- 1Department of Genetics, Harvard Medical School, Blavatnik Institute, Boston, MA, United States
- 2Harvard Medical School, Howard Hughes Medical Institute, Boston, MA, United States
Oenocytes are large secretory cells present in the abdomen of insects known to synthesize very-long-chain fatty acids to produce hydrocarbons and pheromones that mediate courtship behavior in adult flies. In recent years, oenocytes have been implicated in the regulation of energy metabolism. These hepatocyte-like cells accumulate lipid droplets under starvation and can non-autonomously regulate tracheal waterproofing and adipocyte lipid composition. Here, we summarize evidence, mostly from Drosophila, establishing that oenocytes perform liver-like functions. We also compare the functional differences in oenocytes and the fat body, another lipid storage tissue which also performs liver-like functions. Lastly, we examine signaling pathways that regulate oenocyte metabolism derived from other metabolic tissues, as well as oenocyte-derived signals that regulate energy homeostasis.
Introduction
Regulating energy utilization and storage is central to animal physiology and adaptation to environmental challenges. Under conditions of nutrition surplus, glucose is converted to fatty acids, which are then synthesized into triglycerides (TGs) and stored as lipid droplets. Excessive lipid stores can be detrimental and have been associated with various metabolic diseases, such as cardiovascular diseases (CVDs), non-alcoholic fatty liver disease (NAFLD), obesity and insulin resistance, making understanding lipid metabolism of great importance to human health.
The liver is the major detoxifying organ of the body and plays a central role in regulating the metabolism of carbohydrates, proteins and lipids. Moreover, the liver is the major site for glycogen storage and very low-density lipoprotein (VLDL) secretion (1, 2). During starvation, adipocytes undergo lipolysis to produce free fatty acids (FFAs). FFAs are processed by hepatic oxidation to generate ketone bodies in the liver which are then used as fuels for other tissues. If mobilization of FFAs exceeds the rate of lipid oxidation, re-esterification of surplus FFAs to TGs occurs in the liver, leading to an increase in intrahepatic TG content, i.e., steatosis. NAFLD, a common manifestation of the metabolic syndrome, is characterized by steatosis in the absence of starvation. Nonalcoholic hepatic steatosis is present in approximately 25% of the adult population worldwide, and NAFLD is the most common liver disease in Western societies. Thus, understanding how hepatic diseases regulate cellular processes in peripheral organs and how other organs contribute to steatosis is of interest to human metabolic diseases.
Major metabolic and endocrine pathways are conserved in Drosophila, making this model organism well suited to dissect the cellular and molecular mechanisms underlying physiology (3–5). The fly fat body is equivalent to the vertebrate white adipose tissue (WAT), which stores excess fat as TGs. In addition, fly oenocytes, which are similar to hepatocyte cells, are important for mobilizing stored lipids from the fly fat body (6). Like mammals, flies convert excess carbohydrates into TGs through de novo lipogenesis (7, 8). In addition, excess carbohydrates and amino acids can also be processed into UDP-glucose, which fuels glycogen synthesis (9). Regulation of energy storage in flies involves several signaling pathways, including insulin/insulin-like growth factor (IGF) signaling, which is similar to the insulin signaling in mammals (10). However, unlike mammals, there are eight different Drosophila insulin-like peptides (dILPs). Most of these modulate the IGF pathway through a single insulin receptor, InR (10, 11). Under nutrient-deprivation or energy demanding conditions, lipids are released from the fat body through increased lipolysis (12), and are further processed in oenocytes (6, 13). Signaling that regulates catabolism of lipids and carbohydrates include adipokinetic hormone (Akh), which is similar to glucagon in mammals and ecdysone, which antagonizes insulin signaling (14, 15).
In this review, we explore the potential of Drosophila oenocytes as a model for hepatic diseases. We summarize the different roles of oenocytes and the fat body in regulating carbohydrate and lipid metabolism under normal or starved conditions. We also discuss the intricate interplay of oenocytes with other tissues, including the fat body and muscles, in shaping organismal lipid storage.
Oenocytes as the Lipid Metabolizing Center
Oenocytes were originally described as wax-producing cells because histological stains and organic extractions suggested that they contain wax particles or other lipids (16). These unusual cells contain abundant smooth endoplasmic reticulum (ER), which synthesizes lipids, phospholipids, steroids and metabolizes carbohydrates (17). In addition, oenocytes are also highly enriched in peroxisomes (18), the major sites for metabolism of reactive oxidative species (ROS) and β-oxidation of very-long-chain fatty acids. Both smooth ER and peroxisomes are highly enriched in mammalian hepatocytes, highlighting the functional similarities between oenocytes and liver cells in lipid metabolism.
Oenocytes in different insects have been shown to change with the molting cycle (16), prompting investigation of the role of oenocytes in production of the insect hormone ecdysone. The active form of ecdysone, 20-hydroxyecdysone (20E), which is the primary molting hormone, regulates a variety of physiological processes, including metamorphosis, immune response, and reproduction (19–21). In larvae, ecdysone is mainly synthesized in the prothoracic gland from its cholesterol precursor via a set of cytochromes P450 proteins encoded by the “Halloween genes” (22). In adults, ecdysone is mainly but not exclusively synthesized in ovary (23). Ecdysone is converted to 20HE in peripheral tissues, such as the fat body, Malpighian tubules, and midgut. These tissues express shade (shd), which encodes an E-20-monooxygenase that mediates the hydroxylation of ecdysone at carbon 20 (24). Interestingly, two of the ecdysone biosynthesis genes, Phantom and Shadow, are highly expressed in adult oenocytes (25), suggesting that oenocytes participate in ecdysone biosynthesis. Further, manipulation of spidey, which encodes a steroid dehydrogenase, regulates ecdysone metabolite levels (26). Silencing or overexpression of spidey during embryonic development results in pupal lethality, similar to what is observed for mutations in ecdysone signaling pathway genes (26). In addition, oenocyte-specific knockdown of spidey in larvae results in accelerated oxidation of 20HE, while overall 20HE levels remain unchanged. Finally, overexpression of spidey in oenocytes leads to dramatic reduction of 20HE and its catabolic metabolites (26) (Figure 1). In further support of the role of oenocytes in ecdysone biosynthesis, isolated oenocyte-fat body complexes (OEFC) from adult males of the cricket Gryllus bimaculatus have been found to secrete ecdysteroids (32). Moreover, ecdysone has been linked to lipid metabolism in various tissues (33–35), suggesting that oenocytes regulate lipid metabolism by modulating ecdysone levels. It remains to be elucidated whether oenoytes contribute to ecdysone synthesis during adult or larval stages, a question that could be addressed using genetic ablation of the oenocytes. Furthermore, the functional significance of ecdysone in oenocytes physiology and metabolism is of interest.
