
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 26 September 2024
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1441589
This article is part of the Research Topic Towards an Expansion of Sustainable Global Marine Aquaculture View all 13 articles
Understanding the molecular mechanism of melanogenesis in Plectropomus leopardus is important for exploring the pattern of skin colour variation in grouper. The research team conducted a combined transcriptomic and proteomic analysis of P. leopardus skin tissues in red-skinned and black-skinned fish and found that the common differences were reflected in the melanogenesis pathway. Therefore, to further investigate the molecular mechanism of melanogenesis in P. leopardus, the full-length sequences of the erk1/2 and mitf genes were obtained in this study using the RACE technique. Through structure-function analysis and differential expression in different red-skinned and black-skinned P. leopardus tissues, it was found that the MAPK signalling pathway may be involved in skin colour changes in P. leopardus, and when erk1/2 expression was decreased in P. leopardus, mitf expression increased accordingly. On the one hand, through short-term in vivo injection of erk1/2-dsRNA, the optimal interference primer for experimented fish was found to be group D: F2R1(F2: TAATACGACTCACTATAGGGATCAACGACATTCTCAGGGC; R1: TAATACGACTCACTATAGGGTCCATGGAGAAAGTGAAGGG), the optimal injection site was the tail vein, the optimal interference concentration was 5 µg/g, and the duration of the interference effect was 5 days. The results of long-term interference showed that when erk1/2 expression was decreased in P. leopardus, the skin colour of the treats fish then darkened, which indicated that ERK1/2 was involved in the regulation of melanogenesis. On the other hand, in vitro Co-Immunoprecipitation (Co-IP) results showed that there was a direct or indirect interaction between MITF and ERK1/2 proteins. In conclusion, this is the first time that an interaction between ERK1/2 and MITF, which indicated that ERK1/2 was involved in the regulation of melanogenesis through the regulation of MITF in P. leopardus. These results further enrich our understanding of the theoretical basis of the changing pattern of skin colour in P. leopardus and provides a new perspective for exploring the variable skin colouration of coral reef fish.
Plectropomus leopardus, belonging to Perciformes, Epinephelinae (Sugianti and Mujiyanto, 2016), mainly lives in the tropical waters of the Pacific and Indian Oceans, and a small number of them also exist in the east coast and southern waters of Hainan (Yoseda et al., 2008). Because of its rich nutrition and bright colour, P. leopardus is popular among consumers and has broad market prospects (Wang et al., 2011; Sun et al., 2015). However, environmental factors such as changes in light intensity and human activities during artificial culture produced a stress response in P. leopardus, resulting in the gradual darkening or blackening of the skin colour, which directly affects the ornamental and economic value of P. leopardus (Zhao et al., 2016). Recent studies in wild P. leopardus have found the presence of melanomas (Sweet et al., 2012), indicating that P. leopardus can be used as a good model to study the molecular mechanism of melanin metabolic diseases in fish (Lerebours et al., 2016). Exploring the process of melanogenesis can not only enrich our basic understanding of P. leopardus and skin colour research but also has certain scientific value in ecological conservation applications.
The MAPK signalling pathway is involved in the metabolic activities of organisms and plays a crucial role in cell growth and development, migration, differentiation, and apoptosis (Wang et al., 2017). There are three main highly conserved signalling pathways in the MAPK family, namely, ERK1/2, JNK1, and p38MAPK, which function through a tertiary kinase cascade and are inactive in the nonphosphorylated state; the MAPK signalling pathway stays quiescent and is activated upon stimulation by stepwise phosphorylation of MAPK (Zheng et al., 2009). In addition, MITF (microphthalmia-associated transcription factor) is a tissue-specific MAPK substrate found in melanocytes (Wellbrock and Arozarena, 2015) and the level of MAPK pathway activation is critical for MITF abundance and function in melanocytes (Molina et al., 2005). Recently, the MAPK pathway was found to be involved in the biological activities of pigment cells during skin colour formation in fish. By comparing the transcriptome and microbiome of Carassius auratus and Oreochromis mossambicus, researchers found that the MAPK signalling pathway is one of the key pathways involved in the regulation of skin colour formation (Zhu et al., 2016). These transcriptional level and epigenetic studies have provided good insights, but the specific mechanism through which signalling pathways and skin colour variation are related has not been fully elucidated. In particular, the skin colour of P. leopardus is very sensitive to changes in environmental factors, and MAPK signalling may be closely related to the regulation of environmental stress (Jalmi and Sinha, 2015).
The MAPK signalling pathway has been extensively studied in mammalian melanin synthesis, which is involved in the regulation of survival of many pigmented mammalian cells. For example, lipopolysaccharide (LPS) induces melanin production in human melanocytes through activation of the p38MAPK signalling pathway. Meanwhile, it was demonstrated that p38MAPK activation mediated the expression of the mitf and tyrosinase (tyr) genes (Zhou et al., 2021), which in turn induced melanin production, after negative regulation by p38MAPK inhibitors (Ahn et al., 2008). The same results were confirmed in mouse studies, where inhibition of p38MAPK resulted in reduced melanin secretion by melanocytes (Kim et al., 2007). JNK1 was revealed to be primarily involved in the regulation of melanocyte resistance to adversity. Minocycline reduces cell death in cutaneous melanocytes by activating the JNK1 pathway against the threat posed by H2O2 (Song et al., 2008); similarly, endothelin-1 plays a role in protecting human cutaneous melanocytes from UV-induced DNA damage by activating the JNK1 and p38MAPK signalling pathways (von Koschembahr et al., 2015). Interestingly, in contrast to p38MAPK and JNK1, activation of ERK1/2 inhibits melanocyte melanin synthesis. Inhibition of ERK1/2 activity stimulates melanin synthesis (Kim et al., 2002). Abundant evidence suggests that ERK1/2 is closely linked to the vital activities of melanocytes and plays an important role in the melanogenic pathway in mammals, and that this role is closely linked to mitf, a key gene for melanogenesis. Some studies have confirmed that ERK1/2 can directly bind to Ser73, the phosphorylation site of MITF, phosphorylate MITF and thus regulate melanin synthesis (Figure 1) (Wu et al., 2000).
Fish skin colour studies, using the comparative transcriptome approach, in red crucian carp (Zhang et al., 2017), Oreochromis mossambicus (Zhu et al., 2016; Wang et al., 2018)and P. leopardus (Dai et al., 2015) have identified the MAPK signalling pathway as potentially being involved in the regulation of skin colour in fishes. However, the molecular mechanism through which the MAPK signalling pathway regulates melanin synthesis in fish is still not elucidated. The research team conducted a combined transcriptomic and proteomic analysis of skin tissues from red-skinned and black-skinned P. leopardus and found that the differences were reflected in the melanogenesis pathway, which was closely related to the MAPK signalling pathway, and the activation levels of ERK1/2, which differed the most among experimental fish with different skin colours (Wen et al., 2022). Previous pre-experimental results showed that some fish turned black after 24h after RNAi interfered with erk1/2, This study will take these data as an entry point to explore the connection between ERK1/2, MITF and melanocyte differentiation and skin colour changes in P. leopardus by combining in vivo RNAi and in vitro Co-IP experiments as well as integrating these results with those of morphological observations, molecular biology, cell culture, in vivo injection, and protein interactions in our research. These results will open new perspectives for us to investigate the regulatory mechanism of skin colour change in P. leopardus and provide basic information for the in-depth understanding of how skin colour change occurs in coral reef fishes.
The Plectropomus leopardus used in this experiment were placed in a recirculating aerated bucket of water at 28°C in a culture room with a controlled 12:12 h light:dark cycle. A Motic SMZ-168 body vision system was used to differentiate between individuals with different skin colours in the culture population (Wen et al., 2022). We also screened 300 red-coloured and 300 black-coloured individuals, each with a body length of 11 ± 0.5 cm and a body weight of 32 ± 0.5 g, from cultured fish with large variations in skin colour. All the sampling procedures were conducted according the standards and ethical guidelines established by the Animal Ethical Review Committee of Hainan University, Haikou, China.
The transcriptome data were searched for erk1/2 and mitf candidate sequences (Wen et al., 2022). To clone the intermediate fragments of these two genes, PCR primers for erk1/2 and mitf were designed (Supplementary Table S1), total RNA was extracted from P. leopardus skin tissue using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and genomic DNA was removed using a PrimeScriptTM RT kit (Takara, Japan) with gDNA Eraser. An RNA purification kit (Qiagen, Valencia, CA) was used to purify RNA. RNA was tested for integrity and concentration using agarose gel electrophoresis and a Thermo Scientific NanoDrop instrument (Thermo Scientific, USA), respectively. One microgram of total RNA was reverse transcribed into cDNA using the HiScript III RT SμperMix for qPCR kit (Vazyme, Nanjing, China), and the intermediate fragments of the two genes were amplified by polymerase chain reaction (PCR).
