- 1Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai, Thailand
- 2Faculty of Science, Mae Fah Luang University, Chiang Rai, Thailand
- 3Department of Health Sciences, Faculty of Science, University of Mauritius, Reduit, Mauritius
- 4Department of Molecular Biology and Genetic, Sciences and Arts Faculty, Kırşehir Ahi Evran University, Kirsehir, Turkey
- 5Biology Department, Graduate School of Natural and Applied Sciences, Kırşehir Ahi Evran University, Kirsehir, Turkey
- 6Retired, Goa Velha, India
- 7Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 8World Agroforestry Centre, Kunming, China
- 9Honghe Center for Mountain Futures, Kunming Institute of Botany, Kunming, China
- 10Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 11Research Center of Microbial Diversity and Sustainable Utilization, Chiang Mai University, Chiang Mai, Thailand
- 12Academy of Science, The Royal Society of Thailand, Bangkok, Thailand
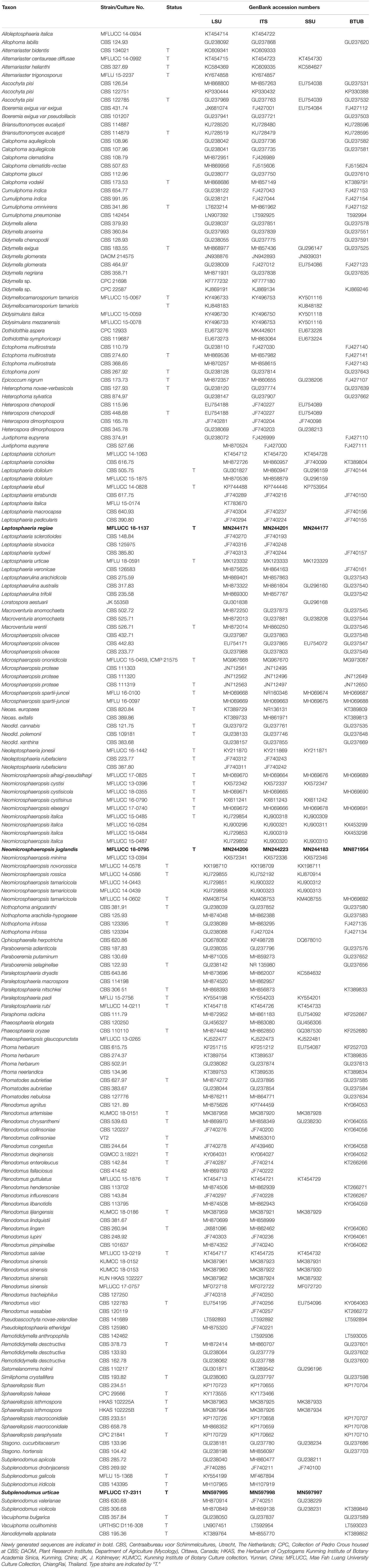
During an ongoing investigation of Ascomycetes from plant substrates, three saprobic species were found from plant substrates. Two new species, Leptosphaeria regiae and Neomicrosphaeropsis juglandis were isolated from dead branches of Juglans regia from Turkey. Another species is introduced herein as Subplenodomus urticae sp. nov within the family Leptosphaeriaceae found on Urtica dioica in Italy. Multigene phylogenies based on combined LSU, ITS, SSU, and β-tubulin DNA sequence data generated from maximum likelihood and MrBayes analyses indicate that Leptosphaeria regiae is related to L. slovacica and forms an independent lineage within the genus Leptosphaeria. Subplenodomus urticae is basal to S. iridicola and its establishment as a new species is strongly supported. Neomicrosphaeropsis juglandis forms a moderately supported lineage in between N. italica and N. elaeagni in the Didymellaceae. Full morphological details are provided herein and phylogenetic relationships of the three new species are also discussed.
Introduction
The bitunicate fungi commonly known as Dothideomycetes is one of the largest group of fungi with a high level of diversity (Zhang et al., 2012; Hyde et al., 2013, 2018, 2019; Tibpromma et al., 2017; Jayasiri et al., 2019). Most of them exist as decomposers, endophytes, epiphytes, fungicolous, lichenized, or lichenicolous fungi in diverse environments (Jeewon et al., 2013, 2017; Phukhamsakda et al., 2016; Doilom et al., 2017; Wanasinghe et al., 2018; Pem et al., 2019a; Phookamsak et al., 2019). They can reproduce either sexually or asexually (Doilom et al., 2014, 2018; Wijayawardene et al., 2014, 2018; Pem et al., 2019c, d). The role of fungi as decomposers is crucial as it helps in recycling of nutrients and releasing enzymes hence maintaining the nutrient compositions of the ecosystem (Hyde and Jeewon, 2003; Hyde et al., 2005; Tang et al., 2005). Decomposers also play vital role in the eco-system such as breakdown of rock to form soils, protection against pathogens and as a food source and alteration of pollutants (Dighton, 2016; Singh et al., 2016). Leptosphaeriaceae is a family in the order Pleosporales (Dothideomycetes, Ascomycota) introduced by Barr (1987) and typified by the genus Leptosphaeria. There are 1,800 epithets of Leptosphaeriaceae recorded in Index Fungorum (2020) with the largest number of species (1,669) occurring in the genus Leptosphaeria and 130 epithets in Mycobank (2020) but most of them lack molecular data. The family Leptosphaeriaceae is characterized by immersed to superficial ascomata, cylindrical to oblong pedicellate asci and reddish brown or yellowish brown, septate ascospores (Hyde et al., 2013; Ariyawansa et al., 2015; Dayarathne et al., 2015; Wanasinghe et al., 2016). Asexual morph are coelomycetous producing phialidic or annellidic conidiogenous cells (Wijayawardene et al., 2018). Leptosphaeriaceae species differ from other closely related families in the Dothideomycetes by the presence of a scleroplectenchymatous peridium. During our ongoing survey, another species resembling those of the asexual genus Neomicrosphaeropsis was observed. The genus Neomicrosphaeropsis was introduced by Thambugala et al. (2016) to accommodate four species namely; N. italica, N. novorossica, N. rossica, and N. tamaricicola. The type species is N. italica and was isolated from dead branches of Tamarix (Tamaricaceae) in Italy. Species of Neomicrosphaeropsis are pathogens or endophytes (Wijayawardene et al., 2017) and are morphologically characterized by hyaline to light brown, aseptate, obovoid to ellipsoidal conidia (Wijayawardene et al., 2018). The genus Neomicrosphaeropsis has been reported to comprise complex species which are morphologically similar but phylogenetically different (Thambugala et al., 2016). Neomicrosphaeropsis resembles species of Microsphaeropsis which is also accommodated in Didymellaceae as well as species of Coniothyrium in having hyaline to light brown, aseptate, obovoid to ellipsoidal, smooth-walled conidia (Verkley et al., 2014; Crous et al., 2019). There are currently 10 epithets in the genus Neomicrosphaeropsis (Index Fungorum, 2020). In this study, we introduce a new asexual species in the genus Neomicrosphaeropsis isolated from stems of Juglans regia (Juglandaceae) in Turkey using multi-gene (LSU, ITS, SSU, and β-tubulin) phylogenetic data. To the best of our knowledge, there have been no fungal species of Leptosphaeriaceae and Didymellaceae associated with Juglans regia in Turkey. We also report on a new species of Leptosphaeriaceae specifically in the genus Subplenodomus found on Urtica dioica from Italy. The aim of this study is to characterize these three fungal isolates in terms of morphology and phylogeny based on multi-gene sequence data.
Materials and Methods
Samples Collection, Morphological Examination, and Isolation
Specimens were collected from dead stems and branches of Juglans regia in the Corum and Kirikkale province of Turkey and on Urtica dioica in the province of Forlì-Cesena (FC) Italy (Figure 1). Samples were stored in Zip-lock bags and returned to the laboratories for examination and description of morphological characters. The specimens were observed under a Motic SMZ 168 series dissecting stereo-microscope. Free hand sections of fungal structures were taken and mounted in water for microscopic study. Photomicrography was carried out using a Canon 750D digital camera fitted to the microscope. Measurements were made with the Tarosoft (R) Image Frame Work software. The images were processed with Adobe Photoshop CS5 v. 12.0 software (Adobe Systems, United States) to illustrate fungal characters using a photoplate. Pure cultures were established from single ascospores/conidia on 2% malt extract agar (MEA; 62 g/L Criterion in distilled water) as described in Vijaykrishna et al. (2004) and Pem et al. (2019d). Cultures were incubated at 25°C for up to 5 weeks and cultural characters were observed and measured after a week and again after 4 weeks following Liu et al. (2014). Holotype specimens are deposited in the herbarium located at Mae Fah Luang University (MFLU) and isotype specimens are deposited at the Kunming Institute of Botany, Academia Sinica Herbarium (HKAS), China. Ex-type living cultures are deposited at the Mae Fah Luang Culture Collection (MFLUCC) and duplicates at the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell cultures (DSMZ). Faces of fungi numbers (Jayasiri et al., 2015) and Index Fungorum number1 are provided.

Figure 1. Geographical coordinates and collecting site of specimen (a,b: Turkey, c: Juglans regia; d,e: Italy, f: Urtica dioica).
