
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol., 25 August 2023
Sec. Systems Microbiology
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1185787
This article is part of the Research TopicEnvironments-Pathogens-The Gut Microbiota and Host DiseasesView all 12 articles
 Patricia Guevara-Ramírez†
Patricia Guevara-Ramírez†
 Santiago Cadena-Ullauri
Santiago Cadena-Ullauri Elius Paz-Cruz
Elius Paz-Cruz Rafael Tamayo-Trujillo
Rafael Tamayo-Trujillo Viviana A. Ruiz-Pozo
Viviana A. Ruiz-Pozo Ana Karina Zambrano*
†
Ana Karina Zambrano*
†
Hematologic neoplasms represent 6.5% of all cancers worldwide. They are characterized by the uncontrolled growth of hematopoietic and lymphoid cells and a decreased immune system efficacy. Pathological conditions in hematologic cancer could disrupt the balance of the gut microbiota, potentially promoting the proliferation of opportunistic pathogens. In this review, we highlight studies that analyzed and described the role of gut microbiota in different types of hematologic diseases. For instance, myeloma is often associated with Pseudomonas aeruginosa and Clostridium leptum, while in leukemias, Streptococcus is the most common genus, and Lachnospiraceae and Ruminococcaceae are less prevalent. Lymphoma exhibits a moderate reduction in microbiota diversity. Moreover, certain factors such as delivery mode, diet, and other environmental factors can alter the diversity of the microbiota, leading to dysbiosis. This dysbiosis may inhibit the immune response and increase susceptibility to cancer. A comprehensive analysis of microbiota-cancer interactions may be useful for disease management and provide valuable information on host-microbiota dynamics, as well as the possible use of microbiota as a distinguishable marker for cancer progression.
Hematologic malignancies are characterized by the uncontrolled growth of hematopoietic and lymphoid cells, resulting in decreased immune system efficacy (Méndez-Ferrer et al., 2020). Hematologic neoplasms account for 6.5% of all cancers worldwide (De Moraes Hungria et al., 2019; Kocarnik et al., 2022). The World Health Organization (WHO) classifies hematologic malignancies based on morphology, immunophenotype, genetics, and clinical features (Khoury et al., 2022). The most common subtypes include leukemia, Hodgkin’s lymphoma (HL), non-Hodgkin’s lymphoma (NHL), and multiple myeloma (MM) (Keykhaei et al., 2021). Hematologic diseases have been associated with genetic factors and alterations of the immune system. However, several studies also suggest a potential correlation between hematologic cancers and alteration in the microbiota. For instance, research shown that the growth of gastric mucosa-associated lymphoid tissue (MALT) lymphoma tumors can be stimulated by signaling antigens released by the bacterium Helicobacter pylori (H. pylori), highlighting a possible link between bacteria and MALT lymphoma (Ferreri et al., 2013; Kuo and Cheng, 2013; Portlock et al., 2015).
The human gut microbiota (GM) is a population of microorganisms, including bacteria, archaea, fungi, protozoa, and viruses, that coexist within the intestinal tract (D’Angelo et al., 2021). Furthermore, these microorganisms produce metabolites such as short-chain fatty acids (SCFAs), which could have anti-carcinogenic properties. The most predominant SCFAs, acetate, propionate, and butyrate, play crucial roles in ion absorption and intestinal motility (Jasiński et al., 2021). In particular, butyrate has been studied for its anti-inflammatory properties (Ubeda et al., 2010; Canani et al., 2011; Zimmerman et al., 2012; Bin et al., 2021). However, conflicting findings suggest that the effects of butyrate on cell proliferation vary, depending on factors such as time, cell type, and concentration; it could either promote or prevent cell proliferation. Nonetheless, it has been proposed that excessive butyrate production following dysbiosis and inflammation may promote tumor proliferation, potentially outweighing its beneficial properties (Donohoe et al., 2012).
Metagenomics and metabolomics analyses have provided valuable insights into the role of intestinal microbiota in malignant neoplasms (Frankel et al., 2017). These studies suggest that pathological conditions in hematologic cancer (HC) can lead to dysbiosis, which is an imbalance of the microbiota (Ahmed et al., 2020; Dutta and Lim, 2020; Tsvetikova and Koshel, 2020; Zheng et al., 2020; Abdelazeem et al., 2021). Imbalances in the microbiota can inhibit the colonization of beneficial probiotic bacteria, promote harmful enteropathogens proliferation, and alter cytokine signaling, thus affecting the immune system (Alexander et al., 2017). In this review, we highlight studies that analyzed the role of GM in different types of hematologic diseases, especially leukemias, lymphomas, and myelomas. Additionally, we describe the factors that can alter the human gut microbiota and its correlation with hematologic cancer predisposition and progression.
Hematologic diseases have been associated with dysbiosis, leading to a limited capacity of the microbiota’s metabolites to modulate inflammatory processes, and disrupting intestinal homeostasis. Understanding the relationship between the host and gut microbiota is crucial. Germ-free mice experiments have shown that certain bacteria, such as Bacteroides and Escherichia spp., could have an immunogenic effect by stimulating the production of immunoglobulin A (IgA) plasmacytes (Moreau et al., 1978; Strauch et al., 2005). The microbiota interacts with the immune system via the intestinal epithelium, which comprises enterocytes, goblet cells, neuroendocrine cells, tuft cells, Paneth cells, and Microfold cells (M cells), plays an essential role in innate immunity and host defense (Allaire et al., 2018).
Peyer’s patches are clusters of lymphoid tissue that line the walls of the small intestine. They contain immune cells such as innate lymphoid cells (ILCs), natural killer (NK) cells, T and B lymphocytes, and M cells (Elemam et al., 2017). Pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) and Nod-like receptors (NLRs), are expressed by both epithelial and immune cells. These receptors can recognize pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) (Rankin et al., 2013). Remarkably, a study in mice suggests that gut microbiota manipulation can modulate cancer immunotherapy by increasing T cells within the tumor microenvironment (Sivan et al., 2015). GM has been linked to immunological response because microorganisms can facilitate the transport of macromolecules and antigens through the gut epithelium.
Moreover, flagellin is the primary component of the bacterial flagellum; it mediates the interaction between the intestinal epithelium and host immunity. Flagellin can be recognized by TLR5, found in B-cells and CD4+ T-cells. Differentiated B-cells produce IgA that neutralizes the pathogen and prevents subsequent infection (Eaves-Pyles et al., 2011; Haiko and Westerlund-Wikström, 2013). Generally, TLRs activation by antigens from the normal gut microbiota signals the inhibition of inflammatory reactions, which is necessary to maintain intestinal homeostasis. NLRs recognize specific microbial molecules and initiate the formation of inflammasomes, which act as sensors for damage-associated patterns (Lavelle et al., 2010; Parlato and Yeretssian, 2014). Thus, immune dysregulation in hematologic diseases could alter the interaction with the microbiota, inhibiting the role of its metabolites and leading to an increased vulnerability to infections and a rise in the severity of hematological cancer.
The interactions between the microbiome and hematologic cancer are influenced by intrinsic and extrinsic factors. Intrinsic factors, such as genetics, immune status, and overall health, can shape both the composition and functionality of the gut microbiota. Genetic variations in host genes can influence the expression of microbial receptors, impacting the colonization and survival of specific microbial species. Immune dysregulation can lead to microbial imbalances contributing to carcinogenesis (Rahman et al., 2022). Extrinsic factors, such as nutrition, lifestyle, drugs, anticancer therapy, and environmental exposures, also influence the gut microbiota. Physical exercise, stress, diet, type of delivery, pollution, and chemicals indirectly impact the gut microbiota through their effects on human physiology and metabolism (Bajinka et al., 2020). Altogether, these variables alter the gut microbial ecosystem, increasing the host’s susceptibility to hematopoietic malignancies (Figure 1; Uribe-Herranz et al., 2021).
