- Department of Digestive Diseases, Xijing Hospital, Fourth Military Medical University, Xi’an, China
Background: The etiological factors of Cholestatic Liver Diseases especially primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC) are not fully illustrated. It has been reported in previous observational studies that gut microbiota are associated with cholestatic liver diseases. However, there is uncertainty regarding the causality of this association. By using Mendelian randomization, this study aimed to examine the causal impact of gut microbiota on cholestatic liver diseases.
Methods: From large-scale genome-wide association studies, genetic instruments for each gut microbiota taxa as well as primary biliary cholangitis and primary sclerosing cholangitis were developed. Subsequently, we conducted a two-sample Mendelian randomization analysis, supplemented by multiple post hoc sensitivity analyses. Additionally, we performed reverse MR analyses to investigate the possibility of the reverse causal association.
Result: This two-sample MR study indicated that the order Bacillales, family Peptostreptococcaceae, family Ruminococcaceae, genus Anaerotruncu was associated with a decreased risk of developing PBC, and that order Selenomonadales, family Bifidobacteriaceae may be factors that increase the risk of PBC. On the other hand, we also identified order Selenomonadales, family Rhodospirillaceae, and genus RuminococcaceaeUCG013 were positively associated with PSC. The order Actinomycetales, family Actinomycetaceae, genus Actinomyces, genus Alloprevotella, genus Barnesiella, and genus Peptococcus were found negative associations with the risk of PSC. The reverse MR analysis demonstrated no statistically significant relationship between PBC, PSC and these specific gut microbial taxa.
Conclusion: Our findings offered novel evidence that the abundance of particular bacteria contributes to the risk of PBC and PSC, which may contribute to more effective approaches to PBC and PSC therapy and prevention.
1 Introduction
Cholestatic liver disease (CLD) refers to a group of disorders in which bile synthesis, secretion, and excretion are compromised for a variety of reasons (1). CLD dominantly includes primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC). PBC, a chronic cholestatic liver disease, is hallmarked by non-suppurative inflammation within the small intrahepatic bile duct (2). PSC, a rare cholestatic liver disease, could result in bile duct fibrosis and strictures. In contrast to PBC, which has a female predominance, the majority of PSC patients are male (3). Up to 80% of PSC patients also suffer from IBD, indicating the involvement of the gut-liver axis in PSC (4). Patients with PBC can effectively control the disease by taking medications such as Ursodeoxycholic Acid, Bezafibrate and Fenofibrate (5–7). However, there is currently no satisfactory treatment for PSC.
Though the exact mechanism underlying PBC and PSC is still not fully illustrated, it is reported that genetics, environment, immune factors, gut microbiota, and individual susceptibility may all contribute to the development of these diseases (4, 8, 9).
Environmental factors are thought to cause PBC and PSC in individuals who are genetically predisposed, resulting in a loss of tolerance to self-antigens. Molecules derived from microbiota can activate the immune system and lead to autoimmune inflammation (10). According to recent research, gut microbiota dysbiosis can affect the immune system leading to autoimmune diseases such as celiac disease, inflammatory bowel disease, and cholestatic liver diseases (11–13). In numerous studies focused on the gut-liver axis, a link has been established between gut microbiota dysbiosis and PBC and PSC pathophysiology (14–16). Research has indicated a significant reduction in the abundance of microbiota in individuals with PBC and PSC compared to healthy controls (17, 18). However, some bacterial genera, such as Haemophilus, Veillonella, Clostridium, and Bifidobacterium, are increased in PBC patients compared to healthy controls (19, 20), while Lactobacillus, Streptococcus, and Veillonella proportions are higher in PSC patients than healthy controls (21).
Nevertheless, it is important to note that observational studies are often influenced by confounding factors. Moreover, the aforementioned studies have examined different populations with varying dietary habits. Therefore, these cross-sectional studies do not allow for definitive conclusions to be drawn.
Mendelian randomization (MR) is an innovative approach to investigate the association between an exposure and a noteworthy outcome (22). Alleles are randomly allocated, according to Mendelian’s laws of inheritance, and genotypes are fixed at conception. Thus, confounders and reverse causality are unlikely to affect the causal relationship. MR analysis exploited common genetic variations to represent a modifiable environmental exposure, which has become widely used to investigate potential causal relationships between environmental exposures and outcomes. In two-sample MR analysis, single-nucleotide polymorphisms (SNPs) can serve as instrumental variables (IVs) to investigate casual associations between exposures and outcomes (23).
In summary, the causality of the associations between the gut microbiota and PBC and PSC remains inconclusive. In this study, we conducted a two-sample Mendelian randomization analysis using comprehensive summary statistics from large-scale genome-wide association studies (GWAS) of gut microbiota (GM), PBC and PSC to conduct this question.
2 Materials and methods
2.1 Study design
As shown in Figure 1, we implemented a two-sample MR to explore casual associations between CLD (specifically PBC and PSC) and gut microbiota.
