
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 24 July 2023
Sec. Terrestrial Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1229828
This article is part of the Research Topic Hazardous Pollutants in Agricultural Soil and Environment View all 15 articles
 Samiksha Joshi1
Samiksha Joshi1 Saurabh Gangola1*
Saurabh Gangola1* Geeta Bhandari2
Geeta Bhandari2 Narendra Singh Bhandari1
Narendra Singh Bhandari1 Deepa Nainwal1
Deepa Nainwal1 Anju Rani3
Anju Rani3 Sumira Malik4,5,6*
Sumira Malik4,5,6* Petr Slama7*
Petr Slama7*The increasing rate of industrialization, anthropogenic, and geological activities have expedited the release of heavy metals (HMs) at higher concentration in environment. HM contamination resulting due to its persistent nature, injudicious use poses a potential threat by causing metal toxicities in humans and animals as well as severe damage to aquatic organisms. Bioremediation is an emerging and reliable solution for mitigation of these contaminants using rhizospheric microorganisms in an environmentally safe manner. The strategies are based on exploiting microbial metabolism and various approaches developed by plant growth promoting bacteria (PGPB) to minimize the toxicity concentration of HM at optimum levels for the environmental clean-up. Rhizospheric bacteria are employed for significant growth of plants in soil contaminated with HM. Exploitation of bacteria possessing plant-beneficial traits as well as metal detoxifying property is an economical and promising approach for bioremediation of HM. Microbial cells exhibit different mechanisms of HM resistance such as active transport, extra cellular barrier, extracellular and intracellular sequestration, and reduction of HM. Tolerance of HM in microorganisms may be chromosomal or plasmid originated. Proteins such as MerT and MerA of mer operon and czcCBA, ArsR, ArsA, ArsD, ArsB, and ArsC genes are responsible for metal detoxification in bacterial cell. This review gives insights about the potential of rhizospheric bacteria in HM removal from various polluted areas. In addition, it also gives deep insights about different mechanism of action expressed by microorganisms for HM detoxification. The dual-purpose use of biological agent as plant growth enhancement and remediation of HM contaminated site is the most significant future prospect of this article.
The term “heavy metals (HMs)” represents a unique group of metals and metalloids existing naturally with high density and atomic weight. Among several HMs contamination of arsenic (As), cadmium (Cd), chromium (Cr), lead (Pb), and mercury (Hg) in environment are considered as highly toxic and are found in terrestrial, aerial, and aquatic eco-systems more than their threshold values (World Health Organization [WHO], 2010; Agency for Toxic Substances and Disease Registry [ATSDR], 2015). Commonly such metals are called as “toxic HMs” or “most problematic HMs” (Rahman and Singh, 2019). HMs are important for the growth of organisms at their optimum desirable concentrations (WHO: 0.001–3 mg/L), however at higher concentrations, these can lead to biotoxicity and have detrimental consequences on human and environmental health (Musa et al., 2013; Gupta et al., 2016). HMs are major industrial effluents, which may subsequently get accumulated in different ecosystems leading to immense threat to the various agro-ecosystems (Cheung and Gu, 2007). HM pollution has now become a crucial matter of environmental concern worldwide due to its non-biodegradability and bioaccumulation in nature (Gautam et al., 2014). Exposure of HMs exhibited severe consequences in humans (like inflammatory, respiratory, and cardiovascular diseases), animals (Engwa et al., 2019), plants (reduced growth rate, photosynthesis, and yield) (Asati et al., 2016), microorganisms (metabolism, growth, and morphology) (Ayangbenro and Babalola, 2017) and aquatic lives (death and reproduction) (Pandey and Madhuri, 2014). Worldwide, there are almost 5 million contaminated sites of soil with HMs concentration exceeding the regulatory levels (Li et al., 2019). Biogeochemical cycles on disruption leads to deposition of HM and other contaminants into aquatic and terrestrial environment like, combustion of fossil fuels, mining, nuclear power plants, industrial effluents, sludges, preservatives including organometallic compounds, dust from smelters, waste from brewery, and distillery units (Zhang et al., 2013; Gangola et al., 2023a). Apart from being toxic in nature, few HMs are also reported as essential micronutrients for efficient plant growth. HMs may also function as cofactor of several important enzymes, required for metabolism of hydrogen, involved in methane biogenesis and acetogenesis.
Metal contamination of food and water has been reported to result into several births related defects like cancer, lesion of skin, impairment of liver and kidney functions, and many more. Millions of people of Argentina, Taiwan, Bangladesh, India, Poland, China, Hungry, Japan, Belgium, North Mexico, Chile, and Mongolia are suffering from health-related issues mentioned above due to metal contamination of ground water (Tseng et al., 2005). Lead, cadmium and mercury have been given second, third and seventh rank, respectively, due to their highly toxic and widespread nature. A huge number of superfund sites were found to be HM polluted (Peters, 1999). HMs are present in different ecosystem naturally or due to anthropogenic activities like smelting of metal ores, fuel and energy production, sludge dumps, mine tailings, agricultural activities, and gas exhaust (Raskin et al., 1994). HMs impose more intensive challenge because of its recalcitrant nature and occurrence in a form that cannot undergo complete degradation but can only be complexed with some compounds or only its chemical form can be altered. Several parameters such as physical, chemical, and ecological characteristics of the polluted sites should be checked for successful achievement of bioremediation (Gu, 2021). Physicochemical and biological methods are used for remediation of sites contaminated with HMs, both of which have their own pros and cons while use of microorganisms as a biological tool is the emerging technology for degradation and remediation of pollutants at contaminated sites (Gu, 2018; Ansari et al., 2023). Microbial bioremediation is a simple, economic, ecofriendly, inexpensive, and efficient approach done for removal and detoxification of toxic pollutants using native microorganisms (Kumar Mishra, 2017; Spain et al., 2021). Removal of pollutant from any contaminated sites by employing microbial system could be achieved maximally by knowing the toxic concentration of pollutant and maintenance energy required by the microbial system (Gao and Gu, 2021). Several bioremediation methods are reported for the degradation of more than 50 pollutants, but very little innovative approach was found (Gu, 2016). Mechanisms involved mainly were biosorption, bio-oxidation and bio mineralization (Jin et al., 2018). Microbial population associated with plant roots are efficient in remediation of polluted soils and promote plant growth by several direct and indirect mechanisms such as siderophore production, phytohormone production, phosphate solubilization, biological N2 fixation, antibiotic production, synthesis of lytic enzymes, etc. (Goswami et al., 2016; El-Meihy et al., 2019; Joshi et al., 2023a). Exploitation of plant growth promoting bacteria (PGPB) having potential of metal detoxification as well as multiple plant growth promoting traits are promising metal bioremediation tool. Catabolic efficiency as well as production of bio surfactant and enzymes by microbes is a novel approach to enhance their remediation efficacy (Le et al., 2017). Plant growth promoting rhizobacteria (PGPR) has shown improvement in metal immobilization or mobilization in HM contaminated soils and plant biomass as they are metal resistant and phytoremediation enhancing agents (Ma et al., 2016). Removal of environmental contaminants that cause ecological imbalance is a global concern. Considering this important, this review addresses microbes as a tool for bioremediation of toxic HMs from different contaminated systems with emphasis on the involved mechanism.
Heavy metals are present in the environment as a result of both naturally occurring pedogenetic processes and human activities such as mining, smelting, electroplating, pesticide use, release of biosolids and phosphate fertilizer (Dixit et al., 2015). HM concentrations are also influenced by weathering of minerals, erosion, volcanic activity, atmospheric deposition, and other natural sources (Fulekar et al., 2009; Sabiha-Javied et al., 2009). Unfortunately, human activity has the potential to interfere with the natural geochemical cycle of metals, causing HMs to accumulate in soil and water. When HM concentrations reach permissible levels, this can pose a risk to human health, as well as to the health of plants, animals, and aquatic life (D’amore et al., 2005). Due to excessive production from anthropogenic and natural sources, movement from mines to areas where humans are more exposed, industrial waste discharge, and increasing bioavailability, HMs end up as pollutants in soil and water. As is a metal that can be found in biosolids, insecticides, ore mining, smelting, and wood preservatives. Electroplating, phosphate fertilizers, plastic stabilizers, paints, and pigments are sources of Cd. Fly ash, steel manufacturing, and tanneries all include Cr. Biosolids, fertilizers, ore mining, pesticides, and smelting are sources of copper. Hg is present in medical waste, coal combustion, and the mining of silver. Ni can be found in surgical equipment, wastewater, culinary appliances, and automotive batteries. Pb was observed in the aerial emissions produced by the burning of used batteries, insecticides, and herbicides.
According to Lombi and Gerzabek (1998), the equation can be used to depict how HMs are distributed/mass balance in the soil’s surroundings:
In the equation, the following variables are included: M for HM, p for parent material, a for atmospheric deposition, f for fertilizer source, ag for agrochemical source, ow for organic waste source, ip for inorganic pollutant, cr for crop removal, and l for losses due to leaching, volatilization, and other processes.
Metals in soil occur in both available and non-available forms to a microorganism (Sposito, 2000) which is directly related to positive and negatively charged salts of the metal. Various factors influencing availability of HMs in soil includes temperature, cation exchange capacity (CEC), redox potential, pH, aeration capacity, organic matter content, microbial activity in the rhizosphere, clay minerals, water quantity, root exudates, hydrous metal oxides, climate, and metal chemical properties (Roane and Pepper, 2000; Fischerová et al., 2006). Lasat (2002) showed bioavailability of HMs to be affected by some bacterial traits such as acidification, synthesis of chelating agents, and altering the redox potential. Metals exist in soluble cationic form under aerobic and oxidized conditions whereas found as insoluble carbonate or sulfides under anaerobic and reduced environment. Moreover, bioavailability of metals in free ionic form increases at acidic pH whereas reduced at high pH because of precipitation. Availability of HMs in soils is observed to be highest for zinc followed by copper, cadmium, and nickel. Metals with high CEC are observed to have reduced toxicity even at their higher concentrations (Roane and Pepper, 2000). Several reports on availability of HMs and their uptake by plants can be essential for predicting the effect of HMs on growth of plant under stressed conditions and population of rhizospheric microbes, as well as assessment of execution of bioremediation techniques for remediation of metal stressed soils. HMs are recalcitrant which affect and alters their toxicity over time. High concentration of HMs negatively affects physiology of microbes whereas also used as important micronutrient for their growth (Ahemad, 2012). Bioavailability of metals decides the possibility of interaction between metal species and bacteria that can occur either to neutralize their toxic effects or to fulfill their metabolic needs. Mostly, naturally existing microbial biomass are most effective and participate actively in detoxification mechanism, but still they are not well characterized or known. This scientific gap is due to lack of knowledge in cultivation of such unculturable microorganisms and continuous dynamic changes in their traits essential for adaptation to the existing environment. Bacteria in rhizospheric region affect HM speciation which further leads to changes in bioavailability of metals. Furthermore, they prevent phytotoxicity in plants by converting bioavailable form of HMs to their non-bioavailable forms in soils (Jing et al., 2007). Bioavailability of metal varies from species to species. Speciation of metals and the resulting bioavailability decides the overall toxicity as well as physiological effects of a metal on biological systems (Knotek-Smith et al., 2003). Nutrient status of the soil also affects the bioavailability of HMs. Several in situ experiments rely on the enrichment of the existing microbial population by adding nutrients such as carbon, nitrogen, phosphorus, etc., called bioaugmentation. Most of the degradation processes are oxidative in which microorganisms harness energy through electron exchange. The availability of the oxygen is the major kinetic obstacle to the aerobic microbes due to low solubility in water.
According to Gao and Gu (2021) at a particular concentration of pollutant or below its toxicity limit, microorganisms require lower energy for their maintenance, isolation, enrichment, and their efficient utilization for biodegradation. HMs are categorized according to their reactivity, effect on biological systems and their target sites. Some of them have vital role (such as zinc, iron, copper, magnesium, and calcium) in biological system whereas others are also reported to have carcinogenic and cytotoxic effects (like mercury, lead, and cadmium). Rapid industrialization, urbanization as well as mining activities have altered biogeochemical cycling which further raised accumulation of HMs in terrestrial, atmosphere, and aquatic eco-system. Such changes may severely affect the biotic communities present on those sites. Various reports showed HM stress on higher plants like cadmium, lead, and mercury inhibiting chlorophyll biosynthesis, respiration as well as oxidative stress which may further cause cellular damage, disturbs ionic balance of the cell, and toxicity in higher organisms (Mithoefer et al., 2004; Han F. X. et al., 2006; Han S. H. et al., 2006). Toxic HMs contaminates groundwater and biota and have negative effects on health of human beings. It is essential to examine the distribution and concentration of toxic HMs in riverine ecosystems (Islam et al., 2018). Major sources of contamination in aquatic system include domestic sewage, industrial effluents, agricultural run-off, and mining operations (Zhuang et al., 2013). Water contamination with HMs is a sensitive environmental issue which adversely affects surface water, ground water, plants, human, animal’s health (Rezania et al., 2016), aquatic organisms (Abrar et al., 2011) as well as alters histopathological tissues of aquatic organisms (Ahmed et al., 2014). HM contaminated water bodies are a severe concern worldwide because of biomagnifications, environmental persistence, bioaccumulation, and toxicity (Rajaei et al., 2012). Contamination of riverine sediments by HMs may cause ecological risk to organisms present in benthic region (Pacle Decena et al., 2018). Soil with more inputs of fertilizers is enriched with HMs. Bioavailability of HMs varies according to physicochemical properties and metal speciation of soil which is further essential for plant uptake. Urban areas soil has been reported with high amount of lead, out of which only up to 85% is bio accessible (Mackay et al., 2013). Non-essential HMs are reported to be hazardous and highly toxic to plants, humans, and animals even at minimum concentrations (Mahboob et al., 2014). Also, few essential HMs may increase risk of toxicity if used at increased concentrations as listed in Table 1. Metals that are toxic, persistent, and bio accumulative are more harmful (DeForest et al., 2007). Several HMs have been referred as teratogenic, mutagenic, and carcinogenic. Cadmium with potential of bioaccumulation and high toxicity resulted into population decline of freshwater mussels (Ngo, 2007).
Undoubtedly, the combination of ecological analysis and chemical data can offer greater understanding of the environmental condition of ecosystems and the transformations that have occurred (Gu, 2014). HMs cannot be subjected to complete degradation, i.e., modification in the nuclear structure does not occur, thus they can only undergo change in their oxidation states which in turn changes their chemical behavior (Sobariu et al., 2017). Change in oxidation may lead to various consequences like solubility of metal increase in water that can be further easily removed by leaching or may decrease its solubility by which it becomes less available for decontamination, may get converted to less toxic forms or can be removed from polluted sites via volatilization (Garbisu and Alkorta, 2003).
