
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 30 May 2018
Sec. Food Microbiology
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01126
Potato is prone to many drastic diseases like potato common scab (CS). As no highly effective methods exist for managing CS, this study explored the possibility of using biological control. Ten bacterial strains were isolated from CS-infected potato tubers from four different locations of Punjab, Pakistan, and identified based on biochemical and molecular analysis. Analysis of 16s rDNA sequences amplified by PCR revealed the isolated bacterial strains to be Streptomyces scabies, S. turgidiscabies and S. stelliscabiei. Pathogenic islands were also confirmed among the isolates after identification of txtAB, nec1, and tomA genes with PCR amplification. One strain isolated from soil was antagonistic to the pathogenic Streptomyces spp., and determined to be Streptomyces A1RT on the basis of 16s rRNA sequencing. A methanolic extract of Streptomyces A1RT contained Isatropolone C, which was purified and structurally determined by 1H- and 13C-NMR, 1H/1H-COSY, HMQC, and HMBC techniques. Streptomyces A1RT also produced the plant growth hormone indole-3-acetic acid (IAA) with a titer of 26 μg ml-1 as confirmed by spectrophotometry and HPLC. In a greenhouse assay, disease severity index was established from 0 to 500. Average disease severity indexes were recorded as 63, 130.5, and 78 for Streptomyces scabies, S. turgidiscabies and S. stelliscabiei, respectively. When Streptomyces A1RT was applied in soil that contained one of these pathogenic isolates, the average disease severity indexes were significantly (P < 0.05) reduced to 11.1, 5.6 and 8.4, respectively. A significant increase in tuber weight and shoot development was also observed with the tubers treated with Streptomyces A1RT. The use of the plant growth-promoting Streptomyces A1RT against potato CS thus provides an alternative strategy to control the disease without affecting environmental, plants, animals and human health.
Common scab (CS) is a recurrent plant disease found all over the world (Loria et al., 1997). It is caused by the Gram positive, filamentous species of Streptomyces in the order of Actinomycetales. Two third of known antibiotics are produced by Actinomycetes and 80% of these antibiotics are produced by Streptomyces spp. (Ahmad et al., 2017; Wang et al., 2017). Under the genus Streptomyce, some species can be plant pathogens, such as Streptomyces scabies, causing CS of potatoes. The main pathogens of CS include Streptomyces scabies, S. acidiscabies, S. turgidiscabies, S. europaeiscabiei and other members, including S. bottropensis, S. stelliscabiei and S. aureofaciens (Lambert and Loria, 1989; Miyajima et al., 1998; Leiminger et al., 2013). These different species are grouped together because of their different prevalence, phenotypic and genotypic characters and they produce similar signs and symptoms to analogous hosts (Loria et al., 1997).
Pathogenic characters of these species are governed by the plant phytotoxin, thaxtomin. The predominant form of thaxtomin produced by S. scabies is thaxtomin A. The biosynthesis of thaxtomin A is prompted by the plant cellobiose and cellotriose (Johnson et al., 2007). Genotypically thaxtomin A production is encoded by the non-ribosomal peptide synthetase genes [txtA and txtB (txtAB)] and cytochrome P-450 type monooxygenase gene (txtC) (Healy et al., 2002). The txtAB genes are pathogenicity determinant, but other genes like nec1 and tomA facilitate or enhance the pathogenicity, although they are not absolutely required (Kreuze et al., 1999; Healy et al., 2000; Flores-González et al., 2008; Barry et al., 2012).
The use of bacterial strains for CS suppression depends upon many factors including the interaction between antagonists and pathogens, environmental conditions, ecological and evolutionary dynamics of antagonistic strains (Kinkel et al., 2012). The use of higher densities of antagonistic Streptomyces are consistent with CS disease suppression (Wanner, 2007) due to the complementarity among antibiotics of Streptomyces. Environmental stress like soil pH, moisture and temperature can be varied in different fields or even within the same field, in the same or different seasons. The effectiveness of same antagonist can be varied among season to seasons in the same or even different fields (Prévost et al., 2006). Some potato cultivars are more sensitive to CS pathogens and they are difficult to control because their adaptiveness to harbor S. scabies is greater as compared to antagonistic strain (Neeno-Eckwall et al., 2001).
There is long-standing interest in the use of plant growth-promoting bacteria as a strategy for sustainable agriculture (Shennan, 2008). The use of natural or manipulated microbial communities which suppress phytopathogens not only has the potential to reduce disease input but also reduce the need of chemical inputs such as pesticides and fungicides (Emmert and Handelsman, 1999). In Nature, plants benefit from symbiotic bacteria that counteract pathogens and thus inhibit the growth of disease in the plant (Lorang et al., 1995). PGRP are focused for use as biological control agent due to beneficial characters which include the production of bioactive compounds (Samac et al., 2003) and secondary metabolites including antibiotics, insecticides, and pesticides (Berdy, 2005). In addition, plant growth-promoting bacteria enhance plant growth by producing phytohormones like auxins (Lee et al., 2004), siderophores (Ryan et al., 2008), and by activating plant defense pathways (Lin et al., 2012).
In the past, microorganisms have shown good potential as plant-growth promoting bacteria to enhance the growth and yield of rice (Lucas et al., 2009), wheat (Majeed et al., 2015), beans (Perez-Montano et al., 2014), maize (Qaisrani et al., 2014), and soya bean (Cassán et al., 2009). But limited data is available for growth-promotion (Abbas et al., 2014) and disease suppression (Kobayashi et al., 2015) in potatoes. Streptomyces spp. have been reported as plant growth promoting bacteria due to beneficial effects in resisting pathogens (Neeno-Eckwall et al., 2001), producing lytic enzymes (Prapagdee et al., 2008), resisting environmental effects (Coombs and Franco, 2003), and secreting siderophores and phytohormones (Berg, 2009; Khamna et al., 2009) which are beneficial in interacting with the plant rhizosphere (Khamna et al., 2009).
In present study, potato CS pathogens were isolated from Punjab, Pakistan. The antagonistic bacterial strains were isolated from a potato field with no CS history over the past 5 years. It was hypothesized that such fields may contain microorganisms which augment plant growth and help in disease suppression in potato crops. With this aim, a study was conducted for isolation and characterization of potato CS pathogens. Sampling was also performed to isolate antagonistic bacterial strains. We described the potential of Streptomyces A1RT as plant growth- promoting and CS disease suppressing in potato crop. To our best knowledge this is the first report of biocontrol against four major CS pathogens.
CS-infected potato tubers were collected from Potato Research Institute (PRI), Sahiwal, Pakistan. Potato samples were shifted to the Department of Microbiology and Molecular Genetics, University of the Punjab Lahore, Pakistan. Samples were graded according to their disease severity as very low, low, moderate, moderately high, high and very high depending upon the percentage 10, 20, 40, 60, 80, and 90% of CS symptoms respectively.
Fourteen potato samples with various CS symptoms were processed. All potato samples were washed with sterile distilled water and then disinfected with 2.8% sodium hypochlorite (NaOCl) solution, followed by rinsing three times with sterile distilled water. The scabby lesions were carefully cut with sterile scalpel and homogenized with 2 mL Tris–HCl (pH 7.2) in a Eppendorf tube followed by incubation for 10 min at 50°C in incubator. The homogenized mixture was further diluted three times with Tris–HCl (pH 7.2). For each aliquot, 100 μl diluted sample was spread on yeast malt extract (YME) agar plates (Meng et al., 2011) and incubated at 28°C for 7 days. Selected single bacterial colonies were spread on another YME agar plate and physical growth characters were recorded for each Streptomyces spp. Ten bacterial isolates having Streptomyces-like growth characters were isolated and purified on additional YME agar plates. These Streptomyces spp. were identified based on typical growth characters, diffusible pigments and spore colors compared with already identified Streptomyces spp. (positive controls kindly provided by Dr. Jurgen Leiminger, Freising, Germany).
For isolation of antagonistic bacterial spp., soil samples were collected from a potato growing field that have had very low or no potato CS history over past 5 years. Antibiotic- producing bacterial spp., especially Streptomyces spp., were selected as potential CS antagonists (Kharel et al., 2005).
