- 1Key Laboratory of New Animal Drug Project of Gansu Province, Lanzhou, China
- 2Key Laboratory of Veterinary Pharmaceutical Development, Ministry of Agriculture, Lanzhou, China
- 3Lanzhou Institute of Husbandry and Pharmaceutical Sciences, Chinese Academy of Agricultural Sciences, Lanzhou, China
A new antibacterial strategy based on inhibiting bacterial quorum sensing (QS) has emerged as a promising method of attenuating bacterial pathogenicity and preventing bacterial resistance to antibiotics. In this study, we screened Echinatin (Ech) with high-efficiency anti-QS from 13 flavonoids through the AI-2 bioluminescence assay. Additionally, crystal violet (CV) staining combined with confocal laser scanning microscopy (CLSM) was used to evaluate the effect of anti-biofilm against Escherichia coli (E. coli). Further, the antibacterial synergistic effect of Ech and marketed antibiotics were measured by broth dilution and Alamar Blue Assay. It was found that Ech interfered with the phenotype of QS, including biofilm formation, exopolysaccharide (EPS) production, and motility, without affecting bacterial growth and metabolic activity. Moreover, qRT-PCR exhibited that Ech significantly reduced the expression of QS-regulated genes (luxS, pfs, lsrB, lsrK, lsrR, flhC, flhD, fliC, csgD, and stx2). More important, Ech with currently marketed colistin antibiotics (including colistin B and colistin E) showed significantly synergistically increased antibacterial activity in overcoming antibiotic resistance of E. coli. In summary, these results suggested the potent anti-QS and novel antibacterial synergist candidate of Ech for treating E. coli infections.
Introduction
As one of the pathogenically versatile bacterial organisms, E. coli can cause various infections, including diarrhea, urinary tract infections, sepsis, and hemolytic-uremic syndrome (Riley, 2020). Pathogenic E. coli causes great economic losses to animal and poultry industries, as well as a serious threat to human health. For example, millions of dollars are lost each year due to Avian pathogenic E. coli (APEC) infections (Lutful Kabir, 2010). In the United States, E. coli O157:H7 is estimated to cause 95,000 illnesses a year (Scallan et al., 2011; Beshearse et al., 2021), and Shiga-producing E. coli (STEC) causes 5,960 infections annually (Hale et al., 2012). Antimicrobial resistance has become a worldwide concern and an increasing threat to human and animal health (Hawkey, 2008). Currently, most antibacterial compounds target the basic physiological processes of bacteria, which increases the likelihood of bacteria developing resistance to multiple drugs (Munguia and Nizet, 2017). Thus, new therapeutic strategies are urgently needed for treating multidrug-resistant pathogen infections.
At present, alternative approaches to antimicrobial therapy have focused on inhibiting the virulence factors of bacterial pathogens (Dickey et al., 2017; Buroni and Chiarelli, 2020; Loubet et al., 2020; Piewngam et al., 2020; Silva et al., 2020). QS is a cellular mechanism mediated by autoinducers, which allows bacteria to organize behavior depending on their density (Ng and Bassler, 2009). Interference with QS systems does not exert selection pressure on bacteria compared with antibiotics thus reducing the emergence and spread of resistant mutants (Sully et al., 2014; Quave et al., 2015; Ning et al., 2021). QS is a process that involves bacteria communicating with signaling molecules called autoinducers (AIs). There are several types of AI molecules, including diffusible signaling factors (DSFs), autoinducer-2 (AI-2), indole, and Acyl-homoserine lactones (AHLs, AI-1), etc. (Guo et al., 2013; Whiteley et al., 2017, 2018). QS systems function based on cell density, which increases the concentration of AI as cell density increases. Upon reaching a certain level of concentration of AI, signaling is activated that modulates the expression of genes related to bacterial physiology, biofilm formation, motility, and virulence (Papenfort and Bassler, 2016). Antibacterial strategies based on inhibiting bacterial QS have emerged as a new promising method of preventing bacterial resistance to antibiotics, as well as inhibiting the expression of virulence factors (Manner and Fallarero, 2018; Wang et al., 2019; Minich et al., 2022).
AI-2 has been considered to be a “universal” signaling molecule involved in bacterial communication at the inter-and intra-species level, which is widely found in Gram-negative and Gram-positive bacteria. For most bacteria, the regulatory function of the AI-2 QS system is mainly reflected in four aspects, including bacterial virulence, biofilm, motility, and other functions (Bassler, 2002; Sturme et al., 2002; Reading and Sperandio, 2006; Choudhary and Schmidt-Dannert, 2010). For example, AI-2 could influence the production of virulence factors and the formation of biofilm in E. coli, Salmonella, and S. suis (Kendall et al., 2007; Ju et al., 2018; Sun et al., 2020; Li et al., 2021). The inhibition of AI-2 can effectively reduce bacterial virulence, biofilm formation, and bacterial resistance, which can be used to replace antibiotics. In this context, it’s reported that plant-derived compounds with diverse structures have been widely investigated as AI-2 QS inhibitors (Karnjana et al., 2020; Li et al., 2021; Meng et al., 2022). Ech, a flavonoid isolated from glycyrrhiza, had antioxidant, antitumor, anti-virus, and other biological activities (Ji et al., 2016; Zhu et al., 2018; Kwak et al., 2019; Hu et al., 2021; Ran et al., 2021). Despite many pharmacologic investigations, there have been no reports on the anti-QS activity of Ech.
AI-2 bioluminescence assay (Vibrio harveyi (V. harveyi) BB170 bioluminescence assay) is the most commonly used method to detect AI-2 signal molecules. The bioluminescence reporter strain V. harveyi BB170 (luxN:: Tn5) is a mutant strain, whose fluorescence is only regulated by AI-2 signal molecule (Bassler et al., 1994, 1997; Surette et al., 1999). Therefore, the inhibitory effect of the inhibitors on E. coli AI-2 production can be judged by the intensity of V. harveyi bioluminescence. Here, based on V. harveyi BB170 bioluminescence assay, 13 natural product compounds that inhibit AI-2 production were screened and evaluated. Notably, we found that one of the compounds, Ech, effectively blocks E. coli QS. Furthermore, the study examined the antibiofilm and antivirulence properties of Ech against E. coli and evaluated the synergistic effects of combining Ech with antibiotics.
Materials and methods
Natural product compounds
Echinatin, Aloeemodin, Loureirin B, Cardamonin, Cynaroside, Artemetin, Neosperidin dihydrochalcone, Hesperetin, and Vitexin used in the study were obtained from Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China). Phloretin was purchased from Macklin Inc. (Shanghai, China). Scutellarin and Acacetin were obtained from MCE (Shanghai, China). Gentisin was obtained from ChemFaces (Wuhan, China). Dimethylsulfoxide (DMSO, Sigma) was used to dissolve all compounds to the concentration of 40 mM.
Bacterial strains and cells
Escherichia coli O157:H7 (ATCC 43895) was purchased from Beina Chuanglian Biotechnology Research Institute (Beijing, China). Clinical strains E. coli O101、E. coli C83654、E. coli O149、E. coli XJ24、E. coli KD-13-1 were isolated and maintained in our laboratory. Luria-Bertani (LB, HuanKai Microbial, Guangdong, China) and Luria-Bertani agar (LA, HuanKai Microbial, Guangdong, China) medium were used to cultivate all E. coli strains. V. harveyi BB170 and V. harveyi BB152 were kindly provided by Researcher Han Xiangan (Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences). AB medium supplemented with 1 mM L-arginine, 1% glycerol, and 10 mM Phosphate buffer (pH = 7.2) for culturing V. harveyi.
Caco-2 cell lines were obtained from ATCC and cultured under standard conditions containing MEM medium (Gibco, Grand Island, NY, United States) supplemented with 20% FBS (Gibco, Grand Island, NY, USA), 1% non-essential amino acids (Gibco, Grand Island, NY, USA), 10 mM HEPES (Solarbio, Beijing, China), 1 mM L-glutamine (Gibco, Grand Island, NY, USA), and 1 mM sodium pyruvate (Gibco, Grand Island, NY, United States).
Anti-QS inhibitor screening
AI-2 bioluminescence assay was used to screen QS inhibitors as described previously (Bassler et al., 1994; Taga and Xavier, 2011) with minor modifications. Briefly, E. coli O157:H7 was cultured for 16 h with 50 μM of natural product compounds and centrifuged at 12000 × g for 5 min. Cell-free supernatant was collected in a 0.22 μm filter. The bioluminescence reporter strain V. harveyi BB170 grew in AB medium to 1.0 ~ 1.1 of OD600nm at 30°C under shaking and then diluted at 1:2500 with fresh AB medium. Twenty micro liter of cell-free supernatant mixed with 180 μl of V. harveyi BB170 culture in black 96-well plates (Jingan, Shanghai, China) and incubated for 3.5 h at 30°C in the dark. A multipurpose microplate reader (Enspire; PerkinElmer, USA) was used to measure bioluminescence. Cell-free supernatants of V. harveyi BB152 overnight cultures were used as control. Further study was conducted on the compound with the highest AI-2 inhibition. We also determined the IC50 for AI-2 inhibition with the selected compound with the same procedure described above.
