
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 10 December 2020
Sec. Microbiotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fmicb.2020.594052
This article is part of the Research Topic Microbial Systems as Paradigms Of Successful And Sustainable Interactions View all 13 articles
Tetrahydrofuran (THF) is a universal solvent widely used in the synthesis of chemicals and pharmaceuticals. As a refractory organic contaminant, it can only be degraded by a small group of microbes. In this study, a thiamine auxotrophic THF-degrading bacterium, Rhodococcus ruber ZM07, was isolated from an enrichment culture H-1. It was cocultured with Escherichia coli K12 (which cannot degrade THF but can produce thiamine) and/or Escherichia coli K12ΔthiE (which can neither degrade THF nor produce thiamine) with or without exogenous thiamine. This study aims to understand the interaction mechanisms between ZM07 and K12. We found that K12 accounted for 30% of the total when cocultured and transferred with ZM07 in thiamine-free systems; in addition, in the three-strain (ZM07, K12, and K12ΔthiE) cocultured system without thiamine, K12ΔthiE disappeared in the 8th transfer, while K12 could still stably exist (the relative abundance remained at approximately 30%). The growth of K12 was significantly inhibited in the thiamine-rich system. Its proportion was almost below 4% after the fourth transfer in both the two-strain (ZM07 and K12) and three-strain (ZM07, K12, and K12ΔthiE) systems; K12ΔthiE’s percentage was higher than K12’s in the three-strain (ZM07, K12, and K12ΔthiE) cocultured system with exogenous thiamine, and both represented only a small proportion (less than 1% by the fourth transfer). The results of the coculture of K12 and K12ΔthiE in thiamine-free medium indicated that intraspecific competition between them may be one of the main reasons for the extinction of K12ΔthiE in the three-strain (ZM07, K12, and K12ΔthiE) system without exogenous thiamine. Furthermore, we found that ZM07 could cooperate with K12 through extracellular metabolites exchanges without physical contact. This study provides novel insight into how microbes cooperate and compete with one another during THF degradation.
As an important solvent, tetrahydrofuran (THF) is widely applied in the chemical industry. According to toxicity tests, THF induces cell proliferation and causes DNA damage, thereby increasing the risk of cancer in laboratory animals (Gamer et al., 2002; Hermida et al., 2006); moreover, severe central nervous system disorders can be triggered by acute exposure to high concentrations of THF (Katahira et al., 1982; Chhabra et al., 1990). In recent years, biodegradation has been universally acknowledged as an ecofriendly remediation strategy for contaminant removal with high degradation rates (Oh et al., 2010). However, a few kinds of THF-degrading microorganisms (including sixteen bacteria and three fungi) have been reported (Bernhardt and Diekmann, 1991; Parales et al., 1994; Kohlweyer et al., 2000; Skinner et al., 2009; Yao et al., 2009; Masuda et al., 2012; Tajima et al., 2012; Sei et al., 2013; Ren et al., 2020). Furthermore, the biodegradation pathway of THF has not been investigated thoroughly (Skinner et al., 2009). One of the most widely accepted THF metabolic pathways is the oxidation pathway, whereby THF is initially oxidized into 2-hydroxytetrahydrofuran (2-OH THF), which then can form γ-butyrolactone or γ-hydroxybutyraldehyde. Both of these compounds can form γ-hydroxybutyrate, which can be further oxidized to succinate and enter the tricarboxylic acid cycle (Bernhardt and Diekmann, 1991; Thiemer et al., 2003; Morenohorn et al., 2005; Tajima et al., 2012).
According to previous studies, bacteria are ubiquitous and coexist to survive in nature (Dubey and Ben-Yehuda, 2011; Schuster et al., 2013; Amin et al., 2015; Bruger and Waters, 2018). Cooperative behaviors among microorganisms are commonplace (Brown and Johnstone, 2001; Velicer, 2003). For microbes, communication and cooperation are essential in their natural lives. Through cooperation, microorganisms can more efficiently perform biological functions, such as foraging, biofilm formation, pathopoiesis, anabolism and biodegradation (Webb et al., 2003; Du et al., 2012; Rakoff-Nahoum et al., 2014; Filkins et al., 2015; Tao et al., 2017). Many crucial genetic functions of cooperators are leaky, some productions are produced by microbes of community and become the “public good,” which represents an important type of cooperation between microorganisms (Morris et al., 2012; Morris, 2015). Not only the producers but also other nearby members can benefit from diffusible substances released into the environment (Van Gestel et al., 2014; Lujan et al., 2015). Cross-feeding (or syntrophy) is another important type of interaction among microorganisms (Sieuwerts et al., 2008; Morris et al., 2013). Metabolites are released into the extracellular environment and then utilized by others as nutrients or energy sources (Mee et al., 2014; Pande et al., 2016). Previous studies found that a number of microorganisms are auxotrophic in nature and rely on external nutrients (i.e., amino acids, vitamins and other cofactors) for growth (D’Souza et al., 2014; Wexler and Goodman, 2017; Liu et al., 2018). Hydrocarbon-degrading bacterial consortia are built by microbial interactions among amino acids and vitamin auxotrophy and their cooperators (Hubalek et al., 2017). However, the question remains as to how auxotrophic microorganisms, especially contaminant-degrading bacteria, perform their functions in microbial communities. In this study, a THF-degrading bacterium, Rhodococcus ruber ZM07, was isolated from a THF-degrading bacterial culture H-1 (Huang et al., 2019). This strain cannot be continuously passaged in basal salt medium (BSM) with THF as the sole carbon source, although it can cooperate with many kinds of other bacteria, such as Escherichia coli K12, to form a stable symbiotic system. Strain ZM07 has been verified as a thiamine auxotroph. Therefore, it is very likely that some of the microorganisms that degrade refractory pollutants, e.g., THF, but have not yet been isolated might also be auxotrophic and depend on other non-degrading microbes to perform their functions in nature. Strain ZM07 was deemed highly suitable for this study, which investigated this interesting phenomenon and the ecological significance of auxotrophic strains.
Cooperative behavior can potentially be exploited by uncooperative cheaters (Fiegna and Velicer, 2003; West et al., 2006), which gain benefits from other cooperators without contributing; thus, they achieve a competitive advantage and can invade the community (Smith, 2001; Velicer, 2003; West et al., 2006). If cheaters are unrestricted in the bacterial community, they may quickly overgrow, ultimately leading to a population collapse of the community (Hardin, 1968). In a previous study, cheaters could help to counteract competition among microorganisms and foster biodiversity in well-mixed media by invading the community of cooperating siderophore producers (Leinweber et al., 2017). However, another study indicated that cheaters could be resisted by the conditional privatization strategy under stressful conditions (Jin et al., 2018). Therefore, it might be important to investigate the behavior pattern of uncooperative organisms in systems. We could deduce the stability of the cooperative community of auxotrophic degrading bacteria and nondegrading bacteria in actual environments, such as in wastewater treatment, by studying the noncooperator K12ΔthiE constructed in this study.
Auxotrophic microorganisms that degrade refractory pollutants depend on other nondegrading microbes to perform their functions in nature. However, the understanding of the interaction mechanisms between degrading and nondegrading microorganisms as well as the ecological significance of auxotrophic strains is limited. In this study, different cooperative and competitive systems were constructed to explore the interspecies and transspecies communications among the thiamine auxotrophic THF-degrading bacterium ZM07 and its cooperator K12 as well as the stability of the cooperative community of these two strains. This research was proposed to investigate the functions of auxotrophic degrading bacteria and nondegrading bacteria in a cooperative system for THF degradation. The proposed model of auxotrophic THF-degrading and non-THF-degrading microorganisms may provide an insight into the interaction mechanisms in multispecies ecosystems.
The strains and plasmids used in this study are listed in Table 1. The THF-degrading strain Rhodococcus ruber ZM07 (collection number CCTCC AB 2019217) was isolated from an enrichment culture designated H-1 (Huang et al., 2019). The non-THF-degrading strain Escherichia coli K12 (Bachmann et al., 1976) and its knockout strain K12ΔthiE (which can neither degrade THF nor produce thiamine) were both cocultured with ZM07, and the latter strain was constructed as described in the Supplementary Methods. The λ-Red recombination plasmid pDK46 (Datsenko and Wanner, 2000) used for thiE disruption was purchased from Youbio Biological Technology Co., Ltd. (Changsha, China). Plasmid pGemT7cat (Król et al., 2010) was used as a template for PCR amplification of the fused chloramphenicol-selectable marker.
The strains were initially cultivated in lysogeny broth (LB) culture medium and then transferred into BSM supplemented with THF as the sole carbon source. One liter of LB contained 10 g tryptone, 5 g yeast extract, and 10 g NaCl, and the initial pH of the media was 7.4. One liter of BSM contained 3.240 g K2HPO4, 1.000 g NaH2PO4⋅H2O, 2.000 g NH4Cl, 0.123 g C6H8NNa3O7, 0.200 g MgSO4⋅7H2O, 0.012 g FeSO4⋅7H2O, 0.003 g MnSO4⋅H2O, 0.003 g ZnSO4⋅7H2O, and 0.001 g CoCl2⋅6H2O (Parales et al., 1994; Huang et al., 2019). For experiments under thiamine-rich conditions, 0.01 mM external thiamine was added to the BSM. All strains used in this study were cultured in 100 mL of BSM with an initial pH of 7.0 at 30°C, referred to the conditions of THF-degrading bacterial culture H-1 (Huang et al., 2019). Most experiments were performed using 500 mL shaking flasks at 160 rpm, while specifically designed two-phase reactors were used in the experiment to verify the interaction mode between strains ZM07 and K12 with a shaking rate of 100 rpm.
(1) To investigate the interaction mechanism between ZM07 and K12, this study used a two-phase reactor that can separate cells but allow extracellular metabolites to pass through. The reactor is divided into two chambers (each 500 mL by volume and containing 100 mL of BSM) by track-etched polyethylene terephthalate (PET) nucleopore membranes (with a pore size distribution at 0.22 μm), which were purchased from Wuwei Kejin Xinfa Technology Co., Ltd. (Gansu, China). The test experiments of the reactor performance are detailed in Supplementary Methods. Strains ZM07 and K12 were inoculated separately into each side of the reactor at a ratio of 1:1 (the initial inoculum size of each strain was OD600 = 0.06). Samples were collected every 24 h for analysis of the biomass (OD600) and determination of the residual THF concentrations.
