
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 19 May 2022
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.825389
This article is part of the Research TopicCarbapenemases in Gram-negative Bacteria: A Global Health Threat and Therapeutic ChallengeView all 19 articles
 Xinhua Luo1†
Xinhua Luo1† Kai Mu2,3†
Kai Mu2,3† Yujie Zhao4
Yujie Zhao4 Jin Zhang1
Jin Zhang1 Ying Qu1
Ying Qu1 Dakang Hu1
Dakang Hu1 Yifan Jia1
Yifan Jia1 Piaopiao Dai1
Piaopiao Dai1 Jian Weng5
Jian Weng5 Dongguo Wang6*†
Dongguo Wang6*† Lianhua Yu1*
Lianhua Yu1*To demonstrate the detailed genetic characteristics of a blaNDM–1-carrying multidrug-resistant Aeromonas caviae strain, the complete genome of the A. caviae strain K433 was sequenced by Illumina HiSeq and Oxford nanopore platforms, and mobile genetic elements associated with antibiotic resistance genes were analyzed by a series of bioinformatics methods. A. caviae K433 which was determined to produce class B carbapenemase, was resistant to most antibiotics tested except amikacin. The genome of K433 consisted of a chromosome cK433 (6,482-kb length) and two plasmids: pK433-qnrS (7.212-kb length) and pK433-NDM (200.855-kb length), the last being the first investigated blaNDM-carrying plasmid from Aeromonas spp. By comparison of the backbone and MDR regions from the plasmids studied, they involved a highly homologous sequence structure. This study provides in-depth genetic insights into the plasmids integrated with blaNDM-carrying genetic elements from Aeromonas spp.
Aeromonas spp. was first recognized as a human pathogen in 1954 when it was isolated from a blood sample (Parker and Shaw, 2011). In the following years, there were more confirmed cases of Aeromonas spp. causing human infections with varying degrees of severity, mainly, gastroenteritis (Parker and Shaw, 2011). Aeromonas spp. is ubiquitous in water, which can form biofilms, and then colonize the water system, drinking water may be a potential source of infection. Aeromonas spp. was mainly found in marine environments and freshwater (Figueira et al., 2011; Martino et al., 2014), and their spread is related to contact and ingestion of contaminated water or food.
In 2018, Aeromonas spp. was investigated in a wastewater treatment plant effluent in Tokyo, Japan, and two strains harboring the blaKPC–2 gene were detected (Sekizuka et al., 2019). Besides KPC, Aeromonas caviae producing VIM was reported in an Israeli hospital in 2014 (Adler et al., 2014), Aeromonas hydrophila carrying GES-24 carbapenemase was discovered in 2018 from a hospitalized patient in Okinawa, Japan (Uechi et al., 2018), and A. caviae from India was confirmed to carry OXA-181-carbapenemase (Anandan et al., 2017). Aeromonas spp. simultaneously harboring blaCTX–M–15, blaSHV–12, blaPER–1, and blaFOX–2, was isolated from Adriatic Sea of Croatia (Maravić et al., 2013). In the past 10 years, carbapenemase-producing bacteria have been isolated from non-human sources, including the aquatic environment. The carbapenemase-producing bacteria from the aquatic environment are particularly susceptible to human activities. Bidirectional movement of the carbapenemase-producing bacteria between the aquatic environment and humans has been occurring all the time (Hammer-Dedet et al., 2020). The fewest members of metallo-beta-lactamases B2 are composed of different species of Aeromonas, such as A. hydrophila, Aeromonas Veronii, and Serratia Fonticola, named CphA, ImiS, and SFH-I in the literature, respectively (Mojica et al., 2022). Just due to the pooling of carbapenemase-producing Aeromonas spp. from water, acquired resistance genes appear from time-to-time in the clinic, which deserves attention.
Although the aforementioned genes involving carbapenemase or other beta-lactamase were reported in Aeromonas spp., blaNDM has not been reported in A. caviae to date. Since the blaNDM gene was first discovered in India in 2009 (Yong et al., 2009), it has rapidly spread all over the world (Dortet et al., 2014). Although blaNDM was originally determined in a Klebsiella pneumoniae plasmid (Yong et al., 2009), it has also been reported in the recent years that blaNDM has been found in the chromosomes of Enterobacteriaceae (Girlich et al., 2015; Shen et al., 2017; Sakamoto et al., 2018; Reynolds et al., 2019; Kong et al., 2020). The strains carrying metallo-beta-lactamases are capable of hydrolyzing all beta-lactam antibiotics except aztreonam, which has raised great concerns worldwide.
In this work, we first discovered a multidrug-resistant A. caviae strain carrying blaNDM–1. The whole genome of the strain was sequenced and the mobile genetic elements of the strain containing drug-resistant genes were thoroughly and genetically studied.
