- 1Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 2Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, Zhanjiang, China
In the present study, a g-type lysozyme was successfully screened and cloned from Pinctada fucata martensii (designated as PmlysG). The cDNA has a length of 973 bp with an open reading frame (ORF) of 769 bp, encoding a protein of 255 amino acids. The PmlysG transcript was detected in multiple tissues by quantitative real-time PCR (qRT-PCR), with the highest expression being in the hepatopancreas. Additionally, the temporal expression of PmlysG mRNA in the hepatopancreas after in vivo stimulation with pathogen-associated molecular patterns (PAMPs), such as lipopolysaccharide (LPS), peptidoglycan (PGN) and polyinosinic acid (PolyI:C) was detected by qRT-PCR. Although PmlysG responded to all three stimulation modes, it rapidly responded to PGN stimulation. Meanwhile, the recombinant protein of g-type lysozyme of P.f. martensii (rPmlysG) was used for antibacterial function analysis, and the results showed that rPmlysG has antibacterial function against Vibrio parahaemolyticus, Aeromonas hydrophila, and Pseudomonas aeruginosa. Overall, these study results suggest that the identified PmlysG participates in the innate immune responses of P.f. martensii against pathogen infection.
Introduction
Lysozyme, belonging to the GH (hydrolytic glycosidase [(β-) glycoside hydrolase) subfamily 22, is a ubiquitous enzyme found in various organisms. It is a key effector molecule of the invertebrate and vertebrate innate immune systems. It can catalyze the hydrolysis of β-1,4-glycosidic linkage between N-acetylmuramic acid and N-acetylglucosamine alternating the sugar residues in the bacterial cell walls and peptidoglycan (PGN), thus inducing bacterial cell lysis (Jollès and Jollès, 1984; Prager and Jollès, 1996). According to the source, structure, and physicochemical properties, lysozymes can be classified into six types: Goose-type-lysozyme (g-type); Invertebrate-type-Iysozyme (i-type); Chicken-type-lysozyme (c-type); Bacterial lysozyme; Plant lysozyme and Bacteriophage lysozyme (Jiménez-Cantizano et al., 2008). Lysozyme exerts a synergistic effect on the cationic antimicrobial peptides exposed to the PGN layer of gram-negative bacteria (Hancock and Scott, 2000).
G-type lysozymes are mostly abundant in poultry eggs and were first identified in an antibacterial peptide of a white bird egg (Canfield and McMurry, 1967). In aquatic organisms, g-type lysozyme was first identified in Paralichthys olivaceus (Hikima et al., 2001). Besides, the g-type lysozyme has been identified in Epinephelus coioides (Yin et al., 2003), Pseudosciaena crocea (Zheng et al., 2007), and Ctenopharyngodon idellus (Ye et al., 2010). As a non-specific immune factor of fish, g-type lysozyme is closely related to the immune function of fish against bacterial infection. It is a natural endogenous antitoxin and helps improve the immunity of the body through its antibacterial, antiviral, and anti-inflammatory activities (Shakoori et al., 2019). For instance, g-type lysozyme significantly increased in different tissues of C. idellus after infection with Aeromonas hydrophila (Ye et al., 2010). Furthermore, the recombinant protein of g-type lysozyme induced by Escherichia coli poses antibacterial activity against various gram-negative and gram-positive bacteria in different environments (Li et al., 2008; Zhang et al., 2012).
Unlike advanced animals, mollusks are typical invertebrates without any unique immune system. In this regard, the innate immune factors play a crucial role in invertebrate immunity (Wang et al., 2013; He et al., 2019; He et al., 2020), including heme-mediated cellular and humoral immune responses (Fan et al., 2022; Lv et al., 2022). The innate immune factors dissolve the invading microorganisms or bacterial tissues using constitutive and inducible antibacterial molecules (Rolff and Siva-Jothy, 2003; He et al., 2018). Shellfish are filter-feeding organisms and are often exposed to various potential pathogens in the aquatic environment. Correspondingly, lysozyme in shellfish helps in pathogen defense due to its antibacterial effect and also helps in ingestion and digestion (Nilsen et al., 1999; Nilsen et al., 2003).
Recent studies have also identified the g-type lysozymes in invertebrates. For instance, the g-type lysozyme has been successfully identified in the adductor muscle of Chlamys farreri with active participation in immune response (Li et al., 2013). Similarly, a study has identified nine SNP sites and three ins-del polymorphic sites in the promoter region of G-type lysozyme of Japanese scallops (Mizuhopecten yessoensis). These mutations are classified into two haplotypes, which are associated with different transcription factor binding sites (He et al., 2012).
Pearl oysters (Pinctada fucata martensii) are one of the primary shellfish reared in the seawater pearls of southern China with high economic value (Wu et al., 2017a; Lu et al., 2022). Nevertheless, several P.f. martensii have died in recent years due to environmental pollution (Qiu et al., 2014; Wu et al., 2017b). Thus, improving disease resistance in P.f. martensii has great significance.
In the present study, the cDNA sequence of goose-type lysozyme in P.f. martensii (PmlysG) was cloned using the rapid amplification of cDNA ends (RACE) technique and the expression of PmlysG in various tissues was analyzed using quantitative real-time PCR (qRT-PCR). Additionally, the PmlysG after treatment with pathogen-associated molecular patterns (PAMPs) were evaluated, including lipopolysaccharide(LPS), PGN, and polyinosinic acid (PolyI:C). Lastly, the recombinant protein of PmlysG (rPmlysG) was used against bacteria in vitro to further analyze the molecule characteristics of g-type lysozyme in P.f. martensii.
Materials and methods
Experimental materials and screening of immune effector molecules
P.f. martensii of approximately 2 years of age, with shell lengths of 5–6 cm, were collected directly from the sea in Chengwu, Zhanjiang, Guangdong Province, China. The P.f. martensii were cultured at 25–27°C in tanks with recirculating seawater for three days prior to experimentation.
The amino acid sequences with antimicrobial properties were obtained from the antimicrobial peptide database (APD3, http://aps.unmc.edu/AP/main.php), PubMed (https://www.ncbi.nlm.nih.gov/pubmed/), and National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). The genome data of P.f. martensii (Accession: PXD006786) (Du et al., 2017)was compared with the AMP database using a local reference. The gene sequences with the highest alignment rate were analyzed using the online blast in the NCBI protein sequence database to predict the types of immune effector molecules collected. A total of 21 sequences were obtained (Table S1). All the 21 sequences are interesting, some sequences have been studied and published in our previous study (He et al., 2020; Liang et al., 2022), other sequences are in study. One of the immune effector molecules identified as PmlysG was selected for follow-up studies in this manuscript.
Characterization and cloning of PmlysG
The PmlysG specific primers for PCR amplification were designed using Primer Premier 5.0 (Table 1). The total RNA was obtained from P.f. martensii hemocytes using the Trizol reagent (Thermo-Fisher Scientific, USA). RNase-free DNase (Promega, USA) was used to avoid DNA contamination. The PCR product integrity was assessed on 1% agarose gel. The RNA concentration was determined based on the OD260/OD280 ratio using a NanoDrop 2000 spectrophotometer (Thermo-Fisher Scientific, USA). The cDNA was prepared using the Reverse Transcriptase M-MLV, following the manufacturer’s protocol. The 3’ and 5’ ends of the PmlysG gene were cloned by RACE using the SMART RACE cDNA Amplification Kit (Clontech).
Bioinformatics analysis
The homologous gene sequence was obtained through BLAST search. The open reading frame (ORF) was identified using the ORF finder. The theoretical isoelectric point and the molecular mass of the predicted amino acid sequence were estimated using ExPASy (https://web.exPasy.org/protparam/). The signal peptide sequence was predicted using the Signalp4.0 server. The transmembrane domain analysis of sequences was conducted using TMHMMServerv 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). The amino acid sequence and functional sites were predicted using SoftBerryPsite. The protein secondary structure was predicted using SOPAM.
The protein sequences of PmlysG and g-type lysozyme from six species (Argopcten irradians, Mytilus galloprovincialis, Azumapcten farreri, Haliotis discus, Mizuhopecten yessoensis, and Physlla acuta) were compared using DNAMAN6.0. Finally, the neighbor-joining phylogenetic tree was constructed using MEGA6.0.
Tissue expression analysis
The relative expression level of PmlysG in various tissues of P.f. martensii was measured by quantitative real-time PCR. The tissues, including mantle, hemocytes, gonads, gills, hepatopancreas, and adductor muscle were obtained from 10 pearl oysters and immediately stored in liquid nitrogen for further use. The total RNA was extracted from the samples according to a previously reported method (He et al., 2020). After obtaining the total RNA from various tissues, it was reverse transcribed into cDNA. Based on the obtained cDNA sequence, a pair of specific primers of PmlysG was designed using Primer Premier 5.0 (Table 1). The quantitative level of PmlysG in each tissue was detected through the premix and lightcyclr96 real-time PCR system (Roche) according to a previously reported method (Liang et al., 2022).