The root of plants provides aeration to the soil and influences the distribution of rhizospheric microbes through soil. The root system is also capable of penetrating the impermeable zones of soils and draw the soluble forms of the organic contaminants. A significant symbiotic relationship between the plant root system and rhizospheric microbes is observed and this relationship has long been exploited for rhizo-remediation (Gangola et al., 2022a). Successful rhizo-remediation is dependent upon several factors such as: root and rhizosphere colonization by microbes, formation of various useful metabolites by the plants, survival, and ecological interactions with other organisms. This novel approach can also be called phytoremediation. The employment of leguminous plants for bioremediation has seen a rise in the recent times due to immense bioremediation ability and capability of biological nitrogen fixation as well as root nodule formation (Pastor et al., 2003; Kamaludeen and Ramasamy, 2008; Gangola et al., 2022b). Rhizospheric microbes have immense potential of nutrient cycling, restoration of soil structure, decontamination of various contaminants, pest management, and enhanced plant growth (Gangola et al., 2023b). Therefore, soil microbes can improve the bioremediation potential of plants and in turn lower the phytotoxicity of the soil contaminants. In symbiotic associations between microbes and plants, plant make carbon source available to the associated microbes which in turn helps in reduction of phytotoxicity of the soil contaminants. In addition, some non-specific associations are also observed in between microbes and plants where metabolic processes of plant stimulate the microbial growth, which in turn degrade the soil pollutants due to their inherent metabolic activity. Plants roots are capable of releasing root exudates and increasing the metal ion solubility. These biochemical processes increase the bioremediation potential of rhizospheric microbes associated with the plant roots and thus is of significance for remediation of heavy-metal pollution soils (Gangola et al., 2021, 2022c). This association also accounts for effective phytoremediation due to potential of microbes to influence bioavailability and solubility of the HM. PGPRs boost up the growth and development as well as improve the metal tolerance by decontaminating the toxic HMs (Tassi et al., 2008).
Bioremediation offers transformation, degradation, or detoxification of hazardous compounds, organic or inorganic waste by natural biological activity of bacteria, fungi, or plants in the environment (Gu and Pan, 2006; Gu and Wang, 2013; Siddiquee et al., 2015; Gu, 2021). This process is less expensive; low technology based and can be done on site as well as it can be improved by addition of nutrient, an electron acceptor, potent microbial population, pH, soil type, and temperature (Vidali, 2001; Gu, 2019). Moreover, alteration of environmental parameters to provide optimum conditions for microbial activity and growth allow bioremediation to be more effective and proceed at a faster rate (Gu, 2003; Su, 2014). Microbes with ability to remediate when applied to the contaminated site results into transformation of toxic compounds via several metabolic reactions (Siddiquee et al., 2015). Efficient biodegradation and remediation of pollutants at contaminated sites can be achieved by the knowledge of microbial biochemical reactions along with its engineering and management (Gu, 2021; Joshi et al., 2023b). Several approaches for bioremediation include bioreactor, biosorption, biostimulation, bioventing, bio filters, composting, bioaugmentation, and land farming. Several reports have demonstrated efficient elimination of HMs or their conversion to benign or less toxic forms using potent microbes (Qazilbash, 2004). Potential of microbes of exploiting available nitrogen or carbon source for their growth and survival is used in bioremediation and thus bacteria utilizes the contaminants as source of nutrient (Tang et al., 2019). Soil microbes in rhizospheric zone are considered as efficient degraders to overcome HM stress. Other than these plants also possess several detoxifying strategies like synthesis of thiols with high ability to take up HMs. MacNaughton et al. (1999) showed bioremediation as a significant technology to detoxify ground water, soils, and coastal water bodies. Interaction of microbes with HMs can occur via different mechanisms as shown in Figure 1, such as biomineralization, biotransformation, bioleaching, biosorption, etc., which further helps in metal bioremediation (Pande et al., 2022). Several investigations as listed in Table 2, revealed that bacterial communities belonging to genus Escherichia, Bacillus, and Mycobacterium isolated from various contaminated sites are found efficient for the removal of HM (Cd, Cr, and Cu) (Jiang et al., 2015). Similarly, they can be remediated by fungal genera, such as Pleurotus, Acremonium, and Fusarium. Li et al. (2019) discovered microbial consortia with high remediation potential of HMs. Composting and Immobilization are used to enhance the rate of remediation by increasing the activity of microorganisms (Poulsen and Bester, 2010; Chen et al., 2015). Rhizobium-legume symbiotic association is a significant bioremediation method for metal contaminated soil and have been exploited for removal of arsenic from soils. Mandal et al. (2011) have reported isolation of arsenate (2.8 mM arsenate) resistant symbiotic bacteria Rhizobium strain (VMA301) from roots of Vigna mungo. This resistant strain showed efficient N2 fixation as well as helped in decontamination of arsenic containing soil. However, a delayed nodulation and reduced nitrogenase activity was also observed in arsenic treated plants. Arsenic was found to accumulate in roots at higher rate than in the root nodules. Although a lot of research has been done on environmental pollutant degradation but still waiting for the need of efficient biodegradation technology (Gu, 2016).
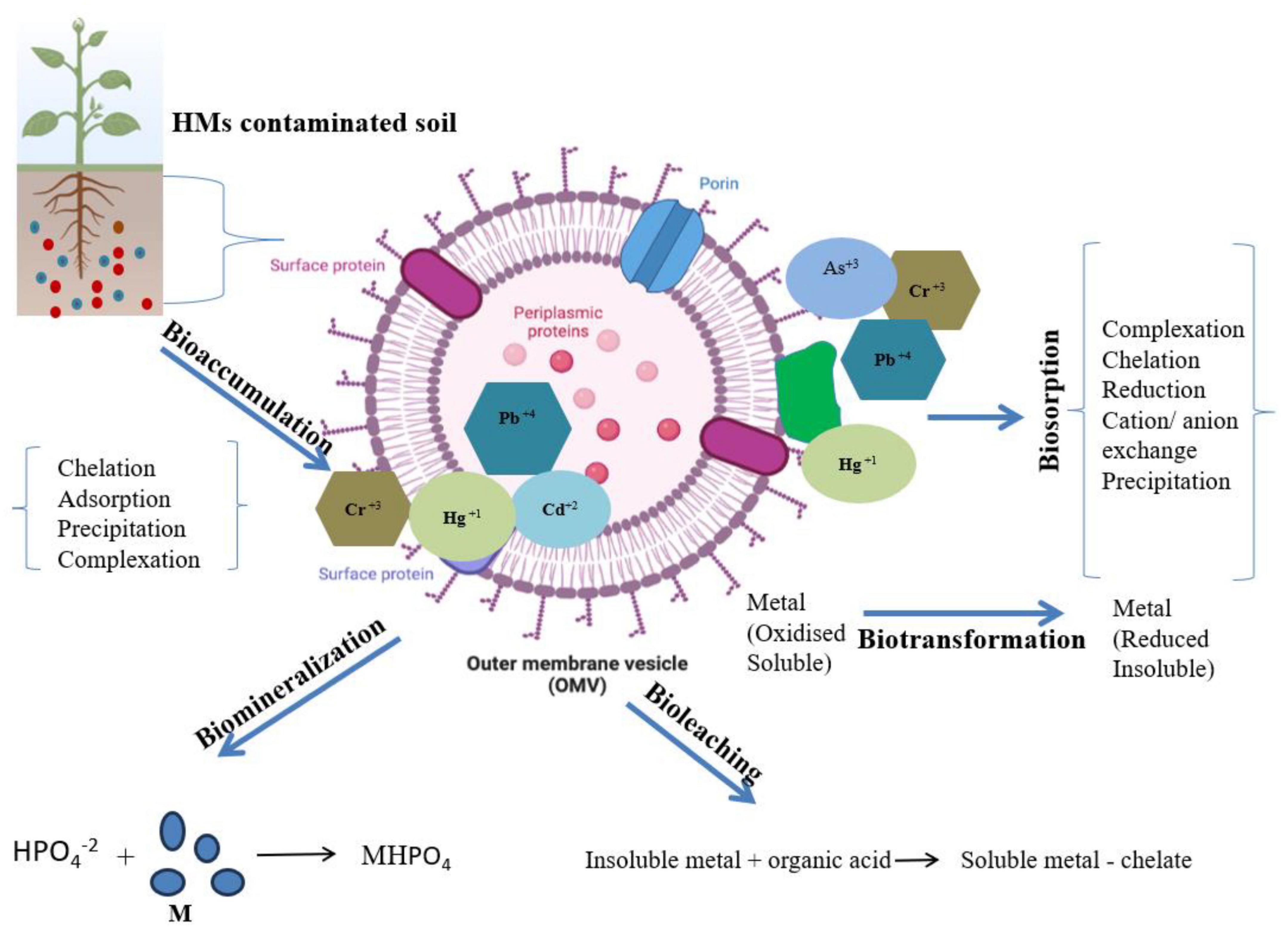
Figure 1. Different strategies such as: bioleaching, biomineralization, biotransformation, biosorption, and bioaccumulation by microbial system for the removal or transformation of toxic HMs from contaminated sites.
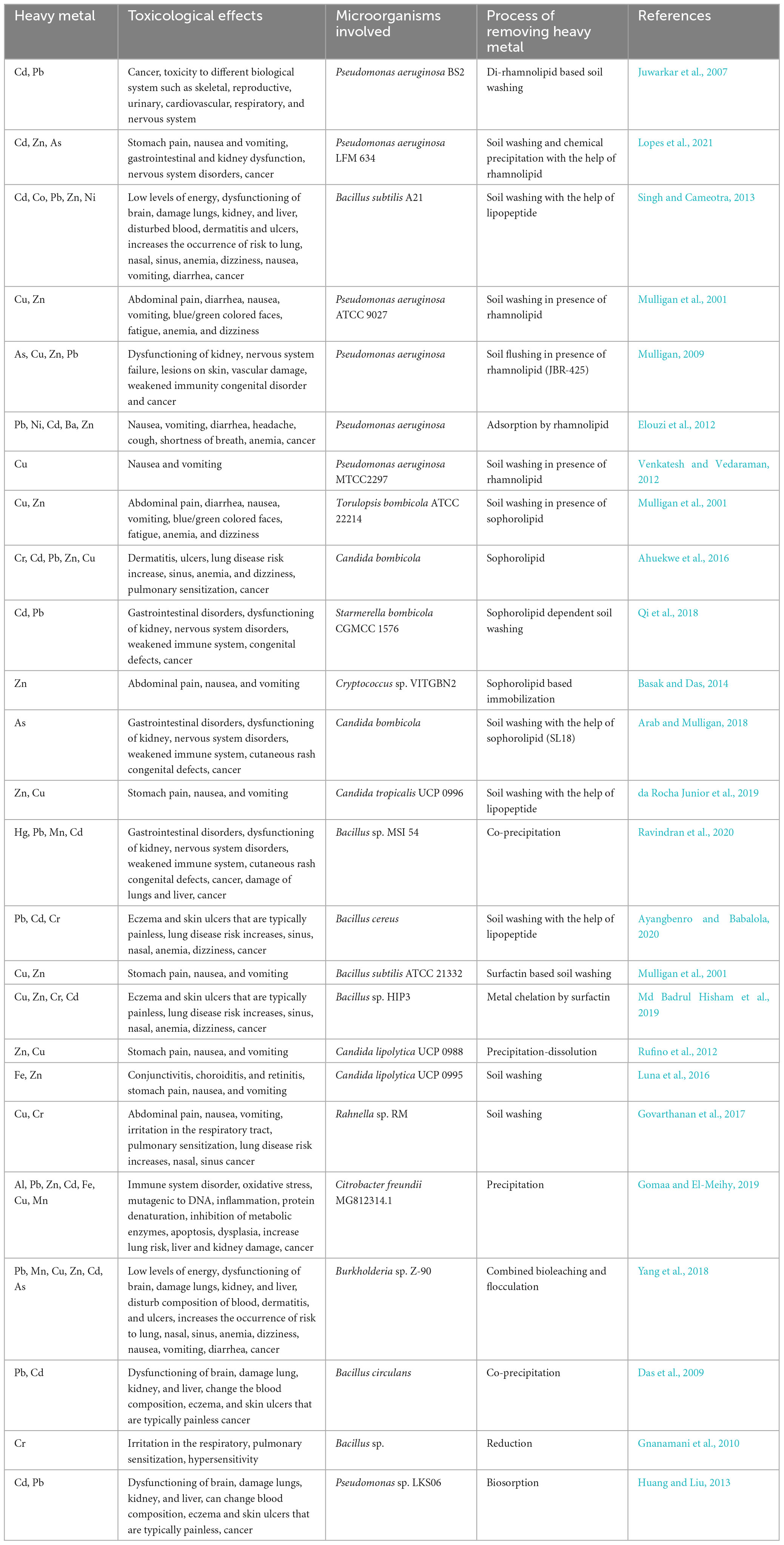
Table 2. Heavy metal mediated toxicological effects and microorganisms involved in the process of removing them from soil.
Biosorption by microorganisms remediates the contaminated sites or HMs through binding with extracellular polymers or adsorption on the cell surface (Gangola et al., 2018). The outer cell shield of microbes carries active groups of compounds which provides sorption ability and further helps in binding of metals. Such linking occurs due to negative potential of active groups present on outer membrane and cationic metal ions. Biosorption is a reversible process which makes it essential for both recovery [for non-toxic and essential HMs such as gold (Au), Zinc (Zn), and copper (Cu)] and removal of HMs. Various studies showed binding of Cr3+, Cd2+, Pb2+, Cu2+, Co2+, Zn2+, and Ni2+ to microbial biomass by sorption at pH between 5 and 7 and get liberated at low pH whereas few metal ions like Ag2+ and Au2+ stay bound to biosorbents even at low pH (Kisielowska et al., 2010). Microorganisms are found as rapid adsorbers of HM ions for example Bacillus sp. showed 60% sorption of its Cu2+ at pH 7.2, at initial phase and reached to adsorption equilibrium within 10 min (He and Tebo, 1998). Several studies demonstrated good binding affinity of microbes via biosorption such as Streptomyces rimosus for iron and lead (Sahmoune, 2019), Staphylococcus hominis strain AMB-2 for cadmium and lead (Rahman et al., 2019), Cronobacter muytjensii KSCAS2 for multiple HMs (Saranya et al., 2018), and Staphylococcus aureus biofilms for U(IV) remediation (Shukla et al., 2020).
