
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Med. , 05 March 2025
Sec. Dermatology
Volume 12 - 2025 | https://doi.org/10.3389/fmed.2025.1476685
The incidence of skin cancer continues to rise due to increased sun exposure and tanning habits, requiring early detection and treatment for favorable outcomes. Skin biopsy is an important diagnostic tool in dermatology and pathology, as it provides a valuable understanding of various skin diseases. Proper handling of skin biopsy specimens is vital to ensure accurate histopathological assessment. Still, the use of light microscopy and immunofluorescence provides a comprehensive approach to evaluating skin biopsy specimens, with each contributing unique information to aid in accurate diagnosis and management. This review highlights the evolution of skin biopsy practices, from traditional techniques to advanced methods incorporating artificial intelligence (AI) and convolutional neural networks. AI technologies enhance diagnostic accuracy and efficiency, aiding in the rapid analysis of skin lesions and biopsies. Despite challenges such as the need for extensively annotated datasets and ethical considerations, AI shows promise in dermatological diagnostics. The future of skin biopsy lies in minimally invasive techniques, liquid biopsies, and integrated pharmacogenomics for personalized medicine.
Skin cancer is one of the most common types of cancer globally, affecting millions of people each year. The primary types are basal cell carcinoma (BCC), squamous cell carcinoma (SCC), and melanoma, with BCC being the most frequent (1). In 2020, non-melanoma skin cancer (NMSC), after lung and prostate cancers, was the third most frequently diagnosed cancer in males worldwide. In fact, NMSC was the first and most common cancer in Northern America, Australia, and New Zealand (2). The rate of skin cancer is increasing because of more sun exposure and tanning habits. Detecting and treating skin cancer early is essential for a positive outcome, but there are obstacles to diagnosis and healthcare accessibility. Raising public awareness and promoting preventive actions, like applying sunscreen and limiting sun exposure, are critical to lowering the risk of skin cancer (3, 4).
Skin biopsies are crucial diagnostic procedures in dermatology, providing valuable information about various skin conditions. Skin biopsy is valuable in distinguishing between different types of skin diseases, including but not limited to bullous pemphigoid, dermatitis herpetiformis, pemphigus vulgaris, epidermolysis bullosa simplex, discoid lupus erythematosus, systemic lupus erythematosus, erythema multiforme, lichen planus actinicus, leukocytoclastic vasculitis, vasculitis, urticarial vasculitis, polymorphous light eruption, demodex follicularis, porphyria, thrombotic thrombocytopenia, vitiligo, and psoriasis. Furthermore, it helps in diagnosing spongiotic patterns such as spongiotic dermatitis, pompholyx, and pityriasis. Recent advances in skin biopsy techniques for diagnosing alopecia have improved precision in identifying underlying causes, particularly in distinguishing between scarring and non-scarring types (5). Key techniques include the use of punch biopsy, ideally taken at a 4 μm thickness to ensure deeper analysis and assessment across all follicle levels (6). Both vertical and horizontal sectioning are commonly used to evaluate different layers of hair structure and associated pathologies (7, 8). Findings such as follicular miniaturization, perifollicular fibrosis, and the ratio of growth phase to resting phase hairs help in diagnosing conditions such as androgenetic alopecia, alopecia areata, and scarring alopecia (9).
A typical skin biopsy involves taking a small tissue sample from the skin and examining it under a microscope to identify abnormalities and diagnose different skin diseases (10). This method provides detailed information about the tissue’s structure and any pathological changes. The combined use of light microscopy (LM) and immunofluorescence (IF) provides a full approach to diagnosing skin biopsies.
LM, through the use of mainly hematoxylin and eosin (H&E), offers detailed views of cellular architecture, which facilitates the identification of general pathological changes. IF techniques, which involve the use of antibodies tagged with fluorescent dyes to detect specific antigens in the tissue, are useful for diagnosing autoimmune and inflammatory diseases, such as lupus erythematosus and pemphigoid, by revealing the presence and distribution of immune deposits in the skin. Combining LM and IF enhances diagnostic accuracy and provides a comprehensive evaluation of skin biopsy specimens (11).
The literature lacks a review that bridges traditional biopsy techniques with modern advancements such as AI and convolutional neural networks (CNNs). This gap leaves a need for a resource that highlights the progression from traditional to modern techniques and discusses their combined potential to enhance diagnostic accuracy. As the incidence of skin cancer continues to rise, there is a pressing need for improved diagnostic tools and techniques. Traditional skin biopsy methods, while effective, can be further enhanced by incorporating modern technologies such as AI and CNNs (12). These technologies promise to increase diagnostic accuracy and efficiency, addressing some limitations of current practices. This review highlights the evolution of skin biopsy practices, from traditional techniques to advanced methods incorporating AI and CNNs.
After obtaining informed consent, the area of the skin to be biopsied is cleaned with an antiseptic solution, and using a dissecting microscope to look for gross indications such as blisters or vesicles, a fresh skin biopsy is bisected into two pieces: one half for light microscopy and the other half for immunofluorescence (13). For light microscopy, the skin biopsy portion is fixed in 10% neutral buffered formalin, processed for histology, sectioned at 3 μm thickness, and stained using the hematoxylin and eosin method. For immunofluorescence, the skin biopsy portion is snap-frozen in liquid nitrogen, sectioned at 5 μm thickness, fixed in cold acetone, and stained with fluorescence isothiocyanate-conjugated antibodies.
IF is a powerful technique used in conjunction with skin biopsy to diagnose various skin diseases. In dermatology, IF is particularly useful for diagnosing autoimmune blistering diseases and other dermatoses (14). When combined with a skin biopsy, IF provides detailed information about the presence and distribution of immune components in the skin (15). Direct IF (DIF) is the most commonly used method for skin biopsies. DIF demonstrates IgG, IgA, and IgM immunoglobulins, and C3 complement in skin biopsies (16). For example, DIF shows IgG and C3 deposits in the epidermis in pemphigus vulgaris, reveals linear IgG and C3 deposits along the basement membrane zone in Bullous Pemphigoid, demonstrates granular IgA deposits at the dermal-epidermal junction in dermatitis herpetiformis, and shows a full band of IgG, IgM, IgA, and C3 at the dermo-epidermal junction in lupus erythematosus (17, 18).
The frozen section procedure for skin biopsy is a quick, intraoperative diagnostic method, often used in surgeries for skin cancer, such as Mohs micrographic surgery (MMS) (19). After the tissue sample is collected, it is rapidly frozen in a cryostat, allowing the tissue structure to be preserved temporarily without formalin fixation. A measure of 5–6 μm thickness is then cut using a cryostat and stained with hematoxylin and eosin, for microscopic examination. Then, the pathologist assesses the tissue’s cellular structures and determines whether cancer cells are present at the margins, which guides the surgeon on whether further excision is needed (20).
