- Laboratory for Molecular Infection Medicine Sweden, Department of Molecular Biology, Umeå Centre for Microbial Research, Umeå University, Umeå, Sweden
Bacteria can colonize virtually any environment on Earth due to their remarkable capacity to detect and respond quickly and adequately to environmental stressors. Vibrio cholerae is a cosmopolitan bacterium that inhabits a vast range of environments. The V. cholerae life cycle comprises diverse environmental and infective stages. The bacterium is found in aquatic ecosystems both under free-living conditions or associated with a wide range of aquatic organisms, and some strains are also capable of causing epidemics in humans. In order to adapt between environments, V. cholerae possesses a versatile metabolism characterized by the rapid cross-regulation of energy-producing pathways. Low oxygen concentration is a key environmental factor that governs V. cholerae physiology. This article reviews the metabolic plasticity that enables V. cholerae to thrive on low oxygen concentrations and its role in environmental and host adaptation.
Introduction
Vibrio cholerae is a Gram-negative facultative anaerobic bacterium that inhabits estuaries, rivers, and other aquatic environments (Reen et al., 2006) and can cause Cholera disease via contaminated water or food. Upon ingestion, the organism colonizes the small intestine (SI), where it secretes the potent cholera toxin (CT). CT is directly responsible for a purging diarrheal illness that can kill an adult in 24 h (Mekalanos et al., 1997). Cholera disease is also dependent on the production of the type IV pilus TCP (Waldor and Mekalanos, 1996; Häse and Mekalanos, 1998), the type VI secretion system T6SS (Fu et al., 2018), and alternative secreted virulence factors including the hemagglutinin/protease (HAP), the multifunctional RTX toxin, and the hemolysin A/cytolysin (VCC) (Hanne and Finkelstein, 1982; Olivier et al., 2007). During its infection, V. cholerae cells are shed in large numbers into the environment through the stool, where they can survive either as free-living cells, by forming biofilms on the chitin surface of crustaceans (Silva and Benitez, 2016) or by colonizing the gut of birds and fish until they are ingested again by humans, thus completing its life cycle (De Haan and Hirst, 2004; Yahr, 2006).
These transitions of V. cholerae from the human gastrointestinal tract (GI) to the environment require a rapid metabolic adaption to a variety of environmental factors, adaptation to varying oxygen concentrations being one of the most significant. The most important requirement of any living organism is to respire oxygen, since its respiration (reduction to water) is the main source of energy for the cell. During aerobic respiration, electron donors such as NAD(P)H or FADH2 are oxidized, and the resulting free electrons are conducted through an electron transport chain (ETC) to the final electron acceptor, oxygen (Figure 1A). ETCs are formed by electron carriers with increasing redox potential embedded within cellular membranes. Typically, these electron transporters are transmembrane proteins, small mobile haem-containing proteins (cytochromes), and redox-active lipids (ubiquinones). The transport of electrons releases free energy that promotes translocation of protons through the membrane, generating an electrochemical gradient (aka. proton motive force, PMF). Dissipation of this proton gradient by an ATP synthase supports phosphorylation of ADP to render energy in the form of ATP. This process is commonly known as oxidative phosphorylation.
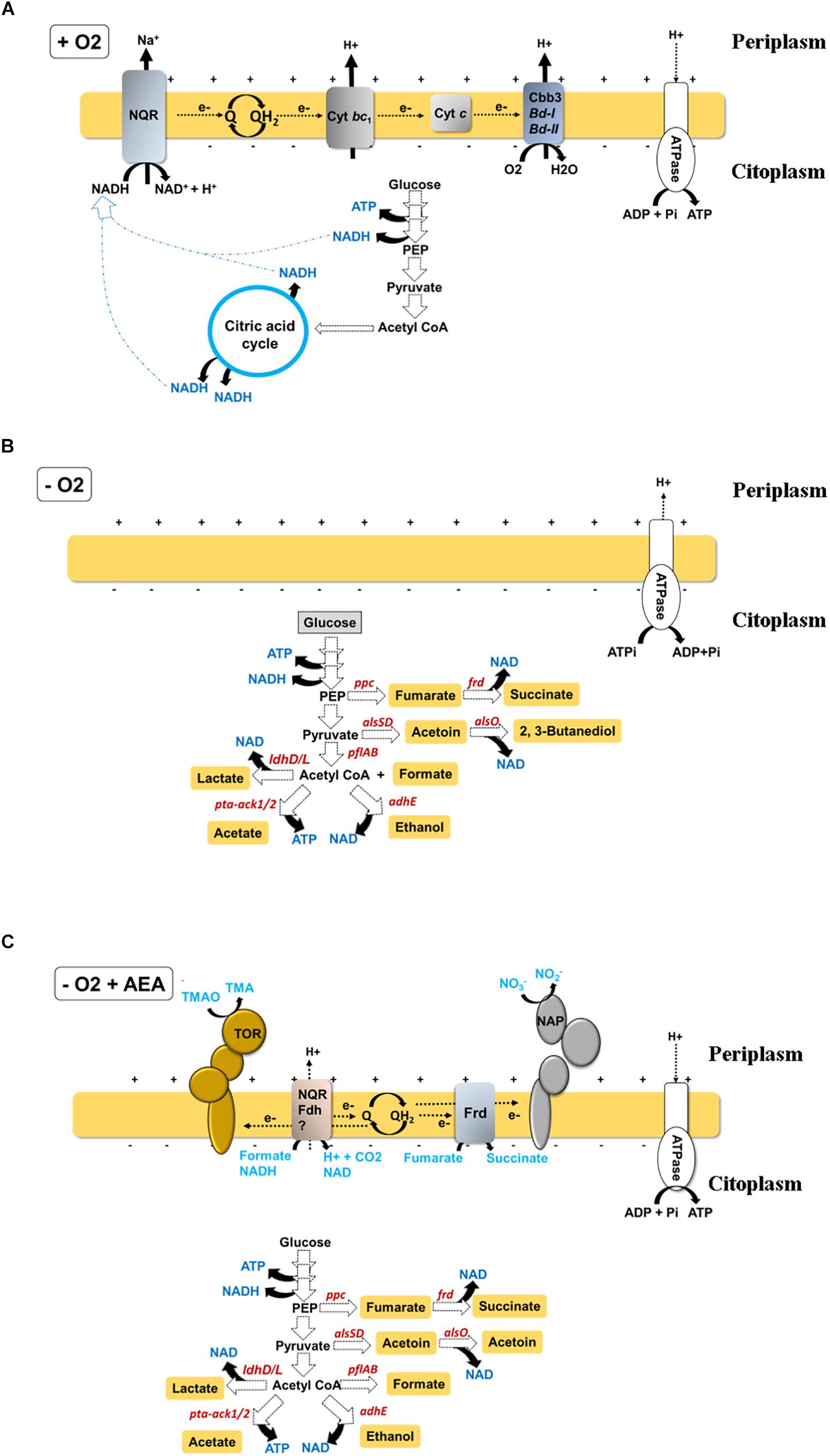
Figure 1. Schematic of redox balance and energy generating pathways in V. cholerae during (A) oxic growth, where V. cholerae obtains ATP through oxidative phosphorylation by respiration of oxygen using an electron transport chain initiated by a Na+-translocating NADH:quinone oxidoreductase (Na+-NQR), (B) hypoxic growth, where V. cholerae obtains energy by fermentation. In this condition, ATP is generated by substrate-level phosphorylation. As in the absence of final electron acceptors respiration is inhibited, the proton motive force (PMF) is established by proton pumping by the ATPase (with ATP consumption), and by sodium transporters (C) hypoxic growth in the presence of AEA, where V. cholerae is able to simultaneously obtain energy by substrate-level phosphorylation during fermentation and respiration of AEA. Represented in each scheme are only components experimentally demonstrated as relevant for the growth of the bacteria. Other components of the respiratory chain, such as alternative NADH dehydrogenases (see Table 1) whose inactivation does not affect V. cholerae growth are not shown.
Although atmospheric oxygen concentration is 20.9%, partial pressure of oxygen in water drops to approximately 1% and fluctuates depending on the respiratory activity of neighboring organisms and the relative distance to the aerial surface (Mortimer, 1981). Furthermore, within the human small intestine, oxygen concentrations descend below 3% (Crompton et al., 1965; He et al., 1999) and are further lowered to a nearly anoxic environment by the action of the commensal microbiota and host metabolism (Byndloss et al., 2017).
To adapt to these shifts in oxygen concentrations, V. cholerae encodes a repertoire of four respiratory oxygen reductases (Heidelberg et al., 2000): three bd-type oxygen reductases that receive electrons directly from the ubiquinol pool (Yang et al., 2008) and one cbb3-type haem-copper oxygen reductase (Hemp et al., 2005) that receives electrons from the membrane-bound bc1 complex. These enzymes display a distinct affinity for oxygen (Esposti et al., 2019); however, biochemical studies on these respiratory complexes in V. cholerae are lacking.
In the total absence of oxygen as electron acceptor, V. cholerae can also grow by respiring (i.e., reducing) a variety of organic and inorganic alternative electron acceptors (AEA), including fumarate, nitrate (NO3–) (Bueno et al., 2018), and trimethylamine N-oxide (TMAO) (Braun and Thöny-Meyer, 2005; Lee et al., 2012). In niches where both O2 and an AEA are absent, V. cholerae has the capacity to grow fermenting diverse carbohydrates such as sucrose, dextrin, maltose, glucose, mannitol, sorbitol, lactose, and starch (Nobechi, 1925; Wang et al., 2009). Fermentation in V. cholerae varies depending on the strain. El Tor N16961 has the capacity to produce 2,3-butanediol as a fermentative neutral end product, avoiding acidification of the medium. In contrast, the classical biotype O395 is unable to synthesize 2,3-butanediol, and hence its viability is compromised during glucose mixed fermentation due to acidification of the medium by synthesis of organic acids (Yoon and Mekalanos, 2006; Lee et al., 2020).
