
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 25 May 2023
Sec. Infectious Agents and Disease
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1171423
 Keyu Wang1†
Keyu Wang1† Meiliang Gong1†
Meiliang Gong1† Sumin Zhao2†
Sumin Zhao2† Chengcai Lai3
Chengcai Lai3 Lingna Zhao4
Lingna Zhao4 Sijie Cheng5
Sijie Cheng5 Min Xia6
Min Xia6 Yuru Li1
Yuru Li1 Kun Wang1
Kun Wang1 Heqiang Sun1
Heqiang Sun1 Pingjun Zhu7
Pingjun Zhu7 Yu Zhou1*
Yu Zhou1* Qiangguo Ao8*
Qiangguo Ao8* Xinli Deng1*
Xinli Deng1*Long noncoding RNAs (lncRNAs) have been associated with a variety of biological activities, including immune responses. However, the function of lncRNAs in antiviral innate immune responses are not fully understood. Here, we identified a novel lncRNA, termed dual function regulating influenza virus (DFRV), elevating in a dose- and time-dependent manner during influenza A virus (IAV) infection, which was dependent on the NFκB signaling pathway. Meanwhile, DFRV was spliced into two transcripts post IAV infection, in which DFRV long suppress the viral replication while DFRV short plays the opposite role. Moreover, DFRV regulates IL-1β and TNF-α via activating several pro-inflammatory signaling cascades, including NFκB, STAT3, PI3K, AKT, ERK1/2 and p38. Besides, DFRV short can inhibit DFRV long expression in a dose-dependent manner. Collectively, our studies reveal that DFRV may act as a potential dual-regulator to preserve innate immune homeostasis in IAV infection.
Influenza is an infectious respiratory disease caused by influenza A and influenza B viruses in humans (Uyeki et al., 2022). Approximately 1 billion infections, 3–5 million cases of serious illness, and 300,000–500,000 deaths are predicted to emerge from annual influenza epidemics, according to the WHO (Krammer et al., 2018). Host innate immunity is a critical barrier that prevents virus reproduction and transmission in new hosts. Both type I interferons [namely, interferon-α (IFNα) and IFNβ) and type III interferons (namely, IFNλ) are essential for the antiviral immune response in epithelial cells (Crotta et al., 2013; Wang et al., 2017). Additionally, several innate immune cells like neutrophils, macrophages, and dendritic cells become activated and secrete pro-inflammatory cytokines (Helft et al., 2012; Tecchio et al., 2014; Pernet et al., 2019), which attracts other innate cells like natural killer cells and pro-inflammatory monocytes to the lung where they can eliminate infected cells and prevent viral infection (Chen et al., 2018; Krammer et al., 2018). Despite the fact that these host reactions to infection are essential for the final viral clearance and activation of adaptive immune responses, while excessive inflammation exacerbates the immunopathology, which is known as “cytokine storm” (Liu, Q. et al., 2016). Although the signaling pathway of antiviral responses has been well characterized, further research is still needed to elucidate the exact mechanism.
Long noncoding RNAs (lncRNAs) are defined as transcripts of more than 200 nucleotides that are not translated into proteins (Kopp and Mendell, 2018; Bridges et al., 2021), and are involved with a variety of biological processes, including cell cycle (Kitagawa et al., 2013), differentiation (Fatica and Bozzoni, 2014; Delas et al., 2017), and metabolism (Sirey et al., 2019), as well as a number of disease (Batista and Chang, 2013; Wang et al., 2013; Huarte, 2015; Li et al., 2021). Recent research indicates that lncRNAs are crucial to the antiviral immune response. For instance, LncRNA PCNAP1 increases tumor growth in hepatocellular carcinoma and affects the replication of the hepatitis B virus (Feng et al., 2019). In vesicular stomatitis virus (VSV) infection, Lnc-Lsm3b effectively competes with viral RNA for binding RIG-I via multibranch loop topologies (Jiang et al., 2018), and Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response (Lin et al., 2019). In influenza virus infection, lncRNA-NRAV promotes IAV replication and pathogenicity by regulating the initial transcription of multiple critical interferon-stimulated genes (ISGs; Ouyang et al., 2014), whereas lncRNA-NEAT1 promotes the expression of antiviral genes such as interleukin-8 (IL-8; Imamura et al., 2014). In a prior study, we identified two lncRNAs called AVAN and IVRPIE, of which AVAN promotes antiviral innate immunity by interacting with TRIM25 and enhancing the transcription of FOXO3a (Lai et al., 2021), while IVRPIE inhibits IAV replication by promoting the expression of IFN and ISGs (Zhao et al., 2020). However, the biological significance and the underlying mechanisms for the lncRNAs in IAV induced immunity and inflammation remain to be further elaborated.
In this study, we defined a novel lncRNA, named DFRV (dual function regulating influenza virus), which plays a dual function in IAV infection. DFRV was dramatically increased in a time-and dose-dependent manner post IAV infection. In addition, DFRV were mainly located in the nucleus, and the rapid amplification of cDNA ends (RACE) data revealed that DFRV has two transcript variants, which showed the opposite function. Moreover, DFRV-long inhibit IAV replication via promoting pro-inflammation signaling pathway, while DFRV-short suppress DFRV-long expression. These results reveal DFRV as a potential dual-regulator to maintain innate immune homeostasis following IAV infection.
The Academy of Military Medical Sciences provided all of the viruses utilized in this investigation. The Influenza A virus A/Beijing/501/2009 (BJ501, H1N1), A/Wisconsin/67/2005(H3N2), A/Anhui/01/2005 (H5N1), A/Hebei/01/2013 (H7N9), B/Ann Arbor/1/66 (IBV) and Sendai virus (SeV) were propagated in embryonated chicken eggs via the allantoic route. Adenovirus was propagated in Vero cells.
MDCK, A549 and THP-1 cells were purchased from ATCC; MDCK cells were cultured in DMEM (Gibco) supplemented with 10% FBS and 100 U/mL penicillin–streptomycin; A549 cells were cultured in DMEM/F-12(1:1) basic culture medium (Gibco) supplemented with 10% FBS and 100 U/mL penicillin–streptomycin; and THP-1 cells were cultured in RPMI-1640 medium (Gibco) supplemented with 10% FBS and 100 U/mL penicillin–streptomycin. All of the cells were cultured at 37°C in a humidified atmosphere containing 5% CO2.
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen). The commercial PrimeScript RT Master Mix (Takara) was used to create cDNA by reverse transcription, which was then subjected to quantitative PCR on the ABI 7500 PCR System (Applied Biosystems) utilizing TB Green® Premix Ex TaqTM II (Tli RNaseH Plus; Takara). qRT-PCR was performed with primer annealing at 50°C for 2 min, an initial denaturation for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C and 30 s at 72°C. Primer pairs (Supplementary Table S1) were designed using Primer Premier Software 5.0 and synthesized by Invitrogen. To make primer efficiency curves, the simple cDNA was diluted for a 5-fold gradient, and 2 μL of each diluted-simple was used as the template to amplify the cDNA with the primers of target gene and internal reference gene respectively, and the melting curve was analyzed at 60–95°C. The results showed that the R2 of each standard curve is 0.99 (example for DFRV primer), and the amplification efficiency of PCR is calculated to be between 90 and 110% (Supplementary Figures 1A,B). The 2−ΔΔCt method was used to calculate expression relative to the internal control, which normalized to GAPDH expression. ABI 7500 SDS software version 1.3 was used to evaluate the quantitative results.
The DFRV gene was cloned into the pcDNA 3.1(+) plasmid. DFRV-specific Antisense oligonucleotides (ASOs), and negative control ASO were synthesized by Integrated DNA Technologies (IDT). The targeting sequences of the ASOs were as follows: ASO Negative Control: 5′-mG*mC*mG*mA*mC*T*A*T*A*C*G*C*G*C*A*mA*mU*mA*mU*mG-3′, ASO1: 5′-mC*mG*mU*mC*mG*T*C*C*A*A*G*C*C*T*T*mC*mC*mC*mC*mA-3′, ASO2: 5′-mU*mU*mG*mA*mC*A*G*C*G*T*A*G*G*C*A*mC*mC*mA*mG*mC-3′, ASO3: 5′-mA*mC*mU*mU*mG*C*C*A*C*G*T*T*A*G*A*mG*mG*mU*mG*mA-3′. In the above ASO targeting sequences, * represents phosphorothioate bonds and m represents 2′OMe (2′-O-methyl base). Cells were transfected with plasmid or ASOs using jetPRIME (Polyplus) according to the manufacturer’s instructions.
Coding potential calculator 2 was used to examine the non-coding potential of DFRV1 (Kang et al., 2017), and s-fold2 (Rennie et al., 2019), MXfold23 (Sato et al., 2021), pKiss4 (Janssen and Giegerich, 2015), RNAshapes5 (Janssen and Giegerich, 2015), RNAfold6 (Dimonte et al., 2021) was used to predict the secondary structure for DFRV.
The manufacturer’s recommendations were followed while separating the cytoplasmic and nuclear fractions using the PARIS Kit Protein and RNA Isolation System (Invitrogen). Briefly, Cell Fractionation Buffer was used to lyse A549 cells on ice for 5–10 min. The lysates were centrifuged 1–5 min at 4°C and 500 × g. A fresh tube was filled with the supernatant cytoplasmic fraction after it had been aspirated from the nuclear pellet. The nuclear pellets were washed Cell Fractionation Buffer and then vigorously lysed with Cell Disruption Buffer using a pipet or vortex. RNA was extracted using TRIzol Reagent and quantitative PCR was performed to detect DFRV expression in cytoplasmic and nuclear fraction.
According to the manufacturer’s instructions, the SMARTerTM RACE cDNA Amplification Kit (Clontech) was used to conduct the 5′ and 3′ RACE analyses. The RACE PCR products were cloned into pMD-19 T vector (Takara) and sequenced. The primers for DFRV RACE were as follows:
5′RACE outer: 5′-GACAGCGTAGGCACCAGCGAGAC-3′,
5′RACE inner: 5′-CTGCTAGCAAGCGACGAATTGCG-3′,
3′RACE outer: 5′-GCGCCAGTTCTTCCTAAAAGGAAAG-3′,
3′RACE inner: 5′-CAATGGTCACCTCTAACGTGGCAAG-3′.
Cytokines levels were measured using an ELISA kit according to the manufacturer’s instructions. Human precoated IL-1β ELISA kit (Cat. NO. 1110122), Human precoated TNF-α ELISA kit (Cat. NO. 1117202) and human IFN-α ELISA kit (Cat. NO. 1110012) were purchased from Dakewe, and human IFN-β ELISA kit (Cat. NO. E-EL-H0085c) were purchased from Elabscience.
