- 1Department of Marine Biology, Leon H. Charney School of Marine Sciences, University of Haifa, Haifa, Israel
- 2The Interuniversity Institute of Marine Sciences, Eilat, Israel
- 3Department of Earth and Environmental Sciences, Ben-Gurion University of the Negev, Be’er Sheva, Israel
Scleractinian corals are evolutionary-successful calcifying marine organisms, which utilize an endo-symbiotic relationship with photosynthetic dinoflagellate algae that supply energy products to their coral hosts. This energy further supports a higher calcification rate during the day in a process known as light enhanced calcification. Although this process has been studied for decades, the mechanisms behind it are still unknown. However, photosynthesis and respiration also cause daily fluctuations in oxygen and pH levels, resulting in the coral facing highly variable conditions. Here we correlated gene expression patterns with the physiological differences along the diel cycle to provide new insights on the daily dynamic processes, including circadian rhythm, calcification, symbiosis, cellular arrangement, metabolism, and energy budget. During daytime, when solar radiation levels are highest, we observed increased calcification rate combined with an extensive up-regulation of genes associated with reactive oxygen species, redox, metabolism, ion transporters, skeletal organic matrix, and mineral formation. During the night, we observed a vast shift toward up-regulation of genes associated with cilia movement, tissue development, cellular movement, antioxidants, protein synthesis, and skeletal organic matrix formation. Our results suggest that light enhanced calcification is related to several processes that occur across the diel cycle; during nighttime, tissue might elevate away from the skeleton, extending the calcifying space area to enable the formation of a new organic framework template. During daytime, the combination of synthesis of acid-rich proteins and a greater flux of ions to the sites of calcification facilitate the conditions for extensive mineral growth.
Introduction
Coral reefs are among the most diverse and productive ecosystems in the ocean and provide substantial economic and cultural resources (Spalding et al., 2001). Stony corals that form the reefs evolved during the Cambrian period (503 Ma) (McFadden et al., 2021) and are one of the first known metazoans to precipitate a calcium carbonate exoskeleton in a biologically mediated process (Knoll, 2003). This process leads to the production of approximately 4 kg calcium carbonate per square meter per year in the oceans (Smith and Kinsey, 1976). Their ecological success is related to endo-photosymbiosis with unicellular dinoflagellate algae of the family Symbiodiniaceae (Stanley, 2006; LaJeunesse et al., 2018) that evolved later, during the Devonian period (383 Ma) (McFadden et al., 2021). As a result of the photosymbiosis, the algae translocate more than 90% of their photosynthetic products to their coral host (Muscatine, 1990; Falkowski et al., 1993).
Due to the presence of the endosymbiont algae and their photosynthetic activities, symbiotic corals are exposed to wide daily variations in intracellular oxygen concentrations and pH levels (Shashar et al., 1993; Kuhl et al., 1995; Al-Horani et al., 2005; Linsmayer et al., 2020). During the daytime, when photosynthesis rates are high, the coral experiences hyperoxia and a higher pH (Kuhl et al., 1995), while during the night, in the absence of photosynthesis, respiration of both host and algae causes hypoxia and low pH (Shashar et al., 1993; Kuhl et al., 1995; Al-Horani et al., 2005; Linsmayer et al., 2020). Oxygen levels have important implications on corals’ energy budget as they determine whether aerobic versus anaerobic metabolic pathways are used, which, in turn, has implications for the efficiency of the energy production (Nelson and Altieri, 2019).
It has long been observed that photosynthesis-enhanced calcification rates occur in symbiotic corals (Yonge, 1931; Kawaguti, 1948), in a process known as “light-enhanced calcification” (LEC) (Vandermeulen and Muscatine, 1974; Barnes and Chalker, 1990; Gattuso et al., 1999). Moya et al. (2006) determined that the endogenous circadian rhythm does not stimulate the LEC phenomenon, instead, it is driven only by light. In addition, Marshall (1996) reported that non-symbiotic tropical corals exhibit similar calcification rates to symbiotic corals suggesting that calcification may be dark-repressed and not light-enhanced. In this context, Colombo-Pallotta et al. (2010) proposed that calcification rates are mainly promoted by translocation of photosynthetic products such as glycerol and oxygen to the mitochondria in order to generate enough ATP to cover the high energy demand for calcification. Therefore, the authors of these studies concluded that light itself does not necessarily stimulate calcification in symbiotic corals (Marshall, 1996), and corals must rely on a different source of energy to support calcification (Mass et al., 2007).
Skeletal formation studies have suggested two steps of mineral formation related to the diel cycle (Gladfelter, 1983; Vago et al., 1997; Cohen and McConnaughey, 2003). During the night, the tissue is elevated from the skeleton, creating a larger calcifying fluid space (Vago et al., 1997), followed by the formation of nano-granule particles at the centers of calcification (CoCs), while during the daytime, aragonite needle-shaped fibers are radiated outward from the newly formed CoCs, rapidly filling the calcifying fluid space (Barnes, 1970, 1972; Gladfelter, 1983; Cohen and McConnaughey, 2003). However, a recent study, which used nano secondary ion mass spectrometry (nano-SIMS), demonstrated that filling of the mineral space occurred during both day and night (Domart-Coulon et al., 2014).
The biomineralization process is associated with a set of macromolecules such as proteins (Constantz and Weiner, 1988), phospholipids (Isa and Okazaki, 1987), and polysaccharides (Cuif et al., 2003; Naggi et al., 2018), which are secreted to the extracellular matrix (ECM) (Peled et al., 2020). Mass et al. (2013) identified a group of coral acid-rich proteins (CARPs) that catalyzed calcium carbonate precipitation in vitro. In addition, the proteome of the skeletal organic matrix contains an assemblage of adhesion and structural proteins as well as CARPs, which together create a framework for the precipitation of aragonite (Drake et al., 2013; Ramos-Silva et al., 2013; Takeuchi et al., 2016; Peled et al., 2020; Mummadisetti et al., 2021). Immunolocalization assays and chemical cross-linking have revealed a non-random spatial arrangement of key matrix proteins in the skeleton of the common symbiotic coral Stylophora pistillata (Mass et al., 2014; Mummadisetti et al., 2021). This arrangement suggests that the organic components are intimately associated with the mineral phase, organized in a highly ordered structure consistent with a diel calcification pattern (Mass et al., 2014).
The main aim of this study was to elucidate the temporal sequence of events throughout the biomineralization reaction of the endosymbiotic coral Stylophora pistillata. We correlated physiological and transcriptomic approaches to better understand the dynamics of diverse genetic processes critical for corals’ development through the diel cycle, including the biomineralization process. Specifically, we correlated the gene expression pattern with calcification rates, dark respiration rates, and tissue biomass at eight time points across the diel cycle.
Materials and Methods
Sample Preparation
Four colonies of Stylophora pistillata were collected from the reef in front of the Interuniversity Institute of marine science in Eilat (IUI; 29°30′16″N, 34°55’7″E) under a permit from the Israeli Natural Parks Authority, permit number 42410/2019. Each colony was fragmented into 40 fragments of 2 cm nubbins, glued onto transparent film paper, and allowed to recover for 3.5 months in an outdoor running seawater system at the IUI. In addition, from each colony, four fragments of 0.5 cm and 2 cm long were prepared for respiration and calcification analyses, respectively. The seawater was continuously pumped by a high-volume water pump (Ebara, stainless steel, 3LSF 50–125/4.0, flow rate ca. 30 m3 h–1) from 30 meters deep on the adjacent reef slope. The seawater inlets are covered by a coarse mesh (2 cm2) to exclude macro marine biota such as fish and large invertebrates. The mesh covers are manually scrubbed clean once a month to maintain water flow. Corals were maintained in ambient light:dark and temperature cycle from February to May 2020. The outdoor system was exposed to the full spectrum ambient sunlight (ca. 1800 μmol m–2 s–1 PAR at midday on a cloudless day). The growth of lateral edges of each nubbin indicated the recovery and new growth of the corals.
Coral Sampling and Time Points
On 10th of May 2020, nubbins from each colony were randomly sampled from the water-system every 3 h: 06:00am, 09:00am, 12:00pm, 15:00pm, 18:00pm, 21:00pm, 00:00am and 03:00am. Light (lux) and temperature were monitored throughout the duration of the experiment (Supplementary Figure 2) using HOBO pendant temperature and light data loggers (Onset, United States). The units of light lux were converted to PAR using a calibration curve generated by the comparison of HOBO and a Full-Spectrum Quantum PAR sensors and PAR meter (Apogee instrument, United States) diel measurements. The sample of 6:00 am took place approximately at sunrise, and 6:00 pm took place at sunset. Two nubbins from three colonies at each time point (total of n = 64) were briefly drip-dried to remove excess seawater and mucus, snap frozen in liquid N2, and stored at –80°C until processed for genetic and physiological analyses. At the same time points, calcification and respiration rates were measured (Supplementary Figure 1).
RNA and DNA Extraction
Total RNA and DNA were extracted using the Zymo DNA/RNA kit (catalog number D7003) following the manufacturer’s protocol. DNAase treatment was performed within the RNA extraction procedures according to the manufacturer’s protocol. RNA concentration and quality were confirmed using a NanoDrop 2000 (Thermo Fisher Scientific, United States) and RNA ScreenTape (Agilent).
Symbiodiniaceae Species Analyses
The internal transcribed spacer (ITS2) of Symbiodiniaceae DNA was amplified using Symbiodiniaceae-specific primers as described by Arif et al. (2014). The amplified fragments were then separated with electrophoresis, using a 1% agarose gel. 10 μL of the PCR products were sent to HyLab (Hy Laboratories Ltd., Israel), where they were subjected to a second PCR using the Access Array tag for Illumina primers (Fluidigm Corporation, United States). Index and adaptor sequences were added as required for sequencing on the Illumina system. Then, samples were purified using AMPure XP beads (Beckman Coulter Inc., United States), and the concentration was determined by Qubit (Thermo Fisher Scientific, United States). Samples were sequenced on the Illumina Miseq using a v2-500 cycle kit to generate 250 × 2, paired-end reads.
RNA Sequencing
RNA libraries were prepared at the Nancy and Stephen Grand Israel National Center for Personalized Medicine Weizmann Institute of Science using the Genomics in-house protocol for mRNA-seq. Briefly, the polyA fraction (mRNA) was purified from 500 ng of total RNA following fragmentation and the generation of double-stranded cDNA. Then, end repair, A base addition, adapter ligation, and PCR amplification steps were performed. The quality of the libraries was evaluated by Qubit (Thermo fisher scientific) and TapeStation (Agilent). Libraries were sequenced using Illumina NovaSeq S1 in 100 cycles on two lanes.
RNA-seq reads (150 paired-end, NovaSeq S1 100 cycle kit) were first cleaned from adapters using Cutadapt (Martin, 2011) and low-quality regions using Trimmomatic (Bolger et al., 2014) and then mapped to the S. pistillata and Symbiodinium microadriaticum genomes. Overall, a median of 29 vs. 0.9 million reads per sample were mapped uniquely to exons of S. pisitllata (NCBI GCF_002571385.1) vs. S. microadriaticum (NCBI GCA_001939145.1), respectively. This was done using STAR (Dobin and Gingeras, 2016). Reference coordinates and annotations of S. pisitllata NCBI were combined with additional annotations (reefgenomics spis v.1.0) using UCSC Liftover tools.
