- 1Helmholtz International Lab for Anti-Infectives, Shandong University-Helmholtz Institute of Biotechnology, State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China
- 2Department of Microbiology, University of Chittagong, Chittagong, Bangladesh
- 3Industrial Microbiology Research Division, BCSIR Chattogram Laboratories, Bangladesh Council of Scientific and Industrial Research (BCSIR), Chattogram, Bangladesh
- 4Chinese Academy of Sciences (CAS) Key Laboratory of Quantitative Engineering Biology, Shenzhen Institute of Synthetic Biology, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
Natural products derived from microorganisms serve as a vital resource of valuable pharmaceuticals and therapeutic agents. Streptomyces is the most ubiquitous bacterial genus in the environments with prolific capability to produce diverse and valuable natural products with significant biological activities in medicine, environments, food industries, and agronomy sectors. However, many natural products remain unexplored among Streptomyces. It is exigent to develop novel antibiotics, agrochemicals, anticancer medicines, etc., due to the fast growth in resistance to antibiotics, cancer chemotherapeutics, and pesticides. This review article focused the natural products secreted by Streptomyces and their function and importance in curing diseases and agriculture. Moreover, it discussed genomic-driven drug discovery strategies and also gave a future perspective for drug development from the Streptomyces.
Introduction
Diversity, morphology, taxonomy, and genetics of Streptomyces
The genus Streptomyces includes spore-forming, filamentous, and Gram-positive bacteria in Actinobacteria phylum (Khadayat et al., 2020; Lee et al., 2020), residing in a variety of environments, such as extreme environments and underexplored habitats, terrestrial and marine regions, symbionts, endophyte, mangroves etc. Over 850 species of Streptomyces have been studied (Kemung et al., 2018; Aryal et al., 2020). A gram of soil is estimated to contain 109 colony-forming units (CFU) of bacteria, with 107 CFU of Actinobacteria (Harir et al., 2018). Streptomyces species can form extensive branching substrate and aerial mycelia (Chater, 2016) and possess complex morphological characteristics with a complex multicellular development starting from the germination of hyphae from spores and ending with the formation of septa with a chain of uninucleated spores.
Streptomyces have an exceptionally high G + C% (>70%) and linear, moderately large genomes (8-10 Mb), regarded as special features among bacteria (Hopwood, 2019). Streptomyces have multiple biosynthetic gene clusters (BGCs) on each genome, which are the source of numerous bioactive compounds with medical or agricultural use (Ward and Allenby, 2018; Nicault et al., 2021).
Necessities of new natural products
Antimicrobial resistance (AMR), one of the most severe public health challenges nowadays, is caused by resistant pathogenic bacteria, as well as fungi, viruses, and parasites (Amann et al., 2019). Every year, it causes deaths of 17 million people worldwide, along with a huge number of children and the elderly (Procópio et al., 2012). Multidrug resistant bacteria, some known as “superbugs,” make the conditions even worse. In the last few decades, the emergence of multidrug resistant bacteria increased devastatingly. The most common multidrug resistant bacteria are Escherichia coli resistant to cephalosporin and fluoroquinolones, Klebsiella pneumoniae to cephalosporin and carbapenems, Staphylococcus aureus to methicillin and vancomycin, Neisseria gonorrhoeae to cephalosporin, non-typhoidal Salmonella to fluoroquinolones and extended spectrum beta lactamase (ESBL), Streptococcus pneumoniae to penicillin, Shigella species to fluoroquinolones, and Mycobacterium tuberculosis to rifampicin, isoniazid, fluoroquinolone, and carbapenemase-producing enterobacteriaceae. The core reason for the recent increase in the antibiotic resistance phenomenon is indiscriminate and irrational use of antibiotics, poor hygiene and sanitation, frequent and self-medication, and poor control system and surveillance. The emergence and spreading of resistance mechanisms among bacteria are faster than the discovery and development of new antibiotics.
In addition, some non-negligible life-threatening diseases that enforce severe death worldwide are awaiting new drugs, including cancers as major causes of death in most of countries (Rotow and Bivona, 2017; Perin et al., 2022) and Alzheimer’s disease, which still has no effective drugs discovered and has caused dementia in 13.2 million patients in China by 2019 and will reach 150 million globally by 2050. In recent 30–40 years, some new infective diseases have fluently emerged to humans, including HIV, SARS, and Zika virus. In the last two and half years, SARS-CoV-2 has been causing serious problems for the human health as a pandemic worldwide. Specific and effective drugs for these emerging diseases have been yet underway (National Institutes of Health, 2007). The agrochemical field is also awaiting new generation of drugs, although limited types of agrochemicals, including avemectin or spinosad and glyphosate-derived compounds, are used widely.
In addition to the possibility of combining failing drugs with other compounds that appear to restore the desired effects (Brown, 2015), screening novel natural products, especially from Streptomyces, offers a viable alternative and an opportunity to improve the efficacy of treatments for these diseases, as an utmost significant solution to lessen this dreadful outcome.
Natural products from Streptomyces
Streptomyces species have been considered as a repository of a diverse range of natural products (Azerang and Sardari, 2017; Genilloud, 2017; Lapaz et al., 2019; Xia et al., 2020) because of their powerful and complex secondary metabolism (Chang et al., 2021). Streptomyces produce around 100,000 antibiotic compounds, which account for 70–80% of all natural bioactive products with pharmacological or agrochemical applications (Bubici, 2018; Harir et al., 2018; Abdel-Razek et al., 2020).
Streptomyces produce a variety of natural products with high structural diversity, including macrolides, tetracyclines, aminoglycosides, glycopeptides, ansamycins, and terpenes. For example, Streptomyces hygroscopicus secretes around 180 metabolites with a wide range of bioactivities.
Streptomyces-derived bioactive natural products have the capability to function as antimicrobial, antiviral, cytotoxic, antitumor, antihypertensive, immunosuppressive, insecticide, antioxidative, plant growth-promoting, and herbicidal agents (Singh and Khajuria, 2018; Nguyen et al., 2020; Salwan and Sharma, 2020; Pham et al., 2021; Sharma et al., 2021). So, the discovered metabolites can be classified into four classes: (i) Compounds with regulatory activities include morphogenic agents, siderophores, and growth factors; (ii) anti-protozoans, antibacterials, antifungals, and antivirals as antagonistic agents; (iii) insecticides, pesticides, and herbicides as agrobiologicals; and (iv) neurological agents, immunomodulators, antitumor agents, and enzyme inhibitors as pharmacological drugs.
Streptomyces produce these compounds as secondary metabolites during the stationary phase, which are not necessarily required for their development or multiplication but can provide a competitive advantage to the organisms. Some of these metabolites aid reproductive bacterial cells by confiscating metals like iron (siderophores), shielding them from UV light (via pigmentation), limiting competition (antibiotics), and allowing them to communicate with other species, mediate important host–microbe and microbe–microbe interactions in mammalian or animal guts, and regulate biosynthetic pathways (Bolourian and Mojtahedi, 2018b; Wang et al., 2019). Furthermore, they play a vital role in soil biodegradation and in humus formation and produce numerous volatile substances, such as geosmin, which is responsible for the feature “wet earth odor.” Here, we discuss the essential products of Streptomyces rendering antibacterial, antifungal, antiviral, anti-immune, and anticancer properties and several important agrochemicals.
Streptomyces-derived bioactive products with antagonistic or pharmacological properties
Antibacterial compounds with high diversity
Antibiotics are the most valuable products of Streptomyces. Actinomycetes produce two-thirds of all microbial antibiotics, with roughly 80% derived from the Streptomyces genus, which are well known for their therapeutic and agricultural uses. The most widely used antibiotics are derived from Streptomyces, from streptomycin first developed from soil isolates (Waksman, 1953) to well-known daptomycin (Figure 1) approved in 2003 by the FDA (Waters and Tadi, 2021). Terrestrial, marine, and estuarine environments are niches for the storage of antimicrobial drugs for Streptomyces. Almost half of the Streptomyces species isolated have been identified as antibiotic producers.
Some popular antibiotics, for example, cephamycin, chloramphenicol, tetracycline, kanamycin, spectinomycin, monensin, and mitomycin C (Figure 1) have been originated from the species of S. clavuligerus, S. venezuelae, S. aureofaciens, S. kanamyceticus, S. spectablis, S. cinnamonensis, and S. lavendulae, respectively (Akintunde, 2014). S. griseus, S. avermitilis, and S. cattleya produce streptomycin, polypeptide avermectin, and fluorometabolites (Figure 1), respectively (Ikeda et al., 1999; Nicault et al., 2021). Other than that, there are also some other species of Streptomyces that can produce a good number of antibiotics. A new strain S. parvulus strain MARS-17, found in the soil of Rajshahi, Bangladesh, secretes actinomycin D (Figure 1; Rahman et al., 2011). A study showed that n-butanol extracts of S. flavogriseus strain ACTK2 exhibit a broad spectrum of antimicrobial activity (Dezfully and Ramanayaka, 2015). S. cheonanensis VUK-A strain secretes two bioactive compounds 2-methyl butyl propyl phthalate and diethyl phthalate (Figure 1) possessing a broad range of antimicrobial activity (Dezfully and Ramanayaka, 2015).
Nowadays, several Streptomyces species are accountable for both medicinal and commercial antibiotics, and they perform magnificently in these contexts. Many antibiotics have been accumulated in one species of Streptomyces (e.g., S. griseus and S. hygroscopicus), as well as the same antibiotics are produced from different Streptomyces species (e.g., streptothricin and actinomycin).
Antibiotics are usually grouped together based on their action mechanism, chemical structure, or spectrum of activity. Antibiotics derived from Streptomyces are categorized according to their principal structural classes. Major types of antibiotics produced by Streptomyces are aminoglycosides (such as gentamicin, streptomycin, tobramycin, neomycin, and kanamycin), anthracyclines (doxorubicin), glycopeptides, β-lactams (monobactams, cephalosporin, and carbapenems), macrolides (clarithromycin, erythromycin, and azithromycin), ansamycins (rifamycin), nucleosides, peptides, polyenes, polyesters, and tetracyclines.
Antibacterial compounds of Streptomyces in extreme environments
Most of bioactive compounds were isolated from conventional terrestrial environments, mentioned as before. Streptomyces isolated from extreme environments also have the capability to generate some antibacterial products, including alkaloids, angucyclines, macrolides, and peptides (Sivalingam et al., 2019). Some strains of natural bioactive antibacterial metabolite secretors are Streptomyces sp. RAUACT-1, S. arenicola, S. griseus, S. nodosus, S. lincolnensis, S. pacifica, and S. pristinaespiralis secreting 1,4-dihydroxy-2-(3-hydroxybutyl)-9,10-anthraquinone, anthracene 9,10 anthracene, arenimycin, frigocyclinone, lajollamycin, lincomycin, mitomycin C, pacificanones A and B, pristinamycin, and rapamycin, respectively. Streptomyces secrete bisanthraquinone, carbomycin, glaciapyrroles, and tirandamycins that also display antibacterial activity (Harir et al., 2018). Recently, thermophilic Streptomyces werraensis MI S.24 3, isolated from a very harsh environment in Egypt, has both antibacterial and cytotoxic properties (Mohamed et al., 2021).
The production of one-of-a-kind polyketides and phenolic compounds by Streptomyces lanatus AR2 with observable bioactivities made it possible for the organism to adapt to harsh environment like salty wetland, yielding useful natural products with biotechnological, pharmaceutical, and medical applications (Riahi et al., 2019).
Marine Streptomyces are considered the largest and an outstanding repository of myriad natural products and antibiotics. Marine Streptomyces are found in different aquaculture, such as in seaweeds, mollusks, fishes, sponges, mangroves, seawater, and sediments (Dharmaraj, 2010).
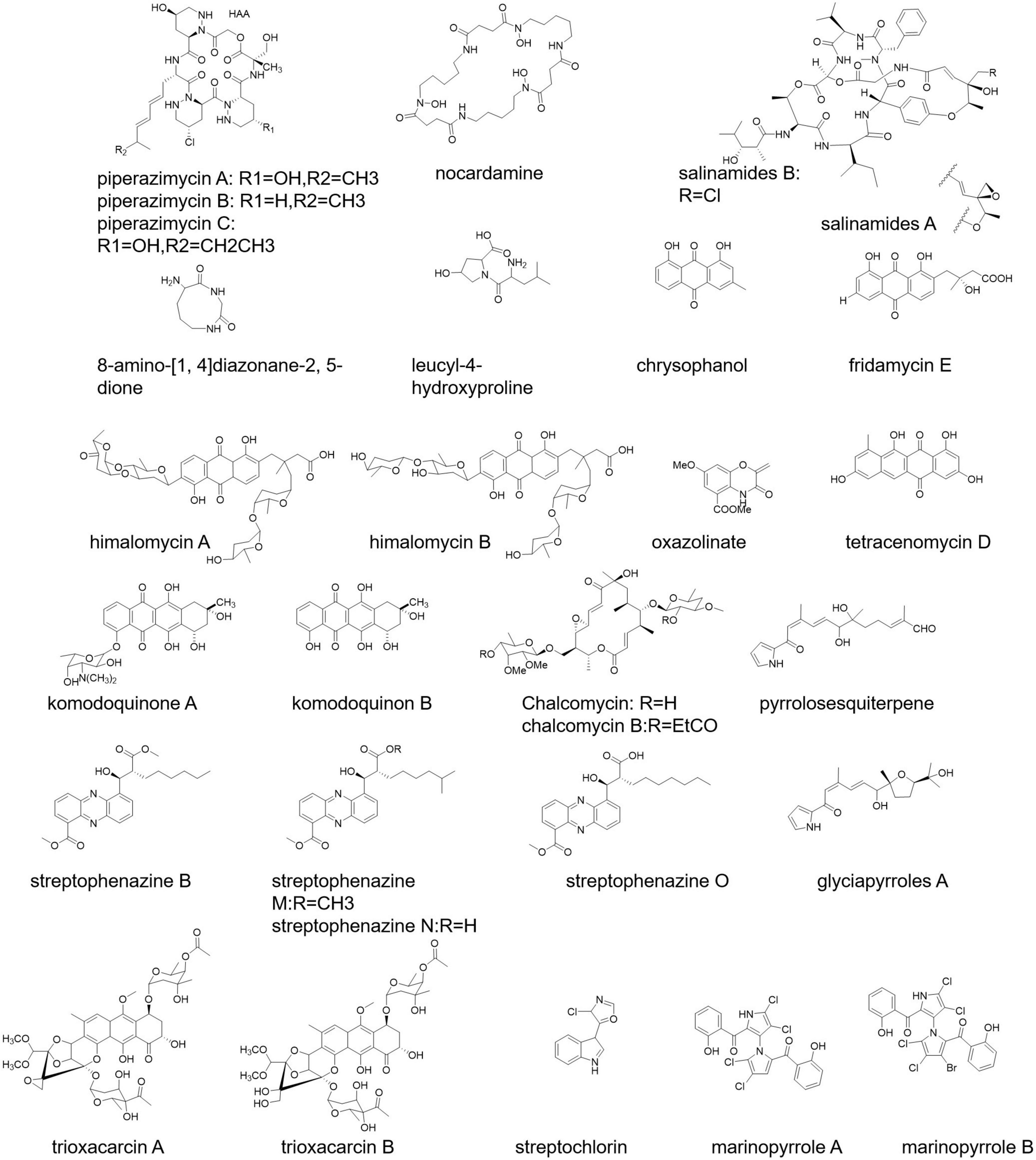
Piperazimycin A-C, allophycocyanin, nocardamine, salinamides A and B, 8-amino-[1,4] diazonane-2, 5-dione, and leucyl-4-hydroxyproline are some of the marine-originated polypeptide antibiotics (Figure 2) isolated from Streptomyces strain KMM7210, Streptomyces strain M097, Streptomyces sp. (from an unidentified marine sponge), Streptomyces sp. CNB-091, and S. acrimycini, respectively (Hernández et al., 2004; Hou et al., 2006; Sobolevskaya et al., 2007; Dharmaraj, 2010).
