
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 September 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.991987
This article is part of the Research Topic Community Series in Plants and Microbial Communities: Diversity, Pathogens and Biological Control, volume II View all 25 articles
Trichoderma spp. are a group of widespread fungi with important applications in many aspects of human life, but they are also pathogens that cause green mold disease on mushrooms. During a survey of mushroom cultivation in Guizhou, China, five strains of Trichoderma from three different localities were isolated from soil in mushroom bags of Hymenopellis raphanipes. The typical morphology of having gregarious, reddish stromata and gregarious phialides and the results of phylogenetic analyses based on a combined dataset of RPB2, TEF, and ITS gene sequences demonstrated that these green-spored Trichoderma belong to a new taxon, Trichoderma hymenopellicola. Pathogenicity tests by covering fungal mycelial blocks or soil mixed with spore suspension in mushroom bags showed similar symptoms to those in the field, and the same fungal pathogen had been observed and re-isolated from these symptoms, which fulfill Koch’s postulates. A primary screening test of nine common fungicides indicated that prochloraz-manganese chloride complex and propiconazole are the top two effective fungicides inhibiting the pathogen, whereas the former was further indicated as a suitable fungicide to control Trichoderma hymenopellicola, with a high inhibition ratio to the pathogen and low toxicity to the mushroom.
Hymenopellis raphanipes (≡ Oudemansiella raphanipes) is a widely cultivated mushroom with edible and medicinal properties. With the increasing scale of cultivation, the annual yield of Hymenopellis raphanipes in China has exceeded 20,000 tons per year, with a corresponding output value of over 350 billion CNY. Meanwhile, various diseases began to appear during the cultivation, whereas researchers have paid more attention on optimizing its growth condition. It has been reported that cobweb disease caused by species of Cladobotryum on Hymenopellis raphanipes is a major problem during mushroom cultivation and can cause great yield losses (Liu et al., 2020; Qin et al., 2021). During our investigation of mushroom cultivation, the green mold disease was found on the soil surface and fruiting bodies of Hymenopellis raphanipes bags from three different localities. This disease has a 30–50% incidence and can cause mushroom bag rot and a decline in yield and quality.
Green mold disease is a common disease that often occurs during mushroom cultivation, with symptom of having green, villiform mycelium on the surface. Trichoderma spp. are the causal agent of green mold disease, which comprises a group of mostly saprobic fungi that are widespread in soil, healthy plants, wood, or other fungi. They are widely used to control fungal pathogens (Hasan et al., 2012; Liu et al., 2012; Abo-Elyousr et al., 2014; Poveda et al., 2019), produce antibiotics, enzymes, and biofuel (Degenkolb et al., 2008; Jun et al., 2011), and bioremediation xenobiotic compounds in water and soil (Katayama and Matsumura, 1993; Harman et al., 2004; Ezzi and Lynch, 2005). On the contrary, some species can cause economic losses in mushroom growing (Samuels et al., 2002; Kim et al., 2012) or infect humans (Kredics et al., 2003; Kuhls et al., 2010). To avoid the negative effects of Trichoderma on humans, fungicides like carbendazim, hexaconazole, metrafenone, prochloraz, tebuconazole, and thiophanate-methyl, can be used to prevent the occurrence and spreading of Trichoderma spp. (Madhavi et al., 2011; Kosanoviæ et al., 2015; Lukoviæ et al., 2021).
Trichoderma includes two groups of species with different colors of ascospores, i.e., hyaline and green ascospores. Green-spored Trichoderma species were first intensively studied by Chaverri and Samuels (2004). Afterward, Jaklitsch and Voglmayr (2015) divided them into six subclades, viz. Ceramicum, Chlorosporum, Harzianum, Helium, Spinulosum, and Strictipile, but this treatment has not been fully accepted by other researchers (Chen and Zhuang, 2017). Recently, Bustamante et al. (2021) conducted multilocus phylogenetic analyses and four DNA-based methods to delimit the Trichoderma species of the Harzianum lineage that comprises most green-spored species.
Morphologically, the new pathogen is characterized by green ascospores and colonies with reddish or yellowish stromata. Phylogenetically, it forms a distinct clade sister to Trichoderma epimyces, T. priscilae, T. purpureum, T. rufobrunneum, and T. tenue. To find a way of controlling the disease, the compatibility of the new pathogen with nine fungicide candidates was tested, and the top two most sensitive fungicides were further applied to strains from all three localities, as well as the mushroom strain. A fungicide that can inhibit the growth of the pathogen and is less toxic to the mushroom would be a suitable agent to prevent the occurrence and spreading of the disease.
Mushroom bags with diseased Hymenopellis raphanipes were collected from mushroom cultivation bases at Jianhe (JH), Shuicheng (SC), and Zhenfeng (ZF) counties in Guizhou, China, during 2019 to 2021. Pathogens were isolated using the spread plate and tissue isolation method resulting in a total of five fungal isolates. Purified cultures were incubated on cornmeal dextrose agar (CMD), potato dextrose agar (PDA), and synthetic low nutrient agar (SNA) plates at 25, 30, and 35°C. Ex-type strains were deposited at the Culture Collection, Department of Plant Pathology, Guizhou University (GUCC). MycoBank number was registered for the new taxon (Crous et al., 2004).
Photographs of fresh stromata were taken using an ultra-depth of field stereomicroscope (digital microscope system Keyence VHX-7000) to illustrate the macrostructures. Sections were made using a stereomicroscope (Leica S9i) and mounted in water or a rehydrated 5% KOH solution. Photomicrographs of perithecia, asci, ascospores, conidiophores, conidia, and phialides were taken with a compound light microscope (Zeiss Scope 5 with color camera AxioCam 208) to observe the morphological characteristics. All measurements of the observed structures were made with ZEN2 (blue edition) software.