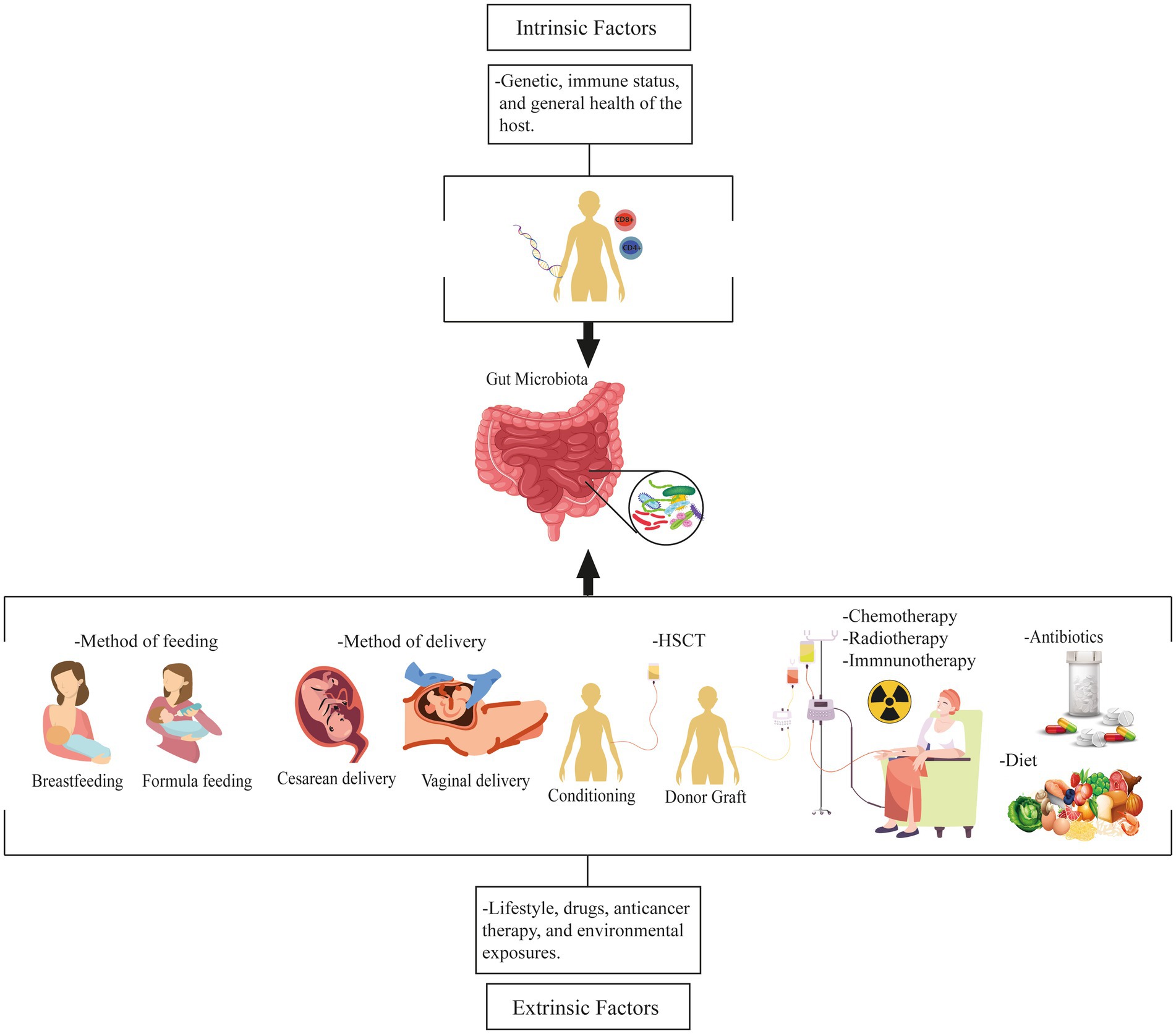
Figure 1. Factors influencing the gut microbiota and its relationship to hematologic malignancies. These factors are intrinsic and extrinsic. Intrinsic factors include genetics, immune status, and overall health, whereas extrinsic factors include nutrition, lifestyle, medications, anticancer therapy, and environmental exposures that affect the gut microbiota. HSCT, Hematopoietic stem cell transplantation.
The type of delivery can influence the diversity of the neonate’s gut microbiota. During vaginal delivery, the neonate is exposed to vaginal, perineal, and fecal flora, with the most abundant bacteria being Lactobacillus, Prevotella, Sneathia (Stiemsma and Michels, 2018), and Gardnerella vaginalis (Chen et al., 2021). Conversely, neonates born by cesarean delivery have distinct intestinal microbiota colonized by skin bacteria, such as Staphylococcus, Corynebacterium, and Propionibacterium (Greenbaum et al., 2018; Sędzikowska and Szablewski, 2021). Research has correlated the type of delivery with a predisposition to the development of hematologic diseases such as leukemia and HL, concluding that cesarean deliveries had higher rates of HC development compared to vaginal delivery (Momen et al., 2014; Greenbaum et al., 2018; Marcoux et al., 2022).
Breastfeeding colonizes the infant’s gut microbiome through contact with the nipple-areola and breast milk microbes. The microbiota of breastfed infants is dominated by Bifidobacterium, Ruminococcus, and Lactobacillus spp. In contrast, bottle-fed infants exhibit a higher prevalence of Proteobacteria, Streptococcus, Bacteroides, Clostridium, Bifidobacterium, and Atopobium in their microbiota. According to numerous studies, breastfeeding is important in lowering the risk of infant leukemia (Ajrouche et al., 2015; Amitay et al., 2016), while formula feeding has been associated with an increased risk of various diseases (Stiemsma and Michels, 2018; Sędzikowska and Szablewski, 2021; Su et al., 2021).
Recent studies have suggested that dietary factors can shape gut microbiota (Alexander et al., 2017; Uribe-Herranz et al., 2021). There are different diet types, depending on the country and the area (rural or urban). Certain diets are characterized by high fat and carbohydrate intake but low fiber, while others are rich in both protein and fiber. The metabolism of these foods can result in the enrichment or elimination of different bacterial populations and lead to the formation of specific metabolites (Koh et al., 2016; Li et al., 2021). Investigations found that fiber (Liu et al., 2015), oligosaccharides (Hosomi et al., 2009), glutamine (Han et al., 2016), and lactoferrin are potentially beneficial molecules during leukemia treatment because they increase the proportions of beneficial commensals (Iyama et al., 2014; Masetti et al., 2021).
The composition of the microbiota is influenced by various factors, including cancer treatments and therapies. One critical factor are medications, such as antibiotics, which can disrupt the balance of the gut microbiota, leading to dysbiosis that may affect cancer treatment outcomes. For example, although antibiotics are commonly administered in hematologic cancer treatment to prevent infections, they can affect bacteria such as Faecalibacterium, Anaerostipes, and Blautia, potentially disrupting the overall gut microbial ecosystem (Dunn et al., 2022; Sochacka-ćwikła et al., 2022).
Furthermore, various anticancer treatments, such as chemotherapy, radiotherapy, and immunotherapy, have a profound impact on the gut microbiota of hematologic cancer patients. Specific chemotherapeutic drugs (cladribine, vidarabine, cisplatin, and gemcitabine) may become less effective against certain bacteria, and could decrease the abundance of beneficial bacteria like Bifidobacterium, Lactobacillus, and Faecalibacterium prausnitzii (F. prausnitzii) while promoting potentially harmful bacteria, such as Escherichia and Enterococcus faecium (Zwielehner et al., 2011; Pflug et al., 2016; Dunn et al., 2022).
Additionally, hematopoietic stem cell transplantation (HSCT) can lead to changes in the microbiota and give rise to complications such as graft-versus-host disease (GVHD). Severe GVHD has been associated with an increased abundance of Enterobacteriaceae, while Clostridia have been linked to anti-inflammatory responses (Hong et al., 2021). Studies have demonstrated shifts in the microbiota during the conditioning stage, with chemotherapeutic agents damaging intestinal epithelial cells and increasing the susceptibility to bacteremia (Shono and van den Brink, 2018; Hong et al., 2021; Ingham et al., 2021; Margolis et al., 2023). The conditioning regimen used before HSCT significantly alters the gut microbiome, surpassing even the effects of the transplant itself (Jørgensen et al., 2022).
In summary, the relationship between microbiota and hematologic cancer is complex and influenced by various factors. Understanding these factors and their impact on the gut microbiota is crucial for developing personalized therapeutic strategies.
Several investigations have evaluated the variations in gut microbiota composition in mouse models and hematologic patients (Figure 2). Moreover, the microbiota composition could change depending on the specific type of hematologic cancer (Supplementary Table S1; Riley et al., 2013; Allegra et al., 2019).
