- 1Applied Mathematics and AI, Moscow Power Engineering Institute, Moscow, Russia
- 2Sber AI Lab, Moscow, Russia
- 3Moscow Institute of Physics and Technology, Dolgoprudny, Russia
- 4LLC SberMedAI, Moscow, Russia
Purpose: A reduced acquisition time positively impacts the patient's comfort and the PET scanner's throughput. AI methods may allow for reducing PET acquisition time without sacrificing image quality. The study aims to compare various neural networks to find the best models for PET denoising.
Methods: Our experiments consider 212 studies (56,908 images) for 7MBq/kg injected activity and evaluate the models using 2D (RMSE, SSIM) and 3D (SUVpeak and SUVmax error for the regions of interest) metrics. We tested 2D and 2.5D ResNet, Unet, SwinIR, 3D MedNeXt, and 3D UX-Net. We have also compared supervised methods with the unsupervised CycleGAN approach.
Results and conclusion: The best model for PET denoising is 3D MedNeXt. It improved SSIM on 38.2% and RMSE on 28.1% in 30-s PET denoising and on 16.9% and 11.4% in 60-s PET denoising when compared to the original 90-s PET reducing at the same time SUVmax discrepancy dispersion.
1 Introduction
Positron emission tomography (PET) is a molecular imaging technique that produces a three-dimensional radiotracer distribution map representing properties of biologic tissues, such as metabolic activity. Many patients undergo more than one PET/CT scan per year. According to the OECD/EU report (42), an average number of PET scans per 1,000 people is 3.3 in EU25 countries, with a maximum value of 10.2 in Denmark. The higher the injected activity, the less noise in the reconstructed images and the more radioactive exposure for a patient and for the healthcare operators.
The task of accelerating a PET/CT scan is to create an algorithm that takes a low-time PET image as input and converts it into an image with diagnostic quality that corresponding to a PET image with a standard exposure time. This task is equivalent to reducing the administered dose of a radio-pharmaceutical. Both of these tasks are noise reduction tasks (54).
Deep learning methods may reduce injected activity or acquisition time by utilizing low-dose (LD)/low-time (LT) and full-dose (FD)/full-time (FT) images (Figure 1) to train models that can predict standard-dose images from LD/LT inputs. A reduced acquisition time positively impacts the patient's comfort or the scanner's throughput, which enables more patients to be scanned daily, lowering costs.

Figure 1. Comparison of denoising methods applied on low-time (30 s) PET reconstruction and SUVmax estimation within the region of interest. Tumor segmentation is done automatically by nnUnet using CT and PET. (A) CT (B) Low-time PET (30 s). (C) Full-time PET (90 s). (D) Reconstructed PET (by SwinIR 5 layers). (E) Reconstructed PET (by MedNeXt small). (F) Reconstructed PET (by Gaussian filter).
The drawback of the recent studies is the need for more comparison between a broad group of methods on a level playing field, especially between supervised and unsupervised methods. Moreover, as methods are tested on different data sets and for various PET time frames, comparing them is complicated. Furthermore, studies differ in the metrics used for denoising quality assessment, for example, some (20, 23, 59, 62) evaluate only image similarity metrics such as RMSE but do not take into account SUV characteristics.
Our study aims to overcome the drawbacks by finding the best backbone and model and comparing the performance of supervised and unsupervised methods for PET denoising. We tested supervised 2D and 2.5D methods [ResNet, Unet, SwinIR (31)] and 3D MedNeXt (49). We have also applied unsupervised pix2pix and CycleGAN with identity and image prior losses (36, 62). We reconstructed FT 90-s/bed position PET from LT PET 30 and 60 s/bed position. Our PET denoising is indicated for use in whole-body PET-CT in patients with primary staging and for assessing the dynamics of those with adenocarcinoma [with a proliferation index of more than 20% (ki67)] and melanomas.
1.1 Related works
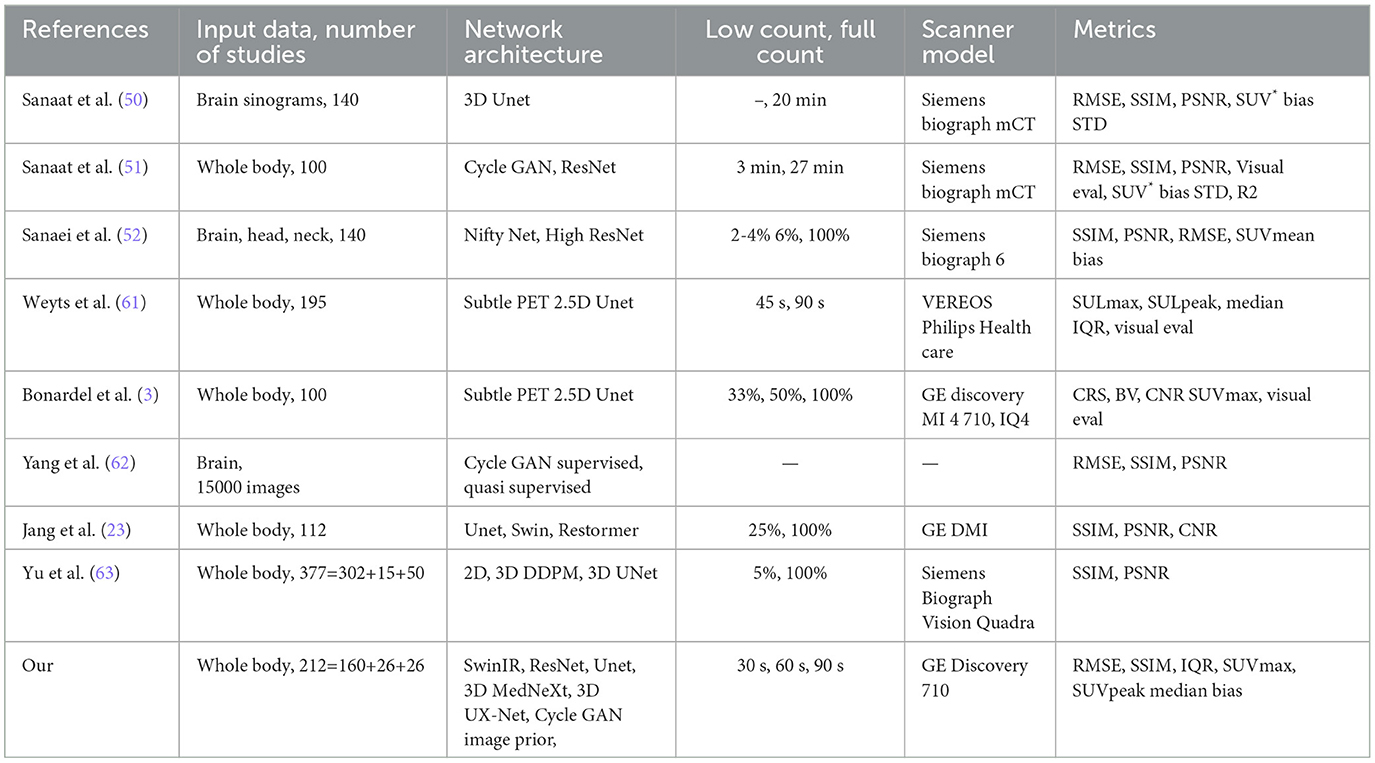
The latest (as of 2024) review on the low-count PET reconstruction are the studies (8, 19). Early studies of PET denoising treated specific parts of the human body, such as the brain and lungs, and used small-size data sets to produce low-quality reconstructions. For example, Gong et al. (16) utilized pretrained VGG19 and perception loss for supervised denoising lungs and brains. One could find the comprehensive overview of methods before 2020 in (50). Table 1 represents the summary of the later studies on PET denoising. In the table, the sign * indicates the SUV metrics where the authors did not specify what kind of SUV—mean, peak, or max they estimated.
An obvious approach for self-supervised PET denoising is to train the model on artificially degraded images (43). CycleGAN is the most popular unsupervised model for PET denoising. The article (28) was the first applied CycleGAN model for whole-body PET denoising. The study (9) also used unsupervised learning. CycleGAN performed better over Unet and Unet GAN in peak signal-to-noise ratio (PSNR) for all human body parts. Sanaat et al. (51) also utilized CycleGAN architecture and demonstrated its performance over ResNet, both trained on 60 studies data set. The ResNet showed, in turn, better results than Unet, which coincides with the results of our experiments. The studies (28, 51) do not reveal the CycleGAN backbone used in their studies; therefore, it remains to be seen if CycleGAN in (28, 51) achieved high performance due to the unsupervised scheme and the adversarial losses or because of difference in the backbone. CycleGAN model applies in different medical image denoising problems such as optical coherence tomography images (39) and low-dose X-ray CT (26, 60). 3D CycleGAN framework with self-attention generates the FC PET image from LC PET with CT aid in the study (29). The study (66) used CycleGAN with Wasserstein loss for stability. Another type of generative models used for PET denoising is diffusion models (17, 45, 46, 57) .
The PET denoising problem is very similar to the PET reconstruction from CT. The study (5) demonstrated that non-contrast CT alone could differentiate regions with different FDG uptake and simulate PET images. To predict three clinical outcomes, using the simulated PET, the article (5) constructed random forest models on the radiomic features. The objective of this experiment was to compare predictive accuracy between the Cycle GAN-simulated and FT PET. ROC AUC for simulated PET achieved to be comparable with ground truth PET—0.59 vs. 0.60, 0.79 vs. 0.82, and 0.62 vs. 0.63. The study (30) denoised CT images by a GAN with the reconstruction loss.
