- 1Key Laboratory of Marine Genetics and Breeding, Ministry of Education, Ocean University of China, Qingdao, China
- 2Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 3Key laboratory of Tropical Aquatic Germplasm of Hainan Province, Sanya Oceanographic Institution, Ocean University of China, Sanya, China
- 4Hainan Yazhou Bay Seed Laboratory, Sanya, China
The habitat occupied by flatfish fry differs considerably in light regime from that of the adult. The diversity of opsins and their expression patterns appear greatest for flatfishes to experience variable light environments. Yet, opsin repertoires and expression patterns in this group of fishes are poorly described. To understand how the visual system has adapted to such changes, we unveil that Japanese flounder (Paralichthys olivaceus) has a visual system adapted to a benthic environment by fine-tuning paralogous opsins (SWS2A and RH2) for wavelength shift and regulated expression. P. olivaceus express five basic opsin genes (M/LWS, SWS1, SWS2, RH1 and RH2) and gene-specific duplications were observed in RH2 and SWS2 paralogues. The expression of the three short-wavelength sensitive genes, SWS2Aα, SWS2Aβ, and SWS2B, is significantly elevated at the benthic stages, especially in SWS2Aβ a striking expression change is observed. The four middle-wavelength sensitive genes exhibit divergent expressions, the expression of RH2A-1 and RH2A-2 increased, while that of RH2B-1 and RH2B-2 decreased significantly from pelagic to benthic stage, especially RH2A-2. At present, changes at a total of 26 sites are known to have modified the λmax of various visual pigments during vertebrate evolution. Thus, these tuning site variations in our P. olivaceus are suspected to cause a green-shift in the λmax of SWS2Aα pigments and blue-shift in that of in RH2A-2. Together, our results suggest that RH2 and SWS2 opsin repertoires serve to optimize visual function under variable light environments by gene family duplications, differential expressions, and maximum absorption wavelength (λmax) variations.
Introduction
Opsins are membrane-bound proteins with seven α-helical structures, belonging to G-protein-coupled receptors (GPCRs) families (Hárosi, 1994; Yokoyama, 1997; Shichida and Imai, 1998). The visual opsins present in photoreceptor cells mainly function as light signal receptors from outer environment using a vitamin-A derived chromophore, 11-cis-retinal. Based on color vision discrimination of spectral differences in photic environments, visual opsins are sorted into five sub-families, namely, “red-sensitive” M/LWS, “green-sensitive” RH2, “blue-sensitive” SWS2, “ultraviolet-sensitive” SWS1, and “dim-light sensitive rhodopsin” RH1 (Yokoyama, 1997; Yokoyama, 2000b; Yokoyama, 2002; Yokoyama, 2008). As the first step of vision, opsins play vital roles in food gathering, communication, predator avoidance, mate selection, and navigation (Hoffmann et al., 2007; Watson et al., 2011; Cortesi et al., 2015).
Visual systems in fish are typically complex because of the remarkable diversity of visual opsins compared with other vertebrates (Rennison et al., 2012; Davies et al., 2015). The reason for this diversity is largely related to the underlying genetic mechanisms, including sequence evolution, lineage-specific tandem duplication, changes in gene expression, gene fusion, and gene mutation (Chinen et al., 2003; Hofmann and Carleton, 2009; Rennison et al., 2012; Nakamura et al., 2013; Cortesi et al., 2015). For example, the interaction of both depth-related variation in opsin sensitivity as well as nuptial colouration (ie sexual selection) that resulted in speciation (Seehausen et al., 2008).
With increasing depth in the ocean the light gets dimmer, the sunlight penetrates the clearest oceanic water to a maximum of 1000 m and long wavelength light and ultraviolet light attenuated most strongly so that, eventually, the light left contains only blue wavelengths close to 480 nm, like dim blue moonlight when observation in the deep water (Land and Osorio, 2011). Therefore, opsin pigments are usually tuned to adapt the peak transmission by wavelength shifts. A292S replacement, an amino acid replacement at site 292 of the rhodopsin protein, causes a blue-shift in RH1 pigments of East African cichlid fishes, providing functional adaptations to the deep-water photic environment (Sugawara et al., 2005). Although only RH1 and RH2 genes, the coelacanth adapt to weak photic environment at depths of 200 m by both pigments short wavelength shifts (Yokoyama, 2000a). The adult elephant shark migrates into shallower river mouth to spawn. Therefore, the retention and duplication of LWS pigments in elephant shark show a novel adaptation to the varying photic environment of the deep and shallow waters encountered by short wavelength shift in LWS1 (Davies et al., 2009).
A great number of teleosts experience dramatic changes in morphology and physiology from larva to juvenile stage (Youson, 1988). Among them, the drastic metamorphic changes from symmetrical to asymmetrical body in flatfishes is one of typical examples. In addition to metamorphic changes, the photic environments inhabited by flatfish are quite divergent throughout their life cycle. Flatfish larvae are pelagic, juveniles inhabit shallow, near-shore waters, and adults are benthic and often descend to depths that exceed 200 m (Iwanicki et al., 2017). It appears that diversity of opsin genes is quite different in flatfish. Nine visual opsin genes have been characterized in turbot (Scophthalmus maximus), eight in barfin flounder (Verasper moseri), while tongue sole (Cynoglossus semilaevis), winter flounder (Pseudopleuronectes americanus) and Atlantic halibut (Hippoglossus hippoglossus) contain five opsin genes (Helvik et al., 2002; Mader and Cameron, 2004; Chen et al., 2014; Kasagi et al., 2015; Figueras et al., 2016). Thus, it appears that diversity of opsins is important for flatfishes to experience variable light environments.
P. olivaceus is an economically important marine species cultivated in China, Japan and Korea. P. olivaceus also undergo metamorphosis process, including 90° rotation in body position and the migration of one eye to the contralateral side, which lead to a transition from the pelagic to the benthic habitat accompanied by changes in the photic environment (Fuiman, 1997; Gibson, 1997; Geffen et al., 2007). However, the molecular mechanism responsible for the adaptation of P. olivaceus to the dim-bottom vision after metamorphosis remains largely unknown. It is thus important to elucidate this evolutionary change, not only at anatomic and physiological levels but also at molecular and genetic levels. The aim of this study was to characterize the visual system of P. olivaceus in the course of their adaptation to various photic environments.
Materials and methods
Ethics statement
This study was approved by the Animal Care and Use Committee of the Centre for Applied Aquatic Genomics at the Ocean University of China.
