
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 21 April 2023
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fmicb.2023.1135579
This article is part of the Research Topic Microbe-Nanoparticle Interactions: A Mechanistic Approach View all 6 articles
Antimicrobial Resistance (AMR) raises a serious concern as it contributes to the global mortality by 5 million deaths per year. The overall impact pertaining to significant membrane changes, through broad spectrum drugs have rendered the bacteria resistant over the years. The economic expenditure due to increasing drug resistance poses a global burden on healthcare community and must be dealt with immediate effect. Nanoparticles (NP) have demonstrated inherent therapeutic potential or can serve as nanocarriers of antibiotics against multidrug resistant (MDR) pathogens. These carriers can mask the antibiotics and help evade the resistance mechanism of the bacteria. The targeted delivery can be fine-tuned through surface functionalization of Nanocarriers using aptamers, antibodies etc. This review covers various molecular mechanisms acquired by resistant bacteria towards membrane modification. Mechanistic insight on ‘NP surface-bacterial membrane’ interactions are crucial in deciding the role of NP as therapeutic. Finally, we highlight the potential accessible membrane targets for designing smart surface-functionalized nanocarriers which can act as bacteria-targeted robots over the existing clinically available antibiotics. As the bacterial strains around us continue to evolve into resistant versions, nanomedicine can offer promising and alternative tools in overcoming AMR.
The emergence of antimicrobial resistance is rendering antibiotics ineffective for an ever-increasing number of infections. World Health Organization (WHO) reported in 2019, the pipeline of 32 new antibiotics against the priority pathogens, amongst which only 6 of them were found to be novel. A study published in The Lancet 2022, suggests 4.95 million mortalities can be attributed to cases concerned with resistant bacterial infections (Murray et al., 2022). Out of which, 1.27 million fatalities were solely due to AMR. Hence, antimicrobial resistance poses a global threat and requires concerted actions with immediate effects by the healthcare community and the policy makers. Bacterial resistance arises mainly due to the lack of proper stewardship of the available drugs. The broad range antimicrobials generally prescribed to treat wide spectrum of bacterial infections and the ease of availability of the drugs became the root cause of drug resistant infections. Till date, the pace of discovery and approval of novel therapeutics is a limiting factor.
WHO enlists a category of fatal pathogens as ESKAPEE which includes Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli species reported to have become MDR pathogens. Broadly, the antibiotic-sensitive strains undergo genomic mutation or acquire antibiotic-resistant genes through horizontal gene transfer. These strains result in pathogenic MDR phenotype and the culprit behind most nosocomial infections. Plenty of clinical data into the mechanisms of antibiotic resistance obtained from isolated pathogenic microorganisms across hospitals is being reported routinely (Wang et al., 2020b; Murray et al., 2022). Hence, it is imperative to thoroughly explore potential targets both at the genotypic and phenotypic level. Membrane-based targets being the first line of defence should be closely investigated, especially in ESKAPEE subgroup as they have the potential to respond towards novel therapeutics and reverse AMR.
Emerging technologies such as nanomedicine have potential as alternative therapeutics in mitigating AMR. Nanoparticles exhibit innate anti-bacterial potential or can serve as a ‘Trojan-horse’ to deliver antibiotics to drug resistant bacteria. Antibiotics administered in bulk can cause tissue-toxicity, but is administered nonetheless in increased doses to mitigate the pathogen and achieve its therapeutic effect. At the physiological level, utilizing nanocarriers as antibiotic-delivery vehicle have shown promising outcomes mainly due to improved pharmacokinetics in terms of lowering the volume of distribution of antibiotic at the site of infection and extending serum half-life due to slow antibiotic release. These nanocarriers can be further improvised by decorating their surface with molecules like antibodies, aptamers etc. specific to targets on the bacterial membrane and reduce the undesirable systemic toxicity in patients.
This review covers the different mechanisms used by the MDR pathogens for membrane modification. In our research for this review article, we came across few articles highlighting the mechanism of bacterial death. Therefore, we have mainly included recent articles which elaborated on the mechanistic insights into bacterial membrane-nanoparticle surface interaction. We believe, this can have implications in establishing the significant role of NPs as an alternative or synergist to antibiotic therapy. Since the bacteria continue to develop resistance to the available antibiotics, nanomedicine can prove to be a boon as a targeted molecular therapy in combating MDR pathogens.
Antimicrobial resistance is the ability of the bacteria to resist the action of an antimicrobial hence making the treatment unsuccessful. The problem is aggravated due to the ability of many pathogenic bacteria to form biofilms and the presence of persister cells in the population. These cells occupy around 1% of the bacterial population in the culture that is in the dormant phase (Keren et al., 2004; Wood et al., 2013). The bacterial resistance mechanisms could be intrinsic, i.e., the inherent nature of some bacterial species to possess functional or structural modifications providing resistance to the antibiotic. Acquired resistance, however, is the resistance developed over time from selection pressure or through horizontal transfer from other bacterial strains. The overall resistance mechanisms can be categorized as: (a) limiting drug uptake, (b) target modification, (c) drug inactivation, and (d) activated efflux pumps (Figure 1).
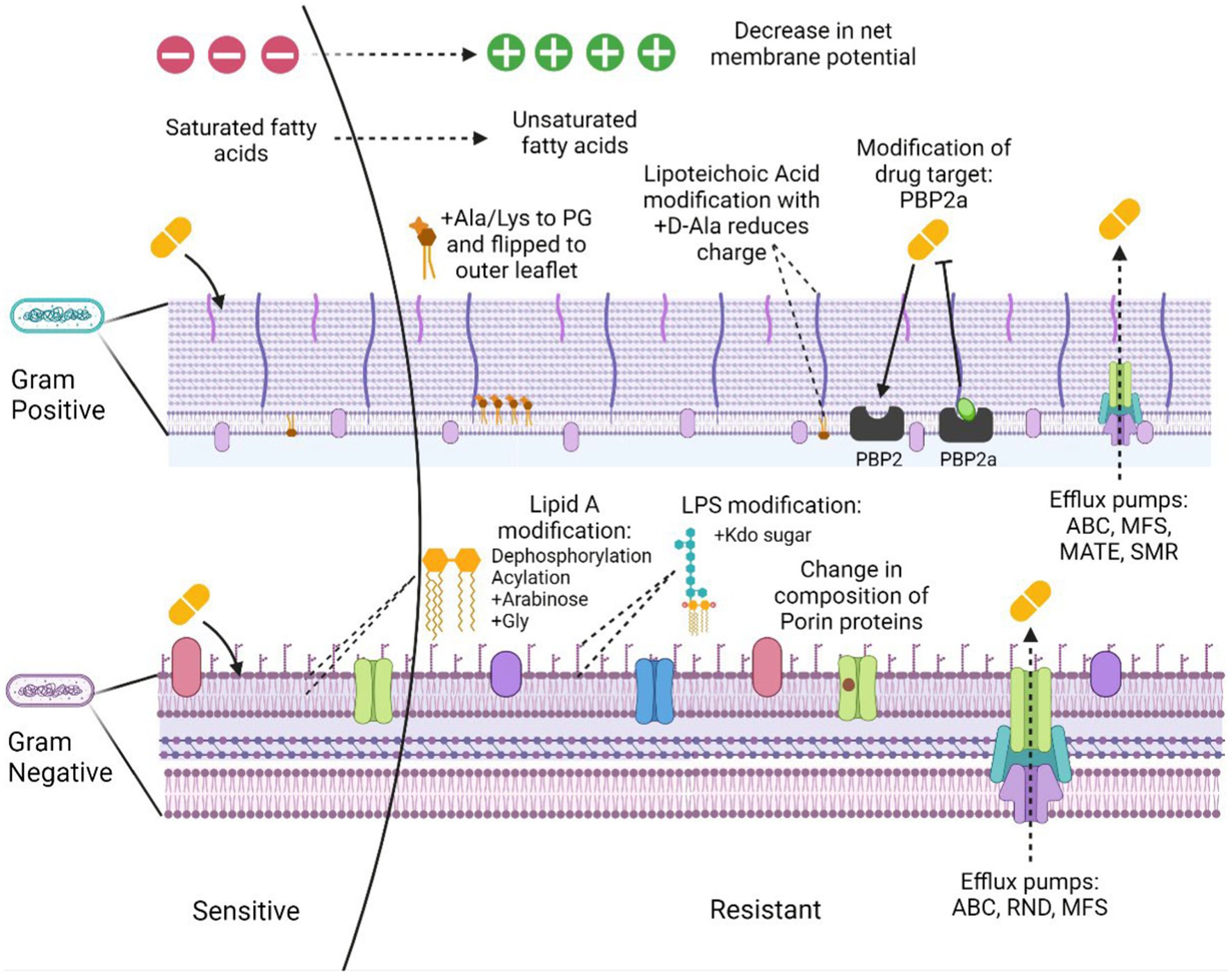
Figure 1. Mechanisms of membrane-based drug resistance in Gram positive and Gram negative bacteria. Figure created in Biorender.
(a) Limiting drug uptake
The limited drug uptake arises due to certain structural components of the Gram-positive and the Gram-negative bacterial cell walls that provide a barrier to the uptake of many antimicrobial agents. The presence of an additional outer layer with increased lipid content in case of mycobacteria, allows the passage of hydrophobic drugs with ease but limits the access of hydrophilic drugs (Lambert, 2002). Gram negative bacteria possess an additional outer membrane that permits the passage of the hydrophilic drugs through β-barrel protein channels called porins (Gill et al., 1998; Blair et al., 2014). Drug uptake through these channels is often restricted by downregulation of these porins. Recently, it was identified that K. pneumoniae mediates resistance against carbapenem, considered as a drug of last resort, via mutations that constrict the outer membrane porins (OmpK35) and (OmpK36; Wong et al., 2019) and restrict uptake of the antibiotic.
(b) Target modification.
Of the most common modifications used by the Gram-positive bacteria towards the beta lactam antibiotics are the variation in the penicillin binding proteins (PBPs). The altered PBP2a protein acquired by methicillin resistance gene (mecA) present on Staphylococcal cassette chromosome mec (SCCmec) mobile genetic element in S. aureus (Reygaert, 2009) confers resistance to methicillin. Vancomycin resistance is acquired through van genes, resulting in the altered metabolites of peptidoglycan synthesis, causing decreased binding to the antibiotic (Beceiro et al., 2013; Cox and Wright, 2013).
(c) Drug inactivation.
Yet another mechanism of resistance is through drug inactivation that occurs in majorly two ways: by complete degradation or by surface modification. Actual degradation of a drug can be done through hydrolyzing enzymes such as beta-lactamase. There are over 7,000 characterized β-lactamases. Carbapenamases such as K. pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM) and oxacillinase (OXA) are known to inactivate multiple antibiotics such as cephalosporins and penicillins apart from carbapenems (Queenan and Bush, 2007). However, tetracycline class of antibiotic can be inactivated through tetX gene (Kumar et al., 2013; Blair et al., 2015). TetX catalyzes resistance through oxygen mediated drug destruction, depending on flavin adenine dinucleotide (FAD) for its monooxygenase activity (Yang et al., 2004). Bacteria also produce enzymes that are capable of altering the drug molecule. Enzymes such as phosphotransferases can modify drugs such as aminoglycosides and macrolides (Golkar et al., 2018).
(d) Activated efflux pumps.
Other category includes the genes encoding efflux pumps, which are present on the bacterial chromosomes. Some of these genes exhibit constitutive expression while others are induced through a stimulus or a specific substrate. The primary purpose of these efflux pumps is to eject out the toxic substances out of the cell (Blair et al., 2014). Some recent studies in resistant Acinetobacter nosocomialis, showed that AdeRS two component system (TCS) confers resistance to tigecycline, eravacycline antibiotics through Ade ATP binding cassette (AdeABC) efflux pump (Lee et al., 2020) therefore, AdeRS system can be a prospective drug target against tigecycline resistance. Similarly, these broad-spectrum antibiotics of last resort have also been targeted in the Gram-positive Staphylococcus species, through a variation in Tet(L) efflux pump leading to a compromise in the effective treatment with these drugs (Wang et al., 2021). Other significant impact of resistance to tetracyclines and ciprofloxacins is through CmeABC efflux pump in Campylobacter jejuni. The cmeA gene was found to be highly expressed in tetracycline resistant isolates hence, conferring resistance (Sharifi et al., 2021). Evolution has paved a way for many efflux pumps to become major mediators of antimicrobial resistance.
Bacterial membranes form an accessible target for majority of the antimicrobials. The outer membrane in Gram-negative bacteria performs a vital role in forming an additional protective layer without compromising on the exchange of components important for sustainability. The modification of the lipid bilayer with certain porin proteins of specific size makes it a permeability barrier. The impact of this lipid bilayer is crucial to the antibiotic susceptibility of the microorganisms. The hydrophilic drugs like beta-lactams, make use of pore formation through porin proteins to breach and enter the cell whereas, the hydrophobic drugs including the macrolides diffuse across the cell. Following are the kinds of membrane alterations responsible for acquiring resistance:
In Gram-negative bacteria like E. coli the overall composition of the membrane can be altered by degrading the existing proteins and incorporating the molecules into new proteins present in the outer membrane. Certain proteases present in the outer membrane such as BepA, DegS, YcaL degrade the damaged proteins (Chang, 2016; Daimon et al., 2017; Soltes et al., 2017). At the level of transcription, porin encoding genes are controlled by two-component systems, non-coding RNAs and other regulatory molecules. These systems work under the regulatory control of environmental stimuli (Pratt et al., 1996; Chen et al., 2004; De La Cruz and Calva, 2010) hence, working at the systemic level. Upon sensing the environmental osmolarity or the ethanol levels, EnvZ-OmpR TCS gets triggered to re-establish the composition of porin proteins present in the outer membrane (Fernández and Hancock, 2012; Chang, 2016; Kenney and Anand, 2020). CpxR member of the envelop stress response also sense the environmental osmolarity or the antimicrobials (Raivio, 2014; Delhaye et al., 2016). Treatment with the drug increases the levels of micC (small non-coding RNA) which work in concert with the cpxR gene to remodel the bacterial membrane against the β-lactams (Dam et al., 2017).