Differential Expression Analysis
Differential Expression (DE) analysis was conducted using DEseq2 (Love et al., 2014) and Voom (Law et al., 2014). In both tools, we considered the time factor and the colony factor since the samples come from three different colonies. Both tools gave comparable results, and we chose to use Voom since it allows testing for differential expression trends (here we tested quadratic trends). Vegan1 NMDS graphs initially gave > 0.1 NMDS stress values; ≤0.1 values are required for a close representation of pairwise dissimilarity between objects in a low dimensional space. In order to obtain lower NMDS stress values, the original read counts were cleaned from the effect of an additional batch factor, using the SVA ComBat-Seq function in R (Leek et al., 2012). SVA results were used only for visualization purpose.
Functional Enrichment Analyses
Functional enrichment analyses were conducted using Goseq based on DEseq2 results and using Clusterprofiler based on Voom expression profiles groups (Young et al., 2010; Yu et al., 2012).
Biomineralization genes were added in the DE table based on three sources (Drake et al., 2013; Zoccola et al., 2015; Peled et al., 2020).
Protein Concentration
Coral tissue of each frozen nubbin was airbrushed using cold 100 mM phosphate-buffered saline (PBS) mixed with 0.1 mM EDTA buffer (pH 7). The tissue slurry was electrically homogenized for 20s (Diax-100, Heidolph, Germany). Total protein concentration was measured using the Bradford assay (BIO-RAD-5000201) following the manufacture protocol (Bradford, 1976). The absorbance of samples was read with triplicate technical replicates, and protein concentration was determined against BSA standards.
Dark Respiration Rates
At each time point, four 0.5 cm nubbins of each colony were transferred directly from the water table to a 24-well glass microplate (Loligo systems, Denmark). Each microplate well (1700 μL) contained one nubbin measured (n = 16) after 30 min of dark incubation. An optical fluorescence oxygen system (PreSens, Germany) was used to quantify oxygen consumption rates inside the sealed wells. Measurements were carried out for 15 min in the dark. Data were recorded with MicroResp™ software (Loligo systems, Denmark). Before measurements, the oxygen sensors were calibrated for approximately 30–45 min against air-saturated seawater (100% oxygen) and a saturated solution of sodium sulfite (zero oxygen), as recommended by the user manual (Loligo systems, Denmark). Dark respiration rates were normalized to the sample protein concentration (mg).
Calcification Rates
Calcification rates were estimated using the total alkalinity anomaly method (TA) (Gran, 1952) with some modifications. Briefly, at each time point, fragments with a similar volume of approximately 1 mL were placed in transparent sealed 4 ml chambers with 0.22 μm filtered seawater and kept in the aquaria system for 3 h to represent the natural light and temperature regime. Water was re-filtered post incubation, and triplicates were measured with Titroline 7000 titrator (SI Analytics, Germany). Endpoint alkalinity in the samples was subtracted from time zero samples (no incubation). Samples were titrated with 0.005M HCl and calibrated by Dickson reference material (Dickson, 1981) for oceanic measurements. Calcification rates [μmol CaCO3 h–1 mg−1] were calculated using the equation described by Schneider and Erez (2006):
where Δ Alk (mg kg–1) is the difference in alkalinity between the beginning and the end of the incubation period, V is the volume (mL), 1.028 is the density of seawater of the northern Gulf of Eilat, T is the duration of incubation (h), and P is the protein concentration (mg).
Results
Symbiodiniaceae Species Analyses
To demonstrate that the observed physiological and molecular changes in the coral host cells are not dependent on photosymbiont species, ITS analysis was performed. A total number of 296,922 ITS2 high-quality sequence reads (mean length = 315 bp) were produced from four colonies. Non-operational taxonomic units (OTUs) of other organisms that are most likely part of the coral holobiome were filtered out. After filtration, only OTUs of Symbiodiniaceae remained. The species identified in all four biological samples was Symbiodinium microadriaticum (formerly Symbiodinium clade A) (98% similarity).
Gene Expression Trends of the Diel Cycle
Our differential gene expression analysis reveals a distinct diel expression pattern of the stony coral S. pistillata. We first searched for diurnal-dependent quadratic-trends of gene expression using Voom (Law et al., 2014), which identifies continuous changes in gene expression along time. Out of 20,102 tested genes, 5,287 genes showed significant time-dependent trends. These genes were grouped into 14 clusters based on the time at maximum/minimum expression (Figure 1). These clusters can be further divided into four time-specific expression pattern groups: (i) morning peak clusters – up-regulation starts during the night, followed by a morning peak of expression (06:00 to 09:00 am) and noon down-regulation (Figure 1, orange box, clusters A, B, C, and D); (ii) noon peak clusters – up-regulation starts during the morning, followed by a noon peak of expression (12:00 to 15:00) and evening down-regulation (Figure 1, red box, clusters E, F, G, and H); (iii) evening peak clusters – up-regulation starts during the noon period, followed by an evening peak of expression (21:00) and nighttime down-regulation (Figure 1, green box, clusters I and J); and (iv) night peak clusters – up-regulation starts during the evening, followed by a night peak of expression (00:00) and morning down-regulation (Figure 1, blue box, clusters K, L, M, and N). Those categories can be further divided to “day expressed genes” (group i, ii, and iii) and “night expressed genes” (group iv).
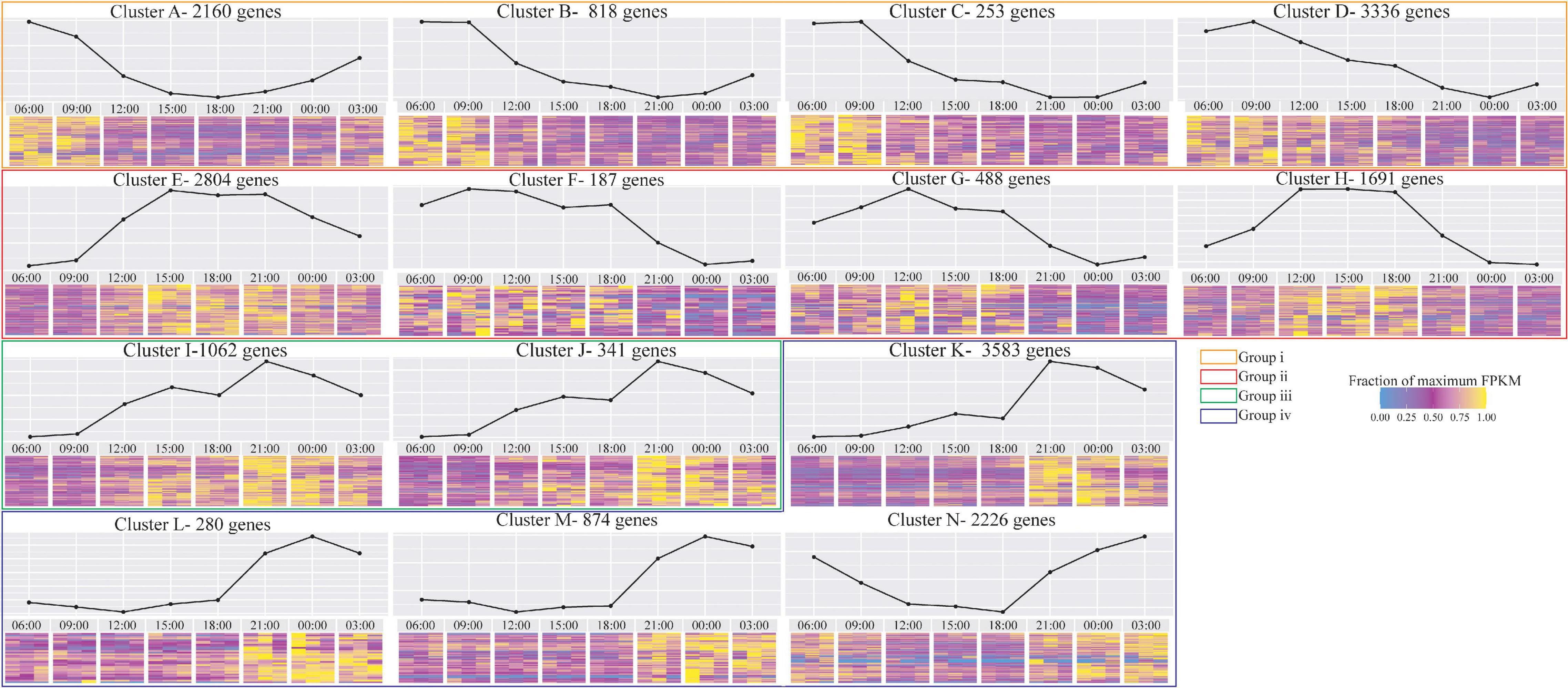
Figure 1. Diurnal trends in gene expression of the coral host, clustered based on minimum/maximum expression peak, based on 5,287 genes showing significant trends using Voom out of 20,102 differentially expressed S. pistillata genes tested. In each cluster, the graph above represents mean Fragments Per Kilobase Million (FPKM) of the genes. The heatmap below represents the fraction of maximum FPKM values of each gene and time-point. Clusters that represent similar expression trends along the diel cycle are framed in colored boxes.
Next, we examined the expression pattern of cryptochrome genes (CRY’s) that are known to be involved in the circadian rhythm signaling proteins in animals (Stanewsky et al., 1998; Lin and Todo, 2005) and as blue-light photoreceptors in plants and corals (Falciatore and Bowler, 2005; Levy et al., 2007). Indeed, cryptochrome 1 (CRY1) and cryptochrome DASH (CRY-DASH) were up-regulated in the morning, followed by a noon peak of expression and evening down-regulation (Figure 1 clusters G and H, respectively).
Functional Enrichment
We next performed Gene Ontology (GO) functional-enrichment based on kyoto encyclopedia of genes and genomes (KEGG) pathway database (Kanehisa et al., 2004) and S. pistillata Trinotate annotations to identify functional terms associated with diel cycle (Figure 2 and Supplementary Table 1). We refer to these enrichment patterns, based on the biological context, as follows.
