- 1School of Life and Environmental Sciences, The University of Sydney, Sydney, NSW, Australia
- 2Sydney Institute of Marine Science, Sydney, NSW, Australia
- 3UWA Oceans Institute and School of Biological Sciences, University of Western Australia, Perth, WA, Australia
- 4Singapore Centre for Environmental Life Sciences Engineering (SCELSE), Nanyang Technological University, Singapore, Singapore
- 5School of Biological, Earth and Environmental Sciences, The University of New South Wales, Sydney, NSW, Australia
- 6Asian School of the Environment, Nanyang Technological University, Singapore, Singapore
- 7Nanyang Environment & Water Research Institute (NEWRI), Nanyang Technological University, Singapore, Singapore
- 8Dipartimento di Biologia, Università di Pisa, Pisa, Italy
Introduction: The collapse of macroalgal habitats is altering the structure of benthic communities on rocky shores globally. Nonetheless, how the loss of canopy-forming macroalgae influences the structure of epilithic microbial communities is yet to be explored.
Methods: Here, we used experimental field manipulations and 16S-rRNA-gene amplicon sequencing to determine the effects of macroalgal loss on the understory bacterial communities and their relationship with epiphytic bacteria on macroalgae. Beds of the fucoid Hormosira banksii were exposed to different levels of disturbance resulting in five treatments: (i) 100% removal of Hormosira individuals, (ii) 50% removal, (iii) no removal, (iv) a procedural control that mimicked the removal process, but no Hormosira was removed and (v) adjacent bare rock. Canopy cover, bacterial communities (epilithic and epiphytic) and benthic macroorganisms were monitored for 16 months.
Results: Results showed that reductions in canopy cover rapidly altered understory bacterial diversity and composition. Hormosira canopies in 50% and 100% removal plots showed signs of recovery over time, but understory epilithic bacterial communities remained distinct throughout the experiment in plots that experienced full Hormosira removal. Changes in bacterial communities were not related to changes in other benthic macroorganisms.
Discussion: These results demonstrate that understory epilithic bacterial communities respond rapidly to environmental disturbances at small scales and these changes can be long-lasting. A deeper knowledge of the ecological role of understory epilithic microbial communities is needed to better understand potential cascading effects of disturbances on the functioning of macroalgal-dominated systems.
1 Introduction
Disturbances play an important role in shaping and maintaining ecological community structure and diversity (Pickett and White, 1985; Fraterrigo & Rusak, 2008; Peters et al., 2011). Terrestrial and marine ecosystems are facing substantial challenges as a result of the recent increase in the frequency and intensity of disturbance events, causing changes in resource availability (Wilson and Tilman, 1993; Peters et al., 2011), genetic diversity (Banks et al., 2013; Coleman et al., 2020) and local environmental conditions (Castorani et al., 2018). In particular, disturbances affecting dominant habitat-forming species can trigger detrimental cascading effects on associated assemblages of species (e.g. understory species) that benefit from the resources and environmental conditions they provide (Brooker et al., 2008; Takolander et al., 2017; Castorani et al., 2018; Narwani et al., 2019). These changes may, in turn, generate stabilizing feedback, negatively affecting the recovery of habitat-formers and, hence, locking the system into an alternative degraded state (Bever et al., 1997; Folke et al., 2004; Hamman and Hawkes, 2013; van der Putten et al., 2013).
On rocky shores, declines of habitat-forming macroalgae due to natural or human disturbances (e.g., decrease in water quality, increased herbivore pressure, storms or trampling) can lead to drastic changes to understory benthic community structure and ecological interactions (Witman, 1987; Benedetti-Cecchi et al., 2001; Marzinelli et al., 2012; Crowe et al., 2013 and Castorani et al., 2018). In particular, nutrient loading and sedimentation rates (Strain et al., 2014) have been demonstrated to promote the shift in dominance from canopy- to turf-forming or invasive algal species (Valentine & Johnson, 2003; Gorman et al., 2009). Over-exploitation of high trophic-level species can also facilitate the formation of barren grounds dominated by encrusting coralline or turfing algae through the release of herbivores from predation (Poore et al., 2012; Filbee-Dexter and Scheibling, 2014; Ling et al., 2015; Coleman & Kennelly, 2019). In many circumstances, these changes are seemingly ‘locked in’ (i.e. achieve hysteresis), though the mechanisms behind this are largely unknown. Bacterial communities are a largely ignored component of understory biotic communities that may be affected by the decline of canopy-forming macroalgae and may, in turn, facilitate such impacts.
On intertidal rocky shores, epilithic bacterial communities grow in dense biopolymer extracellular biofilms that can extend across large areas or as associated epiphytic communities on macroorganisms (Flemming & Wuertz, 2019). Biofilms provide microorganisms an active area of nutrient exchange and suitable environmental conditions (Flemming et al., 2016) and play an important role in resource acquisition (e.g., CO2, NO3-, and prokaryotic-derived vitamins), defence against fouling (Rao et al., 2007), morphological development and recruitment of invertebrates and macroalgae (Joint et al., 2002; Patel et al., 2003; Hadfield, 2011; Pedicini et al., 2023). They also fuel rocky shore food webs (Hawkins and Hartnoll, 1983) by directly supporting grazing species (e.g., gastropods) (Underwood, 1978). Changes in grazer assemblages, in turn, can determine the recruitment, growth and survival rates of macroalgae (Hawkins, 1981 and Coleman et al., 2006). However, changes in the abundance of grazing macrobenthic organisms (Arboleda-Baena et al., 2022), shifts in the availability of biogenic settlement surfaces (e.g., calcified structures or understory algal turfs, Bulleri et al., 2018; Roush & Garcia-Pichel, 2020) and antibacterial activities (Iguchi et al., 1982) may also play an important role in shaping epilithic bacterial biofilms. Canopy forming-macroalgae can also directly influence the adjacent epilithic bacterial community through the provision of resources (e.g., DOC, Elsherbini et al., 2023), protection against environmental factors (Pocklington et al., 2019), release of secondary metabolites (Egan et al., 2013) or a direct transfer of free-living bacteria to the substrata (Bulleri et al., 2018). Thus, changes to understory epilithic bacterial biofilms may have substantial direct and/or indirect impacts on the biodiversity and functioning of intertidal rocky shores.
Hormosira banksii (Turner, Decaisne, 1842; hereafter Hormosira) is a perennial fucoid macroalga that is often the dominant habitat-forming species on intertidal rocky shores of temperate Australasia (Kain et al, 2015; Lilley & Schiel, 2006). Hormosira forms a dense monotypic canopy of several hundred individuals per square meter (Schiel & Taylor, 1999) and directly influences the presence of other algae and invertebrates through the amelioration of environmental variables such as temperature and desiccation (Keough and Quinn, 1998; Schiel and Lilley, 2007). Although Hormosira can tolerate fluctuations in temperature and desiccation (Kain, 2015), it is susceptible to natural and anthropogenic disturbances such as severe storms (Underwood, 1998), poor water quality (Fairweather, 1990; Bellgrove et al., 2010; Cameron et al., 2021), sedimentation and trampling (Povey and Keough, 1991; Keough and Quinn, 1998; Schiel and Taylor, 1999). The recovery of Hormosira following such disturbances is often slow (Bellgrove et al., 2010; Lewis et al., 2021) and dependent on the extent of canopy damage (Underwood, 1998) and subsequent competitive exclusion by coralline turfs (Bellgrove et al., 2010). It is unclear, however, how canopy disturbance influences understory epilithic bacterial communities and, in turn, how such changes regulate macroalgal recovery. Recent studies have shown that human disturbances such as urbanisation and the addition of nutrients alter the structure of epilithic bacterial communities underneath intertidal and subtidal macroalgal canopies (Elsherbini et al., 2023) and, indirectly, macroalgal recruitment (Pedicini et al., 2023).
Here, we performed a manipulative experiment to test the effects of Hormosira canopy disturbance on the understory epilithic bacterial communities. We manipulated the density of Hormosira in the field, resulting in five treatments: (i) 100% removal of Hormosira individuals, (ii) 50% removal, (iii) no removal, (iv) a procedural control that mimicked the removal process, but no Hormosira was removed and (v) adjacent bare rock. We characterised the bacterial communities and benthic understory macroorganisms, and quantified canopy cover over 1.5 years. We examined the recovery of the canopy and changes over time in the structure of microbial, invertebrate and macroalgal communities to test the following hypotheses: (i) that understory epilithic bacterial communities would change after canopy disturbance relative to controls, (ii) that the magnitude of change and rate of recovery would depend on the level of disturbance, with the greatest changes and slowest recovery for assemblages under the full canopy removal treatment. In addition, (iii) we also determined whether observed changes in epilithic bacterial communities were influenced by shifts in other understory macroorganisms such as grazers or sessile species (i.e., coralline algae or algal understory and turfs). Finally, iv) we assessed whether Hormosira plays an important role in the establishment and structuring of the understory epilithic bacterial community via transfer of taxa from its surface.
2 Materials and methods
2.1 Experimental manipulation of Hormosira
This study was carried out on an east-facing flat rocky platform at Coalcliff, New South Wales, Australia (-34.248°, 150.978°) and lasted for 16 months. At this site, thalli of Hormosira were ~15cm in length, forming a bed of 100% canopy cover in interspersed patches of ~150m2. The experiment was setup in October 2018 and consisted of 25 randomly selected 50x50cm plots, ~3m apart, randomly assigned to five experimental treatments that were spatially interspersed on the rocky platform. Disturbance treatments included plots where Hormosira canopy was 1) completely (100% removal, zero density of Hormosira) or 2) partially removed (50% removal and 43.5 ± 3.2 ind/m2 SE of Hormosira density), 3) undisturbed Hormosira control plots (0% removal), 4) procedural control plots where the canopy was disturbed through physically handling and shaking H. bansksii, simulating the manual disturbance done in the disturbance treatments, but not removed (PC, both control treatments with Hormosira density of 87 ± 6.4 ind/m2 SE), and 5) plots with bare rock where Hormosira was not present (n=5 for each treatment).
