
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 21 March 2025
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1525524
Climate change and human activities are altering the Mediterranean marine biodiversity. Monitoring these alterations over time is crucial for assessing the health of coastal environments and preserving local species. However, this monitoring process is resource-intensive, requiring taxonomic experts and significant amounts of time. To address this, we present an automated pipeline that detects, classifies and segments 17 species of Mediterranean fish using YOLOv8, integrated into an underwater stereo vision system capable of real-time inference and selective data storage. The proposed model demonstrates strong performance in detecting, classifying, and segmenting 17 Mediterranean fish species, achieving an mAP50(B) of 0.886 and an mAP50(M) of 0.889.
Oceans are essential for human society, providing invaluable natural resources, supporting diverse ecosystems, and maintaining biodiversity. In an era of significant anthropogenic impact through activities such as fish stocking, shipping, aquaculture, pollution, and habitat modification, which cause considerable ecological and economic damage (Effrosynidis et al., 2020), monitoring changes in marine ecosystems is crucial. This enables biodiversity protection and sustainable management of ocean resources, ensuring their efficient and responsible use.
Underwater marine imagery is a widely used tool for monitoring these changes in ecosystems of interest, as it allows for the study of various important parameters such as the impact of environmental stressors over time, biodiversity assessment, changes in habitat structure or the distribution of marine species and their behaviour at multiple spatial and temporal scales (Aguzzi et al., 2015).
Regarding biodiversity assessment on fish species, recognition is essential for identifying the abundance of species in a specific area, identifying endangered species, and controlling production management, making it a critical aspect of ecosystem management (Alaba et al., 2022).
Traditionally, tasks such as detecting, classifying, segmenting, and counting fish in underwater imagery have relied mainly on manual analysis by expert biologists, a process that can often be resource-intensive and costly. These tasks have been difficult to automate because traditional computer vision techniques do not perform well in underwater conditions, where the background is complex and the shape and texture features of fish are subtle (Siddiqui et al., 2018). Lower-cost approaches have also been developed, including underwater photography competitions and citizen science initiatives (Tiralongo et al., 2021; Roberts et al., 2022).
The integration of technological advances has significantly enhanced the ability to collect larger and higher-quality datasets for marine biodiversity monitoring. However, the accumulation of such vast amounts of data also demands greater processing capabilities, making the synergy between emerging technologies and traditional ecological methods increasingly necessary (McClure et al., 2020). In this context, the integration of new technologies and citizen science may allow scientists to create and process larger volumes of data than possible with conventional method and has the potential to maximise outcomes in ecological monitoring by improving the efficiency of large-scale data analysis (Garcia-Soto et al., 2021).
In recent years, deep learning has revolutionised the field of computer vision, achieving remarkable success in visual recognition and detection tasks. The application of these technologies to aquatic fauna is still in its early stages due to the unique challenges of the underwater environment and difficulties with data collection. However, recent advances have made it possible to automate these processes with fairly good results.
This work presents a deep learning model able to detect, classify and segment seventeen species of Mediterranean fish and its integration in an underwater Stereo Vision System (SVS) that can monitor areas of interest and automatically select the relevant data, avoiding unnecessary storage and the need for a human in the loop.
The remainder of this document is structured as follows. Section 2 reviews the related work on fish detection and segmentation and highlights the main contributions of this work. Section 3 describes the adopted methodology and materials, including the description of the dataset, the neural network architecture used and the hyperparameter study performed. Section 4 presents the experimental results. Section 5 explains the system implementation on an underwater SVS. Finally, Section 6 summarises the main conclusions and presents possible future research lines.
This section provides a summary of the current state of research on deep learning in the field of aquatic animal recognition and detection, as well as an overview of the most significant contributions of this work.
Monitoring fish populations is essential for environmental conservation and the development of sustainable fisheries (Saleh et al., 2022). There is a growing interest in using non-invasive monitoring techniques as biodiversity management tools, as they do not interfere with the ecosystem. Underwater video and digital still cameras are therefore rapidly being adopted by marine scientists and managers as tools for non-destructively quantifying and measuring the relative abundance, cover and size of marine fauna (Salman et al., 2016).
A clear example of the significant potential of imagery as a source of biological information for environmental monitoring is the proliferation of underwater cabled observatories. Notable examples include the Ocean Network Canada (ONC), the European Multidisciplinary Seafloor and Water-Column Observatories (EMSO) and the Expandable Seafloor Observatory (OBSEA) in Catalonia, Spain. Moreover, the use of fixed or mobile underwater cameras for detection purposes is widely spreading (Cui et al., 2020; Lopez-vazquez et al., 2020; Szymak et al., 2020; Coro and Bjerregaard Walsh, 2021). However, the processing of image data within ecological applications is still partly manual, and cabled observatory platforms and their networks currently lack software tools for automated recognition and classification of biologically relevant image content (Lopez-vazquez et al., 2020).
Manual processing of this data is very labour-intensive and time-consuming, which has led to significant efforts to develop automated methods for fish species detection and classification.
Traditionally, classical vision techniques such as Gabor filters or Support Vector Machine (SVM) have been used for underwater fish classification (Hu et al., 2012; Ogunlana et al., 2015; Rathi et al., 2018; Alsmadi et al., 2019). However, advances in artificial intelligence and deep learning over the last decade have enabled the application of these powerful technologies to the field of fish classification. Given the complexity of the underwater environment, deep learning-based methods appear to offer superior generalisation capacity and performance to address the challenges posed by these scenarios (Li J. et al., 2023).
Hybrid methods have also been used, with Qin et al. (2016) proposing a custom convolutional neural network (CNN) with an SVM classifier at the top, which achieved an accuracy of 0.986. Similarly, Deep and Dash (2019) proposed a hybrid CNN framework that used CNN for feature extraction and SVM and K-Nearest Neighbour for classification, achieving an accuracy of 0.988 on the same dataset.
All these approaches can be classified according to the problem they are trying to solve: classification, object detection or instance segmentation approaches.
The goal of the first group is to categorise underwater images based on the specific underwater animal or object depicted. A distinction can be made between works that encompass all fish species into a generic fish class (Lopez-vazquez et al., 2020; Szymak et al., 2020) and those that differentiate between various fish species. In the latter group, Siddiqui et al. employed CNN and SVM to classify seventeen classes of Australian reef fish, achieving a 0.89 accuracy on their own dataset and a 0.967 accuracy on the LifeClef’15 dataset (Siddiqui et al., 2018).
Even though these methods obtained good results, classifying fish from underwater images is only the first step towards an automated ecosystem monitoring pipeline. Typically, in marine environments, many fish can be found in the same image, therefore, an object detection approach becomes more tailored to the problem, as it provides information about which fish species can be found in an image and where they are located.
Consequently, a second group of studies has focussed on applying object detection to fish classification. This computer vision task combines object localisation, which determines the precise locations of objects in an image, and object classification, which assigns them to specific categories. This dual objective allows object detection to provide a richer understanding of visual data compared to simple image classification (Zhao et al., 2019). In the context of fish classification it allows to classify multiple fish in a single image and locate them within the image, typically enclosing each detected specimen with a bounding box.
Initially, works within this group were based on the usage and improvement of convolutional neural networks. A clear example is the work of Dos Santos and Gonçalves (dos Santos and Gonçalves, 2019),, which introduced a CNN based approach to enhance the recognition accuracy of Pantanal fish with similar characteristics. The CNN, comprising three branches for classifying species, family, and order, demonstrated an improvement in accuracy from 0.864 to 0.873 compared to traditional methods. In recent years, the continuous optimisation of deep learning algorithms has led to the development of more sophisticated and powerful network architectures.
At present, object detection algorithms are mainly divided into two types of detectors: one-stage detectors and two-stage detectors, with R-CNN and Faster R-CNN representing two-stage object detectors, and different versions of YOLO representing one-stage object detectors. These networks have become really popular and have been widely adopted. Jalal et al. (2020) proposed a hybrid solution by combining YOLOv3 deep neural network with optical flow and Gaussian mixture models to detect and classify fish in unconstrained underwater videos. Liu et al. (2020) proposed a marine biometric recognition algorithm based on YOLOv3-GAN network and demonstrated its improvement in performance with respect to YOLOv3. Knausgård et al. (2022) presented a deep learning-based approach using YOLOv3 for detection and CNN-SENet for classification was implemented for temperate fish. They obtained a 0.870 mAP50 on the Fish4Knowledge dataset and a 0.837 for the temperate fish dataset, differentiating between four species.