Despite the importance of hypoxic metabolism in human pathogens, experimental studies in laboratories are still carried out in the presence of oxygen, conditions that pathogens will never face. In this review we dissect the metabolic pathways employed by V. cholerae to prevail when oxygen is scarce, with an emphasis on infective processes.
Adaptations of V. Cholerae to Hypoxia
Under standard experimental conditions (i.e., aeration, 37°C, in LB medium), V. cholerae divides roughly every 16–20 min, yielding around 3 × 109 CFU after 12 h of growth. Under these conditions, V. cholerae cells obtain energy in the form of ATP by respiration of oxygen, as eukaryotic mitochondria do (Figure 1A). However, in contrast to eukaryotic cells, V. cholerae is also able to generate energy and maintain its physiological functions in the absence of oxygen using AEA such as TMAO, fumarate and nitrate (Braun and Thöny-Meyer, 2005; Lee et al., 2012; Bueno et al., 2018; Figure 1C). In contrast to other enteric pathogens, V. cholerae cannot obtain energy to grow under hypoxia using DMSO (Braun and Thöny-Meyer, 2005; Lee et al., 2012), tetrathionate, or sulfate as AEA. In the absence of an AEA, V. cholerae still can survive by fermenting internal metabolic electron acceptors (EAs) derived from carbohydrate catabolism, such as pyruvate and acetyl coenzyme A (AcCoA) (Nobechi, 1924; Figure 1B).
In general, there is a hierarchy in the use of AEA, where the most efficient AEAs are reduced (NO3– and TMAO) followed by those that yield less energy (nitrite, DMSO, tetrathionate). Ultimately, in the absence of AEAs, redox reactions are balanced by fermentation, where energy is generated by substrate-level phosphorylation (Unden and Bongaerts, 1997).
Anaerobic Nitrate Respiration
Nitrate is an inorganic ion which is abundant in the environment and the human diet (Lundberg et al., 2004; World Health Organization [WHO], 2011; Weitzberg and Lundberg, 2013). Its respiration by bacteria can generate nitrogen gas (N2) or ammonium as final products. When the final product is N2 this process is known as denitrification, and it is one of the more important processes in the nitrogen cycle since it returns fixed nitrogen to the atmosphere and thereby completes the cycle (Martinez-Espinosa et al., 2011). This reductive process occurs in four steps, beginning with the reduction of NO3– to NO2–, followed by the sequential reduction to the intermediates NO, N2O and finally to N2. The enzymes involved in denitrification are nitrate-, nitrite-, nitric oxide-, and nitrous oxide reductases, encoded by nar/nap, nir/nrf, nor, and nos genes, respectively (Bueno et al., 2012) whose synthesis is highly coordinated since accumulation of some of the nitrogen oxide intermediates, such as nitrite and nitric oxide, is toxic for the bacterial cells. Contrary to the anaerobic respiration of other AEA (e.g., TMAO, fumarate), which use menaquinone (MQ) as linker between NADH dehydrogenase and the terminal reductases, denitrification uses ubiquinone (UQ) (E0′ = +100 mV) to transfer electrons to the nitrate reductase (E0′ = +433 mV) (Unden and Bongaerts, 1997; Table 1).
Despite the fact that anaerobic respiration is preferred over fermentation by most microbes, some enteropathogens such as Escherichia coli, Salmonella typhimurium, Citrobacter rodentium, and V. cholerae, simultaneously perform fermentation and respiration of NO3– (Bueno et al., 2018; Figure 1C). In this scenario, fermentative products may acidify the growth medium and protonate NO2–, the product of NO3– respiration, to nitrous acid HNO2 (Da Silva et al., 2006) which might cross the bacterial membrane to interfere with diverse metabolic functions of the organism (Rowe et al., 1979). Additionally, HNO2 has been suggested to act as an uncoupler of the PMF (Meijer et al., 1979; Sijbesma et al., 1996) and generates a variety of reactive nitrogen species (RNS), including nitric oxide (NO), nitrogen dioxide (NO2), dinitrogen trioxide (N2O3), dinitrogen tetraoxide (N2O4), nitrite (NO2–), S-nitrosothiols, peroxinitrite (ONOO–) and nitrate. This repertoire of RNS may interact with numerous targets within microbial cells, including tyrosine residues of proteins, thiols and metal centers, and nucleotide and membrane lipids (Fang, 1997; Nathan and Shiloh, 2000), thereby inhibiting their biological functions.
The denitrification pathway is generally conserved among enteric pathogens (Arkenberg et al., 2011) but not in V. cholerae, which, despite having a periplasmic nitrate reductase Nap operon (vca0676-80) lacks the nitrite (Nir/Nrf), nitric oxide (Nor), or nitrous oxide (Nos) reductases (Heidelberg et al., 2000). As a consequence, V. cholerae accumulates toxic nitrite in anoxia in the presence of nitrate and so it was believed that it was unable to grow by nitrate respiration (Braun and Thöny-Meyer, 2005). However, it has been demonstrated that this inability was due to membrane potential dissipation by the formation of nitrous acid under fermentative acidifying conditions. Thus, the capacity of V. cholerae to grow by nitrate reduction, as denitrifiers do, was achieved by buffering the pH of the growth medium, minimizing the production of nitrous acid (Bueno et al., 2018). Although in microbiology the capacity to grow is always considered a positive trait, it was found that the growth arrest exerted by nitrite accumulation under low pH also had a relevant function for V. cholerae and other enteric pathogens, as it lead to increased cell viability during fermentative acidification of the media (Bueno et al., 2018; Figure 2B). Since nitrite accumulation is widespread among nitrate reducing bacterial species (Almeida et al., 1995; Bueno et al., 2008; Bergaust et al., 2010; Rowley et al., 2012), these results suggested an unprecedented role for nitrite in bacterial persistence.
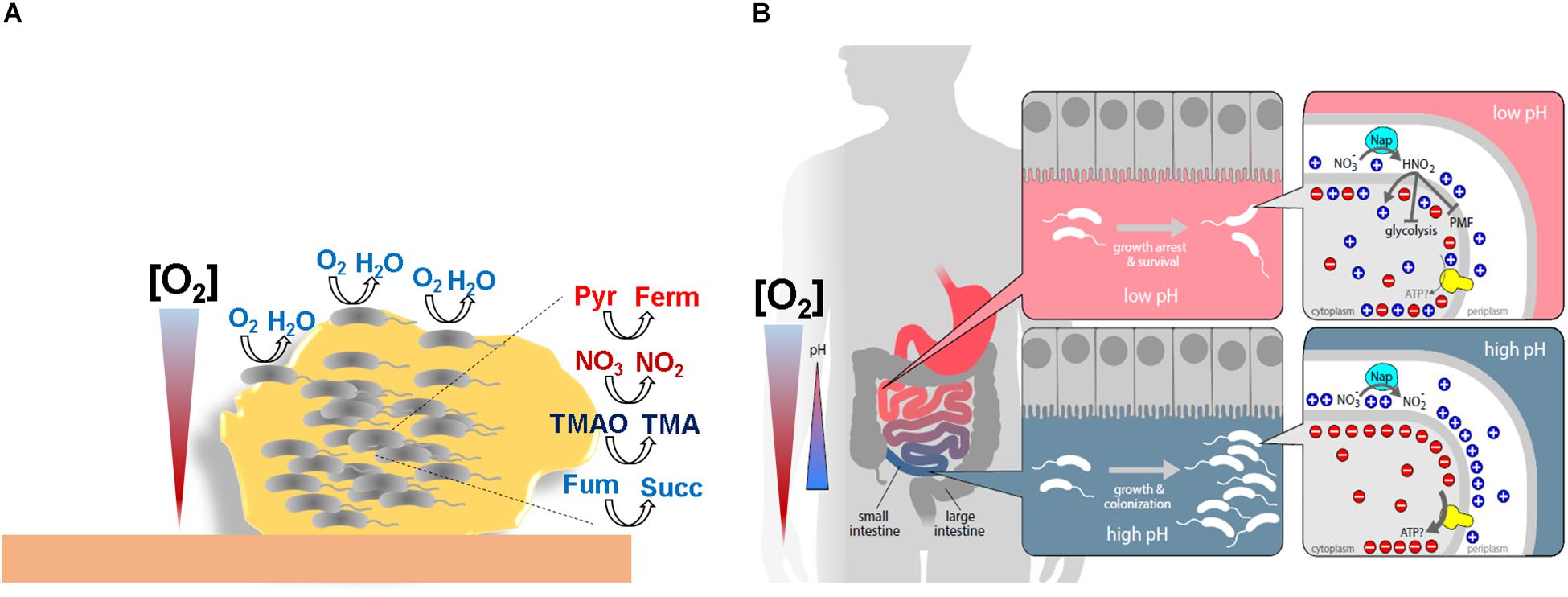
Figure 2. Schematic representations of V. cholerae niches where oxygen concentrations are limited. (A) Within the biofilm bacteria faces different oxygen concentrations. Cells situated in the periphery of the biofilm, where oxygen tensions are higher, will obtain energy through respiration of oxygen. However, cells situated in inner layers, where oxygen concentrations are scarce, will obtain energy through fermentation or/and nitrate, TMAO, fumarate respiration. (B) Human intestine colonization model showing the divergent outcomes during anaerobic nitrate respiration on bacterial expansion dependent on oxygen concentrations and pH. Pyr: pyruvate. Ferm: fermentative products. Fum: fumarate. Succ: succinate.
