
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 April 2018
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.00611
This article is part of the Research Topic Alternative Therapeutics Against Antimicrobial-Resistant Pathogens View all 27 articles
Seaweeds of the intertidal regions are a rich source of surface associated bacteria and are potential source of antimicrobial molecules. In the present study, 77 epiphytic isolates from eight different algae collected from Little Andaman were enumerated. On testing for their antimicrobial activities against certain pathogens twelve isolates showed positive and six of them showed significant antimicrobial inhibition zone against Shigella boydii type 1, Shigella flexneri type 2a, Shigella dysenteriae type 5, Enterotoxigenic Escherichia coli O115, Enteropathogenic E. coli serotype O114, Vibrio cholera; O1 Ogawa, Aeromonas hydrophila, Klebsiella pneumoniae, Staphylococcus aureus. Based on the activity these six isolates (G1C, G2C, G3C, UK, UVAD, and Tor1) were identified by 16S rRNA gene sequence and were found to belong to the phyla Firmicutes and Proteobacteria. Purified antimicrobial compounds obtained from these isolates were identified by GC-MS. Furan derivatives were identified from G2C Pseudomonas stutzeri KJ849834, UVAD Alcanivorax dieselolei KJ849833, UK Vibrio sp. KJ849837, Tor1 Exiguobacterium profundum KJ849838. While 2-Pyrrolidinone, Phenol, 2, 4-bis (1, 1-dimethylethyl) were from G3C Vibrio owensii KJ849836 and (1-Allylcyclopropyl) methanol from the extracts of G1C Bacillus sp. KJ849835. The results of the present study shows that these six potent isolates isolated from the seaweeds are found to be a source of antimicrobial compounds.
Marine eukaryotes such as seaweeds are one of the primary producers which offers nutrient rich environment for microbial communities (Egan et al., 2008; Wahl, 2008). Biofilm forming bacteria isolated from the surface of seaweeds release certain compounds (Zheng et al., 2005) which serve as nutrient supplement for the algae (Croft et al., 2005), such compounds protect the host plant from the fouling communities (Rao et al., 2007). Surface associated marine organisms such as bacteria, fungi, diatoms, larval forms of marine invertebrate’s have been reported to be associated with the thallus of seaweeds (Goecke et al., 2010; Burke et al., 2011; Murthy et al., 2016; Karthick and Mohan, 2017). Such host association particularly epiphytic bacteria are sources of certain natural compounds (Singh et al., 2011; Ali et al., 2012; Martin et al., 2014). Importance of microbial diversity of seaweeds, particularly bacterial genus are highly host specific with novel species, which have emerged from these algal environment (Goecke et al., 2013).
The secondary metabolites produced by these bacteria are highly recognized for their importance in the field of biomedical applications (Armstrong et al., 2001; Kelecom, 2002; Burgess et al., 2003). Antimicrobial activity of the epiphytic bacterial communities from seaweeds have been reported (Kanagasabhapathy et al., 2006, 2008; Vijayalakshmi et al., 2008; Ravisankar et al., 2013; Horta et al., 2014; Martin et al., 2014; Karthick et al., 2015a), similarly anti-diatom activity have also been observed by Kumar et al. (2011). In the Andaman Islands, luxuriant growth of all the three groups of seaweeds are available throughout the year. Some studies on taxonomy of seaweeds have been carried out in this region but information on the epiphytic interaction and its potentiality has not been undertaken. Based on the occurrence of seaweeds in Little Andaman and their bacterial association, the present study has been undertaken to describe the isolation of epiphytic bacterial, screening, optimization, evaluation and identification of potential isolates and their antimicrobial activity against different pathogens as test organisms.
Eight different seaweeds representing all the three groups were handpicked from the intertidal region of Harminder Bay Bridge, Little Andaman, Andaman Islands, India. Among these, six species Gracilaria corticata, Acanthophora spicifera (red algae), Ulva lactuca (green algae), Sargassum swartzii, Turbinaria ornata, and Padina tetrastromatica (brown algae) are common and other two species Mastophora rosea (red algae) and Caulerpa microphysa (green algae) were found to be rare in occurrence in these islands. The collected samples were placed in sterile plastic bags and transported to the laboratory. These were washed thrice with autoclaved seawater to remove loosely bounded epiphytes, sand particles and other attached settlements on the surface of thallus. After rinsing, firmly attached epiphytic bacteria from thallus region were swabbed with sterile cotton buds and these were then swabbed on Zobell marine agar plate (Himedia). Plates were incubated for 5 days at 32°C (Lemos et al., 1985). After incubation, colonies were picked and restreaked for the isolation of individual colonies and the purity of the isolates were checked under the microscope for single morphology. These pure cultures obtained were stored at –20°C in marine broth supplemented with 20% glycerol.
The antagonistic activity of epiphytic bacteria obtained were studied on solid media by cross streaking and double-layer method described Lemos et al. (1985) and agar well diffusion method by Karthick et al. (2013b).
All the 77 isolates were cultured on 100ml marine broth, Luria broth and minimal media by modifying the methodology slightly by decreasing the incubation time and by increasing the temperature for obtaining better results. The culture broth was centrifuged at 10000 rpm for 30 s to remove the cells and cell free broth was extracted thrice with 100 ml of ethyl acetate. All the solvents were removed under reduced pressure at 40°C (Zheng et al., 2005). Crude extracts obtained were stored at –20°C until usage for the antimicrobial assay against targeted pathogens. Sterile media without culture being adjusted to pH 7 were used as control.
All the potential isolates were cultured in inorganic salt medium referred to as minimal medium for the extraction of secondary metabolites (Jafarzade et al., 2013).
Potential cultures were cultivated in 100 ml minimal medium supplemented with 3% NaCl, 1% glucose and 1% yeast extract as carbon and nitrogen sources in a 250 ml Erlenmeyer flask and incubated at 32°C for 24 h in an incubator shaker. Five milliliter of this culture was used as bacterial (Starter) culture (Jafarzade et al., 2013).
1 ml of starter cultures were grown with minimal media supplemented with 3% NaCl, 1% glucose, and 1% yeast extract prepared and inoculated with minimal media supplemented with 0.75% of sodium chloride, 1% of glucose and yeast extract for the production of antimicrobial compounds with various pH levels (6–8) at 32°C for 5 days. After incubation, supernatant was extracted three times with ethyl acetate (EtOAc). The sterile media without the culture adjusted to pH was used as control. The extracts were then tested for antimicrobial activity.
