
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 26 May 2020
Sec. Aquatic Microbiology
Volume 7 - 2020 | https://doi.org/10.3389/fmars.2020.00360
This article is part of the Research Topic Ecology and Physiology of Nitrification View all 15 articles
The Eastern Tropical North Pacific (ETNP) is a large, persistent, and intensifying oxygen minimum zone (OMZ) that accounts for almost half of the total area of global OMZs. Within the OMZ core (∼350–700 m depth), dissolved oxygen is typically near or below the analytical detection limit of modern sensors (∼10 nM). Steep oxygen gradients above and below the OMZ core lead to vertical structuring of microbial communities that also vary between particle-associated (PA) and free-living (FL) size fractions. Here, we use 16S amplicon sequencing (iTags) to analyze the diversity and distribution of prokaryotic populations between FL and PA size fractions and among the range of ambient redox conditions. The hydrographic conditions at our study area were distinct from those previously reported in the ETNP and other OMZs, such as the ETSP. Trace oxygen concentrations (∼0.35 μM) were present throughout the OMZ core at our sampling location. Consequently, nitrite accumulations typically reported for OMZ cores were absent as were sequences for anammox bacteria (Brocadiales genus Candidatus Scalindua), which are commonly found across oxic-anoxic boundaries in other systems. However, ammonia-oxidizing bacteria (AOB) and archaea (AOA) distributions and maximal autotrophic carbon assimilation rates (1.4 μM C d–1) coincided with a pronounced ammonium concentration maximum near the top of the OMZ core. In addition, members of the genus Nitrospina, a dominant nitrite-oxidizing bacterial (NOB) clade were present suggesting that both ammonia and nitrite oxidation occur at trace oxygen concentrations. Analysis of similarity test (ANOSIM) and Non-metric Dimensional Scaling (nMDS) revealed that bacterial and archaeal phylogenetic representations were significantly different between size fractions. Based on ANOSIM and iTag profiles, composition of PA assemblages was less influenced by the prevailing depth-dependent biogeochemical regime than the FL fraction. Based on the presence of AOA, NOB and trace oxygen in the OMZ core we suggest that nitrification is an active process in the nitrogen cycle of this region of the ETNP OMZ.
In response to ongoing climate change and localized human activities, concentrations of dissolved oxygen have been declining in the open ocean and in coastal marine systems (Breitburg et al., 2018). Estimated oxygen loss from the open ocean during the past 60 years exceeds 2% (Schmidtko et al., 2017), creating concerns about the consequences of oxygen-depleted zone expansion (Paulmier and Ruiz-Pino, 2009). Open ocean OMZs form when high surface primary production fuels biological oxygen demand in subsurface waters that exceeds rates of physical ventilation at depth. Oxygen concentrations in OMZ water columns can have steep gradients (oxycline) above and below the oxygen-depleted core creating hypoxic (typically between 2 and ∼90 μM), suboxic (<2 μM) and anoxic (below detection limit (∼10 nM) layers of varying dimensions (Bertagnolli and Stewart, 2018). Oxygen gradients lead to vertical structuring of metazoan and microbial communities and biogeochemical processes along these extensive oxyclines (Belmar et al., 2011).
Some of the highest rates of nitrogen loss have been recorded in OMZs of the Eastern Tropical North Pacific (ETNP) and South Pacific (ETSP) (Callbeck et al., 2017; Penn et al., 2019), the permanently stratified Cariaco Basin (Montes et al., 2013), the Arabian Sea (Ward et al., 2009), and the OMZ of the Benguela upwelling system (Kuypers et al., 2005). In these systems, the microbial processes of canonical denitrification (heterotrophic reduction of nitrate to nitrogen intermediates and often to dinitrogen gas) and anammox (anaerobic ammonium oxidation) lead to nitrogen losses that can potentially limit primary production (Ward et al., 2007). Moreover, oceanic nitrous oxide emissions (a potent greenhouse gas) from microbial denitrification occurring in OMZs is estimated to account for at least one third of global natural nitrous oxide emissions (Naqvi et al., 2010).
The ETNP OMZ is a large, persistent, and intensifying oxygen minimum zone that accounts for almost half of the total area of global OMZs, is located between 0–25°N latitude and 75 and 180°W longitude (Paulmier and Ruiz-Pino, 2009; Schmidtko et al., 2017). Due to their ecological importance, the biogeochemistry and microbial diversity of different ETNP OMZ regions have been studied intensively (e.g., Beman and Carolan, 2013; Duret et al., 2015; Ganesh et al., 2015; Chronopoulou et al., 2017; Pack et al., 2015; Peng et al., 2015). Previous studies report that dissolved oxygen in this OMZ core (∼250–750 m depth) is typically near or below analytical detection limits (∼10 nM) (Tiano et al., 2014; Garcia-Robledo et al., 2017). However, along the northern margin of the ETNP’s OMZ (study site location ∼22°N) oxygen concentrations at 500 m can reach annual averages between 10 and 20 μM (Paulmier and Ruiz-Pino, 2009; Data from World Ocean Atlas 2013)1. During the field campaign reported herein, we measured oxygen in the OMZ core at sufficient concentrations (0.35 μM) to support aerobic microbial processes, such as ammonium and nitrite oxidation, and partially inhibit important anaerobic microbial processes. Aerobic microbial processes have been detected previously in seemingly suboxic or anoxic layers of the ETNP OMZ (Peng et al., 2015; Garcia-Robledo et al., 2017; Penn et al., 2019). However, factors controlling the distribution and activities of specific functional groups of microorganisms in OMZs are not yet fully understood.
The presence of nitrifiers where oxygen is undetectable in an OMZ can be explained by recent shifts in the vertical position of the oxycline due to episodic vertical oxygen ventilation, which can lead to ephemeral trace oxygen levels within OMZ cores (Muller-Karger et al., 2001; Ulloa et al., 2012; Garcia-Robledo et al., 2017). Such transient conditions can be exploited by aerobic or microaerophilic populations, including nitrifiers. Moreover, sinking particles from the epipelagic (aggregated cells, fecal pellets, and complex organic materials) can contain trace levels of oxygen (Ganesh et al., 2014). Thus, oxygen and aerobic microbes can be transported to otherwise anoxic waters, temporarily allowing aerobic metabolisms to occur in association with particles. Particles are known to be hotspots of microbial biogeochemical cycling (Simon et al., 2002; Ganesh et al., 2014) and can support contrasting anaerobic or aerobic microbial processes that are not observed in the free-living state (Alldredge and Cohen, 1987; Wright et al., 2012; Suter et al., 2018).
In the present study, we investigate prokaryotic communities occupying the northern margin of the ETNP’s OMZ and environmental factors that likely influence their vertical distributions using 16S amplicon sequencing (iTags) coupled with multivariate statistics. We examined two size fractions; the free-living (0.2–2.7 μm) fraction, and the particle-associated fraction (>2.7 μm, capturing particles as well as protistan cells) at multiple depths along the oxycline that correspond to distinct redox conditions.
A 28-day oceanographic expedition starting from Manzanillo, Mexico and returning to San Diego, CA was conducted onboard the R/V Sikuliaq, from 19 January to 15 February 2017 (UNOLS cruise number SKQ201701S). Sampling of OMZ waters was conducted off the Baja California Peninsula. Among the 16 stations occupied during this cruise, Station #12 (21.6°N 117.8°W) was the source of all samples analyzed for this study. This station is located at the northern margin of the ETNP OMZ, approximately 350 nautical miles (∼650 km) offshore from the Baja California Peninsula (Figure 1). At this station, hydrographic data were obtained using a conductivity-temperature-depth (CTD) system (Seabird 3 temperature sensor, Seabird nine digital pressure sensor, and a Seabird four conductivity sensor), a Seapoint fluorimeter (range: 0.01 to 125 μg Chla L–1), and a RI.NKO III series optical oxygen sensor (JFE Advantech Co., Ltd.), all mounted on a rosette. The oxygen sensor was calibrated by the two point calibration method using air-bubbled water for 100% DO saturation and a dilute sodium sulfite solution for 0% DO saturation. Water samples for biological and chemical analyses were collected in 10-liter Niskin bottles on the rosette.
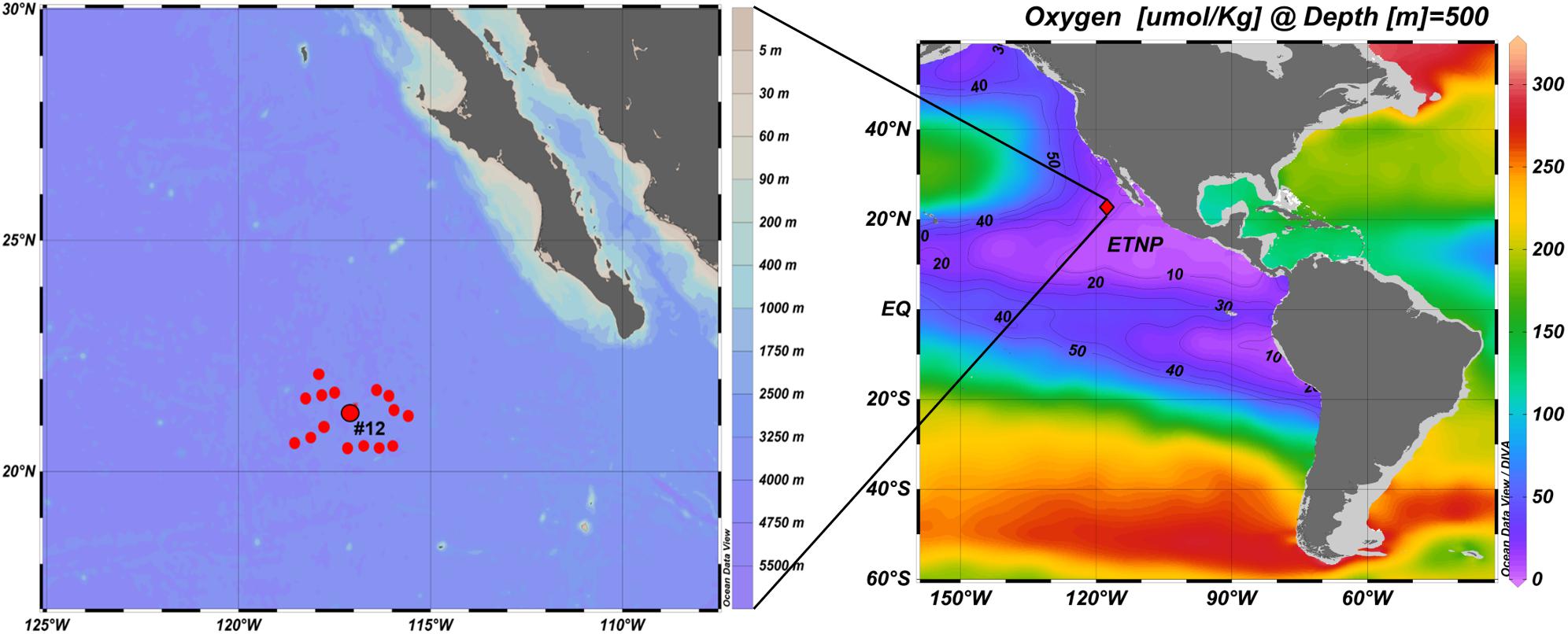
Figure 1. Locations of 16 ETNP stations occupied during the cruise. Station #12 (21.6°N 117.8°W) was the source of the samples analyzed for this study (prepared using Ocean Data View, Schlitzer, R., ODV, https://odv.awi.de, 2018).