Streptomyces sp. 6921 from marine sediments of Mauritius secretes C-glycosides himalomycins, anthraquinones, fridamycin E, and chromophore (Figure 2) possessing strong antibacterial activity (Maskey et al., 2003). S. corchorusii AUBN secretes tetracenomycin D (Figure 2) having cytotoxic activity (Adinarayana et al., 2006). Komodoquinone A, B (Figure 2), anthracycline antibiotics, are produced from the fermentation broth of Streptomyces sp. KS3 showing neurogenic activity (Itoh T. et al., 2003).
Chalcomycin and chalcomycin B (Figure 2) are macrolides isolated from Streptomyces sp. M491 and Streptomyces isolate B7064 (Wu et al., 2006). Streptomyces sp. NPS008187 produced pyrrolosesquiterpenes and glyciapyrroles A (Figure 2) possessing strong antibacterial activity (Macherla et al., 2005). The product streptophenazines (Figure 2) from marine Streptomyces sp. 182SMLY works against a methicillin-resistant S. aureus (Liang et al., 2017). Another example is marinopyrroles (Figure 2; Al-Ansari et al., 2019).
A new tetracycline analog SBR-22, a polyketide, isolated from S. psammoticus exhibits antibacterial activity against methicillin-resistant S. aureus (MRSA) (Sujatha et al., 2005). Trioxacarcin A, B, and C (Figure 2) show anti-plasmodial activity, and these products have been secreted by S. ochraceus and S. bottropensis (Dharmaraj, 2010). Streptochlorin (Figure 2) from the Streptomyces strain 04DH110 possesses antiproliferative activity (Shin et al., 2007).
A strain of Streptomyces olivaceus isolated from a mangrove in Macau, China, was subjected to whole-genome sequencing and MS/MS analysis to determine the strain potential to produce secondary metabolites. According to the findings, the genome of S. olivaceus had a total of 105 gene clusters, and genome mining was able to predict the presence of 53 known secondary metabolites. There were 28 secondary metabolites that were classified as antibiotics and derived from S. olivaceus; however, none of them had been discovered before (Hu et al., 2021).
Antibacterial compounds against biofilm formation in “superbugs”
Biofilm formation is considered to be one of critical factors to determine the toxicity of some pathogenic bacteria, including well-known MRSA. Streptomyces-derived numerous antibiotics or their derivates were used for antibiofilm activity in MRSA. A total of three antibiotics, namely, rifampin, daptomycin, and tigecycline were exploited for the treatment of device-related Staphylococcal biofilm infections such as antimicrobial lock therapy (Hogan et al., 2016). Daptomycin, tigecycline, and rifampin showed better antibiofilm activity against mature biofilms, whereas vancomycin failed to eradicate older biofilms in model experiments (Hogan et al., 2016). Minocycline is also found to be effective compared with vancomycin in inhibiting biofilm formation and eliminating mature biofilms in ica-positive and ica locus-negative MRSA (Chopra et al., 2015).
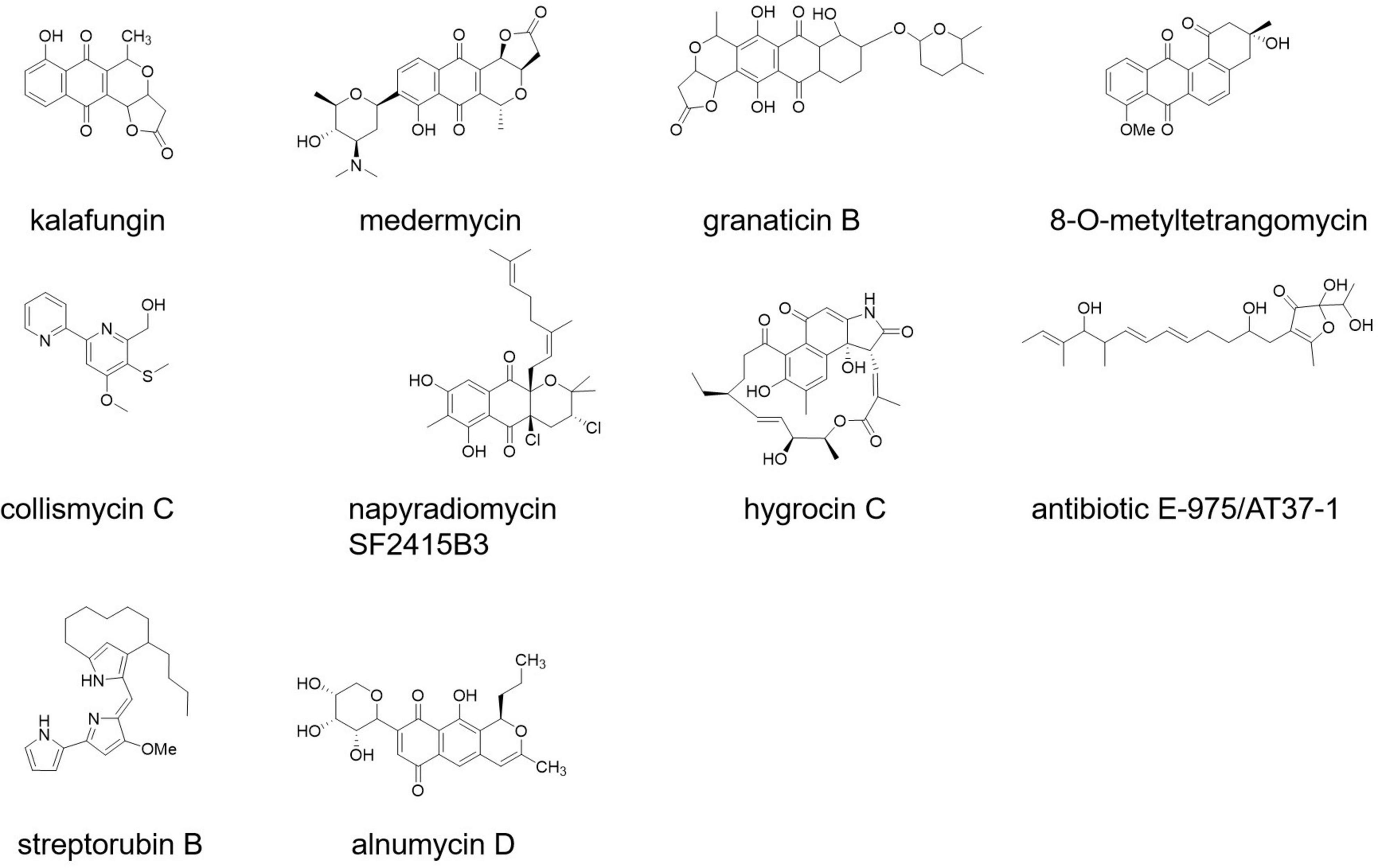
Some newly discovered bioactive compounds from Streptomyces species showed promising antibiofilm activities against S. aureus and MRSA (at the micromolar level), including antibiotic 5812-A/C (Vasilchenko et al., 2020), streptorubin B (Suzuki et al., 2015), alnumycin D, granaticin B, kalafungin, medermycin (Oja et al., 2015), collismycin C, napyradiomycin SF2415B3 (Bauermeister et al., 2019), hygrocin C (Wang et al., 2018), 8-O-metyltetrangomycin (Mary et al., 2021), panglimycin D (Kim et al., 2020), and AT37-1 (Figure 3; Driche et al., 2017).
Antifungal products from Streptomyces
The resurgence of fungal infection is a public health crisis. Secondary metabolites produced by Actinobacteria, particularly members of the Streptomyces genus, are known to have a wide range of biological activities, including antifungal compounds (Siupka et al., 2021).
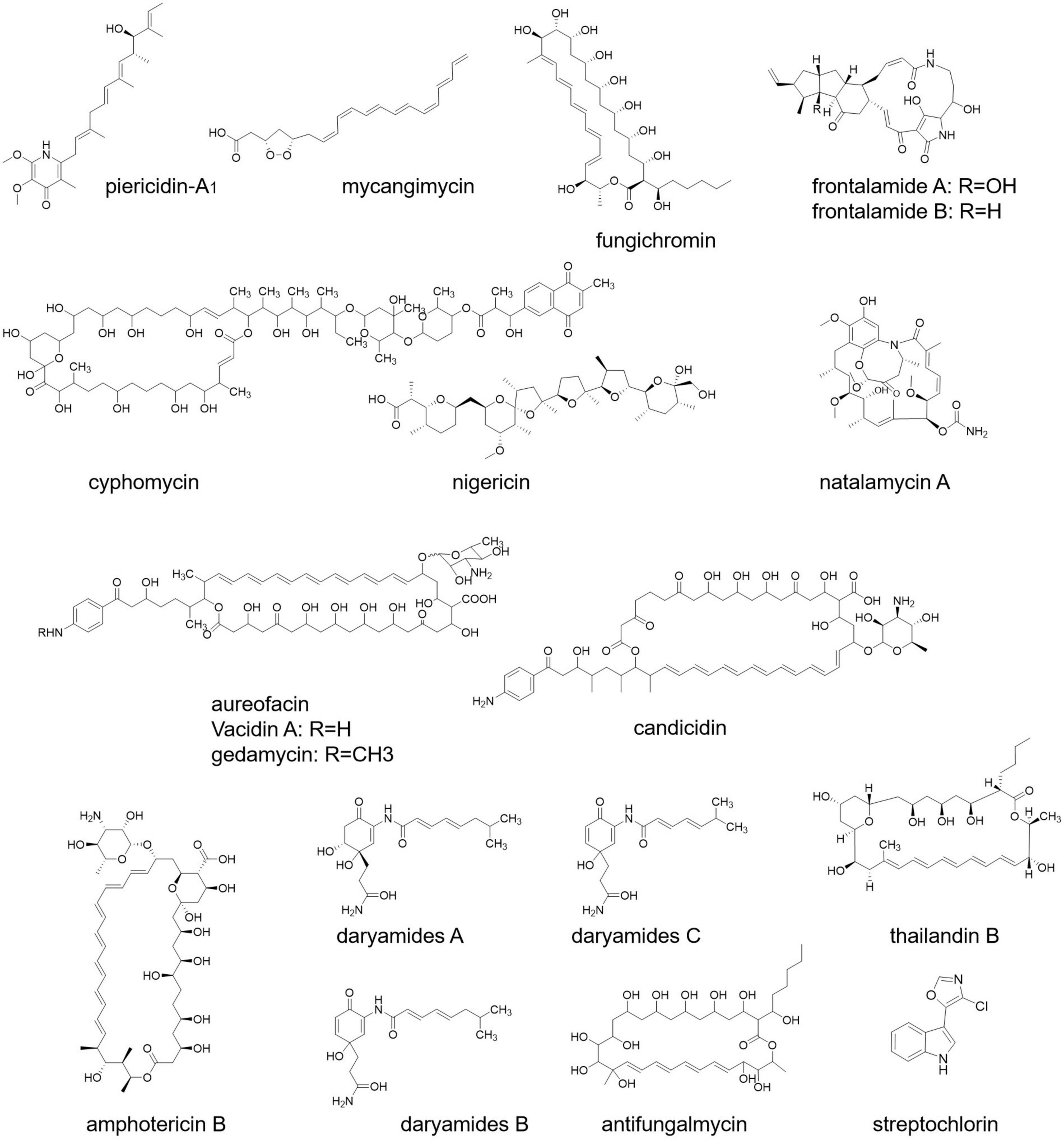
According to a study, Streptomyces sp. ICBG292, isolated from the exoskeleton of Cyphomyrmex workers, produces piericidin-A1 and nigericin (Figure 4), which inhibit the growth of fungal pathogen Escovopsis and also prevent the infection of Leishmania donovani, a human pathogen (Ortega et al., 2019). In another research, it has been found that pathogenic fungus Rhizoctonia solani AG-4 deteriorating the condition of the leaf of cabbage can be treated by S. padanus strain PMS-702. The responsible compound for this fungal inhibition is the polyene macrolide fungichromin (Figure 4), which possesses strong antifungal activity (Shih et al., 2003).
A new molecule named cyphomycin (Figure 4) possesses antifungal activities against multidrug-resistant fungal pathogens (Chevrette et al., 2019). Streptomyces symbiotically with southern pine beetle (Dendroctonus frontalis) secretes the secondary metabolites mycangimycin, frontalamide A, and frontalamide B (Figure 4). Frontalamide possesses antifungal activity and mycangimycin against the growth of the antagonistic fungus Ophiostoma minus (Chevrette et al., 2019). Streptochlorin and natalamycin derived from Streptomyces also exert antifungal activity (Chevrette et al., 2019). Other than that, S. nodosus, S. aureofaciens, S. griseus, S. noursei, S. aureus, and S. diastachromogenes exert antifungal compounds named amphotericin, aureofacin, candicidin (Figure 4), nystatin, oligomycin, and actinomycin D, respectively (Akintunde, 2014).
Daryamide (Figure 4) is another antifungal compound of Streptomyces (Fu et al., 2017). A strain of Streptomyces identified from rhizospheric soils secretes four antifungal compounds, namely, actinomycin, fungichromin, thailandin B, and antifungalmycin (Figure 4; Peng et al., 2020). In a study, it has been shown that S. halstedii K122 exhibits antifungal compounds bafilomycin B1 and C1, which inhibit the growth of fungi Penicillium roqueforti, Aspergillus fumigatus, Paecilomyces variotii, and Mucor hiemalis (Frändberg et al., 2000). In another study, antifungal protein 1 (Afp1) from S. tendae is used to limit fungal growth (Freitas et al., 2005).
Antiviral compounds from Streptomyces
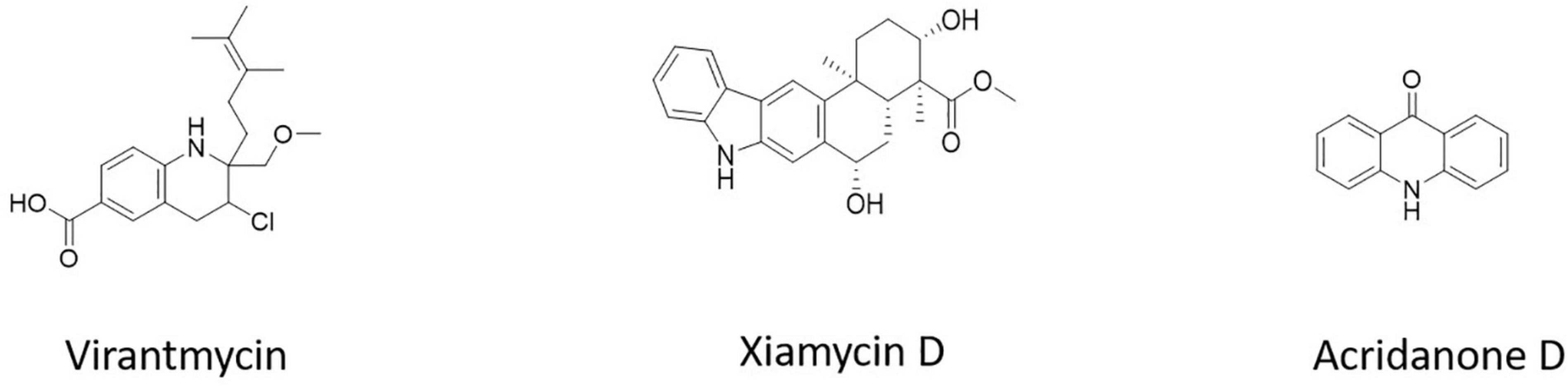
Virantmycin B (1) (Figure 5), isolated from Streptomyces sp. AM-2504, showed activity against the dengue virus (Hirota-Takahata et al., 2017). The bioactive chemical xiamycin D (Figure 5), isolated from Streptomyces sp. (#HK18) culture, had the strongest inhibitory effect on PEDV proliferation (Hayakawa et al., 2013). 9(10H)-acridanone (Figure 5), a secondary metabolite isolated from S. fradiae strain VITMK2, could inhibit the white spot syndrome virus (WSSV) (Hayakawa et al., 2013).
