
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 20 October 2022
Sec. Aquatic Physiology
Volume 9 - 2022 | https://doi.org/10.3389/fmars.2022.1021377
 Litong Yang1†
Litong Yang1† Ying Zhang1†
Ying Zhang1† Huizhen Chen1,2
Huizhen Chen1,2 Qian Gao1,2
Qian Gao1,2 Peng Luo1
Peng Luo1 Jianxing Hu1
Jianxing Hu1 Fanyu Zheng1
Fanyu Zheng1 Yunyu Yuan3
Yunyu Yuan3 Liang Fu3
Liang Fu3 Chang Chen1,4*
Chang Chen1,4*The outbreak of coral-eating Acanthaster spp., commonly known as the Crown-of-Thorn Starfish (CoTS), contributes to a significant proportion of coral loss in the tropical Indo-Pacific region. After the dramatic loss of coral due to their predation, CoTS is expected to face food shortages before coral recovers, which is usually accompanied by the sudden disappearance of its population. To reveal the response of CoTS to starvation stress, we conducted a four-month starvation experiment to investigate the physiological and molecular changes in the stomach tissue by combining the metabolites and enzyme activity measurements with transcriptome analysis. The results showed that the concentrations of primary metabolites and associated enzyme activities, as well as the amount of total antioxidant were not significantly altered between fed and starved CoTS in any case. However, starvation suppressed the expression of the genes involved in glycolysis and citrate cycle, development and movement, but enhanced that of the genes associated with sleep promotion, immunity, lysosome and glucose supply. This suggests that long-term starvation may induce CoTS to enter into a dormancy-like status characterized by reduced unnecessary physical activities for survival, accelerated recycling of nutrients, and enhanced immunity.
Coral reefs are one of the ecosystems with the highest productivity level and can provide essential ecological functions (Moberg and Folke, 1999; Elliff and Silva, 2017). However, in recent years, the coral reef ecosystem has been under severe threat of degradation and destruction due to the increased mortality of reef-building corals (De’ath et al., 2012; Leray et al., 2012; Tkachenko et al., 2020). Among many causes of coral reef decline, the outbreak of the coral-eating Crown-of-Thorn Starfish (CoTS) Acanthaster spp. accounts for 42% of a 27-year coral decline in the Great Barrier Reef (Australia), which is even higher than the well-known coral bleaching (10%) (De’ath et al., 2012). Recently, multiple CoTS outbreaks have been observed in the South China Sea region, causing tremendous coral loss ranging from 43% to 97% (Li et al., 2019; Tkachenko et al., 2020; Heng et al., 2021).
The outbreaks are found to be periodic every 15-20 years in coral reefs worldwide (Pratchett et al., 2017; Li et al., 2019). After 2-5 years of intensive predation on corals, such outbreaks usually end with extremely low coral cover and the disappearance of the majority CoTS population (Saponari et al., 2018; Li et al., 2019; Tkachenko et al., 2020). It was hypothesized that CoTS might migrate for alternative habitat searching (Moran, 1986) or die due to infection by deadly pathogens under starvation stress (Zann et al., 1987; Birkeland and Lucas, 1990). However, none of them is proven to stand since CoTS is not likely to perform long-distance migration under starvation and there is no record of mass death of CoTS in the field (Sigl and Laforsch, 2016; Pratchett et al., 2017; Ling et al., 2020). Moreover, given its relatively long life span (estimated up to 17 years in the field) compared to the short period (2-5 years) with sufficient food (Stump, 1996; Saponari et al., 2018; Li et al., 2019; Tkachenko et al., 2020) and the existing possibility that some CoTS may have survived between the outbreaks (Stump, 1996), CoTS may have developed a survival strategy when facing such prolonged food shortages in the field. However, even with at least 50 years of research on this corallivore starfish, the vast knowledge gap on the well-being and whereabouts of the CoTS population facing food depletion remains to be filled (Pratchett et al., 2017). Thus, a closer investigation on its starvation state could help us to better understand the successful comebacks of CoTS recorded after 10-15 years of coral recovery and provide better solutions for future CoTS population control (Nakamura et al., 2014; Pratchett et al., 2017; Li et al., 2019; Kuo et al., 2022).
To study the response of CoTS to starvation stress, we captured CoTS from the South China Sea and kept them indoors in two groups with or without feeding of corals for four months. At the end of the experiment, the stomach tissues were collected from the survived CoTS for physiological and transcriptomic investigations. The activities of metabolites and enzymes related to energy production and antioxidants were measured in these tissues, and their transcriptomes were analyzed by RNA sequencing (RNA-seq).
Fifty-two adult CoTS were collected from the South China Sea near Tanmen (Hainan, China) on August 2021 during the spawning season and separated gently into two groups in an indoor recirculating aquaculture system (4 x 4 x 1.2 m3) with filtered seawater (temperature = 23.5 ± 0.8°C, salinity = 30.4 ± 2.9 ppt) at Tropical Aquatic Research and Development Center (Hainan, China). To minimize the coral consumption, fed group was constituted of three randomly selected adult CoTS fed with artificially bred corals (mainly Acropora, coral supply was replenished after its complete consumption) (Caballes et al., 2016), while the starved group contained forty-nine CoTS reared without any feeding. After four months, six starfishes that survived starvation were randomly selected as six replicates for the starved group. Together with two out of three CoTS survived with feeding (starved for 0 days, control group), stomach tissues were collected through anatomy (Figure 1). Obtained tissue samples were labeled and fast frozen in liquid nitrogen before being sent to the laboratory (Guangzhou, China) with dry ice and stored at -80°C.
Protein content was determined with BCA Protein Assay Kit (Solarbio, Beijing). Parameters involved in glucose metabolism, including glucose content, pyruvate (PA) content, and pyruvate kinase (PK) activity were measured using commercial kits (Solarbio, Beijing). Antioxidant activity differences between the two groups were determined through superoxide dismutase (SOD) activity, catalase (CAT) activity, and total antioxidant (T-AOC) with commercial kits (Solarbio, Beijing). All data were normalized to wet weight per gram of stomach tissue.
Total RNA from the stomach tissue was extracted with TransZol Up Plus RNA Kit (TransGen Biotech, Beijing) by following the manufacturer’s instructions. RNA quality was evaluated through Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and agarose gel electrophoresis, while RNA concentration was assessed with Qubit 2.0 Fluorometer (Thermo Scientific, USA). Qualified RNA was purified with Oligo (dT) beads and used for cDNA synthesis with NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs, USA), followed by repairing and sequencing using Illumina Novaseq6000 by Gene Denovo Biotechnology Co. (Guangzhou, China).
The obtained data from physiological assessments were processed to Welch’s t-test (the difference is significant if P value < 0.05) and graph illustration using GraphPad Prism software version 9.00 (GraphPad Software, USA). During RNA-seq, reads were filtered by fastp (version 0.18.0) to obtain high-quality data (Chen et al., 2018), including adapter reads, low-quality reads (Q value ≤ 20), and unknown nucleotides (>10%). Clean reads were mapped to the reference genome of CoTS (https://www.ncbi.nlm.nih.gov/bioproject/PRJDB3175/) through HISAT2. 2.4 (Kim et al., 2015), followed by assembling with StringTie v1.3.1 (Pertea et al., 2015; Pertea et al., 2016). Gene abundance was calculated with an FPKM (fragment per kilobase of transcript per million mapped reads) value, which allows the quantification of gene expression abundance and variations among the samples by applying RSEM software (Li and Dewey, 2011). Obtained gene expressions were processed for differential analysis by DESeq2 (Love et al., 2014). Significant DEGs (differentially expressed genes) were selected with the parameter of false discovery rate (FDR) below 0.05 and absolute fold change ≥ 2 in all genes of each sample. To provide a global gene expression pattern and cluster in all DEGs, hierarchical clustering of differential gene expression patterns and was performed through Z-score calculation. To estimate dissimilarity of gene expression patterns between the CoTS individuals from the fed and starved group, a nonmetric multidimensional scaling (NMDS) analysis was performed on the FPKM values of all DEGs by using the vegan package in R (www.R-project.org), a stress value was calculated to evaluate the goodness of fit in the ordination analysis (Clarke, 1993). To analyze the biological properties and functions of sequenced genes, Gene Ontology (GO) analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were conducted. Key genes and pathways selection were based on the enrichment criteria of Q value < 0.05 and the biological processes suspected to alter under starvation stress.