The primary antibodies used in the analysis were anti-NFκB p65, anti-phospho-NFκB p65 (Ser536), anti-STAT3, anti-phospho-STAT3 (Tyr705), anti-PI3K, anti-phospho-PI3K p85 (Tyr458)/p55 (Tyr199), anti-Akt, anti-phospho-Akt (Thr308), anti-p38, anti-phospho-p38 (Thr180/Tyr182), anti-SAPK/JNK, anti-phospho-SAPK/JNK (Thr183/Tyr185), anti-Erk1/2, anti-phospho-Erk1/2 (Thr202/Tyr204), anti-β-actin (13E5) and anti-rabbit IgG were purchased from Cell Signaling Technology, Western Chemiluminescent Horseradish Peroxidase Substrate was purchased from Millipore Corporation, Signal pathway inhibitor including Fludarabine (STAT1, 100 μM, CAT.NO. T1038), SCH772984 (MEK1/ERK1/ERK2, 100 μM, CAT.NO. T6066), MCC950 (NLRP3, 10 μM, CAT.NO. T3701), Ruxolitinib phosphate (JAK1/2, 100 μM, CAT.NO. T3043), SB203580 (p38 MAPK, 100 μM, CAT.NO. T1764), Hydroxychloroquine sulfate (TLR7/9, 100 μM, CAT.NO. T0951) and SP600125 (JNK1/2/3, 10 μM, CAT.NO. T3109) were purchased from TargetMol, PDTC (NFκB, 100 μM, CAT.NO. S3633) were purchased from SELLECK. For signal pathway inhibitor administration, A549 cells were pretreated with the inhibitor for 12 h and the supernatants were removed, followed by BJ501 infection for 24 h.
All of the cells were lysed on ice in RIPA buffer (Solarbio) supplemented with protease and phosphatase inhibitor cocktail (100 ×; Thermo Fisher) for 10 min. A quarter volume of 5 × loading dye was added to the supernatants. Then the mixtures were heated at 95°C and stored at − 80°C. SDS-PAGE was used to separate the samples, and they were then transferred to a nitrocellulose filter membrane (Roche). The membranes were blocked for 1 h while shaking at room temperature with 5% non-fat milk (Becton Dickinson) in 1 × Tris-buffered saline and 0.1% Triton 100. The membranes were incubated with primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies. The band was visualized using the Automatic chemiluminescence imaging system (Tanon). The band intensity was analyzed using Quantity One software.
RNA fluorescence in situ hybridization (RNA FISH) was performed as described previously (Liu, L. et al., 2016). Hybridization was carried out using DNA probe sets (Biosearch Technologies) according to the protocol provided by Biosearch Technologies. Cells were observed on a FV1000 confocal laser microscope (Olympus).
GraphPad Prism version 5.0 (GraphPad Software Inc.) was used to conduct all of the statistical analyses. An ANOVA was used to examine the data for measurements taken at a single time point, and a two-tailed t-test was used to further investigate the findings if a significant difference between the groups was found. Statistically significant was defined as p < 0.05. All experiments were performed in triplicate.
To investigate the functions of host lncRNAs during IAV infection, we analyzed whole transcriptional alterations using RNASeq from patients infected with IAV in the acute stage and their matching recovery-stage (9 months after recovery) in a previous study (GSE108807; Lai et al., 2021). According to the basal expression level and fold change (fold change > 2), 30 lncRNA were chosen to confirm the various expression in A549 cell by qRT-PCR, in which XLOC 010049 was highly elevated and has a high basal expression (Figures 1A,B). Therefore, XLOC 010049, termed DFRV, was chosen for further function studies.
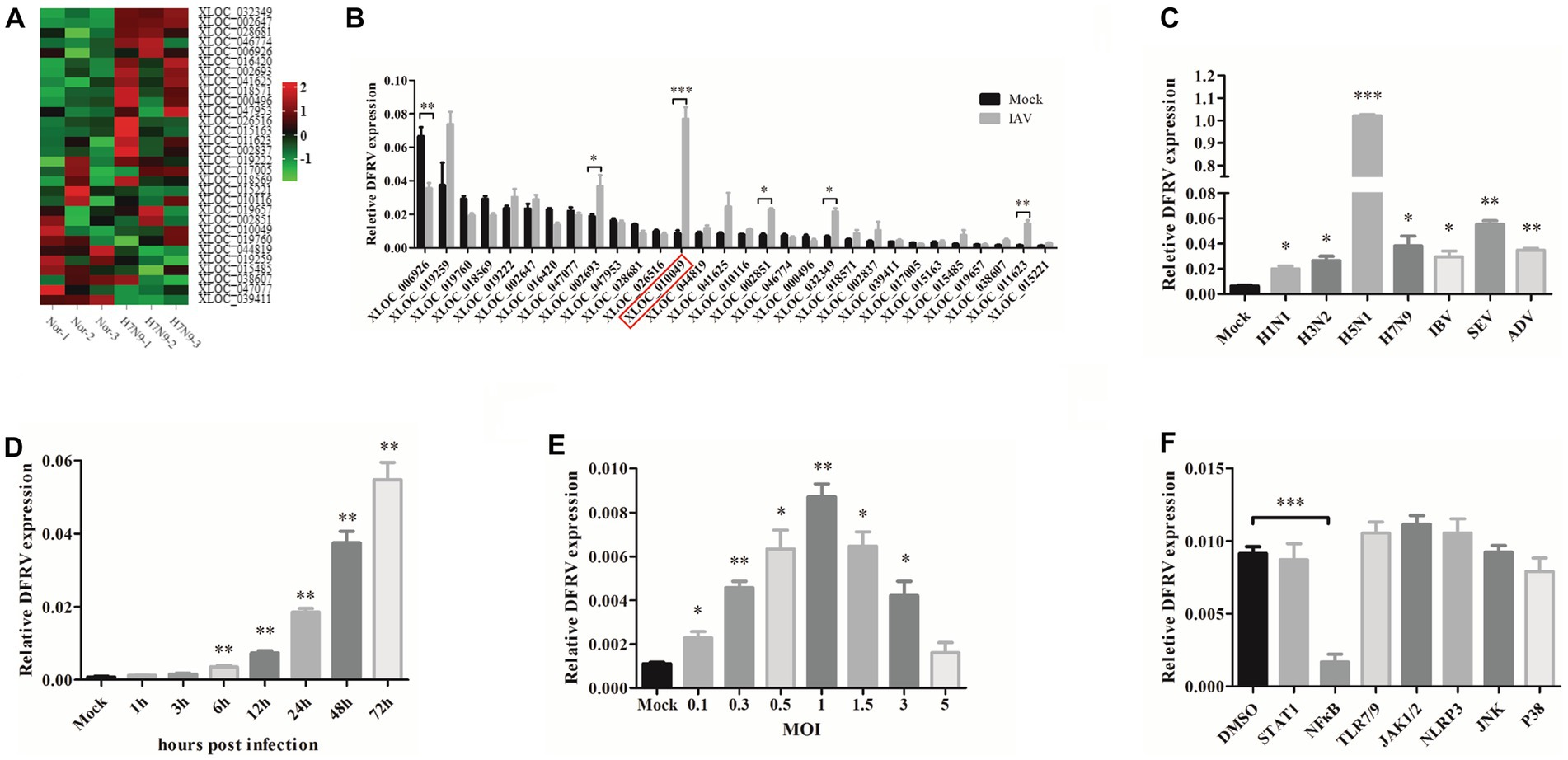
Figure 1. Long noncoding RNA dual function regulating influenza virus (lncRNA DFRV) is significantly elevated in influenza A virus (IAV) infection. (A) Cluster heat map showing 30 selected lncRNAs differentially expressed in IAV-infected patient compared with recovery-stage samples based on RNA-seq data (FC > 2; p < 0.05). (B) The differential expressions of 30 selected lncRNAs were confirmed by RT-PCR in A549 cells stimulated with BJ501 (MOI = 1) or mock for 24 h. DFRV (XLOC_010049) is indicated by red rectangle. (C) A549 cells were infected with H1N1, H3N2, H5N1, H7N9, influenza B virus (IBV), Sendai virus (SEV) and Adenovirus (ADV) at MOI = 1 for 24 h, qRT-PCR was performed to determine the DFRV expression. (D) A549 cells were infected with BJ501 at different multiplicity of infection (MOI) for 24 h, and (E) A549 cells were infected with BJ501 (MOI = 1) for different hours post infection, qRT-PCR was performed to determine the DFRV expression. (F) A549 cells were pretreated with 7 different signaling pathway inhibitors for 12 h followed by BJ501 infection for 24 h. qRT-qPCR was performed to determine the DFRV expression. All of the data are shown as the mean ± SD; n = 3. *p < 0.05; **p < 0.01;***p < 0.001.
Additionally, we discovered that DFRV was increased in A549 and THP-1 cells following influenza virus infection with H1N1, H3N2, H5N1, H7N9, influenza B virus (IBV), other RNA virus (Sendai virus, SEV), and DNA virus (Adenovirus; Figure 1C; Supplementary Figure 2A). Meanwhile, DFRV was upregulated in a dose- and time-dependent manner in A549 cell (Figures 1D,E), and similar result was achieved in the THP-1 cell (Supplementary Figures 2B,C), indicating that it principally functioned in immune cells and lung cells. Moreover, signaling pathway inhibitor was administrated to explore the regulating mechanism of DFRV expression, and we found that DFRV can be inhibited in A549 (Figure 1F) and THP-1 cells (Supplementary Figure 2E) by the kinase inhibitor of NF-κB (PDTC), and the dose-dependent effect of PDTC on inhibiting DFRV expression was validated in A549 cells and THP-1 cells (Supplementary Figures 2D,F). These findings suggest that DFRV was upregulated post virus infection, which was regulated by the NFκB signaling pathway.