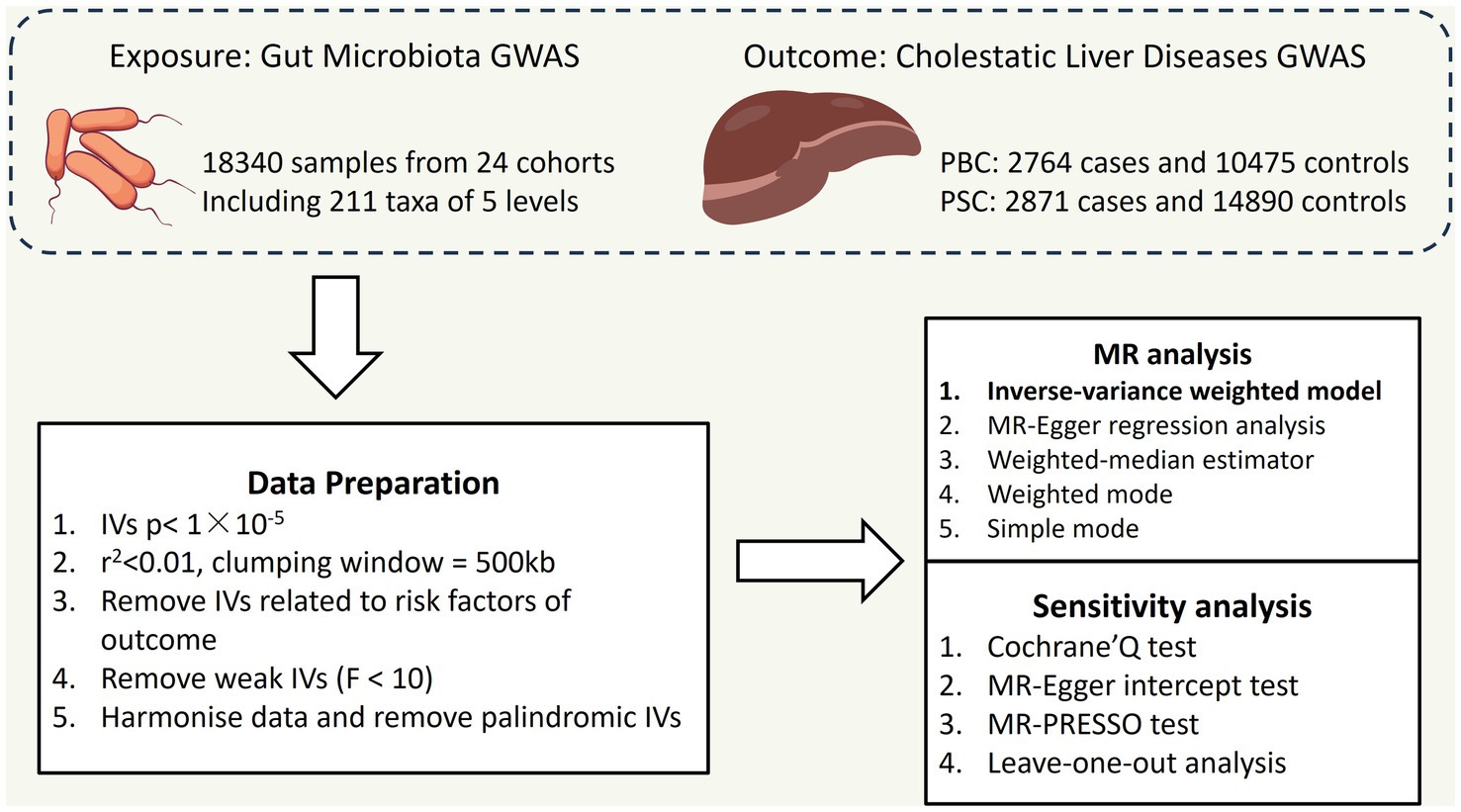
Figure 1. Overall flow chart of this study PBC: primary biliary cholangitis; PSC: primary sclerosing cholangitis; IVs: instrumental variables.
2.2 Data source for exposure
MiBioGen consortium was formed to study the role of human genes in gut microbiota composition. Our study used the latest gut microbiota GWAS data extracted from 18,340 individuals. In this GWAS study, genetic variants associated with 211 GM taxa (9 phyla, 16 classes, 35 families, and 131 genera) were identified. Here is a link to download GWAS summary statistics for GMs.1
2.3 Data source for the outcomes
In two European cohorts, we obtained GWAS summary statistics for PBC and PSC. The PBC GWAS dataset comprises 10,475 controls and 2,764 cases (24), and the PSC GWAS dataset contains 2,871 cases and 14,890 controls (25).
2.4 Identification of IVs
To confirm the causal association of PBC and PSC with the gut microbiota, suitable IVs were chosen by implementing the subsequent quality control measures.
Firstly, we selected the IVs that are strongly correlated with GM taxa. As the initial threshold (p < 5 × 10−8) did not yield a sufficient number of IVS, we opted for a relatively lenient threshold (p < 1 × 10−5) to ensure enough IVs for obtaining robust results. Additionally, linkage disequilibrium (LD) correlation coefficient was set to r2 < 0. 01 and clumping window >500 kb to mitigate LD. Then, palindromic SNPs were removed from the IVs. Lastly, to evaluate weak instrumental bias, we calculated the F statistic of IVs. An F-statistic greater than 10 in MR analyses indicated no weak instrumental bias.
2.5 Statistical methods
Defined as the primary MR method for inferring causality, the inverse variance weighted (IVW) method is an extension of the Wald ratio method (26). In addition to IVW, we also applied four other MR methods: simple mode, weighted median, MR-Egger, and weighted mode. MR Egger’s method could also be used to detect directional pleiotropy (27).
We also conducted several sensitivity analyses to validate the stability of the causal association. We first performed Cochrane’s Q test to evaluate the heterogeneity across all selected SNPs. Additionally, we used MR-PRESSO and the MR-Egger intercept test for detection purposes. To assess the robustness of our results, we performed a leave-one-out analysis. All p < 0.05 was thought to be significant. Reverse MR analysis was employed to confirm the causal direction. It followed similar methods as forward MR, but PBC, PSC was regarded as the exposures, and we extracted SNPs associated with PBC, PSC as the IVs (p < 5 × 10–8).
We performed all analyses in this study using R software (version 4.2.1). We utilized R packages including the “ggplot2,” “TwoSampleMR,” and “MRPRESSO” for our MR study.
3 Results
3.1 Genetic IVs for gut microbiome
There were 2,934 SNPs as IVs linked to 211 GM taxa (9 phyla, 16 classes, 35 families, and 131 genera) in our MR study. The F-values of the selected SNPs ranged from 14.59 to 88.43, indicating a lower risk of weak instrument bias. The detailed information of all SNPs is shown in Supplementary Table 1.
3.2 PBC
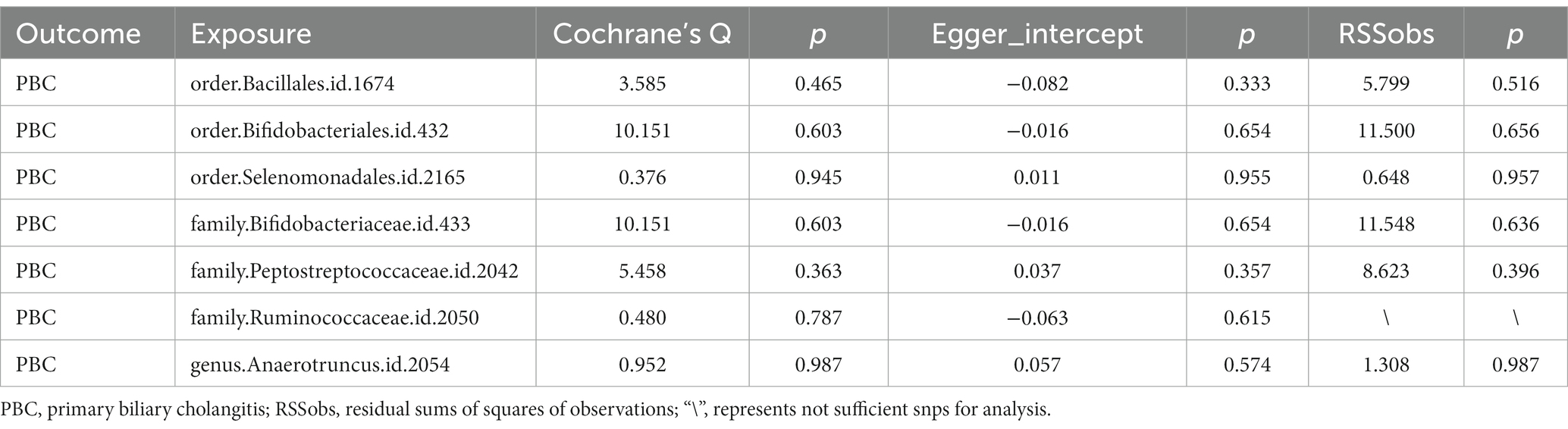
Seven bacterial taxa were identified to be associated with PBC. It was determined that two of these taxa may increase the risk of PBC, specifically containing the order Selenomonadales (IVW OR = 2.13, 95% CI 1.10–4.14, p = 0.026), family Bifidobacteriaceae (IVW OR = 1.40, 95% CI 1.06–1.85, p = 0.019).
