- 1Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology and Business University, Beijing, China
- 2Beijing Engineering and Technology Research Center of Food Additives, Beijing Technology and Business University, Beijing, China
- 3School of Food and Health, Beijing Technology and Business University, Beijing, China
Amino acid metabolism could exert regulatory effects on Monascus pigments (MPs) biosynthesis. In this work, MPs biosynthesis regulated by methionine and S-adenosylmethionine (SAM) was investigated in Monascus purpureus RP2. The results indicated that the addition of methionine in fermentation significantly reduced MPs production by 60–70%, and it induced a higher expression of SAM synthetase Mon2A2272 and consequently led to SAM accumulation. However, the addition of SAM in fermentation promoted MPs production by a maximum of 35%, while over-expression of the gene Mon2A2272 led to a decrease in MPs yield, suggesting that SAM synthetase and SAM were likely to play different regulatory roles in MPs biosynthesis. Furthermore, the gene transcription profile indicated that SAM synthetase expression led to a higher expression of the transcriptional regulatory protein of the MPs biosynthesis gene cluster, while the addition of SAM gave rise to a higher expression of MPs biosynthesis activator and the global regulator LaeA, which probably accounted for changes in MPs production and the mycelium colony morphology of M. purpureus RP2 triggered by methionine and SAM. This work proposed a possible regulation mechanism of MPs biosynthesis by SAM metabolism from methionine. The findings provided a new perspective for a deep understanding of MPs biosynthesis regulation in M. purpureus.
Introduction
Monascus pigments (MPs) are Chinese traditional natural colorants with an application history of thousands of years. Nowadays, they are still widely used in the fields of food, pharmaceutical, cosmetic manufacture, and printing, dyeing, and textile industries (Lin et al., 2008; Mapari et al., 2010; Feng et al., 2012; Patakova, 2013; Dufosse et al., 2014; Mostafa and Abbady, 2014; Srianta et al., 2014; Chen et al., 2015). MPs are a large class of structurally related secondary polyketide metabolites shared with a common azaphilone skeleton produced by Monascus spp. and are traditionally classified as red, orange, and yellow pigments (Chen et al., 2015, Chen W. et al., 2017). It is generally recognized that MPs are biosynthesized via the polyketide synthase (PKS) pathway, and the conserved PKS gene cluster in Monascus is identified to be composed of a dozen gene elements encoding the polyketide synthase, transcription factors, and other functional enzymes (Yang et al., 2014; Chen et al., 2015, Chen W. et al., 2017).
The PKS pathway responsible for MPs biosynthesis is quite complicated and still ambiguous. Continuous studies (Yang et al., 2014; Chen et al., 2015, Chen W. et al., 2017) revealed that MPs biosynthesis started with the esterification of a β-ketoacid from the fatty acid synthase pathway to the chromophore from the PKS pathway, generating the yellow intermediates. The yellow intermediates are then reduced and oxidized into the classical yellow MPs (Monascin and Ankaflavin) and orange MPs (Rubropunctatin and Monascorubrin), respectively. Finally, the classical red MPs (Rubropunctamine and Monascorubramine) are formed by direct integration of the amido group (amines or amino acids) into the orange MPs.
Based on current understanding of the MPs biosynthesis pathway, the amino donors are believed to be directly involved in the red MPs molecule formation (Chen et al., 2015, Chen W. et al., 2017). However, in vitro chemical reactions demonstrated that only 4 of 18 amino donors (arginine, lysine, γ-aminobutyric acid, and ammonia) directly reacted with the orange MPs to form the red MPs (Chen W. et al., 2017), indicating that not all the amino compounds could act as the precursor substrates of MPs. Even though, it is indispensable to take into account the roles of nitrogen sources in MPs biosynthesis. Several previous studies reported that different nitrogen sources supplemented in fermentation medium made a significant impact on the yield and component composition of MPs (Chen and Johns, 1993; Jung et al., 2003, 2005, 2011; Dhale et al., 2011; Hajjaj et al., 2012; Arai et al., 2013; Zhang et al., 2013; Said et al., 2014; Shi et al., 2015; Xiong et al., 2015). Therefore, it is reasonable to conclude that various nitrogen sources could exert regulatory effects on MPs biosynthesis in different ways.
Methionine is an important sulfur-containing amino acid that plays essential roles in multiple biological processes, such as DNA methylation, protein structure, and polyamine synthesis (Brosnan et al., 2007; Cavuoto and Fenech, 2012). Methionine could be transformed into S-adenosylmethionine (SAM) by the catalysis of SAM synthetase (EC 2.5.1.6) (Lieber and Lester, 2002). SAM is the major cellular methyl group donor in living organisms and it is involved in the methylation processes of DNA, RNA, proteins, metabolites, and phospholipids (Lieber and Lester, 2002; Brosnan et al., 2007; Gerke et al., 2012). In microorganisms, SAM is involved in a variety of processes, such as protection of DNA and mRNA integrity, the function of restriction enzymes, regulation of gene expression, efficient translation, cellular differentiation, stress response, and secondary metabolites biosynthesis (Lieber and Lester, 2002; Brosnan et al., 2007; Gerke et al., 2012). It is evident that the metabolism of SAM from methionine serves not only as the nitrogen source supply but also as the regulator of multiple biological processes. In this study, the bidirectional regulatory effects of methionine and SAM on MPs biosynthesis were observed in Monascus purpureus RP2. SAM and SAM synthetase were proved to be the key effector molecules for MPs biosynthesis regulation, which probably interacted with the global regulator LaeA and the PKS gene cluster. These findings provided new insights into the regulation mechanism of MPs biosynthesis in M. purpureus.
