
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 21 May 2024
Sec. Marine Pollution
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1409399
This article is part of the Research Topic Microplastics and Microorganisms in the Environment, Volume II View all 6 articles
Expanded polystyrene (EPS), commonly used as floating material in mariculture, could be a significant source of coastal PS waste. Prolonged floating in seawater leads to the formation of dense biofilms, which could disperse into the oceanic environment and spread to water bodies even the seabed as EPS deteriorates into microplastics. To understand the bacterial community structure in EPS surface biofilms, this study employed high-throughput sequencing technology to analyze the biofilms on EPS foams used in offshore mariculture aside Xiamen. Analysis of 24 biofilm samples from different seasons revealed significant differences in bacterial community structures. However, we have identified the core groups of the EPS biofilm, including Flavobacteriaceae, Rhodobacteraceae, Halomonadaceae, Exiguobacteriaceae, Vibrionaceae, and Pseudoalteromonadaceae. The Halomonadaceae and Exiguobacteriaceae are particularly attracted to the subtropical EPS material. In addition, groups with potential of degrading PS were identified, such as Alcanivorax, Pseudomonas, Acinetobacter, Exiguobacterium, Brevundimonas, and Bacillus, as well as the potential fish pathogen Acinetobacter johnsonii. The microbial biofilms formed on floating EPS demonstrate high diversity and are strongly influenced by environmental conditions. This study contributes to a deeper understanding of the interactions of bacteria in the biofilm with widely used and dispersed floating materials in the oceans.
EPS foam is widely applied in various aspects of daily life due to its low cost, superior and enduring thermal insulation, unique cushioning and shock resistance, as well as its water resistance and anti-aging properties (Derraik, 2002; Andrady and Neal, 2009; Petrella et al., 2020). In 2022, the global annual production of PS and EPS reached 20,815,600 tons (Plastics Europe, 2023). The use of EPS floating rafts in coastal aquaculture, which are often destroyed by storm or discarded after use, has led to an overflow of foam waste in various marine environments, from coastal areas to the open ocean (Mendoza and Jones, 2015; Bond et al., 2018; Turner, 2020; Isobe and Iwasaki, 2022). Meanwhile, EPS is a carrier for potentially pathogenic human bacteria (PHPB), antibiotic-resistant bacteria (ARB) (Naudet et al., 2023), and other pollutants, including toxic compounds such as polychlorinated biphenyls (PCBs), polybrominated diphenyl ethers (PBDEs), polycyclic aromatic hydrocarbons (PAHs), and organochlorine pesticides (DDTs) (Hirai et al., 2011; Zhang et al., 2015; Wang et al., 2018), are raising new concerns, further threatening marine ecological safety and human health.
In marine environments, the initial bacterial communities forming on the surfaces of plastics are influenced by the properties of the plastics such as hydrophobicity, crystallinity, crystal structure, and roughness (Pompilio et al., 2008) and are primarily dominated by γ-Proteobacteria, α-Proteobacteria, and Bacteroidetes that swiftly colonize the plastic surfaces (Lee et al., 2008; Oberbeckmann et al., 2015). However, as the biofilm matures, the influence of these physical characteristics on the bacterial community composition gradually diminishes (Dussud et al., 2018). As EPS waste remains afloat in seawater over time, diverse and active biofilms are formed on its surface. Our recent studies suggest that biofilm microbes may be involved in the degradation and fragmentation of PS plastics (Lv et al., 2023), secreting a variety of enzymes that participate in the process of plastic biodegradation (Howard and McCarthy, 2023). This indicates that the formation of marine plastic biofilms can have an impact on the fragmentation and biodegradation of marine plastics, and potentially influencing global biogeochemical cycles.
There is currently limited knowledge about the composition of bacterial communities in the biofilms of marine floating EPS waste in subtropical mariculture areas, although it has been reported that environmental factors such as temperature, salinity, and light can affect the composition and function of the microbial communities in the biofilms (Zhi et al., 2023). Subtropical estuaries, which are rich in marine biodiversity and ideal marine farms, offer valuable insights into the diversity composition of biofilms on floating plastic wastes to understand their environmental fate and microbial processes involved. In 2022, we described the bacterial diversity in biofilms formed on the surface of EPS, and it was found that seven bacterial families in the community, including Sphingomonadaceae, Rhodanobacteraceae, Rhizobiaceae, Dermacoccaceae, Rhodocyclaceae, Hyphomicrobiaceae, and Methyloligellaceae, were possibly involved in PS degradation (Liu et al., 2023). In order to further verify their degradation potential, nine genera of degrading bacteria were isolated and confirmed to degrade PS, including Novsphingobium, Gordonia, Stappia, Mesobacillus, Alcanivorax, Flexivirga, Cytobacillus, Thioclava and Thalassospira (Liu et al., 2023), indicating the presence of a large number of EPS-biodegrading bacteria in the subtropical coastal region. Additionally, the subtropics have unique climatic conditions, such as intense ultraviolet radiation, and high marine biodiversity, as well as human activities like mariculture; therefore, how they interact with biological processes to affect the distribution, fragmentation, and degradation of EPS, deserve attention.
Xiamen is characterized by a typical subtropical marine climate and its coastal area is influenced by the input from the Jiulong River estuary and human activities. The Xiamen-Kinmen Sea area has always been a hanging culture area for aquaculture species like oysters, with extensive use of plastic floating materials (Figure 1). To understand the bacterial biodiversity of biofilms on EPS plastic, we collected the pieces plastic foams that had washed up on intertidal zones from April to October 2022 and analyzed the plastisphere community composition to evaluate the role of microbes in biodegradation of the plastic waste, which may influence the environmental fate in site or afloat with current.
In this study, EPS foam samples stranded on the beach were collected in April, May, July, and October of 2022. Large pieces of plastic exhibiting visible biofilms were selected (Figure 2), cut into small pieces (approximately 1.5cm × 2cm × 0.5cm), and transferred to 50ml centrifuge tubes. Sterile seawater was added to submerge the foam pieces, and the tubes were then shaken gently for 45 minutes using Vortex-Genie 2. The samples were subsequently centrifuged at 4°C and 7000 r/min for 10 minutes, with the pellets retained for subsequent sequencing experiments. 24 valid samples collected from April to October are described in detail as follows: April (11 samples): Various plastic surface features (pale-yellow a1, golden yellow sponge-like a2, dried film-like a3, pit accumulated with algae and sand a5, foam debris within pits a6, dark flocculent a7, dried algae a8, moist dark a9, sticky bacterial-like a10, orange-yellow a11, and grey adhesions a12); May (3 samples): Plastic surface and worm burrows (sandworm burrows m1-m2 and surface pits m3); July (3 samples): Surface dry, with eroded pits (dried eroded pits j1, dry black flocculent adhesions j2, and red EPS pieces j3); October (7 samples): diversified sampling including green attachments near the surface to worm burrows in the central area (green adhesions near the surface oc1, sandworm burrows near the center oc2, yellow adhesions oc3, surface sandworm burrows oc4, as well as surface features including deep red sunken adhesions oc5, smaller black spot adhesions oc6, and larger black spot adhesions oc7).
