
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 08 February 2022
Sec. Microbiotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.835131
This article is part of the Research Topic Promising and sustainable microbial feedstocks for biotechnological processes View all 7 articles
Corynebacterium glutamicum is used for the million-ton-scale production of amino acids. Valorization of sidestreams from agri- and aqua-culture has focused on the production of biofuels and carboxylic acids. Nitrogen present in various amounts in sidestreams may be valuable for the production of amines, amino acids and other nitrogenous compounds. Metabolic engineering of C. glutamicum for valorization of agri- and aqua-culture sidestreams addresses to bridge this gap. The product portfolio accessible via C. glutamicum fermentation primarily features amino acids and diamines for large-volume markets in addition to various specialty amines. On the one hand, this review covers metabolic engineering of C. glutamicum to efficiently utilize components of various sidestreams. On the other hand, examples of the design and implementation of synthetic pathways not present in native metabolism to produce sought after nitrogenous compounds will be provided. Perspectives and challenges of this concept will be discussed.
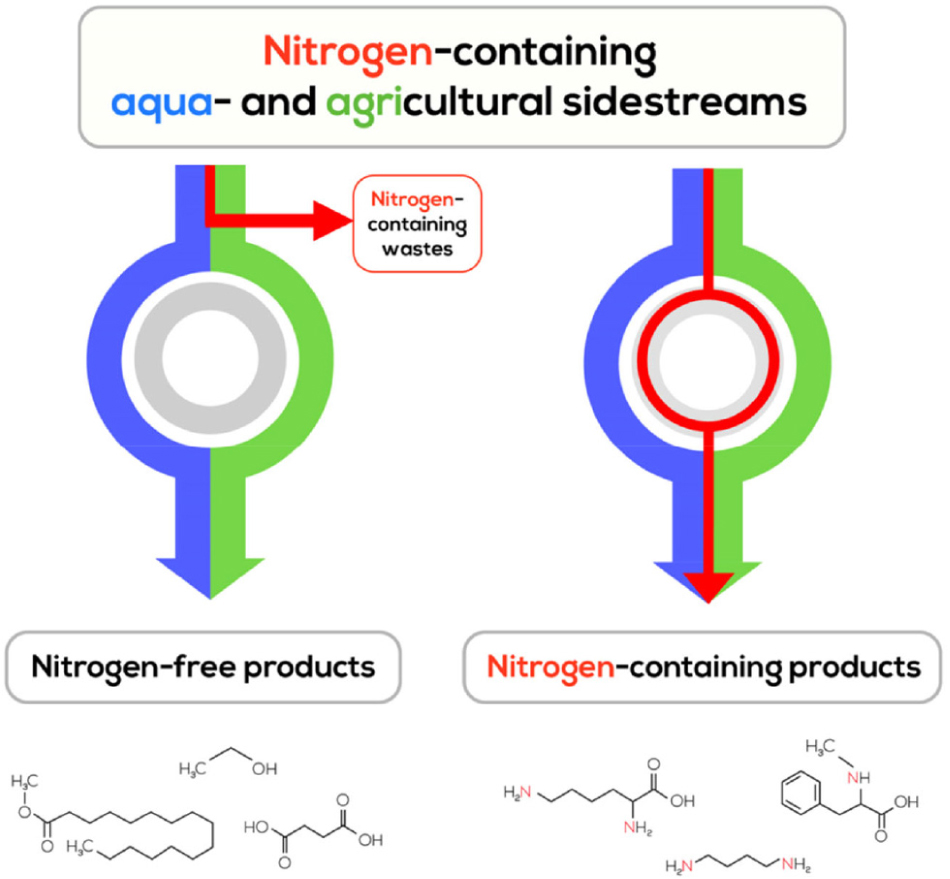
Graphical Abstract. Strategy to valorize non-utilized nitrogen from sidestreams by producing nitrogen-containing products.
In this review, we focus on metabolic engineering of Corynebacterium glutamicum for production of nitrogenous compounds from agri- and aqua-culture sidestreams. To achieve a circular bioeconomy, we need to manage renewable bioresources, reduce dependence on fossil-based and other non-sustainable resources, and avoid food- and feed-competitive raw materials. In that sense, sidestreams from agri- and aqua-culture are valuable resources and a number of approaches for their use have been reviewed previously, e.g., regarding lignin valorization (Wendisch et al., 2018), lignocellulosic feedstocks (Chen et al., 2019) or food wastes (Lee and Wendisch, 2017b). Metabolic engineering and enzyme screening and engineering were instrumental to achieve efficient bioprocesses based on sidestreams or their constituents (Lee and Wendisch, 2017b). The microbes utilizing these sidestreams produce a variety of products. Hitherto, the product scope was narrow, focusing on specific compounds such as organic acids to chemically produce polyesters or alcohols for use as biofuels (Vivek et al., 2021). Strikingly, most of the target compounds lack nitrogen although the aqua- and agricultural sidestreams typically contain a nitrogenous fraction. Thus, the production of nitrogenous value-added compounds from these sidestreams is highly relevant to make use of the full potential of biorefineries in the circular bioeconomy concept.
In this regard, the biotechnological workhorse C. glutamicum is relevant as it is used for the million-ton-scale production of amino acids (Wendisch, 2020) and it has been engineered for production of other nitrogenous compounds such as diamines for large-volume markets in addition to various specialty amines (Mindt et al., 2020). Moreover, a flexible feedstock concept has been realized for this bacterium enabling it to efficiently use various substrates present in aqua- and agri-sidestreams or their respective hydrolyzates. Recent developments, perspectives and challenges of production of nitrogenous compounds from agri- and aqua-culture sidestreams by metabolically engineered C. glutamicum will be discussed.
Biorefineries typically target to production of nitrogen-free value-added products from agri- and aqua-culture sidestreams, but do not make full use of the nitrogen fraction. Wastes from food in general has a distinctive configuration of nearly 30–60 wt% starch, 10–40 wt% lipids, and 5–10 wt% protein (Pleissner and Lin, 2013). Huge amount of protein, from plant and animal derivation, could be a sensuous attractive raw material for the value addition and it has to be better explored for various bioprocessing. Plant and animal parts which were uneatable were removed during the process of harvesting and post harvesting and other nutrient-rich wastes, such as manure and dead-stock all contain high levels of recoverable protein and currently, only a portion of it finds repeated use as animal feeds or animal feed constituent.
Even though the consumable portion of these protein-rich by-products could be used for recuperation of essential amino acids for human consumption, or as is for use in animal feeds, higher value solicitation for inedible and non-essential amino acids can be utilized as a feedstock for protein-based materials and other platform chemicals. Apart from the more specific uses of protein hydrolyzates, the agro-industrial by-products also find value addition through use in biological processes. Many of such products could be ideal media for several microorganisms competent of producing a diversification of bioactive metabolites, antimicrobial compounds and enzymes.
Among the plant-derived additives, grains, such as maize, barley, sorghum, oats, and wheat, and these grain by-products include corn gluten meal, brewers and distiller’s grains, malt sprouts, brewer’s yeast, and wheat mill feed represent a considerable portion (Naik et al., 2010). The inedible and non-essential amino acids obtained from these by-products can supply a feedstock for protein-based plastics, biopesticides or commodity organic compounds (Scott et al., 2007). Similarly, oil production by-products such as oil meals and press cakes from processing oilseeds, like soybean, canola, sunflower seed, linseed, palm kernel and others, are also notable feed ingredients. Some of these oil seeds have high crude protein content such as soybean meal (44.4–53.8%), cotton scotton seed meal (34–44%), castor seed cake (31–36%), canola seed meal (35–37%), pea nut meal (50–51%), etc. Further, various nuts, seeds, and their by-products, such as hulls and seed screenings, legume by-products, such as bean straw meal and hulls also contain high levels of protein (∼20–30%). Hicks and Verbeek (2017) broadly reviewed the typical protein content and global production of some of the protein meals produced from the agricultural industry. Dried ruminant waste (manure), dried poultry waste, dried poultry litter, dried swine waste, undried processed animal waste products, and processed animal waste derivatives are some of the animal waste, which has been used as a feed ingredient. Similarly, fishmeal, dried fish soluble, crab meal, shrimp meal, fish protein concentrate, and other fish by-products are some of the feed ingredients derived from marine origin. The protein content in dried manure ranges from 12 to 18 wt% for cattle, 28 to 48 wt% for poultry, and 22 to 25 wt% for pigs (Chen et al., 2003), making it a supreme source of valuable protein and nutrients.
Slaughterhouse wastes contain, e.g., blood, fat, bones, inedible parts. Protein recovery from inedible tissues is an essential step in valorization of slaughterhouse waste and the general practice is solubilizing the protein in an aqueous medium with the help of heat, chemicals, and enzymes individually or in amalgam followed by work up of the hydrolyzate to recover partially hydrolyzed protein (Adhikari et al., 2018).
As a potential renewable and nitrogen-rich feedstock, tons of shrimp shell waste (counting head, exoskeleton and walking appendages) are generated from the increasing consumption of seafood and most of them is either dumped or land filled. There is massive opportunity to introduce emerging methods and technologies in seafood waste application to produce value-added chemicals and materials. Similarly, seafood waste utilization will create benefits in both environmental and economic aspects. A crustacean shell in general contains 20–40% protein. It is reported that wastes from all the chelipede genotypes of M. vollenhovenii showed advisable protein content between 40.53 ± 0.56% DM and 53.73 ± 0.24% DM (Oyebola and Yildiz, 2016). High levels of protein, encompassing 40–50% dry matter, were found in the shell wastes of shrimp, 44–52% crude protein appeared in shrimp’s discard, and 50.3% crude protein was reported for processing waste powder of M. rosenbergii (Shahidi, 1994). This high level of protein indicates the potential for a wider use of the wastes from the M. vollenhovenii genotypes as proteinaceous food source for humans. Using a reformed chitinase (‘Chit46-CBM3’), (Deng et al., 2020) come out with a one-step shrimp shell processing method. The method could transform 46.5% of chitin in shrimp shells to chitin oligomers by hydrolysis in 12 h which resulted in partial protein release accompanied chitin hydrolysis. The hydrolyzate includes 8.8 g/L chitin oligomers and 11.3 g/L protein and this could support robust microbial growth. Marine wastes extract (MWE), prepared from marine organic wastes, and were used as nitrogen source for sulfate-reducing bacteria (SRB). These acidophiles occur in mine drainage and are exposed to high metal and sulfate concentrations (Dev and Bhattacharya, 2014). The MWE contains 13.95 g/L of nitrogen, and other micronutrients like K, Na, P, S, Ca, Fe, Mg, Mn, Zn, Co, Cu and Ni, and has a C/N ratio of 0.107. Protein pastes were prepared from shell waste through enzymatic treatment using trypsin, papain, pepsin and the average protein content in the protein paste was about 450 g/kg (Chakrabarti, 2002). Similarly, processing waste of prawns can be processed to protein powder mechanically or chemically with sodium hydroxide followed. Relative studies on the composition and amino acid profile, acceptability among consumers and nutritional quality of the protein powders revealed that the product prepared by freeze drying of the press liquor is superior in all aspects studied than the product prepared by mild alkali extraction.
Even though the substantial analyses and nutritional studies have demonstrated that the algal proteins are of high quality and comparable to conventional vegetable proteins, due to high production costs as well as technical hassles, the exploitation of algal protein is still in primary level (Becker, 2007). The use of whole microalgae biomass as a protein source in food and feed is simple and entrenched. However, there is an instant requirement for the development of practicable and robust processes, which can fractionate the microalgae biomass in different value-added products. Access to proteins from microalgae requires cell disruption and extraction. The critical review (Amorim et al., 2020) addresses the current state of the production of microalgae proteins for multifarious applications, and possibilities to concatenate the production of proteins and advanced biofuels.
Extraction of protein from agri-food residues, such as wheat bran, rice bran, brewer’s spent grains, pomegranate seeds, cauliflower leaves, pumpkin seed, and coffee beans has been widely studied (Liu et al., 2016; Phongthai et al., 2016; Xu et al., 2017; Talekar et al., 2018; Wen et al., 2020). During extraction, acid and alkali can hydrolyze proteins into smaller proteins or shorter peptides, which result in lower molecular weight of proteins in the extracts.
Hydrothermal liquefaction (HTL) of high moistured biomass such as microalgae, macroalgae, sludge, manure, and food waste, for the production of bio-oil has been widely concerned worldwide. A process for microalgae is based on extraction under high pressure, temperature and in supercritical water (Yang et al., 2018; Hietala et al., 2019). The high N content (20–40%) of these biomasses poses a challenge since up to 10% N may end up in the bio-oils may result as consequence of the HTL processing parameters (Leng et al., 2020). The nutrients present in the aqueous side fraction (Hu et al., 2017) can be reused in fermentation (Mehrabadi et al., 2016). The bio-oil obtained after HTL contains a large amount of nitrogen compared to that obtained by the solvent extraction method. On an average, total nitrogen (1–18 g/L), ammonium (0.34–12 g/L), and phosphate (0.7–12 g/L) are obtained (Shakya et al., 2017).
Pyrolysis of protein-rich biomass (e.g., algae, sewage sludge, lignocellulosic biomass) also suffer from a high nitrogen (N) content of the resulting bio-oils. The nitrogenous compounds present in these bio-oils are aliphatic and heterocyclic amines, amides and nitriles. Such bio-oil cannot be used as fuel directly since the high N content will induce massive emission of nitrogen oxides during combustion. The review by Leng et al. (2020) summarized the effects of biomass compositions and pyrolysis parameters on the contents of N and the N-containing chemical components in bio-oil.
Due to the increase in global population, there is a need for new sources of protein and thus proteins from agri-food by-products have obtained increasing attention. Although growing, the animal feed market suffers from protein biorefinery by-products and recirculation of nitrogen has been identified as target to revamp the overall process. The newer protein conversion process developed will create value-added chemicals by recycling ammonia and releasing biofuels and high-value amino acids. Use of protein biomass in biogas production or by incineration to generate heat and electricity are limited. The use of agricultural and agro-based-industry wastes and seafood waste utilization will fabricate benefits in both environmental and economic aspects and using them as raw materials can help to diminish the overall production cost and will accord to the recycling of waste as well.