A. caviae strain K433 was isolated from a patient’s sputum in the Taizhou Municipal Hospital affiliated with the Taizhou University of China in 2018. EC600 (highly resistant to rifampicin) and Escherichia coli DH5α were used as hosts for conjugal and plasmid transfers, respectively. Strain K433 was initially identified by Vitek 2. Later, it was confirmed by PCR amplification and sequencing of 16S rRNA with primers: Forward, 5′-AGAGTTTGATCATGGCTCAG-3′; Reverse: 5′-GGTTACCTTGTTACGACTT-3′ (Demarta et al., 1999). Moreover, bacterial species identification was also performed using genome sequence-based average nucleotide identity (ANI) analysis1 (Richter and Rosselló-Móra, 2009).
The activities of class A serine carbapenemase and class B metallo β-lactamase could be suppressed by 3-aminophenyl boronic acid (APB) and ethylenediamine tetra-acetic acid (EDTA) (Pournaras et al., 2013). We chose APB combined with EDTA to detect the carbapenemase of strain K433 according to the previous report (Tsakris et al., 2010).
The interpretation of the results was as follows: (1) if the diameter of the inhibition zone of the imipenem disc with APB solution differs from that of the single-imipenem disc by ≥5 mm, it could be judged that the tested strain produced class A carbapenemase; (2) if the diameter of the inhibition zone of the imipenem disc with EDTA solution differed from that of the single-imipenem disc by ≥5 mm, it might be that the tested strain produced class B carbapenemase; (3) If APB + EDTA were added concurrently, the diameters of the inhibition zone of the imipenem discs with APB + EDTA differed from that of the single-imipenem disc by ≥5 mm, it could be confirmed that the tested strain simultaneously produced class A carbapenemase + class B metallo β-lactamase; (4) if the difference between the inhibition zone diameter of the imipenem disc containing enzyme inhibitor and the single-imipenem disc was less than 5 mm, it could be determined that the bacteria did not produce class A carbapenemase or class B metallo β-lactamase.
The method used for testing bacterial resistance was BioMérieux VITEK2, and the results were determined in accordance with the 2020 Clinical and Laboratory Standards Association (CLSI) guidelines (Clinical and Laboratory Standards Institute [CLSI], 2020).
12 antibiotics, namely, cefepime, aztreonam, imipenem, meropenem, amikacin, ciprofloxacin, levofloxacin, tigecycline, minocycline, tigecycline/clavulanic acid, and piperacillin/tazobactam, were tested. E. coli ATCC 25922 was used as the quality control strain.
Bacterial plasmid DNA of strain K433 was extracted using a plasmid extraction kit (TaKaRa, Dalian, China) in accordance with the manufacturer’s instructions. The plasmid was transferred in an attempt from the A. caviae K433 isolate into EC600 and E. coli DH5a through conjugal transfer and electroporation, respectively. For the selection of transconjugants and/or transformants containing the blaNDM marker, 2 μg/ml imipenem and 1,000 μg/ml rifampicin were used according to specific circumstances.
Genomic DNA was extracted from strain K433 using a Gentra Puregene Yeast/Bact. Kit (Qiagen, Valencia, CA, United States). Libraries were prepared separately using the TruePrepTM DNA Library Prep Kit V2 and the SQU-LSK109 Ligation Sequencing kit. After the preparation of the library was completed, it was separately sequenced on an Illumina HiSeq X Ten platform (Illumina Inc., San Diego, CA, United States) and GridION X5 platform (Oxford Nanopore, United Kingdom). To improve the reliability of data processing, raw data from the HiSeq X Ten platform and the GridION X5 platform were trimmed to obtain the high-quality clean reads (clean data) by Canu v1.8.2. The paired-end short Illumina reads and the long Nanopore reads were ‘‘de novo’’ assembled using Unicycler v0.4.5.3
Open reading frames and pseudogenes were predicted using RAST2.0 (Brettin et al., 2015), BLASTP/BLASTN (Boratyn et al., 2013), UniProtKB/Swiss-Prot (Boutet et al., 2016), and RefSeq databases (O’Leary et al., 2016). Annotation of drug resistance genes, mobile genetic elements, and other features were performed using online databases, such as CARD (Liang et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and the Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN. The genome map was drawn using Inkscape 0.48.1.4
Nucleotide sequence accession numbers for chromosome K433 (ck433), plasmid K433-qnrS (pK433-qnrS), and plasmid K433-NDM (pK433-NDM) were CP084031, OK017455, and OK287926, respectively.
It was collected for comparative analysis between pK433-NDM and related plasmids, including p13ZX28-272, p13ZX28-TC-98, p13ZX36-200, pCP077202, pCP077203, and pCP077204, which nucleotide sequence accession numbers were MN101850, MN101852, MN101853, CP077202, CP077203, and CP077204, respectively.
Through the 16S rRNA sequence and genome sequence-based ANI analysis, strain K433 was identified to be A. caviae eventually. The results of the antimicrobial susceptibility tests on strain K433 were shown in Table 1. Through detection of enzymatic properties, the strain K433 was confirmed to harbor only class B metallo β-lactamase. After bacterial conjugative transfer and electroporation assays, no transconjugant or transformant carrying pK433-NDM could be recovered despite repeated trials.