Fish samples
P. olivaceus samples were collected from Haiyang Yellow Sea Aquatic Product CO., Ltd, Shandong, China. The samples were collected on the basis of the process of metamorphosis including pre-metamorphosis 14 days post hatching, (dph), metamorphic climax (25 dph), and post-metamorphosis (49 dph) in accordance with the migration of eyes (Takashi, 1982). A total of three individuals were collected at each time point. Prior to simple collection for adult fish, all individuals were anesthetized by MS-222 at 30 mg/mL. P. olivaceus eyes were collected from each fish, frozen immediately in liquid nitrogen, and then stored at -80°C until analyzed.
RNA extraction and cDNA synthesis
Total RNA from each sample was extracted separately using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and treated with RNase-free DNase I (TaKaRa, Dalian, China) to eliminate the genomic DNA contamination. cDNA synthesis was performed with 1μg total RNA and random hexamer primers using Reverse Transcriptase M-MLV Kit (TaKaRa, Dalian, China) following the manufacturer’s protocol. The quality and the quantity of RNA were then detected by 1.5% agarose gel electrophoresis and spectrophotometry with NanoPhotometer Pearl (Implen, Munich, Germany).
Expression of visual opsin genes
Specific primer pairs for visual opsin genes were listed in Table 1. Pre-experiments were conducted to confirm the generation of single cDNA PCR products. The 18S rRNA gene was used as internal reference gene. Samples from three individuals were pooled together and performed in triplicate. Then quantitative real-time PCR (qRT-PCR) was performed with 2×SYBR Green qPCR Master Mix (US Everbright Inc.) by LightCycler 480 (Roche, Forrentrasse, Switzerland) at 95°C for 5 min pre-incubation, followed by 45 cycles of 95°C for 15 s and 60°C for 45 s. Relative expression level of genes was calculated by 2-ΔΔCt method.
The qRT-PCR data were subjected to analysis using one-way ANOVA with SPSS 20.0 (IBM, New York, USA). P < 0.05 was considered to indicate statistical significance. All data were expressed as the mean ± standard error of the mean (SEM).
Phylogenetic analysis and synteny analysis
Sequence alignments of a variety of visual opsin genes were generated by ClustalW (Chenna et al., 2003). A Bayesian probabilistic inference method was performed with a Metropolis Markov chain Monte Carlo (MCMC) algorithm using MrBayes 3.2.2 software (Huelsenbeck and Ronquist, 2001; Ronquist et al., 2012). A general time-reversal (GTR) model (Lanave et al., 1984) was used, applying a gamma-distributed rate of variation across all sites with a proportion of invariable sites. Two runs and four chains in parallel were performed for 400000 generations with a chain sample frequency of 10 generations. The first 25% (10000) of all trees generated were discarded as burnin to allow for tree convergence and a low (close to 0.01) standard deviation of split frequencies.
Whole-genome sequences used for synteny analysis were obtained from NCBI and Ensembl. The upstream and downstream genes of RH2 and SWS2 were searched from gff3 files. Then, the figures were drawn by Genomicus.
Results and discussion
Opsin gene duplication and phylogeny
We identified all types of opsin genes, M/LWS, SWS1, SWS2, RH1 and RH2 from both transcriptomes (Accession number SRX500343) and annotated genes (Wang et al., 2014; Shao et al., 2017). The P. olivaceus genome possesses 10 visual pigment genes: one each for SWS1, M/LWS, and RH1, three for SWS2, and four for RH2. All ten P. olivaceus opsin genes were positioned ancestrally to their teleost counterparts within each clade in the phylogenetic tree (Figure S1). The three blue sensitive SWS2 genes reside in a single tandem-array as two distinct clades: SWS2B and SWS2Aα/SWS2Aβ (Figures 1B, F). The four RH2 genes, RH2A-1, RH2A-2, RH2B-1 and RH2B-2, form another highly conserved tandem array as two distinct clades, RH2A1/A2 and RH2B1/B2 (Figures 1A, D). Accession numbers of visual opsin genes for phylogenetic analysis were listed in Table S1.
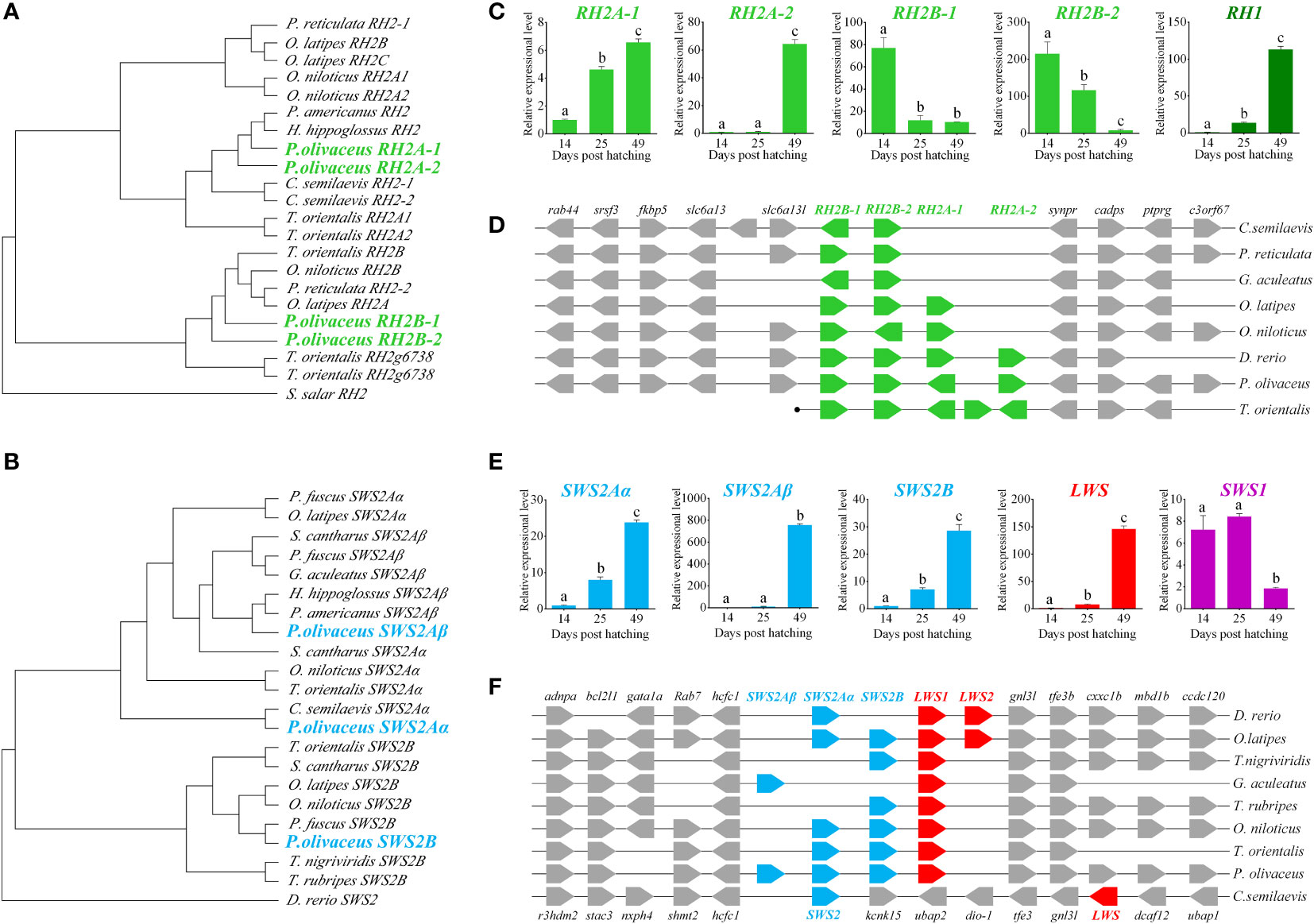
Figure 1 Charactorization of opsin genes in P. olivaceus. (A, B) The phylogenetic analysis of RH2 and SWS2 genes. (C) The expression of RH genes during the process of metamorphosis in P. olivaceus. (D) Syntenic analyses of RH genes in teleosts. (E) The expression of SWS genes during the process of metamorphosis in P. olivaceus. (F) Syntenic analyses of SWS genes in teleosts. Different letters represent significant difference.