100 ml of minimal medium was dispensed into 250 ml Erlenmeyer flasks and sterilized. Yeast extract (1%) and glucose (1%) were filter sterilized and added as nitrogen and carbon sources just prior to inoculation. One milliliter of the starter culture was inoculated into the sterilized medium. Effect of salinity in the production of antimicrobial properties at various concentrations of sodium chloride ranging from 1 to 3% with constant pH of 7 at 32°C for 5 days was experimented. After incubation cell free supernatant was extracted three times with ethyl acetate (EtOAc). Sterile media without the inoculum adjusted with various concentration of sodium chloride was used as control (Jafarzade et al., 2013). The extracts were then tested for antimicrobial activity.
Effect of different concentration (1–3%) of glucose and yeast extract for the production of antimicrobial compound by the epiphytic bacterial isolates was studied using 1 ml of the starter culture inoculated into the minimal medium. Other parameters such as pH 7 and 0.75% sodium chloride were maintained at optimum level during the primary screening at 32°C for 5 days. After incubation supernatant was extracted three times with ethyl acetate (EtOAc). The sterile medium containing glucose, yeast extract and sodium chloride was used as control. The extracts were then tested for antimicrobial activity.
Eighteen bacterial pathogens namely Escherichia coli MTCC 443, Klebsiella pneumoniae MTCC 109, Salmonella typhi MTCC 733, Staphylococcus aureus MTCC 96, Shigella flexneri MTCC 1457, Shigella flexneri type2a 503004, Shigella boydii type 1 NK2379, Shigella sonnei NK4010, Shigella dysenteriae type 5 NK2440, Enterotoxic E. coli serotype 0115, Enteropathogenic E. coli serotype 0114, Shiga toxin producing E. coli serotype O157:H7 VT3, Vibrio fluvialis IDH 02036, Vibrio parahaemolyticus serovar O3: K6 K5030, Vibrio cholera O139, Vibrio cholera 01, Ogawa Eltor, Aeromonas hydrophila IDH1585, Salmonella enterica serovar typhi C6953 and three fungal strains Aspergillus niger, Aspergillus flavus and Rhizopus sp. were used for studying the antibacterial and antifungal assay.
Minimum inhibitory concentration (MIC) of TLC purified metabolites of potential six isolates (G1C, G2C, G3C, UK, UVAD, and Tor1) was tested against Klebsiella pneumoniae and Staphylococcus aureus and it was determined by well diffusion assay. 50 mg of TLC purified extracts was dissolved in 1 ml DMSO. 50 and 100 μl/ml concentration of 50 mg/ml concentration of purified extracts was transferred into the well prepared (9 mm) with well cutter. Gentamicin was used as a positive control and Dimethyl sulfoxide (DMSO) was used as a negative control. Growth inhibition zone formed after the incubation was examined with measuring the diameter (mm) and results were recorded. All the assay was performed in triplicates.
Concentrated fractions were fractioned by Thin Layer Chromatography (TLC) using Silica gel plates with different solvents in a ratio of 2:2:1 ethyl acetate, chloroform and methanol. Bands was scraped from the plates and screened for antimicrobial assay. Active fraction was collected and analyzed by Gas Chromatography and Mass spectrometry GC-MS QP 2010 Shimadzu Corp (Japan). One μl of purified fractioned extract was loaded into the DB-5 Column with Helium as a carrier gas at a flow rate of 1 ml/min. Split Injection mode of the ratio of 1:20 was adopted. Temperature programming was from 75°C for 2 min further increased to 175°C with 15°C/min and then increased up to 280°C at the rate of 5°C/min. Sample run time was maintained upto 10 min. The peaks representing mass to charge ratio characteristic of the antimicrobial fractions were compared with those in the mass spectrum of NIST library identifying the corresponding organic antimicrobial compounds.
Phenotypic characterization of all the seventy seven bacterial isolates were identified following as described in the Bergey’s manual of systemic Bacteriology (Brenner et al., 2005).
Genomic DNA was prepared from the bacterial isolates by following the method of Mohandass et al. (2012). PCR amplification of 16S rRNA gene was conducted in a final volume of 25 μl with the bacterial consensus universal forward and reverse 16S rDNA primers 27F and 1492R (Lane, 1991). The reaction mixture contained 1x PCR buffer (Sigma, United States), 2.5 mM MgCl2, 200 μM DNTP’s, 1U of Taq DNA polymerase, 25 picomol of each forward and reverse oligonucleotide primers and approximately 20 ng of genomic DNA. The amplification profile consisted of an initial denaturation at 94°C for 3 min, followed by 35 cycles at 94°C for 1 min, 55°C for 1 min and 72°C for 1 min. This was followed by a final extension step of 72°C for 5 min. The samples were held at 4°C until further analysis. The PCR products were sequenced by an automated Sequencer (Applied Biosystems, Foster City, CA, United States) at the National Institute of Oceanography, Goa, India. The sequences were submitted to Gen Bank for which accession numbers were assigned.
The PINTAIL program (Ashelford et al., 2005) was used to check chimera formations. The partial 16S rRNA gene sequences of the potential isolates were compared with those available in the public databases. Identification upto the species level was determined by a 16S rDNA sequence similarity of more than 99% with that of the prototype sequence in GenBank. Sequence alignment and comparison were performed using the multiple sequence alignment program Clustal X 1.81 (Thompson et al., 1997). Sequences were edited manually to remove the gaps. Neighbor-joining method was employed to construct the Phylogenetic tree using MEGA4 software (Tamura et al., 2007) and the maximum likelihood method was adopted for calculating the evolutionary distance (Tamura et al., 2004).
Young pure cultures of SG107, 108, 114, 115, 120, and Tor6 were grown on Trypticase Soy Broth Agar (TSBA) for 24 or 48 h at 28°C. The Gas chromatographic analysis of whole cell fatty acid methyl ester (FAME) was performed for further identification and grouping of isolates. FAME extraction were performed using the standard procedures of extraction, purification, and methylation (Sasser, 1990). Fatty acid profiles generated were compared against an inbuilt Sherlock TSBA Library Version 6.0B [S/N 160284] (MIDI Inc., Newark, DE, United States). A similarity index of more than 0.500 was used for clustering of isolates at species level. Cellular fatty acid composition analysis was done at Regional center of Kochi, National Institute of Oceanography.
All the seventy seven cultivable epiphytic bacterial isolates obtained from the thallus of eight different seaweeds were plated on Marine agar. These isolates were purified and based on by phenotypic characterization were assigned to belong to the phylum Firmicutes and Proteobacteria (Table 1). Among these isolates, six of them (G1C, G2C, G3C, UK, UVAD, and Tor1) showed wide range of activities against pathogens with an range of 10–30 mm zone of inhibition and were identified by partial 16S rRNA gene sequences (Table 2). The isolate G1C showed 99% similarity as Bacillus sp., G2C as Pseudomonas stutzeri, G3C and UK were identified as Vibrio owensii and Vibrio sp., respectively. The Isolate UVAD was identified as Alcanivorax dieselolei with 99% similarity and Torl strain was identified with 99.7% similarity as Exiguobacterium profundum (Figure 1).
