- 1Department of Pediatrics, China-Japan Friendship Hospital, Beijing, China
- 2Institute of Clinical Medical Sciences, China-Japan Friendship Hospital, Beijing, China
Objectives: This meta-analysis aimed to test the association of angiotensin-converting enzyme (ACE) gene I/D polymorphism with asthma risk and circulating ACE changes.
Methods: Public literature retrieval, publication selection, and information extraction were completed independently by two investigators. Effect-size values are expressed as odds ratios (ORs) or standardized mean differences (SMDs) with a 95% confidence interval (95% CI).
Results: Nineteen studies (2,888 patients and 9,549 controls) fulfilled the eligibility criteria. Overall investigations demonstrated that ACE gene I/D polymorphism was significantly associated with asthma risk under allelic (OR, 95% CI: 1.26, 1.08 to 1.48), homozygous genotypic (1.50, 1.09 to 2.06), and recessive (1.53, 1.24 to 1.89) models with moderate heterogeneity (I2 statistic: 64% to 79%). Subsidiary investigations recorded that race, matched status, asthma diagnosis, sample size, and age possibly accounted for the existence of significant heterogeneity. Relative to carriers with the II genotype, those with the DD genotype, ID genotype, and the combination of DD and ID genotypes had significantly higher concentrations of circulating ACE (WMD: 3.13, 2.07, and 2.83 U/L, respectively, p < 0.05). Adoption of Mendelian randomization analyses revealed that one unit increment in circulating ACE concentrations was found to be significantly associated with a 1.14-fold increased risk of asthma (95% CI: 1.02 to 4.24).
Conclusion: We provided strong meta-analytical evidence supporting the causal implication of high circulating ACE concentrations in the development of asthma.
Introduction
Asthma is a highly heritable disease. The heritability of childhood asthma reached as high as 82% (1). A long list of asthma-susceptibility genes has been identified (2, 3). However, no consensus exists on which gene actually involves the pathogenesis of asthma, even with genome-wide association studies (4–6). In this regard, the candidate gene approach still represents an alternative strategy (7). Based on a known biological function, a gene can be a candidate to precipitate asthma. Importantly, the gene can be screened to see which mutation actually embodies its function. One such case is the insertion/deletion (I/D) polymorphism in the gene coding angiotensin-converting enzyme (ACE).
The association of ACE gene I/D polymorphism with asthma has been widely studied. For example, the DD genotype of the ACE gene was overrepresented in patients with asthma relative to healthy controls (8). In an early meta-analysis, the DD homozygote carriers had an overall about 60% increased risk of asthma compared with the II + ID carriers, and this risk was more evident in Asians (9). A recent meta-analysis in children indicated that ACE gene I/D polymorphism was associated with a significant risk of asthma (10). Other studies, however, did not reveal any significance between this polymorphism and asthma risk (11–13). The reasons behind this inconsistency are multiple, likely because of differences in origins, baseline characteristics of study participants, diagnosis criteria of asthma, and statistical power to derive significance. To this point, the synthesis of individually underpowered studies with the same research goals can help shed some light on these reasons.
To yield robust evidence, we aimed to perform an updated meta-analysis and test the association of ACE gene I/D polymorphism with asthma risk and changes in circulating ACE concentrations. Meanwhile, heterogeneity sources attributable to inconsistent observations were explored.
Methods
Performance guidelines
This meta-analysis was performed according to the guidelines in the preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement. The PRISMA checklist is shown in Supplementary Table 1, and the PRISMA flow diagram is shown in Figure 1.
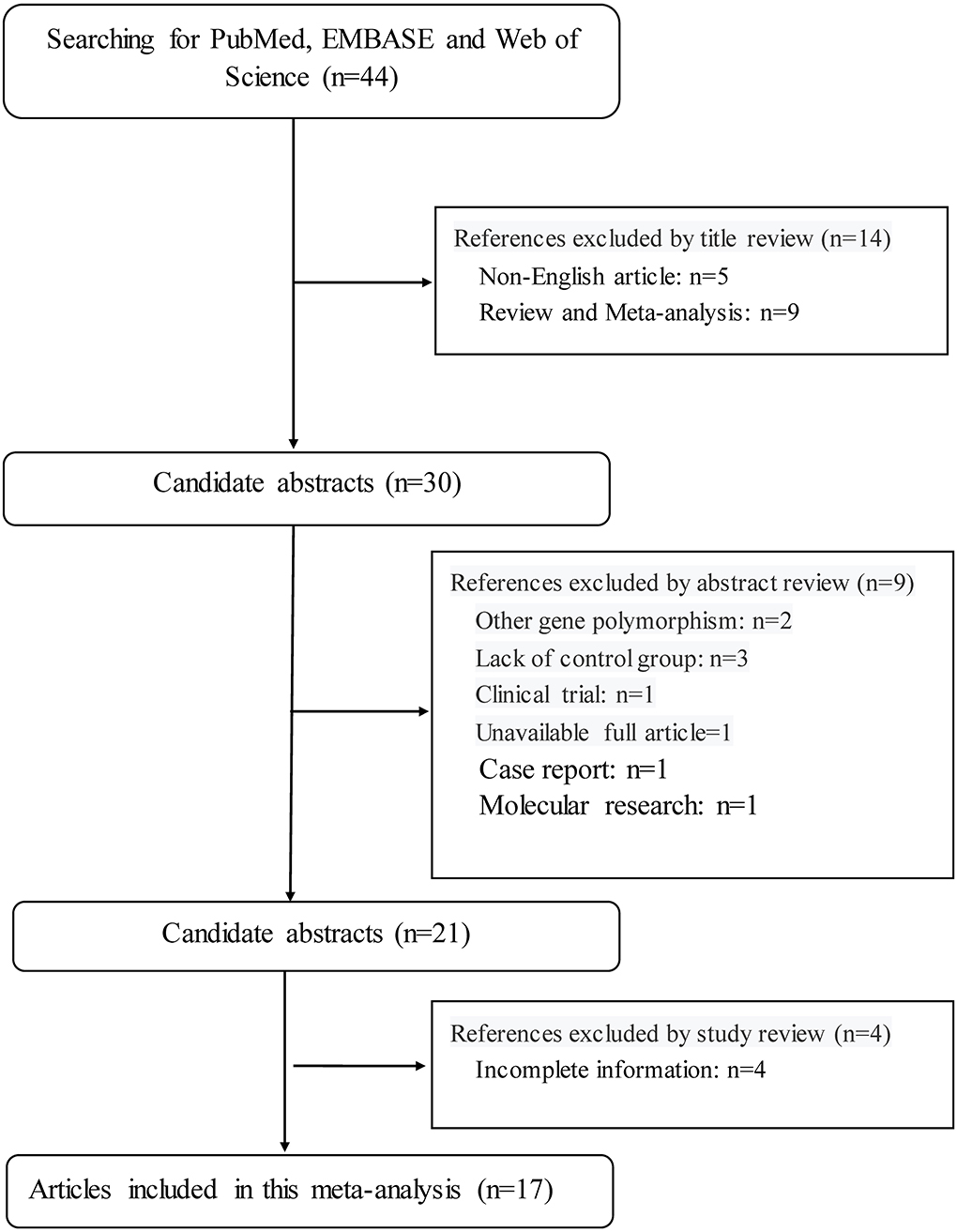
Figure 1. The PRISMA flowchart illustrates the selection process of qualified articles in this meta-analysis.