Opsins are one of the most dynamic gene families in the evolution of vertebrates tightly linked to the adaptive evolution of animal vision system. Teleost opsins are most diversified ones for the successful occupation of habitat ranging from clear shallow rivers to dark deep sea. The phylogenetic results of the three SWS2 and four RH2 opsins in our P. olivaceus revealed both RH2A1/A2 and RH2B1/B2 pairs were apparently derived from additional rounds of species-specific gene duplications, like SWS gene evolution in other percomorph fishes (Cortesi et al., 2015).
Whole-genome and single-gene duplications are important evolutionary mechanisms that provide new genetic raw material for the formation of biological diversification (Ohno, 1970).The multiple rounds of whole-genome duplications are considered to have laid the genomic foundation for the evolutionary status of teleost fishes (Meyer and Van de Peer, 2005). While single-gene duplications are thought more important for adaptive diversification of entire gene families, such as MHC genes (De Souza and Bonilla-Rodriguez, 2007) and opsins in fishes (Hofmann and Carleton, 2009). In the P. olivaceus genome, the copy numbers of RH2 and SWS2 opsins not only differ from the deep ocean-adapted tuna fish (Nakamura et al., 2013), but also from those of tongue sole and turbot. The tongue sole has only one SWS2Aα (with no SWS2Aβ and SWS2B) gene and two RH2 genes, whereas turbot has two SWS2 genes and five RH2 genes with one of them being not tandemly duplicated (Chen et al., 2014; Figueras et al., 2016). Although they experience the same metamorphosis during early development and occupy benthic habitat during adult life, tongue sole usually distributes in river mouth and shallow waters with depths of 5 to15m, flounder prefers depths of 20 to 70m, whereas turbot is most abundant in 30 to100m.
Opsin gene expression
Opsin genes exhibit drastic changes in expression patterns from pelagic larvae to benthic juveniles. The scotopic vision gene RH1 and long-wavelength sensitive gene LWS1 show poor expression before metamorphosis (14 dph) but significantly increased during (25 dph) and after (49 dph) metamorphosis (P < 0.01). The expression of the three short-wavelength sensitive genes, SWS2Aα, SWS2Aβ, and SWS2B, is also significantly elevated at the benthic stages (P < 0.01), especially in SWS2Aβ a striking expression change is observed. The four middle wavelength sensitive pigment genes exhibit divergent expressions, the expression of RH2A-1 and RH2A-2 increases, while that of RH2B-1 and RH2B-2 decreased significantly from pelagic to benthic stage (P < 0.01), especially RH2A-2 (Figures 1C, E).
To date, little has been known about the expressions of opsin genes at different developmental stages of flatfish. Chen et al. (2014) reported in tongue sole that the expression of the scotopic vision RH1 gene and long-wavelength sensitive LWS gene are signifcantly higher in the benthic stages, which are similar in P. olivaceus in this study. They also reported a expressional decrease from pelagic to benthic stages of RH2 gene. However, they did not define which RH2 gene is detected, as there are two RH2 genes in the tongue sole genome. Our result showed obvious expressional diversification of RH2 genes with the two RH2A-1/RH2A-2 decrease while RH2B-1/RH2B-2 increase from pelagic to benthic stages. These observations may suggest that flounder possesses diversed sensitivity of different wavelengths during different life stages, reflecting its adaptation to changed vision environent. Because flatfish experience methamorphosis and undergo an eye migration during this process, it is reasonable to consider that the dynamic expression patterns of RH2A and RH2B genes is possibily caused by temperal and spatial development of different subtypes of opsins during early development, similar to the case of zebrafish (Takechi & Kawamura, 2005) .
Although RH2 and SWS2 belonged to opsin gene family, the functions were different. It has been reported that RH2 was green-sensitive, whereas SWS2 was blue-sensitive (Yokoyama, 1997; Yokoyama, 2000b; Yokoyama, 2002; Yokoyama, 2008). Both RH2 and SWS2 were expanded during evolution P. olivaceus genome. The expanded RH2 might have divergent in function to adapt benthic habitat after metamorphosis. We speculated that RH2B-1/2 maintained the original green-sensitive function. But RH2A-1/2 differentiated shifted wave-length sensitivity. Because, the expression patterns of RH2B-1/2 showed marked decrease after metamorphosis as they are no longer important in benthic deep sea environment, while RH2A-1/2 showed sharply elevated expression patterns.