Second generation feedstocks contain proteins and, thus, nitrogen. During biofuel production nitrogen is not used. However, it can be produced as by-product of biofuels (Huo et al., 2011). Alternatively, agri- and aquaculture sidestreams may be used for production of nitrogenous products such as amino acids, thus, incorporating the nitrogenous fraction of these second generation feedstocks. Several bacteria are used for fermentative production of amino acids, e.g., E. coli and C. glutamicum. One of the latest commercializations of an amino acid production process may be the E. coli process developed by Metabolic Explorer for fermentative production of L-methionine, which was acquired by Evonik (2019). For the use of alternative substrates, the industrially relevant E. coli, C. glutamicum, Pseudomonas, Bacillus, and yeast strains have been engineered to broaden their substrate scope (Wendisch et al., 2016). As compared to E. coli, the range of substrates available for C. glutamicum is narrow, e.g., including lactate, citrate, ethanol, and dicarboxylic acids besides glucose, fructose and sucrose (Polen et al., 2007; Arndt et al., 2008; Georgi et al., 2008; Youn et al., 2009). Metabolic engineering considerably widened the relatively narrow scope of the wild type (Wendisch and Lee, 2020). In the following, the metabolic engineering strategies to enable production of nitrogenous compounds from constituents of agri- and aquaculture sidestreams are detailed (Figure 1) and, where possible, examples for production of nitrogenous compounds by direct utilization of hydrolyzates are given.
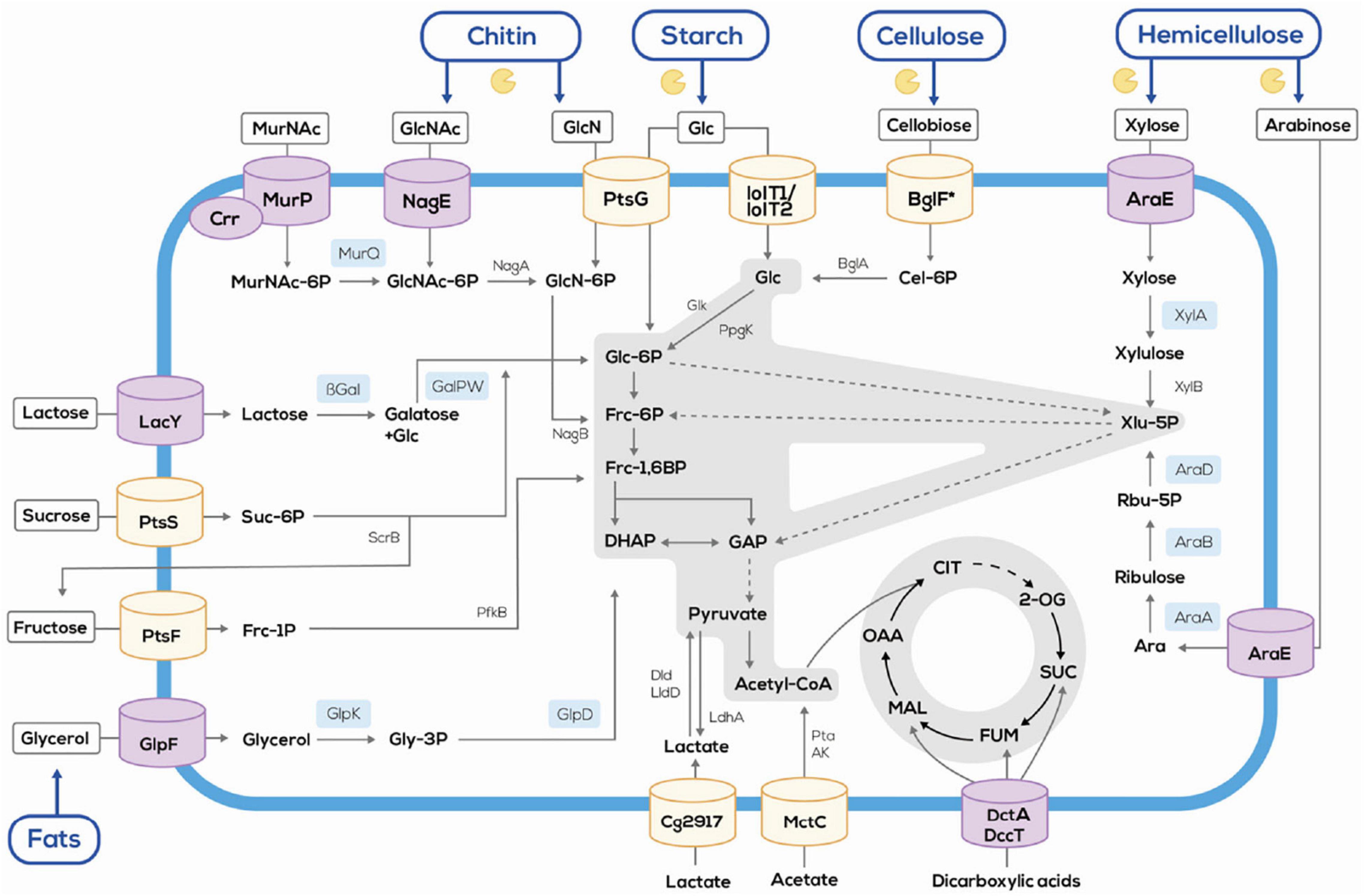
Figure 1. Schematic overview of the flexible feedstock concept established in C. glutamicum by systems metabolic engineering. Orchid cylinders and sky blue boxes indicate heterologous proteins. The symbol and blue lines indicate polysaccharides degradation via heterologous enzymes. AK, acetate kinase; Ara, arabinose; AraA, arabinose isomerase; AraB, ribulokinase; AraD, ribulose 5-phosphate 4-epimerase; AraE, arabinose transporter; βgal, β-galactosidase; BglA, phospho-β-glucosidase; BglF, cellobiose specific PTS; Cel-6-P, cellobiose-6-phosphate; Cg2917, putative L-lactate permease; Crr, glucose-specific PTS system enzyme IIA component; DccT, C4-dicarboxylate divalent anion/sodium symporter-type transporters; DHAP, dihydroxyacetone phosphate; Dld, quinone-dependent L-lactate dehydrogenase; Frc-1,6BP, fructose-1,6-bisphosphate; Frc-6P, fructose-6-phosphate; Frc-1P, fructose-1-phosphate; GalPW, galactose pathway; GAP, glyceraldehyde-3-phosphate; Glc-6P, glucose-6-phosphate; Glc, glucose; GlcN, glucosamine; GlcNAc, N-acetylglucosamine; GlcN-6P, glucosamine-6-phosphate, Glk, ATP dependent glucokinase; GlpD, glycerol-3-phosphate dehydrogenase; GlpF, glycerol facilitator; GlpK, glycerol kinase; Gly-3P, glycerol-3-phosphate; IolT1/IolT2, inositol transporters, also accepting glucose; LacY, lactose permease; LdhA, lactate dehydrogenase; LldD, quinone-dependent L-lactate dehydrogenase; MctC, monocarboxylic acid transporter; MurNAc, N-acetylmuramic acid; MurP, N-acetylmuramic acid PTS permease; MurQ, etherase; NagA, N-acetylglucosamine-6-P deacetylase, NagB, glucosamine-6-P deaminase; NagE, GlcNAc-specific PTS system from Corynebacterium glycinophilum; PfkB, 1-phosphofructokinase; PpgK, polyphosphate dependent glucokinase; Pta, phosphotransacetylase; PtsF, fructose-specific PTS; PtsG, glucose-specific PTS; PtsS, sucrose specific PTS; Rbu-5P, ribulose-5-phosphate; ScrB, sucrose-6-phosphate hydrolase; Suc-6P, sucrose-6-phosphate; Xlu-5P, xylulose-5-phosphate; XylA, xylose isomerase; XylB, xylulokinase.
Fats that are present in sidestreams may be transesterified with methanol or ethanol to yield the biodiesels FAME or FAEE, thus, they have an immediate biofuel application, while the stoichiometric byproduct glycerol accumulates. The C. glutamicum genes for glycerol kinase and glycerol-3-phosphate dehydrogenase are transcriptionally silent. Only upon overexpression of these genes or their respective orthologs from E. coli could the engineered strains grow and produce amino acids from glycerol (Rittmann et al., 2008; Meiswinkel et al., 2013b). The additional expression of the E. coli glycerol facilitator gene glpF increased the growth rate and volumetric productivity. Glycerol was used for production of the following nitrogenous compounds by recombinant C. glutamicum: amino acids L-glutamate, L-lysine, L-ornithine, L-arginine, and the diamine putrescine (Meiswinkel et al., 2013b).
Chitin occurs in cell walls of fungi, the exoskeletons of crustaceans and insects, fish scales, radulae of mollusks, and cephalopod beaks, making it one of the most abundant biopolymers in nature. Technically, chitin is an accessible sidestream of marine fisheries (crustacean shells), mushroom farming and fungal fermentations for the production of enzymes (fungal mycelia). Chitin is made up of glucosamine and N-acetylglucosamine, whereas glucosamine and N-acetylmuramic acid constitute the bacterial cell wall peptidoglycan. The hexosamines serve as sources of carbon, energy and nitrogen, which is relevant in particular for production of nitrogenous compounds such as amino acids. Glucosamine is the only hexosamine utilized by wild-type C. glutamicum. It is taken up into the C. glutamicum cell and phosphorylated using phosphoenolpyruvate by a side activity of the glucose-specific phosphotransferase system PtsG (Uhde et al., 2013). Uptake and phosphorylation of N-acetylglucosamine to N-acetylglucosamine 6-phosphate required expression of the gene for the N-acetylglucosamine PTS (NagE) from C. glycinophilum (Matano et al., 2014). Since the native glucosamine 6-phosphate deaminase gene nagB is repressed by the transcriptional regulator NanR (Uhde et al., 2016), deletion of nanR or plasmid-borne overexpression of nagB efficient resulted in fast growth and production with N-acetylglucosamine (Matano et al., 2014; Uhde et al., 2016). N-Acetylmuramic acid is an N-acetylated sugar acid that makes up half of the polysaccharide content of peptidoglycan. When the E. coli genes murP, crr, and murQ coding for a PTS subunit specific for N-acetylmuramic acid, a general PTS component, and an etherase for cleavage of N-acetylmuramic acid 6-phosphate to yield N-acetylglucosamine 6-phosphate and lactate were expressed, C. glutamicum could utilize N-acetylmuramic acid (Sgobba et al., 2018a). L-Lysine was produced from glucosamine, N-acetylglucosamine as well as N-acetylmuramic acid (Matano et al., 2014; Uhde et al., 2016; Sgobba et al., 2018a).
A synthetic consortium composed of a chitin degrading E. coli strain and a C. glutamicum strain producing nitrogenous compounds has been used to develop chitin-based production (Vortmann et al., 2021). In this proof-of-concept study, the chitin monomer N-acetylglucosamine is hydrolyzed by a lysine-auxotrophic E. coli strain to yield acetate and glucosamine. Acetate is the only carbon source for the engineered E. coli strain, whereas the lysine-secreting C. glutamicum producer strain was engineered to only use glucosamine. Chitin degrading enzymes (chitinase, chitin deacetylase and glucosaminidase) had to be added to the growth media, but they may be synthesized and secreted by the E. coli strain used (Vortmann et al., 2021).
Seaweed has not been fully exploited as sustainable microbial feedstock for the fermentation industry. Algal biomass contains various polysaccharides, e.g., alginate and laminarin in brown macroalga that also contain mannitol. C. glutamicum cannot directly use the polymers, but possesses an arabitol utilization operon. The operon is repressed by the regulatory protein AtlR and can be induced by arabitol, but not by mannitol (Laslo et al., 2012). Strains lacking atlR utilize mannitol via arabitol uptake permease and arabitol dehydrogenase. This concept was taken further by genetic engineering inspired by carbon flux analysis leading to a titer of about 2 g/L L-lysine (Hoffmann et al., 2018). Combining systems metabolic engineering with heterologous fructokinase, a titer of about 76 g/L L-lysine was reached. Acid pre-treated seaweed Durvillaea antarctica or alginate-free extracts of the alga Laminaria digitate were sown to serve as carbon sources (Hoffmann et al., 2018). To make full use of macroalgal biomass C. glutamicum has to be engineered for growth with all major polysaccharide fractions present in brown, green and red algae.
Starch is not a native substrate of C. glutamicum wild type, but upon expression and secretion of α-amylase from Streptomyces L-lysine and L-glutamate were produced from starch (Seibold et al., 2006; Chen et al., 2019). In a concept similar to that described above for chitin, a synthetic E. coli–C. glutamicum consortium has been engineered for growth and production with starch (Sgobba et al., 2018b). The remaining challenges are that starchy biomass does have competing uses as food and feed, that debranching enzymes such as pullulanase have to be secreted or that starch has to be hydrolyzed to access cyclodextrin and branched starch portions (Burgardt et al., 2021).