To define the basic characters of DFRV, 5′ and 3′ rapid amplification of cDNA ends (RACE) were performed. Interestingly, two transcripts (named DFRV long and DFRV short) was identified. Both of the DFRV transcripts share the same 5′ sequences, while the length of 3′ sequences were 128 and 37 nt respectively, and both of which contain a polyadenylated tail (Figure 2A; Supplementary Table S2). To validate the abundance of two transcripts of DFRV, we designed a pair of qPCR primer in 3′ end of DFRV long to detect the expression of DFRV long (Supplementary Figure 3A). The result showed a similar elevating expression in DFRV total and DFRV long (Supplementary Figure 3B), and the expression level of DFRV long was higher than DFRV short, while the significance was only found in mock groups (Supplementary Figure 3C). The human lncRNA gene DFRV is located on chromosome 12q13.13, overlapping with the sense strand of poly (rC) binding protein 2 (PCBP2) gene (Figure 2B). Meanwhile, DFRV had no protein-coding potential, according to coding potential calculator 2 (Figure 2C). Besides, DFRV is primarily found in the nucleus and is identified by subcellular fractionation (Figure 2D), which is validated by RNA-FISH (Supplementary Figure 3D). Furthermore, the S-fold server was used to estimate the secondary structure of the RNA, and the results revealed that DFRV long had one more stem-loop structure at the 3′ end than DFRV short (Figure 2E), as well as in other four secondary structure prediction software (Supplementary Figures 4A–D). Based on the secondary structure data, we hypothesize that two transcripts of DFRV may play separate roles in IAV infection.
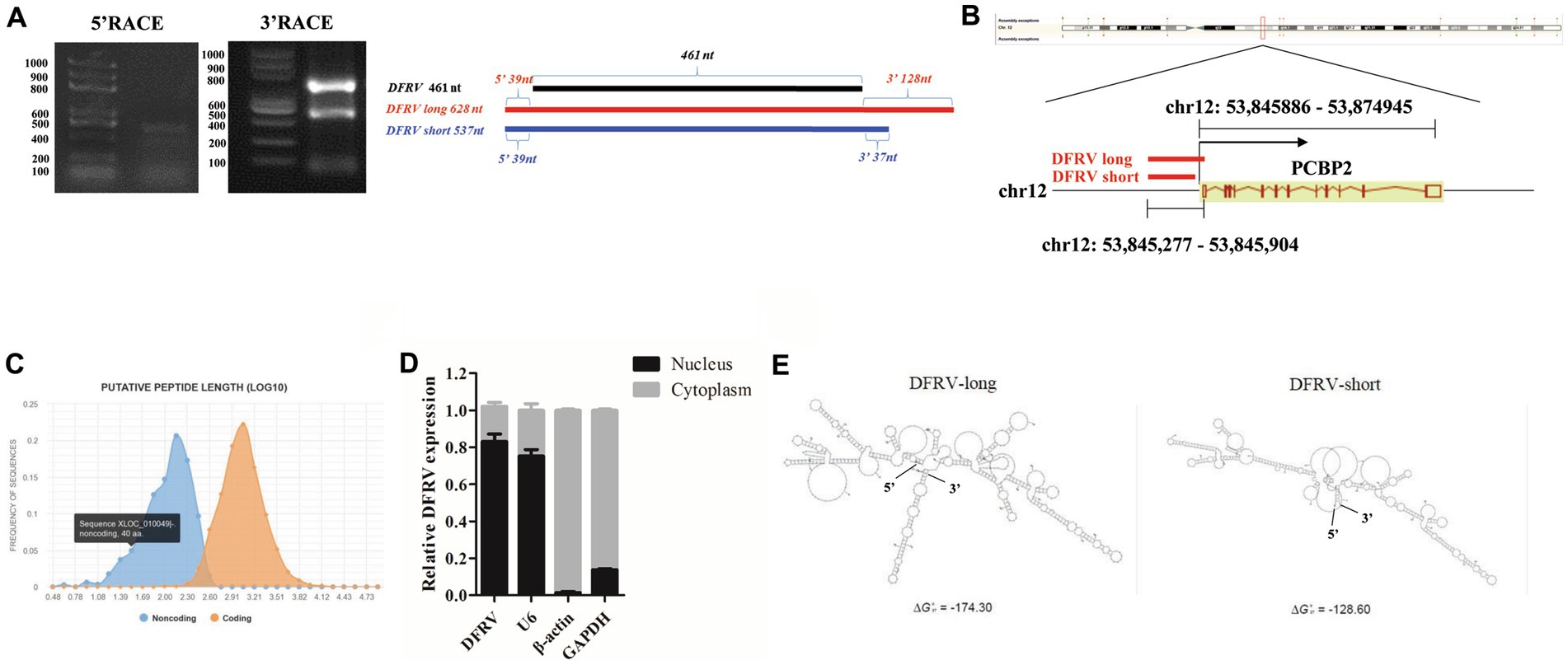
Figure 2. Two transcripts of DFRV have different structures and major located in nucleus. (A) The 5′ and 3′ end sequences of DFRV determined by RACE (left) and schematic diagram showing the RACE products of DFRV (right). (B) Schematic diagram of gene locus of DFRV. (C) Protein-coding potential analysis of DFRV was performed by coding potential calculator 2. (D) Cytoplasmic and nuclear fractions from infected A549 cells were separated, and the expression of DFRV were determined by RT-qPCR. U6 RNA served as nuclear gene expression control, β-actin and GAPDH served as cytoplasmic gene control (n = 3; means ± SEM). (E) Secondary structure predictions of DFRV were performed through S-fold, and the structure with minimum free energy (MEF) of DFRV long (left) and short (right) were exhibited.
To further assess the role of DFRV in IAV infection, A549 cells were transfected with DFRV long plasmid, and the cell viability, virus load, and antiviral related cytokines were measured. The results revealed that DFRV was highly expressed following DFRV long transfection (Figure 3A), while no significant effect was observed in the adjacent gene (PCBP2) by altering DFRV long expression (Figure 3B). Meanwhile, no difference in cell viability was detected using the MTS assay post DFRV long expression (Figure 3C). However, the viral titers were significantly alleviate in DFRV long overexpressing cells than in control cells by qRT-PCR (Figure 3D), and a similar result was achieved by TCID50 assay (Figure 3G). Moreover, both the mRNA level in A549 cells and protein level in cell culture supernatant of the pro-inflammation cytokines, IL-1β and TNF-α, were up-regulated in DFRV long overexpressing cells post IAV infection (Figures 3E,F,H,I), whereas the expression of IFN-α and β were negatively regulated (Supplementary Figures 5A-D). These results imply that DFRV long reduce IAV replication, and that pro-inflammatory cytokines, rather than cell death and IFN production, were responsible for the reduction of virus replication.
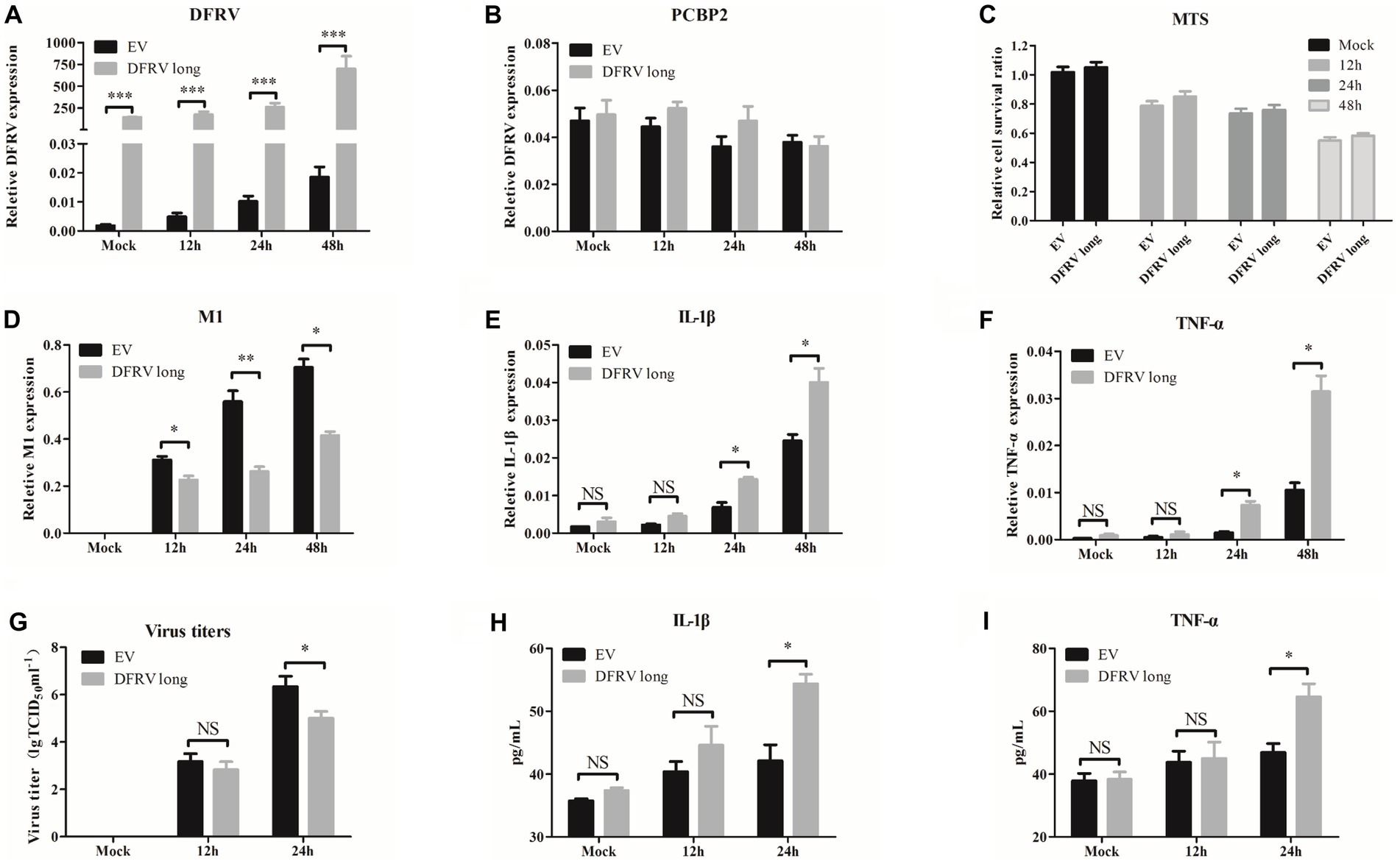
Figure 3. Dual function regulating influenza virus (DFRV) long inhibit viral replication and positively regulate IL-1β and TNF-α in A549 cells. DFRV long were transiently overexpressed in A549 cells for transfected with plasmids for 14 h, and infected with BJ501 (MOI = 1) in 12, 24, and 48 h. The expression level of DFRV (A), PCRP2 (B) was determine by qRT-PCR. The cell viability was determined by MTS (C). IAV titers were, respectively, determined by IAV segment M1 expression by qRT-PCR (D) and TCID50 assay (G). The mRNA expression level of IL-1β (E) and TNF-α (F) was determined by qRT-PCR. A549 culture supernatants were collected to measure the IL-1β (H) and TNF-α (I) expression by ELISA. All of the data are shown as the mean ± SD; n = 3. *p < 0.05; **p < 0.01;***p < 0.001, EV: Empty vector.
