
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 22 July 2022
Sec. Microbe and Virus Interactions with Plants
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.925355
This article is part of the Research Topic Impact of Novel Omic Technologies on Biological Control Against Plant Pathogens View all 11 articles
A correction has been applied to this article in:
Corrigendum: Comparative genomic analysis reveals cellulase plays an important role in the pathogenicity of Setosphaeria turcica f. sp. zeae
Setosphaeria turcica f. sp. zeae and S. turcica f. sp. sorghi, the two formae speciales of S. turcica, cause northern leaf blight disease of corn and sorghum, respectively, and often cause serious economic losses. They have obvious physiological differentiation and show complete host specificity. Host specificity is often closely related to pathogen virulence factors, including secreted protein effectors and secondary metabolites. Genomic sequencing can provide more information for understanding the virulence mechanisms of pathogens. However, the complete genomic sequence of S. turcica f. sp. sorghi has not yet been reported, and no comparative genomic information is available for the two formae speciales. In this study, S. turcica f. sp. zeae was predicted to have fewer secreted proteins, pathogen-host interaction (PHI) genes and carbohydrate-active enzymes (CAZys) than S. turcica f. sp. sorghi. Fifteen and 20 polyketide synthase (PKS) genes were identified in S. turcica f. sp. zeae and S. turcica f. sp. sorghi, respectively, which maintained high homology. There were eight functionally annotated effector protein-encoding genes specifically in S. turcica f. sp. zeae, among which the encoding gene StCEL2 of endo-1, 4-β-D-glucanase, an important component of cellulase, was significantly up-regulated during the interaction process. Finally, gluconolactone inhibited cellulase activity and decreased infection rate and pathogenicity, which indicates that cellulase is essential for maintaining virulence. These findings demonstrate that cellulase plays an important role in the pathogenicity of S. turcica f. sp. zeae. Our results also provide a theoretical basis for future research on the molecular mechanisms underlying the pathogenicity of the two formae speciales and for identifying any associated genes.
Northern leaf blight caused by Setosphaeria turcica is a major disease of gramineous crops and leads to serious yield losses of cereal crops in the world, especially during the growing season when the temperature is moderate (15°C–25°C) and the dew is heavy (Ramathani et al., 2011; Galiano-carneiro and Miedaner, 2017). Under natural conditions, S. turcica infects a broad range of plants, including corn, sorghum, Sudan grasses, and other sorghum species (Robert, 1960; Bhowmik and Prasada, 1970; Martin et al., 2011). Mitra (1923) first reported a clear physiological differentiation of S. turcica, with different formae speciales. S. turcica is classified as S. turcica f. sp. zeae, S. turcica f. sp. Sorghi, and S. turcica f. sp. complexa, based on the pathogen infecting a specific host or group of hosts and producing the typical spots. S. turcica f. sp. zeae can only infect corn, S. turcica f. sp. sorghi is virulent to sorghum and Sudan grasses, whereas S. turcica f. sp. complexa infects more plants (Bergquist and Masias, 1974).
Previous studies on S. turcica mainly focused on strains isolated from corn (S. turcica f. sp. zeae). With the completion of the genome sequencing of S. turcica f. sp. zeae in recent years (Ohm et al., 2012), considerable genetic information became available to understand its infection mechanism and its interaction with corn. At present, molecular-level studies on the pathogenicity of S. turcica f. sp. zeae mainly focus on signal transduction pathways and extracellular secretions of pathogens, such as cell wall degrading enzymes, host-specific toxin, and melanin (Cuq et al., 1993; Degefu et al., 2004; Ni, 2004). Previous studies have shown that HT (from Helminthosporium turcicum) toxin can induce typical symptoms of northern leaf blight in corn. Further, 1, 8-dihydroxynaphthalene (DHN) melanin has been shown to be closely related to pathogenicity (Butler et al., 2001; Nosanchuk and Casadevall, 2003). The secretion of melanin promotes the production of adhesive cells and increases the turgor pressure, which enhances the penetration of S. turcica into the corn tissue (Lagunas-Muñoz et al., 2006). Many genes (StLAC2, StPKS and St4HNR) have been proved to play important roles in the melanin synthesis pathway (Wen et al., 2008; Zhang et al., 2011; Ma et al., 2017). Some oxidoreductases are involved in various physiological metabolic activities of pathogens. Deletion of peroxisomes might interfere with the development of pathogenic fungi, reduce virulence, and decrease the ability to resist plant defense enzymes (Segmüller et al., 2008). However, the virulence factors of S. turcica f. sp. sorghi have not yet been investigated at the molecular level.
The two formae speciales of S. turcica could not be distinguished by morphology and internal transcribed spacer (ITS), but inoculation results showed that S. turcica f. sp. zeae and S. turcica f. sp. sorghi have high specificity on host and have no obvious cross-infection (Tang et al., 2014). Meanwhile, microsatellites had also been used to distinguish S. turcica from corn and sorghum (Nieuwoudt et al., 2018). With the advent of new molecular research techniques, the study of pathogens has remarkably benefited from the information of the genome and the analysis of comparative genomics. A comparative genomic study was conducted on the two pathogens in corn, Ustilago mayais and Sporisorium reilianum; 43 variant regions were identified in the two species. These regions mainly encode secretory effectors and some virulence clusters (Schirawski et al., 2010). The specialized secondary metabolites and small secretory protein effectors of pathogens are closely related to host specificity (Buiate et al., 2017). Alternaria longipes and A. alternata can also cause tobacco brown spot disease, but comparative genomic analysis revealed that A. longipes has more plant-pathogen-associated genes, carbohydrate-active enzymes (CAZys), secreted protein genes and conditionally dispensable chromosomes (Hou et al., 2016). Therefore, exploring the differences in pathogenicity mechanism between the formae speciales of S. turcica and host interaction requires the genomic information of the two formae speciales.
It was reported that pathogens can secrete a large amount of cellulase during the pathogenesis process, which softens the host cell wall leading to faster infection rates and longer disease duration (Wanjiru et al., 2002). In cellulase-inhibited mutants of Erwinia carotovora subsp. carotovora, this process of cell wall softening was significantly reduced in potato tissues (Walker et al., 1994). Owing to the secretion of other cell wall degrading enzymes, the pathogenicity of Cochliobolus carbonum was not affected by the destruction of the CEL1 gene (Sposato et al., 1995). Furthermore, a highly aggressive strain of Phaeosphaeria nodorum secreted more cellulase than a weakly aggressive strain (Lalaoui et al., 2008). The cellulase activity of S. turcica f. sp. zeae was slightly higher than that of S. turcica f. sp. sorghi, and the cellulase genes of S. turcica f. sp. zeae and S. turcica f. sp. sorghi were significantly upregulated at 72 and 36 h after inoculation, respectively (Tang et al., 2015). D-glucono-l, 5-lactone, a mixed inhibitor of cellulase activity in Trichoderma reesei, mainly affects the activity of glucosidase, but also has a inhibitory effect on the activity of exoglucanase, endoglucanase, and related enzymes, and it can induce cellulase gene expression (Reese et al., 1971; Kou et al., 2014).
Obvious host specializations are noted in S. turcica f. sp. zeae and S. turcica f. sp. sorghi. The whole genome sequencing and comparative genomic analysis of the two formae speciales of S. turcica can help in the identification of the relevant pathogenic genes that help in host-specific interactions; the whole genome sequence of S. turcica f. sp. zeae has been published in 2012 (Ohm et al., 2012). In the present study, a strain named GD003 was isolated from sorghum leaves infected with northern leaf blight by using the monospore separation method, and then identified using Koch’s postulates and ITS sequencing of S. turcica f. sp. sorghi. Whole-genome sequencing and gene function annotations revealed important genomic information about S. turcica. Comparative genomic analysis revealed differences in the genomes between the two formae speciales, including secreted proteins, pathogen-host interaction (PHI) genes, CAZys and secondary metabolic pathways. Furthermore, we used gluconolactone to alter the pathogenicity of S. turcica f. sp. zeae and speculated that the cellulase was one of the important reasons for its pathogenicity. The study findings might provide important theoretical information for the pathogenic differentiation mechanism of the formae speciales of S. turcica and provide an effective reference for the prevention of northern leaf blight and genetic breeding of resistant varieties.
Strain GD003 was isolated from sorghum leaves infected with northern leaf blight by using the single-spore isolation method (Gao et al., 2010). The spores were transferred to water agar (WA: 17 g agar and 1 L ddH2O) by tapping the leaves, and then the single spore was directly picked up under low magnification and transferred to potato dextrose agar (PDA: 200 g potato, 20 g glucose, and 17 g agar, and 1 L ddH2O) by using a simple homemade needle. The strain GD003 was deposited in the Institute of Plant Immunology, Shenyang Agricultural University and used to study pathogenic mechanism of the pathogen for 5 years. The strain was incubated under continuous darkness at 25°C.
ITS sequences and the inoculation and were used for the identification of strain GD003. Mycelia were collected from potato dextrose broth, the DNA was separated using the modified CTAB method, and the ITS sequences were amplified using PCR by using primers ITS1 and ITS4 (Gardes and Bruns, 1993; Okori et al., 2004). The amplified product was sequenced, and phylogenetic relationships were analyzed using MEGA4.0 (Tamura et al., 2007) as well as the neighbor-joining (NJ) model. Bootstrap replication was set to 1,000, and the bootstrap value was at the branch node. Sweet sorghum variety LR115 and corn variety Huobai susceptible to northern leaf blight were obtained from Dr. Jiang (Liaoning Academy of Agricultural Sciences, China). Three germinated seeds were sown in pots having 15 cm diameter and cultivated in a greenhouse with a temperature of 21/18°C day/night and light intensities of 35–50 Klux. When the plants grew to the V6 stage, the strain incubated for 2 weeks was added to a small amount of sterile water and filtered through a double-layered gauze to form a suspension of 1 × 106 conidia per milliliters. Tween-20 was added to the prepared spore suspension to a final concentration of 0.1%, and the seedlings were inoculated the spore suspension by using a sprayer; after inoculation, the seedlings were transferred to a plastic shed for 48 h for moistening, and then transferred to a greenhouse. The leaves of plants were inspected for symptoms of infection at 14 days after inoculation.
