- 1Department of Food Science and Engineering, School of Chemistry and Chemical Engineering, Harbin Institute of Technology, Harbin, China
- 2Institute of Microbiology, Heilongjiang Academy of Sciences, Harbin, China
CRISPR-Cas (Clustered regularly interspaced short palindromic repeats-CRISPR associated proteins) loci, which provide a specific immunity against exogenous elements, are hypervariable among distinct prokaryotes. Based on previous researches, this review focuses on concluding systematical genome editing protocols in Streptococcus thermophilus. Firstly, its protocols and optimized conditions in gene editing are introduced. What’s more, classification and diversity analyses of S. thermophilus CRISPR-Cas benefit the further understanding of evolution relationship among Streptococcus. Ability of its foreign segment integration and spacer source analyses also indicate a new direction of phage resistance. Above all, all of these point out its potential to be regarded as another model system other than type II CRISPR-Cas in Streptococcus pyogenes.
Introduction
CRISPR-Cas system was found in thermophilic archaea firstly and used to withstand foreign DNA infection (Garneau et al., 2010). It consists of CRISPR and Cas protein, the former is a piece of regular clusters short-interval palindromes, consisting of several repeat–interval sequences; the latter is a kind of proteins associated with the CRISPRs gene, with nuclease and helicase activity (Chylinski et al., 2014). The mechanism of CRISPR-Cas system is similar to RNA interference. Its immune process includes three stages: adaptation, expression and interference (Choi and Lee, 2016). Moreover, a new classification system mainly including three types was proposed by Makarova et al. (2011). Type II CRISPR system has been widely used as an important gene editing tool. It is also called gRNA-Cas9 complex system, since it mainly functions through gRNA-Cas9 compound, which can not only specifically recognize DNA fragments and PAM but also induce cleavage of DNA double strands at target site.
Streptococcus thermophilus is one of the most valuable lactic acid bacteria, and widely used in production of dairy products. Besides, its CRISPR/Cas system seems to be another popular research area. After long-term immune and evolution processes, S. thermophilus CRISPR/Cas loci present rich diversities. According to the differences of genetic position, repeat sequences and cas genes, they are classified into four types, namely CRISPR1, CRISPR2, CRISPR3 and CRISPR4, which are all included in three types of CRISPR-Cas systems. Furthermore, CRISPR sequences vary from strain to strain because of the space amount differences. Hence, this article focus on the diversities of four CRISPR/Cas and CRISPR function researches, such as sgRNA-Cas9 gene editing technology, evolution relationships and anti-phage activity of S. thermophilus and its potential of becoming another model creature in Type II CRISPR/Cas researches. This work is of help to understand and accelerate researches about environmental adaptability and anti-phage mechanism of S. thermophilus, CRISPR gene editing, comparative genomic studies and evolution of Streptococcus genus and even prokaryotes.
Application of S. thermophilus CRISPR-Cas System for Gene Editing
Compared with the extremely low recombineering efficiency, down to 10-4–10-6 (Sharan et al., 2009), and time-consuming optimization of traditional homologous recombination-based methods, CRISPR system could significantly simplify the mutant construction because of its high editing efficiency (50∼100%) and possibility to select double-crossover events in one step (Chen et al., 2017).
CRISPR-Cas systems start from sequence specific recognition and target DNA loci cleavage and end with fixed mutations through DNA repair mechanism in host cell (Selle and Barrangou, 2015). The process of genome modification mediated by CRISPR-Cas9 is shown in Figure 1. Recently, CRISPR-Cas9 system of S. pyogenes (SpCas9) has been widely applied as a tool in genome modification. However, shortcomings of this system are noticed as its further study and extensive application (Hsu et al., 2013). Thus, researchers focused on the more progressive S. thermophilus CRISPR-Cas9 system (StCas9), which presents lower off-targeting efficiency, improved specificity and smaller Cas9 fragment (Xu et al., 2015; Müller et al., 2016). Its main process includes construction of functional vector containing StCas9 system sequence and expression assay in host cell. Especially, the foremost step is the design of sgRNA, two complementary single-stranded oligo sequences consisting of crRNA and tracrRNA.