Cytotoxicity
Ech’s toxicity was evaluated in Caco-2 cells using the CCK-8 assay. Caco-2 cells (105 cells/mL) were plated in a 96-well plate with 5% CO2 for 24 h at 37°C. The culture medium was then replaced by different concentrations of Ech (6.25, 12.5, 25, 50, 100, 200, 400 μM) for 24 h. After incubation, the plate was incubated at 37°C for an additional hour with 10 μl of CCK-8 (MCE, China). The absorbance was measured at 450 nm using Multiskan Go Reader (Thermo Fisher Scientific, United States).
Growth and metabolic activity
As described previously, the growth ability of E. coli was studied by the broth dilution method (CLSI, 2018; Swetha et al., 2019). In brief, E. coli O157:H7 bacterial suspension (OD600 = 0.01) with various concentration of Ech (6.25, 12.5, 25, 50, 100, 200 μM) was seeded into 96-well plate and incubated for 24 h at 37°C. The Multiskan Go Reader (Thermo Fisher Scientific, United States) was used to measure the absorbance at 600 nm.
Metabolic activity was measured with an Alamar Blue assay (Swetha et al., 2019). E. coli O157:H7 bacterial suspension (OD600 = 0.01) with various concentrations of Ech (6.25, 12.5, 25, 50, 100, 200 μM) was seeded into 12-well plate and incubated for 24 h at 37°C. Cells from each well were harvested at 10000 × g for 5 min and then washed twice using PBS (pH = 7.2). Metabolic activity was measured using Alamar Blue assay according to the manufacturer’s prescribed protocol (Invitrogen™, Thermo Fisher Scientific, USA). PBS containing only AB dye was considered a blank. The metabolic activity was calculated based on the absorbance at 570 nm and 600 nm using the following formula (Swetha et al., 2019):
Eoxi (OD570) – extinction coefficient in oxidized form of AB at 570 nm = 80,586;
Ered (OD570) – extinction coefficient in reduced form of AB at 570 nm = 155,677;
Eoxi (OD600) – extinction coefficient in oxidized form of AB at 600 nm = 117,216;
Ered (OD600) – extinction coefficient in reduced form of AB at 570 nm = 14,652;
B-blank; T-samples.
Biofilm assay
CV staining
The formation of biofilms was assessed using CV staining based on previous study (O’Toole, 2011) with slight modification. In brief, E. coli O157:H7 bacterial suspension (OD600 = 0.01) was cultured in LB medium at the various concentrations of Ech (6.25, 12.5, 25, 50, 100, 200 μM) in a 96-well plate (Corning Costar® 3,599, Corning, NY, United States) for 24 h at 37°C. The plate was washed three times with PBS (pH = 7.2) and then fixed for 1 h at 60°C. Methanol was used to fix the cells and 0.1% CV was used to stain them for 30 min. Next, the CV was rinsed with distilled water and dried under heat. Finally, the CV attached to wells was dissolved in 95% ethanol and then measured the absorbance at 570 nm using Multiskan Go Reader (Thermo Fisher Scientific, United States).
Confocal laser scanning microscopy
Biofilm formation of E. coli was determined using CLSM according to previous study (Zhang et al., 2020) with slight modification. In brief, E. coli O157:H7 bacterial suspension (OD600 = 0.01) supplemented with different concentrations of Ech (12.5, 25, 50 μM) was seeded into a 6-well plate with coverslips and incubated for 24 h at 37°C. The suspensions were removed, and the wells were washed with PBS (pH = 7.2). The biofilm was stained using BacLight Live/Dead viability kit (L7012, Invitrogen™, Thermo Fisher Scientific, United States) according to the procedure and observed by CLSM (Zeiss LSM800, Zeiss, Tokyo, Japan).
EPS production
According to the previous method, Ruthenium Red staining assessed EPS production (Adnan et al., 2020). Cell suspensions (106 CFU/ml) of E. coli O157:H7 and different concentrations of Ech were cultured in a 96-well plate for 24 h at 37°C. The plate was washed with PBS (pH = 7.2), stained with 0.01% ruthenium red (Yuanye, Shanghai, China), and then incubated at 37°C for 1 h. 0.01% ruthenium red was used to fill the wells without biofilm was used as blank. 0.01% ruthenium red was used to fill the wells with biofilm and without Ech was used as a positive control. The absorbance was performed at 450 nm using Multiskan Go Reader after the liquid carrying the residual stain was transferred to new 96-well plates (Thermo Fisher Scientific, USA). EPS inhibition was calculated as follows formula (Adnan et al., 2020):
Whereas:
AB = absorbance of the blank.
AS = absorbance of the sample.
AP = absorbance of the positive control.
Motility assay
According to the previous description, the motility of E.coli was performed (Vikram et al., 2013). Briefly, overnight E. coli O157:H7 was diluted to OD600 = 0.01, and then a semisolid agar media (0.3% LB agar) containing 12.5, 25, and 50 μM of Gin was used for the motility assay. One micro liter of the diluted bacterial solution was inserted into the middle of the plate. DMSO alone was used as the control. Halo zone diameters were measured after incubation for 16–18 h at 37°C to assess motility.
qRT-PCR
qRT-PCR was used to measure Ech’s effect on QS-regulated, biofilm formation, motility, and virulence factor-related genes of E. coli. Escherichia coli O157:H7 was incubated in the 12-wells plate with and without Ech at 37°C for 24 h. Bacterial RNA Kit(Omega, USA) was used to extract total RNA. RNA concentration was determined by NanoDrop OneC spectrophotometer (Thermo Scientific, USA). PrimeScript™ RT reagent Kit with gDNA Eraser (TAKARA Corporation, Japan) was used to reverse transcribe RNA into cDNA. qRT -PCR was analyzed using TB Green® Premix Ex TaqTM II (Tli RNaseH Plus) (TAKARA Corporation, Japan). Based on the 2−∆∆Ct method, the relative changes in gene expression levels were analyzed. The gapA gene was used as an internal control (Hu et al., 2013). This study used the primers listed in Supplementary Table 1.
Antibacterial activity
Antibacterial activity was evaluated based on the previous method (Swetha et al., 2019; Liu et al., 2021) with some modifications. Briefly, E. coli O157:H7 and five clinical strains (E. coli O101, E. coli C83654, E. coli O149, E. coli XJ24, E. coli KD-13-1) bacterial suspensions (OD600 = 0.01) were mixed with antibiotics (1/2 MIC, 1/4 MIC, 1/8 MIC) with or without Ech (50 μM) at 37°C for 16–18 h. Antibacterial effects were assessed by metabolic activity with Alamar Blue assay. Tests were conducted in triplicate.
Statistical analysis
The experiment was repeated three times with three replicates for each treatment, and data represent the mean ± SD. The significance of differences was evaluated with multiple t-tests for two groups or non-parametric one-way ANOVA for multiple groups using GraphPad Prism (GraphPad Prism 8; GraphPad). Where *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Results
Screening of QS inhibitors against Escherichia coli
To identify new QS inhibitors, 13 flavonoid compounds from nature compounds were screened by the V. harveyi BB170 bioluminescence assay (Supplementary Table 2). The results showed that the QS inhibition rates of two compounds Ech and aloeemodin were greater than 70% at 50 μM. The QS inhibition rates of six compounds loureirin B, phloretin, cardamonin, neosperidin dihydrochalcone, cynaroside, and acacetin were between 60 and 70%, and five compounds scutellarin, artemetin, hesperetin, vitexin, and gentiin were less than 50%. In particular, Ech was the better anti-QS activity with an IC50 of 21.67 μM (Figure 1). Therefore, it was focused on during our study.