Dichloromethane extracts (DCME) of Streptomyces sp. demonstrated broad and potent antiviral activity against a variety of influenza viruses, including H1N1 and H3N2 subtypes, and influenza B virus. DCME may interact with the HA2 subunit of influenza A virus (IAV) hemagglutinin (HA) via stopping the fusion process between the viral and host cell membranes, preventing the virus from entering host cells, according to a thorough mode-of-action investigation (Zhao et al., 2019). Streptomyces sp. SMU03 produced an antiviral butanolide [(4S)-4-hydroxy-10-methyl-11-oxo-dodec-2-en-1,4-olide] that has broad and robust activity against influenza viruses, including H1N1 and H3N2 subtypes, and influenza B virus (Li et al., 2018).
Immunostimulant, anti-immunosuppressive, and vasoactive substances from Streptomyces
There are thousands of natural substances that influence immune cell activity or antibody secretion to manage infection and maintain immunological homeostasis and thus regulate the immune system. Immunostimulants are a diverse collection of chemicals that act on the immune system in a non-specific way by activating it, either by upregulating it or by favoring the activity of one of its components (Ortuño-Sahagún et al., 2017). Secondary metabolites from Streptomyces bacteria have a wide range of immunomodulatory activity made a significant contribution to immunomodulatory therapies (Mahmoudi et al., 2016).
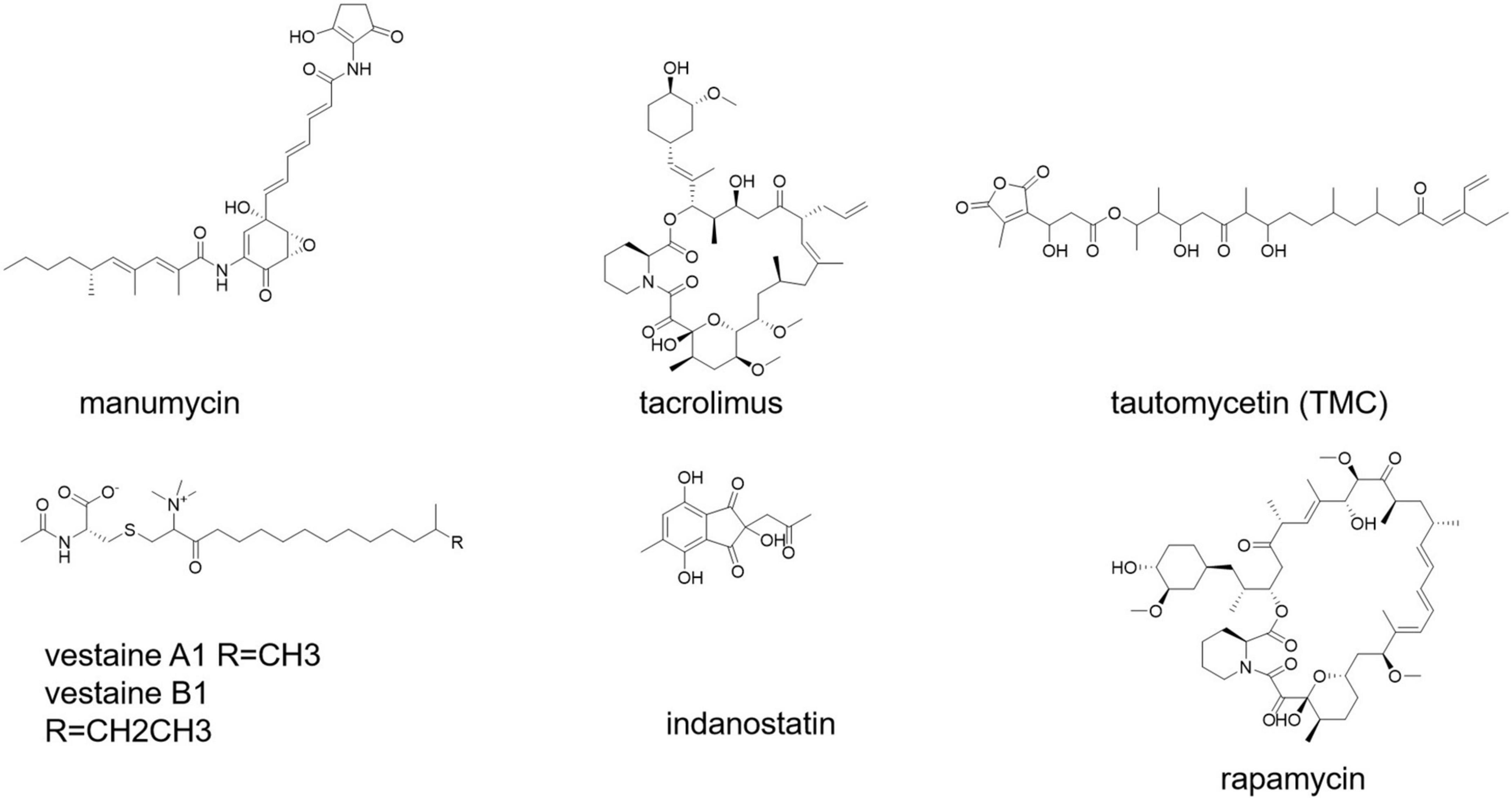
Secondary metabolites from S. calvus have been shown to be effective immunomodulators in human peripheral blood mononuclear cells (PBMCs) (Mahmoudi et al., 2016). Short polyketides generated by several streptomycetes and related bacteria that have strong anti-inflammatory and pro-apoptotic properties are known as manumycin-type compounds (Figure 6; Hrdý et al., 2020).
Immunosuppressants are used to prevent transplanted tissues and organs from being rejected, as well as to treat autoimmune illnesses (Fireman et al., 2004). Antiproliferative/immunosuppressive chemicals produced by Streptomyces bacteria (e.g., rapamycin and tacrolimus) (Figure 6) play an important role in the resistance of certain mucosal cancers (e.g., colon cancer) and the hygiene hypothesis (Bolourian and Mojtahedi, 2018a). Tacrolimus was discovered as a potent immunosuppressant in the strain Streptomyces tsukubaensis 9993T, which was first used in Japan in 1993 to reduce graft rejection after liver transplantation (Barreiro et al., 2012; Muramatsu and Nagai, 2013). Various taxonomically different Streptomyces strains, notably S. tacrolimus ATCC 55098T, have been identified as tacrolimus producers (Kim and Park, 2008; Martínez-Castro et al., 2011).
Streptomyces sp. CK4412 synthesized an activated T-cell-specific immunosuppressive chemical named tautomycetin (TMC) (Figure 6; Choi et al., 2007). TMC has a mechanistically distinct immunosuppressive action that was 100 times greater than that of cyclosporine A (Choi et al., 2017). TMC significantly suppressed breast cancer cell propagation, migration, and invasion, suggesting that it could be used as a possible breast cancer therapeutic alternative (Niu et al., 2013). Rapamycin and tacrolimus are antiproliferative/immunosuppressive drugs found in Streptomyces. S. hygroscopicus and S. tubercidicus probiotics are effective against inflammatory diseases (Bolourian and Mojtahedi, 2018a).
A vasoactive substance is an endogenous agent or pharmaceutical medicine that, by its vasoactivity, or vascular activity, increases or decreases blood pressure and/or heart rate (effect on blood vessels). Streptomyces sp. SANK 63697 produced two vasoactive chemicals, namely, vestaine A1 and vestaine B1 (Figure 6; Hirota-Takahata et al., 2017). Streptomyces sp. RAI20 yielded indanostatin (Figure 6), a neuroprotective compound against glutamate toxicity (Hayakawa et al., 2013).
Anticancer compounds from Streptomyces
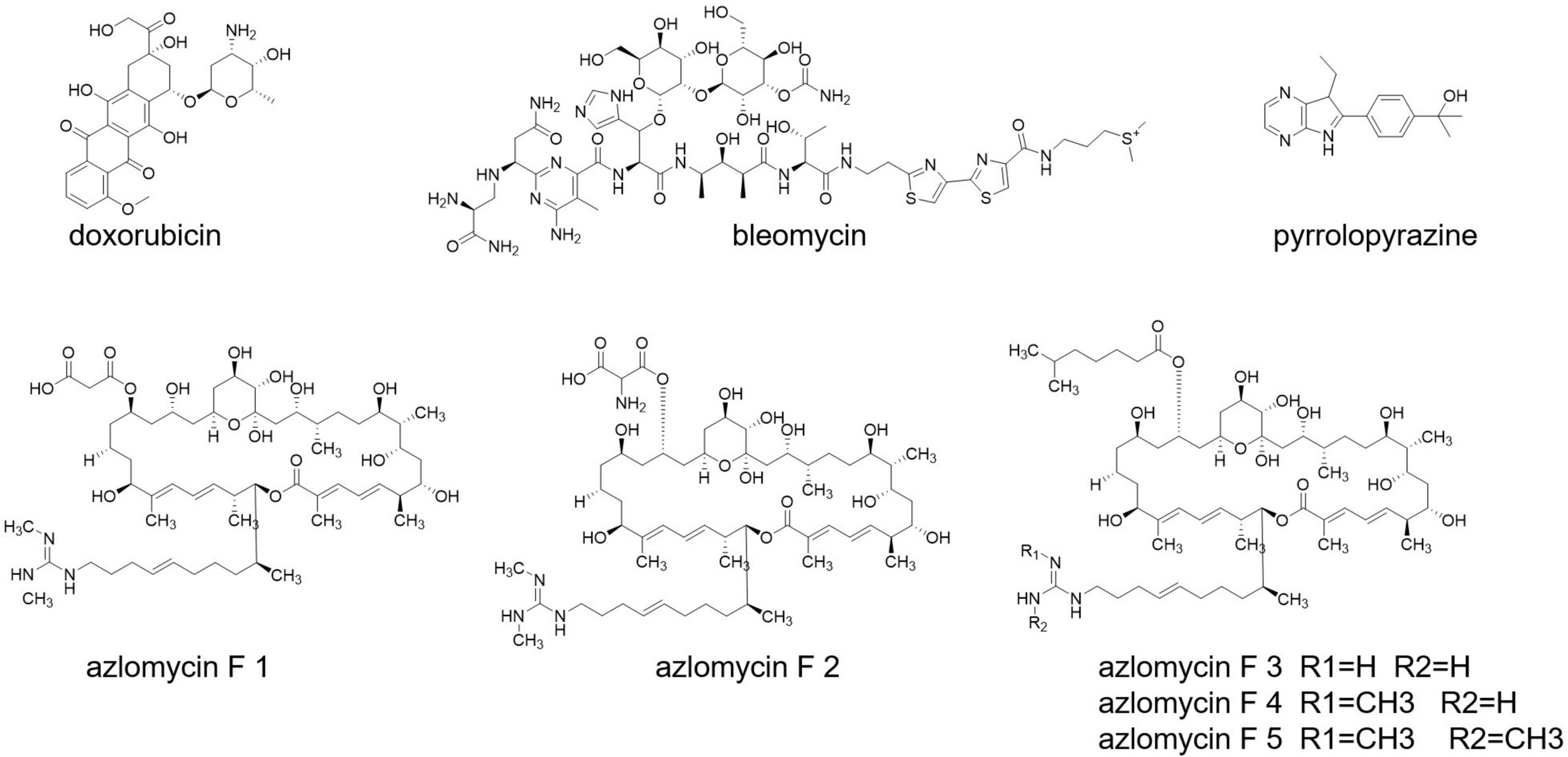
Cancer is still a dreadful disease and causes millions of death every year. Streptomyces also produce some well-known anticancer drugs, such as doxorubicin and bleomycin (Grimm et al., 1994; Du et al., 2000; Figure 7). A research carried out with Malaysian mangrove forest revealed the presence of Malaysia phenolic and pyrrolopyrazine (Figure 7) compounds in Streptomyces having anticancer activities (Azman et al., 2017). Streptomyces sp. 211726, isolated from soil, also exerts anticancer compounds, with seven azalomycin F analogs (Figure 7) and macrocyclic lactones (Tan et al., 2015).
Anticancer activity has been notified from the product selina-4(14),7(11)-diene-8,9-dio from Streptomyces spp. QD518 (Wu et al., 2006). Amorphane and T-muurolol (Figure 7) are terpene antibiotics secreted by Streptomyces sp. M491 (Wu et al., 2007; Ding et al., 2009). T-Muurolol shows moderate cytotoxic activity (Ding et al., 2009). Daryamides A-C (Figure 7) from Streptomyces sp. CNQ-085 has anticancer and anticytotoxic activities (Asolkar et al., 2006). Streptomyces spp. BL-49-58-005 has been isolated from a marine invertebrate, and it secrets indoles A, B having cytotoxic activity.
Streptomyces-derived bioactive compounds with agrobiologic properties
Streptomyces species regulate soil fertility by interacting with a variety of factors and act as nutrient enhancers, as an abundant source of bioactive secondary metabolites used in agriculture (Vurukonda et al., 2018), which are all impacted by chemical pesticides or insecticides with complex structures.
Insecticide, pesticide, and herbicide substances
The S. hydrogenans DH16 extract ethyl acetate had larvicidal and growth-suppressive properties (Kaur et al., 2014). Streptomyces produces avermectins, which have activity against mites, pear psylla, and diamond back moth (Lehman et al., 2004). S. rimosus could be used to reduce pesticide pollution in environments (Khajezadeh et al., 2020). A variety of cellulolytic and hydrolytic enzymes produced by Streptomyces may enter into the plant materials by rupturing the epidermal cell walls and middle lamellae between plant cells (Viaene et al., 2016). Spinosad from actinomyces Saccharopolyspora spinosa was registered by the EPA as a new chemical class of insecticides with a double glycosylated polyketide structure, and it was now accumulated heterologously at the highest yield (70 mg/L) in Streptomyces albus, while in Streptomyces hosts, pathway refactoring led to the production of some novel spinosad derivatives (Song et al., 2020; An et al., 2021).