Search strategies
Using predefined key terms, four electronic databases—PubMed, HuGE Navigator, EMBASE (Excerpt Medica Database), and Web of Science—were searched from literature inception through March 2022. Key terms used for searching possibly eligible publications were formulated using the MeSH (Medical Subject Headings) database and are expressed in the Boolean format, that is (“asthma” or “atopic”) and (“polymorphism” or “SNP” or “variant” or “mutation” or “variation” or “genetic” or “genotype” or “allele”) and (“ACE” or “angiotensin-converting enzyme” or “angiotensin I converting enzyme” or “ACE1” or “DCP” or “DCP1” or “CD143”).
Initial screening of searched publications was restricted to the English language and human beings. The search was also extended to the reference lists of major publications (reviews and meta-analyses) retrieved. The search was independently performed by two investigators (Q. H. and F. Y.), and any difference in numbers was resolved via discussion and a consensus was attained.
Inclusion criteria
Included publications must concurrently meet the following four criteria: (i) class of evidence: case-control design; (ii) outcome: asthma with clear definition; (iii) necessary data: genotype counts (or allele counts in the absence of genotype counts) of ACE gene I/D polymorphism between patients with asthma and controls, or circulating ACE concentrations across I/D genotypes in either patients or controls or both; and (iv) genotype determination: valid methodology.
Exclusion criteria
Publications were excluded if one or more of the following criteria were met: (i) type of publication: review, letter to editor or correspondence, editorial, comment, conference abstract, and case report or series; (ii) publication with duplicate participant samples; (iii) involvement of only cases; (iv) endpoint other than asthma; and (v) unpublished data.
Publication selection
The selection of eligible publications was handled in two steps. First, the title and abstract (if available) were reviewed and removal was applied based on exclusion criteria. After the first round of publication removal, the full text was read and eligibility was checked based on inclusion criteria.
The selection process of eligible publications was completed independently by two investigators (Q. H. and F. Y.), and any divergence was solved by discussion and if necessary by a third investigator (W. N.).
Information extraction
By the use of a uniform data extraction Excel sheet, information from each qualified publication was independently extracted by two investigators (Q. H. and F. Y.), and two Excel sheets were compared by kappa statistics. Any disagreement was solved by rechecking the full text until a consensus was attained.
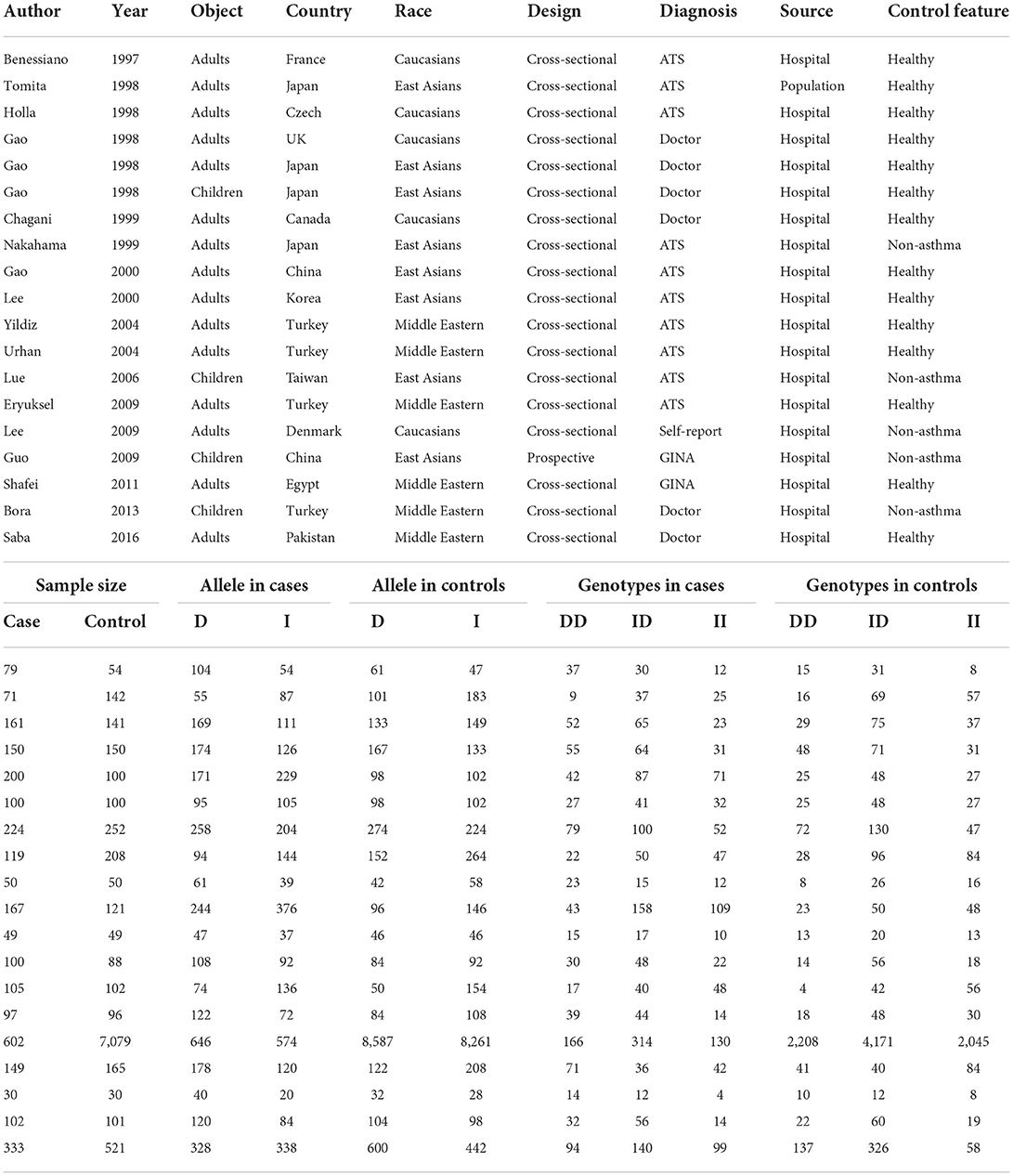
The first author's name, year of publication, country where participants were enrolled, race, sample size, design, source of controls, matched condition, diagnosis of asthma, genotype counts of ACE gene I/D polymorphism, and baseline characteristics of study participants, including age, gender, and ACE concentrations in circulation, were all extracted.
Statistical analyses
STATA software version 14.1 for Windows (Stata Corp, College Station, Texas) was utilized for this meta-analysis. The association of ACE gene I/D polymorphism with the risk of asthma was measured by odds ratio (OR) with a 95% confidence interval (95% CI). Changes in circulating ACE concentrations between genotypes of this polymorphism were denoted by standardized mean difference (SMD) with a 95% CI. Both OR and SMD were derived under the random-effects model, because in the absence of heterogeneity, fixed-effects and random-effects models yield very similar estimates, whereas, in the presence of heterogeneity, the random-effects model is preferred (14). In the case of the significant association of ACE gene I/D polymorphism with asthma risk and ACE changes in circulation, the Mendelian randomization technique was employed to infer the possible causality between circulating ACE and asthma.