A pathogenicity test was conducted by inoculating fungal mycelial blocks and spore suspensions of fungal strains isolated from JH, SC, and ZF on the soil surface of 90-day-old mushroom bags and fruiting bodies of Hymenopellis raphanipes all groups were further incubated at room temperature. PDA blocks and distilled water were used in the control check (CK) to replace the mycelial blocks and spore suspensions. Photographs of bags were taken after 1, 7, and 14 days, respectively, to check if any green mycelium occurred. Then, the fungal pathogen was re-examined and re-isolated from the diseased area to fulfill Koch’s postulates.
An antagonistic experiment was also conducted by inoculating mycelial blocks of the pathogen and mushroom on PDA plates and incubating at room temperature. When the colony of pathogen and mushroom overlapped, photographs of the petri dishes and photomicrographs of the overlapping hyphae were taken to see if there was any interaction between the two species.
Total genomic fungal DNA was extracted by a CwBiotech Plant Genomic DNA Kit (Changping, Beijing, China) following the manufacturer’s protocol. The internal transcribed spacer (ITS) along with the 5.8S ribosomal rDNA, partial translation elongation factor 1-α (TEF), and RNA polymerase II second largest subunit (RPB2) were amplified with the primer pairs ITS5/ITS4, EF1-728F/TEF1LLErev, and fRPB2-5F/fRPB2-7cR, respectively (White et al., 1990; Carbone and Kohn, 1999; Liu et al., 1999; Jaklitsch et al., 2005).
Polymerase chain reaction (PCR) reactions were employed in a 25 μl reaction mixture containing 1.6 μl dNTP mix (2.5 mM/μl), 0.2 μl of Taq polymerase (5 U/μl), 2 μL polymerase buffer (10 × /μl), 1 μl forward and reward primers (10 μM/μl), and 1 μl DNA template. Amplifications were performed in a T100™ Thermal Cycler (BIO-RAD), which was programmed for an initial denaturation at 95°C for 3 min followed by 34 cycles of 1 min at 95°C, 30 s at 55°C and extension at 72°C for 1 min, and a final extension at 72°C for 10 min. PCR products were sequenced by using the same PCR primers used in amplification reactions by Sangon Biotech (Shanghai) Co., Ltd.
Sequences of each gene generated from forward and reverse primers were assembled with BioEdit v.7.2.5 (Hall, 1999), and consensus sequences were then combined with related sequences downloaded from GenBank (Supplementary Table 1). Each gene dataset was aligned separately by Mafft v7.187 (Katoh and Standley, 2013), and manually aligned where necessary. The nucleotide substitution model for each gene was determined by the Bayesian information criterion (BIC) using jModelTest v2.1.6 (Darriba et al., 2012). Phylogenetic trees based on a concatenated dataset of RPB2, TEF, and ITS, generated by SequenceMatrix v1.7.8 (Vaidya et al., 2011), were constructed using maximum likelihood (ML) and Bayesian inference (BI) analyses at the CIPRES Web Portal (Miller et al., 2010). ML was performed using the “RAxML-HPC BlackBox” tool (Stamatakis, 2014). The Markov Chain Monte Carlo (MCMC) algorithm for BI with two parallel runs of four chains was performed using the “MrBayes on XSEDE” tool (Ronquist et al., 2012). Trees were sampled every 100 generations, and runs were stopped automatically when the average standard deviation of split frequencies fell below 0.01. A 50% majority rule consensus tree was summarized after discarding the first 25% of samples. The resulting trees were visualized in FigTree v1.4.3 (Rambaut, 2016).
The type strain of the new collection was first used to conduct primary fungicide sensitivity tests in vitro against four biological fungicides and five chemical fungicides with five different gradient concentrations based on the instructions and our preliminary tests (Table 1). Each fungicide was applied to PDA plates with three replicates, and the same amount of distillation water was applied in CK. Mycelial blocks of 5 mm in diameter from 6-day-old cultures were placed in the center of the plates and incubated at 25°C. Afterward, the diameters of colony (D) were measured after 6-day incubation, and the inhibition ratio (IR = 1–D/DCK) was calculated. Linear regression analysis and the half maximal effective concentration (EC50) were calculated using DPS V18.10, and the significance of the difference was calculated by Duncan’s new multiple range test.
After the primary test, representative strains from the other two collection sites were replicated three times, as well as the mushroom strain, to further test their sensitivity to the top two most effective fungicides.
Both soil inoculating groups of covering mycelial blocks and soil mixed with spore suspension of Trichoderma hymenopellicola exhibited similar symptoms of green mold disease in the field after 7 days (Figures 1D–L), while the control group do not have (Figures 1A–C). The white mycelium can be observed on the surface of mushroom bags after 3–5 days and spread fast, which covered the whole surface of substrate and turned green within 10 days. The rate of Trichoderma hymenopellicola infecting mushroom bags is about 50%, which is similar to its incidence in the field. The same fungal pathogen had been observed and re-isolated from these symptoms, which fulfils Koch’s postulates.