Generally, light of longer wave length was predominant in pelagic habitat, and RH2B-1/2 was sensitive to green. Depth of life stage increased with a change from pelagic to benthic, and from shallow to deep sea habitat. The wavelength also changed as the depth of the seawater increased, with blue and blue-green light of relatively shorter wavelengths becoming predominant in deep environment. The remarkably increased expression of SWS2 paralogues were in consensus with the habitat adaptation requirement.
Tuning site analysis
Some amino acid sequence variations can change the maximum absorption wavelength (λmax) of the visual pigments. At present, changes at a total of 26 sites are known to have modified the λmax of various visual pigments during vertebrate evolution. We examined amino acid sequence variations among the three SWS2 genes and the four RH2 genes for functional implications (Tables S2, S3), which are rather numerous as compared to those of other fish species (Figures 2, 3); some of these sites appear involved in fine-tuning of light sensitivity in blue and green opsins. In P. olivaceus, SWS2 is different from other teleosts at several predicted spectral tuning sites. The amino acids of the three SWS2 members at the two important tuning sites 94 and 97 were all different from one another, and site 86 in SWS2Aβ and 94 in SWS2Aα being specific amino acid changes in the teleosts. Different from most other teleost SWS2 genes that possess S292 and varied 116 (T, L, V or A), P. olivaceus SWS2Aβ possessing M116 and A292 while SWS2Aα possess M116 that are virtually the same with corresponding tuning sites of most RH2 orthologous genes of most other teleosts (Table 2).
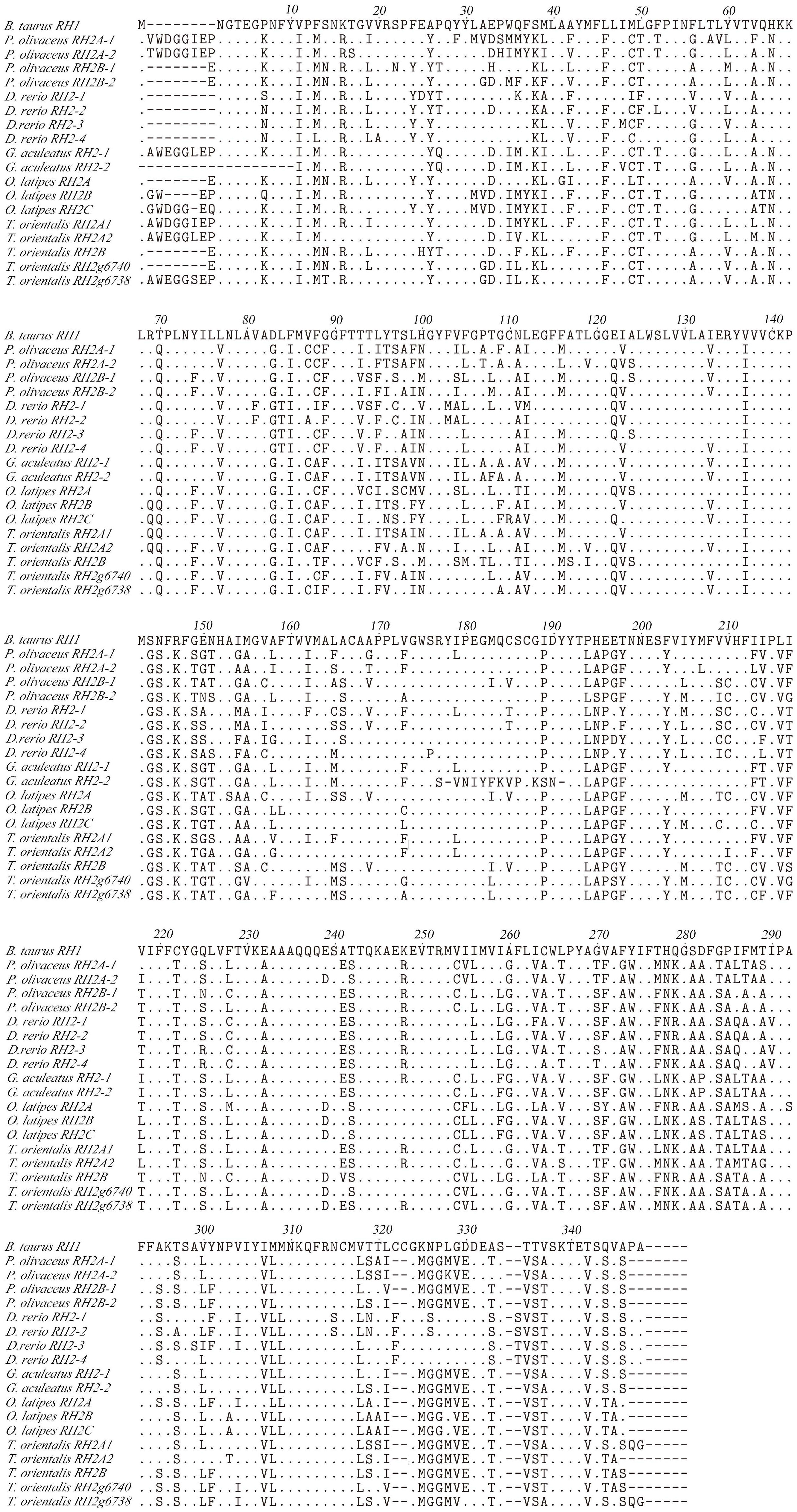
Figure 2 Amino acids alignment of RH2. The dots represented the same amino acid sites compared with B. taurus RH1 sequence among different species. The horizontal lines represented the missing of amino acid sites compared with each other.