To obtain the full-length cDNA sequences of erk1/2 and mitf, 3’ and 5’-RACE-ready cDNAs were prepared according to the instructions in the SMARTer® RACE 5’/3’ Kit (Clontech, USA). cDNA was prepared according to the SMARTer® RACE 5’/3’ Kit (Clontech, USA) instructions, and two pairs of RACE primers (Supplementary Table S1) were designed for nested PCR amplification. cDNAs were separated and purified by agarose gel electrophoresis, ligated into vectors, transformed and sequenced. After splicing the sequences, a pair of gene-specific primers (Supplementary Table S1) was designed based on the terminal sequence of the cDNA for full-length cDNA amplification, and the products were sequenced again to confirm the nucleotide sequence. Finally, the full-length cDNA sequences of erk1/2 and mitf were verified with high-fidelity PrimerSTAR HS DNA polymerase reagent (Takara, Japan).
We retrieved ERK1/2 and MITF protein sequences of different species from GenBank and phylogenetically compared them with those of P. leopardus and aligned them multiple times using ClustalW2. We constructed a bootstrap neighbour-joining (NJ) phylogenetic tree using MEGA 5.0 software and tested branching reliability using bootstrap resampling with 1000 pseudoreplicates.
Total RNA was extracted from skin tissues of red-coloured and black-coloured fish using TRIzol (Invitrogen, 15,596-025) according to the manufacturer’s instructions. Reverse transcription PCR was performed using HiScript® III RT SuperMix for qPCR (+gDNA wiper) (Vazyme™, R123-01). RT-qPCR was performed on a Roche LightCycler 384 real-time PCR system (Applied Roche, Basel, Switzerland) using ChamQ SYBR Color qPCR Master Mix (Vazyme Biotech Co., Piscataway, NJ, China) with the following steps: 40 cycles of initial denaturation at 95°C for 30 s, followed by 95°C for 5 s, 55°C for 30 s, 72°C for 30 s, and a 95°C to 65°C cycle. All reactions were performed in triplicate in a final reaction volume of 10 μL. RT-qPCR primers designed with Primer Express® software Primer5.0 are shown in Supplementary Table S2.
dsRNA for erk1/2 was synthesized in vitro using the TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. We prepared templates for erk1/2 synthesis by amplifying skin tissue in P. leopardus cDNA with the design of four pairs of RNAi primers (Supplementary Tables S3, S4). We measured the concentration of dsRNA at 260 nm using a Thermo Scientific NanoDrop, dsRNA purity and integrity were detected by 1% agarose gel electrophoresis and then stored at -80°C for use.
Preliminary experiments showed that the interference effect was greatest with the group D (F2R1) interfering agent, a dose of 5 μg/g of body weight, and the injection site was tail vein injection. By taking samples on days 1, 2, 3, 4, 5, 6, 7, 9, 11, 13, and 15 after injection and then detecting erk1/2 mRNA expression, it was found that the interference effect was greatest on day 5 after injection. In this study, we randomly divided juvenile experimental fish with an initial body weight of 32 ± 0.5 g into eight groups with eight fish in each group for one, two, three, and four consecutive rounds of RNA interference and injected erk1/2-dsRNA into the tail vein at dose of 5 μg/g of body weight. The control group was injected with an equal volume (300 μL) of DEPC H2O. Twelve tissues were taken (dorsal skin (BSK), abdominal skin (ASK), liver (L), kidney (K), spleen (SP), gill (G), heart (H), intestine (I), eye (E), muscle (M), and brain (BR), and two samples were taken from each group. Microscopic observation of skin tissue was performed before sampling. After sampling, the samples were rapidly frozen in liquid nitrogen and stored at -80°C until use. One sample was used for RT-qPCR analysis of mRNA expression using the primers shown in Supplementary Table S5; the other biological sample was used for the related enzyme activity assay.
BSK and ASK skin tissues obtained from fish in the experiment were homogenized with PBS and then centrifuged at 2500 rpm/min for 10 min to obtain the supernatant. A fish tyrosinase (TYR) enzyme immunoassay kit (Abimat PharmaTech Shanghai Co., Ltd., AB-10122A) and a fish melanin (ML) content immunosorbent assay kit (Shanghai Enzymotec Biotech Co., Ltd., ml025778-96T) were used to determine to tyrosinase activity and the concentration of melanin in the samples, respectively. The tyrosinase activity and melanin content of the samples were calculated from the standard curve.
The BSK of the experimented fish was observed microscopically with a Tipscope microscope (Kenwickis (Wuhan) Technology Co., Ltd., China), and five identical positions were selected at the observation interface for melanocyte counting. This observation occurred after anaesthesia with MS222 and before tissue sampling.
In this study, proteins interacting with ERK1/2 were identified by Co-IP. HEK 293T cells transfected with empty vector were used as the control group, while the ERK1/2 plasmid was transfected into the experimental group. Cells were lysed 24 h after transfection, and Western blotting was performed both before and after Co-IP. The differences in bound proteins in the experimental and control groups were later compared by silver staining. Specific binding was considered to have occurred if a protein-stained band appeared at the same position in the experimental group but not in the control group, while a band at the same position in the treated group and the control group was considered nonspecific binding. The input group was set up for electrophoresis and immunoblotting to determine whether the transfected plasmid was expressed in the cells. FLAG antibody detection before IP (MITF target protein size: 41.5 kD; DIAAN:2064); HIS antibody detection before IP (ERK1/2 target protein size 43.1 kD; CUSABIO: CSB-MA000159); GAPDH antibody detection (GAPDH target protein size 36kD; proteintech:60004-1-Ig).
The erk1/2 and mitf mRNA expression levels were calculated using the comparative CT (2−ΔΔCt) method (Livak and Schmittgen, 2001). All results are expressed as the mean ± standard error of three replicate experiments. The means and standard deviations of three replicate experiments were calculated to confirm homogeneity of variance, and comparisons between means were made using one-way ANOVA. Each data set was analysed using GraphPad Prism 5.0 software (GraphPad software Inc., CA, USA). Multiple comparisons between groups were performed using Tukey B and Duncan’s test, with p < 0.05 being considered a statistically significant difference. SPSS 17 (Chicago, IL, USA) was used for all statistical analyses.
The study results showed that the full-length sequence of P. leopardus erk1/2 was 1449 bp, and the ORF of erk1/2 was 1176 bp, encoding a protein molecule containing 391 amino acids. The length of the obtained 3’ RACE product was 105 bp, containing a typical polyA tail; the length of the obtained 5’ RACE product was 168 bp. The intermediate fragment of the P. leopardus erk1/2 gene, the 3’ RACE and the 5’ RACE base sequences were spliced together using DNAStar software (Supplementary Figure S1A). The gene sequence was uploaded to NCBI (ID: OM831230). DNAMAN multiple sequence comparison analysis revealed that ERK1/2 is a serine/threonine protein kinase that his highly conserved among various different species, and sequence comparison showed that P. leopardus and Epinephelus lanceoletus have up to 93.62% similarity (Supplementary Figure S2A).
The full-length sequence of P. leopardus mitf was 1802 bp, and the ORF of mitf was 1134 bp, encoding a protein molecule containing 377 amino acids. The length of the obtained 3’ RACE product was 560 bp, containing a typical polyA tail; the length of the obtained 5’ RACE product was 108 bp. The base sequences of the intermediate fragment of the P. leopardus mitf gene, the 3’ RACE and the 5’ RACE base sequences were spliced together using DNAStar software (Supplementary Figure S1B), and the gene sequences were uploaded to NCBI (ID: OM914601). DNAMAN multiple sequence comparison analysis revealed that the MITF proteins of P. leopardus and Epinephelus lanceoletus had up to 88.89% similarity (Supplementary Figure S2B).
Phylogenetic analysis of ERK1/2 from representative fish and mammals yielded an NJ phylogenetic tree indicating that ERK1/2 from P. leopardus was highly homologous to that from Epinephelus coioides and Siniperca chuatsi (Supplementary Figure S3A). Analysis of the ERK1/2 amino acid sequence using the signalP4.1 website revealed no signal peptide and indicated that the ERK1/2 protein may be intracellular. Additional analysis using the Compute pl/Mw website revealed that the isoelectric point of ERK1/2 was pH 6.19, and the predicted molecular weight was 44.304 kDa. The hydrophobicity of the ERK1/2 protein was analysed by using ProtScale online software, and the results showed that ERK1/2 is hydrophilic and is a soluble protein.
The NJ phylogenetic tree showed that the MITF of P. leopardus is highly homologous to that of E. lanceoletus (Supplementary Figure S3B). Analysis of the MITF amino acid sequence using the signalP4.1 website revealed no signal peptide and indicated that the MITF protein was probably intracellular. Additional analysis using the Compute pl/Mw website showed that the isoelectric point of MITF was pH 5.92, and the predicted molecular was 41.796 kDa. In addition, MITF protein has relatively good hydrophilicity and is considered to be a soluble protein.
The expression of erk1/2 and mitf in 10 tissues of P. leopardus (skin, liver, spleen, head kidney, intestine, heart, fin, eye, muscle, brain) was examined using RT-qPCR, and the primers are shown in Table S2. Quantitative relative expression was plotted using real-time fluorescence as shown in Figure 2. The results showed that the erk1/2 and mitf genes were constitutively expressed in the skin, liver, spleen, head kidney, intestine, heart, fin, eye, muscle, and brain. The highest level of erk1/2 expression was observed in the fins of P. leopardus and the lowest expression was observed in the heart. mitf genes were most highly expressed in the eyes and the lowest expression was observed in the kidney.
