
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 18 March 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.838392
This article is part of the Research Topic"One Health" Approach For Revealing Reservoirs And Transmission Of Antimicrobial Resistance, Volume IIView all 22 articles
 Rim Abdallah1,2
Rim Abdallah1,2 Edmond Kuete Yimagou1,2
Edmond Kuete Yimagou1,2 Linda Hadjadj1,2
Linda Hadjadj1,2 Oleg Mediannikov1,2
Oleg Mediannikov1,2 Ahmad Ibrahim1,2
Ahmad Ibrahim1,2 Bernard Davoust1,2
Bernard Davoust1,2 Amanda Barciela3
Amanda Barciela3 R. Adriana Hernandez-Aguilar3,4
R. Adriana Hernandez-Aguilar3,4 Georges Diatta5
Georges Diatta5 Cheikh Sokhna2,5
Cheikh Sokhna2,5 Didier Raoult1,2
Didier Raoult1,2 Jean-Marc Rolain1,2
Jean-Marc Rolain1,2 Sophie Alexandra Baron1,2*
Sophie Alexandra Baron1,2*Introduction: The role of wildlife in the transmission of antimicrobial resistant (AMR) is suspected but scarcely reported in current studies. Therefore, we studied the dynamics and prevalence of antibiotic-resistant Enterobacterales in antibiotic-limited areas of Senegal.
Materials and Methods: We collected fecal samples from monkeys and apes (N = 226) and non-fecal environmental samples (N = 113) from Senegal in 2015 and 2019. We grew the samples on selective media, subsequently isolated AMR Enterobacterales, and then sequenced their genomes.
Results: We isolated 72 different Enterobacterales among which we obtained a resistance rate of 65% for colistin (N = 47/72) and 29% for third generation-cephalosporin (C3G) (29%, N = 21/72). Interestingly, almost 46% of our isolates, among Enterobacter sp., Citrobacter cronae and Klebsiella aerogenes, belong to 34 new STs. Moreover, the genes blaCTX–M–15, blaTEM1B, sul2, dfrA14, qnrs, aph(3′′), aph(6), tetA, and tetR harbored within a transposon on the IncY plasmid of ST224 Escherichia coli were transferred and inserted into a ST10 E. coli phage coding region.
Conclusion: Wildlife constitutes a rich, unexplored reservoir of natural microbial diversity, AMR genes and international resistant clones pathogenic in humans. The presence of a transposon that carries AMR genes is intriguing since no antibiotics are used in the non-human primates we studied.
Antimicrobial resistant (AMR) bacteria are widely found in humans, animals, and the environment, which could be all part of the same ecosystem in certain areas (McEwen and Collignon, 2018). AMR genes can be transferred within this ecosystem, and homologous resistance genes could be found in pathogens, normal flora and soil bacteria (Woolhouse et al., 2015). AMR, even in a “One Health” perspective, is considered to be linked to human activity, in hospitals and livestock farms or the artificial environment created by people such as houses and cities. However, this often overlooks the role played by the wild environment, where human activity is lower than in artificial environments created by humans (Chokshi et al., 2019).
According to the World Health Organization (WHO), Africa has one of the largest gap in data on the incidence of AMR (World Health Organization, 2014) and genomic analysis of this AMR bacteria in African wildlife has rarely been explored, to our knowledge (Ramey and Ahlstrom, 2019; Miller et al., 2020). However, it is essential information for understanding the sources of antibiotic resistance, its spread and the factors involved in this spread. In Africa, and more precisely in Senegal, AMR has mainly been studied in relation to human medicine (Dromigny et al., 2005), as well as animal farming (Vounba et al., 2019). High levels of resistance in this country indicate that human hospital outbreaks are reservoirs of extended spectrum β-lactamases (ESBL)-Enterobacterales and are potential sources of colonization and infection (Chereau et al., 2015). The poor hygiene conditions that usually exist in hospitals in low-income countries, limited access to diagnostic tools, and reduced availability of second line antibiotics may promote the horizontal transmission of multidrug-resistant bacteria (Breurec et al., 2016; Bernabé et al., 2017). Other studies have shown this resistance is widespread on farms where animals are raised with the use of antibiotics as growth promoters (Vounba et al., 2019).
In a previous work, we collected samples from wildlife chimpanzees (Pan troglodytes verus) and termites in Senegal (Baron et al., 2021). We isolated enterobacteria carrying multiple antibiotic resistance genes from which we identified international high-risk clones (ST307 and ST147) of carbapenemase-producing Klebsiella pneumoniae. The high similarities between plasmids found in isolates from chimpanzees and termites suggest lateral gene exchanges between these two species. In this work, we pursued by exploring antibiotic resistance in other Enterobacterales isolated from the same specimens. We completed our study by analyzing another collection of samples isolated from Senegal wildlife [green monkeys (Chlrocebus sabaeus), baboons (Papio papio), chimpanzees (Pan troglodytes verus), and non-fecal environmental samples].
The objective of this work was to determine the level of resistance to antibiotics used in humans in Enterobacterales isolated from samples from wildlife in Senegal and to determine the genetic support of this resistance. Interestingly, we identified a transposon in Escherichia coli strains, located on a plasmid and in the chromosome. This observation supports the role of wildlife as a reservoir of resistance genes and as a place of exchange.
Between 26th and 30th of August 2019, 215 non-human primates (NHPs) fecal samples (110 baboons, 95 green monkeys, and 10 chimpanzees) and 113 non-fecal environmental samples (47 termites, 42 soil samples from termite mounds, and 24 samples including fruits, other plant parts, a centipede and a maggot) were collected in four sites in Senegal. The NHPs fecal samples were collected in three sites located in southern Senegal in the Niokolo-Koba National Park: Simenti, Dar Salam, and Niokolo Poste sites. The sample collectors observed the monkeys all day to collect their fresh stool in real time so that they could be sure that it belonged to that NHPs and not to others. The non-fecal environmental samples, with the exception of the centipede and the maggot, were NHP foods, were collected in one site in south-eastern Senegal, the Dindefelo Community Nature Reserve (12°22′01.4′′N, 12°18′00.0′′W), which is located on the border with Guinea Conakry about 35 km from the town of Kedougou. The permission to collect these samples was approved by the Direction des Parcs Nationaux and Direction des Eaux, Forêts, Chasse et de la Conservation des Sols of the Senegal Ministry for the Environment and Sustainable Development (002737/DEFCCS/DGF de la Direction des Eaux, Forêts, Chasses et de la Conservation des Sols du 27/06/2019). The monkey cohort consisted of 95 green monkey samples (Chlrocebus sabaeus) (Linnaeus, 1766) and 110 Guinea baboon samples (Papio papio) (Desmarest, 1820), while the ape cohort consists of 10 chimpanzee samples (Pan troglodytes verus)(Schwarz, 1934) (Primates: Hominidae). It should be noted that the morphological and molecular identifications of the non-fecal environmental samples have previously been reported (Baron et al., 2021).
In addition, fecal samples collected in 2015 from Niokolo Koba National Park, Senegal, were added to this study. This cohort included seven Guinea baboon samples and four green monkey samples. Permission to collect samples was granted by the National Parks Direction and Direction des Eaux, Forêts, Chasse et de la Conservation des Sols of the Senegal Ministry for the Environment and Sustainable Development (001914/DEF/DGF de la Direction des Eaux, Forêts, Chasses et de la Conservation des Sols du 05/06/2016). No other permissions were required, as this research was non-invasive and the collection of the samples did not disrupt the wild fauna. Samples were directly stored at +4°C and cultured 2 days after collection.
