- 1Atlantic Oceanographic and Meteorological Laboratory, Ocean and Atmospheric Research, National Oceanic and Atmospheric Administration, Miami, FL, United States
- 2Cooperative Institute for Marine and Atmospheric Studies, Rosenstiel School for Marine, Atmospheric, & Earth Science, University of Miami, Miami, FL, United States
Within the Gulf of Mexico (GOM), measurements of ocean pH are limited across space and time. This has hindered our ability to robustly monitor and study regional carbon dynamics, inclusive of ocean acidification, over this biogeochemically variable sea. The 2021 launch of Biogeochemical-Argo (BGC-Argo) ocean profiling floats that carry five sensors represented the entry of this particular ocean observing technology into this region. The GOM BGC-Argo floats have vastly increased the number of oxygen, nitrate, pH, chlorophyll-a fluorescence, and particulate backscattering profile observations within the “open GOM” region (>1,000 m water column depth). To circumvent a set of uncertainties associated with the collected sensor pH data, regionally trained neural network algorithms were developed to skillfully predict GOM pH (total scale, in situ temperature and pressure), which served as a secondary QC and sensor performance assessment tool. The GOM neural network pH (GOM-NNpH) algorithms were trained using a selection of climate quality CTD and bottle data (temperature, salinity, oxygen, nitrate, pressure, and location) collected as a part of NOAA’s Gulf of Mexico Ecosystems and Carbon Cruises (GOMECC). Neural network pH estimates were generated using the newly developed GOMNNpH algorithm and two widely used, globally trained neural network algorithms (Empirical Seawater Property Estimation Routines (ESPER) and CArbonate system and Nutrients concentration from hYdrological properties and Oxygen using a Neural-network (CANYON-B)) to compare algorithm performance against validation data. The results demonstrate the advanced skill of the GOM-NNpH in capturing water column variability and robustly reconstructing GOM pH profiles. Using a combination of concurrent float-measured seawater values of pressure, temperature, salinity, and oxygen, a GOM-NNpH algorithm was applied to two years of BGC-Argo float data. Resulting data were used to diagnose the performance of float pH sensors and to generate a time series of neural network estimated pH based on the collected float profiles. These algorithms emphasize the value of regionally-trained neural networks and their utility by the BGC-Argo community. Further, the GOM-NNpH algorithms can also be applied by a variety of users to estimate pH values in the open GOM in the absence of direct pH observations.
1 Introduction
Sustained ocean observations of pH and other ocean carbon system variables are central to monitoring the anthropogenic carbon impacts on our oceans (Weller et al., 2019). As a range of climate-related stressors emerge, inclusive of acidification, warming, and deoxygenation, they will have cascading implications for marine organisms, ecosystems, and services they provide (Doney et al., 2012; Henson et al., 2017). The United Nations (UN), though the World Meteorological Organization and the Intergovernmental Oceanographic Commission, governs the Global Ocean Observing System (GOOS), which is designed to monitor climate impacts through a blend of numerous ocean observing approaches and technologies (ships, fixed moorings, autonomous vehicles and profiling floats) that have varied capabilities (measurement type, sampling resolution, quality) (Chai et al., 2020; Roemmich et al., 2021). GOOS recognizes ocean pH as an Essential Ocean Variable that can be used to characterize the invasion of anthropogenic carbon and climate related ocean changes (GOOS, 2019). While the global coverage of ocean observations has vastly increased over the last several decades, major gaps in ocean biogeochemical observing, and particularly for pH, still exist across the globe (Claustre et al., 2020). The OneArgo vision for launching 1,000 globally distributed Biogeochemical-Argo (BGC-Argo) floats, capable of returning near real-time observations of water column pH, will revolutionize our global-scale understanding of ocean acidification (Roemmich et al., 2021), allowing major advances towards UN and GOOS goals.
Gaps in ocean pH observations can be particularly prevalent in marginal seas, such as the Gulf of Mexico (GOM), which are often not included in large-scale observational campaigns such as GO-SHIP or have observational data that are not publicly available. The absence of open GOM (which we define here as >1,000 m water depth regions) observations was acutely demonstrated following the Deepwater Horizon oil spill where the lack of pre-event environmental data hindered a basic assessment of the oil and dispersant effects ocean biogeochemistry (Passow and Overton, 2021). Intensive efforts to characterize the GOM following this environmental disaster revealed that the GOM is home to one of the most diverse mesopelagic (200-1000 m) ecosystems in the world ocean (Sutton et al., 2017, 2022). Of note, the GOM mid-water ecosystem is regarded as one of the most “hyper-diverse” regions of the global ocean, with tight connections to the epipelagic (0-200 m) and the bathypelagic (>1,000 m) through diel vertical migration and various life stages (Sutton et al., 2020). The general lack of a sustained open GOM time series for variables such as oxygen, nutrients, and inorganic carbon diminishes our capability to establish baselines of biologically-relevant essential ocean variables as well as monitoring climate impacts (Pasqueron de Fommervault et al., 2017; Osborne et al., 2022).
In the GOM, four BGC-Argo floats were deployed in late 2021, representing the first in situ observations of their kind within the region. The suite of sensor packages mounted on the GOM BGC-Argo floats simultaneously measure temperature, salinity, pressure, oxygen, nitrate, pH, chlorophyll-a, and optical backscatter over the upper 2,000 meters of the water column on 10-day cycles. These observations, if sustained, offer the potential to detect in situ long-term change and variability by spatiotemporally characterizing the GOM. The advancement of BGC-Argo sensor technology to observe water column pH now complement ship data, and have the potential to vastly increasing the frequency and spatial coverage of observations not only in the GOM but across the globe. However, ship-based profiles remain the highest quality data source for monitoring ocean carbonate chemistry and serve critical role in float data validation and quality control procedures for the growing global array of BGC-Argo floats.
Publicly available and high-quality observations of carbonate chemistry variables within the GOM, particularly pH in the open GOM, have been highly limited (Osborne et al., 2022). Based on the Global Ocean Data Analysis Project (GLODAP v2.2023) dataset, 19 unique pH profiles collected between July 2007 and August 2017 are available in the >2,000 m domain of the GOM (Lauvset et al., 2023). The World Ocean Atlas database, used to generate the widely used World Ocean Atlas climatologies, contains 35 ship-based pH profiles collected between 1933 and 1974 (Boyer et al., 2016). Collectively, 58 pH shipboard profiles (with potentially some overlap in data) are available in these widely used ocean databases. The majority of these profiles belong to the NOAA’s Gulf of Mexico Ecosystems and Carbon Cruises (GOMECC) program that has been conducted on roughly four-year intervals (July-August 2012, July-August 2017, and September-October 2021). These cruises make up an unparalleled basin-wide “climate quality” carbonate chemistry bottle dataset for the GOM (Wanninkhof et al., 2013; Wanninkhof and Baringer, 2014; Barbero et al., 2019, 2024);. Climate quality refers to analytical uncertainty of approximately 0.003 in pH (Newton et al., 2015), a precision that is only currently achievable by a very limited number of laboratories such as NOAA’s Atlantic Oceanographic and Meteorological Laboratory (AOML), which analyzed samples collected during GOMECC. While not considered climate quality, BGC-Argo float pH profiles vastly build out the historical GOM pH dataset and, at the time of writing, just two years of BGC float operations in the GOM had already yielded 103 pH profiles.
While innovative, cutting edge, and the first of its kind, pH float sensors and handling of sensor data continue to be in development. Measurement of pH via sensor technology is especially challenging in part due to the very low concentrations of hydrogen ions in seawater. A census of the global BGC-Argo array showed that sensor challenges have slowed the growth of the pH dataset produced by the array, though major advancements in sensor lifetime and data quality are underway (Stoer et al., 2023). A recent study in the North Atlantic, showed cases where considerable differences between float data and validation data exist, raising important questions and driving active research on how to best quality control and adjust problematic float pH profiles (Wimart-Rousseau et al., 2024). Moreover, biases in the quality correction of concurrent oxygen profiles, which are used to determine BGC float pH correction factors, can cause significant biases to be propagated into float pH data, particularly at depth (Bushinsky et al., 2024). pH quality control assessments have been challenged by the “pH-pump offset” phenomenon that is observed in some but not all pH-equipped floats and results from the small changes in the flow of water over the sensor, impacting the quality of pH measurements (Johnson et al., 2023). While agreed upon quality control and correction procedures can address many of these challenges, inconsistencies in sensor behavior can at times make correction procedures difficult to implement.
Identifying solutions to these challenges will permit the maximal use of the revolutionary BGC-Argo time series and its ability to answer long-standing ocean carbon questions. pH sensor challenges faced by the particular floats operating in the GOM BGC-Argo array provided an opportunity to develop a series of regionally trained machine learning neural network algorithms to 1) assess float pH sensor performance and 2) as an alternate way to approximate pH using float observations. Due to the strong relationships that exist between ocean physical (i.e., temperature, salinity) and biogeochemical (i.e., oxygen, pH, nitrate) parameters of seawater (e.g., Juranek et al., 2011; Williams et al., 2016; Carter et al., 2018), unobserved ocean variables can often be accurately extrapolated from combinations of measured variables (e.g., Bittig et al., 2018; Carter et al., 2021).
Neural network algorithms have the ability to model complex non-linear relationships between predictors and estimated quantities (Hornik et al., 1989; Tu, 1996) that are often observed in ocean biogeochemistry, particularly carbonate chemistry. Two recently developed, globally trained, and widely used neural networks, Empirical Seawater Property Estimation Routines (ESPER); Carter et al., 2021) and the CArbonate system and Nutrients concentration from hYdrological properties and Oxygen using a Neural-network (CANYON-B; Bittig et al., 2018) are widely used within the BGC-Argo community. ESPER routines published by Carter et al. (2021) include both locally interpolated regressions (ESPER_LIR) and neural network algorithms (ESPER_NN). We chose to assess the performance specifically of neural network routines, therefore for the remainder of this manuscript ESPER refers to ESPER_NN algorithms published by Carter et al. (2021). The 2021 update to the ESPER routines represent the most recently published advances in ocean-based neural network algorithms and are based on quality-controlled observations in the Global Ocean Data Analysis Project (GLODAP) version 2 (GLODAPv2.2020; Olsen et al., 2020). GLODAP v2.2020 includes quality-controlled data from 946 cruises, covering the global ocean from 1972 to 2019. ESPER are a series of feed-forward neural network algorithms that have been trained using combinations of climate-quality observations of ocean physics and biogeochemistry across the global ocean (Carter et al., 2021). CANYON-B (Bittig et al., 2018) is a Bayesian, rather than a plain feed-forward, neural network approach that incorporates uncertainty in its training data. Specifically, local (rather than globally constant) uncertainty values associated with the input data were to train CANYON-B, permitting a more representative output uncertainty, which is likely underestimated by plain feed-forward neural networks (Bittig et al., 2018). Relative to the ESPER routines, the CANYON-B neural network is trained using older and therefore smaller version of GLODAPv2 (Key et al., 2015; Olsen et al., 2016). To date, these estimation algorithms have proven central to the creation of global data products (Carter et al., 2021; Sharp et al., 2022; Ito et al., 2024), to filling observational holes in datasets (Carter et al., 2019; Jiang et al., 2019, 2024), and to adjusting sensor data collected by autonomous sensor platforms (Maurer et al., 2021).
While globally-trained algorithms can be applied to marginal seas, the value of a regionally-trained Mediterranean Sea neural network algorithm demonstrated its enhanced ability to represent peculiar conditions of this region relative to globally trained neural networks (Fourrier et al., 2020). While Carter et al. (2021) intentionally incorporated datasets outside of GLODAPv2.2020 to include marginal seas (such as the GOM), these data were used in ESPER validation rather than training procedures and CANYON-B lacks GOM training and validation data entirely. We assert that it is possible that larger errors would result when applying global algorithms to marginal sea regions due to distinct region-specific physical and biogeochemical dynamics not represented in the training dataset. For the GOM, these dynamics include items such as the Loop Current and mesoscale eddies (McWhorter et al., 2024) and major riverine outflows that reach beyond the continental margins (Gomez et al., 2024). Using the GOMECC cruise dataset, we developed a set of Gulf of Mexico Neural Network pH (GOM-NNpH) algorithms, which are feed-forward algorithms trained using climate quality GOM-collected shipboard bottle data. These algorithms skillfully estimate pH, which use four combinations of predictor variables (latitude, longitude, pressure, temperature, salinity, oxygen, and nitrate). We compare the performance of the GOM-trained neural network to ESPER and CANYON-B to determine the importance of employing a regionally trained neural network. We then apply our GOM-NNpH to assess performance and diagnose pH sensor behavior for the GOM BGC float array.