For DNA extraction, the selected Streptomyces colonies were grown in YME broth for 5 days at 28°C. Each culture was centrifuged and the pellets were subjected to DNA extraction by using the Gene-JET genomic DNA purification kit (Thermo Scientific, United States). The extracted DNA was quantified by absorbance at 260 nm (NanoDrop, Thermo Scientific, United States). High quality DNA (A260/A280 ratio was 1.7) samples were subjected to PCR amplification by using 16S rRNA universal primers (Edwards et al., 1989) for the characterization of the Streptomyces spp. Each PCR reaction contained 1 μL DNA template, 2.5 μL PCR buffer (MgCl2), 1 μL Taq DNA polymerase, 1 μL 2 mM dNTPs, 1 μL 10 pmol of each primer and 18.5 μL H2O to afford a total volume of 25 μl. PCR amplification was performed in BioRad Thermocycler, United States. The PCR reaction was carried out with an initial denaturation at 94°C for 3 min followed by 35 cycles of denaturation at 94°C for 20 s, annealing at 60°C for 30 s, and extension at 72°C for 40 s. Following the cycling, a final extension at 72°C for 3 min was performed.
To identify Streptomyces strains, species-specific primers were used for S. scabies (Lehtonen et al., 2004), S. turgidiscabies, S. acidiscabiei (Tagawa et al., 2008), S. europaeiscabiei, S. stelliscabiei and S. bottropensis (Wanner, 2006). The 16S-23S internal transcribed spacer (ITS) region was amplified by using the ITS-F and ITS-R primer pair (Song et al., 2004). The amplicon was digested with Hpy99I (New England Biolabs) which cut the ITS site at 1629-1633 nucleotide position. The restriction pattern was then visualized by gel electrophoresis. The PCR products were sequenced by GACT, Germany. The 16S rRNA sequences have been submitted to European Nucleotide Database EMBL with the accession numbers KU560917, LN908787 and LN908789.
Polymerase chain reaction primers were designed to identify pathogenicity island (PAI) of the susceptible pathogenic Streptomyces strains. PCR amplification was performed by using txtAB gene, tomA gene (Wanner, 2006) and nec1 gene (Bukhalid et al., 1998). PCR was performed as conditions described above except annealing temperature was set as 48°C, 55°C and 60°C for the txtAB, tomA and nec1 genes, respectively. All the identified genes have been deposited in the NCBI with the accession numbers KX842598-606.
To check the antibacterial activity of the antagonistic Streptomyces spp. against pathogenic CS causing Streptomyces strains, a disk diffusion assay was performed (CLSI, 2015). Twelve Streptomyces spp. were grown separately in YME broth for 5–7 days at 28°C in shaking incubator. An aliquot of 100 μL of pathogenic Streptomyces spp. culture was spread as a lawn on YME agar plates with the help of RattlerTM plating beads (Zymo Research Cooperation, United States). After drying the plates, individual filter paper disks were used to absorb 25 μL of MeOH extract from each antagonistic Streptomyces isolate. The filter disks were then applied to the YME agar plates, which were then placed in incubator at 28°C for 3 days in upright position. After incubation, the inhibition zone around the filter disks were recorded in millimeters (mm). To confirm the antibacterial activity of Isatropolone C containing fractions, the bacterial inhibition assay was performed using Isatropolone C dissolved in methanol.
To determine the potential of antagonistic Streptomyces isolates as plant growth-promoting organisms, colorimetric estimation of IAA was performed as described by Gordon and Weber (1951). Bacterial strains were grown in triplicates for 5–7 days at 28°C, 180 rpm, in ISP-2 liquid medium supplemented with 500 μg mL-1 L-tryptophan (filter sterilized). After incubation, the culture was centrifuged at 14,000 rpm for 15 min. One milliliter of supernatant was mixed with 2 mL Salkowski’s reagent (prepared by adding 1 mL of 0.5 M FeCl2 to 50 mL 35% perchloric acid) and incubated in the dark at room temperature for half an hour. Development of pink color was observed as an indicator of IAA production. IAA estimation was performed by measuring the absorbance at 535 nm (Uvikon 933). A standard curve for IAA was generated and the IAA titer was expressed as μg mL-1.
Indole-3-acetic acid production was also determined by HPLC-DAD-MS. Culture supernatants generated as described above were acidified to pH 2.5 with 1N HCl and extracted twice with an equal volume of ethyl acetate. The organic phase was evaporated in vacuo and the resulting powder redissolved in 5 mL MeOH. HPLC analysis was performed on a Agilent 1100 system equipped with a diode array detector and a quadrupole mass detector. The crude methanolic extract of IAA was diluted 10 times and 20 μL was injected onto a reverse phase XBridge C18 column (3.5 μm, 100 mm × 4.6 mm) equilibrated with 95:5 methanol/water. The extract was resolved isocratically at a flow rate of 0.5 mL min-1. The retention time of IAA was identified by comparison to a standard sample (Sigma-Aldrich).
Antagonistic strains were cultivated in 500 mL shake flasks containing 150 mL TSB medium for 24–48 h at 28°C, 180 rpm. Ten milliliters of this preculture was used to inoculate 150 mL of YME medium and incubated for 3–5 days under the same conditions. After cultivation, the cells were removed by centrifugation and the culture supernatant was extracted twice with 100 mL ethyl acetate. After concentration in vacuo, the extract was dissolved in methanol and used for HPLC-DAD-MS analysis. The HPLC conditions were identical to those used for IAA analysis.
A 100 mL TSB pre-culture of Streptomyces sp. A1RT was used to inoculate 10 L YME, which was then incubated for 3 days at 28°C, 180 rpm. After incubation, the cells were removed by centrifugation and the supernatant extracted twice with an equal volume of ethyl acetate. The solvent was evaporated in vacuo to produce a powder. Acetylation was performed with acetic anhydride and pyridine. One hundred milligrams of the powdered extract was re-suspended in 1 mL pyridine in a twin neck round bottom flask with continuous stirring. The flask was flushed with nitrogen and 100 μL acetic anhydride was added by drops via syringe. The reaction was allowed to proceed for 30 min at room temperature, whereupon the reaction was concentrated in vacuo. The resulting powder was redissolved in 5 mL methanol and applied to an Oasis® HLB20 35 cc cartridge (6 g). The cartridge was eluted with a step gradient of methanol, ranging from 20 to 100%. Each fraction was analyzed by HPLC-DAD-MS. Fractions containing acetylated Isatropolone C were concentrated in vacuo.
Acetylated Isatropolone C was further purified by semi-preparative HPLC (Agilent Technologies) using a Zorbax B-C18 (9.4 mm × 150 mm) pre-column and a Zorbax B-C18 (9.4 mm × 20 mm) main column. A concentrated methanol solution of crude compound was applied to the column and eluted with acetonitrile / 0.5% acetic acid as buffer A and water / 0.5% acetic acid as buffer B at a flow rate of 2 mL min-1. Purified acetylated Isatropolone C was dissolved in CD3OD and analyzed by 1H-NMR (400 MHZ) and 13C-NMR (100 MHZ) on a Bruker DRX-500 spectrometer (Bruker, Karlsruhe, Germany). 2D-NMR (1H/1H-COSY, HMQC, and HMBC) and high resolution MS were also used to confirm the structure of the compound.
The pathogenicity of the isolated Streptomyces toward potato tubers was tested as described by Wanner (2006). S. europaeiscabiei strain G1 was used as a positive control (kindly provided by Dr. Jurgen Leiminger, Freising, Germany). To confirm the normal growth pattern of potato tubers, one control without any bacterial inoculation was also used. The Streptomyces isolates were inoculated in 100 mL YME broth in 250 ml flasks and placed in shaking incubator at 28°C, 180 rpm, for 5–7 days until they attained 106 CFU mL-1 concentration. Each culture was centrifuged at 10,000 rpm for 2 min to collect cells. The cell pellet was re-suspended in 100 mL-1 sterile distilled water to prepare vermiculite. Medium size pots (8 L) were substrate-filled with autoclaved Compo Sana Universal® (Munster Germany). One hundred mL vermiculite inoculum was added and spread equally in each substrate filled pot, in three replicates. Potato tubers were surface sterilized by 0.5% bleach for 10–15 min and washed with sterilized water. The potatoes were twice grown in a greenhouse, once in September 2015 and a second time in March 2016. The average temperature was 22–25°C. Plants were watered as required and continuously monitored for growth. After 3 months the progeny tubers were harvested, weighed, and the length of shoots and roots recorded. The tubers were also scored for disease. Pathogenic Streptomyces spp. were again isolated from infected lesions in harvested tubers to confirm the source of CS.