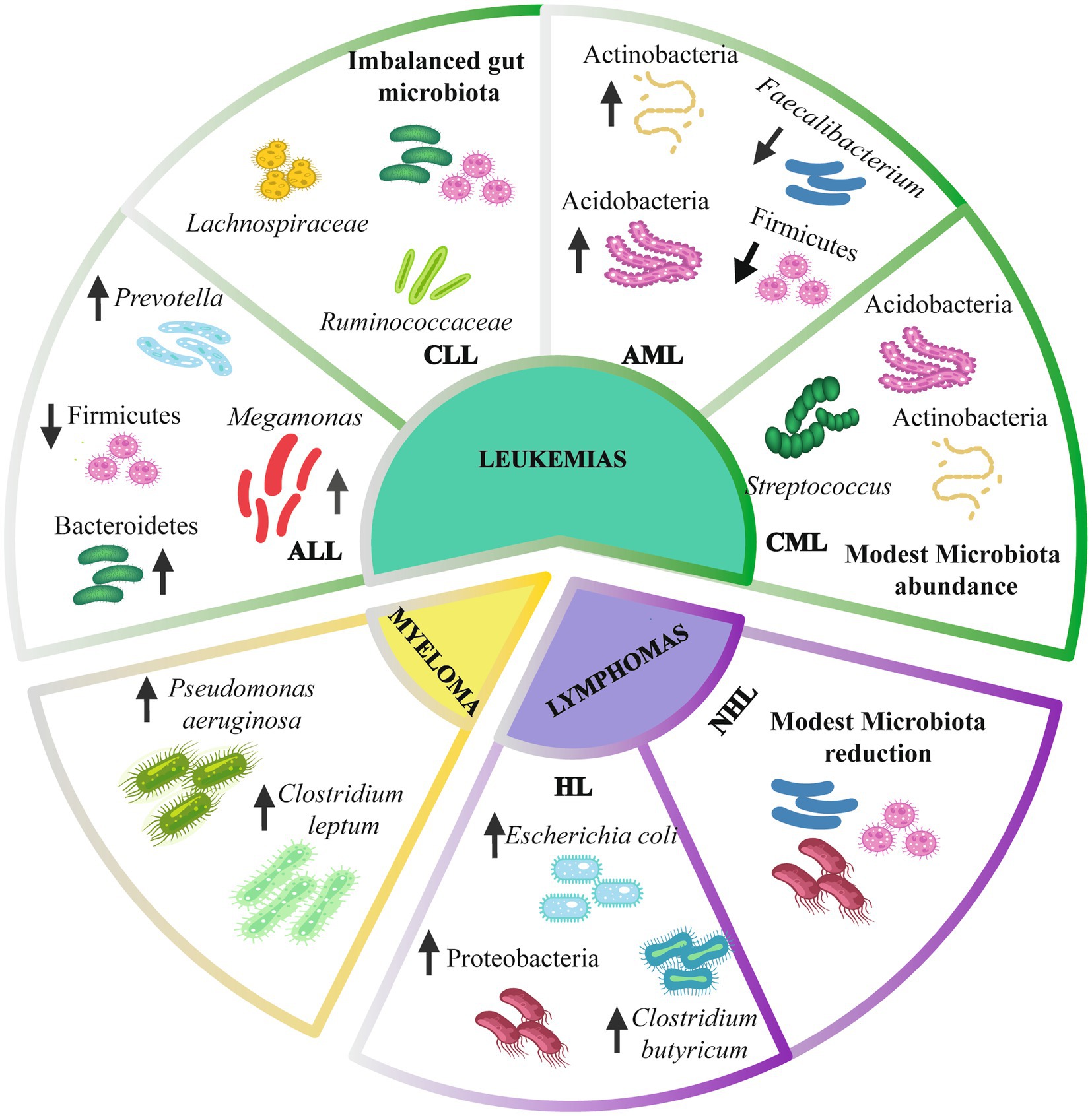
Figure 2. Gut microbiota composition in hematologic cancer. Leukemias have alterations of the intestinal microbiome at the phylum level, including Firmicutes, Bacteroidetes, and Actinobacteria. At the genus level, there is an alteration in Prevotella, Megamonas, Faecalibacterium, and Streptococcus. Lymphomas present a modest reduction of the intestinal microbiota, mainly an increase in Escherichia coli and Clostridium butyricum. Myeloma presents an alteration of Pseudomonas aeruginosa and Clostridium leptum species. Up arrows indicate an increase. Down arrows indicate a decrease. ALL, Acute lymphoblastic leukemia; CLL, Chronic lymphoblastic leukemia; AML, Acute myelogenous leukemia; CML, Chronic myelogenous leukemia; HL, Hodgkin lymphoma; NHL, non-Hodgkin lymphoma.
The role of the gut microbiota in acute lymphoblastic leukemia (ALL) the development remains unclear and is currently under investigation. Reports have identified variations in the GM composition profile in ALL patients compared to a healthy population. Other studies have shown a reduction in the relative abundance of Edwardsiella tarda and Prevotella maculosa in ALL patients, which was positively correlated with interleukin-10 levels (Kostic et al., 2013; Schirmer et al., 2016; Li et al., 2019; Liu et al., 2020).
Another study reported that Faecalibacterium abundance was reduced among ALL patients and negatively correlated with interleukin-6 (IL-6) and C-reactive protein (CRP) (Chua et al., 2017). Similarly, Megamonas was abundant in the gut microbiota of ALL children and correlated with the systemic inflammatory cytokines IL-6 (Sakon et al., 2008; Cozen et al., 2013; Bai et al., 2017; Li et al., 2018; Neisi et al., 2019; Ansari et al., 2021).
Furthermore, NGS analyses have revealed changes in microbiota diversity in ALL individuals, with an increase in Bacteroidetes and a decrease in Firmicutes. These alterations may be detrimental to leukemia patients. The Firmicutes phylum is the principal producer of butyrate (Venegas et al., 2019), which has been shown to have anti-cancer activities (Geng et al., 2021). For instance, researchers reported a significant reduction in butyrate production by the GM. Additionally, they found intestinal barrier damage in leukemia patients, which accelerated lipopolysaccharide (LPS) leakage into the bloodstream (Wang et al., 2022). LPS has been associated with leukemia progression both in vivo and in vitro. Butyrate is produced by certain bacteria such as Eubacterium, Streptococcus, Clostridium, Bacteroides, Roseburia, Coprococcus, Ruminocococcus, and Butyrivibrio (Ramsay et al., 2006; Anshory et al., 2023; Singh et al., 2023). Butyrate can repair the damage in the intestinal barrier, inhibiting LPS leakage and potentially playing a protective role against leukemia progression (Wang et al., 2022).
A common feature of chronic lymphocytic leukemia (CLL) is chronic systemic inflammation, with reports suggesting that dysbiosis may contribute to inflammation (Kawari et al., 2019). In the immune microenvironment of the intestine, T helper 17 cells (Th17) play an important role. Several studies demonstrate that increased levels of Th17 are an unfavorable prognostic factor in CLL. Huang et al. (2020) propose that Prevotella induces Th17 cell production in the mouse colon, highlighting its potential role in intestinal immune system formation (Huang et al., 2020).
Another study found that in patients with CLL, the most abundant bacteria were Bacteroides, Parabacteroides, Prevotella, and Acinetobacter, while there was a depletion of Lachnospiraceae and Ruminococcaceae (Faitová et al., 2022). In contrast, one study reported an increase in the abundance of Firmicutes and a decrease in Bacteroidetes compared to healthy individuals (Kawari et al., 2019).
The decrease in Lachnospiraceae and Ruminococcaceae may have several consequences for leukemia development (Vacca et al., 2020; Masetti et al., 2021). Lachnospiraceae has been associated with resistance to high radiation doses, hematopoiesis restoration, and butyrate-mediated repair of the gastrointestinal system in the host (Ma et al., 2021). Furthermore, studies have reported that the abundance of Lachnospiraceae is correlated with reduced side effects in patients with graft versus host disease (GVHD) (Ma et al., 2021).
Ruminococcus is another bacterium that produces several SCFAs (Mirzaei et al., 2021), and its deficit is associated with disruptions in several signaling pathways (Mirzaei et al., 2021). While the mechanisms of Ruminococcaceae in improving patient outcomes in leukemia are still unknown, there is evidence of increased Ruminococcaceae abundance in patients who achieved complete remission after PD-1 immunotherapy and CAR T-cell therapy (Ma et al., 2021; Zhou et al., 2022). Hence, Ruminococcaceae and its metabolites could improve the diagnosis and treatment of several cancer types.
Researchers have studied the role of gut microbiota in acute myelogenous leukemia (AML) by examining the differences in microbiota with and without treatment. One study published by Wang et al. (2022) reported a decrease in the gut microbiota diversity of AML patients. Moreover, the study found that intestinal damage was correlated with an increase in lipopolysaccharide levels and AML progression. Regarding bacterial species, the authors found that the reduction of Faecalibacterium could be involved in the proliferation and invasion of tumor cells and suppression of apoptosis (Ma et al., 2020; Wang et al., 2022).
Research suggests that most Faecalibacterium strains are associated with energy production for intestinal epithelial cells and the synthesis of metabolites, such as butyric acid, bioactive peptides, and anti-inflammatory substances, which contribute to intestinal health (Zou et al., 2021). Butyric acid modulates signaling pathways by interacting with the proinflammatory nuclear transcription factor NF-kB and inhibiting histone deacetylase (Knudsen et al., 2018). The regulation of metabolites, such as butyrate, could be an alternative for AML therapy development.