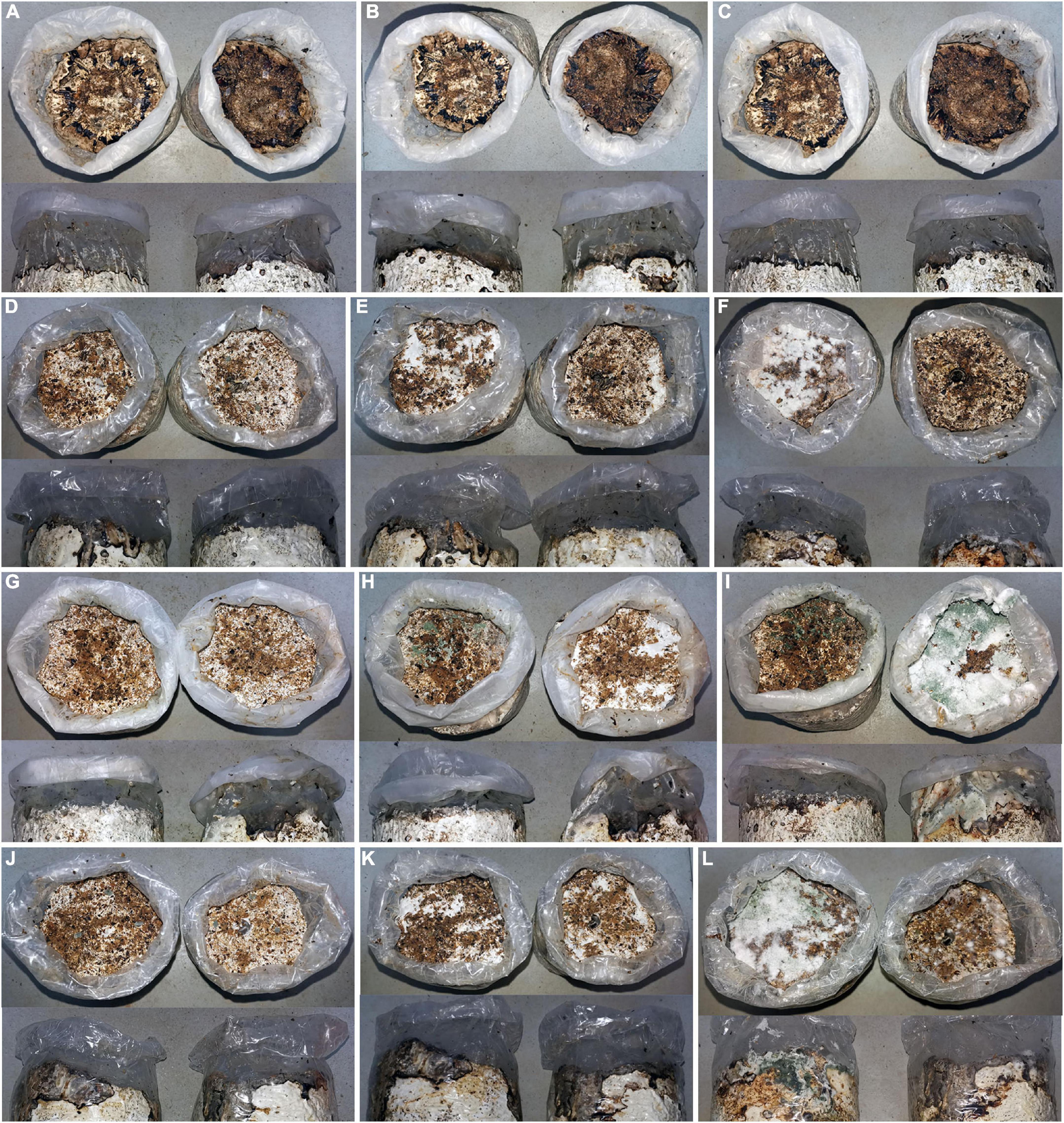
Figure 1. Pathogenicity test of Trichoderma hymenopellicola by covering its mycelial blocks (upper) and soil mixed with spore suspension (lower). (A–C) CK group after 1, 7, and 14 days. (D–F) JH group after 1, 7, and 14 days. (G–I) SC group after 1, 7, and 14 days. (J–L) ZF group after 1, 7, and 14 days.
The result of an antagonistic experiment demonstrated that hyphae of Trichoderma hymenopellicola can cause hyphae of Hymenopellis raphanipes to grow abnormally (Figures 2E,F), and the overlapping part of two colonies does not form a clear boundary (Figures 2A–D). On the contrary, infections were not observed in the fruiting body inoculating group.
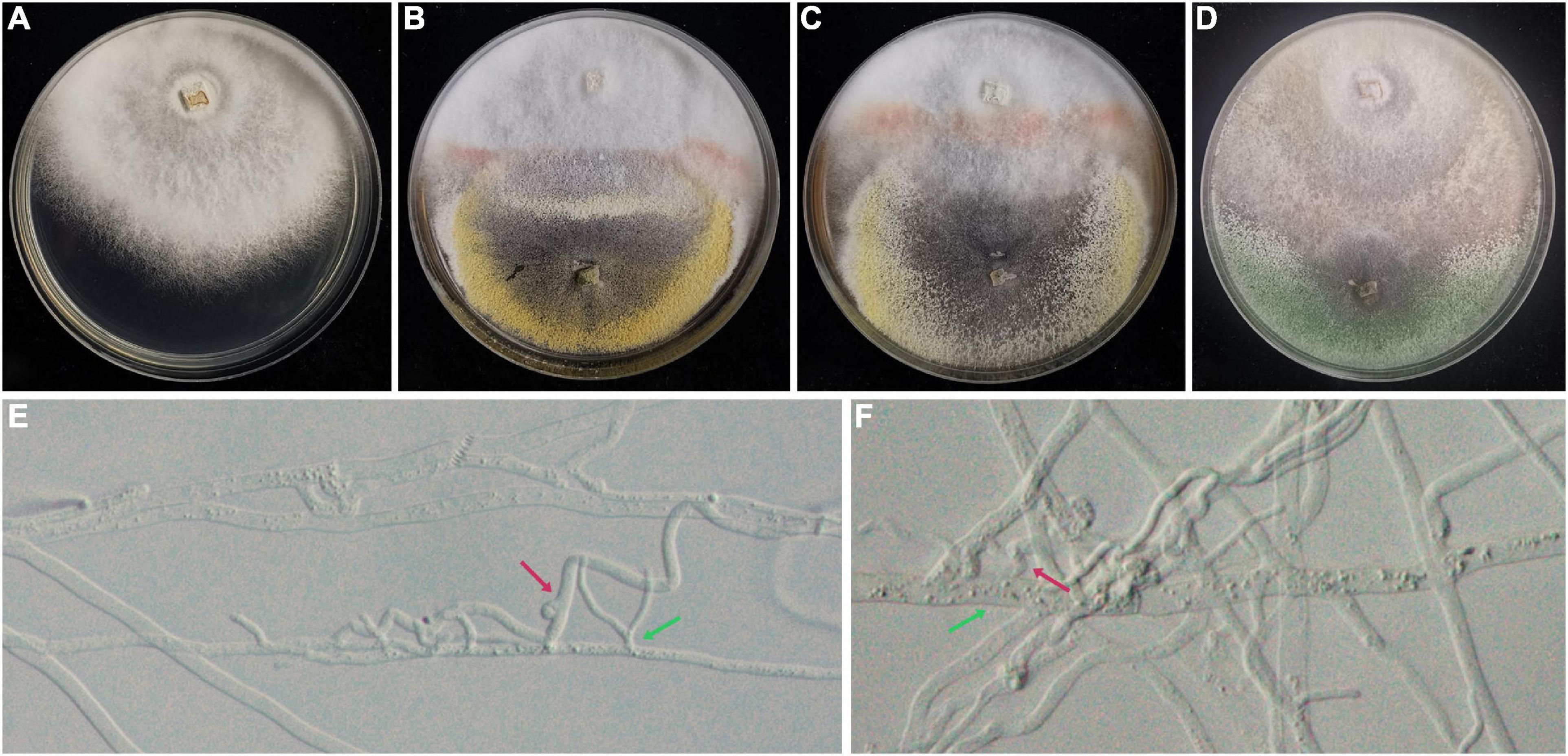
Figure 2. Antagonistic experiment between strains of Trichoderma hymenopellicola and Hymenopellis raphanipes on PDA. (A) Mushroom colony (CK). (B) Mushroom colony with pathogen from JH. (C) Mushroom colony with pathogen from SC. (D) Mushroom colony with pathogen from ZF. (E,F) Hyphae of Trichoderma hymenopellicola (green arrow) and abnormal mushroom hyphae (red arrow).
The concatenated dataset (Supplementary Table 1) consists of 129 strains and 2,922 unambiguously aligned sites (ITS, 609; RPB2, 1024; and TEF, 1289). The best-fit substitution model of each gene is TPM1uf + I + G (RPB2 and TEF) and ITS (TPM2uf + I + G). The RAxML analysis of the combined dataset yielded a best-scoring tree with a final ML optimization likelihood value of –35239.670452. Estimated base frequencies are as follows: A = 0.233134, C = 0.285526, G = 0.253003, and T = 0.228336; substitution rates AC = 1.134637, AG = 4.477934, AT = 1.149518, CG = 1.048786, CT = 6.335323, and GT = 1.000000; proportion of invariable sites I = 0.544721; and gamma distribution shape parameter α = 0.951765. The Bayesian analysis ran 29,64,000 generations before the average standard deviation for split frequencies