
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 15 November 2019
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 10 - 2019 | https://doi.org/10.3389/fmicb.2019.02508
This article is part of the Research Topic Characterization Of Mobile Genetic Elements Associated With Acquired Resistance Mechanisms View all 15 articles
 Qiaoxiang Cheng1†
Qiaoxiang Cheng1† Xiaoyuan Jiang3†
Xiaoyuan Jiang3† Yanan Xu1,2
Yanan Xu1,2 Lingfei Hu3
Lingfei Hu3 Wenbo Luo3
Wenbo Luo3 Zhe Yin3
Zhe Yin3 Huixia Gao2
Huixia Gao2 Wenhui Yang3
Wenhui Yang3 Huiying Yang3
Huiying Yang3 Yuee Zhao3
Yuee Zhao3 Xiaodong Zhao3
Xiaodong Zhao3 Dongsheng Zhou3*
Dongsheng Zhou3* Erhei Dai1,2*
Erhei Dai1,2*A collection of 11 IncC plasmids from China were fully sequenced herein and compared with reference plasmids pR148 and pR55. These 13 plasmids could be assigned into three different subgroups: type 1, type 2, and type 1/2 hybrid. Type 1/2-hybrid plasmids most likely emerged from homologous recombination between type 1 and type 2 plasmids. Different IncC plasmids had evolved to acquire quite different profiles of accessory modules and thus different collections of resistance genes. The accessory resistance modules included not only the blaCMY-carrying region, the ARI-A island, and the ARI-B island, but also various additional kinds of resistance islands such as the blaCTX–M-carrying regions and the MDR regions. Insertion of accessory modules was sometimes accompanied by deletion, inversion, and translocation of surrounding backbone regions. pR148 and pR55 were confirmed to have the most complete backbones for type 1 and type 2, respectively. This was the first report of a blaIMP–8-carrying IncC plasmid, and that of three novel mobile elements: a Tn1696-derived unit transposon Tn6395, a class 2 integron In2-76, and an insertion sequence ISEcl10.
Plasmids of IncA and IncC incompatibility groups have been combined into IncA/C since 1970s, but IncA and IncC groups are essentially compatible (Ambrose et al., 2018) and have significant nucleotide divergence across their backbones (Harmer and Hall, 2015) and, thus, IncA [reference plasmid pRA1 (Fricke et al., 2009)] and IncC should be separated from IncA/C (Harmer et al., 2017). IncC plasmids have two distinct types, namely, type 1 and type 2 (Harmer and Hall, 2014), which are represented by reference plasmids pR148 (Del Castillo et al., 2013), and pR55 (Benoît et al., 2012), respectively. These two types can be distinguished by two genes of substitution (orf1832 in type 1 or orf1847 in type 2, and rhs1 in type 1 or rhs2 in type 2) and two small intergenic insertions (i1 and i2 found in type 2 rather than type 1) (Harmer and Hall, 2014).
A large array of antibiotic resistance genes have been found in IncC plasmids and are clustered in various accessory modules inserted at various sites of the IncC backbones (Harmer and Hall, 2015). IncC plasmids contain at least three hotspots for integration of the blaCMY-carrying region and two antibiotic resistance islands designated ARI-A and ARI-B. The blaCMY-carrying region and ARI-A are frequently found in type 1 plasmids but not type 2; both type 1 and type 2 plasmids carry ARI-B, but this island is not always present in these plasmids (Harmer and Hall, 2015).
Our previous study (Ma et al., 2017) has shown considerable genomic variation in a collection of three type 1 IncC plasmids from China together with pR148. This follow-up study presents the complete nucleotide sequences of 11 new IncC plasmids from China and a further comprehensive genomic comparison of these 11 plasmids together with pR148 and pR55, providing a deeper insight into parallel evolution of IncC plasmids from China.
The 11 isolates (Supplementary Table S1) were recovered from the sputum or blood specimens of 11 different patients with nosocomial infections in 10 Chinese public hospitals from 2012 to 2016. Bacterial species identification was performed using genome sequence-based average nucleotide identity analysis (Richter and Rossello-Mora, 2009).
Plasmid conjugal transfer experiments were carried out with rifampin-resistant Escherichia coli EC600 or sodium azide-resistant E. coli J53 being used as a recipient and each of the QD1501, A2293, and 205880 isolates as a donor. Three milliliters of overnight cultures of each of donor and recipient bacteria were mixed together, harvested, and resuspended in 80 μl of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 hydrophilic nylon membrane filter with a 0.45 μm pore size (Millipore) that was placed on a BHI agar (BD Biosciences) plate and then incubated for mating at 37°C for 12 to 18 h. Bacteria were washed from filter membrane and spotted on Muller-Hinton (MH) agar (BD Biosciences) plates, for selecting an E. coli transconjugant carrying blaNDM (pQD1501-Ct1), blaCTX–M (pA2293-Ct2), or blaCMY (p205880-Ct1/2). 200 μg/ml sodium azide (for J53) or 1000 μg/ml rifampin (for EC600), together with 4 μg/ml imipenem (for blaNDM) or 200 μg/ml ampicillin (for blaCTX–M or blaCMY), was for E. coli transconjugant selection.
Genomic DNA was isolated from each of the above 11 isolates using an UltraClean Microbial Kit (Qiagen, NW, Germany). The genomic DNA of strains 427113, T5282, and 397108 was sequenced from mate-pair libraries with an average insert size of 5 kb (ranging from 2 to 10 kb) using a MiSeq sequencer (Illumina, CA, United States) and DNA contigs were assembled based on their contig coverages using Newbler 2.6 (Nederbragt, 2014). Quality control, removing adapters, and low quality reads were performed using Trimmomatic 0.36 (Bolger et al., 2014). The filtered clean reads were then assembled using Newbler 2.6 (Nederbragt, 2014), followed by extraction of the consensus sequence with CLC Genomics Workbench 3.0 (Qiagen Bioinformatics). Gapfiller V1.11 (Boetzer and Pirovano, 2012) was used for gap closure. For all the other eight isolates, genome sequencing was performed with a sheared DNA library with an average size of 15 kb (ranging from 10 to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, CA, United States), as well as a paired-end library with an average insert size of 400 bp (ranging from 150 to 600 kb) on a HiSeq sequencer (Illumina, CA, United States). The paired-end short Illumina reads were used to correct the long PacBio reads utilizing proovread (Hackl et al., 2014), and then the corrected PacBio reads were assembled de novo utilizing SMARTdenovo1.
Open reading frames (ORFs) and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN (Boratyn et al., 2013) searches against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’Leary et al., 2016). Annotation of resistance genes, mobile elements, and other features was carried out using the online databases including CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.12. Heatmaps were plotted with MeV 4.9.0 (Saeed et al., 2003).
The backbone regions of indicative plasmids were aligned using MUMmer 3.0 (Kurtz et al., 2004). Inference of homologous recombination was performed using ClonalFrameML (Didelot and Wilson, 2015) to remove recombination-associated single-nucleotide polymorphisms (SNPs). A maximum-likelihood tree was constructed from recombination-free SNPs using MEGA7 (Kumar et al., 2016) with a bootstrap iteration of 1000.
Bacterial antimicrobial susceptibility was tested by BioMérieux VITEK 2 and interpreted as per the 2017 Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2017).