Materials and Methods
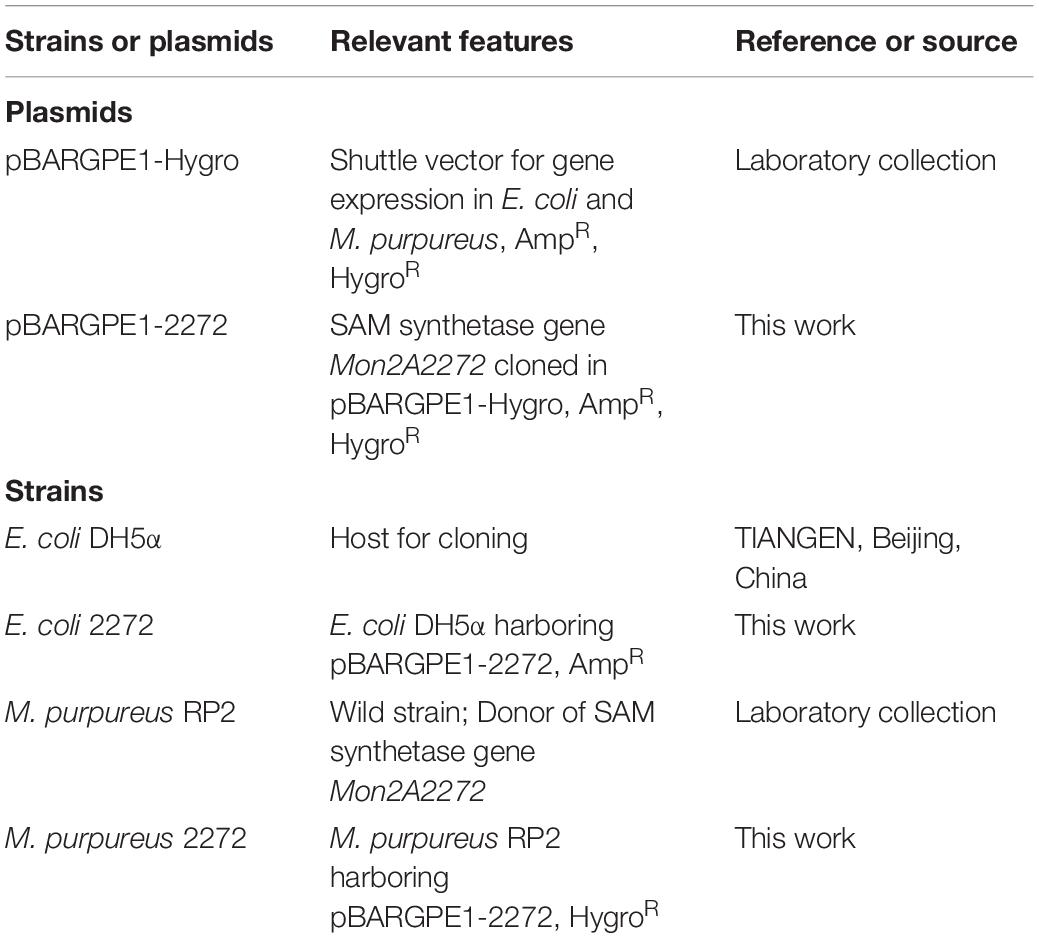
Strains, Plasmids, and Culture Conditions
The strains and plasmids used in this work are listed in Table 1. Escherichia coli strains were grown at 37°C in Luria-Bertani (LB) medium with vigorous shaking at 220 rpm in a shaking incubator with a rotational radius of 10 cm. M. purpureus strains were cultured in Potato Dextrose Agar (PDA) medium at 30°C. When needed, 200 μg/ml ampicillin for E. coli or 20 μg/ml hygromycin B for M. purpureus was added to the medium for recombinant screening.
For MPs production by liquid-state fermentation, M. purpureus was cultured at 30°C in PDA medium for 10 days and the spore suspension was prepared by washing the mycelium with sterile water and collected by filtration with sterile gauze. The spore suspension (10%) was inoculated into seed medium (40 g/L rice flour, 8 g/L peptones, 5 g/L soybean meal, 1 g/L MgSO4⋅7H2O, 2 g/L KH2PO4, and 2 g/L NaNO3) and cultured at 33°C with vigorous shaking at 200 rpm in a shaking incubator with a rotational radius of 10 cm for 48 h. The seed culture (10%) was then inoculated into a fermentation medium (20 g/L glucose, 5 g/L Yeast Nitrogen Base W/O Amino acids, 5 g/L K2HPO4⋅3H2O, 0.5 g/L MgSO4⋅7H2O, 5 g/L KH2PO4, 0.1 g/L CaCl2, 0.03 g/L MnSO4⋅H2O, 0.01 g/L FeSO4⋅7H2O, and 0.01 g/L ZnSO4⋅7H2O) and cultured at 33°C with vigorous shaking at 200 rpm in a shaking incubator with a rotational radius of 10 cm for 12 days. When needed, 3 g/L methionine or 1 g/L SAM was added to the fermentation medium. For colony morphology observation, M. purpureus was cultured at 30°C in the fermentation medium containing agar (20 g/L) for 6–9 days.
Measurement of Monascus Pigments Production
For measurement of MPs production, the fermentation culture was centrifuged at 8,000 × g for 10 min to collect mycelia and medium supernatant, respectively. Samples of the mycelia or medium supernatant were soaked in 50 ml of 70% (v/v) ethanol and incubated in a water bath (60°C) for 1 h to extract MPs. The extraction mixture was centrifuged at 8,000 × g for 10 min to collect the supernatant, which was used to measure MPs concentration by spectrophotometer (OD 410 nm for yellow MPs, OD 468 nm for orange MPs, and OD 505 nm for red MPs).
DNA Manipulation Techniques
Standard DNA manipulation techniques were performed as described by Green and Sambrook (2012). Total RNA from M. purpureus was prepared using the RNAprep Pure Plant Kit (TIANGEN, Beijing, China) following the manufacturer’s instructions. RNA was subjected to reverse transcription to generate cDNA using the Quantscript RT Kit (TIANGEN, Beijing, China) following the manufacturer’s protocol. Quantitative Real-Time PCR (q RT-PCR) was performed using the SuperReal PreMix Plus (SYBR Green) Kit (TIANGEN, Beijing, China) in the CFX96 Touch Real-Time PCR System (Bio-Rad, United States) with the following cycling conditions: 95°C for 2 min, followed by 40 cycles of 94°C for 20 s, 63°C for 45 s, and 60°C for 5 min. The GAPDH gene was used for transcript normalization. All reactions were performed in triplicate. The 2–ΔΔCt method was used to analyze the data, which was corrected for primer efficiencies using the untreated group means as the reference condition (Schmittgen and Livak, 2008; Putt et al., 2017).
DNA amplification and recombinant DNA construction were performed using the Ex Taq DNA Polymerase and DNA Ligation Kit (Takara, Beijing, China) following the manufacturer’s protocol. Plasmid DNA from E. coli was isolated using the High-purity Plasmid Miniprep Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. A standard heat shock transformation method was used to introduce plasmid DNA to E. coli (Green and Sambrook, 2012). Primers used for gene transcription level assay by q RT-PCR and gene cloning by PCR are listed in Supplementary Table 1.
Vector Construction for Over-Expression of the S-Adenosylmethionine Synthetase Gene Mon2A2272
The SAM synthetase gene Mon2A2272 was amplified by PCR from the cDNA derived from the total RNA of M. purpureus RP2 using the specific primers SAMS-F/R designed according to the genomic sequence of M. purpureus RP2. The amplicon was inserted into the shuttle vector pBARGPE1-Hygro for gene expression in E. coli and M. purpureus to construct the recombinant vector pBARGPE1-2272. The DNA ligation mixture was transformed into E. coli DH5α and transformants were screened on LB agar plates containing ampicillin. The recombinant vector was verified by sequencing and alignment analysis using the DNAMAN software package and BLAST Program at NCBI against the GenBank database.
Electroporation of Monascus purpureus
The recombinant vector pBARGPE1-2272 was introduced into M. purpureus RP2 by electroporation as previously described with modifications (Kim et al., 2003; Lakrod et al., 2003). For the preparation of the electrocompetent protoplast of M. purpureus RP2, a fresh spore suspension was prepared as described previously and was spread on a PDA plate and cultured at 30°C for 30–40 h. Mycelia (300 mg) were collected and washed with 1 M MgSO4 and incubated at 30°C for 3 h in 50 ml of enzymolysis buffer (1% snailase, 0.1% cellulase, and 0.3% lysozyme). The enzymolysis solution was filtered with sterile gauze and the spheroplast suspension was collected and centrifuged at 8,000 × g at 4°C for 5 min. The spheroplasts were harvested and washed with cooled sorbitol buffer (1 M) and resuspended in 200 μl of the cooled protoplast buffer (25 mM Tris-HCl, pH 7.5; 25 mM CaCl2, 1.2 M sorbitol).