2 Data and methods
2.1 Biogeochemical-Argo floats
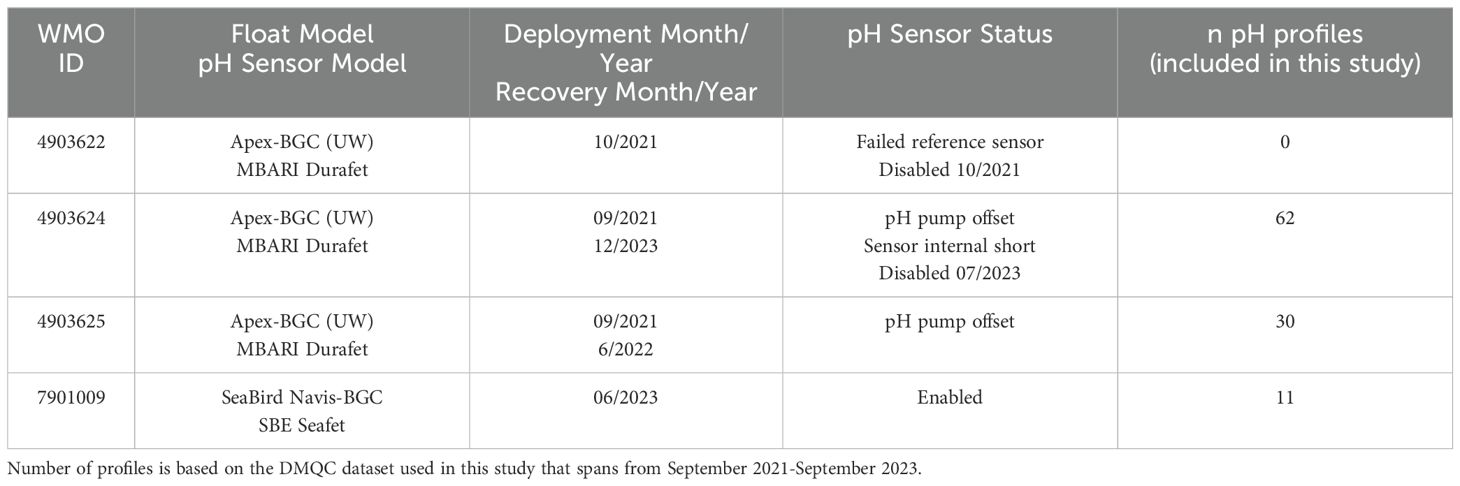
BGC-Argo floats are equipped with five to six sensors including 1) a CTD (conductivity, temperature, and depth), chemical sensors capable of measuring 2) dissolved oxygen, 3) nitrate, 4) pH on the total scale (henceforth pH, measured at in situ temperature and pressure) (Marion et al., 2011; Waters and Millero, 2013), 5) a bio-optical sensor capable of measuring chlorophyll fluorescence, and particulate backscatter (Riser et al., 2018; Bittig et al., 2019; Chai et al., 2020) and 6) a downwelling irradiance sensor capable of measuring light penetration (Organelli et al., 2017). Four GOM BGC-Argo floats were deployed in September-October 2021 and in June of 2023 (see float model and sensor details in Table 1). Generally, on 10-day intervals, the GOM floats collected a vertical profile from 2,000 m during its ascent to the sea surface. Between profiles, the GOM floats have been programmed to park at 1,500 m (rather than 1,000 m standard park depth) where they passively drift between cycles. A total of 205 float profiles have been collected by the GOM array during the first two years of operation (Figure 1). Sensor failures for two floats (Table 1), diagnosed using the transmitted sensor engineering files, resulted in abrupt and dramaticaly drifting pH profiles with clearly unreasonable values (e.g. values of <5). Excluding these profiles, a subset of 103 pH profiles (inclusive of pH profiles effected by pH pump offset) out of the 205 total collected BGC-float profiles have pH data that are included in this analysis.
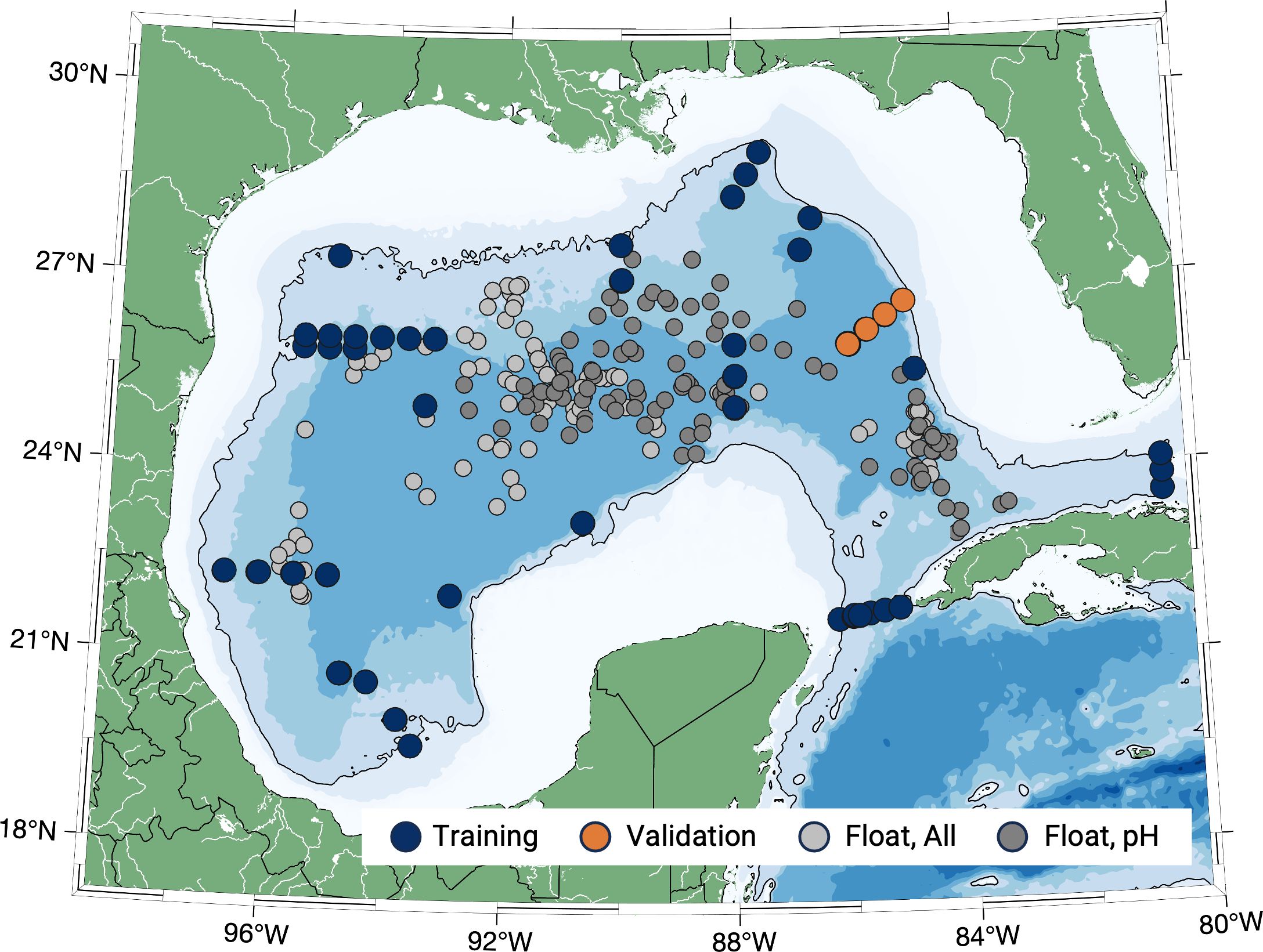
Figure 1. GOMECC stations used to develop the GOM-NNpH algorithms. The neural network training dataset includes the GOMECC bottle profiles shown in navy blue (used for training 70%, validation 15%, and testing 15% of data) and “Tampa Line” GOMECC stations were manually held back and used to independently validate the performance of the neural network in orange. The number of bottle observations used for training and validation of each algorithms are summarized in Table 2. The float profile locations (n = 205) collected between September 2021-October 2023 are shown in gray with the subset of floats with available sensor pH measurements shown in dark gray (n = 103). The black line indicates the 1,000 m isobath, the “open Gulf” water column depth threshold in this study.
Following the delayed mode quality control (DMQC) methodology of Wong et al. (2024), for float CTD and trajectory data, we confirmed that no adjustments for our float CTD data were required. This is unsurprising as typically only 15% of the “core Argo’’ parameters (i.e. temperature and salinity) require adjustment. Conversely, DMQC adjustments are always required for float oxygen, nitrate, and pH sensor data. We determined and applied corrections for float oxygen, nitrate, and pH profiles using the methodology and Sage-O2 and Sage software developed by Maurer et al. (2021). For pH and nitrate corrections, we applied a reference depth of 1480-1520 m and ESPER-NN to estimate local deep reference values. A series of up to 16 ESPER neural network equations were applied within the Sage software and the output with the lowest associated uncertainty is returned and used to calculate and then apply a reference depth offset. Equation 7 used in our study (see Table 2 for inputs), is also used by Sage to in part determine float pH corrections. Following DMQC corrections, reported float sensor accuracy for in-air corrected or World Ocean Atlas climatology corrected oxygen are 1% (Maurer et al., 2021) and 3% (Takeshita et al., 2013) of surface oxygen values, respectively. Oxygen sensor data uncertainty propagate, adding an additional source of uncertainty, when float oxygen data are used as input parameters to derive a deep reference value for nitrate and pH DMQC or algorithm estimated values. Reported BGC float nitrate and pH sensor accuracies are within 0.5 μmol kg-1 190 and 0.007 pH units, respectively (Johnson et al., 2017; Maurer et al., 2021).
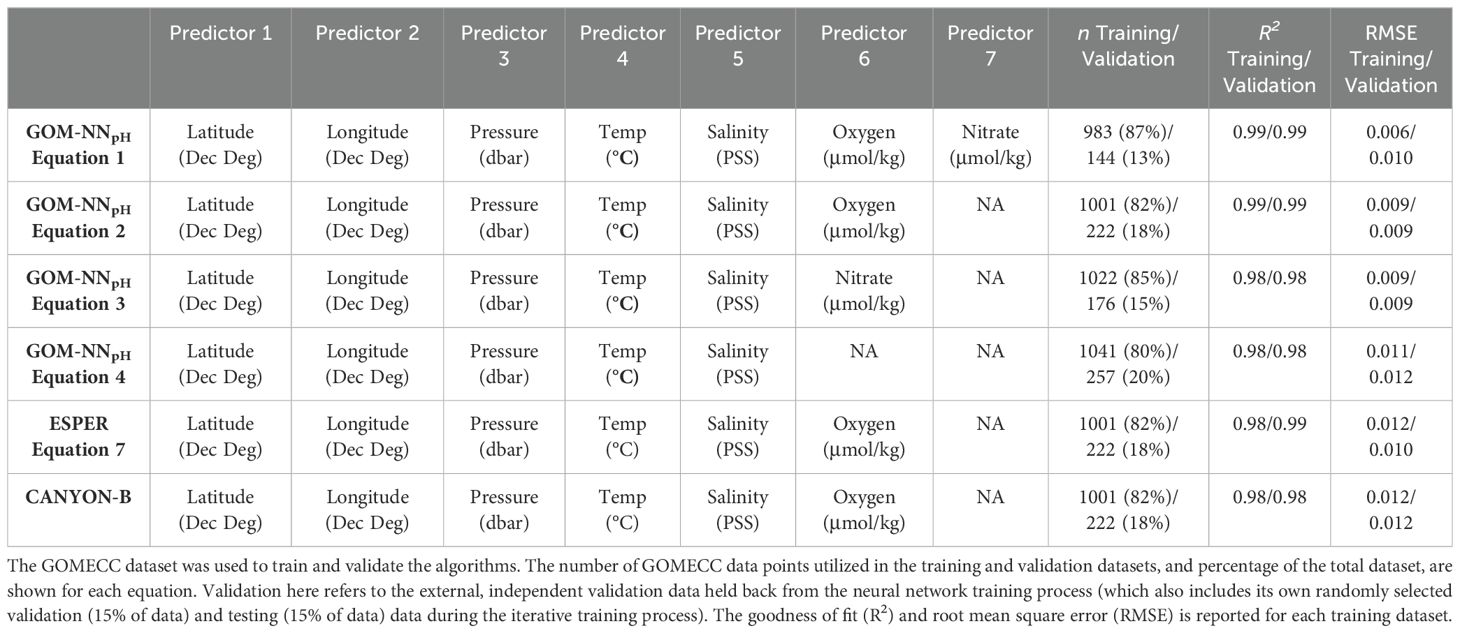
Table 2. The combinations of predictors used to estimate the pH for each of the 4 GOM-NNpH equations.