Several significant DEGs from the two groups were selected from the RNA-seq libraries to conduct qRT-PCR to verify the reliability and confirm the gene expression differences in fed (n = 2) and starved (n = 6) CoTS’s stomach tissue samples. qRT-PCR primers were designed with the Primer-Blast on the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) (Ye et al., 2012) and synthesized by Sangon Biotech Co. (Shanghai, China). Sequences of the primers were listed in Supplementary Table 1.
Previously extracted total RNA was reverse-transcribed into cDNAs with Evo M-MLV RT Mix Kit (with gDNA Clean for qPCR) (Accurate Biology, China) as the template, reacted with the 2 x SYBR® Green Pro Taq HS Premix kit (Accurate Biology, China) on Thermal Cycler Dice® Real Time System III (Takara, Japan). Cytochrome b gene was used as a housekeeping gene for internal standardization (Supplementary Table 1). qRT-PCR reaction was performed with initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s; and a final dissociation stage at 95°C for 15 s, 60°C for 30 s and 95°C for 15 s. To reveal the relative gene expression of fed and starved CoTS tissue under each gene, a comparative CT method (2−ΔΔCT) was applied (Livak and Schmittgen, 2001). Triplicate cDNA sample amplifications were performed for each gene, followed by an unpaired t-test and graph illustration using GraphPad Prism software version 9.00 (GraphPad Software, USA).
The previous study has shown that CoTS is able to survive months-long starvation (Birkeland and Lucas, 1990), so it is believed that it has developed specific strategies to respond to prolonged food supply suspension, which is, however, not elucidated yet. In this study, we found that 18 out of 49 CoTS in the starved group and 2 out of 3 CoTS in the fed group survived after a four-month experiment, resulting in a survival rate of 36.7% and 66.7% for starved and fed CoTS respectively. Although starvation stress greatly decreased the survival rate of the adult CoTS, this result suggests their potent capacity to adjust to long-term food shortages, which is usually expected after their outbreaks in the natural environment. The following results from biochemical and transcriptomic analysis shed some light on the molecular mechanism of how it responds to starvation stress.
Data reliability was firstly ensured through reads quality assessment. Clean reads from the starved group and the fed group were obtained as 42,300, 293 (99.67%) and 52, 458, 674 (99.70%) respectively, and the GC percentage obtained from all samples was 42.62%. The average percentage of number of bases whose quality value was above Q20 and Q30 (correct base recognition rate ≥ 99%, 99.90%) was 97.90% and 93.95% respectively with an average mapped gene ratio of 89.18% compared to the reference genome as shown in Supplementary Table 2.
By applying the criteria (FDR < 0.05, |log2(fold change)| ≥ 1), 1205 DEGs were identified from starved CoTS stomach tissue, including 656 up-regulated and 549 down-regulated genes as shown in Figure 2A. Heatmap of DEGs expression in Figure 2B shows obvious clustering of gene expression patterns of each individual CoTS inside the fed and starved group and the number of DEGs between the two groups was approximately the same. Ordination of the genes as shown in the NMDS analysis result (Figure 2C) indicates a noticeable separation of the DEGs expression of each CoTS samples between the fed and starved group and a close resemblance of gene expression inside the two groups, obtained stress value as 0.0453 (below 0.05) suggests a good fit and a reliable ordination result (Clarke, 1993). To reveal the functions of the identified genes, the KEGG and GO database were used for gene alignments and annotation. The top 10 KEGG enriched pathways (Figure 3A) showed the most significantly enriched pathway was lysosome, followed by antigen processing and presentation pathway, tuberculosis, cell cycle, and so on. In GO analysis (Figure 3B), enrichment results indicated the cellular process, metabolic process, and binding as the top 3 enriched terms were greatly altered by starvation stress.
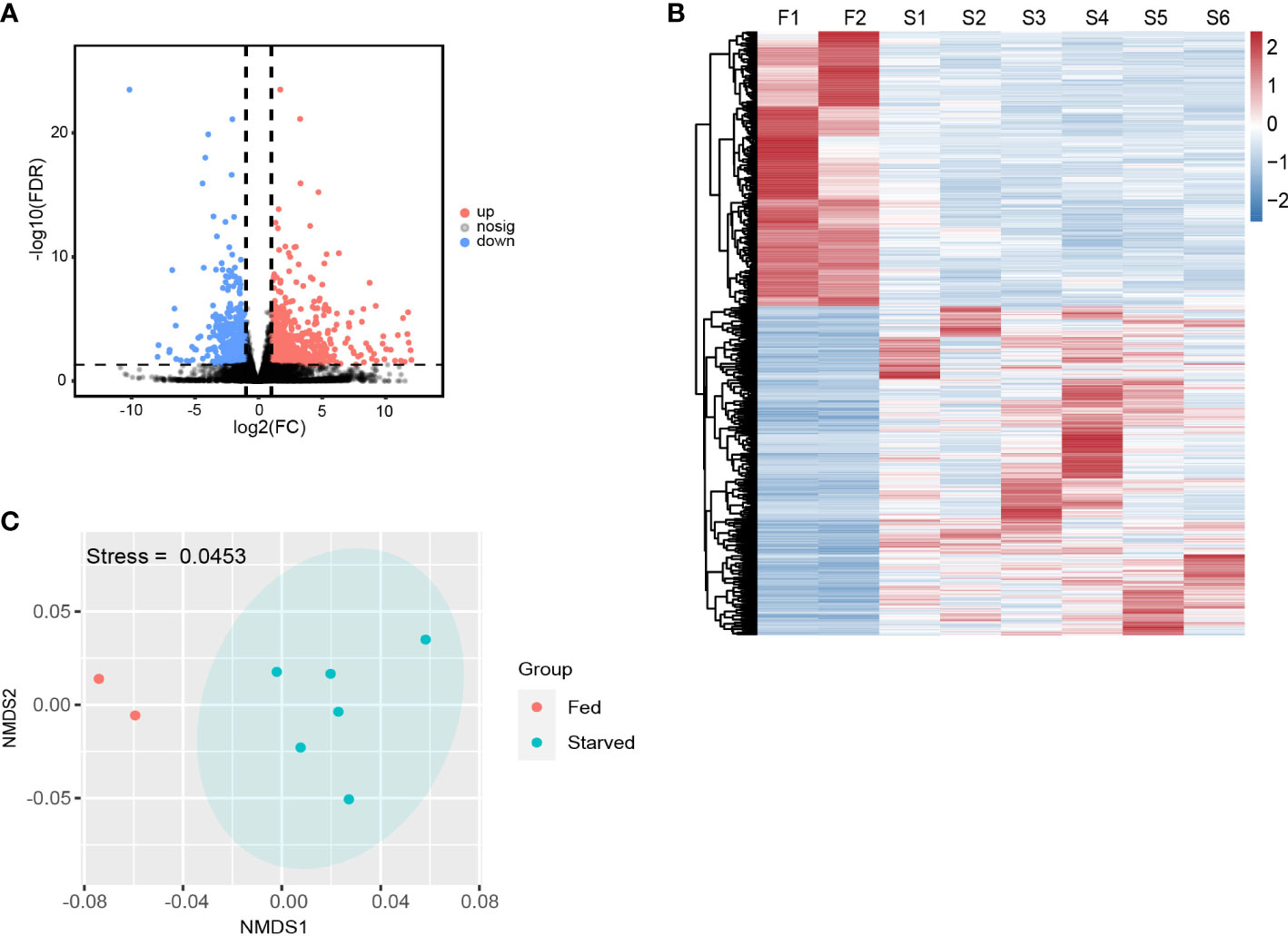
Figure 2 Overall DEGs expressions in starved and fed CoTS. (A) Volcano map of differential genes in transcriptome (FDR < 0.05, |log2(fold change)| ≥ 1). (B) Heatmap of the DEGs in fed and starved CoTS individuals (F = fed CoTS, n = 2; S = starved CoTS, n = 6). (C) Nonmetric multidimensional scaling (NMDS) plot of DEGs among fed and starved CoTS individuals.