DNA Extraction, Amplification, and Sequencing
Isolates were grown on MEA at 16 ± 2°C for 8 weeks. DNA was extracted from fresh fungal mycelium using the DNA extraction kit (E.Z.N.A Fungal DNA Mini Kit, D3390-02, Omega Bio-Tek) following the manufacturer’s protocol. Polymerase Chain Reaction (PCR) was performed to amplify specific gene regions. Primers LR0R and LR5 were used to amplify part of the nuclear ribosomal large subunit 28S rRNA gene (LSU) (Vilgalys and Hester, 1990; Rehner and Samuels, 1994). The Internal transcribed spacer rDNA region (ITS1, 5.8S rDNA, and ITS2) was determined using the primer ITS5/ITS4 and the 18S small subunit ribosomal RNA (SSU) was amplified using NS1/NS4 (White et al., 1990). For the beta-tubulin (TUB) gene, partial cds region, the primers Bt2a/Bt2b was used (Woudenberg et al., 2009). Primer sequences are available at the WASABI (Web Accessible Sequence Analysis for Biological Inference) database at the AFTOL website (aftol.org). Amplification reactions profiles for LSU, ITS, SSU, and β-tubulin gene followed Tang et al. (2007), Wang et al. (2007), and Luo et al. (2017). The analysis of PCR amplification products (amplimers) were performed by the commercial sequencing provider (BGI, Ltd Shenzhen, PR China). Same primer pairs used for amplification process were used for sequencing. The nucleotide sequence data acquired is deposited in GenBank (Table 1). The final alignment and tree are deposited in the TreeBASE repository site2. The recommendations outlined by Jeewon and Hyde (2016) were used to establish the new taxa.
Phylogenetic Analysis
SeqMan v. 7.0.0 (DNASTAR, Madison, WI, United States) was used to assemble consensus sequences. Sequences of closely related strains were recovered from BLAST searches of GenBank3 together with sequences of representative species used by Chen et al. (2015), Thambugala et al. (2016), and Phookamsak et al. (2019) and these are listed in Table 1. Sequences were aligned with online MAFFT v. 7 (Kuraku and Katoh, 2013; Katoh et al., 2019)4. The alignments were checked visually and improved manually where necessary using BioEdit v. 7.0.5.2 (Hall, 1999). Ambiguous regions were excluded from the analyses and gaps were treated as missing data. All novel sequences were deposited in GenBank and the final alignment and tree deposited in TreeBASE2. Phylogenetic analyses were based on maximum likelihood (ML) and Bayesian inference (BI) methods. Maximum likelihood analyses (ML), for single and combined gene alignments included 1,000 bootstrap replicates and was performed using RAxML-HPC2 run on XSEDE (8.2.8) (Stamatakis, 2014) in the CIPRES Science Gateway platform (Miller et al., 2011) using GTR+I+G model of evolution. The final tree was selected among suboptimal trees from each run by comparing likelihood scores with the GTRGAMMA nucleotide substitution model. The best fitting substitution model for each single gene partition and the concatenated data set was determined in MrModeltest 2.3 (Nylander, 2004) for Bayesian inference posterior probabilities (PP). GTR+I+G model was used for each partition or each gene separately, and incorporated into the analysis. The Bayesian inference posterior probabilities (PP) distribution (Zhaxybayeva and Gogarten, 2002) was estimated by Markov Chain Monte Carlo sampling (MCMC) in MrBayes v. 3.2.2 (Ronquist et al., 2012). The MCMC analyses, with six chains were run, started from random tree topology and lasted 1,000,000 generations and sampled every 100 generations (Nylander et al., 2008). The Tracer v. 1.5.0 software program was used to calculate the distribution of log-likelihood scores in order to determine the stationary phase for each search, to check whether extra runs were required to achieve convergence, the stable likelihood plateaus and burn–in value (Drummond et al., 2012). The first 2,000 generations were excluded as burn-in and 10,000 trees were obtained. Maximum likelihood bootstrap values equal or greater than 50% and Bayesian inference posterior probabilities (PP) equal or greater than 0.90 are given in black below or above each node (Figure 1). The phylograms were viewed in FigTree v1.45 and edited using Microsoft PowerPoint 2016.
Results
Taxonomy
Leptosphaeria regiae D. Pem, Selcuk, Jeewon & K.D. Hyde, sp. nov. Figure 2

Figure 2. Leptosphaeria regiae (MFLU 17-0523, holotype). (a) Specimen. (b,c) Appearance of ascomata on host surface. (d) Vertical section through the ascoma. (e) Peridium. (f) Hamathecium. (g–k) Asci. (l–o) Ascospores. (p) Germinating ascospore. (q,r) Culture characteristics on MEA (q: above view; r: reverse view). Scale bars: (b–d) = 200 μm, (e,g,h,k) = 50 μm, (f) = 5 μm, (i,j) = 20 μm, (l–o) = 10 μm.
[urn:lsid:indexfungorum.org:names: 557056]
Facesoffungi Number: FoF 06229
Etymology – Name reflects the host from which the fungus was isolated.
Holotype – MFLU 17-0523
Saprobic on dead stem of Juglans regia. Sexual morph: Ascomata 315–377 μm high, 364–410 μm diam., solitary or gregarious, superficial or semi-immersed on host tissue, visible as black spots on host surface, brown to dark brown. Ostiole apex dark brown to black, ostiolar canal filled with periphyses, papilla not conspicuous. Peridium 41–50 μm wide, comprising two cell types, outer layer composed of large, heavily pigmented, thick-walled cells of textura angularis, inner layer composed of scleroplectenchymatous cells of textura angularis. Hamathecium comprising numerous, 1.4–2.6 μm diam., slime coated, branched, cellular pseudoparaphyses. Asci 99–130 × 9–10 μm ( = 104.8 × 9.9 μm, n = 30), 8-spored, bitunicate, numerous, cylindrical to cylindric-clavate, short pedicellate, apically rounded, with indistinct ocular chamber. Ascospores 15–18 × 6–7 μm ( = 17.1 × 7.1 μm, n = 30), uni to bi-seriate, hyaline brown when immature, becoming yellowish brown to brown at maturity, ellipsoid to broadly fusiform, with rounded to acute ends, slightly clavate, narrow toward the base, 3-septate, constricted at septum, widest above the central septum, smooth-walled. Asexual morph: Undetermined.
Culture characteristics – Colonies on MEA, 17–20 mm diam. after 7 days at 16°C, margin irregular, aerial mycelia thinly hairy, sparse, white and flat; reverse dark brown, white at the margin.
Material examined – TURKEY, The Middle Kizilirmak river basin, Kirşehir province, Kaman district, Savcili small town, 911 m a.s.l., 39° 13′ 684″N, 33° 41′ 034″E, on dead stem of Juglans regia (Juglandaceae), 8 June 2012, Faruk Selcuk (MFLU 17-0523 holotype); ibid. (isotype in HKAS), ex-type living culture MFLUCC 18-1137.
GenBank accession numbers: LSU: MN244171, SSU: MN244177, ITS: MN244201
Notes – The new isolate Leptosphaeria regiae was obtained from dead stem of Juglans regia. In the NCBI BLASTn search of ITS sequence L. regiae has a closest match with L. sclerotioides (Preuss ex Sacc.) de Gruyter et al. (2013) (LP7-MRL) with identities 477/492 (97%) and 1% gaps. In our multigene phylogenetic analysis, L. regiae clusters close to L. slovacica (CBS 125975) with strong bootstrap support (100% ML, 1.00 PP). A comparison of 528 base pairs across the ITS (+5.8S) regions shows 71 (11.9%) base pair differences between L. regiae and L. slovacica. Morphologically, L. regiae differs from L. slovacica in its smaller ascospores (15–18 μm vs. 18–22 μm). We therefore, introduce Leptosphaeria regiae as a new species in the genus Leptosphaeria based on differences in morphology and DNA sequence data.