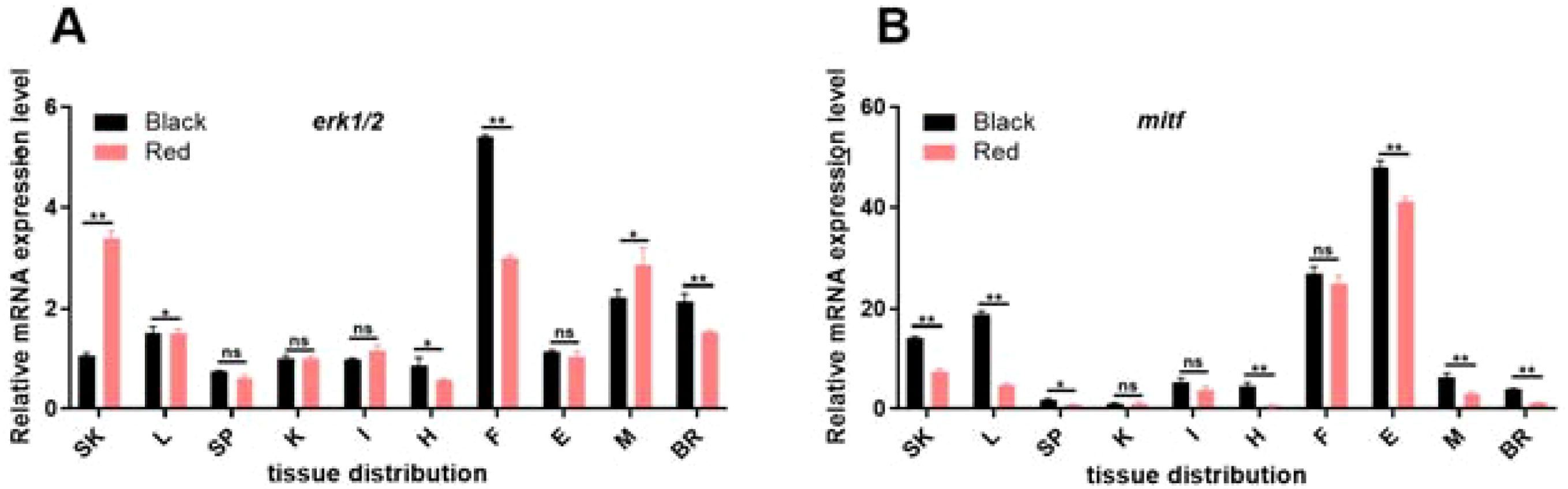
Figure 2. Relative expression of erk1/2 (A) and mitf (B) in tissue species in red-skin and black-skin P. leopardus. **, Indicates significant differences in the gene of tissues with red-skin and black-skin individuals. SK, skin; L, liver; SP, spleen; K, kidney; I, intestines; H, heart; F, fin; E, eye; G, gills; M, musle; BR, brain. Different letters indicate significant differences in gene expression between different experimental groups.
Whether the synthesized dsRNA can have a significant interference effect is determined by the quality of the primers. In this study, four pairs of specific primers were designed, and the T7 promoter was added to the primers. All four pairs of primers can synthesize dsRNA in vitro. As seen from the experimental results shown in Supplementary Figure S4, 24 hours after the injection of erk1/2-dsRNA, the four primer pairs have different degrees of erk1/2 RNA interference in all tissues of P. leopardus. Comparing the differences in erk1/2 mRNA expression in each tissue after injection, the primer pair in group D erk1/2-iF2R1 (743 bp) was selected for subsequent experiments.
The injection site directly affects the interference efficiency of erk1/2-dsRNA. The sites at which fish underwent in vivo injections included the abdominal cavity, dorsal muscle and tail vein. From the experimental results shown in Supplementary Figure S5, it can be seen that the interference effect of abdominal cavity and dorsal muscle injection fluctuated greatly, while the interference effect was most obvious after tail vein injection, so this site was chosen for the subsequent experiments.
Based on the results of previous studies, five dose groups, 0, 1, 5, 10, and 15 μg/g, were designed. Among them, 0 μg/g was the control group, and animals in this group were injected with an equal volume(300μL) of DEPC water. The results shown in Figure 3 reveal that there was no death of experimental fish 24 h after injection, and analysis of erk1/2 mRNA expression in each tissue revealed that the 5 μg/g dose group had the greatest RNA interference effect. Therefore, 5 μg/g was chosen as the subsequent interference dose.
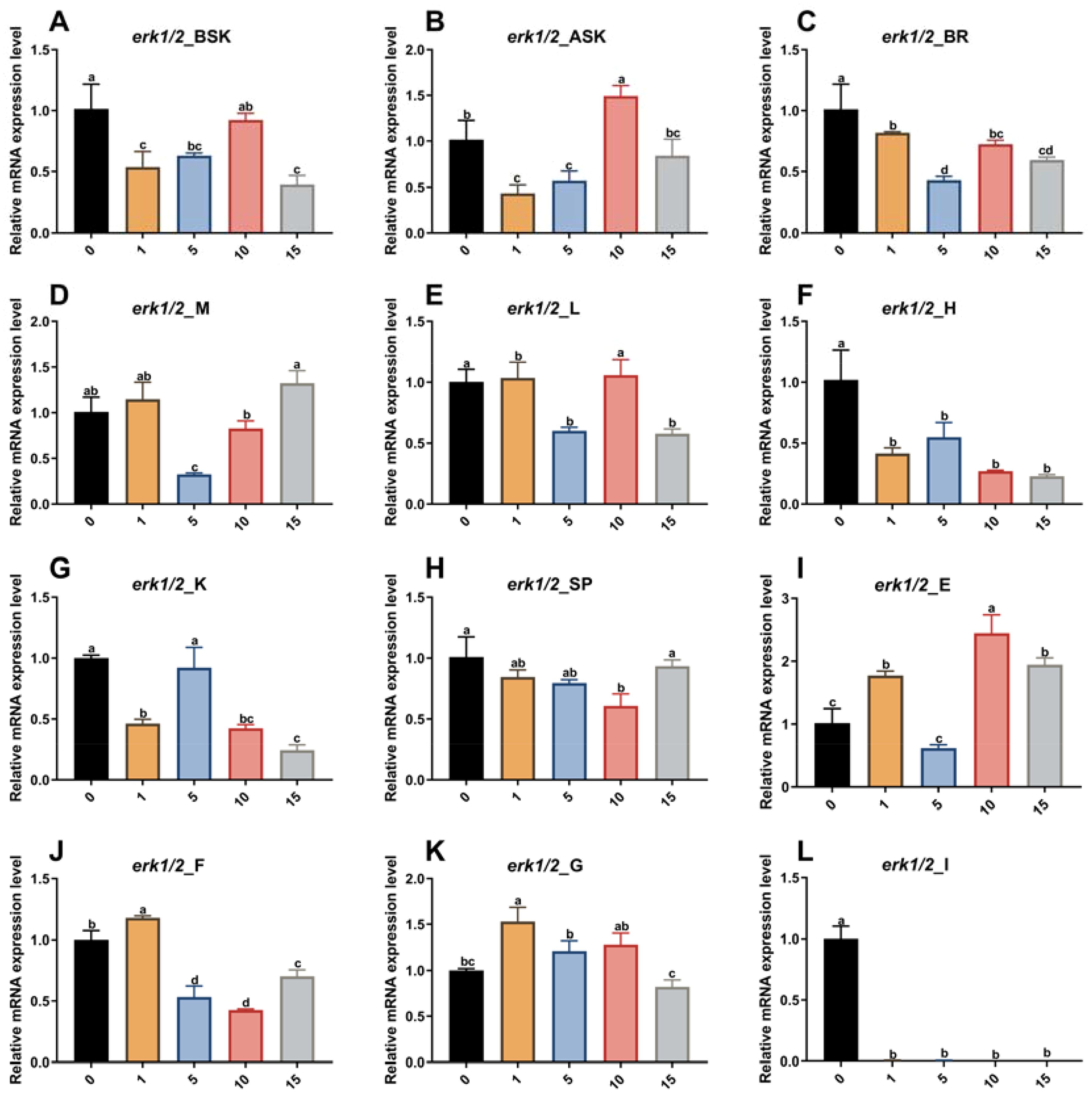
Figure 3. Effect of different doses of erk1/2-dsRNA injected into the tail vein on the relative expression of the erk1/2 gene in various P. leopardus tissues (n=8). 0, 1, 5, 10 and 15 indicate erk1/2 injection concentrations of 0 µg/g (DEPC water), 1 µg/g, 5 µg/g, 10 µg/g and 15 µg/g, respectively. BSK, back skin; ASK, abdominal; BR, brain; M, muscle; L, liver; H, heart; K, kidney; SP, spleen; E, eye; F, fin; G, gills I, intestines. Different letters indicate significant differences in gene expression between different experimental groups.
