
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol. , 11 July 2023
Sec. Evolutionary and Genomic Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1235695
This article is part of the Research Topic Insights on Plant-Associated Microorganisms: Diversity, Systematics and Genomics View all 23 articles
This article is a correction to:
Complete genome sequence of biocontrol strain Bacillus velezensis YC89 and its biocontrol potential against sugarcane red rot
 Linyan Xie1
Linyan Xie1 Lufeng Liu1,2,3
Lufeng Liu1,2,3 Yanju Luo1
Yanju Luo1 Xibing Rao1
Xibing Rao1 Yining Di1,2,3
Yining Di1,2,3 Han Liu1
Han Liu1 Zhenfeng Qian1
Zhenfeng Qian1 Qingqing Shen1
Qingqing Shen1 Lilian He1,3*
Lilian He1,3* Fusheng Li1,3*
Fusheng Li1,3*A corrigendum on
Complete genome sequence of biocontrol strain Bacillus velezensis YC89 and its biocontrol potential against sugarcane red rot
by Xie, L., Liu, L., Luo, Y., Rao, X., Di, Y., Liu, H., Qian, Z., Shen, Q., He, L., and Li, F. (2023). Front. Microbiol. 14:1180474. doi: 10.3389/fmicb.2023.1180474
In the published article, there was an error regarding the affiliations for Lufeng Liu. As well as having affiliation 2, they should also have “1 College of Agronomy and Biotechnology, Yunnan Agricultural University, Kunming, China” and “3 Sugarcane Research Institute, Yunnan Agricultural University, Kunming, China.”
In the published article, there was an error regarding the affiliations for Yining Di. As well as having affiliations 2 and 3, they should also have “1 College of Agronomy and Biotechnology, Yunnan Agricultural University, Kunming, China.”
In the published article, there was an error regarding the affiliations for Lilian He. As well as having affiliation 1, they should also have “3 Sugarcane Research Institute, Yunnan Agricultural University, Kunming, China.”
In the published article, there was an error regarding the affiliations for Fusheng Li. As well as having affiliation 1, they should also have “3 Sugarcane Research Institute, Yunnan Agricultural University, Kunming, China.”
A correction has been made to Results, “3.1. Genomic features and annotation of Bacillus velezensis YC89”, paragraph two. The sentence previously stated:
“Among these pathways, the most represented pathways included carbohydrate metabolism (242 genes, 6.52% of all CDS), followed by amino acid metabolism, and metabolism of cofactors and vitamin pathways (Supplementary Figure S3).”
The corrected sentence appears below:
“Among these pathways, the most represented pathways included carbohydrate metabolism (242 genes, 6.52% of all CDS), followed by amino acid metabolism, and metabolism of cofactors and vitamin pathways (Supplementary Figure S2).”
A correction has been made to Results, “3.2. Identification of strain YC89”, paragraph one. The sentence previously stated:
“These results indicate that YC89 is closely related to B. velezensis FZB42 and B. amyloliquefaciens 6B (Supplementary Figure S3). For an in-depth analysis of the taxonomic status of YC89 strain, a genomewide phylogenomic tree was constructed. The results revealed that strain YC89 formed a close genetic relationship with strain B. velezensis GS-1 (Supplementary Figure S4).”
The corrected sentence appears below:
“These results indicate that YC89 is closely related to B. velezensis FZB42 and B. amyloliquefaciens 6B (Supplementary Figure S3A). For an in-depth analysis of the taxonomic status of YC89 strain, a genome-wide phylogenomic tree was constructed. The results revealed that strain YC89 formed a close genetic relationship with strain B. velezensis GS-1 (Supplementary Figure S3B).”
A correction has been made to Results, “3.8.2. Evaluation of Bacillus velezensis YC89 for biocontrol potential and growth promotion under greenhouse conditions”, paragraph one. The sentence previously stated:
“Collectively, the potted results indicated that the YC89 strain could control the red rot disease of sugarcane and promote the growth of sugarcane plants (Supplementary Figure S5).”
The corrected sentence appears below:
“Collectively, the potted results indicated that the YC89 strain could control the red rot disease of sugarcane and promote the growth of sugarcane plants (Supplementary Figure S4).”
