
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 10 August 2021
Sec. Microbiotechnology
Volume 12 - 2021 | https://doi.org/10.3389/fmicb.2021.724103
 Yuanyuan Yao1,2†
Yuanyuan Yao1,2† Xian Shu1,2†
Xian Shu1,2† Dongdong Wang1
Dongdong Wang1 Wenjie Kan1
Wenjie Kan1 Pengfei Su1
Pengfei Su1 Hao Hu1
Hao Hu1 Xu Chen1
Xu Chen1 Dacheng Wang1
Dacheng Wang1 Shengwei Huang1*
Shengwei Huang1* Lifang Wu1,2*
Lifang Wu1,2*Aflatoxin B1 (AFB1) is the most harmful mycotoxin produced by filamentous fungi and presents a serious threat to human and animal health. Therefore, it is essential to protect humans and animals from AFB1-induced acute and chronic toxicity. In this study, Pseudomonas strain m29 having a high efficiency of AFB1 transformation was isolated from soil. The transformation ratio by m29 was more than 97% within 24 h, and the optimum temperature for transformation was 37°C. Moreover, the AFB1 transforming activity was mainly attributed to the cell-free supernatant of strain m29. The metabolite that plays a crucial role in AFB1 transformation is likely 1,2-dimethylhydrazine or 1,1-dimethylhydrazine, as identified by GC-MS and LC-MS analysis. AFB1 was transformed into a product with molecular formula C17H14O7. To the best of our knowledge, this is the first study of non-enzymatic AFB1 transformation by bacteria. Importantly, this AFB1 transformation mechanism could be universal to various microorganisms.
Aflatoxins are a class of highly toxic secondary metabolites produced mainly by genera of Aspergillus, including Aspergillus flavus, Aspergillus parasiticus, Aspergillus tamari, and Aspergillus nominus, under both field and storage conditions (prefer to 20–35°C and relative humidity above 89%) (Diener and Davis, 1967; Kurtzman et al., 1987). Aflatoxin B1, B2, G1, and G2 are the most common among more than 20 kinds of aflatoxins, of which Aflatoxin B1 (AFB1) is the most toxic and carcinogenic (Gourama and Bullerman, 1995). Aflatoxin contamination of food and feed results in significant economic losses worldwide and seriously threatens human health. Therefore, considerable interest has been focused on finding effective AFB1 detoxification methods for food safety (Strosnider et al., 2006).
Various methods, including several physical and chemical strategies, have been proposed for the removal of aflatoxin contamination to manage the economic losses and health risks caused by the toxin, such as absorption, irradiation, ozone treatment, and sodium bisulfite treatment (Hagler et al., 1983; Diaz et al., 2004; Isman and Biyik, 2009; Kamber et al., 2017). In recent years, researchers have focused on microbial detoxification of aflatoxin due to its unique advantages like minimum loss of product qualities, mild processing conditions, and low cost (Verheecke et al., 2016; Raksha Rao et al., 2017). Over the past decades, several bacterial or fungal strains, such as Rhodococcus erythropolis (Alberts et al., 2006), Bacillus licheniformis (Wang et al., 2018), Pseudomonas aeruginosa (Sangare et al., 2014), Cellulosimicrobium funkei (Sun et al., 2015), and Aspergillus niger (Zhang et al., 2014), have been developed to remove aflatoxins. However, the industrial application of these strains is limited by some obvious disadvantages, such as low transformation efficiency, narrow operating temperature, and unknown removal mechanism.
In addition, there have been few studies on the mechanism of bacterial transformation of AFB1. Almost all studies have shown that bacterial transformation of AFB1 is an enzyme-dependent process. The enzymes responsible for AFB1 transformation have been identified as oxidase, reductase, and peroxidase (Doyle and Marth, 1979; Wu et al., 2015; Adebo et al., 2017). Aflatoxin oxidase (AFO) was identified in 1998 as the first enzyme known to transform AFB1 (Yao et al., 1998). Two F420H2-dependent reductases (FDR-A and FDR-B) from Mycobacterium smegmatis were also reported to catalyze the reduction of the α,β-unsaturated ester moiety of aflatoxins (Taylor et al., 2010). Zhao et al. (2011) found an aflatoxin-transforming enzyme (MADE) from Myxococcus flavus ANSM068 that can remove aflatoxin B1, G1, and M1 from a solution. Wang et al. (2011) studied the conversion of AFB1 to AFB1-8,9-dihydrodiol by manganese peroxidase (MNP) from Phanerochaete sordida YK-624, which effectively eliminated the mutagenic activity of AFB1. However, no data are currently available on the non-enzymatic AFB1 transformation by bacteria.
In this study, Pseudomonas geniculata strain m29 was isolated, and the mechanism of AFB1 transformation by strain m29 was explored. This strain transformed AFB1 through an extracellular and non-enzymatic reaction, and the metabolite responsible for AFB1 transformation was isolated and identified. This study is the first to show the non-enzymatic transformation of AFB1 by bacteria.
Aflatoxin B1 was purchased from Sangon Biotech Co., Ltd. (Shanghai, China) and standard solution was diluted with methanol to prepare an AFB1 stock solution at 25 ppm. Other reagents were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Feed was purchased from Baiyi Feed Technology Co., Ltd. (Liuyang, China). Coumarin medium (CM) was prepared according to the method described by Guan et al. (2008). Nutrient broth (NB) medium was used for liquid cultures of bacteria.
Nine soil samples were collected from several wheat fields in Hefei, Anhui Province, China and screened for strains capable of transforming AFB1. The preliminary screening was conducted according to the method described by Guan et al. (2008) and Raksha Rao et al. (2017). Colonies that grew well on coumarin medium were considered to possess the ability to transform AFB1.
The AFB1 transformation ability of the isolates was determined as follows: 16 isolates were incubated with NB containing 0.5 ppm AFB1 at 37°C overnight in a gyrotary shaker incubator (180 rpm), and uninoculated NB processed similarly served as a control. The residual AFB1 was extracted and detected using high-performance liquid chromatography (HPLC) according to the methods described by Shu et al. (2018). The limit of detection for AFB1 (3σ criterion of blank) is 0.2 ppb.
The genomic DNA of isolate m29 was extracted using an EasyPure Bacteria Genomic DNA Kit (TransGen Biotech Co., Ltd., Beijing, China), and the 16S rRNA gene fragment was amplified using PCR with universal primers (27F and 1492R) and sequenced. The comparison of the obtained sequence with available 16S rRNA gene sequences in the GenBank database was conducted using BLAST program. Five isolates (m6, m36, m29, xls3, and xls8) were collected and identified. Among the monocultures, isolate m29 was selected because of the growth performance and AFB1 degrading activity. Physical and biochemical characterization of isolate m29 was performed according to standard methods (Tindall et al., 2007).
A culture of m29 was inoculated at 1% (v/v) into 10 ml NB medium. AFB1 was added to the culture to achieve the indicated final concentration (0.5 ppm). Strain m29 was incubated at 20, 24, 28, 32, 37, 40, and 42°C for 24 h to determine the effect of temperature on AFB1 transformation. The residual AFB1 in the samples was calculated to determine the optimal degradation temperature. Then, strain m29 was cultured in NB containing AFB1 at the optimal temperature for 72 h, and samples were taken at 0, 3, 6, 12, 24, 48, and 72 h. The residual AFB1 was analyzed according to the method mentioned previously.
The anti-aflatoxigenic effect of strain M29 on A. flavus stain 3.6305 (A. flavus) purchased from the China General Microbiological Culture Collection Center (CGMCC) (aflatoxin producing capacity 422.54 μg/L in liquid culture medium) was determined according to a previously described method (Shu et al., 2018) with minor modification. Briefly, a 200-g pulverized feed containing 5 ml of A. flavus spore suspension (1 × 106 CFU/ml) was inoculated with 15 ml of m29 culture (1 × 108 CFU/ml) at 28°C, and the treatment with NB medium was used as the control. Samples were taken after 15 days to detect AFB1 by HPLC. In addition, the feed 15 days after inoculation with A. flavus was autoclaved at high temperature for 1 h to completely eliminate A. flavus. The obtained AFB1-contaminated feed was inoculated with m29 culture at 28°C, and samples were taken 7 days later to detect AFB1. The treatment with NB medium was used as the control.
Aflatoxin B1 transformation by different components of the m29 culture, including the supernatant, cells, and cell lysate, was determined according to a previously described method (Xie et al., 2019). Isolate m29 was inoculated into NB and cultured at 37°C for 24 h. The m29 culture was centrifuged at 8,000 × g to obtain the supernatant and cells. To obtain cell lysate, the cells were washed twice with phosphate buffer solution (PBS, 0.02 M, pH 7.2) and then disintegrated using an ultrasonic cell disintegrator (Ningbo Xinzhi Instrument Inc., Ningbo, China) for 30 min. After centrifugation at 10,000 × g for 10 min, the supernatant was used as cell lysate. Afterward, AFB1 with a final concentration of 0.5 ppm was treated with the cell-free supernatant, cells and cell lysates obtained above, respectively. The mixtures were incubated at 37°C for 24 h. The residual AFB1 was analyzed according to the method mentioned previously.
