- 1Department of Microbiology, Universidade Federal de Viçosa, Viçosa, Brazil
- 2Department of Nutrition, Universidade Federal de Juiz de Fora, Governador Valadares, Brazil
- 3Department of Biochemistry and Molecular Biology, Universidade Federal de Viçosa, Viçosa, Brazil
- 4Department of Food and Experimental Nutrition, Food Research Center, Universidade de São Paulo, São Paulo, Brazil
Acyl-homoserine lactones (AHLs) are quorum sensing (QS) signaling molecules that mediate cell-to-cell communication in Gram-negative bacteria. Salmonella does not produce AHL, however, it can recognize AHLs produced by other species through SdiA protein modulating important cellular functions. In this work, the influence of the N-dodecanoyl-DL-homoserine lactone (C12-HSL) on glucose consumption, metabolic profile, and gene expression of Salmonella throughout the cultivation time in Tryptic Soy Broth (TSB) under anaerobic conditions was evaluated. Analysis of the supernatant culture in high-performance liquid chromatography (HPLC) revealed lower glucose uptake after 4 and 6 h of the addition of C12-HSL. Gas chromatography-mass spectrometry (GC-MS) based analysis of the intracellular metabolites revealed C12-HSL perturbation in the abundance levels of metabolites related to the metabolic pathways of glycerolipids, purines, amino acids, and aminoacyl-tRNA biosynthesis. The real-time quantitative PCR (RT-qPCR) indicated that Salmonella increase expression of genes associated with nucleoside degradation and quantification of metabolites supported the induction of pentose phosphate pathway to ensure growth under lower glucose consumption. The obtained data suggest an important role of C12-HSL in the optimization of metabolism at a situation of high population densities.
Introduction
Salmonella is a facultative intracellular enteric pathogen that infects both humans and animals (Andino and Hanning, 2015). Gastrointestinal diseases of infectious origin caused by the ingestion of food contaminated by this pathogen constitute a significant public health problem in the world (Santhi et al., 2012). It is estimated that 535,000 cases of invasive non-typhoid Salmonella disease occurred in 2017, causing 77,500 deaths (Stanaway et al., 2019). Among the serotypes involved, Salmonella Enteritidis is one of the most commonly associated (Balasubramanian et al., 2019).
Salmonella is a robust pathogen and has a wide repertoire of strategies to cause infection in the host. These include genes that encode virulence factors, usually grouped in pathogenicity islands (Fookes et al., 2011; Kaur and Jain, 2012; Nieto et al., 2016), as well as, the presence of virulence plasmid pSLT, fimbriae, flagellum, in addition to the capacity to resist the stresses found in the host (Fabrega and Vila, 2013; Runkel et al., 2013). Besides, the ability to adjust the metabolism to adapt to the physical conditions and available nutrients found in the host during infection seems to be fundamental for the success of Salmonella as a pathogen (Dandekar et al., 2015; Rivera-Chávez and Andreas, 2015; Herrero-Fresno and Olsen, 2018). In addition, the regulation of these phenotypes can occur in a synchronized way, through a cellular communication mechanism called quorum sensing (QS).
The QS is a mechanism used by many bacteria that is based on the production of signaling molecules called auto-inducers (AIs). Usually, AIs diffuse or are transported to the environment where they accumulate and, upon reaching a threshold concentration, they bind to intracellular or extracellular receptors regulating the expression of target genes (Fuqua et al., 1994; Keller and Surette, 2006; Hense and Schuster, 2015). In Salmonella, three QS systems have already been described and are mediated by AI-1, AI-2, and AI-3 (Walters and Sperandio, 2006; Simões et al., 2010).
In Gram-negative bacteria, the most studied mechanism of QS is mediated by acyl-homoserine lactone (AHL), an AI-1, synthesized by LuxI protein homologs (Papenfort and Bassler, 2016). Salmonella does not contain the luxI gene in its genome; thus, it does not synthesize AI-1. However, Salmonella can detect and respond to AHLs produced by other species because it encodes a transcriptional regulator homologous to LuxR proteins called SdiA (Michael et al., 2001; Banerjee and Ray, 2016).
Knowing phenotypes regulated by the AI-1 mediated QS system in Salmonella is of interest and could help the comprehension of the pathogen’s virulence. For instance, in Salmonella Typhimurium, SdiA positively regulates the expression of the rck (resistance to complement killing) operon, which codes for pefI, srgD, srgA, srgB, rck, and srgC genes, and it is found in the virulence plasmid pSLT (Ahmer et al., 1998; Soares and Ahmer, 2011; Sabag-Daigle et al., 2012). Also, the addition of 50 nmol L–1 of N-dodecanoyl homoserine lactone (C12-HSL) promoted an increase in the expression of the virulence genes and those involved in biofilm formation in Salmonella Enteritidis PT4 578 (Campos-Galvão et al., 2016). Moreover, the biofilm formed after the addition of C12-HSL is more compact and mature when compared to other AHLs with a lower carbon chain (Campos-Galvão et al., 2016). This could be explained by its higher binding affinity to the SdiA protein when compared to other AHLs through molecular docking studies (Almeida et al., 2016). Besides C12-HSL, other AHLs tested did not influence the growth of Salmonella Enteritidis PT4 578 cultivated in anaerobic Tryptic Soy Broth (TSB) medium at 37°C (Campos-Galvão et al., 2015; Almeida et al., 2017b). Cultivation of Salmonella Enteritidis PT4 578 in anaerobic TSB medium supplemented with C12-HSL also led to alterations in protein abundance and levels of organic acids associated with the early stationary phase of growth (Almeida et al., 2017a). It also increases the expression of thiol-related proteins and thiol, as well as the alteration in the fatty acid profile (Almeida et al., 2018). However, despite the evidence of the association between cell density-dependent gene expression, so far, there is a paucity of information related to the overall impact of AI-1 on the metabolism of Salmonella Enteritidis.
In this work, high-performance liquid chromatography (HPLC), metabolomics based gas chromatography-mass spectrometry (GC-MS) and real-time quantitative PCR (RT-qPCR) were used to quantitatively investigate the metabolic response of Salmonella Enteritidis PT4 578 to the addition of C12-HSL in TSB under anaerobic conditions.
Materials and Methods
Bacterial Strain
The Salmonella enterica serovar Enteritidis PT4 578 (GenBank: 16S ribosomal RNA gene MF066708.1) used in this study was isolated from chicken meat and donated by the Oswaldo Cruz Foundation (FIOCRUZ) Rio de Janeiro, Brazil. The culture was maintained in Luria Bertani broth (LB) (tryptone 1%, yeast extract 0.5% and NaCl 0.4%) with 20% (vol/vol) sterile glycerol at −20°C. All experiments were performed in laboratories with biosafety level 2.
Culture Medium
Cell culture was performed in Tryptic Soy Broth (TSB; Sigma-Aldrich, St. Louis, MO, United States) prepared with CO2 under O2 free conditions, dispensed in anaerobic flasks sealed with a butyl rubber stopper and autoclaved at 121°C for 15 min.
Culture Conditions
Before each experiment, cells were cultured two consecutive times in anaerobic bottles containing 10 mL of anaerobic TSB and incubated for 24 h at 37°C. Then, 0.2 mL were transferred to 20 mL of anaerobic TSB and incubated for 4 h at 37°C. Cells were centrifuged at 12,000 g for 10 min at 4°C (Sorvall, Waltham, MA, United States), and washed twice with 1 mL of saline 0.85% (wt/vol) and, finally, resuspended in 5 mL of saline 0.85%. The optical density of the cells was standardized at 0.1 at 600 nm (OD600nm) using a spectrophotometer (Thermo Fisher Scientific, Vantaa, Uusimaa, Finland) (Almeida et al., 2017b), which corresponds to, approximately, 107 CFU.mL–1. The N-dodecanoyl-DL-homoserine lactone (C12-HSL; PubChem CID:11565426; Fluka, St. Gallen, Buchs, Switzerland) was suspended in acetonitrile (Merck, Darmstadt, Hessen, Germany) at a concentration of 10 mmol.L–1. Three flasks containing 90 mL of anaerobic TSB were supplemented with 500 μL of C12-HSL to a final concentration of 50 nmol.L–1, and the other three flasks were supplemented with the same volume of acetonitrile as a control. The choice of C12-HSL was based on studies showing its increased effect on biofilm formation by Salmonella compared to other AHLs with shorter carbon chains due to its higher affinity to SdiA. The concentration too was based on those same studies (Campos-Galvão et al., 2015, 2016; Almeida et al., 2016, 2017b, 2018). The acetonitrile concentration was less than 1% (vol/vol) in the culture medium since according to Michael et al. (2001), this concentration does not exhibit an effect on the growth and response of Salmonella to AHL. A total of 90 mL of medium was inoculated with an aliquot of 10 mL of the standardized inoculum resulting in a final volume of 100 mL and incubated at 37°C for 36 h. Samples were taken at specific time points of 4, 6, 7, 12, 24, and 36 h.