Despite the considerable success of these algorithms, they continue to face challenges, including those related to occlusions. Some researchers have attempted to address this challenge by employing semantic segmentation or instance segmentation algorithms (Li J. et al., 2023).
Semantic segmentation assigns a label to each pixel in an image based on the object or region it belongs to, allowing for detailed analysis. Instance segmentation goes further by distinguishing individual objects within the same category. In other words, it assigns each pixel to a specific instance of an object, effectively combining object detection and semantic segmentation (Hafiz, 2020).
Therefore, the results obtained from instance segmentation are more informative and can serve as the basis for larger tasks, such as biomass estimation. Consequently, numerous works have been published in recent years focussing on the application of instance segmentation in the domain of fish classification (Álvarez-Ellacuría et al., 2020; Garcia et al., 2020; Abinaya et al., 2022; Muñoz-Benavent et al., 2022; Ubina et al., 2022). However, none of these works present a complete solution. Rather, they either work in a constricted environment or present a solution tailored to a single class of fish.
The Mediterranean Sea is an example of a scarcity of approaches in the field of deep learning for fish detection and instance segmentation (Catalán et al., 2023). Although considerable research has been conducted on tropical fish, there are few published works or models for Mediterranean fish classification. Catalán et al. (2023) investigated the impact of diverse backgrounds, varying labelling practices, image quantity, and selection methods on classification outcomes. Additionally, they compared the performance of FASTER-RCNN and YOLOv5 for Mediterranean fish classification, achieving the most favourable results with YOLOv5, which reached an mAP50 of 0.84 for an object detection task with a generic fish class. Additionally, an mAP50 of 0.42 was achieved for a study distinguishing between sixteen classes, and an F1-score of 0.75 was achieved when distinguishing between eight classes.
In this context, the primary objective of this study is to generate a novel instance segmentation dataset for Mediterranean fish and conduct a hyperparameter study to develop a robust model capable of accurately detecting, localising and segmenting seventeen species of Mediterranean fish in real-world scenarios. Furthermore, the model is adapted and integrated into an underwater SVS, which is deployed to perform real-world tests.
The main contributions of this work are composed of:
1. Generating an open instance segmentation dataset from fish species of the Mediterranean Sea.
2. Obtaining an instance segmentation trained model to detect, classify and segment seventeen species of Mediterranean Fish, outperforming the current best publicly available model.
3. Implementing and integrating the system on an underwater SVS that allows online inference, and testing it in a real-world environment.
This section explains the dataset formation, including its labelling and organisation; and presents the neural network model used, its training procedure, hyperparameter study, and validation metrics.
This subsection explains the formation and management of the dataset used to train and test the deep neural network.
For this study, an instance segmentation dataset comprising thirty-six Mediterranean fish species, containing a total of 4,635 images, has been assembled.
The dataset is composed of images from multiple sources. Firstly, it contains images from two public datasets, already labelled in bounding box form, found on the Roboflow platform (Dwyer et al., 2024): (1) the IMEDEA dataset (Dataset, 2023), that provided images of Mediterranean fish species from the coasts of Spain, as well as from the Sub-Eye Observatory (IMEDEA, 2024); and (2) the OBSEA dataset (2023), that contributes with images from the OBSEA cabled observatory (Nogueras et al., 2010). Secondly, images from the MINKA public database (Minka Observations, 2024) are also incorporated. By gathering images from different cameras, locations and sources a dataset with a variety of backgrounds, light conditions, and resolutions is generated.
Although the dataset comprises thirty-six distinct fish species, only seventeen have sufficient instances to be considered representative and used in this work.
As previously stated, the objective is to create an instance segmentation database. To achieve this, the labels have to be transformed into masks, which is a time-consuming and laborious task. To simplify and optimise this process, a semi-automated approach is adopted, utilising the assistance of the segmentation model, Segment Anything Model (SAM) (Kirillov et al., 2023).
The procedure for each image in the dataset is as follows: for each observation, a bounding box is obtained and used as an input for SAM. Subsequently, a manual review of the masks obtained with SAM is conducted, and they are adapted to the required format of the instance segmentation dataset.
It should be noted that, given the peculiarities of the underwater environment and the anatomy of some fish, the manual review is an important step that cannot be skipped. Some examples of cases when the predictions of SAM need to be corrected are displayed in Figure 1.

Figure 1. Examples of masks inferred by SAM when a bounding box is passed to the model. These selected examples showcase some cases when the model fails to generate correct masks and posterior manual correction is required.
For each image, a list of the fish objects present in the image is included, along with a class ID and the polygon describing the fish mask. Some examples of images of the dataset along with their corresponding labels are displayed in Figure 2.

Figure 2. Examples of images from the dataset and their corresponding labels. The selected examples showcase the diversity of locations and backgrounds present in the dataset.
The dataset is split into a trainval partition composed of 4,173 images (90% of the data) and a test partition composed of 462 images (10% of the data). The dataset distribution in trainval and test is detailed in Table 1.
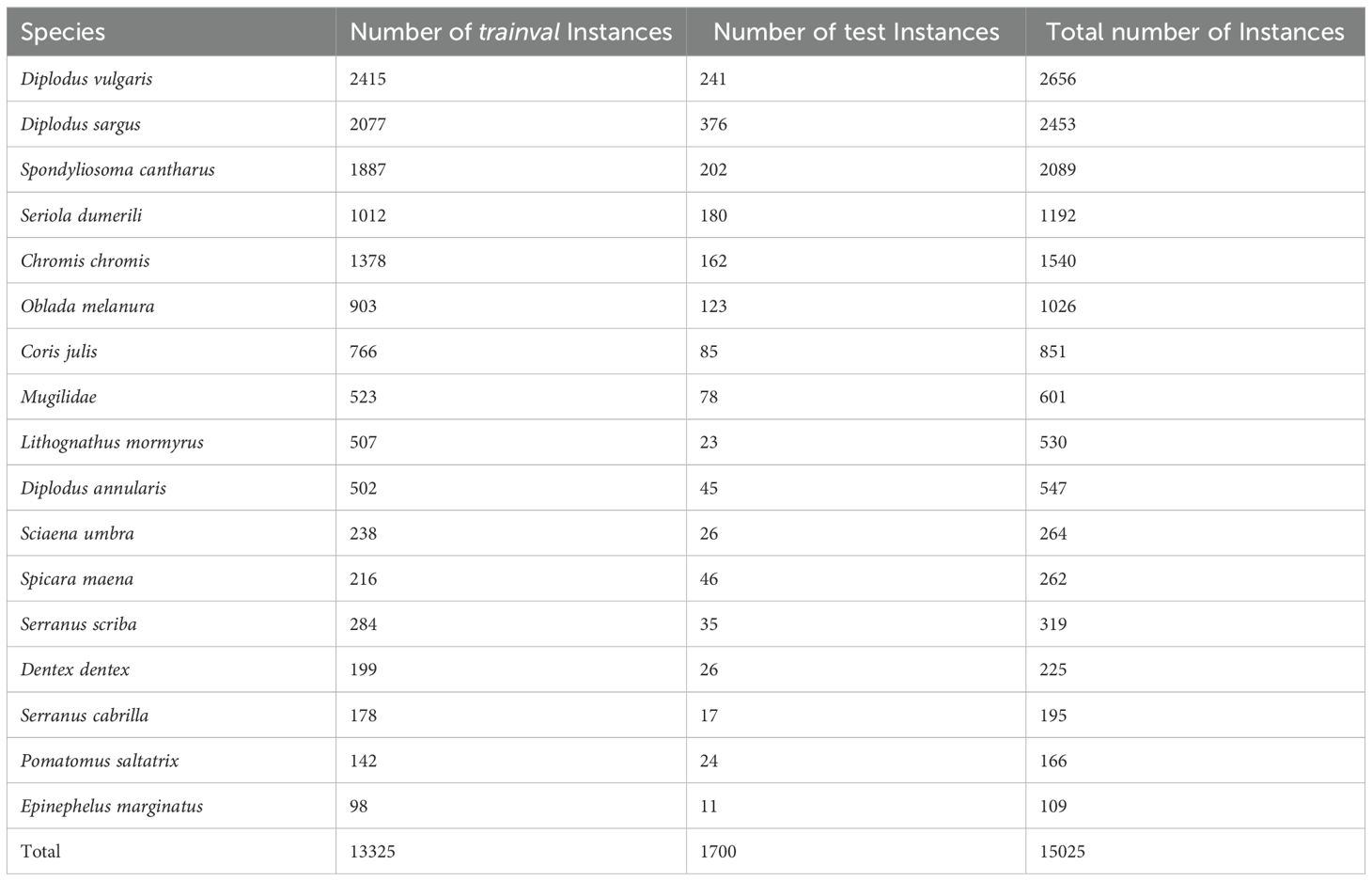
Table 1. Number of instances per species of the dataset categorised into the trainval and test sets, including total counts.