The most popular supervised models [Table 1 in (33)] for PET denoising are ResNet [e.g., (51)] and Unet-style networks [e.g., (53, 55)]. The article (52) used HighResNet, demonstrating that due to PET acquisition's stochastic nature, any LD versions of the PET data would bear complementary/additional information regarding the underlying signal in the standard PET image. This complementary knowledge could improve a deep learning-based denoising framework and [as (52) showed] enhance the quality of FD prediction—PSNR increased from 41.4 to 44.9 due to additional LD images. The study (20) used Swin transformer for FD brain image reconstruction from LC sinograms. The article (23) proposed spatial and channel-wise encoder–decoder transformer—Spatch Transformer that demonstrated better denoising quality over Swin transformer, Restormer, and Unet for 25% low-count PET. An important metric for noise reduction quality is tumor edge preservation. The study (41) showed that CNNs have the same edge preserving quality as bilateral filtering, but it does not provide any quantitative measure of edge preservation.
SubtlePETTM (12) is a commercial product; its official site claims that “SubtlePET is an AI-powered software solution that denoises images conducted in 25% of the original scan duration (e.g., 1 min instead of 4)”. SubtlePET uses multi-slice 2.5D encoder–decoder U-Net (61) optimizing L1 norm and SSIM. The networks were trained with paired low- and high-count PET series from a wide range of patients and from various PET/CT and PET/MR devices (10 General Electric, 5 Siemens, and 2 Philips models). The training data included millions of paired image patches from hundreds of patient scans with multi-slice PET data and data augmentation.
The studies (25, 61, 62) investigated FT 90-s PET reconstruction from LT 30-, 45-, and 60-s images using SubtlePET. The work (61) conducted a study on the efficiency of SubtlePET by comparing denoised LT 45-s PET with FT 90-s PET. The visual analysis revealed a high similarity between FT and reconstructed LT PET. SubtlePET detected 856 lesions for 162 (of 195) patients. Of these, 836 lesions were visualized in both original 90-s PET and denoised 45-s PET, resulting in a lesion concordance rate of 97.7%. The study (3) examined the limits of the SubtlePET denoising algorithm applied to statistically reduced PET raw data from three different last-generation PET scanners compared to the regular acquisition in phantom (spheres) and patient. Enhanced images (PET 33% + SubtlePET) had slightly increased noise compared to PET 100% and could potentially lose information regarding lesion detectability. The PET 100% and PET 50% + SubtlePET were qualitatively comparable regarding the patient data sets. In this case, the SubtlePET algorithm was able to correctly recover the SUVmax values of the lesions and maintain a noise level equivalent to FT images.
The main issue with the studies mentioned above is that it is difficult to compare methods as they use different metrics and data sets. In this study, we have compared various methods, including approaches from other studies and new methods.
The main contribution of the article are
• Our comparative experiments of 2D, 2.5D, and 3D models with different backbones and losses trained and tested in the same conditions on a specially collected data set from clinical practice data showed better performance [Root Mean Square Error (RSME) and Structural Similarity Index] of 3D methods.
• We have trained neural networks of the SwinIR (2D and 2.5), 3D UX-Net (3D), and MedNeXt (3D) architectures. Our tests have shown that MedNeXt is the best at restoring an image from 30 s to 90 s and it is approximately equal to 3D UX-Net, and they solve the problem better than all other tested architectures.
2 Materials and methods
2.1 Whole-body PET data set
We used 212 PET scans from 212 patients obtained during a retrospective study. The data were obtained from scanners calibrated using the NEMA phantom according to the EARL method in June and July 2021. The training subset contains 160 scans with 42,656 images, validation, and test data both consist of 26 scans and 7,126 images, respectively. The patient age is between 21 and 84 years old, and two-thirds of patients are between 49 and 71 years old. The median age is 61; 71% of patients are women. An average height is 1.66 ± STD = 0.09 meters and ranges from 1.47 to 1.94 meters. The weight range is from 34 kg to 150 kg with an average value of 79 ± STD = 18 kg. The body mass index (BMI) is 28.5 ± STD = 6.3.
The number of tumors in the train subset is 521. When excluding 4 patients with more than 50 tumors, the total number of tumors is 139. Of the 160 patients in train subset, 30 do not have any tumors detected. The number of tumors detected in the test set is 74 (for 26 patients, four do not have any tumors), and in the validation set, it is 97 (for another 26, four patients also do not have a tumor). We analyzed the PET scan as a 3D object and, therefore, used all slices to detect the tumor.
There are two ways (5) to simulate low-dose PET—short time frame and decimation. The most common way of decimation is the simulation of a dose reduction by randomized subsampling of PET list-mode data. Another decimation method is randomly sampling the data by a specific factor in each bin of the PET sinogram (50). Short time frames with the corrections taking a shorter amount of time into account will produce images with an SUV uptake similar to the original one. We use short time frame approach in our study collecting PET data with 30, 60 and 90 s/bed positions. All images were collected during the same acquisition session that differs, for example, from studies (50, 51) where the LT images obtained through a separate fast PET acquisition corresponding to the FT scans.
In the data set we used in this study, patients were injected intravenously with 7 MBq/kG [18F]FDG after a 6-h fasting period and blood glucose level testing. The PET data are collected from GE Discovery 710 and were reconstructed using the VPFXS reconstruction method (24). PET frame resolution is 256 × 256. The slice thickness is 3.27 mm, and pixel spacing is 2.73 mm.
The studies are in anonymized DICOM (*.dcm) format, from which one can extract the patient weight, half-life, total dose values and delay Δt between the injection time, and the scan start time for the further SUV calculation.
2.2 Methodology
2.2.1 Problem statement
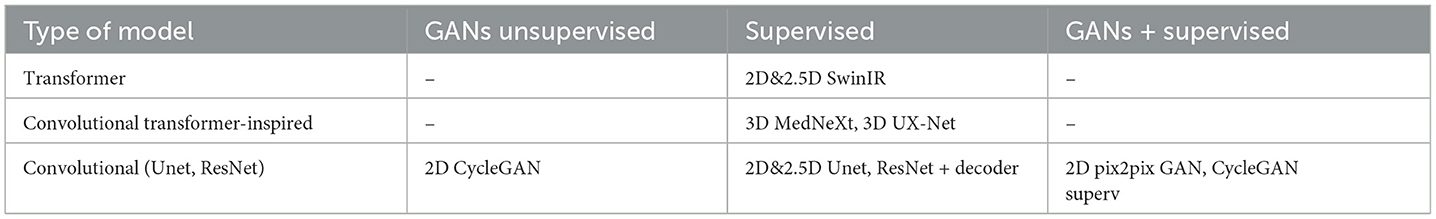
The study aims to assess the quality of the PET denoising for supervised and unsupervised models and find the best model. Unet, ResNet, CycleGAN, pix2pix GAN, SwinIR transformer, MedNeXt, and 3D UX-Net are models to be tested in this research. More details of the network architecture are in the next section.
In academic research, gaussian convolution has been widely recognized as a baseline model for denoising due to its simplicity and proven effectiveness in various denoising tasks. The filter's parameters are crucial for achieving optimal denoising results, and therefore, we optimized them on a validation data set. All models shared the same learning schedule and parameters (with minor differences described in the next section) and, therefore have the same level playing field and could be fairly compared. Table 2 is a systematization of the methods and models used in the study.
2.2.2 Denoising quality assessment
Two metric types describe the quality of denoising: a similarity of 2D PET images and concordance of the tumor's SUV characteristics. The metrics for similarity are SSIM (48) and RMSE:
where FTi is the i-th measurement, is its corresponding prediction, and n is a number of pixels. The SSIM parameters in our study are the same as in scikit-image library. In this report, we used ISSIM=1-SSIM instead of SSIM as ISSIM is more convenient for similar images, and SSIM is higher than 0.9 for most original and denoised PET. We defined relative metrics in the same way as in (50):
the Equation 2 demonstrates the improvement of the denoising method for noised LT image by showcasing the decrease in the discrepancy between the denoised and original image. This equation provides a clear and quantifiable measure of the improvement achieved through the denoising process. The relative ISSIM is defined similarly as Equation 2. The relative metric changes in range from −∞ to 100%. The negative value means that the method has deteriorated the quality of the image, 0%—there are no changes, 100%—the image has been fully denoised and coincides with the original one.