Neomicrosphaeropsis juglandis D. Pem, Selcuk F, Jeewon & K.D. Hyde, sp. nov. Figure 3
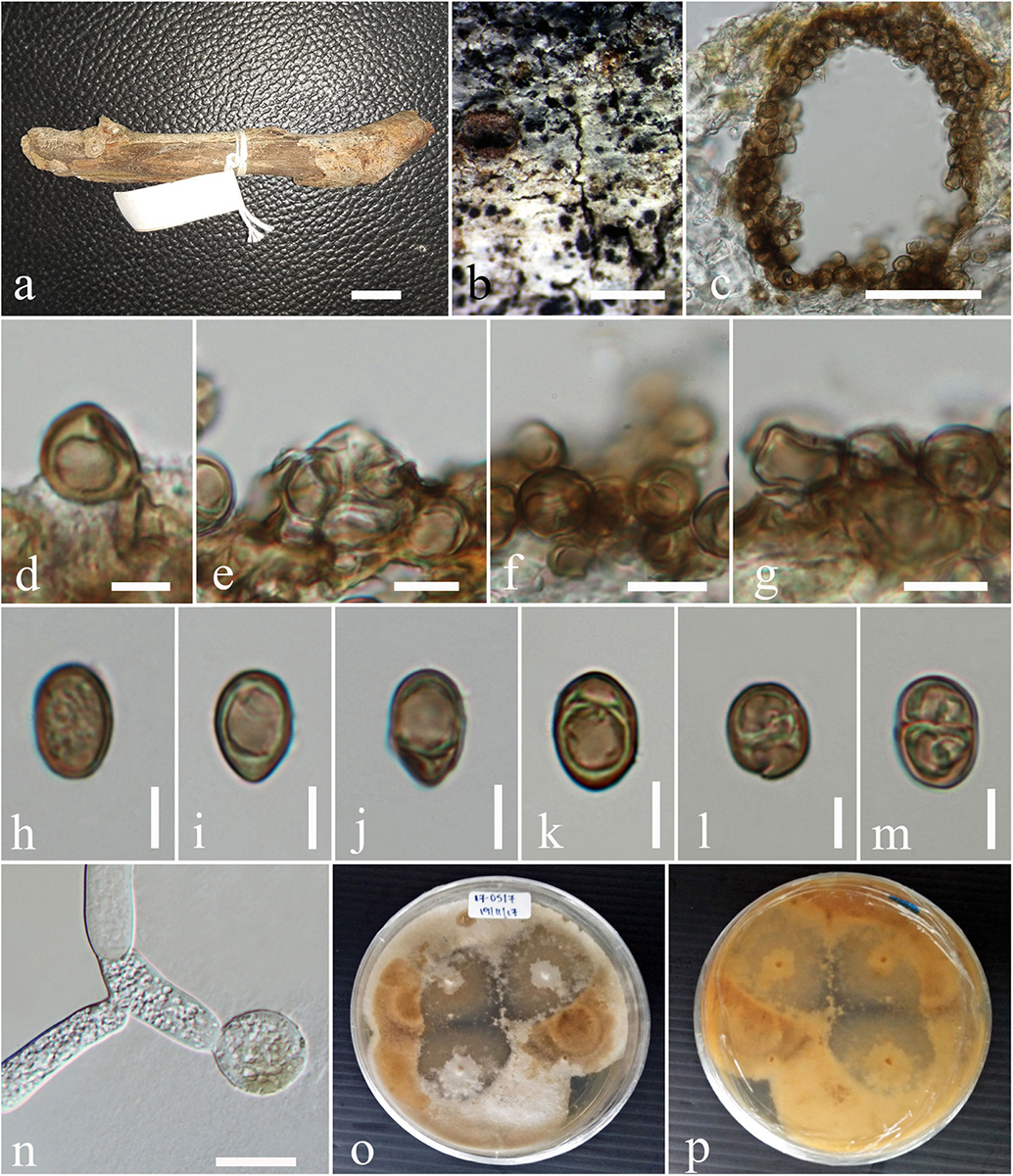
Figure 3. Neomicrosphaeropsis juglandis (MFLU 17-0517, holotype). (a,b) Appearance of conidiomata on host surface. (c) Vertical section through conidioma. (d–g) Conidiogenous cells and developing conidia. (h–m) Conidia. (n) Germinating conidia. (o,p) Culture characters on MEA (o: Above view; p: Reverse view). Scale bars: (a,b) = 1000 μm, (c) = 50 μm, (d) = 5 μm, (e–g) = 10 μm, (h–m) = 5 μm, (n) = 15 μm.
[urn:lsid:indexfungorum.org:names: 556688]
Facesoffungi number– FoF 06211
Etymology – Name reflects the host from which the fungus was isolated
Holotype – MFLU 17-0517
Saprobic on stems of Juglans regia. Sexual morph: Undetermined. Asexual morph: Conidiomata 158–189 μm high × 172–228 μm diam. ( = 170.6 × 202.9 μm, n = 10), pycnidial, scattered, solitary, aggregated or gregarious, immersed, slightly erumpent, black, globose to subglobose, uni- to bi-loculate, non-ostiolate. Conidiomatal wall 10–20 μm wide comprising light to dark brown, thick-walled cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cells enteroblastic, phialidic, light brown, integrated, smooth. Conidia 8–11 × 6–7 μm ( = 9.8 × 6.9 μm, n = 50), yellowish or greenish brown, aseptate, obovoid to ellipsoidal, smooth-walled, sometimes guttulate.
Culture characteristics – Colonies growing on MEA, reaching a diameter of 25 mm after 7 days at 25°C, circular to irregular, flat to slightly raised, mycelium medium sparse, surface initially white, becoming pale saffron to pale white, reverse dark-gray with whitish edge, smooth at surface with entire to slightly filamentous edge, thinly hairy.
Material examined – TURKEY, The Middle Kizilirmak river-basin, Kirikkale province, Delice district, Çerikli small town, 682 ma.s.l., 39° 53′ 689″N, 33° 59′ 769″E, on dead aerial stems of Juglans regia L. (Juglandaceae), 8 August 2012, Faruk Selcuk (MFLU 17–0517, holotype); ibid. (isotype in HKAS), ex-type living culture MFLUCC 18-0795, DSM 109836.
GenBank accession numbers – LSU: MN244206, SSU: MN244183, ITS: MN244223, BTUB: MN871955.
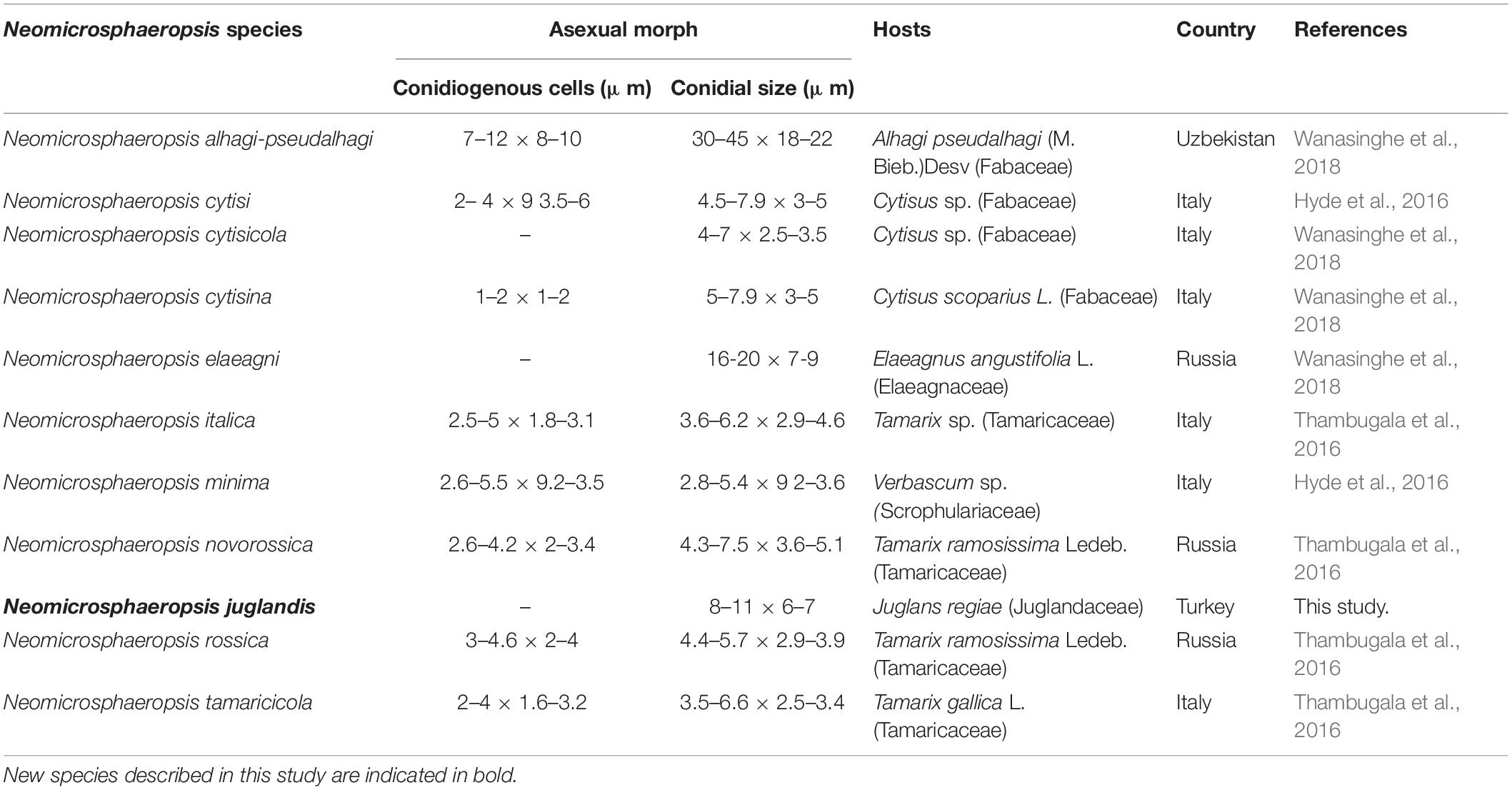
Notes – Our new taxon Neomicrosphaeropsis juglandis is characterized by large, aseptate conidia with a unique yellowish or greenish brown color and measures 8–11 × 6–7 μm, compared to N. italica (3.6–6.2 μm × 2.9–4.6 μm), the type species of the genus Neomicrosphaeropsis. The ITS sequence comparison of N. juglandis with N. tamaricicola, N. italica and N. rossica reveals a difference of 0.8% (5 base pairs difference). However, strong evidence to support N. juglandis as a new species comes from the comparison of the RPB2 gene of our new species, N. juglandis to that of N. italica which shows a pairwise difference of 1.6% as well as that of β-tubulin with N. italica showing a difference of 2.1%. We therefore, introduce N. juglandis as a new species in the genus Neomicrosphaeropsis (Didymellaceae) based on morphological and phylogenetic evidence derived especially from protein coding genes.
