- 1State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 2Guangzhou Laboratory, Guangzhou, China
- 3School of Pharmaceutical Science and Yunnan Key Laboratory of Pharmacology for Natural Products, Kunming Medical University, Kunming, China
- 4Dongguan People’s Hospital, Dongguan, China
- 5The Affiliated Anning First Hospital and Faculty of Life Science and Technology, Kunming University of Science and Technology, Kunming, China
- 6Guangzhou Key Laboratory for Clinical Rapid Diagnosis and Early Warning of Infectious Diseases, Guangzhou, China
- 7State Key Laboratory of Quality Research in Chinese Medicine, Macau University of Science and Technology, Taipa, Macau SAR, China
Background: Acute exacerbation of chronic obstructive pulmonary disease (AECOPD) is associated with high mortality rates. Viral and bacterial coinfection is the primary cause of AECOPD. How coinfection with these microbes influences host inflammatory response and the gut microbiota composition is not entirely understood.
Methods: We developed a mouse model of AECOPD by cigarette smoke exposure and sequential infection with influenza H1N1 virus and non-typeable Haemophilus influenzae (NTHi). Viral and bacterial titer was determined using MDCK cells and chocolate agar plates, respectively. The levels of cytokines, adhesion molecules, and inflammatory cells in the lungs were measured using Bio-Plex and flow cytometry assays. Gut microbiota was analyzed using 16S rRNA gene sequencing. Correlations between cytokines and gut microbiota were determined using Spearman’s rank correlation coefficient test.
Results: Coinfection with H1N1 and NTHi resulted in more severe lung injury, higher mortality, declined lung function in COPD mice. H1N1 enhanced NTHi growth in the lungs, but NTHi had no effect on H1N1. In addition, coinfection increased the levels of cytokines and adhesion molecules, as well as immune cells including total and M1 macrophages, neutrophils, monocytes, NK cells, and CD4 + T cells. In contrast, alveolar macrophages were depleted. Furthermore, coinfection caused a decline in the diversity of gut bacteria. Muribaculaceae, Lactobacillus, Akkermansia, Lachnospiraceae, and Rikenella were further found to be negatively correlated with cytokine levels, whereas Bacteroides was positively correlated.
Conclusion: Coinfection with H1N1 and NTHi causes a deterioration in COPD mice due to increased lung inflammation, which is correlated with dysbiosis of the gut microbiota.
1. Introduction
Acute exacerbation is a catastrophic event during the course of chronic obstructive pulmonary disease (COPD) and contributes to COPD-related mortality (Hillas et al., 2016). In China, COPD patients experience between 0.5 and 3.5 acute exacerbations annually, which accounts for substantial economic burden. Infection is the most common cause of AECOPD, accounting for 70–85% of all cases (Leung et al., 2017). Approximately 50% of these patients have bacterial infections, such as Haemophilus influenzae (NTHi), Streptococcus pneumoniae, and Staphylococcus aureus, while 30% have respiratory virus infections, such as rhinovirus, parainfluenza virus, and influenza virus (Leung et al., 2017). Viruses are more likely to be detected in AECOPD patients with influenza-like symptoms (Biancardi et al., 2016). Bacteria could colonize in lower airways of COPD patients, which may stimulate a secondary infection after viral infection. Coinfection with virus and bacteria is also common in COPD, accounting for 6.5–27% of infection induced-AECOPD (Papi et al., 2006; Hewitt et al., 2016). Patients with AECOPD who had a coinfection had significantly worse lung function and required longer hospitalizations (Papi et al., 2006).
Clinical and animal studies have revealed a synergistic relationship between influenza virus infection and bacterial infection (Jones et al., 1983; Lee et al., 2010; Bosch et al., 2013). Influenza infection can increase susceptibility and severity to secondary bacterial infection (Lee et al., 2010). However, it is still unclear how coinfection influences host response in AECOPD. In addition, there is growing evidence that the gastrointestinal and respiratory tracts are inherently related, and that the interaction between the gut microbiota and host immunity influences disease progression (Budden et al., 2017; Qu et al., 2022). Patients with COPD or viral infection have a disturbed gut microbiota, which worsens pulmonary inflammation and disease severity (Yildiz et al., 2018; Li et al., 2021). Despite good adherence to GOLD Class D recommended anti-microbial therapy, many COPD patients still experience exacerbation, and customized treatment for high frequency exacerbators based on dynamic changes of microbiota is still lacking (Brennan et al., 2022). This limitation highlights the need for a better understanding of the host and microbial response in COPD with influenza and bacterial coinfection. In this study, we developed a COPD mouse model using an oral and nasal cigarette smoking (CS) exposure system, and further induced exacerbation by sequentially infecting the mice with influenza H1N1 virus and NTHi. We also determined viral and bacterial growth, cytokine response, adhesion molecule levels, the presence of inflammatory cells, and gut microbiome.
2. Materials and methods
2.1. Preparation of virus and bacteria
The influenza A/Puerto Rico/8/34 (PR8, H1N1) virus was obtained from the American Type Culture Collection (ATCC) and propagated in the allantoic cavities of 10-day-old embryonated SPF chicken eggs. The virus was tittered in MDCK cells (ATCC) using TCID50 (50% tissue culture infectious dose) assay calculated by Reed and Muench method (Reed and Muench, 1938).
The NTHi ATCC49766, a reference strain, was kindly provided by Professor Zhuo from our laboratory. Due to the infectivity and virulence in mice (Kimura et al., 2020), we selected this strain for studies of dual infection-induced AECOPD. NTHi stocks were recovered in chocolate agar plates containing 0.33 mg/L vancomycin (Guangdong Huankai, China) and grown in brain heart infusion (BHI). 100 μL serial dilutions of NTHi in PBS were inoculated in chocolate agar plates for 24 h in a 37°C, 5% CO2 incubator, after which the colony-forming units (CFU) were determined. All infections experiments were conducted in a biosafety level 2 Plus (BSL-2 +) laboratory following the protocols approved by Guangzhou Medical University.
2.2. Mouse model of coinfection-induced AECOPD
Wild-type C57BL/6N male mice (6–8 weeks old), were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). All the mice were housed in the specific pathogen-free facilities and all experimental protocols were approved by the Animal Care and Use Committee of Kunming Medical University. All mice were allowed to acclimatize for a week prior to cigarette smoke (CS) exposure.
The mice were randomly divided into 5 groups: CS exposure (Group 1), air exposure (Group 2), CS exposure with PR8 infection (Group 3), CS exposure with NTHi infection (Group 4), and CS exposure with PR8 and NTHi coinfection (Group 5). The experimental flow chart was shown in Figure 1. Briefly, group 1, 3, 4, and 5 were exposed to one cigarette (Hongtashan, China) per day for 14 weeks in an oral and nasal exposure system (Beijing Huironghe Technology, China), as previously described (Xiao et al., 2021). Each cigarette yielded 10 mg tar, 11 mg carbon monoxide, and 1.0 mg nicotine. The parameters of exposure were as followed: concentration of oxygen, (20 ± 0.5) %; dilution flow, 10 L/min; air extraction flow, 13 L/min; air humidity, (60 ± 5)%. The cigarette suction parameters were as followed: suction time, 2 s; time interval, 2 s; suction frequency, 10 s; suction volume, 35 mL. At week 14 post-CS exposure, mice from group 3 and 5 were anesthetized with 2.5% isoflurane and intranasally infected with 0.5 LD50 of PR8. After 3 days, group 5 were sequentially challenged with 105 CFU of NTHi. These infectious doses have been established in our previous study of PR8-NTHi coinfection (Wu et al., 2021). At 24 h after NTHi challenge, the invasive lung function was measured. The mice were then sacrificed and the lungs were collected. The whole lungs were weighted and the lung index (lung weight/body weight ratio × 100) was calculated. Left lungs were then rinsed with HBSS containing 2% FBS before flow cytometric analysis of immune cells. Right lungs were homogenized in 1 mL PBS, and 100 μL of homogenate was used for bacterial titer determination. The remaining homogenates were further centrifugated at 3,000 rpm, 4°C, for 10 min, and the resultant supernatant was subjected to measurements of virus titer and cytokine levels. Also, whole lungs were harvested for a hematoxylin and eosin (H and E) staining. In addition, colon luminal content was collected for gut microbiota analysis. In survival study, the body weight and survival of mice were recorded daily for 15 days after PR8 infection.
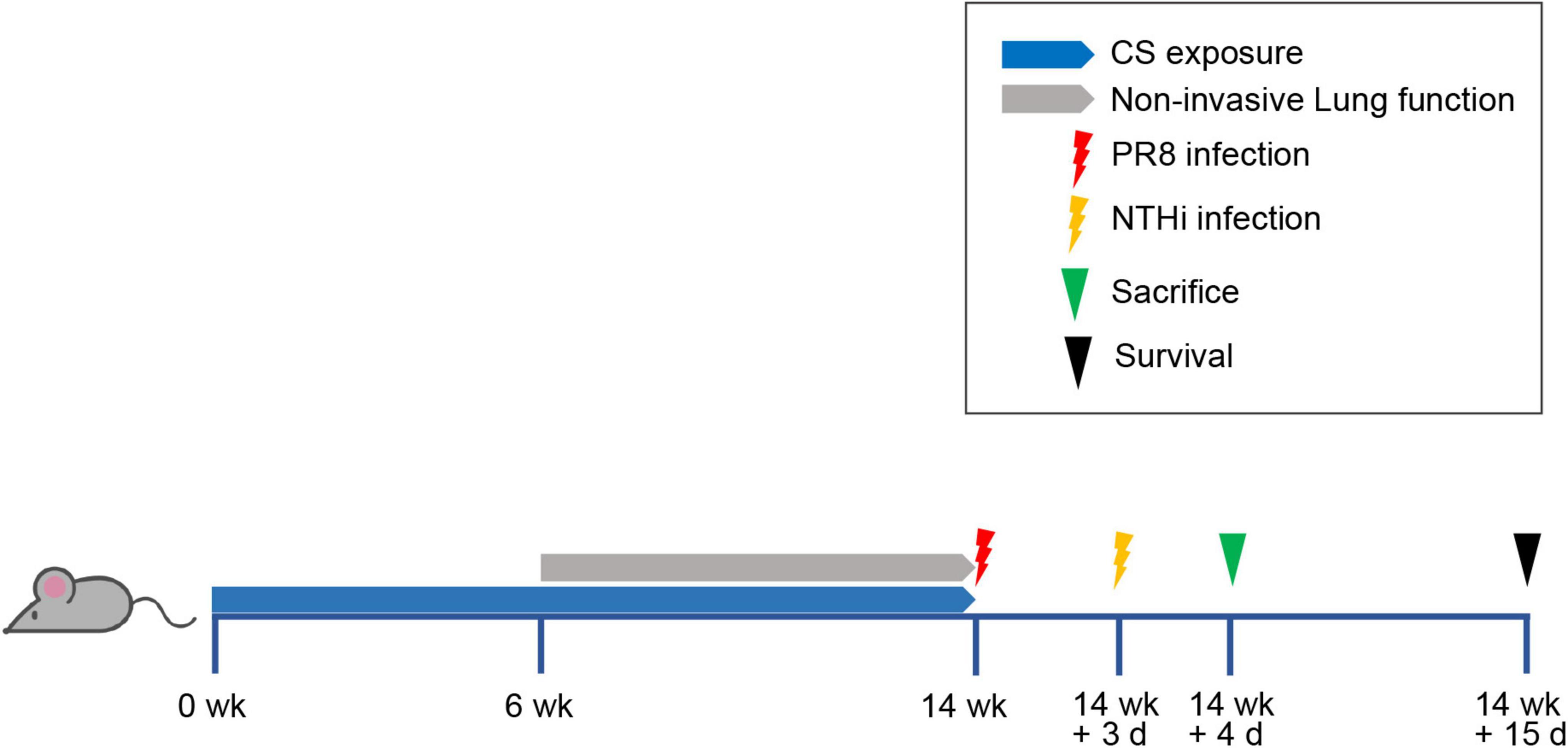
Figure 1. Experimental flow chart. Mice were exposed to one cigarette per day for 14 weeks and then intranasally inoculated with non-lethal dose of PR8 or PBS. At 3 days post-infection (dpi), mice were challenged with NTHi or PBS. After detecting invasive lung function at 24 h post-NTHi challenge, mice were sacrificed and samples were collected for further analysis, including histopathology, lung index, replication of pathogens, inflammatory mediators, immune cells, and gut microbiota. Survival rate was calculated at day 15 after PR8 infection.