The positively charged sugar residues attach to the lipidA molecules present as a component of LPS in the bacterial membrane, thus neutralizing the negative charge conferred by the phosphate groups. Such modifications prevent the binding of cationic antimicrobials leading to resistance. One such example was identified in resistant Gram-negative Salmonella typhimurium, E. coli and P. aeruginosa species, where 4-amino-4-deoxy-l-arabinose (l-Ara4N) moiety having positively charged amine group at C-4 position is attached to the negatively charged phosphate group of lipidA(C-4; Trent et al., 2001; Zhou et al., 2001). The phoPQ TCS in Salmonella transcribes pmrCAB locus conferring polymyxin resistance. This system controls activation of genes responsible for modifying lipidA with Ara4N (Trent et al., 2001; Zhou et al., 2001; Zavascki et al., 2007). A recent study identified pmrAB system in P. aeruginosa that is involved in lipidA modification through Ara4N (Moskowitz et al., 2004). Besides arabinose modification, galactosamine residues were also found to be responsible for developing resistance to the antibiotics. Such as in case of A. baumannii with colistin resistance features a D-galactosamine attachment to lipidA 1-phosphate residue (Pelletier et al., 2013).
Human colonizing pathogens such as Staphylococcus aureus acquires resistance to Cationic antimicrobial peptides (CAMPs) by expressing mprF (multiple peptide resistant factor; Ernst and Peschel, 2011). mprF catalyzes the addition of Ala or Lys residues on phosphatidyl glycerol moieties. This leads to the transition of charge on the phospholipid to positive with the addition of lysine or neutral with alanine. The other domain of mprF is a flippase that flips the peptidoglycan alanine (PG-Ala) and peptidoglycan lysine (PG-Lys) residues to the outer membrane (Ernst et al., 2009; Klein et al., 2009). This translocation of the charged residues reduces the overall negative charge on the membrane and hence, decrease the binding affinity of the cationic antibiotics. The decrease in the affinity of the positively charged membrane disintegrants can also arise from the incorporation of D-alanyl residues mediated via the dlt operon (Neuhaus et al., 1996; Kovács et al., 2006). GraSR, TCS overexpresses the dlt operon system, which is subjected to resistance development in Staphylococcus aureus to anionic daptomycin (Li et al., 2007a; Yang et al., 2009). In a recent study, the resistant Vibrio cholerae to polymyxin B was found to alter lipidA via glycine amino acids on the acyl chain residue of glucosamine (Hankins et al., 2012; Henderson et al., 2014).
LipidA modification with pEtN residue on 1-phosphate, leads to resistance in Gram negative pathogens. This surface change brings upon positive charge to the lipidA while lowering the interaction of positively charged antibiotics to the membrane. This change is brought about by pmrC enzyme which works under the stringent control of pmrAB TCS. Colistin resistance in A. baumannii arises due to the modification of lipidA group by pEtN residues (Pelletier et al., 2013). Additionally, pEtN can also be incorporated on LPS kdo sugar subunits of Gram-negative E. coli and Salmonella species (Kanipes et al., 2001; Raetz et al., 2007).
Efflux pumps are membrane transporters which work to expel out smaller molecules like antibiotics which targets the intracellular organelles/ biomolecules, hence developing resistance to them. The ATP binding cassette (ABC) transporters, and membrane bound efflux pumps, couple the energy of ATP hydrolysis to expel out the substrate. VraFG transporter promotes resistance in S. aureus and Staphylococcus epidermidis species to CAMP (Li et al., 2007a,b). In S. epidermidis expression levels of the homologues of vraFG are high in presence of human cationic antimicrobial peptide human beta defensin 3 (hCAMP hbD3) under regulation of the aps operon (Li et al., 2007b). Whereas S. aureus expresses vraFG system under high levels of hCAMP stress (Li et al., 2007a) resulting in resistance against these CAMPs besides the anionic antimicrobials or neutral peptides (Li et al., 2007a,b). Alternately, these ABC transporters permits the passage of the antimicrobials inside the cell for them to get degraded by the proteases (Li et al., 2007b). Such mechanism was reported in S. typhimurium for melittin / protamine residues.
Remodeling of the acyl group of lipids leads to modification in membrane thickness and fluidity, which aids in developing resistance to various antimicrobial agents as has been seen in bacteria such as Enterococcus faecalis (Aricha et al., 2004; Kumariya et al., 2015; Maria-Neto et al., 2015). Such alterations in the bacterial membrane lipids include modification in the content of unsaturated fatty acids, the length and proportion of the acyl tail that incorporates the branched tail lipids and cyclopropane (Sohlenkamp and Geiger, 2016; López-Lara and Geiger, 2017). Previous studies suggest, that a less branched lipids in the bilayer membrane having a long and saturated chain assists in forming a thick and ordered lipid structure along with slow diffusion of lipids (Filippov et al., 2007; Kučerka et al., 2011; Poger et al., 2014; Marquardt et al., 2016; Levental et al., 2020). Such changes in the lipid composition of bacteria leads to development of resistance against the antimicrobial peptides (AMPs). For example, increasing unsaturated lipid moieties potentially inhibit the construction of daptomycin oligomers followed by varied pore formation by daptomycin (Taylor et al., 2017; Beriashvili et al., 2018). Similarly, Tao et al., 2021 reported that Gram negative bacteria exhibited membrane plasticity and suggested significant impact on glycerolipids in response to colistin treatment (Tao et al., 2021). Apart from modifications in the physio-chemical properties of the membrane, remodeling of acyl tail can occur in response to change in enzymatic pathways which ultimately develops resistance (MacDermott-Opeskin et al., 2022). For example, Hines group reported that pgsA mutation resulted in reduced PG content, aggregation of fatty acids upstream and successive reassembling the profiles of fatty acids (Hines et al., 2017).
The presence of the peptidoglycan layer on the bacterial cell wall, as well as other unique features such as specific ion channels and proteins that are not found on the host cells make the bacterial cell wall a preliminary target to the available therapeutic options. Over the years bacteria have evolved ways to develop resistance to almost every available antimicrobial. It has been anticipated that AMR will emerge to be one of the leading causes of death by 2050 if the problem is not dealt with immediate effects (O’Neill, 2016). To facilitate drug targeting, bacterial membrane provides a platform for directed interaction of a metabolite in the form of a protein, enzyme, or any surface receptor to that of a drug. Table 1 summarizes the list of potential drug targets present in the resistant ESKAPEE pathogens.
In this section, we have tabulated membrane proteins that play a role in the antimicrobial resistance mechanism of ESKAPEE strains. Preference was given to the membrane proteins and the plasmid encoded genes as they can be acquired through horizontal gene transfer. ESKAPEE pathogens account for majority of the nosocomial infections including catheter associated bloodstream and urinary tract infections, with E. faecium being one of them. EfrAB a heterodimeric transporter in E. faecium, confers resistance to antibiotics and biocides. Studies report that toxic substances like ethidium bromide, 4′, 6-diamidino-2-phenylindole (DAPI), doxycycline, novobiocin, doxorubicin amongst others were pumped out using the efflux pump activity of EfrAB (Davis et al., n.d.; Lubelski et al., 2007). Linezolid resistance arises as a result of inhibition of bacterial translation pathway (Diekema and Jones, 2000). A study suggests a plasmid encoding gene, optrA exhibiting resistance to phenicols and oxazolidinones (Wang et al., 2015).
Next, in the ESKAPEE list is Staphylococcus aureus, a superbug known to have developed resistance to nearly all the drugs of last resort. MRSA (Methicillin resistant Staphylococcus aureus) and VRSA (Vancomycin resistant Staphylococcus aureus), emerges as the most pathogenic strains due to the plasmid encoded mecA gene present on the mobile genetic element SCCmec (Jevons, 1961; Katayama et al., 2000; Olsen et al., 2006) and the presence of vanA operon (Arthur et al., 1993; Périchon and Courvalin, 2009) on the bacterial plasmid, respectively. Apart from these Gram-positive commensals, the Gram-negative bacteria have also turned out to be extremely resistant species responsible for high mortality rate worldwide. Through literature analysis, we enlist some membrane bound efflux pumps (Mehmood et al., 2020; Singh et al., 2022) and porin proteins (Chen et al., 2010; Singh et al., 2022) accountable for bacterial virulence and resistance to antibiotics belonging to carbapenem class of antibiotics against drug resistant K. pneumoniae. Further the problem is aggravated due to the biofilm forming capabilities of some of these pathogens. Studies report a few outer membrane proteins to be present at the helm of developing resistance and promoting biofilm formation in strains of Gram negative, A. baumannii (Labrador-Herrera et al., 2020; Nie et al., 2020).
Certain two-component regulatory systems also help in promoting virulence and outer membrane vesicles (Kim et al., 2019); which help the bacteria to adapt to the unfavorable environment conditions. Another harmful microbe, P. aeruginosa reportedly brings about resistance to macrolides and aminoglycosides via membrane proteins MphE, MsrE and a TCS AmgS (Ho-Fung Lau et al., 2013; Chen et al., 2020) respectively and via efflux pumps (mexAB-oprM, mexAB-oprJ; Azimi et al., 2015). Amongst the Enterobacteriaceae species they have conferred resistance through NDM beta-lactamases (Pedersen et al., 2018), mcr genes (Liao et al., 2022) and quinolone resistance genes (Cano et al., 2009). The last in the list is E coli, conferring kanamycin resistance through outer membrane mltA interacting proteinA (MipA) protein, i.e., responsible for interaction with MltA (membrane bound lytic transglycosylase) alongside TolC protein (Zhang et al., 2015). Also, AcrAB efflux pump with the New Delhi metallo beta-lactamase 1 (blaNDM-1) mediated resistance to carbapenems have been reported to be membrane proteins which can lead to potential drug candidates as specific targets (Chetri et al., 2019). Potentially both plasmid encoded and the membrane bound genes confers resistance to the bacterial species therefore, targeting these specific genes might serve to be an important drug candidate in overcoming the multi drug resistance problem.
Nanoparticles (NPs) represent alternatives in addressing AMR as not just drug delivery vehicles but also, they are capable of exerting inherent antimicrobial efficacy. Together they can generate an overall robust synergistic response (Vallet-Regí et al., 2019). Last few decades have seen significant applications in both disease diagnosis and therapeutics (Castillo et al., 2019). In terms of size, nanocarriers can range from 10 nm metal oxides of silver (silver NP or AgNP) and gold (gold NP or AuNP) to polymer-based nanoparticles 200–400 nm. Lipid NPs, nanoemulsions, polymer-lipid hybrid NPs or nanomicelles all represent myriad forms of nanocarriers which although differ in composition, can be utilized towards antimicrobial activity. Antibiotics can be conjugated to inorganic NPs or loaded (entrapped or adsorbed) onto organic NPs like liposomes or nanoemulsion. NPs can be tailor-made keeping in view the route of administration and the ensuing pathophysiological barrier- systemic through intravenous injection, aerosolized NPs for lung delivery, oral delivery for gastrointestinal infections (Kirtane et al., 2021). Nanocarriers can potentially improve the therapeutic index of encapsulated drugs or antibiotics by (1) Sustained or controlled release from nanocarriers and (2) minimizing the systemic drug concentration and consequently adverse effects. NP encapsulated drugs are protected from unwarranted enzymatic degradation or oxidation in vivo compared to their free drug counterparts (Moreno-Sastre et al., 2015).
NPs can exert their antimicrobial activity on the virtue of their surface charge, size and shape or a combination of all of these factors (Makabenta et al., 2020). Nanoparticles can potentially interact and interfere with proteins structure and integrity rupturing cellular membrane causing leakage and cell death. Antimicrobials-loaded nanoparticles, also referred to as “nanobiotics” can affect metabolic processes such as inhibition of transmembrane ATP formation, alter signal transduction, interfere with ribosomal subunits and their functioning, cause mitochondrial dysfunction and DNA damage in pathogens (Chakraborty et al., 2022a). In this review, however, as the cell wall/ cell membrane is the first line of bacterial defence system, the focus will be on the mechanisms of cell death based on interaction between nanoparticle and bacterial membranes.
Several inorganic nanoparticles are well established antimicrobial agents as well as drug delivery vehicles (Giner-Casares et al., 2016). Inorganic nanoparticles are used either in the form of metal like silver (Ag), gold (Au), zinc (Zn) or as metal oxide such as Zinc oxide (ZnO), Titanium oxide (TiO2) etc. Interestingly, akin to antibiotics, metallic nanoparticles can distinguish bacterial cellular system from eukaryotic cellular (mammalian cells) system through bacterial specific transport system and metalloproteins. But unlike antibiotics with specific mechanisms of action (against protein, ion channel, enzyme etc.), metal NPs are more efficient as they can mediate bactericidal activity through multiple mechanism such as membrane integration and damage, inhibition of drug efflux pumps, blocking electron transport, denaturing protein or mimicry and sequestering of endogenous ions (Lemire et al., 2013; Rumyantceva et al., 2019). This can have implications in breakdown of multiple pathways/ cellular components simultaneously leaving little or no time for bacteria to develop resistance mechanisms.
Therefore, metal-based nanoparticles present attractive antibiotic-alternatives against bacteria. Metal NPs such as Ag-NPs (Abbaszadegan et al., 2015), Au-NPs (Li et al., 2020), Cu NPs (Chatterjee et al., 2014; Sánchez-López et al., 2020), ZnO NPs (Kim et al., 2020; Vihodceva et al., 2021), αFe2O3 NPs (Vihodceva et al., 2021), TiO2 NPs (Siwińska-Stefańska et al., 2018) etc. have been reported extensively for their ability to either kill bacteria or to inhibit their proliferation.