The complete nucleotide sequences of plasmids p24845-Ct2, p205880-Ct1/2, pT5282-Ct2, pKpn47-Ct1/2, p11935-Ct1/2, pQD1501-Ct1, p12085-Ct1, pA2293-Ct2, p397108-Ct2, p427113-Ct1/2, and pA1763-Ct2 were submitted to GenBank under the accession numbers MF344572 to MF344574, MN310369, MN310370, MN310375, MN310377, MN310378, MH917284, MH917285, and MG764552, respectively.
Of the 11 IncC plasmids fully sequenced in this work (Table 1 and Figure 1), two and five could be assigned into type 1 and type 2, respectively, based on the presence or absence of orf1832/orf1847, rhs1/rhs2, i1, and i2, while the remaining four, containing orf1832 and rhs1 (characteristic of type 1) plus i2 (characteristic of type 2), were recognized as type 1/2 hybrid. A total of 597 core SNPs (among them 69 were recombination-free) were identified from the backbone regions of these 11 plasmids together with type 1 reference pR148 and type 2 reference pR55. As shown in the phylogenetic tree (Figure 1A) constructed from the 69 recombination-free SNPs, these 13 plasmids could be clustered into three separate clades, corresponding to the above three subtypes. A pairwise sequence comparison using BLASTN showed that these 13 plasmids displayed >99% nucleotide identity across ≥50% of their backbone sequences within each of subgroup IncC plasmids, and had >97% nucleotide identity across ≥47% of their backbone sequences between different subgroups (Figure 1B and Supplementary Table S2). The above results confirmed a parallel diversification and evolution of type 1, type 2, and type 1/2-hybrid IncC plasmids. These 13 plasmids varied in size from about 90 kb to nearly 206 kb with variation in the number of predicted ORFs from 118 to 246, and each plasmid was dissected into the IncC backbone regions and the accessory modules, which were defined as acquired DNA regions associated and bordered with mobile elements (Table 2 and Supplementary Figure S1). These 13 plasmids shared common IncC backbone genes repA, parAB, tra2, dcm2, yacC, int, kfrA, and uvrD.
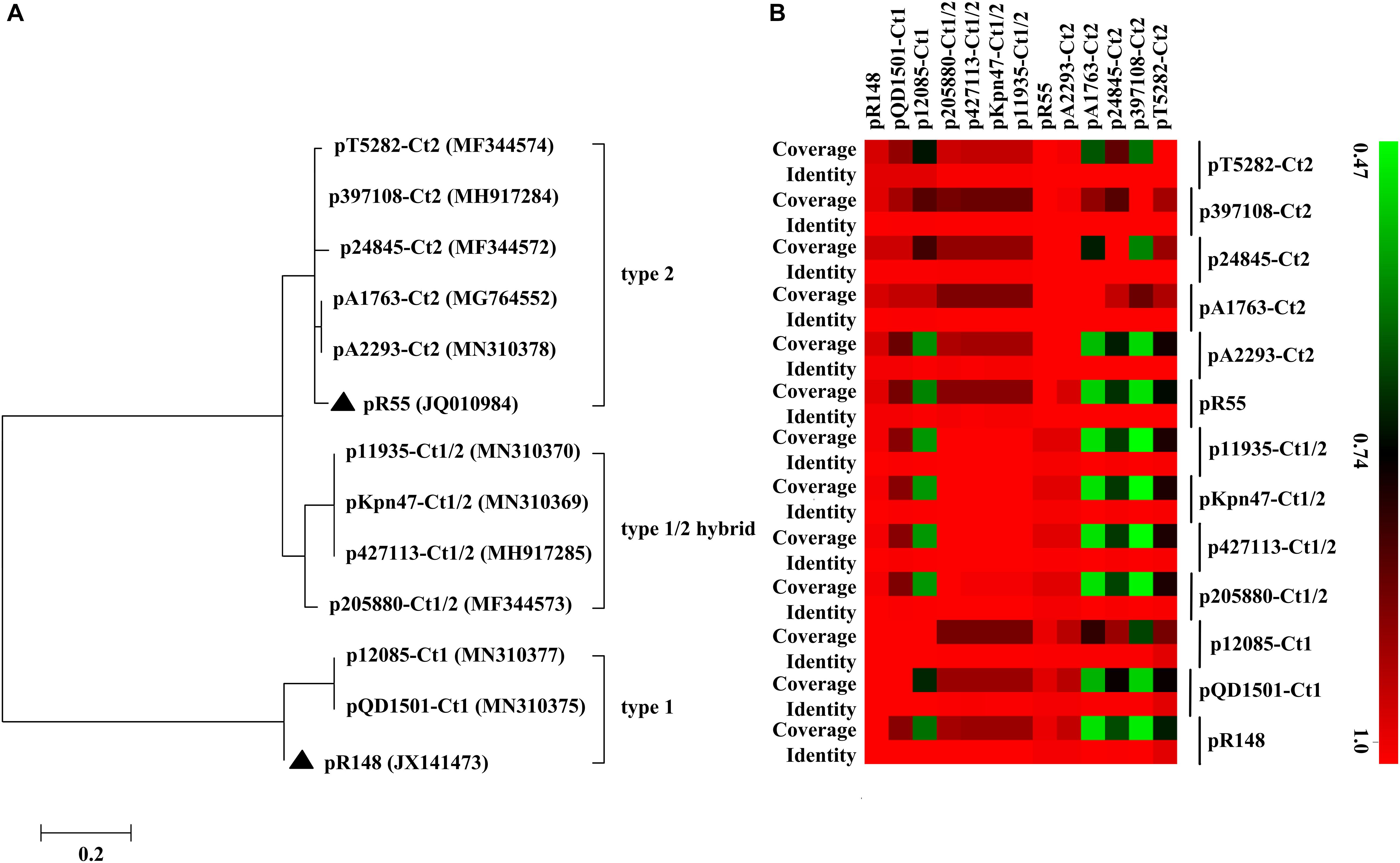
Figure 1. Evolutionary relationships of IncC plasmids. (A) A maximum-likelihood phylogenetic tree. The tree is constructed from recombination-free SNPs of core backbone regions. The degree of support (percentage) for each cluster of associated taxa, as determined by bootstrap analysis, is shown next to each branch. The bar corresponds to the scale of sequence divergence. Triangles indicate the references for type 1 and type 2 IncC plasmids. (B) A heatmap of pairwise comparison of plasmid backbone sequences. The original BLAST coverage and nucleotide identity values are shown in Supplementary Table S2.
