
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 26 May 2022
Sec. Virology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.884044
This article is part of the Research TopicViruses in Agricultural Systems: Interactions with Plants, Insect Pollinators and FungiView all 10 articles
Chlorophyll a/b-binding protein of light-harvesting complex II type 1 like (LHC II-1L) is an essential component of photosynthesis, which mainly maintains the stability of the electron transport chain. However, how the LHC II-1L protein of Fragaria vesca (FvLHC II-1L) affects viral infection remains unclear. In this study, we demonstrated that the movement protein P1 of strawberry vein banding virus (SVBV P1) interacted with FvLHC II-1L in vivo and in vitro by bimolecular fluorescence complementation and pull-down assays. SVBV P1 was co-localized with FvLHC II-1L at the edge of epidermal cells of Nicotiana benthamiana leaves, and FvLHC II-1L protein expression was upregulated in SVBV-infected F. vesca. We also found that FvLHC II-1L effectively promoted SVBV P1 to compensate for the intercellular movement of movement-deficient potato virus X (PVXΔP25) and the systemic movement of movement-deficient cucumber mosaic virus (CMVΔMP). Transient overexpression of FvLHC II-1L and inoculation of an infectious clone of SVBV showed that the course of SVBV infection in F. vesca was accelerated. Collectively, the results showed that SVBV P1 protein can interact with FvLHC II-1L protein, which in turn promotes F. vesca infection by SVBV.
The strawberry vein banding virus (SVBV) is a latent virus that severely harms strawberry plants in major strawberry growing areas worldwide (Ratti et al., 2009; Chen et al., 2016a). In China, SVBV, which is primarily transmitted by aphids or tissue culture seedlings, is mainly distributed in the Sichuan, Hebei, Liaoning, Shandong, and Anhui provinces (Pan et al., 2018; Jiang et al., 2021), where SVBV-infected strawberries display characteristic symptoms, such as weak growth, uneven leaf color, a decrease in the number of creeping stems, and deformed fruits (Frazier, 1974).
Strawberry vein banding virus is a double-stranded DNA virus belonging to the genus Caulimovirus of the Caulimoviridae family. It contains seven open reading frames (ORFs), each of which encode a protein (Pattanaik et al., 2004). ORF I encodes a movement protein (MP) called P1 that can promote the intercellular movement of SVBV, which plays an essential role in the process of viral infection (Rui et al., 2021). SVBV shares similar genome structure with cauliflower mosaic virus (CaMV) belonging to the same genus. According to the proteins encoded by each ORF of CaMV, it was inferred that ORF II, III, IV, and V of SVBV may encode proteins associated with aphid infection, a DNA-binding protein, a coat protein (CP), a reverse transcriptase protein (Pan et al., 2018). P6, encoded by ORF VI, is a multifunctional protein that functions as an RNA silencing suppressor and translation transactivator (Li et al., 2018a), and ORF VII encodes an unknown protein (Schoelz et al., 2016). Our previous study showed that SVBV P1 may regulates intracellular and intercellular movement of SVBV during viral infection (Rui et al., 2021).
After invading the host plants, viruses spread through plasmodesmata (PD) to neighboring cells with the help of MPs (Schoelz et al., 2011). MPs can be transported intracellularly and distantly by binding virus particles, binding viral nucleic acids, inducing the formation of PD tubular structures, and changing the size of the PD aperture (Chen et al., 2000; Lucas, 2006; Park et al., 2014). It has been shown that host-encoded proteins interacting with viral MPs can control the pore size of PD and/or change the localization of viral MP on the PD to affect the process of viral infection (Peiro et al., 2014; Wu and Cheng, 2020). For example, the Alfalfa mosaic virus (AMV) MP can interact with Arabidopsis thaliana patellin 3 (AtPATL3); furthermore, overexpression of AtPATL3 makes AMV MP unable to target PD, thereby affecting the intercellular movement of the virus (Peiro et al., 2014). Similarly, the rice membrane-associated protein remorin (REM1), which interacts with the MP NSvc4 encoded by the rice stripe virus (RSV), is involved in viral infection. Silencing of the NbREM1 gene in Nicotiana benthamiana can reduce the deposition of callose, enhance the permeability of PD, and promote viral infection (Fu et al., 2018). Triple gene block protein 1, encoded by barley stripe mosaic virus (BSMV), can hijack the nucleolar protein fibrillarin 2 to form a viral ribonucleoprotein movement complex, thus promoting viral cell-to-cell movement (Li et al., 2018b). Moreover, Citrus macrophylla miraculin-like protein 2 is capable of hijacking MP P33 of Citrus tristeza virus to form aggregates in the cytoplasm and inhibit the cell-to-cell movement of the virus (Sun et al., 2021). Thus, host factors may interact with MPs to affect the process of viral infection.
In higher plants, photosystem II (PSII) is a large protein complex located on the thylakoid membrane of chloroplasts. It comprises the PSII core complex and light-harvesting complex II (LHC II), which drive water oxidation and are key components of photosynthesis (Huang et al., 2013). LHC II mainly binds to chlorophyll a/b and has three transmembrane domains, generally in the form of small monomers and macromeres (Neilson and Durnford, 2010). LHC II is responsible for transferring the absorbed light energy to the PSII reaction center and maintaining the stability of the PSII electron transport chain (Allen et al., 1981; Nilsson et al., 1997). LHC II not only participates in plant photosynthesis but also regulates plant growth and development. Silencing of AtLhcb1 in Arabidopsis thaliana hindered the formation of LHC II trimers, affected photosynthesis, and caused plant dwarfing (Pietrzykowska et al., 2014); loss-of-function mutations in AtTHF1 decreased the rate of LHC II degradation and delayed leaf yellowing (Huang et al., 2013). Our previous study indicated that SVBV P1 can interact with chlorophyll a/b-binding protein of LHC II type 1 like of Fragaria vesca (FvLHC II-1L), based on the results of yeast two-hybrid (Y2H) assay (Zhang et al., 2019), but the role of FvLHC II-1L in SVBV infection is still largely unknown.
In this study, we used bimolecular fluorescence complementation (BiFC) and pull-down assays to demonstrate that P1 interacts with FvLHC II-1L in vivo and in vitro. Quantitative real-time polymerase chain reaction (RT-qPCR) showed that FvLHC II-1L expression was significantly upregulated following SVBV infection. The subcellular localization assay revealed that SVBV P1 and FvLHC II-1L co-localized at the edge of the plant cell and that FvLHC II-1L was able to promote SVBV P1 aggregation. Furthermore, we showed that FvLHC II-1L promoted SVBV P1 to compensate for the movement of movement-deficient potato virus X (PVXΔP25) and cucumber mosaic virus (CMVΔMP). Overexpression of FvLHC II-1L enhanced SVBV infection in F. vesca.
All plants used in this study were grown in a growth chamber, set at 25°C, under 60% relative humidity and a 16 h light and 8 h dark photoperiod.
Six- to eight-week-old N. benthamiana plants were used for Agrobacterium tumefaciens infiltration, as described previously (Chen et al., 2016b). Equal volumes of individual Agrobacterium cultures (optical density at 600 nm, OD600 = 1.0) were mixed before co-infiltration.
Strawberry plants were infiltrated as described previously (Tian et al., 2015). The roots of strawberry plant seedlings were rinsed with distilled water, and then, whole plants were submerged in A. tumefaciens inoculum containing pTRV1 and pTRV2 or its derivatives (OD600 = 1.0) and placed in a vacuum at 101 kPa atmospheric pressure for 30 s; this procedure was repeated. The treated plants were washed with distilled water and cultured in pots containing nutrient solution, and after 24 h, they were transplanted into pots containing nutrient soil.
The coding sequences of SVBV P1 (GenBank No: X97304.1) and FvLHC II-1L (GenBank No: XM_004303830.2) were cloned into pCV-cYFP and pCV-nYFP, respectively (Li et al., 2018a), to generate cYFP-SVBV P1, nYFP-FvLHC II-1L, nYFP-P1, and cYFP-FvLHC II-1L. The recombinant plasmids were transformed into A. tumefaciens (GV3101) and co-infiltrated into N. benthamiana leaves. The agro-infiltrated leaves were observed using confocal microscopy (Olympus FV1000, Tokyo, Japan) at approximately 3 days post inoculation (dpi).