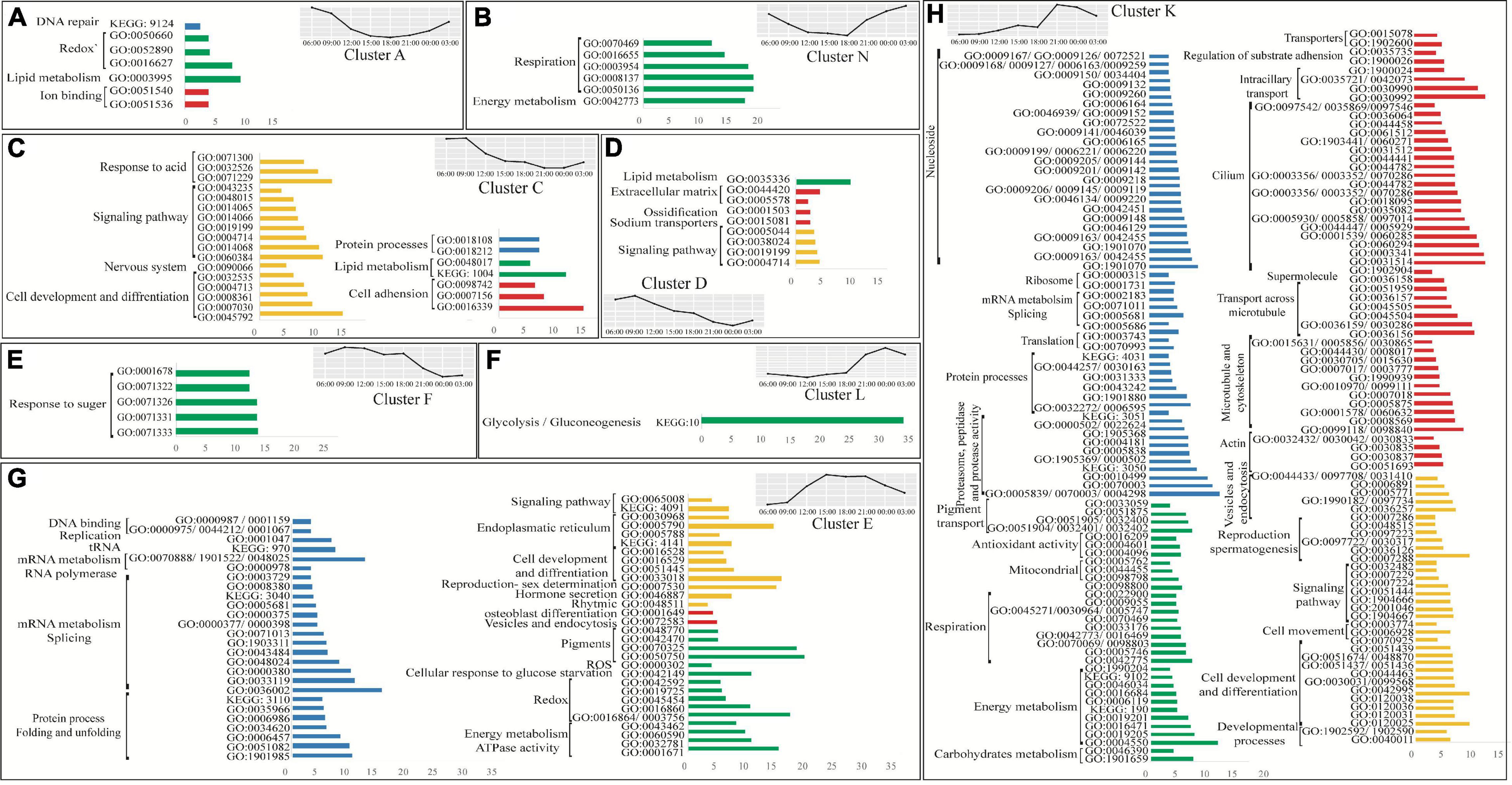
Figure 2. Enriched groups of host genes in the most abundant selected clusters. In each bar plot, the y-axis represents the GO/KEGG ID and the x-axis represents the enrichment factor. The labels represent the GO/KEGG-related term. Groups were filtered by a minimal enrichment factor value of 3 (with some exceptions). Functional terms with comparable enrichment factors and categories have been merged to the same bar. Blue bars represent the genetic code processing (DNA, RNA, and proteins), green bars represent metabolic processes, red bars represent structural terms, and yellow bars represent cellular processes. The full enrichment table is Supplementary Table 1.
Protein Biomass Production
Transcription and translation processes, in which DNA is transcribed to mRNA and eventually to proteins, were found to be dependent on the diel cycle pattern described above. Terms related to DNA and RNA formation such as “DNA binding,” “replication,” “mRNA processes,” “splicing,” “RNA polymerase,” “tRNA,” “protein folding,” and “protein unfoldin” were significantly over-represented during the noontime peak group (pattern group ii, Figure 2G). In contrast, terms related to protein formation such as “ribosomes,” “translation,” “protein processes,” and “proteasome, peptidase and protease activity,” which plays a role in protein degradation, were over-represented only later during the afternoon with a peak in the expression during the evening (pattern group iii, Figure 2H). Upon quantifying the total protein biomass, we found that the total protein biomass at nighttime was about two-fold higher than daytime [Figure 3, one-way ANOVA F (7,14) = 6.0575, followed by Tukey’s post-test (p = 0.00143)].
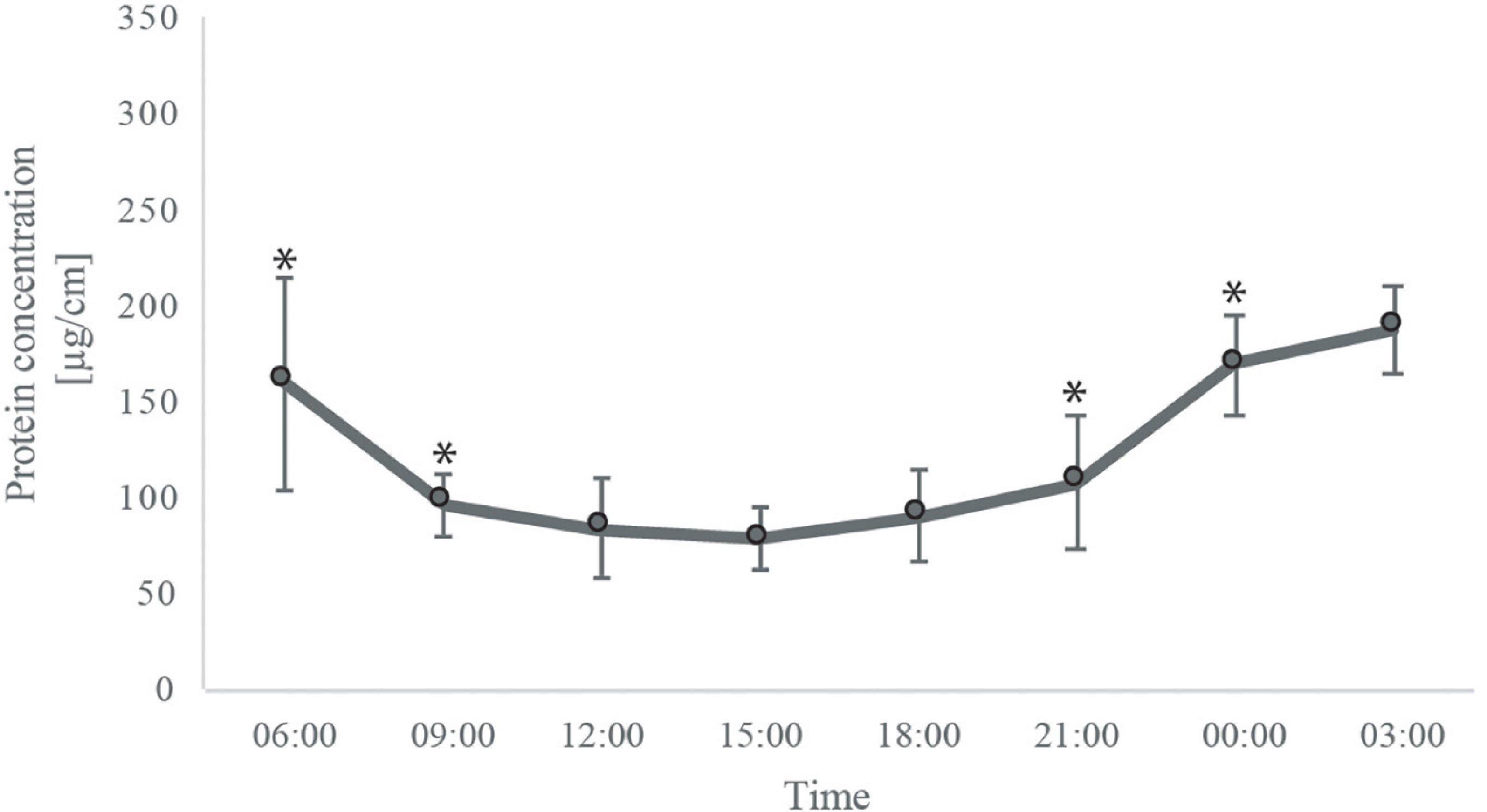
Figure 3. Mean total protein concentration of S. pistillata fragments at each time point. Error bars represent the standard error [one way ANOVA F(7,14) = 6.0575, followed by Tukey’s post-test] (p = 0.0014).
Host Genes Associated With Photosynthesis-Dependent Oxidative Stress
We found that biological processes (BP) like “antioxidant activity” terms are over-represented during the night (Figure 2H), whereas “redox” related terms were over-represented at all time points (Figures 2A,G). “DNA repair” related terms are significantly over-represented during the night and morning (pattern group i, Figure 2A). This seems to reflect the DNA damage and oxidative stress related to high irradiance (Supplementary Figure 2) and high photosynthetic activity during the day (Schneider et al., 2009), as specifically indicated by noontime over-representation of “reactive oxygen species (ROS) activity” (pattern group ii, Figure 2G).
Host Genes Associated With Photosynthetic Metabolites
High photosynthesis rates during the day (Barnes and Chalker, 1990; Schneider et al., 2009) may increase the energy translocation from the symbionts to the host. The combination of this with heterotrophic feeding might be reflected by significant daytime over-representation of host metabolism-related terms. We observed morning over-representation of “lipid metabolism” (pattern group i, Figures 2A,C,D), and noon over-representation of “response to sugars,” “response to glucose starvation” and “energy metabolism and ATPase activity” terms (pattern group ii, Figures 2E,G). In addition, “carbohydrates metabolism” and “glycolysis/gluconeogenesis” terms are over-represented during the night (pattern group iv, Figures 2F,H).
Cellular Respiration
In this study, we measured dark respiration rates and did not find significant differences throughout the diurnal cycle [repeated measured ANOVA F(7,25) = 2.455 (p > 0.05)]. Coral respiration rates follow dark adaptation usually present slight variation compared to post-illumination respiration rates due to the absence of photosynthesis production of oxygen which kinetically limited mitochondrial respiration (Colombo-Pallotta et al., 2010). Moreover, our enrichment analysis indicates that different cellular respiration-related terms are over-represented during all time points (Figure 2). During the morning and noontime, terms involved in “redox” are over-represented (pattern groups i and ii, Figures 2A,G, respectively), while during the night, terms related to “mitochondrial” and “respiration” are over-represented (pattern group iv, Figures 2B,H).
Development and Reproductive Behavior
We found that developmental terms are over-represented throughout the diel cycle. “Signaling pathway” and “cell development and differentiation” are over-represented during the morning, noon, and night (pattern groups i, ii, and iv, Figures 2C,D,G,H, respectively), while other specific developmental terms are time exclusive; “nervous system” terms are morning over-represented (pattern groups i, Figure 2C), “endoplasmic reticulum,” and “rhythmic” terms are noon over-represented (pattern groups ii, Figure 2G), and “cell movement” terms are night over-represented (pattern groups iv, Figure 2H). Furthermore, as the experiment took place during S. pistillata reproductive season (January – September) (Shefy et al., 2018), terms related to “sex determination” (pattern groups ii, Figure 2G) and “spermatogenesis” (pattern groups iv, Figure 2H) were over-represented during the noon and night.
Structural Genes
Enrichment analysis of cellular components (CC) reveals a night over representation of terms that can either play a role in structural components or alternatively in the spermatogenesis process. This includes terms that are related to the formation of cells’ structural machinery and cytoskeleton, including such terms as “actin,” “microtubule,” “supermolecules,” “cilium,” “regulation of substrate adhesions,” transport related genes such as “transporters,” “intraciliary transport,” and “transport across microtubule” (pattern groups iv, Figure 2H). Exception groups that are “ion binding” and “cell adhesion,” “extracellular matrix” and “ossification” terms that are over-represented during the morning (pattern groups i, Figures 2A,C,D), and “osteoblast differentiation” and “vesicles and endocytosis” terms that are over-represented during at noon (pattern groups ii, Figure 2G).