reached 0.00998. The analysis generated 59,282 trees in total, from which 44,462 were sampled after burn-in, and the 99% credible set contains 35,309 trees. Our new strains belong to a distinct clade that is genetically distant from Trichoderma epimyces, T. priscilae, T. purpureum, T. rufobrunneum, and T. tenue, and is divided into three subclades represented by strains from the three localities (Figure 3).
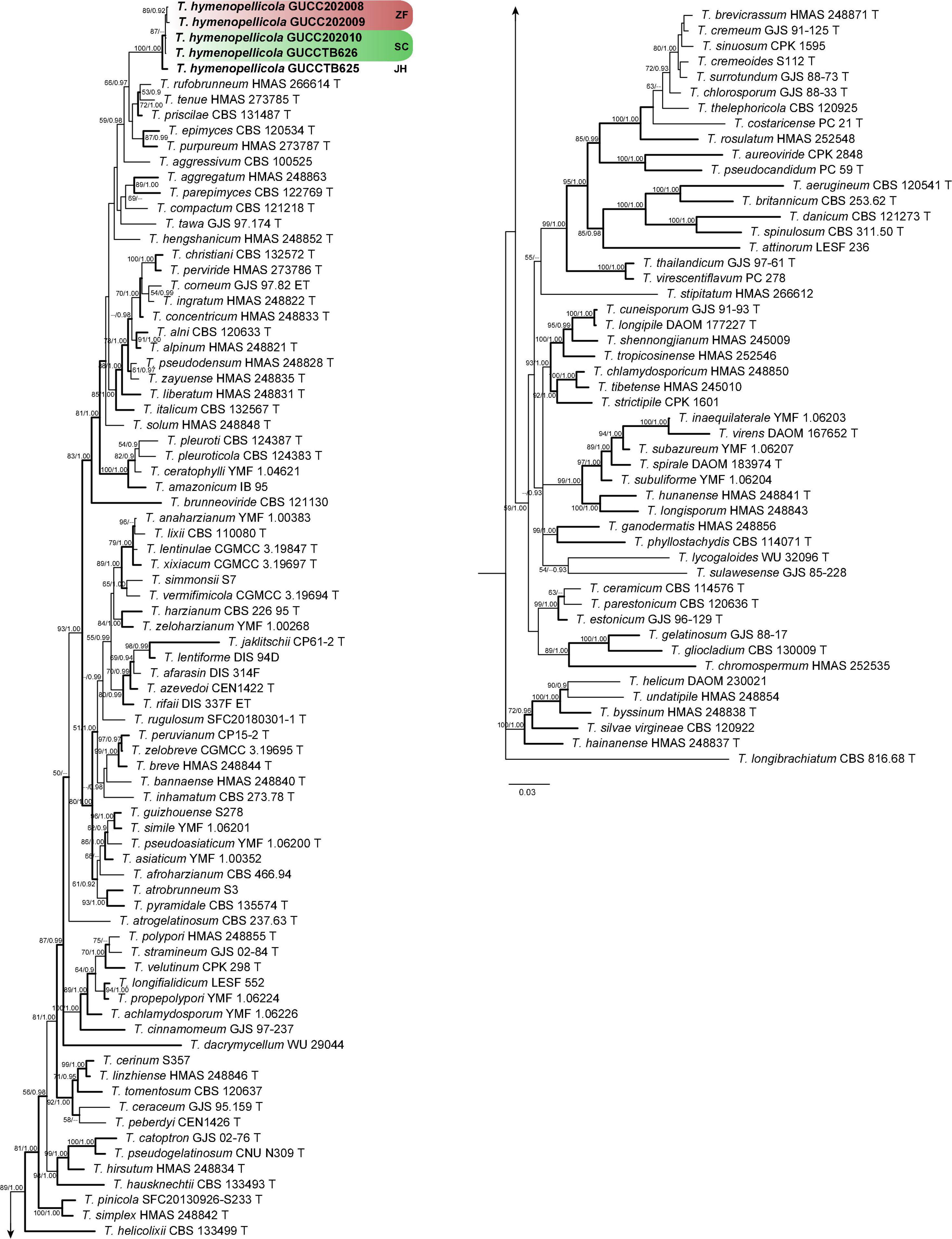
Figure 3. Phylogram generated from Bayesian inference based on combined ITS, RPB2, and TEF sequence data of green-spored Trichoderma species with T. longibrachiatum as the outgroup. Maximum likelihood bootstrap support (BS) above 50% and Bayesian posterior probabilities (PP) above 0.9 are shown at nodes. Clades with strong support (BS ≥ 70, PP ≥ 0.95) are indicate in bold. New sequences obtained from this study are in bold. ‘T’ represents ex-type strains.
Trichoderma hymenopellicola X.Y. Zeng, X.X. Yuan and F.H. Tian, sp. nov. Figure 4.

Figure 4. Trichoderma hymenopellicola (HGUP 202007, GUCC 202010). (A–C) Disease in the field. (D) Fresh stromata. (E) Ostiolar dots on stromata surface. (F) Cortical and subcortical tissue. (G) Cortical and subcortical tissue in section. (H) Ostiole. (I,J) Asci with ascospores. (K) Conidiophores and phialides. (L) Conidia. (M) Cultures on CMD (5 days). (N) Cultures on PDA (5 days). (O) Cultures on SNA (4 days). Scale bars: (D) 1,000 μm, (E,F) 100 μm, (G) 50 μm, (H–K) 20 μm, (L) 10 μm.
MycoBank number: 840876.
Etymology: In reference to its occurrence in Hymenopellis raphanipes bags (Figures 4A–C).
Stromata 1–15 mm in diameter, 1–10 mm thick (n = 10), mostly gregarious, discoid or undulate, with yellowish margin and pale red, depressed center when young, becoming reddish with rugose surface when mature (Figure 4D). Ostiolar dots are umbilicate brown. Rehydrated stromata are larger than dry ones, the surface is