Statistical analysis was performed using SPSS software (IBM SPSS Statistics, Version 21). Data was collected in triplicates and subjected to one-way analysis of variance (ANOVA). Compared means were separated by Duncan’s multiple range test (DMRT). Values of p < 0.05 were considered statistically significant.
Out of ten Streptomyces spp. isolated from CS infected tubers, seven CS-causing Streptomyces spp. were identified based on 16S rRNA analysis by using specific primers designed for different CS species (Supplementary Table S1). Two isolates were identified as S. scabies, four as S. turgidiscabies, and one as S. stelliscabiei. The two S. scabies isolates were distinguished from S. europaeiscabiei by the presence of a Hpy99I restriction enzyme site within the 16S-23S ITS region of the 16S rRNA sequences (Flores-González et al., 2008).
Polymerase chain reaction was used to identify genes in the isolates that encode pathogenicity. Seven out of ten Streptomyces isolates, representing S. scabies, S. turgidiscabies and S. stelliscabiei, were subjected to PCR using primers to amplify txtAB gene. Beside txtAB gene, nec1 and tomA genes were detected by PCR amplification (Figures 1A–C). Neither nec1 nor tomA were detected in the txtAB-negative Streptomyces isolates but both genes were present among all txtAB-positive Streptomyces isolates. Accession numbers are summarized in Supplementary Table S2.
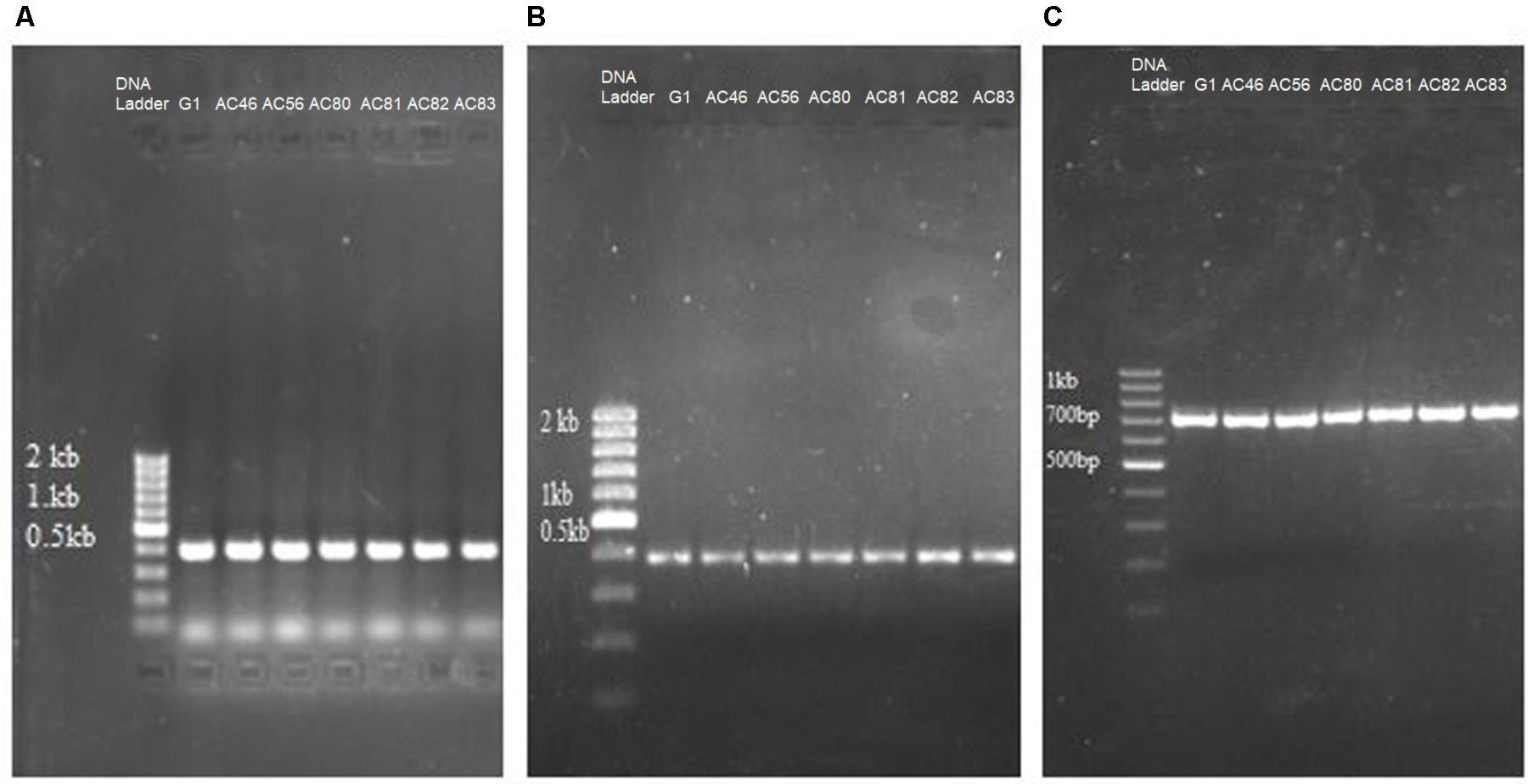
FIGURE 1. PCR detection of genes related to CS pathogenicity in seven Streptomyces isolates. Shown are the agarose electrophoresis gels of PCR reactions amplifying (A) txtAB; (B) nec1; (C) tomA. The name of each isolate (G1, AC46, AC56, AC80, AC81, AC82, AC83) is shown above the respective lanes of the gels.
Streptomyces A1RT, which shows activity against S. scabies (see below), did not test positive for txtAB, nec1 and tomA by PCR, indicating that this strain is non-pathogenic and hence, does not produce thaxtomin A.
Streptomyces strains were isolated from soil, showing activity against S. scabies. Out of twelve antagonistic Streptomyces strains, extracts of seven strains showed bioactivity (inhibition zone ranged from 6 to 26 mm) (Supplementary Table S3). The highest extract activity (26 mm) against S. scabies was derived from Streptomyces A1RT (Supplementary Table S3 and Figure 2). The Streptomyces A1RT extract was also active against S. turgidiscabies and S. stelliscabiei (data not shown).
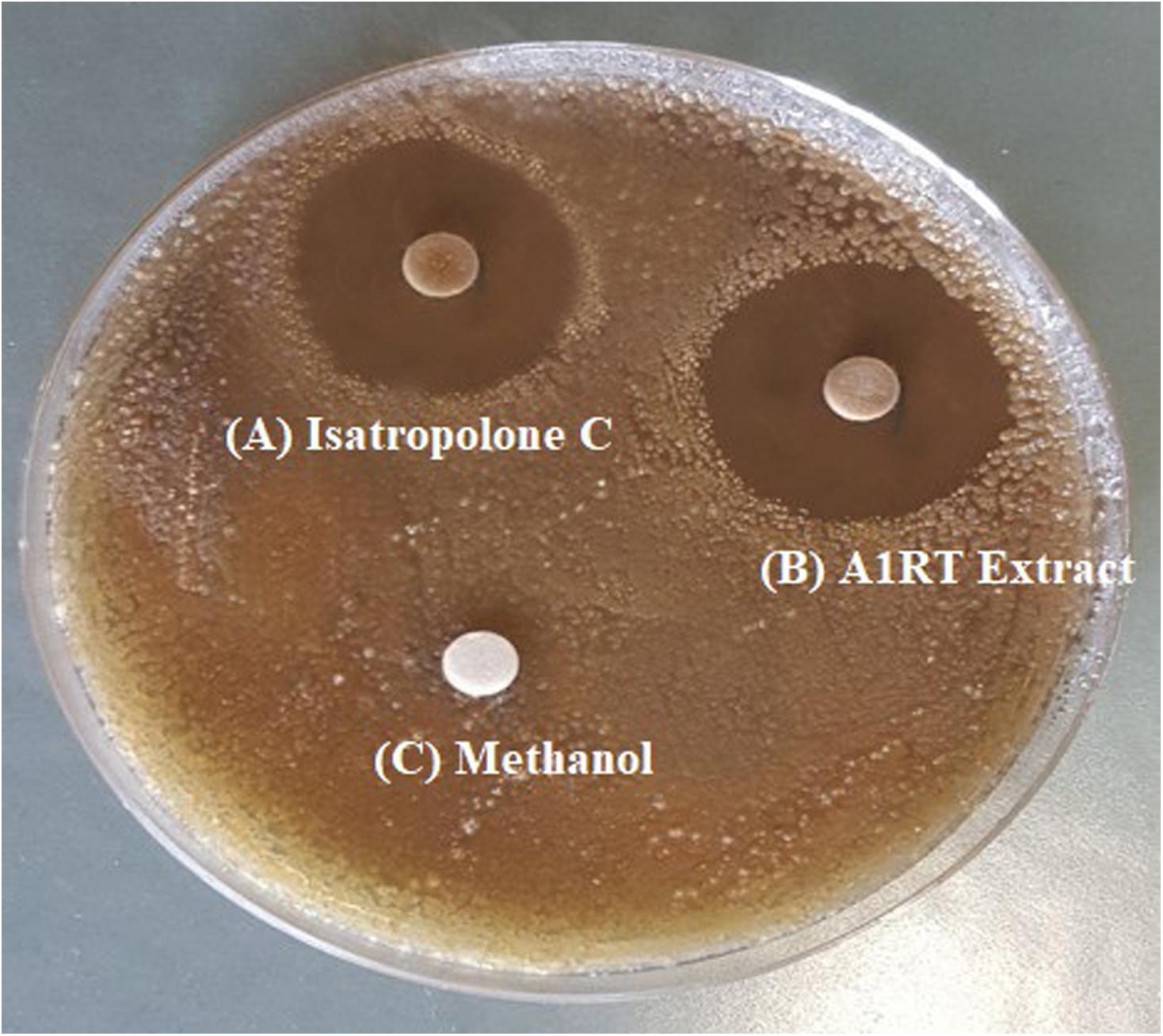
FIGURE 2. Inhibition of S. scabies by disk diffusion assay. S. scabies strain AC46 was grown on a solid medium plate in the presence of filter paper disks containing (A) Purified Isatropolone C dissolved in methanol. (B) Crude extract of Streptomyces A1RT (C) Methanol only.