Several GOM floats demonstrated pH offset, resulting in profile shifts at 1,000 m between >0.01 pH units. The offset manifests at 1,000 m due to the pH sensor being plumbed into the CTD pump line and at this depth the CTD enters into a continuous pumping mode as it increases sampling resolution for the upper half of the profile. The proposed DMQC protocol for adjusting pump offset float profiles (visible offset in pH > 0.01) is to use a shallower reference depth to a value that is above the pump on depth (typically 1,000 m), and to flag the deeper portion of the profile as bad (Johnson et al., 2023). While a correction approach has been proposed for dealing with pump offset profiles, the root cause of the sensor behavior has not been fully characterized (Johnson et al., 2023). Due to magnitude of the offsets observed and the lack of confidence we felt in deriving and applying this correction factor, we chose to not apply a pump offset correction. Rather, these data were included to demonstrate the utility of using neural network approaches to diagnose sensor behavior.
2.2 Neural network algorithm design and equations
The GOM-NNpH algorithm was developed using MATLAB’s deep learning toolbox. The architecture was designed to optimally estimate pH by using inputs of BGC-Argo float data. A neural network, which is a supervised machine learning model, was selected to be trained and applied with GOMECC data as it achieved superior performance over random forest and multiple linear regression models during algorithm development testing. The GOM-NNpH is a feed-forward neural network that contains two hidden layers (Figure 2). A total of four GOM-NNpH algorithms were trained with various combinations of predictors. The number of neurons in each hidden layer corresponds to the number of input parameters utilized in a given algorithm. The predictor variables within the input layer were given varying weights for each neuron in the algorithm design. When training the two-layer networks, data were randomly divided into three subsets: 70% for training, 15% for validation, and 15% for testing that were used during the neural network training process. The Levenberg-Marquardt optimization algorithm (Hagan and Menhaj, 1994), was used in the training process with a performance metric of mean squared error. Each algorithm was trained using 1,000 iterations using the algorithm specific GOMECC dataset.
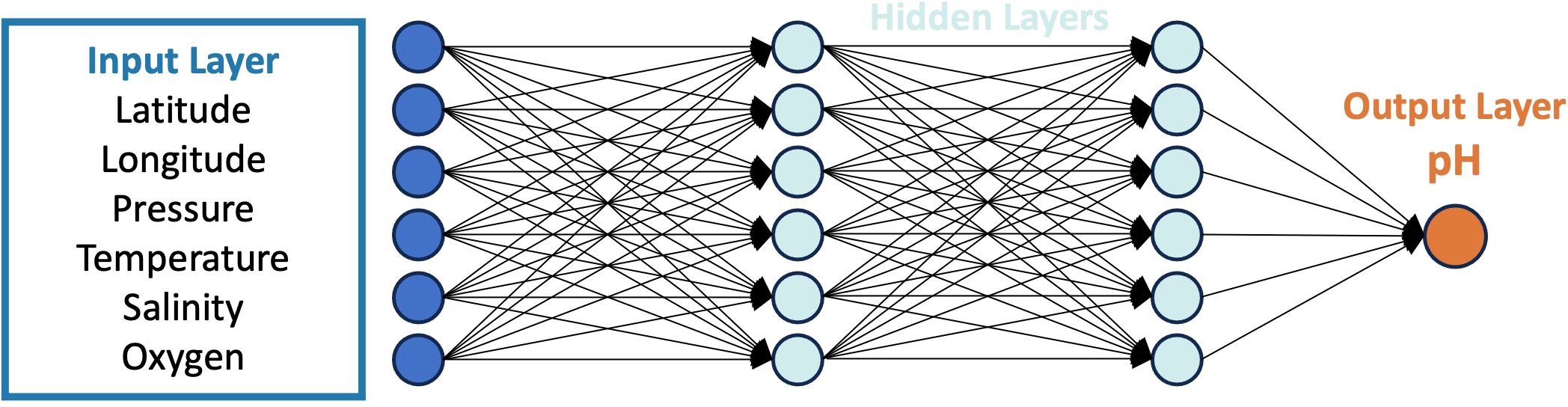
Figure 2. Schematic of one the GOM-NNpH algorithms (Equation 2) design, which differs from the other algorithms only in the number of input variables in the input layer (Table 2). The GOM-NNpH algorithm structures include two hidden layers and one output layer that were run in 1,000 iterations.
2.3 Data source for neural network training and validation
The GOMECC bottle dataset represents the largest source of GOM carbonate chemistry data (Wanninkhof et al., 2013; Wanninkhof and Baringer, 2014; Barbero et al., 2019, 2024), that have equal data quality to the GO-SHIP Program (Sloyan et al., 2019). GOMECC has routinely reoccupied a series of cross shelf transects that terminate in the deep GOM basin, with a growing number of transect locations having been added to the GOMECC program in the GOM since its inception. A subset of the GOMECC data were mined for training and validation of the GOM-NNpH algorithms based on several criteria (Figure 1). First, only GOMECC cruises that included spectrophotometric pH measurements at 25˚C were utilized. This resulted in the exclusion of GOMECC-1 data (2007) where carbonate system variables were measured (Dissolved Inorganic Carbon (DIC), Total Alkalinity (TA), and CO2 fugacity (fCO2) but did not include direct measurements of pH. Second, we incorporated only open GOM stations (minimum of 1,000 m water column) to coincide with the open GOM water masses where BGC-Argo floats are operating. The exclusion of shallower stations eliminates complex carbonate system dynamics that exist along GOM continental margins, particularly the effects of large riverine outflows (e.g., Huang et al., 2015; Osborne et al., 2022), which would likely not be well parameterized by the GOMECC dataset.
We chose to hold back the GOMECC Tampa Line stations (n = 4 profiles), which are located off of the West Florida Shelf (Figure 1, orange symbols). These stations were reoccupied during GOMECC-2 (July-2012), GOMECC-3 (July-2017), and GOMECC-4 (September-2021) and were used as an external, independent validation dataset. Our motive in holding back the Tampa Line bottle data was to test the performance of the GOM-NNpH with a region of the GOM where no training data was used in the algorithm development.
GOMECC data utilized in the GOM-NNpH algorithms include CTD measured temperature, salinity, and pressure, Winkler titration oxygen, colorimetric measurements of nitrate made onboard using an autoanalyzer, and spectrophotometric pH (measured onboard at 25˚C). Following best practices (Hood et al., 2010), calibrants and reference material determined accuracies based on replicates indicated O2 <2 µmol kg-1; Nitrate < 0.1 µmol kg-1; and pH < 0.002. GOMECC calibrated CTD temperature and salinity profile uncertainties are within WOCE standards, 0.002°C and 0.002 PSU, respectively. Shoreside secondary quality control was executed and WOCE quality control flags were applied and only data flagged as ‘good’ or ‘replicate’ were used.
A direct comparison between bottle and float data requires a conversion of bottle spectrophotometric pH, measured onboard at 25°C and ambient atmospheric pressure, to in situ temperature and pressure pH. This conversion was completed using CO2Sys MATLAB with inputs of in situ temperature, salinity, pressure, phosphate, silicate, dissolved inorganic carbon, and pH (Van Heuven et al., 2009). Following the recommendations from Dickson et al. (2007), the constants for carbonic acid K1 and K2 from Lueker et al. (2000), the borate-to-salinity relationship from Lee et al. (2010), the constant for hydrogen fluoride KF from Perez and Fraga (1987), and the constant for hydrogen sulfate KS from Dickson (1990) were used. For the purpose of our study, bottle pH data that has been converted to in situ temperature and pressure are exclusively utilized in our study to permit a direct comparison with sensor pH results.
3 Results
3.1 GOM-NNpH algorithm assessment
The quality of predictions generated by each of the GOM-NNpH algorithms are reported in terms of goodness of fit (R2) and Root Mean Square Error (RMSE) between the measured and neural network predicted values (Table 2; Figure 3). The RMSE is especially useful, as it is in the same units as the original data and therefore can be compared directly to measurement uncertainties. Generally, algorithms with more input predictors tend to perform better when the predictor measurements are high quality (Carter et al., 2021). While Equation 3 and Equation 4 show skill when compared to the training data (Table 2; Figure 3), their performance is relatively weaker than Equations 1 and 2. This result highlights the importance of biogeochemical variables as a predictor of pH. Equation 4, trained using only physical predictors, demonstrates a negative bias in the upper pH range (>8) in the independent Tampa Line validation test. Equations 1 and 2 perform comparably and yielded with the same R2 value (0.99). Equation 1 yields a lower RMSE (0.006 pH units) relative to Equation 2 (0.009 pH units) based on the training dataset (Table 2). Validation results based on the application of Equations 1 and 2 to the GOMECC Tampa Line validation data yields marginally better results for Equation 2 relative to Equation 1 (RMSE 0.001 pH unit difference, Table 2). A paired t-test preformed on the neural network pH variances from the bottle validation (for the available overlapping data n = 144) indicate that the difference between the two model results is significant (p<0.001). Performance of Equation 2 based on the RMSE (Equation 2 = 0.007 pH units versus Equation 1 = 0.010 pH units) and mean absolute error (MAE) (Equation 2 = 0.005 pH units versus Equation 1 = 0.006 pH units) but suggests that Equation 2 performs marginally better when applied to the independent Tampa Line validation dataset. The training of Equation 2 benefited from a larger training and validation dataset, due to greater availability of oxygen relative to nitrate data in the GOMECC data selection. Equation 1 includes validation only from 2017 and 2021 due to no usable nitrate data available along the Tampa line in 2012. For these reasons, combined with the lowest BGC-Argo sensor uncertainties being associated with CTD and oxygen optode data, we chose to apply Equation 2 for to our GOM BGC-Argo dataset for the remained of the detailed assessment performed in our study.
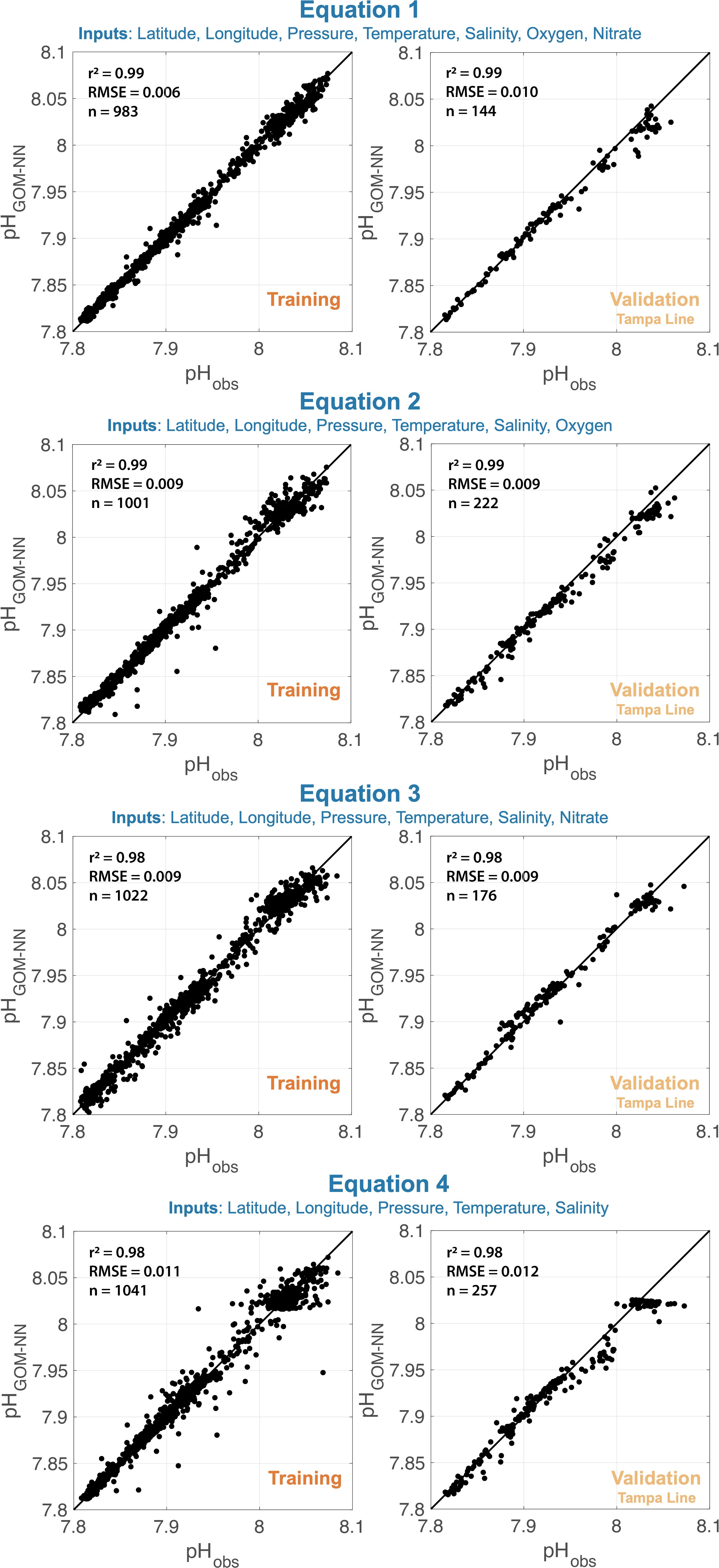
Figure 3. Visualization of the four GOM-NNpH algorithm performances. Performance is assessed by comparing the algorithm estimated pH to; (left) observed GOMECC bottle pH data associated with the 15% of bottle data held back in the neural network training framework and; (right) the observed bottle pH associated with the independent validation dataset (GOMECC Tampa Line, Figure 1 orange circles) that were excluded from the neural network training process entirely.