The epilithic microbial community was sampled during low tides, two (Time 1: 6/11/2018), three (Time 2: 26/11/2018), twenty-eight (Time 3: 19/5/2019) and sixty-one weeks (Time 4: 8/1/2020) after the initial disturbance. Sampling times were chosen to determine short (T1 and T2) and long term (T3 and T4) responses of both Hormosira and the epilithic bacterial community. Due to the harsh environmental conditions of the intertidal, It was expected that monitoring at a short term would provide us with valuable information on the immediate changes of bacterial communities post-disturbance. Comparatively, long-term sampling it was thought that due to Hormosira’s known slow growth rate, potential recovery to produce new canopy cover would only occur after approximately ~ 1 year post disturbance (as seen in Pocklington et al., 2019; Cameron et al., 2021). At each time, an epilithic bacterial sample was taken by swabbing the substratum within a 5x5 cm quadrat at each plot for 30 seconds using a sterile cotton swab (Marzinelli et al., 2015). To ensure that the swabbed area of the substratum was only sampled once during the experiment, each plot was divided into four quarters using the diagonals and the 5x5cm quadrat was placed in each quarter once. In plots where Hormosira was not removed (i.e., control and procedural control) or was only partially removed (50% cover), a second bacterial swab sample was also taken from the surface of Hormosira laminae (similar area as above) to compare understory epilithic and host-associated epiphytic bacterial communities. Prior to swabbing, benthic or algal surfaces were rinsed with 0.22 µm filtered seawater to remove any unattached epibionts. Swabs were stored immediately inside sterile cryogenic tubes and liquid nitrogen, transported to the University of New South Wales, Sydney; and kept at -80°C until DNA extractions were performed. Unsuccessful processing during molecular analysis, due to contamination during DNA extractions and/or low PCR amplification, resulted in different number of samples among times (epilithic: Time 1: n = 25; Time 2: n = 24; Time 3: n = 19; Time 4: n = 19; and epiphytic: Time 1: n = 14; Time 2: n = 12; Time 3: n = 10; Time 4: n = 8).
2.2 Benthic macroorganisms
Following the initial disturbance, the i) percent canopy cover of Hormosira and ii) organisms comprising the benthic understory (i.e., the substratum area that was swabbed) in each plot were monitored through photographic techniques at the same sampling times as the microbial swabs. Hormosira canopy cover was quantified (as plots with 50% density removal might still have high canopy cover) via the online machine learning engine, CoralNet, which performs a semiautomatic point annotation, based on specified classifying labels (Beijbom et al., 2015). These classifying labels were verified through the Collaborative and Automated Tools for Analysis of Marine Imagery classification scheme (CATAMI; v.1.2, Althaus et al., 2015) and included point annotations for seven taxa/groups: Hormosira, calcareous encrusting tube worms (TW), articulated calcareous red algae (ACR), brown laminate macroalgae (BLM), turf algae (TA), grazers (i.e., gastropods), calcareous crustose algae (CCA: healthy and BCCA: bleached) and two abiotic variables, sand and rock (examples of each category can be seen in Figure S1).
2.3 DNA processing and bioinformatics
DNA was randomly extracted from each swab sample using the Powersoil DNA Isolation Kit (Mo Bio Laboratories #12888-100) following the manufacturer guidelines. DNA extracts were stored in a freezer at -20°C until PCR amplification with primers 341F (5’ -CCTACGGGNGGCWGCAG- 3’) and 805R (5’ -GACTACHVGGGTATCTAATCC- 3’) which target the V3-V4 regions of the 16S rRNA gene (Klindworth et al., 2013). Agarose gel electrophoresis, Nanodrop 1000 and the Qubit 2.0 fluorometer (Thermo Fisher Scientific) were used to check the quality and quantity of the amplicons before being sent for sequencing in an Illumina MiSeq 2000 Platform at the Ramaccioti Centre for Genomics.
Gene sequence reads were quality filtered without specified maximum expected errors (as suggested by Prodan et al., 2020) and maximum truncation lengths assigned to forward and reverse reads independently. Trimming parameters for maximum truncation were decided upon inspection of the quality error plots of both reads with any base pairs with a median Q score below 30 removed. Using the untrimmed sequences, the maximum error rates were calculated and included in the DADA2 denoising model. Sample inference was performed using the DADA2 denoising algorithm and all resulting denoised paired reads were merged to form unique amplicon sequence variants (ASV). These unique ASV sequences were used to construct an abundance per sample table and subsequently used to detect and remove chimeric sequences (consensus method; Callahan et al., 2016). An initial taxonomic assignment to the ASV table without chimeric sequences was done using SILVA v. 138.1 (Quast et al., 2013) to increase the detection of chloroplasts and mitochondrial ASV. These were removed from the ASV table, and a second taxonomic assignment was done using the Genome Taxonomy Database, which provides a higher taxonomic resolution (GTDB, Parks et al., 2022). The DADA2 pipeline was performed using R v.4.1.1 and the dada2 package v.1.26 (Callahan et al., 2016). An average of 76.3%± 0.65 SE of total reads was kept at the end of the bioinformatic pipeline.
Two separate data subsets were created from the original bacterial ASV table: (1) samples taken exclusively from biofilms in the benthic substrate (hereafter Dataset 1, 87 samples in five disturbance treatment levels) and (2) samples taken from host-associated epiphytic bacterial communities on the surface of Hormosira and samples from the nearby understory epilithic bacterial community within the same plot (hereafter Dataset 2, 44 samples from treatments only including 50% and 0% Hormosira removal treatments). Statistical analyses of each of the two data subsets were performed separately. Singletons and sequences with low abundance (i.e., <0.01% of the total) were removed and data sets were normalized independently using DESeq2’s median of rations method to account for heterogeneous library sizes (DESeq2, package phyloseq v.1.40.1; Love et al., 2014). Abundance tables for both analyses were independently square-root transformed before statistical analyses (Swift et al., 2023). An ASV accumulation curve was also constructed for each data subset to evaluate the sampling effort to effectively describe the overall bacterial community in the experiment (package vegan v.2.6-4, Oksanen et al., 2013).
2.4 Statistical analyses
2.4.1 Bacterial communities
Alpha diversity indices assessing observed bacterial richness (number of ASV), diversity (Shannon-Wiener index) and evenness (Pielou index) were calculated using the R package phyloseq v.1.44; (Mcmurdie and Holmes, 2013) and analyzed using linear mixed models (LMM). One model, testing for the effects of the different intensity of Hormosira canopy removal included ‘disturbance’ (fixed, 5 levels) and ‘sampling times’ (fixed, crossed, 4 levels) as fixed effects. Plot was included as a random effect in the models to account for repeated measures taken from plots throughout the experiment.
A second model, fitted to examine differences in bacterial communities between the understory and Hormosira surfaces, included ‘disturbance’ (fixed, 2 levels, only including control, undisturbed treatments) and ‘sampling times’ (fixed, crossed, 4 levels) and ‘substratum type’ (fixed, 2 levels: substratum vs Hormosira) and their interaction as fixed effects and the plot as a random effect. All assumptions including linearity, homogeneity of variance and normality were validated suing the R package performance 0.10.4 (Lüdecke et al., 2021). P-values for each model term were calculated using the Anova function in R package car v.3.1-2 using a Satterthwaite approximation method (Luke, 2017) and random effects were inferred through a likelihood ratio test (alpha<0.05; package lmerTest 3.1-3, Kuznetsova et al., 2017). If significant differences in the alpha diversity indices were detected between the fixed factors, post hoc contrasts were calculated (i.e., Tukey HSD; R package emmeans v.1.8.6; Russell, 2022). To further assess the effects of Hormosira removal treatments on the microbial compartment, we also tested for associations between epilithic bacterial alpha diversity indices and Hormosira percentage canopy cover using Pearson’s correlations (R package corrplot v.0.92, Wei and Simko 2021 only for Dataset 1).
Bacterial community composition was visualised using non-metric multidimensional scaling (NMDS) ordination (package vegan; Oksanen et al., 2013). Differences in bacterial community composition were determined through a Permutational Analysis of Variance using Bray-Curtis dissimilarities among samples (PERMANOVA+ v.1.0.5 and Primer-e v.6.1.15) and including the same fixed and random factors as above for both datasets. Multivariate post hoc pairwise tests were performed if significant structural differences were found. Assumptions of equal variance among groups were checked using a permutational analysis of multivariate dispersion using the same program as above.
Comparative analysis of abundance was performed by fitting two multivariate generalized linear models, assuming a negative-binomial distribution (R package mvabund v.4.2.1, Wang et al., 2012) to test the 1) effects of the Hormosira removal treatments across time on the abundance (total abundance: counts) of specific epilithic bacterial ASVs (only Dataset 1), and 2) differences between bacterial communities associated with the surface of Hormosira and understory substrate (Dataset 2, including the fixed factor: ‘substratum type’). For each of the two models, univariate tests were used to identify ASVs that significantly differed in abundance among treatments (R package mvabund, adjusted method for univariate tests; Wang et al., 2012). In some analyses, several dozens of ASVs were found to differ among treatments, so, we focused on bacterial ASVs with the highest average relative abundance and topmost prevalence across all samples in further analyses. For each identified ASV, linear general mixed models and post hoc tests (where significant main effects were found) were performed to determine differences in the relative abundance across fixed factor levels. All model validation, model inferences and multiple pairwise comparisons were done similarly to the alpha diversity index analysis above.
2.4.2 Hormosira canopy cover and other benthic macroorganisms
To determine differences in the canopy cover of Hormosira and other macrobenthic components, a linear model was fitted with these factors: ‘disturbance’ (fixed, 5 levels), ‘sampling times’ (fixed, crossed, 4 levels) and ‘plot’ (random, 28 levels). Model assumptions of normality, homogeneity of variance and influential observations as well as post-hoc contrasts were validated and calculated as above.
A distance-based redundancy analysis (dbRDA) was also included to explore linear relationships between the structure of the biofilm community (i.e., Bray-Curtis dissimilarity indices from the understory epilithic bacterial dataset) and the percentage cover of Hormosira and other macrobenthic components such as calcareous encrusting tube worms, articulate crustose red algae, brown laminate macroalgae, turf algae, grazers, calcareous crustose algae (package vegan, Oksanen et al., 2013). A stepwise model selection based on Akaike information criteria (AIC) was used to determine the best-performing model and predictor variables, which were then added to ordinations to determine linear trends between these predictors and bacterial communities (function envfit, package vegan, Oksanen et al., 2013). Marginal tests were also done to examine relationships with single predictor variables. For these tests, all predictor variables were transformed to reduce skewness (i.e., arcsine transformation) and normalized. No multicollinearity was found between any of the predictor variables (Variance Inflation Factor <2).