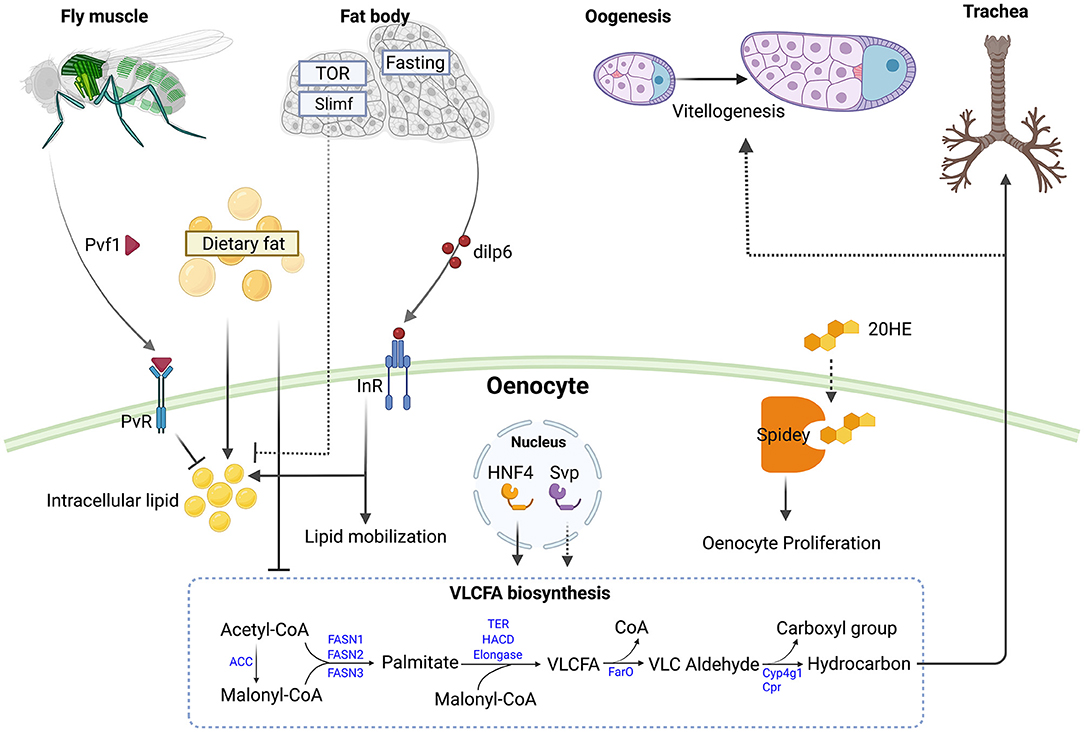
Figure 1. Roles of oenocytes in modulating metabolism and responding to nutritional signal. Muscle derived PDGF- and VEGF-related factor 1 (Pvf1) signals to PDGF- and VEGF-receptor related (PvR) in oenocytes to inhibit lipid accumulation and lipogenesis in adult flies (27). Elevated levels of circulating fats can promote oenocyte steatosis and inhibit very long chain fatty acid biosynthesis (28). Knockdown of target of rapamycin (TOR) or slimfast (slif) in the larval fat body increase steatosis in oenocytes (6). During fasting, the adult fat body produces Insulin-like peptide 6 (Ilp6) which activates the Insulin-like receptor (InR) in oenocytes to promote lipid accumulation and mobilization (13). The nuclear receptors Hepatocyte nuclear factor 4 (Hnf4) and seven up (svp) in adult oenocytes regulate very long chain fatty acid (VLCFA) biosynthesis (26, 29). Knockdown of svp in oenocytes impairs vitellogenesis in ovaries, possibly by reducing hydrocarbon production (30). Spidey in oenocytes can metabolize 20-hydroxyecdysone (20HE) and promotes oenocyte proliferation (26). Larval oenocyte-derived hydrocarbon can waterproof the trachea (31). The VLCFA biosynthesis pathway is depicted with Drosophila genes (in blue) (29). Abbreviations in the pathway: ACC, Acetyl-CoA carboxylase; FASN1-3, Fatty acid synthase 1-3; TER, HACD and Elongase are each encoded by multiple genes; FarO, Fatty acyl-CoA reductase in oenocytes; Cyp4g1, cytochrome P450 4g1; Cpr, Cytochrome P450 reductase. Figure is created with BioRender.com.
In line with the central role in lipid metabolism, oenocytes are also the major sites for VLCFAs synthesis, which are constituents of cellular lipids such as sphingolipids and cuticular hydrocarbons. Oenocyte ablation during larval stages using the GAL4/UAS-reaper system leads to compromised tracheal air-filling and the larvae exhibit hypoxia-induced phenotypes in larvae. Strikingly, oenocyte ablation or knockdown in larvae of genes encoding VLCFA metabolizing enzymes (ACC, KAR, elongase) result in severe tracheal defects, with the tracheal tubes filled with an aqueous solution (31). Interestingly, the spiracle ducts, which are connected to the trachea and correspond to the respiratory openings found on the thorax and abdomen of larvae, are no longer filled with Oil Red O-staining in oenocyte-ablated or Acc mutant larvae (31), suggesting that larval oenocytes might provide VLCFAs that coat the trachea. Alternatively, VLCFAs, which are synthesized from oenocytes, can signal non-autonomously to control lipid metabolism in spiracles, which have been proposed to obtain waterproofing lipids from specialized spiracular gland cells (Figure 1) (16).
In adult flies, cuticular hydrocarbons have been found to provide protection from desiccation and to act as pheromones for sexual communication and modulate longevity (16, 36, 37). Previous studies have suggested that cytoplasmic projections from the oenocytes contact the epidermis and that these cell-to-cell contacts facilitate lipid or lipoprotein transfer from oenocytes to the epidermis (38–42). Adult male and female flies in which oenocytes have been ablated using the GAL4/UAS system, show reduced levels of most of the cuticular hydrocarbons (36). Oenocyte-specific knockdown of genes regulating hydrocarbon production, such as CYP4g1, leads to reduced desiccation resistance (43). Further, mutations in the nuclear receptor Hnf4, which is strongly expressed in oenocytes (6), show reduced expression of genes involved in VLCFA biosynthesis, including KAR, CYP4g1, Cpr and genes encoding elongases; reduced levels of hydrocarbons; and decreased dry starvation resistance (Figure 1) (29). Interestingly, mouse hepatocytes isolated from mice with a mutation in HNF4α, the mammalian ortholog of Hnf4, also show markedly decreased expression of genes that encode elongases, i.e., Elovl3 or Elovl5. Mice mutant for Elovl1 and Elovl4 die shortly after birth from acute dehydration and loss of epidermal hydrophobic barrier function (44, 45), suggesting that the regulation of VLCFA biosynthesis by HNF4 is evolutionarily conserved.