Determination of Growth, pH, and Titratable Acidity
Salmonella growth was done by the drop plate method on Plate Count Agar (PCA, Himedia, Mumbai, Maharashtra, India) (Morton, 2001). The plates were incubated at 37°C for 36 h. Colony counts were manually performed and expressed as log CFU.mL–1. A portion of 2.5 mL of culture was also withdrawn and the pH was measured in potentiometer (Hanna Instruments, Woonsocket, RI, United States) and the titration was performed with 0.1 N sodium hydroxide (NaOH) in the presence of 1% (wt/vol) phenolphthalein. The results of titratable acidity were expressed as eq.L–1 of acids obtained.
Analysis of Glucose Consumption
Glucose consumption was assessed by the quantification of glucose present in the extracellular medium by high-performance liquid chromatography (HPLC). At pre-established times, including initial growth time of 0 h, 2 mL of the culture were removed, immediately frozen in liquid nitrogen, and stored at −80°C until analysis. The sample was centrifuged at 12,000 g for 10 min, filtered on cellulose nitrate membrane and 1 mL of each sample was analyzed in HPLC Dionex Ultimate 3000 (Dionex Corporation, Sunnyvale, CA, United States) coupled to a refractive index Shodex RI-101 (Dionex Corporation, Sunnyvale, CA, United States), maintained at 40°C. Aminex® HPX-87H ion-exchange column 4.6 mm × 300 nm 0.45 μm (Bio-Rad, Hercules, CA, United States), maintained at 45°C and a Micro-Guard Cation H column (Bio-Rad, Hercules, CA, United States) were used (Bento et al., 2015). The mobile phase contained 5 mmol.L–1 sulfuric acid (H2SO4) (Sigma-Aldrich, St. Louis, MO, United States), the flow was 0.7 mL.min–1, and the injection of 20 μL. The HPLC was calibrated with the standard glucose curve, prepared at a final concentration of 25 mmol.L–1.
Analysis of Intracellular Metabolites
Sampling and Quenching of Cell Metabolism
A total of 10 mL was harvested at each time of sampling and immediately added into 15 mL of 60% methanol/water solution at −6°C for quenching of cellular metabolism, frozen in liquid nitrogen, and stored at −80°C until extraction (Winder et al., 2008).
Intracellular Metabolite Extraction
The samples were centrifuged at 12,000 g for 10 min at 4°C and intracellular metabolites were extracted from the quenched cell pellet using 1.5 mL of a mixture containing water, methanol (Sigma-Aldrich, St. Louis, MO, United States) and chloroform (Sigma-Aldrich, St. Louis, MO, United States) at −6°C in the proportions 1:2.5:1 following the protocol described by Vital et al. (2017). In each sample, 60 μL of ribitol (0.2 mg.mL–1 stock in ultrapure water) (Sigma-Aldrich, St. Louis, MO, United States) were added as an internal quantitative standard. The samples were agitated in thermomixer (Eppendorf, Hamburg, Hamburg, Germany) at 950 rpm for 10 min at 4°C. Subsequently, the samples were centrifuged at 11,000 g for 10 min at 4°C, and 1 mL of the supernatant was transferred to a new microtube containing 750 μL of ultrapure water. The samples were homogenized in a vortex and centrifuged at 14,000 g for 15 min at 4°C. The upper phase (polar phase) was collected and fractionated into aliquots in 1.5 mL microtubes. Aliquots of 200 μL were evaporated in a centrifugal vacuum concentrator (Speedvac; Eppendorf, Hamburg, Hamburg, Germany). The samples were stored at −80°C until derivatization.
Chemical Derivatization
The samples were derivatized according to the protocol described by Lisec et al. (2006). In summary, 40 μL of methoxyamine hydrochloride (Sigma-Aldrich, St. Louis, MO, United States) at 20 mg.mL–1 in pure pyridine (Merck, Darmstadt, Hessen, Germany) were added to each tube. The samples were shaken in thermomixer at 950 rpm for 2 h at 37°C. A volume of 70 μL of a solution of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA; Sigma-Aldrich, St. Louis, MO, United States) with 40 μL.mL–1 of a mix of fatty acid methyl esters (FAMEs; Sigma-Aldrich, St. Louis, MO, United States) was added with standards for the retention time. The samples were shaken in thermomixer at 950 rpm for 30 min at 37°C. Finally, 90 μL of each sample were transferred to glass vials suitable for GC-MS analysis.
Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
The samples were analyzed using a system of gas chromatography time-of-flight mass spectrometry (GC-TOF-MS), chromatograph Agilent Technologies 7890A spectrometer Leco, TruTOF® HT TOFMS. The column used was a DB-35ms 30 m × 0.32 mm, 0.25 μm capillary column (Agilent Technologies, Santa Clara, CA, United States). The parameters of injection and analysis were defined according to Lisec et al. (2006). An aliquot of 1 μL of each sample was injected in splitless mode with the injector temperature at 230°C. The flow of helium gas through the capillary column was adjusted to 2 mL.min–1. The temperature remained isothermal for 2 min at 80°C then, increased by 15°C per min up to 330°C and, that temperature was maintained for 6 min.
Data Mining, Data Normalization
The chromatograms had their baseline corrected, and the compilation deconvolution algorithm provided by the software ChromaTOF (LECO, Mönchengladbach, Nordrhein-Westfalen, Germany) was used. The deconvoluted spectra were used to assign the peaks using TagSearch software (Cuadros-Inostroza et al., 2009), and spectral mass libraries of compounds derived from trimethyl silicon (TMS) obtained from the Max Planck Institute for Plant Molecular Physiology1. Areas of peak chromatography were verified and normalized by the peak area corresponding to the internal ribitol standard. The raw and normalized data were submitted to repository Metabolomics Workbench2 under track ID 1977.
Real-Time Quantitative PCR (RT-qPCR)
An RT-qPCR analysis was conducted to check the gene expression of sdiA and selected genes of purine metabolism deoD, deoR, add, and hpt. The culture of Salmonella Enteritidis PT4 was obtained as described in item 2.3. The total RNA was extracted in triplicate with Trizol® Reagent (Invitrogen, Carlsbad, CA, United States) according to the supplier’s instructions. The quality of the RNA was evaluated in agarose gel, and the amount was estimated by measuring the absorbance at 260 nm in Nanodrop (Thermo Fisher Scientific, Waltham, MA, United States). A total of 1 μg was treated with RQ1 DNase I- RNase free (Promega, Madison, WI, United States). Complementary DNA (cDNA) synthesis was performed using 1 μg treated RNA and the ImProm-II Reverse Transcription kit with Random Hexamer Primer according to the manufacturer’s instructions. The program GenScript was used to draw the specifics primers for the gene encoding the SdiA protein and some genes of the purine metabolism deoD, deoR, add, and hpt (Table 1) using the S. enterica subsp. enterica serovar Enteritidis strain P125109 (GenBank: NC_011294.1) genome as a template. The self-annealing loops and dimers formation was evaluated using the Oligo Explorer tool and the primer specificity was verified by electronic PCR. The expressions of endogenous gene 16S (Johny et al., 2017) were used to normalize the data. RT-qPCR was performed with Sybr Green I Master Mix (Promega, Madison, WI, United States) in the 96-well plates in a Bio-Rad C1000 Thermal Cycler under custom thermal cycling conditions (denaturation step at 95°C for 2 min, followed by 40 cycles of a denaturation step at 95°C for 15 s, and an annealing/elongation step at 60°C for 1 min). The efficiency of amplification was determined by running a standard curve for each primer with serial dilutions DNA and calculated with the formula E = (10 (1/–slope) – 1) × 100. The relative standard curve was used to calculate the relative quantity (Rq) values of each sample for each gene (Mendes et al., 2013).
Statistical Analysis
The experiments were carried out in three biological replicates. The data of growth, pH, titratable acidity, glucose consumption, and gene expression were submitted to analysis of variance (ANOVA) and Tukey’s test using the Statistical Analysis System and Genetics Software® (Ferreira, 2014). A p < 0.05 was considered to be statistically significant. The graphs were built on the GraphPad Prism 5.0 program (GraphPad Inc., San Diego, CA, United States). For metabolites, principal component analysis (PCA) was performed with the triplicates of each time using software R. The average values of the triplicates were used to construct the heatmap and dendrogram, using software R. The Pearson correlation analysis was also performed using software R (R Foundation for Statistical Computing, Vienna, Austria) and the metabolites with p < 0.05 were considered significant and used for pathway impact analysis using the Metaboanalyst 2.0 software (Xia Lab, Montreal, QC, Canada; Xia et al., 2009; Xia and Wishart, 2016), where the data of Escherichia coli K12 were used as a metabolic reference map. The metabolites of the pathways most impacted by the addition of C12-HSL were subjected to Tukey’s test using GraphPad Prism 5.0 program. A p < 0.05 was considered to be statistically significant.