The effects of incubation time, temperature, and metal ions on AFB1 transformation were carried out as described by Raksha Rao et al. (2017) with minor modifications. The supernatant was obtained as described previously and exposed to 0.5 ppm AFB1. NB processed similarly served as a control. The mixture was cultured at 37°C for 48 h, and samples were taken at 1, 3, 6, 12, 24, 36, and 48 h. The reaction mixture was incubated at 20, 30, 40, 50, or 60°C for 24 h to study the effect of temperature.
Concentrations of 10 mM Cu2+ (CuSO4), Zn2+ (ZnSO4), Mg2+ (MgCl2), Fe3+ (FeCl3), or Mn2+ (MnCl2) were added to the mixture to study the effects of metal ions on AFB1 transformation, the supernatant without added metal ions served as a control. The residual AFB1 was analyzed according to the method mentioned previously.
The effects of proteinase K and SDS on the AFB1 transformation by the supernatant were studied according to the method described by Guan et al. (2008). The supernatant was treated with a concentration of 1 mg/ml proteinase K and 1% SDS. The residual AFB1 was analyzed according to the HPLC method mentioned previously.
The supernatant was ultra-filtered using a Millipore 8050 ultra-filtration unit according to the method described by Zhou et al. (2012) with small modifications. A volume of 50 ml supernatant was filtered through a 3-kDa NMWL membrane to obtain two fractions: a retentate (volume was adjusted to 50 ml as fraction 1, F1; MW > 3 kDa) and a permeate (MW < 3 kDa). The permeate was further subjected to ultrafiltration through a 1-kDa NMWL membrane to produce a second retentate (volume was adjusted to 50 ml as fraction 2, F2; 1 kDa < MW < 3 kDa) and permeate (fraction 3, F3; MW < 1 kDa). The AFB1 removal efficiency of the three fractions was determined using HPLC.
A 50-ml volume of F3 was thoroughly evaporated in a rotary evaporator with a water aspirator vacuum at a rotation speed of 100 rpm and pressure of 150 mmHg in a water bath held at 45°C (Shanghai Yarong Instrument Inc., Shanghai, China) (Cheng, 2003). Then, the liquid in the collecting flask was removed, and its volume was adjusted to 50 ml (named component 1, C1). The residues in the evaporating flask were re-dissolved in 50 ml of distilled water (named component 2, C2). A 50-ml volume of NB medium was treated similarly (named evap-NB). C1, C2, and evap-NB were incubated with AFB1 at 37°C for 24 h (the final concentration of AFB1 was adjusted to 0.5 ppm) to study the transformation ability of the different components obtained by evaporation. The evap-NB sample containing AFB1 served as the control, and the residual AFB1 was detected using HPLC.
C1 was prepared as described previously and analyzed using the headspace technique coupled with gas chromatography–mass spectrometry (GC-MS; Gotor-Vila et al., 2017). C1 was incubated at 65°C, and the compounds in the headspace were trapped for 40 min. The trapped compounds were desorbed into the GC injection port at 150°C for 3 min. The oven temperature was set at 50°C for 5 min and then programmed to rise from 40 to 100°C at 20°C/min. The transfer line was heated to 250°C, as was the ion source. The helium carrier gas was set at a flow rate of 1.2 ml/min. The mass spectrometer was operated in electron impact mode at 70 eV, with a scanning range of 30/300 m/z. Volatile compounds were tentatively identified by comparing the mass spectra and the retention times with the data system library (NIST 11 MS Library). The evap-NB was performed under the same conditions as the control, and all measurements were collected with three replicates.
LC-MS analysis was performed using the AGILENT-1200HPLC/6520QTOFMS (United States) system with a C18 analytical column (Gemini 150 × 2.0 mm, particle size 3 μm; Phenomenex). A linear gradient of 5–95% acetonitrile (MeCN)–H2O (v/v, 0.1% formic acid) over 15 min was applied to the column, followed by 95 ml MeCN (v/v, 0.1% formic acid) over 5 min with a flow rate of 0.3 ml/min. Mass spectrometry was performed in positive ion mode.
C1, evap-NB, 0.1, and 1% aqueous solution of hydrazine were prepared and treated with AFB1 (20 ppm). The samples were then incubated at 37°C for 24 h. Finally, all samples were directly analyzed using LC-MS without chloroform extraction.
C1 from strains m6, m36, xls3, and xls8 were prepared using the same method as that for m29 mentioned previously. Similarly, C1 from different strains were incubated with 20 ppm AFB1 for 12 h and the products were detected using LC-MS.
All analyses were performed in triplicate, with the values expressed as mean ± SD. The data were analyzed further using ANOVA at a 95% confidence level followed by Tukey’s test (SPSS 19.0; IBM, United States); differences were considered significant when p < 0.05.
In this study, 16 isolates were found to reduce the concentration of AFB1 in NB after a 24-h incubation at 37°C, with different effects (Table 1). Six strains had an AFB1 transformation ratio of more than 85%, of which isolate m29 had the highest transformation ratio of 89.86%. Thus, this isolate was chosen for further study.
Physiological and biochemical characterization showed that isolate m29 is a Gram-negative bacterium (Supplementary Table 1). The 16S rRNA gene sequence and phylogenetic evolution analysis showed that the closest relative of strain m29 is P. geniculata (99% similarity). The resulting sequence was deposited to the GenBank database under the accession number MZ277329. Similarly, m6, m36, xls3, and xls8 were identified as Pantoea rodasii, Pseudomonas taiwanensis, Citrobacter portucalensis, and Shigella sonnei, respectively.
Based on the physiological and biochemical characterization results and 16S rRNA gene sequence analysis, isolate m29 was identified as P. geniculata m29. Several kinds of Pseudomonas have been reported to transform AFB1, such as P. putida (Samuel et al., 2014; Singh and Mehta, 2019) and P. aeruginosa (Sangare et al., 2014). However, this is the first study to report AFB1 transformation by P. geniculata.
Most of the strains that have been reported displayed AFB1 transformation activity do so at a narrow temperature range. For example, A. niger reduces only 25–45% of AFB1 at 20–50°C (Zhang et al., 2014). Interestingly, the AFB1 transformation ratios of P. geniculata m29 were more than 78% over a wide range of temperatures (20–42°C) (Figure 1A). Moreover, the AFB1 transformation ratio reaches a maximum at 37°C, and there was no significant difference between the ratios at 32 and 42°C. A similar result reported by Guan et al. (2008) showed that S. maltophilia 35-3 also presented with the highest AFB1 transformation ratio at 37°C.
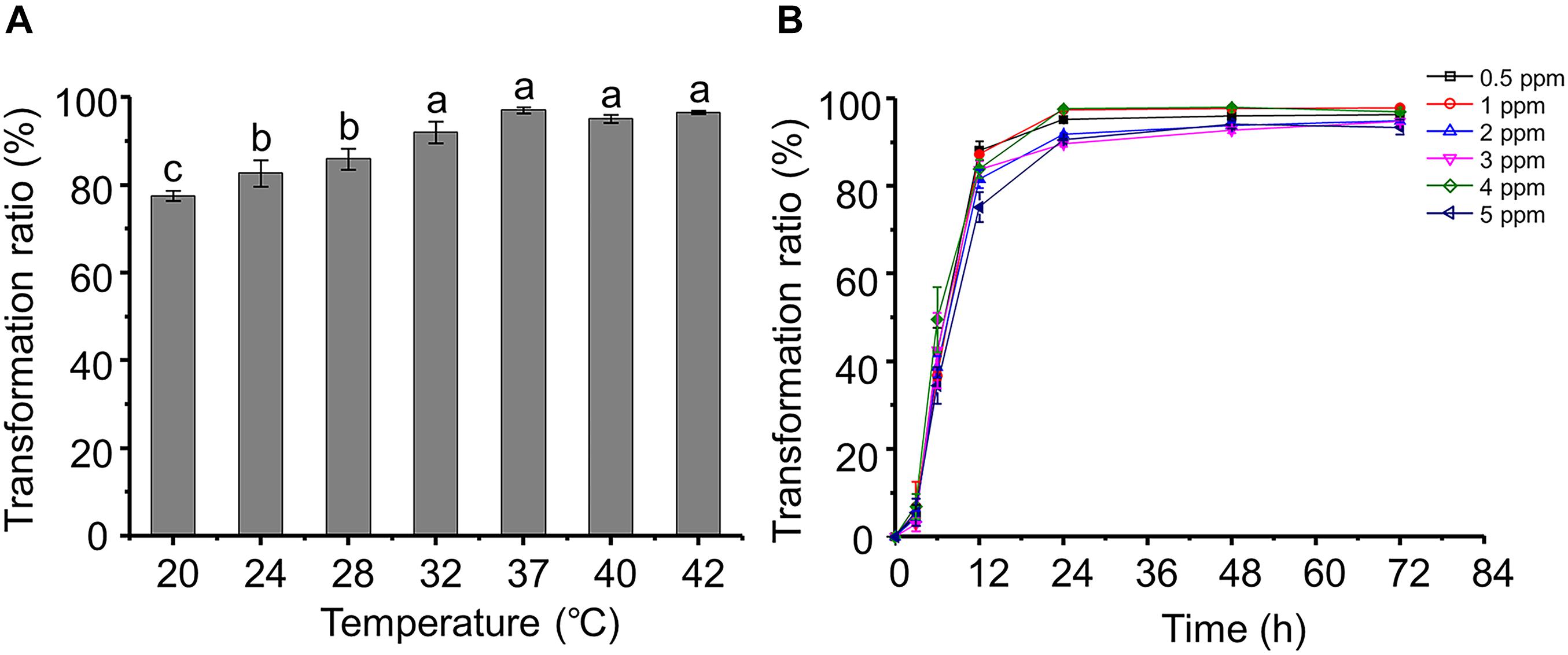
Figure 1. Aflatoxin B1 (AFB1) transformation characteristics of Pseudomonas geniculata m29 (P. geniculata m29). (A) Effect of temperature on AFB1 transformation by P. geniculata m29. (B) Kinetics of AFB1 transformation by P. geniculata m29 at 37°C. Values expressed as mean ± SD, and different letters represent significant difference according to Tukey’s LSD test (p < 0.05).