The PmlysG transcript expression level in the hepatopancreas was quantified in terms of PAMPs, including LPS, PGN, PolyI:C to determine the possible role of PmlysG in immune responses. Healthy P.f. martensii (n = 320) samples were randomly divided into four groups(n = 80 per group), including LPS, PGN, PolyI:C, and PBS (control). The experimental groups (LPS, PGN, and PolyI:C were injected with 100 μL of LPS (10 μg/mL), 100 μL of PGN (10 μg/mL), and 100 μL of PolyI:C (10 μg/mL), respectively. The control group was injected with 100 μL of PBS. The mRNA expression levels of PmlysG were determined at 0, 3, 6, 9, 12,24, 48,72, and 96 hours after stimulation with PAMPs. The relative expression levels of the target genes were determined by the 2^-ΔΔct method (Livak and Schmittgen, 2001), using GAPDH as the reference gene.
Prokaryotic expression and purification
PCR fragment encoding the mature peptide of PmlysG was amplified using the specific primers PmlysGR and PmlysGF. The PCR product digested with M1uI/HindIII was subcloned into pET-28a (+) digested with the same enzymes to obtain the plasmid pET28a-PmlysG. The pET28a-PmlysG compound was verified through restriction enzyme digestion and DNA sequencing.
The recombinant plasmid pET28a-PmlysG was transferred into the BL21 (DE3) competent cells. An excellent single colony was inoculated in 4mL medium containing 50 μg/mL of kanamycin. When the OD reached 0.6 ~ 0.8, the isopropyl-beta-D-thiogalactopyranoside (IPTG)was added to a final concentration of 0.5 mM, and then induced at 15°C and 37°C. The fusion protein was purified using the HisBind purification kit (Pharmacia, Sweden), following the manufacturer’s protocol.
Antimicrobial assay
Several purified proteins were used against Vibrio parahaemolyticus, Aeromonas hydrophila, Pseudomonas aeruginosa, Staphylococcus aureus, Micrococcus luteus, Bacillus subtilis, and Escherichia coli to assess their antibacterial activities. Briefly, each bacterium was cultured in a 2216E liquid medium to the logarithmic growth phase. The bacterial solutions were centrifuged (3000 xg, 10 minutes), washed thrice using 1xPBS, and resuspended in PBS. The purified protein (50 μL, 500 μg/mL) was mixed and incubated with 50 μL of each bacterial suspension at room temperature for 2 hours using PBS as a negative control. The mixture was then incubated at 37°C and the OD600 values at 0, 1, 2, 3, 4, 5,6, 7, 8, 9, 10, 11, and 12 hours were measured using a microplate reader(EnSpire, PerkinElmer). Each experiment was conducted in triplicate.
Transmission electron microscopy
Approximately 200 μL of rPmlysG and 200 μL of bacterial solution in the exponential growth period were mixed at 37 °C for 2 hours using PBS as the control. The mixture was centrifuged at 3000 rpm and room temperature for 10 minutes for deposition and then washed thrice with PBS to remove impurities. Later, 200 μL of 2% sodium phosphotungstate aqueous solution was added to the bacterial suspension, which was then dropped on copper grids. The samples were air-dried after removing the residual water using filter paper for 5 minutes. Finally, the images were observed using a JEM-1230 JEM-1400 (Japan Electronics Corp) microscope under standard operating conditions.
Statistical analysis
All experimental data were analyzed by One-way analysis of variance (ANOVAs) using SPSS 19.0 (IBM, USA). The differences between means were considered significant at P < 0.05 and extremely significant at P < 0.01.
Results
Characterization of PmlysG
As depicted in Figure 1, the PmlysG cDNA is 973 bp long and contains a 3’ untranslated region (UTR) of 121 bp and a 5’ UTR of 84 bp. The PmlysG cDNA includes an ORF of 769 bp, encoding 255 amino acids. The predicted molecular mass of the PmlysG is 27.26 KDa, with an isoelectric point (pI) of 7.27. The highest hydrophobicity continuously appears from position 8, with an index of 2.844. The highest hydrophilicity (hydrophilic protein) appears at position 198, with an index of -2.289 and an overall average coefficient of -0.306. The positively charged residue (Arg + Lys) is 15, and the negatively charged residue (ASP + Glu) is 15, indicating that PmlysG is a neutral charge.
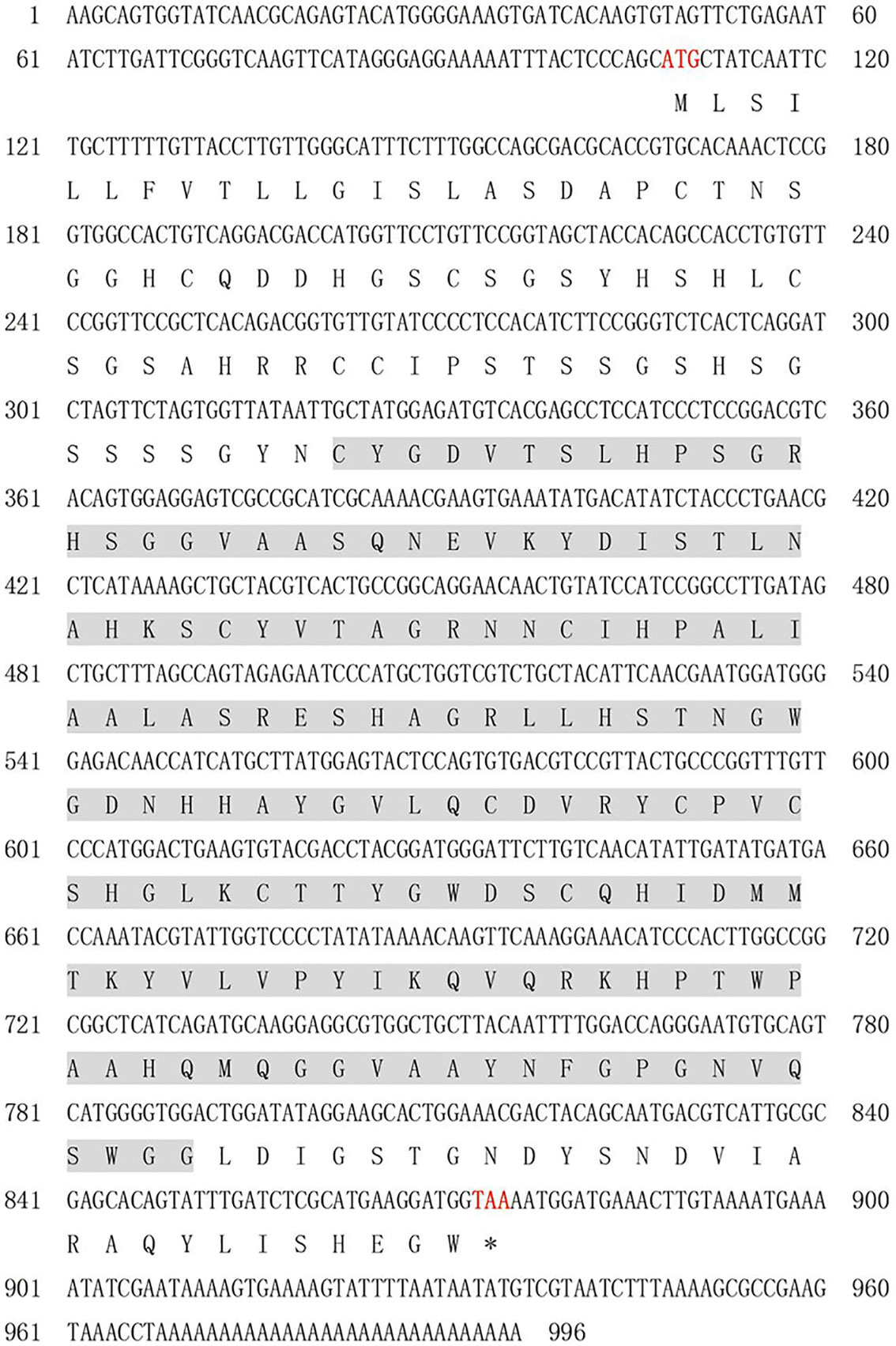
Figure 1 A cDNA sequence and predicted amino acid sequence of PmlysG. Red font indicates start codon and stop codon, the straight line indicates signal peptide, and the shaded portion shows the d153I_ domain.
The amino acids at positions 1-16 have a signal peptide (Supplementary Figure S1). The secondary structure predictions suggested that the active protein mainly contains random coils (51.70%), extended strands (12.94%), α-helices (30.20%), and β-turn (5.49%). The domain analysis identified a single d153I_ domain containing 176 amino acids (Supplementary Figure S2).
Multiple sequence alignment and phylogenic relationships
The alignment of PmlysG with the G-type lysozyme of other species indicated that PmlysG was similar to the g-type lysozyme in Argopecten irradians (60.5%), Mytilus galloprovincialis (58.74%), Azumapecten farreri (57%), and Haliotis discus (45.88%). The consistent similarity among the species reached 55.76% (Figure 2), based on which a phylogenetic tree was then constructed.