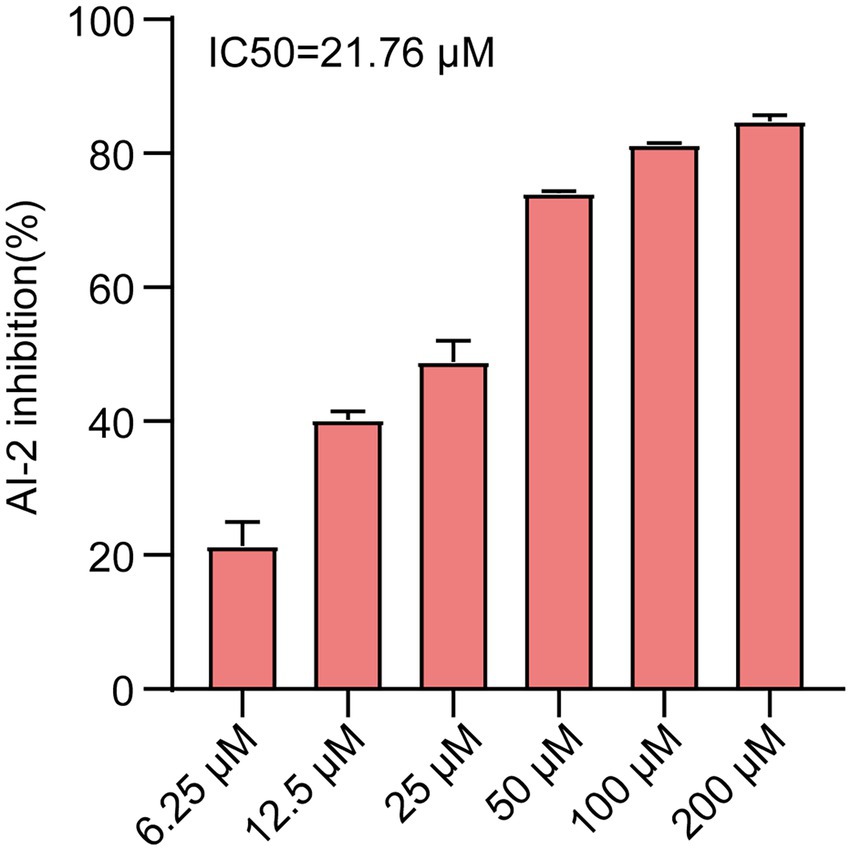
Figure 1. AI-2 inhibition of E. coli was treated with various concentrations of Ech (6.25, 12.5, 25, 50, 100, 200 μM) by AI-2 Bioluminescence assay and IC50 = 21.76 μM. All experiments were carried out in triplicate, and data represent the mean ± SD.
Cytotoxicity of Ech On Caco-2
Ech’s cytotoxicity was assessed in the study to develop it as a safer alternative to antibiotics. The viability of Caco-2 cells was evaluated using CCK-8 assay after treatment with Ech at various concentrations (6.25, 12.5, 25, 50, 100, 200 μM). Compared with the control, Ech is non-toxic to Caco-2 cells at concentrations below 100 μM (Figure 2).
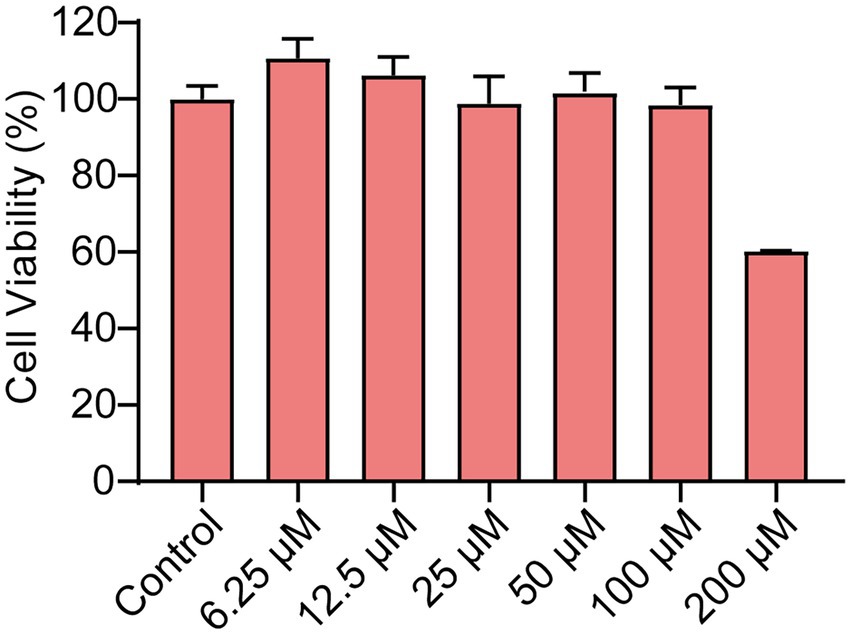
Figure 2. The Cytotoxicity of Ech in Caco-2 cells. The cells were treated with Ech at different concentrations (0, 6.25, 12.5, 25, 50, 100, and 200 μM) for 24 h. Data represent means ± SD of three experiments conducted in triplicate.
Effects of Ech on growth and metabolic activity of Escherichia coli
The effects of Ech on growth and metabolic activity were revealed by the microbroth dilution and Alamar Blue (AB) assay (Figure 3). Results showed that control and Ech treated cells showed no significant differences in the fluorescent intensity of AB dye. In addition, E. coli growth was not significantly different between the Ech treated sample and the control culture. These results showed the non-antibacterial effect of Ech.
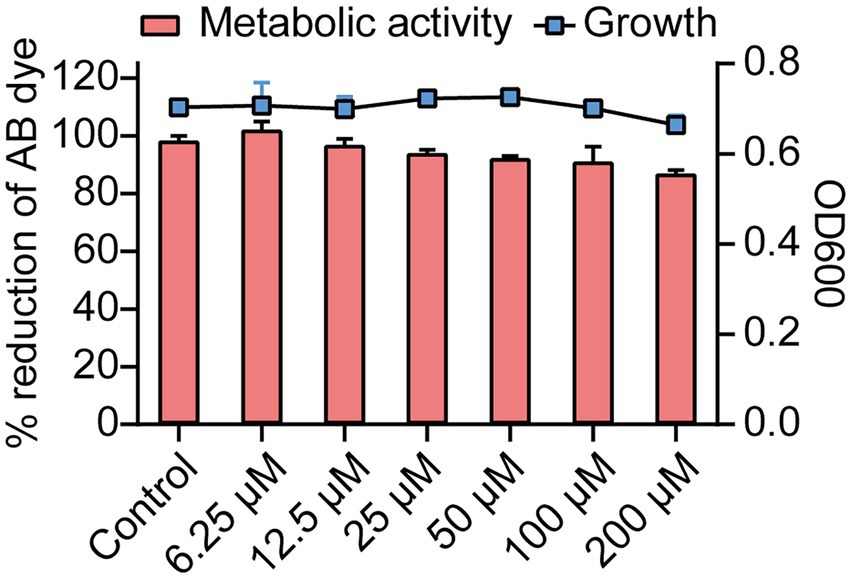
Figure 3. Growth and metabolic activity of E. coli in the presence of Ech. The line graph shows the growth of E. coli with Ech measured using a microbroth dilution assay. The bar graph shows the metabolic activity of E. coli based on the AB assay. Data represent means± SD of three experiments conducted in triplicate.
Effects of Ech on biofilm formation of Escherichia coli
Next, we assessed the effects of QS inhibitor on the formation of biofilm against E. coli by CV staining and CLSM. CV staining showed that Ech significantly inhibited the biofilm formation of E. coli in a dose-dependent manner (Figure 4A). More specifically, inhibition reached 40% at 6.25 μM of inhibitor concentration, while 71.54% inhibition was achieved at 200 μM. In addition, the antibiofilm of Ech was further verified by CLSM. As shown in Figure 4B, green fluorescence (living cells) and red fluorescence (death cells) showed significant decreases with increasing concentration, which proved that Ech could effectively inhibit the adhesion of E. coli and reduce biofilm formation.
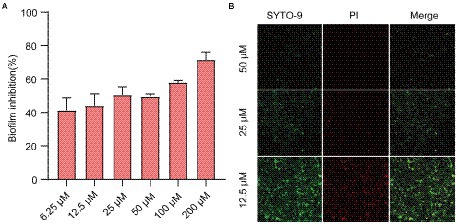
Figure 4. Effects of Ech on E. coli biofilm formation. (A) Biofilm formation inhibition at various concentrations of Ech (6.25, 12.5, 25, 50, 100, and 200 μM) for 24 h by CV staining. (B) Three-dimensional (3D) image of E. coli biofilm with Ech (12.5, 25, and 50 μM) for 24 h by CLSM. Error bars are mean ± SD.
Effects of Ech on EPS production of Escherichia coli
In a biofilm matrix, EPS is among the most critical components (Branda et al., 2005). As was shown in Figure 5, Ech inhibited EPS production dose-dependently. The EPS inhibition reached 50% at the concentration of 100 μM. It was consistent with the results of CV staining. This result further confirms the inhibition of Ech on the formation of biofilm.