Strain K433 carried a 6,482-kb-long chromosome cK433, a 200.855-kb-long plasmid pK433-NDM, and a 7.212-kb-long plasmid pK433-qnrS (Supplementary Table 1). Plasmid pK433-NDM involved the region of blaMOX–6 gene, and a 42.3-kb-long MDR region where blaNDM was inserted (Supplementary Figure 1). Plasmid pK433-qnrS only contained drug-resistance gene qnrS2 (Supplementary Figure 2). All resistance genes were listed in Table 2.
Integrative and mobilizable elements (IMEs) were extremely closely related to the acquisition or loss of bacterial resistance to antibiotics (Bellanger et al., 2014; Delavat et al., 2017). Three IMEs were found on cK433, including IME1, IME2, and IME3 regions (Figure 1).
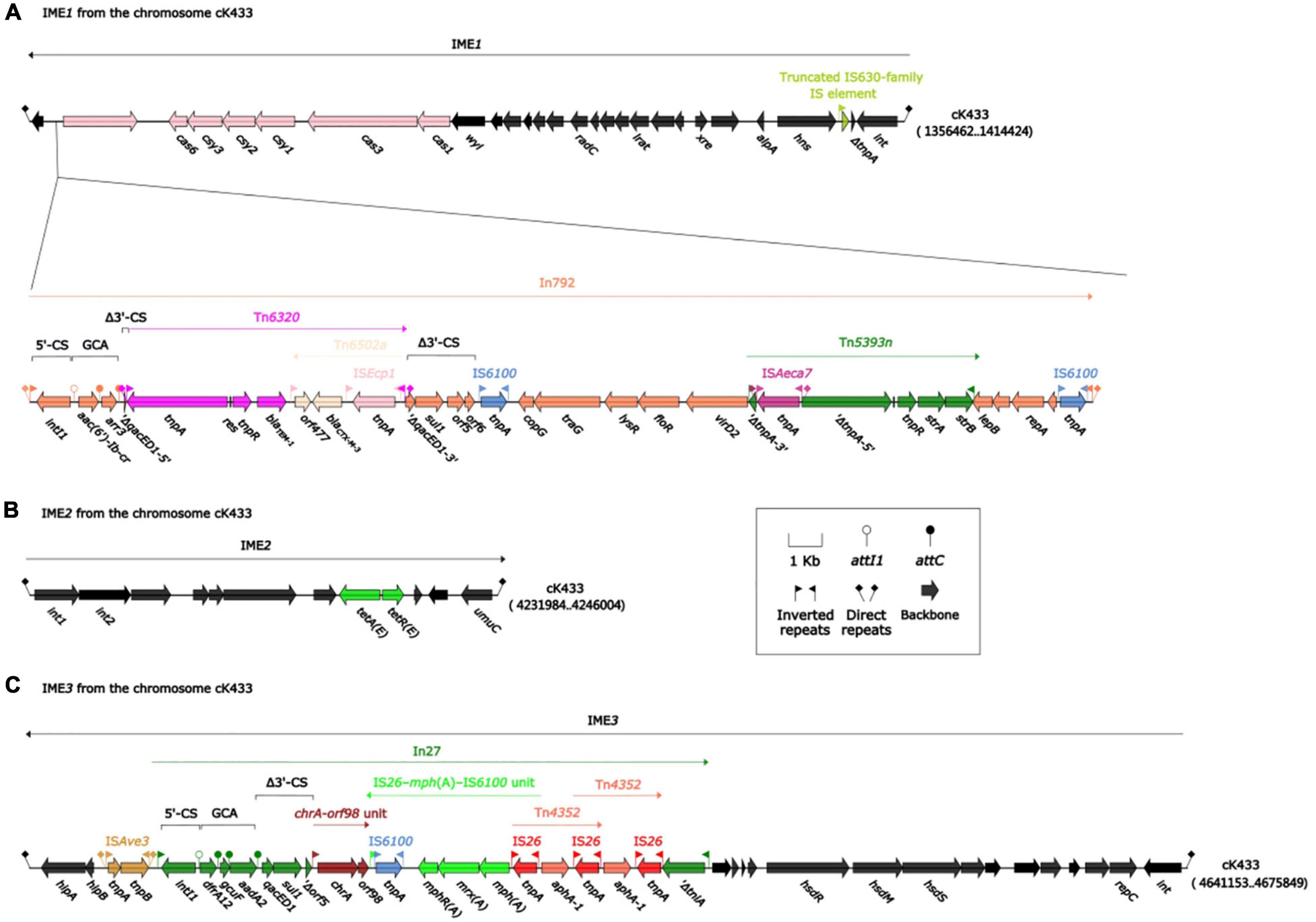
Figure 1. Mobile genetic elements associated with resistant genes on chromosome cK433. IMEs were abbreviation integrative and mobilizable elements. Three IMEs (IME1, IME2, and IME3) were to be discovered on chromosome cK433. (A) IME1, comprising the backbone region, cas-csy module and In792; (B) IME2, consisting of backbone region and tetA-tetR module; (C) IME3, involving the backbone region, ISAve3 and In27.