Pseudomonas geniculata m29 was incubated with different initial AFB1 concentrations (Figure 1B) to study the kinetics of AFB1 transformation. The transformation of AFB1 by strain m29 was a relatively rapid and continuous process. The transformation ratio of AFB1 at 3 h was less than 7%, and AFB1 content rapidly decreased from 3 to 24 h. After this period, the concentration of AFB1 no longer decreased significantly and remained at a very low level.
The transformation ratio of m29 can reach 97.07% at 24 h, which is the highest rate of microbial transformation in the published literature. For example, Streptomyces lividans TK24 can degrade 88% AFB1 after 24 h of incubation and S. aureofaciens ATCC 10762 by 86% (Eshelli et al., 2015). Harkai et al. (2016) observed a reduction of 88.34% for AFB1 by Streptomyces cacaoi subsp. asoensis after 5 days of incubation. Therefore, strain m29 is a more rapid biocatalyst for AFB1 transformation than others reported up to now.
Aflatoxin B1 is mutagenic and harmful to bacteria. Significantly, the bacteria capable of AFB1 biotransformation can tolerate high doses of AFB1. Li et al. (2018) reported that the AFB1 transformation ratio of Candida versatilis CGMCC 3790 decreased when the initial concentration increased from 10 to 55 ng/g. In contrast, it is clear that the AFB1 concentration had no significant effect on the transformation effect of m29, even up to 5 ppm (Figure 1B). Given the high AFB1 transformation efficiency and strong tolerance to AFB1, m29 might be a potential candidate for AFB1 removal in food and feed.
In vitro antagonistic experiments showed that m29 could significantly inhibit the growth of A. flavus (Figure 2). Furthermore, in the feed co-cultured with A. flavus and m29, a 75.40% reduction in AFB1 can be observed after 15 days (Table 2). In addition, 7 days after inoculating the AFB1-containing feed with m29 culture, AFB1 decreased by 47.95%. These results further proved that m29 had good application prospects.
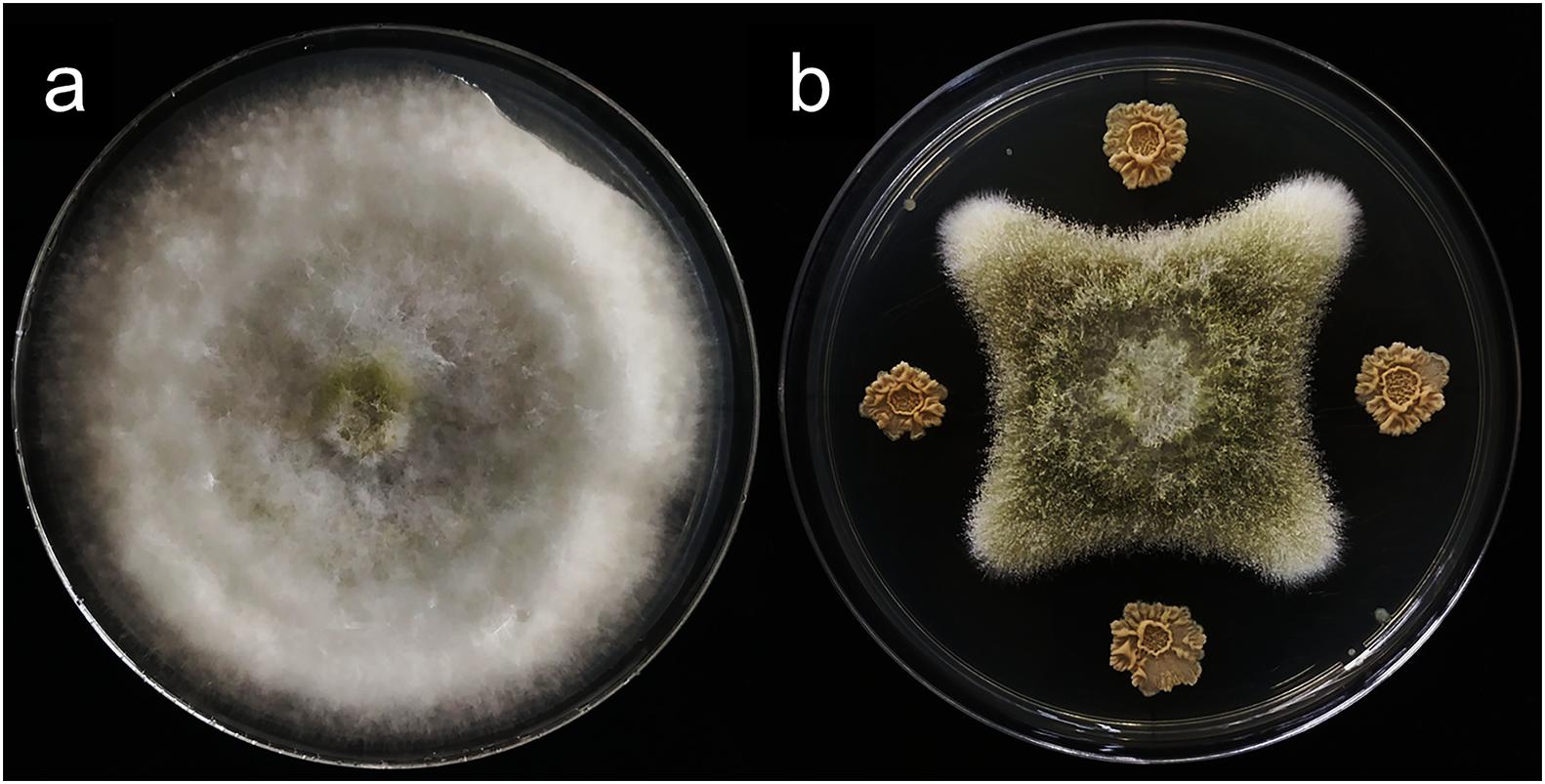
Figure 2. The inhibitory effect of strain m29 on the growth of Aspergillus flavus (A. flavus). (A) A. flavus grown for 7 days; (B) A. flavus was located in the middle of the PDA plate, and m29 was located 3 cm apart away from the center.