2.3. Pulmonary function measurement
Pulmonary function in conscious mice was determined once every 2 weeks from week 6 to 14 after initiation of CS exposure using whole body plethysmography (EMKA Technologies, Canada). The spirometric parameters included frequency (F), tidal volume (TV), expiratory volume (EV), and inspiratory time (Ti).
Invasive lung function of mice was measured by using FlexiVent system (SCIREQ, Canada) according to the manufacturer’s protocol. Briefly, after anesthetization with tribromoethanol (Avertin, 240 mg/kg), mice were tracheotomized, intubated, and placed in the chamber of the FlexiVent system. To maintain muscular relaxation, mice were given intraperitoneal injections of vecuronium bromide (6 mg/kg). Mechanical ventilation was subsequently initiated to measure lung function determined by the forced vital capacity (FVC), the forced expiratory volume in 0.1 s (FEV0.1), respiratory resistance (Rrs), tissue damping (G), and respiratory system compliance (Crs). Each measurement was performed in triplicate. The data were analyzed by the FlexiVent software (SCIREQ, Montréal, QC, Canada).
2.4. Histopathology
The lung histopathology was performed as previously described (Shen et al., 2014). Briefly, the whole lungs of mice were removed and fixed in 4% paraformaldehyde for 24 h. After dehydration and embeddedness, lungs were sectioned to 3 μm thickness and stained with H and E or Masson’s trichrome. The histopathological lesions were observed and recorded under a light microscope. Trichrome staining intensity of 4 randomly selected areas per mouse lung was analyzed using ImageJ software.
2.5. Bacterial and viral titer determination
Haemophilus influenzae were quantified by incubating 100 μL of 10-fold serial dilutions of lung homogenates in chocolate agar plates containing vancomycin, as described above. After centrifugation of the remaining lung homogenates, supernatants were used to determine TCID50 in MDCK cells.
2.6. Cytokine and adhesion molecule measurement
The supernatants of lung homogenates were also used for the measurement of inflammatory cytokines, chemokines, and adhesion molecules. The cytokines and chemokines were determined by using the Luminex ProcartaPlex kit (Invitrogen, Waltham, MA, USA) and the Mouse magnetic Luminex assays kit (R&D, Santa Clara, CA, USA) in a Bio-Plex 200 Multiplex Testing System (Bio-Rad, Hercules, CA, USA) following manufacturer’s instructions. Mouse carcinoembryonic adhesion molecule-1 (CEACAM-1) (MyBioSource, San Diego, CA, Canada), intracellular adhesion molecules-1 (ICAM-1) (R&D), and fibronectin (Fn) (Abnova, Walnut, CA, USA), were determined by ELISA assays.
2.7. Flow cytometric analysis
The collected left lung was digested by the enzyme mixture in the gentleMACS Dissociator (Miltenyi Biotec, Gaithersburg, MD, USA). The resultant digested fluid was allowed to run through a 70-micron cell strainer to obtain a single-cell suspension. The erythrocytes were further lysed in a red blood cell lysis buffer (Biolegend, San Diego, CA, USA). Cells were resuspended with 1 mL FACS buffer (PBS contained 2% FBS and 5 mM EDTA), and the Fc receptors were blocked by anti-mouse CD16/32 (Biolegend, San Diego, CA, USA) at 4°C for 10 min. Cells were then stained with special anti-mouse antibodies, including GhostDye510 (TONBO Biosciences, USA), FITC anti-mouse CD11b (Biolegend, San Diego, CA, USA, Clone M1/70), PerCP-Cy5.5 anti-mouse Ly-6C (Biolegend, San Diego, CA, USA, Clone HK1.4), PE anti-mouse F4/80 (Biolegend, San Diego, CA, USA, Clone BM8), PE-Cy7 anti-mouse CD86 (Biolegend, San Diego, CA, USA, Clone GL-1), APC anti-mouse Ly-6G (Biolegend, San Diego, CA, USA, Clone 1A8), APC-Cy7 anti-mouse CD45 (Biolegend, San Diego, CA, USA, Clone 30-F11), BV421 anti-mouse CD11c (Biolegend, San Diego, CA, USA, Clone N418), BV605 anti-mouse CD206 (Biolegend, San Diego, CA, USA, Clone C068C2), FITC anti-mouse CD4 (Biolegend, San Diego, CA, USA, Clone RM4-5), PerCP-Cy5.5 anti-mouse CD3 (Biolegend, San Diego, CA, USA, Clone 17A2), PE anti-mouse CD19 (Biolegend, San Diego, CA, USA, Clone 6D5), APC anti-mouse CD8 (Biolegend, San Diego, CA, USA, Clone 53-6.7), and BV421 anti-mouse NK1.1 (Biolegend, San Diego, CA, USA, Clone PK136). Flow cytometric analysis of stained cells were performed using a flow cytometer (PARTEC CyFlow Space, Germany). Acquired data were analyzed using FlowJo software.
2.8. Gut microbiota analysis
Colon luminal contents in mice were collected immediately after euthanasia and were rapidly flash-frozen in liquid nitrogen for 30 min before storing at −80°C. Total bacterial DNA was extracted by using a Magnetic soil and stool DNA kit (TIANGEN, China). The V3-V4 region of 16S rRNA gene was amplified using PCR assay with. The amplicon primer sequences were as follows: forward primer 5′-(GTGCCAGCMGCCGCGGTAA)-3′; reverse primer 5′-(GGACTACHVGGGTWTCTAAT)-3′. The libraries were constructed using NEB next ultra-library prep kit (Illumina, USA) and paired-end sequenced on Illumina Novaseq6000 platform (Illumina, USA) of Novogene Co., Ltd. (Beijing, China). The filtering of raw sequence and taxonomic classification were performed as described previously (Li et al., 2020). The diagrams of alpha and beta diversity and relative abundance of microbiota were drawn using the R packages ggplot2 and vegan. The significance of beta diversity based on Weighted UniFrac dissimilarity was tested by analysis of similarities (Anosim) within the R package vegan.
2.9. Statistical analysis
Statistical analysis was performed by using GraphPad Prism 8.0 and SPSS 16.0 software. The differences among groups were compared by using the unpaired two-tailed student’s t-test, welch’s t-test or non-parametric test. Correlations between cytokines and gut microbiota were assessed by using Spearman’s rank correlation. The value of p < 0.05 was considered statistically significant.
3. Results
3.1. Coinfection with PR8 and NTHi results in higher mortality in COPD mice
To replicate COPD in humans, we first developed a mouse model by CS exposure for 14 weeks. Mice exposed to CS had impaired lung function, as indicated by an increase in F and a decrease in Ti, TV, and EV (Figures 2A–D). Besides, CS-exposed mice gained weight significantly slower than control mice (Figure 2E). These results suggested that COPD mice were successfully established. We further developed the AECOPD model by sequentially infecting COPD mice with PR8 and NTHi. Coinfected mice exhibited 100% lethality (p < 0.001), but PR8 or NTHi alone had no effect on mortality, indicating that the combination of two pathogens caused synergistic mortality (Figure 3).
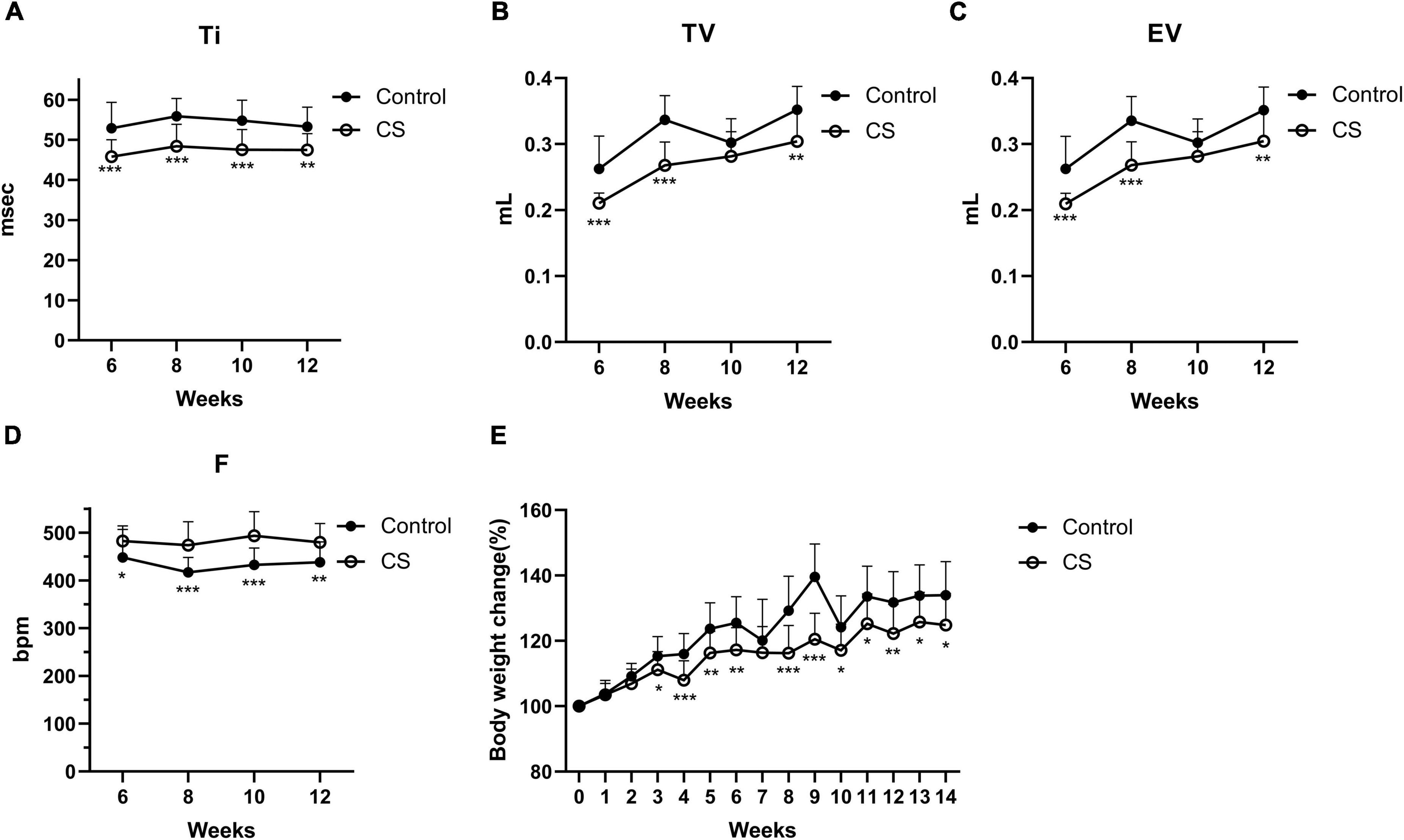
Figure 2. Lung function and body weight changes in conscious mice during CS exposure. Inspiratory time (Ti) (A), tidal volume (TV) (B), expiratory volume (EV) (C), frequency (F) (D), and body weight changes (E) in control (n = 21) and CS-exposed (n = 16) group. Data was expressed as means ± SD. *p < 0.05, **p < 0.01, and ***p < 0.001, compared with air exposure group.