IME1, flanked by a pair of attL/attR (14 bp in length), had a backbone (containing int) with insertion of two accessory modules: 43.9-kb strAB–blaCTX–M–3 region and truncated IS630-family IS element. The 43.9-kb strAB–blaCTX–M–3 region, including In792 [gene cassette array (GCA): aac(6’)-Ib-cr–arr3], was inserted between the orf339 and wyl gene at the left end of the backbone region, and truncated IS630-family IS element was inserted between the hns and orf114 gene at the right end of the backbone region (Figure 1A). Meanwhile, the unit transposon Tn6320 (carrying blaTEM–1 and blaCTX–M–3) was inserted into the qacED1 gene of In792. Tn5393n was inserted between the virD2 and lepB gene at the right end of In792, following, ISAeca7 was inserted into ΔtnpA gene, which was divided into two parts on the left end of Tn5393n, then, two identical IS6100s were inserted between the 3′-CS and the right end of In792, forming the current complex IME1 structure just like “Russian nesting dolls.”
IME2 consisted of the backbone region and tetA-tetR module which was related to tetracycline drug resistance (Figure 1B). IME3 contained the backbone region, ISAve3 and In27 [GCA: dfrA12–gcuF–aadA2] which was truncated by chrA-orf98 unit, IS26-mph(A)-IS6100 unit, and two intersecting Tn4352 (Figure 1C).
According to the BLASTN alignments of the complete sequence of plasmid pK433-NDM in the NCBI GenBank database, we found that the top three plasmids ranked by coverage value were pCP077202 (59%), pCP077203 (26%), and pCP077204 (24%), and their identities were both 100%. These three plasmids (pCP077202, pCP077203, and pCP077204) collected from GenBank all belonged to Aeromonas spp. in the United States and had a close correlation with plasmid pK433-NDM. Plasmids pCP077202, pCP077203, and pCP077204 were from the same strain with 161.381-, 85.67-, and 80.98-kb length, respectively. The sequence composition and structure of pK433-NDM and pCP077203 were highly similar (>95% identity) around the first 35 kb length in the plasmid maintenance region (Figure 2). Both plasmids pK433-NDM and pCP077202 contained the MDR region, in which there was also a high similarity with the composition and structure located on the MDR region upstream and downstream of the plasmid maintenance regions (>95% identity) (Figure 2). The comparison of MDR regions for 42.3 kb long pK433-NDM and 40.2 kb long pCP077202 is illustrated in Figure 3. The composition and structure of the sequence approximate 32 kb long on the left end of plasmid maintenance regions of pK433-NDM (23111.55424) and pCP077204 (5096.36653) were also highly similar (>95% identity). However, no plasmid replication gene was found in plasmid pCP077204 and pCP077202.
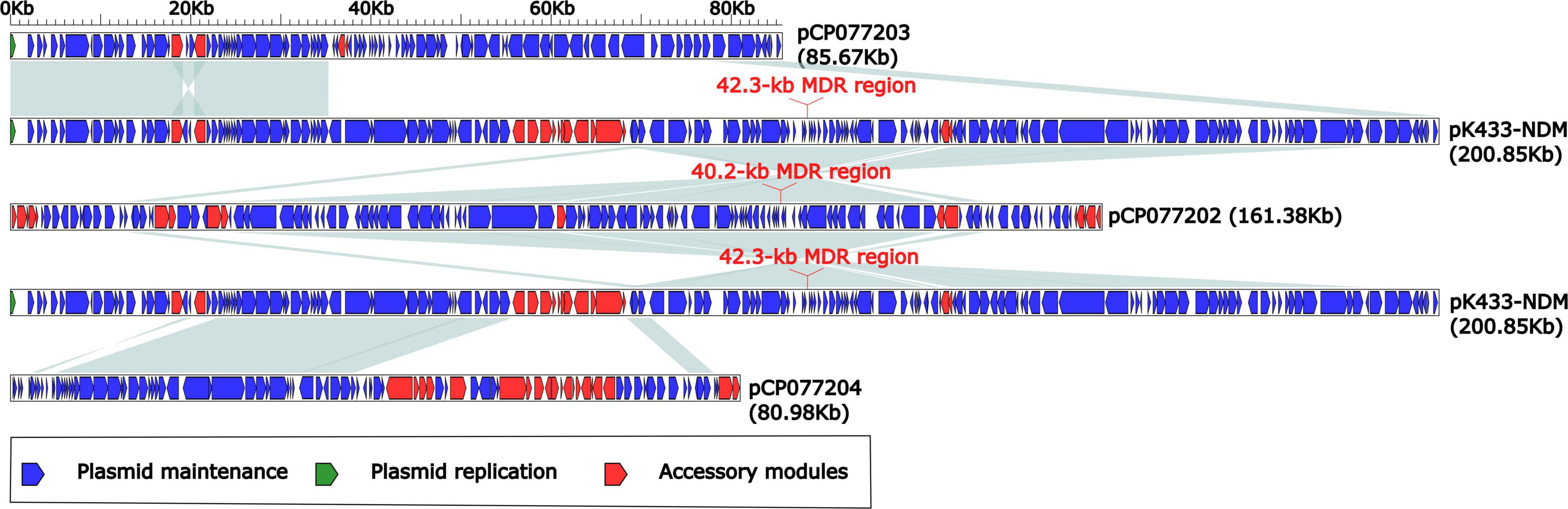
Figure 2. Comparison of plasmids pK433-NDM, pCP077202, pCP077203, and pCP077204. Plasmids pCP077202, pCP077203, and pCP077204 were obtained from GenBank, which came from Aeromonas spp. in the United States. Plasmids pCP077202, pCP077203, and pCP077204 had 161.381, 85.67, and 80.98 kb lengths, respectively. The shadow of light blue represented >95% identity.