Results
pH and Titratable Acidity
The pH and titratable acidity of the growth medium were evaluated to characterize the occurrence of sublethal acid stress and to quantify acidic compounds throughout the cultivation time. Both in the control and treatment with the addition of 50 nmol.L–1 C12-HSL, there was a gradual reduction of pH, and the minimum value reached 5.31 and 5.28 respectively, at 12 h incubation, and at this time the highest values of acid compounds were obtained (Supplementary Table S1). At 4, 6, and 24 h of cultivation, the culture medium with the addition of C12-HSL had slightly higher pH than the control (Supplementary Table S1). However, the difference in titratable acidity was only observed with 24 h of cultivation, and the cultivation of Salmonella with C12-HSL presented a higher titratable acidity (Supplementary Table S1).
Growth and Glucose Consumption
Salmonella growth and glucose consumption in the control or C12-HSL treatment in anaerobic TSB medium for 36 h were evaluated (Figure 1). C12-HSL has no apparent influence on the growth of Salmonella Enteritidis PT4 578 (Figure 1). However, the concentration of glucose in the extracellular medium with the addition of C12-HSL after 4 and 6 h of cultivation was 26 and 29% higher, respectively, indicating a lower consumption of this sugar when compared to the control (Figure 1). Despite the lower glucose consumption in the C12-HSL treatment, numbers of log CFU mL–1 values indicated a population similar to the control (Figure 1). On the other hand, after 7 h of cultivation, there is no difference in glucose uptake (Figure 1).
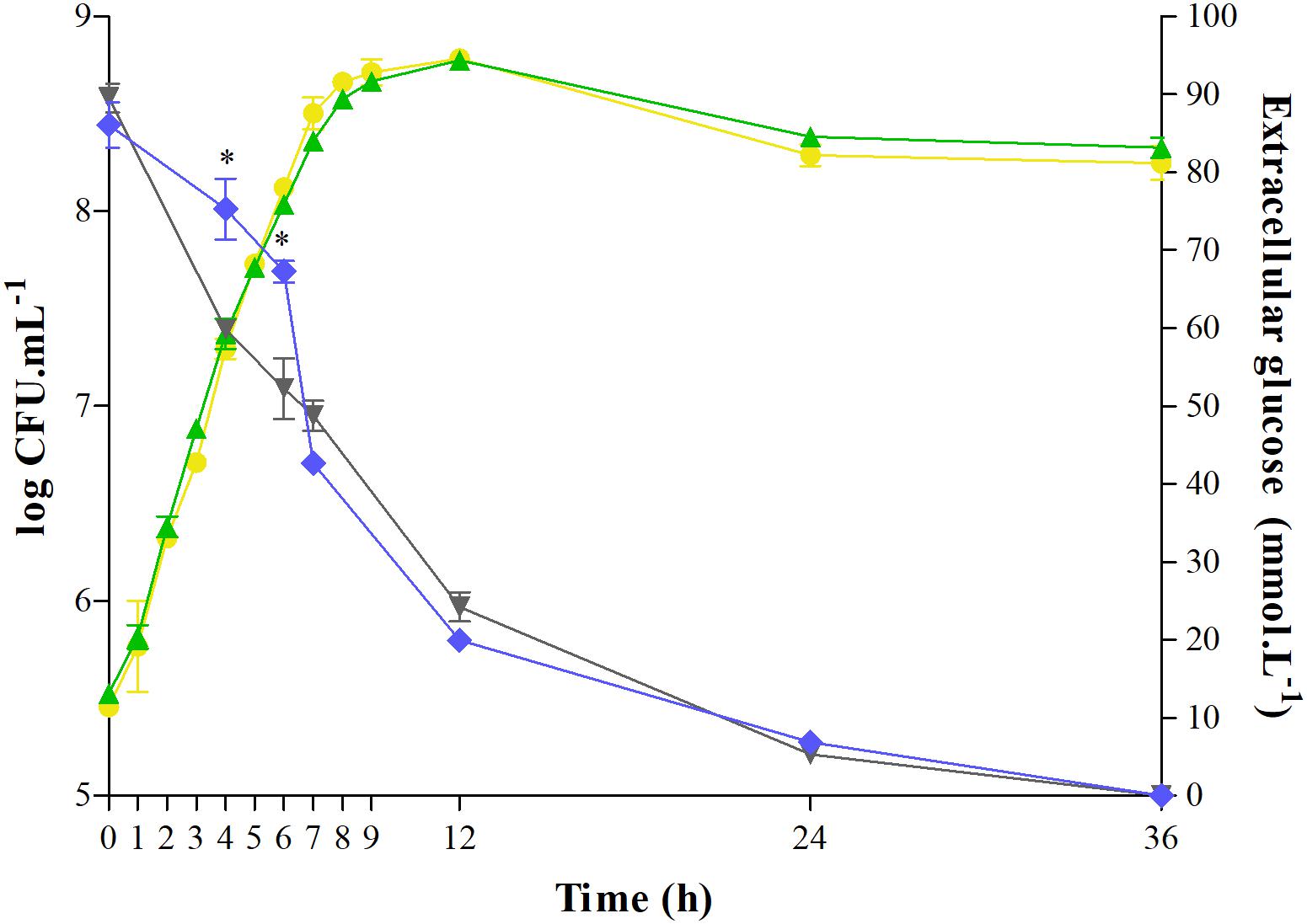
Figure 1. Quantification of extracellular glucose and determination of growth of Salmonella Enteritidis PT4 578 cultivated in anaerobic TSB at 37°C for 36 h with addition of acetonitrile (gray triangle and yellow circle, respectively) or 50 nmol L–1 of C12-HSL (blue diamond and green triangle, respectively). Error bars indicate a standard deviation, and * means significant difference (p < 0.05).
Intracellular Metabolites
The intracellular metabolites were evaluated by GC-TOF-MS. Our results show that the addition of C12-HSL influenced the levels of a large variety of metabolites in Salmonella. We used principal component analysis (PCA) to evaluate the quality of replicates, to understand the data structure, and to detect data patterns. The score plot generated by submitting intracellular metabolites to PCA is demonstrated in Figure 2A. Principal component 1 (PC-1) and PC-2 explained together 88.67% of the data variance (Figure 2A). The variation between the replicates is much lower than the variation between the times, indicating that the chosen methanol concentration of 60% for the metabolic quenching was sufficient to keep the cells intact, avoiding leaks and loss of metabolites being thus, very suitable for the analysis of Salmonella intracellular metabolites. The PCA analysis shows that there is a clear dispersion between the C12-HSL treatment and the control in the time of 4 h and over the growing time the samples tend to cluster, demonstrating that the changes in the metabolic profile caused by C12-HSL tend to reduce over the cultivation time and, from 24 h, no difference is observed (Figure 2A).