Bioleaching process mainly involves complexation, biological dissolution, bio-oxidation (Jin et al., 2018), or conversion of sparingly soluble metal compounds into easily soluble forms which can be removed easily (Kisielowska et al., 2010). Various microorganisms known as potent metal mobilizers produces low molecular weight organic acids such as tartaric acid, oxalic acid, gluconic acid, malic acid, citric acid, and butyric acid for the solubilization of HMs from the insoluble ores and exudation of complexion agents that can easily dissolve HMs from soil particles comprising HM minerals. Application of Acidithiobacillus thiooxidans and Acidithiobacillus ferrooxidans in metal leaching is proven to be effective because of their biochemical attributes as well as high resistance to temperature and pH (Błaszczyk, 2007). Few studies showed that rate of cadmium leaching promoted by providing more nutrients to microbes for increased production of organic acids (Jin et al., 2018). Furthermore, a few genera such as Citrobacter synthesized free inorganic phosphate with high potential to trap toxic metal ions and forming insoluble metal phosphates (Marchenko et al., 2015). Some efficient strains of bacteria like Bacillus licheniformis and Corynebacterium sp. are reported to perform oxidation-reduction reactions and convert the valance of HMs leading to alter their toxicity and mobility (Gavrilescu, 2004). Acidophiles are mainly involved in bioleaching (Srichandan et al., 2014). Arsenic removal by bioleaching using individual and mixed culture of A. ferrooxidans and A. thiooxidans was reported by Zhang and Gu (2007). Bioprecipitation and biocrystallization of HM compounds by microbes results in conversion of metal into form which is less toxic. This occurs mainly due to enzymatic activities (Sklodowska, 2000). Transformation of toxic metal by microbes may occur by several reactions like demethylation, methylation, oxidation, and reduction. For example, Gram-positive bacteria obtained from tannery sewers, transformed highly toxic chromium (VI) to its less toxic form chromium (III) (Kisielowska et al., 2010). Bioaccumulation is a toxic kinetic process in which active metabolism leads to HM uptake of contaminants inside the organism when the rate of absorption of contaminant exceeds its rate of loss (Chojnacka, 2010). Microbes with bioaccumulation potential should have high tolerance to variety of contaminants as well as, superior bio transformational ability to make it non-toxic in nature (Mishra and Malik, 2013). This is not considered as much valuable approach as after certain amount of accumulation it can exert toxic effects to cells. Studies showed removal of HM to be more effective by bioaccumulation as compared to biosorption (Henriques et al., 2015). In an investigation, biomasses of Bacillus megaterium and Bacillus circulans demonstrated efficient removal of toxic form of chromium (VI) at much high rate (32–34.5 mg g–1 dw) by bioaccumulation as compared to biosorption (15.7–39.9 mg g–1 dw) (Srinath et al., 2002). Bioaccumulation potential was shown by Pseudomonas putida 62 BN for cadmium and Bacillus cereus M116 for nickel (II) (Rani and Goel, 2009; Naskar et al., 2020). Microbial transformation results in reduced toxicity and vitalization of HMs. Some reports showed detoxification of arsenic (III) by Acinetobacter sp. as well as Micrococcus sp. and chromium (VI) by Bacillus sp. SFC 500-1E (Tayang and Songachan, 2021). Some of the key bacterial HMs resistant mechanisms are explained.
The cell wall, plasma membrane, and capsule act as an extracellular barrier to stop metal ions from entering the cell (El-Helow et al., 2000). Through ionizable groups like amino, carboxyl, hydroxyl and phosphate on their cell wall or capsule, bacteria can bind metal ions. According to Pardo et al. (2003), this adsorption is a passive process that can even take place in dead bacterial cells. The two phases involved in the accumulation of metal ions by living cells are gradual active transport into the cytoplasm and non-specific initial adsorption by the cell wall (McEldowney, 2000).
Acinetobacter sp. (Irawati et al., 2015), Marinobacter sp. (Bhaskar and Bhosle, 2006), Klebsiella sp. (Mohan et al., 2019), and Enterobacter cloacae (Iyer et al., 2005) are just a few of the bacteria whose capsules have been shown to accumulate metal ions primarily through carboxyl groups of their polysaccharides. Exopolysaccharide (EPS) production by copper-tolerant Pseudomonas aeruginosa strains is twice as high as that of copper-sensitive strains (Kazy et al., 2002). Metal ions, however, can also stop bacteria from producing EPS. Multiple copper-tolerant but deficient in the synthesis of EPS gellan Sphingomonas paucimobilis mutants were found by Richau et al. (1997). The authors hypothesized that because EPS synthesis is a very energy-intensive process, the mutants’ higher copper tolerance was caused by a slower growth rate and the use of stored energy for defense against metal stress.
The majority of HM resistance methods used by bacteria, sometimes referred to as efflux, export metal ions from cells. On chromosomes (Franke et al., 2001; Lee et al., 2007) and plasmids (Nies, 2000), efflux system genetic factors can be discovered. The magnesium transport system allows the entry of cobalt, cadmium, nickel, zinc, and manganese ions to the Ralstonia metallidurans, whereas some metal ions can enter the cell through the essential element uptake systems such as chromate which is transported via the sulfate transport system (Gonzalez Henao and Ghneim-Herrera, 2021). Through ATP hydrolysis or an electrochemical gradient (Mathivanan et al., 2021), metal ions are exported from the cell. Proteins from three families make up efflux systems: P-type ATPases, CDF (cation diffusion facilitator), and RND (resistance, nodulation, cell division). Gram-negative bacteria’s P-type ATPases and CDF proteins move particular substrates across the plasma membrane and into the periplasm. It is important to note that CDF proteins specifically interact with divalent metal ions (Zn2+, Co2+, Ni2+, Cd2+, and Fe2+), whereas P-type ATPases primarily transfer metal ions that have a high affinity for sulfhydryl groups (Cu+/Ag+, Zn2+/Cd2+/Pb2+). RND proteins assemble into transport complexes that carry cations from the periplasm across the plasma membrane (Nies, 2003).
The efflux system that allows the multi-resistant bacterium R. metallidurans CH34 to tolerate copper, cobalt, and zinc ions is encoded by the Czc operon. The system makes use of an electrochemical gradient and is made up of the CzcCB2A efflux complex, which also functions as a cation-proton antiporter and includes the subunits CzcC, CzcB, and RND-protein CzcA. While CzcA might contribute to some degree of HM resistance, CzcC and CzcB are necessary for the efflux system to operate properly (Nies, 2000). With CPx-type ATPases discovered in bacteria like Enterococcus hirae (CopA and CopB), Streptococcus mutans (Vats and Lee, 2001), and Escherichia coli (Rensing et al., 1999), the P-type ATPase family comprises transporters for mono- and divalent metal cations. In bacteria like Staphylococcus aureus and P. putida, P-type ATPases called CadA and ZntA play a role in cadmium tolerance (Oger et al., 2003; Lee et al., 2007).
Certain bacteria can use other mechanisms in addition to efflux systems to resist HMs (Saxena et al., 2002). For instance, the P. putida strain S4 transports copper ions from the cytoplasm and sequesters them in the periplasm using an ATPase efflux mechanism. Another illustration is the ars system, which has 3–5 genes and is found in both Gram-positive and Gram-negative bacteria. The ArsA/ArsB ATPase pump and the ArsC reductase are both encoded by the ars operon. According to Mukhopadhyay et al. (2002), in the first stage, cytoplasmic ArsC arsenate reductase enzymatically converts arsenate to arsenite, which is then exported by the efflux system through the plasma membrane.
Through a procedure known as intracellular sequestration, diverse substances in cell cytoplasm combine metal ions. Two kinds of eukaryotic metal-binding peptides—metallothioneins and phytochelatins—are high in cysteine residues and bind metal ions via sulfhydryl groups (Pinto et al., 2003). For metallothionein synthesis, which is triggered by cadmium and zinc ions, Synechococcus sp. encodes two genes, smtA and smtB (Ybarra and Webb, 1999). Low-molecular-weight proteins with a high cysteine content enable a cadmium-tolerant strain of P. putida to sequester copper, cadmium, and zinc ions intracellularly. According to Ivanova et al. (2002), some marine gamma-proteobacteria synthesize phytochelatin-like low-molecular-weight proteins that are cadmium-inducible. Glutathione is used by Rhizobium leguminosarum cells to sequester cadmium ions intracellularly (Lima et al., 2006).
Metal ion accumulation in the periplasm or outer membrane of microbial cell, or their fusion as insoluble compounds, is referred to as extracellular sequestration. For instance, copper-resistant strains of Pseudomonas syringae produce CopA, CopB, and CopC, which bind copper ions and cause copper to build up in the periplasm or outer membrane, resulting in blue bacterial colonies (Cha and Cooksey, 1991). Similar to how copper-tolerant Pseudomonas pickettii US321 builds up copper ions in the periplasm or outer membrane, resistant strains likely store copper as a complex and transport it into the cytoplasm, while sensitive strains likely build up copper in a toxic free ionic form that damages cells (Gilotra and Srivastava, 1997). Pseudomonas stutzeri AG259, which was isolated from the soil of a silver mine, accumulates silver ions as sulfide complexes on the cell surface or in elemental form in the periplasm due to plasmid-encoded resistance to high concentrations of silver ions in the medium (Haefeli et al., 1984; Slawson et al., 1992; Klaus et al., 1999). Some bacteria release metal ions from the cytoplasm into the periplasm, where they are then trapped. The periplasmic protein SilE of the Salmonella sp. strain selectively binds silver ions, which are then exported by the ATPase pumps SilCBA and SilP (Silver, 2003).
Smirnova (2005) claims that a variety of HM ions, such as chromate, molybdate, and vanadate, are reduced by bacteria from various biological niches. Some bacteria can use metals and metalloids as electron acceptors or donors to produce energy. During anaerobic respiration, bacteria can use oxidized metals as terminal electron acceptors, and enzymatic reduction of metal ions can result in the creation of less hazardous forms of HMs including mercury and chromium (Barkay et al., 2003; Viti et al., 2003).
The mer-operon, which imparts tolerance to mercury by encoding proteins like MerT and MerA, is one of the best-studied mechanisms for metal detoxification (Brown et al., 2002). Divalent mercury ions can enter the cell by the MerT transport protein and are then converted to elemental mercury by the intracellular MerA reductase.
According to Nies (2003), some plasmids or transposons that can be shared by different bacteria through horizontal gene transfer (HGT) contain genetic determinants of HM tolerance. Natural transformations occurring frequently, the isolation of plasmid- and transposons-bearing bacteria from a variety of environments, and the acquisition of new traits by autochthonous microbiota after exposure to plasmid-bearing bacteria are all evidence supporting the role of HGT in bacterial evolution under changing environmental conditions (Coombs and Barkay, 2005).
Bacterial plasmids were where metal resistance mechanisms were first discovered (Summers and Silver, 1972). The Ars operons on the chromosomes of E. coli, P. aeruginosa, and Bacillus subtilis, which structurally mimic plasmid-borne genetic determinants, are examples of metal resistance systems that are similar to those on plasmids (Mukhopadhyay et al., 2002). However, hazardous metal resistance genes are carried on plasmids while important metal ion homeostasis genes are typically found on chromosomes (Bruins et al., 2000; Cervantes et al., 2001).
The mer operon, independent of its position on the chromosome or plasmid, is a well-studied metal tolerance system that is found in diverse types of bacteria, according to Narita et al. (2003) and Reniero et al. (1998). merA, merT, merP, and merR make up the majority of the mer operon, while other genes like merB, merC, merD, merE, merF, and merG may also be present. According to Große et al. (2004), the czc operon on the PMOL30 plasmid of R. metallidurans CH34 encodes resistance to cadmium, zinc, and cobalt ions through the czcCBA genes as well as regulatory, promotor, and unknown functional genes czcN and czcI. According to Mergeay et al. (2003), R. metallidurans CH34 also harbors the PMOL28 mega plasmid, which encodes resistance to Co2+, Ni2+, and chromate as well as tolerance genes for Tl+ and Hg2+ and contains several previously unidentified metal resistance genes. ArsR, ArsA, ArsD, ArsB, and ArsC are just a few of the proteins that are regulated by ars operon and involved in arsenate tolerance, found in many different bacterial groups (Mukhopadhyay et al., 2002). Both Gram-positive (Oger et al., 2003) and Gram-negative (Lee et al., 2007) bacteria have the cop operon, which confers tolerance to copper ions. The cop operon and the cadmium tolerance cad system may differ in structure and location, as observed in various bacterial groups, such as E. hirae and Pseudomonas spp. (Bruins et al., 2000).
Researchers have proposed microbially induced carbonate precipitation (MICP) as a viable approach for environmentally friendly and sustainable bioremediation of metal contaminants (Dhami et al., 2013; Zhang et al., 2018). This mechanism, which involves the secretion of urease by microorganisms, offers a simple and controllable method to rapidly generate substantial amounts of carbonates. When urea is hydrolyzed by urease, CO2 and NH3 are produced, which subsequently react in the solution to produce ammonium, bicarbonate, and hydroxide. As a result, the pH increases (becomes more alkaline), and carbonate ions are formed. Under appropriate conditions of sufficient ionic activity and the presence of divalent cations, carbonate ions can precipitate out of the solution.
According to Yin et al. (2021), cadmium (Cd) and strontium (Sr), along with other metals and radionuclides, can undergo precipitation and create insoluble carbonate minerals of their own by following same pathway. Alternatively, these metals can be co-precipitated with calcium carbonate if the calcifying microorganisms are able to sustain in HM contaminated environment. Certain HM ions, such as Cu2+, Cd2+, Pb2+, Zn2+, and Sr2+, which have ion radii similar to that of Ca2+ can be integrated into the crystal lattice of CaCO3 through isomorphic substitution of Ca2+ or by penetrating crystal interstices or defects (Kim et al., 2021). As a result, the HMs transform from soluble state of ions into insoluble state, effectively prevent their release back into the environment. The wide-ranging application of MICP and its capacity to sequester HMs establish it as a feasible in situ remediation technique for HMs polluted sites. Additionally, its resilience to changes in redox potential in the surrounding environment contributes to its high effectiveness and long-term stability in bioremediation (Lauchnor et al., 2013).
Microorganisms survive via different mechanisms in the presence of HMs to resist metal toxicity as shown in Figure 2. Some commonly used strategies employed by microbes include extrusion, exopolysaccharide secretion, biotransformation, metallothionein as well as enzyme synthesis (Dixit et al., 2015). Furthermore, major mechanisms to resist HMs by microorganism involves several procedures like ion exchange, metal efflux pumps, metal oxidation, electrostatic interaction, methylation, redox process, metal-organic complexion, precipitation, exclusion by permeability barrier, metal ligand degradation, surface complexation, demethylation, metal sequestration, and bio surfactant production. Detoxification of metals by microbes can also be done by extracellular chemical precipitation, volatilization, and valence conversion (Ramasamy et al., 2006; Yang et al., 2015). Microbes have negatively charged groups on their cell surface like sulfonate, hydroxyl, alcohol, phosphoryl, carboxyl, thioether, sulfhydryl, amine, ester, and thiol implicated in metal adsorption (Gavrilescu, 2004). Few studies reported ability of bacteria to accumulate metals intracellularly such as sequestration of zinc, cadmium, and copper ions by cadmium-tolerant E. coli, P. putida strain (Hussain et al., 2022), and R. leguminosarum cells (Pereira et al., 2006).