Calcification
Diurnal measurements of calcification rate revealed an increase in calcification rate with light intensity. However, the peak calcification rate did not match the peak light intensity but was delayed by ∼ 3h [Figure 4A, repeated measured ANOVA F(7,25) = 1289.77 (p < 0.00001), followed by Tukey’s post-test]. Interestingly, the expression pattern of biomineralization “toolkit” genes (Drake et al., 2013; Peled et al., 2020) is time-dependent and follows the diurnal pattern as well. We found that 46 out of 101 biomineralization related genes were DE across the diel cycle (Figure 4 according to their molecular function). Throughout the whole diel cycle, we observed up-regulation of genes that play a role in the adhesion and formation of the protein framework of the skeletal organic matrix (SOM) (Drake et al., 2013; Mummadisetti et al., 2021). Specifically, genes annotated as MAM and LDL receptor and vitellogenin-like represent the morning peak (Figure 4Bi). Growth factor (EGF) and laminin-G domain-containing (EGF LamG), protocadherin 5, and sushi domain-containing proteins represented the noon peak (Figure 4Bii), and phosphopentothenoylcysteine decarboxylase subunit VHS3, CUB domain-containing, vitellogenin, and another protocadherin-like represented the evening peak (Figure 4Biii).
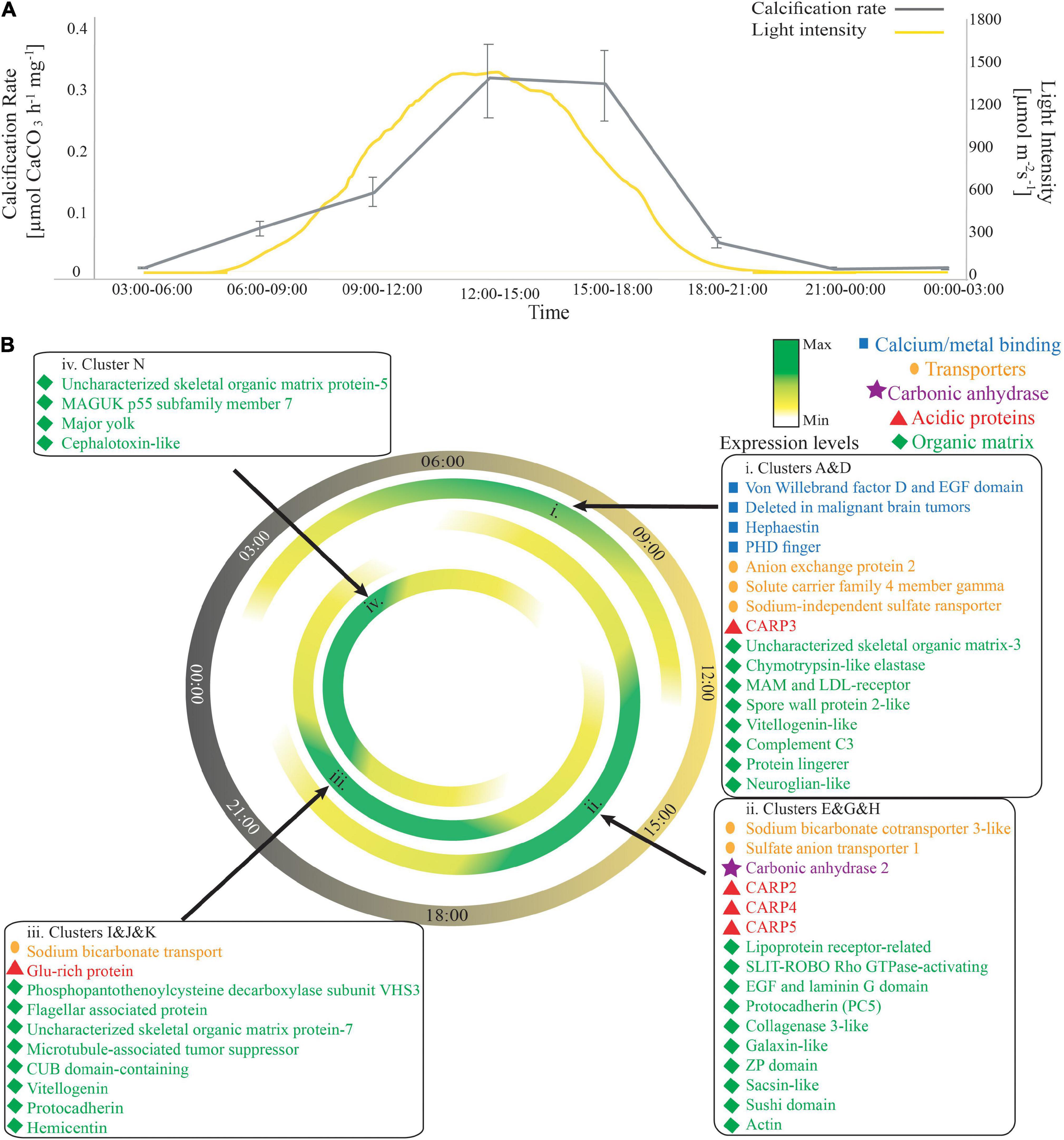
Figure 4. Biomineralization pattern across the diel cycle. (A) Average calcification rate (μmol CaCO3 mg– 1h– 1, gray line) and light intensities (μmol m– 2s– 1, yellow line) during each of the sampling points. Error bars represent the standard error [repeated measured ANOVA F(7,25) = 1289.77 (p < 0.00001), followed by Tukey’s post-test]. (B) The expression pattern of known biomineralization related genes. Each box is indicated with a black arrow to the hour of peak expression. Blue squares represent the calcium/metal binding genes, orange circles represent ion transporters, purple stars represent the carbonic anhydrase enzyme, STPCA2, red triangles represent the CARPs, and the green diamonds represent the other organic matrix genes. The full table with gene accession numbers can be found in Supplementary Table 2.
In contrast, the expression of calcium/metal-binding genes was restricted to specific hours, representing a morning peak in expression (Figure 4Bi). In addition, the acid-rich proteins (CARPs), which have been suggested to control nucleation and growth of the aragonite crystal-like fibers of the skeleton, also represent a unique expression pattern across the diel cycle. CARP3 was highly expressed during the morning (Figure 4Bi), whereas CARPs 2, 4, and 5 were highly expressed during noontime, following the trend of calcification rate (Figure 4A). All these detected CARPs were downregulated during the night. One exception is the glutamic-rich protein (CARP6) that was up-regulated during the afternoon with peak expression during the evening (Figure 4Biii). CARP1 was not differentially expressed throughout the day.
Different bicarbonate transporters were highly expressed in each time point. Solute carrier family 4-member gamma (SLC4γ), which was localized previously to the calicoblastic layer (Zoccola et al., 2015), anion exchange protein 2, and sodium-independent sulfate anion transporter (XM_022925996.1) were highly expressed during the morning (Figure 4Bi); sulfate anion transporter 1 (XP_022800348.1) and sodium bicarbonate cotransporter 3-like (XP_022801462.1) were highly expressed at noon (Figure 4Bii), and another sodium bicarbonate transporter (XP_022783031.1) was highly expressed during the evening (Figure 4Biii). In contrast, the expression of carbonic anhydrase 2 (STPCA2), which catalyzes the reversible hydration of carbon dioxide into bicarbonate and has been localized to both the calicodermis (Bertucci et al., 2010) and the coral skeleton (Moya et al., 2008), was restricted only to daytime, with a peak at noontime, representing a similar trend to the expression pattern of CARPs 2, 4, 5 (Figure 4Bii) and to calcification rates (Figure 4A).
Discussion
In this study, we characterized both physiology and gene expression patterns at eight time points across the diel cycle. Our clustering analysis revealed the dynamics of diverse genetic processes critical for corals’ development through the diel cycle, including circadian rhythm, calcification, symbiosis, cellular arrangement, metabolism, and energy budget. We also found that time of day, which is correlated with light intensities, may trigger some of the processes directly or indirectly.
Circadian Rhythm
The blue-light–sensing photoreceptors CRY genes were first identified in stony corals by Levy et al. (2007), who confirmed that these genes are daylight-dependent and are upregulated during the day. Indeed, in our data, CRY1 and CRY-DASH, which were differentially expressed, represent the same daylight-dependent trend.
Protein Production and Cellular Development
The current central dogma in molecular biology of all living organisms is that DNA is transcribed to RNA which directs protein synthesis; there are many studies that show a positive correlation between differential expression of specific mRNAs and the corresponding proteins (Greenbaum et al., 2002; Lu et al., 2007; Ning et al., 2007). Thus, many studies, including this study, have used transcriptome analysis to examine gene regulation under different conditions. However, there is growing evidence of variations between the expressed mRNA and the corresponding proteins, and for some gene categories, the correlation is stronger than others, resulting in a lag between the transcription and protein synthesis (Gygi et al., 1999; Shankavaram et al., 2007; Gry et al., 2009). This lag can result from a series of intertwining processes of the mRNA prior to translation into functional proteins, also known as the “posttranscriptional” regulation (PTR) (Schaefke et al., 2018). These regulatory processes include processes such as splicing (Berget et al., 1977; Chow et al., 1977), polyadenylation (Elkon et al., 2013; Tian and Manley, 2013), decay (McManus et al., 2015), and translation (Raser and O’shea, 2013; Schaefke et al., 2018).
Our functional enrichment analysis suggests that a lag between the synthesis of mRNA and proteins might occur. We observed that processes related to DNA and RNA formation were over-represented at noon, whereas only later during the evening, processes related to protein formation and PTR (e.g., “mRNA metabolism,” “splicing,” “ribosome,” and “translation” terms, Figure 2) were over-represented. In addition, the protein processes over-represented during the evening correspond with the physiological analysis of the protein biomass, which increased significantly at nighttime.
Furthermore, increases in the protein biomass and the over-representation of “cell development and differentiation,” “developmental processes,” “cell movement,” and “signaling pathway” terms during the night might be related to diverse cellular processes such as cell proliferation, cell migration and tissue vegetative growth to bud new polyps (Duerden, 1905; Kramarsky-winter and Loya, 1996; Bertucci et al., 2015). In addition, night over-representation of structural terms, such as “microtubule and cytoskeleton,” “actin,” “supermolecule” and the “regulation of substrate adhesion” might indicate cytoskeleton rearrangement, which serves as a framework for developing migrating cells (Pollard and Borisy, 1980; Ridley et al., 2003).
Cellular Processes
Light stimulates the photosynthesis process, which results in carbon fixation (Schmitz and Kremer, 1977), and pH and oxygen concentration elevation (Kuhl et al., 1995; Al-Horani et al., 2003a,b). The production of oxygen as a byproduct of the photosynthesis reaction results in hyperoxia (Linsmayer et al., 2020). Oxygen is an unstable molecule, and as a result of oxygen reduction, it produces superoxide (), which is the precursor of most other reactive oxygen species (ROS) (Turrens, 2003; Lesser, 2006; Nelson and Altieri, 2019). A recent study showed high concentrations of superoxide at the coral surfaces both during mid-day and midnight (Zhang et al., 2016), suggesting a constant production of extracellular superoxide during the whole diel cycle (Zhang et al., 2016). In contrast, our GO enrichment results indicate that “ROS” related terms are over-represented during the daytime, with a similar trend to the elevation of light intensities (Figure 2G and Supplementary Figure 2).
In addition, the elevation of the “ROS” related terms requires the elevation of antioxidants activity, which inhibit the ROS accumulation in the cells (Nelson and Altieri, 2019). In contrast, our enrichment analysis indicates that “antioxidant activity” related terms were over-represented only during the night (Figure 2H). This observation could be related to the lag between the transcription and translation due to posttranscriptional regulation processes (Schaefke et al., 2018). Alternatively, as we observed over-representation of terms related to “redox” during all hours (Figures 2A,E), ROS accumulation might be controlled by the entire redox-sensing and signaling networks as suggested previously in plants (Noctor et al., 2018; Farooq et al., 2019). However, further physiology and molecular analyses are required to elucidate the mechanism controlling ROS-related damage. Furthermore, the increase in UV radiation results in DNA damage (Sinha and Häder, 2002; Petruseva et al., 2014), which may be the trigger for the over-representation of “DNA repair” terms during the night as observed here and previously suggested in the Hydra (Barve et al., 2021).