For electroporation of M. purpureus RP2, the purified recombinant plasmid pBARGPE1-2272 (1 μg) and the cooled protoplast suspension (100 μl, 109 protoplasts/ml) were mixed well and transferred to a cooled electroporation cuvette (Bio-Rad, United States) with a 0.2-cm electrode gap. The electroporation cuvette was incubated in an ice bath for 15 min and electroporated in Gene Pulser (Bio-Rad, United States) using the following settings: voltage, 3 kV/cm; capacitance, 25 μF; and resistance, 400 Ω. Following the electric pulse, 1 ml of regeneration broth (Potato Dextrose Broth plus 1.2 M sorbitol) was immediately added to the cuvette and the electroporation mixture was transferred to a sterile 1.5 ml tube and incubated with agitation at 1,000 × g at 30°C for 2 h. The mixture was plated onto PDA plates containing hygromycin B and cultured at 30°C in the dark for 3–6 days. The recombinant strain M. purpureus 2272 was screened and validated by colony PCR of the hygromycin resistance gene using the specific primers Hyg-F/R. Additionally, q RT-PCR was carried out for analysis of the transcriptional level of Mon2A2272 in M. purpureus 2272.
Measurement of S-Adenosylmethionine
For measurement of SAM produced by M. purpureus, the fermentation culture was centrifuged at 8,000 × g for 10 min to collect mycelia and medium supernatant, respectively. The medium supernatant was filtered with a 0.22 μm pore membrane and used for SAM determination. The wet mycelia were dried at −80°C by vacuum freezing and incubated in the solution containing 4 ml of 1.5 M perchloric acid and 4 ml of 1.5 M ammonia water at 4°C for overnight. The mixture was centrifuged at 10,000 × g for 15 min to collect the supernatant for SAM determination. SAM was determined by high-performance liquid chromatography (HPLC) (SHIMADZU, Shanghai, China) as previously described with modifications (Wang et al., 2012). The HPLC analysis conditions were as follows: Inertsil ODS-3 column (4.6 mm × 250 mm, 5 μm, SHIMADZU, Shanghai, China); injection volume 20 μl; oven temperature 30°C; flow rate 1 ml/min; the eluate was monitored at 260 nm. Mobile phase A (10 mM ammonium formate buffer, pH 3.5) and phase B (methanol) were run on an isocratic elution program (95% phase A buffer and 5% phase B buffer). The standard chemicals SAM was purchased from Macklin Biochemical Co., Ltd (Shanghai, China).
Results and Discussion
Addition of Methionine Reduced Monascus Pigments Production and Enhanced S-Adenosylmethionine Synthetase Expression and S-Adenosylmethionine Generation
The fermentation medium containing YNB without amino acids as the nitrogen source was used to evaluate the effect of methionine addition on MPs biosynthesis in M. purpureus RP2. Evaluation of serial gradient concentrations (1–9 g/L) of methionine indicated that 3 g/L was the optimal volume of addition that exerted a significant effect on MPs production (data not shown). Fermentation results showed that the yields of red, orange, and yellow MPs were all significantly reduced by 60–70% during 12 days of liquid fermentation with the addition of 3 g/L methionine (Figure 1). Meanwhile, the bright red, orange, and yellow colors of MPs became much more intense within and around the mycelium colony cultivated without methionine than with methionine in the fermentation agar plates (Figure 2). MPs production was remarkably inhibited in the presence of methionine in both liquid fermentation and plate culture. In addition, it is noteworthy that the morphologies were quite different between the mycelium colonies grown in plates with and without methionine as shown in Figure 2, indicating that the addition of methionine also changed the mycelial development of M. purpureus RP2.
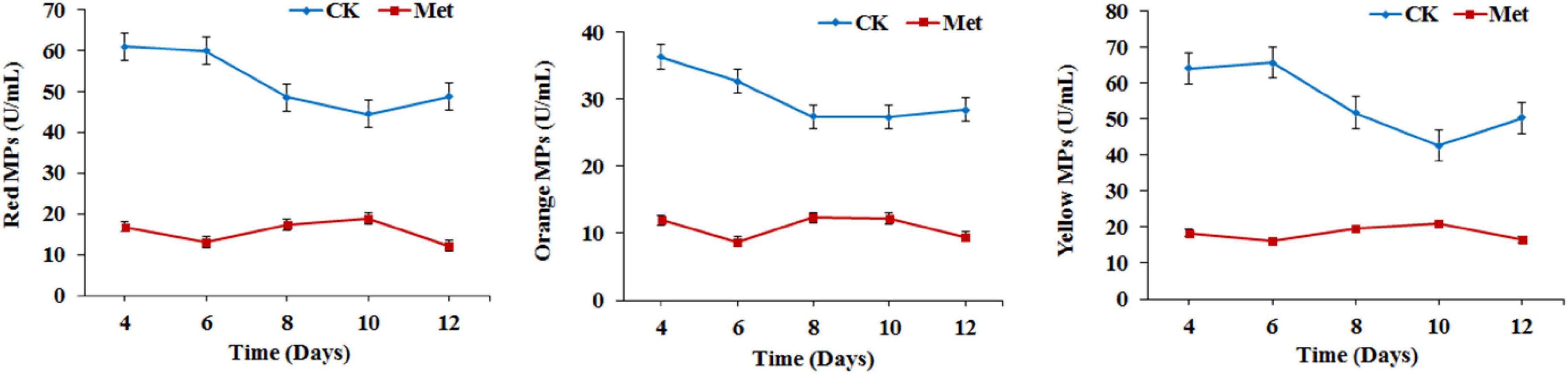
Figure 1. Monascus pigments (MPs) production in Monascus purpureus RP2. CK, fermentation without addition of methionine; Met, fermentation with addition of 3 g/L methionine.