Between-study heterogeneity was measured by the inconsistency index (I2) statistic. I2 denotes the percent of variability observed between studies that are the result of heterogeneity but not a chance observation. Higher I2 indicates a higher likelihood of heterogeneity. An I2 statistic of over 50% is indicative of significant heterogeneity. The sources of heterogeneity were statistical, clinical, and methodological aspects. To track these sources, subsidiary analyses according to pre-specified factors (including sample size, race, matched condition, source of controls, and diagnosis of asthma) and meta-regression analyses of both continuous and categorical factors were carried out. Subgroups involving two or more studies are displayed.
In addition, cumulative analyses were conducted to see the impact of the first publication on subsequent publications and evolution of accumulated estimates over time. Sensitivity analyses were conducted to see the impact of any single study on the overall effect-size estimate by removing an individual study each time to check whether any of these estimates can bias the overall estimates.
Publication bias was assessed by Begg's funnel plot. If the funnel shape is asymmetrically inverted, it suggests a correlation between pooled estimate and study size (publication bias or small study bias). From a statistical aspect, publication bias was measured by Egger's test, which appraises funnel asymmetry with a significance level set at 10%.
Results
Qualified publications
By the use of key terms, searching four public databases yielded 44 publications in the English language. Application of inclusion criteria and exclusion criteria, 17 of 44 publications were qualified for synthesis in this meta-analysis (8, 12, 13, 15–28).
In total, 19 independent studies were isolated from 17 qualified publications, including 2,888 patients with asthma and 9,549 controls, among who the genotypes of ACE gene I/D polymorphism were assayed with validated typing methods. Out of 19 independent studies, 3 (including 431 subjects) provided data on the changes in circulating ACE concentrations across the genotypes of this polymorphism.
Baseline characteristics
The baseline characteristics of qualified studies are shown in Table 1. Thirteen studies were performed among adults, three among children, and one among both.
Overall analyses
The association between ACE gene I/D polymorphism and asthma risk was separately evaluated under allelic, homozygous genotypic, dominant, and recessive models of inheritance. Figure 2 shows the overall forest plots of the four models. This polymorphism was significantly associated with asthma risk in the allelic model, with the D allele corresponding to 1.26 times more likely to have asthma than the I allele (95% CI: 1.08 to 1.48). Significance was also noticed under homozygous genotypic (DD vs. II: OR = 1.50, 95% CI: 1.09 to 2.06) and recessive (DD vs. ID plus II: OR = 1.53, 95% CI: 1.24 to 1.89) models. However, there was no observable association under the dominant model (DD plus DI vs. II: OR = 1.14, 95% CI: 0.87 to 1.48). As of between-study heterogeneity, the I2 statistic ranged from 64 to 79% across the four models of inheritance, suggesting that diversity in effect-size estimates was not due to chance.