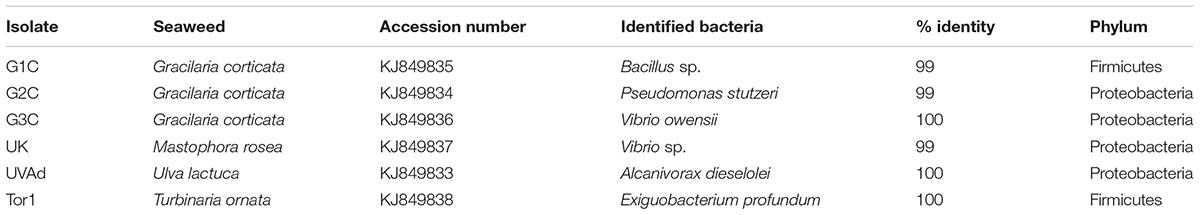
TABLE 2. 16S rRNA gene sequence identity of six potential bacterial isolates obtained from different seaweeds.
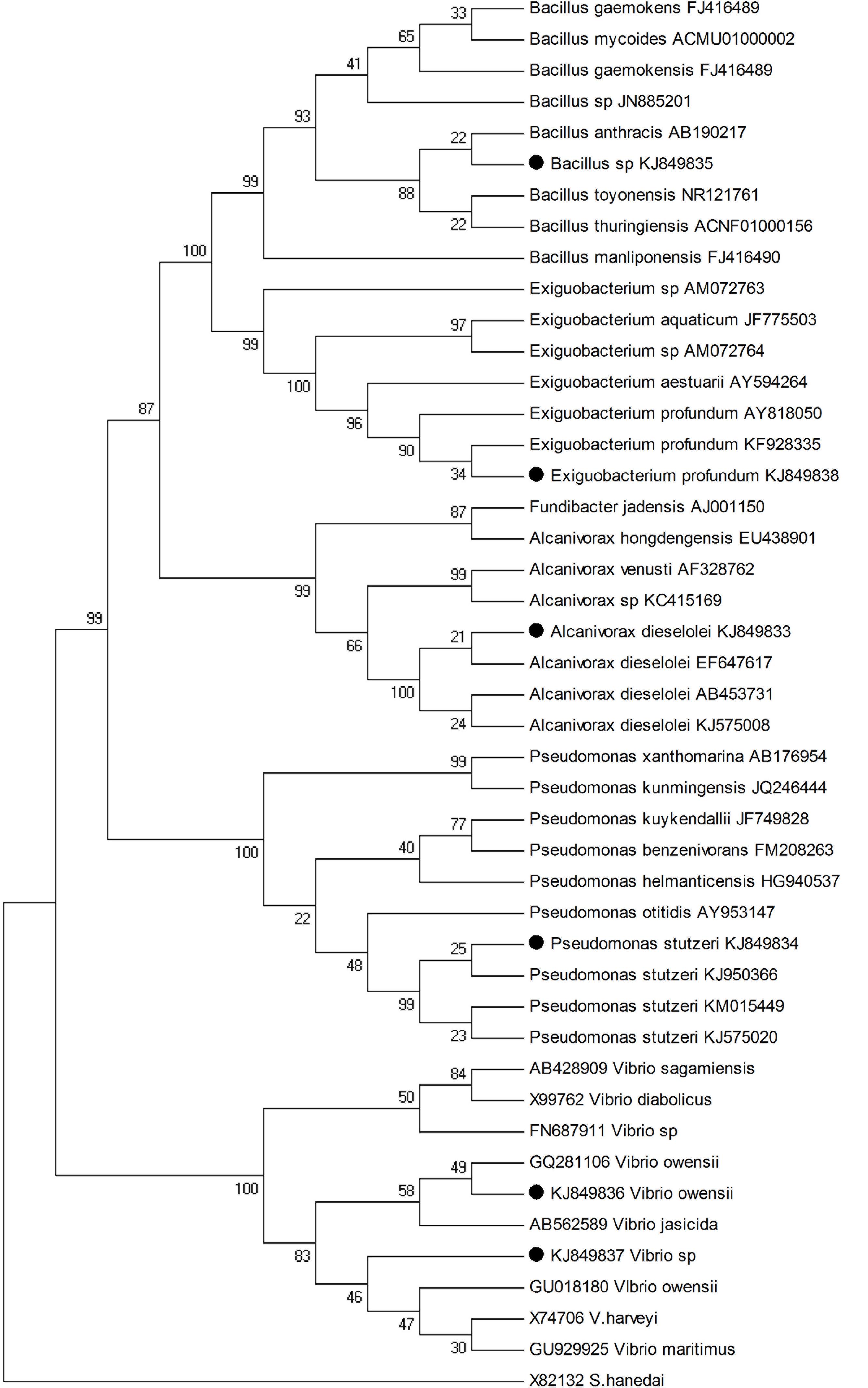
FIGURE 1. Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences showing the relationship of six potential strains with its closest neighbors. Bootstrap values (50%) are shown at branch points in value and S. hanedai X82132 were used as outgroup.
Six other isolates showed moderate to less activity activities against the pathogens with a range of 5–10 mm zone of inhibition and were identified by FAME analysis. Among them three isolates SG107 as Bacillus sp., SG108 Paenibacillus lentimorbus and SG115 as Bacillus sphaericus belonged to phylum Firmicutes. The other three isolates Pantoea agglomerans, SG 120 was Vibrio aestuarianus and TR was identified as Klebsiella pneumoniae ozaena and were assigned to phylum Proteobacteria, SG114 (Table 3).