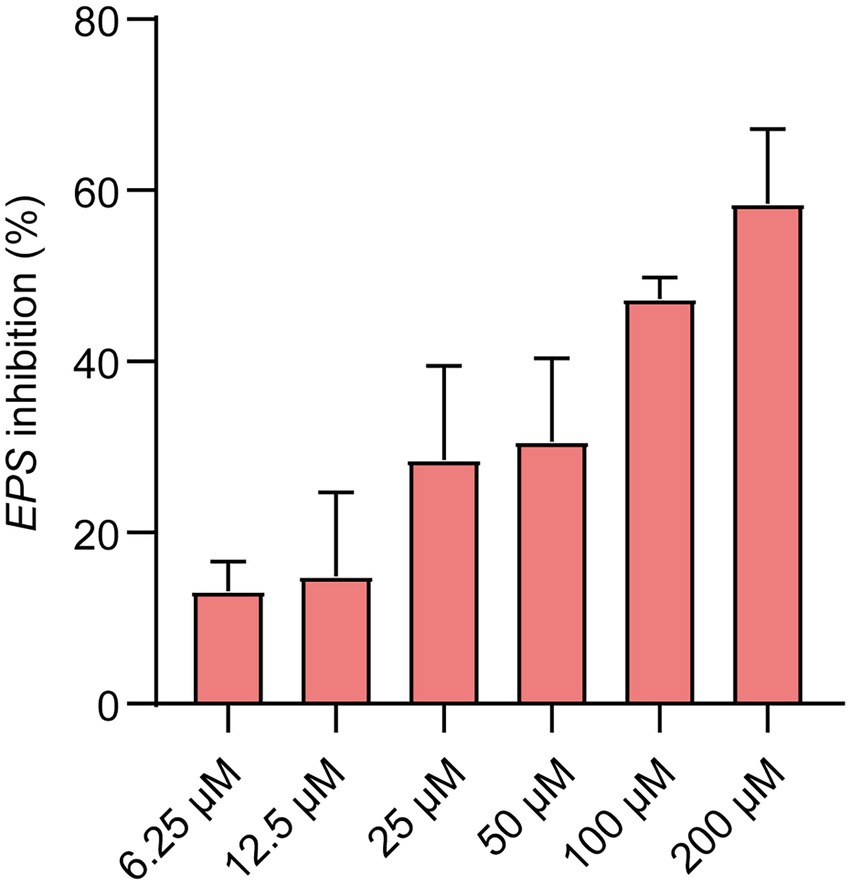
Figure 5. EPS inhibition (%) showed at the various concentrations of Ech (6.25, 12.5, 25, 50, 100, and 200 μM) for 24 h. Data represent means ± SD of three experiments conducted in triplicate.
Effects of Ech on the motility of Escherichia coli
Quorum-sensing controls the motility of E.coli. Figure 6A showed that QS inhibitor Ech reduced the motility in a dose-dependent. Further, we assessed the halo zone quantitatively (Figure 6B). Results showed that Ech significantly inhibited the motility of E. coli in comparison with the control group (p < 0.0001).
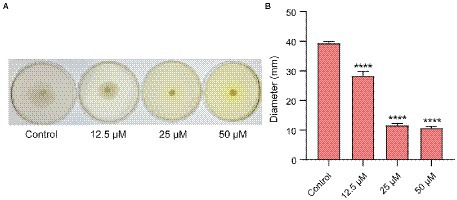
Figure 6. Motility inhibition of E. coli with Ech. (A) Images of motility following incubation with E. coli at the various concentration of Ech (0, 12.5, 25, and 50 μM). (B) Quantitative estimation of motility based on the halo zone’s diameter. **** = p < 0.0001.
Effect of Ech on the expression of QS-regulated genes of Escherichia coli
To further evaluate the potential molecular basis responsible for QS inhibition by Ech, we measured the expression of QS, motility, and biofilm genes by qRT - PCR. Ech reduced the expression of QS-regulated genes like luxS, pfs, lsrB, lsrK, and lsrR (43, 48, 52, 90, and 61% respectively), motility-regulated genes flhC, flhD, and fliC (61, 35, and 53%, respectively), biofilm-regulated genes csgD (52%), and virulence factor-regulated gene stx2 (69%; Figure 7). The result demonstrated that Ech inhibited QS, biofilm formation, motility, and virulence factor production.
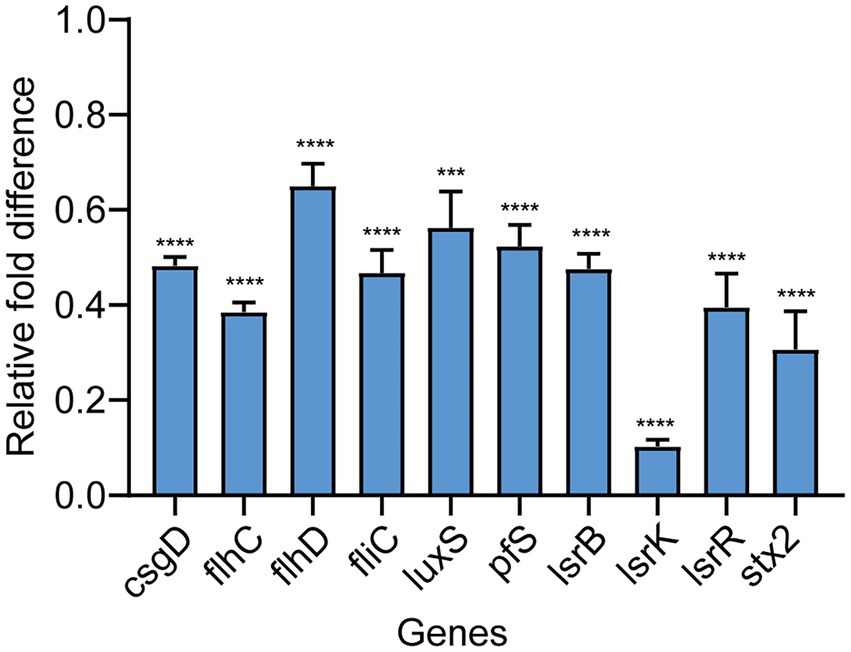
Figure 7. Effects of Ech on expression QS-regulated genes of E. coli. The qRT-PCR results showed significantly different nine genes (csgD, flhC, flhD, fliC, luxS, pfs, lsrB, lsrK, lsrR, and stx2) compared with the control. Data represent means ± SD of three experiments conducted in triplicate.
Synergistic effects of Ech with antibiotics against Escherichia coli
It was reported that antibiotic resistance was closed related to QS. So, we evaluated the synergistic antimicrobial effect of QS inhibitor Ech with conventional antibiotics against E. coli O157:H7 and five clinical isolates of strains (E. coli C83654, E. coli XJ24, E. coli O101, E. coli O149, E. coli KD-13-1). First, we evaluated the synergistic antibacterial effect of Ech and six different antibiotics against E. coli O157:H7. According to Figure 8, QS inhibitor Ech only has synergistic antibacterial activity with colistin antibiotics (1/2 MIC, 1/4 MIC, 1/8 MIC). In addition, the synergistic antibacterial effect of Ech with polymyxin B and polymyxin E on five clinical isolates of E. coli was further verified (Figures 9, 10). These results suggested that QS inhibitor Ech in conjunction with conventional antibiotics could be an efficient therapeutic strategy for inhibiting pathogens like E. coli.
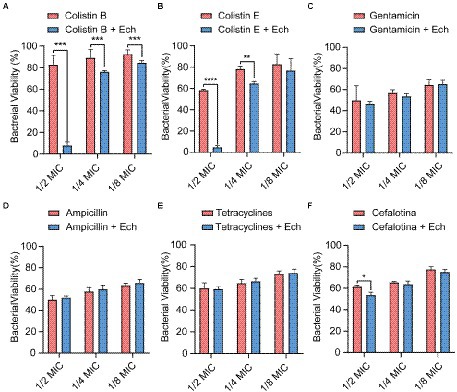
Figure 8. Synergistic effects of Ech (50 μM) with six different antibiotics in E. coli O157:H7 on bacterial viability. Data represent means ± SD of three experiments conducted in triplicate. * = p < 0.05; ** = p < 0.01; *** = p < 0.001;**** = p < 0.0001.
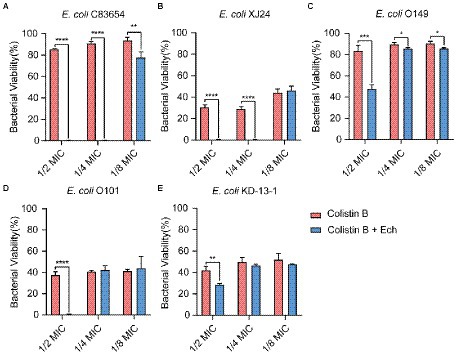
Figure 9. Synergistic effects of Ech (50 μM) with Colistin B in five different E. coli on bacterial viability. E. coli (A) C83654, (B) XJ24, (C) O149, (D) O101, (E) KD-13-1. Data represent means ± SD of three experiments conducted in triplicate. * = p < 0.05; ** = p < 0.01; *** = p < 0.001; **** = p < 0.0001.
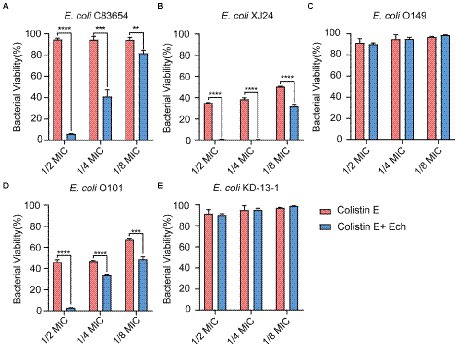
Figure 10. Synergistic effects of Ech (50 μM) with Colistin E in five different E. coli on bacterial viability. E. coli (A) C83654, (B) XJ24, (C) O149, (D) O101, (E) KD-13-1. Data represent means ± SD of three experiments conducted in triplicate. ** = p < 0.01; *** = p < 0.001; **** = p < 0.0001.