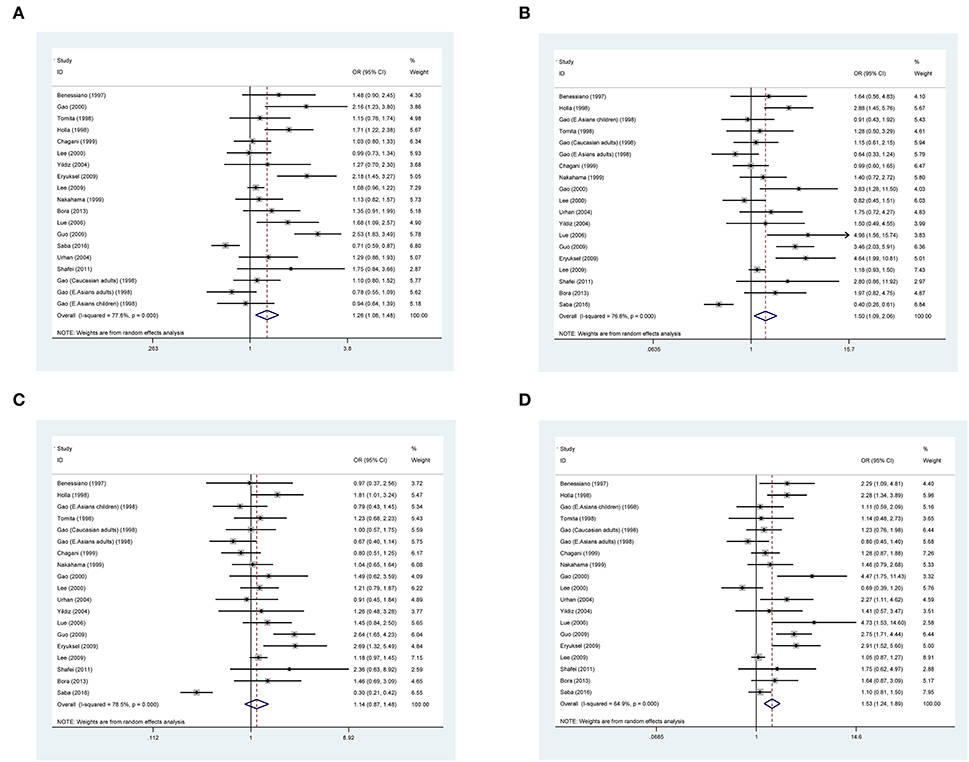
Figure 2. Forest plots of the angiotensin-converting enzyme (ACE) gene insertion/deletion (I/D) polymorphism in association with asthma under allelic (A), homozygous genotypic (B), dominant models (C), and recessive model (D).
In addition, this association was also evaluated under the heterozygous model of inheritance (Supplementary Figure 1), and there was no hint of statistical significance (D/I vs. II: OR = 0.96, 95% CI: 0.74 to 1.25). Considering the opposite effects of allele D and allele I on asthma risk, the heterozygous model is only interrogated in overall analyses.
Cumulative and influential analyses
Supplementary Figures 2, 3 exhibit the cumulative and influential analyses on the association between ACE gene I/D polymorphism and asthma risk, respectively. The cumulative impact over time was stabilized since the year 2000. Influential analyses were conducted by evaluating the impact of each study on the pooled OR via the deletion of one study each time, which revealed no single study impacted the pooled ORs significantly.
Publication bias
Figure 3 shows Begg's funnel plot inspecting the potential satisfaction of publication bias under the allelic model. The Begg's funnel plot seemed asymmetrical by inspection, which was confirmed by Egger's test, with the probability being 0.052.
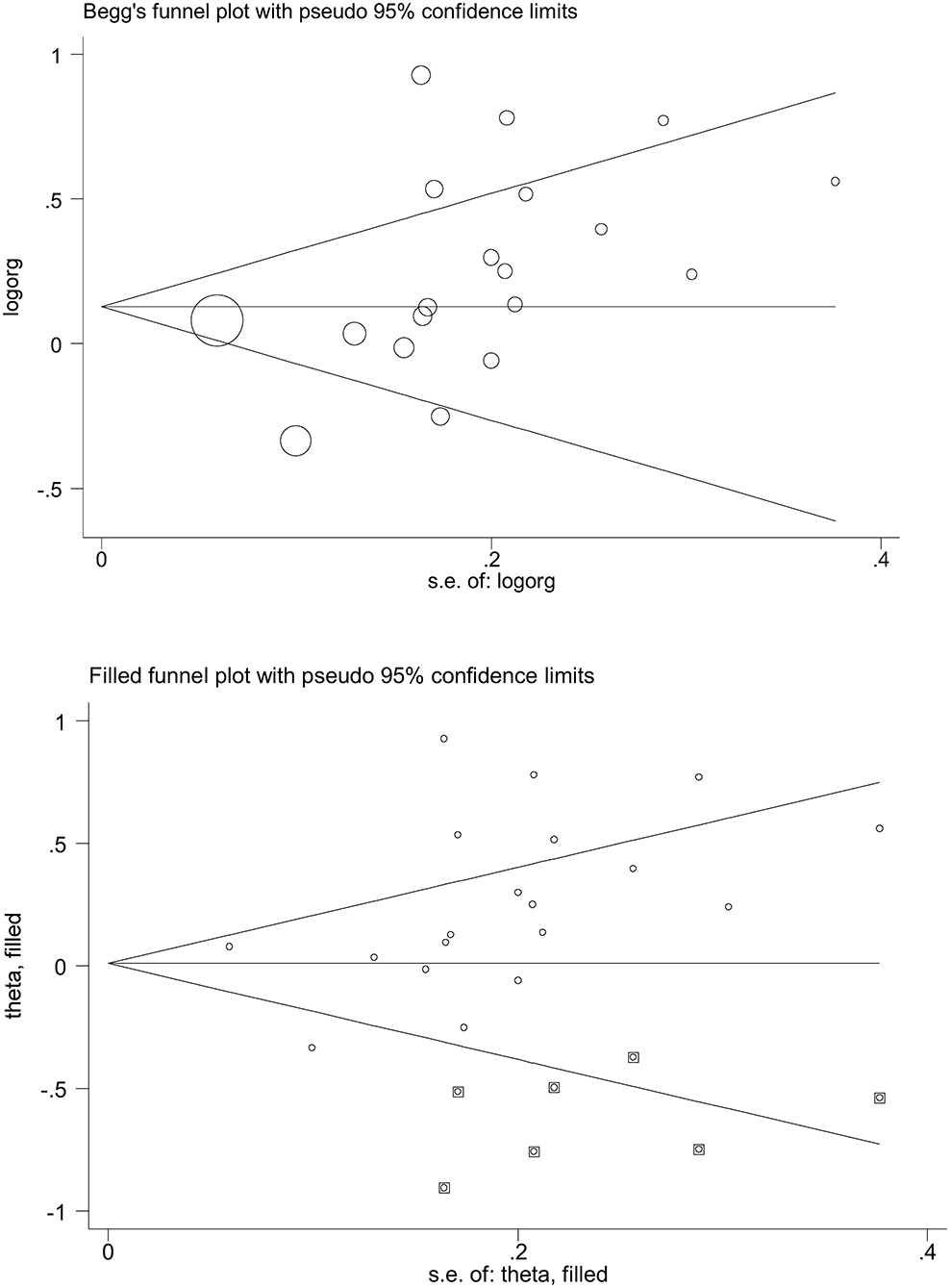
Figure 3. Begg's and filled funnel plots of the angiotensin-converting enzyme (ACE) gene insertion/deletion (I/D) polymorphism in association with asthma under the allelic model.
