
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 November 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.1073243
This article is part of the Research Topic The Discovery of Bioactive Natural Products: The Isolation and Structural Elucidation, Genome Mining, OSMAC Strategy, and Biotransformation View all 15 articles
 Zong-Jie Wang1†
Zong-Jie Wang1† Xiaotong Liu1†
Xiaotong Liu1† Haibo Zhou1†
Haibo Zhou1† Yang Liu1
Yang Liu1 Lin Zhong1,2
Lin Zhong1,2 Xue Wang1
Xue Wang1 Qiang Tu1,2
Qiang Tu1,2 Liujie Huo1
Liujie Huo1 Fu Yan1
Fu Yan1 Lichuan Gu1
Lichuan Gu1 Rolf Müller3
Rolf Müller3 Youming Zhang1,2*
Youming Zhang1,2* Xiaoying Bian1*
Xiaoying Bian1* Xiaokun Xu1*
Xiaokun Xu1*Heterologous expression is an indispensable approach to exploiting natural products from phylogenetically diverse microbial communities. In this study, we constructed a heterologous expression system based on strain Burkholderia thailandensis E264 by deleting efflux pump genes and screening constitutive strong promoters. The biosynthetic gene cluster (BGC) of disorazol from Sorangium cellulosum So ce12 was expressed successfully with this host, and the yield of its product, disorazol F2, rather than A1, was improved to 38.3 mg/L by promoter substitution and insertion. In addition to the disorazol gene cluster, the BGC of rhizoxin from Burkholderia rhizoxinica was also expressed efficiently, whereas no specific peak was detected when shuangdaolide BGC from Streptomyces sp. B59 was transformed into the host. This system provides another option to explore natural products from different phylogenetic taxa.
Natural products and their derivatives are important sources of new drugs (Newman and Cragg, 2020). With the development of next-generation sequencing technologies, an increasing amount of genomic data is available in public databases, and bioinformatic analysis has indicated that most of them are unexplored (Paoli et al., 2022). More than 99% of microbial organisms are currently unculturable (Pace, 1997); therefore, exploiting these treasures has attracted the interest of researchers worldwide. Heterologous expression has been confirmed as an effective strategy for obtaining natural products from slow-growing or even unculturable organisms, as well as poorly explored organisms, due to the lack of efficient genetic manipulation tools (Huo et al., 2019; Paoli et al., 2022).
Disorazols are a class of macrocyclic polyketides discovered in the myxobacterial strain S. cellulosum So ce12 (Jansen, 1994), and the flagship compound, disorazol A1, can inhibit the proliferation of different cancer cell lines at close to picomolar levels by interfering with microtubular dynamics (Elnakady et al., 2004; Menchon et al., 2018). The disorazol gene cluster was identified in So ce12 by transposon mutagenesis (Carvalho et al., 2005; Kopp et al., 2005). It is composed of four clustered genes (disABCD) and unidentified genes encoding tailoring enzymes responsible for epoxidation and methylation, which may lie outside of the biosynthetic gene cluster (BGC). Among them, disABC encode hybrid trans-AT PKS/NRPS megaenzymes, and disD encodes a separate protein that contains an acyl transferase domain and a possible oxidoreductase domain (Figure 1A). In our previous study, the disorazol BGC cloned in a bacterial artificial chromosome library was reconstructed by Red/ET-mediated recombineering (Zhang et al., 1998) and expressed in the model myxobacterium Myxococcus xanthus DK1622, considering the difficulties of culture and genetic manipulation of the original producer (Tu et al., 2016). However, this heterologous host grows relatively slowly and the yield remains quite low (<1 mg/L).
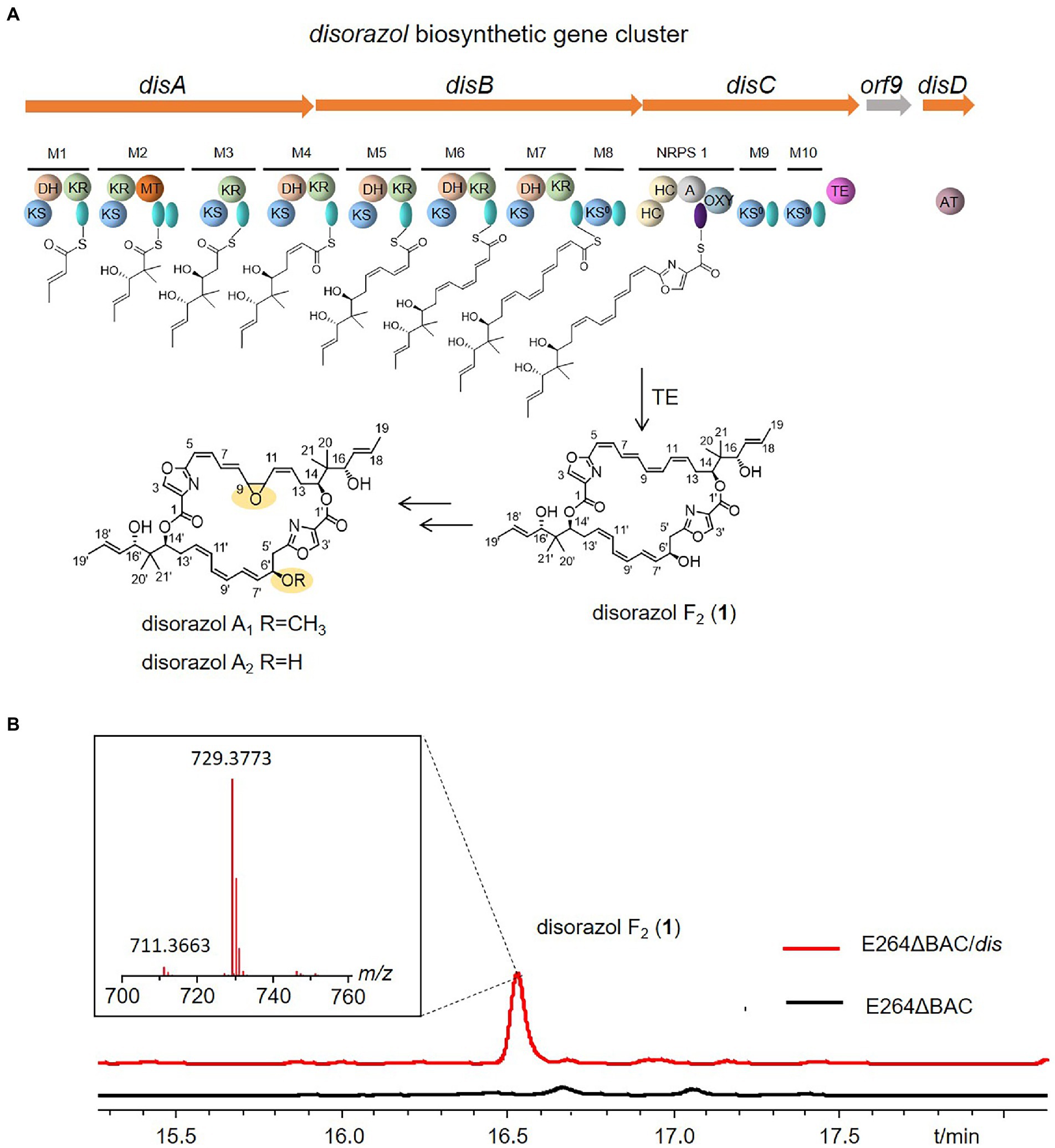
Figure 1. Domain organization of the disorazole biosynthetic gene cluster from Sorangium cellulosum So ce12 (A), and disorazol F2 produced by E264ΔBAC/dis (B). KS: ketosynthase, AT: acyl transferase, green oval: acyl carrier protein (ACP), HC: condensation/heterocyclization, A: adenylation, purple oval: peptidyl carrier protein (PCP), OXY: oxygenase, KR: ketoreductase, DH: dehydratase, MT: C-methyltransferase, TE: thioesterase,