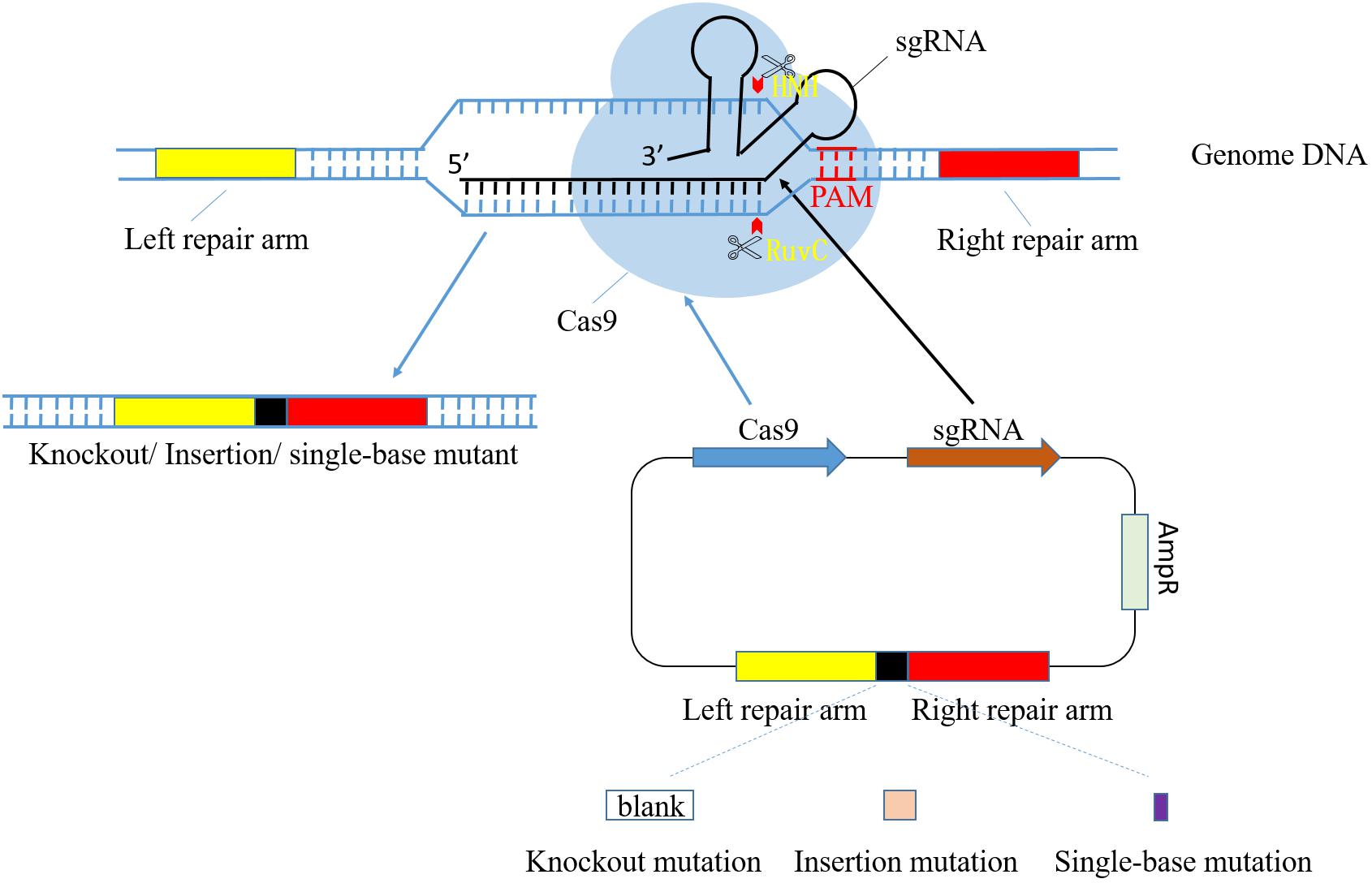
FIGURE 1. Diagram of CRISPR/Cas9 system-mediated gene editing process. pCas9 plasmid consists of cas9 gene, sgRNA, resistance locus and homologous arms. Cas9 and sgRNA gene express in order to identify target DNA and cleave at specific sites in front of PAM. RuvC and HNH are both nuclease domain in Cas9, and they cleave target DNA strand and its antisense strand, respectively. Homologous arm sequences in plasmid are complementary to their counterparts in genome so that the impact target gene fragment can be selected accurately. The whole target DNA is able to be deleted, replaced or added with a piece of new sequence through design of different homologous repair arm.