From Niskin bottles, 2–5 L of water from 10 depths were directly passed through a 2.7 μm hydrophilic glass fiber filter (EMD Millipore, Burlington, MA, United States) and then through an in-line 0.2 μm Millipore® SterivexTM filter (EMD Millipore, Burlington, MA, United States) to collect material for 16S amplicon sequencing (iTags). The >2.7 μm filters primarily captured the particle and protist-associated cells (“PA” fraction) and the 0.2 μm filter primarily captured free-living cells (0.2–2.7 μm = “FL” fraction). The 10 depths sampled targeted different biogeochemical regimes (Table 1); including normoxic conditions below the photic zone (=115 μM O2, “O”), upper oxycline (114–5 μM O2, “UOC”), the OMZ core (0.35–5 μM O2, “OMZc”), lower oxycline (5–50 μM O2, “LOC”), and deep normoxic (>50 μM O2, “DO”) samples. Two biological replicates were processed from each of the 10 sampled depths. Filters were immediately preserved for nucleic acid extraction by adding 1.8 ml sterile DNA lysis buffer (50 mM Tris-HCL, 40 mM EDTA, 0.73 M sucrose) through the inlet of the Sterivex filter using a micropipette. The Sterivex filter was sealed with parafilm. Glass fiber filters were transferred into sterile 4 ml cryovials and were immersed in the lysis buffer. Sterivex and glass fiber filters were stored at −80°C.
Photoautotrophic and chemoautotrophic production were measured using bulk community 13C-bicarbonate incorporation according to Fisher and Haines (1979) and Sambrotto et al. (2015). A CTD cast was performed to obtain the irradiance profile based on the attenuation of PAR just prior to the collection of water for productivity measurements. For photoautotrophic 13C-assimilation, water from five depths (corresponding to 100, 55, 30, 10, and 1.5% of surface irradiance) was dispensed into 2 L transparent polycarbonate (PC) bottles. Incubation bottles were covered with neutral density plastic screen to simulate in situ irradiance levels and were incubated for 24 h in an on-deck incubator. The incubators were continuously flushed with surface seawater to maintain in situ mixed layer temperature (at ∼23°C) which was monitored throughout the experiments. Water for dark 13C-assimilation was collected the same way at six depths (100, 200, 500, 650, 750, and 1500 m) and incubated with 13C-bicarbonate for 24 h in 2 L PC bottles in the dark at in situ temperature. After incubation, particulate material was collected on GF/F filters (previously combusted) under gentle vacuum. Total carbon content and 13C/12C ratios of the samples were measured by a continuous flow isotope ratio mass spectrometer at the UC Davis stable isotope facility2.
Water for nutrients (nitrate, nitrite, phosphate and ammonium) was collected at 20 depths that included normoxic conditions within and below the photic zone, upper and lower oxycline depths, OMZ core, and deep normoxic waters. These samples were analyzed by continuous flow analysis (CFA) at the Instrumental Analysis Laboratory, School of Marine and Atmospheric Sciences, Stony Brook University, following methods of Gordon et al. (1993).
To enumerate total prokaryoplankton abundances, 200 ml samples from 20 depths throughout the water column were preserved with 2% borate-buffered formaldehyde in high density polyethylene (HDPE) bottles and stored at 4°C. Back in the laboratory, subsamples were filtered, and all filters were stained with 1 μg/ml (final concentration) 4′,6-diamidino-2-phenylindole (DAPI) and abundances of DAPI-positive cells were quantified using an epifluorescence microscope with a UV excitation filter set (Porter and Feig, 1980).
For DNA extractions, we used a modification of the Somerville et al. (1988) protocol appropriate for extracting DNA from glass fiber filters. To improve handling and increase the extraction yield, we transferred the glass fiber filters from the 4 ml cryovials that were collected onboard, into 15 ml sterile Falcon tubes. Glass fiber and Sterivex filters were immersed in 40 μl lysozyme solution (50 mM) for 1 h in a 37oC shaker incubator. Lysozyme solution contained 2 mg lysozyme in 40 μl DNA lysis buffer; 50 mM Tris-HCL, 40 mM EDTA, 0.73 M sucrose). After the lysozyme treatment, 100 μl of Proteinase K solution (20 mg/ml) and 100 μl of 20% SDS were added to both types of filters and the tubes were shaken in the incubator for an additional 2 h at 55°C. After the Proteinase K incubation, crude lysates from each glass fiber and Sterivex filter were transferred to separate 15 ml sterile Falcon tubes. Both filter types were rinsed twice with 1 ml lysis buffer and incubated during each rinse for 15 min at 55oC in a shaker incubator. After rinsing, lysate fluids were combined with the crude lysate from their respective Sterivex and glass fiber filters. DNA extraction of the lysates was performed using a phenol-chloroform protocol. Bacterial and archaeal 16S rRNA gene fragments were PCR amplified from the samples using the primers: 515F-Y (5′-GTGYCAGCMGCCGCGGTAA) and 926R (5′-CCGYCAATTYMTTTRAGTTT[GT2]) (Parada et al., 2016). PCR products were sequenced at Georgia Genomics and Bioinformatics Core Facility (U. of Georgia) using llumina MiSeq PE300 and barcoded primers.
Sequence analysis was performed using Quantitative Insights Into Microbial Ecology (QIIME) v1.9.1 software (Caporaso et al., 2010). Raw sequence reads were processed through Trim Galore3, FLASH (Magoč and Salzberg, 2011) and FASTX_Toolkit4 for trimming and removal of low quality/short reads. During quality control, sequences shorter than 250 nucleotides were removed. Operational Taxonomic Unit (OTU) clusters were constructed at 97% similarity with the script pick_otus.py within QIIME (Caporaso et al., 2010) v1.9.1 software and “uclust”. All sequence data generated in this study has been deposited into Sequence Read Archive (SRA) GenBank under the accession numbers SRR10173043-SRR10172999.
The normalized relative abundance table of OTUs present in each sample was obtained using QIIME v1.9.1. Non-Metric Dimensional Scaling (nMDS) and Analysis of Similarities test (ANOSIM) were conducted based on this relative abundance OTU table using R (version 3.2.1).
Bubble plots were created in R (version 3.2.1) to present diversity in each sample at two taxonomic levels (Phylum, Order) using the Vegan package, version 2.3–2. First, OTUs were segregated by FL and PA size fractions (0.2–2.7 μm and >2.7 μm) and analyzed separately. Relative abundances of individual OTUs from replicate biological samples were averaged and then normalized using the Hellinger transformation (Clarke et al., 2013). Only OTUs with >0.1% average relative abundance in at least one sample were included.
nMDS analysis of all samples was performed in R using the same version and package above. Input for these analyses was a Bray-Curtis dissimilarity matrix of the Hellinger-transformed OTU tables. For nMDS analyses, stress values are reported as a measure of goodness of fit. To assess the influence of biotic and abiotic factors on sample ordination, vectors representing contextual data (oxygen, nutrients, DIC assimilation, total prokaryoplankton counts, etc.) were fit to the ordination plots using goodness of fit statistics (r2 and p-value) as indicators for each vector.
Finally, in order to assess if groups of OTUs were significantly different among water samples from different redox regimes (O, UOC, LOC, OMZc, and DO) and size fractions, ANOSIM tests between groups was performed using the dissimilarity matrix as input (Clarke et al., 2013).
The hydrographic conditions at our study area were distinct from those previously reported in the ETNP (Podlaska et al., 2012) and other OMZs, such as the ETSP (Bristow et al., 2016). Oxygen profiles measured in situ from all 16 stations occupied during the cruise, consistently indicated trace oxygen was present in the OMZ core of this region. The minimum oxygen concentrations within the water column varied between 0.28 and 0.38 μM, revealing the presence of trace oxygen even in OMZ core waters (∼350–700 m). In addition, nitrite did not accumulate within the OMZ core at station #12, indicating the presence of microoxic conditions during the period of our cruise. The presence/absence of nitrite accumulation has been used previously to indicate anoxia in OMZs (Ulloa et al., 2012; Peng et al., 2015; Sun et al., 2019).
Vertical profiles of temperature, salinity, and water density (Sigma-t) for our master station (#12) are presented in Figure 2A. The surface layer was well-mixed to 100 m, below which a steep thermocline appears where temperatures dropped from ∼23 to 12°C between 100 and 250 m, and declined more gradually to ∼2°C by 2,500 m. Salinity values were highest in surface waters (34.25 PSU), decreasing down to ∼150 m (33.60 PSU), and then gradually increasing toward a maximum of 34.60 PSU by ∼400 m. In general, seawater density (Sigma-t) increased gradually from 100 to 2,500 m reflecting decreasing temperature and increasing salinity. At station #12, a narrow peak in oxygen concentration was observed below the well-oxygenated mixed layer at 120 m (220 μM O2; Figure 2B), which coincided with a deep chlorophyll a (Chl a) fluorescence maximum at 106 m. A steep oxycline was observed between 200 and ∼350 m, above the OMZ core which extended from 350 to ∼700 m. Below ∼700 m depth, oxygen gradually increased to 69 μM at 2,500 m.
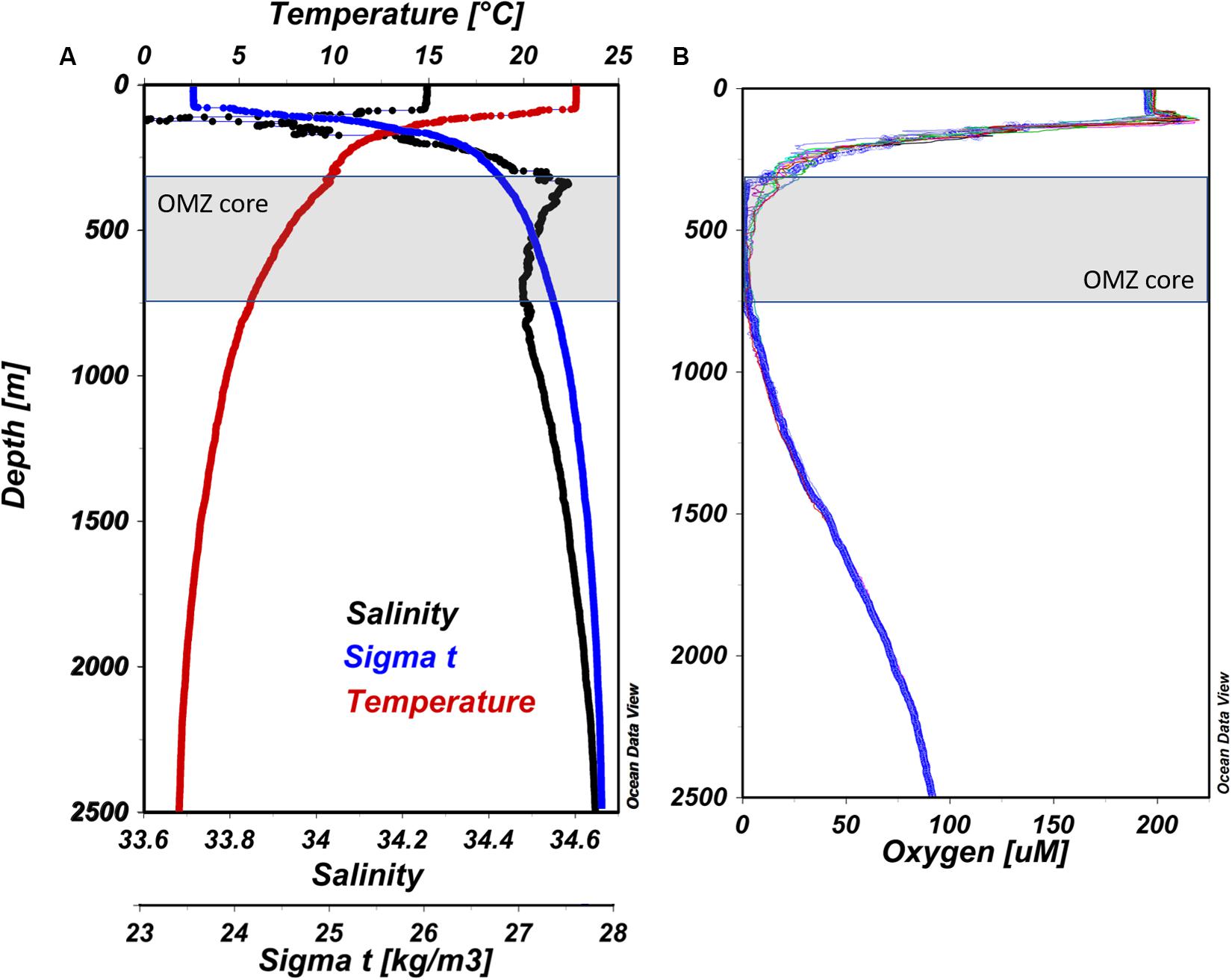
Figure 2. (A) Vertical profiles of CTD sensor data from ETNP station #12. Temperature, salinity, and Sigma-t are presented as red, black, and blue lines, respectively. (B) Vertical profiles of dissolved oxygen concentration from all stations surveyed at the cruise. Blue solid line represent station #12 (prepared using Ocean Data View, Schlitzer, R., ODV, https://odv.awi.de, 2018).