Non-human primates samples were aliquoted and enriched in Tryptone Soy Broth (TSB, BioMérieux, Marcy l’Etoile, France) at 37°C for 72 h. Then, 20 μl was then inoculated on antibiotic-containing media: an LBJMR (Lucie Bardet Jean-Marc Rolain) plate (containing 4 μg/ml colistin and 100 μg/ml vancomycin) (Bardet et al., 2017), MacConkey (BioMérieux) + ertapenem (0.5 μg/ml) and MacConkey + cefotaxime (1 μg/ml). The critical concentrations selected were based on the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (Version 11.0). Inoculation was followed by incubation at 37°C for 24 h. Colonies that grew on the different media were replicated on Columbia agar + 5% sheep blood (BioMérieux) for further analyses. Bacterial identification was performed by Matrix Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) mass spectrometry (Bruker Daltonik, Bremen, Germany), as described previously (Seng et al., 2009). Concerning non-fecal environmental samples, cultures were initially inoculated on MacConkey + cefotaxime and MacConkey + ertapenem and described in a previous study (Baron et al., 2021). Briefly, samples were crushed and put in TSB at 37°C for 72 h, then 20 μl were inoculated on selective media. In this study, we also cultured the non-fecal environmental samples on LBJMR media. Unique and common Enterobacterales strains of NHPs and the non-fecal environmental samples were visualized by Cytoscape program to show interaction between the two ecosystems (Shannon et al., 2003).
Antibiotic Susceptibility Test (AST) were performed using the disk diffusion method following EUCAST recommendations. In case of resistance observed in disk diffusion method, minimum inhibitory concentrations (MIC) of imipenem and ertapenem were determined using the E-test method (BioMérieux), while colistin MIC was determined using the UMIC microdilution method (Biocentric, Bandol, France). The ESBL profile was detected by the observation of a champagne-cork or a keyhole between a third or fourth generation cephalosporin and clavulanic acid (Drieux et al., 2008). The ß-CARBA test (Biorad, Hercules, CA, United States) was performed to identify the strain with carbapenemase activity.
The DNA of enterobacterial isolates was extracted with the BioRobot EZ1 (Qiagen, Courtaboeuf, France) using a commercial EZ1 DNA extraction kit (Qiagen) and quantified by a Qubit assay (Life Technologies, Carlsbad, CA, United States). De novo sequencing was performed using MiSeq technology (Illumina Inc., San Diego, CA, United States) in a paired-end strategy. Libraries were prepared using the Nextera XR DNA sample prep kit (Illumina). Briefly, DNAs were fragmented and tagged with adapters and dual-index barcodes. Then, DNAs were purified using the AMPure XP beads (Beckman Coulter, Inc., Fullerton, CA, United States) and pooled in equimolar concentrations. An Illumina-generated PhiX control libraries was added to the libraries that was then 2 × 250 bp paired-end sequenced on an Illumina Miseq. Genomes have been deposited under the Bioproject number PRJNA738374 on the NCBI Database. The three K. pneumoniae strains (Q1947, Q1948, and Q1945) described previously (Baron et al., 2021) were deposited under genome accession numbers GCA_903166445.1, GCA_903166485.1, and GCA_903166415.1, respectively. They were then assembled using Spades (Galaxy Version 3.12.0 + galaxy1) (Bankevich et al., 2012), and annotated with Prokka (Galaxy Version 1.14.6 + galaxy1) (Seemann, 2014) using the Galaxy platform.1 The detection of AMR genes, plasmids and virulence genes was performed with Abricate (Galaxy Version 1.0.1) using Resfinder, Plasmidfinder and VFDB databases, respectively. The sequence type (ST) of the isolates was determined using the pubMLST2 and Pasteur Institute databases.3 Pangenome analysis was performed using the Roary software (version 3.13.0) while preserving the default settings (95%: the minimum percentage of identity and 99%: a gene needs to be in to be core). The analysis included 51 genomes of sub-Saharan African strains, isolated from human, animal and the environment, that were available in PATRIC database (version 3.6.12) (accession numbers and genome details are available in Supplementary Table 1). We then used Fast tree software (version 2.1.10) to build a maximum likelihood phylogenetic tree with predefined parameters. It was computed using iTOL.4 A core single nucleotide polymorphism (SNP) was performed using snippy (Galaxy Version 4.6.0 + galaxy0) for the mostly related strains. We sequenced the genomes of two strains, Q2160 (ST224) and Q2170 (ST10), using a MinION sequencer (Oxford Nanopore Technologies Inc., Oxford, United Kingdom) and assembled genomes using both Illumina and MinION reads to reconstitute the IncY plasmid and compare it to the transposon of the Q2170 isolate. Bacteriophages were detected using PHASTER (Arndt et al., 2016). The reconstitution of the genetic environment was performed using the Easyfig program (Sullivan et al., 2011).
This experiment was done using the Q2160 E. coli strain containing the IncY plasmid as a donor strain and an acid-resistant E. coli J53 (F- pro Azi r) as the receiver strain. The strains were enriched in TSB for 24 h then mixed in the same tube (1 ml of the donor strain in 9 ml of the receiver strain) for another 24 h. Transconjugants were selected on LB agar, supplemented by 120 μg/mL of sodium acid and 1 μg/mL of cefotaxime (Sennati et al., 2016).
From the 226 NHP samples, we isolated 617 bacteria including 590 Gram negative bacteria (GNB) and 27 Gram positive bacteria (GPB). The detail of the number of bacterial isolates on the different media is shown in Supplementary Table 2 and Figure 1. Among these GNB, 234 were Enterobacterales, of which 29 that are not naturally resistant to the antibiotics in consideration, were studied in this work. Briefly, two of these 29 Enterobacterales grew on MacConkey + ertapenem medium: one E. coli, and one Proteus sp. On MacConkey + cefotaxime media, we isolated 16 Enterobacterales (12 E. coli and four Morganella morganii). Finally, on LBJMR (Lucie Bardet Jean-Marc Rolain) medium, we isolated three E. coli and eight Enterobacter sp. (Three E. cloacae, three E. quasiroggenkampii, one E. asburiae, and one E. bugandensis) (Supplementary Tables 3, 4).