Lignocellulosic hydrolyzates such as spent sulfite liquor or pre-treated Miscanthus biomass have been used to produce nitrogenous compounds with metabolically engineered C. glutamicum strains (Joo et al., 2017; Sinner et al., 2021). Cellulose supported growth upon expression of exogenous genes for cellulases, β-glucanases, and β-glucosidases (Kim et al., 2014; Anusree et al., 2016). The hemicellulosic xylan fraction can be cleaved by endoxylanases and β-xylosidases to yield xylose (Imao et al., 2017). In the isomerase pathway that was transferred to C. glutamicum, xylose is converted to xylulose-5-phosphate, an intermediate of the pentose phosphate pathway, in a two-step pathway. While C. glutamicum possesses a gene for xylulose kinase, heterologous expression of a xylose isomerase was required. Combined overexpression of the genes coding for the respective enzymes allowed for growth (Kawaguchi et al., 2006), production of lysine and astaxanthin (Henke et al., 2018), L-ornithine (Eberhardt et al., 2014), 5-aminovalerate (Jorge et al., 2017b), and sarcosine (Mindt et al., 2019a). Transfer of the Weimberg pathway, which converts xylose to 2-oxoglutarate by five sequential enzyme reactions, supported growth with xylose when the exogenous Caulobacter crescentus genes were expressed (Radek et al., 2014). Later it was shown that it is sufficient to express either the C. crescentus genes encoding xylonate dehydratase or that coding for 2-oxo-3-deoxy-d-xylonate dehydratase (Brüsseler et al., 2019). Side reactions of two native enzymes (IolG and KsaD), a spontaneous lactonization reaction could be coupled to one of the two non-native dehydratases (either XylD or XylX) (Brüsseler et al., 2019). While C. glutamicum utilizes ribose, the other pentose in lignocellulosic hydrolyzates, arabinose, is not a native substrate, but overexpression of the araBAD operon from E. coli led to production of L-glutamate, L-lysine, L-ornithine, and L-arginine, respectively, from arabinose (Schneider et al., 2011). Rice straw hydrolyzates supported amino acid production best when both araBAD and xylAB were overexpressed (Meiswinkel et al., 2013a). The fact that C. glutamicum readily utilizes acetate as sole and combined source of carbon and energy (Gerstmeir et al., 2003) was exploited for production using blends, e.g., of acetate and xylose (Mindt et al., 2019b) or of lignocellulosic acetate (Kiefer et al., 2021).
Corynebacterium glutamicum cannot utilize carbon sources that lack carbon–carbon bonds such as methanol. Methanol can be obtained by reducing carbon dioxide and is considered a valuable carbon source for biotechnological processes (Clomburg et al., 2017). The knowledge about natural methylotrophs that can grow fast and efficiently with methanol (Müller et al., 2015) inspired metabolic engineering of C. glutamicum for methanol utilization. Naturally, C. glutamicum oxidizes methanol to carbon dioxide yielding reduction equivalents. To assimilate methanol into biomass or products such as L-lysine and cadaverine ribulose monophosphate cycle genes from natural methylotrophs had to be expressed in C. glutamicum (Lessmeier et al., 2015). This was demonstrated for production of L-lysine and cadaverine in an isotope-labeling experiment (Lessmeier et al., 2015). Methanol-essential growth was achieved for E. coli (Meyer et al., 2018) and for glutamate-producing C. glutamicum (Tuyishime et al., 2018). Adaptive laboratory evolution led to higher recalcitrance to methanol and helped to identify bottlenecks such as formation of methanethiol as toxic by-product (Hennig et al., 2020; Wang et al., 2020).
Monomethylamine is synthesized chemically from ammonia and methanol, but it also occurs in nature, e.g., due to putrefaction of proteins or by demethylation of methylated L-arginine and L-lysine residues present in histone proteins. Some methylotrophs are able to utilize monomethylamine as sole source of energy, carbon and nitrogen, but C. glutamicum cannot use monomethylamine as growth substrate. However, monomethylamine may serve as co-substrate for the production of N-methylated amines and amino acids by recombinant C. glutamicum strains. In these strains, reductive methylamination of a 2-oxoacid by a side activity of the Pseudomonas putida enzyme DpkA (Muramatsu et al., 2005), which in this bacterium reduces Δ1-piperideine-2-carboxylate to L-pipecolic acid, yields N-methylated amino acids, e.g., pyruvate can be reductively aminated by DpkA using monomethylamine to N-methyl-L-alanine (Mindt et al., 2018). This concept was also applied to reductive methylamination of glyoxylate and phenylpyruvate to yield sarcosine (N-methyl-glycine) and N-methyl-L-phenylpyruvate, respectively (Mindt et al., 2020; Kerbs et al., 2021).
One aspect of metabolically engineered strains relevant for large-scale fermentation processes is genetic stability. The metabolic engineering approaches listed in Table 1 comprise both chromosomal modifications and plasmid-borne gene expression. The more common rolling circle-replicating plasmids are less stabile than theta-type replicating plasmids and shifting from rolling circle-replicating to theta-type replicating plasmids may improve productivity as shown, e.g., for production of the diamine cadaverine by B. methanolicus (Irla et al., 2016). However, regarding genetic stability, chromosomal changes introduced into the genome of the production host remains the gold standard of genetic engineering.
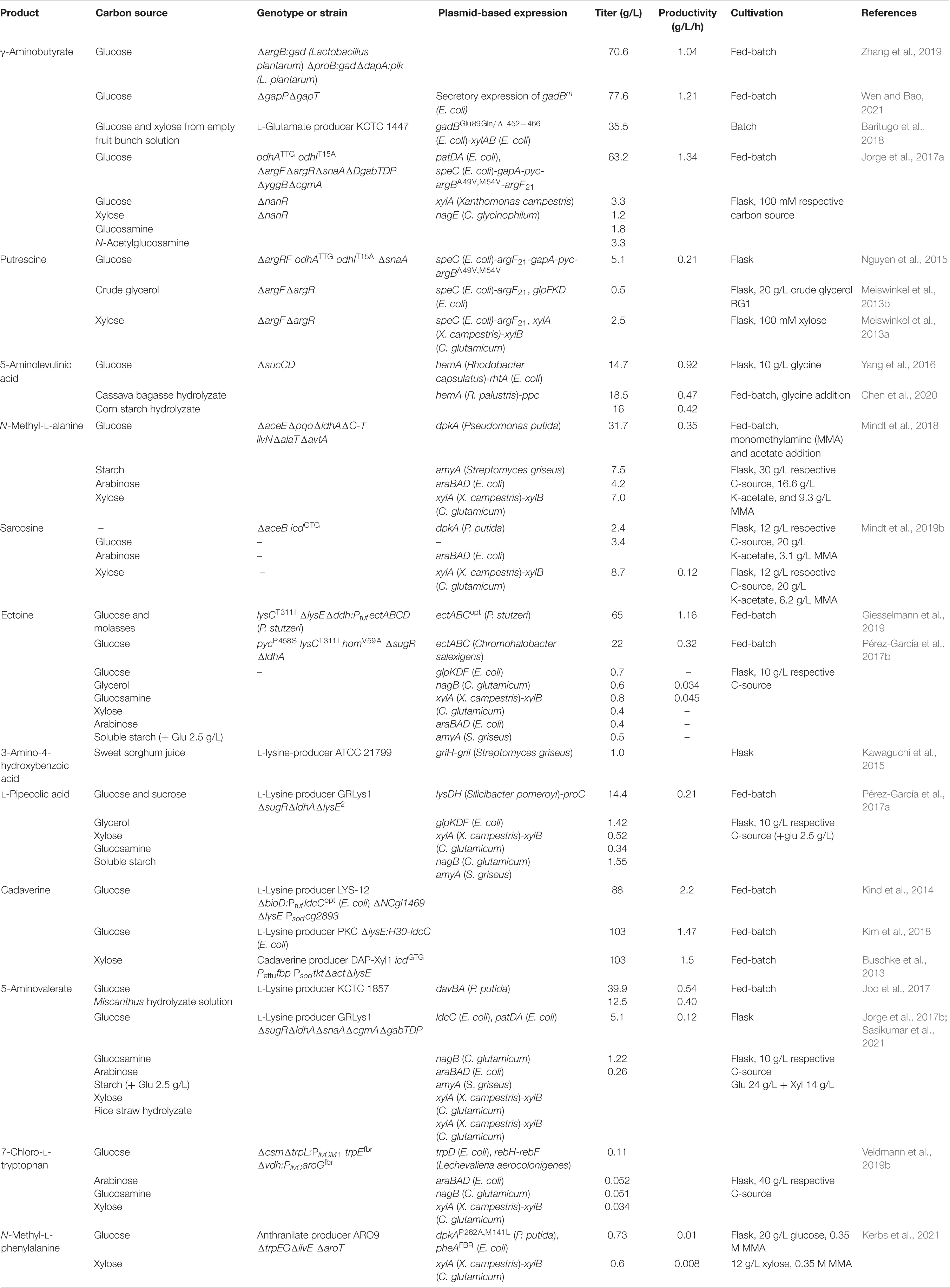
Table 1. Summary of fermentative production of nitrogenous compounds by engineered C. glutamicum from renewable carbon sources.
Notably, all C. glutamicum strains rely on ammonium salts or urea to provide nitrogen for sidestream-based production of nitrogenous compounds. This is due to the fact that the bacterium does not secrete proteases or peptidases. Only access to amino sugars has been established for C. glutamicum using exogenous genes. Monomethylamine (s. above) that is present in aquaculture sidestreams can be used for production N-alkylated amino acids by C. glutamicum strains that use it to reductively aminate 2-oxo acids. Thus, we predict that future metabolic engineering of C. glutamicum may include secretion of proteases and peptidases to access proteins and peptides and other polymeric nitrogen-containing substrates as nitrogen sources, thus, enlarging the currently available nitrogen substrate scope (ammonium, urea, some amino sugars, some alkylamines).
Diverse nitrogenous compounds have been produced by systems metabolic engineering of C. glutamicum through extending and intercepting metabolic pathways derived from glycolysis, TCA cycle, aspartate-family pathways, and the shikimate pathway (Figure 2). The combined metabolic networks of reconstituted synthetic pathways with utilization pathways for alternative carbon sources have accelerated sustainable production of nitrogenous compounds from agri- and aqua-culture sidestreams (Baritugo et al., 2018; Wolf et al., 2021). Here, the product portfolio accessible via C. glutamicum fermentation primarily features amino acids, halogenated amino acids, diamines, and alkylamines (Wendisch and Lee, 2020; Table 1). Besides, examples of the design and implementation of synthetic pathways not present in native metabolism to produce sought after nitrogenous compounds will be provided.
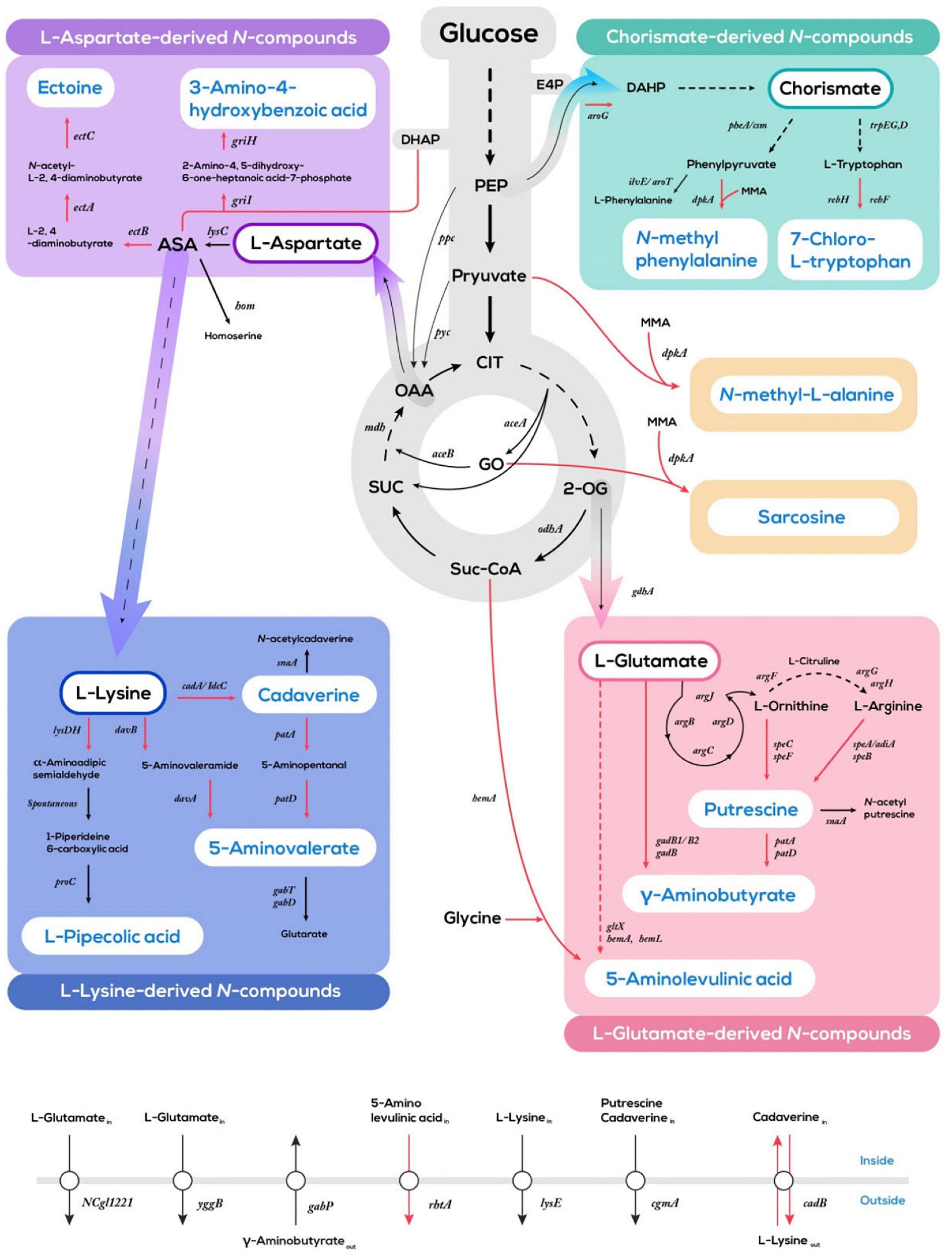
Figure 2. Schematic overview of systems metabolic engineering of C. glutamicum for the production of nitrogenous compounds from glucose. Red lines indicate artificial synthetic pathways; black colored lines indicate endogenous pathways. Multi-step reactions are represented by dotted lines. E4P, erythrose 4-phosphate; DHAP, dihydroxyacetone phosphate; DAHP, 3-deoxy-D-arabinoheptulosonate 7-phosphate; PEP, phosphoenolpyruvate; CIT, citrate; 2-OG, 2-oxoglutarate; Suc-CoA, succinyl-CoA; SUC, succinate; OAA, oxaloacetate; GO, glyoxylate; ASA, L-aspartate 4-semialdehyde; MMA, monomethylamine.