The well-mixed surface layer (100 m) in this region is subject to high rates of primary production that influence nutrient concentrations. Phosphate and nitrate concentrations (Figures 3A,B) were lowest in the surface layer (0 – ∼100 m), and increased steeply from 150 to 250 m, followed by more gradual increases down to ∼900 m. Nitrite distributions were distinct from other nutrients, exhibiting maximum concentration at 150 m (∼1 μM N) and secondary peaks at 25 and 250 m (Figure 3B). Ammonium was only detected at 150 m (∼0.7 μM N) and 500 m (0.4 μM N; Figure 3A).
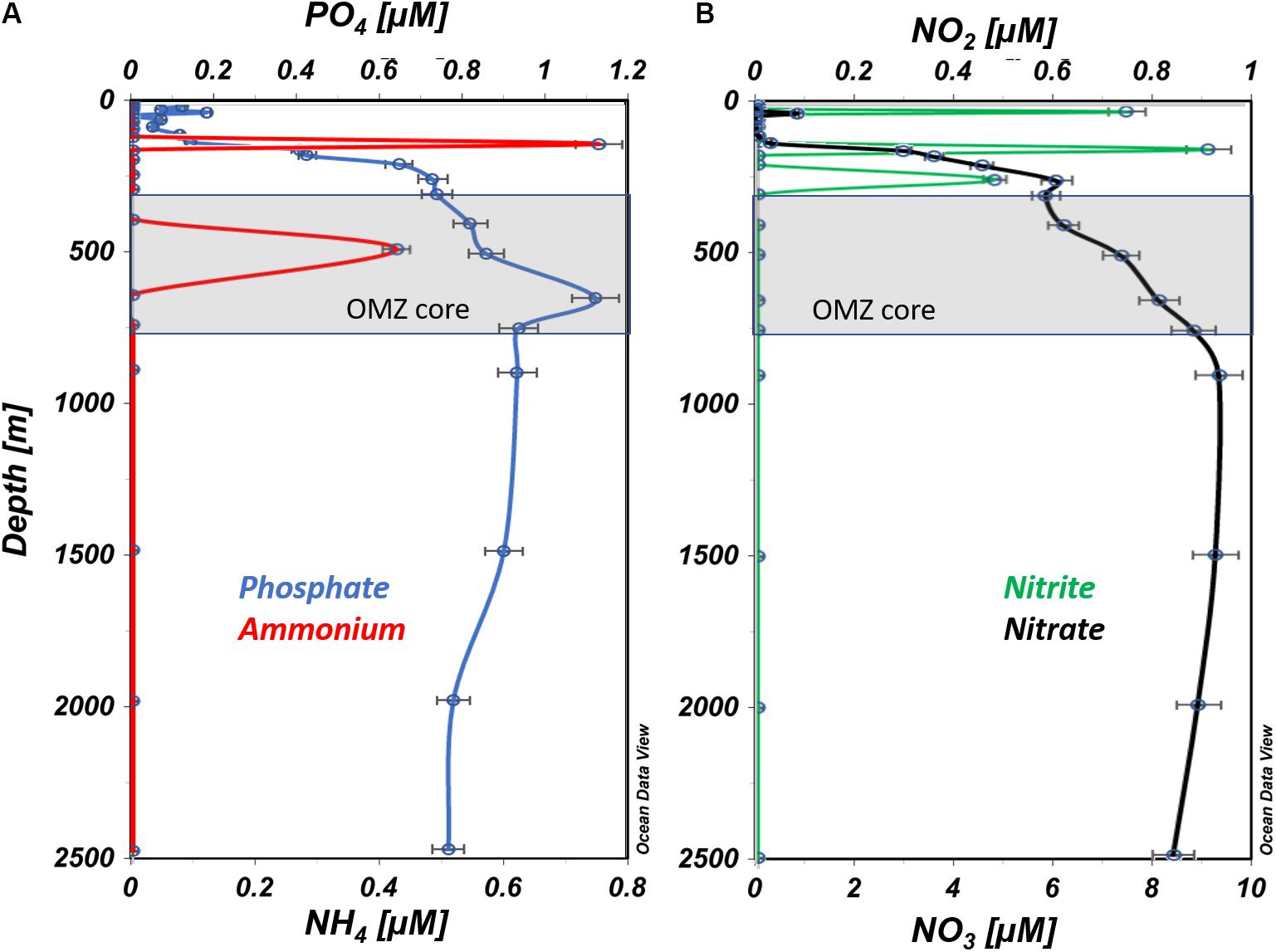
Figure 3. Vertical profiles of nutrient concentrations from ETNP station #12. (A) Ammonium and phosphate are presented in blue and red, respectively. (B) Nitrate and nitrite are presented in black and green, respectively (prepared using Ocean Data View, Schlitzer, R., ODV, https://odv.awi.de, 2018).
Biological variables measured at station #12 suggest intense microbial activity just above the upper oxycline (100–150 m) down into the OMZ core (500 m). Dark and light dissolved inorganic carbon assimilation (DIC assimilation), fluorescence, a proxy of Chl a indicative of phytoplankton biomass, and total prokaryoplankton concentrations are shown in Figure 4. A very pronounced fluorescence peak (∼0.7 relative fluorescence units) was observed at 106 m and corresponded with maximum representation of OTUs belonging to the cyanobacterial Subsection I (Prochlorococcus). The highest prokaryotic abundances were measured in surface waters (6.8 × 108 cells L–1; Figure 4). In general, cell concentrations decreased with depth, but secondary peaks were observed coincident with the 106 m fluorescence peak and just above the OMZ core (300 m).
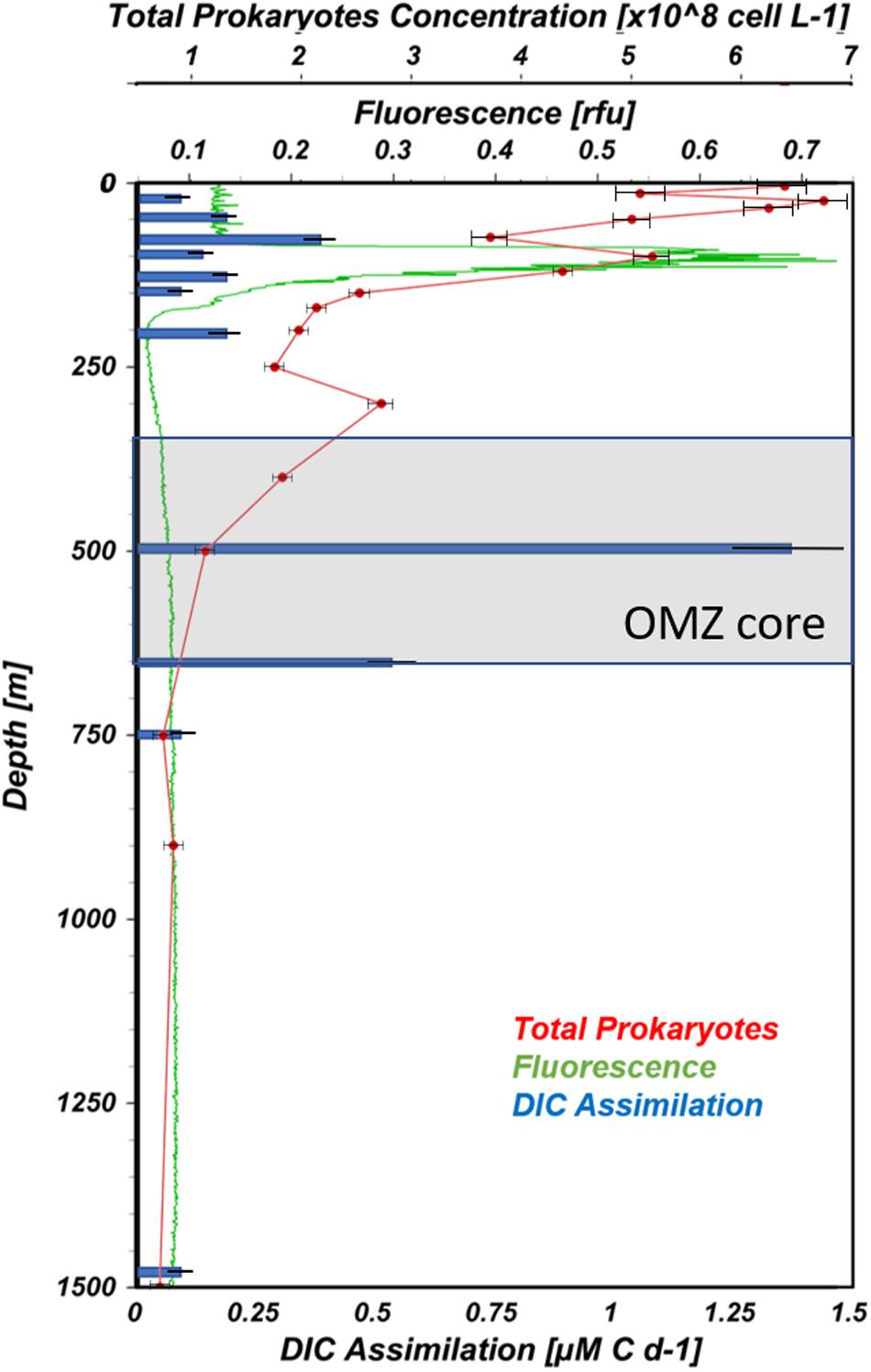
Figure 4. Vertical profiles of biological variables from ETNP station #12. Total prokaryoplankton, fluorescence, and DIC assimilation are presented as red, green and blue lines, respectively (prepared using Ocean Data View, Schlitzer, R., ODV, https://odv.awi.de, 2018).
The maximum DIC assimilation (1.4 μM C d–1) was observed in the OMZ core at 500 m, coinciding with one of the ammonium concentrations peaks (Figures 3B, 4). Similar to the present study, Podlaska et al. (2012) reported maximum DIC assimilation rates varying between 0.02 and 6.4 μM C d–1 among four ETNP stations 7° south of the current study. These rates are considerably faster than those reported for nitrifiers in normoxic regions, such as in the mesopelagic Atlantic Ocean, 0.03–10.4 nM C d–1 (Pachiadaki et al., 2017) or South China Sea and the Western Pacific Ocean, 0.008–1.31 nM C d–1 (Zhang et al., 2020). The disparity between these putatively nitrifier-driven rates may simply be explained by ammonium distributions. NH4+ concentrations in the ETNP studies varied between 400 and 2500 nM-N with depth-dependent peaks (Podlaska et al., 2012; this study). Whereas at those normoxic sites, NH4+ concentrations were only on the order of 30–50 nM-N through the depths sampled (Zhang et al., 2020). At two of Podlaska et al.’s (2012) stations, they showed that 30 μM NH4+ amendments could stimulate DIC assimilation by 1.5 to 33-fold among samples from four to six depths. Observed stimulations suggest nitrifying activity in this system is ammonium limited.