In the published article, there were errors in Figures 1–6 as published. The order of images was incorrect, such that the captions appeared alongside the wrong images. The corrected Figures 1–6 and their captions appear below.
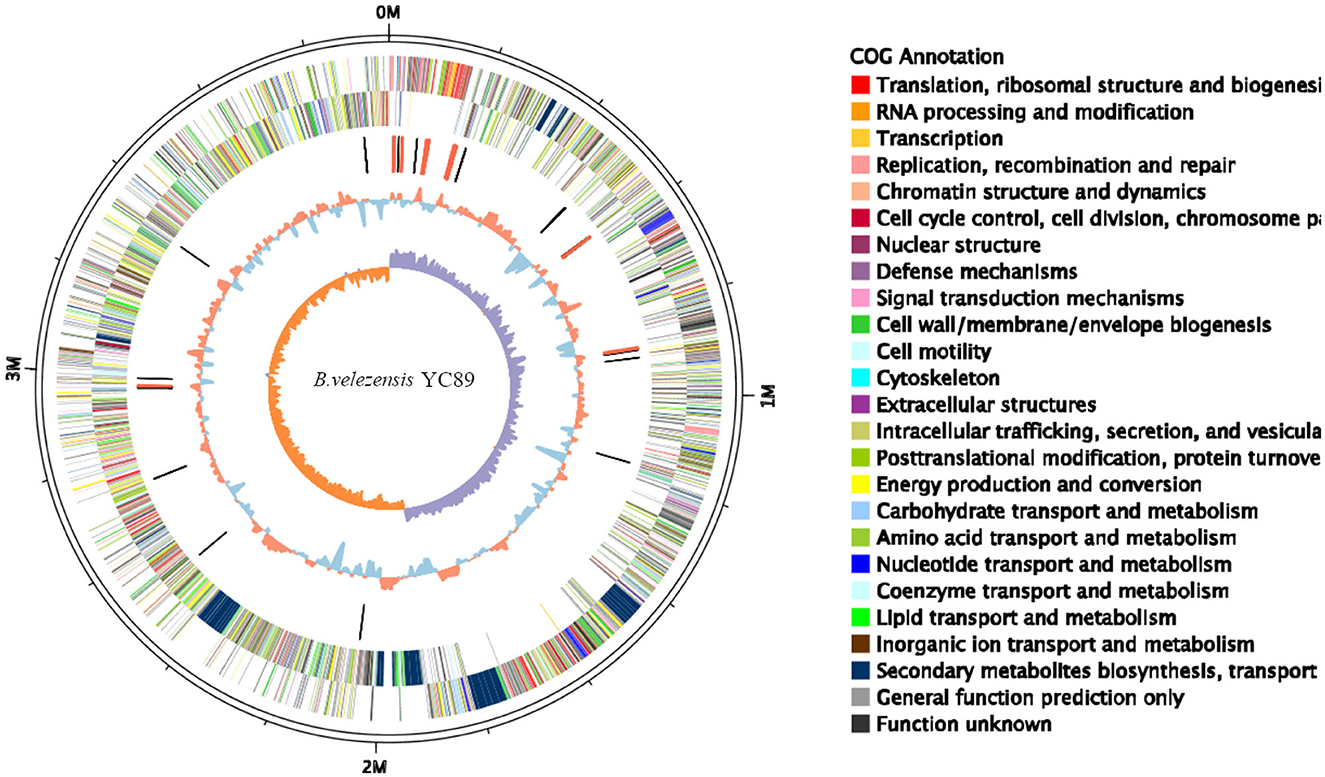
Figure 1. Circular genome map of Bacillus velezensis YC89. From the innermost to the outermost: ring 1 GC skew positive (Orange) and negative (purple); ring 2 for GC content, Orange is greater than the average, blue is less than the average; ring 3 for distribution of rRNAs (red) and tRNAs (black); ring 4 COG classifications of protein-coding genes on the reverse strand; ring 5 COG classifications of protein-coding genes on the forward strand; ring 6 for genome size (black line).