After P. leopardus was injected with erk1/2-dsRNA at a dose of 5 μg/g, erk1/2 mRNA expression decreased in all tissues of the experimental group by varying degrees from days 1 to 15, which indicated that the RNA interference was successful. The results of this experiment are shown in Figure 4. The expression of erk1/2 mRNA reached its lowest level on day 5 after injection of erk1/2-dsRNA. Therefore, in the subsequent long-term interference experiment, the injection was performed every 5 days for a total of 20 days.
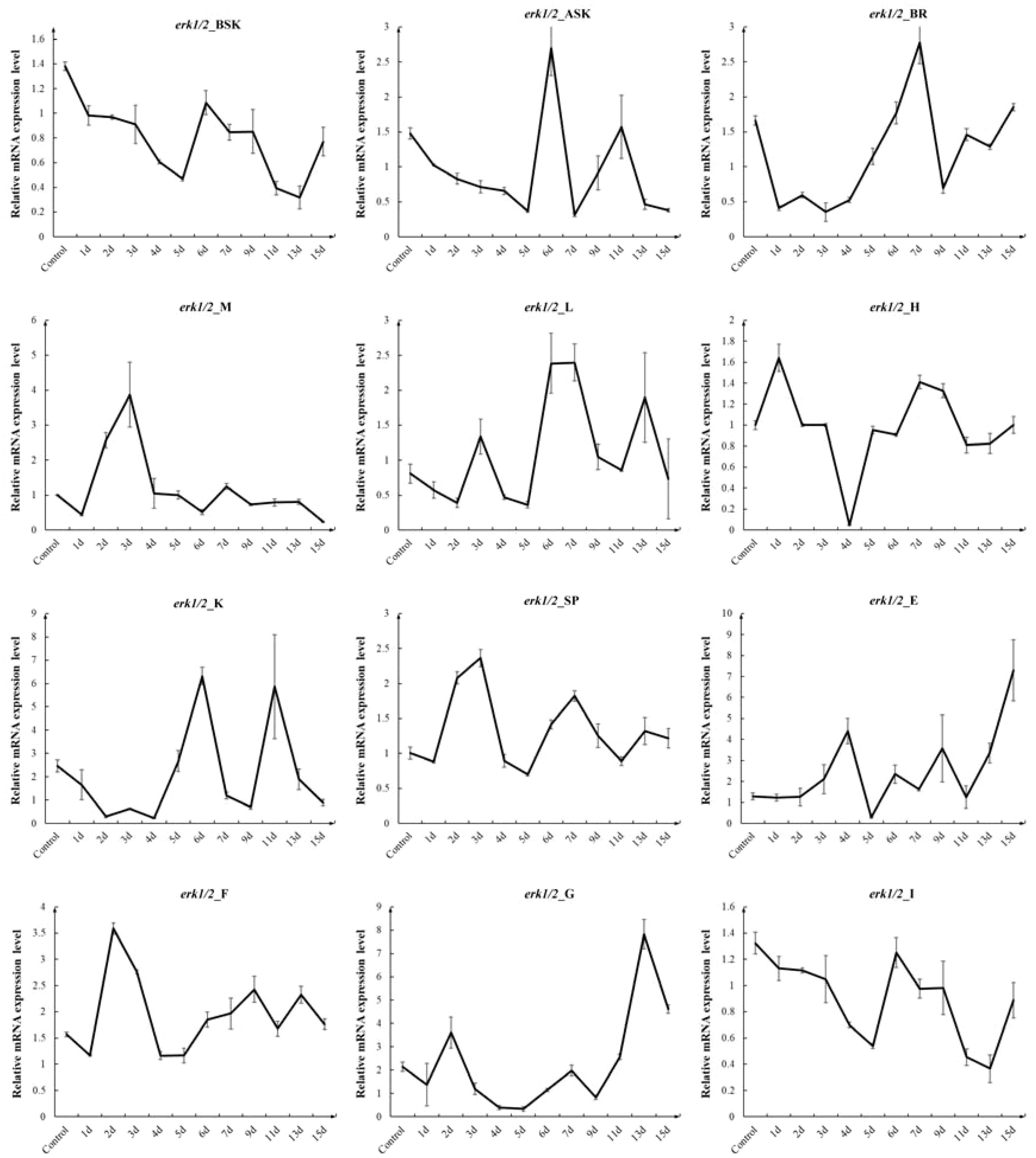
Figure 4. Effect of injection of erk1/2-dsRNA from the tail vein at 5 µg/g for different times on the relative expression of the erk1/2 gene in various tissues of P. leopardus (n=8). 1d, 2d, 3d, 4d, 5d, 6d, 7d, 9d, 11d, 13d, 15d denote 1, 2, 3, 4, 5, 6, 7, 9, 11, 13, 15 days after injection. BSK, back skin; ASK, abdominal; BR, brain; M, muscle; L, liver; H, heart; K, kidney; SP, spleen; E, eye; F, fin; G, gills I, intestines.
The experimental results in Figure 5A show that four consecutive injections of erk1/2-dsRNA resulted in changes in erk1/2 mRNA expression in all P. leopardus tissues, with an overall decreasing trend, suggesting successful interference with the expression of erk1/2 mRNA. In addition, Figure 5B shows that the expression of mitf and kitα mRNA (Nassar and Tan, 2020) (The mutational landscape of mucosal melanoma), which are related to melanogenesis, was elevated, while the expression of dopachrome tautomerase (dct) mRNA was decreased (Zhou et al., 2021)(Epigenetic regulation of melanogenesis).
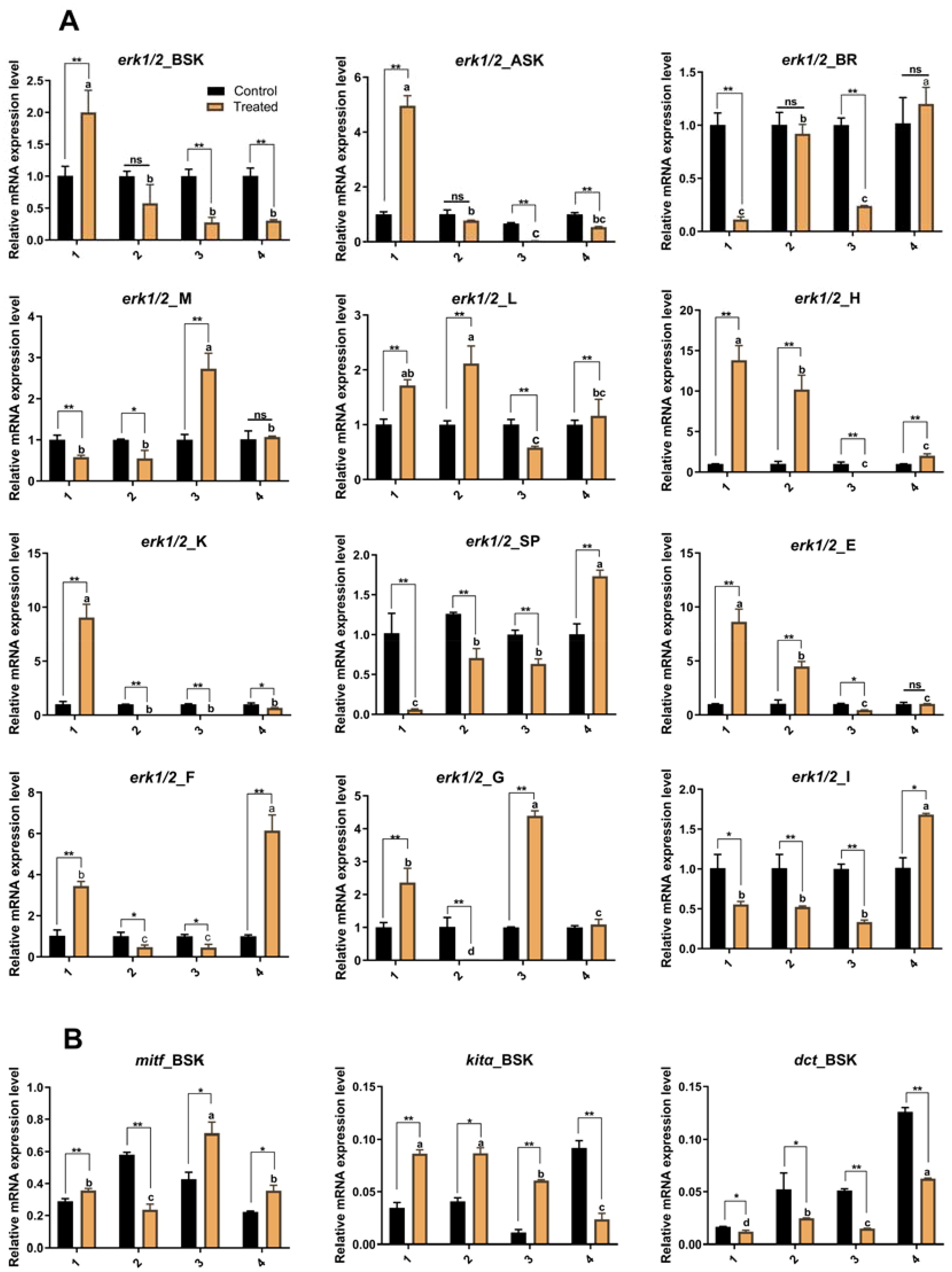
Figure 5. Effect of continuous injection of erk1/2-dsRNA on gene expression in P. leopardus. (A) The effect of continuous erk1/2-dsRNA injection on the erk1/2 gene in various tissues of P. leopardus; (B) The effect of continuous erk1/2-dsRNA injection on the expression of mitf, kitα, and dct genes in the dorsal skin of P. leopardus. 1, 2, 3, and 4 indicate the number of injections. BSK, back skin; ASK, abdominal; BR, brain; M, muscle; L, liver; H, heart; K, kidney; SP, spleen; E, eye; F, fin; G, gills I, intestines. **, Indicates significant differences in gene expression between different control and experimental groups. Different letters indicate significant differences in gene expression between different experimental groups.
