- 1Department of Burn and Plastic Surgery, First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
- 2Department of Dermatology, First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
- 3Department of Plastic and Reconstructive Surgery, Guangdong Second Provincial General Hospital, Guangzhou, China
Objective: Skin fibrosis is a lesion in the dermis causing to itching, pain, and psychological stress. The gut microbiome plays as an essential role in skin diseases developments. We conducted a Mendelian randomization study to determine the causal association between the gut microbiome and skin fibrosis.
Methods: We retrieved valid instrumental variables from the genome-wide association study (GWAS) files of the gut microbiome (n = 18,340) conducted by the MiBioGen consortium. Skin fibrosis-associated data were downloaded from the GWAS Catalog. Subsequently, a two-sample Mendelian randomization (MR) analysis was performed to determine whether the gut microbiome was related to skin fibrosis. A reverse MR analysis was also performed on the bacterial traits which were causally associated with skin fibrosis in the forward MR analysis. In addition, we performed an MR-Pleiotropy Residual Sum and Outlier analysis to remove outliers and a sensitivity analysis to verify our results.
Results: According to the inverse variance-weighted estimation, we identified that ten bacterial traits (Class Actinobacteria, Class Bacteroidia, family Bifidobacteriaceae, family Rikenellaceae, genus Lachnospiraceae (UCG004 group), genus Ruminococcaceae (UCG013 group), order Bacteroidales, order Bifidobacteriales, genus Peptococcus and genus Victivallis) were negatively correlated with skin fibrosis while five bacterial traits (genus Olsenella, genus Oscillospira, genus Turicibacter, genus Lachnospiraceae (NK4A136group), and genus Sellimonas) were positively correlated. No results were obtained from reverse MR analysis. No significant heterogeneity or horizontal pleiotropy was observed in MR analysis.
Objective conclusion: There is a causal association between the gut microbiome and skin fibrosis, indicating the existence of a gut-skin axis. This provides a new breakthrough point for mechanistic and clinical studies of skin fibrosis.
1 Introduction
Skin fibrosis is an imbalance between extracellular matrix (ECM) synthesis and dermal degradation in the dermis. The remodelling process often leads to a loss of physiological architecture and skin malfunction including keloids, hypertrophic scars, systemic sclerosis, and dystrophic epidermolysis. Skin fibrosis is usually caused by surgery, burns and trauma and causes disfiguration, limitation of movement, or even significant psychological distress, such as keloids (1, 2). Keloids are common dermal skin fibrotic diseases. Owing to its aggressive proliferation, it is considered as benign skin tumour that harms the patient’s physiological and psychological health (3, 4). Keloids are refractory to cure, and their recurrence rate is over 45%. There are various treatment options, including intralesional steroid injections and surgical resection combined with postoperative radiation (4, 5). Therefore, it is important to explore the aetiology and pathogenic processes of skin fibrosis to provide novel insights into its treatment.
Many microbial communities reside in the intestine and affect human health and disease (6, 7). For example, the gut microbiome has been reported as a risk or preventive factor that can indirectly affect the response to immunotherapy in cancers (6, 8). The gut microbiome also influences the maintenance of maternal and foetal health during pregnancy (9). Recently, researchers have found that the gut microbiome plays a significant role in a wide variety of skin disorders, such as acne vulgaris, and reported the existence of a gut-skin axis (10, 11). However, the relationship between the gut microbiome and skin fibrosis remains unclear. Probing this association can be helpful for the treatment and clarification of the mechanism of skin fibrosis.
Unfortunately, the gut microbiome is associated with confounding factors, such as age, lifestyle, and living environment. Controlling these confounding biases inherent is difficult in the observational studies (12, 13). The advent of Mendelian randomization (MR) reduces the effect of these biases (14, 15). As Genome-wide association studies (GWAS) have greatly improved, we can observe the genetic susceptibility to the gut microbiome and skin fibrosis (16, 17). MR can be considered a new method for exploring the relationship between the gut microbiome and skin fibrosis (8, 13, 18).
In this study, we used two-sample MR analysis, an extension of the MR method, to analyse GWAS summary statistics from MiBioGen and the GWAS Catalog. Based on these results, we aimed to identify the role of the gut microbiome in skin fibrosis and propose new treatment strategies.