Figure 3. Comparison of MDR regions from plasmids pK433-NDM, pCO077202, pKP-14-6-NDM-1, p13ZX28-272, p13ZX36-200, and p13ZX28-TC-98. pK433-NDM and pCO077202 came from Aeromonas spp. pKP-14-6-NDM-1 was isolated from Klebsiella pneumoniae, and p13ZX28-272, p13ZX36-200, and p13ZX28-TC-98 were all achieved from Escherichia coli.
All the aforementioned plasmids except pK433-NDM were obtained from GenBank. pKP-14-6-NDM-1 was isolated from K. pneumoniae and p13ZX28-272, p13ZX36-200, and p13ZX28-TC-98 were all achieved from E. coli. The coverage and identity of the MDR region from aforementioned plasmids were listed in Supplementary Table 2. Compared with MDR regions from plasmids pK433-NDM and pCP077202, it seemed that In37 [Variable region 1 (VR1) containing aacA4cr, blaOXA–1, and catB1 and VR2 containing blaPER–1] of MDR region from pCP077202 was replaced by In384 [VR1 containing dfrA12, VR2 containing blaNDM–1 and bleMBL, and VR3 containing blaOXA] of MDR region from pK433-NDM, and the remaining regions had a high degree of identity (>95%) with MDR region from pK433-NDM. (Figure 3). However, the MDR region from pK433-NDM carried the blaNDM–1 gene located in the truncated composite transposon Tn125, while the MDR region from pCP077202 did not, which was the significant difference between them. Compared with MDR regions from pK433-NDM and pK-14-6-NDM-1, both involved blaNDM gene and were highly consistent with aaC2-tmrB region and In384 (>95% identity), and also ΔTn21, chrA-orf98 unit and IS26-mph(A)-IS6100 unit (>95% identity). Compared with MDR regions from pK-14-6-NDM-1 and p13ZX28-272, the regions containing blaNDM gene showed high consistency except for the insertion type of integron (In469 from p13ZX28-272 replaced by In384 from pK-14-6-NDM-1). Compared with p13ZX28-272, p13ZX36-200, and p13ZX28-TC-98, MDR regions of pK-14-6-NDM-1 and p13ZX28-272 showed the highest identity (>95%) but revealed different coverage, which was listed in Supplementary Table 2. Interestingly, In469 and In384 in pK433-NDM, pK-14-6-NDM-1, p13ZX28-272, p13ZX36-200, and p13ZX28-TC-98 all contained the identical ISCR1 and ΔTn125 structure, which suggested ISCR1 prompted the accumulation of ΔTn125 between these plasmids.
Various types of antibiotic resistance genes have been discovered over and over again in Aeromonas spp. from nature, which is commonly resistant to quinolone and β-lactam drugs (Piotrowska et al., 2017). It is very likely that Aeromonas spp. is naturally an important repository of acquired β-lactamase genes from wastewater or sludge, which was to be found in plentiful genes harboring classes A, B, C, and D β-lactamase (Piotrowska et al., 2017). However, only a tiny amount of class B carbapenemases were found, such as AsbM1, IMP-19, VIM, ImiS, ImiH, and CphA (Janda and Abbott, 2010; Piotrowska et al., 2017). So far, NDM had never been reported. To our knowledge, this is the first study involving NDM carbapenemase from an A. caviae strain (K433), which was isolated from inpatient’s source with multidrug-resistance in our hospital. This study not only provided the first evidence of nosocomial infection and colonization of an NDM-producing A. caviae, but also revealed the strong transmission ability of NDM.