At least seven major modular differences were identified across the whole plasmid sequences (Figure 2 and Supplementary Figure S2). First, two small regions (dsbA–orf429, and orf858–orf192/orf189–nuc) displayed <95% nucleotide identity between all the three type 1 plasmids and all the 10 type 1/2-hybrid and type 2 plasmids, for which a gene substitution (orf192 in the former three, and orf189 in the later ten) occurred. Second, the ARI-B island or the ΔTn6292–ΔIS26–IS26 region was inserted into the backbone dcm1 region of all the 10 type 1/2-hybrid and type 2 plasmids, leading to a 4.5- to 11.0-kb deletion within the dcm1 region of these 10 plasmids expect for pR55. Third, the three type 1 plasmids had a backbone gene orf489 showing <90% nucleotide identity to all the other plasmids; ISKpn18 was inserted at a site within the backbone gene orf501 in p397108-Ct2, resulting in a 60.1-kb deletion (containing the whole tra1 region and the dcm3 region). Fourth, compared to the prototype tra1 region as observed in pR148 and additional two plasmids, various insertions of accessory modules were found in the other plasmids: (i) Tn6538a or Tn6538b or Tn6538c or the blaCMY–6–blaNDM–1 region was inserted downstream of tivF1 of all the seven type 1 and type 1/2-hybrid plasmids except for pR148, resulting in a 48.1-kb deletion in p12085-Ct1; (ii) the insertion of ISEhe3 into orf672 of pT5282-Ct2 or that of blaCTX–M–3 region into rlx of pA1763-Ct2 led to a 24.5-kb or 57.4-kb deletion, respectively; and (iii) ISKpn18 plus IS1R was inserted into tivF6 of pKpn47-Ct1/2 or p11935-Ct1/2, while the MDR (multidrug resistance) region was inserted into the same gene of p427113-Ct1/2 or p24845-Ct2, leading to a 35.5-kb deletion in p24845-Ct2. Fifth, the backbone dcm3 region was completely or partially lost in five plasmids, which was caused by accessory module insertion or other unknown reasons. Sixth, the ARI-A island was inserted into the backbone gene orf240 of all the seven type 1 and type 1/2-hybrid plasmids except for p12085-Ct1, while ISKpn52:IS30 plus ISAs25 was inserted into a type 2 plasmid pA2293-Ct2. Seventh, Tn6187, IS100kyp and Tn6558 were inserted at different sites around the backbone ter gene of pR55, pA2293-Ct2, and p397108-Ct2, respectively; additionally, three separate insertions of the MDR region, the blaCTX–M–14–blaIMP–8 region, and the ISEhe3 element into the backbone ter–to–tivF3 region made its segmentation in pT5282-Ct2.
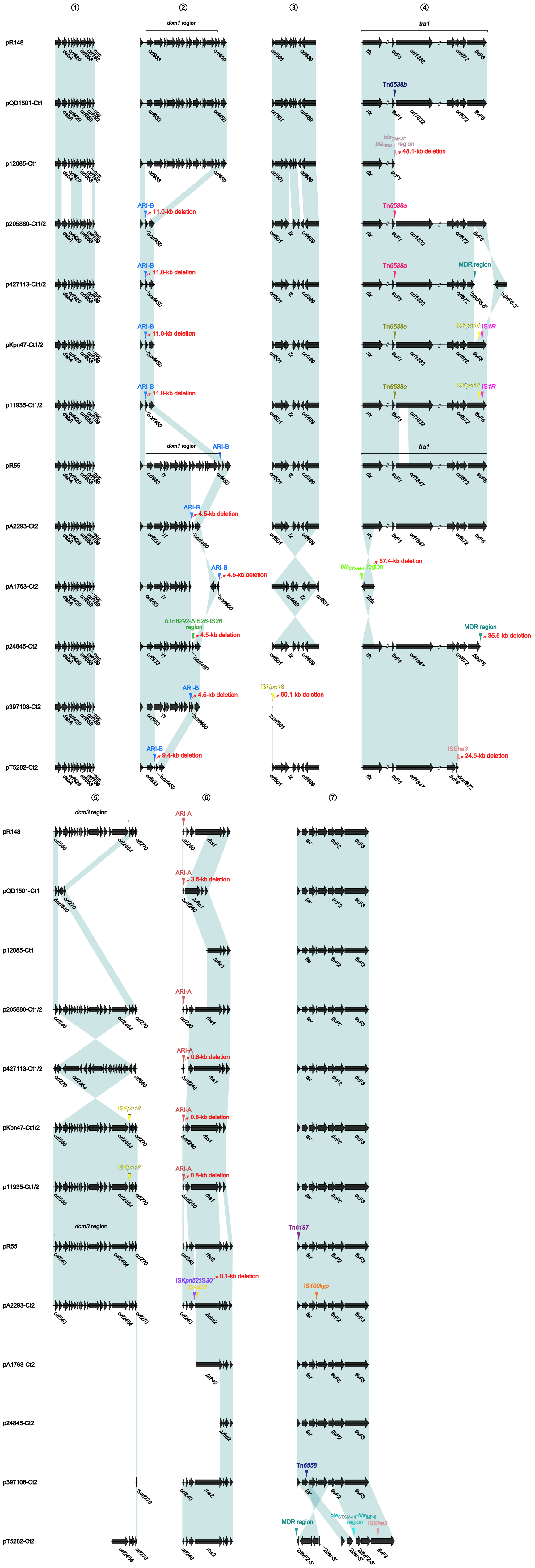
Figure 2. Major modular differences among IncC plasmids. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on function classification. Shading denotes regions of homology (≥95% nucleotide identity).
As found in all the seven type 1 and type 1/2-hybrid plasmids except for p12085-Ct1 (type 1), the ARI-A islands (Figure 3) were identified as Tn1696 (Partridge et al., 2001) derivatives. The Tn1696 derivatives in pR148 and p205880-Ct1/2 were recognized as intact unit transposons Tn6358 (Ma et al., 2017) and Tn6395, respectively, which differed from Tn1696 mainly by insertion of In834 (Ma et al., 2017) and In1212 instead of In4 into a primary tnpAR–mer structure. The Tn1696 derivatives in the other four plasmids could not be discriminated as intact transposons due to 3′-terminal truncations, which were in some cases accompanied with insertion of additional resistance modules: blaNDM–1-carrying ΔTn125 (Poirel et al., 2012) in pQD1501-Ct1 and strAB-carrying ΔTn6029 (Cain et al., 2010) in p427113-Ct1/2.
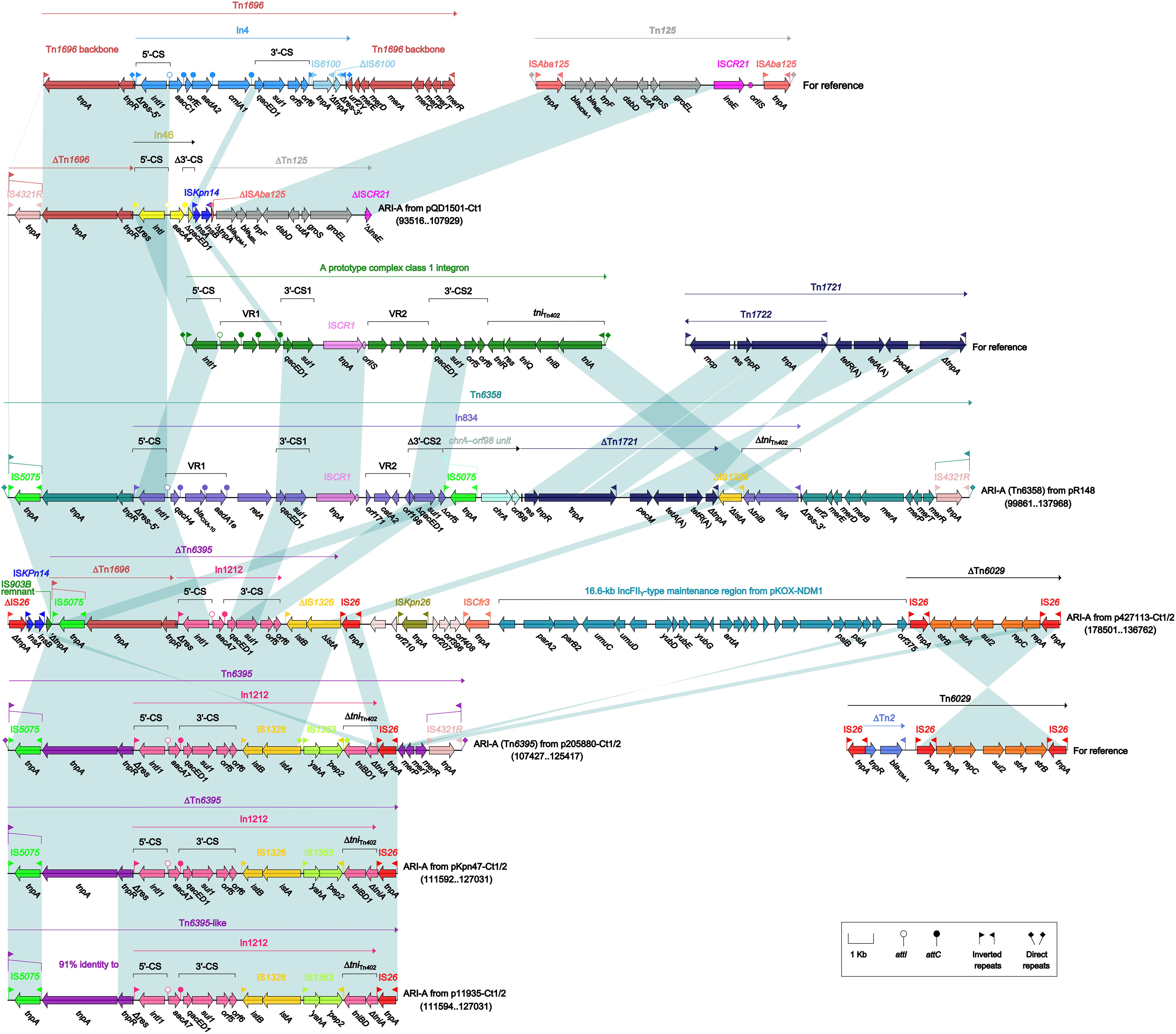
Figure 3. Organization of ARI-A islands and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within corresponding plasmids. The accession numbers of Tn1696 (Partridge et al., 2001), Tn125 (Poirel et al., 2012), Tn1721 (Allmeier et al., 1992), and Tn6029 (Cain et al., 2010) for reference are U12338, JN872328, X61367, and HQ840942, respectively.