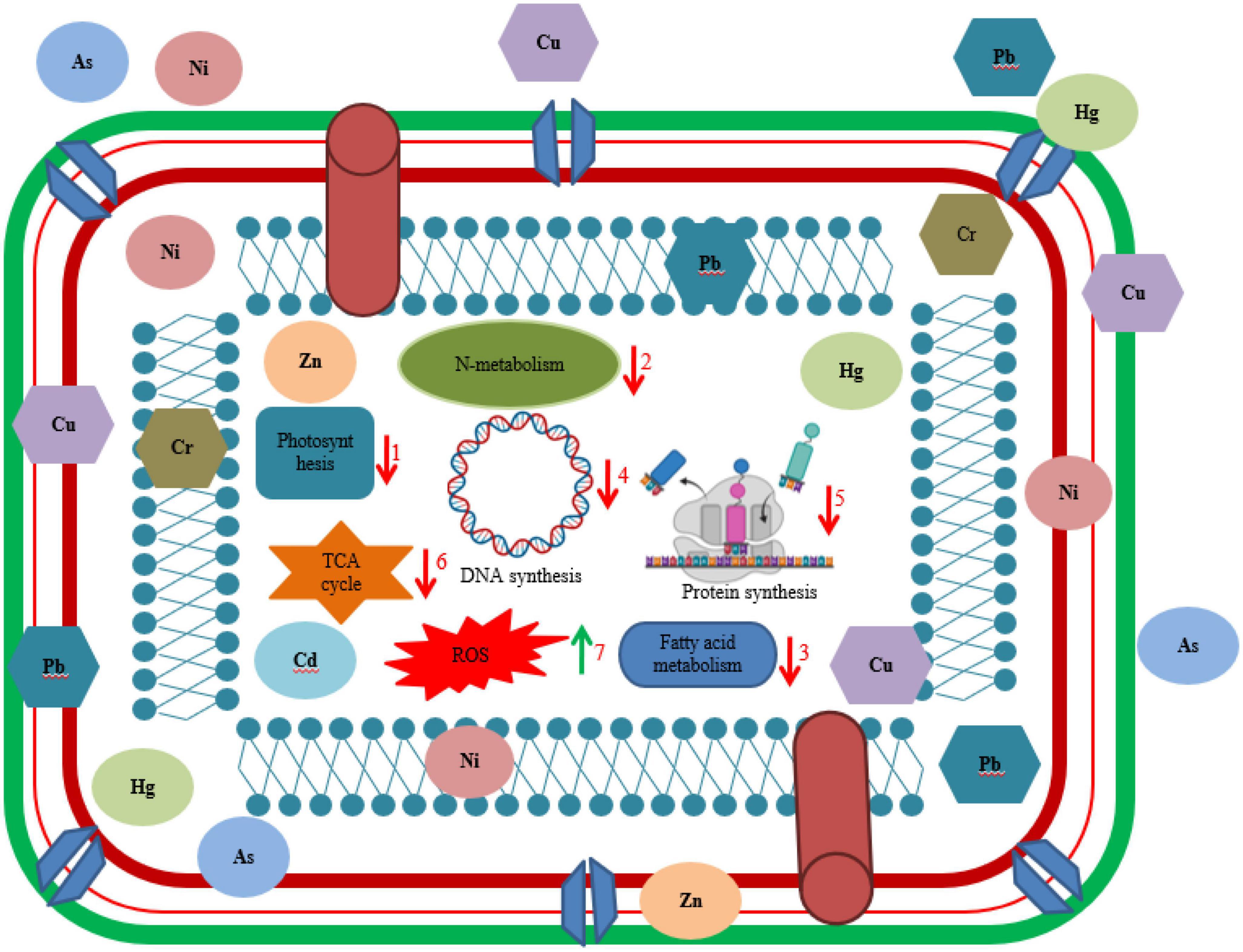
Figure 2. Illustration of interaction of living cell with heavy metals. Different biochemical dysfunctioning of cell occurred due to toxicity of heavy metal such as (1) decrease in rate of photosynthesis by inhibition of photosynthetic enzymes and chlorophyll biosynthesis, (2) significant decrease in nitrogen metabolism, (3) decreased rate of fatty acid metabolism, (4) disruption of DNA synthesis, (5) inhibition of protein synthesis, (6) negatively affecting key enzymes in TCA cycle, and (7) overproduction of reactive oxygen species (ROS).
Accumulation of metal ions extracellularly occurs by complexation as insoluble compounds or via cellular components in the periplasm. Several species belonging to Geobacter and Desulfuromonas are efficient to convert extremely hazardous metals to their benign forms. Some studies showing extracellular sequestration of HMs includes removal of copper ions by copper-resistant P. syringae, zinc ions by Synechocystis PCC 6803 strain, reduction of lethal forms of Manganese (IV), Uranium (VI), and Chromium (VI) to their non-toxic forms Manganese (II), Uranium (IV), and Chromium (III) by application of Geobacter metallireducens, a strict anaerobe (Igiri et al., 2018). Removal of cadmium (Cd) ions by Klebsiella planticola and P. aeruginosa as well as lead by the Vibrio harveyi strain via precipitation has been stated by several researchers (Sharma et al., 2000; Wang et al., 2002; Mire et al., 2004). Non-viable cells of Brevibacterium sp., P. putida, and Bacillus sp., showed effective biosorption of numerous HM ions (Green-Ruiz, 2006). Methylation by microbes play a crucial role in remediation of HMs. Methylated substances are frequently explosive; for example, Escherichia spp., Pseudomonas spp., Clostridium spp., and Bacillus spp., can bio methylate mercury (II) to gaseous methyl mercury. In polluted topsoil, bio methylation of arsenic, lead, and selenium to gaseous arsines, dimethyl lead and dimethyl selenide was observed, respectively (Ramasamy et al., 2006).
Bioaccumulation and biosorptive abilities of metal-resistant bacteria are used to remediate HM-contaminated environments (Kim et al., 2015). Several properties of bacteria such as, high surface-to-volume ratios, wide distribution, capacity to thrive in controlled environments, active chemisorption sites, size, and resistance to environmental conditions contribute to their robust biosorption potentiality (Srivastava et al., 2015; Mosa et al., 2016).
Bacteria and fungi are also reported to produces organic acids as a natural chelating agent for HMs (Seneviratne et al., 2017). Gluconic, acetic, oxalic, and malic acids are the most commonly reported organic acids for HM solubilization (Ullah et al., 2015; Gube, 2016). Out of single, consortium or immobilized forms of bacterial culture consortiums are observed to be more stable, metabolically superior, survive longer for metal biosorption and suited more for field use (Gu and Berry, 1991; Kader et al., 2007). Some studies demonstrating remediation of HM using bacteria includes removal of chromium (Cr) by consortia of Acinetobacter sp. and Arthrobacter sp. (De et al., 2008), cadmium, lead, and chromium removal in tannery effluent by mixed culture of B. megaterium, Penicillium sp., B. subtilis, and Aspergillus niger (Abioye et al., 2018), Pb removal by Micrococcus luteus (Puyen et al., 2012), removal of Cr6+, Cu, and Ni utilizing zeolite immobilized Desulfovibrio desulfuricans (Kim et al., 2015).
Biofilms are also proven as effective bioremediating agent, biological stabilizer as well as possess high tolerance against toxic compounds at lethal quantities. Biofilms act as biosorbents or secrete exo-polymeric substances having surfactant or emulsifier properties for remediation of HMs (El-Masry et al., 2004). Several issues faced due to metallic stress includes reduced soil microbial activity as well as crop output (Ahemad, 2012), oxidative stress, modification, and loss of protein functionality leading to growth impairment, browning of roots, photosystems inactivation, and chlorosis in plants (Shaw et al., 2004; Göhre and Paszkowski, 2006; Seth et al., 2008). PGPB are well known to have adapted mechanisms for removal of metal pollutants like oxidation–reduction, biosorption, complexation, and precipitation (Muñoz et al., 2006; Singh et al., 2010). The bacterial response to a specific HM in the cleanup of metal-contaminated locations is of significant use (Hemambika et al., 2011). Various studies reported use of PGPB as bioremediating agents such as remediation of copper toxicity by Azospirillum lipoferum (UAP154 and UAP40), R. leguminosarum CPMex46 and Pseudomonas fluorescens Avm, in alfalfa Medicago sativa seeds (Carrillo-Castañeda et al., 2005); nickel toxicity by Microbacterium arabinogalactanolyticum, Sphingomonas macrogoltabidus, and Microbacterium liquefaciens in Alyssum murale plants (Abou-Shanab et al., 2003). Furthermore, another study showed that hydroxamate siderophores produced by PGPB strains chelates HMs present in rhizospheric region of plants leading to inhibition of free radical formation and prevention of oxidative damage to plants (Dimkpa et al., 2009). Tripathi et al. (2005) reported growth promotion as well as inhibition of lead and cadmium toxicity by P. putida in Phaseolus vulgaris. Similarly, inoculation with Pseudomonas sp. Ps29C and B. megaterium Bm4C showed plant growth promotion and low nickel toxicity in Brassica juncea (Rajkumar and Freitas, 2008). Barzanti et al. (2007) found that bacteria aided plant growth under nickel stress, which is consistent with previous observations. In all, these investigations clearly demonstrated the potential of PGPB to improve plant biomass under HM stress. As a result, using metal detoxifying PGPB in conjunction with other plant growth-promoting activities can significantly improve the efficiency of the entire remediation process (Glick, 2012).
Rhizoremediation involves crucial role of rhizomicrobial population in detoxification process of HM in contaminated soils (Kuiper et al., 2004). These microbes exhibit high metabolic activity around the roots of plants. Bacterial population dominating HMs stressed sites includes Pseudomonas, Rhizobia, Arthrobacter, and Bacillus (Pires et al., 2017). One of the most well-known symbiotic associations found between legume and rhizobia is capable to remediate HM toxicity and improve the condition of contaminated soils (Checcucci et al., 2017). They have the potential to transform a wide range of metals and alter metal dissolution, toxicity, speciation, mobility, and degradation in soil (Gadd, 2010). Several reviews are available on metal–microbe’s interaction by Giller et al. (2009), Khan (2005), Gadd (2010), and Kong and Glick (2017). Interaction between microorganisms and metals in the rhizosphere is highly specific and influenced by physico-chemical properties of soil, soil type, metabolic activity, concentration, type of metal species, and microbial diversity.
Globalization and the technological improvement is achieving its height day-by-day for the convenience to the humankind. But its use is not done at an optimal level or judicially by the generation due to which its toxic components accumulating exponentially in the environment are affecting human health as well as agroecosystems by causing toxicity. Bioremediation using indigenous microorganisms from contaminated sites seems prudent, for exploiting their true potential. Several strategies including, oxidation, reduction, condensation, hydrolysis, isomerization, chelation, precipitation, complexation, immobilization, adsorption, bioaccumulation, and production of biosurfactant are expressed by microbial system for the removal or transformation of toxic HM into their non-toxic state in the contaminated sites. Preventive measures must be taken to avoid use of metal containing pesticides in agriculture and proper monitoring must be done before releasing metal containing effluents into water bodies. For future prospects, the use of genetically modified microorganisms could remove HM efficiently from the contaminated sites. Hence need to develop some other biological based techniques which efficiently work at contaminated sites and transformation of research from laboratory level to field or industrial level is also required.
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abioye, O. P., Oyewole, O. A., Oyeleke, S. B., Adeyemi, M. O., and Orukotan, A. A. (2018). Biosorption of lead, chromium and cadmium in tannery effluent using indigenous microorganisms. Braz. J. Biol. Sci. 5, 25–32. doi: 10.21472/bjbs.050903
Abou-Shanab, R. A., Angle, J. S., Delorme, T. A., Chaney, R. L., Van Berkum, P., Moawad, H., et al. (2003). Rhizobacterial effects on nickel extraction from soil and uptake by Alyssum murale. New Phytol. 158, 219–224. doi: 10.1046/j.1469-8137.2003.00721.x
Abrar, M., Hussain, Z., Akif, M., Sok, K., Muhammad, A., Khan, A., et al. (2011). Textile effluents and their contribution towards aquatic pollution in the Kabul River (Pakistan). J. Chem. Soc. Pak. 24, 106–111.
Adrees, M., Ali, S., Rizwan, M., Ibrahim, M., Abbas, F., Farid, M., et al. (2015). The effect of excess copper on growth and physiology of important food crops: A review. Environ. Sci. Pollut. Res. 22, 8148–8162. doi: 10.1007/s11356-015-4496-5
Agency for Toxic Substances and Disease Registry [ATSDR], (2015). Priority list of hazardous substances. Available online at: http://www.atsdr.cdc.gov/spl/index.html#modalIdString_myTable2015
Ahemad, M. (2012). Implication of bacterial resistance against heavy metals in bioremediation: A review. IIOAB J. 3, 39–46.
Ahemad, M. (2015). Enhancing phytoremediation of chromium-stressed soils through plant-growth-promoting bacteria. J. Genet. Eng. Biotechnol. 13, 51–58. doi: 10.1016/j.jgeb.2015.02.001
Ahmed, M. K., Parvin, E., Islam, M. M., Akter, M. S., Khan, S., and Al-Mamun, M. H. (2014). Lead-and cadmium-induced histopathological changes in gill, kidney and liver tissue of freshwater climbing perch Anabas testudineus (Bloch, 1792). Chem. Ecol. 30, 532–540. doi: 10.1080/02757540.2014.889123
Ahuekwe, E. F., Okoli, B. E., Stanley, H. O., and Kinigoma, B. (2016). “Evaluation of hydrocarbon emulsification and heavy metal detoxification potentials of sophorolipid biosurfactants produced from waste substrates using yeast and mushroom,” in SPE African health, safety, security, environment, and social responsibility conference and exhibition, (Richardson, TX: OnePetro), doi: 10.2118/183578-MS
Amorim, S. S., Ruas, F. A. D., Barboza, N. R., de Oliveira Neves, V. G., Leao, V. A., and Guerra-Sa, R. (2018). Manganese (Mn2+) tolerance and biosorption by Meyerozyma guilliermondii and Meyerozyma caribbica strains. J. Environ. Chem. Eng. 6, 4538–4545. doi: 10.1016/j.jece.2018.06.061
Anjum, S. A., Tanveer, M., Hussain, S., Shahzad, B., Ashraf, U., Fahad, S., et al. (2016). Osmoregulation and antioxidant production in maize under combined cadmium and arsenic stress. Environ. Sci. Pollut. Res. 23, 11864–11875. doi: 10.1007/s11356-016-6382-1
Ansari, F., Ahmad, A., and Rafatullah, M. (2023). Review on bioremediation technologies of polycyclic aromatic hydrocarbons (PAHs) from soil: Mechanisms and future perspective. Int. Biodeterior. Biodegrad. 179:105582. doi: 10.1016/j.ibiod.2023.105582
Arab, F., and Mulligan, C. N. (2018). An eco-friendly method for heavy metal removal from mine tailings. Environ. Sci. Pollut. Res. 25, 16202–16216. doi: 10.1007/s11356-018-1770-3
Asati, A., Pichhode, M., and Nikhil, K. (2016). Effect of heavy metals on plants: An overview. Int. J. Appl. Innov. Eng. Manag. 5, 56–66.