Consisted with the function in DFRV long, overexpression of DFRV short had no effect on the PCBP2 expression or cell viability (Figures 4A–C). Additionally, contrary to the DFRV long function, DFRV short overexpression exacerbates the virus replication (Figures 4D,G), and both mRNA and protein levels of IL-1β and TNF-α were significantly lower than control cells (Figures 4E,F,H,I). However, overexpression of DFRV short still inhibits the IFN expression as compared to empty vector group (Supplementary Figures 5E–H). Thus, the opposing effects of DFRV short and long in virus replication could be attributed to their distinct impacts on pro-inflammation cytokines.
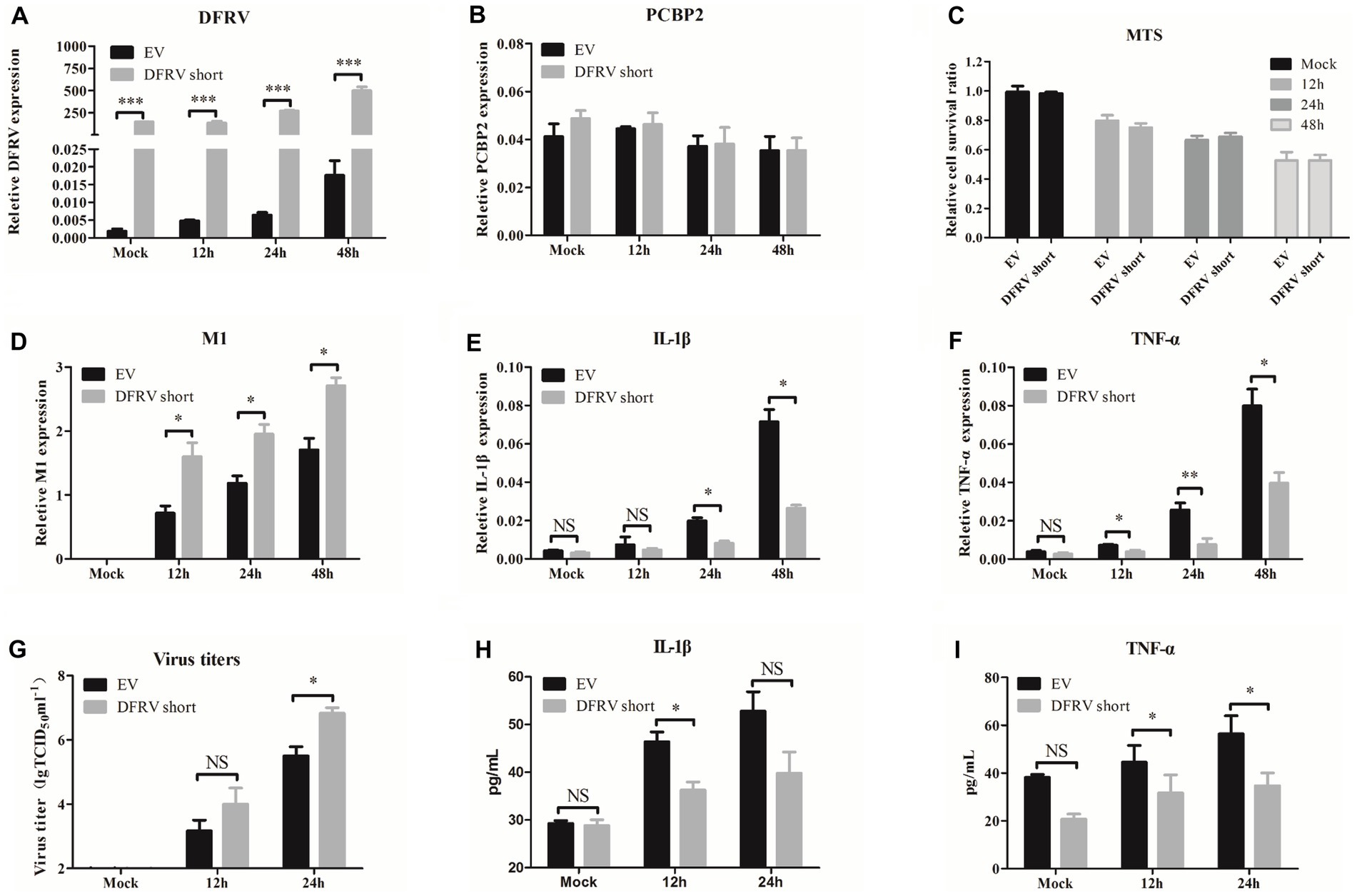
Figure 4. Dual function regulating influenza virus (DFRV) short promote viral replication and negatively regulate IL-1β and TNF-α in A549 cells. DFRV short were transiently overexpressed in A549 cells for transfected with plasmids for 14 h, and infected with BJ501 (MOI = 1) in 12, 24 and 48 h. The expression level of DFRV (A), PCRP2 (B) was determine by qRT-PCR. The cell viability was determined by MTS (C). IAV titers were, respectively, determined by IAV segment M1 expression by qRT-PCR (D) and TCID50 assay (G). The mRNA expression level of IL-1β (E) and TNF-α (F) was determined by qRT-PCR. A549 culture supernatants were collected to measure the IL-1β (H) and TNF-α (I) expression by ELISA. All of the data are shown as the mean ± SD; n = 3. *p < 0.05; **p < 0.01;***p < 0.001, EV: Empty vector.
To further investigate the function of the two transcripts, we designed 3 DFRV specific Antisense Oligonucleotides (ASO). However, only the last 91 nt in the 3′ end of the two transcripts are different and highly conservative, implying that the target of all 3 ASOs are the common sequence of two transcripts (Supplementary Figure 6A), in which ASO3 achieves the best knockdown efficiency (Supplementary Figure 6B). Following that, A549 cells were transfected with ASO3 to downregulate DFRV expression (Figure 5A), and the effect of DFRV downregulation on IAV viral replication or cytokine expression was examined as previously described. There was no significant difference in PCBP2 expression or cell viability (Figures 5B,C), as well as virus load after two transcripts were knocked down (Figures 5D,G). Contrary to DFRV short overexpression, the mRNA expression level of IL-1β and TNF-α was higher in ASO-transfected cells compared to control cells (Figures 5E,F), while no significance was observed in the protein level of cell culture supernatants (Figures 5H,I). Moreover, ASO3 reversed the downregulation of IFN caused by DFRV overexpression (Supplementary Figure 6C–F). Besides, co-transfection with DFRV long and short showed a satisfactory efficiency in A549 cells (Supplementary Figure 6G), and co-transfection with DFRV long and short had no significant effect on virus load and inflammatory response (Supplementary Figures 6H,I), while IFN-α and -β was significant suppressed post 24 h IAV infection (Supplementary Figures 6 K,L).
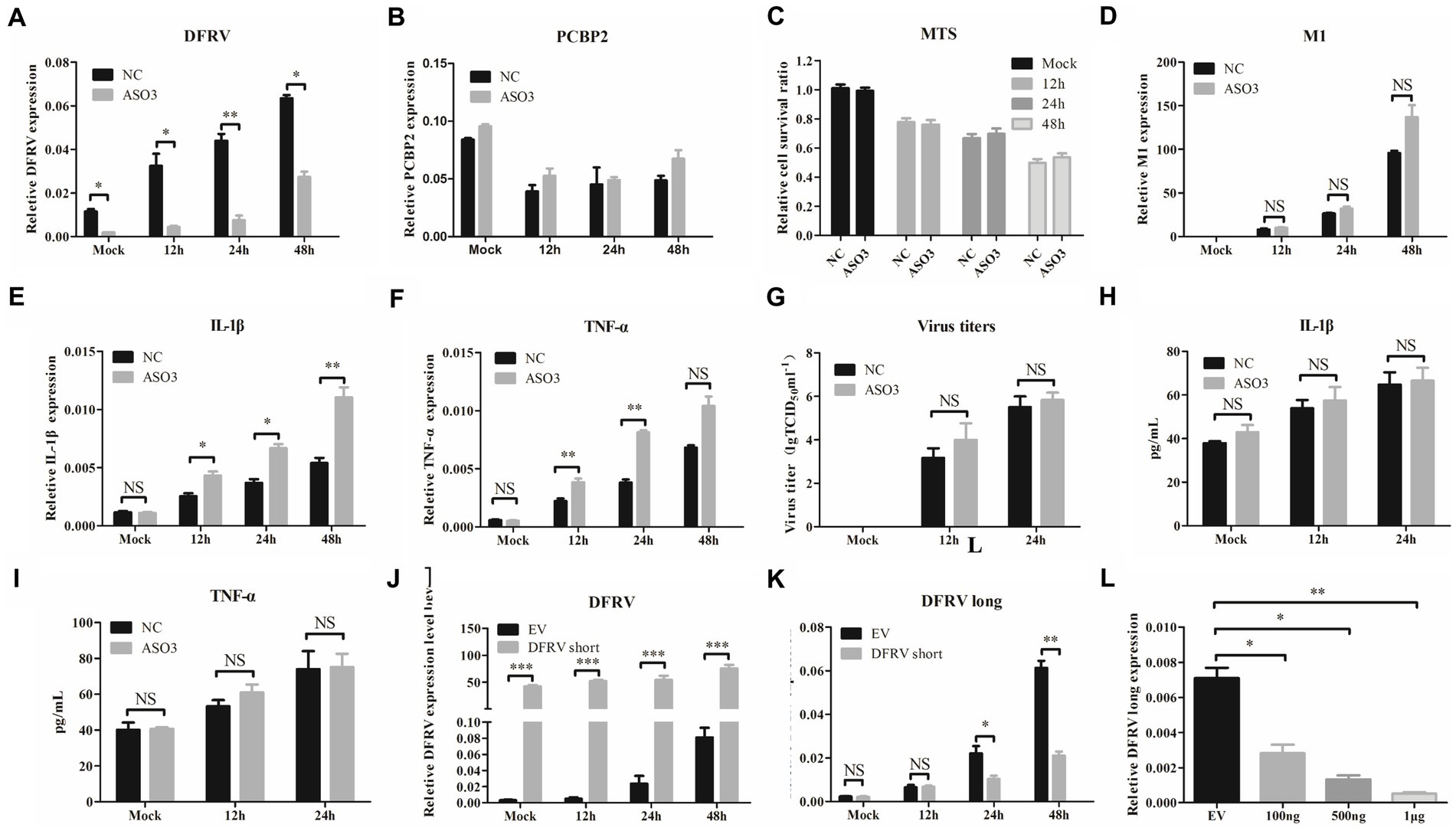
Figure 5. Dual function regulating influenza virus (DFRV) long might play a preferential role in antiviral immune response. DFRV-related ASOs were designed and transfected to A549 cells for 14 h to knockdown the DFRV expression, and infected with BJ501 (MOI = 1) in 12, 24, and 48 h. The expression level of DFRV (A), PCRP2 (B) was determine by qRT-PCR. The cell viability was determined by Cell Proliferation Assay (MTS) (C). IAV titers were, respectively, determined by IAV segment M1 expression by qRT-PCR (D) and TCID50 assay (G). The mRNA expression level of IL-1β (E) and TNF-α (F) was determined by qRT-PCR. A549 culture supernatants were collected to measure the IL-1β (H) and TNF-α (I) expression by ELISA. A549 cells were transfected with DFRV short plasmids for 14 h, and infected with BJ501 (MOI = 1) in 12, 24 and 48 h, and DFRV expression level (J), DFRV long expression level (K) was determine by qRT-PCR. (L) A549 cells were transfected with DFRV short plasmids in different copies for 14 h, and the expression level of DFRV long was determine by qRT-PCR.