The full-length FvLHC II-1L or cyan fluorescent protein (CFP) respectively was inserted between the BamHI/SalI restriction sites within the pCAM1307-3*Flag based expression vector to produce pCAM1307-FvLHC II-1L-3*Flag (FvLHC II-1L-Flag) or pCAM1307-CFP-3*Flag (CFP-Flag). To detect the SVBV P1–FvLHC II-1L interaction, a pull-down assay was conducted using protein extracts of FvLHC II-1L-Flag from infiltrated N. benthamiana leaves at 3 dpi. The full-length cDNAs of SVBV P1 were amplified and inserted into the pHMTc. The plasmid pHMTc was kindly supplied by Prof. Xiaorong Tao (Nanjing agricultural University, China). The construct pHMTc-SVBV P1 was transformed into Escherichia coli BL21 (DE3) cells. Total protein extracts were incubated with the maltose-binding protein (MBP)-SVBV P1 fusion proteins. The MBP pull-down assay was performed as previously described (Zhu et al., 2017).
For the subcellular localization study, SVBV P1 and FvLHC II-1L were inserted into a modified pCAM2300 containing green fluorescent protein (GFP) or red fluorescent protein (RFP), respectively, and then transformed into A. tumefaciens (Feng et al., 2013). The cultures were co-infiltrated with N. benthamiana. After 3 days, the infiltrated leaves were imaged using a confocal microscope (Olympus FV1000, Tokyo, Japan). Excitation wavelength was 488 nm for GFP, 514 nm for yellow fluorescent protein (YFP), 561 nm for RFP, and 640 nm for chloroplast.
Total RNA was extracted from agro-infiltrated leaves using the Omini Plant RNA Plant Kit (CWBIO, Beijing, China). cDNA was synthesized using PrimeScript™ RT reagent kit (TaKaRa, Tokyo, Japan). RT-qPCR was performed as described previously (Li et al., 2019).
Total protein was extracted from agro-infiltrated leaves using RIPA lysis buffer II (Sangon Biotech, Shanghai, China). After electrophoresis, the gel was placed in Coomassie Brilliant Blue stain (CBB, BeyoBlue™ Coomassie Blue Super Fast Staining Solution; Beyotime, Shanghai, China), soaked for 1 h, then decolorized with pure water. Western blotting was performed as previously described (Tian et al., 2014; Li et al., 2018a). Total protein was separated via sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, then separately incubated using GFP mouse monoclonal antibody, c-Myc mouse monoclonal antibody, DYKDDDDK (Flag) mouse monoclonal antibody, MBP mouse monoclonal antibody (TransGen, Beijing, China), FvLHC II-1L rabbit polyclonal antibody, SVBV P1 rabbit polyclonal antibody, and then incubated using goat anti-rabbit IgG (H + L), HRP Conjugate and goat anti-mouse IgG (H + L), HRP Conjugate (TransGen, Beijing, China) with dilutions at 1:5,000 was used in immunoblot analysis. The detection signal was visualized using the EasySee Western Blot Kit (Transgene Biotech, Beijing, China).
For potato virus X movement complementation experiments, a movement-deficient PVX-GFP construct (PVXΔP25-GFP) and construct P25, expressing the PVX P25 protein, were kindly supplied by Prof. Fei Yan (Ningbo University, China). The segment of FvLHC II-1L fused Myc-tag in the 5-termius was inserted between the BamHI/SalI restriction sites within the pBin438 based expression vector to produce pBin438-Myc-FvLHC II-1L. The full-length SVBV P1 was inserted between the BamHI/SalI restriction sites within the pBin438 based expression vector to produce pBin438-SVBV P1, and the assays were performed as previously described (Yan et al., 2012; Rui et al., 2021). The GFP fluorescence of the infiltrated leaves was observed using a confocal microscope (Olympus FV1000, Tokyo, Japan).
The CMV complementation assay was performed as previously described (Shen et al., 2014). The full-length SVBV P1 was inserted between the NcoI/XbaI restriction sites within the pCB301-CMV RNA3ΔMP based expression vector to produce pCB301-CMV RNA3ΔMP-SVBV P1. The plasmid pCB301-CMV RNA3ΔMP was kindly supplied by Prof. Xiaorong Tao (Nanjing agriculture University, China). A. tumefaciens containing pCB301-CMV RNA3 or pCB301-CMV RNA3ΔMP-SVBV P1 was mixed with A. tumefaciens containing pCB301-CMV RNA1 and RNA2 in a 1:1:1 ratio.
The full-length ORF of FvLHC II-1L was inserted behind the CP of the TRV2 vector as previously described (Tian et al., 2014). pTRV1 and pTRV2 or its derivatives were transformed, in order, into A. tumefaciens and co-transfected into F. vesca using a vacuum-infiltration method, as previously described (Tian et al., 2015).
Strawberry vein banding virus-infected strawberry leaves were harvested at 35 dpi. Total DNA was extracted from the strawberry leaves using the CTAB method. Southern blotting was performed as previously described (Feng et al., 2016). SVBV DNA accumulation in SVBV-infected plants was detected by digoxigenin-labeled probes from the 5-termius of SVBV CP gene using DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Basel, Switzerland) according to manufacturer instructions.
The full-length FvLHC II-1L was inserted between the XbaI/HindIII restriction sites within the pCAM1307-Myc based expression vector to produce pCAM1307-Myc-FvLHC II-1L. The plasmid was transformed into A. tumefaciens strain LBA4404. The transgenic N. benthamiana plants of T0 generation were obtained by leaf disc transformation method. The homozygous FvLHCII-1L transgenic N. benthamiana was obtained after screening. RT-qPCR and Western blot analyzes were used to detect the expression of FvLHC II-11L. The primers used for vector construction are listed in Supplementary Table S1.
To examine how SVBV P1 causes plant disease during viral infection, we screened a F. vesca cDNA library using the Y2H assay to identify the host proteins involved. Consequently, FvLHC II-1L was noted to interact with SVBV P1 (Zhang et al., 2019). To further verify the interaction between SVBV P1 and FvLHC II-1L, a pull-down experiment was performed. The results showed that MBP-SVBV P1 specifically pulled down the FvLHC II-1L-Flag protein (Figure 1A). SVBV P1 was absent in the cyan fluorescent protein (CFP)-Flag pulldown products, and FvLHC II-1L-Flag was not able to pull down MBP (Figure 1A), indicating that SVBV P1 can interact with FvLHC II-1L in vitro. Furthermore, a BiFC assay was performed to examine the interaction between SVBV P1 and FvLHC II-1L in vivo using N. benthamiana. As shown in Figure 1B, when cYFP-SVBV P1 and nYFP-FvLHC II-1l were co-expressed in N. benthamiana leaf cells, strong YFP fluorescence was observed, similar to the co-infiltration with nYFP-4A and cYFP-P2 (positive control). As expected, no fluorescence could be observed in N. benthamiana leaf epidermal cells co-infiltrated with cYFP-SVBV P1 and nYFP or nYFP-FvLHC II-1L and cYFP (negative controls; Figure 1B), indicating that SVBV P1 can interact with FvLHC II-1L in vivo. Collectively, these results suggest that SVBV P1 interacts with FvLHC II-1L in vitro and in vivo.

Figure 1. Strawberry vein banding virus (SVBV) P1 interacts with chlorohyll a/b-binding protein of light-harvesting complex II type 1 like of Fragaria vesca (FvLHC II-1L) in vitro and in vivo. (A) Interaction between SVBV P1 and FvLHC II-1L was elucidated using an in vitro pull-down assay. Maltose-binding protein (MBP)-SVBV P1 or MBP was expressed in, and purified from, Escherichia coli BL21(DE3). FvLHC II-1L-Flag or cyan fluorescent protein (CFP)-Flag was expressed in Nicotiana benthamiana leaves and harvested at 3 days post inoculation (dpi). Purified MBP-SVBV P1 or MBP was mixed with plant extract of FvLHC II-1L-Flag or CFP-Flag and pulled down with Flag beads. (B) Interaction between SVBV P1 and FvLHC II-1L was examined using an in vivo bimolecular fluorescence complementation (BiFC) assay. N. benthamiana leaves were co-agroinfiltrated with the following combinations of plasmids: cYFP-SVBV P1 and nYFP-FvLHC II-1L, nYFP-4A and cYFP-P2 (positive control), cYFP-SVBV P1 and nYFP, and nYFP-FvLHC II-1L and cYFP (negative controls). Yellow fluorescent protein (YFP) fluorescence was observed via confocal microscopy at 3 dpi. Bar scale = 100 μm.
