
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 28 July 2022
Sec. Food Microbiology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.915721
This article is part of the Research TopicFrom Traditional to Modern: Progress of Molds and Yeasts in Fermented-food Production, Volume IIView all 12 articles
The biosynthesis of penicillin G (PG) is compartmentalized, and the transportation of the end and intermediate products, and substrates (precursors) such as L-cysteine (L-Cys), L-valine (L-Val) and phenylacetic acid (PAA) requires traversing membrane barriers. However, the transportation system of PAA as a side chain of PG are unclear yet. To discover ABC transporters (ABCTs) involved in the transportation of PAA, the expression levels of 38 ABCT genes in the genome of Monascus ruber M7, culturing with and without PAA, were examined, and found that one abct gene, namely abct31, was considerably up-regulated with PAA, indicating that abct31 may be relative with PAA transportation. Furthermore the disruption of abct31 was carried out, and the effects of two PG substrate's amino acids (L-Cys and L-Val), PAA and some other weak acids on the morphologies and production of secondary metabolites (SMs) of Δabct31 and M. ruber M7, were performed through feeding experiments. The results revealed that L-Cys, L-Val and PAA substantially impacted the morphologies and SMs production of Δabct31 and M. ruber M7. The UPLC-MS/MS analysis findings demonstrated that Δabct31 did not interrupt the synthesis of PG in M. ruber M7. According to the results, it suggests that abct31 is involved in the resistance and detoxification of the weak acids, including the PAA in M. ruber M7.
In view of the present antibiotic resistance crises and the slowing rate of antibiotic discovery, there has been a worldwide push to look into the large quantity of unexplored natural product sources for novel antibiotics or their precursors (Seiple et al., 2016). Even though β-lactam antibiotics have been present since the 1920s, they remained the most commonly utilized antibiotic class. Meanwhile, new routes for biosynthesis of β-lactam are continually being found (Gaudelli et al., 2015). In China and some other Asian regions, Monascus spp. are popular as conventional edible fungi. Their fermented products, namely red yeast rice, red mold rice (RMR), Anka, or Hongqu, have been widely utilized in China as folk cures and culinary colorants for 2,000 years (Lee et al., 2010; Lee and Pan, 2012; Guo et al., 2019). Researchers have publicized that Monascus spp. can create both useful and harmful secondary metabolites (SMs), such as Monascus pigments, aminobutyric acid, dimurumic acid, and monacolin K, and a typical type of mycotoxin, which is known as citrinin (Ozcengiz and Demain, 2013; Yuliana et al., 2017; Chen and Pan, 2019). Recently, Chen (2015) found a probable gene cluster having liability for β-lactam synthesis in the genome of M. ruber M7, and one β-lactam biosynthesis gene was identified in the gene cluster (Ramzan et al., 2019). M. purpureus was also proved to produce γ-lactam according to Wei et al. (2017).
Many microorganisms, including filamentous fungi, actinomycetes, and Gram-negative bacteria, may synthesize several types of β-lactam antibiotics (Lobanovska and Pilla, 2017). During the biosynthetic pathway of the β-lactam antibiotics, the non-ribosomal peptide synthetase initially condenses the three precursor amino acids, L-α-aminoadipic acid, L-cysteine and L-valine, to form the basic nucleus of β-lactam antibiotic, isopenicillin N (IPN) with hydrophilic feature (Fernández-Aguado et al., 2014). The IPN fabrication is taken place in the cytoplasm of β-lactam producers (Barreiro and García-Estrada, 2019). Then, the conversion of IPN is taken place into penicillin G (PG) in the peroxisomal matrix of β-lactam producers (Van Den Berg et al., 2008; Ávalos et al., 2014). During this process, an hydrophilic α-aminoadipyl side chain, phenylacetic acid (PAA), is catalyzed into a hydrophobic phenyl acetyl CoA of IPN by the IPN acyl transferase (García-Estrada et al., 2008). The entry of the side chain of PG, PAA, into the cells takes place via passive diffusion. And acidification of cytosol is caused by the influx of PAA. After entering the cells, PAA is dissociated and releases protons. Due to the acidification of cytoplasm, enzymatic activities are inhibited, which causes the ultimate death of the cell (Weber et al., 2012). Nevertheless, cytoplasmic acidification is unlikely to be the only cause of mild acid toxicity. The more hydrophobic sorbic acid, for example, may impair the organization of the membrane, leading to oxidative stress due to a lesser functioning of the respiratory chain, and so toxicity could be a multidimensional phenomenon. So, during the biosynthesis of penicillin, the shipping of intermediates and their precursors across the cellular membranes is an important matter (Martín et al., 2005). Several transporters are not fully explored (Fernández-Aguado et al., 2013, 2014). According to their abilities to transport diverse organic composites, the transporters are classified into the extrusion of multidrug and toxic compounds, small multidrug resistance, ATP-binding cassette (ABC) superfamily, major facilitator superfamily, and so on (Martín et al., 2005).
Binding and hydrolyzing ATP to transport the substrates across the lipid bilayer is the characteristic ability of the ABC transporter family (Wilkens, 2015). The well-studied ABC exporters in prokaryotes and eukaryotes play a central role in life activities. There have been expansive investigations of their association with many human diseases (Fletcher and Mullins, 2010; Tang et al., 2021) and resistance to multiple drugs (Al Shawi et al., 2011). Weber et al. (2012) found that PAA in higher concentrations could be toxic to the cells, and the proliferation of the common ancestor of the recent penicillin-producing strains, P. chrysogenum Wisconsin 54-1255, is not possible at a high concentration (18 mM) of PAA. The high penicillin-producing strain P. chrysogenum DS17690, on the other hand, is generally insensitive to such PAA doses, showing that DS17690 cells have established PAA resistance mechanisms. Likewise, a weak acid extrusion system has been identified in Saccharomyces cerevisiae. PDR12 is an ABC transporter from S. cerevisiae that prevents cytoplasmic acidification and transports metabolites of yeast metabolism out of the cells (Hazelwood et al., 2006; Zhang et al., 2021).
Though several features of the traffic systems of β-lactam antibiotics need to be investigated further, it has been thought that the involvement of the transmembrane transporter proteins plays a role in their synthesis. As a result, more researches about the involvement of transporters in the β-lactam antibiotic gene cluster are required. Moreover, it has been strongly predicted that the ABC transporters encoding genes are present in the gene clusters. In the present study, the expression and identification of all ABC transporters present in the genome of M. ruber M7 for the biosynthesis of the β-lactam antibiotics have been investigated after being grown in the presence and absence of PAA. In the presence of PAA, the results revealed one ABC transporter (ABCT31), whose role in β-lactam antibiotic production was explored via abct31-deletion. Furthermore, compared to M. ruber M7, the resistance of the mutant to the weak acid toxicity was assessed at various concentrations with the presence and absence of PAA.
The Δabct31 mutant was created using the M. ruber M7 strain (CCAM 070120, Culture Collection of State Key Laboratory of Agricultural Microbiology, < city>Wuhan < /city>, China), which can produce Monascus pigments and citrinin but not monacolin K. (Chen and Hu, 2005). Czapek yeast extract agar (CYA), potato dextrose agar (PDA), glycerol nitrate agar (G25N 25%), and malt extract agar (MA) media were used for the phenotypic analyses (He et al., 2013). On PDA, resistance markers hygromycin B (30 μg/mL) and neomycin (15 μg/mL) were used for transformants screening. PDA slants of strains were prepared and kept at 28°C.
The strains were cultured on the PDA plates with cellophane covers. Extraction of the genomic DNA of the grown strains from their mycelia was done using the cetyltrimethylammonium bromide method by Shao et al. (2009).
The method described by Liu et al. (2014) was followed to conduct the qRT-PCR using the SLAN Fluorescence Quantitative Detection System from Wuhan Good Biotechnology Co., Ltd., (Wuhan, Hubei, China).
Table 1 lists the primer pairs utilized in the current investigation. The abct31gene was amplified by PCR using M. ruber M7 genomic DNA under the subsequent conditions. The first denaturation step was performed for 5 min at 94°C, preceded by 35 amplification cycles at 94°C for 30 s, at 58°C for 30 s, and for 1 min at 72°C. However, the T100 Thermal Cycler was used at the end during the extension phase at 72°C for 10 min (Bio-Rad, Hercules, CA, USA) (Chen et al., 2012a,b). SoftBerry's FGENESH software (https://linuxl.softberry.com/berry.phtml) was used to anticipate the ABCT31 amino acid sequences. The Pfam 27.0 program (http://pfam.xfam.org/) was used to examine the ABCT31 functional regions. While the BLASTP program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was utilized to interpret the homology of the presumed amino acid sequences of ABCT31. Moreover, the PSIPREDv3.2 server (http://bioi nf.cs.ucl.ac.uk/psip red/) (Buchan et al., 2013; Waterhouse et al., 2018) and RaptorX server (http://rapt orx.uchi cago.edu/) (Wang et al., 2016b) were employed, respectively, to explain the predictions of genes for secondary structural elements. Similarly, from the PSIPRED server, HMMTOP, MEMSAT3, and MEMSAT-SVM programs were used to describe membrane helix and topology predictions (Pandey et al., 2018). The Deep Loc-1.0 server (https://services.healthtech.dtu.dk/service.php?DeepLoc-1.0) was used to determine the likely subcellular protein localization (Emanuelsson et al., 2000). Predictions of all genes ontology domains for the probable gene term annotations were done using the FFPred 3 program (http://bioinf.cs.ucl.ac.uk/psipred/) (Pandey et al., 2018).
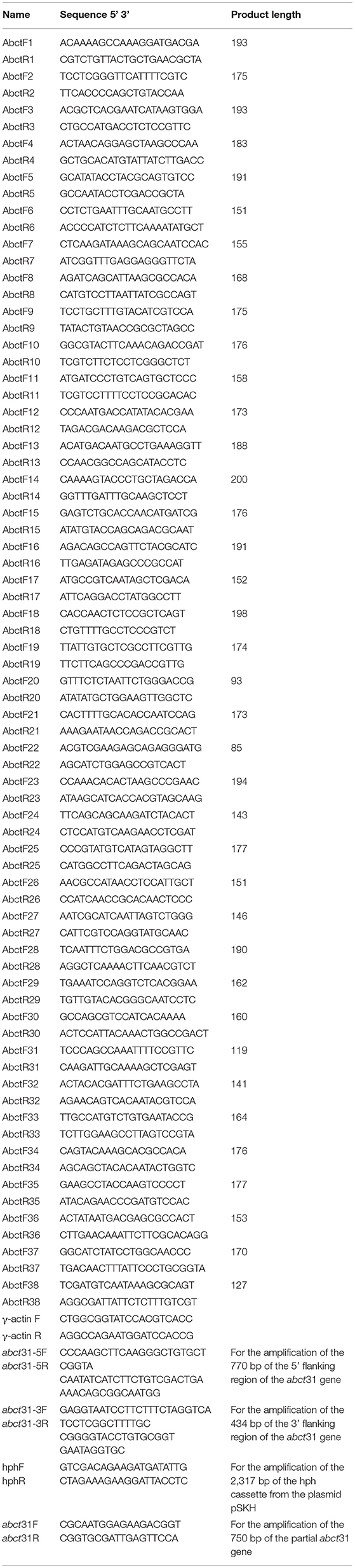
Table 1. Primers for all ABC transporter genes in M. ruber M7 and used in the construction of deletion cassettes for the gene abct31.
The approach of Shao et al. (2009) was used to delete the targeted gene abct31. The construction of the cassette for gene disruption (5'UTR-hph-3'UTR) was carried out with a double-joint PCR procedure. The primer pairs are listed in Table 1, whereas Figure 1A demonstrates the schematic illustration (Wang et al., 2011). For the development of the deletion strain (Δabct31), construction of the Agrobacterium tumefaciens cells expressing the disruption vector (pC- abct31) of abct31 was done, and its co-cultivation was carried out with M. ruber M7.