The ARI-B islands (Figure 4) were found in all the 10 type 2 and type 1/2-hybrid plasmids except for p24845-Ct2 (type 2), and all of them contained a sul2 gene that was originated from either GIsul2 (Nigro and Hall, 2011) or Tn6029 (Cain et al., 2010), because various sul2-carrying remnants of GIsul2 or Tn6029 were found in these ARI-B islands. In general, these ARI-B islands acquired dramatically different collection of resistance modules, which contained genes resistance to older (such as dfrA, strAB, tetA, and floR) and newer (such as blaCTX–M, armA, msr, and mph) antibiotics, and thus varied considerably in size with a highly mosaic nature. The ΔTn6292–ΔIS26–IS26 region (Figure 4), which did not contain any of resistance genes, was inserted at the site specific for ARI-B insertion.
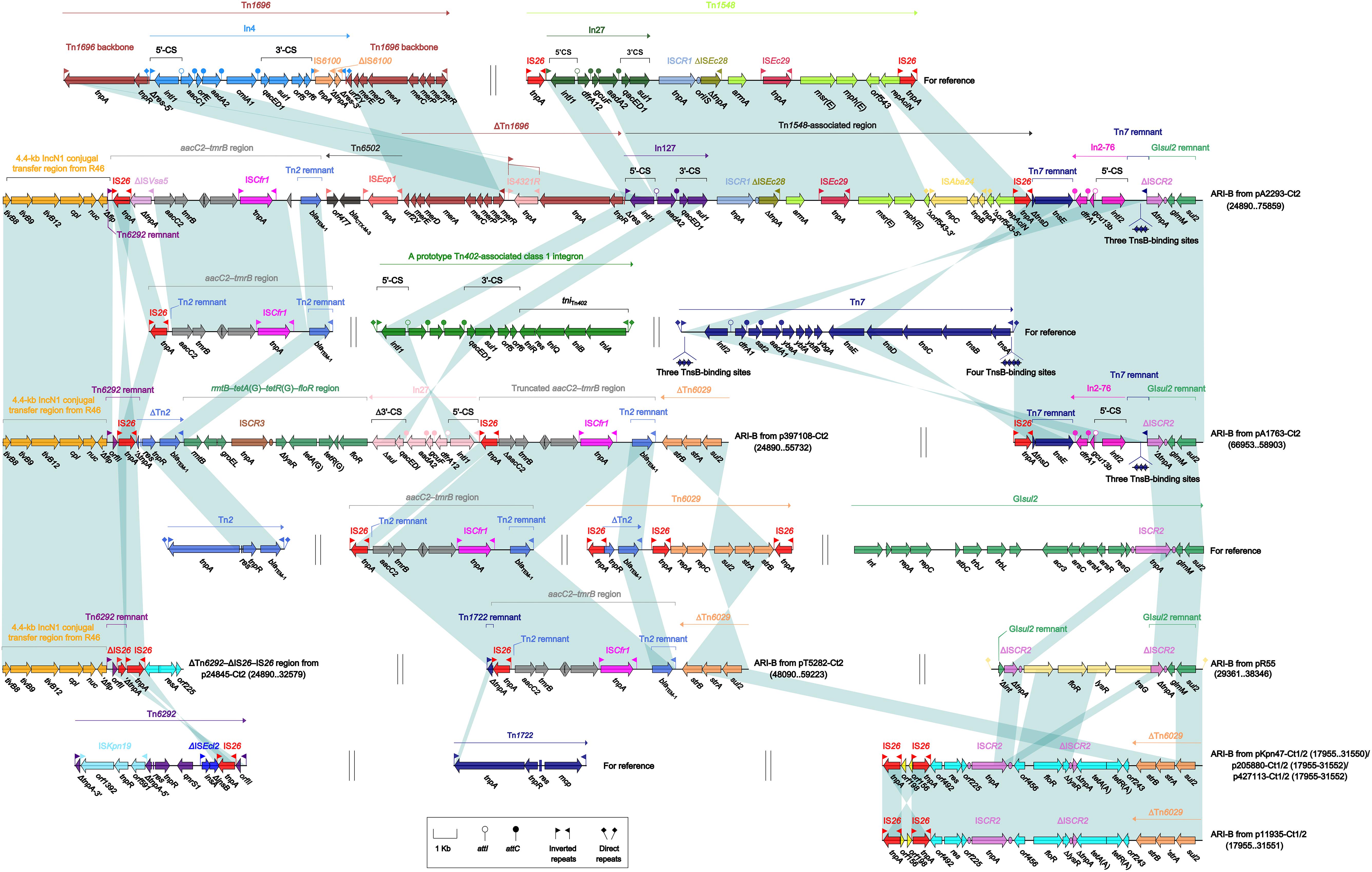
Figure 4. Organization of ARI-B islands and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements, and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within corresponding plasmids. The accession numbers of Tn1696 (Partridge et al., 2001), Tn1548 (Galimand et al., 2005), aacC2–tmrB region (Partridge et al., 2012), Tn7 (see Zhan et al., 2018, for gene organization), Tn2 (Bailey et al., 2011), Tn6029 (Cain et al., 2010), GIsul2 (Nigro and Hall, 2011), Tn6292 (Wei et al., 2016), and Tn1722 (Allmeier et al., 1992) for reference are U12338, AF550415, JX101693, KX117211, HM749967, HQ840942, AE014073, KU886034, and X61367, respectively.