Corynebacterium glutamicum traditionally employed for commercial production of L-glutamate is an ideal platform strain for the production of various nitrogenous compounds derived from L-glutamate, including γ-aminobutyrate (Takahashi et al., 2012), L-ornithine (Jensen and Wendisch, 2013; Jensen et al., 2015), L-arginine (Jensen et al., 2015), putrescine (Schneider et al., 2012), and 5-aminolevulinic acid (Ramzi et al., 2015; Figure 2 and Table 1). Additionally, the intermediates in glycolysis and TCA cycle can be converted to nitrogenous compounds including 5-aminolevulinate (Yang et al., 2016), N-methyl-L-alanine (Mindt et al., 2018), and sarcosine (Mindt et al., 2019b) by introducing synthetic pathways and/or intercepting unnecessary pathways into C. glutamicum.
γ-Aminobutyrate (GABA), one of the non-proteinogenic ω-amino acids, functions as a primary inhibitory neurotransmitter in animals and has several therapeutic activities, including anti-anxiety, anti-depression, anti-schizophrenia, analgesics, diuresis, and hypotension (Zhang et al., 2014; Shi et al., 2017). In addition, butyrolactam is derived from GABA by ring closure and is used to synthesize the biodegradable polyamide (PA, nylon) PA 4 by ring-opening condensation (Baritugo et al., 2018).
As the conversion of L-glutamate to GABA is mediated by pyridoxal phosphate (PLP)-dependent glutamate decarboxylase (GAD; EC 4.1.1.15), C. glutamicum with a superior L-glutamate pool has been employed for efficient GABA production from renewable sources. Pioneering studies for GABA production using C. glutamicum were performed by heterologous expression of gadB from E. coli (Takahashi et al., 2012), gadB1/gadB2 from Lactobacillus brevis (Shi and Li, 2011), and gad from Lactobacillus plantarum (Zhang et al., 2014). Improving GAD activities from L. brevis and E. coli with broadening active pH range to neutral was obtained by protein engineering, yielding increase of GABA production in C. glutamicum (Choi et al., 2015). With these features, Baritugo et al. (2018) achieved 35.5 g/L of GABA at bath fermentation using empty fruit bunch solution containing glucose and xylose by recombinant C. glutamicum, in which E. coli gadBm (Glu89Gln/Δ452–466) and xylAB genes encoding xylose isomerase and xylulokinase were expressed under control of the promoter H36. Due to the requirement of PLP as a cofactor of GAD and absence of pyridoxal kinase in C. glutamicum, an engineered strain was accomplished by expression of plk encoding pyridoxal kinase and gad from L. plantarum, together with blocking of byproducts formation pathways (Zhang et al., 2014). In fed-batch cultures, 70.6 g/L of GABA were accumulated using glucose after 70 h via a two-stage pH control. Increase of intracellular pool of L-glutamate for GABA production was attained by deletion of pknG encoding serine/threonine protein kinase (Okai et al., 2014), overexpression of ppc or deletion of mdh (Shi et al., 2017), or deletion of NCgl1221 encoding L-glutamate exporter (Cho et al., 2017). Another approach to enhance GABA production was conducted by secretion of GAD enzyme and blocking GABA import and degradation (ΔgapPΔgapT), whereby extracellular accumulated L-glutamate was directly converted to GABA (Wen and Bao, 2021). Consequently, 77.6 g/L of GABA with a yield of 0.44 g/g were accumulated on fed-batch fermentation. A new route for GABA production via the growth-associated conversion of putrescine to GABA was established by heterologous expression of patA coding for putrescine transaminase and patD coding for γ-aminobutyraldehyde dehydrogenase from E. coli (Jorge et al., 2016). Improved production of GABA was taken by blocking by-product formation of N-acetylputrescine (ΔsnaA), avoiding export of L-glutamate and putrescine (ΔyggBΔcgmA), and increasing precursor supply by reducing 2-oxoglutarate dehydrogenase activity (odhATTGodhIT15A) (Jorge et al., 2017a). The engineered strain enabled production of 63.2 g/L of GABA with the highest volumetric productivity up to 1.34 g/L/h. Moreover, expression of xylA from Xanthomonas campestris, deletion of nanR, and deletion of nanR and expression of nagE from Corynebacterium glycinophilum, respectively, led to production of GABA using sustainable resources xylose, glucosamine, and N-acetyl-glucosamine. Of these, GABA titer and productivity from N-acetyl-glucosamine were higher than those from D-glucose.
Polyamides are polymers in which the repeating building blocks, amine and carboxylic acid or lactams, are linked by amide bonds (Kind et al., 2010). Commercially available PA 6 and PA 6,10 are typically produced via chemical routes from petroleum-based chemicals. Due to shortage of fossil resources and environmental concerns, bio-based production of polyamides using sustainable resources has attracted considerable interest. Putrescine (1,4-diaminobutane) is a 4-carbon aliphatic diamine and a commodity building block for the synthesis of bio-based PA 4,6 and PA 4,10 (Baritugo et al., 2018).
Corynebacterium glutamicum has a competitive potential for putrescine production due to its innate abilities, including overproduction of the main precursor L-glutamate, remarkable tolerance to high-concentrations of putrescine, and no putrescine degradation routes (Schneider and Wendisch, 2010). Putrescine production by C. glutamicum was first evaluated by recruiting two alternative routes via the ODC (ornithine decarboxylase) and ADC (arginine decarboxylase) pathways from E. coli (Schneider and Wendisch, 2010). As a result, the putrescine yield in C. glutamicum implanted the ODC pathway was 40-fold higher than that provided with the ADC pathway. Putrescine production was further enhanced by following strategies: (i) inactivation of L-arginine repressor by deletion of argR, (ii) fine-tuning argF expression encoding ornithine transcarbamoylase through modulation of transcription and translation efficiencies, (iii) reducing 2-oxoglutarate dehydrogenase activity by chromosomal changes odhATTG and odhIT15A, (iv) increasing precursor supply flux by expression of gapA and pyc genes, (v) deregulation of ArgB by introducing mutated argB encoding feedback-resistant N-acetylglutamate kinase, (vi) blocking accumulation of by-product N-acetylputrescine by deletion of snaA (Nguyen et al., 2015). Expression of speC from E. coli in engineered C. glutamicum harboring genetic properties described above led to production of 5.1 g/L putrescine with a productivity of 0.21 g/L/h in baffled flasks. Another strategy to improve putrescine production was first attempted by construction of ornithine overproducing strain, followed by plasmid-mediated expression of speC from E. coli resulted in production of putrescine with a yield on glucose of 0.17 g/g (Jensen et al., 2015). Putrescine production from crude glycerol (Meiswinkel et al., 2013b), hemicellulosic xylose (Meiswinkel et al., 2013a), and glucosamine (Uhde et al., 2013) as alternative carbon sources has been demonstrated by expression of glpFKD from E. coli, xylA from X. campestris and xylB from C. glutamicum, and nagB from C. glutamicum, respectively, in engineered strains.
5-Aminolevulinic acid (5-ALA) is a 5-carbon non-proteinogenic δ-amino acid and is a major precursor of tetrapyrroles, including porphyrin, heme, chlorophyll, and vitamin B12 (Chen et al., 2020). 5-ALA can be used in medicine, agriculture, and feed additive as photodynamic therapy, a potential bio-stimulant for crop production, and a feed amino acid for livestock rearing.
Two kinds of the artificial pathways were introduced for 5-ALA production in C. glutamicum. The C5 pathway comprised three enzymatic steps converting L-glutamate to 5-ALA via L-glutamyl-tRNA and L-glutamate-1-semialdehyde, and corresponding enzymes are glutamyl-tRNA synthetase, glutamyl-tRNA reductase, and glutamate-1-semialdehyde aminotransferase encoded by gltX, hemA, and hemL (Ramzi et al., 2015). 5-ALA-producing strain has been engineered by expression of a mutated hemA from Salmonella typhimurium and hemL from E. coli, together with flux redistribution of TCA cycle by mutation of OdhIT14A/T15A and introduction of threonine/homoserine exporter RhtA from E. coli (Ramzi et al., 2015; Ko et al., 2019). Under ethambutol induction condition, 5-ALA production reached to 2.9 g/L in shaking incubator. The C4 pathway was reconstituted by expression of different sources of hemA encoding 5-aminolevulinic acid synthase by which succinyl-CoA and glycine are condensed to form 5-ALA (Feng et al., 2016; Yang et al., 2016; Chen et al., 2020). Of these, expression of hemA from Rhodobacter capsulatus and rhtA encoding threonine/homoserine exporter from E. coli in C. glutamicum (ΔsucCD) showed the highest titer of 5-ALA (14.7 g/L) from D-glucose and glycine in fed-batch fermentation (Yang et al., 2016). Similarly, optimized expression of hemA from R. palustris as well as additional expression of ppc gene led to 16.3 g/L of 5-ALA from D-glucose and glycine (Chen et al., 2020). Use of cassava bagasse hydrolyzate and corn starch hydrolyzate as cheap raw materials resulted in accumulation of 18.5 g/L and 16 g/L of 5-ALA, respectively, whereas use of beet molasses gave rise to serious suppression of 5-ALA production.
N-Methylated amino acids are important building blocks for peptide-based drugs such as vancomycin, actinomycin D, cyclosporine A, and N-methylated tubulysin and received increasing interest in the pharmaceutical industry due to improved pharmacokinetic properties (Mindt et al., 2018; Wendisch and Lee, 2020). Since chemical syntheses of N-methylated amino acids are often limited by low yield, incomplete stereoselectivity, and side reactions, microbial and biocatalytic approaches for their production may offer cleaner and greener alternatives over chemical methods (Muramatsu et al., 2005; Mindt et al., 2020).
Typically, N-methyl amino acid dehydrogenase/imine reductase DpkA from P. putida can catalyze the reductive alkylamination of 2-oxo acids to yield the corresponding N-alkylamino acids (Mihara et al., 2005). As a proof-of-concept, fermentative production of N-methyl-L-alanine (NMeAla), one of representative N-alkylamino acids, was obtained by introduction of dpkA from P. putida in pyruvate-overproducing C. glutamicum, which generated 31.7 g/L of NMeAla from D-glucose and monomethylamine (MMA) in fed-batch fermentation (Mindt et al., 2018). In addition, sustainable production of NMeAla from alternative feedstocks was achieved by extending metabolic pathways to utilize starch, arabinose, or xylose.
Sarcosine (N-methylglycine) is ubiquitous in biological materials and occurs as an intermediate in choline and glycine metabolism (Mindt et al., 2019b). It is used in cosmetic formulations such as hair conditioners and surfactant cleansers and has clinical significance for the treatment of schizophrenia and depression (Hyslop et al., 2019).
Due to its catalytic efficiency and broad substrate specificity, DpkA from P. putida is able to catalyze the formation of sarcosine from glyoxylate as substrate for N-alkylation in the presence of MMA. Accumulation of glyoxylate in C. glutamicum resulted from deletion of aceB encoding malate synthase and the start codon exchange of gene icdGTG encoding isocitrate dehydrogenase (Mindt et al., 2019b). Then optimized expression of dpkA in the constructed strains enabled production of 2.4 g/L, 2.7 g/L, and 3.4 g/L of sarcosine, respectively, from glucose, lignocellulosic xylose and arabinose supplemented with potassium acetate. Besides, 8.7 g/L of sarcosine with a productivity of 0.12 g/L/h were achieved in a xylose-based optimized culture, demonstrating that sarcosine production from xylose and arabinose outperforms glucose-based production. The mutation of DpkA (DpkAF117L) in the substrate binding site led to increased specific activity toward glyoxylate and MMA as substrates and allowed faster production of sarcosine (0.16 g/L/h) compared to the control enzyme from xylose-based fermentation process (Mindt et al., 2019b).
In engineered C. glutamicum strains, the coupled metabolic networks for L-lysine biosynthesis via L-aspartate and utilization of several alternative carbon sources have been extended not only by synthetic pathways via L-aspartate 4-semialdehyde for production of ectoine (Becker et al., 2013) and 3-amino-4-hydroxybenzoic acid (Kawaguchi et al., 2015), but also L-lysine degradation pathways for production of L-lysine-derived nitrogenous compounds, such as L-pipecolic acid (Pérez-García et al., 2016, 2019), cadaverine (Kim et al., 2018), and 5-aminovalerate (Rohles et al., 2016; Figure 2 and Table 1).
The cyclic amino acid ectoine (1,4,5,6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid) is a representative compatible solute that is wide-spread among halophilic and halotolerant eubacteria (Liu et al., 2021). It not only functions as osmoprotectant, biofunctional stabilizer, skin protector, and potential drugs for diseases but also is widely used in cosmetic and medical industries (Liu et al., 2021). Since ectoine production in the halophilic microbes through ‘bacterial milking’ process causes corrosive damage to devices, its production process under low salt conditions is crucial for economical production (Wolf et al., 2021). In this sense, C. glutamicum has emerged as a prominent strain for ectoine production.