To overcome this problem, the strains from Burkholderiales with high GC content and abundant secondary metabolites (Kunakom and Eustáquio, 2019) can be used to express the disorazol gene cluster, which was inspired by the success of employing Schlegelella brevitalea DSM 7029 (Tang et al., 2019) as an efficient heterologous host to produce myxobacteria-derived epothilones, vioprolides and myxochelins (Bian et al., 2017; Yan et al., 2018; Liu J. et al., 2021). Burkholderia thailandensis E264, isolated from rice fields in Thailand, is a low virulence member of genus Burkholderia compared with B. pseudomallei, and B. mallei. Although a few infection cases caused by B. thailandensis in immunocompromised patients were reported (Brett et al., 1998; Glass et al., 2006; Chang et al., 2017), it is extensively studied as a model organism of the factors controlling virulence or as a producer of secondary metabolites, such as the metabolite bactobolin produced by a hybrid PKS-NRPS gene cluster (Duerkop et al., 2009; Seyedsayamdost et al., 2010), thailandamide biosynthesized through PKS (Nguyen et al., 2008), and the acyldepsitripeptide histone deacetylase inhibitor burkholdac catalyzed by NRPS (Biggins et al., 2011). Abundant secondary metabolites indicated that it could provide abundant substrates for expressing heterologous BGCs. In addition to possessing huge biosynthetic potential, strain E264 also has a fast growth rate, which makes it more suitable as a heterologous host (Mao et al., 2017; Liu et al., 2022). However, as a member of the genus Burkholderia, the intrinsic multidrug-resistance of the wild-type strain E264 makes genetic manipulation difficult, which partially results from the resistance-nodulation-division family efflux pumps (Biot et al., 2011, 2013).
In this study, we first deleted multidrug-resistance genes and obtained several antibiotic-sensitive mutants. The native potent promoters in strain E264 were screened by transcriptome analysis and evaluated in vivo by cloning them into a promoterless luciferase reporter vector. Selected promoters were used to improve the yields of disorazol in this heterologous host. Finally, the heterologous expression system was used to express the gene clusters from other phylogenetic taxa. The BGC of rhizoxin cloned by Red/ET-mediated direct cloning (Fu et al., 2012) was expressed successfully, whereas no specific peak was detected in the fermentation of the heterologous host containing shuangdaolide BGC cloned by direct cloning from Streptomyces sp. B59.
During the course of expressing the disorazol gene cluster, B. thailandensis E264 showed better performance than the well-studied host DSM 7029 (Bian et al., 2017; Yan et al., 2018; Liu J. et al., 2021) in terms of growth rate and production. To facilitate genetic manipulation, three efflux pumps in strain E264, AmrAB-OprA (abbreviated as A), BpeAB-OprB (B), and BpeEF-OprC (C) (Biot et al., 2011, 2013), were inactivated by homologous recombination combined with pheS counterselection mediated seamless gene deletions (Barrett et al., 2008), and mutants with one, two, or three pump knockouts were obtained. Antibiotic susceptibility determined by the disc diffusion test was shown in tables S4 and S5. For most antibiotics, the mutant with one pump knockout (E264ΔA, E264ΔB, or E264ΔC) showed improved antibiotic sensitivity compared to the wild-type, suggesting that they are involved in multidrug resistance. Therefore, the mutant with three pump knockouts (E264ΔBAC) was constructed and used to express gene clusters or characterize promoters in subsequent experiments.
Compared with the wild-type, E264ΔBAC was more sensitive to the tested antibiotics, except ampicillin (Figure 2A, Table 1). The minimum inhibitory concentrations (MIC) of the tested antibiotics against mutant E264ΔBAC were much lower than the concentrations used to select transformants (Supplementary Table S6). The MIC value (<7.5 μg/ml) of kanamycin against the mutant was much lower than the concentration (300 μg/ml) used to select transformants of the wild type (Kang et al., 2011). The MIC values of tetracycline, chloramphenicol, and erythromycin are lower than 0.6, 1.9, and 3.1 μg/ml, respectively. The antibiotic-sensitive mutant E264ΔBAC facilitates the genetic engineering of this potential heterologous host with low-concentration antibiotics as a selectable marker. In addition, the growth rate of the mutant E264ΔBAC was not affected by engineering, and the growth curve of the mutant was similar to that of the wild-type strain E264, except that the maximum OD600 was slightly lower (Figure 2B).
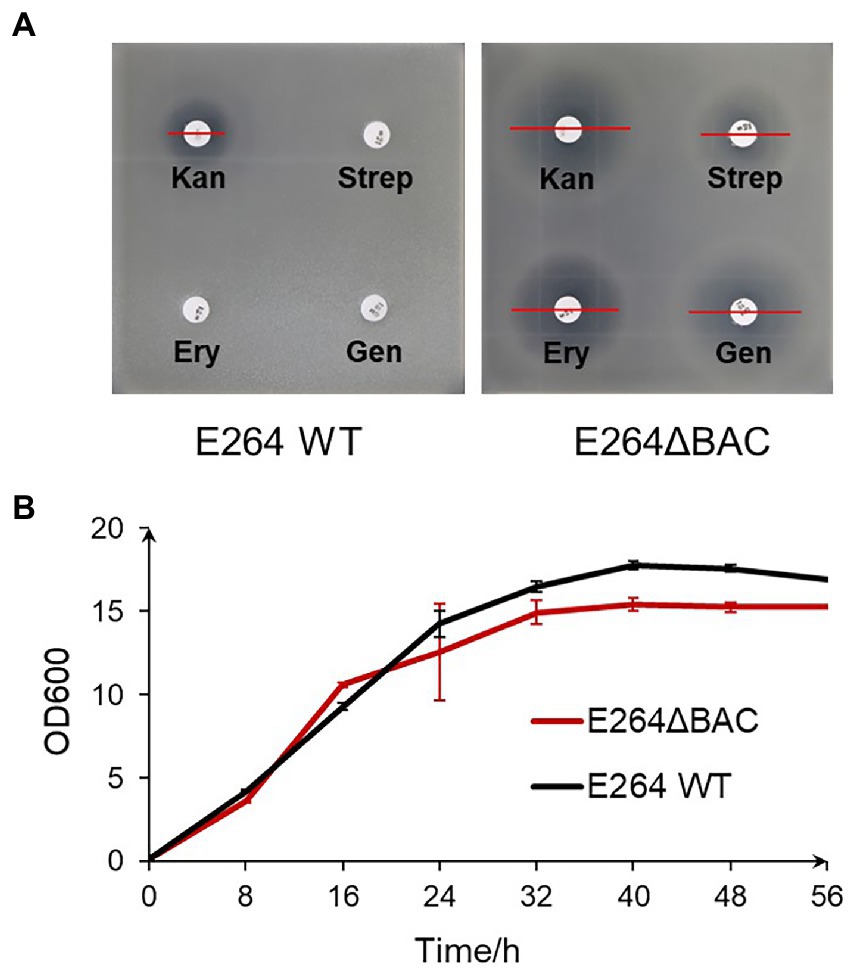
Figure 2. Characteristics of engineered strain E264. (A) Antibiotic sensibility test of mutant E264ΔBAC compared with wild-type strain E264. Kan, kanamycin 30 μg/disc; Strep, streptomycin 10 μg/disc; Ery, erythromycin 15 μg/disc; Gen, gentamicin 10 μg/disc. (B) Growth curves of both wild-type strain E264 and mutant E264ΔBAC.