2.2.3 SUV error estimation
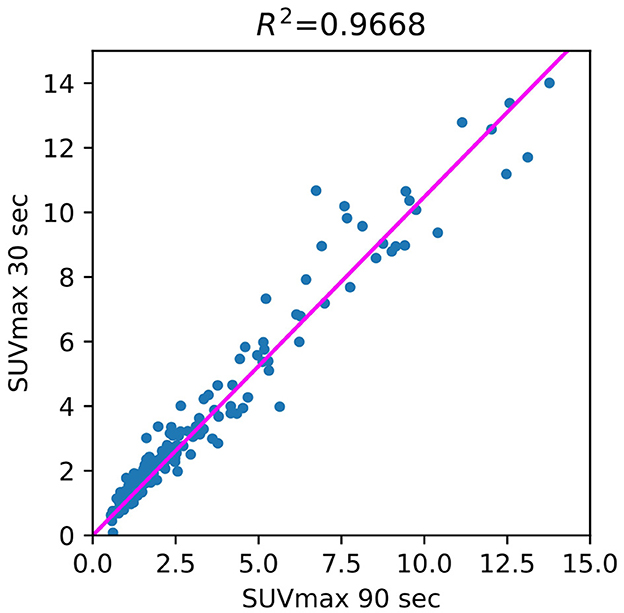
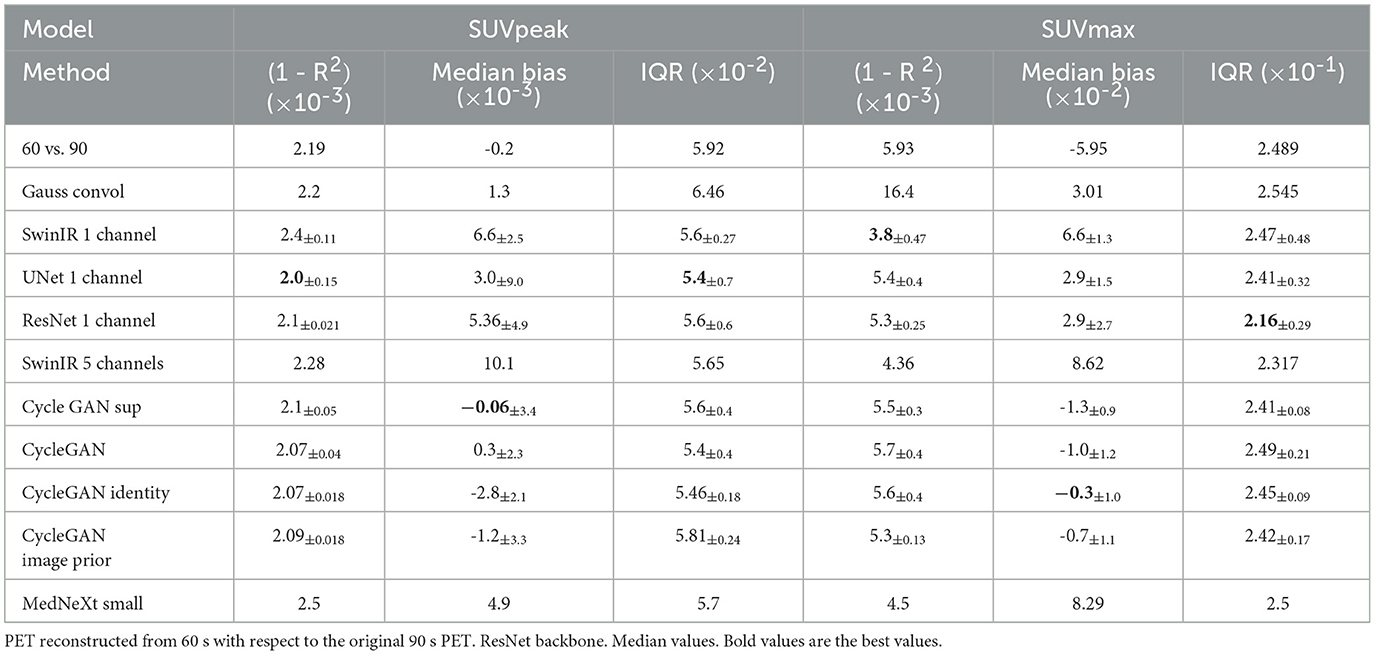
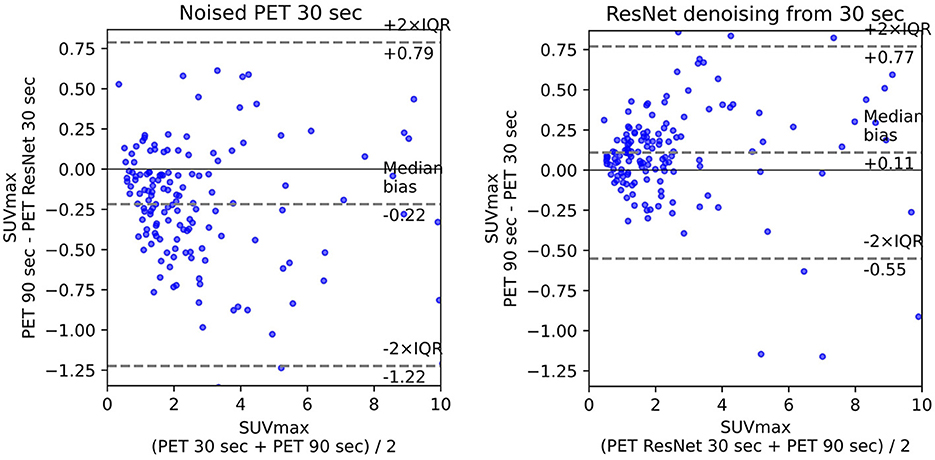
The use of standard uptake value (SUV) is now commonplace (14) in clinical FDG-PET/CT oncology imaging and has a specific role in assessing patient response to cancer therapy. SUVmean, SUVpeak (58), and SUVmax are the values commonly used in PET studies. There are many ways to estimate the correlation between pairs of SUV values for the FT original PET and denoised PET reconstructed from LT. The most common are bias and STD (Figure 6) in terms of Bland–Altman plots (25, 29, 61) and R2 (Figure 2).
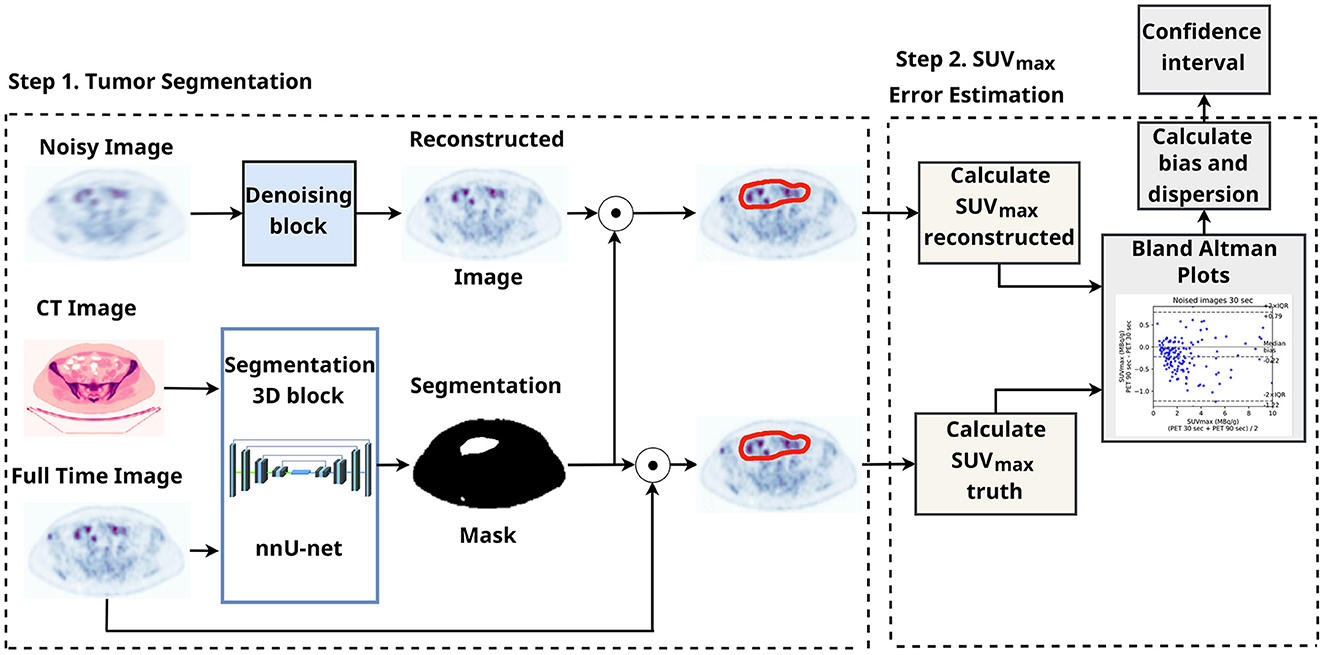
CT is available along with the ground truth PET and could enhance the quality of the tumor segmentation. Instead of employing a radiologist for the malignant tumor detection, we segmented tumors automatically in 3D with the help of nnUnet (21). The pretrained weights are the same as in the AutoPET competition baseline (15). The nnUnet neural network manipulated two channels (PET & CT) input with 400 × 400 resolution. Figure 1 demonstrates tumor segmentation and SUVmax estimation for the region of interest. The CT and PET images are to be resized as they have 512 × 512 and 256 × 256 resolution.
Figure 3 illustrates the SUV confidence interval estimation scheme. After the nnUnet segmentation, cc3d library1 extracts 3D connected components and separates different tumors. We excluded from the study tumors with a maximum length of less than 7 mm and an average SUV of less than 0.5. Bland–Altman plot is a standard instrument of data similarity evaluation in biomedical research. The plot operates with the region of interest SUV for original and denoised PET. The Bland–Altman plot's bias and dispersion are indicators of denoising quality and are used in the latest step of the scheme in Figure 3 for the confidence interval assessment. The total number of tumors in validation and test data is 171.
2.3 Neural network implementation and training details
2.3.1 2D convolutional networks
Unet and ResNet models, pix2pix, and CycleGAN are based on the pytorch implementation of CycleGAN.2 The model parameter numbers are in Table 3. The number of channels in the bottleneck for both models is 64. The Unet model has 54.4 million parameters, the ResNet served as encoder has 11.4 mil. parameters itself, and the decoder has 0.37 mil parameters—11.8 mil parameters in total. The decoder exploits transposed convolutions and does not have skip connections.
SwinIR (31) integrates the advantages of both CNN and transformer. On the one hand, CNN has the advantage of processing images of a large size due to the local attention mechanism. On the other hand, it has the benefit of the transformer to model long-range dependency with the shifted window (34). SwinIR exceeded state-of-the-art CNN performance in denoising and JPEG compression artifact reduction. We implemented code from the official SwinIR repository.3
SwinIR consists of three modules, namely, shallow feature extraction, deep feature extraction, and high-quality image reconstruction modules. The shallow feature extraction module uses a convolution layer to extract shallow features directly transmitted to the reconstruction module to preserve low-frequency information.
The deep feature extraction module mainly comprises residual Swin Transformer blocks, each utilizing several Swin Transformer layers for local attention and cross-window interaction. In addition, Liang et al. (31) added a convolution layer at the end of the block for feature enhancement and used a residual connection. In the end, shallow and deep features are fused in the reconstruction module for high-quality image reconstruction. The patch size of SwinIR in our training is 32; the window size is 8.