The twelve strains were also analyzed for their ability to produce IAA. The best producer, Streptomyces A1RT, produced IAA at a concentration of 26 μg mL-1 after 4 days of incubation at 28°C. Production was confirmed by HPLC analysis of the extract (Figure 3).
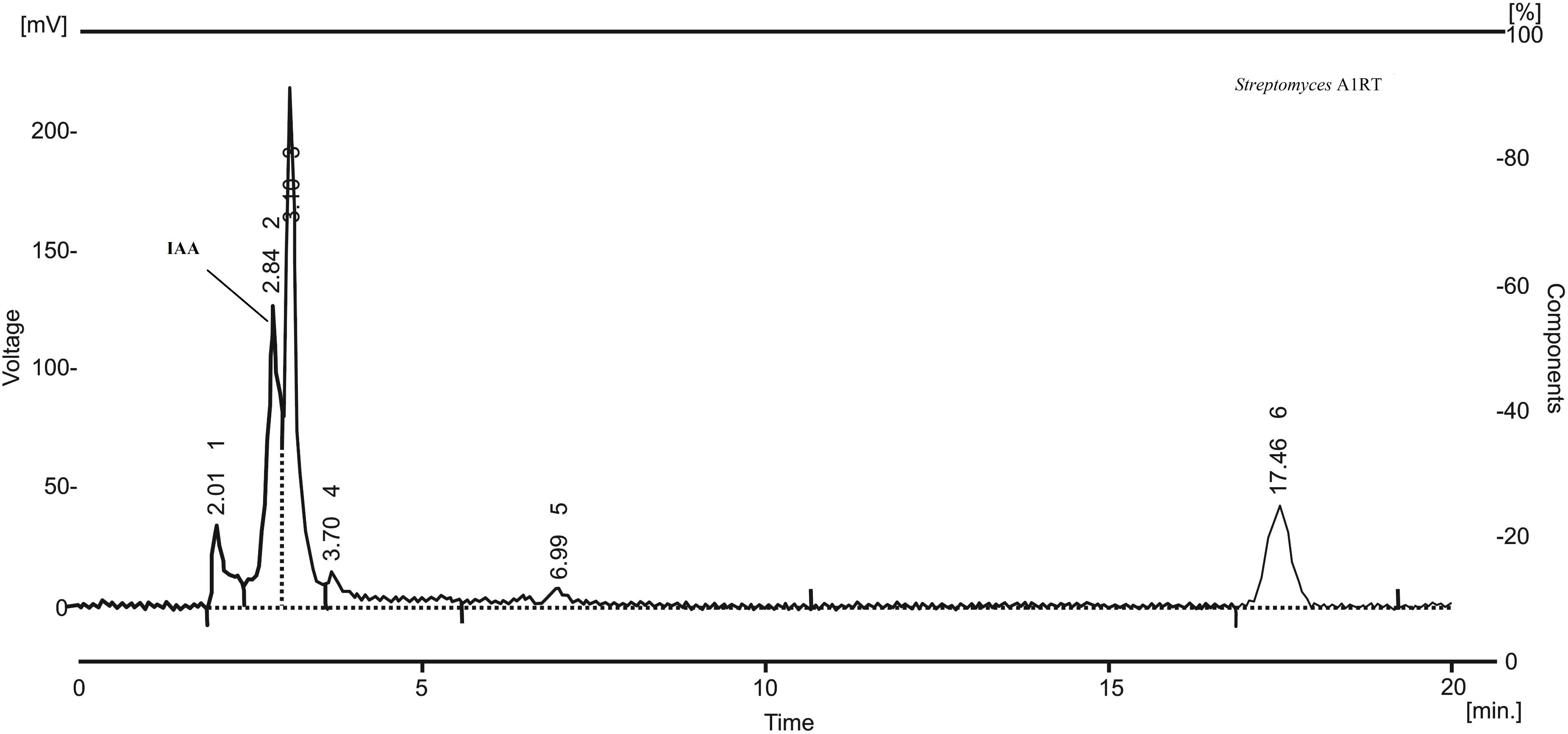
FIGURE 3. HPLC chromatogram of an extract from Streptomyces A1RT. The peak for indole-3-acetic acid (IAA) is shown. The retention time of the peak is identical to a standard sample of indole-3-acetic acid.
One prominent metabolite produced by Streptomyces A1RT was selected for purification and structural characterization. Through the targeted mass fractionation, a dark yellow to orange amorphous powder was isolated which showed antibiotic activity. The molecular formula was predicted to be C24H24O10 by high resolution MS (Supplementary Figure S1) based on the observed molecular ion [M-H]-; with m/z 471.1302 (calcd. for C24H24O10, m/z 471.1302), which corresponds to a hydrogen deficiency index of 13.
Due to the instability of this compound during purification, the crude extracts of the fermentation broth were subjected to global acetylation to protect putative reactive hydroxy groups in the molecule (Yu et al., 2012). A monoacylated molecule with m/z 513.2 ([M-H]-) was detected in the extract, which was isolated through a sequence of chromatographic steps to yield 5 mg of purified compound. Single and 2-dimensional NMR spectroscopic data (Supplementary Table S4 and Supplementary Figures S3–S7) for the compound was consistent with a O-acylated derivative of the recently described Isatropolone C (Figure 4) (Cai et al., 2017). The absorbance spectrum (Supplementary Figure S2) revealed distinct maxima at 300–350 nm and closely resembled the spectrum reported for Isatropolone A (Cai et al., 2017).
Three Streptomyces isolates (AC46, AC56, and AC80) produced CS lesions on surface of potatoes while no observable CS symptoms were detected for tubers infested with combination of pathogens and antagonistic bacterial strain (Figure 5). The CS symptoms ranged from superficial to deep pitted scars and protruding CS lesions. In order to calculat disease severity, disease severity (DS) index was established ranging from 0 to 500 as described by Wanner (2009). All tested organisms having DS index higher than 20 were considered pathogenic (Wanner, 2009). The DS index of test strains were ranged from 102, 57, 107 and 58 for S. europaeiscabiei strain G1, AC46, AC56 and AC80, respectively. The DS index was significantly (p < 0.05) reduced to 14.7, 1.8, 4.7 and 1.4 for strains G1, AC46, AC56 and AC80, respectively when used in combination with Streptomyces A1RT (Figure 6). The DS index was 0 for tuber exposed to Streptomyces A1RT and 2.3 for non-infested tubers.
