- 1Department of Infectious Diseases, The First Affiliated Hospital of Xi'an Jiaotong University, Xi'an, China
- 2State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, College of Medicine, The First Affiliated Hospital, Zhejiang University, Hangzhou, China
In the past decades, many studies have focused on aging because of our pursuit of longevity. With lifespans extended, the regenerative capacity of the liver gradually declines due to the existence of aging. This is partially due to the unique microenvironment in the aged liver, which affects a series of physiological processes. In this review, we summarize the related researches in the last decade and try to highlight the aging-related alterations in the aged liver.
Introduction
During aging, the liver undergoes a series of degenerative changes. Briefly, it presents a progressive decrease in functional liver mass, thus reducing its functional reserve, making it more difficult to maintain homeostasis and vulnerable to external stress or damage (1). Till now, the mechanisms underlying liver aging still remain unclear. As we known, the main causes of aging are DNA damage, telomeres shortening, epigenic alterations, and impairment of proteostasis (2). The aged liver is usually accompanied with failure of regeneration, metabolic dysfunction, redox imbalance, and development of chronic or malignant liver diseases (3–6). The impairment of regenerative capacity in the aged liver is affected by both intracellular factors and extracellular factors (7). Intriguingly, we may be able to recover their regenerative capacity via changing a microenvironment for the senescent hepatocytes (8). The aging-related alterations in the liver form a unique microenvironment and affect a series of physiological processes. Moreover, this unique microenvironment may function as a vital role that causes the liver to become susceptible to chronic diseases or tumors (9). For instance, it affects the fate of hepatocytes and promotes neoplastic development (10). Moreover, hepatocytes in this microenvironment are more susceptible to ischemia/reperfusion (I/R) injury (11–13). Of particular interest is the way to effectively eliminate the effects of aging and reverse the unique aging microenvironment in the aged liver. As reviewed by Xu et al., modulation of autophagy could function as an effective strategy for reverse of the aged liver (14). Autophagy mainly functions as a cytoprective role in liver diseases. Modulation of autophagy could markedly alleviate aging-related liver injury, promote liver regeneration, block I/R induced injury, and reverse the aging microenvironment in the aged liver (14–17).
Hallmarks of Hepatic Aging Process
Over the past years, our understanding of the aging process experiences a great advance. Of notable interest is calling for accurate hallmarks of aging in different organisms and in different organs. In 2013, Lopez-Otin et al. reviewed and concluded 9 cellular hallmarks of aging: genomic instability, epigenetic alterations, loss of proteostasis, telomere attrition, mitochondrial dysfunction, deregulated nutrient sensing, cellular senescence, stem cell exhaustion, and altered intercellular communication (18). Aging related changes in the aged liver occur at genomic, epigenetic, molecular, cellular and sub-cellular levels. Moreover, the aging process in the liver undergoes all the cellular hallmarks, these changes occur in various cells in the liver (parenchymal cells and non-parenchymal cells) and extracellular matrix (ECM) (Figure 1), and finally lead to impairment of liver function (19–21). In a recently published review, Hunt et al. enumerated the hallmarks of the aging process in the liver. Besides the hallmarks reviewed by Lopez-Otin et al., they also covered response to endoplasmic reticulum (ER) stress and aging human liver into discussion (18). Recent years, long non-coding RNAs and microRNAs have gained a lot of interest, and they are considered to be involved in the aging process, especially in the metabolic modulation and intercellular communication (22–25). Moreover, Barbosa et al. also highlighted the relevance of autophagy activity to the hallmarks of aging (26).
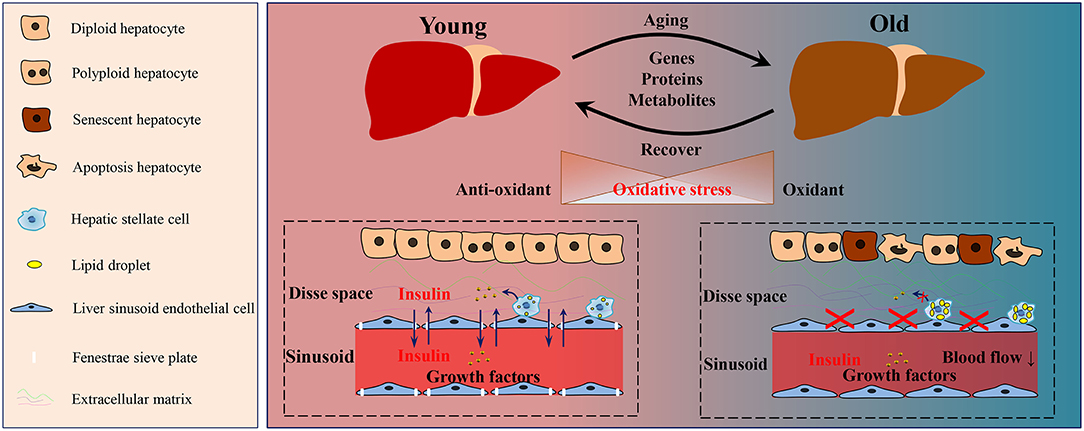
Figure 1. Schematic diagram of the aging-related changes in the liver. During the aging process of the liver, a series of alterations was observed in genes, proteins and metabolites. The oxidative homeostasis was disrupted, thus lead to oxidative stress. During aging, the number of polyploid, senescent, and apoptotic hepatocytes increased. Hepatic stellate cells in the aged liver contained more lipid droplets and released less growth factors than those in the young liver. The defenestration liver sinusoid endothelial cells occurred during aging, and thus disrupted the substance exchange, such as insulin and growth factors. Moreover, more extracellular matrix was deposited in Disse space in the aged liver.