2.3.2 GANs
Pix2Pix and CycleGAN models use PatchGAN (22) with 2.8 million parameters. ResNet, Unet, and CycleGAN models predict the difference between noised and denoised images. The SwinIR model has this difference built into its architecture, like in (38). The Pix2Pix GAN discriminator also used image difference to distinguish between noised and denoised PET. This simple approach applied for the PET denoising improved the results significantly but was used before only in the transformer-based model for CT denoising.
L1 loss is used in all models (except for unsupervised CycleGAN and SwinIR) to optimize the similarity between denoised LT and FT images. SwinIR uses Charbonnier loss (6). Pix2pix GAN also uses Euclidean adversarial loss. In the original CycleGAN paper (67), identity mapping loss
helps preserve the color of the input painting. The loss prevents the network from denoise the FT image and vice versa. Park et al. (47) claims that more weights for the cycle consistency loss and identity loss made the CycleGAN model translate the blood-pool image close to the actual bone image. We will investigate the influence of identity loss on PET denoising.
CycleGAN is an unsupervised method. Therefore, its usage is beneficial if there is a lot of unpaired data in both domains. However, getting paired data with different PET acquisition times is an ordinary task that could be done automatically without any additional action on a patient. The study (62) showed that the use of the additional supervised reconstruction loss (4) in CycleGAN makes the training stable and considerably improves PSNR and SSIM
We used supervised CycleGAN as an upper boundary for ISSIM and RMSE metrics that unsupervised CycleGAN could achieve with the image prior loss. We also studied its effect on SUVmax error. We trained CycleGAN with identity. Zhu et al. (67) and image prior (60) losses in addition to adversarial and cycle consistency losses. Image prior loss
is based on the assumption of similarity between LT noised and FT original PET slices. It performs a regularization over CycleGAN generators, preventing them from generating denoised PET images that are very different from the original one.
2.3.3 2.5D and 3D methods
The 2.5D models have a similar architecture as their 2D counterparts. The difference is in the number of input channels (2, 44). To predict the ith slice denoising, we used k adjacent slices—2k+1 slices in total. These slices were fed into our models as 2k+1 channel images. The increase in input channel number slightly raised GPU memory and time consumption. We trained the 2.5D model in the same manner as we trained the 2D model. The only difference is that we used only the central channel of SwinIR output as the result.
We employed novel 3D ConvNeXt-like architectures, namely, MedNeXt (49) small (5.6 million parameters), and 3D UX-Net (27). These models draw inspiration from the transformer architecture, but it is a pure convolutional model. The authors of MedNeXt adopted the general design of ConvNeXt block for 3D-UNet-like architecture. The MedNext architercture utilizes depthwise convolution, GroupNorm, and Expansion and Compression layers. The novelty of the model is the usage of residiual inverted bottlenecks in place of regular up and downsampling blocks as well as the technique of iteratively increasing kernel size. We fed a 32 PET slices chunk into the input of the network.
2.3.4 Training details
The training parameters are in Table 3. The hyperparameter tuning is done on the validation data set by maximizing SSIM with optuna library. SSIM is preferrable over L1 and RMSE as it, more than other metrics, coincides with human perception and makes denoised PET look similar to the original (48).
We considered identity and image prior loss coefficients between 0 and 30 and weight decay for Unet in the 0.001–0.2. The ISSIM dependence of image prior loss coefficient looks the same as in (60). The quality of denoising is stable to the identity loss coefficient but could deteriorate up to 20% of its value when choosing a coefficient higher than 18.
The augmentations used in training are horizontal and vertical flips. The augmentations did not significantly improve metrics for ResNet, but they made the training process more stable. In contrast, CycleGAN with ResNet backbone metrics slightly dropped when trained with augmented images. The Unet performance improved significantly after applying augmentations but still lagged behind ResNet; this could be partly due to overfitting as Unet has more parameters than ResNet.
Adam is an optimizer for the training process. Unet was trained with weight decay = 0.002 to prevent overfitting improving relative ISSIM from 27.8% to 29.0%. The usage of dropout has a similar effect. The learning rate was chosen individually to achieve the best performance for each model. We trained supervised methods and pix2pix GANs with ResNet backbones using a cos learning rate schedule, max lr = 0.0002 for 35 epochs. CycleGAN training includes 30 epochs with a constant learning rate of 0.0001, which is linearly reduced to zero for the following 15 epochs. The Optuna library helped to fit the optimal learning rate schedule for SwinIR.
We trained models with batch size 32 except CycleGAN.The original CycleGAN (67) used batch size 1. Unlike the original work in the recent study (37), the batch size that generates the best PSNR value is 64, using the initial learning rate. The experiments demonstrated that the batch size does not have to be 1 or 4, but it depends on the data set size and the problem type. Therefore, CycleGAN was trained with batch size 16.
3 Results
For 7 MBq/kg injected activity and 90-s FT, the 3D methods have shown exceptional results in enhancing SSIM and MSE metrics, surpassing 2D methods. The 2.5D approach showed superiority over 2D counterparts for SwinIR only. For 60-s PET, SwinIR 2.5D has slightly surpassed 3D methods. When evaluating the SUV error, 3D methods demonstrated comparable results to the 2D methods. Unlike in the study (65), adding CT information as a second channel, along with PET, did not improve the quality of denoising process. The examples of denoised images are in Figures 4, 5.
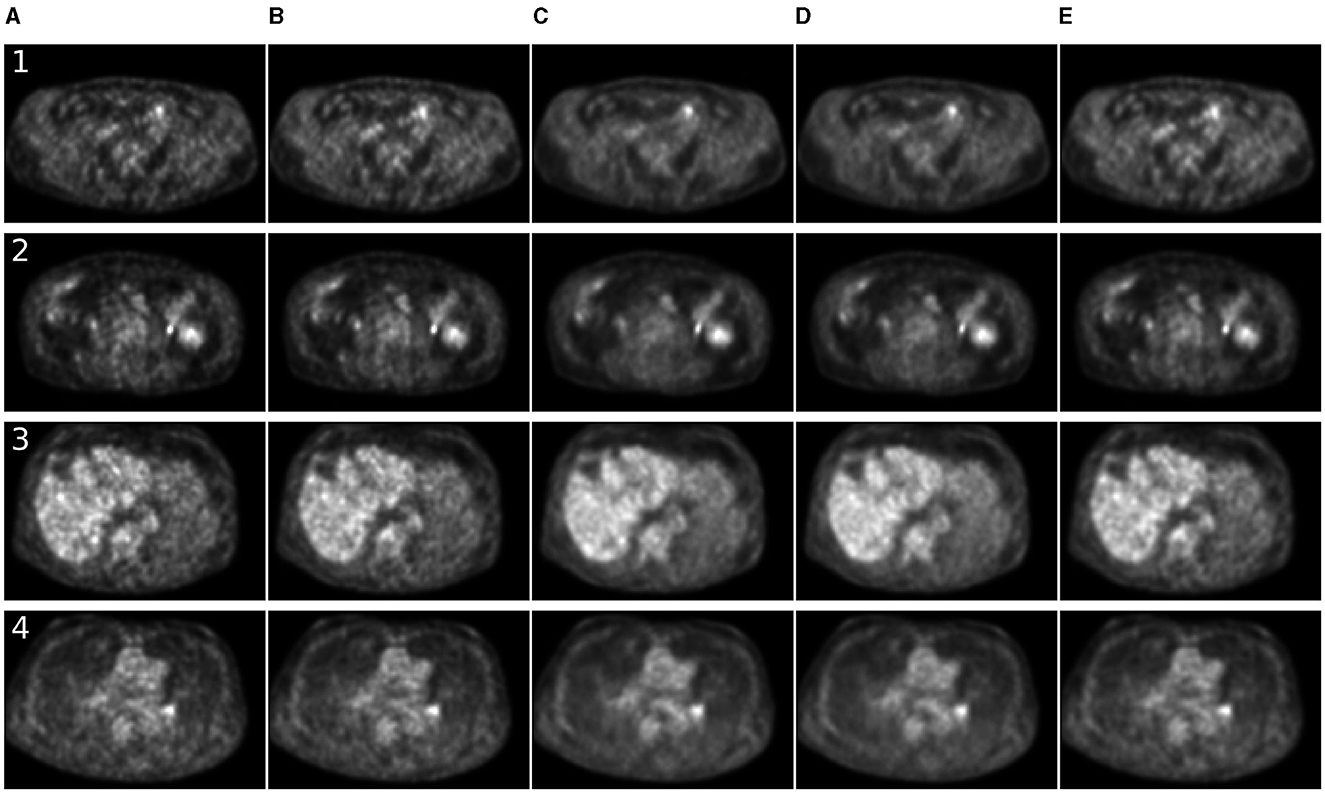
Figure 4. Comparison of denoising methods applied on low-time (30 s) PET reconstruction. (A) Low-time PET (30 s). (B) Full-time PET (90 s). (C) Reconstructed PET (by SwinIR 5 channels). (D) Reconstructed PET (by MedNeXt small). (E) Reconstructed PET (by Gaussian filter).