This section presents the selected neural network for classifying and segmenting Mediterranean fish.
YOLO is an open-source, state-of-the-art family of deep learning models widely used for object detection and instance segmentation tasks. Originally introduced by Redmon et al. in 2016 (Redmon et al., 2016), YOLO revolutionised object detection as a single-shot object detector, outperforming other networks like FASTER-RCNN (Ren et al., 2017) in terms of speed, gaining significant popularity in the field. Over time, numerous improvements to the original network have been proposed, resulting in the development of new YOLO versions such as YOLOv3, YOLOv4, YOLOv5, YOLOv6, YOLOv7, YOLOv8, YOLOv9 and YOLOv10 (Redmon and Farhadi, 2018; Bochkovskiy et al., 2020; Jocher, 2020; Wang et al., 2022; Jocher et al., 2023; Li C. et al., 2023).
The v8 version, YOLOv8, has been selected. YOLOv8 incorporates several architectural improvements over previous YOLO versions, including an anchor-free design and mosaic augmentation, which enhance its generalisation capabilities. Additionally, it offers a range of different-sized versions, providing scalability and adaptability to meet the requirements of specific tasks. Among these size options, the large version (YOLOv8l) has been chosen for its balance between high performance and manageable computational demands.
In addition to size scalability, YOLOv8 exhibits task modularity, allowing for variants tailored to specific tasks such as classification, object detection, and instance segmentation. For this work, which focuses on fish segmentation, the segmentation variant YOLOv8l-seg has been selected due to its state-of-the-art performance, speed, and efficiency. The selected model generates masks and confidence percentages for the detected objects as depicted in Figure 3.
A hyperparameter study is conducted to optimise network performance. This involves determining a set of parameters and tuning them. However, not all possible combinations are considered, as the study would require an unassimilable amount of time and resources. Consequently, an ablation study is performed. With this approach, when a hyperparameter is being tuned, all other hyperparameters are held constant. The performance of the network for the different values of the current hyperparameter is then compared, and the best one is selected and fixed before moving on to the next hyperparameter. This method allows for systematic and efficient identification of the optimal hyperparameter settings within the available resources.
The considered hyperparameters are:
● Input image size: defines the resolution of images fed into the network. Adjusting the input image size can influence the performance and inference speed. Higher-resolution images can capture more detail but also require more computation. This parameter is considered for tuning as most of the images are larger than the default size and contain small fish. A significant downscaling can convert these small background fish into noise.
● Data augmentation: consists of applying transformations to the input data (e.g., rotations, translations, or crops) to increase the variety of the data and reduce overfitting. This helps the model generalise unseen data better. While data augmentation is a widely used technique with well-documented benefits, the question of which specific types are most effective remains unresolved. Three options for data augmentation are studied: no data augmentation, the default data augmentation settings, and a custom selection of augmentations, which will henceforth be referred to as custom data augmentation. The main difference between the default and the custom augmentation is the incorporation of copy-paste augmentation and a reduction in the mosaic probability. The rationale for these modifications is that mosaic augmentation can crop or resize a fish, potentially reducing its size, whereas preserving the entire fish and its original size in some images is a desired outcome. By preserving the entire fish and its original size in certain images, and incorporating copy-paste augmentation to address occlusions and enhance background generalisation, the custom approach aims to improve model performance.
● Learning rate: controls the pace at which the network learns by modifying its training step. It influences the convergence speed and stability of the training process. A learning rate that is too high can cause the training to diverge, while a learning rate that is too low can result in slow convergence.
● Class loss weight: specifies the importance of the class prediction loss in the total loss function. Adjusting the class loss weight balances the contribution of classification errors relative to other types of errors, such as localisation and confidence errors. Proper tuning of this weight can improve the ability of the model to accurately classify objects. Some preliminary work suggests that increasing the class loss weight can increase the performance of the model in cases where classification is important (DS & AI Solutions, 2023), as is the case of this work.
● Optimiser: adjusts the parameters of the model to minimise the loss function during training. Common optimisers include Stochastic Gradient Descent (SGD), Adam, and AdamW. Each optimiser has its mechanism for updating weights and can significantly impact the training efficiency and model performance. For each optimiser, the recommended learning rate for YOLOv8-seg is used.
The summary of the tested values for each hyperparameter is shown in Table 2. Each training procedure is conducted with a batch size of seven and the weights are initialised to the pre-trained weights used for the COCO dataset (Lin et al., 2014). The training procedures are carried out in a computer with the following specifications — processor: Intel i9-129000k, RAM: 64 GB, GPU: NVIDIA GeForce RTX 4090.
To validate and assess the robustness of the network hyperparameter selection process, a 5-fold cross-validation is conducted. This 5-fold cross-validation involves dividing the trainval partition of the dataset into five distinct folds. Subsequently, five distinct training procedures are conducted for each hyperparameter combination, with a different fold serving as the validation data and the remaining four folds serving as the training data for each one. Finally, the hyperparameter combination that achieves the best average results is selected, and the best combination of parameters is evaluated over the test partition to test the robustness of the model. Figure 4 illustrates this process.
Typically, deep learning models are evaluated using Precision and Recall metrics, which are calculated based on the number of True Positives (TP), False Positives (FP), and False Negatives (FN), as described in Equations 1 and 2.
To determine whether a prediction is a True Positive (TP) or a False Positive (FP), a localisation metric known as Intersection Over Union (IoU) is employed. When the IoU between a prediction and the ground truth label exceeds a specified threshold (typically 0.5), the prediction is classified as a TP; otherwise, it is classified as an FP. The IoU is calculated using Equation 3:
where the Area of Overlap is the intersection area between the predicted and ground truth boxes or masks, and the Area of Union is the total area covered by the predicted and ground truth boxes or masks.
However, neither Precision nor Recall alone provide a complete picture of model performance. To offer a more comprehensive metric for evaluation, mAP at 50% IoU (mAP50) is used. In the context of YOLOv8-seg, the mAP50 metric is calculated for both bounding boxes (mAP50(B)) and masks (mAP50(M)) to evaluate detection and segmentation accuracy, respectively. This is described by Equations 4 and 5, where AP represents the average precision, which is calculated as the area under the precision-recall curve and N is the number of classes.
The hyperparameter combination that obtains the best results with the 5-fold cross-validation process is selected and set for all the variants of the model presented in this work.
To avoid the loss of valuable information and the inability to classify fish that do not belong to the seventeen studied classes, an experiment is carried out to assess the impact of introducing a generic fish class. As stated in Section 3.1, the dataset contains several labelled fish with insufficient instances to form new classes. These instances have been excluded from the training of the 17-class model and treated as background; however, for this experiment, they are mapped into a generic fish class. The objective of this experiment is to ascertain whether including a generic fish class, in addition to the 17 species, can facilitate a more comprehensive understanding and improve the ability of the system to classify fish not belonging to the seventeen known classes.
Additionally, another experiment is conducted by training a single-class fish segmentation model mapping all species in the dataset into a single generic fish class. This approach enhances the ability of the model to generalise across various habitats and species, providing broader and more adaptable information. This model is evaluated for its generalisation capability using the DeepFish segmentation dataset (Saleh et al., 2020), which includes 310 images of tropical fish from Australia.