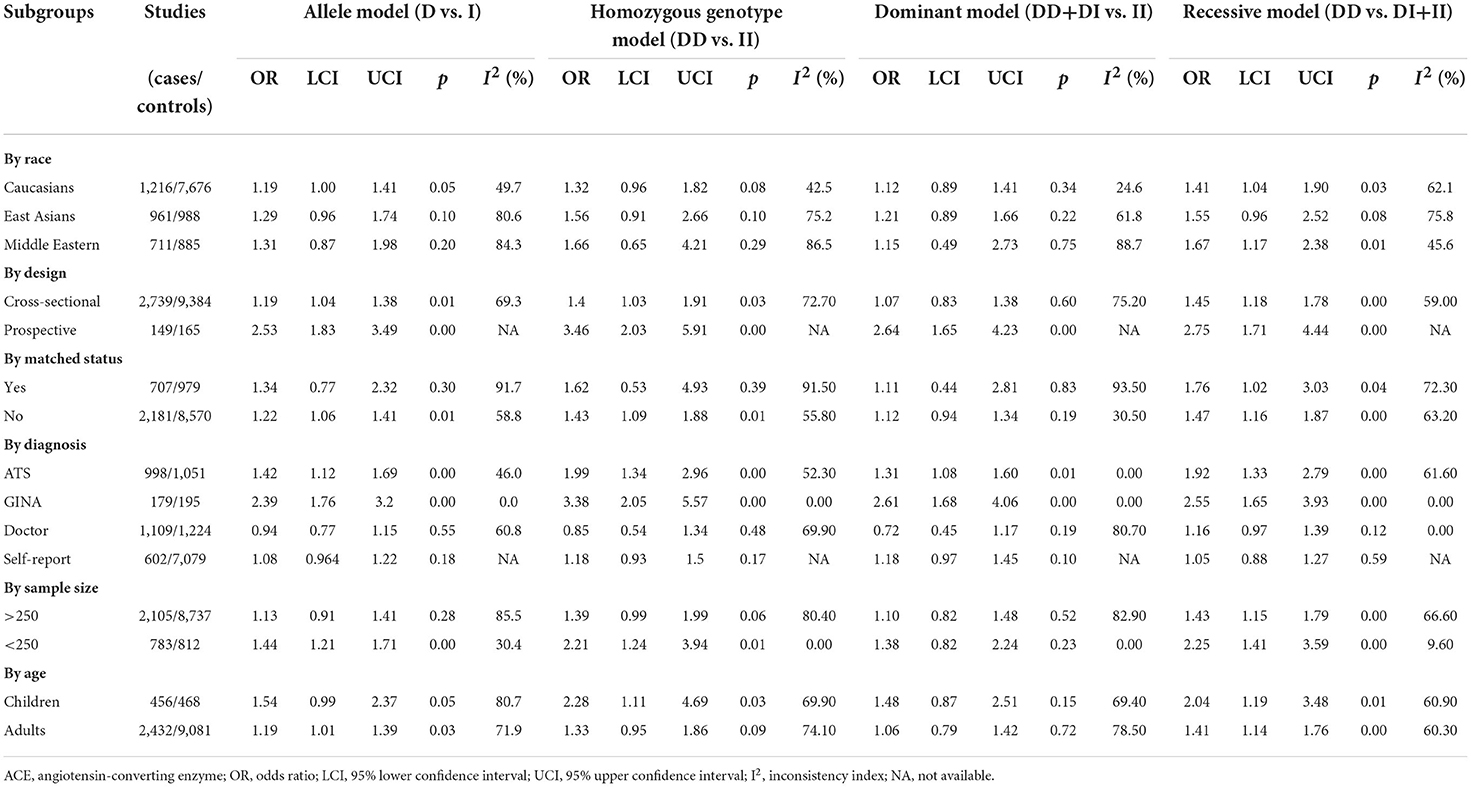
Subgroup analyses
As summarized in Table 2, the association between ACE gene I/D polymorphism and asthma risk was examined upon stratification by several potential factors on a categorical scale under the four models of inheritance.