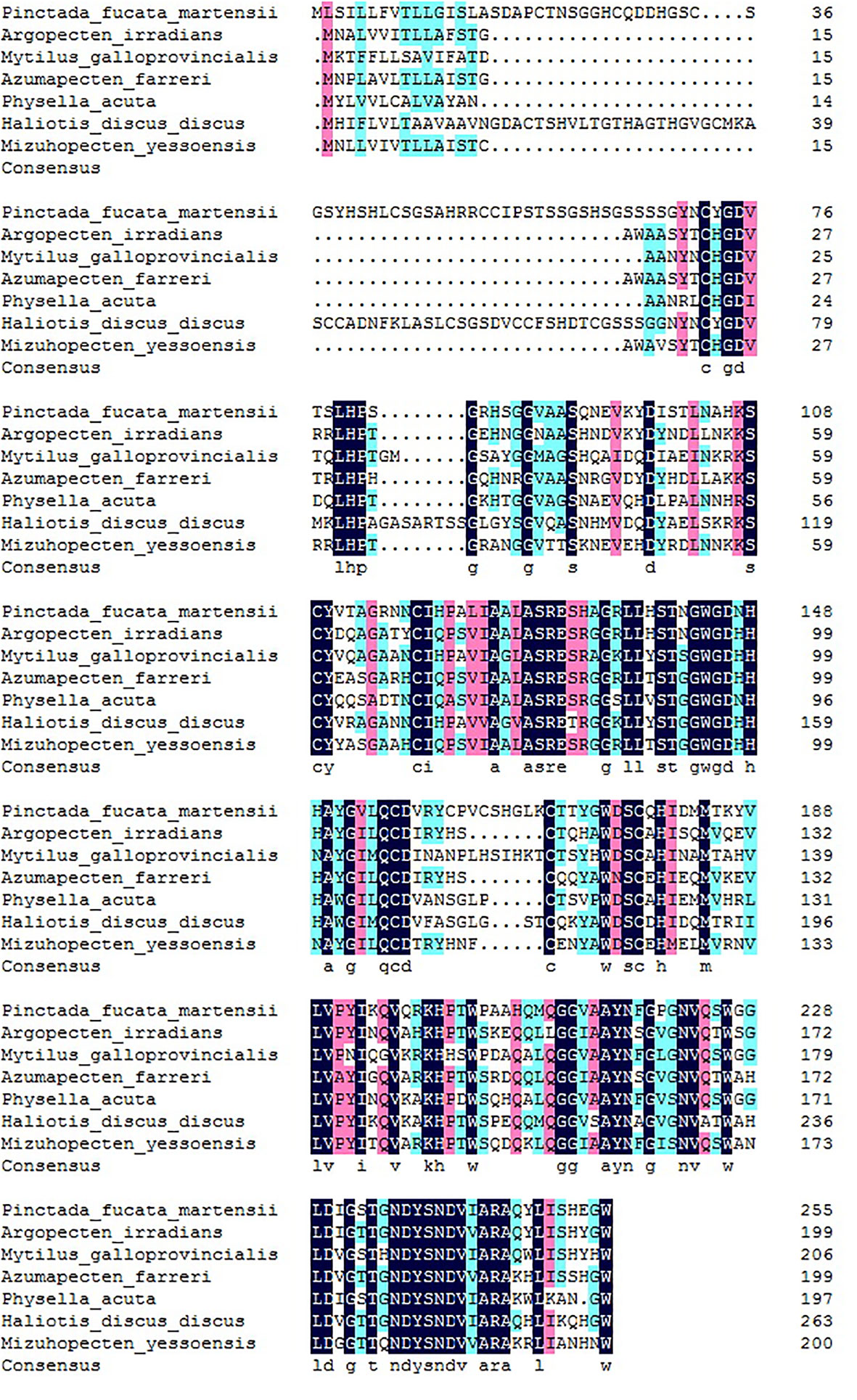
Figure 2 PmlysG Homology alignment and the coincident amino acids. Light blue and pink indicate that the similarity is more than 75% and 50%, respectively. Navy blue: Consensus amino acids.
The phylogenetic tree classification of PmlysG showed that invertebrates and vertebrates formed a separate large branch. P.f. martensii shared the closest genetic relationship with Argopecten irradians, Azumapecten farreri, and Mizuhopecten yessoensis (Figure 3).
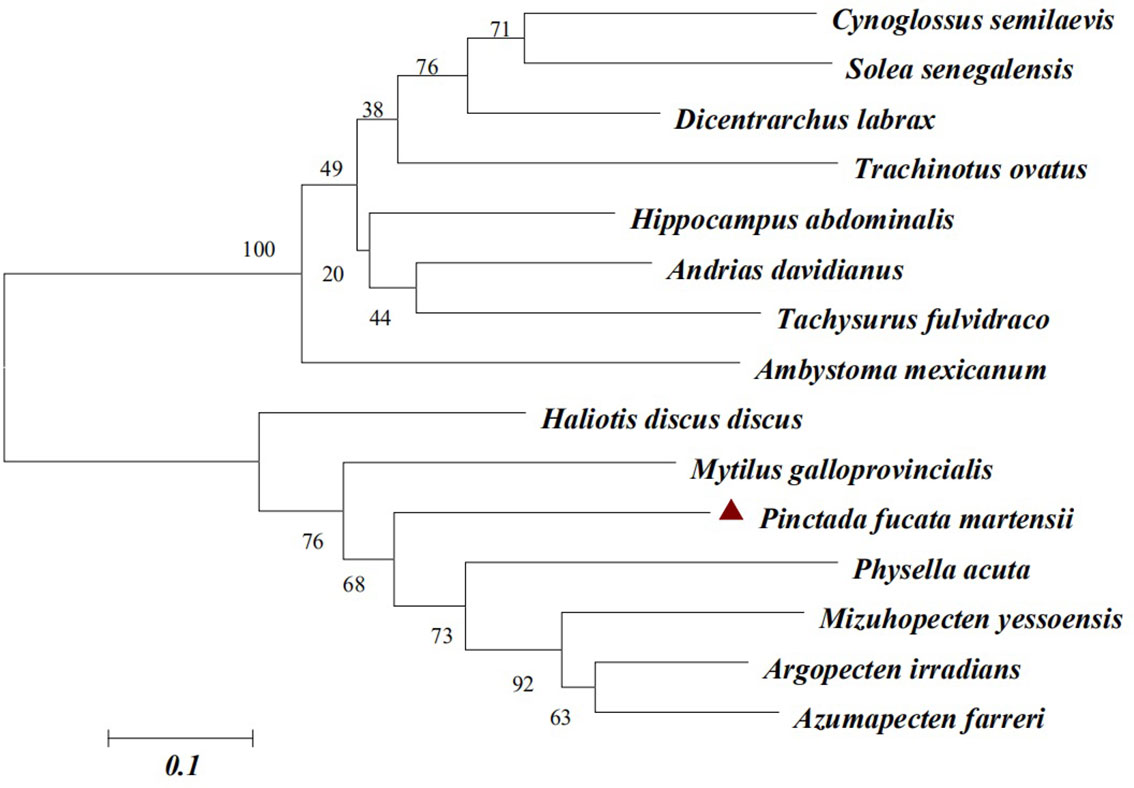
Figure 3 PmlysG Phylogenetic tree construction (Neighbor-joining method). GenBank accession numbers for some species are as follows: Cynoglossus semilaevis (AEQ19605.1), Solea senegalensi s(CCA95109.1), Dicentrarchus labrax (AIE45885.1), Tachysurus fulvidraco (ALD84262.1), Ambystoma mexicanum (AEQ98812.1), Haliotis discus (AGQ50335.1), Mytilus galloprovincialis (AFQ35865.1), Physella acuta (ADV36303.1), Mizuhopecten yessoensis (AEY77130.1), Argopecten irradians (AAX09979.1), Azumapecten farreri (ABB53641.1).
Quantitative analysis of PmlysG in different tissues
The expression levels of PmlysG in the hepatopancreas, gonads, hemocytes, gill, adductor muscle, and mantle were analyzed using qRT PCR under normal physiological conditions. The PmlysG mRNA was ubiquitously expressed in various tissues, with significantly high expression in the hepatopancreas (P < 0.01) (Figure 4).