(2) Coculture of ZM07 and K12: Strains were cultivated in LB, harvested by centrifugation (7,000 × g, 10 min), and washed with BSM three times. The resulting cells were resuspended with BSM (OD600 = 3); subsequently, 100 mL of BSM with 20 mM THF was inoculated with 1 mL of strain ZM07 and 1 mL of strain K12 to generate a cell ratio of 1:1 for coculture as the first transfer. The corresponding monoculture systems were incubated with the same inoculum of ZM07 and 1 mL of BSM. Subsequently, the coculture and monoculture were transferred (OD600 = 6 and 1 mL of cell suspension) in the same way for the second and third transfers. Samples were collected every 24 h for analysis of the biomass (OD600) and determination of the residual THF concentrations.
(3) Coculture of K12 and K12ΔthiE: To test the survivability of K12ΔthiE in the thiamine-limited and thiamine-rich media used in this study, we cocultured K12 wild type and K12ΔthiE in BSM supplemented with 20 mM succinate as the sole carbon source. K12 and K12ΔthiE were inoculated with a cell ratio of 1:1 either supplemented with (0.01 mM) thiamine or without thiamine. We used the method described above to passage coculture strains for 8 transfers every 12 h, and the initial cell densities and cell ratios of K12 and K12ΔthiE are shown in Supplementary Table S1. Samples were collected at the end of cultivation of each transfer for analysis of biomass (OD600) and relative abundance.
(4) Coculture of ZM07, K12, and K12ΔthiE: To assess the synergetic and competitive relationships in the three-strain (ZM07, K12, and K12ΔthiE) and two-strain (ZM07 and K12, ZM07 and K12ΔthiE) communities during THF degradation with or without extra thiamine, we conducted experiments on three different combinations of the three strains (ZM07 and K12, ZM07 and K12ΔthiE, ZM07 and K12 and K12ΔthiE) with sufficient (0.01 mM) thiamine or without thiamine. The initial inoculum size and strain ratio of the three strains in every experiment are shown in Supplementary Table S1. Each combination with thiamine was transferred to fresh medium every 3 days, and 2 mL samples were collected at 48 h for analysis of the biomass (OD600) and determination of the residual THF concentrations and relative strain abundances. Analogously, the cultures without additional thiamine were transferred every 4 days, and samples were collected at 72 h for analysis of the biomass (OD600) and determination of the residual THF concentrations and relative strain abundances.
The relative abundances of ZM07, K12, and K12ΔthiE were determined by quantitative PCR (qPCR). Genomic DNA of the bacterial consortium was isolated with an E.Z.N.A® Bacterial DNA Kit (Omega Bio-Tek) according to the manufacturer’s instructions. Primers for the qPCR assay are shown in Supplementary Table S2. The qPCR reactions were carried out in a volume of 20 μL containing 10 μL of TB GreenTM Premix Ex TaqTM (TaKaRa, Dalian, China), 0.4 μL each of forward and reverse gene-specific primers (10 μM), 0.4 μL of DNA (50–100 ng/μL) and 8.8 μL of ddH2O. The following conditions were used for PCR: 95°C for 2 min, followed by 40 cycles of 10 s at 95°C, 20 s at 58°C and 20 s at 72°C. Three independent DNA samples were assayed, and the 2–ΔΔCT method was used to calculate the gene abundance level of the strains (Livak and Schmittgen, 2001).
The metabolic pathways of strains ZM07 and K12 were analyzed using the method described in previous research (Li et al., 2018). The whole-genome sequencing results of strains ZM07 and K12 were annotated by the Kyoto Encyclopedia of Genes and Genomes (KEGG1). The draft genome sequence data of strain ZM07 have been included in the National Center for Biotechnology Information (NCBI), accession number JACVXT000000000, and strain K12 genome sequence data can also be found in NCBI (GenBank accession number NC_000913.3). We then compared the results to find differences and annotated them in KEGG Mapper with different colors (pathways that are unique in ZM07 and K12 are colored green and blue, respectively, while pathways harbored by both strains are colored brown). We searched for the anabolic pathways that were completely present only in the K12 strain.
Experiments were conducted to verify that strain ZM07 is a thiamine auxotroph. Different amounts of thiamine were added to the third transfer of ZM07 (the strain could barely grow in BSM with 40 mM THF as the sole carbon source after continuous passage culture). Furthermore, strain ZM07 was cocultured with K12 and K12ΔthiE with 40 mM THF, respectively, for two transfers. Samples were collected 72 h after each transfer for analysis of the biomass (OD600) and determination of the residual THF concentrations.
To verify whether trace wild-type K12 can form a stable interaction system with ZM07, we added 0.5% (OD600 = 0.0003) K12 wild type to ZM07 culture in the first transfer and passaged the culture for 8 transfers. In addition, we added 0.5% of the K12 wild type to the third transfer of the ZM07 culture, which was cultivated for 4 days alone with almost no growth. In other tests, 0.5% K12ΔthiE was added to the ZM07 and K12 coculture in the first and third transfers and passaged for 8 transfers to determine how trace K12ΔthiE behaves when invading the ZM07 and K12 system. The initial cell densities and cell ratios of the three strains in every experiment are shown in Supplementary Table S1. Each combination was transferred to fresh medium every 4 days, and samples were collected at 72 h for analysis of the biomass (OD600) and determination of the residual THF concentrations and relative strain abundances.
Strain ZM07 in the first transfer was cultured in 100 mL of BSM supplemented with 20 mM THF for 1, 2, 3, and 4 days. Then, the supernatant of ZM07, which was obtained using a 0.22 μm vacuum bottle filter (BIOFIL, China), was used to cultivate strain K12 (the initial inoculum size of K12 was OD600 = 0.06) for 3 days. Furthermore, THF, 2-OH THF, γ-butyrolactone and succinate (three intermediate metabolites of THF) were chosen to test the ability of strain K12 to use these compounds (5 mM each). Strain K12 was cultured in 100 mL of BSM at 30°C and 160 rpm, with samples collected after 12, 24, and 72 h to measure the OD600. Furthermore, the effects of THF, 2-OH THF and γ-butyrolactone on the growth and morphology of K12 and K12ΔthiE were also studied as described in Supplementary Methods.
The cultures were centrifuged at 10,000 × g for 10 min, and 500 μL of the supernatant was subjected to THF concentration determination using gas chromatograph (GC-2014C; SHIMADZU, Japan). The temperatures of the injector, oven, and detector were set to 250, 160, and 250°C, respectively. The degradation ratio was calculated as described in our previous study (Huang et al., 2019). The THF intermediate metabolites produced by strain ZM07 were detected as described in Supplementary Methods. The biomass was monitored by recording the OD600 using a UV spectrometer (UV-3100PC; MAPADA; Shanghai). Succinate detection is shown below: Samples of extracellular supernatants and intracellular extracts were derivatized using pyridine and N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) and analyzed by gas chromatography-mass spectrometry (GC-MS). Each experiment was performed in triplicate. P-values for all assays were determined using a two-tailed Student’s t-test.
The THF-degrading strain Rhodococcus ruber ZM07 was isolated from an enrichment culture H-1 (Huang et al., 2019), and cannot be subcultured serially in BSM with THF as the sole carbon source. Before carrying out experiments, monoculture growths of strains ZM07, K12, and K12ΔthiE in BSM (not supplemented with THF or any other carbon sources) with or without thiamine were tested. The results showed that these three strains cannot grow in BSM without THF or any other carbon sources (Supplementary Figure S1). Monoculture experiments showed that ZM07 degraded 20 mM THF completely within 3 days in the first transfer, while its THF-degrading ability was inhibited when its growth rate slowed in the second transfer; furthermore, its growth was inhibited when using THF in the third transfer (Figure 1A). We speculated that intracellular thiamine, which may still remain during the first transfer, supported the growth of strain ZM07 in the starting phase. In contrast, when cocultured with strain K12, ZM07 was able to proliferate well and continually degrade THF during passaging (Figure 1B).
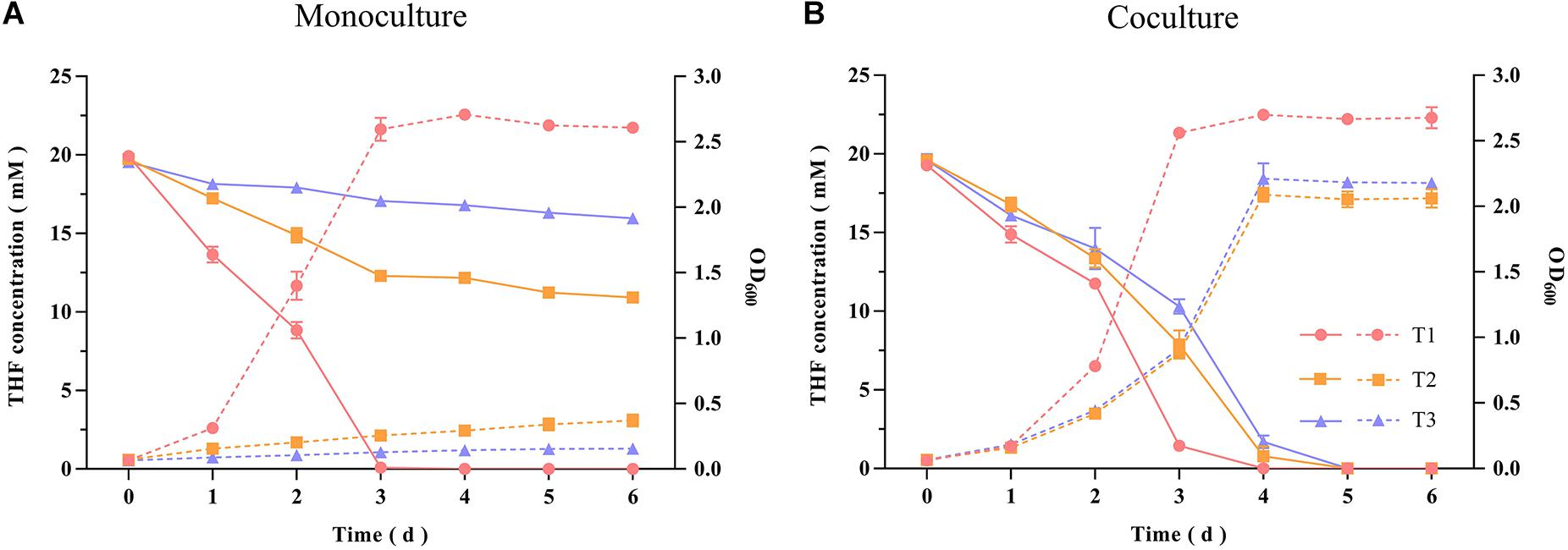
Figure 1. THF degradation and growth curves of strain ZM07 monocultured (A) and cocultured with strain K12 (B) in the first, second and third transfers. The solid lines represent THF concentration curves; the dotted lines represent growth curves (OD600); the circles represent the first transfer; the squares represent the second transfer; and the triangles represent the third transfer. Error bars: SD from three independent replicates, which may be smaller than the marker.