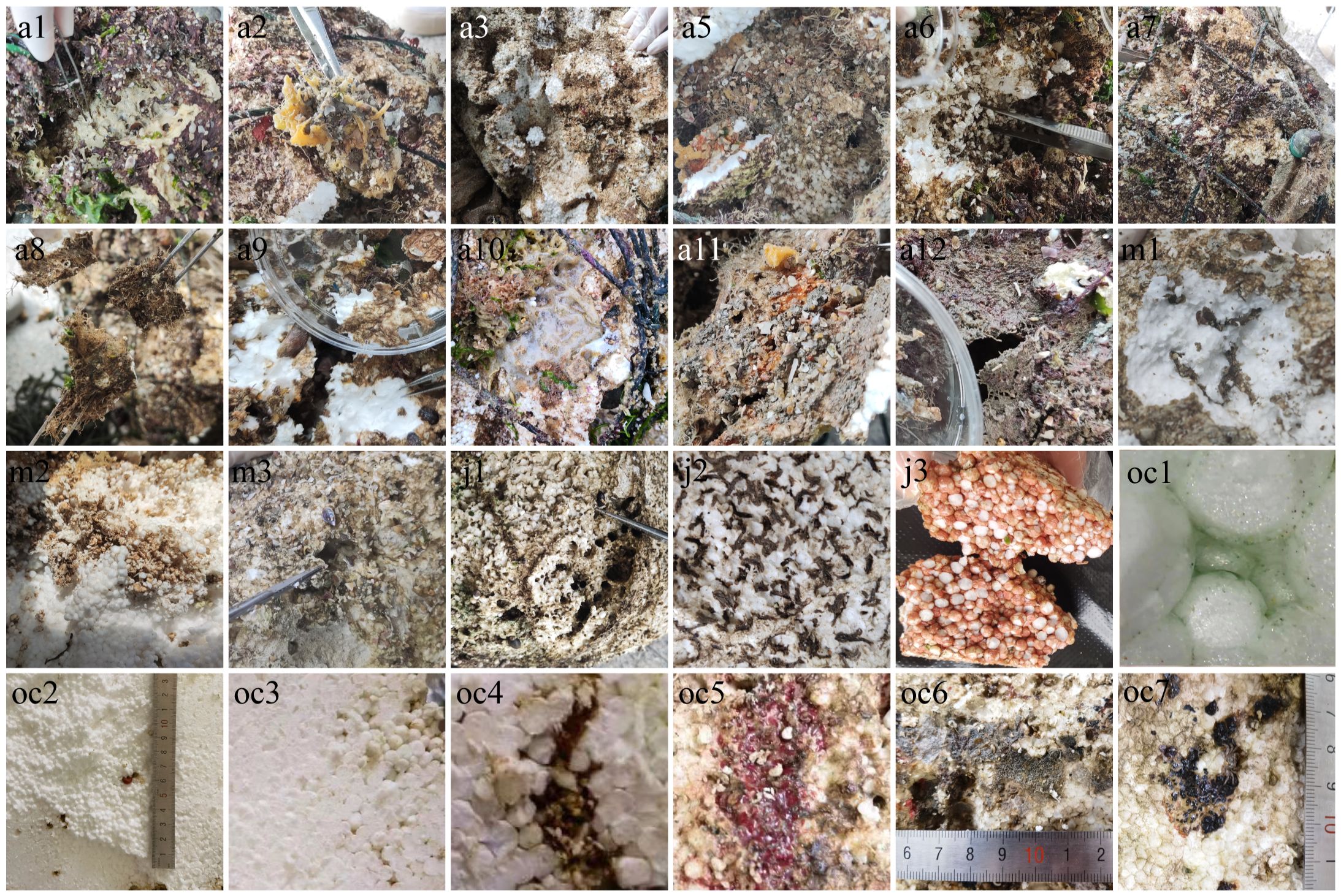
Figure 2 Sample live image. “a” stands for April, from a1 to a3 and a5 to a12, with a total of 11 samples. “m” stands for May, from m1 to m3, a total of three. “j” stands for July, from j1 to j3, a total of three. “oc” stands for October, from oc1 to oc7, a total of seven.
Commercial Power Soil DNA Isolation Kits (MoBio, USA) were used to extract DNA from processed samples according to the manufacturer’s instructions. The concentration and purity of the DNA extractions were checked by 1% agarose gel electrophoresis and a spectrophotometer (Nanodrop, ND-2000, USA), and stored at -80°C before amplification. We performed full-length 16S sequencing on samples collected before May and V3-V4 variable region sequencing on samples collected after July. The reason is that we found both sequencing methods provided useful information for comparing species abundance and diversity levels during May and July.
The extracted DNA served as a template for PCR amplification of the full-length 16S rRNA gene using barcoded primers 27F (5’-AGRGTTYGATYMTGGCTCAG-3’) and 1492R (5’-RGYTACCTTGTTACGACTT-3’) (Weisburg et al., 1991) for the April and May samples. The PCR reaction system comprised 4 μL of 5×FastPfu buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of forward primer (5 μM), 0.8 μL of reverse primer (5 μM), 0.4 μL of FastPfu polymerase, 0.2 μL of BSA, 10 ng of template DNA, and was adjusted to 20 μL with distilled water. Each sample had three replicates. The amplification program was as follows: an initial denaturation at 95°C for 3 min, followed by 27 cycles of denaturation at 95°C for 30s, annealing at 60°C for 30s, extension at 72°C for 30s, and a final extension at 72°C for 10 min, then held at 4°C (PCR instrument: T100 Thermal Cycler PCR, USA). The PCR products were checked by 2% agarose gel electrophoresis, purified with magnetic beads, and quantified using Qubit 4.0 (Thermo Fisher Scientific, USA). SMRTbell prep kit 3.0 was used for the library construction, which includes DNA damage repair, end repair, and adapter ligation. Sequencing was performed using the PacBio Sequel II System. HiFi reads for subsequent data analysis were generated from the sequenced subreads using SMRT-Link v11.0’s CCS mode.
The extracted DNA served as a template for PCR amplification of the V3-V4 variable regions of the 16S rRNA gene using barcoded primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) (Liu et al., 2016) for the July and October samples. The PCR reaction system comprised 4 μL of 5×TransStart FastPfu buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of forward primer (5 μM), 0.8 μL of reverse primer (5 μM), 0.4 μL of Trans Start FastPfu DNA polymerase, 10 ng of template DNA, and adjusted to 20 μL with distilled water. The amplification program was an initial denaturation at 95°C for 3 minutes, followed by 27 cycles of denaturation at 95°C for 30s, annealing at 55°C for 30s, extension at 72°C for 30s, and a final extension at 72°C for 10 minutes, then held at 4°C (PCR instrument: T100 Thermal Cycler, BIO-RAD, USA). The PCR products were purified using a PCR Clean-Up Kit and quantified using Qubit 4.0 (Thermo Fisher Scientific, USA). NEXTFLEX Rapid DNA-Seq Kit was used to construct the library, which includes steps like adapter ligation, bead-based size selection to remove adapter dimers, PCR amplification for library enrichment, and bead-based purification to recover the final library. Sequencing was conducted on the Illumina Miseq platform.
For the April and May samples, data for each sample was differentiated based on Barcode sequences, length-filtered, and orientation-corrected to retain bacterial sequences of 1000-1800 bp. Using UPARSE (Edgar, 2013) (http://drive5.com/uparse/, version 11), sequences were clustered into OTUs at 97% similarity and chimeras were removed. To minimize the effect of sequencing depth on subsequent alpha and beta diversity analysis, all sample sequences were normalized to 2287, with the average sequence coverage (Good’s coverage) 96.64%. OTUs were taxonomically annotated using the RDP classifier (Wang et al., 2007) (http://rdp.cme.msu.edu/, version 11.5) against the Silva 16S rRNA gene database (v138) with a confidence threshold of 70%, and the community composition of each sample was tallied at different taxonomic levels.
For the July and October samples, raw paired-end sequencing sequences were quality-controlled using fastp (Chen et al., 2018) (https://github.com/OpenGene/fastp, version 0.19.6) and merged using FLASH (Magoč and Salzberg, 2011) (http://www.cbcb.umd.edu/software/flash, version 1.2.11). Post-quality control and assembly, optimized sequences were denoised using the DADA2 plugin in the Qiime2 workflow. Sequences after DADA2 denoising are known as ASVs (Amplicon Sequence Variants). Similar to the April and May samples, sequences were normalized to 2287 to mitigate sequencing depth effects on further diversity analyses, reaching an average sequence coverage of 96.82%. The blast classifier in Qiime2 was used to perform taxonomic analysis of ASVs based on the Silva 16S rRNA gene database (v138).