The biosynthesis of ectoine from L-aspartate 4-semialdehyde involves three enzymes, namely, L-2,4-diaminobutyrate aminotransferase (EctB), L-2,4-diaminobutyrate acetyltransferase (EctA), and ectoine synthase (EctC). The recombinant strain for ectoine production was implemented by expression of codon-optimized ectABCD from Pseudomonas stutzeri in a strain with lysCT311I mutation followed by inactivation of L-lysine exporter (Becker et al., 2013). The resulting strain yielded 4.5 g/L of ectoine under low-salt conditions after 16 h cultivation. Subsequently, C. glutamicum that contained an optimized heterologous ectoine pathway with a fine-tuned monocistronic expression of ectB, ectA, and ectC genes produced 65 g/L ectoine on a glucose-molasses medium within 56 h in a fed-batch process (Giesselmann et al., 2019). On the other hand, Pérez-García et al. (2017b) developed an ectoine-producing C. glutamicum from L-lysine-producing basal strain (pycP458S lysCT311I homV59A), because L-lysine and ectoine share L-aspartate 4-semialdehyde as a same precursor. The strategies comprised expression of ectABC from Chromohalobacter salexigens, deletion of sugR encoding DeoR-type regulator, and deletion of ldhA encoding lactate dehydrogenase. 22 g/L of ectoine and 0.32 g/L/h of productivity were obtained in a fed-batch culture. Additionally, C. glutamicum strains utilizing the several carbon sources, i.e., glycerol, glucosamine, xylose, arabinose, and soluble starch, were achieved by introduction of glpKDF from E. coli, nagB from C. glutamicum, xylA from X. campestris and xylB from C. glutamicum, araBAD from E. coli, and amyA from Streptomyces griseus, respectively. Among tested alternatives carbon sources, the highest ectoine yield was obtained using glucosamine, providing both carbon and nitrogen sources for C. glutamicum.
3-Amino-4-hydroxybenzoic acid (3,4-AHBA) is a benzene derivative that acts as a precursor for a variety of secondary metabolites produced by Streptomyces and for the synthesis of polybenzoxazole, a thermostable bioplastic (Suzuki et al., 2006; Kawaguchi et al., 2015). Unlike most aromatic compounds synthesized via the shikimate pathway, it is formed from L-aspartate 4-semialdehyde and dihydroxyacetone phosphate by the activities of GriI and GriH. A 3,4-AHBA-producing C. glutamicum was derived from L-lysine-producing C. glutamicum by transformation of a plasmid expressing griH and griI genes from S. griseus (Kawaguchi et al., 2015). Owing to fermentative properties of C. glutamicum utilizing sucrose, the engineered strain led to accumulation of 1.0 g/L 3,4-AHBA from sweet sorghum juice, an alternative feedstock for sugar cane molasses, which harbors fermentable sugars and amino acids as a nitrogen source. Moreover, this novel pathway has guided an alternative route for the biosynthesis of a benzene ring from C3 and C4 metabolites independent of the shikimate pathway.
The cyclic α-amino acid L-pipecolic acid (piperidine 2-carboxylic acid, L-PA) is a critical regulator of inducible plant immunity and an important precursor of immunosuppressants, antitumor agent, or peptide antibiotic (Pérez-García et al., 2016, 2019).
The production of L-pipecolic acid via L-lysine was attempted by introduction of lysDH encoding lysine 6-dehydrogenase from Silicibacter pomeroyi combined with endogenous expression of proC encoding pyrroline 5-carboxylate reductase in the L-lysine-producing C. glutamicum (GRLys1ΔsugRΔldhA) (Pérez-García et al., 2016). This allowed production of L-PA to a titer of 1.8 g/L with a productivity 0.04 g/L/h. Transport engineering of C. glutamicum by abolishing L-lysine export resulted in production of 3.9 g/L of L-PA without accumulation of L-lysine in shake flask cultivation (Pérez-García et al., 2017a). The resulting strain produced 14.4 g/L of L-PA with a productivity 0.21 g/L/h by fed-batch cultivation using glucose and sucrose as carbon sources. Finally, heterologous expression of relevant genes (described above) for utilization of alternative carbon sources, i.e., glycerol, xylose, glucosamine, and soluble starch, resulted in production of L-PA from these sustainable resources.
Cadaverine (1,5-diaminopentane) is a 5-carbon aliphatic diamine and can be used as a platform chemical for the synthesis of bio-polyamide PA 5,4 and PA 5,10 (Buschke et al., 2013).
Cadaverine is formed from L-lysine catalyzed by lysine decarboxylases (E.C. 4.1.1.18). The production of cadaverine was accomplished in C. glutamicum with a high intracellular L-lysine pool through extending L-lysine pathway by optimal expression of several sources of lysine decarboxylase genes, including cadA and ldcC from E. coli and ldc from Hafnia alvei (Mimitsuka et al., 2007; Kind et al., 2010; Li et al., 2014; Oh et al., 2015). Improvement of cadaverine production was implemented with the following strategies. These include blocking the accumulation of acetylated cadaverine (ΔsnaA) (Nguyen et al., 2015), together with engineering of cellular transport processes, such as deletion of L-lysine exporter (lysE), overexpression of a putative putrescine and cadaverine exporter gene (cgmA, cg2893) (Kind et al., 2011; Lubitz et al., 2016), and overexpression of cadB encoding cadaverine-lysine antiporter from E. coli (Li et al., 2014). The breakthrough works to boost cadaverine yield were done from L-lysine-overproducing strains in accordance with the strategies described above by Kind et al. (2014) and Kim et al. (2018). The resulting strains, respectively, allowed titers of 88 g/L and 103 g/L of cadaverine with productivities of 2.2 g/L/h and 1.47 g/L/h in fed-batch processes. The sustainable production of cadaverine in C. glutamicum was performed by using alternative renewable sources, e.g., soluble starch (Tateno et al., 2009), the hemicellulose sugar xylose (Buschke et al., 2011), xylooligosaccharides (Imao et al., 2017), methanol (Lessmeier et al., 2015), and cellobiose (Ikeda et al., 2013). The secretory expression of genes encoding α-amylase from Streptococcus bovis, β−glucosidase from Thermobifida fusca, and β−xylosidase from B. subtilis with PorH anchor protein, respectively, in engineered C. glutamicum strains enabled efficient utilization of soluble starch, cellobiose, and xylooligosaccharides, respectively, and sustainable production of cadaverine. Methanol is also a sustainable carbon source for microbial fermentations. Introduction of methanol oxidation and formaldehyde assimilation pathways as well as blocking formaldehyde oxidation (ΔaldΔfadH) in non-methylotrophic C. glutamicum led to cell growth in the presence of methanol and a partial conversion of methanol to cadaverine (Lessmeier et al., 2015). The overall approach for enhanced production of cadaverine from xylose in C. glutamicum was performed based on an integrated analysis of fluxome and transcriptome and in silico pathway modeling (Buschke et al., 2013). Systems metabolic engineering of C. glutamicum targeted to genes related with pentose phosphate pathway, TCA cycle, and by-products formation enabled efficient production of 103 g/L of cadaverine from xylose with a product yield of 32%.
5-Aminovalerate (5-AVA, 5-aminopentanoic acid) is a non-proteinogenic 5-carbon ω-amino acid and is an attractive building block for polymer synthesis (Rohles et al., 2016; Shin et al., 2016). 5-AVA can be converted to δ-valerolactam via a cyclization reaction followed by synthesis of bio-based PA 6,5 from purified δ-valerolactam with ε-caprolactam (Park et al., 2014).
As C. glutamicum is a well-established industrial L-lysine producer, the metabolic pathway from L-lysine toward 5-AVA was reconstructed by expressing davB and davA coding for lysine 2-monooxygenase and 5-aminovalereramidase from Pseudomonas putida in several tested L-lysine overproducing strains (Rohles et al., 2016; Shin et al., 2016; Joo et al., 2017). Of these, Joo et al. (2017) achieved 40 g/L of 5-AVA in fed-batch culture from D-glucose. In addition, they examined the feasibility of 5-AVA production (12.5 g/L) from lignocellulosic raw material, a Miscanthus hydrolyzate solution. Meanwhile, Rohles et al. (2016) demonstrated that accumulation of the by-products L-lysine and glutarate can be minimized by deleting the lysE gene and a putative gabT gene, yielding 28 g/L AVA with a productivity of 0.9 g/L/h from L-glucose and beet molasses as carbon sources in fed-batch fermentation. The lysine 2-monooxygenase in the synthetic pathway requires oxygen for activity, which often may limit production performance during fermentation. In this regard, an alternative pathway comprising ldcC, patA, and patD genes encoding lysine decarboxylase, putrescine transaminase, and γ-aminobutyraldehyde dehydrogenase, respectively, from E. coli was constructed in C. glutamicum combined with blocking the accumulation of the by-products cadaverine, N-acetylcadaverine, and glutarate (Jorge et al., 2017b). The resulting strain enabled production of 5.1 g/L 5-AVA with a productivity of 0.12 g/L/h from D-glucose in shake flasks. Also, 5-AVA was produced from the alternative feedstocks starch, glucosamine, xylose, arabinose, and rice straw hydrolyzate (Jorge et al., 2017b; Sasikumar et al., 2021).
A wide range of aromatic compounds produced from petroleum-based feedstocks and extraction of plants are currently being explored by microbial fermentation (Lee and Wendisch, 2017a). Due to the presence of multiple metabolic pathways that degrade and assimilate various aromatic compounds and its inherently higher tolerance to toxic aromatics, C. glutamicum is considered a prominent strain for the production of nitrogenous aromatic compounds, such as halogenated L-tryptophan (Veldmann et al., 2019a,b), N-methyl-L-phenylalanine (Kerbs et al., 2021), 4-aminobenzoate, anthranilate, N-methylanthranilate (Walter et al., 2020), indole-3-actic acid (Kim et al., 2019), and violacein (Wendisch and Lee, 2020; Figure 2). Recent advances in metabolic engineering of C. glutamicum with respect to the sustainable production of nitrogenous aromatic compounds from alternative feedstocks are listed in Table 1.
Halogenated aromatics are important building blocks or intermediates of bioactive compounds associated with the agrochemical, chemical, and pharmaceutical industries (Payne et al., 2015; Veldmann et al., 2019a,b). Generally, the chemical halogenation processes of arenes often suffer from poor regioselectivity and require harsh reaction conditions. It was known that flavin-dependent halogenase overcomes these issues and catalyzes the halogenation of arene compounds with high regioselectivity (Payne et al., 2015). Of these, 7-chloro-L-tryptophan (7-Cl-Trp) is a precursor of the alkaloid antibiotic rebeccamycin and the antifungal pyrrolnitrin (Veldmann et al., 2019b). Recently, enzymatic halogenation of L-tryptophan was established by utilizing immobilized tryptophan 7-halogenase with cofactor regeneration system (Frese and Sewald, 2015).
Fermentative production of halogenated amino acid 7-Cl-Trp from renewable sources and chloride salts was realized by Veldmann et al. (2019b) in an L-tryptophan-producing C. glutamicum. First, to provide a precursor, L-tryptophan-producing C. glutamicum was metabolically engineered by introducing genes encoding feedback-resistant enzymes AroG and TrpE and blocking carbon flow to L-tyrosine and L-phenylalanine as well as by plasmid-mediated expression of E. coli trpD. Finally, overexpression of the genes encoding tryptophan 7-halogeanse RebH and flavin reductase RebF from Lechevalieria aerocolonigenes led to chlorination of L-tryptophan to 7-Cl-Trp (108 mg/L). For access of C. glutamicum to renewable feedstocks, araBAD, xylAB, and nagB genes, respectively, were transformed to C. glutamicum, resulting in efficient utilization of arabinose, xylose, and glucosamine and production of 7-Cl-Trp in flasks cultivation.
Many bioactive peptides harbor N-methylated amino acids, e.g., N-methyl-L-alanine, N-methyl-L-glutamate, sarcosine, and N-methyl-L-phenylalanine (NMePhe). In particular, NMePhe-rich peptides can be considered as promising blood–brain barrier shuttle candidates for improving brain drug delivery of small neurodrugs (Malakoutikhah et al., 2010).
De novo production of NMePhe in C. glutamicum was based on reductive methylamination of phenylpyruvate by an engineered mutant DpkAP262A, M141L, which exhibited similar catalytic efficiencies for phenylpyruvate and pyruvate (Kerbs et al., 2021). To provide phenylpyruvate pool, the carbon flux into L-tryptophan and L-phenylalanine was blocked by removing trpEG, ilvE, and aroT genes in genome of anthranilate-producing C. glutamicum. Upon expression of dpkAP262A, M141L and pheAFBR encoding feedback-resistant chorismate mutase/prephenate dehydratase from E. coli, the engineered stain produced 0.73 g/L of NMePhe from glucose-based fermentation. When genes involved in xylose-utilizing enzymes were expressed in the engineered strain, an NMePhe titer of 0.6 g/L with a productivity of 0.008 g/L/h was reached from xylose-based fermentation.
To better make use of sidestreams from aqua- and agriculture for fermentative processes with engineered C. glutamicum strains producing nitrogen-containing value-added compounds we foresee the need for strain optimization and process intensification. ALE has already been applied to improve growth and production with methanol (Hennig et al., 2020; Wang et al., 2020) and has led to a new pathway of fructose utilization (Krahn et al., 2021). Moreover, volumetric productivities for 5-aminovalerate and glutarate have been enhanced by ALE approaches (Haupka et al., 2020; Prell et al., 2021). ALE will facilitate and accelerate the access to various (nitrogen-containing) feedstocks overcoming recalcitrance of the (polymeric) substrate, toxicity of their breakdown products or contaminants they contain.
Hydrolyzates such as spent sulfite liquor (SSL) are challenging to use since they may contain inhibitors and differ between batches. Online off-gas analysis and particle filtering for simultaneous process state and parameter estimation have recently been used to judge performance of a single C. glutamicum strain during SSL bioprocessing (Sinner et al., 2021). Furthermore, synthetic consortia have been developed for division of labor between different strains, e.g., various C. glutamicum strains or C. glutamicum and E. coli strains production based on either starch, chitin or SSL (Sgobba et al., 2018b; Pérez-García et al., 2021; Vortmann et al., 2021). SSL bioprocessing for production of riboflavin was improved in fed-batch fermentations based on a inoculation strategy using strains that either utilized glucose, glucose + mannose, glucose + xylose or all three sugars (Pérez-García et al., 2021). Besides using consortia of independent strains of the same species, a mutualistic synthetic E. coli/C. glutamicum consortium produced lysine, cadaverine and L-pipecolic acid from starch (Sgobba et al., 2018b). The lysine producing, starch-negative C. glutamicum strain produced the nitrogenous target compounds from glucose liberated by the lysine auxotrophic, starch hydrolyzing E. coli strain that functioned as substrate-converter. Despite the enormous potential that such synthetic microbial consortia have for biotechnological applications, fundamental research questions remain to be answered and process control strategies such as Kalman filtering have to be developed (Tuveri et al., 2021). In this respect, the single-cell behavior has to be studied by dynamic microfluidic single-cell cultivation (dMSCC) technology. As a first application, dMSCC was used for lysine production by a consortium consisting of a lysine auxotrophic strain and a strain producing lysine when induced with IPTG. Notably, the use photocaged IPTG with/without deprotection by light demonstrated at the single-cell level that the lysine auxotrophic strain depended on the lysine producing strain (Burmeister et al., 2021). While insight gained by these approaches still needs to be transferred to real-world fermentative processes, we foresee important contributions of microbial consortia to the field.