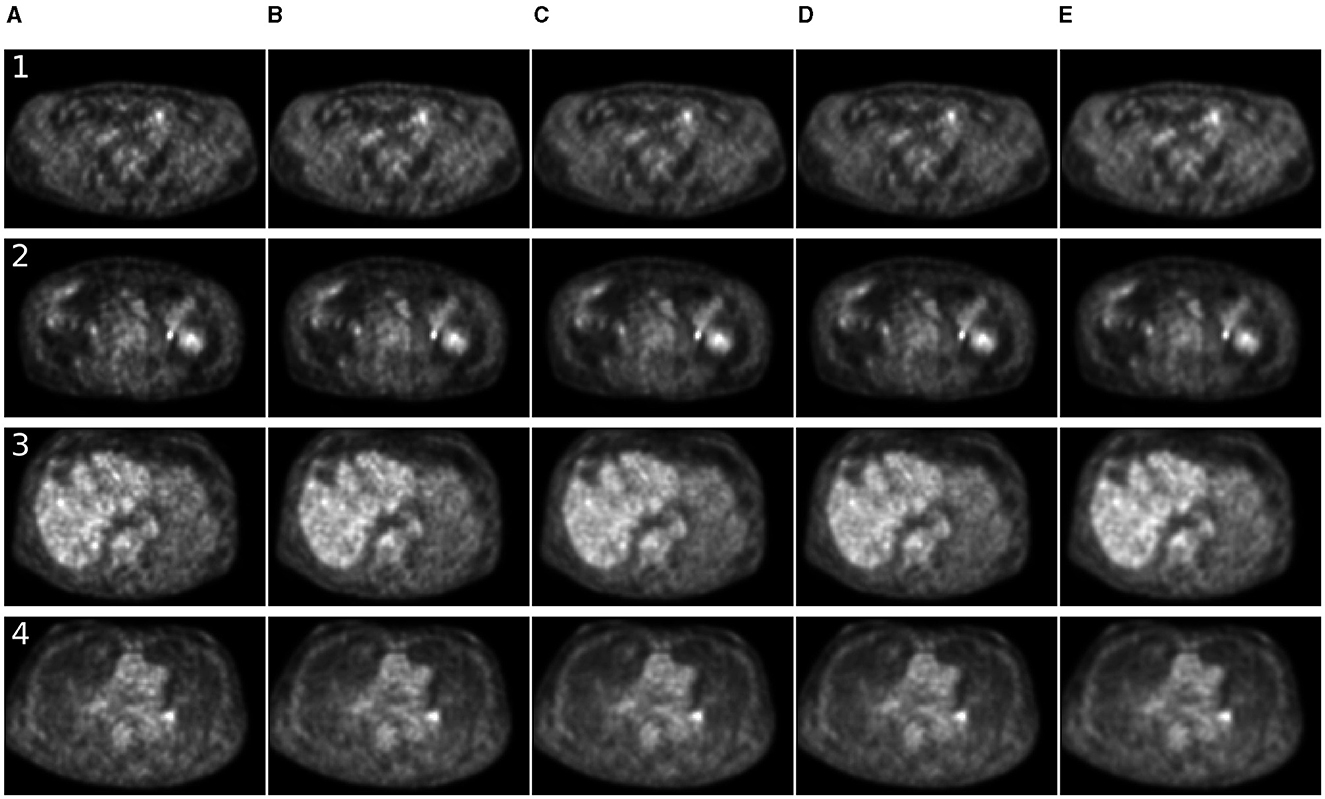
Figure 5. Comparison of denoising methods applied on low-time (60 s) PET reconstruction. (A) Low-time PET (60 s). (B) Full-time PET (90 s). (C) Reconstructed PET (by SwinIR 5 channels). (D) Reconstructed PET (by MedNeXt small). (E) Reconstructed PET (by Gaussian filter).
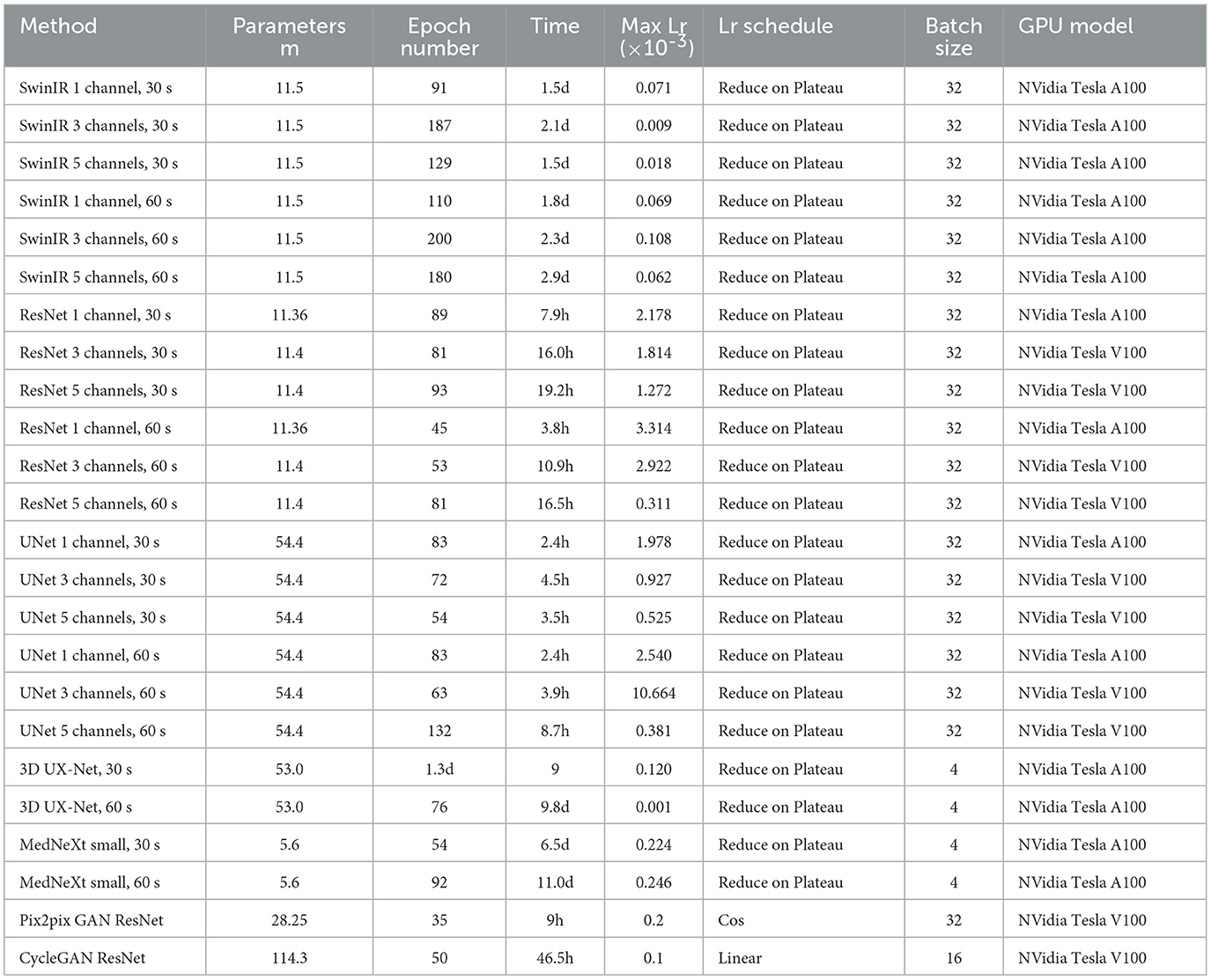
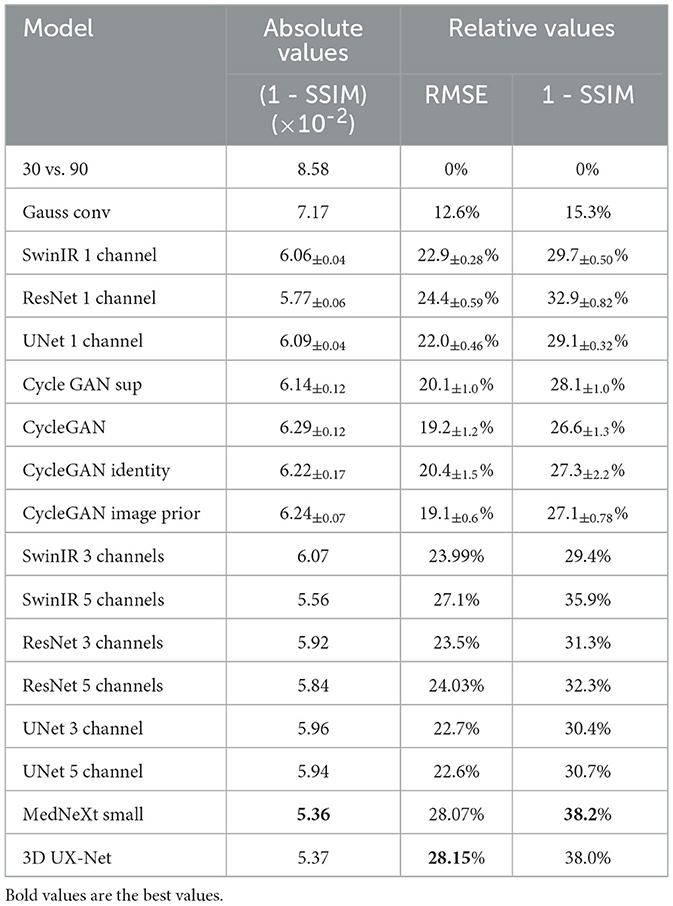
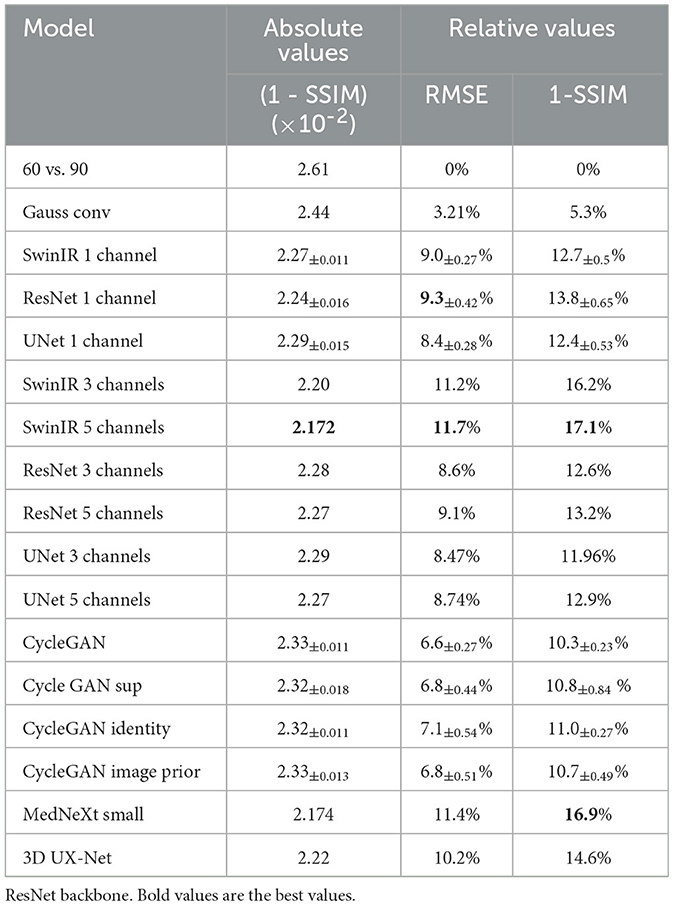
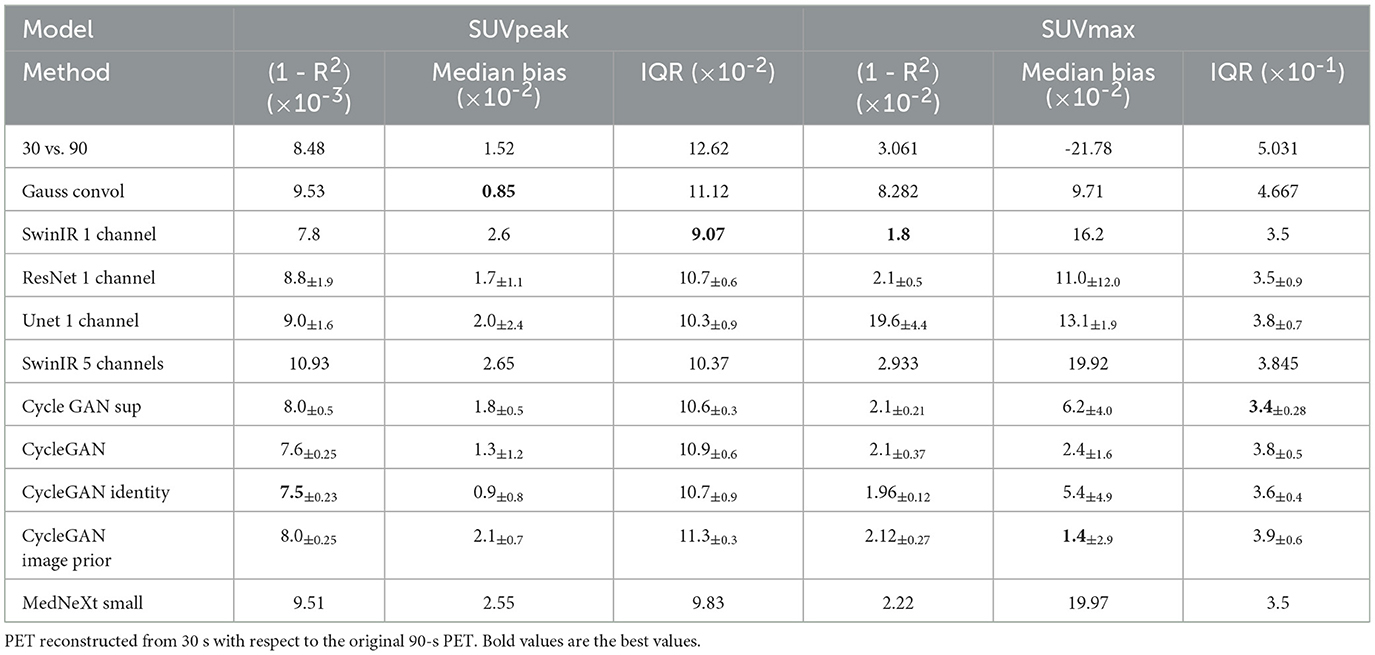
The ISSIM and RMSE metrics for 30 sec and 60 sec PET are in Tables 4, 5, respectively. If the value in a Table has ±, it means that the network we trained independently five times to estimate the confidence intervals (95% confidence level). Tables 6, 7 represent discrepancy [Median bias and Interquartile Range (IQR) values for the Bland–Altman plot, Figure 6] in SUVpeak and SUVmax estimation. We preferred median and IQR over mean and STD like (61) did as these metrics are more robust to the outliers.
The prediction of the difference between noised and denoised images rather than the prediction of the denoised image itself significantly (twice as much for ISSIM metric) improved the quality of denoising as it is easy for the network to produce noise rather than images (7).
3.1 Comparison of 2D and 2.5D methods
ResNet has the best image similarity metrics among 2D methods for both weakly noised (60 s) and strongly noised (30 s) PET. SwinIR and Unet follow it. The quality of SwinIR PET restoration is better than that of Unet. At the same time, Jang et al. (23) demonstrates better Swin performance over Unet on 25% low-count data. This fact indicates that convolutional layers of SwinIR for shallow feature extraction improve reconstruction quality compared to the pure Swin transformer architecture.
The 2.5D approach did not improve the denoising quality for ResNet but significantly improved it for SwinIR with five layers (±2 slices around the slice of interest). This could have happened due to differences in Transformer and convolution architectures. Transformers are better at mixing information between channels. SwinIR with five layers have a similar performance as 3D MedNext for 60-s PET in SSIM and MSE but lagged significantly behind in SUVmax estimation.
3.2 Comparison of GAN performance
ResNet backbone with convolutional decoder without skip connection outperformed Unet and SwinIR in all cases, that is why in Table 4 and the following tables, we presented the results for CycleGAN and Pix2pix GAN for the ResNet backbone only. Pix2Pix GAN (ResNet + PatchGAN discriminator) without distance loss produces realistic denoised images with low metrics. The SSIM steadily improves while the distance loss coefficient increases and reaches its plateau when the GAN degrades to a simple supervised model, as distance loss outweighs adversarial losses. Our Pix2Pix GAN did not show a higher quality over other methods as 3D CVT-GAN in (64), or BiC-GAN (13) for the brain's PET synthesis.
As the original Pix2Pix paper (22) mentioned, the random input z does not change the GAN results, and the model is indeed deterministic. We concluded that the Pix2Pix GAN model is inappropriate for the denoising problem as adversarial loss improves image appearance instead of SSIM. So, it produces realistic images that are far from the original images. For that reason, we have not included pix2pix GAN metrics in Tables 4–7. We trained PixPix GAN with the coefficient by distance loss of 10.0 as it does not outweigh the adversarial loss.