The improved CTAB method was used to extract genomic DNA from GD003, a sorghum-specific strain of S. turcica. After DNA was qualified by electrophoresis, two DNA libraries were constructed, of which 350 bp small fragment library was sequenced at paired-end by HiSeq PE150 and 20 kb SMRT Bell library was sequenced at single-molecule by PacBio RSII. Sequencing was performed at the Beijing Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The low quality reads were filtered by the SMRT Link v5.0.1 (Li et al., 2010) and the filtered reads were assemblied to generate contigs. The relationship between the contigs were determined by SOAPdenovo2 (Luo et al., 2012) to obtain the final assembly results that reflecting the basic conditions of the sample genome, including total data, GC content, read coverage depth, and mass value distribution.
The genome sequences of S. turcica f. sp. zeae Et28A was deposited at joint genome institute (JGI) with project ID 401988. The open reading frames across the genome were predicted and filtered using Augustus software (Stanke et al., 2006), and the number, total length, average length, and proportion of encoding genes were recorded. The gene function annotation was mainly based on the comparison of protein sequences, and the local comparison tool BlastP (Gao et al., 2011) was used for homology matching with the annotation results on GenBank, gene ontology (GO; Ashburner et al., 2000), kyoto encyclopedia of genes and genomes (KEGG; Kanehisa et al., 2004), cluster of orthologous groups (COG; Tatusov et al., 2003), non-redundant protein sequence (NR; Li et al., 2002), transporter classification database (TCDB; Milton et al., 2009), Swiss-Prot (Bairoch and Apweiler, 2000), PHI (Urban et al., 2015) and CAZy (Cantarel et al., 2009) databases to obtain the corresponding functional annotation information. SignalP (Petersen et al., 2011) was used to analyze the N-terminal signal peptide of encoded proteins, TMHMM (Krogh et al., 2001) was used for transmembrane structure prediction, and TargetP (Emanuelsson et al., 2007) was used to predict the subcellular localization of encoded proteins. Proteins located extracellularly, with signal peptide and lacking transmembrane domains, were defined as secreted proteins. Further, effectors in secreted proteins were screened by EffectorP (Sperschneider et al., 2016).
The key genes for secondary metabolite syntheses were identified using antiSMASH v4.0.2 program (Medema et al., 2011), especially polyketide synthase (PKS) coding genes. The MEGA4.0 (Tamura et al., 2007) was used to compare the protein domains encoded by PKSs of two formae speciales and other plant pathogenic fungi, and to construct phylogenetic tree, including Bipolaris maydis T-toxin synthesis related to PKS1 (GenBank accession number: AAB08104), Fusarium graminearum zearalenone synthesis related to PKS4 (GenBank accession number: ABB90283), Aspergillus nidulans locastatin synthesis related to LovF (GenBank accession number: AAD34559), F. verticillioides fumonisin synthesis related to Fum1p (GenBank accession number: AAD43562), A. steynii ochratoxin synthesis related to PKS (GenBank accession number: AHZ61902), and A. alternate ACT-toxin synthesis related to ACTTS3 (GenBank accession number: BAJ14522), as well as genes closely related to melanin synthesis such as B. oryzae PKS1 (GenBank accession number: BAD22832), S. turcicia PKS (GenBank accession number: AEE68981), Ascochyta rabiei PKS1 (GenBank accession number: ACS74449), Podospora anserine PKS1 (GenBank accession number: CDP25014), Colletotrichum lagenarium PKS1 (GenBank accession number: BAA18956), A. fumigatus A1b1p (GenBank accession number: ACJ13039), Ceratocystis resinifera PKS1 (GenBank accession number: AAO60166).
The mycelium disk of strain Et28A with diameter of 1 cm was inoculated onto corn leaves in vitro, and 50–100 mg of the leaves under the disk were cut with RNA-free scissors at different infection periods (0, 24, 48, 72, and 96 h), wrapped in tin foil and immediately frozen in liquid nitrogen for 10 min, transferred to −80°C for storage. Total cellular RNA was isolated using an Ultrapure RNA Kit (CWBIO, Beijing, China), and then cDNA was synthesized using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time; TaKaRa, Tokyo, Japan). The reaction mixture contained 10 μl of TB Green Premix Ex Taq II (Tli RhaseH Plus; TaKaRa, Tokyo, Japan), 1 μl of forward primer, 1 μl of reverse primer, 2 μl of cDNA, and 6 μl of ddH2O. The reactions were performed in the CFX-96 system (BioRad, Hercules, CA, United States), and all samples were tested in triplicate and repeated twice. All reaction conditions were performed as follows: initial denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s and annealing at 60°C for 30 s. The cycle threshold (Ct) values were analyzed using CFX Manager and relative expression levels of functionally annotated effector protein-coding genes specific for S. turcica f. sp. zeae were calculated at each period according to the 2-△△Ct method.
The gluconolactone solution was sterilized and cooled, and then added to a PDA medium under sterile conditions to final concentrations of 0.2, 0.4, and 0.8% (w/v). Five millimeter agar disks containing mycelium of strain Et28A were transferred to the medium with an inoculation needle. Then the side of the hyphae was pressed down to the medium and one agar disk was placed in the center of each dish. Each treatment was repeated five times and cultured at 25°C for 5 days. The gluconolactone solution was replaced by sterile water for the control group. For each treatment group, the colony diameter was measured then the growth inhibition rate was calculated (Quiroga et al., 2001).
To analyze the effect of gluconolactone on cellulase activity, preparation of the crude enzyme solution was slightly modified based on the methods of Lee and Blackburn (1975). The agar disks containing mycelium of strain Et28A that were cultured on the PDA were added to Czaper liquid culture medium (2 g KNO3, 0.5 g KCl, 0.01 g FeSO4, 1 g K2HPO4, 0.5 g MgSO4, 10 g sodium carboxymethyl cellulose, and 1 L ddH2O) both with and without gluconolactone (0.2, 0.4, and 0.8%; w/v). Then, nine disks were inoculated in 150 ml medium and were shaken for 1 h per day (120 rpm), incubated for 15 days at 25°C in the dark, and then filtered through sterile double gauze (22 mesh). The filtrate was centrifuged at 4°C and 10,000 g for 20 min and the crude enzyme solution was the supernatant. Cellulase activity was determined based on the method described by Eveleigh et al. (2009). Briefly, 1 ml of 1% sodium carboxymethyl cellulose in 0.1 M citrate buffer (pH 4.5) and 0.5 ml extracted crude enzyme solution were placed in a test tube and then incubated in a 50°C water bath for 30 min. After the reaction mixture was cooled, 3 ml of 3, 5-dinitrosalicylic acid reagent was added, and the solution was heated to 100°C for 5 min. The absorbance at 540 nm of the reaction mixture after appropriate dilution was measured with a spectrophotometer. A cellulase activity unit (U) was defined as the amount of enzyme required to catalyze the reaction to produce 1 μmol of reducing sugar per min under specific conditions. All enzyme activity assays were repeated three times. The protein content was determined using the Coomassie brilliant blue G250 staining method (Bradford, 1976).
Analysis of expression levels was used to measure the effect of gluconolactone on endo-1, 4-β-D-glucanohydrolase encoding gene A2464. The agar disks containing the mycelium of S. turcica f. sp. zeae Et28A that were cultured on the PDA were inoculated in vitro on corn leaves at the 6–8 leaf stage. One hundred microliter gluconolactone solutions (0.2 and 0.4%, w/v) were added to the edge of the disks each day, and the control group was treated with sterile water. The expression levels of A2464 were determined at different infection periods (0, 6, 12, 24, 36, 48, 72, and 96 h). All samples were tested in triplicate and repeated twice. Finally, the infection rate and pathogenicity were determined and the inoculation method was described above. Pathogenicity was determined after culturing for 72 h in the dark at 25°C for moistening. Then it was stained with trypan blue, dehydrated with saturated chloral hydrate, and rinsed with sterile water before being placed under a microscope to observe the infection efficiency of S. turcica f. sp. zeae Et28A. Each treatment was repeated three times.
All statistical tests were calculated in SPSS Statistics 19 software. Data were represented as means ± standard error of at least three repeated experiments. p < 0.05 was defined as a statistically significant difference.
Phylogenetic tree of the ITS sequences showed that GD003, S. turcica f. sp. zeae strain QDY1307 (GenBank accession number: KJ922736.1) and S. turcica f. sp. sorghi strain LLG1302 (GenBank accession number: KJ922728.1) were in the same branch (only four differential bases; Figure 1A). The ITS sequences do not distinguish the two formae speciales of S. turcica. Within 14 days after inoculation, this GD003 strain formed a typical long spindle lesion on sorghum leaves, while no visible reaction was evident on corn leaves (Figure 1B), so the pathogenicity tests identified strain GD003 as S. turcica f. sp. sorghi.

Figure 1. Identification of GD003 based on ITS sequences and pathogenicity. (A) Phylogenetic analysis of ITS sequences of GD003. The numbers in parentheses indicate the accession numbers of ITS sequences of the species in GenBank. (B) Pathogenicity of GD003 strain on sorghum and corn. (a) Typical lesions of GD003 on sorghum. (b) No visible reaction of GD003 on corn.
After electrophoresis, 159.60 ng/ml DNA yielded OD260/280 of 1.87 and OD260/230 of 2.24; the fragment size was mainly distributed above 30 K, and the genome was slightly broken, which met the requirements for single-sequencing database creation. A total of 7.82 Gb of reads were obtained by sequencing the genome of S. turcica f. sp. sorghi GD003 (depth: 177×), including 938,546 reads. The length of the sequence N50 was 11,965 bp, and the average sequencing quality value was 0.86. The genome assembly revealed 22 contigs (Supplementary Fasta 1) with a total length of 44,063,561 bp and a GC content of 50.7%. The scatter diagram of S. turcica f. sp. sorghi GD003 genomic GC-depth was mostly concentrated in the range of 40%–60% (Supplementary Figure 1).
A total of 10,428 protein-coding genes (Supplementary Fasta 2) were predicted in the genome of S. turcica f. sp. sorghi GD003, accounting for 35.47% of the total length of the genome sequence, and the average length of the coding genes was 1,499 bp. In contrast, only 8,276 protein-coding genes (Supplementary Fasta 3) were found in the genome of S. turcica f. sp. zeae Et28A, accounting for 26.86% of the total length of the genomic sequence, and the average length of the coding genes was 1,396 bp (Table 1). From the gene distribution map, the most abundant S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A genes were found to be concentrated in the region of 2,500 bp or more, including 1,431 and 882 genes, respectively.