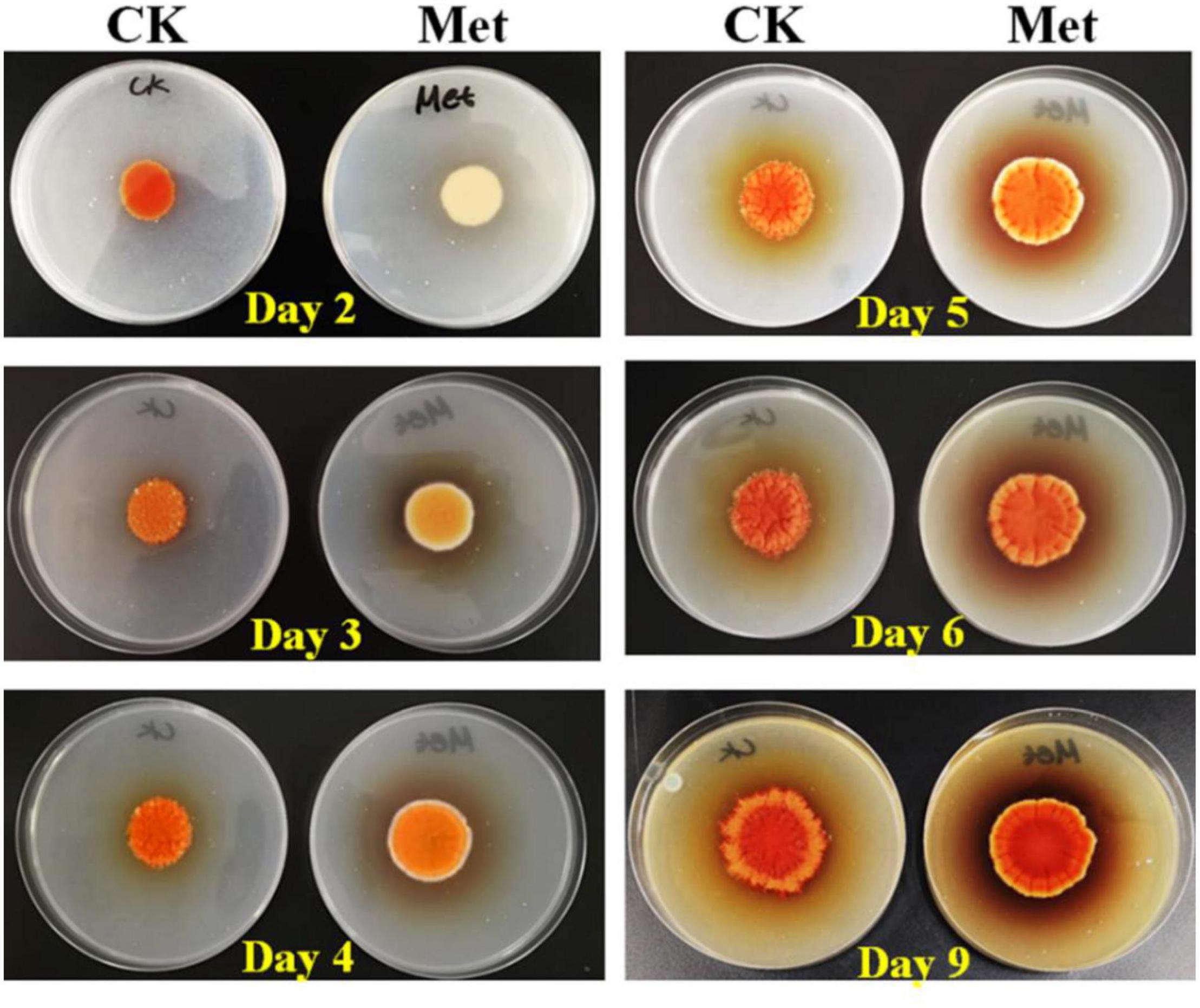
Figure 2. Cultivation of M. purpureus RP2 in fermentation agar plates. CK, fermentation without addition of methionine; Met, fermentation with addition of 3 g/L methionine.
To figure out the reason that methionine inhibited MPs production, the metabolism of methionine was investigated in M. purpureus RP2. Generally, methionine could be transformed to SAM by the catalysis of SAM synthetase and the derivative would act as the major cellular methyl donor and precursor to organic radicals involved in a variety of important biological processes (Lieber and Lester, 2002; Brosnan et al., 2007). Unsurprisingly, it was detected that the concentration of intracellular SAM in M. purpureus RP2 increased by 100–200 μg/ml in fermentation with methionine compared with that without methionine. In addition, q RT-PCR analysis also showed that the transcriptional level of the SAM synthetase gene Mon2A2272 in M. purpureus RP2 at the beginning of fermentation was about 12-fold higher with methionine than without methionine, which could account for SAM generation. Therefore, it is reasonable to conclude that the addition of methionine in the fermentation medium induced SAM synthetase Mon2A2272 expression and consequently enhanced SAM generation, directly or indirectly leading to a significant reduction in MPs production in M. purpureus RP2. However, it is still questionable whether SAM synthetase Mon2A2272 expressed at a higher level or SAM accumulation acted as the effector that affected MP production.
Addition of S-Adenosylmethionine Enhanced Monascus Pigments Production
As the addition of methionine led to SAM generation and MP production reduction in M. purpureus RP2, it is essential to reveal the causal connection between SAM accumulation and MP production. The fermentation with the addition of 1 g/L SAM was conducted to investigate the effect of high-level SAM on MP production in M. purpureus RP2. Unexpectedly, a maximum 35% increase in MPs production was detected in fermentation with SAM in comparison with that without SAM (Figure 3). The fermentation agar plate cultivation also showed that a brighter yellow or orange halo was observed around the mycelium colony with SAM (Figure 4), which was in accordance with MPs yield determination. Moreover, the addition of SAM also altered the morphology of the mycelium colony (Figure 4), but it is a different pattern from that derived from the addition of methionine (Figure 2). In addition, though the addition of SAM resulted in a higher accumulation of SAM in M. purpureus RP2 after 3 days of fermentation as shown in Figure 5, q RT-PCR assay indicated that extra SAM did not affect the expression of the SAM synthetase gene Mon2A2272 (Figure 6). It suggested that SAM individually played the regulatory role in MP production and mycelium development in M. purpureus RP2.
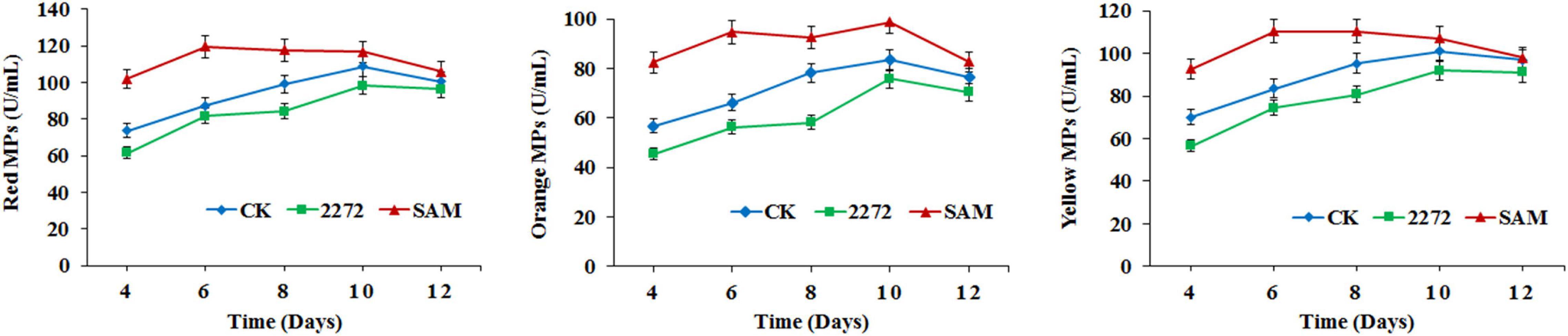
Figure 3. Monascus pigments production in M. purpureus RP2 and M. purpureus 2272. CK, M. purpureus RP2 fermentation without addition of amino acid; 2272, M. purpureus 2272 fermentation without addition of amino acid; (SAM), M. purpureus RP2 fermentation with addition of 1 g/L SAM.