Regulation of the reduction of NO3– to NO2– has been comprehensively studied in E. coli. The transcriptional control of this process has been attributed to three systems: (i) Fnr transcriptional factor (fumarate nitrate reductase regulator), and the two-component systems (TCS): (ii) NarX-NarL, and (iii) NarQ-NarP (Rabin and Stewart, 1992; Noriega et al., 2010). Fnr is the transcriptional factor implicated in sensing external O2 concentrations and inducing the expression of anoxic lifestyle genes., such as the fermentative and the nitrate reduction nap/nar genes (Melville and Gunsalus, 1996; Lamberg and Kiley, 2000; Stewart, 2003; Browning et al., 2004; Crack et al., 2004). While NarXL in E. coli is a specific system for nitrate response, NarQP can sense nitrate, nitrite, and other signals (Lee et al., 1999; Noriega et al., 2010). The presence in V. cholerae of a homologous system to the E. coli NarQP suggests that the overall responses to nitrate and nitrogen oxides intermediates derived from its reduction might be entirely governed by this two-component system.
Anaerobic TMAO Respiration
TMAO is a small organic molecule abundant in the environment, aquatic animals and humans. In aquatic animals it supports homeostasis during changes in hydrostatic pressure and salinity (Pang et al., 1977; Zerbst-Boroffka et al., 2005). In humans, TMAO is mostly generated from trimethylamine (TMA) oxidation, which is a catabolic product of choline, carnitine, and phosphatidylcholine by the gut microbiota (Koeth et al., 2013).
When oxygen is scarce, TMAO can substitute for oxygen as the final electron acceptor in the V. cholerae respiratory chain to produce energy during its reduction to TMA (Figure 1C) by the TMAO reductase complex. TMAO reductase is encoded by the tor operon and consists of TorA, the TMAO reductase enzyme; TorC, a c-type cytochrome; and TorD, a specific chaperone for TorA (Silvestro et al., 1989; Pommier et al., 1998; Ansaldi et al., 1999). Since the redox potential of TMAO is lower than that of NO3– (E0′ = +130 mV vs +433 mV), menaquinone (E0′ = −74 mV), a quinone with lower redox potential, replaces ubiquinone as the adapter molecule for the electron transfer to TMAO (Table 1). Interestingly, aerobic expression of Tor in E. coli was demonstrated to be a stochastic bet-hedging strategy to avoid sudden anoxic entrapment (Carey et al., 2018). Whether such regulation of Tor is also present in V. cholerae is currently unknown.
One unique feature of anaerobic TMAO respiration in V. cholerae is that, while the presence of TMAO supports a rapid increase of biomass during the first 4 h of incubation, cell viability is largely impaired by a ppGpp-dependent response once stationary phase is reached (Oh et al., 2014). Interestingly, cholera toxin (CT) is produced during TMAO respiration (Childers and Klose, 2007; Lee et al., 2012), however, the CT inducer does not seem to be the respiration of TMAO itself but rather an intermediate product such as ROS followed by the ppGpp-dependent stringent response (Lee et al., 2012; Oh et al., 2014). In fact, addition of H2O2 in the presence of TMAO also enhanced the synthesis of CT. These results might suggest that TMAO-dependent CT production in V. cholerae may be linked with the stress conditions experienced by the pathogen while colonizing the host and thus it might aid the development of the infection.
Expression of the torCAD operon is controlled by a three-component system composed by the proteins TorT, TorR and TorS (Baraquet et al., 2006; Moore and Hendrickson, 2012). When there is TMAO in the medium, the periplasmic sensor TorT interacts with the membrane-bound protein TorS and this phosphorylates TorR, which ultimately induces the expression of torCAD. In contrast to other alternative respiratory systems, expression of the Tor system is not induced by the absence of oxygen in E. coli (Ansaldi et al., 2007) and it is independent from the global hypoxia regulators (i.e., FNR and ArcBA) (Pascal et al., 1984; Lynch and Lin, 1996) or from AEA signals such as nitrate (Pascal et al., 1984; Ansaldi et al., 2007). In addition to TorSR, TMAO respiration in V. cholerae is also governed by the catabolite repression protein (CRP), impairing TMAO reduction when glucose is available under anaerobic conditions. It has been proposed that the depletion of intracellular cAMP under high glucose concentrations leads to the inactivation of CRP and, as a consequence, expression of the TMAO reductase TorA is abrogated (Oh et al., 2015).
Despite that dimethyl sulfoxide (DMSO) is an organic compound analogous to TMAO, and it is also serves as final electron acceptor in some bacterial species (McCrindle et al., 2005), V. cholerae is unable to use it for anaerobic respiration (Braun and Thöny-Meyer, 2005; Lee et al., 2012).
Anaerobic Fumarate Respiration
Fumarate is an intermediate in the tricarboxylic acid (TCA) cycle, which can also replace the role of oxygen as respiratory final electron acceptor in V. cholerae (Figure 1C; Braun and Thöny-Meyer, 2005; Lee et al., 2012). Although fumarate presents a very low redox potential (E0′ = 30 mV), it supports robust anaerobic growth in V. cholerae (Braun and Thöny-Meyer, 2005; Lee et al., 2012) and as for TMAO respiration, menaquinone is used as the ETC adaptor for fumarate (Table 1). In aerobiosis, fumarate is generated from succinate by the TCA cycle enzyme succinate dehydrogenase (Sdh), but in the absence of oxygen, the reverse reaction can be catalyzed by the fumarate reductase (Frd), hence allowing the resulting fumarate to be used as the AEA.
The fumarate reductase in V. cholerae is a membrane-bound complex composed of four polypeptides designated FrdA, FrdB, FrdC, and FrdD (VC2656-2659) (Heidelberg et al., 2000). The catalytic FrdAB components face the inner side of the cytoplasmic membrane, where fumarate is reduced to succinate (Lemire et al., 1982). Despite being the most extensive form of anaerobic respiration, regulation of fumarate respiration in V. cholerae, and its importance for colonization during infection, is not yet known. In E.coli, FNR and Arc control the expression of the frdABCD operon during hypoxic conditions (Jones and Gunsalus, 1987; Iuchi and Lin, 1988; Park et al., 1995) but if NO3– is present in the environment, expression of the frd operon is suppressed by the Nar two-component regulatory system (Gunsalus, 1992). Therefore, the response to nitrate is dominant over anoxia in modulating the expression of the fumarate respiration machinery (Jones and Gunsalus, 1985, Gunsalus, 1987). In addition, exogenous fumarate also regulates the frdABCD operon via the DcuSR regulatory two-component system (Zientz et al., 1998). Since TMAO does not appear to control fumarate reductase gene expression it was suggested that E. coli does not display a hierarchical use of TMAO over fumarate (Jones and Gunsalus, 1987).
Fermentative Metabolism
When both oxygen and an AEA are absent in the environment, V. cholerae is still able to generate energy and maintain its physiological functions by inducing the fermentative pathway (Figure 1B). In comparison to the AEA-dependent pathways, fermentation is the lowest energy generating pathway where energy is produced by substrate-level phosphorylation and pyridine nucleotides replace quinones as intermediate electron drivers.
Fermentation can be divided into two parts: first, glucose oxidation produces NADH and pyruvate, and second, pyruvate is reduced and NAD+ regenerated. Although there is only a one pathway for the first step, there are multiple alternatives for the second, which produces lactate, acetate, ethanol, formate and 2,3-Butanediol (Yoon and Mekalanos, 2006; Hawver et al., 2016; Bueno et al., 2018).
While reduction of pyruvate to lactate and ethanol provides a mechanism for NAD+ regeneration, the acetate branch generates energy in the form of ATP (Figure 1B). The lactate dehydrogenase enzyme (Ldh) catalyzes the reduction of pyruvate to lactate. Although inactivation of E. coli Ldh does not have any effect on hypoxic growth in vitro (Mat-Jan et al., 1989), the role of the V. cholerae lactate dehydrogenase LdhA is still unknown.
AcCoA is a vital metabolic intermediate for any living organism (Pietrocola et al., 2015). When oxygen is present, the so-called pyruvate dehydrogenase complex (PDC) transforms pyruvate to AcCoA with the generation of NADH, however, under hypoxia, the high NADH/NAD+ ratio inactivates the PDH complex (Hansen and Henning, 1966). Under these conditions, pyruvate will then be converted to AcCoA by the oxygen sensitive PflAB pyruvate formate-lyase complex (PFL), with the co-production of formate, which is a common electron donor during anaerobic nitrate respiration and the substrate for dihydrogen production (Cole and Wimpenny, 1968; Garland et al., 1975). In contrast to the oxidation of pyruvate to AcCoA in the presence of oxygen, AcCoA generation under anaerobiosis does not generate reducing equivalents. Thus, formate production during fermentation is the best suited reaction for balancing cellular redox status. Given its toxicity, formate is rapidly pumped out into the bacterial periplasm by the transporter FocA where it is used as electron donor by the FDH-N or FDH-O formate dehydrogenases for nitrate or oxygen respiration, respectively. This process is coupled with the generation of PMF (Clark, 1989; Sawers, 2005). Under hypoxic conditions, inactivation of PFL in E. coli reduces energy production and as consequence, the bacterial growth yield is impaired. The bacterium is able to counter this defect by the coupled induction of the glycolytic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and lactate dehydrogenase, to increase cellular ATP and dissipate the excess of NADH produced by GAPDH, respectively (Zhu and Shimizu, 2004). The importance of this pathway in the hypoxic metabolism of V. cholerae also remains unknown.
The second NAD+ regenerating pathway consists of the reduction of AcCoA to acetaldehyde and this to ethanol by the alcohol dehydrogenase (AdhE) protein (Clark, 1989). Inactivation of AdhE in V. cholerae renders strains incapable of growing under fermentative conditions with glucose as the sole carbon source (Bueno et al., 2018), suggesting that NAD+ regeneration cannot be compensated by other NAD+ producing branches such as reduction of pyruvate to lactate. Interestingly, addition of an external electron acceptor such as nitrate relieves the redox and energetic constraints supporting V. cholerae growth (Bueno et al., 2018).
