
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Microbiol. , 01 May 2017
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 8 - 2017 | https://doi.org/10.3389/fmicb.2017.00760
This article is part of the Research Topic The search for biological active agent(s) from actinobacteria View all 25 articles
Deep sea is a unique and extreme environment. It is a hot spot for hunting marine actinomycetes resources and secondary metabolites. The novel deep sea actinomycete species reported from 2006 to 2016 including 21 species under 13 genera with the maximum number from Microbacterium, followed by Dermacoccus, Streptomyces and Verrucosispora, and one novel species for the other 9 genera. Eight genera of actinomycetes were reported to produce secondary metabolites, among which Streptomyces is the richest producer. Most of the compounds produced by the deep sea actinomycetes presented antimicrobial and anti-cancer cell activities. Gene clusters related to biosynthesis of desotamide, heronamide, and lobophorin have been identified from the deep sea derived Streptomyces.
The search and discovery of novel microbes that produce new secondary metabolites can be expected to remain significant in the race against new and emerging diseases and antibiotic resistant pathogens (Berdy, 2012; Manivasagan et al., 2013). Actinomycetes are widely distributed in various marine habitats, ranging from sea sand (Hong et al., 2008), mangrove sediments (Hong et al., 2009; Hong, 2013; Azman et al., 2015), sea water (Zhang L. et al., 2012), coastal sediments (Yu et al., 2015), and deep sea sediments (Zhang et al., 2015; Chen et al., 2016). The increasing number of literature on marine actinomycetes strongly supported the view that marine environments including deep sea are significant sources for search and discovery of both diverse actinomycetes resources and secondary metabolites (Skropeta and Wei, 2014; Xu et al., 2014).
Deep sea habitats show extreme variations in available nutrients, light, oxygen concentration, pressure, salinity, and temperature. Deep-sea organisms have developed unique biochemical metabolic and physiological capabilities, which not only ensure their survival in this habitat but also provide potential for the production of novel metabolites absent in terrestrial microorganisms (Fenical, 1993; Bull et al., 2000; Skropeta and Wei, 2014). Through molecular ecology studies, actinobacterial operational taxonomic units (OTUs) have been identified from deep sea sediments. Most of those foreseeably exhibit novel species, genera and families (Stach and Bull, 2005; Chen et al., 2016). Diverse species of actinomycetes cultured from the deep seafloor surface, including the deepest sea sediment samples from the Mariana Trench, have shown great biosynthetic capacities and thus a potent source of novel natural products (Pathom-aree et al., 2006d; Abdel-Mageed et al., 2010). With the breakthrough of technological barriers associated with deep sea actinobacteria isolation strategies, such as sample collection and cultivation under standard laboratory conditions, more and more deep sea actinobacteria and their natural products have been identified. Here we review the recent progress on deep sea actinomycetes and their metabolites from literature during year 2006–2016.
The vast oceans cover 70% of the world's surface, with 95% greater than 1,000 m deep. Deep sea environments are divided into the bathyal zone (depths between 200 and 2,000 m), the abyssal (depths between 2,000 and 6,000 m) and the hadal zone (depths below 6,000 m) (Harino et al., 2009). Below sea level pressure is increased by depth, thereby in the deepest part of the trenches, the pressure varying from 10 atm at the shelf- slope interface to >1,000 atm. At bathyal depths temperatures taper off rapidly with increasing depth to 2°C. Deep-sea species must adjust their biochemical processes to survive in low temperatures, because the cold reduces chemical reaction rates. Oxygen concentration drops along with the depth, oxygen-minimum layer in mid-water, usually between 300 and 1,000 m depth. Light intensity decreases exponentially with depth in the water column. No photosynthetically useful light reaches the sea floor below about 250 m (Thistle, 2003).
Start at about 200 m depth, the deep sea is characterized by high pressure, low temperature, lack of light and variable salinity and oxygen concentration (Das et al., 2006), at the shelf break, where a clear change of fauna from shallow to deep water is observed (Thistle, 2003). According to Haefner (2003), in cold deep sea mud the diversity of life can be remarkably high with species richness rivaling that of tropical rain forest. Studying the species level of microbial diversity, finding a large number of rare species which more than half of them considered as new species and more than 95% is unidentified, furthermore the expanding of biodiversity reach to the 5,000 m in depth to abyssal which the peak amount of species at the depths of 3,000 m and beyond (Skropeta, 2008). On earth abyssal hills are the most abundant of biomass, but on wider abyss the ecological impact of the habitat heterogeneity is largely unexplored (Durden et al., 2015).
However, so far only a few actinomycetes have been isolated from deep sea. It is because of technological barriers associated with isolation strategies. Therefore, we are in the pace to develop efficient cultivation methods to recover the actinobacteria population from extreme deep sea habitats. To achieve the task, firstly collection of samples from deep sea plays a pivotal role. In recent years several advancements have been developed in the context of sample collection from deep sea such as modified sediment grab and designer-built bounce corer (Fenical and Jensen, 2006), remote-operated submarine vehicle (Pathom-aree et al., 2006d), neuston sampling devices (Hakvåg et al., 2008), multi-core sampler (Xu et al., 2009), gravity or piston cores (D'Hondt et al., 2009), and untethered coring device (Prieto-Davó et al., 2013).