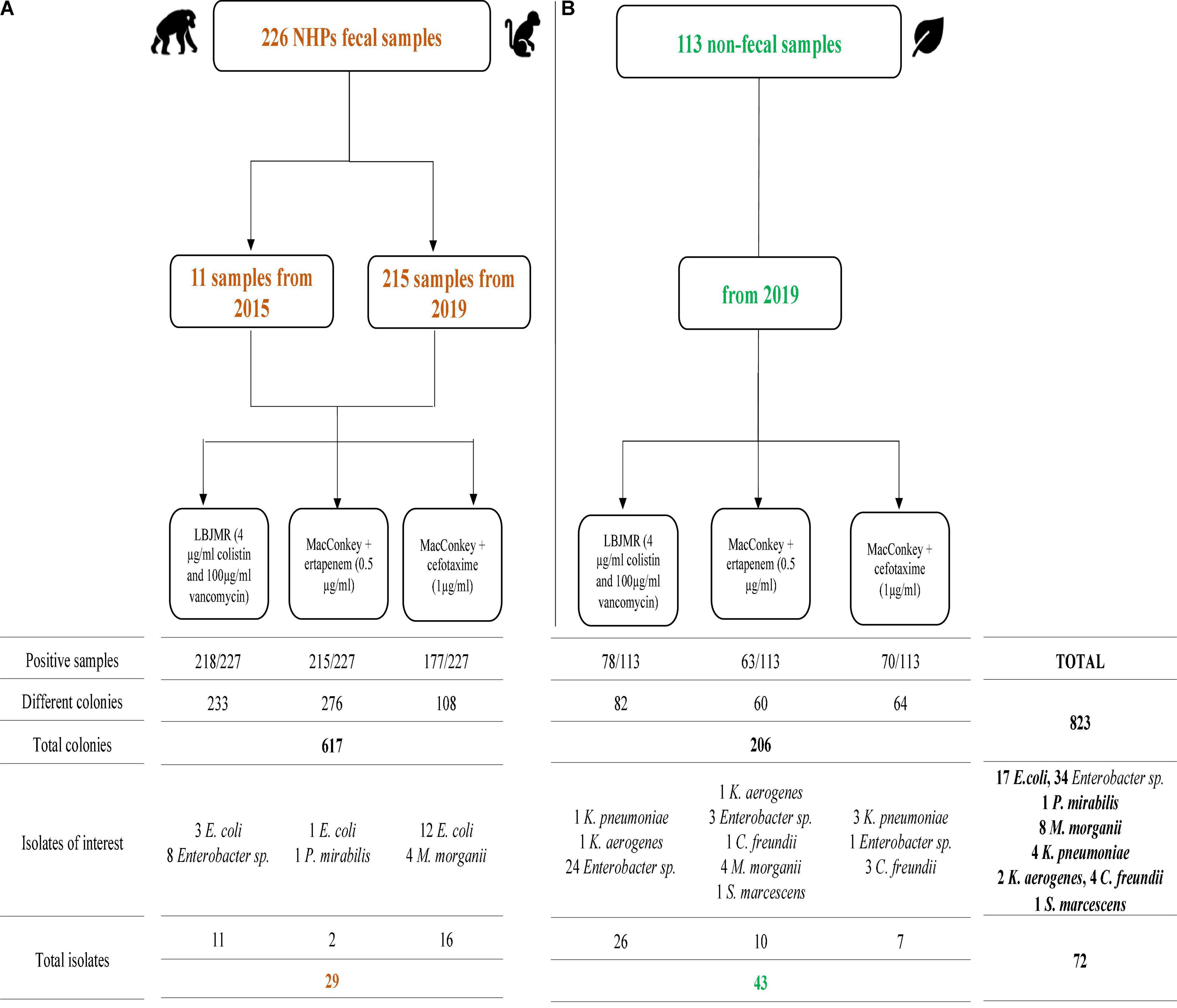
Figure 1. Summary of positive non-human primates (NHPs) fecal samples (A) and non-fecal environmental samples (B) cultures on different selective media. Isolates of interest are those that were sequenced.
From the 113 non-fecal environmental samples, we isolated 206 bacteria including 201 GNB and 5 GPB. The detail of the number of bacterial isolates on the different media is shown in Supplementary Table 5 and Figure 1. Among the GNB, 84 were Enterobacterales, of which 43 non-naturally resistant were studied in this work. Briefly, ten Enterobacterales grew on MacConkey + ertapenem, including four M. morganii, three Enterobacter sp. (one E. quasiroggenkampii, one E. cloacae, and one E. hormaechei), one Citrobacter cronae, one Klebsiella aerogenes, and one Serratia marcescens. On MacConkey + cefotaxime, we isolated three K. pneumoniae, two C. cronae, and two Enterobacter sp. (one E. quasiroggenkampii and one E. hormaechei). Finally, on LBJMR, 25 Enterobacter sp. (nine E. cloacae, eight E. quasiroggenkampii, four E. bugandensis, two E. roggenkampii, and two E. sichuanensis), one K. aerogenes and one K. pneumoniae were isolated. Identical strains from the same origin are reported once. It should be noted that culture results from non-fecal environmental samples on MacConkey + ertapenem, MacConkey + cefotaxime, and MacConkey media alone have previously been described (Baron et al., 2021), but the strains of interest were sequenced for this study. Specific and common species between samples were represented in Figure 2.
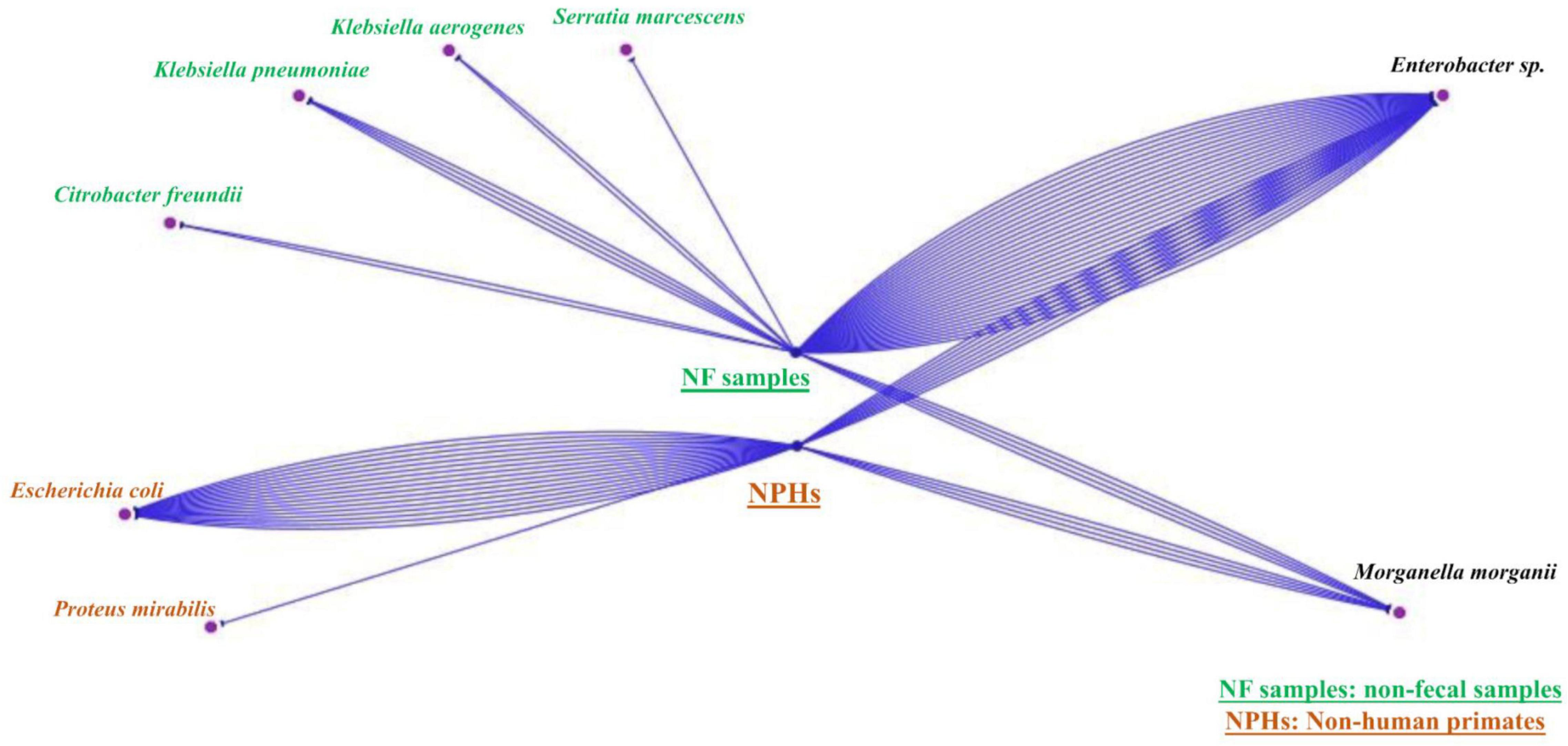
Figure 2. Simple view of the Cytoscape visualizing unique and common species of the two samples’ types (NHPs and non-feacal samples). The blue lines represent the number of strains found each type. There are strains that belong just to non-fecal environmental samples, others that belong just to NHPs and strains that can be found in both of them.