On the contrary, 4 taxa including order Bacillales (IVW OR = 0.75, 95%CI 0.58–0.95, p = 0.035), family Peptostreptococcaceae (IVW OR = 0.65, 95% CI 0.43–0.98, p = 0.037), family Ruminococcaceae (IVW OR 0.33, 95% CI 0.15–0.72, p = 0.005) and genus Anaerotruncu (IVW OR 0.59, 95% CI 0.37–0.95, p = 0.28) are identified as having negative associations with PBC, and may causally reduce the risk of PBC (Figure 2). Other results are shown in Supplementary Table 2.
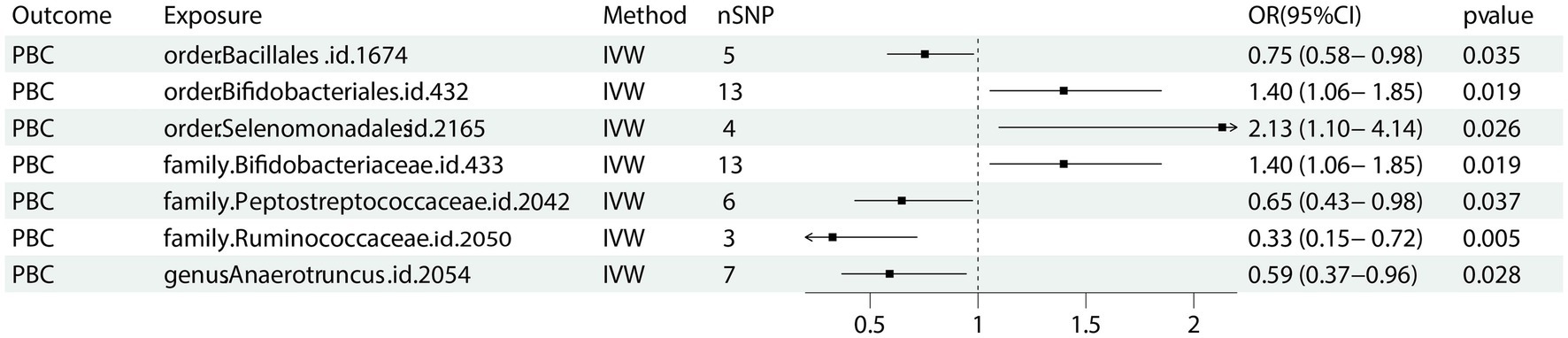
Figure 2. Forest plot of GM taxa associated with PBC identified by IVW method. PBC: primary biliary cholangitis; nSNP, number of the single nucleotide polymorphisms; IVW: inverse variance weighted method.
The Cochrane’s Q test, the MR-Egger intercept test, and the MR-PRESSO test did not indicate any obvious heterogeneity in selected SNPs (Table 1) and showed that there is no pleiotropy or outliers (p > 0.05). Examination of forest plots and scatter plots was conducted (Supplementary Figures 1, 2). Finally, the leave-one-out method confirms our main results’ robustness (Figure 3).
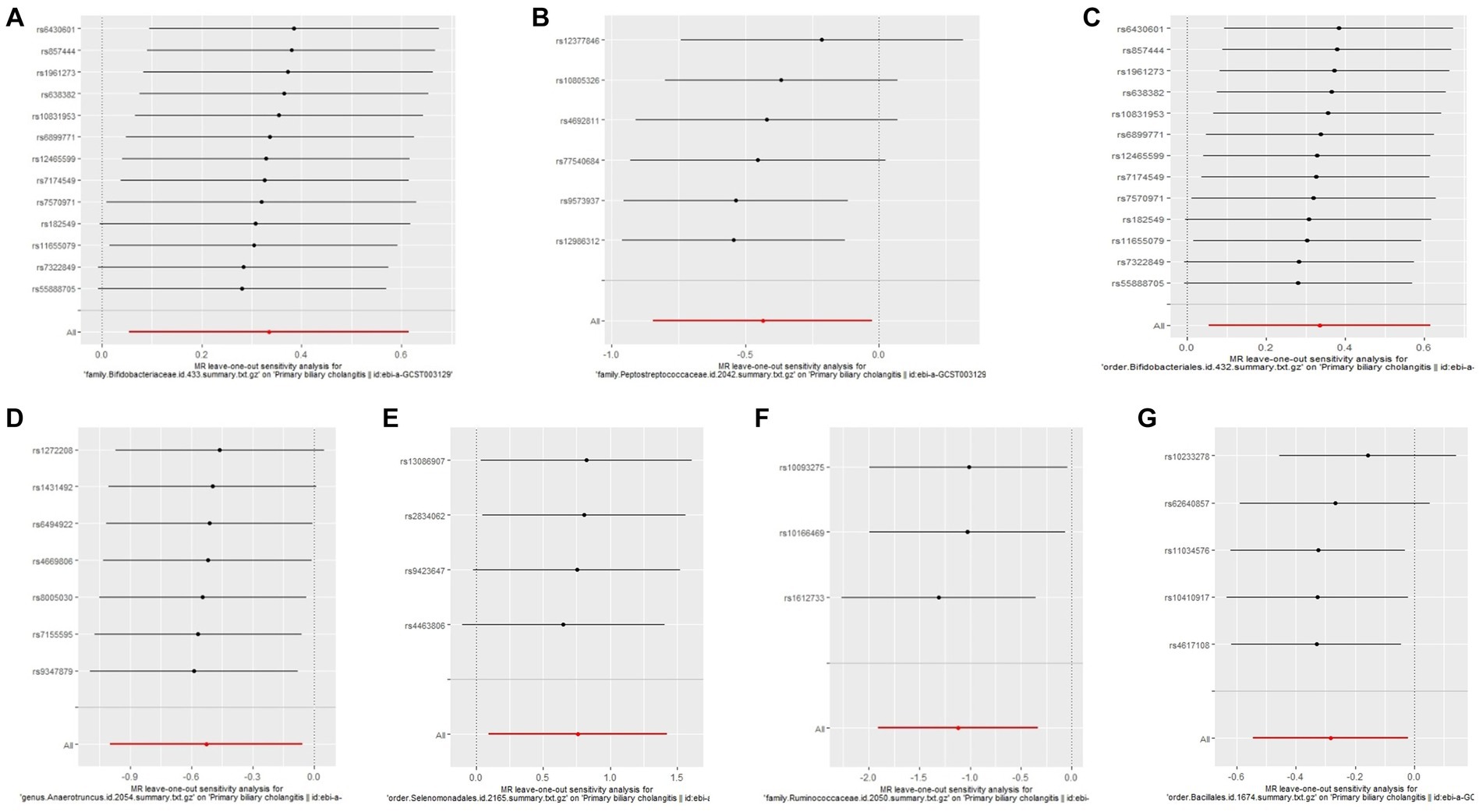