Accordingly, we suspected that the THF-degrading bacterium ZM07 may not be able to produce some growth factors that are critical to its growth. To verify this hypothesis, we plotted a comparative metabolic pathway network for ZM07 and K12 with the pathways harbored by the different strains, which are colored differently (Supplementary Figure S2). The map of thiamine metabolism supports our hypothesis since the thiamine synthesis pathway is incomplete in strain ZM07 but intact in strain K12 (Figure 2). As an essential growth factor, thiamine pyrophosphate is produced through the condensation reaction of 4-amino-5-hydroxymethyl pyrimidine pyrophosphate and 4-methyl-5-(beta-hydroxyethyl)-thiazole monophosphate (Begley et al., 1999). As shown in Figure 2, strain ZM07 lacks the genes thiF, thiH, and thiI (highlighted by the purple boxes), which are required for 4-methyl-5-(beta-hydroxyethyl)-thiazole synthesis (Webb et al., 1997; Dorrestein et al., 2004; Martinez-Gomez et al., 2004).
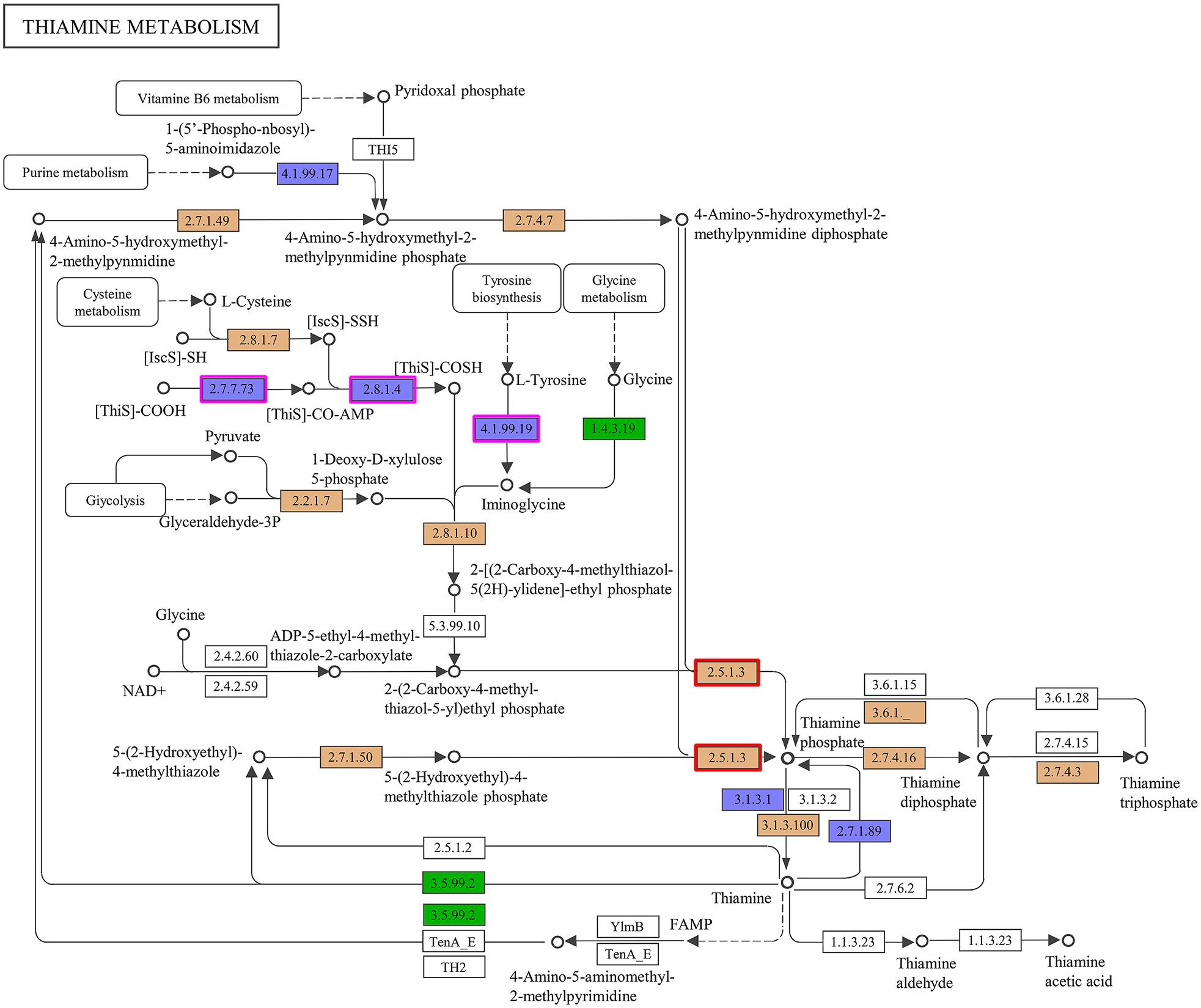
Figure 2. Comparison of the KEGG metabolic pathways reconstructed from the complete genomes of ZM07 and K12. Here, thiamine metabolism (map00730) is shown. The ZM07-specific pathways are colored in green; the K12-specific pathways are colored in blue; shared pathways are colored in brown; and pathways present in neither ZM07 nor K12 are colored in white. The genes for thiF (No. 2.7.7.73), thiH (No. 4.1.99.19), and thiI (No. 2.8.1.4) are highlighted by purple boxes, and the gene for thiE (No. 2.5.1.3) is highlighted by red boxes.
To further verify our hypothesis, we performed two experiments. First, we cocultured the wild-type strain K12 and the thiE (highlighted by the red boxes in Figure 2)-deficient strain K12ΔthiE separately with strain ZM07. The thiE gene encodes thiamine phosphate synthase (ThiE), which is responsible for linking the thiazole and pyrimidine moieties of thiamine monophosphate (TMP) generated in respective metabolic pathways (Chiu et al., 1999; Jurgenson et al., 2009). The results showed that strain ZM07 could grow and degrade THF normally when cocultured and transferred with K12 wild type (Supplementary Figure S3D), while it could not restore its growth and THF degradation ability when cocultured with K12ΔthiE (which is very obvious after the second transfer) (Supplementary Figure S3E). The coculture of ZM07 and K12ΔthiE (basically no growth) quickly became cloudy with additional thiamine on the 1st day, with a final OD600 of 2.541 ± 0.056 (data not shown), while the growth of coculture without additional thiamine was very slow (Supplementary Figure S4A). Further experiments were performed to confirm that THF-degrading bacterium ZM07 is indeed a thiamine auxotroph strain by adding thiamine directly to the culture medium. The results showed that when supplied with more than 10–5 mM thiamine, strain ZM07 grew well and degraded THF normally (Supplementary Figure S5). Nevertheless, the growth and THF-degrading ability of ZM07 were still inhibited when thiamine concentrations were below 10–5 mM (Supplementary Figure S5). The results indicated that THF-degrading bacterium ZM07 cannot grow alone with THF as the sole carbon source since it is a thiamine auxotroph strain.
To determine the interaction mode between strains ZM07 and K12, the metabolic intermediates of THF were detected and identified at different cultivation time of strain ZM07 when utilizing 20 mM THF as substrates firstly. The results showed that no metabolites were detected in extracellular supernatants (Supplementary Figure S6A), while one peak that appeared at retention times of 6.816 min were observed in intracellular extracts (Supplementary Figure S6B). According to the retention times of standards, the sources of the peak was identified as γ-butyrolactone. Furthermore, succinate was identified in both of the samples of extracellular supernatants and intracellular extracts (Supplementary Figures S6D,E). To investigate whether K12 could utilize the intermediate metabolites of THF degraded by ZM07 in the cocultured system, we used THF and its intermediate metabolites (2-OH THF, γ-butyrolactone and succinate) as carbon sources to cultivate strain K12. The results showed that K12 could utilize only succinate to survive, with the highest OD600 value reaching 0.305 ± 0.002, while it could not utilize THF, 2-OH THF, or γ-butyrolactone as sole carbon sources (Supplementary Table S3). The effects of several different concentrations (0, 1.25, 2.5, and 5 mM) of THF, 2-OH THF and γ-butyrolactone on growth of K12 and its mutant were tested. The results showed that different THF, 2-OH THF, and γ-butyrolactone concentrations (0, 1.25, 2.5, and 5 mM) did not significantly influence the biomass of K12 or K12ΔthiE (Supplementary Figure S7). Otherwise, the transmission electron microscopy (TEM) images of two strains morphology after exposuring the cell to these three compounds indicated that there are no obvious changes of both two strains exposing to 5 mM THF, 2-OH THF, and γ-butyrolactone (Supplementary Figure S8).
In addition, we used culture supernatants of ZM07 (cultured for 1, 2, 3, and 4 days) to culture strain K12 (Supplementary Table S4), and it showed no growth probably because THF intermediate metabolites in the supernatants of strain ZM07, which could be used by strain K12, are easily accessible carbon sources and cannot be accumulated, hence the instantaneous concentration is too low to support the growth of K12. Finally, two-phase reactors were used to explore whether physical contact was needed to mediate the interaction between these two species. The test experiments of the reactor performance indicated that the membrane can separate cells and allow only extracellular metabolite exchange (Supplementary Figures S9, S10). The results showed that the separately cultured strain K12 could help ZM07 grow and degrade THF (Figures 3A,B). A comparison with the contacting coculture of ZM07 and K12 showed that THF was degraded even faster in the group of the non-contacting coculture (Figure 3B), which indicated that there might be some cooperation and competition mechanisms in these two strains. The highest OD600 value of K12 reached 0.313 ± 0.026 (Figure 3C), indicating that physical contact is not a prerequisite for the interaction between strains ZM07 and K12. Separately cultured strain K12 could grow well when continuous supplying with THF intermediate metabolites as carbon sources from ZM07 through membrane, while it was not growing with discontinuous supplies cultured in the supernatants of strain ZM07. Thus, the exchange of extracellular metabolites is one way these two strains interact.