The tyrosinase (TYR) activity and melanin content of the BSK and ASK of P. leopardus were increased after four consecutive injections of erk1/2-dsRNA, but the difference was not significant (Supplementary Figure S7). However, elevated TYR activity inclines the organism to the true melanogenesis pathway (Hoekstra et al., 2006; Ito and Wakamatsu, 2003). which is consistent with these findings.
The experimental results show that observation of P. leopardus 24 h after the injection of erk1/2-dsRNA, reveals a significant change in the local skin colour at the injection site (Wen et al., 2022); Phenotypic observation of experimental fish after long-term RNAi revealed that the skin colour of treated fish was darker than control fish (Figure 6A), and the number of melanocytes in the experimental group was significantly higher than that in the control group under the same viewing angle (Figure 6B).
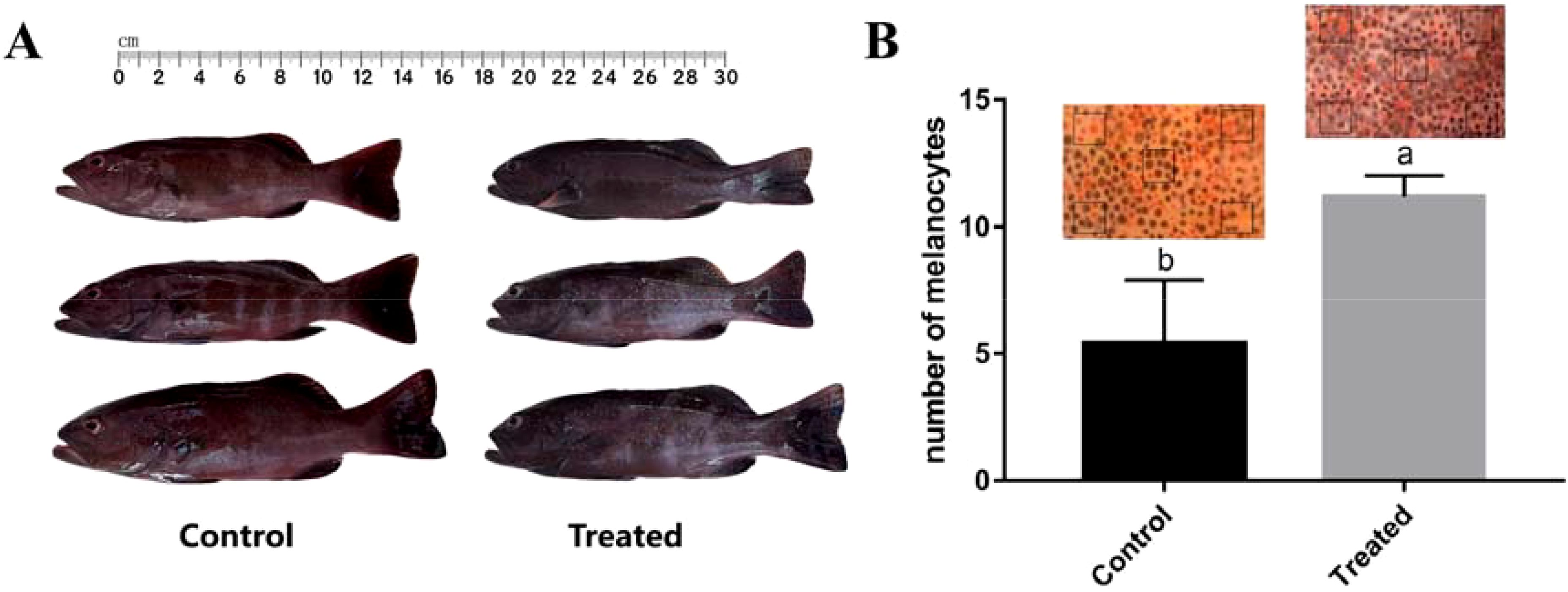
Figure 6. Effects of continuous erk1/2-dsRNA injection on phenotypes in P. leopardus (n=8). (A) Observation of variation in skin-colour after long-term RNAi; (B) Microscopic observation and statistics of variation in body-colour at the same viewing angle. The statistical area is the boxed portion of the figure, with 5 tails for each of the experimental and control groups counted separately. Different letters indicate significant differences in number of melanocytes between different experimental groups.
After silver staining the gel scanning map results showed that the target proteins were labelled according to their positional size (Figure 7). Before Co-IP, Figure 7A shows that the FLAG antibody could detect the MITF signal after normal type exposure for 2 min; the His antibody could detect the ERK1/2 target signal after ultrasensitive type exposure for 20 min; and the GADPH antibody could detect the internal reference signal, indicating that the MITF, ERK1/2, and GAPDH proteins can be normally expressed in HEK 293T cells. After Co-IP, Figure 7B shows that the MITF target signal was detected in the Co-IP group after exposure to FLAG antibody, and the ERK1/2 target signal was detected in the Co-IP group after exposure to HIS antibody for 10 min. The two target proteins were still detectable after Co-IP, which indicates that MITF and ERK1/2 have a mutual binding relationship in P. leopardus.
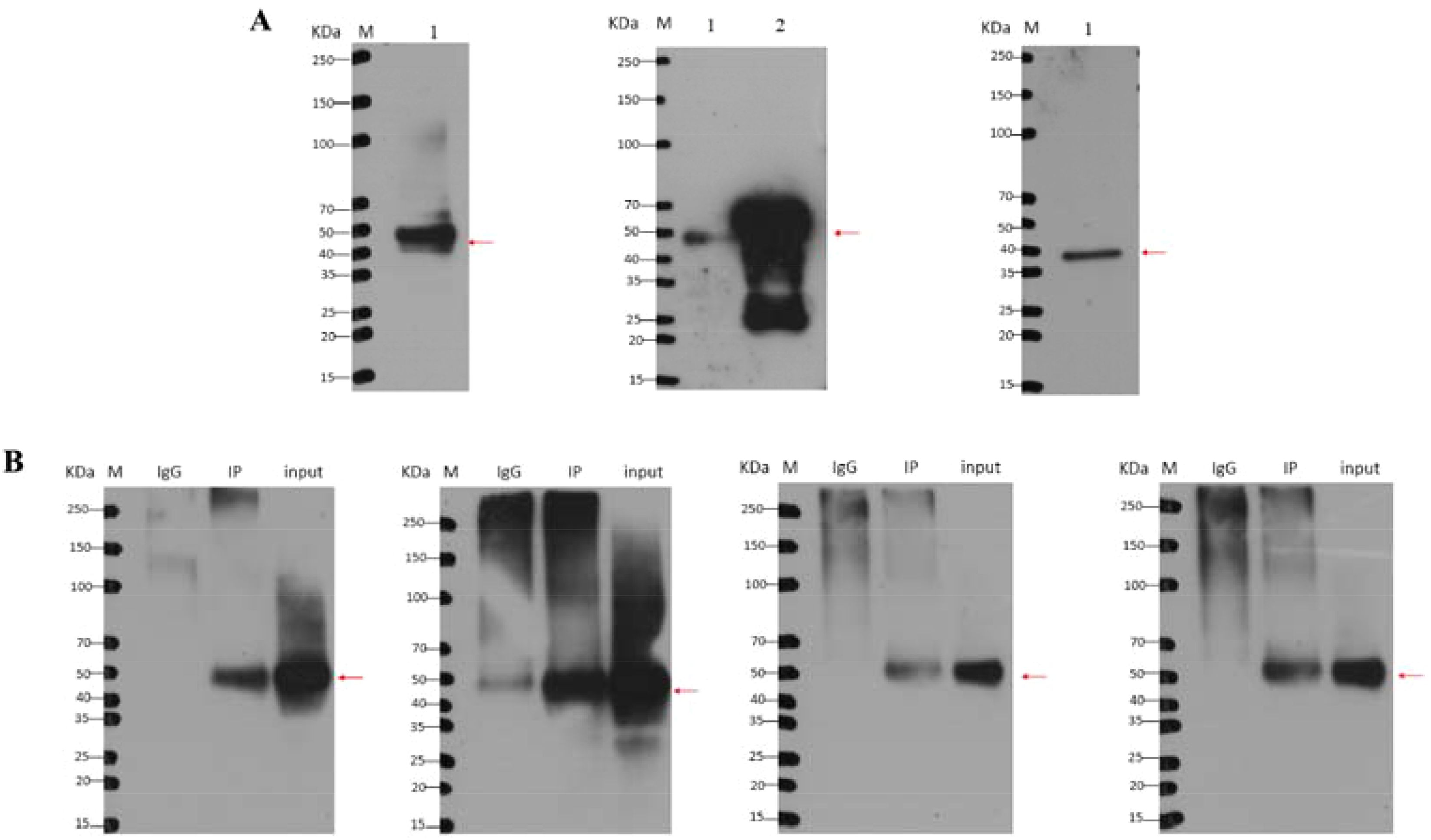
Figure 7. Western blot detection before and after Co-IP. (A) Western blot detection before Co-IP; (B) Western blot detection after Co-IP. M, marker; 1, 293T cotransfected cells; 2, positive control. The target proteins are labelled according to their position and size in the figure.