To explore the interaction between the two transcripts of DFRV, we determined the expression level of DFRV long in cells transfected with DFRV short (Figure 5J). Interestingly, the results revealed that DFRV short overexpression significantly suppresses DFRV long expression post IAV infection (Figure 5K). In addition, DFRV short significantly inhibit the DFRV long expression in a dose-dependent manner (Figure 5L).
To elucidate the mechanism of the altered expression in IL-1β and TNF-α induced by DFRV, we examined the pro-inflammatory signaling pathway after DFRV overexpression. As expected, prominent phosphorylation of several pro-inflammatory signaling pathways such as NFκB, MAPKs including ERK1/2 and p38 MAPK, STAT3, PI3K and AKT were observed at 24 h after the DFRV long overexpression, in contrast, these pro-inflammatory signaling phosphorylation were weaker in DFRV short overexpression group, and the altering signaling phosphorylation of ASO were accompanied with pro-inflammatory cytokine expression (Figures 6A–G). Collectively, DFRV long may play a dual role in the antivirals immune response by activating pro-inflammatory signaling pathways.
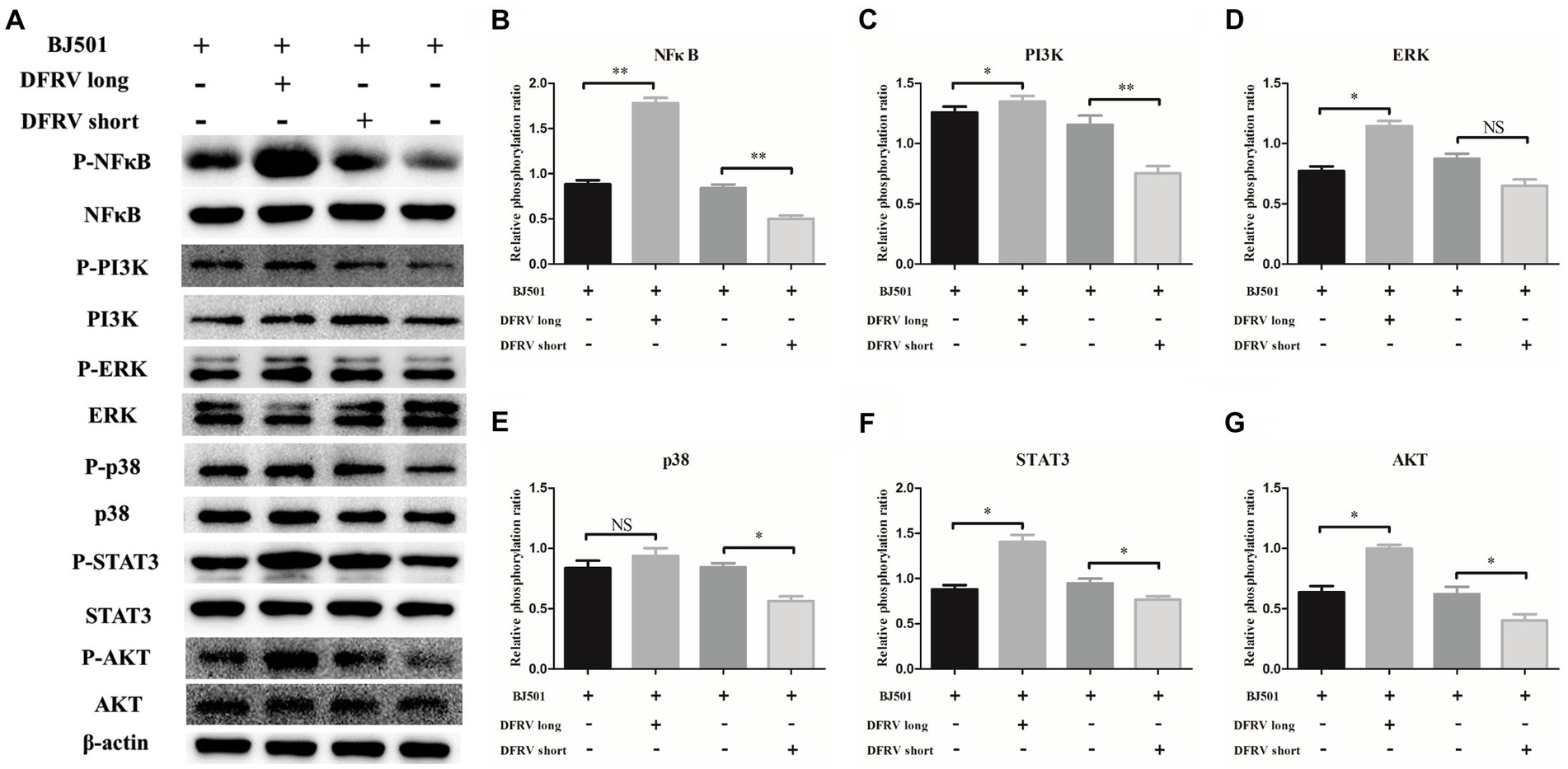
Figure 6. Dual function regulating influenza virus (DFRV) regulate the inflammatory response via several pro-inflammatory signaling pathways. (A) Western blot analysis of pro-inflammatory signaling in A549 cells transfected with DFRV long, short and ASO upon BJ501 (MOI = 1) infection for 24 h, β-actin were used as a loading control. (B-G) The phosphorylation ratio of NFκB (B), PI3K (C), ERK (D), p38 (E), STAT3 (F) and AKT (G) was calculated. All of the data are shown as the mean ± SD; n = 3. *p < 0.05; **p < 0.01; ***p < 0.001, EV: Empty vector.
LncRNA plays an important role in various biological processes, and emerging research confirms the significance lncRNAs in innate immunity and host–virus interactions (Fortes and Morris, 2016; Basavappa et al., 2019; Liu et al., 2019). It has been demonstrated that a number of lncRNAs regulate the identification by innate immune cells and the transcription of gene expression programs such as NEAT1, MAHAT, OASL-IT1 (Morchikh et al., 2017; Wang et al., 2021; Liu et al., 2022). Here, we discover a novel lncRNA termed DFRV that was significantly up-regulated during IAV infection. DFRV was split into two transcripts, each of which plays an opposing role in antiviral immunity. Moreover, DFRV long suppress viral replication by promoting the expression of IL-1β and TNF-α, and pro-inflammatory signaling pathways including NFκB, STAT3, PI3K-AKT, ERK and p38 were elevated in this biological process. Besides, DFRV long serves as a key component in antivirals immune response, while DFRV short regulates the expression of DFRV long (Figure 7).
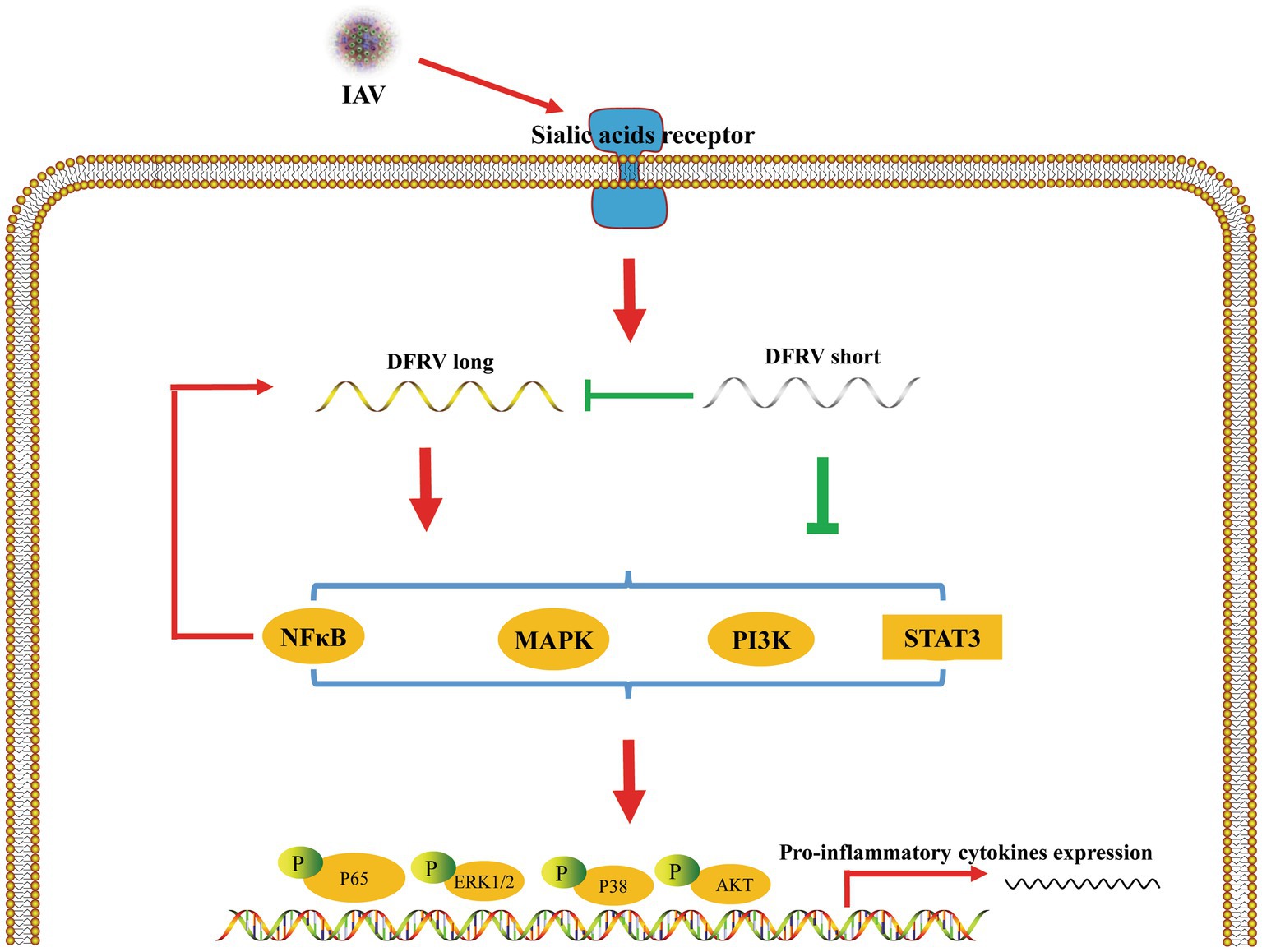
Figure 7. Schematic of the mechanisms by which DFRV regulates antivirus innate immune responses. IAV infection induced DFRV up-regulation, which is regulated by NFκB signaling pathway. DFRV is spliced to DFRV long and short post IAV infection, in which DFRV short inhibit the expression of DFRV long, and DFRV regulate pro-inflammatory cytokines expression via MAPKs, NFκB, STAT3, PI3K-AKT signaling pathways.