It is crucial to cultivate deep sea actinomyetes under standard laboratory conditions. There are several factors that influence the isolation, such as pre-treatment of dry heat (Shin et al., 2008), media composition (Luo et al., 2011; Pan et al., 2013; Song et al., 2015), dilution factor (Pathom-aree et al., 2006a), seawater requirement (Song et al., 2015), artificial seawater (Pan et al., 2013; Pesic et al., 2013) and incubation time (Song et al., 2015). It has also shown the addition of different antibiotics on selective media can inhibit the growth of fungal and bacterial contamination in order to enhance the actinomycetes growth similar to those used in isolation of actinomycetes from terrestrial sample. Long term freeze storage of deep sea sediment samples at −80°C has shown to prevent the growth of fast-growing bacteria which in results enhance the actinomycetes population (Ulanova and Goo, 2015). For the initial isolation of Streptomyces, cultivation temperatures have also influenced the recovery from deep sea sediment samples. Optimal growth temperature generally ranging from 25 to 30°C for successful cultivation of deep sea actinomycetes (Jeong et al., 2006; Luo et al., 2011; Pesic et al., 2013).
Heat pre-treatment procedures have been used effectively for the selective isolation of members of several actinomycete taxa and also inhibited growth of bacterial and fungal colonies. Moreover, actinomycete spores and hyphae are more sensitive to wet than dry heat hence relatively low temperature regimes are used to pretreat water and soil suspensions. Although heat pretreatment procedures decrease the ratio of bacteria to actinomycetes on isolation plates, the numbers of actinomycetes may also be reduced (Williams et al., 1972; Pathom-aree et al., 2006a,b,c,d). Pathom-aree et al., isolated actinomycetes from Norwegian fjord sediments support that the numbers of actinomycetes were reduced when used heat pretreatment for isolation; fewer actinomycetes were isolated on selective media inoculated with suspensions treated at 55°C as opposed to 50°C. Similarly, higher counts were generally recorded on isolation plates seeded with non-heat pretreated suspensions (Pathom-aree et al., 2006d).
For the other method, Jensen et al., 2005 used dry and stamp method for isolation actinomycetes from tropical Pacific Ocean and found that using this method for isolation of actinomycetes showed good recovery of 44%. In addition, Ulanova and Goo (2015) found that the majority of actinomycete-like colonies were also isolated using dry stamping technique from subseafloor sediments at the Nankai and Okinawa Troughs.
Novel actinomycete species isolated from deep sea environment between 2006 and 2016, have yielded an impressive array of novel species with the highest number found at depths of abyssal zone and deeper. Different media has been used by researchers (Table 1). It is worth to be noticed that long time culturing and low temperature were employed for some of the novel isolates (Table 1). Only one novel Microbacterium marinum was obtained by pretreatment at 55°C, 6 min, others were from none heat pretreated samples (Table 1). The novel deep sea actinomycete species including 21 species under 13 genera with the maximum number from Microbacterium (n = 4), followed by Dermacoccus (n = 3), Streptomyces (n = 3) and Verrucosispora (n = 2), and one novel species for each of the other 9 genera (Table 1).
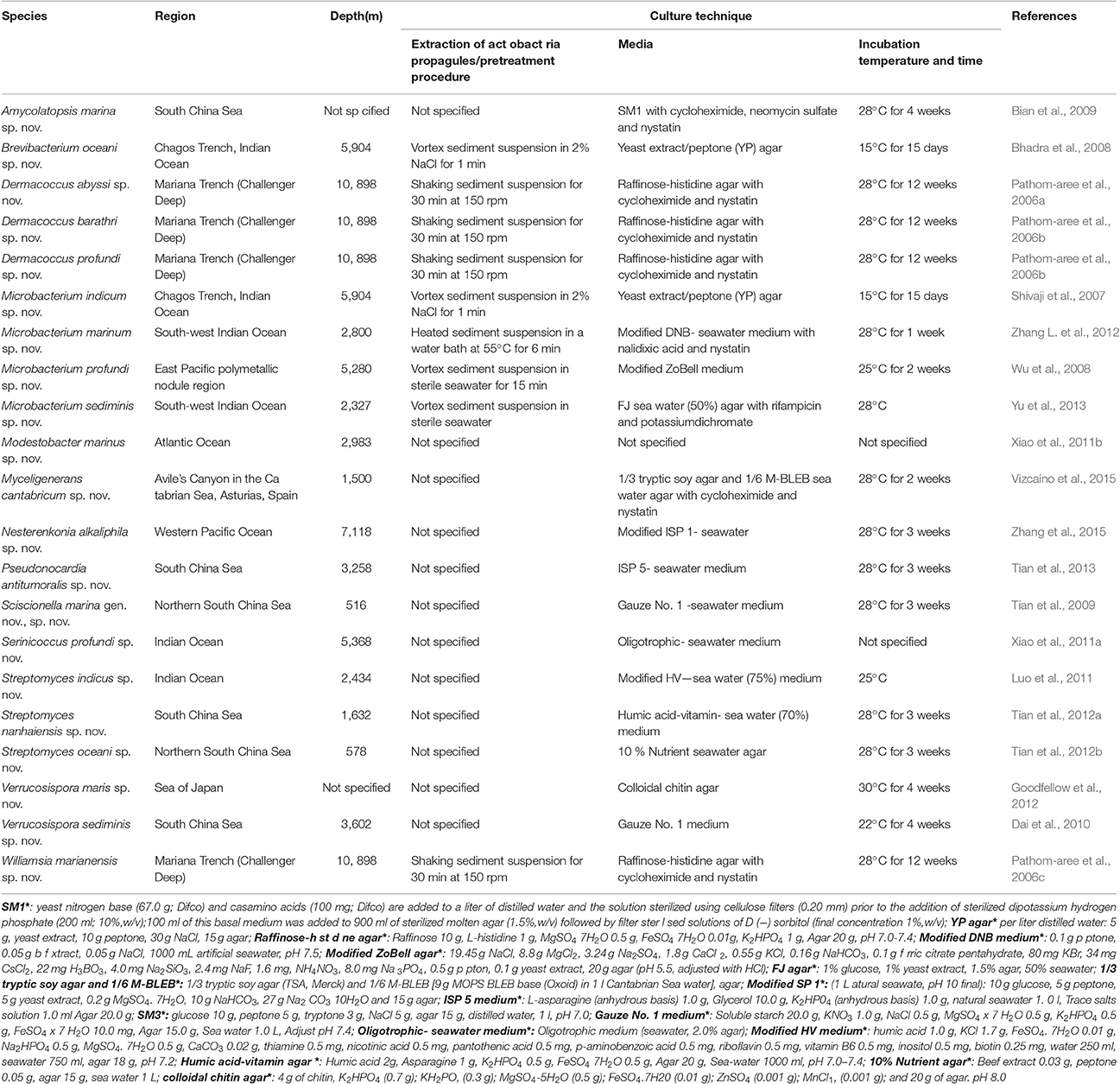
Table 1. New actinomycetes species (n = 21) isolated from deep sea environment between 2006 and 2015.