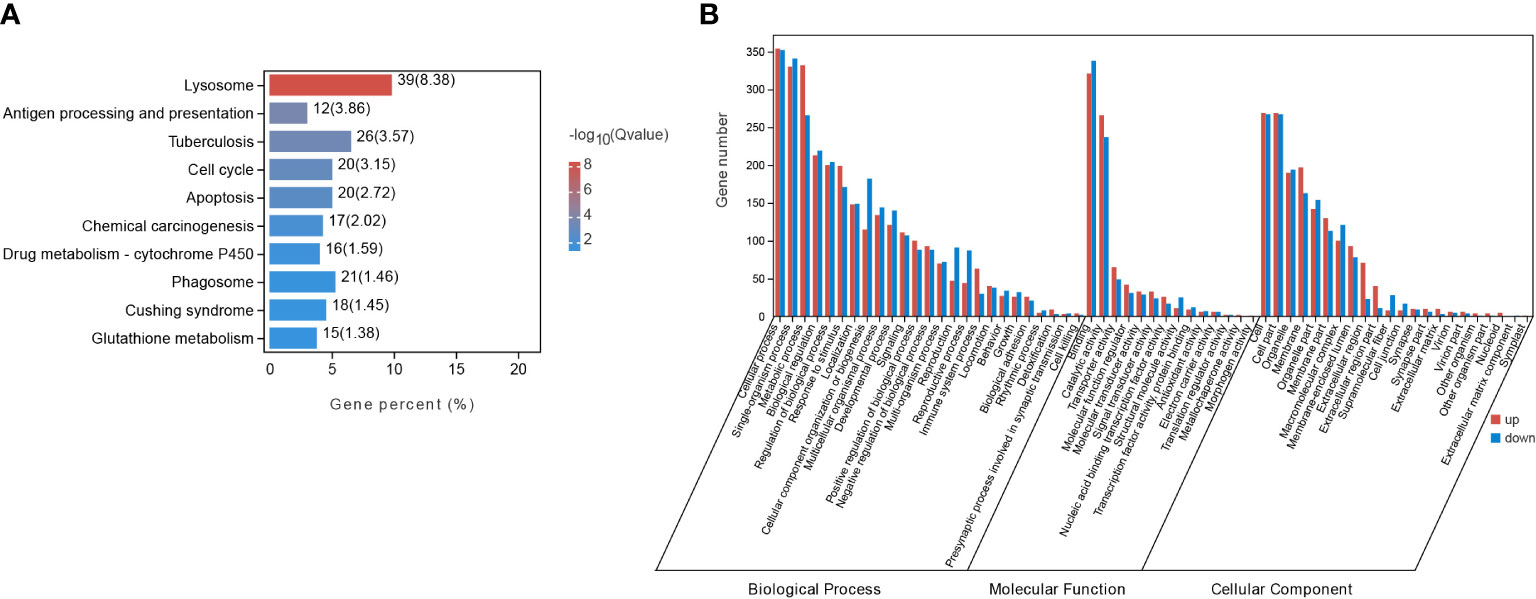
Figure 3 DEGs enrichment analysis in starved and fed CoTS. (A) Top 10 of KEGG enriched pathways. (B) Histogram of GO enriched genes in transcriptomes.
As shown in Figure 4, 12 selected genes altered in metabolism, immunity, development processes, sleep, and muscle construction were processed with qRT-PCR verification. Compared to the fed group, gene expression for uncharacterized protein LOC110982408 (named SLPUP1 in the present study) in sleep; glycine N-methyltransferase-like (GNMT) in amino acid metabolism; uncharacterized protein LOC110984484 (named LYS in the present study), legumain-like isoform X1 (LGMN) and cathepsin L1-like (LCP1) in immunity; cyclin-dependent kinase inhibitor 1B-like (named CCB in the present study) in cell cycle were all significantly upregulated compared to the expression level of fed CoTS samples, while the expression for myosin heavy chain and striated muscle-like (MYH16) and actin, muscle (CYIA) favor muscle construction; amine oxidase [flavin-containing] B-like isoform X1(MAOB) in amino acid metabolism; retinal dehydrogenase 2-like (ALDH1A2) in carbohydrates metabolism; G1/S-specific cyclin-D2-like (CCND2) and calmodulin-1-like isoform X1 (CAM1) in cell cycle were significantly downregulated. Similar gene expression trends were confirmed by comparing the qRT-PCR results with the RNA-seq data, indicating the RNA-seq results were reliable for further analysis.
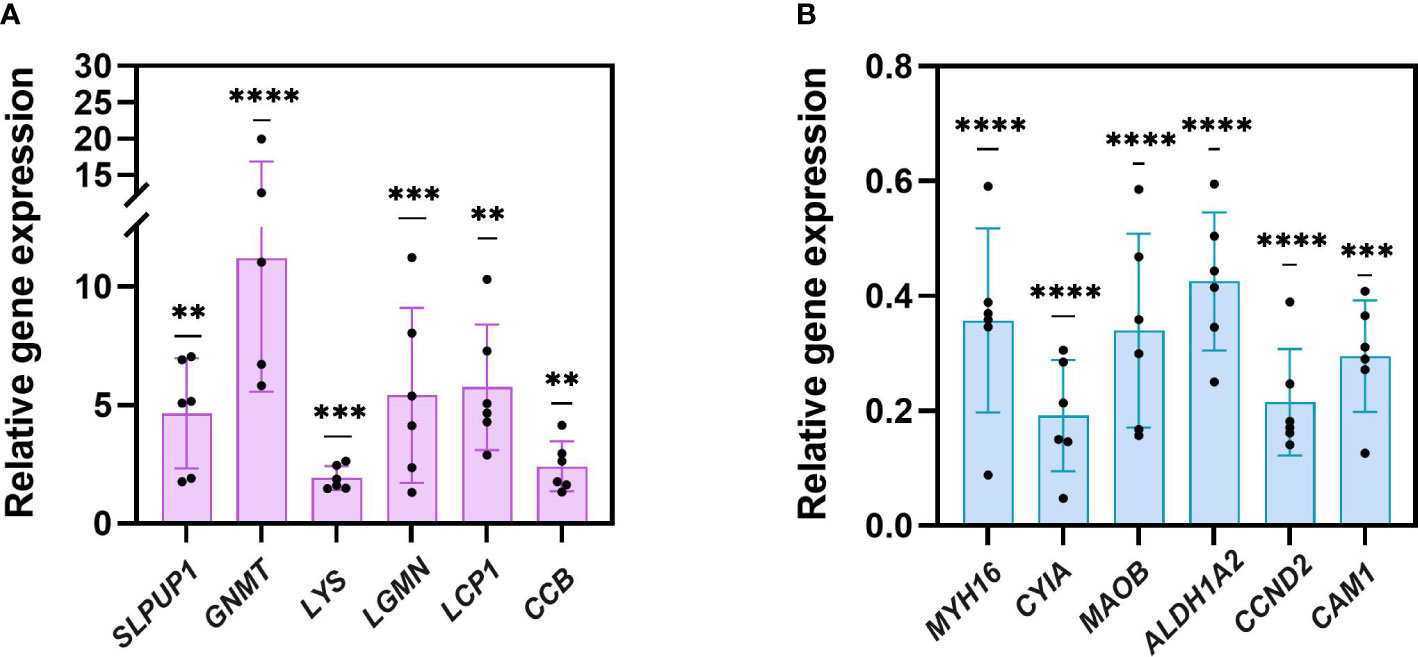
Figure 4 qRT-PCR verification of 12 genes. (A) six upregulated genes. (B) six downregulated genes (**p < 0.005, ***p< 0.001, ****p < 0.0001).