It is worth noticing that race, matched status, asthma diagnosis, sample size, and age were possible sources of between-study heterogeneity, particularly under the recessive model. For example, the mutation of ACE gene I/D polymorphism was associated with the significant risk of asthma in Caucasians under allelic and recessive models, with the odds reaching 1.19 (95% CI: 1.00 to 1.41) and 1.41 (95% CI: 1.04 to 1.90), respectively, and no significance was observed in East Asians, irrespective of the models of inheritance.
The majority of subgroups showed improved between-study heterogeneity by pooling studies with homogeneous characteristics of interest.
Meta-regression analyses
An alternative way to explore sources of between-study heterogeneity is to perform meta-regression analyses. By regressing age, gender, asthma severity, race, matched status, asthma diagnosis, sample size, and study design (Supplementary Table 2), no hints of significance were seen at a significance level of 5%.
Genotype–phenotype analyses
The relationship between ACE gene I/D polymorphism and circulating ACE concentrations is shown in Table 3. Relative to carriers with the II genotype, those with the DD genotype, ID genotype, and the combined DD and ID genotypes showed significantly higher concentrations of circulating ACE (WMD: 3.13, 2.07, and 2.83 U/L, respectively, p < 0.05 for all).
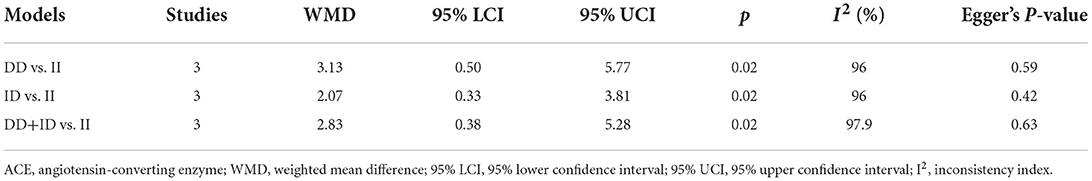
Table 3. Mean changes in circulating ACE concentrations between carriers of different genotypes of ACE gene I/D polymorphism.
Mendelian randomization analyses
Given the significance observed in both genotype-disease and genotype–phenotype analyses, the Mendelian randomization technique was utilized to infer the possible causal association between circulating ACE and asthma risk. Under the assumptions of the Mendelian randomization technique and by use of ACE gene I/D polymorphism as an instrumental variable, one unit increment in circulating ACE concentrations was found to be significantly associated with a 1.14-fold increased risk of asthma (95% CI: 1.02 to 4.24) under the homozygous genotypic model.
Discussion
The aim of this meta-analysis was to test the association of ACE gene I/D polymorphism with asthma risk and circulating ACE changes as well as explore sources for heterogeneity in the English literature. Through a comprehensive pooling of 19 independent studies involving 2,888 cases and 9,549 controls, this polymorphism was associated with a significant risk of asthma and changes in circulating ACE concentrations. Importantly, further adoption of the Mendelian randomization technique revealed that genetically increased ACE concentrations were causally associated with an increased risk of asthma. To the best of our knowledge, this is thus far the first meta-analytical evidence concerning the causal relation between circulating ACE and asthma risk in the literature.
It is well-known that asthma is a chronic pulmonary disease characterized by intermittent and reversible airflow obstruction (29). The exact etiology of asthma currently remains elusive; however, there is convincing evidence that asthma is a highly inheritable disease (1, 30). To shed light on the genetic profiles of asthma and seek reasons attributable to the inconsistency of previous individual studies, we, in this meta-analysis, aimed to test the association of ACE gene I/D polymorphism with asthma risk, as well as with circulating ACE concentrations. Our genotype-disease and genotype–phenotype analyses showed that carriers of the mutant DD homozygote had a 50% increased risk of asthma and 3.13 U/L increased concentrations of circulating ACE relative to the wild II homozygote. Under the assumptions of the Mendelian randomization technique, it is expected that circulating ACE may be a causal risk factor for the development of asthma. The implication of circulating ACE in asthma is biologically plausible. There is evidence that ACE expressed in the lungs plays a key role in the pathogenesis of bronchial asthma. It is because ACE can mediate the proliferation of smooth vascular muscle cells (31), which affects aggregation and adhesion of platelets and monocytes and consequently leads to excessive bronchiectasis (32). The biological mechanism behind the causal implication of circulating ACE in asthma is not clear at present (33). It is reasonable to speculate that if involved, the mutation of I/D polymorphism can alter the expression of ACE in circulation or tissues, which triggers the development of asthma.
It is also worth noticing that according to our subsidiary analyses, differences in race, matched status, asthma diagnosis, sample size, and age might account for previously diverging findings of individual studies. Taking race as an example, we found that the susceptibility of ACE gene I/D polymorphism to asthma was race-dependent, with significance observed in Caucasians but not in East Asians, in agreement with the findings of previous studies (16, 18, 19, 21, 34). Indeed, asthma is a multifactorial disease to which genetic, environmental, and lifestyle-related factors contribute jointly (35). For feasibility reasons, it is recommended to construct a list of candidate genetic determinants for asthma in each racial group. Moreover, diagnostic criteria for asthma can also confound the association between ACE gene I/D polymorphism and asthma risk. In this meta-analysis, significance was observed in studies based on ATS and GINA criteria, whereas there was no observable significance in studies with self-reported asthma. To derive a reliable estimate, it is important to diagnose asthma formally. Differing from the observations of subsidiary analyses, we failed to reveal any statistical significance in meta-regression analyses, an alternative method to explore sources of between-study heterogeneity. It is of practical importance to bear in mind that meta-regression analyses, albeit enabling covariates in either continuous or categorical format to be regressed, do not have the methodological rigor of a properly designed study that is intended to test the effect of these covariates formally (36).
Another important finding is the obvious changes in circulating ACE between genotypes of ACE gene I/D polymorphism. Considering the fact that the I/D polymorphism is located in the 16th intron, it is unlikely to be functional at the transcription level. There is a possibility that this polymorphism is strongly linked to another functional locus in either the promoter or exon or 3′-untranslated region of the ACE gene that is responsible for the regulation of circulating ACE concentrations. Further genomic and functional explorations of the ACE gene are encouraged.