Four different blaCMY-carrying regions (Figure 5) were found in all the seven type 1 and type 1/2-hybrid plasmids except for pR148 and were manifested as three intact ISEcp1-based transposition units Tn6538a (p205880-Ct1/2 and p427113-Ct1/2), Tn6538b (pQD1501-Ct1), and Tn6538c (pKpn47-Ct1/2 and p11935-Ct1/2), and the blaCMY–6–blaNDM–1 region (p12085-Ct1). Tn6538a, Tn6538b, and Tn6538c were highly similar to one another but differed by substitution of blaCMY–2 with blaCMY–6 or by insertion of IS1R into ISEcp1. The blaCMY–6–blaNDM–1 region was a combination of ΔTn6538b carrying blaCMY–6, ISKpn14, and ΔTn125 containing blaNDM–1.
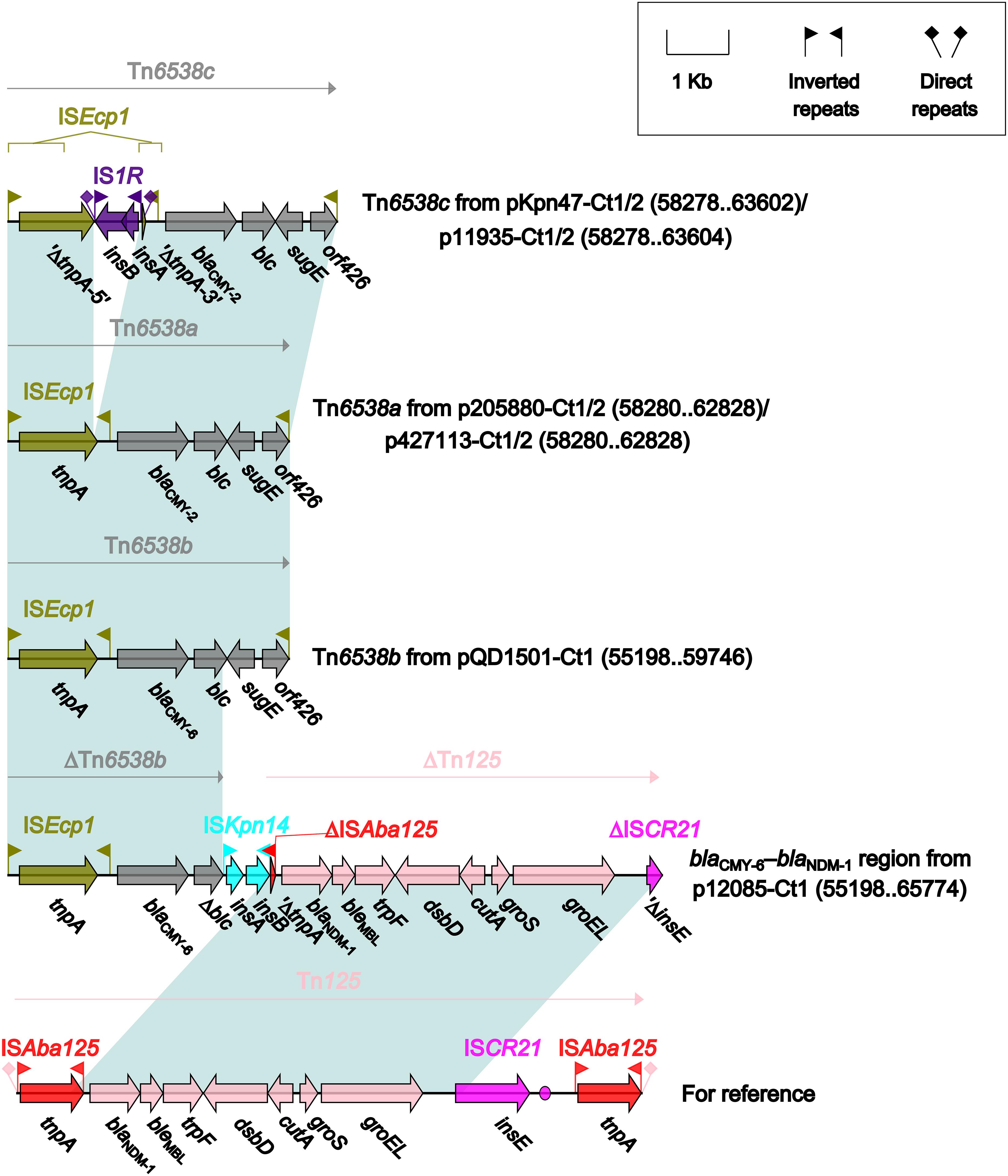
Figure 5. Organization of blaCMY-carrying regions and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within corresponding plasmids. The accession number of Tn125 (Poirel et al., 2012) for reference is JN872328.
Four different blaCTX–M-carrying regions were found in four type 2 plasmids: Tn6558 (p397108-Ct2), the blaCTX–M–14–blaIMP–8 region (pT5282-Ct2), and the blaCTX–M–3 region (pA1763-Ct2) (Figure 6), as well as the ARI-B island (pA2293-Ct2) (Figure 4). Insertion of a truncated version of Tn6503, which was an ISEcp1-based, blaCTX–M–14-carrying transposition unit (Feng et al., 2015), into the cryptic unit transposon Tn1722 generated Tn6558, while the blaCTX–M–14–blaIMP–8 region was resulted from three separate insertions of Tn6503, In655 (carrying blaIMP–8), and IS26 into Tn1722. The blaCTX–M–3 region in pA1763-Ct2 was highly similar to a partial region of the ARI-B island in pA2293-Ct2, and both carried the ISEcp1-based blaCTX–M–3-carrying transposition unit Tn6502 (Liang et al., 2014).