Several techniques are used to obtain skin biopsy specimens, each with its indications: Punch biopsy, where a circular blade is used to remove a cylindrical core of skin, including the epidermis, dermis, and superficial subcutis. It is commonly used to diagnose inflammatory and neoplastic skin conditions. Shave biopsy, where a scalpel or razor blade is used to shave off a superficial layer of skin. This method is suitable for lesions confined to the epidermis, such as warts and superficial basal cell carcinomas. Excisional biopsy, where the entire lesion is removed with a margin of normal skin. It is used for small, suspicious lesions where complete removal is necessary. An incisional biopsy, where a portion of the lesion is removed, is often used for larger lesions or when a diagnosis cannot be made with less invasive methods (21). Figure 1 shows the different types of skin biopsies.
AI is a branch of computer science focused on creating systems capable of performing tasks that typically require human intelligence. These tasks include learning, reasoning, problem-solving, understanding language, and recognizing patterns (22). AI technologies, such as machine learning and neural networks, enable computers to learn from data and improve over time. AI is widely used in various fields, including healthcare, helping to automate processes, enhance decision-making, and solve complex problems. It enhances the ability to diagnose, treat, and manage diseases (23). In dermatology, AI, particularly through technologies such as CNNs, is used to analyze images of skin lesions, helping to detect conditions such as skin cancer with high accuracy (24). AI assists dermatologists by providing quick and precise analyses, reducing the time needed for diagnosis, and improving patient outcomes.
CNNs are a specialized type of AI designed to process and analyze visual data. It mimics the human brain’s way of recognizing patterns and features in images, making it particularly effective for tasks such as image classification, object detection, and facial recognition (25). CNNs are widely used in various fields, including healthcare, where they assist in diagnosing medical images, as well as in technology for applications such as self-driving cars and image search engines. Their ability to learn from large sets of labeled data allows them to identify complex patterns and provide accurate predictions. There are different types of CNN architectures available, such as NASNet-Large, AlexNet, Inception-ResNet-v2, Inception-v3, ResNet-50, SqueezeNet, and Vgg19, and (26, 27). In dermatology, CNNs are used to help diagnose skin conditions by examining images of skin lesions (28, 29). They can differentiate between various types of skin cancers such as melanoma and basal cell carcinoma, with high accuracy (30). By learning from thousands of labeled images, CNNs can assist dermatologists in making quicker and more accurate diagnoses, improving patient care and outcomes (31). However, a key concern with AI is bias caused by a lack of diverse skin tones in training data. Research shows that many AI systems are mainly trained on images of lighter skin, making them less accurate at diagnosing conditions in people with darker skin (32–34). A review, which identified that only 20% (14/70) about race and 10% (7/70) about skin color, reported that bias may contribute to higher rates of false positives and false negatives, thus affecting patient outcomes and contributing to healthcare differences (35). A recent systemic review has also reported that the majority of the studies reviewed were obtained from light skin and therefore provide insufficient evidence to comment on the overall accuracy of AI models for darker skin types. They concluded that the lack of diversity in studies is likely caused by the shortage of available datasets (36).
The excitement regarding AI in dermatology began in 2017 when a study compared the diagnostic performance of an AI-powered network with that of 21 board-certified dermatologists in evaluating biopsy-proven clinical images of benign and skin cancers. The findings showed that the AI system demonstrated diagnostic accuracy equal to that of human experts, achieving a level of competence comparable to that of experienced dermatologists (24).
Not widely applied but several studies have shown that direct image analysis is a reality for accurate classification of routine diagnoses for skin biopsies (37–40). In fact, one of the common uses of AI in skin biopsy is diagnosis. Basal cell carcinoma (BCC) showed a sensitivity of 98.23% and a specificity of 98.51% using CNNs from 1,255 whole-slide images (41). Another study revealed that CNNs achieved an accuracy of 99.5% for nodular BCC, 99.3% for dermal nevus (DN), and 100.0% for seborrheic keratosis (SK) (42). Another interesting finding regarding the AI is that CNNs, which were trained using 595 histopathologic images of melanomas and nevi, were classified by an expert dermato-histopathologist. When tested with an additional 100 images, the CNNs showed a discordance rate of only 19% compared to the histopathologist’s classifications (43). This rate is similar to the discordance between human pathologists, which is reported in the literature to be 25–26% (44, 45).
A total of 1,377 patches of healthy tissue and 2,141 patches of melanoma were assessed in the training/validation set, while 791 patches of healthy tissue and 1,122 patches of pathological tissue were evaluated in the test dataset. The CNN findings showed 95.7% sensitivity, 97.7% specificity, and 96.5% accuracy when compared with the dermatopathologist results (46). In a retrospective study, which reviewed 225,230 pathological patches cut from 79 formalin-fixed paraffin-embedded pathological slides from 73 patients (55 non-malignant eyelid nevus slides from 55 patients and 24 malignant melanoma slides from 18 patients), H&E stained whole-slide images (WSIs) and compared with 7 board-certified pathologists, the CNN findings showed 91.4% accuracy, 91% sensitivity, and 92.8% specificity (47).
In comparison with 95 human experts, of whom 62 were board-certified dermatologists, the CNNs, which were trained on 7,895 dermoscopic and 5,829 close-up images of lesions excised at a primary skin cancer clinic, showed a higher accuracy rate than those experts in diagnosis of common malignant cases such as basal cell carcinoma, actinic keratoses or Bowen disease, and squamous cell carcinoma or keratoacanthoma but did not reach the accuracy of human experts in rare malignant non-pigmented lesions such as amelanotic melanoma and benign non-pigmented lesions (48). It is important to note that amelanotic melanoma is not easy to diagnose, even for experts (49). Another systematic review of 39 studies for the detection of NMSC found that the AI overall diagnostic accuracy, in comparison with histopathologic diagnosis, was high and ranged from 72 to 100% (50). In a training set of 1,629 images (743 malignant lip, 886 benign lip diseases), the findings showed that CNNs were equivalent to the dermatologists and superior to the non-dermatologists in classifying malignancy (51). On a set of 1,417 images from 308 regions of interest on skin histopathology slides, where the presence or absence of basal cell carcinoma needs to be determined, the findings showed that deep learning architectures had a 91.4% accuracy in comparison with histopathology (52).