This section presents the performance results obtained from the hyperparameter study, examining the influence of each hyperparameter on the results. Additionally, it details the performance of a model trained with the optimised hyperparameters when evaluated against the test set.
Following the selection of the YOLOv8l-seg architecture, a hyperparameter study is conducted to efficiently identify optimal hyperparameter settings given resource constraints using an ablation study, as detailed in Section 3.3. Table 3 presents the results of this hyperparameter study, obtained using 5-fold cross-validation.
First, two different options for image size are studied. It can be observed that increasing the input image size has a significant impact on the model performance. This is not unexpected and can be explained by the dataset containing numerous images with small labelled fish in the background. If the reduction of the images is too significant, as is observed when the input size is set to the default value of 640, these instances are difficult to detect. This introduces noise into the model, which may affect its performance.
Secondly, the impact of data augmentation techniques is evaluated. As anticipated, data augmentation significantly enhances the generalisation capacity of the model. Moreover, it can be observed that the custom augmentation improves the performance of the model by 1% compared to the default data augmentation settings of YOLOv8l-seg.
Following, the SGD, Adam and AdamW optimisers are evaluated. For each optimiser, the recommended learning rate by YOLOv8 is selected. The best result is obtained using the SGD optimiser.
For the learning rate study, it is determined that for the SGD optimiser, the default learning rate value of 0.01 yielded the best results.
Finally, the class loss weight is modified. While the improvement is minor, results show that setting the class loss value to 8 reached the best results.
The combination of hyperparameters that obtained the best results in the hyperparameter study (image_size = 1280, lr = 0.01, custom data augmentations, SGD optimiser, cls_loss = 8) yielded an mAP50(B) of 0.886 and an mAP50(M) of 0.887. This configuration was then evaluated on the test partition, obtaining an mAP50(B) of 0.886 and an mAP50(M) of 0.890, demonstrating its good performance on unseen data.
As far as we are aware, the resulting models achieve superior performance compared to the only publicly available model for Mediterranean fish species classification by Catalan et al (Catalán et al., 2023), which reported an mAP(B) of 0.42 for sixteen classes and an F1-score of 0.75 for eight classes. The trained model presented in this work achieved an F1-score of 0.853 for seventeen species. Despite the differences in the datasets and classes, it is believed that this new model demonstrates enhanced performance by offering a greater number of classes and providing more informative outputs, as YOLOv8l-seg produces instance masks. To the best of our knowledge, this model is the most effective publicly available model for Mediterranean fish species classification and segmentation.
The confusion matrix in Figure 5 illustrates that the model effectively distinguishes between fish species. The most confusions occur between fish with similar characteristics, particularly within the Diplodus genus. The primary weakness of the model is its occasional failure to detect some fish instances, which may be due to the challenge of identifying small fish against complex backgrounds.
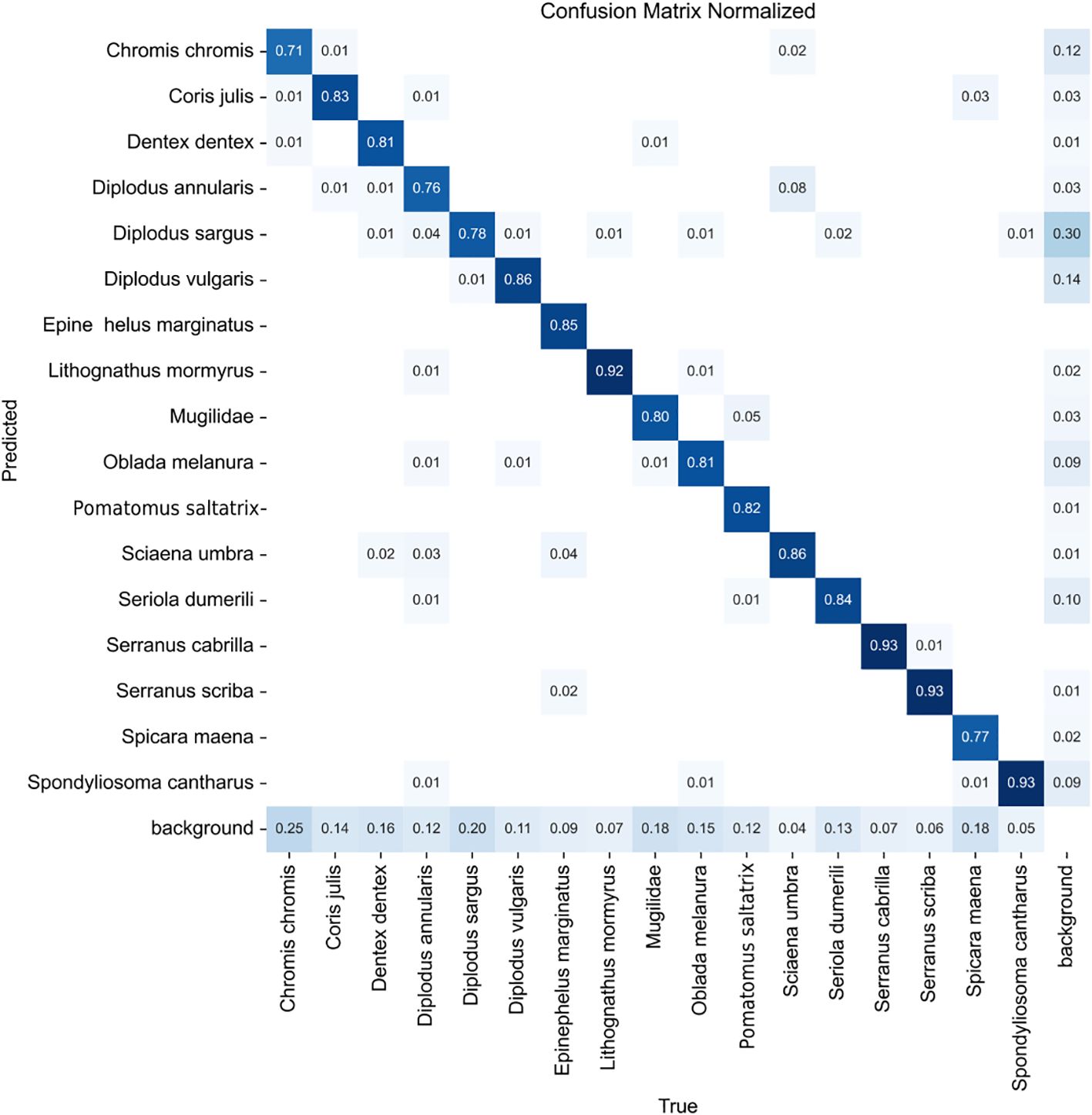
Figure 5. Confusion matrix obtained when evaluating the model trained with the best hyperparameters on the test partition.
To address the issue of missed detections, a new training process that introduces a generic fish class is considered. This class includes individuals from species lacking sufficient instances to be considered distinctive classes in previous experiments. This approach provides insights for future training procedures, specifically regarding whether to include species with a small number of instances as a generic fish class or to exclude them from labelling, allowing the model to consider them as background.
Table 4 illustrates that this approach enables the model to offer additional information without sacrificing overall performance. Although the generic fish class shows relatively low performance, it does not affect the accuracy of the existing class detections and even enhances the results for some of them. As displayed in Figure 6, errors stem from missed or confused detections rather than FPs. The primary confusion occurs with the Diplodus sargus class, likely because many specimens in the generic class are tagged as Diplodus sp., indicating an unclear genus. Consequently, some misclassifications may actually be correct.