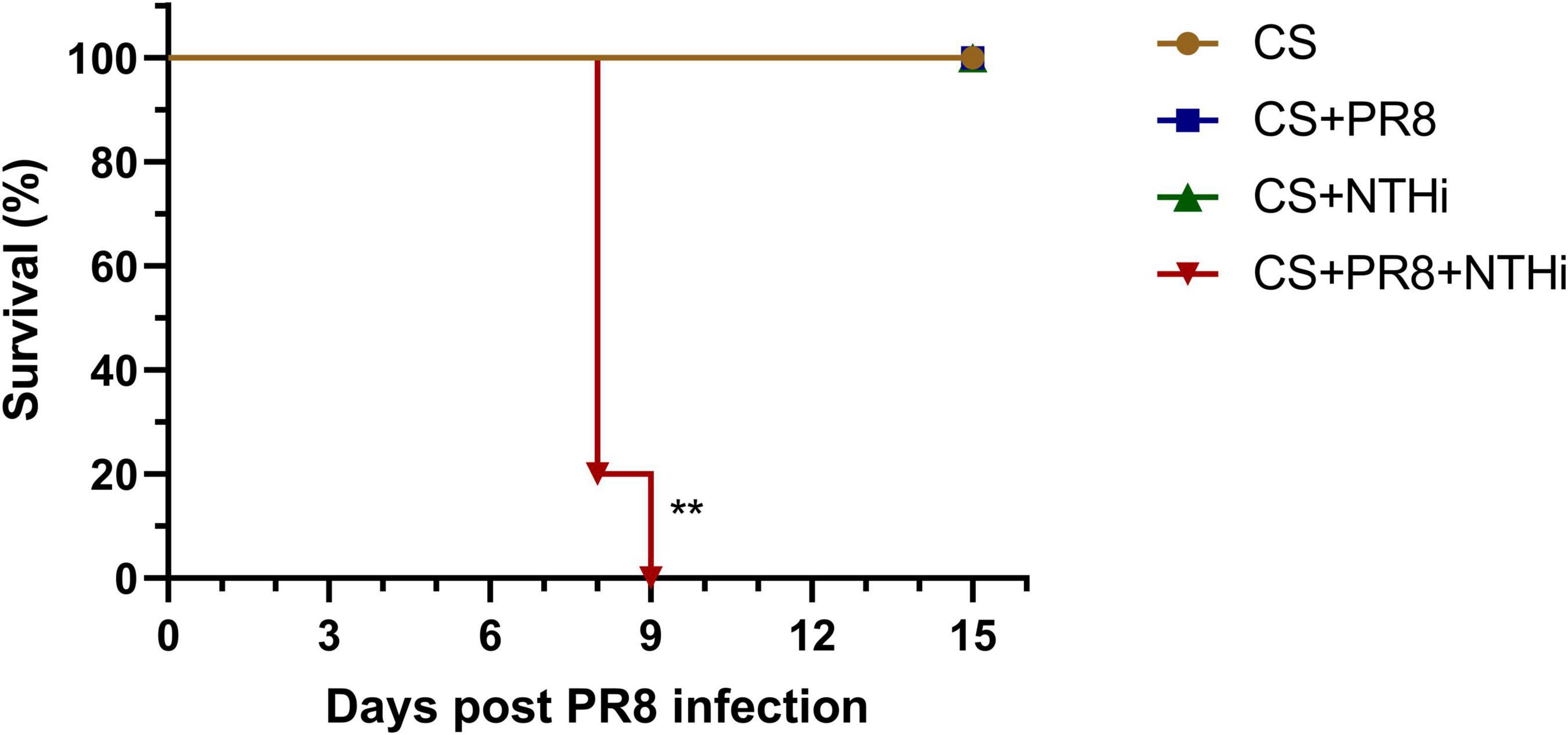
Figure 3. Lethal synergistic effect of PR8-NTHi coinfection in COPD mice. Three days following intranasal infection with 1 LD50 of PR8, COPD mice were challenged with 105 CFU of NTHi. Survival (n = 5) was recorded for 15 days. **p < 0.01 compared with CS-exposed group.
3.2. Coinfection results in declined lung function in COPD mice
As expected, CS exposure significantly impaired lung function, as shown by a decline in FEV0.1/FVC and Crs, and a increase in Rrs and G compared to control mice (Figures 4A–D), but PR8 or NTHi alone did not result in further impairment (Figures 4A–D). Interestingly, coinfection deteriorated the impairment of lung function in COPD mice (Figures 4A–D).
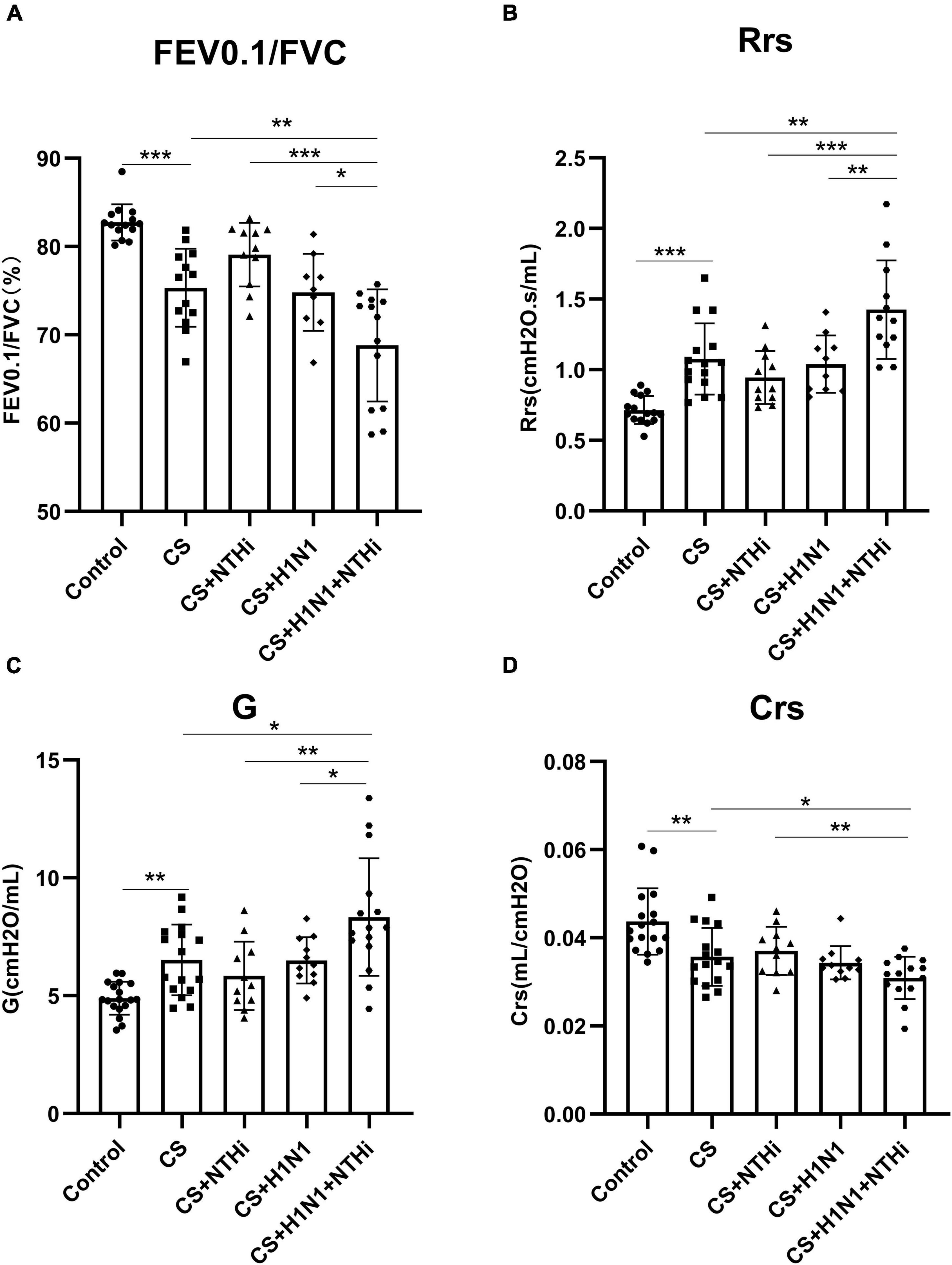
Figure 4. Coinfection with PR8 and NTHi impaired invasive lung function in AECOPD mice. At 24 h after NTHi challenge, invasive lung function was measured by forced expiratory volume in 0.1 s/forced vital capacity (FEV0.1/FVC) (A), respiratory resistance (Rrs) (B), tissue damping (G) (C), and respiratory system compliance (Crs) (D). Data are expressed as mean ± SD (n = 10–15). *p < 0.05, **p < 0.01, and ***p < 0.001.
3.3. PR8 enhances NTHi growth in lungs of COPD mice
The combination of PR8 and NTHi resulted in significant higher levels of NTHi load in mice lungs at 24 h post-NTHi challenge (Figure 5A), but the PR8 titer in coinfected mice did not differ from that in PR8-infected mice (Figure 5B).