Several researches demonstrate that StCas9 shows similar cleavage activity as SpCas9 and feasibility of gene editing in human, plants and microorganisms (Xu et al., 2015; Müller et al., 2016; Steinert et al., 2016). One of the essential parts in StCas9 system is Cas9 protein. As the undertaker of the cutting function, Cas9 is composed of nuclease (NUC) and a recognition (REC) lobe (Anders et al., 2014; Nishimasu et al., 2014). The former, including NUC, RuvC like nuclease and PI domains, is mainly responsible for PAM-specific cutting of target DNA double-strand. And the latter is responsible for recognition of sgRNA-target DNA complex (Chen et al., 2017). StCas9 system includes St1Cas9 and St3Cas9, which represent S. thermophilus CRISPR-Cas9 system in CRISPR1 and CRISPR3 loci, respectively. St1Cas9 and St3Cas9 recognize 5′-NNAGAAW and 5′-NGGNG, respectively (Garneau et al., 2010; Magadán et al., 2012). Hence, the target efficiency shown between them is slightly different. St3Cas9 presents higher target efficiency than St1Cas9 (Müller et al., 2016). However, both of them can be used in genome modification according to existing researches (Xu et al., 2015; Müller et al., 2016; Steinert et al., 2016).
Furthermore, there are several pivotal factors affecting activities and editing efficiency of StCas9 system. Firstly, sgRNA, consisting of crRNA (CRISPR RNA) and tracrRNA (trans-activating RNA), owns its typical stem-loop structure, which could enhance the interaction between Cas9 and sgRNA (Jinek et al., 2014; Nishimasu et al., 2014). But stem and ring has influence on its activity. For instance, stem-loop structure in 3′ end of tracrRNA plays vital roles in activity of StCas9. And its internal bulges are essential for the functions of StCas9 while the change of side loop sequences has no effect (Xu et al., 2015). Of note, crRNA is the designed spacer acting on target DNA strand to provide specific targeting. tracrRNA is responsible for Cas9 combination and its 3′ region is vital to StCas9 activity (Wiedenheft et al., 2012; Xu et al., 2015). Furthermore the dosage of Cas9 and sgRNA is closely related to off-target efficiency of StCas9 system (Hsu et al., 2013). And truncated tracrRNA could improve its targeting efficiency (Pattanayak et al., 2013). In addition, PAM and its nearby bases are crucial for specific recognition of target fragments. Especially, G in PAM second base is essential for activity of StCas9. In conclusion, StCas9 system can not only be widely used in gene editing but also present more progressive application prospects.
Moreover, a spacer-based S. thermophilus CRISPR array assembling method has been reported and its assembling steps have been shown in the previous study (Guo et al., 2015). Thus this can be used as a breakthrough of convenient multiplex targeting (Cong et al., 2013). At first, tracrRNA cassette, cas9 fragment and CRISPR leader sequence together with CRISPR terminator should be obtained by PCR amplifying. Then amplification products are assembled into a proper vector with particular restriction endonucleases sites. As for multiplex targeting, the strategy of DR (direct repeat)-Spacer guidance is adopted instead of sgRNA guidance (Guo et al., 2015). Proper primers should be synthesized for amplifying of interested sequence containing needed DR arrays and specific spacers firstly. Particularly, when designing primers, consideration of retaining corresponding restriction sites is necessary. Through directly annealing oligonucleotides, the first DR array can be simply assembled into primary CRISPR vector (Xu et al., 2015). Next, primers are used to extend and assemble subsequent spacers and DR sequences into CRISPR locus to form an artificial Spacer-DR structure (Gou et al., 2007). Varieties of specific vectors aiming at different target genes can be reconstructed by this way. Finally, this recombinant plasmid is transferred into recipient cells to obtain target mutants, and phenotype observation and sequencing are carried out to assay its editing efficiency.
Harnessing catalytically inactive Cas9, whose endonucleolytic activity has been eliminated but identification and binding functions are still retained (Qi et al., 2013), together with certain transcription factors preventing or facilitating the banding of RNA polymerase (RNAP) and its promoter, an efficient transcription regulation can be achieved (Bikard et al., 2013). An ideal dCas9 (dead Cas9) could be constructed through mutating D10A and H840A in RuvC and HNH domains respectively (Jinek et al., 2012). Of note, the usual dCas9 is demonstrated to block transcription elongation through competitive inhibition with RNAP, while researchers have converted it into a transcriptional activator by linking it withω subunit of RNAP covalently (Dove and Hochschild, 1998; Bikard et al., 2013). A pool of specific spacers could be assembled into constructed vector simultaneously via Golden Gate assembly. Then, dCas9 system would present a pretty high activation in transcriptional regulation (Chen et al., 2017).