Antifungal substances
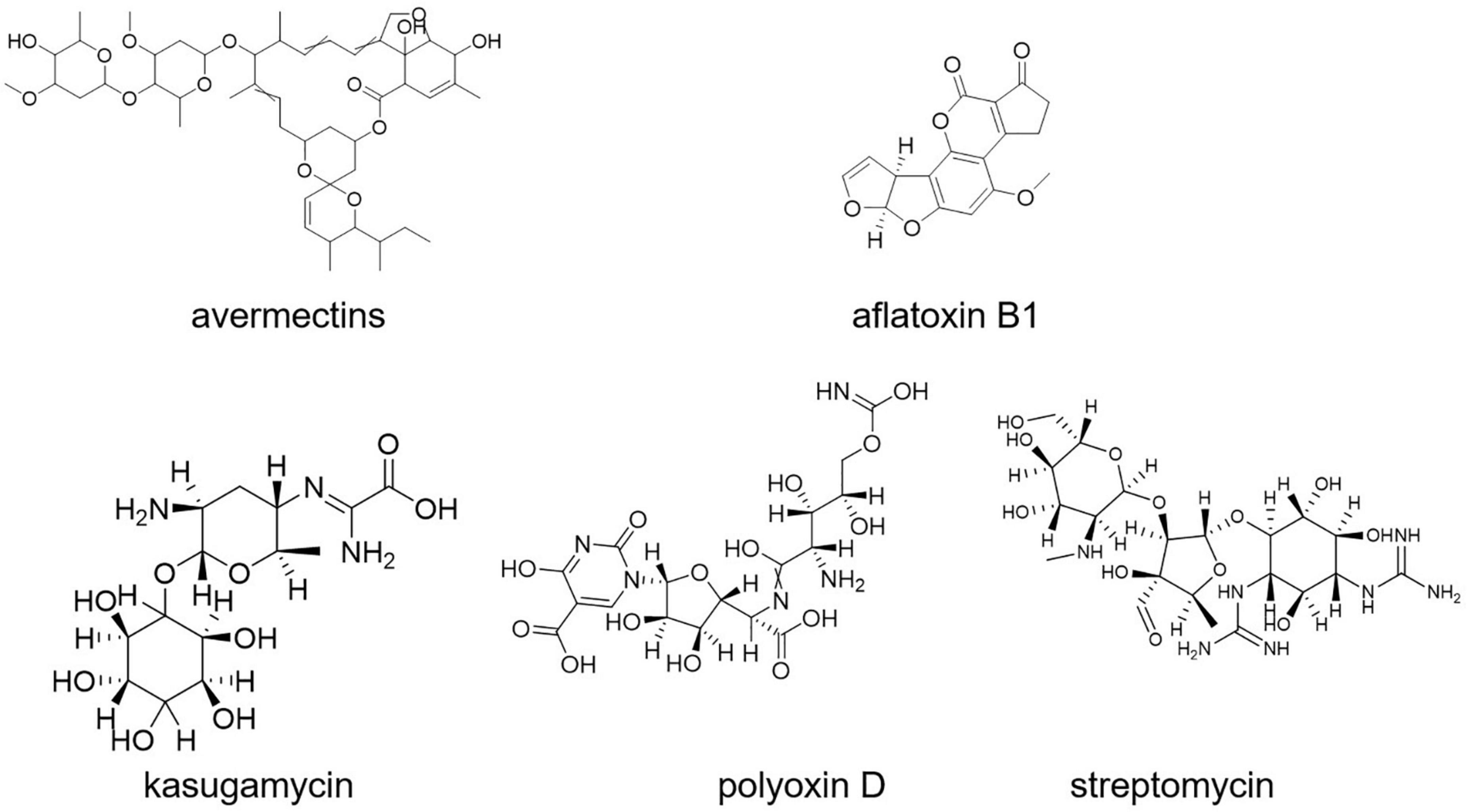
Aflatoxin B1 contamination is controlled with S. roseolus (Caceres et al., 2018). Similarly, S. yanglinensis could inhibit the formation of Aspergillus flavus mycelium in protecting food and feed from aflatoxin contamination (Shakeel et al., 2018; Lyu et al., 2020). S. griseoviridis K61 (Mycostop) was used for the biocontrol of black scorch on date palms caused by Ceratocystis radicicola and soilborne diseases, namely, Fusarium oxysporum sp. lycopersici (FOL) and verticillium of tomato (Suleman et al., 2002; Minuto et al., 2006). A total of four different strains of Streptomyces (viz., S. violaceusniger YCED9, S. saraceticus KH400, S. lydicus WYEC108, and S. griseoviridis K61) were used for biocontrol of fungal and bacterial soil diseases in nine (ix) different formulations. Kasugamycin, streptomycin, and polyoxin D (Figure 8) are three purified metabolites produced by Streptomyces sp. that are used as foliar fungicides and bactericides (Rey and Dumas, 2017). The synthesis of a thiopeptide that stops Fusarium cell wall biosynthesis could be linked to the disease-suppressive actions of Streptomyces strains, according to genome mining and mutant analysis (Cha et al., 2016; Rey and Dumas, 2017).
Streptomyces S4-7 was found to be resistant to Fusarium wilt disease (Kim et al., 2017). Culture extracts of Streptomyces globisporus JK-1 could inhibit Magnaporthe oryzae more effectively than tricyclazole, a commonly used chemical fungicide for the control of the rice blast fungus (Newitt et al., 2019). Kasugamycin (an antibiotic derived from S. kasugaensis, with brand name Kasumin in Japan) is used to prevent rice blast disease. Rice plants with transgenic expression of the S. griseus chitinase-encoding gene chiC showed improved resistance to the rice blast fungus Magnaporthe grisea (Itoh Y. et al., 2003).
Natural products from endophytic Streptomyces species
Streptomyces species are unique evolutionally because they lie between fungi and bacteria. Endophytic Streptomyces species appear asymptomatically in animal or human hosts by participating in the physiological process in the animal body or guts. Most of endophytic Streptomyces from plants are involved in the rhizophagy cycle and improve plant growth, reduce oxidative stress, and suppress the growth of competitive plants (Hallmann et al., 1997; Bacon and White, 2000). Here, we mentioned some different endophytic Streptomyces species that can produce bioactive natural products.
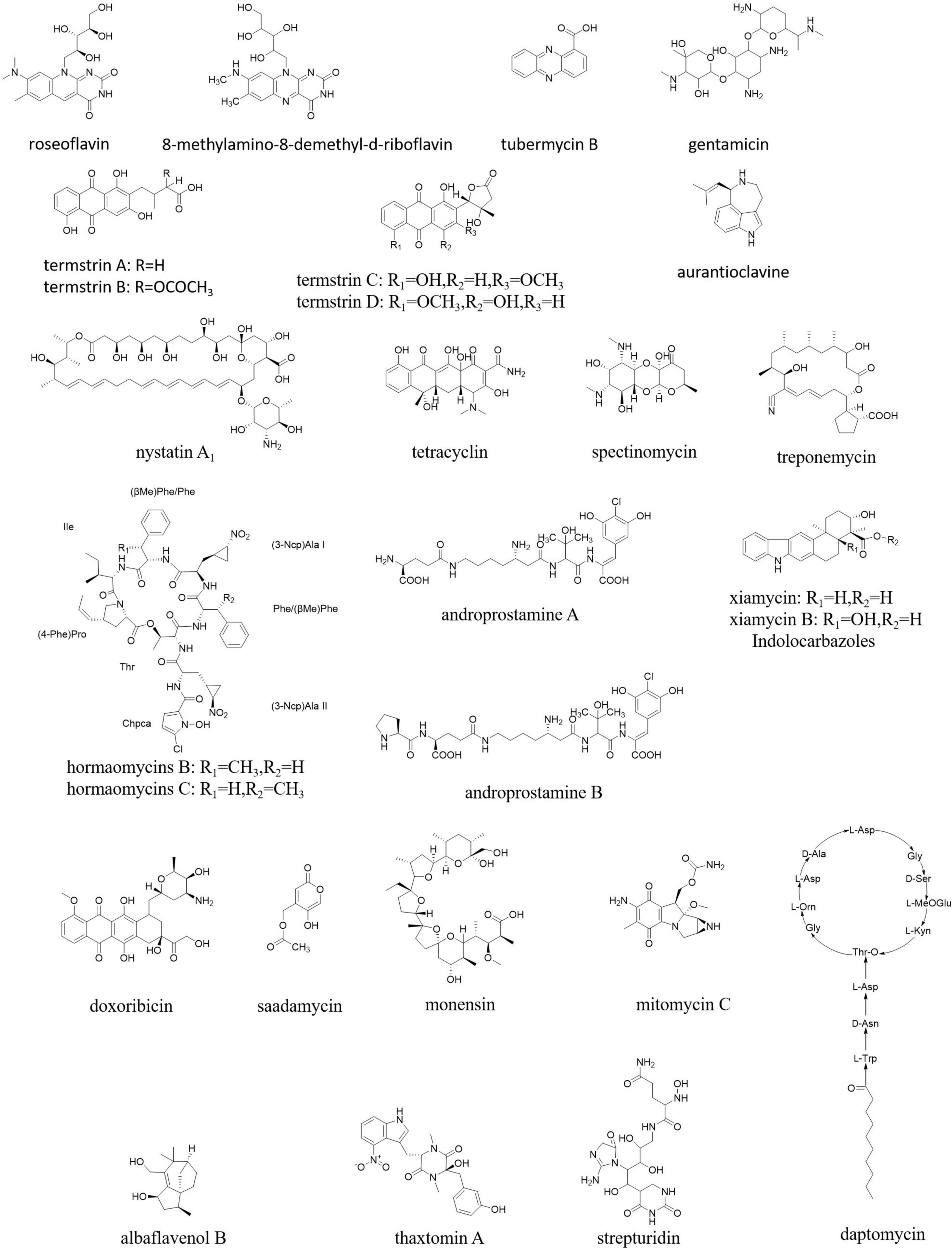
A diverse group of pharmaceutical compounds (Figure 9) have been isolated from different endophytic Streptomyces species and have a wide range of activities, such as antibacterial, antifungal, antimalarial, anticancer, and larvicidal, including nystatin, munumbicin, hormaomycin, tetracycline, coronamycin, spectinomycin, treponemycin, androprostamines, indolocarbazoles, doxorubicin, anthracycline, daptomycin, monensin, mitomycin C, saadamycin, strepturidin, thaxtomin A, xiamycin, and albaflavenol B (Ayswaria et al., 2020).
Although Streptomyces in mammal guts are less prevalent than those in soils, among a limited number of Streptomyces identified as “old friends” in human guts, a species showed activity against colon cancer. So, it was deduced that such Streptomyces able to produce antiproliferatives/immunosuppressants could reduce colon cancer directly while residing in human guts as possible “old friends” (Bolourian and Mojtahedi, 2018b). A high number of natural products were identified from Streptomyces in the animal guts, especially in the termite guts. For example, roseoflavin and 8-methylamino-8-demethyl-d-riboflavin from S. davaonensis living in the body surface of the queen of Odontotermes formosanus strongly inhibited Bacillus subtilis and S. aureus (Zhou et al., 2021). Termstrins A, B, C, and D (anthraquinone derivatives) were produced by Streptomyces sp. BYF63 from termite guts; among them, termstrins A and C showed potent antibacterial activities against S. aureus (Zhang et al., 2020). A polyene-producing Streptomyces spp. from the fungus-growing termite Macrotermes barneyi showed strong bioactivity against the antagonistic fungus Xylaria (Li J. et al., 2021).
Overall, 12 isolates from 65 endophytic Streptomyces sp. were found to have antibacterial activity against penicillin-resistant S. aureus (Passari et al., 2017). An endophytic actinomycete isolated from an ethnomedicinal plant Streptomyces kebangsaanensis sp. nov. produced tubermycin B (phenazine-1-carboxylic acid) with antibacterial activity (Tan et al., 2013). Streptomyces sp. SUK was proved to produce non-toxic antibiotics, such as aurantioclavine and gentamicin. Endophytic Streptomyces spp. from maize had antibacterial and antifungal activities against many pathogens (de Araújo et al., 2000).
Endophytic Streptomyces spp. SUK 12 and SUK 48 extracts were found to be shown anti-plasmodial activity (Ahmad et al., 2021). A Streptomyces strain producing munumbicins A, B, C, and D was reported to inhibit malarial infection (Castillo et al., 2002). Shimizu et al. (2004) revealed antitumor agent actinomycin X2-producing endophytic S. galbus R-5 (Shimizu et al., 2004). Tanvir et al. (2014) reported the larvicidal effect of endophytic Streptomyces spp.
Many endophytic Streptomyces species could produce amylases and cellulases, especially extracellular amylases being widely used in the textile, distilling, food, and brewing industries. For example, endophytic alkali-thermotolerant S. gulbargensis DAS 131 was reported to produce amylases (Syed et al., 2009).
Some endophytic Streptomyces species were reported to accumulate phytohormones, antibiotics, and siderophores used to improve the fitness of the plants. Overall, three Streptomyces species, namely, Streptomyces fulvoviolaceus, Streptomyces caelestis, and Streptomyces coelicolor, showed strong activity against one or more pathogenic fungi, including Phytophthora erythroseptica, Pythium ultimum, Sclerotinia sclerotiorum, Mycosphaerella fijiensis, and Rhizoctonia solani (Zin et al., 2007). Streptomyces SUK 25 produced pharmaceutically active compounds (PhACs) in submerged fermentation using response surface methodology (RSM) as a tool for optimization (Al-Shaibani et al., 2021).
Some endophytic Streptomyces are able to produce biosurfactants. Streptomyces sp. DPUA1566 could produce biosurfactant production using corn steep liquor substrates (Santos et al., 2019). Marine endophytic Streptomyces spp. NRC50 and NRC51 were able to accumulate prodigiosin-like compounds (red pigments) using agrowaste materials as substrates (El-Bondkly et al., 2012). Some volatile organic products with antifungal activity from Streptomyces setonii WY228 could control black spot disease of sweet potato (Gong et al., 2022).
Action modes of bioactive compounds from Streptomyces species
Different bioactive natural products from Streptomyces exert their activities via different mechanisms.
Action modes of antibacterial compounds
Most of the antibiotics act upon some specific parts of bacteria and stop the cellular processes or cellular synthesis. The specific targets include inhibition of (1) deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) synthesis (DNA topoisomerase or RNA polymerase), (2) protein synthesis (ribosome), or (3) cell wall (peptidoglycan) synthesis (Figure 10).