It has been extensively investigated how IAV infection cause symptoms in humans. Through the binding of HA to either 2, 3- or 2, 6-linked sialic acids, IAV predominantly targets airway and alveolar epithelial cells (Kobasa et al., 2004; Chutinimitkul et al., 2010; van Riel et al., 2010; Te Velthuis and Fodor, 2016). The innate immune system serves as an effective first line of defense against influenza viruses and is crucial in limiting the anatomic spread of IAV (White et al., 2008; Iwasaki and Pillai, 2014; Wang et al., 2018).Various pattern recognition receptors (PRRs) identify the viral RNA present in infected cells as foreign, activating intracellular immune pathways that result in the release of type I interferons (IFNs), pro-inflammatory cytokines, and chemokines (Durbin et al., 2013; Pothlichet et al., 2013; Iwasaki and Pillai, 2014). In this study, 30 lncRNA were chosen to confirm the differential expression in A549 cell by qRT-PCR, in which XLOC 010049 was highly elevated and has a high basal expression. However, these data were different from RNA-seq. For the RNA-seq, we used neutrophil samples from patients infected with the influenza A (H7N9) virus in the acute stage and their matched recovery-stage samples. Because our primary concern was influenza-induced acute lung injury, differential expression were validated in A549 cells. Inconsistencies in sequencing and validation results may have been exacerbated by differences in sample types (Consortium S M-I, 2014). Simultaneously, discordant information such as the patient’s age and previous disease may lead to contradictory differential expression results (Aird et al., 2011). Meanwhile, we searched the RNA-seq database and found in GSE61517 (Fabozzi et al., 2018), DFRV was significantly increased in BEAS-2B cells 24 h hours post three H3N2 viruses, including two seasonal IAV/H3N2 strains, Brisbane/10/07 and Perth/16/09 (reference strains for past vaccine seasons) and the well-characterized laboratory strain Udorn/307/72, which is consistent with our findings. In our study, DFRV was dramatically up-regulated in A549 and THP-1 cells post IAV infection in a dose- and time-dependent manner, which is dependent on NFκB signaling activation. Besides, the results showed that THP-1 cells were more sensitive to IAV infection than A549 cells as an immune cell, while several studies have shown that IAV replication is more significant in A549 cells than in THP-1 cells (van Riel et al., 2011; Li et al., 2018). Therefore, we chose A549 cells for the following function study.
Furthermore, two transcripts of DFRV were identified by RACE. Both of DFRV long and short were elevated post IAV infection, in which DFRV long was significantly higher than DFRV short without IAV infection, while no significance were found post IAV infection, which indicate a higher up-regulation of DFRV short than DFRV long post IAV infection. Previous research demonstrated that the varied roles of lncRNAs are based on their susceptibility to fold into thermodynamically stable secondary and higher-order structures (Novikova et al., 2012; Mercer and Mattick, 2013). For example, structural alterations in lncRNA H19 influence regulatory functions and serve as optimal targets for further in-depth investigations into their molecular interactions (Martens et al., 2021). Likewise, HOTAIR is composed of four separate structural modules, two of which precisely correspond to predicted protein-binding domains (Somarowthu et al., 2015), and a proper folding structure of the MEG3 RNA molecule is crucial for its biological functions (Zhang et al., 2010). Our findings demonstrated that, in comparison to the DFRV long, the DFRV short lacks a stem-loop structure. Thus, we further explore the role of two transcripts of DFRV. Interestingly, our results showed a diametrically opposed role for DFRV long and short in virus replications.
Because lncRNAs have been shown to affect the expression of neighboring genes, and it has been proposed that many other lncRNAs can also function as local regulators (Engreitz et al., 2016), we investigated the effect of DFRV on the neighboring gene PCBP2 and the result indicated no significant difference, suggesting that the antiviral response of DFRV is independent of PCBP2. Hypercytokinemia, also known as cytokine storm, which is usually present in patients with severe IAV infection and is closely associated to disease severity and may be a predictor of disease progression and mortality (Iwasaki and Medzhitov, 2011; Gui and Chen, 2021). However, cell death and inflammation do not always harm the host; moderate cell death and inflammation can facilitate the elimination of viruses (Kuriakose et al., 2016; Zheng et al., 2020). Hence, we determined whether DFRV had any influence on the activation of cell viability, pro-inflammatory cytokines and IFNs. The results showed there was no significant difference in cell viability levels in A549 cells transfected with DFRV long, short and ASO, while DFRV long positively regulates IL-1β and TNF-α production at both the mRNA and protein levels, whereas DFRV short inhibit those expression. Meanwhile, both DFRV long and short negatively regulate the IFN-α and -β expression. Collectively, these results reveal that DFRV regulate the pro-inflammatory cytokine stimulation in response to IAV replication.
The initiation of an inflammatory response induced by viral infection is dependent on several signaling cascades, including NFκB, JAK–STAT, and MAPKs (Guo et al., 2012; Ding et al., 2017; Li et al., 2020). The NFκB signaling pathway is activated during IAV infection and regulated the expression of important pro-inflammatory cytokines such as IL-1, IL-6, IL-8 and TNF-α (White et al., 2008). STAT3 is a significant transcription factor that is essential for expression of a variety of pro-inflammatory and immune response genes (Pandey et al., 2022; Yao et al., 2022), and a recent study reported that miR-4,485 regulates the production of cytokines by targeting STAT3-PI3K-AKT signaling pathway, thus alleviating H1N1-induced injury in A549 cells (Guo et al., 2020). Furthermore, the dysregulation of pro-inflammatory mediators caused by low-or high-pathogenic influenza has been linked to MAPKs, such as ERK1/2 and p38 MAPK (Kother et al., 2014; Dai et al., 2018; Zhou et al., 2021). To test whether DFRV stimulates these signaling, phosphorylation of NFκB, STAT3, PI3K, AKT, ERK1/2 and p38 was examined by western blotting. As expected, significant difference in the levels of these signaling was observed among the infected DFRV long and short-overexpressing or ASO-transfected cells, which were in consisted with pro-inflammatory cytokine expression. In combination with the major location of DFRV in nucleus, our results further support the idea that DFRV play important roles in antiviral immune response dependent on pro-inflammatory signaling cascades.
It has been acknowledged that the diverse functions of RNA transcripts in human diseases may be interpreted by the distinct structures that result from various splicing modalities (Huang et al., 2018). Previous research has shown that MBNL3 induces the inclusion of exon 4 in the lncRNA-PXN-AS1, and the two major isoforms of lncRNA-PXN-AS1 have distinct effects on Paxillin (PXN; Yuan et al., 2017). LncRNAs and circRNAs are two different types of transcriptional products of the genome, while differential splicing of the same gene can result in the formation of either lncRNAs or circRNAs, even different lncRNAs or circRNAs can be produced from a single gene due to splicing of different exons and introns (Huang et al., 2018). Meanwhile, IAV infection also triggers a widespread program of alternative splicing of host genes, which is a crucial process in the viral life cycle (Thompson and Lynch, 2019; Thompson et al., 2020; Esparza et al., 2022), and a large number of infection-induced splicing processes generate proteins critical for IAV replication (Thompson et al., 2020). Thus, more in depth studies are required to understand the mechanism by triggering alternative splicing to produce two DFRV transcriptions in IAV infection.
However, there are still some limitations in our study. Firstly, because only the last 91 nt in the 3′ end of the two transcripts are different and highly conservative, we cannot design a primer or ASO target for the DFRV short, which limits us to further investigate the interaction between the two transcript of DFRV. Secondly, although mouse genome contains the DFRV homolog sequence, we did not succeed in altering mouse DFRV expression, which remains to be a barrier to in vivo studies. Besides, lncRNAs have been reported to regulate gene expression in different ways, including by targeting chromatin modifiers (Herman et al., 2022), acting in cis or in trans (Chen, 2016), interacting with proteins, RNAs, and DNAs (Wu et al., 2017), acting as sponges to sequester RNA-binding proteins (RBPs) and microRNAs (Paraskevopoulou and Hatzigeorgiou, 2016) and so on. Further studies focusing on the interaction factor of DFRV is necessary to elucidate the mechanism of the regulating roles of DFRV in antiviral immunity.
In conclusion, we discovered a novel lncRNA, DFRV, which was elevated in IAV infection and was spliced into two transcripts with the opposite function in viral replication. Moreover, the antiviral functions of DFRV up-regulates IL-1β and TNF-α is dependent on several pro-inflammatory signaling cascades. The precise mechanism of DRFV in antiviral activities needs to be further determined.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/geo/, GSE108807.