KEGG enrichment analysis indicates the most significantly altered pathways was in immunity, evidenced by the three immunity-related pathways among the Top 10 of KEGG pathways (Figure 3A), including 39 DEGs enriched in lysosome with the smallest Q value (Figure 3A, Supplementary Table 3.1), 12 DEGs in antigen processing and presentation, and 21 DEGs in phagosome as listed in Supplementary Table 3.1. Other immunity-related DEGs were found enriched in toll-like receptors (TLRs) signaling pathway (7 genes), complement and coagulation cascade pathway (5 genes) (Supplementary Table 3.1), all of which together revealed a more complete immune responses of CoTS under starvation.
Unlike vertebrates, starfishes as invertebrates only have innate immune responses (Chiaramonte and Russo, 2015). TLRs play an essential role in the innate immune responses in starfish that trigger transcription of immune functional genes to induce phagocytosis and produce immune effectors (Smith et al., 2010). As shown in Figure 5A, BPI as the function protein for lipopolysaccharide-binding was upregulated, presumably leading to more efficient reorganization of the pathogen. As such, the upregulated gene expression in TRAF6 and TBK1 compose of a highly conserved cascade, resulting in higher expression of the nuclear factor NFKB1, which activates phagocytosis and multiple immune effectors such as inflammatory cytokines (Betancur et al., 2017). When developed phagosomes find and fuse with lysosomes to eliminate pathogens and apoptotic cells to maintain cellular homeostasis, the upregulation of V-ATPase encoding gene VHAAC39-1 (Figure 5A) would lead to a higher activity of phagolysosome in starved CoTS (Lee et al., 2020; Lancaster et al., 2021). Due to its essential role in phagosome and lysosome lumen acidification that lowers the pH and activates hydrolytic enzymes, its upregulation suggests starved CoTS enhanced their digestion ability for prevention and resource usage (Lancaster et al., 2021; Nguyen and Yates, 2021). Expression of cathepsin genes (CTSA, CTSB, CTSC, CTSD, CTSL, CTSZ and LCP1) in starved CoTS were significantly increased (Figure 5A; Supplementary Table 3.1). Cathepsins can not only favor the antigen processing and presentation process to cause destruction of the phagocytosed microbes inside the lysosomes (Delaissé et al., 1991; Stinchcombe et al., 2004; Zhang et al., 2019), the released ones from secretory lysosomes can also assist microbe elimination extracellularly (Pollard and Borisy, 2003; Nair et al., 2005). Thus, the upregulation of the cathepsin genes in starved CoTS further suggested ability for pathogen removal was enhanced. In addition, starved CoTS showed an upregulation of complement and coagulation cascade activity (Supplementary Table 3.1), which allows pathogen uptake and facilitates phagocytosis to eliminate non-self-molecules, evidenced by the increased gene expression of complement factor B (C2, MCT7 in Figure 5A), which could increase the complement C3 production and furthermore activate phagocytosis process (Lubbers et al., 2017; Wahltinez et al., 2020).
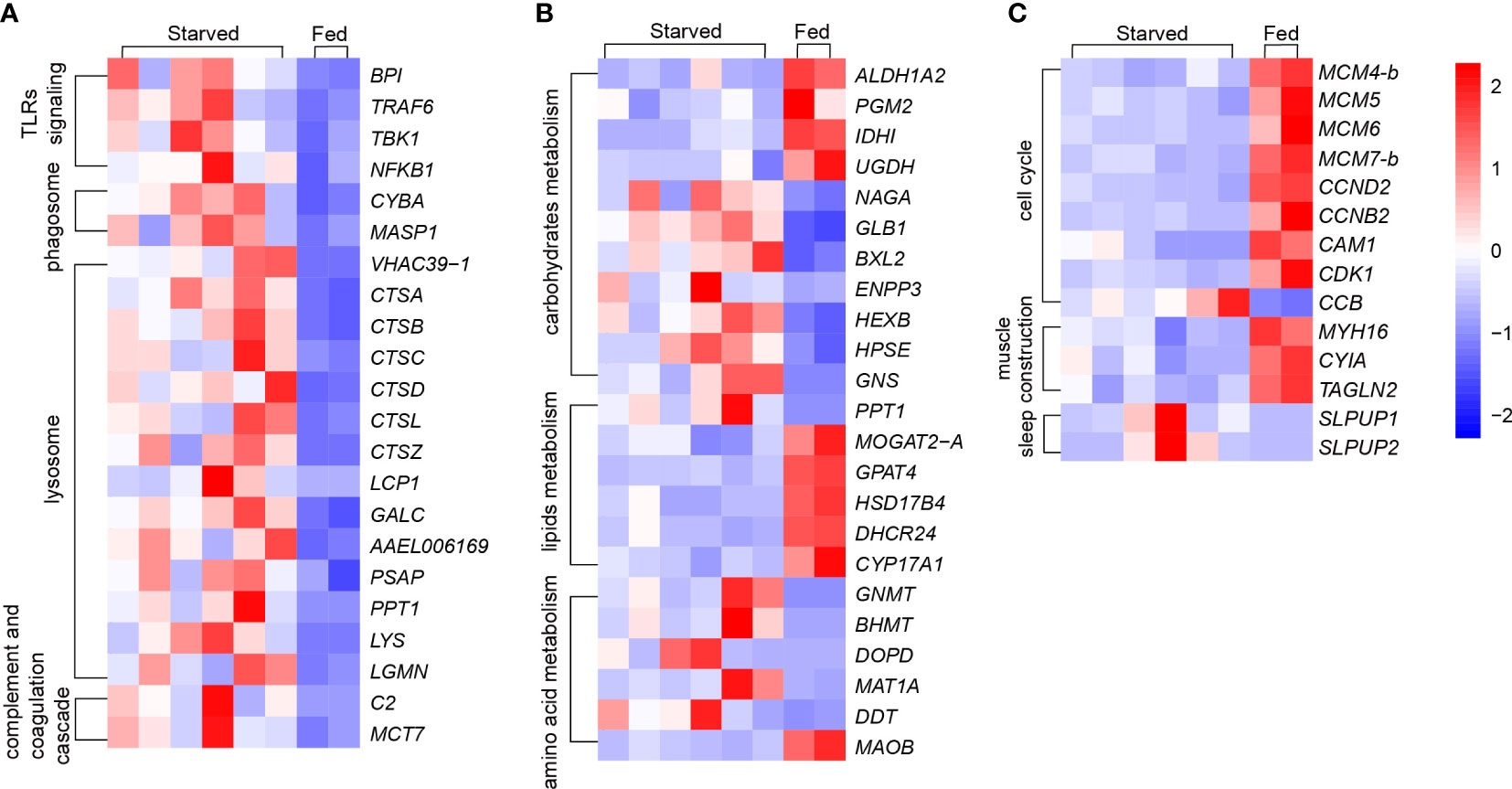
Figure 5 Changes of key genes under starvation in CoTS. (A) Immune responses: Toll-like receptors (TLRs) signing, phagosome, lysosome and complement and coagulation cascade. (B) Metabolism responses of carbohydrates, lipids and amino acids. (C) Other key changes of cell cycle, muscle construction and sleep.
In addition, CAT, SOD and T-AOC activities in echinoderms have been reported to increase under environmental stresses such as high temperature, high salinity and food depletion, which are essential for eliminating excess intracellular ROS (Liu et al., 2016). However, from the antioxidant activity assessments, average SOD activity was obtained as 93.4 U/g and 76.3 U/g for starved and fed CoTS respectively, CAT activity was 15166.0 U/g and 18666.5 U/g for starved and fed CoTS respectively, and the T-AOC amount was 6.1 µmol/g and 4.03 µmol/g for starved and fed CoTS respectively, statistical analyses suggest the difference in antioxidative capability caused by starvation was not significant in this study (Figure 6). Such results could on the one hand be due to the limited number of samples tested. On the other hand, the enzymatic activity may vary in tested individuals as such differences have been found in different animal genders (Crago and Klaper, 2011), both of which are recommended improvements for further experiment.