Figure 3. Leave one out analysis of the MR results of GM taxa associated with PBC. (A) family. Bifidobacteriaceae.id.433, (B) family.Peptostreptococcaceae.id.2042, (C) order.Bifidobacteriales.id.432, (D) genus.Anaerotruncus.id.2054, (E) order.Selenomonadales.id.2165, (F) family.Ruminococcaceae.id.2050, (G) order.Bacillales.id.1674.
3.3 PSC
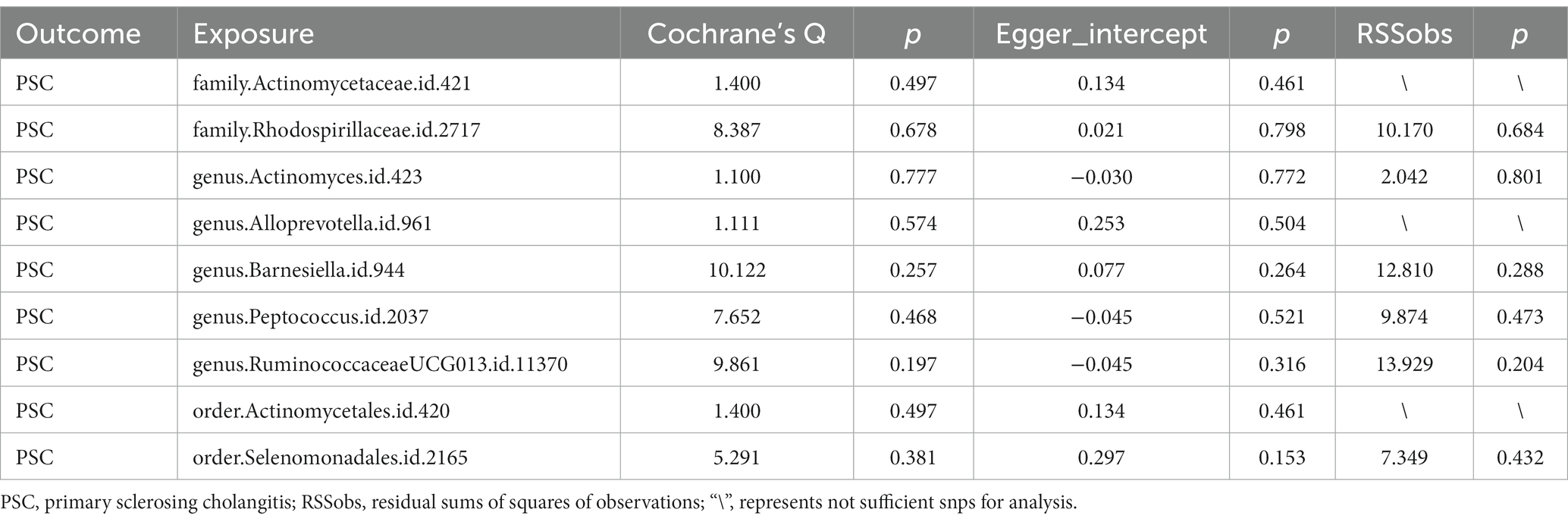
Nine bacterial traits were found to be associated with PSC, specifically order Selenomonadales (IVW OR 1.72, 95% CI 1.00–2.93, p = 0.048), family Rhodospirillaceae (IVW OR 1.30, 95% CI 1.01–2.68, p = 0.042) and genus RuminococcaceaeUCG013 (IVW OR 1.63, 95% CI 1.04–2.57, p = 0.034) were positively causally associated with PSC.
As for order Actinomycetales (IVW OR 0.59, 95% CI 0.36–0.98, p = 0.042), family Actinomycetaceae (IVW OR 1.72, 95% CI 0.36–0.98 p = 0.042), genus Actinomyces (IVW OR 0.62, 95% CI 0.42–0.90, p = 0.012), genus alloprevotella (IVW OR 0.68, 95% CI 0.50–0.94, p = 0.018), genus Barnesiella (IVW OR 0.63, 95% CI 0.42–0.95, p = 0.027) as well as genus Peptococcus (IVW OR 0.79, 95% CI 0.63–0.99, p = 0.041) were found negative association with the risk of PSC (Figure 4) Other results are shown in Supplementary Table 3.