Subplenodomus urticae D. Pem, Camporesi, Jeewon & K.D. Hyde, sp. nov. Figure 4
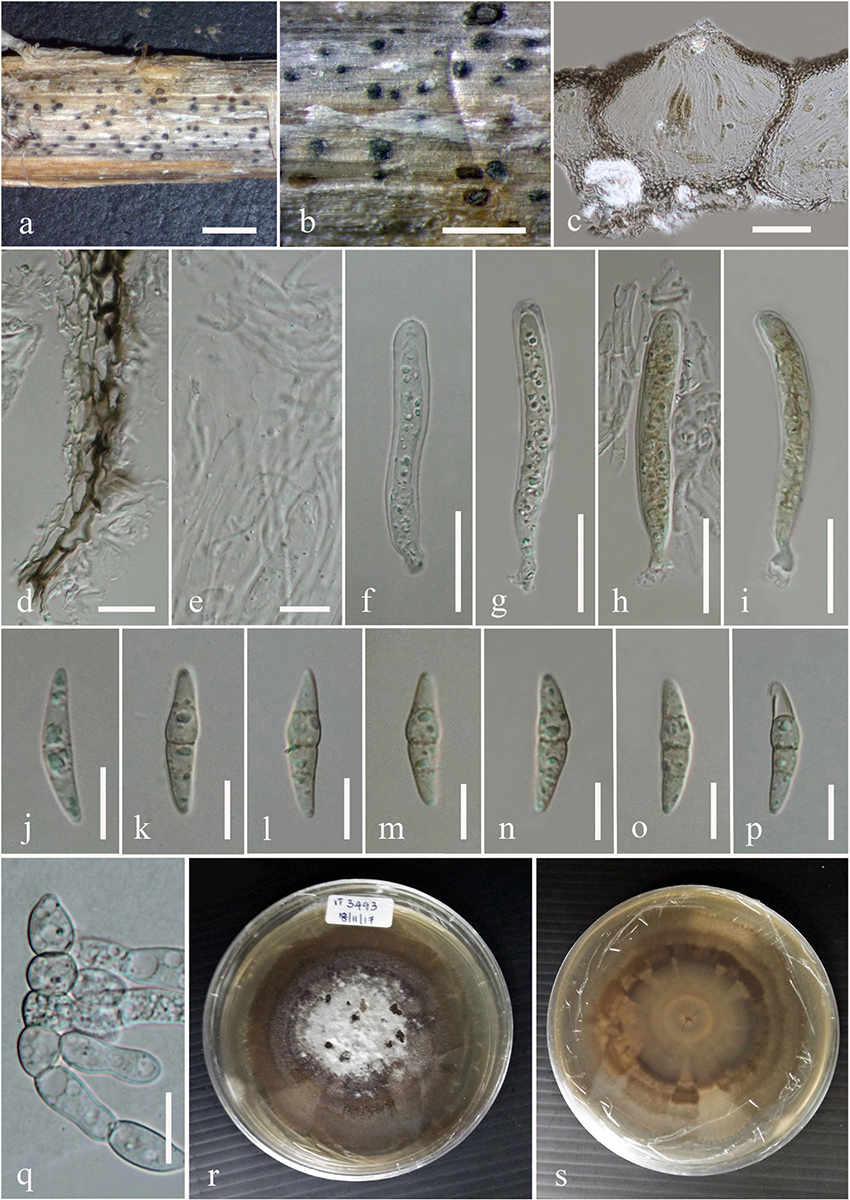
Figure 4. Subplenodomus urticae (MFLU 17-1694, holotype). (a,b) Appearance of ascomata on host surface. (c) Vertical section through the ascoma. (d) Peridium. (e) Hamathecium. (f–i) Asci. (j–p) Ascospores. (q) Germinating ascospore. (r,s) Culture characteristics on MEA (r: above view; s: reverse view). Scale bars: (b–d) = 200 μm, (e,g,h,k) = 50 μm, (f) = 5 μm, (i,j) = 20 μm, (l–o) = 10 μm.
[urn:lsid:indexfungorum.org:names: 557057]
Facesoffungi Number: FoF 06855
Etymology – Name reflects the host from which the fungus was isolated
Holotype – MFLU 17-1694
Saprobic on dead stem of Urtica dioica. Sexual morph: Ascomata 98–162 μm high, 111–200 μm diam., solitary or gregarious, superficial or semi-immersed on host tissue, visible as black spots on host surface, dark brown to black, papillate. Ostiole 29–32 × 48–66 μm, smooth, ostiolar canal filled with periphyses. Peridium 10–28 μm wide, comprising two cell types, outer layer composed of large, heavily pigmented, thick-walled cells of textura angularis, inner layer composed of hyaline cells of textura prismatica. Hamathecium comprising numerous, long, 1.7–2.4 μm ( = 2.2 μm, n = 50) wide, broad, transversely septate, branched, cellular pseudoparaphyses. Asci 44–65 × 7.3–10.7 μm ( = 58.4 × 8.9 μm, n = 30), 8-spored, bitunicate, numerous, cylindrical to cylindric-clavate, short pedicellate, apically rounded, with indistinct ocular chamber. Ascospores 19–24 × 4.4–5.6 μm ( = 21.9 × 5.2 μm, n = 30), overlapping biseriate, hyaline when immature, becoming yellowish brown to brown at maturity, ellipsoidal to broadly fusiform, tapering at the ends, 3–septate, constricted at septum, widest at second septum, smooth-walled. Asexual morph: Undetermined.
Culture characteristics – Circular, surface rough, entire edge, in the middle powdery, on the edge thinly hairy, margin well-defined and slightly radiating, white and slightly raised in the middle, greenish gray at the edge; reverse white in the middle, strongly radiating, cracking the media, greenish-gray at the edges.
Material examined – ITALY, near Balze – Verghereto [province of Forlì-Cesena (FC)], on stem of Urtica dioica (Urticaceae), 19 September 2017, Erio Camporesi (MFLU 17-1694, holotype), ibid. (isotype in HKAS), ex-type living culture MFLUCC 17-2311.
GenBank accession numbers – LSU: MN597995, SSU: MN597997, ITS: MN597998.
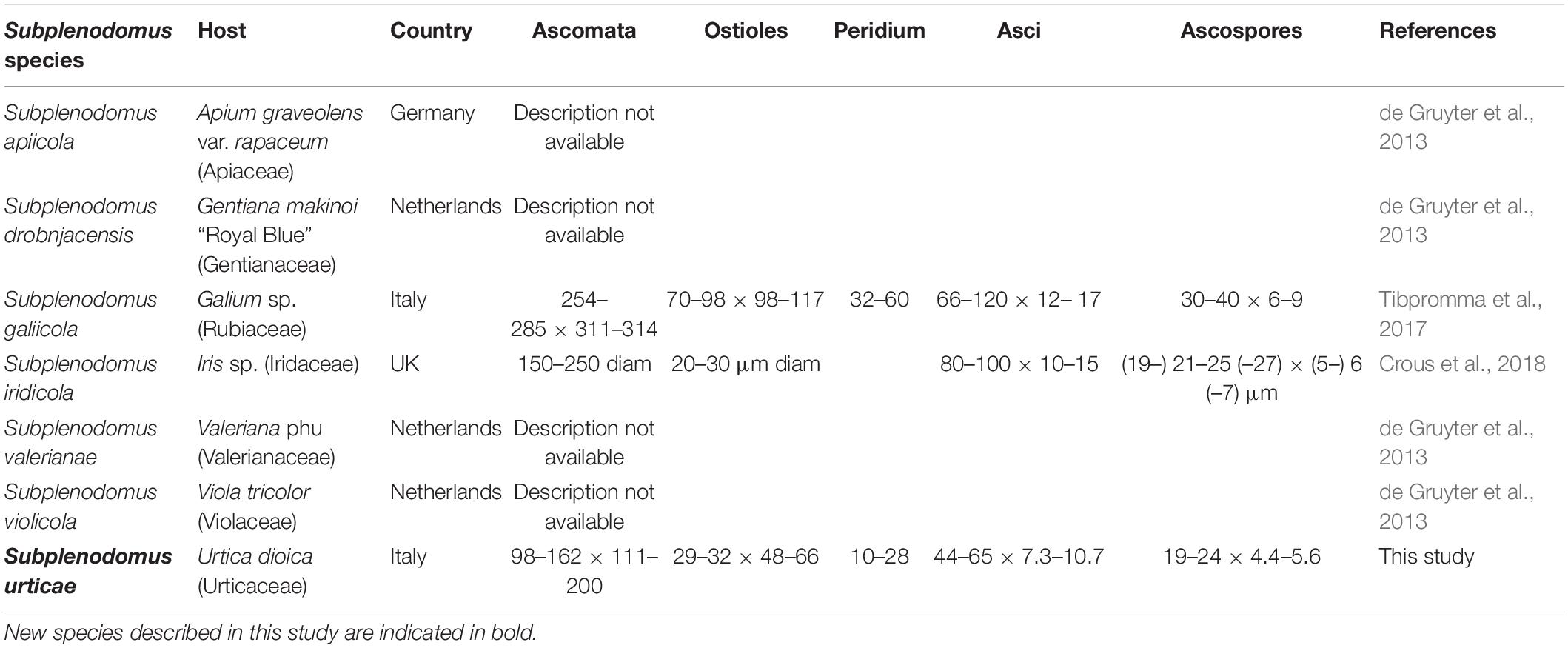
Notes – Subplenodomus urticae was collected from dead stem of Urtica dioica. Morphologically, the present collection matches the description of S. galiicola and S. iridicola in having broad cylindrical, with club-shaped pedicel. However, S. urticae is distinct from S. galiicola in having smaller ascomata (98–162 × 111–200 μm vs. 254–285 × 311–314 μm), smaller ostiole (29–32 × 48–66 μm vs. 70–98 × 98–117 μm), shorter asci (44–65 × 7.3–10.7 μm vs. 66–120 × 12–17 μm) and smaller ascospores (19–24 × 4.4–5.6 μm vs. 30–40 × 6–9 μm. Phylogenetically, S. urticae forms an independent lineage distinct from S. iridicola and other Subplenodomus species. Subplenodomus urticae differs from S. iridicola in having shorter asci (44–65 × 7.3–10.7 μm vs. 80–100 × 10–15 μm) and shorter ascospores (19–24 × 4.4–5.6 μm vs. 21–25 × 6–7 μm). Other species of Subplenodomus are in their asexual morph and thus cannot be compared. A comparison of 528 ITS (+5.8S) nucleotides between S. urticae and S. galiicola shows 57 (9.5%) base pair difference while that of S. urticae and S. iridicola shows 69 (13.1%) base pair difference. Thus, a new taxon is introduced as S. urticae based on the recommendations provided by Jeewon and Hyde (2016).