To investigate the subcellular localization of SVBV P1 and FvLHC II-1L, SVBV P1 and FvLHC II-1L were fused to the C-terminus of GFP, respectively, to obtain SVBV P1-GFP and FvLHC II-1L-GFP. The constructs were then transformed into A. tumefaciens and infiltrated into N. benthamiana leaves, as described previously (Rui et al., 2021). GFP fluorescence was observed in the cytoplasm and cell periphery of SVBV P1-GFP-infiltrated N. benthamiana leaves, and GFP fluorescence was observed in chloroplasts of FvLHC II-1L-GFP-infiltrated N. benthamiana leaves (Figure 2A). To determine whether FvLHC II-1L affects the localization of SVBV P1, we co-expressed SVBV P1-GFP and FvLHC II-1L-RFP in N. benthamiana leaves using the agroinfiltration method. The results showed that SVBV P1-GFP and FvLHC II-1L-RFP were co-localized at the edge of N. benthamiana epidermal cells, and GFP fluorescence converged at the edge of cells (Figure 2B). These results suggest that FvLHC II-1L promotes SVBV P1 aggregation at the periphery of plant cells.
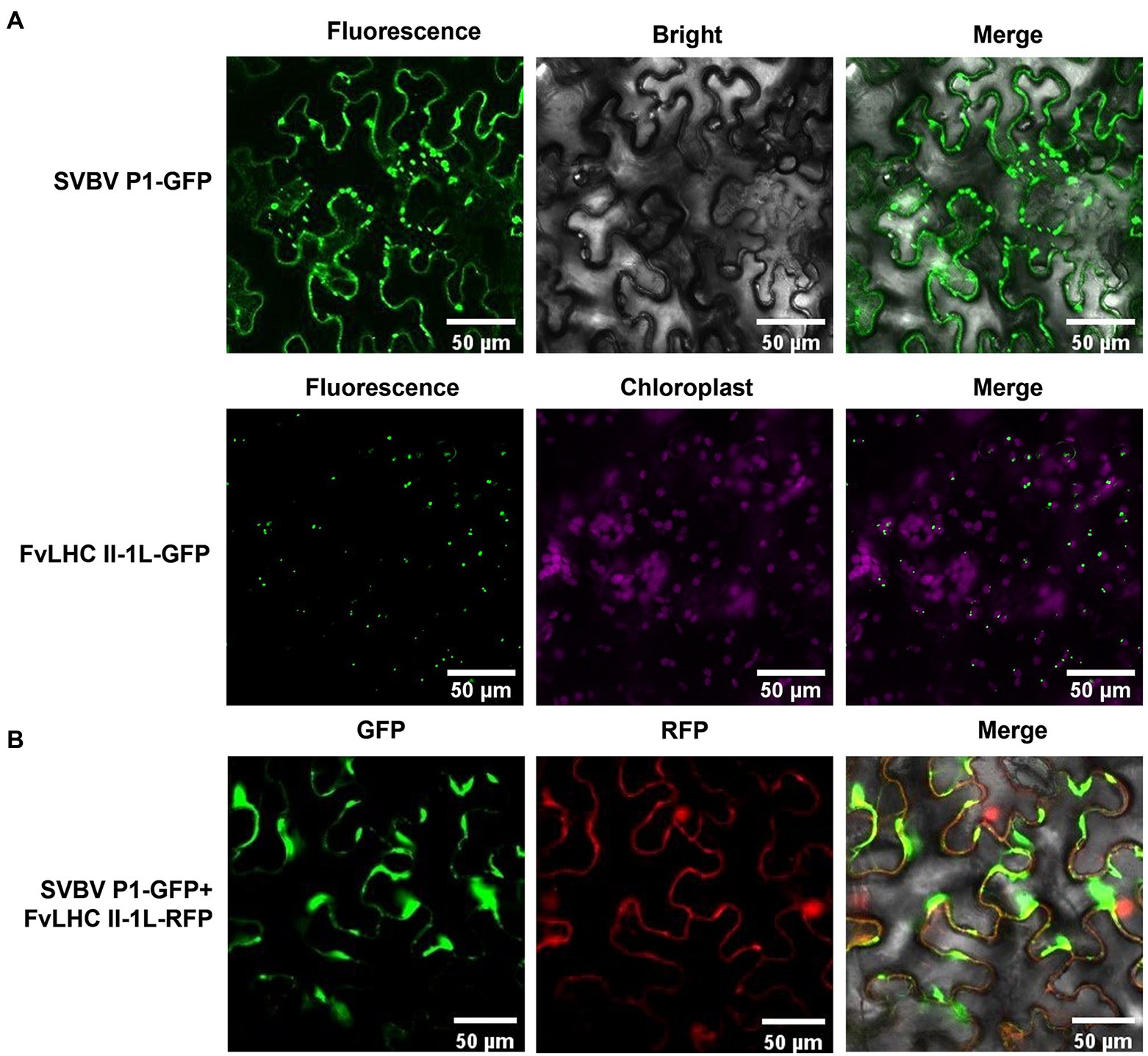
Figure 2. Subcellular localization of SVBV P1 and FvLHC II-1L. (A) Subcellular localization of SVBV P1 and FvLHC II-1L in N. benthamiana leaves. N. benthamiana leaves were infiltrated with the indicated vector combinations. Green fluorescent protein (GFP) was observed at 3 dpi. (B) The co-localization of SVBV P1 and FvLHC II-1L in N. benthamiana leaves. The fluorescence was observed via confocal microscopy at 3 dpi. Bar scale = 50 μm.
To investigate the response of FvLHC II-1L to SVBV infection, healthy F. vesca was inoculated with A. tumefaciens containing the infectious clone of SVBV using the vacuum-infiltration method, as described previously (Tian et al., 2015). Compared with the mock control, F. vesca inoculated with the infectious clone of SVBV showed vein banding symptoms at 35 dpi (Figure 3A). Next, leaves with vein banding symptoms were collected from infected plants and subjected to RT-qPCR analysis. As shown in Figure 3B, the expression of FvLHC II-1L was significantly upregulated by approximately 3.5-fold following SVBV infection. This result suggests that SVBV can upregulate the transcript levels of FvLHC II-1L in F. vesca.

Figure 3. Expression of FvLHC II-1L is upregulated by SVBV in infected F. vesca. (A) Phenotype of mock-inoculated or SVBV-inoculated F. vesca. (B) The transcript levels of FvLHC II-1L in mock-inoculated and SVBV-infected leaves of F. vesca were determined using quantitative real-time polymerase chain reaction (RT-qPCR) analysis. Error bars indicate the mean ± standard deviation of three replicates. A two-sample unequal variance directional t-test was used to test the significance of the difference (*p < 0.05).
Protein P1 of strawberry vein banding virus has been shown to complement the cell-to-cell movement of movement-defective PVX (Rui et al., 2021). To determine whether FvLHC II-1L affects the function of SVBV P1, we performed a complement assay. A. tumefaciens containing pBin438, P25, pBin438-SVBV P1, and pBin438-MYC-FvLHC II-1L were co-infiltrated with movement-defective PVX (PVXΔP25-GFP) into N. benthamiana leaves. At 3 dpi, strong GFP fluorescence was observed in infiltrated and adjacent cells of N. benthamiana leaves co-expressing pBin438-FvLHC II-1L and pBin438-SVBV P1, which was stronger than that observed in N. benthamiana leaves infiltrated with SVBV P1 alone (Figure 4A). N. benthamiana leaves infiltrated with pBin438 or P25 were used as negative and positive controls, respectively (Figure 4A). The accumulation of GFP protein in N. benthamiana leaves (Figure 4A) was also determined by western blot analysis. As expected, higher GFP protein accumulation was observed in N. benthamiana leaves co-expressing FvLHC II-1L and SVBV P1, compared to N. benthamiana leaves infiltrated with pBin438, P25, or pBin438-SVBV P1 (Figure 4B).