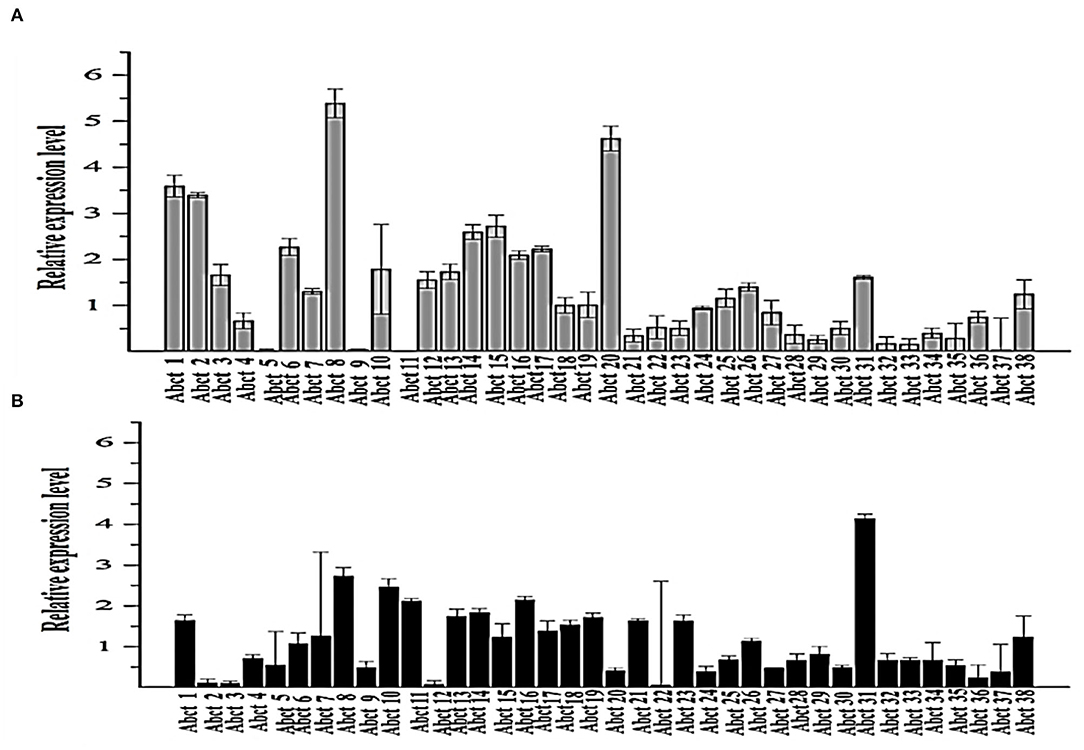
Figure 1. Quantitative real-time PCR analysis of the expression of all ABC transporter genes in the M. ruber M7 toward PAA. (A) The comparative expression of all ABC transporter genes in M. ruber M7 without PAA; (B) A comparative expression of all ABC transporter genes in M. ruber M7 with PAA. M. ruber M7 was cultivated and incubated at 28°C for 7 d in the presence (black bar) and absence (gray bar) of PAA at 10 mM. The values were represented as average values ± sd.
The abct31 gene deletion strain was further confirmed utilizing the DIG-High Prime DNA Labeling & Detection Starter kit's method by applying PCR and Southern blot method (Roche, Mannheim, Germany). The restriction enzyme XbaI was used to digest the DNA of the putative Δabct31 strain and M. ruber M7, respectively. As shown in Table 1, the respective primer pairs hphF/hphR and abct31F/abct31R were used to amplify the hph gene (probe 2) and abct31 gene (probe 1) by the PCR. Finally, probes 1 and 2 confirmed the Δabct31 strain.
In order to study the phenotypic characteristics, colonial and microbiological features of parental (M. ruber M7) and Δabct31strains were inoculated on PDA, G25N, MA, and CYA media plates and incubated at 28°C for 15 days (Chen et al., 2017a).
Estimation of the biomasses of M. ruber M7 and Δabct31 was accomplished by the gravimetric method. The collection of mycelia on PDA plates was done following the drying at 60°C until the constant weights were obtained. Three replicates were used to compute the mean biomasses (Wang et al., 2016a).
The PG was extracted from the respective M. ruber M7 (wild-type) and Δabct31 by the previously defined method (Ramzan et al., 2019).
The previously reported method (Ullán et al., 2008; Ramzan et al., 2019) was used for the analysis of intra- and extracellular PG contents.
The mass profile of the extract was produced using electrospray ionization (ESI) coupled Acquity TQD tandem quadruple mass spectrometer (Waters, Manchester, UK). The conditions for the ESI-MS to detect MS were set according to Ramzan et al. (2019). The presence of PG metabolites in the extract was confirmed using a PG standard with a purity of more than 98.0 percent (Sigma-Aldrich) (Lopes et al., 2013).
The feeding of amino acids was used to investigate the transportation role of the abct31 gene (Yang et al., 2012). For this purpose, Δabct31 and M. ruber M7 were separately grown on PDA with 2 mM concentrations of PG pathway amino acids such as D-Val, L-Cys and PAA, respectively. Then, phenotypic characteristics, biomasses, and PG synthesis were analyzed.
The feeding of mutants and M. ruber M7 on PDA with the addition of some weak acids including PAA, acetic acid, adipic acid and sorbic acid was used to examine the transportation mechanism of the abct31 gene (Weber et al., 2012). This procedure was carried out according to the method described by Weber et al. (2012) with slight modification such as PAA (mM): 50, 62.5, 75, 100, and 125; acetic acid (mM): 5, 10, 15, 20, and 25; adipic acid (mM): 12.5, 25, 37.5, and 50; and sorbic acid (mM): 2, 2.75, 3.5, and 4.25 concentration. The phenotypical features and relative expression of abct31 gene in M. ruber M7 were analyzed.
All of the experiments were carried out in triplicate. The statistical studies were performed using the Statistics 8.1 program (Analytical Software, USA). After performing the Tukey test, a P < 0.05 was taken as statistically significant, and a P < 0.01 was considered highly significant.
To understand the proper mechanism of the abct31 gene as a transporter in the translocation of intermediates metabolites of the PG pathway, M. ruber M7 was cultivated for 7 days at 10 mM in the +PAA (presence) or –PAA (absence). The determination of the transcript levels of all ABC transporter genes in the genome of M. ruber M7 was done while the γ-actin gene used as a reference gene (Figure 1).
The ABC transporter expression levels have been noted in M. ruber M7 with or without PAA. The experiment results revealed a notable upsurge in the expression of abct31 (Figure 1B) in the presence of PAA in M. ruber M7, whereas without the PAA (Figure 1A), the expression of abct31 was considerably lower. In the +PAA/–PAA, all other ABC transporter genes displayed no expression or did not show a distinct variation in their transcript levels in M. ruber M7 (Figures 1A,B).
The ABCT31 protein sequence from M. ruber M7 was analyzed using NCBI's blast tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and homologous aligning of the abct31 with 27 distinct ABC transporters from other genomes as well as identification of different conserved domains were discovered (Table 2). Softberry's FGENESH software was also used to predict the 1,257 amino acid sequence. abct31 corresponds to the ABC transporter family, according to a database search using the Pfam 27.0 tool (http://pfam.xfam.org/). The expression of an ABC-membrane family C domain (PLN03130, pfam00005) represents hydrophobic transmembrane helices, (Schulz and Kolukisaoglu, 2006), and a key component of ABC transport proteins and its recognition is conserved by the low E values (Figure 2).
Although atomic resolution crystal structures of various soluble proteins have been discovered, equivalent advancement has not been reported for transporter proteins. The key challenge is the severe exertion in crystallizing them due to flexibility in their conformation. As a result, developing a homology-based protein model is a viable option (Gao et al., 2021). The ABCT31's final 3D structure (Figure 3A), which was created using the SWISS-MODEL software and a template-based homology model, was visualized using Procheck. The basic layout of all ABC transporters is the same, with two transmembrane domains (TMDs) and two nucleotide-binding domains (NBDs). The structural analysis has revealed that ABCT31 protein performed as an importer and possessed two TMDs marked as TMD1 and TMD2, similarly to NBDs grouped into NBD1 and NBD2 in the three-dimensional structure presented in Figure 3A. In ABCT31, protein is fused between TMDs and NBDs. The ABC transporter's fundamental unit comprises all of these domains. The ABC signature motifs, Walker A (P-loop) and Walker B, which are prevalent but not limited to ABC transporters, are used to classify the NBD.
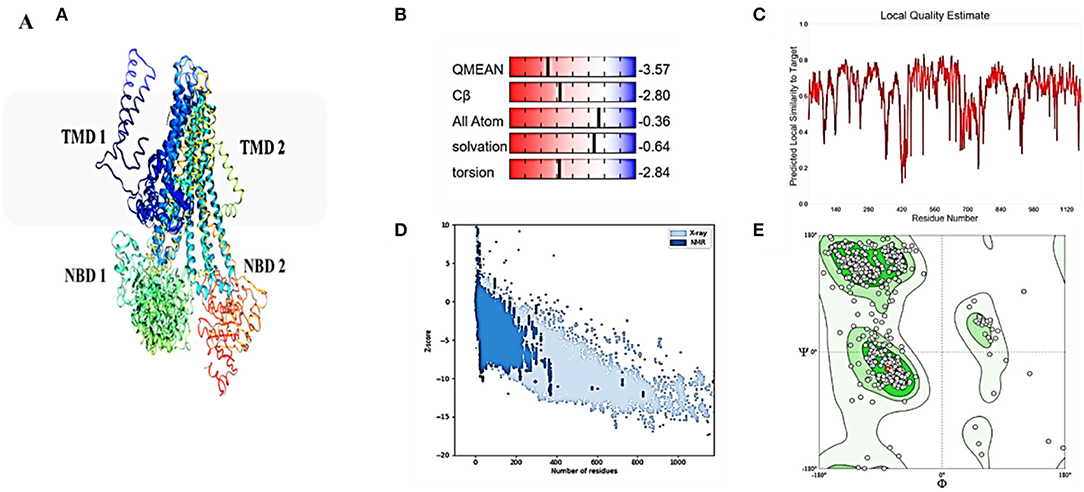
Figure 3. Homology model and checking results for ABCT31 protein. Verification of homology modeling (A) Homology model of ABCT31 protein TMD, Transmembrane domain; NBD, Nucleotide-binding domains; (B) Plot displaying the model's normalized Q mean score; (C) Plot displaying the model's local quality; (D) Z score value of the model developed; (E) Ramachandran plot validating the energy-minimized model's backbone dihedral angles.
The assemblage, dynamics, surface characteristics, and thermodynamics of inorganic, polymeric, and biological systems have been studied using molecular modeling approaches. The target protein 3D framework has been developed using the SWISS-MODEL software and template-based homology modeling. The findings suggest that the ABCT31 protein seems to have a homological similarity with the 6pza 1 (ATP-binding cassette, sub-family C member 8). The highest template for homology-based modeling for ABCT31 was reported as having a sequence identical score of 33.17 percent.
The QMEAN-Z score was found to be 3.57 in the context of the global score by QMEAN, indicating the relatively strong quality of the model (<4) (Figure 3B). The model quality determinant is the Z score value representing the structure's total energy. The observed Z score value of ABCT31 was noted as −10.34, as shown in Figure 3D, which falls in the range normally found in comparable protein chains in the PDB, demonstrating the structure's dependability. The energy plot presented in Figure 3E depicts the quality of the local model by graphing energies as a function of the position of amino acids in sequence, with +ve values corresponding to problematic or incorrect regions of a model in general. The residues further validated the projected model's consistency with negative energy (Kulkarni and Devarumath, 2014).
