
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 05 October 2021
Sec. Microbial Symbioses
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.739572
This article is part of the Research Topic Nutritional Physiology and Gut Microbiome View all 21 articles
Poor utilization efficiency of plant protein diets always leads to intestinal barrier dysfunction and growth inhibition in animals. Probiotics have shown promise in improving growth performance and gut health of the host. However, obtaining the host-beneficial probiotic from thousands of bacterial phylotypes is challenging. Here, four intestinal autochthonous bacteria were isolated from fast-growing bullfrog after a 60-day feeding on a soybean meal (SM)-based diet. Another feeding trial was conducted to evaluate the effects of supplementing these strains in an SM-based diet on growth, nutrient digestibility, immunity, and gut health of bullfrog. A high-SM basal diet was used as a non-supplemented control group (NC), and four other diets were prepared by supplementing the basal diet with 1 × 107 CFU/g of Bacillus siamensis, Bacillus tequilensis (BT), Bacillus velezensis, and Lactococcus lactis (LL). Results showed that weight gain, feed efficiency, nitrogen retention, and apparent digestibility coefficients of dry matter and protein were significantly higher in the LL group compared with the NC group (p < 0.05). Furthermore, compared with the NC group, both BT and LL groups showed markedly higher jejunal protease and amylase activities, serum complement 4 and immunoglobulin M levels, jejunal muscularis thickness (p < 0.05), and up-regulated expression of il-10 and zo-1 genes (p < 0.05). High-throughput sequencing revealed higher abundances of Bacillus and Cetobacterium in BT and LL groups, respectively, accompanied with decreased abundances of Enterobacter and Escherichia–Shigella. Besides, KEGG pathways related to metabolisms were significantly enhanced by the LL diet relative to the NC diet (p < 0.05). Overall, the beneficial effects of two frog-derived probiotics were determined: supplementation of L. lactis in SM-based diet promoted growth and nutrient digestibility; both B. tequilensis and L. lactis supplementation improved immune response and intestinal barrier function of bullfrogs.
Plant proteins are widely used as substitutes for expensive animal-derived protein sources in aquaculture and livestock sectors (Li Z.C. et al., 2017; Naylor et al., 2021). However, due to their general drawbacks such as low palatability, imbalanced amino acid profile, and presence of anti-nutritional factors (Liu et al., 2019; Peng et al., 2019; Yao et al., 2019), an excess proportion of plant proteins in diets usually induce immune dysfunction, gut inflammation, and subsequent growth inhibition in animals (Sahlmann et al., 2013; Wang et al., 2020).
Recently, probiotics have garnered significant attraction for disease prevention and growth promotion in aquaculture (Melo-Bolivar et al., 2021). Efficacy of probiotics is dictated by genetic, nutritional, and environmental factors, and the origin of the probiotic strains, accordingly some probiotics are only effective in specific animals (Sun et al., 2009). To avoid potentially harmful effects on the host and endogenous microbiota, intestinal autochthonous bacteria have more advantages over allochthonous bacteria in colonizing host’s intestinal mucosa and exerting physiological effects (Ringø et al., 2018). In addition, it has been reported that bacteria attached to the intestinal epithelial surfaces are more likely to be the real autochthonous bacteria (Denev et al., 2009). Bullfrog (Lithobates catesbeianus) has become one of the most economically valuable farmed amphibians worldwide. The evaluation of the cultivable indigenous microbiota from bullfrog specimens has already been performed as well as the study of the beneficial properties of some bacterial groups to advance in the design of probiotics to control both bacterial and fungal diseases (Montel Mendoza et al., 2012; Niederle et al., 2019). Some in vivo studies proposed the use of native or commercial lactic acid bacteria as potential probiotics for bullfrog hatcheries (de Clarla Dias et al., 2010; Pereira et al., 2016, 2018). However, information about the application of autochthonous probiotics isolated from intestinal mucosa of bullfrogs is scarce.
Growth rate of animals is determined by a series of factors such as ambient temperature, stocking density, available nutrients, etc. (Baltz et al., 1998; Nielsen, 2012). In addition, increasing studies showed that gut microbiota is deeply involved in various host physiological processes (Gao et al., 2018). Thus, it can be regarded as an “internal factor,” which confers multifaceted effects on the host. Remarkable growth difference usually occurs in farmed animals in breeding industry although they might be reared under the same environmental and dietary conditions. Sun et al. (2009) found that gut microbial composition differs in fast-growing (FG) and slow-growing (SG) groupers where higher abundances of potential pathogens were recorded in SG individuals. Similarly, Gui et al. (2017) reported that gut microbiota of FG chicken showed higher diversity and richness compared with the slow- and medium-growing chicken. Furthermore, a research on sea cucumber (Apostichopus japonicus) revealed that the differences in Actinobacteria abundance might be associated with the remarkable difference in body weight (Sha et al., 2016). These studies indicated that the growth of animals is closely linked to the gut microbiota. Although gut microbiota has long been researched through traditional and molecular techniques, information on intestinal microbiota–host crosstalk is scarce. Thus, recognition of the host-beneficial probiotics from thousands of bacterial phylotypes is challenging. Besides, as diet plays a key role in animals’ gut community, whether some sort of bacterial species enriched by a specific diet could in turn contribute to utilization of the same diet is uncertain.
Soybean meal (SM) has been widely used as a typical plant protein source in aquafeed. Previous studies conducted in our laboratory showed that high-SM diet caused intestine inflammation in bullfrog referred to as SM-induced enteritis (SBMIE), accompanied with poor growth and feed utilization (Ding et al., 2019; Yang et al., 2019). In the present study, initially a high-SM diet was fed to bullfrogs for 60 days. Presumably, the FG individuals might harbor a more favorable gut microbiota, which could adapt to the gut environment established by the SM-based diet, and this in turn contributes to the host’s growth. Then, the dominant gut bacterial species enriched in FG bullfrogs were isolated. A 58-day feeding trial was conducted to investigate the effects of supplementing these bacterial strains in a high-SM diet on growth performance, feed utilization, nutrient digestibility, immune function, and gut health of bullfrog.
A high-SM diet (Supplementary Table 1) was used to feed 108 bullfrogs of similar size (26.14 ± 0.21 g) for 60 days. After the feeding trial, bullfrogs were euthanized by destroying the spinal cord with a pin and weighed (Wang et al., 2020). Then, 12 heaviest bullfrogs and 12 lightest bullfrogs were sampled and grouped into FG and SG groups, respectively. The gut brush border membranes (mucosa) of bullfrogs from two groups were scraped with a coverslip on ice under sterile conditions, and then moved into sterile tubes. The mucosa samples were homogenized in normal saline solution (NSS, 0.7% NaCl). After gradient dilution, each dilution was evenly spread onto three different plates in triplicate, including nutrient agar (pH 7.3 ± 0.1), Man Rogosa Sharpe (pH 6.5 ± 0.2) agar, and Bacillus agar (pH 7.0 ± 0.2; Hopebio Technology Co. Ltd, China). The plates were incubated in normoxic incubator (33∘C for 24 h) for growth of aerobic and facultative anaerobic bacteria. Then, the bacterial colonies were divided into different types based on the colony characteristics of shape, structure, size, opacity, and color, and the colonies of each recognizable type was counted for determining number and occurrence rate [(the number of plates containing the strain)/(the number of all plates) × 100]. Then, three to five representatives of each colony type were streaked on corresponding plates repeatedly until pure cultures were obtained. Finally, a total of 35 representative isolates with different types were successfully isolated from agar plates, and the isolates with higher occurrence rates and numbers in the plates of the FG group compared with those of the SG group (Table 1) were picked up for gene sequencing. The bacterial DNA was extracted using bacterial genome DNA extraction kits (SBS Genetech, China) and sent for sequence analysis of the 16S rRNA gene by Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China). The 16S rRNA sequences were blast in Ezbiocloud to determine genetic homology.
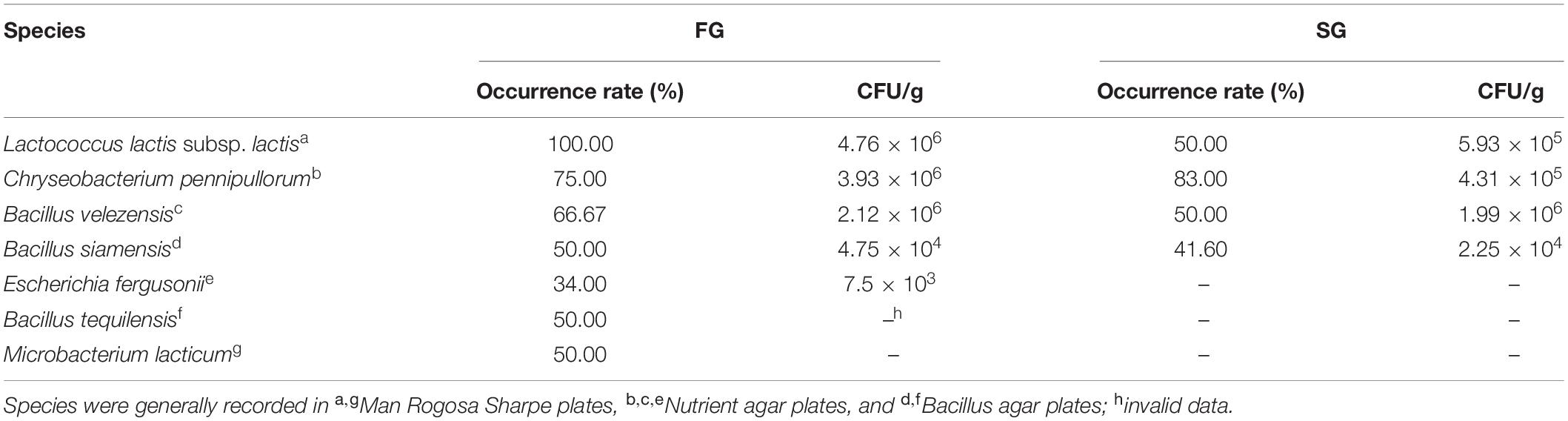
Table 1. Species with a higher occurrence rate or number in plates of fast-growing (FG) group compared with those of slow-growing (SG) group.
Bacillus species and a lactic acid bacterium were cultured in nutrient broth or MRS broth overnight. These cultures were then centrifuged at 4,000 × g for 5 min, and the bacterial pellets were suspended in sterile NSS after washing twice with NSS. The number of alive bacterial cells in the suspensions was determined by plate-counting method (Balestra and Misaghi, 1997). A basal diet (Supplementary Table 1) containing 55% SM was produced following the protocol described by Li et al. (2019). Four treated diets were prepared by supplementing the basal diet with 1 × 107 CFU/g feed of bacterial suspension of Bacillus siamensis (BS diet), Bacillus tequilensis (BT diet), Bacillus velezensis (BV diet), and Lactococcus lactis (LL diet). The non-supplemented control diet (NC diet) was supplemented with equivalent sterile NSS, then all diets were encapsulated with a mixture of fish oil and soybean oil and dried in the shade. To ensure the viability of bacterial cells, diets were prepared every 2 weeks and stored at −20∘C until used.