Asgher, M., Khan, M. I. R., Anjum, N. A., and Khan, N. A. (2015). Minimising toxicity of cadmium in plants—role of plant growth regulators. Protoplasma 252, 399–413. doi: 10.1007/s00709-014-0710-4
Ayangbenro, A. S., and Babalola, O. O. (2017). A new strategy for heavy metal polluted environments: A review of microbial biosorbents. Int. J. Environ. Health Res. 14:94. doi: 10.3390/ijerph14010094
Ayangbenro, A. S., and Babalola, O. O. (2020). Genomic analysis of Bacillus cereus NWUAB01 and its heavy metal removal from polluted soil. Sci. Rep. 10:19660. doi: 10.1038/s41598-020-75170-x
Bachman, G. R., and Miller, W. B. (1995). Iron chelate inducible iron/manganese toxicity in zonal geranium. J. Plant Nutr. 18, 1917–1929. doi: 10.1080/01904169509365033
Balestrasse, K. B., Benavides, M. P., Gallego, S. M., and Tomaro, M. L. (2003). Effect of cadmium stress on nitrogen metabolism in nodules and roots of soybean plants. Funct. Plant Biol. 30, 57–64. doi: 10.1071/FP02074
Barkay, T., Miller, S. M., and Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27, 355–384. doi: 10.1016/S0168-6445(03)00046-9
Barzanti, R., Ozino, F., Bazzicalupo, M., Gabbrielli, R., Galardi, F., Gonnelli, C., et al. (2007). Isolation and characterization of endophytic bacteria from the nickel hyperaccumulator plant Alyssum bertolonii. Microb. Ecol. 53, 306–316. doi: 10.1007/s00248-006-9164-3
Basak, G., and Das, N. (2014). Characterization of sophorolipid biosurfactant produced by Cryptococcus sp. VITGBN2 and its application on Zn(II) removal from electroplating wastewater. J. Environ. Biol. 35, 1087–94.
Bhaskar, P. V., and Bhosle, N. B. (2006). Bacterial extracellular polymeric substance (EPS): A carrier of heavy metals in the marine food-chain. Environ. Int. 32, 191–198. doi: 10.1016/j.envint.2005.08.010
Brown, N. L., Shih, Y. C., Leang, C., Glendinning, K. J., Hobman, J. L., and Wilson, J. R. (2002). Mercury transport and resistance. Biochem. Soc. Trans. 30, 715–718. doi: 10.1042/bst0300715
Bruins, M. R., Kapil, S., and Oehme, F. W. (2000). Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 45, 198–207. doi: 10.1006/eesa.1999.1860
Cakmak, I. (2000). Tansley Review No. 111 Possible roles of zinc in protecting plant cells from damage by reactive oxygen species. New Phytol. 146, 185–205. doi: 10.1046/j.1469-8137.2000.00630.x
Cargnelutti, D., Tabaldi, L. A., Spanevello, R. M., de Oliveira Jucoski, G., Battisti, V., Redin, M., et al. (2006). Mercury toxicity induces oxidative stress in growing cucumber seedlings. Chemosphere 65, 999–1006. doi: 10.1016/j.chemosphere.2006.03.037
Carrillo-Castañeda, G., Muñoz, J. J., and Peralta-Videa, J. R. (2005). A spectrophotometric method to determine the siderophore production by strains of fluorescent Pseudomonas in the presence of copper and iron. Microchem. J. 81, 35–40. doi: 10.1016/j.microc.2005.01.018
Cervantes, C., Campos-García, J., Devars, S., Gutiérrez-Corona, F., Loza-Tavera, H., Torres-Guzmán, J. C., et al. (2001). Interactions of chromium with microorganisms and plants. FEMS Microbiol. Rev. 25, 335–347. doi: 10.1111/j.1574-6976.2001.tb00581.x
Cha, J. S., and Cooksey, D. A. (1991). Copper resistance in Pseudomonas syringae mediated by periplasmic and outer membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 88, 8915–8919. doi: 10.1073/pnas.88.20.8915
Chatterjee, C., Gopal, R., and Dube, B. K. (2006). Physiological and biochemical responses of French bean to excess cobalt. J. Plant Nutr. 29, 127–136. doi: 10.1080/01904160500416513
Checcucci, A., Bazzicalupo, M., and Mengoni, A. (2017). “Exploiting nitrogen-fixing rhizobial symbionts genetic resources for improving phytoremediation of contaminated soils,” in Enhancing cleanup of environmental pollutants, eds N. Anjum, S. Gill, and N. Tuteja (Cham: Springer), 275–288. doi: 10.1007/978-3-319-55426-6_13
Chen, M., Xu, P., Zeng, G., Yang, C., Huang, D., and Zhang, J. (2015). Bioremediation of soils contaminated with polycyclic aromatic hydrocarbons, petroleum, pesticides, chlorophenols and heavy metals by composting: Applications, microbes and future research needs. Biotechnol. Adv. 33, 745–755. doi: 10.1016/j.biotechadv.2015.05.003
Cheung, K. H., and Gu, J. D. (2007). Mechanism of hexavalent chromium detoxification by microorganisms and bioremediation application potential: A review. Int. Biodeterior. Biodegr. 59, 8–15. doi: 10.1016/j.ibiod.2006.05.002
Chojnacka, K. (2010). Biosorption and bioaccumulation–the prospects for practical applications. Environ. Int. 36, 299–307. doi: 10.1016/j.envint.2009.12.001
Coombs, J. M., and Barkay, T. (2005). New findings on evolution of metal homeostasis genes: Evidence from comparative genome analysis of bacteria and archaea. Appl. Environ. Microbiol. 71, 7083–7091. doi: 10.1128/AEM.71.11.7083-7091.2005
da Rocha Junior, R. B., Meira, H. M., Almeida, D. G., Rufino, R. D., Luna, J. M., et al. (2019). Application of a low-cost biosurfactant in heavy metal remediation processes. Biodegradation 30, 215–233. doi: 10.1007/s10532-018-9833-1
D’amore, J. J., Al-Abed, S. R., Scheckel, K. G., and Ryan, J. A. (2005). Methods for speciation of metals in soils: A review. J. Environ. Qual. 34, 1707–1745. doi: 10.2134/jeq2004.0014
Das, P., Mukherjee, S., and Sen, R. (2009). Biosurfactant of marine origin exhibiting heavy metal remediation properties. Bioresour. Technol. 100, 4887–4890. doi: 10.1016/j.biortech.2009.05.028
De, J., Ramaiah, N., and Vardanyan, L. (2008). Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar. Biotechnol. 10, 471–477. doi: 10.1007/s10126-008-9083-z
DeForest, D. K., Brix, K. V., and Adams, W. J. (2007). Assessing metal bioaccumulation in aquatic environments: The inverse relationship between bioaccumulation factors, trophic transfer factors and exposure concentration. Aquat. Toxicol. 84, 236–246. doi: 10.1016/j.aquatox.2007.02.022
Dhami, N. K., Reddy, M. S., and Mukherjee, A. (2013). Biomineralization of calcium carbonates and their engineered applications: A review. Front. Microbiol. 4:314. doi: 10.3389/fmicb.2013.00314/full
Dhankhar, R., Sainger, P. A., and Sainger, M. (2012). Phytoextraction of zinc: Physiological and molecular mechanism. Soil Sediment Contam. Int. J. 21, 115–133. doi: 10.1080/15320383.2012.636983
Dimkpa, C. O., Merten, D., Svatoš, A., Büchel, G., and Kothe, E. (2009). Siderophores mediate reduced and increased uptake of cadmium by Streptomyces tendae F4 and sunflower (Helianthus annuus), respectively. J. Appl. Microbiol. 107, 1687–1696. doi: 10.1111/j.1365-2672.2009.04355.x
Dixit, R., Malaviya, D., Pandiyan, K., Singh, U. B., Sahu, A., Shukla, R., et al. (2015). Bioremediation of heavy metals from soil and aquatic environment: An overview of principles and criteria of fundamental processes. Sustainability 7, 2189–2212. doi: 10.3390/su7022189
El-Helow, E. R., Sabry, S. A., and Amer, R. M. (2000). Cadmium biosorption by a cadmium resistant strain of Bacillus thuringiensis: Regulation and optimization of cell surface affinity for metal cations. Biometals 13, 273–80. doi: 10.1023/A:1009291931258
El-Masry, M. H., El-Bestawy, E., and El-Adl, N. I. (2004). Bioremediation of vegetable oil and grease from polluted wastewater using a sand biofilm system. World J. Microbiol. Biotechnol. 20, 551–557. doi: 10.1023/B:WIBI.0000043162.17813.17
El-Meihy, R. M., Abou-Aly, H. E., Youssef, A. M., Tewfike, T. A., and El-Alkshar, E. A. (2019). Efficiency of heavy metals-tolerant plant growth promoting bacteria for alleviating heavy metals toxicity on sorghum. Environ. Exp. Bot. 162, 295–301. doi: 10.1016/j.envexpbot.2019.03.005
Elouzi, A. A., Akasha, A. A., Elgerbi, A. M., El-Baseir, M., and El Gammudi, B. A. (2012). Removal of heavy metals contamination by bio-surfactants (Rhamnolipids). J. Chem. Pharm. Res. 4, 4337–4341.
Engwa, G. A., Ferdinand, P. U., Nwalo, F. N., and Unachukwu, M. N. (2019). “Mechanism and health effects of heavy metal toxicity in humans,” in Poisoning in the modern world-new tricks for an old dog, eds O. Karcioglu and B. Arsian (London: IntechOpen Limited), 70–90.
Fashola, M. O., Ngole-Jeme, V. M., and Babalola, O. O. (2016). Heavy metal pollution from gold mines: Environmental effects and bacterial strategies for resistance. Int. J. Environ. Res. Public Health 13:1047. doi: 10.3390/ijerph13111047
Fischerová, Z., Tlustoš, P., Száková, J., and Šichorová, K. (2006). A comparison of phytoremediation capability of selected plant species for given trace elements. Environ. Pollut. 144, 93–100. doi: 10.1016/j.envpol.2006.01.005
Fontes, R. L. F., and Cox, F. R. (1998). Zinc toxicity in soybean grown at high iron concentration in nutrient solution. J. Plant Nutr. 21, 1723–1730. doi: 10.1080/01904169809365517
Foy, C. D., Chaney, R. T., and White, M. C. (1978). The physiology of metal toxicity in plants. Annu. Rev. Plant Physiol. 29, 511–566. doi: 10.1146/annurev.pp.29.060178.002455
Foy, C. D., Weil, R. R., and Coradetti, C. A. (1995). Differential manganese tolerances of cotton genotypes in nutrient solution. J. Plant Nutr. 18, 685–706. doi: 10.1080/01904169509364931
Franke, S., Grass, G., and Nies, D. H. (2001). The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology 147, 965–972. doi: 10.1099/00221287-147-4-965
Fulekar, M. H., Singh, A., and Bhaduri, A. M. (2009). Genetic engineering strategies for enhancing phytoremediation of heavy metals. Afr. J. Biotechnol. 8, 529–535.
Gadd, G. M. (2010). Metals, minerals and microbes: Geomicrobiology and bioremediation. Microbiology 156, 609–643. doi: 10.1099/mic.0.037143-0
Gangola, S., Bhatt, P., Joshi, S., Bhandari, N. S., Kumar, S., Prakash, O., et al. (2022a). “Isolation, Enrichment, and characterization of Fungi for the degradation of Organic Contaminants,” in Mycoremediation protocols, eds D. Udayanga, P. Bhatt, D. Manamgoda, and J. M. Saez (New York, NY: Springer), 1–11. doi: 10.1007/978-1-0716-2006-9_1
Gangola, S., Bhatt, P., Kumar, A. J., Bhandari, G., Joshi, S., Punetha, A., et al. (2022b). Biotechnological tools to elucidate the mechanism of pesticide degradation in the environment. Chemosphere 296:133916. doi: 10.1016/j.chemosphere.2022.133916
Gangola, S., Sharma, A., Joshi, S., Bhandari, G., Prakash, O., Govarthanan, M., et al. (2022c). Novel mechanism and degradation kinetics of pesticides mixture using Bacillus sp. strain 3C in contaminated sites. Pestic. Biochem. Phy. 181:104996. doi: 10.1016/j.pestbp.2021.104996
Gangola, S., Bhandari, G., Joshi, S., Sharma, A., Simsek, H., and Bhatt, P. (2023a). Esterase and ALDH dehydrogenase-based pesticide degradation by Bacillus brevis 1B from a contaminated environment. Environ. Res. 232:116332. doi: 10.1016/j.envres.2023.116332
Gangola, S., Joshi, S., Bhandari, G., Bhatt, P., Kumar, S., Bhandari, N. S., et al. (2023b). “Remediation of heavy metals by rhizospheric bacteria and their mechanism of detoxification,” in Advanced microbial technology for sustainable agriculture and environment, eds S. Gangola, S. Kumar, S. Joshi, and P. Bhatt (Cambridge, MA: Academic Press), 31–46. doi: 10.1016/B978-0-323-95090-9.00005-4
Gangola, S., Joshi, S., Kumar, S., Sharma, B., and Sharma, A. (2021). Differential proteomic analysis under pesticides stress and normal conditions in Bacillus cereus 2D. PLoS One 16:e0253106. doi: 10.1371/journal.pone.0253106
Gangola, S., Sharma, A., Bhatt, P., Khati, P., and Chaudhary, P. (2018). Presence of esterase and laccase in Bacillus subtilis facilitates biodegradation and detoxification of cypermethrin. Sci. Rep. 8:12755. doi: 10.1038/s41598-018-31082-5
Gao, L., and Gu, J. D. (2021). A new unified conceptual framework involving maintenance energy, metabolism and toxicity for research on degradation of organic pollutants. Int. Biodeterior. Biodegrad. 162:105253. doi: 10.1016/j.ibiod.2021.105253
Garbisu, C., and Alkorta, I. (2003). Basic concepts on heavy metal soil bioremediation. Eur. J. Min. Process. Environ. Protect. 3, 58–66.
Gautam, R. K., Sharma, S. K., Mahiya, S., and Chattopadhyaya, M. C. (2014). “Contamination of heavy metals in aquatic media: Transport, toxicity and technologies for remediation,” in Heavy metals in water: Presence, removal and safety, ed. S. K. Sharma (London: Royal Society of Chemistry), 1–24. doi: 10.1039/9781782620174-00001
Gavrilescu, M. (2004). Removal of heavy metals from the environment by biosorption. Eng. Life Sci. 4, 219–232. doi: 10.1002/elsc.200420026
Giller, K. E., Witter, E., and McGrath, S. P. (2009). Heavy metals and soil microbes. Soil Biol. Biochem. 41, 2031–2037. doi: 10.1016/j.soilbio.2009.04.026
Gilotra, U., and Srivastava, S. (1997). Plasmid-encoded sequestration of copper by Pseudomonas pickettii strain US321. Curr. Microbiol. 34, 378–381. doi: 10.1007/s002849900199
Glick, B. R. (2012). Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012:963401. doi: 10.6064/2012/963401
Gnanamani, A., Kavitha, V., Radhakrishnan, N., Rajakumar, G. S., Sekaran, G., and Mandal, A. B. (2010). Microbial products (biosurfactant and extracellular chromate reductase) of marine microorganism are the potential agents reduce the oxidative stress induced by toxic heavy metals. Colloids Surf. B Biointerfaces 79, 334–339. doi: 10.1016/j.colsurfb.2010.04.007
Göhre, V., and Paszkowski, U. (2006). Contribution of the arbuscular mycorrhizal symbiosis to heavy metal phytoremediation. Planta 223, 1115–1122.