Another important use of AI in dermatology is with onychomycosis, which is best demonstrated by the periodic-acid-Schiff (PAS) staining method in comparison with other methods such as direct microscopy using potassium hydroxide staining, fluorescence optical preparation, and culture (53, 54). PAS has high specificity, is not expensive, and the detection of tinea is high if present in a high number (55). However, if the fungi are present in a small number, histopathologic detection is time-consuming and the risk of missing fungi is high (56). Subsequently, this might result in a delay in diagnosis and repeating preparation and analysis (57). The literature shows that the sensitivity of detecting fungi using histopathologic evaluation has been reported to range between 80 and 85% (58). A study, that used CNNs with 664 corresponding H&E- and PAS-stained histologic whole-slide images (WSIs) of human nail plates from four different laboratories, showed a sensitivity and specificity of 93 and 77%, respectively. Their study demonstrated comparable sensitivity to that of the 11 board-certified dermatopathologists (56). Another similar study reported that CNNs showed 94.1% sensitivity and 98% specificity, for a dataset of 528 whole-slide images of nail samples for onychomycosis (59). Another interesting finding of AI with microorganisms is that the CNN model in a dataset of 1,819 thick smear images from 150 patients showed effectiveness in discriminating between positive (parasitic) and negative image patches with 93.46% accuracy, 92.59% sensitivity, 94.33% specificity, 94.25% precision, and 92.74% negative predictive value (60). A recent systemic review on the use of AI in skin disease diagnosis in primary care settings showed that CNN has a sensitivity ranging from 58 to 96.1%, with accuracies varying from 41 to 93% (61). Other studies have used AI in research in dermatopathology (62, 63). These findings highlight the AI potential to enhance diagnostic accuracy and efficiency in dermatopathology.
Artificial intelligence (AI) applications in frozen section analysis are advancing to assist pathologists in rapidly evaluating slides, identifying cancerous cells, and distinguishing them from healthy tissue, which enhances diagnostic speed and accuracy in the operating room setting. Several studies using AI and deep learning models have been valuable tools in enhancing the assessment of MMS slides for skin cancer detection with high sensitivity and specificity rates (64, 65). A review of selected recent articles on the application of artificial intelligence in diagnosing and managing skin diseases is shown in Table 1.
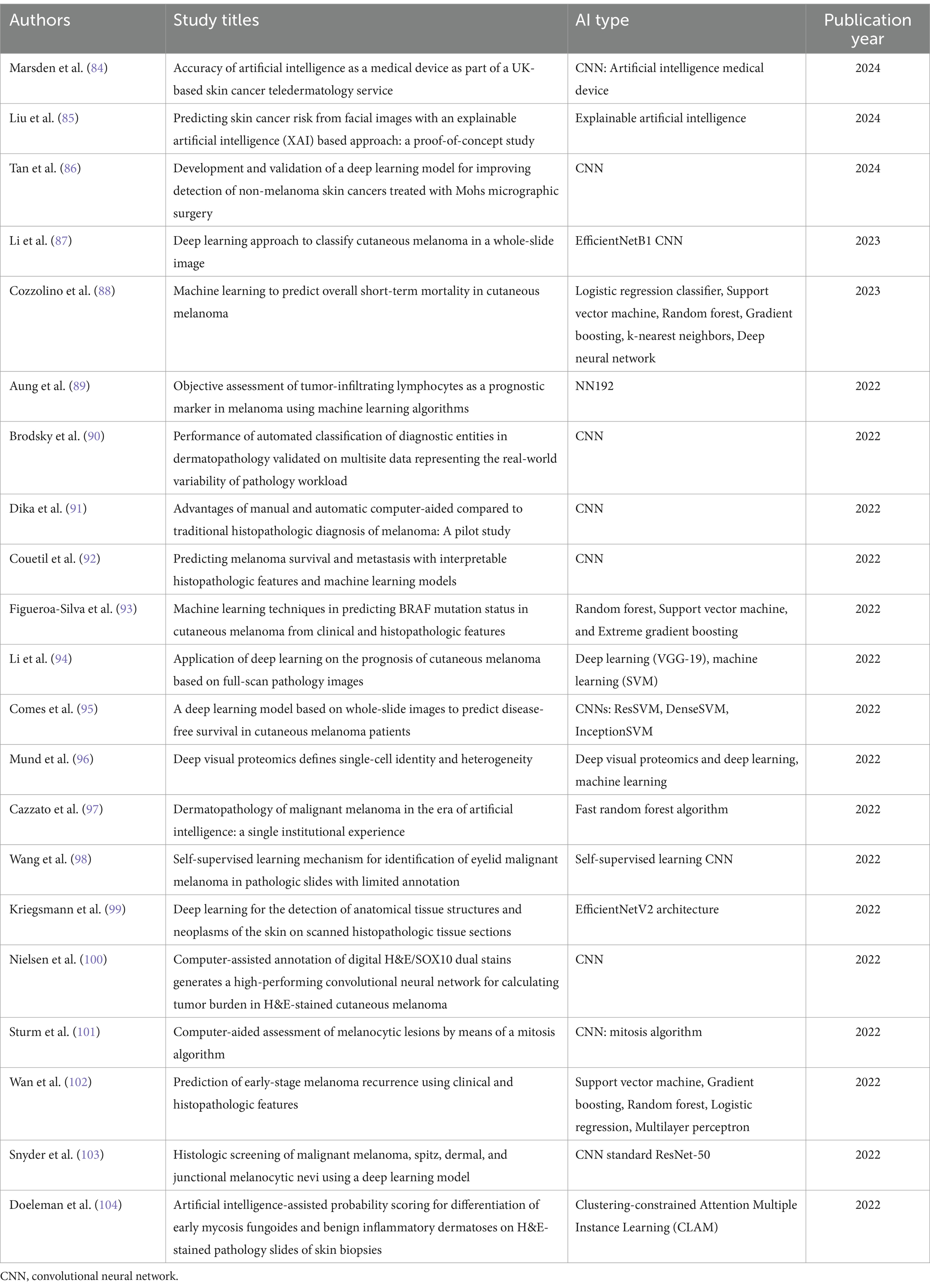
Table 1. Review of selected recent articles on the application of artificial intelligence in diagnosing and managing skin diseases.
AI assists in the interpretation of biopsy samples. AI serves as a valuable tool for pathologists, offering second opinions and highlighting areas of concern within biopsy samples (40). AI can reduce diagnostic errors by providing consistent and objective analysis, minimizing inter- and intra-observer variability (66). AI can process large volumes of biopsy samples quickly, reducing turnaround times and enabling faster diagnosis and treatment initiation. AI can be used to predict disease outcomes and responses to treatment based on biopsy findings and other patient data, aiding in personalized medicine and tailored therapeutic strategies (67).