The relationship between the MAPK signalling pathway and colour has been abundantly studied in mammals and fish, but the mechanism of regulation in fish is not clear. In this study, the full-length P. leopardus erk1/2 gene sequence was cloned, and the erk1/2 gene does not exist as an isoform in this species. The complete cDNAs of erk1 and erk2 were cloned in the early 1990s (Boulton et al., 1990, 1991). After comparison, it was found that the ERK1 and ERK2 proteins share approximately 83% amino acid homology (Supplementary Figures SI, S2) (Boulton et al., 1990). The phylogenetic tree suggests that in P. leopardus ERK1/2 is a MAPK3 (Supplementary Figure S3), which has important implications for our understanding of the ERK pathway. RT-qPCR was used to investigate the differences in the tissue distribution of P. leopardus erk1/2 in red-skinned and black-skinned fish, and the results showed that erk1/2 showed a constitutive distribution in all of the examined tissues in all experimental fishes, with higher expression observed in skin, fins, muscle, and brain (Figure 2), and when erk1/2 expression was decreased in P. leopardus, mitf expression increased accordingly. This result is similar to the erk1/2 expression pattern in other vertebrates, but there is a significant difference in expression in other species, which is similar to the distribution of erk1/2 in mammals (Boulton et al., 1990), suggesting that erk1/2 may be widely involved in a variety of essential processes in organisms (Schmitt et al., 2019). ERK1/2 expression in skin tissues was significantly lower in experimental fish with black skin than that in fish with red skin. Related studies in mammals have confirmed that ERK1/2 directly binds to Ser73, the phosphorylation site of MITF, phosphorylates MITF and thus regulates melanin synthesis (Wu et al., 2000). In mammals, the ERK signalling pathway is one of the major regulators of a variety of biological activities, including cell differentiation and proliferation (Yang, 2020). The results of this study suggest that the ERK1/2 protein is a very conserved serine/threonine protein kinase, and the erk1/2 gene of P. leopardus may have a direct or indirect regulatory role in skin colour, which is a critical life process in pigment cells and ultimately exerts a role in the regulation of the number, migration, morphology, and function of pigment cells.
In addition, pigment cells in fish skin tissues develop by differentiation from pigmentoblasts in the neural crest, which are devoid of pigment precursors, and the role of the mitf gene in this process should not be underestimated; mitf is not only a prominent marker gene for melanin stem cells and is mainly involved in the regulation of melanocyte differentiation but also plays a role in regulating the differentiation of nonmelanocyte cells (Johnson et al., 2011). The current study shows that the expression of the mitf gene is relatively high in tissues with an abundant number of melanocytes. For example, Zhang et al. found that the skin colour of Carassius auratus was able to change from grey to red, and mitf mRNA expression was reduced accordingly (Zhang et al., 2017). Expression of the mitf gene was also significantly higher in skin tissues in black-white Cyprinus carpio than in golden, tricolour, and red-white Cyprinus carpio (Liu et al., 2015). This phenomenon is not unique to fish, and similar results have been observed in terrestrial vertebrates (Xin et al., 2018; Zhang et al., 2016). In the present study, the expression of the mitf gene in various tissues of P. leopardus was similar to the above findings. The mitf gene was highly expressed in the dorsal skin, eye and fin tissues of P. leopardus (Figure 2), and this result verifies that the mitf gene regulates melanin synthesis in the organism. When the mitf gene is expressed at high levels in an organism, it will favour melanin production in that individual (Steunou et al., 2013); if the gene is expressed at a low levels, it will inhibit or reduce melanin production in the organism (Wu et al., 2021). Combining the erk1/2 results from this study and those from related studies in mammals, we hypothesize that in P. leopardus, erk1/2 may be involved in the regulation of melanogenesis through the regulation of mitf.
The erk1/2 gene plays an important role in the formation of skin colour in mammals and fish (Luo et al., 2013), but there are still few studies on erk1/2 loss-of-function in fish. In RNAi experiments, the dsRNAs synthesized with different primer sequences had different in effects. In this study, group D (erk1/2iF2R1, 743 bp) had the best RNA interference effect (Supplementary Figure S4), indicating that the nucleic acid sequence of erk1/2-dsRNA (group D) was an efficient RNAi target, which was consistent with the findings of the researchers (Wang et al., 2013). Then, equal doses of erk1/2-dsRNA were injected into three different sites, namely, the abdominal cavity, the back muscle, and the tail vein, and analysis of erk1/2 mRNA expression in each tissue of the experimental fish showed that tail vein injection had the best interference effect (Supplementary Figure S5). This is likely because it is possible to transport the injected interference sequence throughout the body through the fish blood circulation system allowing the sequence to produce its full RNA interfering effect (Bu, 2019). Therefore, based on the results of this experiment and the specifications of the experimented fish, tail vein injection is proposed for subsequent experiments. In addition to the specific dsRNA sequence used, and the injection site both directly affecting the efficacy of RNAi, the dsRNA injection dose also has an effect (Ge et al., 2020; Tan et al., 2020). The results of this study showed that the experimental fish injected with 5 μg/g dsRNA had the best interference effect (Figure 3), and the erk1/2 RNA interference effect was not completely positively correlated with the dsRNA injection dose. In recent years, an interference dose of 4-5 μg/g has mostly been used for RNAi in crustaceans, and the interference effect is obvious at this dose. After one injection of erk1/2-dsRNA (5 μg/g), the expression of erk1/2 mRNA decreased and reached the lowest level 5 days post-injection in most tissues, and the interference effect was most stable at this time point (Figure 4). In addition, fish skin colour is coregulated by many genes, and some researchers found that the mRNA expression of the slc7a11 gene was decreased after siRNA treatment, resulting in a significant downregulation of the expression of genes related to melanin formation, such as mitf in rabbit skin fibroblasts (Yang et al., 2018). In this study, we found that the expression level of erk1/2 mRNA in experimental fish could be significantly reduced by injecting erk1/2-dsRNA after long-term interference was completed, and the expression levels of mitf and kitα mRNA in dorsal skin tissues were significantly increased following erk1/2 interference (Figure 6), suggesting that erk1/2 has a negative regulatory effect on mitf and other genes related to skin colour formation in fish. Previous findings suggest that erk1/2 has a regulatory role in fish melanogenesis, which is possibly related to the synthesis of tyrosine proteases. In melanosomes, tyrosinase acts as a catalyst in the first two stages of the melanogenesis mechanism, and overexpression of tyrosinase leads to overproduction of melanin and pathological disorders such as hyperpigmentation (Hearing and Jiménez, 1987). Measurements of tyrosinase activity and melanin content in the DSK and ASK of P. leopardus in this study showed that erk1/2 expression was suppressed, and melanin content increased along with the TYR activity involved in melanin synthesis (Supplementary Figure S6), which promoted melanogenesis (Chung et al., 2019). It has also been shown that long-term RNA interference can lead to significant phenotypic changes in tissues or organs (Tan et al., 2020; Ventura et al., 2009), and it is clear that fewer injections can reduce the damage caused to experimental fish. In the present study, the number of melanocytes in the skin of P. leopardus increased significantly after several consecutive injections, and the fish skin darkened significantly (Figure 7). In conclusion, the results of gene expression, enzyme activity and phenotypic observation suggest that ERK1/2 may be involved in melanogenesis in P. leopardus through the regulation of MITF, but the specific mechanism through which these proteins interact still needs further study.