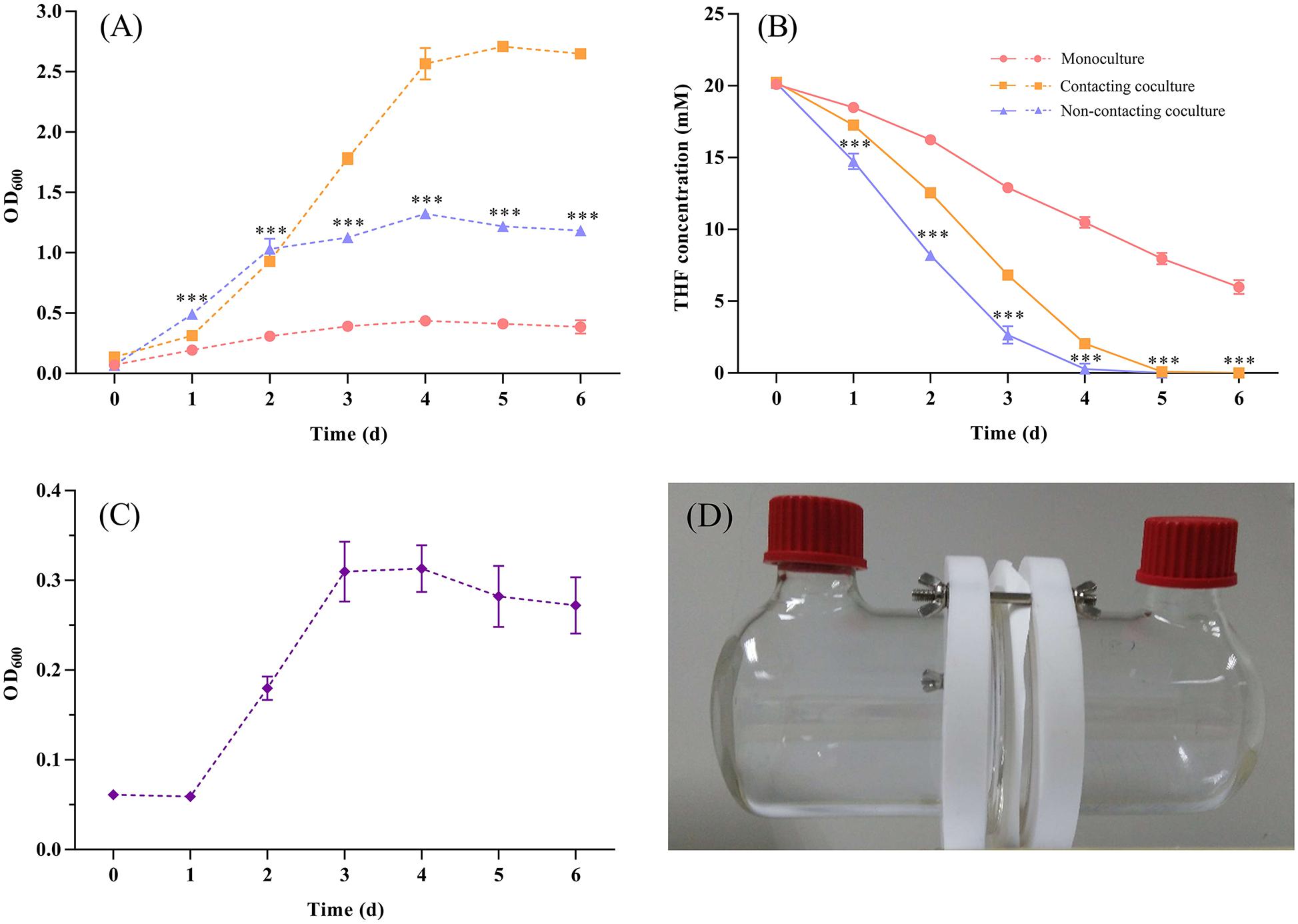
Figure 3. (A) Growth curves of strain ZM07 monocultured (pink line) and cocultured with K12 under the non-contact condition (blue line) in two-phase reactors and the growth curve of the two strains cocultured under the contact condition (yellow line). (B) THF degradation curves of strain ZM07 in the three cases described above. (C) Growth curve of strain K12 cocultured with ZM07 under the non-contact condition in a two-phase reactor. (D) Two-phase reactor used to separate the cultured strains in this study. The P-value indicates a significant difference between monocultured strain ZM07 and strain ZM07 cocultured with K12 in two-phase reactors and was determined using Student’s t-test (n = 3, ∗∗∗p < 0.001). Error bars: SD from three independent replicates, which may be smaller than the marker.
Succinate, one of the intermediate metabolites of THF, was used as a carbon source to explore the intraspecific competition between wild-type K12 and its knockout strain. The results showed that under thiamine-limited conditions, strain K12 wild type outcompeted strain K12ΔthiE. The proportion of K12ΔthiE was drastically reduced during passage and disappeared almost entirely by the fourth transfer (Figure 4A), while its proportion remained stable in the coculture system under thiamine-rich conditions. Moreover, the composition of K12ΔthiE and K12 remained stable (at a ratio of approximately 6:4) during passage with sufficient thiamine (Figure 4B). However, there was no difference in the biomass between K12 and K12ΔthiE with or without thiamine in the first transfer (Supplementary Figures S11A,B). Furthermore, strain K12ΔthiE cannot be continuously passaged in BSM without thiamine, and the final biomass of 1 day of cultivation between K12 and the coculture of K12 and K12ΔthiE showed no difference (Supplementary Figure S11C). In conclusion, K12ΔthiE has obvious disadvantages in thiamine-limited medium. However, additional thiamine can significantly increase the competitiveness of K12ΔthiE against its wild-type relative.
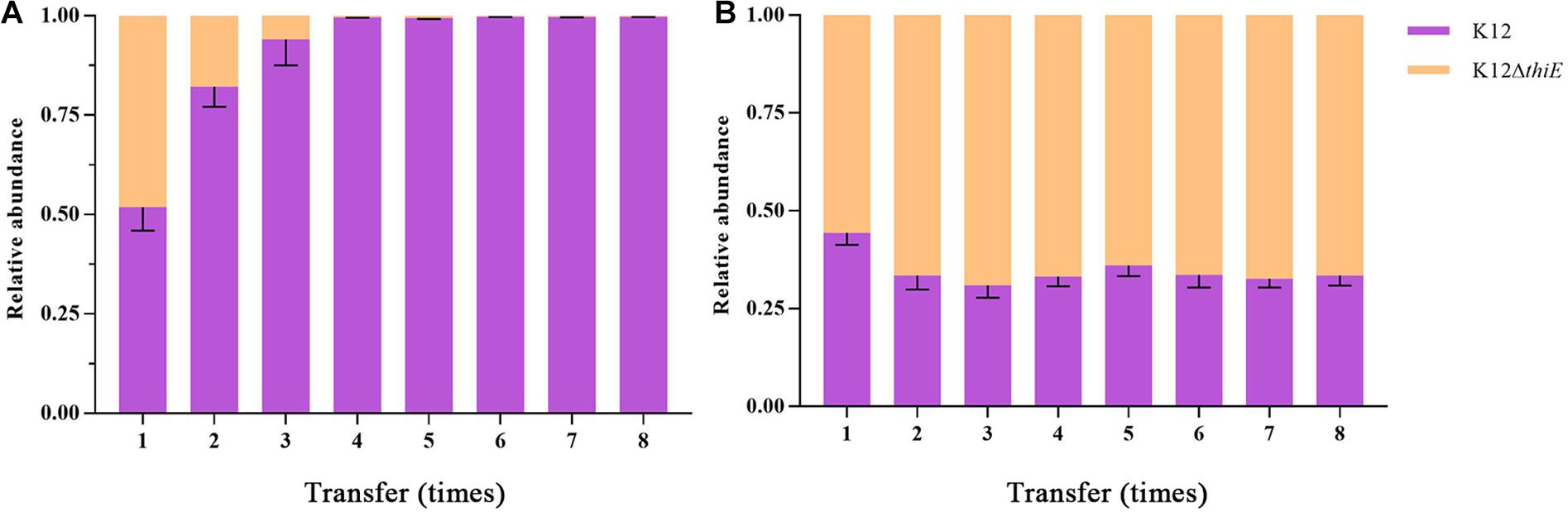
Figure 4. Relative abundances of K12 and K12ΔthiE cocultured under 20 mM succinate without (A) or with (B) 0.01 mM thiamine across different transfers. Error bars: SD from three independent replicates.
In thiamine-limited medium, experiments revealed that the relative abundance of K12 remained at approximately 30% in both the two-strain system (ZM07 and K12) and the three-strain system (ZM07, K12, and K12ΔthiE) during passage without additional thiamine (Figures 5A,B). In the invasion experiments, a trace amount (the initial inoculum size of the strain was OD600 = 0.0003, and same as below) of K12 was added to the ZM07 culture in the 1st transfer, and the results showed that the relative abundances of ZM07 and K12 finally became similar to those of the coculture of ZM07 and K12 without additional thiamine (Figure 5A) during passage (Figure 5C). In addition, the addition of trace amounts of K12 to the third transfer of the ZM07 culture that was cultivated for 4 days alone with almost no growth could easily restore the growth and THF-degrading ability of ZM07 (Supplementary Figure S4B). These results indicated that strain K12 could help auxotrophic strain ZM07 rapidly regain its growth and THF degradation abilities. Then, the results for the coculture of ZM07, K12, and K12ΔthiE showed that K12ΔthiE could not continuously exist in the symbiotic system composed of ZM07 and K12 in thiamine-limited medium and instead disappeared after the 8th transfer (Figure 5B). In the invasion experiments, when trace amounts of K12ΔthiE were added to the ZM07 and K12 system in the 1st transfer, K12ΔthiE disappeared by the 8th transfer during passage (Figure 5D). Additionally, to eliminate the influence of intracellular thiamine, which may still remain during the first two transfers, experiments were performed in which trace amounts of K12ΔthiE were added to the ZM07 and K12 system in the 3rd transfer; however, K12ΔthiE disappeared even faster (Supplementary Figure S12). These results indicated that K12ΔthiE cannot invade the symbiotic system of strains ZM07 and K12.