Bullfrogs were obtained from a commercial farm and transported to the fisheries laboratory of Jimei University (Xiamen, China). Before starting the experiment, bullfrogs were reared in an indoor aquarium (π × 160 × 80 cm) supplied with 4–6 cm of freshwater. All bullfrogs were fed the basal diet twice daily for 3 weeks to acclimate them to experimental conditions. Then, a total of 195 disease-free bullfrogs with homogenous size (initial mean body weight 29.61 ± 0.28 g) were equally divided into five groups with triplicates per group (13 bullfrogs per tank), and they were allocated into fifteen 10-L tanks. Bullfrogs within three tanks were randomly assigned to each dietary treatment and hand-fed to apparent satiation twice daily (8:00 and 18:00) for 58 days. After each feeding, the uneaten feeds were siphoned out and the water in each tank was entirely renewed with fresh water. Feces samples in each tank were collected as described by Lin et al. (2020) over the last 2 weeks for digestibility analysis. During the feeding period, a 12 h light/12 h dark photoperiod was maintained by fluorescent lamp, air temperature ranged from 28 to 32∘C, and water temperature ranged from 27 to 31∘C.
At the end of the feeding trial, bullfrogs were fasted for 24 h and euthanized by destroying the spinal cord with a pin. The number and total weight of bullfrogs in each tank were recorded for analyses of survival and final body weight. Three bullfrogs per tank were randomly sampled and frozen at −20∘C for body composition analysis. The abdomen of 10 additional bullfrogs in each tank was opened immediately with sterile scissors. Blood samples of the aforementioned bullfrogs were collected from ductus arteriosus using a sterile syringe, transported into sterile tubes, and kept at 4∘C overnight. Then, serum was separated after centrifugation (3,000 × g, 10 min, 4∘C) and stored at −80∘C for further analyses. Liver and hind legs were dissected from three bullfrogs per tank and weighed for calculations of hepatosomatic index (HSI) and hind leg index (HLI). The jejunum samples were collected from two bullfrogs per tank and fixed in Bouin’s solution for morphology analysis. Jejunum samples were collected from three bullfrogs per tank and kept at −80∘C for tissue RNA extraction and analysis of enzyme activity. Jejunum samples of two bullfrogs per tank were sampled and pooled for intestinal microbiota analysis.
The crude protein, crude lipid, moisture, and ash contents in diet, whole body, muscle, and feces samples were analyzed according to the standard method of AOAC (2002). Moisture content was estimated by drying in an oven at 105∘C until a constant weight was reached. Crude protein (N × 6.25) was determined by the Dumas method (Gerhardt, Germany). Crude lipid content was quantified by ether extraction, and ash content was measured by the combustion method in a muffle furnace at 550∘C for 8 h. The mineral element content of samples was determined by inductively coupled plasma atomic emission spectroscopy (ICP-OES; Leeman, United States).
Intestine samples were homogenized in 10 volumes (w/v) of NSS and centrifuged at 3,000 × g for 10 min. Then, the homogenized solution was collected and stored at 4∘C. The protease activity was measured by Folin-phenol method (Lowry et al., 1951). One unit of protease activity was defined as the amount of the hydrolysis of casein that liberated 1 μg of tyrosine per minute. Lipase and amylase activities were quantified using commercial kits (Nanjing Jiancheng Biological Company, China). One unit of lipase activity was defined as the amount of enzyme that hydrolyzes 1 μmol substrate per minute at 37°C. One unit of amylase activity was defined as the amount of protein that hydrolyzed 10 mg starch per 30 min at 37°C.
Serum D-lactate concentration, complement 3 (C3), complement 4 (C4), and IgM levels were measured by competition method according to Syedbasha et al. (2016) using amphibian ELISA kits from Nanjing Jiancheng Biological Company (Nanjing, China). Serum diamine oxidase (DAO) and lysozyme (LZM) activities were determined by colorimetric method using commercial assay kits (Nanjing Jiancheng Bioengineering Institute, China). One unit of LZM activity was defined as the amount of enzyme needed to decrease absorbance at a rate of 0.001 min–1⋅ml–1 at 37∘C, and one unit of DAO activity was defined as 1 mmol ammonia formed per minute per milliliter of serum at 37∘C.
Total RNA was extracted from jejunum of bullfrogs using FastPure Tissue Total RNA Isolation Kit (Vazyme Biotech Co., Ltd, China). The purity of the total RNA was analyzed in 1% agarose gel electrophoresis, and the concentration of the total RNA was quantitated using NanoDrop One Ultramicro spectrophotometer (Thermo Fisher, United States). Total RNA was reversely transcribed to cDNA by TransScript ALL-in-one First-Strand cDNA Synthesis Kit (TransGen Biotech Co., Ltd., China). Quantitative PCR was performed on ABI StepOne Plus (Thermal Cycler, United States). The primers of the gene (Supplementary Table 2) and PCR amplification program were designed as our previous research on bullfrogs (Ding et al., 2019; Lin et al., 2021). The relative expression levels of genes were calculated using 2–ΔΔCt method.
Jejunal samples were stained with H&E following the standard histological procedures conducted by Servicebio Biotechnology Co., Ltd. (Wuhan, China). The micrographs were observed with a light microscope (Leica DM5500B, Germany), and Image J software was used for morphometric analysis.
Bacterial DNA was extracted from bullfrog jejunum using HiPure Soil DNA Kits (Magen, China). The DNA yield was detected using Nanodrop 2000 (Thermo Scientific, United States). The amplicons of 16S rRNA gene V3–V4 region were extracted from 2% agarose gel, purified by the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, United States), and quantified using ABI StepOnePlus Real-Time PCR System (Life Technologies, United States). Then, the purified amplicons were paired-end sequenced on Illumina MiSeq PE 250 (Gene Denovo Biotechnology Co., Ltd., China).
Noisy sequences of raw tags were filtered by QIIME software to obtain high-quality clean tags. Clean tags were searched against the reference database to perform reference-based chimera checking using UCHIME algorithm. All chimeric tags were removed and finally obtained effective tags were used for further analysis. The effective tags were clustered into operational taxonomic units (OTUs) of ≥97% similarity using UPARSE software. The tag sequence with the highest abundance was selected as a representative sequence within each cluster. Venn analysis and principal coordinates analysis (PCoA) based on Jaccard distance matrix were performed in R project package. The representative sequences were classified into organisms using RDP classifier based on SILVA database with the confidence threshold value of 80%. The abundance statistics of each taxonomy were visualized using Krona. The KEGG pathway analysis of the OTUs was inferred using PICRUSt2 (Douglas et al., 2020). Analysis of function difference between groups was calculated by Welch’s t-test in R project package. The raw reads were uploaded to NCBI Sequence Read Archive database (accession number PRJNA747862).
Weight gain (WG, %) = (W1 − W0)/W0 × 100
Feeding rate (FR, %/day) = WD/(W1/2 + W0/2)/t × 100
Feed efficiency (FE) = (W1 − W0)/WD
Nitrogen retention ratio (NRR, %) = (W1 × P1 − W0 × P0)/(WD × PD) × 100
Survival (%) = N1/N0 × 100
Hepatosomatic index (%) = liver weight/W1 × 100
Hind leg index (%) = hind leg weight/W1 × 100
Apparent digestibility coefficients (ADCs, %) = (1 − F0/D0 × (DY/FY)) × 100
Where W1 and W0 are the mean final and initial wet body weights, respectively; WD is dry feed intake; t is feeding days; PD, P0, and P1 are the concentrations of crude protein in the diet, initial body, and final body, respectively; N0 and N1 are the initial and final number of bullfrogs, respectively; DY and FY are the concentration of yttrium in diet and feces, respectively; and D0 and F0 are the quantities of compositions in diets and feces, respectively.
The homogeneity of variances of data was tested before further analysis. The significance of difference between the FG group and the SG group was analyzed by Student’s t-test using SPSS 22.0. One-way ANOVA followed by Tukey multiple comparison test was used to identify differences among NC, BT, BV, BS, and LL groups. The values are presented as mean ± SEM and the differences were considered significant at p < 0.05.
After the 60-day feeding on the SM-based diet, the FG group showed significantly higher WG than the SG group (p < 0.05; Supplementary Figure 1). Some species enriched in the gut of the FG group, but scarce in the gut of the SG group, and the species with higher occurrence rate and numbers in the FG group compared with the SG group are listed in Table 1. Three Bacillus species (B. siamensis, B. tequilensis, and B. velezensis) and a lactic acid bacterium (L. lactis subsp. lactis) were then selected for the subsequent feeding trial.
Compared with the NC group, WG was significantly increased in bullfrogs fed the LL diet (p < 0.05), and intermediary WG values were observed in BS and BT groups (p > 0.05; Table 2). No marked difference was found in FR between the NC group and each treated group (p > 0.05). LL supplementation led to a remarkable increment of FE compared with the other groups (p < 0.05). Moreover, the highest NRR was found in the LL group, which was significantly higher than that in NC and BV groups (p < 0.05). No significant difference was observed in survival, HSI, and HLI among all groups (p > 0.05).
Bullfrogs fed the LL diet exhibited markedly higher whole-body protein content than both NC and BV groups (p < 0.05; Table 3). Whole-body moisture content significantly decreased in the LL group compared with that in the BV group (p < 0.05). Higher protein but lower moisture contents were found in the muscle of the BT group compared with that of the BV group (p < 0.05). No significant changes were found in whole-body lipid and ash, and muscle lipid contents (p > 0.05).
Apparent digestibility coefficients of dry matter and crude protein were significantly increased in the LL group compared with those in NC, BS, and BV groups (p < 0.05; Figures 1A,B). There were no marked changes in ADCs of calcium and phosphorus among all groups (p > 0.05; Figures 1C,D). In addition, LL and BT groups exhibited profoundly higher jejunal protease and amylase activities than NC and BV groups (p < 0.05; Figures 2A,C). Bacteria supplementation did not significantly affect jejunal lipase activity (p > 0.05; Figure 2B).