3.2 Comparison GOM-NNpH with globally trained neural networks
We assessed the performance of the GOM-NNpH relative to two globally trained algorithms that include a vastly larger training and validation dataset (10,000’s to 100,000’s data points) (Bittig et al., 2018; Carter et al., 2021). We compare our GOM-NNpH Equation 2 to ESPER Equation 7 and CANYON-B (which has parallel input predictors) using the GOMECC Tampa Line data (Figure 1) that was held back from the GOM-NNpH training process. Our statistical results suggest a similar robustness in performance across the algorithms, with a slightly better RMSE reported for the GOM-NNpH algorithm (0.009 pH units) relative to ESPER (0.010 pH units) and CANYON-B (0.012 pH units) (Figure 4). The MAE for the GOM-NNpH is also marginally lower (0.007 pH units) relative to ESPER (0.008 pH units) and CANYON-B (0.009 pH units) (Figure 4).
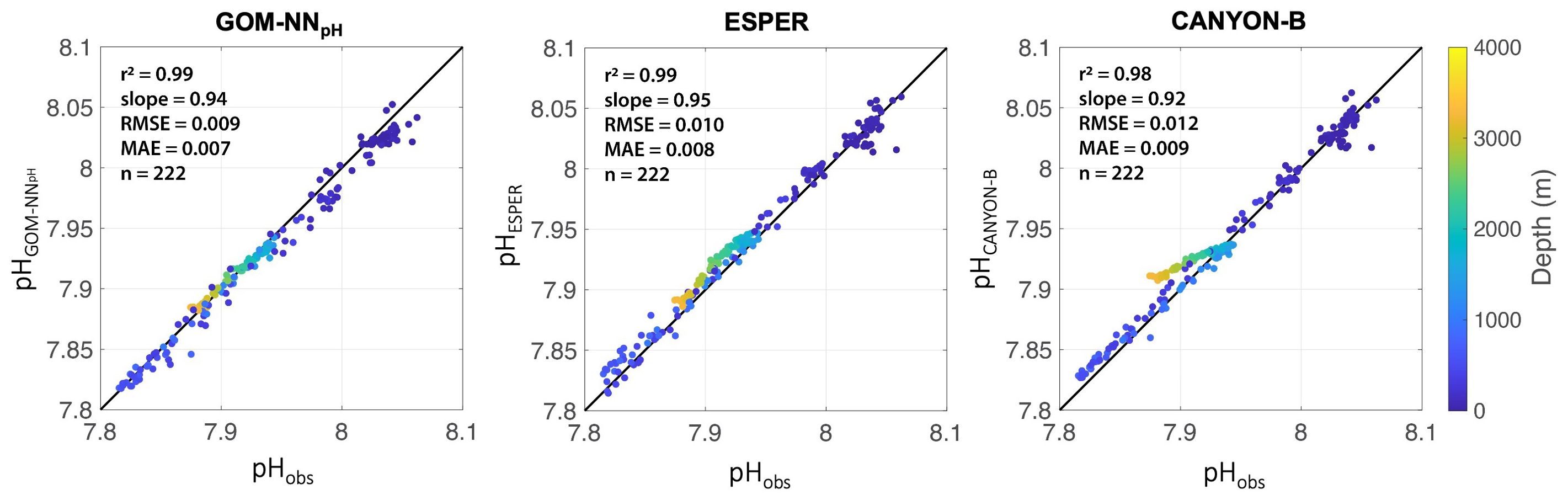
Figure 4. (Left) ESPER Equation 7, (center) GOM-NNpH Equation 2, and (right)CANYON-B neural network performance based on the GOMECC Tampa Line observations that were held back (Figure 1 orange symbols) from the GOM-NNpH training process (n observations = 222). Colors correspond to the measurement depth (m).
Visualizing the algorithm variations from the observed validation data for the two algorithms in depth space, shows that there are notable differences among the GOM-NNpH, ESPER, and CANYON-B outputs (Figure 5). While the RSME is similar, the pattered biases in ESPER- and CANYON-B-generated estimates are considerable compared GOM-NNpH. In the deep portion of the water column >1500 m, ESPER and CANYON-B both produce a notable positive bias, that result in pH values that are as much as 0.01 and 0.03 pH units higher than the GOM observations (Figure 5). ESPER and CANYON-B demonstrate spurious behavior within the pH minima (typically ~600 m within the GOM) that also results in a distinctive positive bias. Differences between GOM-NNpH algorithm outputs and observations are greatest at the surface, ranging from -0.025 to 0.01 (excluding outliers), which is on par with the variations observed for ESPER and CAYON-B at the surface (Figure 5).
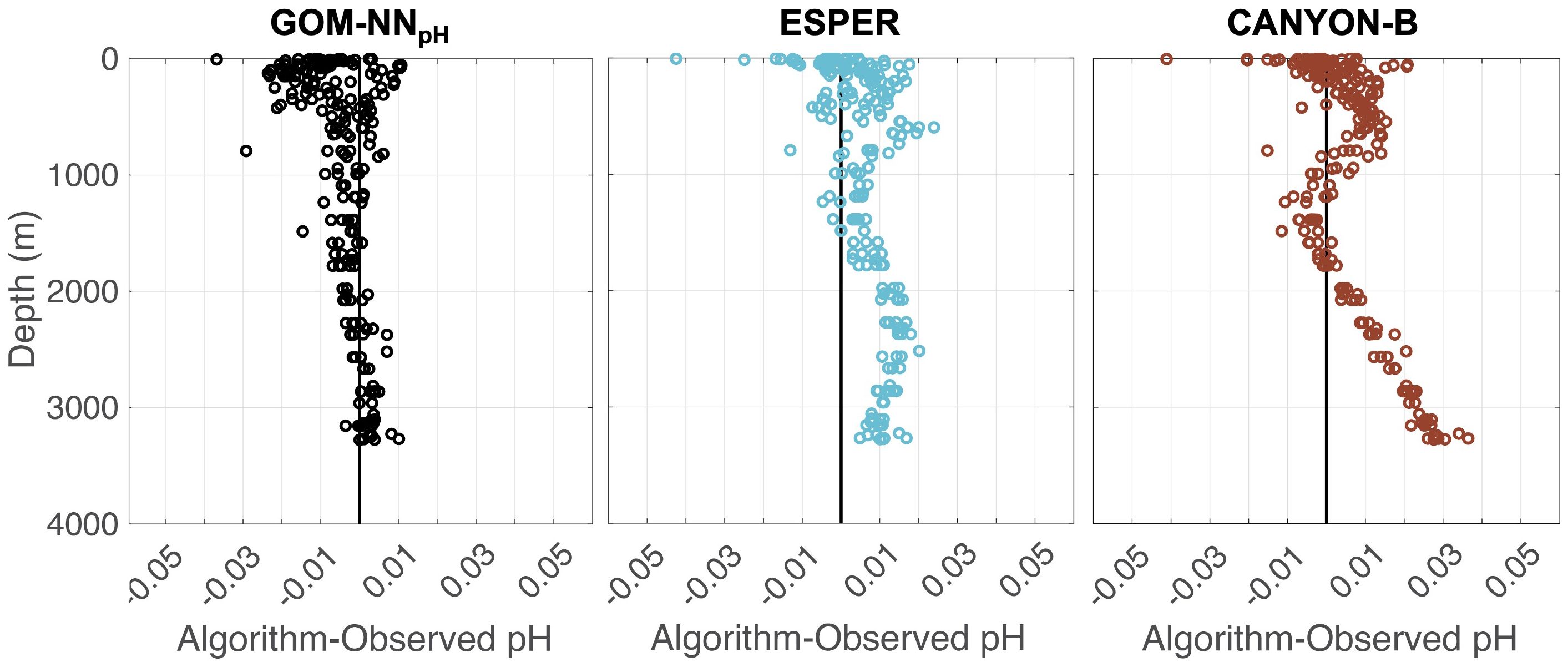
Figure 5. Comparison of ESPER Equation 7 (left), GOM-NNpH Equation 2 (center), and CANYON-B (right) algorithm-estimated pH minus bottle validation pH. This bottle comparison is applied only to the GOMECC Tampa Line bottle data (Figure 1, orange circles) that were excluded from the GOM-NNpH training process.
3.3 Test application of algorithms to Biogeochemical-Argo float data
Shipboard bottle data provides an independent, high quality validation data source that can be used to quantify the differences between sensor and neural network algorithm pH estimates. A direct comparison of these datasets is possible for the first float profile when a CTD cast is collected at the time of deployment. For the neural network assessment, outputs generated by GOM-NNpH Equation 2, ESPER Equation 7, CANYON-B, which each utilize latitude, longitude, pressure, temperature, salinity, and oxygen, were employed. Comparisons using depth-equivalent float and discrete bottle data, referred to as “matchups”, are reported following the approach described by Johnson et al. (2017).
There are important sensor-bottle matchup caveats to recognize. Float sensors can undergo significant drift due to sensor conditioning during the ~1-5 profile collections, with the greatest offsets often associated with the first collected profile. Johnson et al. (2017) attempted to overcome this limitation by matching bottle data collected at the time of deployment with the second profile collected by the float ~10 days later. Further, it is possible that depth-derived matchups that have temporal offsets that may be influenced by internal waves that would produce offsets between the bottle validation data that are the result of natural variability. It is possible to overcome the influence of internal waves by producing matchups based on density rather than depth, however we have chosen to report depth-based matchups in order to compare directly to previously published results (Johnson et al., 2017; Maurer et al., 2021). With these caveats in mind, the matchup comparison is the only means of comparing equivalent sensor, neural network, with bottle validation data.
CTD validation casts collected at the approximate site and time of float deployment for BGC-Argo floats 4903624, 4903625, and 7901009 are compared to the first and second profiles collected by each float (Figure 6; Table 3). Note that BGC-Argo float 7901009 (Navis float) only reached 1,150 m during its first profile, as Navis BGC-Argo floats undergo an auto-ballasting process that enables the float to reach its full profiling depth of 2,000 m over the first few cycles of operation (Figure 6). Less than 24 hours elapses between the start time of the matchup CTD and the transmission time of the first float profile at the end of the profile collection, which takes on average ~6 hours to collect. The second float profile was collected approximately 10 days following the first profile. Figure 6 demonstrates the significant differences between the first and second float sensor pH profiles collected 10 days apart by float 4903625 that are a result of natural variability. Based on a satellite-based assessment (see method described in McWhorter et al., 2024), the first profile and validation cast for float 4903625 are collected in GOM Common Water and over the next 10 days, this float drifted into the Loop Current where profile two was collected. This demonstrates that major biogeochemical variability can exists over small spatial scales in the GOM, necessitating that matchups compare contemporaneous data collections when possible.