After obtaining the mutant, the plasmid p15A-dis (Supplementary Figure S1; Tu et al., 2016) was introduced into the mutant E264ΔBAC via conjugation mediated by the donor strain Escherichia coli WM3064 (Dehio and Meyer, 1997). A specific peak at m/z 729.4 was observed in the crude extract of the mutant with the disorazol gene cluster, and the area of the specific peak produced by mutant E264ΔBAC was much higher than that of S. brevitalea DSM 7029, which we used for heterologous expression of cis-PKS-derived epothilone (Figure 1B and Supplementary Figure S2; Bian et al., 2017), suggesting that E264 is more suitable for expressing this trans-AT PKS gene cluster. However, the molecular weight of the product (1) was different from that of disorazol A1 (Figure 1B). The molecular formula of 1 was predicted to be C42H52N2O9 based on HR-ESIMS (m/z 729.3773 [M + H]+, calcd 729.3746; Figure 1B), which is consistent with disorazol F2. Furthermore, the 1H and 13C NMR spectra of 1 confirmed that the product is disorazol F2 (1) (Supplementary Table S7, Supplementary Figures S3 and S4; Jansen, 1994). Compared to disorazol A1, the epoxide at C-9/C-10 in 1 was replaced by a double bond, and the hydroxyl group at C-6′ was not methylated (Figure 1A).
The heterologous expression of disorazol in a phylogenetically distant relative of the myxobacterial strain S. cellulosum could exclude the influence of homologous genes in the previously reported myxobacterial host M. xanthus DK1622 (Tu et al., 2016) and facilitate the elucidation of its biosynthetic pathway. The orf9 encoding a ubiquinone biosynthesis protein Coq4 is not necessary for disorazol production (Tu et al., 2016). The product produced by the heterologous host B. thailandensis E264 is disorazol F2 rather than A1 or A2 produced by original producer So ce12 or myxobacterial host DK1622, indicating that the epoxidation and methylation of disorazols may be catalyzed by enzymes encoded by genes located outside of the gene cluster. Another debate regarding disorazols is the installation of a hydroxyl group at C-6. Theoretically, the DH7 domain could catalyze the elimination of hydroxyl groups at C-6 and C-6′ and form symmetrical macrodiolide products; however, most natural disorazols are unsymmetrical owing to the presence of the hydroxyl group at C-6′. The product obtained from E264ΔBAC::Ptet-dis is disorazol F2, an unsymmetrical macrodiolide, which suggests that the hydroxyl group at C-6′ is not installed during post-PKS modification.
Initially, the yield of disorazol F2 (1) was less than 0.4 mg/L. To improve the yield of F2 (1), we first characterized potent native promoters in E264. Fifty native promoters with high expression levels at different phases of growth in M9 broth were selected based on the data of transcriptome analysis. However, the data of transcriptome may be affected by sample processing procedures, such as liquid nitrogen freezing, we further characterized the promoters in vivo by a luciferase assay to obtain a series of potent constitutive promoters of E264 (Supplementary Figure S5, Supplementary Table S8; Ouyang et al., 2020). To avoid possible positional effects, an attB site, the attachment site of site-specific recombination, was integrated into the locus of operon BpeEF-OprC, resulting in the mutant E264ΔBAC::attB. The potent promoters P11 (promoter of a hypothetical protein), P17 (promoter of a radical SAM protein), P33 (promoter of a hypothetical protein), P35 (promoter of a peroxiredoxin), P44 (promoter of a membrane protein), and P46 (promoter of a DUF4148 domain-containing protein) were employed to promote the expression of the disorazol gene cluster, resulting in a significant increase in yield (Figure 3A). Among them, the optimal promoter, P46, resulted in an approximately 23-fold increase in the average yield of disorazol F2 (1) to 9.3 mg/L. Promoters P17 and P44 showed similar effects, which led to an obvious increase in yield to 7.5 and 8.6 mg/L, respectively. Other promoters also improved the yields by 1.5-fold to 6.5-fold. To further improve the yield, promoters P46, P44, P17, and P46 were inserted upstream of genes disA, disB, disC, and disD using the RedEx method (Supplementary Figure S6; Song et al., 2020), which generated mutants 1P (P46), 2P (P46 + P44), 3P (P46 + P44 + P17), and 4P (P46 + P44 + P17 + P46; Figure 3B). The yield of disorazol F2 (1) showed a positive correlation with the number of promoters, which reached to 38.3 mg/L in mutant 4P-dis, 96-fold higher than that of E264ΔBAC::Ptet-dis. The significantly improved efficiency of heterologous expression encouraged us to express BGCs from other phylogenetical taxa.