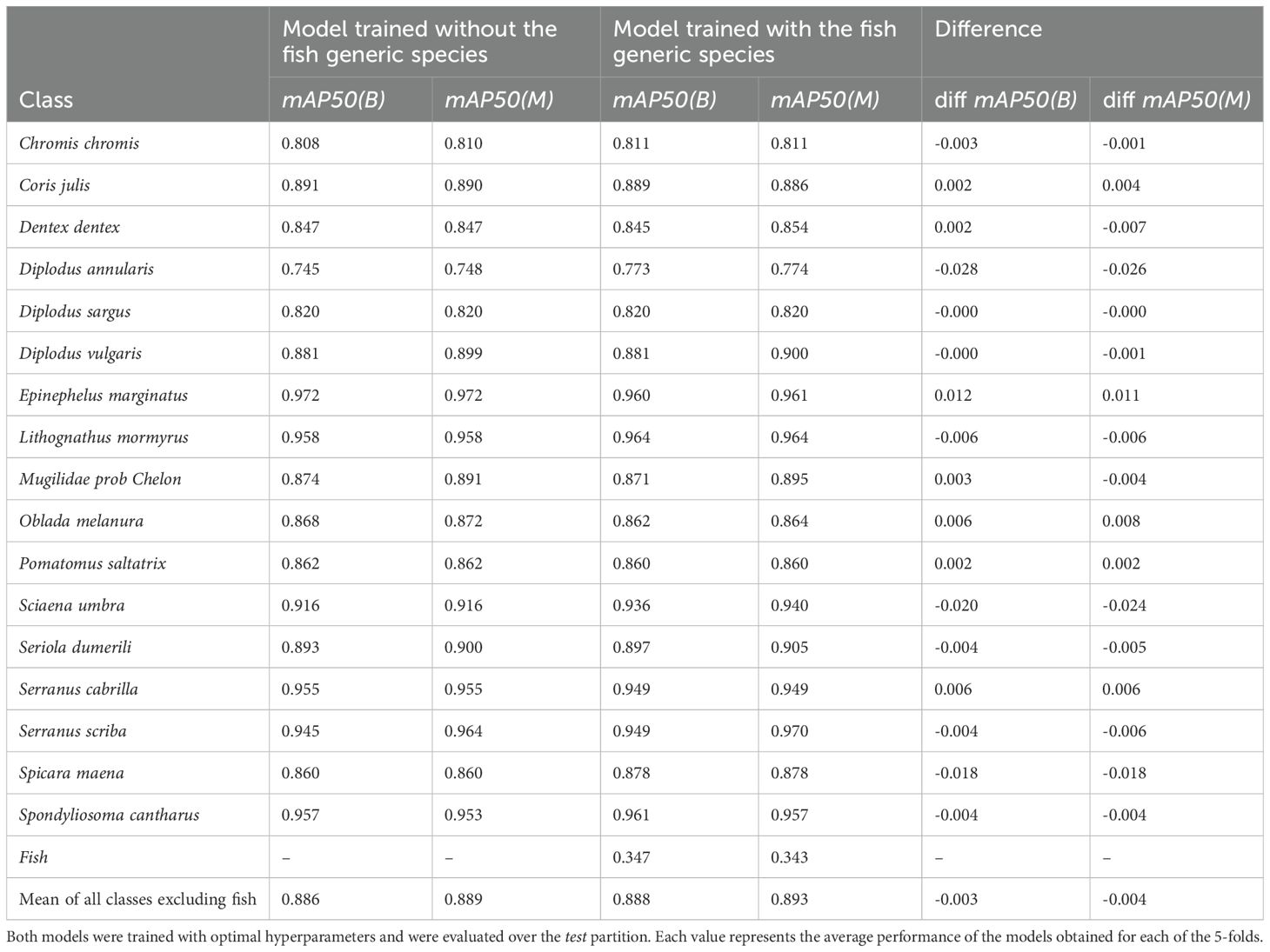
Table 4. Comparison of the mean results per class when including a fish generic class and when excluding it.
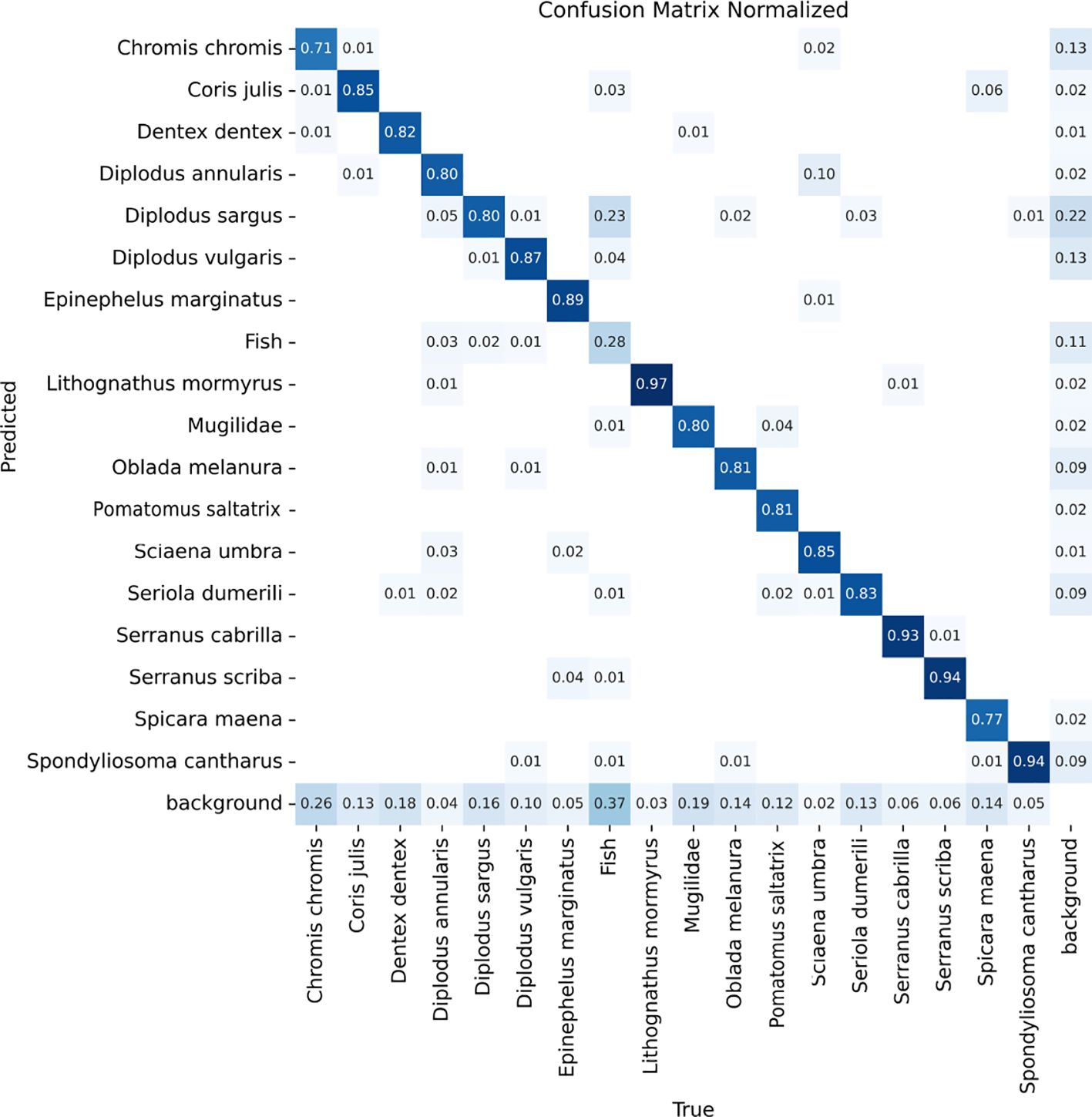
Figure 6. Confusion matrix obtained when evaluating the model trained with the best hyperparameters on the test partition including the fish generic class.
Furthermore, a single-class classification network is also trained by remapping every species of the dataset into a generic fish class to obtain a more general model. The best hyperparameter combination is used to train this single-class model which achieved an mAP50(B) of 0.915 and an mAP50(M) of 0.920. Table 5 shows a comparison between the results obtained for a generic fish species with our dataset and the Deepfish dataset. Even though the fish in the latter dataset are tropical Australian fish, which may differ in morphology and features from those in the Mediterranean, the model sustains its high performance. This demonstrates its robustness and ability to function effectively across different habitats and environments.