2 Materials and methods
2.1 Design of study
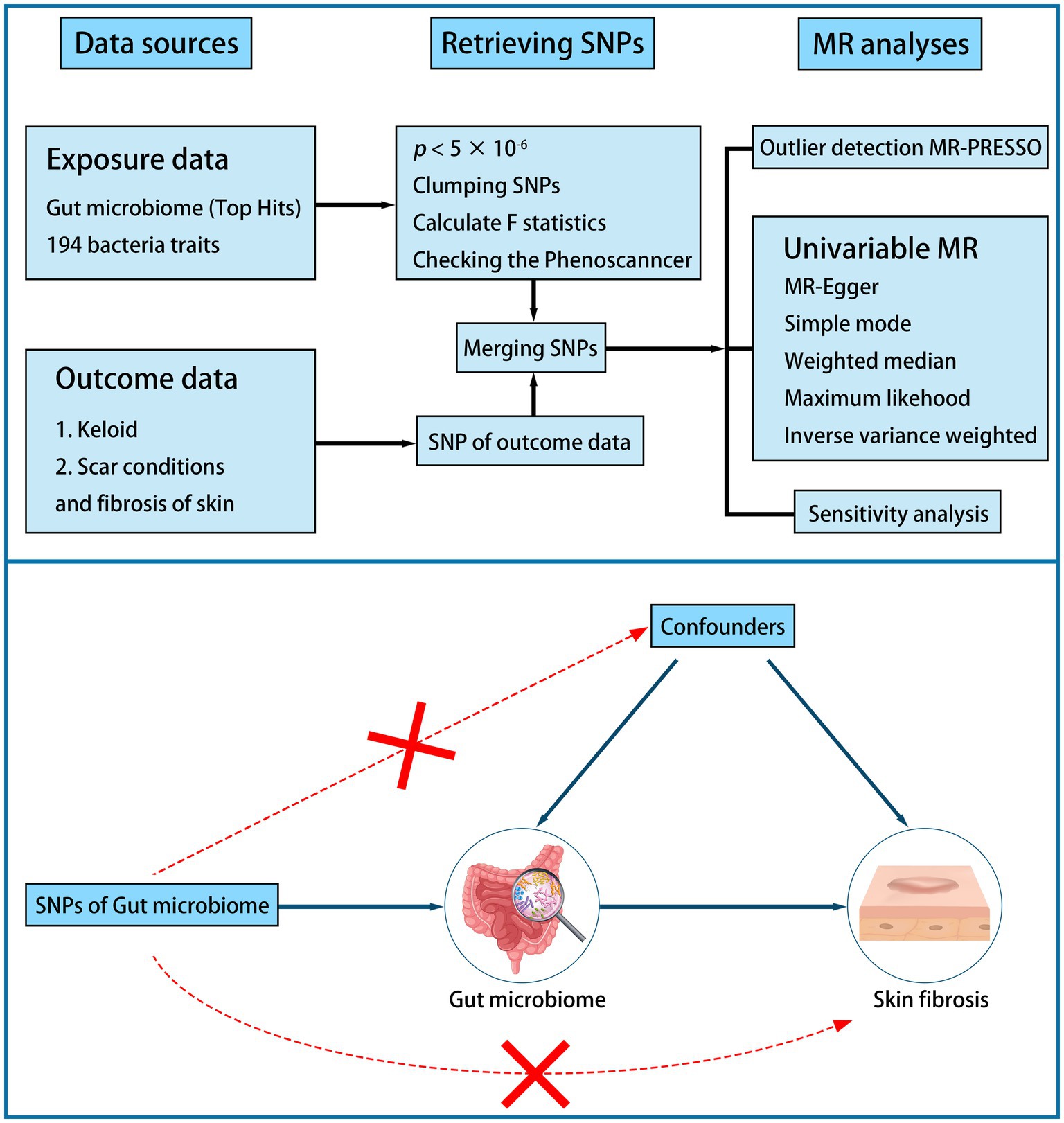
A two-sample MR study was performed to examine the causal association between the gut microbiome and skin fibrosis (Figure 1). First, we identified a valid instrumental variable (IV). IVs must be associated with exposure but not with confounders. IVs must relate to outcomes only through exposure. Second, the appropriate IVs were subjected to MR analysis. Sensitivity analysis was conducted to verify these results (18, 19).
2.2 Data preparation
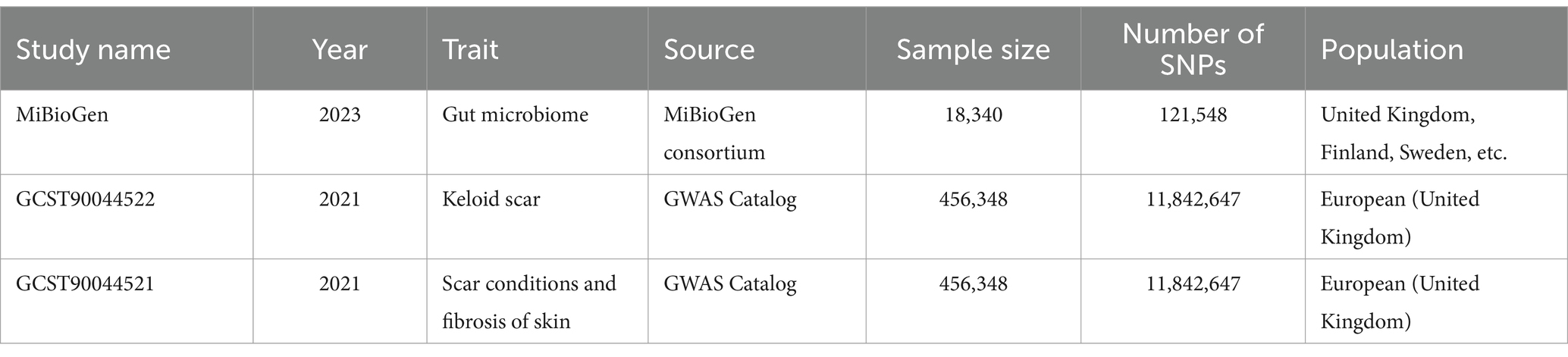
Human gut microbiome GWAS statistics were retrieved from the MiBioGen Consortium, an international project on genome-wide genotypes, and 16S faecal microbiome data (17, 20). In total, 194 bacterial traits were identified in the dataset. In this project, 18,340 individuals from 24 cohorts were included, of which 13,266 were of European ancestry. Thus, participants of European ancestry were selected to reduce the influence of ethnicity. We obtained GWAS data for keloids from GCST90044522. This study included 201 cases and 456,147 controls of European descent. In addition, summary genetic data for scar conditions and skin fibrosis were downloaded from GCST90044521 including 1,887 cases and 454,461 controls (Table 1) (21). The detail of gut microbiome was also shown (Supplementary Table S1).
2.3 Single nucleotide polymorphisms selection
We selected appropriate SNPs as IVs to reduce biases and improve the credibility of the results. The selection criteria were as follows: (1) the SNP was associated with the gut microbiome (p < 5 × 10−6); (2) the SNP was independent (r2 < 0.01 and clump distance >10,000 kb, EUR population reference); (3) it was highly correlated to exposure traits (the F-statistic >10); (4) palindromic SNPs were harmonized in exposure and outcome data; and (5) the SNPs associated with potential risk factors, such as inflammatory factor, were deleted based on PhenoScanner V2 (5, 16, 18, 22, 23).
2.4 Statistical analyses
Before the MR analysis, we performed an MR-Pleiotropy Residual Sum and Outlier (MR-PRESSO) analysis to identify and remove outliers.