smooth, and they become purple in 5% KOH (Figure 4E). Cortical layer comprising thick-walled, brown cells with textura angularis (Figure 4F). Perithecia 185–208 × 124–179 μm (n = 20), flask-shaped or subglobose, crowded (Figure 4G). Peridium 9–14 μm thick at sides, 13–22 μm thick at base (n = 30), light brown. Ostioles 87–102 × 33–44 μm (n = 30) (Figure 4H). Asci 87–98 × 5–6 μm (n = 30), including a 9–19 μm long stipe, 16-spored, cylindrical, hyaline (Figures 4I,J). Ascospores 5–7 × 3–5 μm (n = 50), 1-seriate, ellipsoid to globose, green, and verrucose.
Growth optimum at 25°C and no growth at 35°C on all media. On CMD after 72 h 49–50 mm, mycelium covering the plate after 4 days (Figure 4M). On PDA after 72 h 45–49 mm, mycelium covering the plate after 5 days (Figure 4N). Colony hyaline, greenish, yellowish, or pink when old, dense, circular, margin well defined and stellate due to parallel, aggregated surface hyphae. Aerial hyphae numerous, thin, and complexly branched, forming radial and circular strands. Conidiophores 1–3 level are branched and tapered at the tips (Figure 4K). Phialides are mostly gregarious, subfusiform, or cylindrical. Conidia are subglobose, smooth, hyaline, and scar indistinct, with no or few minute guttules (Figure 4L). On SNA, mycelium had covered the plate for 72 h (Figure 4O). Colonies are hyaline, thin, and circular. Aerial hyphae are scant and thin.
China, Guizhou, Liupanshui, Shuicheng district, on soil surfaces of Hymenopellis raphanipes bags, March 2021, X-XY (holotype HGUP20071; ex-type culture GUCC202010; culture GUCCTB626). China, Guizhou, Southeast Guizhou Autonomous prefecture, Jianhe County, on the soil surface of Hymenopellis raphanipes bags, July 2021, X-XY (GUCCTB625). China, Guizhou, Southwest Guizhou Autonomous prefecture, Zhenfeng County, on the soil surface of Hymenopellis raphanipes bags, January 2020, X-XY (cultures GUCC202008 and GUCC202009).