Different numbers of genes in the two formae speciales genomes were annotated in each functional database (Table 2). By comparing S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A, 704 and 521 secreted proteins were predicted, respectively (Supplementary Figure 2), containing 161 and 137 effectors, which were required for the pathogens to act directly or indirectly on the hosts. The findings suggested that the S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A could directly secrete 42 and 33 small cysteine-rich proteins (SCRPs; the number of amino acids is less than or equal to 200, and cysteine content is 4% or more), respectively, of which 30 SCRPs existed in both formae speciales.
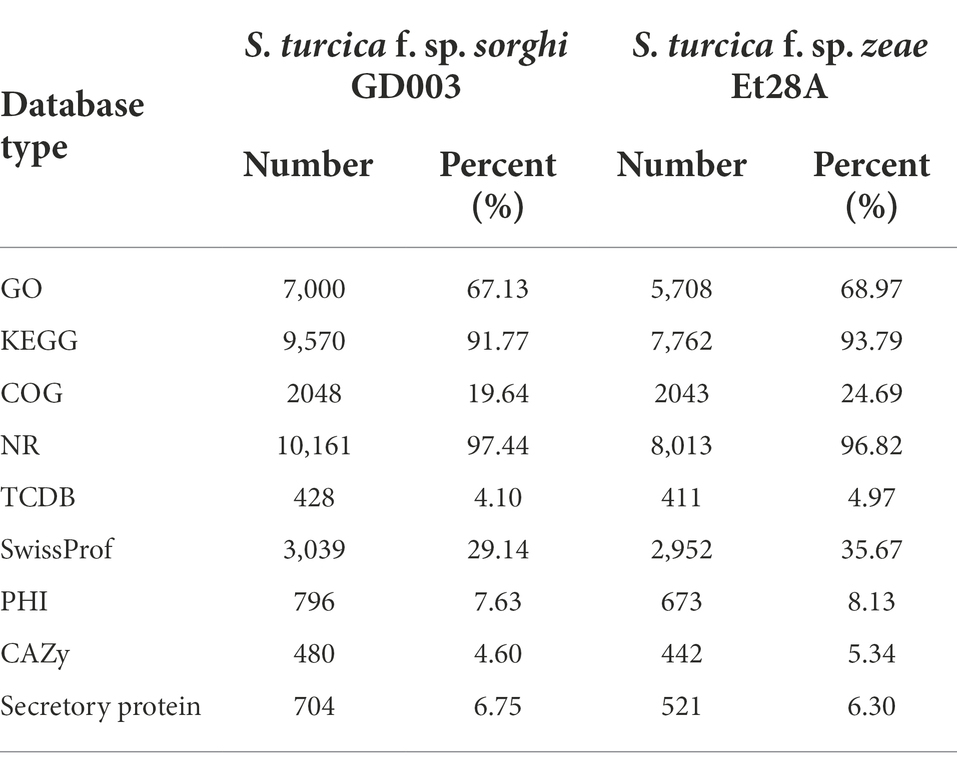
Table 2. Number of genes annotated by different functional databases in Setosphaeria turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A genomes.
In the present study, 796 and 673 PHI genes were detected in S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A, covering 609 and 539 PHI accessions, respectively, but they had seven types of phenotypic mutations (Supplementary Table 1). The PHI information of 655 genes was identical in the two formae speciales. Among 141 PHI genes unique to S. turcica f. sp. sorghi GD003, excluding 81 genes that did not affect the pathogenicity, phenotypic mutants of 45 genes (accession number: PHI139, PHI323, PHI339, PHI3837, PHI3865, PHI4992, etc.) had reduced virulence. Further searches revealed that the secreted proteins of S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A contained 62 and 54 PHI related genes, respectively, of which 51 PHI genes were homologous and 11 PHI genes were specific in S. turcica f. sp. sorghi GD003.
Blastp alignment was performed using the genomically encoded proteins and CAZy database, and 480 and 442 CAZys were identified from S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A, respectively; the related CAZys were mainly involved in carbohydrate degradation, modification, and biosynthesis (Figure 2). The most common CAZys were glycoside hydrolases (GHs) containing 224 and 216 genes, respectively, and the remaining were auxiliary activities (AAs), glycosyltransferases (GTs), carbohydrate binding modules (CBMs), carbohydrate esterases (CEs) and polysaccharide lyases (PLs). Further analysis found that 216 (30.68%) and 178 (34.17%) CAZy genes were identified in the secreted proteins of S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A, respectively, and most of these genes were associated with GHs in the subfamily classification (Supplementary Table 2).
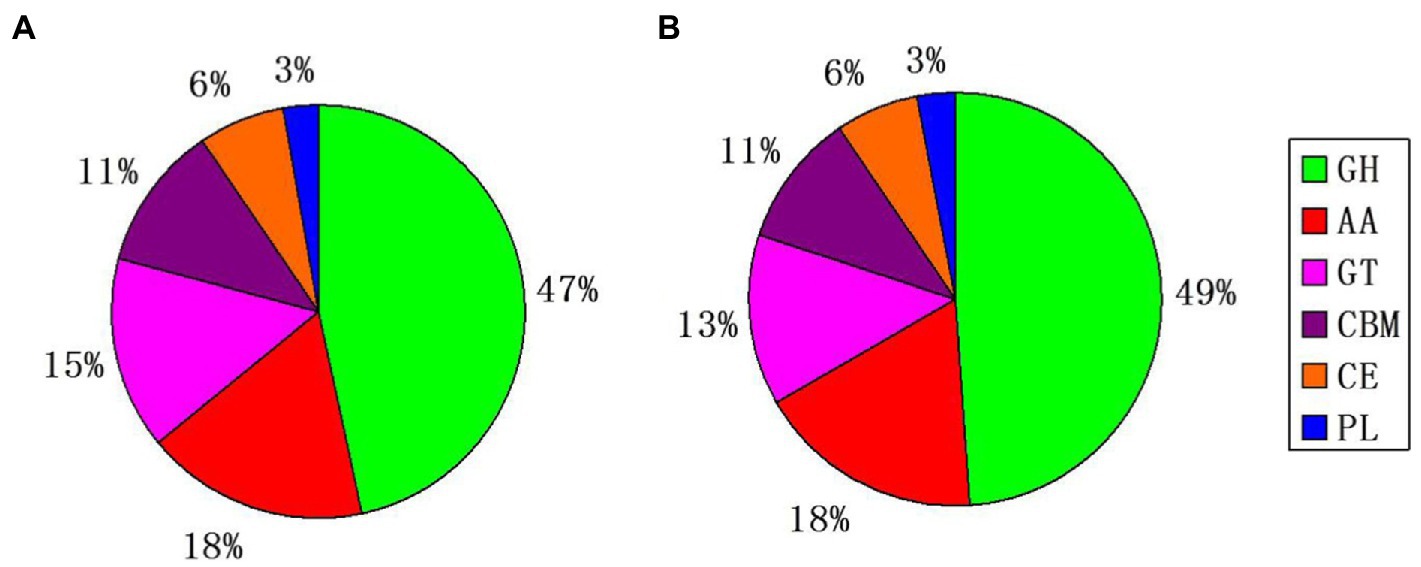
Figure 2. Classification and comparison of the two formae speciales by using whole genome CAZy database. (A) Setosphaeria turcica f. sp. sorghi GD003. (B) S. turcica f. sp. zeae Et28A. GH, glycoside hydrolase; AA, auxiliary activity; GT, glycosyltransferase; CBM, carbohydrate-binding module; CE, carbohydrate esterase, and PL, polysaccharide lyase.
Further, 20 and 15 PKSs were predicted for secondary metabolic gene clusters in S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A genomes, respectively (Supplementary Table 3). The PKS genes of the two formae speciales were mainly divided into two types (Figure 3A). The core domain of type I consisted of ketoacyl synthase (KS), acyltransferase (AT) and dehydratase (DH), including 16 S. turcica f. sp. sorghi GD003 PKSs, 12 S. turcica f. sp. zeae Et28A PKSs, and 6 other PKSs related to the synthesis of phytopathogenic mycotoxins. The other type of core domain was KS + AT, including 4 S. turcica f. sp. sorghi GD003 PKSs, 3 S. turcica f. sp. zeae Et28A PKSs, and 7 known melanin synthesis-related PKSs from phytopathogenic fungi. Further analysis of PKSs associated with melanin synthesis revealed that the two formae speciales shared the same core domain, including one KS, one AT, two acyl carrier proteins (ACPs) and one thioesterase (TE), and both coding sequences were 99.48% similar to the known S. turcica PKS (GenBank accession number: AEE68981; Figure 3B).