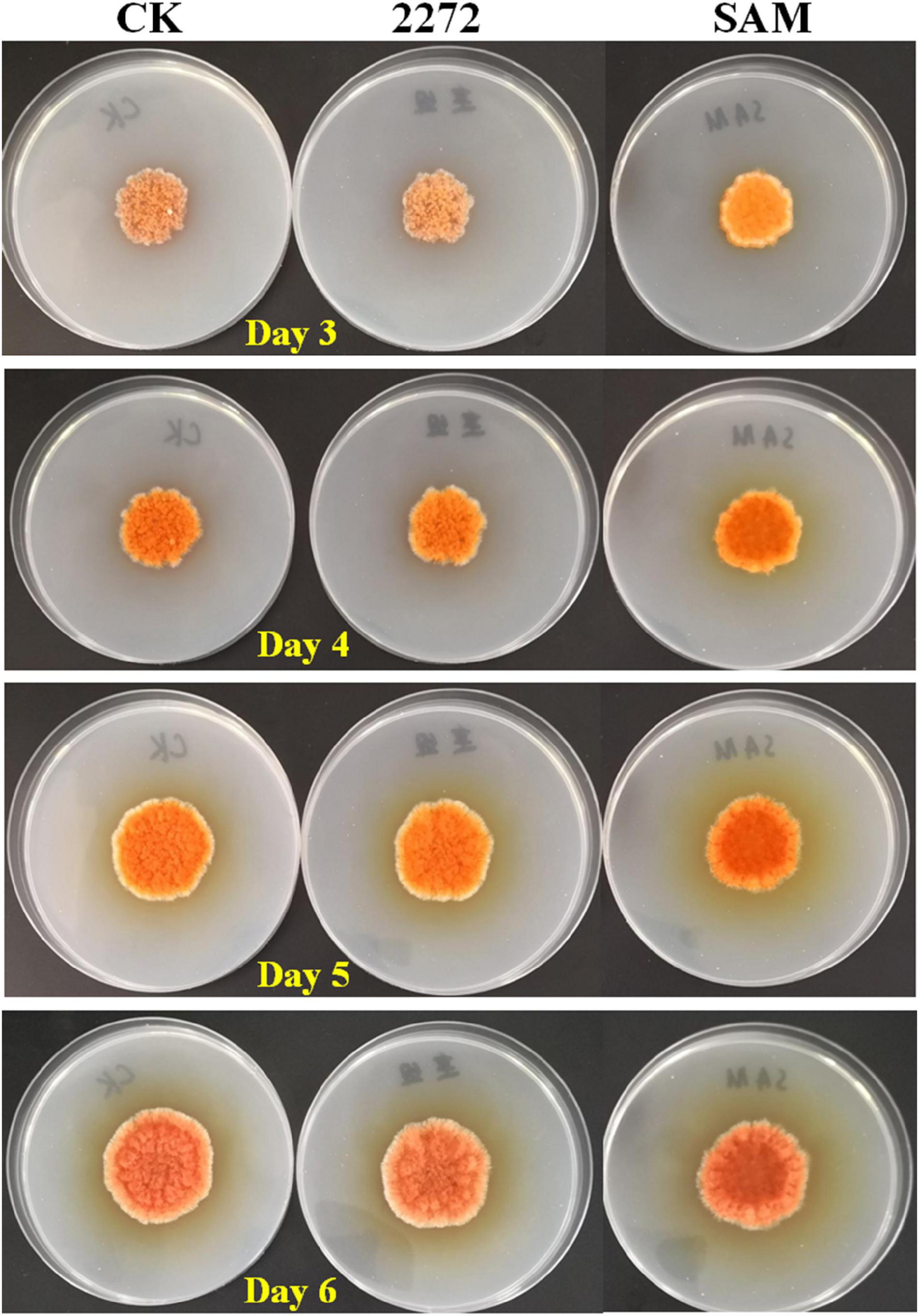
Figure 4. Cultivation of M. purpureus RP2 and M. purpureus 2272 in fermentation agar plates. CK, M. purpureus RP2 fermentation without addition of amino acid; 2272, M. purpureus 2272 fermentation without addition of amino acid; SAM, M. purpureus RP2 fermentation with addition of 1 g/L SAM.
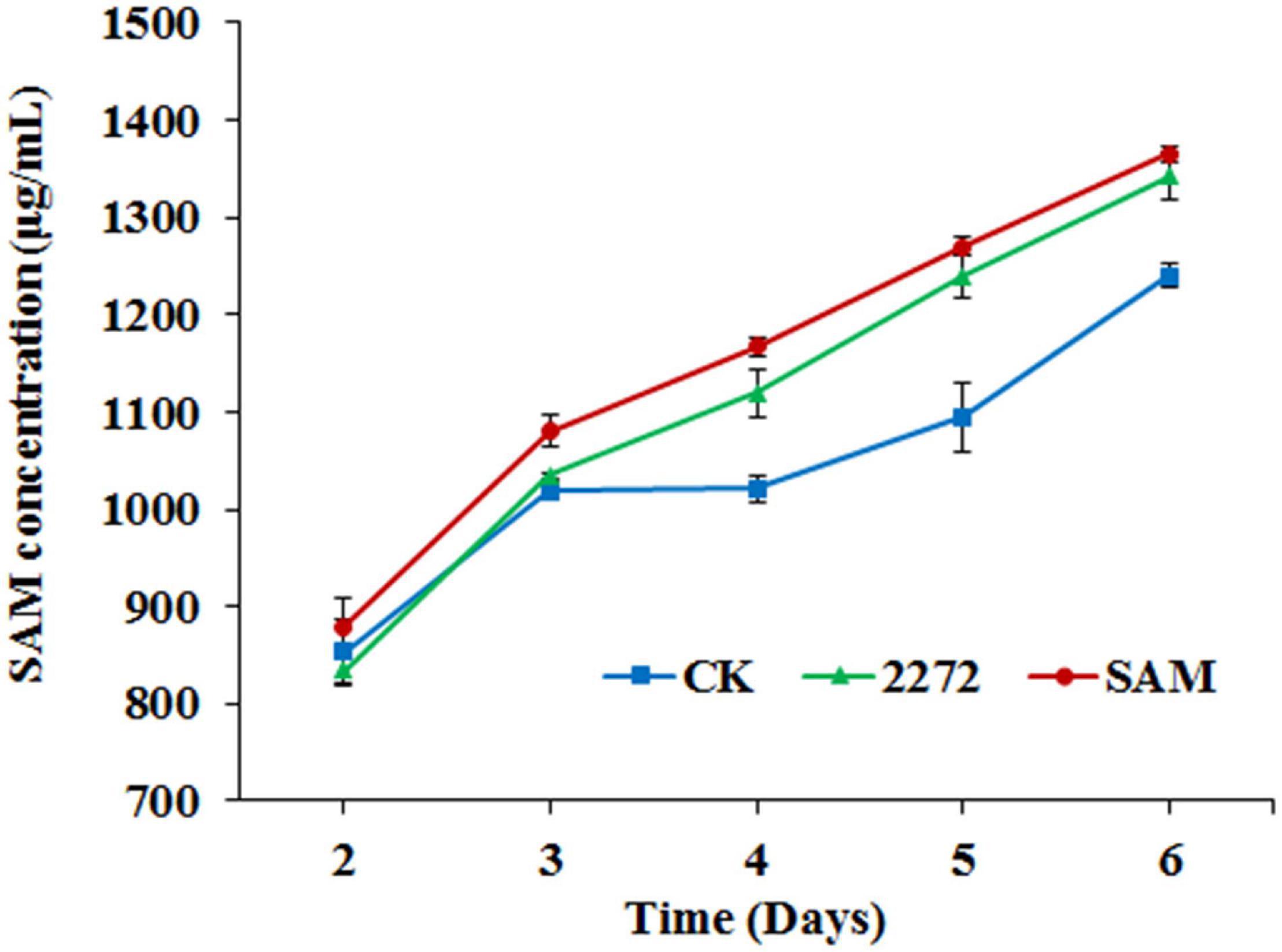
Figure 5. S-adenosylmethionine accumulation in the fermentation broth of M. purpureus RP2 and M. purpureus 2272. CK, M. purpureus RP2 fermentation without addition of amino acid; 2272, M. purpureus 2272 fermentation without addition of amino acid; SAM, M. purpureus RP2 fermentation with the addition of 1 g/L SAM.