Biorefinery processes typically operate under sterile conditions with the associated costs for, e.g., autoclaving. Phosphite (also name phosphonate), which is used as organic fertilizer and is non-toxic to humans and animals, does not support growth of C. glutamicum as source of phosphorus. Transfer of phosphite dehydrogenase from Pseudomonas stutzeri to a lysine producing C. glutamicum strain enabled growth in non-autoclaved growth medium that contained phosphite instead of phosphate (Lei et al., 2021). While lysine was produced under non-sterile conditions, this approach can only be applied at larger scale using sidestreams from aqua- and agriculture that lack phosphate or require addition of a phosphorus source.
VW, KN, and J-HL wrote, revised, and edited the manuscript. All authors contributed to the article and approved the submitted version.
We acknowledge the financial support of the German Research Foundation (DFG) and the Open Access Publication Fund of Bielefeld University for the article processing charge. VW gratefully acknowledges support by the state of North Rhine-Westphalia (NRW) and the “European Regional Development Fund (EFRE),” Project “Cluster Industrial Biotechnology (CLIB) Kompetenzzentrum Biotechnologie (CKB)” (34.EFRE-0300095/1703FI04), and by the ERA CoBioBlue project SIDESTREAM (031B0950A). J-HL acknowledges support by Basic Science Research Program through the National Research Foundation of Korea (NRF-2018R1D1A1B07047207). KN acknowledges the grant from DBT, New Delhi and BMBF, Germany. The funding bodies had no role in the design of the study or the collection, analysis, or interpretation of data or in writing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We gratefully acknowledge excellent support by You-Lim Lee for the design of figures.
Adhikari, B., Chae, M., and Bressler, D. (2018). Utilization of slaughterhouse waste in value-added applications: recent advances in the development of wood adhesives. Polymers 10:176. doi: 10.3390/polym10020176
Amorim, M. L., Soares, J., Coimbra, J. S. D. R., Leite, M. D. O., Albino, L. F. T., and Martins, M. A. (2020). Microalgae proteins: production, separation, isolation, quantification, and application in food and feed. Crit. Rev. Food Sci. Nutr. 61, 1976–2002. doi: 10.1080/10408398.2020.1768046
Anusree, M., Wendisch, V. F., and Nampoothiri, K. M. (2016). Co-expression of endoglucanase and beta-glucosidase in Corynebacterium glutamicum DM1729 towards direct lysine fermentation from cellulose. Bioresour. Technol. 213, 239–244. doi: 10.1016/j.biortech.2016.03.019
Arndt, A., Auchter, M., Ishige, T., Wendisch, V. F., and Eikmanns, B. J. (2008). Ethanol catabolism in Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 15, 222–233. doi: 10.1159/000107370
Baritugo, K. A., Kim, H. T., David, Y., Khang, T. U., Hyun, S. M., Kang, K. H., et al. (2018). Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum strains from empty fruit bunch biosugar solution. Microb. Cell Fact. 17:129. doi: 10.1186/s12934-018-0977-9
Becker, E. W. (2007). Micro-algae as a source of protein. Biotechnol. Adv. 25, 207–210. doi: 10.1016/j.biotechadv.2006.11.002
Becker, J., Schafer, R., Kohlstedt, M., Harder, B. J., Borchert, N. S., Stöveken, N., et al. (2013). Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb. Cell Fact. 12:110. doi: 10.1186/1475-2859-12-110
Brüsseler, C., Spath, A., Sokolowsky, S., and Marienhagen, J. (2019). Alone at last! - heterologous expression of a single gene is sufficient for establishing the five-step Weimberg pathway in Corynebacterium glutamicum. Metab. Eng. Commun. 9:e00090. doi: 10.1016/j.mec.2019.e00090
Burgardt, A., Prell, C., and Wendisch, V. F. (2021). Utilization of a wheat sidestream for 5-aminovalerate production in Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 9:732271. doi: 10.3389/fbioe.2021.732271
Burmeister, A., Akhtar, Q., Hollmann, L., Tenhaef, N., Hilgers, F., Hogenkamp, F., et al. (2021). (Optochemical) control of synthetic microbial coculture interactions on a microcolony level. ACS Synth. Biol. 10, 1308–1319. doi: 10.1021/acssynbio.0c00382
Buschke, N., Becker, J., Schafer, R., Kiefer, P., Biedendieck, R., and Wittmann, C. (2013). Systems metabolic engineering of xylose-utilizing Corynebacterium glutamicum for production of 1,5-diaminopentane. Biotechnol. J. 8, 557–570. doi: 10.1002/biot.201200367
Buschke, N., Schroder, H., and Wittmann, C. (2011). Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol. J. 6, 306–317.
Chakrabarti, R. (2002). Carotenoprotein from tropical brown shrimp shell waste by enzymatic process. Food Biotechnol. 16, 81–90.
Chen, J., Wang, Y., Guo, X., Rao, D., Zhou, W., Zheng, P., et al. (2020). Efficient bioproduction of 5-aminolevulinic acid, a promising biostimulant and nutrient, from renewable bioresources by engineered Corynebacterium glutamicum. Biotechnol. Biofuels 13:41. doi: 10.1186/s13068-020-01685-0
Chen, S., Liao, W., Liu, C., Wen, Z., Kincaid, R. L., Harrison, J. H., et al. (2003). Value-Added Chemicals from Animal Manure. Washington, DC: Pacific Northwest National Laboratory.
Chen, Z., Liu, G., Zhang, J., and Bao, J. (2019). A preliminary study on L-lysine fermentation from lignocellulose feedstock and techno-economic evaluation. Bioresour. Technol. 271, 196–201. doi: 10.1016/j.biortech.2018.09.098
Cho, J. S., Choi, K. R., Prabowo, C. P. S., Shin, J. H., Yang, D., Jang, J., et al. (2017). CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab. Eng. 42, 157–167. doi: 10.1016/j.ymben.2017.06.010
Choi, J. W., Yim, S. S., Lee, S. H., Kang, T. J., Park, S. J., and Jeong, K. J. (2015). Enhanced production of gamma-aminobutyrate (GABA) in recombinant Corynebacterium glutamicum by expressing glutamate decarboxylase active in expanded pH range. Microb. Cell Fact. 14:21. doi: 10.1186/s12934-015-0205-9
Clomburg, J. M., Crumbley, A. M., and Gonzalez, R. (2017). Industrial biomanufacturing: the future of chemical production. Science 355:aag0804. doi: 10.1126/science.aag0804
Deng, J.-J., Zhang, M.-S., Li, Z.-W., Lu, D.-L., Mao, H.-H., Zhu, M.-J., et al. (2020). One-step processing of shrimp shell waste with a chitinase fused to a carbohydrate-binding module. Green Chem. 22, 6862–6873.
Dev, S., and Bhattacharya, J. (2014). Use of marine waste extract as a nitrogen source for biological sulfate reduction: development of a suitable alternative. Mine Water Environ. 33, 362–371.
Eberhardt, D., Jensen, J. V., and Wendisch, V. F. (2014). L-citrulline production by metabolically engineered Corynebacterium glutamicum from glucose and alternative carbon sources. AMB Express 4:85. doi: 10.1186/s13568-014-0085-0
Evonik (2019). Evonik. Available Online at: https://corporate.evonik.com/en/media/press_releases/pages/article.aspx?articleId=106336 [accessed March 3, 2019].
Feng, L., Zhang, Y., Fu, J., Mao, Y., Chen, T., Zhao, X., et al. (2016). Metabolic engineering of Corynebacterium glutamicum for efficient production of 5-aminolevulinic acid. Biotechnol. Bioeng. 113, 1284–1293.
Frese, M., and Sewald, N. (2015). Enzymatic halogenation of tryptophan on a gram scale. Angew. Chem. Int. Ed Engl. 54, 298–301. doi: 10.1002/anie.201408561
Georgi, T., Engels, V., and Wendisch, V. F. (2008). Regulation of L-lactate utilization by the FadR-type regulator LldR of Corynebacterium glutamicum. J. Bacteriol. 190, 963–971. doi: 10.1128/JB.01147-07
Gerstmeir, R., Wendisch, V. F., Schnicke, S., Ruan, H., Farwick, M., Reinscheid, D., et al. (2003). Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104, 99–122.
Giesselmann, G., Dietrich, D., Jungmann, L., Kohlstedt, M., Jeon, E. J., Yim, S. S., et al. (2019). Metabolic engineering of Corynebacterium glutamicum for high-level ectoine production: design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway. Biotechnol. J. 14:e1800417. doi: 10.1002/biot.201800417
Haupka, C., Delepine, B., Irla, M., Heux, S., and Wendisch, V. F. (2020). Flux enforcement for fermentative production of 5-aminovalerate and glutarate by Corynebacterium glutamicum. Catalysts 10:1065.
Henke, N. A., Wiebe, D., Pérez-García, F., Peters-Wendisch, P., and Wendisch, V. F. (2018). Coproduction of cell-bound and secreted value-added compounds: simultaneous production of carotenoids and amino acids by Corynebacterium glutamicum. Bioresour. Technol. 247, 744–752. doi: 10.1016/j.biortech.2017.09.167
Hennig, G., Haupka, C., Brito, L. F., Ruckert, C., Cahoreau, E., Heux, S., et al. (2020). Methanol-essential growth of Corynebacterium glutamicum: adaptive laboratory evolution overcomes limitation due to methanethiol assimilation pathway. Int. J. Mol. Sci. 21:3617. doi: 10.3390/ijms21103617
Hicks, T. M., and Verbeek, C. J. R. (2017). Chapter 1 - protein-rich by-products: production statistics, legislative restrictions, and management options. Protein Byproducts 2016, 1–18.
Hietala, D. C., Godwin, C. M., Cardinale, B. J., and Savage, P. E. (2019). The independent and coupled effects of feedstock characteristics and reaction conditions on biocrude production by hydrothermal liquefaction. Appl. Energy 235, 714–728.
Hoffmann, S. L., Jungmann, L., Schiefelbein, S., Peyriga, L., Cahoreau, E., Portais, J. C., et al. (2018). Lysine production from the sugar alcohol mannitol: design of the cell factory Corynebacterium glutamicum SEA-3 through integrated analysis and engineering of metabolic pathway fluxes. Metab. Eng. 47, 475–487. doi: 10.1016/j.ymben.2018.04.019
Hu, Y., Feng, S., Yuan, Z., Xu, C. C., and Bassi, A. (2017). Investigation of aqueous phase recycling for improving bio-crude oil yield in hydrothermal liquefaction of algae. Bioresour. Technol. 239, 151–159. doi: 10.1016/j.biortech.2017.05.033
Huo, Y. X., Cho, K. M., Rivera, J. G., Monte, E., Shen, C. R., Yan, Y., et al. (2011). Conversion of proteins into biofuels by engineering nitrogen flux. Nat. Biotechnol. 29, 346–351. doi: 10.1038/nbt.1789
Hyslop, J. F., Lovelock, S. L., Watson, A. J. B., Sutton, P. W., and Roiban, G. D. (2019). N-Alkyl-alpha-amino acids in nature and their biocatalytic preparation. J. Biotechnol. 293, 56–65. doi: 10.1016/j.jbiotec.2019.01.006
Ikeda, N., Miyamoto, M., Adachi, N., Nakano, M., Tanaka, T., and Kondo, A. (2013). Direct cadaverine production from cellobiose using beta-glucosidase displaying Escherichia coli. AMB Express 3:67. doi: 10.1186/2191-0855-3-67
Imao, K., Konishi, R., Kishida, M., Hirata, Y., Segawa, S., Adachi, N., et al. (2017). 1,5-Diaminopentane production from xylooligosaccharides using metabolically engineered Corynebacterium glutamicum displaying beta-xylosidase on the cell surface. Bioresour. Technol. 245, 1684–1691. doi: 10.1016/j.biortech.2017.05.135
Irla, M., Heggeset, T. M., Naerdal, I., Paul, L., Haugen, T., Le, S. B., et al. (2016). Genome-based genetic tool development for Bacillus methanolicus: theta- and rolling circle-replicating plasmids for inducible gene expression and application to methanol-based cadaverine production. Front. Microbiol. 7:1481. doi: 10.3389/fmicb.2016.01481
Jensen, J. V., Eberhardt, D., and Wendisch, V. F. (2015). Modular pathway engineering of Corynebacterium glutamicum for production of the glutamate-derived compounds ornithine, proline, putrescine, citrulline, and arginine. J. Biotechnol. 214, 85–94. doi: 10.1016/j.jbiotec.2015.09.017
Jensen, J. V., and Wendisch, V. F. (2013). Ornithine cyclodeaminase-based proline production by Corynebacterium glutamicum. Microb. Cell Fact. 12:63. doi: 10.1186/1475-2859-12-63
Joo, J. C., Oh, Y. H., Yu, J. H., Hyun, S. M., Khang, T. U., Kang, K. H., et al. (2017). Production of 5-aminovaleric acid in recombinant Corynebacterium glutamicum strains from a Miscanthus hydrolysate solution prepared by a newly developed Miscanthus hydrolysis process. Bioresour. Technol. 245, 1692–1700. doi: 10.1016/j.biortech.2017.05.131
Jorge, J. M., Leggewie, C., and Wendisch, V. F. (2016). A new metabolic route for the production of gamma-aminobutyric acid by Corynebacterium glutamicum from glucose. Amino Acids 48, 2519–2531. doi: 10.1007/s00726-016-2272-6
Jorge, J. M., Nguyen, A. Q., Pérez-García, F., Kind, S., and Wendisch, V. F. (2017a). Improved fermentative production of gamma-aminobutyric acid via the putrescine route: systems metabolic engineering for production from glucose, amino sugars, and xylose. Biotechnol. Bioeng. 114, 862–873. doi: 10.1002/bit.26211
Jorge, J. M. P., Pérez-García, F., and Wendisch, V. F. (2017b). A new metabolic route for the fermentative production of 5-aminovalerate from glucose and alternative carbon sources. Bioresour. Technol. 245, 1701–1709. doi: 10.1016/j.biortech.2017.04.108
Kawaguchi, H., Sasaki, K., Uematsu, K., Tsuge, Y., Teramura, H., Okai, N., et al. (2015). 3-Amino-4-hydroxybenzoic acid production from sweet sorghum juice by recombinant Corynebacterium glutamicum. Bioresour. Technol. 198, 410–417. doi: 10.1016/j.biortech.2015.09.024
Kawaguchi, H., Vertes, A. A., Okino, S., Inui, M., and Yukawa, H. (2006). Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72, 3418–3428. doi: 10.1128/AEM.72.5.3418-3428.2006
Kerbs, A., Mindt, M., Schwardmann, L., and Wendisch, V. F. (2021). Sustainable production of N-methylphenylalanine by reductive methylamination of phenylpyruvate using engineered Corynebacterium glutamicum. Microorganisms 9:824. doi: 10.3390/microorganisms9040824
Kiefer, D., Merkel, M., Lilge, L., Hausmann, R., and Henkel, M. (2021). High cell density cultivation of Corynebacterium glutamicum on bio-based lignocellulosic acetate using pH-coupled online feeding control. Bioresour. Technol. 340:125666. doi: 10.1016/j.biortech.2021.125666
Kim, H. T., Baritugo, K.-A., Oh, Y. H., Hyun, S. M., Khang, T. U., Kang, K. H., et al. (2018). Metabolic engineering of Corynebacterium glutamicum for the high-level production of cadaverine that can be used for the synthesis of biopolyamide 510. ACS Sustain. Chem. Eng. 6, 5296–5305.