Accumulation of ammonium often results from remineralization of sinking organic material that reaches depth from more productive surface waters (Lam et al., 2011; Beman and Popp, 2012). Alternatively, ammonium enrichments can be generated by excretion from vertically migratory mesozooplankton. In fact, during our cruise Wishner et al. (2018) conducted mesozooplankton surveys using a 1-m2 MOCNESS net and observed an abundance peak of the copepod, Lucicutia hulsemannae, between 500 and 600 m. It is likely that the ammonium enrichment observed at 500m during the current study combined with the presence of trace oxygen fueled chemoautotrophic production by nitrifiers. In the photic zone, a smaller peak of DIC assimilation was observed at 90 m (0.38 μM C d–1) just above the Chl a peak where OTUs affiliated with the cyanobacterial Subsection I (Prochlorococcus) were most highly represented.
Microbial phylogenetic composition, cell abundances, depth distributions, and activities can vary widely among size-fractionated samples in OMZs (Ganesh et al., 2014; Suter et al., 2018). Ganesh et al. (2014) observed distinct phylogenetic compositions of PA and FL assemblages in the ETSP. These differences suggested that many PA prokaryoplankton taxa are specialists favoring biogeochemical conditions in the detritosphere and prefer surface attachment, association with protists, or growth in biofilms. Furthermore, particles that have been colonized in surface waters and sink through the OMZ likely contain prokaryotic assemblages phylogenetically distinct from the surrounding waters from which they are collected (Suter et al., 2018).
In the present study, two size fractions (0.2–2.7 μm and >2.7 μm) of the microbial community from each depth were analyzed. ANOSIM tests results (R value = 0.67, p < 0.001) revealed that bacterial and archaeal taxonomic representation was significantly different between the FL and PA size fractions. Supplementary Figure 1 shows the nMDS for all samples based on the relative abundances of 463 OTUs, clearly showing community differences among the size fractions (FL and PA).
Size partitioning of prokaryote assemblages is evident in the average relative abundances of bacterial and archaeal iTags at the phylum taxonomic level (Figure 5). Of the 17 main groups with average relative abundances above 0.1% in at least one sample, only Proteobacteria and Thaumarchaeota were abundant in both size fractions and under all redox conditions. The top three most abundant bacterial classes were alpha-, gamma- and delta-proteobacteria with alpha-proteobacteria dominating bacterial inventories at all depths. Signatures of cyanobacteria and/or chloroplasts were only abundant in the surface waters in both size fractions. Marinimicrobia (SAR 406) and Euryarchaeota were abundant throughout the water column in the FL fraction, while Planctomycetes and Bacteroidetes were also abundant throughout all depths in the PA fraction. Firmicutes were most highly represented in the PA fraction at 150 and 500 m depth.
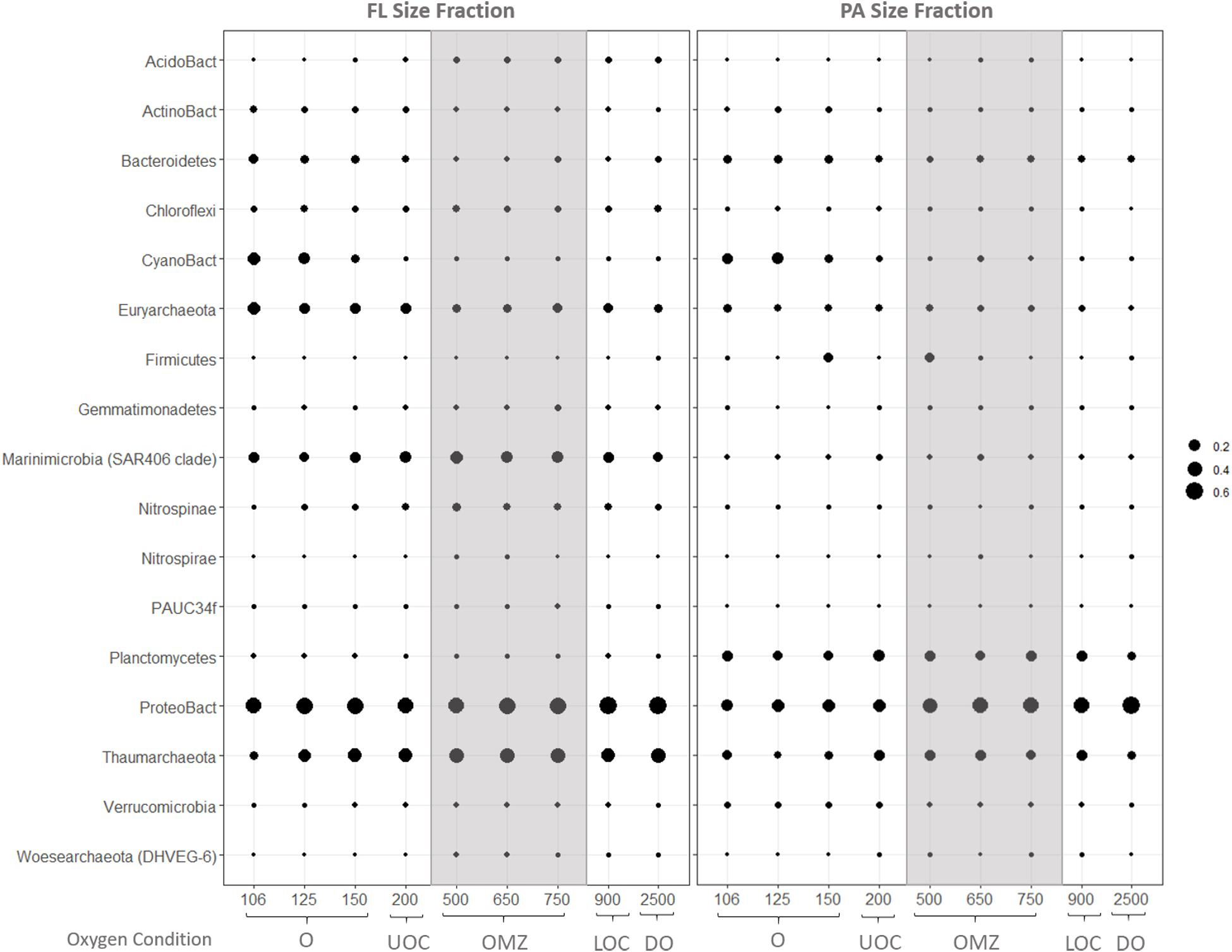
Figure 5. Average relative abundances of iTags in the FL (left) and PA (right) size fractions at coarse phylogenetic resolution. Samples in each panel are organized from left to right by depth, spanning from the oxic (O), upper oxycline (UOC), oxygen minimum zone core (OMZc), lower oxycline (LOC) and deep oxic (DO) depths. Bubbles are the average relative abundances.
Anaerobacillus sp. (Bacillaceae) comprised 75% of the OTUs belonging to the Firmicutes in the PA fraction at 150 m (O) and 500 m (OMZc). The four known species within this genus are strictly anaerobic, heterotrophic, diazotrophic bacteria that possess the nifH gene which is a widely sequenced marker gene for nitrogen fixation (Zavarzina et al., 2009; Wang et al., 2015). Although our study lacks metagenomic and metatranscriptomic data on nifH, we hypothesize nitrogen fixation is possibly carried out by particle-associated Anaerobacillus species at 150 and 500 m, the NH4 maxima. This highlights the potential importance of particle-associated microorganisms in OMZ water columns and can offer an explanation for how anaerobic nitrogen fixation can occur at the same depths as microaerophilic nitrifiers that oxidize ammonium to nitrite.
We postulate that the ammonium peak observed at 500 m supports ammonium oxidation (the first step of nitrification) if AOB, AOA and trace oxygen are present. In fact, our iTag data reveal an abundant, well-described AOA, Marine Group I (Thaumarchaeota) OTU in the FL and PA fractions of OMZ core (Figures 6, 7) coincident with the 500 m ammonium maximum (0.4 μM) and oxygen minimum (0.35 μM). Marine Group I were also abundant in the UOC and DO in both size fractions in our station. BLASTn analysis revealed many of our Marine Group I SSU signatures were affiliated with uncultured archaeon clones L4, L5, and L6 (100% query coverage; 97.1% percentage ID and e-value 0). Zou et al. (2019) determined that these clones are affiliated with AOA, providing indirect evidence that our Marine Group I signatures are also largely AOAs. Other phylogenetic studies from oxygen minimum zones and seasonally stratified anoxic basins reveal that Marine Group I is well represented in the oxycline and is believed to be a major driver of nitrification in the oceans (Pitcher et al., 2011; Swan et al., 2014). Studies of the Chilean OMZ showed that AOA and NOB could be responsible for ammonium and nitrite oxidation rates between 0.16 and 1 μM N d–1 at very low oxygen levels (5–30 nM) if oxygen fluxes are sufficient (Ward et al., 1989; Molina et al., 2010; Bristow et al., 2016). Elsewhere in the ETNP, Peng et al. (2015) reported that AOA dominated ammonia oxidation over AOB within the oxycline. Likewise, Podlaska et al. (2012) suggested that ammonium and nitrite oxidation were important in the upper oxycline, OMZ core, and deep waters at their ETNP stations, based on metabolic activity and nitrogen species profiles. Considering that our iTag library revealed AOA in the OMZ core and that nitrification can occur at trace oxygen concentrations, we posit that ammonium oxidation was an active process in the OMZ core at this site in the ETNP.
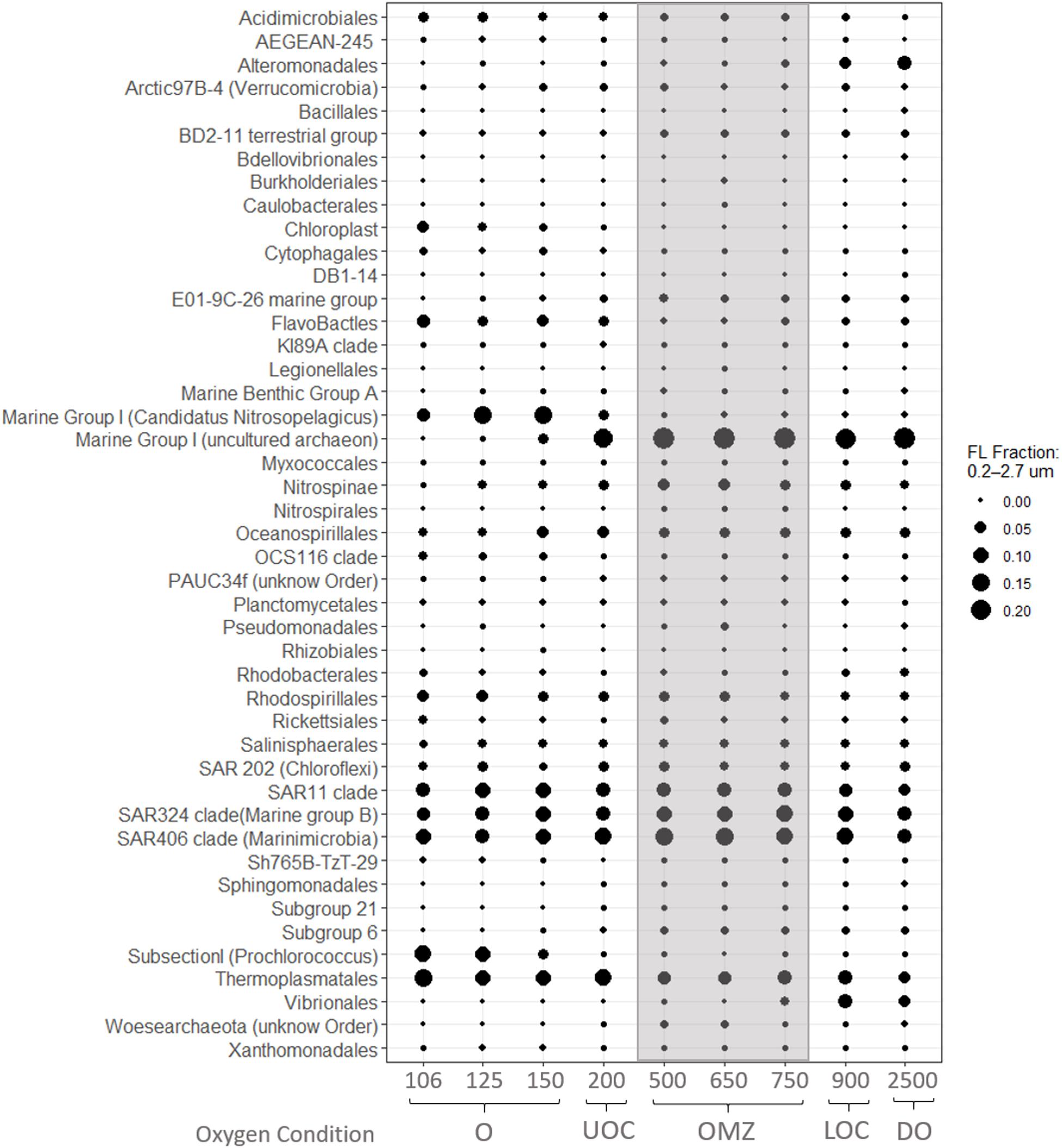
Figure 6. Average relative abundances of iTags at taxonomic level of Order from the 0.2–2.7 μm (FL) size fraction. Samples in each panel are organized from left to right by depth, spanning the oxic (O), Upper oxycline (UOC), oxygen minimum zone core (OMZc), Lower Oxycline (LOC) and deep oxic (DO) depths. Bubbles are the average relative abundances in the FL fraction.