Aging Liver: Suitable Animal Models
To fully understand the aging-related alterations in liver, suitable animal models are deemed necessary. Generally, these animal models can be divided into: naturally aging models, accelerated aging models, mimetic aging models, and transgenic aging models. Naturally aging models, especially aging rodents (mice aged ≥ 12 months, rats aged ≥ 16 months), are widely used in aging research because of their similarity of characteristics with human aging process. They have been applied in most of the studies related to aging liver in the last decade. Moreover, researchers used the senescence-accelerated mouse (SAMP10) to study the underlying mechanism of the aging liver (27, 28). In addition, mimetic aging models are also widely applied, which included D-galactose, radiation, β-amyloid, aluminum chloride induced aging models. Of these, D-galactose mimics naturally aging via inducing mitochondrial dysfunction and oxidative stress, increasing inflammation and apoptosis in the liver. Radiation induced aging models are usually used in cancer treatment (radiotherapy) related aging researches (29). Application of β-amyloid and aluminum chloride induces brain disorders, and these models are usually applied in Alzheimer's Disease related studies (30, 31). In recent years, transgenic aging models gain more and more attention with the development of gene editing technologies. Nevertheless, these animal models can only partially reproduce the typical pathological and biochemical alterations of aging.
Aging Related Alterations in the Liver
Currently, many researchers have reported the mechanisms of aging and its effects on the liver. In 2013, Capel et al. found that aging-related genes in the aged liver were mainly related to xenobiotic and fatty acid metabolism, retinoid X receptor function, and oxidative stress (32). In another microarray based study in the liver of aged rats, various genes involved in lipid metabolism and cell growth were downregulated (33). In 2015, White et al. revealed the main aging related alterations were involved in immune response, metabolic processes, RNA modification, and cell activation (34). Bochkis et al. showed that the inflammatory regulators (NFκB, IRF3, TLR4, et al.) upregulated related genes in the aged livers, nucleosome occupancy increased with age in mammalian liver (35). Sato et al. revealed that the aging process induced reprogramming of circadian rhythms in the liver. They showed that hepatic metabolic processes, such as protein acetylation and Nicotinamide adenine dinucleotide (NAD+) metabolism, were critically involved in the circadian change and remodel (36). In 2019, Chen et al. reported the transcriptional profiling of aging liver in Yana pigs (37). They showed that the up-regulated genes in pig aging liver were mainly involved in immune response, while the down-regulated genes were mainly enriched for metabolism (34). In another research, Bacalini et al. identified aging correlated genes enriched in the following biological processes: inflammatory response, interferon gamma response, and allograft rejection (38).
It is well known that oxidative stress plays a key role in the process of aging. Considering mitochondria and peroxisomes are important sources of reactive oxygen species (ROS), several studies have characterized aging related changes in liver peroxisomes and mitochondria using proteomics technology. In 2011, Amelina et al. identified 8 differentially expressed proteins of young (10-week-old) vs. old (18-month-old) in mouse liver peroxisomes (39). Five proteins were significantly up-regulated in the aged liver: epoxide hydrolase 2, 3-ketoacyl-CoA thiolase A, peroxisomal sarcosine oxidase, peroxisomal 2,4-dienoyl-CoA reductase, and prohibitin-2. And 3 proteins were down-regulated: pancreatic alpha-amylase, cytochrome c oxidase subunit 6C, and Cytochrome b-c1 complex subunit 2. These differentially expressed proteins mainly involved in the following processes: detoxification of xenobiotics, peroxisomal-oxidation, and production of ROS (39). In 2011, Musicco et al. identified differentially expressed hepatic mitochondrial proteins between old (28-month-old) and adult (12-month-old) rats. These differentially expressed proteins were involved in several mitochondrial metabolic pathways, such as pyruvate dehydrogenase complex, tricarboxylic acid cycle, and oxidative phosphorylation system chain (40). In another study, Bakala et al. revealed 16 differentially expressed hepatic mitochondrial proteins between young (3-month-old) and old (20-month-old) rats. They also found that most of these proteins were enzymes related to metabolic processes including fatty acid β-oxidation reaction, oxidative phosphorylation system chain complex I/V components, and tricarboxylic acid cycle (41). Moreover, to deeply reveal the aging-related organ specific function deterioration, Ori et al. identified the molecular alterations in the liver in young (6-month-old) and old (24-month-old) rats using an integrated approach (42). In this study, the researchers revealed several aged-related changes: altered translation and protein abundance, protein re-localization, alternative splicing, and altered protein phosphorylation. The liver proteome homeostasis was affected during aging, and the majority of these alterations were due to transcriptional regulation. The age-specific impacts on the liver were tightly related to the liver function: small molecule metabolism, monosaccharide metabolism, angiogenesis, protein degradation, complement activation, and antigen processing (42). To further investigate the molecular networks of longevity, Heinze et al. conducted a cross-species comparison between the long-lived naked mole-rats and shorter-lived guinea pigs (43). They revealed that 30 hepatic proteins were associated with sustaining longevity and most of these proteins were linked to lipid or fatty acid, and xenobiotic metabolism (43). Taken together, all of the above studies advance our understanding of aging-related changes in liver proteins that are significantly associated with organ function. However, we have to recognize there are several limitations to these studies. Firstly, different animal models are utilized in these studies. Some researchers choose mice as animal models, while others choose rats. In addition, they selected animals of different ages for the aging study. Secondly, the number of biological replicates is limited. Thus, these above findings need to be further validated.