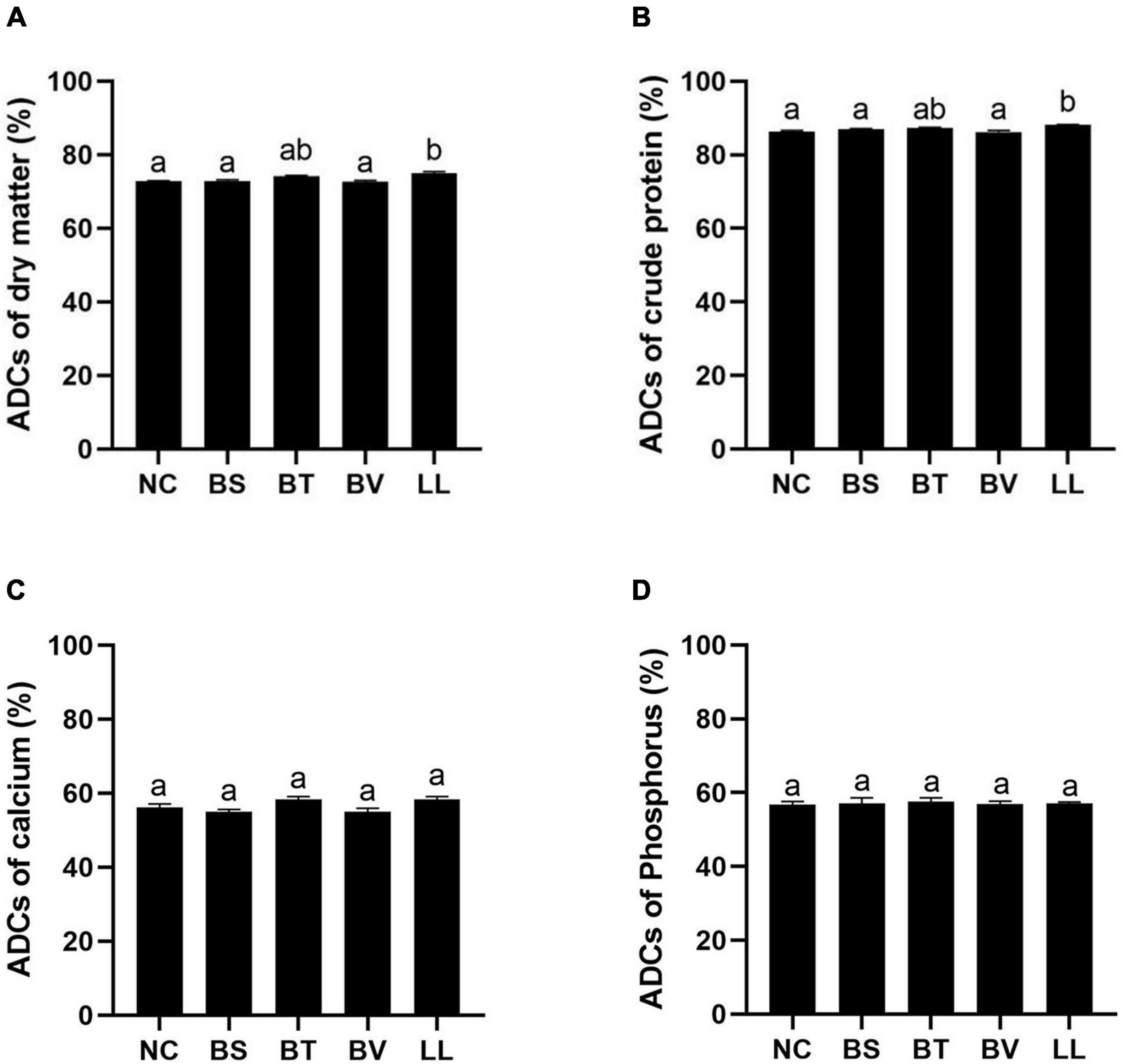
Figure 1. Apparent digestibility coefficients (ADCs) of nutrients: (A) dry matter, (B) protein, (C) calcium, and (D) phosphorus. Bars with different letters were significantly different (p < 0.05). NC, control diet; BS, control diet containing 1 × 107 CFU/g of Bacillus siamensis; BT, control diet containing 1 × 107 CFU/g of Bacillus tequilensis; BV, control diet containing 1 × 107 CFU/g of Bacillus velezensis; and LL, control diet containing 1 × 107 CFU/g of Lactococcus lactis.
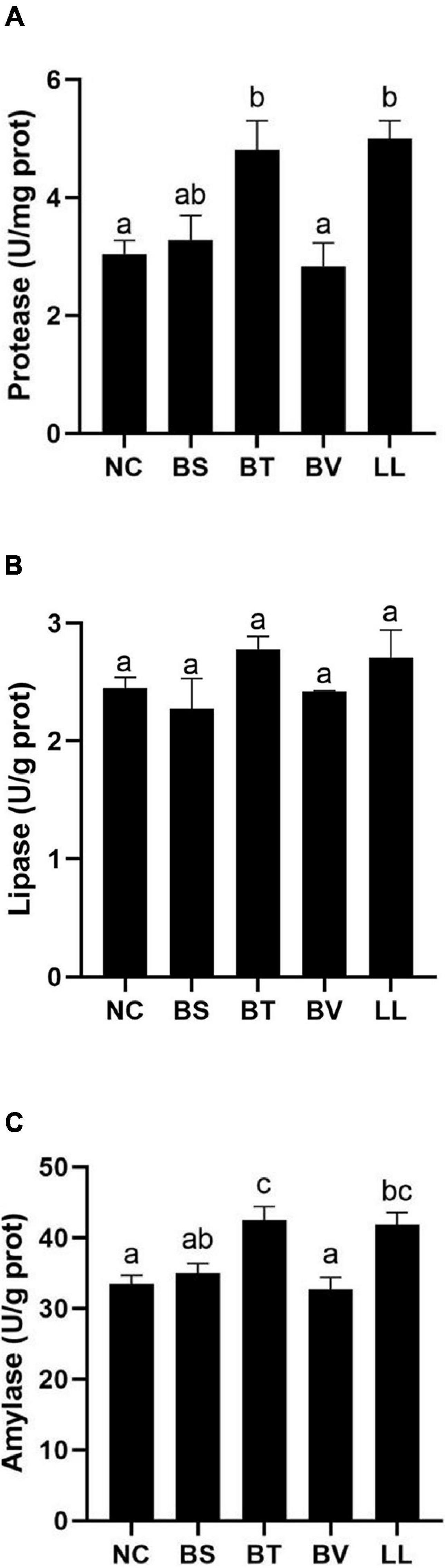
Figure 2. Digestive enzymes activity in jejunum of bullfrog: (A) protease, (B) lipase, and (C) amylase. Bars with different letters were significantly different (p < 0.05). NC, control diet; BS, control diet containing 1 × 107 CFU/g of Bacillus siamensis; BT, control diet containing 1 × 107 CFU/g of Bacillus tequilensis; BV, control diet containing 1 × 107 CFU/g of Bacillus velezensis; and LL, control diet containing 1 × 107 CFU/g of Lactococcus lactis.
Bullfrogs fed LL diet showed significantly higher serum LZM activity and C3 level compared with the NC group (p < 0.05; Table 4). Moreover, serum C4 and IgM levels were remarkably increased in both BT and LL groups relative to NC and BV groups (p < 0.05). BS and BV diets did not affect the aforementioned immune parameters in bullfrogs (p > 0.05).
A typical SBMIE phenomenon was detected in the jejunal epithelium of NC, BS, and BV groups, including shrunken mucosal folds, narrow muscularis, and partial separation of tissue (Figure 3A). Conversely, the mucosal folds in the LL group were longer and curlier, accompanied by more branches. Besides, no obvious separation of tissue or degenerative changes were found in the BT and LL groups. Morphometric analysis of jejunum showed increased number (p < 0.05) and height (p > 0.05) of mucosal folds in the LL group compared with the NC group (Table 5). Furthermore, muscularis thickness in BT and LL groups was significantly higher than that in the other groups (p < 0.05). There was no significant difference in mucosal fold width and lamina propria thickness among groups (p > 0.05). In addition, the mRNA expression of tight junction protein zo-1 (p < 0.05) and occludin (p > 0.05) increased in BT and LL groups compared with the NC group (Figures 3B,C). Moreover, compared with the NC group, BT and LL diets resulted in a significant reduction of serum DAO activity (p < 0.05; Figure 3D). However, no significant difference was found in serum D-lactate concentration among all groups (p > 0.05; Figure 3E).
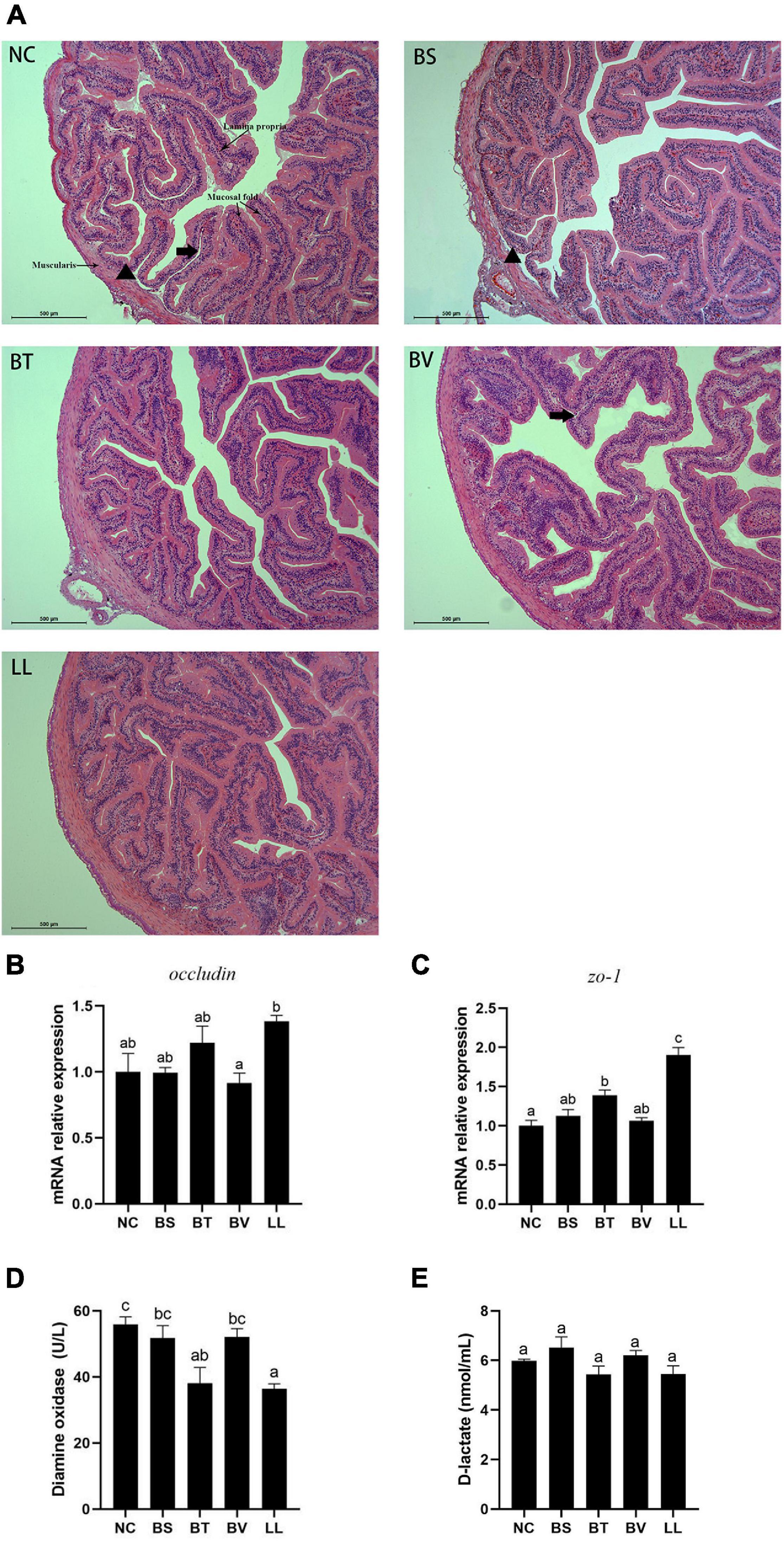
Figure 3. (A) Jejunal morphology of bullfrog using H&E staining. The arrow pointed to the separation between lamina propria and mucosa. The triangle pointed to the separation between submucosa and muscularis. (B,C) Expression of tight junction proteins genes (occludin and zo-1) in jejunum of bullfrog. (D,E) Intestinal epithelial permeability indicators (diamine oxidase and D-lactate) in serum of bullfrog. Bars with different letters were significantly different (p < 0.05). zo, zonula occludens; NC, control diet; BS, control diet containing 1 × 107 CFU/g of Bacillus siamensis; BT, control diet containing 1 × 107 CFU/g of Bacillus tequilensis; BV, control diet containing 1 × 107 CFU/g of Bacillus velezensis; and LL, control diet containing 1 × 107 CFU/g of Lactococcus lactis.