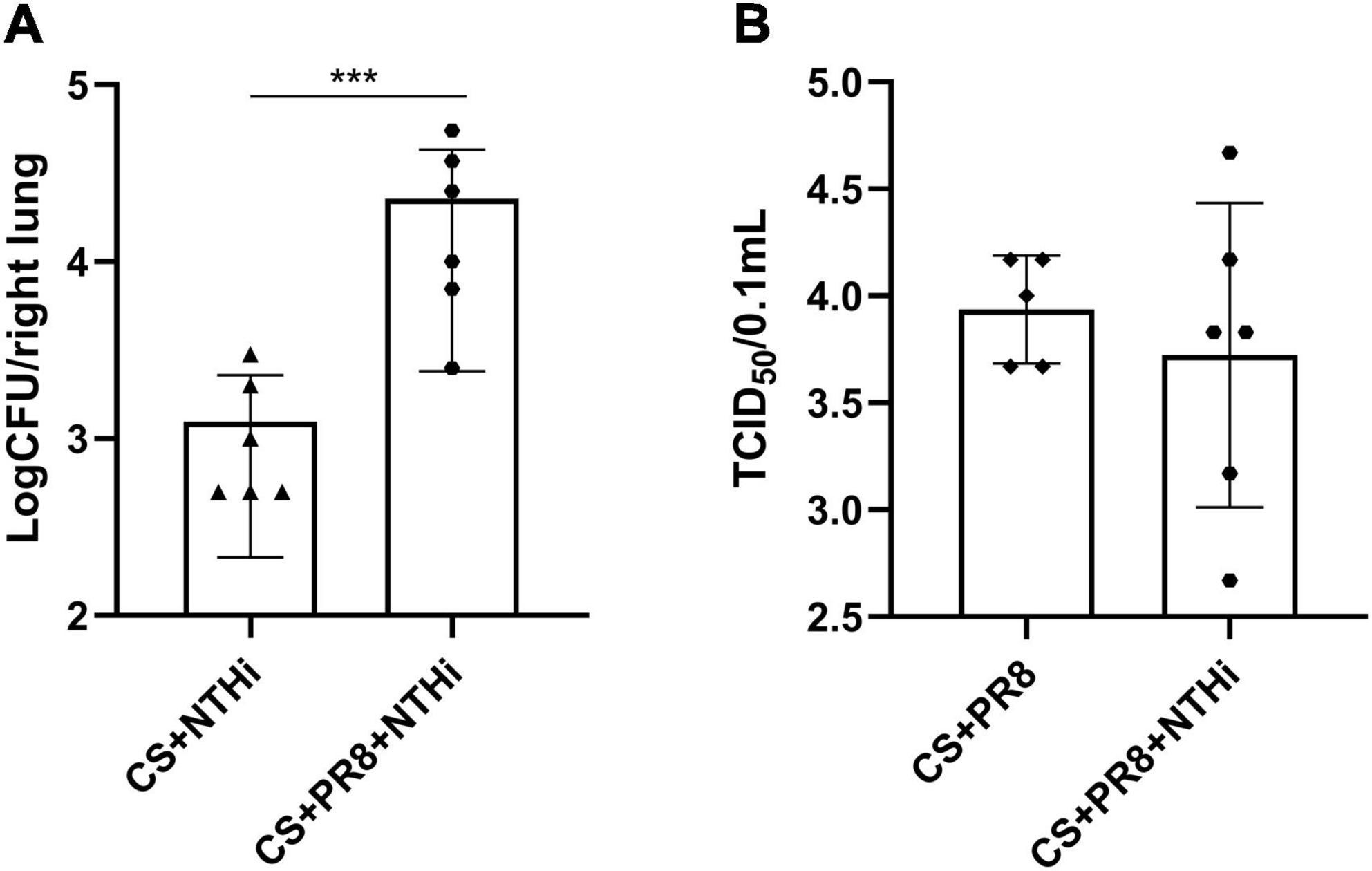
Figure 5. Replication of PR8 and NTHi in AECOPD mice. At 24 h post-NTHi infection, lung homogenates were added to chocolate agar plates containing vancomycin to determine bacterial CFU (A), and the supernatants of lung homogenates were added to MDCK cells to determine viral TCID50 (B). Data are expressed as mean ± SD (n = 6). ***p < 0.001.
3.4. Coinfection aggravates lung injury in COPD mice
Mice exposed to CS had emphysema-like pathological changes, including bronchial wall thickening, alveolar wall thinning and destruction, and alveolar enlargement below the pleura, as compared with control group (Figures 6A–D). Mild inflammation was observed in COPD mice infected with NTHi, as shown by the recruitment of neutrophils into bronchial lumen (Figures 6E, F). In PR8-infected COPD mice, lymphocytic inflammation was present in peribronchial alveolar (Figures 6G, H). In addition, coinfected mice had more severe lung injury, as evidenced by infiltration of neutrophils into bronchiolar cavity, profuse hemorrhage and edema, and destruction of ciliated columnar epithelium (Figures 6I, J). In accordance with histopathological findings, a single pathogen resulted in a higher lung index than CS exposure alone, and coinfection resulted in the highest lung index (Figure 6K). In addition, mild collagen deposition was observed in smooth muscle layer of trachea and lung blood vessels of normal control mice (Figures 6L, Q). Compared to control group, mice exposed to CS exhibited bronchial wall thickening and significantly increased collagen deposition (Figures 6M, Q). Collagen expression was not significantly different between the CS, CS + NTHi, and CS + PR8 groups (Figures 6M, N, O, Q), but it was more extensive and significant in coinfected mice (Figures 6P, Q). These findings indicate that coinfection significantly increases the collagen deposition in lungs of COPD mice.
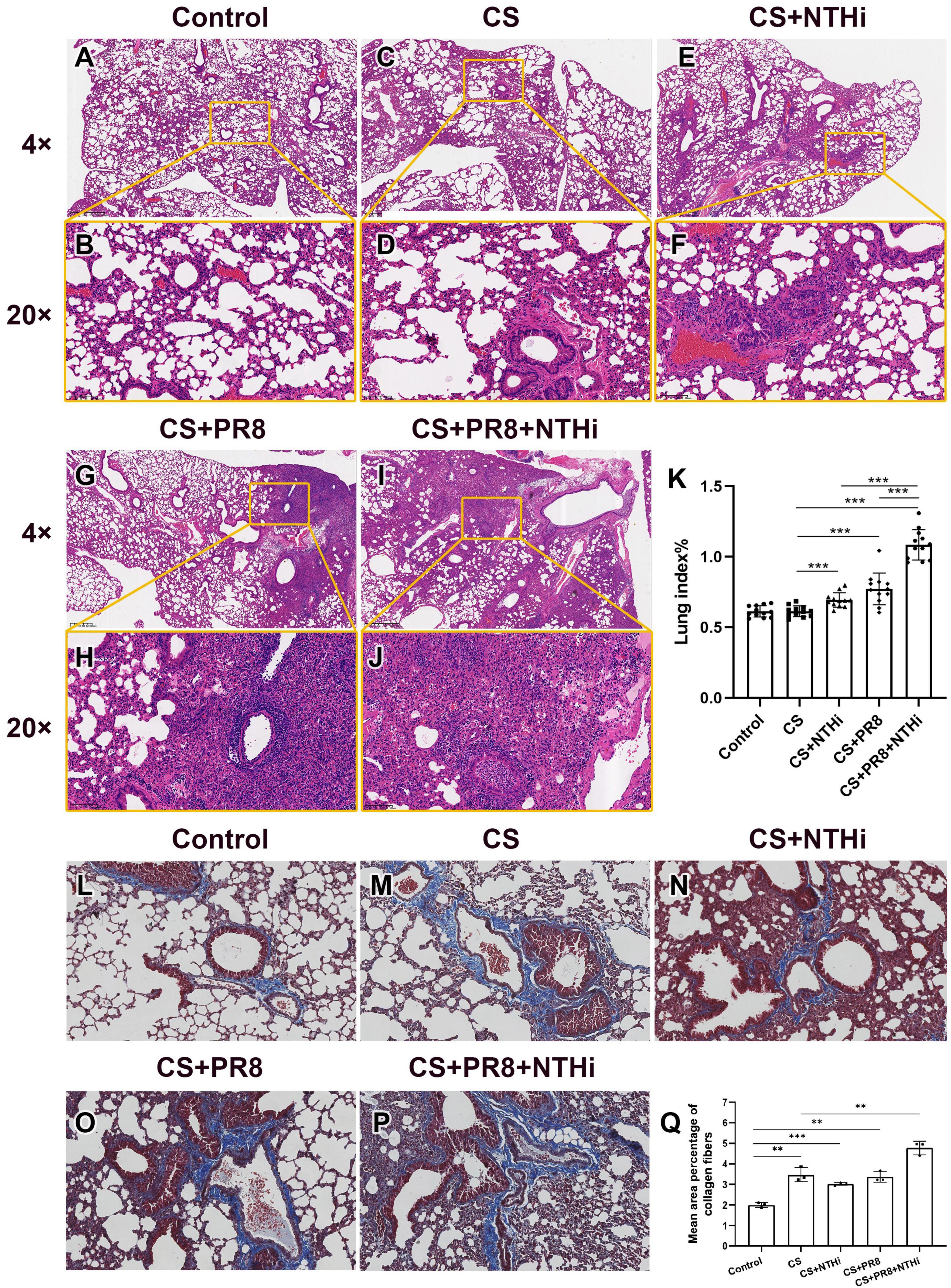
Figure 6. Coinfection aggravates lung injury in AECOPD mice. Pathological changes of control group (A,B), CS-exposed group (C,D), CS + NTHi group (E,F), CS + PR8 group (G,H), and CS + PR8 + NTHi group (I,J) were observed in H and E-stained sections. Original magnification, × 4 and × 20. Lung index was calculated as lung weight/body weight × 100 (K). Collagen fibers of control (L), CS-exposed (M), CS + NTHi (N), CS + PR8 (O), and CS + PR8 + NTHi (P) groups were stained with Masson’s trichrome and quantified using ImageJ (Q). Data are representative images (n = 3 in each group) or expressed as mean ± SD (n = 12 for lung index, n = 3 for percentage of collagen fibers). **p < 0.01 and ***p < 0.001.
3.5. Coinfection results in increased inflammation in lungs of COPD mice
Since pathological inflammation was pronounced in AECOPD mice, we further determined the expression levels of inflammatory cytokines and immune cells in the lungs. NTHi alone induced increased levels of cytokines, including IL-1β, IL-6, TNF-α, IL-22, IL-17, KC, and MIG, as compared with non-infected COPD mice (Figures 7B–H). Besides, PR8 infection alone also triggered significantly higher expression of inflammatory cytokines and chemokines, including CRP, IL-1β, IL-6, TNF-α IL-22, IL-17, KC, and MIG (Figures 7A–H). Furthermore, coinfected mice had significant higher levels of CRP, IL-1β, IL-6, TNF-α, IL-22 and KC as compared with PR8 or NTHi alone (Figures 7A–E, G). We also found that the levels of the adhesion molecules, including ICAM-1 and CEACAM-1, as well as the extracellular matrix Fn, were significantly higher in coinfected mice than the CS group (Figures 7I–K).
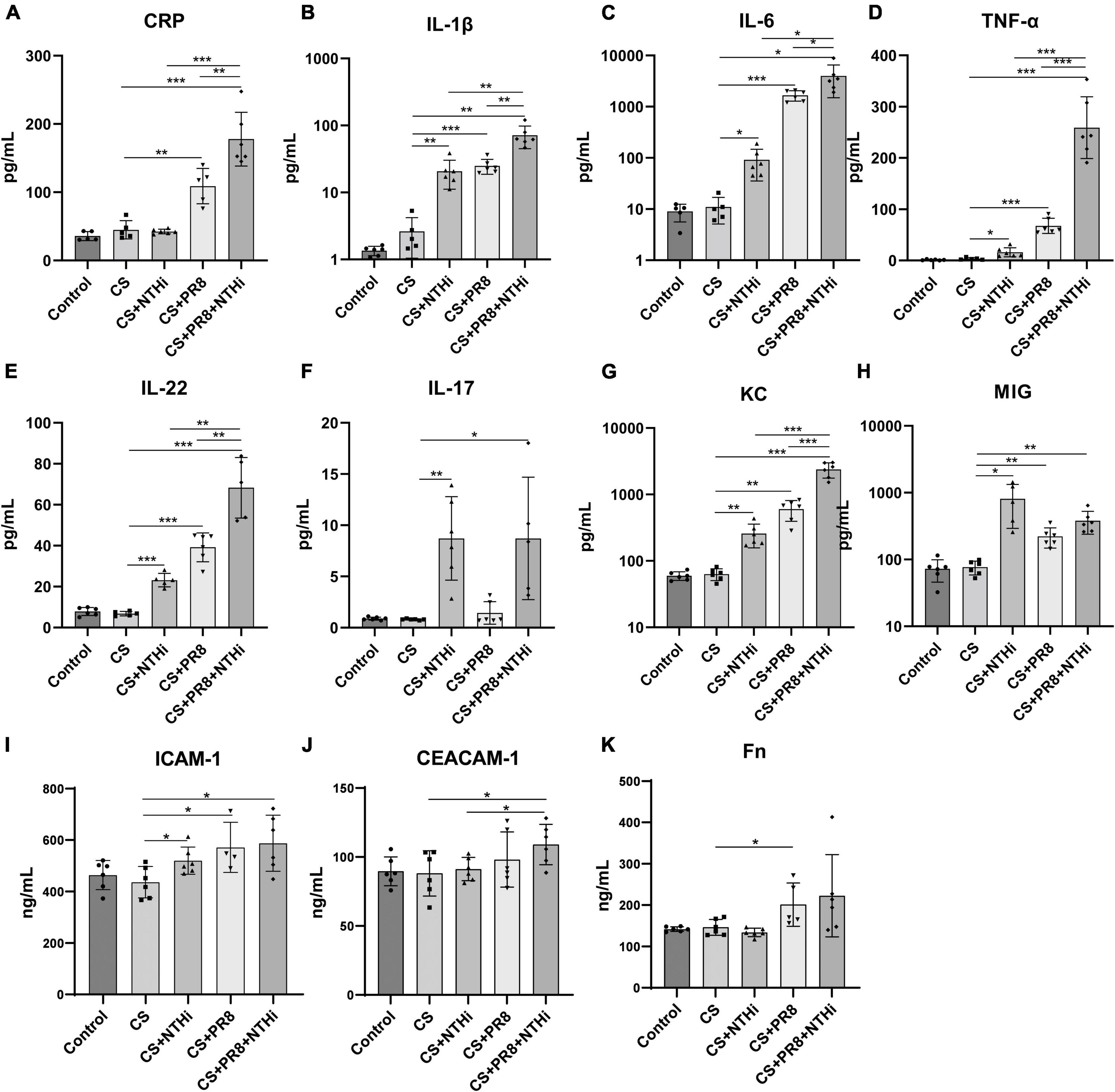
Figure 7. Cytokine and adhesion molecule expression in lung homogenates of AECOPD mice. The levels of CRP (A), IL-1β (B), IL-6 (C), TNF-α (D), IL-22 (E), IL-17 (F), KC (G), MIG (H), ICAM-1 (I), CEACAM-1 (J), and Fn (K) were measured by the Bio-Plex Mouse Cytokines assay or ELISA. Data are expressed as mean ± SD (n = 6 in each group). *p < 0.05, **p < 0.01, and ***p < 0.001.