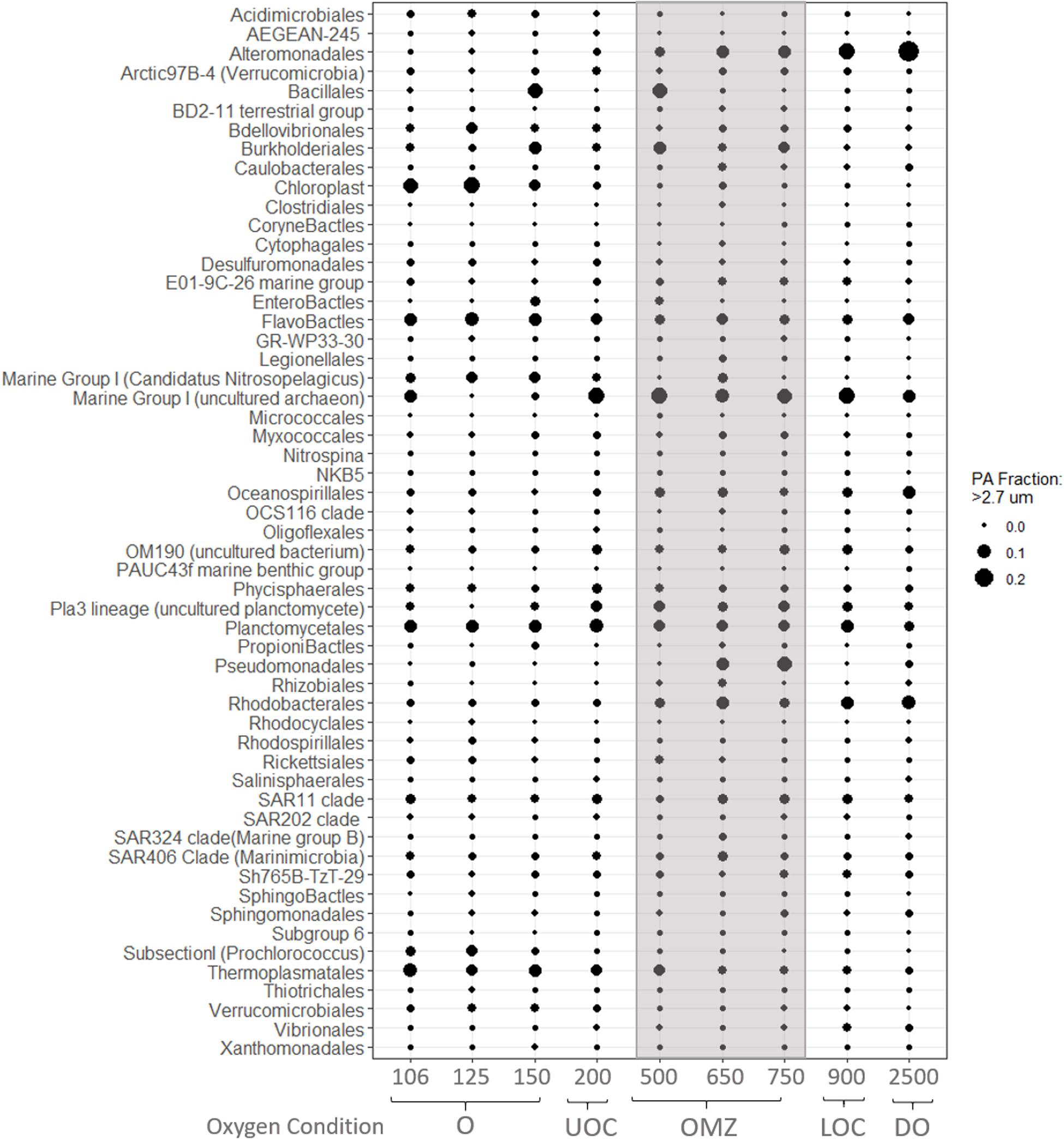
Figure 7. Average relative abundances of iTags at the taxonomic level of Order from the PA fraction. Samples in each panel are organized from left to right by depth, spanning from samples in the oxic (O), upper oxycline (UOC), oxygen minimum zone core depleted (OMZc), lower oxycline (LOC) and deep oxic (DO) depths. Bubbles are the average relative abundances in the PA fraction.
Similarly, based on a sharp decrease in nitrite concentrations below 150 m (Figure 3) and evidence of nitrite-oxidizing taxa in our iTAG libraries, we hypothesize that nitrite oxidation must be an active process where traces of oxygen are present. Four genera are recognized to be responsible for nitrite oxidation in the ocean; Nitrobacter (alpha-proteobacteria), Nitrococcus (gamma-proteobacteria), Nitrospina (Nitrospinae) and Nitrospira (Nitrospirae). However, several studies report the last two as the most common groups in the open ocean (Levipan et al., 2014; Pachiadaki et al., 2017; Sun et al., 2019). We did not recover any Nitrococcus or Nitrobacter sequences in our iTag libraries. Nitrospina sp. was the dominant NOB taxon in the oxic layer (150 m), UOC, OMZ core and deep waters, appearing primarily in the FL fraction (Figure 6), but also in the PA fraction (Figure 7) to a lesser extent. Although Nitrospira OTUs were found in the oxic layer and OMZ core their average relative abundances were less than 0.1% in all samples (FL and PA fractions).
Nitrospinae is a cosmopolitan chemoautotrophic bacterial clade present in major oxygenated mesopelagic water masses (e.g., Pachiadaki et al., 2017). Although, the type species for the Nitrospinae phylum (Nitrospina gracilis) was described to be strictly aerobic (Lücker et al., 2013), it has been found to be present and active in OMZ locations and in an anoxic basin (Rodriguez-Mora et al., 2013; Bristow et al., 2016; Sun et al., 2019). Suter et al. (2018) found a relatively high abundance of Nitrospinae along the oxycline of the Cariaco Basin, where oxygen concentrations declined from ∼120 to 0.4 μM, but they were absent in anoxic and euxinic layers, suggesting an adaptation to low but non-zero oxygen conditions. Studies in coastal habitats and in open ocean waters showed that abundances and distributions of Nitrospinae correlated with those of AOA, implicating metabolic interdependence between members of these clades (Mincer et al., 2007; Santoro et al., 2010). Furthermore, Nitrospina gracilis’ genome encodes for a cytochrome cbb3-type terminal oxidase with high O2 affinity and for nitrite and ammonia transporters that enable Nitrospina to sustain respiration and assimilatory nitrogen uptake in low-oxygen environments (Lücker et al., 2013). If the Nitrospinae detected at our station are similarly adapted, then it is plausible that these aerobes are actively oxidizing nitrite in the OMZ core where oxygen concentrations average 0.35 μM O2. This proposition is also consistent with evidence for microaerophilic respiration (nitrite oxidation) at functionally anoxic depths from the Peruvian OMZs and ETNP (Kalvelage et al., 2015; Sun et al., 2017). Thus, we suggest that coupled ammonia and nitrite oxidations likely play an important role in nitrogen cycling in the OMZ core at this location when traces of oxygen are present.
Distributions of bacterial and archaeal OTUs appeared to be strongly influenced by vertical oxygen distributions. ANOSIM tests were performed to compare taxonomic compositions of both size fractions and of waters from oxic (O), upper oxycline (UOC), oxygen minimum zone core (OMZc), lower oxycline (LOC) and deep oxic (DO) layers. For the FL fraction, the ANOSIM R value (0.80, p < 0.001) suggests a strong dissimilarity among prokaryotic assemblages along the redox gradient. nMDS ordination of OTUs in the FL fraction (Figure 8A) reveals that groups living in well-oxygenated surface waters (O) are significantly different from those in underlying layers. The goodness of fit (r2 and p-values) of each environmental variable vector in the ordination space for the FL prokaryoplankton assemblage shows that OTU distributions are strongly influenced by oxygen (r2 = 0.85; p < 0.001), nitrite (r2 = 0.92; p < 0.001), and ammonium (r2 = 0.91; p < 0.001) concentrations.
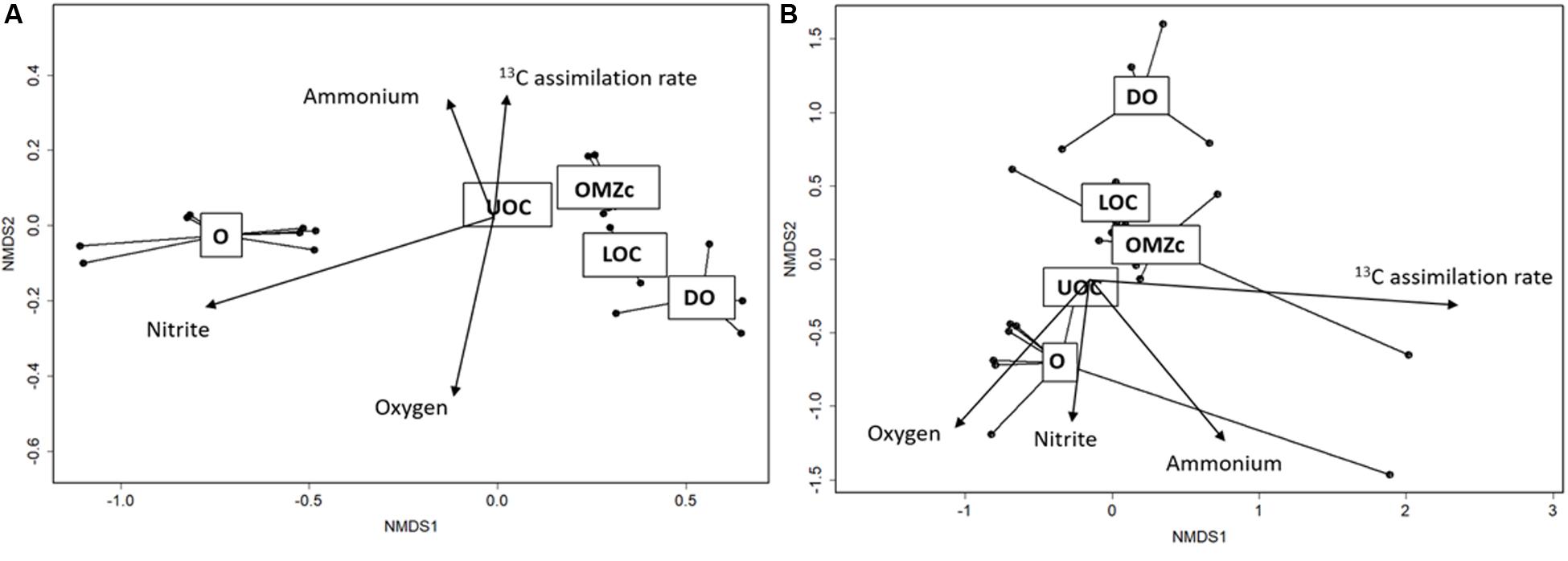
Figure 8. (A) Ordination (nMDS) plot of OTUs in the FL fraction. Stress for this analysis was 4%. Overlain vectors correspond to O2, nitrate, nitrite concentrations and 13C assimilation. r2 and p-values of each vector are: O2: r2 = 0.85; p < 0.00001, nitrate: r2 = 0.93; p < 0.00001, nitrite: r2 = 0.16; p < 0.130 and 13C assimilation: r2: = 0.30; p < 0.017. (B) Ordination (nMDS) plot of OTUs in the PA size fraction. Stress for this analysis was 8%. r2 and p-values of each vector are: O2: r2 = 0.32; p < 0.001, nitrite: r2 = 0.50; p < 0.001, ammonium: r2 = 0.16; p < 0.128 and 13C assimilation rate: r2 = 0.25; p < 0.005. Dots represent each of the 26 samples. Samples in the ordination space segregated by oxygen conditions (O = oxic, UOC = upper oxycline, OMZc = oxygen minimum zone core, LOC = lower oxycline and DO = deep oxic).