In addition, metabolites are markedly altered during aging. Metabolomics technology enables us to fully understand the dynamic changes of metabolites, qualitatively and quantitatively (44). In 2013, Smiljanic et al. revealed aging-related cholesterol metabolism alterations in the liver of aged rats, and found that aging induced elevation of 2 cholesterol precursors: lanosterol and lathosterol (45). In another study, Pagliassotti et al. identified several metabolites that reduced in the liver of aged mice: pyruvate, lactate, nicotinamide, UDP-N-acetylglucosamine and UDP-N-acetylgalactosamine (46). Of these, UDP-N-acetylglucosamine was associated with longevity and protein quality control (47). Sato et al. quantitatively measured NAD+ and metabolites participated in its biosynthetic pathway (36). The authors revealed that nicotinamide and ADP ribose were significantly altered during aging and identified NAD+ metabolism as a core liver specific circadian metabolic pathway of aging. The improvement of NAD+ availability increased sirtuin 1 activity and rescued protein acetylation, thereby reversed the aging process (36). In 2019, Ando et al. found that ether-linked diacylglycerols accumulated in the liver of aged mice, which may associate with cell arrest and apoptosis (48). Wesley et al. reported the liver specific elevation of B-alanine and uridine, and the decrease of NAD and formate in aged rats (49).
Hepatic Cells of the Aging Liver
In the aged liver, number of hepatocytes decreases, and remnant hepatocytes experience an autonomous decline in the regenerative capacity (50). Moreover, the aging related hepatic structural changes form a hypoxia condition, which induces higher glucose production and elevation of phosphoenol pyruvate carboxykinase, a gluconeogenesis-regulating enzyme. This process is regulated by hepatocyte nuclear factor 4 α, and targeting its expression can reverse the aging related effects on gluconeogenesis (51). In addition, apoptotic, senescent and polyploidy hepatocytes accumulate in the aged liver (52–54). Intriguingly, targeted approaches that modulating the level of apoptosis can block the aging effects (52). The senescent hepatocytes are typically characterized by accumulation of DNA damage, activation of tumor suppressor pathways (p53, p16ink4a, and C/EBPα) (53, 55, 56). These senescent cells remain metabolically active, but no longer able to proliferate. They present with the senescence-associated secretory phenotype and affect their neighboring cells, e.g., induce macrophage migration, immune cell recruitment, and modulation of ECM (52, 57). For polyploidy hepatocytes, its role in aging liver still remains controversial. Polyploidy is also considered as a feature of the aging process, and several hypotheses have been proposed to explain their function and significance (58, 59). Some studies indicated that hepatocyte polyploidy is a terminally differentiated state (58). It is suggested that the polyploidy state restricted proliferation of hepatocytes (60). Reversal of hepatocyte ploidy can reverse the aging-related function disorder (61). Oppositely, other studies reported that hepatocyte polyploidy may enhance liver function, retains the ability to divide and proliferate, and even contributes to hepatocyte turnover during aging (58, 62, 63). Verma et al. found that hepatocytes and cholangiocytes in liver donors over a wide range showed no telomere shortening (64). However, Aini et al. observed telomere shortening in hepatocytes in human liver allografts (54). Thus, it is still debated about the contribution of telomere shortening to senescent hepatocytes (20). Intriguingly, Lin et al. identified a novel subset of hepatocytes with high levels of telomerase that repopulated the liver (65). This emerging view may enable us to re-consider the role of telomerase in senescent hepatocytes. Moreover, continuous cell proliferation could reactive telomerase, reverse senescence and polyploid in hepatocytes during aging (53). In addition, Bacalini et al. reported DNA methylation of 6 age-related differentially methylated positions (ELOVL2 island, MACROD1 island, CYP1B1 island, CCNJ island, ZIC1 island, ZIC1 shore) in primary human hepatocytes (38). IFN-α and its related signaling elevate in aging hepatocytes (66). During aging, hepatocytes highly express activin A and p15INK4b, which are thought related to decreased proliferation and increased apoptosis (67). Ultrastructural changes of aged hepatocytes were characterized by cytoplasmic vacuolization, hypertrophy, and changes in density of the nuclei and mitochondria (68). Moreover, somatic mutations were accumulated with age in human hepatocytes (69). However, post-translational modification of histone (H3K9me3/H3K14ac modification) was decreased, which subsequently lead to gene alteration (70). Liver sinusoidal endothelial cells (LESCs) are critically involved in managing the liver microenvironment. However, LSECs in the aged liver are gradually thickened and undergo typical age-related changes (defenestration or pseudocapillarization) (71–73). Pseudocapillarization of the sinusoid lining delays liver regeneration in the aged liver via disturbing endothelium-dependent processes, decreasing hepatosinusoidal blood flow, impairing platelet adhesion to sinusoid, and preventing growth factors to reach target cells (74). The old LESCs present with reduced endocytic capacity and low responsive to 2,5-Dimethoxy-4-Iodoamphetamine (71, 75). In addition, aging-related changes in LESCs lead to hepatic insulin resistance and influence glucose homeostasis, consistent with the reduced transendothelial transfer activity of insulin (76). Moreover, aging LESCs leads to up-regulation of cell adhesion molecules, thus induces an accumulation of CD68 macrophages and neutrophils. Together with the up-regulation of p16 and fibrinogen 2, these alterations induced by aging LESCs finally lead to pro-inflammatory phenotype (77). However, these changes may be reversed by targeting aging-related defenestration (78). HSCs reside around LESCs and their activation is partially regulated by LESCs (79). Thus, the above mentioned aging-related LESCs changes also have an impact on HSCs. In general, HSCs are activated and transdifferentiate into myofibroblasts (α-SMA+) upon injury. Aging is related to the hyperplasia of HSCs. During aging, it exists an increase in the number and diameter of lipid droplets in HSCs (80). Telomere attrition, a hallmark of the aging liver, also occurs in HSCs (64). In addition, integrin α5/β1 decreases in HSCs and thereby reduces the levels of hepatocyte growth factor in the aged liver, thus impairs liver regeneration (81). Moreover, production of ECM components by HSCs, such as laminin, is impaired in aged mice (81, 82). Thus, aging-related HSCs alterations have a vital influence on ECM remodeling. As we know, there is a close association between HSCs and liver progenitor cells (LPCs) (83), and HSCs may act as positive regulators of LPCs. LPCs, or called oval cells, are critically involved in maintaining the liver homeostasis. However, they are rarely observed in normal conditions. LPCs activation exists in various liver diseases, such as viral hepatitis, liver cirrhosis, and liver cancer (84, 85). Upon injury, LPCs activate, proliferate, and accumulate around the portal vein or central vein, and this process is termed ductual reaction (86). However, their activation decreases with age. As reported, Thy-1 (+) LPCs declined in aged donor, which might responsible for impaired liver regeneration (87). Qian et al. revealed that oval cell in aged mice showed low response to 3,5-diethoxycarbonyl-1,4-dihydrocollidine induced liver injury, due to the down-regulation of laminin (82).