Phylogenetic Analyses
In the multi-locus phylogeny inferred from the combined dataset of LSU, ITS, SSU, and β-tubulin, several well-supported clades can be recognized which are used for the delimitation of the 10 genera namely, Plenodomus, Alternariaster, Sphaerellopsis, Leptosphaeria, Alloleptosphaeria, Pseudoleptosphaeria, Subplenodomus, Paraleptosphaeria, and Heterospora (Figure 5). The genus Plenodomus forms a well-supported clade within the family Leptosphaeriaceae and comprised 26 strains as well as the type species Plenodomus lingam (Tode: Fr.) Höhn. The genus Alternariaster also forms a well-supported clade sister to the Plenodomus clade and included four strains namely, A. trigonosporus, A. centaureae-diffusae, A. bidentis along with A. helianthi, the type species of the genus Alternariaster. Sphaerellopsis strains together with the type strain, Sphaerellopsis filum (Biv.) B. Sutton clustered in a distinct clade, fully supported in all analyses (89% ML, 1.00 PP). Leptosphaeria sensu stricto forms a well-supported clade in the family Leptosphaeriaceae comprising L. doliolum strains, the type species, strains of 12 other species along with the new species Leptosphaeria regiae (MFLUCC 18-1137). Our new taxon, L. regiae is close to L. slovacica (CBS 125975). The monotypic genera Alloleptosphaeria and Pseudoleptosphaeria form a distinct clade sister to each other and close to the genus Neoleptosphaeria which comprise three strains with N. rubefaciens as type species. Our new taxon, Subplenodomus urticae constitutes an independent lineage and fits within the genus Subplenodomus. The genus Paraleptosphaeria forms a well-supported monophyletic clade (89% ML, 1.00 PP) with four strains together with the type species Paraleptosphaeria nitschkei. The new species Neomicrosphaeropsis juglandis clusters in the family Didymellaceae with moderate support (85% ML, 1.00 PP). The multigene analyses show that N. juglandis (MFLUCC 18-0795) is phylogenetically related to species in the genus Neomicrosphaeropsis in particular to N. italica (Figure 5). We therefore, describe the three taxa as new based on the recommendations outlined by Jeewon and Hyde (2016).
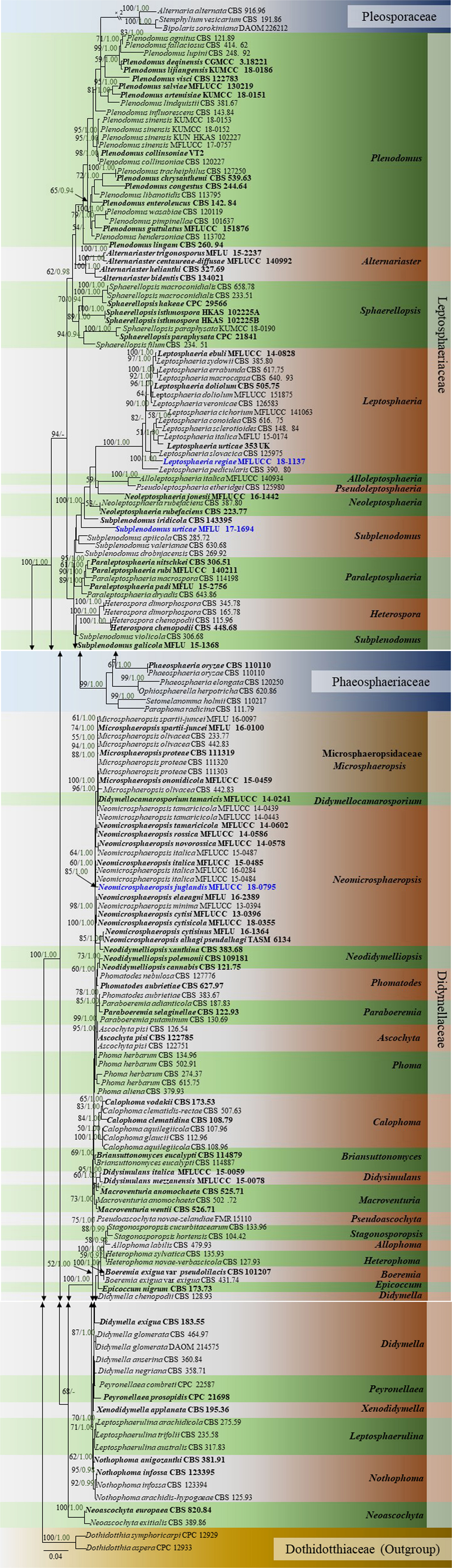
Figure 5. Phylogram generated from maximum likelihood and Bayesian analysis based on combined LSU, ITS, SSU, and β-tubulin sequence data retrieved from GenBank. Related sequences were referred to Chen et al. (2015), Thambugala et al. (2016), and Phookamsak et al. (2019). One hundred and sixty-nine different taxa are included in the combined genes sequence analyses which comprised 2528 characters (LSU: 1–899, ITS: 900–1510, SSU: 1511–2515, β-tubulin: 2516–2878) including gaps. Dothidotthia symphoricarpi (CPC 12929) and Neodothidotthia negundinicola (CPC 12933) are used as the out-group taxa. Maximum likelihood (ML) analysis was conducted in the CIPRES Science Gateway V.3.3. The best sorting RAxML tree with a final likelihood value of -25154.560152 is presented. Estimated base frequencies were as follows: A = 0.242800, C = 0.228484, G = 0.270168, T = 0.258548; substitution rates AC = 1.463454, AG = 3.304206, AT = 1.844561, CG = 0.804481, CT = 7.244176, GT = 1.000000; gamma distribution shape parameter α = 0.629245; proportion of invariant 0.688986. ML bootstrap values ≥50% are given as the first set of numbers and approximate likelihood-ratio test (aLRT) ≥0.90 values as the second set of numbers above the nodes. Voucher/strain numbers are given after the taxon names, the one from type material are indicated in bold face. The new taxa are given in bold and blue. The bar length indicates the number of nucleotide substitutions per site.
Discussion
Several recent papers have described saprobic fungi from the class Dothideomycetes from different hosts across the world (Pem et al., 2018, 2019a,b; Hyde et al., 2019, 2020; Phookamsak et al., 2018, 2019). This study reports on three species that are new to science collected from Turkey and Italy. They are Leptosphaeria regiae, Subplenodomus urticae from the family Leptopshaeriaceae and Neomicrosphaeropsis juglandis from the family Didymellaceae. Both Leptosphaeriaceae and Didymellaceae are highly diverse family with more than hundred species discovered during the last 10 years.