As expected for surface waters, recovered sequences in the FL fraction were dominated by OTUs of cyanobacteria mainly affiliated with Prochlorococcus. Other notable OTUs present in the FL fraction from the surface waters included the autotroph Candidatus Nitrosopelagicus (Thaumarchaeota), and OTUs from the heterotrophic clade of Flavobacteriales (Figure 6). Both Candidatus Nitrosopelagicus and Flavobacteriales (Figure 6) were abundant in our oxic water samples and co-occurred with the higher nitrite and ammonium concentrations observed near the base of the euphotic zone (150 m) (Figure 3).
Thaumarchaeota are the major ammonia oxidizers in the oceans that aerobically oxidize ammonia to nitrite. The presence of Candidatus Nitrosopelagicus in the FL fraction in the oxic layer may indicate active ammonia oxidation with concomitant production of nitrite near the base of the euphotic zone. Highly represented Flavobacteriales in the FL fraction of the oxic layer is consistent with observations from other studies in the ETNP OMZ (Fuchsman et al., 2017). The presence of Flavobacteriales in epipelagic waters of the seasonal deep OMZ in the Gulf of Alaska was reported to be a response to phytoplankton blooms (Muck et al., 2019). Other studies have shown that increased abundances of Flavobacteriales in epipelagic waters are significantly related with phytoplankton-derived polysaccharides and decomposition of algal-derived organic matter (Teeling et al., 2012; Taylor et al., 2014).
Members of the Vibrionales and Alteromonadales were highly represented in the deep oxic FL samples, but not in the surface water libraries (Figure 6). Specific ecotypes of Alteromonadales in deep waters are considered to have an important ecological role in denitrification and in the degradation of recalcitrant compounds in sinking particles found at meso- and bathypelagic depths (Martinez et al., 2008). We hypothesize that they play a similar role in our 900 and 2,500 m samples. Within the Vibrionaceae family, Vibrio and Photobacterium were the most highly represented genera. Some Vibrio and Photobacterium species can be free-living, while others are bioluminescent symbionts occupying light organs of deep-sea fishes (Haygood, 1990). The Vibrionales whose signatures we detected at 900 and 2,500 m could be symbionts of deep-sea fishes or free-living bacteria.
In the FL fraction, SAR11, SAR324 (Marine Group B) and SAR406 (Marinimicrobia) clades were abundant throughout the water column (Figure 6). Wright et al. (2012) reported that OTUs representing the SAR11 and SAR324 clades dominated suboxic and anoxic environments in other OMZs and suggested that members of these clades could have overlapping habitat preferences. Within single-cell amplified genomes, Tsementzi et al. (2016) identified SAR11 lineages that carry adaptions for low oxygen. These include genes for respiratory nitrate reductases (nar) that represented ∼40% of the nar transcripts identified in the ETNP OMZ off Mexico. The vertical distribution of these SAR11 nar transcripts was similar to that reported by Ganesh et al. (2015), showing a nar expression peak in the OMZ core. Furthermore, phylogenetic analysis revealed affiliation with facultative aerobic gamma-proteobacteria, Deferribacteres and Geobacter sp. (delta-proteobacteria), all of which are known nitrate-reducers (Tsementzi et al., 2016). Marinimicrobia and SAR324 clades identified in the Northeastern subarctic Pacific OMZ and the seasonally anoxic Saanich Inlet have been reported to commonly encode for key enzymes in denitrification, i.e., nitrate reductases (nar), nitrous oxide reductases (nos), and nitrite reductases (nir) genes (Hawley et al., 2017). While observed high relative abundances of SAR11, SAR324 (Marine Group B), and SAR406 (Marinimicrobia) at all depths is not definitive evidence of their specific activity, we hypothesize that these clades may be contributing to denitrification at this site.
The PA size fraction also showed vertical structuring of prokaryotic assemblages along the redox gradient. The goodness of fit (r2 and p-values) of each vector representing contextual variables in the ordination space with PA prokaryoplankton assemblage showed that OTU distributions had significant, but weaker correlations than those observed for the FL fraction with oxygen (r2 = 0.32, p < 0.001), nitrite (r2 = 0.50, p < 0.001), ammonium (r2 = 0.50, p < 0.001). The ANOSIM R value (0.26) suggests that biogeochemical conditions exert selective pressures on both FL and PA assemblages, but the pressures on PA were weaker. The observed weaker vertical zonation of PA assemblage composition may simply be explained by sinking particles being colonized in surface waters, where most form, and sinking downward with shallow water assemblages. Alternatively, it may be indicative of particles from different depths sharing greater similarities in their chemical microenvironments than experienced in the surrounding waters by the FL fraction. Support for these hypotheses will be evaluated below.
Not unexpectedly, the PA fraction yielded sequences of chloroplasts from larger eukaryotic species, such as Eucampia Antarctica, and several uncultured Prymnesiophytes (C19847, C5574). Also, Enterobacteriales and Firmicutes of the order Bacilliales were highly represented in oxic waters (150 m; Figure 7). The PA fraction in the OMZc and LOC samples was dominated by iTags from Rhizobiales, Burkholderiales and Pseudomonadales, members of which are well-known chemoorganotrophic denitrifiers (Figure 7). The relative abundances of Rhodobacteriales, Oceanospirillales, and Marine Group I (Thaumarchaeota) OTUs increased from the UOC and reached a maximum in the OMZc and deep oxic waters (Figure 7). Alteromonadales was another group that appeared to prefer conditions in the PA fraction of OMZc, LOC and deep oxic samples, with maximum iTag representation at 2,500 m (Figure 8).
Planctomycetes were abundant in the PA fraction under all redox conditions and largely absent in the FL fraction. Of the 491 OTUs recovered in this study, 10% belonged to this group. The two most numerically important classes within the Planctomycetes phylum were the Phycisphaerae (13 OTUs) and Planctomycetacia (23 OTUs). Blastopirellula, Gemmata, Planctomyces, and Rhodopirellula were the most abundant genera. These genera include heterotrophic, non-annamox species that can cope with low oxygen concentrations, but are obligate aerobes (Woebken et al., 2007).
Planctomycetes are widely distributed in marine environments with a prominent role in anaerobic ammonia oxidation. However, anammox Planctomycetes are only found within the genetically and metabolically distinct order, Brocadiales (Woebken et al., 2007; Fuerst and Sagulenko, 2011). The metabolic and genetic differences between anammox and non-anammox Planctomycetes involve the absence of C1 metabolic genes, absence of almost any planctomycete-specific genes (e.g., sulfatases that breakdown of sulfated polysaccharides and are involved in carbon recycling), and absence of paralogous gene families (probably involved in internal compartmentalization) in anammox planctomycetes (Woebken et al., 2007). Notably, no sequences of anammox bacteria belonging to “Candidatus Brocadiales’ order or genus “Candidatus Scalindua” were recovered in our survey. This clade, however, is commonly reported from other ETNP locations (e.g., Rush et al., 2012; Ganesh et al., 2018) and from other oxygen-depleted systems, such as the ETSP (Stevens and Ulloa, 2008) or anoxic basins (Cernadas-Martín et al., 2017). While the absence of proof is not proof of absence, failure to recover “Candidatus Scalindua” sequences in our iTag libraries suggests either that this group was rare at this location in the ETNP during the sampled period or that our vertical sampling resolution was insufficient to capture a narrow anammox layer.
We hypothesize that denitrification dominated N2 production over anammox at the master station during our study period. Our iTag data reveal the presence of well-known denitrifiers in both size fractions and the complete absence of anammox bacterial sequences. Generally, higher rates of anammox (N2 production) are observed in anoxic, non-sulfidic systems, but denitrification dominates where hydrogen sulfide is produced (Thamdrup, 2012). We speculate that hydrogen sulfide production at this ETNP site is negligible because OTUs belonging to sulfur cycling organisms were not recovered at relative abundances exceeding 0.1% of total sequences in any library. Episodic input of new organic matter to the ETSP OMZ was determined to favor production of N2O and N2 via denitrification, with anammox bacteria oxidizing ammonium at a slower rate (Dalsgaard et al., 2012). It is possible that prior to our cruise an oxygen ventilation event occurred, leading to ephemeral trace oxygen levels and introduction of new organic matter within the OMZ core. Some denitrifying microorganism are known to be facultative aerobes, having the ability to colonize biogenic particles in oxic waters. As these particles sink into the OMZ and organic matter is remineralized, localized anoxia can develop within particles when microbial respiration exceeds diffusive O2 influx. Such conditions enable facultative aerobic microbes to occupy different niches within particles, conducting a range of anaerobic processes, including denitrification (Arístegui et al., 2009). Trace oxygen present in the OMZ core at the master station may have been sufficient to inhibit anammox bacteria leaving denitrification as the sole pathway for the production of N2O and N2.
The ability to denitrify is broadly polyphyletic within the domain Bacteria and therefore, denitrification genes (e.g., nar, nor, nir, and nos) are present in a range of organisms with widely different denitrification regulatory phenotypes (Bergaust et al., 2011). Dalsgaard et al. (2014) suggested that denitrification in OMZs results from individual processes catalyzed by different groups of coexisting microorganisms. This suggests that a functional group of diverse denitrifying taxa can develop an ecological “bet hedging” strategy in which an array of different ecophysiotypes are poised to respond to transient changes in redox conditions. Similar to Dalsgaard et al.’s (2014) proposal for the anoxic OMZ off Chile, we hypothesize that “bet hedging” strategies were operative at our master ETNP station. However, this hypothesis requires further investigation.
Organization of prokaryoplankton assemblages in response to biogeochemical gradients will depend upon how dependent, adaptable, or sensitive different groups are to any of a myriad of environmental variables. Our statistical analyses of iTag profiles revealed that PA assemblages were less influenced by the biogeochemical regime of surrounding waters than FL fraction assemblages. Particles are known to offer unique biogeochemical and physical microenvironments (Alldredge and Cohen, 1987; Ganesh et al., 2014). However, the degree to which PA assemblage composition changes among depths in OMZs in response to biogeochemical conditions in ambient waters remains an open question (Ganesh et al., 2014). Our results suggest that surrounding biogeochemical conditions in the ETNP’s OMZ do, in fact, influence observed structure of PA assemblages, i.e., shallow oxic, oxycline, core, and deep oxic assemblages were distinct from one another (Figures 7, 8B). If most sinking particles were formed in surface waters and microbial assemblages were essentially established there, then PA assemblages among depths would likely exhibit a higher similarity than evident in Figure 8B. Alternatively, particles may be generated at different depths by aggregation processes or release by vertically migrating zooplankton (Simon et al., 2002; Wishner et al., 2018). For slowly sinking particles generated in shallow waters, chance collisions with microorganisms and colonization by chemotactic attraction may be sufficiently frequent to shift assemblage composition as particles sink further into the OMZ (Stocker, 2012). In these cases, PA assemblages are likely to be dissimilar among depths.