Metal nanoparticles can be synthesized following sol–gel process, chemical vapor deposition (CVD) as bottom-up approach. Top-down approach include pyrolysis for synthesis of bimetallic NPs (Arora et al., 2020) or thermolysis for metal-polymer nanocomposites (Pal Singh Chauhan et al., 2019). Chemical reduction of metal salts is the most routinely employed technique for synthesis of metal NPs. Either strong reductants such as Sodium borohydride (NaBH4) and sodium hydrophosphite (NaH2PO4) or mild reductants such as plant extracts can be used as reducing agents for chemical or biological synthesis of NPs, respectively, (Kharissova et al., 2019). Furthermore, surfactants can be mixed to coat NP surface and to prevent aggregation. Examples of metal ions reducing to NPs include both noble and non-noble metals such as silver, gold, cobalt, nickel, copper and lead (Reverberi et al., 2016).
Inorganic NPs can be utilized either alone or with appropriate antibiotics for both additive or synergistic effect for pronounced bactericidal activity (Hari et al., 2014; Haji et al., 2022). Mahsa et al. conjugated antibiotic Vancomycin, which is a known inhibitor of cell wall synthesis to Ag NPs and evaluated against E. coli (MDR), S. aureus (Methicillin and vancomycin intermediate), S. epidermidis (MDR), P. aeruginosa (MDR) and E. faecalis (MDR, vancomycin resistant; Esmaeillou et al., 2017). Vancomycin-capped Ag NP treatment led to lower minimum inhibitory concentration (MIC) than free Vancomycin and exhibited overall stronger anti-bacterial activity against Gram positive than Gram negative bacteria. The authors suggested a synergistic effect of both Vancomycin and Ag NP in inhibiting cell wall synthesis and its structural disintegration.
Nanoparticles demonstrating robust properties in terms of small size (Suttiponparnit et al., 2011), high surface area to volume ratio and ease of preparation are suitable for theranostic purpose (Dadfar et al., 2019; Erkey and Türk, 2021). Due to having significant bactericidal activity, metal nanoparticle-based treatment of bacterial infections has attracted the interest of researchers to explore and understand it extensively (Yuan et al., 2017; Sánchez-López et al., 2020). While bactericidal activity is desirable, the widespread use of metal NPs is limited by its unwarranted accumulation and toxicity towards human tissues at high concentrations (Duval et al., 2019).
Organic nanoparticles referred to as polymeric or lipid-based nanoparticles are synthesized using natural or synthetic polymer, lipids, excipients as carrier constituents and acts as multifunctional delivery vehicles for antibiotics. Usage of organic NPs is growing exponentially due to their ability to deliver vast array of cargoes including- drugs, genes, peptides and other bioactive molecules. Additionally, organic nanoparticles are better than their inorganic counterparts owing to superior properties in terms of biocompatibility, biodegradability, enhanced cargo delivery with minimal systemic toxicity. Polymer-based nanocarrier can be further classified into polyester, polyamides etc. and have been known for effective delivery of antibacterial drug as well to targeted cells via pathway such as endocytosis, adsorption, ligand-receptor or contact-release etc. (Huh and Kwon, 2011; Ranjan et al., 2012). Methods of synthesis such as solvent diffusion, polymer precipitation, emulsion polymerization have been covered extensively for different class of organic nanomedicine such as lipid-based nanoparticles (Wang et al., 2020a), solid-lipid nanoparticles (Wang et al., 2020a), polymeric (Spirescu et al., 2021), lipid polymer hybrid nanocarriers (Shah et al., 2022).
There are various amphiphiles (Mehta et al., 2021), lipids (Fischer, 2020) and polymers (Qiu et al., 2020) which are known for exhibiting inherent antibacterial activity. This is especially important as the outer layer of nanocarrier is the first point-of-contact while encountering bacteria and can be tailor-made depending on target- in this case Gram-positive, Gram-negative or Mycobacterial cell wall. This can further enhance the functional activity of the nanoparticles. For example, Sarah et al., (Richards et al., 2018) recently showed that the polymer poly(dimethylaminoethyl methacrylate; PDMAEMA) having different degree of polymerization can act as antibacterial agent against E. coli and M. tuberculosis (model organism of TB). Mechanistic analysis further showed that in E. coli PDMAEMA can lead to cell membrane disruption followed by cell death whereas in M. tuberculosis it showed bacteriostatic effect.
In another recent work, Luo et al. prepared and characterized dialdehyde nanocrystalline cellulose (DNC) and evaluated in S. aureus (ATCC 6538P), S. epidermidis (ATCC 12228) and S. pneumoniae (ATCC 6305; Luo et al., 2021). Strong antibacterial activity against MRSA correlated with higher aldehyde content in DNC as observed in vitro. The bioactive nanomaterial significantly reduced MRSA infection on the skin of mice model, exhibited excellent skin compatibility, low cytotoxicity and no acute oral toxicity. The underlying mechanism was suggested to be disruption of membrane protein and leakage of cellular content. Harnessing such bioactive polymers (Wang et al., 2022b) or NP components can render nanocarrier additional bactericidal properties.
Cell wall is the outermost cellular component present in bacterial and archaeal species (Pasquina-Lemonche et al., 2020). The primary structural component of the cell wall comprises of peptidoglycan, which essentially provides structural integrity and acts as constraint to the integral turgor (Turner et al., 2014). Peptidoglycan is a macromolecule made of glycan units, crosslinked by peptide chain and therefore is a well-established crucial target of multiple antibiotics (Rojas et al., 2018). Several studies reported interaction of NPs with outer cell wall component led to breakdown of structural integrity of microbe followed by cell leakage and death.
In this context, shape of nanocarrier plays crucial role in its interaction with bacterial surface or biofilm. In a recent study (Acharya et al., 2018), shape dependent physical mutilation and lethal damage was observed in different bacterial species by sphere-shaped in contrast to rod-shaped silver NPs. AgNP-sp. showed enhanced antibacterial activity when exposed to bacterial strains- E. coli (MIC = 190 μg/ml), and S. aureus (MIC = 190 μg/ml), B. subtilis AST5-2 (MIC = 195 μg/ml), P. aeruginosa AL2-14B (MIC = 188 μg/ml) and K. pneumoniae AWD5 (MIC = 184 μg/ml). K. pneumoniae exhibiting the lowest MIC was further analysed using Field emission scanning electron microscopy (FESEM) and showed silver NP interaction with cell wall as the cause of bacterial death. The same group recently reported comparative evaluation of antibacterial activity of nanospheres with nanorods, nanotriangles and nanohexagons against Gram positive and Gram negative bacteria (Acharya et al., 2021). The authors attributed highest bactericidal effect in nanospheres owing to maximum release of silver ions over nanoparticles of other shapes.
Detailed analysis suggested that besides vancomycin, Ag NPs interacts with cell wall and rupture its architecture leading to cell death. In a similar work, instead of metal NP, chitosan-based polymeric NPs were formulated encapsulating ampicillin and tested against S. aureus strains (ATCC25923, ATCC29213, and ATCC43300; Ciro et al., 2019). It was observed that the positively charged chitosan in the outer NP layer interacted electrostatically with outer surface of bacteria specifically with lipoteichoic acid and ruptured its integrity followed by cell wall that led to loss of cellular integrity and cell death (Caudill et al., 2020).
LPS is the outer most layer present in Gram negative bacteria. Functionally it imparts protection from foreign invaders and is involved in immune modulation. Cytoplasmic division requires the synthesis and transport of millions of new LPS molecules continuously (Clairfeuille et al., 2020). LPS comprises of lipid A membrane-anchor, core oligosaccharide and O-antigen (Shrivastava and Chng, 2019; Clairfeuille et al., 2020). Due to its immunostimulatory properties, LPS has been traditionally considered as potential target for antibiotics like polymyxin (colistin; Van Langevelde et al., 1998; Cano et al., 2009). Metals used in their nanoparticle form allow its absolute interaction with the LPS components and being smaller in size, are easy to bypass through channel proteins. Furthermore, the negative charge on bacterial LPS allows its electrostatic interactions with metal-based NPs and can disrupt membrane integrity (Beyth et al., 2015). Hence, these ionic interaction potentially generate oxidative stress via free radicals leading to cell membrane disruption followed by cell death (Lee et al., 2019; Mammari et al., 2022).
Khan et al. (2013) and Ansari et al. (2014a) studied the antibacterial activity of Ag NPs on 80 Gram negative clinical isolates of E. coli samples and investigated the mechanism of action behind therapeutic effect of NP. This investigation was confirmed by using Attenuated Total Reflectance-Fourier transform infrared (ATR-FTIR) spectroscopy. It was observed that IR spectra of membrane-based lipid polysaccharide (LPS) and Phosphoethanolamine (PE) changes significantly after exposure to Ag NPs. Moreover, it was also found that on one hand, the interaction of the O-antigen of LPS with Ag NPs leads to formation of hydrogen bond, on the other side, exposure of Ag NPs to membrane PE led to disruption of its phosphodiester bonds into phosphomonoesters and ultimately results in highly distorted alkyl chain.
Another similar study has shown that exposure of Al2O3 NPs to clinical isolates of E. coli significantly inhibited the proliferation of bacterial population. Investigation of antibacterial mechanism as observed by ATR-FTIR, suggested that interaction of membrane phospholipid especially L-α-Phosphoethanolamine (PE) and O-antigen of LPS with Al2O3 NPs led to the disruption of cellular integrity. Additionally, formation of H-bond, interfering with membrane ligands led to change in the amphiphilicity of membrane and hence caused cell leaking and cell death (Ansari et al., 2014b; Figure 2).
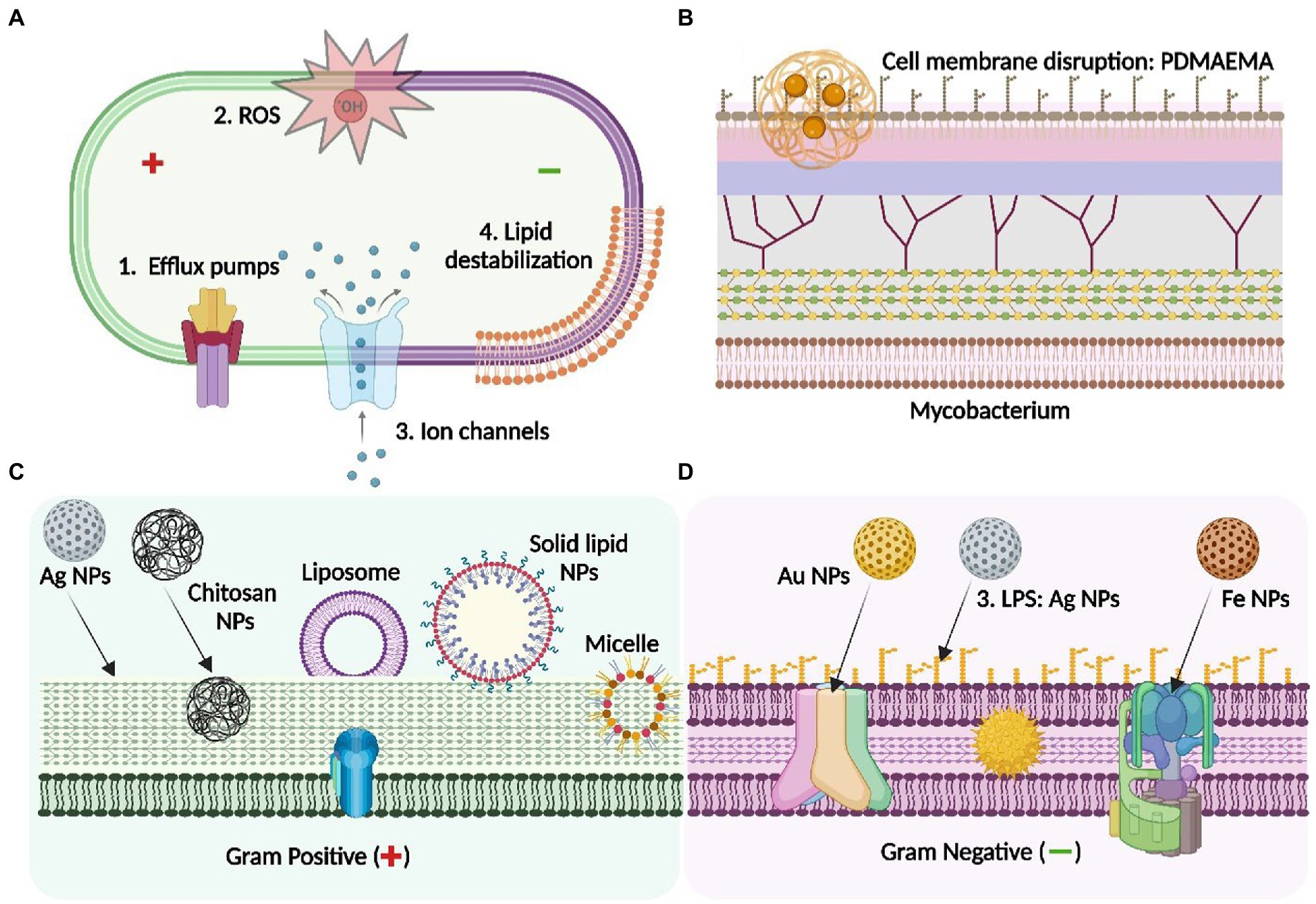
Figure 2. Antibacterial mechanisms of different class of nanoparticle: Mechanistic insight into the nanoparticle interaction with bacterial membrane to overcome AMR. (A) depicts the general membrane targets of the NPs through the activity of efflux pumps, reactive oxygen species (ROS), ion channels and membrane lipid destabilisation serving as antibacterial agents: (B) depicts the mycobacterial membrane and the interaction of PDMAEMA [poly(dimethylaminoethyl) methacrylate] polymeric NPs with the LPS layer of the mycobacterium, leading to cell membrane disruption; (C) represents the precise membrane interaction of the NPs with the Gram- negative bacteria- Ag NPs interact with LPS whereas AuNPs and Fe NPs binds with different targets through specific membrane transport proteins including the porin channels and the F0-F1 ATPase respectively; (D) shows the specific targets of different types of NPs on the Gram-positive bacterial membrane-Ag NPs, chitosan (polymeric) NPs, solid-lipid NPs: These interact with peptidoglycan layer of the cell envelop to exert antimicrobial activity. Others, including micelle or liposome-based NPs serves as a carrier to enhance the penetration of the antibiotic by interacting with the thick peptidoglycan of the bacterial membrane. Figure created in Biorender.