Acetate is generated in two steps, AcCoA produced in glycolysis is converted to acetyl-phosphate by the phosphotransferase PTA, and subsequently, the acetate kinase Ack generates acetate. Inactivation of this fermentative branch in E. coli provokes a remarkable perturbation of the bacterial fermentative profile under hypoxia which results in impaired bacterial growth rates and synthesis of fermentative intermediates, which is countered by lactate overflow (Castaño-Cerezo et al., 2009). In contrast to E. coli, V. cholerae presents two copies of ack, ack1, and ack2. Despite that the function of Ack remains unknown in V. cholerae, this seeming functional redundancy suggests the particular importance that this fermentative branch might have for the hypoxic lifestyle in V. cholerae.
2,3-Butanediol is produced by V. cholerae as well as by a few enterobacteria such as Serratia, Klebsiella, or Enterobacter species (White, 2000). Production of this compound benefits the cell since it decreases the production of fermentative organic acids, thus reducing the otherwise detrimental acidification of the medium. In V. cholerae, fermentation varies depending on the strain. While the El Tor biotype strain N16961 strain produces 2,3-butanediol, the classical biotype strain O395 is unable to synthesize this compound, and as a result its viability is compromised during glucose mixed fermentation (Yoon and Mekalanos, 2006). Interestingly, although V. cholerae biotype strain N16961 synthesizes 2,3-butanediol (Yoon and Mekalanos, 2006), we have observed that this compound does not control the drop of pH due to fermentation under strict anaerobic conditions in the biotype strain C6706 (Bueno et al., 2018). The presence of oxygen in the mentioned studies (Yoon and Mekalanos, 2006) might suggest that 2,3-Butanediol production in V. cholerae is reduced under hypoxic conditions, suggesting potential benefits of accumulating fermentative organic acids during anaerobic respiration.
In E. coli, fermentative metabolism is controlled by the FNR (fumarate, nitrate respiration) transcriptional regulator (Gunsalus, 1992; Becker et al., 1996) and the ArcBA (aerobic respiration control) two-component regulatory system (Spiro and Guest, 1991; Sawers and Suppmann, 1992; Gunsalus and Park, 1994; Guest et al., 1996; Georgellis et al., 1999; Levanon et al., 2005; Shalel-Levanon et al., 2005; Unden et al., 1995). However, expression of the adhE gene is governed by the Fis (Factor for inversion stimulation) and Cra (Catabolite repressor activator) proteins and the cellular NADH/NAD+ ratio (Membrillo-Hernández and Lin, 1999). The presence of homologous systems to FNR/ArcBA in V. cholerae might suggest their implication in regulating fermentative metabolism, however, their relevance for hypoxic metabolism is still unknown.
Despite the evident benefits of fermentative metabolism in facultative anaerobes, one drawback is the acidification of the growth medium by the production of organic acids. Fermentative enterobacteria, and especially V. cholerae, grow optimally at an alkaline pH. Hence, it was speculated that fermentative acidification to pH 4,5 impaired bacterial growth rate and viability. Surprisingly, we have found that fermentative acidification combined with nitrite production drives V. cholerae into a growth-arrested persistent mode. Interestingly, this survival mechanism is spread among diverse enteric pathogens underscoring a potential strategy to adapt to the hypoxic intestine during infection (Bueno et al., 2018). From an ecological and evolutionary point of view, these findings provide insights into why some bacteria retain certain processes that in principle can be considered undesirable under certain conditions. In addition to the role of toxic products such as nitrous acid during fermentative acidification and nitrate reduction, they could also be used by bacteria as metabolic weapons against their hosts during infection.
Relevance of Hypoxic Metabolism in V. Cholerae Niches
The life cycle of V. cholerae includes infective and environmental stages where V. cholerae accumulates and forms polymicrobial aggregates such as sludge, films, mats, flocs, or biofilms (Faruque et al., 2006; Islam et al., 2007; Tamayo et al., 2010). Biofilms present a dense structure that together with high bacterial respiratory activity results in an abrupt drop in their internal oxygen concentration (Xu et al., 1998; Walters et al., 2003; Werner et al., 2004; Jo et al., 2017; Karampatzakis et al., 2017). In addition to biofilms, V. cholerae faces low oxygen tensions and anoxia due to the oxidative metabolism of commensal microorganisms and host colonocytes in the human gut during infection (Zheng et al., 2015; Byndloss et al., 2017; Chun et al., 2017). Under such conditions, energy generation by canonical oxygen respiratory systems will not proceed and the bacterium must re-program its metabolism to obtain energy while keeping up its cellular redox status. V. cholerae might achieve this by (i) exchanging common oxygen respiratory terminal oxidases with oxidases that have higher affinity for oxygen (Heidelberg et al., 2000), (ii) using fermentative pathways, or (iii) using an AEA (Bueno et al., 2018; Figure 2A). Indeed, it has been demonstrated experimentally that biofilm formation in Pseudomonas aeruginosa, Streptococcus aureus, Escherichia coli, and Bacillus subtilis induces hypoxia related enzymes such as high-oxygen-affinity terminal oxidases (Jo et al., 2017), anaerobic ribonucleotide reductases (RNR) (Crespo et al., 2016), anaerobic threonine fermentation (Létoffé et al., 2017), mixed fermentative enzymes (Chen et al., 2015), and anaerobic nitrate respiration (Yoon et al., 2004), which are crucial for the development and maturation of the biofilm.
Although the relevance of hypoxic metabolism for V. cholerae biofilm development and turnover has not yet been investigated, induction of hypoxic related pathways in V. cholerae was identified by transcriptomic studies during colonization experiments using the mouse and rabbit infection models (Table 2; Merrell et al., 2002; Bina et al., 2003; Xu et al., 2003; Yoon and Mekalanos, 2006; Schild et al., 2007; Mandlik et al., 2011; Minato et al., 2014; Oh et al., 2014; Bueno et al., 2018).
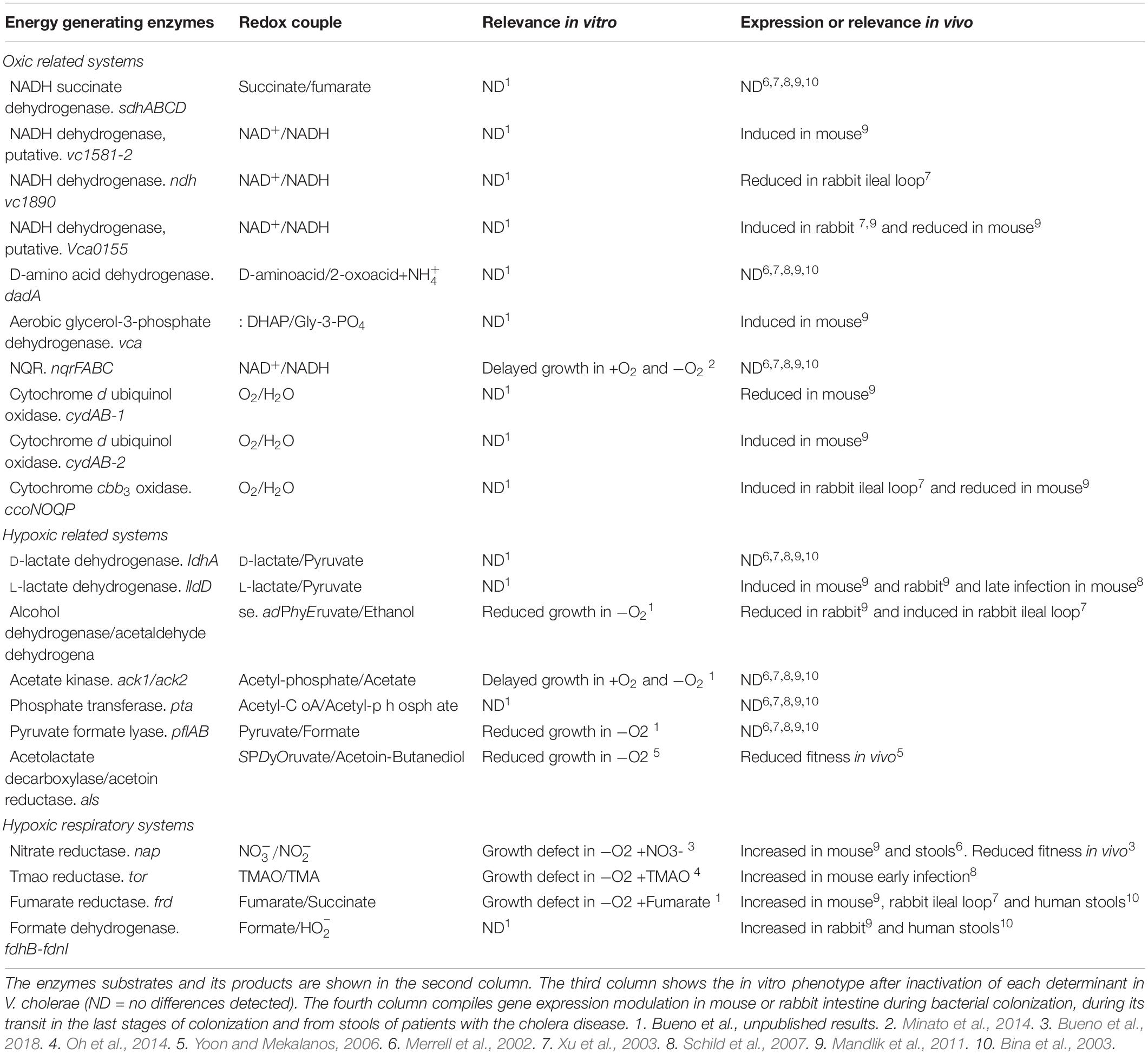
Table 2. Determinants supporting V. cholerae energetic metabolism under oxic, hypoxic and hypoxic respiratory conditions.