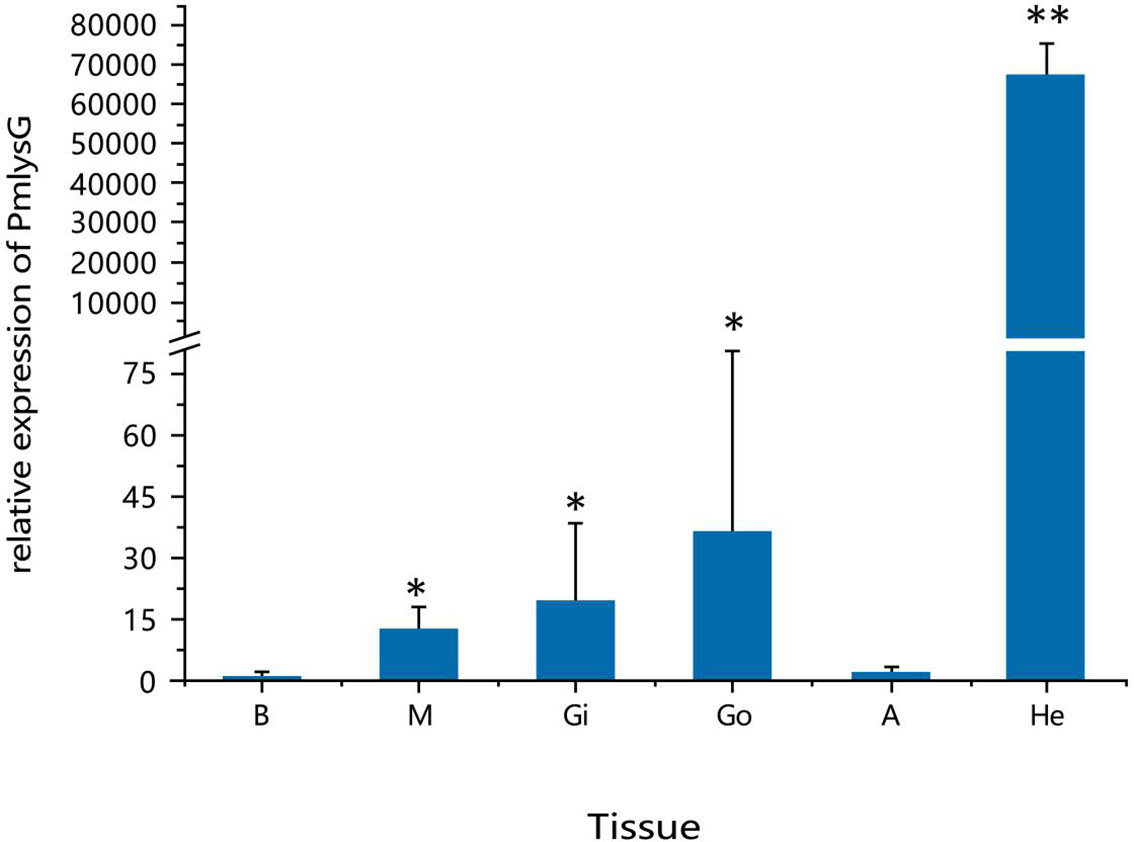
Figure 4 Quantitative analysis of PmlysG gene expression. He: Hepatopancreas, Gi: Gill, Go: Gonad, M: Mantle, H: hemocytes, A: Adductor muscle. *, P < 0.05; **, P < 0.01.
The mRNA expression of PmlysG after PAMPs stimulation
Compared with the control, the PmlysG mRNA expression rapidly increased in the hepatopancreas at 6 hours after LPS stimulation, reaching the maximum level at 72 hours (P<0.01), then it quickly returned to the normal level at 96 hours (Figure 5A). Moreover, PmlysG mRNA began to increase at 3 hours after PGN stimulation, reaching the maximum level at 6 hours (P < 0.01), then decreased at 12 hours, increased at 24 hours, gradually decreased at 48 hours, and finally returned to the normal level (Figure 5B). However, after PolyI:C stimulation, PmlysG mRNA did not change in the first 12 hours, but began to rise at 24 and 48 hours, reaching a maximum at 72 hours (P < 0.05), then began to decline after 72 hours (Figure 5C)
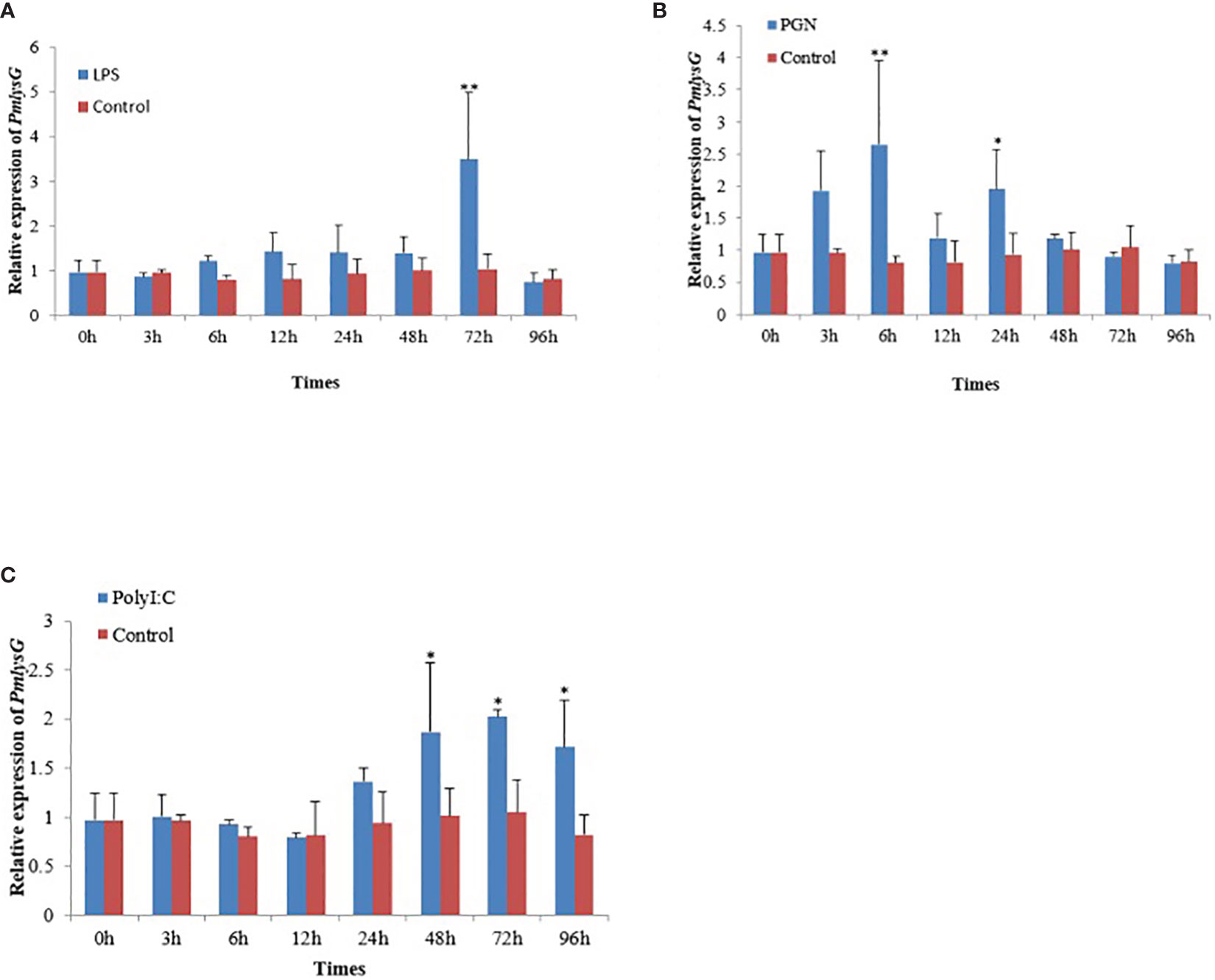
Figure 5 The PmlysG mRNA expression level in the hepatopancrease of P.f martensii after PAMPs stimulation. (A) LPS stimulation. (B) PGN stimulation, (C) PolyI: C stimulation. The asterisks *, P < 0.05, **, and P < 0.01 indicate significant differences between the control and stimulation groups. Vertical bars represent the mean ± SD (n=5).
Induced expression and purification of the PmlysG
The digested PmlysG gene fragment was about 1570 bp (Figure 6A). The successful construction of vector pET28-PmlysG was verified through sequencing. Further validation showed that 26.52 KDa protein was present in the recombinant bacteria but not in the negative control. Moreover, no target bands were found in lanes 3 and 4 of the supernatant at 15 °C and 37 °C. However, Clear target bands appeared in lanes 1 and 2 (whole bacteria), 5 and 6 (precipitation) of the experimental groups at 15 °C and 37 °C (Figure 6B). Western blot further verified the 27 kDa clear single staining bands, and the positions of the lanes were consistent with the above positions. Further analysis showed that the recombinant protein induced at 15 °C (lane 5) was brighter than the one induced at 37 °C (lane 6) (Figure 6C). These results indicate that the product is mainly expressed as an inclusion. Since the recombinant protein was expressed as an inclusion body, it was dialyzed, renatured, and transferred to a soluble buffer (PBS, 10% glycerol, 1 mL arginine, pH 7.4).