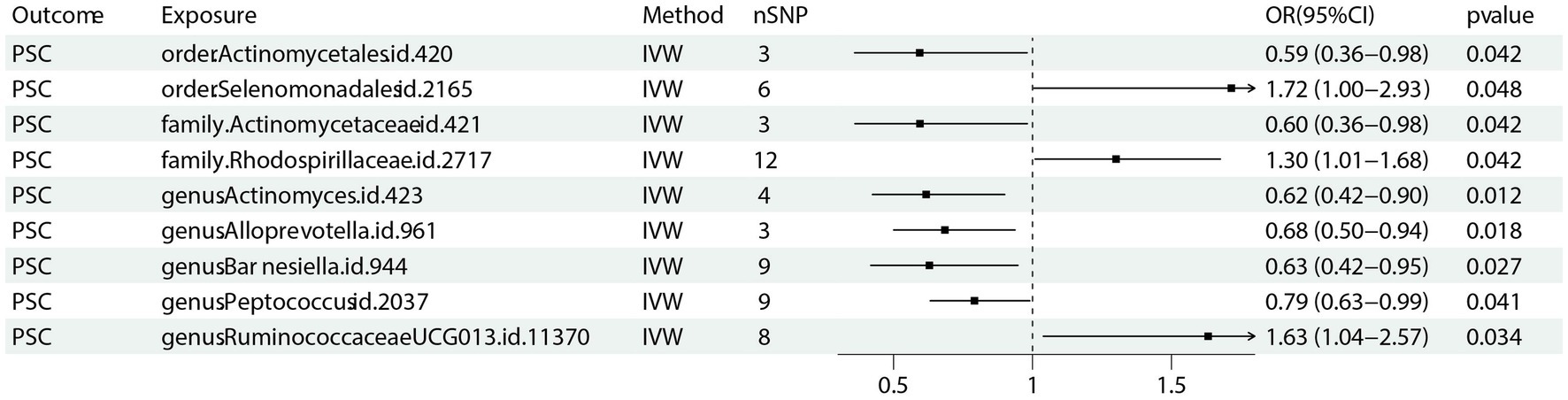
Figure 4. Forest plot of GM taxa associated with PSC identified by IVW method. PSC: primary sclerosing cholangitis; nSNP, number of the single nucleotide polymorphisms; IVW: inverse variance weighted method.
Through Cochran’s Q test, we detected no heterogeneity (p > 0.05). All p-values of the MR-PRESSO test and the MR-egger interpret test were > 0.05, indicating the absence of outliers or pleiotropy (Table 2). We then examined the forest plot and scatter plot (Supplementary Figures 3, 4). Finally, the robustness of our primary findings was validated using the leave-one-out method (Figure 5).
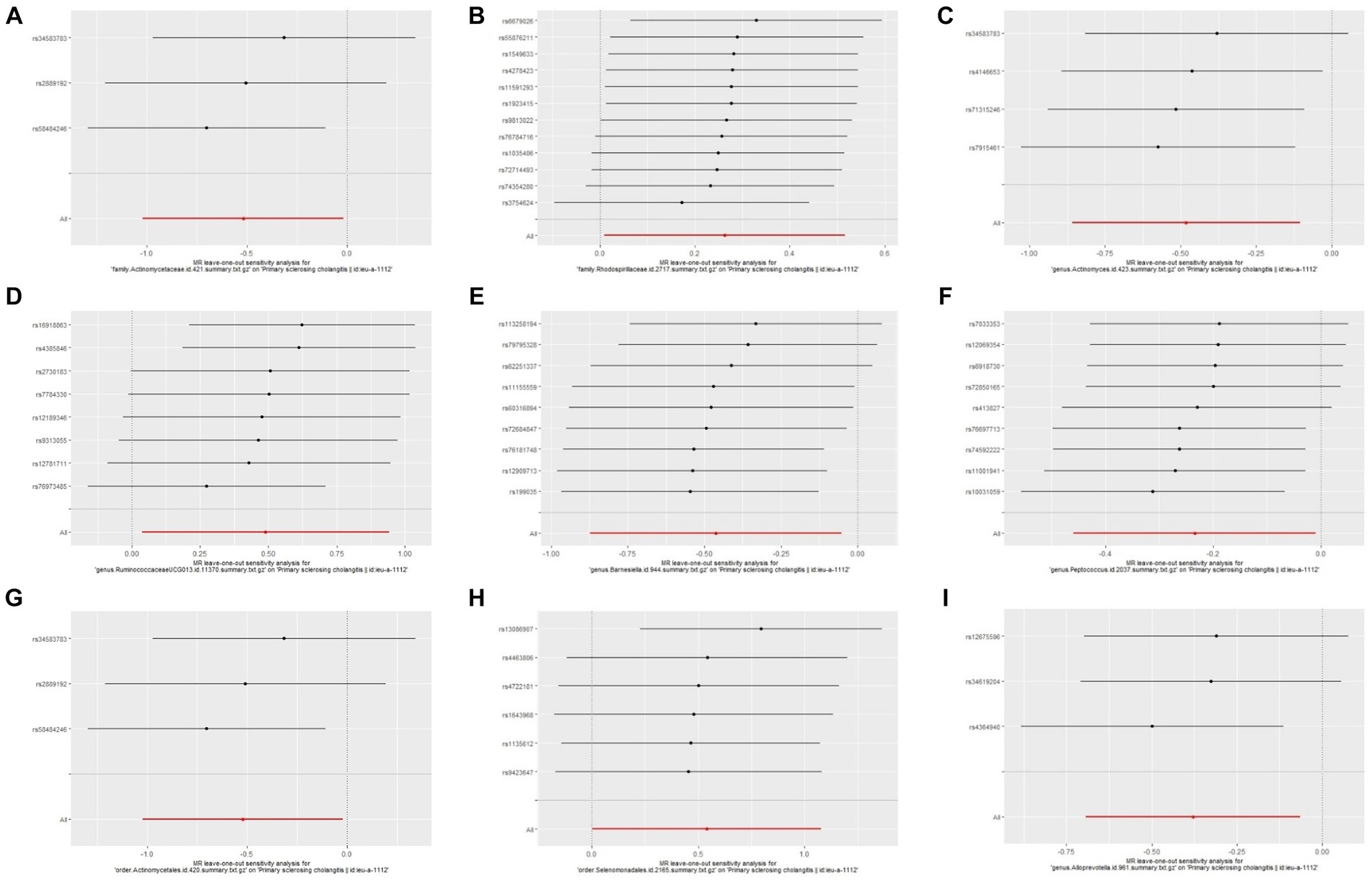
Figure 5. Leave one out analysis of the MR results of GM taxa associated with PSC. (A) family.Actinomycetaceae.id.421, (B) family.Rhodospirillaceae.id.2717, (C) genus.Actinomyces.id.423, (D) genus.RuminococcaceaeUCG013.id.11370, (E) genus.Barnesiella.id.944, (F) genus.Peptococcus.id.2037, (G) order.Actinomycetales.id.420, (H) order.Selenomonadales.id.2165, (I) genus.Alloprevotella.id.961.
3.4 Reverse Mendelian randomization
A reverse MR analysis was utilized through the IVW method to explore the potential causal association between PBC, PSC, and these specific gut microbial taxa. The data presented in Supplementary Table 4 did not show any significant reverse causal association between PBC, PSC and these specific gut microbial taxa.