We performed MR analysis using the inverse variance-weighted (IVW), MR–Egger, weighted median, simple mode, and weighted mode methods. We chose the IVW as the primary method because it can provide more robust estimates in a broader set of scenarios. Other methods serve as complementary methods (8, 18, 24).
To verify our study, we performed a sensitivity analysis using Cochran’s test. Heterogeneity was tested using Cochran’s Q test. If the Q statistic is p < 5 × 10−2, it indicates the presence of heterogeneity (25, 26). We tested for the pleiotropic effect using the MR–Egger regression intercept. There was no horizontal pleiotropy of the study when p > 5 × 10−2 (24). Furthermore, a leave-one-out analysis was used to assess the horizontal pleiotropy. Finally, reverse MR analysis was performed using the same methods and settings (8, 13).
Statistical analyses were performed using R version 4.3.0, and the MR analysis was performed using the TwosampleMR package (version 0.5.7) (27).
3 Results
3.1 IVs associated with the gut microbiome
1,384 SNPs were retrieved from 194 bacterial traits, including phyla, classes, orders, families, and genus levels by the genera. There was no bias in weak IVs (all F > 17). Additional details are provided in Supplementary Table S2.
3.2 Causal effects of the gut microbiome on skin fibrosis
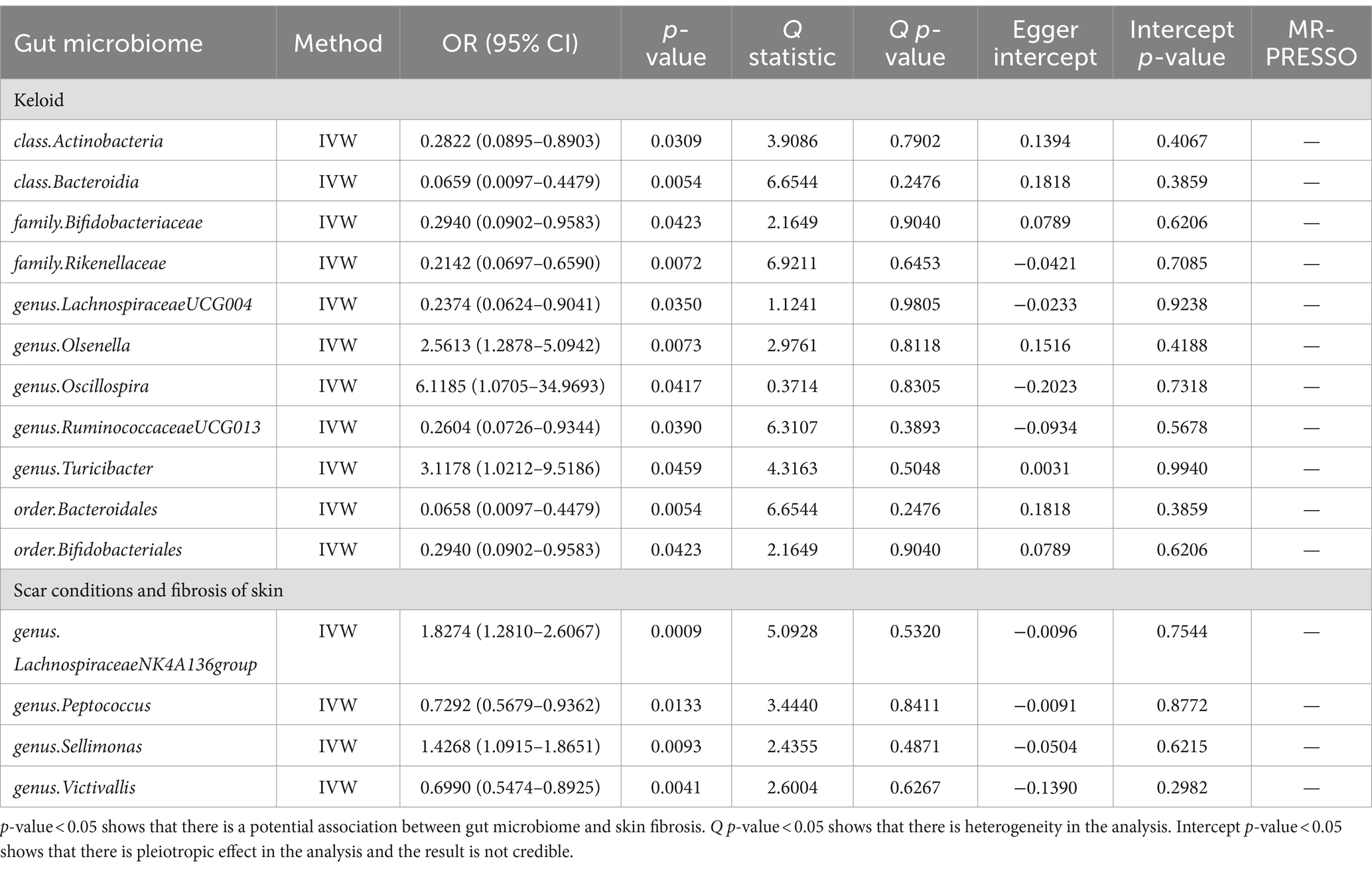
We performed an MR analysis of 194 bacterial traits (Supplementary Table S3). Most of these factors were not associated with skin fibrosis. However, 15 bacterial features were potentially associated with skin fibrosis. The odds ratio (OR) of Class Actinobacteria was 0.2822, while the 95% confidence interval (CI) was 0.0895–0.8903, and the p value was 3.09 × 10−2. The OR of Class Bacteroidia was 0.0659, the 95% CI was 0.0097–0.4479, and the p value was 5.4 × 10−3. The OR of family Bifidobacteriaceae was 0.2940, while the 95% CI was 0.0902–0.9583, and the p value was 4.23 × 10−2. The OR of family Rikenellaceae was 0.2142, the 95% CI was 0.0697–0.6590 and the p