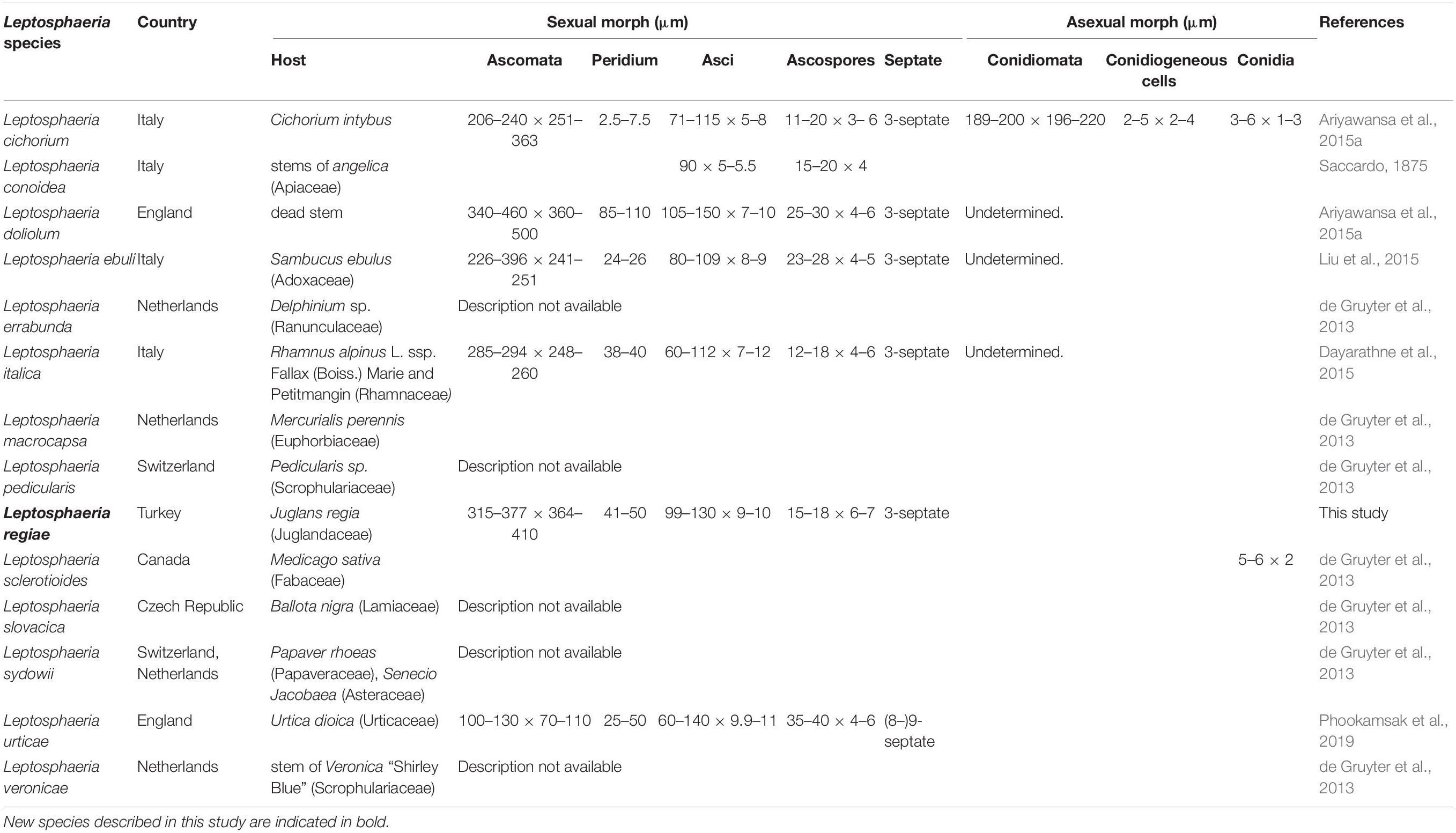
Leptosphaeria regiae forms a distinct lineage basal to L. slovacica (CBS 125975). Phylogeny recovered herein depict a close association of Leptosphaeria regiae to L. slovacica but the affinities of the latter with other species is obscure. Despite a close phylogenetic link, these two species are morphologically different. Leptosphaeria regiae differs from L. slovacica in having relatively shorter ascospores (15–18 μm vs. 18–22 μm) (de Gruyter et al., 2013). Leptosphaeria pedicularis is in its asexual state characterized by black globose perithecia, short ostiole and hyaline cylindrical conidia and cannot be morphologically compared to L. regiae. Comparison of the ITS sequences of these species shows 6.7% (L. regiae vs. L. slovacica) and 7.3% (L. regiae vs. L. pedicularis) nucleotides differences, respectively. Leptosphaeria regiae seems to be most closely allied to L. cichorium by resemblance of general morphological features such as superficial or semi-immersed globose to subglobose ascomata, peridium of schleroplectenchymatous cells and yellowish brown, fusoid, 3-septate ascospores (Ariyawansa et al., 2015). However, L. regiae has longer asci (99–130 × 9–10 μm vs. 71–115 × 5–8 μm) compared to L. cichorium. A pairwise comparison of 523 ITS (+5.8S) sequence data reveals 34 (6.8%) base pair differences between L. regiae and L. cichorium which confirms the two species as distinct. Leptosphaeria regiae also resembles L. italica in sharing superficial or semi-immersed globose ascomata and fusiform ascospores while the former differs in having longer asci (99–130 × 9–10 μm vs. 60–112 μm × 7–12 μm) (Dayarathne et al., 2015). Furthermore, L. italica occurs on Rhamnus alpinus in the Province of Forlì-Cesena in Italy while L. regiae was found on Juglans regia in the Çorum province of Turkey and they are phylogenetically apart (Figure 5). It is also worth to compare the new species L. regiae to that of L. doliolum, the type species of the genus Leptosphaeria. The former has shorter asci (99–130 × 9–10 μm vs. 105–150 × 7–10 μm) and smaller ascospores (15–18 × 6–7 μm vs. 25–30 × 4–6 μm) compared to L. doliolum which was recorded on dead stem in England (Shearer et al., 1990; Liu et al., 2015). Comparison of ITS sequence data between L. regiae and L. doliolum shows 4.4% base pair differences and these two species are far apart in the phylogenetic tree. Likewise, our new species L. regiae also bears morphological resemblance to L. urticae in having cylindric-clavate, short pedicellate asci but markedly differs from L. urticae in having shorter ascospores (15–18 × 6–7 μm vs. 35–40 × 4–6 μm) and in the number of ascospore septa (3 vs. 8–9) (Phookamsak et al., 2019). Moreover, the multi-locus phylogenetic study demonstrates that both species could be clearly differentiated with 25 (5.1%) ITS nucleotides differences between them. Leptosphaeria regiae also shares similarities to L. ebuli in having cylindrical to cylindric-clavate, short pedicellate asci. However, L. regiae has larger ascomata (315–377 × 364–410 μm vs. 226–396 × 241–251 μm) and shorter ascospores (15–18 × 6–7 vs. 23–28 × 4–5 μm) compared to L. ebuli (Liu et al., 2015). Leptosphaeria regiae and L. ebuli are phylogenetically distant and ITS DNA sequence comparison reveals 28 (5.9%) base pair differences. There are 605 estimated species in the genus Leptosphaeria but only 15 species have DNA sequence data (Species Fungorum, 2020). A synopsis of all recognized species having molecular data are provided in Table 2.
There are six morphological species in the genus Subplenodomus (Species Fungorum, 2020) and all six species are described based on DNA sequence data (Tibpromma et al., 2017; Crous et al., 2018). The new species Subplenodomus urticae is morphologically similar to S. iridicola in sharing superficial or semi-immersed black ascomata and cylindrical asci with club-shaped pedicel but distinct in that the latter was described from Iris sp. (Iridaceae) from UK and has larger ascospores (21–25 × 5–7 μm vs. 19–24 × 4.4–5.6 μm) (Crous et al., 2018). A synopsis of Subplenodomus species is provided in Table 3. Phylogenetically, S. urticae clusters in the genus Subplenodomus basal to S. iridicola. Subplenodomus violicola is the type species of Subplenodomus and was established by de Gruyter et al. (2013). Since then, five additional species have been described in the genus. Subplenodomus urticae differs from S. iridicola by 13.1% nucleotide differences in the ITS regions. In our multi-gene analysis, the affinities of Subplenodomus corroborates those reported by previous studies (Schoch et al., 2009; Zhang et al., 2012; Crous et al., 2018; Phookamsak et al., 2019). Subplenodomus apiicola, S. drobnjacensis, S. valerianae, and S. violicola all produce pycnidia with an elongated neck. The pycnidial wall is pseudoparenchymatous. The new species S. urticae is unique and well-distinct among all the species reported in the genus Subplenodomus. Subplenodomus urticae is the first Subplenodomus species reported from Urtica dioica (Urticaceae) and is unique in having shorter cylindrical asci as well as ascospores compared to other species of Subplenodomus.
Our new species Neomicrosphaeropsis juglandis is an independent lineage close to N. italica. Among the several genes regions analyzed, it was noted that β-tubulin DNA sequence data generated relatively well-resolved topologies to support intergeneric relationships within the Didymellaceae and particularly in connection with Neomicrosphaeropsis (data not shown). Our new taxon is an addition to Neomicrosphaeropsis and is also the first record of the genus on Juglans regia in Turkey. A synopsis of the asexual morph of existing species of Neomicrosphaeropsis is provided in Table 4. Our new taxon is unique in that it produces larger aseptate conidia compared to other Neomicrosphaeropsis species and has been reported from a different host. Among the phenotypically diverse species, the genus Neomicrosphaeropsis as well as Didymellocamarosporium in Didymellaceae produce pigmented, muriform spores (Hyde et al., 2016; Thambugala et al., 2016). One interesting finding is also that Neomicrosphaeropsis cytisi, N. cytisicola and N. cytisina are morphologically similar with conidial measurement ranging between 4–7.9 × 2.5–3.5 μm and all of them were isolated from Cytisus sp. (Fabaceae). The authors differentiate the two species based on size and form of conidiomata. However, it is highly probable that N. cytisi, N. Cytisicola, and N. cytisina are all same species as the ITS sequences of the three species are same (no base pair difference). RPB2 comparison of N. cytisi and N. cytisicola shows only three base pair difference across 1089 nucleotides examined. No TEF and BTUB sequences are available for the three species for comparison. Likewise, N. tamaricicola, N. rossica, N. Novorossica, and N. italica have been isolated from Tamarix species and are morphologically similar with conidial measurements ranging from 3.5–6.6 × 2.5–4.6 μm. Comparison of RPB2 and BTUB gene sequences between N. italica and N. tamaricola shows no base pair difference. However, comparison of TEF gene sequences between N. italica to N. rossica and N. tamaricicola shows 7 or 1.0% and 9 or 1.3% base pair differences, respectively, while there was no base pair differences between N. rossica and N. tamaricicola. It can be possible that some of these species are the same and need to be synonymised in future studies. In our phylogenetic analyses, N. minima is closely related to N. cytisi with strong bootstrap support. Comparison of available ITS and TEF sequences of N. minima and N. cytisi reveals zero base pair difference, however, there are some slight differences in conidial sizes (N. minima: 2.8–5.4 × 9.2–3.6 μm vs. N. cytisi: 4.5–7.9 × 3–5 μm). Neomicrosphaeropsis minima also differs from N. cytisi in conidiomatal size (N. minima: 60–80 μm diam., 60–95 μm high vs. N. cytisi: 75–155 μm diam., 75–130 μm high) and were isolated from different hosts. Whether these species are distinct and merit a specific taxonomic rank warrant further investigations given that minor conidial and conidiomatal size differences could vary under cultural and host conditions. Recollecting and sequencing of more coniothyrium-like and more fungi similar to Neomicrosphaeropsis from different geographical regions are also essential to clarify the placement of species and to infer relationships in Neomicrosphaeropsis and Neomicrosphaeropsis-like genera within the Didymellaceae (Pleosporales).