Besides non-immune cells mentioned above, immune cells are also play important roles in maintaining liver function. Aged liver exhibits increased immune cell infiltration, such as macrophages, T-cells, B-cells, NK cells, and neutrophils, which is accompanied by a high inflammatory status (88). The increased immune cell infiltration and high inflammatory status in the aged liver were thought to be a detrimental factor of liver regeneration. Till now, our understanding of the infiltrated immune cells in aged liver is still superficial. During aging, neutrophils are recruited by p16Ink4a-expressing cells and induce oxidative damage telomeres in non-immune cells (89). Intriguingly, modulation of the immune cell infiltration in aged liver, such as depletion of macrophages and NK cells could significantly leads to improved regeneration (66). Nevertheless, aging leads to decline of macrophage function. Briefly, aging leads to impaired autophagy and phagocytosis, dysregulated secretion of pro-inflammatory mediators, alterations in cell morphology and distribution, and changes in epigenetic signature (90).
Extracellular Matrix of the Aging Liver
The ECM provides structural support and reside environment for liver cells. In recent years, the ECM gains a lot of attention and some of its novel functions in regenerative medicine have been uncovered (91, 92). The role of the ECM in aging and longevity has been reviewed (93, 94). Major aging-induced modifications of the ECM are glycation, fragmentation and carbamylation (94). Karsdal et al. outlined the aging-related changes in the turnover of the ECM by detecting 15 serum biomarkers, which provided a new perspective for the therapy of chronic liver disease (95, 96). Mahmoud et al. observed increased ECM deposit in the Disse space in the livers of old rats by transmission electron microscopy (68). Delire et al. showed that the liver of old mice with impaired ECM remodeling capacity was more susceptible to carbon tetrachloride induced fibrosis, owing to a reduction of the chemokine (C-X-C motif) ligand 9 and matrix metallo-proteinases-13 axis (19).
Till now, the researches referred to the aging-related alterations of liver ECM are limited. The ECM is important for not only structural support, and also for maintaining liver function during aging. Aging induces impairment of ECM remodeling, making the liver more susceptible to injury. However, to fully understand the role of the ECM in the aging liver, we still need more experimental work.
Concluding Remarks and Future Directions
Liver aging is a complicated process with a series of organ-specific alterations, from genes, proteins to metabolites. These alterations form a unique microenvironment in the aged liver and have an impact on liver functions. Elimination of the aging effects and reversal of the aging microenvironment are beneficial to the reparative capacity of the aged liver. In the future, the development of drugs or anti-aging vaccines to treat aging-related liver disorders will be urgently needed. Modulation of autophagy is considered as a promising “rejuvenation” strategy for the aging liver. However, how to effectively and appropriately modulate autophagy without harm is still not available. Meanwhile, novel therapeutic targets for reversing the hepatic aging microenvironment are hotly pursued. Hopefully, we will develop effective anti-aging strategies based on novel therapeutic targets in the future.
Author Contributions
JL and YZ designed the work. YZ, YY, and QL searched the databases and listed the related literature. YZ and YY drafted the manuscript. JL provided critical revision of the manuscript. All authors approved the final version.
Funding
We thank the support from the Institutional Foundation of the First Affiliated Hospital of Xi'an Jiaotong University (2021QN-05) and the National Natural Science Fund (No. 81700559).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Kim IH, Kisseleva T, Brenner AD. Aging and liver disease. Curr Opin Gastroenterol. (2015) 31:184–91. doi: 10.1097/MOG.0000000000000176
2. Pinto C, Ninfole E, Gaggiano L, Benedetti A, Marzioni M, Maroni M. Aging and the biological response to liver injury. Semin Liver Dis. (2020) 40:225–32. doi: 10.1055/s-0039-3402033
3. Schmucker DL. Age-related changes in liver structure and function: implications for disease? Exp Gerontol. (2005) 40:650–9. doi: 10.1016/j.exger.2005.06.009
4. Timchenko NA. Aging and liver regeneration. Trends Endocrinol Metab. (2009) 20:171–6. doi: 10.1016/j.tem.2009.01.005
5. Huang YL, Shen ZQ, Huang CH, Lin CH, Tsai FT. Cisd2 slows down liver aging and attenuates age-related metabolic dysfunction in male mice. Aging Cell. (2021) 20:e13523. doi: 10.1111/acel.13523
6. Bellanti F, Vendemiale G. The aging liver: redox biology and liver regeneration. Antioxid Redox Signal. (2021) 35:832–847. doi: 10.1089/ars.2021.0048