Gomaa, E. Z., and El-Meihy, R. M. (2019). Bacterial biosurfactant from Citrobacter freundii MG812314.1 as a bioremoval tool of heavy metals from wastewater. Bull. Natl. Res. Cent. 43, 1–14. doi: 10.1186/s42269-019-0088-8
Gonzalez Henao, S., and Ghneim-Herrera, T. (2021). Heavy metals in soils and the remediation potential of bacteria associated with the plant microbiome. Front. Environ. Sci. 9:604216. doi: 10.3389/fenvs.2021.604216
Goswami, D., Thakker, J. N., and Dhandhukia, P. C. (2016). Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2:1127500. doi: 10.1080/23311932.2015.1127500
Govarthanan, M., Mythili, R., Selvankumar, T., Kamala-Kannan, S., Choi, D., and Chang, Y. C. (2017). Isolation and characterization of a biosurfactant-producing heavy metal resistant Rahnella sp. RM isolated from chromium-contaminated soil. Biotechnol. Bioprocess Eng. 22, 186–194. doi: 10.1007/s12257-016-0652-0
Green-Ruiz, C. (2006). Mercury (II) removal from aqueous solutions by nonviable Bacillus sp. from a tropical estuary. Bioresour. Technol. 97, 1907–1911. doi: 10.1016/j.biortech.2005.08.014
Große, C., Anton, A., Hoffmann, T., Franke, S., Schleuder, G., and Nies, D. H. (2004). Identification of a regulatory pathway that controls the heavy-metal resistance system Czc via promoter czcNp in Ralstonia metallidurans. Arch. Microbiol. 182, 109–118. doi: 10.1007/s00203-004-0670-8
Gu, J. D. (2003). Microbiological deterioration and degradation of synthetic polymeric materials: Recent research advances. Int. Biodeterior. Biodegrad. 52, 69–91. doi: 10.1016/S0964-8305(02)00177-4
Gu, J. D. (2014). Assessment of ecosystem health and ecotoxicology through chemical analysis and modeling. Ecotoxicology 23, 475–479. doi: 10.1007/s10646-014-1206-x
Gu, J. D. (2016). Biodegradation testing: So many tests but very little new innovation. Appl. Environ. Biotechnol. 1, 92–95. doi: 10.26789/AEB.2016.01.007
Gu, J. D. (2018). Bioremediation of toxic metals and metalloids for cleaning up from soils and sediments. Appl. Environ. Biotechnol. 3, 48–51. doi: 10.26789/AEB.2018.02.006
Gu, J. D. (2019). Microbial ecotoxicology as an emerging research subject. Appl. Environ. Biotechnol. 1, 1–4.
Gu, J. D. (2021). On environmental biotechnology of bioremediation. Appl. Environ. Biotechnol. 5, 3–8. doi: 10.26789/AEB.2019.01.001
Gu, J. D., and Berry, D. F. (1991). Degradation of substituted indoles by an indole-degrading methanogenic consortium. Appl. Environ. Microbiol. 57, 2622–2627. doi: 10.1128/aem.57.9.2622-2627.1991
Gu, J. D., and Pan, L. (2006). Comparing the growth characteristics of three bacteria involved in degrading rubbers. J. Polym. Environ. 14, 273–279. doi: 10.1007/s10924-006-0016-5
Gu, J. D., and Wang, Y. (2013). A new era for geomicrobial ecotoxicology in environmental science research. Int. Biodeterior. Biodegr. 85, 345–346. doi: 10.1016/j.ibiod.2012.06.024
Gube, M. (2016). “Fungal molecular response to heavy metal stress,” in Biochemistry and molecular biology, ed. D. Hoffmeister (Cham: Springer), 47–68. doi: 10.1007/978-3-319-27790-5_4
Gunes, A., Pilbeam, D. J., and Inal, A. (2009). Effect of arsenic–phosphorus interaction on arsenic-induced oxidative stress in chickpea plants. Plant Soil 314, 211–220. doi: 10.1007/s11104-008-9719-9
Guo, J., Dai, X., Xu, W., and Ma, M. (2008). Overexpressing GSH1 and AsPCS1 simultaneously increases the tolerance and accumulation of cadmium and arsenic in Arabidopsis thaliana. Chemosphere 72, 1020–1026. doi: 10.1016/j.chemosphere.2008.04.018
Gupta, A., Joia, J., Sood, A., Sood, R., Sidhu, C., and Kaur, G. (2016). Microbes as potential tool for remediation of heavy metals: A review. J. Microb. Biochem. Technol. 8, 364–372. doi: 10.4172/1948-5948.1000310
Habiba, U., Ali, S., Farid, M., Shakoor, M. B., Rizwan, M., Ibrahim, M., et al. (2015). EDTA enhanced plant growth, antioxidant defense system, and phytoextraction of copper by Brassica napus L. Environ. Sci. Pollut. Res. 22, 1534–1544. doi: 10.1007/s11356-014-3431-5
Haefeli, C., Franklin, C., and Hardy, K. (1984). Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine. J. Bacteriol. 158, 389–392. doi: 10.1128/jb.158.1.389-392.1984
Han, F. X., Su, Y., Monts, D. L., Waggoner, C. A., and Plodinec, M. J. (2006). Binding, distribution, and plant uptake of mercury in a soil from Oak Ridge, Tennessee, USA. Sci. Total Environ. 368, 753–768. doi: 10.1016/j.scitotenv.2006.02.026
Han, S. H., Lee, J. C., Oh, C. Y., and Kim, P. G. (2006). Alleviation of Cd toxicity by composted sewage sludge in Cd-treated Schmidt birch (Betula schmidtii) seedlings. Chemosphere 65, 541–546. doi: 10.1016/j.chemosphere.2006.02.049
He, L. M., and Tebo, B. M. (1998). Surface charge properties of and Cu (II) adsorption by spores of the marine Bacillus sp. strain SG-1. Appl. Environ. Microbiol. 64, 1123–1129. doi: 10.1128/AEM.64.3.1123-1129.1998
Hemambika, B., Rani, M. J., and Kannan, V. R. (2011). Biosorption of heavy metals by immobilized and dead fungal cells: A comparative assessment. J. Ecol. Nat. Environ. 3, 168–175.
Henriques, B., Rocha, L. S., Lopes, C. B., Figueira, P., Monteiro, R. J., Duarte, A. D. C., et al. (2015). Study on bioaccumulation and biosorption of mercury by living marine macroalgae: Prospecting for a new remediation biotechnology applied to saline waters. J. Chem. Eng. 281, 759–770. doi: 10.1016/j.cej.2015.07.013
Huang, W., and Liu, Z. M. (2013). Biosorption of Cd (II)/Pb (II) from aqueous solution by biosurfactant-producing bacteria: Isotherm kinetic characteristic and mechanism studies. Colloids Surf. B Biointerfaces 105, 113–119. doi: 10.1016/j.colsurfb.2012.12.040
Hussain, S., Khan, M., Sheikh, T. M. M., Mumtaz, M. Z., Chohan, T. A., Shamim, S., et al. (2022). Zinc essentiality, toxicity, and its bacterial bioremediation: A comprehensive insight. Front. Microbiol. 13:900740. doi: 10.3389/fmicb.2022.900740
Igiri, B. E., Okoduwa, S. I., Idoko, G. O., Akabuogu, E. P., Adeyi, A. O., and Ejiogu, I. K. (2018). Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. J. Toxicol. 2018:2568038. doi: 10.1155/2018/2568038
Irawati, W., Parhusip, A. J., and Sopiah, N. (2015). Heavy metals biosorption by copper resistant bacteria of Acinetobacter Sp. IrC2. Microbiol. Indonesia 9, 163–170. doi: 10.5454/mi.9.4.4
Islam, M. S., Hossain, M. B., Matin, A., and Sarker, M. S. I. (2018). Assessment of heavy metal pollution, distribution and source apportionment in the sediment from Feni River estuary, Bangladesh. Chemosphere 202, 25–32. doi: 10.1016/j.chemosphere.2018.03.077
Ivanova, E. P., Kurilenko, V. V., Kurilenko, A. V., Gorshkova, N. M., Shubin, F. N., Nicolau, D. V., et al. (2002). Tolerance to cadmium of free-living and associated with marine animals and eelgrass marine gamma-proteobacteria. Curr. Microbiol. 44, 357–362. doi: 10.1007/s00284-001-0017-5
Iyer, A., Mody, K., and Jha, B. (2005). Biosorption of heavy metals by a marine bacterium. Mar. Pollut. Bull. 50, 340–343. doi: 10.1016/j.marpolbul.2004.11.012
Jiang, Y., Yang, K., Wang, H., Shang, Y., and Yang, X. (2015). Characteristics of phenol degradation in saline conditions of a halophilic strain JS3 isolated from industrial activated sludge. Mar. Pollut. Bull. 99, 230–234. doi: 10.1016/j.marpolbul.2015.07.021
Jin, Y., Luan, Y., Ning, Y., and Wang, L. (2018). Effects and mechanisms of microbial remediation of heavy metals in soil: A critical review. Appl. Sci. 8:1336. doi: 10.3390/app8081336
Jing, Y. D., He, Z. L., and Yang, X. E. (2007). Role of soil rhizobacteria in phytoremediation of heavy metal contaminated soils. J. Zhejiang Univ. Sci. B. 8, 192–207. doi: 10.1631/jzus.2007.B0192
Joshi, S., Gangola, S., Jaggi, V., and Sahgal, M. (2023a). Functional characterization and molecular fingerprinting of potential phosphate solubilizing bacterial candidates from Shisham rhizosphere. Sci. Rep. 13:7003. doi: 10.1038/s41598-023-33217-9
Joshi, S., Gangola, S., Rani, A., Sahgal, M., Tewari, S., Bhandari, N. S., et al. (2023b). “Recent molecular and omics approaches to study rhizosphere functioning,” in Advanced microbial technology for sustainable agriculture and environment, eds S. Gangola, S. Kumar, S. Joshi, and P. Bhatt (Cambridge, MA: Academic Press), 1–13. doi: 10.1016/B978-0-323-95090-9.00009-1
Juwarkar, A. A., Nair, A., Dubey, K. V., Singh, S. K., and Devotta, S. (2007). Biosurfactant technology for remediation of cadmium and lead contaminated soils. Chemosphere 68, 1996–2002. doi: 10.1016/j.chemosphere.2007.02.027
Kader, J., Sannasi, P., Othman, O., Ismail, B. S., and Salmijah, S. (2007). Removal of Cr (VI) from aqueous solutions by growing and non-growing populations of environmental bacterial consortia. Glob. J. Environ. Res. 1, 12–17.
Kamaludeen, S. P. B., and Ramasamy, K. (2008). Rhizoremediation of metals: Harnessing microbial communities. Indian J. Microbiol. 48, 80–88. doi: 10.1007/s12088-008-0008-3
Kazy, S. K., Sar, P., Singh, S. P., Sen, A. K., and D’souza, S. (2002). Extracellular polysaccharides of a copper-sensitive and a copper-resistant Pseudomonas aeruginosa strain: Synthesis, chemical nature and copper binding. World J. Microbiol. Biotechnol. 18, 583–588. doi: 10.1023/A:1016354713289
Khan, A. G. (2005). Role of soil microbes in the rhizospheres of plants growing on trace metal contaminated soils in phytoremediation. J. Trace. Elem. Med. Biol. 18, 355–364. doi: 10.1016/j.jtemb.2005.02.006
Khan, M. I. R., Iqbal, N., Masood, A., Mobin, M., Anjum, N. A., and Khan, N. A. (2016). Modulation and significance of nitrogen and sulfur metabolism in cadmium challenged plants. Plant Growth Regul. 78, 1–11. doi: 10.1007/s10725-015-0071-9
Kim, I. H., Choi, J. H., Joo, J. O., Kim, Y. K., Choi, J. W., and Oh, B. K. (2015). Development of a microbe-zeolite carrier for the effective elimination of heavy metals from seawater. J. Microbiol. Biotechnol. 25, 1542–1546. doi: 10.4014/jmb.1504.04067
Kim, Y., Kwon, S., and Roh, Y. (2021). Effect of divalent cations (Cu, Zn, Pb, Cd, and Sr) on microbially induced calcium carbonate precipitation and mineralogical properties. Front. Microbiol. 12:763. doi: 10.3389/fmicb.2021.646748
Kisielowska, E., Hołda, A., and Niedoba, T. (2010). Removal of heavy metals from coal medium with application of biotechnological methods. Górnictwoi Geoinżynieria 34, 93–104.