WHAT IF was used to assess the packing environment for residues of the modeled ABCT31 to the experimental structures. An equal or < -5.0 score indicates poor structural packing (Singh et al., 2016; Stitou et al., 2021). Compared with the template X-ray structure (Figure 3C), the produced structure has similar packing scores, with only a few residues having poor packing, as their score values have been shown (−1.074) lesser than −5.0. The ABCT31's stereochemical and energetic features were evaluated using a Ramachandran plot, which revealed that 1.3 percent of residues (15) in the disallowed/outlier region, 5.6 percent of residues (66) in the allowed region, and 93.1 percent of residues (1094) in the most favored zones, of the RAMPAGE server (Figure 3E). The statistics in the preferred and allowed regions and the low proportion in the outlier show the acceptability of the abct31's Ramachandran plot. The PROCHECK goodness factor (G-factor) revealed important information about the distances of covalent and overall bond angles. However, after the analysis of the G-factor for the modeled ABCT31 was > 0.502, indicating that the quality of the projected model was extremely high. The G-factor and the overall Ramachandran plot properties ensured the ABCT31structure's quality (Table 3). The overall quality factor of ABCT31 was 88.1651, according to an ERRAT analysis. The quality of the 3D protein model generated in this work is confirmed by the findings acquired from multiple quality evaluation servers.
As described in section Model Validation and Homology Modeling, the NBDs possess some conserved motifs, which play an important role in the hydrolysis and binding of ATP in the opening and closing mechanism of the TMDs'. NBDs dominate the subunits of ABC, and these might be divided into two constituent domains: the conserved Walker A motif or P-loop (GXXGXGK(S/T), which is contained in the catalytic core domain, Walker B motif (ϕϕϕϕD, ϕ represents the hydrophobic residue), switch region (also known as Q-loop and H-motif) together with an α-helical domain that is more diverse on a structural basis, and it contains the signature motif of ABC (LSXGQXXXXX). The nucleotide state affects the comparative alignment of the catalytic domains and helix. The ABC subunits were packed together in a “head-to-tail” configuration in an intact transporter so that one subunit's P loop is positioned toward the other subunit's signature. In the present ABC protein, the sequence of Walker A (GPIGSGKTS), Walker B (IVLLDD) ABC transporter signature (LSRGQKQRIA), and the three-loop structures Q (FCDQ), D (SGLD), and H (VLVTHAN) was also identified and located, respectively, as shown in Figure 4A.
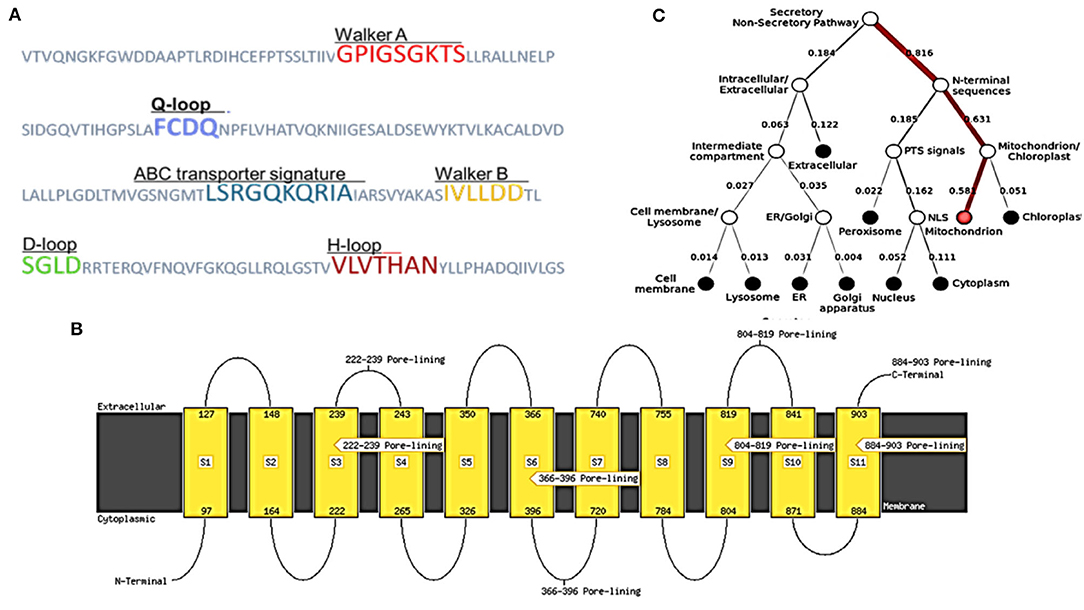
Figure 4. Structural features and location analysis of the ABCT31 protein. (A) Features loop and walker of the abct31 protein; (B) Topology analysis of the ABCT31 protein; (C) Location of abct31 gene in M. ruber M7.
Low E values (Figure 1) determined by the CDD software indicated that most amino acids in the ABC-MRP domains (accession: cd03250 and cd03244) are substantially retained in all homologs. The extremely preserved amino acid motifs are contained in the NBD of the ABC-MRP domain and play a role in generating the required energy, which is ultimately used for powering the transport mechanism (Ghilarov et al., 2021). The Walker A [GXXGXGK(S/T)] and Walker B (hhhhDE, where h represents the hydrophobic amino acid) motifs were discovered to be ATP-binding motifs inside the NBD (Figure 4A). The establishment of formally extensive connections with an ATP molecule's phosphate group is done by the Walker A motif. Similarly, the coordination of water and Mg2+ at the catalytic site is done by the Asp of the Walker B motif.
In contrast, the catalytic glutamate is essential for the hydrolysis of ATP (Zaitseva et al., 2005). Likewise, a conserved Gln plays an important function in coordinating the ATP molecule. This is located between the Walker sequences in the Q-loop. The identification of the conserved D-loop and switch or H-loop as additional motifs was done downstream of the Walker B motif and, respectively, termed as Asp and His. These coordinate the γ-phosphate through the collaboration of either D-loop (water molecule) or H-loop (direct hydrogen binding) (George and Jones, 2012). The Walker sequences of each NBD bearing the conserved amino acid sequence LSGGQ were also identified. This discovered subdomain is also implicated in ATP binding and is termed a helical subdomain or an ABC signature motif (Zaitseva et al., 2005; Chen et al., 2017b).
MEMSAT SVM is a support vector machine (SVM) oriented predictor, which is very dependable software that can also predict pore-lining residues. MEMSAT 3, HMMTOP, and MEMSAT SVM are transmembrane protein topological predicting tools. With a total entropy of 18.8083, these programs indicated 11 transmembrane helices (Figure 4B). The illustrated transmembrane helices have a high number of hydrophobic amino acids, and the MEMSAT SVM software was used to represent the final interpreted topology of this transmembrane protein, as shown in Figure 4B. It also discovered four pore-lining helices with a pore stoichiometry of 1, containing residues from 222 to 239, 366 to 396, 804 to 819, and 884 to 903. It has been observed that the transmembrane domain area of transporter proteins forming transport channels is mostly made up of membrane-spanning alpha-helices with bends and kinks, giving significant structural variation that is necessary for the channel's activity (Moussatova et al., 2008; Nugent and Jones, 2012). The occurrence of transmembrane with hydrophobic regions revealed its membrane-embedded architecture and participation in transmembrane transportation, which can be confirmed further through functional and structural analysis.
FFPred predicted the biological function as transmembrane transport with 0.907 probability based on gene ontology. Molecular function analysis predicted transmembrane transporter activity as 0.963 and substrate-specific activity of transmembrane transporter as 0.769. However, the cellular component prediction was predicted as an intrinsic or integral part of the membrane with a probability of 1.0. Deeploc server identified that abct31 protein is a membrane type with 0.9973 scores while the soluble scoring is 0.0027. It also predicted that it is located in the cell membrane with a score of 0.378 (Figure 4C).
Structural information, outcomes of GO prediction, and domain analysis altogether recommend the localization and function of ABCT31 protein membrane in the transportation of substrates through transmembrane, especially the transportation of phenylacetic acid is needed to be studied further through the 3D structure analysis. However, another trusted online method known as the RaptorX-binding site prediction tool identified PA as the potential ligand and discovered two binding sites regulated by residues G487 P483, I484, G485, S486, K488, and T489 (Figure 5).
To examine the role of abct31 in vivo, the development of the abct31 gene disruption strain (Δabct31) was done by using a method (Shao et al., 2009; Liu et al., 2014, 2019). The amplification of the upstream 5' flanking region and downstream 3' flanking region (Figure 6B) was done from the genomic DNA of M. ruber M7 (schematic presentation in Figure 6A). At the same time, the pKSH vector was used to amplify the hph gene fragment (Figure 6C). All amplified fragments were purified by gel electrophoresis and a gel purification kit (Figure 6D). During the next step, the disruption cassette was ligated with the linearized pCAMBIA3300 vector by T4 ligase to make the knock-out vector (pC- abct31) of abct31. The successful ligation was further confirmed by double digestion of pC-abct31 (Lane 1-2); two bands, 8.0 kb by pCAMBIA3300 and 3.3 kb of disruption cassette, are presented in Figure 6E.
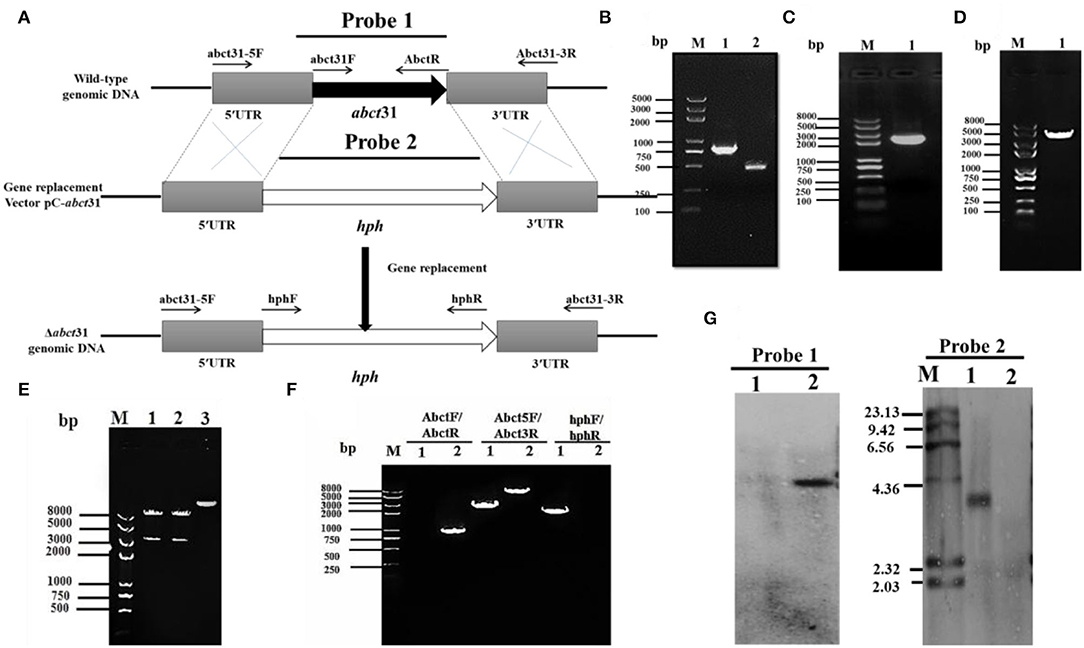
Figure 6. Construction and verification of Δabct31 strain. (A) Pictorial presentation for the homologous recombination approach to create abct31 disruption strains. (B) M: Trans 2k II Marker; Lane1: PCR product of 5' franking region (770 bp); Lane 2: PCR product of 3' franking (434 bp). (C) M: Trans 2k plus II Marker; Lane 1: hph (2,137 bp). (D) M: Trans 2k plus II Marker; Lane 1: PCR product of recombinant fragment of 5'UTR, hph and 3'UTR (3341 bp). (E) M: Trans 2k plus II Marker; Lane1: Restriction enzyme digestion analysis of vector pC-abct31 (KpnI/HindIII). (F) Validation of abct31 homologous recombination events. M: Trans 2k plus II Marker; Lane 1, the abct31 strain; Lane 2, the wild-type strain (M7). Different distinct bands were obtained by PCR amplification for selected pair of primer. (G) Southern hybridization analysis. Lane 1, Xba1 digested genomic DNA of Δabct31; lane 2, Xba1 digested genomic DNA of M7 respectively; M: λDNA/HindIII marker; Probe 1: abct31 gene; Probe 2: hph gene.