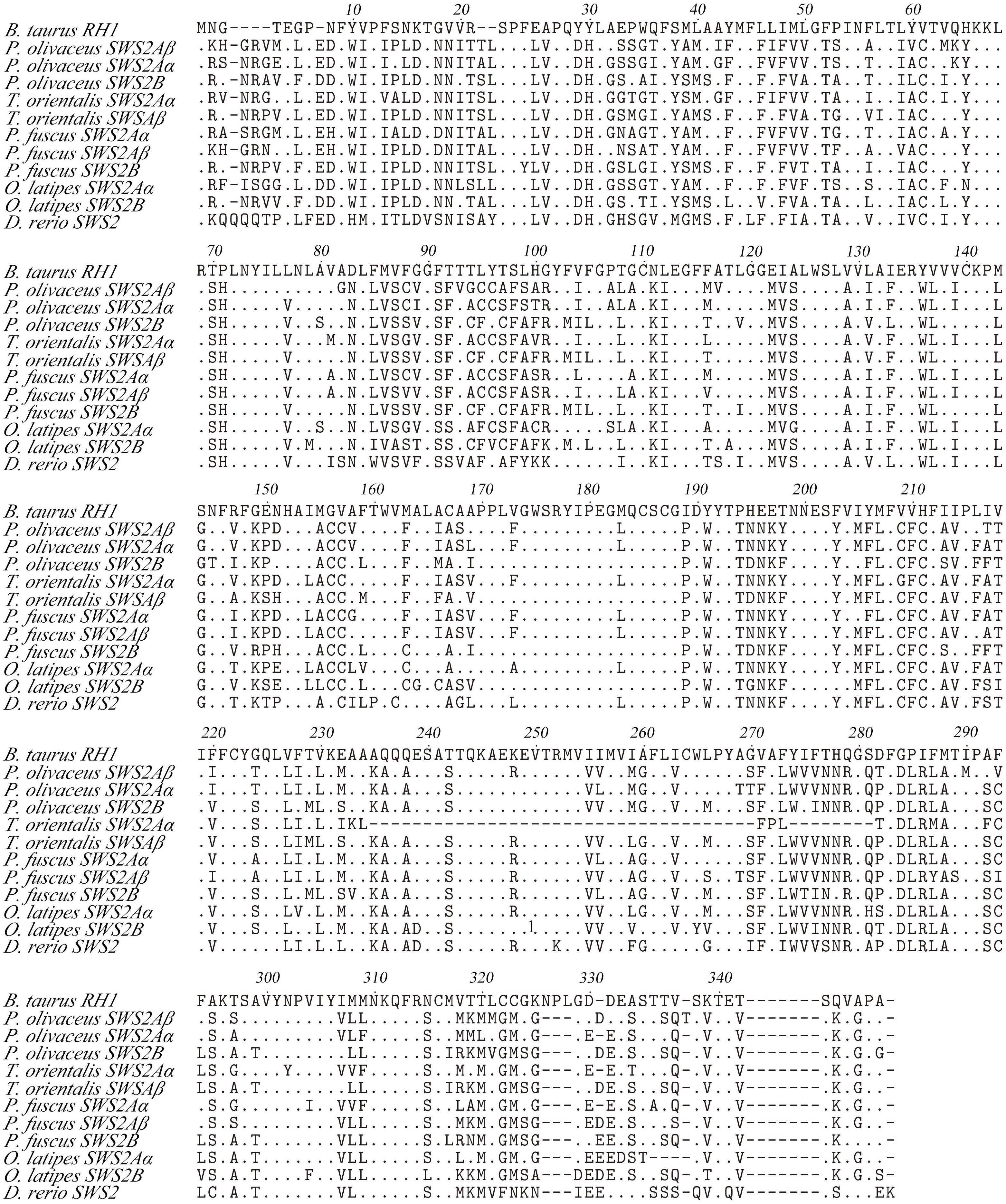
Figure 3 Amino acids alignment of SWS2. The dots represented the same amino acid sites compared with B. taurus RH1 sequence among different species. The horizontal lines represented the missing of amino acid sites compared with each other.
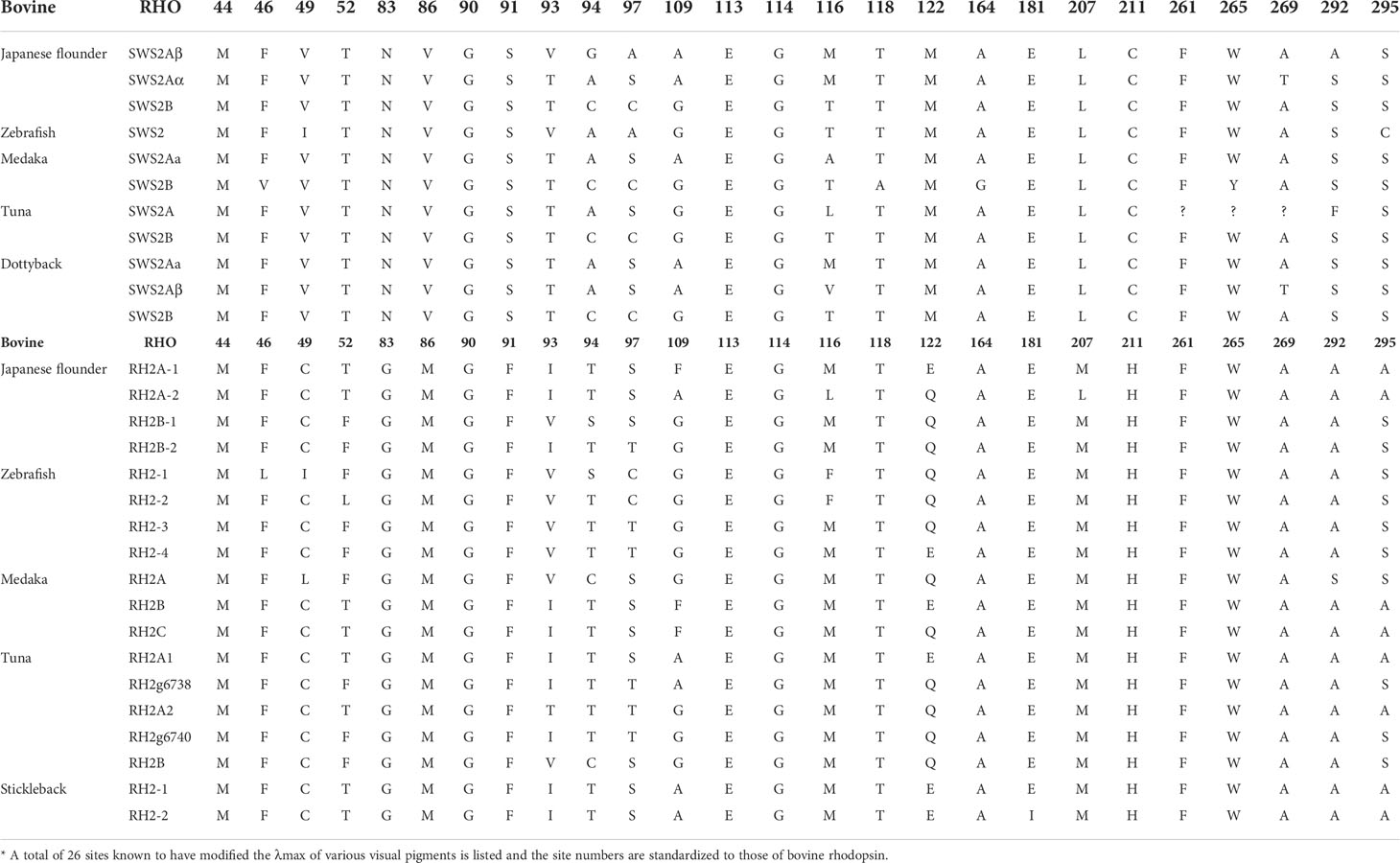
Table 2 Comparison of the representative amino acid sites involved in the light sensitivity of SWS2 and RH2 opsins among several teleosts*.