Discussion
Microbial infections continue to pose a serious problem because of antibiotic resistance, making alternative therapies imperative. QS not only regulates different pathogenic processes like virulence production (Defoirdt, 2018), biofilm formation (Rickard et al., 2006; Irie and Parsek, 2008), and antibiotic sensitivity (Stenvang et al., 2016; Liu et al., 2021) but also does not result in the development of resistance (Sully et al., 2014; Quave et al., 2015; Ning et al., 2021). It has emerged as the potential target for fighting antibiotic resistance (Suga and Smith, 2003; Geske et al., 2005; Rasmussen and Givskov, 2006). In the present study, we identified that Ech, a novel QS inhibitor selected from 13 different flavonoids, did not inhibit E. coli growth and metabolic activity but affected QS-regulated processes like biofilm formation, EPS production, motility, and virulence factor production. Furthermore, we demonstrated the synergistic effect of Ech combination with colistin B and colistin E against E. coli O157:H7 and five clinical isolates of strains. In short, we concluded that Ech presents excellent anti-QS and synergistic antibacterial effects against E. coli.
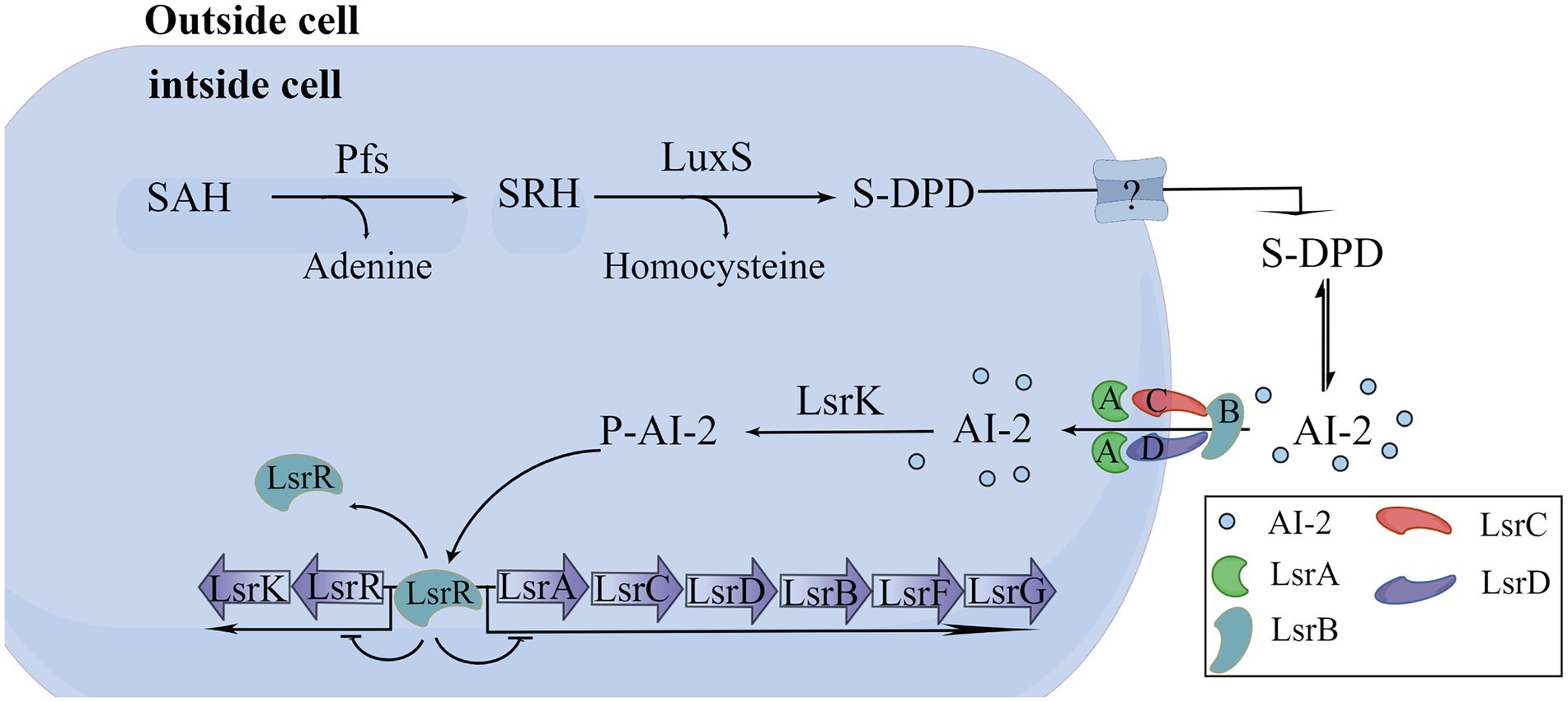
At present, the detection methods of signal molecule AI-2 mainly include V. harveyi BB170 bioluminescence assay, LuxP-based fluorescence resonance energy transfer (LuxP-FRET) reporting system assay, QS engineering protein detection assay, Gas Chromatography-Mass Spectrometer (GC–MS) assay, high-performance liquid chromatography with tandem mass spectrometric (HPLC-MS/MS) assay, and high-performance liquid chromatography with fluorescence detection (HPLC-FLD) assay (Yan et al., 2016). Among them, V harveyi BB170 bioluminescence assay is the most commonly used method at present. In this study, we screened Ech with high-efficiency anti-QS from 13 different flavonoids through the AI-2 bioluminescence assay. In addition, we proved that Ech had no cell toxicity in Caco-2 in the effective dose and adverse effects on bacterial growth and metabolic activity. Further, Ech also significantly decreased the expression of QS-regulated genes, including luxS, pfs, lsrB, lsrK, and lsrR. Several studies have shown that AI-2 regulates the formation of biofilm, the production of virulence factors, motility, and resistance of bacteria. So, pathogen virulence could be reduced by inhibiting AI-2 production (Ma et al., 2017; Sedlmayer et al., 2021; Jiang et al., 2022). LuxS and pfs are responsible for the synthesis of AI-2 with S-adenosylhomocysteine (SAH) as substrate (Figure 11; Zhao et al., 2018; Wu et al., 2019). Similarly, it has been reported that luxS and pfs deletion mutants of E. coli inhibit AI-2 production, biofilm formation, and motility (Yang et al., 2014; Wang et al., 2016; Zuberi et al., 2017). Therefore, the luxS and pfs have been considered important therapeutic drug targets, and many inhibitors against luxS and pfs have been investigated (Shivaprasad et al., 2021; Sharifi and Nayeri Fasaei, 2022). The lsr system is liable for AI-2 detection, uptake, and signal transduction in E. coli (Pereira et al., 2013). LsrR is an inhibitor of the lsr system. There is very little extracellular AI-2 during the early stages of bacterial growth. LsrR is active and inhibits the transcription of the lsr system. LsrK phosphorylates uptake of AI-2 as it accumulates in the extracellular space. The binding of phospho-AI-2 to lsrR results in the initiation of transport by the lsr system. LsrB is an AI-2 receptor responsible for the internalization of AI-2 in E. coli (Figure 11). Similarly, lsrR, lsrK, and lsrB deletion mutant has been reported to interfere with signal transduction and inhibit biofilm, motility, and pathogenicity (Li et al., 2007; Jani et al., 2017; Laganenka and Sourjik, 2018; Zuo et al., 2019). So, the lsrR, lsrK, and lsrB have been considered important therapeutic drug targets. Many inhibitors against lsrR, lsrK, and lsrB have been investigated (Peng et al., 2018; Gatta et al., 2019; Stotani et al., 2019; Gatta et al., 2020). Our results suggested that Ech could interfere with the AI-2 synthesis, secretion, or transport through their effects on luxS, pfs, lsrK, or lsrB.
The biofilm is an aggregate of microorganisms where microorganisms are adhered to the substrate (e.g., stainless, glass, meats, and vegetables) and encapsulated within a self-produced matrix of EPS, proteins, and extracellular DNA (eDNA) (Hobley et al., 2015; Flemming et al., 2016; Karygianni et al., 2020). EPS can separate colonies and form a pathway for transporting metabolites and nutrients. In addition, EPS can maintain the biofilm’s structure and contribute to bacteria colonization on the non-biological surface (Fan et al., 2022). Curli fimbriae and cellulose, whose products are regulated by csgD, are common EPS components found in bacterial biofilms (Dieltjens et al., 2020). CsgD stimulates the formation of E. coli biofilms by simultaneously activating the expression of the curli- encoding genes csg operon and inhibiting negative affect factors such as yagS and pepD (Brombacher et al., 2003). Otherwise, csgD directly inhibits flagellum formation of fliE and fliEFGH operon, thereby regulating biofilm formation and cell motility by inhibiting flagellum formation and rotation. This study showed that Ech inhibited EPS production in a dose-dependent manner by Ruthenium Red stain. Further, Ech also significantly decreased csgD expression (Figure 7). The results demonstrated that Ech reduced EPS production by inhibiting QS.