Drosophila Oenocytes as an Emerging Model for Liver Function
Studies of oenocytes focused on their role in hydrocarbon synthesis until Gutierrez et al. showed that larval oenocytes can store and process lipids under starvation, a function analogous to what occurs in the mammalian liver. Under fed conditions, Drosophila larvae store lipids in the midgut epithelial cells and the fat body (6, 46). However, after 14 h of fasting, lipid droplets can no longer be detected in the fat body and the midgut, but intense Oil Red O staining persists in the oenocytes (6). Observation of this starvation induced steatosis suggested that larval oenocytes have hepatocyte-like functions. Further, although the progenitors of larval and adult oenocytes are different (16), Chatterjee et al. showed that like larval oenocytes, adult oenocytes also exhibits starvation-induced steatosis and that this process is mediated by fat body secreted Drosophila insulin-like peptide 6 (dILP6) (13). Under acute fasting response, adult female flies accumulate lipid droplets in the oenocytes, although the level of this steatosis is mild and more heterogenous as compared to what is observed in larvae. In starvation conditions, several genes with catabolic and gluconeogenic functions in hepatocytes are also highly induced in oenocytes, including amylase proximal (amy-P) and phosphoenolpyruvate carboxykinase (pepck), which catalyzes the rate-limiting step in gluconeogenesis (13).
In addition to gluconeogenic genes, which are induced in oenocytes in response to starvation response, oenocytes are also enriched with genes involved in ketogenesis, long-chain fatty acid metabolism, and peroxisomal function, all of which are also enriched in the mammalian liver. Huang et al. performed oenocyte-specific translatomic profiling using the RiboTag sequencing approach to explore this. By comparing oenocyte RiboTag data with previously published fly whole body transcriptome datasets, these authors identified genes enriched in adult oenocytes (25). Further, by comparing these genes to genes enriched in mammalian liver, Huang et al. identified several commonly enriched genes. One of them is HMG-CoA synthase (Hmgs in flies and HMGCS1/2 in humans), which encodes the key enzyme involved in ketogenesis and cholesterol biosynthesis. Others include HMG-CoA lyase (CG10399 in flies and HMGCL in humans) and D-β-hydroxybutyrate dehydrogenase (shroud in flies and BDH1 in humans), which encode key enzymes involved in ketogenesis. These observations suggest that oenocytes may be the primary site for ketogenesis in flies. Our current understanding of ketone body metabolism in insects is limited to locusts. Bailey et al. showed that the hemolymph of the locust Schistocerca gregaria contains appreciable levels of acetoacetate and at least a small amount of 3-hydroxybutyrate (47). The levels of acetoacetate, but not of 3-hydroxybutyrate, increase during starvation and flight. This is different from mammals, in which 3-hydroxybutyrate is the dominating ketone body. It seems possible that Drosophila oenocytes produce alternative ketone bodies, including acetoacetate, under flight or starved condition.
Genes involved in the synthesis of VLCFAs and microsomal fatty acid elongation are also highly enriched in oenocytes and the liver (25). These include very-long-chain 3-ketoacyl-coA synthase (CG18609 in flies and ELOVL2 in humans), which catalyzes the first step of VLCFA synthesis in smooth ER (25), and several key genes involved in the production of cuticular hydrocarbons, such as Cyp4g1, Cpr, FarO (16, 28, 29, 41). Notably, the role of the mammalian liver in synthesizing hydrocarbons is unclear, although it has been shown that VLCFA genes (ELOVL2 and ELOVL6) are enriched in this organ. Fibroblast growth factor 21 (branchless in fly and FGF21 in human), is also a key hormonal factor that is enriched in both oenocytes and the liver (25). FGF21 is an important metabolic regulator that has anti-diabetic properties in humans (48). Several studies have shown that FGF21 stimulates fatty acids oxidation, ketone body production and inhibits lipogenesis (49, 50). Interestingly, exogenously provided FGF21 increases longevity and stress tolerance in female silkworms (Bombyx mori), possibly through activated AMPK, FoxO, and sirtuins (51), suggesting that insects can be used as animal models for evaluating the pharmaceutical effects of FGF21. The role of FGF21 in fly oenocytes remains to be characterized and may provide an excellent model to further decipher the role of FGF21 in oenocyte/liver function.
Comparison of the Roles of Oenocytes and the Fat Body With Liver Functions
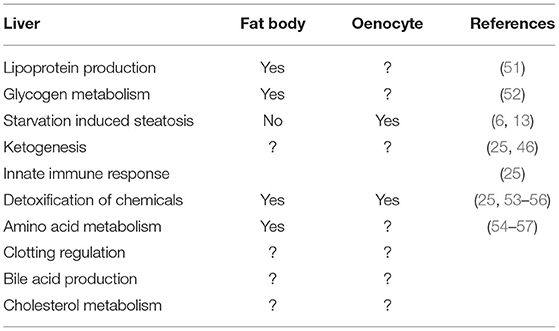
In addition to oenocytes, the fat body has also been regarded as a liver-like tissue in Drosophila, given its roles in nutrient sensing, glycogen storage, detoxification, the immune response, and lipid storage (3). Interestingly, comparison of oenocyte- and fat body-enriched genes revealed that there was very little overlap between these two tissues (25), suggesting that they perform distinct biological functions. Indeed, Gene Ontology (GO) analysis revealed an enrichment of genes involved in carboxylic acid and amino acid metabolism in the fat body, whereas oenocytes showed an enrichment for genes involved in fatty acid elongation, biosynthesis, xenobiotic metabolism and peroxisomal function (25). See a summary of comparison of liver function with the fat body and the oenocyte in Table 1.
In mammals, the liver and gut are the two primary organs for the secretion of lipoproteins, which deliver lipids and sterols to peripheral tissues. Apolipoprotein B (ApoB) is the primary apolipoprotein that scaffolds chylomicrons and very low-density lipoproteins (VLDL), which are secreted by the gut and liver, respectively. In Drosophila and other insects, lipophorins (Lpp) are the major lipoproteins similar to apoB-containing lipoproteins in mammals (58). Lpp scaffolding apolipoproteins are the highly conserved apolipophorins (apoLpp) (59). In addition, the two Drosophila lipoprotein receptors (LpR1 and LpR2), homologous to LDL receptors in mammals (60, 61), can promote Lpp uptake. A BLAST search against human apoB yields four Drosophila genes: apolipophorin (apolpp), microsomal triacylglycerol transfer protein (Mtp), apolipoprotein lipid transfer particle (Apoltp) and crossveinless d (cv-d) (58). Drosophila larvae hemolymph contains three circulating lipoproteins: Lpp, LTP, and Cv-d. Among them, Lpp is the major lipid carrier as more than 95% of hemolymph lipids co-fractionate with it (58). Interestingly, the larval fat body is the primary tissue that secretes Lpp, as fat body-specific Lpp knockdown strongly reduces the level of circulating Lpp and LTP in the hemolymph. Based on this, the fat body performs liver-like function regulating lipid transport through secreted lipoproteins. It remains to be determined whether and how oenocytes regulate lipoprotein circulation in the hemolymph, as four apoB genes are also expressed in oenocytes. In addition, oenocyte-specific over-expression of the lipogenic genes fatty acid synthase 1 and 3 (fasn1 and fasn3) can increase lipid droplet size in the fat body, suggesting that lipids generated from oenocytes are transported to the fat body, possibly through lipoproteins (27).