The Alpha diversity indices such as Chao 1 and Shannon index were calculated using mothur (Schloss et al., 2009) software (http://www.mothur.org/wiki/Calculators), and rarefaction curves were also constructed using R language tools. Further, Principal Coordinates Analysis (PCoA) and Non-metric Multidimensional Scaling (NMDS) analysis based on the Bray-Curtis algorithm, and Hierarchical clustering analysis using the average linkage method was performed on all samples to examine the similarity of microbial community structures. ANOSIM was used to test if the differences in microbial community structures between sample groups were significant. Additionally, community bar charts, pie charts, Venn diagrams, and heatmaps were plotted using R language (version 3.3.1).
The sequences of this study were submitted to the China National Center for Bioinformation (CNCB) Genomics Database (GSA), with the database accession numbers CRA013490 and CRA013487.
Sobs rarefaction curves were used to assess the sequencing data. Although the sobs curves were not parallel to the x-axis, they began to plateau (Supplementary Figure 1), indicating that the amount of sequencing data was reasonable and that increasing the data volume would only yield a few additional species.
The richness index (Chao/Ace), diversity index (Shannon/Simpson), and sequencing depth index (Good’s coverage) of the samples are shown in Table 1. They demonstrated the changes in bacterial community diversity within the EPS biofilms across different months. Using the Simpson and Chao1 indices as measures of diversity, we found that the average Simpson index values for the samples from April, May, and October were 0.04, 0.05, and 0.04 respectively, indicating no significant differences in bacterial community diversity between these months. Similarly, the average Chao1 index values also showed consistency, being 320, 351, and 350 for the same months, further confirming the similarity in the bacterial communities in the EPS of these three periods. In contrast, the July samples had a significantly higher average Simpson index value (0.13) than those of April, May, and October, while their average Chao1 index value (184) was lower than those of the aforementioned months, suggesting a lower richness of bacterial communities within the biofilms in July. These results reflect the trends in the variability of the bacterial communities in the EPS biofilms under different environmental conditions throughout the months. The average coverage of all samples in the four groups was over 94.62%, indicating that the sequencing depth of this study covered the microbial composition in the environmental samples sufficiently. Specially, the top three samples with the highest bacterial richness and diversity indices are all sandworm burrow samples (oc4, m1, and m2). This indicates that the surface samples have lower bacterial richness and diversity compared to the sandworm burrow samples.
The microbial community composition of EPS samples collected for four months was analyzed for β-diversity to explore the similarities and differences. The results of Hierarchical clustering analysis (Figure 3A) showed that the sandworm burrow samples (m1, m2, oc2 and oc4) in May and October clustered together, while samples from April and July separated respectively, indicating notable differences in bacterial community composition in EPS biofilms among different months, while microbial compositions of sandworm burrow samples were similar across months. Similar results were observed in PCoA (Figure 3B) and NMDS analyses (R=0.9065, P=0.001) (Figure 3C) based on Bray-Curtis, which reflect the degree of differences among samples through the distances between points. The PCoA and NMDS plots revealed no overlap between April samples and others, with close distances within April samples, while July samples were further apart from samples of other months. The samples from October and May overlapped, with sandworm burrow samples (oc2 and oc4) showing the closest resemblance to May samples. ANOSIM results (Figure 3D) confirmed significant differences in bacterial community structures among different months (R=0.9065, P=0.001), with the largest within-group variation observed in May and the smallest in April. A Venn diagram (Figure 3E) showed nearly half of the genera were shared in the sandworm burrow samples of May and October (133 genera, accounting for 47.67%), suggesting that sampling time had a minimal impact on the microbial community composition of sandworm burrows.

Figure 3 Beta diversity (A–C) and Analysis of similarities (D) at genus level of bacteria at EPS biofilms of April, May, July and October, and Venn diagram in genus level of clamworm channel samples (E). (A) The length between branches represents the distance between samples, and different colors represent different parts. (B, C) X-axis and Y-axis represent the two selected spindles, and percentage represents the value of the explanation degree of the spindles to the difference in the composition of bacteria in different parts. The scale of X axis and Y axis is relative distance, which has no practical significance. Each point represents a sample, and points of the same color come from the same group. (D) The corresponding box Between represents the distance value of the difference between the groups, and the remaining boxes represent the difference distance value within the group; The Y-axis scale indicates the magnitude of the distance value.
Bacterial taxa from all samples were identified, belonging to 44 phyla, 125 classes. Bacterial phyla with an abundance greater than 5% in all samples are illustrated in Figure 4A, mainly including Proteobacteria, Bacteroidota, Actinobacteriota, Firmicutes, and Cyanobacteria, which are commonly reported as dominant bacterial groups colonizing plastic surfaces. Within the communities, the phylum Proteobacteria attained a notable average abundance of 46.02%, emerging as a significantly dominant group with respective average relative abundances of 65.11%, 50.27%, 44.49%, and 38.53% during the months of May, October, July, and April. Following closely was the phylum Bacteroidota, which on average occupied 18.84% of the total sample abundances. Notably, its presence reached a peak frequency of 32.95% in the April sample set, markedly surpassing other months (ranging from 4.45% to 8.72%), thereby revealing its distinctive distribution pattern. In comparison with other leading phyla, Actinobacteriota sustained an average relative abundance of 7.09% across all samples. Data points in July (12.66%), April (8.17%), October (5.48%), and May (1.31%) further disclosed the temporal relative distribution trends of this taxa. Moreover, Firmicutes exhibited a substantial relative abundance of 26.22% in July samples, and Cyanobacteria accentuated their presence with a proportion of 15.49% in the October samples. The relative abundances of both phyla plummeted to nearly zero in other monthly samples, highlighting their variable abundances across different time frames. Other phyla had lower relative abundance across samples.