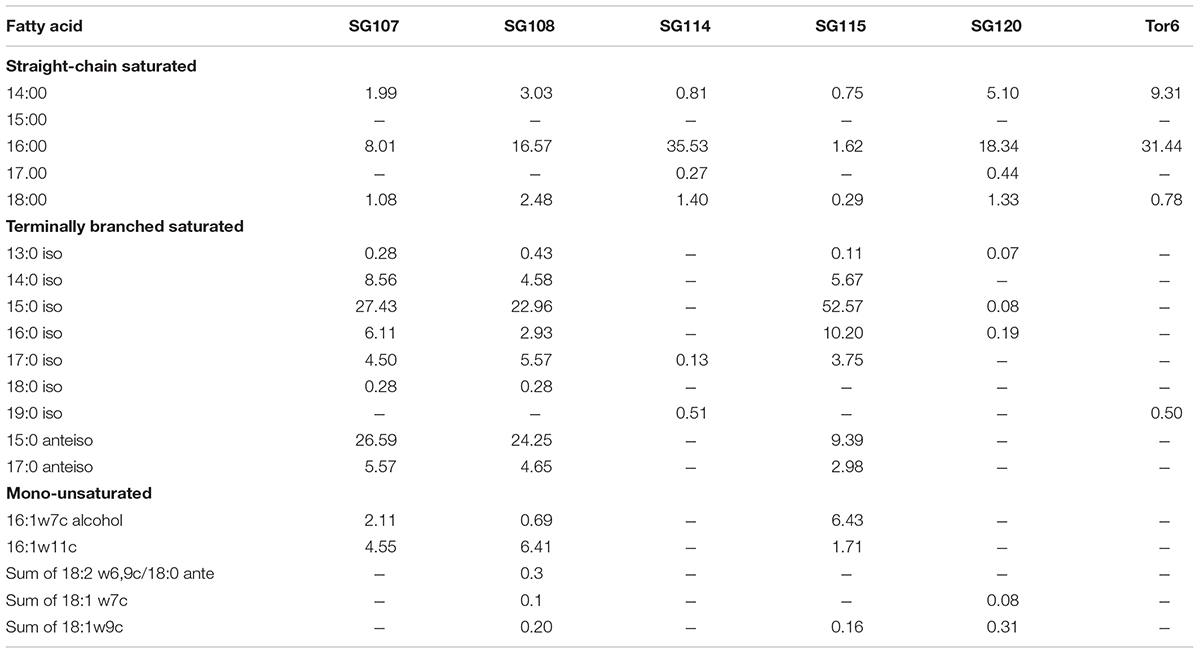
TABLE 3. Comparison of cellular fatty acid composition of 6 moderate activity identified from brown algae (1) SG107 Bacillus sp, (2) SG108 Paenibacillus lentimorbus, (3) SG114 Pantoea agglomerans, (4) SG115 Bacillus sphaericus, (5) SG120 Vibrio aestuarianus, (6) Tor6 Klebsiella pneumoniae ozaenae.
Antimicrobial activity for all the 77 bacterial isolates were tested by adopting three different methods (Agar overlay, cross streaking and agar well diffusion technique) against 21 pathogens. Among these larger zones of inhibition were observed in agar well diffusion assay and this assay was chosen for further antimicrobial activity test. All the isolates were cultured in three different media (marine broth, luria broth and minimal medium) among the medium used minimal media showed broad range of antimicrobial activity (Table 4). Based on the preliminary activity only six potential isolates (G1C, G2C, G3C, UK, UVAD, and Tor1) were optimized in minimal medium for the production of antimicrobial compounds.
Isolate G1C showed strong inhibitory activity against Shigella boydii (31 mm), Enterotoxigenic E. coli (28 mm), Enteropathogenic E. coli and Aeromonas hydrophila (23 mm) and this higher inhibition zones were obtained with minimal media supplemented with 1 and 2% Sodium chloride, 1% of glucose and yeast extract with pH in the range of 7–8. Isolate G2C was more effective against Shigatoxin E. coli (26 mm), Aeromonas hydrophila and Salmonella typhi (24 mm) observed from minimal medium containing only 1% of sodium chloride, glucose and yeast extract with pH in the range of 6–7. Isolate G3C was effective against Salmonella enterica serovar typhimurium (26 mm), Vibrio cholerae Eltor and Shigella dysenteriae (24 mm) and antimicrobial activity was observed with minimal media with 1 and 2% of sodium chloride, glucose and 1% of yeast extract with pH in the range of 6–7. Isolate TOR1 exhibited broad range of antibacterial activity against Klebsiella pneumoniae, Shigella dysenteriae (31 mm), Shigella sonnei (25 mm). UVAD isolate showed maximum zone of inhibitory activity against Staphylococcus aureus (30 mm), Salmonella enterica serovar typhimurium (29 mm) and Shigella dysenteriae (27 mm) and isolate UK displayed maximum zone of inhibition against Aeromonas hydrophila (29 mm), Shigella flexneri 2A and Shigella flexneri (24 mm), respectively. All these six bacterial isolates showed maximum inhibitory activity against most of the tested pathogens (Table 4).
Cell free supernatant of the six potential isolates (G1C, G2C, G3C, UK, UVAD, and Tor1) that showed antimicrobial activity were purified by TLC. Results of minimum inhibitory concentration of TLC purified metabolites of all the six potential isolates at 50 mg/mL concentration diluted in 50 and 100 μl/ml concentration showed the positive results against K. pneumoniae and S. aureus. 50 μl/ml concentration of potential extracts exhibited inhibitory activity against the tested pathogens. Maximum zone of inhibition 10 mm was measured against K. pneumoniae by UK, UVAD, Tor1, G1C, and G3C extract at 100 μl concentration. 9 mm zone of inhibition was measured against S. aureus by 100 μl concentration of G2C, G3C, UK, and Tor1. Based on the inhibitory growth of bacteria at 50 μl concentration of purified metabolites, it can be concluded that minimum inhibitory concentration was observed at 2.5 mg/mL. Based on the activity purified compounds obtained from these six isolates were characterized and identified. Furan derivatives were found to be present in four of the isolates namely G2C Pseudomonas stutzeri, Tor1 Exiguobacterium profundum. UVAD Alcanivorax dieselolei and UK Vibrio sp. While 2-Pyrrolidinone, Phenol, 2, 4-bis (1, 1-dimethylethyl) were identified from the isolate G3C Vibrio owensii and (1-Allylcyclopropyl) methanol from G1C Bacillus sp.
Seaweed biomass were found in large quantities in both intertidal and subtidal regions of all the regions of Andaman Island and in Little Andaman’s (Karthick et al., 2013a,b). Good hemolytic activity in certain seaweeds of these Island has been reported recently (Punnam Chander et al., 2014). Besides these studies Karthick et al. (2015b) had reported on antimicrobial activity of certain seaweeds against pathogenic bacterial and fungal stains. Several authors suggested that macro algal associated bacteria were found to be an efficient producer of antimicrobial compounds (Burgess et al., 1999; Lee et al., 2006; Kanagasabhapathy et al., 2008; Karthick et al., 2015a; Ismail et al., 2016). On the other hand, certain brown algae also produced biologically active compounds which inhibited the settlement of bacterial colonies on the thallus (Nagayama et al., 2002).
In the present study it was observed that higher number of epiphytic bacteria were isolated from brown and red algae, certainly the proportion of higher isolates were from brown rather than green and red algae. On surface colonization non-pigmented bacterial isolates were found dominant in most of the seaweeds used in this study. Epiphytic bacteria from marine macro algae have been well studied in reference to their ecological importance with host organisms (Croft et al., 2005) with a dominance of Gram-negative bacteria. Similarly in the present study 46 Gram-negative bacterial isolates were isolated in comparison to 31 being Gram-positive. Bacteria belonging to genus the Bacillus were dominant with 20 isolates followed by other genus such as Vibrio, Aeromonas, and Pseudomonas.