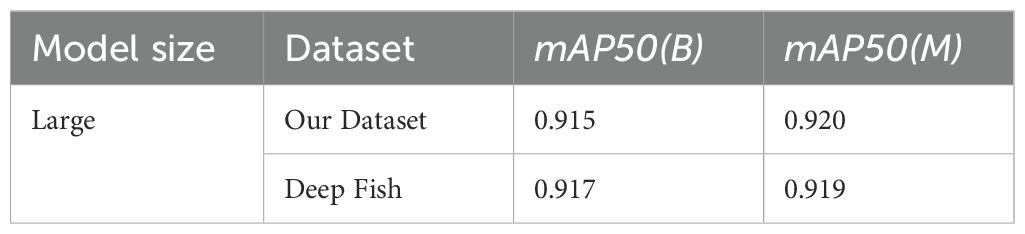
Table 5. Performance comparison of generic fish segmentation large model evaluated on our dataset versus the DeepFish dataset.
Once the best hyperparameters are selected, they are used to train a model, which is then integrated into an SVS, as illustrated in Figure 7. The SVS is equipped with two USB3 Chameleon3 cameras; an Intel Nuc Pro12 running on Ubuntu 20.04 and Robot Operating System (ROS); two LED lights, which are controlled by an Arduino Nano; and powered directly by a high-capacity 14,8V 70 Ah Li-Ion battery. Detailed descriptions of the system can be found in the work of Alfaro-Dufour et al. (2024).
The SVS is subject to power consumption constraints, necessitating the utilisation of a lightweight network. Thus, reducing the model size is essential for enabling efficient online processing.
Real-time operation is crucial, making inference time a key factor. YOLOv8-seg offers a range of models with different sizes, providing the necessary scalability and adaptability for specific application requirements. Inference time for every YOLOv8-seg model was measured on the SVS computer and are detailed in Table 6. The YOLOv8-seg nano (YOLOv8n-seg) model was the fastest, with a processing time of 1.348 seconds per image, allowing for a frame rate of 0.74 frames per second. As a result, the YOLOv8n-seg model is chosen for integration into the underwater SVS.
A periodic process has been implemented to reduce power consumption. This process activates the camera and the acquisition and processing pipeline based on a predefined cycle that includes both acquisition and idle periods. When initiated, the process starts an image capture sequence. Each image captured is then inferred by the trained model. If a fish is detected with a confidence level above a predefined threshold, the image is stored. In addition, detection data is logged, including the time stamp, number of detections, their respective classes and confidence levels. This approach eliminates the need to store large amounts of video and allows continuous, autonomous monitoring of fish presence throughout the deployment. Figure 8 summarises this process.
Given that the SVS is designed to be integrated into a larger monitoring system which may include other marine robots, the network has been integrated into the Robot Operating System (ROS) (Quigley et al., 2009). This framework is a standard in robotics due to its versatility and comprehensive collection of tools and libraries that facilitate the implementation of new code and functionalities. Firstly, the images captured by the camera are retrieved, rectified, fed into the processing unit, and processed by the neural network, which generates predictions of fish instances present in the images. The identified fishes and their masks are published back into ROS for access by other robots, sensors, or actuators.
Field tests have been conducted to test the overall system in real-world conditions. The SVS has been deployed in different locations on the coast of Spain. Figure 9 shows the deployed SVS during field tests. The SVS operated in automatic mode, capturing images at 2-minute intervals, within 10-minute periods. The online inference node identified fish species and published the results as ROS topics. Figure 10 shows an example of the output resulting from the online inference performed during the field tests. Additionally, illustrating the results of online inference conducted both during day and night are available in the Supplementary Material.
To evaluate the performance of the model when working with images obtained by the SVS and assess its generalisation capability, some of the images recorded during field tests have been manually labelled to serve as an independent test set. During real-world experiments, only four fish species from the studied species were found in the recordings. The independent test comprises a total of 98 images including these four fish species. Table 7 describes the detailed number of instances per class.
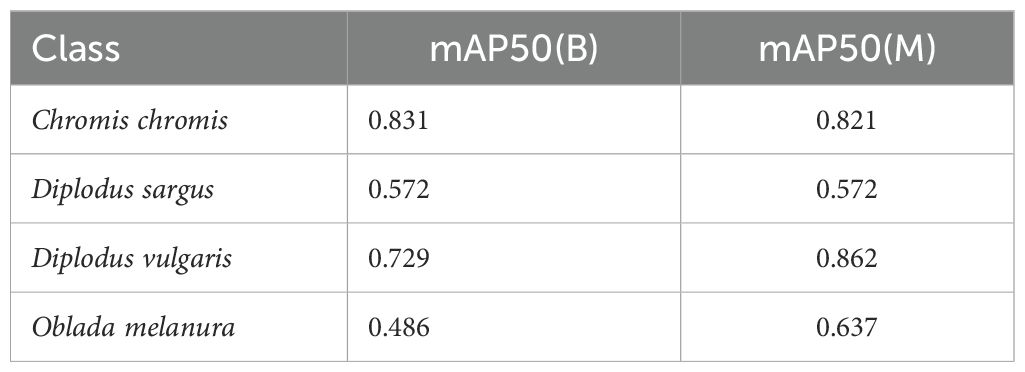
Table 7. Description of the independent test set conformed from images obtained during the field experiments.
Table 8 presents the per-class metrics for the detected species. Although a decrease in performance is observable, this is likely attributable to the field test images comprising numerous tiny fishes, frequently obscured by the background, and featuring classes with highly similar morphology and characteristics. Additionally, it is important to acknowledge that, as previously stated, the nano model is being employed due to due to constraints imposed by online inference times. This model offers inferior performance compared to the large model.
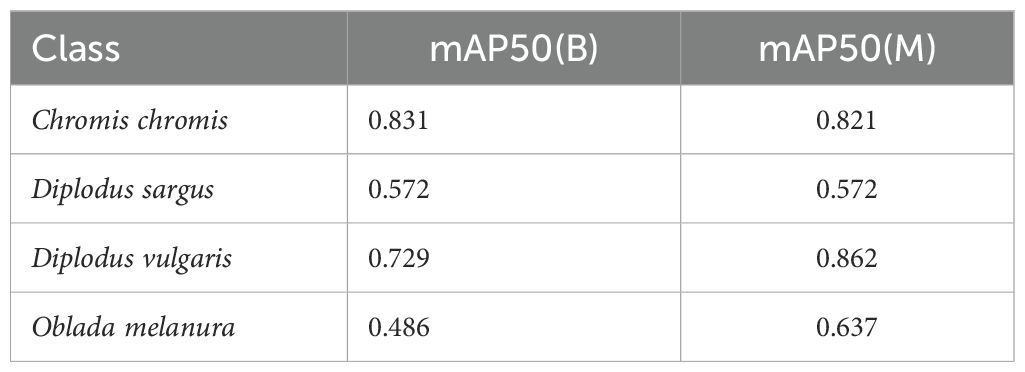
Table 8. Test results obtained when evaluating the model on the independent test obtained during the field tests with the underwater SVS.
This paper presents a new instance segmentation dataset for Mediterranean fish and a deep learning model to automate the process of detecting, classifying, and segmenting seventeen Mediterranean fish species. As far as we are aware, the newly trained model achieved superior performance concerning the best publicly available model, as evidenced by an mAP50(B) and mAP50(M) of 0.886 and 0.889, respectively. The trained model and the data used in this study are available in this Zenodo repository (Muntaner et al., 2024).
A hyperparameter study for YOLOv8l-seg is presented, and questions such as whether the inclusion of a generic fish class benefits the system have been investigated. Although the generic fish class exhibited relatively lower performance, it did not introduce noise into the model and enhanced the performance for some of the classes. The primary errors observed were associated with missed or confused detections rather than FPs, indicating the robustness of the classification system.
Furthermore, the training and integration of a lightweight version of the network into an SVS have enabled real-time processing, achieving inference times suitable for online applications. This advancement facilitates autonomous and continuous monitoring of fish presence, highlighting the utility of the model in real-world underwater environments.
While the model demonstrates strong performance, its real-world deployment also highlights certain limitations. Challenges such as variations in species appearance due to changing lighting and water conditions, as well as the detection of small or distant specimens, require further investigation. Future work will focus on increasing its robustness, reliability, and adaptability to diverse underwater conditions. Nonetheless, the results demonstrate that, despite these challenges, the system performs well in real-world environments and can serve as a powerful tool for reducing the cost of data acquisition and processing for researchers.