Compared with the NC group, BT and LL supplementation led to up-regulated expression of anti-inflammatory cytokines including il-10 (p < 0.05) and il-4 (p > 0.05) genes (Figures 4A,B). No significant difference was found in the expression of pro-inflammatory cytokines (il-17 and tnf-α genes) between the NC group and each of the treated groups (p > 0.05; Figures 4C,D).
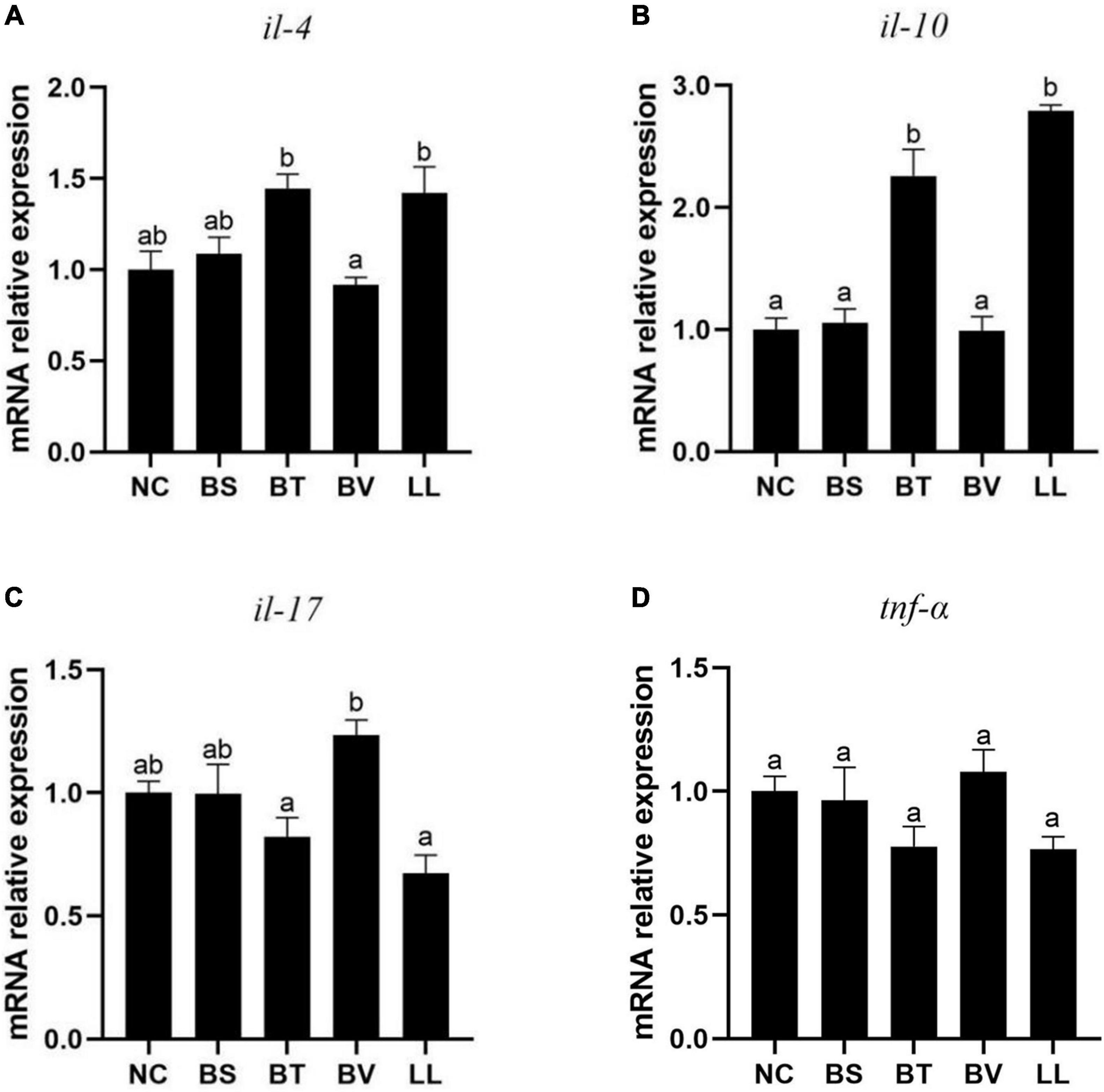
Figure 4. Expression of jejunal inflammatory cytokines of bullfrog, including (A) il-4, (B) il-10, (C) il-17, and (D) tnf-α. Bars with different letters were significantly different (p < 0.05). il, interleukin; tnf, tumor necrosis factor; NC, control diet; BS, control diet containing 1 × 107 CFU/g of Bacillus siamensis; BT, control diet containing 1 × 107 CFU/g of Bacillus tequilensis; BV, control diet containing 1 × 107 CFU/g of Bacillus velezensis; and LL, control diet containing 1 × 107 CFU/g of Lactococcus lactis.
A total of 2,160,089 effective tags were obtained from five groups, with an average of 108,004 ± 1,024 per sample. The resulting sequences were clustered into OTUs at 97% sequence identity. Thus, a total of 2,457 OTUs were found in five groups, with an average of 123 ± 5 per sample. Venn diagram of bacterial communities showed that the core OTUs of all groups was 45, and total OTUs of NC, BS, BT, BV, and LL groups were 199, 108, 140, 95, and 98, respectively, (Figure 5A). PCoA plot based on Jaccard distance matrix was executed to indicate the similarity of intestinal microbial communities among experimental groups. Some data points of NC, BS, and BV groups had overlap regions (Figure 5B), which were distinct from those of BT and LL groups.
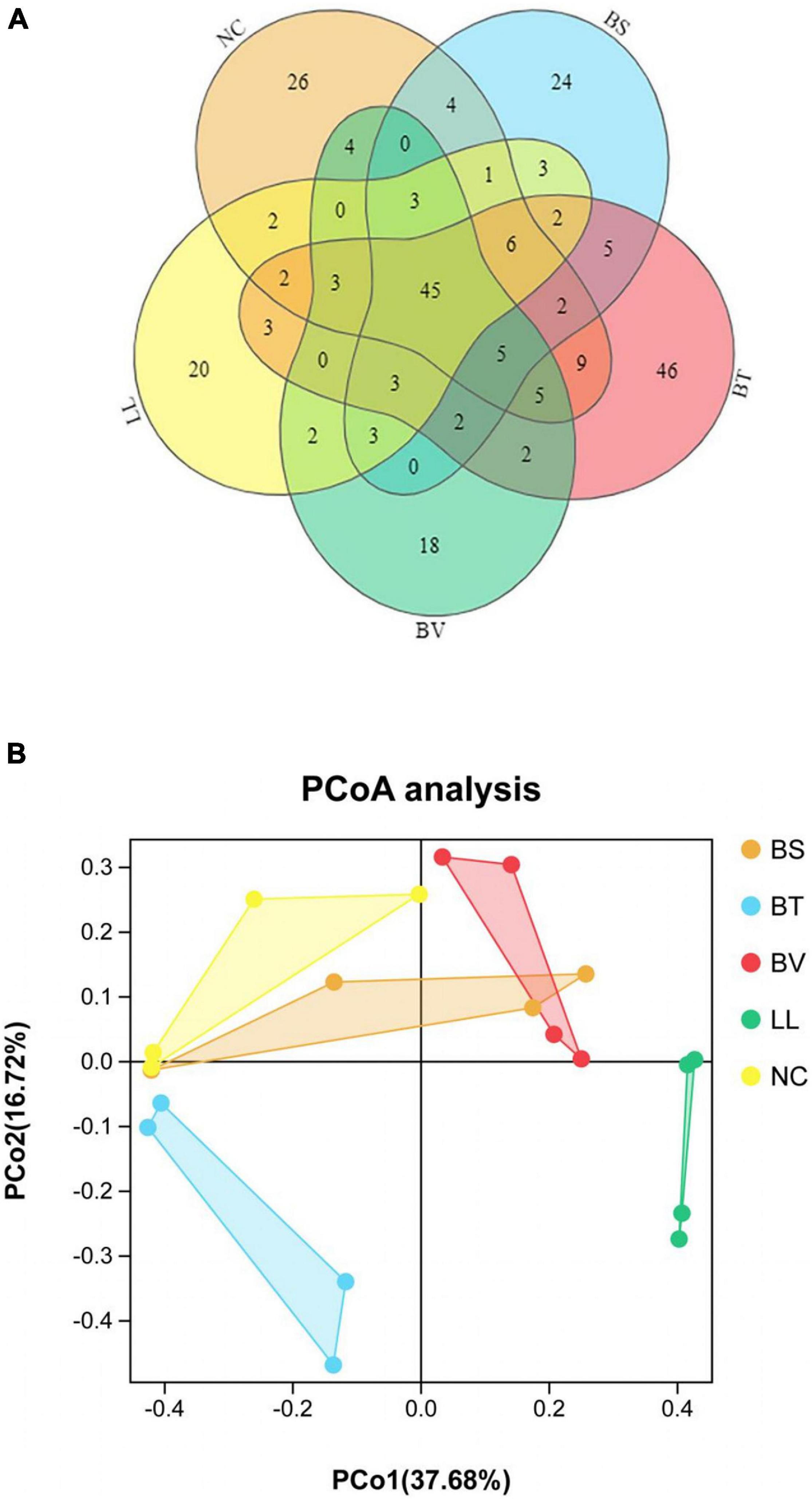
Figure 5. (A) Venn diagram and (B) principal component analysis (PCoA) based on Jaccard distance matrix of jejunal microbiota. NC, control diet; BS, control diet containing 1 × 107 CFU/g of Bacillus siamensis; BT, control diet containing 1 × 107 CFU/g of Bacillus tequilensis; BV, control diet containing 1 × 107 CFU/g of Bacillus velezensis; and LL, control diet containing 1 × 107 CFU/g of Lactococcus lactis.
In general, Proteobacteria, Fusobacteria, and Firmicutes were the three most dominant bacterial phyla in the jejunum of bullfrog (Figure 6A), and their relative abundance was different among experimental groups. The relative abundance of Proteobacteria (the dominant phylum) in NC, BS, BT, and BV groups was 81.84, 61.75, 55.91, and 70.94%, respectively, whereas the dominant phylum in the LL group was Fusobacteria (67.57%). The LL group showed significantly lower abundance of Proteobacteria and higher abundance of Fusobacteria than the NC group (p < 0.05; Figure 6B). The highest abundance of Firmicutes was found in the BT group (41.82%) which significantly differed from that of BS, BV, and LL groups (p < 0.05). The major genera in the NC group were Enterobacter (50.27%), Plesiomonas (12.10%), and Escherichia–Shigella (6.59%; Figure 7A), and the dominant genera in treated groups were Enterobacter (BS, 48.62%), Bacillus (BT, 33.86%), Escherichia–Shigella (BV, 26.53%), and Cetobacterium (LL, 67.20%). The LL group exhibited significantly lower abundance of Enterobacter and higher abundance of Cetobacterium compared with the NC group (p < 0.05; Figure 7B). Moreover, the highest abundance of Bacillus and Escherichia–Shigella were recorded in the BT and BV groups, respectively.