As technology advances, there is a growing potential to enhance the diagnostic abilities of skin biopsy through innovative techniques and tools. Future developments aim to improve the precision, efficiency, and comprehensiveness of skin biopsies, finally benefiting patient care. Regarding innovations in biopsy techniques, there have been significant advancements in minimally invasive procedures and liquid biopsy methods. Microneedle biopsy involves the use of microneedle arrays to collect small amounts of tissue or interstitial fluid with less discomfort and scarring for patients (68, 69). Laser-assisted biopsy uses lasers to precisely target and excise tissue samples, minimizing damage to surrounding areas and enhancing accuracy while reducing healing times (70). Additionally, research into liquid biopsy focuses on identifying circulating biomarkers, such as cell-free DNA, RNA, and proteins, which can provide diagnostic information from a blood sample and potentially reduce the need for traditional tissue biopsies (71). Due to advances in multi-omics such as genomics, transcriptomics, and in genomic analyses such as next-generation sequencing, the identification of many gene mutations has been explored, as with BRAF, NRAS, and c-KIT in melanoma (72–74). Regarding personalized medicine and precision dermatology genomic and molecular profiling by integrating these data with skin biopsy results to gain a comprehensive understanding of individual patient conditions. This approach can guide targeted therapies and improve treatment outcomes (75, 76). Pharmacogenomics, the study of how genes affect a person’s response to drugs, is increasingly being used in skin biopsy analysis to modify treatments and improve outcomes. This approach allows for personalized medicine, where genomic and molecular profiling of biopsy samples can reveal specific genetic variations that influence drug efficacy and safety (77). For instance, understanding a patient’s unique genetic makeup can help dermatologists select the most effective therapies and minimize adverse effects, optimizing the treatment of skin cancers and other dermatological conditions (78, 79). As technology evolves, the integration of pharmacogenomics with skin biopsies holds promise for more precise and effective dermatological care.
Despite the prevalence of skin lesions, scientists face challenges in obtaining annotated training and skin images as skin disease images are still insufficient (80). One critical challenge with CNNs is their need for a large amount of data; the quality and size of the image dataset are essential for effective CNN training and validation (81). Another important drawback is that experts in computer science, biomedical, and medicine are insufficient (82). Furthermore, gaining acceptance from healthcare professionals requires addressing concerns about reliability and transparency. Another major issue with AI is determining who is responsible for any diagnostic errors made by the AI. As well as other moral and ethical issues related to the use of AI. In addition, there are many kinds of skin diseases (83). It might be difficult for AI to identify all specific skin diseases. Furthermore, as AI models trained mostly on lighter skin tones may have lower accuracy for darker skin, this might raise concerns about diagnostic differences (36).
Histopathological diagnosis of skin biopsy is still the gold standard method for skin diseases. However, the integration of traditional histopathological techniques such as light microscopy and immunofluorescence with advanced technologies such as artificial intelligence and convolutional neural networks enhances diagnostic accuracy and efficiency. Innovations in biopsy techniques, including minimally invasive procedures and liquid biopsies, should further improve patient outcomes by reducing discomfort and providing comprehensive diagnostic information. The application of pharmacogenomics in skin biopsy analysis facilitates personalized medicine by modifying treatments based on individual genetic profiles. Despite the advancements, challenges remain, in particular, in obtaining high-quality annotated images for training AI models and addressing ethical and responsibility concerns associated with AI diagnostics. Overall, the review highlights the evolution and future potential of skin biopsy practices in improving dermatological care.
NA: Conceptualization, Formal analysis, Methodology, Resources, Writing – original draft, Writing – review & editing. MA: Formal analysis, Investigation, Methodology, Resources, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors extend their sincere appreciation to Ms. Nooran AlHamdani for her exceptional effort in designing and illustrating Figure 1 included in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Perera, E, Gnaneswaran, N, Staines, C, Win, AK, and Sinclair, R. Incidence and prevalence of non-melanoma skin cancer in Australia: a systematic review. Australas J Dermatol. (2015) 56:258–67. doi: 10.1111/ajd.12282
2. Ferlay, J, Colombet, M, Soerjomataram, I, Parkin, DM, Piñeros, M, Znaor, A, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. (2021) 149:778–89. doi: 10.1002/ijc.33588
3. Saladi, RN, and Persaud, AN. The causes of skin cancer: a comprehensive review. Drugs Today. (2005) 41:37–54. doi: 10.1358/dot.2005.41.1.875777
4. Narayanan, DL, Saladi, RN, and Fox, JL. Ultraviolet radiation and skin cancer. Int J Dermatol. (2010) 49:978–86. doi: 10.1111/j.1365-4632.2010.04474.x
5. Villablanca, S, Fischer, C, García-García, SC, Mascaró-Galy, JM, and Ferrando, J. Primary scarring alopecia: clinical-pathological review of 72 cases and review of the literature. Skin Appendage Disord. (2017) 3:132–43. doi: 10.1159/000467395