7. Pibiri M. Liver regeneration in aged mice: new insights. Aging. (2018) 10:1801–24. doi: 10.18632/aging.101524
8. Liu L, Yannam GR, Nishikawa T, Yamamoto T, Basma H, Ito R, et al. The microenvironment in hepatocyte regeneration and function in rats with advanced cirrhosis. Hepatology. (2012) 55:1529–39. doi: 10.1002/hep.24815
9. Kim IH, Xu J, Liu X, Koyama Y, Ma HY, Diggle K, et al. Aging increases the susceptibility of hepatic inflammation, liver fibrosis and aging in response to high-fat diet in mice. Age. (2016) 38:291–302. doi: 10.1007/s11357-016-9938-6
10. Marongiu F, Serra MP, Doratiotto S, Sini M, Fanti M, Cadoni E, et al. Aging promotes neoplastic disease through effects on the tissue microenvironment. Aging. (2016) 8:3390–99. doi: 10.18632/aging.101128
11. Wang JH, Ahn IS, Fischer TD, Byeon JI, Dunn WA, Behrns KE, et al. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology. (2011) 141:2188–99.e6. doi: 10.1053/j.gastro.2011.08.005
12. Chun SK, Lee S, Flores-Toro J, U RY, Yang MJ, Go KL, et al. Loss of sirtuin 1 and mitofusin 2 contributes to enhanced ischemia/reperfusion injury in aged livers. Aging Cell. (2018) 17:e12761. doi: 10.1111/acel.12761
13. Bi J, Yang L, Wang T, Zhang J, Li T, Ren Y, et al. Irisin improves autophagy of aged hepatocytes via increasing telomerase activity in liver injury. Oxid Med Cell Longev. (2020) 2020:6946037. doi: 10.1155/2020/6946037
14. Xu F, Tautenhahn HM, Dirsch O, Dahmen U. Modulation of autophagy: a novel “rejuvenation” strategy for the aging liver. Oxid Med Cell Longev. (2021) 2021:6611126. doi: 10.1155/2021/6611126
15. Xu X, Hueckstaedt LK, Ren J. Deficiency of insulin-like growth factor 1 attenuates aging-induced changes in hepatic function: role of autophagy. J Hepatol. (2013) 59:308–17. doi: 10.1016/j.jhep.2013.03.037
16. Liu A, Guo E, Yang J, Yang Y, Liu S, Jiang X, et al. Young plasma reverses age-dependent alterations in hepatic function through the restoration of autophagy. Aging Cell. (2018) 17:e12708. doi: 10.1111/acel.12708
17. Liu A, Yang J, Hu Q, Dirsch O, Dahmen U, Zhang C, et al. Young plasma attenuates age-dependent liver ischemia reperfusion injury. FASEB J. (2019) 33:3063–73. doi: 10.1096/fj.201801234R
18. Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. (2013) 153:1194–217. doi: 10.1016/j.cell.2013.05.039
19. Delire B, Lebrun V, Selvais C, Henriet P, Bertrand A, Horsmans Y, et al. Aging enhances liver fibrotic response in mice through hampering extracellular matrix remodeling. Aging. (2016) 9:98–113. doi: 10.18632/aging.101124
20. Hunt NJ, Kang SWS, Lockwood GP, Le Couteur DG, Cogger CV. Hallmarks of aging in the liver. Comput Struct Biotechnol J. (2019) 17:1151–61. doi: 10.1016/j.csbj.2019.07.021
21. Allaire M, Gilgenkrantz H. The aged liver: beyond cellular senescence. Clin Res Hepatol Gastroenterol. (2020) 44:6–11. doi: 10.1016/j.clinre.2019.07.011
22. Morsiani C, Bacalini MG, Santoro A, Garagnani P, Collura S, D'Errico A, et al. The peculiar aging of human liver: a geroscience perspective within transplant context. Ageing Res Rev. (2019) 51:24–34. doi: 10.1016/j.arr.2019.02.002
23. Victoria B, Nunez Lopez YO, Masternak MM. MicroRNAs and the metabolic hallmarks of aging. Mol Cell Endocrinol. (2017) 455:131–47. doi: 10.1016/j.mce.2016.12.021
24. Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. Long noncoding RNAs (lncRNAs) and the molecular hallmarks of aging. Aging. (2014) 6:992–1009. doi: 10.18632/aging.100710
25. Capri M, Olivieri F, Lanzarini C, Remondini D, Borelli V, Lazzarini R, et al. Identification of miR-31-5p, miR-141-3p, miR-200c-3p, and GLT1 as human liver aging markers sensitive to donor-recipient age-mismatch in transplants. Aging Cell. (2017) 16:262–72. doi: 10.1111/acel.12549
26. Barbosa MC, Grosso RA, Fader MC. Hallmarks of aging: an autophagic perspective. Front Endocrinol. (2018) 9:790. doi: 10.3389/fendo.2018.00790
27. Honma T, Yanaka M, Tsuduki T, Ikeda I. Increased lipid accumulation in liver and white adipose tissue in aging in the SAMP10 mouse. J Nutr Sci Vitaminol. (2011) 57:123–9. doi: 10.3177/jnsv.57.123
28. Honma T, Shinohara N, Ito J, Kijima R, Sugawara S, Arai T, et al. High-fat diet intake accelerates aging, increases expression of Hsd11b1, and promotes lipid accumulation in liver of SAMP10 mouse. Biogerontology. (2012) 13:93–103. doi: 10.1007/s10522-011-9363-2
29. Zhu W, Zhang X, Yu M, Lin B, Yu C. Radiation-induced liver injury and hepatocyte senescence. Cell Death Discovery. (2021) 7:244. doi: 10.1038/s41420-021-00634-6
30. Wong-Guerra M, Montano-Peguero Y, Ramírez-Sánchez J, Jiménez-Martin J, Fonseca-Fonseca LA, Hernández-Enseñat D, et al. JM-20 treatment prevents neuronal damage and memory impairment induced by aluminum chloride in rats. Neurotoxicology. (2021) 87:70–85. doi: 10.1016/j.neuro.2021.08.017