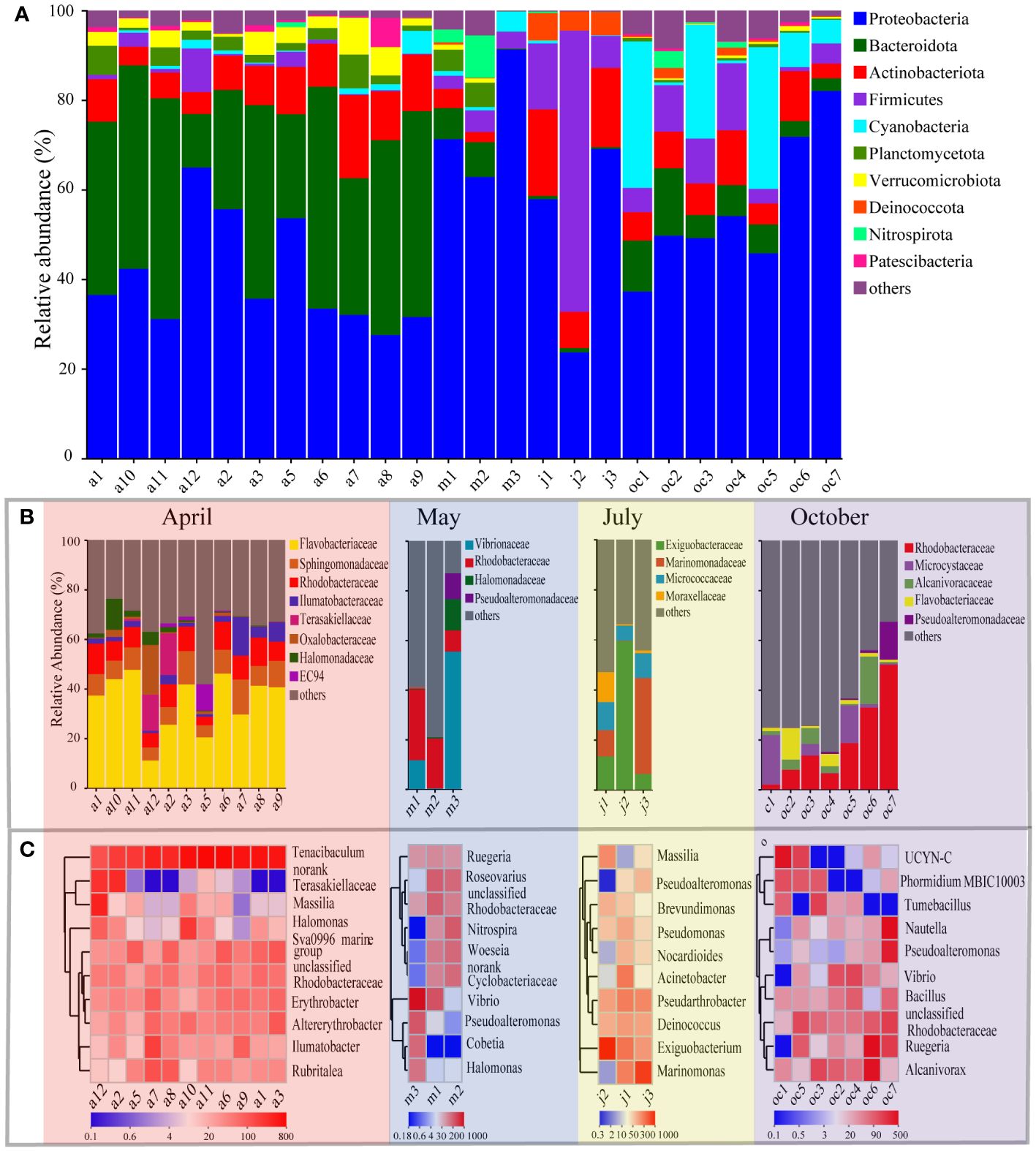
Figure 4 The dominant species of bacteria in EPS biofilms at phylum (A), family (B) and genus (C) level. (A, B) The horizontal coordinate is the sample name, the vertical coordinate is the proportion of species in the sample, the different colors of the column represent different species, and the length of the column represents the size of the proportion of the species. (C) The horizontal coordinate is the sample name, and the vertical coordinate is the species name. The variation of the abundance of different species in the sample is shown through the color gradient of the color block. The value represented by the color gradient is shown on the downside of the figure.
At the family level, bacterial community composition was analyzed, focusing on taxa that exhibited a relative abundance of more than 10% in at least one sample. As shown in Figure 4B, Flavobacteriaceae, Sphingomonadaceae, Rhodobacteraceae, Ilumatobacteraceae, Terasakiellaceae, Oxalobacteraceae, Halomonadaceae, and EC94 were the dominant families in April. Their respective average relative abundances were 35.03%, 8.91%, 8.78%, 3.95%, 2.96%, 2.49%, 2.36%, and 1.29%, with Flavobacteriaceae being notably dominant in the April samples. In May (Figure 4B), the community structure shifted, with the principal families being Vibrionaceae, Halomonadaceae, Pseudoalteromonadaceae, and Rhodobacteraceae, having average relative abundances of 22.47%, 4.44%, 1.76%, and 1.90% respectively. In this instance, Vibrionaceae emerged as the dominant family in the May sample group. The July samples (Figure 4B) showed a prevalence of Exiguobacteraceae, Marinomonadaceae, Micrococcaceae, and Moraxellaceae, with respective average relative abundances of 26.39%, 16.35%, 9.00%, and 4.57%. Throughout this month, Exiguobacteraceae stood out as the most widespread family. In the October EPS samples (Figure 4B), the bacterial community was primarily composed of Rhodobacteraceae, Alcanivoracaceae, Microcystaceae, Pseudoalteromonadaceae, and Flavobacteriaceae, with respective average relative abundances of 18.81%, 5.04%, 5.95%, 2.57%, and 3.40%. Rhodobacteraceae were identified as the leading family. Notably, Flavobacteriaceae, Rhodobacteraceae, Halomonadaceae, and Pseudoalteromonadaceae were consistently present in the sample groups across several months. Specifically, Flavobacteriaceae were detected in the April and October samples; Rhodobacteraceae recurred in the April, May, and October sample groups; Halomonadaceae appeared in both April and May; Pseudoalteromonadaceae were observed in the May and October samples. We classified these recurring groups as core groups in this study, signifying their importance and stability.
At the genus level, the dominant members in each mouth group were indicated by the top 10 abundant taxa were shown in Figure 4C. Different seasons possess various dominant members. As the Beta diversity described in Figure 3. (A, B, C), the bacterial composition was similar within a sampling group in general. For example, in April, all 11 samples contained bacterial communities with the dominant genera in common, including Tenacibaculum, Sva0996 marine group, unclassified Rhodobacteraceae, Erythrobacter, Altererythrobacter, Ilumatobacter, and Ruegeria, with exceptions of unclassified Terasakiellaceae and Massilia that dominated in sample a12, and Halomonas in a10 in addition to the most predominant member Tenacibaculum that dominated nearly all the 11 samples of April. However, Tenacibaculum was not predominant in other samples of May to October. In May, the samples showed unique bacterial commotions compared to April, but much resembled to samples in October. The dominant genera at the genus level in two samples of May (m1 and m2) were Roseovarius, an unclassified Rhodobacteraceae, an unclassified Cyclobacteriaceae, Woeseia, and Nitrospira, while the genera in sample m3 were different, which absolutely dominated by Vibrio, Pseudoalteromonas, Cobetia, and Halomonas. Among them, Vibrio shared as the most predominant member of the two samples (m1 and m3), while Rugeria occurred in all May samples. In addition, the unclassified Rhodobacteraceae occurred commonly as in April and Oct. In July samples the dominant genera were noted for Marinomonas, Exiguobacterium, Deniococcus and Pseudarthrobacter in addition to Massilia, Pseudoalteromonas, Acinetobacter. The first two alternatively dominated the July bacterial communities, while Deniococcus and Pseudarthrobacter occurred in all July samples as a sub-dominant member. Deniococcus, Exiguobacterium, and Pseudarthrobacter all belonged to Terrabacteria group. In October samples, the unclassified Rhodobacteraceae, Ruegeria, Pseudoalteromonas and Vibrio reoccurred as one of the dominant members in most samples. In addition, some unique bacteria occurred such as Alcanivorax and Bacillus occurred in all October samples. Moreover, Phormidium of Oscillatoriaceae, Tumebacillus of Alicyclobacillaceae, and Nautella of Roseobacteraceae were also present in some October samples. Additionally, a N2-fixing cyanobacterial phylotype named UCYN-C, previously detected in tropical Atlantic Ocean at a water depth (200 m) of high salinity, low temperature, and nutrients, was dominant in our two samples.