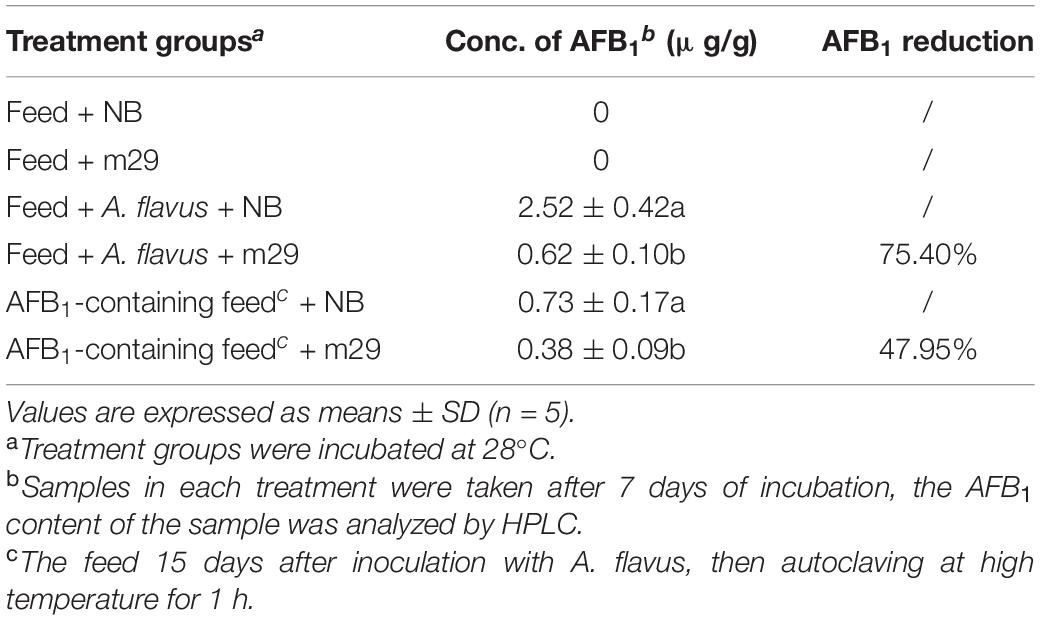
Table 2. Aflatoxin B1 (AFB1) inhibition in feed containing co-cultures of Pseudomonas geniculata m29 and Aspergillus flavus.
The AFB1 transformation ratio of the supernatant reached 80% after a 24-h incubation, compared with 46.38 and 20.69% of cells and cell lysate, respectively, (Figure 3), suggesting that the supernatant played a major role in AFB1 transformation and the absorption capacity of the cell walls only plays a small role in AFB1 removal. It seems that the removal of AFB1 by m29 was caused mainly by a metabolite secreted out of the cells, which is in accordance with findings reported for in Bacillus subtilis (Xia et al., 2017). AFB1 removal by a cell-free supernatant can overcome the disadvantage of using whole cultures that may damage the taste and nutrition of a product (Adebo et al., 2017; Shu et al., 2018). Interestingly, there was no significant change in the AFB1 conversion capacity of the supernatant treated at 121°C for 20 min, suggesting that the substances responsible for AFB1 in the supernatant may be small molecule compounds or heat-resistant proteins. Similar results were reported by Sangare et al. (2014). In addition, the AFB1 transformation ratio of heated cell lysate was significantly reduced, which indicated that intracellular heat-labile components (probably enzymes) also play an important role in AFB1 transformation by m29. Similarly, Li et al. (2018) reported that C. versatilis CGMCC 3790 transforms AFB1 through intracellular heat-labile enzymes.
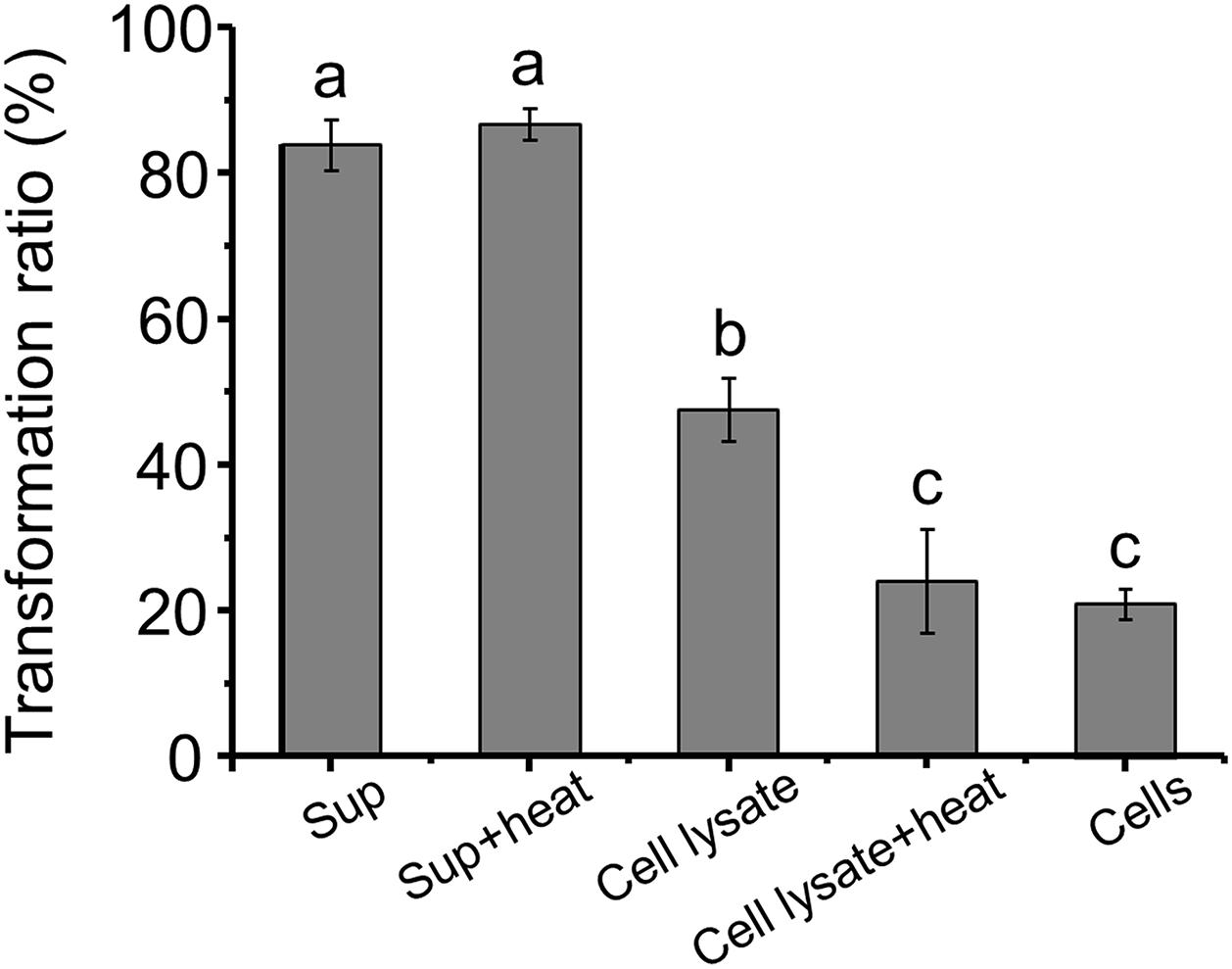
Figure 3. Aflatoxin B1 (AFB1) transformation by the supernatant, cells and cell lysate of m29 at 37°C after 24 h of incubation. The initial concentration of AFB1 was 0.5 ppm. Sup: supernatant; Sup + heat: supernatant treated at 121°C for 20 min; Cell lysate + heat: cell lysate treated at 121°C for 20 min. Values expressed as mean ± SD, and different letters represent significant difference according to Tukey’s LSD test (p < 0.05).
The dynamics of AFB1 transformation by the cell-free supernatant are shown in Figure 4A. It seems that AFB1 transformation by the supernatant of m29 is a relatively rapid process. Most of the transformation occurs within 12 h, and the transformation ratio reached 51.49% after only 1 h of incubation, which is faster than previously reported in other bacteria. For example, Alberts et al. (2006) reported that supernatant of R. erythropolis transformed 68.2% of AFB1 after 72 h of incubation. Similarly, Song et al. (2019) reported that P. aeruginosa M19 removed only 32.8% of AFB1 in the initial 6 h, and 80% of AFB1 was reduced after 144 h of incubation.