A total of 72 Enterobacterales were isolated from NHPs and non-fecal environmental samples. We confirmed the phenotypic resistance of these isolates by antibiotic susceptibility tests (AST) (Supplementary Table 4). Sixteen isolates were confirmed to be resistant to cefotaxime (N = 16/339 samples, 5%), two resistant to ertapenem (N = 2/340 samples, 0.6%), and 37 resistant to colistin (N = 37/339 samples, 11%). The ß-carba-test was negative for the two ertapenem-resistant isolates. These antibiotic-resistant isolates were isolated from different NHP species and the environment. In chimpanzees, we isolated one ESBL-producing E. coli and six colistin-resistant Enterobacter sp. In green monkeys (Chlorocebus sabaeus), we identified seven E. coli and one Morganella morganii which produced ESBL, and one colistin-resistant Enterobacter cloacae. Four ESBL-producing and two colistin-resistant E. coli as well as one Enterobacter asburiae were found in Guinea baboons (Papio papio). From the non-fecal environmental samples, we isolated two ertapenem-resistant M. morganii, 27 colistin-resistant Enterobacterales including 25 Enterobacter sp., one K. pneumoniae and one K. aerogenes and three ESBL-producing K. pneumoniae.
Of these 72 Enterobacterales, 23 had a multidrug resistant profile (MDR) (Magiorakos et al., 2012) and were resistant to four or more antibiotic families, 46 to fewer than four antibiotic families, and three had no resistance. Resistance to third generation cephalosporins (3GC) was 29% (N = 21/72) whereas colistin resistance represented 65% (47/72) of the total strains. 3GC-resistant isolates, had co-resistance with tetracycline family (52%, N = 11/21) (doxycycline), fluoroquinolone (43%, N = 9) (ciprofloxacin) and trimethoprim-sulfamethoxazole (62%, N = 13). Resistance to amoxicillin was found in 75% of the E. coli strains (N = 12/16).
We sequenced the genome of the 72 Enterobacterales and studied clonal population and AMR genes. Details of the assembly results are provided in Supplementary Table 4. The population distribution of Enterobacterales was mainly represented by new sequence types (STs), indicating the existence of a population specific to the ecosystem analyzed but we found clonal complexes that are associated with human infections for E. coli and K. pneumoniae. Of the 16 E. coli, six belonged to ST10, two to ST224, and nine to other unique STs (ST212, ST202, ST196, ST469, ST2803, ST642, ST10648, ST3580, and ST6611). For K. pneumoniae, three belonged to ST307 and had previously been described (Baron et al., 2021), but one had a new ST5460. Of the 37 Enterobacter sp. sequenced, only three strains belonged to a known ST (ST1084- Q3805, ST113- Q2141, and ST565-Q2153), while the remaining 34 isolates belonged to 29 new different STs. Interestingly, one group was containing strains belonged to the chimpanzee and non-fecal environmental samples. It consisted of 11 STs (ST1545, ST1547, ST1548, ST1549, ST1552, ST1554, ST1563, ST1565, ST1566, ST1567, and ST1570) combining strains of the environment origin (N = 7) and strains from chimpanzees (N = 4). Finally, two K. aerogenes and two C. cronae from the non-fecal environmental samples belonged to new STs ST225 and ST226 for K. aerogenes and ST574 and ST575 for C. cronae (Supplementary Table 4).
We noticed that 53 of the 72 Enterobacterales had four or more resistance genes, some had the same association of genes, such as E. coli and K. pneumoniae (Supplementary Table 4 and Table 1). We found a CTX-M-15 type β-lactamases in 12 E. coli from NHPs, three ST307 K. pneumoniae isolated from the environment (Figure 3). Four of the 12 E. coli (2 ST224, 1 ST202, and ST469) harbored an IncY plasmid of 91,959 bp length (GC% = 52.06%) carrying resistance genes to beta-lactam (blaCTX–M–15, blaTEM1B), sulfonamide (sul2, dfrA14), fluroquinolones (qnrs), tetracycline (tetA, tetR) and aminosides [aph(3′′), aph(6)]. The plasmid IncY was non conjugative, did not have a complete conjugative apparatus, and conjugation to an E. coli J53 failed on three attempts. It was found in three green monkey and one baboon samples (Figure 3). A Tn2 transposon of 30,340 bp (GC% = 52.05%) containing the same AMR genes found on the IncY plasmid [blaCTX–M–15, blaTEM1B, sul2, dfrA14, qnrs1, aph(3′′), aph(6), tetA, and tetR] was observed in the chromosome of four of the six ST10 isolates (Q2156, Q2170, Q2165, and Q2157). These four strains differed from 1 to 14 SNPs in their core genome suggesting they had the same origin (Figure 3). Interestingly, this transposon was similar to that present in the plasmid IncY (Figure 4) but was inserted in a SfII-like prophage (KC736978) of 35.5 bp (GC% = 50.23%) in the chromosome of the ST10 isolate (Q2170). On the 5′–3′ locus, an insertion sequence (IS)26 inserted in an integrase (INT) of the phage, causing a deletion of 8 bp in the integrase sequence, shifting the reading frame and leading to the synthesis of 187 amino acid residues of the integrase protein against 216 amino acid residues in the reference phage. On the 3′–5′ locus, the IS26 inserted before the tail fiber assembly protein (TfaE), leading to the truncation of a hypothetical protein located between the integrase (INT) and the TfaE protein. Interestingly, the intact SFII-like prophage was found in a β-lactamases susceptible E. coli isolate (Q3820) that was only resistant to colistin (Figure 4). Finally, the four other E. coli isolates carrying CTX-M-15 enzymes belonged to different STs [two ST10 (Q2157, Q2164), one ST3580 (Q2155), and one ST212 (Q3821)], had no IncY plasmid and did not have the same transposon. Three of them carried one to five plasmids (IncF, Col, and IncB plasmids, details in Supplementary Table 4), but the location of blaCTX–M–15 on the plasmids could not be identified in these genomes. The conjugation experiment showed negative results as no growth has been detected for a J53 E. coli receiving the cefotaxime resistant gene.