The numbers of novel microbial metabolites from deep sea sediment samples have been increasing, especially from deep sea streptomycetes. Eight genera of actinomycetes were reported to produce secondary metabolites, among which Streptomyces is the richest producer (Table 2). Earlier culture dependent studies strongly suggested that Streptomyces species are present in considerable number in deep sea sediment samples (Jensen et al., 2005; Pathom-aree et al., 2006d). In addition several novel species of deep sea derived Streptomyces strains with distinct metabolites have been reported which indicates deep sea Streptomyces are really worth in the context of novel natural products discovery (Pan et al., 2015; Song et al., 2015).
The deepest sea sediment samples from the Mariana Trench have been shown to possess great biosynthetic capacities. Seven dermacozines A–G were reported from the actinobacteria Dermacoccus abyssi sp. nov., strains MT1.1 and MT1.2 isolated from Mariana Trench sediment collected at a depth of 10 898 m. Dermacozines F and G displayed moderate cytotoxic activity against the leukemia cell line K562 with IC50 values of 9 and 7 mM, respectively, whereas dermacozine C also exhibited high radical scavenger activity with an IC50 value of 8.4 mM (Abdel-Mageed et al., 2010).
In recent years, South China Sea has been emerging as a potentially abundant source of novel species/genera of marine actinomycetes. Some bioactive compounds, such as pseudonocardians A-C, grincamycins B-F, and abyssomicins J-L were reported. Natural products derived from deep sea actinomycetes discovery have displayed a wide range of bioactivities, such as antitumor, antimicrobial, antifouling, and anti-fibrotic activities (Table 2).
Lobophorins H and I together with three known analogs, O-β-kijanosyl-(1 → 17)-kijanolide, lobophorins B and F were yielded by Streptomyces sp. 12A35, isolated from a deep sea sediment sample collected at a depth of 2,134 m in South China Sea (Pan et al., 2013). While, lobophorins E and F, along with two known analogs lobophorins A and B were discovered from the products of the deep sea Streptomyces sp. SCSIO 01127, was isolated from sample collected at a depth of 1,350 m in the South China Sea (Niu et al., 2011). The gene cluster involved in biosynthesis of lobophorin was the first type I PKS gene cluster identified from the deep sea derived Streptomyces. Three glycosyltransferases (GTs) LobG1-LobG3 genes-inactivation mutants yielded five different glycosylated metabolites, and the result suggested that LobG3 as an iterative GT to attach two L-digitoxoses (Li et al., 2013). Desotamides B, C and D together with a known desotamide A were obtained from deep sea derived Streptomyces scopuliridis SCSIO ZJ46, recovered from sediment sample collected at a depth of 3,536 m in the South China Sea (Song et al., 2014). A 39 kb gene cluster governing the biosynthesis of the anti-infective desotamides has been isolated from the strain. Desotamides A and B and a new desotamide G have been obtained by heterologous expression of desotamide gene cluster in Streptomyces coelicolor M1152 (Li et al., 2015).
Heronamides D, E, and Fare discovered from the products of Streptomyces sp. SCSIO 03032, which was isolated from deep sea sediment sample collected at a depth of 3,412 m in the Bay of Bengal, Indian Ocean (Zhang W. et al., 2014). The gene cluster governing the biosynthesis of heronamide has been isolated from strain SCSIO 03032. The gene inactivation study confirmed that P450 enzyme encode HerO as an 8-hydroxylase for tailoring heronamide biosynthesis. Feeding experiments with labeled small carboxylic acid molecules confirmed the migrated double bonds in the conjugated diene-containing side chain of heronamides (Zhu et al., 2015).
Marformycins A-F were obtained from fermentation broth of deep sea sediment-derived Streptomyces drozdowiczii SCSIO 1014, which was isolated from sample collected at a depth of 1,396 m in South China Sea. All compounds exerted selective anti-microbial activity against Micrococcus luteus, Propionibacterium acnes, and P. granulosum. Marformycins A-E displayed inhibitory activity against M. luteus with MICs of 0.25, 4.0, 0.25, 0.063, and 4.0 μg/mL, respectively, while they did not displayed any cytotoxicity (Liu D. et al., 2015). It is suggested that these compounds may be used as promising candidatures for anti-infective drug leads. The gene cluster that responsible for the biosynthesis of marformycin is about 45 kb in size and has been identified from strain SCSIO 10141. The gene inactivation studies indicated that three NRPS proteins MfnC, MfnD, MfnE, a free adenylation (A) enzyme MfnK, and a free peptidyl carrier protein (PCP) MfnL were essential for the generation of the marformycin core scaffold. Further, MfnN was found to use an intact cyclodepsipeptide intermediate as its substrate (Liu J. et al., 2015).