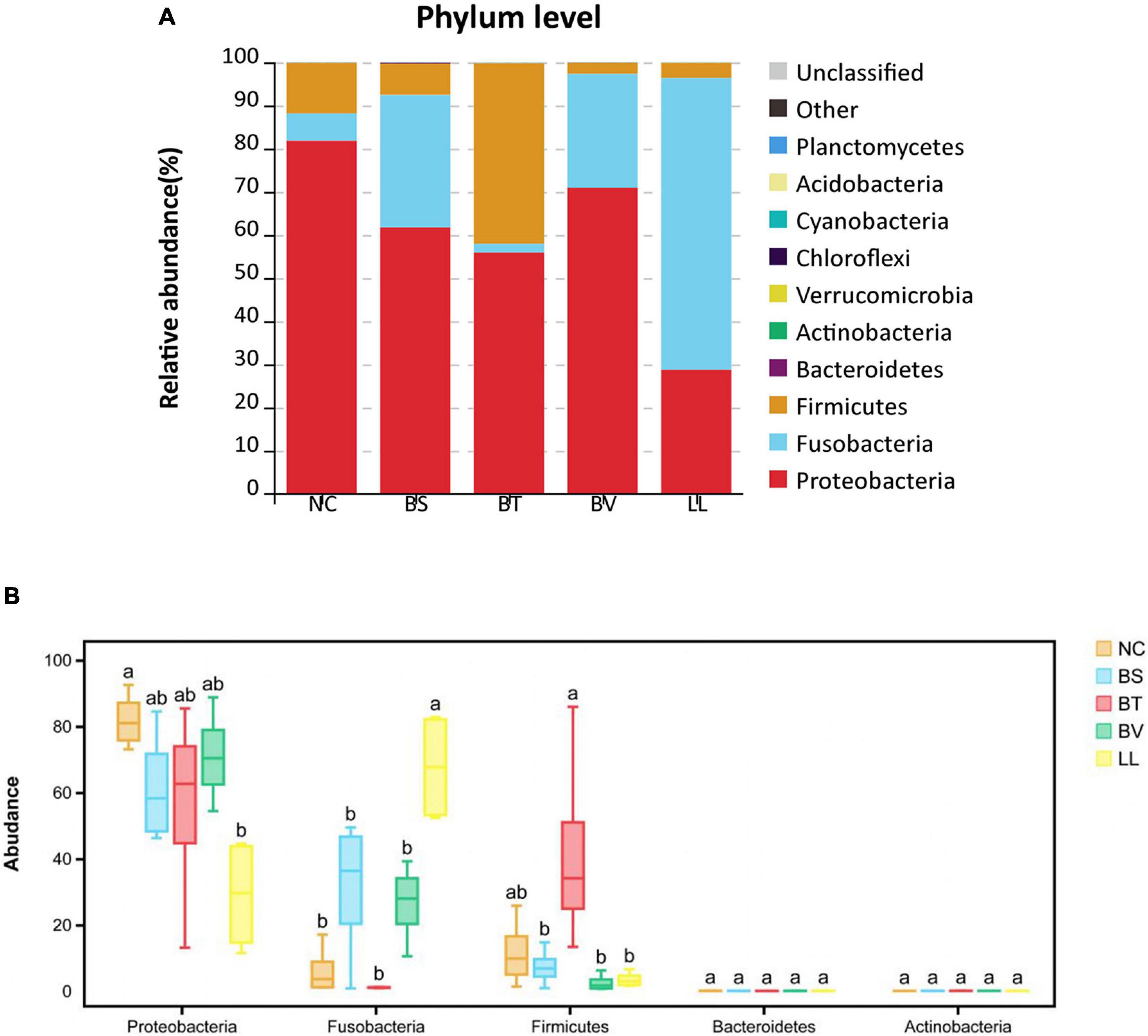
Figure 6. The relative abundance of major bacterial phyla in (A) barplot and (B) boxplot. Bars with different letters were significantly different (p < 0.05). NC, control diet; BS, control diet containing 1 × 107 CFU/g of Bacillus siamensis; BT, control diet containing 1 × 107 CFU/g of Bacillus tequilensis; BV, control diet containing 1 × 107 CFU/g of Bacillus velezensis; and LL, control diet containing 1 × 107 CFU/g of Lactococcus lactis.
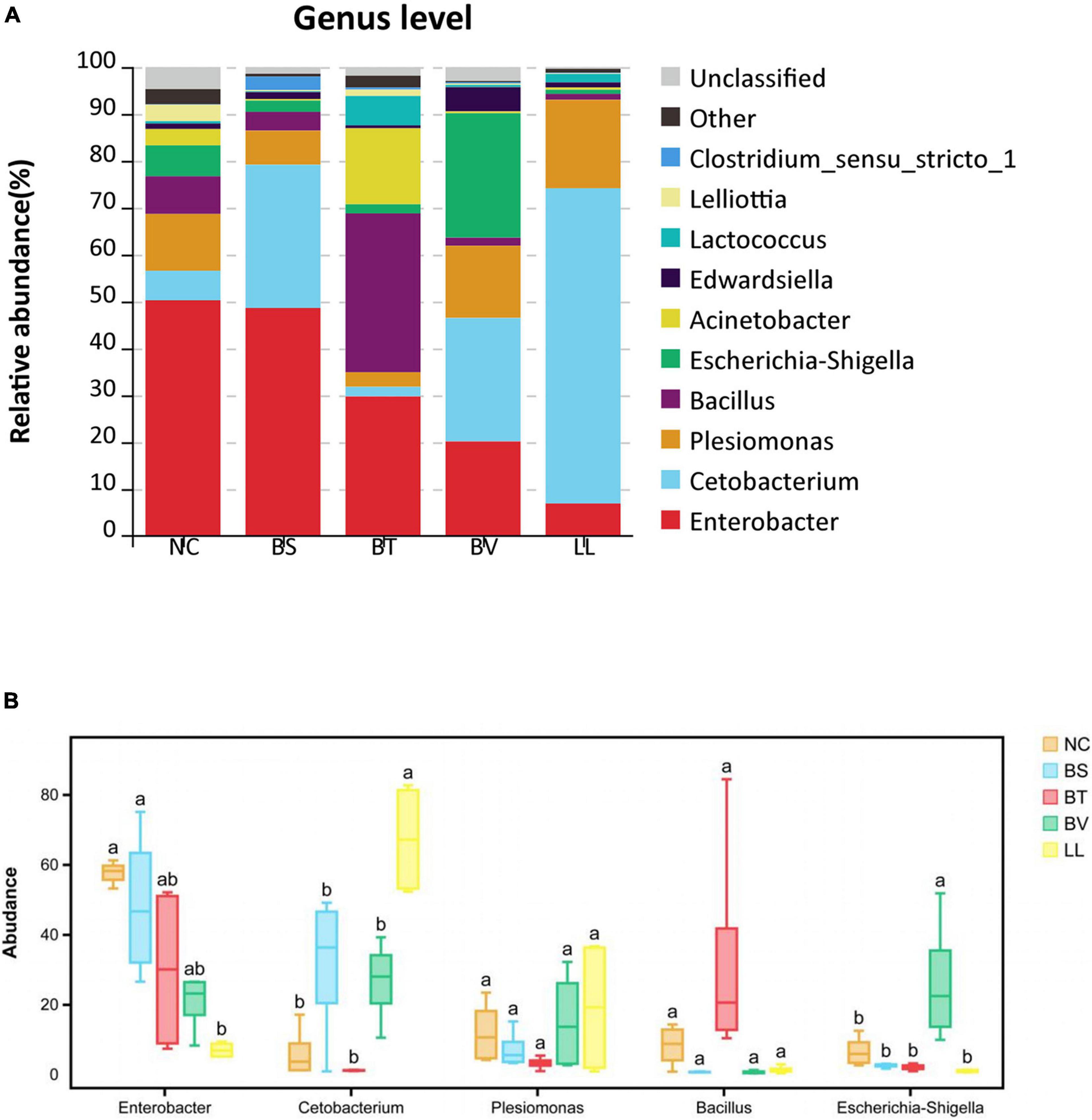
Figure 7. The relative abundance of major bacterial genera in (A) barplot and (B) boxplot. Bars with different letters were significantly different (p < 0.05). NC, control diet; BS, control diet containing 1 × 107 CFU/g of Bacillus siamensis; BT, control diet containing 1 × 107 CFU/g of Bacillus tequilensis; BV, control diet containing 1 × 107 CFU/g of Bacillus velezensis; and LL, control diet containing 1 × 107 CFU/g of Lactococcus lactis.
Predicted functional analysis of intestinal microbiota by PICRUSt2 revealed the top 20 level-2 KO groups. The KEGG pathways enriched in BS and BV groups were similar to that in the NC group (Figure 8A), whereas BT and LL groups showed a higher abundance of partial KEGG pathways than the NC group. Lipid metabolism was markedly enhanced by the BT diet compared with the NC diet (p < 0.05; Figure 8B). Moreover, 14 KEGG pathways including carbohydrate metabolism, metabolism of cofactors and vitamins, metabolism of terpenoids and polyketides, metabolism of other amino acids, energy metabolism, replication and repair, folding, sorting and degradation, glycan biosynthesis and metabolism, membrane transport, translation, biosynthesis of other secondary metabolites, nucleotide metabolism, cell growth and death, and transcription were significantly enriched in the LL group compared with the NC group (p < 0.05; Figure 8C).