Venn diagrams and pie charts showed shared genera over four months (Figure 5), and the common colonizing taxa on the EPS surface were 47, which account for 12.08% of the total relative abundance of species of genus (Figure 5A). In the Pie chart (Figure 5B), the main taxa were Vibrio (11.6%), Exiguobacterium (10.34%), unclassified Rhodobacteraceae (8.87%), Erythrobacter (5.85%), Sva0996 marine group (5.41%), Altererythrobacter (4.48%), Pseudoalteromonas (4.29%), Ilumatobacter (4.07%), Bacillus (4.00%), and Roseovarius (3.98%), others with less than 2% abundance were combined. Utilizing the Kruskal-Wallis H test combined with the false discovery rate (FDR) method for adjusting P-values for multiple comparisons, we further assessed the significant differences among common colonizing taxa on the EPS surface across sample groups from different months (Figure 5C). Confidence intervals were calculated using the “Tukey-Kramer” method at a 95% confidence level. The differences in average relative abundance of common bacterial groups across sample groups from different months were calculated. In the July samples, the relative abundance of Exiguobacterium and Acinetobacter was significantly higher than in other months. Vibrio and Roseovarius had a notably higher relative abundance in the May samples than in other months. The Sva0996 marine group, Altererythrobacter, Ilumatobacter, and norank Rhizobiaceae were more abundant in the April samples compared to other months. Similar to the results shown in Figure 4C, unclassified Rhodobacteraceae had a significantly lower relative abundance in the July samples compared to other months. Pseudoalteromonas, Erythrobacter, Bacillus, Nitrospira, unclassified Sphingomonadaceae, Methyloceanibacter, and Pleurocapsa did not show significant differences in relative abundance among the four monthly samples.
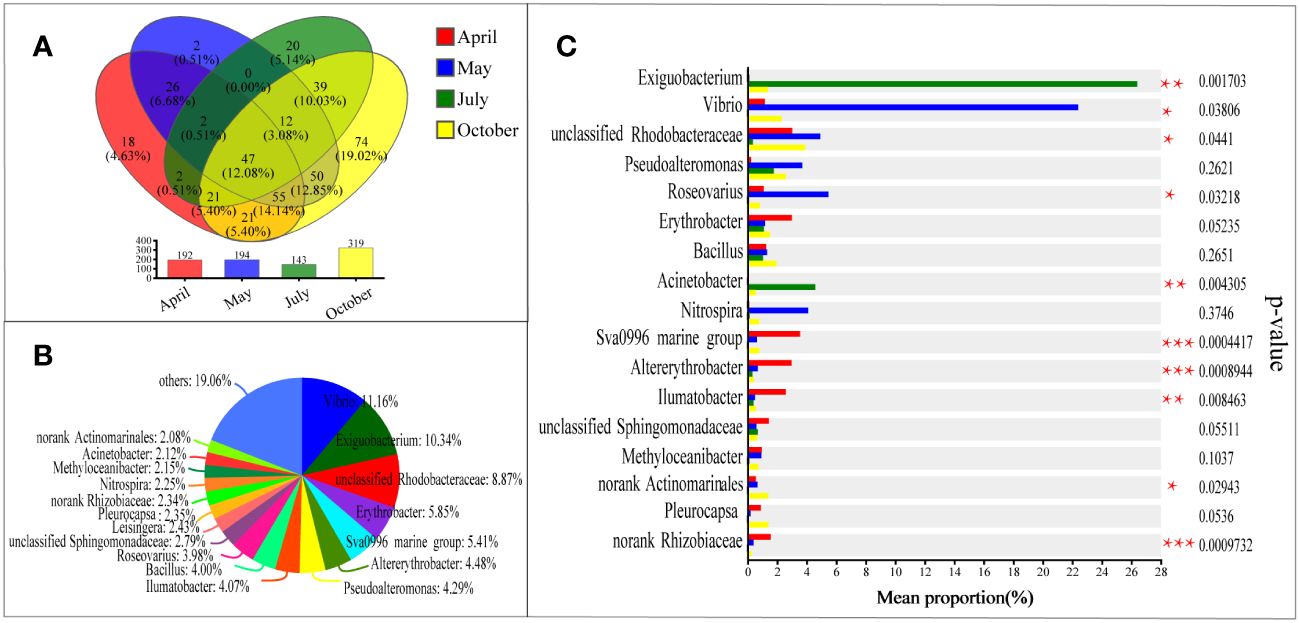
Figure 5 Venn diagram (A) of bacteria at sample of April, May, July and October, long term colonization microbial community pie plot (B) and bar plot (C) at the level of genus. (A) Different colors represent different groups, overlapping numbers represent the number of species common to multiple groups, and non-overlapping numbers represent the number of species unique to the corresponding group. (B) The distribution of common genus in different groups. Different colors represent different genus. Pie area represents the percentage of the number of genus in the total genus. (C) The difference in average relative abundance of the same species between different groups, and whether the difference is significant (P-value, asterisk means significant difference) are marked. *: P<0.05; **: P<0.01; ***: P<0.001.
Plastic debris provides a unique ecological niche for microorganisms in the ocean, demonstrating the potential to select and colonize different microbial communities (Zettler et al., 2013; De Tender et al., 2015; Bryant et al., 2016). A comparison of the community structures of environmental EPS biofilms found significant differences(P<0.001) among the four-month sample groups, especially the community structures of the July samples, which showed the most significant differences compared to others. Probably, temperature is one of the key environment factors affecting the microbial community structure of the biofilms in this study. The average temperature range for April, May, July, and October 2022 is 17-26°C, 19-25°C, 25-34°C, and 21-29°C, respectively, with the average temperature in Xiamen in July being significantly higher than other months (Xie, 2023)(Supplementary Figure 2). However, the microbial community structure of inside EPS of worm burrows, isolated from the external environment, is not easily affected by external environments, as evidenced by the stability of the microbial community structures of worm tunnel samples collected in May and October (Figure 3).
The dominant bacterial groups in the EPS biofilms of this study included Flavobacteriaceae, Rhodobacteraceae, Halomonadaceae, Exiguobacteriaceae, Vibrionaceae, and Pseudoalteromonadaceae (Figure 4B). The survey found that Flavobacteriaceae, Rhodobacteraceae, Vibrionaceae, and Pseudoalteromonadaceae also appeared as core groups on PS biofilms in temperate, subtropical, and tropical areas in other studies (Oberbeckmann et al., 2017; Frère et al., 2018; Kesy et al., 2019; Kumar et al., 2021; Tu et al., 2021; Xie et al., 2021; Zhang et al., 2022), while Halomonadaceae and Exiguobacteriaceae were particularly attracted to the subtropical EPS debris in this study (Figure 4B). Studies have shown that the presence of Rhodobacteraceae is associated with hard surface substrates for attachment, whether on PS plastic or glass surfaces, Rhodobacteraceae are the primary colonizers (Hoellein et al., 2014; Oberbeckmann et al., 2016). In this study, predominant members of Flavobacteriaceae such as Tenacibaculum and members of Pseudoalteromonadaceae such as Pseudoalteromonas have shown the ability to adhere to plastics and form biofilms (Egan et al., 2002; Nowlan et al., 2020; Hansen et al., 2021; Nowlan et al., 2021). Members of the Vibrionaceae, particularly Vibrio, are commonly found in plastic biofilm communities in marine environments (Xie et al., 2021; Zhang et al., 2022) and can form stable biofilms on plastic surfaces (Hansen et al., 2021). The formation of biofilms contributes to the biodegradation of plastics (Mor and Sivan, 2008), but no PS-degrading bacteria have been found. Halomonadaceae, the largest halophilic family, could degrade organic pollutants such as crude oil (Hassanshahian et al., 2012) and phenol (Bonfá et al., 2013), recently we found Halomonas titanicae can degrade PS and lead to microplastic and nanoplastic generation (Lv et al., 2024). Exiguobacteriaceae includes PS-degrading groups, with members such as Exiguobacterium undae DR14 and Exiguobacterium sibiricum DR11 identified as being capable of degrading PS (Chauhan et al., 2018). Consistently, a PS-degrading bacterium Exiguobacterium sp. YT2 was isolated from the terrestrial gut of the EPS-chewing, which removed PS mass by 7.4% in 60 days (Yang et al., 2015). Therefore, Halomonadaceae and Exiguobacteriaceae identified in this study may be the key species involved in PS degradation.