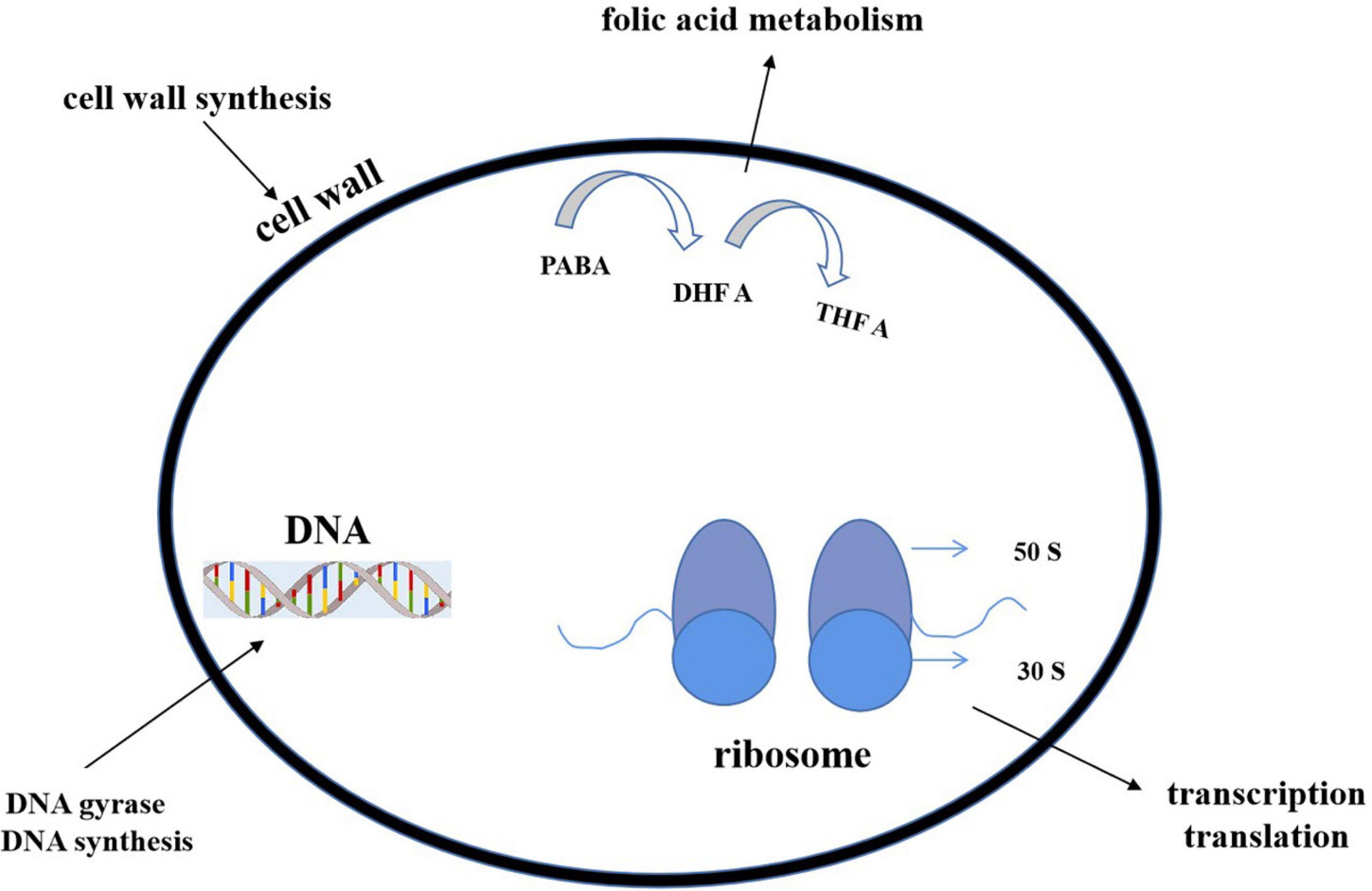
Figure 10. Action modes of antibacterial compounds. By targeting the bacterial cell wall/cell membrane, antibiotics inhibit bacterial growth. The synthesis of nucleic acids and proteins are two further targets. The latter is a function of ribosomes, which are nucleoprotein complexes made up of a small and large subunit (30 S and 50 S in bacteria, as shown in the figure). Antibiotics can also operate as antimetabolites by blocking folate metabolism (and thus DNA synthesis) through a pathway that includes para aminobenzoic acid (PABA) and two folic acid precursors, dihydrofolic acid (DHF) and tetrahydrofolic acid (THFA) (THF). Antibiotics can stop DNA gyrase from changing the shape of DNA, which is important for replication and transcription.
Cell wall biosynthesis inhibition
Generally, the β-lactam group include penicillin, cephalosporin, and vancomycin. The first two antibiotics inhibit the cell wall precursor–peptidoglycan crosslink. A serine hydroxyl group attacks β-lactam rings and produces an acyl intermediate, which hydrolyzes very slowly, terminates cell wall synthesis, and lessens the number of reproducing cells (Walsh, 2003). Vancomycin binds with the network of strong hydrogen bonding and stops the cell wall generation process (Rossiter et al., 2017). Lipid II is an essential intermediate in peptidoglycan biosynthesis in bacteria. Some peptides (like some lanthiopeptides) could bind to lipid II to impair cell wall synthesis. Meanwhile, such binding also caused the formation of a polymerized complex, leading to perforation on cell membranes and electrolyte outflow (Li C. et al., 2021).
Inhibition of protein synthesis
Erythromycin, oxazolidinones including linezolid, and tetracycline, come under this category. Erythromycin and linezolid attack the larger 50S subunit of the ribosome (Levy and Marshall, 2004; Ruuskanen and Järvinen, 2014). Tetracycline binds the smaller 30S subunit and blocks the translation process (Rossiter et al., 2017).
Inhibition of DNA or RNA synthesis (DNA topoisomerase or RNA polymerase)
DNA and RNA replication have been blocked by rifampicin, quinolones, ciprofloxacin, etc. (Kohanski et al., 2010). They interfere with the DNA gyrase, unwind the both strands of DNA, bind with the cleavage site, and stop the process of further of DNA synthesis (Rossiter et al., 2017).
Action modes of antifungal compounds
Fungal cell membranes have a unique sterol, ergosterol, differing from mammalian cell membranes (cholesterol). The chitin structured fungal cell wall is absent in human cell. Thus, for some selective antifungal agents, fungal cell walls may be considered to be prime targets (Sudhir et al., 2010).
Piericidin A1 showed antitumor/anti-insect activity by competitive binding to the mitochondrial complex I (Li et al., 2019). It was also found to have the most potent inhibitory activity against Xanthomonas oryzae pv. oryzicola and Penicillium decumbens (Shang et al., 2018; Li et al., 2019). Piericidin A and glucopiericidin A are also potential quorum sensing inhibitors (QSIs) that suppress the expression of Eca virulence genes of plant pathogen Erwinia carotovora subsp. atroseptica (Eca), implying that they could be used as soft rot disease control agents on potato tubers (Kang et al., 2016).
Nigericin can promote an exchange of K+ for H+, which changes the ion gradient across membranes involved in energetic metabolism and thus inhibits the growth of the Leishmania parasite (Ortega et al., 2019).
Fungichromin may inhibit the biofilm formation of C. albicans by downregulating the expression of the genes (ALS1, ALS3, HWP1, HYR1, EFG1, CPH1, and BCR1) (An et al., 2016). Streptochlorin mechanism of action for antifungal activity is unknown; however, a few studies have shown that it can inhibit the activity of monoamine oxidase (Jia et al., 2018).
Another antifungal agent, AFP1, as a novel member of the bg-crystallin superfamily, could change fungal morphogenesis. It was observed to bind to crab shell chitin, chitosan (deacetylated chitin), and the cell walls of the fungus P. variotii (Campos-Olivas et al., 2001).
Action modes of antiviral compounds
Viruses are obligate intracellular parasites and have no cell walls or cell membranes. They do not have metabolic machinery of their own and have to use hosts’ enzymes. Targeting the viruses themselves and the host cell components are the two main techniques for developing antiviral drugs.
Antiviral compounds by inhibiting virus attachment, entrance, uncoating, genome replication, and inhibiting some viral enzymes (DNA polymerase, protease, nucleotide reverse transcriptase, and integrase) are some antiviral medications that specifically target viruses. Protease inhibitors including ritonavir, atazanavir, and darunavir; viral DNA polymerase inhibitors including acyclovir, tenofovir, valganciclovir, and valacyclovir; and integrase inhibitors (such as raltegravir) are among the top 200 drugs by sales in the 2010s. But, many viral infections still lack effective antiviral treatment.
Xiamycin D showed inhibition against PEDV RNA expression, nucleocapsid protein synthesis, and also lowering RNA levels. Its activity is associated with the GP2 spike protein, a key regulator in the viral entry step (Kim et al., 2016).
Some known antiviral drugs were found to enforce the cell resistance to a virus (interferons), block viral attachment, diffusion into the host cells (amantadine), or viral deproteinization process in the cell (amantadine). They also lower cell metabolisms to inhibit viral nucleic acid synthesis. Other antiviral agents, including butanolide, could prevent viral entrance into host cells by blocking the fusogenic process of viral attachment proteins, such as hemagglutinin (HA) of influenza A virus (IAV) (Li et al., 2018).
Action modes of insecticide, pesticide, and herbicide substances
Chemicals known as “insecticides” are put to use to get rid of insects either by killing or by preventing them from carrying out harmful or unwanted behaviors. Based on the difference in structure and mode of action, insecticides are classified into organophosphates, pyrethroids, triazines, carbamates, and organochlorines. Many insecticides act upon the insect’s nervous system (e.g., cholinesterase inhibition), while others act as growth regulators or endotoxins.
Organophosphate pesticides could weaken acetylcholinesterase, leading to cholinergic toxicity. Carbamate or phosphate pesticides were found to disrupt the equilibrium between acetylcholine synthesis and release, on the one hand, and its hydrolysis, on the other hand, by inhibiting acetylcholinesterase and cause the accumulation of acetylcholine at the synaptic level, resulting in prolonged activation of cholinergic receptors.
Herbicides can result in deformities in plants via blocking cell division and photosynthesis or by mimicking natural plant growth hormones. The potential effects of herbicides are strictly dependent on their toxic mode of action and way of application.
Strategies for new natural product discovery from Streptomyces
Although many compounds with biological activity were isolated from Streptomyces, only two new classes of antibiotics were introduced in the clinic in the last 30 years. On the other hand, microbial resistance to existing antibiotics is increasing day by day. It is a matter of great concern that humanity may return to the pre-antibiotic era. Antibiotic resistance (AR) is widely considered to be the next global pandemic.
General consideration for screening new natural products from Streptomyces
A large quantity of Streptomyces strains having capability to produce bioactive compounds remain unexplored (Genilloud, 2014). Nowadays, new drug discovery has become more difficult, but demands for novel antibiotics in the hospitals have pushed drug screening from nature to be resumed. Some new Streptomyces strains have been discovered from new environments. Marine environments have emerged as potential sources during the last decade, as well as soils of deserts, mountain, and icy places. Symbiotic relationships have attracted attentions of the scientific community. Another important perspective is paying attention to natural growth conditions.
After the availability of whole-genome sequencing technique, many cryptic or silent biosynthetic gene clusters (BGCs) in Streptomyces were revealed. Among 20–50 biosynthetic pathways for natural products in every Streptomyces strain, only a limited number of products have been identified. It still remains a huge challenge to activate these pathways to discover new secondary metabolites from Streptomyces (Supplementary Figure S1).
In laboratory/industry, culture conditions for Streptomyces growth differ greatly from natural conditions. In natural habitats of Streptomyces, natural product production may remain dormant as these substances are related to stress conditions or defense against niche competitors. But most of BGCs are not expressed or slightly expressed under laboratory conditions.
In addition, in laboratory conditions, a majority (>90%) of Streptomyces are hard to culture or are even non-culturable. This problem could be solved by using the functional metagenomics approach.
Currently, elicitors are used to manipulate Streptomyces. Elicitors can induce differentiation of Streptomyces and trigger cryptic antibiotic pathways. Secondary metabolism could also be conditioned by differentiation. The mycelium morphology and different life cycle stages are important for antibiotic production. To improve active cryptic biosynthetic gene clusters and secondary metabolite production, different complementary strategies have been developed, either to improve the screening of new activities or to improve the production of known molecules.
It has been hypothesized that cryptic antibiotic gene clusters that have been inactive may need to be reactivated. Some newer methods were used to make it achievable, such as by adding signaling molecule N-acetylglucosamine to the medium for cultivating S. hygroscopicus, S. collinus, S. clavuligerus, S. griseus, and S. venezuelae, resulting in the activation of antibiotic-producing genes (Rigali et al., 2008). The inclusion of bacterial hormones gamma-butyrolactones (γ-butyrolactones) produced similar results. Alternatively, it was found that adding sub-inhibitory concentrations of lincomycin into the medium caused production of chemicals that are not formed on lincomycin-free medium (Imai et al., 2015).
Aside from the new methods already described, there have been reports of other recent ways used to encourage the identification of new Streptomyces compounds. There have also been reports of innovative co-transformation strategies being used in Streptomyces sp. (Kallifidas et al., 2012).
Promoter engineering was developed to activate silent BGCs. The Clustered Regularly Interspaced Short Palindromic Repeats-Cas9 (CRISPR-Cas9) gene editing tool was effectively employed to add the kasO promoter (97 base pair) into S. roseosporus NRRL 15998. This promoter region then allowed the silent aurR1 gene cluster to be activated, resulting in the synthesis of auroramycin when expressed (Kallifidas et al., 2012).
Despite the fact that whole-genome sequencing was used to identify the gene clusters, heterologous expression of these gene clusters typically goes unnoticed. Scientists employed environmental DNA clones (eDNA), specifically those expressing PKS-type transcription factors, and transplanted them into S. albus with the equivalent minimal PKS containing clone. Tetarimycin was eventually produced and isolated in S. albus using this approach (Kallifidas et al., 2012).
Genomic-driven natural product discovery from Streptomyces
Recent data suggest that genome mining of large-scale genomic datasets reveals the potential of secretion of various natural products from microbes (Doroghazi et al., 2014; Ju et al., 2015; Pan et al., 2017) with the possibilities of the production of antibiotics too (Bérdy, 2012; Ward and Allenby, 2018). Genome mining is a process that helps perceive the biosynthetic pathway for production of natural compounds (Zhao et al., 2019). This process is dependent on biosynthetic gene clusters (BGCs) for natural products (Medema et al., 2015).
By genome mining approaches in silico, it was revealed that Streptomyces genomes have 25–70 BGCs, but only a small fraction of these bioactive compounds is synthesized in the laboratory exploiting culture methods (Belknap et al., 2020). Furthermore, genome mining enables the finding of untapped biosynthetic pathways from uncultivated Streptomyces hiding in a variety of underexplored habitats, as well as uncharacterized biosynthetic pathways contained in the genomes of cultured Streptomyces (Niu, 2018). In this case, knowledge about evolutionary relationships, BGC diversity, and distribution patterns of BGCs are very important. However, it is not easy to analyze the distribution of BGCs in case of Streptomyces as these species as prolific producers contain at least 1,000 genomes and 20–70 BGCs on each genome (Ward and Allenby, 2018). A recent study found a cluster of Streptomyces BGC by genome mining that have antitumor properties (Belknap et al., 2020). In another study, genome mining is used to identify hidden diversity of clusters of Streptomyces having ecological impact (Ward and Allenby, 2018). In S. coelicolor genome, cryptic biosynthetic gene clusters were identified by genome mining (Challis, 2014). antiSMASH,1 and MIBiG databases2 contain reference BGCs. In antiSMASH database, among 39 Streptomyces genomes, 1346 BGCs are found. Some BGCs possess conserved regions for production of ectoine, osmotic compatible solute, hopene for prokaryotic membrane stability, siderophores like desferrioxime for iron uptake, melanin and spore pigments, carotenoids like isorenieratene, butyrolactones, geosmin, and methylisoborneol. Chlortetracycline, antimycin, albaflavenone, and ectoine are some of the antibiotics from the BGC clusters by Streptomyces (Ward and Allenby, 2018).
Next, with the development of synthetic biology-inspired new strategies, including TAR or Red/ET strategies, large whole BGCs could be directly cloned and expressed successfully in heterologous hosts to produce compounds silent in the native cells (Abbasi et al., 2020), and some Streptomyces still act as good candidates as hosts, such as S. albus and S. coelicolor. De novo reconstruction of new BGCs or engineering of pathways is nowadays available and expressed in heterologous chassis cells, allowing more rapid finding new compounds (Alam et al., 2021).
It is unequivocally true that genome mining is a promising area that emphasizes on the biosynthetic pathways of microbes, facilitating the production of antibiotics. Nowadays, there is no alternative to genome mining to quench the thirst of finding new antibiotics.