The key liver-like characteristic of the fat body is glycogen storage and utilization. In humans, glycogen is primarily stored in the liver (~100 g) and in skeletal muscles (~500 g). However, human muscles do not show major decreases in glycogen during fasting (52, 62), and only liver stored glycogen contributes to the release of glucose into the blood, specifically during fasting. Thus, the liver is viewed as a “glucostat” that maintains circulating sugar levels (63). Net hepatic glycogen synthesis is one of the major direct effects of insulin on hepatocytes and an important mechanism for suppression of hepatic glucose production (64). During fasting, the pancreas secretes glucagon to initiate a cascade of kinase activity that leads to release of glucose from stored glycogen via glycogenolysis. Similarly, in fly larvae, glycogen metabolism in the fat body plays a crucial role in the maintenance of circulating sugars under fasting conditions. One difference is that in Drosophila, glycogen from the fat body is converted to glucose, as well as trehalose, a form of nonreducing disaccharide primarily present in the insect hemolymph (65). The concentration of circulating trehalose concentration in third instar larvae is ~25 ug/ul, vs. ~0.1 ug/ul of glucose (66). In flies, excess energy from food, can result from feeding of flies on a high sugar diet, induces the expression of glycogen synthase (glyS), which promotes glycogen levels (10). Interestingly, unlike the action of glucagon in the mammalian liver, fasting-induced glycogen breakdown in the fly fat body is not regulated by AKH (Glucagon-like homolog in flies) (65). Instead, glycogen mobilization in the fat body is regulated by a decrease in sugar availability (65) and depends on glycogen phosphorylase (GlyP) (9), which increases in activity during larval development and remains high during pupal-adult development. In addition to GlyP, glycogen autophagy is also involved in glycogen breakdown in larval muscles (53). In adult flies, glycogen is stored mostly in the flight muscles, the fat body and oocytes. Whether oenocytes contribute to glycogen storage and mobilization is currently unclear and deserves further investigation.
Detoxification of toxic substances from the human body is mainly carried out by the liver. Detoxification are performed by phase I and phase II drug metabolizing enzymes (DMEs), as well as phase III transporters (54). Phase I DMEs consist of the cytochrome P450 (CYP) microsomal enzymes which are abundant in the liver, gastrointestinal tract, lung, and kidney. Phase II metabolizing and conjugating enzymes consist of superfamily of enzymes, including glutathione S-transferases (GSTs) (55–57). Interestingly, RiboTag analysis revealed that the microsomal GST Mgstl is highly enriched in Drosophila oenocytes (25). Further, transcriptome analysis in the yellow fever mosquito Aedes aegypti also showed that pupal oenocytes highly express cytochrome P450 genes (67), suggesting that oenocytes are responsible for detoxifications in insects. In addition to oenocytes, the fat body can also perform detoxification. For example, phenobarbital (present in insecticides) administration induced Cyp6a2 expression in the Drosophila midgut and the pericuticular fat bodies (68). Also, biochemical analysis demonstrated that the fat body of the black swallowtail capillaries can metabolize linear furanocoumarins bergapten and xanthotoxin (69). Altogether, these data indicate that insect fat body and oenocytes can perform detoxification, analogous to the mammalian liver.
The liver, along with small intestines, kidneys and muscle are important organs for amino acid metabolism. The liver is the primary organ responsible for amino acids catabolism with the exception of branched amino acids. The amine group is separated and converted into urea, which is released into the blood and the remaining carbon group can be used for gluconeogenesis and ketogenesis (70). In honeybee (Apis mellifera) larvae, amino acids are synthesized into Hexamerins, which are storage proteins secreted by the fat body made of six polypeptide subunits. Hexamerins provide amino acids to other tissues during development and disappear during adulthood (71). In Drosophila larval storage proteins (Lsp) play a similar role (72). The fat body is also an important sensor for amino acids. Dietary amino acids are sensed by target of rapamycin (TOR)/regulatory associated protein of TOR (RAPTOR) in the Drosophila larval fat body, which can remotely control dILP release from the brain, thereby regulating systemic growth (73). In addition, an amino acid transporter, slimfast, can regulate TOR signaling in the fat body and regulates phosphatidyl-inositol 3-kinase (PI3K) signaling in peripheral tissues (74). Together, these results suggest that the insect fat body functions as a sensor and regulator that couples nutritional status to growth, through a humoral mechanism. However, whether insect fat body or oenocytes participate in amino acid catabolism and how they contribute to glycogen or carbohydrate metabolism remains unclear.
Regulation of Fat Body and Oenocytes Metabolism Under Starvation
Perhaps the most distinct feature of oenocytes is their ability to accumulate lipids under starvation (6). Oenocytes are also important for the survival of the adult flies, as flies lacking oenocytes have increased sensitivity to starvation (13). Thus, oenocytes may process lipids or carbohydrates generated from the fat body to provide energy under starvation. In line with this, oenocyte ablation in larvae blocks fat body TAG depletion following starvation, suggesting a defect in lipid mobilization (6). Chatterjee et al. performed RNA-seq profiling under starvation conditions and showed that oenocytes containing carcasses had elevated gene expression in carbohydrate metabolism, the oxidation-reduction process, and amine metabolism. In contrast, the expression of genes involved in the defense response, chorion-containing eggshell formation, and proteolysis was increased in the fat body (13). Consistent with these observations, the classic catabolic and gluconeogenic genes amylase proximal (amy-p) and phosphoenolpyruvate carboxykinase (pepck) are induced in starved oenocytes. In addition, lipophorin receptor 2 (lpr2), which is responsible for capturing lipids in the hemolymph, was also induced in oenocytes but not in the fat body (13). Altogether, these observations suggest that oenocytes and the fat body have different physiological response and that oenocytes might process lipids released from fat bodies under starvation.
Roles of the Fat Body and Oenocytes in Immune Responses
Hepatocytes play an important role in controlling innate immunity via production of pattern-recognition receptors (PRRs) and pathogen associated molecular patterns (PAMPs) (75). As is the case for the mammalian liver, fly fat body and oenocytes express genes involved in immunity (25). Interestingly, the two innate immunity pathways (Toll and Imd) are differentially enriched in oenocytes vs. the fat body. Specifically, genes in the Imd pathway are enriched in oenocytes whereas Toll pathway genes are enriched in the fat body. The Toll pathway controls resistance to Gram-positive bacterial infections and fungal infections, and the Imd pathway controls resistance to Gram-negative bacterial infections (76). Downstream of Toll signaling are the NF-κB transcription factors: dorsal and DIF, whose immune-regulated functions are conserved in mammals (76). Relish, another NF-κB protein, is regulated by the Imd pathway and controls the expression of most of the Drosophila antimicrobial peptides (AMPs) (77). Further, many downstream effectors of Relish are highly enriched in oenocytes, including DptA, DptB, CecC, Dro and MTK. On the other hand, genes regulated by Toll signaling, including Tl, PGRP-SA, GNBP3 and modSP, are preferentially expressed in the fat body (25).