The discovery of novel actinomycete taxa with unique metabolic activity from deep sea samples, and novel compounds with the greatest biogenic, metabolic diversity and biological activities clearly illustrate that indigenous deep sea actinomycetes indeed exist in the oceans and are an important source of novel secondary metabolites. Other function of deep sea actinobacteria is also interesting such as oil degradation and biosurfactant production (Wang et al., 2014). It is worth to be noticed that no heat pretreatment, dry and stamp method and low temperature incubation were more productive for actinomycetes isolation from some deep sea samples. With the development of culture independent techniques, more productive strategy of strain isolation guided by the deep sea actinomycetes distribution or direct cloning and heterologous express the functional genes could be approached.
MK contribute the introduction, deep sea environment and biodiversity, actinomycete cultivation, novel taxa, and Table 1. PS contribute sample collection, Table 2 and biosynthesis of secondary metabolites from deep sea streptomycetes. KH and ZD conceived the idea and revised the whole manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The work described here was partially supported by the EU FP7 project PharmaSea (312184).
Abdel-Mageed, W. M., Milne, B. F., Wagner, M., Schumacher, M., Sandor, P., Pathom-aree, W., et al. (2010). Dermacozines, a new phenazine family from deep-sea dermacocci isolated from a Mariana Trench sediment. Org. Biomol. Chem. 8, 2352–2362. doi: 10.1039/c001445a
Azman, A. S., Othman, I., Velu, S. S., Cha, K. G., and Lee, L. H. (2015). Mangrove rare actinobacteria: taxonomy, natural compound, and discovery of bioactivity. Front. Microbiol. 6:856. doi: 10.3389/fmicb.2015.00856
Berdy, J. (2012). Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. 65, 385–395. doi: 10.1038/ja.2012.27
Bhadra, B., Raghukumar, C., Pindi, P. K., and Shivaji, S. (2008). Brevibacterium oceani sp. nov., isolated from deep-sea sediment of the Chagos Trench, Indian ocean. Int. J. Syst. Evol. Microbiol. 58, 57–60. doi: 10.1099/ijs.0.64869-0
Bian, J., Li, Y., Wang, J., Song, F. H., Liu, M., Dai, H. Q., et al. (2009). Amycolatopsis marina sp. nov., an actinomycete isolated from an ocean sediment. Int. J. Syst. Evol. Microbiol. 59, 477–481. doi: 10.1099/ijs.0.000026-0
Bull, A. T., Ward, A. C., and Goodfellow, M. (2000). Search and discovery strategies for biotechnology: the paradigm shift. Microbiol. Mol. Biol. Rev. 64, 573–606. doi: 10.1128/MMBR.64.3.573-606.2000
Chen, P., Zhang, L., Guo, X., Dai, X., Liu, L., Xi, L., et al. (2016). Diversity, biogeography, and biodegradation potential of actinobacteria in the deep-sea sediments along the deep sea sediments along the Southwest Indian Ridge. Front. Microbiol. 7:1340. doi: 10.3389/fmicb.2016.01340
D'Hondt, S., Spivack, A., Pockalny, R., Ferdelman, T., Fischer, J., Kallmeyer, J., et al. (2009). Sub seafloor sedimentary life in the South Pacific Gyre. Proc. Natl. Acad. Sci. U.S.A. 106, 11651–11656. doi: 10.1073/pnas.0811793106
Dai, H. Q., Wang, J., Xin, Y. H., Pei, G., Tang, S. K., Ren, B., et al. (2010). Verrucosispora sediminis sp. nov., a cyclodipeptide- producing actinomycete from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 60, 1807–1812. doi: 10.1099/ijs.0.017053-0
Das, S., Lyla, P., and Khan, S. A. (2006). Marine microbial diversity and ecology: importance and future perspectives. Curr. Sci. 90, 1325–1335.
Durden, J. M., Bett, B. J., Jones, D. O. B., Huvenne, V. A. I., and Ruhl, H. A. (2015). Abyssal hills hidden source of increased habitat heterogeneity, benthic megafaunal biomass and diversity in the deep sea. Prog. Oceanogr. 137, 209–218. doi: 10.1016/j.pocean.2015.06.006
Fenical, W. (1993). Chemical studies of marine bacteria: developing a new resource. Chem. Rev. 93, 1673–1683. doi: 10.1021/cr00021a001
Fenical, W., and Jensen, P. R. (2006). Developing a new resource for drug discovery: marine actinomycete bacteria. Nat. Chem. Biol. 2, 666–673. doi: 10.1038/nchembio841
Gärtner, A., Ohlendorf, B., Schulz, D., Zinecker, H., Wiese, J., and Imhoff, J. F. (2011). Levantilides A and B, 20-membered Macrolides from a Micromonospora strain isolated from the Mediterranean deep sea sediment. Mar. Drugs 9, 98–108. doi: 10.3390/md9010098
Gaudêncio, S. P., MacMillan, J. B., Jensen, P. R., and Fenical, W. (2008). Ammosamides A and B new cytotoxic alkaloids isolated from a marine Streptomyces sp. Planta Med. 74:PB172. doi: 10.1055/s-0028-1084516
Goodfellow, M., Stach, J. E., Brown, R., Bonda, A. N., Jones, A. L., Mexson, J., et al. (2012). Verrucosispora maris sp. nov., a novel deep-sea actinomycete isolated from a marine sediment which produces abyssomicins. Antonie van Leeuwenhoek 101, 185–193. doi: 10.1007/s10482-011-9651-5
Haefner, B. (2003). Drugs from the deep: marine natural products as drug candidates. Drug Discov. Today 8, 536–544. doi: 10.1016/S1359-6446(03)02713-2
