- 1Tianjin Key Laboratory of Animal and Plant Resistance, Tianjin Key Laboratory of Conservation and Utilization of Animal Diversity, Tianjin Normal University, Tianjin, China
- 2School of Marine Science and Technology, Tianjin University, Tianjin, China
- 3College of Marine and Environmental Sciences, Tianjin University of Science and Technology, Tianjin, China
Regular oxygen depletion is occurring every summer in the depression area of the Bohai Sea. The community structure and potential functions of microbes in expanding marine hypoxic area are of great importance due to their roles in biogeochemical cycling. In this study, the diversity and distribution of bacteria based on 16S rRNA gene in sediment along an inshore-offshore transect across the oxygen-depletion area in the Bohai Sea was investigated in June, July and August of 2018 by employing high-throughput sequencing. Results revealed that the bacteria community was dominated by Proteobacteria (42.67%), Actinobacteria (14.13%), Chloroflexi (13.02%), Acidobacteria (8.01%), and Bacteroidetes (6.30%). During oxygen depletion, the bacteria community from inshore site A3 subjected to dramatic variation from June to August, but the composition tended to be stable in sites from the depression area along the transect. Distinct switch of bacteria from aerobic to anaerobic group was observed when the DO concentration <4.2 mg/L, typically represented by dominance of Anaerolineaceae in August sample. Further, KEGG prediction by PICRUSt confirmed the variations by showing significant difference in functional pathways, especially the nitrogen metabolisms, before and after DO depletion (p < 0.05). These variations could be influenced by depth, NO2– concentration and DO availability based on RDA analysis. The details in diversity and composition of bacteria under continually observation provide insights into both instant and long-term response of bacteria community to oxygen depletion, and the distinct functional switch under this process expands our knowledge on the metabolic character of bacteria in worldwide hypoxia areas.
Introduction
As a vital part of marine ecosystem, sediments are impacted by geological, hydrodynamical, physicochemical and biological processes (Köster and Meyer-Reil, 2001) and acting as both source and sink of adsorbed nutrients (Zhou et al., 2017), heavy metals (Dou et al., 2013), and persistent hydrophobic organics (Lofrano et al., 2017). The microbes that living in the sediments are responsible for various biogeochemical transformations and organic matter decomposition by means of uptake and release of chemicals (Petro et al., 2017). Due to excessive anthropogenic nitrogen inputs resulted from application of synthetic nitrogenous fertilizers and industry activity, nitrogen pollution in coastal ecosystem has led to deleterious ecological changes (Canfield et al., 2010; Cui et al., 2013). The negative effects including eutrophication and harmful algal blooms (Anderson et al., 2002; Bricker et al., 2008), expanding hypoxia and anoxic zones (Diaz and Rosenberg, 2008; Paulmier and Ruiz-Pino, 2009; Breitburg et al., 2018) are increasing in high frequency. Additionally, as a key environmental factor, dissolved oxygen (DO) affects biochemical processes of marine organisms and also acts as one of the vital drivers in the global carbon and nitrogen cycling (Voss et al., 2013; Mahaffey et al., 2020). Bacteria is an important contributor to the net loss of fixed nitrogen in oxygen minimum zones (OMZ) of oceans (Lam et al., 2009; Ward et al., 2009; Stramma et al., 2020). At the same time, some autotrophic denitrifying bacteria and anaerobic ammonium oxidizing (anammox) bacteria are also involved in the fixation of carbon dioxide (CO2) by activating electron acceptors: the former fixes CO2 while reducing sulfur, forming compounds with nitrate; the latter fixes CO2 while oxidizing ammonium plus nitrite to dinitrogen, both of which are under anaerobic environment (Koeve and Kähler, 2010).
Certain bacterial community are sensitive enough to be an indicator when inferring bacteria changes under hypoxic environment (Wright et al., 2012), such as Proteobacteria, Bacteroidetes, Actinobacteria, and Planctomycetes. Some members belonging to the suboxic-related phyla can use electron acceptors as an alternative to oxygen, by which mechanisms driving changes in the bacterial community to response oxygen depletion or totally lack of dissolved oxygen. Therefore, the community composition of bacteria and unique biogeochemical cycling processes in hypoxic area have been extensively studied in recent years (Beman and Carolan, 2013; Li et al., 2014; Fuchsman et al., 2019; Sen et al., 2021). Certain bacteria have been reported to be adapted for low-oxygen environments, such as SAR11 bacteria, NC10 bacteria and Protochlorococcus (Füssel et al., 2012; Padilla et al., 2016), which are related to ocean hypoxia and nitrogen loss and generally distributed in the core hypoxic area with high abundance (Thiele et al., 2019) or intensive activity (Tsementzi et al., 2016). In addition, some investigations have determined the metabolic basis for SAR11 and confirmed their role as a model to study the carbon and nitrogen cycles in the OMZs (Tsementzi et al., 2016; Bertagnolli and Stewart, 2018; Ruiz-Perez et al., 2021). However, direct clues on the effects of oxygen depletion on bacterial diversity and metabolism are still limited. Therefore, to explore the response mechanism of the functional bacterial groups in hypoxia environment is of great importance.
It is a common method to achieve information about bacteria diversity and abundance by profiling the 16S-rRNA gene. Moreover, examining the functions of bacterial communities is critical for estimating and understanding of how bacterial communities respond to the changes of ecosystem (Ward et al., 2017; Escales et al., 2019). Recent investigations on functional potential prediction of bacteria by employing PICRUSt2 in different environments, including soils (Toole et al., 2021), seawater (Xue et al., 2021), marine sediment (Su et al., 2018) are in help to decipher the detailed function variations successfully. In addition, benefiting from the function prediction of bacteria from the 16S rRNA gene sequences solely, diverse unknown microbes could be identified phylogenetically (Tsaban et al., 2021). With the unique contribution of each clade to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways are developed, PICRUSt2 prediction is increasingly applied in microbial ecology studies.
Bohai Sea, with an area of 77,000 km2 and the average water depth of 18 m, is semi-enclosed and located in northern China. It is characterized by a long water exchange half-life as much as 17–21 months (Hao et al., 2002; Luo et al., 2021). Due to the long-term impact of industry and agriculture, heavy metal pollution and excessive inorganic nitrogen has been accumulated in the Bohai Sea (Duan and Li, 2017; Mu et al., 2017), which led to severe eutrophication and the formation of oxygen-deficient zone in the central Bohai Sea (Zhao et al., 2017). Microbes are suffering from severe ecological pollution but also argued to be key for remediation since the denitrification bacteria could remove excessive nitrogen from marine environment. Based on current study on microbial composition, Proteobacteria and Bacteroidetes are widely distributed in Bohai Sea sediments (Wang et al., 2013, 2014; Li et al., 2017), with temperature and DO are the key impacting factors. Moreover, other environmental vital drivers such as salinity (Lv et al., 2016) and pH (Zhang et al., 2017) have been proved to determine bacterial communities also. However, to the best of our knowledge, the detailed information on bacteria diversity in the coastal Bohai Sea sediments remain limited, and none of the above studies put emphasize on the variation of bacterial communities under hypoxia in sediment of Bohai Sea.
In this study, the diversity and distribution of bacteria along one transect covering inshore and oxygen-depleted offshore area of Bohai Sea were investigated from June to August, 2018. By employing 16S rRNA gene high-throughput sequencing, in combination with metagenomic analysis, bacteria diversity, distribution, phylogeny and functions during oxygen depletion was evaluated and predicted. Furthermore, key environmental factors driving the bacterial communities were discussed. All of which provide insights into the relationship between oxygen depletion and relating microbial functions.
Materials and Methods
Sample Collection
Samples were collected in four cruises carried out in June, July, early and late August of 2018, respectively, in the Bohai Sea along one inshore-offshore transect (A3–A7:39.38°–39.69°N, 119.63° –120.55°E) (Figure 1), details of sampling site and date were listed in Supplementary Table 1. The sediment samples were collected in stainless steel 0.1 m2 GyO’Hara box corer, and the top 1–2 cm surface sediment were gently scraped off before subsampling were made after homogenization. All the samples were stored in sterilized 60 ml centrifuge tubes and frozen in liquid nitrogen immediately. It is worth to note that samples were attached to dry ice during transportation and then stored at –80°C until DNA extraction.
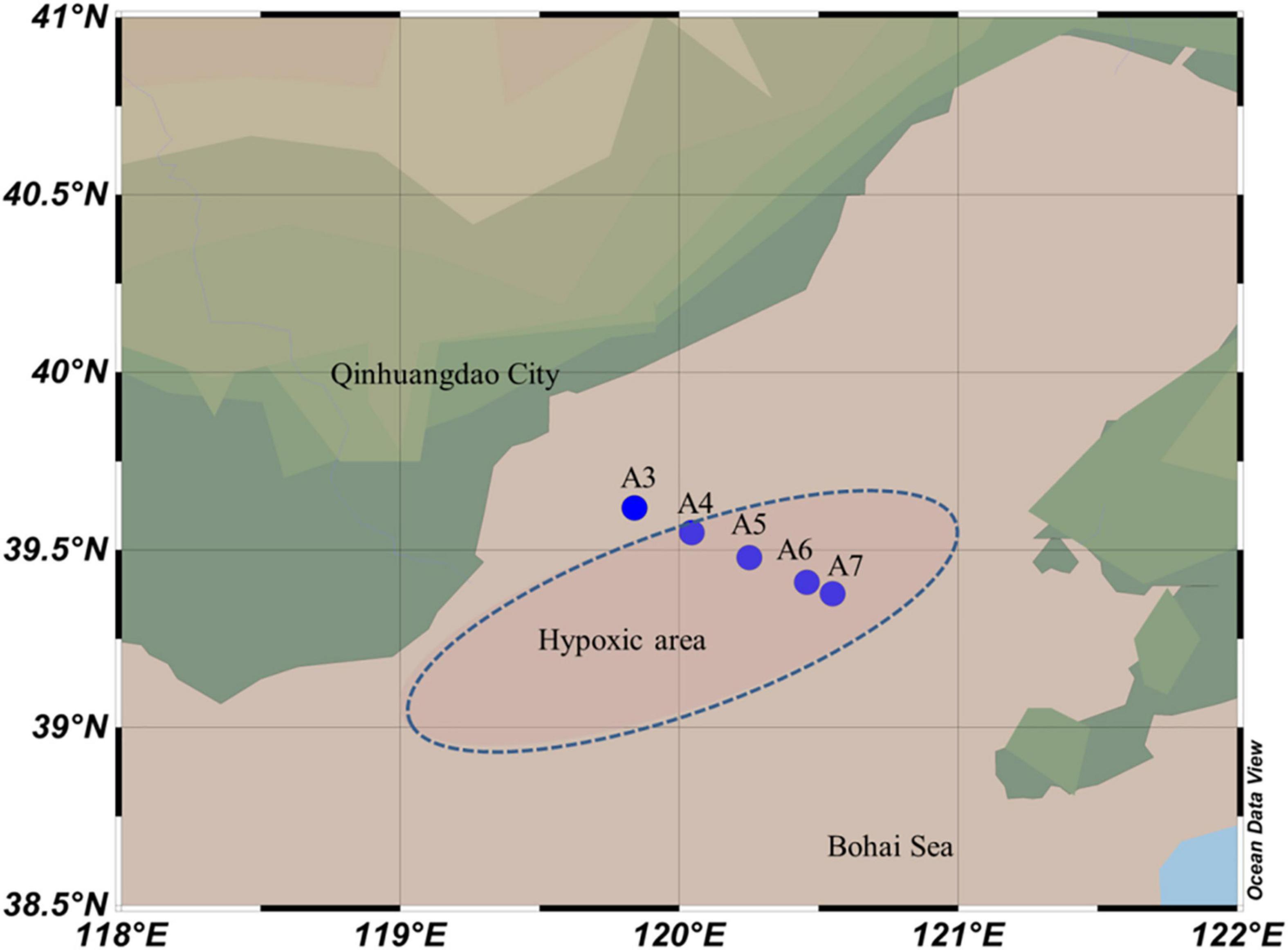
Figure 1. Map showing the sampling sites in Bohai Sea along inshore—offshore transect. The blue dash line oval showing the reported hypoxic area.