The six putative deleted (Δabct31) strains obtained by Agrobacterium-mediated T-DNA transformation to M. ruber M7 as a recipient were also identified and authenticated by PCR analyses. One of the mutants was subjected to further PCR verification, and the results are as shown in Figure 6F. The 3 pairs of primers Abct31-5F/Abct31-3R, Abct31F/Abct31R, and hphF/hphR (Table 1) used for the validation of the homologous recombination event. Lane 2 for the M7 genomic DNA and Lane 1 for the Δabct31 strain, the primer pair Abct5F/Abct3R 3.3 kb and 6.0 kb band, were amplified, respectively. In the case of ORF prime pair Abct31F/Abct31R, nothing was amplified from the Δabct31 strain while the 800 bp band amplified M7 genomic DNA. Controversially, the hphF/hphR primer pair 2.1 kb band appeared in Lane 1 (Δabct31 strain), and nothing was seen in Lane 2. The above PCR results are demonstrated in Figure 6F, the main differences in genomic DNA of Δabct31 strain and Monascus ruber (wild type) strain.
Furthermore, performing the transformant Southern hybridization verification, the results are shown in Figure 6G; with Probe 2 (Table 1) as a probe, a single hybridization band appeared in the transformant Δabct31 and nothing appeared in M7 DNA. However, ORF (Table 1) was used as a Probe 1; no hybridization band appeared for Δabct31 strain. Hybridization bands indicate that the abct31 gene in the mutant has been successfully replaced by the single copy of the hph resistance gene.
Many genes involved in the manufacture of pigments, antibiotics, herbicides, and other secondary metabolites have been cloned in the previous decade. The biosynthetic genes for cephalosporin, penicillin, cephamycins, and penicillin are grouped in clusters, as are the biosynthetic genes for other secondary metabolites and antibiotics (Ullán et al., 2002). Homologous recombination (HR) is the core tool in DNA repair that is efficiently used for gene knockout from any part of DNA (Aguilera and Gómez-González, 2008). This study shows that HR is an extremely useful tool for gene removal in filamentous fungi. The abct31 encodes the ABC transporter in the β-Lactam gene cluster of M7. The Δabct31 (deletion mutant) of this gene is created by using the gene disruption cassette (5'UTR-hph-3'UTR). In this cassette, the hph is used as a resistant gene for the screening of the mutant. The cassette's 5'UTR (770 bp) and 3'UTR (434 bp) fragments are amplified from the upstream and downstream regions of the gene, respectively. Further verification of the gene's role in abct31 β-lactam gene cluster of M7 has been conducted by evaluating the relative expression level toward weak organic acid.
The phenotypical structures were observed to explore the morphological development differences between the Δabct31 strain as related to M. ruber M7. Both deletion and wild-type strains were inoculated on four different media, including PDA, G25N, CYA, and MA, and cultivated at 28°C for 15 days. The colony size, color, and other morphological properties observed (Figure 7A) are commonly applied to investigate the morphological characteristics of Monascus strains (Li et al., 2010, 2014).
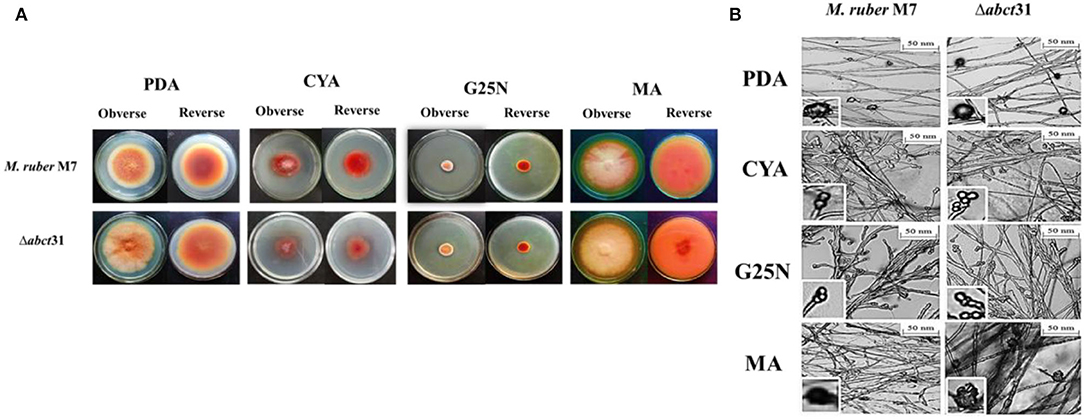
Figure 7. Morphological comparison of M. ruber M7and Δabct31. (A) Colony morphology of M. ruber M7 and Δabct31 on PDA, CYA, G25N, MA plates, and cultured at 28°C for 15 days; (B) Cleistothecia and conidia development in M. ruber M7and Δabct31 inoculated on PDA, CYA, G25N, MA plates, and cultured at 28°C for 8 days.
While seeing the results of M. ruber M7 and abct31 deletion strain (Δabct31), it was observed that the colonies of Δabct31 normally grew when these were compared with colonies of wild-type M. ruber M7 for all media (Figure 7A). However, there was a substantial difference between the strains on PDA and MA medium plates in terms of phenotypic analyses, such as colony appearance, colony edges, the size of PDA, colony diameter, and growth rate (Figure 7A). But when it comes to CYA, Δabct31's colony color was darker than M. ruber M7's. There were slight changes in the colony color and size of Δabct31 when compared with M. ruber M7 in G25N.
From Figure 7B, it can be seen that the Δabct31 mutant can produce a cleistothecium compared with M. ruber M7 on PDA and MA. There was no change observed in the cleistothecia growth pattern and size. However, the conidia structure can be observed in CYA and G25N. In both media, the conidia were observed in normal shape and size. It was clearly noted that no difference was found in the quality and quantity of cleistothecium and conidium (Figure 7B). As a result, there were no differences in cleistothecia and conidia's overall development and phenotypic characteristics between the Δabct31 (mutant strain) and M. ruber M7 (wild strain).
As well as considering the growth and structure of the hyphae in mutant Δabct31 strain remained unchanged compared with the control of M. ruber M7. There were no structural abnormalities found in mycelia regarding the diameter and shape. Moreover, the branching pattern and growth of the mycelia also typically look normal in spreading form, as shown in Figure 7B.
Initially, the morphological changes in the development of the colony and pigments of the M. ruber M7 and Δabct31 strains were explored against PAA, L-cysteine, and D-valine at 2 mM concentration (Figure 8A). The effect on the development of conidia and cleistothecia was also perceived (Figure 8B).
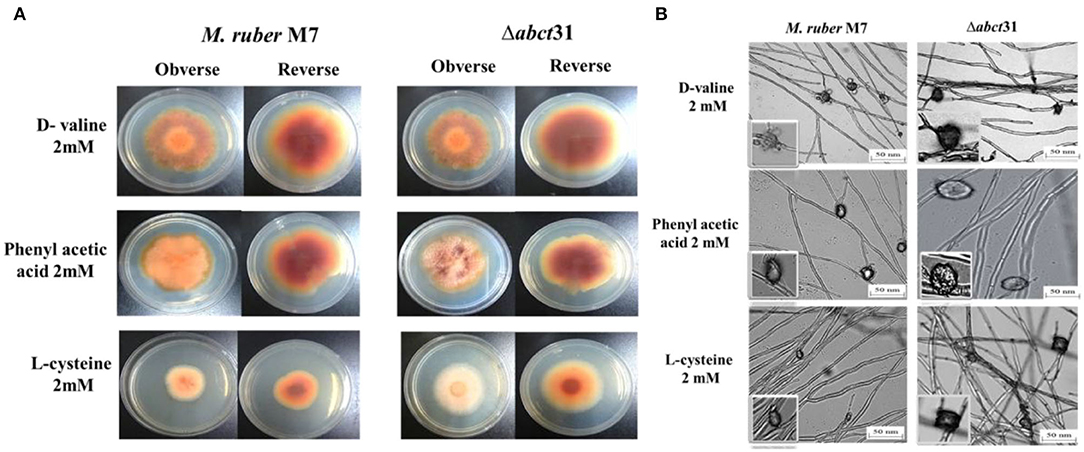
Figure 8. Morphological comparison of M. ruber M7and Δabct31 to evaluate the sensitivity toward pathway amino acid supplementation. (A) Colony morphology of M. ruber M7 and Δabct31 inoculated on PDA plates supplemented with D-valine, phenylacetic acid, and L-cysteine at 2 mM concentrations for each cultured at 28°C for 15 days. (B) Cleistothecia and conidia development in M. ruber M7 and Δabct31 cultured on PDA plates which supplemented with D-valine, phenylacetic acid, and L-cysteine at 2 mM concentrations for each and cultured at 28°C for 10 days.
Whereas seeing the outcomes of M. ruber M7 and Δabct31, a substantial difference was detected related to the phenotypic characteristics, such as the size and appearance of the colony and the colony growth rate and diameter. Considering the Δabct31 on PDA supplemented with PAA, the colony size exhibits resistance to growth, the colony color is clearly reddish, and colony edges are dissimilar from the M. ruber M7 on the PDA-PAA plate. Similarly, from colony diameter, the Δabct31 displayed less sensitivity toward L-Cys supplementation. Moreover, colony color is a pale orange on the PDA-L-Cys feed plate for Δabct31 when compared with wild-type M. ruber M7 and colony diameter is higher than wild type (Figure 8A). On the PDA-D-Val plate, the Δabct31 mutant colony color and appearance look similar to M. ruber M7 (Figure 8A). Hence, Δabct31 has a strong sensitivity toward PAA feeding.
From the results shown in Figure 8B, it can be seen that the Δabct31 can produce a cleistothecium compared with M. ruber M7 on PDA for the supplementation of all pathway precursors. It was clearly noted that there is no difference in the quantity of cleistothecium for all precursors. Furthermore, when comparing the mutant strains to the wild strain for D-Val and L-Cys, there were no differences in general development and phenotypic of cleistothecia. However, in the case of PAA feeding, the cleistothecia size increased compared to the wild type. Besides, considering the growth and structure of the hyphae in mutants Δabct31 strains looks normal and parallel to the control of M. ruber M7 for all precursors. No structural defects such as weakening or inflammation were found in the hyphae of the mycelium by supplementation of precursors.
Moreover, the branching pattern and growth of the mycelia were also typically scattered, as presented in Figure 8B. Hence, the feeding of pathway precursors such as D-Val, and L-Cys at 2 mM has no effect on the Δabct31 regarding microscopic structures. In contrast, Δabct31 exhibits a sensitivity toward PAA feeding regarding cleistothecia size.
The biomasses (dry mycelium) of the M. ruber M7 and Δabct31 were appraised employing the gravimetric process. All strains were uniformly spread on PDA plates covered with cellophane-sheet without and with supplementation of the phenylacetic acid, L-cysteine, and D-valine at 2 mM concentration, cultivated at 28°C. The mycelia were collected at a specific time interval and then dried in an oven at 60°C until constant weights were obtained. The average weight of biomass was calculated and presented in Figure 9.