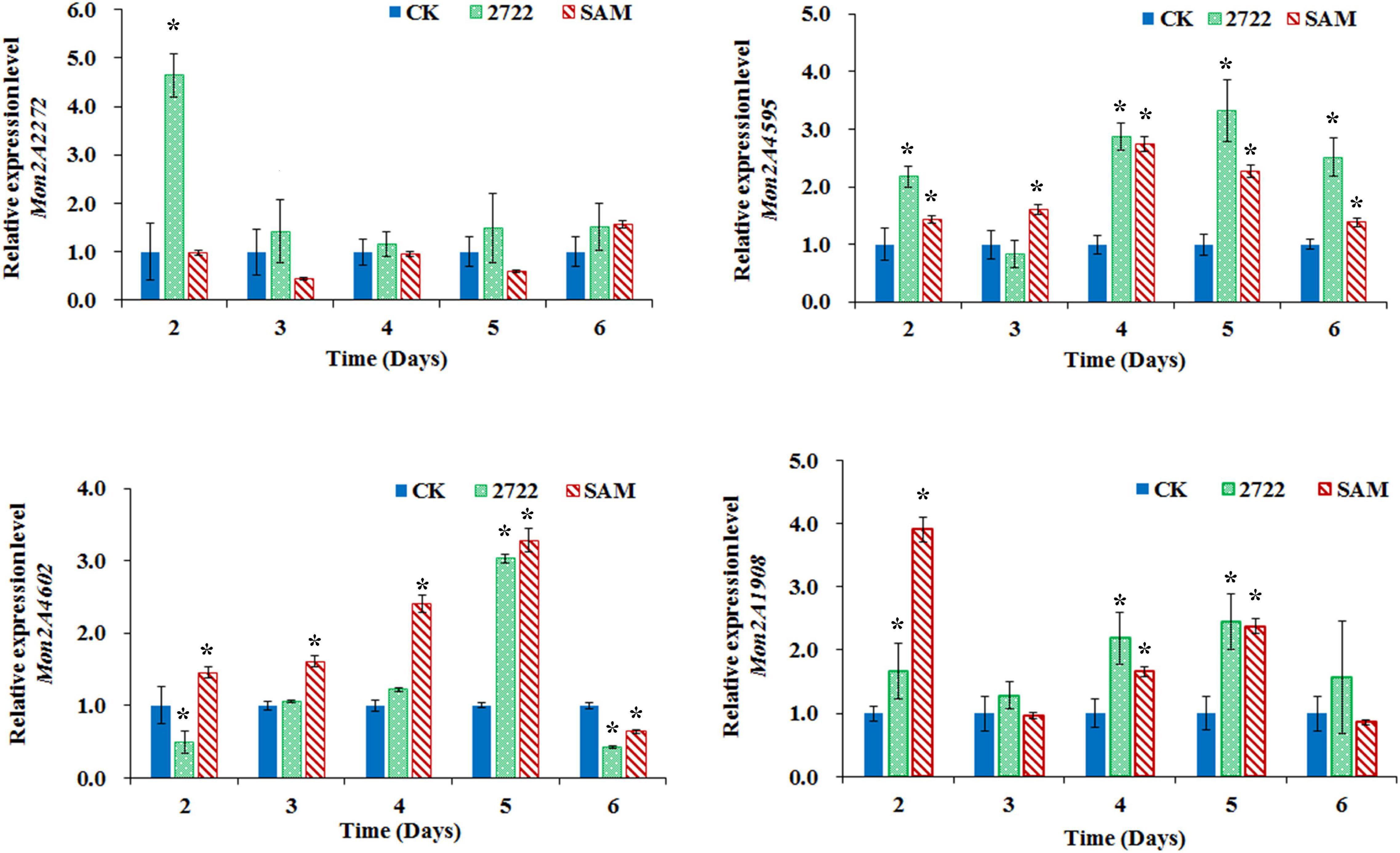
Figure 6. Gene transcriptional level assay in M. purpureus RP2 and M. purpureus 2272. CK, M. purpureus RP2 fermentation without addition of amino acid; 2272, M. purpureus 2272 fermentation without addition of amino acid; SAM, M. purpureus RP2 fermentation with the addition of 1 g/L SAM; Bars with an asterisk are significantly different (p < 0.05).
S-Adenosylmethionine Synthetase Mon2A2272 Over-Expression Enhanced S-Adenosylmethionine Generation and Reduced Monascus Pigments Production
As previously reported, the addition of methionine induced SAM synthetase Mon2A2272 expression, enhanced SAM generation, and inhibited MPs production. SAM synthetase or SAM was suspected to be the effector of the reduction in MP production. However, when a high concentration of SAM was added in fermentation, MP production increased as shown in Figures 3, 4. Hence, the causal connection between the high expression level of SAM synthetase and MPs production was further investigated by the construction of the recombinant strain M. purpureus 2272 with over-expression of the SAM synthetase gene Mon2A2272. The SAM synthetase gene Mon2A2272 cloned from M. purpureus RP2 was 1,161 bp in size and encoded a protein containing 386 amino acid residues, which shared a 100% homology with the SAM synthase from M. purpureus (GenBank Accession No. TQB68059.1). The q RT-PCR analysis showed that the transcriptional level of the SAM synthetase gene Mon2A2272 in M. purpureus 2272 was about 4.5-fold higher than that in the wild strain M. purpureus RP2 on day 2 of fermentation (Figure 6). Consequently, much more SAM was generated and accumulated in the fermentation broth in M. purpureus 2272 than that in M. purpureus RP2 after 3 days of fermentation (Figure 5). In addition, the yields of red, orange, and yellow MPs all decreased in M. purpureus 2272 in comparison with M. purpureus RP2 during 12 days of fermentation (Figure 3). Morphology observation showed no obvious difference between the mycelium colonies of M. purpureus 2272 and M. purpureus RP2 (Figure 4), indicating that expression of the SAM synthetase gene Mon2A2272 did not affect the mycelial development. The results gave a probable explanation for MPs reduction by the addition of methionine in fermentation. SAM synthetase expression likely exerted the negative regulation on MPs production in M. purpureus RP2.