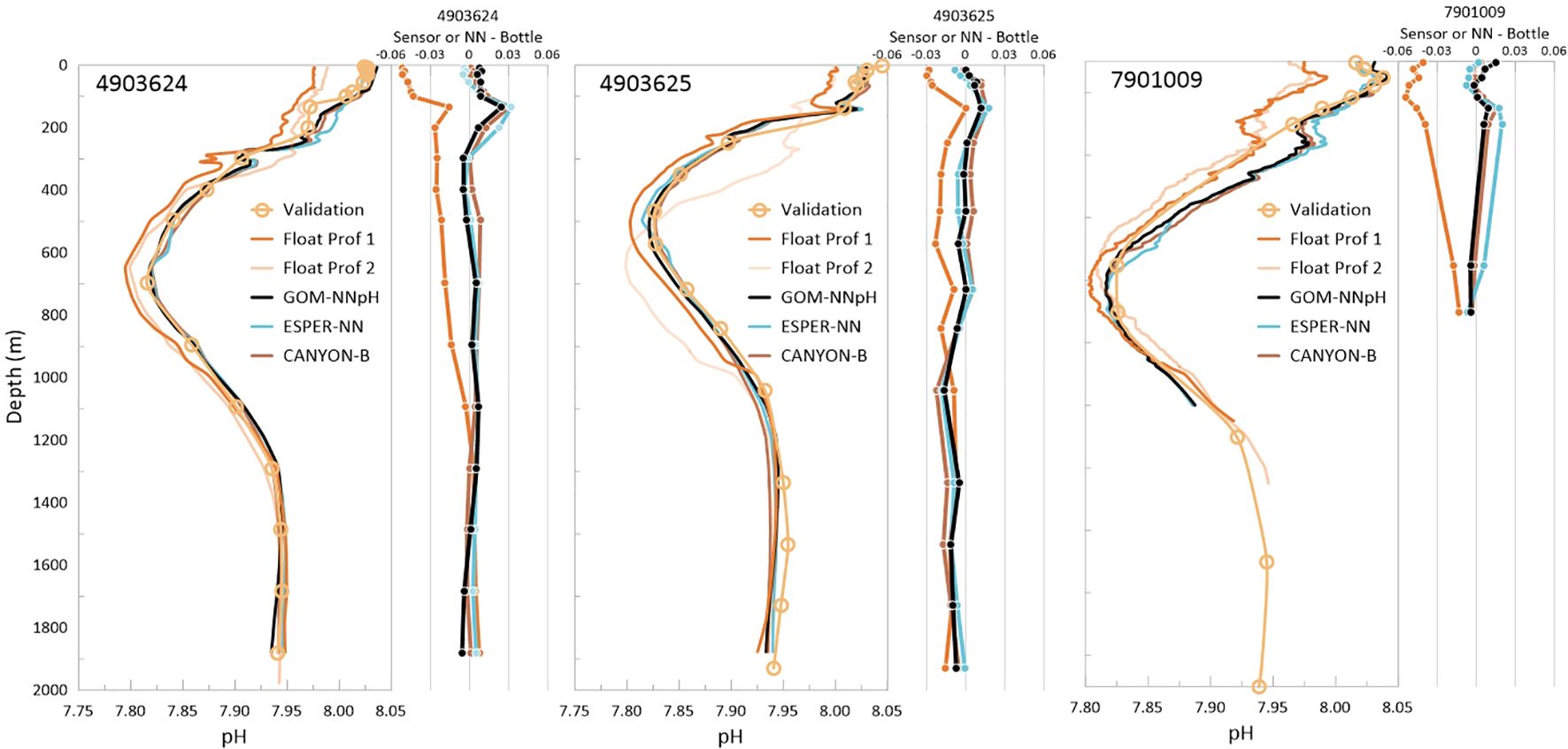
Figure 6. Comparison of bottle validation data collected at the time and location of the BGC-Argo float’s first and second profile for floats 4903624 (left), 4903625 (center), and 7901009 (right). Bottle matchup GOM-NNpH (Equation 2), ESPER (Equation 7), and CANYON-B algorithm estimated pH are also shown. Right panels within each figure show algorithm or sensor pH data minus the bottle data.

Table 3. Statistical results of the float sensor and GOM-NNpH depth matchup with bottle validation data.
The matchup results show that the neural network pH results match the validation more closely than sensor pH. The largest matchup differences are associated with the float sensor data in the upper 1000 meters of the water column, which appear to increase with decreasing pressure (Figure 6). Across the full depth range for all GOM float sensor matchups, a median offset of -0.022 pH units is observed (Table 3). This is significantly higher than the 0.002 median offset determined in a bottle matchup analysis of globally distributed BGC-Argo floats (Maurer et al., 2021). Recall the caveat that sensor conditioning that can occur during the first several float profile collections may be exacerbating these bottle-sensor offsets. The convergence of all datasets at depth with the validation data suggest that sensor conditioning is not the only potential influence on surface value offsets observed in the sensor dataset. An additional comparison of the sensor pH differences from the three neural network pH estimates for the bottle validation dataset further illustrates a positively correlated pressure-dependent offset in the sensor pH dataset (Figure 7).
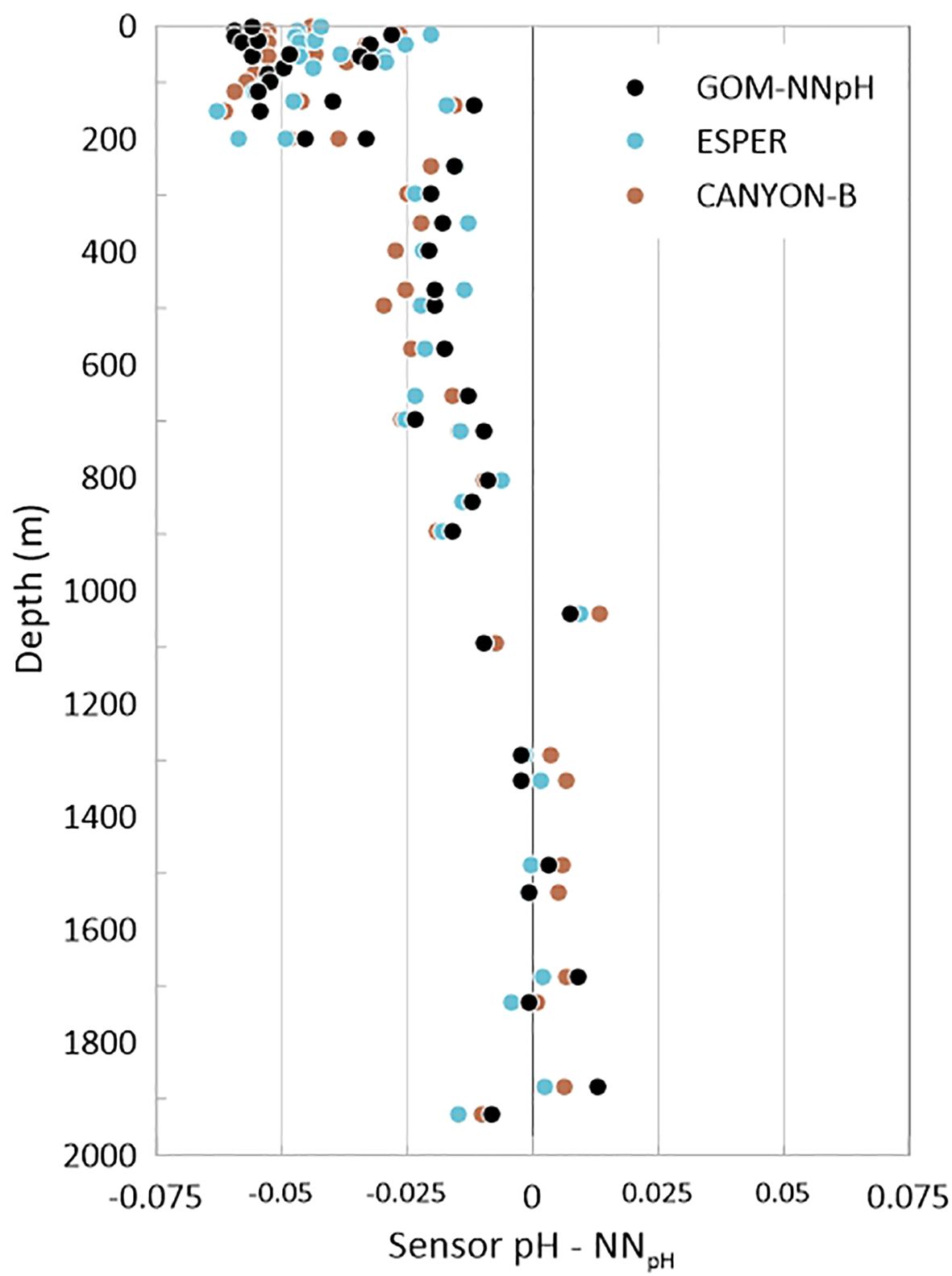
Figure 7. Comparison of sensor pH differences from GOM-NNpH (Equation 2), ESPER (Equation 7), and CANYON-B algorithm estimated pH. This comparison is done using the compiled bottle validation matchup data associated with the first collected float profiles for floats 4903624, 4903625, 7901009.
Two of the GOM Apex floats, 4903622 and 4903625, demonstrated pH-pump offset, evidenced by abrupt shifts in pH values between the ~950 and ~1000 dbar (Figure 8). Based on a visual assessment and comparison with bottle matchup and GOM-NNpH estimates, we approximate that the pH pump offset causes offsets in the first float profiles of -0.014 pH units for 4903624 and 0.021 pH units for 4903625. Application of these corrections across the upper half of the profile vastly improves the matchup RMSE for float 4903625 (RMSE 0.009 pH units), however it only moderately improves the RMSE for 4903624 (RMSE 0.020 pH units). For both floats, but especially for 4903624, corrected profiles continue to show considerable offsets from the validation data and neural network results (Figure 8). And float 7901009, which is unaffected by pH pump-offset, also appears to show inversely related pressure-dependent differences (Figure 6).
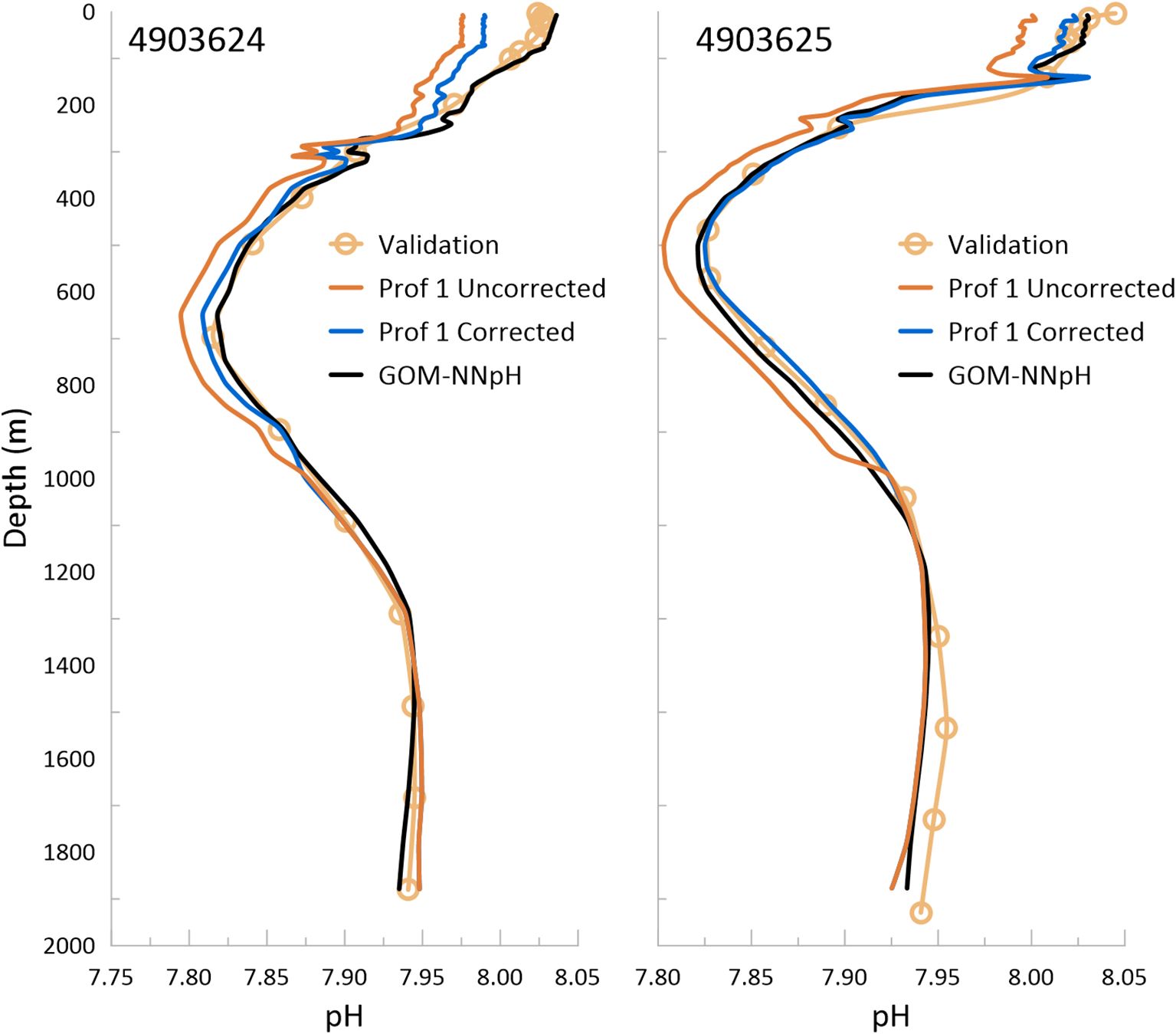
Figure 8. Based on a visual comparison with the bottle validation and GOM-NNpH results, a pH pump-offset correction factor was estimated for float 4903624 (-0.014) and float 4903625 (-0.021). The uncorrected and corrected profile 1 for each float are shown here in comparison to the bottle validation and GOM-NNpH.