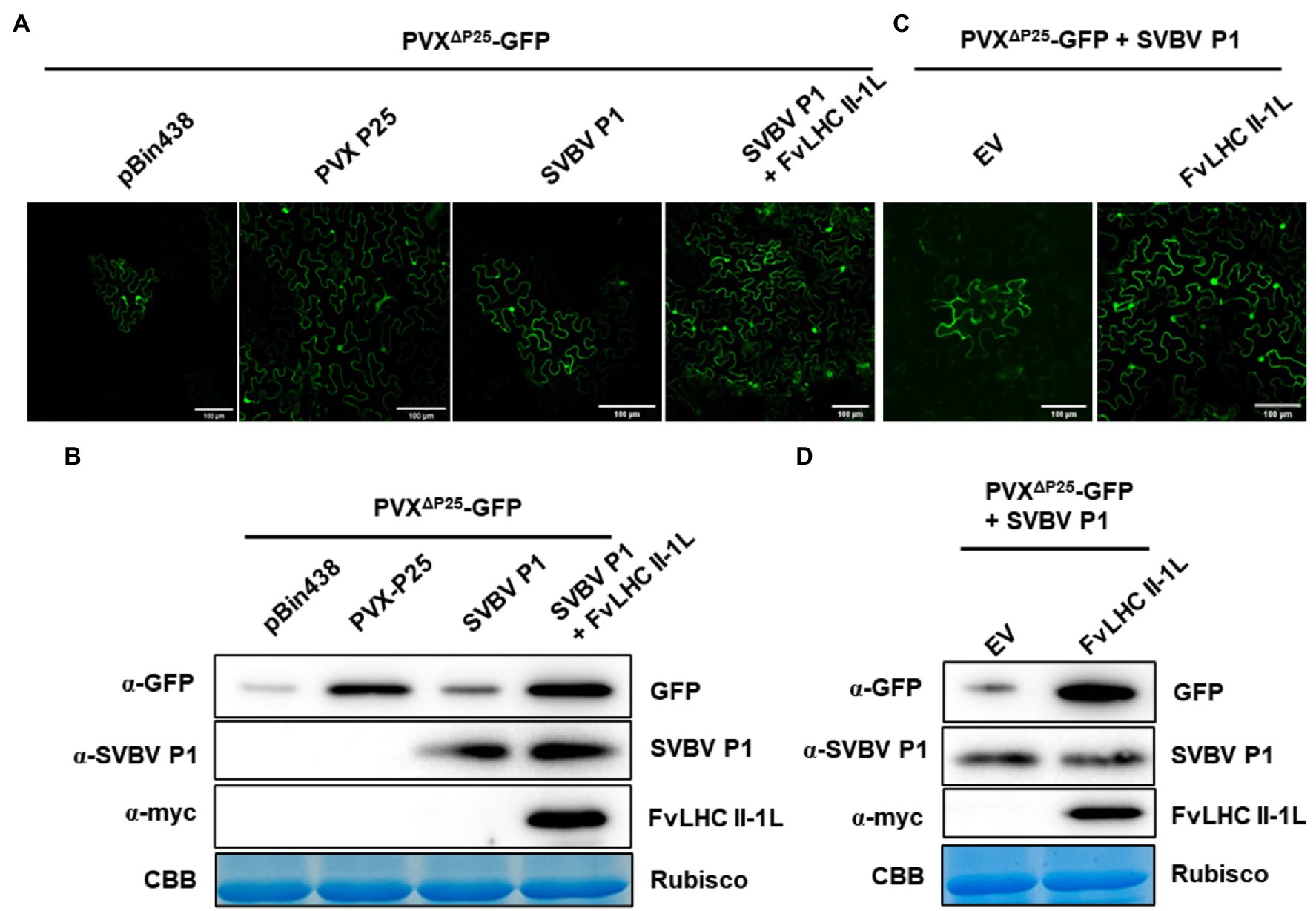
Figure 4. FvLHC II-1L assists SVBV P1 in complementing movement-defective potato virus X (PVX). (A) FvLHC II-1L promotes SVBV P1 to complement movement-defective PVX (PVXΔP25-GFP) in N. benthamiana. PVXΔP25-GFP was co-infiltrated with pBin438 empty vector, P25, SVBV P1, or SVBV P1 and FvLHC II-1L in N. benthamiana. GFP fluorescence was observed at 3 dpi. Bar scale = 100 μm. (B) Western blot analysis of GFP, SVBV P1, or FvLHC II-1L protein accumulation in infiltrated leaves at 3 dpi. α-GFP, α-SVBV P1, and α-myc antibodies were used to detect the accumulation of GFP, SVBV P1, and FvLHC II-1L, respectively. Coomassie Brilliant Blue-stained RuBisCO was used as the loading control. (C) FvLHC II-1L promotes SVBV P1 to complement PVXΔP25-GFP in FvLHC II-1L transgenic N. benthamiana. PVXΔP25-GFP was co-expressed with SVBV P1 in empty vector (EV) or FvLHC II-1L transgenic N. benthamiana. GFP fluorescence was observed at 3 dpi. (D) The accumulation of GFP, SVBV P1, and FvLHC II-1L proteins in infiltrated leaves was detected via western blotting at 3 dpi.
To further understand the role of FvLHC II-1L, with respect to SVBV P1 complementing PVXΔP25-GFP, transgenic N. benthamiana plants overexpressing MYC-tagged FvLHC II-1L (p1307-MYC-FvLHC II-1L) were generated. The mRNA and protein levels of FvLHC II-1L in transgenic plants were quantified via RT-qPCR and western blotting, respectively (Supplementary Figure S1). Then, A. tumefaciens containing pBin438-SVBV P1 and PVXΔP25-GFP was co-infiltrated into FvLHC II-1L transgenic N. benthamiana leaves. As shown in Figure 4C, the area of GFP fluorescence in FvLHC II-1L transgenic plant leaves was noticeably larger than that in empty vector transgenic N. benthamiana plant leaves. Western blot analysis revealed a greater accumulation of GFP protein in FvLHC II-1L transgenic plant leaves, compared to leaves of the empty vector control plant (Figure 4D). Collectively, these results suggest that FvLHC II-1L can assist SVBV P1 in complementing movement-defective PVX.
To investigate whether SVBV P1 can complement the systemic movement of CMVΔMP, the recombinant plasmid CMVΔMP-SVBV P1 was constructed. The construct was then transformed into A. tumefaciens, followed by infiltration into N. benthamiana seedlings. After 8 days, compared with the CMV plant (positive control), more mild leaf crimping was observed in systemic leaves of N. benthamiana seedlings infiltrated with CMVΔMP-SVBV P1, while the systemic leaves of CMVΔMP-erGFP-infected N. benthamiana seedlings showed no symptoms (Figure 5A). As shown in Figure 5B, the accumulation of CMV CP was detected in CMVΔMP-SVBV P1-infected N. benthamiana systemic leaves but not in CMVΔMP-erGFP-infected N. benthamiana systemic leaves.