Data Availability Statement
The datasets generated for this study can be found in the sequences retrieved from the 28S rRNA, Internal transcribed spacer rDNA region (ITS1, 5.8S rDNA, and ITS2), 18S rRNA and beta-tubulin (TUB) gene sequencing and were deposited at GenBank-NCBI under the nucleotide accession number as follows: MN244171, MN244201, MN244177 (Leptosphaeria regiae), MN244206, MN244223, MN244183, MN871954 (Neomicrosphaeropsis juglandis), MN597995, MN597998, MN597997 (Subplenodomus urticae).
Author Contributions
DP, RJ, and KH designed the study. FS did the sample collections. DP and RJ were involved in the phylogenetic analyses. SL contributed to the research funds. All authors contributed to the article and approved the submitted version.
Funding
This research work was partially supported by the Chiang Mai University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
DP thanks the Mae Fah Luang University for supporting studies in Dothideomycetes fungi. MD thanks the 5th batch of Postdoctoral Orientation Training Personnel in Yunnan Province (Grant No. Y934283261) and the 64th batch of China Postdoctoral Science Foundation (Grant No. Y913082271). RJ thanks the University of Mauritius for research support. Mr. Erio Camporesi is acknowledged for his support in sample collection. KH thanks Chiang Mai University for the award of a Visiting Professor scholarship. SL thanks Chiang Mai University for research support. DP is also grateful to the project entitled Comparison of Diversity and Biogeographical distribution of Ascomycetous fungi from two protected areas in Turkey and Thailand (TÜBİTAK-NSTDA).
Footnotes
- ^ http://www.indexfungorum.org
- ^ http://www.treebase.org/
- ^ http://www.ncbi.nlm.nih.gov
- ^ http://mafft.cbrc.jp/alignment/server/index.html
- ^ http://tree.bio.ed.ac.uk/software/figtree/
References
Ariyawansa, H. A., Phukhamsakda, C., Thambugala, K. M., Bulgakov, T. S., Wanasinghe, D. N., Perera, R. H., et al. (2015). Revision and phylogeny of Leptosphaeriaceae. Fungal Divers. 74, 19–51. doi: 10.1007/s13225-015-0349-2
Chen, Q., Jiang, J. R., Zhang, G. Z., Cai, L., and Crous, P. W. (2015). Resolving the Phoma enigma. Stud. Mycol. 82, 137–217. doi: 10.1016/j.simyco.2015.10.003
Crous, P. W., Schumacher, R. K., Akulov, A., Thangavel, R., and Hernandez-Restrepo, M. (2019). New and interesting fungi. 2. Fungal Sys. Evol. 3, 57–124. doi: 10.3114/fuse.2019.03.06
Crous, P. W., Schumacher, R. K., Wingfield, M. J., Akulov, A., Denman, S., Roux, J., et al. (2018). New and interesting fungi. 1. Fungal Sys. Evol. 1, 169–215.
Dayarathne, M. C., Phookamsak, R., Ariyawansa, H. A., Jones, E. B. G., Camporesi, E., and Hyde, K. D. (2015). Phylogenetic and morphological appraisal of Leptosphaeria italica sp. nov. (Leptosphaeriaceae, Pleosporales) from Italy. Mycosphere 6, 634–642. doi: 10.5943/mycosphere/6/5/13
de Gruyter, J., Woudenberg, J. H. C., Aveskamp, M. M., Verkley, G. J. M., Groenewald, J. Z., and Crous, P. W. (2013). Redisposition of phoma-like anamorphs in Pleosporales. Stud. Mycol. 75, 1–36. doi: 10.3114/sim0004
Dighton, J. (2016). Fungi in Ecosystem Processes, Vol. 31. Boca Raton, FL: CRC Press, doi: 10.1201/9781315371528
Doilom, M., Dissanayake, A. J., Wanasinghe, D. N., Boonmee, S., Liu, J. K., Bhat, D. J., et al. (2017). Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 82, 107–182. doi: 10.1007/s13225-016-0368-7
Doilom, M., Hyde, K. D., Phookamsak, R., Dai, D. Q., Tang, L. Z., Hongsanan, S., et al. (2018). Mycosphere notes 225-274: types of some genera of Ascomycota. Mycosphere 9, 647–754. doi: 10.5943/mycosphere/9/4/3
Doilom, M., Shuttleworth, L. A., Roux, J., Chukeatirote, E., and Kevin, H. D. (2014). Barriopsis tectonae sp. nov. a new species of Botryosphaeriaceae from Tectona grandis (teak) in Thailand. Phytotaxa 176, 81–91.
Drummond, A., Suchard, M. A., Xie, D., and Rambaut, A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 22, 1185–1192. doi: 10.1093/molbev/mss075
Hall, T. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hyde, K. D., Cai, L., and Jeewon, R. (2005). “Tropical Fungi,” in The Fungal Community: Its Organization and Role in the Ecosystem, 3rd Edn, eds J. Dighton, P. Oudemans, and J. White (Boca Raton, FL: Taylor & Francis), 93–115.
Hyde, K. D., Chaiwan, N., Norphanphoun, C., Boonmee, S., Camporesi, E., Chethana, K. W. T., et al. (2018). Mycosphere notes 169–224. Mycosphere 9, 271–430. doi: 10.5943/mycosphere/9/2/8
Hyde, K. D., de Silva, N. I., Jeewon, R., Bhat, D. J., Phookamsak, R., Doilom, M., et al. (2020). AJOM new records and collections of fungi: 1-100. Asian J. Mycol. 3, 22–294.
Hyde, K. D., Hongsanan, S., Jeewon, R., Bhat, J. D., and McKenzie, E. H. C. (2016). Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 80, 1–270. doi: 10.1007/s13225-016-0373-x
Hyde, K. D., and Jeewon, R. (2003). “Physiological studies and molecular diversity of freshwater lignicolous fungi,” in Freshwater Mycology, eds C. K. M. Tsui and K. D. Hyde (Hong Kong: Hong Kong University Press), 173–193.
Hyde, K. D., Jones, E. B. G., Liu, J. K., Ariyawansa, H., Boehm, E., Boonmee, S., et al. (2013). Families of Dothideomycetes. Fungal Divers. 63, 1–313.
Hyde, K. D., Tennakoon, D. S., Jeewon, R., Bhat, J. D., Maharachchikumbura, S. S. N., Rossi, W., et al. (2019). Fungal diversity notes 1036-1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 96, 1–242. doi: 10.1007/s13225-019-00429-2
Index Fungorum (2020). Available online at: http://www.indexfungorum.org/names/Names.asp (accessed February 26 2019).
Jayasiri, S. C., Hyde, K. D., Ariyawansa, H. A., Bhat, J., Buyck, B., Cai, L., et al. (2015). The faces of fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 74, 3–18. doi: 10.1007/s13225-015-0351-8
Jayasiri, S. C., Hyde, K. D., Jones, E. B. G., McKenzie, E. H. C., Jeewon, R., Phillips, A. J. L., et al. (2019). Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 10, 1–186. doi: 10.5943/mycosphere/10/1/1
Jeewon, R., and Hyde, K. D. (2016). Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7, 1669–1677. doi: 10.5943/mycosphere/7/11/4
Jeewon, R., Ittoo, J., Mahadeb, D., Jaufeerally-Fakim, Y., Hong Kai, W., and Ai-Rong, L. (2013). DNA based identification and phylogenetic characterization of endophytic and saprobic fungi from Antidesma madagascariense, a medicinal plant in Mauritius. J. Mycol. 2013:781914. doi: 10.1155/2013/781914
Jeewon, R., Wanasinghe, D. N., Rampadaruth, S., Puchooa, D., Zhou, L., and Liu, A. (2017). Nomenclatural and identification pitfalls of endophytic mycota based on DNA sequence analyses of ribosomal and protein genes phylogenetic markers: a taxonomic dead end? Mycosphere 8, 1802–1817. doi: 10.5943/mycosphere/8/10/7
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kuraku, Z., and Katoh, N. (2013). aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 41, W22–W28.
Liu, J. K., Hyde, K. D., Jones, E. B. G., Ariyawansa, H. A., Bhat, J. D., Boonmee, S., et al. (2015). Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 72, 1–197.
Liu, J. K., Phookamsak, R., Dai, D. Q., Tanaka, K., Jones, E. B. G., Xu, J. C., et al. (2014). Roussoellaceae, a new pleosporalean family to accommodate the genera Neoroussoella gen. nov., Roussoella and Roussoellopsis. Phytotaxa 181, 1–33.
Luo, Z. L., Bhat, J. D., Jeewon, R., Boonmee, S., Bao, D. F., Zhao, Y. C., et al. (2017). Molecular phylogeny and morphological characterization of asexual fungi (Tubeufiaceae) from freshwater habitats in Yunnan, China. Cryptogamie Mycol. 38, 1–28. doi: 10.7872/crym/v38.iss1.2017.27
Miller, M., Pfeiffer, W., and Schwartz, T. (2011). “CIPRES science gateway: a community resource for phylogenetic analyses,” in Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, July 18- 21, Salt Lake City, UT.