Physic-Chemical Parameters
At each site, the salinity, dissolved oxygen (DO), pH, and temperature of seawater was recorded in situ by employing the SeaBird model SBE9 conductivity-temperature-depth (CTD) recorder (Sea-Bird Electronics). The pore water from sediment cores were obtained by centrifuging at 6,000 r/min and poisoned by HgCl2 before analysis. Concentration of ammonium, nitrite and nitrate were determined by using a AA3 nutrient Auto Analyzer.
DNA Extraction and High-Throughput Sequencing
The DNA extraction and PCR amplification were following the published procedures (Wang et al., 2019) with replacement of primer sets targeting the V3, V4 region of 16S rRNA gene. Phylogenetically diagnostic sequences were amplified using the universal primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Dai et al., 2016). High-throughput sequencing was performed using Illumina MiSeq system (Illumina MiSeq, United States).
Statistical Analysis
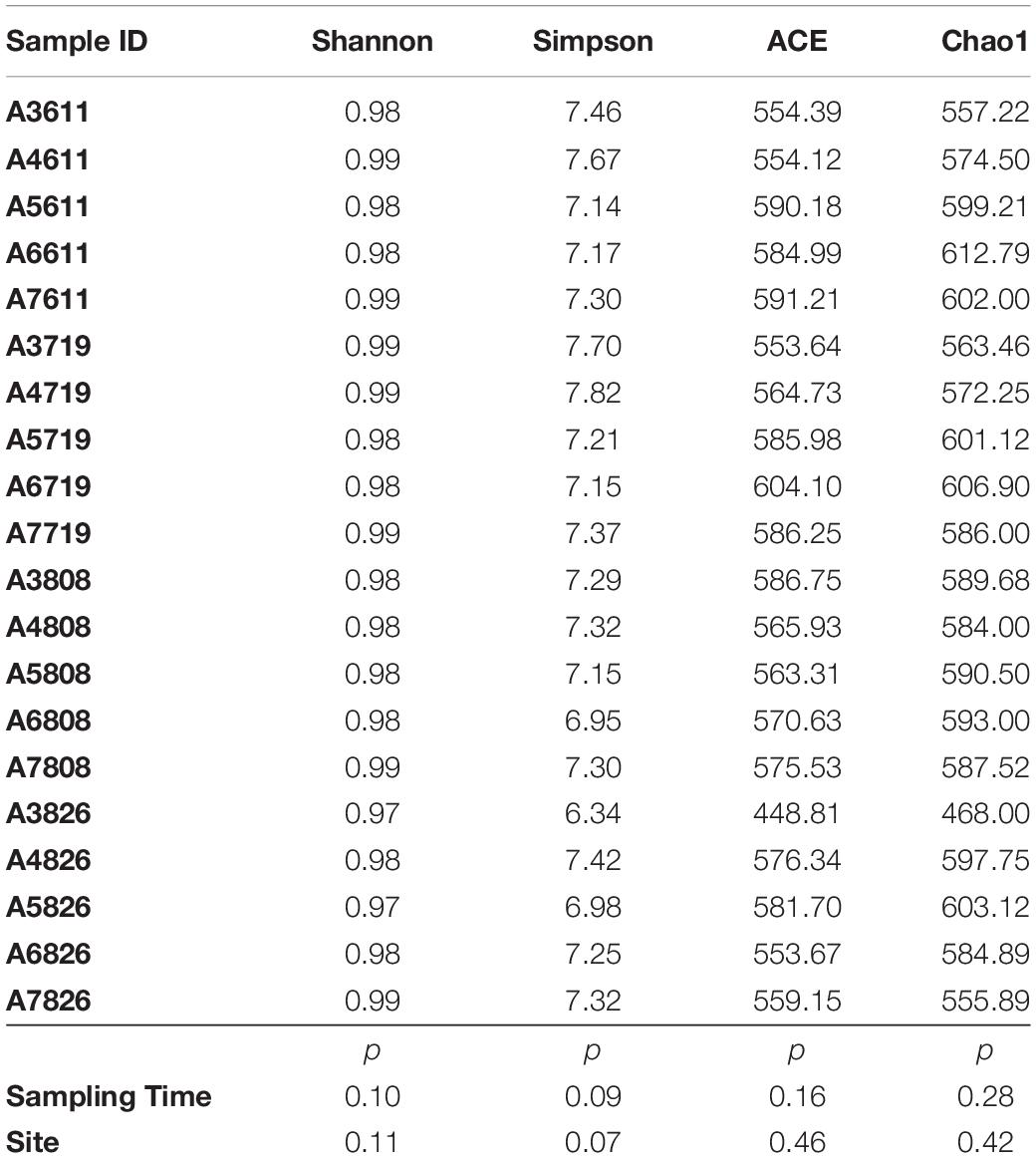
The OTUs (operational taxonomic units) were defined by 97% identity for taxonomy assignment using Uclust method (Edgar, 2010). Rarefaction analysis was performed for all the OTUs using Mothur (version v.1.30) and R with the aim to ensure the amount of the sequences was as reasonable as possible (Supplementary Figure 1). Mothur software was then used to evaluate the species richness and alpha diversity index of the samples (Shannon, Simpson, ACE and Chao 1). Beta diversity at phylum level was calculated between the five sites and four sampling time using non-metric multidimensional scaling analysis (NMDS) based on binary-jaccard distance using the R (3.6.3). Phylogenetic analysis of the representative sequences from dominant OTU (top 18 OTUs of 16S rRNA gene) was conducted with the software of MEGA7.0. Neighbor-joining tree of the representative OTUs and the reference sequences from NCBI GenBank was established with 1,000 replicates of bootstrap value. Bacterial community dynamics at generic level with eight environmental variables (Temperature, salinity, pH, DO, NO3–, NO2–, NH4+) were analyzed using canonical correspondence analysis (CANOCO v.5.02) and mapped with R language vegan package. Significance test was conducted using one-way ANOVA by employing SPSS 17.0.
Functional Prediction of Bacteria by PICRUSt
Functional potential of microbial communities in sediment samples were predicted using PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) (Langille et al., 2013). PICRUSt is a bioinformatics tool that predicts the functional composition of a metagenome using 16S rRNA gene sequences and a reference genome database. Sequences used for PICRUSt prediction were clustered into OTUs (97% similarity) against the Greengenes 13.5 database using QIIME2 software (version 2.9.1). Then the rarefied OTU table was used for normalization of 16S rRNA gene copy numbers. The normalized OTU table and the KEGG database were compared to obtain different function files for each sample. Finally, PICRUSt was used to obtain information at three pathway levels. The KEGG orthology (KO) genes that involved in nitrogen transformation in the KEGG database were compared with the PICRUSt function prediction results, and the KO related to nitrogen metabolism with the gene copy numbers in the respective sample was selected for analysis.
Nucleotide Sequences Accession Numbers
The annotated nucleotide sequences of 16S rRNA gene were deposited to NCBI under accession numbers SRR17253936-SRR17253955.
Results
Biogeochemical Characteristics of Bohai Sea
In total, 20 sediment samples were collected from 5 sites and 3 months along inshore-offshore transect covering oxygen depletion area (Supplementary Table 1). The hydrological parameters (except depth and NO3–) showed significant variation before and after DO depletion (p < 0.05, Supplementary Table 1), but no significance was observed among sites within each sampling time. As shown in Supplementary Figure 2, both DO and salinity decreased in August, especially in the reported oxygen depletion area (sites A4–A7 in this study). At the same time, concentration of NO2– and NH4+ were also accumulated in August. The variation of NO3– concentrations was not significant in sediment across sampling time, and the highest value occurred in August A5 site, which was also core of the oxygen-depletion zone.
Community Composition of Bacteria During Oxygen Depletion
By applying high-throughput sequencing targeting V3, V4 region of bacterial 16S rRNA gene, 1,198,861 reads and 77,560 clean reads were achieved from 20 samples. Based on 97% similarity, 8,032 OTUs were identified by reference from NCBI. A rarefaction curve was plotted with 34,070 sequences for each sample (Supplementary Figure 1). The community composition revealed by 16S rRNA gene at phylum level and genus level was shown in Figure 2. In general, Proteobacteria dominated by 42.67% among all the detected communities, followed by Actinobacteria (14.13%), Chloroflexi (13.02%), Acidobacteria (8.01%), Bacteroidetes (6.30%), and Gemmatimonadetes (2.95%). Phylum of Proteobacteria was mainly composed of classes Gammaproteobacteria, Deltaproteobacteria, and Alphaproteobacteria, among which, Gammaproteobacteria obviously was the most dominant class (19.12%). The relative proportion of every phylum within each sample was changing between sampling sites and time. Proteobacteria was most abundant in A3826 (56.75%), but overall, they tend to distribute in offshore sites (Figure 2B). From A3 to A5, Chloroflexi was less than Actinobacteria in June and July, but greater than Actinobacteria when time turned to August. But this variation was observed in the offshore samples A6 and A7. With their location in the reported core hypoxia area, the relative abundance of these phylum kept constant along the four sampling times. Phylum Firmicutes was only abundant in A3 and A4 sites before the oxygen began to deplete. For the dominant phylum, strong variation was observed in inshore sites, and their relative composition tend to be stable in the offshore and core hypoxia sites.