Previously report suggested that the λmax values of P. olivaceus SWS2β was 465.6 nm, which was categorized into blue-sensitive opsin (Kasagi et al., 2018). According to the results in bluefin killifish (Yokoyama et al., 2007), the amino acid compositions of tuning sites 44, 46, 94, 97, 109, 116, 118, 265, and 292 of the SWS2A genes tend to shift the λmax value. But because almost all RH2 pigments have M116, and they are associated with the pigments with green-shifted λmax values, the site changes in our P. olivaceus are suspected to cause a green-shift in the λmax of SWS2Aαpigments. When compared with the wavelength shifts caused by site-directed mutations in other species, the λmax value of SWS2B were found shifting toward purple (shorter wavelength-sensitive) by all tuning site changes, especially site 94C, 97C.
Similarly, amino acid variations at sites 83, 116, 207 and 292 have been reported to be the tuning sites affecting λmax value in RH2 pigments (Yokoyama et al., 1999). The sequences of P. olivaceus RH2 are also different from other teleosts at some of the predicted spectral tuning sites (Matsumoto et al., 2006; Hofmann and Carleton, 2009; Flamarique et al., 2013; Nakamura et al., 2013; Cortesi et al., 2015). The P. olivaceus RH2A-1, RH2B-1 and RH2B-2 were similar in tuning site variations, whereas RH2A-2 pigment contained distinct amino acid changes. Most fish RH2 contain an M116 or F116 and an M207, but the P. olivaceus RH2A-2 possessed an L116 and an L207 (Table 2). The only known L207 was found in L. chalumnae RH2Alc (Yokoyama et al., 1999). It is interesting to notice that all the reported SWS2 pigments in teleosts have an L207. Comparing the RH2 and SWS2 sequences, the L116 and L207 mutations are suspected to cause a blue-shift in the λmax of in P. olivaceus RH2A-2 pigment.
Because the adult flatfish lives in deep benthic environment where blue and blue-green lights are predominant, the identified tuning site mutations resulting blue-shift in the λmax of RH2A-2 as well as those resulting green-shift in the λmax of SWS2A all reflected the evolutionary results of flounder genome to adapt the dim-bottom vision habitat (Figure 4).
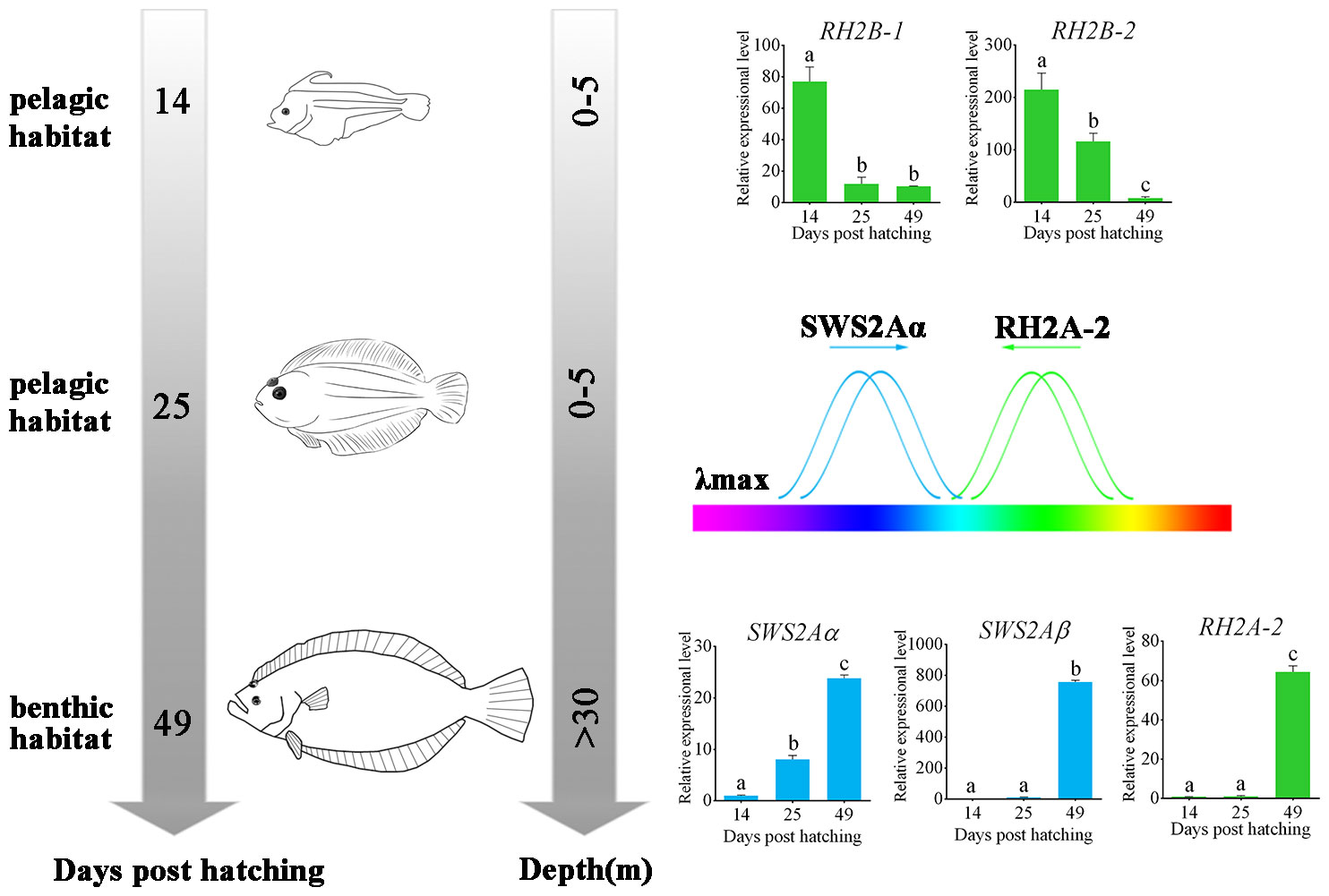
Figure 4 Schematic of metamorphosis and adapting benthic habitat of P. olivaceus. The wavelength shift by both RH2 and SWS2 opsins made P. olivaceus possess a broader wavelength adaptation from green to purple-blue lights. Different letters represent significant difference.