4 Discussion
In this two-sample Mendelian Randomization study, we found that 7 bacterial taxa were associated with PBC, and 9 bacterial taxa were associated with PSC.
Previous article reported (28) that Order Selenomonadales, Order Bifidobacteriales, Genus Lachnospiraceae_UCG_004, Family Peptostreptococcaceae, and Family Ruminococcaceae were related to PBC, which have been validated in our study. In addition, our results also found that order Bacillales and genus Anaerotruncu were potential risk factor for PBC. These results expand previous research and provide a foundation for further research on PBC, and we also compared the differences between PBC and PSC.
We identified order Selenomonadales as a protective factor in both PBC and PSC. Selenomonadales are anaerobic bacteria that typically have a curved or bent shape. They contribute to the formation and function of complex gut microbiota. These bacteria can utilize various carbon sources such as glucose, lactose, and cellulose to produce organic acids and gases through fermentation (29).
It was also found that the family Peptostreptococcaceae is a protective factor for PBC, while the genus Peptococcus has a similar protective effect on PSC. These results indicate that certain gut microbiota might play a common role in the occurrence of PBC and PSC.
The family Peptostreptococcaceae belongs to the phylum Firmicutes (30). This family includes the genera Peptostreptococcus, Finegoldia, and Anaerococcus. Bacteria in the family Peptostreptococcaceae are typically anaerobic organisms and can be found in the digestive tract, skin, and other body surfaces of humans and animals, as well as in soil and water environments.
There are also some bacteria that play different roles in PBC and PSC. For example, the family Ruminococcaceae plays a protective role in PBC, while the genus RuminococcaceaeUCG013 increases the risk of PSC. These results suggest that different bacteria within the same family may also have different effects.
Family Ruminococcaceae family are usually anaerobic organisms. These bacteria are able to utilize cellulose and other components of plant cell walls to produce organic acids and gases through fermentation, providing energy and nutrients to the host. These bacteria in the human gut are associated with intestinal health and metabolism (31).
Due to Mendelian randomization analysis using GM GWAS data, different levels of bacteria, such as genus, family, and order, may extract the same SNPs, which results in different levels of bacteria having the same effect. For example, order Bifidobacteriales and family Bifidobacteriaceae both extract 13 SNPs, and the results indicated the same protective effect on PBC. What’s more, although the number of extracted SNPs differed, order Actinomycetales, family Actinomycetaceae, and genus Actinomyces are all having protective effects on PSC.
Bifidobacterium is generally regarded as probiotics and possesses numerous advantages, such as facilitating food digestion, synthesizing vitamins, and augmenting immune system functionality (32). However, our findings provide evidence that the family Bifidobacteriaceae may elevate the risk of PBC, which is consistence with previous research which identified Bifidobacterium is increased in PBC patients (20).
Actinomycetales are widely present in natural environments, including soil, water bodies, and plant surfaces. They can also survive in the bodies of humans and other animals, such as in the oral cavity, intestines, and skin. Actinomycetales have the ability to produce some important enzymes and bioactive substances (33). For example, they can produce cellulases, proteases, and acid phosphatases, as well as antioxidants and anti-tumor substances.
We found that specific bacterial features are causally related to the risk of PBC and PSC. The underlying mechanism of the influence of bacteria features on PBC and PSC has been extensively studied. Metabolites, especially short-chain fatty acids (SCFAs), are one of the most crucial factors (34, 35), with butyric acid, propionic acid, and acetic acid being the predominant constituents. A critical function of SCFAs is to act as signaling molecules that regulate the immune system, cellular growth, and metabolic activity of the host (36).
Butyrate, an essential metabolite derived from the GM, contributes to maintaining the integrity gut barrier by supplying energy to colonic epithelial cells. Additionally, it modulates genes associated with the circadian clock, thereby performing its anti-inflammatory function (37). Moreover, it can modulate T cell proliferation and regulate the activation of B cells that produce IL-10 and/or IL-17 (38). These cytokines will aggravate the inflammation of bile duct cells in patients with PBC and PSC and worsen bile stasis. There is also evidence that bacterial-derived peptides induce CD8+ T cell clonal expansion (39, 40), which are the main effector cells causing bile duct damage in patients with PBC and PSC (41).
Bile acids also play an important role in the pathogenesis of PBC and PSC. Bile acids not only play a role in digesting food, but also serve as important messengers for liver and intestinal communication.
Firstly, bile acids have a direct bactericidal effect and have an inhibitory effect on the growth of gut microbiota. In addition, bile acids regulate the composition of gut microbiota by regulating farnesoid X receptors. Finally, bile acids can also serve as raw materials for gut microbiota to promote the proliferation of some bacteria (42, 43). Dysbiosis of the GM, characterized by a decrease in microbial diversity and changes in specific bacterial species, may be linked to an elevated risk of developing PSC and PBC (44–46).
Based on our Mendelian randomization study, we have found a reasonable correlation between the gut microbiota taxa, PBC, and PSC. Some probiotics and their metabolites can restore the ecological balance of gut microbiota, repair the intestinal mucosal barrier and regulate systemic immune function. Our research can provide a foundation for further research (47). Future studies are required to better understand the mechanisms behind this prevalent disease and identify potential therapeutic targets.
Our study has some limitations: (a) Since the normally used threshold (p < 5 × 10−8) did not yield enough IVs, we set a lenient threshold (p < 1 × 10−5). (b) It’s difficult to determine whether some specific species are related to the outcome since most GM studies using 16S rRNA permit resolution at the genus level. (c) We refrained from conducting multiple corrections in our study. However, it is worth mentioning that the rigorous application of a multiple-testing correction may be excessively conservative and could potentially miss out on partially potential GM taxa that are causally correlated to CLD. Hence, we made the decision not to incorporate multiple correlations. Furthermore, it is important to note that the Bonferroni correction has the possibility of generating false negative results.
5 Conclusion
Our findings offer novel evidence that supports the causal influence of particular bacterial abundance on the risk of PBC and PSC. The GM is anticipated to be a promising treatment and prevention target for PBC and PSC.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
JY: Data curation, Investigation, Writing – original draft. GM: Investigation, Methodology, Writing – original draft. KW: Software, Writing – original draft. HY: Software, Writing – original draft. SJ: Data curation, Methodology, Writing – original draft. QF: Methodology, Validation, Writing – original draft. XZ: Supervision, Writing – review & editing. GG: Supervision, Visualization, Writing – review & editing. YH: Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (No. 82270551 and 2023KJXX-026).