6. Waśkiel, A, Rakowska, A, Sikora, M, Olszewska, M, and Rudnicka, L. Trichoscopy of alopecia areata: an update. J Dermatol. (2018) 45:692–700. doi: 10.1111/1346-8138.14283
7. Knopp, E. The scalp biopsy for hair loss and its interpretation. Semin Cutan Med Surg. (2015) 34:57–66. doi: 10.12788/j.sder.2015.0144
8. Sperling, LC. The role of the scalp biopsy in the evaluation of alopecia. J Am Acad Dermatol. (2023) 89:S16–9. doi: 10.1016/j.jaad.2023.05.047
9. Rambwawasvika, H. Alopecia types, current and future treatment. J Dermatol Cosmetol. (2021) 5:93–9. doi: 10.15406/jdc.2021.05.00190
10. Nischal, U, Nischal, KC, and Khopkar, U. Techniques of skin biopsy and practical considerations. J Cutan Aesthet Surg. (2008) 1:107–11. doi: 10.4103/0974-2077.44174
11. Elston, DM, Stratman, EJ, and Miller, SJ. Skin biopsy: biopsy issues in specific diseases. J Am Acad Dermatol. (2016) 74:1–16. doi: 10.1016/j.jaad.2015.06.033
12. Reddy, S, Shaheed, A, and Patel, R. Artificial intelligence in Dermoscopy: enhancing diagnosis to distinguish benign and malignant skin lesions. Cureus. (2024) 16:e54656. doi: 10.7759/cureus.54656
13. Chaidemenos, GC, Maltezos, E, Chrysomallis, F, Chrysomallis, F, Kouskoukis, K, Kapetis, E, et al. Value of routine diagnostic criteria of bullous pemphigoid. Int J Dermatol. (1998) 37:206–10. doi: 10.1046/j.1365-4362.1998.00271.x
14. Tintle, SJ, Cruse, AR, Brodell, RT, and Duong, B. Classic findings, mimickers, and distinguishing features in primary blistering skin disease. Arch Pathol Lab Med. (2020) 144:136–47. doi: 10.5858/arpa.2019-0175-RA
15. Valencia-Guerrero, A, Deng, A, Dresser, K, Bouliane, G, and Cornejo, KM. The value of direct immunofluorescence on proteinase-digested formalin-fixed paraffin-embedded skin biopsies. Am J Dermatopathol. (2018) 40:111–7. doi: 10.1097/DAD.0000000000000934
16. Mysorekar, VV, Sumathy, TK, and Shyam Prasad, AL. Role of direct immunofluorescence in dermatological disorders. Indian Dermatol Online J. (2015) 6:172–80. doi: 10.4103/2229-5178.156386
17. Pfaltz, K, Mertz, K, Rose, C, Scheidegger, P, Pfaltz, M, and Kempf, W. C3d immunohistochemistry on formalin-fixed tissue is a valuable tool in the diagnosis of bullous pemphigoid of the skin. J Cutan Pathol. (2010) 37:654–8. doi: 10.1111/j.1600-0560.2009.01450.x
18. Fijałkowska, A, Kądziela, M, and Żebrowska, A. The spectrum of cutaneous manifestations in lupus erythematosus: a comprehensive review. J Clin Med. (2024) 13:2419. doi: 10.3390/jcm13082419
19. Gunter, GR. Weber a frozen section analysis in the management of skin cancers. Ann Plast Surg. (1999) 43:156–60. doi: 10.1097/00000637-199943020-00009
20. Smith-Zagone, MJ, and Schwartz, MR. Frozen section of skin specimens. Arch Pathol Lab Med. (2005) 129:1536–43. doi: 10.5858/2005-129-1536-FSOSS
21. Alguire, PC, and Mathes, BM. Skin biopsy techniques for the internist. J Gen Intern Med. (1998) 13:46–54. doi: 10.1046/j.1525-1497.1998.00009.x
22. Kaul, V, Enslin, S, and Gross, SA. History of artificial intelligence in medicine. Gastrointest Endosc. (2020) 92:807–12. doi: 10.1016/j.gie.2020.06.040
23. Miller, DD, and Brown, EW. Artificial intelligence in medical practice: the question to the answer? Am J Med. (2018) 131:129–33. doi: 10.1016/j.amjmed.2017.10.035
24. Esteva, A, Kuprel, B, Novoa, RA, Ko, J, Swetter, SM, Blau, HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. (2017) 542:115–8. doi: 10.1038/nature21056
25. Litjens, G, Kooi, T, Bejnordi, BE, Setio, AA, Ciompi, F, Ghafoorian, M, et al. A survey on deep learning in medical image analysis. Med Image Anal. (2017) 42:60–88. doi: 10.1016/j.media.2017.07.005
26. Khosravi, P, Kazemi, E, Imielinski, M, Elemento, O, and Hajirasouliha, I. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine. (2018) 27:317–28. doi: 10.1016/j.ebiom.2017.12.026
27. Han, T, Liu, C, Yang, W, and Jiang, D. Learning transferable features in deep convolutional neural networks for diagnosing unseen machine conditions. ISA Trans. (2019) 93:341–53. doi: 10.1016/j.isatra.2019.03.017
28. Hu, Z, Tang, J, Wang, Z, Zhang, K, Zhang, L, and Sun, Q. Deep learning for image-based cancer detection and diagnosis—a survey. Pattern Recogn. (2018) 83:134–49. doi: 10.1016/j.patcog.2018.05.014
29. Fujisawa, Y, Otomo, Y, Ogata, Y, Nakamura, Y, Okiyama, N, Ohara, K, et al. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br J Dermatol. (2019) 180:373–81. doi: 10.1111/bjd.16924
30. Tschandl, P, Codella, N, Akay, BN, Argenziano, G, Braun, RP, Cabo, H, et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol. (2019) 20:938–47. doi: 10.1016/S1470-2045(19)30333-X
31. Dildar, M, Akram, S, Irfan, M, Khan, HU, Ramzan, M, Mahmood, AR, et al. Skin cancer detection: a review using deep learning techniques. Int J Environ Res Public Health. (2021) 18:5479. doi: 10.3390/ijerph18105479
32. Adamson, AS, and Smith, A. Machine learning and health care disparities in dermatology. JAMA Dermatol. (2018) 154:1247–8. doi: 10.1001/jamadermatol.2018.2348
33. Han, SS, Kim, MS, Lim, W, Park, GH, Park, I, and Chang, SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol. (2018) 138:1529–38. doi: 10.1016/j.jid.2018.01.028
34. Liu, Y, Jain, A, Eng, C, Way, DH, Lee, K, Bui, P, et al. A deep learning system for differential diagnosis of skin diseases. Nat Med. (2020) 26:900–8. doi: 10.1038/s41591-020-0842-3
35. Daneshjou, R, Smith, MP, Sun, MD, Rotemberg, V, and Zou, J. Lack of transparency and potential bias in artificial intelligence data sets and algorithms: a scoping review. JAMA Dermatol. (2021) 157:1362–9. doi: 10.1001/jamadermatol.2021.3129
36. Liu, Y, Primiero, CA, Kulkarni, V, Soyer, HP, and Betz-Stablein, B. Artificial intelligence for the classification of pigmented skin lesions in populations with skin of color: a systematic review. Dermatology. (2023) 239:499–513. doi: 10.1159/000530225