The over-representation of “cilium” related terms during nighttime might be related to water exchange on the coral surface or to heterotrophic feeding behavior. In corals, cilium activity was found to be associated with heterotrophic feeding, which occurs mostly during nighttime (Yonge, 1930; Coles, 1961; Porter, 1974). However, as the tentacles of Stylophora pistillata are continuously expanded, and feeding behavior can also occur during daytime (Levy et al., 2003), cilium activity during the night might be an attempt to reduce the hypoxic conditions. We observed constant respiration rates throughout the diel cycle; however, during the night, photosynthesis decreases, resulting in hypoxic conditions (Linsmayer et al., 2020). A recent study showed that motile cilia cover the entire outer surface of the coral and can stir an approximately 2 mm thick layer of water from the coral surface (Shapiro et al., 2014). In addition, the authors showed that in ambient conditions, these vortices control the exchange of nutrients and oxygen between the coral and its environment (Shapiro et al., 2014). Nevertheless, as cilia have diverse functions and locations in the coral tissue (Levy et al., 2021; Tambutté et al., 2021), further analysis is required to distinguish between the different roles and functions of the various cilium-related genes.
Metabolism
The observation of over-representation of “lipid metabolism” terms during the day was previously reported in the coral Acropora millepora as well (Bertucci et al., 2015). As cnidarians cannot synthesize sterols (Tarrant et al., 2009), they must acquire it from external sources such as predation or photosynthesis products (Von Holt and Von Holt, 1968; Crossland et al., 1980). Crossland et al. (1980) suggested that lipids are synthesized at the endosymbionts’ chloroplasts and then transferred to host tissues. Moreover, other studies identified lipid bodies located in the host cells containing symbionts (Kellogg and Patton, 1983; Patton and Burris, 1983), and it has been suggested that lipid bodies in the host cells have a diel rhythmic pattern, with an increasing density and size of lipid bodies during the daytime hours (Chen et al., 2012).
Glucose is considered to be the major translocated metabolite from the symbionts to the host cells (Gordon and Leggat, 2010). The over-representation of the terms related to “response to sugar” during daytime observed here may therefore reflect the effect of the photosynthetic diel cycle (Schneider et al., 2009). Previously, it has been reported that glucose transporters are up-regulated during the daytime (Bertucci et al., 2015; Ruiz-Jones and Palumbi, 2015). In our data, we did not observe over-representation in glucose transporters terms; however, “carbohydrates metabolism” and “glycolysis/gluconeogenesis” terms are night enriched. A similar pattern was observed in plants where energy synthesis occurs during the day and is regulated during nighttime (Kojima et al., 2007; Smeekens et al., 2010; Lastdrager et al., 2014). Therefore, we suggest that sugar transport occurs during the day, and the coral host cells’ cellular processes of energy consumption and metabolism occur predominately during nighttime.
Biomineralization
The majority of CaCO3 which forms in the ocean is precipitated by photosynthetic organisms or their hosts (Ries, 2010). Coccolithophores perform direct photosynthesis (Balch et al., 1992), while foraminifera and scleractinian corals utilize energy indirectly via the establishment of symbiosis with intracellular algae (Gattuso et al., 1999). Although LEC has been observed in corals (Kawaguti, 1948; Moya et al., 2006; Schneider et al., 2009), the effects of light on the calcification mechanism remain poorly understood. In accordance with former studies, we found that calcification rates are highly correlated with light intensity. Although we did not measure photosynthesis in this experiment, it is well documented that photosynthesis and calcification rates follow the light intensity pattern, with a slight lag from the peak in photosynthesis to the peak in calcification (Barnes and Chalker, 1990; Mass et al., 2007; Schneider et al., 2009). Previous studies referred to the enhancement of calcification rate as a photosynthesis-driven process due to the elevation of oxygen and glycerol concentration (Colombo-Pallotta et al., 2010). However, other studies have shown that while photosynthesis is partially activated over a wide range of wavelengths, LEC occurs specifically in a narrow waveband of the blue spectrum, when oxygen production is reduced (Wijgerde et al., 2012; Cohen et al., 2016). Therefore, the authors suggested that blue light receptors are involved in LEC (Cohen et al., 2016), as previous have shown blue light to be the cue for circadian rhythm (Levy et al., 2007; Mason et al., 2012). In this study, we did not perform any light manipulation, and therefore, we cannot rule out any of the suggested mechanisms for LEC.
Using in vivo laser measurements Vago et al. (1997) suggested that during the night, the tissue is elevated from the skeleton to increase the calcifying fluid space for deposition of nano-granule particles at the CoC’s, followed by daytime needle-shaped fibers space filling in the newly formed CoC’s (Gladfelter, 1983; Cohen and McConnaughey, 2003). It has been suggested that the first process is not light-dependent while the latter is driven by light (Barnes and Crossland, 1980). To enable the tissue elevation from the skeleton at night, the tissue must move and rearrange. Indeed, as described above, we observed an increase in the protein biomass and over-representation of developmental and structural related terms during the night. This may indicate that the tissue has elevated from the skeleton and serves as a framework for the newly developed skeleton. However, the spatial and temporal dynamics of cellular development and movement are still unclear, and further study is required to address it.
Recent studies have suggested that the SOM, which includes proteins (Drake et al., 2013; Ramos-Silva et al., 2013; Takeuchi et al., 2016; Peled et al., 2020), polysaccharides (Cuif et al., 2008; Naggi et al., 2018) and other macromolecules, spatially interacts with and creates an organic framework which controls the biomineral deposition, shape, size and three-dimensional structure (Drake et al., 2013; Peled et al., 2020; Mummadisetti et al., 2021). For example, acid-rich proteins, which are negatively charged, have the ability to interact with calcium ions providing points for nucleation (Addadi and Weiner, 1985). Our RNA-seq data reveal that some of these highly acidic genes, including CARP1, are not differentially expressed across the whole diel cycle, which may indicate a constant framework might form the space-filling fibers during the whole diel cycle, as demonstrated previously using nano-SIM (Domart-Coulon et al., 2014). While the up-regulation of CARPs 2, 3, 4, and 5 during the day supports rapid calcification during the daytime. CARPs 3, 4, and 5 are aspartic-rich proteins, which have been localized in the mineral aragonite fibers (Mass et al., 2014) and were suggested to have similar roles in mineral nucleation and calcium binding/concentrating during the biomineralization process (Drake et al., 2013; Mass et al., 2014). In our study, CARP3 was the first of these genes to be up-regulated and expressed during the morning. Previous in vitro studies have demonstrated that CARP3 can promote the precipitation of Mg-calcite (Gavriel et al., 2018). In addition, the initial mineral phase observed in newly settled primary polyps is nascent Mg-calcite (Neder et al., 2019). Therefore, CARP3 might play a role in the daily mineral initiation and CaCO3 polymorph alteration. In addition, CARP 4 and 5, which were up-regulated during the daytime, belong to a highly acidic subfamily of proteins that is well conserved across the Order Scleractinia (Zaquin et al., 2021) and are characterized by two acidic regions (Drake et al., 2013; Mass et al., 2013) suggested to be a template for calcium carbonate nucleation or growth (Drake et al., 2013).
Interestingly, the glutamic-rich proteins CARP2 and 6 were up-regulated during daytime and nighttime, respectively. Glutamic-rich proteins were found to stabilize amorphous calcium carbonate (ACC) in other marine biomineralizing organisms (Aizenberg et al., 1996, 2002).
In addition, glutamic-rich proteins have been correlated with ACC (Akiva et al., 2018), and CARP2 was located in S. pistillata CoCs (Mass et al., 2014), which are enriched with ACC (Devol et al., 2015; Mass et al., 2017; Von Euw et al., 2017). Although CoCs have been suggested to be formed during the night (Vago et al., 1997), we suggest that CoCs may form during the whole diel cycle.
Additionally, the expression of proteins with calcium/metal binding domains is limited to the morning peak cluster. One of these proteins contains a von Willebrand factor (vWF) domain. The vWF domain is part of a large family of adhesion glycoproteins known to stabilize calcium binding structures (Jakobi et al., 2011) and to contribute to the formation of the organic framework of the calcifying space (Mummadisetti et al., 2021). In addition, this protein and CARP2 were up-regulated during larval development of Pocilopora acuta (Mass et al., 2016), where ACC was detected (Akiva et al., 2018), suggesting a role for these proteins in the formation of the initial ACC phase.
Another aspect that might influence the variation in the rate of skeleton formation across the diel cycle is ion supply and concentration mechanisms in the calcifying fluid space. Previous studies have demonstrated that ion supply and concentration involves a combination of active bicarbonate transporters (Zoccola et al., 2015), endocytosis of Ca2+ rich vesicles (Neder et al., 2019; Ganot et al., 2020), and passive paracellular transports (Tambutte et al., 2011). This results in elevation of the pH and aragonite saturation state (Ωarag) in the calcifying fluid with respect to the surrounding seawater (Venn et al., 2011; Comeau et al., 2017; Sevilgen et al., 2019). Our data indicate that during the day, there is an over-representation of vesicle-related terms (Figure 2G). In addition, most bicarbonate transporters, including SLC4γ that have been localized to the calicoblastic cells (Zoccola et al., 2015), are up-regulated in the day. The combination of the above might indicate that during the day, a greater flux of Ca2+ and carbonate ions are transported to the calcifying fluid, promoting the induction of mineral deposition. Furthermore, the enzyme carbonic anhydrase 2 (STPCA2), which catalyzes the hydration of carbon dioxide into bicarbonate and was found in the skeleton (Drake et al., 2013) is also up-regulated in the daytime. Therefore, the Ωarag elevation driven by greater flux of ions may potentially enhance the rate of skeleton deposition during daylight hours.
Conclusion
Reef-building corals exhibit complex rhythmic responses to diurnal, lunar, and annual changes. The diel cycle is highly influenced by light due to the presence of photosynthetic endosymbionts that have a profound physiochemical influence on the intracellular environment. The present data demonstrate the daily dynamics of several important processes of coral biology such as circadian, photosynthesis, metabolism, cellular arrangement, and calcification.
Based on our findings, we propose the following diel mechanism which includes calcification, cellular arrangement, metabolism, and energy budget (Figure 5). First, the production of glucose and lipid bodies during the day might be related to photosynthesis together with predation throughout the day. These products are later utilized by the host for different cellular processes, such as cellular development, transcription and translation, respiration, ciliary movement, and calcification. In addition, ROS related genes react with the free oxygen species to reduce oxidative damages. During the nighttime, when the host and the symbionts are respiring, and there is no oxygen producing photosynthesis, cilia movement might also aid with reducing the hypoxic conditions, and antioxidants deal with ROS accumulated in the tissue during daytime.