Nutrients and Tissues Regulating Lipid Metabolism in Oenocytes
Emerging evidence suggests that there is a close interaction between the fat body and the oenocytes in both larvae and adults. In larvae, the fat body consists of free-floating fat cells that are physically associated with the oenocytes (6, 78). In adults, fat body cells are tightly attached to the oenocytes (79). The fat body was the first tissue described to non-autonomously regulate oenocyte metabolism, as lipid accumulation in oenocytes depends on fat body nutritional sensors (6). Knockdown of slimfast, a fat body-specific amino acid transporter, causes lipid droplet accumulation in the oenocytes. Further, inhibition of TOR activity, following overexpression of TSC1 and TSC2 in the fat body, leads to a marked increase of steatosis in oenocytes. Similarly, inhibiting the phosphatidylinositol-3 kinase pathway through overexpression of PTEN also leads to severe steatosis in oenocytes, suggesting that oenocytes are regulated by the fat body nutritional status, either in response to fluctuating amino acid levels or through a TOR-dependent signaling peptide secreted by the fat body or by an intermediate tissue such as the intestine (Figure 1).
In addition to amino acids, oenocytes also respond to dietary lipids and fatty acids. When fed under high dietary lipids during larval stages, adult flies produce less pheromones (28), indicating that oenocytes VLCFA biosynthesis function is hindered in response to an increased level of circulating lipid (Figure 1). Further, overexpression of the Brummer (Bmm) lipase in the fat body induces steatosis in oenocytes under fed conditions. Conversely, overexpression of Lsd2 in the fat body under starved conditions reduces steatosis in oenocytes (6). Altogether, these results suggest that oenocytes develop steatosis when circulating levels of FFAs increase, which happens during starvation or under high fat diet treatment. This is consistent with what is observed in in vertebrates, in which either high fat diet or high levels of lipid uptake in hepatocytes all contributes to NAFLD pathogenesis (80–82).
In addition to nutritional signals, oenocytes are also regulated –by ligands received from other tissues under various conditions. The expression of unpaired 2 (upd2), which encodes the first adipokine discovered in response to nutrients, is induced by dietary sugar and nutrients (83). Interestingly, oenocytes show abnormal steatosis in fed upd2 mutants (83), suggesting that oenocytes might be regulated by fat-body derived upd2 in response to dietary nutrients. Unlike other tissues, such as the fat body, oenocytes exhibit increased levels of insulin activity when starved, as detected by the increased membrane localization of a PI3Kinase reporter (13, 84). Under starvation, the level of dILP6 is elevated in the adult fat body (85), and overexpression of dILP6 in the fat body can induce oenocyte insulin/IGF signaling (IIS) and steatosis in adult females (13). To the contrary, fat body-specific knockdown of dILP6 reduces the level of starvation-induced steatosis in oenocytes (13). Altogether, these data suggest that under starvation, the fat body signals to the oenocytes to increase lipid mobilization and carbohydrate metabolism via dILP6 (Figure 1).
Oenocyte metabolism can also be regulated by a PDGF/VEGF-like ligand from the muscle. Ghosh et al. demonstrated that muscle-specific knockdown of pvf1 leads to severe lipid accumulation in adipose tissue and oenocytes by increasing de novo lipogenesis (27). Single-nuclei RNA-sequencing (snRNA-seq) revealed that PDGF- and VEGF-receptor related (Pvr), which encodes the PVf1 receptor, is highly enriched in oenocytes. Further, this muscle-to-oenocyte signaling was found to inhibit rapid lipid expansion in newly eclosed flies. Altogether, these analyses suggest that Pvf1 acts as a myokine that suppresses oenocyte lipid synthesis (Figure 1). Interestingly, in vertebrates, VEGF-A and VEGF-B have been shown to be stored and released by muscles (86, 87). Furthermore, it has also been demonstrated that VEGF levels are lower in NAFLD patients than in healthy controls (88), and that a variety of myokines can reduce insulin resistance and fat accumulation in the liver (89). Thus, the Drosophila muscle-to-oenocyte model might be useful for discovery of new therapeutics for treating NAFLD.
Crosstalk of Oenocytes With Other Tissues in Shaping Organismal Metabolism
Considering the role of oenocytes in metabolism, it is not surprising that many studies in which the function of oenocytes has been perturbed report observing defects in the metabolism of other tissues. For example, loss of oenocytes cytochrome P450, Cyp4g1, leads to an elevated oleic acid/stearic acid ratio, that is specific to TGs and not phospholipids, suggesting an altered fat body lipid composition and metabolism (6). Additionally, oenocyte-specific inhibition of TOR, following overexpression of TSC1 and TSC2 can induce lipogenesis and increase lipid storage in the fat body (27). Further, oenocyte ablation or knockdown of genes encoding VLCFA metabolizing enzymes in larvae leads to severe tracheal defects and altered spiracle lipid metabolism (31). In addition, in a genome-wide RNAi screen in Drosophila for obesity-causing genes, many genes were identified that regulate whole-body fat content in an oenocyte-dependent manner (90). Interesting targets included inflammation-related genes and genes regulating ubiquitination, including TNF-receptor-associated factor 4 (Traf4), the interleukin enhancer-binding factor (ILF2), and the ubiquitin-conjugating enzyme (UBE2N) (90). These data suggest a role for immune regulatory networks and the ubiquitination in regulating fat storage in oenocytes. Oenocytes can also regulate vitellogenesis during oogenesis, as oenocyte-specific knock down of the seven up (svp), which encodes a nuclear receptor, can increase vitellogenic follicle egg chamber death (30). Moreover, svp is a known oenocyte-specific modulator of cuticle lipids, raising the possibility that female oenocytes regulate vitellogenic follicles through the production of hydrocarbons (Figure 1).
Conclusions
Major advances have been made in recent years in understanding the role of insect oenocytes in metabolism and physiology, and many fascinating research areas remain unexplored. One largely ignored yet important area is to delineate the differences and similarities between oenocyte function in Drosophila larvae and adults. Understanding such differences will be important for choosing the most appropriate stage to model a specific human disease. Another area of interest is the synthesis of VLCFAs and their derivatives in oenocytes, as this in turn affects not only hydrocarbon synthesis but also lipid metabolism in trachea, fat body and oocytes. Such non-autonomous regulation remains largely obscure and may be mediated through metabolites or signaling pathway activities. Finally, it is highly likely that additional oenocyte functions and crosstalk with additional tissues remain to be identified. For example, the role of oenocytes in immunity and bacterial defense, as suggested by the expression of IMD signaling pathway genes, need to be clarified. The role of oenocytes in drug detoxification as well as amino acids metabolism remains elusive. In addition, whether oenocyte metabolic functions regulate aspects of fly behavior, other than mating, remains to be explored.