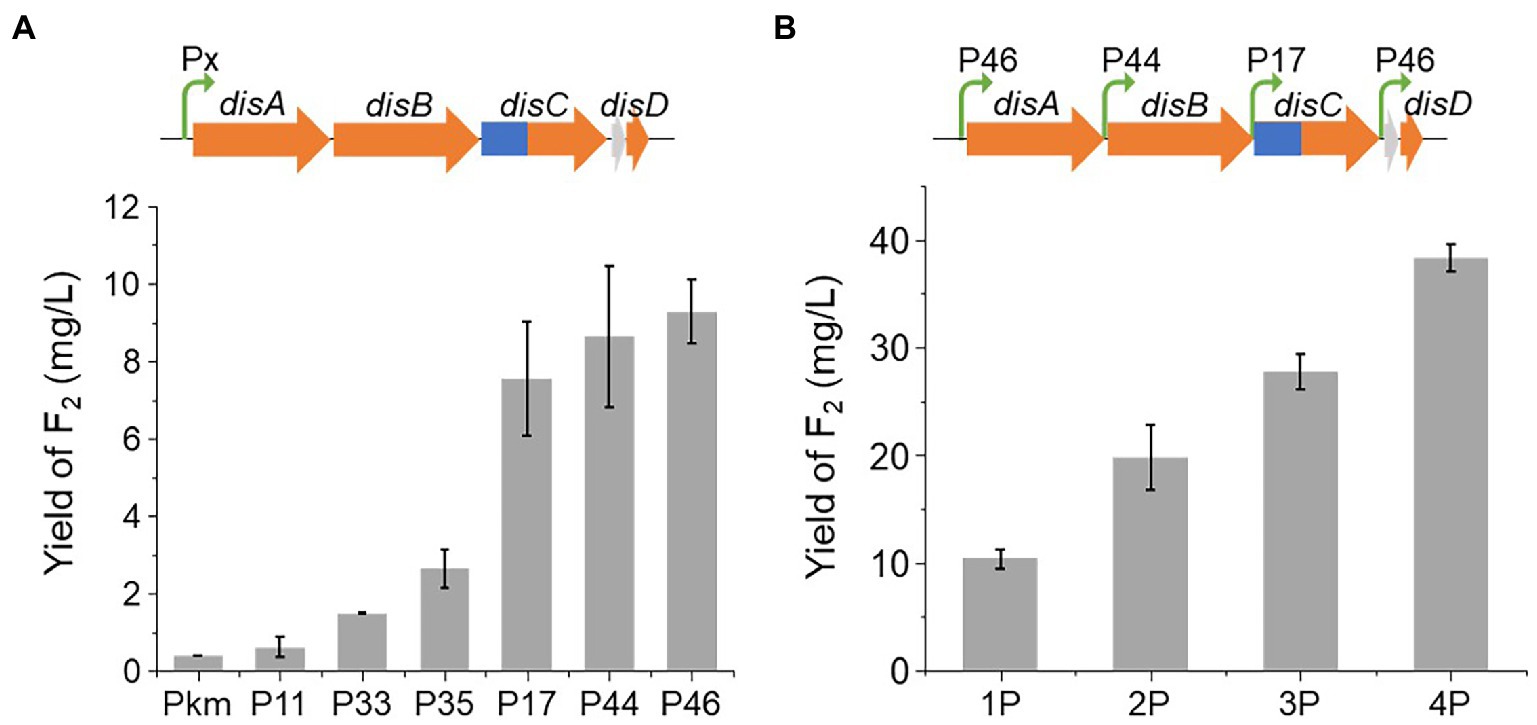
Figure 3. The yield of disorazol F2 produced by E264ΔBAC with different promoter strategies. (A) The yields of F2 (1) in E264ΔBAC::attB/dis from cluster expression promoted by different promoters (Px). (B) The yields of F2 (1) in E264ΔBAC::attB/dis driven by different numbers of promoters.
In addition to the gene cluster from myxabacteria, the BGCs of rhizoxin (rhi) from Burkholderia rhizoxinica (Partida-Martinez and Hertweck, 2007) and shuangdaolide (sdl; Supplementary Figure S7) from Streptomyces sp. B59 (Liu Y. et al., 2021) were transferred to E264ΔBAC::attB. No specific peak was detected in the crude extract of E264ΔBAC::attB/sdl, whereas a series of specific peaks were observed in the fermentation product of E264ΔBAC::attB/rhi, whose molecular weights were consistent with the reported rhizoxins (Figure 4). Specific peaks a and b with a high proportion were detected at m/z 628.3 with tR = 13.4 min and at m/z 642.3 with tR = 14.8 min. Based on HRMS, the molecular formula of the compound in peak a was established as C35H49NO9 (m/z 628.3483 [M + H]+, calcd 628.3482), which is identical to that of rhizoxins M1, Z2, and S2; peak b may contain rhizoxin M2 or Z2 (C36H51NO9, m/z 642.3636 [M + H]+, calcd 642.3637). In addition to the major peaks, we also observed some specific peaks in a relatively low proportion of the fermentation product of E264ΔBAC::attB/rhi. Their MS/MS spectra were similar to peaks a and b, and HRMS suggested that they were also related to known rhizoxins (Supplementary Table S9). The BGC of rhizoxin from closely related species was expressed successfully in the host based on strain E264, and the potential of this heterologous expression system still needs to be further developed.
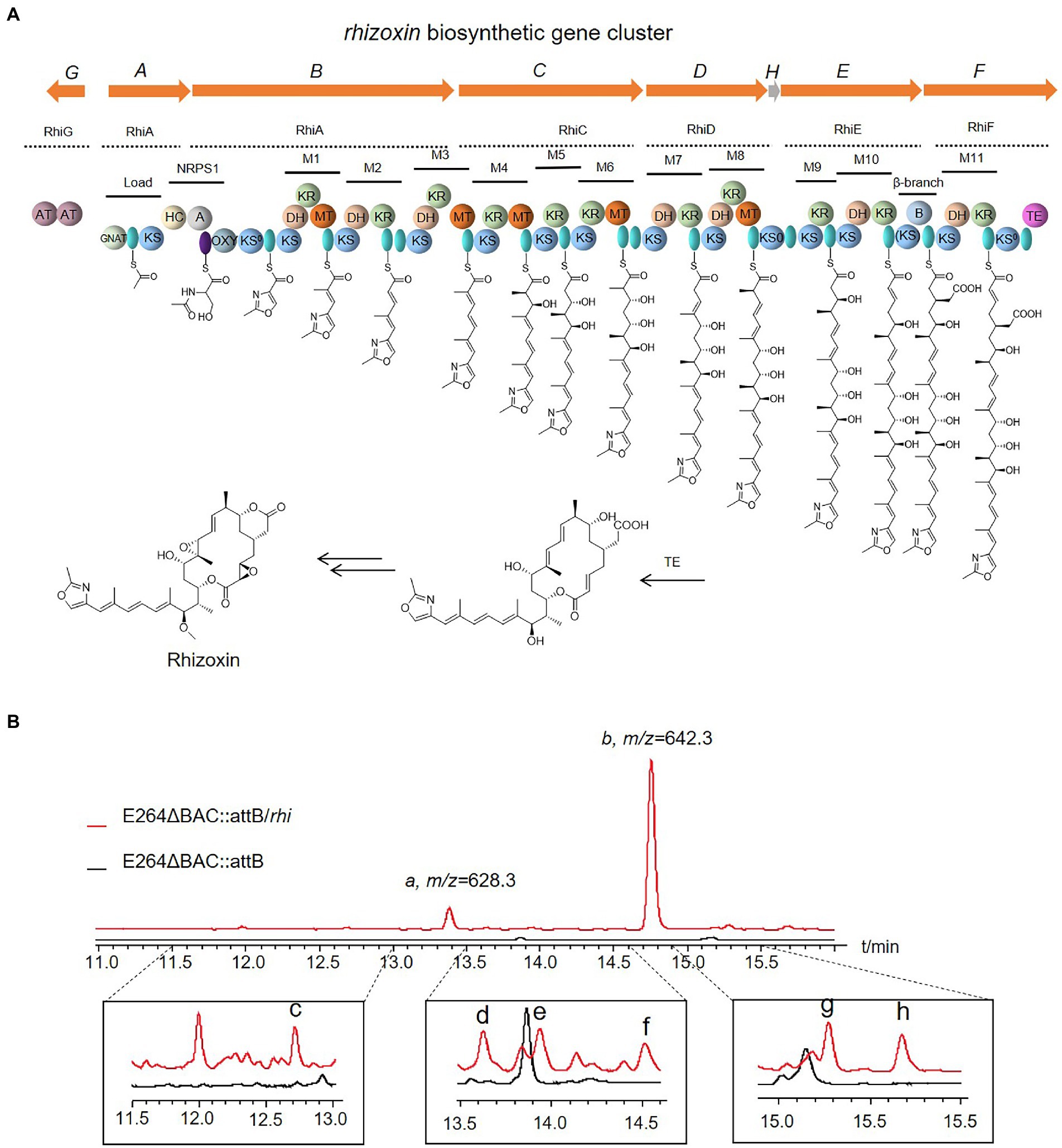
Figure 4. Organization of the rhizoxin (rhi) biosynthesis gene cluster and model for rhizoxin biosynthesis (A), and the fermentation products of E264ΔBAC::attB/rhi (B). GNAT: N-acetyltransferase; KS: ketosynthase, AT: acyl transferase, green oval: ACP, HC: condensation/heterocyclization, A: adenylation, purple oval: PCP, OXY: oxygenase, KR: ketoreductase, DH: dehydratase, MT: C-methyltransferase, TE: thioesterase, B: unknown domain possibly involved in β-branching.