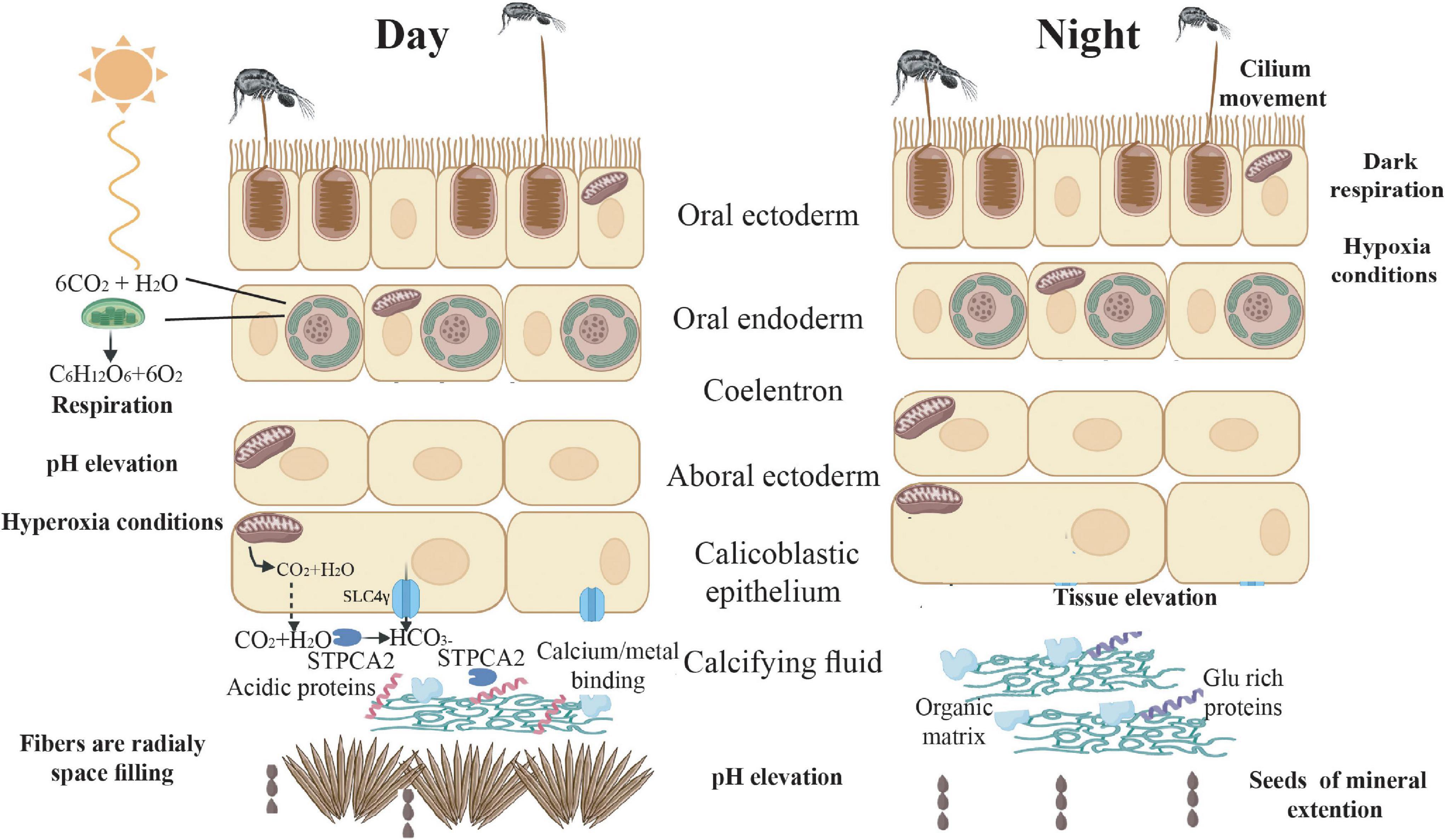
Figure 5. Suggested mechanism of gene expression and physiology across the diel cycle. During daytime, energy production increases, due to photosynthesis and predation, and ROS react with free oxygen reducing ROS damage. In addition, calcification rate increases due to synthesis of acidic proteins (CARP’s) that play a role in mineral nucleation and growth. Furthermore, the high flux of ions to the sites of calcification via transporters, modulate the pH to facilitate an environment favorable for mineral deposition. During the night, cilia movements may alleviate hypoxic conditions. Cellular development and movement might lead to tissue elevation from the skeleton, followed by the new organic framework template for mineral deposition.
In terms of light enhanced calcification, our results support the idea of tissue extension during nighttime, that may elevate tissue from the skeleton, increasing the calcifying space allowing a new organic framework template to be formed. During the daytime, there is a greater flux of ions to the sites of calcification and up-regulation of ion binding domains and some CARPs. The combination of the above leads to an enhanced calcification rate and the formation of needle-shaped aragonite fibers that grow rapidly and fill the space between the extended CoC’s gaps.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA725620.
Author Contributions
MN and TM designed the experiment. MN carried out the experiment, the physiology, the molecular analyses, and wrote the draft manuscript. RS and GA performed the alkalinity measurement. AM performed the RNAseq analysis. RS, AM, GA, and TM contributed to the writing and improving of the manuscript. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Funding
This work has received funding from the European Research Council under the European Union’s Horizon 2020 Research and Innovation Programme (Grant Agreement No. 755876).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Maya Lalzar from the Bioinformatics Core Unit, the University of Haifa for fruitful discussions and suggested improvements. We thank Shani Levy, Jessica Bellworthy, and Jeana Drake for reading the manuscript and suggesting improvements. We thank the Crown Genomics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine at the Weizmann Institute of Science for transcriptome sequencing. Computations presented in this work were performed on the Hive computer cluster at the University of Haifa.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.745171/full#supplementary-material
Footnotes
References
Addadi, L., and Weiner, S. (1985). Interactions between acidic proteins and crystals: stereochemical requirements in biomineralization. Proc. Natl. Acad. Sci. U S A. 82, 4110–4114. doi: 10.1073/pnas.82.12.4110
Aizenberg, J., Addadi, L., Weiner, S., and Lambert, G. (1996). Stabilization of amorphous calcium carbonate by specialized macromolecules in biological and synthetic precipitates. Adv. Mater. 8, 222–226. doi: 10.1002/adma.19960080307
Aizenberg, J., Lambert, G., Weiner, S., and Addadi, L. (2002). Factors involved in the formation of amorphous and crystalline calcium carbonate: a study of an ascidian skeleton. J. Am. Chem. Soc. 124, 32–39. doi: 10.1021/ja016990l
Akiva, A., Neder, M., Kahil, K., Gavriel, R., Pinkas, I., Goobes, G., and Mass, T. (2018). Minerals in the pre-settled coral Stylophora pistillata crystallize via protein and ion changes. Nat. Commun. 9:1880. doi: 10.1038/s41467-018-04285-7
Al-Horani, F. A., Al-Moghrabi, S. M., and de Beer, D. (2003a). Microsensor study of photosynthesis and calcification in the scleractinian coral, Galaxea fascicularis: active internal carbon cycle. J. Exp. Mar. Biol. Ecol. 288, 1–15.
Al-Horani, F. A., Al-Moghrabi, S. M., and de Beer, D. (2003b). The mechanism of calcification and its relation to photosynthesis and respiration in the scleractinian coral Galaxea fascicularis. Mar. Biol. 142, 419–426. doi: 10.1007/s00227-002-0981-8
Al-Horani, F. A., Ferdelman, T., Al-Moghrabi, S. M., and De Beer, D. (2005). Spatial distribution of calcification and photosynthesis in the scleractinian coral Galaxea fascicularis. Coral Reefs 24, 173–180. doi: 10.1007/s00338-004-0461-3
Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C. J., Lajeunesse, T. C., et al. (2014). Assessing symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418–4433. doi: 10.1111/mec.12869
Balch, W. M., Holligan, P. M., and Kilpatrick, K. A. (1992). Calcification, photosynthesis and growth of the bloom-forming coccolithophore, Emiliania huxleyi. Cont. Shelf Res. 12, 1353–1374.
Barnes, D. J. (1970). Coral skeletons: an explanation of their growth and structure. Science 170, 1305–1308. doi: 10.1126/science.170.3964.1305
Barnes, D. J. (1972). The structure and formation of growth-ridges in scleractinian coral skeletons. Proc. R. Soc. Lond. B. Biol. Sci. 182, 331–350. doi: 10.1098/rspb.1972.0083
Barnes, D. J., and Crossland, C. J. (1980). Diurnal and seasonal variations in the growth of a staghorn coral measured by time-lapse photography. Limnol. Oceanogr. 25, 1113–1117. doi: 10.4319/lo.1980.25.6.1113
Barnes, D. J., and Chalker, B. E. (1990). “Calcification and photosynthesis in reef-building corals and algae,” in Ecosystems of the World. (Ed.), Z. Dubinsky (Amsterdam: Elsevier).
Barve, A., Galande, A. A., Ghaskadbi, S. S., and Ghaskadbi, S. (2021). DNA repair repertoire of the enigmatic Hydra. Front. Genet. 12:670695. doi: 10.3389/fgene.2021.670695
Berget, S. M., Moore, C., and Sharp, P. A. (1977). Spliced segments at the 5’ terminus of adenovirus 2 late mRNA. Proc. Natl. Acad. Sci. U S A. 74, 3171–3175. doi: 10.1073/pnas.74.8.3171
Bertucci, A., Forêt, S., Ball, E. E., and Miller, D. J. (2015). Transcriptomic differences between day and night in Acropora millepora provide new insights into metabolite exchange and light-enhanced calcification in corals. Molec. Ecol. 24, 4489–4504. doi: 10.1111/mec.13328
Bertucci, A., Zoccola, D., Tambutté, S., Vullo, D., and Supuran, C. T. (2010). Carbonic anhydrase activators. the first activation study of a coral secretory isoform with amino acids and amines. Bioorg. Med. Chem. 18, 2300–2303. doi: 10.1016/j.bmc.2010.01.059
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bradford, M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chen, W. N. U., Kang, H. J., Weis, V. M., Mayfield, A. B., Jiang, P. L., Fang, L. S., et al. (2012). Diel rhythmicity of lipid-body formation in a coral-Symbiodinium endosymbiosis. Coral Reefs 31, 521–534. doi: 10.1007/s00338-011-0868-6
Chow, L. T., Roberts, J. M., Lewis, J. B., and Broker, T. R. (1977). A map of cytoplasmic RNA transcripts from lytic adenovirus type 2, determined by electron microscopy of RNA:DNA hybrid. Cell 11, 819–836. doi: 10.1016/0092-867490294-X
Cohen, A. L., and McConnaughey, T. A. (2003). Geochemical perspectives on coral mineralization. Rev. Mineral. Geochem. 54, 151–187. doi: 10.2113/0540151
Cohen, I., Dubinsky, Z., and Erez, J. (2016). Light enhanced calcification in hermatypic corals: new insights from light spectral responses. Front. Mar. Sci. 2:122. doi: 10.3389/fmars.2015.00122
Coles, S. L. (1961). Estimates of feeding and respiration for three scleractinian corals. Limnol. Oceanogr. 14, 949–953. doi: 10.4319/lo.1969.14.6.0949
Colombo-Pallotta, M. F., Rodríguez-Román, A., and Iglesias-Prieto, R. (2010). Calcification in bleached and unbleached Montastraea faveolata: evaluating the role of oxygen and glycerol. Coral Reefs 29, 899–907. doi: 10.1007/s00338-010-0638-x
Comeau, S., Carpenter, R. C., Edmunds, P. J., Comeau, S., Tambutte, E., Tambutte, S., et al. (2017). Coral calcifying fluid pH is modulated by seawater carbonate chemistry not solely seawater pH. Proc. R. Soc. B. 284:20161669. doi: 10.6084/m9.figshare.c.3649919
Constantz, B., and Weiner, S. (1988). Acidic macromolecules associated with the mineral phase of scleractinian coral skeletons. J. Exp. Zool. 248, 253–258. doi: 10.1002/jez.1402480302
Crossland, C. J., Barnes, D. J., and Borowitzka, M. A. (1980). Diurnal lipid and mucus production in coral. Mar. Biol. 60, 81–90.
Cuif, J. P., Dauphin, Y., Doucet, J., Salome, M., and Susini, J. (2003). XANES mapping of organic sulfate in three scleractinian coral skeletons. Geochim. Cosmochim. Acta 67, 75–83.