Bacterial membrane is considered as one of the most prominent targets for any anti-bacterial drug or antibiotic as the outer structure acts as a physical barrier and mediator between outer environment and inner cellular components. The membrane proteins and their components including transport protein (Li et al., 2016), anchor proteins (Sun et al., 2021), structural proteins (Lyu et al., 2022), efflux proteins (Soto, 2013; Sun et al., 2014; Du et al., 2018) and ion channels (Prindle et al., 2015) play essential role towards cell physiology and metabolic regulation. Outer leaflet of plasma membrane has architecture of LPS while inner leaflets incorporate mixture of approximately 25 phospholipid type and various protein complexes too. Many metals in nanoparticulate form can exert their antimicrobial activity by destabilizing these membrane proteins (Rudramurthy et al., 2016).
Hamida et al. (2020) showed that biogenic Ag NPs cause the bacterial cell death via the generation of ROS and experimental result concluded that exposure of Ag NPs show biofilm inhibition and virulence activities in MRSA. Mechanical analysis suggested that disruption of membrane proteins mediated via several mechanisms including generation of free radicals leading to oxidative stress, lipid peroxidation interferes with fluidity and stability of membrane leading to cell death. In another recent study, Jiang et al. (2022) exhibited that when nanoparticle-pinched polymer brushes (NPPBs) consisting chemically inert silica nanospheres (covalently grafted with hydrophilic polymer brushes) were exposed to Gram negative strains including E. coli, tobramycin and gentamycin-resistant P. aeruginosa PA14, and Gram Positive S. aureus and S. aureus MU50 (methicillin, oxacillin, and vancomycin-resistant S. aureus), it induced pore formation in the membrane followed by cell death (Nikaido, 2009). Although it was suggested that smaller sized NPPBPs had more intensified antibacterial potential against both Gram positive and Gram negative bacteria compared to larger NPPBs (dsilica > 50 nm).
Gabrielyan et al. (2019) noted antibacterial effect of Fe3O4 NPs on two resistant strains of Gram negative bacteria- ampicillin resistant E. coli DH5α-pUC18 and kanamycin resistant E. coli pARG-25 strains, respectively, and further evaluated the mechanism of action. It was observed that Fe3O4 NPs, although in a concentration dependent manner, significantly started reducing the bacterial growth and increased the latent lag phase at the concentration of 50 μg/ml and exhibited maximum inhibition at concentration of 250 μg/ml. Although kanamycin resistant E. coli pARG-25 was found to be more susceptible than ampicillin resistant E. coli DH5α-pUC18 but exposing 100 μg/ml of Fe3O4NPs to E. coli DH5α-pUC18 either in the absence or presence of antibiotic ampicillin or Kanamycin decreased the cell viability by 5- to 7-fold, respectively. They further determined H+ flux and H+ membrane conductance as an indicator of bacterial membrane function. The production of H2, interestingly suggested that Fe3O4 decreased energy dependent H+ efflux by E. coli DH5α-pUC18 by ~1.2 and ~1.5-fold, respectively, when grown in the absence and presence of ampicillin. Interestingly, exposure of Fe3O4 to E. coli grown in the absence of antibiotics led to ~1.7-fold decrease in H+ flux in the presence of N,N-Dicyclohexylcarbodiimide (DCCD), inhibitor of F0F1 ATPase (Vardanyan et al., 2015). Besides this, approximately ~1.2-fold H+ conductance of E. coli was enhanced, in the absence of ampicillin while in the presence of ampicillin ~1.9 fold increased when compared to control. Lastly, they estimated the H2 production, and observed that besides reducing the redox potential (Eh) to negative value, H2 production was also reduced (Poladyan et al., 2012; Sargsyan et al., 2016).
Trchounian et al. (2017) has reported that H2 production is directly related to the membrane associated formate hydrogen lyase (FHL) complexes that functions to split formate into H2 and CO2. Post exposure to Fe3O4 (in absence of antibiotic), it was observed that H2 production was decreased ~1.2-fold as compared to control. Through this work, including decrease in H+ fluxes in the absence of FoF1 ATPase author concluded that Fe3O4 NPs potentially interact with membrane associated proteins including FHL, membrane transport channel protein etc. which ultimately distort the membrane integrity by enhancing the membrane permeability and leads to cell death.
Beside targeting global membrane proteins, NPs can also exert bactericidal effect upon interaction with particular protein present in the membrane. We found very few studies to have evaluated specific NP-membrane protein interaction, and these have been included in this review. Efflux pumps are transport proteins which essentially play role in extrusion of substances from inner to exterior environment. In bacterial system, at least 6 different drug efflux pumps have been identified that contribute towards the efflux pathways (Du et al., 2018). Among them, there is one ATP-binding cassette (ABC) which directly utilises ATP as an energy source to drive transport, while rest type of efflux pump known as secondary active transporters including major facilitator superfamily (MFS), the multidrug and toxin extrusion (MATE) family, the small multidrug resistance (SMR) family, the resistance-nodulation-cell division (RND) superfamily and the proteobacterial antimicrobial compound efflux (PACE) etc. empowered by electrochemical energy generated in membrane ion transport gradient (Hassan et al., 2013, 2015).
Interestingly, NPs can easily bypass efflux pumps by two mechanisms – (1) act as Trojan horse and deliver antibiotics intracellularly and (2) interact with efflux pump causing irreversible blockage. Several studies have reported effective antibacterial activity when bioactive molecule(s) delivered by encapsulating into nanoparticles. For example, Padwal et al. (2014) and Padwal et al. (2015) studied the antibacterial effect of iron oxide functionalized by polyacrylic acid in the presence of rifampicin antibiotic (PLA-MNP). Exposing Mycobacterium smegmatis to the combination of rifampicin and PLA-MNP led to 4-fold reduction in bacterial proliferation as compared to either of the treatments. Further mechanistic analysis suggested that beside enhanced cell permeability, efflux pump disruption was responsible.
In another study, it was observed that Copper Nanoparticles (Cu NPs) exhibited bactericidal activity by inhibiting norA pump through generation of Cu which directly blocked the efflux pump (Ashajyothi et al., 2016). Similarly, Au NPs functionalized with pyrimidine showed antibacterial activity by sequestering ions such as magnesium and calcium present in the cell. Further analysis suggested that use of vancomycin and Au NPs together led the inhibition of E. coli and P. aeruginosa proliferation by directly targeting efflux pump (Zhao et al., 2010).
In another study, Christena et al. (2015) showed that Cu NPs have potential to act as antibiofilm inhibitor as well as the efflux pump inhibitor. Study revealed that exposure to Cu NPs (0.065 mM) exhibited remarkable inhibition in wild type strains of both S. aureus and P. aeruginosa and less but significant efflux inhibitory effect against MRSA and drug resistant mutant strains of S. aureus. Exposure to and increased concentration of 0.13 mM Cu NPs led to significant inhibition of biofilm formed by S. aureus and P. aeruginosa. Efflux pump inhibition and antibacterial effect of Cu NPs is suggested partly by particles’ effects and ionic effects, respectively. In support of their hypothesis on efflux pump inhibition, the authors performed cartwheel assay, real time efflux and membrane permeable related studies.
In another recent study, it was shown that the magnetite nanoparticles (MNP) coated with polyacrylic acid (PAA) treated together with rifampicin (a first line anti-TB drug) exhibited synergistic effect including 4-fold growth inhibition of M. smegmatis mc2 155 and 3-fold enhanced intracellular aggregation of rifampicin. Through mechanistic study, it was concluded that MNP-PAA formulation blocked the efflux system of mycobacterium (Padwal et al., 2014). These studies suggest that combination of inorganic NPs together with antibiotics can be a promising strategy in blocking efflux pump inhibitors and deliver antibiotic dose within pathogens at therapeutic levels.
Ion channels are the membrane component that play vital role in maintaining physiological balance and homeostatic condition. Different types of ion-specific channels have been identified based on voltage gated and non-voltage gated. Membrane ion channels like Na+, K+ and Cl− are well characterized at structural and functional level (Booth et al., 2003). Altogether, ion channel functions include uptake of particular ions and small molecules etc. (Booth et al., 2003). Upregulation of membrane ion channels have been reported in MDR bacteria (Nikaido, 2009). Interestingly, several studies have suggested good affinity of NPs towards ion channels either through ionic or electrostatic interaction (Yin et al., 2019).
Au NPs functionalized with pyrimidine (Au-DAPT) 4,6-Diamino-2-pyrimidinethiol experimented clinical isolates, MDR E. coli and MDR P. aeruginosa hospitals in China showed effective bacterial cell death. Further, mechanistic analysis suggested that Au-DAPT interact with outer membrane vesicles (OMV) especially sequester and chelate the ions Mg2+ and Ca2+ which caused disturbance in membrane potential, enhancement in cell membrane permeability and thereby cell death (Zhao et al., 2010). Similarly, Song et al., found that exposing Chlorohexidine nanoemulsion against MRSA caused K+, Mg2+ ions leakage which increased the membrane electrical conductivity and thereby exhibited strong ability to damage membrane proteins and increased overall cellular permeability (Song et al., 2016).
ROS is a stress response wherein reactive oxygen species generated using photosensitizers (photosensitive compounds/ metals) which interacts with bacterial biomolecules such as lipids, fatty acids, proteins, DNA, ribosomes etc. eventually leading to cell death (Hong et al., 2019; Li et al., 2021). Although microbes possess antioxidant system to maintain or neutralize the cellular ROS response but an excessive ROS generation can potentially disrupt cellular homeostasis. Nanomaterials having potential to interact with biomolecules, proteins or other macromolecules generate ROS through lipid peroxidation, generation of free radical (1O2), superoxide radicals (O2−), and hydroxyl radicals (.OH). Nanomaterials especially certain metal-based NPs (such as Copper, Tellurium, Titanium) are known to cause ROS development owing to pro-oxidant functional group which allow more ultra-reactive surface, secondly, the involvement of transition metal ion nanoparticles providing multivalent sites for interaction and after that, post-internalisation interaction of NPs with cellular components (Chakraborty et al., 2022b).
Several studies have shown promising antibacterial activity due to ROS generation. Alhadrami and Shoudri (2020) studied the effect of TiO2 NPs on MRSA, E. coli, and P. aeruginosa. The authors observed effective ROS generation with TiO2 due to interaction with membrane lipid and proteins leading to cell death. Similarly, Pramanik et al. (2012) showed that exposure of Copper Iodide (CuI) to Bacillus subtilis (ATCC 6633), Shigella dysenteriae (ATCC 12039) and E. coli DHF 5α (ATCC 10536) cause ROS generation. Further, mechanistic analysis concluded that interaction of Copper Iodide with various amine groups present in the membrane biomolecules and others led to generation of ROS causing cell death. In another study, Sarwar et al. (2016) evaluated the activity of ~10 nm ZnO NPs on two biotypes of O-serotype of V. cholera -Classical and El Tor, with the latter being more susceptible in biofilm and planktonic forms. Further analysis showed enhanced membrane fluidity and polarization along with protein leakage. This distortion of membrane integrity was caused by ROS-mediated free radical generation and its interaction with membrane lipids, proteins, fatty acids followed by cell death (Sarwar et al., 2016). In another recent study, Morena et al. (2021) showed the generation of ROS due to exposure of Hybrid Tellurium Lignin Nanoparticles (TeLigNPs) in S. aureus (ATCC 25923), E. coli (ATCC 25922), and P. aeruginosa (ATCC 10145). Further mechanistic analysis suggested interaction of active surface of TeLigNPs with hydrophilic surface of bacterial membrane. The NPs further integrated with the outer membrane causing lipid peroxidation, generation of highly reactive short aldehyde chain which diffused intracellularly to oxidize amine and thiol groups of proteins thereby affecting the cellular homeostasis and causing cell death (Vermaas et al., 2019).
ROS generation using photosensitizer and light under particular wavelength has successfully translated as research field referred to as antibacterial photodynamic therapy (aPDT). Recently, Yan et al. (2021) designed ultra-thin hollow silica-chitosan NPs and loaded with photosensitizer Ce6. In comparison to free Ce6, its nanoparticle formulation led to stronger antibacterial activity against S. aureus, enhanced adherence and obliteration of mature S. aureus biofilms and 81% decline in biomass. In another work, non-toxic plant pigment chlorophyll was demonstrated to exhibit excellent photodynamic bactericidal properties (Liu et al., 2022). Additionally, if combined with antibiotics, aPDT can show synergistic effects as demonstrated by Liu et al. (Wang et al., 2022a) with gentamycin (antibiotic) and Toluidine (photosensitizer) in terms of growth inhibition in MDR S. aureus. The authors also reported the treatment to be effective in burn-infected mice by reducing number of bacteria colonizing the wound, decline in inflammatory markers and promoting wound healing process. Co-encapsulating photosensitizers with antibiotics with NPs for synergistic effect should be explored for nanomedicine-based aPDT (Table 2).
Overall, nanomedicine has shown promising results in terms of bench-to-bedside research for cancer therapy and these innovations were responsible for early FDA approvals for Doxorubicin-loaded liposome (Doxil®) and Paclitaxel-loaded albumin nanoparticles (Abraxane®) in early 1990s. However, approvals for antimicrobial nanomedicine are few and far between till date. While silver nanoparticles have made it to marketed products from antibacterial creams to face masks, only a handful other NPs are approved as antimicrobial agents, most of which belong to liposomes. Liposomes represent small- (SUVs) or large-unilamellar vesicles (LUVs) composed of phospholipid bilayers with the ability to entrap both hydrophilic and hydrophobic compounds and ease of surface functionalization (Ferreira et al., 2021). They present the most widely researched and FDA approved nanomedicine (Table 3) due to their stability, fusablity with bacterial membrane (fusogenic liposomes) and enhanced biological performance in terms of controlled antibiotic delivery due to prolonged plasma circulation. Besides systemic administration, in one of the recent clinical trials, liposomal amikacin for inhalation (LAI) for nebulizer-based inhalation in Mycobacterium abcessus infected Cystic Fibrosis patients showed promising results (Caimmi et al., 2018).