Ravisankar et al. (2013) observed that the surface of the brown algae Hypnea valentiae and Padina tetrastromatica contained more number of non-pigmented bacterial colonies which are similar to our studies wherein 10 isolates were obtained from Padina tetrastromatica. Similar observations were observed in Tunisian waters, where 17 isolates were obtained from the thallus of Ulva intestinalis (Ali et al., 2010) and 10 isolates were reported from Ulva lactuca in Fiji waters, of which majority of the isolates were efficient antimicrobial producers (Kumar et al., 2011).
In the present study twelve isolates (15.7%) of the total 77 isolated exhibited antimicrobial activity and six isolates showed broad spectrum of activity against both bacterial and fungal pathogens. Similarly (Jayanth et al., 2002) isolated 14.52% of associated bacteria from the red algae Gracilaria with antagonistic properties against certain human pathogens. On the other hand 11% of associated bacteria isolated from seaweeds were reported to have antagonistic nature against Bacillus subtilis, E. coli, S. aureus, Agrobacterium tumefaciens, and Saccharomyces cerevisiae (Zheng et al., 2005).
The 16S rRNA sequences of bacterial isolates obtained from the surface of green algae Ulva australis and Delisea pulchra, belonged to the representative’s classes of Alpha and Gammaproteobacteria and interestingly Actinobacteria, Firmicutes, and Bacteroidetes were observed as antimicrobial producers (Penesyan et al., 2009). Ali et al. (2012) on 16S rRNA sequence of the isolates obtained from the surface of coralline red algae Jania rubens found them belong to the group Proteobacteria. Similar observation made by Singh et al. (2015) also highlighted that bacterial isolates belonged to the order Bacillales, Pseudomonadales, Alteromonadales, and Vibrionales were dominant in green algae Ulva lactuca, U. fasciata, and red algae Gracilaria corticata and G. dura. In this study also it’s evident that all the 77 bacterial isolates were closely related to the phylum Proteobacteria and Firmicutes. These findings substantiate that these groups are more specific to the macro algal surface. Similarly species belonging to the genera Bacillus and Vibrio were found to be strong antimicrobial producers colonizing more on the surface of seaweeds.
Genus Bacillus predominantly colonizes on the surface of marine niche and several studies have been reported Bacillus from different marine sources particularly associated with brown algae (Thakur and Anil, 2000) and from the thallus surface of different red algae (Kanagasabhapathy et al., 2008). Apart from their association with seaweeds Bacillus were previously isolated from sediments and seaweeds with antimicrobial properties (Prieto et al., 2012). So far, more than 800 metabolites have been reported with various biological activities from the Bacillus genera. Recently, cell free supernatant extracted from Bacillus associated with a nematode were found to be very effective against multidrug resistant Staphylococcus aureus (Susilowati et al., 2015). As observed in the present study one potential isolate G1C identified as Bacillus sp. showed remarkable activity against most of the tested pathogens, in particular against toxin producing pathogens like Shigella boydii, Enterotoxigenic E. coli, Shigatoxin E. coli Enteropathogenic E. coli, and Aeromonas hydrophila etc. Similarly SG107 Bacillus sp. and SG115 Bacillus sphaericus obtained from the brown algae Sargassum swartzii also showed moderate to less activity against few pathogens.
Vibrios being truly marine and they are widespread in various marine niches and are known to produce secondary metabolites for their survival. Earlier genus Vibrio sp., Pseudomonas sp., and Bacillus pumilus were reported to be a probiotic bacteria used in aquaculture (Hill et al., 2009). Kanagasabhapathy et al. (2008) reported that Vibrio strain isolated from red algae showed certain biological activities. In the present study, two potential Vibrio isolates GC3 Vibrio owensii and UK Vibrio sp. were obtained from the surface of red algae Gracilaria corticata and Mastophora rosea, respectively, and these isolates exhibited wider range of antimicrobial activity against most of the tested pathogens like Salmonella typhi, Shigella dysenteriae, Vibrio cholerae, and Staphylococcus aureus. Similarly, Pawar et al. (2015) extracted antibacterial compounds from marine Vibrio sp. which were found to be active against numerous pathogens.
In our study of 14 bacterial isolates were obtained green algae Ulva lactuca among these one isolate UVAD Alcanivorax dieselolei was found to possess higher range of antimicrobial activity. Previously this species Alcanivorax dieselolei has been reported to be isolated from the deep sea sediment involved in degrading alcanes (Liu and Shao, 2005), and petroleum products (Brito et al., 2006). Ali et al. (2010) reported that two epiphytic bacteria obtained from green alga U. intestinalis showed potent antimicrobial activity. These studies suggest that green algae Ulva species attracts novel bacterial colonization on their surface with potential microbial communities and these isolates produce various compounds to protect the host from the predators and other micro and macro fouling colonization.
Pseudomonas stutzeri has been reported to have wide range of biological activity by the production of secondary metabolites. Previously Pseudomonas stutzeri isolated from fish gut exhibited antimicrobial activity (Uzair et al., 2008), hydrocarbon degradation (Vazquez et al., 2009), and reported as uncommon opportunistic pathogen (Park et al., 2013), controlling biofilm formation (Wu et al., 2016). In this study Pseudomonas stutzeri isolated from the red algae Gracilaria corticata produced antimicrobial compounds which showing potent activity against numerous toxin producing pathogens S. aureus, Shigella boydii, S. flexneri 2A, S. dysenteriae, K. pneumoniae, Et. E. coli, St. E. coli, V. cholerae Eltor, A. hydrophila.
Earlier Exiguobacterium sp. showed antimicrobial properties (Shatila et al., 2013) and this bacterium also known to produce antifouling compound and thus protected the host organisms from fouling communities (Jain et al., 2013). In the present study Tor1 Exiguobacterium profundum isolate obtained from Turbinaria ornata, showed antibacterial activity against clinical pathogens S. aureus, Shigella boydii, S. flexneri, S. flexneri 2A, S. dysenteriae, K. pneumoniae, Et. E. coli, and St. E. coli. The same genus was identified in earlier studies from different seaweeds occurring in different geographical locations showing various biological activities. In the present study 6 potential isolates obtained from seaweeds were found to be good antimicrobial producers. The same genus was identified in earlier studies from different seaweeds occurring in different geographical locations showing various biological activities (Table 5).