Hakvåg, S., Fjaervik, E., Josefsen, K. D., Ian, E., Ellingsen, T. E., and Zotchev, S. B. (2008). Characterization of Streptomyces spp. isolated from the sea surface microlayer in the Trondheim Fjord, Norway. Mar. Drugs 6, 620–635. doi: 10.3390/md6040620
Harino, H., Arai, T., Ohji, M., and Miyazaki, N. (2009). “Organotin contamination in deep sea environment,” in Ecotoxicology of Antifouling Biocides, eds T. Arai, H. Harino, M. Ohji, and W.J. Langston (New York, NY: Springer), 95–97. doi: 10.1007/978-4-431-85709-9_6
Hohmann, C., Schneider, K., Brunter, C., Irran, E., Nicholson, G., Bull, A. T., et al. (2009). Caboxamycin, a new antibiotic of the benzoxazole family produced by the deep-sea strain Streptomyces sp. NTK 937. J. Antibiot. 62, 99–104. doi: 10.1038/ja.2008.24
Hong, K. (2013). Actinomycetes from mangrove and their secondary metabolites. Acta Microbiol. Sin. 53, 1131–1141
Hong, K., Gao, A. H., Xie, Q. Y., Gao, H., Zhuang, L., Lin, H. P., et al. (2009). Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 7, 24–44. doi: 10.3390/md7010024
Hong, S. G., Lee, Y. K., Yim, J. H., Chun, J., and Lee, H. K. (2008). Sanguibacter antarcticus sp. nov., isolated from Antarctic sea sand. Int. J. Syst. Evol. Microbiol. 58, 50–52. doi: 10.1099/ijs.0.65031-0
Huang, H. B., Yang, T. T., Ren, X. M., Liu, J., Song, Y. X., Sun, A. J., et al. (2012). Cytotoxic Angucycline class glycosides from the deep sea actinomycete Streptomyces lusitanus SCSIO LR32. J. Nat. Prod. 75, 202–208. doi: 10.1021/np2008335
Huang, H. B., Yao, Y. L., He, Z. X., Yang, T. T., Ma, J. Y., Tian, X. P., et al. (2011). Antimalarial β-carboline andindolactam alkaloids from Marinactinospora thermotolerans a deep sea isolate. J. Nat. Prod. 74, 2122–2127. doi: 10.1021/np200399t
Igarashi, Y., Ikeda, M., Miyanaga, S., Kasai, H., Shizuri, Y., and Matsuura, N. (2015). Two butenolides with PPARα agonistic activity from a marine-derived Streptomyces. J. Antibiot. 68, 345–347. doi: 10.1038/ja.2014.151
Jensen, P. R., Mincer, T. J., Williams, P. G., and Fenical, W. (2005). Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek 87, 43–48. doi: 10.1007/s10482-004-6540-1
Jeong, S. Y., Shin, H. J., Kim, T. S., Lee, H. S., Park, S. K., and Kim, H. M. (2006). Streptokordin, a new cytotoxic compound of the methylpyridine class from a marine-derived Streptomyces sp. KORDI-3238. J. Antibiot. 59, 234–240. doi: 10.1038/ja.2006.33
Komaki, H., Ichikawa, N., Hosoyama, A., Fujita, N., and Igarashi, Y. (2015). Draft genome sequence of marine-derived Streptomyces sp. TP-A0873, a producer of a Pyrrolizidine alkaloid bohemamine. Genome Announc. 3, e00008–e00015. doi: 10.1128/genomea.00008-15
Li, Q., Song, Y., Qin, X., Zhang, X., Sun, A., and Ju, J. (2015). Identification of the biosynthetic gene cluster for the anti-infective desotamides and production of a new analogue in a heterologous host. J. Nat. Prod. 78, 944–948. doi: 10.1021/acs.jnatprod.5b00009
Li, S., Tian, X., Niu, S., Zhang, W., Chen, Y., Zhang, H., et al. (2011). Pseudonocardians AC, new Diazaanthraquinone derivatives from a deap sea actinomycete Pseudonocardia sp. SCSIO 01299. Mar. Drugs 9, 1428–1439. doi: 10.3390/md9081428
Li, S., Xiao, J., Zhu, Y., Zhang, G., Yang, C., Zhang, H., et al. (2013). Dissecting glycosylation steps in lobophorin biosynthesis implies an iterative glycosyltransferase. Org Lett. 15, 1374–1377. doi: 10.1021/ol400342e
Li, X., Dobretsov, S., Xu, Y., Xiao, X., Hung, O. X., and Qian, P. Y. (2006). Antifouling diketopiperazines produced by a deep-sea bacterium, Streptomyces fungicidicus. Biofouling 22, 201–208. doi: 10.1080/08927010600780771
Liu, D., Lin, H., Proksch, P., Tang, X., Shao, Z., and Lin, W. (2015). Microbacterins A and B, new peptaibols from the deep sea actinomycete Microbacterium sediminis sp. nov. YLB-01(T). Org Lett. 17, 1220–1223. doi: 10.1021/acs.orglett.5b00172
Liu, J., Wang, B., Li, H., Xie, Y., Li, Q., Qin, X., et al. (2015). Biosynthesis of the antiinfective marformycins featuring pre-NRPS assembly line N-formylation and Omethylation and post-assembly line C-hydroxylation chemistries. Org Lett. 17, 1509–1512. doi: 10.1021/acs.orglett.5b00389
Liu, N., Shang, F., Xi, L. J., and Huang, Y. (2013). Tetroazolemycins A and B, two new oxazole-thiazole siderophores from deep-sea Streptomyces olivaceus FXJ8.012. Mar. Drugs 11, 1524–1533. doi: 10.3390/md11051524
Luo, M. H., Tang, G. L., Jub, J. H., Lua, L. C., and Huang, H. B. (2015). A new diketopiperazine derivative from a deep sea-derived Streptomyces sp. SCSIO 04496. Nat. Prod. Res. 30, 1–6. doi: 10.1080/14786419.2015.1045509
Luo, Y., Xiao, J., Wang, Y., Xu, J., Xie, S., and Xu, J. (2011). Streptomyces indicussp. nov., an actinomycete isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 61, 27126. doi: 10.1099/ijs.0.029389-0
Manivasagan, P., Venkatesan, J., Sivakumar, K., and Kim, S. K. (2013). Marine actinobacterial metabolites: current status and future perspectives. Microbiol. Res. 168, 311–332. doi: 10.1016/j.micres.2013.02.002