Limitations
Several limitations should be acknowledged in this meta-analysis. First, this meta-analysis synthesized evidence from publications written in the English language, and selection bias cannot be excluded. Second, all included studies are cross-sectionally designed; however, causality was inferred by means of the Mendelian randomization technique. Third, only a few features were commonly provided by the majority of included studies, and it is expected that more features are needed to examine their potential confounding impact on between-study heterogeneity. Fourth, there was moderate evidence of publication bias, which might limit the generalizability of our findings.
Conclusion
Taken together, we, for the first time, provided systematic evidence supporting the causal implication of high circulating ACE in the development of asthma by means of the Mendelian randomization technique. Further experimental studies are needed to determine the culprit genetic loci in the ACE gene that can simultaneously regulate circulating ACE concentrations and precipitate the onset and progression of asthma.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
QZ and WN planned and designed the study, and directed its implementation. QH and FY contributed to data acquisition. QH, YH, and BP conducted statistical analyses. QH and WN wrote and revised the manuscript. All authors read and approved the final manuscript prior to submission.
Funding
This work was financially supported by the CAMS Innovation Fund for Medical Sciences (2021-12M-C&T-B-089) and the National Natural Science Foundation of China (Grant number: 81970042).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.941944/full#supplementary-material
References
1. Ullemar V, Magnusson PKE, Lundholm C, Zettergren A, Melen E, Lichtenstein P, et al. Heritability and confirmation of genetic association studies for childhood asthma in twins. Allergy. (2016) 71:230–8. doi: 10.1111/all.12783
2. Bansal M, Garg M, Agrawal A. Advances in asthma genetics. Adv Genet. (2021) 107:1–32. doi: 10.1016/bs.adgen.2020.11.001
3. Ober C. Asthma genetics in the post-GWAS era. Ann Am Thorac Soc. (2016) 13(Suppl. 1):S85–90. doi: 10.1513/AnnalsATS.201507-459MG
4. Hirota T, Takahashi A, Kubo M, Tatsuhiko T, Tomita K, Doi D, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. (2011) 43:893–6. doi: 10.1038/ng.887
5. Bonnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. (2014) 46:51–5. doi: 10.1038/ng.2830
6. Anantharaman R, Andiappan AK, Nikanth PP, Suri BK, Wang DY, Chew FT. Genome-wide association study identifies PERLD1 as asthma candidate gene. BMC Med Genet. (2011) 12:170. doi: 10.1186/1471-2350-12-170
7. Raita Y, Zhu Z, Camargo CA, Freishtat RJ, Ngo D, Liang L, et al. Relationship of soluble interleukin-6 receptors with asthma: a mendelian randomization study. Front Med. (2021) 8:665057. doi: 10.3389/fmed.2021.665057
8. Benessiano J, Crestani B, Mestari F, Klouche W, Neukirch F, Hacein-Bey S, et al. High frequency of a deletion polymorphism of the angiotensin-converting enzyme gene in asthma. J Allergy Clin Immunol. (1997) 99:53–7. doi: 10.1016/S0091-6749(97)70300-2
9. Zhang YG, Li XB, Zhang J, Huang J, He C, Deng Y, et al. The I/D polymorphism of angiotensin-converting enzyme gene and asthma risk: a meta-analysis. Allergy. (2011) 66:197–205. doi: 10.1111/j.1398-9995.2010.02438.x
10. Shao Z, Jin H, Sun H, Dong C, Xu B, Zha L. Angiotensin-converting enzyme insertion/deletion polymorphism and susceptibility to pediatric asthma: a meta-analysis. J Renin Angiotensin Aldosterone Syst. (2020) 21:1470320320923475. doi: 10.1177/1470320320923475
11. Rad IA, Bagheri M, Rahimi-Rad MH. Deletion allele of the ACE gene is not a risk factor for asthma predisposition. Pneumologia. (2011) 60:208–12. doi: 10.1081/jas-120026073
12. Yildiz P, Oflaz H, Cine N, Genchallac H, Erginel-Unaltuna N, Yildiz A, et al. Endothelial dysfunction in patients with asthma: the role of polymorphisms of ACE and endothelial NOS genes. J Asthma. (2004) 41:159–66. doi: 10.1081/JAS-120026073
13. Gao PS, Mao XQ, Kawai M, Enomoto T, Sasaki S, Shaldon SR, et al. Lack of association between ACE gene polymorphisms and atopy and asthma in British and Japanese populations. Clin Genet. (1998) 54:245–7. doi: 10.1111/j.1399-0004.1998.tb04294.x
14. Cohn LD, Becker BJ. How meta-analysis increases statistical power. Psychol Methods.(2003) 8:243–53. doi: 10.1037/1082-989X.8.3.243
15. Urhan M, Degirmenci I, Harmanci E, Gunes HV, Metintas M, Basaran A. High frequency of DD polymorphism of the angiotensin-converting enzyme gene in Turkish asthmatic patients. Allergy Asthma Proc. (2004) 25:243–7.