All the results let us conclude that P. olivaceus have adapted the benthic vision environment by: 1. gene-specific duplications in both RH2 and SWS2 genes, 2. tuning site mutations for wavelength shift by both RH2 and SWS2 opsins that make P. olivaceus possess a broader wavelength adaptation from green to purple-blue lights; 3. an expressional modification of different RH2 and SWS2 paralogues to adapt either pelagic or benthic environments.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by the Animal Care and Use Committee of the Centre for Applied Aquatic Genomics at the Ocean University of China. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
ZRZ conducted the experiments, data analyses and drafted the manuscript. YZL and WZ conducted data analyses and collected the materials. XXD provided technical support and participated in discussion. JXL contributed to project conception, drafted and finalized the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2020YFD0901002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1019660/full#supplementary-material
References
Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., et al. (2003). Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31 (13), 3497–3500. doi: 10.1093/nar/gkg500
Chen S., Zhang G., Shao C., Huang Q., Liu G., Zhang P., et al. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46, 253. doi: 10.1038/ng.2890
Chinen A., Hamaoka T., Yamada Y., Kawamura S. (2003). Gene duplication and spectral diversification of cone visual pigments of zebrafish. Genetics 163 (2), 663–675. doi: 10.1093/genetics/163.2.663
Cortesi F., Musilová Z., Stieb S. M., Hart N. S., Siebeck U. E., Malmstrøm M., et al. (2015). Ancestral duplications and highly dynamic opsin gene evolution in percomorph fishes. Proc. Natl. Acad. Sci. U.S.A. 112 (5), 1493–1498. doi: 10.1073/pnas.1417803112
Davies W. L., Carvalho L. S., Tay B. H., Brenner S., Hunt D. M., Venkatesh B. (2009). Into the blue: gene duplication and loss underlie color vision adaptations in a deep-sea chimaera, the elephant shark Callorhinchus milii. Genome Res. 19 (3), 415–426. doi: 10.1101/gr.084509.108
Davies W. I., Tamai T. K., Zheng L., Fu J. K., Rihel J., Foster R. G., et al. (2015). An extended family of novel vertebrate photopigments is widely expressed and displays a diversity of function. Genome Res. 25 (11), 1666–1679. doi: 10.1101/gr.189886.115
De Souza P., Bonilla-Rodriguez G. O. (2007). Fish hemoglobins. Braz. J. Med. Biol. Res. 40 (6), 769–778. doi: 10.1590/s0100-879x2007000600004
Figueras A., Robledo D., Corvelo A., Hermida M., Pereiro P., Rubiolo J. A., et al. (2016). Whole genome sequencing of turbot (Scophthalmus maximus; pleuronectiformes): a fish adapted to demersal life. DNA Res. 23 (3), 181–192. doi: 10.1093/dnares/dsw007
Flamarique I. N., Cheng C. L., Bergstrom C., Reimchen T. E. (2013). Pronounced heritable variation and limited phenotypic plasticity in visual pigments and opsin expression of threespine stickleback photoreceptors. J. Exp. Biol. 216 (4), 656–667. doi: 10.1242/jeb.078840
Fuiman L. A. (1997). What can flatfish ontogenies tell us about pelagic and benthic lifestyles? J. Sea. Res. 7 (3), 257–267. doi: 10.1016/S1385-1101(97)00013-0
Geffen A., van der Veer H., Nash R. (2007). The cost of metamorphosis in flatfishes. J. Sea. Res. 58 (1), 35–45. doi: 10.1016/j.seares.2007.02.004
Gibson R. (1997). Behaviour and the distribution of flatfishes. J. Sea. Res. 37 (3), 241–256. doi: 10.1016/S1385-1101(97)00019-1
Hárosi F. I. (1994). An analysis of two spectral properties of vertebrate visual pigments. Vision Res. 34 (11), 1359–1367. doi: 10.1016/0042-6989(94)90134-1
Helvik J. V., Drivenes Ø., NÆSs T. H., Fjose A., Seo H.-C. (2002). Molecular cloning and characterization of five opsin genes from the marine flatfish Atlantic halibut (Hippoglossus hippoglossus). Vis. Neurosci. 18 (5), 767–780. doi: 10.1017/S095252380118510X
Hoffmann M., Tripathi N., Henz S. R., Lindholm A. K., Weigel D., Breden F., et al. (2007). Opsin gene duplication and diversification in the guppy, a model for sexual selection. Proc. Biol. Sci. 274 (1606), 33–42. doi: 10.1098/rspb.2006.3707
Hofmann C. M., Carleton K. L. (2009). Gene duplication and differential gene expression play an important role in the diversification of visual pigments in fish. Integr. Comp. Biol. 49 (6), 630–643. doi: 10.1093/icb/icp079
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17 (8), 754–755. doi: 10.1093/bioinformatics/17.8.754
Iwanicki T. W., Novales Flamarique I., Ausiό J., Morris E., Taylor J. S. (2017). Fine-tuning light sensitivity in the starry flounder (Platichthys stellatus) retina: Regional variation in photoreceptor cell morphology and opsin gene expression. J. Comp. Neurol. 525 (10), 2328–2342. doi: 10.1002/cne.24205
Kasagi S., Mizusawa K., Murakami N., Andoh T., Furufuji S., Kawamura S., et al. (2015). Molecular and functional characterization of opsins in barfin flounder (Verasper moseri). Gene 556 (2), 182–191. doi: 10.1016/j.gene.2014.11.054
Kasagi S., Mizusawa K., Takahashi A. (2018). Green-shifting of SWS2A opsin sensitivity and loss of function of RH2-a opsin in flounders, genus verasper. Ecol. Evol. 8 (2), 1399–1410. doi: 10.1002/ece3.3745