We performed motility experiments in a semi-solid agar medium to determine whether Ech affects bacterial motility. The results showed that Ech inhibited bacterial motility in a dose-dependent manner. Additionally, flagellum-regulated genes such as flhC, flhD, and fliC were also significantly down-regulated by Ech. FlhDC is an activator of the flagellum regulatory cascade, which regulates flagellum synthesis and motility (Guttenplan and Kearns, 2013). FliC, as a flagellum filament structural protein, is involved in the pathogenesis of infection, the production of biofilm, and motility (Chaban et al., 2015). The results demonstrated that Ech reduced flagellum motility by inhibiting QS.
Substances that constitute bacterial virulence are called virulence factors including invasiveness and virulence. Among the six major pathotypes of E. coli, Shiga toxin-producing E. coli is the most prevalent (Moreau et al., 2021). Shiga toxin is one of the major virulence factors of Shiga toxin-producing E. coli O157: H7. It can induce the formation of A/E lesions, and cause HUS if it enters the circulatory system (Kordbacheh et al., 2019). In humans, the stx2 gene codes for the Stx2 which is associated with more severe diseases caused by Shiga toxins, which play an essential role in the pathogenesis of E. coli O157:H7 (Sheng et al., 2016). In the present student, we found that the virulence factor regulatory gene stx2 was downregulated by 69%. The results demonstrated that Ech reduced virulence factor production by inhibiting QS.
Multiple antibiotic resistance mechanisms have evolved due to the extensive use of antibiotics during clinical treatment. The primary resistance mechanisms are to passivate antibiotics by eliminating antibiotics by efflux pump system, chemical modifications, and target gene modification (Zhao et al., 2020). Meanwhile, the biofilms formed by many pathogens result in strong resistance (Rajput et al., 2018). QS system can reduce antibiotic resistance by regulating biofilm formation (Zhao et al., 2020). In this study, we found that combined QS inhibitor Ech and colistin antibiotics could play a synergistic antibacterial effect. The results suggested that combining antibiotics with anti-QS compounds appears to be an effective therapeutic strategy for treating pathogen infections.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
Y-BB wrote the manuscript and participated in some experiments. M-YS, L-YW, and Y-TB performed part of the experiments. W-WW, X-ZZ, and BL revised the manuscript. J-YZ and W-WW directed the project and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the research on the National Natural Science Foundation of China, grant number 32102727–the earmarked fund for CARS, grant number CARS-37–Innovation Project of Chinese Academy of Agricultural Sciences, grant number 25-LZIHPS-05.
Acknowledgments
We thank Researcher Xiangan Han for providing the strain Vibrio harveyi BB170 and Vibrio harveyi BB152. Additionally, we are grateful for the suggestions made by reviewers.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1003692/full#supplementary-material
References
Adnan, M., Patel, M., Deshpande, S., Alreshidi, M., Siddiqui, A. J., Reddy, M. N., et al. (2020). Effect of adiantum philippense extract on biofilm formation, adhesion with its antibacterial activities against foodborne pathogens, and characterization of bioactive metabolites: an in vitro-in silico approach. Front. Microbiol. 11:823. doi: 10.3389/fmicb.2020.00823
Bassler, B. L. (2002). Small talk. Cell-to-cell communication in bacteria. Cells 109, 421–424. doi: 10.1016/s0092-8674(02)00749-3
Bassler, B. L., Greenberg, E. P., and Stevens, A. M. (1997). Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179, 4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997
Bassler, B. L., Wright, M., and Silverman, M. R. (1994). Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13, 273–286. doi: 10.1111/j.1365-2958.1994.tb00422.x
Beshearse, E., Bruce, B. B., Nane, G. F., Cooke, R. M., Aspinall, W., Hald, T., et al. (2021). Attribution of illnesses transmitted by food and water to comprehensive transmission pathways using structured expert judgment. United States Emerg. Infect. Dis. 27, 182–195. doi: 10.3201/eid2701.200316
Branda, S. S., Vik, S., Friedman, L., and Kolter, R. (2005). Biofilms: the matrix revisited. Trends Microbiol. 13, 20–26. doi: 10.1016/j.tim.2004.11.006
Brombacher, E., Dorel, C., Zehnder, A. J. B., and Landini, P. (2003). The curli biosynthesis regulator csgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149, 2847–2857. doi: 10.1099/mic.0.26306-0
Buroni, S., and Chiarelli, L. R. (2020). Antivirulence compounds: a future direction to overcome antibiotic resistance? Future Microbiol. 15, 299–301. doi: 10.2217/fmb-2019-0294
Chaban, B., Hughes, H. V., and Beeby, M. (2015). The flagellum in bacterial pathogens: for motility and a whole lot more. Semin. Cell Dev. Biol. 46, 91–103. doi: 10.1016/j.semcdb.2015.10.032
Choudhary, S., and Schmidt-Dannert, C. (2010). Applications of quorum sensing in biotechnology. Appl. Microbiol. Biotechnol. 86, 1267–1279. doi: 10.1007/s00253-010-2521-7
CLSI (2018). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically 11th ed. (Wayne, PA: Clinical and Laboratory Standards Institute).
Defoirdt, T. (2018). Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 26, 313–328. doi: 10.1016/j.tim.2017.10.005
Dickey, S. W., Cheung, G. Y. C., and Otto, M. (2017). Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 16, 457–471. doi: 10.1038/nrd.2017.23
Dieltjens, L., Appermans, K., Lissens, M., Lories, B., Kim, W., Van der Eycken, E. V., et al. (2020). Inhibiting bacterial cooperation is an evolutionarily robust anti-biofilm strategy. Nat. Commun. 11:107. doi: 10.1038/s41467-019-13660-x
Fan, Q., Zuo, J., Wang, H., Grenier, D., Yi, L., and Wang, Y. (2022). Contribution of quorum sensing to virulence and antibiotic resistance in zoonotic bacteria. Biotechnol. Adv. 59:107965. doi: 10.1016/j.biotechadv.2022.107965
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Gatta, V., Ilina, P., Porter, A., McElroy, S., and Tammela, P. (2019). Targeting quorum sensing: high-throughput screening to identify novel lsrK inhibitors. Int. J. Mol. Sci. 20:3112. doi: 10.3390/ijms20123112
Gatta, V., Tomašič, T., Ilaš, J., Zidar, N., Peterlin Mašič, L., Barančoková, M., et al. (2020). A new cell-based AI-2-mediated quorum sensing interference assay in screening of lsrK-targeted inhibitors. Chembiochem 21, 1918–1922. doi: 10.1002/cbic.201900773
Geske, G. D., Wezeman, R. J., Siegel, A. P., and Blackwell, H. E. (2005). Small molecule inhibitors of bacterial quorum sensing and biofilm formation. J. Am. Chem. Soc. 127, 12762–12763. doi: 10.1021/ja0530321
Guo, M., Gamby, S., Zheng, Y., and Sintim, H. O. (2013). Small molecule inhibitors of AI-2 signaling in bacteria: state-of-the-art and future perspectives for anti-quorum sensing agents. Int. J. Mol. Sci. 14, 17694–17728. doi: 10.3390/ijms140917694
Guttenplan, S. B., and Kearns, D. B. (2013). Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 37, 849–871. doi: 10.1111/1574-6976.12018
Hale, C. R., Scallan, E., Cronquist, A. B., Dunn, J., Smith, K., Robinson, T., et al. (2012). Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin. Infect. Dis. 54, S472–S479. doi: 10.1093/cid/cis051
Hawkey, P. M. (2008). The growing burden of antimicrobial resistance. J. Antimicrob. Chemother. 62, i1–i9. doi: 10.1093/jac/dkn241
Hobley, L., Harkins, C., MacPhee, C. E., and Stanley-Wall, N. R. (2015). Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 39, 649–669. doi: 10.1093/femsre/fuv015