16. Tomita H, Sato S, Matsuda R, Ogisu N, Mori T, Niimi T, et al. Genetic polymorphism of the angiotensin-converting enzyme (ACE) in asthmatic patients. Respir Med. (1998) 92:1305–10. doi: 10.1016/S0954-6111(98)90134-2
17. Saba N, Yusuf O, Rehman S, Munir S, Ahmad leS, Mansoor A, et al. An angiotensin I-converting enzyme insertion/detion polymorphism is associated with Pakistani asthmatic cases and controls. J Biosci. (2016) 41:439–44. doi: 10.1007/s12038-016-9617-x
18. Nakahama H, Obata K, Nakajima T, Nakamura H, Kitada O, Sugita M, et al. Renin-angiotensin system component gene polymorphism in Japanese bronchial asthma patients. J Asthma. (1999) 36:187–93. doi: 10.3109/02770909909056316
19. Lee YC, Cheon KT, Lee HB, Kim W, Rhee YK, Kim DS. Gene polymorphisms of endothelial nitric oxide synthase and angiotensin-converting enzyme in patients with asthma. Allergy. (2000) 55:959–63. doi: 10.1034/j.1398-9995.2000.00724.x
20. Lee J, Nordestgaard BG, Dahl M. Elevated ACE activity is not associated with asthma, COPD, and COPD co-morbidity. Respir Med. (2009) 103:1286–92. doi: 10.1016/j.rmed.2009.04.003
21. Hollá L, Văsku A, Znojil V, Sisková L, Vácha J. Association of 3 gene polymorphisms with atopic diseases. J Allergy Clin Immunol. (1999) 103:702–8. doi: 10.1016/S0091-6749(99)70246-0
22. Gao J, Lin Y, Xiao Y, Xu K, Xu W, Zhu Y, et al. Polymorphism of angiotensin-converting enzyme gene and genetic susceptibility to asthma with familial aggregation. Chin Med Sci J. (2000) 15:24–8.
23. Eryüksel E, Ceyhan BB, Bircan R, Avşar M, Cirakoglu B. Angiotensin converting enzyme gene polymorphism in Turkish asthmatic patients. J Asthma. (2009) 46:335–8. doi: 10.1080/02770900802660972
24. El-Shafei MS, Farres MN, Shahin RY. Evaluation of angiotensin converting enzyme gene polymorphism and susceptibility to bronchial asthma among Egyptians. Allergol Immunopathol. (2012) 40:275–80. doi: 10.1016/j.aller.2011.05.010
25. Chagani T, Pare PD, Zhu S, Weir TD, Bai TR, Behbehani NA, et al. Prevalence of tumor necrosis factor-alpha and angiotensin converting enzyme polymorphisms in mild/moderate and fatal/near-fatal asthma. Am J Respir Crit Care Med. (1999) 160:278–82. doi: 10.1164/ajrccm.160.1.9808032
26. Lue KH, Ku MS, Li C, Sun HL, Lee HS, Chou MC. ACE gene polymorphism might disclose why some Taiwanese children with allergic rhinitis develop asthma symptoms but others do not. Pediatr Allergy Immunol. (2006) 17:508–13. doi: 10.1111/j.1399-3038.2006.00452.x
27. Guo S, Zhang JH, Yan YD, Ding YF, Sheng JY. Association between renin-angiotensin system gene polymorphism and recurrent wheezing in Chinese children: a 4-year follow-up study. J Int Med Res. (2009) 37:351–8. doi: 10.1177/147323000903700209
28. Bora E, Soylar R, Ayyildiz ZA, Uzuner N, Bozkaya OG, Ercal D, et al. Plasminogen activator inhibitor-1 and angiotensin converting enzyme gene polymorphisms in Turkish asthmatic children. Allergol Immunopathol. (2013) 41:11–6. doi: 10.1016/j.aller.2011.12.003
29. Bush A. Pathophysiological mechanisms of asthma. Front Pediatr. (2019) 7:68. doi: 10.3389/fped.2019.00068
30. Wang TN, Chao YY, Wang TH, ChenCJ, Ko YC. Familial risk of asthma among adolescents and their relatives in Taiwan. J Asthma. (2001) 38:485–94. doi: 10.1081/JAS-100105869
31. Tong Y, Ye C, Ren XS, Qiu Y, Zang YH, Xiong XQ, et al. Exosome-mediated transfer of ACE (angiotensin-converting enzyme) from adventitial fibroblasts of spontaneously hypertensive rats promotes vascular smooth muscle cell migration. Hypertension. (2018) 72:881–8. doi: 10.1161/HYPERTENSIONAHA.118.11375
32. Rechkina. Models of gen-gene interaction in determining the severity of bronchial asthma in children. Am J Intern Med. (2020) 8:182–91. doi: 10.11648/j.ajim.20200804.17
33. Krynytska I. The association of angiotensin-converting enzyme gene insertion/deletion polymorphism with bronchial asthma. Pol Merkur Lekarski. (2021) XLIX:442–4.
34. Ding QL, Sun SF, Cao C, Deng ZC. Association between angiotensin-converting enzyme I/D polymorphism and asthma risk: a meta-analysis involving 11,897 subjects. J Asthma. (2012) 49:557–62. doi: 10.3109/02770903.2012.685540
35. Tan LD, Alismail A, Ariue B. Asthma guidelines: comparison of the National Heart, Lung, and Blood Institute Expert Panel Report 4 with Global Initiative for Asthma 2021. Curr Opin Pulm Med. (2022) 28:234–44. doi: 10.1097/MCP.0000000000000867
Keywords: angiotensin-converting enzyme, asthma, meta-analysis, polymorphism, Mendelian randomization
Citation: Hui Q, Hao Y, Ye F, Pang B, Niu W and Zhang Q (2022) Genetically high angiotensin-converting enzyme concentrations causally increase asthma risk: A meta-analysis using Mendelian randomization. Front. Med. 9:941944. doi: 10.3389/fmed.2022.941944
Received: 12 May 2022; Accepted: 15 September 2022;
Published: 07 November 2022.
Edited by:
Yi Liu, Shandong Provincial Hospital, ChinaReviewed by:
Jia Mi, Binzhou Medical University, ChinaTadesse Bekele Tafesse, Haramaya University, Ethiopia
Copyright © 2022 Hui, Hao, Ye, Pang, Niu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenquan Niu, bml1d2VucXVhbkB6cnloeXkuY29tLmNu; Qi Zhang, emhhbmdxaWtleWFuQDE2My5jb20=
 Qin Hui1
Qin Hui1 Fang Ye
Fang Ye Bo Pang
Bo Pang Wenquan Niu
Wenquan Niu Qi Zhang
Qi Zhang