Owing to the low yield of the original producer So ce12, the cytotoxicity of disorazol F2 has never been reported in previous studies. In this study, the cytotoxicity of disorazol F2 (1) was evaluated against three human cancer cell lines (HepG2, HCT116, and A549; Table 2). Although one-two orders of magnitude weaker than epoxide disorazols A1 and A2 (Tu et al., 2016), disorazol F2 still exhibited potent activity against the tested cell lines with IC50 values ranging from 0.25 to 3.11 nM, one to three orders of magnitude stronger than clinically used doxorubicin or vincristine. The cytotoxicity assay compared with previously reported data (Irschik et al., 1995; Elnakady et al., 2004; Hopkins and Wipf, 2009; Tu et al., 2016) suggested that the activity of disorazols is affected by the epoxide at C-9/C-10, which suggests the tailoring enzyme of the disorazol pathway is still a significant task.
Disorazols are a family of natural products with nanomolar cytotoxicity. However, the difficulties in culture and genetic manipulation of the original producer have limited further study. The heterologous expression system developed in this study efficiently overcomes these problems. This provides a powerful tool for illuminating this fantastic pathway. Heterologous expression in strain E264 indicated that the enzymes catalyzed epoxidation and methylation outside of the gene cluster. The gene cluster of disorazol also possesses some interesting characteristics unusual in classical PKS, such as a missing loading domain in module 1 and repeated ACP domains in module 2, as well as these ‘redundant’ domains in genes disB and disC. These interesting points may be elucidated in the near future by expressing domain (s) inactivation or deletion of the disorazol gene cluster with an efficient heterologous host.
This heterologous expression system also provides a platform for producing unnatural disorazol by PKS engineering. Natural products are designed to help producers occupy an ecological niche rather than to help people cure diseases. Therefore, it is effective to improve drug properties of them by increasing structural diversity (Floss, 2006). We believe that an increasing number of disorazol derivatives will be generated using this platform, which will promote drug development in this fantastic family.
Finally, myxobacteria are becoming an important source of natural products for drug discovery and still possess enormous biosynthetic potential (Weissman and Müller, 2010; Bader et al., 2020). The gene cluster of epothilone, a marketed anti-cancer drug isolated from Sorangium cellulosum, was successfully expressed in strain DSM 7029 with higher yields after multiple medium optimization and genome engineering (Bian et al., 2017; Yu et al., 2020). The heterologous expression system based on strain E264 provides another choice for the exploration of natural products from myxobacteria.
In addition to the advantages mentioned above, some problems must be addressed in this heterologous expression system. First, although strain E264 have been extensively studied in lab, as a class II opportunistic pathogen, potential risks need to be evaluated to ensure safety during large-scale fermentation. Additional measures, such as appropriate containment, are needed to ensure safety, which will significantly increase costs in industrial production. To overcome this problem, the construction of attenuated mutants through genetic engineering may be feasible. Secondly, this study obtained disorazol F2 rather than A1, which might leave the real “natural products” in the process of expressing other gene clusters. This problem needs to be addressed in other systems. Finally, the expression of shuangdaolide in the host failed, which may have resulted from their phylogenetic relationships. This result indicates that various hosts are required to express gene clusters from different taxa.
In summary, we developed an efficient heterologous expression system based on streamlined B. thailandensis E264, in which the disorazol gene cluster was expressed successfully. The yield of the heterologous product, disorazol F2, was improved 96-fold using promoter substitution and insertion. The expression of the rhizoxin gene cluster from Burkholderia rhizoxina was also successful, whereas no specific peak was detected in the fermentation of the host containing the shuangdaolide gene cluster. Furthermore, the safety and compatibility of the heterologous expression system should be improved.
Burkholderia thailandensis E264 was purchased from German collection of microorganisms and cell cultures GmbH (DSMZ) and was cultured in low-salt Luria-Bertani medium (1% tryptone, 0.5% yeast extract, 0.1% NaCl) at 37°C. The markerless mutants of strain E264 were established as described in section construction of deletion mutants. Escherichia coli GB05-red was used for mediating homologous recombination between a linear and circular DNA molecular (LCHR) while E. coli GB05-dir mediating linear plus linear recombination (LLHR). Escherichia coli GB05-dir-gyrA462 or GB05-red-gyrA462 were used for constructing plasmids with counterselectable marker CcdB. Escherichia coli WM3064 was employed as a conjugal donor (Dehio and Meyer, 1997). Unless otherwise specified, the strains of Escherichia coli were cultured under the same conditions with strain E264. Tetracycline (Tet) was added to a final concentration of 50 μg/ml for selective growth of wild type strain of B. thailandensis E264. For mutant E264ΔBAC, antibiotics were added to final concentrations of 30 μg/ml kanamycin (Kan) or 5 μg/ml gentamicin (Genta) as required, for strains of E. coli, the concentrations were 30 μg/ml Kan, 100 μg/ml ampicillin (Amp) or 15 μg/ml chloramphenicol (Cm).