Additionally, semi-supervised learning techniques will be utilised to augment the number of classes without the need for extensive manual labelling. Expanding the taxonomy will facilitate a more comprehensive understanding of the underwater ecosystem, contributing to more detailed and accurate monitoring efforts.
Moreover, efforts will be made to reduce the computational load of the system. The objective is to achieve a reduction in power consumption and inference times by optimising the architecture of the network. This will enhance the efficiency of the system, making it more suitable for prolonged autonomous deployment in energy-constrained environments.
Finally, it is planned to conduct more diverse and extended field tests to gather comprehensive data that can be used to evaluate and refine the performance of the model in real-world scenarios.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://zenodo.org/records/13120727?token=eyJhbGciOiJIUzUxMiJ9.eyJpZCI6ImU3ZTRlOTQ3LTYyNzAtNDcyYy04MjJhLWIwN2U2ZTEwZmVkZSIsImRhdGEiOnt9LCJyYW5kb20iOiIxZDcwNDE2NjMwZTljNWNiNDZhNzkxYjYzZDc5ZjkzOSJ9.5m4PbN5J-fB1oubvtCu5NshPNL9ZEu0VAiqKMg4Qc32W142fc5JrihCQ_GrmPwJtbamE6ZsSwLgzIAs1nNzkZg.
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because the data used was publicly available.
CM: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. AN: Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. MM: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YG: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. Caterina Muntaner-Gonzalez was supported by the Consejería de Educación y Universidades del Gobierno de las Illes Balears under the contract FPU-2022-010-C CAIB 2022. This work is partially supported by ERDF A way of making Europe, by Grant PLEC2021-007525/AEI/10.13039/501100011033 funded by the Agencia Estatal de Investigación, under Next Generation EU/PRTR. This work has been partially sponsored and promoted by the Comunitat Autònoma de les Illes Balears through the Direcció General de Recerca, Innovació i Transformació Digital and the Conselleria de Economia, Hisenda i Innovació via Plans complementaris del Pla de Recuperació, Transformació i Resiliència (EU/PRTR-C17. I1.). This work has been partially sponsored and promoted by the Comunitat Autonòma de les Illes Balears through the Conselleria d’Educació I Universitats and by the European Union - Next Generation EU (BIO/002A.1 and BIO/022B.1). The authors are grateful for the support received from Grant PID2020-115332RB-C33 funded by MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe”.
The authors would like to thank IMEDEA and OBSEA for their support in helping us build the dataset for this study. We also appreciate the Roboflow and Minka communities for sharing data, which contributed to the development of our research. We would like to thank the Spanish Navy, especially its naval base in the Port of Soller (Spain), for its collaboration and for the loan of its facilities, which have been key to carrying out the experimental tasks of this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The views and opinions expressed are solely those of the author or authors, and do not necessarily reflect those of the Conselleria d’Educació i Universitats, the European Union or the European Commission. Therefore, none of these organizations shall not be held liable.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1525524/full#supplementary-material
Abinaya N. S., Susan D., Sidharthan R. K. (2022). Deep learning-based segmental analysis of fish for biomass estimation in an occulted environment. Computers and Electronics in Agriculture. 197, 106985. doi: 10.1016/j.compag.2022.106985
Aguzzi J., Doya C., Tecchio S., De Leo F., Azzurro E., Costa C., et al. (2015). Coastal observatories for monitoring of fish behaviour and their responses to environmental changes. Rev. Fish Biol. Fisheries 25, 463–483. doi: 10.1007/s11160-015-9387-9
Alaba S. Y., Nabi M., Shah C., Prior J., Campbell M. D., Wallace F., et al. (2022). Class-aware fish species recognition using deep learning for an imbalanced dataset. Sensors 22, 8268. doi: 10.3390/s22218268
Alfaro-Dufour E., Muntaner-González C., Martorell-Torres A., Oliver-Codina G. (2024). “Lanty: A deep sea stereo vision system,” in 2024 IEEE International Conference on Industrial Technology (ICIT) (Bristol, United Kingdom: IEEE), 1–6. doi: 10.1109/ICIT58233.2024.10540725
Alsmadi M. K., Tayfour M., Alkhasawneh R. A., Badawi U., Almarashdeh I., Haddad F. (2019). Robust feature extraction methods for general fish classification. Int. J. Electrical Comput. Eng. (2088-8708) 9, 5192–5204. doi: 10.11591/ijece.v9i6.pp5192-5204
Álvarez-Ellacuría A., Palmer M., Catalán I. A., Lisani J. L. (2020). Image-based, unsupervised estimation of fish size from commercial landings using deep learning. ICES J. Mar. Sci. 77, 1330–1339. doi: 10.1093/icesjms/fsz216
Bochkovskiy A., Wang C. Y., Liao H. Y. M. (2020). Yolov4: Optimal speed and accuracy of object detection. arXiv preprint arXiv:2004.10934. doi: 10.48550/arXiv.2004.10934
Catalán I. A., Álvarez-Ellacuría A., Lisani J. L., Sánchez J., Vizoso G., Heinrichs-Maquilón A. E., et al. (2023). Automatic detection and classification of coastal Mediterranean fish from underwater images: Good practices for robust training. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1151758
Coro G., Bjerregaard Walsh M. (2021). An intelligent and cost-effective remote underwater video device for fish size monitoring. Ecol. Inf. 63, 101311. doi: 10.1016/j.ecoinf.2021.101311
Cui S., Zhou Y., Wang Y., Zhai L. (2020). Fish detection using deep learning. Appl. Comput. Intell. Soft Comput. 2020, 3738108. doi: 10.1155/2020/3738108
dataset O. (2023). Merged + filtered dataset. Available online at: https://universe.roboflow.com/fish-od-w8vfm/merged-filtered (Accessed August 6, 2024).
Dataset I. (2023). Imedea dataset. Available online at: https://universe.roboflow.com/fish-od-w8vfm/imedea (Accessed August 6, 2024).
Deep B. V., Dash R. (2019). “Underwater fish species recognition using deep learning techniques,” in 2019 6th International Conference on Signal Processing and Integrated Networks, SPIN 2019 (Noida, India: IEEE), 665–669. doi: 10.1109/SPIN.2019.8711657
dos Santos A. A., Gonçalves W. N. (2019). Improving pantanal fish species recognition through taxonomic ranks in convolutional neural networks. Ecol. Inf. 53, 100977. doi: 10.1016/j.ecoinf.2019.100977
Dwyer B., Nelson J., Hansen T., et al. (2024). Roboflow (version 1.0). Available online at: https://roboflow.com.computervision (Accessed March 14, 2025).
Effrosynidis D., Tsikliras A., Arampatzis A., Sylaios G. (2020). Species distribution modelling via feature engineering and machine learning for pelagic fishes in the mediterranean sea. Appl. Sci. 10, 8900. doi: 10.3390/app10248900
Garcia R., Prados R., Quintana J., Tempelaar A., Gracias N., Rosen S., et al. (2020). Automatic segmentation of fish using deep learning with application to fish size measurement. ICES J. Mar. Sci. 77, 1354–1366. doi: 10.1093/icesjms/fsz186
Garcia-Soto C., Seys J. J., Zielinski O., Busch J. A., Luna S., Baez J. C., et al. (2021). Marine citizen science: Current state in europe and new technological developments. Front. Mar. Sci. 8, 621472. doi: 10.3389/fmars.2021.621472
Hafiz A. M. (2020). Bhat GM. A survey on instance segmentation: state of the art. Int. J. Multimedia Inf. Retrieval 9, 171–189. doi: 10.1007/s13735-020-00195-x
Hu J., Li D., Duan Q., Han Y., Chen G., Si X. (2012). Fish species classification by color, texture and multi-class support vector machine using computer vision. Comput. Electron. Agric. 88, 133–140. doi: 10.1016/j.compag.2012.07.008
IMEDEA (2024). Sub-eye: Underwater observatory – port d’andratx underwater cabled coastal observatory (Accessed August 6, 2024).