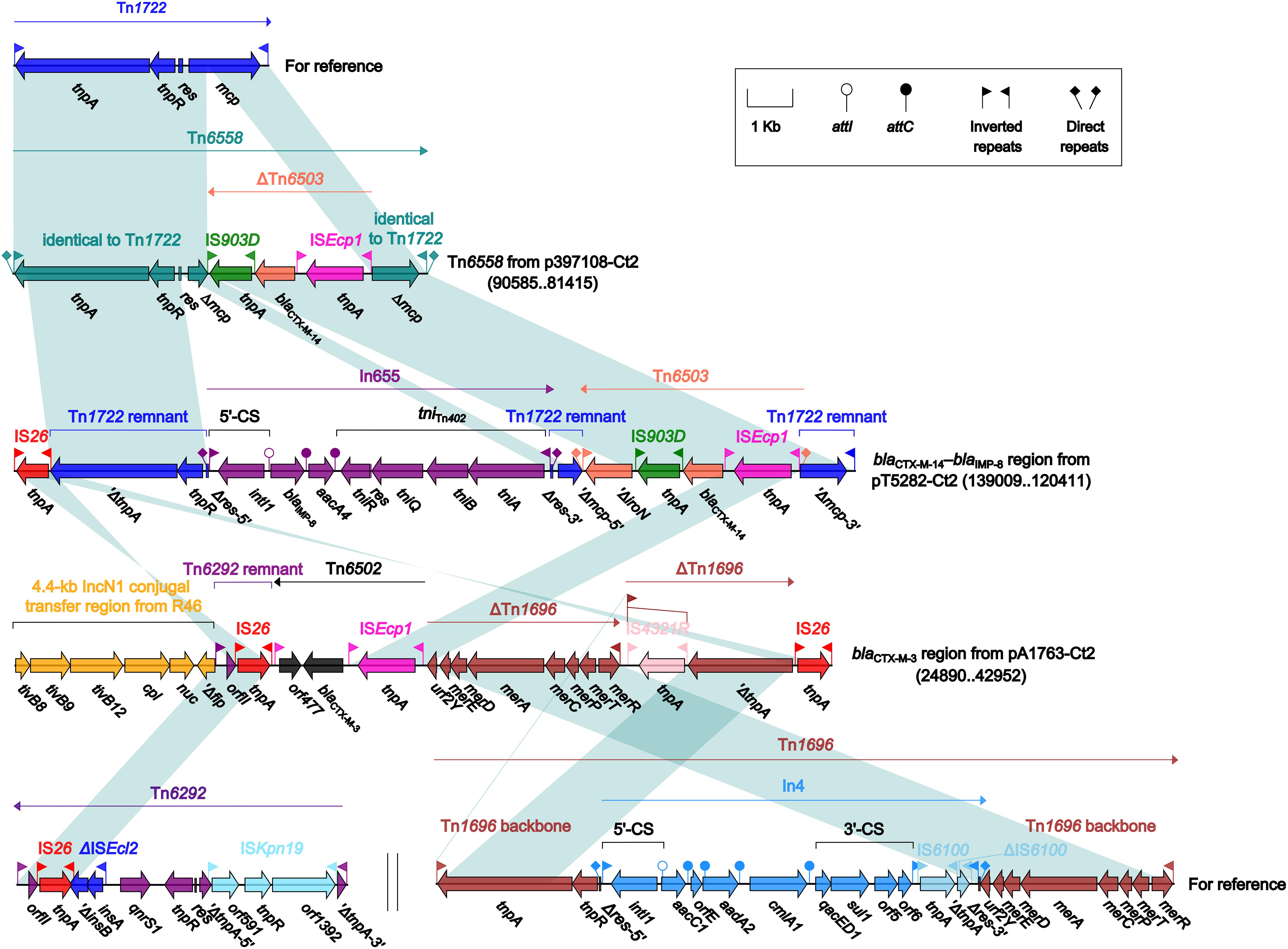
Figure 6. Organization of blaCTX–M-carrying regions and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within corresponding plasmids. The accession numbers of Tn1722 (Allmeier et al., 1992), Tn6292 (Wei et al., 2016) and Tn1696 (Partridge et al., 2001) for reference are X61367, KU886034 and U12338, respectively.
Four different MDR regions (Figure 7) were found in four plasmids: Tn6187 (Benoît et al., 2012) in pR55, and three MDR regions in p427113-Ct1/2, pT5282-Ct2, and p24845-Ct2. As similar to the above ARI-B islands, different MDR regions were assembled from different collections of resistance units [especially including ΔTn6296 carrying blaKPC–2, IS26–mph(A)–mrx–mphR(A)–IS6100 unit, In207, In384, and rmtB–tetA(G)–tetR(G)–floR region] via homologous or non-homologous recombination.
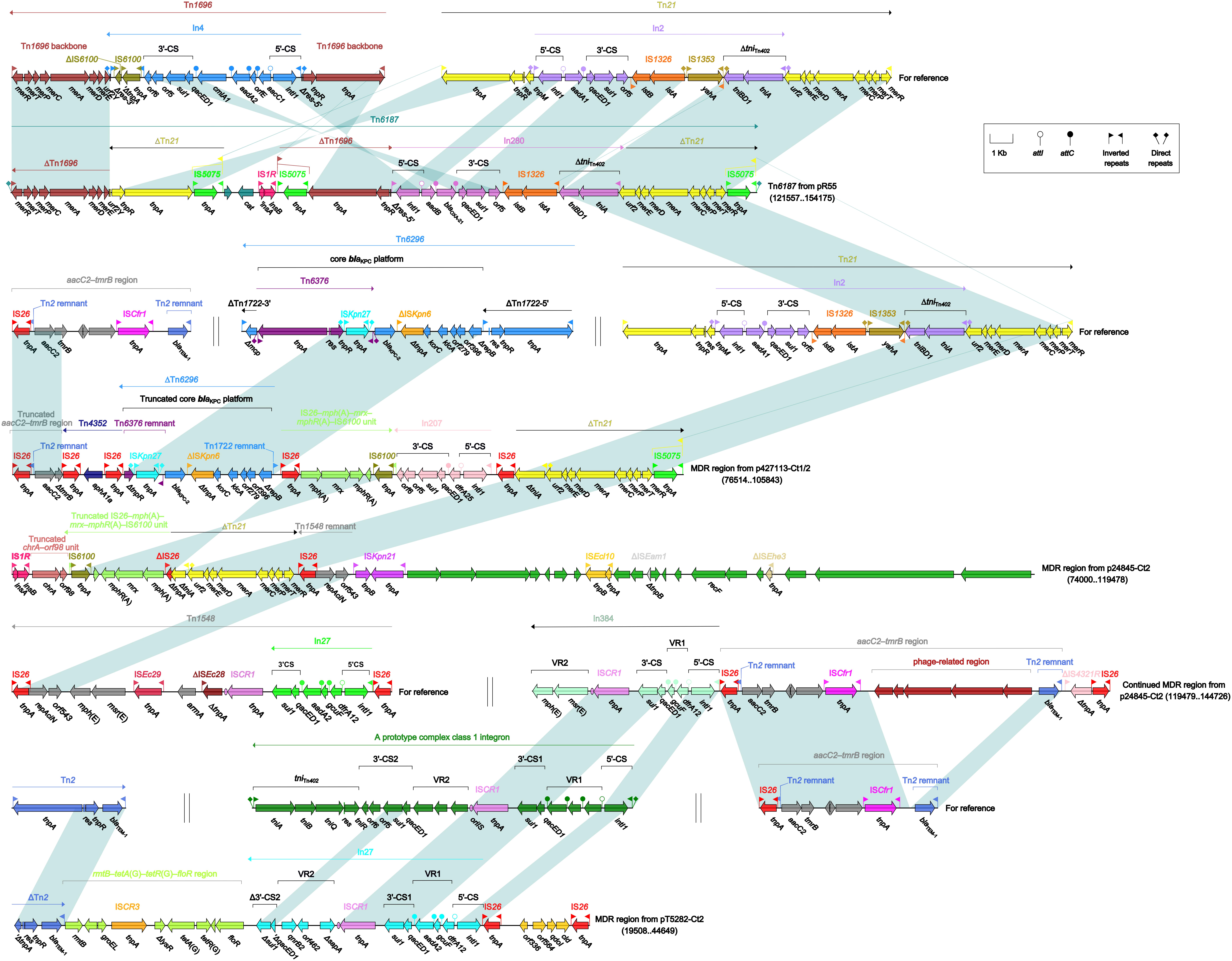
Figure 7. Organization of MDR regions and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within corresponding plasmids. The accession number of Tn1696 (Partridge et al., 2001), Tn21 (Partridge et al., 2001), Tn1548 (Galimand et al., 2005), Tn2 (Bailey et al., 2011), aacC2–tmrB region (Partridge et al., 2012) and Tn6296 (Jiang et al., 2010) for reference are U12338, AF071413, AF550415, HM749967, JX101693 and FJ628167, respectively.