In addition, bacterial groups that included previously reported PS-degrading bacteria were identified as the dominant groups in this study, such as Alcanivorax, Pseudomonas, Acinetobacter, Brevundimonas, and Bacillus (Figure 4C). Bacillus is the only group whose relative abundance is less affected by sampling season (Figure 5). Bacillus is a common plastic-degrading group and can degrade various plastics, which has been mainly isolated from terrestrial soil environments (Kiatkamjornwong et al., 1999; Galgali et al., 2002; Oikawa et al., 2003; Atiq et al., 2010; Asmita et al., 2015; Mohan et al., 2016). Recently, Bacillus cereus isolated from the mangrove of the Malaysian Peninsula was found to have the ability to degrade PS plastics (Auta et al., 2017). Bacillus paralicheniformis G1 isolated from deep sea sediments also has the ability to degrade PS (Ganesh Kumar et al., 2021). Additionally, bacteria Alcanivorax, Acinetobacter and Brevundimonas have been as a marine PS-degrading bacterium by our group (Liu et al., 2023; Lv et al., 2024; Zhao et al., 2024), as well as in soil environments and insect guts (Galgali et al., 2002; Oikawa et al., 2003; Ward et al., 2006; Atiq et al., 2010; Asmita et al., 2015; Mohan et al., 2016; Sekhar et al., 2016; Kim et al., 2020; Lee et al., 2020; Wang et al., 2020; Kim et al., 2021)prior to this study (Figure 4C). Pseudomonas traditionally considered to be degraders of PS found only in soil environments and insect gut, have recently been discovered to dominate in marine PS biofilms (Kumar et al., 2021; Bae and Yoo, 2022; Shi et al., 2022). This finding suggests that Pseudomonas originating from the marine may also possess a vast potential for PS degradation. Therefore, we speculate that in present case, they are prevalent PS-degrading bacteria not limited to terrestrial environments but also widespread in the marine plastics. However, whether these dominant groups derived from marine possess the ability to degrade PS still needs to be further studied and confirmed with pure cultures.
This study has detected several bacteria with potential pathogenicity, including those harmful to marine organisms, rise concerns. For example, Tenacibaculum, reported as an essential pathogen in global aquaculture (Satyam et al., 2023), in addition to some species of Vibrio (Silva et al., 2019) and Acinetobacter (Noel et al., 2022). To determine if they are pathogenic, we constructed a phylogenetic tree with the OTUs predominant in our dataset (Figure 6). The phylogenetic tree result of the 16S rRNA gene found that the dominant OTU6757 Acinetobacter is close to Acinetobacter johnsonii (Figure 6A), which has potential pathogenicity for fish (Kozińska et al., 2014). Tenacibaculum (OTU7310 and OTU7274) is close to Tenacibactulum pelagium on the evolutionary tree (Figure 6B), which is far from the species that causes fish diseases (Satyam et al., 2023). Similarly, OTU7016 and OTU5409, which belong to the Vibrio, are close to Vibrio azureus and Vibrio japonicus in the evolutionary tree (Figure 6C), and there have been no reports of related pathogenicity (Hoff et al., 2020).

Figure 6 Construct of phylogenetic relationship tree of bacterial OTU based on 16S rRNA gene sequences shown the position of OTU within the genus. (A) the position of OTU6757 within the genus Acinetobacter. (B) the position of OTU7274 and OTU7310 within the genus Tenacibaculum. (C) the position of OTU5409 and OTU7016 within the genus Vibrio. Bootstrap values indicated on the node with numbers using the neighbor-joining analysis of expressed as percentages of 1000 replications. Sequence divergence represented by score bar at 0.005 nucleotide substitution.
The EPS plastic biofilm in the Xiamen subtropical marine area is mainly constituted by Flavobacteriaceae, Rhodobacteraceae, Halomonadaceae, Exiguobacteriaceae, Vibrionaceae, and Pseudoalteromonadaceae. Compared with other subtropical environment PS biofilms, Halomonadaceae and Exiguobacteriaceae are particularly enriched here, which may play an important role in EPS biofilm formation. In addition, dominant bacterial groups with PS degradation potential ability were found, such as Alcanivorax, Pseudomonas, Acinetobacter, Exiguobacterium, Brevundimonas, and Bacillus, as well as the potential fish pathogen Acinetobacter johnsonii. These results provide a reference for the further study of plastic-degrading bacteria in the subtropical marine environment.
The sequences of this study were submitted to the China National Center for Bioinformation (CNCB) Genomics Database (GSA), with the database accession numbers CRA013490 and CRA013487.
BZ: Data curation, Formal Analysis, Investigation, Software, Visualization, Writing–original draft, Writing–review & editing. RL: Methodology, Writing–review & editing. HX: Validation, Visualization, Writing–original draft. SZ: Writing–review & editing. JW: Writing review & editing. ZS: Conception, Funding acquisition, Methodology, Resources, Supervision, Writing–original draft, Writing–review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Projects of the National Science Foundation of China (No. 91851203 and 42030412), the COMRA Program (No. DY-XZ-04 and DY135-B2-01), Scientific Research Foundation of Third Institute of Oceanography (No. 2019021), Xiamen Ocean Economic Innovation and Development Demonstration Project (No. 16PZP001SF16).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1409399/full#supplementary-material
Andrady A. L., Neal M. A. (2009). Applications and societal benefits of plastics. Philos. Trans. R. Soc Lond. Ser. B: Biol. Sci. 364, 1977–1984. doi: 10.1098/rstb.2008.0304
Asmita K., Shubhamsingh T., Tejashree S. (2015). Isolation of plastic degrading microorganisms from soil samples collected at various locations in mumbai, India. Proc. Environ. Sci. 4, 77-85.
Atiq N., Ahmed S., Ali M., Andleeb S., Ahmad B., Robson G. (2010). Isolation and identification of polystyrene biodegrading bacteria from soil.Afr. J. Microbiol. Res. 4, 1537–1541.