value was 7.2 × 10−3. The OR of genus Lachnospiraceae (UCG004 group) was 0.2374, the 95% CI was 0.0624–0.9041 and the p value was 3.5 × 10−2. The OR of genus Olsenella was 2.5613, the 95% CI was 1.2878–5.0942 and the p value was 7.3 × 10−3. The OR of genus Oscillospira was 6.1185, the 95% CI was 1.0705–34.9693 and the p value was 4.17 × 10−2. The OR of genus Ruminococcaceae (UCG013 group) was 0.2604, while the 95% CI was 0.0726–0.9344 and the p value was 3.9 × 10−2. The OR of genus Turicibacter was 3.1178, while the 95% CI was 1.0212–9.5186 and the p value was 4.59 × 10−2. The OR of order Bacteroidales was 0.0658, while the 95% CI was 0.0097–0.4479 and the p value was 5.4 × 10−3. The OR of order Bifidobacteriales was 0.2940, while the 95% CI was 0.0902–0.9583 and the p value was 4.23 × 10−2. These were causally associated with keloids. Moreover, the OR of genus Lachnospiraceae (NK4A136group) was 1.8274, while the 95% CI was 1.2810–2.6067 and the p value was 9 × 10−4. The OR of genus Peptococcus was 0.7292, while the 95% CI was 0.5679–0.9362 and the p value was 1.33 × 10−2. The OR of genus Sellimonas was 1.4268, the 95% CI was 1.0915–1.8651 and the p value was 9.3 × 10−3. The OR of genus Victivallis was 0.6990, while the 95% CI was 0.5474–0.8925 and the p value was 4.1 × 10−3. The OR > 1 means that the gut microbiome was positively associated with skin fibrosis while the OR < 1 means negatively associated with skin fibrosis. Besides, p < 0.05 means the result is statistically significant (Figure 2). These results indicate that the gut can affect skin disease, supporting the existence of the gut-skin axis (Figure 3). Refer to single nucleotide polymorphisms annotator (SNIPA), the genes associated with SNP of these bacterial trait were exhibited in Supplementary Table S4.
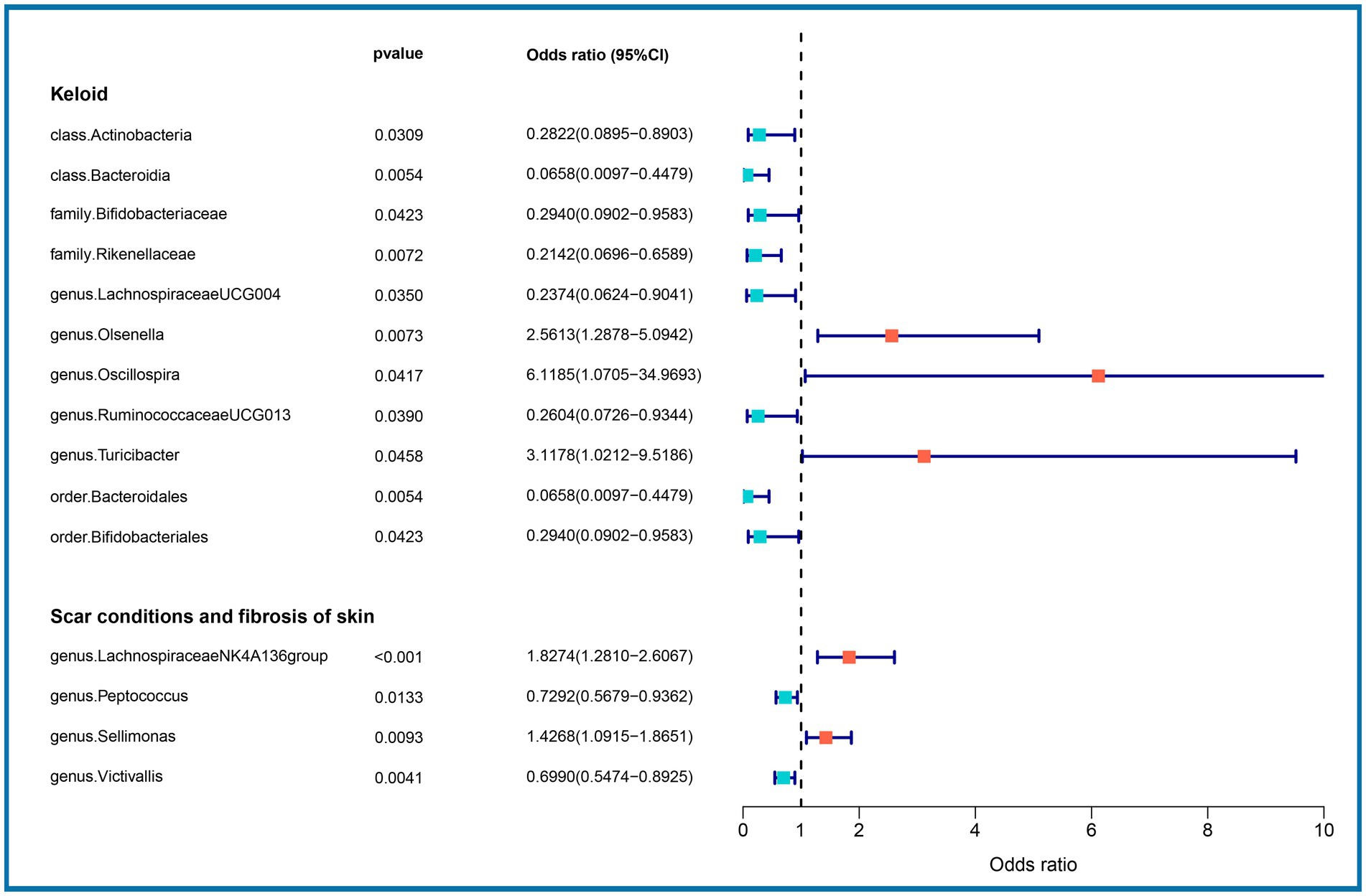
Figure 2. Association between gut microbiome (15 bacterial features of MiBioGen consortium) and skin fibrosis (GCST90044521 & GCST90044522) (p < 5 × 10−6). Color code = “#48D1CC,” “#FA8072” and “#FF0000.”