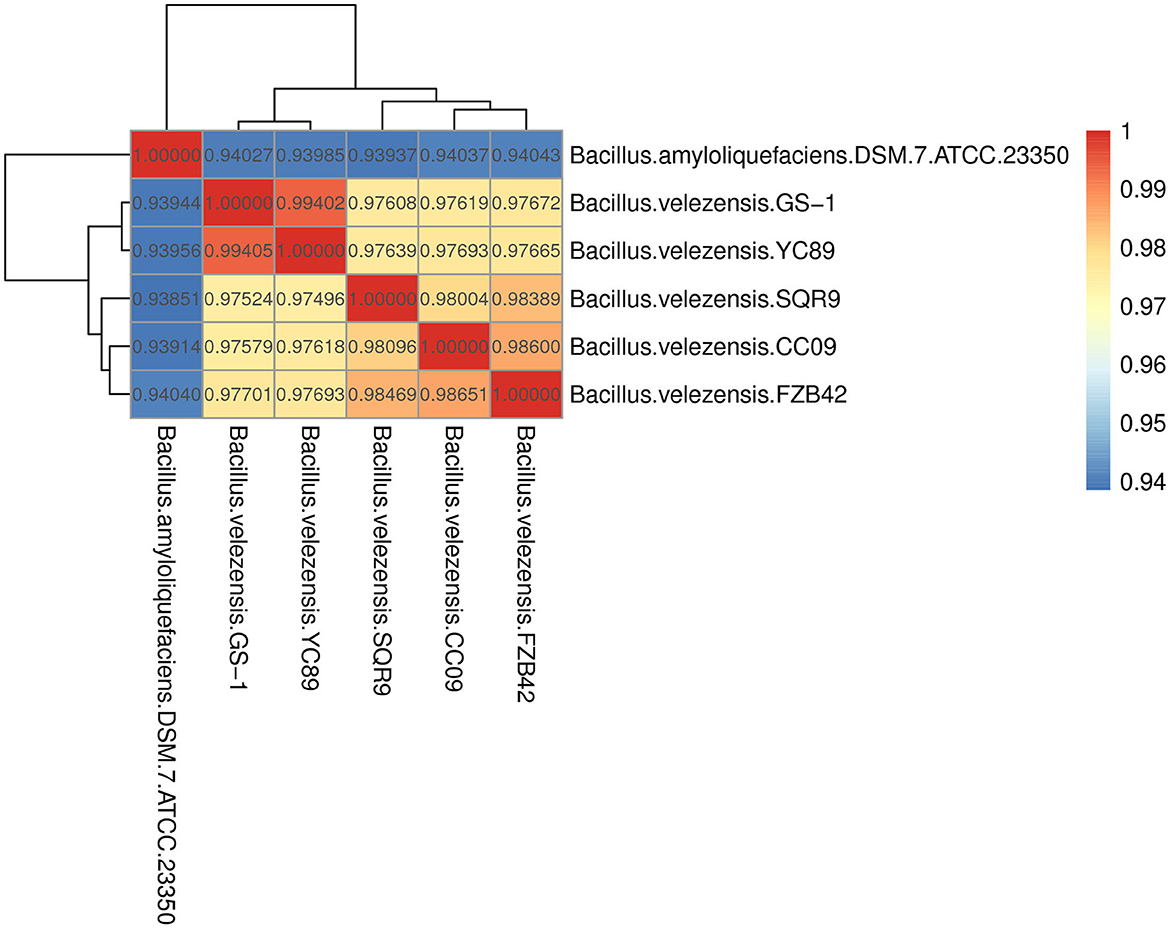
Figure 3. Heatmap of pairwise average nucleotide identity (ANI) values for whole genomes of YC89 and five other Bacillus species.