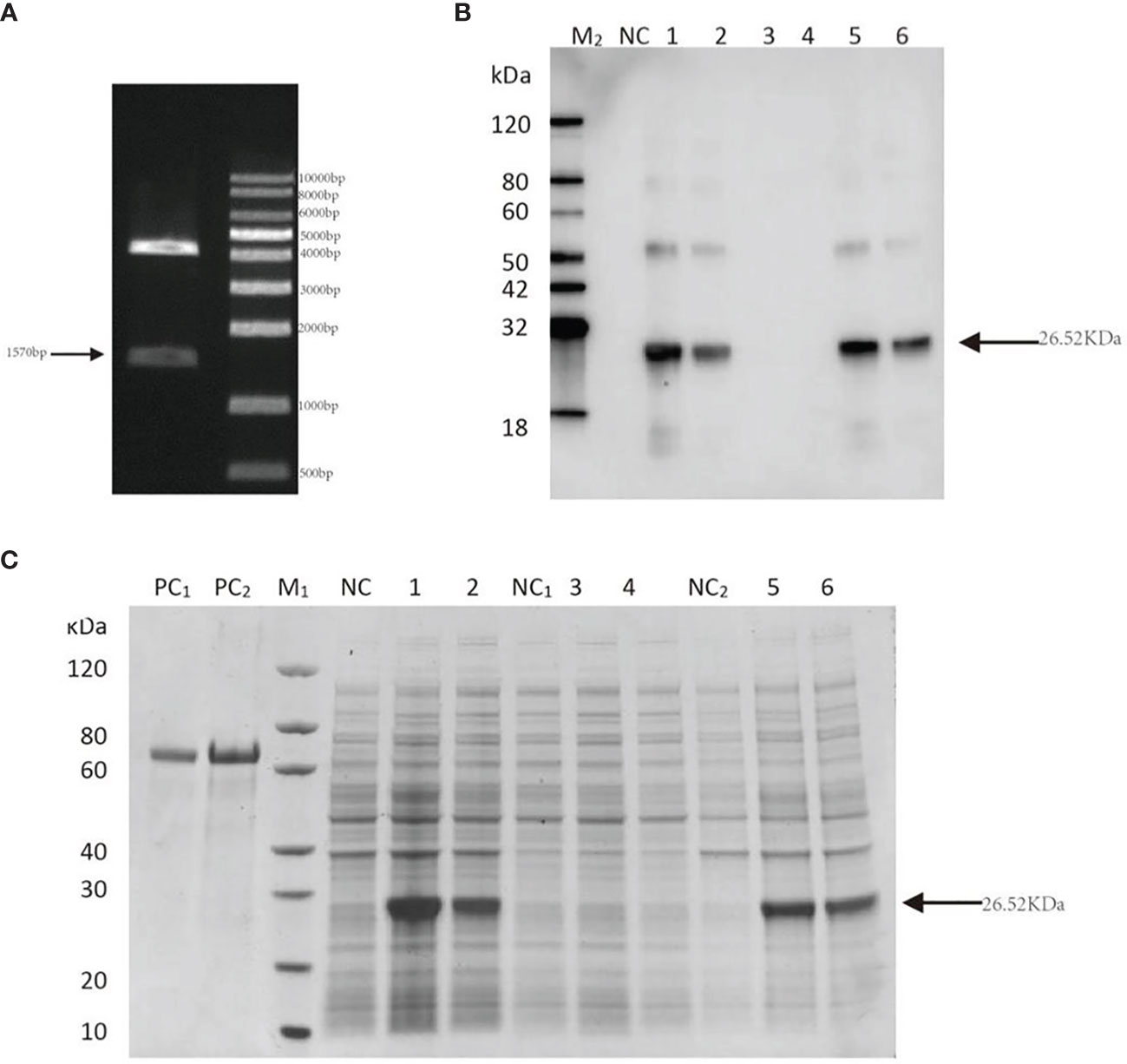
Figure 6 Construction of PmlysG recombinant protein. (A) Verification of recombinant plasmid of PmlysG 1: Bands of interest M: Maker. (B) SDS-PAGE (C) Western Blot. PC1: BSA (1 μg); PC2: BSA (2 μg); M1,M2: Marker; NC(whole bacteria): Cell lysate without induction; 1: Cell lysate with induction at 15°C for 16 hours; 2: Cell lysate with induction at 37°C for 4 hours; NC1(supernatant): Supernatant of cell lysate without induction; 3: Supernatant of cell lysate with induction at 15°C for 16 hours; 4: Supernatant of cell lysate with induction at 37°C for 4 hours; NC2 (precipitation): Debris of cell lysate without induction; 5: Debris of cell lysate with induction at 15°C for 16 hours; 6: Debris of cell lysate with induction at 37°C for 4 hours.
Antimicrobial activity
Seven bacteria, including three gram-positive bacteria and four gram-negative bacteria, were determined in the bacterial inhibition experiment. The purified protein (500 μg/mL) significantly inhibited the growth of gram-negative species (V. parahaemolyticus, A. hydrophila, and P. aeruginosa) (P<0.05) (Figure 7), but exhibited little effect on the growth of S. aureus, M. luteus, B. subtilis, and E. coli (Supplementary Figure S3).
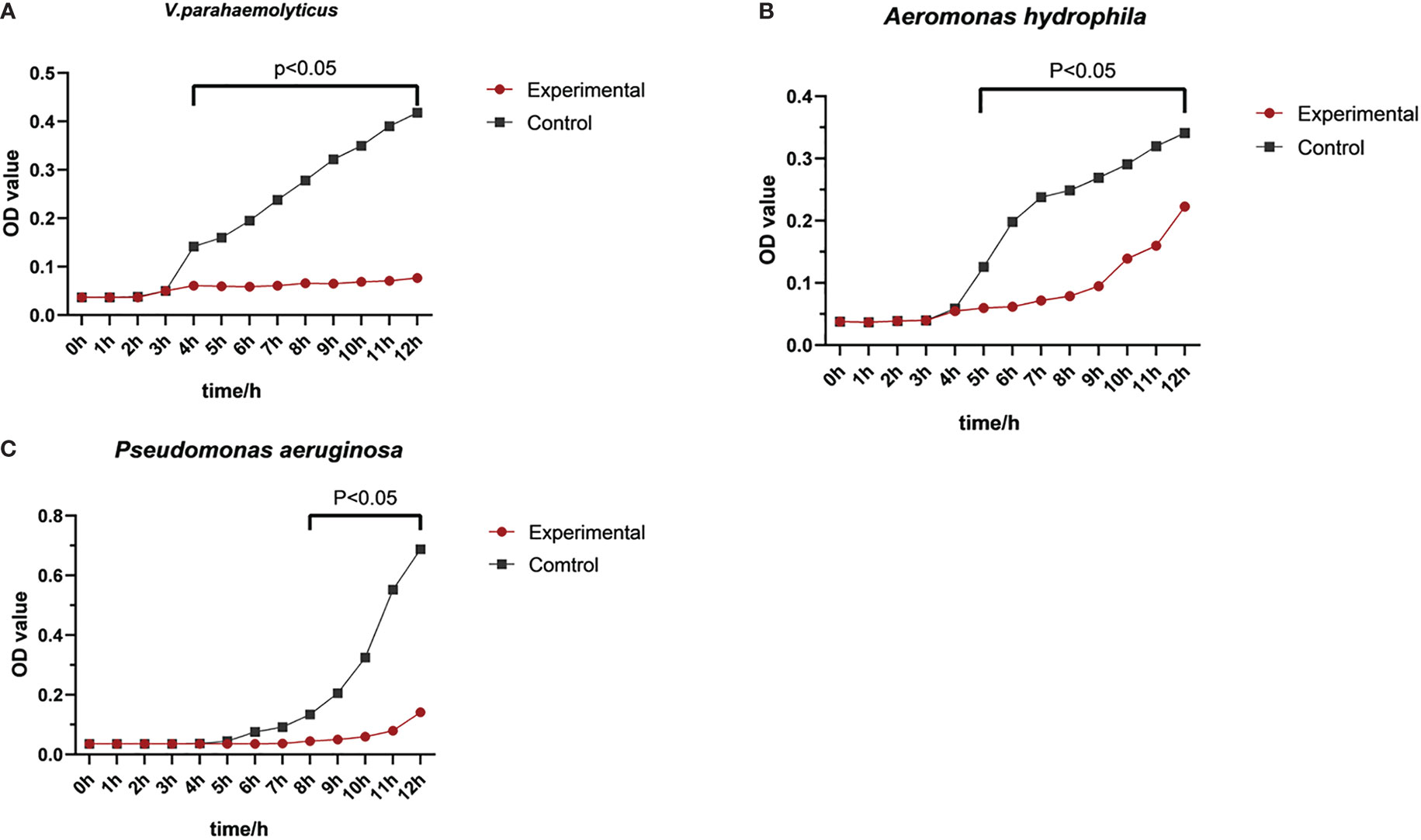
Figure 7 Antibacterial activity of rPmlysG. Control group: PBS. Experimental groups; (A) V. parahaemolyticus challenge; (B) A hydrophila challenge; (C) Pseudomonas aeruginosa challenge.