Other ‘nanobiotics’ in clinical use include formulations of mupirocin, gentamicin and polymyxin B. Recently, during the second wave of Covid, patients recovered from long Covid developed secondary infection due to the fungus Mucormycosis (Black fungus). Amphotericin is an anti-fungal agent against mucormycosis but is highly toxic to nervous system and is administered as Ambisome®- liposomes encapsulating Amphotericin. There was a global shortage of the liposome due to increase in number of patients with Mucormycosis. Hence, it is imperative to develop antibacterial nanomedicine as humanity wades through the surge of AMR.
Mechanistic insights into membrane interaction with NP can yield potential targets for specific action only to bacteria without affecting human cells. Development of such nanoparticles for active delivery can further improve antibiotic pharmacokinetics. There is need to develop effective strategy to counter life threatening situation such as non-response to antibiotics or sepsis. Administering nanoparticles which can block efflux pump inhibitors in such critical situations can be specifically helpful as they can further facilitate antibiotic retention and bacterial killing of resistant strains. Antimicrobial nanomedicine, especially metal-based NPs have shown quick response in killing bacteria upon interaction with bacterial membrane. In general, translational research on nanocarrier-based antibiotic delivery should be promoted as it can yield faster antibacterial response at lower antibiotic doses. Additionally, it prevents undesirable systemic exposure of antibiotics to non-target human tissues thereby reducing the development of microbial resistance.
Generation of toxic by-products following treatment with nanoparticles is a major roadblock in advancement of nanomedicine. Toxicitiy of Ag, ZnO, and CuO NPs and the ensuing toxicity in organs such as spleen, liver, lungs, bone marrow and colon owing to tissue accumulation have been reviewed previously (Liu et al., 2020). Others have reported neuro- and nephrotoxicity with Al2O3 and CuO NPs, respectively, due to DNA damage and oxidative stress (Ivask et al., 2014; Liu et al., 2020). Metal-NPs toxicity can be mitigated by optimizing and administering minimum dose or combination with antibiotics to obtain synergistic effects. Additionally, coating with aptamers, antibody can improve therapeutic effect by targeting specifically to bacteria. Alternatively, organic NPs formulated with biodegradable carrier moieties such as polymers, lipids or amphiphiles can alleviate toxicity issues (Fischer, 2020; Qiu et al., 2020; Mehta et al., 2021).
Besides addressing toxicity and improving pharmacokinetics in vivo, the clinical translatability of nanomedicine in terms of batch-to-batch reproducibility, contaminant-less synthesis and storage stability is pertinent. Improved quality control guidelines for industry-grade nanomaterials and intellectual property regulations can further bring nano-based products through clinical trials to consumers.
As newer pathogenic strains emerge, development of alternative antimicrobial therapy is imminent. Using nanoparticles as a vehicle for delivering therapeutic agents such as antibiotics or antimicrobial peptides not only enhances its delivery efficiency but also reduces drug toxicity, improves bioavailability and specificity. Nanoparticles have shown ability to penetrate biofilms and can also act as slow-release reservoirs of antibiotics. However, most studies are restricted to in vitro bactericidal activity without evaluation of mechanism of action. Comprehensive studies are required to understand the mechanism of interaction of different classes of nanoparticles with bacterial membrane. Research should focus on narrowing down the generic NP-membrane interaction mechanisms, as the nanoparticle interaction can differ depending on the structural differences of Gram negative, Gram positive or Mycobacterium membranes.
Furthermore, optimizing nanoparticle formulations for physical and chemical stability, batch-to-batch reproducibility, quality control and scale up capabilities are paramount to move towards in vivo testing. This gap needs to be further abridged by additional studies of ‘antibacterial nanomedicine’ on mice model of bacterial infection and biodistribution profile to aid its clinical translatability. Various robust animal models of skin, lung, gut and systemic infections exist and must be utilized to further characterize and test antimicrobial nanomedicine.
In this review, we highlighted on the bacterial membrane where most of the AMR-related alterations occur at genomic or functional level. This can have direct implications in the interaction of nanoparticles at the bacterial membrane interface. Although few, we also reviewed research articles that elucidate mechanism of NP-membrane interactions. Identification and characterization of potential bacterial membrane targets specifically in the drug resistant strains can help design NPs towards targeted therapy. Surface conjugation with aptamers or antibodies can provide specificity and further reduce the dose of antibiotic required for antibacterial action. With few clinically approved nanomedicine, there is scope for major research in utilizing the potential of nanotechnology as an alternative to antibiotics.
MSe and MSi: conceptualization, writing—review and editing, and supervision. SM and SG: writing—original draft preparation. All authors contributed to the article and approved the submitted version.
Figures were created using Biorender platform (www.biorender.com/, accessed in 2022).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbaszadegan, A., Ghahramani, Y., Gholami, A., Hemmateenejad, B., Dorostkar, S., Nabavizadeh, M., et al. (2015). The effect of charge at the surface of silver nanoparticles on antimicrobial activity against Gram-positive and Gram-negative bacteria: a preliminary study. J. Nanomater. 2015, 1–8. doi: 10.1155/2015/720654
Acharya, D., Pandey, P., and Mohanta, B. (2021). A comparative study on the antibacterial activity of different shaped silver nanoparticles. Chem. Pap. 75, 4907–4915. doi: 10.1007/s11696-021-01722-8
Acharya, D., Singha, K. M., Pandey, P., Mohanta, B., Rajkumari, J., and Singha, L. P. (2018). Shape dependent physical mutilation and lethal effects of silver nanoparticles on bacteria. Sci. Report. 8, 201–211. doi: 10.1038/s41598-017-18590-6
Alhadrami, H. A., and Shoudri, R. A. (2020). Titanium oxide (TiO2) nanoparticles for treatment of wound infection. J. Pure Appl. Microbiol. 15, 437–451. doi: 10.20944/PREPRINTS202011.0307.V1
Ansari, M. A., Khan, H. M., Khan, A. A., Ahmad, M. K., Mahdi, A. A., Pal, R., et al. (2014a). Interaction of silver nanoparticles with Escherichia coli and their cell envelope biomolecules. J. Basic Microbiol. 54, 905–915. doi: 10.1002/jobm.201300457
Ansari, M. A., Khan, H. M., Khan, A. A., Cameotra, S. S., Saquib, Q., and Musarrat, J. (2014b). Interaction of Al2O3 nanoparticles with Escherichia coli and their cell envelope biomolecules. J. Appl. Microbiol. 116, 772–783. doi: 10.1111/JAM.12423
Aricha, B., Fishov, I., Cohen, Z., Sikron, N., Pesakhov, S., Khozin-Goldberg, I., et al. (2004). Differences in membrane fluidity and fatty acid composition between phenotypic variants of Streptococcus pneumoniae. J. Bacteriol. 186, 4638–4644. doi: 10.1128/JB.186.14.4638-4644.2004/ASSET/53947252-5AFF-4C09-80A5-E6776D5C359D/ASSETS/GRAPHIC/ZJB0140438480002.JPEG
Arora, N., Thangavelu, K., and Karanikolos, G. N. (2020). Bimetallic nanoparticles for antimicrobial applications. Front. Chem. 8:412. doi: 10.3389/FCHEM.2020.00412/BIBTEX
Arthur, M., Molinas, C., Depardieu, F., and Courvalin, P. (1993). Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175, 117–127. doi: 10.1128/jb.175.1.117-127.1993
Ashajyothi, C., Harish, K. H., Dubey, N., and Chandrakanth, R. K. (2016). Antibiofilm activity of biogenic copper and zinc oxide nanoparticles-antimicrobials collegiate against multiple drug resistant bacteria: a nanoscale approach. J. Nanostructure Chem. 6, 329–341. doi: 10.1007/s40097-016-0205-2
Azimi, L., Namvar,, Brahimzade, A. E., Jamali, S., Lari, A. R., Astega,, Bijari, A., Lari, A. R., and Astega,. (2015). Relative expression of efflux pumps in multi drug resistant pseudomonas aeruginosa. Roum. Arch. Microbiol. Immunol. 74, 86–90.
Barros, C. H. N., Hiebner, D. W., Fulaz, S., Vitale, S., Quinn, L., and Casey, E. (2021). Synthesis and self-assembly of curcumin-modified amphiphilic polymeric micelles with antibacterial activity. J. Nanobiotechnol. 19, 1–15. doi: 10.1186/S12951-021-00851-2/FIGURES/8
Beceiro, A., Tomás, M., and Bou, G. (2013). Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 26, 185–230. doi: 10.1128/CMR.00059-12
Beriashvili, D., Taylor, R., Kralt, B., Abu Mazen, N., Taylor, S. D., and Palmer, M. (2018). Mechanistic studies on the effect of membrane lipid acyl chain composition on daptomycin pore formation. Chem. Phys. Lipids 216, 73–79. doi: 10.1016/j.chemphyslip.2018.09.015
Beyth, N., Houri-Haddad, Y., Domb, A., Khan, W., and Hazan, R. (2015). Alternative antimicrobial approach: nano-antimicrobial materials. Evid. Based Complement. Alternat. Med. 2015:246012. doi: 10.1155/2015/246012
Blair, J. M. A., Richmond, G. E., and Piddock, L. J. V. (2014). Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 9, 1165–1177. doi: 10.2217/fmb.14.66
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. V. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Booth, I. R., Edwards, M. D., and Miller, S. (2003). Bacterial ion channels. Biochemistry 42, 10045–10053. doi: 10.1021/bi034953w
Caimmi, D., Martocq, N., Trioleyre, D., Guinet, C., Godreuil, S., Daniel, T., et al. (2018). Positive effect of liposomal amikacin for inhalation on Mycobacterium abcessus in cystic fibrosis patients. Open Forum Infect. Dis. 5:ofy034. doi: 10.1093/ofid/ofy034
Cano, M. E., Rodríguez-Martínez, J. M., Agüero, J., Pascual, A., Calvo, J., García-Lobo, J. M., et al. (2009). Detection of plasmid-mediated quinolone resistance genes in clinical isolates of Enterobacter spp. in Spain. J. Clin. Microbiol. 47, 2033–2039. doi: 10.1128/JCM.02229-08
Castillo, R. R., Lozano, D., González, B., Manzano, M., Izquierdo-Barba, I., and Vallet-Regí, M. (2019). Advances in mesoporous silica nanoparticles for targeted stimuli-responsive drug delivery: an update 16, 415–439. doi: 10.1080/17425247.2019.1598375
Caudill, E. R., Hernandez, R. T., Johnson, K. P., O’Rourke, J. T., Zhu, L., Haynes, C. L., et al. (2020). Wall teichoic acids govern cationic gold nanoparticle interaction with Gram-positive bacterial cell walls. Chem. Sci. 11, 4106–4118. doi: 10.1039/C9SC05436G
Chakraborty, N., Jha, D., Roy, I., Kumar, P., Gaurav, S. S., Marimuthu, K., et al. (2022a). Nanobiotics against antimicrobial resistance: harnessing the power of nanoscale materials and technologies. J. Nanobiotechnol. 20, 1–25. doi: 10.1186/S12951-022-01573-9
Chakraborty, S., Prakash, P., Shah, J., Mayya, C., Singh, S., Ranganathan, R., et al. (2022b). CuO nanoparticles as copper-ion reservoirs for elesclomol-mediated intracellular oxidative stress: implications for anticancer therapies. ACS Appl. Nano Mater. 5, 1607–1620. doi: 10.1021/ACSANM.1C04350/SUPPL_FILE/AN1C04350_SI_005.MP4
Chang, Z. (2016). The function of the DegP (HtrA) protein: protease versus chaperone. IUBMB Life 68, 904–907. doi: 10.1002/iub.1561
Chatterjee, A. K., Chakraborty, R., and Basu, T. (2014). Mechanism of antibacterial activity of copper nanoparticles. Nanotechnology 25:135101. doi: 10.1088/0957-4484/25/13/135101
Chen, Q., Lu, W., Zhou, D., Zheng, G., Liu, H., Qian, C., et al. (2020). Characterization of two macrolide resistance-related genes in multidrug-resistant Pseudomonas aeruginosa isolates. Pol. J. Microbiol. 69:349. doi: 10.33073/pjm-2020-038
Chen, J. H., Siu, L. K., Fung, C. P., Lin, J. C., Yeh, K. M., Chen, T. L., et al. (2010). Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J. Antimicrob. Chemother. 65, 986–990. doi: 10.1093/jac/dkq056
Chen, S., Zhang, A., Blyn, L. B., and Storz, G. (2004). MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 186, 6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004
Chetri, S., Bhowmik, D., Dhar, D., Chakravarty, A., and Bhattacharjee, A. (2019). Effect of concentration gradient carbapenem exposure on expression of blaNDM-1 and acrA in carbapenem resistant Escherichia coli. Infect. Genet. Evol. 73, 332–336. doi: 10.1016/j.meegid.2019.05.024
Christena, L. R., Mangalagowri, V., Pradheeba, P., Ahmed, K. B. A., Shalini, B. I. S., Vidyalakshmi, M., et al. (2015). Copper nanoparticles as an efflux pump inhibitor to tackle drug resistant bacteria. RSC Adv. 5, 12899–12909. doi: 10.1039/C4RA15382K
Ciro, Y., Rojas, J., Oñate-Garzon, J., and Salamanca, C. H. (2019). Synthesis, characterisation and biological evaluation of ampicillin-chitosan-polyanion nanoparticles produced by ionic gelation and polyelectrolyte complexation assisted by high-intensity sonication. Polymers 11:1758. doi: 10.3390/POLYM11111758
Clairfeuille, T., Buchholz, K. R., Li, Q., Verschueren, E., Liu, P., Sangaraju, D., et al. (2020, 2020). Structure of the essential inner membrane lipopolysaccharide–PbgA complex. Nature 584, 479–483. doi: 10.1038/s41586-020-2597-x
Cox, G., and Wright, G. D. (2013). Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 303, 287–292. doi: 10.1016/j.ijmm.2013.02.009
Dadfar, S. M., Roemhild, K., Drude, N. I., von Stillfried, S., Knüchel, R., Kiessling, F., et al. (2019). Iron oxide nanoparticles: diagnostic, therapeutic and theranostic applications. Adv. Drug Deliv. Rev. 138, 302–325. doi: 10.1016/j.addr.2019.01.005
Daimon, Y., Iwama-Masui, C., Tanaka, Y., Shiota, T., Suzuki, T., Miyazaki, R., et al. (2017). The TPR domain of BepA is required for productive interaction with substrate proteins and the β-barrel assembly machinery complex. Mol. Microbiol. 106, 760–776. doi: 10.1111/mmi.13844
Dam, S., Pagès, J. M., and Masi, M. (2017, 2017). Dual regulation of the small RNA MicC and the quiescent Porin OmpN in response to antibiotic stress in Escherichia coli. Antibiotics 6:33. doi: 10.3390/antibiotics6040033
Davis, D. R., McAlpine, J. B., Pazoles, C. J., Talbot, M. K., Alder, E. A., White, C., et al. (n.d.). Enterococcus faecalis multi-drug resistance transporters: Application for antibiotic discovery. J. Mol. Microbiol. Biotechnol. 3, 179–184.