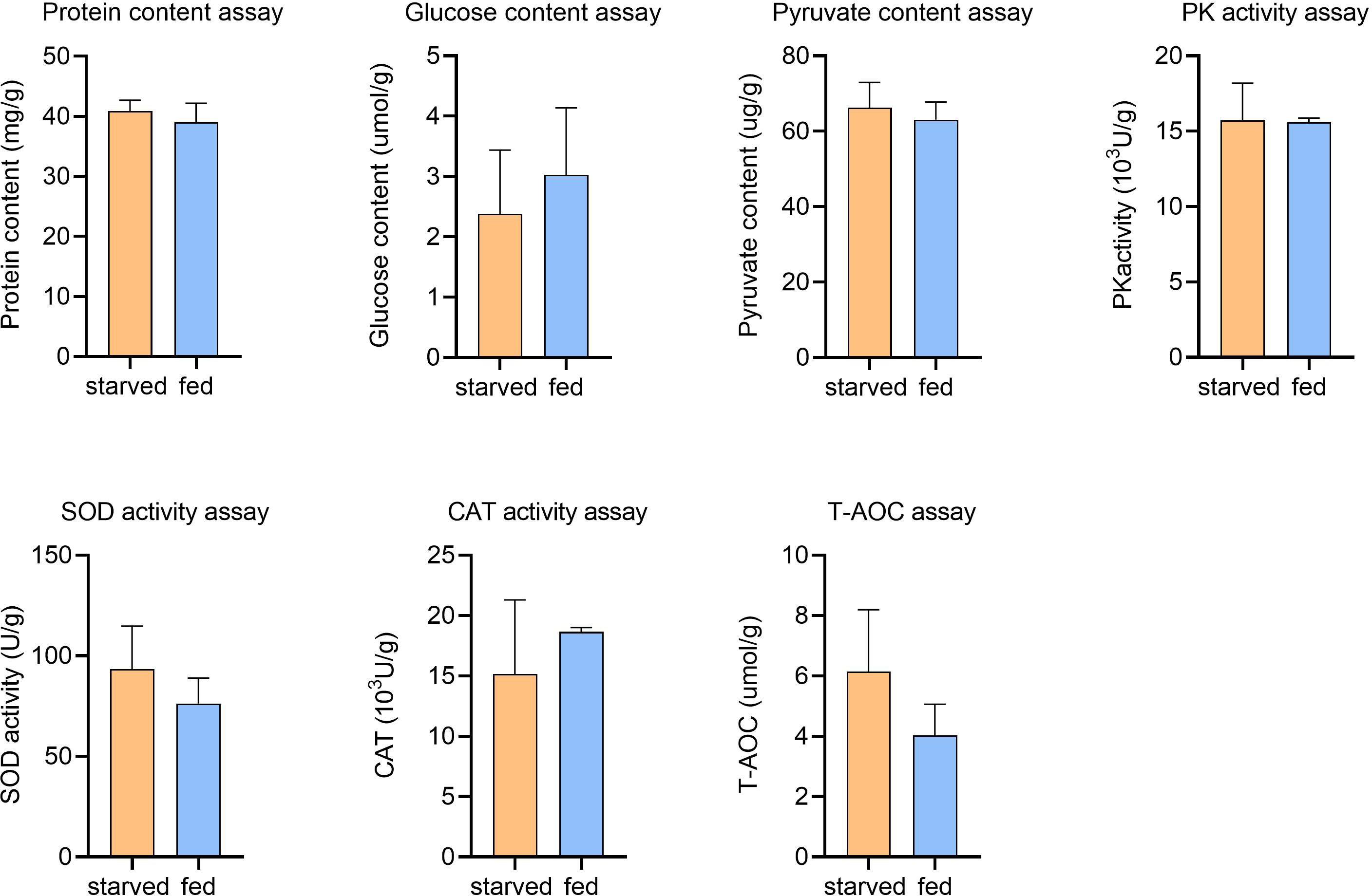
Figure 6 Physiological assays for starved and fed CoTS stomach tissue. Means ± S.E., nstarved = 6, nfed = 2.
Alive animals always demand a continuing supply of energy, either exogenous or endogenous energy input. When the exogenous energy input suspends, endogenous physiological fuel stores are expected to function as energy suppliers (McCue, 2010). As the available amount of energy is limited, and the energy expenditure in immune responses was suspected to increase in starved CoTS, which may cause species-specific energy suppression and reallocation, allowing species to wisely respond to starvation stress to extend survival time by reducing unnecessary activities and remaining essential processes for survival (McCue, 2010).
Starting with the physiological assessments, a negligible difference in the average glucose level was found in the starved CoTS (2.4 µmol/g) compared to fed CoTS (3.0 µmol/g), while the average PA content was 66.3 µg/g and 63.1 µg/g, PK activity was obtained as 1572.4 U/g and 1559.3 U/g from starved and fed CoTS respectively. Such results suggest in the present study, no significant changes in glucose concentrations were found between starved and fed CoTS, nor were changes in PA and PK activities critical for energy production (Figure 6), implying that CoTS may have developed alternative pathways to maintain the relative balance of glucose utilization and production, which is well documented at the transcriptome level. DEGs analysis implies a decreased glucose utilization activity in starved CoTS, evidenced by the 4 downregulated enzyme-coding genes in glucose utilization (e.g. ALDH1A2 in Figure 5B) as listed in Supplementary Table 3.2. In addition, acetyl-CoA might be in shortage as the gene expression of its utilizing enzymes (e.g. IDH1) was significantly decreased, resulting in suppressed citrate cycle. However, the upregulated expression of 4 genes in galactose metabolism (e.g. NAGA and GLB1 in Figure 5B), 5 genes in starch and sucrose metabolism (e.g. BXL2, ENPP3 in Figure 5B) and 5 genes in glycan degradation (e.g. HEXB, HPSE, and GNS in Figure 5B) pathways promotes glucose production (Supplementary Table 3.2). This phenomenon was also reported in aestivating sea cucumber, where starch and sucrose are hydrolyzed to produce glucose in response to starvation (Yang et al., 2021). Thus, such a shift could be a conserved strategy in starved echinoderms.
Lipids are usually considered the primary endogenous fuel in starved animals when faced with glucose depletion (McCue, 2010), alterations occurred in lipid metabolism were investigated through analysis of DEGs associated with lipid metabolism KEGG pathways. As shown in Figure 5B, PPT1 upregulated in starved CoTS stomach tissue, which was enriched in pathways of fatty acid metabolism and lysosome (Supplementary Table 3.2). Functioning as the catalyser of the thioester cleavage reactions in the hydrolysis of long chain fatty acyl CoAs (Hellsten et al., 1996), PPT1 may favour the degradation of substrates in lysosomes and the maintenance of cortical neurons (Hellsten et al., 1996; Yun et al., 2020). MOGAT2-A catalyses the synthesis of diacylglycerol downregulated in starved CoTS, which may suggest a lower absorption of fat occurred in starved CoTS stomach tissues (Cao et al., 2004). Downregulation of GPAT4 was found in starved CoTS, which impacts triacylglycerol synthesis during development and regulates glucose as well as lipid homeostasis (Yu et al., 2018). The downregulation of the HSD17B4 in starved CoTS stomach tissue may reflect a potential deficiency of D-bifunctional protein, which acts as the catalyser of multiple steps of beta-oxidation of very long chain fatty acids, and its downregulation was proven to break the lipid homeostasis in the nervous system and gonads, which eventually cause psychomotor retardation and infertility in mammalian (Lieber et al., 2014). In addition, steroid biosynthesis may be downregulated in starved CoTS as revealed by the significant downregulation of genes like CYP17A1 and DHCR24 in their stomach tissue as shown in Figure 5B. Although the exact role or mechanism of the genes remains unclear in starfish, CYP17A1 was found to promote development in invertebrate, such as gonad maturation (Thitiphuree et al., 2019), while DHCR24 functions as cell-protective protein for ROS scavenger and anti-apoptosis (Lu et al., 2012). As discussed above, starved CoTS seem to tighten up the lipid metabolism control for energy absorption and utilization, while loosening the control of activities in development and cell protection.