Reported firstly in 2009, NDM-1 has caused a major public health problem because of its high resistance profile to carbapenems and its global prevalence (Yong et al., 2009). To date, 40 variants of NDM carbapenemases have been reported.5 The bacterial strains harboring blaNDM exhibited significantly increased MICs for carbapenems, cephalosporins, penicillins, ticarcilin/clavulanic acid, and piperacillin/tazobactam except for aztreonam, just as shown in the susceptibility test of A. caviae K433 (Table 1). Strain K433 was resistant to almost all the antibiotics (including imipenem, meropenem, and tigecycline) except amikacin. Among the reported mechanisms of tigecycline resistance, the bacterial efflux pump system plays a major role. The overexpression of characteristic efflux pumps AdeABC, AdeFGH, and AdeIJK, together with the deletion and mutation of the two-component regulatory systems adeR and adeS, can lead to tigecycline resistance (Nguyen et al., 2014). In addition, the reasons for the decreased sensitivity to tigecycline include the inactivation of tigecycline by the modification enzyme Tet(X), the alteration of the cell membrane permeability because of the mutation of the plsC gene, and the decreased affinity between tigecycline and the ribosome due to the mutation of the rpsJ gene, etc. (Beabout et al., 2015). Recently, studies have reported that tigecycline resistance can be transmitted in bacteria by conjugation of plasmids carrying resistance genes (Partridge et al., 2018). In this study, we have not detected the tet(X) gene or other tigecycline resistance genes in strain K433. The possible mechanisms of tigecycline resistance in K433 were the overexpression of bacterial efflux pump system or/and the altering of cell membrane permeability, etc.
After high-throughput sequencing, it was determined that A. caviae K433 carried two plasmids: (pK433-NDM and pK433-qnrS), and one 6,482-kb-long chromosome cK433, carrying three IMEs: (IME1, IME2, and IME3) (Figure 1). IMEs and ICEs (integrative and conjugative elements) (Botelho and Schulenburg, 2021) are two different types of mobile genetic elements. They are often integrated into bacterial chromosomes to prompt the spread of resistance genes. IMEs cannot be self-transmitted, and they move between cells with the help of other conjugative elements that encode proteins involving in the complete conjugation function. IMEs usually have attL, int, rlx, oriT, and attR, but do not contain conjugative transfer genes (Luo et al., 2021). As for other properties of chromosome cK433, further study is needed.
It was utterly different between pK433-NDM and pK433-qnrS. Plasmid pK433-qnrS (7212 kb in length) had only qnrS2-repC-repA-mob gene cassettes (Supplementary Figure 2), while plasmid pK433-NDM (200.855 kb in length) possessed the backbone, including plasmid maintenance and replication regions, and variable regions: 42.3-kb MDR region, blaMOX–6 region, ISAeme19 and ISAS17 (Supplementary Figure 1). We speculated that such a length of plasmid and the complex structure of the MDR region may result in the failures of plasmid conjugative transfer and electroporation experiment for pK433-NDM. There were six units or modules in the MDR region from pK433-NDM, revealing Tn2, ΔTn21, IS26-mph(A)-IS6100 unit, chA-orf98 unit, In384, and aaC2-tmrB region (Figure 3). The biggest differences between the MDR regions from pCP077202 and pK433-NDM were that In384 from pK433-NDM replaced the position of In37 from pCP077202, and, In37 involved 2 variable regions (VR), In384 contained 3 variable regions, then, VR2 carried ΔTn125 with blaNDM (Figure 3). Such a complex plasmid structure would greatly enhance the resistance to the drugs, such as carbapenems, cephalosporins, and penicillins (Table 1). Except that the MDR region was somewhat comparable, the backbone regions of plasmids: pCP077202, pCP077203, and pCP077204 which came from the same strain were more or less identified with the pK433-NDM, but there were some repeat backbone regions between the pCP077202, pCP077203, and pCP077204 (Figure 2). In general, these plasmids from different Aeromonas spp. had more similar structures and compositions despite of coming from different countries, different times, and even different races (Figure 2). As for the comparative analysis of the MDR regions from the pK433-NDM, p13ZX28-272, p13ZX28TC-98, pKP14-6-NDM-1, and p13ZX36-200, we found that the MDR regions from different plasmids almost had the identical structure, harboring ΔTn125 with blaNDM gene. It suggested that after the In384 carrying ΔTn125 with blaNDM gene was replaced by the In469 which also carried the ΔTn125 with blaNDM gene, it might be evolved even more epidemic; simultaneously, we also speculated that part of the plasmid structure and composition of A. caviae cK433 might come from other popular plasmids, and there was a potential risk of transmission, which must be actively prevented.
This study characterized the genome structure and constitution of the blaNDM-carrying multidrug-resistant A. caviae strain K433. Plasmids pK433-NDM and pK433-qnrS and chromosome cK433 were discovered. In total, three drug-resistant-gene-associated IMEs (IME1, IME2, and IME3) were inserted into complex gene structures, including integrons, transposons, and other mobile genetic modules or units, and studied in cK433. Four plasmids: pK433-NDM, pCP077202, pCP077203, and pCP077204 were compared with the backbone and MDR regions. It showed a highly homologous sequence structure between pK433-NDM and plasmids from the same strain: pCP077202, pCP077203, and pCP077204, in the backbone regions. It also indicated a highly homologous sequence structure between the MDR regions of pK433-NDM, pCP077202, pKP-14-6-NDM-1, p13ZX28-272, p13ZX36-200, and p13ZX28-TC-98. This study would provide a further theoretical basis for genetic evolution for plasmids involving blaNDM-carrying genetic elements from Aeromonas spp.