De La Cruz, M. Á., and Calva, E. (2010). The complexities of porin genetic regulation. J. Mol. Microbiol. Biotechnol. 18, 24–36. doi: 10.1159/000274309
Delhaye, A., Collet, J. F., and Laloux, G. (2016). Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. MBio, 7: e00047–e16. doi: 10.1128/mBio.00047-16
Diekema, D. J., and Jones, R. N. (2000). Oxazolidinones: a review. Drugs 59, 7–16. doi: 10.2165/00003495-200059010-00002
Doualla-Bell, F., Boyd, D. A., Savard, P., Yousfi, K., Bernaquez, I., Wong, S., et al. (2021). Analysis of an IncR plasmid carrying blaNDM-1 linked to an azithromycin resistance region in Enterobacter hormaechei involved in an outbreak in Quebec. Microbiol. Spectr. 9:e0199821. doi: 10.1128/spectrum.01998-21
Du, D., Wang-Kan, X., Neuberger, A., van Veen, H. W., Pos, K. M., Piddock, L. J. V., et al. (2018, 2018). Multidrug efflux pumps: structure, function and regulation. Nat. Rev. Microbiol. 16, 523–539. doi: 10.1038/s41579-018-0048-6
Duval, R. E., Gouyau, J., and Lamouroux, E. (2019). Limitations of recent studies dealing with the antibacterial properties of silver nanoparticles: fact and opinion. Nano 9. doi: 10.3390/nano9121775
Erkey, C., and Türk, M. (2021). Synthesis of nanostructured composites of metals by supercritical deposition (SCD). Supercrit. Fluid Sci. Technol. 8, 129–209. doi: 10.1016/B978-0-444-64089-5.00001-9
Ernst, C. M., and Peschel, A. (2011). Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol. Microbiol. 80, 290–299. doi: 10.1111/j.1365-2958.2011.07576.x
Ernst, C. M., Staubitz, P., Mishra, N. N., Yang, S. J., Hornig, G., Kalbacher, H., et al. (2009). The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660. doi: 10.1371/journal.ppat.1000660
Esmaeillou, M., Zarrini, G., Rezaee, M. A., Mojarrad, J. S., and Bahadori, A. (2017). Vancomycin capped with silver nanoparticles as an antibacterial agent against multi-drug resistance bacteria. Adv. Pharm. Bull. 7:479. doi: 10.15171/apb.2017.058
Fernández, L., and Hancock, R. E. W. (2012). Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681. doi: 10.1128/CMR.00043-12
Ferreira, M., Ogren, M., Dias, J. N. R., Silva, M., Gil, S., Tavares, L., et al. (2021). Liposomes as antibiotic delivery systems: a promising nanotechnological strategy against antimicrobial resistance. Molecules 26:2047. doi: 10.3390/molecules26072047
Filippov, A., Orädd, G., and Lindblom, G. (2007). Domain formation in model membranes studied by pulsed-field gradient-NMR: the role of lipid polyunsaturation. Biophys. J. 93, 3182–3190. doi: 10.1529/biophysj.107.111534
Fischer, C. L. (2020). Antimicrobial activity of host-derived lipids. Antibiotics 9:75. doi: 10.3390/antibiotics9020075
Gabrielyan, L., Hakobyan, L., Hovhannisyan, A., and Trchounian, A. (2019). Effects of iron oxide (Fe3O4) nanoparticles on Escherichia coli antibiotic-resistant strains. J. Appl. Microbiol. c–1116. doi: 10.1111/jam.14214
Gill, M. J., Simjee, S., Al-Hattawi, K., Robertson, B. D., Easmon, C. S. F., and Ison, C. A. (1998). Gonococcal resistance to beta-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus. Antimicrob. Agents Chemother. 42, 2799–2803. doi: 10.1128/AAC.42.11.2799
Giner-Casares, J. J., Henriksen-Lacey, M., García, I., and Liz-Marzán, L. M. (2016). Plasmonic surfaces for cell growth and retrieval triggered by near-infrared light. undefined 55, 974–978. doi: 10.1002/ANIE.201509025
Golkar, T., Zielinski, M., and Berghuis, A. M. (2018). Look and outlook on enzyme-mediated macrolide resistance. Front. Microbiol. 9:1942. doi: 10.3389/FMICB.2018.01942/BIBTEX
Haji, S. H., Ali, F. A., and Aka, S. T. H. (2022). Synergistic antibacterial activity of silver nanoparticles biosynthesized by carbapenem-resistant Gram-negative bacilli. Sci. Report. 12, 1–13. doi: 10.1038/s41598-022-19698-0
Hamida, R. S., Ali, M. A., Goda, D. A., Khalil, M. I., and Al-Zaban, M. I. (2020). Novel biogenic silver nanoparticle-induced reactive oxygen species inhibit the biofilm formation and virulence activities of methicillin-resistant Staphylococcus aureus (MRSA) strain. Front. Bioeng. Biotechnol. 8, 433.
Hankins, J. V., Madsen, J. A., Giles, D. K., Brodbelt, J. S., and Trent, M. S. (2012). Amino acid addition to vibrio cholerae LPS establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria. Proc. Natl. Acad. Sci. U. S. A. 109, 8722–8727. doi: 10.1073/pnas.1201313109
Hari, N., Thomas, T. K., and Nair, A. J. (2014). Comparative study on the synergistic action of differentially synthesized silver nanoparticles with β-cephem antibiotics and chloramphenicol. J. Nanosci. 2014, 1–8. doi: 10.1155/2014/201482
Hassan, K. A., Jackson, S. M., Penesyan, A., Patching, S. G., Tetu, S. G., Eijkelkamp, B. A., et al. (2013). Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. U. S. A. 110, 20254–20259. doi: 10.1073/pnas.1317052110
Hassan, K. A., Liu, Q., Henderson, P. J. F., and Paulsen, I. T. (2015). Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. MBio 6:e01982–14. doi: 10.1128/mBio.01982-14
Henderson, J. C., Fage, C. D., Cannon, J. R., Brodbelt, J. S., Keatinge-Clay, A. T., and Trent, M. S. (2014). Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases. ACS Chem. Biol. 9, 2382–2392. doi: 10.1021/cb500438x
Hines, K. M., Waalkes, A., Penewit, K., Holmes, E. A., Salipante, S. J., Werth, B. J., et al. (2017). Characterization of the mechanisms of daptomycin resistance among gram-positive bacterial pathogens by multidimensional lipidomics. mSphere 2:e00492–17. doi: 10.1128/mSphere.00492-17
Ho-Fung Lau, C., Fraud, S., Jones, M., Peterson, S. N., and Poole, K. (2013). Mutational activation of the AmgRS two-component system in aminoglycoside-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57:2243. doi: 10.1128/AAC.00170-13
Hong, Y., Zeng, J., Wang, X., Drlica, K., and Zhao, X. (2019). Post-stress bacterial cell death mediated by reactive oxygen species. Proc. Natl. Acad. Sci. U. S. A. 116, 10064–10071. doi: 10.1073/pnas.1901730116
Huh, A. J., and Kwon, Y. J. (2011). “Nanoantibiotics”: a new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release 156, 128–145. doi: 10.1016/j.jconrel.2011.07.002
Ivask, A., Juganson, K., Bondarenko, O., Mortimer, M., Aruoja, V., Kasemets, K., et al. (2014). Mechanisms of toxic action of Ag, ZnO and CuO nanoparticles to selected ecotoxicological test organisms and mammalian cells in vitro: a comparative review. Nanotoxicology 8, 57–71. doi: 10.3109/17435390.2013.855831
Jiang, Y., Zheng, W., Tran, K., Kamilar, E., Bariwal, J., Ma, H., et al. (2022). Hydrophilic nanoparticles that kill bacteria while sparing mammalian cells reveal the antibiotic role of nanostructures. Nat. Commun. 131:1–17. doi: 10.1038/s41467-021-27193-9
Jourghanian, P., Ghaffari, S., Ardjmand, M., Haghighat, S., and Mohammadnejad, M. (2016). Sustained release curcumin loaded solid lipid nanoparticles. Adv. Pharm. Bull. 6, 17–21. doi: 10.15171/APB.2016.004
Kanipes, M. I., Lin, S., Cotter, R. J., and Raetz, C. R. H. (2001). Ca2+-induced phosphoethanolamine transfer to the outer 3-deoxy-D-manno-octulosonic acid moiety of Escherichia coli lipopolysaccharide. A novel membrane enzyme dependent upon phosphatidylethanolamine. J. Biol. Chem. 276, 1156–1163. doi: 10.1074/jbc.M009019200
Katayama, Y., Ito, T., and Hiramatsu, K. (2000). A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44, 1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000
Kenney, L. J., and Anand, G. S. (2020). EnvZ/OmpR two-component signaling: an archetype system that can function noncanonically. EcoSal Plus 9. doi: 10.1128/ecosalplus.ESP-0001-2019
Keren, I., Kaldalu, N., Spoering, A., Wang, Y., and Lewis, K. (2004). Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18. doi: 10.1016/S0378-1097(03)00856-5
Khan, M. J., Husain, Q., and Ansari, (2013). Polyaniline-assisted silver nanoparticles: A novel support for the immobilization of α-amylase. Appl. Microbiol. Biotechnol. 97, 1513–1522. doi: 10.1007/S00253-012-4384-6/METRICS
Kharissova, O. V., Kharisov, B. I., González, C. M. O., Méndez, Y. P., and López, I. (2019). Greener synthesis of chemical compounds and materials. R. Soc. Open Sciy 01:191378. doi: 10.1098/rsos.191378
Kim, S. Y., Kim, M. H., Kim, S. I., Son, J. H., Kim, S., Lee, Y. C., et al. (2019). The sensor kinase BfmS controls production of outer membrane vesicles in Acinetobacter baumannii. BMC Microbiol. 19:301. doi: 10.1186/s12866-019-1679-0
Kim, I., Viswanathan, K., Kasi, G., Sadeghi, K., Thanakkasaranee, S., and Seo, J. (2020). Preparation and characterization of positively surface charged zinc oxide nanoparticles against bacterial pathogens. Microb. Pathog. 149:104290. doi: 10.1016/j.micpath.2020.104290
Kirtane, A. R., Verma, M., Karandikar, P., Furin, J., Langer, R., and Traverso, G. (2021). Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 16, 369–384. doi: 10.1038/s41565-021-00866-8
Klein, S., Lorenzo, C., Hoffmann, S., Walther, J. M., Storbeck, S., Piekarski, T., et al. (2009). Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol. Microbiol. 71, 551–565. doi: 10.1111/j.1365-2958.2008.06562.x
Kovács, M., Halfmann, A., Fedtke, I., Heintz, M., Peschel, A., Vollmer, W., et al. (2006). A functional dlt operon, encoding proteins required for incorporation of D-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188, 5797–5805. doi: 10.1128/JB.00336-06
Kučerka, N., Nieh, M. P., and Katsaras, J. (2011). Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. Biochim. Biophys. Acta Biomembr. 1808, 2761–2771. doi: 10.1016/j.bbamem.2011.07.022
Kulshrestha, S., Qayyum, S., and Khan, A. U. (2017). Antibiofilm efficacy of green synthesized graphene oxide-silver nanocomposite using Lagerstroemia speciosa floral extract: a comparative study on inhibition of gram-positive and gram-negative biofilms. Microb. Pathog. 103, 167–177. doi: 10.1016/j.micpath.2016.12.022
Kumar, S., Mukherjee, M. M., and Varela, M. F. (2013). Modulation of bacterial multidrug resistance efflux pumps of the major facilitator superfamily. Int. J. Bacteriol. 2013, 1–15. doi: 10.1155/2013/204141
Kumariya, R., Sood, S. K., Rajput, Y. S., Saini, N., and Garsa, A. K. (2015). Increased membrane surface positive charge and altered membrane fluidity leads to cationic antimicrobial peptide resistance in Enterococcus faecalis. Biochim. Biophys. Acta Biomembr. 1848, 1367–1375. doi: 10.1016/j.bbamem.2015.03.007
Labrador-Herrera, G., Pérez-Pulido, A. J., Álvarez-Marín, R., Casimiro-Soriguer, C. S., Cebrero-Cangueiro, T., Morán-Barrio, J., et al. (2020). Virulence role of the outer membrane protein CarO in carbapenem-resistant Acinetobacter baumannii. 11, 1727–1737. doi: 10.1080/21505594.2020.1855912
Lambert, P. A. (2002). Cellular impermeability and uptake of biocides and antibiotics in gram-positive bacteria and mycobacteria. Symp. Ser. Soc. Appl. Microbiol., 31:46S–54S. doi: 10.1046/j.1365-2672.92.5s1.7.x
Lavilla Lerma, L., Benomar, N., Sánchez Valenzuela, A., Muñoz, C., Del, C., Gálvez, A., et al. (2014). Role of EfrAB efflux pump in biocide tolerance and antibiotic resistance of Enterococcus faecalis and Enterococcus faecium isolated from traditional fermented foods and the effect of EDTA as EfrAB inhibitor. Food Microbiol. 44, 249–257. doi: 10.1016/j.fm.2014.06.009