3 Results
3.1 Response of epilithic bacterial communities to Hormosira removal
A total of 6,418 microbial ASVs across 81 samples were included in the analysis and sampling effort was found to be adequate for this community (Figure S2A). All alpha diversity indices show a significant effect of disturbance treatment, independent of sampling time (Figure 1 and Table S1, Richness, F4,76 = 15.68, p<0.001; Shannon-Wiener diversity: F4,76 = 24.2, p<0.001; evenness: F4,76 = 20.95, p<0.001). Control plots (0% removal and PC) had higher bacterial diversity (p<0.032) and evenness values (p<0.010) compared to plots where Hormosira had been completely cleared. However, there was no difference between these controls and the 50% removal treatments (see post hoc tests in Table S2). Bacterial richness (decrease of 59 ± 2.7% SE; p<0.008), diversity (decrease of 27 ± 2.2% SE; p<0.001) and evenness (decrease of 15 ± 1.8% SE; p<0.023) was lower in bare rock plots compared to all other disturbance treatments (Figure 1 and post hoc tests in Table S2). Bacterial richness, diversity and evenness were positively correlated with canopy cover of Hormosira (Figure 2).
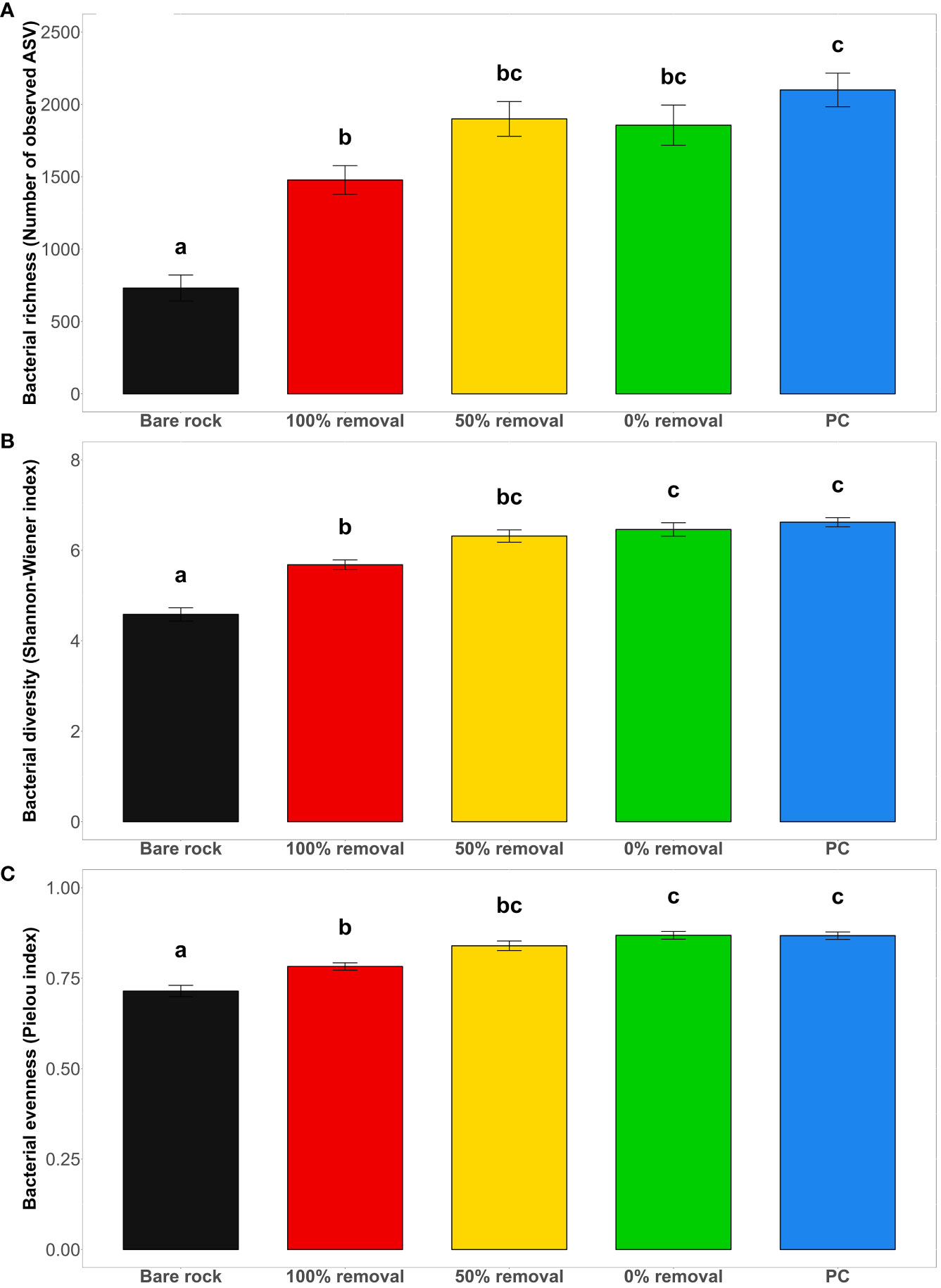
Figure 1 Effect of Hormosira disturbance treatments (Bare rock (n=18), 100% removal (n=17), 50% removal (n=18) and undisturbed control (0% removal, n=20), and procedural control (n=14)) on benthic epilithic bacterial alpha diversity indices (mean ± SE) including bacterial (A) richness, (B) diversity (Shannon-Wiener Index) and (C) evenness (Pielou Index). Sampling times (T1-T4) were pooled within disturbance treatment levels for each alpha diversity metric as the interaction term was not significant (sampling time x disturbance treatment, F4,76 = 1.76, p>0.081, Table S1). Different letters indicate significant differences between treatments obtained from post hoc tests (p<0.05).
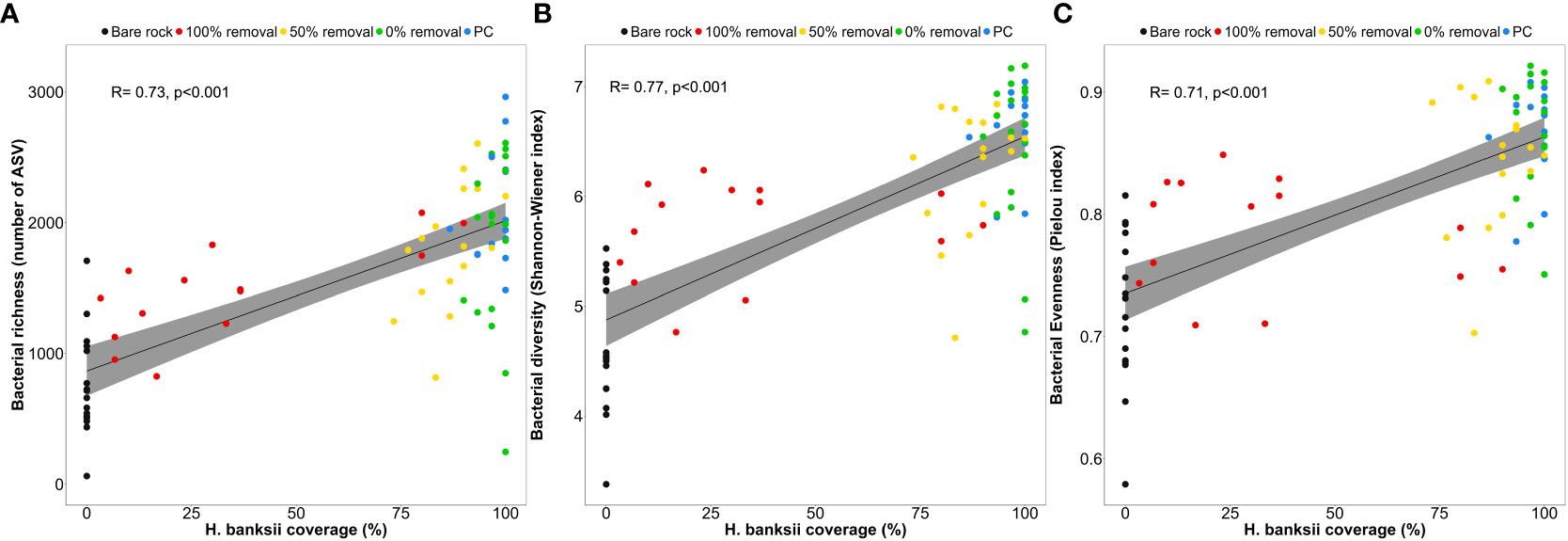
Figure 2 Relationship of % of Hormosira canopy cover on benthic bacterial alpha diversity indices including bacterial (A) richness, (B) diversity (Shannon-Wiener Index) and (C) evenness (Pielou Index); and evaluated across disturbance treatments (bare rock [n=18], 100% removal [n=17], 50% removal [n=18] and undisturbed control [0% removal, n=20], and procedural control [n=14]) and including all sampling times. Grey area represents the calculated confidence interval for each correlation (95%).
There was an interaction between disturbance and sampling time on the structure (i.e. composition and relative abundance of ASVs) of understory epilithic bacterial communities (Figure 3 and Table S3, PERMANOVA, F12,69 = 1.22, p<0.001). Structural intragroup variability among disturbance treatments was found (PERMDISP, Table S3: F4,76 = 3.35m, p=0.035), but only between the procedural controls and other disturbance treatments (see post hoc tests Table S4). Thus, this structural intragroup variability was not found to be a driver of the differences found with the PERMANOVA. The epilithic bacterial community structure on bare rock plots consistently differed from all other treatments at all sampling times, except T3, when they did not differ from plots with 50% removal and procedural controls (Figure 3 and Table S5). Bacterial community structure in 100% Hormosira removal plots was distinct from those on bare rock and other treatments at most sampling times (Figure 3, post hoc tests in Table S5), except for T3 when they did not differ from 50% removal and controls (p>0.09). No differences in epilithic bacterial community structure were found at any time point between control and 50% removal plots (Figure 3 and Table S5).