Phylogenetically, our new collections cluster with Trichoderma aggressivum, T. epimyces, T. priscilae, T. purpureum, T. rufobrunneum, and T. tenue in the Harzianum lineage with high posterior probability (Figure 3), but with at least 2% (8/607 nucleotides, 5 gaps) difference in ITS, 4% (32/903 nucleotides, no gaps) difference in RPB2, and 5% (27/575 nucleotides, 5 gaps) difference in TEF. Morphologically, our new collections are most similar to T. epimyces in the size of stromata, perithecia, asci, and ascospores but have deeper color of stromata and ascospores, less pigment on media, and faster growth rate on media (Jaklitsch, 2009). The difference in our collections with T. epimyces is more than 2% (11/574 nucleotides, 3 gaps) in ITS, 4% (33/933 nucleotides, no gaps) in RPB2, and 6% (32/571 nucleotides, 6 gaps) in TEF.
The compatibility of Trichoderma hymenopellicola GUCC202010 to the nine fungicide candidates with regression equations is listed in Table 2, among which prochloraz-manganese chloride complex and propiconazole are the top two effective fungicides inhibiting the mycelium of Trichoderma hymenopellicola with EC50 less than 0.05 mg/L, while Osthole is found to be the best biological fungicide (Table 2).
The compatibility from high to low of strains from all three localities to the prochloraz-manganese chloride complex and propiconazole is SC, ZF, and JH, respectively (Table 3). The toxicity of these two chemicals to Hymenopellis raphanipes strains is also listed in Table 3. The results showed that the prochloraz-manganese chloride complex is a suitable chemical agent to control Trichoderma hymenopellicola, with a high inhibition ratio for the pathogen and low toxicity to the mushroom.
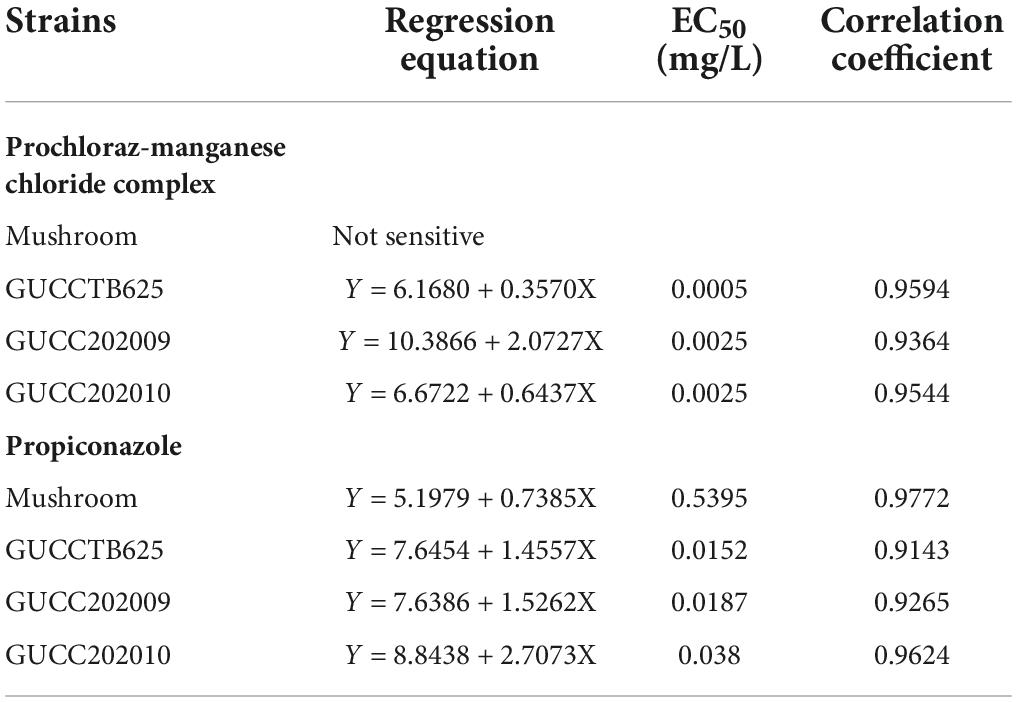
Table 3. Sensitivity of mushroom strain and Trichoderma strains to prochloraz-manganese chloride complex and propiconazole.
Hymenopellis raphanipes was first described by Berkeley (1850) in India and is widely distributed in China (Hao et al., 2016). It had been previously misidentified as H. furfuracea, H. radicata, Termitomyces fuliginosus, or T. badius, until Hao et al. (2016) clarified its taxonomic placement based on both morphology and phylogeny. In recent years, cultivation scales of Hymenopellis raphanipes in China have increased rapidly, accompanying a series of diseases resulting in great yield losses, such as cobweb disease (Liu et al., 2020; Qin et al., 2021). Green mold disease is also a very common disease in mushroom cultivation, typical of superficial, green, and villiform mycelium. It can be either a competitive disease that antagonizes the normal growth of mushrooms or an infectious disease that causes the fruiting bodies or mycelium of mushrooms to grow abnormally, or both (Bian, 2013). In our study, only inoculations of soil showed the typical symptoms of green mold disease, while infections were not observed in the fruiting body inoculating experiment. However, the hyphae of the pathogen can affect the growth of mushroom mycelium, indicating that the infection is susceptible at the early stages of mushroom cultivation. In addition, the color of Trichoderma hymenopellicola colonies on PDA plates can also be yellow or pink, besides green in the field, which may be the result of environmental changes.