37. Jiang, YQ, Xiong, JH, Li, HY, Yang, XH, Yu, WT, Gao, M, et al. Recognizing basal cell carcinoma on smartphone-captured digital histopathology images with a deep neural network. Br J Dermatol. (2020) 182:754–62. doi: 10.1111/bjd.18026
38. Hart, SN, Flotte, W, Norgan, AP, Shah, KK, Buchan, ZR, Mounajjed, T, et al. Classification of melanocytic lesions in selected and whole-slide images via convolutional neural networks. J Pathol Inform. (2019) 10:5. doi: 10.4103/jpi.jpi_32_18
39. Ianni, JD, Soans, RE, Sankarapandian, S, Chamarthi, RV, Ayyagari, D, Olsen, TG, et al. Tailored for real-world: a whole slide image classification system validated on uncurated multi-site data emulating the prospective pathology workload. Sci Rep. (2020) 10:3217. doi: 10.1038/s41598-020-59985-2
40. Kulkarni, PM, Robinson, EJ, Pradhan, SJ, Gartrell-Corrado, RD, Rohr, BR, Trager, MH, et al. Deep learning based on standard H&E images of primary melanoma tumors identifies patients at risk for visceral recurrence and death. Clin Cancer Res. (2020) 26:1126–34. doi: 10.1158/1078-0432.CCR-19-1495
41. Duschner, N, Baguer, DO, Schmidt, M, Griewank, KG, Hadaschik, E, Hetzer, S, et al. Applying an artificial intelligence deep learning approach to routine dermatopathological diagnosis of basal cell carcinoma. J Dtsch Dermatol Ges. (2023) 21:1329–37. doi: 10.1111/ddg.15180
42. Olsen, TG, Jackson, BH, Feeser, TA, Kent, MN, Moad, JC, Krishnamurthy, S, et al. Diagnostic performance of deep learning algorithms applied to three common diagnoses in dermatopathology. J Pathol Inform. (2018) 9:32. doi: 10.4103/jpi.jpi_31_18
43. Hekler, A, Utikal, JS, Enk, AH, Berking, C, Klode, J, Schadendorf, D, et al. Pathologist-level classification of histopathological melanoma images with deep neural networks. Eur J Cancer. (2019) 115:79–83. doi: 10.1016/j.ejca.2019.04.021
44. Corona, R, Mele, A, Amini, M, De Rosa, G, Coppola, G, Piccardi, P, et al. Interobserver variability on the histopathologic diagnosis of cutaneous melanoma and other pigmented skin lesions. J Clin Oncol. (1996) 14:1218–23. doi: 10.1200/JCO.1996.14.4.1218
45. Lodha, S, Saggar, S, Celebi, JT, and Silvers, DN. Discordance in the histopathologic diagnosis of difficult melanocytic neoplasms in the clinical setting. J Cutan Pathol. (2008) 35:349–52. doi: 10.1111/j.1600-0560.2007.00970.x
46. De Logu, F, Ugolini, F, Maio, V, Simi, S, Cossu, A, Massi, D, et al. Recognition of cutaneous melanoma on digitized histopathological slides via artificial intelligence algorithm. Front Oncol. (2020) 10:1559. doi: 10.3389/fonc.2020.01559
47. Wang, L, Ding, L, Liu, Z, Sun, L, Chen, L, Jia, R, et al. Automated identification of malignancy in whole-slide pathological images: identification of eyelid malignant melanoma in gigapixel pathological slides using deep learning. Br J Ophthalmol. (2020) 104:318–23. doi: 10.1136/bjophthalmol-2018-313706
48. Tschandl, P, Rosendahl, C, Akay, BN, Argenziano, G, Blum, A, Braun, RP, et al. Expert-level diagnosis of nonpigmented skin cancer by combined convolutional neural networks. JAMA Dermatol. (2019) 155:58–65. doi: 10.1001/jamadermatol.2018.4378
49. Menzies, SW, Kreusch, J, Byth, K, Pizzichetta, MA, Marghoob, A, Braun, R, et al. Dermoscopic evaluation of amelanotic and hypomelanotic melanoma. Arch Dermatol. (2008) 144:1120–7. doi: 10.1001/archderm.144.9.1120
50. Marka, A, Carter, JB, Toto, E, and Hassanpour, S. Automated detection of nonmelanoma skin cancer using digital images: a systematic review. BMC Med Imaging. (2019) 19:21. doi: 10.1186/s12880-019-0307-7
51. Cho, SI, Sun, S, Mun, JH, Kim, C, Kim, SY, Cho, S, et al. Dermatologist-level classification of malignant lip diseases using a deep convolutional neural network. Br J Dermatol. (2020) 182:1388–94. doi: 10.1111/bjd.18459
52. Cruz-Roa, AA, Arevalo, O, Madabhushi, A, and González, FA. A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection. Med Image Comput Comput Assist Interv. (2013) 16:403–10. doi: 10.1007/978-3-642-40763-5_50
53. Velasquez-Agudelo, V, and Cardona-Arias, JA. Meta-analysis of the utility of culture, biopsy, and direct KOH examination for the diagnosis of onychomycosis. BMC Infect Dis. (2017) 17:166. doi: 10.1186/s12879-017-2258-3
54. Helfen, M, Wagenpfeil, S, Vogt, T, and Muller, CS. Neglect of the histological diagnostics of onychomycosis—the best would be so easy. J Dtsch Dermatol Ges. (2021) 19:885–8. doi: 10.1111/ddg.14382_g
55. Reisberger, EM, Abels, C, Landthaler, M, and Szeimies, RM. Histopathological diagnosis of onychomycosis by periodic acid-Schiff-stained nail clippings. Br J Dermatol. (2003) 148:749–54. doi: 10.1046/j.1365-2133.2003.05029.x
56. Jansen, P, Creosteanu, A, Matyas, V, Dilling, A, Pina, A, Saggini, A, et al. Deep learning assisted diagnosis of onychomycosis on whole-slide images. J Fungi. (2022) 8:912. doi: 10.3390/jof8090912
57. Lilly, KK, Koshnick, RL, Grill, JP, Khalil, ZM, Nelson, DB, and Warshaw, EM. Cost-effectiveness of diagnostic tests for toenail onychomycosis: a repeated-measure, single-blinded, cross-sectional evaluation of 7 diagnostic tests. J Am Acad Dermatol. (2006) 55:620–6. doi: 10.1016/j.jaad.2006.03.033
58. Lawry, MA, Haneke, E, Strobeck, K, Martin, S, Zimmer, B, and Romano, PS. Methods for diagnosing onychomycosis: a comparative study and review of the literature. Arch Dermatol. (2000) 136:1112–6. doi: 10.1001/archderm.136.9.1112
59. Decroos, F, Springenberg, S, Lang, T, Päpper, M, Zapf, A, Metze, D, et al. A deep learning approach for histopathological diagnosis of onychomycosis: not inferior to analogue diagnosis by histopathologists. Acta Derm Venereol. (2021) 101:10.2340/00015555-3893. doi: 10.2340/00015555-3893
60. Yang, F, Poostchi, M, Yu, H, Zhou, Z, Silamut, K, Yu, J, et al. Deep learning for smartphone-based malaria parasite detection in thick blood smears. IEEE J Biomed Health Inform. (2020) 24:1427–38. doi: 10.1109/JBHI.2019.2939121