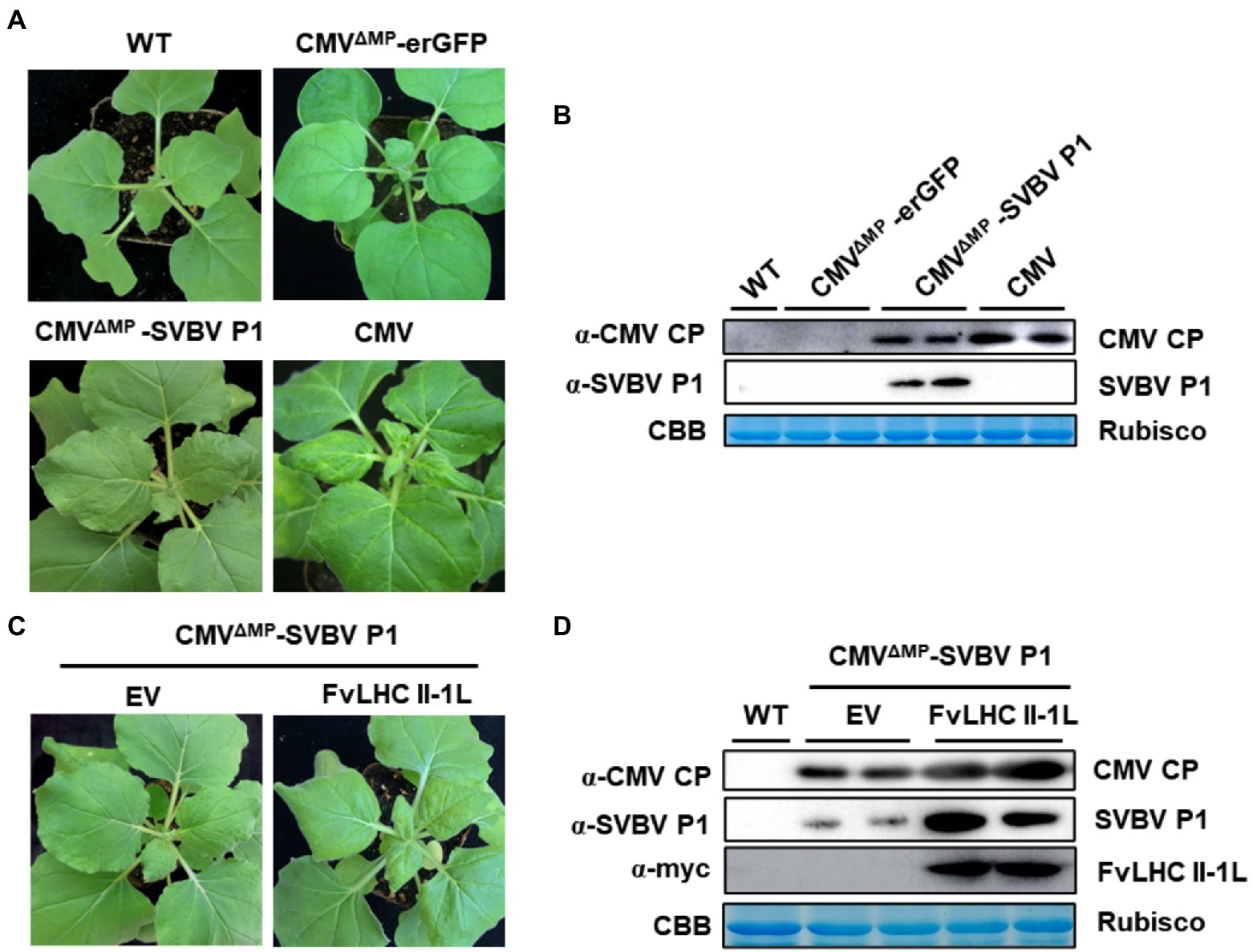
Figure 5. FvLHC II-1L assists SVBV P1 in complementing movement-defective cucumber mosaic virus (CMV). (A) Systemic symptoms of wild-type N. benthamiana plants agroinfiltrated with CMV, CMVΔMP-erGFP, or CMVΔMP-SVBV P1 at 8 dpi. WT indicates non-infiltrated plants that served as the negative control. (B) Accumulation of CMV coat protein (CP) and SVBV P1 in systemically infected leaves was detected via western blotting. (C) Systemic symptoms of empty vector (EV) or FvLHC II-1L transgenic N. benthamiana plants agroinfiltrated with CMVΔMP–SVBV P1 at 8 dpi. (D) Accumulation of CMV CP, SVBV P1, and FvLHC II-1L in systemically infected leaves was detected via western blotting.
To determine whether FvLHC II-1L promotes SVBV P1 to complement the movement of CMVΔMP, empty vector and FvLHC II-1L transgenic N. benthamiana leaves were infiltrated with CMVΔMP-SVBV P1. Compared with the empty vector plants, more severe symptoms were observed in the systemic leaves of CMVΔMP-SVBV P1-infected FvLHC II-1L transgenic N. benthamiana (Figure 5C). As expected, CMV CP and SVBV P1 protein accumulation was higher in FvLHC II-1L plants (Figure 5D). These results suggest that SVBV P1 was able to complement the systemic movement of CMVΔMP, and FvLHC II-1L can enhance the complement function of SVBV P1.
To further clarify the role of FvLHC II-1L in the infection progression of SVBV in F. vesca plants, we used the TRV system to overexpress’in F. vesca seedlings before inoculation with SVBV. At 15 dpi, systemic leaves of F. vesca seedlings were sampled and subjected to RT-qPCR and western blot analyses. The results showed that the transcript levels of FvLHC II-1L and the accumulation of CP-FvLHC II-1L fusion protein in F. vesca seedlings infiltrated with pTRV-FvLHC II-1L were higher than those in the seedlings infiltrated with TRV:00 (negative control; Figure 6A).
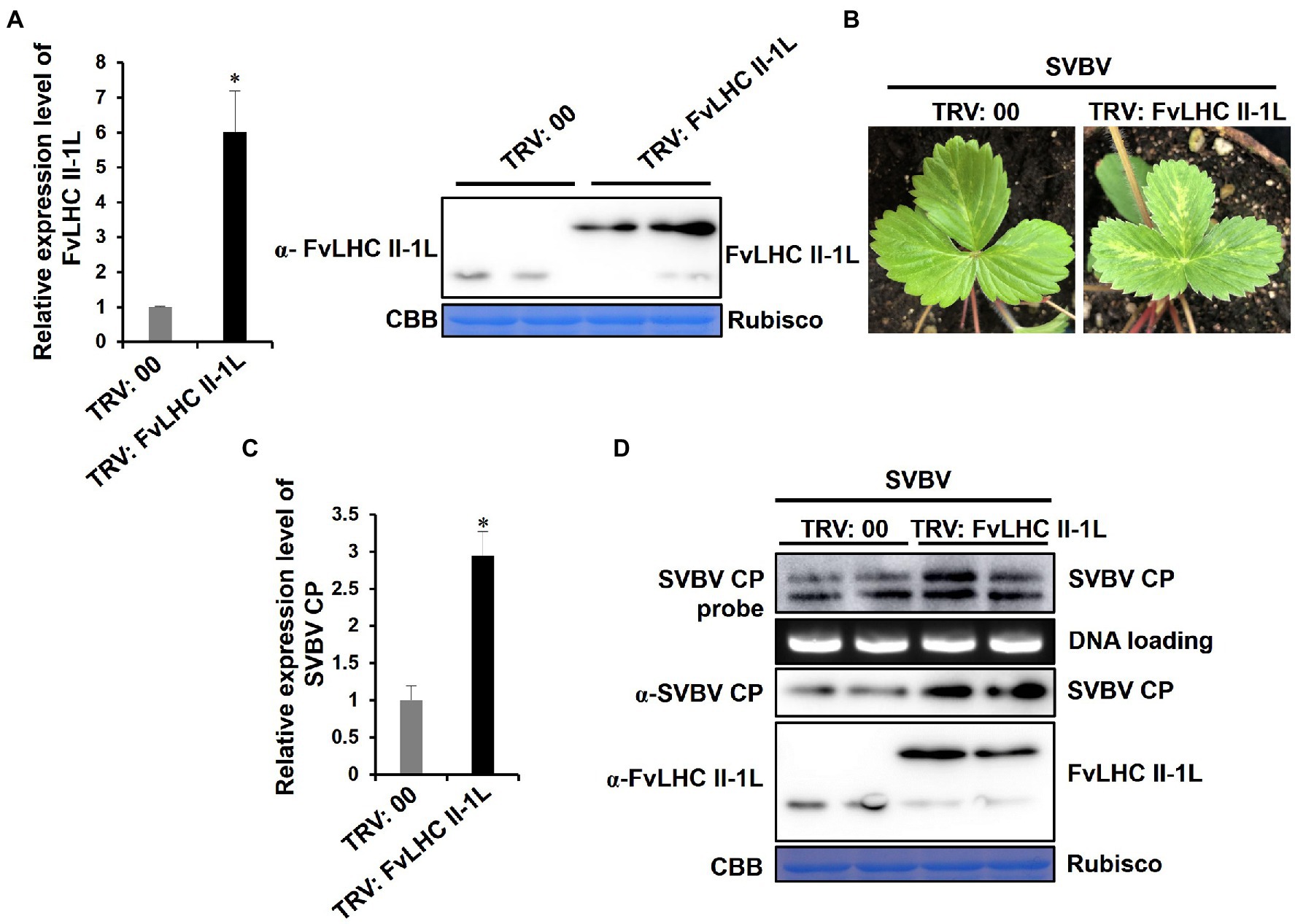
Figure 6. Overexpression of FvLHC II-1L promotes F. vesca infection by SVBV. (A) The transcript levels of FvLHC II-1L and the accumulation of FvLHC II-1L protein in TRV:00-infected or TRV:FvLHC II-1L-infected leaves of F. vesca were determined via RT-qPCR analysis and Western blot at 15 dpi. TRV:00 indicates plants infiltrated with tobacco rattle virus (TRV) empty vector, serving as the negative control. (B) Systemic symptoms of TRV:00 or TRV:FvLHC II-1L pre-infected plants inoculated with SVBV via vacuum-infiltration. The phenotype of SVBV infection was monitored and photographed at 35 days post SVBV inoculation. (C) The transcript levels of SVBV CP in systemically infected leaves of TRV:00 and TRV:FvLHC II-1L pre-infected F. vesca were quantified using RT-qPCR analysis. (D) Accumulation of SVBV CP and FvLHC II-1L proteins and SVBV CP DNA, in systemically infected leaves, was detected via Western and Southern blotting, respectively.