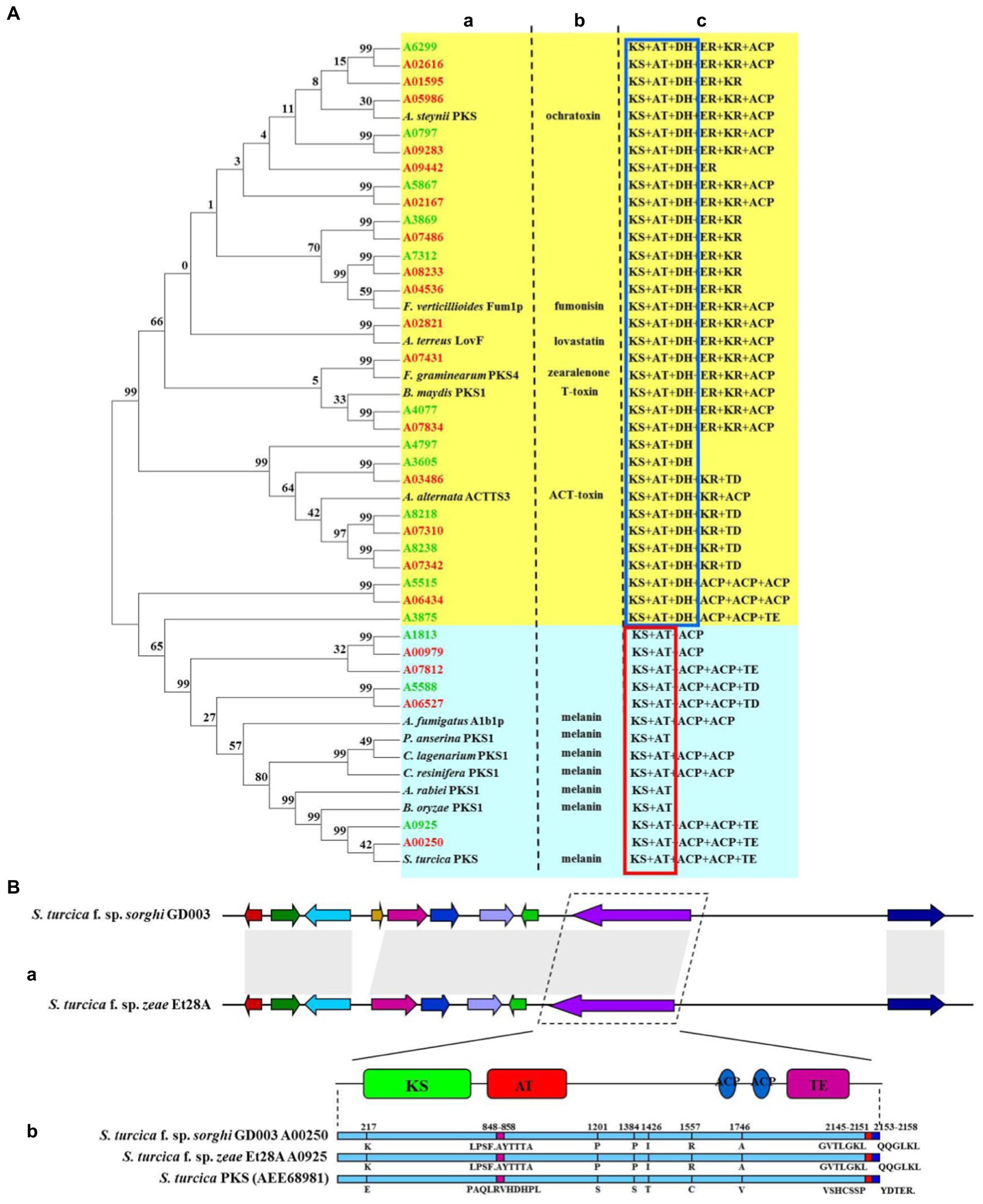
Figure 3. Comparative analysis of polyketide synthase (PKS) key genes of two formae speciales of Setosphaeria turcica. (A) Phylogeny and domain comparison of PKSs from Setosphaeria turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A with that from other known plant pathogenic fungi. (a) Phylogenetic analysis of proteins encoded by PKS genes. (b) PKS-related toxin and melanin from other known phytopathogenic fungi. (c) PKS-associated protein domains. KS, ketoacyl synthase; AT, acyltransferase; DH, dehydratase; ER, enoylreductase; KR, ketoacyl reductase; ACP, acyl carrier protein; TE, thioesterase; and TD, tudor domain. Words marked in green and red indicate gene names for S. turcica f. sp. zeae Et28A and S. turcica f. sp. sorghi GD003, respectively. The red and blue boxes indicate common domains. (B) Comparison of gene clusters and core gene-coding sequences of melanin synthesis in S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A genomes. (a) Melanin synthesis gene clusters of the two formae speciales. (b) Differences in protein sequence of the melanin synthesis core gene from known S. turcica.
In our study, 21 effector protein-coding genes were found specifically in S. turcica f. sp. zeae Et28A, and 13 of them were defined as encoding hypothetical proteins. 18S rRNA was used as a reference gene, primer sequences of eight functionally annotated effector protein-encoding genes specific to S. turcica f. sp. zeae were designed (Table 3). Compared with the 0 h control, only A1078 was downregulated after inoculation, while the expression levels of A0353, A2199, A2464, A3017, A6166, and A8125 were significantly upregulated. The expression level of A2464 after 72 h of inoculation increased by more than 150-fold compared with that before (Figure 4).
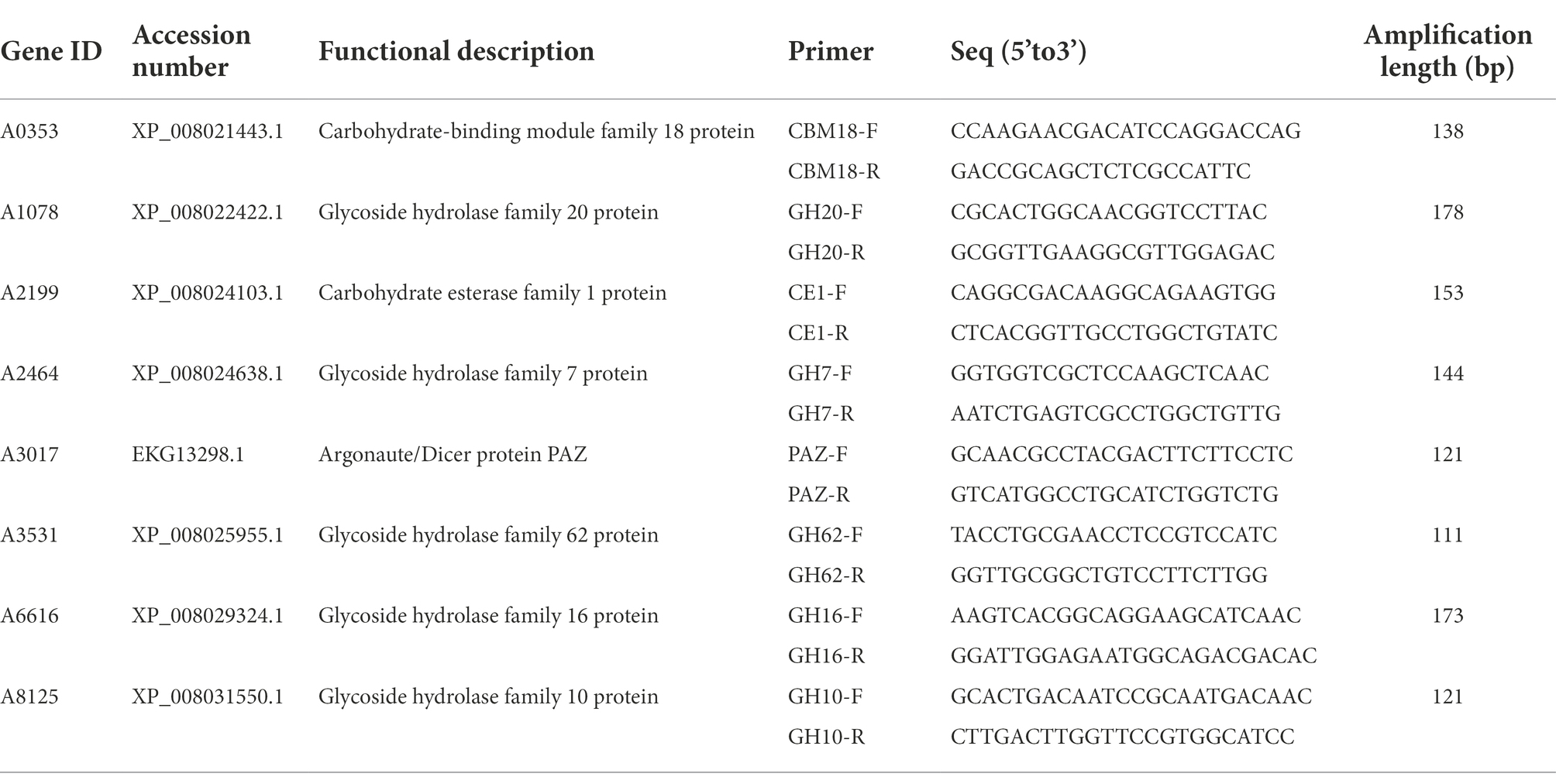
Table 3. Primer sequences and amplification lengths of functionally annotated effector protein-coding genes in Setosphaeria turcica f. sp. zeae.
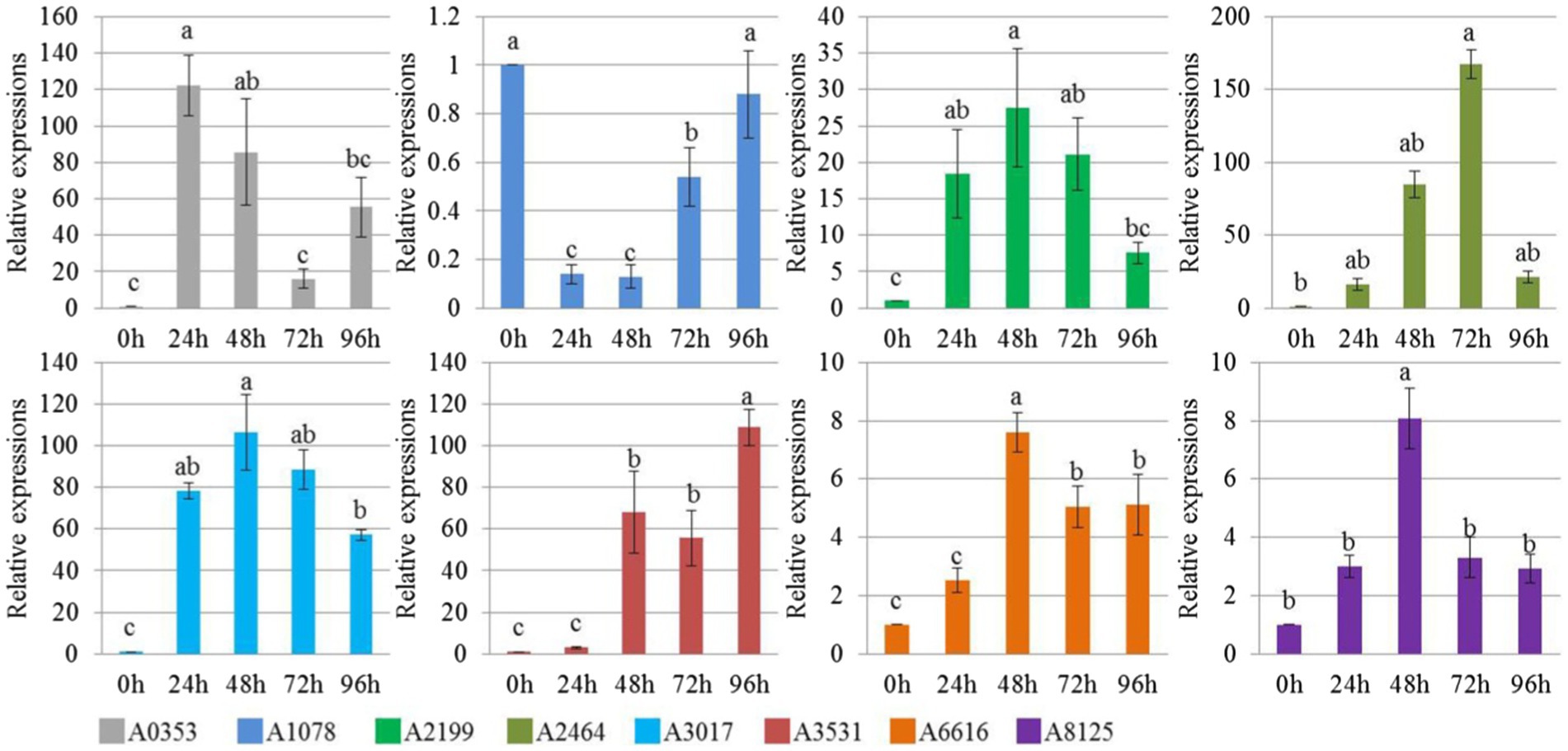
Figure 4. Relative expression level of Setosphaeria turcica f. sp. zeae specific effector coding genes at different infection periods postinoculation. Error bars represent means ± SE of three repeated experiments (n = 3). Different letters indicate significant differences (p < 0.05). A0353 encodes carbohydrate binding module family 18 protein; A1078 encodes glycoside hydrolase family 20 protein; A2199 encodes carbohydrate esterase family 1 protein; A2464 encodes glycoside hydrolase family 7 protein; A3017 encodes Argonaute/Dicer protein PAZ; A3531 encodes glycoside hydrolase family 62 protein; A6616 encodes glycoside hydrolase family 16 protein; A8125 encodes glycoside hydrolase family 10 protein.