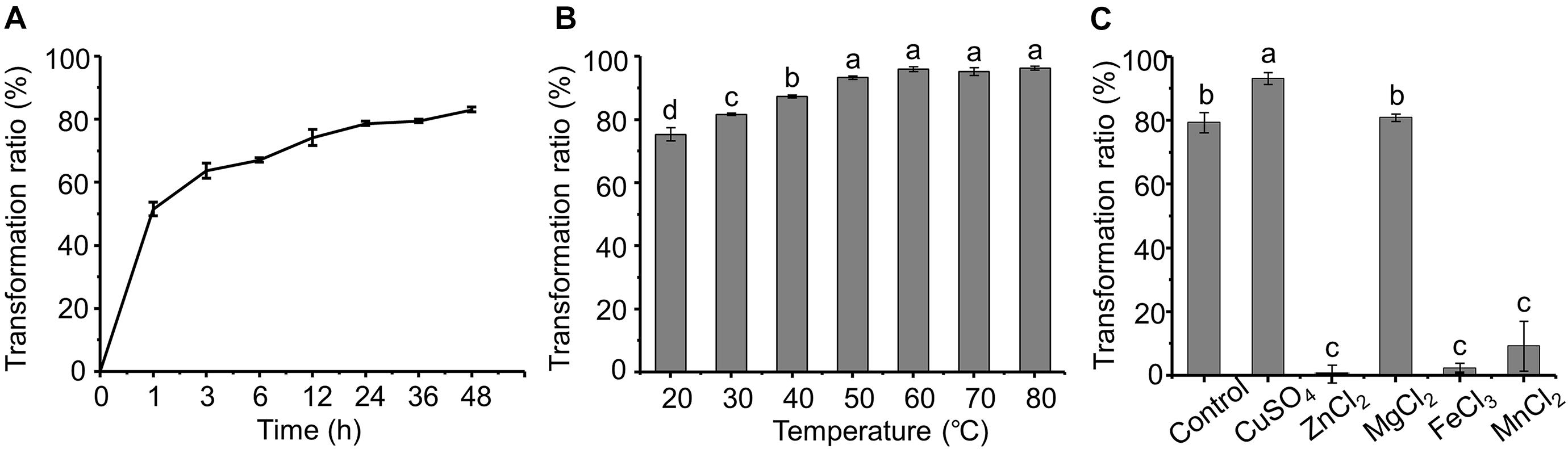
Figure 4. Effect of time, temperature and metal ions on aflatoxin B1 (AFB1) transformation by supernatant of m29. (A) Effect of incubation time; (B) effect of temperature; (C) effect of metal ions. Values expressed as mean ± SD, and different letters represent significant difference according to Tukey’s LSD test (p < 0.05).
Furthermore, the AFB1 transformation ratio of the supernatant increased with temperature, and the transformation ratio reached 93.37% after incubating at 50°C for 24 h (Figure 4B). It is worth noting that even with incubation at 60°C for 24 h, the AFB1 transformation ratio by the supernatant was not significantly affected, indicating that m29 transformed AFB1 through a heat-resistant enzyme or other metabolite. Similarly, the AFB1 transformation ratio of the cell-free supernatant of Fusarium sp. WCQ3361 had no significant change at a wide range of temperatures from 0 to 90°C (Wang et al., 2017). The excellent thermal stability means that m29 can stably and efficiently remove AFB1 in different applications.
The effect of metal ions on the AFB1 transformation ability of the supernatant is shown in Figure 4C. Cu2+ can stimulate AFB1 transformation, while Zn2+, Fe3+, and Mn2+ notably inhibited the transformation ability of the m29 supernatant. These results indicate that Cu2+ may change the structure of the AFB1-transforming metabolite in the supernatant and activate its activity. The activation effect of Cu2+ and inhibition effect of Zn2+ and Fe3+ are in agreement with a study of the AFB1 transformation ability of the culture supernatant of B. licheniformis CFR1 (Raksha Rao et al., 2017).
The AFB1 transformation ability of the supernatant was not affected by proteinase K, while SDS can significantly reduce the transformation ability (Figure 5A), indicating that AFB1 might be transformed by the supernatant of m29 in a non-enzymatic manner. For instance, chemicals such as 1% sodium bisulfite, sodium hydroxide, and aqueous ammonia transform more than 80% of AFB1 after 24 h (Moerck et al., 1980).
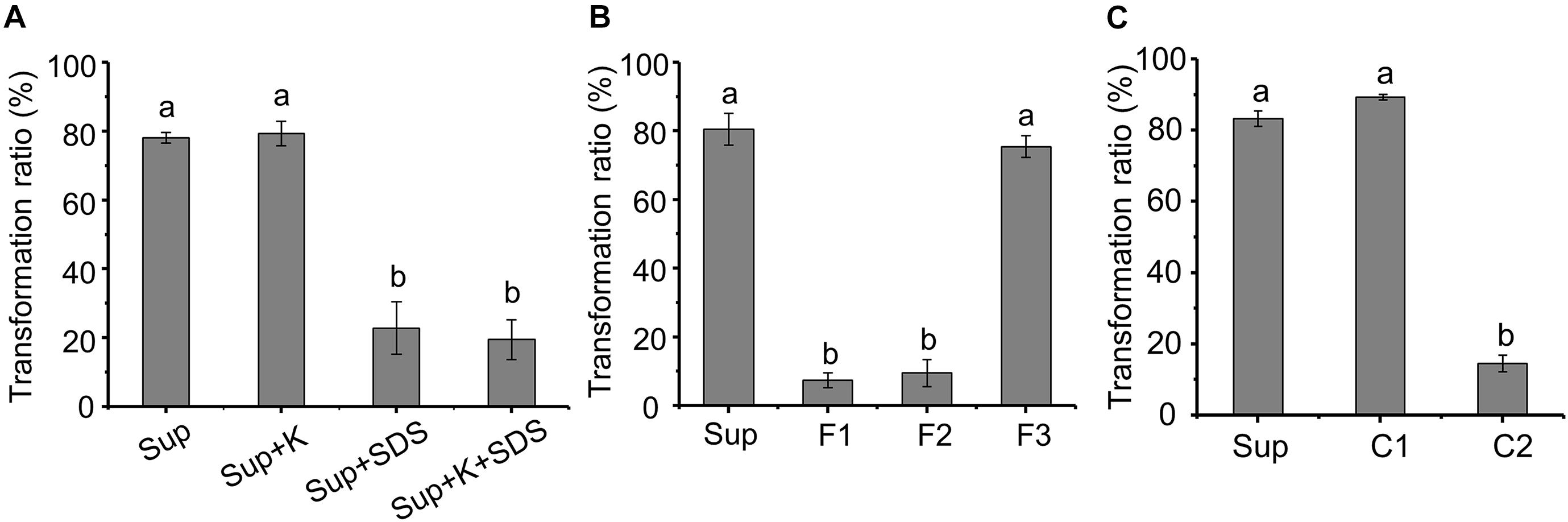
Figure 5. Preliminary analysis of the metabolite transforming aflatoxin B1 (AFB1). (A) Effect of protein K and SDS on AFB1 transformation by supernatant. Sup: supernatant; Sup + K: proteinase K treated supernatant; Sup + SDS: SDS treated supernatant; Sup + K + SDS: proteinase K and SDS treated supernatant. (B) AFB1 transformation by three fractions obtained from supernatant by ultrafiltration. F1, F2, and F3 correspond to samples >3, 1–3, or <1 kDa, respectively. (C) AFB1 transformation by component obtained from rotary evaporator. Values expressed as mean ± SD, and different letters represent significant difference according to Tukey’s LSD test (p < 0.05).
To identify the metabolite responsible for AFB1 transformation, the molecular weight (MW) of the metabolite in the supernatant that transforms AFB1 was preliminarily determined using ultrafiltration (Figure 5B). The AFB1 transformation ability of F1 and F2 was extremely low, while the AFB1 transformation ability of F3 was equal to the untreated supernatant, which indicates that the MW of the AFB1-transforming metabolite in the supernatant is lower than 1 kDa. It is very likely that small molecules (less than 1 kDa) or short peptides are responsible for the transformation of AFB1.
Components C1 and C2 were obtained from F3 by rotary evaporation, in which the volatile compounds were retained in C1. Figure 5C shows that the AFB1 transformation ratio of C1 was 89.25%, while the AFB1 transformation ratio of C2 was extremely low (<15%). Furthermore, C1 treated at different temperatures for 1 h were used to transform AFB1, and the AFB1 transformation ability of C1 treated at 60 and 90°C decreased by 24.12 and 95.34%, respectively, (Supplementary Figure 2). These results suggest that the primary AFB1-transforming metabolite produced by m29 is volatile, and AFB1 transformation is an extracellular, non-enzymatic reaction. To our knowledge, this is the first report that a volatile compound mediates the microbial transformation of AFB1. A similar study conducted by Diniz et al. (2002) reported that the sulfate-reducing bacterium D. alaskensis can produce hydrogen sulfide to reduce and decolorize azo dye, which is also an extracellular and non-enzymatic reaction.