Kim, S. J., Hyeon, J. E., Jeon, S. D., Choi, G. W., and Han, S. O. (2014). Bi-functional cellulases complexes displayed on the cell surface of Corynebacterium glutamicum increase hydrolysis of lignocelluloses at elevated temperature. Enzyme Microb. Technol. 66, 67–73. doi: 10.1016/j.enzmictec.2014.08.010
Kim, Y. M., Kwak, M. H., Kim, H. S., and Lee, J. H. (2019). Production of indole-3-acetate in Corynebacterium glutamicum by heterologous expression of the indole-3-pyruvate pathway genes. Microbiol. Biotechnol. Lett. 47, 242–249. doi: 10.4014/mbl.1901.01013
Kind, S., Jeong, W. K., Schroder, H., and Wittmann, C. (2010). Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab. Eng. 12, 341–351. doi: 10.1016/j.ymben.2010.03.005
Kind, S., Kreye, S., and Wittmann, C. (2011). Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab. Eng. 13, 617–627. doi: 10.1016/j.ymben.2011.07.006
Kind, S., Neubauer, S., Becker, J., Yamamoto, M., Volkert, M., Abendroth, G., et al. (2014). From zero to hero - production of bio-based nylon from renewable resources using engineered Corynebacterium glutamicum. Metab. Eng. 25, 113–123. doi: 10.1016/j.ymben.2014.05.007
Ko, Y. J., You, S. K., Kim, M., Lee, E., Shin, S. K., Park, H. M., et al. (2019). Enhanced production of 5-aminolevulinic acid via flux redistribution of TCA cycle toward L-glutamate in Corynebacterium glutamicum. Biotechnol. Bioprocess Eng. 24, 915–923.
Krahn, I., Bonder, D., Torregrosa-Barragan, L., Stoppel, D., Krause, J. P., Rosenfeldt, N., et al. (2021). Evolving a new efficient mode of fructose utilization for improved bioproduction in Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 9:669093. doi: 10.3389/fbioe.2021.669093
Laslo, T., Von Zaluskowski, P., Gabris, C., Lodd, E., Rückert, C., Dangel, P., et al. (2012). Arabitol metabolism of Corynebacterium glutamicum and its regulation by AtlR. J. Bacteriol. 194, 941–955. doi: 10.1128/JB.06064-11
Lee, J. H., and Wendisch, V. F. (2017a). Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J. Biotechnol. 257, 211–221. doi: 10.1016/j.jbiotec.2016.11.016
Lee, J. H., and Wendisch, V. F. (2017b). Production of amino acids - genetic and metabolic engineering approaches. Bioresour. Technol. 245, 1575–1587. doi: 10.1016/j.biortech.2017.05.065
Lei, M., Peng, X., Sun, W., Zhang, D., Wang, Z., Yang, Z., et al. (2021). Nonsterile L-lysine fermentation using engineered phosphite-grown Corynebacterium glutamicum. ACS Omega 6, 10160–10167. doi: 10.1021/acsomega.1c00226
Leng, L., Zhang, W., Peng, H., Li, H., Jiang, S., and Huang, H. (2020). Nitrogen in bio-oil produced from hydrothermal liquefaction of biomass: a review. Chem. Eng. J. 401:126030.
Lessmeier, L., Pfeifenschneider, J., Carnicer, M., Heux, S., Portais, J. C., and Wendisch, V. F. (2015). Production of carbon-13-labeled cadaverine by engineered Corynebacterium glutamicum using carbon-13-labeled methanol as co-substrate. Appl. Microbiol. Biotechnol. 99, 10163–10176. doi: 10.1007/s00253-015-6906-5
Li, M., Li, D., Huang, Y., Liu, M., Wang, H., Tang, Q., et al. (2014). Improving the secretion of cadaverine in Corynebacterium glutamicum by cadaverine-lysine antiporter. J. Ind. Microbiol. Biotechnol. 41, 701–709. doi: 10.1007/s10295-014-1409-4
Liu, M., Liu, H., Shi, M., Jiang, M., Li, L., and Zheng, Y. (2021). Microbial production of ectoine and hydroxyectoine as high-value chemicals. Microb. Cell Fact. 20:76. doi: 10.1186/s12934-021-01567-6
Liu, R. L., Yu, P., Ge, X. L., Bai, X. F., Li, X. Q., and Fu, Q. (2016). Establishment of an aqueous PEG 200-based deep eutectic solvent extraction and enrichment method for pumpkin (Cucurbita moschata) seed protein. Food Anal. Methods 10, 1669–1680.
Lubitz, D., Jorge, J. M., Pérez-García, F., Taniguchi, H., and Wendisch, V. F. (2016). Roles of export genes cgmA and lysE for the production of L-arginine and L-citrulline by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 100, 8465–8474.
Malakoutikhah, M., Prades, R., Teixido, M., and Giralt, E. (2010). N-methyl phenylalanine-rich peptides as highly versatile blood-brain barrier shuttles. J. Med. Chem. 53, 2354–2363. doi: 10.1021/jm901654x
Matano, C., Uhde, A., Youn, J. W., Maeda, T., Clermont, L., Marin, K., et al. (2014). Engineering of Corynebacterium glutamicum for growth and L-lysine and lycopene production from N-acetyl-glucosamine. Appl. Microbiol. Biotechnol. 98, 5633–5643. doi: 10.1007/s00253-014-5676-9
Mehrabadi, A., Craggs, R., and Farid, M. M. (2016). Biodiesel production potential of wastewater treatment high rate algal pond biomass. Bioresour. Technol. 221, 222–233. doi: 10.1016/j.biortech.2016.09.028
Meiswinkel, T. M., Gopinath, V., Lindner, S. N., Nampoothiri, K. M., and Wendisch, V. F. (2013a). Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb. Biotechnol. 6, 131–140. doi: 10.1111/1751-7915.12001
Meiswinkel, T. M., Rittmann, D., Lindner, S. N., and Wendisch, V. F. (2013b). Crude glycerol-based production of amino acids and putrescine by Corynebacterium glutamicum. Bioresour. Technol. 145, 254–258. doi: 10.1016/j.biortech.2013.02.053
Meyer, F., Keller, P., Hartl, J., Groninger, O. G., Kiefer, P., and Vorholt, J. A. (2018). Methanol-essential growth of Escherichia coli. Nat. Commun. 9:1508. doi: 10.1038/s41467-018-03937-y
Mihara, H., Muramatsu, H., Kakutani, R., Yasuda, M., Ueda, M., Kurihara, T., et al. (2005). N-methyl-L-amino acid dehydrogenase from Pseudomonas putida. A novel member of an unusual NAD(P)-dependent oxidoreductase superfamily. FEBS J. 272, 1117–1123. doi: 10.1111/j.1742-4658.2004.04541.x
Mimitsuka, T., Sawai, H., Hatsu, M., and Yamada, K. (2007). Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci. Biotechnol. Biochem. 71, 2130–2135.
Mindt, M., Hannibal, S., Heuser, M., Risse, J. M., Sasikumar, K., Nampoothiri, K. M., et al. (2019a). Fermentative production of N-alkylated glycine derivatives by recombinant Corynebacterium glutamicum using a mutant of imine reductase DpkA from Pseudomonas putida. Front. Bioeng. Biotechnol. 7:232. doi: 10.3389/fbioe.2019.00232
Mindt, M., Heuser, M., and Wendisch, V. F. (2019b). Xylose as preferred substrate for sarcosine production by recombinant Corynebacterium glutamicum. Bioresour. Technol. 281, 135–142. doi: 10.1016/j.biortech.2019.02.084
Mindt, M., Risse, J. M., Gruss, H., Sewald, N., Eikmanns, B. J., and Wendisch, V. F. (2018). One-step process for production of N-methylated amino acids from sugars and methylamine using recombinant Corynebacterium glutamicum as biocatalyst. Sci. Rep. 8:12895. doi: 10.1038/s41598-018-31309-5
Mindt, M., Walter, T., Kugler, P., and Wendisch, V. F. (2020). Microbial engineering for production of N-functionalized amino acids and amines. Biotechnol. J. 15:1900451. doi: 10.1002/biot.201900451
Müller, J. E., Heggeset, T. M., Wendisch, V. F., Vorholt, J. A., and Brautaset, T. (2015). Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol. Appl. Microbiol. Biotechnol. 99, 535–551. doi: 10.1007/s00253-014-6224-3
Muramatsu, H., Mihara, H., Kakutani, R., Yasuda, M., Ueda, M., Kurihara, T., et al. (2005). The putative malate/lactate dehydrogenase from Pseudomonas putida is an NADPH-dependent delta1-piperideine-2-carboxylate/delta1-pyrroline-2-carboxylate reductase involved in the catabolism of D-lysine and D-proline. J. Biol. Chem. 280, 5329–5335. doi: 10.1074/jbc.M411918200
Naik, S. N., Goud, V. V., Rout, P. K., and Dalai, A. K. (2010). Production of first and second generation biofuels: a comprehensive review. Renew. Sustain. Energy Rev. 14, 578–597.
Nguyen, A. Q., Schneider, J., and Wendisch, V. F. (2015). Elimination of polyamine N-acetylation and regulatory engineering improved putrescine production by Corynebacterium glutamicum. J. Biotechnol. 201, 75–85. doi: 10.1016/j.jbiotec.2014.10.035
Oh, Y. H., Choi, J. W., Kim, E. Y., Song, B. K., Jeong, K. J., Park, K., et al. (2015). Construction of synthetic promoter-based expression cassettes for the production of cadaverine in recombinant Corynebacterium glutamicum. Appl. Biochem. Biotechnol. 176, 2065–2075. doi: 10.1007/s12010-015-1701-4
Okai, N., Takahashi, C., Hatada, K., Ogino, C., and Kondo, A. (2014). Disruption of pknG enhances production of gamma-aminobutyric acid by Corynebacterium glutamicum expressing glutamate decarboxylase. AMB Express 4:20. doi: 10.1186/s13568-014-0020-4
Oyebola, O. O., and Yildiz, F. (2016). Nutritional quality of wastes emanating from processing of genotypes of freshwater Prawn, Macrobrachium vollenhovenii. Cogent Food Agric. 2:1268287.
Park, S. J., Oh, Y. H., Noh, W., Kim, H. Y., Shin, J. H., Lee, E. G., et al. (2014). High-level conversion of L-lysine into 5-aminovalerate that can be used for nylon 6,5 synthesis. Biotechnol. J. 9, 1322–1328. doi: 10.1002/biot.201400156
Payne, J. T., Poor, C. B., and Lewis, J. C. (2015). Directed evolution of RebH for site-selective halogenation of large biologically active molecules. Angew. Chem. Int. Ed Engl. 54, 4226–4230. doi: 10.1002/anie.201411901
Pérez-García, F., Brito, L. F., and Wendisch, V. F. (2019). Function of L-pipecolic acid as compatible solute in Corynebacterium glutamicum as basis for its production under hyperosmolar conditions. Front. Microbiol. 10:340. doi: 10.3389/fmicb.2019.00340
Pérez-García, F., Burgardt, A., Kallman, D. R., Wendisch, V. F., and Bar, N. (2021). Dynamic co-cultivation process of Corynebacterium glutamicum strains for the fermentative production of riboflavin. Fermentation 7:11.
Pérez-García, F., Peters-Wendisch, P., and Wendisch, V. F. (2016). Engineering Corynebacterium glutamicum for fast production of L-lysine and L-pipecolic acid. Appl. Microbiol. Biotechnol. 100, 8075–8090.
Pérez-García, F., Risse, J. M., Friehs, K., and Wendisch, V. F. (2017a). Fermentative production of L-pipecolic acid from glucose and alternative carbon sources. Biotechnol. J. 12:1600646. doi: 10.1002/biot.201600646
Pérez-García, F., Ziert, C., Risse, J. M., and Wendisch, V. F. (2017b). Improved fermentative production of the compatible solute ectoine by Corynebacterium glutamicum from glucose and alternative carbon sources. J. Biotechnol. 258, 59–68. doi: 10.1016/j.jbiotec.2017.04.039
Phongthai, S., Lim, S.-T., and Rawdkuen, S. (2016). Optimization of microwave-assisted extraction of rice bran protein and its hydrolysates properties. J. Cereal Sci. 70, 146–154.