Provenance of the particles captured in each of our PA samples cannot be determined with available data. Furthermore, we have no information on prokaryotes associated with protists captured in our PA size fraction. Therefore, we cannot determine to what degree differences in PA assemblages observed through the water column are driven by particle sources/quality, protist-prokaryotic associations, colonization in transit, or both ambient and particle biogeochemical regimes. Clearly, further focused studies are necessary to assess the relative contributions of these variables to prokaryote assemblage composition in size-fractionated samples.
Our study site in the ETNP was characterized by a steep vertical oxygen concentration gradient that strongly influenced prokaryotic assemblage composition. Bacterial and archaeal OTUs were strongly segregated between the FL and PA size fractions at all depths sampled, with environmental conditions having a stronger influence on the FL fraction. In contrast to other ETNP OMZ studies, our location on the northern margin had a suboxic core (350–700 m), maintaining low oxygen concentrations (0.35–2.30 μM) and exhibiting very limited nitrite accumulation only in the upper oxycline. Prokaryoplankton assemblages within the core were distinct from those previously reported from other regions in the ETNP and the ETSP OMZs, and from truly anoxic water columns, such as the Cariaco Basin and Black Sea. Three major groups; SUP 05, artic96BD-19 (both gamma-proteobacteria), and Candidatus Scalindua (Brocadiales), commonly abundant in OMZs and anoxic environments, were completely undetected in our iTag libraries (Stevens and Ulloa, 2008; Podlaska et al., 2012; Beman and Carolan, 2013; Cernadas-Martín et al., 2017; Suter et al., 2018). While taxon abundance and activity are not always linked, scarcity of these key players at this site suggests chemoautotrophic oxidation of sulfur and anammox were not important processes at the time of sampling this site.
We detected the presence of ammonia-oxidizing archaea in the OMZ core which suggests that aerobic ammonium oxidation occurs at low oxygen concentrations (<2 μM). We hypothesize that at the time of sampling oxygen supply to the OMZ core was sufficient to support ammonium and nitrite oxidation and to inhibit other important anaerobic processes. Microaerophilic nitrifiers may play an important role in nitrogen cycling in some regions of the ETNP OMZ. Furthermore, the growing ETNP literature illustrates that distributions of hydrographic and geochemical properties within this OMZ vary geographically and temporally. Accounting for this variability is crucial to understanding how the expansion of the OMZ regions will affect future marine environments.
All iTag sequence data generated in this study have been deposited into Sequences Read Archive (SRA) GenBank under the accession numbers SRR10173043–SRR10172999.
All authors participated in experimental design. LM and PM collected samples, performed experiments, and conducted the data analysis. LM took the lead on manuscript writing with contributions from all authors.
This project resulted from a cruise of opportunity and was not expressly funded by any particular project. Projects which contributed to travel and sequencing costs had no funds allocated to publication costs and expired over a year ago. As I am a very active member of the Frontiers in Marine Science Editorial Board. I was granted one free submission and now request a waiver of the Article Processing Charges for this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the cruise’s chief scientists, Drs. Karen Wishner and Brad Seibel, for their invitation to participate in the cruise and for providing the logistics necessary to transport materials and equipment used during the expedition. We also thank Elizabeth Suter and Maria Pachiadaki for their inputs in the manuscript and data analysis. Thanks to the captain and crews of the R/V Sikuliaq (University of Alaska). This communication benefited greatly from insightful suggestions provided by our reviewers (JB and XS). This work was partially supported by the National Science Foundation (NSF) grants (OCE-1336082 to VE and OCE-1335436 and OCE-1259110 to GT).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00360/full#supplementary-material
Alldredge, A., and Cohen, Y. (1987). Can microscale chemical patches persist in the sea? microelectrode study of marine snow, fecal pellets. Science 235, 689–691. doi: 10.1126/science.235.4789.689
Arístegui, J., Gasol, J. M., Duarte, C. M., and Herndl, G. J. (2009). Microbial oceanography of the dark ocean’s pelagic realm. Limnol. Oceanogr. 54, 1501–1529. doi: 10.4319/lo.2009.54.5.1501
Belmar, L., Molina, V., and Ulloa, O. (2011). Abundance and phylogenetic identity of archaeoplankton in the permanent oxygen minimum zone of the eastern tropical South Pacific. FEMS Microbiol. Ecol. 78, 314–326. doi: 10.1111/j.1574-6941.2011.01159.x
Beman, J. M., and Carolan, M. T. (2013). Deoxygenation alters bacterial diversity and community composition in the ocean’s largest oxygen minimum zone. Nat. Commun. 4:2705. doi: 10.1038/ncomms3705
Beman, M., and Popp, B. (2012). Quantification of ammonia oxidation rates and ammonia-oxidizing archaea and bacteria at high resolution in the Gulf of of California and Eastern Tropical North Pacific Ocean. Limnol. Oceanogr. 57, 711–726. doi: 10.4319/lo.2012.57.3.0711
Bergaust, L., Bakken, L. R., and Frostegård, Å. (2011). Denitrification regulatory phenotype, a new term for the characterization of denitrifying bacteria. Biochem. Soc. Trans. 39, 207–212. doi: 10.1042/BST0390207
Bertagnolli, A. D., and Stewart, F. J. (2018). Microbial niches in marine oxygen minimum zones. Nat. Rev. Microbiol. 16, 723–729. doi: 10.1038/s41579-018-0087-z
Breitburg, D., Levin, L. A., Oschlies, A., Grégoire, M., Chavez, F. P., and Conley, D. J. (2018). Declining oxygen in the global ocean and coastal waters. Science 359:eaam7240. doi: 10.1126/science.aam7240
Bristow, L. A., Dalsgaard, T., Tiano, L., Mills, D. B., Bertagnolli, A. D., Wright, J. J., et al. (2016). Ammonium and nitrite oxidation at nanomolar oxygen concentrations in oxygen minimum zone waters. Proc. Natl. Acad. Sci. U.S.A. 113, 10601–10606. doi: 10.1073/pnas.1600359113
Callbeck, C. M., Lavic, G., Stramma, L., Kuypers, M. M., and Bristow, L. A. (2017). Enhanced nitrogen loss by eddy-induced vertical transport in the offshore Peruvian oxygen minimum zone. PLoS One 12:e0170059. doi: 10.1371/journal.pone.0170059
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336.
Cernadas-Martín, S., Suter, E. A., Scranton, M. I., Astor, Y., and Taylor, G. T. (2017). Aerobic and anaerobic ammonium oxidizers in the Cariaco Basin: distributions of major taxa and nitrogen species across the redoxcline. Aquat. Microb. Ecol. 79, 31–48. doi: 10.3354/ame01817
Chronopoulou, P., Shelley, F., Pritchard, W. J., Maanoja, S. T., and Trimmer, M. (2017). Origin and fate of methane in the Eastern Tropical North Pacific oxygen minimum zone. Nat. Publ. Gr. 11, 1386–1399. doi: 10.1038/ismej.2017.6
Clarke, K., Gorley, R. N., Somerrfield, P. J., and Warwick, R. M. (2013). Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, 3nd Edn. Plymouth: PRIMER-E.
Dalsgaard, A., Thamdrup, B., Farias, L., and Revsbech, N. P. (2012). Anammox and denitrification in the oxygen minimum zone of the Eastern South Pacific. Limnol. Oceanogr. 57, 1331–1346. doi: 10.4319/lo.2012.57.5.1331
Dalsgaard, T., Stewart, F. J., Thamdrup, B., De Brabandere, L., Revsbech, N. P., Ulloa, O., et al. (2014). Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen minimum zone off northern Chile. mBio 5:e01966-14. doi: 10.1128/mBio.01966-14
Duret, M., Pachiadaki, M. G., Sarode, N., Christaki, U., and Edgcomb, V. P. (2015). Size-fractionated diversity of eukaryotic microbial communities in the Eastern Tropical North Paciic oxygen minimum zone. FEMS Microbiol. Ecol. 91:fiv037. doi: 10.1093/femsec/fiv037
Fisher, T., and Haines, E. (1979). A comment on the calculation for stable isotopes1 of atom percent enrichment. Limnol. Oceanogr. 24, 593–595. doi: 10.4319/lo.1979.24.3.0593
Fuchsman, C. A., Devol, A. H., Saunders, J. K., McKay, C., and Rocap, G. (2017). Niche partitioning of the N Cycling microbial community of an offshore oxygen deficient zone. Front. Microbiol. 8:2384. doi: 10.3389/fmicb.2017.02384
Fuerst, J., and Sagulenko, E. (2011). Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat. Rev. Microbiol. 9, 403–413. doi: 10.1038/nrmicro2578
Ganesh, S., Bertagnolli, A. D., Bristow, L. A., Padilla, C. C., Blackwood, N., Aldunate, M., et al. (2018). Single cell genomic and transcriptomic evidence for the use of alternative nitrogen substrates by anammox bacteria. ISME J. 12, 2706–2722. doi: 10.1038/s41396-018-0223-9
Ganesh, S., Bristow, L. A., Larsen, M., Sarode, N., Thamdrup, B., and Stewart, F. J. (2015). Size-fraction partitioning of community gene transcription and nitrogen metabolism in a marine oxygen minimum zone. ISME J. 9, 2682–2696. doi: 10.1038/ismej.2015.44
Ganesh, S., Parris, D. J., DeLong, E. F., and Stewart, F. J. (2014). Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 8, 187–211. doi: 10.1038/ismej.2013.144
Garcia-Robledo, E., Padilla, C. C., Aldunate, M., Stewart, F. J., Ulloa, O., Paulmier, A., et al. (2017). Cryptic oxygen cycling in anoxic marine zones. Proc. Natl. Acad. Sci. U.S.A. 114, 8319–8324. doi: 10.1073/pnas.1619844114
Gordon, L., Jennings, J. Jr., Ross, A., and Krest, J. (1993). A Suggested Protocol for Continuous Flow Automated Analysis of Seawater Nutrients (Phosphate, Nitrate, Nitrite and Silicic Acid). WOCE Operation Manual 91-1. Washington, DC: US WHP Office, 1989–1991.
Hawley, A. K., Nobu, M. K., Wright, J. J., Durno, W. E., Morgan-lang, C., Sage, B., et al. (2017). Diverse Marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat. Commun. 8:1507. doi: 10.1038/s41467-017-01376-9
Haygood, M. G. (1990). Relationship of the luminous bacterial symbiont of the Caribbean flashlight fish, Kryptophanaron alfredi (family Anomalopidae) to other luminous bacteria based on bacterial luciferase (luxA) genes. Arch. Microbiol. 154, 496–497.