The visual inspection approach using profile 1 relies on the availability of the bottle data, which are limited by the potential for sensor conditioning-related variations. Therefore, exclusive use of bottle validation data to identify a float pump-offset correction factors is an imperfect approach. Further, applying a blanket correction across the full collection of float profiles is unreliable, as the pump-offset may not be stable over time. Ideally, profile specific pH-pump offsets are determined, which can be generated, for example, using neural network simulations of pH. Sensor and neural network profiles can be directly compared in an attempt to diagnose sensor behavior and potentially quantify an appropriate offset. However, quantifying the propagation of uncertainty and a general assessment of various correction approaches is still needed. The current standard for correcting pH pump-offset, which suggests the shoaling of the reference depth to <1000 m (above the pump on depth) (Johnson et al., 2023), can introduce other uncertainties during the DMQC process. Specifically in the GOM, biogeochemical variability related to the Loop Current can be induced at 1000 m, therefore this cannot be assumed to be a stable reference depth. For this reason, we choose to not publish a pH pump-corrected dataset associated with our floats as to not publish data we are not confident in.
The non-pH pump-offset corrected BGC-Argo sensor data-bottle validation matchups, collected in association with the first float profile, are summarized in Table 3. A total n = 43 bottle matchups are available across our dataset. GOM-NNpH pH estimates were generated to directly compare NN-bottle to the float-bottle matchup results. Based on the mean, there are highly significant differences (t-test p <0.005) between the algorithm matchup compared to the BGC-Argo float pH sensor matchup. Matchup offsets are consistently greater in the BGC-Argo pH sensor dataset, which are visualized in Figure 9, demonstrating the considerable float negative pH bias in the upper 1,000 m of the water column observed for all three floats, which becomes larger with decreasing pressure.
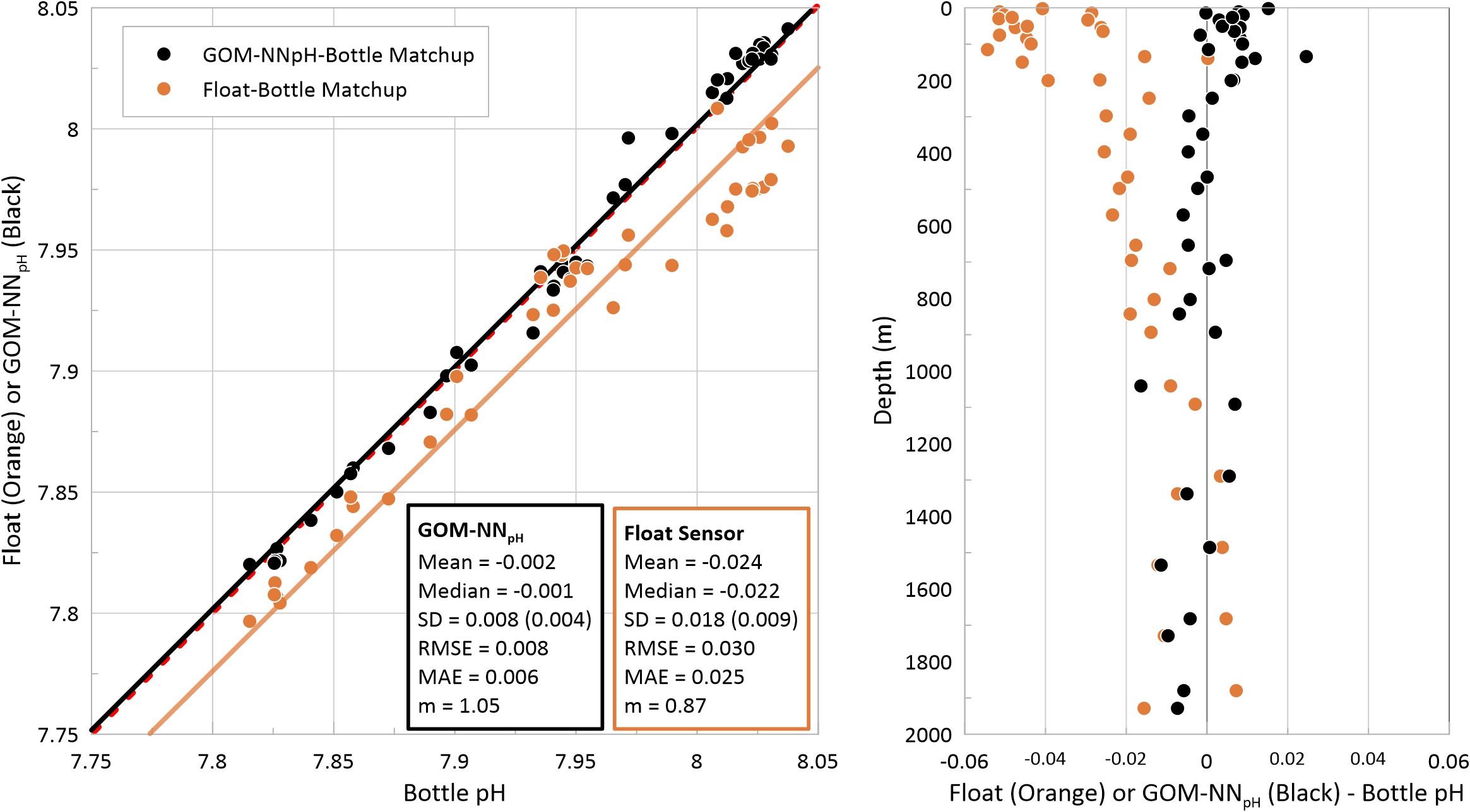
Figure 9. (Left) Matchup of float sensor pH (orange) and GOM-NNpH (black) with depth equivalent shipboard bottle validation pH (n=43 per matchup). The dashed red line represents the 1:1 and the solid orange and black lines are the simple linear fit of the float and GOM-NNpH data, respectively. Statistics for matchups are reported across the full matchup dataset here, and broken out per float in Table 3. (Right) Matchup differences, shown as float sensor pH (orange) or GOM-NNpH (black) minus bottle matchup pH, plotted in depth space. Note that pH pump-offsets are not corrected in the float sensor dataset, therefore 2 out of 3 floats have a bias in the upper 1000 of the water column.
Based on the bottle matchup assessment for the full GOM BGG float array, we report a MAE of 0.025 and RMSE of 0.030 pH units (Table 3). Previously, the pH sensor uncertainty across the large BGC-Argo arrays has been reported as half of one standard deviation based on bottle-float matchups (Maurer et al., 2021). This hinges on the assumption that half of the variability between a bottle validation cast and the first float profile is the result of natural ocean variability (Johnson et al., 2017). Based on the error approach reported in Maurer et al. (2021), the GOM array yields a 0.009 pH unit uncertainty, which is on par with the reported uncertainty of 0.007 from a meta-analysis of globally distributed BGC-Argo floats (Johnson et al., 2017; Maurer et al., 2021). Overall, the GOM-NNpH algorithm yields the most robust matchups, with an RMSE of 0.008 pH units and an adjusted uncertainty (half of one standard deviation) of 0.004 (Table 3).
We further assess performance of the GOM BGC-Argo pH sensors by calculating mean pH profiles for all sensor pH profiles and GOM-NNpH profile estimates per float (Figure 10). Mean pH profiles represent averaged values across a series of binned depths for all cycles collected over the lifetime of a given float. This assessment ingests a much larger dataset (n profiles = 103, n data points = 11870 per data type) relative to the currently available matchup validation dataset (n profiles = 3, n data points = 43 per data type) and overcomes the caveat that validation matchups with the first float profiles can be hindered by sensor conditioning that can yield magnified sensor biases during the first few float cycles of float operation.
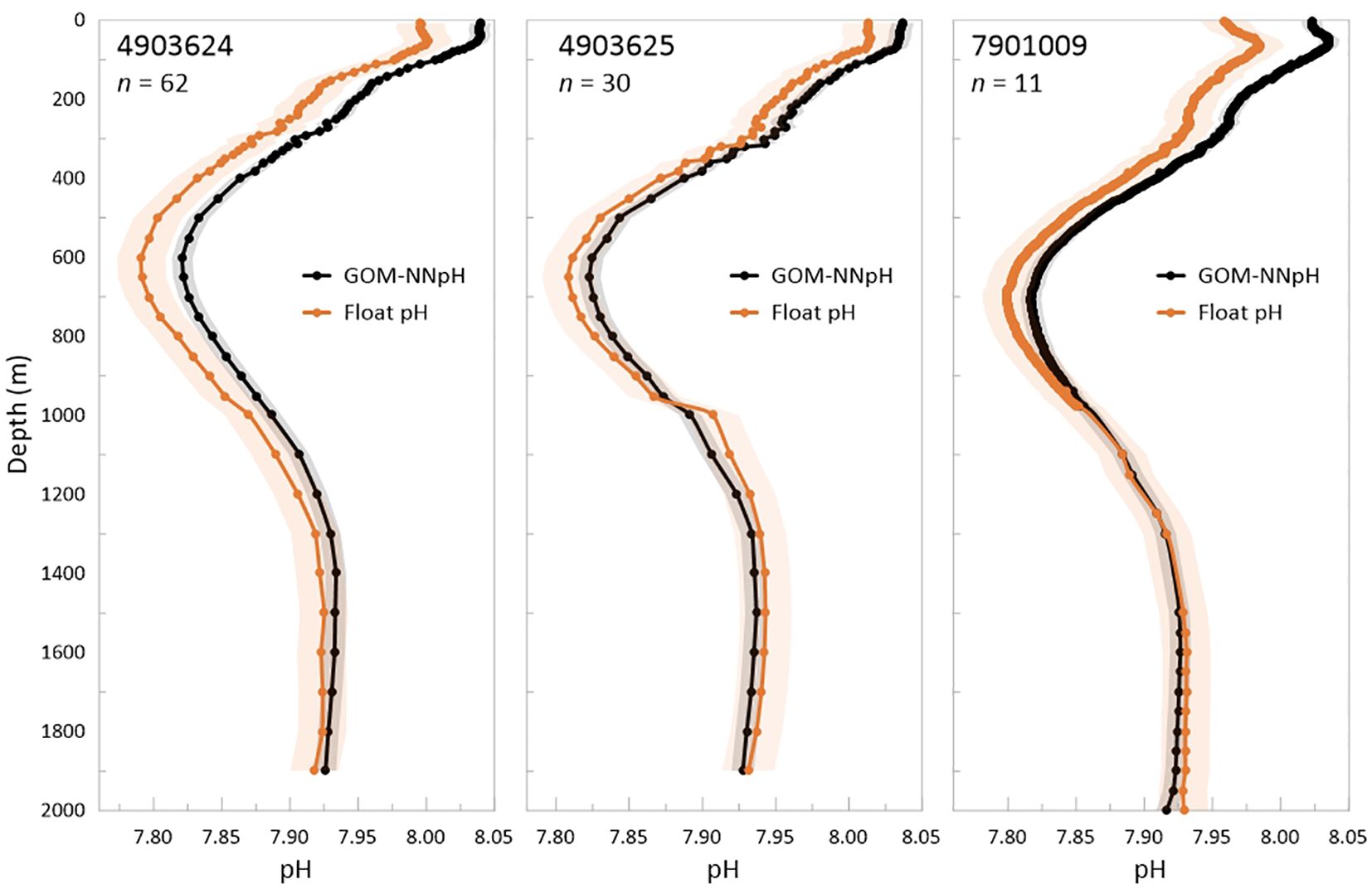
Figure 10. Comparison of the mean profiles per float generated for measured float pH sensor and GOM-NNpH estimated pH. Circle symbols indicate sample depths, which vary between Apex-BGC and Navis float models. Data shown are mean profiles per data type over the lifetime of a float (n indicates the number of profiles that were averaged to generate a mean profile per float). Mean profiles are generated only for floats and cycles where reasonable float sensor pH measurements were available, The standard deviation of the mean is shown as shaded area for each data type based on the statistical results of the GOM array-wide validation matchup generated in this study.