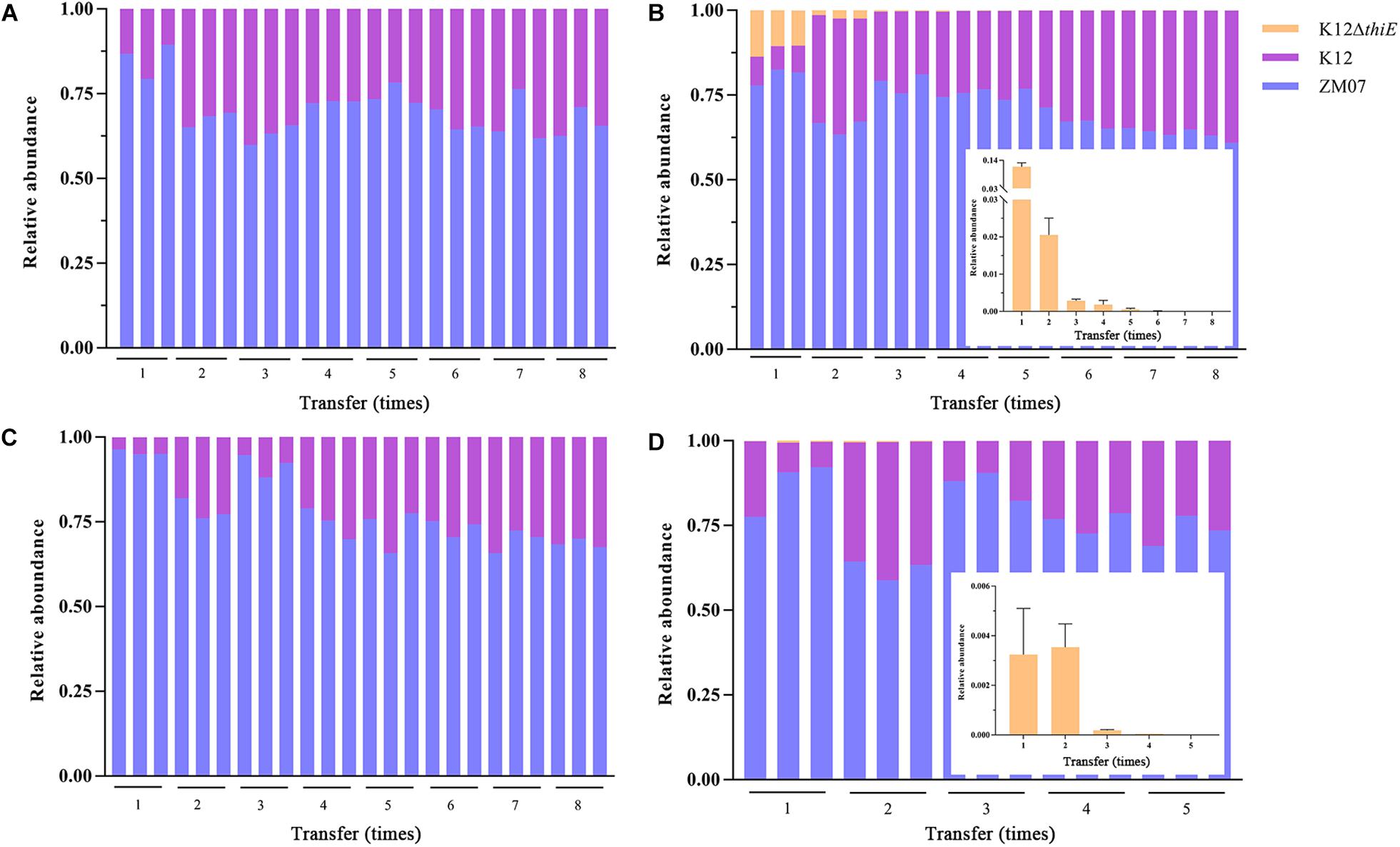
Figure 5. Relative abundances of ZM07, K12, and K12ΔthiE in a two-strain system (ZM07 and K12) (A) and in a three-strain system (B) across different transfers without thiamine. Relative abundances of three strains in a two-strain system (ZM07 and trace amount of K12) (C) and in a three-strain system (ZM07, K12 and trace amount of K12ΔthiE) (D) across different transfers. The plots within (B,D) show details of the relative abundance of K12ΔthiE in corresponding experiments. Three independent replicates were performed for each group. Error bars: SD from three independent replicates.
In thiamine-rich medium, the results showed that additional thiamine drastically increased the advantage of ZM07 and decreased the proportion of K12 in the coculture of ZM07 and K12 (Figure 6A), which indicated that strain K12 was significantly inhibited by additional thiamine. Moreover, despite occupying only a small proportion of the cocultured systems, strain K12ΔthiE existed throughout the whole transfer process in the two-strain system of ZM07 and K12ΔthiE (Figure 6B) or in the three-strain system of ZM07, K12, and K12ΔthiE (Figure 6C) with additional thiamine, which differed from its fate in thiamine-limited systems (Figure 5B). Furthermore, the proportion of K12 in the cocultured system of ZM07 and K12 showed no significant differences from the proportion of K12ΔthiE in the cocultured system of ZM07 and K12ΔthiE in thiamine-rich medium (Figure 6D); however, K12ΔthiE grew better than K12 in the three-strain system with additional thiamine (Figure 6E), which was similar to the results of the intraspecific competition experiment of K12 and K12ΔthiE (Figure 4B). The results indicated that additional thiamine could increase the competitiveness of K12ΔthiE against its wild-type relative. Additionally, the THF degradation ratio and the total biomass of the systems showed no significant difference among different groups (monoculture of ZM07; two-strain coculture of ZM07 and K12, or ZM07 and K12ΔthiE; and three-strain coculture of ZM07, K12, and K12ΔthiE) in thiamine-rich medium (Supplementary Figure S13), and all combinations of these three strains mentioned above were able to completely degrade 20 mM THF within 4 days. We can conclude that exogenous thiamine might eliminate the effect of K12 and K12ΔthiE on the THF degradation ability of strain ZM07 in the cocultured systems.
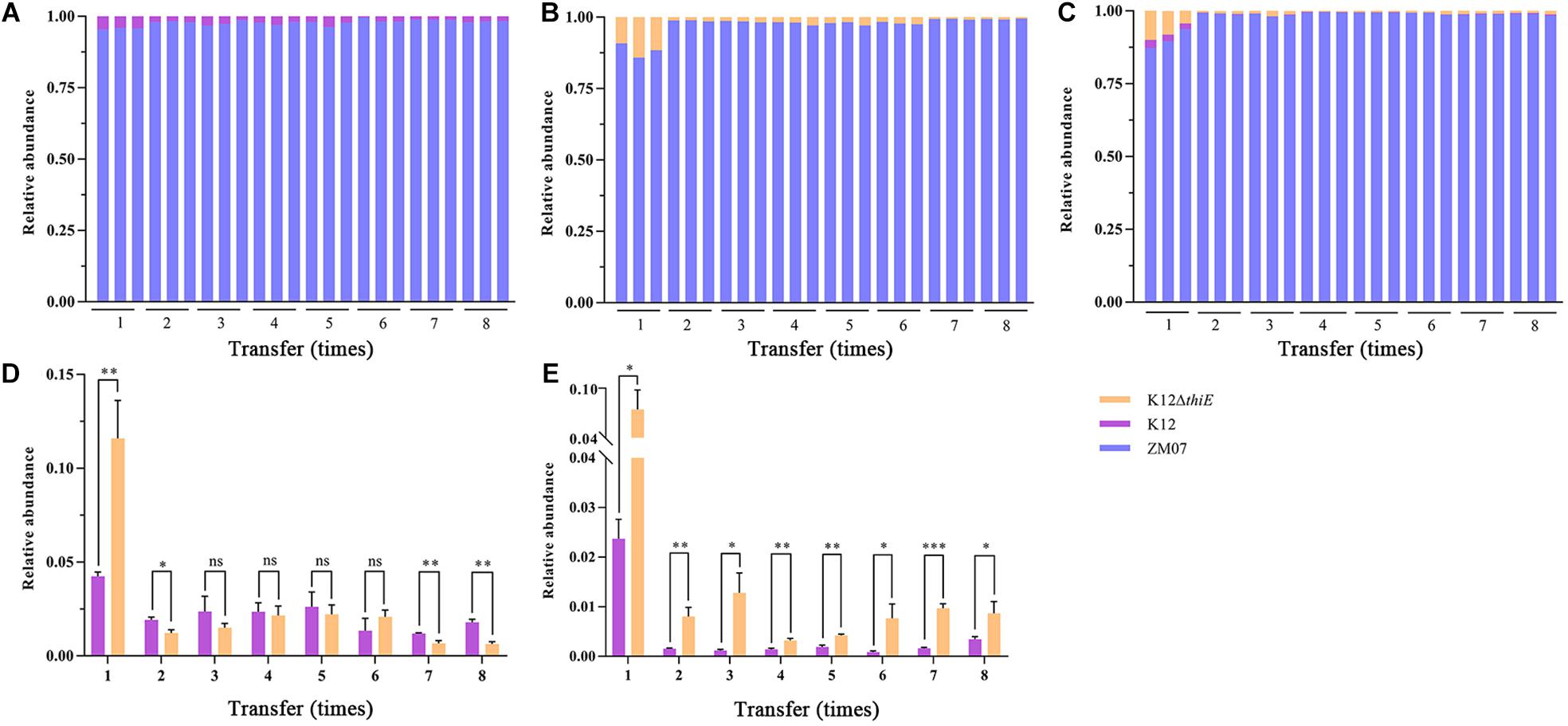
Figure 6. Relative abundances of ZM07, K12, and K12ΔthiE in two-strain systems (ZM07 and K12) (A), (ZM07 and K12ΔthiE) (B) and a three-strain system (C) in different transfers with 0.01 mM thiamine. (D) Relative abundances of K12 and K12ΔthiE in the above experiments in two-strain systems (the relative abundance of strain K12 in a cocultured system of ZM07 and K12 in (A) and the relative abundance of strain K12ΔthiE in a cocultured system of ZM07 and K12ΔthiE in panel (B) in different transfers with 0.01 mM thiamine. (E) Relative abundances of K12 and K12ΔthiE in the above experiments in the three-strain system (ZM07, K12, and K12ΔthiE in (C) in different transfers with 0.01 mM thiamine. Three independent replicates were performed for each group. Error bars: SD from three independent replicates. The P value indicates statistical significance between K12 and K12ΔthiE determined using Student’s t-test (n = 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Auxotrophic strains rely on external nutrients for growth, and they are also very likely to benefit their cooperators, which facilitates the establishment of a stable interaction system between them. Rhodococci have a wide range of degradation spectra (Martínková et al., 2009), compatibility with foreign genes (Larkin et al., 2006), and effective degradability; therefore, they have enormous application potential for the treatment of environmental pollutants. Thiamine auxotrophic microorganisms are ubiquitous in the Rhodococcus genus (Hummel et al., 1987; Denome et al., 1994). The Rhodococcus ruber strain ZM07 used in this study is a natural thiamine auxotrophic THF-degrading strain. Compared with other strains in the enrichment culture H-1, strain ZM07 seems to have greater survival advantages in nutrient-poor environments with high THF concentrations (Huang et al., 2019). Therefore, during the long-term evolution process, why have many species of Rhodococcus genus lost thiamine synthesis ability, even though it is extremely important for their survival and they cannot live independently without a supply of thiamine provided by other microbes? The “Black Queen Hypothesis” and streamlining theory might provide a reference for this topic; according to these theories, genome reduction might result in higher fitness and optimized resource allocation (Morris et al., 2012; Wolf and Koonin, 2013; Batut et al., 2014; Giovannoni et al., 2014; Otero-Bravo et al., 2018; Zengler and Zaramela, 2018). Prochlorococcus mutants gradually lost the ability to reduce HOOH in water; however, they can still enjoy the benefits of decomposing H2O2 because other microorganisms in the community still have this function (Morris et al., 2012). Accordingly, we conjecture that “landlords,” such as the strain ZM07 and Prochlorococcus mutants, may play vital roles in the microbial community. Their lost functions can be compensated for by “leakage” from synthesis-capable organisms since thiamine, a public good, is a readily available substance in the environment. In this manner, the auxotrophic functional bacterium ZM07 may be able to degrade THF more effectively and easily by forming a stable symbiotic system with non-THF-degrading bacteria in nature.