We further performed flow cytometric analysis of inflammatory cells, including macrophages (CD45 + /CD11b + /F4/80 + /Ly6G-), M1 macrophages (CD45 + /CD11b + /F4/80 + /Ly6G-/CD86 +), M2 macrophages (CD45 + /CD11b + /F4/80 + /Ly6G-/CD206 +), alveolar macrophages (CD45 + F4/80 + CD11c + CD11b-), neutrophils (CD45 + /Ly6C + /Ly6G + /CD11b +), monocytes (CD45 + Ly6C + Ly6G-CD11b +), NK cells (NK1.1 + CD3-), T cells (CD45 + /CD3 +), CD4 + T cells (CD45 + /CD3 + /CD4 + /CD8-), CD8 + T cells (CD45 + /CD3 + /CD4-/CD8 +), and B cells (CD45 + /CD3-/CD19 +). COPD mice infected with NTHi alone had significantly increased M1 and M2 macrophages but decreased alveolar macrophages as compared with non-infected COPD mice (Figures 8B, C, E). PR8 infection alone resulted in an increase in the number of M1 and M2 macrophages, neutrophils, monocytes, and NK cells in the infected COPD mice (Figures 8B, C, F–H). In coinfected mice, total macrophages, M1 macrophages, neutrophils, monocytes, and NK cells were significantly increased (Figure 8A, B, F–H), whereas the alveolar macrophages were significantly decreased (Figure 8E). We also showed that coinfection resulted in a significant increase in the ratio of M1/M2 (Figure 8D). Regarding adaptive immune cells, coinfection significantly reduced the levels of CD4 + T cells and B cells (Figures 8J, L), but had no effect on total number of T cells or CD8 + T cells (Figures 8I, K).
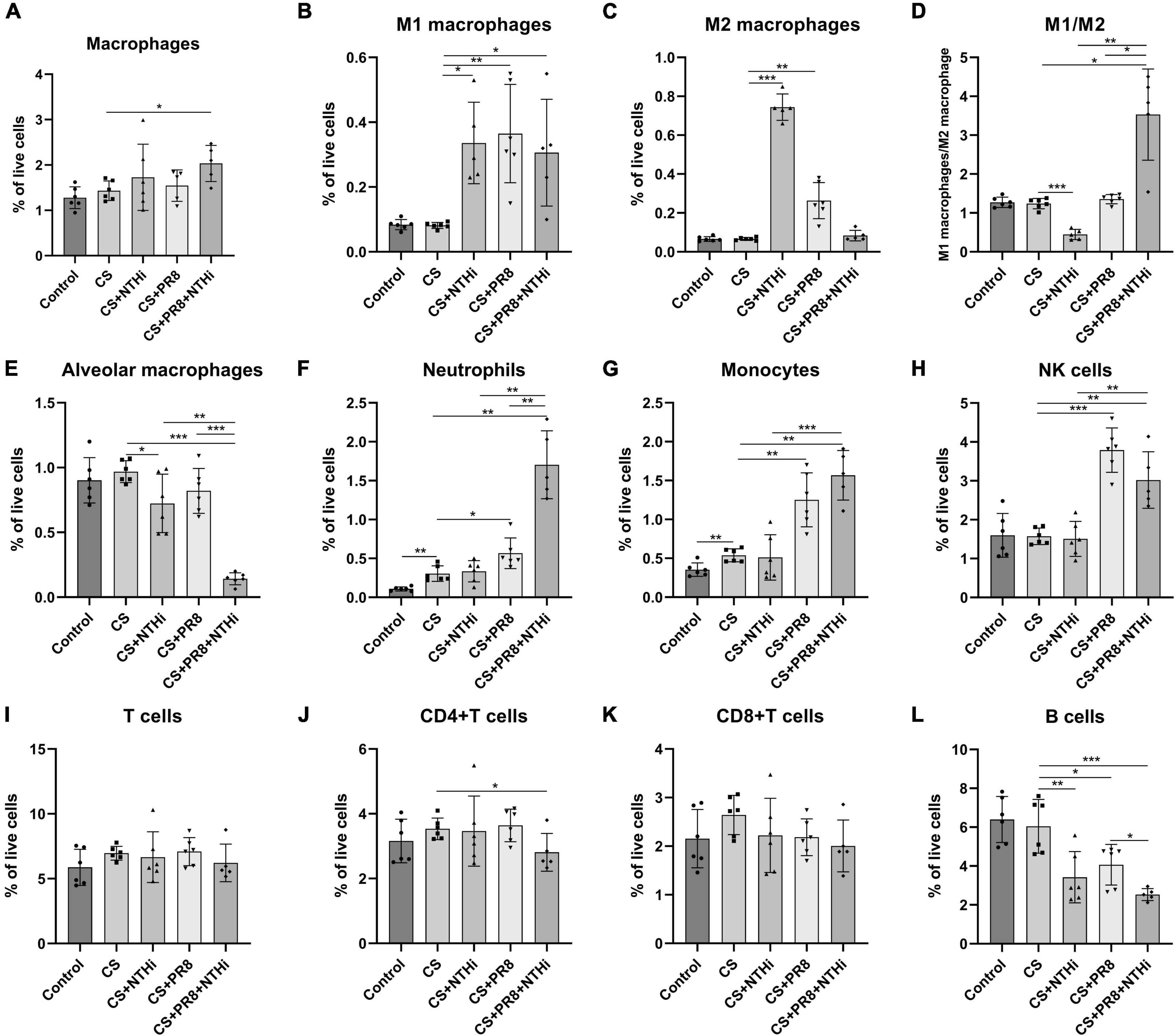
Figure 8. Flow cytometric analysis of innate and adaptive immune cells in AECOPD mice lungs. Following digestion of lungs, the collected cells were analyzed by flow cytometry for monocyte-derived macrophages (A), M1 macrophages (B), M2 macrophages (C), M1 macrophages to M2 macrophages ratio (D), alveolar macrophages (E), neutrophils (F), monocytes (G), NK cells (H), T cells (I), CD4 + T cells (J), CD8 + T cells (K), and B cells (L). Data are expressed as mean ± SD (n = 6). *p < 0.05, **p < 0.01, and ***p < 0.001.
3.6. Coinfection results in the gut microbiota dysbiosis in COPD mice
It has been reported that exposure to CS or biofuels disrupts the gut microbiota of COPD mice, which may influence disease progression (Li et al., 2021). Infection with IAV can also alter the gut microbiota, which increases susceptibility to viral infection and bacterial superinfection, exacerbates lung injury, and even increases mortality (Yildiz et al., 2018). However, the alteration of gut microbiota in AECOPD individuals remained unknown. Hence, we sequenced the 16S rRNA gene of colon contents to further investigate the effect of coinfection on the gut microbiota in COPD mice. After removing low-quantity data, 1,810,199 effective sequence reads from 30 samples were utilized for analysis and 9,403 operational taxonomic units (OTUs) were identified. The number of OTUs was significantly higher in mice exposed to CS than in the control group, but COPD mice infected with NTHi or PR8 alone had a slightly lower number of OTUs, and the decrease was even more pronounced in coinfection (Figure 9A). Chao 1 indices also showed a similar pattern of decline in alpha diversity (Figure 9B). In addition, PCoA results revealed varying degrees of shift across all groups, with the most significant shifts occurring between CS and CPN (p = 0.0149) and between CN and CPN (p = 0.0049) (Figure 9C).