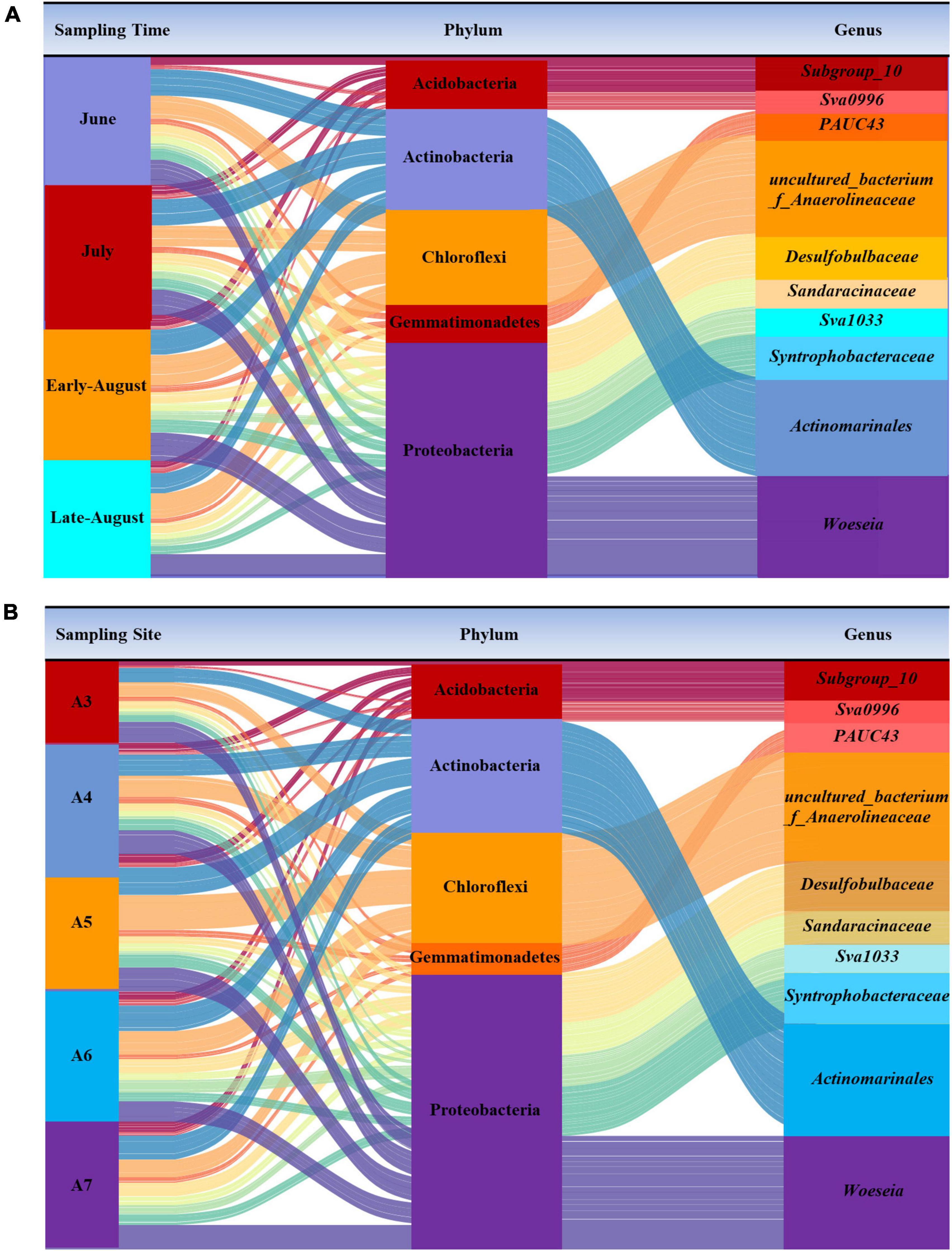
Figure 2. The community composition of dominant bacteria at phylum and genus level from each sampling time (A) and site (B).
Switch of bacteria was observed after oxygen depletion. Members of class Acidimicrobiia under phylum Actinobacteria were dominant in June and July, genus Woeseia (9.61%) of Proteobacteria was dominant and evenly distributed at all sites across all sampling time, and their dominance was replaced by the anaerobic bacteria Anaerolineaceae that belonging to Chloroflexi by 12.5% in August (Figure 2A). In addition, specific genera including Amphiplicatus, Ellin6067, Dokdonella, Defluviimonas from Proteobacteria, OLB12, and Rikenella from Bacteroidetes, Lactobacillus from Firmicutes and Mucispirillum from Deferribacteres, most of which were aero-tolerant or anaerobic bacteria, were also detected as a result of oxygen depletion with first occurrence in August samples.
The spatial distribution of bacteria was evaluated by non-metric multidimensional scaling (NMDS) analysis based on the binary-jaccard method (Figure 3). Samples from the same sites were clustered together, while samples from different sampling time were separately distributed. Among which, samples from A6 and A7 overlapped with each other, which suggested the high similarity of community composition of bacteria in A6 and A7.
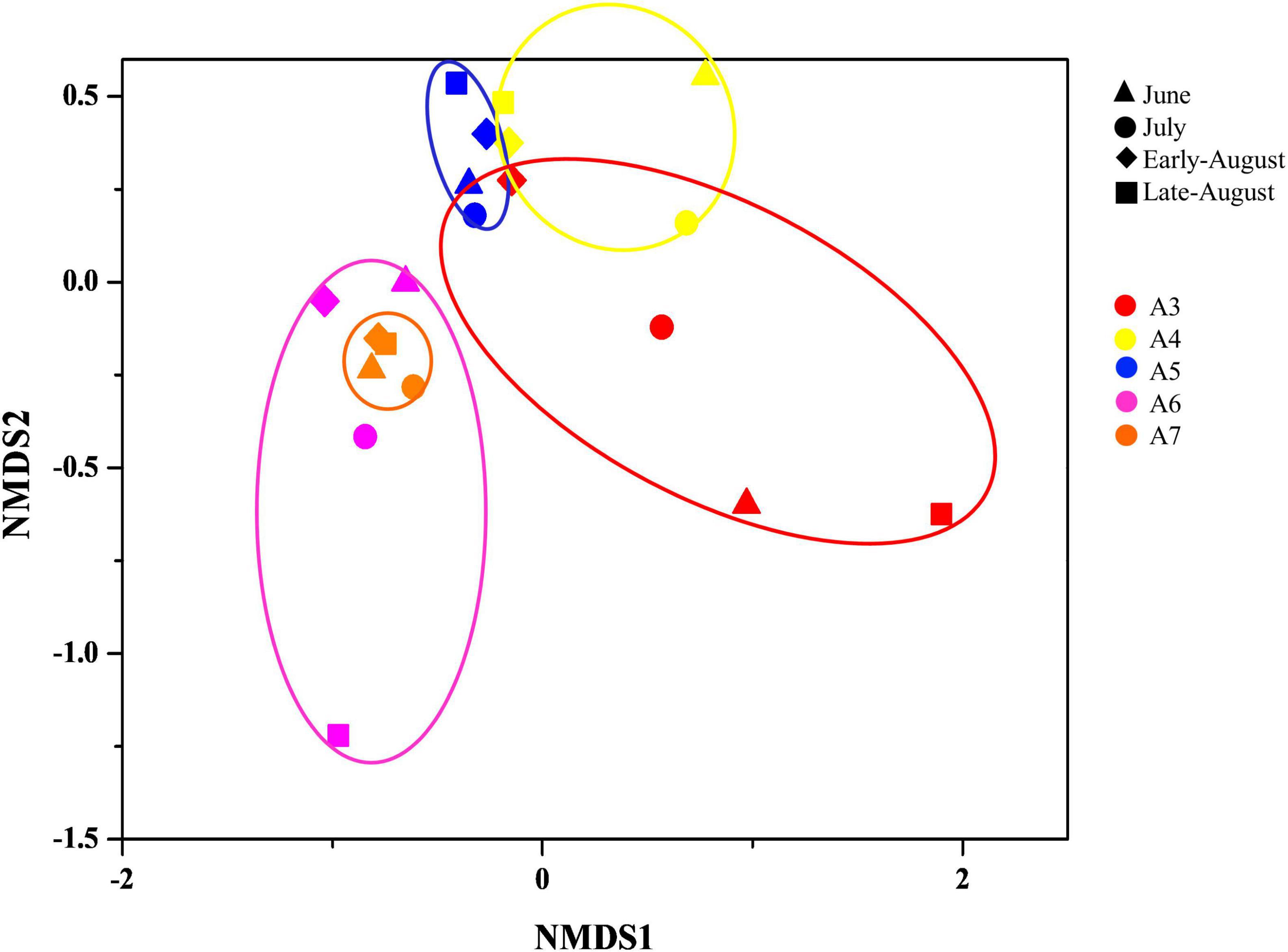
Figure 3. Non-metric multidimensional scaling (NMDS) plot depicting phylum-level bacteria composition of each sample from different sampling time and site. (Axis define 2D space that allows the best spatial representation of sample distance, based on binary-jaccard distance with stress = 0.154. The points in different color represent sampling sites, different symbol represent sampling time, and the distance between points represents the degree of difference).
Spatiotemporal Variations of Alpha Diversity
ACE and Chao 1 indices showed that the greatest diversity of 16S rRNA gene was observed at the offshore site A6 in June and July, while the lowest value occurred at the nearest inshore site A3 in Late-August. The diversity indices decreased slightly in the reported core oxygen depletion sites when oxygen began depleted, but decreased sharply in site A3, which subjected to low DO only in this study. The indices of Shannon and Simpson showed consistent trend with ACE and Chao1. Spatially, the diversity of the 16S rRNA gene was higher at offshore sites than the inshore; temporally, higher diversity was appeared in June, and decreased in August. As shown in Table 1, the differences observed was not significant between sites or sampling times (p > 0.05).