Figure 3. Existence of gut-skin axis supported by a two-sample Mendelian randomization (15 bacterial features of MiBioGen consortium). Color code = “#48D1CC,” “#FA8072” and “#FF0000.”
In addition, we used the traditional GWAS significance threshold (p < 5 × 10−8) as the selection criterion for the IVs. However few SNPs remained, and no significant results were observed in Supplementary Table S5.
In the reverse MR analysis, we found that there were not enough SNPs that were limited by the size of the gut microbiome. Using the selection criteria, no IVs were found in the skin fibrosis and gut microbiome data simultaneously.
3.3 Sensitivity analysis
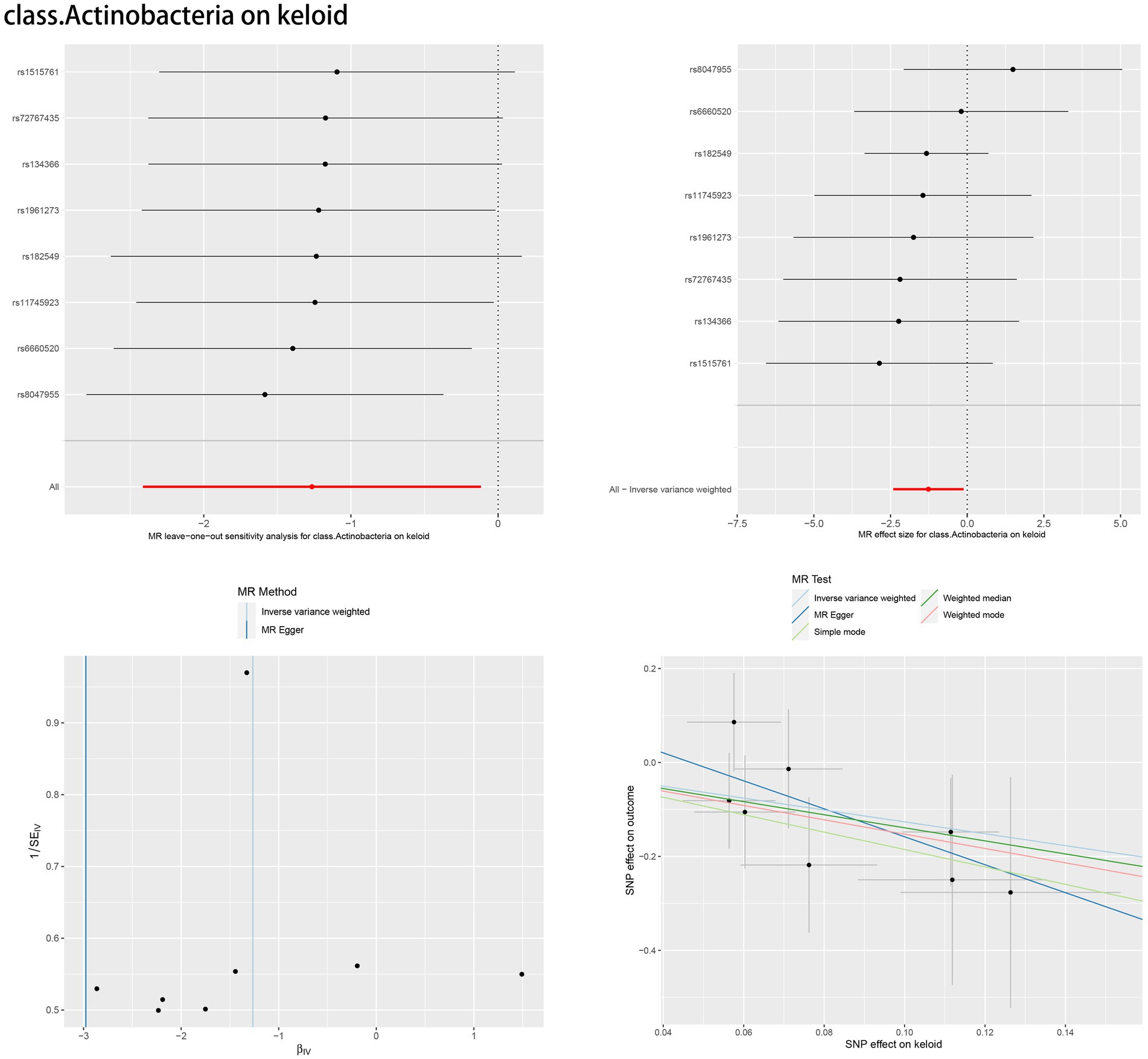
According to Cochran’s Q test and MR–Egger regression intercept analysis, there was no significant heterogeneity or directional horizontal pleiotropy. Furthermore, the results of MR-PRESSO showed no outliers (Table 2). Leave-one-out analysis was performed to verify the MR results. Forest plots, scatter plots and funnel plots for the causal association between the gut microbiome and skin fibrosis are also shown (Figure 4, Supplementary Figures S1–S14).
4 Discussion
In the occurrence of skin disease, researchers observe microbial dysbiosis both in the gut and skin (10, 28). For example, the microbiological composition of healthy skin is balanced with proper quantities of human and microbial antimicrobial peptides (AMPs). AMPs are a secretory product of epithelial and immune cells regulated by the Toll-like receptor (TLR) pathway. AMPs can maintain intestinal homeostasis and prevent the entry of gut bacteria into the bloodstream. When TLR pathways are inhibited, gut allostasis occurs. Dysbiosis of the gut microbiome influences the conversion of complex polysaccharides into vitamins and short-chain fatty acids (SCFAs) to improve the integrity of the gut barrier (10). They may also affect nitric oxide (NO) production and influence blood flow through the denitrification pathway (29). Impairment of intestinal epithelial cells leads to reduce production of AMPs and immunoglobulin A (IgA), which exacerbates gut microbial dysbiosis. As a result, gut bacteria enter the bloodstream through the gut wall and then reach the skin, causing dysbiosis of the skin microbiome and tissue inflammation (10, 11, 30).