Lee, Y. T., Chen, H. Y., Yang, Y. S., Chou, Y. C., Chang, T. Y., Hsu, W. J., et al. (2020). AdeABC efflux pump controlled by AdeRS two component system conferring resistance to tigecycline, omadacycline and eravacycline in clinical carbapenem resistant Acinetobacter nosocomialis. Front. Microbiol. 11:584789. doi: 10.3389/fmicb.2020.584789
Lee, N. Y., Ko, W. C., and Hsueh, P. R. (2019). Nanoparticles in the treatment of infections caused by multidrug-resistant organisms. Front. Pharmacol. 10:1153. doi: 10.3389/FPHAR.2019.01153/BIBTEX
Lemire, J. A., Harrison, J. J., and Turner, R. J. (2013). Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 11, 371–384. doi: 10.1038/nrmicro3028
Levental, I., Levental, K. R., and Heberle, F. A. (2020). Lipid rafts: controversies resolved, mysteries remain. Trends Cell Biol. 30, 341–353. doi: 10.1016/j.tcb.2020.01.009
Li, M., Cha, D. J., Lai, Y., Villaruz, A. E., Sturdevant, D. E., and Otto, M. (2007a). The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66, 1136–1147. doi: 10.1111/J.1365-2958.2007.05986.X
Li, M., Lai, Y., Villaruz, A. E., Cha, D. J., Sturdevant, D. E., and Otto, M. (2007b). Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. U. S. A. 104, 9469–9474. doi: 10.1073/PNAS.0702159104
Li, Z., Xu, J., Jiang, T., Ge, Y., Liu, P., Zhang, M., et al. (2016). Overexpression of transport proteins improves the production of 5-aminovalerate from l-lysine in Escherichia coli. Sci. Report. 6, 1–8. doi: 10.1038/srep30884
Li, Y., Zhen, J., Tian, Q., Shen, C., Zhang, L., Yang, K., et al. (2020). One step synthesis of positively charged gold nanoclusters as effective antimicrobial nanoagents against multidrug-resistant bacteria and biofilms. J. Colloid Interface Sci. 569, 235–243. doi: 10.1016/j.jcis.2020.02.084
Li, H., Zhou, X., Huang, Y., Liao, B., Cheng, L., and Ren, B. (2021). Reactive oxygen species in pathogen clearance: the killing mechanisms, the adaption response, and the side effects. Front. Microbiol. 11:3610. doi: 10.3389/FMICB.2020.622534/BIBTEX
Liao, W., Cui, Y., Quan, J., Zhao, D., Han, X., Shi, Q., et al. (2022). High prevalence of colistin resistance and mcr-9/10 genes in Enterobacter spp. in a tertiary hospital over a decade. Int. J. Antimicrob. Agents 59:106573. doi: 10.1016/j.ijantimicag.2022.106573
Liu, Y., Busscher, H. J., Zhao, B., Li, Y., Zhang, Z., Van Der Mei, H. C., et al. (2016). Surface-adaptive, antimicrobially loaded, micellar nanocarriers with enhanced penetration and killing efficiency in staphylococcal biofilms. ACS Nano 10, 4779–4789. doi: 10.1021/acsnano.6b01370
Liu, X., Liu, S., Mai, B., Su, X., Guo, X., Chang, Y., et al. (2022). Synergistic gentamicin-photodynamic therapy against resistant bacteria in burn wound infections. Photodiagn. Photodyn. Ther. 39:103034. doi: 10.1016/J.PDPDT.2022.103034
Liu, H., Zhang, W., Fang, Y., Yang, H., Tian, L., Li, K., et al. (2020). Neurotoxicity of aluminum oxide nanoparticles and their mechanistic role in dopaminergic neuron injury involving p53-related pathways. J. Hazard. Mater. 392:122312. doi: 10.1016/j.jhazmat.2020.122312
López-Lara, I. M., and Geiger, O. (2017). Bacterial lipid diversity. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1287–1299. doi: 10.1016/J.BBALIP.2016.10.007
Lubelski, J., Konings, W. N., and Driessen, A. J. M. (2007). Distribution and physiology of ABC-type transporters contributing to multidrug resistance in bacteria. Microbiol. Mol. Biol. Rev. 71, 463–476. doi: 10.1128/MMBR.00001-07
Luo, H., Lan, H., Cha, R., Yu, X., Gao, P., Zhang, P., et al. (2021). Dialdehyde nanocrystalline cellulose as antibiotic substitutes against multidrug-resistant bacteria. ACS Appl. Mater. Interfaces 13, 33802–33811. doi: 10.1021/acsami.1c06308
Lyu, J., Liu, C., Zhang, T., Schrecke, S., Elam, N. P., Packianathan, C., et al. (2022). Structural basis for lipid and copper regulation of the ABC transporter MsbA. Nat. Commun. 13, 1–11. doi: 10.1038/s41467-022-34905-2
MacDermott-Opeskin, H. I., Gupta, V., and O’Mara, M. L. (2022). Lipid-mediated antimicrobial resistance: a phantom menace or a new hope? Biophys. Rev. 14, 145–162. doi: 10.1007/s12551-021-00912-8
Makabenta, J. M. V., Nabawy, A., Li, C. H., Schmidt-Malan, S., Patel, R., and Rotello, V. M. (2020). Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat. Rev. Microbiol. 19, 23–36. doi: 10.1038/S41579-020-0420-1
Mammari, N., Lamouroux, E., Boudier, A., and Duval, R. E. (2022). Current knowledge on the oxidative-stress-mediated antimicrobial properties of metal-based nanoparticles. Microorganisms 10:437. doi: 10.3390/microorganisms10020437
Maria-Neto, S., De Almeida, K. C., Macedo, M. L. R., and Franco, O. L. (2015). Understanding bacterial resistance to antimicrobial peptides: from the surface to deep inside. Biochim. Biophys. Acta Biomembr. 1848, 3078–3088. doi: 10.1016/j.bbamem.2015.02.017
Marquardt, D., Heberle, F. A., Greathouse, D. V., Koeppe, R. E., Standaert, R. F., Van Oosten, B. J., et al. (2016). Lipid bilayer thickness determines cholesterol’s location in model membranes. Soft Matter 12, 9417–9428. doi: 10.1039/C6SM01777K
Mehmood, A., Naseer, S., Ali, A., Fatimah, H., Rehman, S., and Kiani, A. (2020). Identification of novel vaccine candidates against carbapenem resistant Klebsiella pneumoniae: a systematic reverse proteomic approach. Comput. Biol. Chem. 89:107380. doi: 10.1016/j.compbiolchem.2020.107380
Mehta, D., Saini, V., Aggarwal, B., Khan, A., and Bajaj, A. (2021). Unlocking the bacterial membrane as a therapeutic target for next-generation antimicrobial amphiphiles. Mol. Asp. Med. 81:100999. doi: 10.1016/j.mam.2021.100999
Morena, A. G., Bassegoda, A., Hoyo, J., and Tzanov, T. (2021). Hybrid tellurium-lignin nanoparticles with enhanced antibacterial properties. ACS Appl. Mater. Interfaces 13, 14885–14893. doi: 10.1021/ACSAMI.0C22301/ASSET/IMAGES/LARGE/AM0C22301_0008.JPEG
Moreno-Sastre, M., Pastor, M., Salomon, C. J., Esquisabel, A., and Pedraz, J. L. (2015). Pulmonary drug delivery: a review on nanocarriers for antibacterial chemotherapy. J. Antimicrob. Chemother. 70, 2945–2955. doi: 10.1093/jac/dkv192
Moskowitz, S. M., Ernst, R. K., and Miller, S. I. (2004). PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186, 575–579. doi: 10.1128/JB.186.2.575-579.2004
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Neuhaus, F. C., Heaton, M. P., Debabov, D. V., and Zhang, Q. (1996). The dlt operon in the biosynthesis of D-alanyl-lipoteichoic acid in Lactobacillus casei. Microb. Drug Resist. 2, 77–84. doi: 10.1089/mdr.1996.2.77
Nie, D., Hu, Y., Chen, Z., Li, M., Hou, Z., Luo, X., et al. (2020). Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. Sci. 27:26. doi: 10.1186/s12929-020-0617-7
Nikaido, H. (2009). Multidrug resistance in bacteria. Annu. Rev. Biochem. 78:119. doi: 10.1146/annurev.biochem.78.082907.145923
O’Neill, Jim (2016). Tackling drug-resistant infections globally: Final report and recommendations the review on antimicrobial resistance chaired.
Olsen, J. E., Christensen, H., and Aarestrup, F. M. (2006). Diversity and evolution of blaZ from Staphylococcus aureus and coagulase-negative staphylococci. J. Antimicrob. Chemother. 57, 450–460. doi: 10.1093/jac/dki492
Padwal, P., Bandyopadhyaya, R., and Mehra, S. (2014). Polyacrylic acid-coated iron oxide nanoparticles for targeting drug resistance in mycobacteria. Langmuir 30, 15266–15276. doi: 10.1021/LA503808D/SUPPL_FILE/LA503808D_SI_001.PDF
Padwal, P., Bandyopadhyaya, R., and Mehra, S. (2015). Biocompatible citric acid-coated iron oxide nanoparticles to enhance the activity of first-line anti-TB drugs in Mycobacterium smegmatis. J. Chem. Technol. Biotechnol. 90, 1773–1781. doi: 10.1002/jctb.4766
Pal Singh Chauhan, N., Yazdanpanah, A., and Mozafari, M. (2019). Polymer-metal nanocomposites with antimicrobial activity. Biocidal Polym. 4:83–106. doi: 10.1515/9783110639131-004/HTML
Pasquina-Lemonche, L., Burns, J., Turner, R. D., Kumar, S., Tank, R., Mullin, N., et al. (2020). The architecture of the gram-positive bacterial cell wall. Nature 582, 294–297. doi: 10.1038/s41586-020-2236-6
Pedersen, T., Sekyere, J. O., Govinden, U., Moodley, K., Sivertsen, A., Samuelsen, Ø., et al. (2018). Spread of plasmid-encoded NDM-1 and GES-5 carbapenemases among extensively drug-resistant and pandrug-resistant clinical enterobacteriaceae in Durban, South Africa. Antimicrob. Agents Chemother. 62:e02178–17. doi: 10.1128/AAC.02178-17
Pelletier, M. R., Casella, L. G., Jones, J. W., Adams, M. D., Zurawski, D. V., Hazlett, K. R. O., et al. (2013). Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob. Agents Chemother. 57, 4831–4840. doi: 10.1128/AAC.00865-13
Périchon, B., and Courvalin, P. (2009). VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53, 4580–4587. doi: 10.1128/AAC.00346-09
Poger, D., Caron, B., and Mark, A. E. (2014). Effect of methyl-branched fatty acids on the structure of lipid bilayers. J. Phys. Chem. B 118, 13838–13848. doi: 10.1021/jp503910r
Poladyan, A., Avagyan, A., Vassilian, A., and Trchounian, A. (2012). Oxidative and reductive routes of glycerol and glucose fermentation by Escherichia coli batch cultures and their regulation by oxidizing and reducing reagents at different pHs. Curr. Microbiol. 66, 49–55. doi: 10.1007/S00284-012-0240-2
Pramanik, A., Laha, D., Bhattacharya, D., Pramanik, P., and Karmakar, P. (2012). A novel study of antibacterial activity of copper iodide nanoparticle mediated by DNA and membrane damage. Colloids Surf. B: Biointerfaces 96, 50–55. doi: 10.1016/j.colsurfb.2012.03.021
Pratt, L. A., Hsing, W., Gibson, K. E., and Silhavy, T. J. (1996). From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20, 911–917. doi: 10.1111/j.1365-2958.1996.tb02532.x
Prindle, A., Liu, J., Asally, M., Ly, S., Garcia-Ojalvo, J., and Süel, G. M. (2015). Ion channels enable electrical communication in bacterial communities. Nature 527, 59–63. doi: 10.1038/nature15709
Qiu, H., Si, Z., Luo, Y., Feng, P., Wu, X., Hou, W., et al. (2020). The mechanisms and the applications of antibacterial polymers in surface modification on medical devices. Front. Bioeng. Biotechnol. 8:910. doi: 10.3389/FBIOE.2020.00910/BIBTEX
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/CMR.00001-07
Raetz, C. R. H., Reynolds, C. M., Trent, M. S., and Bishop, R. E. (2007). Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329. doi: 10.1146/annurev.biochem.76.010307.145803
Raivio, T. L. (2014). Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim. Biophys. Acta 1843, 1529–1541. doi: 10.1016/J.BBAMCR.2013.10.018
Ranjan, A., Pothayee, N., Seleem, M. N., Boyle, S. M., Kasimanickam, R., Riffle, J. S., et al. (2012). Nanomedicine for intracellular therapy. FEMS Microbiol. Lett. 332, 1–9. doi: 10.1111/j.1574-6968.2012.02566.x
Reverberi, A. P., Kuznetsov, N. T., Meshalkin, V. P., Salerno, M., and Fabiano, B. (2016). Systematical analysis of chemical methods in metal nanoparticles synthesis. Theor. Found. Chem. Eng. 50, 59–66. doi: 10.1134/S0040579516010127
Reygaert, W. (2009). Methicillin-resistant Staphylococcus aureus (MRSA): molecular aspects of antimicrobial resistance and virulence. Clin. Lab. Sci. 22, 115–119.