31. Oyeleke MB, Owoyele VB. Saponins and flavonoids from Bacopa floribunda plant extract exhibit antioxidant and anti-inflammatory effects on amyloid beta 1-42-induced Alzheimer's disease in BALB/c mice. J Ethnopharmacol. (2022) 288:114997. doi: 10.1016/j.jep.2022.114997
32. Capel F, Rolland-Valognes G, Dacquet C, Brun M, Lonchampt M, Ktorza A, et al. Analysis of sterol-regulatory element-binding protein 1c target genes in mouse liver during aging and high-fat diet. J Nutrigenet Nutrigenomics. (2013) 6:107–22. doi: 10.1159/000350751
33. Chishti MA, Kaya N, Binbakheet AB, Al-Mohanna F, Goyns MH, Colak D. Induction of cell proliferation in old rat liver can reset certain gene expression levels characteristic of old liver to those associated with young liver. Age. (2013) 35:719–32. doi: 10.1007/s11357-012-9404-z
34. White RR, Milholland B, MacRae SL, Lin M, Zheng D, Vijg J. Comprehensive transcriptional landscape of aging mouse liver. BMC Genomics. (2015) 16:899. doi: 10.1186/s12864-015-2061-8
35. Bochkis IM, Przybylski D, Chen J, Regev A. Changes in nucleosome occupancy associated with metabolic alterations in aged mammalian liver. Cell Rep. (2014) 9:996–1006. doi: 10.1016/j.celrep.2014.09.048
36. Sato S, Solanas G, Peixoto FO, Bee L, Symeonidi A, Schmidt MS, et al. Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell. (2017) 170:664–77.e11. doi: 10.1016/j.cell.2017.07.042
37. Chen J, Zou Q, Lv D, Raza MA, Wang X, Chen Y, et al. Comprehensive transcriptional profiling of aging porcine liver. PeerJ. (2019) 7:e6949. doi: 10.7717/peerj.6949
38. Bacalini MG, Franceschi C, Gentilini D, Ravaioli F, Zhou X, Remondini D, et al. Molecular aging of human liver: an epigenetic/transcriptomic signature. J Gerontol A Biol Sci Med Sci. (2019) 74:1–8. doi: 10.1093/gerona/gly048
39. Amelina H, Sjodin MO, Bergquist J, Cristobal S. Quantitative subproteomic analysis of age-related changes in mouse liver peroxisomes by iTRAQ LC-MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci. (2011) 879:3393–400. doi: 10.1016/j.jchromb.2011.08.044
40. Musicco C, Capelli V, Pesce V, Timperio AM, Calvani M, Mosconi L, et al. Rat liver mitochondrial proteome: changes associated with aging and acetyl-L-carnitine treatment. J Proteomics. (2011) 74:2536–47. doi: 10.1016/j.jprot.2011.05.041
41. Bakala H, Ladouce R, Baraibar MA, Friguet B. Differential expression and glycative damage affect specific mitochondrial proteins with aging in rat liver. Biochim Biophys Acta. (2013) 1832:2057–67. doi: 10.1016/j.bbadis.2013.07.015
42. Ori A, Toyama BH, Harris MS, Bock T, Iskar M, Bork P, et al. Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. (2015) 1:224–37. doi: 10.1016/j.cels.2015.08.012
43. Heinze I, Bens M, Calzia E, Holtze S, Dakhovnik O, Sahm A, et al. Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging. BMC Biol. (2018) 16:82. doi: 10.1186/s12915-018-0547-y
44. Zhao Y, Chen E, Huang K, Xie Z, Zhang S, Wu D, et al. Dynamic alterations of plasma metabolites in the progression of liver regeneration after partial hepatectomy. J Proteome Res. (2020) 19:174–85. doi: 10.1021/acs.jproteome.9b00493
45. Smiljanic K, Vanmierlo T, Djordjevic AM, Perovic M, Loncarevic-Vasiljkovic N, Tesic V, et al. Aging induces tissue-specific changes in cholesterol metabolism in rat brain and liver. Lipids. (2013) 48:1069–77. doi: 10.1007/s11745-013-3836-9
46. Pagliassotti MJ, Estrada AL, Hudson WM, Wei Y, Wang D, Seals DR, et al. Trehalose supplementation reduces hepatic endoplasmic reticulum stress and inflammatory signaling in old mice. J Nutr Biochem. (2017) 45:15–23. doi: 10.1016/j.jnutbio.2017.02.022
47. Denzel MS, Storm NJ, Gutschmidt A, Baddi R, Hinze Y, Jarosch E, et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell. (2014) 156:1167–78. doi: 10.1016/j.cell.2014.01.061
48. Ando A, Oka M, Satomi Y. Deoxysphingolipids and ether-linked diacylglycerols accumulate in the tissues of aged mice. Cell Biosci. (2019) 9:61. doi: 10.1186/s13578-019-0324-9
49. Wesley UV, Bhute VJ, Hatcher JF, Palecek SP, Dempsey JR. Local and systemic metabolic alterations in brain, plasma, and liver of rats in response to aging and ischemic stroke, as detected by nuclear magnetic resonance (NMR) spectroscopy. Neurochem Int. (2019) 127:113–24. doi: 10.1016/j.neuint.2019.01.025
50. Serra MP, Marongiu F, Marongiu M, Contini A, Laconi E. Cell-autonomous decrease in proliferative competitiveness of the aged hepatocyte. J Hepatol. (2015) 62:1341–8. doi: 10.1016/j.jhep.2015.01.015
51. Park EY, Lee CH, Lee EK, Kim JH, Cova A, Lee SK, et al. HNF4alpha contributes to glucose formation in aged rat hepatocytes. Exp Gerontol. (2013) 48:1518–25. doi: 10.1016/j.exger.2013.10.011
52. Zhong HH, Hu SJ, Yu B, Jiang SS, Zhang J, Luo D, et al. Apoptosis in the aging liver. Oncotarget. (2017) 8:102640–52. doi: 10.18632/oncotarget.21123
53. Wang MJ, Chen F, Li JX, Liu CC, Zhang HB, Xia Y, et al. Reversal of hepatocyte senescence after continuous in vivo cell proliferation. Hepatology. (2014) 60:349–61. doi: 10.1002/hep.27094