Functional Prediction of Bacteria by PICRUSt
Community prediction using PICRUSt was performed to determine the function of the observed bacteria. Based on the KEGG database, 6 categories of biological metabolic pathways were obtained, including metabolism, genetic information processing, environmental information processing, cellular processes, human diseases and organismal systems. Metabolism, genetic information processing and environmental information processing appeared to be the primary components, accounting for 51.88, 20.22, and 11.12% of all components, respectively. An analysis on the secondary functional levels of predicted genes identified 39 sub-functions, with relative abundance greater than 1% are shown in Figure 4A. A large proportion of the microbial functions were found in carbohydrate metabolism (9.76%), amino acid metabolism (9.63%), membrane transport (9.40%), energy metabolism (9.11%), replication and repair (8.78%), translation (6.09%), metabolism of cofactors and vitamins (5.37%), and nucleotide metabolism (4.23%). The results showed that the relative abundance of bacteria in the carbohydrate metabolism and membrane transport was higher in the nearshore station (DO sufficient area). The abundance of bacteria in relatively offshore sites (DO depletion area) was high in other functional pathways that account for a relatively large proportion.
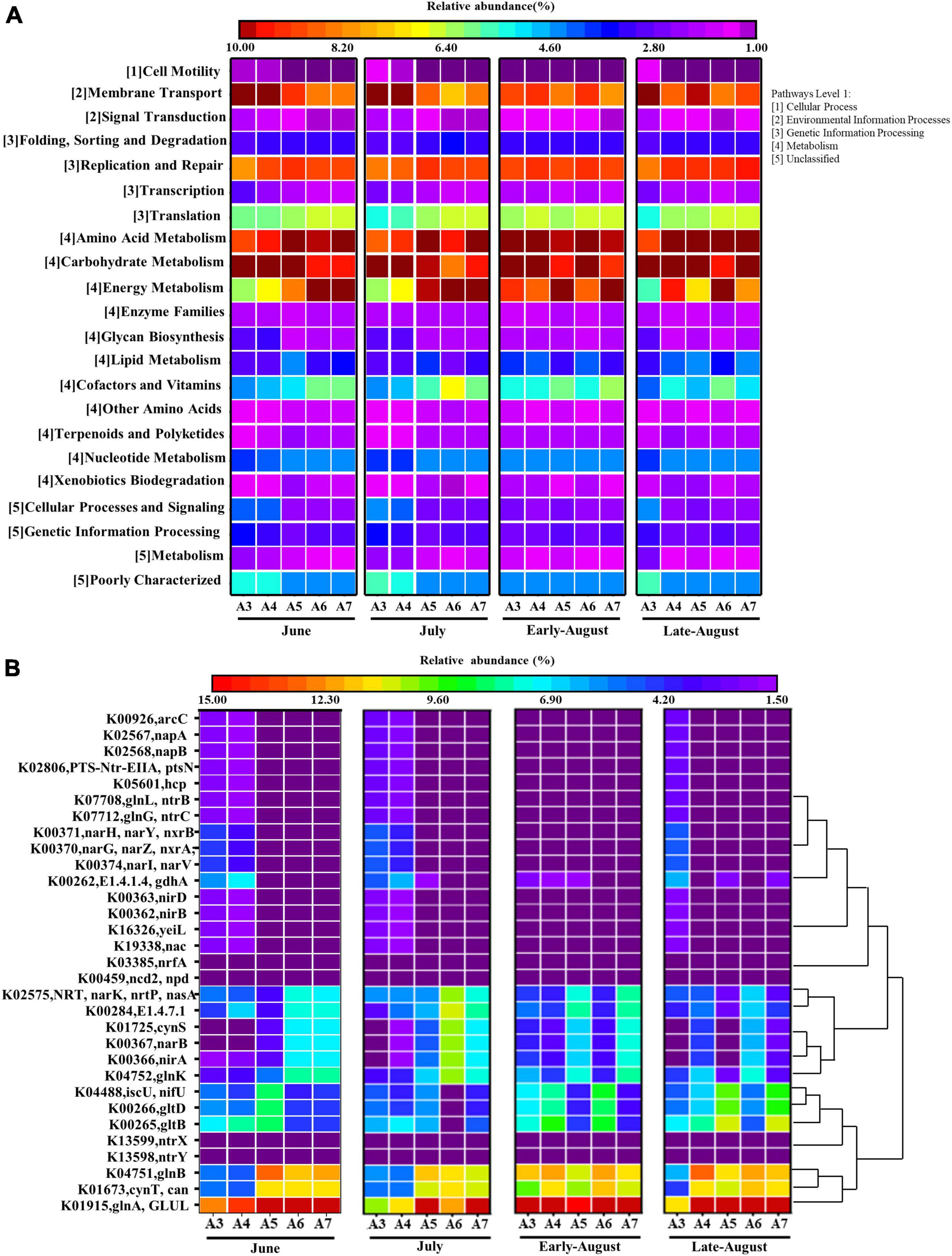
Figure 4. Heatmap profiles showing the functional categories (KEGG level 2) of bacteria communities predicted by PICRUSt analysis (A) and nitrogen metabolism-related KO based on predicted gene copy (B) in each sampling time and site. (Rows represent the 22 KEGG orthology (KO) functions, columns represent the twenty samples, and the color intensity in the heatmap represents the relative abundance (%) of the functional genes. Pathways are displayed on the y axis, the number (1–5) in front of pathway names correspond to the number (1–5) of Pathways Level 1 shown on the right).
To gain more insight into nitrogen metabolism, the functional genes that involved in nitrogen metabolism were analyzed further and 31 KOs relating to nitrogen metabolism with the corresponding gene copy numbers in the sample were selected (Figure 4B). Results showed that a majority of the genes were significantly differentiated among the sites (p < 0.05) (Supplementary Table 2). KO predictions K00366 (nirA), K00367 (narB), and K02575 (narK), which were associated with denitrification pathway, were highly abundant in oxygen-depleted sites for each month. Surprisingly, other denitrification related genes were abundant in nearshore sites, including K00371 (narH), K00370 (narG), K00374 (narI), K00363 (nirD), K00362 (nirB), where the content of DO was not depleted, but the dominance was weak. Additionally, during the transition period between non-hypoxia and hypoxia (sample from July), the expression of functional genes related to denitrification decreased significantly in sites A3 and A4 (p < 0.05) (Supplementary Table 2), which were only slightly recovered in August. The KOs responsible for nitrogen fixation were found to be abundant in offshore sites and August samples (DO-depleted areas) for the following genes: K01915 (glnA), K04751 (glnB), and K04752 (glnK), while differences in expression of functional genes were not significant in these samples.
Environmental Drivers on Community Compositions of Bacteria
RDA (redundancy analysis) was performed to find out the relationship between the environmental parameters and bacterial structure (Figure 5). Identified phylum along with 8 environmental variables (Depth, Temperature, salinity, pH, NO3–, NO2–, NH4+, DO) were analyzed using CANOCO. The length of an environmental parameter arrow in the sorting diagram indicated the strength of the relationship of that variable with community composition. For the first two RDA dimensions, 53.57 and 72.50% of the environmental variables were explained for the total variance, and the results showing that depth (p < 0.01), DO and NO2– (p < 0.05) were the key impact factors shaping the community composition encoded by 16S rRNA gene. NO2– and temperature were positively correlated with the samples in site A7, indicating the microbial communities from this site were significantly influenced by these two parameters. Depth and NO3– were positively correlated with samples from A5 and A6, and DO mainly influenced samples from A3 and A4. Further, a triplot revealed that Chloroflexi, Acidobacteria were positively correlated with NO3– and depth. The most abundant phylum, Proteobacteria was highly influenced by NO2– and temperature. In general, the concentration of nitrogen was key to the bacterial communities in the studied area. These results suggested that the distribution of bacteria was affected by multiple environmental factors rather than a single parameter.
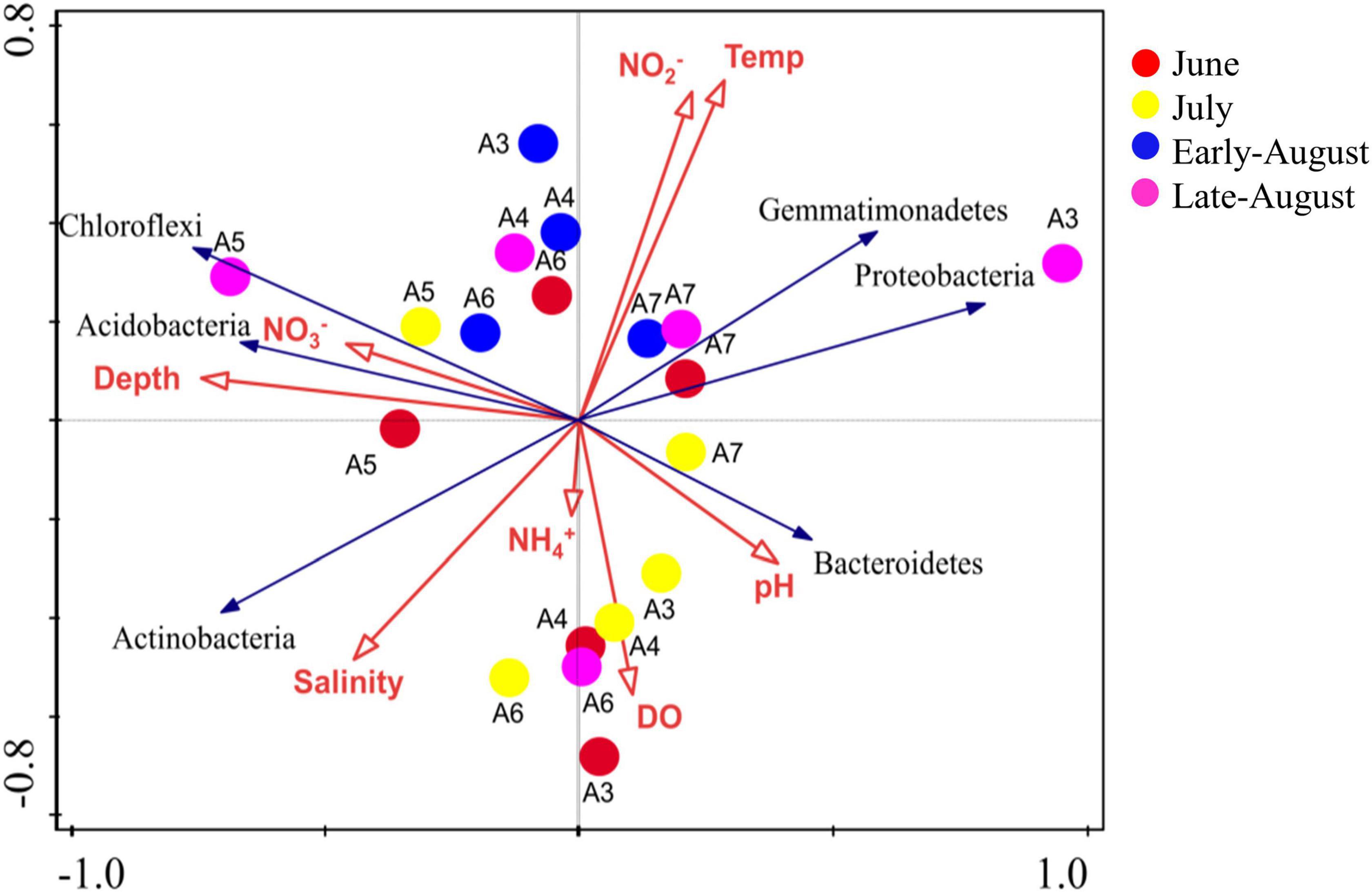
Figure 5. Redundancy analysis (RDA) shows the relationship between environmental variables and the bacteria communities.