According to research, chronic myelogenous leukemia (CML) patients have a higher abundance of Actinobacteria, Acidobacteria, and Chloroflexi, as well as a decreased abundance of Tenericutes. Furthermore, studies have described an increase in the levels of the Streptococcus genus in patients with CML compared to control patients (Yu et al., 2021). Several studies suggest an association between Streptococcus bacteria and an increase in the proinflammatory cytokine interferon γ (Bagheri et al., 2022). Streptococcus is essential in the sugar fermentation process, producing lactic acid as the main compound, which could have implications for CML progression (van den Bogert et al., 2013). Therefore, an imbalance of microbiota components could lead to proinflammatory responses, potentially triggering carcinogenesis (Liu et al., 2021).
An increased Streptococcus abundance may have a deleterious effect on leukemias, whereas the Actinobacteria abundance may help to decrease the adverse effects. Research has shown that the Actinobacteria phylum may benefit acute leukemia patients, as it is positively associated with Allo-HSCT immunotherapy (Ma et al., 2021) and exhibits antioxidant activities (Almuhayawi et al., 2021). Several Actinobacteria metabolites, such as indolocarbazoles, isoprenoids, non-ribosomal peptides, anthracyclines, macrolides, and enediynes, exhibit antioxidant and antitumoral properties. These metabolites have shown cytotoxic activity against cancer cell lines by reducing cyclooxygenase and lipoxygenase activity (Zhou et al., 2017; Almuhayawi et al., 2021). Cyclooxygenase is involved in prostaglandin synthesis, which promotes the proliferation of leukemia cells and the production of reactive oxygen species, while lipoxygenase catalyzes the production of hydroxyl eicosatetraenoic acids and leukotrienes, contributing to apoptosis suppression and the stimulation of tumor cell proliferation (Almuhayawi et al., 2021).
Understanding the correlation between gut microbiota, adaptive and innate immunity, and diseases like Hodgkin’s lymphoma is essential. Yuan et al. (2021) characterized the gut microbiota of 25 untreated individuals with diffuse large B cell lymphoma. Compared to the control group, the authors observed a higher abundance of Proteobacteria at the phylum level, as well as Escherichia coli (E. coli) and Clostridium butyricum (C. butyricum) species.
Various analyses have suggested that an increased prevalence of the bacterial phylum Proteobacteria could serve as a potential marker for an unstable microbial community (Shin et al., 2015; Tang et al., 2019) and be associated with B-cell differentiation (Yuan et al., 2021). Unlike most microbes, which are strict anaerobes, Proteobacteria are frequently facultatively or obligate anaerobic, enabling them to tolerate a wide range of toxic conditions.
On the other hand, E. coli produces colibactin and cytolethal-distending toxins, which have been associated with DNA breaks in epithelial cells, promoting genetic mutations and contributing to tumor formation. E. coli plays a crucial role in lymphoproliferative processes and infections by primarily colonizing the mucosal layer of the gastrointestinal tract, where it can contribute to chronic inflammation. Inflammation can persist due to these bacteria’ immune evasion strategies, including blocking TLR-4 signaling, NF-κB activity, and proinflammatory cytokines production in cells (Olson et al., 2014; Conway and Cohen, 2015; Rolhion and Chassaing, 2016).
Moreover, C. butyricum, a bacterium that produces butyrate and acetate, has been studied for its potential therapeutic use in dysbiosis-related diseases (Li et al., 2022). C. butyricum can also slow tumor growth by modulating Wnt/β-catenin signaling, which leads to decreased proliferation, and increased apoptosis (Tomita et al., 2022).
MALT lymphoma has been associated with a Helicobacter pylori infection, which could be involved in tumorigenesis and a chronic inflammatory response (Wotherspoon et al., 1991; O’Rourke, 2008; Saito et al., 2012; Moleiro et al., 2016). A retrospective study by Moleiro et al. (2016) showed that H. pylori eradication therapy could be effective for complete remission in patients (Moleiro et al., 2016).
Recent findings have shown an association between gut microbiota and MM (Lax et al., 2014; Alkharabsheh et al., 2020; Shapiro et al., 2021). Zhang et al. (2019) found that Pseudomonas aeruginosa and Clostridium leptum (C. leptum) were more abundant in MM patients. Moreover, higher levels of C. leptum were observed in MM patients with advanced stages of the disease. Pseudomonas aeruginosa can cause bacterial infections, while C. leptum is involved in the intestinal glucose metabolism pathway. Therefore, further research on these bacteria is critical for a better understanding of their roles (Zhang et al., 2019).
Clostridium leptum regulates glucose concentration in the intestinal microenvironment by producing butyrate through the pyruvate and acetyl-coenzyme A pathway. Butyrate plays a role in increasing regulatory T cells and suppressing interleukin 17 (IL-17) (Linares and Hermouet, 2022). For instance, Calcinotto et al. (2018) showed that a lack of IL-17 in MM mice, or treatment with antibiotics or antibodies that block IL-17/IL-17R interactions, leads toa delay in MM progression. The study identified Prevotella heparinolytica as the causal bacteria for IL-17 proliferation (Calcinotto et al., 2018). Therefore, the presence of butyrate-producing bacteria in the intestinal microbiota of MM patients is positively correlated with higher rates of minimal residual disease (MRD) negativity (Brevi et al., 2022).
Furthermore, Pianko et al. (2019) analyzed the microbiota composition of MRD in MM patients and found that MRD-negative treatment response was associated with a higher abundance of Eubacterium hallii and F. prausnitzii. Eubacterium hallii produces propionate, while F. prausnitzii produces butyrate. Both metabolites modulate immunity through autoinflammatory functions (Pianko et al., 2019).
The evidence presented in this mini-review underscores the role of specific microorganisms in the progression of hematologic diseases, given that microbiota imbalances have been found in all types of HC. Each type of HC —myeloma, lymphoma, and leukemia— exhibits distinct microbiota characteristics. Myeloma is characterized by an increased abundance of Pseudomonas aeruginosa and Clostridium leptum; lymphoma is associated with a higher proportion of E. coli and C. butyricum, while leukemia is marked by a decrease in Lachnospiraceae and Ruminococcaceae. These bacteria interact with immune cells in the epithelial tissue through their antigens or by secreting metabolites, potentially influencing the tumor environment. While these findings offer valuable insights, it is crucial to acknowledge that other factors and mechanisms may also contribute to cancer progression, warranting further investigation of the role and interactions of the gut microbiota with the tumor environment (Arthur et al., 2017). Notably, gut microbiota modulation may play a significant role in immune and treatment outcomes (Matson et al., 2018).
Microbiota modulation can be influenced by various factors, which may increase the risk of cancer development (De Agüero et al., 2016). Early interactions between the newborn, the mother, and the environment, such as the delivery and feeding methods, play a pivotal role in shaping the microbial microenvironment and long-term cancer susceptibility. Additionally, diet represents a critical factor that can be modified to prevent an imbalance of beneficial bacteria. Microbial food fermentation produces primary metabolites that can have either beneficial or detrimental effects on the host. Ongoing large-scale clinical trials are actively evaluating the efficacy of microbiota modulation, including dietary interventions and intratumoral injection of engineered bacteria (Sepich-Poore et al., 2021), as potential therapies for hematologic malignancies.
A comprehensive analysis of the microbiota concerning cancer may support disease management and deepen our understanding of host-microbial evolution. It also holds promise in exploring the microbiota as a distinguishable marker for cancer progression (Kalia et al., 2022). Fecal microbiome transplantation (FMT) is an alternative for restoring healthy microbiota in patients with hematologic diseases (Zheng et al., 2020). However, the characteristics of a healthy microbiome remain undefined, which leads to ongoing evaluation of FMT’s effectiveness in treating hematologic cancer, along with challenges like optimizing fecal processing and ensuring patient safety.
One of the main limitations of this research is that it relies on cross-sectional studies, limiting the capacity to establish a cause-effect relationship between microbiota and HC. Therefore, conducting longitudinal studies that measure the microbiota at different time points is essential for gain a comprehensive understanding of this interaction (Vogtmann and Goedert, 2016; Hou et al., 2022). There are other limitations, such as small sample sizes, ethnic bias, and the absence of control groups or disease staging in some studies. Moreover, technical limitations are also present as different techniques were used to identify microorganisms, resulting in the inability to capture the full complexity of the intestinal microbiota, potentially missing rare or less abundant species.
Furthermore, variations in the microbiome across different geographical regions should be considered. Characterizing microbiotas from diverse areas is essential to identify their primary composition. Moreover, it is crucial to carefully account for confounding factors such as diet, medication use, and the environment, as they could significantly impact the composition of the microbiota and its association with cancer progression (Fontana et al., 2019; Dwiyanto et al., 2021).