Nachtigall, J., Schneider, K., Bruntner, C., Bull, A. T., Goodfellow, M., and Zinecker, H. (2011). Benzoxacystol, a benzoxazine-type enzyme inhibitor from the deep-sea strain Streptomyces sp. NTK 935. J. Antibiot. 64, 453–457. doi: 10.1038/ja.2011.26
Niu, S., Li, S. M., Chen, Y., Tian, X. P., Zhang, H. B., Zhang, G. T., et al. (2011). Lobophorins E and F, new spirotetronate antibiotics from a South China sea-derived Streptomyces sp. SCSIO 01127. J. Antibiot. 64, 711–716. doi: 10.1038/ja.2011.78
Pan, H. Q., Yu, S. Y., Song, C. F., Wang, N., Hua, H. M., Hu, J. C., et al. (2015). Identification and characterization of the antifungal substances of a novel Streptomyces cavourensis NA4. J. Ind. Microbiol. Biotechnol. 25, 353–357. doi: 10.4014/jmb.1407.07025
Pan, H. Q., Zhang, S. Y., Wang, N., Li, Z. L., Hua, H. M., Hu, J. C., et al. (2013). New spirotetronate antibiotics lobophorins H and I from a South China sea derived Streptomyces sp. 12A35. Mar. Drugs 11, 3891–3901. doi: 10.3390/md11103891
Pathom-aree, W., Nogi, Y., Sutcliffe, L. C., Ward, A. C., Horikoshi, K., Bull, A. T., et al. (2006a). Dermacoccus abyssi sp. nov., a piezotolerant actinomycete isolated from the Mariana Trench. Dermacoccus abyssi sp. nov., a piezotolerant actinomycete isolated from the Mariana Trench. Int. J. Syst. Evol. Microbiol. 56 1233–1237. doi: 10.1099/ijs.0.64133-0
Pathom-aree, W., Nogi, Y., Sutcliffe, L. C., Ward, A. C., Horikoshi, K., Bull, A. T., et al. (2006b). Dermacoccus barathri sp. nov. and Dermacoccus profundi sp. nov., novel actinomycetes isolated from deep-sea mud of the Mariana Trench. Int. J. Syst. Evol. Microbiol. 56, 2303–2307. doi: 10.1099/ijs.0.64250-0
Pathom-aree, W., Nogi, Y., Sutcliffe, L. C., Ward, A. C., Horikoshi, K., Bull, A. T., et al. (2006c). Williamsia marianensis sp. nov., a novel actinomycete isolated from the Mariana Trench. Int. J. Syst. Evol. Microbiol. 56, 1123–1126. doi: 10.1099/ijs.0.64132-0
Pathom-aree, W., Stach, J. E., Ward, A. C., Horikoshi, K., Bull, A. T., and Goodfellow, M. (2006d). Diversity of actinomycetes isolated from Challenger deep sediment (10,898 m) from the Mariana Trench. Extremophiles 10, 181–189. doi: 10.1007/s00792-005-0482-z
Pesic, A., Baumann, H. I., Kleinschmidt, K., Ensle, P., Wiese, J., Süssmuth, R. D., et al. (2013). Champacyclin, a new cyclic octapeptide from Streptomyces strain C42 isolated from the Baltic Sea. Mar Drugs11, 4834–4857. doi: 10.3390/md11124834
Prieto-Davó, A., Villarreal-Gómez, L. J., Forschner-Dancause, S., Bull, A. T., Stach, J. E., Smith, C., et al. (2013). Targeted search for actinomycetes from nearshore and deep sea marine sediments. FEMS Microbiol Ecol. 84, 510–518. doi: 10.1111/1574-6941.12082
Raju, R., Piggott, A. M., Conte, M., Tnimov, Z., Alexandrov, K., and Capon, R. J. (2010). Nocardiopsins: new FKBP12-binding macrolide polyketides from an Australian marinederived actinomycete, Nocardiopsis sp. Chem. Eur. J. 16, 3194–3200. doi: 10.1002/chem.200902933
Shin, H. J., Kim, T. S., Lee, H. S., Park, J. Y., Choi, I. K., and Kwon, H. J. (2008). Streptopyrrolidine, an angiogenesis inhibitor from a marine-derived Streptomyces sp. KORDI-3973. Phytochemistry 69, 2363–2366. doi: 10.1016/j.phytochem.2008.05.020
Shivaji, S., Bhadra, B., Rao, R. S., Chaturvedi, P., Pindi, P. K., and Raghukumar, C. (2007). Microbacterium indicum sp. nov., isolated from a deep-sea sediment sample from the Chagos Trench, Indian Ocean. Int. J. Syst. Evol. Microbiol. 57, 1819–1822. doi: 10.1099/ijs.0.64782-0
Skropeta, D. (2008). Deep-sea natural products. Nat. Prod. Rep. 25, 1131–1166. doi: 10.1039/b808743a
Skropeta, D., and Wei, L. (2014). Recent advances in deep-sea natural products. Nat. Prod. Rep. 31, 999–1025. doi: 10.1039/C3NP70118B
Song, Y., Huang, H. B., Chen, Y. C., Ding, J. C., Zhang, Y., Sun, A., et al. (2013). Cytotoxic and antibacterial Marfuraquinocins from the deep South China sea-derived Streptomyces niveus SCSIO 3406. J. Nat. Prod. 76, 2263–2268. doi: 10.1021/np4006025
Song, Y., Li, Q., Liu, X., Chen, Y., Zhang, Y., Sun, A., et al. (2014). Cyclic hexapeptides from the deep south China sea-derived Streptomyces scopuliridis SCSIO ZJ46 active against pathogenic gram-positive bacteria. J. Nat. Prod. 77, 1937–1941. doi: 10.1021/np500399v
Song, Y., Liu, G., Li, J., Huang, H., Zhang, X., Zhang, H., et al. (2015). Cytotoxic and antibacterial Angucycline- and Prodigiosin- analogues from the deep-sea derived Streptomyces sp. SCSIO 11594. Mar. Drugs 13, 1304–1316. doi: 10.3390/md13031304
Stach, J. E. M., and Bull, A. T. (2005). Estimating and comparing the diversity of marine actinobacteria. Antonie van Leeuwenhoek 87, 3–9. doi: 10.1007/s10482-004-6524-1
Sun, Y., Takada, K., Nogi, Y., Okada, S., and Matsunaga, S. (2014). Lower homologues of ahpatinin, aspartic protease inhibitors, from a marine Streptomyces sp. J. Nat. Prod. 77, 1749–1752. doi: 10.1021/np500337m
Thistle, D. (2003). “The deep-sea floor: an overview,” in Ecosystems of the Deep Ocean, ed P. A. Tyler (Amsterdam: Elsevier), 5–39.