The Diversity of S. thermophilus CRISPR Loci and Their Evolution Relationship
According to CRISPR sequence information of several known genome strains published on CRISPR database1, there are more than one type CRISPR loci in most S. thermophilus strains. The main four CRISPR types can be distinguished by distinct DR and distribution or composition of cas genes. Supplementary Table S1 lists an overview of some basic CRISPR repeat-spacer informations. Moreover, CRISPR-Cas distribution in S. thermophilus ND07 is shown in Supplementary Figure S1 and these four loci all belong to known CRISPR-Cas systems.
CRISPR1 and CRISPR3 are attributed to Csn Type II-A and both CRISPR loci play crucial roles in adaptation and interference stages (Magadán et al., 2012). The distribution and architecture of cas sequences in CRISPR1 and CRISPR3 loci are relatively conservative, with the same amount of cas genes in upstream of their repeat-spacer regions. However, the sequence similarities between them are low, which present 33.6% and 41.3% identity in cas1 and cas2, respectively (Horvath et al., 2008). In addition, they all show strong abilities of new spacers integrating, especially CRISPR1. Several evidences demonstrate this conclusion. Through vitro experiments, S. thermophilus can naturally obtain spacers from bacteriophages and plasmids DNA into its CRISPR1 and CRISPR3 (Garneau et al., 2010). And there are more spacers in CRISPR1 locus universally. This characteristic can not only provide intensive identifications of exogenous DNA but also do good to the process of spacer integration. Furthermore, CRISPR1 is most widely distributed in S. thermophilus.
CRISPR2/Cas, included in Csm Type III-A system, is far more different in both repeat sequences and distribution of cas genes, with 27.78% identity in DR sequences and cas genes located at both sides of repeat-spacer region. Besides, as for CRISPR2 locus, high proportion of cas but low percentage of repeat-spacer region result in its poor ability of integrating new spacers. Distribution of CRISPR loci shows that CRISPR2 is probably a piece of degenerated fragment originated from gram-positive ancestor (Horvath et al., 2008).
CRISPR4, belonging to Cse Type I-E system, exists only in few strains. With only 28 nt repeat sequences and multiple cas modules distributed upstream the repeat-spacer region, CRISPR4 undergoes an unique transcription process and plays distinct roles during phage resistance. There is no novel spacer addition when foreign DNA invading, but significant up-regulated expression of gene encoding Cascade complex can be observed, especially cas7 (Young et al., 2012). Thus, CRISPR4 mainly functions through antiviral defense mediated by Cascade superfamily. In addition, its cas2 stop codon coincides with partial sequence of the repeat element (Carte et al., 2014).
It is found that distribution of Cas1 and Cas2 has intensive influences on acquisition of invading sequences. They are able to form Cas1–Cas2-protospacer DNA complex when selecting specific foreign DNA (Wang et al., 2015). And both of them retain among all CRISPR-Cas systems and present conservations, which can be applied in comparative genomic researches (Bolotin et al., 2005). It has been demonstrated that Cas1–Cas2 complex acts as integrase to integrate novel spacers into CRISPR locus (Rollie et al., 2015). In addition, new spacers are prone to integrate into the 5′ end (leading end) of repeat-spacer region, while vestigial spacers are apt to be deleted from its 3′ end (tail end) (Deveau et al., 2008). Thus, the CRISPR tail end exhibits high homologies between different strains while the leading end shows high variability. And the deletion and integration process can occur simultaneously.
Study demonstrated that consistency between CRISPR and Cas protein is relatively high in same type (Horvath et al., 2008). This observation indicates coevolution and cooperation relationships between them. And there are significant homologies of several Cas protein among different species, such as Cas7 and Cas9, which all play major roles in defending foreign DNA (Young et al., 2012). It is found that CRISPR3 is universal in most Streptococcus species while CRISPR1 and CRISPR4 exist only in few species (Horvath et al., 2008). Through analysis of spacer sequences and cas genes, species exposed to similar surroundings with evolution relationship can be found out.