Regarding fermentative metabolism, expression of genes encoding the fermentative enzymes L-lactate dehydrogenase (L-LdhA), acetolactate decarboxylase/acetoin reductase (AlsSDO) and alcohol dehydrogenase AdhE were induced during V. cholerae colonization (Table 2; Xu et al., 2003; Yoon and Mekalanos, 2006; Schild et al., 2007; Mandlik et al., 2011). The prominent induction of the two branches which dissipate redox potential during hypoxic growth through the generation of lactate and ethanol might suggest that during infection V. cholerae experiences a remarkable redox constraint, probably due to the low oxygen concentrations prevalent in the overpopulated intestines. The als operon, implicated in the synthesis of 2,3-butanediol, has a role in maintaining the pH at neutral levels during fermentative processes (Yoon and Mekalanos, 2006). Therefore, production 2,3-butanediol by certain V. cholerae strains (e.g., N16961) underscores the importance of countering the acidifying fermentative processes during hypoxic growth in vivo. Similarly to fermentative metabolism, bacterial respiratory pathways also seem to be modulated during infective processes. Thus, expression of V. cholerae respiratory enzymes such as nitrate (Nap), TMAO (Tor), and fumarate reductases (Frd) were also induced during infection experiments in mice, and consistently, inactivation of Nap and Tor respiratory systems reduced V. cholerae fitness during intestinal colonization in mice (Table 2; Oh et al., 2014; Bueno et al., 2018). Interestingly, expression of the nitrate reductase and fumarate reductases was also enhanced in human stool samples, suggesting a possible role of these respiratory enzymes in the dissemination and transmission of V. cholerae (Table 2; Merrell et al., 2002; Bina et al., 2003).
In addition to the activation of hypoxic metabolism to maintain bacterial energy when oxygen is scarce, low oxygen concentrations could be a niche specific signal (e.g., indicating a host niche), able to trigger virulence programs in pathogens. Indeed, induction of the toxin-coregulated pilus (TCP) was detected when V. cholerae cultures were subjected to oxygen deprivation (Marrero et al., 2009), suggesting that hypoxia might be a key signal used by the pathogen to determine niche localization. In addition, induction of TCP was also found to be dependent on low oxygen concentrations by reduction of the cysteine-containing regulatory protein OhrR in concert with AphB (Liu et al., 2011, 2016). Although anoxia itself seemed not to induce the synthesis of the CT in V. cholerae (Krishnan et al., 2004; Marrero et al., 2009), CT was highly stimulated by the presence of TMAO as final electron acceptor (Lee et al., 2012; Oh et al., 2014). Interestingly, TMAO respiration during host colonization has only been shown to be relevant in the enteropathogen V. cholerae.
The importance of hypoxic metabolism in infection has also been reported in other pathogenic bacteria. In Brucella suis for example, persistence within mouse macrophages is mediated by the induction of the high-oxygen-affinity terminal oxidase cbb3 (Jiménez De Bagüés et al., 2007). The fermentative pathway was also found to be induced in pathogenic bacteria during infection (Beckham et al., 2014; Luong et al., 2015). Similarly, the anaerobic respiration of nitrate (Winter et al., 2013; Lopez et al., 2015), tetrathionate (Winter et al., 2010), 1,2-propanediol, and fumarate (Ge et al., 2000; Jones et al., 2007) was induced during infection, and enhanced pathogen survival and persistence within the host.
Altogether, this experimental evidence suggests that hypoxic metabolism plays an important role in a variety of bacterial pathogens under both free-living conditions and during infective processes. However, whether induction of hypoxic metabolism is strictly only required for balancing bacterial energetics or if it has also a direct role in bacterial pathogenicity during infective stages (e.g., synthesis of bacterial effectors and toxins) still needs to be addressed.
Conclusion and Future Perspectives
Human infectious diseases caused by bacterial pathogens remain a global concern for public health care systems which result in millions of deaths per year worldwide. Oxygen concentration is low in most tissues within the host. Therefore, the standard laboratory aerobic culture of bacteria is undoubtedly far from mimicking the real scenario within the host and will result in misleading findings in understanding the molecular mechanisms used by pathogenic bacteria to cause diseases. Thus, considering that the hypoxic metabolic state is the prevalent physiological condition of pathogens during infection, and adopting adequate methodologies for their studies is a must in order to solve new enigmas in host-pathogen interactions.
Despite our deep knowledge of microbial fermentative and respiratory pathways, a comprehensive understanding of the crosstalk among hypoxic processes is still missing. In the environment or within the host, several types of fermentable carbon sources and respiratory terminal acceptors are simultaneously available, and hence, understanding pathogen responses while co-metabolizing different hypoxic substrates is an important question that remains to be addressed. In addition, with the advent of novel microscopy techniques, flow cytometry and microfluidic devices it will be possible to visualize the dynamics of those systems in real-time.
Regardless of the importance of studying the behavior of clonal bacterial populations, it has been demonstrated that small sub-populations can present altered behavior within the community (Veening et al., 2008; Lycus et al., 2018). Thus, understanding hypoxic regulation at single cell level represents another missing piece in better understanding population responses. In fact, in nature there are multiple cases of metabolic cooperation and competition between organisms co-inhabiting a niche. However, how hypoxic metabolic products influence or modulate the host’s microbiota and immune system during infection is unknown. Hence, given that many enteropathogens accumulate nitrite, it is possible that nitrite may be instrumental for pathogenic bacteria to compete against neighboring commensals and/or modulate the host immune system during infection. Future research combining omic data with single cell mechanistic insights will shed light on how regulation of hypoxic metabolism is orchestrated both in vitro and in vivo.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
EB research is supported by the Kempe Foundation. EB and VP are thankful to the Swedish Research Council, Laboratory for Molecular Infection Medicine Sweden (MIMS) and UCMR for the financial support. Research in the Cava lab is supported by MIMS, the Knut and Alice Wallenberg Foundation (KAW), the Swedish Research Council, and the Kempe Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Michael Gilmore for proofreading the manuscript.
References
Almeida, J. S., Júlio, S. M., Reis, M. A. M., and Carrondo, M. J. T. (1995). Nitrite inhibition of denitrification by Pseudomonas fluorescens. Biotechnol. Bioeng. 46, 194–201. doi: 10.1002/bit.260460303
Ansaldi, M., Bordi, C., Lepelletier, M., and Méjean, V. (1999). TorC apocytochrome negatively autoregulates the trimethylamine N-oxide (TMAO) reductase operon in Escherichia coli. Mol. Microbiol. 33, 284–295. doi: 10.1046/j.1365-2958.1999.01468.x
Ansaldi, M., Théraulaz, L., Baraquet, C., Panis, G., and Méjean, V. (2007). Aerobic TMAO respiration in Escherichia coli. Mol. Microbiol. 66, 484–494. doi: 10.1111/j.1365-2958.2007.05936.x
Arkenberg, A., Runkel, S., Richardson, D. J., and Rowley, G. (2011). The production and detoxification of a potent cytotoxin, nitric oxide, by pathogenic enteric bacteria. Biochem. Soc. Trans. 39, 1876–1879. doi: 10.1042/BST20110716
Baraquet, C., Théraulaz, L., Guiral, M., Lafitte, D., Méjean, V., and Jourlin-Castelli, C. (2006). TorT, a member of a new periplasmic binding protein family, triggers induction of the Tor respiratory system upon trimethylamine N-oxide electron-acceptor binding in Escherichia coli. J. Biol. Chem. 281, 38189–38199. doi: 10.1074/jbc.M604321200
Becker, S., Holighaus, G., Gabrielczyk, T., and Unden, G. (1996). O2 as the regulatory signal for FNR-dependent gene regulation in Escherichia coli. J. Bacteriol. 178, 4515–4521. doi: 10.1128/jb.178.15.4515-4521.1996
Beckham, K. S. H., Connolly, J. P. R., Ritchie, J. M., Wang, D., Gawthorne, J. A., Tahoun, A., et al. (2014). The metabolic enzyme AdhE controls the virulence of Escherichia coliO157: H7. Mol. Microbiol. 93, 199–211. doi: 10.1111/mmi.12651
Bergaust, L., Mao, Y., Bakken, L. R., and Frostegård, Å (2010). Denitrification response patterns during the transition to anoxic respiration and posttranscriptional effects of suboptimal ph on nitrogen oxide reductase in paracoccus denitrificans. Appl. Environ. Microbiol. 76, 6387–6396. doi: 10.1128/AEM.00608-610
Bina, J., Zhu, J., Dziejman, M., Faruque, S., Calderwood, S., and Mekalanos, J. (2003). ToxR regulon of Vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. U.S.A. 100, 2801–2806. doi: 10.1073/pnas.2628026100
Braun, M., and Thöny-Meyer, L. (2005). Cytochrome c maturation and the physiological role of c-type cytochromes in Vibrio cholerae. J. Bacteriol. 187, 5996–6004. doi: 10.1128/JB.187.17.5996-6004.2005
Browning, D. F., Cole, J. A., and Busby, S. J. W. (2004). Transcription activation by remodelling of a nucleoprotein assembly: the role of NarL at the FNR-dependent Escherichia coli nirpromoter. Mol. Microbiol. 53, 203–215. doi: 10.1111/j.1365-2958.2004.04104.x
Bueno, E., Bedmar, E. J., Richardson, D. J., and Delgado, M. J. (2008). Role of Bradyrhizobium japonicum cytochrome c550 in nitrite and nitrate respiration. FEMS Microbiol. Lett. 279, 188–194. doi: 10.1111/j.1574-6968.2007.01034.x
Bueno, E., Mesa, S., Bedmar, E. J., Richardson, D. J., and Delgado, M. J. (2012). Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid. Redox Signal. 16, 819–852. doi: 10.1089/ars.2011.4051