According to previous studies, cooperation through public good is ubiquitous among microbial members in nature (Crespi, 2001; West et al., 2006). In this study, we proposed a contact-independent interaction mode between ZM07 and K12. Based on the results of two-phase reactors, these two strains’ cooperation relies on extracellular metabolic interactions, which do not require physical contact between strains (Figure 3). During THF degradation, cooperator K12 provides thiamine as a public good to ZM07, which is critical to growth and could pass through the membrane without difficulty (Supplementary Figure S10). Through the results of THF intermediate metabolites detection and using abilities of K12, THF-degrading bacterium ZM07 most likely provided available metabolites of THF that are easily used, such as succinate for K12 (Supplementary Table S3 and Supplementary Figure S6). In addition, the highest OD600 value of ZM07 cultured separately with K12 in two-phase reactors reached only 1.323 ± 0.022 (Figure 3A), while it could reach up to 2.708 ± 0.012 with normal growth in the first transfer (Figure 1A). The low public goods exchange rate between the two strains might represent an important reason for its poor growth. Alternatively, there might be some other contact-dependent interaction mechanisms between these two strains that need to be further studied. However, cooperators (who produce public goods) would generate a metabolic cost, while all the microorganisms in the community benefit from it, regardless of whether they are producers (Brockhurst et al., 2010; Popat et al., 2012). Hence, such cooperative systems can easily be invaded by cheaters. The invasion of cheaters usually causes low proportions of cooperators who contribute public goods to maintain growth stability, consequently damaging the interests of the cooperative system (Dunny et al., 2008). In the cocultured system of ZM07, K12, and K12ΔthiE in thiamine-limited medium, the non-cooperator K12ΔthiE did not cause the entire community to crash; instead, it gradually disappeared during the passages (Figure 5B), indicating that the symbiotic system of strains ZM07 and K12 was stable and K12ΔthiE could not invade it. The reasons for the success of the cooperative system that resists the non-cooperator might be as follows. First, intraspecific competition experiments between the K12 wild type and defective strain showed that under thiamine-limited conditions, K12ΔthiE faced fierce intraspecific competition and was inhibited during coculture with K12 (Figure 4). Intraspecific competition experiments indicate that one of the reasons for the failure of K12ΔthiE is the insufficient thiamine secreted by K12 to the culture medium. A previous study showed that thiamine tends to be stored intracellularly rather than secreted extracellularly (Schyns et al., 2005), which also supports our conjectures. Furthermore, the results showed that the growth of strain ZM07 was inhibited by the shortage of thiamine in the community (Supplementary Figure S5) and that the degrading bacterial strain ZM07 cannot keep the entire community functioning with such low activity in the thiamine-limited medium. At the beginning of inoculation, strain ZM07 lacked thiamine, strain K12 lacked a carbon source, and both strains depended on each other to grow; hence, it was impossible to eliminate either of the two strains through rapid proliferation, which eventually might result in a stable symbiotic system. Experiments using trace amounts of K12 to rapidly restore the growth and THF-degrading ability of ZM07 (Figure 5C) also demonstrated the stability of the community of ZM07 and K12. In the cocultured system, strain ZM07 might have stronger thiamine utilization ability than K12ΔthiE, and since it can degrade THF, strain ZM07 could gain some preferential access to the broken-down products. These two reasons might explain why strain ZM07 shows a faster growth rate than K12ΔthiE, which might be an important factor in resisting non-cooperator invasion. In the fierce competition for dual nutrients (carbon source and thiamine), the non-cooperator could not find a sufficient niche in the coculture system without additional thiamine. Moreover, the dual pressures of carbon deficiency and thiamine deficiency might have accelerated K12ΔthiE extinction.
In this study, the addition of thiamine weakened the advantage of K12, whether it was cocultured with ZM07 alone or cocultured with both ZM07 and K12ΔthiE, and the relative abundance of K12 was decreased to a very low level in the cocultured systems (Figures 6A,C). Otherwise, no difference occurred in the proportions of K12 and K12ΔthiE when they were cocultured separately with ZM07 in thiamine-rich medium (Figure 6D), indicating that adding thiamine eliminates the advantage of the K12 wild type, which can only attain its position when needed by ZM07 in a cocultured system. Why could the cooperative system recognize the role of K12 and inhibit it under thiamine-rich conditions? We speculate that the THF-degrading bacterium ZM07 might reduce the secretion of intermediate metabolites of THF when thiamine is easily available. Conversely, strain ZM07 has to maintain its THF-degrading ability through a metabolite cross-feeding interaction with K12 under thiamine-limited conditions. Furthermore, K12ΔthiE grew better than K12 in the three-strain system with additional thiamine (Figure 6E), indicating that there is a metabolic cost for thiamine synthesis as discussed above. In the three-strain communities in this study, the non-cooperator K12ΔthiE disappeared by the 8th transfer in thiamine-limited medium (Figure 5B) but remained in the medium supplemented with thiamine (Figure 6C), indicating that the additional thiamine might slow down the elimination of K12ΔthiE in cooperative communities. Accordingly, in the kingdom of the THF degradation system, cooperators may utilize thiamine as “rent” that they exchange for “food” (carbon sources) from the “landlord,” the THF-degrading bacterium. Non-cooperators who are unable to provide “rent” are likely to be gradually eliminated.
Based on the above results, we propose a model depicting the succession process of these three strains during passaging in the communities. In a cocultured system, K12 provides thiamine, an essential growth factor, as a public good to thiamine auxotrophic bacteria ZM07 and K12ΔthiE; THF-degrading bacterium ZM07 degrades THF (the sole carbon source in the cocultured system) and provides easily accessible intermediates (succinate and other downstream products) to the cooperator strain K12 and the non-cooperator K12ΔthiE as carbon sources. Eventually, the non-cooperator K12ΔthiE (which can neither degrade THF nor produce thiamine) disappears during passaging, indicating that non-cooperator strain K12ΔthiE cannot invade the symbiotic ZM07 and K12 system (Figure 7). In thiamine-rich medium, both K12 and K12ΔthiE represented only a small proportion of the coculture system, while ZM07 became dominant (Figure 7). Based on the observed interactions between strains ZM07 and K12, “rent” must be paid to maintain a place in the symbiotic system of THF degradation; non-cooperators who cannot pay the “rent” might be excluded by the cooperative system. However, thiamine auxotrophic THF-degrading bacterium ZM07 might not need cooperators when surrounded by sufficient thiamine. Taken together, we show that cooperation and competition mechanisms exist in THF multispecies ecosystems to maintain the stability of communities. Auxotrophic degrading bacteria and their cooperators can play an important role in species coexistence in microbial communities.