Anti-Phage Activity Provided by CRISPR/Cas System
Recently, studies about acquired immunization provided by CRISPR/Cas system imply an effective approach to protect S. thermophilus from phages attack (Barrangou et al., 2007; Deveau et al., 2008). S. thermophilus phages, belonging to Siphoviridae family, have adverse effects on S. thermophilus growth and reproduction and lead to huge economic losses in dairy industry consequently. S. thermophilus phages are classified into two groups named cos and pac type according to their DNA packaging mechanism (Marrec et al., 1997). DT1, Sfi19, Sfi21 and 7201 belong to cos-type, whereas O1205, Sfi11, and 2972 are pac-type phages (Lévesque et al., 2005). They are all common bacteriophages with complete genome sequenced and present high homologies, which would improve anti-phage ability of CRISPR/Cas system.
The CRISPR-Cas system builds an ‘indestructible wall’ in bacteria, functioning via interfering and cutting off exogenous genes (Choi and Lee, 2016). When upon exposure to phage, host strains integrate short sequences from invading phages as spacers. In addition, new spacer addition is only detected in CRISPR1 and CRISPR3 (Paezespino et al., 2015). And neither CRISPR2 nor CRISPR4 acquired any novel DR sequence although CRISPR4/Cas presented biochemical activities in vitro (Sinkunas et al., 2013). After transcription of CRISPR locus containing added DR sequences and further processing, the single stranded crRNA with a second structure combines with Cas or Cascade complex to form CRISPR effect nucleoprotein complex (crRNP). The crRNP functions as endonuclease guided by crRNA. Eventually, the host is immune to the re-invading phage through sequence-specific cleavage of phage DNA.
The homologies between CRISPR spacer sequences and existing sequences in the GenBank database are analyzed (Supplementary Table S2). Firstly, most spacers are homologous with S. thermophilus bacteriophages genome. In general, one spacer is homologous with several phages and multiple spacers could resist the same phage. Intriguingly, most spacers in CRISPR2 are homologous with S. thermophilus bacteriophage 73, whereas spacers in other CRISPR loci show homologies with several phages, such as bacteriophage Sfi19, Sfi21, 7201, TP-J34 and 20617 etc., especially 20617, which is homologous with the most spacers. And researchers reported that even part of introns coexisting in S. thermophilus phages share a putative common ancestor (Lévesque et al., 2005). Secondly, the tail end spacers are inclined to be less homologous with exogenous DNA, so that they play less important roles than the 5′ end one. Similar short homologous fragments are found between S. thermophilus bacteriophages and other Streptococcus phages. By the way, there are some spacers homologous with plasmids. It is reported that fragments from plasmids are added as spacers in S. thermophilus CRISPR loci (Garneau et al., 2010). This provides novel methods to eliminate dissemination of antibiotic-resistance gene.
In addition, researches focusing on coevolution between hosts and phages propose that after exposure to phage invasion, not only do new spacers add to S. thermophilus CRISPR loci but also mutations happen in phage genome (Barrangou et al., 2007). And more newly added spacers originate from the coding strand of host genome. It can be presumed that this results from specific PAM motifs, which determine the location of integrated fragments in phage genome. Novel spacers are apt to play more important roles on resisting phages. Of note, spacer deletion has been detected in bacteriophage-insensitive S. thermophilus mutants (BIMs) during phage resistance process. And the deletion always occurs at adjacent position with addition, implying possibility that deletion happens on account of homologous recombination between DR sequences (Deveau et al., 2008).
Potential of Being Another Model System in CRISPR/Cas Researches
Streptococcus thermophilus is a common probiotic strain used in diary industry. Researchers focusing on its CRISPR system have established complete and systematic knowledge hierarchy about classification, diversity and evolution of its CRISPR loci (Carte et al., 2014; Wu et al., 2014). Lots of evidences indicate that S. thermophilus CRISPR has potential of being a characteristic model in CRISPR-Cas systems. Moreover, S. thermophilus is closely related to S. pyogenes, whose type II CRISPR/Cas has been regarded as an important model system. S. thermophilus CRISPR/Cas9 is used to mediate genome editing in eukaryote (Xu et al., 2015; Fujii et al., 2016). Furthermore, compared to S. pyogenes, S. thermophilus has higher biosecurities and is readily available (Ferretti et al., 2001).