At 15 dpi, TRV:00-infiltrated or TRV:FvLHC II-1L-infiltrated plants were infiltrated with the infectious clone of SVBV. After 35 days, the systemic symptoms of TRV:FvLHC II-1L-infiltrated plants were more severe than those of TRV:00-infiltrated plants (Figure 6B). As expected, RT-qPCR, western blotting, and Southern blotting showed that the levels of SVBV CP mRNA transcripts, SVBV CP protein, and DNA accumulation were significantly increased in TRV:FvLHC II-IL plants (Figures 6C,D). These results indicated that FvLHC II-1L promoted F. vesca infection by SVBV.
To infect host plants, plant viruses have to move between cells to achieve local infection, and then move further, over longer distances through the phloem vascular bundles to achieve systematic infection. Both these processes are related to MPs encoded by viruses (Zhou et al., 2019). Not only is SVBV P1 essential for SVBV intercellular movement but it can also complement the movement function of heterologous viruses and promote the intercellular movement of movement-deficient PVX (Rui et al., 2021). In this study, we found that compared with the movement-deficient CMV, the movement-deficient CMV inserted with SVBV P1 systematically moved to the upper leaves via long-distance movement, indicating that SVBV P1 complemented the movement-deficient CMV (Figures 5A,B). This complement function is not unique to SVBV P1. The MP Nsvc4 encoded by RSV can complement cell-to-cell movement of movement-deficient PVX (Fu et al., 2018). The MP NSm, encoded by tomato spotted wilt virus, can complement cell-to-cell and long-distance movement of movement-deficient tobacco mosaic virus (TMV) and CMV (Li et al., 2009). The MPs encoded by some Rhabdoviruses can complement the intercellular movement of movement-deficient tomato mosaic virus and PVX (Zhou et al., 2019). Thus, the functions of MPs of different viruses are complementary.
During viral infection, many proteins encoded by plant viruses can interact with chloroplast proteins and affect the course of disease development (Bhat et al., 2013; Mathioudakis et al., 2013; Cheng et al., 2021). The replicase protein of TMV interacts with the chloroplast ATPase γ subunit AtpC to block ATP synthesis, decrease photosynthate accumulation, and aggravate viral symptoms (Bhat et al., 2013). The replicase protein of tobacco vein banding mosaic virus interacts with the chloroplast 50S ribosomal protein large submit 1 to promote the accumulation of viral replicase and improve viral replication (Cheng et al., 2021). Pepino mosaic virus (PepMV) MP P26 interacts with tomato chloroplast catalase 1 (CAT1) to improve the activity of CAT1. Silencing of the CAT1 gene of N. benthamiana resulted in a significant decrease in RNA accumulation of PepMV and inhibition of systemic infection by the virus (Mathioudakis et al., 2013). These results suggest that host chloroplast proteins have a close association with plant viral infection. In the present study, the FvLHC II-1L protein, which is another chloroplast protein responsible for maintaining the stability of the electron transport chain of PS II, is an important regulatory factor of photosynthesis and participates in normal plant growth and development. In a previous study, we screened the FvLHC II-1L protein from the cDNA library of F. vesca using SVBV P1 as the bait protein (Zhang et al., 2019). Here, FvLHC II-1L was shown to interact with SVBV P1 both in vitro and in vivo (Figure 1). Further experiments showed that the transcription of FvLHC II-1L was upregulated during SVBV infection, suggesting that FvLHC II-1L may be involved in viral infection (Figure 3). This finding is consistent with those of previous studies showing that chloroplast proteins frequently have important roles in facilitating viral infection (Cheng et al., 2021).
Furthermore, we noted that SVBV P1 and FvLHC II-1L were co-located at the edge of cells, wherein SVBV P1 formed irregular aggregates (Figure 2). FvLHC II-1L not only assists SVBV P1 to accelerate the intercellular movement of the PVX transport-deficient mutant but also promotes SVBV P1 to complement the systemic movement of the CMV transport-deficient mutant (Figures 4, 5). It has been reported that the MP P3N-PIPO of turnip mosaic virus (TuMV) interacts with Arabidopsis thaliana plasma membrane-associated Ca2+ binding protein 1 to expand the diameter of PD and promote viral infection (Vijayapalani et al., 2012). The PVX MP TGB12K interacts with tobacco TIP1 protein, which can act on β-1,3-glucanase and increase the diameter of PD, thereby promoting intercellular transport of PVX (Fridborg et al., 2003). The P25 protein of PVX interacts with the chloroplast ferridoxin 1 (FD1) protein to reduce callose deposition in PD and enhance viral infection (Yang et al., 2020). Rice chloroplast FD1 has previously been demonstrated to be an important component of the photosynthetic electron transport chain that can affect plant growth and development. Therefore, it is reasonable to speculate that FvLHC II-1L functions as a key protein in the photosynthetic electron transport chain of F. vesca, which can interact with SVBV P1 and co-locate with it in cell walls, decrease callose deposition, increase the diameter of PD, and promote viral intercellular and systemic movement. Additionally, in the current study, we showed that overexpression of FvLHC II-1L accelerated the course of SVBV infection (Figure 6).