In conclusion, this mini review emphasizes the crucial role of specific microorganisms in hematologic cancer progression and highlights the significance of modulating the microbiota in immune responses and treatment outcomes. However, further research is essential to explore and comprehend the complexities of interactions between the gut microbiota and the tumor environment. Such studies are crucial for the development of targeted and effective microbiota-focused anticancer strategies, holding great promise for the future of hematologic cancer treatments.
PG-R and AZ: conceptualization and writing – review and editing. SC-U: writing – original draft. EP-C, RT-T, and VR-P: investigation. AZ: supervision. All authors contributed to the article and approved the submitted version.
The publication of this article will be funded by Universidad UTE-Ecuador. The funder had no role in the study design, bibliographic analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1185787/full#supplementary-material
Abdelazeem, K. N. M., Kalo, M. Z., Hammer, S. B., and Lang, F. (2021). The gut microbiota metabolite urolithin a inhibits NF – κB activation in LPS stimulated BMDMs. Sci. Rep. 11, 1–16. doi: 10.1038/s41598-021-86514-6
Ahmed, N., Ghannoum, M., Gallogly, M., and De Lima, M. (2020). Influence of gut microbiome on multiple myeloma: Friend or foe? J. Immunother. Cancer, 8–10. doi: 10.1136/jitc-2020-000576
Ajrouche, R., Rudant, J., Orsi, L., Petit, A., Baruchel, A., Lambilliotte, A., et al. (2015). Childhood acute lymphoblastic leukaemia and indicators of early immune stimulation: the Estelle study (SFCE). Br. J. Cancer 112, 1017–1026. doi: 10.1038/bjc.2015.53
Alexander, J. L., Wilson, I. D., Teare, J., Marchesi, J. R., Nicholson, J. K., and Kinross, J. M. (2017). Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 14, 356–365. doi: 10.1038/nrgastro.2017.20
Alkharabsheh, O., Sidiqi, M. H., Aljama, M. A., Gertz, M. A., and Frankel, A. E. (2020). The human microbiota in multiple myeloma and proteasome inhibitors. Acta Haematol. 143, 118–123. doi: 10.1159/000500976
Allaire, J. M., Crowley, S. M., Law, H. T., Chang, S. Y., Ko, H. J., and Vallance, B. A. (2018). The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 39, 677–696. doi: 10.1016/j.it.2018.04.002
Allegra, A., Innao, V., Allegra, A. G., Ettari, R., Pugliese, M., Pulvirenti, N., et al. (2019). Role of the microbiota in hematologic malignancies. Neth. J. Med. 77, 67–80.
Almuhayawi, M. S., Mohamed, M. S. M., Abdel-Mawgoud, M., Selim, S., Al Jaouni, S. K., and Abdelgawad, H. (2021). Bioactive potential of several actinobacteria isolated from microbiologically barely explored desert habitat, Saudi Arabia. Biology (Basel) 10, 1–22. doi: 10.3390/biology10030235
Amitay, E. L., Dubnov Raz, G., and Keinan-Boker, L. (2016). Breastfeeding, other early life exposures and childhood Leukemia and lymphoma. Nutr. Cancer 68, 968–977. doi: 10.1080/01635581.2016.1190020
Ansari, A., Bose, S., You, Y., Park, S., and Kim, Y. (2021). Molecular mechanism of microbiota metabolites in preterm birth: pathological and therapeutic insights. Int. J. Mol. Sci. 22, 1–20. doi: 10.3390/ijms22158145
Anshory, M., Effendi, R. M. R. A., Kalim, H., Dwiyana, R. F., Suwarsa, O., Nijsten, T. E. C., et al. (2023). Butyrate properties in immune-related diseases: friend or foe? Fermentation 9, 1–19. doi: 10.3390/fermentation9030205
Arthur, J. C., Gharaibeh, R. Z., Mühlbauer, M., and Chanona, E. P. (2017). Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Janelle Nat. Commun. 176, 139–148. doi: 10.1038/ncomms5724
Bagheri, Z., Moeinzadeh, L., and Razmkhah, M. (2022). Roles of microbiota in cancer: from tumor development to treatment. J. Oncol. 2022:3845104. doi: 10.1155/2022/3845104
Bai, L., Zhou, P., Li, D., and Ju, X. (2017). Changes in the gastrointestinal microbiota of children with acute lymphoblastic leukaemia and its association with antibiotics in the short term. J. Med. Microbiol. 66, 1297–1307. doi: 10.1099/jmm.0.000568
Bajinka, O., Tan, Y., Abdelhalim, K. A., Özdemir, G., and Qiu, X. (2020). Extrinsic factors influencing gut microbes, the immediate consequences and restoring eubiosis. AMB Express. 10:1066. doi: 10.1186/s13568-020-01066-8
Bin, Z. L., Zhang, Y. C., Huang, H. H., and Lin, J. (2021). Prospects for clinical applications of butyrate-producing bacteria. World J. Clin. Pediatr. 10, 84–92. doi: 10.5409/wjcp.v10.i5.84
Brevi, A., Cogrossi, L. L., Lorenzoni, M., Mattorre, B., and Bellone, M. (2022). The insider: impact of the gut microbiota on cancer immunity and response to therapies in multiple myeloma. Front. Immunol. 13, 1–11. doi: 10.3389/fimmu.2022.845422
Calcinotto, A., Brevi, A., Chesi, M., Ferrarese, R., Perez, L. G., Grioni, M., et al. (2018). Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. doi: 10.1038/s41467-018-07305-8
Canani, R. B., Di, C. M., Leone, L., Pedata, M., Meli, R., and Calignano, A. (2011). Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 17, 1519–1528. doi: 10.3748/wjg.v17.i12.1519
Chen, X., Lu, Y., Chen, T., and Li, R. (2021). The female vaginal microbiome in health and bacterial vaginosis. Front. Cell. Infect. Microbiol. 11, 1–15. doi: 10.3389/fcimb.2021.631972
Conway, T., and Cohen, P. S. (2015). Commenal and pathogenic E. coli metabolism in the gut. Microbiol Spectr. 3:10. doi: 10.1128/microbiolspec.MBP-0006-2014
Chua, L. L., Rajasuriar, R., Azanan, M. S., Abdullah, N. K., Tang, M. S., Lee, S. C., et al. (2017). Reduced microbial diversity in adult survivors of childhood acute lymphoblastic leukemia and microbial associations with increased immune activation. Microbiome 5:250. doi: 10.1186/s40168-017-0250-1
Cozen, W., Yu, G., Gail, M. H., Ridaura, V. K., Nathwani, B. N., Hwang, A. E., et al. (2013). Fecal microbiota diversity in survivors of adolescent/young adult Hodgkin lymphoma: a study of twins. Br. J. Cancer 108:1163. doi: 10.1038/bjc.2013.60
D’Angelo, C. R., Sudakaran, S., and Callander, N. S. (2021). Clinical effects and applications of the gut microbiome in hematologic malignancies. Cancer 127, 679–687. doi: 10.1002/cncr.33400
De Agüero, M. G., Ganal-Vonarburg, S. C., Fuhrer, T., Rupp, S., Uchimura, Y., Li, H., et al. (2016). The maternal microbiota drives early postnatal innate immune development. Science (80-) 351, 1296–1302. doi: 10.1126/science.aad2571
De Moraes Hungria, V. T., Chiattone, C., Pavlovsky, M., Abenoza, L. M., Agreda, G. P., Armenta, J., et al. (2019). Epidemiology of hematologic malignancies in real-world settings: findings from the hemato-oncology Latin america observational registry study. J. Glob. Oncol. 2019, 1–19. doi: 10.1200/JGO.19.00025
Donohoe, D. R., Collins, L. B., Wali, A., Bigler, R., Sun, W., and Bultman, S. J. (2012). The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 48, 612–626. doi: 10.1016/j.molcel.2012.08.033
Dunn, K. A., MacDonald, T., Rodrigues, G. J., Forbrigger, Z., Bielawski, J. P., Langille, M. G. I., et al. (2022). Antibiotic and antifungal use in pediatric leukemia and lymphoma patients are associated with increasing opportunistic pathogens and decreasing bacteria responsible for activities that enhance colonic defense. Front. Cell. Infect. Microbiol. 12, 1–19. doi: 10.3389/fcimb.2022.924707