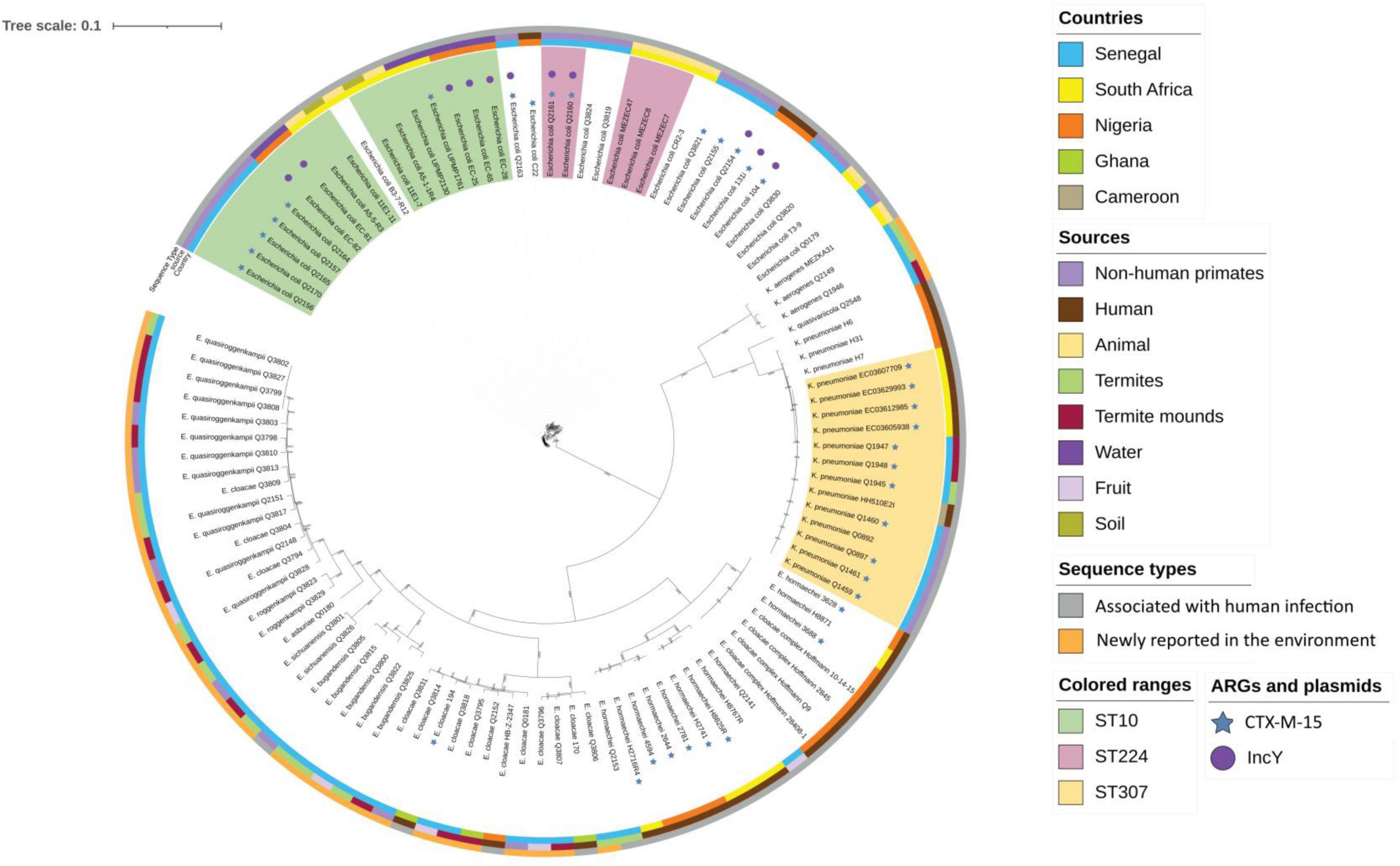
Figure 3. Maximum likelihood phylogenetic tree representation of the 9 Klebsiella spp., 16 E. coli, and 37 Enterobacter spp. strains compared with strains from different origin of the Sub-Saharan region. The tree was generated and annotated with the iTOL tool (https://itol.embl.de/tree/).
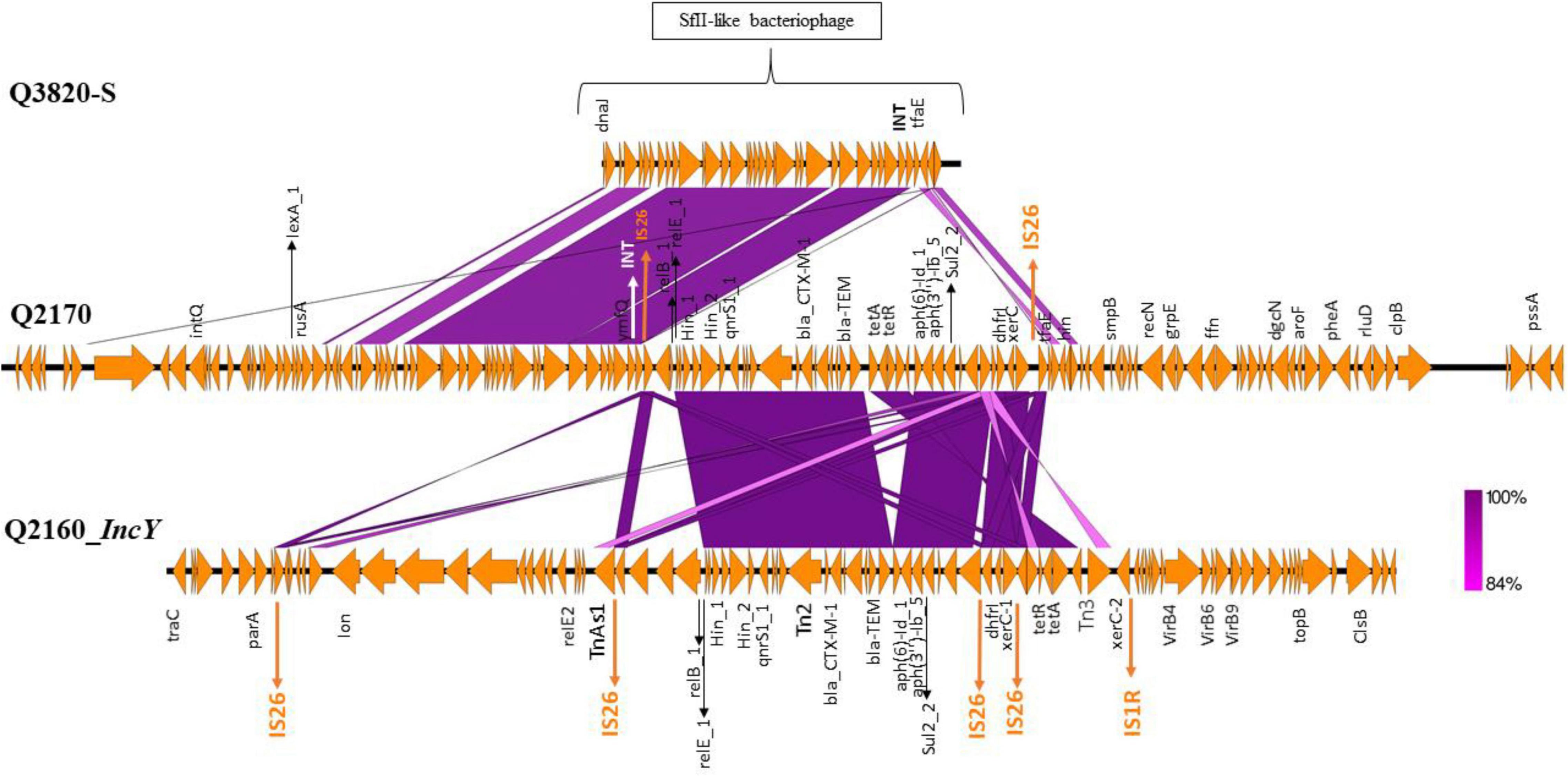
Figure 4. Genomic comparison of the SFII-like bacteriophage region of the 3GC-susceptible Escherichia coli strain (Q3820), the transposon integrated in the SFII-like prophage region of the ST10 3GC-resistant E. coli(Q2170) and the IncY plasmid sequence harboring the same transposon of the ST224 E. coli (Q2160). This comparison shows the putative transfer of resistance genes between these two strains. The figure was made using the Easyfig program.