Chloroplasts are not only involved in photosynthesis but also have a close association with salicylic acid and other resistance pathways that are activated by the accumulation of reactive oxygen species (ROS; Nomura et al., 2012; Chan et al., 2016; Serrano et al., 2016). The C4 protein encoded by tomato yellow leaf curve virus can move from the cell membrane to the chloroplast during tobacco infection and can bind to the calcium sensor protein of chloroplasts to inhibit resistance signal transduction (Medina-Puche et al., 2020). The γb protein, encoded by BSMV, interacts with chloroplast NADPH-dependent thioredoxin reductase C (NTRC) to inhibit NTRC-mediated antioxidant defense and promote viral infection (Wang et al., 2021). Similarly, the VPg protein of TuMV interacts with the NADH dehydrogenase-like complex M subunit (NdhM) of N. benthamiana chloroplasts to prevent export from the nucleus and inhibit the NdhM-mediated defense response in the host (Zhai et al., 2021). FvLHC II-1L is the core protein of the light-harvesting complex of PS II in F. vesca. We speculated that the overexpression of FvLHC II-1L would result in chloroplasts absorbing excess light energy, leading to disruption of the balance between ROS production and cleanup and initiation of ROS-dependent resistance signal pathways. Interaction of SVBV P1 with FvLHC II-1L likely reduces the levels of FvLHC II-1L in chloroplasts, leading to a reduction in the intensity of photosynthesis, and subsequently, the reduction of ROS accumulation, inhibition of immune response, and promotion of viral infection.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
XZ, LJ, and TJ conceived and designed the study. SX, XZ, KX, and ZW performed the experiments and data analysis. SX, XZ, and KX wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the Grants from the National Natural Science Foundation of China (nos. 32072386 and 31801700), the Anhui Provincial Key Research and Development Plan (no. 202004a06020013), the Zhejiang Basic Public Welfare Research Project of China (no. LGN19C140002), and the Postdoctoral Science Foundation of Anhui Province (no. 2019B360).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Xiaorong Tao (Nanjing Agricultural University, China) for providing the system of movement-defective CMV. We also thank Fei Yan (Ningbo University, China) for providing the wild-type PVX-GFP and the movement-deficient PVXΔP25.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.884044/full#supplementary-material
A. tumefaciens, Agrobacterium tumefaciens; AMV, Alfalfa mosaic virus; BiFC, Bimolecular fluorescence complementation; BSMV, Barley stripe mosaic virus; CMVΔMP, Movement-defective cucumber mosaic virus; CAT1, chloroplast catalase 1; cDNA, Complementary DNA; CP, Coat protein; dpi, Days post inoculation; erGFP, Endoplasmic reticulum-targeted green fluorescent protein; EV, Empty vector; FD1, Ferridoxin 1 protein; F. vesca, Fragaria vesca; FvLHC II-1L, Chlorophyll a/b-binding protein of light-harvesting complex II type 1 like of Fragaria vesca; GFP, Green fluorescent protein; LHC II, Light-harvesting complex II; MP, Movement protein; N. benthamiana, Nicotiana benthamiana; NdhM, NADH dehydrogenase-like complex M subunit; NTRC, NADPH-dependent thioredoxin reductase C; PD, Plasmodesmata; PepMV, Pepino mosaic virus; PSII, Photosystem II; PVXΔP25-GFP, Movement-deficient potato virus X green fluorescent protein; RT-qPCR, Quantitative real-time polymerase chain reaction; REM1, Remorin protein; RFP, Red fluorescent protein; ROS, Reactive oxygen species; RSV, Rice stripe virus; SVBV, Strawberry vein banding virus; SVBV P1, Movement protein P1 of strawberry vein banding virus; TMV, Tobacco mosaic virus; TRV, Tobacco rattle virus; YFP, Yellow fluorescent protein; Y2H, Yeast two-hybrid.
Allen, J. F., Bennett, J., Steinback, K. E., and Arntzen, C. J. (1981). Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature 291, 25–29. doi: 10.1038/291025a0
Bhat, S., Folimonova, S. Y., Cole, A. B., Ballard, K. D., Lei, Z., Watson, B. S., et al. (2013). Influence of host chloroplast proteins on tobacco mosaic virus accumulation and intercellular movement. Plant Physiol. 161, 134–147. doi: 10.1104/pp.112.207860
Chan, K. X., Phua, S. Y., Crisp, P., McQuinn, R., and Pogson, B. J. (2016). Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53. doi: 10.1146/annurev-arplant-043015-111854
Chen, M. H., Sheng, J., Hind, G., Handa, A. K., and Citovsky, V. (2000). Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 19, 913–920. doi: 10.1093/emboj/19.5.913
Chen, J., Zhang, H., Feng, M., Zuo, D., Hu, Y., and Jiang, T. (2016a). Transcriptome analysis of woodland strawberry (Fragaria vesca) response to the infection by strawberry vein banding virus (SVBV). Virol. J. 13:128. doi: 10.1186/s12985-016-0584-5
Chen, X., Zhu, M., Jiang, L., Zhao, W., Li, J., Wu, J., et al. (2016b). A multilayered regulatory mechanism for the autoinhibition and activation of a plant CC-NB-LRR resistance protein with an extra N-terminal domain. New Phytol. 212, 161–175. doi: 10.1111/nph.14013
Cheng, D. J., Xu, X. J., Yan, Z. Y., Tettey, C. K., Fang, L., Yang, G. L., et al. (2021). The chloroplast ribosomal protein large subunit 1 interacts with viral polymerase and promotes virus infection. Plant Physiol. 187, 174–186. doi: 10.1093/plphys/kiab249
Feng, Z., Chen, X., Bao, Y., Dong, J., Zhang, Z., and Tao, X. (2013). Nucleocapsid of tomato spotted wilt tospovirus forms mobile particles that traffic on an actin/endoplasmic reticulum network driven by myosin XI-K. New Phytol. 200, 1212–1224. doi: 10.1111/nph.12447
Feng, M., Zhang, H., Pan, Y., Hu, Y., Chen, J., Zuo, D., et al. (2016). Complete nucleotide sequence of strawberry vein banding virus Chinese isolate and infectivity of its full-length DNA clone. Virol. J. 13:164. doi: 10.1186/s12985-016-0624-1
Frazier, N. W. (1974). Detection of graft-transmissible diseases in strawberry by a modified leaf grafting technique. Plant Dis. Rep. 58, 203–207.
Fridborg, I., Grainger, J., Page, A., Coleman, M., Findlay, K., and Angell, S. (2003). TIP, a novel host factor linking callose degradation with the cell-to-cell movement of potato virus X. Mol. Plant Microbe Interact. 16, 132–140. doi: 10.1094/MPMI.2003.16.2.132
Fu, S., Xu, Y., Li, C., Li, Y., Wu, J., and Zhou, X. (2018). Rice stripe virus interferes with S-acylation of Remorin and induces its autophagic degradation to facilitate virus infection. Mol. Plant 11, 269–287. doi: 10.1016/j.molp.2017.11.011
Huang, W., Chen, Q., Zhu, Y., Hu, F., Zhang, L., Ma, Z., et al. (2013). Arabidopsis thylakoid formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol. Plant 6, 1673–1691. doi: 10.1093/mp/sst069
Jiang, L., Li, S., Jiang, X., and Jiang, T. (2021). Complete genome sequence of two strawberry vein banding virus isolates from China. Biocell 45, 1695–1702. doi: 10.32604/biocell.2021.015250
Li, S., Hu, Y., Jiang, L., Rui, P., Zhao, Q., Feng, J., et al. (2018a). Strawberry vein banding virus P6 protein is a translation trans-activator and its activity can be suppressed by FveIF3g. Viruses 10:717. doi: 10.3390/v10120717
Li, W., Lewandowski, D. J., Hilf, M. E., and Adkins, S. (2009). Identification of domains of the tomato spotted wilt virus NSm protein involved in tubule formation, movement and symptomatology. Virology 390, 110–121. doi: 10.1016/j.virol.2009.04.027
Li, P., Liu, C., Deng, W. H., Yao, D. M., Pan, L. L., Li, Y. Q., et al. (2019). Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 15:e1007607. doi: 10.1371/journal.ppat.1007607