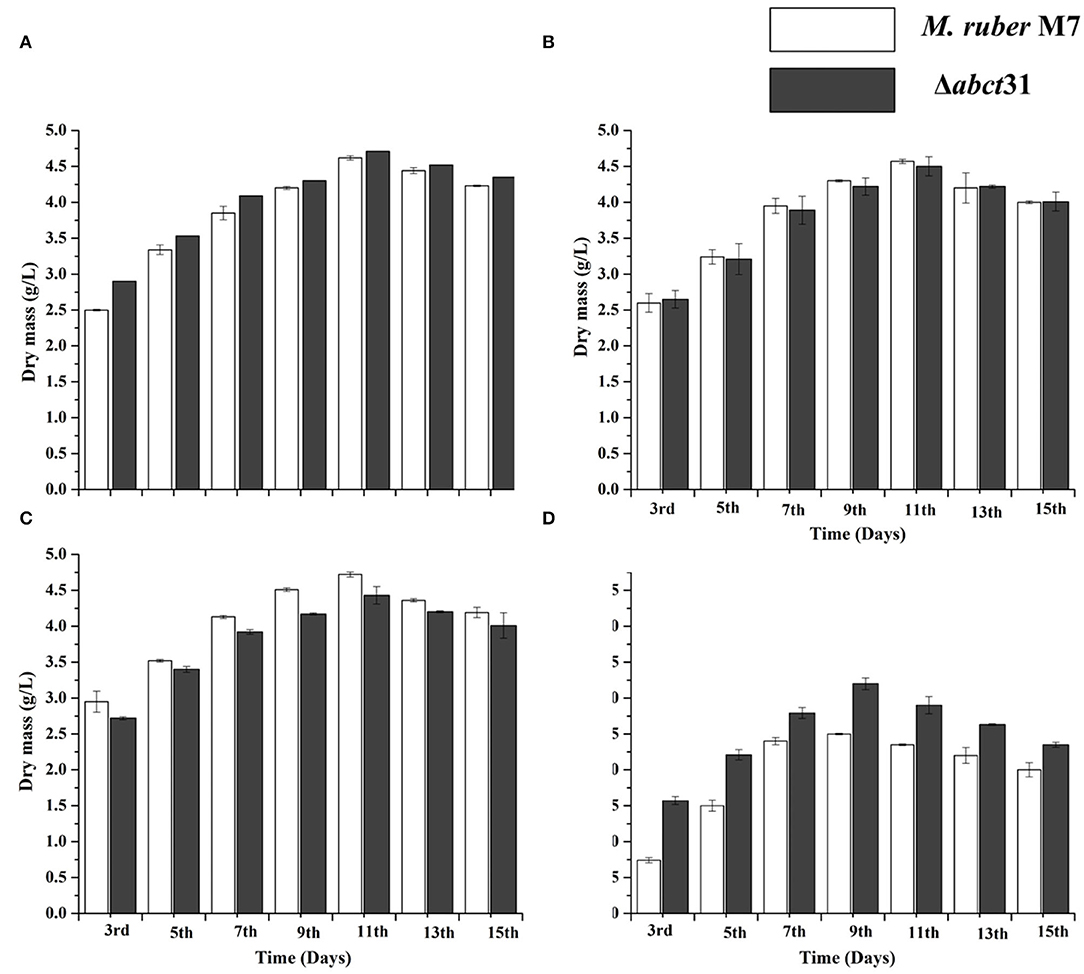
Figure 9. Comparison of biomass (dry cell weight) of the M. ruber M7 and Δabct31 against pathway amino acid. (A) PDB medium without amino acid (control); (B) PDB medium with D-valine; (C) PDB medium with phenylacetic acid; (D) PDB medium with L-cysteine, at 2 mM concentrations and incubated at 28°C without agitation. The bar representing the mean of triplicate values and error bars show standard deviation.
The samples of biomass of M. ruber M7 and Δabct31 cultured on PDA and incubated at 28°C for 15 days were collected on alternate days for the 3rd−15th day for biomass comparison. The mean of the triplicate data is represented by the bar, while the error bars illustrate the standard deviation. By weighing the weight of the dried mycelia, the biomass (dry mass) of M. ruber M7 (control) to Δabct31 strain was determined. The data shown in Figure 9A revealed a substantial (p < 0.05) increase in biomass values for both strains up to the 15th day without supplementation. However, the highest dry cell weight was found on the 11th day, and onwards, the inclination in biomass was observed.
The biomass results presented in Figures 9B–D reveal that among the precursors such as D-Val, PAA, and L-Cys feeding fermentation, a variation in biomass production was observed for mutant Δabct31 when compared with the wild type (M. ruber M7). However, mycelium production starts to increase after the 3rd day of fermentation. The results are presented in Figure 9B; for the D-Val, feeding fermentation showed a non-significant reduction in the biomass for Δabct31 when compared with the M. ruber M7, up to 11th d; for PAA feeding (Figure 9C), the biomass for Δabct31 was significantly lower when compared with the M. ruber M7 until the 15th d, and significantly (p < 0.01) higher biomass was observed in the L-Cys feeding case for Δabct31 when compared with M. ruber M7 (Figures 5–9D). However, the results presented in Figures 9B,C exhibited a significant (p < 0.05) increase in the biomass quantity for Δabct31 and M. ruber M7 up to 11th d for D-Val and PAA, at 9th d of L-Cys supplementation, shown in Figure 9D, and a clear reduction in the biomass value was shown in L-Cys < PAA < D-Val precursors. Overall, in the L-Cys feeding experiment, biomass production value was the lowest compared to other precursors.
Monascus spp. can yield numerous secondary metabolites (Chen et al., 2017b), especially Monascus pigments (MPs) such as red (Monascorubramine and Rubropunctamine), orange (Monascorubrin and Rubropunctatin), and yellow (Ankaflavin and Monascin). To check the effect of Δabct31 gene deletion on MPs, the MPs profile was detected without and with supplementation of the phenylacetic acid, L-cysteine, and D-valine, at 2 mM. Table 4 presents the MPs' average results. The production of the overall pigment rose significantly (p < 0.01) from days 3 to 15 for all strains.
When any strain is grown in a liquid media, it produces metabolites and transports them to growth media via a well-defined transport system. After the fermentation of Δ abct31 and wild-type strains in PDB, both extracellular and intracellular contents of pigments were observed. The overall means of extracellular and intracellular MPs were calculated. The results (Table 4) showed that MPs slightly increased in Δabct31 for control and D-Val, PAA, and L-Cys precursor feeding, during PDB fermentation duration from 3rd to 15th d. The slight increase of MPs of Δabct31 might be due to their role in transporting intermediates. This may be due to the side chain amino acid level increased in the medium; more substrates are available for the pigment production.
The HPLC was used to study the involvement of the abct31 gene for penicillin production in M. ruber M7 and deletion (Δabct31). Furthermore, the role of the feeding of precursors, i.e., D-Val, PAA, and L-Cys at 2 mM on PG production in both strains was also observed. The samples collected on the 7th day, in addition to the treated samples, were analyzed by HPLC for PG estimation.
In the Δabct31 strains, the PG level was observed in intracellular and extracellular. However, the HPLC results showed that these strains could produce the PG at the intracellular level and in the extracellular medium for the 7th day (Figure 10). But the concentration of PG is significantly decreased in the extra- and intracellular part of the mutant when compared with M. ruber M7. The average yield of PG in transformants' (Δabct31) intracellular portion is calculated as 3.2 ± 0.08 μg/g, when compared with the parental strain's yield (Figure 10A). However, the extracellular results showed a similar pattern for PG production Δabct31, yielding the PG, 2.9 ± 0.21 μg/g when compared with the parental strain (Figure 10A).
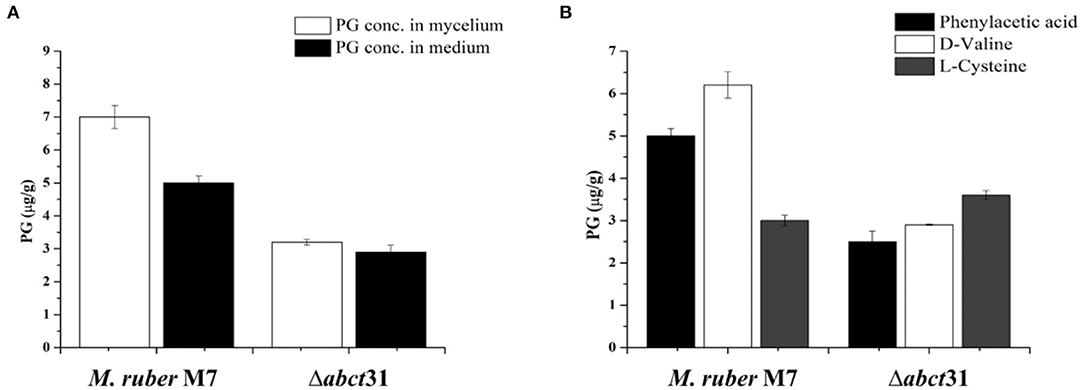
Figure 10. Comparison of PG concentration of the wild-type strain and the Δabct31 transformants on the 7th day. (A) PG concentration in mycelium and medium; (B) Comparison of PG concentration affected by supplementation of D-valine, phenylacetic acid, and L-cysteine at 2 mM on 7th day. The bar representing the mean of triplicate values and error bars show standard deviation.
From Figure 10B, it has been observed that the individual feeding of D-Val, PAA, and L-Cys at 2 mM concentration has a substantial impact on the M. ruber M7. The highest PG concentration was 6.20 ± 0.17 μg/g observed for the D-Val feeding, in contrast to the L-Cys the lowest PG level (3.23 ± 0.12 μg/g). However, for Δabct31 mutants, the lowest PG production of 2.50 ± 0.24 μg/g in Δabct31 mutants has been detected for PAA feeding. While among these precursors feeding in Δabct31, the highest PG level was observed for L-Cys 3.64 ± 0.10 μg/g, presented in Figure 10B. From all results of feeding experiments, it has been concluded that Δabct31 has the highest sensitivity toward PAA precursor.
In order to investigate further the difference in the metabolic profile between M. ruber M7 and mutant (Δabct31), ultra-performance liquid chromatography (UPLC) was used for more elaboration of the HPLC results. The results are shown in Figure 11. The penicillin G spectrum is shown in Figure 11C. As shown in Figures 11A,B, the peak appearance for penicillin G took place at around 3.8 min for sample filtrates of M. ruber M7 and Δabct31, respectively. The UV-Vis spectra confirm the presence of PG in both strains; however, the overall peak area was reduced. Hence, abct31 might be involved in the transportation of intermediate (IPN and PAA) compounds to produce PG in M. ruber M7. For additional validation about the function of the abct31 gene regarding the PG production, UPLC-MS/MS has been performed.
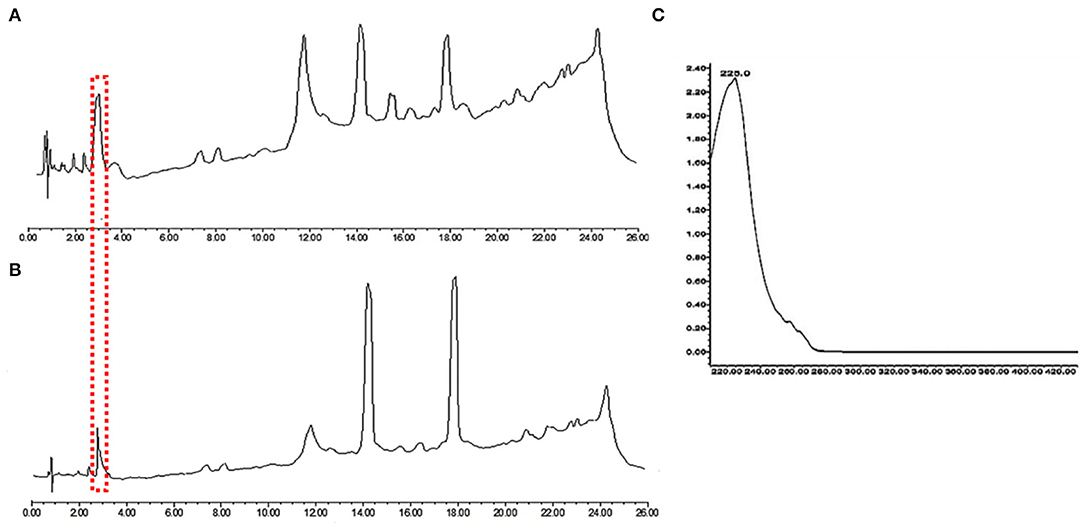
Figure 11. Comparison of eluted products of M. ruber M7 and Δabct31 transformant for solid-state fermentation on rice. (A) Metabolic profile of M. ruber M7 at 250 nm; (B) Metabolic profile of Δabct31 at 250 nm; (C) UV-Vis spectrum of benzyl penicillin (PG) which is shown by the arrows in metabolic profiles.