Kitao, M., Lei, T. T., and Koike, T. (1997). Effects of manganese toxicity on photosynthesis of white birch (Betula platyphylla var. japonica) seedlings. Physiol. Plant. 101, 249–256. doi: 10.1111/j.1399-3054.1997.tb0099
Klaus, T., Joerger, R., Olsson, E., and Granqvist, C. G. (1999). Silver-based crystalline nanoparticles, microbially fabricated. Proc. Natl. Acad. Sci. U.S.A. 96, 13611–13614. doi: 10.1073/pnas.96.24.1361
Knotek-Smith, H. M., Deobald, L. A., Ederer, M., and Crawford, D. L. (2003). Cadmium stress studies: Media development, enrichment, consortia analysis, and environmental relevance. Biometals 16, 251–261. doi: 10.1023/A:1020617013927
Kong, Z., and Glick, B. R. (2017). The role of plant growth-promoting bacteria in metal phytoremediation. Adv. Microb. Physiol. 71, 97–132. doi: 10.1016/bs.ampbs.2017.04.001
Kuiper, I., Lagendijk, E. L., Bloemberg, G. V., and Lugtenberg, B. J. (2004). Rhizoremediation: A beneficial plant-microbe interaction. Mol. Plant Microbe Interact. 17, 6–15. doi: 10.1094/MPMI.2004.17.1.6
Kumar Mishra, G. (2017). Microbes in heavy metal remediation: A review on current trends and patents. Recent Pat. Biotechnol. 11, 188–196. doi: 10.2174/1872208311666170120121025
Kumar, S., Dubey, R. S., Tripathi, R. D., Chakrabarty, D., and Trivedi, P. K. (2015). Omics and biotechnology of arsenic stress and detoxification in plants: Current updates and prospective. Environ. Int. 74, 221–230. doi: 10.1016/j.envint.2014.10.019
Lasat, M. M. (2002). Phytoextraction of toxic metals, a review of biological mechanisms. J. Environ. Qual. 31, 109–120. doi: 10.2134/jeq2002.1090
Lauchnor, E. G., Schultz, L. N., Bugni, S., Mitchell, A. C., Cunningham, A. B., and Gerlach, R. (2013). Bacterially induced calcium carbonate precipitation and strontium coprecipitation in a porous media flow system. Environ. Sci. Technol. 47, 1557–1564. doi: 10.1021/es304240y
Le, T. T., Son, M. H., Nam, I. H., Yoon, H., Kang, Y. G., and Chang, Y. S. (2017). Transformation of hexabromocyclododecane in contaminated soil in association with microbial diversity. J. Hazard. Mater. 325, 82–89. doi: 10.1016/j.jhazmat.2016.11.058
Lee, S., Kim, Y. Y., Lee, Y., and An, G. (2007). Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 145, 831–842. doi: 10.1104/pp.107.102236
Li, C., Zhou, K., Qin, W., Tian, C., Qi, M., Yan, X., et al. (2019). A review on heavy metals contamination in soil: Effects, sources, and remediation techniques. Soil Sediment Contam. 28, 380–394. doi: 10.1080/15320383.2019.1592108
Lima, A. I. G., Corticeiro, S. C., and de Almeida Paula Figueira, E. M. (2006). Glutathione-mediated cadmium sequestration in Rhizobium leguminosarum. Enzyme Microb. Technol. 39, 763–769. doi: 10.1016/j.enzmictec.2005.12.009
Lombi, E., and Gerzabek, M. H. (1998). Determination of mobile heavy metal fraction in soil: Results of a pot experiment with sewage sludge. Commun. Soil Sci. Plant Anal. 29, 2545–2556. doi: 10.1080/00103629809370133
Lopes, C. S. C., Teixeira, D. B., Braz, B. F., Santelli, R. E., de Castilho, L. V. A., Gomez, J. G. C., et al. (2021). Application of rhamnolipid surfactant for remediation of toxic metals of long-and short-term contamination sites. Int. J. Environ. Sci. Technol. 18, 575–588. doi: 10.1007/s13762-020-02889-5
Luna, J. M., Rufino, R. D., and Sarubbo, L. A. (2016). Biosurfactant from Candida sphaerica UCP0995 exhibiting heavy metal remediation properties. Process Saf. Environ. Prot. 102, 558–566. doi: 10.1016/j.psep.2016.05.010
Ma, Y., Rajkumar, M., Zhang, C., and Freitas, H. (2016). Beneficial role of bacterial endophytes in heavy metal phytoremediation. J. Environ. Manage. 174, 14–25. doi: 10.1016/j.jenvman.2016.02.047
Mackay, A. K., Taylor, M. P., Munksgaard, N. C., Hudson-Edwards, K. A., and Burn-Nunes, L. (2013). Identification of environmental lead sources and pathways in a mining and smelting town: Mount Isa, Australia. Environ. Pollut. 180, 304–311. doi: 10.1016/j.envpol.2013.05.007
MacNaughton, S. J., Stephen, J. R., Venosa, A. D., Davis, G. A., Chang, Y. J., and White, D. C. (1999). Microbial population changes during bioremediation of an experimental oil spill. Appl. Environ. Microbiol. 65, 3566–3574. doi: 10.1128/AEM.65.8.3566-3574.1999
Mahboob, S., Al-Balwai, H. F. A., Al-Misned, F., and Ahmad, Z. (2014). Investigation on the genotoxicity of mercuric chloride to freshwater Clarias gariepinus. Pak. Vet. J. 34, 100–103.
Malik, A. (2004). Metal bioremediation through growing cells. Environ. Int. 30, 261–278. doi: 10.1016/j.envint.2003.08.001
Mandal, S. M., Gouri, S. S., De, D., Das, B. K., Mondal, K. C., and Pati, B. R. (2011). Effect of arsenic on nodulation and nitrogen fixation of blackgram (Vigna mungo). Indian J. Microbiol. 51, 44–47. doi: 10.1007/s12088-011-0080-y
Marchenko, A. M., Pshinko, G. N., Demchenko, V. Y., and Goncharuk, V. V. (2015). Leaching heavy metal from deposits of heavy metals with bacteria oxidizing elemental sulphur. J. Water Chem. Technol. 37, 311–316. doi: 10.3103/S1063455X15060090
Mathivanan, K., Chandirika, J. U., Vinothkanna, A., Yin, H., Liu, X., and Meng, D. (2021). Bacterial adaptive strategies to cope with metal toxicity in the contaminated environment–A review. Ecotoxicol. Environ. Saf. 226:112863. doi: 10.1016/j.ecoenv.2021.112863
McEldowney, S. (2000). The impact of surface attachment on cadmium accumulation by Pseudomonas fluorescens H2. FEMS Microbiol. Ecol. 33, 121–128. doi: 10.1111/j.1574-6941.2000.tb00734.x
Md Badrul Hisham, N. H., Ibrahim, M. F., Ramli, N., and Abd-Aziz, S. (2019). Production of biosurfactant produced from used cooking oil by Bacillus sp. HIP3 for heavy metals removal. Molecules 24:2617. doi: 10.3390/molecules24142617
Mergeay, M., Monchy, S., Vallaeys, T., Auquier, V., Benotmane, A., Bertin, P., et al. (2003). Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: Towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27, 385–410. doi: 10.1016/S0168-6445(03)00045-7
Mire, C. E., Tourjee, J. A., O’Brien, W. F., Ramanujachary, K. V., and Hecht, G. B. (2004). Lead precipitation by Vibrio harveyi: Evidence for novel quorum-sensing interactions. Appl. Environ. Microbiol. 70, 855–864. doi: 10.1128/AEM.70.2.855-864.2004
Mishra, A., and Malik, A. (2013). Recent advances in microbial metal bioaccumulation. Crit. Rev. Environ. Sci. Technol. 43, 1162–1222. doi: 10.1080/10934529.2011.627044
Mithoefer, A., Schulze, B., and Boland, W. (2004). Biotic and heavy metal stress in plants: Evidence for common signals. FEBS Lett. 566, 1–5. doi: 10.1016/j.febslet.2004.04.011
Mohan, A. K., Martis, S., Chiplunkar, S., Kamath, S., Goveas, L. C., and Rao, C. V. (2019). Heavy metal tolerance of Klebsiella pneumoniae Kpn555 isolated from coffee pulp waste. Borneo J. Resour. Sci. Technol. 9, 101–106.
Mosa, K. A., Saadoun, I., Kumar, K., Helmy, M., and Dhankher, O. P. (2016). Potential biotechnological strategies for the cleanup of heavy metals and metalloids. Front. Plant Sci. 7:303. doi: 10.3389/fpls.2016.00303
Mukhopadhyay, R., Rosen, B. P., Phung, L. T., and Silver, S. (2002). Microbial arsenic: From geocycles to genes and enzymes. FEMS Microbiol. Rev. 26, 311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x
Mulligan, C. N. (2009). Recent advances in the environmental applications of biosurfactants. Curr. Opin. Colloid Interface Sci. 14, 372–378. doi: 10.1016/j.cocis.2009.06.005
Mulligan, C. N., Yong, R. N., and Gibbs, B. F. (2001). Heavy metal removal from sediments by biosurfactants. J. Hazard. Mater. 85, 111–125. doi: 10.1016/S0304-3894(01)00224-2
Muñoz, R., Alvarez, M. T., Muñoz, A., Terrazas, E., Guieysse, B., and Mattiasson, B. (2006). Sequential removal of heavy metals ions and organic pollutants using an algal-bacterial consortium. Chemosphere 63, 903–911. doi: 10.1016/j.chemosphere.2005.09.062
Musa, O. K., Shaibu, M. M., and Kudamnya, E. A. (2013). Heavy metal concentration in groundwater around Obajana and its environs, Kogi State, North Central Nigeria. Am. Int. J. Contemp. Res. 3, 170–177.
Nakazawa, R., Kameda, Y., Ito, T., Ogita, Y., Michihata, R., and Takenaga, H. (2004). Selection and characterization of nickel-tolerant tobacco cells. Biol. Plant. 48, 497–502. doi: 10.1023/B:BIOP.0000047143.23646.18
Narita, M., Chiba, K., Nishizawa, H., Ishii, H., Huang, C. C., Kawabata, Z. I., et al. (2003). Diversity of mercury resistance determinants among Bacillus strains isolated from sediment of Minamata Bay. FEMS Microbiol. Lett. 223, 73–82. doi: 10.1016/S0378-1097(03)00325-2
Naskar, A., Majumder, R., and Goswami, M. (2020). Bioaccumulation of Ni (II) on growing cells of Bacillus sp.: Response surface modeling and mechanistic insight. Environ. Technol. Innov. 20:101057. doi: 10.1016/j.eti.2020.101057
Ngo, T. L. P. (2007). The affect of water and soil environment to vegetable quality. Vietnam J. Agric. Rural Dev. 17, 15–20.
Nies, D. H. (2000). Heavy metal-resistant bacteria as extremophiles: Molecular physiology and biotechnological use of Ralstonia sp. CH34. Extremophiles 4, 77–82. doi: 10.1007/s007920050140
Nies, D. H. (2003). Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339. doi: 10.1016/S0168-6445(03)00048-2
Oger, C., Mahillon, J., and Petit, F. (2003). Distribution and diversity of a cadmium resistance (cadA) determinant and occurrence of IS 257 insertion sequences in Staphylococcal bacteria isolated from a contaminated estuary (Seine, France). FEMS Microbiol. Ecol. 43, 173–183. doi: 10.1111/j.1574-6941.2003.tb01056.x
Pacle Decena, S. C., Sanita Arguelles, M., and Liporada Robel, L. (2018). Assessing heavy metal contamination in surface sediments in an urban river in the Philippines. Pol. J. Environ. Stud. 27, 1983–95. doi: 10.15244/pjoes/75204
Pande, V., Pandey, S. C., Sati, D., Bhatt, P., and Samant, M. (2022). Microbial interventions in bioremediation of heavy metal contaminants in agroecosystem. Front. Microbiol. 13:824084. doi: 10.3389/fmicb.2022.824084
Pandey, G., and Madhuri, S. (2014). Heavy metals causing toxicity in animals and fishes. Res. J. Anim. Vet. Fish. Sci. 2, 17–23.
Pardo, R., Herguedas, M., Barrado, E., and Vega, M. (2003). Biosorption of cadmium, copper, lead and zinc by inactive biomass of Pseudomonas putida. Anal. Bioanal. Chem. 376, 26–32. doi: 10.1007/s00216-003-1843-z
Pastor, J., Hernández, A. J., Prieto, N., and Fernández-Pascual, M. (2003). Accumulating behaviour of Lupinus albus L. growing in a normal and a decalcified calcic luvisol polluted with Zn. J. Plant Physiol. 160, 1457–1465. doi: 10.1078/0176-1617-01007
Peralta-Videa, J. R., Lopez, M. L., Narayan, M., Saupe, G., and Gardea-Torresdey, J. (2009). The biochemistry of environmental heavy metal uptake by plants: Implications for the food chain. Int. J. Biochem. Cell Biol. 41, 1665–1677. doi: 10.1016/j.biocel.2009.03.005
Pereira, S. I. A., Lima, A. I. G., and Figueira, E. M. D. A. P. (2006). Heavy metal toxicity in Rhizobium leguminosarum biovar viciae isolated from soils subjected to different sources of heavy-metal contamination: Effects on protein expression. Appl. Soil Ecol. 33, 286–293. doi: 10.1016/j.apsoil.2005.10.002
Peters, R. W. (1999). Chelant extraction of heavy metals from contaminated soils. J. Hazard. Mater. 66, 151–210.
Pinto, E., Sigaud-kutner, T. C., Leitao, M. A., Okamoto, O. K., Morse, D., and Colepicolo, P. (2003). Heavy metal–induced oxidative stress in algae 1. J. Phycol. 39, 1008–1018. doi: 10.1111/j.0022-3646.2003.02-193.x
Pires, C., Franco, A. R., Pereira, S. I., Henriques, I., Correia, A., Magan, N., et al. (2017). Metal (loid)-contaminated soils as a source of culturable heterotrophic aerobic bacteria for remediation applications. Geomicrobiol. J. 34, 760–768. doi: 10.1080/01490451.2016.1261968
Poulsen, T. G., and Bester, K. (2010). Organic micropollutant degradation in sewage sludge during composting under thermophilic conditions. Environ. Sci. Technol. 44, 5086–5091. doi: 10.1021/es9038243
Puyen, Z. M., Villagrasa, E., Maldonado, J., Diestra, E., Esteve, I., and Solé, A. (2012). Biosorption of lead and copper by heavy-metal tolerant Micrococcus luteus DE2008. Bioresour. Technol. 126, 233–237. doi: 10.1016/j.biortech.2012.09.036
Qazilbash, A. A. (2004). Isolation and characterization of heavy metal tolerant biota from industrially polluted soils and their role in bioremediation. Islamabad: Quaid-i-Azam University.
Qi, X., Xu, X., Zhong, C., Jiang, T., Wei, W., and Song, X. (2018). Removal of cadmium and lead from contaminated soils using sophorolipids from fermentation culture of Starmerella bombicola CGMCC 1576 fermentation. Int. J. Environ. Res. Public Health 15:2334. doi: 10.3390/ijerph15112334
Rahman, Z., and Singh, V. P. (2019). The relative impact of toxic heavy metals (THMs)(arsenic (As), cadmium (Cd), chromium (Cr)(VI), mercury (Hg), and lead (Pb)) on the total environment: An overview. Environ. Monit. Assess. 191, 1–21. doi: 10.1007/s10661-019-7528-7
Rahman, Z., Thomas, L., and Singh, V. P. (2019). Biosorption of heavy metals by a lead (Pb) resistant bacterium, Staphylococcus hominis strain AMB-2. J. Basic Microbiol. 59, 477–486. doi: 10.1002/jobm.201900024
Rajaei, G., Mansouri, B., Jahantigh, H., and Hamidian, A. H. (2012). Metal concentrations in the water of Chah nimeh reservoirs in Zabol, Iran. Bull. Environ. Contam. Toxicol. 89, 495–500. doi: 10.1007/s00128-012-0738-0
Rajkumar, M., and Freitas, H. (2008). Effects of inoculation of plant-growth promoting bacteria on Ni uptake by Indian mustard. Bioresour. Technol. 99, 3491–3498. doi: 10.1016/j.biortech.2007.07.046
Ramasamy, V., Murugesan, S., Mullainathan, S., and Chaparro, M. A. E. (2006). Magnetic characterization of recently excavated sediments of Cauvery river, Tamilnadu, India. Pollut. Res. 25:357.
Rani, A., and Goel, R. (2009). “Strategies for crop improvement in contaminated soils using metal-tolerant bioinoculants,” in Microbial strategies for crop improvement, eds M. Khan, A. Zaidi, and J. Musarrat (Berlin: Springer), 85–104. doi: 10.1007/978-3-642-01979-1_5
Raskin, I., Kumar, P. N., Dushenkov, S., and Salt, D. E. (1994). Bioconcentration of heavy metals by plants. Curr. Opin. Biotechnol. 5, 285–290.