In earlier studies Furan derivatives were reported to have antimicrobial properties (Kirilmis et al., 2009; Joshi et al., 2010; Ramasamy and Balasubramanian, 2012), cytotoxic agent (Wang et al., 2008) and were observed to have a wide range of biological activities like antiproliferative, antiviral, antifungal, immunosuppressive, anti-platelet, anti-oxidative, insecticidal, anti-inflammatory, anti-feedant, and cancer preventative activity (Venkateshwarlu et al., 2013). In our present study we have identified Furan compounds from four potential isolates (G2C, UVAD, Tor1, and UK). Apart from antimicrobial properties, these compounds are being used for other pharmacological properties (Bober et al., 2012). In this study G3C Vibrio owensii produced antimicrobial compounds such as 2-Pyrrolidinone, Phenol, 2, 4-bisdimetyl ethyl)-ester and Pyrrolo [1,2-a] pyrazine-1,4-dione. Earlier, these compounds were reported to have antimicrobial properties (Sutariya et al., 2012; Khatiwora et al., 2013; Padmavati et al., 2014; Dhanya et al., 2016). Marine Vibrio sp. is highly capable of producing Phenol, pyrrolo [1,2-a]pyrazine-1,4-dione, Pyrrolidinone derivative compounds containing pharmacological properties (Pawar et al., 2015). Based on earlier findings and as observed in the present study Vibrios are efficient producer of Phenol and Pyrrolidinone derivatives. In conclusion based on the findings of the present study, the compounds produced from six potential isolates (G1C, G2C, G3C, UK, UVAD, and Tor1) having effective antimicrobial properties, could be further studied for other activities. These isolates could prove to be potential candidates for the production of novel antimicrobial compounds in order to control the pathogens.
PK designed the work, performed all the experiments, analyzed and wrote the manuscript. RM contributed the designation of research work and evaluated the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are greatful to Dr. C. Mohandass, Chief Scientist, National Institute of Oceanography (NIO), Goa for sequencing studies, Dr. Anas Abdulaziz, Scientist, Marine Microbial Reference Facility (NIO) RC, Kochi, Dr. R. Babu Rajendran, Professor, Department of Environmental Biotechnology, Bharathidasan University for GC-MS facilities. The authors also thank personally to their colleagues Dr. K. N. Murthy, Dr. C. H. Ramesh, Dr. Sumantha Narayana for their support during field collection and Pondicherry University authorities for providing the facilities.
Ali, A. I., Bour, M. E., Ktari, L., Bolhuis, H., Ahmed, A., Boudabbous, A., et al. (2012). Jania rubens-associated bacteria: molecular identification and antimicrobial activity. J. Appl. Phycol. 24, 525–534. doi: 10.1007/s10811-011-9758-0
Ali, I. B., Ktari, L., Bolhuis, H., Boudabbous, A., Stal, L. J., and Bour, M. E. L. (2010). Ulva intestinalis associated bacteria: molecular identification and antimicrobial potential. Rap. Comm. Int. Mer. Medit. 39:372.
Armstrong, E., Yan, L., Boyd, K. G., Wright, P. C., and Burgess, J. G. (2001). The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461, 37–40. doi: 10.1023/A:1012756913566
Ashelford, K. E., Chuzhanova, N. A., Fry, J. C., Jones, A. J., and Weightman, A. J. (2005). At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71, 7724–7736. doi: 10.1128/AEM.71.12.7724-7736.2005
Bober, L., Kawczak, P., and Baczek, T. (2012). Pharmacological classification and activity evaluation of furan and thiophene amide derivatives applying semi-empirical ab initio molecular modeling methods. Int. J. Mol. Sci. 13, 6665–6678. doi: 10.3390/ijms13066665
Brenner, D. J., Krieg, N. R., and Staley, J. T. (eds). (2005). Bergeys Manual of Systematic Bacteriology. New York, NY: Springer.
Brito, E. M., Guyoneaud, R., Urizza, M., Ranchou-Peyruse, A., Verbaere, A., Crapez, M. C., et al. (2006). Characterization of hydrocarbonoclastic bacterial communities from mangrove sediments in Guanabara Bay, Brazil. Res. Microbiol. 157, 752–762. doi: 10.1016/j.resmic.2006.03.005
Burgess, J. G., Boyd, K. G., Armstrong, E., Jiang, Z., Yan, L., Berggren, M., et al. (2003). The development of a marine natural product-based antifouling paint. Biofouling 19, 197–205. doi: 10.1080/0892701031000061778
Burgess, J. G., Mearns-Spragg, A., Jordan, E. M., Bregu, M., and Boyd, K. G. (1999). Microbial antagonism: a neglected avenue of natural products research. J. Biotechnol. 70, 27–32. doi: 10.1016/S0168-1656(99)00054-1
Burke, C., Thomas, T., Lewis, M., Steinberg, P., and Kjelleberg, S. (2011). Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 5, 590–600. doi: 10.1038/ismej.2010.164
Croft, M. T., Lawrence, A. D., Raux-Deery, E., Warren, M. J., and Smith, A. G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Dhanya, K. I., Swati, V. I., Vanka, K. S., and Osborne, W. J. (2016). Antimicrobial activity of Ulva reticulata and its endophytes. J. Ocean Univ. China 15, 363–369.
Egan, S., Thomas, T., and Kjelleberg, S. (2008). Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opin. Microbiol. 11, 219–225. doi: 10.1016/j.mib.2008.04.001
Goecke, F., Labes, A., Wiese, J., and Imhoff, J. F. (2010). Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 409, 267–299. doi: 10.3354/meps08607
Goecke, F., Thiel, V., Wiese, J., Labes, A., and Imhoff, J. (2013). Algae as an important environment for bacteria – phylogenetic relationships among new bacterial species isolated from algae. Phycologia 52, 14–24. doi: 10.2216/12-24.1
Harder, T., Dobretsov, S., and Qian, P.-Y. (2004). Waterborne polar macromolecules act as algal antifoulants in the seaweed Ulva reticulata. Mar. Ecol. Prog. Ser. 274, 131–141. doi: 10.3354/meps274133
Hill, J. E., Baiano, J. C. F., and Barnes, A. C. (2009). Isolation of a novel strain of Bacillus pumilus from penaeid shrimp that is inhibitory against marine pathogens. J. Fish Dis. 32, 1007–1016. doi: 10.1111/j.1365-2761.2009.01084.x
Horta, A., Pinteus, S., Alves, C., Fino, N., Silva, J., Fernandez, S., et al. (2014). Antioxidant and antimicrobial potential of the Bifurcaria bifurcata epiphytic bacteria. Mar. Drugs 12, 1676–1689. doi: 10.3390/md12031676
Ismail, A., Ktari, L., Ahmed, M., Bolhuis, H., Boudabbous, A., Stal, L. J., et al. (2016). Antimicrobial activities of bacteria associated with the brown alga Padina pavonica. Front. Microbiol. 7:1072. doi: 10.3389/fmicb.2016.01072
Jafarzade, M., Shayesteh, F., Usup, G., and Ahmad, A. (2013). Influence of culture conditions and medium composition on the production of antibacterial compounds by marine Serratia sp. WPRA3. J. Microbiol. 51, 373–379. doi: 10.1007/s12275-013-2440-2
Jain, S. C., Sivakumar, A. J., and Pandian, P. M. (2013). Surface associated bacteria of marine algae in Kovalam beach, Chennai, had screened for its antifouling activity. Indian J. Geomarine Sci. 42, 498–502.