Since C1 is the main component with an AFB1 transformation ability, the active metabolite in C1 was analyzed using HPLC. However, no new chromatographic peak was observed in C1 at 190–800 nm (Supplementary Figure 3). The headspace coupled with GC-MS was used to analyze the active metabolite with an AFB1 transformation ability. The gas chromatogram of evap-NB (control) was shown in Figure 6A and the Figure 6B indicated the mass spectra of peak in Figure 6A at 2.99 min. The peak at 2.99 min (Figure 6C) is a putative AFB1-transforming metabolite, and its GC mass spectra are shown in Figure 6D. Comparison with the data system library indicates that the AFB1-transforming metabolites might be hydrazine compounds, such as 1,2-dimethylhydrazine and 1,1-dimethylhydrazine. Furthermore, the signal at m/z 61 can only be detected in C1 by LC-MS (Figure 6E), and the HR-ESIMS spectra of this compound are shown in Supplementary Figure 4, which confirmed that the MW of this compound was 60 g/mol. Here, due to the lack of standards for 1,2-dimethylhydrazine and 1,1-dimethylhydrazine, a 0.1% aqueous solution of hydrazine was used to transform AFB1. As shown in Supplementary Figure 5, the AFB1 transformation product of C1 was the same as that of a 0.1% aqueous solution of hydrazine. In conclusion, these results suggest that the AFB1-transforming metabolite of m29 is likely to be 1,2-dimethylhydrazine or 1,1-dimethylhydrazine (Figure 6F). In addition to m29, a variety of microorganisms have been reported to produce various hydrazine-containing compounds, such as katorazone from Streptomyces sp. IFM 11299, gyromitrins from Gyromitra esculenta, and spinamycin from Streptomyces albospinus (Le Goff and Ouazzani, 2014).
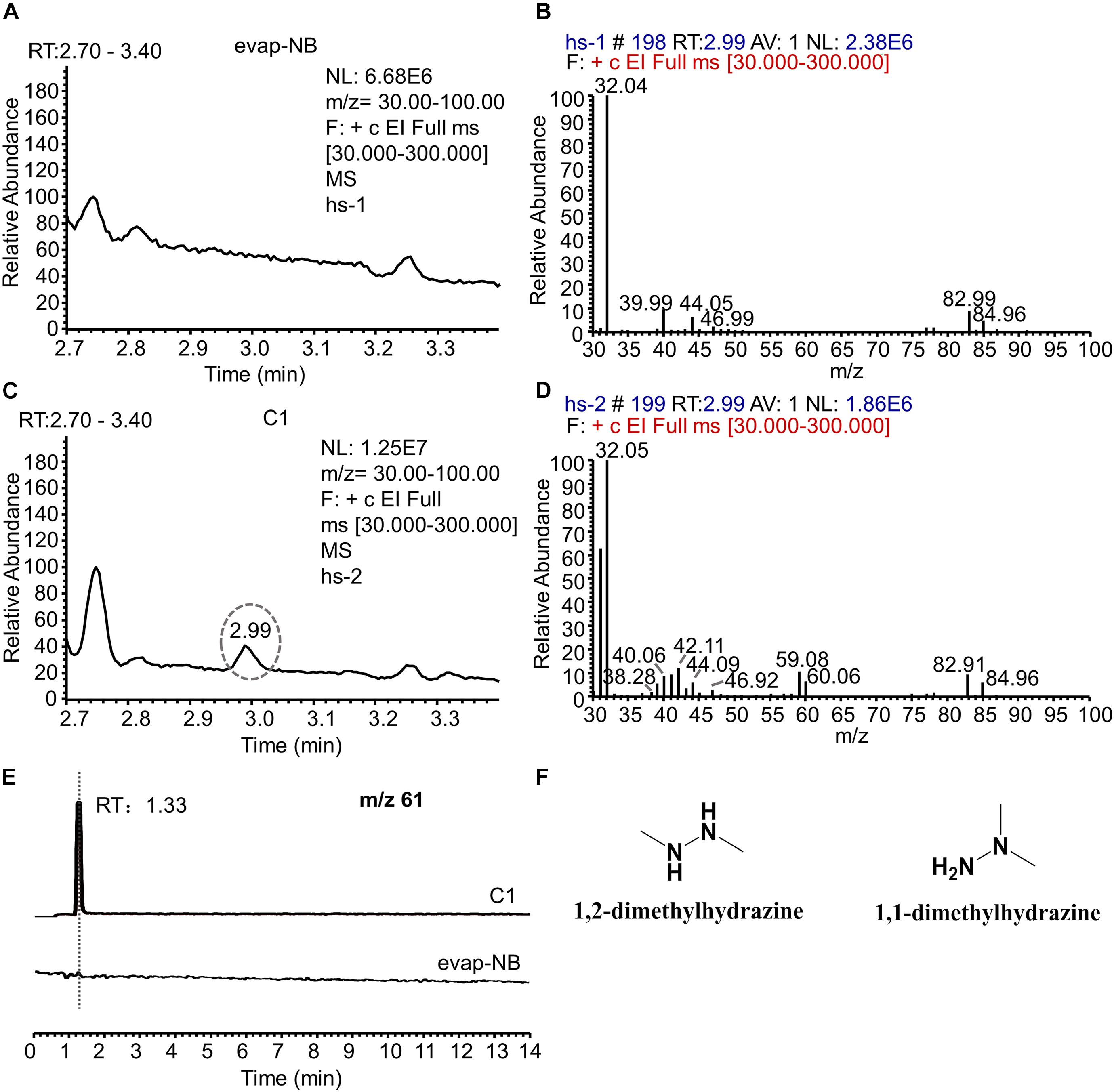
Figure 6. GC-MS and LC-MS analysis of evap-NB and C1. (A) Gas chromatogram of evap-NB. (B) Mass spectra of peak in chromatogram A at 2.99 min. (C) Gas chromatogram of C1; (D) mass spectra of peak in chromatogram C at 2.99 min. (E) Single ion monitoring of m/z 61. (F) Structure of 1,2-dimethylhydrazine and 1,1-dimethylhydrazine. GC-MS, gas chromatography–mass spectrometry; LC-MS, liquid chromatography–mass spectrometry.
After co-incubation with m29 culture and AFB1, chloroform was used to extract AFB1 and the transformation product; the structure of the product was further determined by LC-MS (Supplementary Figure 6). However, no transformation product was found, and similar results have been reported by other researchers (Farzaneh et al., 2012; Sangare et al., 2014; Raksha Rao et al., 2017; Xia et al., 2017; Shu et al., 2018). It speculated that the chemical properties of AFB1 transformation products are different from those of AFB1, making them difficult to be detected (Alberts et al., 2006).
C1 was incubated with 20 ppm AFB1 for 24 h, and the transformation product was directly analyzed by LC-MS without extraction using chloroform (Figure 7A) to investigate further the identity of the transformation product. A transformation product with a MW of 330 g/mol (18 units more that of AFB1) was observed. The UV and MS data of AFB1 and the AFB1 transformation product are shown in Supplementary Figures 7, 8. To further determine the structure of the AFB1 transformation product, coumarin was reacted with C1, and the product 3 with a MW of 164 g/mol (18 units more that of coumarin) was observed (Figure 7B and Supplementary Figures 9,10). Therefore, the lactone rings of AFB1 and coumarin react with C1, rather than the carbonyl group on the five-membered ring of AFB1. The structure of transformation product and speculative reaction mechanism is shown in Figure 7C. The same transformation product with unclarified mechanism has also been reported, which was less toxic than AFB1 (Qiu et al., 2021). Interestingly, 1% hydrazine could convert AFB1 into product two with a MW of 326 g/mol (Supplementary Figure 11), which was speculated to be the product of the reaction of the carbonyl group of AFB1 with hydrazine. The UV spectra, MS spectra, and speculative structure of product two are shown in Supplementary Figure 11. The findings also imply that the 1,2-dimethylhydrazine or 1,1-dimethylhydrazine content in C1 might be very low, resulting in the absence of product two in the reaction between AFB1 and C1.