Mycobank (2020). Mycobank Database: Fungal Databases, Nomenclature & Species Banks. Available online at: http://www.mycobank.org/ (Accessed 26 February 2020)Google Scholar
Nylander, J. A. A. (2004). MrModeltest Version 2. Program Distributed by the Author. Uppsala: Evolutionary Biology Centre, Uppsala University.
Nylander, J. A. A., Wilgenbusch, J. C., Warren, D. L., and Swofford, D. L. (2008). AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583. doi: 10.1093/bioinformatics/btm388
Pem, D., Gafforov, Y., Jeewon, R., Hongsanan, S., Promputtha, I., Doilom, M., et al. (2018). Multigene phylogeny coupled with morphological characterization reveal two new species of Holmiella and taxonomic insights within patellariaceae. Cryptogamie Mycol. 39, 193–209. doi: 10.7872/crym/v39.iss2.2018.193
Pem, D., Hyde, K. D., Doilom, M., Camporesi, E., Hongsanan, S., Rampadarath, S., et al. (2019a). Multigene phylogenetic analyses to establish new Valsaria species and taxonomic significance of spore ornamentation. PLoS One 14:e0217982. doi: 10.1371/journal.pone.0217982
Pem, D., Jeewon, R., Bhat, D. J., Doilom, M., Boonmee, S., Hongsanan, S., et al. (2019b). Mycosphere Notes 275–324: a morphotaxonomic revision and typification of obscure Dothideomycetes genera (Incertae sedis). Mycosphere 10, 1115–1246. doi: 10.5943/mycosphere/10/1/22
Pem, D., Jeewon, R., Bulgakov, T., Gafforov, Y., Hongsanan, S., Phookamsak, R., et al. (2019c). Taxonomy and molecular phylogeny of Thyrostroma ephedricola sp. nov. (Dothidotthiaceae) and proposal for Thyrostroma jaczewskii comb. nov. Phytotaxa 416, 243–256. doi: 10.11646/phytotaxa.416.4.3
Pem, D., Jeewon, R., Gafforov, Y., Hongsanan, S., Phukhamsakda, C., Promputtha, I., et al. (2019d). Melanocamarosporioides ugamica gen. et sp. nov., a novel member of the family Melanommataceae from Uzbekistan. Mycol. Progress 18, 471–481. doi: 10.1007/s11557-018-1448-8
Phookamsak, R., Hyde, K. D., Jeewon, R., Bhat, J. D., Jones, E. B. G., Maharachchikumbura, S. S. N., et al. (2019). Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95, 1–273. doi: 10.1007/s13225-019-00421-w
Phookamsak, R., Lu, Y. Z., Hyde, K. D., Jeewon, R., Li, J., Doilom, M., et al. (2018). Phylogenetic characterization of two novel Kamalomyces species in Tubeufiaceae (Tubeufiales). Mycol. Progress 17, 647–660. doi: 10.1007/s11557-017-1365-2
Phukhamsakda, C., Hongsanan, S., Ryberg, M., Ariyawansa, H. A., Chomnunti, P., Bahkali, A. H., et al. (2016). The evolution of Massarineae with Longipedicellataceae fam. nov. Mycosphere 7, 1713–1731. doi: 10.5943/mycosphere/7/11/7
Rehner, S. A., and Samuels, G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98, 625–634. doi: 10.1016/s0953-7562(09)80409-7
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Saccardo, P. A. (1875). Fungi veneti novi vel critici. II Nuovo Giornale Botanico Italiano. 7, 299–329.
Schoch, C. L., Crous, P. W., Groenewald, J. Z., Boehm, E. W. A., Burgess, T. I., de Gruyter, J., et al. (2009). A class–wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 64, 1–15. doi: 10.3114/sim.2009.64.01
Shearer, C. A., Crane, J. L., and Chandra, R. K. R. (1990). Studies in Leptosphaeria. Lectotypification of Sphaeria doliolum. Mycologia 82, 496–500. doi: 10.1080/00275514.1990.12025913
Singh, R., Rani, A., Kumar, P., Shukla, G., and Kumar, A. (2016). The decomposer microorganisms in the environment and their succession of substrates. IJESRT 5:7. doi: 10.5281/zenodo.58637
Species Fungorum (2020). CABI Databases. Available online at: http://www.speciesfungorum.org/ (Accessed January, 2020).
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tang, A. M. C., Jeewon, R., and Hyde, K. D. (2005). Successional patterns of microfungi in fallen leaves of Castanopsis fissa (Fagaceae) in Hong Kong forest. Can. J. Microbiol. 51, 967–974. doi: 10.1139/w05-086
Tang, A. M. C., Jeewon, R., and Hyde, K. D. (2007). Phylogenetic utility of protein (RPB2, B-tubulin) and ribosomal (18S, 28S) gene sequences in the systematics of Sordariomycetes (Ascomycota, Fungi). Antonie van Leeuwenhoek 91, 327–349. doi: 10.1007/s10482-006-9120-8
Thambugala, K. M., Daranagama, D. A., Phillips, A. J. L., Bulgakov, T. S., Bhat, D. J., Camporesi, E., et al. (2016). Microfungi on Tamarix. Fungal Divers. 82, 239–306. doi: 10.1007/s13225-016-0371-z
Tibpromma, S., Hyde, K. D., Jeewon, R., Maharachchikumbura, S. S. N., Liu, J. K., Bhat, J. D., et al. (2017). Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 83, 1–261. doi: 10.1007/s13225-017-0378-0
Verkley, G. J. M., Dukik, K., Renfurm, R., Göker, M., and Stielow, J. B. (2014). Novel genera and species of coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 32, 25–51. doi: 10.3767/003158514X679191
Vijaykrishna, D., Mostert, L., Jeewon, R., Hyde, K. D., and Crous, P. W. (2004). Pleurostomosphora, an anamorph of Pleurostoma (Calosphaeriales), a new anamorph genus morphologically similar to Philaphora. Stud. Mycol. 50, 387–398.
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wanasinghe, D. N., Camporesi, E., and Hu, D. M. (2016). Neoleptosphaeria jonesii sp nov., a novel saprobic sexual species, in Leptosphaeriaceae. Mycosphere 7, 1368–1377. doi: 10.5943/mycosphere/7/9/10
Wanasinghe, D. N., Phukhamsakda, C., Hyde, K. D., Jeewon, R., Lee, H. B., Jones, E. B. G., et al. (2018). Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 89, 1–236. doi: 10.1007/s13225-018-0395-7
Wang, H. K., Aproot, A., Crous, P. W., Hyde, K. D., and Jeewon, R. (2007). The polyphyletic nature of Pleosporales: an example from Massariosphaeria based on ribosomal DNA and RPB2 gene phylogenies. Fungal Biol. 111, 1268–1276. doi: 10.1016/j.mycres.2007.08.014
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, CA: Academic Press), 315–322. doi: 10.1016/b978-0-12-372180-8.50042-1
Wijayawardene, N. N., Crous, P. W., Kirk, P. M., Hawksworth, D. L., Boonmee, S., Braun, U., et al. (2014). Naming and outline of Dothideomycetes-2014 including proposals for the protection or suppression of generic names. Fungal Divers. 69, 1–55. doi: 10.1007/s13225-014-0309-2
Wijayawardene, N. N., Hyde, K. D., Lumbsch, H. T., Liu, J. K., Maharachchikumbura, S. S. N., Ekanayaka, A. H., et al. (2018). Outline of Ascomycota: 2017. Fungal Divers. 88, 167–263. doi: 10.1007/s13225-018-0394-8
Wijayawardene, N. N., Hyde, K. D., Rajeshkumar, K. C., Hawksworth, D. L., Madrid, H., and Kirk, P. M. (2017). Notes for genera: ascomycota. Fungal Divers. 86, 1–594. doi: 10.1007/s13225-017-0386-0
Woudenberg, J. H. C., Aveskamp, M. M., de Gruyter, J., Spiers, A. G., and Crous, P. W. (2009). Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia 22, 56–62. doi: 10.1016/j.mycres.2007.01.009
Zhang, Y., Crous, P. W., Schoch, C. L., and Hyde, K. D. (2012). Pleosporales. Fungal Divers. 53, 1–221. doi: 10.1007/s13225-011-0117-x
Keywords: 3 new species, asexual fungi, Dothideomycetes, morphology, phylogeny
Citation: Pem D, Jeewon R, Selcuk F, Ulukapi M, Bhat J, Doilom M, Lumyong S and Hyde KD (2020) Ribosomal and Protein Gene Phylogeny Reveals Novel Saprobic Fungal Species From Juglans regia and Urtica dioica. Front. Microbiol. 11:1303. doi: 10.3389/fmicb.2020.01303
Received: 04 January 2020; Accepted: 22 May 2020;
Published: 30 June 2020.
Edited by:
Baokai Cui, Beijing Forestry University, ChinaReviewed by:
Sinang Hongsanan, Shenzhen University, ChinaFeng-Yan Bai, Institute of Microbiology (CAS), China
Copyright © 2020 Pem, Jeewon, Selcuk, Ulukapi, Bhat, Doilom, Lumyong and Hyde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saisamorn Lumyong, c2Nib2kwMDlAZ21haWwuY29t
 Dhandevi Pem
Dhandevi Pem Rajesh Jeewon
Rajesh Jeewon Faruk Selcuk
Faruk Selcuk Merve Ulukapi5
Merve Ulukapi5 Mingkwan Doilom
Mingkwan Doilom Saisamorn Lumyong
Saisamorn Lumyong Kevin D. Hyde
Kevin D. Hyde