Author Contributions
KH drafted and revised the manuscript. YL and NP helped to draft and revise the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Stephanie Mohr and Nirmalya Chatterjee for their critical comments of the manuscript. We would like to acknowledge the support from the American Heart Association (AHA) Postdoctoral Fellowship granted to KH (ID: 826844), and from Sigrid Jusélius Foundation and the Finnish Cultural Foundation (Suomen Kulttuurirahasto) granted to YL. NP is an investigator of the Howard Hughes Medical Institute.
References
1. Gonzalez JT, Fuchs CJ, Betts JA, van Loon LJ. Liver glycogen metabolism during and after prolonged endurance-type exercise. Am J Physiol Endocrinol Metab. (2016) 311:E543–53. doi: 10.1152/ajpendo.00232.2016
2. Tiwari S, Siddiqi SA. Intracellular trafficking and secretion of VLDL. Arterioscler Thromb Vasc Biol. (2012) 32:1079–86. doi: 10.1161/ATVBAHA.111.241471
3. Droujinine IA, Perrimon N. Interorgan communication pathways in physiology: focus on drosophila. Annu Rev Genet. (2016) 50:539–70. doi: 10.1146/annurev-genet-121415-122024
4. E.Bier.. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. (2005) 6:9–23. doi: 10.1038/nrg1503
5. Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. (2001) 11:1114–25. doi: 10.1101/gr.169101
6. Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. (2007) 445:275–80. doi: 10.1038/nature05382
7. Bailey E. Biochemistry of Insect Flight, Insect Biochemistry and Function. Springer. (1975) p. 89–176. doi: 10.1007/978-1-4899-3204-4_2
8. Heier C, Kuhnlein RP. Triacylglycerol metabolism in drosophila melanogaster. Genetics. (2018) 210:1163–84. doi: 10.1534/genetics.118.301583
9. J.Steele.. Glycogen phosphorylase in insects. Insect Biochem. (1982) 12:131–47. doi: 10.1016/0020-1790(82)90001-4
10. Chatterjee N, Perrimon N. What fuels the fly: energy metabolism in drosophila and its application to the study of obesity and diabetes. Sci Adv. (2021) 7:eabg4336. doi: 10.1126/sciadv.abg4336
11. Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. (2001) 11:213–21. doi: 10.1016/S0960-9822(01)00068-9
12. Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, Müller G, et al. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. (2005) 1:323–30. doi: 10.1016/j.cmet.2005.04.003
13. Chatterjee D, Katewa SD, Qi Y, Jackson SA, Kapahi P, Jasper H. Control of metabolic adaptation to fasting by dILP6-induced insulin signaling in Drosophila oenocytes. Proc Natl Acad Sci U S A. (2014) 111:17959–64. doi: 10.1073/pnas.1409241111
14. Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. (2004) 7:179–92. doi: 10.1016/j.devcel.2004.07.005
15. Colombani J, Bianchini L, Layalle S, Pondeville E, Dauphin-Villemant C, Antoniewski C, et al. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. (2005) 310:667–70. doi: 10.1126/science.1119432
16. Makki R, Cinnamon E, Gould AP. The development and functions of oenocytes. Annu Rev Entomol. (2014) 59:405–25. doi: 10.1146/annurev-ento-011613-162056
17. Cole L, Kramer PR. Chapter 1.1 - The Cell. In: Cole L, Kramer PR, editors. Human Physiology, Biochemistry and Basic Medicine. Boston: Academic Press. (2016) p. 3–10. doi: 10.1016/B978-0-12-803699-0.00001-3
18. Huang K, Miao T, Chang K, Kim J, Kang P, Jiang Q, et al. Impaired peroxisomal import in Drosophila oenocytes causes cardiac dysfunction by inducing upd3 as a peroxikine. Nat Commun. (2020) 11:2943. doi: 10.1038/s41467-020-16781-w
19. Yamanaka N, Rewitz KF, O'Connor MB. Ecdysone control of developmental transitions: lessons from drosophila research. Annu Rev Entomol. (2013) 58:497–516. doi: 10.1146/annurev-ento-120811-153608
20. Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, et al. Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J Exp Biol. (2008) 211:2712–24. doi: 10.1242/jeb.014878
21. Ahmed SMH, Maldera JA, Krunic D, Paiva-Silva GO, Pénalva C, Teleman AA, et al. Fitness trade-offs incurred by ovary-to-gut steroid signalling in Drosophila. Nature. (2020) 584:415–9. doi: 10.1038/s41586-020-2462-y
22. L.I. Gilbert. Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol Cell Endocrinol. (2004) 215:1–10. doi: 10.1016/j.mce.2003.11.003
23. Lafont R, Dauphin-Villemant C, Warren JT, Rees H. 4 - Ecdysteroid Chemistry and Biochemistry. In: Gilbert LI, editors. Insect Endocrinology. San Diego: Academic Press. (2012) p. 106–176. doi: 10.1016/B978-0-12-384749-2.10004-4
24. Petryk A, Warren JT, Marqués G, Jarcho MP, Gilbert LI, Kahler J, et al. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc Nat Acad Sci. (2003) 100:13773–8. doi: 10.1073/pnas.2336088100
25. Huang K, Chen W, Zhu F, Li PW, Kapahi P, Bai H. RiboTag translatomic profiling of Drosophila oenocytes under aging and induced oxidative stress. BMC Genomics. (2019) 20:50. doi: 10.1186/s12864-018-5404-4
26. Chiang YN, Tan KJ, Chung H, Lavrynenko O, Shevchenko A, Yew JY. Steroid hormone signaling is essential for pheromone production and oenocyte survival. PLoS Genet. (2016) 12:e1006126. doi: 10.1371/journal.pgen.1006126
27. Ghosh AC, Tattikota SG, Liu Y, Comjean A, Hu Y,V, Barrera, et al. Drosophila PDGF/VEGF signaling from muscles to hepatocyte-like cells protects against obesity. Elife. (2020) 9:e56969. doi: 10.7554/eLife.56969
28. Wicker-Thomas C, Garrido D, Bontonou G, Napal L, Mazuras N, Denis B, et al. Flexible origin of hydrocarbon/pheromone precursors in Drosophila melanogaster. J Lipid Res. (2015) 56:2094–101. doi: 10.1194/jlr.M060368
29. Storelli G, Nam HJ, Simcox J, Villanueva CJ, Thummel CS. Drosophila HNF4 directs a switch in lipid metabolism that supports the transition to adulthood. Dev Cell. 48 (2019) 200–14 e6. doi: 10.1016/j.devcel.2018.11.030