At a concentration of 0.4% (w/v), hyphal growth was significantly inhibited. When the concentration increased to 0.8% (w/v), the inhibition rate reached 46.12%, which indicated that gluconolactone inhibited the hyphal growth of S. turcica f. sp. zeae Et28A (Figure 5A). The cellulase activity of S. turcica f. sp. zeae Et28A was significantly inhibited by different concentrations of gluconolactone (p < 0.05), and the effect increased with increasing concentration (Figure 5B). Despite different gluconolactone concentrations (0.2 and 0.4%, w/v), StCEL2 gene expression showed a consistent trend, all peaked at 72 h. StCEL2 gene expression level increased with increasing gluconolactone concentration during the same infection period (Figure 5C). Differently-treated pathogens could invade the host and caused corn leaf lesions after 72 h of inoculation. However, the number of invasion sites observed in the control group was significantly higher than that in the gluconolactone treatment group. Furthermore, the number of invasion sites decreased with increasing concentration (Figure 5D), indicating that gluconolactone affected the infection and pathogenicity of S. turcica f. sp. zeae Et28A.
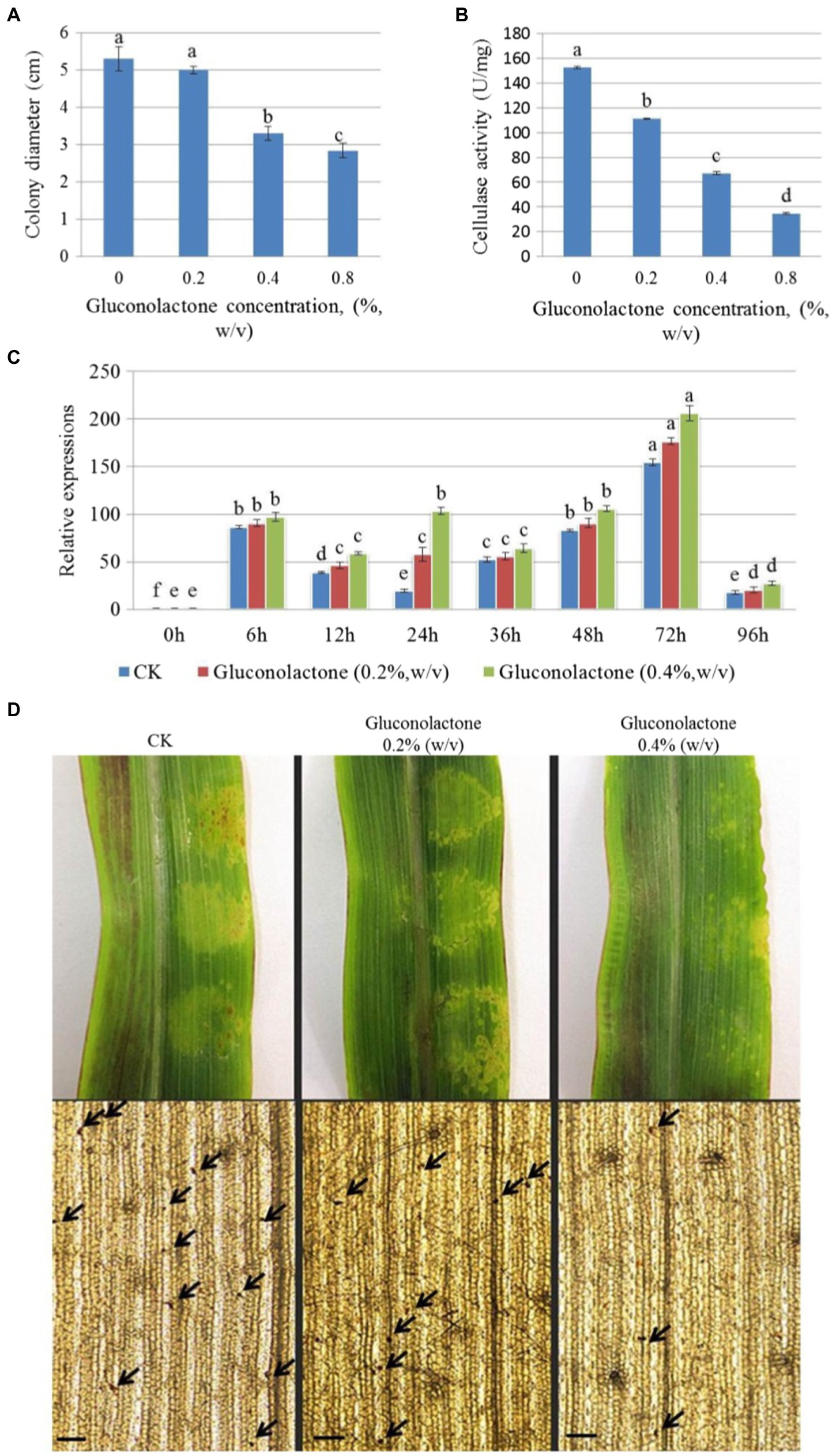
Figure 5. Effect of gluconolactone on Setosphaeria turcica f. sp. zeae. (A) Effect of gluconolactone on colony diameter. Error bars are presented as means ± SE (n = 5). Different letters indicate significant differences (p < 0.05). (B) Effect of gluconolactone on cellulase activity. Error bars indicate means ± SE (n = 3). (C) Effect of glucolactone on the expression level of endo-1, 4-β-D-glucanase encoding gene StCEL2. Data represent means ± SE (n = 3). (D) Effect of gluconolactone on the pathogenicity and infection rate. Arrow positions are the infection dots. Scale bars are equal to 50 μm.
Previous studies have shown that the variability of ITS sequences among different formae speciales of the same fungus is limited. For example, the ITS and EF-alpha elongation factor analyses cannot identify the formae speciales of F. oxysporum (Zhang et al., 2013). Since the ITS information of GD003, S. turcica f. sp. zeae strain QDY1307 (GenBank accession number: KJ922736.1), and S. turcica f. sp. sorghi strain LLG1302 (GenBank accession number: KJ922728.1) reveal a difference of only four bases, it proves once again that ITS could not identify the formae speciales of S. turcica. Because of the obvious host-specificity between the formae speciales (Mitra, 1923), the inoculation of sorghum leaves showed a clear feature of northern leaf blight. Finally, GD003 was identified as S. turcica f. sp. sorghi and named S. turcica f. sp. sorghi GD003.
In this study, we comprehensively reported the first genome information of S. turcica f. sp. sorghi and compared it with the published genomic data of S. turcica f. sp. zeae Et28A (JGI ID: 401988), which can provide a reference for revealing the pathogenic mechanism of S. turcica. Different strains of the same species also have large differences in genomic structure and encoded proteins (Condon et al., 2013). Several random amplified polymorphic DNA haplotypes uniquely present in sorghum strains of S. turcica were not observed in strains collected from corn (Borchardt et al., 1998; Ferguson and Carson, 2004). Genetic differences were confirmed in two formae speciales of S. turcica by universally primed polymerase chain reaction (Tang et al., 2014). The genome size of S. turcica f. sp. zeae GD003 (44.06 Mb) is greater than that of S. turcica f. sp. sorghi Et28A (43.01 Mb), which may be caused by the pressure of host selection (Thrall et al., 2002). Changes of GC content were speculated to prompt Curvularia lunata to mutate more frequently in virulence differentiation (Gao et al., 2014). The GC content of the same species shows a concentrated distribution in the sequencing depth profile (Forsdyke, 1996). Concentration of most of the points in the distribution map in a narrow range (40%–60%) indicates no pollution in the assembly results.