Regarding other Enterobacterales, three C. cronae genomes harbored a fluroquinolone-resistance gene (qnrB12) as well as the chromosomal beta-lactamase blaCMY –98_1. In the 37 Enterobacter spp., we identified four different chromosomal ampC genes, namely blaCMH–3–1 (N = 10), blaACT–6–1 (N = 10), blaCMG–1 (N = 14), and blaMIR–1 (N = 2). None of these strains had an inducible expression of their AmpC β-lactamase with a phenotypic resistance to 3 GC. In the eight M. morganii genomes isolated from green monkeys and non-fecal environmental samples, we found the ampC β-lactamase (blaMOR–2, blaDHA12), conferring resistance to 3 GC. The summary of antibiotic resistance genes in these strains is presented in Table 1. The blaCMG gene shows 98.34% similarity with blaACT–62 suggesting that they could be allelic variants of the same AmpC gene.
We compared the genomes of E. coli, K. pneumonia, and Enterobacter spp. isolated in our work with 51 genomes from different origin isolated in sub-Saharan Africa (Figure 3). Enterobacter isolates clustered together, far from the human isolates which were consistent with the ST results. E. coli and K. pneumoniae isolates were more related with human isolates but their core genome differed by more than 20,000 SNPs suggesting there were not directly related. We have also included in this analysis two strains of E. coli (131i and 104) that were isolated from animals use for human consumption in Nigeria (Sharma et al., 2020) that were interesting because they carried an IncY plasmid, the antibiotic resistance genes included in the transposon [blaCTX–M–15, blaTEM1B, sul2, dfrA14, qnrs1, aph(3′′), aph(6), tetA and tetR] and a SfII-like prophage. However, these isolates did not cluster with isolates that contained the transposon (Q2154, Q2156, Q2160, Q2161, Q2163, Q2165, Q2157, and Q2170). Unfortunately, we were not able to find the antibiotic resistance genes location in these two isolates.
Microbial diversity in natural environments is considered to be the largest unexplored reservoir of biodiversity on earth. Many studies have described bacterial diversity in primates, but few of them have focused on their genomic characteristics. This explains the large number of new STs of Enterobacter sp., C. cronae, and K. aerogenes found in our study. E. coli [ST10 (Mohsin et al., 2017) and ST224 (Cao et al., 2014)] and K. pneumoniae [ST307 and ST147 (Peirano et al., 2020)] found in our previous study (Baron et al., 2021) are reported to cause human infections. Although our newly described STs have not yet been found in humans and transmission cannot be excluded.
Interestingly, in our study, 11 green monkeys, three baboons and one chimpanzee carried an Enterobacterales with a MDR profile, often an ESBL profile. This MDR profile was also found in the Enterobacterales of nine non-fecal environmental samples showing resistance to 3GC, tetracycline and fluoroquinolones. Moreover, the rate of resistance was relatively high for a cohort far from massive antibiotic exposure, especially for colistin but also for 3GC (29%, N = 21/72). Moreover, 18% (N = 13/72) of the isolated strains were resistant to tetracycline and 18% (N = 13/72) were resistant to sulphonamides. According to the WHO report published in 2014 highlighting resistance levels to the most widely-used antibiotics, fluoroquinolones and 3GC antibiotics are known to be less used in Africa, where the resistance rate is lower than in Europe (Bassoum et al., 2019). Our results show that 13% (N = 9/72) of the isolates were fluoroquinolone-resistant and 29% (21/72) were C3G-resistant. These phenotypic results were confirmed genotypically, where AMR genes were found to be consistent with isolate antibiotic resistance. 3GC-resistance is often co-presented in this study with tetracycline, fluoroquinolone and sulphonamide resistance, conferring a MDR profile for the strain, which was demonstrated in the IncY plasmid that carries all the AMR genes conferring resistance to these antibiotics at the same time. In contrast, mobile colistin resistance (mcr) genes were not found and further analysis will be needed in order to decipher the resistance mechanisms of Enterobacter sp.
The IncY plasmid has been found in the environment (Ma et al., 2021), including in water samples (Moremi et al., 2016) and in animals such as camels in Tunisia (Saidani et al., 2019). Although it could be mobilizable in silico because of the current flanking sequence surrounding this DNA element, the conjugation experimental attempt to mobilize it has failed. This plasmid is often associated with ESBL-resistance genes, especially blaCTX–M–15 (Rasheed et al., 2020). Here, AMR genes clustered in this plasmid within a transposon [blaCTX–M–15, blaTEM1B, sul2, dfrA14, qnrs1_1, aph(3′′), aph(6), tetA, and tetR] in four E. coli strains, and were surrounded by IS26 from each side. Interestingly, we showed that this transposon was inserted in a SfII-like phage on the chromosome of a ST10 isolate (Figure 4) leading to nucleotides deletion that modified the prophage integrase (INT) amino-acid sequence from a side and truncated a hypothetical protein from the other side. Phages are believed to play a critical role in the horizontal transfer of bacterial genes, including antibiotic resistance genes (Kondo et al., 2021). The SfII phage is known to play a role in virulence spread (Mavris et al., 1997). This finding demonstrates that intermediate steps between transposons and the genomes of the phage can also be found and the recombination site of a prophage is vulnerable and can receive mobile elements (Brown-Jaque et al., 2015). Some studies have shown the integration of the transposon next to the recombination site of a phage (Huang et al., 2017) but none has shown the insertion of the transposon inside the phage to our knowledge. We were interested in the two E. coli strains that were isolated from poultry in Nigeria (Sharma et al., 2020) and contained the same phage, plasmid and antibiotic resistance genes that our E. coli isolates but we were unable to verify if they have the same transposon and its location. These findings, however, suggest a circulation of these mobile elements in bacteria from animals in Africa.