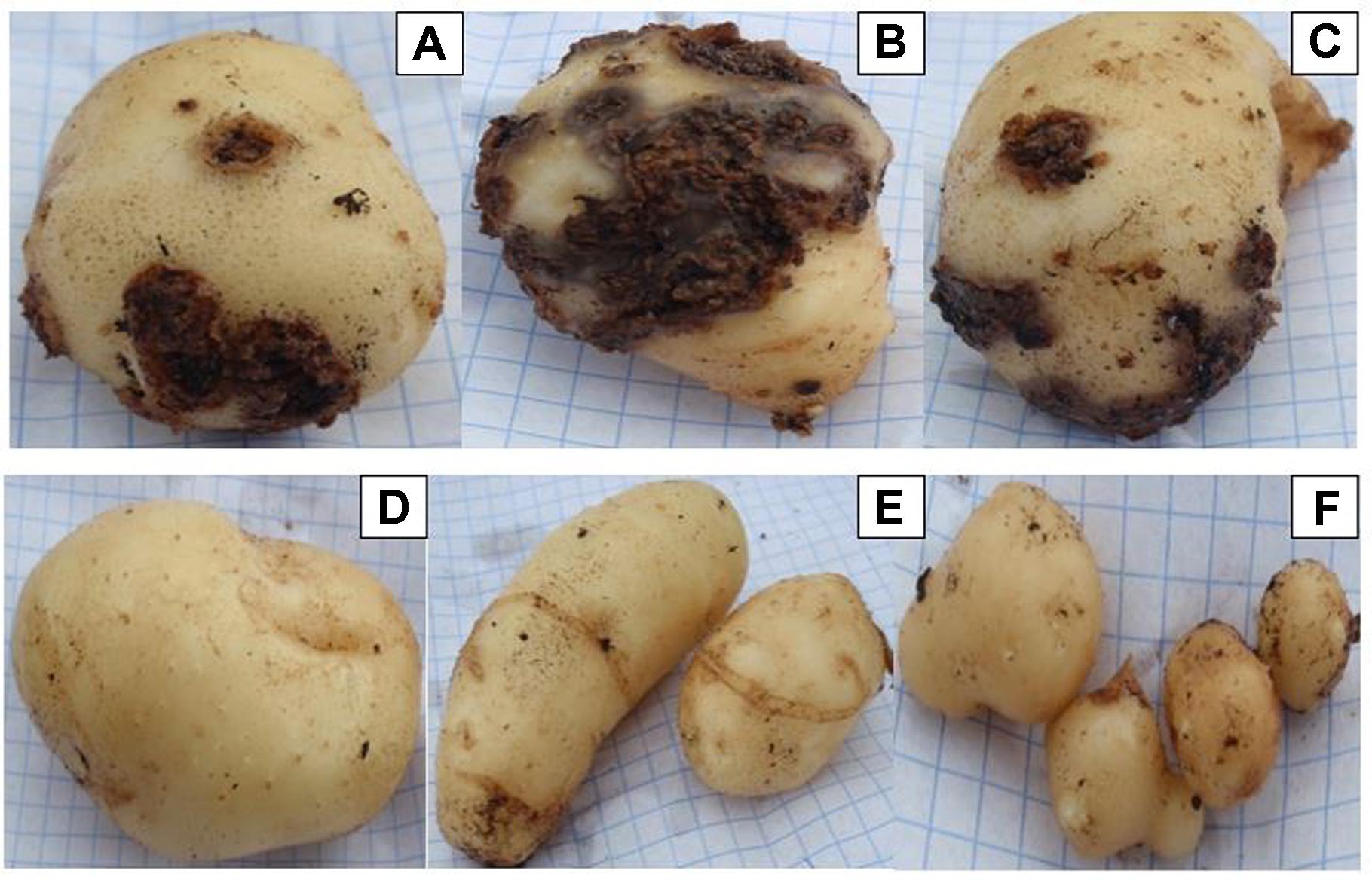
FIGURE 5. Potato tubers harvested from potting soil treated with Streptomyces strains (A) AC46; (B) AC56; (C) AC80; (D) A1RT+AC46; (E) A1RT+AC56; and (F) A1RT+AC80.
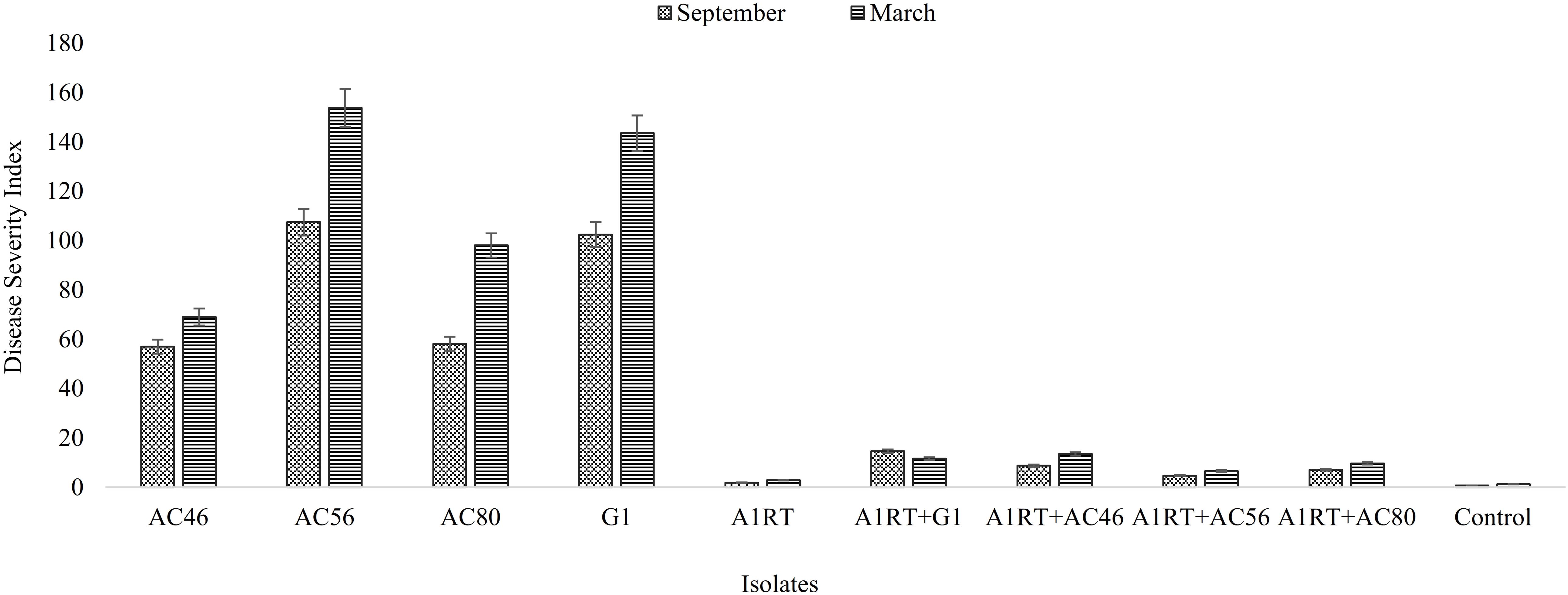
FIGURE 6. Graphical representation of disease severity index calculated after harvesting. Error bars represent standard error. The results shown in triplicates (p < 0.05).
Growth promotion potential was assessed in terms of relative gain in tuber weight, root and shoot development to the plants exposed to Streptomyces A1RT as compared to the plants exposed to CS pathogens only. For example, when tubers inoculated with CS pathogens, average tuber weight was measured as 40 g which significantly (p < 0.05) increased up to 60 g when used in combination with Streptomyces A1RT. Similarly, an average 5–10 cm increase in shoot and root length (Figure 7) was observed in plants inoculated with pathogens and Streptomyces A1RT as compared to plants exposed to CS pathogens only. However, not a significant difference (p > 0.05) was observed between experiments conducted in March and September.