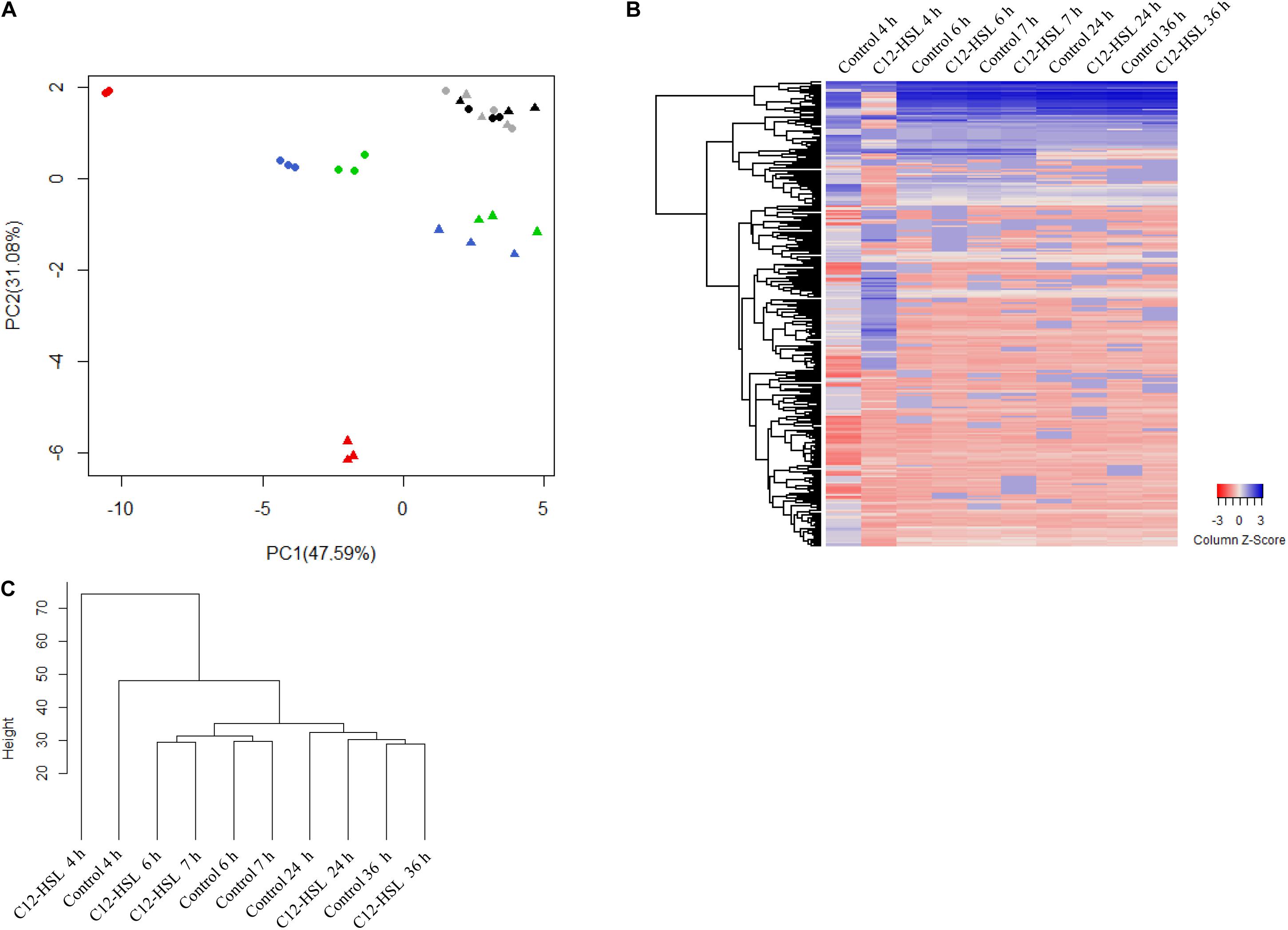
Figure 2. Principal component analysis (PCA) score plot of metabolites from Salmonella Enteritidis PT4 578 anaerobically cultivated in TSB at 37°C with addition of acetonitrile (filled triangle) or 50 nmol L–1 of C12-HSL (filled circle). The colors represent the times of cultivation 4 h (red), 6 h (blue), 7 h (green), 24 h (gray), and 36 h (black) (A). Heatmap of the relative levels of each metabolite identified by GC-MS over time. Each row corresponds to a unique metabolite and each column at the mean of the triplicate values. The color scale ranges from red (low abundance) to blue (high abundance) (B). Dendrogram of the mean values of the triplicates at each time. The height of the arms is proportional to the difference in the abundance profile of the metabolites (C).
The heatmap (Figure 2B) shows, in a global way, changes in the relative levels of each metabolite over time. The profile in the time of 4 h, in the C12-HSL treatment, has more variations, and this observation is confirmed by the dendrogram resulting from cluster analysis by agglomerative hierarchical methods (Figure 2C).
Based on uncorrelated metabolites by Pearson’s analysis, the metabolic pathway impact revealed the distinct perturbation of C12-HSL in a total of seven pathways (Figure 3). Among these pathways, four are related to amino acid metabolism, and the others correspond to the aminoacyl-tRNA biosynthesis, metabolism of glycerolipid and purines (Figure 3 and Supplementary Table S2).
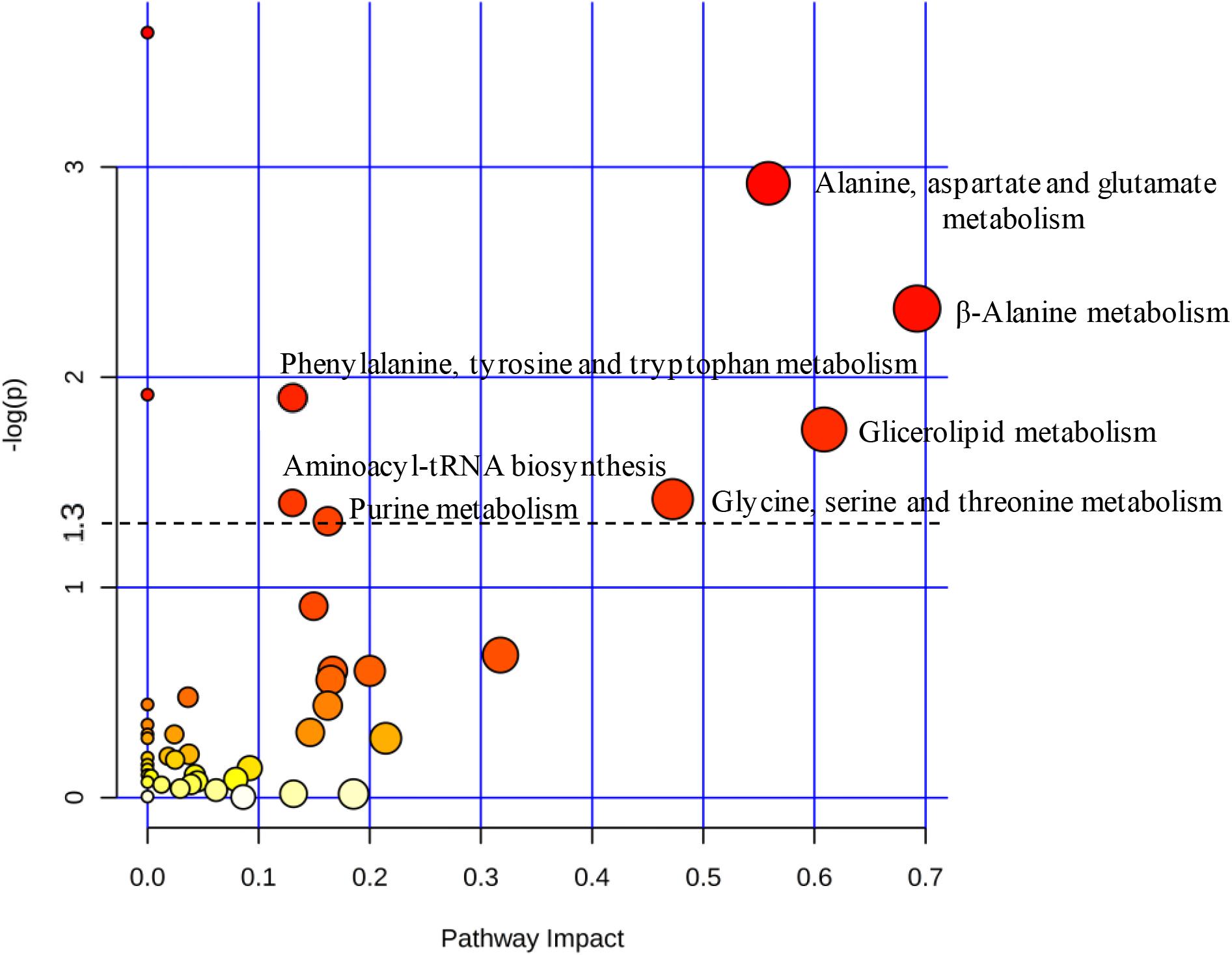
Figure 3. The pathway impact analysis using MetaboAnalyst 2.0. Metabolic pathways with values –log (p) ≥ 1.3 (p ≤ 0.05) were considered significant (above dashed line). The node color is based on its p-value and the scale of significance goes from white (highest p-value) to red (lowest p-value). The node radius is determined based on their pathway impact values.
After 4 h of cultivation the levels of cyclic guanosine monophosphate (3’, 5’-Cyclic GMP), cyclic adenosine monophosphate (3’, 5’-Cyclic AMP) and the first nucleotide formed during the purine synthesis, the inosine monophosphate (IMP) decrease in comparison to the control (Supplementary Figure S1 and Supplementary Table S2). There was also a reduction in levels of nucleosides guanosine, xanthosine, and adenosine, but an increase in hypoxanthine and inosine bases levels (Supplementary Figure S1 and Supplementary Table S2). Also, there was an increase in deoxynucleoside levels, deoxyguanine, and deoxyadenosine (Supplementary Figure S1 and Supplementary Table S2).
The individual analysis of some identified metabolites and belonging to the glycolytic pathway and pentose phosphate pathway (PPP), showed that there is a change in their levels due to the addition of C12-HSL (Figure 4). The reduction of ribose-5-phosphate (ribose-5-P) levels may represent greater interconversion between its isomer ribulose-5-phosphate (ribulose-5-P) and increased activation of the non-oxidative branch of PPP (Figure 5).