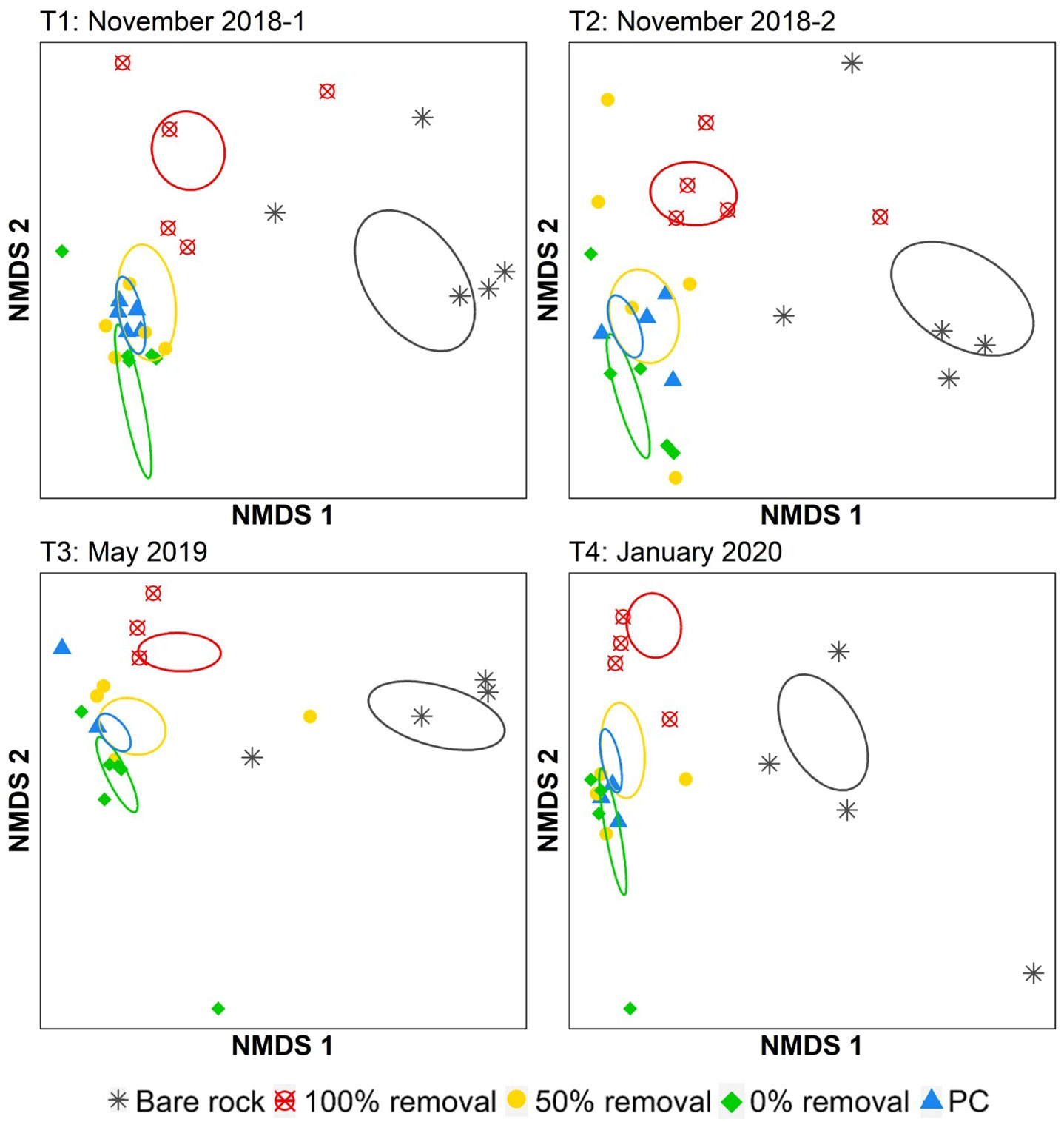
Figure 3 NMDS ordination of understory epilithic bacterial community structure calculated from Bray-Curtis dissimilarities (stress, T1 = 0.06, T2 = 0.09, T3 = 0.08 and T4 = 0.08) across all treatments for each time point. Ellipses shows the standard error of the centroids calculated from the mean centroid of each treatment category. PC, Procedural control.
A differential abundance analysis was used to identify epilithic bacterial ASVs that differed in abundance between Hormosira disturbance treatments and across time. For this first model, 40 bacterial ASV were identified to be affected by these factors, but only the 10 most abundant (0.06%< and >0.005%) and prevalent ASVs (>34% present across all samples) were selected. Results from this analysis show that soon after the removal of Hormosira (T1, November 2018-1), treatments with bare rock and 100% removal had similar abundances of bacterial groups such as Verrucomicrobiae (i.e., genus Rubritalea) and Bacteroidia (i.e., genera Winogradskyella, Olleya and an unidentified ASV of the family Flavobacteriaceae), but with lower relative abundance compared to other treatments (Figure S3). After T1, all of these ASV decreased in abundance across the following sampling times regardless of the Hormosira disturbance treatment, while other groups had a more complex temporal abundance pattern. For example, one ASV assigned to the genus Vibrio and one to unidentified Pirellulales, had a higher abundance in the control group compared to other removal treatments, but this pattern was not constant through time (Figure S4). Comparatively, another ASV assigned to the genus Vibrio, a Cetobacterium and an unidentified Phormidiaceae (Cyanobacteria), had a higher abundance in 100% Hormosira removal plots than in those assigned to other treatments, but only at some sampling times (Figure S5).
There were differences in epilithic bacterial community structure over time for treatments with 0% and 50% removal, as well as procedural controls (PERMANOVA, Table S3 and post hoc tests, Table S7). The differential abundance analysis focusing on the most prevalent ASV, showed a similar pattern with some groups increasing in the last two sampling times across all treatments, except bare rock (e.g., Unidentified genus of Pirelullales, Figure S4) while other ASV decreased substantially in the latter time points (e.g., genus Olleya, unidentified genus of Flavobacteriales, genus UBA9320 and one Vibrio, Figures S3–5). Relative abundances of the ten selected ASV remained constant through time and generally with lower abundances in bare rock compared to other Hormosira removal treatments.
3.2 Relationships between experimentally induced changes in Hormosira cover, invertebrate abundance and epilithic bacterial communities
The effect of the experimental disturbance to the canopy Hormosira changes through time (Figure S6 and Table S8; interaction, F12,76 = 19.79, p<0.001). After the initial disturbance, a lower canopy cover was predictably found in the plots with complete removal of Hormosira and bare rock compared to other disturbance treatments, a trend that continued throughout the experiment (Figure S6A and see post hoc tests in Table S9, overall t ratio<-3.04, p<0.03). However, treatments with complete removal of Hormosira experienced a rapid recovery in Hormosira cover from the time of disturbance to the end of the experiment (increase of 68 ± 8% cover from T1) (Figure S6B and post hoc tests in Table S10B, t ratio< -4.47, p<0.001). No differences in canopy cover were found between plots with 50% removal and both control treatments at any stage of the experiment (Figure S5A and Tables S9–10).
There were no significant differences in the covers of other understory macrobenthic components across disturbance treatments, although values of most groups (BLM, ACR, TW, TA and CCA) were generally lower in bare rock plots (Figures S7–9). The exception to this pattern were grazers, which were observed to have higher coverage values in bare rock plots compared to other treatments, and the amount of sand coverage which was higher in plots with 50% and 0% Hormosira removal compared to bare rock (Figures S7A–B). The dbRDA, marginal test and vector fit determined that Hormosira canopy cover was the main and most consistent driver of the structure of the understory epilithic bacterial communities across all sampling times albeit explaining a small amount of the total variation (Figure 4, and Table S11; marginal tests: p<0.015; envfit: r2 >0.82). Other understory benthic components such as rocky substrate, turfing algae, grazers, tube worms, brown laminate macroalgae, bleached and unbleached CCA, were found to influence the overall structure of the epilithic bacterial community across more than one sampling time (marginal tests: p<0.040) but to a lesser magnitude (Table S11, envfit: r2<0.43, p<0.006) or limited to specific sampling times (e.g., CCA, T3:May 2019, marginal tests: p=0.001 and envfit: r2 = 0.86, p=0.02).
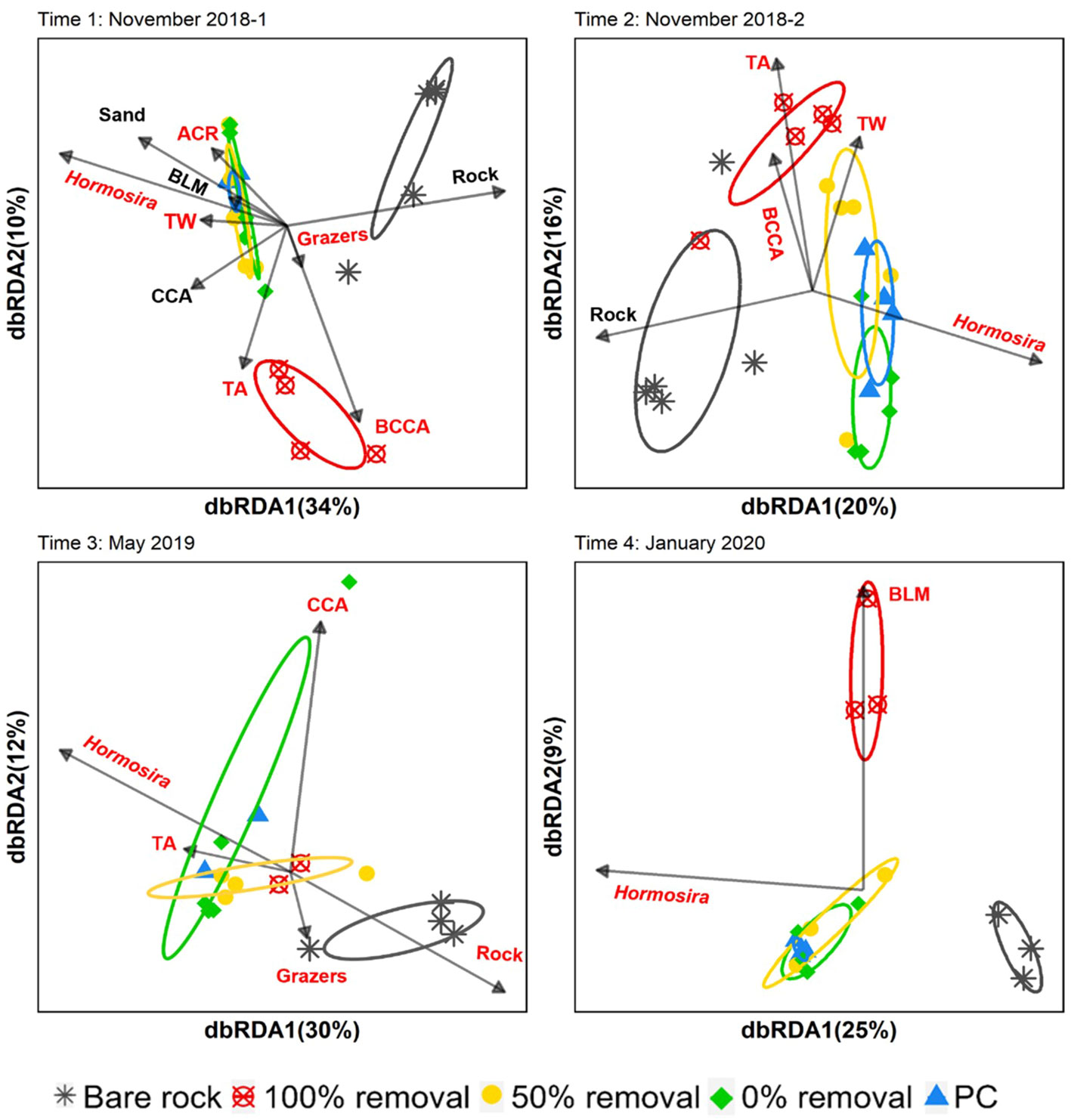
Figure 4 Distance-based redundancy analysis (dbRDA) ordination showing the relationship between the percent canopy cover and the benthic communities (Grazers, tubeworms [TW], crustose coralline algae [CCA], bleached crustose coralline algae [BCCA], sand, rock, turf algae [TA], brown laminate macroalgae [BLM] and articulate crustose red algae [ACR]), and the structure of the understory epilithic bacterial community (Bray-Curtis dissimilarities based on sqrt-transformed data). Ellipses shows the standard error of the centroids calculated from the mean centroid each treatment category. Arrows represent the linear relationship between each predictor variable and the community structure. Significant marginal tests are shown in red.