Protein or amino acid metabolism is usually considered the last resort of fuel for energy production when animals face critical lipid levels (McCue, 2010). It was suggested that some animals tend to recycle endogenous proteins, either to reduce the protein requirements or to reduce the net protein loss under starvation stress (McCue, 2010). According to the protein level test (Figure 6), average protein contents were 40.8 mg/g for starved CoTS and 39.1 mg/g for fed CoTS, such negligible difference between the two groups implying a homeostasis state may be achieved in starved CoTS. The previously mentioned cathepsin genes (e.g. CTSB) under immunity may also favor protein recycling through its significant upregulation. According to Lu et al. (2011) and Zhou et al. (2014), they as proteinases can promote autolysis by digesting connective tissues in echinoderms. Such genes that favor nutrient recycling were found not only phagolysosome-related pathways but also involved in metabolism, indicating the phagolysosome in starved CoTS might have become the nutrients processing center through exogenous pathogen digestion and endogenous recycling. Metabolism of multifunctional amino acids such as glycine was enhanced (e.g. GNMT, BHMT in Figure 5B), which plays an important role in regulating glucose level, antioxidant activity, and immunity (Takahashi et al., 2016; Hughey et al., 2018), all of which help reduce the impact of the starvation stress.
Starved CoTS was found to downregulate several functional gene expressions related to self-development. As shown in Figure 5C, expression levels for genes that enriched in cell cycle and DNA replication pathways (e.g. CCND2) were suppressed. In addition, expression of CAM1 dramatically reduced in starved CoTS. As an important Ca2+ regulator, downregulation of CAM1 may slow cell cycle phases and cellular proliferation (Berchtold and Villalobo, 2014). The cyclin-dependent kinase inhibitor 1B (named as CCB in the present study) that inhibits cell cycle was greatly upregulated. Such alterations indicate the cell cycle might be arrested in starved CoTS, which is a phenomenon also found in both invertebrates and vertebrates (Wu and Storey, 2012; Zhu et al., 2016). As reported by Zhu et al. (2016), the arrest of the cell cycle is an energy-saving action in response to the hypometabolic state of aestivating sea cucumbers. In addition, hibernating squirrels were found to dramatically decrease the ATP-expensive functional activities, which caused the cell cycle arrest (Wu and Storey, 2012). Other functional processes such as steroid biosynthesis as mentioned before was found significantly suppressed, which have been proven to make a profound impact on reproduction, development, and more in echinoderms (Köhler et al., 2007; Li et al., 2018). For instance, homologous of identified sex hormones, and corticosteroids and their receptors have been proven to have endocrine actions in starfish (Mita et al., 2011; Caballes and Pratchett, 2017). Other genes that promote energy consumption activities such as CoA utilization, vision development and neuromodulation were downregulated (e.g. ALDH1A2, ACAA2 and MAOB in Figure 5B) (Mondovì, 2017; Cho et al., 2021). The upregulation of MAT1A (Figure 5B) that causing degradation of the multifunctional amino acids implies the potential suppression of the biological processes such as catalyzation, regulation, and binding in starved CoTS (Marino and Gladyshev, 2010; Brosnan and Brosnan, 2020).
Sleep is a widely conserved process in the animal kingdom, especially in starved animals, sleep has been reported as a survival strategy for energy conservation and repairing (Roth et al., 2010; Menezes et al., 2020). Similarly in echinoderms, Li et al. (2018) reported that clock-related genes Egr1 and Klf2 are the key triggers for extended sleep, which lead to aestivation in sea cucumber A. japonicus. In this study, several genes related to sleep were annotated by the GO database, including the two most significantly altered genes, named SLPUP1 and SLPUP2. Over 200-fold change in upregulation of gene expression was found in SLPUP genes (Supplementary Table 3.3, Figure 5C), which share a conserved domain with the SLEEPLESS protein (SSS) (Lu et al., 2020) that was identified as a conserved sleep-promoting factor from Drosophila to mammals (Koh et al., 2008; Wu et al., 2014). The upregulation of SSS could lead to several phenotypes including decreased motility and activity-rest cycles (Ruan et al., 2017). Thus, it is likely that SLPUP genes play an important role in extending sleep and reducing motility in starved CoTS for energy saving and prolonged survival. The reduction motility of starved CoTS could be evidenced by the significantly downregulated muscle-related genes such as MYH16, CYIA, and TAGLN2 (Figure 5C), encoding for myosin heavy chain, actin (muscle), and myophilin-like protein respectively. Since the normal level of interaction of those motor proteins is essential for muscle contraction and further movement, such great downregulation would lead to a relatively immobile state in animals (Wells et al., 1996; Dominguez and Holmes, 2011). Thus, CoTS could suppress these processes as an altered energy expenditure strategy in response to starvation.
Compared to endotherms, ectotherms such as sea stars appear to have a broader tolerance to starvation due to their relatively low energy requirements (McCue, 2010). The results of the present study suggest that CoTS have developed a strategy to cope with months-long starvation characterized by minimizing non-survival related energy-consuming activities, increasing material recycling, and enhancing immunity. The present study reveals for the first time the survival strategy of CoTS under prolonged starvation stress at the molecular level, which will help us better understand the mechanism of CoTS outbreaks.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. Datasets supporting RNA sequencing data of this article have been deposited in the NCBI SRA under accession code PRJNA868810.
YL and ZY equally conducted the experiments as well as data analysis, and YL wrote the manuscript. CH and GQ helped to analyze transcriptome data. LP critically reviewed the manuscript. HJ, ZF, YY, FL collected and reared CoTS, CC provided funding, designed the experiment, analyzed the data and revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded in part by The National Key Research and Development Program of China (2020YFD0901100), The National Natural Science Foundation of China (31872605).
We thank the Tropical Aquatic Research and Development Center (Hainan, China) for providing facilities to rear the starfishes. We are grateful to Guangzhou Gene Denovo Biotechnology Co., Ltd for assisting in sequencing and bioinformatics analysis. We thank the data archive support from South China Sea Ocean Data Center (http://data.scsio.ac.cn, accessed on 16th July 2022), National science & Technology infrastructure of China (http://www.geodata.cn, accessed on 16th July 2022).
Authors YY and LF were employed by Sansha Track Ocean Coral Reef Conservation Research Institute Co., Ltd.,.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1021377/full#supplementary-material
Berchtold M. W., Villalobo A. (2014). The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim. Biophys. Acta 1843 (2), 398–435. doi: 10.1016/j.bbamcr.2013.10.021
Betancur P. A., Abraham B. J., Yiu Y. Y., Willingham S. B., Khameneh F., Zarnegar M., et al. (2017). A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat. Commun. 8 (1), 14802. doi: 10.1038/ncomms14802
Birkeland C., Lucas J. (1990). Acanthaster planci: major management problem of coral reefs (Boca Raton, FL, USA: CRC press).