The skin performs its functions and undergoes constant renewal during homeostasis. Gut microbial dysbiosis can cause skin allostasis (11, 31). For example, the dysbiosis of Firmicutes and Bacteroides alters the serological cytokine levels and promotes inflammation, leading to the development of acne vulgaris (31). This interaction is mainly mediated by the immune system (10, 32). Additionally, bacterial metabolites, such as butyrate, are related to the integrity of the epithelial barrier which engages to protect the skin (10, 30, 33, 34). Intact skin is crucial for maintaining homeostasis (35). This intricate interaction is known as the gut-skin axis. However, there are few reports on the role of gut-skin axis in skin fibrosis (28). Thus, we designed an MR analysis to determine the impact of the gut microbiome on the skin.
In this study, we evaluated the casual association between the abundance of specific bacterial signatures and the risk of skin fibrosis. Ten bacterial traits showed protective effects against skin fibrosis: Class Actinobacteria, Class Bacteroidia, family Bifidobacteriaceae, family Rikenellaceae, genus Lachnospiraceae (UCG004 group), genus Ruminococcaceae (UCG013 group), order Bacteroidales, order Bifidobacteriales, genus Peptococcus, and genus Victivallis. The genus Olsenella, genus Oscillospira, genus Turicibacter, genus Lachnospiraceae (NK4A136group), and genus Sellimonas are risk factors for skin fibrosis. In a randomised clinical pilot trial, participants consecutively consumed milk containing family Bifidobacteriaceae twice a day for 8 weeks. Compared with the pre-intake period, researchers found that the relative abundance levels of family Bifidobacteriaceae were significantly increased, and the skin condition of participants improved (36). Genus Lachnospiraceae (UCG004 group) acts as probiotics and increases the production of SCFAs including butyric acid for skin homeostasis (33, 37). Sodium butyrate dampens the profibrotic response induced by TGF-β1 in human dermal fibroblasts (38). A study reported higher abundance levels of genus Turicibacter in patients with systemic lupus erythematosus than in healthy individuals (39). These results are consistent with those of our study and support the existence of a gut-skin axis. Besides, Order Bacteroidales and genus Olsenella are associated with checkpoint blockade immunotherapy in melanoma (40, 41). The mechanism underlying other bacterial traits in the gut-skin axis remains to be elucidated.
There are currently no effective treatments for skin fibrosis (2). With the development of research on gut microbiome and skin fibrosis, we can elucidate the mechanisms of skin fibrosis and explore new therapeutic targets. These include the intake of probiotics, transplantation of the faecal microbiome, dietary modification, and drug–microbiome combination treatment (42–45). Relative abundances of gut microbiome were regularly detected in the suspected population. Before the disease onset, early screening and diagnosis can be performed by engineering the gut microbiome and restoring intestinal homeostasis. In the context of skin fibrosis, we propose novel and effective therapeutic strategies based on changes in specific gut microbiome abundance levels. Based on these levels, we can evaluate the effects of treatment and adjust therapeutic strategies for precise treatment. After treatment, diet can be leveraged to optimise SCFA production, maintain a healthy gut-skin axis, and reduce the risk of recurrence (46). To achieve this goal, we not only need appropriate data analysis to probe the causal association between the gut microbiome and skin fibrosis but also require a large number of rigorous clinical trials for validation.
Our study has some limitations. First, the results might not be entirely applicable to individuals of non-European descent because almost all samples are European, and only a few gut microbiome samples are from other races (17, 20, 21). Second, there might be a possibility of identifying false-positive findings owing to the absence of no Bonferroni correction. However, the results with IVW-derived p values less than 0.05 should also be treated cautiously (14). Third, the GWAS data for the gut microbiome were coordinated using by 16S rRNA gene sequencing, and the lowest taxonomic level was the genus. As a result, it is difficult to estimate the relationships between specific strains or species and skin fibrosis (17, 18). Additionally, we cannot exclude a reverse causal association between the gut microbiome and skin fibrosis because of the relatively small size of the gut microbiome. We hope that future GWAS data of the gut microbiome will be more sufficient, and we can perform analyses between a specific species and skin fibrosis in both European and non-European populations to reduce bias and improve universality.
5 Conclusion
The MR analysis revealed a causal association between the gut microbiome and skin fibrosis, supporting the existence of the gut-skin axis. When confronted with skin fibrosis, we also need to be cautious of the condition of the gut microbiome. Abundance of the gut microbiome can be a potential biomarker for early diagnosis and treatment of skin fibrosis. The role of the gut-skin axis in skin fibrosis requires further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
ZZ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. ZX: Conceptualization, Investigation, Software, Writing – original draft, Writing – review & editing. DL: Conceptualization, Investigation, Software, Writing – original draft, Writing – review & editing. YR: Writing – review & editing. ZH: Writing – review & editing. RY: Funding acquisition, Writing – review & editing. YD: Writing – review & editing. XC: Project administration, Supervision, Writing – review & editing. BT: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Nos. 81871565, 82072181, 82103706, and 82272273).
Acknowledgments
The authors express their gratitude to the participants and investigators of the MiBioGen consortium, the GWAS Catalog, PhenoScanner V2 and SNIPA.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1380938/full#supplementary-material.