30. Weaver LN, Drummond-Barbosa D. The nuclear receptor seven up functions in adipocytes and oenocytes to control distinct steps of Drosophila oogenesis. Dev Biol. (2019) 456:179–89. doi: 10.1016/j.ydbio.2019.08.015
31. Parvy J-P, Napal L, Rubin T, Poidevin M, Perrin L, Wicker-Thomas C, et al. Drosophila melanogaster Acetyl-CoA-Carboxylase Sustains a Fatty Acid–Dependent Remote Signal to Waterproof the Respiratory System. PLoS Genet. (2012) 8:e1002925. doi: 10.1371/journal.pgen.1002925
32. Römer F, Bressel H-U. Secretion and metabolism of ecdysteroids by oenocyte-fat body complexes (OEFC) in adult males of Gryllus bimaculatus DEG (Insecta). Zeitschrift für Naturforschung C. (1994) 49:871–80. doi: 10.1515/znc-1994-11-1224
33. Matthew Sieber H, Allan C. Spradling steroid signaling establishes a female metabolic state and regulates SREBP to control oocyte lipid accumulation current biology. Curr Biol. (2015) 25:993–1004. doi: 10.1016/j.cub.2015.02.019
34. Carney GE, Bender M. The Drosophila ecdysone receptor (EcR) gene is required maternally for normal oogenesis. Genetics. (2000) 154:1203–11. doi: 10.1093/genetics/154.3.1203
35. Delanoue R, Slaidina M, Léopold P. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in drosophila fat cells. Dev Cell. (2010) 18:1012–21. doi: 10.1016/j.devcel.2010.05.007
36. Billeter JC, Atallah J, Krupp JJ, Millar JG, Levine JD. Specialized cells tag sexual and species identity in Drosophila melanogaster. Nature. (2009) 461:987–91. doi: 10.1038/nature08495
37. Stefana MI, Driscoll PC, Obata F, Pengelly AR, Newell CL, MacRae JI, et al. Developmental diet regulates Drosophila lifespan via lipid autotoxins. Nat Commun. (2017) 8:1384. doi: 10.1038/s41467-017-01740-9
38. Whitten JM. The post-embryonic development of the tracheal system in drosophila melanogaster. J Cell Sci. (1957) 123–50. doi: 10.1242/jcs.s3-98.41.123
39. E.Rinterknecht. Cuticulogenesis correlated with ultrastructural changes in oenocytes and epidermal cells in the late cockroach embryo. Tissue Cell. (1985) 17:723–43. doi: 10.1016/0040-8166(85)90007-2
40. Wigglesworth VB Memoirs: Memoirs: The Physiology of the Cuticle and of Ecdysis in Rhodnius prolixus (Triatomidae Hemiptera); Hemiptera); with special reference to the function of the oenocytes and of the dermal glands. J Cell Sci. (1933) s2-76:269–318. doi: 10.1242/jcs.s2-76.302.269
41. V.B. Wigglesworth. The source of lipids and polyphenols for the insect cuticle: The role of fat body, oenocytes and oenocytoids. Tissue Cell. (1988) 20:919–32. doi: 10.1016/0040-8166(88)90033-X
42. Wolfe LS The The deposition of the third instar larval cuticle of Calliphora erythrocephala. J Cell Sci. (1954) s3-95:49–66. doi: 10.1242/jcs.s3-95.29.49
43. Qiu Y, Tittiger C, Wicker-Thomas C, Le Goff G, Young S, Wajnberg E, et al. An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc Natl Acad Sci U S A. (2012) 109:14858–63. doi: 10.1073/pnas.1208650109
44. Sassa T, Ohno Y, Suzuki S, Nomura T, Nishioka C, Kashiwagi T, et al. Impaired epidermal permeability barrier in mice lacking elovl1, the gene responsible for very-long-chain fatty acid production. Mol Cell Biol. (2013) 33:2787–96. doi: 10.1128/MCB.00192-13
45. Cameron DJ, Tong Z, Yang Z, Kaminoh J, Kamiyah S, Chen H, et al. Essential role of Elovl4 in very long chain fatty acid synthesis, skin permeability barrier function, neonatal survival. Int J Biol Sci. (2007) 3:111–9. doi: 10.7150/ijbs.3.111
46. Butterworth FM, Bodenstein D, King RC. Adipose tissue of drosophila melanogaster. i an experimental study of larval fat body. J Exp Zool. (1965) 158:141–53. doi: 10.1002/jez.1401580203
47. Bailey EJ, Horne JA, Izatt ME, Hill L. Concentrations of acetoacetate and 3-hydroxybutyrate in pigeon blood and desert locust haemolymph. Life Sci II. (1971) 10:1415–9. doi: 10.1016/0024-3205(71)90351-1
48. Tezze C, Romanello V, Sandri M. FGF21 as modulator of metabolism in health and disease. Front Physiol. (2019) 10:419. doi: 10.3389/fphys.2019.00419
49. Fisher FM, Maratos-Flier E. Understanding the physiology of FGF21. Annu Rev Physiol. (2016) 78:223–41. doi: 10.1146/annurev-physiol-021115-105339
50. Staiger H, Keuper M, Berti L, Hrabe de AngelisM, Haring HU. Fibroblast growth factor 21-metabolic role in mice and men. Endocr Rev. (2017) 38:468–88. doi: 10.1210/er.2017-00016
51. Song JB, Hao KG, Chen X, Zhang YH, Cheng ZL, Mao S, et al. Fibroblast growth factor 21 prolongs lifespan and improves stress tolerance in the silkworm, Bombyx mori. Ann Transl Med. (2020) 8:220. doi: 10.21037/atm.2020.01.18
52. Vendelbo MH, Clasen BF, Treebak JT, Moller L, Krusenstjerna-Hafstrom T, Madsen M, et al. Insulin resistance after a 72-h fast is associated with impaired AS160 phosphorylation and accumulation of lipid and glycogen in human skeletal muscle. Am J Physiol Endocrinol Metab. (2012) 302:E190–200. doi: 10.1152/ajpendo.00207.2011
53. Zirin J, Nieuwenhuis J, Perrimon N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol. (2013) 11:e1001708. doi: 10.1371/journal.pbio.1001708
54. Xu C, Li CY, Kong AN. Induction of phase I, II and III drug metabolism/transport by xenobiotics. Arch Pharm Res. (2005) 28:249–68. doi: 10.1007/BF02977789
55. Moscow JA, Dixon KH. Glutathione-related enzymes, glutathione and multidrug resistance. Cytotechnology. (1993) 12:155–70. doi: 10.1007/BF00744663
56. Schilter B, Turesky RJ, Juillerat M, Honegger P, Guigoz Y. Phase I and phase II xenobiotic reactions and metabolism of the food-borne carcinogen 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in aggregating liver cell cultures. Biochem Pharmacol. (1993) 45:1087–96. doi: 10.1016/0006-2952(93)90253-S