Brosnan M. E., Brosnan J. T. (2020). Histidine metabolism and function. J. Nutr. 150 (Supplement_1), 2570S–2575S. doi: 10.1093/jn/nxaa079
Caballes C. F., Pratchett M. S. (2017). Environmental and biological cues for spawning in the crown-of-thorns starfish. PloS One 12 (3), e0173964. doi: 10.1371/journal.pone.0173964
Caballes C. F., Pratchett M. S., Kerr A. M., Rivera-Posada J. A. (2016). The role of maternal nutrition on oocyte size and quality, with respect to early larval development in the coral-eating starfish, Acanthaster planci. PloS One 11 (6), e0158007. doi: 10.1371/journal.pone.0158007
Cao J., Hawkins E., Brozinick J., Liu X., Zhang H., Burn P., et al. (2004). A predominant role of acyl-CoA: monoacylglycerol acyltransferase-2 in dietary fat absorption implicated by tissue distribution, subcellular localization, and up-regulation by high fat diet. J. Biol. Chem. 279 (18), 18878–18886. doi: 10.1074/jbc.m313272200
Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi: 10.1093/bioinformatics/bty560
Chiaramonte M., Russo R. (2015). The echinoderm innate humoral immune response. Ital. J. Zool. 82 (3), 300–308. doi: 10.1080/11250003.2015.1061615
Cho K., Lee S.-M., Heo J., Kwon Y. M., Chung D., Yu W.-J., et al. (2021). Retinaldehyde dehydrogenase inhibition-related adverse outcome pathway: Potential risk of retinoic acid synthesis inhibition during embryogenesis. Toxins 13 (11), 739. doi: 10.3390/toxins13110739
Clarke K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18 (1), 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Crago J., Klaper R. D. (2011). Influence of gender, feeding regimen, and exposure duration on gene expression associated with xenobiotic metabolism in fathead minnows (Pimephales promelas). Comp. Biochem. Physiol. Part C.: Toxicol. Pharmacol. 154 (3), 208–212. doi: 10.1016/j.cbpc.2011.05.016
De’ath G., Fabricius K. E., Sweatman H., Puotinen M. (2012). The 27–year decline of coral cover on the great barrier reef and its causes. PNAS 109 (44), 17995–17999. doi: 10.1073/pnas.1208909109
Delaissé J. M., Ledent P., Vaes G. (1991). Collagenolytic cysteine proteinases of bone tissue. cathepsin b, (pro)cathepsin l and a cathepsin l-like 70 kDa proteinase. Biochem. J. 279 (1), 167–174. doi: 10.1042/bj2790167
Dominguez R., Holmes K. C. (2011). Actin structure and function. Annu. Rev. Biophysics 40, 169–186. doi: 10.1146/annurev-biophys-042910-155359
Elliff C. I., Silva I. R. (2017). Coral reefs as the first line of defense: Shoreline protection in face of climate change. Mar. Environ. Res. 127, 148–154. doi: 10.1016/j.marenvres.2017.03.007
Hellsten E., Vesa J., Olkkonen V. M., Jalanko A., Peltonen L. (1996). Human palmitoyl protein thioesterase: evidence for lysosomal targeting of the enzyme and disturbed cellular routing in infantile neuronal ceroid lipofuscinosis. EMBO J. 15 (19), 5240–5245. doi: 10.1002/j.1460-2075.1996.tb00909.x
Heng W. K., Ho M.-J., Kuo C.-Y., Huang Y.-Y., Ko C.-Y., Jeng M.-S., et al. (2021). Crown-of-thorns starfish outbreak at Taiping island (Itu aba), spratlys, south China Sea. Bull. Mar. Sci. 98 (1), 101–102. doi: 10.5343/bms.2021.0030
Hughey C. C., Trefts E., Bracy D. P., James F. D., Donahue E. P., Wasserman D. H. (2018). Glycine n-methyltransferase deletion in mice diverts carbon flux from gluconeogenesis to pathways that utilize excess methionine cycle intermediates. J. Biol. Chem. 293 (30), 11944–11954. doi: 10.1074/jbc.ra118.002568
Kim D., Langmead B., Salzberg S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Koh K., Joiner W. J., Wu M. N., Yue Z., Smith C. J., Sehgal A. (2008). Identification of SLEEPLESS, a sleep-promoting factor. Science 321, 5887, 372–376. doi: 10.1126/science.1155942
Köhler H.-R., Kloas W., Schirling M., Lutz I., Reye A. L., Langen J.-S., et al. (2007). Sex steroid receptor evolution and signalling in aquatic invertebrates. Ecotoxicology 16 (1), 131–143. doi: 10.1007/s10646-006-0111-3
Kuo C.-Y., Ho M.-J., Heng W. K., Huang Y.-Y., Ko C.-Y., Jiang G.-C., et al. (2022). What is for dessert? crown-of-thorns starfish feeds on non-scleractinian anthozoans at Taiping island (Itu aba), spratlys, south China Sea. Mar. Biodivers. 52 (1):2. doi: 10.1007/s12526-021-01240-6
Lancaster C. E., Fountain A., Dayam R. M., Somerville E., Sheth J., Jacobelli V., et al. (2021). Phagosome resolution regenerates lysosomes and maintains the degradative capacity in phagocytes. J. Cell Biol. 220 (9):e202005072. doi: 10.1083/jcb.202005072
Lee H.-J., Woo Y., Hahn T.-W., Jung Y. M., Jung Y.-J. (2020). Formation and maturation of the phagosome: A key mechanism in innate immunity against intracellular bacterial infection. Microorganisms 8 (9), 1298. doi: 10.3390/microorganisms8091298
Leray M., Béraud M., Anker A., Chancerelle Y., Mills S. C. (2012). Acanthaster planci outbreak: Decline in coral health, coral size structure modification and consequences for obligate decapod assemblages. PloS One 7 (4), e35456. doi: 10.1371/journal.pone.0035456
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12 (1), 323. doi: 10.1186/1471-2105-12-323
Lieber D. S., Hershman S. G., Slate N. G., Calvo S. E., Sims K. B., Schmahmann J. D., et al. (2014). Next generation sequencing with copy number variant detection expands the phenotypic spectrum of HSD17B4-deficiency. BMC Med. Genet. 15 (30), 30. doi: 10.1186/1471-2350-15-30
Ling S. D., Cowan Z. L., Boada J., Flukes E. B., Pratchett M. S. (2020). Homing behaviour by destructive crown-of-thorns starfish is triggered by local availability of coral prey. Proc. R. Soc. B.: Biol. Sci. 287 (1938), 20201341. doi: 10.1098/rspb.2020.1341
Liu S. L., Zhou Y., Ru X. S., Zhang M. Z., Cao X. B., Yang H. S. (2016). Differences in immune function and metabolites between aestivating and non-aestivating Apostichopus japonicus. Aquaculture 459, 36–42. doi: 10.1016/j.aquaculture.2016.03.029
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Δ Δ C) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li Y., Wang R., Xun X., Wang J., Bao L., Thimmappa R., et al. (2018). Sea Cucumber genome provides insights into saponin biosynthesis and aestivation regulation. Cell Discovery 4 (1), 29. doi: 10.1038/s41421-018-0030-5
Li Y., Wu Z., Liang J., Chen S., Zhao J. (2019). Analysis on the outbreak period and cause of Acanthaster planci in xisha islands in recent 15 years. Chin. Sci. Bull. 64 (33), 3478–3484. doi: 10.1360/TB-2019-0152
Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 1–21. doi: 10.1186/s13059-014-0550-8
Lubbers R., van Essen M. F., van Kooten C., Trouw L. A. (2017). Production of complement components by cells of the immune system. Clin. Exp. Immunol. 188 (2), 183–194. doi: 10.1111/cei.12952
Lu X., Li Y., Liu J., Cao X., Wang X., Wang D., et al. (2012). The membrane topological analysis of 3β-hydroxysteroid-Δ24 reductase (DHCR24) on endoplasmic reticulum. J. Mol. Endocrinol. 48 (1), 1–9. doi: 10.1530/jme-11-0132
Lu P., Takai K., Weaver V. M., Werb Z. (2011). Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harbor Perspect. Biol. 3 (12), a005058–a005058. doi: 10.1101/cshperspect.a005058
Lu S., Wang J., Chitsaz F., Derbyshire M. K., Geer R. C., Gonzales N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48 (D1), D265–D268. doi: 10.1093/nar/gkz991
Marino S. M., Gladyshev V. N. (2010). Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 404 (5), 902–916. doi: 10.1016/j.jmb.2010.09.027
McCue M. D. (2010). Starvation physiology: Reviewing the different strategies animals use to survive a common challenge. Comp. Biochem. Physiol. Part A.: Mol. Integr. Physiol. 156 (1), 1–18. doi: 10.1016/j.cbpa.2010.01.002