References
1. Condorelli, AG, El Hachem, M, Zambruno, G, Nystrom, A, Candi, E, and Castiglia, D. Notch-Ing up knowledge on molecular mechanisms of skin fibrosis: focus on the multifaceted notch Signalling pathway. J Biomed Sci. (2021) 28:36. doi: 10.1186/s12929-021-00732-8
2. Griffin, MF, Huber, J, Evan, FJ, Quarto, N, and Longaker, MT. The role of Wnt signaling in skin fibrosis. Med Res Rev. (2022) 42:615–28. doi: 10.1002/med.21853
3. Lv, W, Ren, Y, Hou, K, Hu, W, Yi, Y, Xiong, M, et al. Epigenetic modification mechanisms involved in keloid: current status and prospect. Clin Epigenetics. (2020) 12:183. doi: 10.1186/s13148-020-00981-8
4. Frech, FS, Hernandez, L, Urbonas, R, Zaken, GA, Dreyfuss, I, and Nouri, K. Hypertrophic scars and keloids: advances in treatment and review of established therapies. Am J Clin Dermatol. (2023) 24:225–45. doi: 10.1007/s40257-022-00744-6
5. Huang, C, and Ogawa, R. Systemic factors that shape cutaneous pathological scarring. FASEB J. (2020) 34:13171–84. doi: 10.1096/fj.202001157R
6. Yachida, S, Mizutani, S, Shiroma, H, Shiba, S, Nakajima, T, Sakamoto, T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. (2019) 25:968–76. doi: 10.1038/s41591-019-0458-7
7. Sorboni, SG, Moghaddam, HS, Jafarzadeh-Esfehani, R, and Soleimanpour, S. A comprehensive review on the role of the gut microbiome in human neurological disorders. Clin Microbiol Rev. (2022) 35:e0033820. doi: 10.1128/CMR.00338-20
8. Long, Y, Tang, L, Zhou, Y, Zhao, S, and Zhu, H. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med. (2023) 21:66. doi: 10.1186/s12916-023-02761-6
9. Di Simone, N, Santamaria Ortiz, A, Specchia, M, Tersigni, C, Villa, P, Gasbarrini, A, et al. Recent insights on the maternal microbiota: impact on pregnancy outcomes. Front Immunol. (2020) 11:528202. doi: 10.3389/fimmu.2020.528202
10. Mahmud, MR, Akter, S, Tamanna, SK, Mazumder, L, Esti, IZ, Banerjee, S, et al. Impact of gut microbiome on skin health: gut-skin Axis observed through the lenses of therapeutics and skin diseases. Gut Microbes. (2022) 14:2096995. doi: 10.1080/19490976.2022.2096995
11. De Pessemier, B, Grine, L, Debaere, M, Maes, A, Paetzold, B, and Callewaert, C. Gut-skin axis: current knowledge of the interrelationship between microbial Dysbiosis and skin conditions. Microorganisms. (2021) 9:20353. doi: 10.3390/microorganisms9020353
12. Rinninella, E, Raoul, P, Cintoni, M, Franceschi, F, Miggiano, GAD, Gasbarrini, A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. (2019) 7:10014. doi: 10.3390/microorganisms7010014
13. Li, P, Wang, H, Guo, L, Gou, X, Chen, G, Lin, D, et al. Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. (2022) 20:443. doi: 10.1186/s12916-022-02657-x
14. Chen, X, Kong, J, Pan, J, Huang, K, Zhou, W, Diao, X, et al. Kidney damage causally affects the brain cortical structure: a Mendelian randomization study. EBioMedicine. (2021) 72:103592. doi: 10.1016/j.ebiom.2021.103592
15. Davey Smith, G, and Hemani, G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. (2014) 23:R89–98. doi: 10.1093/hmg/ddu328
16. Distler, JHW, Gyorfi, AH, Ramanujam, M, Whitfield, ML, Konigshoff, M, and Lafyatis, R. Shared and distinct mechanisms of fibrosis. Nat Rev Rheumatol. (2019) 15:705–30. doi: 10.1038/s41584-019-0322-7
17. Kurilshikov, A, Medina-Gomez, C, Bacigalupe, R, Radjabzadeh, D, Wang, J, Demirkan, A, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. (2021) 53:156–65. doi: 10.1038/s41588-020-00763-1
18. Meng, C, Deng, P, Miao, R, Tang, H, Li, Y, Wang, J, et al. Gut microbiome and risk of Ischaemic stroke: a comprehensive Mendelian randomization study. Eur J Prev Cardiol. (2023) 30:613–20. doi: 10.1093/eurjpc/zwad052
19. Sekula, P, Del Greco, MF, Pattaro, C, and Kottgen, A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. (2016) 27:3253–65. doi: 10.1681/ASN.2016010098
20. van der Velde, KJ, Imhann, F, Charbon, B, Pang, C, van Enckevort, D, Slofstra, M, et al. Molgenis research: advanced bioinformatics data software for non-Bioinformaticians. Bioinformatics. (2019) 35:1076–8. doi: 10.1093/bioinformatics/bty742
21. Jiang, L, Zheng, Z, Fang, H, and Yang, J. A generalized linear mixed model association tool for biobank-scale data. Nat Genet. (2021) 53:1616–21. doi: 10.1038/s41588-021-00954-4
22. Kamat, MA, Blackshaw, JA, Young, R, Surendran, P, Burgess, S, Danesh, J, et al. Phenoscanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics. (2019) 35:4851–3. doi: 10.1093/bioinformatics/btz469
23. Burgess, S, and Thompson, SGCollaboration CCG. Avoiding Bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036
24. Bowden, J, Davey Smith, G, and Burgess, S. Mendelian randomization with invalid instruments: effect estimation and Bias detection through egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
25. Greco, MF, Minelli, C, Sheehan, NA, and Thompson, JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. (2015) 34:2926–40. doi: 10.1002/sim.6522
26. Bowden, J, Del Greco, MF, Minelli, C, Zhao, Q, Lawlor, DA, Sheehan, NA, et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the Nome assumption. Int J Epidemiol. (2019) 48:728–42. doi: 10.1093/ije/dyy258