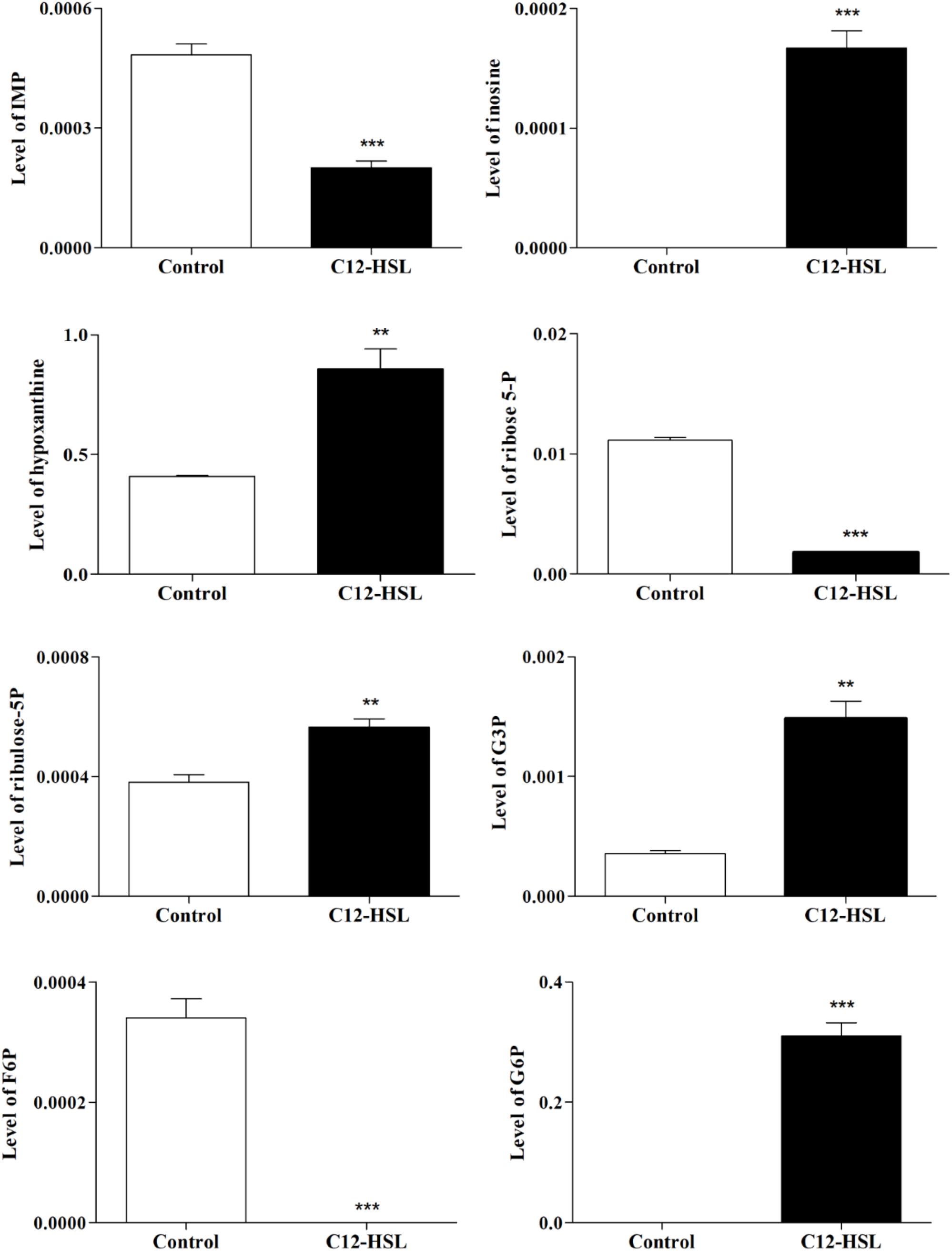
Figure 4. The relative levels of metabolites belonging to the glycolytic pathway and the PPP identified after 4 h of cultivation with addition of acetonitrile (white bar) or 50 nmol L–1 of C12-HSL treatment (black bar). Error bars indicate the standard error, and values that are significantly different to the control by Tukey’s test are indicated; **p < 0.01; ***p < 0.001. PPP, pentose phosphate pathway; IMP, inosine monophosphate; G3P, glyceraldehyde-3-phosphate; F6P, fructose-6- phosphate; G6P, glucose-6-phosphate; -P, -phosphate.
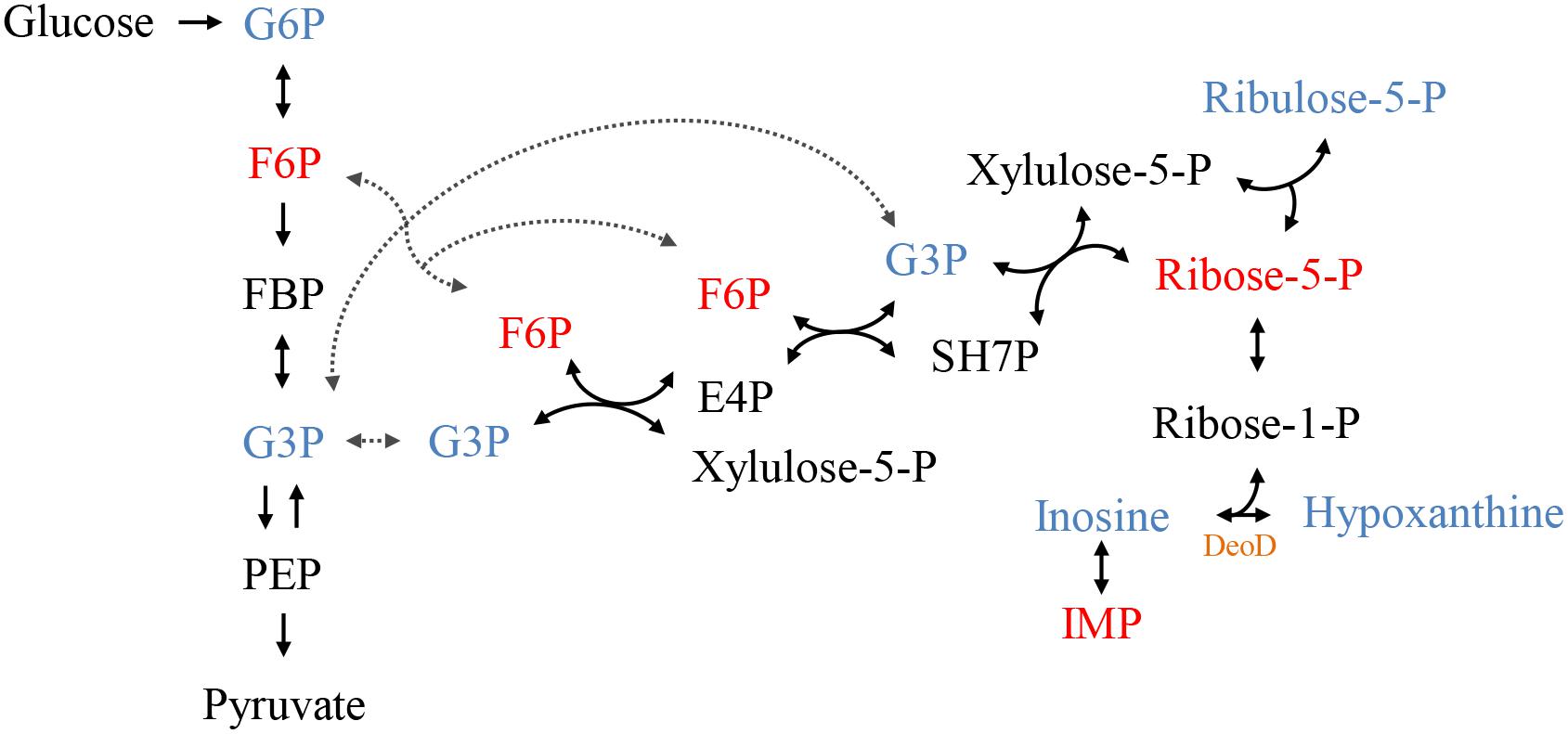
Figure 5. The connection between the glycolytic pathway and the pentose phosphate pathway (PPP). The colors represent the metabolites identified by GC-MS, blue represents increased levels and red, reduced levels compared to control. In black metabolites that have not been identified but complete the pathway. In orange highlighted the protein DeoD, whose gene expression was evaluated in this work. PPP, pentose phosphate pathway; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6- bisphosphate; G3P, glyceraldehyde-3-phosphate; PEP, phosphoenolpyruvate; SH7P, sedoheptulose-7-phosphate; E4P, erythrose 4-phosphate; IMP, inosine monophosphate; -P, -phosphate.