Hu, Y., Liu, M., Qin, H., Lin, H., An, X., Shi, Z., et al. (2021). Artemether, Artesunate, Arteannuin B, Echinatin, Licochalcone B and Andrographolide effectively inhibit SARS-CoV-2 and related viruses in vitro. Front. Cell. Infect. Microbiol. 11:680127. doi: 10.3389/fcimb.2021.680127
Hu, J., Wang, B., Fang, X., Means, W. J., McCormick, R. J., Gomelsky, M., et al. (2013). C-di-GMP signaling regulates E. coli O157:H7 adhesion to colonic epithelium. Vet. Microbiol. 164, 344–351. doi: 10.1016/j.vetmic.2013.02.023
Irie, Y., and Parsek, M. R. (2008). Quorum sensing and microbial biofilms. Curr. Top. Microbiol. Immunol. 322, 67–84. doi: 10.1007/978-3-540-75418-3_4
Jani, S., Seely, A. L., Peabody, V. G., Jayaraman, A., and Manson, M. D. (2017). Chemotaxis to self-generated AI-2 promotes biofilm formation in Escherichia coli. Microbiology 163, 1778–1790. doi: 10.1099/mic.0.000567
Ji, S., Li, Z., Song, W., Wang, Y., Liang, W., Li, K., et al. (2016). Bioactive constituents of glycyrrhiza uralensis (licorice): discovery of the effective components of a traditional herbal medicine. J. Nat. Prod. 79, 281–292. doi: 10.1021/acs.jnatprod.5b00877
Jiang, K., Xu, Y., Yuan, B., Yue, Y., Zhao, M., Luo, R., et al. (2022). Effect of autoinducer-2 quorum sensing inhibitor on interspecies quorum sensing. Front. Microbiol. 13:791802. doi: 10.3389/fmicb.2022.791802
Ju, X. Y., Li, J. J., Zhu, M. J., Lu, Z. X., Lv, F. X., Zhu, X. Y., et al. (2018). Effect of the luxS gene on biofilm formation and antibiotic resistance by salmonella serovar Dublin. Food Res. Int. 107, 385–393. doi: 10.1016/j.foodres.2018.02.039
Karnjana, K., Nobsathian, S., Soowannayan, C., Zhao, W., Tang, Y. J., and Wongprasert, K. (2020). Purification and evaluation of N-benzyl cinnamamide from red seaweed gracilaria fisheri as an inhibitor of Vibrio harveyi AI-2 quorum sensing. Mar. Drugs 18:80. doi: 10.3390/md18020080
Karygianni, L., Ren, Z., Koo, H., and Thurnheer, T. (2020). Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 28, 668–681. doi: 10.1016/j.tim.2020.03.016
Kendall, M. M., Rasko, D. A., and Sperandio, V. (2007). Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect. Immun. 75, 4875–4884. doi: 10.1128/iai.00550-07
Kordbacheh, E., Nazarian, S., Hajizadeh, A., Fasihi-Ramandi, M., and Fathi, J. (2019). Recombinant HcpA-EspA-Tir-Stx2B chimeric protein induces immunity against attachment and toxicity of Escherichia coli O157:H7. Microb. Pathog. 129, 176–182. doi: 10.1016/j.micpath.2019.02.004
Kwak, A. W., Choi, J. S., Lee, M. H., Oh, H. N., Cho, S. S., Yoon, G., et al. (2019). Retrochalcone Echinatin triggers apoptosis of esophageal squamous cell carcinoma via ROS- and ER stress-mediated signaling pathways. Molecules 24:4055. doi: 10.3390/molecules24224055
Laganenka, L., and Sourjik, V. (2018). Autoinducer 2-dependent Escherichia coli biofilm formation is enhanced in a dual-species coculture. Appl. Environ. Microbiol. 84, e02638–02617. doi: 10.1128/aem.02638-17
Li, J., Attila, C., Wang, L., Wood, T. K., Valdes, J. J., and Bentley, W. E. (2007). Quorum sensing in Escherichia coli is signaled by AI-2/lsrR: effects on small RNA and biofilm architecture. J. Bacteriol. 189, 6011–6020. doi: 10.1128/JB.00014-07
Li, J. P., Fan, Q. Y., Jin, M. Y., Mao, C. L., Zhang, H., Zhang, X. L., et al. (2021). Paeoniflorin reduce luxS/AI-2 system-controlled biofilm formation and virulence in Streptococcus suis. Virulence 12, 3062–3073. doi: 10.1080/21505594.2021.2010398
Liu, J., Hou, J. S., Chang, Y. Q., Peng, L. J., Zhang, X. Y., Miao, Z. Y., et al. (2021). New pqs quorum sensing system inhibitor as an antibacterial synergist against multidrug-resistant Pseudomonas aeruginosa. J. Med. Chem. 65, 688–709. doi: 10.1021/acs.jmedchem.1c01781
Loubet, P., Ranfaing, J., Dinh, A., Dunyach-Remy, C., Bernard, L., Bruyère, F., et al. (2020). Alternative therapeutic options to antibiotics for the treatment of urinary tract infections. Front. Microbiol. 11:1509. doi: 10.3389/fmicb.2020.01509
Lutful Kabir, S. M. (2010). Avian Colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. Int. J. Environ. Res. Public Health 7, 89–114. doi: 10.3390/ijerph7010089
Ma, Y. P., Hao, L., Ke, H., Liang, Z. L., Ma, J. Y., Liu, Z. X., et al. (2017). LuxS/AI-2 in Streptococcus agalactiae reveals a key role in acid tolerance and virulence. Res. Vet. Sci. 115, 501–507. doi: 10.1016/j.rvsc.2017.07.032
Manner, S., and Fallarero, A. (2018). Screening of natural product derivatives identifies two structurally related flavonoids as potent quorum sensing inhibitors against gram-negative bacteria. Int. J. Mol. Sci. 19:1346. doi: 10.3390/ijms19051346
Meng, F., Zhang, F., Chen, Q., Yang, M., Yang, Y., Li, X., et al. (2022). Virtual screening and in vitro experimental verification of LuxS inhibitors from natural products for lactobacillus reuteri. Biomed. Pharmacother. 147:112521. doi: 10.1016/j.biopha.2021.112521
Minich, A., Levarski, Z., Mikulášová, M., Straka, M., Liptáková, A., and Stuchlík, S. (2022). Complex analysis of vanillin and syringic acid as natural antimicrobial agents against Staphylococcus epidermidis biofilms. Int. J. Mol. Sci. 23:1816. doi: 10.3390/ijms23031816
Moreau, M. R., Kudva, I. T., Katani, R., Cote, R., Li, L., Arthur, T. M., et al. (2021). Nonfimbrial adhesin mutants reveal divergent Escherichia coli O157:H7 adherence mechanisms on human and cattle epithelial cells. Int. J. Microbiol. 2021:8868151. doi: 10.1155/2021/8868151
Munguia, J., and Nizet, V. (2017). Pharmacological targeting of the host-pathogen interaction: alternatives to classical antibiotics to combat drug-resistant superbugs. Trends Pharmacol. Sci. 38, 473–488. doi: 10.1016/j.tips.2017.02.003
Ng, W. L., and Bassler, B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43, 197–222. doi: 10.1146/annurev-genet-102108-134304
Ning, Q., Wang, D., and You, J. (2021). Joint effects of antibiotics and quorum sensing inhibitors on resistance development in bacteria. Environ Sci Process Impacts 23, 995–1005. doi: 10.1039/d1em00047k
O’Toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 47:2437. doi: 10.3791/2437
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Peng, L. Y., Yuan, M., Cui, Z. Q., Wu, Z. M., Yu, Z. J., Song, K., et al. (2018). Rutin inhibits quorum sensing, biofilm formation and virulence genes in avian pathogenic Escherichia coli. Microb. Pathog. 119, 54–59. doi: 10.1016/j.micpath.2018.04.007
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Piewngam, P., Chiou, J., Chatterjee, P., and Otto, M. (2020). Alternative approaches to treat bacterial infections: targeting quorum-sensing. Expert Rev. Anti-Infect. Ther. 18, 499–510. doi: 10.1080/14787210.2020.1750951
Quave, C. L., Lyles, J. T., Kavanaugh, J. S., Nelson, K., Parlet, C. P., Crosby, H. A., et al. (2015). Castanea sativa (European chestnut) leaf extracts rich in ursene and oleanane derivatives block Staphylococcus aureus virulence and pathogenesis without detectable resistance. PLoS One 10:e0136486. doi: 10.1371/journal.pone.0136486
Rajput, A., Thakur, A., Sharma, S., and Kumar, M. (2018). ABiofilm: a resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res. 46, D894–d900. doi: 10.1093/nar/gkx1157
Ran, H., Liu, H., and Wu, P. (2021). Echinatin mitigates H2O2-induced oxidative damage and apoptosis in lens epithelial cells via the Nrf2/HO-1 pathway. Adv. Clin. Exp. Med. 30, 1195–1203. doi: 10.17219/acem/139130
Rasmussen, T. B., and Givskov, M. (2006). Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 296, 149–161. doi: 10.1016/j.ijmm.2006.02.005