3.3 Comparison between epilithic and epiphytic bacterial biofilms
A total of 4519 bacterial ASVs across 44 samples were included in the second dataset that focused on a comparison between host associated epiphytic bacteria in Hormosira and the understory epilithic bacterial communities. These ASVs provided a robust representation of the sampled communities (Figure S2B).
There was a significant interaction among disturbance, sampling time and substratum type on bacterial richness (Figure S10A and Table S12, F3,40 = 3.13, p=0.05). Pairwise contrasts showed that bacterial richness was higher on rocky substrata than on the surface of Hormosira, consistently across sampling times and disturbance levels (summary of pairwise comparisons in Table S13). Likewise, bacterial diversity (F1,42 = 116.33, p< 0.001) and evenness (F1,42 = 67.35, p<0.001) were higher on rocky substrata than on Hormosira (Figures S10B, C and Table S12), but they did not vary with time or disturbance level.
Disturbance (F1,42 = 1.45, p<0.001), sampling time (F3,40 = 3.08, p<0.001) and substratum type (F1,42 = 31.26, p<0.001) independently influenced the structure of the whole bacterial community (PERMANOVA, Figure 5 and Table S14), despite variability among samples within each substratum types (Figure 5 and Table S14, PERMIDISP, F=66.76, df=1 and p=0.001). A total of 630 ASVs were found to differ significantly in relative abundance between the understory substrata and Hormosira surface samples. Analysis of the most abundant and prevalent bacterial ASVs revealed that the surface of Hormosira had higher relative abundances of the classes Bacteroidia (i.e., identified genera: Maribacter, Haliscomenobacter, Unidentified Saprospiraceae and Unidentified Bacteroidia), Verrucomicrobiae (i.e., genus Rubritalea), Alphaproteobacteria (i.e., genera Sulfitobacter and Octadecabacter), Gammaproteobacteria (i.e., Unidentified genus) and Aciidomicrobia (i.e., MedAcidi-G1) compared to the adjacent understory substrata (Figure S11). A full list of bacterial genera that differed between rocky substrata and Hormosira samples is presented in Table S15.
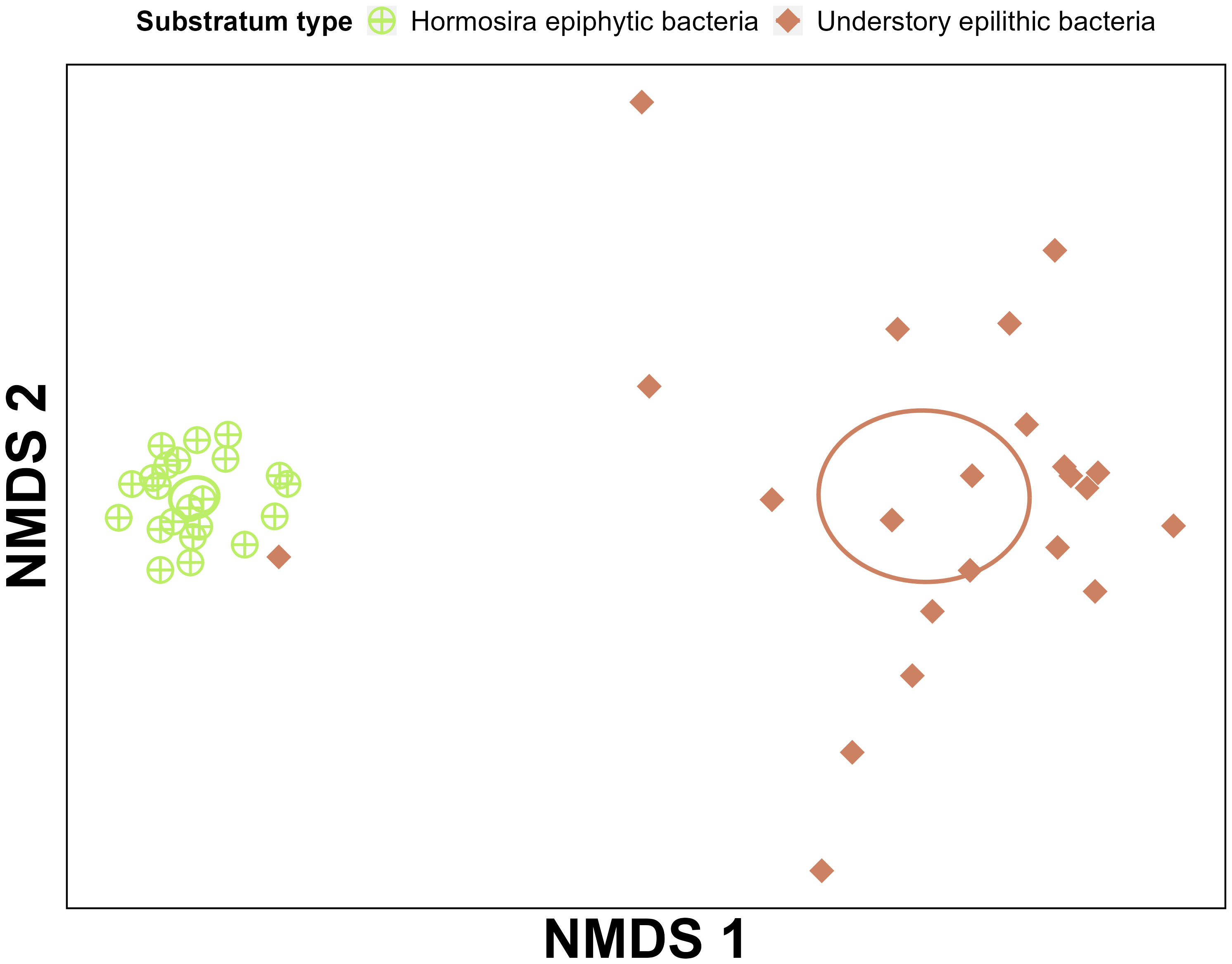
Figure 5 NMDS ordination comparing the epiphytic bacterial community structure associated to the surface of Hormosira and the understory epilithic biofilms. Ordination includes only data from treatments with 0% and 50% Hormosira removal and were calculated from Bray-Curtis dissimilarities (stress = 0.09). Ellipses shows the standard error of the centroids calculated from the mean centroid each treatment category.
4 Discussion
4.1 Disturbance of the canopy alters understory epilithic bacterial communities
Understory epilithic microbial communities are key components of biodiversity in seaweed-dominated systems, influencing the recruitment of seaweeds, invertebrates and regulating species interactions (Underwood, 1978; Pedicini et al., 2023; and Hawkins & Hartnoll, 1983). Understanding how and under which conditions these microbial changes may cascade through the system is critical to better predict and manage responses to environmental disturbances. Here, we found that bacterial changes were not related with the assemblage of understory macroorganisms, suggesting a direct effect of the macroalgae canopy removal.
Full, but not partial, removal of the habitat-forming seaweed Hormosira banksii had a strong impact on canopy cover and caused changes in the understory epilithic bacterial communities that persisted for ~1.5 years, despite the appearance of new Hormosira recruits. These fully cleared plots had the lowest bacterial richness (20-30% lower), diversity (10-14% lower), evenness (7-10% lower) and a different community structure compared to partially cleared (50% density) or undisturbed plots. Hormosira may facilitate the recruitment and development of macroalgae, including conspecifics, and other benthic macroorganisms through stress amelioration and a nursery effect given by its canopy cover (Cameron et al., 2021; Lewis et al., 2021), and likely have a similar effect on the bacterial communities beneath them. Complete removal of canopies may have influenced the structure of the epilithic bacterial community through a variety of mechanisms including through changes on the transfer rates of nutrients (e.g.DOC, labile sugars or hydrophobic molecules), allelochemical agents (e.g. antibacterial agents) or free-living microorganisms from the diffusive boundary layer of the blade or holdfast to the understory substratum (Pfister et al., 2019; Elsherbini et al., 2023).
In contrast to fully cleared plots, partial reductions of Hormosira density (i.e., 50% removal) had little effect on the understory epilithic bacterial communities. In these plots, canopy cover was still relatively high throughout the experiment (above 73% of cover) and probably offered a similar benthic understory environment to the canopy on undisturbed plots. Thus, a moderate thinning of canopies would have limited effects on the structure of the epilithic bacterial community since it would not significantly modify their chemically mediated effects nor their influence on environmental conditions. As shown in other systems (e.g., Cline et al., 2014 or Butitta et al., 2017), shifts between alternative states are often regulated by critical thresholds. Although very few studies have investigated transitions between alternative states on intertidal rocky shores, Rindi et al. (2018) showed that the shift in dominance from canopy to turf-forming macroalgae on a Mediterranean rocky shore was initiated by exceeding a reduction of 75% of the canopy cover. The responses of the microbial community to the total and 50% removal of Hormosira, generating covers of ~10% and ~73%, respectively, may suggest that the decline of Hormosira could cause marked changes to epilithic bacterial communities only when exceeding a critical threshold of cover.