Secreted protein is a generic term for a class of proteins that are produced by cells at specific times and conditions and transported extracellularly, which is often directly related to PHI, is a candidate effector, and is more likely to exhibit population differences during natural selection (Klein et al., 1996). Therefore, studies on secreted proteins might help understand host specificity issues in PHIs. The secreted proteins related to pathogenicity are mainly avirulence genes of pathogens, products of pathogenic genes, and related regulatory proteins (Rep, 2005), such as cell wall degrading enzymes can reduce or even overcome the host’s barrier to pathogen infection (Brito et al., 2006), elicitor substances can induce pathogenic responses in host plants (Kamoun et al., 1998), and some of the secreted proteins can also degrade the antifungal toxin produced by host plants to facilitate the progression of infection (Sperschneider et al., 2016). In this study, more secreted proteins were predicted in S. turcica f. sp. sorghi GD003. Many phytopathogenic fungi can also directly secrete SCRPs, which have a close relationship with the mechanism of pathogenesis (Rep et al., 2004). These small molecular proteins can act as virulence effector proteins and have carbohydrate-binding activity; they can directly play a role by interfering with host cell signal transduction or inhibiting host PAMP-triggered immunity responses (Marcet-Houben et al., 2012). Twelve SCRPs unique to S. turcica f. sp. sorghi GD003 were all uncharacterized proteins, and further studies are warranted to determine the function of SCRPs specific to the two formae speciales in PHI.
The genes involved in the interaction between pathogens and hosts play a crucial role in pathogenesis; their products are directly involved in the adaptation and response of pathogens to the host-infecting environment, and the secreted elicitors can directly induce the host plants to express the symptom (Barrett et al., 2009). The PHI database integrates pathogen-related genes to different hosts such as animals, plants, and microorganisms and is widely used to investigate plant pathogen genomes and genes implicated in virulence (Winnenburg et al., 2006; Urban et al., 2015). In this study, 45 genes with phenotypic mutations with reduced pathogenicity were specifically present in S. turcica f. sp. sorghi GD003, such as PHI139 was required for Cryptococcus neoformans to maintain virulence (Chang and Kwon-chung, 1999), the loss of PHI339 significantly reduced the pathogenicity of C. lindemuthianum (Siriputthaiwan et al., 2005), and PHI4992 was required for Candida albicans biofilm formation in vitro and in vivo (Desai et al., 2015). Differences in these PHI-related genes might lead to differences in pathogenicity between the two formae speciales. Significantly, there were 11 PHI genes specific for the secreted protein encoding genes of S. turcica f. sp. sorghi GD003, the knockout of PHI323 significantly reduced the virulence of Verticillium fungicola (Amey et al., 2003), and PHI3865 was required for Penicillium expansum to cause blue mold rot (Barad et al., 2012), while the related genes PHI569, PHI2849, and PHI6126 reported in Fusarium did not affect their pathogenicity.
The plant pathogenic fungi CAZys play a crucial role in degrading plant cell walls, breaking through host passive defense systems, and establishing PHI relationships (Cantarel et al., 2009). The enzymes encoded by the GH, CE, and PL family genes play a role in depolymerizing cell walls (Walton, 1994), and they had only slight differences between the two formae speciales. Considering the errors in gene sequencing and energy prediction, the CAZy species and quantity of the two formae speciales could basically be thought to be consistent at the genome level. Further, 32 and 26 CBM family genes were found in the S. turcica f. sp. sorghi GD003 and S. turcica f. sp. zeae Et28A genomes, respectively; the modules of approximately 40 residues in the family genes were unique to the fungi and played a key role in cellulose degradation in plant cell walls (Quentin et al., 2002). Comparison of CAZy annotation results in secreted proteins of the two formae speciales revealed that the content of GHs in S. turcica f. sp. sorghi GD003 was significantly higher than that in S. turcica f. sp. zeae Et28A, which might suggest that the former has stronger pathogenic ability than the latter.
Among the secondary metabolites of plant pathogens, melanin and toxin are the two key pathogenic factors in S. turcica. The synthesis of these two virulence substances is mainly mediated by PKSs. PKS1 had been successfully cloned in B. oryzae and C. resinifera and was found to affect the synthesis of melanin and reduce pathogenicity (Moriwaki et al., 2004; Tanguay et al., 2006). StPKS of S. turcica f. sp. zeae was shown to play a role in the DHN melanin synthesis pathway, and its decreased expression reduced melanin production (Zhang et al., 2011). Moreover, S. turcica f. sp. zeae Et28A had an additional betaenone biosynthetic gene cluster unlike in S. turcica f. sp. sorghi GD003; this cluster acted as a phytotoxin that inhibited multiple protein kinases (Patrick and Heimbrook, 1996) and caused significant growth inhibition of Beta vulgaris (Haraguchi et al., 1983). This study was the first to identify the core domain of the PKS genes related to melanin and toxin biosynthesis of the two formae speciales of S. turcica, and it was found that they had high homology with PKS genes of other pathogenic fungi (common domain of toxin-synthesized PKS genes: KS + AT + DH; common domain of melanin-synthesized PKS genes: KS + AT). Differences in key genes involved in secondary metabolite synthesis have less effect on pathogenic differentiation of the two formae speciales of S. turcica.
The determination of the host range in plant pathogens is often closely related to the fungal effectors (Baroncelli et al., 2016). A total of 346 candidate effectors in S. turcica were identified by time-course RNAseq, and SIX13-like proteins of S. turcica isolated from corn and sorghum were demonstrated to have host-specific polymorphisms (Human et al., 2020). In this study, we first excluded the influence of shared effector protein coding genes of pathogens on host specificity, and only analyzed the expression of specific effector protein-coding genes in S. turcica f. sp. zeae during the interaction with corn. In the future, further verification of the functions of differential genes is required. During the interaction between plants and pathogens, the activity of hydrolase is conducive to the invasion of the pathogen and the expansion of the disease course. The hydrolytic enzymes related to pathogenicity mainly include cellulase, hemicellulase, pectinase, xylanase, etc. (van den Brink and de Vries, 2011). The significant up-regulation of α-L-arabinofuranosidase encoding gene-A3531 (targeting xylan in plant fibers) during the infection process once again proves that many pathogenic related genes are simultaneously expressed in the interaction, and the time and level of expression determine the pathogenic level of the pathogen to the host (Kim et al., 2016).
Cellulose, an important component of plant cell walls, has a stable structure, which functions effectively to resist pathogen invasion and exogenous stress (Hu et al., 2018). Many cell wall degrading enzymes produced by pathogens cooperate to degrade host cell walls and infect the host (Cooper et al., 1988). Novo et al. (2006) found high levels of activity for endo-1, 4-β-D-glucanase, and β-1, 4-D-glucosidase in invasive V. dahliae strains. Furthermore, highly invasive P. nodorum strains can produce more cellulase (Lalaoui et al., 2008). The cellulase activity of S. turcica f. sp. zeae was previously reported to be slightly higher than that of S. turcica f. sp. sorghi, and it was noted that differences in cell wall degrading enzymes might be one of the reasons for its pathogenic specialization (Tang et al., 2015). The protein encoded by A2464 was annotated as a member of the seventh family of GHs with endo-1, 4-β-D-glucanohydrolase activity (EC 3.2.1.4). This is an important component of cellulase gene and might be a cause of the pathogenicity of S. turcica f. sp. zeae Et28A. Therefore, we selected the highest expressed S. turcica f. sp. zeae specific cellulase gene (StCEL2) for analysis. In this study, we used gluconolactone to treat S. turcica f. sp. zeae and analyzed its inhibitory effect on cellulase activity, and our results are consistent with those of Holtzapple et al. (1990). Interestingly, the expression of StCEL2, an endo-1, 4-β-D-glucanase coding gene, was upregulated, and increased with increasing concentration of gluconolactone. This is probably due to gluconolactone inhibiting cellulase activity, and the pathogen taking advantage of this stress to invade the host, inducing the expression of related genes (Kou et al., 2014). In summary, gluconolactone can reduce infection rate and pathogenicity by inhibiting the cellulase activity of S. turcica f. sp. zeae. Furthermore, methods such as gene knockout will be applied to reveal the reasons of two formae speciales infect specific hosts. These results provide useful information for understanding the mechanism of infection and pathogenic differentiation of S. turcica f. sp. zeae.
In this study, we reported the genome sequence of S. turcica f. sp. sorghi and compared it with the number of genes of S. turcica f. sp. zeae in each functional database. Because of the obvious host specificity of the two formae speciales, we focused on the differences in the coding genes of secreted proteins and secondary metabolites, and pointed out the expression levels of specific effector protein-coding genes in S. turcica f. sp. zeae at different infection periods. Furthermore, the close relationship between cellulase and pathogenicity of S. turcica f. sp. zeae was determined by the inhibitory effect of enzyme activity, and it was clear that cellulase was one of the important factors of its pathogenicity. In summary, our results provide a novel ideas for studying the interaction between pathogens and the host, and lay a strong foundation for further knockout of cellulase genes and mining of pathogenicity-related genes. Obviously, our data improve the understanding of important pathogens of S. turcica, increase the genomic information of S. turcica f. sp. sorghi, and contribute to the study of pathogenic mechanisms.
The raw sequence reads of Setosphaeria turcica f. sp. sorghi have been deposited in GeneBank under the accession number PRJNA860778.
ZM, BL, and ZG conceived and designed the experiments. ZM, YH, ZZ, and XL performed the experiments and analyzed the data. ZM drafted the manuscript, which was critically revised by YX. All authors contributed to the article and approved the submitted version.