Acknowledgments
The authors want to thank all researchers who shared publicly available GWAS summary data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2024.1342119/full#supplementary-material
Footnotes
References
1. Ibrahim, SH, Kamath, BM, Loomes, KM, and Karpen, SJ. Cholestatic liver diseases of genetic etiology: advances and controversies. Hepatol Baltim Md. (2022) 75:1627–46. doi: 10.1002/hep.32437
2. Lindor, KD, Bowlus, CL, Boyer, J, Levy, C, and Mayo, M. Primary biliary cholangitis: 2018 practice guidance from the American Association for the Study of Liver Diseases. Hepatology. (2019) 69:394–19. doi: 10.1002/hep.30145
3. Eaton, JE, Talwalkar, JA, Lazaridis, KN, Gores, GJ, and Lindor, KD. Pathogenesis of primary sclerosing cholangitis and advances in diagnosis and management. Gastroenterology. (2013) 145:521–36. doi: 10.1053/j.gastro.2013.06.052
4. Karlsen, TH, Folseraas, T, Thorburn, D, and Vesterhus, M. Primary sclerosing cholangitis - a comprehensive review. J Hepatol. (2017) 67:1298–23. doi: 10.1016/j.jhep.2017.07.022
5. Levy, C, Kendrick, S, Bowlus, CL, Tanaka, A, Jones, D, Kremer, AE, et al. GLIMMER: a randomized phase 2b dose-ranging trial of Linerixibat in primary biliary cholangitis patients with pruritus. Clin Gastroenterol Hepatol. (2022) 21:10217:1902–1912.e13. doi: 10.1016/j.cgh.2022.10.032
6. Bowlus, CL, Galambos, MR, Aspinall, RJ, Hirschfield, GM, Jones, DEJ, Dörffel, Y, et al. A phase II, randomized, open-label, 52-week study of seladelpar in patients with primary biliary cholangitis. J Hepatol. (2022) 77:353–64. doi: 10.1016/j.jhep.2022.02.033
7. de Veer, RC, van Hooff, MC, Corpechot, C, Thorburn, D, Invernizzi, P, Lammers, WJ, et al. Ursodeoxycholic acid treatment-induced GLOBE score changes are associated with liver transplantation-free survival in patients with primary biliary cholangitis. Am J Gastroenterol. (2022) 118:1196–03. doi: 10.14309/ajg.0000000000002128
8. Leung, KK, Deeb, M, and Hirschfield, GM. Review article: pathophysiology and management of primary biliary cholangitis. Aliment Pharmacol Ther. (2020) 52:1150–64. doi: 10.1111/apt.16023
9. Hirschfield, GM, Beuers, U, Corpechot, C, Invernizzi, P, Jones, D, Marzioni, M, et al. EASL clinical practice guidelines: the diagnosis and management of patients with primary biliary cholangitis. J Hepatol. (2017) 67:145–72. doi: 10.1016/j.jhep.2017.03.022
10. Hov, JR, and Karlsen, TH. The microbiota and the gut-liver axis in primary sclerosing cholangitis. Nat Rev Gastroenterol Hepatol. (2023) 20:135–54. doi: 10.1038/s41575-022-00690-y
11. Zeng, Y, Cao, S, and Yang, H. Roles of gut microbiome in epilepsy risk: a Mendelian randomization study. Front Microbiol. (2023) 14:1115014. doi: 10.3389/fmicb.2023.1115014
12. Li, T, Feng, Y, Wang, C, Shi, T, Abudurexiti, A, Zhang, M, et al. Assessment of causal associations among gut microbiota, metabolites, and celiac disease: a bidirectional Mendelian randomization study. Front Microbiol. (2023) 14:1087622. doi: 10.3389/fmicb.2023.1087622
13. Liu, B, Ye, D, Yang, H, Song, J, Sun, X, Mao, Y, et al. Two-sample Mendelian randomization analysis investigates causal associations between gut microbial genera and inflammatory bowel disease, and specificity causal associations in ulcerative colitis or Crohn’s disease. Front Immunol. (2022) 13:921546. doi: 10.3389/fimmu.2022.921546
14. Fuchs, CD, and Trauner, M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat Rev Gastroenterol Hepatol. (2022) 19:432–50. doi: 10.1038/s41575-021-00566-7
15. Ley, RE, Peterson, DA, and Gordon, JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. (2006) 124:837–48. doi: 10.1016/j.cell.2006.02.017
16. González-Regueiro, JA, Moreno-Castañeda, L, Uribe, M, and Chávez-Tapia, NC. The role of bile acids in glucose metabolism and their relation with diabetes. Ann Hepatol. (2017) 16:S21–6. doi: 10.5604/01.3001.0010.5672
17. Tang, R, Wei, Y, Li, Y, Chen, W, Chen, H, Wang, Q, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. (2018) 67:534–41. doi: 10.1136/gutjnl-2016-313332
18. Kummen, M, Holm, K, Anmarkrud, JA, Nygård, S, Vesterhus, M, Høivik, ML, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. (2017) 66:611–9. doi: 10.1136/gutjnl-2015-310500
19. Abe, K, Takahashi, A, Fujita, M, Imaizumi, H, Hayashi, M, Okai, K, et al. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS One. (2018) 13:e0198757. doi: 10.1371/journal.pone.0198757
20. Lv, LX, Fang, DQ, Shi, D, Chen, DY, Yan, R, Zhu, YX, et al. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol. (2016) 18:2272–86. doi: 10.1111/1462-2920.13401
21. Sabino, J, Vieira-Silva, S, Machiels, K, Joossens, M, Falony, G, Ballet, V, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut. (2016) 65:1681–9. doi: 10.1136/gutjnl-2015-311004
22. Swanson, SA, Tiemeier, H, Ikram, MA, and Hernán, MA. Nature as a Trialist?: deconstructing the analogy between Mendelian randomization and randomized trials. Epidemiol Camb Mass. (2017) 28:653–9. doi: 10.1097/EDE.0000000000000699
23. Lawlor, DA, Harbord, RM, Sterne, JAC, Timpson, N, and Davey, SG. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. (2008) 27:1133–63. doi: 10.1002/sim.3034
24. Cordell, HJ, Han, Y, Mells, GF, Li, Y, Hirschfield, GM, Greene, CS, et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun. (2015) 6:8019. doi: 10.1038/ncomms9019