KW, CL, and XD contributed to conception and design of the study. SZ, CL, LZ, and SC performed the experiment. MG, MX, and HS organized the database. KW, MG, LZ, SC, and KW performed the statistical analysis. KW and MG wrote the first draft of the manuscript. PZ, YZ, and QA reviewed and revised the manuscript. YZ, QA, and XD supervised the study. KW and XD take primary responsibility for communication with the journal and editorial office during the submission process, throughout peer review and during publication. All authors contributed to the article and approved the submitted version.
This research was funded by “National Natural Science Foundation of China, grant number 82000004 and 81771700” and “Beijing Natural Science Foundation, grant number 7232150.”
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1171423/full#supplementary-material
1. ^http://cpc2.cbi.pku.edu.cn
2. ^http://sfold.wadsworth.org/
3. ^http://www.dna.bio.keio.ac.jp/mxfold2/
4. ^https://bibiserv.cebitec.uni-bielefeld.de/pkiss?id=pkiss_view_webservive
5. ^https://bibiserv.cebitec.uni-bielefeld.de/rnashapes?id=rnashapes_view_Webservice
6. ^http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi
Aird, D., Ross, M. G., Chen, W. S., Danielsson, M., Fennell, T., Russ, C., et al. (2011). Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12:R18. doi: 10.1186/gb-2011-12-2-r18
Basavappa, M., Cherry, S., and Henao-Mejia, J. (2019). Long noncoding RNAs and the regulation of innate immunity and host-virus interactions. J. Leukoc. Biol. 106, 83–93. doi: 10.1002/JLB.3MIR0918-354R
Batista, P. J., and Chang, H. Y. (2013). Long noncoding RNAs: cellular address codes in development and disease. Cells 152, 1298–1307. doi: 10.1016/j.cell.2013.02.012
Bridges, M. C., Daulagala, A. C., and Kourtidis, A. (2021). LNCcation: lncRNA localization and function. J. Cell Biol. 220:e202009045. doi: 10.1083/jcb.202009045
Chen, L. L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41, 761–772. doi: 10.1016/j.tibs.2016.07.003
Chen, X., Liu, S., Goraya, M. U., Maarouf, M., Huang, S., and Chen, J. L. (2018). Host immune response to influenza a virus infection. Front. Immunol. 9:320. doi: 10.3389/fimmu.2018.00320
Chutinimitkul, S., Herfst, S., Steel, J., Lowen, A. C., Ye, J., Van Riel, D., et al. (2010). Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A (H1N1) virus affects receptor binding. J. Virol. 84, 11802–11813. doi: 10.1128/JVI.01136-10
Consortium S M-I (2014). A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the sequencing quality control consortium. Nat. Biotechnol. 32, 903–914. doi: 10.1038/nbt.2957
Crotta, S., Davidson, S., Mahlakoiv, T., Desmet, C. J., Buckwalter, M. R., Albert, M. L., et al. (2013). Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog. 9:e1003773. doi: 10.1371/journal.ppat.1003773
Dai, J., Gu, L., Su, Y., Wang, Q., Zhao, Y., Chen, X., et al. (2018). Inhibition of curcumin on influenza a virus infection and influenzal pneumonia via oxidative stress, TLR2/4, p38/JNK MAPK and NF-kappaB pathways. Int. Immunopharmacol. 54, 177–187. doi: 10.1016/j.intimp.2017.11.009
Delas, M. J., Sabin, L. R., Dolzhenko, E., Knott, S. R., Munera Maravilla, E., Jackson, B. T., et al. (2017). lncRNA requirements for mouse acute myeloid leukemia and normal differentiation. elife 6:e25607. doi: 10.7554/eLife.25607
Dimonte, S., Fabeni, L., Pellegrino, M., and Aquaro, S. (2021). Specific synonymous mutations tightly correlate with HIV-1 co-receptor usage and differentially affect the secondary structure of HIV-1 Env RNA. Acta Virol. 65, 173–180. doi: 10.4149/av_2021_211
Ding, Y., Chen, L., Wu, W., Yang, J., Yang, Z., and Liu, S. (2017). Andrographolide inhibits influenza a virus-induced inflammation in a murine model through NF-kappaB and JAK-STAT signaling pathway. Microbes Infect. 19, 605–615. doi: 10.1016/j.micinf.2017.08.009
Durbin, R. K., Kotenko, S. V., and Durbin, J. E. (2013). Interferon induction and function at the mucosal surface. Immunol. Rev. 255, 25–39. doi: 10.1111/imr.12101
Engreitz, J. M., Haines, J. E., Perez, E. M., Munson, G., Chen, J., Kane, M., et al. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. doi: 10.1038/nature20149
Esparza, M., Bhat, P., and Fontoura, B. M. (2022). Viral-host interactions during splicing and nuclear export of influenza virus mRNAs. Curr. Opin. Virol. 55:101254. doi: 10.1016/j.coviro.2022.101254
Fabozzi, G., Oler, A. J., Liu, P., Chen, Y., Mindaye, S., Dolan, M. A., et al. (2018). Strand-specific dual RNA sequencing of bronchial epithelial cells infected with influenza a/H3N2 viruses reveals splicing of gene segment 6 and novel host-virus interactions. J. Virol. 92:e00518–18. doi: 10.1128/JVI.00518-18
Fatica, A., and Bozzoni, I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21. doi: 10.1038/nrg3606
Feng, J., Yang, G., Liu, Y., Gao, Y., Zhao, M., Bu, Y., et al. (2019). LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics 9, 5227–5245. doi: 10.7150/thno.34273
Fortes, P., and Morris, K. V. (2016). Long noncoding RNAs in viral infections. Virus Res. 212, 1–11. doi: 10.1016/j.virusres.2015.10.002
Gui, R., and Chen, Q. (2021). Molecular events involved in influenza a virus-induced cell death. Front. Microbiol. 12:797789. doi: 10.3389/fmicb.2021.797789
Guo, W., Liu, W., Chen, G., Hong, S., Qian, C., Xie, N., et al. (2012). Water-soluble andrographolide sulfonate exerts anti-sepsis action in mice through down-regulating p38 MAPK, STAT3 and NF-kappaB pathways. Int. Immunopharmacol. 14, 613–619. doi: 10.1016/j.intimp.2012.09.002
Guo, L., Wang, Q., and Zhang, D. (2020). MicroRNA-4485 ameliorates severe influenza pneumonia via inhibition of the STAT3/PI3K/AKT signaling pathway. Oncol. Lett. 20:215. doi: 10.3892/ol.2020.12078
Helft, J., Manicassamy, B., Guermonprez, P., Hashimoto, D., Silvin, A., Agudo, J., et al. (2012). Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J. Clin. Invest. 122, 4037–4047. doi: 10.1172/JCI60659
Herman, A. B., Tsitsipatis, D., and Gorospe, M. (2022). Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell 82, 2252–2266. doi: 10.1016/j.molcel.2022.05.027
Huang, M. S., Zhu, T., Li, L., Xie, P., Li, X., Zhou, H. H., et al. (2018). LncRNAs and CircRNAs from the same gene: masterpieces of RNA splicing. Cancer Lett. 415, 49–57. doi: 10.1016/j.canlet.2017.11.034
Huarte, M. (2015). The emerging role of lncRNAs in cancer. Nat. Med. 21, 1253–1261. doi: 10.1038/nm.3981
Imamura, K., Imamachi, N., Akizuki, G., Kumakura, M., Kawaguchi, A., Nagata, K., et al. (2014). Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 53, 393–406. doi: 10.1016/j.molcel.2014.01.009
Iwasaki, A., and Medzhitov, R. (2011). A new shield for a cytokine storm. Cells 146, 861–862. doi: 10.1016/j.cell.2011.08.027
Iwasaki, A., and Pillai, P. S. (2014). Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–328. doi: 10.1038/nri3665
Janssen, S., and Giegerich, R. (2015). The RNA shapes studio. Bioinformatics 31, 423–425. doi: 10.1093/bioinformatics/btu649
Jiang, M., Zhang, S., Yang, Z., Lin, H., Zhu, J., Liu, L., et al. (2018). Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cells 173, 906–919.e13. doi: 10.1016/j.cell.2018.03.064
Kang, Y. J., Yang, D. C., Kong, L., Hou, M., Meng, Y. Q., Wei, L., et al. (2017). CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16. doi: 10.1093/nar/gkx428
Kitagawa, M., Kitagawa, K., Kotake, Y., Niida, H., and Ohhata, T. (2013). Cell cycle regulation by long non-coding RNAs. Cell. Mol. Life Sci. 70, 4785–4794. doi: 10.1007/s00018-013-1423-0
Kobasa, D., Takada, A., Shinya, K., Hatta, M., Halfmann, P., Theriault, S., et al. (2004). Enhanced virulence of influenza a viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431, 703–707. doi: 10.1038/nature02951
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cells 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Kother, K., Nordhoff, C., Masemann, D., Varga, G., Bream, J. H., Gaestel, M., et al. (2014). MAPKAP kinase 3 suppresses Ifng gene expression and attenuates NK cell cytotoxicity and Th1 CD4 T-cell development upon influenza a virus infection. FASEB J. 28, 4235–4246. doi: 10.1096/fj.14-249599
Krammer, F., Smith, G. J. D., Fouchier, R. A. M., Peiris, M., Kedzierska, K., Doherty, P. C., et al. (2018). Influenza. Nat. Rev. Dis. Primers. 4:3. doi: 10.1038/s41572-018-0002-y
Kuriakose, T., Man, S. M., Malireddi, R. K., Karki, R., Kesavardhana, S., Place, D. E., et al. (2016). ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol 1:aag2045. doi: 10.1126/sciimmunol.aag2045
Lai, C., Liu, L., Liu, Q., Wang, K., Cheng, S., Zhao, L., et al. (2021). Long noncoding RNA AVAN promotes antiviral innate immunity by interacting with TRIM25 and enhancing the transcription of FOXO3a. Cell Death Differ. 28, 2900–2915. doi: 10.1038/s41418-021-00791-2
Li, J., Jie, X., Liang, X., Chen, Z., Xie, P., Pan, X., et al. (2020). Sinensetin suppresses influenza a virus-triggered inflammation through inhibition of NF-kappaB and MAPKs signalings. BMC Complement Med. Ther. 20:135. doi: 10.1186/s12906-020-02918-3