Unsupervised CycleGAN provides weaker image similarity metrics than supervised. The optimal coefficient for the identity loss is 2.2, and the image prior loss is 9.2. For the distance loss in supervised CycleGAN, we used the same coefficient as for image prior loss was 9.2. For the reduced 30% data set, the impact of the image prior loss is the same as in (60). However for the full-size data set, the image prior loss effect is less pronounced. The reason is that for the small-size data set, the image prior loss works as regularization and prevents overfitting. For example, the size of the data set in (60) is only 906 CT images with 512 × 512 resolution.
The image prior loss improves the quality and stability of the reconstruction. The identity loss has a more profound effect on the SSIM and RMSE metrics than the image prior loss for the weakly noised PET (Table 5). We used the same coefficient for image prior loss as for the distance loss in supervised CycleGAN - 9.2. Supervised CycleGAN image similarity metrics lie between supervised and unsupervised methods but have the advantage of CycleGAN estimating SUVmax with lower bias and dispersion. Our results contradict (28) where CycleGAN outperformed supervised Unet and Unet GAN. That could stem from the small size of the data set used in (28), or the reason is the usage in CycleGAN backbone other than Unet.
4 Discussion
The results of experiments showed 3D MedNext is the best model for PET denoising, though 2.5D SwinIR slightly outperforms it in MSE for 60-s PET. The supervised methods produce reconstructed PET with positive SUVmax bias (Figure 6) because they flatten the signals too much. On the other hand, CycleGAN family methods have predominantly negative or around zero bias such as SubtlePET algorithm (61), which is an advantage of unsupervised methods.
The SSIM metric was an optimization goal for the network, so the model having the lowest ISSIM and RMSE results does not necessarily produce the best SUV reconstruction for tumors. In (52), the SUVmean bias was not improved by HighResNet (LD contains only 6% of the FD) even though the PSNR and SSIM of the reconstructed image were better than the LD. There is also a high variation in SUVs for the same model trained several times.
Due to biological or technological factors, SUV may significantly differ from one measurement to another (1, 11, 32). For example, technological factors include inter-scanner variability, image reconstruction, processing parameter changes, and calibration error between a scanner and a dose calibrator. An example of biological factors are respiratory motion (18) like cardiac motion, and body motion. The LT denoising is acceptable if its error on SUV values is smaller than SUV variations from different FT measurements.
We did use SUVmax and SUVpeak measurements as these are metrics commonly used by clinicians for follow-up FDG-PET/CT scans and therapy response evaluation (40). The studies (4, 35, 56) reported high ΔSUVmax between two FDG acquisitions. One should consider the details of these experiments to compare them with the results of our study. The uncertainty estimation of the PET reconstruction (10) is also an important topic for the future research.
The recent work (11) conducted the following experiment. The six phantom spheres of 10–37 mm diameters were filled with the concentration 20.04 MBq/ml. The FT 150-s mode was divided into subsets of shorter frames varying from 4 to 30 s. The SUVmax monotonically increases with sphere's diameter. The ratio of the standard SUVmax deviation to its average value for 30-s PET is approximately 15% for a 10 mm sphere and the confidence interval length achieves up to 0.5 kBq/ml. The experiment (11) does not take into account biological and most of technical factors (1), so the final SUVmax discrepancy between two PET could achieve higher values. The aforementioned estimation of SUVmax discrepancy for the same tumor between two PET acquisitions shows that SUVmax denoising error for the 30- and 60-s PET achieved in our study lies in the acceptable range.