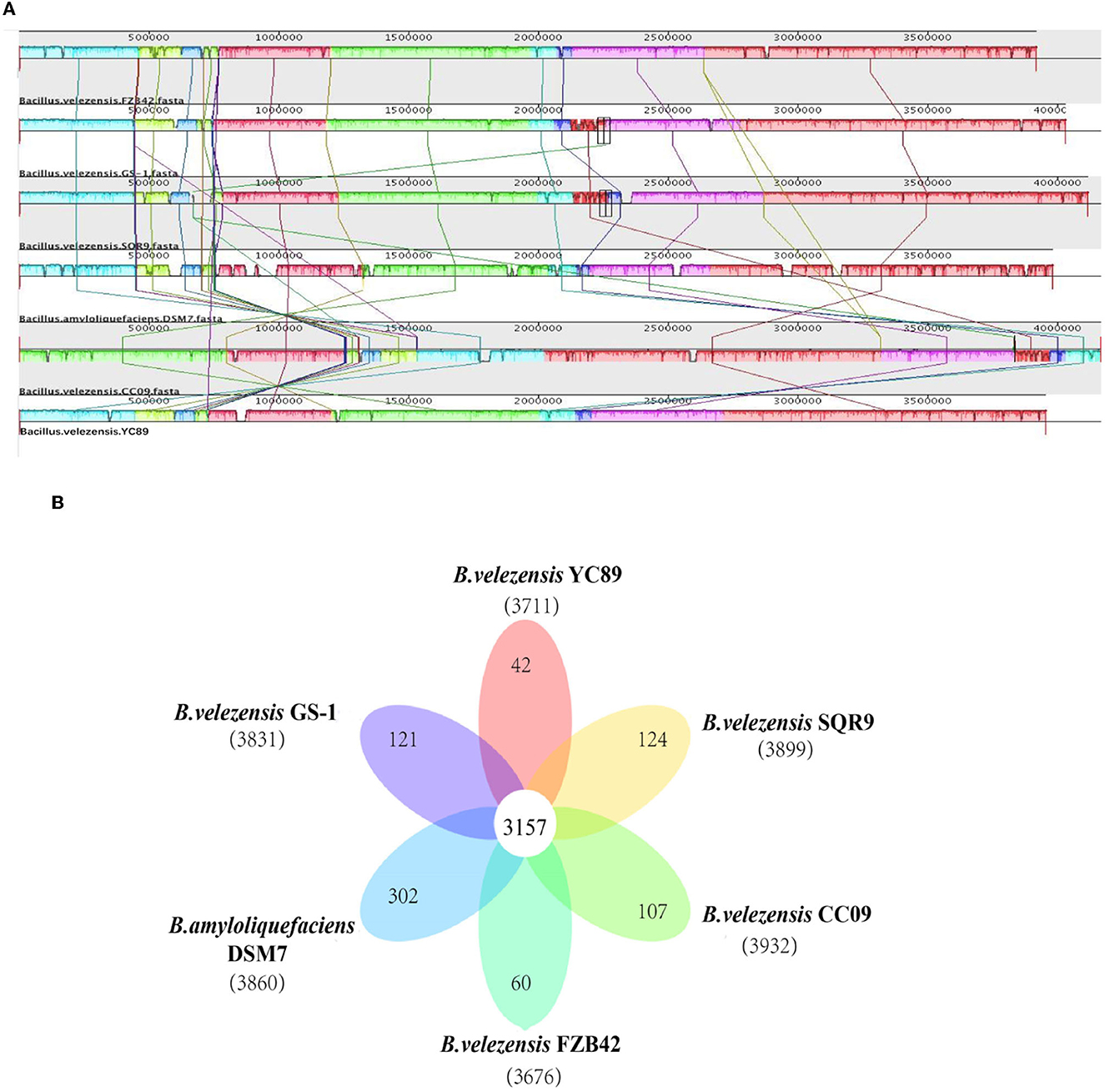
Figure 4. Comparison of B. velezensis YC89 genome sequences against five other Bacillus species genome sequences. (A) Synteny analysis of B. velezensis YC89, B. velezensis FZB42, B. velezensis CC09, B. velezensis SQR9, B. velezensis GS-1 and B. amyloliquefaciens DSM7 genomes. Pairwise alignments of the genomes were generated using MAUVE. The genome of strain YC89 was used as the reference genome. Boxes with the same color indicate syntenic regions. Boxes below the horizontal strain line indicate inverted regions. Rearrangements are shown and by colored lines. Scale is in nucleotides. (B) Venn diagram showing the number of clusters of orthologous genes shared and unique genes.