25. The UK-PSC ConsortiumThe International IBD Genetics ConsortiumThe International PSC Study GroupJi, SG, Juran, BD, Mucha, S, et al. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat Genet. (2017) 49:269–73. doi: 10.1038/ng.3745
26. Pagoni, P, Dimou, NL, Murphy, N, and Stergiakouli, E. Using Mendelian randomisation to assess causality in observational studies. Evid Based Ment Health. (2019) 22:67–71. doi: 10.1136/ebmental-2019-300085
27. Bowden, J, Davey Smith, G, Haycock, PC, and Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
28. Zhang, J, Wu, G, Tang, Y, Liu, H, Ge, X, Peng, R, et al. Causal associations between gut microbiota and primary biliary cholangitis: a bidirectional two-sample Mendelian randomization study. Front Microbiol. (2023) 14:1273024. doi: 10.3389/fmicb.2023.1273024
29. Marchandin, H, Teyssier, C, Campos, J, Jean-Pierre, H, Roger, F, Gay, B, et al. Negativicoccus succinicivorans gen. Nov., sp. nov., isolated from human clinical samples, emended description of the family Veillonellaceae and description of Negativicutes classis nov., Selenomonadales Ord. Nov. and Acidaminococcaceae fam. Nov. in the bacterial phylum Firmicutes. Int J Syst Evol Microbiol. (2010) 60:1271–9. doi: 10.1099/ijs.0.013102-0
30. Aas, JA, Paster, BJ, Stokes, LN, Olsen, I, and Dewhirst, FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. (2005) 43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005
31. Feng, J, Ma, H, Huang, Y, Li, J, and Li, W. Ruminococcaceae_UCG-013 promotes obesity resistance in mice. Biomedicine. (2022) 10:3272. doi: 10.3390/biomedicines10123272
32. Laursen, MF, Sakanaka, M, von Burg, N, Mörbe, U, Andersen, D, Moll, JM, et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat Microbiol. (2021) 6:1367–82. doi: 10.1038/s41564-021-00970-4
33. Lechevalier, HA, and Lechevalier, MP. Biology of actinomycetes. Annu Rev Microbiol. (1967) 21:71–00. doi: 10.1146/annurev.mi.21.100167.000443
34. Klaassen, CD, and Cui, JY. Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab Dispos Biol Fate Chem. (2015) 43:1505–21. doi: 10.1124/dmd.115.065698
35. Tabibian, JH, O'Hara, SP, Trussoni, CE, Tietz, PS, Splinter, PL, Mounajjed, T, et al. Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology. (2016) 63:185–96. doi: 10.1002/hep.27927
36. Parada Venegas, D, de la Fuente, MK, Landskron, G, González, MJ, Quera, R, Dijkstra, G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol. (2019) 10:277. doi: 10.3389/fimmu.2019.00277
37. Stilling, RM, van de Wouw, M, Clarke, G, Stanton, C, Dinan, TG, and Cryan, JF. The neuropharmacology of butyrate: the bread and butter of the microbiota-gut-brain axis? Neurochem Int. (2016) 99:110–32. doi: 10.1016/j.neuint.2016.06.011
38. Huang, X, Chen, Y, Zhang, F, Yang, Q, and Zhang, G. Peripheral Th17/Treg cell-mediated immunity imbalance in allergic rhinitis patients. Braz J Otorhinolaryngol. (2014) 80:152–5. doi: 10.5935/1808-8694.20140031
39. Wick, MJ, and Ljunggren, HG. Processing of bacterial antigens for peptide presentation on MHC class I molecules. Immunol Rev. (1999) 172:153–62. doi: 10.1111/j.1600-065x.1999.tb01363.x
40. Rodríguez, T, Pérez, O, Ugrinovic, S, Bracho, G, and Mastroeni, P. Bacterial derived proteoliposome as ideal delivery system and cellular adjuvant. Vaccine. (2006) 24:S24–5. doi: 10.1016/j.vaccine.2005.01.106
41. Itoh, A, Adams, D, Huang, W, Wu, Y, Kachapati, K, Bednar, KJ, et al. Enoxacin up-regulates MicroRNA biogenesis and Down-regulates cytotoxic CD8 T-cell function in autoimmune cholangitis. Hepatol Baltim Md. (2021) 74:835–46. doi: 10.1002/hep.31724
42. Watanabe, M, Fukiya, S, and Yokota, A. Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J Lipid Res. (2017) 58:1143–52. doi: 10.1194/jlr.M075143
43. Gadaleta, RM, van Erpecum, KJ, Oldenburg, B, Willemsen, ECL, Renooij, W, Murzilli, S, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. (2011) 60:463–72. doi: 10.1136/gut.2010.212159
44. Cordell, HJ, Fryett, JJ, Ueno, K, Darlay, R, Aiba, Y, Hitomi, Y, et al. An international genome-wide meta-analysis of primary biliary cholangitis: novel risk loci and candidate drugs. J Hepatol. (2021) 75:572–81. doi: 10.1016/j.jhep.2021.04.055
45. Schneider, KM, Candels, LS, Hov, JR, Myllys, M, Hassan, R, Schneider, CV, et al. Gut microbiota depletion exacerbates cholestatic liver injury via loss of FXR signalling. Nat Metab. (2021) 3:1228–41. doi: 10.1038/s42255-021-00452-1
46. Terziroli Beretta-Piccoli, B, Mieli-Vergani, G, and Vergani, D. HLA, gut microbiome and hepatic autoimmunity. Front Immunol. (2022) 13:980768. doi: 10.3389/fimmu.2022.980768
Keywords: primary sclerosing cholangitis, primary biliary cholangitis, Mendelian randomization, cholestatic liver diseases, gut microbiota
Citation: Yang J, Ma G, Wang K, Yang H, Jiang S, Fan Q, Zhou X, Guo G and Han Y (2024) Causal associations between gut microbiota and Cholestatic liver diseases: a Mendelian randomization study. Front. Med. 11:1342119. doi: 10.3389/fmed.2024.1342119
Edited by:
Monica Barone, University of Bologna, ItalyReviewed by:
Xingshun Qi, General Hospital of Northern Theater Command, ChinaAkihiko Oka, Shimane University, Japan
Copyright © 2024 Yang, Ma, Wang, Yang, Jiang, Fan, Zhou, Guo and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guanya Guo, Z3VvZ3VhbnlhQDEyNi5jb20=; Ying Han, aGFueWluZzFAZm1tdS5lZHUuY24=
†These authors have contributed equally to this work
 Jiaqi Yang
Jiaqi Yang Gang Ma†
Gang Ma† Kemei Wang
Kemei Wang Ying Han
Ying Han