Trichoderma hymenopellicola is the first report that Trichoderma spp. cause green mold disease on Hymenopellis raphanipes. The new species has gregarious, larger, reddish stromata, gregarious phialides, and a fast-growing rate on media, with a preference of growing on SNA. The current classification system of Trichoderma relies on the phylogeny, as most species were isolated from environments, such as soil, and lack sexual morphs (Chen and Zhuang, 2017; Zheng et al., 2021). However, the recognition of subclades is empirical and does not have a compatible standard, and the TEF sequence data are not all from the same region. In this study, we included 128 Trichoderma species of the Harzianum lineage and green-spored species according to previous studies to better interpret their phylogenetic relationship (Zhu and Zhuang, 2015; Chen and Zhuang, 2017; Bustamante et al., 2021; Zheng et al., 2021). The topology of phylogenetic trees based on single gene and concatenated genes is similar, except that Trichoderma aggressivum does not cluster in our new collection in the tree generated from the RPB2 dataset. In the future, more studies with full morphological illustration and description are needed for further clarifying the interspecific relationship and fast identification of Trichoderma species.
Fungicides, such as carbendazim, hexaconazole, metrafenone, prochloraz, tebuconazole, and thiophanate-methyl, have been used to prevent the occurrence and spreading of Trichoderma spp., whereas prochloraz is the most effective one with an EC50 less than 0.4 mg/L (Madhavi et al., 2011; Kosanoviæ et al., 2015; Lukoviæ et al., 2021). In our study, we tested its manganese chloride complex, which is much more effective than prochloraz itself with an EC50 less than 0.005 mg/L and is non-toxic to the mushroom. As both an infectious and competitive disease, species of Trichoderma grow much faster than mushrooms, and infection at the early stage will cause no fruiting in mushroom bags. As a result, control of the green-mold disease should be done as early as possible during cultivation.
In this study, we reported the first green mold disease on Hymenopellis raphanipes. Its causal agent was further confirmed as a new species, Trichoderma hymenopellicola sp. nov., based on morphology and phylogeny. The new pathogen can infect the mycelium of Hymenopellis raphanipes at an early stage and be competitive in the field. However, it can be controlled by applying prochloraz-manganese chloride complex, and this fungicide has no effect on the growth of Hymenopellis raphanipes. The results of this study provide essential information for future prevention and control of green mold diseases on Hymenopellis raphanipes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MZ330754-MZ330756, ON074580, ON074583, ON088661-ON088664, ON102005-ON102008, and ON102011.
X-YZ and F-HT: designing, analyzing, and writing. X-XY, K-QP, Y-TP, T-JT, and NW: collecting specimens and conducting experiments. All authors contributed to the article and approved the submitted version.
This study was funded by the following projects: the National Natural Science Foundation of China, NSFC No. 32260044; Guizhou Science and Technology Department, Science and Technology Support Project QKH (2021) general 199; Natural Science Special Research Fund of Guizhou University, Special Post 2021(25); and Department of Education of Guizhou Province, Natural Science Research Projects (2021)054.
The authors declare that the diseased mushroom bags used in the study were provided by Guizhou Kangqunyuan Biotechnology Co., Ltd.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.991987/full#supplementary-material
Abo-Elyousr, K. A., Abdel-Hafez, S. I., and Abdel-Rahim, I. R. (2014). Isolation of Trichoderma and evaluation of their antagonistic potential against Alternaria porri. J. Phytopathol. 162, 567–574. doi: 10.1111/jph.12228
Berkeley, M. J. (1850). Decades of fungi. Decades XXV. to XXX. Sikkim Himalaya fungi, collected by Dr. J.D. Hooker. Hookers J. Bot. Kew Gard. Miscellany 2, 42–51.
Bian, Y. (2013). Research progression infectious and competitive mycelial diseases of edible mushrooms. Acta Edulis Fungi 20, 1–7.
Bustamante, D. E., Calderon, M. S., Leiva, S., Mendoza, J. E., Arce, M., and Oliva, M. (2021). Three new species of Trichoderma in the Harzianum and Longibrachiatum lineages from Peruvian cacao crop soils based on an integrative approach. Mycologia 113, 1056–1072. doi: 10.1080/00275514.2021.1917243
Carbone, I., and Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Chaverri, P., and Samuels, G. J. (2004). Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): species with green ascospores. Stud. Mycol. 48, 1–116.
Chen, K., and Zhuang, W. Y. (2017). Discovery from a large-scaled survey of Trichoderma in soil of China. Sci. Rep. 7:9090. doi: 10.1038/s41598-017-07807-3
Crous, P. W., Gams, W., Stalpers, J. A., Robert, V., and Stegehuis, G. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Stud. Mycol. 50, 19–22.