54. Aini W, Miyagawa-Hayashino A, Tsuruyama T, Hashimoto S, Sumiyoshi S, Ozeki M, et al. Telomere shortening and karyotypic alterations in hepatocytes in long-term transplanted human liver allografts. Transpl Int. (2012) 25:956–66. doi: 10.1111/j.1432-2277.2012.01523.x
55. Wang C, Jurk D, Maddick M, Nelson G, Martin-Ruiz C, von Zglinicki T. DNA damage response and cellular senescence in tissues of aging mice. Aging Cell. (2009) 8:311–23. doi: 10.1111/j.1474-9726.2009.00481.x
56. Nguyen P, Valanejad L, Cast A, Wright M, Garcia JM, El-Serag HB, et al. Elimination of age-associated hepatic steatosis and correction of aging phenotype by inhibition of cdk4-C/EBPalpha-p300 axis. Cell Rep. (2018) 24:1597–609. doi: 10.1016/j.celrep.2018.07.014
57. Engelmann C, Tacke F. The potential role of cellular senescence in non-alcoholic fatty liver disease. Int J Mol Sci. (2022) 23:652. doi: 10.3390/ijms23020652
58. Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol. (2013) 24:347–56. doi: 10.1016/j.semcdb.2013.01.003
59. Tormos AM, Taléns-Visconti R, Sastre J. Regulation of cytokinesis and its clinical significance. Crit Rev Clin Lab Sci. (2015) 52:159–67. doi: 10.3109/10408363.2015.1012191
60. Wilkinson PD, Delgado ER, Alencastro F, Leek MP, Roy N, Weirich MP, et al. The polyploid state restricts hepatocyte proliferation and liver regeneration in mice. Hepatology. (2019) 69:1242–58. doi: 10.1002/hep.30286
61. Liu Q, Chen F, Yang T, Su J, Song S, Fu ZR, et al. Aged-related function disorder of liver is reversed after exposing to young milieu via conversion of hepatocyte ploidy. Aging Dis. (2021) 12:1238–51. doi: 10.14336/AD.2020.1227
62. Wang N, Hao F, Shi Y, Wang J. The controversial role of polyploidy in hepatocellular carcinoma. Onco Targets Ther. (2021) 14:5335–44. doi: 10.2147/OTT.S340435
63. Matsumoto T, Wakefield L, Grompe M. The significance of polyploid hepatocytes during aging process. Cell Mol Gastroenterol Hepatol. (2021) 11:1347–9. doi: 10.1016/j.jcmgh.2020.12.011
64. Verma S, Tachtatzis P, Penrhyn-Lowe S, Scarpini C, Jurk D, Von Zglinicki T, et al. Sustained telomere length in hepatocytes and cholangiocytes with increasing age in normal liver. Hepatology. (2012) 56:1510–20. doi: 10.1002/hep.25787
65. Lin S, Nascimento EM, Gajera CR, Chen L, Neuhofer P, Garbuzov A, et al. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature. (2018) 556:244–8. doi: 10.1038/s41586-018-0004-7
66. Singh P, Goode T, Dean A, Awad SS, Darlington JG. Elevated interferon gamma signaling contributes to impaired regeneration in the aged liver. J Gerontol A Biol Sci Med Sci. (2011) 66:944–56. doi: 10.1093/gerona/glr094
67. Menthena A, Koehler CI, Sandhu JS, Yovchev MI, Hurston E, Shafritz DA, et al. Activin A, p15INK4b signaling, and cell competition promote stem/progenitor cell repopulation of livers in aging rats. Gastroenterology. (2011) 140:1009–20. doi: 10.1053/j.gastro.2010.12.003
68. Mahmoud YI, Hegazy GH. Ginger and alpha lipoic acid ameliorate age-related ultrastructural changes in rat liver. Biotech Histochem. (2016) 91:86–95. doi: 10.3109/10520295.2015.1076578
69. Brazhnik K, Sun S, Alani O, Kinkhabwala M, Wolkoff AW, Maslov AY, et al. Single-cell analysis reveals different age-related somatic mutation profiles between stem and differentiated cells in human liver. Sci Adv. (2020) 6:eaax2659. doi: 10.1126/sciadv.aax2659
70. Price AJ, Manjegowda MC, Kain J, Anandh S, Bochkis MI. Hdac3, Setdb1, and Kap1 mark H3K9me3/H3K14ac bivalent regions in young and aged liver. Aging Cell. (2020) 19:e13092. doi: 10.1111/acel.13092
71. Simon-Santamaria J, Malovic I, Warren A, Oteiza A, Le Couteur D, Smedsrod B, et al. Age-related changes in scavenger receptor-mediated endocytosis in rat liver sinusoidal endothelial cells. J Gerontol A Biol Sci Med Sci. (2010) 65:951–60. doi: 10.1093/gerona/glq108
72. Hilmer SN, Cogger VC, Fraser R, McLean AJ, Sullivan D, Le Couteur GD. Age-related changes in the hepatic sinusoidal endothelium impede lipoprotein transfer in the rat. Hepatology. (2005) 42:1349–54. doi: 10.1002/hep.20937
73. Hunt NJ, McCourt PAG, Le Couteur DG, Cogger CV. Novel targets for delaying aging: the importance of the liver and advances in drug delivery. Adv Drug Deliv Rev. (2018) 135:39–49. doi: 10.1016/j.addr.2018.09.006
74. Furrer K, Rickenbacher A, Tian Y, Jochum W, Bittermann AG, Kach A, et al. Serotonin reverts age-related capillarization and failure of regeneration in the liver through a VEGF-dependent pathway. Proc Natl Acad Sci U S A. (2011) 108:2945–50. doi: 10.1073/pnas.1012531108
75. Cogger VC, Mitchell SJ, Warren A, de Cabo R, Le Couteur GD. Age-related loss of responsiveness to 2,5-dimethoxy-4-iodoamphetamine in liver sinusoidal endothelial cells. J Gerontol A Biol Sci Med Sci. (2014) 69:514–8. doi: 10.1093/gerona/glt124