Tian, X. P., Long, L. J., Li, S. M., Zhang, J., Xu, Y., He, J., et al. (2013). Pseudonocardia antitumoralis sp. nov., a deoxynyboquinone-producing actinomycete isolated from a deep-sea sediment. Int. J. Syst. Evol. Microbiol. 63, 893–899. doi: 10.1099/ijs.0.037135-0
Tian, X. P., Long, L. J., Wang, F. Z., Xu, Y., Li, J., Zhang, J., et al. (2012a). Streptomyces nanhaiensis sp. nov., a marine streptomycete isolated from a deep-sea sediment. Int. J. Syst. Evol. Microbiol. 62, 864–868. doi: 10.1099/ijs.0.031591-0
Tian, X. P., Xu, Y., Zhang, J., Li, J., Chen, Z., Kim, C. J., et al. (2012b). Streptomyces oceani sp. nov., a new obligate marine actinomycete isolated from a deep-sea sample of seep authigenic carbonate nodule in South China Sea. Antonie van Leeuwenhoek 102, 335–343. doi: 10.1007/s10482-012-9743-x
Tian, X. P., Zhi, X. Y., Qiu, Y. Q., Zhang, Y. Q., Tang, S. K., Xu, L. H., et al. (2009). Sciscionella marina gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China Sea. Int. J. Syst. Evol. Microbiol. 59, 222–228. doi: 10.1099/ijs.0.001982-0
Ulanova, D., and Goo, K. S. (2015). Diversity of actinomycetes isolated from subseafloor sediments after prolonged low-temperature storage. Folia Microbiol. 60, 211–216. doi: 10.1007/s12223-014-0361-z
Um, S., Kim, Y. J., Kwon, H., Wen, H., Kim, S. H., Kwon, H. C., et al. (2013). Sungsanpin, a lasso peptide from a deep-sea streptomycete. J. Nat. Prod. 76, 873–879. doi: 10.1021/np300902g
Vizcaíno, A. S., González, V., Branã, A. F., Molina, A., Acunã, J. L., Garcıá, L. A., et al. (2015). Myceligenerans cantabricum sp. nov., a barotolerant actinobacterium isolated from a deep cold-water coral. Int. J. Syst. Evol. Microbiol. 65, 1328–1334. doi: 10.1099/ijs.0.000107
Wagner, M., Abdel-Mageed, W. M., Ebel, R., Bull, A. T., Goodfellow, M., Fiedler, H. P., et al. (2014). Dermacozines H−J isolated from a deep-sea strain of Dermacoccus abyssi from Mariana Trench sediments. J. Nat. Prod. 77, 416–420. doi: 10.1021/np400952d
Wang, Q., Song, F. H., Xiao, X., Huang, P., Li, L., Monte, A., et al. (2013). Abyssomicins from the south China sea deep sea sediment Verrucosispora sp.: natural thioether michael addition adducts as antitubercular prodrugs. Angew. Chem. Int. Ed. Engl. 52, 1231–1234. doi: 10.1002/anie.201208801
Wang, W., Cai, B., and Shao, Z. (2014). Oil degradation and biosurfactant production by the deep sea bacterium Dietziamaris As-13-3. Front. Microbiol. 5:711. doi: 10.3389/fmicb.2014.00711
Williams, S. T., Shameemullah, M., Watson, E. T., and Mayfield, C. I. (1972). Studies on the ecology of actinomycetes in soil. VI. The influence of moisture tension on growth and survival. Soil Biol. Biochem. 4, 215–225. doi: 10.1016/0038-0717(72)90014-4
Wu, Y. H., Wu, M., Wang, C. S., Wang, X. G., Yang, J. Y., Oren, A., et al. (2008). Microbacterium profundi sp. nov., isolated from deep-sea sediment of polymetallic nodule environments. Int. J. Syst. Evol. Microbiol. 58, 2930–2934. doi: 10.1099/ijs.0.2008/000455-0
Xiao, J., Luo, Y., Xie, S., and Xu, J. (2011a). Serinicoccus profundi sp. nov., an actinomycete isolated from deep-sea sediment, and emended description of the genus Serinicoccus. Int. J. Syst. Evol. Microbiol. 61, 16–19. doi: 10.1099/ijs.0.019976-0
Xiao, J., Luo, Y., Xu, J., Xie, S., and Xu, J. (2011b). Modestobacter marinussp. nov., a psychrotolerant actinobacterium from deep-sea sediment, and emended description of the genus Modestobacter. Int. J. Syst. Evol. Microbiol. 61, 1710–1714. doi: 10.1099/ijs.0.023085-0
Xu, D. B., Ye, W. W., Han, Y., Deng, Z. X., and Hong, K. (2014). Natural products from mangrove actinomycetes. Mar. Drugs 12, 2590–2613. doi: 10.3390/md12052590