Plasmids pQD1501-Ct1, pA2293-Ct2, and p205880-Ct1/2 were arbitrarily selected as the representatives of type 1, type 2, and type 1/2 hybrid, respectively, and could be transferred from the wild-type isolates into J53 or EC600 through conjugation, generating E. coli transconjugants QD1501-NDM-J53, A2293-CTXM-J53, and 205880-CMY-EC600, respectively. The self-transferable nature was consistent with the presence of two complete sets (tra1 and tra2) of conjugal transfer genes in each plasmid, although Tn6538a or Tn6538b was inserted at an intergenic site in tra1 of p205880-Ct1/2 or pQD1501-Ct1, respectively. All the three wild-type isolates QD1501, A2293, and 205880 and one transconjugant QD1501-NDM-J53 were resistant to imipenem with minimum inhibitory concentration (MIC) values ≥16 (due to production of NDM or KPC enzyme) but the other two transconjugants A2293-CTXM-J53 and 205880-CMY-EC600 remained susceptible to imipenem. All the above strains were resistant to cefuroxime with MIC values ≥32 (due to production of NDM or CTX-M or CMY enzyme).
Comparative genomics analysis of the collection of IncC plasmids (n = 13 in this study) provides a deeper understanding of diversification of IncC plasmids. Different IncC plasmids have evolved to acquire very different profiles of accessory modules and thus different collections of resistance genes (Figure 8). Insertion of accessory modules is often accompanied by deletion of surrounding backbone regions. Type 1/2-hybrid IncC plasmids most likely emerged from homologous recombination between type 1 and type 2 plasmids. In addition to using the presence or absence of signature sequences orf1832/orf1847, rhs1/rhs2, i1, and i2, a phylogenetic analysis using recombination-free SNPs of core backbone regions can steadily discriminate type 1/2 hybrid from type 1 and type 2. As shown previously (Ma et al., 2017) and herein, additional resistance islands (e.g., the MDR regions and the blaCTX–M-carrying regions in this study) can be found in type 1, type 2, and type 1/2-hybrid plasmids.
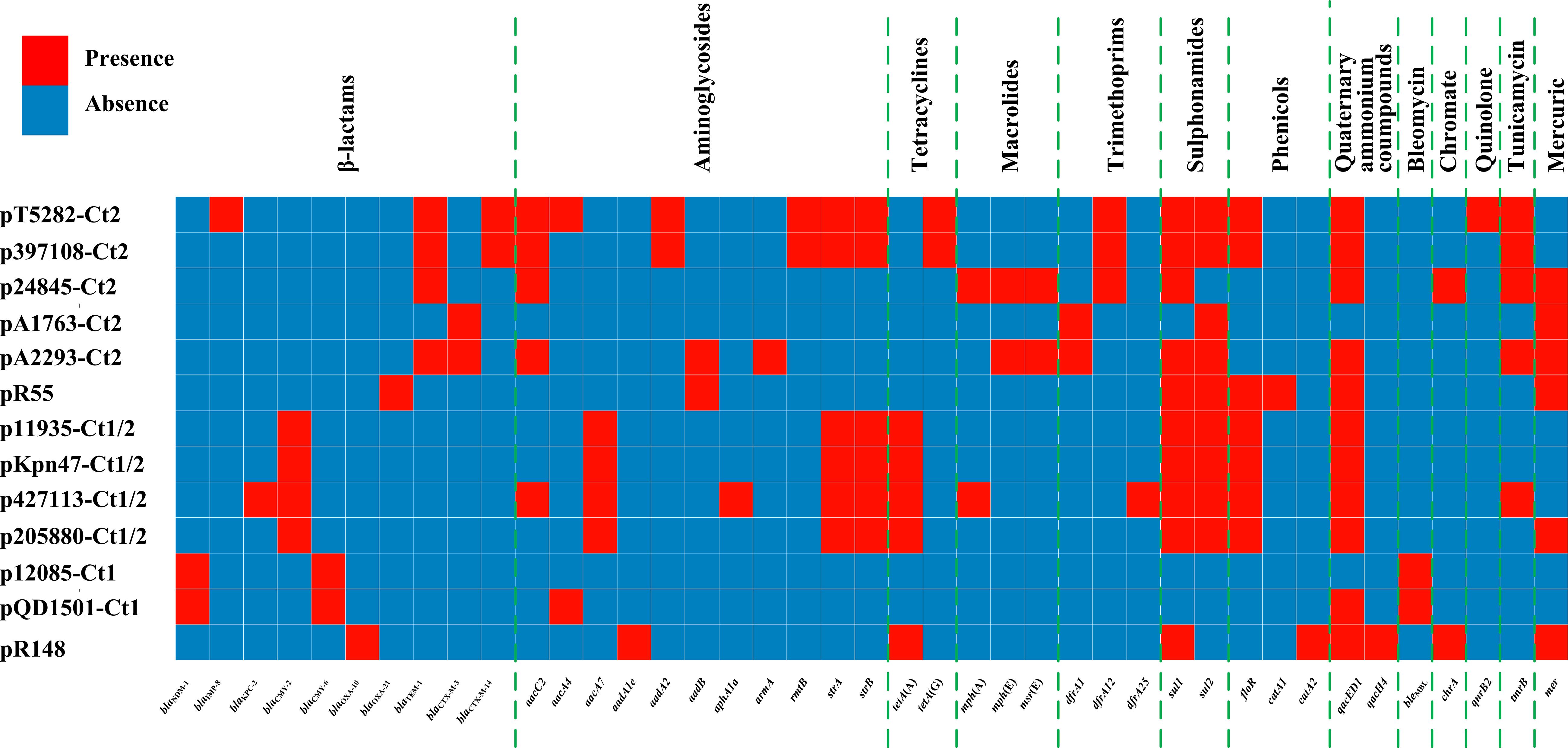
Figure 8. A heatmap of prevalence of resistance genes in IncC plasmids. The original data are shown in Supplementary Table S3.
The IncC reference plasmids pR148 (166 kb in length, 195 ORFs) and pR55 (171 kb in length, 203 ORFs) had the most complete backbones for type 1 and type 2, respectively. By contrast, the type 1 plasmid p12085-Ct1 and the type 2 plasmids pA1763-Ct2 and p397108-Ct2 harbored extremely simple backbones due to deletion of large backbone regions. In addition to insertion of accessory modules and deletion of backbone regions, large inversion (such as those in p427113-Ct1/2 and pA1763-Ct2) and translocation (such as those in pT5282-Ct2) events occurred within backbone regions during genomic diversification of IncC plasmids (Ma et al., 2017; Supplementary Figure S2).
The carbapenemase genes blaNDM–1, blaKPC–2, and blaIMP–8 were identified in pQD1501-Ct1 and p12085-Ct1, p427113-Ct1/2, and pT5282-Ct2, respectively. This is the first report of a blaIMP–8-carrying IncC plasmid. This study presents three novel mobile elements [firstly identified in this study, including Tn6395 (p205880-Ct1/2), In2-76 (pA1763-Ct2), and ISEcl10 (p24845-Ct2)] and additionally six newly named mobile elements [firstly designated in this study but with previously determined sequences, including Tn6538a (p205880-Ct1/2 and p427113-Ct1/2), Tn6538b (pQD1501-Ct1), Tn6538c (pKpn47-Ct1/2 and p11935-Ct1/2), Tn6502 (pA2293-Ct2 and pA1763-Ct2), Tn6503 (pT5282-Ct2), and ISKpn52 (pA2293-Ct2)].
The datasets generated for this study can be found in the complete nucleotide sequences of plasmids p24845-Ct2, p205880-Ct1/2, pT5282-Ct2, pKpn47-Ct1/2, p11935-Ct1/2, pQD1501-Ct1, p12085-Ct1, pA2293-Ct2, p397108-Ct2, p427113-Ct1/2, and pA1763-Ct2 were submitted to GenBank under the accession numbers MF344572 to MF344574, MN310369, MN310370, MN310375, MN310377, MN310378, MH917284, MH917285, and MG764552, respectively.