Most of the epilithic bacterial groups on understory substrata are common in coastal waters (Freitas et al., 2012; Handley & Lloyd, 2013), sediments (e.g. genus Rubritalea, Lee et al., 2016) and below macroalgal canopies in temperate rocky systems (e.g. class Bacteroidia, Elsherbini et al., 2023). Fully cleared plots were characterized by an overall decrease in the relative abundance of ASVs assigned to the class Bacteroidia (i.e., genera from the family Flavobacteriaceae including Winogradskyella, Olleya, and an unidentified genus), Verrucomicrobiae (genus Rubritalea), Planctomycetes (unidentified genus of the order Pirellulales) and Gammaproteobacteria (genus Vibrio) to values more similar to those found in bare rock plots. The only exceptions to this pattern were the higher relative abundances of a Vibrio (three weeks post-disturbance) and a Cyanobacteria (seven months post-disturbance; ASV assigned to the family Phormidiaceae) and the lower abundance of a Cetobacterium in fully cleared plots compared to bare rocks. However, the fluctuations in relative abundance among predominant epilithic bacterial groups were not consistent over time. This can be attributed to the influence of seasonal environmental factors, such as temperature and nutrient concentration, which likely exert their effects with a certain degree of stochasticity (Antunes et al., 2019; and Caruso, 2020).
The functional relevance of prevalent bacterial groups that had lower relative abundances in cleared plots throughout the experiment is still understudied (Dogs et al., 2017). However, it is likely that most of these groups play an important role in carbon cycling as copiothrophic bacteria (e.g., family Flavobacteriaceae; Elsherbini et al., 2023 or the genus Vibrio, Nelson et al., 2013) and their abundance may be reflective of the organic enrichment produced by Hormosira on the understory substratum (Anderson, 2016). Future research is needed to further understand the functional implications that the loss these macroalgae may have on intertidal rocky systems.
4.2 Canopy recovery and understory bacterial communities
Changes in the bacterial communities caused by the experimental reduction of Hormosira density and cover did not appear to affect the subsequent recovery of the macroalgae, which occurred through the recruitment of juveniles. Indeed, Hormosira cover in fully cleared plots increased to an average of 83 ± SE 2.7% after ~16 months since the start of the experiment (Figure S6, a total increase of 75 ± SE 6% of coverage across all 0% density plots), but the space in the plots was mainly occupied by juvenile individuals which did not form a true canopy. The relatively slow recovery of a full canopy by Hormosira post-disturbance is consistent with other experimental studies, with recovery to full adult size (>80 mm) expected to occur within 2 to 5 years although this is tightly linked to the type and magnitude of the disturbance (Keough and Quinn, 1998; Underwood, 1998; Schiel and Taylor, 1999; Lilley and Schiel, 2006; Pocklington et al., 2019; Cameron et al., 2021; Lewis et al., 2021). This may explain why bacterial communities in these plots remained different throughout the entire experiment. However, it could also be predicted that the nursery effects of Hormosira will be restored following the growth of these juvenile sporophytes, possibly facilitating the recovery of the structure of the original microbial biofilm (Park et al., 2011).
Recent studies on epilithic bacterial communities have shown strong effects of urbanisation on their composition, which in turn negatively influenced recruitment of habitat-forming macroalgae (Pedicini et al., 2023). In our study, however, Hormosira recruitment was observed across all cleared plots suggesting that observed changes in the understory bacteria did not affect recruitment of Hormosira. This could be due to the persistence of a potentially “core” component of the microbial community in the remnant biofilm which may play a critical role in influencing the rate of Hormosira recruitment. A list of the most prevalent bacterial ASVs (frequency> 95%) across disturbance treatments (0, 50 and 100% Hormosira removal) and sampling times is given in Table S16. These prevalent bacterial taxa could potentially represent a part of this “core” bacterial community; however, this finding still remains speculative. The prevalence of these taxa might not be directly linked to a functional role that is essential to Hormosira recruitment or survival after a disturbance, but rather, a process of colonization occurring homogenously across the intertidal rocky habitat (Burke et al., 2011 and discussed further below).
4.3 Differences between epiphytic and epilithic bacterial biofilms
Little is known about the mechanisms and factors that influence the establishment and structure of understory epilithic bacterial communities, for example whether bacteria that colonize the substratum are sourced from the water column or from nearby macro-organisms such as the canopy-forming macroalgae. Here, Hormosira-associated epiphytic bacterial communities were found to be mostly distinct and less diverse compared to understory epilithic bacterial communities, suggesting that few components of the understory microbial communities are supplied by Hormosira. Many recent studies have found that epiphytic microbes associated with macroalgae differ from the surrounding environment such as water and rocks (Lachnit et al., 2011; Roth-Schulze et al., 2016; Weigel et al., 2022). Genera such as Rubritalea, Maribacter, Octadecabacter and Haliscomenobacter were enriched on Hormosira and have also been found associated with other subtidal and intertidal macroalgae where they play an important role in algal growth, nutrition and resistance to stress (Lin et al., 2018; Quigley et al., 2018; Samo et al., 2018; Capistrant-Fossa et al., 2021) The structure of epyphtic bacterial communities could thus be determined by selective processes driven by Hormosira (e.g., production of secondary metabolites by Hormosira, Egan et al., 2013 and Roth-Schulze et al., 2016) It has also been suggested that the key level at which to address the assembly of bacterial communities may not be at a taxonomic (i.e., ASV) level but rather at the level of core functional genes (Burke et al., 2011). In this model, bacterial communities may originate from the water column (e.g., via local colonization by incoming tidal waterflows) and communities are subsequently shaped by neutral/random processes and competition with other microbes, retaining a core residual function independent of taxa (e.g. competitive lottery model, Burke et al., 2011). Functional genes associated with macroalgal recruitment have been found in many bacterial taxa, for example the N-acylhomoserine lactone gene (AHL) that has been associated with enhanced settlement of Ulva zoospores through quorum sensing (Joint et al., 2002). Characterisation of core functional genes in the epiphytic versus epilithic communities may thus provide an interesting avenue for future research.
5 Conclusion
Disturbances and declines of macroalgal canopies occurring globally are likely to have consequences for benthic epilithic bacterial communities (Pedicini et al., 2023). In this study, we were able to determine strong effects of Hormosira canopy removal on bacterial communities associated with the understory substrata, but little evidence that other macrobenthic components, including macroalgae and mobile invertebrates, played an important role in shaping these communities. Plots with complete removal of Hormosira maintained a distinct bacterial community from that found on adjacent bare rock and could provide a suitable biofilm that supports new algal recruitment and development. However, macroalgal recovery after disruption can be slow, as it may rely upon extant biofilms and nursing effects of nearby or regrowing canopy cover to produce favourable conditions. Whether larger-scale or subsequent disturbances would further impact this ability to recover remains to be explored. Future work needs to focus on how long changes in the understory bacterial community remain after disruption of canopies by major disturbance events and whether induced changes in specific bacterial taxa translate into functional changes that could substantially impair the resilience of the system.
Data availability statement
The data presented in the study are deposited in the Dryad repository, accession number https://doi.org/10.5061/dryad.g1jwstqww.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SG: Conceptualization, Formal Analysis, Visualization, Writing – original draft, Writing – review & editing. GW: Conceptualization, Formal Analysis, Visualization, Writing – original draft, Writing – review & editing. HT: Conceptualization, Investigation, Writing – review & editing, Writing – original draft. KL: Conceptualization, Investigation, Writing – review & editing, Writing – original draft. MM-P: Conceptualization, Writing – review & editing, Writing – original draft. FL: Conceptualization, Writing – review & editing, Writing – original draft. SK: Funding acquisition, Resources, Writing – original draft, Writing – review & editing. FB: Conceptualization, Writing – original draft, Writing – review & editing. PS: Conceptualization, Funding acquisition, Resources, Writing – review & editing. EM: Conceptualization, Funding acquisition, Resources, Writing – review & editing, Investigation, Methodology, Project administration, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Australian Research Council funds to EM, PS and SK (DP180104041) and the Nanyang Technological University Undergraduate Research Experience on Campus to HT.