The genomic DNA (gDNA) of strain E264 was isolated using phenol – chloroform – isoamyl alcohol mixture (Wang et al., 2020). To obtain markerless mutants, genetic tools developed by Barrett et al were modified and applied in this study (Barrett et al., 2008). Briefly, about 1 kb regions upstream and downstream of targeted genes amplified by PCR using PrimeSTAR® HS DNA polymerase with GC buffer were cloned into vector pBR322-amp-tet-pheS (linearized with restriction enzymes XabI and NcoI) by strain GB05-dir. The plasmids with homology arms were introduced into strain E264 by natural transformation (Garcia, 2017), and correct clones verified by colony PCR were cultured in LSLB without antibiotic for 12 h to eliminate selection marker. The overnight cultures were centrifuged and washed twice with liquid M9G medium (M9 medium of Sangon, Shanghai supplemented with 20 mM Glucose), then 50 μl cell suspensions were plated on M9G plates supplementing with 0.1% 4-Chloro-D, L-phenylalanine (Sigma-Aldrich) to screen the recombinants with selection marker deletion. Markerless mutants were further verified by colony PCR. To generate mutant E264ΔBAC::attB, the sequence of attB (GGGTGCCAGGGCGTGCCCTTGGGCTCCCCGGGCGCGTA) was inserted between the homology arms of pBR322-Amp-Tet-pheS-oprC through oligonucleotide synthesis, and the plasmid was introduced into mutant E264ΔBA followed by selection and counterselection successively.
To make test plates, the overnight cultures (200 μl) of strain E264 or E264ΔBAC were mixed with 50 ml melted LSLB agar medium. Then, the paper discs containing different antibiotics (HangweiTM, Hangzhou) were placed on the media. The plates were cultured at 37°C overnight to measure the diameter of inhibition zone.
The amp-ccdB cassette flanked with two restriction enzyme NdeI sites and about 40 bp homology arms was amplified from plasmid p15A-amp-ccdB using primers amp-ccdB-P11-F/R (Wang et al., 2018), the PCR product and plasmid p15A-genta-int-attP-P11-firefly (Ouyang et al., 2020) were electroporated into E. coli GB05-red-gyrA462 expressed recombinases Redα/Redβ, which resulted in plasmid p15A-genta-int-attP-amp-ccdB-firefly. Colonies growth on plates supplemented with Amp and Genta were transferred into 1.8 ml fresh LB broth with appropriate antibiotics and incubated at 37°C overnight. The plasmid DNA was extracted from the overnight culture and confirmed by restriction analysis.
Promoter Px was amplified using primers PX-F/R with 40 bp homology arms, and plasmid p15A-genta-int-attP-amp-ccdB-firefly was linearized with restriction endocuclease NdeI. The products of PCR and restriction enzyme digestion were electroporated into E. coli GB05-dir expressed recombinases RecE/RecT, which resulted in plasmid p15A-genta-int-attP-Px-firefly.
Plasmids with different promoters located upstream of luciferase gene were introduced into E264ΔBAC::attB by conjugation mediated by WM3064 (Dehio and Meyer, 1997), The expression levels of report gene in transformants were detected by Luciferase Assay System of Promega Corporation. Briefly, 20 μl cell lysates prepared according the instruction were mixed with 100 μl of Luciferase Assay Reagent, and the light produced by the mixture was quantified by GloMaxTM 96 Microplate Luminometer of Promega Corporation.
Cloning vector BAC-cm flanked with PacI/HpaI sites and homology arms was amplified by PCR from plasmid pBeloBAC11 (digested with BamHI and HindIII) using primers bac-dis-F/R. The products of PCR and plasmid p15A-dis (Tu et al., 2016) linearized with XbaI were electroporated into E. coli GB05-dir, which resulted in plasmid pBAC-cm-dis.
Plasmid p15A-genta-int-attP-Px-firefly was linearized with MfeI, then cassette p15A-genta-int-attP-Px with homology arms was amplified by PCR using primers Px-dis-F/R. The PCR product and linearized plasmid pBAC-cm-dis (digested with PacI and HpaI) were assembled by LLHR mediated by GB05-dir, which resulted in plasmid p15A-genta-int-attP-Px-dis.
To construct plasmids with multiple promoters and improve the yield of disorazol, the strategy of RedEx was employed (Song et al., 2020). Breifly, plasmid p15A-genta-int-attP-P46-dis was electroporated into E. coli GB05-red-gyrA462, then the cassette 2P-amp-ccdB, dis (17985–18044)-PacI-amp-ccdB-PacI-dis (18015–18044)-P44-dis (18053–18110; the first nucleotide of disA was designated as 1), generated by fusion PCR was electroporated into E. coli GB-red-gyrA462 with plasmid p15A-genta-int-attP-P46-dis. The cassette 2P-amp-ccdB was inserted upstream of disB through LCHR mediated by recombinases Redα/Redβ which resulted in plasmid p15A-genta-int-attP-2P-dis-amp-ccdB. The plasmid p15A-genta-int-attP-2P-dis-amp-ccdB was digested with PmeI, and 100 ng linearized plasmid DNA was treated with 0.2 U T4pol (New England BioLabs) in a 20 μl reaction at 25°C for 30 min, 75°C for 20 min, 50°C for 20 min and then held at 4°C in a thermocycler. The reaction mixture was electroporated into E. coli GB2005 cells after desalting treatment. The recombinant p15A-genta-int-attP-2P-dis (2 promoters) with promoter P44 inserting upstream disB was identified with KpnI/PmeI restriction analysis.
Utilizing the strategy of RedEX, promoter P17 was inserted upstream of gene disC and P46 was inserted upstream of gene disD successively, which resulted in plasmid p15A-genta-int-attP-3P-dis (3 promoters) and p15A-genta-int-attP-4P-dis (4 promoters), respectively.
Single colonies were inoculated into 2 ml Eppendorf tubes containing 1.8 ml LB broth supplemented with appropriate antibiotics and incubated at 37°C overnight with shaking at 900 rpm. The seed cultures (1%, v/v) were transferred into 50 ml M9 broth in 250 ml Erlenmeyer flask and incubated at 30°C with shaking at 200 rpm for 24 h before 2% XAD-16 resin was added, then the incubation was continued for another 48 h. The resin was collected by centrifugation and resuspended with 50 ml of methanol. The mixtures were shaken at 30°C, 200 rpm for 2 h. The methanol was removed by evaporation in vacuo, and residues were dissolved with 1 ml of methanol. After filtering with 0.22 μm membrane, the crude extracts were analyzed by UPLC-MS (UltiMate 3,000 UPLC system combined with Bruker amazon SL Ion Trap mass spectrometer). The C18 column (2.1 × 100 mm. 2.2 μm, Thermo) was utilized to analyze the crude extracts at a 0.3 ml/min flow rate using the following program: 0–3 min 5% solvent B; 3–22 min, 5 95% with linear gradient; then, 22–25 min, 5% solvent B (Solvent A, Milli Q water supplemented with 0.1% formic acid; Solvent B, acetonitrile supplemented with 0.1% formic acid). Mass spectra were acquired in positive ion mode.
The crude extract from 10 L culture of E264ΔBAC::Ptet/dis was fractionated with Sephadex LH-20 column (GE Healthcare) chromatography using MeOH as a mobile phase. Fractions containing F2 (1) were combined and further purified by semipreparative reverse-phase HPLC (Agilent ZORBAX SB-C18 column, 250 × 9.4 mm, 5 μm; gradient elution 0–2 min, 64% ACN; 2–17 min, 64–90% ACN; 17–22 min, 95% ACN, 22–25 min, 64% ACN; 2.5 ml/min). Finally, 1.5 mg of disorazol F2 (1) was obtained with retention time at 15.1 min.