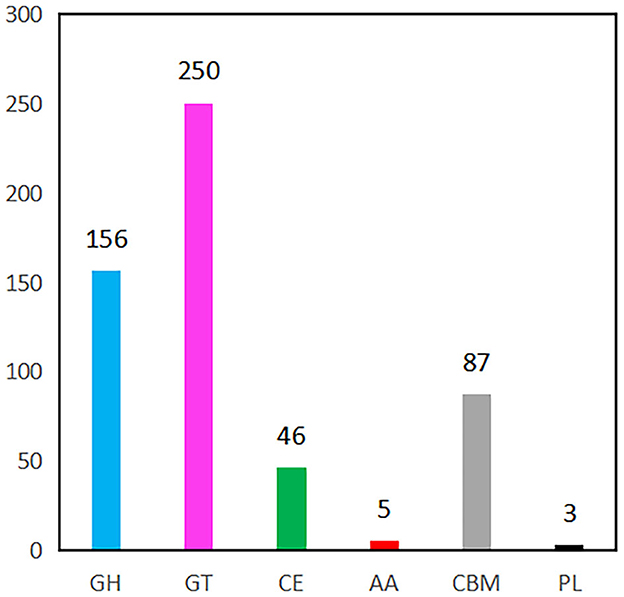
Figure 5. CAZymes gene classification in YC89 genome. GH, Glycoside Hydrolases; GT, Glycosyl Transferases, CE, Carbohydrate Esterases, AA, Auxiliary Activities, CBM, Carbohydrate-Binding Modules; PL, Polysaccharide Lyase.
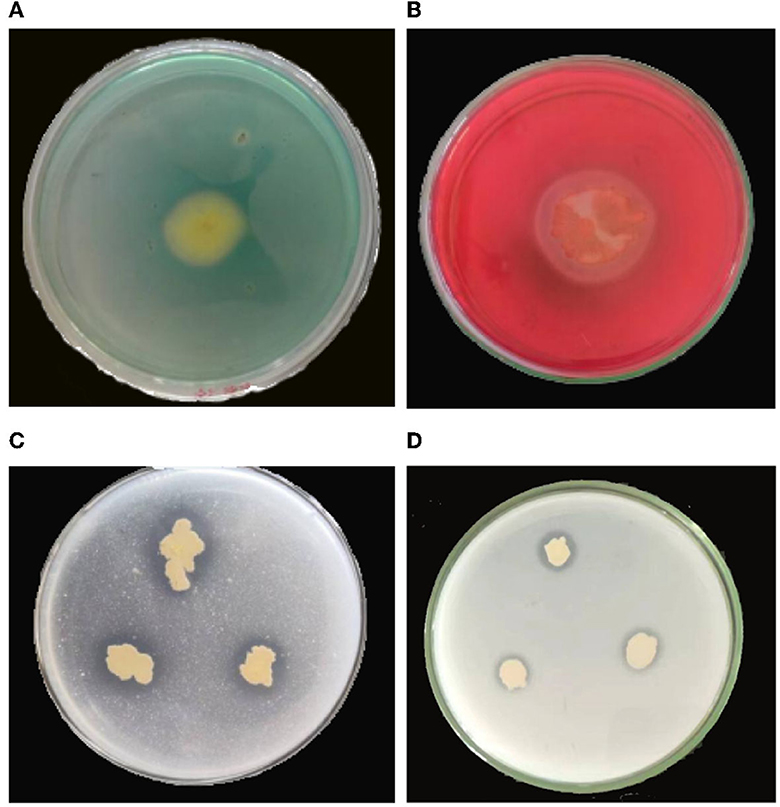
Figure 6. Determination of biological activity of B. velezensis YC89. (A): siderophore, (B): cellulase, (C): Monkina, (D): NBRIP.
The authors apologize for these errors and state that they do not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Bacillus velezensis, biocontrol, sugarcane red rot, genome sequencing, biocontrol mechanism
Citation: Xie L, Liu L, Luo Y, Rao X, Di Y, Liu H, Qian Z, Shen Q, He L and Li F (2023) Corrigendum: Complete genome sequence of biocontrol strain Bacillus velezensis YC89 and its biocontrol potential against sugarcane red rot. Front. Microbiol. 14:1235695. doi: 10.3389/fmicb.2023.1235695
Received: 06 June 2023; Accepted: 30 June 2023;
Published: 11 July 2023.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2023 Xie, Liu, Luo, Rao, Di, Liu, Qian, Shen, He and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fusheng Li, bGZzODEwQHNpbmEuY29t; Lilian He, aGVsaWxpYW45MDVAc29odS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.