To investigate the metabolites, a mass profile of M. ruber and Δabct31 was developed using the rice (solid-state fermentation) filtrate. Because the filtrate revealed a wider range of biological compounds, Figure 12 shows the findings of the footprinting of the numerous metabolites found in M. ruber M7.
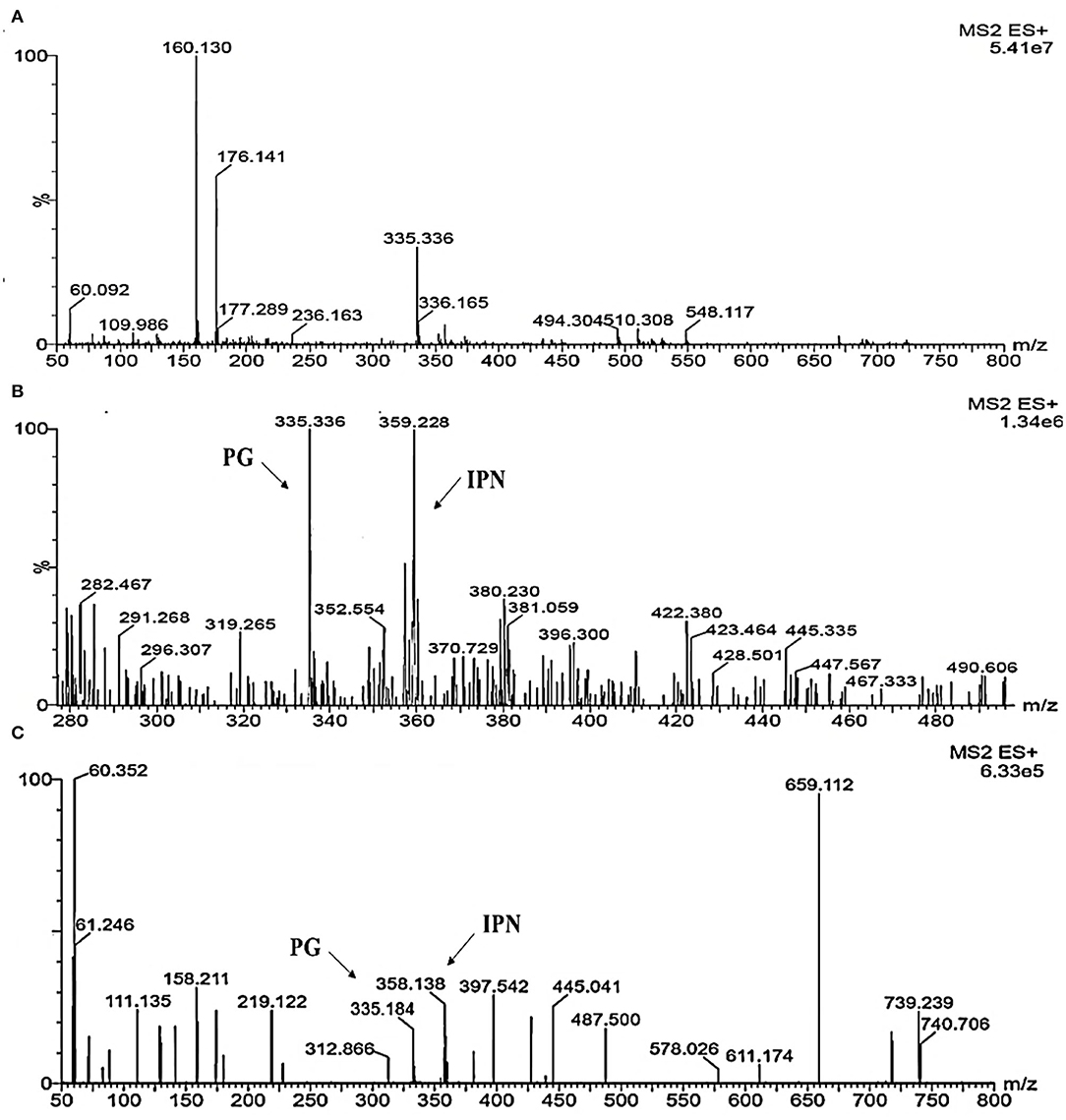
Figure 12. Detection of β-metabolites detected M. ruber M7 and Δabct31 transformants by UPLC-MS/MS. UPLC-MS/MS spectrogram of the M. ruber M7 and Δabct31 for solid-state fermented on rice displays that the atomic mass of ion (m/z 335.336) matched well with the standard penicillin, the molecular weight for isopenicillin N is 359.3 g/mol, in spectrogram ion m/z 359.00 representing the isopenicillin N peak. (A) Spectra in A penicillin G (PG) standard with the mass 335.336; (B) A pattern of fragmentation of M. ruber M7; (C) A pattern of fragmentation of Δabct31.
According to Figure 12, molecular masses associated with β-Lactam such as PG and their intermediates IPN were discovered in the culture filtrate. As a result, PG and IPN in the extract M. ruber M7 were also observed in the UPLC-MS/MS study. The existence of the 335.336 m/z peaks for PG and 395 m/z for isopenicillin N in the crude extract of M7 (Figure 12B) and alike fragmentation design present in the penicillin G standard (Figure 12A) has confirmed the identity of this metabolite. Moreover, as Figure 12C shows that the PG and IPN peaks were also detected in Δabct31.
From the above results, the presence of the IPN peak in Figure 12C demonstrated that abct31 might be responsible for the transportation of intermediate, phenylacetic acid. For further clarification, the sensitivity toward weak acids has also been evaluated.
The toxicity of PAA with weak acids was tested for Δabct31 and in the M. ruber M7 since the Δabct31 gene was substantially expressed when the parental strain was cultured in the presence of PAA. Herein, the spores of M. ruber M7 and Δabct31 were plated on PDA with the supplementation of increased concentrations of sorbic acid, acetic acid, and adipic acid at pH 6.2. Further analyses were done for the colony morphologies and relative expression.
To evaluate the difference in the colonies between Δabct31 and M. ruber M7, both of them were inoculated on individual PDA plates supplemented with sorbic acid at different concentrations (0, 1, 2.5, 5, 7.5, 10 mM) with acetic acid (0, 0.5, 0.75, 1, 1.25, 1.5 mM) concentrations and also with adipic acid at different concentration levels (0, 1, 2, 3 mM) and then incubated for 12 d at 28°C; the colony sizes, color, and other morphological properties were observed (Figure 13).
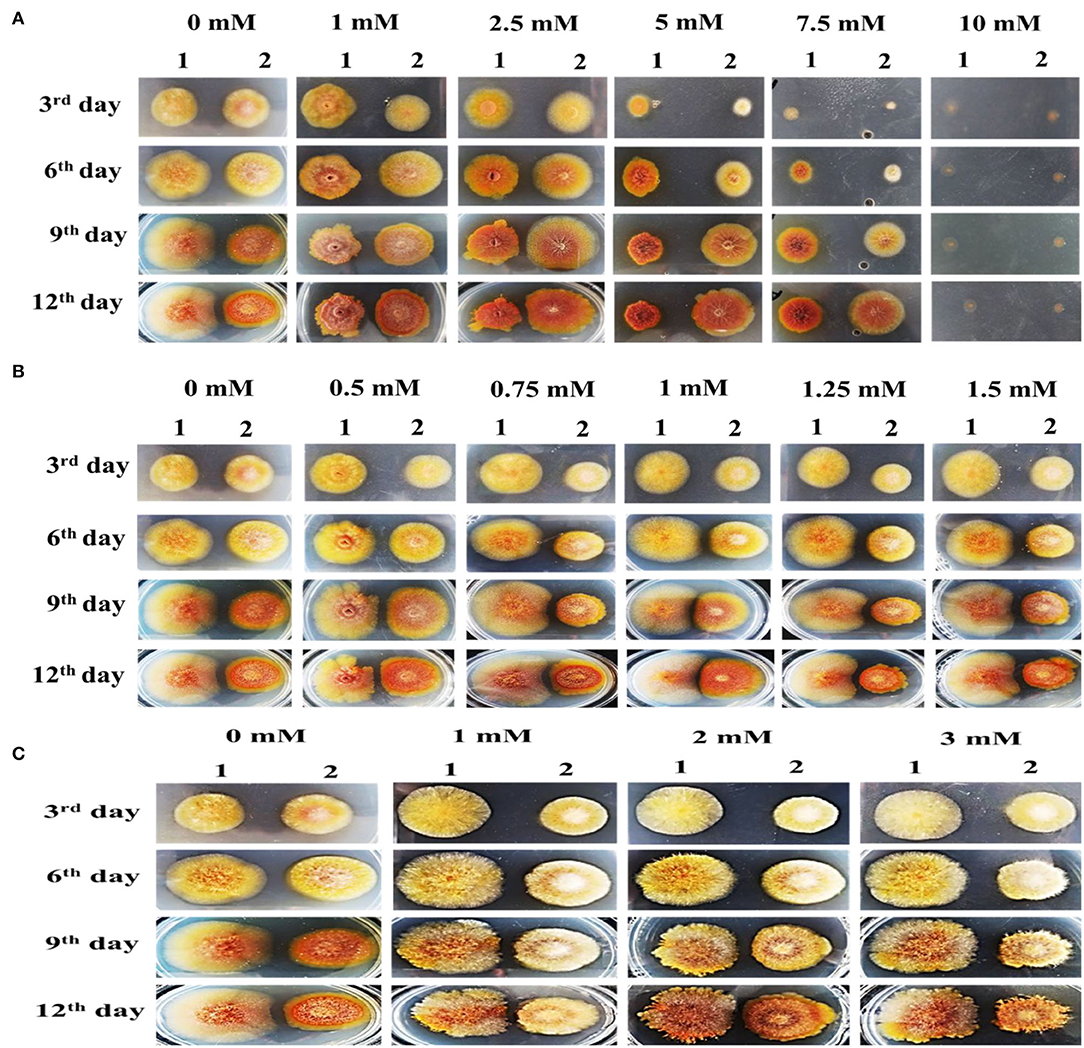
Figure 13. Colony morphology of M. ruber M7 and Δabct31 for the feeding of weak acids. The morphologies of M. ruber M7 (1st vertical row) and Δabct31 (2nd vertical row) colonies on PDA plates supplemented with weak acid at 28°C for 12d. (A) Colony morphology of M. ruber M7 and Δabct31 for the feeding of sorbic acids at 0, 1, 2.5, 5, 7.5, and 10 mM concentration, respectively; (B) Colony morphology of M. ruber M7 and Δabct31 for the feeding of acetic acids at 0, 0.5, 0.75, 1, 1.25, 1.5 mM concentration, respectively; (C) Colony morphology of M. ruber M7 and Δabct31 for the feeding of adipic acids at 0, 1, 2, and 3 mM concentration, respectively.
The results in Figure 13A have shown that Δabct31 colonial sizes were bigger than those of M7 for all tested concentrations of sorbic acid, and Δabct31 colonial colors were a little bit light when compared with M. ruber M7 when the concentrations of sorbic acid were (1, 2.5, 5, 7.5 mM). Although the colonial sizes of M. ruber M7 were reduced as the concentrations of sorbic acid were increased, the colonial colors seemed reddish. The colonial edges were dissimilar from the controlled growth of M. ruber M7. Similarly, from colonial diameters, it was observed that Δabct31 displayed less sensitivity for sorbic acid supplementation. At the highest concentration of sorbic acid 10 mM, both strains did not show any growth up to 12d (Figure 13A).