Pleissner, D., and Lin, C. S. K. (2013). Valorisation of food waste in biotechnological processes. Sustain. Chem. Process. 1:21.
Polen, T., Schluesener, D., Poetsch, A., Bott, M., and Wendisch, V. F. (2007). Characterization of citrate utilization in Corynebacterium glutamicum by transcriptome and proteome analysis. FEMS Microbiol. Lett. 273, 109–119. doi: 10.1111/j.1574-6968.2007.00793.x
Prell, C., Busche, T., Ruckert, C., Nolte, L., Brandenbusch, C., and Wendisch, V. F. (2021). Adaptive laboratory evolution accelerated glutarate production by Corynebacterium glutamicum. Microb. Cell Fact. 20:97. doi: 10.1186/s12934-021-01586-3
Radek, A., Krumbach, K., Gatgens, J., Wendisch, V. F., Wiechert, W., Bott, M., et al. (2014). Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of D-xylose containing substrates. J. Biotechnol. 192(Pt A), 156–160. doi: 10.1016/j.jbiotec.2014.09.026
Ramzi, A. B., Hyeon, J. E., Kim, S. W., Park, C., and Han, S. O. (2015). 5-Aminolevulinic acid production in engineered Corynebacterium glutamicum via C5 biosynthesis pathway. Enzyme Microb. Technol. 81, 1–7. doi: 10.1016/j.enzmictec.2015.07.004
Rittmann, D., Lindner, S. N., and Wendisch, V. F. (2008). Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl. Environ. Microbiol. 74, 6216–6222. doi: 10.1128/AEM.00963-08
Rohles, C. M., Giesselmann, G., Kohlstedt, M., Wittmann, C., and Becker, J. (2016). Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate. Microb. Cell Fact. 15:154. doi: 10.1186/s12934-016-0553-0
Sasikumar, K., Hannibal, S., Wendisch, V. F., and Nampoothiri, K. M. (2021). Production of biopolyamide precursors 5-amino valeric acid and putrescine from rice straw hydrolysate by engineered Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 9:635509. doi: 10.3389/fbioe.2021.635509
Schneider, J., Eberhardt, D., and Wendisch, V. F. (2012). Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system. Appl. Microbiol. Biotechnol. 95, 169–178. doi: 10.1007/s00253-012-3956-9
Schneider, J., Niermann, K., and Wendisch, V. F. (2011). Production of the amino acids L-glutamate, L-lysine, L-ornithine and L-arginine from arabinose by recombinant Corynebacterium glutamicum. J. Biotechnol. 154, 191–198. doi: 10.1016/j.jbiotec.2010.07.009
Schneider, J., and Wendisch, V. F. (2010). Putrescine production by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 88, 859–868. doi: 10.1007/s00253-010-2778-x
Scott, E., Peter, F., and Sanders, J. (2007). Biomass in the manufacture of industrial products–the use of proteins and amino acids. Appl. Microbiol. Biotechnol. 75, 751–762. doi: 10.1007/s00253-007-0932-x
Seibold, G., Auchter, M., Berens, S., Kalinowski, J., and Eikmanns, B. J. (2006). Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J. Biotechnol. 124, 381–391. doi: 10.1016/j.jbiotec.2005.12.027
Sgobba, E., Blöbaum, L., and Wendisch, V. F. (2018a). Production of food and feed additives from non-food-competing feedstocks: valorizing N-acetylmuramic acid for amino acid and carotenoid fermentation with Corynebacterium glutamicum. Front. Microbiol. 9:2046. doi: 10.3389/fmicb.2018.02046
Sgobba, E., Stumpf, A. K., Vortmann, M., Jagmann, N., Krehenbrink, M., Dirks-Hofmeister, M. E., et al. (2018b). Synthetic Escherichia coli-Corynebacterium glutamicum consortia for L-lysine production from starch and sucrose. Bioresour. Technol. 260, 302–310. doi: 10.1016/j.biortech.2018.03.113
Shahidi, F. (1994). “Flavor of meat and meat products—an overview,” in Flavor of Meat and Meat Products, ed. F. Shahidi (Boston, MA: Springer), 1–3.
Shakya, R., Adhikari, S., Mahadevan, R., Shanmugam, S. R., Nam, H., Hassan, E. B., et al. (2017). Influence of biochemical composition during hydrothermal liquefaction of algae on product yields and fuel properties. Bioresour. Technol. 243, 1112–1120.
Shi, F., and Li, Y. (2011). Synthesis of gamma-aminobutyric acid by expressing Lactobacillus brevis-derived glutamate decarboxylase in the Corynebacterium glutamicum strain ATCC 13032. Biotechnol. Lett. 33, 2469–2474. doi: 10.1007/s10529-011-0723-4
Shi, F., Zhang, M., and Li, Y. (2017). Overexpression of ppc or deletion of mdh for improving production of γ-aminobutyric acid in recombinant Corynebacterium glutamicum. World J. Microbiol. Biotechnol. 33:122.
Shin, J. H., Park, S. H., Oh, Y. H., Choi, J. W., Lee, M. H., Cho, J. S., et al. (2016). Metabolic engineering of Corynebacterium glutamicum for enhanced production of 5-aminovaleric acid. Microb. Cell Fact. 15:174.
Sinner, P., Stiegler, M., Herwig, C., and Kager, J. (2021). Noninvasive online monitoring of Corynebacterium glutamicum fed-batch bioprocesses subject to spent sulfite liquor raw material uncertainty. Bioresour. Technol. 321:124395. doi: 10.1016/j.biortech.2020.124395
Suzuki, H., Ohnishi, Y., Furusho, Y., Sakuda, S., and Horinouchi, S. (2006). Novel benzene ring biosynthesis from C(3) and C(4) primary metabolites by two enzymes. J. Biol. Chem. 281, 36944–36951. doi: 10.1074/jbc.M608103200
Takahashi, C., Shirakawa, J., Tsuchidate, T., Okai, N., Hatada, K., Nakayama, H., et al. (2012). Robust production of gamma-amino butyric acid using recombinant Corynebacterium glutamicum expressing glutamate decarboxylase from Escherichia coli. Enzyme Microb. Technol. 51, 171–176. doi: 10.1016/j.enzmictec.2012.05.010
Talekar, S., Patti, A. F., Singh, R., Vijayraghavan, R., and Arora, A. (2018). From waste to wealth: high recovery of nutraceuticals from pomegranate seed waste using a green extraction process. Ind. Crops Prod. 112, 790–802.
Tateno, T., Okada, Y., Tsuchidate, T., Tanaka, T., Fukuda, H., and Kondo, A. (2009). Direct production of cadaverine from soluble starch using Corynebacterium glutamicum coexpressing alpha-amylase and lysine decarboxylase. Appl. Microbiol. Biotechnol. 82, 115–121.
Tuveri, A., Pérez-García, F., Lira-Parada, P. A., Imsland, L., and Bar, N. (2021). Sensor fusion based on extended and unscented kalman filter for bioprocess monitoring. J. Process Control 106, 195–207. doi: 10.1016/j.jprocont.2021.09.005
Tuyishime, P., Wang, Y., Fan, L., Zhang, Q., Li, Q., Zheng, P., et al. (2018). Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production. Metab. Eng. 49, 220–231. doi: 10.1016/j.ymben.2018.07.011
Uhde, A., Brühl, N., Goldbeck, O., Matano, C., Gurow, O., Rückert, C., et al. (2016). Transcription of sialic acid catabolism genes in Corynebacterium glutamicum is subject to catabolite repression and control by the transcriptional repressor NanR. J. Bacteriol. 198, 2204–2218. doi: 10.1128/JB.00820-15
Uhde, A., Youn, J. W., Maeda, T., Clermont, L., Matano, C., Krämer, R., et al. (2013). Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 97, 1679–1687. doi: 10.1007/s00253-012-4313-8
Veldmann, K. H., Dachwitz, S., Risse, J. M., Lee, J. H., Sewald, N., and Wendisch, V. F. (2019a). Bromination of L-tryptophan in a fermentative process with Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 7:219. doi: 10.3389/fbioe.2019.00219
Veldmann, K. H., Minges, H., Sewald, N., Lee, J. H., and Wendisch, V. F. (2019b). Metabolic engineering of Corynebacterium glutamicum for the fermentative production of halogenated tryptophan. J. Biotechnol. 291, 7–16. doi: 10.1016/j.jbiotec.2018.12.008
Vivek, N., Hazeena, S. H., Alphy, M. P., Kumar, V., Magdouli, S., Sindhu, R., et al. (2021). Recent advances in microbial biosynthesis of C3-C5 diols: genetics and process engineering approaches. Bioresour. Technol. 322:124527. doi: 10.1016/j.biortech.2020.124527
Vortmann, M., Stumpf, A. K., Sgobba, E., Dirks-Hofmeister, M. E., Krehenbrink, M., Wendisch, V. F., et al. (2021). A bottom-up approach towards a bacterial consortium for the biotechnological conversion of chitin to L-lysine. Appl. Microbiol. Biotechnol. 105, 1547–1561. doi: 10.1007/s00253-021-11112-5
Walter, T., Al Medani, N., Burgardt, A., Cankar, K., Ferrer, L., Kerbs, A., et al. (2020). Fermentative N-methylanthranilate production by engineered Corynebacterium glutamicum. Microorganisms 8:866.
Wang, Y., Fan, L., Tuyishime, P., Liu, J., Zhang, K., Gao, N., et al. (2020). Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum. Commun. Biol. 3:217. doi: 10.1038/s42003-020-0954-9
Wen, J., and Bao, J. (2021). Improved fermentative gamma-aminobutyric acid production by secretory expression of glutamate decarboxylase by Corynebacterium glutamicum. J. Biotechnol. 331, 19–25. doi: 10.1016/j.jbiotec.2021.03.003
Wen, L., Álvarez, C., Zhang, Z., Poojary, M. M., Lund, M. N., Sun, D.-W., et al. (2020). Optimisation and characterisation of protein extraction from coffee silverskin assisted by ultrasound or microwave techniques. Biomass Convers. Biorefin. 11, 1575–1585.
Wendisch, V. F. (2020). Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 58, 17–34. doi: 10.1016/j.ymben.2019.03.008
Wendisch, V. F., Brito, L. F., Gil Lopez, M., Hennig, G., Pfeifenschneider, J., Sgobba, E., et al. (2016). The flexible feedstock concept in industrial biotechnology: metabolic engineering of Escherichia coli, Corynebacterium glutamicum, Pseudomonas, Bacillus and yeast strains for access to alternative carbon sources. J. Biotechnol. 234, 139–157. doi: 10.1016/j.jbiotec.2016.07.022
Wendisch, V. F., and Lee, J. H. (2020). Metabolic Engineering in Corynebacterium glutamicum. Cham: Springer.
Wendisch, V. F., Mindt, M., and Pérez-García, F. (2018). Biotechnological production of mono- and diamines using bacteria: recent progress, applications, and perspectives. Appl. Microbiol. Biotechnol. 102, 3583–3594. doi: 10.1007/s00253-018-8890-z
Wolf, S., Becker, J., Tsuge, Y., Kawaguchi, H., Kondo, A., Marienhagen, J., et al. (2021). Advances in metabolic engineering of Corynebacterium glutamicum to produce high-value active ingredients for food, feed, human health, and well-being. Essays Biochem. 65, 197–212. doi: 10.1042/EBC20200134
Xu, Y., Li, Y., Bao, T., Zheng, X., Chen, W., and Wang, J. (2017). A recyclable protein resource derived from cauliflower by-products: potential biological activities of protein hydrolysates. Food Chem. 221, 114–122. doi: 10.1016/j.foodchem.2016.10.053
Yang, J., He, Q., Niu, H., Corscadden, K., and Astatkie, T. (2018). Hydrothermal liquefaction of biomass model components for product yield prediction and reaction pathways exploration. Appl. Energy 228, 1618–1628.
Yang, P., Liu, W., Cheng, X., Wang, J., Wang, Q., and Qi, Q. (2016). A new strategy for production of 5-aminolevulinic acid in recombinant Corynebacterium glutamicum with high yield. Appl. Environ. Microbiol. 82, 2709–2717. doi: 10.1128/AEM.00224-16
Youn, J. W., Jolkver, E., Kramer, R., Marin, K., and Wendisch, V. F. (2009). Characterization of the dicarboxylate transporter DctA in Corynebacterium glutamicum. J. Bacteriol. 191, 5480–5488. doi: 10.1128/JB.00640-09
Zhang, B., Gao, G., Chu, X. H., and Ye, B. C. (2019). Metabolic engineering of Corynebacterium glutamicum S9114 to enhance the production of L-ornithine driven by glucose and xylose. Bioresour. Technol. 284, 204–213. doi: 10.1016/j.biortech.2019.03.122
Keywords: nitrogen valorization, agriculture sidestreams, aquaculture sidestreams, Corynebacterium, biorefinery, circular bioeconomy, metabolic engineering
Citation: Wendisch VF, Nampoothiri KM and Lee J-H (2022) Metabolic Engineering for Valorization of Agri- and Aqua-Culture Sidestreams for Production of Nitrogenous Compounds by Corynebacterium glutamicum. Front. Microbiol. 13:835131. doi: 10.3389/fmicb.2022.835131
Received: 14 December 2021; Accepted: 13 January 2022;
Published: 08 February 2022.
Edited by:
João M. P. Jorge, Universidade Nova de Lisboa, PortugalReviewed by:
Xiaoqiang Ma, Singapore-MIT Alliance for Research and Technology (SMART), SingaporeCopyright © 2022 Wendisch, Nampoothiri and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Volker F. Wendisch, dm9sa2VyLndlbmRpc2NoQHVuaS1iaWVsZWZlbGQuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.