Jayanth, K., Jeyasekaran, G., and Shakila, R. J. (2002). Isolation of marine bacteria, antagonistic to human pathogens. Ind. J. Mar. Sci. 31, 39–44.
Joshi, S. D., Ashwini, J., and Gadaginamath, G. S. (2010). Synthesis and antimicrobial evaluation of some new pyrrolylnaphtho [2, 1-b] furan derivatives. Ind. J. Pharm. Educ. Res. 44, 148–155.
Kanagasabhapathy, M., Sasaki, H., Haldar, S., Yamasaki, S., and Nagata, S. (2006). Antibacterial activities of marine epibiotic bacteria isolated from brown algae of Japan. Ann. Microbiol. 56, 167–173. doi: 10.1007/BF03175000
Kanagasabhapathy, M., Sasaki, H., and Nagata, S. (2008). Phylogenetic identification of epibiotic bacteria possessing antimicrobial activities isolated from red algal species of Japan. World J. Microbiol. Biotechnol. 24, 2315–2321. doi: 10.1007/s11274-008-9746-y
Karthick, P., and Mohan, P. M. (2017). Associated epiphytic faunal distribution on brown algae Sargassum wightii Greville ex J.Agardh, in the environments, off South Andaman. J. Terr. Mar. Res. 1, 41–44.
Karthick, P., Mohanraju, R., Murthy, K. N., and Ramesh, C. H. (2013a). Seaweed potential of Little Andaman, India. Seaweed Res. Utilin. 35, 17–21.
Karthick, P., Mohanraju, R., Ramesh, C. H., Murthy, K. N., and Narayana, S. (2013b). Distribution and diversity of seaweeds in North and South Andaman Island. Seaweed Res. Utilin. 35, 8–16.
Karthick, P., Mohanraju, R., Mohandass, C., and Rajasabapathy, R. (2015a). Antimicrobial activity of Serratia sp isolated from the coralline red algae Amphiroa anceps. Indian J. Geomarine Sci. 44, 1857–1866.
Karthick, P., Mohanraju, R., Murthy, K. N., and Ramesh, C. H. (2015b). Antibacterial activity of seaweeds collected from South Andaman, India. J. Algal Biomass Util. 6, 33–36.
Kelecom, A. (2002). Secondary metabolites from marine microorganisms. An. Acad. Bras. Cienc. 74, 151–170. doi: 10.1590/S0001-37652002000100012
Khatiwora, E., Adsula, V. B., Kulkarni, M., Deshpande, N. R., and Kashalkar, R. V. (2013). Isolation and characterization of substituted dibutyl phthalate from Ipomoea carnea stem. Der Pharma Chemica 5, 5–10.
Kirilmis, C., Koca, M., Servi, S., and Gur, S. (2009). Synthesis and antimicrobial activity of dinaphtho[2,1-b]furan-2-yl-methanone and their oxime derivatives. Turkish J. Chem. 33, 375–384.
Kumar, V., Rao, D., Thomas, T., Kjelleberg, S., and Egan, S. (2011). Antidiatom and antibacterial activity of epiphytic bacteria isolated from Ulva lactuca in tropical waters. World J. Microbiol. Biotechnol. 27, 1543–1549. doi: 10.1007/s11274-010-0606-1
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley & Sons Inc), 115–175.
Lee, Y. K., Jung, H. J., and Lee, H. K. (2006). Marine bacteria associated with the Korean brown alga, Undaria pinnatifida. J. Microbiol. 44, 694–698.
Lemos, M. L., Toranzo, A. E., and Barja, J. L. (1985). Antibiotic activity of epiphytic bacteria isolated from intertidal seaweeds. Microbial. Ecol. 11, 149–163. doi: 10.1007/BF02010487
Liu, C., and Shao, Z. (2005). Alcanivorax dieselolei sp. nov., a novel alkane-degrading bacterium isolated from sea water and deep-sea sediment. Int. J. Syst Evol. Microbiol. 55, 1181–1186. doi: 10.1099/ijs.0.63443-0
Martin, M., Portetelle, D., Michel, G., and Vandenbol, M. (2014). Microorganisms living on macroalgae: diversity, interactions, and biotechnological applications. Appl Microbiol. Biotechnol. 98, 2917–2935. doi: 10.1007/s00253-014-5557-2
Mohandass, C., Rajasabapathy, R., Ravindran, C., Colaco, A., and Meena, R. M. (2012). Bacterial diversity and their adaptations in the shallow water hydrothermal vent at D. Joao de Castro Seamount (DJCS), Azores, Portugal. Cah. Biol. Mar. 53, 65–76.
Murthy, K. N., Mohanraju, R., Karthick, P., and Ramesh, C. H. (2016). Phenotypic and molecular characterization of epiphytic Vibrios from the marine macro algae of Andaman Islands, India. Indian J. Mar. Sci. 45, 304–309.
Nagayama, K., Iwamura, Y., Shibata, T., Hirayama, I., and Nakamura, T. (2002). Bactericidal activity of phlorotannins from the brown alga Ecklonia kurome. J. Antimicrob. Chemother. 50, 889–893. doi: 10.1093/jac/dkf222
Padmavati, A. R., Abinaya, B., and Pandian, S. K. (2014). Phenol, 2,4-bis(1,1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens. Biofouling 30, 1111–1122. doi: 10.1080/08927014.2014.972386
Park, S. W., Back, J. H., Lee, S. W., Song, J. H., Shin, C. H., Kim, G. E., et al. (2013). Successful antibiotic treatment of Pseudomonas stutzeri-induced peritonitis without peritoneal dialysis catheter removal in continuous ambulatory peritoneal dialysis. Kidney Res. Clin. Pract. 32, 81–83. doi: 10.1016/j.krcp.2013.04.004
Pawar, R., Mohandass, C., Sivaperumal, E., Sabu, E., Rajasabapathy, R., and Jagtap, T. (2015). Epiphytic marine pigmented bacteria: a prospective source of antioxidants. Braz. J. Microbiol. 46, 29–39. doi: 10.1590/S1517-838246120130353
Penesyan, A., Marshall-Jones, Z., Holmstrom, C., Kjelleberg, S., and Egan, S. (2009). Antimicrobial activity observed among cultured marine epiphytic bacteria reflects their potential as a source of new drugs. FEMS Microbiol. Ecol. 69, 113–124. doi: 10.1111/j.1574-6941.2009.00688.x
Prieto, M., Sullivan, L. O., McLoughlin, P., Hughes, H., O’Connor, P. M., and Gardiner, G. E. (2012). Assessment of the bacteriocinogenic potential of marine bacteria reveals lichenicidin production by seaweed-derived Bacillus spp. Mar. Drugs 10, 2280–2299. doi: 10.3390/md10102280
Punnam Chander, M., Sachithanandam, V., and Vijayachari, P. (2014). Antimicrobial and hemolytic activity of seaweed Padina gymnospora from South Andaman, Andaman and Nicobar Islands of India. Int. J. Curr. Microbiol. App. Sci. 3, 364–369.