Acknowledgments
We thank Garance Ronot, Juliette Prothon, Renske Jongen for field assistance and Madeleine Langley for DNA extractions and PCRs.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1264797/full#supplementary-material
References
Althaus F., Hill N., Ferrari R., Edwards L., Przeslawski R., Schönberg C. H. L., et al. (2015). A standardised vocabulary for identifying benthic biota and substrata from underwater imagery: the CATAMI classification scheme. PloS One 10 (10), e0141039. doi: 10.1371/journal.pone.0141039
Anderson O. R. (2016). Marine and estuarine natural microbial biofilms: ecological and biogeochemical dimensions. Aims Microbiol. 2 (3), 304–331. doi: 10.3934/microbiol.2016.3.304
Antunes J., Leão P., Vasconcelos V. (2019). Marine biofilms: diversity of communities and of chemical cues. Environ. Microbiol. Rep. 11 (3), 287–305. doi: 10.1111/1758-2229.12694
Arboleda-Baena C., Pareja C. B., Pla I., Logares R., de la Iglesia R., Navarrete S. A. (2022). Hidden interactions in the intertidal rocky shore: variation in pedal mucus microbiota among marine grazers that feed on epilithic biofilm communities. PeerJ 10, e13642. doi: 10.7717/peerj.13642
Banks S. C., Cary G. J., Smith A. L., Davies I. D., Driscoll D. A., Gill A. M., et al. (2013). How does ecological disturbance influence genetic diversity? Trends Ecol. Evol. 28 (11), 670–679. doi: 10.1016/j.tree.2013.08.005
Beijbom O., Edmunds P. J., Roelfsema C., Smith J., Kline D. I., Neal B. P., et al. (2015). Towards automated annotation of benthic survey images: Variability of human experts and operational modes of automation. PloS One 10 (7), e0130312. doi: 10.1371/journal.pone.0130312
Bellgrove A., McKenzie P. F., McKenzie J. L., Sfiligoj B. J. (2010). Restoration of the habitat-forming fucoid alga Hormosira banksii at effluent-affected sites: competitive exclusion by coralline turfs. Mar. Ecol. Prog. Ser. 419, 47–56. doi: 10.3354/meps08843
Benedetti-Cecchi L., Pannacciulli F., Bulleri F., Moschella P. S., Airoldi L., Relini G., et al. (2001). Predicting the consequences of anthropogenic disturbance: large-scale effects of loss of canopy algae on rocky shores. Mar. Ecol. Prog. Ser. 214, 137–150. doi: 10.3354/meps214137
Bever J. D., Westover K. M., Antonovics J. (1997). Incorporating the soil community into plant population dynamics: the utility of the feedback approach. J. Ecol. 85, 561–573. doi: 10.2307/2960528
Brooker R. W., Maestre F. T., Callaway R. M., Lortie C. L., Cavieres L. A., Kunstler G., et al. (2008). Facilitation in plant communities: the past, the present, and the future. J. Ecol. 96 (1), 18–34. doi: 10.1111/j.1365-2745.2007.01295.x
Bulleri F., Thiault L., Mills S. C., Nugues M. M., Eckert E. M., Corno G., et al. (2018). Erect macroalgae influence epilithic bacterial assemblages and reduce coral recruitment. Mar. Ecol. Prog. Ser. 597, 65–77. doi: 10.3354/meps12583
Burke C., Steinberg P., Rusch D., Kjelleberg S., Thomas T. (2011). Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. 108 (34), 14288–14293. doi: 10.1073/pnas.1101591108
Butitta V. L., Carpenter S. R., Loken L. C., Pace M. L., Stanley E. H. (2017). Spatial early warning signals in a lake manipulation. Ecosphere 8 (10), e01941. doi: 10.1002/ecs2.1941
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13 (7), 581–583. doi: 10.1038/nmeth.3869
Cameron H., Amor M. D., Bellgrove A. (2021). Barriers to restoration: Pollution alters nurse effects for an ecosystem engineer. J. Appl. Ecol. 58 (12), 2783–2796. doi: 10.1111/1365-2664.14010
Capistrant-Fossa K. A., Morrison H. G., Engelen A. H., Quigley C. T. C., Morozov A., Serrão E. A., et al. (2021). The microbiome of the habitat-forming brown alga Fucus vesiculosus (Phaeophyceae) has similar cross-Atlantic structure that reflects past and present drivers1. J. Phycol. 57 (6), 1681–1698. doi: 10.1111/jpy.13194
Caruso G. (2020). Microbial colonization in marine environments: overview of current knowledge and emerging research topics. J. Mar. Sci. Eng. 8 (2), 78. doi: 10.3390/jmse8020078
Castorani M. C. N., Reed D. C., Miller R. J. (2018). Loss of foundation species: Disturbance frequency outweighs severity in structuring kelp forest communities. Ecology 99 (11), 2442–2454. doi: 10.1002/ecy.2485
Cline T. J., Seekell D. A., Carpenter S. R., Pace M. L., Hodgson J. R., Kitchell J. F., et al. (2014). Early warnings of regime shifts: evaluation of spatial indicators from a whole-ecosystem experiment. Ecosphere 5 (8), 1–13. doi: 10.1890/ES13-00398.1
Coleman M. A., Kennelly S. J. (2019). Microscopic assemblages in kelp forests and urchin barrens. Aquat. Bot. 154, 66–71. doi: 10.1016/j.aquabot.2019.01.005
Coleman M. A., Minne A. J., Vranken S., Wernberg T. (2020). Genetic tropicalisation following a marine heatwave. Sci. Rep. 10 (1), 12726. doi: 10.1038/s41598-020-69665-w
Coleman R. A., Underwood A. J., Benedetti-Cecchi L., Åberg P., Arenas F., Arrontes J., et al. (2006). A continental scale evaluation of the role of limpet grazing on rocky shores. Oecologia 147, 556–564. doi: 10.1007/s00442-005-0296-9
Crowe T. P., Cusson M., Bulleri F., Davoult D., Arenas F., Aspden R., et al. (2013). Large-scale variation in combined impacts of canopy loss and disturbance on community structure and ecosystem functioning. PloS One 8 (6), e66238. doi: 10.1371/journal.pone.0066238
Dogs M., Wemheuer B., Wolter L., Bergen N., Daniel R., Simon M., et al. (2017). Rhodobacteraceae on the marine brown alga Fucus spiralis are abundant and show physiological adaptation to an epiphytic lifestyle. Systematic Appl. Microbiol. 40 (6), 370–382. doi: 10.1016/j.syapm.2017.05.006
Egan S., Harder T., Burke C., Steinberg P., Kjelleberg S., Thomas T. (2013). The seaweed holobiont: Understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 37 (3), 462–476. doi: 10.1111/1574-6976.12011
Elsherbini J., Corzett C., Ravaglioli C., Tamburello L., Polz M., Bulleri F. (2023). Epilithic bacterial assemblages on subtidal rocky reefs: variation among alternative habitats at ambient and enhanced nutrient levels. Microbial Ecol., 86, 1–13. doi: 10.1007/s00248-023-02174-1
Fairweather P. G. (1990). Sewage and the biota on seashores: assessment of impact in relation to natural variability. Environ. Monit. Assess. 14, 197–210. doi: 10.1007/BF00677916
Filbee-Dexter K., Scheibling R. E. (2014). Sea urchin barrens as alternative stable states of collapsed kelp ecosystems. Mar. Ecol. Prog. Ser. 495, 1–25. doi: 10.3354/meps10573
Flemming H. C., Wingender J., Szewzyk U., Steinberg P., Rice S. A., Kjelleberg S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14 (9), 563–575. doi: 10.1038/nrmicro.2016.94
Flemming H. C., Wuertz S. (2019). Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17 (4), 247–260. doi: 10.1038/s41579-019-0158-9
Folke C., Carpenter S., Walker B., Scheffer M., Elmqvist T., Gunderson L., et al. (2004). Regime shifts, resilience, and biodiversity in ecosystem management. Annu. Rev. Ecol. Evol. Syst. 35, 557–581. doi: 10.1146/annurev.ecolsys.35.021103.105711
Fraterrigo J. M., Rusak J. A. (2008). Disturbance-driven changes in the variability of ecological patterns and processes. Ecol. Lett. 11 (7), 756–770. doi: 10.1111/j.1461-0248.2008.01191.x
Freitas S., Hatosy S., Fuhrman J. A., Huse S. M., Mark Welch D. B., Sogin M. L., et al. (2012). Global distribution and diversity of marine Verrucomicrobia. ISME J. 6 (8), 1499–1505. doi: 10.1038/ismej.2012.3
Gorman D., Russell B. D., Connell S. D. (2009). Land-to-sea connectivity: linking human-derived terrestrial subsidies to subtidal habitat change on open rocky coasts. Ecol. Appl. 19 (5), 1114–1126. doi: 10.1890/08-0831.1
Hadfield M. G. (2011). Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Annu. Rev. Mar. Sci. 3, 453–470. doi: 10.1146/annurev-marine-120709-142753
Hamman S. T., Hawkes C. V. (2013). Biogeochemical and microbial legacies of non-native grasses can affect restoration success. Restor. Ecol. 21 (1), 58–66. doi: 10.1111/j.1526-100X.2011.00856.x
Handley K., Lloyd J. (2013). Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by Marinobacter species. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00136
Hawkins S. J. (1981). The influence of season and barnacles on the algal colonization of Patella vulgata exclusion areas. J. Mar. Biol. Assoc. United Kingdom 61 (1), 1–15. doi: 10.1017/S0025315400045872
Hawkins S. J., Hartnoll R. G. (1983). Grazing of intertidal algae by marine invertebrates. Oceanogr. Mar. biol.: an Annu. Rev. 21, 195–282. Available at: https://api.semanticscholar.org/CorpusID:87723063.
Iguchi S. M., Aikawa T., Matsumoto J. J. (1982). Antibacterial activity of snail mucus mucin. Comp. Biochem. Physiol. Part A: Physiol. 72 (3), 571–574. doi: 10.1016/0300-9629(82)90123-2
Joint I., Tait K., Callow M. E., Callow J. A., Milton D., Williams P., et al. (2002). Cell-to-cell communication across the prokaryote-eukaryote boundary. Science 298 (5596), 1207–1207. doi: 10.1126/science.1077075
Kain J. M. (2015). Hormosira banksii (Phaeophyceae): A tough survivor in the harsh conditions of high intertidal pools in southeast Australia. Eur. J. Phycol. 50 (4), 408–421. doi: 10.1080/09670262.2015.1075594
Keough M. J., Quinn G. P. (1998). Effects of periodic disturbances from trampling onrocky intertidal algal beds. Ecol. Appl. 8 (1), 141–161. doi: 10.1890/1051-0761(1998)008[0141:EOPDFT]2.0.CO;2
Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41 (1), e1. doi: 10.1093/nar/gks808
Kuznetsova A., Brockhoff P. B., Christensen R. H. B. (2017). lmerTest package: tests in linear mixed effects models. J. Stat. Software 82 (13), 1–26. doi: 10.18637/jss.v082.i13
Lachnit T., Meske D., Wahl M., Harder T., Schmitz R. (2011). Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13 (3), 655–665. doi: 10.1111/j.1462-2920.2010.02371.x
Lee Y. M., Kim M.-K., Ahn D. H., Kim H.-W., Park H., Shin S. C. (2016). Comparative analysis of lacinutrix genomes and their association with bacterial habitat. PloS One 11 (2), e0148889. doi: 10.1371/journal.pone.0148889
Lewis R. D., Johnson C. R., Wright J. T. (2021). Demography of the intertidal fucoid hormosira banksii: importance of recruitment to local abundance. J. Phycol. 57 (2), 664–676. doi: 10.1111/jpy.13124
Lilley S. A., Schiel D. R. (2006). Community effects following the deletion of a habitat-forming alga from rocky marine shores. Oecologia 148 (4), 672–681. doi: 10.1007/s00442-006-0411-6
Lin J. D., Lemay M. A., Parfrey L. W. (2018). Diverse bacteria utilize alginate within the microbiome of the giant kelp macrocystis pyrifera. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01914
Ling S. D., Scheibling R. E., Rassweiler A., Johnson C. R., Shears N., Connell S. D., et al. (2015). Global regime shift dynamics of catastrophic sea urchin overgrazing. Philos. Trans. R. Soc. B: Biol. Sci. 370 (1659), 20130269. doi: 10.1098/rstb.2013.0269
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Lüdecke D., Ben-Shachar M. S., Patil I., Waggoner P., Makowski D. (2021). performance: An R package for assessment, comparison and testing of statistical models. J. Open Source Software 6 (60). doi: 10.21105/joss.03139
Luke S. G. (2017). Evaluating significance in linear mixed-effects models in R. Behav. Res. Methods 49 (4), 1494–1502. doi: 10.3758/s13428-016-0809-y
Marzinelli E. M., Burrows M. T., Jackson A. C., Mayer-Pinto M. (2012). Positive and negative effects of habitat-forming algae on survival, growth and intra-specific competition of limpets. PloS One 7 (12), e51601. doi: 10.1371/journal.pone.0051601
Marzinelli E. M., Campbell A. H., Zozaya Valdes E., Vergés A., Nielsen S., Wernberg T., et al. (2015). Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol. 17 (10), 4078–4088. doi: 10.1111/1462-2920.12972
Mcmurdie P. J., Holmes S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8 (4), e61217. doi: 10.1371/journal.pone.0061217
Narwani A., Reyes M., Pereira A. L., Penson H., Dennis S. R., Derrer S., et al. (2019). Interactive effects of foundation species on ecosystem functioning and stability in response to disturbance. Proc. R. Soc. B: Biol. Sci. 286 (1913), 20191857. doi: 10.1098/rspb.2019.1857
Nelson C. E., Goldberg S. J., Wegley Kelly L., Haas A. F., Smith J. E., Rohwer F., et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 7 (5), 962–979. doi: 10.1038/ismej.2012.161
Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’hara R. B., et al. (2013). Package ‘vegan’. Community ecology package, version, Vol. 2. 1–295. Available at: https://github.com/vegandevs/vegan.