Metagenomic approaches for natural product discovery from Streptomyces
Metagenomic mining is another effective and practical method for finding natural product biosynthesis pathways. This stage entails collecting uncultured microorganisms from ambient samples, creating a genome library, and then performing function-based or sequence-based screening on the metagenomic library (Alam et al., 2021). However, various hurdles must be overcome before their full biosynthetic potential may be realized. One example is the selection of the vector and host in metagenome mining. BAC vectors, which can hold large DNA inserts harboring complete BGCs, are preferred for metagenomic library construction over cosmid and fosmid vectors. For example, S. albus is evaluated for its color phenotypic, whereas sequence-based screening relies on sequence tags. Then, using phylogenetic analysis or PCR-based screening, the clones of interest or BCGs are selected, allowing for faster chemical diversity screening and the discovery of novel antibiotic compounds (Niu, 2018). However, isolating DNA fragments of suitable size from eDNA remains difficult. In addition, E. coli and Streptomyces spp. are the most typical hosts utilized for library development. Taking into account the metabolic variety of environmental samples, hosts in addition to E. coli and actinomycetes need additional investigation.
In total, seven effective epoxyketone proteasome inhibitors and two unusual tryptophan dimer natural products were identified by screening soil samples with conserved sequence tags (Chang et al., 2015; Owen et al., 2015). To enrich the library of scaffold-specific antibacterial products, environmental samples are screened in the presence of a specific antibiotic. This is based on the idea that antibiotic producers must develop a self-defense mechanism. Pekiskomycin, a glycopeptide antibacterial drug with a unique peptide structure, was discovered using PCR and phylogenetic analysis, which help screen potential producers for chemical diversity (Thaker et al., 2013). The S. albus metagenomic library led to the discovery of metatricycloene, a novel natural product (Iqbal et al., 2016). It was fascinating that S. albus acted as heterologous host to express cadasides (CDE) gene cluster from a soil eDNA sample (Wu et al., 2019).
Conclusion and future perspectives
Antibiotic resistance is spreading faster than we can develop new antimicrobials and techniques to counteract it. Antibiotic-resistant diseases demand novel drugs. Multidrug resistance is a serious difficulty in treating infectious illnesses, both in the community and in hospitals, and restricts the therapy of bacterial infections. Antibiotic discovery rates decrease drastically after the “golden age” of the 1940s to the 1960s. Streptomyces is a promising natural antibiotic manufacturer and bioactive chemical source used to find and make new drugs. Numerous Streptomyces species include BGCs that encode chemicals with diverse activity, making them valuable.
However, identifying new antibiotics in Streptomyces is difficult because of rediscovering known antibiotics. To avoid rediscovering the same molecules, new multidisciplinary approaches are needed for resuming natural products screening from Streptomyces. To improve Streptomyces screening, multidisciplinary techniques that integrate methodologies will be critical to unlock hidden pathways and discovering new bioactive chemicals. Biosynthetic pathways from previously unknown Streptomyces are now accessible with advances in genomics-driven technologies. It is simple to apply metagenome, pan-genome, and genome mining to these filamentous Streptomyces bacteria.
Bioinformatics, heterologous expression, synthetic biology, mass spectrometry (MS), and nuclear magnetic resonance (NMR)-based metabolomics are needed to unravel the intricacy of metabolic pathways in understudied Streptomyces. Undoubtedly, advancements in these methods, especially in synthetic biology and artificial intelligence used in bioinformatics fields, will enhance our knowledge of the variety and distribution of these biosynthetic pathways and optimize the biosynthetic potential of chemical weapons against diseases resistant to antimicrobials.
Author contributions
AL conceived the concept and funds, supervised the work, and validated the results. KA, AM, and SS wrote the original draft of manuscript. KA, JH, and Y-MZ conducted software. SI, CS, and YW conducted validation. YZ contributed to visualization, writing, and data analysis. All authors read and approved the manuscript.
Funding
This study was supported by the National Key R&D Program of China (2019YFA0905700), the National Natural Science Foundation of China (32170038 and 32270088), and the Open Project Program of the State Key Laboratory of Bio-based Material and Green Papermaking (KF201825) and the 111 Project (B16030).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.968053/full#supplementary-material
Supplementary Figure 1 | General procedures for screening bioactive compounds from Streptomyces.
Footnotes
References
Abbasi, M. N., Fu, J., Bian, X., Wang, H., Zhang, Y., and Li, A. (2020). Recombineering for genetic engineering of natural product biosynthetic pathways. Trends Biotechnol. 38, 715–728.
Abdel-Razek, A. S., El-Naggar, M. E., Allam, A., Morsy, O. M., and Othman, S. I. (2020). Microbial natural products in drug discovery. Processes 8:470.
Adinarayana, G., Venkateshan, M. R., Bapiraju, V., Sujatha, P., Premkumar, J., Ellaiah, P., et al. (2006). Cytotoxic compounds from the marine actinobacterium Streptomyces corchorusii AUBN 1/7 1. Russ. J. Bioorg. Chem. 32, 295–300.
Ahmad, S. J., Zin, N. M., Mazlan, N. W., Baharum, S. N., Baba, M. S., and Lau, Y. L. (2021). Metabolite profiling of endophytic Streptomyces spp. and its antiplasmodial potential. PeerJ 9:e10816. doi: 10.7717/peerj.10816
Akintunde, O. G. (2014). Production of an Antibiotic-like Activity by Streptomyces sp. COUK1 under Different Growth Conditions. Ph.D. thesis. Johnson City, TN: East Tennessee State University.
Alam, K., Abbasi, M. N., Hao, J., Zhang, Y., and Li, A. (2021). Strategies for natural products discovery from uncultured microorganisms. Molecules 26:2977.
Al-Ansari, M., Alkubaisi, N., Vijayaragavan, P., and Murugan, K. (2019). Antimicrobial potential of Streptomyces sp. to the Gram positive and Gram negative pathogens. J. Infect. Public Health 12, 861–866. doi: 10.1016/j.jiph.2019.05.016
Al-Shaibani, M. M., Radin Mohamed, R. M. S., Zin, N. M., Al-Gheethi, A., Al-Sahari, M., and El Enshasy, H. A. (2021). Enhanced pharmaceutically active compounds productivity from Streptomyces SUK 25: Optimization, characterization, mechanism and techno-economic analysis. Molecules 26:2510. doi: 10.3390/molecules26092510
Amann, S., Neef, K., and Kohl, S. (2019). Antimicrobial resistance (AMR). Eur. J. Hosp. Pharm. 26, 175–177.
An, D., Wang, X., Li, J., Jiang, S., Ma, X., Zhang, H., et al. (2016). The activity of fungichromin against the formation of Candida albicans biofilm. Biol. Pharm. Bull. 39, 1948–1954. doi: 10.1248/bpb.b16-00380
An, Z., Tao, H., Wang, Y., Xia, B., Zou, Y., Fu, S., et al. (2021). Increasing the heterologous production of spinosad in Streptomyces albus J1074 by regulating biosynthesis of its polyketide skeleton. Synth. Syst. Biotechnol. 6, 292–301. doi: 10.1016/j.synbio.2021.09.008
Aryal, S., Neupane, L., Adhikari, R., Regmi, B., Koirala, N., and Joshi, D. R. (2020). Novel Streptomyces Sp. Reported in 2018: A Meta-Analysis. AntiInfect. Agents 19, 2–13.
Asolkar, R. N., Jensen, P. R., Kauffman, C. A., and Fenical, W. (2006). Daryamides A–C, weakly cytotoxic polyketides from a marine-derived actinomycete of the genus Streptomyces strain CNQ-085. J. Nat. Prod. 69, 1756–1759. doi: 10.1021/np0603828
Ayswaria, R., Vasu, V., and Krishna, R. (2020). Diverse endophytic Streptomyces species with dynamic metabolites and their meritorious applications: A critical review. Crit. Rev. Microbiol. 46, 750–758. doi: 10.1080/1040841X.2020.1828816
Azerang, P., and Sardari, S. (2017). Bioactive compound produced from Actinomycetes-Streptomyces. Chemistry 151, 1507–1523.
Azman, A.-S., Othman, I., Fang, C.-M., Chan, K.-G., Goh, B.-H., and Lee, L.-H. (2017). Antibacterial, anticancer and neuroprotective activities of rare actinobacteria from mangrove forest soils. Indian J. Microbiol. 57, 177–187. doi: 10.1007/s12088-016-0627-z
Bacon, C. W., and White, J. F. Jr. (2000). “Physiological adaptations in the evolution of endophytism in the Clavicipitaceae,” in Microbial Endophytes, eds C. W. Bacon and J. F. White (Boca Raton, FL: CRC Press), 251–276.
Barreiro, C., Prieto, C., Sola-Landa, A., Solera, E., Martínez-Castro, M., Pérez-Redondo, R., et al. (2012). Draft genome of Streptomyces tsukubaensis NRRL 18488, the producer of the clinically important immunosuppressant tacrolimus (FK506). J. Bacteriol. 194, 3756–3757. doi: 10.1128/JB.00692-12
Bauermeister, A., Pereira, F., Grilo, I. R., Godinho, C. C., Paulino, M., Almeida, V., et al. (2019). Intra-clade metabolomic profiling of MAR4 Streptomyces from the Macaronesia Atlantic region reveals a source of anti-biofilm metabolites. Environ. Microbiol. 21, 1099–1112. doi: 10.1111/1462-2920.14529
Belknap, K. C., Park, C. J., Barth, B. M., and Andam, C. P. (2020). Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 10:2003. doi: 10.1038/s41598-020-58904-9
Bérdy, J. (2012). Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 65, 385–395.
Bolourian, A., and Mojtahedi, Z. (2018b). Streptomyces, shared microbiome member of soil and gut, as ‘old friends’ against colon cancer. FEMS Microbiol. Ecol. 94:fiy120. doi: 10.1093/femsec/fiy120
Bolourian, A., and Mojtahedi, Z. (2018a). Immunosuppressants produced by Streptomyces: Evolution, hygiene hypothesis, tumour rapalog resistance and probiotics. Environ. Microbiol. Rep. 10, 123–126. doi: 10.1111/1758-2229.12617
Brown, D. (2015). Antibiotic resistance breakers: Can repurposed drugs fill the antibiotic discovery void? Nat. Rev. Drug Discov. 14, 821–832. doi: 10.1038/nrd4675
Caceres, I., Snini, S. P., Puel, O., and Mathieu, F. (2018). Streptomyces roseolus, a promising biocontrol agent against Aspergillus flavus, the main aflatoxin B1 producer. Toxins 10:442. doi: 10.3390/toxins10110442
Campos-Olivas, R., Hörr, I., Bormann, C., Jung, G., and Gronenborn, A. M. (2001). Solution structure, backbone dynamics and chitin binding of the anti-fungal protein from Streptomyces tendae TÜ901. J. Mol. Biol. 308, 765–782. doi: 10.1006/jmbi.2001.4622
Castillo, U. F., Strobel, G. A., Ford, E. J., Hess, W. M., Porter, H., Jensen, J. B., et al. (2002). Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscansaa. Microbiology 148, 2675–2685.
Cha, J.-Y., Han, S., Hong, H.-J., Cho, H., Kim, D., Kwon, Y., et al. (2016). Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 10, 119–129. doi: 10.1038/ismej.2015.95
Challis, G. L. (2014). Exploitation of the Streptomyces coelicolor A3 (2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 41, 219–232. doi: 10.1007/s10295-013-1383-2
Chang, F.-Y., Ternei, M. A., Calle, P. Y., and Brady, S. F. (2015). Targeted metagenomics: Finding rare tryptophan dimer natural products in the environment. J. Am. Chem. Soc. 137, 6044–6052.
Chang, T.-L., Huang, T.-W., Wang, Y.-X., Liu, C.-P., Kirby, R., Chu, C.-M., et al. (2021). An Actinobacterial Isolate, Streptomyces sp. YX44, produces broad-spectrum antibiotics that strongly inhibit Staphylococcus aureus. Microorganisms 9:630. doi: 10.3390/microorganisms9030630
Chevrette, M. G., Carlson, C. M., Ortega, H. E., Thomas, C., Ananiev, G. E., Barns, K. J., et al. (2019). The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 10:516. doi: 10.1038/s41467-019-08438-0
Choi, S.-S., Hur, Y.-A., Sherman, D. H., and Kim, E.-S. (2007). Isolation of the biosynthetic gene cluster for tautomycetin, a linear polyketide T cell-specific immunomodulator from Streptomyces sp. CK4412. Microbiology 153, 1095–1102. doi: 10.1099/mic.0.2006/003194-0
Choi, S.-S., Nah, H.-J., Pyeon, H., and Kim, E.-S. (2017). Biosynthesis, regulation, and engineering of a linear polyketide tautomycetin: A novel immunosuppressant in Streptomyces sp. CK4412. J. Ind. Microbiol. Biotechnol. 44, 555–561. doi: 10.1007/s10295-016-1847-2
Chopra, S., Harjai, K., and Chhibber, S. (2015). Antibiotic susceptibility of ica-positive and ica-negative MRSA in different phases of biofilm growth. J. Antibiot. 68, 15–22. doi: 10.1038/ja.2014.96
de Araújo, J. M., da Silva, A. C., and Azevedo, J. L. (2000). Isolation of endophytic actinomycetes from roots and leaves of maize (Zea mays L.). Braz. Arch. Biol. Technol. 43, 447–451.
Dezfully, N. K., and Ramanayaka, J. G. (2015). Isolation, identification and evaluation of antimicrobial activity of Streptomyces flavogriseus, strain ACTK2 from soil sample of Kodagu, Karnataka State (India). Jundishapur J. Microbiol. 8:e15107. doi: 10.5812/jjm.15107
Dharmaraj, S. (2010). Marine Streptomyces as a novel source of bioactive substances. World J. Microbiol. Biotechnol. 26, 2123–2139.
Ding, L., Pfoh, R., Ruhl, S., Qin, S., and Laatsch, H. (2009). T-muurolol sesquiterpenes from the marine Streptomyces sp. M491 and revision of the configuration of previously reported amorphanes. J. Nat. Prod. 72, 99–101. doi: 10.1021/np8006843
Doroghazi, J. R., Albright, J. C., Goering, A. W., Ju, K.-S., Haines, R. R., Tchalukov, K. A., et al. (2014). A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968. doi: 10.1038/nchembio.1659
Driche, E. H., Sabaou, N., Bijani, C., Zitouni, A., Pont, F., Mathieu, F., et al. (2017). Streptomyces sp. AT37 isolated from a Saharan soil produces a furanone derivative active against multidrug-resistant Staphylococcus aureus. World J. Microbiol. Biotechnol. 33:105. doi: 10.1007/s11274-017-2265-y
Du, L., Sánchez, C., Chen, M., Edwards, D. J., and Shen, B. (2000). The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 7, 623–642. doi: 10.1016/s1074-5521(00)00011-9
El-Bondkly, A., El-Gendy, M., and Bassyouni, R. H. (2012). Overproduction and biological activity of prodigiosin-like pigments from recombinant fusant of endophytic marine Streptomyces species. Antonie Van Leeuwenhoek 102, 719–734. doi: 10.1007/s10482-012-9772-5
Fireman, M., DiMartini, A. F., Armstrong, S. C., and Cozza, K. L. (2004). Immunosuppressants. Psychosomatics 45, 354–360.
Frändberg, E., Petersson, C., Lundgren, L. N., and Schnürer, J. (2000). Streptomyces halstedii K122 produces the antifungal compounds bafilomycin B1 and C1. Can. J. Microbiol. 46, 753–758.