Richards, S. J., Isufi, K., Wilkins, L. E., Lipecki, J., Fullam, E., and Gibson, M. I. (2018). Multivalent antimicrobial polymer nanoparticles target mycobacteria and Gram-negative bacteria by distinct mechanisms. Biomacromolecules 19, 256–264. doi: 10.1021/acs.biomac.7b01561
Rojas, E. R., Billings, G., Odermatt, P. D., Auer, G. K., Zhu, L., Miguel, A., et al. (2018). The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 559, 617–621. doi: 10.1038/s41586-018-0344-3
Rudramurthy, G. R., Swamy, M. K., Sinniah, U. R., and Ghasemzadeh, A. (2016). Nanoparticles: alternatives against drug-resistant pathogenic microbes. Molecules 21:836. doi: 10.3390/molecules21070836
Rumyantceva, V., Rumyantceva, V., Koshel, E., and Vinogradov, V. (2019). Biocide-conjugated magnetite nanoparticles as an advanced platform for biofilm treatment. Ther. Deliv. 10, 241–250. doi: 10.4155/TDE-2019-0011
Sánchez-López, E., Gomes, D., Esteruelas, G., Bonilla, L., Lopez-Machado, A. L., Galindo, R., et al. (2020). Metal-based nanoparticles as antimicrobial agents: an overview. Nano 10:292. doi: 10.3390/NANO10020292
Sargsyan, H., Trchounian, K., Gabrielyan, L., and Trchounian, A. (2016). Novel approach of ethanol waste utilization: biohydrogen production by mixed cultures of dark- and photo-fermentative bacteria using distillers grains. Int. J. Hydrog. Energy 41, 2377–2382. doi: 10.1016/J.IJHYDENE.2015.11.082
Sarwar, S., Chakraborti, S., Bera, S., Sheikh, I. A., Hoque, K. M., and Chakrabarti, P. (2016). The antimicrobial activity of ZnO nanoparticles against Vibrio cholerae: variation in response depends on biotype. Nanomed. Nanotechnol. Biol. Med. 12, 1499–1509. doi: 10.1016/J.NANO.2016.02.006
Shah, S., Famta, P., Raghuvanshi, R. S., Singh, S. B., and Srivastava, S. (2022). Lipid polymer hybrid nanocarriers: insights into synthesis aspects, characterization, release mechanisms, surface functionalization and potential implications. Colloid Inter. Sci. Commun. 46:100570. doi: 10.1016/j.colcom.2021.100570
Sharifi, S., Bakhshi, B., and Najar-peerayeh, S. (2021). Significant contribution of the CmeABC efflux pump in high-level resistance to ciprofloxacin and tetracycline in Campylobacter jejuni and Campylobacter coli clinical isolates. Ann. Clin. Microbiol. Antimicrob. 20:36. doi: 10.1186/s12941-021-00439-6
Shawa, M., Furuta, Y., Mulenga, G., Mubanga, M., Mulenga, E., Zorigt, T., et al. (2021). Novel chromosomal insertions of ISEcp1-blaCTX-M-15 and diverse antimicrobial resistance genes in Zambian clinical isolates of Enterobacter cloacae and Escherichia coli. Antimicrob. Resist. Infect. Control 10:79. doi: 10.1186/s13756-021-00941-8
Shrivastava, R., and Chng, S. S. (2019). Lipid trafficking across the Gram-negative cell envelope. J. Biol. Chem. 294, 14175–14184. doi: 10.1074/jbc.AW119.008139
Singh, S., Wilksch, J. J., Dunstan, R. A., Mularski, A., Wang, N., Hocking, D., et al. (2022). LPS O antigen plays a key role in Klebsiella pneumoniae capsule retention. Microbiol. Spectr. 10:e0151721. doi: 10.1128/spectrum.01517-21
Siwińska-Stefańska, K., Kubiak, A., Piasecki, A., Goscianska, J., Nowaczyk, G., Jurga, S., et al. (2018). TiO2-ZnO binary oxide systems: comprehensive characterization and tests of photocatalytic activity. Materials 11:841. doi: 10.3390/ma11050841
Sohlenkamp, C., and Geiger, O. (2016). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159. doi: 10.1093/femsre/fuv008
Soltes, G. R., Martin, N. R., Park, E., Sutterlin, H. A., and Silhavy, T. J. (2017). Distinctive roles for periplasmic proteases in the maintenance of essential outer membrane protein assembly. J. Bacteriol. 199:e00418–17. doi: 10.1128/JB.00418-17
Song, Z., Sun, H., Yang, Y., Jing, H., Yang, L., Tong, Y., et al. (2016). Enhanced efficacy and anti-biofilm activity of novel nanoemulsions against skin burn wound multi-drug resistant MRSA infections. Nanomedicine 12, 1543–1555. doi: 10.1016/J.NANO.2016.01.015
Soto, S. M. (2013). Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4:223. doi: 10.4161/VIRU.23724
Spirescu, V. A., Chircov, C., Grumezescu, A. M., and Andronescu, E. (2021). Polymeric nanoparticles for antimicrobial therapies: an up-to-date overview. Polymers 13, 1–27. doi: 10.3390/POLYM13050724
Sun, Y. J., Bai, F., Luo, A. C., Zhuang, X. Y., Lin, T. S., Sung, Y. C., et al. (2021). Probing bacterial cell wall growth by tracing wall-anchored protein complexes. Nat. Commun. 12, 1–9. doi: 10.1038/s41467-021-22483-8
Sun, J., Deng, Z., and Yan, A. (2014). Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 453, 254–267. doi: 10.1016/j.bbrc.2014.05.090
Suttiponparnit, K., Jiang, J., Sahu, M., Suvachittanont, S., Charinpanitkul, T., and Biswas, P. (2011). Role of surface area, primary particle size, and crystal phase on titanium dioxide nanoparticle dispersion properties. Nanoscale Res. Lett. 6, 1–8. doi: 10.1007/S11671-010-9772-1/FIGURES/7
Tanvir, F., Yaqub, A., Tanvir, S., and Anderson, W. A. (2017). Colorimetric enumeration of bacterial contamination in water based on β-galactosidase gold nanoshell activity. Enzym. Microb. Technol. 99, 49–56. doi: 10.1016/j.enzmictec.2017.01.006
Tao, Y., Acket, S., Beaumont, E., Galez, H., Duma, L., and Rossez, Y. (2021). Colistin treatment affects lipid composition of Acinetobacter baumannii. Antibiotics 10:528. doi: 10.3390/antibiotics10050528
Taylor, R., Beriashvili, D., Taylor, S., and Palmer, M. (2017). Daptomycin pore formation is restricted by lipid acyl chain composition. ACS Infect. Dis. 3, 797–801. doi: 10.1021/acsinfecdis.7b00138
Trchounian, K., Poladyan, A., and Trchounian, A. (2017). Enhancement of Escherichia coli bacterial biomass and hydrogen production by some heavy metal ions and their mixtures during glycerol vs glucose fermentation at a relatively wide range of pH. Int. J. Hydrog. Energy 42, 6590–6597. doi: 10.1016/J.IJHYDENE.2017.02.003
Trent, M. S., Ribeiro, A. A., Lin, S., Cotter, R. J., and Raetz, C. R. H. (2001). An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid a: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276, 43122–43131. doi: 10.1074/jbc.M106961200
Turner, R. D., Vollmer, W., and Foster, S. J. (2014). Different walls for rods and balls: the diversity of peptidoglycan. Mol. Microbiol. 91, 862–874. doi: 10.1111/mmi.12513
Vallet-Regí, M., González, B., and Izquierdo-Barba, I. (2019). Nanomaterials as promising alternative in the infection treatment. Int. J. Mol. Sci. 20:3806. doi: 10.3390/ijms20153806
Van Langevelde, P., Kwappenberg, K. M. C., Groeneveld, P. H. P., Mattie, H., and Van Dissel, J. T. (1998). Antibiotic-induced lipopolysaccharide (LPS) release from Salmonella typhi: delay between killing by ceftazidime and imipenem and release of LPS. Antimicrob. Agents Chemother. 42:739. doi: 10.1128/AAC.42.4.739
Vardanyan, Z., Gevorkyan, V., Ananyan, M., Vardapetyan, H., and Trchounian, A. (2015). Effects of various heavy metal nanoparticles on Enterococcus hirae and Escherichia coli growth and proton-coupled membrane transport. J. Nanobiotechnol. 13:69. doi: 10.1186/s12951-015-0131-3
Vermaas, J. V., Dixon, R. A., Chen, F., Mansfield, S. D., Boerjan, W., Ralph, J., et al. (2019). Passive membrane transport of lignin-related compounds. Proc. Natl. Acad. Sci. U. S. A. 116, 23117–23123. doi: 10.1073/pnas.1904643116
Vihodceva, S., Šutka, A., Sihtmäe, M., Rosenberg, M., Otsus, M., Kurvet, I., et al. (2021). Antibacterial activity of positively and negatively charged hematite (α-Fe2O3) nanoparticles to Escherichia coli, Staphylococcus aureus and Vibrio fischeri. Nano 11:652. doi: 10.3390/nano11030652
Wang, Z., Geng, H., Nie, C., and Xing, C. (2022b). Conjugated polymers for combatting antimicrobial resistance. Chin. J. Chem. 40, 759–772. doi: 10.1002/CJOC.202100692
Wang, N., Li, D., Schwarz, S., Qin, S., Yao, H., and Du, X.-D. (2021). Novel Tet(L) efflux pump variants conferring resistance to tigecycline and eravacycline in Staphylococcus Spp. Microbiol. Spectr. 9:e0131021. doi: 10.1128/Spectrum.01310-21
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
Wang, D. Y., van der Mei, H. C., Ren, Y., Busscher, H. J., and Shi, L. (2020a). Lipid-based antimicrobial delivery-Systems for the treatment of bacterial infections. Front. Chem. 7:872. doi: 10.3389/FCHEM.2019.00872/BIBTEX
Wang, M., Wang, W., Niu, Y., Liu, T., Li, L., Zhang, M., et al. (2020b). A clinical extensively-drug resistant (XDR) Escherichia coli and role of its β-lactamase genes. Front. Microbiol. 11:3178. doi: 10.3389/FMICB.2020.590357/BIBTEX
Wang, G., Yang, C., Shan, M., Jia, H., Zhang, S., Chen, X., et al. (2022a). Synergistic poly(lactic acid) antibacterial surface combining superhydrophobicity for antiadhesion and chlorophyll for photodynamic therapy. Langmuir 38, 8987–8998. doi: 10.1021/ACS.LANGMUIR.2C01377/SUPPL_FILE/LA2C01377_SI_004.MP4
Wong, J. L. C., Romano, M., Kerry, L. E., Kwong, H. S., Low, W. W., Brett, S. J., et al. (2019). OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo. Nat. Commun. 10, 1–10. doi: 10.1038/s41467-019-11756-y
Wood, T. K., Knabel, S. J., and Kwan, B. W. (2013). Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 79, 7116–7121. doi: 10.1128/AEM.02636-13
Yan, C., Shao, X., Shu, Q., Teng, Y., Qiao, Y., Guan, P., et al. (2021). Chitosan modified ultra-thin hollow nanoparticles for photosensitizer loading and enhancing photodynamic antibacterial activities. Int. J. Biol. Macromol. 186, 839–848. doi: 10.1016/j.ijbiomac.2021.07.078
Yang, S. J., Kreiswirth, B. N., Sakoulas, G., Yeaman, M. R., Xiong, Y. Q., Sawa, A., et al. (2009). Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. J. Infect. Dis. 200, 1916–1920. doi: 10.1086/648473
Yang, W., Moore, I. F., Koteva, K. P., Bareich, D. C., Hughes, D. W., and Wright, G. D. (2004). TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 279, 52346–52352. doi: 10.1074/jbc.M409573200
Yin, S., Liu, J., Kang, Y., Lin, Y., Li, D., and Shao, L. (2019). Interactions of nanomaterials with ion channels and related mechanisms. Br. J. Pharmacol. 176:3754. doi: 10.1111/bph.14792
Yuan, Y. G., Peng, Q. L., and Gurunathan, S. (2017). Effects of silver nanoparticles on multiple drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa from mastitis-infected goats: an alternative approach for antimicrobial therapy. Int. J. Mol. Sci. 18:569. doi: 10.3390/ijms18030569
Zavascki, A. P., Goldani, L. Z., Li, J., and Nation, R. L. (2007). Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60, 1206–1215. doi: 10.1093/jac/dkm357
Zhang, D. F., Li, H., Lin, X. M., and Peng, X. X. (2015). Outer membrane proteomics of kanamycin-resistant Escherichia coli identified MipA as a novel antibiotic resistance-related protein. FEMS Microbiol. Lett. 362:74. doi: 10.1093/FEMSLE/FNV074
Zhao, Y., Tian, Y., Cui, Y., Liu, W., Ma, W., and Jiang, X. (2010). Small molecule-capped gold nanoparticles as potent antibacterial agents that target gram-negative bacteria. J. Am. Chem. Soc. 132, 12349–12356. doi: 10.1021/JA1028843/SUPPL_FILE/JA1028843_SI_001.PDF
Zhou, Z., Ribeiro, A. A., Lin, S., Cotter, R. J., Miller, S. I., and Raetz, C. R. H. (2001). Lipid a modifications in polymyxin-resistant Salmonella typhimurium: PMRA-dependent 4-amino-4-deoxy-L-arabinose, and phosphoethanolamine incorporation. J. Biol. Chem. 276, 43111–43121. doi: 10.1074/jbc.M106960200
Keywords: nanomedicine, AMR, antibiotic, membrane, resistant
Citation: Modi SK, Gaur S, Sengupta M and Singh MS (2023) Mechanistic insights into nanoparticle surface-bacterial membrane interactions in overcoming antibiotic resistance. Front. Microbiol. 14:1135579. doi: 10.3389/fmicb.2023.1135579
Received: 01 January 2023; Accepted: 30 March 2023;
Published: 21 April 2023.
Edited by:
Madhuree Kumari, Indian Institute of Science (IISc), IndiaReviewed by:
Liping Wang, Nanjing Agricultural University, ChinaCopyright © 2023 Modi, Gaur, Sengupta and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manu Smriti Singh, bWFudS5zaW5naEBiZW5uZXR0LmVkdS5pbg==; bWFubjI1ODVAZ21haWwuY29t; Mrittika Sengupta, bXJpdHRpa2Euc2VuZ3VwdGFAYmVubmV0dC5lZHUuaW4=; bXJpdHRpa2FzZW5ndXB0YUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.