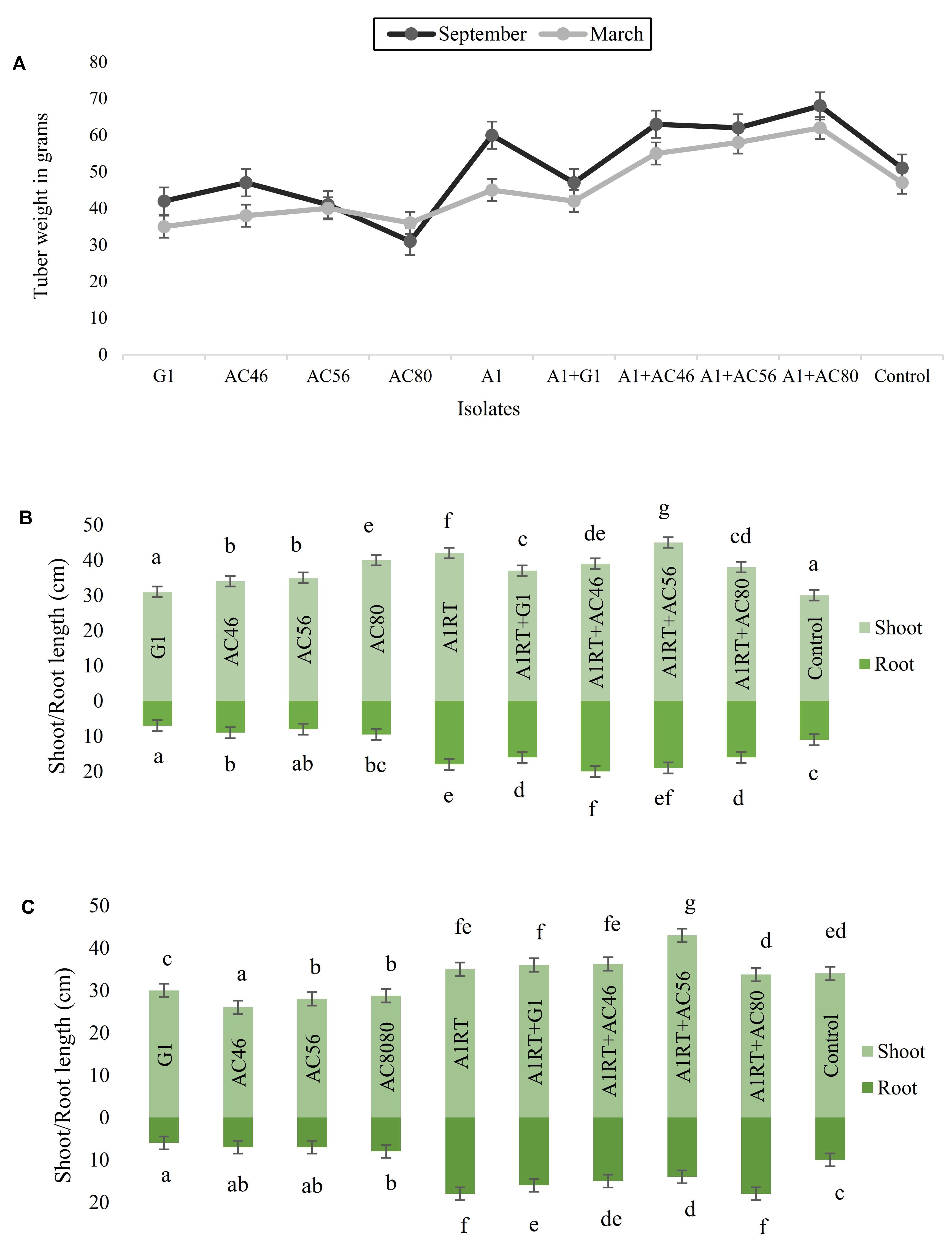
FIGURE 7. Graphical representation of plant growth promoting effects (A): Increase in tuber growth was seen when using Streptomyces A1RT with different CS pathogens. (B) Shoot and root growth in September (C) Shoot and root growth in March. Error bars representing mean ± S.E of triplicates. The letters above the bars indicate significant differences between treatments calculated by Duncan’s multiple range test (DMRT).
The history of controlling soil borne pathogens by using Streptomyces spp. as biocontrol agents is more than 50 years old. In addition to nutrient competition, Streptomyces spp. have multiple mechanisms which can be used to control soil pathogens, including production of antibiotics, degradative enzymes, and nitrous oxide (Cohen and Mazzola, 2006; Mahmoudi et al., 2011). Among them, disease control by producing antibiotics remained an active and most promising strategy. Additionally, Streptomyces spp. have potential as plant growth-promoting microorganisms to enhance carbon and nitrogen resources and efficiently competing in the rhizosphere (Schlatter et al., 2008). Therefore, the ultimate benefit of using plant growth promoting bacteria is not only in disease suppression or enhancement of plant growth factors, but also development of sustainable agricultural practices. This latter factor is especially important in developing countries like Pakistan where almost 250 kg nitrogen and 150 kg each of phosphorus and potash is used per hectare for potato production (Naqqash et al., 2016). To this end, we report the isolation of the strain Streptomyces A1RT from field suppressive soil where CS disease symptoms were negligible over the period of 5 years.
CS disease-promoting bacteria are an evolving threat to potato crops. Although S. scabies is the best known causative agent of CS disease, it was reported that S. turgidiscabies also has the pathogenicity-related genes that encode the production of the phytotoxin thaxtomin A. Although S. turgidiscabies shows high sequence similarity with S. scabies (Kers et al., 2005), both species are different (Miyajima et al., 1998). Moreover, S. turgidiscabies was reported as more of a CS threat in terms of growth characters, virulence, and ability to grow even in humid environmental conditions (Hiltunen et al., 2009). Such features gave rise to the hypothesis that S. turgidiscabies may be evolving as an emerging plant pathogen and have gained decisive pathogenic factors from S. scabies (Loria et al., 2006). Previous data also suggest that due to the versatility and adaptability of S. turgidiscabies to different environmental conditions, it is possible that S. turgidiscabies will displace S. scabies from the environment in the future. Therefore, it is important to find antagonistic organisms that act against this emerging CS pathogen. In past studies, bacterial species such as Bacillus spp. (Han et al., 2005), Pseudomonas spp. (Arseneault et al., 2013), S. albidoflavus (Hayashida et al., 1989) and S. diastatochromogenes (Neeno-Eckwall et al., 2001) have been used as biological control of potato CS. Likewise, Streptomyces spp. have been used as biological control agents against S. scabies (Prévost et al., 2006) and S. turgidiscabies (Hiltunen et al., 2009). However, to our knowledge Streptomyces A1RT is the first example of a biological control that is active against S. scabies and S. turgidiscabies, but also the CS pathogens S. europaeiscabiei and S. stelliscabiei.
It is known that the plant growth-promoting hormone IAA plays a crucial role in germination rate and the development of roots and shoots (Fatima et al., 2009). In this study we demonstrate that Streptomyces A1RT has the ability to produce high levels of IAA. Accordingly, potato tubers treated with Streptomyces A1RT developed better in terms of roots elongation, number of shoots, and tuber weight. Therefore, Streptomyces A1RT is novel in its ability to not only suppress multiple CS pathogens but also promote potato tuber growth.
Greenhouse trials performed by using pathogenic CS strains indicated the CS disease was suppressed in pots when Streptomyces A1RT was present. This may be due to the production of an antibiotic compound by Streptomyces A1RT. Disk diffusion assays clearly indicated that a highly active compound (or compounds) are produced by Streptomyces A1RT. A prominent compound produced by Streptomyces A1RT was purified and spectroscopically shown to be Isatropolone C. Isatropolone C was recently discovered as a metabolite of Streptomyces Gö66 and was shown to have anti-parasitic activity (Cai et al., 2017). In our study, we report that an Isatropolone C containing extract shows high antibiotic activity.
In summary, a major goal of 21st century is to develop an environmentally friendly and sustainable agriculture. This study has identified the novel strain Streptomyces A1RT as a promising and effective method for biological control of multiple CS pathogens, while simultaneously promoting plant growth.
AS conducted all experimental work. ZL assisted with data anlaysis. SZ and JZ performed chromatographic steps and NMR analysis. DZ and AB assisted with experimental design and writing the manuscript.