Expression of sdiA and Genes Related to the Nucleotide Salvage Pathway by RT-qPCR
We performed an RT-qPCR analysis to evaluate sdiA expression and whether changes in purine metabolism were related to the expression of genes that could be related to the accumulation of hypoxanthine. For this, the expression of purine nucleoside phosphorylase (deoD), deoxyribose operon repressor (deoR), adenosine deaminase (add), and hypoxanthine phosphoribosyltransferase (hpt) genes were analyzed. The expression of sdiA gene was the same in the control and C12-HSL treatment in both conditions, the expression varied overtime of cultivation, being higher in the early stages (Figure 6A). Interestingly, deoD, which performs the reversible conversion of nucleosides to free bases and pentose-1-phosphate had the expression positively altered in the C12-HSL treatment (Figure 6B). The deoD gene is a distal gene of the operon deoCABD that is controlled by the deoR repressor, down-regulated in the C12-HSL treatment (Figure 6B). The add which encodes the protein that catalyzes the irreversible conversion of adenosine to inosine and hpt, which encodes the protein that catalyzes the conversion of nucleosides to nucleotides were also down-regulated (Figure 6B).
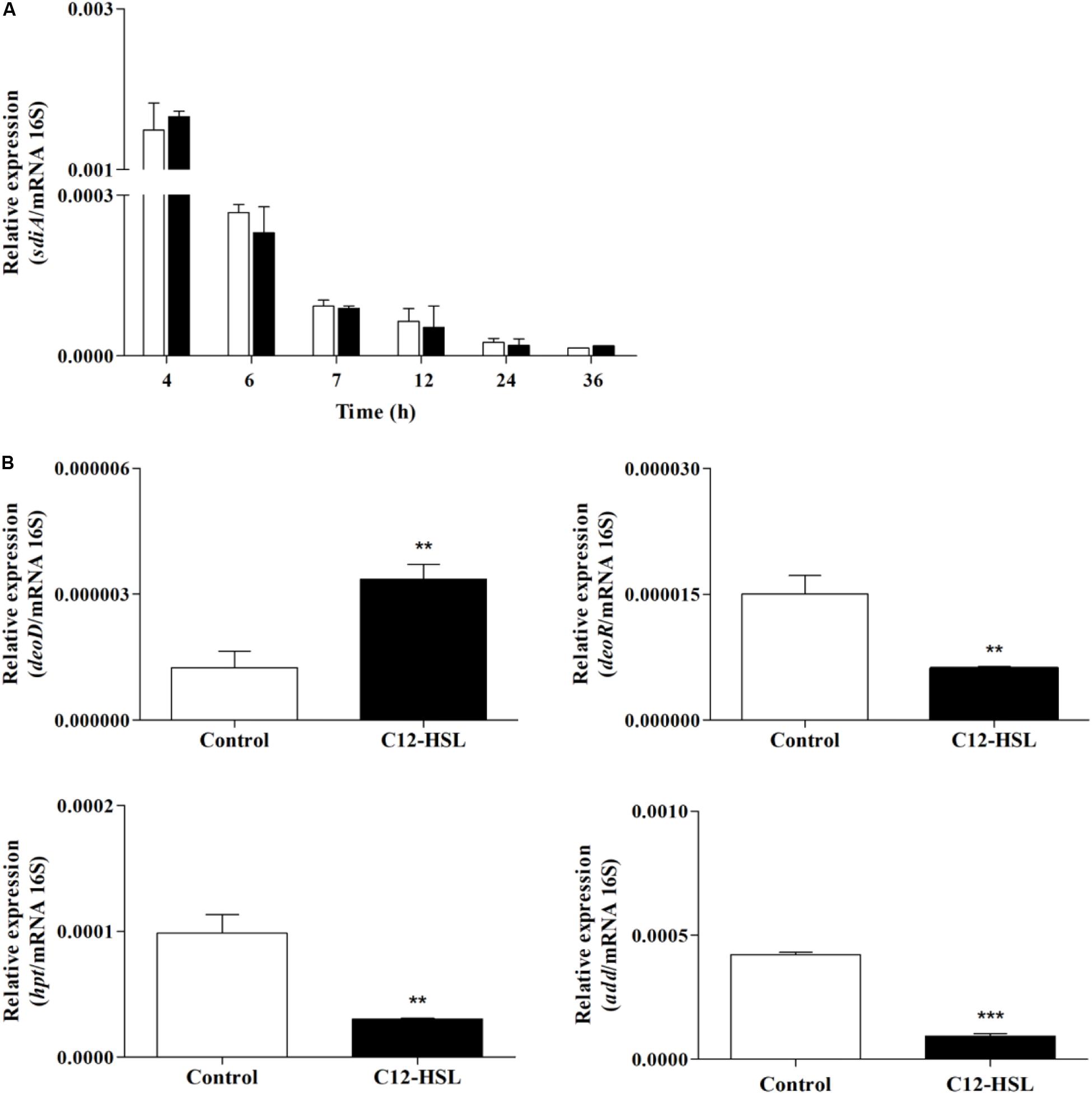
Figure 6. The relative gene expression of sdiA from Salmonella Enteritidis PT4 578 anaerobically cultivated in TSB at 37°C with addition of acetonitrile (white bar) or 50 nmol L–1 of C12-HSL (black bar) (A). The relative gene expression of deoD, deoR, add and hpt from Salmonella Enteritidis PT4 578 after 4 h of cultivation in anaerobic TSB at 37°C in presence of acetonitrile (white bar) or 50 nmol L–1 of C12-HSL (black bar) (B). Error bars indicate the standard deviation and values that are significantly different to the control by Tukey’s test are indicated; **p < 0.01; ***p < 0.001.
Discussion
In this study, we characterized the metabolic response of Salmonella to the addition of C12-HSL under anaerobic conditions. Analysis of pH values demonstrates the occurrence of acid sublethal stress, resulting from the activation of the mixed acid fermentation pathway widely used by enterobacteria under anaerobic conditions. The growth of Salmonella was not altered by the addition of C12-HSL corroborating that observed by Campos-Galvão et al. (2015) and Almeida et al. (2017b) and the lower glucose consumption in the early times suggest that Salmonella can modulate and optimize its metabolism. For this, opting for other favorable metabolic reactions to meet carbon demands in the presence of the signaling molecule, even with glucose availability. This may be a strategy in overcrowded conditions, mimicked by the high concentration of C12-HSL. This phenotype has also been observed in other Gram-negative bacteria. Analyzes based on RNAseq of strains of the rice pathogen Burkholderia glumae mutated in the N-octanoyl-DL-homoserine lactone (C8-HSL) synthase gene (tofI), cognitive receptor (tofR), and transcriptional regulator (qsmR) showed significantly increased expression of ptsI, a representative gene in the sugar phosphotransferase (PTS) system, compared to the wild type (An et al., 2014). Moreover, by nuclear magnetic resonance spectroscopy, it was shown that the transport levels of D-glucose-1-[13C] were significantly higher in the QS mutants than in the wild type. The higher uptake of glucose by the QS mutants in B. glumae indicates that the AI-1 QS system in this pathogen acts as a single metabolic brake when the cells are at a high population (An et al., 2014). Similar behavior was observed in the etiological agent of bubonic plague Yersinia pestis, in which RNAseq analyses demonstrated that the ptsI gene was downregulated in the wild-type compared to an AHL-null strain during the logarithmic growth phase (LaRock et al., 2013). The equal glucose absorption levels after 7 h of cultivation (Figure 1) can be justified by a 1.57-fold increase in the abundance of PtsI protein (phosphoenolpyruvate protein phosphotransferase) in Salmonella Enteritidis PT4 578 at the same time and cultivation in TSB anaerobic medium supplemented with C12-HSL reported by Almeida et al. (2017a).
Interestingly, the addition of C12-HSL resulted in changes in the metabolic profile during the early stages of growth, 4, 6, and 7 h and after these times, the metabolic profile was not significantly altered (Figure 2). This may be related to sdiA expression, which is higher in the early times (Figure 6A). Just like Salmonella, in enterohemorrhagic E. coli (EHEC) AHLs regulate SdiA at the post-transcriptional level, and the expression of sdiA is not affected by the signal molecule (Nguyen et al., 2015). The growth phase, as well as the accumulation of signaling molecules in the medium, were also determinant for the regulation of population density-dependent genes in Burkholderia thailandensis and Pseudomonas aeruginosa (Schuster et al., 2003; Wagner et al., 2003; Majerczyk et al., 2014).
According to the results presented in the PCA and the dendrogram, 4 h is the time that presents the biggest differences between control and C12-HSL treatment, highlighting the pathway of purine metabolism. After 4 h of cultivation, there was also the difference between glucose absorption. Since just deoD had its expression increased after 4 h, we hypothesized that instead of using bases to form nucleotides, in the presence of AHL, Salmonella can break nucleosides to release ribose-1-phosphate that could enter the PPP, generating fructose-6-phosphate and glyceraldehyde-3-phosphate that can follow glycolytic pathway or be converted to hexose and channeled into glycolysis (Figure 5). This reaction could be an interesting alternative both for obtaining energy and for producing glucose, which can be used to form other important cellular structures, thus support the growth with lower consumption of glucose.