Through homology comparison showed in Figure 2, it is found that repeat sequences in S. thermophilus CRISPR3 and S. pyogenes type II CRISPR-Cas loci share an identity of 88.89% (32/36). And there are much more spacers included in S. thermophilus CRISPR3/Cas locus. In addition, both CRISPR-Cas systems show the same structure, with four cas genes including cas9, cas1, cas2 and csn2 located upstream and repeat-spacer sequences located downstream the loci. Furthermore, length of each cas gene is identical and the four pieces of cas sequences present relatively high homology. This is related to their analogous functions. And many studies also demonstrate powerful abilities of S. thermophilus type II CRISPR-Cas in integrating exogenous DNA (Deveau et al., 2008). Of note, the common S. pyogenes CRISPR system uses NGG as its PAM loci, while S. themophilus Cas9 is able to recognize longer PAM. This characteristic imparts St1Cas9 and St3Cas9 with higher targeting efficiency and sequence specificity.
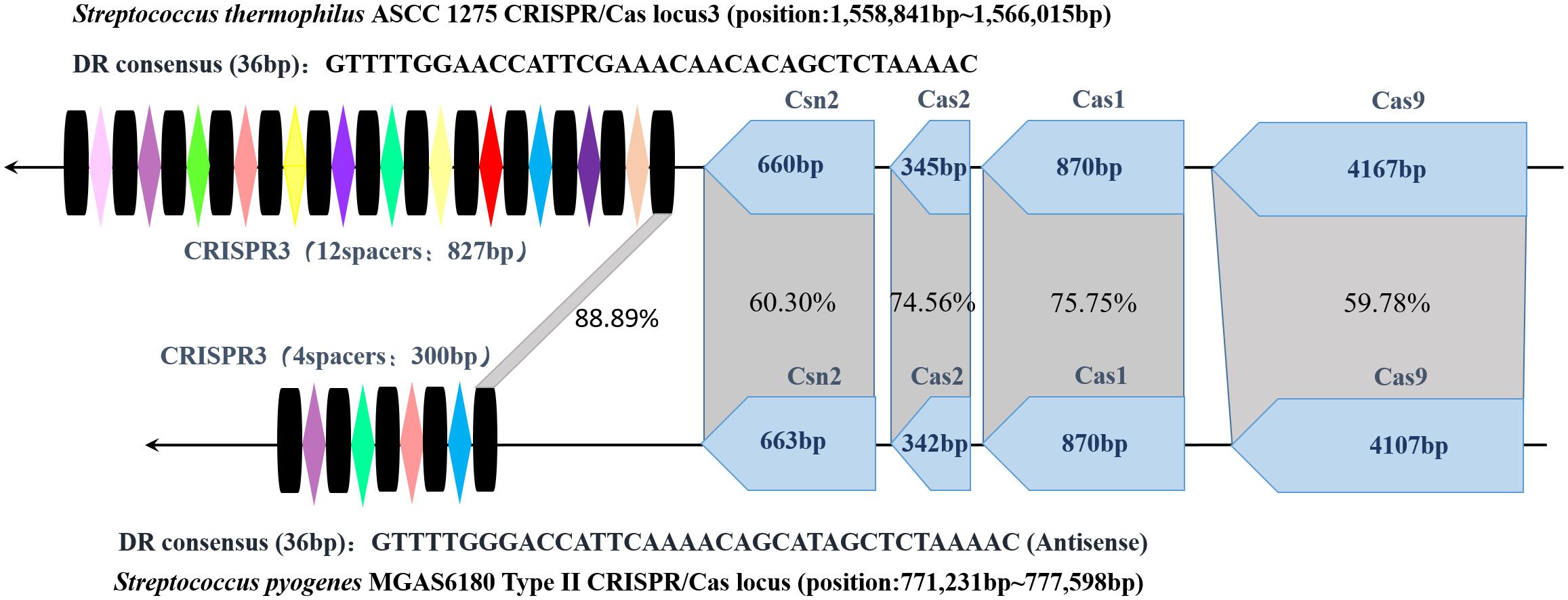
FIGURE 2. Structure of S. thermophilus CRISPR3 and S. pyogenes type II CRISPR-Cas loci and their homology comparison. The spacers and repeat sequences are represented by colored rhombi and black columns, respectively. The homology percentage between four Cas protein genes and repeat sequences are indicated in the gray shading. (S. thermophilus ASCC 1275 NCBI Reference Sequence: NZ_CP006819.1 and S. pyogenes NCBI Reference Sequence: NC_007296.1).