Bueno, E., Sit, B., Waldor, M. K., and Cava, F. (2018). Anaerobic nitrate reduction divergently governs population expansion of the enteropathogen Vibrio cholerae. Nat. Microbiol. 3:1346. doi: 10.1038/s41564-018-0253-0
Byndloss, M. X., Olsan, E. E., Rivera-chávez, F., Tiffany, C. R., Cevallos, S. A., Lokken, K. L., et al. (2017). Microbiota-activated PPAR- g signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 575, 570–575. doi: 10.1126/science.aam9949
Carey, J. N., Mettert, E. L., Roggiani, M., Myers, K. S., Kiley, P. J., Goulian, M., et al. (2018). Regulated stochasticity in a bacterial signaling network permits tolerance to a rapid environmental article regulated stochasticity in a bacterial signaling network permits tolerance to a rapid environmental change. Cell 173, 196.e14–207.e14. doi: 10.1016/j.cell.2018.02.005
Castaño-Cerezo, S., Pastor, J. M., Renilla, S., Bernal, V., Iborra, J. L., and Cánovas, M. (2009). An insight into the role of phosphotransacetylase (pta) and the acetate/acetyl-CoA node in Escherichia coli. Microb. Cell Fact. 8, 1–19. doi: 10.1186/1475-2859-8-54
Chen, Y., Gozz, K., Yan, F., and Chai, Y. (2015). Acetic acid acts as a volatile signal to stimulate bacterial biofilm formation. mBio 6, 406–408. doi: 10.1128/mBio.00392-315
Childers, B. M., and Klose, K. E. (2007). Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol. 2, 335–344. doi: 10.2217/17460913.2.3.335
Chun, C., Zheng, L., and Colgan, S. P. (2017). Tissue metabolism and host-microbial interactions in the intestinal mucosa. Free Radic. Biol. Med. 105, 86–92. doi: 10.1016/j.freeradbiomed.2016.09.027
Clark, D. P. (1989). The fermentation pathways of Escherichia coli. FEMS Microbiol. Lett. 63, 223–234. doi: 10.1016/0378-1097(89)90132-90138
Cole, J. A., and Wimpenny, J. W. T. (1968). Metabolic pathways for nitrate reduction in Escherichia coli. BBA Bioenerg. 162, 39–48. doi: 10.1016/0005-2728(68)90212-90210
Crack, J., Green, J., and Thomson, A. (2004). Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR). J. Biol. Chem. 279, 9278–9286. doi: 10.1074/jbc.m309878200
Crespo, A., Pedraz, L., Astola, J., and Torrents, E. (2016). Pseudomonas aeruginosa exhibits deficient biofilm formation in the absence of class II and III ribonucleotide reductases due to hindered anaerobic growth. Front. Microbiol. 7:688. doi: 10.3389/fmicb.2016.00688
Crompton, D. W., Shrimpton, D. H., and Silver, I. A. (1965). Measurements of the oxygen tension in the lumen of the small intestine of the domestic duck. J. Exp. Biol. 43, 473–478.
Da Silva, G., Kennedy, E. M., and Dlugogorski, B. Z. (2006). Ab initio procedure for aqueous-phase pKa calculation: the acidity of nitrous acid. J. Phys. Chem. A 110, 11371–11376. doi: 10.1021/jp0639243
De Haan, L., and Hirst, T. R. (2004). Cholera toxin: a paradigm for multi-functional engagement of cellular mechanisms (Review). Mol. Membr. Biol. 21, 77–92. doi: 10.1080/09687680410001663267
Esposti, M. D., Mentel, M., Martin, W., and Sousa, F. L. (2019). Oxygen reductases in alphaproteobacterial genomes: physiological evolution from low to high oxygen environments. Front. Microbiol. 10:499. doi: 10.3389/fmicb.2019.00499
Fang, F. C. (1997). Perspectives series: host / pathogen interactions. Mechanisms of nitric oxide-related antimicrobial activity. J. Clin. Invest. 99, 2818–2825. doi: 10.1172/jci119473
Faruque, S. M., Biswas, K., Udden, S. M. N., Ahmad, Q. S., Sack, D. A., Nair, G. B., et al. (2006). Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U.S.A. 103, 6350–6355. doi: 10.1073/pnas.0601277103
Fu, Y., Ho, B. T., and Mekalanos, J. J. (2018). Tracking Vibrio cholerae cell-cell interactions during infection reveals bacterial population dynamics within intestinal microenvironments. Cell Host Microbe 23, 274.e2–281.e2. doi: 10.1016/j.chom.2017.12.006
Garland, P. B., Downie, J. A., and Haddock, B. A. (1975). Proton translocation and respiratory nitrate reductase of Escherichia coli. Biochem. J. 152, 547–559. doi: 10.1042/bj1520547
Ge, Z., Feng, Y., Dangler, C. A., Xu, S., Taylor, N. S., and Fox, J. G. (2000). Fumarate reductase is essential for Helicobacter pylori colonization of the mouse stomach. Microb. Pathog. 29, 279–287. doi: 10.1006/mpat.2000.0391
Georgellis, D., Kwon, O., and Lin, E. C. C. (1999). Amplification of signaling activity of the arc two-component system of Escherichia coli by anaerobic metabolites. An in vitro study with different protein modules. J. Biol. Chem. 274, 35950–35954. doi: 10.1074/jbc.274.50.35950
Guest, J. R., Green, J., Irvine, A. S., and Spiro, S. (1996). “The FNR Modulon and FNR-regulated gene expression,” in Regulation of Gene Expression in Escherichia coli, (Boston, MA: Springer), 317–342. doi: 10.1007/978-1-4684-8601-8_16
Gunsalus, R. P. (1992). Control of electron flow in Escherichia coli: coordinated transcription of respiratory pathway genes. J. Bacteriol. 174, 7069–7074. doi: 10.1128/jb.174.22.7069-7074.1992
Gunsalus, R. P., and Park, S. J. (1994). Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res. Microbiol. 145, 437–450. doi: 10.1016/0923-2508(94)90092-90092
Hanne, L. F., and Finkelstein, R. A. (1982). Characterization and distribution of the hemagglutinins produced by Vibrio cholerae. Infect. Immun. 36, 209–214. doi: 10.1128/iai.36.1.209-214.1982
Hansen, R. G., and Henning, U. (1966). Regulation of pyruvate dehydrogenase activity in Escherichia coli K12. BBA Enzymol. Biol. Oxid. 122, 355–358. doi: 10.1016/0926-6593(66)90076-90072
Häse, C. C., and Mekalanos, J. J. (1998). TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 95, 730–734. doi: 10.1073/pnas.95.2.730
Hawver, L. A., Giulietti, J. M., Baleja, J. D., and Ng, W.-L. (2016). Quorum sensing coordinates cooperative expression of pyruvate. mBio 7, 1–11. doi: 10.1128/mBio.01863-16
He, G., Shankar, R. A., Chzhan, M., Samouilov, A., Kuppusamy, P., and Zweier, J. L. (1999). Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. U.S.A. 96, 4586–4591. doi: 10.1073/pnas.96.8.4586
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
Hemp, J., Christian, C., Barquera, B., Gennis, R. B., and Martínez, T. J. (2005). Helix switching of a key active-site residue in the cytochrome cbb 3 oxidases. Biochemistry 44, 10766–10775. doi: 10.1021/bi050464f
Islam, M. S., Rahman, M. Z., Shafiqu, M., Islam, M. S. K., Sack, D. A., Jahid, M. I. K., et al. (2007). Biofilm acts as a microenvironment for plankton-associated Vibrio cholerae in the aquatic environment of Bangladesh. Microbiol. Immunol. 51, 369–379. doi: 10.1111/j.1348-0421.2007.tb03924.x
Iuchi, S., and Lin, E. C. C. (1988). areA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. U.S.A. 85, 1888–1892. doi: 10.1073/pnas.85.6.1888
Jiménez De Bagüés, M. P., Loisel-Meyer, S., Liautard, J. P., and Jubier-Maurin, V. (2007). Different roles of the two high-oxygen-affinity terminal oxidases of Brucella suis: cytochrome c oxidase, but not ubiquinol oxidase, is required for persistence in mice. Infect. Immun. 75, 531–535. doi: 10.1128/IAI.01185-06
Jo, J., Cortez, K. L., Cornell, W. C., Price-Whelan, A., and Dietrich, L. E. P. (2017). An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. eLife 6, 1–30. doi: 10.7554/eLife.30205
Jones, H. M., and Gunsalus, R. P. (1985). Transcription of the Escherichia coli fumarate reductase genes (frdABCD) and their coordinate regulation by oxygen, nitrate, and fumarate. J. Bacteriol. 164, 1100–1109. doi: 10.1128/jb.164.3.1100-1109.1985
Jones, H. M., and Gunsalus, R. P. (1987). Regulation of Escherichia coli fumarate reductase (frdABCD) operon expression by respiratory electron acceptors and the fnr gene product. J. Bacteriol. 169, 3340–3349. doi: 10.1128/jb.169.7.3340-3349.1987
Jones, S. A., Chowdhury, F. Z., Fabich, A. J., Anderson, A., Schreiner, D. M., House, A. L., et al. (2007). Respiration of Escherichia coli in the mouse intestine. Infect. Immun. 75, 4891–4899. doi: 10.1128/IAI.00484-07
Karampatzakis, A., Sankaran, J., Kandaswamy, K., Rice, S. A., Cohen, Y., and Wohland, T. (2017). Measurement of oxygen concentrations in bacterial biofilms using transient state monitoring by single plane illumination microscopy. Biomed. Phys. Eng. Express 3:035020. doi: 10.1088/2057-1976/aa6db7
Koeth, R. A., Wang, Z., Levison, B. S., Buffa, J. A., Org, E., Sheehy, B. T., et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585. doi: 10.1038/nm.3145
Krishnan, H. H., Ghosh, A., Paul, K., and Chowdhury, R. (2004). Effect of anaerobiosis on expression of virulence factors in Vibrio cholerae. Infect. Immun. 72, 3961–3967. doi: 10.1128/IAI.72.7.3961-3967.2004
Lamberg, K. E., and Kiley, P. J. (2000). FNR-dependent activation of the class II dmsA and narG promoters of Escherichia coli requires FNR-activating regions 1 and 3. Mol. Microbiol. 38, 817–827. doi: 10.1046/j.1365-2958.2000.02172.x
Lee, A. I., Delgado, A., and Gunsalus, R. P. (1999). Signal-dependent phosphorylation of the membrane-bound NarX two- component sensor-transmitter protein of Escherichia coli: nitrate elicits a superior anion ligand response compared to nitrite. J. Bacteriol. 181, 5309–5316. doi: 10.1128/jb.181.17.5309-5316.1999
Lee, D., Kim, E. J., Baek, Y., Lee, J., Youngbae, Y., Nair, G. B., et al. (2020). Alterations in glucose metabolism in Vibrio cholerae serogroup O1 El Tor biotype strains. Sci. Rep. 10:308. doi: 10.1038/s41598-019-57093-57094
Lee, K.-M., Park, Y., Bari, W., Yoon, M. Y., Go, J., Kim, S. C., et al. (2012). Activation of cholera toxin production by anaerobic respiration of trimethylamine N-oxide in Vibrio cholerae. J. Biol. Chem. 287:39742. doi: 10.1074/jbc.M112.394932
Lee, S. H., Hava, D. L., Waldor, M. K., and Camilli, A. (1999). Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99, 625–634. doi: 10.1016/s0092-8674(00)81551-2
Lemire, B. D., Robinson, J. J., and Weiner, J. H. (1982). Identification of membrane anchor polypeptides of Escherichia coli fumarate reductase. J. Bacteriol. 152, 1126–1131.