Discussion
Switch of Bacteria Community in Response to Oxygen Depletion
The ocean OMZs were found extending globally, typical OMZ had been reported from the Indian Ocean (Arabian Sea, the Bay of Bengal), the Pacific Ocean and the Atlantic (Namibian upwelling) (Stramma et al., 2008; Wright et al., 2012; Schmidtko et al., 2017). Under the increasing pressure of anthropogenic activity and climate change, oxygen depletion zone began to occur in central Bohai Sea (Zhai et al., 2012, 2020; Wei et al., 2019), with a regular occurrence every August ranging from 1.07 to 3.60 mg/L (bottom layer water) in the past 10 years. In our monthly investigation from June to August covering marginal and central Bohai Sea in 2018, although the disturbance by typhoon Yagi (landed on August 11 and continued until August 16, 2018) braked the layers and vertical mixing increased the DO content of the reported oxygen-depleted area, the gradually depletion of DO was also observed from July to August, and the lowest value was 4.2 mg/L in the reported hypoxic sites.
With key roles in metabolism played by marine microbes, dissolved oxygen had been confirmed to be an important impacting factor and further influence the microbial biogeochemical cycling in marine sediment (Durbin and Teske, 2012; Spietz et al., 2015; Lipsewers et al., 2017). The instant variations of bacteria composition in Bohai Sea sediment during the depletion of oxygen were revealed by our monthly investigations. Distinct switch of dominant groups, such as the continuous change of dominant groups from Acidimicrobiia in June, Woeseiaceae in July and Anaerolineae in August (Figure 2A), revealed the specific response of bacteria to oxygen depletion. Anaerolineae could thrive in anaerobic, organic carbon-enriched environments, and in turn, eutrophic and anoxic conditions in the marine sediment favored the proliferation of this group of bacteria (Moncada et al., 2019). Anaerolineae had been reported in the middle and deeper layers of the hypoxic coral reef sand (Red Sea), further indicating their preference to the oxygen depletion environment (Schöttner et al., 2011; Andreote et al., 2012), which was also in accordance with our study by showing dominance only in oxygen-depleted August samples. Switch of bacteria under depletion of oxygen was also reflected by the occurrence of the anaerobic species, including Rikenella (Abe et al., 2012), Lactobacillus (Marcial-Coba et al., 2018), and Mucispirillum (Herp et al., 2019). Although they were rare species, their instant response to oxygen depletion in our study were evidences for their potential ecological role in Bohai Sea. Furthermore, switch of bacteria also included some strictly aerobic species, such as Amphiplicatus, which was also a thermophilic bacterium (Zhen-Li et al., 2014). It could be suspected that the high temperature rather than DO facilitated their occurrence in August in inshore sites. Additionally, combined with the results from functional prediction, genes involved in denitrification were abundant in DO sufficient sites. Ellin6067 had been reported as a group of significant ammonia oxidizing bacteria (Podlesnaya et al., 2020) and could be inhibited by activity when DO content was low (Huang et al., 2018). The only occurrence of both Amphiplicatus and Ellin6067 in August in our study indicated that oxygen might not be a limiting factor but could be an indicator for their distribution in marine environment. Moreover, the distribution and diversity of bacteria was also influenced as a function of sediment annum or impacted by seasonality (Laverock et al., 2014; Ebah et al., 2016; Huang et al., 2018). Although in general the abundance and diversity of bacteria in June and July were higher than in August, the difference was not significant (p > 0.05), possibly because the short interval for our sampling time limited the transport and sedimentation of sediment since it was an inter-annual or seasonal event.
Diversity and Distribution of Bacteria in Bohai Sea Sediment
Bacteria in marine sediments had been widely studied, Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria and Bacteroidetes were revealed to be the main component in marine environment (Kerfahi et al., 2014; Walsh et al., 2016; Hoshino et al., 2020; Wang et al., 2020; Peng et al., 2021). But generally, and also obviously, the dominant groups varied due to the influence of geographical locations and environmental factors (Antunes et al., 2020). Compared to changes in relative abundance on a sampling time scale, Proteobacteria, Actinobacteria and Chloroflexi were evenly distributed among the studied sites, however, Acidobacteria and Bacteroidetes varied along the transect (Figure 2B). Acidobacteria, that were dominated in nearshore sites (A3 and A4), had been revealed to be involved in nitrate reduction process (Kielak et al., 2016). This may also be the reason for the high abundance of genes that mediate the denitrification process in nearshore sites (Figure 4B). Bacteroidetes had been reported to be preponderant in nutrient-rich waters (Gomes et al., 2019), which also in support of our results that the abundance of Bacteroidetes was higher in the nutrient-rich sites regardless of the sampling time. Most of the sequences assigned to phylum Actinobacteria belonged to class Acidimicrobiia, and two most representative OTUs were classified as uncultured Actinobacterium (previously classified as OM1 clade), an eosinophilic family (Goodfellow et al., 2012; Chen et al., 2016). This group of bacteria was usually abundant in high salinity and pH sediments, and related to the capability of metabolizing hydrocarbon compounds (Queiroz et al., 2020). Their great abundance in the studied sites might contribute to the degradation of severe oil pollution in the coast of Bohai Sea (Liu et al., 2016; Yu et al., 2018). Woeseia, which belonging to Proteobacteria and most abundant in the inshore sediments of the Bohai Sea, had the capability for both aerobic and anaerobic growth (Du et al., 2016). For oxygen-independent respiration, Woeseia may take part in a truncated denitrification pathway which including periplasmic dissimilatory nitrite and the membrane-bound nitric oxide reductase by employing NirS and NorB gene, respectively (Mußmann et al., 2017). This ubiquitous and abundant group might substantially contribute to nitrous oxide emissions from coastal sediments due to its truncated denitrification which revealed by our nitrogen gene function prediction results that the inshore sediments had more functional genes which playing role in denitrification. In addition, it proved that the role of coastal sediments in nitrogen loss made more contribution than assumed, and may account for a large part of the global oceanic powerful emission of greenhouse gas nitrous oxide.
Aiming to figure out the origin or the phylogenetic distance of bacteria in Bohai Sea sediment, with special focus on the oxygen depletion sites, the phylogenetic analysis was further performed with dominant sequences (top 18 OTUs) and the reference sequences from GenBank by constructing Neighbor-joining tree. Results revealed that bacteria from Bohai Sea oxygen depletion sites had a diverse phylogenetic relationship with bacteria in typical OMZs, such as the Yangtze River estuary, the Gulf of Mexico, the Arabian Sea, the west coast of the Indian Ocean, the North Indian Ocean, and the Bay of Bengal (Figure 6). Moreover, novel hypoxic-adapted species in the sediments of Bohai Sea were also predicted, such as OTU64, OTU82, which were grouped with reported OMZ species but formed a unique clade in the phylogenetic tree. Anaerolineae and SAR11, represented by OTU45, OTU62, respectively, clustered together with most of the sequences from OMZs. As mentioned above, Anaerolineae was strictly anaerobic, which was a typical and specific bacterium in the Bohai Sea oxygen depletion area. While for SAR11 members, which were group of aerobic Alphaproteobacteria, had the ability to assimilate nitrogen-rich organic compounds by allocating surrounding resources when experiences nitrogen stress (Smith et al., 2013). Therefore, this group of bacteria could function as a specific object for research on nitrogen cycle in hypoxic zone. Moreover, due to the close geographic distance, a high degree of kinship of Bohai Sea sediment with the sediment sequences in Jiaozhou Bay, the Yellow Sea and the South China Sea were revealed from the phylogenetic tree.