27. Hemani, G, Tilling, K, and Davey, SG. Orienting the causal relationship between imprecisely measured traits using Gwas summary data. PLoS Genet. (2017) 13:e1007081. doi: 10.1371/journal.pgen.1007081
28. Kim, S, Park, HJ, and Lee, SI. The microbiome in systemic sclerosis: pathophysiology and therapeutic potential. Int J Mol Sci. (2022) 23:16154. doi: 10.3390/ijms232416154
29. Koch, CD, Gladwin, MT, Freeman, BA, Lundberg, JO, Weitzberg, E, and Morris, A. Enterosalivary nitrate metabolism and the microbiome: intersection of microbial metabolism, nitric oxide and diet in cardiac and pulmonary vascular health. Free Radic Biol Med. (2017) 105:48–67. doi: 10.1016/j.freeradbiomed.2016.12.015
30. Lee, HJ, and Kim, M. Skin barrier function and the microbiome. Int J Mol Sci. (2022) 23:113071. doi: 10.3390/ijms232113071
31. Polkowska-Pruszynska, B, Gerkowicz, A, and Krasowska, D. The gut microbiome alterations in allergic and inflammatory skin diseases—an update. J Eur Acad Dermatol Venereol. (2020) 34:455–64. doi: 10.1111/jdv.15951
32. Salem, I, Ramser, A, Isham, N, and Ghannoum, MA. The gut microbiome as a major regulator of the gut-skin axis. Front Microbiol. (2018) 9:1459. doi: 10.3389/fmicb.2018.01459
33. Stec, A, Sikora, M, Maciejewska, M, Paralusz-Stec, K, Michalska, M, Sikorska, E, et al. Bacterial metabolites: a link between gut microbiota and dermatological diseases. Int J Mol Sci. (2023) 24:43494. doi: 10.3390/ijms24043494
34. Chen, J, and Vitetta, L. The role of butyrate in attenuating pathobiont-induced hyperinflammation. Immune Netw. (2020) 20:e15. doi: 10.4110/in.2020.20.e15
35. Akdis, CA . Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat Rev Immunol. (2021) 21:739–51. doi: 10.1038/s41577-021-00538-7
36. Nagino, T, Kaga, C, Kano, M, Masuoka, N, Anbe, M, Moriyama, K, et al. Effects of fermented soymilk with Lactobacillus Casei Shirota on skin condition and the gut microbiota: a randomised clinical pilot trial. Benef Microbes. (2018) 9:209–18. doi: 10.3920/BM2017.0091
37. Gao, Y, Hou, L, Hu, M, Li, D, Tian, Z, Wen, W, et al. Effects of Bacillus Subtilis Bsnk-5-fermented soymilk on the gut microbiota by in vitro fecal fermentation. Food Secur. (2022) 11:13501. doi: 10.3390/foods11213501
38. Park, HJ, Jeong, OY, Chun, SH, Cheon, YH, Kim, M, Kim, S, et al. Butyrate improves skin/lung fibrosis and intestinal Dysbiosis in bleomycin-induced mouse models. Int J Mol Sci. (2021) 22:52765. doi: 10.3390/ijms22052765
39. Zhao, Y, Zhang, B, Liu, C, and Ren, G. Method for accurate diagnose of lupus erythematosus skin lesions based on microbial Rdna sequencing. Saudi J Biol Sci. (2020) 27:2111–5. doi: 10.1016/j.sjbs.2020.05.035
40. Vetizou, M, Pitt, JM, Daillere, R, Lepage, P, Waldschmitt, N, Flament, C, et al. Anticancer immunotherapy by Ctla-4 blockade relies on the gut microbiota. Science. (2015) 350:1079–84. doi: 10.1126/science.aad1329
41. Mager, LF, Burkhard, R, Pett, N, Cooke, NCA, Brown, K, Ramay, H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. (2020) 369:1481–9. doi: 10.1126/science.abc3421
42. Raheem, A, Liang, L, Zhang, G, and Cui, S. Modulatory effects of probiotics during pathogenic infections with emphasis on immune regulation. Front Immunol. (2021) 12:616713. doi: 10.3389/fimmu.2021.616713
43. Fretheim, H, Chung, BK, Didriksen, H, Baekkevold, ES, Midtvedt, O, Brunborg, C, et al. Fecal microbiota transplantation in systemic sclerosis: a double-blind, placebo-controlled randomized pilot trial. PLoS One. (2020) 15:e0232739. doi: 10.1371/journal.pone.0232739
44. Fritsch, J, Garces, L, Quintero, MA, Pignac-Kobinger, J, Santander, AM, Fernandez, I, et al. Low-fat, high-Fiber diet reduces markers of inflammation and Dysbiosis and improves quality of life in patients with ulcerative colitis. Clin Gastroenterol Hepatol. (2021) 19:1189–1199.e30. doi: 10.1016/j.cgh.2020.05.026
45. Zhang, X, Han, Y, Huang, W, Jin, M, and Gao, Z. The influence of the gut microbiota on the bioavailability of Oral drugs. Acta Pharm Sin B. (2021) 11:1789–812. doi: 10.1016/j.apsb.2020.09.013
Keywords: gut microbiome, skin fibrosis, Mendelian randomization, gut-skin axis, single nucleotide polymorphisms
Citation: Zhao Z, Xu Z, Lv D, Rong Y, Hu Z, Yin R, Dong Y, Cao X and Tang B (2024) Impact of the gut microbiome on skin fibrosis: a Mendelian randomization study. Front. Med. 11:1380938. doi: 10.3389/fmed.2024.1380938
Edited by:
Monica Barone, University of Bologna, ItalyReviewed by:
Sankarasubramanian Jagadesan, University of Nebraska Medical Center, United StatesMangala Hegde, Indian Institute of Technology Guwahati, India
Mudasir Rashid, Howard University Hospital, United States
Copyright © 2024 Zhao, Xu, Lv, Rong, Hu, Yin, Dong, Cao and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoling Cao, Q2FveGxpbmczQG1haWwuc3lzdS5lZHUuY24=; Bing Tang, dGFuZ2JpbmdAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
 Zirui Zhao
Zirui Zhao Zhongye Xu
Zhongye Xu Dongming Lv
Dongming Lv Yanchao Rong1
Yanchao Rong1 Zhicheng Hu
Zhicheng Hu Yunxian Dong
Yunxian Dong Bing Tang
Bing Tang