Consistent with the validation matchups, the mean profile comparisons between sensor pH and GOM-NNpH estimated pH demonstrate the highest differences between datasets over the upper 1,000 m. For floats 4903624 and 4903625 this is in part due to an uncorrected pH pump-offset. While application of a tentative pH pump-offset correction for float 4903625 improves the sensor-GOM-NNpH alignment, a considerable pressure dependent difference remains for float 4903624 and is also present for float 7901009. Figure 10 illustrates the particularly prominent pH pump-offset for float 4903625 and observable but smaller for float 4903624. The results of the mean profile comparison qualitatively confirm the observations made based on the significantly smaller validation dataset used in the matchup assessment.
4 Discussion and concluding statements
Neural network approaches for estimating non-linear relationships between various ocean biogeochemical parameters are capable of skillfully producing ocean datasets, specifically demonstrated here for the carbonate chemistry parameter, pH. The GOM is a biogeochemically variable region due to influences including, but likely not limited to, riverine inputs and the Loop Current and associated eddies (Gomez et al., 2024; Le Hénaff et al., 2012; Le Hénaff et al., 2023). Because of these regional influences, GOM biogeochemistry cannot be optimally parameterized by neural networks trained exclusively with open-ocean datasets. The development of a series of GOM-trained neural network algorithms capable of estimating pH within the open GOM basin, represents an advancement in machine learning approaches used to study biogeochemistry in this large marginal sea. Here we offer four new neural network routines (Table 2; Figure 3), trained with the largest, high-quality shipboard carbonate chemistry dataset that is publicly available for the region. These algorithms can be applied to GOM datasets using various combinations of temperature, salinity, oxygen, and nitrate observations to estimate open GOM pH (>1,000 m water depths).
A comparison of the newly developed GOM-NNpH algorithms to two widely used, globally trained neural network algorithms, ESPER and CANYON-B, demonstrate the increased skill of the GOM-trained algorithm for capturing carbonate system variable within the region. We conclude that the globally trained neural networks, designed to fit global-scale variability, lack neurons associated with regionally-specific relationships due to them being sacrificed in the training process. While ESPER and CANYON-B produce reasonable values for the GOM, particularly based on statistical results, these algorithms show patterned biases across the water column (Figure 5). Our results for the GOM, similar to Fourrier et al.’s (2020) findings for the Mediterranean Sea, suggest considerable value of regional neural network routines.
For our application to the GOM BGC-Argo array, we employed GOM-NNpH Equation 2 (inputs: latitude, longitude, pressure, temperature, salinity, and oxygen), based on this algorithm being trained and validated with the largest available biogeochemical dataset and its utilization of float temperature, salinity, and oxygen data. Argo mounted CTDs and oxygen optodes have the longest history of field deployments to date, and by extension the greatest sensor and QC protocol development, and are associated with the lowest measurement uncertainties. Compared to a meta-analysis of globally deployed BGC-Argo floats that reported a pH sensor uncertainty of 0.007 pH units, we report an uncertainty of 0.009 (RMSE 0.030 pH units) for the GOM BGC-Argo float array (Table 3). Visualization of the float pH with validation data demonstrates considerable, differences for the GOM array floats that we connect to pH pump-offset and apparent pressure-dependent effects. Despite the presence of pH pump-offsets in two out of three of our floats, we visually approximate a correction based on a comparison to bottle and GOM-NNpH outputs, but choose to not apply this correction to our full float dataset due to our lack of methodological confidence we have in doing so. While preliminary protocols for corrections exist, the mechanism driving pH-pump offset is currently not well understood and proposed correction protocols present challenges for the GOM. Further, despite the tentative pH pump-offsets corrections applied to our dataset, considerable differences between float pH and bottle pH remained.
A comparison of GOM-NNpH (Equation 2) to the GOM validation dataset demonstrates robust performance, resulting in uncertainty of 0.004 pH units (RMSE 0.008 pH units) (Table 3). Prompted by the application to a new and growing dataset of BGC-Argo float observations within the region, the GOM-NNpH algorithms serve two functions: 1) to skillfully estimate pH profiles using combinations of (non-pH) float sensor data in the event of a failed sensor and 2) as a secondary QC method to compare the pH values from floats with working pH sensors.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://github.com/AOML-BGC-Argo/GOM-pH-NN/blob/main/GOM_NN_Eqn1_training.m; https://usgodae.org/pub/outgoing/argo/ or https://data-argo.ifremer.fr/; https://www.ncei.noaa.gov/access/ocean-carbon-acidification-data-system-portal/.
Author contributions
EO: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. Y-YX: Data curation, Formal analysis, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. MS: Data curation, Software, Writing – review & editing. JM: Formal analysis, Writing – original draft, Writing – review & editing. LB: Data curation, Formal analysis, Funding acquisition, Writing – original draft, Writing – review & editing. RW: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Acidification Program (OAP), ROR #02bfn4816, under project #17928. Support for this analysis was provided by NOAA’s Atlantic Oceanographic and Meteorological Laboratory. MS and LB are supported under the auspices of the Cooperative Institute for Marine and Atmospheric Studies (CIMAS), a cooperative institute of the University of Miami and NOAA (agreement NA20OAR4320472). This research was carried out in part with support from the National Oceanic and Atmospheric Administration’s Ocean Acidification Program (OAP), ROR #02bfn4816, under project #17928.
Acknowledgments
Thank you to our partners at the University of Washington Applied Physics Laboratory who have played an important role in standing up the AOML Biogeochemical-Argo program. Thank you to our partners at Monterey Bay Aquarium Research Institute who continue to lead the charge in developing, testing, and improving the Biogeochemical-Argo float technology and dataset. Thank you to Brandon Navarro who assisted with delayed mode quality control assessment of the float CTD data. Thank you to Brendan Carter who provided useful comments during the development of this study and manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Barbero L., Pierrot D., Wanninkhof R., Baringer M. O., Byrne R. H., Langdon C., et al. (2019). Dissolved inorganic carbon, total alkalinity, nutrients, and other variables collected from CTD profile, discrete bottle, and surface underway observations using CTD, Niskin bottle, flow-through pump, and other instruments from NOAA Ship Ronald H. Brown in the GOM of Mexico, Southeastern coast of the United States, and Mexican and Cuban coasts during the third GOM of Mexico and East Coast Carbon (GOMECC-3) Cruise from 2017-07-18 to 2017-08-20 (NCEI Accession 0188978) (Asheville, North Carolina USA: NOAA National Centers for Environmental Information). doi: 10.25921/yy5k-dw60
Barbero L., Stefanick A., Hooper J., Wanninkhof R., Baringer M. O., et al. (2024). Surface underway and surface discrete measurements of dissolved inorganic carbon (DIC), total alkalinity (TA), water temperature, salinity, partial pressure of CO2 (pCO2), pH(total scale) nutrients and other parameters obtained onboard the NOAA ship Ronald H. Brown during the GOMECC-4 cruise in the GOM of Mexico and North Atlantic Ocean from 2021-09-13 to 2021-10-21 (NCEI Accession 0286818) (Asheville, North Carolina USA: NOAA National Centers for Environmental Information). Available at: https://www.ncei.noaa.gov/archive/accession/0286818.
Bittig H. C., Maurer T. L., Plant J. N., Schmechtig C., Wong A. P.S., Claustre H., et al. (2019). A BGC-argo guide: planning, deployment, data handling and usage. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00502
Bittig H. C., Steinhoff T., Claustre H., Fiedler B., Williams N. L., Sauzède R., et al. (2018). An alternative to static climatologies: robust estimation of open ocean CO2 variables and nutrient concentrations from T, S, and O2 data using bayesian neural networks. Front. Mar. Sci. 5. doi: 10.3389/fmars.2018.00328
Boyer T. P., Antonov J. I., Baranova O. K., Coleman C., García H. E., Grodsky A., et al. (2016). NCEI Standard Product: World Ocean Database (WOD) (NOAA National Centers for Environmental Information). Available online at: https://www.ncei.noaa.gov/archive/accession/NCEI-WOD (Accessed 2024-03-29).
Bushinsky S. M., Nachod Z., Fassbender A. J., Tamsitt V. M., Takeshita Y., William N. (2024). Offset between profiling float and shipboard oxygen observations at depth imparts bias on float pH and derived pCO2. ESS Open Archive. doi: 10.22541/essoar.171199408.87526758/v1
Carter B. R., Bittig H. C., Fassbender A. J., Sharp J. D., Takeshita Y., Xu Y.-Y., et al. (2021). New and updated global empirical seawater property estimation routines. Limnol. Oceanogr. Methods / ASLO 19, 785–809. doi: 10.1002/lom3.v19.12
Carter B. R., Feely R. A., Williams N. L., Dickson A. G., Fong M. B., Takeshita Y. (2018). Updated methods for global locally interpolated estimation of alkalinity, pH, and nitrate. Limnol. Oceanogr. Methods 16, 119–131. doi: 10.1002/lom3.10232
Carter B. R., Williams N. L., Evans W., Fassbender A. J., Barbero L., Hauri C., et al. (2019). Time of detection as a metric for prioritizing between climate observation quality, frequency, and duration. Geophys. Res. Lett. 46, 3853–3861. doi: 10.1029/2018GL080773
Chai F., Johnson K. S., Claustre H., Xing X., Wang Y., Boss E., et al. (2020). Monitoring ocean biogeochemistry with autonomous platforms. Nat. Rev. Earth Environ. 1, 315–326. doi: 10.1038/s43017-020-0053-y
Claustre H., Johnson K. S., Takeshita Y. (2020). Observing the global ocean with biogeochemical-argo. Annu. Rev. Mar. Sci. 12. doi: 10.1146/annurev-marine-010419-010956
Dickson A. G. (1990). Standard potential of the reaction: and and the standard acidity constant of the ion HSO4– in synthetic sea water from 273.15 to 318.15 K. J. Chem. Thermodynam. 22, 113–127. doi: 10.1016/0021-9614(90)90074-Z
Dickson A. G., Sabine C. L., Christian J. R. Eds. (2007). Guide to Best Practices for Ocean CO2 Measurements. PICES Special Publication 3, 191 pp. Available online at: https://repository.oceanbestpractices.org/bitstream/handle/11329/249/Guide_all_in_one.pdf?sequence=5
Doney S. C., Ruckelshaus M., Emmett Duffy J., Barry J. P., Chan F., English C. A., et al. (2012). Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Fourrier M., Coppola L., Claustre H., D’Ortenzio F., Sauzède R., Gattuso J.-P. (2020). A regional neural network approach to estimate water-column nutrient concentrations and carbonate system variables in the Mediterranean Sea: CANYON-MED. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00620
Gomez F. A., Wanninkhof R., Barbero L., Lee S.-K. (2024). Mississippi river chemistry impacts on the interannual variability of Aragonite saturation state in the northern GOM of Mexico. J. Geophys. Res. Oceans 129. doi: 10.1029/2023jc020436
Hagan M. T., Menhaj M. B. (1994). Training feedforward networks with the marquardt algorithm. IEEE Trans. Neural Networks 5, 989–993. doi: 10.1109/72.329697
Henson S. A., Beaulieu C., Ilyina T., John J. G., Long M., Séférian R., et al. (2017). Rapid emergence of climate change in environmental drivers of marine ecosystems. Nat. Commun. 8, 14682. doi: 10.1038/ncomms14682
Hood E. M., Sabine C. L., Sloyan B. M. (2010).GO-SHIP Repeat Hydrography Manual: A Collection of Expert Reports and Guidelines. Available online at: http://www.go-ship.org/HydroMan.html (Accessed March 31, 2024).