61. Escalé-Besa, A, Vidal-Alaball, J, Miró Catalina, Q, Gracia, VHG, Marin-Gomez, FX, and Fuster-Casanovas, A. The use of artificial intelligence for skin disease diagnosis in primary care settings: a systematic review. Healthcare. (2024) 12:1192. doi: 10.3390/healthcare12121192
62. Fisher, HM, Hoehndorf, R, Bazelato, BS, Dadras, SS, King, LE Jr, Gkoutos, GV, et al. DermO; an ontology for the description of dermatologic disease. J Biomed Semantics. (2016) 7:38. doi: 10.1186/s13326-016-0085-x
63. Lott, JP, Boudreau, DM, Barnhill, RL, Weinstock, MA, Knopp, E, Piepkorn, MW, et al. Population-based analysis of histologically confirmed melanocytic proliferations using natural language processing. JAMA Dermatol. (2018) 154:24–9. doi: 10.1001/jamadermatol.2017.4060
64. Campanella, G, Nehal, KS, Lee, EH, Rossi, A, Possum, B, Manuel, G, et al. A deep learning algorithm with high sensitivity for the detection of basal cell carcinoma in Mohs micrographic surgery frozen sections. J Am Acad Dermatol. (2021) 85:1285–6. doi: 10.1016/j.jaad.2020.09.012
65. Geijs, DJ, Hillen, LM, Dooper, S, Winnepenninckx, V, Varra, V, Carr, DR, et al. Weakly-supervised classification of Mohs surgical sections using artificial intelligence. Mod Pathol. (2024) 38:100653. doi: 10.1016/j.modpat.2024.100653
66. Kiran, N, Sapna, F, Kiran, F, Kumar, D, Raja, F, Shiwlani, S, et al. Digital pathology: transforming diagnosis in the digital age. Cureus. (2023) 15:e44620. doi: 10.7759/cureus.44620
67. Li, Z, Koban, KC, Schenck, TL, Giunta, RE, Li, Q, and Sun, Y. Artificial intelligence in dermatology image analysis: current developments and future trends. J Clin Med. (2022) 11:6826. doi: 10.3390/jcm11226826
68. Kislevitz, M, Wamsley, C, Bartels, M, Lu, KB, Li, X, Pinch, S, et al. Clinical translation of scarless 0.33-mm core microbiopsy for molecular evaluation of human skin. Aesthet Surg J. (2021) 41:NP1710–20. doi: 10.1093/asj/sjaa332
69. Pozner, JN, Kilmer, SL, Geronemus, RG, Jack, M, Burns, JA, and Kaminer, MS. Cytrellis: a novel microcoring technology for scarless skin removal: summary of three prospective clinical trials. Plast Reconstr Surg Glob Open. (2021) 9:e3905. doi: 10.1097/GOX.0000000000003905
70. Raj Kirit, EP, Sivuni, A, Ponugupati, S, and Gold, MH. Efficacy and safety of triple wavelength laser hair reduction in skin types IV to V. J Cosmet Dermatol. (2021) 20:1117–23. doi: 10.1111/jocd.13995
71. Zaporozhchenko, IA, Ponomaryova, AA, Rykova, EY, and Laktionov, PP. The potential of circulating cell-free RNA as a cancer biomarker: challenges and opportunities. Expert Rev Mol Diagn. (2018) 18:133–45. doi: 10.1080/14737159.2018.1425143
72. Garbe, C, Amaral, T, Peris, K, Hauschild, A, Arenberger, P, Basset-Seguin, N, et al. European consensus-based interdisciplinary guideline for melanoma. Part 1: diagnostics: update 2022. Eur J Cancer. (2022) 170:236–55. doi: 10.1016/j.ejca.2022.03.008
73. Wang, H, Dang, T, Feng, J, Wu, W, He, L, and Yang, J. Identification of differentially methylated genes for severe acne by genome-wide DNA methylation and gene expression analysis. Epigenetics. (2023) 18:2199373. doi: 10.1080/15592294.2023.2199373
74. Yang, TT, Yu, S, Ke, CK, and Cheng, ST. The genomic landscape of melanoma and its therapeutic implications. Genes. (2023) 14:1021. doi: 10.3390/genes14051021
75. Wu, J, Fang, Z, Liu, T, Hu, W, Wu, Y, and Li, S. Maximizing the utility of transcriptomics data in inflammatory skin diseases. Front Immunol. (2021) 12:761890. doi: 10.3389/fimmu.2021.761890
76. Theocharidis, G, Tekkela, S, Veves, A, McGrath, JA, and Onoufriadis, A. Single-cell transcriptomics in human skin research: available technologies, technical considerations and disease applications. Exp Dermatol. (2022) 31:655–73. doi: 10.1111/exd.14547
77. Wilbaux, M, Yang, S, Jullion, A, Demanse, D, Porta, DG, Myers, A, et al. Integration of pharmacokinetics, pharmacodynamics, safety, and efficacy into model-informed dose selection in oncology first-in-human study: a case of roblitinib (FGF401). Clin Pharmacol Ther. (2022) 112:1329–39. doi: 10.1002/cpt.2752
78. Guin, D, Rani, J, Singh, P, Grover, S, Bora, S, Talwar, P, et al. Global text mining and development of Pharmacogenomic knowledge resource for precision medicine. Front Pharmacol. (2019) 10:839. doi: 10.3389/fphar.2019.00839
79. Su, M, Zhang, Z, Zhou, L, Han, C, Huang, C, and Nice, EC. Proteomics, personalized medicine and cancer. Cancers. (2021) 13:2512. doi: 10.3390/cancers13112512
80. Strzelecki, M, Kociołek, M, Strąkowska, M, Kozłowski, M, Grzybowski, A, and Szczypiński, PM. Artificial intelligence in the detection of skin cancer: state of the art. Clin Dermatol. (2024) 42:280–95. doi: 10.1016/j.clindermatol.2023.12.022
81. Gao, J, Jiang, Q, Zhou, B, and Chen, D. Convolutional neural networks for computer aided detection or diagnosis in medical image analysis: an overview. Math Biosci Eng. (2019) 16:6536–61. doi: 10.3934/mbe.2019326
82. Li, CX, Shen, CB, Xue, K, Shen, X, Jing, Y, Wang, ZY, et al. Artificial intelligence in dermatology: past, present, and future. Chin Med J. (2019) 132:2017–20. doi: 10.1097/CM9.0000000000000372
83. Demers, AA, Nugent, Z, Mihalcioiu, C, Wiseman, MC, and Kliewer, EV. Trends of nonmelanoma skin cancer from 1960 through 2000 in a Canadian population. J Am Acad Dermatol. (2005) 53:320–8. doi: 10.1016/j.jaad.2005.03.043
84. Marsden, H, Kemos, P, Venzi, M, Noy, M, Maheswaran, S, Francis, N, et al. Accuracy of an artificial intelligence as a medical device as part of a UK-based skin cancer teledermatology service. Front Med. (2024) 11:1302363. doi: 10.3389/fmed.2024.1302363