Transmission electron microscope observation
The morphological characteristics of V. parahaemolyticus and A. hydrophila after interacting with rPmlysG were visualized using TEM to assess the antibacterial mechanism of g-type lysozyme in P.f. martensii. The exterior of the control V. parahaemolyticus (Figures 8A-C) was clear, and the contents were dense and uniform. The middle of V. parahaemolyticus shrank after rPmlysG treatment (shown by an arrow in Figures 8D, E). Meanwhile, plasmolysis was observed (shown by the arrow in Figure 8F), indicating that the cell wall collapse led to the release of local contents from the cell. The control group of A. hydrophila is shown in Figures 8G–I. The cell appeared thin with a clear edge and uniform content, whereas the bacterial cell wall was irregular in the experiment group of Aeromonas hydrophila (Figure 8J). Further magnification revealed that these two cell walls were dissolved, and the contents were lost in the hollow state (shown by an arrow in Figures 8K–L).
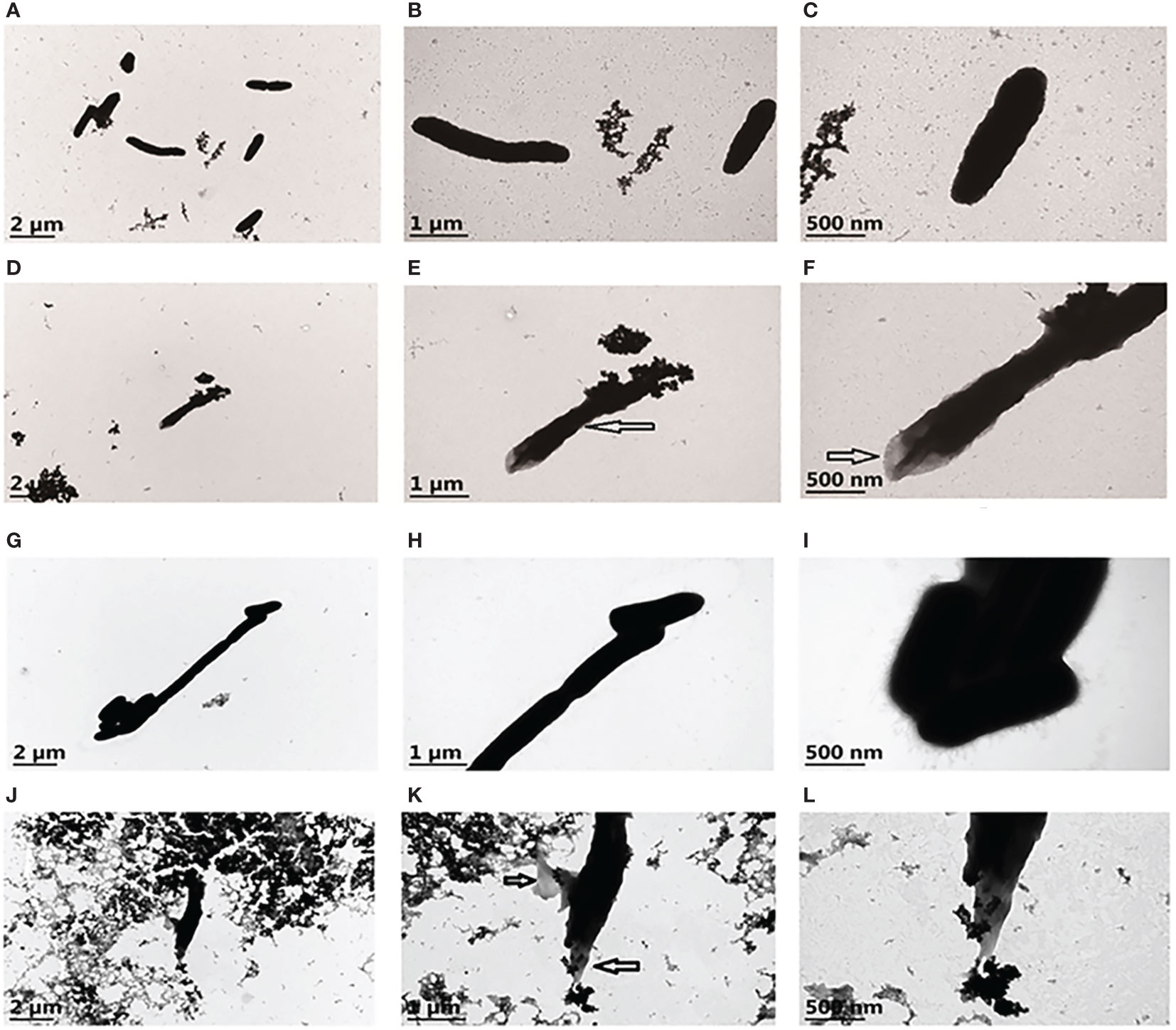
Figure 8 Transmission Electron Microscopy of V. parahaemolyticus and A. hydrophila untreated and treated with rPmlysG. (A) The control group of V. parahaemolyticus untreated with rPmlysG; (B, C) represent enlarged views of A; (D) V. parahaemolyticus action with rPmlysG; (E, F) represent enlarged views of D; (G) The control group of A. hydrophila untreated with rPmlysG; (H, I) represent enlarged views of G; (J) A. hydrophila action with rPmlysG; (K, L) represent enlarged views of J.
Discussion
Innate immunity is the first line of defense in marine invertebrates against pathogenic attacks (Wang et al., 2018; Cao et al., 2019). Lysozyme is a vital component of innate immunity and has been detected in body fluids and tissues of many bivalve scallops (McHenery and Birkbeck, 1982; Matsumoto et al., 2006). Besides, the g-type lysozymes have also been identified in vertebrates and some mollusks (Zhao et al., 2007). Therefore, studying the structure and immune characteristics of g-type lysozymes in P.f. martensii is of great significance. In this study, a g-type lysozyme gene, encoding a polypeptide with 255 amino acids, was isolated from P.f. martensii. Sequence analysis showed that the PmlysG gene contain a d153I_domain. Following the alignment of domain structure and NCBI prediction, the protein was deduced to be a g-type lysozyme. The physical and chemical properties analysis revealed that PmlysG formed a signal peptide at positions 1-16 amino acids, indicating that PmlysG is a secretory protein. This was consistent with the description of Chlamys farreri (Zhao et al., 2007) and gastropod Oncomelania (Zhang et al., 2012). All these species had a signal peptide, indicating the presence of secretory proteins (Irwin and Gong, 2003). It is likely that activation of PmlysG expression is a host immune defense directed against general bacterial invasion. Moreover, there was no transmembrane structure, indicating that the protein was not a transmembrane protein. In contrast, the signal peptide is not common in the g-type lysozymes of fish. Only a few fish g-type lysozymes have secretion signals, such as the g-type lysozymes from Salmo sala (Kyomuhendo et al., 2007), but lacks the disulfide bond and N-terminal signal peptide, which is consistent with the lysozymes from C. idellus (Yin et al., 2003), Scophthalmus maximus (Zhao et al., 2011), and Epinephelus coioides (Wei et al., 2014). PmlysG has three conserved catalytic sites (Glu, Asp, Asp), the critical involvement of Glu and Asp in the catalytic activity, also indicate that the Glu residue has pivotal role in the structural stability of G-type lysozyme (Hirakawa et al., 2008). It can promote G-type lysozyme catalyze the hydrolysis of the β-1,4-glycosidic linkage between N-acetylmuramic acid and N-acetylglucosamine alternating sugar residues in the bacterial cell walls and peptidoglycan. The crystal structures of GEL, free and complexed with (GlcNAc)3, revealed that the three-dimensional position of Glu73 in GEL is analogous to those of Glu35 in HEL and Glu11 in T4L, which are believed to act as a general acid to donate a proton to the glycosidic bond, thereby facilitating bond cleavage (Weaver et al., 1995; Kawamura et al., 2006).
Multiple sequence alignments showed that the g-type lysozymes of PmlysG had high similarity with other species, with the highest similarity in Argopecten irradians (60.5%), indicating that PmlysG maintains high conservatism during evolution. Evolutionary analysis showed that P.f. martensii could cluster with Argopcten irradians, Azumapecten farreri, and Mizuhopecten yessoensis, consistent with the relationship of species evolution.
Therefore, assessing the functional characteristics of PmlysG is highly necessitated. Herein, the qRT-PCR analysis showed that PmlysG mRNA was expressed in all tissues. Moreover, the mRNA expression was the highest in hepatopancreas compared with other tissues. Previous studies have detected lysozyme activity in the hepatopancreas of several marine bivalves. The mRNA expression level of Meretrix meretrix was higher in the hepatopancreas than in other tissues (Yue et al., 2011). The high expression level of PmlysG mRNA in the hepatopancreas indicate that it is the primary site for the synthesis of g-type lysozymes. These results confirm that PmlysG is a digestive enzyme that can protect digestive organs from bacterial attacks. Digestive lysozymes may have a potentially low pH and isoelectric points and higher resistance to proteases (Callewaert and Michiels, 2010).