Jalal A., Salman A., Mian A., Shortis M., Shafait F. (2020). Fish detection and species classification in underwater environments using deep learning with temporal information. Ecol. Inf. 57, 101088. doi: 10.1016/j.ecoinf.2020.101088
Kirillov A., Mintun E., Ravi N., Mao H., Rolland C., Gustafson L., et al. (2023). Segment anything. In Proceedings of the IEEE/CVF international conference on computer vision. pp. 4015–4026. doi: 10.48550/arXiv.2304.02643
Knausgård K. M., Wiklund A., Sørdalen T. K., Halvorsen K. T., Kleiven A. R., Jiao L., et al. (2022). Temperate fish detection and classification: a deep learning based approach. Appl. Intell. 52, 6988–7001. doi: 10.1007/s10489-020-02154-9
Li C., Li L., Geng Y., Jiang H., Cheng M., Zhang B., et al. (2023). Yolov6 v3.0: A full-scale reloading. doi: 10.48550/arXiv.2301.05586
Li J., Xu W., Deng L., Xiao Y., Han Z., Zheng H. (2023). Deep learning for visual recognition and detection of aquatic animals: A review. Rev. Aquacult. 15, 409–433. doi: 10.1111/raq.12726
Lin T. Y., Maire M., Belongie S., Hays J., Perona P., Ramanan D., et al. (2014). “Microsoft coco: Common objects in context,” in Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, September 6-12, 2014, Proceedings, Part V 13 (Zurich, Switzerland: Springer), 740–755. doi: 10.1007/978-3-319-10602-1_48
Liu P., Yang H., Fu J. (2020). “Marine biometric recognition algorithm based on yolov3-gan network,” in MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, South Korea, January 5–8, 2020, Proceedings, Part I 26 (Daejeon, South Korea: Springer), 581–592. doi: 10.1007/978-3-030-37731-1_47
Lopez-vazquez V., Lopez-guede J. M., Marini S., Fanelli E., Johnsen E., Aguzzi J. (2020). Video image enhancement and machine learning pipeline for underwater animal detection and classification at cabled observatories. Sens. (Switzerland) 20 (3), 726. doi: 10.3390/s20030726
McClure E. C., Sievers M., Brown C. J., Buelow C. A., Ditria E. M., Hayes M. A., et al. (2020). Artificial intelligence meets citizen science to supercharge ecological monitoring. Patterns 1 1 (7), 100109. doi: 10.1016/j.patter.2020.100109
Muñoz-Benavent P., Martínez-Peiró J., Andreu-García G., Puig-Pons V., Espinosa V., Pérez-Arjona I., et al. (2022). Impact evaluation of deep learning on image segmentation for automatic bluefin tuna sizing. Aquacult. Eng. 99, 102299. doi: 10.1016/j.aquaeng.2022.102299
Muntaner C., Nadal-Martínez A., Martin-Abadal M., Gonzalez-Cid Y. (2024). Automatic deep learningbased pipeline for mediterranean fish segmentation dataset. doi: 10.5281/zenodo.13120727
Nogueras M., del Rio J., Cadena J., Sorribas J., Artero C., Dañobeitia J., et al. (2010). “Obsea an oceanographic seafloor observatory,” in 2010 IEEE International Symposium on Industrial Electronics (Bari, Italy: IEEE), 488–492. doi: 10.1109/ISIE.2010.5637675
Ogunlana S., Olabode O., Oluwadare S., Iwasokun G. (2015). Fish classification using support vector machine. Afr. J. Comput. ICT 8, 75–82.
Qin H., Li X., Liang J., Peng Y., Zhang C. (2016). DeepFish: Accurate underwater live fish recognition with a deep architecture. Neurocomputing 187, 49–58. doi: 10.1016/j.neucom.2015.10.122
Quigley M., Conley K., Gerkey B., Faust J., Foote T., Leibs J., et al. (2009). “Ros: an open-source robot operating system,” in ICRA workshop on open source software, vol. vol. 3. (Kobe, Japan: IEEE), 5.
Rathi D., Jain S., Indu S. (2018). “Underwater fish species classification using convolutional neural network and deep learning,” in 2017 9th International Conference on Advances in Pattern Recognition, ICAPR 2017 (Bangalore, India: IEEE), 344–349. doi: 10.1109/ICAPR.2017.8593044
Redmon J., Divvala S., Girshick R., Farhadi A. (2016). “You only look once: Unified, real-time object detection,” in Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (Las Vegas, NV, USA: IEEE), 779–788.
Redmon J., Farhadi A. (2018). Yolov3: An incremental improvement. arXiv preprint arXiv:1804.02767. doi: 10.48550/arXiv.1804.02767
Ren S., He K., Girshick R., Sun J. (2017). Faster r-cnn: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 39, 1137–1149. doi: 10.1109/TPAMI.2016.2577031
Roberts C. J., Vergés A., Callaghan C. T., Poore A. G. (2022). Many cameras make light work: opportunistic photographs of rare species in inaturalist complement structured surveys of reef fish to better understand species richness. Biodivers. Conserv. 31, 1407–1425. doi: 10.1007/s10531-022-02398-6
Saleh A., Laradji I. H., Konovalov D. A., Bradley M., Vazquez D., Sheaves M. (2020). A realistic fish-habitat dataset to evaluate algorithms for underwater visual analysis. Sci. Rep. 10, 14671. doi: 10.1038/s41598-020-71639-x
Saleh A., Sheaves M., Rahimi Azghadi M. (2022). Computer vision and deep learning for fish classification in underwater habitats: A survey. Fish Fisheries 23, 977–999. doi: 10.1111/faf.12666
Salman A., Jalal A., Shafait F., Mian A., Shortis M., Seager J., et al. (2016). Fish species classification in unconstrained underwater environments based on deep learning. Limnol. Oceanogr.: Methods 14, 570–585. doi: 10.1002/lom3.10113
Siddiqui S. A., Salman A., Malik M. I., Shafait F., Mian A., Shortis M. R., et al. (2018). Automatic fish species classification in underwater videos: Exploiting pre-trained deep neural network models to compensate for limited labelled data. ICES J. Mar. Sci. 75, 374–389. doi: 10.1093/icesjms/fsx109
Szymak P., Piskur P., Naus K. (2020). The effectiveness of using a pretrained deep learning neural networks for object classification in underwater video. Remote Sens. 12, 1–19. doi: 10.3390/RS12183020
Tiralongo F., La Mesa G., De Mendoza F. P., Massari F., Azzurro E. (2021). Underwater photo contests to complement coastal fish inventories: Results from two mediterranean marine protected areas. Mediterr. Mar. Sci. 22, 436–445. doi: 10.12681/mms.26176
Ubina N. A., Cheng S. C., Chang C. C., Cai S. Y., Lan H. Y., Lu H. Y. (2022). Intelligent underwater stereo camera design for fish metric estimation using reliable object matching. IEEE Access 10, 74605–74619. doi: 10.1109/ACCESS.2022.3185753
Wang C. Y., Bochkovskiy A., Liao H. Y. M. (2022). YOLOv7: Trainable bag-of-freebies sets new stateof-the-art for real-time object detectors. arXiv preprint arXiv:2207.02696. doi: 10.48550/arXiv.2207.02696
Keywords: deep learning, instance segmentation, fish classification, Mediterranean fish, ecosystem monitoring
Citation: Muntaner-Gonzalez C, Nadal-Martínez A, Martin-Abadal M and Gonzalez-Cid Y (2025) Automatic deep learning-based pipeline for Mediterranean fish segmentation. Front. Mar. Sci. 12:1525524. doi: 10.3389/fmars.2025.1525524
Received: 11 November 2024; Accepted: 24 February 2025;
Published: 21 March 2025.
Edited by:
Fabio Fiorentino, National Research Council (CNR), ItalyReviewed by:
Francesco Tiralongo, University of Catania, ItalyCopyright © 2025 Muntaner-Gonzalez, Nadal-Martínez, Martin-Abadal and Gonzalez-Cid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caterina Muntaner-Gonzalez, Yy5tdW50YW5lckB1aWIuY2F0
†ORCID: Caterina Muntaner-Gonzalez, orcid.org/0009-0003-9091-7970
Antonio Nadal-Martinez, orcid.org/0009-0003-1782-3433
Miguel Martin-Abadal, orcid.org/0000-0003-1925-7378
Yolanda Gonzalez-Cid, orcid.org/0000-0001-6155-2288
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.