The standard curve of disorazol F2 (1) was established by measuring the area of absorption spectrum at UV 280 nm containing 0.05, 0.1, 0.2, 0.5 and 1.0 mg/ml disorazol F2 (1). The yield of disorazol F2 (1) was determined by reference to the standard curve.
The datasets presented in this study can be found in online repositories. The accession number for the RNA-Seq data in this paper is GEO: PRJNA889572.
XX, XB, YZ, and Z-JW: designed research. XX, Z-JW and XL: performed research. HZ, YL, LZ, XW, QT, LH, FY, LG, RM, XB, and YZ: analyzed data. Z-JW, XX, HZ, and XB: wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2021YFC2100500, 2019YFA0905700, 2019YFA09004000), the National Natural Science Foundation of China (32161133013, 32070060, 32100052, and 32170038), the Shandong Provincial Natural Science Foundation of China (ZR2019JQ11, ZR2019ZD18, and ZR2019ZD30), the 111 project (B16030), Youth Interdisciplinary Innovative Research Group of SDU (2020QNQT009), and the China Postdoctoral Science Foundation (2020M682169).
The authors thank Haiyan Sui, Xiangmei Ren, Zhifeng Li, Jingyao Qu, and Jing Zhu of the State Key Laboratory of Microbial Technology of Shandong University for their help and guidance within NMR, LC, and HPLC-HRMS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1073243/full#supplementary-material
Bader, C. D., Panter, F., and Müller, R. (2020). In depth natural product discovery—myxobacterial strains that provided multiple secondary metabolites. Biotechnol. Adv. 39:107480. doi: 10.1016/j.biotechadv.2019.107480
Barrett, A. R., Kang, Y., Inamasu, K. S., Son, M. S., Vukovich, J. M., and Hoang, T. T. (2008). Genetic tools for allelic replacement in Burkholderia species. Appl. Environ. Microbiol. 74, 4498–4508. doi: 10.1128/AEM.00531-08
Bian, X., Tang, B., Yu, Y., Tu, Q., Gross, F., Wang, H., et al. (2017). Heterologous production and yield improvement of epothilones in Burkholderiales strain DSM 7029. ACS Chem. Biol. 12, 1805–1812. doi: 10.1021/acschembio.7b00097
Biggins, J. B., Gleber, C. D., and Brady, S. F. (2011). Acyldepsipeptide HDAC inhibitor production induced in Burkholderia thailandensis. Org. Lett. 13, 1536–1539. doi: 10.1021/ol200225v
Biot, F. V., Lopez, M. M., Poyot, T., Neulat-Ripoll, F., Lignon, S., Caclard, A., et al. (2013). Interplay between three RND efflux pumps in doxycycline-selected strains of Burkholderia thailandensis. PLoS One 8:e84068. doi: 10.1371/journal.pone.0084068
Biot, F. V., Valade, E., Garnotel, E., Chevalier, J., Villard, C., Thibault, F. M., et al. (2011). Involvement of the efflux pumps in chloramphenicol selected strains of Burkholderia thailandensis: proteomic and mechanistic evidence. PLoS One 6:e16892. doi: 10.1371/journal.pone.0016892
Brett, P. J., Deshazer, D., and Woods, D. E. (1998). Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48 Pt 1, 317–320. doi: 10.1099/00207713-48-1-317
Carvalho, R., Reid, R., Viswanathan, N., Gramajo, H., and Julien, B. (2005). The biosynthetic genes for disorazoles, potent cytotoxic compounds that disrupt microtubule formation. Gene 359, 91–98. doi: 10.1016/j.gene.2005.06.003
Chang, K., Luo, J., Xu, H., Li, M., Zhang, F., Li, J., et al. (2017). Human infection with Burkholderia thailandensis, China, 2013. Emerg. Infect. Dis. 23, 1416–1418. doi: 10.3201/eid2308.170048
Dehio, C., and Meyer, M. (1997). Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. bacteriol. 179, 538–540. doi: 10.1128/jb.179.2.538-540.1997
Duerkop, B. A., Varga, J., Chandler, J. R., Peterson, S. B., Herman, J. P., Churchill, M. E. A., et al. (2009). Quorum-sensing control of antibiotic synthesis in Burkholderia thailandensis. J. Bacteriol. 191, 3909–3918. doi: 10.1128/JB.00200-09
Elnakady, Y. A., Sasse, F., Lunsdorf, H., and Reichenbach, H. (2004). Disorazol A1, a highly effective antimitotic agent acting on tubulin polymerization and inducing apoptosis in mammalian cells. Biochem. Pharmacol. 67, 927–935. doi: 10.1016/j.bcp.2003.10.029
Floss, H. G. (2006). Combinatorial biosynthesis-potential and problems. J. Biotechnol. 124, 242–257. doi: 10.1016/j.jbiotec.2005.12.001
Fu, J., Bian, X., Hu, S., Wang, H., Huang, F., Seibert, P. M., et al. (2012). Full-length rec E enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat. Biotechnol. 30, 440–446. doi: 10.1038/nbt.2183
Garcia, E. (2017). Burkholderia thailandensis: genetic manipulation. Curr. Protoc. Microbiol. 45:4C.2.1-4C.2.15. doi: 10.1002/cpmc.27
Glass, M. B., Gee, J. E., Steigerwalt, A. G., Cavuoti, D., Barton, T., Hardy, R. D., et al. (2006). Pneumonia and septicemia caused by Burkholderia thailandensis in the United States. J. Clin. Microbio. 44, 4601–4604. doi: 10.1128/jcm.01585-06
Hopkins, C. D., and Wipf, P. (2009). Isolation, biology and chemistry of the disorazoles: new anti-cancer macrodiolides. Nat. Prod. Rep. 26, 585–601. doi: 10.1039/b813799b
Huo, L., Hug, J. J., Fu, C., Bian, X., Zhang, Y., and Müller, R. (2019). Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 36, 1412–1436. doi: 10.1039/c8np00091c
Irschik, H., Jansen, R., Gerth, K., Hofle, G., and Reichenbach, H. (1995). Disorazol a, an efficient inhibitor of eukaryotic organisms isolated from myxobacteria. J. Antibiot. (Tokyo) 48, 31–35. doi: 10.7164/antibiotics.48.31
Jansen, R. (1994). Disorazoles, highly cytotoxic metabolites from the sorangicin-producing bacterium Sorangium cellulosum, strain So ce 12. Liebigs. Ann. Chem. 1994, 759–773. doi: 10.1002/jlac.199419940802
Kang, Y., Norris, M. H., Wilcox, B. A., Tuanyok, A., Keim, P. S., and Hoang, T. T. (2011). Knockout and pullout recombineering for naturally transformable Burkholderia thailandensis and Burkholderia pseudomallei. Nat. Protoc. 6, 1085–1104. doi: 10.1038/nprot.2011.346