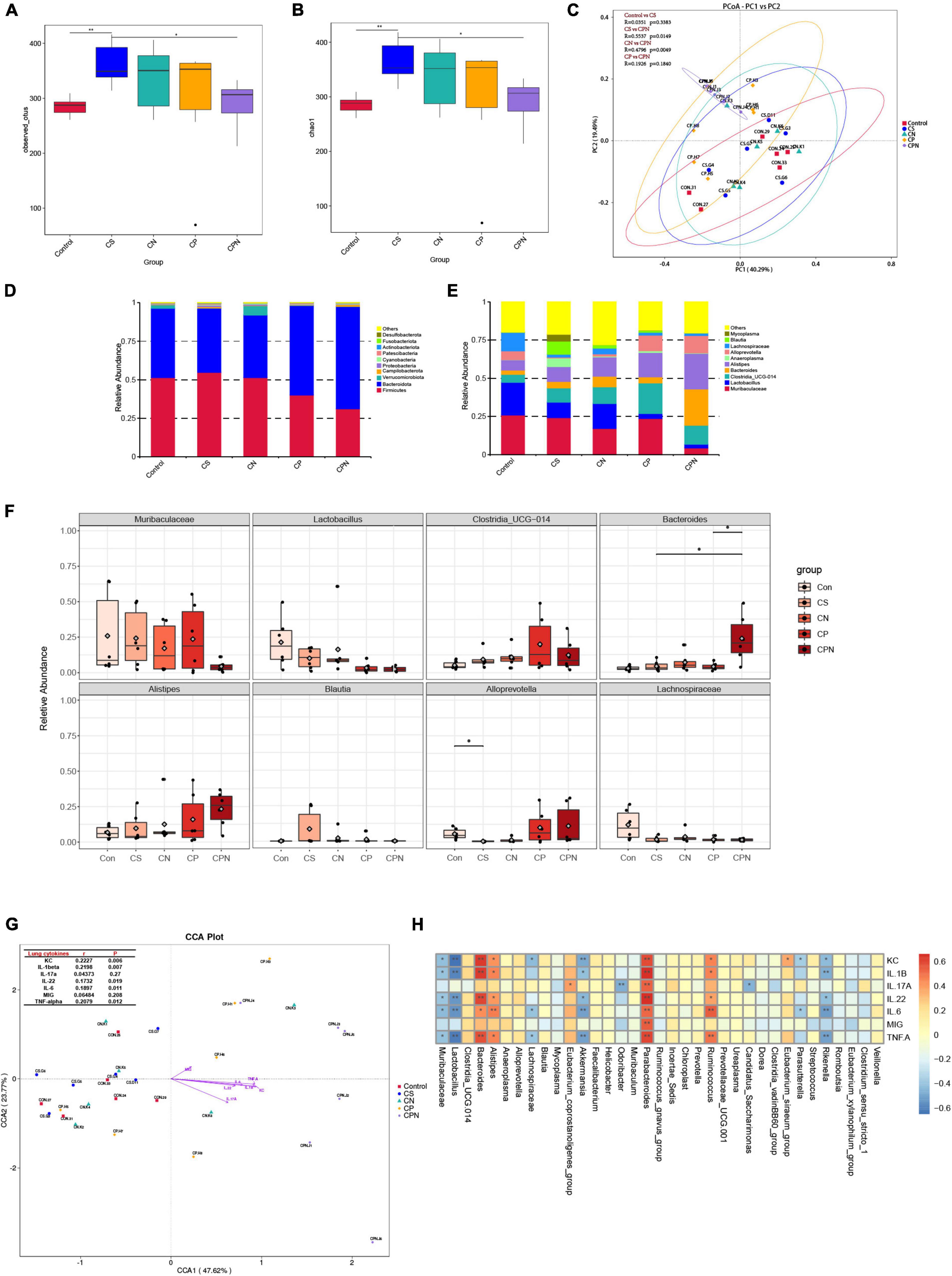
Figure 9. Coinfection alters gut microbiota in COPD mice. Alpha diversity indices, including observed OTUs (A) and Chao 1 (B), for control group, CS group, CS + NTHi group (CN), CS + PR8 group (CP), and CS + PR8 + NTHi group (CPN). (C) PCoA of weighted UniFrac distances, with R and p-values calculated by Anosim. The average relative abundance of the gut microbiota at phylum (D) and genus (E) levels analyzed by Bar-plot. (F) Comparison of the relative abundance at the genus levels. (G) Correlations between cytokines and gut microbiota identified by (CCA), with mental test displayed on the lop-left corner. (H) Correlation between each bacteria genus and cytokine analyzed by spearman correlation tests. *p < 0.05 and **p < 0.01.
We then analyzed the taxonomic level of microbial communities that may have contributed to the changes in alpha and beta diversity. At the phylum level, all colon contents contained four major bacterial phyla: Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria. Mice exposed to CS for 14 weeks had a greater relative abundance of Actinobacteria, which was decreased when COPD mice were infected with NTHi (p < 0.05) or PR8 (p > 0.05) (Figure 9D). COPD mice with coinfection exhibited more pronounced decreases in Actinobacteria and Firmicutes, and an increase in Bacteroidetes (Figure 9D). In comparison to COPD mice, coinfected mice had lower abundance of Muribaculaceae, Lactobacillus, and Lachnospiraceae, and higher abundance of Bacteroides (Figures 9E, F).
Inflammatory cytokine production induced by acute viral respiratory infections is an important cause of gut microbiota changes (Sencio et al., 2021). We further analyzed the correlation between gut microbiota and the expression of lung cytokines. The Mantel test revealed a positive correlation between the diversity of gut microbiota and multiple inflammatory cytokines, including KC, IL-1β, IL-6, and TNF-α, which were significantly elevated in coinfected COPD mice (Figure 9G). We then investigated the association between lung cytokines and individual microbes. Interestingly. the abundance of commensal bacteria, including Muribaculaceae, Lactobacillus, Akkermansia, Lachnospiraceae, and Rikenella, were negatively correlated with most of cytokines. On the contrary, the higher levels of lung cytokines were correlated with increased abundance of Bacteroides, Alistipes, Parabacteroides, and Ruminococcus (Figure 9H), but the last three genus did not significantly differ among all groups.
4. Discussion
Respiratory viral and/or bacterial infection are the most frequently cited causes of AECOPD, and coinfection with these pathogens accounts for 25% of severe AECOPD; this percentage is likely to increase as more sensitive diagnostic tests are developed (Papi et al., 2006). Coinfection is associated with longer hospitalization and more severe lung injury in COPD patients (Papi et al., 2006). However, it is unknown precisely how viral and bacterial coinfection affects the progression of disease during an exacerbation. By exposing mice to cigarette smoke followed by viral and bacterial coinfection, it is advantageous to replicate the development and progression of AECOPD, investigate the interaction between two pathogens, and explore potential therapeutic interventions. In this study, we demonstrated that COPD mice succumbed when exposed to both influenza virus and NTHi, and that influenza virus markedly increased the host susceptibility to NTHi replication, whereas NTHi had no effect on influenza virus growth, which was consistent with our previous findings in coinfected mice without cigarette smoke exposure (Wu et al., 2021). These results suggest that a prior infection with PR8 could promote NTHi growing in the lungs of COPD mice, and the uncontrolled growth of NTHi may be one of the causes of lethality in COPD mice. It has also been reported that influenza infection increases susceptibility to and decreases elimination of other bacteria, including S. aureus and S. pneumoniae (McCullers and Rehg, 2002; Zhao et al., 2021). In addition, our AECOPD mice model not only had pathologic features of viral and bacterial pneumonia, which was consistent with our previous finding in coinfected normal mice (Wu et al., 2021), but also had emphysema, which resulted in decreased lung function.
At present, there are numerous mechanisms underlying the increased susceptibility to infections due to exposure to cigarette smoking. Firstly, cigarette exposure dampens anti-infective mechanism, such as antiviral signaling (Modestou et al., 2010; Duffney et al., 2018), pulmonary pathogen clearance (Sussan et al., 2015), and pathogen-dependent endocytosis (Duffney et al., 2020). In addition, viral infection may compromise the epithelial barrier, alter the mucosal surface environment, degrade antimicrobial peptides, and expose adhesion receptors, such as ICAM-1, CEACAM-1, platelet-activating factor receptor (PAFr), and Fn, thereby facilitating bacterial adhesion and growth (McCullers and Rehg, 2002; Bosch et al., 2013; Su et al., 2018). Therefore, higher levels of ICAM-1 and CEACAM-1 in our coinfected mice may contribute to enhanced NTHi growth.
Acute exacerbations of COPD are episodes of worsening respiratory symptoms that are always associated with increased airway and system inflammation (Wedzicha and Seemungal, 2007). Airway neutrophilia is a common feature of COPD patients and correlated with airway obstruction, decline in FEV1, and development of emphysema (Jasper et al., 2019). Although neutrophils traps pathogens and control overwhelming infections by casting neutrophil extracellular traps (NETs), massive formation of NETs may result in respiratory epithelial and endothelial cell death and accelerates disease progression, which is commonly seen in COPD (Trivedi et al., 2021). In addition, the increased migration of neutrophils in the lungs of COPD patients was frequently accompanied by decreased migration accuracy, resulting in increased “bystander” tissue damage caused by proteinase released from these cells (Sapey et al., 2011; Tregay et al., 2019). Therefore, the increased neutrophils in our CS-exposed mice and its further elevation following coinfection might contribute to the severity of AECOPD in mice. Oxidative stress-induced impairment of alveolar macrophage (AM) function reduces phagocytosis of bacteria and efferocytosis of apoptotic cells, which has been linked to increased susceptibility to AECOPD (Vlahos and Bozinovski, 2014). Although CS exposure did not affect the number of macrophages, M1 and M2 macrophages, or alveolar macrophages in our model, it is possible that the capability of macrophages to eliminate invading microbes was impaired. Our results also showed that a single NTHi or PR8 infection reduced alveolar macrophages by 25 and 15%, respectively, when compared to CS mice without infection, and that alveolar macrophages were further reduced when these two microbes were combined (Figure 8E). This was comparable to the findings of Ghoneim et al. (2013), who demonstrated that coinfection during the AM depletion phase led to significant body weight loss and mortality, and that the depletion of alveolar macrophages during influenza virus infection promoted bacterial superinfections. Macrophage can be polarized into classically activated (M1) and alternatively activated (M2) macrophages. M1 macrophages play an important role in the early stages of inflammation by phagocytosing and eliminating foreign pathogens as well as producing pro-inflammatory cytokines (Huang et al., 2018). In contrast, M2 macrophages are associated with inflammation resolution and tissue repair by reducing levels of pro-inflammatory cytokines in the cellular space (Huang et al., 2018). Balanced polarization of M1/M2 macrophage determines the lung inflammation or injury. At the stage of severe infection and inflammation, macrophages first polarized to M1 phenotype by producing TNF-α, IL-1β, IL-12, and IL-23 against the stimulus l (Shapouri-Moghaddam et al., 2018). Our findings followed a similar pattern, with coinfection causing higher M1/M2 ratio and release of TNF-α and IL-1β. Although total T cells and CD8 + T cells were not significantly different between groups, CD4 + T cells were significantly diminished in our coinfected mice compared to CS-exposed mice without infection. It has been demonstrated that CD4 + T cells play a role in the development and progression of AECOPD, and that a decrease in CD4 + T cells may be one of the contributing factors to the deterioration of COPD (Xue et al., 2022). In addition, multiple subsets of CD4 + T cells, such as regulatory T cells (Tregs), T helper 17 cells (Th17), type 1 T helper (Th1) cells, and Th2 cells, have been implicated in the pathogenesis of AECOPD (Guan et al., 2018; Wei and Sheng Li, 2018), which needs to be investigated further in our study.
Respiratory infections could result in an impaired microbiota phenotype (De Steenhuijsen Piters et al., 2016; Ferreira et al., 2020). Furthermore, it is believed that gut microbiota may increase gut permeability, allowing bacteria and toxins to enter the circulatory system and exacerbate the systemic inflammation (Mizutani et al., 2022). Our and other studies have demonstrated that altered gut microbiota composition correlated with higher levels of inflammatory cytokines (Yeoh et al., 2021; Mizutani et al., 2022). Clinical studies have demonstrated that the gut microbiota of COPD patients differs from that of healthy individuals, with fewer Bacteroides and more Firmicutes (Li et al., 2021; Chiu et al., 2022), which is similar to our finding in COPD mice. In contrast, viral infection could result in an increase in the level of Bacteroides and a decreased level of Firmicutes (Al Khatib et al., 2021), which is also similar to our finding in AECOPD mice. It has also been suggested that dysbiosis of Bacteroidetes and Firmicutes is a potential inflammatory trigger (Li et al., 2021). Changes in the Firmicutes/Bacteroidetes (F/B) ratio caused inflammatory bowel disease (IBD), and single or combined use of probiotic from the phylum Firmicutes were effective in restoring the gut microbial balance by influencing the F/B ratio, thereby alleviating intestinal inflammation (Stojanov et al., 2020). Actinobacteria have been found to colonize on the surface of respiratory mucosa of healthy individuals (Rigauts et al., 2022). We also revealed a dynamic pattern of Actinobacteria that increased with CS exposure but decreased after infection with PR8, NTHi, or both microbes. This could be explained by Li et al. (2022) who demonstrated a negative correlation between Actinobacteria and the frequency of AECOPD. Lachnospiraceae are the most common Firmicutes families that have been shown to decrease in patients with IBD and ulcerative colitis (UC) (Nagao-Kitamoto and Kamada, 2017; Vacca et al., 2020). This finding was consistent with our result in AECOPD mice. A study led by Chen found that NLRP12 could promote beneficial strains of Lachnospiraceae, thereby reducing intestinal inflammation (Chen et al., 2017). Bacteroides spp. are “friendly” commensals when they are in the gut, but when they are located elsewhere in the body, they tend to transform into opportunistic pathogens (Zafar and Saier, 2021). Bacteroides level increased in the gut of our coinfected mice, and this was positively correlated with cytokine expression in the lungs. It was unknown whether Bacteroides moved from the gut to the lung and became pathogenic. In our study, coinfected COPD mice also had decreased level of Muribaculaceae in the guts, which were negatively correlated with pulmonary inflammatory cytokines. This bacteria has been shown to be effective in the treatment of IAV infection (Zelaya et al., 2016). Lactobacillus was found to be negatively correlated with most of cytokines in our coinfected mice, and oral administration of heat-killed or plant-derived Lactobacillus was effective in protecting against IAV infection by enhancing T-cell factor production and IFN response (Kobayashi et al., 2011; Waki et al., 2014). According to clinical studies, Lactobacillus was also the most commonly used probiotic that reduced the risk of viral respiratory tract infections (Shi et al., 2021). Therefore, dysbiosis of the gut microbiota correlates with deterioration in gut and lung inflammation, and development of mono- or poly-microbial intervention may prevent the progression of gut and lung injury.