85. Liu, X, Sangers, TE, Nijsten, T, Kayser, M, Pardo, LM, Wolvius, EB, et al. Predicting skin cancer risk from facial images with an explainable artificial intelligence (XAI) based approach: a proof-of-concept study. EClinicalMedicine. (2024) 71:102550. doi: 10.1016/j.eclinm.2024.102550
86. Tan, E, Lim, S, Lamont, D, Epstein, R, Lim, D, and Lin, F. Development and validation of a deep learning model for improving detection of nonmelanoma skin cancers treated with Mohs micrographic surgery. JAAD Int. (2024) 14:39–47. doi: 10.1016/j.jdin.2023.10.007
87. Li, M, Abe, M, Nakano, S, and Tsuneki, M. Deep learning approach to classify cutaneous melanoma in a whole slide image. Cancers (Basel). (2023) 15:1907. doi: 10.3390/cancers15061907
88. Cozzolino, C, Buja, A, Rugge, M, Miatton, A, Zorzi, M, Vecchiato, A, et al. Machine learning to predict overall short-term mortality in cutaneous melanoma. Discov Oncol. (2023) 14:13. doi: 10.1007/s12672-023-00622-5
89. Aung, TN, Shafi, S, Wilmott, JS, Nourmohammadi, S, Vathiotis, I, Gavrielatou, N, et al. Objective assessment of tumor infiltrating lymphocytes as a prognostic marker in melanoma using machine learning algorithms. EBioMedicine. (2022) 82:104143. doi: 10.1016/j.ebiom.2022.104143
90. Brodsky, V, Levine, L, Solans, EP, Dola, S, Chervony, L, and Polak, S. Performance of automated classification of diagnostic entities in Dermatopathology validated on multisite data representing the real-world variability of pathology workload. Arch Pathol Lab Med. (2023) 147:1093–8. doi: 10.5858/arpa.2021-0550-OA
91. Dika, E, Curti, N, Giampieri, E, Veronesi, G, Misciali, C, Ricci, C, et al. Advantages of manual and automatic computer-aided compared to traditional histopathological diagnosis of melanoma: a pilot study. Pathol Res Pract. (2022) 237:154014. doi: 10.1016/j.prp.2022.154014
92. Couetil, J, Liu, Z, Huang, K, Zhang, J, and Alomari, AK. Predicting melanoma survival and metastasis with interpretable histopathological features and machine learning models. Front Med. (2023) 9:1029227. doi: 10.3389/fmed.2022.1029227
93. Figueroa-Silva, O, Pastur Romay, LA, Viruez Roca, RD, Rojas, MDSY, and Suárez-Peñaranda, JM. Machine learning techniques in predicting BRAF mutation status in cutaneous melanoma from clinical and histopathologic features. Appl Immunohistochem Mol Morphol. (2022) 30:674–80. doi: 10.1097/PAI.0000000000001075
94. Li, A, Li, X, Li, W, Yu, X, Qi, M, and Li, D. Application of deep learning on the prognosis of cutaneous melanoma based on full scan pathology images. Biomed Res Int. (2022) 2022:4864485. doi: 10.1155/2022/4864485
95. Comes, M, Fucci, L, Mele, F, Bove, S, Cristofaro, C, Risi, D, et al. A deep learning model based on whole slide images to predict disease-free survival in cutaneous melanoma patients. Sci Rep. (2022) 12:20366. doi: 10.1038/s41598-022-24315-1
96. Mund, A, Coscia, F, Kriston, A, Hollandi, R, Kovács, F, Brunner, AD, et al. Deep visual proteomics defines single-cell identity and heterogeneity. Nat Biotechnol. (2022) 40:1231–40. doi: 10.1038/s41587-022-01302-5
97. Cazzato, G, Massaro, A, Colagrande, A, Lettini, T, Cicco, S, Parente, P, et al. Dermatopathology of malignant melanoma in the era of artificial intelligence: a single institutional experience. Diagnostics. (2022) 12:1972. doi: 10.3390/diagnostics12081972
98. Wang, L, Jiang, Z, Shao, A, Liu, Z, Gu, R, Ge, R, et al. Self-supervised learning mechanism for identification of eyelid malignant melanoma in pathologic slides with limited annotation. Front Med. (2022) 9:976467. doi: 10.3389/fmed.2022.976467
99. Kriegsmann, K, Lobers, F, Zgorzelski, C, Kriegsmann, J, Janßen, C, Meliß, RR, et al. Deep learning for the detection of anatomical tissue structures and neoplasms of the skin on scanned histopathological tissue sections. Front Oncol. (2022) 12:1022967. doi: 10.3389/fonc.2022.1022967
100. Nielsen, PS, Georgsen, JB, Vinding, MS, Østergaard, LR, and Steiniche, T. Computer-assisted annotation of Digital H&E/SOX10 dual stains generates high-performing convolutional neural network for calculating tumor burden in H&E-stained cutaneous melanoma. Int J Environ Res Public Health. (2022) 19:14327. doi: 10.3390/ijerph192114327
101. Sturm, B, Creytens, D, Smits, J, Ooms, AHAG, Eijken, E, Kurpershoek, E, et al. Computer-aided assessment of melanocytic lesions by means of a mitosis algorithm. Diagnostics. (2022) 12:436. doi: 10.3390/diagnostics12020436
102. Wan, G, Nguyen, N, Liu, F, DeSimone, M, Leung, B, Rajeh, A, et al. Prediction of early-stage melanoma recurrence using clinical and histopathologic features. NPJ Precis Oncol. (2022) 6:79. doi: 10.1038/s41698-022-00321-4
103. Snyder, AN, Zhang, D, Dreesen, SL, Baltimore, CA, Lopez-Garcia, DR, Akers, JY, et al. Histologic screening of malignant melanoma, Spitz, dermal and junctional melanocytic nevi using a deep learning model. Am J Dermatopathol. (2022) 44:650–7. doi: 10.1097/DAD.0000000000002232
104. Doeleman, T, Westerbeek, D, Jansen, P, Hondelink, LM, He, J, Kers, J, et al. Artificial intelligence-assisted probability scoring for differentiation of early mycosis fungoides and benign inflammatory dermatoses on H&E stained pathology slides of skin biopsies. Eur J Cancer. (2022) 173:S11. doi: 10.1016/S0959-8049(22)00572-X
Keywords: skin biopsy, light microscopy, immunofluorescence, artificial intelligence, convolutional neural networks
Citation: Alwahaibi N and Alwahaibi M (2025) Mini review on skin biopsy: traditional and modern techniques. Front. Med. 12:1476685. doi: 10.3389/fmed.2025.1476685
Received: 06 August 2024; Accepted: 18 February 2025;
Published: 05 March 2025.
Edited by:
George Kroumpouzos, Brown University, United StatesReviewed by:
Kevinn Eddy, Calder Biosciences, Inc., United StatesCopyright © 2025 Alwahaibi and Alwahaibi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nasar Alwahaibi, bmFzYXJAc3F1LmVkdS5vbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.