S-Adenosylmethionine and S-Adenosylmethionine Synthetase Mon2A2272 Both Regulated Monascus Pigments Biosynthesis Gene Cluster Expression
The conserved PKS gene cluster was responsible for MPs biosynthesis in Monascus (Yang et al., 2014; Chen et al., 2015, Chen W. et al., 2017). A classical PKS gene cluster was detected in M. purpureus RP2 and it consisted of 15 structural genes encoding functional enzymes and two regulatory elements Mon2A4595 and Mon2A4602 (Supplementary Figure 2). Mon2A4595 encoded the transcriptional regulatory protein and shared a 99% homology with the transcription factor MpigI (GenBank Accession No. APZ73944.1) from Monascus ruber M7; and Mon2A4602 encoded the pigment biosynthesis activator and shared a 97% homology with the pigment biosynthesis activator PigR (GenBank Accession No. AGL44390.1) from M. ruber M7. Yang et al. (2014) reported that most genes in the PKS cluster including the two regulatory elements were expressed at higher levels under high pigment production conditions in M. purpureus YY-1 and Chen D. et al. (2017) reported that expression of the transcriptional regulatory protein in the PKS cluster was upregulated when pigment biosynthesis was induced by blue light in M. purpureus M9. Hence, the MPs biosynthesis regulatory elements Mon2A4595 and Mon2A4602 were selected for gene transcriptional level analysis by qRT-PCR during fermentation. As shown in Figure 6, the transcriptional levels of Mon2A4595 and Mon2A4602 both increased in M. purpureus RP2 with addition of SAM and in M. purpureus 2272 with an expression of SAM synthetase Mon2A2272 during the early 6 days of fermentation when MPs was synthesized in large amounts. Generally, SAM synthetase Mon2A2272 expression led to a higher expression of the transcriptional regulatory protein Mon2A4595, while the addition of SAM gave rise to a higher expression of the pigment biosynthesis activator Mon2A4602. Furthermore, the addition of SAM in fermentation resulted in higher expressions of most functional enzymes encoding genes in the PKS cluster in comparison with that without SAM addition and with Mon2A2272 expression (Supplementary Figure 3). This could probably explain the difference in MP production yield between M. purpureus RP2 with and without the addition of SAM and M. purpureus 2272 with the expression of SAM synthetase Mon2A2272. It suggested that SAM and SAM synthetase both could affect the expression of the PKS gene cluster by the regulatory elements to regulate MPs biosynthesis.
Furthermore, the global regulator, Mon2A1908 that shared a 100% homology with the global regulator of sporulation and secondary metabolism LaeA (GenBank Accession No. AIY63188.1) from M. ruber M7 was subjected to transcription analysis in response to the addition of SAM and expression of SAM synthetase Mon2A2272. It is observed that the transcriptional level of Mon2A1908 was about 4-fold higher in M. purpureus RP2 with the addition of SAM than that in M. purpureus RP2 without SAM and M. purpureus 2272 at the beginning of fermentation (Figure 6). The biosynthesis of many secondary metabolites in filamentous fungi is regulated by global transcriptional regulators, such as LaeA (Lind et al., 2018). In Aspergillus species, the global regulatory proteins VeA and LaeA have been shown to control both development and secondary metabolism (Calvo and Cary, 2015). Liu et al. (2016) reported that inactivation of LaeA in M. ruber drastically reduced the production of multiple secondary metabolites, such as Monascus pigments and citrinin and also resulted in the formation of an abnormal colony phenotype with abundant aerial hyphae. As previously reported, the addition of SAM enhanced MPs production and altered the morphology of the mycelium colony of M. purpureus RP2, it is reasonable to conclude that SAM probably regulates MPs biosynthesis and the mycelial development by interaction with the global regulator Mon2A1908.
Secondary metabolites biosynthesis coupled with asexual and sexual development in Aspergillus fungi is regulated by various environmental signals, such as temperature, pH, light, and carbon or nitrogen sources (Calvo et al., 2002; Calvo and Cary, 2015; Liu et al., 2016; Lind et al., 2018). In addition, it is controlled by a complex global network involved multiple regulators at the cellular level (Calvo et al., 2002; Calvo and Cary, 2015; Lind et al., 2018). This work proposed a possible bi-directional control mechanism of MPs biosynthesis by SAM metabolism from methionine in M. purpureus RP2. It is interesting to note that methionine could induce expression of SAM synthetase and SAM generation and exhibit the inhibited effect on MPs production, while high-level SAM inversely enhanced MPs biosynthesis. It is possible that the dynamic change in SAM synthetase expression level and the pool of SAM would be important signals of the regulatory network for MPs biosynthesis. Nevertheless, it is indispensable to dig out more details on the molecule components and their interactions involved in the regulatory network triggered by methionine and SAM. The findings in this work would provide a new perspective for a deep understanding of MPs biosynthesis regulation in M. purpureus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
SY conceived and designed the experiments, wrote the manuscript, contributed to the manuscript revision, financial support, and supervision. DY, YZ, and BH performed the experimental work and data analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (31401669), Joint Program of Beijing Natural Science Foundation and Beijing Municipal Education Commission (KZ201910011014), Support Project of High-level Teachers in Beijing Municipal Universities (IDHT20180506), and the Talent Training Quality Construction-First Class Professional Construction (PXM2019-014213-000010).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.921540/full#supplementary-material
References
Arai, T., Koganei, K., Umemura, S., Kojima, R., Kato, J., Kasumi, T., et al. (2013). Importance of the ammonia assimilation by Penicillium purpurogenum in amino derivative Monascus pigment. PP-V, production. AMB Exp. 3:19. doi: 10.1186/2191-0855-3-19
Brosnan, J. T., Brosnan, M. E., Bertolo, R., and Brunton, J. A. (2007). Methionine: A metabolically unique amino acid. Livest. Sci. 112, 2–7. doi: 10.1016/j.livsci.2007.07.005
Calvo, A. M., and Cary, J. W. (2015). Association of fungal secondary metabolism and sclerotial biology. Front. Microbiol. 6:62. doi: 10.3389/fmicb.2015.00062
Calvo, A. M., Wilson, R. A., Bok, J. W., and Keller, N. P. (2002). Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 66, 447–459. doi: 10.1128/MMBR.66.3.447-459.2002
Cavuoto, P., and Fenech, M. F. (2012). A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat Rev. 38, 726–736. doi: 10.1016/j.ctrv.2012.01.004
Chen, D., Chen, M., Wu, S., Li, Z., Yang, H., and Wang, C. (2017). The molecular mechanisms of Monascus purpureus M9 responses to blue light based on the transcriptome analysis. Sci. Rep. 7:5537. doi: 10.1038/s41598-017-05990-x
Chen, M. H., and Johns, M. R. (1993). Effect of pH and nitrogen source on pigment production by Monascus purpureus. Appl. Microbiol. Biot. 40, 132–138. doi: 10.1007/BF00170441
Chen, W., Chen, R., Liu, Q., He, Y., He, K., Ding, X., et al. (2017). Orange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 8, 4917–4925. doi: 10.1039/c7sc00475c
Chen, W., He, Y., Zhou, Y., Shao, Y., and Chen, F. (2015). Edible filamentous fungi from the species Monascus: early traditional fermentations, modern molecular biology, and future genomics. Compr. Rev. Food Sci. F. 14, 555–567. doi: 10.1111/1541-4337.12145
Dhale, M. A., Puttananjaiah, M. K. H., Sukumaran, U. K., and Govindaswamy, V. (2011). Production of Monascus purpureus pigments; influenced by amidase and acid protease activity. J. Food Biochem. 35, 1231–1241. doi: 10.1111/j.1745-4514.2010.00447.x
Dufosse, L., Fouillaud, M., Caro, Y., Mapari, S. A., and Sutthiwong, N. (2014). Filamentous fungi are large-scale producers of pigments and colorants for the food industry. Curr. Opin. Biotech. 26, 56–61. doi: 10.1016/j.copbio.2013.09.007
Feng, Y., Shao, Y., and Chen, F. (2012). Monascus pigments. Appl. Microbiol. Biot. 96, 1421–1440. doi: 10.1007/s00253-012-4504-3
Gerke, J., Bayram, Ö, and Braus, G. H. (2012). Fungal S-adenosylmethionine synthetase and the control of development and secondary metabolism in Aspergillus nidulans. Fungal Genet. Biol. 49, 443–454. doi: 10.1016/j.fgb.2012.04.003
Green, M., and Sambrook, J. (2012). Molecular Cloning: A Laboratory Manual, edition no.4. New York, NY: Cold Spring Harbor Laboratory Press.