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Degenkolb, T., Dhren, H. V., Nielsen, K. F., Samuels, G. J., and Brückner, H. (2008). Recent advances and future prospects in peptaibiotics, hydrophobin, and mycotoxin research, and their importance for chemotaxonomy of Trichoderma and Hypocrea. Chem. Biodivers. 5, 671–680. doi: 10.1002/cbdv.200890064
Ezzi, M. I., and Lynch, J. M. (2005). Biodegradation of cyanide by Trichoderma spp. and Fusarium spp. Enzyme Microb. Technol. 36, 849–854. doi: 10.1016/j.enzmictec.2004.03.030
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hao, Y. J., Zhao, Q., Wang, S. X., and Yang, Z. L. (2016). What is the radicate Oudemansiella cultivated in China? Phytotaxa 286, 1–12. doi: 10.11646/phytotaxa.286.1.1
Harman, G., Lorito, M., and Lynch, J. (2004). Uses of Trichoderma spp. to alleviate or remediate soil and water pollution. Adv. Appl. Microbiol. 56, 313–330. doi: 10.1016/S0065-2164(04)56010-0
Hasan, M. M., Rahman, S., Kim, G. H., Abdallah, E., and Oh, D. H. (2012). Antagonistic potentiality of Trichoderma harzianum towards seed-borne fungal pathogens of winter wheat cv. Protiva in vitro and in vivo. J. Microbiol. Biotechnol. 22, 585–591. doi: 10.4014/jmb.1107.07063
Jaklitsch, W. M. (2009). European species of Hypocrea part I. The green-spored species. Stud. Mycol. 63, 1–91. doi: 10.3114/sim.2009.63.01
Jaklitsch, W. M., Komon, M., Kubicek, C. P., and Druzhinina, I. S. (2005). Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97, 1365–1378. doi: 10.1080/15572536.2006.11832743
Jaklitsch, W. M., and Voglmayr, H. (2015). Biodiversity of Trichoderma (Hypocreaceae) in Southern Europe and Macaronesia. Stud. Mycol. 80, 1–87. doi: 10.1016/j.simyco.2014.11.001
Jun, H., Kieselbach, T., and Jönsson, L. J. (2011). Enzyme production by filamentous fungi: analysis of the secretome of Trichoderma reesei grown on unconventional carbon source. Microb. Cell Fact. 10, 1–10. doi: 10.1186/1475-2859-10-68
Katayama, A., and Matsumura, F. (1993). Degradation of organochlorine pesticides, particularly endosulfan, by Trichoderma harzianum. Environ. Toxicol. Chem. 12, 1059–1065. doi: 10.1002/etc.5620120612
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, C. S., Park, M. S., Kim, S. C., Maekawa, N., and Yu, S. H. (2012). Identification of Trichoderma, a competitor of shiitake mushroom (Lentinula edodes), and competition between Lentinula edodes and Trichoderma species in Korea. Plant Pathol. J. 28, 137–148. doi: 10.5423/PPJ.2012.28.2.137
Kosanoviæ, D., Potoènik, I., Vukojeviæ, J., Stajiæ, M., Rekanoviæ, E., Stepanoviæ, M., et al. (2015). Fungicide sensitivity of Trichoderma spp. from Agaricus bisporus farms in Serbia. J. Environ. Sci. Health B 50, 607–613. doi: 10.1080/03601234.2015.1028849
Kredics, L., Antal, Z., Dóczi, I., Manczinger, L., Kevei, F., and Nagy, E. (2003). Clinical importance of the genus Trichoderma. Acta Microbiol. Immunol. Hungar. 50, 105–117. doi: 10.1556/AMicr.50.2003.2-3.1
Kuhls, K., Lieckfeldt, E., Börner, T., and Guého, E. (2010). Molecular reidentification of human pathogenic Trichoderma isolates as Trichoderma longibrachiatum and Trichoderma citrinoviride. Med. Mycol. 37, 25–33. doi: 10.1111/j.1365-280X.1999.00197.x
Liu, G. L., Li, H., Gong, N., Deng, H. C., and Zhang, M. (2020). Identification of a pathogenic fungus causing cobweb disease in Oudemansiella raphanipes by morphological characteristics and phylogenetic analysis. Acta Edulis Fungi 27:30.
Liu, M., Liu, J., and Wang, W. M. (2012). Isolation and functional analysis of Thmfs1, the first major facilitator superfamily transporter from the biocontrol fungus Trichoderma harzianum. Biotechnol. Lett. 34, 1857–1862. doi: 10.1007/s10529-012-0972-x
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Lukoviæ, J., Milijaševiæ-Marèiæ, S., Hatvani, L., Kredics, L., Szûcs, A., Vágvölgyi, C., et al. (2021). Sensitivity of Trichoderma strains from edible mushrooms to the fungicides prochloraz and metrafenone. J. Environ. Sci. Health B 56, 54–63. doi: 10.1080/03601234.2020.1838821
Madhavi, G. B., Bhattiprolu, S. L., and Reddy, V. B. (2011). Compatibility of biocontrol agent Trichoderma viride with various pesticides. J. Hortic. Sci. 6, 71–73.
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE), (Piscataway, NJ: IEEE), 1–8. doi: 10.1109/GCE.2010.5676129
Poveda, J., Hermosa, R., Monte, E., and Nicolás, C. (2019). Trichoderma harzianum favours the access of arbuscular mycorrhizal fungi to non-host Brassicaceae roots and increases plant productivity. Sci. Rep. 9, 1–11. doi: 10.1038/s41598-019-48269-z
Qin, W., Li, J., Zeng, Z., Wang, S., and Liu, Y. (2021). First report of cobweb disease in Oudemansiella raphanipes caused by Cladobotryum varium in Beijing, China. Plant Dis. [Epub ahead of print]. doi: 10.1094/PDIS-02-21-0265-PDN
Rambaut, A. (2016). FigTree version 1.4.3 [Online]. Available: https://github.com/rambaut/figtree/releases/tag/v1.4.3 (accessed July 2022).
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Samuels, G. J., Dodd, S. L., Gams, W., Castlebury, L. A., and Petrini, O. (2002). Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 94, 146–170. doi: 10.1080/15572536.2003.11833257
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Zheng, H., Qiao, M., Lv, Y. F., Du, X., Zhang, K. Q., and Yu, Z. F. (2021). New species of trichoderma isolated as endophytes and saprobes from Southwest China. J. Fungi 7:467. doi: 10.3390/jof7060467
Keywords: fungicide, Hypocreaceae, novel taxa, pathogen, phylogeny, taxonomy
Citation: Zeng X-Y, Yuan X-X, Peng K-Q, Pan Y-T, Tan T-J, Wu N and Tian F-H (2022) Taxonomy and control of Trichoderma hymenopellicola sp. nov. responsible for the first green mold disease on Hymenopellis raphanipes. Front. Microbiol. 13:991987. doi: 10.3389/fmicb.2022.991987
Received: 12 July 2022; Accepted: 06 September 2022;
Published: 29 September 2022.
Edited by:
Amin Uddin Mridha, University of Chittagong, BangladeshReviewed by:
Malkhan Singh Gurjar, Indian Agricultural Research Institute (ICAR), IndiaCopyright © 2022 Zeng, Yuan, Peng, Pan, Tan, Wu and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng-Hua Tian, Zmh0aWFuQGd6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.