Menezes L., de Moraes D. A., Ribeiro-Silva N., Silva S. M. A., Suchecki D., Luz J. (2020). Chronic REM sleep restriction in young rats increases energy expenditure with no change in food intake. Exp. Physiol. 105 (8), 1339–1348. doi: 10.1113/EP088474
Mita M., Yamamoto K., Nakamura M., Nagahama Y. (2011). Hormonal action of relaxin-like gonad-stimulating substance (GSS) on starfish ovaries in growing and fully grown states. Gen. Comp. Endocrinol. 172 (1), 85–89. doi: 10.1016/j.ygcen.2011.01.014
Moberg F., Folke C. (1999). Ecological goods and services of coral reef ecosystems. Ecol. Econom. 29 (2), 215–233. doi: 10.1016/S0921-8009(99)00009-9
Moran P. J. (1986). The acanthaster phenomenon. Oceanogr. Mar. Biol. Ann. Rev. 24, 379–480. doi: 10.1201/9781482267266
Nair S. V., Del Valle H., Gross P. S., Terwilliger D. P., Smith L. C. (2005). Macroarray analysis of coelomocyte gene expression in response to LPS in the sea urchin. identification of unexpected immune diversity in an invertebrate. Physiol. Genomics 22 (1), 33–47. doi: 10.1152/physiolgenomics.00052.2005
Nakamura M., Okaji K., Higa Y., Yamakawa E., Mitarai S. (2014). Spatial and temporal population dynamics of the crown-of-thorns starfish, Acanthaster planci, over a 24-year period along the central west coast of Okinawa island, Japan. Mar. Biol. 161 (11), 2521–2530. doi: 10.1007/s00227-014-2524-5
Nguyen J. A., Yates R. M. (2021). Better together: Current insights into phagosome-lysosome fusion. Front. Immunol. 12. doi: 10.3389/fimmu.2021.636078
Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 11 (9), 1650–1667. doi: 10.1038/nprot.2016.095
Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T., Salzberg S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Pollard T. D., Borisy G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112 (4), 453–465. doi: 10.1016/S0092-8674(03)00120-X
Pratchett M., Caballes C., Wilmes J., Matthews S., Mellin C., Sweatman H., et al. (2017). Thirty years of research on crown-of-Thorns starfish 1986–2016): Scientific advances and emerging opportunities. Diversity 9 (4), 41. doi: 10.3390/d9040041
Roth T. C., Rattenborg N. C., Pravosudov V. V. (2010). The ecological relevance of sleep: the trade-off between sleep, memory and energy conservation. Philos. Trans. R. Soc. B.: Biol. Sci. 365 (1542), 945–959. doi: 10.1098/rstb.2009.0209
Ruan H., Ueda A., Xing X., Wan X., Strub B., Mukai S., et al. (2017). Generation and characterization of new alleles of quiver (qvr) that encodes an extracellular modulator of the shaker potassium channel. Neurogenet 31 (4), 325–336. doi: 10.1080/01677063.2017.1393076
Saponari L., Montalbetti E., Galli P., Strona G., Seveso D., Dehnert I., et al. (2018). Monitoring and assessing a 2-year outbreak of the corallivorous seastar Acanthaster planci in ari atoll, republic of Maldives. Environ. Monit. Assess. 190 (6), 344. doi: 10.1007/s10661-018-6661-z
Sigl R., Laforsch C. (2016). The influence of water currents on movement patterns on sand in the crown-of-Thorns seastar (Acanthaster cf. solaris). Diversity 8 (4), 25. doi: 10.3390/d8040025
Smith L. C., Ghosh J., Buckley K. M., Clow L. A., Dheilly N. M., Haug T., et al. (2010). Echinoderm immunity. Adv Exp Med Biol, 708, 260–301. doi: 10.1007/978-1-4419-8059-5_14.
Stinchcombe J., Bossi G., Griffiths G. M. (2004). Linking albinism and immunity: The secrets of secretory lysosomes. Science 305 (5680), 55–59. doi: 10.1126/science.1095291
Stump R. (1996). An investigation to describe the population dynamics of acanthaster planci (L.) around lizard island, cairns section, great barrier reef marine park (Townsville, Qld, Australia: CRC Reef Research Cent).
Takahashi T., Sasaki K., Somfai T., Nagai T., Manabe N., Edashige K. (2016). N, n-dimethylglycine decreases oxidative stress and improves in vitro development of bovine embryos. J. Reprod. Dev. 62 (2), 209–212. doi: 10.1262/jrd.2015-149
Thitiphuree T., Nagasawa K., Osada M. (2019). Molecular identification of steroidogenesis-related genes in scallops and their potential roles in gametogenesis. J. Steroid Biochem. Mol. Biol. 186, 22–33. doi: 10.1016/j.jsbmb.2018.09.004
Tkachenko K. S., Huan N. H., Thanh N. H., Britayev T. A. (2020). Extensive coral reef decline in nha trang bay, Vietnam: Acanthaster planci outbreak: the final event in a sequence of chronic disturbances. Mar. Freshw. Res. 72 (2), 186–199. doi: 10.1071/MF20005
Wahltinez S. J., Newton A. L., Harms C. A., Lahner L. L., Stacy N. I. (2020). Coelomic fluid evaluation in pisaster ochraceus affected by Sea star wasting syndrome: Evidence of osmodysregulation, calcium homeostasis derangement, and coelomocyte responses. Front. Vet. Sci. 7. doi: 10.3389/fvets.2020.00131
Wells L., Edwards K. A., Bernstein S. I. (1996). Myosin heavy chain isoforms regulate muscle function but not myofibril assembly. EMBO J. 15 (17), 4454–4459. doi: 10.1002/j.1460-2075.1996.tb00822.x
Wu M., Robinson J., Joiner W. (2014). SLEEPLESS is a bifunctional regulator of excitability and cholinergic synaptic transmission. Curr. Biol. 24 (6), 621–629. doi: 10.1016/j.cub.2014.02.026
Wu C.-W., Storey K. B. (2012). Pattern of cellular quiescence over the hibernation cycle in liver of thirteen-lined ground squirrels. Cell Cycle 11 (9), 1714–1726. doi: 10.4161/cc.19799
Yang Q., Zhang X., Lu Z., Huang R., Tran N. T., Wu J., et al. (2021). Transcriptome and metabolome analyses of Sea cucumbers Apostichopus japonicus in southern China during the summer aestivation period. J. Ocean Univ. China 20 (1), 198–212. doi: 10.1007/s11802-021-4482-0
Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13 (1), 1–11. doi: 10.1186/1471-2105-13-134
Yu J., Loh K., Song Z.-Y., Yang H.-Q., Zhang Y., Lin S. (2018). Update on glycerol-3-phosphate acyltransferases: the roles in the development of insulin resistance. Nutr. Diabetes 8 (34), 34. doi: 10.1038/s41387-018-0045-x
Yun H. R., Jo Y. H., Kim J., Nguyen N. N. Y., Shin Y., Kim S. S., et al. (2020). Palmitoyl protein thioesterase 1 is essential for myogenic autophagy of C2C12 skeletal myoblast. Front. Physiol. 11. doi: 10.3389/fphys.2020.569221
Zann L., Brodie J., Berryman C., Naqasima M. (1987). Recruitment, ecology, growth and behavior of juvenile Acanthaster planci (L.)(Echinodermata: Asteroidea). Bull. Mar. Sci. 41 (2), 561–575.
Zhang N., Gao P., Yin B., Li J., Wu T., Kuang Y., et al. (2019). Cathepsin l promotes secretory IgA response by participating in antigen presentation pathways during Mycoplasma hyopneumoniae infection. PloS One 14 (4), e0215408. doi: 10.1371/journal.pone.0215408
Zhou D.-Y., Chang X.-N., Bao S.-S., Song L., Zhu B.-W., Dong X.-P., et al. (2014). Purification and partial characterisation of a cathepsin l-like proteinase from sea cucumber (Stichopus japonicus) and its tissue distribution in body wall. Food Chem. 158, 192–199. doi: 10.1016/j.foodchem.2014.02.105
Keywords: Acanthaster spp., crown-of-thorn starfish, starvation, physiological, transcriptomes
Citation: Yang L, Zhang Y, Chen H, Gao Q, Luo P, Hu J, Zheng F, Yuan Y, Fu L and Chen C (2022) Physiological and transcriptomic responses to starvation in the corallivorous crown-of-thorn starfish. Front. Mar. Sci. 9:1021377. doi: 10.3389/fmars.2022.1021377
Received: 18 August 2022; Accepted: 03 October 2022;
Published: 20 October 2022.
Edited by:
Shinya Shikina, National Taiwan Ocean University, TaiwanCopyright © 2022 Yang, Zhang, Chen, Gao, Luo, Hu, Zheng, Yuan, Fu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Chen, Y2hlbi5jaGFuZ0BzY3Npby5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.