It is a matter of discussion on which metric—SUVmax error or visual similarity should be given priority. Image similarity metrics provide visual information that can help doctors determine whether a tumor is malignant or benign. Image comparison also helps clinicians assess treatment efficacy by comparing pre-treatment images with post-treatment ones to measure any changes due to therapy intervention. On the other hand, SUVmax error provides quantitative data regarding how much a tumor has reduced in size after treatment interventions have been applied; this allows physicians an objective way of evaluating treatments' effectiveness without relying solely on subjective visual assessments from image comparisons alone.
5 Conclusion
PET denoising may allow for reducing an injected dose or increasing the scanner's throughput. We reconstructed PET with a reduced acquisition time of 30 and 60 s and compared it with the original full-time 90-s PET for 7MBq/kg injected activity. The AI models reduced PET denoising for 38% (100%–restoration to original image) in the SSIM metric for 30-s PET and for 17% for 60-s PET. The SUVmax discrepancy for the 30- and 60-s PET achieved in our study lies in the acceptable range.
We trained and tested MedNeXt, 3D UX-Net, SwinIR, Unet, ResNet, and CycleGAN with ResNet backbone and different auxiliary losses. The 3D MedNeXt approach has shown the best results in enhancing SSIM and MSE metrics. The supervised denoising methods have significantly better RMSE and ISSIM than unsupervised ones. This result differs from previous studies claiming that CycleGAN surpasses Unet and ResNet. The ResNet reconstructs PET images with the lowest RMSE and ISSIM, outperforming 2D SwinIR and Unet, but lags significantly behind 2.5 SwinIR with five channels. Supervised CycleGAN achieved the lowest SUVmax error after PET denoising. The SUVmax error of the reconstructed PET is comparable with the reproducibility error due to biological or technological factors. Adding CT information to PET does not improve denoising quality.
Data availability statement
The datasets presented in this article are not readily available because the data that support the findings of this study are available from PET Moscow region, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request. Requests to access the datasets should be directed to Ivan Kruzhilov, aXNrcnV6aGlsb3YmI3gwMDA0MDtzYmVyYmFuay5ydQ==.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants' legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
SK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. IK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. LV: Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. ES: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing. VK: Conceptualization, Data curation, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We would like to express our gratitude to Alexander Nikolaev for his valuable assistance in addressing medical-related issues.
Conflict of interest
ES was employed by LLC SberMedAI.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://github.com/seung-lab/connected-components-3d/
References
1. Adams MC, Turkington TG, Wilson JM, Wong TZ, A. systematic review of the factors affecting accuracy of SUV measurements. Am J Roentgenol. (2010) 195:310–20. doi: 10.2214/AJR.10.4923
2. Avesta A, Hossain S, Lin M, Aboian M, Krumholz HM, Aneja S. Comparing 3D, 2.5D, and 2D approaches to brain image auto-segmentation. Bioengineering. (2023) 10:181. doi: 10.3390/bioengineering10020181
3. Bonardel G, Dupont A, Decazes P, Queneau M, Modzelewski R, Coulot J, et al. Clinical and phantom validation of a deep learning based denoising algorithm for F-18-FDG PET images from lower detection counting in comparison with the standard acquisition. EJNMMI Phys. (2022) 9:1–23. doi: 10.1186/s40658-022-00465-z
4. Burger IA, Huser DM, Burger C, von Schulthess GK, Buck A. Repeatability of FDG quantification in tumor imaging: averaged SUVs are superior to SUVmax. Nucl Med Biol. (2012) 39:666–70. doi: 10.1016/j.nucmedbio.2011.11.002
5. Chandrashekar A, Handa A, Ward J, Grau V, Lee R. A deep learning pipeline to simulate fluorodeoxyglucose (FDG) uptake in head and neck cancers using non-contrast CT images without the administration of radioactive tracer. Insights Imaging. (2022) 13:1–10. doi: 10.1186/s13244-022-01161-3
6. Charbonnier P, Blanc-Feraud L, Aubert G, Barlaud M. Two deterministic half-quadratic regularization algorithms for computed imaging. In: Proceedings of 1st International Conference on Image Processing. Austin, TX: IEEE (1994). p. 168–172.
7. Chen J, Chen J, Chao H, Yang M. Image blind denoising with generative adversarial network based noise modeling. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Salt Lake City, UT: IEEE (2018). p. 3155–64.
8. Chen KT, Zaharchuk G. Image synthesis for low-count PET acquisitions: lower dose, shorter time. In: Biomedical Image Synthesis and Simulation. London: Elsevier (2022). p. 369–391.
9. Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, et al. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging. (2019) 46:2780–9. doi: 10.1007/s00259-019-04468-4
10. Cui J, Xie Y, Joshi AA, Gong K, Kim K, Son YD, et al. PET denoising and uncertainty estimation based on NVAE model using quantile regression loss. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2022: 25th International Conference, Singapore, September 18-22, 2022, Proceedings, Part IV. Cham: Springer (2022). p. 173–183.
11. De Luca GM, Habraken JB. Method to determine the statistical technical variability of SUV metrics. EJNMMI Phys. (2022) 9:40. doi: 10.1186/s40658-022-00470-2
12. De Summa M, Ruggiero MR, Spinosa S, Iachetti G, Esposito S, Annunziata S, et al. Denoising approaches by SubtlePET artificial intelligence in positron emission tomography (PET) for clinical routine application. Clini Transl Imag. (2024) 12:1–10. doi: 10.1007/s40336-024-00625-4
13. Fei Y, Zu C, Jiao Z, Wu X, Zhou J, Shen D, et al. Classification-Aided High-Quality PET Image Synthesis via Bidirectional Contrastive GAN with Shared Information Maximization. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2022: 25th International Conference, Singapore, September 18-22, 2022, Proceedings, Part VI. Cham: Springer (2022). p. 527–537.
14. Fletcher J, Kinahan P. PET/CT standardized uptake values (SUVs) in clinical practice and assessing response to therapy. NIH Public Access. (2010) 31:496–505. doi: 10.1053/j.sult.2010.10.001
15. Gatidis S, Hepp T, Früh M, La Fougère C, Nikolaou K, Pfannenberg C, et al. A whole-body FDG-PET/CT dataset with manually annotated tumor lesions. Sci Data. (2022) 9:1–7. doi: 10.1038/s41597-022-01718-3
16. Gong K, Guan J, Liu CC Qi J. PET image denoising using a deep neural network through fine tuning. IEEE Trans Radiat Plasma Med Sci. (2018) 3:153–61. doi: 10.1109/TRPMS.2018.2877644
17. Gong K, Johnson K, El Fakhri G, Li Q, Pan T, PET. image denoising based on denoising diffusion probabilistic model. Eur J Nucl Med Mol Imaging. (2024) 51:358–68. doi: 10.1007/s00259-023-06417-8
18. Guo X, Zhou B, Pigg D, Spottiswoode B, Casey ME, Liu C, et al. Unsupervised inter-frame motion correction for whole-body dynamic PET using convolutional long short-term memory in a convolutional neural network. Med Image Anal. (2022) 80:102524. doi: 10.1016/j.media.2022.102524
19. Hashimoto F, Onishi Y, Ote K, Tashima H, Reader AJ, Yamaya T. Deep learning-based PET image denoising and reconstruction: a review. Radiol Phys Technol. (2024) 17:24–46. doi: 10.1007/s12194-024-00780-3
20. Hu R, Liu H. TransEM: residual Swin-transformer based regularized PET image reconstruction. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2022: 25th International Conference, Singapore, September 18-22, 2022, Proceedings, Part IV. Singapore: Springer (2022). p. 184–193.
21. Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods. (2021) 18:203–11. doi: 10.1038/s41592-020-01008-z
22. Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. Honolulu, HI: IEEE (2017). p. 1125–34.
23. Jang SI, Pan T, Li Y, Heidari P, Chen J, Li Q, et al. Spach Transformer: Spatial and channel-wise transformer based on local and global self-attentions for PET image denoising. IEEE Trans Med Imaging. (2023). doi: 10.1109/TMI.2023.3336237
24. Kaalep A, Sera T, Rijnsdorp S, Yaqub M, Talsma A, Lodge MA, et al. Feasibility of state of the art PET/CT systems performance harmonisation. Eur J Nucl Med Mol Imaging. (2018) 45:1344–61. doi: 10.1007/s00259-018-3977-4
25. Katsari K, Penna D, Arena V, Polverari G, Ianniello A, Italiano D, et al. Artificial intelligence for reduced dose 18F-FDG PET examinations: a real-world deployment through a standardized framework and business case assessment. EJNMMI Phys. (2021) 8:1–15. doi: 10.1186/s40658-021-00374-7
26. Kwon T, Ye JC. Cycle-free cyclegan using invertible generator for unsupervised low-dose CT denoising. IEEE Trans Computat Imag. (2021) 7:1354–68. doi: 10.1109/TCI.2021.3129369
27. Lee HH, Bao S, Huo Y, Landman BA. 3d ux-net: A large kernel volumetric convnet modernizing hierarchical transformer for medical image segmentation. arXiv [preprint] arXiv:220915076. (2022).
28. Lei Y, Dong X, Wang T, Higgins K, Liu T, Curran WJ, et al. Whole-body PET estimation from low count statistics using cycle-consistent generative adversarial networks. Phys Med Biol. (2019) 64:215017. doi: 10.1088/1361-6560/ab4891
29. Lei Y, Wang T, Dong X, Higgins K, Liu T, Curran WJ, et al. Low dose PET imaging with CT-aided cycle-consistent adversarial networks. In: Medical Imaging 2020: Physics of Medical Imaging. California: SPIE (2020). p. 1043–1049.
30. Li Y, Zhang K, Shi W, Miao Y, Jiang Z. A novel medical image denoising method based on conditional generative adversarial network. Comput Math Methods Med. (2021) 2021:9974017. doi: 10.1155/2021/9974017
31. Liang J, Cao J, Sun G, Zhang K, Van Gool L, Timofte R. SwinIR: image restoration using swin transformer. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. Montreal, BC: IEEE (2021). p. 1833–1844.