This project was supported by the National Key Technology Research and Development Program of China (2018YFD0300307, 2017YFD0300704, and 2016YFD0300704).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.925355/full#supplementary-material
Amey, R. C., Mills, P. R., Bailey, A., and Foster, G. D. (2003). Investigating the role of a Verticillium fungicola β-1, 6-glucanase during infection of Agaricus bisporus using targeted gene disruption. Fungal Genet. Biol. 39, 264–275. doi: 10.1016/S1087-1845(03)00061-6
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bairoch, A., and Apweiler, R. (2000). The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28, 45–48. doi: 10.1093/nar/28.1.45
Barad, S., Horowitz, S. B., Moscovitz, O., Lichter, A., Sherman, A., and Prusky, D. (2012). A Penicillium expansum glucose oxidase-encoding gene, GOX2, is essential for gluconic acid production and acidification during colonization of deciduous fruit. Mol. Plant Microbe Interact. 25, 779–788. doi: 10.1094/MPMI-01-12-0002
Baroncelli, R., Amby, D. B., Zapparata, A., Sarrocco, S., Vannacci, G., Le Floch, G., et al. (2016). Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genomics 17:555. doi: 10.1186/s12864-016-2917-6
Barrett, L. G., Kniskern, J. M., Bodenhausen, N., Zhang, W., and Bergelson, J. (2009). Continua of specificity and virulence in plant host-pathogen interactions: causes and consequences. New Phytol. 183, 513–529. doi: 10.1111/j.1469-8137.2009.02927.x
Bergquist, R. R., and Masias, O. R. (1974). Physiologic specialization in Trichometasphaeria turcica f. sp. zeae and Trichometasphaeria turcica f. sp. sorghi in Hawaii. Phytopathology 64, 645–649. doi: 10.1094/Phyto-64-645
Bhowmik, T. P., and Prasada, R. (1970). Physiologic specialization in Helminthosporium turcicum Pass. from India. J. Phytopathol. 68, 84–87. doi: 10.1111/j.1439-0434.1970.tb02491.x
Borchardt, D. S., Welz, H. G., and Geiger, H. H. (1998). Molecular marker analysis of European Setosphaeria turcica populations. Eur. J. Plant Pathol. 104, 611–617. doi: 10.1023/A:1008641920356
Bradford, M. M. (1976). A rapid method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brito, N., Espino, J. J., and Gonzalez, C. (2006). Endo-β-1, 4-xylanase xyn11A required for virulence in Botrytis cinerea. Mol. Plant Microbe Interact. 19, 25–32. doi: 10.1094/MPMI-19-0025
Buiate, E. A. S., Xavier, K. V., Moore, N., Torres, M. F., Farman, M. L., Schardl, C. L., et al. (2017). A comparative genomic analysis of putative pathogenicity genes in the host-specific sibling species Colletotrichum graminicola and Colletotrichum sublineola. BMC Genomics 18:67. doi: 10.1186/s12864-016-3457-9
Butler, M. J., Day, A. W., Henson, J. M., and Money, N. P. (2001). Pathogenic properties of fungal melanins. Mycologia 93, 1–8. doi: 10.1080/00275514.2001.12061273
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, 233–238. doi: 10.1093/nar/gkn663
Chang, Y. C., and Kwon-chung, K. J. (1999). Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J. Bacteriol. 181, 5636–5643. doi: 10.1128/JB.181.18.5636-5643.1999
Condon, B. J., Leng, Y., Wu, D., Bushley, K. E., Ohm, R. A., Otillar, R., et al. (2013). Comparative genome structure, secondary metabolite, and effector coding capacity across Cochliobolus pathogens. PLoS Genet. 9:e1003233. doi: 10.1371/journal.pgen.1003233
Cooper, R. M., Longman, D., Campbell, A., Henry, M., and Lees, P. E. (1988). Enzymatic adaptation of cereal pathogens to monocotyledonous primary wall. Physiol. Mol. Plant Pathol. 32, 33–47. doi: 10.1016/S0885-5765(88)80004-3
Cuq, F., Herrmanngorline, S., Klaebe, A., Rossignol, M., and Petitprez, M. (1993). Monocerin in Exserohilum turcicum isolates from maize and a study of its phytotoxicity. Phytochemistry 34, 1265–1270. doi: 10.1016/0031-9422(91)80013-Q
Degefu, Y., Lohtander, K., and Paulin, L. (2004). Expression patterns and phylogenetic analysis of two xylanase genes (htxyl1 and htxyl2) from Helminthosporium turcicum, the cause of northern leaf blight of maize. Biochimie 86, 83–90. doi: 10.1016/j.biochi.2004.01.001
Desai, J. V., Cheng, S. J., Ying, T., Nguyen, M. H., Clancy, C. J., Lanni, F., et al. (2015). Coordination of Candida albicans invasion and infection functions by phosphoglycerol phosphatase Rhr2. Pathogens 4, 573–589. doi: 10.3390/pathogens4030573
Emanuelsson, O., Brunak, S., Von, H. G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Eveleigh, D. E., Mandels, M., Androtti, R., and Roche, C. (2009). Mesurement of saccharifying cellulases. Biotechnol. Biofuels 6, 21–23. doi: 10.1186/1754-6834-2-21
Ferguson, L. M., and Carson, M. L. (2004). Spatial diversity of Setosphaeria turcica sampled from the eastern United States. Phytopathology 94, 892–900. doi: 10.1094/PHYTO.2004.94.8.892
Forsdyke, D. R. (1996). Different biological species “broadcast” their DNAs at different (G+C)% “wavelengths”. J. Theor. Biol. 178, 405–417. doi: 10.1006/jtbi.1996.0038
Galiano-carneiro, A. L., and Miedaner, T. (2017). Genetics of resistance and pathogenicity in the maize/Setosphaeria turcica pathosystem and implications for breeding. Front. Plant Sci. 8, 1490. doi: 10.3389/fpls.2017.01490
Gao, J. X., Gao, Z. G., Zhang, X. F., Zhang, S., and Chen, L. (2010). A simple and feasible method for single-spore isolation of Exserohilum turcicum. Microbiology 37, 1548–1550. doi: 10.13344/j.microbiol.china.2010.10.023
Gao, Q., Jin, K., Ying, S. H., Zhang, Y., Xiao, G., Shang, Y., et al. (2011). Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7:e1001264. doi: 10.1371/journal.pgen.1001264
Gao, S. G., Li, Y. Q., Gao, J. X., Suo, Y. J., Fu, K. H., Li, Y. Y., et al. (2014). Genome sequence and virulence variation-related transcriptome profiles of Curvularia lunata, an important maize pathogenic fungus. BMC Genomics 15:627. doi: 10.1186/1471-2164-15-627
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity of basidiomycetes–application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Haraguchi, T., Oguro, M., Nagano, H., Ichihara, A., and Sakamura, S. (1983). Specific inhibitors of eukaryotic DNA synthesis and DNA polymerase α, 3-deoxyaphidicolin and aphidicolin-17-monoacetate. Nucleic Acids Res. 11, 1197–1209. doi: 10.1093/nar/11.4.1197
Holtzapple, M., Cognata, M., Shu, Y., and Hendrickson, C. (1990). Inhibition of Trichoderma reesei cellulase by sugars and solvents. Biotechnol. Bioeng. 36, 275–287. doi: 10.1002/bit.260360310
Hou, Y. J., Ma, X., Wan, W. T., Long, N., Zhang, J., Tan, Y. T., et al. (2016). Comparative genomics of pathogens causing brown spot disease of tobacco: Alternaria longipes and Alternaria alternata. PLoS One 11:e0155258. doi: 10.1371/journal.pone.0155258
Hu, H. Z., Zhang, R., Tao, Z. S., Li, X. K., Li, Y. Y., Huang, J. F., et al. (2018). Cellulose synthase mutants distinctively affect cell growth and cell wall integrity for plant biomass production in Arabidopsis. Plant Cell Physiol. 59, 1144–1157. doi: 10.1093/pcp/pcy050
Human, M. P., Berger, D. K., and Crampton, B. G. (2020). Time-course RNAseq reveals Exserohilum turcicum effectors and pathogenicity determinants. Front. Microbiol. 11:360. doi: 10.3389/fmicb.2020.00360
Kamoun, S., van West, P., Vleeshouwers, V. G., de Groot, K. E., and Govers, F. (1998). Resistance of Nicotiana benthamiana to Phytophthora infestans is mediated by the recognition of the elicitor protein INF1. Plant Cell 10, 1413–1426. doi: 10.1105/tpc.10.9.1413
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, 277–280. doi: 10.1093/nar/gkh063
Kim, S., Cho, Y. J., Song, E. S., Lee, S. H., Kim, J. G., and Kang, L. W. (2016). Time-resolved pathogenic gene expression analysis of the plant pathogen Xanthomonas oryzae pv. Oryzae. BMC Genomics 17:345. doi: 10.1186/s12864-016-2657-7
Klein, R. D., Gu, Q., Goddard, A., and Rosenthal, A. (1996). Selection for genes encoding secreted proteins and receptors. Proc. Natl. Acad. Sci. U. S. A. 93, 7108–7113. doi: 10.1073/pnas.93.14.7108
Kou, Y. B., Xu, J. T., Cao, Y. L., Lv, X. X., Zhao, G. L., Chen, G. J., et al. (2014). Gluconolactone induces cellulase gene expression in cellulolytic filamentous fungus Trichoderma reesei. RSC Adv. 4, 36057. doi: 10.1039/C4RA06731B
Krogh, A., Larsson, B., Von, H. G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Lagunas-Muñoz, V. H., Cabrera-Valladares, N., Bolívar, F., Gosset, G., and Martínez, A. (2006). Optimum melanin production using recombinant Escherichia coli. J. Appl. Microbiol. 101, 1002–1008. doi: 10.1111/j.1365-2672.2006.03013.x
Lalaoui, F., Halama, P., Dumortier, V., and Paul, B. (2008). Cell wall-degrading enzymes produced in vitro by isolates of Phaeosphaeria nodorum differing in aggressiveness. Plant Pathol. 49, 727–733. doi: 10.1046/j.1365-3059.2000.00491.x
Lee, B. H., and Blackburn, T. H. (1975). Cellulase production by a thermophilic Clostridium species. Appl. Microbiol. 30, 346–353. doi: 10.1128/am.30.3.346-353.1975
Li, W. Z., Jaroszewski, L., and Godzik, A. (2002). Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics 18, 77–82. doi: 10.1093/bioinformatics/18.1.77
Li, R. Q., Zhu, H. M., Ruan, J., Qian, W. B., Fang, X. D., Shi, Z. B., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Luo, R., Liu, B., Xie, Y., Li, Z., Huang, W., Yuan, J., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1:18. doi: 10.1186/2047-217x-1-18
Ma, S. X., Cao, K. K., Liu, N., Meng, C., Cao, Z. Y., Dai, D. Q., et al. (2017). The StLAC2 gene is required for cell wall integrity, DHN-melanin synthesis and the pathogenicity of Setosphaeria turcica. Fungal Biol. 121, 6–7. doi: 10.1016/j.funbio.2017.04.003
Marcet-Houben, M., Ballester, A. R., de la Fuente, B., Harries, E., Marcos, J. F., González-Candelas, L., et al. (2012). Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics 13, 646. doi: 10.1186/1471-2164-13-646
Martin, T., Biruma, M., Fridborg, I., Okori, P., and Dixelius, C. (2011). A highly conserved NB-LRR encoding gene cluster effective against Setosphaeria turcica in sorghum. BMC Plant Biol. 11, 151. doi: 10.1186/1471-2229-11-151
Medema, M. H., Blin, K., Cimermancic, P., de Jager, V., Zakrzewski, P., Fischbach, M. A., et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39, W339–W346. doi: 10.1093/nar/gkr466
Milton, H. S. Jr., Yen, M. R., Noto, K., Tamang, D. G., and Elkan, C. (2009). The transporter classification database: recent advances. Nucleic Acids Res. 37, 274–278. doi: 10.1093/nar/gkn862
Mitra, M. (1923).Helminthosporium spp. on cereals and sugarcane in India. Part I. Diseases of Zeamays and Sorghum vulgare caused by species of Helminthosporium. India Dep. Agric. Mem. Bot. Ser. 11, 219–242.