Antimicrobial resistant in humans is a current issue linked to the overuse of antibiotics in clinical, veterinary and agricultural practices, which has been considered to be the main selective pressure for AMR strains and AMR gene dissemination since the 1950s (Thaller et al., 2010). However, the literature is rich in cases reporting the emergence of AMR in the absence of antibiotic use, as is the case for colistin resistance (Olaitan et al., 2016). Other factors, such as anthropological and socioeconomic factors (Collignon et al., 2018) and cross-resistance with other antibiotics may be involved (Napier et al., 2013). The “One Health” approach has emerged several years ago (Thamlikitkul et al., 2015), and puts the issue of AMR in a more global context. This approach has already been well studied for farm animals but remains poorly studied for wild animals (Diallo et al., 2020). The study of NHPs is even more interesting since they are phylogenetically close to humans and may be colonized or infected by species that are close or identical to those that are pathogenic to humans (Wolfe et al., 1998). Since NHPs live in areas with different protection status and human impact, they are a good model for studying the phenomenon of AMR, especially since they also live in groups and display behaviors similar to our own.
Our study shows that AMR can be found in wildlife and that this emergence, in places where human selection and pressure is reduced, is a public health concern. The wild environment is not well studied and hosts a large, undescribed microbial diversity. Transfer of several AMR genes via transposon in plasmids and via integrated prophages should be further explored to understand this dynamic in natural environments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the Direction des Parcs Nationaux and Direction des Eaux, Forêts, Chasse et de la Conservation des Sols of the Senegal Ministry for the Environment and Sustainable Development.
SB, J-MR, and DR designed the study, drafted, and revised the manuscript. OM, BD, AB, GD, and CS collected the samples. RA drafted the manuscript. RA, EK, LH, and AI performed microbiology analyses. RH-A facilitated sample collection and revised the manuscript. All authors have read and approved the final manuscript.
This work was supported by the French Government under the “Investissements d’Avenir” (Investments for the Future) programme managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03) and by the IHU Mediterranean Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Mouna Hamel for her committed assistance in the laboratory. We are grateful to the Direction des Parcs Nationaux and Direction des Eaux, Forêts, Chasse et de la Conservation des Sols of the Senegal Ministry for the Environment and Sustainable Development for their support. We also thank the MLST and Pasteur Institute databases for helping us submit the new ST profiles. We thank Mamadou Samba Sylla from the Jane Goodall Institute Spain and Senegal for help with sample collection.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.838392/full#supplementary-material
Supplementary Tables 1 | List of reference bacterial genomes used for phylogenetic comparison (Figure 3).
Supplementary Table 2 | List of bacterial isolates in NHPs fecal samples on different media.
Supplementary Table 3 | Summary of the Enterobacterales isolated on selective media (Source, date and localisation were presented).
Supplementary Table 4 | Characteristics summary of Enterobacterales isolated on selective media. Antibiotic susceptibility tests to provide resistance profile and genomic analysis results (antibiotic resistance genes, plasmid and virulence genes as well as allelic profile) were presented.
Supplementary Table 5 | List of bacterial isolates contained in non-fecal environmental samples on different media.
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bardet, L., Le Page, S., Leangapichart, T., and Rolain, J.-M. (2017). LBJMR medium: a new polyvalent culture medium for isolating and selecting vancomycin and colistin-resistant bacteria. BMC Microbiol. 17:220. doi: 10.1186/s12866-017-1128-x
Baron, S. A., Mediannikov, O., Abdallah, R., Kuete Yimagou, E., Medkour, H., Dubourg, G., et al. (2021). Multidrug-resistant Klebsiella pneumoniae clones from wild chimpanzees and termites in senegal. Antimicrob. Agents Chemother. 65:e0255720. doi: 10.1128/AAC.02557-20
Bassoum, O., Lèye, M. M. M., Sougou, N. M., Diongue, M., Niang, K., Tine, J. A. D., et al. (2019). Practices about antibiotic use among urban residents: a cross-sectional survey in rufisque, Senegal. Central Afr. J. Public Health 5:1. doi: 10.11648/j.cajph.20190501.11
Bernabé, K. J., Langendorf, C., Ford, N., Ronat, J.-B., and Murphy, R. A. (2017). Antimicrobial resistance in West Africa: a systematic review and meta-analysis. Int. J. Antimicrob. Agents 50, 629–639. doi: 10.1016/j.ijantimicag.2017.07.002
Breurec, S., Bouchiat, C., Sire, J.-M., Moquet, O., Bercion, R., Cisse, M. F., et al. (2016). High third-generation cephalosporin resistant Enterobacteriaceae prevalence rate among neonatal infections in Dakar, Senegal. BMC Infect. Dis. 16:587. doi: 10.1186/s12879-016-1935-y
Brown-Jaque, M., Calero-Cáceres, W., and Muniesa, M. (2015). Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid 79, 1–7. doi: 10.1016/j.plasmid.2015.01.001
Cao, X., Zhang, Z., Shen, H., Ning, M., Chen, J., Wei, H., et al. (2014). Genotypic characteristics of multidrug-resistant Escherichia coli isolates associated with urinary tract infections. APMIS 122, 1088–1095. doi: 10.1111/apm.12260
Chereau, F., Herindrainy, P., Garin, B., Huynh, B.-T., Randrianirina, F., Padget, M., et al. (2015). Colonization of extended-spectrum-β-lactamase- and NDM-1-Producing Enterobacteriaceae among Pregnant Women in the Community in a Low-Income Country: a potential reservoir for transmission of multiresistant Enterobacteriaceae to Neonates. Antimicrob. Agents Chemother. 59, 3652–3655. doi: 10.1128/AAC.00029-15
Chokshi, A., Sifri, Z., Cennimo, D., and Horng, H. (2019). Global contributors to antibiotic resistance. J. Glob. Infect. Dis. 11, 36–42. doi: 10.4103/jgid.jgid_110_18
Collignon, P., Beggs, J. J., Walsh, T. R., Gandra, S., and Laxminarayan, R. (2018). Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet. Health 2:e398–e405. doi: 10.1016/S2542-5196(18)30186-4
Diallo, O. O., Baron, S. A., Abat, C., Colson, P., Chaudet, H., and Rolain, J.-M. (2020). Antibiotic resistance surveillance systems: a review. J. Glob. Antimicrob. Resist. 23, 430–438. doi: 10.1016/j.jgar.2020.10.009
Drieux, L., Brossier, F., Sougakoff, W., and Jarlier, V. (2008). Phenotypic detection of extended-spectrum β-lactamase production in Enterobacteriaceae: review and bench guide. Clin. Microbiol. Infect. 14, 90–103. doi: 10.1111/j.1469-0691.2007.01846.x