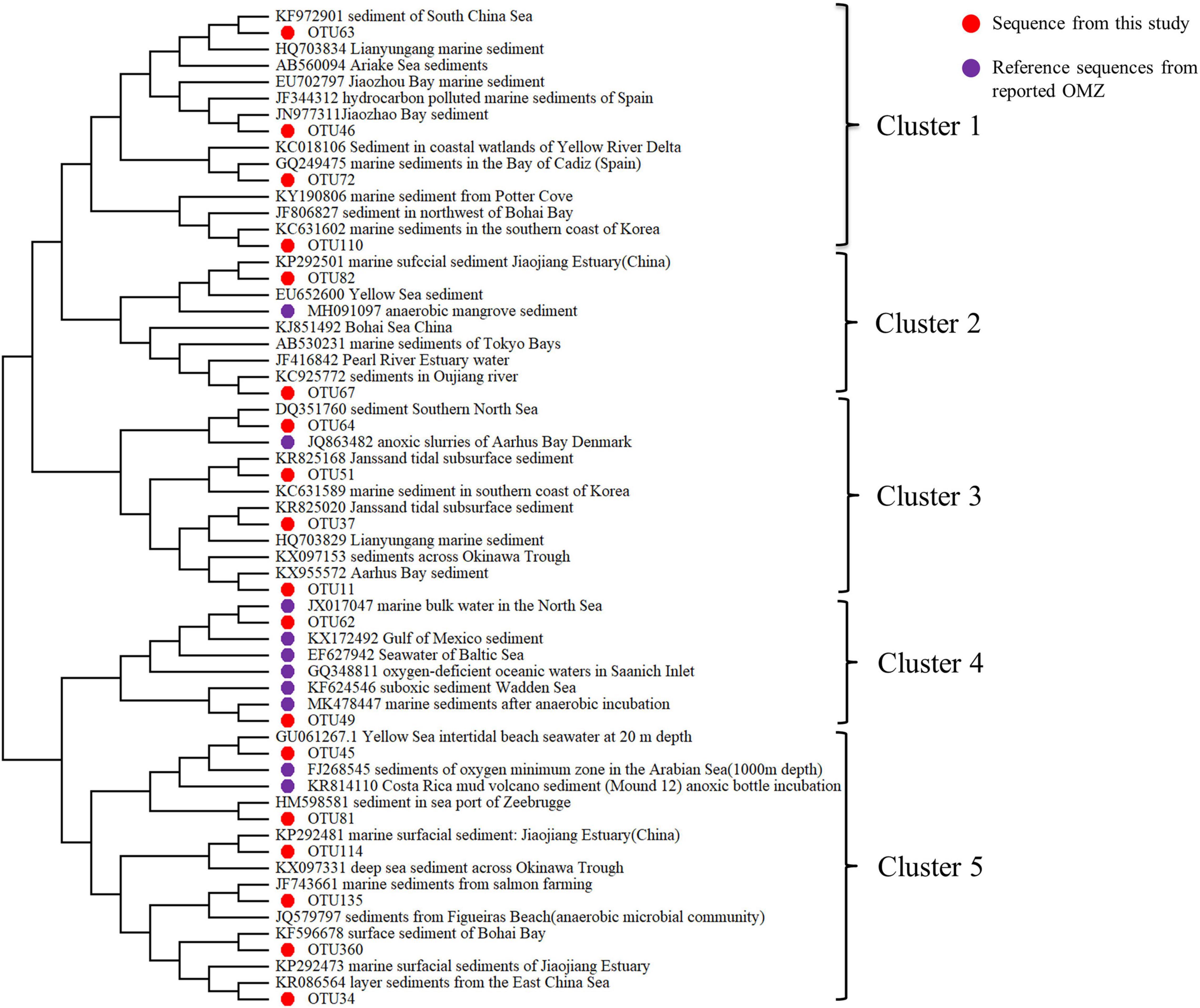
Figure 6. Neighbor-joining phylogenetic tree of the dominant OTUs (Top 18) in the sediment samples of Bohai Sea (Bootstrap values were 1,000 replicates).
Environmental Factors Shaping the Community Composition
As early as decades ago, a number of studies supported the view that different depth would be associated with changes in sediment bacterial community composition (Urakawa et al., 2000; Haglund et al., 2003). At present, Huang et al. (2018) mentioned that nitrification bacteria were highly abundant in shallow sediments, which may explain our result that high abundant functional gene related to nitrification were observed in offshore sites where sampling depth was shallower. Additionally, NO2– played a key role in shaping bacterial community. As an important substrate in the nitrogen cycle, nitrite participates in the nxr-mediated NO2– to NO3– nitrification process and denitrification pathway mediated by nir which reducing NO2– to NO (Zumft, 1997). Nitrite contents and sampling depth may make influence on nitrogen transforming bacteria, especially the nitrifier and denitrifier communities. Furthermore, a recent study found that the nitrogen transforming microbial communities in the hypoxia area and non-hypoxic area of the Yangtze River Estuary are different (Wu et al., 2019), which was highly consistent with our study, that denitrifiers prefer non-hypoxia area and genes involved in nitrification dominant in oxygen depletion area. Dissolved oxygen content substantially influenced bacterial structure and potential function with the discovery and expansion of the global ocean hypoxic zone.
Conclusion
During the occurrence of oxygen depletion from June to August, distinct community switch of bacteria was observed in the sediment of Bohai Sea. Members of class Acidimicrobiia and genus Woeseia affiliating with Gammaproteobacteria dominated before oxygen depletion, while Anaerolineae affiliating with Chloroflexi was absolutely dominant in August when DO < 4.2 mg/L. Occurrence of anaerobic rare species after oxygen depletion indicated their potential roles in Bohai Sea sediment. Spatially, Proteobacteria distributed evenly from inshore to offshore regardless of sampling time, however, relative abundance of Actinobacteria and Bacteroidetes varied among inshore sites but tended to be stable in offshore sites. Firmicutes was only abundant in nearshore and oxygen sufficient samples. KEGG pathways prediction by bacterial community showed significant variation before and after DO depletion (p < 0.05). the Predicted functional genes involved in nitrogen metabolism revealed abundant denitrification pathways in both oxygen rich (inshore) and depleted samples (offshore), while nitrogen fixation pathway was only abundant in oxygen-depleted sites. Local environmental factors including depth, NO2– together with DO play important roles in shaping bacterial community (p < 0.05). Variations in diversity and distribution of bacteria communities over oxygen depletion in the sediment of Bohai Sea provided detailed insights into bacterial response to world expending OMZs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/PRJNA789710, SRR17253936-SRR17253955.
Author Contributions
JW: conceptualization. XG: experimental operation. XG and YL: writing—original draft preparation. JW and GS: writing—review, editing, and funding acquisition. GS and LZ: field sampling. All authors read and approved the final manuscript.
Funding
This study was supported by the Natural Science Foundation of Tianjin City (5KK19004), the National Natural Science Foundation of China (52070143, 42076033, and 41506182), and the Doctoral Foundation of Tianjin Normal University (043135202-XB1714).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the crews for their help and cooperation during the cruises.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.833513/full#supplementary-material
References
Abe, K., Ueki, A., Ohtaki, Y., Kaku, N., Watanabe, K., and Ueki, K. (2012). Anaerocella delicata gen. nov., sp. nov., a strictly anaerobic bacterium in the phylum Bacteroidetes isolated from a methanogenic reactor of cattle farms. J. Gen. Appl. Microbiol. 58, 405–412. doi: 10.2323/jgam.58.405
Anderson, D. M., Glibert, P. M., and Burkholder, J. M. (2002). Harmful algal blooms and eutrophication: nutrient sources, composition, and consequences. Estuaries 25, 704–726. doi: 10.1007/BF02804901
Andreote, F. D., Jiménez, D. J., Chaves, D., Dias, A. C. F., Luvizotto, D. M., Dini-Andreote, F., et al. (2012). The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS One 7:e38600. doi: 10.1371/journal.pone.0038600
Antunes, J. T., Sousa, A. G., Azevedo, J., Rego, A., Leão, P. N., and Vasconcelos, V. (2020). Distinct temporal succession of bacterial communities in early marine biofilms in a portuguese atlantic port. Front. Microbiol. 11:1938. doi: 10.3389/fmicb.2020.01938
Beman, J. M., and Carolan, M. T. (2013). Deoxygenation alters bacterial diversity and community composition in the ocean’s largest oxygen minimum zone. Nat. Commun. 4:2705. doi: 10.1038/ncomms3705
Bertagnolli, A. D., and Stewart, F. J. (2018). Microbial niches in marine oxygen minimum zones. Nat. Rev. Microbiol. 16, 723–729. doi: 10.1038/s41579-018-0087-z
Breitburg, D., Levin, L. A., Oschlies, A., Grégoire, M., Chavez, F. P., Conley, D. J., et al. (2018). Declining oxygen in the global ocean and coastal waters. Science 359:6371. doi: 10.1126/science.aam7240
Bricker, S. B., Longstaff, B., Dennison, W., Jones, A., Boicourt, K., Wicks, C., et al. (2008). Effects of nutrient enrichment in the nation’s estuaries: a decade of change. Harmful Algae 8, 21–32. doi: 10.1016/j.hal.2008.08.028
Canfield, D. E., Glazer, A. N., and Falkowski, P. G. (2010). The evolution and future of Earth’s nitrogen cycle. Science 330, 192–196. doi: 10.1126/science.1186120
Chen, P., Zhang, L., Guo, X., Dai, X., Liu, L., Xi, L., et al. (2016). Diversity, biogeography, and biodegradation potential of actinobacteria in the deep-sea sediments along the southwest Indian ridge. Front. Microbiol. 7:1340. doi: 10.3389/fmicb.2016.01340
Cui, S., Shi, Y., Groffman, P. M., Schlesinger, W. H., and Zhu, Y. G. (2013). Centennial-scale analysis of the creation and fate of reactive nitrogen in China (1910–2010). Proc. Natl. Acad. Sci. U S A. 110, 2052–2057. doi: 10.1073/pnas.1221638110
Dai, T., Zhang, Y., Tang, Y., Bai, Y., Tao, Y., Huang, B., et al. (2016). Identifying the key taxonomic categories that characterize microbial community diversity using full-scale classification: a case study of microbial communities in the sediments of Hangzhou Bay. FEMS Microbiol. Ecol. 92:fiw150. doi: 10.1093/femsec/fiw150
Diaz, R. J., and Rosenberg, R. (2008). Spreading dead zones and consequences for marine ecosystems. Science 321, 926–929. doi: 10.1126/science.1156401
Dou, Y., Li, J., Zhao, J., Hu, B., and Yang, S. (2013). Distribution, enrichment and source of heavy metals in surface sediments of the eastern Beibu Bay, South China Sea. Mar. Pollut. Bull. 67, 137–145. doi: 10.1016/j.marpolbul.2012.11.022
Du, Z. J., Wang, Z. J., Zhao, J. X., and Chen, G. J. (2016). Woeseia oceani gen. nov., sp. nov., a chemoheterotrophic member of the order Chromatiales, and proposal of Woeseiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 107–112. doi: 10.1099/ijsem.0.000683
Duan, X., and Li, Y. (2017). Distributions and sources of heavy metals in sediments of the Bohai Sea, China: a review. Environ. Sci. Pollut. Res. 24, 24753–24764. doi: 10.1007/s11356-017-0330-6
Durbin, A. M., and Teske, A. (2012). Archaea in organic-lean and organic-rich marine subsurface sediments: an environmental gradient reflected in distinct phylogenetic lineages. Front. Microbiol. 3:168. doi: 10.3389/fmicb.2012.00168
Ebah, E., Tersagh, I., and Okpokwasili, G. C. (2016). Studies on seasonal variation and effect of heavy metal pollution on microbial load of marine sediment. Am. J. Mar. Sci. 4, 4–10.