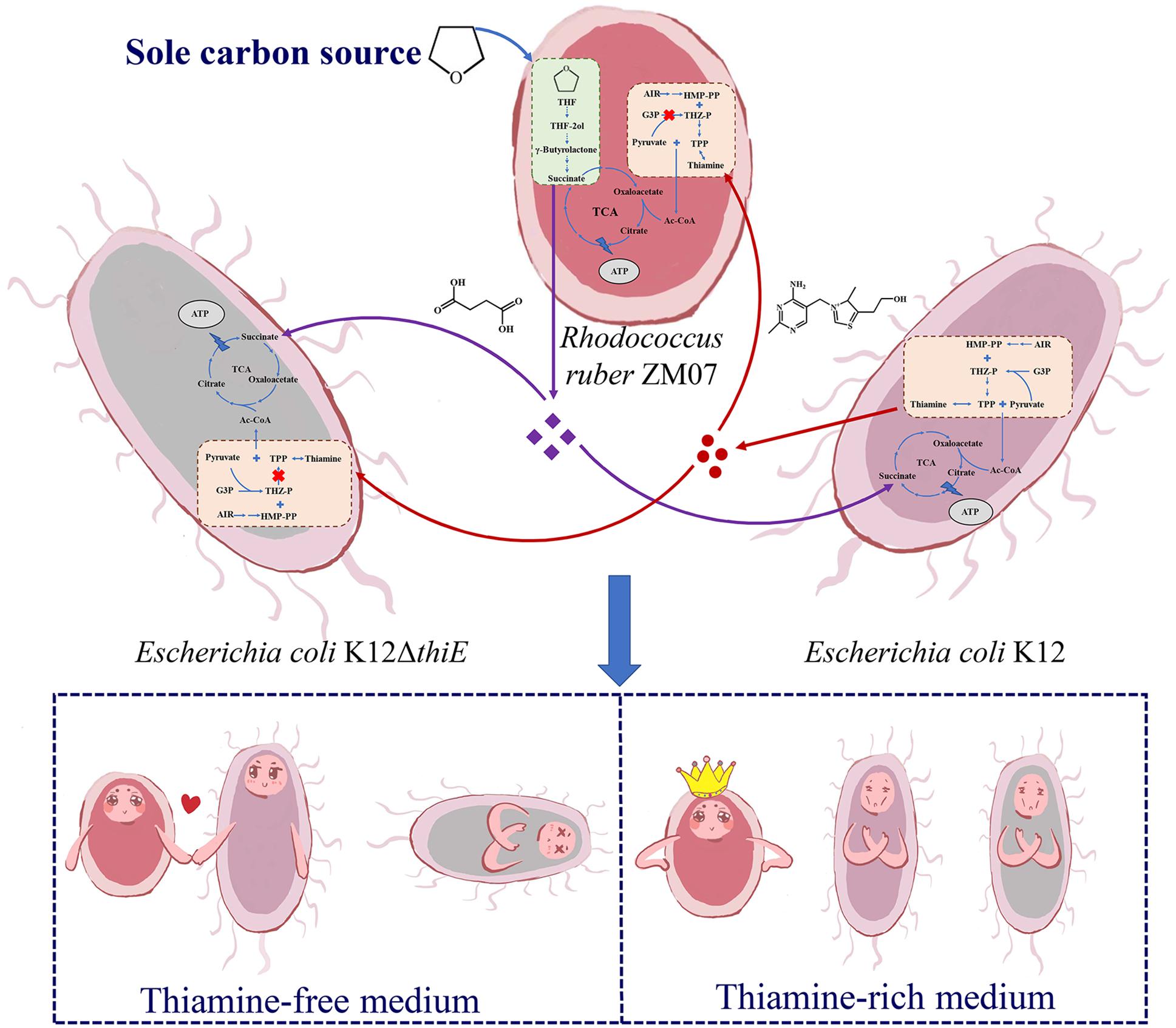
Figure 7. Proposed schematic depiction of the interaction mechanism among strains ZM07, K12, and K12ΔthiE during THF degradation. Strain K12 provides thiamine (red circles) to ZM07 and K12ΔthiE, and strain ZM07 might provide succinate or other downstream products (purple diamonds) to K12 and K12ΔthiE as a carbon source by degrading THF. In thiamine-free medium, ZM07 and K12 formed a stable symbiotic system, and K12ΔthiE disappeared during passaging; in thiamine-rich media, both K12 and K12ΔthiE represented only a small proportion of the coculture system.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
ZL and HH conceived and designed the experiments. HH and YL performed the experiments. HH, MQ, and YL analyzed the data. HH, HW, XW, and YQ wrote the manuscript. All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
This work was financially supported by the National Natural Science Foundation of China (Nos. 41630637 and 31370151).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This is a short text to acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.594052/full#supplementary-material
Amin, S., Hmelo, L., Van Tol, H., Durham, B., Carlson, L., Heal, K., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522, 98–101. doi: 10.1038/nature14488
Bachmann, B. J., Low, K. B., and Taylor, A. L. (1976). Recalibrated linkage map of Escherichia coli K-12. Bacteriol. Rev. 40, 116–167. doi: 10.1128/mmbr.40.1.116-167.1976
Batut, B., Knibbe, C., Marais, G., and Daubin, V. (2014). Reductive genome evolution at both ends of the bacterial population size spectrum. Nat. Rev. Microbiol. 12, 841–850. doi: 10.1038/nrmicro3331
Begley, T. P., Downs, D. M., Ealick, S. E., Mclafferty, F. W., Van Loon, A. P., Taylor, S., et al. (1999). Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171, 293–300. doi: 10.1007/s002030050713
Bernhardt, D., and Diekmann, H. (1991). Degradation of dioxane, tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Appl. Microbiol. Biotechnol. 36, 120–123. doi: 10.1007/BF00164711
Brockhurst, M. A., Habets, M. G., Libberton, B., Buckling, A., and Gardner, A. (2010). Ecological drivers of the evolution of public-goods cooperation in bacteria. Ecology 91, 334–340. doi: 10.1890/09-0293.1
Brown, S. P., and Johnstone, R. A. (2001). Cooperation in the dark: signalling and collective action in quorum-sensing bacteria. Proc. R. Soc. Lond. B 268, 961–965. doi: 10.1098/rspb.2001.1609
Bruger, E. L., and Waters, C. M. (2018). Maximizing growth yield and dispersal via quorum sensing promotes cooperation in Vibrio bacteria. Appl. Environ. Microbiol. 84:e00402-18. doi: 10.1128/AEM.00402-18
Chhabra, R. S., Elwell, M. R., Chou, B. J., Miller, R. A., and Renne, R. A. (1990). Subchronic toxicity of tetrahydrofuran vapors in rats and mice. Toxicol. Sci. 14, 338–345. doi: 10.1016/0272-0590(90)90213-4
Chiu, H.-J., Reddick, J. J., Begley, T. P., and Ealick, S. E. (1999). Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 Å resolution. Biochemistry 38, 6460–6470. doi: 10.1021/bi982903z
Crespi, B. J. (2001). The evolution of social behavior in microorganisms. Trends Ecol. & Evol. 16, 178–183. doi: 10.1016/S0169-5347(01)02115-2
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A 97, 6640–6645. doi: 10.1073/pnas.120163297
Denome, S. A., Oldfield, C., Nash, L. J., and Young, K. D. (1994). Characterization of the desulfurization genes from Rhodococcus sp. strain IGTS8. J. bacteriol. 176, 6707–6716. doi: 10.1128/jb.176.21.6707-6716.1994
Dorrestein, P. C., Zhai, H., Mclafferty, F. W., and Begley, T. P. (2004). The biosynthesis of the thiazole phosphate moiety of thiamin: the sulfur transfer mediated by the sulfur carrier protein ThiS. Chem. Biol. 11, 1373–1381. doi: 10.1016/j.chembiol.2004.08.009
D’Souza, G., Waschina, S., Pande, S., Bohl, K., Kaleta, C., and Kost, C. (2014). Less is more: selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68, 2559–2570. doi: 10.1111/evo.12468
Du, J., Zhou, J., Xue, J., Song, H., and Yuan, Y. (2012). Metabolomic profiling elucidates community dynamics of the Ketogulonicigenium vulgare–Bacillus megaterium consortium. Metabolomics 8, 960–973. doi: 10.1007/s11306-011-0392-2
Dubey, G. P., and Ben-Yehuda, S. (2011). Intercellular nanotubes mediate bacterial communication. Cell 144, 590–600. doi: 10.1016/j.cell.2011.01.015
Dunny, G. M., Brickman, T. J., and Dworkin, M. (2008). Multicellular behavior in bacteria: communication, cooperation, competition and cheating. Bioessays 30, 296–298. doi: 10.1002/bies.20740
Fiegna, F., and Velicer, G. J. (2003). Competitive fates of bacterial social parasites: persistence and self–induced extinction of Myxococcus xanthus cheaters. Proc. R. Soc. Lond. B 270, 1527–1534. doi: 10.1098/rspb.2003.2387
Filkins, L. M., Graber, J. A., Olson, D. G., Dolben, E. L., Lynd, L. R., Bhuju, S., et al. (2015). Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J. Bacteriol. 197, 2252–2264. doi: 10.1128/JB.00059-15
Gamer, A. O., Jaeckh, R., Leibold, E., Kaufmann, W., Gembardt, C., Bahnemann, R., et al. (2002). Investigations on cell proliferation and enzyme induction in male rat kidney and female mouse liver caused by tetrahydrofuran. Toxicol. Sci. 70, 140–149. doi: 10.1093/toxsci/70.1.140
Giovannoni, S. J., Thrash, J. C., and Temperton, B. (2014). Implications of streamlining theory for microbial ecology. ISME J. 8, 1553–1565. doi: 10.1038/ismej.2014.60
Hardin, G. (1968). The tragedy of the commons. Science 162, 1243–1248. doi: 10.1126/science.162.3859.1243
Hermida, S. A. S., Possari, E. P. M., Souza, D. B., Campos, I. P. D. A., Gomes, O. F., Mascio, P. D., et al. (2006). 2’-Deoxyguanosine, 2’-deoxycytidine, and 2’-deoxyadenosine adducts resulting from the reaction of tetrahydrofuran with DNA bases. Chem. Res. Toxicol. 19, 927–936. doi: 10.1021/tx060033d
Huang, H., Yu, H., Qi, M., Liu, Z., Wang, H., and Lu, Z. (2019). Enrichment and characterization of a highly efficient tetrahydrofuran-degrading bacterial culture. Biodegradation 30, 467–479. doi: 10.1007/s10532-019-09888-5
Hubalek, V., Buck, M., Tan, B., Foght, J., Wendeberg, A., Berry, D., et al. (2017). Vitamin and amino acid auxotrophy in anaerobic consortia operating under methanogenic conditions. mSystems 2:e00038-17. doi: 10.1128/mSystems.00038-17
Hummel, W., Schütte, H., Schmidt, E., Wandrey, C., and Kula, M.-R. (1987). Isolation of L-phenylalanine dehydrogenase from Rhodococcus sp. M4 and its application for the production of L-phenylalanine. Appl. Microbiol. Biotechnol. 26, 409–416. doi: 10.1007/BF00253523
Jin, Z., Li, J., Ni, L., Zhang, R., Xia, A., and Jin, F. (2018). Conditional privatization of a public siderophore enables Pseudomonas aeruginosa to resist cheater invasion. Nat. Commun. 9:1383. doi: 10.1038/s41467-018-03791-y
Jurgenson, C. T., Begley, T. P., and Ealick, S. E. (2009). The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78, 569–603. doi: 10.1146/annurev.biochem.78.072407.102340
Katahira, T., Teramoto, K., and Horiguchi, S. (1982). Experimental studies on the acute toxicity of tetrahydrofuran in animals. Sangyo lgaku 24:373. doi: 10.1539/joh1959.24.373
Kohlweyer, U., Thiemer, B., Schrader, T., and Andreesen, J. R. (2000). Tetrahydrofuran degradation by a newly isolated culture of Pseudonocardia sp. strain K1. FEMS Microbiol. Lett. 186, 301–306. doi: 10.1111/j.1574-6968.2000.tb09121.x
Król, J. E., Rogers, L. M., Krone, S. M., and Top, E. M. (2010). Dual reporter system for in situ detection of plasmid transfer under aerobic and anaerobic conditions. Appl. Environ. Microbiol. 76, 4553–4556. doi: 10.1128/AEM.00226-10
Larkin, M. J., Kulakov, L. A., and Allen, C. C. (2006). Biodegradation by members of the genus Rhodococcus: biochemistry, physiology, and genetic adaptation. Adv. Appl. Microbiol. 59, 1–29. doi: 10.1016/S0065-2164(06)59001-X
Leinweber, A., Inglis, R. F., and Kümmerli, R. (2017). Cheating fosters species co-existence in well-mixed bacterial communities. ISME J. 11, 1179–1188. doi: 10.1038/ismej.2016.195