Lanave C., Preparata G., Sacone C., Serio G. (1984). A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20 (1), 86–93. doi: 10.1007/BF02101990
Land M. F., Osorio D. C. (2011). Marine optics: Dark disguise. Curr. Biol. 21 (22), R918–R920. doi: 10.1016/j.cub.2011.10.009
Mader M. M., Cameron D. A. (2004). Photoreceptor differentiation during retinal development, growth, and regeneration in a metamorphic vertebrate. J. Neurosci. 24 (50), 11463–11472. doi: 10.1523/jneurosci.3343-04.2004
Matsumoto Y., Fukamachi S., Mitani H., Kawamura S. (2006). Functional characterization of visual opsin repertoire in medaka (Oryzias latipes). Gene 371 (2), 268–278. doi: 10.1016/j.gene.2005.12.005
Meyer A., Van de Peer Y. (2005). From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27 (9), 937–945. doi: 10.1002/bies.20293
Nakamura Y., Mori K., Saitoh K., Oshima K., Mekuchi M., Sugaya T., et al. (2013). Evolutionary changes of multiple visual pigment genes in the complete genome of pacific bluefin tuna. Proc. .Natl. Acad. Sci. U.S.A. 110 (27), 11061–11066. doi: 10.1073/pnas.1302051110
Ohno S. (1970). “Duplication for the sake of producing more of the same,” in Evolution by gene duplication (Springer, Berlin, Heidelberg), 59–65. doi: 10.1007/978-3-642-86659-3_11
Rennison D. J., Owens G. L., Taylor J. S. (2012). Opsin gene duplication and divergence in ray-finned fish. Mol. Phylogenet. Evol. 62 (3), 986–1008. doi: 10.1016/j.ympev.2011.11.030
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a Large model space. Syst. Biol. 61 (3), 539–542. doi: 10.1093/sysbio/sys029
Seehausen O., Terai Y., Magalhaes I. S., Carleton K. L., Mrosso H. D. J., Miyagi R., et al. (2008). Speciation through sensory drive in cichlid fish. Nature 455, 620. doi: 10.1038/nature07285
Shao C., Bao B., Xie Z., Chen X., Li B., Jia X., et al. (2017). The genome and transcriptome of Japanese flounder provide insights into flatfish asymmetry. Nat. Genet. 49 (1), 119–124. doi: 10.1038/ng
Shichida Y., Imai H. (1998). Visual pigment: G-protein-coupled receptor for light signals. Cell. Mol. Life Sci. 54 (12), 1299–1315. doi: 10.1007/s000180050256
Sugawara T., Terai Y., Imai H., Turner G. F., Koblmüller S., Sturmbauer C., et al. (2005). Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep-water cichlids from lakes Tanganyika and Malawi. Proc. Natl. Acad. Sci. U.S.A. 102 (15), 5448–5453. doi: 10.1073/pnas.0405302102
Takashi M. (1982). The early life history of a flounder paralichthys olivaceus. Nippon. Suisan. Gakk. 48, 1581–1588. doi: 10.2331/suisan.48.1581
Takechi M., Kawamura S. (2005). Temporal and spatial changes in the expression pattern of multiple red and green subtype opsin genes during zebrafish development. J. Exp. Biol. 208 (7), 1337–1345. doi: 10.1242/jeb.01532
Wang W., Wang J., You F., Ma L., Yang X., Gao J., et al. (2014). Detection of alternative splice and gene duplication by RNA sequencing in Japanese flounder, Paralichthys olivaceus. G3 (Bethesda). 4 (12), 2419–2424. doi: 10.1534/g3.114.012138
Watson C. T., Gray S. M., Hoffmann M., Lubieniecki K. P., Joy J. B., Sandkam B. A., et al. (2011). Gene duplication and divergence of long wavelength-sensitive opsin genes in the guppy, poecilia reticulata. J. Mo. Evol. 72 (2), 240–252. doi: 10.1007/s00239-010-9426-z
Yokoyama S. (1997). Molecular genetic basis of adaptive selection: examples from color vision in vertebrates. Annu. Rev. Genet. 31 (1), 315–336. doi: 10.1146/annurev.genet.31.1.315
Yokoyama S. (2000a). Color vision of the coelacanth (Latimeria chalumnae) and adaptive evolution of rhodopsin (RH1) and rhodopsin-like (RH2) pigments. J. Hered. 91 (3), 215–220. doi: 10.1093/jhered/91.3.215
Yokoyama S. (2000b). Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye. Res. 19 (4), 385–419. doi: 10.1016/S1350-9462(00)00002-1
Yokoyama S. (2002). Molecular evolution of color vision in vertebrates. Gene 300 (1), 69–78. doi: 10.1016/S0378-1119(02)00845-4
Yokoyama S. (2008). Evolution of dim-light and color vision pigments. Annu. Rev. Genomics Hum. Genet. 9 (1), 259–282. doi: 10.1146/annurev.genom.9.081307.164228
Yokoyama S., Takenaka N., Blow N. (2007). A novel spectral tuning in the short wavelength-sensitive (SWS1 and SWS2) pigments of bluefin killifish (Lucania goodei). Gene 396 (1), 196–202. doi: 10.1016/j.gene.2007.03.019
Yokoyama S., Zhang H., Radlwimmer F. B., Blow N. S. (1999). Adaptive evolution of color vision of the comoran coelacanth (Latimeria chalumnae). Proc. Natl. Acad. Sci. U.S.A. 96 (11), 6279–6284. doi: 10.1073/pnas.96.11.6279
Keywords: paralichthys olivaceus, visual opsins, gene duplication, spectral tuning, adaptive evolution
Citation: Zhang Z, Liu Y, Zhang W, Du X and Liu J (2022) Benthic visual adaptation by fine-tuning light sensitivity in Japanese flounder (Paralichthys olivaceus). Front. Mar. Sci. 9:1019660. doi: 10.3389/fmars.2022.1019660
Received: 15 August 2022; Accepted: 21 September 2022;
Published: 06 October 2022.
Edited by:
Xiaotong Wang, Ludong University, ChinaCopyright © 2022 Zhang, Liu, Zhang, Du and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinxiang Liu, bGl1amlueGlhbmdAb3VjLmVkdS5jbg==
 Zhengrui Zhang
Zhengrui Zhang Yuezhong Liu1
Yuezhong Liu1 Xinxin Du
Xinxin Du Jinxiang Liu
Jinxiang Liu