32. Lindholm H, Staaf J, Jacobsson H, Brolin F, Hatherly R, Sânchez-Crespo A. Repeatability of the maximum standard uptake value (SUVmax) in FDG PET. Mol Imag Radionucl Ther. (2014) 23:16. doi: 10.4274/Mirt.76376
33. Liu J, Malekzadeh M, Mirian N, Song TA, Liu C, Dutta J. Artificial intelligence-based image enhancement in pet imaging: noise reduction and resolution enhancement. PET Clin. (2021) 16:553–76. doi: 10.1016/j.cpet.2021.06.005
34. Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, et al. Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. Montreal, QC: IEEE (2021). p. 10012–22. doi: 10.1109/ICCV48922.2021.00986
35. Lodge MA, Chaudhry MA, Wahl RL. Noise considerations for PET quantification using maximum and peak standardized uptake value. J Nucl Med. (2012) 53:1041–7. doi: 10.2967/jnumed.111.101733
36. Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, et al. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol. (2019) 64:165019. doi: 10.1088/1361-6560/ab3242
37. Lupión M, Sanjuan J, Ortigosa P. Using a Multi-GPU node to accelerate the training of Pix2Pix neural networks. J Supercomput. (2022) 78:12224–41. doi: 10.1007/s11227-022-04354-1
38. Luthra A, Sulakhe H, Mittal T, Iyer A, Yadav SK. Eformer: edge enhancement based transformer for medical image denoising. arXiv (preprint)abs/2109.08044 (2021).
39. Manakov I, Rohm M, Kern C, Schworm B, Kortuem K, Tresp V. Noise as domain shift: denoising medical images by unpaired image translation. In: Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data. Cham: Springer (2019). p. 3–10.
40. Massaro A, Cittadin S, Milan E, Tamiso L, Pavan L, Secchiero C, et al. Reliability of SUVmax vs. SUVmean in FDG PET/CT. J Nucl Med. (2009) 50:2121.
41. Maus J, Nikulin P, Hofheinz F, Petr J, Braune A, Kotzerke J, et al. Deep learning based bilateral filtering for edge-preserving denoising of respiratory-gated PET. EJNMMI physics. (2024) 11:58. doi: 10.1186/s40658-024-00661-z
42. OECD EU. Health at a Glance: Europe 2020: State of Health in the EU Cycle. Paris: OECD Publishing. (2020).
43. Onishi Y, Hashimoto F, Ote K, Matsubara K, Ibaraki M. Self-supervised pre-training for deep image prior-based robust pet image denoising. IEEE Trans Radiat Plasma Med Sci. (2023) 8:348–56. doi: 10.1109/TRPMS.2023.3280907
44. Ottesen JA Yi D, Tong E, Iv M, Latysheva A, Saxhaug C, et al. 2.5 D and 3D segmentation of brain metastases with deep learning on multinational MRI data. Front Neuroinform. (2023) 16:1056068. doi: 10.3389/fninf.2022.1056068
45. Pan S, Abouei E, Peng J, Qian J, Wynne JF, Wang T, et al. Full-dose whole-body PET synthesis from low-dose PET using high-efficiency denoising diffusion probabilistic model: PET consistency model. Med Phys. (2024). doi: 10.1117/12.3006565
46. Pan S, Abouei E, Peng J, Qian J, Wynne JF, Wang T, et al. Full-dose PET synthesis from low-dose PET using 2D high efficiency denoising diffusion probabilistic model. In: Medical Imaging 2024: Clinical and Biomedical Imaging. California: SPIE (2024). p. 428–435.
47. Park KS, Cho SG, Kim J, Song HC. The effect of weights for cycle-consistency loss and identity loss on blood-pool image to bone image translation with CycleGAN. J Nucl Med. (2021) 62:1189.
48. Renieblas GP, Nogués AT, González AM, León NG, Del Castillo EG. Structural similarity index family for image quality assessment in radiological images. J Med Imag. (2017) 4:035501. doi: 10.1117/1.JMI.4.3.035501
49. Roy S, Koehler G, Ulrich C, Baumgartner M, Petersen J, Isensee F, et al. Mednext: transformer-driven scaling of convnets for medical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer (2023). p. 405–415.
50. Sanaat A, Arabi H, Mainta I, Garibotto V, Zaidi H. Projection space implementation of deep learning-guided low-dose brain PET imaging improves performance over implementation in image space. J Nucl Med. (2020) 61:1388–96. doi: 10.2967/jnumed.119.239327
51. Sanaat A, Shiri I, Arabi H, Mainta I, Nkoulou R, Zaidi H. Deep learning-assisted ultra-fast/low-dose whole-body PET/CT imaging. Eur J Nucl Med Mol Imaging. (2021) 48:2405–15. doi: 10.1007/s00259-020-05167-1
52. Sanaei B, Faghihi R, Arabi H, Zaidi H. Does prior knowledge in the form of multiple low-dose PET images (at different dose levels) improve standard-dose PET prediction? In: 2021 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC). Piscataway, NJ: IEEE (2021). p. 1–3.
53. Sano A, Nishio T, Masuda T, Karasawa K. Denoising PET images for proton therapy using a residual U-net. Biomed Phys Eng Express. (2021) 7:025014. doi: 10.1088/2057-1976/abe33c
54. Schaefferkoetter J, Nai YH, Reilhac A, Townsend DW, Eriksson L, Conti M. Low dose positron emission tomography emulation from decimated high statistics: a clinical validation study. Med Phys. (2019) 46:2638–45. doi: 10.1002/mp.13517
55. Schaefferkoetter J, Yan J, Ortega C, Sertic A, Lechtman E, Eshet Y, et al. Convolutional neural networks for improving image quality with noisy PET data. EJNMMI Res. (2020) 10:1–11. doi: 10.1186/s13550-020-00695-1
56. Schwartz J, Humm J, Gonen M, Kalaigian H, Schoder H, Larson S, et al. Repeatability of SUV measurements in serial PET. Med Phys. (2011) 38:2629–38. doi: 10.1118/1.3578604
57. Shen C, Yang Z, Zhang Y. PET image denoising with score-based diffusion probabilistic models. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer (2023). p. 270–278.
58. Sher A, Lacoeuille F, Fosse P, Vervueren L, Cahouet-Vannier A, Dabli D, et al. For avid glucose tumors, the SUV peak is the most reliable parameter for [18F] FDG-PET/CT quantification, regardless of acquisition time. EJNMMI Res. (2016) 6:1–6. doi: 10.1186/s13550-016-0177-8
59. Spuhler K, Serrano-Sosa M, Cattell R, DeLorenzo C, Huang C. Full-count PET recovery from low-count image using a dilated convolutional neural network. Med Phys. (2020) 47:4928–38. doi: 10.1002/mp.14402
60. Tang C, Li J, Wang L, Li Z, Jiang L, Cai A, et al. Unpaired low-dose CT denoising network based on cycle-consistent generative adversarial network with prior image information. Comput Math Methods Med. (2019) 2019:8639825. doi: 10.1155/2019/8639825
61. Weyts K, Lasnon C, Ciappuccini R, Lequesne J, Corroyer-Dulmont A, Quak E, et al. Artificial intelligence-based PET denoising could allow a two-fold reduction in [18F] FDG PET acquisition time in digital PET/CT. Eur J Nucl Med Mol Imaging. (2022) 49:3750–60. doi: 10.1007/s00259-022-05800-1
62. Yang G, Li C, Yao Y, Wang G, Teng Y. Quasi-supervised learning for super-resolution PET. Comput Med Imaging Grap. (2024) 113:102351. doi: 10.1016/j.compmedimag.2024.102351
63. Yu B, Dong Y, Gong K. Whole-body PET image denoising based on 3D denoising diffusion probabilistic model. J Nucl Med. (2024) 51:358–68.
64. Zeng P, Zhou L, Zu C, Zeng X, Jiao Z, Wu X, et al. 3D CVT-GAN: a 3D convolutional vision transformer-GAN for PET reconstruction. In: Medical Image Computing and Computer Assisted Intervention-MICCAI 2022: 25th International Conference, Singapore, September 18-22, 2022, Proceedings, Part VI. Cham: Springer (2022). p. 516–526.
65. Zhang Q, Hu Y, Zhou C, Zhao Y, Zhang N, Zhou Y, et al. Reducing pediatric total-body PET/CT imaging scan time with multimodal artificial intelligence technology. EJNMMI Phys. (2024) 11:1. doi: 10.1186/s40658-023-00605-z
66. Zhou L, Schaefferkoetter JD, Tham IW, Huang G, Yan J. Supervised learning with cyclegan for low-dose FDG PET image denoising. Med Image Anal. (2020) 65:101770. doi: 10.1016/j.media.2020.101770
Keywords: artificial intelligence, positron emission tomography, SUV, noise reduction, MedNeXt, SwinIR
Citation: Kruzhilov I, Kudin S, Vetoshkin L, Sokolova E and Kokh V (2024) Whole-body PET image denoising for reduced acquisition time. Front. Med. 11:1415058. doi: 10.3389/fmed.2024.1415058
Received: 09 April 2024; Accepted: 30 August 2024;
Published: 30 September 2024.
Edited by:
Lorenzo Faggioni, University of Pisa, ItalyReviewed by:
Daniele Panetta, National Research Council (CNR), ItalyMichel Hesse, Cliniques Universitaires Saint-Luc, Belgium
Copyright © 2024 Kruzhilov, Kudin, Vetoshkin, Sokolova and Kokh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ivan Kruzhilov, aXNrcnV6aGlsb3YmI3gwMDA0MDtzYmVyYmFuay5ydQ==
†These authors have contributed equally to this work and share first authorship
 Ivan Kruzhilov1,2*†
Ivan Kruzhilov1,2*† Stepan Kudin
Stepan Kudin