Li, Q., Lin, F., Yang, C., Wang, J., Lin, Y., Shen, M., et al. (2018). A large-scale comparative metagenomic study reveals the functional interactions in six bloom-forming microcystis-epibiont communities. Front. Microbiol. 9:746. doi: 10.3389/fmicb.2018.00746
Liu, Y.-F., Galzerani, D. D., Mbadinga, S. M., Zaramela, L. S., Gu, J.-D., Mu, B.-Z., et al. (2018). Metabolic capability and in situ activity of microorganisms in an oil reservoir. Microbiome 6:5. doi: 10.1186/s40168-017-0392-1
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lujan, A. M., Gomez, P. S., and Buckling, A. (2015). Siderophore cooperation of the bacterium Pseudomonas fluorescens in soil. Biol. Lett. 11:20140934. doi: 10.1098/rsbl.2014.0934
Martinez-Gomez, N. C., Robers, M., and Downs, D. M. (2004). Mutational analysis of ThiH, a member of the radical S-adenosylmethionine (AdoMet) protein superfamily. J. Biol. Chem. 279, 40505–40510. doi: 10.1074/jbc.M403985200
Martínková, L., Uhnáková, B., Pátek, M., Nešvera, J., and Křen, V. (2009). Biodegradation potential of the genus Rhodococcus. Environ. Int. 35, 162–177. doi: 10.1016/j.envint.2008.07.018
Masuda, H., Mcclay, K., Steffan, R. J., and Zylstra, G. J. (2012). Biodegradation of tetrahydrofuran and 1,4-dioxane by soluble diiron monooxygenase in Pseudonocardia sp. strain ENV478. J. Mol. Microbiol. Biotechnol. 22, 312–316. doi: 10.1159/000343817
Mee, M. T., Collins, J. J., Church, G. M., and Wang, H. H. (2014). Syntrophic exchange in synthetic microbial communities. Proc. Natl. Acad. Sci. U.S.A. 111:201405641. doi: 10.1073/pnas.1405641111
Morenohorn, M., Garbe, L. A., Tressl, R., and Gorisch, H. (2005). Transient accumulation of γ-butyrolactone during degradation of bis(4-chloro-n-butyl) ether by diethylether-grown Rhodococcus sp. strain DTB. Appl. Microbiol. Biotechnol. 69, 335–340. doi: 10.1007/s00253-005-1984-4
Morris, B. E. L., Henneberger, R., Huber, H., and Moissleichinger, C. (2013). Microbial syntrophy: interaction for the common good. FEMS Microbiol. Rev. 37, 384–406. doi: 10.1111/1574-6976.12019
Morris, J. J. (2015). Black Queen evolution: the role of leakiness in structuring microbial communities. Trends Genet. 31, 475–482. doi: 10.1016/j.tig.2015.05.004
Morris, J. J., Lenski, R. E., and Zinser, E. R. (2012). The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3:e00036-12. doi: 10.1128/mBio.00036-12
Oh, S., Kim, J. R., Premier, G. C., Lee, T., Kim, C., and Sloan, W. T. (2010). Sustainable wastewater treatment: how might microbial fuel cells contribute. Biotechnol. Adv. 28, 871–881. doi: 10.1016/j.biotechadv.2010.07.008
Otero-Bravo, A., Goffredi, S., and Sabree, Z. L. (2018). Cladogenesis and genomic streamlining in extracellular endosymbionts of tropical stink bugs. Genome Biol. Evol. 10, 680–693. doi: 10.1093/gbe/evy033
Pande, S., Kaftan, F., Lang, S. N., Svatos, A., Germerodt, S., and Kost, C. (2016). Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. 10, 1413–1423. doi: 10.1038/ismej.2015.212
Parales, R., Adamus, J., White, N., and May, H. (1994). Degradation of 1, 4-dioxane by an actinomycete in pure culture. Appl. Environ. Microbiol. 60, 4527–4530. doi: 10.1128/aem.60.12.4527-4530.1994
Popat, R., Crusz, S. A., Messina, M., Williams, P., West, S. A., and Diggle, S. P. (2012). Quorum-sensing and cheating in bacterial biofilms. Pro. R. Soc. Lond. B 279, 4765–4771. doi: 10.1098/rspb.2012.1976
Rakoff-Nahoum, S., Coyne, M., and Comstock, L. (2014). An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 24, 40–49. doi: 10.1016/j.cub.2013.10.077
Ren, H., Li, H., Wang, H., Huang, H., and Lu, Z. (2020). Biodegradation of tetrahydrofuran by the newly isolated filamentous fungus Pseudallescheria boydii ZM01. Microorganisms 8:1190. doi: 10.3390/microorganisms8081190
Schuster, M., Joseph Sexton, D., Diggle, S. P., and Peter Greenberg, E. (2013). Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67, 43–63. doi: 10.1146/annurev-micro-092412-155635
Schyns, G., Potot, S., Geng, Y., Barbosa, T. M., Henriques, A., and Perkins, J. B. (2005). Isolation and characterization of new thiamine-deregulated mutants of Bacillus subtilis. J. Bacteriol. 187, 8127–8136. doi: 10.1128/JB.187.23.8127-8136.2005
Sei, K., Miyagaki, K., Kakinoki, T., Fukugasako, K., Inoue, D., and Ike, M. (2013). Isolation and characterization of bacterial strains that have high ability to degrade 1,4-dioxane as a sole carbon and energy source. Biodegradation 24, 665–674. doi: 10.1007/s10532-012-9614-1
Sieuwerts, S., De Bok, F. A. M., Hugenholtz, J., and Vlieg, J. E. T. V. H. (2008). Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl. Environ. Microbiol. 74, 4997–5007. doi: 10.1128/AEM.00113-08
Skinner, K., Cuiffetti, L., and Hyman, M. R. (2009). Metabolism and cometabolism of cyclic ethers by a filamentous fungus, a Graphium sp. Appl. Environ. Microbiol. 75, 5514–5522. doi: 10.1128/AEM.00078-09
Smith, J. (2001). The social evolution of bacterial pathogenesis. Proc. R. Soc. Lond. B 268, 61–69. doi: 10.1098/rspb.2000.1330
Tajima, T., Hayashida, N., Matsumura, R., Omura, A., Nakashimada, Y., and Kato, J. (2012). Isolation and characterization of tetrahydrofuran-degrading Rhodococcus aetherivorans strain M8. Process. Biochem. 47, 1665–1669. doi: 10.1016/j.procbio.2011.08.009
Tao, K., Liu, X., Chen, X., Hu, X., Cao, L., and Yuan, X. (2017). Biodegradation of crude oil by a defined co-culture of indigenous bacterial consortium and exogenous Bacillus subtilis. Bioresour. Technol. 224, 327–332. doi: 10.1016/j.biortech.2016.10.073
Thiemer, B., Andreesen, J. R., and Schrader, T. (2003). Cloning and characterization of a gene cluster involved in tetrahydrofuran degradation in Pseudonocardia sp. strain K1. Arch. Microbiol. 179, 266–277. doi: 10.1007/s00203-003-0526-7
Van Gestel, J., Weissing, F. J., Kuipers, O. P., and Kovacs, A. T. (2014). Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 8, 2069–2079. doi: 10.1038/ismej.2014.52
Velicer, G. J. (2003). Social strife in the microbial world. Trends Microbiol. 11, 330–337. doi: 10.1016/S0966-842X(03)00152-5
Webb, E., Claas, K., and Downs, D. M. (1997). Characterization of thiI, a new gene involved in thiazole biosynthesis in Salmonella typhimurium. J. Bacteriol. 179, 4399–4402. doi: 10.1128/jb.179.13.4399-4402.1997
Webb, J. S., Givskov, M., and Kjelleberg, S. (2003). Bacterial biofilms: prokaryotic adventures in multicellularity. Curr. Opin. Microbiol. 6, 578–585. doi: 10.1016/j.mib.2003.10.014
West, S. A., Griffin, A. S., Gardner, A., and Diggle, S. P. (2006). Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607. doi: 10.1038/nrmicro1461
Wexler, A. G., and Goodman, A. L. (2017). An insider’s perspective: bacteroides as a window into the microbiome. Nat. Microbiol. 2:17026. doi: 10.1038/nmicrobiol.2017.26
Wolf, Y. I., and Koonin, E. V. (2013). Genome reduction as the dominant mode of evolution. Bioessays 35, 829–837. doi: 10.1002/bies.201300037
Yao, Y., Lv, Z., Min, H., Lv, Z., and Jiao, H. (2009). Isolation, identification and characterization of a novel Rhodococcus sp. strain in biodegradation of tetrahydrofuran and its medium optimization using sequential statistics-based experimental designs. Bioresour. Technol. 100, 2762–2769. doi: 10.1016/j.biortech.2009.01.006
Keywords: tetrahydrofuran degradation, thiamine auxotroph, cooperation, interaction mechanism, Rhodococcus ruber ZM07, Escherichia coli K12
Citation: Huang H, Qi M, Liu Y, Wang H, Wang X, Qiu Y and Lu Z (2020) Thiamine-Mediated Cooperation Between Auxotrophic Rhodococcus ruber ZM07 and Escherichia coli K12 Drives Efficient Tetrahydrofuran Degradation. Front. Microbiol. 11:594052. doi: 10.3389/fmicb.2020.594052
Received: 17 August 2020; Accepted: 16 November 2020;
Published: 10 December 2020.
Edited by:
Obulisamy Parthiba Karthikeyan, University of Houston, United StatesReviewed by:
Dhiraj Kumar Chaudhary, Korea University, South KoreaCopyright © 2020 Huang, Qi, Liu, Wang, Wang, Qiu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenmei Lu, bHpoZW5tZWlAemp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.