Auta H. S., Emenike C. U., Fauziah S. H. (2017). Screening of bacillus strains isolated from mangrove ecosystems in peninsular Malaysia for microplastic degradation. Environ. pollut. 231, 1552–1559. doi: 10.1016/j.envpol.2017.09.043
Bae S., Yoo K. (2022). Microplastic contamination and microbial colonization in coastal area of busan city, korea. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.1030476
Bond T., Ferrandiz-Mas V., Felipe-Sotelo M., van Sebille E. (2018). The occurrence and degradation of aquatic plastic litter based on polymer physicochemical properties: A review. Crit. Rev. Environ. Sci. Technol. 48, 685–722. doi: 10.1080/10643389.2018.1483155
Bonfá M. R., Grossman M. J., Piubeli F., Mellado E., Durrant L. R. (2013). Phenol degradation by halophilic bacteria isolated from hypersaline environments. Biodegradation 24, 699–709. doi: 10.1007/s10532-012-9617-y
Bryant J. A., Clemente T. M., Viviani D. A., Fong A. A., Thomas K. A., Kemp P., et al. (2016). Diversity and activity of communities inhabiting plastic debris in the north pacific gyre. mSystems 1, 3. doi: 10.1128/mSystems.00024-16
Chauhan D., Agrawal G., Deshmukh S., Roy S. S., Priyadarshini R. (2018). Biofilm formation by exiguobacterium sp. Dr11 and dr14 alter polystyrene surface properties and initiate biodegradation. Rsc. Advances. 8, 837590–837599. doi: 10.1039/c8ra06448b
Chen S., Zhou Y., Chen Y and Gu J. (2018). Fastp: An ultra-fast all-in-one fastq preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Derraik J. G. B. (2002). The pollution of the marine environment by plastic debris: A review. Mar. pollut. Bull. 44, 842–852. doi: 10.1016/S0025-326X(02)00220-5
De Tender C. A., Devriese L. I., Haegeman A., Maes S., Ruttink T and Dawyndt P. (2015). Bacterial community profiling of plastic litter in the belgian part of the north sea. Environ. Sci. Technol. 49, 9629–9638. doi: 10.1021/acs.est.5b01093
Dussud C., Hudec C., George M., Fabre P., Higgs P., Bruzaud S., et al. (2018). Colonization of non-biodegradable and biodegradable plastics by marine microorganisms. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01571
Edgar R. C. (2013). Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Egan S., James S., Holmström C and Kjelleberg S. (2002). Correlation between pigmentation and antifouling compounds produced by pseudoalteromonas tunicata. Environ. Microbiol. 4, 433–442. doi: 10.1046/j.1462-2920.2002.00322.x
Frère L., Maignien L., Chalopin M., Huvet A., Rinnert E., Morrison H., et al. (2018). Microplastic bacterial communities in the bay of brest: Influence of polymer type and size. Environ. pollut. 242, 614–625. doi: 10.1016/j.envpol.2018.07.023
Galgali P., Varma A. J., Puntambekar U. S., Gokhale D. V. (2002). Towards biodegradable polyolefins: Strategy of anchoring minute quantities of monosaccharides and disaccharides onto functionalized polystyrene, and their effect on facilitating polymer biodegradation. Chem. Commun. 23, 2884–2885. doi: 10.1039/b209254a
Ganesh Kumar A., Hinduja M., Sujitha K., Nivedha Rajan N., Dharani G. (2021). Biodegradation of polystyrene by deep-sea bacillus paralicheniformis g1 and genome analysis. Sci. Total Environ. 774, 145002. doi: 10.1016/j.scitotenv.2021.145002
Hansen J., Melchiorsen J., Ciacotich N., Gram L., Sonnenschein E. C. (2021). Effect of polymer type on the colonization of plastic pellets by marine bacteria. FEMS. Microbiol. Lett. 368, 5. doi: 10.1093/femsle/fnab026
Hassanshahian M., Emtiazi G., Cappello S. (2012). Isolation and characterization of crude-oil-degrading bacteria from the persian gulf and the caspian sea. Mar. pollut. Bull. 64, 7–12. doi: 10.1016/j.marpolbul.2011.11.006
Hirai H., Takada H., Ogata Y., Yamashita R., Mizukawa K., Saha M., et al. (2011). Organic micropollutants in marine plastics debris from the open ocean and remote and urban beaches. Mar. pollut. Bull. 62, 1683–1692. doi: 10.1016/j.marpolbul.2011.06.004
Hoellein T., Rojas M., Pink A., Gasior J., Kelly J. (2014). Anthropogenic litter in urban freshwater ecosystems: Distribution and microbial interactions. PloS One 9, e98485. doi: 10.1371/journal.pone.0098485
Hoff J., Daniel B., Stukenberg D., Thuronyi B. W., Waldminghaus T., Fritz G. (2020). Vibrio natriegens: An ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ. Microbiol. 22, 4394–4408. doi: 10.1111/1462-2920.15128
Howard S. A., McCarthy R. R. (2023). Modulating biofilm can potentiate activity of novel plastic-degrading enzymes. npj. Biofilms. Microbiomes. 9, 72. doi: 10.1038/s41522-023-00440-1
Isobe A., Iwasaki S. (2022). The fate of missing ocean plastics: Are they just a marine environmental problem? Sci. Total. Environ. 825, 153935. doi: 10.1016/j.scitotenv.2022.153935
Kesy K., Oberbeckmann S., Kreikemeyer B., Labrenz M. (2019). Spatial environmental heterogeneity determines young biofilm assemblages on microplastics in baltic sea mesocosms. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01665
Kiatkamjornwong S., Sonsuk M., Wittayapichet S., Prasassarakich P., Vejjanukroh P.-C. (1999). Degradation of styrene-g-cassava starch filled polystyrene plastics. Polym. Degrad. Stab. 66, 323–335. doi: 10.1016/S0141-3910(99)00082-8
Kim H. W., Jo J. H., Kim Y. B., Le T. K., Cho C. W., Yun C. H., et al. (2021). Biodegradation of polystyrene by bacteria from the soil in common environments. J. Hazard. Mater. 416, 126239. doi: 10.1016/j.jhazmat.2021.126239
Kim H. R., Lee H. M., Yu H. C., Jeon E., Lee S., Li J., et al. (2020). Biodegradation of polystyrene by pseudomonas sp. Isolated from the gut of superworms (larvae of zophobas atratus). Environ. Sci. Technol. 54, 6987–6996. doi: 10.1021/acs.est.0c01495
Kozińska A., Paździor E., Pękala A and Niemczuk W. (2014). Acinetobacter johnsonii and acinetobacter lwoffii - the emerging fish pathogens. Bull. Vet. Inst. Pulawy. 58, 193–199. doi: 10.2478/bvip-2014-0029
Kumar R., Pandit P., Kumar D., Patel Z., Pandya L., Kumar M., et al. (2021). Landfill microbiome harbour plastic degrading genes: A metagenomic study of solid waste dumping site of gujarat, India. Sci. Total. Environ. 779, 146184. doi: 10.1016/j.scitotenv.2021.146184
Lee H. M., Kim H. R., Jeon E., Yu H. C., Lee S., Li J., et al. (2020). Evaluation of the biodegradation efficiency of four various types of plastics by pseudomonas aeruginosa isolated from the gut extract of superworms. Microorganisms 8, 9. doi: 10.3390/microorganisms8091341
Lee J. W., Nam J. H., Kim Y. H., Lee K. H., Lee D. H. (2008). Bacterial communities in the initial stage of marine biofilm formation on artificial surfaces. J. Microbiol. 46, 174–182. doi: 10.1007/s12275-008-0032-3
Liu C., Zhao D., Ma W., Guo Y., Wang A., Wang Q., et al. (2016). Denitrifying sulfide removal process on high-salinity wastewaters in the presence of halomonas sp. Appl. Environ. Microbiol. 100, 1421–1426. doi: 10.1007/s00253-015-7039-6
Liu R., Zhao S., Zhang B., Li G., Fu X., Yan P., et al. (2023). Biodegradation of polystyrene (ps) by marine bacteria in mangrove ecosystem. J. Hazard. Mater. 442, 130056. doi: 10.1016/j.jhazmat.2022.130056
Lv S., Cui K., Zhao S., Li Y., Liu R., Hu R., et al. (2023). Continuous generation and release of microplastics and nanoplastics from polystyrene by plastic-degrading marine bacteria. J. Hazard. Mater. 465, 133339. doi: 10.1016/j.jhazmat.2023.133339
Lv S., Cui K., Zhao S., Li Y., Liu R., Hu R., et al. (2024). Continuous generation and release of microplastics and nanoplastics from polystyrene by plastic-degrading marine bacteria. J. Hazard. Mater. 465, 133339. doi: 10.1016/j.jhazmat.2023.133339
Magoč T., Salzberg S. L. (2011). Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Mendoza L. M. R., Jones P. R. (2015). Characterisation of microplastics and toxic chemicals extracted from microplastic samples from the north pacific gyre. Environ. Chem. 12, 611–617. doi: 10.1071/EN14236
Mohan A. J., Sekhar V. C., Bhaskar T., Nampoothiri K. M. (2016). Microbial assisted high impact polystyrene (hips) degradation. Bioresour. Technol. 213, 204–207. doi: 10.1016/j.biortech.2016.03.021
Mor R., Sivan A. (2008). Biofilm formation and partial biodegradation of polystyrene by the actinomycete rhodococcus ruber: Biodegradation of polystyrene. Biodegradation 19, 851–858. doi: 10.1007/s10532-008-9188-0
Naudet J., d’Orbcastel E. R., Bouvier T., Godreuil S., Dyall S., Bouvy S., et al. (2023). Identifying macroplastic pathobiomes and antibiotic resistance in a subtropical fish farm. Mar. pollut. Bull. 194, 115267. doi: 10.1016/j.marpolbul.2023.115267
Noel H. R., Petrey J. R., Palmer L. D. (2022). Mobile genetic elements in acinetobacter antibiotic-resistance acquisition and dissemination. Ann. N. Y. Acad. Sci. 1518, 166–182. doi: 10.1111/nyas.14918
Nowlan J. P., Britney S. R., Lumsden J. S., Russell S. (2021). Experimental induction of tenacibaculosis in atlantic salmon (salmo salar l.) using tenacibaculum maritimum, t. Dicentrarchi, and t. Finnmarkense. Pathogens 10, 11. doi: 10.3390/pathogens10111439
Nowlan J. P., Lumsden J. S., Russell S. (2020). Advancements in characterizing tenacibaculum infections in Canada. Pathogens 9, 12. doi: 10.3390/pathogens9121029
Oberbeckmann S., Kreikemeyer B., Labrenz M. (2017). Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02709
Oberbeckmann S., Löder M. G. J., Labrenz M. (2015). Marine microplastic-associated biofilms - a review. Environ. Chem. 12, 551–562. doi: 10.1071/EN15069
Oberbeckmann S., Osborn A. M., Duhaime M. B. (2016). Microbes on a bottle: Substrate, season and geography influence community composition of microbes colonizing marine plastic debris. PloS One 11, e0159289. doi: 10.1371/journal.pone.0159289
Oikawa E., Linn K. T., Endo T., Oikawa T., Ishibashi Y. (2003). Isolation and characterization of polystyrene degrading microorganisms for zero emission treatment of expanded polystyrene. Environ. Eng. Res. 40, 373–379. doi: 10.11532/proes1992.40.373
Petrella A., Di Mundo R., Notarnicola M. (2020). Recycled expanded polystyrene as lightweight aggregate for environmentally sustainable cement conglomerates. Materials 13, 4. doi: 10.3390/ma13040988
Plastics Europe (2023) Plastics–the fast facts 2023. Available at: https://plasticseurope.org/knowledge-hub/plastics-the-fast-facts-2023/ (Accessed March 1, 2024]).