Li, Z., Zhang, Y., Jiang, Z., Jin, X., Zhang, K., Wang, X., et al. (2018b). Hijacking of the nucleolar protein fibrillarin by TGB1 is required for cell-to-cell movement of barley stripe mosaic virus. Mol. Plant Pathol. 19, 1222–1237. doi: 10.1111/mpp.12612
Lucas, W. J. (2006). Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344, 169–184. doi: 10.1016/j.virol.2005.09.026
Mathioudakis, M. M., Veiga, R. S., Canto, T., Medina, V., Mossialos, D., Makris, A. M., et al. (2013). Pepino mosaic virus triple gene block protein 1 (TGBp1) interacts with and increases tomato catalase 1 activity to enhance virus accumulation. Mol. Plant Pathol. 14, 589–601. doi: 10.1111/mpp.12034
Medina-Puche, L., Tan, H., Dogra, V., Wu, M., Rosas-Diaz, T., Wang, L., et al. (2020). A defense pathway linking plasma membrane and chloroplasts and co-opted by pathogens. Cell 182, 1109–1124.e25. doi: 10.1016/j.cell.2020.07.020
Neilson, J. A., and Durnford, D. G. (2010). Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth. Res. 106, 57–71. doi: 10.1007/s11120-010-9576-2
Nilsson, A., Stys, D., Drakenberg, T., Spangfort, M. D., Forsén, S., and Allen, J. F. (1997). Phosphorylation controls the three-dimensional structure of plant light harvesting complex II. J. Biol. Chem. 272, 18350–18357. doi: 10.1074/jbc.272.29.18350
Nomura, H., Komori, T., Uemura, S., Kanda, Y., Shimotani, K., Nakai, K., et al. (2012). Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3:926. doi: 10.1038/ncomms1926
Pan, Y., Zhou, X., Li, S., Feng, M., Shi, M., Zuo, D., et al. (2018). Strawberry vein banding virus P6 protein intracellular transport and an important domain identifcation. J. Integr. Agr. 17, 2031–2041. doi: 10.1016/S2095-3119(18)61978-1
Park, M. R., Jeong, R. D., and Kim, K. H. (2014). Understanding the intracellular trafficking and intercellular transport of potexviruses in their host plants. Front. Plant Sci. 5:60. doi: 10.3389/fpls.2014.00060
Pattanaik, S., Dey, N., Bhattacharyya, S., and Maiti, I. B. (2004). Isolation of full-length transcript promoter from the strawberry vein banding virus (svbv) and expression analysis by protoplasts transient assays and in transgenic plants. Plant Sci. 167, 427–438. doi: 10.1016/j.plantsci.2004.04.011
Peiro, A., Izquierdo-Garcia, A. C., Sanchez-Navarro, J. A., Pallas, V., Mulet, J. M., and Aparicio, F. (2014). Patellins 3 and 6, two members of the plant Patellin family, interact with the movement protein of alfalfa mosaic virus and interfere with viral movement. Mol. Plant Pathol. 15, 881–891. doi: 10.1111/mpp.12146
Pietrzykowska, M., Suorsa, M., Semchonok, D. A., Tikkanen, M., Boekema, E. J., Aro, E. M., et al. (2014). The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 26, 3646–3660. doi: 10.1105/tpc.114.127373
Ratti, C., Pisi, A., Rubies Autonell, C., Babini, A., and Vicchi, V. (2009). First report of strawberry vein banding virus on strawberry in Italy. Plant Dis. 93:675. doi: 10.1094/PDIS-93-6-0675A
Rui, P., Wang, Z., Shan, W., Xia, W., Zhou, X., Yang, L., et al. (2021). P1 of strawberry vein banding virus is a multilocalized protein that functions as a movement protein and interacts with the coat protein. J. Integr. Agr. 21, 1071–1083. doi: 10.1016/S2095-3119(21)63711-5 [Epub ahead of print].
Schoelz, J. E., Angel, C. A., Nelson, R. S., and Leisner, S. M. (2016). A model for intracellular movement of cauliflower mosaic virus: the concept of the mobile virion factory. J. Exp. Bot. 67, 2039–2048. doi: 10.1093/jxb/erv520
Schoelz, J. E., Harries, P. A., and Nelson, R. S. (2011). Intracellular transport of plant viruses: finding the door out of the cell. Mol. Plant 4, 813–831. doi: 10.1093/mp/ssr070
Serrano, I., Audran, C., and Rivas, S. (2016). Chloroplasts at work during plant innate immunity. J. Exp. Bot. 67, 3845–3854. doi: 10.1093/jxb/erw088
Shen, Y., Zhao, X., Yao, M., Li, C., Miriam, K., Zhang, X., et al. (2014). A versatile complementation assay for cell-to-cell and long distance movements by cucumber mosaic virus based agro-infiltration. Virus Res. 190, 25–33. doi: 10.1016/j.virusres.2014.06.013
Sun, Y. D., Zhang, L., and Folimonova, S. Y. (2021). Citrus miraculin-like protein hijacks a viral movement-related p33 protein and induces cellular oxidative stress in defence against citrus tristeza virus. Plant Biotechnol. J. 19, 977–991. doi: 10.1111/pbi.13523
Tian, J., Cheng, L., Han, Z., and Yao, Y. (2015). Tobacco rattle virus mediated gene silencing in strawberry plants. Plant Cell Tiss. Org. 120, 1131–1138. doi: 10.1007/s11240-014-0669-z
Tian, J., Pei, H., Zhang, S., Chen, J., Chen, W., Yang, R., et al. (2014). TRV-GFP: a modified tobacco rattle virus vector for efficient and visualizable analysis of gene function. J. Exp. Bot. 65, 311–322. doi: 10.1093/jxb/ert381
Vijayapalani, P., Maeshima, M., Nagasaki-Takekuchi, N., and Miller, W. A. (2012). Interaction of the trans-frame potyvirus protein P3NPIPO with host protein PCaP1 facilitates potyvirus movement. PLoS Pathog. 8:e1002639. doi: 10.1371/journal.ppat.1002639
Wang, X., Jiang, Z., Yue, N., Jin, X., Zhang, X., Li, Z., et al. (2021). Barley stripe mosaic virus γb protein disrupts chloroplast antioxidant defenses to optimize viral replication. EMBO J. 40:e107660. doi: 10.15252/embj.2021107660
Wu, X., and Cheng, X. (2020). Intercellular movement of plant RNA viruses: targeting replication complexes to the plasmodesma for both accuracy and efficiency. Traffic 21, 725–736. doi: 10.1111/tra.12768
Yan, F., Lu, Y., Lin, L., Zheng, H., and Chen, J. (2012). The ability of PVX p25 to form RL structures in plant cells is necessary for its function in movement, but not for its suppression of RNA silencing. PLoS One 7:e43242. doi: 10.1371/journal.pone.0043242
Yang, X., Lu, Y., Wang, F., Chen, Y., Tian, Y., Jiang, L., et al. (2020). Involvement of the chloroplast gene ferredoxin 1 in multiple responses of Nicotiana benthamiana to potato virus X infection. J. Exp. Bot. 71, 2142–2156. doi: 10.1093/jxb/erz565
Zhai, Y., Yuan, Q., Qiu, S., Li, S., Li, M., Zheng, H., et al. (2021). Turnip mosaic virus impairs perinuclear chloroplast clustering to facilitate viral infection. Plant Cell Environ. 44, 3681–3699. doi: 10.1111/pce.14157
Zhang, X. X., Jiang, X. Z., Li, S., Jiang, L., and Jiang, T. (2019). Screening of the host factors of woodland strawberry interacting with P1 of strawberry vein banding virus by yeast two-hybrid system. Acta Phytopathol. Sin. 49, 56–63. doi: 10.13926/j.cnki.apps.000362
Zhou, X., Lin, W., Sun, K., Wang, S., Zhou, X., Jackson, A. O., et al. (2019). Specificity of plant Rhabdovirus cell-to-cell movement. J. Virol. 93, e00296–e00219. doi: 10.1128/JVI.00296-19
Keywords: strawberry vein banding virus, intercellular movement, FvLHC II-1L protein, P1 protein, viral infection
Citation: Xu S, Zhang X, Xu K, Wang Z, Zhou X, Jiang L and Jiang T (2022) Strawberry Vein Banding Virus Movement Protein P1 Interacts With Light-Harvesting Complex II Type 1 Like of Fragaria vesca to Promote Viral Infection. Front. Microbiol. 13:884044. doi: 10.3389/fmicb.2022.884044
Received: 25 February 2022; Accepted: 06 May 2022;
Published: 26 May 2022.
Edited by:
Sean Michael Prager, University of Saskatchewan, CanadaReviewed by:
Nobumitsu Sasaki, Tokyo University of Agriculture and Technology, JapanCopyright © 2022 Xu, Zhang, Xu, Wang, Zhou, Jiang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tong Jiang, amlhbmd0b25nNDY1MEBzaW5hLmNvbQ==; Lei Jiang, amlhbmdsZWkwNjJ4QGFoYXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.