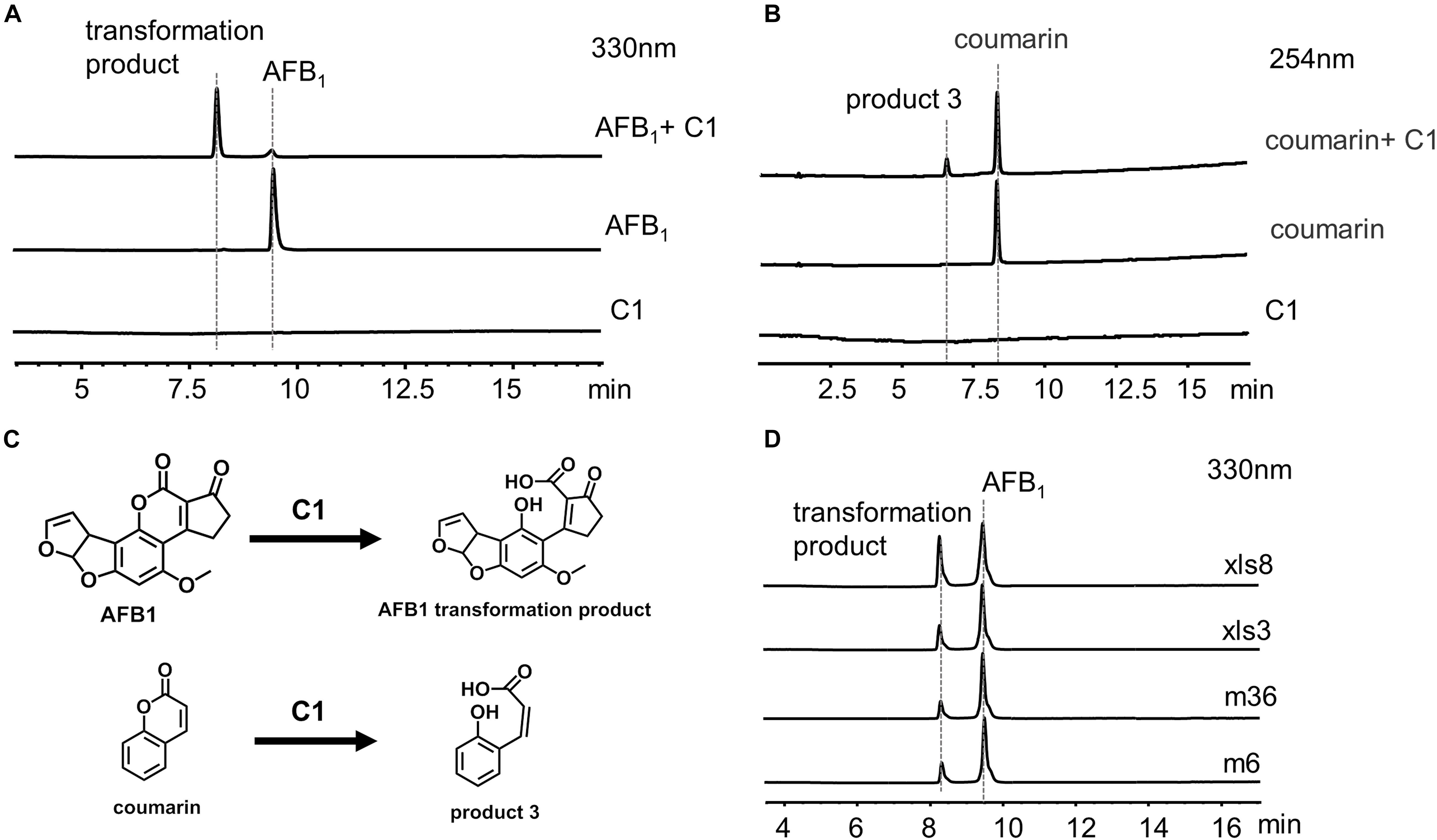
Figure 7. C1 reaction with aflatoxin B1 (AFB1) and coumarin. (A) HPLC analysis of the reaction between C1 and AFB1. (B) HPLC analysis of the reaction between C1 and coumarin. (C) Speculative reaction mechanism of C1. (D) AFB1 was transformed by C1 from different strains. AFB1 was transformed by C1 from four different strains into the same transformation product as m29. HPLC, high-performance liquid chromatography.
We hypothesized that the AFB1 transformation mechanism might be widespread in a variety of bacteria. Therefore, C1 from four different strains (m6, m36, xls3, and xls8) were used to transform AFB1 (Figure 7D). AFB1 was transformed into the same product produced by m29 by C1 of four different strains. These results suggest that the transformation of AFB1 by C1 may be the first step in a general AFB1 detoxification strategy for bacteria. The subsequent AFB1 transformation process may require the further involvement of intracellular enzymes.
In summary, Pseudomonas strain m29 that can efficiently transform AFB1 was isolated. GC-MS and LC-MS analysis indicate that the transformation process is extracellular and non-enzymatic and mainly depends on the hydrazine compound produced during the growth of bacteria. This is the first study to report the non-enzymatic AFB1 transformation by bacteria. In addition, the structure of the AFB1 transformation product was preliminarily identified. It is worth noting that the transformation mechanism of AFB1 may be widespread in a variety of bacteria, indicating that we should also pay attention to the important role of microbial non-enzymatic transformation in the treatment of aflatoxin contamination. Indeed, future studies are needed to elucidate the AFB1 transformation mechanism, and explore the possible use of the P. geniculata m29 in food and feed.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
YY, XS, and SH thoroughly discussed and designed this study. YY and XS were in charge of the investigation, data collection, and article writing. DoW analyzed the biological samples. WK, PS, XC, and DaW helped with data curation and formal analysis. LW provided funding and managed the project. All authors contributed to the article and approved the submitted version.
This work was supported by the Natural Science Foundation of Anhui Province (2008085MC60) and the Key Program of 13th Five-Year Plan, Hefei Institutes of Physical Science, Chinese Academy of Sciences (No. kp-2019-21).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the staff members at Omics-laboratory of the Biotechnology Center for Anhui Agricultural University in Hefei, China for providing technical support in mass data collection and the other valuable discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.724103/full#supplementary-material
Adebo, O. A., Njobeh, P. B., Gbashi, S., Nwinyi, O. C., and Mavumengwana, V. (2017). Review on microbial degradation of aflatoxins. Crit. Rev. Food Sci. Nutr. 57, 3208–3217. doi: 10.1080/10408398.2015.1106440
Alberts, J. F., Engelbrecht, Y., Steyn, P. S., Holzapfel, W. H., and van Zyl, W. H. (2006). Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int. J. Food Microbiol. 109, 121–126. doi: 10.1016/j.ijfoodmicro.2006.01.019
Cheng, C. C. (2003). Recovery of polycyclic aromatic hydrocarbons during solvent evaporation with a rotary evaporator. Polycyclic Aromat. Compd. 23, 315–325. doi: 10.1080/10406630308048
Diaz, D. E., Hagler, W. M., Blackwelder, J. T., Eve, J. A., Hopkins, B. A., Anderson, K. L., et al. (2004). Aflatoxin Binders II: reduction of aflatoxin M1 in milk by sequestering agents of cows consuming aflatoxin in feed. Mycopathologia 157, 233–241. doi: 10.1023/b:myco.0000020587.93872.59
Diener, U. L., and Davis, N. D. (1967). Limiting temperature and relative humidity for growth and production of aflatoxin and free fatty acids by Aspergillus flavus in sterile peanuts. J. Am. Oil Chem. Soc. 44, 259–263. doi: 10.1007/BF02639271
Diniz, P. E., Lopes, A. T., Lino, A. R., and Serralheiro, M. L. (2002). Anaerobic reduction of a sulfonated azo dye, Congo Red, by sulfate-reducing bacteria. Appl. Biochem. Biotechnol. 97, 147–163. doi: 10.1385/abab:97:3:147
Doyle, M. P., and Marth, E. H. (1979). Peroxidase activity in mycelia of Aspergillus parasiticus that degrade aflatoxin. Eur. J. Appl. Microbiol. Biotechnol. 7, 211–217. doi: 10.1007/BF00505027
Eshelli, M., Harvey, L., Edrada-Ebel, R., and McNeil, B. (2015). Metabolomics of the bio-degradation process of aflatoxin B1 by Actinomycetes at an initial pH of 6.0. Toxin 7, 439–456. doi: 10.3390/toxins7020439
Farzaneh, M., Shi, Z.-Q., Ghassempour, A., Sedaghat, N., Ahmadzadeh, M., Mirabolfathy, M., et al. (2012). Aflatoxin B1 degradation by Bacillus subtilis UTBSP1 isolated from pistachio nuts of Iran. Food Control 23, 100–106. doi: 10.1016/j.foodcont.2011.06.018
Gotor-Vila, A., Teixidó, N., Di Francesco, A., Usall, J., Ugolini, L., Torres, R., et al. (2017). Antifungal effect of volatile organic compounds produced by Bacillus amyloliquefaciens CPA-8 against fruit pathogen decays of cherry. Food Microbiol. 64, 219–225. doi: 10.1016/j.fm.2017.01.006
Gourama, H., and Bullerman, L. B. (1995). Aspergillus flavus and Aspergillus parasiticus: aflatoxigenic fungi of concern in foods and feeds: a review. J. Food Prot. 58, 1395–1404. doi: 10.4315/0362-028X-58.12.1395
Guan, S., Ji, C., Zhou, T., Li, J., Ma, Q., and Niu, T. (2008). Aflatoxin B1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int. J. Mol. Sci. 9, 1489–1503. doi: 10.3390/ijms9081489
Hagler, W. M. J., Hutchins, J. E., and Hamilton, P. B. (1983). Destruction of Aflatoxin B1 with sodium bisulfite: isolation of the major product aflatoxin B1S. J. Food Prot. 46, 295–300. doi: 10.4315/0362-028X-46.4.295
Harkai, P., Szabó, I., Cserháti, M., Krifaton, C., Risa, A., Radó, J., et al. (2016). Biodegradation of aflatoxin-B1 and zearalenone by Streptomyces sp. collection. Int. Biodeterior. Biodegrad. 108, 48–56. doi: 10.1016/j.ibiod.2015.12.007
Isman, B., and Biyik, H. (2009). THE AFLATOXIN CONTAMINATION OF FIG FRUITS IN AYDIN CITY (TURKEY). J. Food Saf. 29, 318–330. doi: 10.1111/j.1745-4565.2009.00159.x
Kamber, U., Gülbaz, G., Aksu, P., and Doğan, A. (2017). Detoxification of Aflatoxin B1 in Red Pepper (Capsicum annuum L.) by ozone treatment and its effect on microbiological and sensory quality. J. Food Process. Preserv. 41:e13102. doi: 10.1111/jfpp.13102