Xu, Y., Li, H., Li, X., Xiao, X., and Qian, P. Y. (2009). Inhibitory effects of a branched – chain fatty acid on larval settlement of the polychaete hydroides elegans. Mar. Biotechnol. (NY). 11, 495–504. doi: 10.1007/s10126-008-9161-2
Yang, X. W., Peng, K., Liu, Z., Zhang, G. Y., Li, J., Wang, N., et al. (2013a). Strepsesquitriol, a rearranged zizaane-type sesquiterpenoid from the deep-sea-derived actinomycete Streptomyces sp. SCSIO 10355. J. Nat. Prod. 76, 2360–2363. doi: 10.1021/np400923c
Yang, X. W., Zhang, G. Y., Ying, J. X., Yang, B., Zhou, X. F., Steinmetz, A., et al. (2013b). Isolation, characterization, and bioactivity evaluation of 3-((6-methylpyrazin-2yl)methyl)- 1H-indole, a new alkaloid from a deep-sea-derived actinomycete Serinicoccus profundi sp. nov. Mar. Drugs 11, 33–39. doi: 10.3390/md11010033
You, Z. Y., Wang, Y. H., Zhang, Z. G., Xu, M. J., Xie, S. J., Han, T. S., et al. (2013). Identification of two novel anti-Fibrotic benzopyran compounds produced by engineered strains derived from Streptomyces xiamenensis M1-94P that originated from deep-sea sediments. Mar. Drugs 11, 4035–4049. doi: 10.3390/md11104035
Yu, J., Zhang, L., Liu, Q., Qi, X. H., Ji, Y., and Kim, B. S. (2015). Isolation and characterization of actinobacteria from Yalujiang coastal wetland, North China. Asian Pac. J. Trop. Biomed. 5, 555–560. doi: 10.1016/j.apjtb.2015.04.007
Yu, L., Lai, Q., Yi, Z., Zhang, L., Huang, Y., Gu, L., et al. (2013). Microbacterium sediminis sp. nov., a psychrotolerant, thermotolerant, halotolerant and alkalitolerant actinomycete isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 63, 25–30. doi: 10.1099/ijs.0.029652-0
Zhang, G., Zhang, Y., Yin, X., and Wang, S. (2015). Nesterenkonia alkaliphila sp. nov., an alkaliphilic, halotolerant actinobacteria isolated from the western Pacific Ocean. Int. J. Syst. Evol. Microbiol. 65, 516–521. doi: 10.1099/ijs.0.065623-0
Zhang, L., Xi, L., Ruan, J., and Huang, Y. (2012). Microbacterium marinum sp. nov., isolated from deep-sea water. Syst. Appl. Microbiol. 35, 81–85. doi: 10.1016/j.syapm.2011.11.004
Zhang, Q., Li, S., Chen, Y., Tian, X., Zhang, X., Zhang, G., et al. (2013). New diketopiperazine derivatives from a deep-sea-derived Nocardiopsis alba SCSIO 03039. J. Antibiot. 66, 31–36. doi: 10.1038/ja.2012.88
Zhang, W. J., Liu, Z., Li, S. M., Yang, T. T., Zhang, Q. B., Ma, L., et al. (2012). Spiroindimicins A-D: new Bisindole alkaloids from a deep-sea-derived actinomycete. Org. Lett. 14, 3364–3367. doi: 10.1021/ol301343n
Zhang, W. J., Ma, L., Li, S. M., Liu, Z., Chen, Y. C., et al. (2014). Indimicins A-E, Bisindole alkaloids from the deep-sea-derived Streptomyces sp. SCSIO 03032. J. Nat. Prod. 77, 1887–1892. doi: 10.1021/np500362p
Zhang, W., Li, S., Zhu, Y., Chen, Y., Chen, Y., Zhang, H., et al. (2014). Heronamides D-F, polyketide macrolactams from the deep-sea-derived Streptomyces sp. SCSIO 03032. J. Nat. Prod. 77, 388–391. doi: 10.1021/np400665a
Zhou, X., Huang, H. B., Li, J., Song, Y. X., Jiang, R. W., Liu, J., et al. (2014). New antiinfective cycloheptadepsipeptide congeners and absolute stereochemistry from the deep sea-derived Streptomyces drozdowiczii SCSIO 10141. Tetrahedron 70, 7795–7801. doi: 10.1016/j.tet.2014.02.007
Keywords: deep sea, actinomycetes, bioactive natural products, biosynthesis, novel species
Citation: Kamjam M, Sivalingam P, Deng Z and Hong K (2017) Deep Sea Actinomycetes and Their Secondary Metabolites. Front. Microbiol. 8:760. doi: 10.3389/fmicb.2017.00760
Received: 30 December 2016; Accepted: 12 April 2017;
Published: 01 May 2017.
Edited by:
Learn-Han Lee, Monash University Malaysia, MalaysiaReviewed by:
Joachim Wink, Helmholtz Centre for Infection Research, GermanyCopyright © 2017 Kamjam, Sivalingam, Deng and Hong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kui Hong, a3VpaG9uZzMxQHdodS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.