Dromigny, J. A., Nabeth, P., Juergens-Behr, A., and Perrier-Gros-Claude, J. D. (2005). Risk factors for antibiotic-resistant Escherichia coli isolated from community-acquired urinary tract infections in Dakar, Senegal. J. Antimicrob. Chemother. 56, 236–239. doi: 10.1093/jac/dki158
Huang, W., Wang, G., Sebra, R., Zhuge, J., Yin, C., Aguero-Rosenfeld, M. E., et al. (2017). Emergence and Evolution of multidrug-resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M Integrated in the Chromosome. J. Antimicrob. Chemother. 61:e00076-17. doi: 10.1128/AAC.00076-17
Kondo, K., Kawano, M., and Sugai, M. (2021). Distribution of antimicrobial resistance and virulence genes within the prophage-associated regions in nosocomial pathogens. mSphere 6:e00452-21. doi: 10.1128/mSphere.00452-21
Ma, Y., Chen, J., Fong, K., Nadya, S., Allen, K., Laing, C., et al. (2021). Antibiotic Resistance in Shiga Toxigenic Escherichia coli Isolates from Surface Waters and Sediments in a Mixed Use Urban Agricultural Landscape. Antibiotics (Basel) 10:237. doi: 10.3390/antibiotics10030237
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Mavris, M., Manning, P. A., and Morona, R. (1997). Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol. Microbiol. 26, 939–950. doi: 10.1046/j.1365-2958.1997.6301997.x
McEwen, S. A., and Collignon, P. J. (2018). Antimicrobial resistance: a one health perspective. Microbiol. Spectr. 6:2017. doi: 10.1128/microbiolspec.ARBA-0009-2017
Miller, E. A., Ponder, J. B., Willette, M., Johnson, T. J., and VanderWaal, K. L. (2020). Merging metagenomics and spatial epidemiology to understand the distribution of antimicrobial resistance genes from Enterobacteriaceae in wild owls. Appl. Environ. Microbiol. 86:e00571-20. doi: 10.1128/AEM.00571-20
Mohsin, J., Pál, T., Petersen, J. E., Darwish, D., Ghazawi, A., Ashraf, T., et al. (2017). Plasmid-mediated colistin resistance gene mcr-1 in an Escherichia coli ST10 bloodstream isolate in the sultanate of Oman. Microbial. Drug Resist. 24, 278–282. doi: 10.1089/mdr.2017.0131
Moremi, N., Manda, E. V., Falgenhauer, L., Ghosh, H., Imirzalioglu, C., Matee, M., et al. (2016). Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front. Microbiol. 7:1862. doi: 10.3389/fmicb.2016.01862
Napier, B. A., Burd, E. M., Satola, S. W., Cagle, S. M., Ray, S. M., McGann, P., et al. (2013). Clinical use of colistin induces cross-resistance to host antimicrobials in Acinetobacter baumannii. mBio 4:e00021-13. doi: 10.1128/mBio.00021-13
Olaitan, A. O., Morand, S., and Rolain, J.-M. (2016). Emergence of colistin-resistant bacteria in humans without colistin usage: a new worry and cause for vigilance. Int. J. Antimicrob. Agents 47, 1–3. doi: 10.1016/j.ijantimicag.2015.11.009
Peirano, G., Chen, L., Kreiswirth, B. N., and Pitout, J. D. D. (2020). Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob. Agents Chemother. 64:e01148-20. doi: 10.1128/AAC.01148-20
Ramey, A. M., and Ahlstrom, C. A. (2019). Antibiotic resistant bacteria in wildlife: perspectives on trends, acquisition and dissemination, data gaps, and future directions. J. Wildlife Dis. 56, 1–15. doi: 10.7589/2019-04-099
Rasheed, F., Saeed, M., Alikhan, N.-F., Baker, D., Khurshid, M., Ainsworth, E. V., et al. (2020). Emergence of Resistance to fluoroquinolones and third-generation cephalosporins in Salmonella Typhi in Lahore, Pakistan. Microorganisms 8:1336. doi: 10.3390/microorganisms8091336
Saidani, M., Messadi, L., Mefteh, J., Chaouechi, A., Soudani, A., Selmi, R., et al. (2019). Various Inc-type plasmids and lineages of Escherichia coli and Klebsiella pneumoniae spreading blaCTX-M-15, blaCTX-M-1 and mcr-1 genes in camels in Tunisia. J. Glob. Antimicrob. Resist. 19, 280–283. doi: 10.1016/j.jgar.2019.05.007
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seng, P., Drancourt, M., Gouriet, F., La Scola, B., Fournier, P.-E., Rolain, J. M., et al. (2009). Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 49, 543–551. doi: 10.1086/600885
Sennati, S., Riccobono, E., Di Pilato, V., Villagran, A. L., Pallecchi, L., Bartoloni, A., et al. (2016). pHN7A8-related multiresistance plasmids (blaCTX-M-65, fosA3 and rmtB) detected in clinical isolates of Klebsiella pneumoniae from Bolivia: intercontinental plasmid dissemination? J. Antimicrob. Chemother. 71, 1732–1734. doi: 10.1093/jac/dkv506
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, P., Gupta, S. K., Adenipekun, E. O., Barrett, J. B., Hiott, L. M., Woodley, T. A., et al. (2020). Genome analysis of multidrug-resistant Escherichia coli isolated from poultry in Nigeria. Foodborne Pathog. Dis. 17, 1–7. doi: 10.1089/fpd.2019.2659
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Thaller, M. C., Migliore, L., Marquez, C., Tapia, W., Cedeño, V., Rossolini, G. M., et al. (2010). Tracking acquired antibiotic resistance in commensal bacteria of galápagos land iguanas: no man, no resistance. PLoS One 5:e8989. doi: 10.1371/journal.pone.0008989
Thamlikitkul, V., Rattanaumpawan, P., Boonyasiri, A., Pumsuwan, V., Judaeng, T., Tiengrim, S., et al. (2015). Thailand antimicrobial resistance containment and prevention program. J. Glob. Antimicrob. Resist. 3, 290–294. doi: 10.1016/j.jgar.2015.09.003
Vounba, P., Arsenault, J., Bada-Alambédji, R., and Fairbrother, J. M. (2019). Prevalence of antimicrobial resistance and potential pathogenicity, and possible spread of third generation cephalosporin resistance, in Escherichia coli isolated from healthy chicken farms in the region of Dakar, Senegal. PLoS One 14:e0214304. doi: 10.1371/journal.pone.0214304
Wolfe, N. D., Escalante, A. A., Karesh, W. B., Kilbourn, A., Spielman, A., and Lal, A. A. (1998). Wild primate populations in emerging infectious disease research: the missing link? Emerg. Infect. Dis. J. 4, 149–158. doi: 10.3201/eid0402.980202
Woolhouse, M., Ward, M., van Bunnik, B., and Farrar, J. (2015). Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:20140083. doi: 10.1098/rstb.2014.0083
Keywords: antibiotic resistance, Enterobacterales, primates, environment, Senegal
Citation: Abdallah R, Kuete Yimagou E, Hadjadj L, Mediannikov O, Ibrahim A, Davoust B, Barciela A, Hernandez-Aguilar RA, Diatta G, Sokhna C, Raoult D, Rolain JM and Baron SA (2022) Population Diversity of Antibiotic Resistant Enterobacterales in Samples From Wildlife Origin in Senegal: Identification of a Multidrug Resistance Transposon Carrying blaCTX–M–15 in Escherichia coli. Front. Microbiol. 13:838392. doi: 10.3389/fmicb.2022.838392
Received: 17 December 2021; Accepted: 18 February 2022;
Published: 18 March 2022.
Edited by:
Teresa M. Coque, Ramón y Cajal Institute for Health Research, SpainReviewed by:
Angela Novais, University of Porto, PortugalCopyright © 2022 Abdallah, Kuete Yimagou, Hadjadj, Mediannikov, Ibrahim, Davoust, Barciela, Hernandez-Aguilar, Diatta, Sokhna, Raoult, Rolain and Baron. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophie Alexandra Baron, c29waGllLmJhcm9uQGFwLWhtLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.