This study uses the clinical bacterial isolates obtained from the Chinese public hospitals as listed in Supplementary Table S1. The local legislation did not require the study to be reviewed or approved by an ethics committee, because the bacterial isolates involved in this study were part of the routine hospital laboratory procedures. The research involving biohazards and all related procedures were approved by the Biosafety Committee of the Beijing Institute of Microbiology and Epidemiology.
ED and DZ conceived the study and designed the experimental procedures. QC, XJ, ZY, YX, LH, and HG performed the experiments. QC, WL, XJ, DZ, and ED analyzed the data. WY, HY, YZ, and XZ provided the reagents and materials. DZ, QC, and ED wrote the manuscript.
This work was supported by the National Science and Technology Major Project (2018ZX10733402) of China and the Key Project Plan of Medical Science Research in Hebei Province (Grant No. 20181072).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02508/full#supplementary-material
Allmeier, H., Cresnar, B., Greck, M., and Schmitt, R. (1992). Complete nucleotide sequence of Tn1721: gene organization and a novel gene product with features of a chemotaxis protein. Gene 111, 11–20. doi: 10.1016/0378-1119(92)90597-i
Ambrose, S. J., Harmer, C. J., and Hall, R. M. (2018). Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid 96-97, 7–12. doi: 10.1016/j.plasmid.2018.02.002
Bailey, J. K., Pinyon, J. L., Anantham, S., and Hall, R. M. (2011). Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 66, 745–751. doi: 10.1093/jac/dkq529
Benoît, D., David, B., Gregory, D., Karine, P., Axel, C., and Mulvey, M. R. (2012). Complete nucleotide sequence of the multidrug resistance IncA/C plasmid pR55 from Klebsiella pneumoniae isolated in 1969. J. Antimicrob. Chemother. 67, 2354–2360. doi: 10.1093/jac/dks251
Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with gapfiller. Genome Biol. 13:R56. doi: 10.1186/gb-2012-13-6-r56
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the uniprot knowledgebase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the rast algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Cain, A., Liu, X., Djordjevic, S., and Hall, R. (2010). Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica Serovar typhimurium from Australian animals. Microb. Drug Resist. 16, 197–202. doi: 10.1089/mdr.2010.0042
Del Castillo, C. S., Jun-Ichi, H., Ho-Bin, J., Seong-Won, N., Tae-Sung, J., Janenuj, W., et al. (2013). Comparative sequence analysis of a multidrug-resistant plasmid from aeromonas hydrophila. Antimicrob. Agents Chemother. 57, 120–129. doi: 10.1128/AAC.01239-12
Didelot, X., and Wilson, D. J. (2015). ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 11:e1004041. doi: 10.1371/journal.pcbi.1004041
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Feng, J., Qiu, Y., Yin, Z., Chen, W., Yang, H., Yang, W., et al. (2015). Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J Antimicrob. Chemother. 70, 2987–2991. doi: 10.1093/jac/dkv232
Fricke, W. F., Welch, T. J., Mcdermott, P. F., Mammel, M. K., Leclerc, J. E., White, D. G., et al. (2009). Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191, 4750–4757. doi: 10.1128/JB.00189-09
Galimand, M., Sabtcheva, S., Courvalin, P., and Lambert, T. (2005). Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 49, 2949–2953. doi: 10.1128/aac.49.7.2949-2953.2005
Hackl, T., Hedrich, R., Schultz, J., and Forster, F. (2014). Proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Harmer, C. J., and Hall, R. M. (2014). pRMH760, a precursor of A/C? plasmids carrying blaCMY and blaNDM genes. Microb. Drug Resist. 20, 416–423. doi: 10.1089/mdr.2014.0012
Harmer, C. J., and Hall, R. M. (2015). The A to Z of A/C plasmids. Plasmid 80, 63–82. doi: 10.1016/j.plasmid.2015.04.003
Harmer, C. J., Hamidian, M., and Hall, R. M. (2017). pIP40a, a type 1 IncC plasmid from 1969 carries the integrative element GI sul2 and a novel class II mercury resistance transposon. Plasmid 92, 17–25. doi: 10.1016/j.plasmid.2017.05.004
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiang, Y., Yu, D. L., Wei, Z. Q., Shen, P., Zhou, Z. H., and Yu, Y. S. (2010). Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC–2, blaDHA–1, qnrB4, and armA. Antimicrob. Agents Chemother. 54, 3967–3969. doi: 10.1128/AAC.00137-10
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
Liang, C., Hongyan, H., Chavda, K. D., Shulong, Z., Renkun, L., Hui, L., et al. (2014). Complete sequence of a KPC-producing IncN multidrug-resistant plasmid from an epidemic Escherichia coli sequence type 131 strain in China. Antimicrob. Agents Chemother. 58, 2422–2425. doi: 10.1128/AAC.02587-13
Ma, L., Yin, Z., Zhang, D., Zhan, Z., Wang, Q., Duan, X., et al. (2017). Comparative genomics of type 1 IncC plasmids from China. Future Microbiol. 12, 1511–1522. doi: 10.2217/fmb-2017-0072
Moura, A., Soares, M., Pereira, C., Leitão, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Nederbragt, A. J. (2014). On the middle ground between open source and commercial software - the case of the newbler program. Genome Biol. 15:113. doi: 10.1186/gb4173
Nigro, S. J., and Hall, R. M. (2011). GIsul2, a genomic island carrying the sul2 sulphonamide resistance gene and the small mobile element CR2 found in the Enterobacter cloacae subspecies cloacae type strain ATCC 13047 from 1890, Shigella flexneri ATCC 700930 from 1954 and Acinetobacter baumannii ATCC 17978 from 1951. J. Antimicrob. Chemother. 66, 2175–2176. doi: 10.1093/jac/dkr230
O’Leary, N. A., Wright, M. W., Rodney, B. J., Stacy, C., Diana, H., Rich, M. V., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Partridge, S., Brown, H., Stokes, H., and Hall, R. (2001). Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45, 1263–1270. doi: 10.1128/aac.45.4.1263-1270.2001
Partridge, S. R., Ginn, A. N., Paulsen, I. T., and Iredell, J. R. (2012). pEl1573 carrying blaIMP–4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob. Agents Chemother. 56, 6029–6032. doi: 10.1128/AAC.01189-12
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., and Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089. doi: 10.1128/AAC.05620-11
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. doi: 10.2144/03342mt01
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, 32–36.
Wei, F., Zhou, D., Qian, W., Luo, W., Zhang, D., Qiang, S., et al. (2016). Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 6:33419. doi: 10.1038/srep33419
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: plasmids, IncC, multidrug resistance, resistance islands, Tn6395, In2-76, ISEcl10
Citation: Cheng Q, Jiang X, Xu Y, Hu L, Luo W, Yin Z, Gao H, Yang W, Yang H, Zhao Y, Zhao X, Zhou D and Dai E (2019) Type 1, 2, and 1/2-Hybrid IncC Plasmids From China. Front. Microbiol. 10:2508. doi: 10.3389/fmicb.2019.02508
Received: 09 September 2019; Accepted: 18 October 2019;
Published: 15 November 2019.
Edited by:
Costas C. Papagiannitsis, University of Thessaly, GreeceReviewed by:
Christopher John Harmer, The University of Sydney, AustraliaCopyright © 2019 Cheng, Jiang, Xu, Hu, Luo, Yin, Gao, Yang, Yang, Zhao, Zhao, Zhou and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t; Erhei Dai, ZGFpZWgyMDA4QDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.