The original contributions presented in the study are publicly available. This data can be found here: PRJNA765169, OK287926, and OK017455.
LY and DW conceptualized and designed the study. All authors participated and acquired the data. XL, KM, LY, and DW analyzed and interpreted the data. XL and KM drafted the manuscript. LY and DW critically revised the manuscript. All authors read and approved the final manuscript.
This work was supported by the Foundation of the Public Welfare Program of Natural Science Foundation of Zhejiang (LGF19H200006) and the Zhejiang Health Department of China (2020KY364).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.825389/full#supplementary-material
Adler, A., Assous, M. V., Paikin, S., Shulman, A., Miller-Roll, T., Hillel, S., et al. (2014). Emergence of VIM-producing Aeromonas caviae in Israeli hospitals. J. Antimicrob. Chemother. 69, 1211–1214. doi: 10.1093/jac/dkt505
Anandan, S., Gopi, R., Devanga Ragupathi, N. K., Muthuirulandi Sethuvel, D. P., Gunasekaran, P., Walia, K., et al. (2017). First report of bla(OXA-181)-mediated carbapenem resistance in Aeromonas caviae in association with pKP3-A: threat for rapid dissemination. J. Glob. Antimicrob. Resist. 10, 310–314. doi: 10.1016/j.jgar.2017.07.006
Beabout, K., Hammerstrom, T. G., Perez, A. M., Magalhães, B. F., Prater, A. G., Clements, T. P., et al. (2015). The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 59, 5561–5566. doi: 10.1128/aac.00547-15
Bellanger, X., Payot, S., Leblond-Bourget, N., and Guédon, G. (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 38, 720–760. doi: 10.1111/1574-6976.12058
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Botelho, J., and Schulenburg, H. (2021). The role of integrative and conjugative elements in antibiotic resistance evolution. Trends Microbiol. 29, 8–18. doi: 10.1016/j.tim.2020.05.011
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/swiss-prot, the manually annotated section of the uniprot knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Clinical and Laboratory Standards Institute [CLSI] (2020). Performance Standards for Antimicrobial Susceptibility Testing. 30 th ed. CLSI Supplement M100. Wayne, PA: CLSI.
Delavat, F., Miyazaki, R., Carraro, N., Pradervand, N., and van der Meer, J. R. (2017). The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 41, 512–537. doi: 10.1093/femsre/fux008
Demarta, A., Tonolla, M., Caminada, A. P., Ruggeri, N., and Peduzzi, R. (1999). Signature region within the 16S rDNA sequences of Aeromonas popoffii. FEMS Microbiol. Lett. 172, 239–246. doi: 10.1111/j.1574-6968.1999.tb13474.x
Dortet, L., Poirel, L., and Nordmann, P. (2014). Worldwide dissemination of the NDM-type carbapenemases in Gram-negative bacteria. Biomed Res. Int. 2014:249856. doi: 10.1155/2014/249856
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Figueira, V., Vaz-Moreira, I., Silva, M., and Manaia, C. M. (2011). Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 45, 5599–5611. doi: 10.1016/j.watres.2011.08.021
Girlich, D., Dortet, L., Poirel, L., and Nordmann, P. (2015). Integration of the blaNDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-PmPEL) in a Proteus mirabilis clinical isolate. J. Antimicrob. Chemother. 70, 98–102. doi: 10.1093/jac/dku371
Hammer-Dedet, F., Jumas-Bilak, E., and Licznar-Fajardo, P. (2020). The hydric environment: a hub for clinically relevant carbapenemase encoding genes. Antibiotics 9:699. doi: 10.3390/antibiotics9100699
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/cmr.00039-09
Kong, L. H., Xiang, R., Wang, Y. L., Wu, S. K., Lei, C. W., Kang, Z. Z., et al. (2020). Integration of the blaNDM-1 carbapenemase gene into a novel SXT/R391 integrative and conjugative element in Proteus vulgaris. J. Antimicrob. Chemother. 75, 1439–1442. doi: 10.1093/jac/dkaa068
Liang, Q., Yin, Z., Zhao, Y., Liang, L., Feng, J., Zhan, Z., et al. (2017). Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int. J. Antimicrob. Agents 49, 709–718. doi: 10.1016/j.ijantimicag.2017.01.021
Luo, X., Yin, Z., Zeng, L., Hu, L., Jiang, X., Jing, Y., et al. (2021). Chromosomal integration of huge and complex bla (NDM)-carrying genetic elements in Enterobacteriaceae. Front. Cell Infect. Microbiol. 11:690799. doi: 10.3389/fcimb.2021.690799