Li, C., Ni, Y. Q., Xu, H., Xiang, Q. Y., Zhao, Y., Zhan, J. K., et al. (2021). Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct. Target. Ther. 6:383. doi: 10.1038/s41392-021-00779-x
Li, J., Zhang, K., Chen, Q., Zhang, X., Sun, Y., Bi, Y., et al. (2018). Three amino acid substitutions in the NS1 protein change the virus replication of H5N1 influenza virus in human cells. Virology 519, 64–73. doi: 10.1016/j.virol.2018.04.004
Lin, H., Jiang, M., Liu, L., Yang, Z., Ma, Z., Liu, S., et al. (2019). The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 20, 812–823. doi: 10.1038/s41590-019-0379-0
Liu, Y., Liu, X., Bai, J., Sun, Y., Nauwynck, H., Wang, X., et al. (2022). A new long noncoding RNA, MAHAT, inhibits replication of porcine reproductive and respiratory syndrome virus by recruiting DDX6 to bind to ZNF34 and promote an innate immune response. J. Virol. 96:e0115422. doi: 10.1128/jvi.01154-22
Liu, S., Liu, X., Li, J., Zhou, H., Carr, M. J., Zhang, Z., et al. (2019). Long noncoding RNAs: novel regulators of virus-host interactions. Rev. Med. Virol. 29:e2046. doi: 10.1002/rmv.2046
Liu, L., Yue, H., Liu, Q., Yuan, J., Li, J., Wei, G., et al. (2016). LncRNA MT1JP functions as a tumor suppressor by interacting with TIAR to modulate the p53 pathway. Oncotarget 7, 15787–15800. doi: 10.18632/oncotarget.7487
Liu, Q., Zhou, Y. H., and Yang, Z. Q. (2016). The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell. Mol. Immunol. 13, 3–10. doi: 10.1038/cmi.2015.74
Martens, L., Ruhle, F., Witten, A., Meder, B., Katus, H. A., Arbustini, E., et al. (2021). A genetic variant alters the secondary structure of the lncRNA H19 and is associated with dilated cardiomyopathy. RNA Biol. 18, 409–415. doi: 10.1080/15476286.2021.1952756
Mercer, T. R., and Mattick, J. S. (2013). Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307. doi: 10.1038/nsmb.2480
Morchikh, M., Cribier, A., Raffel, R., Amraoui, S., Cau, J., Severac, D., et al. (2017). HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell 67, 387–399.e5. doi: 10.1016/j.molcel.2017.06.020
Novikova, I. V., Hennelly, S. P., and Sanbonmatsu, K. Y. (2012). Sizing up long non-coding RNAs: do lncRNAs have secondary and tertiary structure? BioArchitecture 2, 189–199. doi: 10.4161/bioa.22592
Ouyang, J., Zhu, X., Chen, Y., Wei, H., Chen, Q., Chi, X., et al. (2014). NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 16, 616–626. doi: 10.1016/j.chom.2014.10.001
Pandey, P., Al Rumaih, Z., Kels, M. J. T., Ng, E., Kc, R., Chaudhri, G., et al. (2022). Targeting ectromelia virus and TNF/NF-kappaB or STAT3 signaling for effective treatment of viral pneumonia. Proc. Natl. Acad. Sci. U. S. A. 119:e2112725119. doi: 10.1073/pnas.2112725119
Paraskevopoulou, M. D., and Hatzigeorgiou, A. G. (2016). Analyzing MiRNA-LncRNA interactions. Methods Mol. Biol. 1402, 271–286. doi: 10.1007/978-1-4939-3378-5_21
Pernet, E., Downey, J., Vinh, D. C., Powell, W. S., and Divangahi, M. (2019). Leukotriene B(4)-type I interferon axis regulates macrophage-mediated disease tolerance to influenza infection. Nat. Microbiol. 4, 1389–1400. doi: 10.1038/s41564-019-0444-3
Pothlichet, J., Meunier, I., Davis, B. K., Ting, J. P., Skamene, E., Von Messling, V., et al. (2013). Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza a virus infected cells. PLoS Pathog. 9:e1003256. doi: 10.1371/journal.ppat.1003256
Rennie, W., Kanoria, S., Liu, C., Carmack, C. S., Lu, J., and Ding, Y. (2019). Sfold tools for MicroRNA target prediction. Methods Mol. Biol. 1970, 31–42. doi: 10.1007/978-1-4939-9207-2_3
Sato, K., Akiyama, M., and Sakakibara, Y. (2021). RNA secondary structure prediction using deep learning with thermodynamic integration. Nat. Commun. 12:941. doi: 10.1038/s41467-021-21194-4
Sirey, T. M., Roberts, K., Haerty, W., Bedoya-Reina, O., Rogatti-Granados, S., Tan, J. Y., et al. (2019). The long non-coding RNA Cerox1 is a post transcriptional regulator of mitochondrial complex I catalytic activity. elife 8:e45051. doi: 10.7554/eLife.45051
Somarowthu, S., Legiewicz, M., Chillon, I., Marcia, M., Liu, F., and Pyle, A. M. (2015). HOTAIR forms an intricate and modular secondary structure. Mol. Cell 58, 353–361. doi: 10.1016/j.molcel.2015.03.006
Te Velthuis, A. J., and Fodor, E. (2016). Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 14, 479–493. doi: 10.1038/nrmicro.2016.87
Tecchio, C., Micheletti, A., and Cassatella, M. A. (2014). Neutrophil-derived cytokines: facts beyond expression. Front. Immunol. 5:508. doi: 10.3389/fimmu.2014.00508
Thompson, M. G., Dittmar, M., Mallory, M. J., Bhat, P., Ferretti, M. B., Fontoura, B. M., et al. (2020). Viral-induced alternative splicing of host genes promotes influenza replication. elife 9:e55500. doi: 10.7554/eLife.55500
Thompson, M. G., and Lynch, K. W. (2019). Functional and mechanistic interplay of host and viral alternative splicing regulation during influenza infection. Cold Spring Harb. Symp. Quant. Biol. 84, 123–131. doi: 10.1101/sqb.2019.84.039040
Uyeki, T. M., Hui, D. S., Zambon, M., Wentworth, D. E., and Monto, A. S. (2022). Influenza. Lancet 400, 693–706. doi: 10.1016/S0140-6736(22)00982-5
Van Riel, D., Den Bakker, M. A., Leijten, L. M., Chutinimitkul, S., Munster, V. J., De Wit, E., et al. (2010). Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am. J. Pathol. 176, 1614–1618. doi: 10.2353/ajpath.2010.090949
Van Riel, D., Leijten, L. M., Van Der Eerden, M., Hoogsteden, H. C., Boven, L. A., Lambrecht, B. N., et al. (2011). Highly pathogenic avian influenza virus H5N1 infects alveolar macrophages without virus production or excessive TNF-alpha induction. PLoS Pathog. 7:e1002099. doi: 10.1371/journal.ppat.1002099
Wang, Y., Chen, L., Chen, B., Li, X., Kang, J., Fan, K., et al. (2013). Mammalian ncRNA-disease repository: a global view of ncRNA-mediated disease network. Cell Death Dis. 4:e765. doi: 10.1038/cddis.2013.292
Wang, Y., Huo, Z., Lin, Q., Lin, Y., Chen, C., Huang, Y., et al. (2021). Positive feedback loop of long noncoding RNA OASL-IT1 and innate immune response restricts the replication of Zika virus in epithelial A549 cells. J. Innate Immun. 13, 179–193. doi: 10.1159/000513606
Wang, K., Lai, C., Gu, H., Zhao, L., Xia, M., Yang, P., et al. (2017). miR-194 inhibits innate antiviral immunity by targeting FGF2 in influenza H1N1 virus infection. Front. Microbiol. 8:2187. doi: 10.3389/fmicb.2017.02187
Wang, K., Lai, C., Li, T., Wang, C., Wang, W., Ni, B., et al. (2018). Basic fibroblast growth factor protects against influenza a virus-induced acute lung injury by recruiting neutrophils. J. Mol. Cell Biol. 10, 573–585. doi: 10.1093/jmcb/mjx047
White, M. R., Doss, M., Boland, P., Tecle, T., and Hartshorn, K. L. (2008). Innate immunity to influenza virus: implications for future therapy. Expert. Rev. Clin. Immunol. 4, 497–514. doi: 10.1586/1744666X.4.4.497
Wu, H., Yang, L., and Chen, L. L. (2017). The diversity of long noncoding RNAs and their generation. Trends Genet. 33, 540–552. doi: 10.1016/j.tig.2017.05.004
Yao, D., Bao, L., Li, F., Liu, B., Wu, X., Hu, Z., et al. (2022). H1N1 influenza virus dose dependent induction of dysregulated innate immune responses and STAT1/3 activation are associated with pulmonary immunopathological damage. Virulence 13, 1558–1572. doi: 10.1080/21505594.2022.2120951
Yuan, J. H., Liu, X. N., Wang, T. T., Pan, W., Tao, Q. F., Zhou, W. P., et al. (2017). The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 19, 820–832. doi: 10.1038/ncb3538
Zhang, X., Rice, K., Wang, Y., Chen, W., Zhong, Y., Nakayama, Y., et al. (2010). Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology 151, 939–947. doi: 10.1210/en.2009-0657
Zhao, L., Xia, M., Wang, K., Lai, C., Fan, H., Gu, H., et al. (2020). A long non-coding RNA IVRPIE promotes host antiviral immune responses through regulating interferon beta1 and ISG expression. Front. Microbiol. 11:260. doi: 10.3389/fmicb.2020.00260
Zheng, M., Karki, R., Vogel, P., and Kanneganti, T. D. (2020). Caspase-6 is a key regulator of innate immunity, Inflammasome activation, and host defense. Cells 181, 674–687.e13. doi: 10.1016/j.cell.2020.03.040
Keywords: long noncoding RNA, influenza A virus, innate immune, proinflammatory signaling, viral replication
Citation: Wang K, Gong M, Zhao S, Lai C, Zhao L, Cheng S, Xia M, Li Y, Wang K, Sun H, Zhu P, Zhou Y, Ao Q and Deng X (2023) A novel lncRNA DFRV plays a dual function in influenza A virus infection. Front. Microbiol. 14:1171423. doi: 10.3389/fmicb.2023.1171423
Received: 22 February 2023; Accepted: 27 April 2023;
Published: 25 May 2023.
Edited by:
Svetlana Khaiboullina, University of Nevada, United StatesReviewed by:
Adam Wesley Whisnant, Julius Maximilian University of Würzburg, GermanyCopyright © 2023 Wang, Gong, Zhao, Lai, Zhao, Cheng, Xia, Li, Wang, Sun, Zhu, Zhou, Ao and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Zhou, emhvdXk0MjdAMTYzLmNvbQ==; Qiangguo Ao, YW9xZzMwMUAxMjYuY29t; Xinli Deng, YmpkeGxAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.