Lung microbiota is variable and distinct of those from guts (Dumas et al., 2018), and nasally infection with microbes could directly alter the lung microbiota. Bacteria colonization is observed in stable COPD patients, and respiratory viral and/or bacterial infection further leads to the development of AECOPD. It is interesting that AECOPD may be caused by new strains of bacteria rather than new families or genera; specifically, new strains of Haemophilus influenzae were implicated (Brennan et al., 2022). Patients with stable COPD were reported to have Firmicutes (31.63%), Bacteroidetes (28.94%), and Proteobacteria (19.68%) in lungs, whereas those with AECOPD had a dominant bacterial phylum of Proteobacteria (30.29%), Firmicutes (29.85%), and Bacteroidetes (14.02%) (Russo et al., 2022). In a Chinese cohort study, Streptococcus was found to be the most predominant genus in sputum of AECOPD patients (Wang et al., 2020). Pasteurellaceae, Fusobacterium, Solobacterium, Haemophilus, Atopobium, Corynebacterium, and Streptococcus were also found to be enriched in the sputum microbiomes of eosinophilic AECOPD (Wang et al., 2020). By sampling endotracheal aspirates from AECOPD individuals, Firmicutes and Proteobacteria was also found to be the most abundant bacterial phyla, and Streptococcus was the most abundant bacterial species (Sun et al., 2020). In an emphysema mouse model, elastase induces a decline in microbiota richness and diversity, with Pseudomonas and Lactobacillus genera increasing and Prevotella decreasing (Wang et al., 2017). IAV causes subtle, transient changes in the microbial composition of the lower respiratory tract of mice, with Lactobacillus dominating (Yildiz et al., 2018). Mice infected with S. pneumoniae after 6 months of exposure to cigarette smoke had decreased pulmonary microbiota diversity and increased Lactobacillaceae levels in the upper respiratory tract (Hilty et al., 2020). However, it remains unclear how changes in the composition of lung microbiome in COPD mice with NTHi infection or IAV-NTHi coinfection. Furthermore, the underlying mechanism driving these changes in lung microbiota observed in patients and animals should be investigated further in our coinfection model.
5. Conclusion
Coinfection with H1N1 and NTHi increases disease severity in COPD mice due to increased lung inflammation, which is correlated with a dysbiosis of gut microbiota.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/supplementary material.
Ethics statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Kunming Medical University.
Author contributions
Z-FY, W-MY, and X-SX were the principal investigators who conceived the scientific idea. XW, BL, CX, K-LM, H-XZ, D-YZ, SC, Y-CW, R-FC, H-MJ, L-PC, L-ZD, and P-FX carried out the experiments. JZ was responsible for pathological examine. XW and R-FL contributed to the experimental design, data analysis, and article writing. Z-SL and R-FL reviewed and revised the manuscript. All authors have read and agreed to the submitted version of the manuscript.
Funding
This work was supported by the National TCM Interdisciplinary Innovation team project (Grant Number ZYYCXTU-D-202201), National Natural Science Foundation of China (Grant Number 81872765), Science Research Project of the Guangdong Province (Grant Number 2022A0505020031), Guangzhou Science and Technology Planning Project (Grant Number 202102100003), Basic and Applied Basic Research Foundation of Guangdong Province (Grant Number 2020A1515111155), Guangdong Provincial Department of Natural Resources project [Grant Number GDNRC(2021)51], Emergency Key Program of Guangzhou Laboratory (Grant Number EKPG21-06), Macao Science and Technology Development Fund (Grant Number 0172/2019/A3), and the Youth Lift Project of China Association for Science and Technology (Grant Number 2020-2022QNRC003).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al Khatib, H. A., Mathew, S., Smatti, M. K., Eltai, N. O., Pathan, S. A., Al Thani, A. A., et al. (2021). Profiling of intestinal microbiota in patients infected with respiratory influenza A and B viruses. Pathogens 10:761. doi: 10.3390/pathogens10060761
Biancardi, E., Fennell, M., Rawlinson, W., and Thomas, P. S. (2016). Viruses are frequently present as the infecting agent in acute exacerbations of chronic obstructive pulmonary disease in patients presenting to hospital. Intern. Med. J. 46, 1160–1165. doi: 10.1111/imj.13213
Bosch, A. A. T. M., Biesbroek, G., Trzcinski, K., Sanders, E. A. M., and Bogaert, D. (2013). Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 9:e1003057. doi: 10.1371/journal.ppat.1003057
Brennan, M., McDonnell, M. J., Harrison, M. J., Duignan, N., O’Regan, A., Murphy, D. M., et al. (2022). Antimicrobial therapies for prevention of recurrent acute exacerbations of COPD (AECOPD): Beyond the guidelines. Respir. Res. 23:58. doi: 10.1186/s12931-022-01947-5
Budden, K. F., Gellatly, S. L., Wood, D. L. A., Cooper, M. A., Morrison, M., Hugenholtz, P., et al. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15, 55–63. doi: 10.1038/nrmicro.2016.142
Chen, L., Wilson, J. E., Koenigsknecht, M. J., Chou, W. C., Montgomery, S. A., Truax, A. D., et al. (2017). NLRP12 attenuates colon inflammation by maintaining colonic microbial diversity and promoting protective commensal bacterial growth. Nat. Immunol. 18, 541–551. doi: 10.1038/ni.3690
Chiu, Y. C., Lee, S. W., Liu, C. W., Lan, T. Y., and Wu, L. S. (2022). Relationship between gut microbiota and lung function decline in patients with chronic obstructive pulmonary disease: A 1-year follow-up study. Respir. Res. 23:10. doi: 10.1186/s12931-022-01928-8
De Steenhuijsen Piters, W. A., Heinonen, S., Hasrat, R., Bunsow, E., Smith, B., Suarez-Arrabal, M. C., et al. (2016). Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am. J. Respir. Crit. Care. Med. 194, 1104–1115. doi: 10.1164/rccm.201602-0220OC
Duffney, P. F., Embong, A. K., McGuire, C. C., Thatcher, T. H., Phipps, R. P., and Sime, P. J. (2020). Cigarette smoke increases susceptibility to infection in lung epithelial cells by upregulating caveolin-dependent endocytosis. PLoS One 15:e0232102. doi: 10.1371/journal.pone.0232102
Duffney, P. F., Mccarthy, C. E., Nogales, A., Thatcher, T. H., Martinez-Sobrido, L., Phipps, R. P., et al. (2018). Cigarette smoke dampens antiviral signaling in small airway epithelial cells by disrupting TLR3 cleavage. Am. J. Physiol. Lung. Cell. Mol. Physiol. 314, L505–L513. doi: 10.1152/ajplung.00406.2017
Dumas, A., Bernard, L., Poquet, Y., Lugo-Villarino, G., and Neyrolles, O. (2018). The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell Microbiol. 20:e12966. doi: 10.1111/cmi.12966
Ferreira, C., Viana, S. D., and Reis, F. (2020). Is gut microbiota dysbiosis a predictor of increased susceptibility to poor outcome of COVID-19 patients? An update. Microorganisms 9:53. doi: 10.3390/microorganisms9010053
Ghoneim, H. E., Thomas, P. G., and McCullers, J. A. (2013). Depletion of alveolar macrophages during influenza infection facilitates bacterial superinfections. J. Immunol. 191, 1250–1259. doi: 10.4049/jimmunol.1300014
Guan, X., Lu, Y., Wang, G., Gibson, P., Chen, F., Fang, K., et al. (2018). The role of regulatory T cell in nontypeable Haemophilus influenzae-induced acute exacerbation of chronic obstructive pulmonary disease. Med. Inflamm. 2018:8387150. doi: 10.1155/2018/8387150
Hewitt, R., Farne, H., Ritchie, A., Luke, E., Johnston, S. L., and Mallia, P. (2016). The role of viral infections in exacerbations of chronic obstructive pulmonary disease and asthma. Ther. Adv. Respir. Dis. 10, 158–174. doi: 10.1177/1753465815618113
Hillas, G., Perlikos, F., and Tzanakis, N. (2016). Acute exacerbation of COPD: Is it the “stroke of the lungs”? Int. J. Chron. Obstruct. Pulmon. Dis. 11, 1579–1586. doi: 10.2147/COPD.S106160
Hilty, M., Wuthrich, T. M., Godel, A., Adelfio, R., Aebi, S., Burgener, S. S., et al. (2020). Chronic cigarette smoke exposure and pneumococcal infection induce oropharyngeal microbiota dysbiosis and contribute to long-lasting lung damage in mice. Microb. Genom. 6:mgen000485. doi: 10.1099/mgen.0.000485
Huang, X. F., Xiu, H. Q., Zhang, S. F., and Zhang, G. S. (2018). The role of macrophages in the pathogenesis of ALI/ARDS. Med. Inflamm. 2018:1264913. doi: 10.1155/2018/1264913
Jasper, A. E., McIver, W. J., Sapey, E., and Walton, G. M. (2019). Understanding the role of neutrophils in chronic inflammatory airway disease. F1000Research 8:F1000FacultyRev–557. doi: 10.12688/f1000research.18411.1
Jones, W. T., Menna, J. H., and Wennerstrom, D. E. (1983). Lethal synergism induced in mice by influenza type A virus and type Ia group B streptococci. Infect. Immun. 41, 618–623. doi: 10.1128/iai.41.2.618-623.1983
Kimura, G., Nishimoto, Y., Nakaoki, T., Ito, K., and Kizawa, Y. (2020). Establishment of nontypeable Haemophilus influenzae airway infection murine model. Eur. Respir. J. 56:2322. doi: 10.1183/13993003.congress-2020.2322
Kobayashi, N., Saito, T., Uematsu, T., Kishi, K., Toba, M., Kohda, N., et al. (2011). Oral administration of heat-killed Lactobacillus pentosus strain b240 augments protection against influenza virus infection in mice. Int. Immunopharmacol. 11, 199–203. doi: 10.1016/j.intimp.2010.11.019
Lee, L. N., Dias, P., Han, D., Yoon, S., Shea, A., Zakharov, V., et al. (2010). A mouse model of lethal synergism between influenza virus and Haemophilus influenzae. Am. J. Pathol. 176, 800–811. doi: 10.2353/ajpath.2010.090596