After LPS stimulation, the mRNA expression level of PmlysG reached a maximum at 72 hours. After PGN stimulation, the mRNA expression level of PmlysG reached a maximum at 6 hours, then increased for the second time at 48 hours. After PolyI:C stimulation, the mRNA expression level of PmlysG reached a maximum at 72 hours. Although the three stimulation modes mediated immune response, they had different effects with time. For instance, the mRNA expression was the highest at 72 hours after LPS and PolyI:C stimulation, suggesting that PmlysG might be less involved in the immune response of pathogens containing LPS and PolyI:C in the early stage. Similarly, the mRNA expression was maximum at 6 hours after PGN stimulation, indicating that PmlysG might quickly participate in the immune defense of PGN model pathogens. These results were consistent with the g-type lysozymes of other species stimulated with PAMPs. For instance, PalysG identified in Physa acuta could sustain high expression in the hepatopancreas within 8-48 hours after LPS stimulation (Guo and He, 2014). CalysG identified in Carassius auratus could sustain high expression in the hepatopancreas within 6-48 hours after LPS stimulation (Wang et al., 2016). These results show that PmlysG has various biological functions, and it is a multifunctional molecule essential for digestive function and host immune response.
The rPmlysG was used to identify the antibacterial properties. Several studies have indicated that some g-type lysozymes could inhibit microbial growth. For instance, a g-type lysozyme isolated from Chlamys farreri (Zhao et al., 2007) (rCFlysG) can inhibit V. parahaemolyticus, V. splendidus, and V. anguillarum. Similarly, a g-type lysozyme of salmon can inhibit the growth of Aeromonas hydrophila (Zhang et al., 2018). The recombinant g-type lysozyme of grass carp can inhibit A. hydrophila, V. parahaemolyticus, and Bacillus cereus (Ye et al., 2010; Yang et al., 2016). Additionally, rPmlysG showed potent lytic activity against V. parahaemolyticus, A. hydrophila, and Pseudomonas aeruginosa. However, the activity was less for gram-positive bacteria. Studies have reported that the antimicrobial action occurs through the hydrolysis of β-1,4-glycosidic linkage between N-acetylmuramic acid and N-acetylglucosamine alternating the sugar residues in the PGN of bacterial cell walls, thereby inducing bacterial cell lysis (Mohapatra et al., 2019). In this study, rPmlysG damaged the cell wall of V. parahaemolyticus and A. hydrophila, inducing the release of bacterial contents. This attack pattern was consistent with the Chitosan-lysozyme nanoparticles (CS-Lys-NPs) (Wu et al., 2017c). This experiment provides insights into immunological research and disease control in P.f. martensii.
Conclusion
In this study, a g-type lysozyme gene was successfully cloned from P.f. martensii. The amino acid sequence of PmlysG shared a high similarity with other known shellfish. The mRNA expression analysis indicated that PmlysG was highly expressed in the hepatopancreas. PAMPs stimulation induced an immune response in the body, and rPmlysG exhibited significant antibacterial activity. Overall, these results confirm that the g-type lysozyme plays a key role in the innate immunity of P.f. martensii.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ZG: Investigation and designed the study, Analyzed all data, Writing - Original draft preparation. CS: Analyzed all sequencing data. HL: Conceptualization, Methodology, Writing- Reviewing and Editing. MZ: Prepared the samples, Analyzed all sequencing data. BL: Conclusion. BZ: Prepared the samples, Formal analysis. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants from the National Natural Science Foundation of China (No. 31472306), Guangdong Natural Science Foundation of China (No. 2021A1515010962), Science and technology Special Fund of Guangdong Province (2021A05250), Special Fund for Harbor Construction and Fishery Industry Development of Guangdong Province (No. A201608B15) and Sustainable Development Project of Shenzhen Science and Technology Program (KCXFZ20211020165547010).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1012323/full#supplementary-material
References
Callewaert L., Michiels C. W. (2010). Lysozymes in the animal kingdom. J. Biosci. 35 (1), 127–160. doi: 10.1007/s12038-010-0015-5
Canfield R. E., McMurry S. (1967). Purification and characterization of a lysozyme from goose egg white. Biochem. Biophys. Res. Commun. 26 (1), 38–42. doi: 10.1016/0006-291x(67)90249-5
Cao Y., Yang S., Feng C., Zhan W., Zheng Z., Wang Q., et al. (2019). Evolution and function analysis of interleukin-17 gene from Pinctada fucata martensii. Fish Shellfish Immunol. 88, 102–110. doi: 10.1016/j.fsi.2019.02.044
Du X., Fan G., Jiao Y., Zhang H., Guo X., Huang R., et al. (2017). The pearl oyster pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. Gigascience 6 (8), 1–12. doi: 10.1093/gigascience/gix059
Fan S., Wang W., Li J., Cao W., Li Q., Wu S., et al. (2022). The truncated MyD88s negatively regulates TLR2 signal on expression of IL17-1 in oyster Crassostrea gigas. Dev. Comp. Immunol. 133, 104446. doi: 10.1016/j.dci.2022.104446
Guo Y., He H. (2014). Identification and characterization of a goose-type lysozyme from sewage snail Physa acuta. Fish Shellfish Immunol. 39 (2), 321–325. doi: 10.1016/j.fsi.2014.05.029
Hancock R. E., Scott M. G. (2000). The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. U.S.A. 97 (16), 8856–8861. doi: 10.1073/pnas.97.16.8856
He J., Liang. H., Wu. Y., Deng Y. (2018). Gene cloning and tissue expression analysis of adapter protein 368 CIKS from Pinctada fucata martensii martensii. J. Guangdong Ocean Univ. 38 (4), 369 1–7. doi: 10.3969/j.issn.1673-9159.2018.04.001
He J., Liang H., Zhu J., Fang X. (2019). Separation, identification and gene expression analysis of PmAMP-1 from Pinctada fucata martensii. Fish Shellfish Immunol. 92, 728–735. doi: 10.1016/j.fsi.2019.07.002
He J., Shen C., Liang H., Fang X., Lu J. (2020). Antimicrobial properties and immune-related gene expression of a c-type lectin isolated from Pinctada fucata martensii. Fish Shellfish Immunol. 105, 330–340. doi: 10.1016/j.fsi.2020.07.017
He C., Yu H., Liu W., Su H., Shan Z., Bao X., et al. (2012). A goose-type lysozyme gene in Japanese scallop (Mizuhopecten yessoensis): cDNA cloning, mRNA expression and promoter sequence analysis. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 162 (1), 34–43. doi: 10.1016/j.cbpb.2012.02.002
Hikima J.-i., Minagawa S., Hirono I., Aoki T. (2001). Molecular cloning, expression and evolution of the Japanese flounder goose-type lysozyme gene, and the lytic activity of its recombinant protein. Biochim. Biophys. Acta (BBA) - Gene Structure Expression 1520 (1), 35–44. doi: 10.1016/S0167-4781(01)00248-2
Hirakawa H., Ochi A., Kawahara Y., Kawamura S., Torikata T., Kuhara S. (2008). Catalytic reaction mechanism of goose egg-white lysozyme by molecular modelling of enzyme-substrate complex. J. Biochem. 144 (6), 753–761. doi: 10.1093/jb/mvn133
Irwin D. M., Gong Z. M. (2003). Molecular evolution of vertebrate goose-type lysozyme genes. J. Mol. Evol. 56 (2), 234–242. doi: 10.1007/s00239-002-2396-z
Jiménez-Cantizano R. M., Infante C., Martin-Antonio B., Ponce M., Hachero I., Navas J. I., et al. (2008). Molecular characterization, phylogeny, and expression of c-type and g-type lysozymes in brill (Scophthalmus rhombus). Fish Shellfish Immunol. 25 (1), 57–65. doi: 10.1016/j.fsi.2007.12.009
Jollès P., Jollès J. (1984). What’s new in lysozyme research? always a model system, today as yesterday. Mol. Cell Biochem. 63 (2), 165–189. doi: 10.1007/bf00285225
Kawamura S., Ohno K., Ohkuma M., Chijiiwa Y., Torikata T. (2006). Experimental verification of the crucial roles of Glu73 in the catalytic activity and structural stability of goose type lysozyme. J. Biochem. 140 (1), 75–85. doi: 10.1093/jb/mvj125
Kyomuhendo P., Myrnes B., Nilsen I. W. (2007). A cold-active salmon goose-type lysozyme with high heat tolerance. Cell. Mol. Life Sci. CMLS 64 (21), 2841–2847. doi: 10.1007/s00018-007-7372-8