The observation in Figure 13B shows that when compared with wild-type M. ruber M7, the colonial sizes of Δabct31 on PDA supplemented with acetic acid were smaller for higher concentrations, such as 0.75, 1, 1.25, and 1.5 mM, and the colonial color was darker than M. ruber M7. In the M7 strain, colonial size exhibited resistance to growth, and the colonial color was clearly lighter on the PDA-acetic acid plate at higher concentrations of 1–1.5 mM. In comparison, at a lower concentration of acetic acid (0.5 mM), the colonial size looked similar in contrast to higher concentrations. Hence, the Δabct31 strain exhibited acetic acid supplementation sensitivity at higher concentrations than M. ruber M7 (Figure 13B).
The results shown in Figure 13C illustrate that Δabct31 colonial sizes were smaller than those of M. ruber M7 for all tested concentrations of adipic acid. The Δabct31 colonial colors remained pale yellow up to the 9th day when compared with M. ruber M7 when the tested concentration of adipic acid was 1–3 mM used. The colonial color of M. ruber M7 strain also considerably changed to control the colonial growth of M. ruber M7. On the 12th day, the colonial color's profile of Δabct31 strain changed for 2 and 3 mM concentration and the colonial colors turned to orange. As well as considering the colonial edges looked more irregular, the compacted growth of mycelium in the colonies could be observed when compared with the M. ruber M7, which were typically spread (Figure 13C).
So, in the following experiments, 7.5 mM sorbic acid, 1.5 mM acetic acid, and 3 mM adipic acid were used to check the abct31 gene response.
The effect of sorbic acid, adipic acid, and acetic acid at 7.5, 1.5, and 3 mM, respectively, on the expression level of the abct31 gene in M. ruber M7 with and without PAA at 10 mM has been investigated. The results are revealed in Figure 14.
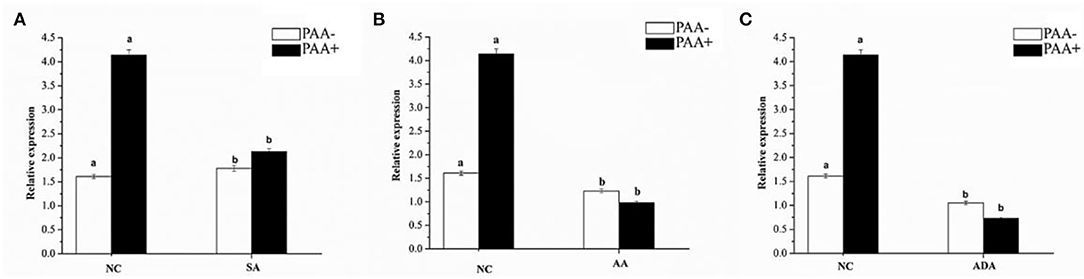
Figure 14. Expression of abct31 in M. ruber M7 to weak acid supplementation with or without PAA. NC was presented in the control group in which M. ruber M7 was cultivated on PDA without sorbic acid. SA presented the sorbic acid group in which M. ruber M7 was cultivated on a PDA with (A) Sorbic acid at 7.5 Mm; (B) Acetic acid at 1.5 Mm; (C) Adipic acid at 3 mM. Both groups were incubated at 28°C for 7d in the presence (black vertical bar) and absence (empty vertical bar) of PAA at 10 mM.
From Figure 14, it has been observed that abct31 in M. ruber M7 exhibited a high transcription level with PAA when compared with the absence of PAA for all weak acids. Although the sorbic acid at 7.5 mM, acetic acid at 1.5 mM, and adipic acid at 3 mM significantly decreased the transcriptional level of abct31 in the absence of PAA, all the weak acids toxicity caused a highly significant (p < 0.01) decrease in the expression level of abct31 in the presence of PAA. Overall, sorbic acid at 7.5 mM causes the abct31 expression level more in the presence of PAA, whereas acetic acid at 1.5 mM and adipic acid at 3 mM cause more expression levels in the absence of PAA.
The findings of this work demonstrate that knocking out the ABC transporter abct31 in M. ruber M7 does not stop the formation of β-lactams, indicating that abct31 does not appear to play a direct part in this process. Nevertheless, when cells develop in +PAA, abct31 expression increases dramatically, indicating more functioning abct31 transport proteins in the cell. To see the effect on gene expression, we needed a high concentration of PAA in our PAA challenge assessments. Furthermore, when compared with its other β-lactam generating strains P. chrysogenum, the abct31 gene expression was higher in M. ruber M7 strains carrying only one copy of the penicillin biosynthetic gene cluster. Altogether, our findings indicate that abct31 expression is influenced by the quantity of its inducer, PAA. When the abct31 gene was expressed, cells became more sensitive to PAA and sorbic acid, accompanied by a drop in abct31 transcript levels when the cells were subjected to these substances.
The side-chain precursor PAA, which is transferred from the cytosol to the peroxisomal matrix and is encoded by the paaT gene in P. chrysogenum, has recently been examined. It encodes a drug/H + antiporter with 12 TMSs, one of which is found in the membrane of peroxisomes as discovered using fluoresce targeted microscopy (Fernández-Aguado et al., 2013, 2014; Martín et al., 2013).
Many other weak acids, such as acetic and adipic acids, were shown to have no effect on the expression of abct31. As a result, they act as inducers through an as-yet-unidentified mechanism. It is possible that abct31 is not the only transporter engaged in exporting PAA. Expressions of the all-other residual ABC transporters were unaffected in the Δabct31 strain cultured in the presence of PAA (unpublished data), indicating that no additional candidates for PAA export were discovered. Transporters from the major facilitator superfamily (MFS) may play a role in the residual low acid resistance potential. As a result of these findings, abct31 appears to play the same role as an ATP-dependent extrusion system for the weak acids to protect cells against these substances. These findings are backed by the theory that an ATP-dependent exporter is involved in the active secretion of the PAA (Xu et al., 2017).
According to our findings, it can be concluded that M. ruber M7 depends on at least two separate detoxifying mechanisms to deal with the PAA during fermentation. These mechanisms might be the conversion of PAA enzymatically into the PG and the active extrusion of PAA from the cell. However, the abct31 transporter is expressed in strains (deletion) with low penicillin G production rates, allowing a detoxifying process that also relies on the ATP-dependent extrusion of PAA.
The outcomes of our study suggest that ABCT31 is the prime ABC transporter, primarily involved in this process of β-lactam synthesis. Additionally, based on these findings, it might be concluded that M. ruber depends upon two separate detoxifying mechanisms to deal with the higher PPA concentrations in the fermentation broth. These mechanisms could be the extrusion of PAA from the cell and then converting PAA into penicillin G by using enzymatic activity. Because of the residuals of PAA, the ABCT31 transporter is expressed and permits a detoxifying pathway. The detoxification mechanism also relies on the extrusion of PAA, which is ATP-dependent extrusion. Understanding the mechanisms governing the intake of these side-chain precursors aid in the clarification of the penicillin biosynthetic pathway. Still, it will also be beneficial economically for the β-lactams producers.
The data presented in the study are deposited in MetaboLights repository, accession number MTBLS4681.
RR has designed and carried out the present research work, conducted experiments, analyzed the data, and written the present manuscript. MV helped in doing the experiments and gave technical guidance. FC provided a place in the laboratory and gave access to the lab facilities for experimentation and funds for the present work. All authors contributed to the article and approved the submitted version.
This work was supported by the Major Program of the National Natural Science Foundation of China (Nos. 31730068 and 31330059 to FC) and the National Key Research and Development Program of China (No. 2018YFD0400404 to FC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguilera, A., and Gómez-González, B. (2008). Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 9, 204–217. doi: 10.1038/nrg2268
Al Shawi, A., Rasul, A., Khan, M., Iqbal, F., and Tonghui, M. (2011). Eupatilin: a flavonoid compound isolated from the artemisia plant, induces apoptosis and G2/M phase cell cycle arrest in human melanoma A375 cells. African J. Pharm. Pharmacol. 5, 582–588. doi: 10.5897/AJPP11.079
Ávalos, J., Díaz-Sánchez, V., García-Martínez, Jorge Castrillo, M., Ruger-Herreros, M., and Limón, M. C. (2014). Biosynthesis and molecular genetics of fungal secondary metabolites. Fungal Biol. 2, 67–79. doi: 10.1007/978-1-4939-1191-2
Barreiro, C., and García-Estrada, C. (2019). Proteomics and Penicillium chrysogenum: unveiling the secrets behind penicillin production. J. Proteomics. 198, 119–131. doi: 10.1016/j.jprot.2018.11.006
Buchan, D. W. A., Minneci, F., Nugent, T. C. O., Bryson, K., and Jones, D. T. (2013). Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 41, 349–357. doi: 10.1093/nar/gkt381
Chen, C.-L., and Pan, T.-M. (2019). Beneficial effects of Monascus purpureus NTU 568-fermented products on cholesterol in vivo and clinical trial: a review. Integr. Clin. Med. 3, 1–3. doi: 10.15761/ICM.1000161
Chen, F., and Hu, X. (2005). Study on red fermented rice with high concentration of monacolin K and low concentration of citrinin. Int. J. Food Microbiol. 103, 331–337. doi: 10.1016/j.ijfoodmicro.2005.03.002
Chen, G., Bei, Q., Huang, T., and Wu, Z. (2017a). Tracking of pigment accumulation and secretion in extractive fermentation of Monascus anka GIM 3.592. Microb. Cell Fact. 16, 1–13. doi: 10.1186/s12934-017-0786-6
Chen, W. (2015). Insights Into Biological Characteristics Of Monascus Ruber M7 by Genomics Approaches. (Doctor thesis), Huazhong Agricultural University, Wuhan, Hubei, China.
Chen, W., Chen, R., Liu, Q., He, Y., He, K., Ding, X., et al. (2017b). Orange, red, yellow: biosynthesis of azaphilone pigments in monascus fungi. Chem. Sci. 8, 4917–4925. doi: 10.1039/C7SC00475C
Chen, W., Xie, T., Shao, Y., and Chen, F. (2012a). Genomic characteristics comparisons of 12 food-related filamentous fungi in tRNA gene set, codon usage and amino acid composition. Gene. 497, 116–124. doi: 10.1016/j.gene.2012.01.016
Chen, W., Xie, T., Shao, Y., and Chen, F. (2012b). Phylogenomic relationships between amylolytic enzymes from 85 strains of fungi. PLoS ONE 7, e49679. doi: 10.1371/journal.pone.0049679
Emanuelsson, O., Nielsen, H., Brunak, S., and Von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Fernández-Aguado, M., Martín, J. F., Rodríguez-Castro, R., García-Estrada, C., Albillos, S. M., Teijeira, F., et al. (2014). New insights into the isopenicillin N transport in Penicillium chrysogenum. Metab. Eng. 22, 89–103. doi: 10.1016/j.ymben.2014.01.004
Fernández-Aguado, M., Ullán, R. V., Teijeira, F., Rodríguez-Castro, R., and Martín, J. F. (2013). The transport of phenylacetic acid across the peroxisomal membrane is mediated by the PaaT protein in Penicillium chrysogenum. Appl. Microbiol. Biotechnol. 97, 3073–3084. doi: 10.1007/s00253-012-4425-1
Fletcher, D. A., and Mullins, R. D. (2010). Cell mechanics and the cytoskeleton. Nature 463, 485–492. doi: 10.1038/nature08908
Gao, S., Yu, S., and Yao, S. (2021). An efficient protein homology detection approach based on seq2seq model and ranking. Biotechnol. Biotechnol. Equip. 35, 633–640. doi: 10.1080/13102818.2021.1892522
García-Estrada, C., Vaca, I., Fierro, F., Sjollema, K., Veenhuis, M., and Martín, J. F. (2008). The unprocessed preprotein form IATC103S of the isopenicillin N acyltransferase is transported inside peroxisomes and regulates its self-processing. Fungal Genet. Biol. 45, 1043–1055. doi: 10.1016/j.fgb.2008.03.005
Gaudelli, N. M., Long, D. H., and Townsend, C. A. (2015). β-Lactam formation by a non-ribosomal peptide synthetase during antibiotic biosynthesis. Nature 520, 383–387. doi: 10.1038/nature14100
George, A. M., and Jones, P. M. (2012). Perspectives on the structure-function of ABC transporters: the Switch and Constant Contact Models. Prog. Biophys. Mol. Biol. 109, 95–107. doi: 10.1016/j.pbiomolbio.2012.06.003
Ghilarov, D., Inaba-Inoue, S., Stepien, P., Qu, F., Michalczyk, E., Pakosz, Z., et al. (2021). Molecular mechanism of SbmA, a promiscuous transporter exploited by antimicrobial peptides. Sci. Adv. 7, eabj5363. doi: 10.1126/sciadv.abj5363
Guo, X., Li, Y., Zhang, R., Yu, J., Ma, X., Chen, M., et al. (2019). Transcriptional regulation contributes more to Monascus pigments diversity in different strains than to DNA sequence variation. World J. Microbiol. Biotechnol. 35, 1–13. doi: 10.1007/s11274-018-2566-9
Hazelwood, L. A., Tai, S. L., Boer, V. M., De Winde, J. H., Pronk, J. T., and Daran, J. M. (2006). A new physiological role for Pdr12p in Saccharomyces cerevisiae: export of aromatic and branched-chain organic acids produced in amino acid catabolism. FEMS Yeast Res. 6, 937–945. doi: 10.1111/j.1567-1364.2006.00094.x
He, Y., Liu, Q., Shao, Y., and Chen, F. (2013). ku70 and ku80 null mutants improve the gene targeting frequency in Monascus ruber M7. Appl. Microbiol. Biotechnol. 97, 4965–4976. doi: 10.1007/s00253-013-4851-8
Kulkarni, P. A., and Devarumath, R. M. (2014). In silico 3D-structure prediction of SsMYB2R: a novel MYB transcription factor from Saccharum spontaneum. Indian J. Biotechnol. 13, 437–447.