Furthermore there are only two types of CRISPR-Cas loci in most S. pyogenes strains genome, including type II and type I-C. Whereas S. thermophilus CRISPR-Cas owns more abundant diversities, including Csn type II-A, type I-E and type III-A. In terms of type II-A system, both CRISPR1 and CRISPR3 are endowed with high exogenous DNA resistance activity. Thus, S. thermophilus CRISPR-Cas system can not only be another characteristic model but also perform better.
Conclusion
Streptococcus thermophilus CRISPR-Cas system presents rich diversities and excellent abilities of exogenous DNA integration. Plenty of profitable characteristics contribute to its bright application prospects. Owing to the existence of RNA-guided Cas9 nucleases, S. thermophilus CRISPR-Cas9 can be used as an efficient genome modification tool with the help of designed sgRNA. In addition, diversity and related evolution relationship provide a unique perspective to classification and evolution researches in Streptococcus and other related species. Furthermore, phage infection test in vitro together with homology analysis of spacers lay foundation for studies about anti-phage effect of CRISPR-Cas system. Future development of CRISPR-Cas will benefit more from the unfavorable factors that affect its actions and expansion of its application range.
Author Contributions
YC developed the ideas presented in this manuscript. MH collected the literature and wrote the manuscript. YC and XQ professionally approved the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (Grant Nos. 31471712 and 31371827).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00257/full#supplementary-material
Footnotes
References
Anders, C., Niewoehner, O., Duerst, A., and Jinek, M. (2014). Structural basis of pam-dependent target DNA recognition by the cas9 endonuclease. Nature 513, 569–573. doi: 10.1038/nature13579
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437. doi: 10.1093/nar/gkt520
Bolotin, A., Quinquis, B., Sorokin, A., and Ehrlich, S. D. (2005). Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561. doi: 10.1099/mic.0.28048-0
Carte, J., Christopher, R. T., Smith, J. T., Olson, S., Barrangou, R., Moineau, S., et al. (2014). The three major types of CRISPR-Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol. Microbiol. 93, 98–112. doi: 10.1111/mmi.12644
Chen, W., Zhang, Y., Yeo, W. S., Bae, T., and Ji, Q. (2017). Rapid and efficient genome editing in Staphylococcus aureus by using an engineered CRISPR/Cas9 system. J. Am. Chem. Soc. 139, 3790–3795. doi: 10.1021/jacs.6b13317
Choi, K. R., and Lee, S. Y. (2016). CRISPR technologies for bacterial systems: current achievements and future directions. Biotechnol. Adv. 34, 1180–1209. doi: 10.1016/j.biotechadv.2016.08.002
Chylinski, K., Makarova, K. S., Charpentier, E., and Koonin, E. V. (2014). Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 42, 6091–6105. doi: 10.1093/nar/gku241
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Deveau, H., Barrangou, R., Garneau, J. E., Labonté, J., Fremaux, C., Boyaval, P., et al. (2008). Phage Response to CRISPR-Encoded Resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400. doi: 10.1128/JB.01412-07
Dove, S. L., and Hochschild, A. (1998). Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 12, 745–754.
Ferretti, J. J., Mcshan, W. M., Ajdic, D., Savic, D. J., Savic, G., Lyon, K., et al. (2001). Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U.S.A. 98, 4658–4663. doi: 10.1073/pnas.071559398
Fujii, W., Kakuta, S., Yoshioka, S., Kyuwa, S., Sugiura, K., and Naito, K. (2016). Zygote-mediated generation of genome-modified mice using Streptococcus thermophilus 1 -derived CRISPR/Cas system. Biochem. Biophys. Res. Commun. 477, 473–476. doi: 10.1016/j.bbrc.2016.06.070
Garneau, J. E., Dupuis, M., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71. doi: 10.1038/nature09523
Gou, D., Zhang, H., Baviskar, P. S., and Liu, L. (2007). Primer extension-based method for the generation of a siRNA/miRNA expression vector. Physiol. Genomics 31, 554–562. doi: 10.1152/physiolgenomics.00005.2007
Guo, L., Xu, K., Liu, Z., Zhang, C., Xin, Y., and Zhang, Z. (2015). Assembling the Streptococcus thermophilus clustered regularly interspaced short palindromic repeats (CRISPR) array for multiplex DNA targeting. Anal. Biochem. 478, 131–133. doi: 10.1016/j.ab.2015.02.028
Horvath, P., Romero, D. A., Coûté-Monvoisin, A. C., Richards, M., Deveau, H., Moineau, S., et al. (2008). Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190, 1401–1412. doi: 10.1128/JB.01415-07
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi: 10.1038/nbt.2647
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jinek, M., Jiang, F., Taylor, D. W., Sternberg, S. H., Kaya, E., Ma, E., et al. (2014). Structures of cas9 endonucleases reveal RNA-mediated conformational activation. Science 343:1247997. doi: 10.1126/science.1247997
Lévesque, C., Duplessis, M., Labonté, J., Labrie, S., Fremaux, C., Tremblay, D., et al. (2005). Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl. Environ. Microbiol. 71, 4057–4068. doi: 10.1128/AEM.71.7.4057-4068.2005
Magadán, A. H., Dupuis, M., Villion, M., and Moineau, S. (2012). Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One 7:e40913. doi: 10.1371/journal.pone.0040913
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. doi: 10.1038/nrmicro2577
Marrec, C. L., Sinderen, D. V., Walsh, L., Stanley, E., Vlegels, E., Moineau, S., et al. (1997). Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 63, 3246–3253.