This research was supported by the government of Pakistan and by the University of Freiburg. We gratefully acknowledge the Robert S. McNamara World Bank fellowship and Higher Education Commission of Pakistan (IRSIP-1-8/ HEC/HRD/2016/5915) for providing funding and visiting opportunity to Germany.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Khaled Attia Shaaban Mahmoud (University of Kentucky, United States) for helping to elucidate the NMR structure and Dr. Jurgen Leiminger (Freising, Germany) for providing technical support and positive strains. We also thank to Dr. Jianjun Hao (University of Maine, United States) for reviewing the manuscript. Thanks to University of the Punjab, Lahore, Pakistan and University of Freiburg, Germany, for providing access to NMR and lab facilities to conduct all research work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01126/full#supplementary-material
Abbas, M. T., Hamza, M. A., Youssef, H. H., Youssef, G. H., Fayez, M., Monib, M., et al. (2014). Bio-preparates support the productivity of potato plants grown under desert farming conditions of north Sinai: five years of field trials. J. Adv. Res. 5, 41–48. doi: 10.1016/j.jare.2012.11.004
Ahmad, M. S., El-Gendy, A. O., Ahmed, R. R., Hassan, H. M., El-Kabbany, H. M., and Merdash, A. G. (2017). Exploring the antimicrobial and antitumor potentials of Streptomyces sp. AGM12-1 Isolated from Egyptian Soil. Front. Microbiol. 8:438. doi: 10.3389/fmicb.2017.00438
Arseneault, T., Goyer, C., and Filion, M. (2013). Phenazine production by Pseudomonas sp. LBUM223 contributes to the biological control of potato common scab. Phytopathology 103, 995–1000. doi: 10.1094/phyto-01-13-0022-r
Barry, S. M., Kers, J. A., Johnson, E. G., Song, L., Aston, P. R., Patel, B., et al. (2012). Cytochrome P450-catalyzed L-tryptophan nitration in thaxtomin phytotoxin biosynthesis. Nat. Chem. Biol. 8, 814–816. doi: 10.1038/nchembio.1048
Berg, G. (2009). Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Bukhalid, R. A., Chung, S. Y., and Loria, R. (1998). nec1, a gene conferring a necrogenic phenotype, is conserved in plant-pathogenic Streptomyces spp. and linked to a transposase pseudogene. Mol. Plant Microbe Interact. 11, 960–967. doi: 10.1094/mpmi.1998.11.10.960
Cai, X., Shi, Y. M., Pohlmann, N., Revermann, O., Bahner, I., Pidot, S. J., et al. (2017). Structure and biosynthesis of isatropolones, bioactive amine-scavenging fluorescent natural products from Streptomyces Go66. Angew. Chem. Int. Ed. Engl. 56, 4945–4949. doi: 10.1002/anie.201701223
Cassán, F., Perrig, D., Sgroy, V., Masciarelli, O., Penna, C., and Luna, V. (2009). Azospirillum brasilense Az39 and Bradyrhizobium japonicum E109, inoculated singly or in combination, promote seed germination and early seedling growth in corn (Zea mays L.) and soybean (Glycine max L.). Eur. J. Soil Biol. 45, 28–35. doi: 10.1016/j.ejsobi.2008.08.005
Cohen, M. F., and Mazzola, M. (2006). Resident bacteria, nitric oxide emission and particle size modulate the effect of Brassica napus seed meal on disease incited by Rhizoctonia solani and Pythium spp. Plant Soil 286, 75–86. doi: 10.1007/s11104-006-9027-1
Coombs, J. T., and Franco, C. M. (2003). Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 69, 5603–5608. doi: 10.1128/AEM.69.9.5603-5608.2003
Edwards, U., Rogall, T., Blocker, H., Emde, M., and Bottger, E. C. (1989). Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853. doi: 10.1093/nar/17.19.7843
Emmert, E. A., and Handelsman, J. (1999). Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol. Lett. 171, 1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x
Fatima, Z., Saleemi, M., Zia, M., Sultan, T., Aslam, M., Rehman, R., et al. (2009). Antifungal activity of plant growth-promoting rhizobacteria isolates against Rhizoctonia solani in wheat. Afr. J. Biotechnol. 8, 219–225. doi: 10.1111/1751-7915.12158
Flores-González, R., Velasco, I., and Montes, F. (2008). Detection and characterization of Streptomyces causing potato common scab in Western Europe. Plant Pathol. 57, 162–169. doi: 10.1111/j.1365-3059.2007.01734.x
Gordon, S. A., and Weber, R. P. (1951). Colorimetric estimation of indoleacetic acid. Plant Physiol. 26, 192–195. doi: 10.1104/pp.26.1.192
Han, J. S., Cheng, J. H., Yoon, T. M., Song, J., Rajkarnikar, A., Kim, W. G., et al. (2005). Biological control agent of common scab disease by antagonistic strain Bacillus sp. sunhua. J. Appl. Microbiol. 99, 213–221. doi: 10.1111/j.1365-2672.2005.02614.x
Hayashida, S., Choi, M.-Y., Nanri, N., Yokoyama, M., and Uematsu, T. (1989). Control of potato common scab with an antibiotic biofertilizer produced from swine feces containing Streptomyces albidoflavus CH-33. Agric. Biol. Chem. 53, 349–354. doi: 10.1080/00021369.1989.10869326
Healy, F. G., Krasnoff, S. B., Wach, M., Gibson, D. M., and Loria, R. (2002). Involvement of a cytochrome P450 monooxygenase in thaxtomin a biosynthesis by Streptomyces acidiscabies. J. Bacteriol. 184, 2019–2029. doi: 10.1128/jb.184.7.2019-2029.2002
Healy, F. G., Wach, M., Krasnoff, S. B., Gibson, D. M., and Loria, R. (2000). The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol. Microbiol. 38, 794–804. doi: 10.1046/j.1365-2958.2000.02170.x
Hiltunen, L. H., Ojanperä, T., Kortemaa, H., Richter, E., Lehtonen, M. J., and Valkonen, J. P. T. (2009). Interactions and biocontrol of pathogenic Streptomyces strains co-occurring in potato scab lesions. J. Appl. Microbiol. 106, 199–212. doi: 10.1111/j.1365-2672.2008.03992.x
Johnson, E. G., Joshi, M. V., Gibson, D. M., and Loria, R. (2007). Cello-oligosaccharides released from host plants induce pathogenicity in scab-causing Streptomyces species. Physiol. Mol. Plant Pathol. 71, 18–25. doi: 10.1016/j.pmpp.2007.09.003
Kers, J. A., Cameron, K. D., Joshi, M. V., Bukhalid, R. A., Morello, J. E., Wach, M. J., et al. (2005). A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol. Microbiol. 55, 1025–1033. doi: 10.1111/j.1365-2958.2004.04461.x
Khamna, S., Yokota, A., and Lumyong, S. (2009). Actinomycetes isolated from medicinal plant rhizosphere soils: diversity and screening of antifungal compounds, indole-3-acetic acid and siderophore production. World J. Microbiol. Biotechnol. 25, 649–655. doi: 10.1007/s11274-008-9933-x
Kharel, M. K., Shepherd, M. D., Nybo, S. E., Smith, M. L., Bosserman, M. A., and Rohr, J. (2005). Isolation of Streptomyces species from soil. Curr. Protoc. Microbiol. Chapter 10:Unit 10E.4.