Létoffé, S., Chalabaev, S., Dugay, J., Stressmann, F., Audrain, B., Portais, J. C., et al. (2017). Biofilm microenvironment induces a widespread adaptive amino-acid fermentation pathway conferring strong fitness advantage in Escherichia coli. PLoS Genet. 13:e1006800. doi: 10.1371/journal.pgen.1006800
Levanon, S. S., San, K. Y., and Bennett, G. N. (2005). Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol. Bioeng. 89, 556–564. doi: 10.1002/bit.20381
Liu, Z., Wang, H., Zhou, Z., Kan, B., Goulian, M., and Zhu, J. (2016). Differential thiol-based switches jump-start Vibrio cholerae pathogenesis article differential thiol-based switches jump-start Vibrio cholerae pathogenesis. CellReports 14, 347–354. doi: 10.1016/j.celrep.2015.12.038
Liu, Z., Yang, M., Peterfreund, G. L., Tsou, A. M., Selamoglu, N., Daldal, F., et al. (2011). Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc. Natl. Acad. Sci. U.S.A. 108, 810–815. doi: 10.1073/pnas.1014640108
Lopez, C. A., Rivera-Chávez, F., Byndloss, M. X., and Bäumler, A. J. (2015). The periplasmic nitrate reductase NapABC supports luminal growth of Salmonella enterica serovar typhimurium during colitis. Infect. Immun. 83, 3470–3478. doi: 10.1128/IAI.00351-315
Lundberg, J. O., Weitzberg, E., Cole, J. A., and Benjamin, N. (2004). Nitrate, bacteria and human health. Nat. Rev. Microbiol. 2, 593–602. doi: 10.1038/nrmicro929
Luong, T. T., Kim, E. H., Bak, J. P., Nguyen, C. T., Choi, S., Briles, D. E., et al. (2015). Ethanol-induced alcohol dehydrogenase E (AdhE) potentiates pneumolysin in Streptococcus pneumoniae. Infect. Immun. 83, 108–119. doi: 10.1128/IAI.02434-2414
Lycus, P., Soriano-Laguna, M. J., Kjos, M., Richardson, D. J., Gates, A. J., Milligan, D. A., et al. (2018). A bet-hedging strategy for denitrifying bacteria curtails their release of N2O. Proc. Natl. Acad. Sci. U.S.A. 115, 11820–11825. doi: 10.1073/pnas.1805000115
Lynch, A. S., and Lin, E. C. C., eds. (1996). Regulation of aerobic and anaerobic metabolism by the arc system,” in Regulation of Gene Expression in Escherichia coli (Springer: Boston, MA). doi: 10.1007/978-1-4684-8601-8_18
Mandlik, A., Livny, J., Robins, W. P., Ritchie, J. M., and Mekalanos, J. J. (2011). Resource RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10, 165–174. doi: 10.1016/j.chom.2011.07.007
Marrero, K., Sánchez, A., Rodríguez-Ulloa, A., González, L. J., Castellanos-Serra, L., Paz-Lago, D., et al. (2009). Anaerobic growth promotes synthesis of colonization factors encoded at the Vibrio pathogenicity island in Vibrio cholerae El Tor. Res. Microbiol. 160, 48–56. doi: 10.1016/j.resmic.2008.10.005
Martinez-Espinosa, R. M., Cole, J. A., Richardson, D. J., and Watmough, N. J. (2011). Enzymology and ecology of the nitrogen cycle. Biochem. Soc. Trans. 39, 175–178. doi: 10.1042/bst0390175
Mat-Jan, F., Alam, K. Y., and Clark, D. P. (1989). Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J. Bacteriol. 171, 342–348. doi: 10.1128/jb.171.1.342-348.1989
McCrindle, S. L., Kappler, U., and McEwan, A. G. (2005). Microbial dimethylsulfoxide and trimethylamine-N-oxide respiration. Adv. Microb. Physiol. 50, 147–198.
Meijer, E. M., Zwaan, J. W., Stouthamer, A. H., and Wever, R. (1979). Anaerobic respiration and energy conservation in Paracoccus denitrficans. Eur. J. Biochem. 96, 69–76. doi: 10.1111/j.1432-1033.1979.tb13014.x
Mekalanos, J. J., Rubin, E. J., and Waldor, M. K. (1997). Cholera: molecular basis for emergence and pathogenesis. FEMS Immunol. Med. Microbiol. 18, 241–248. doi: 10.1016/S0928-8244(97)00054-0
Melville, S. B., and Gunsalus, R. P. (1996). Isolation of an oxygen-sensitive FNR protein of Escherichia coli: interaction at activator and repressor sites of FNR-controlled genes. Proc. Natl. Acad. Sci. U.S.A. 93, 1226–1231. doi: 10.1073/pnas.93.3.1226
Membrillo-Hernández, J., and Lin, E. C. C. (1999). Regulation of expression of the adhE gene, encoding ethanol oxidoreductase in Escherichia coli: transcription from a downstream promoter and regulation by Fnr and RpoS. J. Bacteriol. 181, 7571–7579. doi: 10.1128/jb.181.24.7571-7579.1999
Merrell, D. S., Qadri, F., Cohen, M. B., Calderwood, S. B., Schoolnik, G. K., Dolganov, N. A., et al. (2002). Host-induced epidemic spread of the cholera bacterium. Nature 417, 642–645. doi: 10.1038/nature00778
Minato, Y., Fassio, S. R., Reddekopp, R. L., and Häse, C. C. (2014). Inhibition of the sodium-translocating NADH-ubiquinone oxidoreductase [Na+-NQR] decreases cholera toxin production in Vibrio cholerae O1 at the late exponential growth phase. Microb. Pathog. 66, 36–39. doi: 10.1016/j.micpath.2013.12.002
Moore, J. O., and Hendrickson, W. A. (2012). An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure 20, 729–741. doi: 10.1016/j.str.2012.02.021
Mortimer, C. H. (1981). The oxygen content of air-saturated fresh waters over ranges of temperature and atmospheric pressure of limnological interest. SIL Commun. 22, 1–23. doi: 10.1080/05384680.1981.11904000
Nathan, C., and Shiloh, M. U. (2000). Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 97, 8841–8848. doi: 10.1073/pnas.97.16.8841
Noriega, C. E., Lin, H. Y., Chen, L. L., Williams, S. B., and Stewart, V. (2010). Asymmetric cross-regulation between the nitrate-responsive NarX-NarL and NarQ-NarP two-component regulatory systems from Escherichia coli K-12. Mol. Microbiol. 75, 394–412. doi: 10.1111/j.1365-2958.2009.06987.x
Nobechi, K. (1925). Contributions to the knowledge of Vibrio cholerae I. Fermentation of carbohydrates and polyatomic alcohols By Vibrio cholerae. J. Bacteriol. 10, 197–215. doi: 10.1128/jb.10.3.197-215.1925
Oh, Y. T., Lee, K. M., Bari, W., Kim, H. Y., Kim, H. J., and Yoon, S. S. (2015). Cholera toxin production induced upon anaerobic respiration is suppressed by glucose fermentation in Vibrio cholera. J. Microbiol. Biotechnol. 26, 627–636. doi: 10.4014/jmb.1512.12039
Oh, Y. T., Park, Y., Yoon, M. Y., Bari, W., Go, J., Min, K. B., et al. (2014). Cholera toxin production during anaerobic trimethylamine N-oxide respiration is mediated by stringent response in Vibrio cholerae. J. Biol. Chem. 289, 13232–13242. doi: 10.1074/jbc.M113.540088
Olivier, V., Salzman, N. H., and Fullner Satchell, K. J. (2007). Prolonged colonization of mice by Vibrio cholerae El Tor O1 depends on accessory toxins. Infect. Immun. 75, 5043–5051. doi: 10.1128/IAI.00508-507
Pang, P. K. T., Griffith, R. W., and Atz, J. W. (1977). Osmoregulation in elasmobranchs. Integr. Comp. Biol. 17, 365–377. doi: 10.1093/icb/17.2.365
Park, S., Tseng, C., and Gunsaius, R. P. (1995). Regulation of succinate dehydrogenase (sdhCDAB) operon expression in Escherichia coii in response to and anaerobiosis: role of ArcA and Fnr. Mol. Microbiol. 15, 473–482.