Kurtzman, C. P., Horn, B. W., and Hesseltine, C. W. (1987). Aspergillus nomius, a new aflatoxin-producing species related to Aspergillus flavus and Aspergillus tamarii. Antonie Van Leeuwenhoek 53, 147–158. doi: 10.1007/BF00393843
Le Goff, G., and Ouazzani, J. (2014). Natural hydrazine-containing compounds: biosynthesis, isolation, biological activities and synthesis. Bioorg. Med. Chem. 22, 6529–6544. doi: 10.1016/j.bmc.2014.10.011
Li, J., Huang, J., Jin, Y., Wu, C., Shen, D., Zhang, S., et al. (2018). Mechanism and kinetics of degrading aflatoxin B1 by salt tolerant Candida versatilis CGMCC 3790. J. Hazard. Mater. 359, 382–387. doi: 10.1016/j.jhazmat.2018.05.053
Moerck, K. E., Mc, E. P., Wohlman, A., and Hilton, B. W. (1980). Aflatoxin destruction in corn using sodium bisulfite. sodium hydroxide and aqueous ammonia. J. Food Prot. 43, 571–574. doi: 10.4315/0362-028X-43.7.571
Qiu, T., Wang, H., Yang, Y., Yu, J., Ji, J., Sun, J., et al. (2021). Exploration of biodegradation mechanism by AFB1-degrading strain Aspergillus niger FS10 and its metabolic feedback. Food Control 121:107609. doi: 10.1016/j.foodcont.2020.107609
Raksha Rao, K., Vipin, A. V., Hariprasad, P., Anu Appaiah, K. A., and Venkateswaran, G. (2017). Biological detoxification of Aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 71, 234–241. doi: 10.1016/j.foodcont.2016.06.040
Samuel, M. S., Sivaramakrishna, A., and Mehta, A. (2014). Degradation and detoxification of aflatoxin B1 by Pseudomonas putida. Int. Biodeterior. Biodegrad. 86, 202–209. doi: 10.1016/j.ibiod.2013.08.026
Sangare, L., Zhao, Y., Folly, Y. M., Chang, J., Li, J., Selvaraj, J. N., et al. (2014). Aflatoxin B1 degradation by a Pseudomonas strain. Toxins 6, 3028–3040. doi: 10.3390/toxins6103028
Shu, X., Wang, Y., Zhou, Q., Li, M., Hu, H., Ma, Y., et al. (2018). Biological degradation of aflatoxin B1 by cell-free extracts of Bacillus velezensis DY3108 with Broad PH stability and excellent thermostability. Toxins 10:330. doi: 10.3390/toxins10080330
Singh, J., and Mehta, A. (2019). Protein-mediated degradation of aflatoxin B1 by Pseudomonas putida. Braz. J. Microbiol. 50, 1031–1039. doi: 10.1007/s42770-019-00134-x
Song, J., Zhang, S., Xie, Y., and Li, Q. (2019). Purification and characteristics of an aflatoxin B1 degradation enzyme isolated from Pseudomonas aeruginosa. FEMS Microbiol. Lett. 366:fnz034. doi: 10.1093/femsle/fnz034
Strosnider, H., Azziz-Baumgartner, E., Banziger, M., Bhat, R. V., Breiman, R., Brune, M. N., et al. (2006). Workgroup report: public health strategies for reducing aflatoxin exposure in developing countries. Environ. Health Perspect. 114, 1898–1903. doi: 10.1289/ehp.9302
Sun, L. H., Zhang, N. Y., Sun, R. R., Gao, X., Gu, C., Krumm, C. S., et al. (2015). A novel strain of Cellulosimicrobium funkei can biologically detoxify aflatoxin B1 in ducklings. Microb. Biotechnol. 8, 490–498. doi: 10.1111/1751-7915.12244
Taylor, M. C., Jackson, C. J., Tattersall, D. B., French, N., Peat, T. S., Newman, J., et al. (2010). Identification and characterization of two families of F420H2-dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 78, 561–575. doi: 10.1111/j.1365-2958.2010.07356.x
Tindall, B. J., Sikorski, J., Smibert, R. A., and Krieg, N. R. (2007). “Phenotypic characterization and the principles of comparative systematics,” in Methods for General and Molecular Microbiology, eds C. A. Reddy, T. J. Beveridge, J. A. Breznak, G. A. Marzluf, T. M. Schmidt, and L. R. Snyder (Washington: ASM Press), 330–393.
Verheecke, C., Liboz, T., and Mathieu, F. (2016). Microbial degradation of aflatoxin B1: current status and future advances. Int. J. Food Microbiol. 237, 1–9. doi: 10.1016/j.ijfoodmicro.2016.07.028
Wang, C., Li, Z., Wang, H., Qiu, H., Zhang, M., Li, S., et al. (2017). Rapid biodegradation of aflatoxin B1 by metabolites of Fusarium sp. WCQ3361 with broad working temperature range and excellent thermostability. J. Sci. Food Agric. 97, 1342–1348. doi: 10.1002/jsfa.7872
Wang, J., Ogata, M., Hirai, H., and Kawagishi, H. (2011). Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 314, 164–169. doi: 10.1111/j.1574-6968.2010.02158.x
Wang, Y., Zhang, H., Yan, H., Yin, C., Liu, Y., Xu, Q., et al. (2018). Effective biodegradation of Aflatoxin B1 Using the Bacillus licheniformis (BL010) Strain. Toxins 10:497. doi: 10.3390/toxins10120497
Wu, Y. Z., Lu, F. P., Jiang, H. L., Tan, C. P., Yao, D. S., Xie, C. F., et al. (2015). The furofuran-ring selectivity, hydrogen peroxide-production and low Km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food Chem. Toxicol. 76, 125–131. doi: 10.1016/j.fct.2014.12.004
Xia, X., Zhang, Y., Li, M., Garba, B., Zhang, Q., Wang, Y., et al. (2017). Isolation and characterization of a Bacillus subtilis strain with aflatoxin B1 biodegradation capability. Food Control 75, 92–98. doi: 10.1016/j.foodcont.2016.12.036
Xie, Y., Wang, W., and Zhang, S. (2019). Purification and identification of an aflatoxin B1 degradation enzyme from Pantoea sp. T6. Toxicon 157, 35–42. doi: 10.1016/j.toxicon.2018.11.290
Yao, D. S., Liang, R., Liu, D. L., Gu, L. Q., Ma, L., and Chen, W. Q. (1998). Screening of the fungus whose multienzyme system has catalytic detoxification activity towards aflatoxin B1 (Part I). Ann. N. Y. Acad. Sci. 864, 579–585. doi: 10.1111/j.1749-6632.1998.tb10385.x
Zhang, W., Xue, B., Li, M., Mu, Y., Chen, Z., Li, J., et al. (2014). Screening a strain of Aspergillus niger and optimization of fermentation conditions for degradation of aflatoxin B1. Toxins 6, 3157–3172. doi: 10.3390/toxins6113157
Zhao, L. H., Guan, S., Gao, X., Ma, Q. G., Lei, Y. P., Bai, X. M., et al. (2011). Preparation, purification and characteristics of an aflatoxin degradation enzyme from Myxococcus fulvus ANSM068. J. Appl. Microbiol. 110, 147–155. doi: 10.1111/j.1365-2672.2010.04867.x
Keywords: aflatoxin B1, Pseudomonas geniculata m29, transformation, non-enzymatic, LC-MS analysis
Citation: Yao Y, Shu X, Wang D, Kan W, Su P, Hu H, Chen X, Wang D, Huang S and Wu L (2021) Non-enzymatic Transformation of Aflatoxin B1 by Pseudomonas geniculata m29. Front. Microbiol. 12:724103. doi: 10.3389/fmicb.2021.724103
Received: 12 June 2021; Accepted: 12 July 2021;
Published: 10 August 2021.
Edited by:
Lei Chen, Tianjin University, ChinaReviewed by:
Alka Mehta, School of Biosciences and Technology, Vellore Institute of Technology, IndiaCopyright © 2021 Yao, Shu, Wang, Kan, Su, Hu, Chen, Wang, Huang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengwei Huang, c3dodWFuZ0BpcHAuYWMuY24=; Lifang Wu, bGZ3dUBpcHAuYWMuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.