Dutta, D., and Lim, S. H. (2020). Bidirectional interaction between intestinal microbiome and cancer: opportunities for therapeutic interventions. Biomark Res. 8, 1–15. doi: 10.1186/s40364-020-00211-6
Dwiyanto, J., Hussain, M. H., Reidpath, D., Ong, K. S., Qasim, A., Lee, S. W. H., et al. (2021). Ethnicity influences the gut microbiota of individuals sharing a geographical location: a cross-sectional study from a middle-income country. Sci. Rep. 11, 1–10. doi: 10.1038/s41598-021-82311-3
Eaves-Pyles, T., Bu, H. F., di Tan, X., Cong, Y., Patel, J., Davey, R. A., et al. (2011). Luminal-applied flagellin is internalized by polarized intestinal epithelial cells and elicits immune responses via the TLR5 dependent mechanism. PLoS One 6:24869. doi: 10.1371/journal.pone.0024869
Elemam, N. M., Hannawi, S., and Maghazachi, A. A. (2017). Innate lymphoid cells (ILCs) as mediators of inflammation, release of cytokines and lytic molecules. Toxins 9, 1–18. doi: 10.3390/toxins9120398
Faitová, T., Svanberg, R., Da Cunha-Bang, C., Ilett, E. E., Jørgensen, M., Noguera-Julian, M., et al. (2022). The gut microbiome in patients with chronic lymphocytic leukemia. Haematologica 107, 2238–2243. doi: 10.3324/haematol.2021.280455
Ferreri, A. J. M., Govi, S., and Ponzoni, M. (2013). Marginal zone lymphomas and infectious agents. Semin. Cancer Biol. 23, 431–440. doi: 10.1016/j.semcancer.2013.09.004
Fontana, A., Panebianco, C., Picchianti-Diamanti, A., Laganà, B., Cavalieri, D., Potenza, A., et al. (2019). Gut microbiota profiles differ among individuals depending on their region of origin: an Italian pilot study. Int. J. Environ. Res. Public Health 16:4065. doi: 10.3390/ijerph16214065
Frankel, A. E., Coughlin, L. A., Kim, J., Froehlich, T. W., Xie, Y., Frenkel, E. P., et al. (2017). Metagenomic shotgun sequencing and unbiased Metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia (United States) 19, 848–855. doi: 10.1016/j.neo.2017.08.004
Geng, H. W., Yin, F. Y., Zhang, Z. F., Gong, X., and Yang, Y. (2021). Butyrate suppresses glucose metabolism of colorectal cancer cells via GPR109a-AKT Signaling pathway and enhances chemotherapy. Front. Mol. Biosci. 8, 1–9. doi: 10.3389/fmolb.2021.634874
Greenbaum, S., Sheiner, E., Wainstock, T., Segal, I., Ben-Harush, M., Sergienko, R., et al. (2018). Cesarean delivery and childhood malignancies: a single-Center, population-based cohort study. J. Pediatr. 197, 292–296.e3. doi: 10.1016/j.jpeds.2017.12.049
Haiko, J., and Westerlund-Wikström, B. (2013). The role of the bacterial flagellum in adhesion and virulence. Biology (Basel) 2, 1242–1267. doi: 10.3390/biology2041242
Han, Y., Zhang, F., Wang, J., Zhu, Y., Dai, J., Bu, Y., et al. (2016). Application of glutamine-enriched nutrition therapy in childhood acute lymphoblastic leukemia. Nutr. J. 15, 1–8. doi: 10.1186/s12937-016-0187-4
Hong, T., Wang, R., Wang, X., Yang, S., Wang, W., Gao, Q., et al. (2021). Interplay between the intestinal microbiota and acute graft-versus-host disease: experimental evidence and clinical significance. Front. Immunol. 12, 1–16. doi: 10.3389/fimmu.2021.644982
Hosomi, O., Misawa, Y., Takeya, A., Matahira, Y., Sugahara, K., Kubohara, Y., et al. (2009). Novel oligosaccharide has suppressive activity against human leukemia cell proliferation. Glycoconj. J. 26, 189–198. doi: 10.1007/s10719-008-9175-z
Hou, K., Wu, Z. X., Chen, X. Y., Wang, J. Q., Zhang, D., Xiao, C., et al. (2022). Microbiota in health and diseases. Signal Transduct. Target. Ther. 7:381. doi: 10.1038/s41392-022-01241-2
Huang, Y., Tang, J., Cai, Z., Zhou, K., Chang, L., Bai, Y., et al. (2020). Prevotella induces the production of Th17 cells in the colon of mice. J Immunol Res 2020:9607328. doi: 10.1155/2020/9607328
Ingham, A. C., Kielsen, K., Mordhorst, H., Ifversen, M., Aarestrup, F. M., Müller, K. G., et al. (2021). Microbiota long-term dynamics and prediction of acute graft-versus-host disease in pediatric allogeneic stem cell transplantation. Microbiome 9, 1–28. doi: 10.1101/2021.02.19.21252040
Iyama, S., Sato, T., Tatsumi, H., Hashimoto, A., Tatekoshi, A., Kamihara, Y., et al. (2014). Efficacy of enteral supplementation enriched with glutamine, fiber, and oligosaccharide on mucosal injury following hematopoietic stem cell transplantation. Case Rep. Oncol. 7, 692–699. doi: 10.1159/000368714
Jasiński, M., Biliński, J., and Basak, G. W. (2021). The role of the gut microbiome in pathogenesis, biology, and treatment of plasma cell Dyscrasias. Front. Oncol. 11, 1–10. doi: 10.3389/fonc.2021.741376
Jørgensen, M., Nørgaard, J. C., Ilett, E. E., Marandi, R. Z., Noguera-Julian, M., Paredes, R., et al. (2022). Metabolic potential of the gut microbiome is significantly impacted by conditioning regimen in allogeneic hematopoietic stem cell transplantation recipients. Int. J. Mol. Sci. 23:1115. doi: 10.3390/ijms231911115
Kalia, V. C., Patel, S. K. S., Cho, B. K., Wood, T. K., and Lee, J. K. (2022). Emerging applications of bacteria as antitumor agents. Semin. Cancer Biol. 2020, 1014–1025. doi: 10.1016/j.semcancer.2021.05.012
Kawari, M., Akhtar, M., Sager, M., Basbous, Z., Baydoun, I., Kabanja, J., et al. (2019). Alterations of gut microbiome in untreated chronic lymphocytic Leukemia (CLL); future therapeutic potentials. Blood 134:5455. doi: 10.1182/blood-2019-121643
Keykhaei, M., Masinaei, M., Mohammadi, E., Azadnajafabad, S., Rezaei, N., Saeedi Moghaddam, S., et al. (2021). A global, regional, and national survey on burden and quality of care index (QCI) of hematologic malignancies; global burden of disease systematic analysis 1990–2017. Exp. Hematol. Oncol. 10, 1–15. doi: 10.1186/s40164-021-00198-2
Khoury, J. D., Solary, E., Abla, O., Akkari, Y., Alaggio, R., Apperley, J. F., et al. (2022). The 5th edition of the World Health Organization classification of Haematolymphoid tumours: Myeloid and histiocytic/dendritic neoplasms. Leukemia 36, 1703–1719. doi: 10.1038/s41375-022-01613-1
Knudsen, K. E. B., Lærke, H. N., Hedemann, M. S., Nielsen, T. S., Ingerslev, A. K., Nielsen, D. S. G., et al. (2018). Impact of diet-modulated butyrate production on intestinal barrier function and inflammation. Nutrients 10:1499. doi: 10.3390/nu10101499
Kocarnik, J. M., Compton, K., Dean, F. E., Fu, W., Gaw, B. L., Harvey, J. D., et al. (2022). Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019 a systematic analysis for the global burden of disease study 2019. JAMA Oncol. 8, 420–444. doi: 10.1001/jamaoncol.2021.6987
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cells 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Kuo, S. H., and Cheng, A. L. (2013). Helicobacter pylori and mucosa-associated lymphoid tissue: what’s new. Hematol. Am. Soc. Hematol. Educ. Program 2013, 109–117. doi: 10.1182/asheducation-2013.1.109
Lavelle, E. C., Murphy, C., O’Neill, L. A. J., and Creagh, E. M. (2010). The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 3, 17–28. doi: 10.1038/mi.2009.124
Lax, S., Smith, D. P., Hampton-Marcell, J., Owens, S. M., Handley, K. M., Scott, N. M., et al. (2014). Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345, 1048–1052. doi: 10.1126/science.1254529
Li, W., Deng, X., and Chen, T. (2021). Exploring the modulatory effects of gut microbiota in anti-cancer therapy. Front. Oncol. 11, 1–11. doi: 10.3389/fonc.2021.644454