Pascal, M. C., Burini, J. F., and Chippaux, M. (1984). Regulation of the trimethylamine N-oxide (TMAO) reductase in Escherichia coli: analysis of tor::Mu d1 operon fusion. MGG Mol. Gen. Genet. 195, 351–355. doi: 10.1007/BF00332770
Pietrocola, F., Galluzzi, L., Bravo-San Pedro, J. M., Madeo, F., and Kroemer, G. (2015). Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821. doi: 10.1016/j.cmet.2015.05.014
Pommier, J., Méjean, V., Giordano, G., and Iobbi-Nivol, C. (1998). TorD, a cytoplasmic chaperone that interacts with the unfolded trimethylamine N-oxide reductase enzyme (TorA) in Escherichia coli. J. Biol. Chem. 273, 16615–16620. doi: 10.1074/jbc.273.26.16615
Rabin, R. S., and Stewart, V. (1992). Either of two functionally redundant sensor proteins, NarX and NarQ, is sufficient for nitrate regulation in Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 89, 8419–8423. doi: 10.1073/pnas.89.18.8419
Reen, F. J., Almagro-Moreno, S., Ussery, D., and Boyd, E. F. (2006). The genomic code: inferring Vibrionaceae niche specialization. Nat. Rev. Microbiol. 4, 697–704. doi: 10.1038/nrmicro1476
Rowe, J. J., Yarbrough, Y. M., Rake, J. B., and Eagon, R. G. (1979). Nitrite inhibition of aerobic bacteria. Curr. Microbiol. 2, 51–54. doi: 10.11150/kansenshogakuzasshi1970.53.704
Rowley, G., Hensen, D., Felgate, H., Arkenberg, A., Appia-Ayme, C., Prior, K., et al. (2012). Resolving the contributions of the membrane-bound and periplasmic nitrate reductase systems to nitric oxide and nitrous oxide production in Salmonella enterica serovar Typhimurium. Biochem. J. 441, 755–762. doi: 10.1042/BJ20110971
Sawers, G., and Suppmann, B. (1992). Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J. Bacteriol. 174, 3474–3478. doi: 10.1128/jb.174.11.3474-3478.1992
Sawers, R. G. (2005). Formate and its role in hydrogen production in Escherichia coli. Biochem. Soc. Trans. 33, 42–46. doi: 10.1042/BST0330042
Schild, S., Tamayo, R., Nelson, E. J., Qadri, F., Calderwood, S. B., and Camilli, A. (2007). Article genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2, 264–277. doi: 10.1016/j.chom.2007.09.004
Shalel-Levanon, S., San, K. Y., and Bennett, G. N. (2005). Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol. Bioeng. 92, 147–159. doi: 10.1002/bit.20583
Sijbesma, W. F. H., Almeida, J. S., Reis, M. A. M., and Santos, H. (1996). Uncoupling effect of nitrite during denitrification by Pseudomonas fluorescens: an in vivo 31 P-NMR study. Biotechnol. Bioeng. 52, 176–182.
Silva, A. J., and Benitez, J. A. (2016). Vibrio cholerae biofilms and cholera pathogenesis. PLoS Negl. Trop. Dis. 10:e004330. doi: 10.1371/journal.pntd.0004330
Silvestro, A., Pommier, J., Pascal, M.-C., and Giordano, G. (1989). The inducible trimethylamine N-oxide reductase of Escherichia coli K12: its localization and inducers. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 999, 208–216. doi: 10.1016/0167-4838(89)90220-90223
Spiro, S., and Guest, J. R. (1991). Adaptive responses to oxygen limitation in Escherichia coli. Trends Biochem. Sci. 16, 310–314.
Stewart, V. J. (2003). Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 31, 1–10.
Tamayo, R., Patimalla, B., and Camilli, A. (2010). Growth in a biofilm induces a hyperinfectious phenotype in Vibrio cholerae. Infect. Immun. 78, 3560–3569. doi: 10.1128/IAI.00048-10
Unden, G., Becker, S., Bongaerts, J., Holighaus, G., Schirawski, J., and Six, S. (1995). O2-Sensing and O2-dependent gene regulation in facultatively anaerobic bacteria. Arch. Microbiol. 164, 81–90. doi: 10.1007/BF02525312
Unden, G., and Bongaerts, J. (1997). Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. BBA Bioenerg. 1320, 217–234. doi: 10.1016/S0005-2728(97)00034-30
Veening, J.-W., Smits, W. K., and Kuipers, O. P. (2008). Bistability, epigenetics, and bet-hedging in bacteria. Annu. Rev. Microbiol. 62, 193–210. doi: 10.1146/annurev.micro.62.081307.163002
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. doi: 10.1126/science.272.5270.1910
Walters, M. C. III, Roe, F., Bugnicourt, A., Franklin, M. J., and Stewart, P. S. (2003). Contributions of antibiotic penetration, oxygen limitation. Antimicrob. Agents Chemother. 47, 317–323. doi: 10.1128/AAC.47.1.317
Wang, R., Zhang, H., Qiu, H., and Gao, S. (2009). Proteins involved in difference of sorbitol fermentation rates of the toxigenic and nontoxigenic Vibrio cholerae El Tor strains revealed by comparative proteome analysis. BMC Microbiol. 9:135. doi: 10.1186/1471-2180-9-135
Weitzberg, E., and Lundberg, J. O. (2013). Novel aspects of dietary nitrate and human health. Annu. Rev. Nutr. 33, 129–159. doi: 10.1146/annurev-nutr-071812-161159
Werner, E., Roe, F., Bugnicourt, A., Franklin, M. J., Heydorn, A., Pitts, B., et al. (2004). Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70, 6188–6196. doi: 10.1128/AEM.70.10.6188
White, D. (2000). The Physiology and Biochemistry of Prokaryotes. New York, NY: Oxford University Press.
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Winter, S. E., Winter, M. G., Xavier, M. N., Thiennimitr, P., Poon, V., Keestra, A. M., et al. (2013). Host-derived nitrate boosts growth. Science 339, 708–711. doi: 10.1126/science.1232467
World Health Organization [WHO] (2011). Nitrate and Nitrite in Drinking-Water. Geneva: World Heal. Organ.
Xu, K. D., Stewart, P. S., Xia, F., Huang, C. T., and McFeters, G. A. (1998). Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64, 4035–4039.
Xu, Q., Dziejman, M., and Mekalanos, J. J. (2003). Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U.S.A. 100, 1286–1291. doi: 10.1073/pnas.0337479100
Yahr, T. L. (2006). A critical new pathway for toxin secretion? N. Engl. J. Med. 355, 1171–1172. doi: 10.1056/NEJMcibr063931
Yang, K., Borisov, V. B., Konstantinov, A. A., and Gennis, R. B. (2008). The fully oxidized form of the cytochrome bd quinol oxidase from E. coli does not participate in the catalytic cycle: direct evidence from rapid kinetics studies. FEBS Lett. 582, 3705–3709. doi: 10.1016/j.febslet.2008.09.038
Yoon, S. S., Hennigan, R. F., Hilliard, G. M., Ochsner, U. A., Parvatiyar, K., Kamani, M. C., et al. (2004). Pseudomonas aeruginosa anaerobic respiration in biofilms. Dev. Cell 3, 593–603. doi: 10.1016/s1534-5807(02)00295-292
Yoon, S. S., and Mekalanos, J. J. (2006). 2,3-Butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect. Immun. 74, 6547–6556. doi: 10.1128/IAI.00695-06
Zerbst-Boroffka, I., Kamaltynow, R. M., Harjes, S., Kinne-Saffran, E., and Gross, J. (2005). TMAO and other organic osmolytes in the muscles of amphipods (Crustacea) from shallow and deep water of Lake Baikal. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 142, 58–64. doi: 10.1016/j.cbpa.2005.07.008
Zheng, L., Kelly, C. J., and Colgan, S. P. (2015). Physiologic hypoxia and oxygen homeostasis in the healthy intestine. a review in the theme: cellular responses to hypoxia. Am. J. Physiol. Physiol. 309, C350–C360. doi: 10.1152/ajpcell.00191.2015
Zhu, J., and Shimizu, K. (2004). The effect of pfl gene knockout on the metabolism for optically pure D-lactate production by Escherichia coli. Appl. Microbiol. Biotechnol. 64, 367–375. doi: 10.1007/s00253-003-1499-1499
Keywords: Vibrio cholerae, enteropathogen, respiration, nitrate, fumarate, TMAO, fermentation, fitness
Citation: Bueno E, Pinedo V and Cava F (2020) Adaptation of Vibrio cholerae to Hypoxic Environments. Front. Microbiol. 11:739. doi: 10.3389/fmicb.2020.00739
Received: 20 January 2020; Accepted: 30 March 2020;
Published: 29 April 2020.
Edited by:
Jennifer M. Ritchie, University of Surrey, United KingdomReviewed by:
Jun Zhu, University of Pennsylvania, United StatesJyl S. Matson, The University of Toledo, United States
Copyright © 2020 Bueno, Pinedo and Cava. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emilio Bueno, ZW1pbGlvLmJ1ZW5vQHVtdS5zZQ==; Felipe Cava, ZmVsaXBlLmNhdmFAdW11LnNl
 Emilio Bueno
Emilio Bueno Víctor Pinedo
Víctor Pinedo Felipe Cava*
Felipe Cava*