Kinkel, L. L., Schlatter, D. C., Bakker, M. G., and Arenz, B. E. (2012). Streptomyces competition and co-evolution in relation to plant disease suppression. Res. Microbiol. 163, 490–499. doi: 10.1016/j.resmic.2012.07.005
Kobayashi, Y. O., Kobayashi, A., Maeda, M., Someya, N., and Takenaka, S. (2015). Biological control of potato scab and antibiosis by antagonistic Streptomyces sp. WoRs-501. J. Gen. Plant Pathol. 81, 439–448. doi: 10.1007/s10327-015-0614-y
Kreuze, J. F., Suomalainen, S., Paulin, L., and Valkonen, J. P. (1999). Phylogenetic analysis of 16S rRNA genes and PCR analysis of the nec1 Gene from Streptomyces spp. Causing Common Scab, Pitted Scab, and Netted Scab in Finland. Phytopathology 89, 462–469. doi: 10.1094/phyto.1999.89.6.462
Lambert, D. H., and Loria, R. (1989). Streptomyces scabies sp. nov., nom. rev.† Int. J. Syst. Evol. Microbiol. 39, 387–392. doi: 10.1099/00207713-39-4-387
Lee, S., Flores-Encarnación, M., Contreras-Zentella, M., Garcia-Flores, L., Escamilla, J. E., and Kennedy, C. (2004). Indole-3-acetic acid biosynthesis is deficient in Gluconacetobacter diazotrophicus strains with mutations in cytochrome c biogenesis genes. J. Bacteriol. 186, 5384–5391. doi: 10.1128/JB.186.16.5384-5391.2004
Lehtonen, M. J., Rantala, H., Kreuze, J. F., Bång, H., Kuisma, L., Koski, P., et al. (2004). Occurrence and survival of potato scab pathogens (Streptomyces species) on tuber lesions: quick diagnosis based on a PCR-based assay. Plant Pathol. 53, 280–287. doi: 10.1111/j.0032-0862.2004.01009.x
Leiminger, J., Frank, M., Wenk, C., Poschenrieder, G., Kellermann, A., and Schwarzfischer, A. (2013). Distribution and characterization of Streptomyces species causing potato common scab in Germany. Plant Pathol. 62, 611–623. doi: 10.1111/j.1365-3059.2012.02659.x
Lin, L., Ge, H. M., Yan, T., Qin, Y. H., and Tan, R. X. (2012). Thaxtomin A-deficient endophytic Streptomyces sp. enhances plant disease resistance to pathogenic Streptomyces scabies. Planta 236, 1849–1861. doi: 10.1007/s00425-012-1741-8
Lorang, J., Liu, D., Anderson, N., and Schottel, J. (1995). Identification of potato scab inducing and suppressive species of Streptomyces. Phytopathology 85, 261–268. doi: 10.1094/Phyto-85-261
Loria, R., Bukhalid, R. A., Fry, B. A., and King, R. R. (1997). Plant pathogenicity in the genus Streptomyces. Plant Dis. 81, 836–846. doi: 10.1094/PDIS.1997.81.8.836
Loria, R., Kers, J., and Joshi, M. (2006). Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol 44, 469–487. doi: 10.1146/annurev.phyto.44.032905.091147
Lucas, J., Ramos, B., Montes, F., Ojeda, J., Megías, M., and Gutierrez Mañero, F. J. (2009). Use of two PGPR strains in the integrated management of blast disease in rice (Oryza sativa) in Southern Spain. Field Crops Res. 114, 404–410. doi: 10.1016/j.fcr.2009.09.013
Mahmoudi, E., Tabatabaei, B. E. S., and Venturi, V. (2011). Virulence attenuation of Pectobacterium carotovorum using N-Acyl-homoserine lactone degrading bacteria isolated from potato rhizosphere. Plant Pathol. J. 27, 242–248. doi: 10.5423/PPJ.2011.27.3.242
Majeed, A., Abbasi, M. K., Hameed, S., Imran, A., and Rahim, N. (2015). Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front. Microbiol. 6:198. doi: 10.3389/fmicb.2015.00198
Meng, Q., Yin, J., Rosenzweig, N., Douches, D., and Hao, J. J. (2011). Culture-based assessment of microbial communities in soil suppressive to potato common scab. Plant Dis. 96, 712–717. doi: 10.1094/PDIS-05-11-0441
Miyajima, K., Tanaka, F., Takeuchi, T., and Kuninaga, S. (1998). Streptomyces turgidiscabies sp. nov. Int. J. Syst. Evol. Microbiol. 48, 495–502. doi: 10.1099/00207713-48-2-495
Naqqash, T., Hameed, S., Imran, A., Hanif, M. K., Majeed, A., and van Elsas, J. D. (2016). Differential response of potato toward inoculation with taxonomically diverse plant growth promoting rhizobacteria. Front. Plant Sci. 7:144. doi: 10.3389/fpls.2016.00144
Neeno-Eckwall, E. C., Kinkel, L. L., and Schottel, J. L. (2001). Competition and antibiosis in the biological control of potato scab. Can. J. Microbiol. 47, 332–340. doi: 10.1139/w01-010
Perez-Montano, F., Alias-Villegas, C., Bellogin, R. A., del Cerro, P., Espuny, M. R., Jimenez-Guerrero, I., et al. (2014). Plant growth promotion in cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol. Res. 169, 325–336. doi: 10.1016/j.micres.2013.09.011
Prapagdee, B., Kuekulvong, C., and Mongkolsuk, S. (2008). Antifungal potential of extracellular metabolites produced by Streptomyces hygroscopicus against phytopathogenic fungi. Int. J. Biol. Sci. 4, 330–337. doi: 10.7150/ijbs.4.330
Prévost, K., Couture, G., Shipley, B., Brzezinski, R., and Beaulieu, C. (2006). Effect of chitosan and a biocontrol streptomycete on field and potato tuber bacterial communities. BioControl 51, 533–546. doi: 10.1007/s10526-005-4240-z
Qaisrani, M. M., Mirza, M. S., Zaheer, A., and Malik, K. A. (2014). Isolation and identification by 16s rRNA sequence analysis of achromobacter, azospirillum and rhodococcus strains from the rhizosphere of maize and screening for the beneficial effect on plant growth. Pak. J. Agric. Sci. 51, 91–99.
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Samac, D. A., Willert, A. M., McBride, M. J., and Kinkel, L. L. (2003). Effects of antibiotic-producing Streptomyces on nodulation and leaf spot in alfalfa. Appl. Soil Ecol. 22, 55–66. doi: 10.1016/S0929-1393(02)00109-9
Schlatter, D., Fubuh, A., Xiao, K., Hernandez, D., Hobbie, S., and Kinkel, L. (2008). Resource amendments influence density and competitive phenotypes of streptomyces in soil. Microb. Ecol. 57, 413–420. doi: 10.1007/s00248-008-9433-4
Shennan, C. (2008). Biotic interactions, ecological knowledge and agriculture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 717–739. doi: 10.1098/rstb.2007.2180
Song, J., Lee, S.-C., Kang, J.-W., Baek, H.-J., and Suh, J.-W. (2004). Phylogenetic analysis of Streptomyces spp. isolated from potato scab lesions in Korea on the basis of 16S rRNA gene and 16S–23S rDNA internally transcribed spacer sequences. Int. J. Syst. Evol. Microbiol. 54, 203–209. doi: 10.1099/ijs.0.02624-0
Tagawa, M., Tamaki, H., Manome, A., Koyama, O., and Kamagata, Y. (2008). Development of a genotyping method for potato scab pathogens based on multiplex PCR. Biosci. Biotechnol. Biochem. 72, 2324–2334. doi: 10.1271/bbb.80234
Wang, D., Wang, C., Gui, P., Liu, H., Khalaf, S. M. H., Elsayed, E. A., et al. (2017). Identification, bioactivity, and productivity of actinomycins from the marine-derived Streptomyces heliomycini. Front. Microbiol. 8:1147. doi: 10.3389/fmicb.2017.01147
Wanner, L. A. (2006). A survey of genetic variation in streptomyces isolates causing potato common scab in the United States. Phytopathology 96, 1363–1371. doi: 10.1094/phyto-96-1363
Wanner, L. A. (2007). A new strain of Streptomyces causing common scab in potato. Plant Dis. 91, 352–359. doi: 10.1094/pdis-91-4-0352
Wanner, L. A. (2009). A patchwork of Streptomyces species isolated from potato common scab lesions in North America. Am. J. Potato Res. 86, 247–264. doi: 10.1007/s12230-009-9078-y
CLSI (2015). Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Disk Susceptibility Tests, 12th Edn. Wayne, PA: CLSI.
Keywords: Streptomyces scabies, Streptomyces turgidiscabies, Streptomyces stelliscabiei, Streptomyces europaeiscabiei, biological control, Isatropolone, Pakistan
Citation: Sarwar A, Latif Z, Zhang S, Zhu J, Zechel DL and Bechthold A (2018) Biological Control of Potato Common Scab With Rare Isatropolone C Compound Produced by Plant Growth Promoting Streptomyces A1RT. Front. Microbiol. 9:1126. doi: 10.3389/fmicb.2018.01126
Received: 04 January 2018; Accepted: 14 May 2018;
Published: 30 May 2018.
Edited by:
Abd El-Latif Hesham, Assiut University, EgyptReviewed by:
Learn-Han Lee, Monash University Malaysia, MalaysiaCopyright © 2018 Sarwar, Latif, Zhang, Zhu, Zechel and Bechthold. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Bechthold, YW5kcmVhcy5iZWNodGhvbGRAcGhhcm1hemllLnVuaS1mcmVpYnVyZy5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.