Freitas, D. A., Leclerc, S., Miyoshi, A., Oliveira, S. C., Sommer, P. S. M., Rodrigues, L., et al. (2005). Secretion of Streptomyces tendae antifungal protein 1 by Lactococcus lactis. Braz. J. Med. Biol. Res. 38, 1585–1592. doi: 10.1590/s0100-879x2005001100004
Fu, P., La, S., and MacMillan, J. B. (2017). Daryamide analogues from a marine-derived Streptomyces species. J. Nat. Prod. 80, 1096–1101. doi: 10.1021/acs.jnatprod.7b00011
Genilloud, O. (2014). The re-emerging role of microbial natural products in antibiotic discovery. Antonie Van Leeuwenhoek 106, 173–188. doi: 10.1007/s10482-014-0204-6
Genilloud, O. (2017). Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 34, 1203–1232. doi: 10.1039/c7np00026j
Gong, Y., Liu, J.-Q., Xu, M.-J., Zhang, C.-M., Gao, J., Li, C.-G., et al. (2022). Antifungal volatile organic compounds from Streptomyces setonii WY228 control black spot disease of sweet potato. Appl. Environ. Microbiol. 88, e2317–e2321. doi: 10.1128/aem.02317-21
Grimm, A., Madduri, K., Ali, A., and Hutchinson, C. R. (1994). Characterization of the Streptomyces peucetius ATCC 29050 genes encoding doxorubicin polyketide synthase. Gene 151, 1–10. doi: 10.1016/0378-1119(94)90625-4
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W. F., and Kloepper, J. W. (1997). Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43, 895–914.
Harir, M., Bendif, H., Bellahcene, M., Fortas, Z., and Pogni, R. (2018). Streptomyces secondary metabolites. Basic Biol. Appl. Actinobacteria 6, 99–122.
Hayakawa, Y., Kobayashi, T., and Izawa, M. (2013). Indanostatin, a new neuroprotective compound from Streptomyces sp. J. Antibiot. 66, 731–733. doi: 10.1038/ja.2013.80
Hernández, I. L. C., Macedo, M. L., Berlinck, R. G. S., Ferreira, A. G., and Godinho, M. J. L. (2004). Dipeptide metabolites from the marine derived bacterium Streptomyces acrymicini. J. Braz. Chem. Soc. 15, 441–444.
Hirota-Takahata, Y., Kurosawa, E., Ishimoto, Y., Iwadate, Y., Kizuka, M., Chiba, J., et al. (2017). Vestaines, novel vasoactive compounds, isolated from Streptomyces sp. SANK 63697. J. Antibiot. 70, 179–186. doi: 10.1038/ja.2016.98
Hogan, S., Zapotoczna, M., Stevens, N. T., Humphreys, H., O’Gara, J. P., and O’Neill, E. (2016). In vitro approach for identification of the most effective agents for antimicrobial lock therapy in the treatment of intravascular catheter-related infections caused by Staphylococcus aureus. Antimicrob. Agents Chemother. 60, 2923–2931. doi: 10.1128/AAC.02885-15
Hou, Y.-H., Qin, S., Li, Y.-X., Li, F.-C., Xia, H.-Z., and Zhao, F.-Q. (2006). Heterologous expression and purification of recombinant allophycocyanin in marine Streptomyces sp. isolate M097. World J. Microbiol. Biotechnol. 22, 525–529.
Hrdý, J., Súkeníková, L., Petrásková, P., Novotná, O., Kahoun, D., Petříček, M., et al. (2020). Inhibition of pro-inflammatory cytokines by metabolites of Streptomycetes—A potential alternative to current anti-inflammatory drugs?. Microorganisms 8:621. doi: 10.3390/microorganisms8050621
Hu, D., Lee, S. M.-Y., Li, K., and Mok, K. M. (2021). Secondary metabolite production potential of mangrove-derived Streptomyces olivaceus. Mar. Drugs 19:332. doi: 10.3390/md19060332
Ikeda, H., Nonomiya, T., Usami, M., Ohta, T., and Ōmura, S. (1999). Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc. Natl. Acad. Sci. U. S. A. 96, 9509–9514. doi: 10.1073/pnas.96.17.9509
Imai, Y., Sato, S., Tanaka, Y., Ochi, K., and Hosaka, T. (2015). Lincomycin at subinhibitory concentrations potentiates secondary metabolite production by Streptomyces spp. Appl. Environ. Microbiol. 81, 3869–3879. doi: 10.1128/AEM.04214-14
Iqbal, H. A., Low-Beinart, L., Obiajulu, J. U., and Brady, S. F. (2016). Natural product discovery through improved functional metagenomics in Streptomyces. J. Am. Chem. Soc. 138, 9341–9344. doi: 10.1021/jacs.6b02921
Itoh, T., Kinoshita, M., Aoki, S., and Kobayashi, M. (2003). Komodoquinone A, a novel neuritogenic anthracycline, from marine Streptomyces sp. KS3. J. Nat. Prod. 66, 1373–1377. doi: 10.1021/np030212k
Itoh, Y., Takahashi, K., Takizawa, H., Nikaidou, N., Tanaka, H., Nishihashi, H., et al. (2003). Family 19 chitinase of Streptomyces griseus HUT6037 increases plant resistance to the fungal disease. Biosci. Biotechnol. Biochem. 67, 847–855. doi: 10.1271/bbb.67.847
Jia, C.-Y., Xu, L.-Y., Yu, X., Ding, Y.-B., Jin, B., Zhang, M.-Z., et al. (2018). An efficient synthesis and antifungal evaluation of natural product streptochlorin and its analogues. Fitoterapia 125, 106–110. doi: 10.1016/j.fitote.2017.12.017
Ju, K.-S., Gao, J., Doroghazi, J. R., Wang, K.-K. A., Thibodeaux, C. J., Li, S., et al. (2015). Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc. Natl. Acad. Sci. U. S. A. 112, 12175–12180. doi: 10.1073/pnas.1500873112
Kallifidas, D., Kang, H.-S., and Brady, S. F. (2012). Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J. Am. Chem. Soc. 134, 19552–19555. doi: 10.1021/ja3093828
Kang, J. E., Han, J. W., Jeon, B. J., and Kim, B. S. (2016). Efficacies of quorum sensing inhibitors, piericidin A and glucopiericidin A, produced by Streptomyces xanthocidicus KPP01532 for the control of potato soft rot caused by Erwinia carotovora subsp. atroseptica. Microbiol. Res. 184, 32–41. doi: 10.1016/j.micres.2015.12.005
Kaur, T., Vasudev, A., Sohal, S. K., and Manhas, R. K. (2014). Insecticidal and growth inhibitory potential of Streptomyces hydrogenans DH16 on major pest of India, S podoptera litura (Fab.)(Lepidoptera: Noctuidae). BMC Microbiol. 14:227. doi: 10.1186/s12866-014-0227-1
Kemung, H. M., Tan, L. T.-H., Khan, T. M., Chan, K.-G., Pusparajah, P., Goh, B.-H., et al. (2018). Streptomyces as a prominent resource of future anti-MRSA drugs. Front. Microbiol. 9:2221. doi: 10.3389/fmicb.2018.02221
Khadayat, K., Sherpa, D. D., Malla, K. P., Shrestha, S., Rana, N., Marasini, B. P., et al. (2020). Molecular identification and antimicrobial potential of streptomyces species from nepalese soil. Int. J. Microbiol. 2020:8817467. doi: 10.1155/2020/8817467
Khajezadeh, M., Abbaszadeh-Goudarzi, K., Pourghadamyari, H., and Kafilzadeh, F. (2020). A newly isolated Streptomyces rimosus strain capable of degrading deltamethrin as a pesticide in agricultural soil. J. Basic Microbiol. 60, 435–443. doi: 10.1002/jobm.201900263
Kim, H. S., and Park, Y. I. (2008). Isolation and identification of a novel microorganism producing the immunosuppressant tacrolimus. J. Biosci. Bioeng. 105, 418–421. doi: 10.1263/jbb.105.418
Kim, J.-S., Kang, N. J., Kwak, Y.-S., and Lee, C. (2017). Investigation of genetic diversity of Fusarium oxysporum f. sp. fragariae using PCR-RFLP. Plant Pathol. J. 33:140. doi: 10.5423/PPJ.FT.01.2017.0011
Kim, J. W., Kwon, Y., Bang, S., Kwon, H. E., Park, S., Lee, Y., et al. (2020). Unusual bridged angucyclinones and potent anticancer compounds from Streptomyces bulli GJA1. Org. Biomol. Chem. 18, 8443–8449. doi: 10.1039/d0ob01851a
Kim, S.-H., Ha, T.-K.-Q., Oh, W. K., Shin, J., and Oh, D.-C. (2016). Antiviral indolosesquiterpenoid xiamycins C–E from a halophilic actinomycete. J. Nat. Prod. 79, 51–58. doi: 10.1021/acs.jnatprod.5b00634
Kohanski, M. A., Dwyer, D. J., and Collins, J. J. (2010). How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 8, 423–435.
Lapaz, M. I., Cisneros, E. J., Pianzzola, M. J., and Francis, I. M. (2019). Exploring the exceptional properties of Streptomyces: A hands-on discovery of natural products. Am. Biol. Teach. 81, 658–664.
Lee, N., Hwang, S., Kim, J., Cho, S., Palsson, B., and Cho, B.-K. (2020). Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 18:1548–1556. doi: 10.1016/j.csbj.2020.06.024
Lehman, L. J., Orjala, J. E., Manker, D. C., Jimenez, D. R., Baum, N. A., and Marrone, P. G. (2004). Streptomyces Strain with Insecticidal Activity and Method of Using As An Insecticide. U.S. Patent No. 6,682,925. Washington, DC: U.S. Patent and Trademark Office.
Levy, S. B., and Marshall, B. (2004). Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 10, S122–S129.
Li, C., Alam, K., Zhao, Y., Hao, J., Yang, Q., Zhang, Y., et al. (2021). Mining and biosynthesis of bioactive lanthipeptides from microorganisms. Front. Bioeng. Biotechnol. 9:692466. doi: 10.3389/fbioe.2021.692466
Li, J., Sang, M., Jiang, Y., Wei, J., Shen, Y., Huang, Q., et al. (2021). Polyene-producing Streptomyces spp. From the fungus-growing termite macrotermes barneyi exhibit high inhibitory activity against the antagonistic fungus Xylaria. Front. Microbiol. 12:649962. doi: 10.3389/fmicb.2021.649962
Li, F., Chen, D., Lu, S., Yang, G., Zhang, X., Chen, Z., et al. (2018). Anti-influenza A viral butenolide from Streptomyces sp. Smu03 inhabiting the intestine of Elephas maximus. Viruses 10:356. doi: 10.3390/v10070356
Li, Y., Kong, L., Shen, J., Wang, Q., Liu, Q., Yang, W., et al. (2019). Characterization of the positive SARP family regulator PieR for improving piericidin A1 production in Streptomyces piomogeues var. Hangzhouwanensis. Synth. Syst. Biotechnol. 4, 16–24. doi: 10.1016/j.synbio.2018.12.002
Liang, Y., Chen, L., Ye, X., Anjum, K., Lian, X.-Y., and Zhang, Z. (2017). New streptophenazines from marine Streptomyces sp. 182SMLY. Nat. Prod. Res. 31, 411–417. doi: 10.1080/14786419.2016.1169419
Lyu, A., Yang, L., Wu, M., Zhang, J., and Li, G. (2020). High efficacy of the volatile organic compounds of Streptomyces yanglinensis 3-10 in suppression of Aspergillus contamination on peanut kernels. Front. Microbiol. 11:142. doi: 10.3389/fmicb.2020.00142
Macherla, V. R., Liu, J., Bellows, C., Teisan, S., Nicholson, B., Lam, K. S., et al. (2005). Glaciapyrroles A, B, and C, pyrrolosesquiterpenes from a Streptomyces sp. isolated from an Alaskan marine sediment. J. Nat. Prod. 68, 780–783. doi: 10.1021/np049597c
Mahmoudi, F., Baradaran, B., Dehnad, A., Shanehbandi, D., Mohamed Khosroshahi, L., and Aghapour, M. (2016). The immunomodulatory activity of secondary metabolites isolated from Streptomyces calvus on human peripheral blood mononuclear cells. Br. J. Biomed. Sci. 73, 97–103. doi: 10.1080/09674845.2016.1188476
Martínez-Castro, M., Barreiro, C., Romero, F., Fernandez-Chimeno, R. I., and Martín, J. F. (2011). Streptomyces tacrolimicus sp. nov., a low producer of the immunosuppressant tacrolimus (FK506). Int. J. Syst. Evol. Microbiol. 61, 1084–1088. doi: 10.1099/ijs.0.024273-0
Mary, T. R. J., Kannan, R. R., Iniyan, A. M., Ramachandran, D., and Vincent, S. G. P. (2021). Cell wall distraction and biofilm inhibition of marine Streptomyces derived angucycline in methicillin resistant Staphylococcus aureus. Microb. Pathog. 150:104712. doi: 10.1016/j.micpath.2020.104712
Maskey, R. P., Helmke, E., and Laatsch, H. (2003). Himalomycin A and B: Isolation and structure elucidation of new fridamycin type antibiotics from a marine Streptomyces isolate. J. Antibiot. 56, 942–949. doi: 10.7164/antibiotics.56.942
Medema, M. H., Kottmann, R., Yilmaz, P., Cummings, M., Biggins, J. B., Blin, K., et al. (2015). Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 11, 625–631.
Minuto, A., Spadaro, D., Garibaldi, A., and Gullino, M. L. (2006). Control of soilborne pathogens of tomato using a commercial formulation of Streptomyces griseoviridis and solarization. Crop Prot. 25, 468–475.
Mohamed, H., Hassane, A., Rawway, M., El-Sayed, M., Gomaa, A. E.-R., Abdul-Raouf, U., et al. (2021). Antibacterial and cytotoxic potency of thermophilic Streptomyces werraensis MI-S. 24-3 isolated from an Egyptian extreme environment. Arch. Microbiol. 203, 4961–4972. doi: 10.1007/s00203-021-02487-0
Muramatsu, H., and Nagai, K. (2013). Streptomyces tsukubensis sp. nov., a producer of the immunosuppressant tacrolimus. J. Antibiot. 66, 251–254. doi: 10.1038/ja.2012.116
National Institutes of Health (2007). Understanding Emerging and Re-Emerging Infectious Diseases. Bethesda, MD: National Institutes Health.
Newitt, J. T., Prudence, S. M. M., Hutchings, M. I., and Worsley, S. F. (2019). Biocontrol of cereal crop diseases using streptomycetes. Pathogens 8:78. doi: 10.3390/pathogens8020078
Nguyen, C. T., Dhakal, D., Pham, V. T. T., Nguyen, H. T., and Sohng, J.-K. (2020). Recent advances in strategies for activation and discovery/characterization of cryptic biosynthetic gene clusters in Streptomyces. Microorganisms 8:616. doi: 10.3390/microorganisms8040616
Nicault, M., Zaiter, A., Dumarcay, S., Chaimbault, P., Gelhaye, E., Leblond, P., et al. (2021). Elicitation of Antimicrobial Active Compounds by Streptomyces-Fungus Co-Cultures. Microorganisms 9:178. doi: 10.3390/microorganisms9010178
Niu, G. (2018). Genomics-driven natural product discovery in actinomycetes. Trends Biotechnol. 36, 238–241.