Li, Z., Quan, G., Jiang, X., Yang, Y., Ding, X., Zhang, D., et al. (2018). Effects of metabolites derived from gut microbiota and hosts on pathogens. Front. Cell. Infect. Microbiol. 8:314. doi: 10.3389/fcimb.2018.00314
Li, H., Sun, B., Ning, X., Jiang, S., and Sun, L. (2019). A comparative analysis of Edwardsiella tarda-induced transcriptome profiles in RAW264.7 cells reveals new insights into the strategy of bacterial immune evasion. Int. J. Mol. Sci. 20:151. doi: 10.3390/ijms21010151
Li, J., Zhang, A. H., Wu, F. F., and Wang, X. J. (2022). Alterations in the gut microbiota and their metabolites in colorectal cancer: recent Progress and future prospects. Front. Oncol. 12, 1–14. doi: 10.3389/fonc.2022.841552
Linares, M., and Hermouet, S. (2022). Editorial: the role of microorganisms in multiple myeloma. Front. Immunol. 13, 1–4. doi: 10.3389/fimmu.2022.960829
Liu, P., Holman, C. D. J., Jin, J., and Zhang, M. (2015). Diet and risk of adult leukemia: a multicenter case–control study in China. Cancer Causes Control 26, 1141–1151. doi: 10.1007/s10552-015-0608-2
Liu, X., Zou, Y., Ruan, M., Chang, L., Chen, X., Wang, S., et al. (2020). Pediatric acute lymphoblastic Leukemia patients exhibit distinctive alterations in the gut microbiota. Front. Cell. Infect. Microbiol. 10:558799. doi: 10.3389/fcimb.2020.618747
Liu, X., Zou, Y., Zhang, Y., Liu, L., Duan, Y., Zhang, A., et al. (2021). Characteristics in gut microbiome is associated with chemotherapy-induced pneumonia in pediatric acute lymphoblastic leukemia. BMC Cancer 21, 1–9. doi: 10.1186/s12885-021-08917-y
Ma, T., Chen, Y., Li, L. J., and Zhang, L. S. (2021). Opportunities and challenges for gut microbiota in acute Leukemia. Front. Oncol. 11, 1–10. doi: 10.3389/fonc.2021.692951
Ma, J., Sun, L., Liu, Y., Ren, H., Shen, Y., Bi, F., et al. (2020). Alter between gut bacteria and blood metabolites and the anti-tumor effects of Faecalibacterium prausnitzii in breast cancer. BMC Microbiol. 20:82. doi: 10.1186/s12866-020-01739-1
Marcoux, S., Soullane, S., Lee, G. E., and Auger, N. (2022). Association between caesarean birth and childhood cancer: an age-lagged approach. Acta Paediatr. 2022, 1–8. doi: 10.1111/apa.16335
Margolis, E. B., Alfaro, G. M., Sun, Y., Dallas, R. H., Allison, K. J., Ferrolino, J., et al. (2023). Microbiota predict infections and acute graft-versus-host disease after Pediatric allogeneic hematopoietic stem cell transplantation. J. Infect. Dis. :jiad190. doi: 10.1093/infdis/jiad190
Masetti, R., Muratore, E., Leardini, D., Zama, D., Turroni, S., Brigidi, P., et al. (2021). Gut microbiome in pediatric acute leukemia: from predisposition to cure. Blood Adv. 5, 4619–4629. doi: 10.1182/bloodadvances.2021005129
Matson, V., Fessler, J., Bao, R., Chongsuwat, T., Zha, Y., Alegre, M. L., et al. (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (80-) 359, 104–108. doi: 10.1126/science.aao3290
Méndez-Ferrer, S., Bonnet, D., Steensma, D. P., Hasserjian, R. P., Ghobrial, I. M., Gribben, J. G., et al. (2020). Bone marrow niches in haematological malignancies. Nat. Rev. Cancer 20, 285–298. doi: 10.1038/s41568-020-0245-2
Mirzaei, R., Afaghi, A., Babakhani, S., Sohrabi, M. R., Hosseini-Fard, S. R., Babolhavaeji, K., et al. (2021). Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 139, 111619–111618. doi: 10.1016/j.biopha.2021.111619
Moleiro, J., Ferreira, S., Lage, P., and Dias, P. A. (2016). Gastric malt lymphoma: analysis of a series of consecutive patients over 20 years. United Eur. Gastroenterol. J. 4, 395–402. doi: 10.1177/2050640615612934
Momen, N. C., Olsen, J., Gissler, M., Cnattingius, S., and Li, J. (2014). Delivery by caesarean section and childhood cancer: a nationwide follow-up study in three countries. BJOG An. Int. J. Obstet. Gynaecol. 121, 1343–1350. doi: 10.1111/1471-0528.12667
Moreau, M. C., Ducluzeau, R., Guy-Grand, D., and Muller, M. C. (1978). Increase in the population of duodenal immunoglobulin a plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect. Immun. 21, 532–539. doi: 10.1128/iai.21.2.532-539.1978
Neisi, Z., Ansari-Asl, Z., Jafarinejad-Farsangi, S., Tarzi, M. E., Sedaghat, T., and Nobakht, V. (2019). Synthesis, characterization and biocompatibility of polypyrrole/cu(II) metal-organic framework nanocomposites. Colloids Surfaces B-Biointerfaces 178, 365–376. doi: 10.1016/j.colsurfb.2019.03.032
O’Rourke, J. L. (2008). Gene expression profiling in helicobacter-induced MALT lymphoma with reference to antigen drive and protective immunization. J. Gastroenterol. Hepatol. 23, S151–S156. doi: 10.1111/j.1440-1746.2008.05553.x
Olson, D., Yacoub, A. T., Gjini, A. D., Domingo, G., and Greene, J. N. (2014). Escherichia coli: an important pathogen in patients with hematologic malignancies. Mediterr J. Hematol. Infect. Dis. 6:e2014068. doi: 10.4084/MJHID.2014.068
Parlato, M., and Yeretssian, G. (2014). NOD-like receptors in intestinal homeostasis and epithelial tissue repair. Int. J. Mol. Sci. 15, 9594–9627. doi: 10.3390/ijms15069594
Pflug, N., Kluth, S., Vehreschild, J. J., Bahlo, J., Tacke, D., Biehl, L., et al. (2016). Efficacy of antineoplastic treatment is associated with the use of antibiotics that modulate intestinal microbiota. Onco. Targets. Ther. 5:399. doi: 10.1080/2162402X.2016.1150399
Pianko, M. J., Devlin, S. M., Littmann, E. R., Chansakul, A., Mastey, D., Salcedo, M., et al. (2019). Minimal residual disease negativity in multiple myeloma is associated with intestinal microbiota composition. Blood Adv. 3, 2040–2044. doi: 10.1182/bloodadvances.2019032276
Portlock, C. S., Hamlin, P. A., Gerecitano, J. F., Noy, A., Palomba, M. L., Walkley, J., et al. (2015). A positive prospective trial of antibiotic therapy in advanced stage, Non-Bulky Indolent Lymphoma. Tumor Microenviron. Ther. 2, 14–18. doi: 10.1515/tumor-2015-0001
Rahman, M. M., Islam, R., Shohag, S., Ahasan, T., Sarkar, N., Khan, H., et al. (2022). Microbiome in cancer: role in carcinogenesis and impact in therapeutic strategies. Biomed. Pharmacother. 149:112898. doi: 10.1016/j.biopha.2022.112898
Ramsay, A. G., Scott, K. P., Martin, J. C., Rincon, M. T., and Flint, H. J. (2006). Cell-associated α-amylases of butyrate-producing Firmicute bacteria from the human colon. Microbiology 152, 3281–3290. doi: 10.1099/mic.0.29233-0
Rankin, L., Groom, J., Mielke, L. A., Seillet, C., and Belz, G. T. (2013). Diversity, function, and transcriptional regulation of gut innate lymphocytes. Front. Immunol. 4, 1–15. doi: 10.3389/fonc.2013.00011
Riley, D. R., Sieber, K. B., Robinson, K. M., White, J. R., Ganesan, A., Nourbakhsh, S., et al. (2013). Bacteria-human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Comput. Biol. 9:e1003107. doi: 10.1371/journal.pcbi.1003107
Rolhion, N., and Chassaing, B. (2016). When pathogenic bacteria meet the intestinal microbiota. Philos. Trans. R Soc. B Biol. Sci. 371:504. doi: 10.1098/rstb.2015.0504
Saito, Y., Suzuki, H., Tsugawa, H., Imaeda, H., Matsuzaki, J., Hirata, K., et al. (2012). Overexpression of miR-142-5p and miR-155 in gastric mucosa-associated lymphoid tissue (MALT) lymphoma resistant to Helicobacter pylori eradication. PLoS One 7:e47396. doi: 10.1371/journal.pone.0047396