Studies in vertebrates suggest that ERK1/2 may interact with MITF, and activation of MITF promotes melanogenesis by increasing the expression of melanin-related enzymes (Jimenez-Cervantes et al., 1994; Tsukamoto et al., 1992). Studies have shown that MAPK family proteins, including p38MAPK, ERK1/2, and JNK1, are activated by numerous extracellular stimuli and play key roles in melanin synthesis (Peng et al., 2014). For example, phosphorylated ERK inhibits MITF expression and thus reduces melanin synthesis, while phosphorylation of p38MAPK and JNK1 can increase melanin synthesis by activating MITF (Kang et al., 2015). Kim et al. found that UV-B induced apoptosis in human melanocytes, which correspondingly increased the phosphorylation levels of JNK1 and p38MAPK but transiently inactivated ERK1/2 kinases (Kim et al., 2007). These studies suggest that the three members of the MAPK pathway, JNK1, p38MAPK, and ERK1/2, are mediated through the regulation of key genes for melanin synthesis, which in turn participate in the life process of melanocytes, ultimately exerting regulatory effects on melanocyte number, migration, morphology, and function. The results of the preliminary study showed that erk1/2 was significantly expressed in the brains of P. leopardus with different skin colours, suggesting that erk1/2 may have a direct or indirect regulatory role in skin colour, participate in the life process of pigment cells, and ultimately play a role in the regulation of the number, migration, morphology, and function of pigment cells. Currently, the mechanism of interaction between the different proteins in the process of pigment cell formation is poorly understood and needs to be investigated more fully. In this study, the results of immunoprecipitation indicate that P. leopardus ERK1/2 and MITF proteins are normally expressed in HEK 293T cells (Figure 7A). Moreover, MITF and ERK1/2 target proteins could still be detected after Co-IP (Figure 7B), indicating that the two proteins can bind to each other in P. leopardus. Wu et al. found that ERK1/2 can bind to Ser73, the phosphorylation site of MITF, which phosphorylates MITF and then regulates melanin synthesis (Wu et al., 2000), which is consistent with the results of this study, suggesting that MITF and ERK1/2 bind to each other and are involved in the regulation of melanogenesis in P. leopardus (Figure 1). In addition, the reciprocal relationship between ERK1/2 and MITF was confirmed for the first time in coral reef fishes, which provides technical and theoretical support for more in-depth investigation of the mechanism of skin colour formation in fishes, but further studies are still needed to reveal the reasons for the variation in skin colour in fishes.
In summary, the full-length erk1/2 and mitf genes were cloned and it was found that these genes were differentially expressed in with red-skinned and black-skinned P. leopardus fish, suggesting that erk1/2 and mitf may be involved in melanogenesis. Injection in vivo experiments revealed that ERK1/2 can be involved in regulating melanogenesis in P. leopardus. Co-IP in vitro shows that there was a direct or indirect interaction between MITF and ERK1/2 proteins. Therefore, this study the study demonstrates that ERK1/2 regulates melanogenesis through the mediation of MITF in P. leopardus. Which is the first study showing that ERK1/2 interacts with MITF to regulate melanin synthesis in coral reef fishes. The results of this study further enrich our understanding of the changing pattern of skin colour in P. leopardus and provide a new perspective for exploring the variable skin colour of coral reef fishes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal studies were approved by a Sanya Nanfan Research Institute of Hainan University, Hainan Aquaculture Breeding Engineering Research Center, Hainan Academician Team Innovation Center, Hainan University, Haikou 570228, China. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
MY: Conceptualization, Writing – original draft. JH: Investigation, Writing – review & editing. DZ: Software, Writing – review & editing. HT: Methodology, Software, Writing – review & editing. JCL: Methodology, Writing – review & editing. JL: Supervision, Writing – review & editing. XW: Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32202901); Key R&D Project in Hainan (ZDYF2023XDNY046); The Innovation Center of Hainan University (XTCX2022NYC16).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1441589/full#supplementary-material
Ahn J. H., Jin S. H., Kang H. Y. (2008). LPS induces melanogenesis through p38 MAPK activation in human melanocytes. Arch. Dermatol. Res. 300, 325–329. doi: 10.1007/s00403-008-0863-0
Boulton T. G., Nye S. H., Robbins D. J., Ip N. Y., Radzlejewska E., Morgenbesser S. D., et al. (1991). ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65, 663–675. doi: 10.1016/0092-8674(91)90098-J
Boulton T. G., Yancopoulos G. D., Gregory J. S., Slaughter C., Moomaw C., Hsu J., et al. (1990). An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science 249, 64–67. doi: 10.1126/science.2164259
Bu H. (2019). Study on the RNA innterference of slc7a11 Gene in Red Tilapia (Nanjing: Nanjing Agricultural University).
Chung Y. C., Kim M.-J., Kang E. Y., Kim Y. B., Kim B. S., Park S.-M., et al. (2019). Anti-melanogenic effects of hydroxyectoine via mitf inhibition by jnk, p38, and akt pathways in b16f10 melanoma cells. Natural Product. Commun. 14, 1934578X19858523. doi: 10.1177/1934578X19858523
Dai P., Huan P., Wang H., Lu X., Liu B. (2015). Characterization of a long-chain fatty acid-CoA ligase 1 gene and association between its SNPs and growth traits in the clam Meretrix meretrix. Gene 566, 194–200. doi: 10.1016/j.gene.2015.04.047
Ge H.-L., Tan K., Shi L.-L., Sun R., Wang W.-M., Li Y.-H. (2020). Comparison of effects of dsRNA and siRNA RNA interference on insulin-like androgenic gland gene (IAG) in red swamp crayfish Procambarus clarkii. Gene 752, 144783. doi: 10.1016/j.gene.2020.144783
Hearing V. J., Jiménez M. (1987). Mammalian tyrosinase-the critical regulatory control point in melanocyte pigmentation. Int. J. Biochem. 19, 1141–1147. doi: 10.1016/0020-711X(87)90095-4
Hoekstra R., Fekkes D., Pepplinkhuizen L., Loonen A. J. M., Tuinier S., Verhoeven W. M. A. (2006). Nitric oxide and neopterin in bipolar affective disorder. Neuropsychobiology 54, 75–81. doi: 10.1159/000096042
Ito S., Wakamatsu K. (2003). Quantitative analysis of eumelanin and pheomelanin in humans, mice, and other animals: a comparative review. Pigment. Cell Res. 16, 523–531. doi: 10.1034/j.1600-0749.2003.00072.x
Jalmi S. K., Sinha A. K. (2015). ROS mediated MAPK signaling in abiotic and biotic stress-striking similarities and differences. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00769
Jimenez-Cervantes C., Solano F., Kobayashi T., Urabe K., Hearing V. J., Lozano J. A., et al. (1994). A new enzymatic function in the melanogenic pathway. The 5, 6-dihydroxyindole-2-carboxylic acid oxidase activity of tyrosinase-related protein-1 (TRP1). J. Biol. Chem. 269, 17993–18000. doi: 10.1007/BF00006249
Johnson S. L., Nguyen A. N., Lister J. A. (2011). Mitfa is required at multiple stages of melanocyte differentiation but not to establish the melanocyte stem cell. Dev. Biol. 350, 405–413. doi: 10.1016/j.ydbio.2010.12.004
Kang S. J., Choi B. R., Lee E. K., Kim S. H., Yi H. Y., Park H. R., et al. (2015). Inhibitory effect of dried pomegranate concentration powder on melanogenesis in B16F10 melanoma cells; involvement of p38 and PKA signaling pathways. Int. J. Mol. Sci. 16, 24219–24242. doi: 10.3390/ijms161024219
Kim D. S., Kim S. Y., Chung J. H., Kim K. H., Eun H. C., Park K. C. (2002). Delayed ERK activation by ceramide reduces melanin synthesis in human melanocytes. Cell. Signalling. 14, 779–785. doi: 10.1016/S0898-6568(02)00024-4
Kim D.-S., Park S.-H., Kwon S.-B., Na J.-I., Huh C.-H., Park K.-C. (2007). Additive effects of heat and p38 mapk inhibitor treatment on melanin synthesis. Arch. Pharmacal. Res. 30, 581–586. doi: 10.1007/BF02977652
Lerebours A., Chapman E. C., Sweet M. J., Heupel M. R., Rotchell J. M. (2016). Molecular changes in skin pigmented lesions of the coral trout Plectropomus leopardus. Mar. Environ. Res. 120, 130–135. doi: 10.1016/j.marenvres.2016.07.009