Niu, M., Sun, Y., Liu, X., Tang, L., and Qiu, R. (2013). Tautomycetin induces apoptosis by inactivating Akt through a PP1-independent signaling pathway in human breast cancer cells. J. Pharmacol. Sci. 121, 17–24. doi: 10.1254/jphs.12206fp
Oja, T., San Martin Galindo, P., Taguchi, T., Manner, S., Vuorela, P. M., Ichinose, K., et al. (2015). Effective antibiofilm polyketides against Staphylococcus aureus from the pyranonaphthoquinone biosynthetic pathways of Streptomyces species. Antimicrob. Agents Chemother. 59, 6046–6052. doi: 10.1128/AAC.00991-15
Ortega, H. E., Ferreira, L. L. G., Melo, W. G. P., Oliveira, A. L. L., Alvarenga, R. F. R., Lopes, N. P., et al. (2019). Antifungal compounds from Streptomyces associated with attine ants also inhibit Leishmania donovani. PLoS Negl. Trop. Dis. 13:e0007643. doi: 10.1371/journal.pntd.0007643
Ortuño-Sahagún, D., Zänker, K., Rawat, A. K. S., Kaveri, S. V., and Hegde, P. (2017). Natural immunomodulators. J. Immunol. Res. 2017:7529408.
Owen, J. G., Charlop-Powers, Z., Smith, A. G., Ternei, M. A., Calle, P. Y., Reddy, B. V. B., et al. (2015). Multiplexed metagenome mining using short DNA sequence tags facilitates targeted discovery of epoxyketone proteasome inhibitors. Proc. Natl. Acad. Sci. U. S. A. 112, 4221–4226. doi: 10.1073/pnas.1501124112
Pan, G., Xu, Z., Guo, Z., Ma, M., Yang, D., Zhou, H., et al. (2017). Discovery of the leinamycin family of natural products by mining actinobacterial genomes. Proc. Natl. Acad. Sci. U. S. A. 114, E11131–E11140. doi: 10.1073/pnas.1716245115
Passari, A. K., Mishra, V. K., Singh, G., Singh, P., Kumar, B., Gupta, V. K., et al. (2017). Insights into the functionality of endophytic actinobacteria with a focus on their biosynthetic potential and secondary metabolites production. Sci. Rep. 7:11809.
Peng, F., Zhang, M.-Y., Hou, S.-Y., Chen, J., Wu, Y.-Y., and Zhang, Y.-X. (2020). Insights into Streptomyces spp. isolated from the rhizospheric soil of Panax notoginseng: Isolation, antimicrobial activity and biosynthetic potential for polyketides and non-ribosomal peptides. BMC Microbiol. 20:143. doi: 10.1186/s12866-020-01832-5
Perin, J., Mulick, A., Yeung, D., Villavicencio, F., Lopez, G., Strong, K. L., et al. (2022). Global, regional, and national causes of under-5 mortality in 2000–19: An updated systematic analysis with implications for the Sustainable Development Goals. Lancet Child Adolesc. Health 6, 106–115. doi: 10.1016/S2352-4642(21)00311-4
Pham, V. T. T., Nguyen, C. T., Dhakal, D., Nguyen, H. T., Kim, T.-S., and Sohng, J. K. (2021). Recent Advances in the Heterologous Biosynthesis of Natural Products from Streptomyces. Appl. Sci. 11:1851.
Procópio, R. E., Silva, I. R., Martins, M. K., Azevedo, J. L., and Araújo, J. M. (2012). Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16, 466–471.
Rahman, M., Islam, M. Z., Islam, M., and Ul, A. (2011). Antibacterial activities of Actinomycete isolates collected from soils of Rajshahi, Bangladesh. Biotechnol. Res. Int. 2011:6. doi: 10.4061/2011/857925
Rey, T., and Dumas, B. (2017). Plenty is no plague: Streptomyces symbiosis with crops. Trends Plant Sci. 22, 30–37. doi: 10.1016/j.tplants.2016.10.008
Riahi, K., Hosni, K., Raies, A., and Oliveira, R. (2019). Unique secondary metabolites of a Streptomyces strain isolated from extreme salty wetland show antioxidant and antibacterial activities. J. Appl. Microbiol. 127, 1727–1740. doi: 10.1111/jam.14428
Rigali, S., Titgemeyer, F., Barends, S., Mulder, S., Thomae, A. W., Hopwood, D. A., et al. (2008). Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9, 670–675.
Rossiter, S. E., Fletcher, M. H., and Wuest, W. M. (2017). Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 117, 12415– 12474.
Rotow, J., and Bivona, T. G. (2017). Understanding and targeting resistance mechanisms in NSCLC. Nat. Rev. Cancer 17, 637–658.
Ruuskanen, O., and Järvinen, A. (2014). Editorial commentary: What is the real role of respiratory viruses in severe community-acquired pneumonia? Clin. Infect. Dis. 59, 71–73. doi: 10.1093/cid/ciu242
Salwan, R., and Sharma, V. (2020). Bioactive compounds of Streptomyces: Biosynthesis to applications. Stud. Nat. Prod. Chem. 64, 467–491.
Santos, E. F., Teixeira, M. F. S., Converti, A., Porto, A. L. F., and Sarubbo, L. A. (2019). Production of a new lipoprotein biosurfactant by Streptomyces sp. DPUA1566 isolated from lichens collected in the Brazilian Amazon using agroindustry wastes. Biocatal. Agric. Biotechnol. 17, 142–150.
Shakeel, Q., Lyu, A., Zhang, J., Wu, M., Li, G., Hsiang, T., et al. (2018). Biocontrol of Aspergillus flavus on peanut kernels using Streptomyces yanglinensis 3-10. Front. Microbiol. 9:1049. doi: 10.3389/fmicb.2018.01049
Shang, N.-N., Zhang, Z., Huang, J.-P., Wang, L., Luo, J., Yang, J., et al. (2018). Glycosylated piericidins from an endophytic Streptomyces with cytotoxicity and antimicrobial activity. J. Antibiot. 71, 672–676. doi: 10.1038/s41429-018-0051-1
Sharma, V., Kaur, R., and Salwan, R. (2021). Streptomyces: Host for refactoring of diverse bioactive secondary metabolites. 3 Biotech 11:340. doi: 10.1007/s13205-021-02872-y
Shih, H.-D., Liu, Y.-C., Hsu, F.-L., Mulabagal, V., Dodda, R., and Huang, J.-W. (2003). Fungichromin: A substance from Streptomyces padanus with inhibitory effects on Rhizoctonia solani. J. Agric. Food Chem. 51, 95–99. doi: 10.1021/jf025879b
Shimizu, M., Igarashi, Y., Furumai, T., Onaka, H., and Kunoh, H. (2004). Identification of endophytic Streptomyces sp. R-5 and analysis of its antimicrobial metabolites. J. Gen. Plant Pathol. 70, 66–68.
Shin, H.-J., Jeong, H.-S., Lee, H.-S., Park, S.-K., Kim, H.-M., and Kwon, H.-J. (2007). Isolation and structure determination of streptochlorin, an antiproliferative agent from a marine-derived Streptomyces sp. 04DH110. J. Microbiol. Biotechnol. 17, 1403–1406.
Singh, S., and Khajuria, R. (2018). New and Future Developments in Microbial Biotechnology and Bioengineering. Amsterdam: Elsevier.
Siupka, P., Hansen, F. T., Schier, A., Rocco, S., Sřrensen, T., and Piotrowska-Seget, Z. (2021). Antifungal activity and biosynthetic potential of new Streptomyces sp. MW-W600-10 strain isolated from coal mine water. Int. J. Mol. Sci. 22:7441. doi: 10.3390/ijms22147441
Sivalingam, P., Hong, K., Pote, J., and Prabakar, K. (2019). Extreme environment Streptomyces: Potential sources for new antibacterial and anticancer drug leads?. Int. J. Microbiol. 2019:5283948 doi: 10.1155/2019/5283948
Sobolevskaya, M. P., Denisenko, V. A., Moiseenko, A. S., Shevchenko, L. S., Menzorova, N. I., Sibirtsev, Y. T., et al. (2007). Bioactive metabolites of the marine actinobacterium Streptomyces sp. KMM 7210. Russ. Chem. Bull. 56, 838–840.
Song, C., Luan, J., Li, R., Jiang, C., Hou, Y., Cui, Q., et al. (2020). RedEx: A method for seamless DNA insertion and deletion in large multimodular polyketide synthase gene clusters. Nucleic Acids Res. 48:e130. doi: 10.1093/nar/gkaa956
Sudhir, P. V., Ranjan, D. S., and Ganpat, A. N. (2010). Antifungal agents, WO2009025733. Expert Opin. Ther. Pat. 20, 137–143. doi: 10.1517/13543770903338768
Sujatha, P., Raju, K. B., and Ramana, T. (2005). Studies on a new marine streptomycete BT-408 producing polyketide antibiotic SBR-22 effective against methicillin resistant Staphylococcus aureus. Microbiol. Res. 160, 119–126. doi: 10.1016/j.micres.2004.10.006
Suleman, P., Al-Musallam, A., and Menezes, C. A. (2002). The effect of biofungicide Mycostop on Ceratocystis radicicola, the causal agent of black scorch on date palm. Biocontrol 47, 207–216.
Suzuki, N., Ohtaguro, N., Yoshida, Y., Hirai, M., Matsuo, H., Yamada, Y., et al. (2015). A compound inhibits biofilm formation of Staphylococcus aureus from Streptomyces. Biol. Pharm. Bull. 38, 889–892. doi: 10.1248/bpb.b15-00053
Syed, D. G., Agasar, D., and Pandey, A. (2009). Production and partial purification of α-amylase from a novel isolate Streptomyces gulbargensis. J. Ind. Microbiol. Biotechnol. 36, 189–194. doi: 10.1007/s10295-008-0484-9
Tan, G. Y. A., Franco, C. M. M., Edrada-Ebel, R., Latip, J., and Zin, N. M. (2013). Streptomyces kebangsaanensis sp. nov., an endophytic actinomycete isolated from an ethnomedicinal plant, which produces phenazine-1-carboxylic acid. Int. J. Syst. Evol. Microbiol. 63, 3733–3738. doi: 10.1099/ijs.0.047878-0
Tan, L. T.-H., Ser, H.-L., Yin, W.-F., Chan, K.-G., Lee, L.-H., and Goh, B.-H. (2015). Investigation of antioxidative and anticancer potentials of Streptomyces sp. MUM256 isolated from Malaysia mangrove soil. Front. Microbiol. 6:1316. doi: 10.3389/fmicb.2015.01316
Tanvir, R., Sajid, I., and Hasnain, S. (2014). Larvicidal potential of Asteraceae family endophytic actinomycetes against Culex quinquefasciatus mosquito larvae. Nat. Prod. Res. 28, 2048–2052. doi: 10.1080/14786419.2014.919579
Thaker, M. N., Wang, W., Spanogiannopoulos, P., Waglechner, N., King, A. M., Medina, R., et al. (2013). Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 31, 922–927.
Vasilchenko, A. S., Julian, W. T., Lapchinskaya, O. A., Katrukha, G. S., Sadykova, V. S., and Rogozhin, E. A. (2020). A novel peptide antibiotic produced by Streptomyces roseoflavus strain INA-Ac-5812 with directed activity against Gram-positive bacteria. Front. Microbiol. 11:556063. doi: 10.3389/fmicb.2020.556063
Viaene, T., Langendries, S., Beirinckx, S., Maes, M., and Goormachtig, S. (2016). Streptomyces as a plant’s best friend?. FEMS Microbiol. Ecol. 92:fiw119. doi: 10.1093/femsec/fiw119
Vurukonda, S. S. K. P., Giovanardi, D., and Stefani, E. (2018). Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19:952. doi: 10.3390/ijms19040952
Waksman, S. A. (1953). Streptomycin: Background, isolation, properties, and utilization. Science 118, 259–266.
Wang, J., Nong, X.-H., Amin, M., and Qi, S.-H. (2018). Hygrocin C from marine-derived Streptomyces sp. SCSGAA 0027 inhibits biofilm formation in Bacillus amyloliquefaciens SCSGAB0082 isolated from South China Sea gorgonian. Appl. Microbiol. Biotechnol. 102, 1417–1427. doi: 10.1007/s00253-017-8672-z
Wang, L., Ravichandran, V., Yin, Y., Yin, J., and Zhang, Y. (2019). Natural products from mammalian gut microbiota. Trends Biotechnol. 37, 492–504.
Ward, A. C., and Allenby, N. E. E. (2018). Genome mining for the search and discovery of bioactive compounds: The Streptomyces paradigm. FEMS Microbiol. Lett. 365:fny240. doi: 10.1093/femsle/fny240
Wu, C., Shang, Z., Lemetre, C., Ternei, M. A., and Brady, S. F. (2019). Cadasides, calcium-dependent acidic lipopeptides from the soil metagenome that are active against multidrug-resistant bacteria. J. Am. Chem. Soc. 141, 3910–3919. doi: 10.1021/jacs.8b12087
Wu, S. J., Fotso, S., Li, F., Qin, S., Kelter, G., Fiebig, H. H., et al. (2006). N-Carboxamido-staurosporine and Selina-4 (14), 7 (11)-diene-8, 9-diol, New Metabolites from a Marine Streptomyces sp. J. Antibiot. 59, 331–337. doi: 10.1038/ja.2006.46
Wu, S. J., Fotso, S., Li, F., Qin, S., and Laatsch, H. (2007). Amorphane sesquiterpenes from a marine Streptomyces sp., 1. J. Nat. Prod. 70, 304–306.
Xia, H., Li, X., Li, Z., Zhan, X., Mao, X., and Li, Y. (2020). The application of regulatory cascades in Streptomyces: Yield enhancement and metabolite mining. Front. Microbiol. 11:406. doi: 10.3389/fmicb.2020.00406
Zhang, L., Song, T., Wu, J., Zhang, S., Yin, C., Huang, F., et al. (2020). Antibacterial and cytotoxic metabolites of termite-associated Streptomyces sp. BYF63. J. Antibiot. 73, 766–771. doi: 10.1038/s41429-020-0334-1
Zhao, Q., Wang, L., and Luo, Y. (2019). Recent advances in natural products exploitation in Streptomyces via synthetic biology. Eng. Life Sci. 19, 452–462. doi: 10.1002/elsc.201800137
Zhou, L.-F., Wu, J., Li, S., Li, Q., Jin, L.-P., Yin, C.-P., et al. (2021). Antibacterial potential of termite-associated Streptomyces spp. ACS Omega 6, 4329–4334. doi: 10.1021/acsomega.0c05580
Keywords: Streptomyces, natural products, bioactive compounds, genome mining, metagenomics
Citation: Alam K, Mazumder A, Sikdar S, Zhao Y-M, Hao J, Song C, Wang Y, Sarkar R, Islam S, Zhang Y and Li A (2022) Streptomyces: The biofactory of secondary metabolites. Front. Microbiol. 13:968053. doi: 10.3389/fmicb.2022.968053
Received: 13 June 2022; Accepted: 15 August 2022;
Published: 29 September 2022.
Edited by:
Crtomir Podlipnik, University of Ljubljana, SloveniaReviewed by:
Noraziah Binti Mohamad Zin, National University of Malaysia, MalaysiaAnil Kumar Pinnaka, Institute of Microbial Technology (CSIR), India
Copyright © 2022 Alam, Mazumder, Sikdar, Zhao, Hao, Song, Wang, Sarkar, Islam, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aiying Li, YXlsaUBzZHUuZWR1LmNu; Saiful Islam, c2FpZnVsY3RnQGJjc2lyLmdvdi5iZA==; Youming Zhang, emhhbmd5b3VtaW5nQHNkdS5lZHUuY24=
 Khorshed Alam1
Khorshed Alam1 Suranjana Sikdar
Suranjana Sikdar Yi-Ming Zhao
Yi-Ming Zhao Jinfang Hao
Jinfang Hao Youming Zhang
Youming Zhang Aiying Li
Aiying Li