Kopp, M., Irschik, H., Pradella, S., and Müller, R. (2005). Production of the tubulin destabilizer disorazol in Sorangium cellulosum: biosynthetic machinery and regulatory genes. Chembiochem 6, 1277–1286. doi: 10.1002/cbic.200400459
Kunakom, S., and Eustáquio, A. S. (2019). Burkholderia as a source of natural products. J. Nat. Prod. 82, 2018–2037. doi: 10.1021/acs.jnatprod.8b01068
Liu, J., Wang, X., Dai, G., Zhang, Y., and Bian, X. (2022). Microbial chassis engineering drives heterologous production of complex secondary metabolites. Biotechnol. Adv. 59:107966. doi: 10.1016/j.biotechadv.2022.107966
Liu, Y., Zhou, H., Shen, Q., Dai, G., Yan, F., Li, X., et al. (2021). Discovery of polycyclic macrolide shuangdaolides by heterologous expression of a cryptic trans-AT PKS gene cluster. Org. Lett. 23, 6967–6971. doi: 10.1021/acs.orglett.1c02589
Liu, J., Zhou, H., Yang, Z., Wang, X., Chen, H., Zhong, L., et al. (2021). Rational construction of genome-reduced Burkholderiales chassis facilitates efficient heterologous production of natural products from proteobacteria. Nat. Commun. 12:4347. doi: 10.1038/s41467-021-24645-0
Mao, D., Bushin, L. B., Moon, K., Wu, Y., and Seyedsayamdost, M. R. (2017). Discovery of scm R as a global regulator of secondary metabolism and virulence in Burkholderia thailandensis E264. Proc. Natl. Acad. Sci. U. S. A. 114, E2920–e2928. doi: 10.1073/pnas.1619529114
Menchon, G., Prota, A. E., Lucena-Agell, D., Bucher, P., Jansen, R., Irschik, H., et al. (2018). A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin. Nat. Commun. 9:2106. doi: 10.1038/s41467-018-04535-8
Newman, D. J., and Cragg, G. M. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803. doi: 10.1021/acs.jnatprod.9b01285
Nguyen, T., Ishida, K., Jenke-Kodama, H., Dittmann, E., Gurgui, C., Hochmuth, T., et al. (2008). Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat. Biotechnol. 26, 225–233. doi: 10.1038/nbt1379
Ouyang, Q., Wang, X., Zhang, N., Zhong, L., Liu, J., Ding, X., et al. (2020). Promoter screening facilitates heterologous production of complex secondary metabolites in Burkholderiales strains. ACS Synth. Biol. 9, 457–460. doi: 10.1021/acssynbio.9b00459
Pace, N. R. (1997). A molecular view of microbial diversity and the biosphere. Science 276, 734–740. doi: 10.1126/science.276.5313.734
Paoli, L., Ruscheweyh, H.-J., Forneris, C. C., Hubrich, F., Kautsar, S., Bhushan, A., et al. (2022). Biosynthetic potential of the global ocean microbiome. Nature 607, 111–118. doi: 10.1038/s41586-022-04862-3
Partida-Martinez, L. P., and Hertweck, C. (2007). A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. Chembiochem. 8, 41–45. doi: 10.1002/cbic.200600393
Seyedsayamdost, M. R., Chandler, J. R., Blodgett, J. A. V., Lima, P. S., Duerkop, B. A., Oinuma, K.-I., et al. (2010). Quorum-sensing-regulated bactobolin production by Burkholderia thailandensis E264. Org. Lett. 12, 716–719. doi: 10.1021/ol902751x
Song, C., Luan, J., Li, R., Jiang, C., and Wang, H. (2020). RedEx: a method for seamless DNA insertion and deletion in large multimodular polyketide synthase gene clusters. Nucleic Acids Res. 48:e130. doi: 10.1093/nar/gkaa956
Tang, B., Yu, Y., Liang, J., Zhang, Y., Bian, X., Zhi, X., et al. (2019). Reclassification of 'Polyangium brachysporum' DSM 7029 as Schlegelella brevitalea sp. nov. Int. J. Syst. Evol. Microbiol. 69, 2877–2883. doi: 10.1099/ijsem.0.003571
Tu, Q., Herrmann, J., Hu, S., Raju, R., Bian, X., Zhang, Y., et al. (2016). Genetic engineering and heterologous expression of the disorazol biosynthetic gene cluster via red/ET recombineering. Sci. Rep-UK 6:21066. doi: 10.1038/srep21066
Wang, H., Li, Z., Jia, R., Yin, J., Li, A., Xia, L., et al. (2018). ExoCET: exonuclease in vitro assembly combined with RecET recombination for highly efficient direct DNA cloning from complex genomes. Nucleic Acids Res. 46:e28. doi: 10.1093/nar/gkx1249
Wang, Z. J., Zhou, H., Zhong, G., Huo, L., Tang, Y. J., Zhang, Y., et al. (2020). Genome mining and biosynthesis of primary amine-acylated desferrioxamines in a marine gliding bacterium. Org. Lett. 22, 939–943. doi: 10.1021/acs.orglett.9b04490
Weissman, K. J., and Müller, R. (2010). Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat. Prod. Rep. 27, 1276–1295. doi: 10.1039/c001260m
Yan, F., Auerbach, D., Chai, Y., Keller, L., Tu, Q., Hüttel, S., et al. (2018). Biosynthesis and heterologous production of vioprolides: rational biosynthetic engineering and unprecedented 4-methylazetidinecarboxylic acid formation. Angew. Chem. Int. Ed. Engl. 57, 8754–8759. doi: 10.1002/anie.201802479
Yu, Y., Wang, H., Tang, B., Liang, J., Zhang, L., Wang, H., et al. (2020). Reassembly of the biosynthetic gene cluster enables high epothilone yield in engineered Schlegelella brevitalea. ACS Syn. Bio. 9, 2009–2022. doi: 10.1021/acssynbio.0c00100
Keywords: Burkholderia, chassis, heterologous expression, disorazol, rhizoxin
Citation: Wang Z-J, Liu X, Zhou H, Liu Y, Zhong L, Wang X, Tu Q, Huo L, Yan F, Gu L, Müller R, Zhang Y, Bian X and Xu X (2022) Engineering of Burkholderia thailandensis strain E264 serves as a chassis for expression of complex specialized metabolites. Front. Microbiol. 13:1073243. doi: 10.3389/fmicb.2022.1073243
Received: 18 October 2022; Accepted: 28 October 2022;
Published: 17 November 2022.
Edited by:
Dewu Zhang, Chinese Academy of Medical Sciences, ChinaReviewed by:
Xianpu Ni, Shenyang Pharmaceutical University, ChinaCopyright © 2022 Wang, Liu, Zhou, Liu, Zhong, Wang, Tu, Huo, Yan, Gu, Müller, Zhang, Bian and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youming Zhang, emhhbmd5b3VtaW5nQHNkdS5lZHUuY24=; Xiaoying Bian, YmlhbnhpYW95aW5nQHNkdS5lZHUuY24=; Xiaokun Xu, eHV4aWFva3VuMzEzQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.