Cuif, J. P., Dauphin, Y., Farre, B., Nehrke, G., Nouet, J., and Salomé, M. (2008). Distribution of sulphated polysaccharides within calcareous biominerals suggests a widely shared two-step crystallization process for the microstructural growth units. Mineral. Maga. 72, 233–237. doi: 10.1180/minmag.2008.072.1.233
Devol, R. T., Sun, C. Y., Marcus, M. A., Coppersmith, S. N., Myneni, S. C. B., and Gilbert, P. U. P. A. (2015). Nanoscale transforming mineral phases in fresh nacre. J. Am. Chem. Soc. 137, 13325–13333. doi: 10.1021/jacs.5b07931
Dickson, A. G. (1981). An exact definition of total alkalinity and a procedure for the estimation of alkalinity and total inorganic carbon from titration data. Deep Sea Res. Part I Oceanogr. Res. Pap. 28, 609–623.
Dobin, A., and Gingeras, T. R. (2016). Optimizing RNA-Seq mapping with STAR. Methods Mol. Biol. 1415, 245–262. doi: 10.1007/978-1-4939-3572-7_13
Domart-Coulon, I., Stolarski, J., Brahmi, C., Gutner-Hoch, E., Janiszewska, K., Shemesh, A., et al. (2014). Simultaneous extension of both basic microstructural components in scleractinian coral skeleton during night and daytime, visualized by in situ 86Sr pulse labeling. J. Struc. Biol. 185, 79–88. doi: 10.1016/j.jsb.2013.10.012
Drake, J., Mass, T., Haramaty, L., Zelzion, E., Bhattacharya, D., and Falkowski, P. (2013). Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc. Natl. Acad. Sci. U S A. 110, 3788–3793. doi: 10.1073/pnas
Duerden, J. E. (1905). Recent results on the morphology and development of coral polyps. Smith Misc. Coll. 47, 93–111
Elkon, R., Ugalde, A. P., and Agami, R. (2013). Alternative cleavage and polyadenylation: exten, regulation and function. Nat. Rev. Genet. 14, 496–506 doi: 10.1038/nrg3482
Falciatore, A., and Bowler, C. (2005). The evolution and function of blue and red light photoreceptors. Curr. Top. Dev. Biol. 68, 317–350.
Falkowski, P. G., Dubinsky, Z., Muscatine, L., and McCloskey, L. (1993). Population control in symbiotic corals - Ammonium ions and organic materials maintain the density of zooxanthellae. BioScience 43, 606–611. doi: 10.2307/1312147
Farooq, M. A., Niazi, A. K., Akhtar, J., Saifullah Farooq, M., Souri, Z., Karimi, N., et al. (2019). Acquiring control: the evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 141, 353–369. doi: 10.1016/j.plaphy.2019.04.039
Ganot, P., Tambutté, E., Caminiti-Segonds, N., Toullec, G., Allemand, D., and Tambutté, S. (2020). Ubiquitous macropinocytosis in anthozoans. eLife 9:e50022. doi: 10.7554/eLife.50022
Gattuso, J. P., Allemand, D., and Frankignoulle, M. (1999). Photosynthesis and calcification at cellular, organismal and community levels in coral reefs: a review on interactions and control by carbonate chemistry. Am. Zool. 39, 160–183. doi: 10.1093/icb/39.1.160
Gavriel, R., Nadav-Tsubery, M., Glick, Y., Yarmolenko, A., Kofman, R., Keinan-Adamsky, K., et al. (2018). The coral protein CARP3 acts from a disordered mineral surface film to divert aragonite crystallization in favor of Mg-Calcite. Adv. Func. Mater. 28:1707321. doi: 10.1002/adfm.201707321
Gladfelter, E. H. (1983). Skeletal development in Acropora cervicornis: II. diel patterns of calcium carbonate accretion. Coral Reefs 2, 91–100. doi: 10.1007/BF02395279
Gordon, B. R., and Leggat, W. (2010). Symbiodinium - invertebrate symbioses and the role of metabolomics. Mar. Drugs 8, 2546–2568. doi: 10.3390/md8102546
Gran, G. (1952). Determination of the equivalence point in potentiometric titrations. Part II. The Analyst 77, 661–671.
Greenbaum, D., Jansen, R., and Gerstein, M. (2002). Analysis of mRNA expression and protein abundance data: an approach for the comparison of the enrichment of features in the cellular population of proteins and transcripts. Bioinformatics 18, 585–596. doi: 10.1093/bioinformatics/18.4.585
Gry, M., Rimini, R., Strömberg, S., Asplund, A., Pontén, F., Uhlén, M., et al. (2009). Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genom. 10:365. doi: 10.1186/1471-2164-10-365
Gygi, S. P., Rochon, Y., Franza, B. R., and Aebersold, R. (1999). Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730. doi: 10.1128/mcb.19.3.1720
Isa, Y., and Okazaki, M. (1987). Some observations on the Ca2+- binding phospholipid from scleractinian coral skeletons. Comp. Biochem. Physiol. B. 87, 507–512.
Jakobi, A. J., Mashaghi, A., Tans, S. J., and Huizinga, E. G. (2011). Calcium modulates force sensing by the von willebrand factor A2 domain. Nat. Commun. 2:1. doi: 10.1038/ncomms1385
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, 277–280. doi: 10.1093/nar/gkh063
Kawaguti, S. (1948). The effect of light on calcium deposition in corals. Bull. Oceanogr. Inst. Taiwan 4, 65–70.
Kellogg, R. B., and Patton, J. S. (1983). Lipid droplets, medium of energy exchange in the symbiotic anemone Condylactis gigantea: a model coral polyp. Mar. Biol. 75, 137–149. doi: 10.1007/BF00405996
Knoll, A. A. H. (2003). Biomineralization and evolutionary history. Rev. Mineral. Geochem. 54, 329–356. doi: 10.2113/0540329
Kojima, H., Suzuki, T., Kato, T., Enomoto, K. I., Sato, S., Kato, T., Tabata, S., et al. (2007). Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 49, 1053–1063. doi: 10.1111/j.1365-313X.2006.03016.x
Kramarsky-winter, A. E., and Loya, Y. (1996). Regeneration versus budding in fungiid corals: a trade-off regeneration versus budding in fungiid corals: a trade-off. Mar. Ecol. Prog. Ser. 134, 179–185. doi: 10.3354/meps134179
Kuhl, M., Cohen, Y., Dalsgaard, T., Jorgensen, B. B., and Revsbech, N. P. (1995). Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied with microsensors for O2, pH and light. Mar. Eco. P. Ser. 117, 159–177. doi: 10.3354/meps117159
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580.e6. doi: 10.1016/j.cub.2018.07.008
Lastdrager, J., Hanson, J., and Smeekens, S. (2014). Sugar signals and the control of plant growth and development. J. Exp. Bot. 65, 799–807. doi: 10.1093/jxb/ert474
Law, C. W., Chen, Y., Shi, W., and Smyth, G. K. (2014). Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29. doi: 10.1186/gb-2014-15-2-r29
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E., and Storey, J. D. (2012). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. doi: 10.1093/bioinformatics/bts034
Lesser, M. P. (2006). Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278.
Levy, O., Appelbaum, L., Leggat, W., Gothlif, Y., Hayward, D. C., and Miller, D. J. (2007). Light responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 318, 467–471. doi: 10.1126/science.1145432
Levy, O., Dubinsky, Z., and Achituv, Y. (2003). Photobehavior of stony corals: responses to light spectra and intensity. J. Exper. Biol. 206, 4041–4049. doi: 10.1242/jeb.00622
Levy, S., Elek, A., Grau-Bové, X., Menéndez-Bravo, S., Iglesias, M., Tanay, A., et al. (2021). A stony coral cell atlas illuminates the molecular and cellular basis of coral symbiosis, calcification, and immunity. Cell 184, 2973–2298. doi: 10.1016/j.cell.2021.04.005
Linsmayer, L. B., Deheyn, D. D., Tomanek, L., and Tresguerres, M. (2020). Dynamic regulation of coral energy metabolism throughout the diel cycle. Sci. Rep. 10:19881. doi: 10.1038/s41598-020-76828-2
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lu, P., Vogel, C., Wang, R., Yao, X., and Marcotte, E. M. (2007). Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124. doi: 10.1038/nbt1270
Marshall, A. T. (1996). Calcification in hermatypic and ahermatypic corals. Science 271, 637–639. doi: 10.1126/science.271.5249.637
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10. doi: 10.14806/ej.17.1.200
Mason, B., Schmale, M., Gibbs, P., Miller, M. W., Wang, Q., Levay, K., et al. (2012). Evidence for multiple phototransduction pathways in a reef-building coral. PLoS One 7:e50371. doi: 10.1371/journal.pone.0050371
Mass, T., Drake, J. L., Haramaty, L., Kim, J. D., Zelzion, E., Bhattacharya, D., et al. (2013). Cloning and characterization of four novel coral acid-rich proteins that precipitate carbonates in vitro. Curr. Biol. 23, 1126–1131. doi: 10.1016/j.cub.2013.05.007
Mass, T., Drake, J. L., Peters, E. C., Jiang, W., and Falkowski, P. G. (2014). Immunolocalization of skeletal matrix proteins in tissue and mineral of the coral Stylophora pistillata. Proc. Natl. Acad. Sci. U S A. 111, 12728–12733. doi: 10.1073/pnas.1408621111
Mass, T., Einbinder, S., Brokovich, E., Shashar, N., Vago, R., Erez, J., et al. (2007). Photoacclimation of Stylophora pistillata to light extremes: metabolism and calcification. Mar. Ecol. Prog. Ser. 334, 93–102. doi: 10.3354/meps334093
Mass, T., Giuffre, A. J., Sun, C. -Y., Stifler, C. A., Frazier, M. J., Neder, M., et al. (2017). Amorphous calcium carbonate particles form coral skeletons. Proc. Natl. Acad. Sci. U S A. 114, 7670–7678. doi: 10.1073/pnas.1707890114
Mass, T., Putnam, H. M., Drake, J. L., Zelzion, E., Gates, R. D., Bhattacharya, D., et al. (2016). Temporal and spatial expression patterns of biomineralization proteins during early development in the stony coral Pocillopora damicornis. Proc. R. Soc. B. 283:1829. doi: 10.1098/rspb.2016.0322
McFadden, C. S., Quattrini, A. M., Brugler, M. R., Cowman, P. F., Dueñas, L. F., Kitahara, M. V., et al. (2021). Phylogenomics, origin, and diversification of anthozoans (Phylum cnidaria). Sys. Biol. 70, 635–647. doi: 10.1093/sysbio/syaa103
McManus, J., Cheng, Z., and Vogel, C. (2015). Next-generation analysis of gene expression regulation-comparing the roles of synthesis and degradation. Mol. BioSyst. 11, 2680–2689. doi: 10.1039/c5mb00310e
Moya, A., Tambutté, S., Bertucci, A., Tambutté, E., Lotto, S., Vullo, D., et al. (2008). Carbonic anhydrase in the scleractinian coral Stylophora pistillata: characterization, localization, and role in biomineralization. J. Biol. Chem. 283, 25475–25484. doi: 10.1074/jbc.M804726200
Moya, A., Tambutté, S., Tambutté, E., Zoccola, D., Caminiti, N., and Allemand, D. (2006). Study of calcification during a daily cycle of the coral Stylophora pistillata: implications for ‘light-enhanced calcification.’ J. Exp. Biol. 2, 3413–3419. doi: 10.1242/jeb.02382
Mummadisetti, M. P., Drake, J. L., and Falkowski, P. G. (2021). The spatial network of skeletal proteins in a stony coral. J. R. Soc. Interface 18:20200859. doi: 10.1098/rsif.2020.0859
Muscatine, L. (1990). The role of symbiotic algae in carbon and energy flux in reef corals. Coral Reefs 25, 75–87.