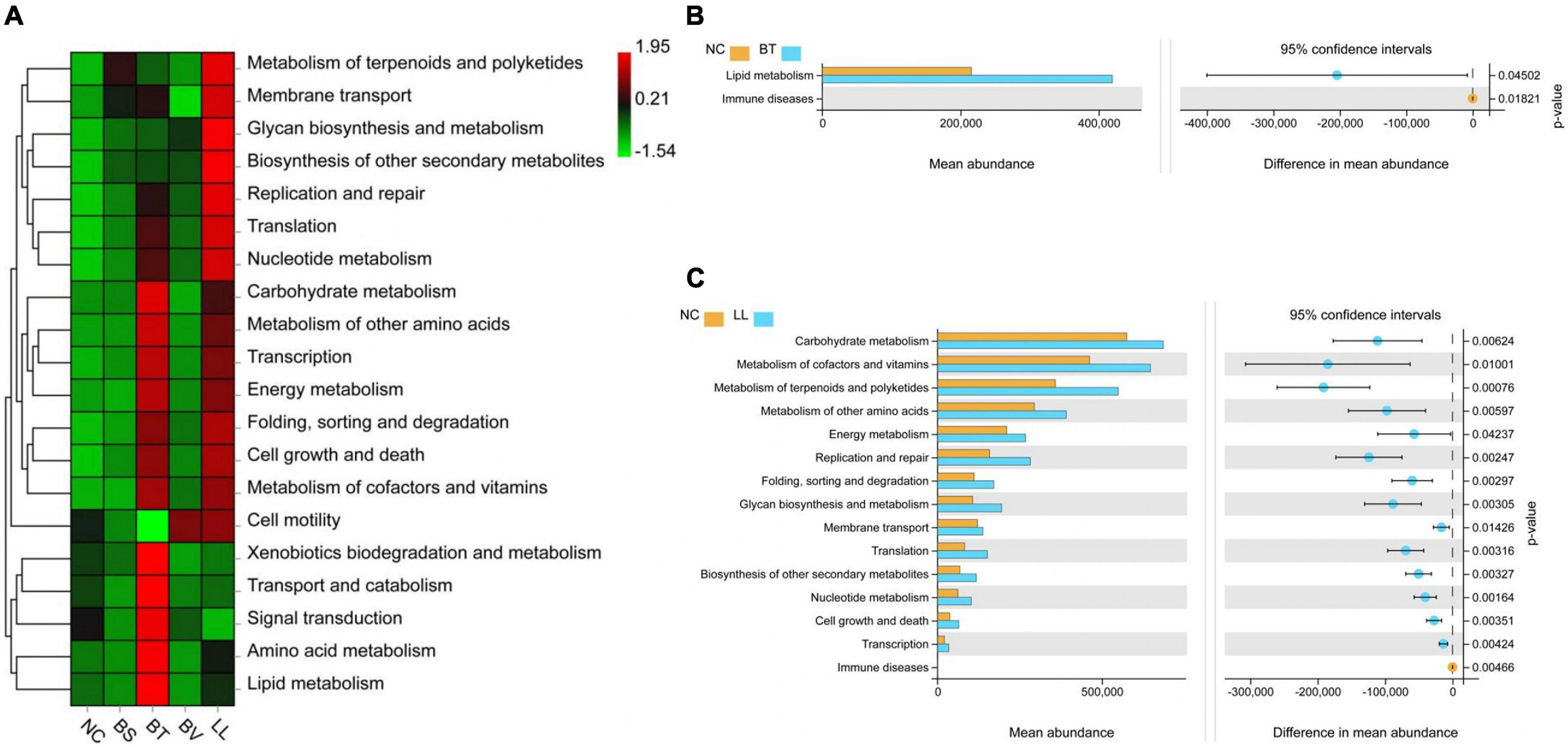
Figure 8. Predicted functional profiles of jejunal microbiota using PICRUSt2 at level 2. (A) The heat map analysis among all groups. (B,C) Analysis of function difference between NC group and BT (or LL) group. Differences were calculated by Welch’s t-test (p < 0.05). NC, control diet; BS, control diet containing 1 × 107 CFU/g of Bacillus siamensis; BT, control diet containing 1 × 107 CFU/g of Bacillus tequilensis; BV, control diet containing 1 × 107 CFU/g of Bacillus velezensis; and LL, control diet containing 1 × 107 CFU/g of Lactococcus lactis.
In the current study, remarkable growth differences were observed among bullfrogs that received SM-based diet for 60 days and reared under the same conditions. Several representative bacterial strains enriched in FG bullfrogs were isolated by standard spread plate method. Presumably, those strains could be adaptive to the gut environment established by a long-term feeding on plant-based diet and might be associated with the growth of bullfrogs. Then, three Bacillus species (B. siamensis, B. tequilensis, and B. velezensis) and a lactic acid bacterium (L. lactis) were selected as feed additives since the two bacterial categories were among the most common probiotics used in breeding production and experimental research.
In livestock industry and aquaculture, it is challenging to improve the feed utilization of plant-sourced proteins and alleviate their adverse effects on animals. Bacillus spp. and lactic acid bacteria are among the most widely used probiotics which exert diverse host-beneficial properties including growth promotion (Yu et al., 2016), immune modulation (Sun et al., 2012), mucosal barrier repair (Lin et al., 2020), disease resistance, etc. (Xia et al., 2019). In the current study, L. lactis supplementation improved growth performance, feed efficiency, and nitrogen retention of bullfrogs, and the increase in nitrogen retention was consistent with the trend in whole-body protein content, which agrees with our previous bullfrog study (Lin et al., 2021). This result indicated that the protein synthesis in bullfrogs might be stimulated by L. lactis treatment. Similarly, Xia et al. (2018) showed that dietary supplementation of L. lactis JCM5805 led to improved weight gain and feed utilization in Nile tilapia (Oreochromis niloticus). Also, Li C. et al. (2017) reported enhanced growth of sea cucumber by dietary L. lactis LH8 application. However, several researches indicated the lack of growth-promoting effect of Bacillus spp. (Reda and Selim, 2015; Yi et al., 2018). In the current study, the three tested Bacillus strains (B. siamensis, B. tequilensis, and B. velezensis) had a slight effect on growth of bullfrog, which might be associated with their antibiosis activity, action mechanism, application dosage, and the complex microbiota–host crosstalk (Sahu et al., 2008).
Humoral immunity is a part of innate immune system in aquatic animals, which consisted of complement, LZM, phagocytosis, etc. (Jia et al., 2016). IgM is the key molecule involved in systemic immunity and immune response mechanisms (Liu et al., 2021). C3 and C4 participate in complement pathways, which are crucial to the elimination of pathogens (Alexander and Ingram, 1992). Excessive levels of dietary plant-protein diets increase the risk of immune homeostasis disruption in animals, especially in aquatic animals (Sahlmann et al., 2013). Bacillus species and lactic acid bacteria have been proven to stimulate the immune system and maintain the immune homeostasis in animals (Ringø et al., 2018). In this study, L. lactis and B. tequilensis diets resulted in elevated serum LZM, IgM, and complement levels in bullfrog. Similar results have also been found in olive flounder (Paralichthys olivaceus) through dietary supplementation of L. lactis I2 (Heo et al., 2013) and in grouper by L. lactis application (Sun et al., 2012). Overall, the current study indicated that the immune response of bullfrog might be improved by dietary supplementation of B. tequilensis and L. lactis.
Intestine morphology is widely used for assessing the intestine development. In the present study, bullfrogs fed the high-SM diet showed a typical SBMIE phenomenon in the jejunum: few and shrunken mucosal folds, as well as narrow muscularis. These degenerative changes might be associated with the inflammatory reaction (Yu et al., 2021). Besides, plant-protein-based diet was proven to induce mucosa damage and infiltration of leukocytes in the lamina propria (Li Y. et al., 2017). In this context, partial separations of tissue were detected in the jejunum of NC, BS, and BV groups. Probiotics have been reported to promote intestinal barrier function through modulation of cytokine production, mucus secretion, macrophage activation, etc. (Anderson et al., 2012). Thus, making use of probiotics conferred a compelling strategy to alleviate adverse impacts caused by plant-protein diet in the gut. For example, Navarrete et al. (2013) reported that dietary L. lactis supplementation aided in the restoration of intestinal morphology impaired by SM-based diet in Atlantic salmon (Salmo salar). Furthermore, a bullfrog study showed an increase in villus height and thickness by B. subtilis application (Lin et al., 2020). Similarly, in the current study, LL and BT diets alleviated the intestinal structural damage caused by high-SM diet. The integrity of intestinal structure is associated with intestinal epithelial permeability (Anderson et al., 2012). Serum DAO activity and D-lactate concentration are two well-established markers for the intestinal mucosal damage (Anderson et al., 2012). In the present study, decreased serum DAO activity indicated the improvement in intestinal integrity and permeability. In addition, intestinal tight junction protein (zo-1 and occludin) were also important components contributing to intestinal physical barrier function (Anderson et al., 2012), and their mRNA expression were up-regulated in bullfrogs fed BT or LL diets as expected. Generally, the results of intestinal morphology, gut permeability, and tight junction proteins collectively indicated that both B. tequilensis and L. lactis aided in maintaining the intestinal barrier functions and preventing SBMIE.
The intestine epithelium is the main site of nutrient digestion and absorption, the longer and thicker intestinal mucosal folds corresponded to enhanced absorptive surface area (Standen et al., 2016), which could be partially responsible for the increased feed efficiency determined in LL and BT groups. Besides, some probiotics could alter intestinal motility and stimulate the digestive enzyme production, leading to greater digestion efficiency (Anderson et al., 2012), as reported in gibel carp (Carassius auratus gibelio) following B. coagulans supplementation (Yu et al., 2016), and in Pacific white shrimp after L. plantarum application (Zheng et al., 2017). Consistently, in the current study, both jejunal protease and amylase activities of bullfrog were increased by both B. tequilensis and L. lactis diets, while feed digestibility was increased by L. lactis treatment. It has been reported that multiple non-digestible ingredients, including oligosaccharides, disaccharide components, and sugar alcohols, were the main substrates for probiotic growth, and many of these ingredients could be fermented by specific bacteria to short-chain fatty acids, which also conferred health benefits for animals (Sahu et al., 2008). Overall, it could be concluded that intestinal digestive and absorptive functions of bullfrog were improved by both B. tequilensis and L. lactis supplementation.
The intestinal microbiota is closely tied to the host’s physiological function, nutrient metabolism, and immune homeostasis (Gao et al., 2018; Xia et al., 2018). Diet intake constitutes a pivotal determinant of the compositions of trillions of microorganisms residing in the gut. In the present study, high-SM diet led to the dominance of Proteobacteria, Firmicutes, and Fusobacteria in the jejunum of bullfrog, which was consistent with previous studies on bullfrog (Wang et al., 2020) and freshwater fish species (Desai et al., 2012; Ni et al., 2012). Reportedly, intestinal microbiota disorder was one of the characteristics of enteritis including SBMIE (Merrifield et al., 2010). In the current study, Enterobacter or Escherichia–Shigella were the major bacterial genera in NC, BS, and BV groups, whereas most bacterial species affiliated to the two genera had been determined as pathogenic bacteria and associated with the inflammatory disorders of animals (Hung et al., 2015; Cao et al., 2017). These results indicated that the long-term SM-based diet might establish favorable conditions for colonization of pathogenic bacteria in bullfrog gut, which might be the main factor that caused intestinal structural damage and microbiota disorder. Probiotics are widely used to modulate the host’s intestinal microbial community; their modes of action include the inhibition of pathogens’ growth by competition for nutrients and adhesion sites, secretion of antibacterial peptides, etc. (Rengpipat et al., 1998; Verschuere et al., 2000). To effectively confer health benefits on the host, it is vitally important for probiotics to adhere and colonize on intestinal mucosa (Alander et al., 1999). In the current study, the BT-fed bullfrogs exhibited an increased abundance of Bacillus and decreased abundance of potential pathogens including Enterobacter, Plesiomonas, and Escherichia–Shigella. However, B. siamensis and B. velezensis diets had a weak effect on the abundance of Bacillus in bullfrog jejunum; it suggested that the ability of Bacillus species to colonize the intestine varied from strain to strain. Interestingly, the intestinal microbiota in the LL-fed bullfrog was characterized by a bloom of Cetobacterium. It was reported that the main components of L. lactis were glycerol phosphate, teichoic acid, and some polysaccharides (Vinogradov et al., 2018), which might serve as substrates for Cetobacterium growth. However, the concrete mechanism of the symbiotic relationship among microorganisms needs to be further studied. Cetobacterium was able to produce vitamin B12 and short-chain fatty acids by fermenting carbohydrates and peptides, and thereby promoting the nutrients’ utilization (Finegold et al., 2003; Tsuchiya et al., 2008; Li et al., 2015), as confirmed by the fact that increased nutrient digestibility was obtained in bullfrogs fed L. lactis diet.