Müller, M., Lee, C. M., Gasiunas, G., Davis, T. H., Cradick, T. J., Siksnys, V., et al. (2016). Streptococcus thermophilus crispr-cas9 systems enable specific editing of the human genome. Mol. Ther. 24, 636–644. doi: 10.1038/mt.2015.218
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. doi: 10.1016/j.cell.2014.02.001
Paezespino, D., Sharon, I., Morovic, W., Stahl, B., Thomas, B. C., Barrangou, R., et al. (2015). CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. mBio 6:e00262-15. doi: 10.1128/mBio.00262-15
Pattanayak, V., Lin, S., Guilinger, J. P., Ma, E., Doudna, J. A., and Liu, D. R. (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31, 839–843. doi: 10.1038/nbt.2673
Qi, L., Larson, M., Gilbert, L., Doudna, J., Weissman, J., Arkin, A., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022
Rollie, C., Schneider, S., Brinkmann, A. S., Bolt, E. L., and White, M. F. (2015). Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife 4:e08716. doi: 10.7554/eLife.08716
Selle, K., and Barrangou, R. (2015). Harnessing CRISPR–Cas systems for bacterial genome editing. Trends Microbiol. 23, 225–232. doi: 10.1016/j.tim.2015.01.008
Sharan, S. K., Thomason, L. C., Kuznetsov, S. G., and Court, D. L. (2009). Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223. doi: 10.1038/nprot.2008.227
Sinkunas, T., Gasiunas, G., Waghmare, S. P., Dickman, M. J., Barrangou, R., Horvath, P., et al. (2013). In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J. 32, 385–394. doi: 10.1038/emboj.2012.352
Steinert, J., Schiml, S., Fauser, F., and Puchta, H. (2016). Highly efficient heritable plant genome engineering using cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 84, 1295–1305. doi: 10.1111/tpj.13078
Wang, J., Li, J., Zhao, H., Sheng, G., Wang, M., Yin, M., et al. (2015). Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163, 840–853. doi: 10.1016/j.cell.2015.10.008
Wiedenheft, B., Sternberg, S. H., and Doudna, J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338. doi: 10.1038/nature10886
Wu, Q., Tun, H. M., Leung, C. C., and Shah, N. P. (2014). Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275. Sci. Rep. 4:4974. doi: 10.1038/srep04974
Xu, K., Ren, C., Liu, Z., Zhang, T., Zhang, T., Li, D., et al. (2015). Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell. Mol. Life Sci. 72, 383–399. doi: 10.1007/s00018-014-1679-z
Keywords: Streptococcus thermophilus, CRISPR-Cas, gene editing, diversity, model system
Citation: Hao M, Cui Y and Qu X (2018) Analysis of CRISPR-Cas System in Streptococcus thermophilus and Its Application. Front. Microbiol. 9:257. doi: 10.3389/fmicb.2018.00257
Received: 26 June 2017; Accepted: 31 January 2018;
Published: 20 February 2018.
Edited by:
Carlos Alberto Moreira-Filho, Faculdade de Medicina, Universidade de São Paulo, BrazilReviewed by:
Jiangxin Wang, Shenzhen University, ChinaGeraldo Aleixo Passos, Departamento de Genética, Universidade de São Paulo, Brazil
Copyright © 2018 Hao, Cui and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhua Cui, eWhjdWlAaGl0LmVkdS5jbg==
 Mengyuan Hao
Mengyuan Hao Yanhua Cui
Yanhua Cui Xiaojun Qu
Xiaojun Qu