Park S. R., Kang Y. H., Choi C. G. (2011). Biofilm: A crucial factor affecting the settlement of seaweed on intertidal rocky surfaces. Estuarine Coast. Shelf Sci. 91 (1), 163–167. doi: 10.1016/j.ecss.2010.10.013
Parks D. H., ChuvoChina M., Rinke C., Mussig A. J., Chaumeil P.-A., Hugenholtz P. (2022). GTDB: An ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 50 (D1), D785–D794. doi: 10.1093/nar/gkab776
Patel P., Callow M. E., Joint I., Callow J. A. (2003). Specificity in the settlement–modifying response of bacterial biofilms towards zoospores of the marine alga Enteromorpha. Environ. Microbiol. 5 (5), 338–349. doi: 10.1046/j.1462-2920.2003.00407.x
Pedicini L., Vannini C., Rindi F., Ravaglioli C., Bertocci I., Bulleri F. (2023). Variations in epilithic microbial biofilm composition and recruitment of a canopy-forming alga between pristine and urban rocky shores. Mar. Environ. Res., 188, 106035. doi: 10.1016/j.marenvres.2023.106035
Peters D. P. C., Lugo A. E., Chapin F. S., Pickett S. T. A., Duniway M., Rocha A. V., et al. (2011). Cross-system comparisons elucidate disturbance complexities and generalities. Ecosphere 2 (81), 1–26. doi: 10.1890/ES11-00115.1
Pfister C. A., Altabet M. A., Weigel B. L. (2019). Kelp beds and their local effects on seawater chemistry, productivity, and microbial communities. Ecology 100 (10), e02798. doi: 10.1002/ecy.2798
Pickett S. T. A., White P. S. (1985). The ecology of natural disturbance and patch dynamics (Academic Press). doi: 10.1016/C2009-0-02952-3
Pocklington J. B., Keough M. J., O’Hara T. D., Bellgrove A. (2019). The influence of canopy cover on the ecological function of A key autogenic ecosystem engineer. Diversity 11 (5), 79. doi: 10.3390/d11050079
Poore A. G., Campbell A. H., Coleman R. A., Edgar G. J., Jormalainen V., Reynolds P. L., et al. (2012). Global patterns in the impact of marine herbivores on benthic primary producers. Ecol. Lett. 15 (8), 912–922. doi: 10.1111/j.1461-0248.2012.01804.x
Povey A., Keough M. J. (1991). Effects of trampling on plant and animal populations on rocky shores [Hormosira mats]. Oikos 61 (3), 355–368. doi: 10.2307/3545243
Prodan A., Tremaroli V., Brolin H., Zwinderman A. H., Nieuwdorp M., Levin E. (2020). Comparing bioinformatic pipelines for microbial 16S rRNA amplicon sequencing. PloS One 15 (1), e0227434. doi: 10.1371/journal.pone.0227434
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41 (D1), D590–D596. doi: 10.1093/nar/gks1219
Quigley C. T. C., Morrison H. G., Mendonça I. R., Brawley S. H. (2018). A common garden experiment with Porphyra umbilicalis (Rhodophyta) evaluates methods to study spatial differences in the macroalgal microbiome. J. Phycol. 54 (5), 653–664. doi: 10.1111/jpy.12763
Rao D., Webb J. S., Holmström C., Case R., Low A., Steinberg P., et al. (2007). Low densities of epiphytic bacteria from the marine alga Ulva australis inhibit settlement of fouling organisms. Appl. Environ. Microbiol. 73 (24), 7844–7852. doi: 10.1128/AEM.01543-07
Rindi L., Dal Bello M., Benedetti-Cecchi L. (2018). Experimental evidence of spatial signatures of approaching regime shifts in macroalgal canopies. Ecology 99, 1709–1715. doi: 10.1002/ecy.2391
Roth-Schulze A. J., Zozaya-Valdés E., Steinberg P. D., Thomas T. (2016). Partitioning of functional and taxonomic diversity in surface-associated microbial communities. Environ. Microbiol. 18 (12), 4391–4402. doi: 10.1111/1462-2920.13325
Roush D., Garcia-Pichel F. (2020). Succession and colonization dynamics of endolithic phototrophs within intertidal carbonates. Microorganisms 8 (2), 214. doi: 10.3390/microorganisms8020214
Russell V.L. (2022). emmeans: Estimated Marginal Means, aka Least-Squares Means (R package version 1.7.4-1). Available at: https://CRAN.R-project.org/package=emmeans.
Samo T. J., Kimbrel J. A., Nilson D. J., Pett-Ridge J., Weber P. K., Mayali X. (2018). Attachment between heterotrophic bacteria and microalgae influences symbiotic microscale interactions. Environ. Microbiol. 20 (12), 4385–4400. doi: 10.1111/1462-2920.14357
Schiel D. R., Lilley S. A. (2007). Gradients of disturbance to an algal canopy and the modification of an intertidal community. Mar. Ecol. Prog. Ser. 339, 1–11. doi: 10.3354/meps339001
Schiel D. R., Taylor D. I. (1999). Effects of trampling on a rocky intertidal algal assemblage in southern New Zealand. J. Exp. Mar. Biol. Ecol. 235 (2), 213–235. doi: 10.1016/S0022-0981(98)00170-1
Strain E. M., Thomson R. J., Micheli F., Mancuso F. P., Airoldi L. (2014). Identifying the interacting roles of stressors in driving the global loss of canopy-forming to mat-forming algae in marine ecosystems. Global Change Biol. 20 (11), 3300–3312. doi: 10.1111/gcb.12619
Swift D., Cresswell K., Johnson R., Stilianoudakis S., Wei X. (2023). A review of normalization and differential abundance methods for microbiome counts data. Wiley Interdiscip. Reviews: Comput. Stat 15 (1), e1586. doi: 10.1002/wics.1586
Takolander A., Cabeza M., Leskinen E. (2017). Climate change can cause complex responses in Baltic Sea macroalgae: A systematic review. J. Sea Res. 123, 16–29. doi: 10.1016/j.seares.2017.03.007
Underwood A. J. (1978). An experimental evaluation of competition between three species of intertidal prosobranch gastropods. Oecologia 33, 185–202. doi: 10.1007/BF00344847
Underwood A. J. (1998). Grazing and disturbance: An experimental analysis of patchiness in recovery from a severe storm by the intertidal alga Hormosira banksii on rocky shores in New South Wales. J. Exp. Mar. Biol. Ecol. 231 (2), 291–306. doi: 10.1016/S0022-0981(98)00091-4
Valentine J. P., Johnson C. R. (2003). Establishment of the introduced kelp Undaria pinnatifida in Tasmania depends on disturbance to native algal assemblages. J. Exp. Mar. Biol. Ecol. 295, 63–90. doi: 10.1016/S0022-0981(03)00272-7
Van der Putten W. H., Bardgett R. D., Bever J. D., Bezemer T. M., Fukami T., Kardol P., et al. (2013). Plant-soil feedbacks: the past, the present and future challenges. J. Ecol. 101, 265–276. doi: 10.1111/1365-2745
Wang Y. I., Naumann U., Wright S. T., Warton D. I. (2012). mvabund–an R package for model-based analysis of multivariate abundance data. Methods Ecol. Evol. 3 (3), 471–474. doi: 10.1111/j.2041-210X.2012.00190.x
Wei T., Simko V. (2021) R package 'corrplot': Visualization of a Correlation Matrix (Version 0.92). Available at: https://github.com/taiyun/corrplot.
Weigel B. L., Miranda K. K., Fogarty E. C., Watson A. R., Pfister C. A. (2022). Functional insights into the kelp microbiome from metagenome-assembled genomes. MSystems 7 (3), e01422–e01421. doi: 10.1128/msystems.01422-21
Wilson S. D., Tilman D. (1993). Plant competition and resource availability in response to disturbance and fertilization. Ecology 74 (2), 599–611. doi: 10.2307/1939319
Keywords: bacteria, understory, seaweed, Hormosira banksii, disturbance, rocky shore, microbiome
Citation: Vadillo Gonzalez S, Wood G, Tiong HYR, Lema KA, Mayer-Pinto M, Lauro FM, Kjelleberg S, Bulleri F, Steinberg PD and Marzinelli EM (2023) Effect of seaweed canopy disturbance on understory microbial communities on rocky shores. Front. Mar. Sci. 10:1264797. doi: 10.3389/fmars.2023.1264797
Received: 21 July 2023; Accepted: 31 October 2023;
Published: 20 November 2023.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Tania Aires, University of Algarve, PortugalGianmaria Califano, University of Lisbon, Portugal
Copyright © 2023 Vadillo Gonzalez, Wood, Tiong, Lema, Mayer-Pinto, Lauro, Kjelleberg, Bulleri, Steinberg and Marzinelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian Vadillo Gonzalez, c2ViYXN0aWFuLnZhZGlsbG9Ac3lkbmV5LmVkdS5hdQ==; Georgina Wood, Z2VvcmdlLndvb2RAdXdhLmVkdS5hdQ==
†These authors have contributed equally to this work and share first authorship
 Sebastian Vadillo Gonzalez
Sebastian Vadillo Gonzalez Georgina Wood
Georgina Wood Hui Yi Regine Tiong4
Hui Yi Regine Tiong4 Kimberley A. Lema
Kimberley A. Lema Mariana Mayer-Pinto
Mariana Mayer-Pinto Federico M. Lauro
Federico M. Lauro Staffan Kjelleberg
Staffan Kjelleberg Fabio Bulleri
Fabio Bulleri Peter D. Steinberg
Peter D. Steinberg Ezequiel M. Marzinelli
Ezequiel M. Marzinelli