Maravić, A., Skoèibušić, M., Samanić, I., Fredotović, Z., Cvjetan, S., Jutronić, M., et al. (2013). Aeromonas spp. simultaneously harbouring bla(CTX-M-15), bla(SHV-12), bla(PER-1) and bla(FOX-2), in wild-growing Mediterranean mussel (Mytilus galloprovincialis) from Adriatic Sea Croatia. Int. J. Food Microbiol. 166, 301–308. doi: 10.1016/j.ijfoodmicro.2013.07.010
Martino, M. E., Fasolato, L., Montemurro, F., Novelli, E., and Cardazzo, B. (2014). Aeromonas spp.: ubiquitous or specialized bugs? Environ. Microbiol. 16, 1005–1018. doi: 10.1111/1462-2920.12215
Mojica, M. F., Rossi, M. A., Vila, A. J., and Bonomo, R. A. (2022). The urgent need for metallo-β-lactamase inhibitors: an unattended global threat. Lancet Infect. Dis. 22, e28–e34. doi: 10.1016/s1473-3099(20)30868-9
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nguyen, F., Starosta, A. L., Arenz, S., Sohmen, D., Dönhöfer, A., and Wilson, D. N. (2014). Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 395, 559–575. doi: 10.1515/hsz-2013-0292
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Parker, J. L., and Shaw, J. G. (2011). Aeromonas spp. clinical microbiology and disease. J. Infect. 62, 109–118. doi: 10.1016/j.jinf.2010.12.003
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31, e88–e17. doi: 10.1128/cmr.00088-17
Piotrowska, M., Przygodzińska, D., Matyjewicz, K., and Popowska, M. (2017). Occurrence and variety of β-lactamase genes among Aeromonas spp. isolated from urban wastewater treatment plant. Front. Microbiol. 8:863. doi: 10.3389/fmicb.2017.00863
Pournaras, S., Zarkotou, O., Poulou, A., Kristo, I., Vrioni, G., Themeli-Digalaki, K., et al. (2013). A combined disk test for direct differentiation of carbapenemase-producing enterobacteriaceae in surveillance rectal swabs. J. Clin. Microbiol. 51, 2986–2990. doi: 10.1128/jcm.00901-13
Reynolds, M. E., Phan, H. T. T., George, S., Hubbard, A. T. M., Stoesser, N., Maciuca, I. E., et al. (2019). Occurrence and characterization of Escherichia coli ST410 co-harbouring blaNDM-5, blaCMY-42 and blaTEM-190 in a dog from the UK. J Antimicrob Chemother 74, 1207–1211. doi: 10.1093/jac/dkz017
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Roberts, A. P., Chandler, M., Courvalin, P., Guédon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Sakamoto, N., Akeda, Y., Sugawara, Y., Takeuchi, D., Motooka, D., Yamamoto, N., et al. (2018). Genomic characterization of carbapenemase-producing Klebsiella pneumoniae with chromosomally carried bla (NDM-1). Antimicrob. Agents Chemother. 62, e1520–e1518. doi: 10.1128/aac.01520-18
Sekizuka, T., Inamine, Y., Segawa, T., Hashino, M., Yatsu, K., and Kuroda, M. (2019). Potential KPC-2 carbapenemase reservoir of environmental Aeromonas hydrophila and Aeromonas caviae isolates from the effluent of an urban wastewater treatment plant in Japan. Environ. Microbiol. Rep. 11, 589–597. doi: 10.1111/1758-2229.12772
Shen, P., Yi, M., Fu, Y., Ruan, Z., Du, X., Yu, Y., et al. (2017). Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J. Clin. Microbiol. 55, 199–205. doi: 10.1128/jcm.01581-16
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Tsakris, A., Poulou, A., Pournaras, S., Voulgari, E., Vrioni, G., Themeli-Digalaki, K., et al. (2010). A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J. Antimicrob. Chemother. 65, 1664–1671. doi: 10.1093/jac/dkq210
Uechi, K., Tada, T., Sawachi, Y., Hishinuma, T., Takaesu, R., Nakama, M., et al. (2018). A carbapenem-resistant clinical isolate of Aeromonas hydrophila in Japan harbouring an acquired gene encoding GES-24 β-lactamase. J. Med. Microbiol. 67, 1535–1537. doi: 10.1099/jmm.0.000842
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/aac.00774-09
Keywords: Aeromonas spp., IMEs, mobile genetic elements, blaNDM, multidrug resistance
Citation: Luo X, Mu K, Zhao Y, Zhang J, Qu Y, Hu D, Jia Y, Dai P, Weng J, Wang D and Yu L (2022) Emergence of blaNDM– 1-Carrying Aeromonas caviae K433 Isolated From Patient With Community-Acquired Pneumonia. Front. Microbiol. 13:825389. doi: 10.3389/fmicb.2022.825389
Received: 30 November 2021; Accepted: 16 February 2022;
Published: 19 May 2022.
Edited by:
Mary Marquart, University of Mississippi Medical Center, United StatesReviewed by:
Milena Dropa, University of São Paulo, BrazilCopyright © 2022 Luo, Mu, Zhao, Zhang, Qu, Hu, Jia, Dai, Weng, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lianhua Yu, eXVsaWFuaHVhNjRAMTI2LmNvbQ==; Dongguo Wang, d2RndHpzQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.