Kalvelage, T., Lavik, G., Jensen, M. M., Revsbech, N. P., Löscher, C., Schunck, H., et al. (2015). Aerobic microbial respiration in oceanic oxygen minimum zones. PLoS One 10:e0133526. doi: 10.1371/journal.pone.0133526
Kuypers, M. M., Lavik, G., Woebken, D., Schmid, M., Fuchs, B. M., Amann, R., et al. (2005). Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc. Nat. Acad. Sci. U.S.A. 102, 6478–6483. doi: 10.1073/pnas.0502088102
Lam, P., Jensen, M., Kopck, A., Lettmann, K., Plancherel, Y., Lavik, G., et al. (2011). Origin and fate of the secondary nitrite maximum in the Arabian Sea Biogeosciences Origin and fate of the secondary nitrite maximum in the Arabian Sea. Biogeosciences 8, 1565–1577. doi: 10.5194/bg-8-1565-2011
Levipan, H. A., Molina, V., and Fernandez, C. (2014). Nitrospina-like bacteria are the main drivers of nitrite oxidation in the seasonal upwelling area of the Eastern South Pacific (Central Chile 36°S). Environ. Microbiol. Rep. 6, 565–573. doi: 10.1111/1758-2229.12158
Lücker, S., Nowka, B., Rattei, T., Spieck, E., and Daims, H. (2013). The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front. Microbiol. 4:27. doi: 10.3389/fmicb.2013.00027
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Martinez, E. I., Martin, A. B., D’Auria, G., Mira, A., Ferriera, S., Johnson, J., et al. (2008). Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2, 1194–1212. doi: 10.1038/ismej.2008.74
Mincer, T. J., Church, M. J., Taylor, L. T., Preston, C., Karl, D. M., and Delong, E. F. (2007). Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9, 1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x
Molina, V. L., Belmar, L., and Ulloa, O. (2010). High diversity of ammonia-oxidizing archaea in permanent and seasonal oxygen-deficient waters of the eastern South Pacific. Environ. Microbiol. 12, 2450–2465. doi: 10.1111/j.1462-2920.2010.02218.x
Montes, E., Altabet, M. A., Muller-Karger, F. E., Scranton, M. I., Thunell, R., Benitez-Nelson, C., et al. (2013). Biogenic nitrogen gas production at the oxic-anoxic interface in the Cariaco Basin, Venezuela. Biogeosciences 10, 267–279. doi: 10.5194/bg-10-267-2013
Muck, S., De Corte, D., Clifford, E. L., Bayer, B., Herndl, G. J., and Sintes, E. (2019). Niche Differentiation of aerobic and anaerobic ammonia oxidizers in a high latitude deep oxygen minimum zone. Front. Microbiol. 10:2141. doi: 10.3389/fmicb.2019.02141
Muller-Karger, F., Varela, R., Thunell, R., Scranton, M., Bohrer, R., Taylor, G., et al. (2001). Annual cycle of primary production in the Cariaco Basin: response to upwelling and implications for vertical export. J. Geophys. Res. 106, 4527–4542. doi: 10.1029/1999JC000291
Naqvi, S. W. A., Bange, H. W., Farías, L., Monteiro, P. M. S., Scranton, M. I., and Zhang, J. (2010). Marine hypoxia/anoxia as a source of CH4 and N2O. Biogeosciences 7, 2159–2190. doi: 10.5194/bg-7-2159-2010
Pachiadaki, M. G., Sintes, E., Bergauer, K., Brown, J. M., Record, N. R., Swan, et al. (2017). Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation. Science 358, 1046–1051. doi: 10.1126/science.aan8260
Pack, M. A., Heintz, M. B., Reeburgh, W. S., Trumbore, S. E., Valentine, D. L., Xu, X., et al. (2015). Methane oxidationin the eastern tropical North PacificOcean water column. J. Geophys. Res. Biogeosci. 120, 1078–1092. doi: 10.1002/2014JG002900
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Paulmier, A., and Ruiz-Pino, D. (2009). Oxygen minimum zones (OMZs) in the modern ocean. Prog. Oceanogr. 80, 113–128. doi: 10.1016/j.pocean.2008.08.001
Peng, X., Fuchsman, C. A., Jayakumar, A., Oleynik, S., Martens-Habbena, W., Devol, A. H., et al. (2015). Ammonia and nitrite oxidation in the Eastern Tropical North Pacific. Glob. Biogeochem. Cycles 29, 2034–2049. doi: 10.1002/2015gb005278
Penn, J., Weber, T., Chang, B. T., and Deutsch, C. (2019). Microbial ecosystems dynamics drive fluctuating nitrogen lost in marine anoxic zones. Proc. Nat. Acad. Sci. U.S.A. 116, 7220–7225. doi: 10.1073/pnas.1818014116
Pitcher, A., Villanueva, L., Hopmans, E. C., Schouten, S., Reichart, G. J., and Sinninghe Damsté, J. S. (2011). Niche segregation of ammonia-oxidizing archaea and anammox bacteria in the Arabian Sea oxygen minimum zone. ISME J. 5, 1896–1904. doi: 10.1038/ismej.2011.60
Podlaska, A., Wakeham, S. G., Fanning, K. A., and Taylor, G. T. (2012). Deep-Sea Research I Microbial community structure and productivity in the oxygen minimum zone of the eastern tropical North Pacific. Deep Res. Part I 66, 77–89. doi: 10.1016/j.dsr.2012.04.002
Porter, K. G., and Feig, Y. S. (1980). The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25, 943–948. doi: 10.4319/lo.1980.25.5.0943
Rodriguez-Mora, M. J., Madrid, V. M., Taylor, G. T., Scranton, M., and Chistoserdov, A. (2013). Bacterial community composition in a large marine anoxic basin: a Cariaco Basin time-series survey. FEMS Microbiol. Ecol. 84, 625–639. doi: 10.1111/1574-6941.12094
Rush, D., Wakeham, S. G., Hopmans, E. C., Schouten, S., and Sinninghe Damsté, J. S. (2012). Biomarker evidence for anammox in the oxygen minimum zone of the Eastern Tropical North Pacific. Org. Geochem. 53, 80–87. doi: 10.1016/j.orggeochem.2012.02.005
Sambrotto, R. N., Burdloff, D., and McKee, K. (2015). Spatial and year-to-year patterns in new and primary productivity in sea ice melt regions of the eastern Bering Sea. Deep Res. Part II Top. Stud. Oceanogr. 134, 86–99. doi: 10.1016/j.dsr2.2015.07.011
Santoro, A. E., Casciotti, K. L., and Francis, C. A. (2010). Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ. Microbiol. 12, 1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x
Schmidtko, S., Stramma, L., and Visbeck, M. (2017). Decline in global oceanic oxygen content during the past five decades. Nature 542, 335–339. doi: 10.1038/nature21399
Simon, M., Grossart, H., Schweitzer, B., and Ploug, H. (2002). Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28, 175–211. doi: 10.3354/ame028175
Somerville, C, Knight, I. T., Straube, W. L., and Colwell, R. R. (1988). Simple, rapid method for direct isolation of nucleic acids from aquatic environments. App. Environ. Microbiol. 55, 548–554.
Stevens, H., and Ulloa, O. (2008). Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ. Microbiol. 10, 1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x
Stocker, R. (2012). Marine microbes see a sea of gradients. Science 338, 628–633. doi: 10.1126/science.1208929
Sun, X., Ji, Q., Jayakumar, A., and Ward, B. B. (2017). Dependence of nitrite oxidation on nitrite and oxygen in low-oxygen seawater. J. Geophys. Res. Lett. 44, 7883–7891. doi: 10.1002/2017GL074355
Sun, X., Kop, L., Lau, M., Frank, J., Jayakumar, A., Lucker, S., et al. (2019). Uncultured Nitrospina-like species are major nitrite oxidizing bacteria in oxygen minimum zones. ISME J. 13, 2391–2402. doi: 10.1038/s41396-019-0443-7
Suter, E. A., Pachiadaki, M., Taylor, G. T., Astor, Y., and Edgcomb, V. P. (2018). Free-living chemoautotrophic and particle-attached heterotrophic prokaryotes dominate microbial assemblages along a pelagic redox gradient. Environ. Microbiol. 20, 693–712. doi: 10.1111/1462-2920.13997
Swan, B. K., Chaffin, M. D., Martinez-Garcia, M., Morrison, H. G., Field, E. K., and Poulton, N. J. (2014). Genomic and metabolic diversity of Marine Group I Thaumarchaeota in the mesopelagic of two subtropical gyres. PLoS One 9:e95380. doi: 10.1371/journal.pone.0095380
Taylor, J. D., Cottingham, S. D., Billinge, J., and Cunliffe, M. (2014). Seasonal microbial community dynamics correlate with phytoplankton-derived polysaccharides in surface coastal waters. ISME J. 8, 245–248. doi: 10.1038/ismej.2013.178
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Thamdrup, B. (2012). New pathways and processes in the global nitrogen cycle. Annu. Rev. Ecol. Evol. Syst. 43, 407–428. doi: 10.1146/annurev-ecolsys-102710-145048
Tiano, L., Garcia-Robledo, E., Dalsgaard, T., Devol, A. H., Ward, B. B., Ulloa, O., et al. (2014). Oxygen distribution and aerobic respiration in the north and south eastern tropical Pacific oxygen minimum zones. Deep Res. Part I Oceanogr. Res. Pap. 94, 173–183. doi: 10.1016/j.dsr.2014.10.001
Tsementzi, D., Wu, J., Deutsch, S., Nath, S., Rodriguez, L. M., Burns, A. S., et al. (2016). SAR11 bacteria linked to ocean anoxia and nitrogen loss. Nature 536, 179–183. doi: 10.1038/nature19068
Ulloa, O., Canfield, D. E., DeLong, E. F., Letelier, R. M., and Stewart, F. J. (2012). Microbial oceanography of anoxic oxygen minimum zones. Proc. Natl. Acad. Sci. U.S.A. 109, 15996–16003. doi: 10.1073/pnas.1205009109
Wang, J. P., Liu, B., Liu, G. H., Ge, C. B., Chen, Q. Q., Zhu, Y. J., et al. (2015). Genome sequence of Anaerobacillus macyae JMM-4T (DSM 16346), the first genomic information of the newly established genus Anaerobacillus. GenomeA 3, 4–5. doi: 10.1128/genomeA.00922-15
Ward, B., Capone, D. G., and Zehr, J. P. (2007). What’s new in the nitrogen cycle? Oceanogrphy 20, 101–109.
Ward, B. B., Devol, A. H., Rich, J. J., Chang, B. X., Bulow, S. E., Naik, H., et al. (2009). Denitrification as the dominant nitrogen loss process in the Arabian Sea. Nat. Lett. 461, 78–81. doi: 10.1038/nature08276
Ward, B. B., Glover, H. E., and Lipschultz, F. (1989). Chemoautotrophic activity and nitrification in the oxygen minimum zone off Peru. Deep Sea Res. Part A 36, 1031–1051. doi: 10.1016/0198-0149(89)90076-9
Wishner, K. F., Seibel, B. A., Roman, C., Deutsch, C., Outram, D., Shaw, C. T., et al. (2018). Ocean deoxygenation and zooplankton: very small oxygen differences matter. Sci. Adv. 4:eaau5180. doi: 10.1126/sciadv.aau5180
Woebken, D., Teeling, H., Wecker, P., Dumitriu, A., Kostadinov, I., DeLong, E. F., et al. (2007). Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. ISME J. 11, 419–435. doi: 10.1038/ismej.2007.63
Wright, J. J., Konwar, K. M., and Hallam, S. J. (2012). Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–394. doi: 10.1038/nrmicro2778
Zavarzina, D. G., Tourova, T. P., Kolganova, T. V., Boulygina, E. S., and Zhilina, T. N. (2009). Description of Anaerobacillus alkalilacustre gen. nov., sp. nov.-Strictly anaerobic diazotrophic bacillus isolated from soda lake and transfer of Bacillus arseniciselenatis, Bacillus macyae, and Bacillus alkalidiazotrophicus to Anaerobacillus as the new combinations A. arseniciselenatis comb. nov., A. macyae comb. nov., and A. alkalidiazotrophicus comb. nov. Microbiology 78, 723–773.
Zhang, Y., Qin, W., Hou, L., Zakem, E. J., Wan, Z., Zhao, Z., et al. (2020). Nitrifier adaptation to low energy flux controls inventory of reduced nitrogen in the dark ocean. Proc. Natl. Acad. Sci. U.S.A. 117, 4823–4830. doi: 10.1073/pnas.1912367117
Keywords: eastern tropical north pacific, oxygen minimum zones, prokaryoplankton community structure, oxygen gradients, nitrification
Citation: Medina Faull L, Mara P, Taylor GT and Edgcomb VP (2020) Imprint of Trace Dissolved Oxygen on Prokaryoplankton Community Structure in an Oxygen Minimum Zone. Front. Mar. Sci. 7:360. doi: 10.3389/fmars.2020.00360
Received: 13 January 2020; Accepted: 28 April 2020;
Published: 26 May 2020.
Edited by:
Sebastian Lücker, Radboud University Nijmegen, NetherlandsReviewed by:
J. Michael Beman, University of California, Merced, United StatesCopyright © 2020 Medina Faull, Mara, Taylor and Edgcomb. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gordon T. Taylor, Z29yZG9uLnRheWxvckBzdG9ueWJyb29rLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.