Naggi, A., Torri, G., Iacomini, M., Colombo Castelli, G., Reggi, M., Fermani, S., et al. (2018). Structure and function of stony coral intraskeletal polysaccharides. ACS Omega 3, 2895–2901. doi: 10.1021/acsomega.7b02053
Neder, M., Laissue, P. P., Akiva, A., Akkaynak, D., Albéric, M., Spaeker, O., et al. (2019). Mineral formation in the primary polyps of pocilloporoid corals. Acta Biomater. 96, 631–645. doi: 10.1016/j.actbio.2019.07.016
Nelson, H. R., and Altieri, A. H. (2019). Oxygen: the universal currency on coral reefs. Coral Reefs 38, 177–198. doi: 10.1007/s00338-019-01765-0
Ning, F., Drinnenberg, I., Kelso, J., Wu, J. R., Pääbo, S., Zeng, R., et al. (2007). Comparison of protein and mRNA expression evolution in humans and chimpanzees. PLoS One 2:e216. doi: 10.1371/journal.pone.000021
Noctor, G., Reichheld, J. P., and Foyer, C. H. (2018). ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. 80, 3–12. doi: 10.1016/j.semcdb.2017.07.013
Patton, J. S., and Burris, J. E. (1983). Lipid synthesis and extrusion by freshly isolated zooxanthellae (Symbiotic algae). Mar. Biol. 75, 131–136. doi: 10.1007/BF00405995
Peled, Y., Drake, J. L., Malik, A., Almuly, R., Lalzar, M., Morgenstern, D., et al. (2020). Optimization of skeletal protein preparation for LC–MS/MS sequencing yields additional coral skeletal proteins in Stylophora pistillata. BMC Mater. 2:8. doi: 10.1186/s42833-020-00014-x
Petruseva, I. O., Evdokimov, A. N., and Lavrik, O. I. (2014). Molecular mechanism of global genome nucleotide excision repair. Acta Nat. 6, 23–34. doi: 10.32607/20758251-2014-6-1-23-34
Pollard, T. D., and Borisy, G. (1980). Cellular motility riven by assembly and disassembly of actin filaments. Cell 14, 363–366. doi: 10.1007/BF02073506
Porter, J. W. (1974). “Zooplankton feeding by the Caribbean reef-building coral Montastrea cavernosa,” in Proceedings of the 2nd International Coral Reef Symposium. (Brisbane: The Great Barrier Reef Committee).
Ramos-Silva, P., Kaandorp, J., Huisman, L., Marie, B., Zanella-Cléon, I., Guichard, N., et al. (2013). The skeletal proteome of the coral Acropora millepora: the evolution of calcification by co-option and domain shuffling. Mol. Biol. Evol. 30, 2099–2112. doi: 10.1093/molbev/mst109
Raser, J. M., and O’shea, E. K. (2013). Noise in gene expression: origins, consequences, and control. Science 309, 2010–2013. doi: 10.1126/science.1105891
Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Mark, H., Borisy, G., et al. (2003). Cell migration: integrating signals from front to back. Science 302, 1704–1709. doi: 10.1126/science.1092053
Ries, J. B. (2010). Review: geological and experimental evidence for secular variation in seawater Mg/Ca (calcite-aragonite seas) and its effects on marine biological calcification. Biogeosciences 7, 2795–2849. doi: 10.5194/bg-7-2795-2010
Ruiz-Jones, L. J., and Palumbi, S. R. (2015). Transcriptome-wide changes in coral gene expression at noon and midnight under field conditions. Biol. Bull. 228, 227–241. doi: 10.1086/BBLv228n3p227
Schaefke, B., Sun, W., Li, Y. S., Fang, L., and Chen, W. (2018). The evolution of posttranscriptional regulation. Wiley Interdisci. Rev. RNA 9:e1485. doi: 10.1002/wrna.1485
Schmitz, K., and Kremer, B. P. (1977). Carbon fixation and analysis of assimilates in a coral-dinoflagellate symbiosis. Mar. Biol. 42, 305–313. doi: 10.1007/BF00402192
Schneider, K., and Erez, J. (2006). The effect of carbonate chemistry on calcification and photosynthesis in the hermatypic coral Acropora eurystoma. Limnol. Oceanogr. 51, 1284–1293. doi: 10.4319/lo.2006.51.3.1284
Schneider, K., Levy, O., Dubinsky, Z., and Erez, J. (2009). In situ diel cycles of photosynthesis and calcification in hermatypic corals. Limnol. Oceanogr. 54, 1995–2002. doi: 10.4319/lo.2009.54.6.1995
Sevilgen, D. S., Venn, A. A., Hu, M. Y., Tambutté, E., De Beer, D., Planas-Bielsa, V., et al. (2019). Full in vivo characterization of carbonate chemistry at the site of calcification in corals. Sci. Adv. 5:7447. doi: 10.1126/sciadv.aau7447
Shankavaram, U. T., Reinhold, W. C., Nishizuka, S., Major, S., Morita, D., Chary, K. K., et al. (2007). Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol. Cancer Ther. 6, 820–832.
Shapiro, O. H., Fernandez, V. I., Garren, M., Guasto, J. S., Debaillon-Vesque, F. P., Kramarsky-Winter, E., et al. (2014). Vortical ciliary flows actively enhance mass transport in reef corals. Proc. Natl. Acad. Sci. U S A. 111, 13391–13396. doi: 10.1073/pnas.1323094111
Shashar, N., Cohen, Y., and Loya, Y. (1993). Extreme diel fluctuations of oxygen in diffusive boundary layers surrounding stony corals. Biol. Bull. 185, 455–461. doi: 10.2307/1542485
Shefy, D., Shashar, N., and Rinkevich, B. (2018). The reproduction of the Red Sea coral Stylophora pistillata from Eilat: 4-decade perspective. Mar. Biol. 165:27. doi: 10.1007/s00227-017-3280-0
Sinha, R. P., and Häder, D. P. (2002). UV-induced DNA damage and repair: a review. Photochem. Photobiol. Sci. 1, 225–236. doi: 10.1039/b201230h
Smeekens, S., Ma, J., Hanson, J., and Rolland, F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 273–278. doi: 10.1016/j.pbi.2009.12.002
Smith, S. V., and Kinsey, D. W. (1976). Calcium carbonate production, coral reef growth, and sea level change. Science 194, 937–939. doi: 10.1126/science.194.4268.937
Spalding, M., Ravilious, C., and Green, E. P. (2001). World Atlas of Coral Reefs. Univ of California Press: Berkeley, CA.
Stanewsky, R., Kaneko, M., Emery, P., Beretta, B., Wager-Smith, K., Kay, S. A., et al. (1998). The cry(b) mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692.
Stanley, G. D. J. (2006). Photosymbiosis and the evolution of modern coral reefs. Science 312, 857–858. doi: 10.1126/science.1123701
Takeuchi, T., Yamada, L., Shinzato, C., and Sawada, H. (2016). Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PLoS One 11:e0156424. doi: 10.1371/journal.pone.0156424
Tambutté, E., Ganot, P., Venn, A. A., and Tambutté, S. (2021). A role for primary cilia in coral calcification? Cell Tissue Res. 383, 1093–1102. doi: 10.1007/s00441-020-03343-1
Tambutte, S., Segonds, N., Zoccola, D., Venn, A., Erez, J., and Allemand, D. (2011). Calcein labelling and electrophysiology: insights on coral tissue permeability and calcification. R. Soc. 279, 19–27. doi: 10.1098/rspb.2011.0733
Tarrant, A. M., Reitzel, A. M., Blomquist, C. H., Haller, F., Tokarz, J., and Adamski, J. (2009). Steroid metabolism in cnidarians: insights from Nematostella vectensis. Mol. Cell 301, 27–36. doi: 10.1016/j.mce.2008.09.037
Tian, B., and Manley, J. L. (2013). Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 38, 312–320. doi: 10.1016/j.tibs.2013.03.005
Turrens, J. F. (2003). Mitochondrial formation of reactive oxygen species. J. Physiol. 552, 335–344. doi: 10.1113/jphysiol.2003.049478
Vago, R., Gill, E., and Collingwood, J. C. (1997). laser measurments of coral growth. Nature 386, 30–31.
Vandermeulen, J. H., and Muscatine, L. (1974). “Influence of symbiotic algae on calcification in reef corals: critique and progress report,” in Symbiosis in the Sea. (Ed.) W. B. Vernberg (Columbia: University of South Carolina).
Venn, A., Tambutte, E., Holcomb, M., Allemand, D., and Tambutte, S. (2011). Live tissue imaging shows reef corals elevate pH under their calcifying tissue relative to seawater. PLoS One 6:e20013. doi: 10.1371/journal.pone.0020013
Von Euw, S., Zhang, Q., Manichev, V., Murali, N., Gross, J., Feldman, L. C., et al. (2017). Biological control of aragonite formation in stony corals. Science 356, 933–938. doi: 10.1126/science.aam6371
Von Holt, C., and Von Holt, M. (1968). Transfer of photosynthetic products from zooxanthellae to coelenterate hosts. Comp. Biochem. Physiol. 24, 73–81.
Wijgerde, T., Henkemans, P., and Osinga, R. (2012). Effects of irradiance and light spectrum on growth of the scleractinian coral Galaxea fascicularis- applicability of LEP and LED lighting to coral aquaculture. Aquaculture 344–349, 188–193. doi: 10.1016/j.aquaculture.2012.03.025
Yonge, C. M. (1930). Studies on the physiology of corals. in feeding mechanisms and food. Sci. Rep. 1, 13–57
Yonge, C. M. (1931). The significance of the relationship between corals and zooxanthellae. Nature 128, 309–311.
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Yu, G., Wang, L. -G., Han, Y., and He, Q. -Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Zaquin, T., Malik, A., Drake, J. L., Putnam, H. M., and Mass, T. (2021). Evolution of protein-mediated biomineralization in scleractinian corals. Front. Genet. 12:618517. doi: 10.3389/fgene.2021.618517
Zhang, T., Diaz, J. M., Brighi, C., Parsons, R. J., McNally, S., Apprill, A., et al. (2016). Dark production of extracellular superoxide by the coral Porites astreoides and representative symbionts. Front. Mar. Sci. 3:232. doi: 10.3389/fmars.2016.00232
Keywords: coral biomineralization, light enhanced calcification, symbiosis, photosynthesis, metabolism, RNAseq
Citation: Neder M, Saar R, Malik A, Antler G and Mass T (2022) New Insights on the Diurnal Mechanism of Calcification in the Stony Coral, Stylophora pistillata. Front. Mar. Sci. 8:745171. doi: 10.3389/fmars.2021.745171
Received: 21 July 2021; Accepted: 29 November 2021;
Published: 05 January 2022.
Edited by:
Eric Jeremy Hochberg, Bermuda Institute of Ocean Sciences, BermudaReviewed by:
Susana Enríquez, National Autonomous University of Mexico, MexicoJoshua Patterson, University of Florida, United States
Copyright © 2022 Neder, Saar, Malik, Antler and Mass. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maayan Neder, bWFheWFuc2xrQGdtYWlsLmNvbQ==; Tali Mass, dG1hc3NAdW5pdi5oYWlmYS5hYy5pbA==
 Maayan Neder
Maayan Neder Raoul Saar2
Raoul Saar2 Assaf Malik
Assaf Malik Gilad Antler
Gilad Antler Tali Mass
Tali Mass