Sakon, H., Nagai, F., Morotomi, M., and Tanaka, R. (2008). Sutterella parvirubra sp. nov. and Megamonas funiformis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 58, 970–975. doi: 10.3389/fcimb.2018.00314
Schirmer, M., Smeekens, S. P., Vlamakis, H., Jaeger, M., Oosting, M., Franzosa, E. A., et al. (2016). Linking the human gut microbiome to inflammatory cytokine production capacity. Cells 167:1897. doi: 10.1016/j.cell.2016.11.046
Sędzikowska, A., and Szablewski, L. (2021). Human gut microbiota in health and selected cancers. Int. J. Mol. Sci. 22:3440. doi: 10.3390/ijms222413440
Sepich-Poore, G. D., Zitvogel, L., Straussman, R., Hasty, J., Wargo, J. A., and Knight, R. (2021). The microbiome and human cancer. Science (80-) 371:eabc4552. doi: 10.1126/science.abc4552
Shapiro, Y. N., Peppercorn, J. M., Yee, A. J., Branagan, A. R., Raje, N. S., and Donnell, E. K. O. (2021). Lifestyle considerations in multiple myeloma. Blood. Cancer J. 11:172. doi: 10.1038/s41408-021-00560-x
Shin, N. R., Whon, T. W., and Bae, J. W. (2015). Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503. doi: 10.1016/j.tibtech.2015.06.011
Shono, Y., and van den Brink, M. R. M. (2018). Gut microbiota injury in allogeneic haematopoietic stem cell transplantation. Nat. Rev. Cancer 18, 283–295. doi: 10.1038/nrc.2018.10
Singh, V., Lee, G. D., Son, H. W., Koh, H., Kim, E. S., Unno, T., et al. (2023). Butyrate producers, “the sentinel of gut”: their intestinal significance with and beyond butyrate, and prospective use as microbial therapeutics. Front. Microbiol. 13:3836. doi: 10.3389/fmicb.2022.1103836
Sivan, A., Corrales, L., Hubert, N., Williams, J. B., Aquino-Michaels, K., Earley, Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science (80-) 350, 1084–1089. doi: 10.1126/science.aac4255
Sochacka-ćwikła, A., Mączyński, M., and Regiec, A. (2022). FDA-approved drugs for hematological malignancies—the last decade review. Cancers (Basel) 14:87. doi: 10.3390/cancers14010087
Stiemsma, L. T., and Michels, K. B. (2018). The role of the microbiome in the developmental origins of health and disease. Pediatrics 141:2437. doi: 10.1542/peds.2017-2437
Strauch, U. G., Obermeier, F., Grunwald, N., Gürster, S., Dunger, N., Schultz, M., et al. (2005). Influence of intestinal bacteria on induction of regulatory T cells: lessons from a transfer model of colitis. Gut 54, 1546–1552. doi: 10.1136/gut.2004.059451
Su, Q., Sun, X., Zhu, L., Yan, Q., Zheng, P., Mao, Y., et al. (2021). Breastfeeding and the risk of childhood cancer: a systematic review and dose-response meta-analysis. BMC Med. 19:1950. doi: 10.1186/s12916-021-01950-5
Tang, W., Zhu, G., Shi, Q., Yang, S., Ma, T., Mishra, S. K., et al. (2019). Characterizing the microbiota in gastrointestinal tract segments of Rhabdophis subminiatus: dynamic changes and functional predictions. Microbiology 8, 1–18. doi: 10.1002/mbo3.789
Tomita, Y., Goto, Y., Sakata, S., Imamura, K., Minemura, A., Oka, K., et al. (2022). Clostridium butyricum therapy restores the decreased efficacy of immune checkpoint blockade in lung cancer patients receiving proton pump inhibitors. Oncoimmunology 11:1010. doi: 10.1080/2162402X.2022.2081010
Tsvetikova, S. A., and Koshel, E. I. (2020). Microbiota and cancer: host cellular mechanisms activated by gut microbial metabolites. Int. J. Med. Microbiol. 310:151425. doi: 10.1016/j.ijmm.2020.151425
Ubeda, C., Taur, Y., Jenq, R. R., Equinda, M. J., Son, T., Samstein, M., et al. (2010). Vancomycin-resistant enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120, 4332–4341. doi: 10.1172/JCI43918
Uribe-Herranz, M., Klein-González, N., Rodríguez-Lobato, L. G., Juan, M., and de Larrea, C. F. (2021). Gut microbiota influence in hematological malignancies: from genesis to cure. Int. J. Mol. Sci. 22, 1–27. doi: 10.3390/ijms22031026
Vacca, M., Celano, G., Calabrese, F. M., Portincasa, P., Gobbetti, M., and De Angelis, M. (2020). The controversial role of human gut lachnospiraceae. Microorganisms 8, 1–25. doi: 10.3390/microorganisms8040573
van den Bogert, B., Erkus, O., Boekhorst, J., de Goffau, M., Smid, E. J., Zoetendal, E. G., et al. (2013). Diversity of human small intestinal streptococcus and Veillonella populations. FEMS Microbiol. Ecol. 85, 376–388. doi: 10.1111/1574-6941.12127
Venegas, D. P., De La Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:1486. doi: 10.3389/fimmu.2019.01486
Vogtmann, E., and Goedert, J. J. (2016). Epidemiologic studies of the human microbiome and cancer. Br. J. Cancer 114, 237–242. doi: 10.1038/bjc.2015.465
Wang, R., Yang, X., Liu, J., Zhong, F., Zhang, C., Chen, Y., et al. (2022). Gut microbiota regulates acute myeloid leukaemia via alteration of intestinal barrier function mediated by butyrate. Nat. Commun. 13, 1–18. doi: 10.1038/s41467-022-30240-8
Wotherspoon, A. C., Ortiz-Hidalgo, C., Falzon, M. R., and Isaacson, P. G. (1991). Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 338, 1175–1176.
Yu, D., Yu, X., Ye, A., Xu, C., Li, X., Geng, W., et al. (2021). Profiling of gut microbial dysbiosis in adults with myeloid leukemia. FEBS Open Bio. 11, 2050–2059. doi: 10.1002/2211-5463.13193
Yuan, L., Wang, W., Zhang, W., Zhang, Y., Wei, C., Li, J., et al. (2021). Gut microbiota in untreated diffuse large B cell lymphoma patients. Front. Microbiol. 12:696. doi: 10.3389/fmicb.2021.646361
Zhang, B., Gu, J., Liu, J., Huang, B., and Li, J. (2019). Fecal microbiota taxonomic shifts in Chinese multiple myeloma patients analyzed by quantitative Polimerase chain reaction (QPCR) and 16S rRNA high-throughput sequencing. Med. Sci. Monit. 25:8269. doi: 10.12659/MSM.919988
Zheng, D., Liwinski, T., and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7
Zhou, Y. J., Zhao, D. D., Liu, H., Chen, H. T., Li, J. J., Mu, X. Q., et al. (2017). Cancer killers in the human gut microbiota: diverse phylogeny and broad spectra. Oncotarget 8, 49574–49591. doi: 10.18632/oncotarget.17319
Zhou, Y., Zhou, C., and Zhang, A. (2022). Gut microbiota in acute leukemia: current evidence and future directions. Front. Microbiol. 13, 1–14. doi: 10.3389/fmicb.2022.1045497
Zimmerman, M. A., Singh, N., Martin, P. M., Thangaraju, M., Ganapathy, V., Waller, J. L., et al. (2012). Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am. J. Physiol. 302:2011. doi: 10.1152/ajpgi.00543.2011
Zou, Y., Lin, X., Xue, W., Tuo, L., Chen, M. S., Chen, X. H., et al. (2021). Characterization and description of Faecalibacterium butyricigenerans sp. nov. and F. longum sp. nov., isolated from human faeces. Sci. Rep. 11, 1–13. doi: 10.1038/s41598-021-90786-3
Keywords: hematologic cancer, leukemia, lymphoma, microbiota, multiple myeloma
Citation: Guevara-Ramírez P, Cadena-Ullauri S, Paz-Cruz E, Tamayo-Trujillo R, Ruiz-Pozo VA and Zambrano AK (2023) Role of the gut microbiota in hematologic cancer. Front. Microbiol. 14:1185787. doi: 10.3389/fmicb.2023.1185787
Received: 14 March 2023; Accepted: 11 August 2023;
Published: 25 August 2023.
Edited by:
Jinbo Xiong, Ningbo University, ChinaReviewed by:
Dulcenombre Gomez-Garre, Hospital Clinico San Carlos, SpainCopyright © 2023 Guevara-Ramírez, Cadena-Ullauri, Paz-Cruz, Tamayo-Trujillo, Ruiz-Pozo and Zambrano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana Karina Zambrano, YW5hemFtYnJhbm8xN0Bob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.