As an added metabolic “organ” of the host, intestinal microbiota participates in a series of host metabolism steps (Backhed et al., 2004; Zhang et al., 2014). In the current study, analyses of predicted functions of gut microbial communities showed that the pathway of lipid metabolism was stimulated by B. tequilensis diet. Zhou et al. (2016) reported that a Bacillus strain contributed to decompose cholesterol into coprostanol, and reduced lipid accumulation by stimulating fecal-lipid and bile-acid output. Besides, pathways of metabolisms, including that of carbohydrate, amino acids, energy, cofactors, and vitamins, were significantly enhanced by L. lactis diet, and they might be attributed to the effects of Cetobacterium on fermenting carbohydrates and peptides as mentioned earlier. Excessive levels of dietary carbohydrate induced metabolic load and stress responses in aquatic animals since most of them were generally considered to be glucose intolerant and poor at utilizing carbohydrates (Qiao et al., 2016; Boonanuntanasarn et al., 2018; Gao et al., 2018). Thus, it is of great significance that gut microbes aid the host in the utilization of carbohydrate and non-digestible ingredients. Collectively, after feeding the SM-based diet supplemented with L. lactis for 58 days, the changes in intestinal structure and digestive function, as well as the alterations in gut microbiota, collaborated to promote the gut health, resulting in improved feed utilization and growth performance of bullfrog.
The beneficial effects of two frog-derived probiotics were determined. Dietary supplementation of L. lactis significantly promoted feed utilization and growth performance of bullfrog, and both L. lactis and B. tequilensis supplementation improved the immune response and alleviated enteritis caused by the high-SM diet. These beneficial effects could be attributed to the improvement in intestinal epithelial integrity and digestive function, as well as the alterations in gut microbiota.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GenBank MZ573378, MZ573379, MZ573380, and MZ573381; BioProject PRJNA747862.
The animal study was reviewed and approved by The Committee on the Ethics of Animal Experiments of Jimei University.
LW and CZ designed the study. ZW performed the experiment, analyzed data, and wrote the manuscript. KL and KS participated in the experiment design and gave valuable advice. XL and SR contributed to revision of the manuscript. All authors have read and approved the final version of this article.
This study was supported by National Natural Science Foundation of China (Grant No. 31602172) and the Science, Technology Leading Project of Fujian Province (Grant No. 2017N0021). SR was supported by NAZV project (Grant No. QK1710310).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.739572/full#supplementary-material
Alander, M., Satokari, R., and Korpela, R. (1999). Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Environ. Microbiol. 65, 351–354. doi: 10.1128/aem.65.1.351-354.1999
Alexander, J. B., and Ingram, G. A. (1992). Noncellular nonspecific defence mechanisms of fish. Ann. Rev. Fish Dis. 2, 249–279. doi: 10.1016/0959-8030(92)90066-7
Anderson, R. C., Dalziel, J. E., Gopal, P. K., Bassett, S., Ellis, A., and Roy, N. C. (2012). “The role of intestinal barrier function in early life in the development of colitis,” in Colitis, ed. M. Fukata (London: InTechOpen), 1–30.
AOAC (2002). Official Methods of Analysis of the Association of Analytical Chemistry, 17th Edn. Arlington: AOAC.
Backhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101, 15718–15723. doi: 10.1073/pnas.0407076101
Balestra, G. M., and Misaghi, I. J. (1997). Increasing the efficiency of the plate counting method for estimating bacterial diversity. J. Microbiol. Meth. 30, 111–117. doi: 10.1016/s0167-7012(97)00056-0
Baltz, D. M., Fleeger, J. W., Rakocinski, C. F., and McCall, J. N. (1998). Food, density, and microhabitat: factors affecting growth and recruitment potential of juvenile saltmarsh fishes. Environ. Biol. Fish. 53, 89–103. doi: 10.1023/a:1007471724608
Boonanuntanasarn, S., Jangprai, A., Kumkhong, S., Plagnes-Juan, E., Veron, V., Burel, C., et al. (2018). Adaptation of Nile tilapia (Oreochromis niloticus) to different levels of dietary carbohydrates: new insights from a long term nutritional study. Aquaculture 496, 58–65.
Cao, H., An, J., Ou, R., Lu, L., Ai, X., and Yang, Y. (2017). Enterobacter aerogenes: an emerging pathogen for enteritis in farmed channel catfish Ictalurus punctatus. Isr. J. Aquacult. Bamid. 69, 1–7.
de Clarla Dias, D., De Stefani, M. V., Ferreira, C. M., Franca, F. M., Tavares Ranzani-Paiva, M. J., and Santos, A. A. (2010). Haematologic and immunologic parameters of bullfrogs, Lithobates catesbeianus, fed probiotics. Aquac. Res. 41, 1064–1071.
Denev, S., Staykov, Y., Moutafchieva, R., and Beev, G. (2009). Microbial ecology of the gastrointestinal tract of fish and the potential application of probiotics and prebiotics in finfish aquaculture. Int. Aquatic Res. 1, 1–29. doi: 10.1002/9781118897263.ch1
Desai, A. R., Links, M. G., Collins, S. A., Mansfield, G. S., and Hill, J. E. (2012). Effects of plant-based diets on the distal gut microbiome of rainbow trout (Oncorhynchus mykiss). Aquaculture 35, 134–142. doi: 10.1016/j.aquaculture.2012.04.005
Ding, L., Wang, L., Lu, K. L., Song, K., and Zhang, C. X. (2019). Effects of replacement of fish meal with soybean meal on growth performance, digestive enzyme activity and intestinal health of Rana catesbeiana. Freshw. Fish. 49, 69–75.
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Finegold, S. M., Vaisanen, M. L., Molitoris, D. R., Tomzynski, T. J., Song, Y., Liu, C., et al. (2003). Cetobacterium somerae sp. nov. from human feces and emended description of the genus Cetobacterium. Syst. Appl. Microbiol. 26, 177–181. doi: 10.1078/072320203322346010
Gao, S., Pan, L. Q., Huang, F., Song, M. S., and Zhang, M. Y. (2018). Metagenomic insights into the structure and function of intestinal microbiota of the farmed Pacific white shrimp (Litopenaeus vannamei). Aquaculture 499, 109–118. doi: 10.1016/j.aquaculture.2018.09.026
Gui, G., Xiao, Y., Dai, B., Chen, X., Yang, H. L., and Xu, E. (2017). Alteration of intestinal archaeal community composition in broilers with different growth rates. Anim. Husb. Feed Sci. 9, 335–359.
Heo, W. S., Kim, Y. R., Kim, E. Y., Bai, S. C., and Kong, I. S. (2013). Effects of dietary probiotic, Lactococcus lactis subsp. lactis I2, supplementation on the growth and immune response of olive flounder (Paralichthys olivaceus). Aquaculture 376, 20–24. doi: 10.1016/j.aquaculture.2012.11.009
Hung, L. D., Hirayama, M., Ly, B. M., and Hori, K. (2015). Biological activity, cDNA cloning and primary structure of lectin KSA-2 from the cultivated red alga Kappaphycus striatum (Schmitz) Doty ex Silva. Phytochem. Lett. 14, 99–105. doi: 10.1016/j.phytol.2015.09.012
Jia, R., Liu, B. L., Han, C., Huang, B., and Lei, J. L. (2016). The physiological performance and immune response of juvenile turbot (Scophthalmus maximus) to nitrite exposure. Comp. Biochem. Phys. C 18, 40–46. doi: 10.1016/j.cbpc.2016.01.002
Li, C., Ren, Y. C., Jiang, S. H., Zhou, S., Zhao, J. S., Wang, R. J., et al. (2017). Effects of dietary supplementation of four strains of lactic acid bacteria on growth, immune-related response and genes expression of the juvenile sea cucumber Apostichopus japonicus Selenka. Fish Shellfish Immun. 74, 69–75. doi: 10.1016/j.fsi.2017.12.037
Li, T. T., Long, M., Gatesoupe, F. J., Zhang, Q., Li, A. H., and Gong, X. N. (2015). Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb. Ecol. 69, 25–36. doi: 10.1007/s00248-014-0480-8
Li, X., Rahimnejad, S., Wang, L., Lu, K., Song, K., and Zhang, C. (2019). Substituting fish meal with housefly (Musca domestica) maggot meal in diets for bullfrog Rana (Lithobates) catesbeiana: effects on growth, digestive enzymes activity, antioxidant capacity and gut health. Aquaculture 499, 295–305. doi: 10.1016/j.aquaculture.2018.09.053
Li, Y., Hu, H., Liu, J., Yang, P., Zhang, Y., Ai, Q., et al. (2017). Dietary soya allergen β-conglycinin induces intestinal inflammatory reactions, serum-specific antibody response and growth reduction in a carnivorous fish species, turbot Scophthalmus maximus L. Aquac. Res. 48, 4022–4037. doi: 10.1111/are.13224
Li, Z. C., Li, Y. K., Lv, Z. Q., Liu, H., Zhao, J. B., Noblet, J., et al. (2017). Net energy of corn, soybean meal and rapeseed meal in growing pigs. J. Anim. Sci. Biotechno. 8:44.
Lin, J. B., Zeng, Q. H., Zhang, C. X., Song, K., Lu, K. L., Wang, L., et al. (2020). Effects of Bacillus subtilis supplementation in soybean meal-based diets on growth performance, diet digestibility and gut health in bullfrog Lithobates catesbeianus. Aquac. Rep. 16:100273. doi: 10.1016/j.aqrep.2020.100273
Lin, J. B., Zhang, C. X., Lu, K. L., Song, K., and Wang, L. (2021). Effect of guanidinoacetic acid and betaine supplementation in soybean meal-based diets on growth performance, muscle energy metabolism and methionine utilization in the bullfrog Lithobates catesbeianus. Aquaculture 533:736167. doi: 10.1016/j.aquaculture.2020.736167
Liu, T., Zhang, G. G., Feng, Y., Kong, C., Ayisi, C. L., Huang, X. X., et al. (2019). Dietary soybean antigen impairs growth and health through stress-induced non-specific immune responses in Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immun. 84, 124–129. doi: 10.1016/j.fsi.2018.09.062
Liu, Z. N., Liao, L. Y., Chen, Q., Lin, S. Q., and Wu, Y. J. (2021). Effects of Hericium erinaceus polysaccharide on immunity and apoptosis of the main immune organs in Muscovy duck reovirus-infected ducklings. Int. J. Biol. Macromol. 171, 448–456. doi: 10.1016/j.ijbiomac.2020.12.222
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951). Protein measurement with the folin phenol reagent. J. Biol. Chem. 193, 265–275. doi: 10.1016/s0021-9258(19)52451-6
Melo-Bolivar, J. F., Ruiz Pardo, R. Y., Hume, M. E., and Villamil Diaz, L. M. (2021). Multistrain probiotics use in main commercially cultured freshwater fish: a systematic review of evidence. Rev. Aquacult. 13, 1758–1780. doi: 10.1111/raq.12543
Merrifield, D. L., Dimitroglou, A., Bradley, G., Baker, R. T., and Davies, S. J. (2010). Soybean meal alters autochthonous microbial populations, microvilli morphology and compromises intestinal enterocyte integrity of rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis. 32, 755–766. doi: 10.1111/j.1365-2761.2009.01052.x
Montel Mendoza, G., Pasteris, S. E., Ale, C. E., Otero, M. C., Buehler, M. I., and Fatima Nader-Macias, M. E. (2012). Cultivable microbiota of Lithobates catesbeianus and advances in the selection of lactic acid bacteria as biological control agents in raniculture. Res. Vet. Sci. 93, 1160–1167.