Pompilio A., Piccolomini R., Picciani C., D’Antonio D., Savini V., Di Bonaventura G. (2008). Factors associated with adherence to and biofilm formation on polystyrene by stenotrophomonas maltophilia: The role of cell surface hydrophobicity and motility. FEMS. Microbiol. Lett. 287, 41–47. doi: 10.1111/fml.2008.287.issue-1
Satyam R., Ahmad S., Raza K. (2023). Comparative genomic assessment of members of genus tenacibaculum: An exploratory study. Mol. Genet. 298, 979–993. doi: 10.1007/s00438-023-02031-3
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Sekhar V. C., Nampoothiri K. M., Mohan A. J., Nair N. R., Bhaskar T., Pandey A. (2016). Microbial degradation of high impact polystyrene (hips), an e-plastic with decabromodiphenyl oxide and antimony trioxide. J. Hazard. Mater. 318, 347–354. doi: 10.1016/j.jhazmat.2016.07.008
Shi Y., Wang S., Wang H., Li Z., Cai J., Han Q., et al. (2022). Diversity and succession of microbial communities on typical microplastics in xincun bay, a long-term mariculture tropical lagoon. Oceanol. Hydrobiol. Stud. 51, 10–22. doi: 10.26881/oahs-2022.1.02
Silva M. M., Maldonado G. C., Castro R. O., de Sá Felizardo J., Cardoso R. P., Anjos R. M. D., et al. (2019). Dispersal of potentially pathogenic bacteria by plastic debris in guanabara bay, rj, Brazil. Mar. pollut. Bull. 141, 561–568. doi: 10.1016/j.marpolbul.2019.02.064
Tu C., Liu Y., Li L., Li Y., Vogts A., Luo Y., et al. (2021). Structural and functional characteristics of microplastic associated biofilms in response to temporal dynamics and polymer types. Bull. Environ. Contam. Toxicol. 107, 633–639. doi: 10.1007/s00128-021-03333-1
Turner A. (2020). Foamed polystyrene in the marine environment: Sources, additives, transport, behavior, and impacts. Environ. Sci. Technol. 54, 10411–10420. doi: 10.1021/acs.est.0c03221
Wang F., Wong C. S., Chen D., Lu X., Wang F and Zeng E. Y. (2018). Interaction of toxic chemicals with microplastics: A critical review. Water Res. 139, 208–219. doi: 10.1016/j.watres.2018.04.003
Wang Q., Garrity G. M., Tiedje JM and Cole J. R. (2007). Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang Z., Xin X., Shi X., Zhang Y. (2020). A polystyrene-degrading acinetobacter bacterium isolated from the larvae of tribolium castaneum. Sci. Total Environ. 726, 138564. doi: 10.1016/j.scitotenv.2020.138564
Ward P. G., Goff M., Donner M., Kaminsky W., O’Connor K. E. (2006). A two step chemo-biotechnological conversion of polystyrene to a biodegradable thermoplastic. Environ. Sci. Technol. 40, 2433–2437. doi: 10.1021/es0517668
Weisburg W. G., Barns S. M., Pelletier DA and Lane D. J. (1991). 16s ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Xie L. (2023) Monthly temperature statistics in xiamen in 2022. Available at: https://www.tianqi24.com/xiamen/history2022.html (Accessed March 1, 2024).
Xie H., Chen J., Feng L., He L., Zhou C., Hong P., et al. (2021). Chemotaxis-selective colonization of mangrove rhizosphere microbes on nine different microplastics. Sci. Total. Environ. 752, 142223. doi: 10.1016/j.scitotenv.2020.142223
Yang Y., Yang J., Wu W. M., Zhao J., Song Y., Gao L., et al. (2015). Biodegradation and mineralization of polystyrene by plastic-eating mealworms: Part 2. Role of gut microorganisms. Environ. Sci. Technol. 49, 12087–12093. doi: 10.1021/acs.est.5b02663
Zettler E. R., Mincer T. J., Amaral-Zettler L. A. (2013). Life in the “plastisphere”: Microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Zhang S. J., Zeng Y. H., Zhu J. M., Cai Z. H., Zhou J. (2022). The structure and assembly mechanisms of plastisphere microbial community in natural marine environment. J. Hazard. Mater. 421, 126780-126793. doi: 10.1016/j.jhazmat.2021.126780
Zhang W. W., Ma X. D., Zhang Z. F., Wang Y., Wang J. Y., Wang J., et al. (2015). Persistent organic pollutants carried on plastic resin pellets from two beaches in China. Mar. pollut. Bull. 99, 28–34. doi: 10.1016/j.marpolbul.2015.08.002
Zhao S., Liu R., Lv S., Zhang B., Wang J., Shao Z. (2024). Polystyrene (ps)-eating marine benthic polychaete and associated gut microbiome dominated by polystyrene-degrading bacteria. Ear. Environ. Commun. 5, 162-174. doi: 10.1038/s43247-024-01318-6
Keywords: plastic waste, expanded polystyrene, biofilms, bacterial community, subtropical mariculture
Citation: Zhang B, Liu R, Xu H, Zhao S, Wang J and Shao Z (2024) Bacterial diversity in the biofilms on mariculture polystyrene foam at Xiamen’s coast. Front. Mar. Sci. 11:1409399. doi: 10.3389/fmars.2024.1409399
Received: 01 April 2024; Accepted: 02 May 2024;
Published: 21 May 2024.
Edited by:
Hans Uwe Dahms, Kaohsiung Medical University, TaiwanReviewed by:
Tan Suet May Amelia, University of Malaysia Terengganu, MalaysiaCopyright © 2024 Zhang, Liu, Xu, Zhao, Wang and Shao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongze Shao, c2hhb3p6QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.