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Escales, A., Hale, L., Voordeckers, J., Yang, Y. F., Firestone, M., Alvarez-Cohen, L., et al. (2019). Microbial functional diversity: from concepts to applications. Ecol. Evol. 9, 12000–12016. doi: 10.1002/ece3.5670
Fuchsman, C. A., Palevsky, H. I., Widner, B., Duffy, M., Carlson, M. C., Neibauer, J. A., et al. (2019). Cyanobacteria and cyanophage contributions to carbon and nitrogen cycling in an oligotrophic oxygen-deficient zone. ISME J. 13, 2714–2726. doi: 10.1038/s41396-019-0452-6
Füssel, J., Lam, P., Lavik, G., Jensen, M. M., Holtappels, M., Günter, M., et al. (2012). Nitrite oxidation in the Namibian oxygen minimum zone. ISME J. 6, 1200–1209. doi: 10.1038/ismej.2011.178
Gomes, J., Khandeparker, R., Meena, R. M., and Ramaiah, N. (2019). Bacterial community composition markedly altered by coastal hypoxia. Indian J. Microbiol. 59, 200–208. doi: 10.1007/s12088-019-00790-5
Goodfellow, M., Kämpfer, P., Busse, H. J., Trujillo, M. E., Suzuki, K. I., Ludwig, W., et al. (2012). Bergey’s Manual® of Systematic Bacteriology: Volume Five the Actinobacteria, Part A. New York, NY: Springer. doi: 10.1007/978-0-387-68233-4
Haglund, A. L., Lantz, P., Törnblom, E., and Tranvik, L. (2003). Depth distribution of active bacteria and bacterial activity in lake sediment. FEMS Microbiol. Ecol. 46, 31–38. doi: 10.1016/S0168-6496(03)00190-9
Hao, W., Tian, T., and Feng, Z. (2002). Numerical study on the water exchange of the Bohai Sea: simulation of the half-life time by dispersion model. J. Ocean Univ. China 32, 519–525.
Herp, S., Brugiroux, S., Garzetti, D., Ring, D., Jochum, L. M., Beutler, M., et al. (2019). Mucispirillum schaedleri antagonizes salmonella virulence to protect mice against colitis. Cell Host Microbe 25, 681–694. doi: 10.1016/j.chom.2019.03.004
Hoshino, T., Doi, H., Uramoto, G. I., Wörmer, L., Adhikari, R. R., Xiao, N., et al. (2020). Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. U S A. 117, 27587–27597. doi: 10.1073/pnas.1919139117
Huang, S., Chen, C., and Jaffé, P. R. (2018). Seasonal distribution of nitrifiers and denitrifiers in urban river sediments affected by agricultural activities. Sci. Total Environ. 642, 1282–1291. doi: 10.1016/j.scitotenv.2018.06.116
Kerfahi, D., Hall-Spencer, J. M., Tripathi, B. M., Milazzo, M., Lee, J., and Adams, J. M. (2014). Shallow water marine sediment bacterial community shifts along a natural CO2 gradient in the Mediterranean Sea off Vulcano. Italy. Microb. Ecol. 67, 819–828. doi: 10.1007/s00248-014-0368-7
Kielak, A. M., Barreto, C. C., Kowalchuk, G. A., van Veen, J. A., and Kuramae, E. E. (2016). The ecology of Acidobacteria: moving beyond genes and genomes. Front. Microbiol. 7:744. doi: 10.3389/fmicb.2016.00744
Koeve, W., and Kähler, P. (2010). Heterotrophic denitrification vs. autotrophic anammox–quantifying collateral effects on the oceanic carbon cycle. Biogeosciences 7, 2327–2337. doi: 10.5194/bg-7-2327-2010
Köster, M., and Meyer-Reil, L. A. (2001). Characterization of carbon and microbial biomass pools in shallow water coastal sediments of the southern Baltic Sea (Nordrügensche Bodden). Mar. Ecol. Prog. Ser. 214, 25–41. doi: 10.3354/meps214025
Lam, P., Lavik, G., Jensen, M. M., van de Vossenberg, J., Schmid, M., Woebken, D., et al. (2009). Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. U S A. 106, 4752–4757. doi: 10.1073/pnas.0812444106
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Laverock, B., Tait, K., Gilbert, J. A., Osborn, A. M., and Widdicombe, S. (2014). Impacts of bioturbation on temporal variation in bacterial and archaeal nitrogen-cycling gene abundance in coastal sediments. Environ. Microbiol. Rep. 6, 113–121. doi: 10.1111/1758-2229.12115
Li, J., Li, N., Li, F., Zou, T., Yu, S., Wang, Y., et al. (2014). Spatial diversity of bacterioplankton communities in surface water of northern South China Sea. PLoS One 9:e113014. doi: 10.1371/journal.pone.0113014
Li, Y., Wu, C., Zhou, M., Wang, E. T., Zhang, Z., Liu, W., et al. (2017). Diversity of cultivable protease-producing bacteria in Laizhou bay sediments, Bohai Sea, China. Front. Microbiol. 8:405. doi: 10.3389/fmicb.2017.00405
Lipsewers, Y. A., Vasquez-Cardenas, D., Seitaj, D., Schauer, R., Hidalgo-Martinez, S., Sinninghe Damsté, J. S., et al. (2017). Impact of seasonal hypoxia on activity and community structure of chemolithoautotrophic bacteria in a coastal sediment. Appl. Environ. Microbiol. 83:e03517-16. doi: 10.1128/AEM.03517-16
Liu, X., Guo, M., Wang, Y., Yu, X., Guo, J., Tang, C., et al. (2016). Assessing pollution-related effects of oil spills from ships in the Chinese Bohai Sea. Mar. Pollut. Bull. 110, 194–202. doi: 10.1016/j.marpolbul.2016.06.062
Lofrano, G., Libralato, G., Minetto, D., De Gisi, S., Todaro, F., Conte, B., et al. (2017). In situ remediation of contaminated marinesediment: an overview. Environ. Sci. Pollut. Res. 24, 5189–5206. doi: 10.1007/s11356-016-8281-x
Luo, C., Lin, L., Shi, J., Liu, Z., Cai, Z., Guo, X., et al. (2021). Seasonal variations in the water residence time in the Bohai Sea using 3D hydrodynamic model study and the adjoint method. Ocean Dyn. 71, 157–173. doi: 10.1007/s10236-020-01438-5
Lv, X., Ma, B., Yu, J., Chang, S. X., Xu, J., Li, Y., et al. (2016). Bacterial community structure and function shift along a successional series of tidal flats in the Yellow River Delta. Sci. Rep. 6:36550. doi: 10.1038/srep36550
Mahaffey, C., Palmer, M., Greenwood, N., and Sharples, J. (2020). Impacts of climate change on dissolved oxygen concentration relevant to the coastal and marine environment around the UK. MCCIP Sci. Rev. 2002, 31–53.
Marcial-Coba, M. S., Cieplak, T., Cahú, T. B., Blennow, A., Knøchel, S., and Nielsen, D. S. (2018). Viability of microencapsulated Akkermansia muciniphila and Lactobacillus plantarum during freeze-drying, storage and in vitro simulated upper gastrointestinal tract passage. Food Funct. 9, 5868–5879. doi: 10.1039/C8FO01331D
Moncada, C., Hassenrück, C., Gärdes, A., and Conaco, C. (2019). Microbial community composition of sediments influenced by intensive mariculture activity. FEMS Microbiol. Ecol. 95:fiz006. doi: 10.1093/femsec/fiz006
Mu, D., Yuan, D., Feng, H., Xing, F., Teo, F. Y., and Li, S. (2017). Nutrient fluxes across sediment-water interface in Bohai Bay Coastal Zone. China. Mar. Pollut. Bull. 114, 705–714. doi: 10.1016/j.marpolbul.2016.10.056
Mußmann, M., Pjevac, P., Krüger, K., and Dyksma, S. (2017). Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J. 11, 1276–1281. doi: 10.1038/ismej.2016.185
Padilla, C. C., Bristow, L. A., Sarode, N., Garcia-Robledo, E., Ramírez, E. G., Benson, C. R., et al. (2016). NC10 bacteria in marine oxygen minimum zones. ISME J. 10, 2067–2071. doi: 10.1038/ismej.2015.262
Paulmier, A., and Ruiz-Pino, D. (2009). Oxygen minimum zones (OMZs) in the modern ocean. Prog. Oceanogr. 80, 113–128. doi: 10.1016/j.pocean.2008.08.001
Peng, X., Su, H., Cai, R., and Han, Y. (2021). Wide-bound salt tolerance of the inocula from marine sediment and their specific microbial community. Environ. Res. 197:111119. doi: 10.1016/j.envres.2021.111119
Petro, C., Starnawski, P., Schramm, A., and Kjeldsen, K. U. (2017). Microbial community assembly in marine sediments. Aquat. Microb. Ecol. 79, 177–195. doi: 10.3354/ame01826
Podlesnaya, G. V., Krasnopeev, A. Y., Potapov, S. A., Tikhonova, I. V., Shtykova, Y. R., Suslova, M. Y., et al. (2020). Diversity of nitrifying bacteria in microbial communities from water and epilithic biofilms of the Lake Baikal littoral zone. Limnol. Freshw. Biol. 3, 1008–1010. doi: 10.31951/2658-3518-2020-A-4-1008
Queiroz, L. L., Bendia, A. G., Duarte, R. T. D., das Graças, D. A., da Silva, A. L. D. C., Nakayama, C. R., et al. (2020). Bacterial diversity in deep-sea sediments under influence of asphalt seep at the São Paulo Plateau. Antonie Van Leeuwenhoek 113, 707–717. doi: 10.1007/s10482-020-01384-8
Ruiz-Perez, C. A., Bertagnolli, A. D., Tsementzi, D., Woyke, T., Stewart, F. J., and Konstantinidis, K. T. (2021). Description of Candidatus Mesopelagibacter carboxydoxydans and Candidatus Anoxipelagibacter denitrificans: Nitrate-reducing SAR11 genera that dominate mesopelagic and anoxic marine zones. Syst. Appl. Microbiol. 44:126185. doi: 10.1016/j.syapm.2021.126185
Schmidtko, S., Stramma, L., and Visbeck, M. (2017). Decline in global oceanic oxygen content during the past five decades. Nature 542, 335–339. doi: 10.1038/nature21399