Liu J. H., Wen S., Luo C., Zhang Y. Q., Tao M., Wang D. W., et al. (2015). Involvement of the mitfa gene in the development of pigment cell in Japanese ornamental (Koi) carp (Cyprinus carpio L.). Genet. Mol. Res. 14, 2775–2784. doi: 10.4238/2015.March.31.7
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo z., Jiang m., Zhou y., Wen s., Wang m., Liu s., et al. (2013). The expression of erk1/2 in tissues and embryos of the Triploid fish. Life Sci. Res. 17, 11–13 + 42. doi: 10.16605/j.cnki.1007-7847.2013.01.002
Molina D. M., Grewal S., Bardwell L. (2005). Characterization of an ERK-binding domain in microphthalmia-associated transcription factor and differential inhibition of ERK2-mediated substrate phosphorylation. J. Biol. Chem. 280, 42051–42060. doi: 10.1074/jbc.M510590200
Nassar K. W., Tan A. C. (2020). The mutational landscape of mucosal melanoma. Semin. Cancer Biol. 61, 139–148. doi: 10.1016/j.semcancer.2019.09.013
Peng H.-Y., Lin C.-C., Wang H.-Y., Shih Y., Chou S.-T. (2014). The melanogenesis alteration effects of Achillea millefolium L. essential oil and linalyl acetate: involvement of oxidative stress and the JNK and ERK signaling pathways in melanoma cells. PloS One 9, e95186. doi: 10.1371/journal.pone.0095186
Schmitt M., Sinnberg T., Nalpas N. C., Maass A., Schittek B., Macek B. (2019). Quantitative proteomics links the intermediate filament nestin to resistance to targeted BRAF inhibition in melanoma cells. Mol. Cell. Proteomics 18, 1096–1109. doi: 10.1074/mcp.RA119.001302
Song X., Xu A., Pan W., Wallin B., Kivlin R., Lu S., et al. (2008). Minocycline protects melanocytes against H2O2-inducedcell death via JNK and p38 MAPK pathways. Int. J. Mol. Med. 22, 9–16. doi: 10.3892/ijmm.22.1.9
Steunou A.-L., Ducoux-Petit M., Lazar I., Monsarrat B., Erard M., Muller C., et al. (2013). Identification of the hypoxia-inducible factor 2α Nuclear interactome in melanoma cells reveals master proteins involved in melanoma development. Mol. Cell. Proteomics 12, 736–748. doi: 10.1074/mcp.M112.020727
Sugianti Y., Mujiyanto M. (2016). Biodiversitas hayati ikan karang di perairan Taman Nasional Karimunjawa Jepara. BAWAL. Widya. Riset. Perikanan. Tangkap. 5, 23–31. doi: 10.15578/bawal.5.1.2013.23-31
Sun Z., Xia S., Feng S., Zhang Z., Rahman M. M., Rajkumar M., et al. (2015). Effects of water temperature on survival, growth, digestive enzyme activities, and body composition of the leopard coral grouper Plectropomus leopardus. Fisheries. ence. 1, 107–112. doi: 10.1007/s12562-014-0832-9
Sweet M., Kirkham N., Bendall M., Currey L., Bythell J., Heupel M. (2012). Evidence of melanoma in wild marine fish populations. 7(8):e41989. doi: 10.1371/journal.pone.0041989
Tan K., Li Y., Zhou M., Wang W. (2020). siRNA knockdown of MrIR induces sex reversal in Macrobrachium rosenbergii. Aquaculture 523, 735172. doi: 10.1016/j.aquaculture.2020.735172
Tsukamoto K., Jackson I. J., Urabe K., Montague P. M., Hearing V. J. (1992). A second tyrosinase-related protein, TRP-2, is a melanogenic enzyme termed DOPAchrome tautomerase. EMBO J. 11, 519–526. doi: 10.1002/j.1460-2075.1992.tb05082.x
Ventura T., Manor R., Aflalo E. D., Weil S., Raviv S., Glazer L., et al. (2009). Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 150, 1278–1286. Available at: https://doi.org/2020071612270331300.
von Koschembahr A. M., Swope V. B., Starner R. J., Abdel-Malek Z. A. (2015). Endothelin-1 protects human melanocytes from UV-induced DNA damage by activating JNK and p38 signalling pathways. Exp. Dermatol. 24, 269–274. doi: 10.1111/exd.12638
Wang R., Qi Z., Zhang X., Wang F., Wang Y., Fu S., et al. (2011). Biological characteristics and Artificial Cultivation Techniques of Plectropomus leopardus. China Fisheries. 2011(4), 33–34. CNKI: SUN : SICA.0.2011-04-020.. doi: 10.3969/j.issn.1002-6681.2011.04.017
Wang T., Wen H., Yang J., Yin H., Liao J., He Y., et al. (2017). Research progress on the relationship between osteoarthritis and MAPK signaling pathway. Chin. J. Ethnomed. Ethnopharmacy. 26, 27–29 + 33. CNKI: SUN : MZMJ.0.2017-19-010. doi: CNKI:SUN:MZMJ.0.2017-19-010
Wang J., Wu M., Wang B., Han Z. (2013). Comparison of the RNA interference effects triggered by dsRNA and siRNA in Tribolium castaneum. Pest Manage. Sci. 69, 781–786. doi: 10.1002/ps.3432
Wang L., Zhu W., Dong Z., Song F., Dong J., Fu J. (2018). Comparative microRNA-seq analysis depicts candidate miRNAs involved in skin color differentiation in red tilapia. Int. J. Mol. Sci. 19, 1209. doi: 10.3390/ijms19041209
Wellbrock C., Arozarena I. (2015). Microphthalmia-associated transcription factor in melanoma development and MAP-kinase pathway targeted therapy. Pigment. Cell Melanoma. Res. 28, 390–406. doi: 10.1111/pcmr.12370
Wen X., Yang M., Zhou K., Huang J., Fan X., Zhang W., et al. (2022). Transcriptomic and proteomic analyses reveal the common and unique pathway (s) underlying different skin colors of leopard coral grouper (Plectropomus leopardus). J. Proteomics 266, 104671. doi: 10.1016/J.JPROT.2022.104671
Wu M., Hemesath T. J., Takemoto C. M., Horstmann M. A., Wells A. G., Price E. R., et al. (2000). c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 14, 301–312. doi: 10.1101/gad.14.3.301
Wu S., Huang J., Li Y., Zhang Q., Pan Y., Wang X. (2021). Cloning and expression analysis of body colour-related gene mitfa in Rainbow Trout (Oncorhynchus mykiss). J. Agric. Biotechnol. 29, 753–763. doi: 10.3969/j.issn.1674-7968.2021.04.013
Xin Q., Li L., Miao Z., Zhang L., Liu Y., Zhu Z., et al. (2018). Expression of MITF gene in Putian Black Duck (Anas anas domesticus) and its association with melanin deposition. J. Agric. Biotechnol. 26, 1928–1937. doi: 10.3969/j.issn.1674-7968.2018.11.012
Yang M. (2020). Molecular cloning and characterization of a cDNA encoding extracellular signal-regulated kinase (ERK) from blood clam Tegillarca granosa (Xiamen: Third Institute Of Oceanography,Ministry of Natural Resources).
Yang N., Mu L. I. N., Zhao B., Wang M., Hu S., Zhao B. I. N., et al. (2018). RNAi-mediated SLC7A11 knockdown inhibits melanogenesis-related genes expression in rabbit skin fibroblasts. J. Genet. 97, 463–468. doi: 10.1007/s12041-018-0945-5
Yoseda K., Yamamoto K., Asami K., Chimura M., Hashimoto K., Kosaka S. (2008). Influence of light intensity on feeding, growth, and early survival of leopard coral grouper (Plectropomus leopardus) larvae under mass-scale rearing conditions. Aquaculture 279, 55–62. doi: 10.1016/j.aquaculture.2008.04.002
Zhang Y., Liu J., Fu W., Xu W., Zhang H., Chen S., et al. (2017). Comparative Transcriptome and DNA methylation analyses of the molecular mechanisms underlying skin color variations in Crucian carp (Carassius carassius L.). BMC Genet. 18, 1–12. doi: 10.1186/s12863-017-0564-9
Zhang H., Xu C., Song X., Zhou N., Zhang L., Xing X., et al. (2016). Progress in melanocyte regulation of microphthalmia-associated transcription factors. Anim. Husbandry. Vet. Med. 48, 113–117. CNKI: SUN : XMYS.0.2016-08-030. doi: CNKI:SUN:XMYS.0.2016-08-030
Zhao N., Zhou B., Li Y., Zhang J., Ma J., Yu X. (2016). Effects of light color on growth, skin color, and physiological indices of juvenile Plectropomus leopardus in a recirculating aquaculture system. J. Fishery. Sci. China 23, 976–984. doi: 10.3724/SP.J.1118.2016.15438
Zheng S., Zheping H., Zhang D. D., Edward M. R. (2009). Phosphorylation of nrf2 at multiple sites by MAP kinases has a limited contribution in modulating the nrf2-dependent antioxidant response. PloS One 4, e6588–. doi: 10.1371/journal.pone.0006588
Zhou S., Zeng H., Huang J., Lei L., Tong X., Li S., et al. (2021). Epigenetic regulation of melanogenesis. Ageing Res. Rev. 69, 101349. doi: 10.1016/j.arr.2021.101349
Keywords: Plectropomus leopardus, ERK1/2, skin colour, MITF, melanogenesis
Citation: Yang M, Huang J, Zheng D, Tang H, Liu J, Luo J and Wen X (2024) ERK1/2 regulates melanin synthesis in fish: a case study on a colourful variety, leopard coral grouper (Plectropomus leopardus). Front. Mar. Sci. 11:1441589. doi: 10.3389/fmars.2024.1441589
Received: 31 May 2024; Accepted: 28 August 2024;
Published: 26 September 2024.
Edited by:
Ida Grong Aursand, SINTEF Ocean, NorwayReviewed by:
Liang Guo, Hunan Normal University, ChinaCopyright © 2024 Yang, Huang, Zheng, Tang, Liu, Luo and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Wen, d2VueGluZmlzaEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.