Hornik K., Stinchcombe M., White H. (1989). Multilayer feedforward networks are universal approximators. Neural Netw. 2, 359–366. doi: 10.1016/0893-6080(89)90020-8
Huang W.-J., Cai W.-J., Wang Y., Lohrenz S. E., Murrell M. C. (2015). The carbon dioxide system on the Mississippi River-dominated continental shelf in the northern GOM of Mexico: 1. Distribution and air-sea CO2 flux. J. Geophys. Res. Oceans 120, 1429–1445. doi: 10.1002/2014JC010498
Ito T., Cervania A., Cross K., Ainchwar S., Delawalla S. (2024). Mapping dissolved oxygen concentrations by combining shipboard and argo observations using machine learning algorithms. J. Geophys. Res.: Mach. Learn. Comput. 1. doi: 10.1029/2024JH000272
Jiang L.-Q., Boyer T. P., Paver C. R., Yoo H., Reagan J. R., Alin S. R., et al. (2024). Climatological distribution of ocean acidification indicators along the north American ocean margins. doi: 10.5194/essd-2024-59
Jiang L.-Q., Carter B. R., Feely R. A., Lauvset S. K., Olsen A. (2019). Surface ocean pH and buffer capacity: past, present and future. Sci. Rep. 9, 18624. doi: 10.1038/s41598-019-55039-4
Johnson K. S., Maurer T. L., Plant J. N., Takeshita Y. (2023). BGC-Argo quality control manual for pH. doi: 10.13155/97828
Johnson K. S., Plant J. N., Coletti L. J. (2017). Biogeochemical sensor performance in the SOCCOM profiling float array. J. Geophys. Res.: Oceans 122, 6416–6436. doi: 10.1002/2017JC012838
Juranek L. W., Feely R. A., Gilbert D., Freeland H., Miller L. A. (2011). Real-time estimation of pH and aragonite saturation state from Argo profiling floats: Prospects for an autonomous carbon observing strategy. Geophys. Res. Lett. 38. doi: 10.1029/2011gl048580
Key R. M., Olsen A., van Heuven S., Lauvset S. K., Velo A., Lin X., et al. (2015). Global Ocean Data Analysis Project, Version 2 (GLODAPv2). doi: 10.3334/CDIAC/OTG.NDP093_GLODAPv2
Lauvset S. K., Lange N., Tanhua T., Bittig H. C., Olsen A., Kozyr A., et al. (2023). Global Ocean Data Analysis Project version 2.2023 (GLODAPv2.2023) (NCEI Accession 0283442) (NOAA National Centers for Environmental Information) (Accessed 2024-03-29).
Le Hénaff M., Kourafalou V. H., Morel Y., Srinivasan A. (2012). Simulating the dynamics and intensification of cyclonic Loop Current Frontal Eddies in the Gulf of Mexico. J. Geophys. Res. doi: 10.1029/2011JC007279
Le Hénaff M., Kourafalou V. H., Androulidakis Y., Ntaganou N., Kang H. (2023). Influence of the Caribbean Sea eddy field on Loop Current predictions. Front. Mar. Sci. 10, 1129402. doi: 10.3389/fmars.2023.1129402
Lee K., Kim T.-W., Byrne R. H., Millero F. J., Feely R. A., Liu Y.-M. (2010). The universal ratio of boron to chlorinity for the North Pacific and North Atlantic oceans. Geochim. Cosmochim. Acta 74 (6), 1801–1811. doi: 10.1016/j.gca.2009.12.027
Lueker T. J., Dickson A. G., Keeling C. D. (2000). Ocean pCO2 calculated from dissolved inorganic carbon, alkalinity, and equations for K1 and K2 : validation based on laboratory measurements of CO2 in gas and seawater at equilibrium. Mar. Chem. 70, 105–119.
Marion G. M., Millero F. J., Camões M. F., Spitzer P., Feistel R., Chen C.-T. A. (2011). pH of seawater. Mar. Chem. 126, 89–96. doi: 10.1016/j.marchem.2011.04.002
Maurer T. L., Plant J. N., Johnson K. S. (2021). Delayed-mode quality control of oxygen, nitrate, and pH data on SOCCOM biogeochemical profiling floats. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.683207
McWhorter J. K., Roman-Stork H. L., Le Hénaff M., Frenzel H., Johnston M. A., Cornec M., et al. (2024). Mesoscale eddies influence coral reef environments in the northwest GOM of Mexico. J. Geophys. Res. Oceans 129. doi: 10.1029/2023jc020821
Newton J. A., Feely R. A., Jewett E. B., Williamson P., Mathis J. (2015). Global Ocean Acidification Observing Network: Requirements and Governance Plan. Second Edition, GOA-ON Available online at: http://www.goa-on.org/docs/GOA-ON_plan_print.pdf.
Olsen A., Key R. M., Heuven S., van Lauvset S. K., Velo A., Lin X., et al. (2016). The Global Ocean Data Analysis Project version 2 (GLODAPv2) – an internally consistent data product for the world ocean. Earth Syst. Sci. Data 8, 297–323. doi: 10.5194/essd-8-297-2016
Olsen A., Lange N., Key R. M., Tanhua T., Bittig H. C., Kozyr A., et al. (2020). Global ocean data analysis project version 2.2020 (GLODAPv2.2020) (NCEI accession 0210813). NOAA Natl. Centers Environ. Inf. doi: 10.25921/2C8H-SA89
Organelli E., Claustre H., Bricaud A., Barbieux M., Uitz J., D’Ortenzio F., et al. (2017). Bio-optical anomalies in the world’s oceans: An investigation on the diffuse attenuation coefficients for downward irradiance derived from Biogeochemical Argo float measurements. J. Geophys. Res. Oceans 122, 3543–3564. doi: 10.1002/2016jc012629
Osborne E., Hu X., Hall E. R., Yates K., Vreeland-Dawson J., Shamberger K., et al. (2022). Ocean acidification in the gulf of Mexico: drivers, impacts, and unknowns. Prog. Oceanogr. 209, 102882. doi: 10.1016/j.pocean.2022.102882
Pasqueron de Fommervault O., Perez-Brunius P., Damien P., Camacho-Ibar V. F., Sheinbaum J. (2017). Temporal variability of chlorophyll distribution in the gulf of Mexico: bio-optical data from profiling floats. Biogeosciences 14, 5647–5662. doi: 10.5194/bg-14-5647-2017
Passow U., Overton E. B. (2021). The complexity of spills: the fate of the deepwater horizon oil. Annu. Rev. Mar. Sci. 13, 109–136. doi: 10.1146/annurev-marine-032320-095153
Perez F. F., Fraga F. (1987). Association constant of fluoride and hydrogen ions in seawater. Mar. Chem. 21, 161–168. doi: 10.1016/0304-4203(87)90036-3
Riser S. C., Swift D., Drucker R. (2018). Profiling floats in SOCCOM: technical capabilities for studying the southern ocean. J. Geophys. Res. C: Oceans 123, 4055–4735. doi: 10.1002/2017JC013419
Roemmich D., Scripps Institution of Oceanography, Talley L., Zilberman N., Osborne E., Johnson K., et al. (2021). The technological, scientific, and sociological revolution of global subsurface ocean observing. Oceanography 2–8. doi: 10.5670/oceanog.2021.supplement.02-02
Sharp J. D., Fassbender A. J., Carter B. R., Johnson G. C., Schultz C., Dunne J. P. (2022). GOBAI-O2: temporally and spatially resolved fields of ocean interior dissolved oxygen over nearly two decades. doi: 10.5194/essd-2022-308
Sloyan B. M., Wanninkhof R., Kramp M., Johnson G. C., Talley L. D., Tanhua T., et al. (2019). The global ocean ship-based hydrographic investigations program (GO-SHIP): A platform for integrated multidisciplinary ocean science. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00445
Stoer A. C., Takeshita Y., Maurer T. L., Demeaux C. B., Bittig H. C., Boss E., et al. (2023). A census of quality-controlled biogeochemical-argo float measurements. Front. Mar. Sci. 10. doi: 10.3389/fmars.2023.1233289
Sutton T. T., Clark M. R., Dunn D. C., Halpin P. N., Rogers A. D., Guinotte J., et al. (2017). A global biogeographic classification of the mesopelagic zone. Deep Sea Res. Part I: Oceanogr. Res. Papers 126, 85–102. doi: 10.1016/j.dsr.2017.05.006
Sutton T. T., Frank T., Judkins H., Romero I. C. (2020). “As GOM Oil Extraction Goes Deeper, Who Is at Risk? Community Structure, Distribution, and Connectivity of the Deep-Pelagic Fauna,” in Scenarios and Responses to Future Deep Oil Spills: Fighting the Next War. Eds. Murawski S. A., Ainsworth C. H., Gilbert S., Hollander D. J., Paris C. B., Schlüter M., Wetzel D. L. (Springer International Publishing, Cham), 403–418.
Sutton T. T., Milligan R. J., Daly K., Boswell K. M., Cook A. B., Cornic M., et al. (2022). The open-ocean gulf of Mexico after deepwater horizon: synthesis of a decade of research. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.753391
Takeshita Y., Martz T. R., Johnson K. S., Plant J. N., Gilbert D., Riser S. C., et al. (2013). A climatology-based quality control procedure for profiling float oxygen data. J. Geophys. Res. Oceans 118, 5640–5650. doi: 10.1002/jgrc.20399
Tu J. V. (1996). Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J. Clin. Epidemiol. 49, 1225–1231. doi: 10.1016/S0895-4356(96)00002-9
Van Heuven S., Pierrot D., Rae J. W. B., Wallace D. W. R. (2009). MATLAB program developed for CO2 system calculations, carbon dioxide information analysis center, Oak Ridge National Laboratory (Oak Ridge, Tennessee: U.S. Department of Energy).
Wanninkhof R., Baringer M. O. (2014). Dissolved oxygen, temperature, salinity, and other variables measured from profile observations using CTD and other instruments from NOAA Ship Ronald H. Brown in the East Coast of the United States and GOM of Mexico during the second GOM of Mexico and East Coast Carbon (GOMECC-2) Cruise from 2012-07-24 to 2012-08-13 (NCEI Accession 0117943) (Asheville, North Carolina USA: NOAA National Centers for Environmental Information). doi: 10.7289/v59w0cds
Wanninkhof R., Zhang J.-Z., Baringer M. O., Langdon C., Cai W.-J., Salisbury, et al. (2013). Partial pressure (or fugacity) of carbon dioxide, dissolved inorganic carbon, alkalinity, temperature, salinity and other variables collected from Surface underway, discrete sample and profile observations using Alkalinity titrator, Barometric pressure sensor and other instruments from NOAA Ship RONALD H. BROWN in the Coastal Waters of Florida, Gray's Reef National Marine Sanctuary and others from 2007-05-11 to 2007-08-04 (NCEI Accession 0083633) (Asheville, North Carolina USA: NOAA National Centers for Environmental Information). doi: 10.3334/cdiac/otg.clivar_nacp_east_coast_cruise_2007
Waters J. F., Millero F. J. (2013). The free proton concentration scale for seawater pH. Mar. Chem. 149, 8–22. doi: 10.1016/j.marchem.2012.11.003
Weller R. A., Baker D.J., Glackin M. M., Roberts S. J., Schmitt R. W., Twigg E. S., et al. (2019). The challenge of sustaining ocean observations. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00105
Williams N. L., Juranek L. W., Johnson K. S., Feely R. A., Riser S. C., Talley L. D., et al. (2016). Empirical algorithms to estimate water column pH in the Southern Ocean. Geophys. Res. Lett. 43, 3415–3422. doi: 10.1002/2016gl068539
Wimart-Rousseau C., Steinhoff T., Klein B., Bittig H., Körtzinger A. (2024). Technical note: assessment of float pH data quality control methods – a case study in the subpolar Northwest Atlantic ocean. Biogeosciences 21, 1191–12115. doi: 10.5194/bg-21-1191-2024
Keywords: Biogeochemical-Argo, neural network, pH, Gulf of Mexico, carbon cycling
Citation: Osborne E, Xu Y-Y, Soden M, McWhorter J, Barbero L and Wanninkhof R (2024) A neural network algorithm for quantifying seawater pH using Biogeochemical-Argo floats in the open Gulf of Mexico. Front. Mar. Sci. 11:1468909. doi: 10.3389/fmars.2024.1468909
Received: 22 July 2024; Accepted: 14 October 2024;
Published: 18 November 2024.
Edited by:
Catherine Marie Schmechtig, UMS3455 Observatoire des sciences de l’Univers Paris-Centre Ecce Terra (ECCE TERRA), FranceReviewed by:
Cathy Wimart-Rousseau, Helmholtz Association of German Research Centres (HZ), GermanyNicolas Metzl, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2024 Osborne, Xu, Soden, McWhorter, Barbero and Wanninkhof. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emily Osborne, ZW1pbHkub3Nib3JuZUBub2FhLmdvdg==
†These authors have contributed equally to this work and share first authorship
 Emily Osborne
Emily Osborne Yuan-Yuan Xu
Yuan-Yuan Xu Madison Soden
Madison Soden Jennifer McWhorter
Jennifer McWhorter Leticia Barbero
Leticia Barbero Rik Wanninkhof
Rik Wanninkhof