Schöttner, S., Pfitzner, B., Grünke, S., Rasheed, M., Wild, C., and Ramette, A. (2011). Drivers of bacterial diversity dynamics in permeable carbonate and silicate coral reef sands from the Red Sea. Environ. Microbiol. 13, 1815–1826. doi: 10.1111/j.1462-2920.2011.02494.x
Sen, K., Bai, M., Sen, B., and Wang, G. (2021). Disentangling the structure and function of mycoplankton communities in the context of marine environmental heterogeneity. Sci. Total Environ. 766:142635. doi: 10.1016/j.scitotenv.2020.142635
Smith, D. P., Thrash, J. C., Nicora, C. D., Lipton, M. S., Burnum-Johnson, K. E., Carini, P., et al. (2013). Proteomic and transcriptomic analyses of “Candidatus Pelagibacter ubique” describe the first PII-independent response to nitrogen limitation in a free-living Alphaproteobacterium. mBio 4:e00133-12. doi: 10.1128/mBio.00133-12
Spietz, R. L., Williams, C. M., Rocap, G., and Horner-Devine, M. C. (2015). A dissolved oxygen threshold for shifts in bacterial community structure in a seasonally hypoxic estuary. PLoS One 10:e0135731. doi: 10.1371/journal.pone.0135731
Stramma, L., Johnson, G. C., Sprintall, J., and Mohrholz, V. (2008). Expanding oxygen-minimum zones in the tropical oceans. Science 320, 655–658. doi: 10.1126/science.1153847
Stramma, L., Schmidtko, S., Bograd, S. J., Ono, T., Ross, T., Sasano, D., et al. (2020). Trends and decadal oscillations of oxygen and nutrients at 50 to 300 m depth in the equatorial and North Pacific. Biogeosciences 17, 813–831. doi: 10.5194/bg-17-813-2020
Su, Z., Dai, T., Tang, Y., Tao, Y., Huang, B., Mu, Q., et al. (2018). Sediment bacterial community structures and their predicted functions implied the impacts from natural processes and anthropogenic activities in coastal area. Mar. Pollut. Bull. 131, 481–495. doi: 10.1016/j.marpolbul.2018.04.052
Thiele, S., Basse, A., Becker, J. W., Lipski, A., Iversen, M. H., and Mollenhauer, G. (2019). Microbial communities in the nepheloid layers and hypoxic zones of the Canary current upwelling system. MicrobiologyOpen 8:e00705. doi: 10.1002/mbo3.705
Toole, D. R., Zhao, J., Martens-Habbena, W., and Strauss, S. L. (2021). Bacterial functional prediction tools detect but underestimate metabolic diversity compared to shotgun metagenomics in southwest Florida soils. Appl. Soil Ecol. 168:104129. doi: 10.1016/j.apsoil.2021.104129
Tsaban, T., Stupp, D., Sherill-Rofe, D., Bloch, I., Sharon, E., Schueler-Furman, O., et al. (2021). CladeOScope: functional interactions through the prism of clade-wise co-evolution. NAR Genom Bioinform. 3:lqab024. doi: 10.1093/nargab/lqab024
Tsementzi, D., Wu, J., Deutsch, S., Nath, S., Rodriguez-R, L. M., Burns, A. S., et al. (2016). SAR11 bacteria linked to ocean anoxia and nitrogen loss. Nature 536, 179–183. doi: 10.1038/nature19068
Urakawa, H., Yoshida, T., Nishimura, M., and Ohwada, K. (2000). Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16S rDNA-based approaches and quinone profiling. Environ. Microbiol. 2, 542–554. doi: 10.1046/j.1462-2920.2000.00137.x
Voss, M., Bange, H. W., Dippner, J. W., Middelburg, J. J., Montoya, J. P., and Ward, B. (2013). The marine nitrogen cycle: recent discoveries, uncertainties and the potential relevance of climate change. Philos. T. R. Soc. B. 368:20130121. doi: 10.1098/rstb.2013.0121
Walsh, E. A., Kirkpatrick, J. B., Rutherford, S. D., Smith, D. C., Sogin, M., and D’Hondt, S. (2016). Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 10, 979–989. doi: 10.1038/ismej.2015.175
Wang, J., Kan, J., Qian, G., Chen, J., Xia, Z., Zhang, X., et al. (2019). Denitrification and anammox: understanding nitrogen loss from Yangtze Estuary to the east China sea (ECS). Environ. Pollut. 252, 1659–1670. doi: 10.1016/j.envpol.2019.06.025
Wang, L., Liu, L., Zheng, B., Zhu, Y., and Wang, X. (2013). Analysis of the bacterial community in the two typical intertidal sediments of Bohai Bay, China by pyrosequencing. Mar. Pollut. Bull. 72, 181–187. doi: 10.1016/j.marpolbul.2013.04.005
Wang, L., Zheng, B., Nan, B., and Hu, P. (2014). Diversity of bacterial community and detection of nirS-and nirK-encoding denitrifying bacteria in sandy intertidal sediments along Laizhou Bay of Bohai Sea. China. Mar. Pollut. Bull. 88, 215–223. doi: 10.1016/j.marpolbul.2014.09.002
Wang, M., Noor, S., Huan, R., Liu, C., Li, J., Shi, Q., et al. (2020). Comparison of the diversity of cultured and total bacterial communities in marine sediment using culture-dependent and sequencing methods. Peer J. 8:e10060. doi: 10.7717/peerj.10060
Ward, B. B., Devol, A. H., Rich, J. J., Chang, B. X., Bulow, S. E., Naik, H., et al. (2009). Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nature 461, 78–81. doi: 10.1038/nature08276
Ward, C. S., Yung, C. M., Davis, K. M., Blinebry, S. K., Williams, T. C., Johnson, Z. I., et al. (2017). Annual community patterns are driven by seasonal switching between closely related marine bacteria. ISME J. 11, 1412–1422. doi: 10.1038/ismej.2017.4
Wei, Q., Wang, B., Yao, Q., Xue, L., Sun, J., Xin, M., et al. (2019). Spatiotemporal variations in the summer hypoxia in the Bohai Sea (China) and controlling mechanisms. Mar. Pollut. Bull. 138, 125–134. doi: 10.1016/j.marpolbul.2018.11.041
Wright, J. J., Konwar, K. M., and Hallam, S. J. (2012). Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–394. doi: 10.1038/nrmicro2778
Wu, D. M., Dai, Q. P., Liu, X. Z., Fan, Y. P., and Wang, J. X. (2019). Comparison of bacterial community structure and potential functions in hypoxic and non-hypoxic zones of the Changjiang Estuary. PLoS One 14:e0217431. doi: 10.1371/journal.pone.0217431
Xue, J., Shi, K., Chen, C., Bai, Y., Cui, Q., Li, N., et al. (2021). Evaluation of response of dynamics change in bioaugmentation process in diesel-polluted seawater via high-throughput sequencing: degradation characteristic, community structure, functional genes. J. Hazard. Mater. 403:123569. doi: 10.1016/j.jhazmat.2020.123569
Yu, F., Xue, S., Zhao, Y., and Chen, G. (2018). Risk assessment of oil spills in the Chinese Bohai Sea for prevention and readiness. Mar. Pollut. Bull. 135, 915–922. doi: 10.1016/j.marpolbul.2018.07.029
Zhai, W. D., Zheng, L. W., Li, C. L., Xiong, T. Q., and Wang, S. Y. (2020). Changing Nutrients, Dissolved Oxygen and Carbonate System in the Bohai and Yellow Seas, China. Singapore: Springer. doi: 10.1007/978-981-15-4886-4_8
Zhai, W., Zhao, H., Zheng, N., and Xu, Y. (2012). Coastal acidification in summer bottom oxygen-depleted waters in northwestern-northern Bohai Sea from June to August in 2011. Chin. Sci. Bull. 57, 1062–1068. doi: 10.1007/s11434-011-4949-2
Zhang, H., Zheng, S., Ding, J., Wang, O., and Liu, F. (2017). Spatial variation in bacterial community in natural wetland-river-sea ecosystems. J. Basic Microbiol. 57, 536–546. doi: 10.1002/jobm.201700041
Zhao, H. D., Kao, S. J., Zhai, W. D., Zang, K. P., Zheng, N., Xu, X. M., et al. (2017). Effects of stratification, organic matter remineralization and bathymetry on summertime oxygen distribution in the Bohai Sea. China. Cont. Shelf Res. 134, 15–25. doi: 10.1016/j.csr.2016.12.004
Zhen-Li, Z., Xin-Qi, Z., Nan, W., Wen-Wu, Z., Xu-Fen, Z., Yi, C., et al. (2014). Amphiplicatus metriothermophilus gen. nov., sp. nov., a thermotolerant alphaproteobacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 64, 2805–2811. doi: 10.1099/ijs.0.062471-0
Zhou, F., Gao, X., Zhang, Y., Yuan, H., Song, J., Liu, K., et al. (2017). Potential mobility of inorganic nutrients and its controls at the sediment-water interface in the main path of Kuroshio Current off eastern Taiwan. Mar. Pollut. Bull. 119, 270–276. doi: 10.1016/j.marpolbul.2017.04.002
Keywords: Bohai Sea, sediment, oxygen depletion, 16S rRNA, community switch, function prediction
Citation: Guo X, Song G, Li Y, Zhao L and Wang J (2022) Switch of Bacteria Community Under Oxygen Depletion in Sediment of Bohai Sea. Front. Mar. Sci. 9:833513. doi: 10.3389/fmars.2022.833513
Received: 11 December 2021; Accepted: 06 January 2022;
Published: 10 February 2022.
Edited by:
Meilin Wu, South China Sea Institute of Oceanology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Hualong Wang, Ocean University of China, ChinaLei Yan, Heilongjiang Bayi Agricultural University, China
Copyright © 2022 Guo, Song, Li, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Wang, andhbmdfaGt1QDE2My5jb20=
 Xiaoxiao Guo
Xiaoxiao Guo Guisheng Song
Guisheng Song Yanying Li
Yanying Li Liang Zhao
Liang Zhao Jing Wang
Jing Wang