Hajjaj, H., François, J. M., Goma, G., and Blanc, P. J. (2012). Effect of amino acids on red pigments and citrinin production in Monascus ruber. J. Food Sci. 77, M156–M159. doi: 10.1111/j.1750-3841.2011.02579.x
Jung, H., Choe, D., Nam, K. Y., Cho, K. H., and Shin, C. S. (2011). Degradation patterns and stability predictions of the original reds and amino acid derivatives of Monascus pigments. Eur. Food Res. Technol. 232, 621–629. doi: 10.1007/s00217-011-1427-7
Jung, H., Kim, C., Kim, K., and Shin, C. S. (2003). Color characteristics of Monascus pigments derived by fermentation with various amino acids. J. Agric. Food Chem. 51, 1302–1306. doi: 10.1021/jf0209387
Jung, H., Kim, C., and Shin, C. S. (2005). Enhanced photostability of Monascus pigments derived with various amino acids via fermentation. J. Agric. Food Chem. 53, 7108–7114. doi: 10.1021/jf0510283
Kim, J. G., Choi, Y. D., Chang, Y. J., and Kim, S. U. (2003). Genetic transformation of Monascus purpureus DSM1379. Biotechnol. Lett. 25, 1509–1514. doi: 10.1023/a:1025438701383
Lakrod, K., Chaisrisook, C., and Skinner, D. Z. (2003). Expression of pigmentation genes following electroporation of albino Monascus purpureus. J. Ind. Microbiol. Biotechnol. 30, 369–374. doi: 10.1007/s10295-003-0058-9
Lieber, C. S., and Lester, P. (2002). S-adenosylmethionine: molecular, biological, and clinical aspects – an introduction. Am. J. Clin. Nutr. 76, 1148S–1150S. doi: 10.1093/ajcn/76/5.1148S
Lin, Y. L., Wang, T. H., Lee, M. H., and Su, N. W. (2008). Biologically active components and nutraceuticals in the Monascus-fermented rice: a review. Appl. Microbiol. Biot. 77, 965–973. doi: 10.1007/s00253-007-1256-6
Lind, A. L., Lim, F. Y., Soukup, A. A., Keller, N. P., and Rokas, A. (2018). An LaeA- and BrlA-dependent cellular network governs tissue-specific secondary metabolism in the human pathogen Aspergillus fumigatus. mSphere 3, e00050–18. doi: 10.1128/mSphere.00050-18
Liu, Q., Cai, L., Shao, Y., Zhou, Y., Li, M., Wang, X., et al. (2016). Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biol. 120, 297–305. doi: 10.1016/j.funbio.2015.10.008
Mapari, S. A., Thrane, U., and Meyer, A. S. (2010). Fungal polyketide azaphilone pigments as future natural food colorants? Trends Biotechnol. 28, 300–307. doi: 10.1016/j.tibtech.2010.03.004
Mostafa, M. E., and Abbady, M. S. (2014). Secondary metabolites and bioactivity of the Monascus pigments review article. Global J. Biotech. Biochem. 9, 1–13. doi: 10.5829/idosi.gjbb.2014.9.1.8268
Patakova, P. (2013). Monascus secondary metabolites: production and biological activity. J. Ind. Microbiol. Biot. 40, 169–181. doi: 10.1007/s10295-012-1216-8
Putt, K. K., Pei, R. S., White, H. M., and Bolling, B. W. (2017). Yogurt inhibits intestinal barrier dysfunction in Caco-2 cells by increasing tight junctions. Food Funct. 8, 406–414. doi: 10.1039/c6fo01592a
Said, F. M., Brooks, J., and Chisti, Y. (2014). Optimal C:N ratio for the production of red pigments by Monascus ruber. World J. Microbiol. Biotechnol. 30, 2471–2479. doi: 10.1007/s11274-014-1672-6
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shi, K., Song, D., Chen, G., Pistolozzi, M., Wu, Z., and Quan, L. (2015). Controlling composition and color characteristics of Monascus pigments by pH and nitrogen sources in submerged fermentation. J. Biosci. Bioeng. 120, 145–154. doi: 10.1016/j.jbiosc.2015.01.001
Srianta, I., Ristiarini, S., Nugerahani, I., Sen, S. K., Zhang, B. B., Xu, G. R., et al. (2014). Recent research and development of Monascus fermentation products. Int. Food Res. J. 21, 1–12.
Wang, Y., Wang, D., Wei, G., and Shao, N. (2012). Enhanced co-production of S-adenosylmethionine and glutathione by an ATP-oriented amino acid addition strategy. Bioresour. Technol. 107, 19–24. doi: 10.1016/j.biortech.2011.12.030
Xiong, X., Zhang, X., Wu, Z., and Wang, Z. (2015). Coupled aminophilic reaction and directed metabolic channeling to red Monascus pigments by extractive fermentation in nonionic surfactant micelle aqueous solution. Proc. Biochem. 50, 180–187. doi: 10.1016/j.procbio.2014.12.002
Yang, Y., Liu, B., Du, X., Li, P., Liang, B., Cheng, X., et al. (2014). Complete genome sequence and transcriptomics analyses reveal pigment biosynthesis and regulatory mechanisms in an industrial strain Monascus purpureus YY-1. Sci. Rep. 5, 8331–8331. doi: 10.1038/srep08331
Keywords: methionine, S-adenosylmethionine, SAM synthetase, Monascus pigments, Monascus purpureus
Citation: Yin S, Yang D, Zhu Y and Huang B (2022) Methionine and S-Adenosylmethionine Regulate Monascus Pigments Biosynthesis in Monascus purpureus. Front. Microbiol. 13:921540. doi: 10.3389/fmicb.2022.921540
Received: 16 April 2022; Accepted: 17 May 2022;
Published: 14 June 2022.
Edited by:
Jun Liu, Central South University Forestry and Technology, ChinaReviewed by:
Fusheng Chen, Huazhong Agricultural University, ChinaChuannan Long, Jiangxi Science and Technology Normal University, China
Copyright © 2022 Yin, Yang, Zhu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sheng Yin, eWluc2hlbmdAYnRidS5lZHUuY24=
 Sheng Yin
Sheng Yin Dongmei Yang3
Dongmei Yang3