Reading, N. C., and Sperandio, V. (2006). Quorum sensing: the many languages of bacteria. FEMS Microbiol. Lett. 254, 1–11. doi: 10.1111/j.1574-6968.2005.00001.x
Rickard, A. H., Palmer, R. J., Blehert, D. S., Campagna, S. R., Semmelhack, M. F., Egland, P. G., et al. (2006). Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60, 1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x
Riley, L. W. (2020). Distinguishing pathovars from nonpathovars: Escherichia coli. Microbiol. Spectr. 8, 1–23. doi: 10.1128/microbiolspec.AME-0014-2020
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.p11101
Sedlmayer, F., Woischnig, A. K., Unterreiner, V., Fuchs, F., Baeschlin, D., Khanna, N., et al. (2021). 5-fluorouracil blocks quorum-sensing of biofilm-embedded methicillin-resistant Staphylococcus aureus in mice. Nucleic Acids Res. 49:e73. doi: 10.1093/nar/gkab251
Sharifi, A., and Nayeri Fasaei, B. (2022). Selected plant essential oils inhibit biofilm formation and luxS- and pfs-mediated quorum sensing by Escherichia coli O157:H7. Lett. Appl. Microbiol. 74, 916–923. doi: 10.1111/lam.13673
Sheng, L., Rasco, B., and Zhu, M. J. (2016). Cinnamon oil inhibits Shiga toxin type 2 phage induction and Shiga toxin type 2 production in Escherichia coli O157:H7. Appl. Environ. Microbiol. 82, 6531–6540. doi: 10.1128/aem.01702-16
Shivaprasad, D. P., Taneja, N. K., Lakra, A., and Sachdev, D. (2021). In vitro and in situ abrogation of biofilm formation in E. coli by vitamin C through ROS generation, disruption of quorum sensing and exopolysaccharide production. Food Chem. 341:128171. doi: 10.1016/j.foodchem.2020.128171
Silva, D. R., Sardi, J. D. C. O., Pitangui, N. D. S., Roque, S. M., Silva, A. C. B. D., and Rosalen, P. L. (2020). Probiotics as an alternative antimicrobial therapy: current reality and future directions. J. Funct. Foods 73:104080. doi: 10.1016/j.jff.2020.104080
Stenvang, M., Dueholm, M. S., Vad, B. S., Seviour, T., Zeng, G., Geifman-Shochat, S., et al. (2016). Epigallocatechin gallate remodels overexpressed functional amyloids in Pseudomonas aeruginosa and increases biofilm susceptibility to antibiotic treatment. J. Biol. Chem. 291, 26540–26553. doi: 10.1074/jbc.M116.739953
Stotani, S., Gatta, V., Medarametla, P., Padmanaban, M., Karawajczyk, A., Giordanetto, F., et al. (2019). DPD-inspired discovery of novel lsrK kinase inhibitors: an opportunity to fight antimicrobial resistance. J. Med. Chem. 62, 2720–2737. doi: 10.1021/acs.jmedchem.9b00025
Sturme, M. H., Kleerebezem, M., Nakayama, J., Akkermans, A. D., Vaugha, E. E., and de Vos, W. M. (2002). Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 81, 233–243. doi: 10.1023/a:1020522919555
Suga, H., and Smith, K. M. (2003). Molecular mechanisms of bacterial quorum sensing as a new drug target. Curr. Opin. Chem. Biol. 7, 586–591. doi: 10.1016/j.cbpa.2003.08.001
Sully, E. K., Malachowa, N., Elmore, B. O., Alexander, S. M., Femling, J. K., Gray, B. M., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174. doi: 10.1371/journal.ppat.1004174
Sun, Y., Li, Y., Luo, Q., Huang, J., Chen, J., Zhang, R., et al. (2020). LuxS/AI-2 quorum sensing system in Edwardsiella piscicida promotes its biofilm formation and pathogenicity. Infect. Immun. 88, e00907–00919. doi: 10.1128/iai.00907-19
Surette, M. G., Miller, M. B., and Bassler, B. L. (1999). Quorum sensing in Escherichia coli, salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. U. S. A. 96, 1639–1644. doi: 10.1073/pnas.96.4.1639
Swetha, T. K., Pooranachithra, M., Subramenium, G. A., Divya, V., Balamurugan, K., and Pandian, S. K. (2019). Umbelliferone impedes biofilm formation and virulence of methicillin-resistant Staphylococcus epidermidis via impairment of initial attachment and intercellular adhesion. Front. Cell. Infect. Microbiol. 9:357. doi: 10.3389/fcimb.2019.00357
Taga, M. E., and Xavier, K. B. (2011). Methods for analysis of bacterial autoinducer-2 production. Curr. Protoc. Microbiol. Chapter 1:Unit1C.1. doi: 10.1002/9780471729259.mc01c01s23
Vikram, A., Jayaprakasha, G. K., Uckoo, R. M., and Patil, B. S. (2013). Inhibition of Escherichia coli O157:H7 motility and biofilm by β-sitosterol glucoside. Biochim. Biophys. Acta 1830, 5219–5228. doi: 10.1016/j.bbagen.2013.07.022
Wang, W., Huang, X., Yang, H., Niu, X., Li, D., Yang, C., et al. (2019). Antibacterial activity and anti-quorum sensing mediated phenotype in response to essential oil from melaleuca bracteata leaves. Int. J. Mol. Sci. 20:5696. doi: 10.3390/ijms20225696
Wang, X., Li, S., Lu, X., Hu, P., Chen, H., Li, Z., et al. (2016). Rapid method of luxS and pfs gene inactivation in enterotoxigenic Escherichia coli and the effect on biofilm formation. Mol. Med. Rep. 13, 257–264. doi: 10.3892/mmr.2015.4532
Whiteley, M., Diggle, S. P., and Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624
Whiteley, M., Diggle, S. P., and Greenberg, E. P. (2018). Corrigendum: Progress in and promise of bacterial quorum sensing research. Nature 555:126. doi: 10.1038/nature25977
Wu, S., Liu, J., Liu, C., Yang, A., and Qiao, J. (2019). Quorum sensing for population-level control of bacteria and potential therapeutic applications. Cell. Mol. Life Sci. 77, 1319–1343. doi: 10.1007/s00018-019-03326-8
Yan, C., Li, B., Gu, Y., Jia, Y., Liu, Y., and He, Y. (2016). Methods for the determination of autoinducer-2—a review. J. Microbiol. China 43, 1333–1338. doi: 10.13344/j.microbiol.china.150514
Yang, Y., Zhou, M., Hou, H., Zhu, J., Yao, F., Zhang, X., et al. (2014). Quorum-sensing gene luxS regulates flagella expression and Shiga-like toxin production in F18ab Escherichia coli. Can. J. Microbiol. 60, 355–361. doi: 10.1139/cjm-2014-0178
Zhang, P., Xu, C., Zhou, X., Qi, R., Liu, L., Lv, F., et al. (2020). Cationic conjugated polymers for enhancing beneficial bacteria adhesion and biofilm formation in gut microbiota. Colloids Surf. B Biointerfaces 188:110815. doi: 10.1016/j.colsurfb.2020.110815
Zhao, J., Quan, C., Jin, L., and Chen, M. (2018). Production, detection and application perspectives of quorum sensing autoinducer-2 in bacteria. J. Biotechnol. 268, 53–60. doi: 10.1016/j.jbiotec.2018.01.009
Zhao, X., Yu, Z., and Ding, T. (2020). Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms 8:425. doi: 10.3390/microorganisms8030425
Zhu, G., Ma, S., Li, X., Zhang, P., Tang, L., Cao, L., et al. (2018). The effect of ethanol extract of Glycyrrhiza uralensis on the voltage-gated sodium channel subtype 1.4. J. Pharmacol. Sci. 136, 57–65. doi: 10.1016/j.jphs.2017.11.008
Zuberi, A., Misba, L., and Khan, A. U. (2017). CRISPR interference (CRISPRi) inhibition of luxS gene expression in E. coli: an approach to inhibit biofilm. Front. Cell. Infect. Microbiol. 7:214. doi: 10.3389/fcimb.2017.00214
Keywords: Escherichia coli, quorum sensing, inhibitor, Echinatin, biofilm, EPS, motility, antibacterial synergist
Citation: Bai Y-B, Shi M-Y, Wang W-W, Wu L-Y, Bai Y-T, Li B, Zhou X-Z and Zhang J-Y (2022) Novel quorum sensing inhibitor Echinatin as an antibacterial synergist against Escherichia coli. Front. Microbiol. 13:1003692. doi: 10.3389/fmicb.2022.1003692
Edited by:
Huancai Lin, Sun Yat-sen University, ChinaReviewed by:
Adline Princy Solomon, SASTRA University, IndiaKhristina Judan Cruz, Central Luzon State University, Philippines
Krassimira Hristova, Marquette University, United States
Copyright © 2022 Bai, Shi, Wang, Wu, Bai, Li, Zhou and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji-Yu Zhang, aW5memp5QHNpbmEuY29t
 Yu-Bin Bai
Yu-Bin Bai Meng-Yan Shi
Meng-Yan Shi Wei-Wei Wang
Wei-Wei Wang Ling-Yu Wu
Ling-Yu Wu Yu-Ting Bai
Yu-Ting Bai Bing Li
Bing Li Xu-Zheng Zhou
Xu-Zheng Zhou Ji-Yu Zhang
Ji-Yu Zhang