Lee, B. H., and Pan, T. M. (2012). Benefit of monascus-fermented products for hypertension prevention: a review. Appl. Microbiol. Biotechnol. 94, 1151–1161. doi: 10.1007/s00253-012-4076-2
Lee, Y. M., Li, H., Hong, J., Cho, H. Y., Bae, K. S., Kim, M. A., et al. (2010). Bioactive metabolites from the sponge-derived fungus Aspergillus versicolor. Arch. Pharm. Res. 33, 231–235. doi: 10.1007/s12272-010-0207-4
Li, L., He, L., Lai, Y., Shao, Y., and Chen, F. (2014). Cloning and functional analysis of the Gβ gene Mgb1 and the Gγ gene Mgg1 in Monascus ruber. J. Microbiol. 52, 35–43. doi: 10.1007/s12275-014-3072-x
Li, L., Shao, Y., Li, Q., Yang, S., and Chen, F. (2010). Identification of Mga1, a G-protein α-subunit gene involved in regulating citrinin and pigment production in Monascus ruber M7. FEMS Microbiol. Lett. 308, 108–114. doi: 10.1111/j.1574-6968.2010.01992.x
Liu, J., Lei, M., Zhou, Y., and Chen, F. (2019). A comprehensive analysis of the small GTPases Ypt7 involved in the regulation of fungal development and secondary metabolism in monascus ruber M7. Front. Microbiol. 10, 452. doi: 10.3389/fmicb.2019.00452
Liu, Q., Xie, N., He, Y., Wang, L., Shao, Y., Zhao, H., et al. (2014). MpigE, a gene involved in pigment biosynthesis in Monascus ruber M7. Appl. Microbiol. Biotechnol. 98, 285–296. doi: 10.1007/s00253-013-5289-8
Lobanovska, M., and Pilla, G. (2017). Penicillin's discovery and antibiotic resistance: lessons for the future? Yale J. Biol. Med. 90, 135–145.
Lopes, F. C., Tichota, D. M., Sauter, I. P., Meira, S. M. M., Segalin, J., Rott, M. B., et al. (2013). Active metabolites produced by Penicillium chrysogenum IFL1 growing on agro-industrial residues. Ann. Microbiol. 63, 771–778. doi: 10.1007/s13213-012-0532-6
Martín, J. F., Casqueiro, J., and Liras, P. (2005). Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr. Opin. Microbiol. 8, 282–293. doi: 10.1016/j.mib.2005.04.009
Martín, J. F., García-Estrada, C., and Ullán, R. V. (2013). Transport of substrates into peroxisomes: the paradigm of β-lactam biosynthetic intermediates. Biomol. Concepts 4, 197–211. doi: 10.1515/bmc-2012-0048
Moussatova, A., Kandt, C., O'Mara, M. L., and Tieleman, D. P. (2008). ATP-binding cassette transporters in Escherichia coli. Biochim. Biophys. Acta - Biomembr. 1778, 1757–1771. doi: 10.1016/j.bbamem.2008.06.009
Nugent, T., and Jones, D. T. (2012). Membrane protein structural bioinformatics. J. Struct. Biol. 179, 327–337. doi: 10.1016/j.jsb.2011.10.008
Ozcengiz, G., and Demain, A. L. (2013). Recent advances in the biosynthesis of penicillins, cephalosporins and clavams and its regulation. Biotechnol. Adv. 31, 287–311. doi: 10.1016/j.biotechadv.2012.12.001
Pandey, V., Krishnan, V., Basak, N., Marathe, A., Thimmegowda, V., Dahuja, A., et al. (2018). Molecular modeling and in silico characterization of GmABCC5: a phytate transporter and potential target for low-phytate crops. 3 Biotech. 8, 1–16. doi: 10.1007/s13205-017-1053-6
Ramzan, R., Safiullah Virk, M., Muhammad, Z., Ahmed, A. M. M., Yuan, X., and Chen, F. (2019). Genetic modification of mfsT gene stimulating the putative penicillin production in Monascus ruber M7 and exhibiting the sensitivity towards precursor amino acids of penicillin pathway. Microorganisms 7, 1–22. doi: 10.3390/microorganisms7100390
Schulz, B., and Kolukisaoglu, H. Ü. (2006). Genomics of plant ABC transporters: The alphabet of photosynthetic life forms or just holes in membranes? FEBS Lett. 580, 1010–1016. doi: 10.1016/j.febslet.2006.01.002
Seiple, I. B., Zhang, Z., Jakubec, P., Langlois-Mercier, A., Wright, P. M., Hog, D. T., et al. (2016). A platform for the discovery of new macrolide antibiotics. Nature 533, 338–345. doi: 10.1038/nature17967
Shao, Y., Ding, Y., Zhao, Y., Yang, S., Xie, B., and Chen, F. (2009). Characteristic analysis of transformants in T-DNA mutation library of Monascus ruber. World J. Microbiol. Biotechnol. 25, 989–995. doi: 10.1007/s11274-009-9977-6
Singh, R., Singh, S., and Pandey, P. N. (2016). In-silico analysis of Sirt2 from Schistosoma mansoni: Structures, conformations, and interactions with inhibitors. J. Biomol. Struct. Dyn. 34, 1042–1051. doi: 10.1080/07391102.2015.1065205
Stitou, M., Toufik, H., Bouachrine, M., and Lamchouri, F. (2021). Quantitative structure–activity relationships analysis, homology modeling, docking and molecular dynamics studies of triterpenoid saponins as Kirsten rat sarcoma inhibitors. J. Biomol. Struct. Dyn. 39, 152–170. doi: 10.1080/07391102.2019.1707122
Tang, K., Xin, Y., Li, K., Chen, X., and Tan, Y. (2021). Cell cytoskeleton and stiffness are mechanical indicators of organotropism in breast cancer. Biology 10, 1–14. doi: 10.3390/biology10040259
Ullán, R., Liu, G., Casqueiro, J., Gutiérrez, S., Bañuelos, O., and Martín, J. (2002). The cefT gene of Acremonium chrysogenum C10 encodes a putative multidrug efflux pump protein that significantly increases cephalosporin C production. Mol. Genet. Genomics. 267, 673–683. doi: 10.1007/s00438-002-0702-5
Ullán, R. V., Teijeira, F., and Martín, J. F. (2008). Expression of the Acremonium chrysogenum cefT gene in Penicillum chrysogenum indicates that it encodes an hydrophilic β-lactam transporter. Curr. Genet. 54, 153–161. doi: 10.1007/s00294-008-0207-9
Van Den Berg, M. A., Albang, R., Albermann, K., Badger, J. H., Daran, J. M. M., et al. (2008). Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 26, 1161–1168. doi: 10.1038/nbt.1498
Wang, L., Dai, Y., Chen, W., Shao, Y., and Chen, F. (2016a). Effects of light intensity and color on the biomass, extracellular red pigment, and citrinin production of monascus ruber. J. Agric. Food Chem. 50, 9506–9514. doi: 10.1021/acs.jafc.6b04056
Wang, L., Wang, W., and Xu, G. (2011). Promotion of monacolin K production by Agrobacterium tumefaciens-mediated transformation in Monascus albidus 9901. Curr. Microbiol. 62, 501–507. doi: 10.1007/s00284-010-9735-x
Wang, S., Li, W., Liu, S., and Xu, J. (2016b). RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 44, 430–435. doi: 10.1093/nar/gkw306
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, 296–303. doi: 10.1093/nar/gky427
Weber, S. S., Kovalchuk, A., Bovenberg, R. A. L., and Driessen, A. J. M. (2012). The ABC transporter ABC40 encodes a phenylacetic acid export system in Penicillium chrysogenum. Fungal Genet. Biol. 49, 915–921. doi: 10.1016/j.fgb.2012.09.003
Wei, W., Lin, S., Chen, M., Liu, T., Wang, A., Li, J., et al. (2017). Monascustin, an unusual γ-lactam from red yeast rice. J. Nat. Prod. 80, 201–204. doi: 10.1021/acs.jnatprod.6b00493
Wilkens, S. (2015). Structure and mechanism of ABC transporters. F1000Prime Rep. 7, 14. doi: 10.12703/P7-14
Xu, Y., Seelig, A., and Bernèche, S. (2017). Unidirectional transport mechanism in an ATP dependent exporter. ACS Cent. Sci. 3, 250–258. doi: 10.1021/acscentsci.7b00068
Yang, J., Xu, X., and Liu, G. (2012). Amplification of an MFS transporter encoding gene penT significantly stimulates penicillin production and enhances the sensitivity of Penicillium chrysogenum to phenylacetic acid. J. Genet. Genomics. 39, 593–602. doi: 10.1016/j.jgg.2012.08.004
Yuliana, A., Singgih, M., Julianti, E., and Blanc, P. J. (2017). Derivates of azaphilone Monascus pigments. Biocatal. Agric. Biotechnol. 9, 183–194. doi: 10.1016/j.bcab.2016.12.014
Zaitseva, J., Jenewein, S., Jumpertz, T., Holland, I. B., and Schmitt, L. (2005). H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 24, 1901–1910. doi: 10.1038/sj.emboj.7600657
Keywords: ABC transporter, secondary metabolites, phenylacetic acid, amino acid, weak acid
Citation: Ramzan R, Virk MS and Chen F (2022) The ABCT31 Transporter Regulates the Export System of Phenylacetic Acid as a Side-Chain Precursor of Penicillin G in Monascus ruber M7. Front. Microbiol. 13:915721. doi: 10.3389/fmicb.2022.915721
Received: 08 April 2022; Accepted: 16 May 2022;
Published: 28 July 2022.
Edited by:
Xucong Lv, Fuzhou University, ChinaReviewed by:
Chan Zhang, Beijing Technology and Business University, ChinaCopyright © 2022 Ramzan, Virk and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fusheng Chen, Y2hlbmZzQG1haWwuaHphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.