76. Mohamad M, Mitchell SJ, Wu LE, White MY, Cordwell SJ, Mach J, et al. Ultrastructure of the liver microcirculation influences hepatic and systemic insulin activity and provides a mechanism for age-related insulin resistance. Aging Cell. (2016) 15:706–15. doi: 10.1111/acel.12481
77. Wan Y, Li X, Slevin E, Harrison K, Li T, Zhang Y, et al. Endothelial dysfunction in pathological processes of chronic liver disease during aging. FASEB J. (2022) 36:e22125. doi: 10.1096/fj.202101426R
78. Hunt NJ, Lockwood GP, Warren A, Mao H, McCourt PAG, Le Couteur DG, et al. Manipulating fenestrations in young and old liver sinusoidal endothelial cells. Am J Physiol Gastrointest Liver Physiol. (2019) 316:G144–54. doi: 10.1152/ajpgi.00179.2018
79. Deleve LD, Wang X, Guo Y. Sinusoidal endothelial cells prevent rat stellate cell activation and promote reversion to quiescence. Hepatology. (2008) 48:920–30. doi: 10.1002/hep.22351
80. Warren A, Cogger VC, Fraser R, Deleve LD, McCuskey RS, Le Couteur GD. The effects of old age on hepatic stellate cells. Curr Gerontol Geriatr Res. (2011) 2011:439835. doi: 10.1155/2011/439835
81. Rohn F, Kordes C, Buschmann T, Reichert D, Wammers M, Poschmann G, et al. Impaired integrin alpha5 /beta1 -mediated hepatocyte growth factor release by stellate cells of the aged liver. Aging Cell. (2020) 19:e13131. doi: 10.1111/acel.13131
82. Qian L, Zhang H, Gu Y, Li D, He S, Wang H, et al. Reduced production of laminin by hepatic stellate cells contributes to impairment in oval cell response to liver injury in aged mice. Aging. (2018) 10:3713–35. doi: 10.18632/aging.101665
83. Zhang W, Chen XP, Zhang WG, Zhang F, Xiang S, Dong HH, et al. Hepatic non-parenchymal cells and extracellular matrix participate in oval cell-mediated liver regeneration. World J Gastroenterol. (2009) 15:552–60. doi: 10.3748/wjg.15.552
84. Dai Z, Song G, Balakrishnan A, Yang T, Yuan Q, Mobus S, et al. Growth differentiation factor 11 attenuates liver fibrosis via expansion of liver progenitor cells. Gut. (2019) 69:1104–15. doi: 10.1136/gutjnl-2019-318812
85. Ko S, Russell JO, Molina LM, Monga PS. Liver progenitors and adult cell plasticity in hepatic injury and repair: knowns and unknowns. Annu Rev Pathol. (2020) 15:23–50. doi: 10.1146/annurev-pathmechdis-012419-032824
86. Fausto N. Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells. Hepatology. (2004) 39:1477–87. doi: 10.1002/hep.20214
87. Ono Y, Kawachi S, Hayashida T, Wakui M, Tanabe M, Itano O, et al. The influence of donor age on liver regeneration and hepatic progenitor cell populations. Surgery. (2011) 150:154–61. doi: 10.1016/j.surg.2011.05.004
88. Singh P, Coskun ZZ, Goode C, Dean A, Thompson-Snipes L, Darlington G. Lymphoid neogenesis and immune infiltration in aged liver. Hepatology. (2008) 47:1680–90. doi: 10.1002/hep.22224
89. Lagnado A, Leslie J, Ruchaud-Sparagano MH, Victorelli S, Hirsova P, Ogrodnik M, et al. Neutrophils induce paracrine telomere dysfunction and senescence in ROS-dependent manner. EMBO J. (2021) 40:e106048. doi: 10.15252/embj.2020106048
90. Stahl EC, Haschak MJ, Popovic B, Brown NB. Macrophages in the aging liver and age-related liver disease. Front Immunol. (2018) 9:2795. doi: 10.3389/fimmu.2018.02795
91. Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. (2017) 547:179–84. doi: 10.1038/nature22978
92. Hammond JS, Gilbert TW, Howard D, Zaitoun A, Michalopoulos G, Shakesheff KM, et al. Scaffolds containing growth factors and extracellular matrix induce hepatocyte proliferation and cell migration in normal and regenerating rat liver. J Hepatol. (2011) 54:279–87. doi: 10.1016/j.jhep.2010.06.040
93. Robert L, Labat-Robert J. Longevity and aging: role of genes and of the extracellular matrix. Biogerontology. (2015) 16:125–9. doi: 10.1007/s10522-014-9544-x
94. Birch HL. Extracellular matrix and ageing. Subcell Biochem. (2018) 90:169–90. doi: 10.1007/978-981-13-2835-0_7
95. Karsdal MA, Genovese F, Madsen EA, Manon-Jensen T, Schuppan D. Collagen and tissue turnover as a function of age: implications for fibrosis. J Hepatol. (2016) 64:103–9. doi: 10.1016/j.jhep.2015.08.014
Keywords: aging, microenvironment, regeneration, hepatic cells, extracellular matrix
Citation: Zhao Y, Yang Y, Li Q and Li J (2022) Understanding the Unique Microenvironment in the Aging Liver. Front. Med. 9:842024. doi: 10.3389/fmed.2022.842024
Received: 23 December 2021; Accepted: 31 January 2022;
Published: 24 February 2022.
Edited by:
Juan Pablo Arab, Pontificia Universidad Católica de Chile, ChileReviewed by:
Daniel Cabrera, Pontificia Universidad Católica de Chile, ChileJorge Arnold, Pontificia Universidad Católica de Chile, Chile
Copyright © 2022 Zhao, Yang, Li and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianzhou Li, bGlqaWFuemhvdTE5ODZAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Yalei Zhao
Yalei Zhao Ya Yang2†
Ya Yang2† Jianzhou Li
Jianzhou Li