Liang H., Zhang M., Shen C., He J., Lu J., Guo Z. (2022). Cloning and functional analysis of a trypsin-like serine protease from Pinctada fucata martensii. Fish Shellfish Immunol. 126, 327–335. doi: 10.1016/j.fsi.2022.05.058
Li H., Parisi M.-G., Toubiana M., Cammarata M., Roch P. (2008). Lysozyme gene expression and hemocyte behaviour in the Mediterranean mussel, mytilus galloprovincialis, after injection of various bacteria or temperature stresses. Fish Shellfish Immunol. 25 (1), 143–152. doi: 10.1016/j.fsi.2008.04.001
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Li L., Zhao J., Wang L., Qiu L., Song L. (2013). Genomic organization, polymorphisms and molecular evolution of the goose-type lysozyme gene from zhikong scallop Chlamys farreri. Gene 513 (1), 40–52. doi: 10.1016/j.gene.2012.10.080
Lu J., Zhang M., Liang H., Shen C., Zhang B., Liang B., et al. (2022). Comparative proteomics and transcriptomics illustrate the allograft-induced stress response in the pearl oyster (Pinctada fucata martensii). Fish Shellfish Immunol. 121 (74), 85. doi: 10.1016/j.fsi.2021.12.055
Lv X., Yang W., Guo Z., Wu W., Li Y., Yan X., et al. (2022). CgHMGB1 functions as a broad-spectrum recognition molecule to induce the expressions of CgIL17-5 and Cgdefh2 via MAPK or NF-κB signaling pathway in Crassostrea gigas. Int. J. Biol. Macromol. 211, 289–300. doi: 10.1016/j.ijbiomac.2022.04.166
Matsumoto T., Nakamura A. M., Takahashi K. G. (2006). Cloning of cDNAs and hybridization analysis of lysozymes from two oyster species, Crassostrea gigas and Ostrea edulis. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 145 (3), 325–330. doi: 10.1016/j.cbpb.2006.08.003
McHenery J. G., Birkbeck T. H. (1982). ). characterization of the lysozyme of Mytilus edulis (L). Comp. Biochem. Physiol. B 71 (4), 583–589. doi: 10.1016/0305-0491(82)90466-7
Mohapatra A., Parida S., Mohanty J., Sahoo P. K. (2019). Identification and functional characterization of a g-type lysozyme gene of Labeo rohita, an Indian major carp species. Dev. Comp. Immunol. 92, 87–98. doi: 10.1016/j.dci.2018.11.004
Nilsen I. W., Myrnes B., Edvardsen R. B., Chourrout D. (2003). Urochordates carry multiple genes for goose-type lysozyme and no genes for chicken- or invertebrate-type lysozymes. Cell. Mol. Life Sci. CMLS 60 (10), 2210–2218. doi: 10.1007/s00018-003-3252-z
Nilsen I. W., Overbø K., Sandsdalen E., Sandaker E., Sletten K., Myrnes B. (1999). Protein purification and gene isolation of chlamysin, a cold-active lysozyme-like enzyme with antibacterial activity. FEBS Lett. 464 (3), 153–158. doi: 10.1016/s0014-5793(99)01693-2
Prager E. M., Jollès P. (1996). Animal lysozymes c and g: an overview. Exs 75, 9–31. doi: 10.1007/978-3-0348-9225-4_2
Qiu Y., Lu H., Zhu J., Chen X., Wang A., Wang Y. (2014). Characterization of novel EST-SSR markers and their correlations with growth and nacreous secretion traits in the pearl oyster Pinctada martensii (Dunker). Aquaculture 420-421, S92–S97. doi: 10.1016/j.aquaculture.2013.09.040
Rolff J., Siva-Jothy M. T. (2003). Invertebrate ecological immunology. Science 301 (5632), 472–475. doi: 10.1126/science.1080623
Shakoori M., Hoseinifar S. H., Paknejad H., Jafari V., Safari R., Van Doan H., et al. (2019). Enrichment of rainbow trout (Oncorhynchus mykiss) fingerlings diet with microbial lysozyme: Effects on growth performance, serum and skin mucus immune parameters. Fish Shellfish Immunol. 86, 480–485. doi: 10.1016/j.fsi.2018.11.077
Wang L., Qiu L., Zhou Z., Song L. (2013). Research progress on the mollusc immunity in China. Dev. Comp. Immunol. 39 (1), 2–10. doi: 10.1016/j.dci.2012.06.014
Wang L., Song X., Song L. (2018). The oyster immunity. Dev. Comp. Immunol. 80, 99–118. doi: 10.1016/j.dci.2017.05.025
Wang M., Zhao X., Kong X., Wang L., Jiao D., Zhang H. (2016). Molecular characterization and expressing analysis of the c-type and g-type lysozymes in qihe crucian carp Carassius auratus. Fish Shellfish Immunol. 52, 210–220. doi: 10.1016/j.fsi.2016.03.040
Weaver L. H., Grütter M. G., Matthews B. W. (1995). The refined structures of goose lysozyme and its complex with a bound trisaccharide show that the “goose-type” lysozymes lack a catalytic aspartate residue. J. Mol. Biol. 245 (1), 54–68. doi: 10.1016/s0022-2836(95)80038-7
Wei S., Huang Y., Huang X., Cai J., Wei J., Li P., et al. (2014). Molecular cloning and characterization of a new G-type lysozyme gene (Ec-lysG) in orange-spotted grouper, epinephelus coioides. Dev. Comp. Immunol. 46 (2), 401–412. doi: 10.1016/j.dci.2014.05.006
Wu Y., Guo Z., Liang H. (2017c). Clone and expression analysis of PmLec-8 gene from Pinctada fucata martensii. J. Guangdong Ocean Univ. 37 (04), 1–7. doi: 10.3969/j.issn.1673-9159.2017.04.001
Wu Y., Liang H., Wang Z., Lei Q., Xia L. (2017b). A novel toll-like receptor from the pearl oyster pinctada fucata martensii is induced in response to stress. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 214, 19–26. doi: 10.1016/j.cbpb.2017.08.006
Wu T., Wu C., Fu S., Wang L., Yuan C., Chen S., et al. (2017a). Integration of lysozyme into chitosan nanoparticles for improving antibacterial activity. Carbohydr. Polymers 155, 192–200. doi: 10.1016/j.carbpol.2016.08.076
Yang Y., Yu H., Li H., Wang A. (2016). Transcriptome profiling of grass carp (Ctenopharyngodon idellus) infected with aeromonas hydrophila. Fish Shellfish Immunol. 51, 329–336. doi: 10.1016/j.fsi.2016.02.035
Ye X., Zhang L., Tian Y., Tan A., Bai J., Li S. (2010). Identification and expression analysis of the g-type and c-type lysozymes in grass carp Ctenopharyngodon idellus. Dev. Comp. Immunol. 34 (5), 501–509. doi: 10.1016/j.dci.2009.12.009
Yin Z. X., He J. G., Deng W. X., Chan S. M. (2003). Molecular cloning, expression of orange-spotted grouper goose-type lysozyme cDNA, and lytic activity of its recombinant protein. Dis. Aquat. Organ. 55 (2), 117–123. doi: 10.3354/dao055117
Yue X., Liu B., Xue Q. (2011). An i-type lysozyme from the Asiatic hard clam meretrix meretrix potentially functioning in host immunity. Fish Shellfish Immunol. 30 (2), 550–558. doi: 10.1016/j.fsi.2010.11.022
Zhang C., Zhang J., Liu M., Huang M. (2018). Molecular cloning, expression and antibacterial activity of goose-type lysozyme gene in Microptenus salmoides. Fish Shellfish Immunol. 82, 9–16. doi: 10.1016/j.fsi.2018.07.058
Zhang S. H., Zhu D. D., Chang M. X., Zhao Q. P., Jiao R., Huang B., et al. (2012). Three goose-type lysozymes in the gastropod Oncomelania hupensis: cDNA sequences and lytic activity of recombinant proteins. Dev. Comp. Immunol. 36 (1), 241–246. doi: 10.1016/j.dci.2011.06.014
Zhao J., Song L., Li C., Zou H., Ni D., Wang W., et al. (2007). Molecular cloning of an invertebrate goose-type lysozyme gene from Chlamys farreri, and lytic activity of the recombinant protein. Mol. Immunol. 44 (6), 1198–1208. doi: 10.1016/j.molimm.2006.06.008
Zhao L., Sun J.-s., Sun L. (2011). The g-type lysozyme of scophthalmus maximus has a broad substrate spectrum and is involved in the immune response against bacterial infection. Fish Shellfish Immunol. 30 (2), 630–637. doi: 10.1016/j.fsi.2010.12.012
Keywords: Pinctada fucata martensii, g-type lysozyme, expression analysis, antibacterial activity, quantitative real-time PCR
Citation: Guo Z, Shen C, Liang H, Zhang M, Liang B and Zhang B (2022) Immune characterization and expression analysis of a goose-type lysozyme gene from Pinctada fucata martensii. Front. Mar. Sci. 9:1012323. doi: 10.3389/fmars.2022.1012323
Received: 05 August 2022; Accepted: 28 September 2022;
Published: 13 October 2022.
Edited by:
Maria Angeles Esteban, University of Murcia, SpainReviewed by:
Jingguang Wei, South China Agricultural University, ChinaZhi Liao, Zhejiang Ocean University, China
Copyright © 2022 Guo, Shen, Liang, Zhang, Liang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiying Liang, empsaWFuZ2h5QDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
 Zhijie Guo
Zhijie Guo Chenghao Shen
Chenghao Shen Haiying Liang
Haiying Liang Meizhen Zhang1
Meizhen Zhang1