Leung, J. M., Tiew, P. Y., Mac Aogain, M., Budden, K. F., Yong, V. F., Thomas, S. S., et al. (2017). The role of acute and chronic respiratory colonization and infections in the pathogenesis of COPD. Respirology 22, 634–650. doi: 10.1111/resp.13032
Li, N., Dai, Z., Wang, Z., Deng, Z., Zhang, J., Pu, J., et al. (2021). Gut microbiota dysbiosis contributes to the development of chronic obstructive pulmonary disease. Respir. Res. 22:274. doi: 10.1186/s12931-021-01872-z
Li, N., Yang, Z., Liao, B., Pan, T., Pu, J., Hao, B., et al. (2020). Chronic exposure to ambient particulate matter induces gut microbial dysbiosis in a rat COPD model. Respir. Res. 21:271. doi: 10.1186/s12931-020-01529-3
Li, W., Wang, B., Tan, M., Song, X., Xie, S., and Wang, C. (2022). Analysis of sputum microbial metagenome in COPD based on exacerbation frequency and lung function: A case control study. Respir. Res. 23:321. doi: 10.1186/s12931-022-02246-9
McCullers, J. A., and Rehg, J. E. (2002). Lethal synergism between influenza virus and Streptococcus pneumoniae: Characterization of a mouse model and the role of platelet-activating factor receptor. J. Infect. Dis. 186, 341–350. doi: 10.1086/341462
Mizutani, T., Ishizaka, A., Koga, M., Ikeuchi, K., Saito, M., Adachi, E., et al. (2022). Correlation analysis between gut microbiota alterations and the cytokine response in patients with coronavirus disease during hospitalization. Microbiol. Spectr. 10:e0168921. doi: 10.1128/spectrum.01689-21
Modestou, M. A., Manzel, L. J., El-Mahdy, S., and Look, D. C. (2010). Inhibition of IFN-gamma-dependent antiviral airway epithelial defense by cigarette smoke. Respir. Res. 11:64. doi: 10.1186/1465-9921-11-64
Nagao-Kitamoto, H., and Kamada, N. (2017). Host-microbial cross-talk in inflammatory bowel disease. Immune Netw. 17, 1–12. doi: 10.4110/in.2017.17.1.1
Papi, A., Bellettato, C. M., Braccioni, F., Romagnoli, M., Casolari, P., Caramori, G., et al. (2006). Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am. J. Respir. Crit. Care. Med. 173, 1114–1121. doi: 10.1164/rccm.200506-859OC
Qu, L., Cheng, Q., Wang, Y., Mu, H., and Zhang, Y. (2022). COPD and gut-lung axis: How microbiota and host inflammasome influence COPD and related therapeutics. Front. Microbiol. 13:868086. doi: 10.3389/fmicb.2022.868086
Reed, L. J., and Muench, H. A. (1938). A simple method for estimating fifty percent endpoints. Am. J. Epidemiol. 27, 493–497. doi: 10.1093/oxfordjournals.aje.a118408
Rigauts, C., Aizawa, J., Taylor, S. L., Rogers, G. B., Govaerts, M., Cos, P., et al. (2022). R othia mucilaginosa is an anti-inflammatory bacterium in the respiratory tract of patients with chronic lung disease. Eur. Respir. J. 59:2101293. doi: 10.1183/13993003.01293-2021
Russo, C., Colaianni, V., Ielo, G., Valle, M. S., Spicuzza, L., and Malaguarnera, L. (2022). Impact of lung microbiota on COPD. Biomedicines 10:1337. doi: 10.3390/biomedicines10061337
Sapey, E., Stockley, J. A., Greenwood, H., Ahmad, A., Bayley, D., Lord, J. M., et al. (2011). Behavioral and structural differences in migrating peripheral neutrophils from patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care. Med. 183, 1176–1186. doi: 10.1164/rccm.201008-1285OC
Sencio, V., Machado, M. G., and Trottein, F. (2021). The lung-gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 14, 296–304. doi: 10.1038/s41385-020-00361-8
Shapouri-Moghaddam, A., Mohammadian, S., Vazini, H., Taghadosi, M., Esmaeili, S. A., Mardani, F., et al. (2018). Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 233, 6425–6440. doi: 10.1002/jcp.26429
Shen, L. L., Liu, Y. N., Shen, H. J., Wen, C., Jia, Y. L., Dong, X. W., et al. (2014). Inhalation of glycopyrronium inhibits cigarette smoke-induced acute lung inflammation in a murine model of COPD. Int. Immunopharmacol. 18, 358–364. doi: 10.1016/j.intimp.2013.12.021
Shi, H. Y., Zhu, X., Li, W. L., Mak, J. W. Y., Wong, S. H., Zhu, S. T., et al. (2021). Modulation of gut microbiota protects against viral respiratory tract infections: A systematic review of animal and clinical studies. Eur. J. Nutr. 60, 4151–4174. doi: 10.1007/s00394-021-02519-x
Stojanov, S., Berlec, A., and Strukelj, B. (2020). The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms. 8:1715. doi: 10.3390/microorganisms8111715
Su, Y. C., Jalalvand, F., Thegerstrom, J., and Riesbeck, K. (2018). The interplay between immune response and bacterial infection in COPD: Focus upon non-typeable Haemophilus influenzae. Front. Immunol. 9:2530. doi: 10.3389/fimmu.2018.02530
Sun, Z., Zhu, Q. L., Shen, Y., Yan, T., and Zhou, X. (2020). Dynamic changes of gut and lung microorganisms during chronic obstructive pulmonary disease exacerbations. Kaohsiung J. Med. Sci. 36, 107–113. doi: 10.1002/kjm2.12147
Sussan, T. E., Gajghate, S., Thimmulappa, R. K., Ma, J., Kim, J. H., Sudini, K., et al. (2015). Exposure to electronic cigarettes impairs pulmonary anti-bacterial and anti-viral defenses in a mouse model. PLoS One 10:e0116861. doi: 10.1371/journal.pone.0116861
Tregay, N., Begg, M., Cahn, A., Farahi, N., Povey, K., Madhavan, S., et al. (2019). Use of autologous (99m) technetium-labelled neutrophils to quantify lung neutrophil clearance in COPD. Thorax 74, 659–666. doi: 10.1136/thoraxjnl-2018-212509
Trivedi, A., Khan, M. A., Bade, G., and Talwar, A. (2021). Orchestration of neutrophil extracellular traps (Nets), a unique innate immune function during chronic obstructive pulmonary disease (COPD) development. Biomedicines 9:53. doi: 10.3390/biomedicines9010053
Vacca, M., Celano, G., Calabrese, F. M., Portincasa, P., Gobbetti, M., and De Angelis, M. (2020). The controversial role of human gut lachnospiraceae. Microorganisms 8:573. doi: 10.3390/microorganisms8040573
Vlahos, R., and Bozinovski, S. (2014). Role of alveolar macrophages in chronic obstructive pulmonary disease. Front. Immunol. 5:435. doi: 10.3389/fimmu.2014.00435
Waki, N., Yajima, N., Suganuma, H., Buddle, B. M., Luo, D., Heiser, A., et al. (2014). Oral administration of Lactobacillus brevis KB290 to mice alleviates clinical symptoms following influenza virus infection. Lett. Appl. Microbiol. 58, 87–93. doi: 10.1111/lam.12160
Wang, J., Chai, J., Sun, L., Zhao, J., and Chang, C. (2020). The sputum microbiome associated with different sub-types of AECOPD in a Chinese cohort. BMC Infect. Dis. 20:610. doi: 10.1186/s12879-020-05313-y
Wang, L., Hao, K., Yang, T., and Wang, C. (2017). Role of the lung microbiome in the pathogenesis of chronic obstructive pulmonary disease. Chin. Med. J. 130, 2107–2111. doi: 10.4103/0366-6999.211452
Wedzicha, J. A., and Seemungal, T. A. R. (2007). COPD exacerbations: Defining their cause and prevention. Lancet 370, 786–796. doi: 10.1016/S0140-6736(07)61382-8
Wei, B., and Sheng Li, C. (2018). Changes in Th1/Th2-producing cytokines during acute exacerbation chronic obstructive pulmonary disease. J. Int. Med. Res. 46, 3890–3902. doi: 10.1177/0300060518781642
Wu, X., Li, R. F., Weng, Y. C., Zhou, H. X., Jiang, H. M., Zhao, J., et al. (2021). Correlation of adhesion molecules and non-typeable haemophilus influenzae growth in a mice coinfected model of acute inflammation. Microbes Infect. 23:104839. doi: 10.1016/j.micinf.2021.104839
Xiao, C., Cheng, S., Li, R., Wang, Y., Zeng, D., Jiang, H., et al. (2021). Isoforskolin alleviates AECOPD by improving pulmonary function and attenuating inflammation which involves downregulation of Th17/IL-17A and NF-kappaB/NLRP3. Front. Pharmacol. 12:721273. doi: 10.3389/fphar.2021.721273
Xue, W., Ma, J., Li, Y., and Xie, C. (2022). Role of CD(4) (+) T and CD(8) (+) T lymphocytes-mediated cellular immunity in pathogenesis of chronic obstructive pulmonary disease. J. Immunol. Res. 2022:1429213. doi: 10.1155/2022/1429213
Yeoh, Y. K., Zuo, T., Lui, G. C., Zhang, F., Liu, Q., Li, A. Y., et al. (2021). Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70, 698–706. doi: 10.1136/gutjnl-2020-323020
Yildiz, S., Mazel-Sanchez, B., Kandasamy, M., Manicassamy, B., and Schmolke, M. (2018). Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome 6:9. doi: 10.1186/s40168-017-0386-z
Zafar, H., and Saier, M. H. Jr. (2021). Gut Bacteroides species in health and disease. Gut Microbes 13, 1–20. doi: 10.1080/19490976.2020.1848158
Zelaya, H., Alvarez, S., Kitazawa, H., and Villena, J. (2016). Respiratory antiviral immunity and immunobiotics: Beneficial effects on inflammation-coagulation interaction during influenza virus infection. Front. Immunol. 7:633. doi: 10.3389/fimmu.2016.00633
Keywords: COPD, influenza, influenzae, mice, inflammation, microbiota
Citation: Wu X, Li R-F, Lin Z-S, Xiao C, Liu B, Mai K-L, Zhou H-X, Zeng D-Y, Cheng S, Weng Y-C, Zhao J, Chen R-F, Jiang H-M, Chen L-P, Deng L-Z, Xie P-F, Yang W-M, Xia X-S and Yang Z-F (2023) Coinfection with influenza virus and non-typeable Haemophilus influenzae aggregates inflammatory lung injury and alters gut microbiota in COPD mice. Front. Microbiol. 14:1137369. doi: 10.3389/fmicb.2023.1137369
Received: 04 January 2023; Accepted: 15 March 2023;
Published: 30 March 2023.
Edited by:
Johnan A. R. Kaleeba, National Cancer Institute (NIH), United StatesReviewed by:
Zhang Wang, South China Normal University, ChinaAdrian G. Rosas-Taraco, Autónoma University of Nuevo León, Mexico
Copyright © 2023 Wu, Li, Lin, Xiao, Liu, Mai, Zhou, Zeng, Cheng, Weng, Zhao, Chen, Jiang, Chen, Deng, Xie, Yang, Xia and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zi-Feng Yang, jeffyah@163.com; Wei-Min Yang, ywmbessie@yeah.net; Xue-Shan Xia, oliverxia2000@aliyun.com
†These authors have contributed equally to this work
 Xiao Wu
Xiao Wu