Ravindran, A., Sajayan, A., Priyadharshini, G. B., Selvin, J., and Kiran, G. S. (2020). Revealing the efficacy of thermostable biosurfactant in heavy metal bioremediation and surface treatment in vegetables. Front. Microbiol. 11:222. doi: 10.3389/fmicb.2020.00222
Reniero, D., Mozzon, E., Galli, E., and Barbieri, P. (1998). Two aberrant mercury resistance transposons in the Pseudomonas stutzeri plasmid pPB. Gene 208, 37–42. doi: 10.1016/S0378-1119(97)00641-0
Rensing, C., Ghosh, M., and Rosen, B. P. (1999). Families of soft-metal-ion-transporting ATPases. J. Bacteriol. 181, 5891–5897. doi: 10.1128/jb.181.19.5891-5897.1999
Rezania, S., Taib, S. M., Din, M. F. M., Dahalan, F. A., and Kamyab, H. (2016). Comprehensive review on phytotechnology: Heavy metals removal by diverse aquatic plants species from wastewater. J. Hazard. Mater. 318, 587–599. doi: 10.1016/j.jhazmat.2016.07.053
Richau, J. A., Choquenet, D., Fialho, A. M., and Sá-Correia, I. (1997). Emergence of Cu++-tolerant mutants defective in gellan synthesis in Cu++-stressed cultures of Sphingomonas paucimobilis. Res. Microbiol. 148, 251–261. doi: 10.1016/S0923-2508(97)85245-X
Roane, T. M., and Pepper, I. L. (2000). “Microorganisms and metal pollution,” in Environmental Microbiology, eds R. Maier, I. Pepper, and C. Gerba (London: Academic Press), 403–423.
Rufino, R. D., Luna, J. M., Campos-Takaki, G. M., Ferreira, S. R., and Sarubbo, L. A. (2012). Application of the biosurfactant produced by Candida lipolytica in the remediation of heavy metals. Chem. Eng. 27, 61–66. doi: 10.3303/CET1227011
Sabiha-Javied, Mehmood, T., Chaudhry, M. M., Tufail, M., and Irfan, N. (2009). Heavy metal pollution from phosphate rock used for the production of fertilizer in Pakistan. Microchem. J. 91, 94–99.
Sahmoune, M. N. (2019). Evaluation of thermodynamic parameters for adsorption of heavy metals by green adsorbents. Environ. Chem. Lett. 17, 697–704.
Sankarammal, M., Thatheyus, A., and Ramya, D. (2014). Bioremoval of cadmium using Pseudomonas fluorescens. Open J. Water. Pollut. Treat. 1, 92–100. doi: 10.15764/WPT.2014.02010
Saranya, K., Sundaramanickam, A., Shekhar, S., Meena, M., Sathishkumar, R. S., and Balasubramanian, T. (2018). Biosorption of multi-heavy metals by coral associated phosphate solubilising bacteria Cronobacter muytjensii KSCAS2. J. Environ. Manag. 222, 396–401. doi: 10.1016/j.jenvman.2018.05.083
Saxena, D., Joshi, N., and Srivastava, S. (2002). Mechanism of copper resistance in a copper mine isolate Pseudomonas putida strain S4. Curr. Microbiol. 45, 0410–0414. doi: 10.1007/s00284-002-3787-5
Sayqal, A., and Ahmed, O. B. (2021). Advances in heavy metal bioremediation: An overview. Appl. Bionics Biomech. 2021:1609149.
Seneviratne, M., Seneviratne, G., Madawala, H. M. S. P., and Vithanage, M. (2017). “Role of rhizospheric microbes in heavy metal uptake by plants,” in Agro-environmental sustainability, eds J. Singh and G. Seneviratne (Cham: Springer), 147–163. doi: 10.1007/978-3-319-49727-3_8
Seth, C. S., Chaturvedi, P. K., and Misra, V. (2008). The role of phytochelatins and antioxidants in tolerance to Cd accumulation in Brassica juncea L. Ecotoxicol. Environ. Saf. 71, 76–85. doi: 10.1016/j.ecoenv.2007.10.030
Sethy, S. K., and Ghosh, S. (2013). Effect of heavy metals on germination of seeds. Nat. Sci. Biol. Med. 4, 272–275.
Sharma, P. K., Balkwill, D. L., Frenkel, A., and Vairavamurthy, M. A. (2000). A new Klebsiella planticola strain (Cd-1) grows anaerobically at high cadmium concentrations and precipitates cadmium sulfide. Appl. Environ. Microbiol. 66, 3083–3087. doi: 10.1128/AEM.66.7.3083-3087.2000
Shaw, B. P., Sahu, S. K., and Mishra, R. K. (2004). “Heavy metal induced oxidative damage in terrestrial plants,” in Heavy metal stress in plants, ed. M. N. V. Prasad (Berlin: Springer), 84–126. doi: 10.1007/978-3-662-07743-6_4
Shukla, S. K., Hariharan, S., and Rao, T. S. (2020). Uranium bioremediation by acid phosphatase activity of Staphylococcus aureus biofilms: Can a foe turn a friend? J. Hazard. Mater. 384:121316. doi: 10.1016/j.jhazmat.2019.121316
Siddiquee, S., Rovina, K., Azad, S. A., Naher, L., Suryani, S., and Chaikaew, P. (2015). Heavy metal contaminants removal from wastewater using the potential filamentous fungi biomass: A review. J. Microb. Biochem. Technol. 7, 384–395. doi: 10.4172/1948-5948.1000243
Silver, S. (2003). Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 27, 341–353. doi: 10.1016/S0168-6445(03)00047-0
Singh, A. K., and Cameotra, S. S. (2013). Efficiency of lipopeptide biosurfactants in removal of petroleum hydrocarbons and heavy metals from contaminated soil. Environ. Sci. Pollut. Res. 20, 7367–7376. doi: 10.1007/s11356-013-1752-4
Singh, R., Singh, D. P., Kumar, N., Bhargava, S. K., and Barman, S. C. (2010). Accumulation and translocation of heavy metals in soil and plants from fly ash contaminated area. J. Environ. Biol. 31, 421–430.
Sklodowska, A. (2000). Biologiczne metody lugowania metali ciezkich-biohydrometalurgia. Postêpy Mmikrobiologii 39, 73–89.
Slawson, R. M., Trevors, J. T., and Lee, H. (1992). Silver accumulation and resistance in Pseudomonas stutzeri. Arch. Microbiol. 158, 398–404. doi: 10.1007/BF00276299
Smirnova, G. F. (2005). Distribution of bacteria resistant to oxygen-containing anions-xenobiotics. Mikrobiol. Z. 67, 11–18.
Sobariu, D. L., Fertu, D. I., Diaconu, M., Pavel, L. V., Hlihor, R., Drãgoi, E. N., et al. (2017). Rhizobacteria and plant symbiosis in heavy metal uptake and its implications for soil bioremediation. New Biotechnol. 39, 125–134. doi: 10.1016/j.nbt.2016.09.002
Spain, O., Plöhn, M., and Funk, C. (2021). The cell wall of green microalgae and its role in heavy metal removal. Physiol. Plant. 173, 526–535. doi: 10.1111/ppl.13405
Sposito, F. G. (2000). “The chemistry of soils,” in Environmental microbiology, eds R. Maier, I. Pepper, and C. Gerba (London: Academic press), 406.
Srichandan, H., Pathak, A., Singh, S., Blight, K., Kim, D. J., and Lee, S. W. (2014). Sequential leaching of metals from spent refinery catalyst in bioleaching–bioleaching and bioleaching–chemical leaching reactor: Comparative study. Hydrometallurgy 150, 130–143. doi: 10.1016/j.hydromet.2014.09.019
Srinath, T., Verma, T., Ramteke, P. W., and Garg, S. K. (2002). Chromium (VI) biosorption and bioaccumulation by chromate resistant bacteria. Chemosphere 48, 427–435. doi: 10.1016/S0045-6535(02)00089-9
Srivastava, S., Agrawal, S. B., and Mondal, M. K. (2015). A review on progress of heavy metal removal using adsorbents of microbial and plant origin. Environ. Sci. Pollut. Res. 22, 15386–15415. doi: 10.1007/s11356-015-5278-9
Su, C. (2014). A review on heavy metal contamination in the soil worldwide: Situation, impact and remediation techniques. Environ. Skeptics Critics 3, 24–38.
Summers, A. O., and Silver, S. (1972). Mercury resistance in a plasmid-bearing strain of Escherichia coli. J. Bacteriol. 112, 1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972
Tang, J., Zhang, J., Ren, L., Zhou, Y., Gao, J., Luo, L., et al. (2019). Diagnosis of soil contamination using microbiological indices: A review on heavy metal pollution. J. Environ. Manag. 242, 121–130. doi: 10.1016/j.jenvman.2019.04.061
Tassi, E., Pouget, J., Petruzzelli, G., and Barbafieri, M. (2008). The effects of exogenous plant growth regulators in the phytoextraction of heavy metals. Chemosphere 71, 66–73. doi: 10.1016/j.chemosphere.2007.10.027
Tayang, A., and Songachan, L. S. (2021). Microbial bioremediation of heavy metals. Curr. Sci. 120, 1013–1025. doi: 10.18520/cs/v120/i6/1013-1025
Tripathi, M., Munot, H. P., Shouche, Y., Meyer, J. M., and Goel, R. (2005). Isolation and functional characterization of siderophore-producing lead-and cadmium-resistant Pseudomonas putida KNP9. Curr. Microbiol. 50, 233–237. doi: 10.1007/s00284-004-4459-4
Tripathi, R. D., Srivastava, S., Mishra, S., Singh, N., Tuli, R., Gupta, D. K., et al. (2007). Arsenic hazards: Strategies for tolerance and remediation by plants. Trends Biotechnol. 25, 158–165. doi: 10.1016/j.tibtech.2007.02.003
Tseng, C. H., Huang, Y. K., Huang, Y. L., Chung, C. J., Yang, M. H., Chen, C. J., et al. (2005). Arsenic exposure, urinary arsenic speciation, and peripheral vascular disease in blackfoot disease-hyperendemic villages in Taiwan. Toxicol. Appl. Pharmacol. 206, 299–308. doi: 10.1016/j.taap.2004.11.022
Ullah, A., Mushtaq, H., Ali, H., Munis, M. F. H., Javed, M. T., and Chaudhary, H. J. (2015). Diazotrophs-assisted phytoremediation of heavy metals: A novel approach. Environ. Sci. Pollut. Res. 22, 2505–2514. doi: 10.1007/s11356-014-3699-5
Vats, N., and Lee, S. F. (2001). Characterization of a copper-transport operon, copYAZ, from Streptococcus mutans. Microbiol. 147, 653–662. doi: 10.1099/00221287-147-3-653
Venkatesh, N. M., and Vedaraman, N. (2012). Remediation of soil contaminated with copper using rhamnolipids produced from Pseudomonas aeruginosa MTCC 2297 using waste frying rice bran oil. Ann. Microbiol. 62, 85–91. doi: 10.1007/s13213-011-0230-9
Vidali, M. (2001). Bioremediation. an overview. Pure Appl. Chem. 73, 1163–1172. doi: 10.1351/pac200173071163
Vikram, A., Johri, T., and Tandon, P. K. (2011). Effect of chromium (IV) on growth and metabolism of Spinacia oleracea (Spinach) plants. Res. Environ. Life Sci. 4, 119–124.
Viti, C., Pace, A., and Giovannetti, L. (2003). Characterization of Cr (VI)-resistant bacteria isolated from chromium-contaminated soil by tannery activity. Curr. Microbiol. 46, 1–5. doi: 10.1007/s00284-002-3800-z
Wang, C. L., Ozuna, S. C., Clark, D. S., and Keasling, J. D. (2002). A deep-sea hydrothermal vent isolate, Pseudomonas aeruginosa CW961, requires thiosulfate for Cd2+ tolerance and precipitation. Biotechnol. Lett. 24, 637–641. doi: 10.1023/A:1015043324584
World Health Organization [WHO], (2010). Preventing disease through healthy environments. Action is needed on chemicals of major public health concern. Geneva: World Health Organization.
Yang, T., Chen, M. L., and Wang, J. H. (2015). Genetic and chemical modification of cells for selective separation and analysis of heavy metals of biological or environmental significance. Trends Anal. Chem. 66, 90–102. doi: 10.1016/j.trac.2014.11.016
Yang, Z., Shi, W., Yang, W., Liang, L., Yao, W., Chai, L., et al. (2018). Combination of bioleaching by gross bacterial biosurfactants and flocculation: A potential remediation for the heavy metal contaminated soils. Chemosphere 206, 83–91. doi: 10.1016/j.chemosphere.2018.04.166
Ybarra, G. R., and Webb, R. (1999). Effects of divalent metal cations and resistance mechanisms of the cyanobacterium Synechococcus sp. strain PCC 7942. J. Hazard. Subst. Res. 2:1. doi: 10.4148/1090-7025.1011
Yin, T., Lin, H., Dong, Y., Li, B., He, Y., Liu, C., et al. (2021). A novel constructed carbonate-mineralized functional bacterial consortium for high-efficiency cadmium biomineralization. J. Hazard. Mater. 401:123269. doi: 10.1016/j.jhazmat.2020.123269
Zhang, W. M., and Gu, S. F. (2007). Catalytic effect of activated carbon on bioleaching of low-grade primary copper sulflde ores. Trans. Nonferr. Mrtal. Soc. China 17, 1123–1127. doi: 10.1016/S1003-6326(07)60236-2
Zhang, W., Ju, Y., Zong, Y., Qi, H., and Zhao, K. (2018). In situ real-time study on dynamics of microbially induced calcium carbonate precipitation at a single-cell level. Environ. Sci. Technol. 52, 9266–9276. doi: 10.1021/acs.est.8b02660
Zhang, X., Wang, H., He, L., Lu, K., Sarmah, A., Li, J., et al. (2013). Using biochar for remediation of soils contaminated with heavy metals and organic pollutants. Environ. Sci. Pollut. Res. 20, 8472–8483. doi: 10.1007/s11356-013-1659-0
Zhou, Z. S., Huang, S. Q., Guo, K., Mehta, S. K., Zhang, P. C., and Yang, Z. M. (2007). Metabolic adaptations to mercury-induced oxidative stress in roots of Medicago sativa L. J. Inorg. Biochem. 101, 1–9. doi: 10.1016/j.jinorgbio.2006.05.011
Keywords: toxicity, heavy metals, detoxification, rhizospheric, bioremediation
Citation: Joshi S, Gangola S, Bhandari G, Bhandari NS, Nainwal D, Rani A, Malik S and Slama P (2023) Rhizospheric bacteria: the key to sustainable heavy metal detoxification strategies. Front. Microbiol. 14:1229828. doi: 10.3389/fmicb.2023.1229828
Received: 27 May 2023; Accepted: 10 July 2023;
Published: 24 July 2023.
Edited by:
Deep Chandra Suyal, Vidyadayini Institute of Science, Management and Technology, IndiaReviewed by:
Om Prakash, Ministry of Ayush, IndiaCopyright © 2023 Joshi, Gangola, Bhandari, Bhandari, Nainwal, Rani, Malik and Slama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saurabh Gangola, c2FpbmRzYXVyYWJoQGdtYWlsLmNvbQ==; Sumira Malik, c21hbGlrQHJuYy5hbWl0eS5lZHU=; Petr Slama, cGV0ci5zbGFtYUBtZW5kZWx1LmN6
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.