Proteomic analysis of the adaptive response to anaerobic conditions of Salmonella Typhimurium cultured in nutrient broth demonstrated that the level of protein DeoD and two enzymes transketolase (TktA) and transaldolase (TalB), which are part of the non-oxidative reactions of the PPP, were induced anaerobically (Encheva et al., 2009). Moreover, the TalB protein had its abundance increased in the presence of C12-HSL after 7 h of cultivation in anaerobiosis (Almeida et al., 2017a). Another protein that has drastically increased its expression when Salmonella has grown in Luria-Bertani (LB) broth under anaerobic conditions is Tsx, which encodes a porin that facilitates the import of nucleosides (Bucarey et al., 2005). The expression of the tsx gene is controlled by two differentially regulated promoters; one of these promoters is down-regulated by the DeoR repressor (Bremer et al., 1988; Bucarey et al., 2006). Based on these data, we can infer that with the addition of C12-HSL and consequent negative regulation of the deoR, can alleviate the repression of the promoter that regulates the expression of Tsx, increasing the synthesis of porins and consequently, the increase of nucleoside internalization.
In P. aeruginosa, AHL signals positively regulate a nucleoside hydrolase (Nuh) which does the hydrolytic degradation of purine nucleotides to nucleobase and ribose (Heurlier et al., 2005). The significance of the QS control of Nuh is not fully understood, but it is added that it is a way of using the products generated during the synthesis of methylthioadenosine, a byproduct of AHL biosynthesis (Heurlier et al., 2006).
Although the significance of QS control in purine metabolism of Salmonella is not fully understood, the variation in expression and accumulation of metabolites provides a clear-cut metabolic phenotype that is under QS control, and this work is the first to provide evidence that the reason is for optimization of metabolism.
Conclusion
Although some studies have already investigated the mechanism of QS in Salmonella, this is the first study that focused quantitatively on the response of Salmonella Enteritidis to AI-1 under anaerobic conditions. The results show that the addition of C12-HSL changes the metabolic profile and reduces glucose absorption by Salmonella Enteritidis under anaerobic conditions. However, despite the reduction of glucose consumption, the growth is not affected, suggesting that Salmonella can use alternative pathways that make pentose moiety of exogenous nucleosides available as sources of carbon and optimize its metabolism in conditions of high population density. The changes in the metabolic profile accentuated in the initial stages of growth corroborate with the literature data and may be related to the highest expression of the response protein, SdiA. Given the limited number of studies on the processes regulated by QS, specifically AI-1, in Salmonella, this work brings new information on the influence of this signaling molecule on the metabolism of the pathogen most frequently reported in outbreaks involving the consumption of contaminated foods. Besides, it reinforces the role of QS in the synchronization of microbial metabolism to changes that can occur in a super crowded environment.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
DC, FA, TM, and MV: conceptualization and experiment design. DC, FA, TM, AA, and NV: data curation. MV and TM: funding acquisition. DC, FA, AA, and NV: performing the experiments. MV: project administration and supervision. DC: validation. DC, FA, TM, UP, and MV: writing original draft. All authors contributed to the article and approved the submitted version.
Funding
DC was supported by a fellowship from “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior” (CAPES) and this research was supported by the “Conselho Nacional de Desenvolvimento Científico e Tecnológico” (CNPq) and the Bill and Melinda Gates Foundation (OPP1193112).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the “Núcleo de Análise de Biomoléculas” (Nubiomol) and the “Diretoria de Tecnologia de Informação” (DTI) at the “Universidade Federal de Viçosa” for the equipments and software used in the analyses of metabolites.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01459/full#supplementary-material
Footnotes
References
Ahmer, B. M. M., van Reeuwijk, J., Timmers, C. D., Valentine, P. J., and Heffron, F. (1998). Salmonella Typhimurium encodes an SdiA homolog, a putative quorum sensor of the LuxR family, that regulates genes on the virulence plasmid. J. Bacteriol. 180, 1185–1193. doi: 10.1128/JB.180.5.1185-1193.1998
Almeida, F. A., Carneiro, D. G., Mendes, T. A. O., Barros, E., Pinto, U. M., Oliveira, L. L., et al. (2018). N-dodecanoyl-homoserine lactone influences the levels of thiol and proteins related to oxidation-reduction process in Salmonella. PLoS One 13:204673. doi: 10.1371/journal.pone.0204673
Almeida, F. A., Pimentel-Filho, N. J., Carrijo, L. C., Bento, C. B. P., Baracat-Pereira, M. C., Pinto, U. M., et al. (2017a). Acyl homoserine lactone changes the abundance of proteins and the levels of organic acids associated with stationary phase in Salmonella Enteritidis. Microb. Pathog. 102, 148–159. doi: 10.1016/j.micpath.2016.11.027
Almeida, F. A., Pimentel-Filho, N. J., Pinto, U. M., Mantovani, H. C., Oliveira, L. L., and Vanetti, M. C. D. (2017b). Acyl homoserine lactone-based quorum sensing stimulates biofilm formation by Salmonella Enteritidis in anaerobic conditions. Arch. Microbiol. 199, 475–486. doi: 10.1007/s00203-016-1313-6
Almeida, F. A., Pinto, U. M., and Vanetti, M. C. D. (2016). Novel insights from molecular docking of SdiA from Salmonella Enteritidis and Escherichia coli with quorum sensing and quorum quenching molecules. Microb. Pathog. 99, 178–190. doi: 10.1016/j.micpath.2016.08.024
An, J. H., Goo, E., Kim, H., Seo, Y.-S., and Hwang, I. (2014). Bacterial quorum sensing and metabolic slowing in a cooperative population. Proc. Natl. Acad. Sci. U.S.A. 111, 14912–14917. doi: 10.1073/pnas.1412431111
Andino, A., and Hanning, I. (2015). Salmonella enterica: survival, colonization, and virulence differences among serovars. Sci. World J. 2015, 1–16. doi: 10.1155/2015/520179
Balasubramanian, R., Im, J., Lee, J. S., Jeon, H. J., Mogeni, O. D., Kim, J. H., et al. (2019). The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccines Immunother. 15, 1421–1426. doi: 10.1080/21645515.2018.1504717
Banerjee, G., and Ray, A. K. (2016). The talking language in some major Gram-negative bacteria. Arch. Microbiol. 198, 489–499. doi: 10.1007/s00203-016-1220-x
Bento, C. B. P., Azevedo, A. C., Detmann, E., and Mantovani, H. C. (2015). Biochemical and genetic diversity of carbohydrate-fermenting and obligate amino acid-fermenting hyper-ammonia-producing bacteria from Nellore steers fed tropical forages and supplemented with casein. BMC Microbiol. 15:28. doi: 10.1186/s12866-015-0369-369
Bremer, E., Gerlach, P., and Middendorf, A. (1988). Double negative and positive control of tsx expression in Escherichia coli. J. Bacteriol. 170, 108–116. doi: 10.1128/jb.170.1.108-116.1988
Bucarey, S. A., Villagra, N. A., Fuentes, J. A., and Mora, G. C. (2006). The cotranscribed Salmonella enterica sv. Typhi tsx and impX genes encode opposing nucleoside-specific import and export proteins. Genetics 173, 25–34. doi: 10.1534/genetics.105.054700
Bucarey, S. A., Villagra, N. A., Martinic, M. P., Trombert, A. N., Santiviago, C. A., Maulén, N. P., et al. (2005). The Salmonella enterica serovar Typhi tsx gene, encoding a nucleoside-specific porin, is essential for prototrophic growth in the absence of nucleosides. Infect. Immun. 73, 6210–6219. doi: 10.1128/IAI.73.10.6210
Campos-Galvão, M. E. M., Leite, T. D. S., Ribon, A. O. B., Araújo, E. F., and Vanetti, M. C. D. (2015). A new repertoire of informations about the quorum sensing system in Salmonella enterica serovar Enteritidis PT4. Genet. Mol. Res. 14, 4068–4084. doi: 10.4238/2015.April.27.22
Campos-Galvão, M. E. M., Ribon, A. O. B., Araújo, E. F., and Vanetti, M. C. D. (2016). Changes in the Salmonella enterica Enteritidis phenotypes in presence of acyl-homoserine lactone quorum sensing signals. J. Basic Microbiol. 56, 493–501. doi: 10.1002/jobm.201500471