Moriwaki, A., Kihara, J., Kobayashi, T., Tokunaga, T., Arase, S., and Honda, Y. (2004). Insertional mutagenesis and characterization of a polyketide synthase gene (PKS1) required for melanin biosynthesis in Bipolaris oryzae. FEMS Microbiol. Lett. 238, 1–8. doi: 10.1016/j.femsle.2004.07.007
Ni, L. N. (2004). Isolation and identification of a high-melanin-producting bacterium. Microbiology 31, 55–59. doi: 10.13344/j.microbiol.china.2004.01.013
Nieuwoudt, A., Human, M. P., Craven, M., and Crampton, B. G. (2018). Genetic differentiation in populations of Exserohilum turcicum from maize and sorghum in South Africa. Plant Pathol. 67, 1483–1491. doi: 10.1111/ppa.12858
Nosanchuk, J. D., and Casadevall, A. (2003). The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 5, 203–223. doi: 10.1046/j.1462-5814.2003.00268.x
Novo, M., Pomar, F., Gayoso, C., and Merino, F. (2006). Cellulase activity in isolates of Verticillium dahliae differing in aggressiveness. Plant Dis. 90, 155–160. doi: 10.1094/PD-90-0155
Ohm, R. A., Feau, N., Henrissat, B., Schoch, C. L., Horwitz, B. A., Barry, K. W., et al. (2012). Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen Dothideomycetes fungi. PLoS Genet. 8:e1003037. doi: 10.1371/journal.ppat.1003037
Okori, P., Rubaihayo, P. R., Ekwamu, A., Fahleson, J., and Dixelius, C. (2004). Genetic characterization of Cercospora sorghi from cultivated and wild sorghum and its relationship to other Cercospora fungi. Phytopathology 94, 743–750. doi: 10.1094/PHYTO.2004.94.7.743
Patrick, D. R., and Heimbrook, D. C. (1996). Protein kinase inhibitors for the treatment of cancer. Drug Discov. Today 1, 325–330. doi: 10.1016/1359-6446(96)10030-1
Petersen, T. N., Brunak, S., Von, H. G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Quentin, M., Ebbelaar, M., Derksen, J., Mariani, C., and van der Valk, H. (2002). Description of a cellulose-binding domain and a linker sequence from aspergillus fungi. Appl. Microbiol. Biotechnol. 58, 658–662. doi: 10.1007/s00253-002-0937-4
Quiroga, E. N., Sampietro, A. R., and Vattuone, M. A. (2001). Screening antifungal activities of selected medicinal plants. J. Ethnopharmacol. 74, 89–96. doi: 10.1016/S0378-8741(00)00350-0
Ramathani, I., Biruma, M., Martin, T., Dixelius, C., and Okari, P. (2011). Disease severity, incidence and races of Setosphaeria turcica on sorghum in Uganda. Eur. J. Plant Pathol. 131, 383–392. doi: 10.1007/s10658-011-9815-1
Reese, E. T., Parrish, F. W., and Ettlinger, M. (1971). Nojirimycin and D-glucono-l, 5-lactone as inhibitors of carbohydrases. Carbohydr. Res. 18, 381–388. doi: 10.1016/S0008-6215(00)80274-8
Rep, M. (2005). Small proteins of plant-pathogenic fungi secreted during host colonization. FEMS Microbiol. Lett. 253, 19–27. doi: 10.1016/j.femsle.2005.09.014
Rep, M., van der Does, H. C., Meiger, M., van Wijk, R., Houterman, P. M., Dekker, H. L., et al. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53, 1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x
Robert, A. L. (1960). Physiologic specialization in Helminthosporium turcicum. Phytopathology 50, 217–220. doi: 10.1111/j.1399-3054.1960.tb08108.x
Schirawski, J., Mannhaupt, G., Münch, K., Breport, T., Schipper, K., Doehlemann, G., et al. (2010). Pathogenicity determinants in smut fungi revealed by genome comparison. Science 330, 1546–1548. doi: 10.1126/science.1195330
Segmüller, N., Kokkelink, L., Giesbert, S., Odinius, D., Kan, J. V., and Tudzynski, P. (2008). NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol. Plant Microbe Interact. 21, 808–819. doi: 10.1094/MPMI-21-6-0808
Siriputthaiwan, P., Jauneau, A., Herbert, C., Garcin, D., and Dumas, B. (2005). Functional analysis of CLPT1, a Rab/GTPase required for protein secretion and pathogenesis in the plant fungal pathogen Colletotrichum lindemuthianum. J. Cell Sci. 118, 323–329. doi: 10.1242/jcs.01616
Sperschneider, J., Gardiner, D. M., Dodds, P. N., Tini, F., Covarelli, L., Singh, K. B., et al. (2016). EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol. 210, 743–761. doi: 10.1111/nph.13794
Sposato, P., Ahn, J. H., and Walton, J. D. (1995). Characterization and disruption of a gene in the maize pathogen Cochliobolus carbonum encoding a cellulase lacking a cellulose binding domain and hinge region. Mol. Plant Microbe Interact. 8, 602–609. doi: 10.1094/MPMI-8-0602
Stanke, M., Tzvetkova, A., and Morgenstern, B. (2006). AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 7, 1–8. doi: 10.1186/gb-2006-7-s1-s11
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tang, L., Gao, Z. G., Yang, R. X., Yao, Y., and Liu, X. (2015). Activity analysis and gene expression of cell wall degradation enzymes in Setosphaeria turcica f. sp. zeae and S. turcica f. sp. sorghi. Acta Phytophylacica Sin. 42, 935–941. doi: 10.13802/j.cnki.zwbhxb.2015.06.011
Tang, L., Gao, Z. G., Yao, Y., and Liu, X. (2014). Identification and genetic diversity of formae speciales of Setosphaeria turcica in China. Plant Dis. 99, 482–487. doi: 10.1094/pdis-06-14-0570-re
Tanguay, P., Loppnau, P., Morin, C., Bernier, L., and Breuil, C. (2006). A spontaneous albino mutant of Ceratocystis resinifera results from a point mutation in the polyketide synthase gene, PKS1. Can. J. Microbiol. 52, 501–507. doi: 10.1139/w05-150
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Eiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinf. 4, 41. doi: 10.1186/1471-2105-4-41
Thrall, P. H., Burdon, J. J., and Young, A. (2002). Variation in resistance and virulence among demes of a plant host–pathogen metapopulation. J. Ecol. 89, 736–748. doi: 10.1046/j.0022-0477.2001.00597.x
Urban, M., Irvine, A. G., Cuzick, A., and Hammond-Kosack, K. E. (2015). Using the pathogen-host interactions database (PHI-base) to investigate plant pathogen genomes and genes implicated in virulence. Front. Plant Sci. 6, 605. doi: 10.3389/fpls.2015.00605
van den Brink, J., and de Vries, R. P. (2011). Fungal enzyme sets for plant polysaccharide degradation. Appl. Microbiol. Biotechnol. 91, 1477–1492. doi: 10.1007/s00253-011-3473-2
Walker, D. S., Reeves, P. J., and Salmond, G. P. C. (1994). The major secreted cellulase, CelV, of Erwinia carotovora subsp. carotovora is an improtant soft rot virulence factor. Mol. Plant Microbe Interact. 7, 425–431. doi: 10.1094/MPMI-7-0425
Walton, J. D. (1994). Deconstructing the cell wall. Plant Physiol. 104, 1113–1118. doi: 10.1104/pp.104.4.1113
Wanjiru, W. M., Kang, Z. S., and Buchenauer, H. (2002). Importance of cell wall degrading enzymes produced by Fusarium graminearum during infection of wheat heads. Eur. J. Plant Pathol. 108, 803–810. doi: 10.1023/A:1020847216155
Wen, L. L., Cao, Z. Y., and Dong, J. G. (2008). Cloning and promoter prediction of melanin biosynthesis gene 4HNR in Setosphaeria turcica. Acta Phytopathol. Sin. 38, 641–644. doi: 10.13926/j.cnki.apps.2008.06.008
Winnenburg, R., Baldwin, T. K., Urban, M., Rawlings, C., Köhler, J., and Hammond-Kosack, K. E. (2006). PHI-base: a new database for pathogen host interactions. Nucleic Acids Res. 34, D459–D464. doi: 10.1093/nar/gkj047
Zhang, X., Cao, Z. Y., Liu, S. W., Guo, L. Y., and Dong, J. G. (2011). Functional analysis of polyketide synthase gene StPKS in Setosphaeria turcica. Sci. Agric. Sin. 44, 1603–1609. doi: 10.3864/j.issn.0578-1752.2011.08.008
Keywords: Setosphaeria turcica, formae speciales, pathogenicity, genome sequencing, gene function, comparative genome, cellulase activity
Citation: Ma Z, Huang Y, Zhang Z, Liu X, Xuan Y, Liu B and Gao Z (2022) Comparative genomic analysis reveals cellulase plays an important role in the pathogenicity of Setosphaeria turcica f. sp. zeae. Front. Microbiol. 13:925355. doi: 10.3389/fmicb.2022.925355
Received: 26 May 2022; Accepted: 29 June 2022;
Published: 22 July 2022.
Edited by:
Boqiang Li, Institute of Botany (CAS), ChinaReviewed by:
Yuli Dai, Fujian Academy of Agricultural Sciences, ChinaCopyright © 2022 Ma, Huang, Zhang, Liu, Xuan, Liu and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Liu, bGl1Ym80NTUyQDEyNi5jb20=; Zenggui Gao, Z2FvemVuZ2d1aUBvdXRsb29rLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.