Navarrete, P., Fuentes, P., Fuente, L. D. L., Barros, L., Magne, F., Opazo, R., et al. (2013). Short-term effects of dietary soybean meal and lactic acid bacteria on the intestinal morphology and microbiota of Atlantic salmon (Salmo salar). Aquacult. Nutr. 19, 827–836. doi: 10.1111/anu.12047
Naylor, R. L., Hardy, R. W., Buschmann, A. H., Bush, S. R., Cao, L., Klinger, D. H., et al. (2021). A 20-year retrospective review of global aquaculture. Nature 591, 551–563. doi: 10.1038/s41586-021-03308-6
Ni, J. J., Yu, Y. H., Zhang, T. L., and Gao, L. (2012). Comparison of intestinal bacterial communities in grass carp, Ctenopharyngodon idellus, from two different habitats. Chin. J. Oceanol. Limn. 30, 757–765. doi: 10.1007/s00343-012-1287-4
Niederle, M. V., Bosch, J., Ale, C. E., Nader-Macias, M. E., Aristimuno Ficoseco, C., Toledo, L. F., et al. (2019). Skin-associated lactic acid bacteria from North American bullfrogs as potential control agents of Batrachochytrium dendrobatidis. PLoS One 14:e0223020. doi: 10.1371/journal.pone.0223020
Nielsen, B. L. (2012). Effects of ambient temperature and early open-field response on the behaviour, feed intake and growth of fast- and slow-growing broiler strains. Animal 6, 1460–1468. doi: 10.1017/s1751731112000353
Peng, C., Tang, X., Shu, Y., He, M., Xia, X., Zhang, Y., et al. (2019). Effects of 7S and 11S on the intestine of weaned piglets after injection and oral administration of soybean antigen protein. Anim. Sci. J. 90, 393–400. doi: 10.1111/asj.13130
Pereira, S. A., Jeronimo, G. T., da Costa Marchiori, N., de Oliveira, H. M., Owatari, M. S., Jesus, G. F. A., et al. (2016). Autochthonous probiotic Lactobacillus sp. in the diet of bullfrog tadpoles Lithobates catesbeianus improves weight gain, feed conversion and gut microbiota. Aquacult. Nutr. 23, 910–916. doi: 10.1111/anu.12458
Pereira, S. A., Jeronimo, G. T., Marchiori, N. C., Oliveira, H. M., Jesus, G. F. A., Schmidt, E. C., et al. (2018). Tadpoles fed supplemented diet with probiotic bacterium isolated from the intestinal tract of bullfrog Lithobates catesbeianus: haematology, cell activity and electron microscopy. Microb. Pathog. 114, 255–263. doi: 10.1016/j.micpath.2017.11.033
Qiao, F., Liu, Y. K., Sun, Y. H., Wang, X. D., and Zhang, M. L. (2016). Influence of different dietary carbohydrate sources on the growth and intestinal microbiota of Litopenaeus vannamei at low salinity. Aquac. Nutr. 23, 444–542. doi: 10.1111/anu.12412
Reda, R. M., and Selim, K. M. (2015). Evaluation of Bacillus amyloliquefaciens on the growth performance, intestinal morphology, hematology and body composition of Nile tilapia, Oreochromis niloticus. Aquacult. Int. 23, 203–217. doi: 10.1007/s10499-014-9809-z
Rengpipat, S., Phianphak, W., Piyatiratitivorakul, S., and Menasveta, P. (1998). Effects of a probiotic bacterium on black tiger shrimp Penaeus monodon survival and growth. Aquaculture 167, 301–313. doi: 10.1016/s0044-8486(98)00305-6
Ringø, E., Hoseinifar, S. H., Ghosh, K., Doan, H. V., Beck, B. R., and Song, S. K. (2018). Lactic acid bacteria in finfish—an update. Front. Microbiol. 9:1818. doi: 10.3389/fmicb.2018.01818
Sahlmann, C., Sutherland, B. J. G., Kortner, T. M., Koop, B. F., Krogdahl, Å, and Bakke, A. M. (2013). Early response of gene expression in the distal intestine of Atlantic salmon (Salmo salar L.) during the development of soybean meal induced enteritis. Fish. Shellfish Immun. 34, 599–609. doi: 10.1016/j.fsi.2012.11.031
Sahu, M. K., Swarnakumar, N. S., Sivakumar, K., Thangaradjou, T., and Kannan, L. (2008). Probiotics in aquaculture: importance and future perspectives. Indian J. Microbiol. 48, 299–308. doi: 10.1007/s12088-008-0024-3
Sha, Y., Liu, M., Wang, B., Jiang, K., Sun, G., and Wang, L. (2016). Gut bacterial diversity of farmed sea cucumbers Apostichopus japonicus with different growth rates. Microbiology 85, 109–115. doi: 10.1134/s0026261716010112
Standen, B. T., Peggs, D. L., Rawling, M. D., Foey, A., Davies, S. J., Santos, G. A., et al. (2016). Dietary administration of a commercial mixed-species probiotic improves growth performance and modulates the intestinal immunity of tilapia, Oreochromis niloticus. Fish Shellfish Immun. 49, 427–435. doi: 10.1016/j.fsi.2015.11.037
Sun, Y. Z., Yang, H. L., Ma, R. L., and Song, K. (2012). Effect of Lactococcus lactis and Enterococcus faecium on growth performance, digestive enzymes and immune response of grouper Epinephelus coioides. Aquacult. Nutr. 18, 281–289. doi: 10.1111/j.1365-2095.2011.00894.x
Sun, Y., Yang, H., Ling, Z., Chang, J., and Ye, J. (2009). Gut microbiota of fast and slow growing grouper Epinephelus coioides. Afr. J. Microbiol. Res. 3, 713–720.
Syedbasha, M., Linnik, J., Santer, D., O’Shea, D., Barakat, K., Joyce, M., et al. (2016). An ELISA based binding and competition method to rapidly determine ligand-receptor interactions. J. Vis. Exp. 109:e53575.
Tsuchiya, C., Sakata, T., and Sugita, H. (2008). Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 46, 43–48.
Verschuere, L., Rombaut, G., Sorgeloos, P., and Verstraete, W. (2000). Probiotic bacteria as biological control agents in aquaculture. Microbiol. Mol. Biol. Rev. 64, 655–671. doi: 10.1128/mmbr.64.4.655-671.2000
Vinogradov, E., Sadovskaya, I., Courtin, P., Kulakauska, S., Grard, T., Mahonyet, J., et al. (2018). Determination of the cell wall polysaccharide and teichoic acid structures from Lactococcus lactis IL1403. Carbohydr. Res. 462, 39–44. doi: 10.1016/j.carres.2018.04.002
Wang, L., Wang, J., Lu, K. L., Song, K., and Rahimnejad, S. (2020). Total replacement of fish meal with soybean meal in diets for bullfrog (Lithobates catesbeianus): effects on growth performance and gut microbial composition. Aquaculture 524:735236. doi: 10.1016/j.aquaculture.2020.735236
Xia, Y., Gao, J. M., Wang, M., Lu, M. X., Chen, G., Gao, F. Y., et al. (2019). Effects of Lactococcus lactis subsp. lactis JCM5805 on colonization dynamics of gut microbiota and regulation of immunity in early ontogenetic stages of tilapia. Fish Shellfish Immun. 86, 53–63. doi: 10.1016/j.fsi.2018.11.022
Xia, Y., Lu, M. X., Chen, G., Cao, J. M., Gao, F. Y., Wang, M., et al. (2018). Effects of dietary Lactobacillus rhamnosus JCM1136 and Lactococcus lactis subsp. lactis JCM5805 on the growth, intestinal microbiota, morphology, immune response and disease resistance of juvenile Nile tilapia, Oreochromis niloticus. Fish Shellfish Immun. 76, 368–379. doi: 10.1016/j.fsi.2018.03.020
Yang, M. Q., Rahimnejad, S., Zhang, C. X., Song, K., Lu, K. L., and Wang, L. (2019). Histomorphology of gastrointestinal tract in bullfrog Rana (Lithobates) catesbeiana and evaluation of the changes induced by a soybean meal-based diet. Aquacult. Res. 51, 164–174. doi: 10.1111/are.14361
Yao, J. T., Kong, C., Hua, X. M., Yang, J. F., Liu, T., Wang, G., et al. (2019). T1R1 expression in obscure puffer (Takifugu fasciatus) is associated with effect of dietary soybean antigenic protein on intestinal health. Aquaculture 501, 202–212. doi: 10.1016/j.aquaculture.2018.11.010
Yi, Y. L., Zhang, Z. H., Zhao, F., Liu, H., and Wang, G. X. (2018). Probiotic potential of Bacillus velezensis JW: antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immun. 78, 322–330. doi: 10.1016/j.fsi.2018.04.055
Yu, G., Liu, Y., Ou, W., Dai, J., Ai, Q., Zhang, W., et al. (2021). The protective role of daidzein in intestinal health of turbot (Scophthalmus maximus L.) fed soybean meal-based diets. Sci. Rep. 11:3352. doi: 10.1038/s41598-021-82866-1
Yu, Y. B., Lv, F., Wang, C. H., Wang, A. M., Zhao, W. H., and Yang, W. P. (2016). Effects of Bacillus coagulans on growth performance, disease resistance, and HSP70 gene expression in Juvenile gibel carp, Carassius auratus gibelio. J. World Aquacult. Soc. 47, 729–740. doi: 10.1111/jwas.12308
Zhang, M. L., Chekan, J. R., Dodd, D., Hong, P. Y., and Cann, I. (2014). Xylan utilization in human gut commensal bacteria is orchestrated by unique modular organization of polysaccharide-degrading enzymes. Proc. Natl. Acad. Sci. U.S.A. 111, E3708–E3717.
Zheng, X. T., Duan, Y. F., Dong, H. B., and Zhang, J. S. (2017). Effects of dietary Lactobacillus plantarum on growth performance, digestive enzymes and gut morphology of Litopenaeus vannamei. Probiotics. Antimicro. 10, 504–510. doi: 10.1007/s12602-017-9300-z
Keywords: Lithobates catesbeianus, autochthonous bacteria, gut structure, gut microbiota, soybean meal-based diet
Citation: Wang Z, Zhang C, Lu K, Song K, Li X, Wang L and Rahimnejad S (2021) Effects of Supplementing Intestinal Autochthonous Bacteria in Plant-Based Diets on Growth, Nutrient Digestibility, and Gut Health of Bullfrogs (Lithobates catesbeianus). Front. Microbiol. 12:739572. doi: 10.3389/fmicb.2021.739572
Received: 11 July 2021; Accepted: 10 September 2021;
Published: 05 October 2021.
Edited by:
Jia Yin, Hunan Normal University, ChinaReviewed by:
Sun Yun, Hainan University, ChinaCopyright © 2021 Wang, Zhang, Lu, Song, Li, Wang and Rahimnejad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ling Wang, bGluZ3dhbmdAam11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.