Cuadros-Inostroza, Á, Caldana, C., Redestig, H., Kusano, M., Lisec, J., Peña-Cortés, H., et al. (2009). TargetSearch - a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinform. 10:428. doi: 10.1186/1471-2105-10-428
Dandekar, T., Fieselmann, A., Fischer, E., Popp, J., Hensel, M., and Noster, J. (2015). Salmonella - how a metabolic generalist adopts an intracellular lifestyle during infection. Front. Cell. Infect. Microbiol. 4:191. doi: 10.3389/fcimb.2014.00191
Encheva, V., Shah, H. N., and Gharbia, S. E. (2009). Proteomic analysis of the adaptive response of Salmonella enterica serovar Typhimurium to growth under anaerobic conditions. Microbiology 155, 2429–2441. doi: 10.1099/mic.0.026138-26130
Fabrega, A., and Vila, J. (2013). Salmonella enterica serovar Typhimurium skills to succeed in the host: virulence and regulation. Clin. Microbiol. Rev. 26, 308–341. doi: 10.1128/CMR.00066-12
Ferreira, D. F. (2014). Sisvar: a guide for its bootstrap procedures in multiple comparisons. Cienc. Agrotecnol. 38, 109–112. doi: 10.1590/S1413-70542014000200001
Fookes, M., Schroeder, G. N., Langridge, G. C., Blondel, C. J., Mammina, C., Connor, T. R., et al. (2011). Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 7:e1002191. doi: 10.1371/journal.ppat.1002191
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Hense, B. A., and Schuster, M. (2015). Core principles of bacterial autoinducer systems. Microbiol. Mol. Biol. Rev. 79, 153–169. doi: 10.1128/MMBR.00024-14
Herrero-Fresno, A., and Olsen, J. E. (2018). Salmonella Typhimurium metabolism affects virulence in the host - A mini-review. Food Microbiol. 71, 98–110. doi: 10.1016/j.fm.2017.04.016
Heurlier, K., Dénervaud, V., and Haas, D. (2006). Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296, 93–102. doi: 10.1016/j.ijmm.2006.01.043
Heurlier, K., Dénervaud, V., Haenni, M., Guy, L., Krishnapillai, V., and Haas, D. (2005). Quorum-sensing-negative (lasR) mutants of Pseudomonas aeruginosa avoid cell lysis and death. J. Bacteriol. 187, 4875–4883. doi: 10.1128/JB.187.14.4875
Johny, A. K., Frye, J. G., Donoghue, A., Donoghue, D. J., Porwollik, S., McClelland, M., et al. (2017). Gene expression response of Salmonella enterica serotype Enteritidis phage type 8 to subinhibitory concentrations of the plant-derived compounds trans -cinnamaldehyde and eugenol. Nat. Protoc. 8, 1–10. doi: 10.3389/fmicb.2017.01828
Kaur, J., and Jain, S. K. (2012). Role of antigens and virulence factors of Salmonella enterica serovar Typhi in its pathogenesis. Microbiol. Res. 167, 199–210. doi: 10.1016/j.micres.2011.08.001
Keller, L., and Surette, M. G. (2006). Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4, 249–258. doi: 10.1038/nrmicro1383
LaRock, C. N., Yu, J., Horswill, A. R., Parsek, M. R., and Minion, F. C. (2013). Transcriptome analysis of acetyl-homoserine lactone-based quorum sensing regulation in Yersinia pestis. PLoS One 8:e62337. doi: 10.1371/journal.pone.0062337
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., and Fernie, A. R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 1, 387–396. doi: 10.1038/nprot.2006.59
Majerczyk, C., Brittnacher, M., Jacobs, M., Armour, C. D., Radey, M., Schneider, E., et al. (2014). Global Analysis of the Burkholderia thailandensis quorum sensing-controlled regulon. J. Bacteriol. 196, 1412–1424. doi: 10.1128/JB.01405-1413
Mendes, T. A. O., Cunha, J. L. R., Lourdes, R. A., Luiz, G. F. R., Lemos, L. D., Santos, A. R. R., et al. (2013). Identification of strain-specific B-cell epitopes in Trypanosoma cruzi using genome-scale epitope prediction and high-throughput immunoscreening with peptide arrays. PLoS One 7:e2524. doi: 10.1371/journal.pntd.0002524
Michael, B., Smith, J. N., Swift, S., Heffron, F., and Ahmer, B. M. M. (2001). SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183, 5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001
Morton, R. D. (2001). Aerobic plate count. compendium of methods for the microbiological examination of foods. Am. Public Health Assoc. 4, 63–67.
Nguyen, Y., Nguyen, N. X., Rogers, J. L., Liao, J., Macmillan, J. B., and Jiang, Y. (2015). Structural and mechanistic roles of novel chemical ligands on the SdiA quorum-sensing transcription regulator. mBio 6, 1–10. doi: 10.1128/mBio.02429-2414
Nieto, P. A., Pardo-Roa, C., Salazar-Echegarai, F. J., Tobar, H. E., Coronado-Arrázola, I., Riedel, C. A., et al. (2016). New insights about excisable pathogenicity islands in Salmonella and their contribution to virulence. Microb. Infect. 18, 302–309. doi: 10.1016/j.micinf.2016.02.001
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 14, 576–588. doi: 10.1038/nrmicro.2016.89
Rivera-Chávez, F., and Andreas, J. B. (2015). The pyromaniac inside you: Salmonella metabolism in the host gut. Annu. Rev. Microbiol. 69, 31–48. doi: 10.1146/annurev-micro-091014-104108
Runkel, S., Wells, H. C., and Rowley, G. (2013). Living with stress. Adv. Appl. Microbiol. 83, 87–144. doi: 10.1016/B978-0-12-407678-5.00003-9
Sabag-Daigle, A., Soares, J. A., Smith, J. N., Elmasry, M. E., and Ahmer, B. M. M. (2012). The acyl-homoserine lactone receptor, SdiA, of Escherichia coli and Salmonella enterica serovar Typhimurium does not respond to indole. Appl. Environ. Microbiol. 78, 5424–5431. doi: 10.1128/AEM.00046-12
Santhi, L. P., Sunkoji, S., Siddiram, S., and Sanghai, S. S. (2012). Patent research in Salmonellosis prevention. Food Res. Int. 45, 809–818. doi: 10.1016/j.foodres.2011.11.006
Schuster, M., Lostroh, C. P., Ogi, T., and Greenberg, E. P. (2003). Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185, 2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003
Simões, M., Simões, L. C., and Vieira, M. J. (2010). A review of current and emergent biofilm control strategies. LWT Food Sci. Technol. 43, 573–583. doi: 10.1016/j.lwt.2009.12.008
Soares, J. A., and Ahmer, B. M. M. (2011). Detection of acyl-homoserine lactones by Escherichia and Salmonella. Curr. Opin. Microbiol. 14, 188–193. doi: 10.1016/j.mib.2011.01.006
Stanaway, J. D., Parisi, A., Sarkar, K., Blacker, B. F., Reiner, R. C., Hay, S. I., et al. (2019). The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the global burden of disease study 2017. Lancet Infect. Dis. 19, 1312–1324. doi: 10.1016/S1473-3099(19)30418-30419
Vital, C. E., Giordano, A., Soares, E. A., Williams, T. C. R., Mesquita, R. O., Vidigal, P. M. P., et al. (2017). An integrative overview of the molecular and physiological responses of sugarcane under drought conditions. Plant Mol. Biol. 94, 577–594. doi: 10.1007/s11103-017-0611-y
Wagner, V. E., Bushnell, D., Passador, L., Brooks, A. I., and Iglewski, B. H. (2003). Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185, 2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003
Walters, M., and Sperandio, V. (2006). Quorum sensing in Escherichia coli and Salmonella. Int. J. Med. Microbiol. 296, 125–131. doi: 10.1016/j.ijmm.2006.01.041
Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., et al. (2008). Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Anal. Chem. 80, 2939–2948. doi: 10.1021/ac7023409
Xia, J., Psychogios, N., Young, N., and Wishart, D. S. (2009). MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 37, W652–W660. doi: 10.1093/nar/gkp356
Keywords: quorum sensing, autoinducer, growth stage, glucose uptake, metabolic pathway
Citation: Carneiro DG, Almeida FA, Aguilar AP, Vieira NM, Pinto UM, Mendes TAO and Vanetti MCD (2020) Salmonella enterica Optimizes Metabolism After Addition of Acyl-Homoserine Lactone Under Anaerobic Conditions. Front. Microbiol. 11:1459. doi: 10.3389/fmicb.2020.01459
Received: 19 November 2019; Accepted: 04 June 2020;
Published: 28 July 2020.
Edited by:
Ulrike Kappler, The University of Queensland, AustraliaReviewed by:
Mark Radosevich, The University of Tennessee, Knoxville, United StatesZaixiang Lou, Jiangnan University, China
Adam Schikora, Julius Kühn-Institut, Germany
Copyright © 2020 Carneiro, Almeida, Aguilar, Vieira, Pinto, Mendes and Vanetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiago A. O. Mendes, tiagoaomendes@ufv.br; Maria Cristina D. Vanetti, mvanetti@ufv.br
 Deisy G. Carneiro
Deisy G. Carneiro Felipe A. Almeida
Felipe A. Almeida Ananda P. Aguilar
Ananda P. Aguilar Nívea M. Vieira
Nívea M. Vieira Uelinton M. Pinto
Uelinton M. Pinto Tiago A. O. Mendes
Tiago A. O. Mendes Maria Cristina D. Vanetti
Maria Cristina D. Vanetti