Ramasamy, M., and Balasubramanian, U. (2012). Identification of bioactive compounds and antimicrobial activity of marine clam Anadara granosa (LINN.). Int. J. Sci. Nat. 3, 263–266.
Rao, D., Webb, J. S., Holmstrom, C., Case, R., Low, A., Steinberg, P., et al. (2007). Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 73, 7844–7852. doi: 10.1128/AEM.01543-07
Ravisankar, A., Gnanambal, M. E., and Sundaram, L. R. (2013). A newly isolated Pseudomonas sp., epibiotic on the seaweed, Padina tetrastromatica, off Southeastern Coast of India, reveals antibacterial action. Appl. Biochem. Biotechnol. 171, 1968–1985. doi: 10.1007/s12010-013-0473-y
Sasser, M. (1990). “Identification of bacteria through fatty acid analysis,” in Methods in Phytobacteriology, eds Z. Klement, K. Rudolph, and D. C. Sands (Budapest: Akademiai Kiado).
Shatila, F., Yusef, H., and Beirut, H. H. (2013). Pigment production by Exiguobacterium aurantiacum FH, a novel Lebanese isolate. Int. J. Curr. Microbiol. App. Sci. 2, 176–191.
Singh, R. P., Baghel, R. S., Reddy, C. R. K., and Jha, B. (2015). Effect of quorum sensing signals produced by seaweed-associated bacteria on carpospore liberation from Gracilaria dura. Front. Plant Sci. 6:117. doi: 10.3389/fpls.2015.00117
Singh, R. P., Mantri, V. A., Reddy, C. R. K., and Jha, B. (2011). Isolation of seaweed-associated bacteria and their morphogenesis-inducing capability in axenic cultures of the green alga Ulva fasciata. Aquat. Biol. 12, 13–21. doi: 10.3354/ab00312
Susilowati, R., Sabdono, A., and Widowati, I. (2015). Isolation and characterization of bacteria associated with brown algae Sargassum spp. from Panjang Island and their antibacterial activities. Procedia Environ. Sci. 23, 240–246. doi: 10.1016/j.proenv.2015.01.036
Sutariya, S. D., Parmar, K. A., and Kharadi, G. J. (2012). Synthesis, spectral studies and antibacterial screening of some novel derivatives of 2-pyrrolidinones based on Schiff base. Der Chem. Sin. 3, 854–859.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Tamura, K., Nei, M., and Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U.S.A. 101, 11030–11035. doi: 10.1073/pnas.0404206101
Thakur, N. L., and Anil, A. C. (2000). Antibacterial activity of the sponge Ircinia ramosa: importance of its surface-associated bacteria. J. Chem. Ecol. 26, 57–71. doi: 10.1023/A:1005485310488
Thompson, J. D., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. doi: 10.1093/nar/25.24.4876
Uzair, B., Ahmed, N., and Edwards, D. H. (2008). The isolation, purification and biological activity of a novel antibacterial compound produced by Pseudomonas stutzeri. FEMS Microbiol. Lett. 279, 243–250. doi: 10.1111/j.1574-6968.2007.01036.x
Vazquez, S., Nogales, B., Ruberto, L., Hernandez, E., Christie-Oleza, J., Lo Balbo, A., et al. (2009). Bacterial community dynamics during bioremediation of diesel oil-contaminated Antarctic soil. Microb. Ecol. 57, 598–610. doi: 10.1007/s00248-008-9420-9
Venkateshwarlu, T., Ravindernath, A., and Chennapragada, K. P. (2013). Synthesis and antimicrobial activity of novel benzo[b]furan derivatives. Der Pharma Chemica 5, 229–234.
Vijayalakshmi, S., Santhanaramasamy, M., Murugesh, S., and Murugan, A. (2008). Isolation and screening of marine associated bacteria from Tamil Nadu, Southeast coast of India for potential antibacterial activity. Ann. Microbiol. 58, 605–609. doi: 10.1007/BF03175564
Wahl, M. (2008). Ecological lever and interface ecology: epibiosis modulates the interactions between host and environment. Biofouling 24, 427–438. doi: 10.1080/08927010802339772
Wang, J., Zhang, H., Yang, X., Zhou, Y., Wang, H., and Bai, H. (2008). HS071, a new furan-type cytotoxic metabolite from Streptomyces sp. HS-HY-071. J. Antibiot. 61, 623–626. doi: 10.1038/ja.2008.82
Wiese, J., Thiel, V., Nagel, K., Staufenberger, T., and Imhoff, J. F. (2009). Diversity of antibiotic-active bacteria associated with the brown alga Laminaria saccharina from the Baltic Sea. Mar. Biotechnol. 11, 287–300. doi: 10.1007/s10126-008-9143-4
Wu, S., Liu, G., Jin, W., Xiu, P., and Sun, C. (2016). Antibiofilm and anti-Infection of a marine bacterial exopolysaccharide against Pseudomonas aeruginosa. Front. Microbiol. 7:102. doi: 10.3389/fmicb.2016.00102
Keywords: Alcanivorax dieselolei, Little Andaman, Furan, Gracilaria corticata, seaweeds
Citation: Karthick P and Mohanraju R (2018) Antimicrobial Potential of Epiphytic Bacteria Associated With Seaweeds of Little Andaman, India. Front. Microbiol. 9:611. doi: 10.3389/fmicb.2018.00611
Received: 30 June 2017; Accepted: 16 March 2018;
Published: 04 April 2018.
Edited by:
Noton Kumar Dutta, Johns Hopkins University, United StatesReviewed by:
Vinay Kumar, Savitribai Phule Pune University, IndiaCopyright © 2018 Karthick and Mohanraju. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Perumal Karthick, a2FydGhpY2ttaWNyb2Jlc0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.