57. Tew KD, Ronai Z. GST function in drug and stress response. Drug Resist Updat. (1999) 2:143–7. doi: 10.1054/drup.1999.0086
58. Palm W, Sampaio JL, Brankatschk M, Carvalho M, Mahmoud A, Shevchenko A, et al. Lipoproteins in Drosophila melanogaster—assembly, function, and influence on tissue lipid composition. PLoS Genet. (2012) 8:e1002828. doi: 10.1371/journal.pgen.1002828
59. Smolenaars MM, Madsen O, Rodenburg KW, Van der Horst DJ. Molecular diversity and evolution of the large lipid transfer protein superfamily. J Lipid Res. (2007) 48:489–502. doi: 10.1194/jlr.R600028-JLR200
60. and JH Bock HH. Lipoprotein receptors in the nervous system. Annu Rev Biochem. (2002) 71:405-434. doi: 10.1146/annurev.biochem.71.110601.135342
61. Khaliullina H, Panáková D, Eugster C, Riedel F, Carvalho M, Eaton S. Patched regulates Smoothened trafficking using lipoprotein-derived lipids. Development. (2009) 136:4111–21. doi: 10.1242/dev.041392
62. Nieman DC, Carlson KA, Brandstater ME, Naegele RT, Blankenship JW. Running endurance in 27-h-fasted humans. J Appl Physiol. (1985) 63:2502–9. doi: 10.1152/jappl.1987.63.6.2502
63. Jensen J, Rustad PI, Kolnes AJ, Lai YC. The role of skeletal muscle glycogen breakdown for regulation of insulin sensitivity by exercise. Front Physiol. (2011) 2:112. doi: 10.3389/fphys.2011.00112
64. Petersen MC, Vatner DF, Shulman GI. Regulation of hepatic glucose metabolism in health and disease. Nat Rev Endocrinol. (2017) 13:572–87. doi: 10.1038/nrendo.2017.80
65. Yamada T, Habara O, Kubo H, Nishimura T. Fat body glycogen serves as a metabolic safeguard for the maintenance of sugar levels in Drosophila. Development. (2018) 145. doi: 10.1242/dev.158865
66. Matsushita R, Nishimura T. Trehalose metabolism confers developmental robustness and stability in Drosophila by regulating glucose homeostasis. Communicat Biol. (2020) 3:170. doi: 10.1038/s42003-020-0889-1
67. Martins GF, Ramalho-Ortigão JM, Lobo NF, Severson DW, McDowell MA, Pimenta PF. Insights into the transcriptome of oenocytes from Aedes aegypti pupae. Mem Inst Oswaldo Cruz. (2011) 106:308–15. doi: 10.1590/S0074-02762011000300009
68. Brun A, Cuany A, Le Mouel T, Berge J, Amichot M. Inducibility of the Drosophila melanogaster cytochrome P450 gene, CYP6A2, by phenobarbital in insecticide susceptible or resistant strains. Insect Biochem Mol Biol. (1996) 26:697–703. doi: 10.1016/S0965-1748(96)00036-7
69. Petersen RA, Zangerl AR, Berenbaum MR, Schuler MA. Expression of CYP6B1 and CYP6B3 cytochrome P450 monooxygenases and furanocoumarin metabolism in different tissues of Papilio polyxenes (Lepidoptera: Papilionidae). Insect Biochem Mol Biol. (2001) 31:679–90. doi: 10.1016/S0965-1748(00)00174-0
70. Blanco, Blanco G. Chapter 16 - amino acid metabolism. In: Blanco A, Blanco G, editors. Medical Biochemistry. Cambridge, MA: Academic Press (2017) p. 367–99. doi: 10.1016/B978-0-12-803550-4.00016-1
71. Martins JR, Bitondi MMG. The HEX 110 hexamerin is a cytoplasmic and nucleolar protein in the ovaries of apis mellifera. PLoS ONE. (2016) 11:e0151035. doi: 10.1371/journal.pone.0151035
72. Wolfe J, Akam ME, Roberts DB. Biochemical and immunological studies on larval serum protein 1, the major haemolymph protein of Drosophila melanogaster third-instar larvae. Eur J Biochem. (1977) 79:47–53. doi: 10.1111/j.1432-1033.1977.tb11782.x
73. Géminard C, Rulifson EJ, Léopold P. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. (2009) 10:199–207. doi: 10.1016/j.cmet.2009.08.002
74. Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Léopold P. A Nutrient sensor mechanism controls drosophila growth. Cell. (2003) 114:739–49. doi: 10.1016/S0092-8674(03)00713-X
75. Gao B, Jeong WI, Tian Z. Liver: An organ with predominant innate immunity. Hepatology. (2008) 47:729–36. doi: 10.1002/hep.22034
76. Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. (2002) 3:121–6. doi: 10.1038/ni0202-121
77. Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci U S A. (1996) 93:10343–7. doi: 10.1073/pnas.93.19.10343
78. Aguila JR, Suszko J, Gibbs AG, Hoshizaki DK. The role of larval fat cells in adult Drosophila melanogaster. J Exp Biology. (2007) 210:956–63. doi: 10.1242/jeb.001586
79. Krupp JJ, Levine JD. Dissection of oenocytes from adult Drosophila melanogaster. J Vis Exp. (2010) 2242. doi: 10.3791/2242
80. Velázquez KT, Enos RT, Bader JE, Sougiannis AT, Carson MS, Chatzistamou I, et al. Prolonged high-fat-diet feeding promotes non-alcoholic fatty liver disease and alters gut microbiota in mice. World J Hepatol. (2019) 11:619–37. doi: 10.4254/wjh.v11.i8.619
81. Doege H, Baillie RA, Ortegon AM, Tsang B, Wu Q, Punreddy S, et al. Targeted deletion of FATP5 reveals multiple functions in liver metabolism: alterations in hepatic lipid homeostasis. Gastroenterology. (2006) 130:1245–58. doi: 10.1053/j.gastro.2006.02.006
82. Falcon A, Doege H, Fluitt A, Tsang B, Watson N, Kay MA, et al. FATP2 is a hepatic fatty acid transporter and peroxisomal very long-chain acyl-CoA synthetase. Am J Physiol Endocrinol Metab. (2010) 299:E384–93. doi: 10.1152/ajpendo.00226.2010
83. Rajan A, Perrimon N. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell. (2012) 151:123–37. doi: 10.1016/j.cell.2012.08.019
84. Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. (2002) 2:239–49. doi: 10.1016/S1534-5807(02)00117-X
85. Bai H, Kang P, Tatar M. Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell. (2012) 11:978–85. doi: 10.1111/acel.12000
86. Boström P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. (2012) 481:463–8. doi: 10.1038/nature10777
87. Vind BF, Pehmøller C, Treebak JT, Birk JB, Hey-Mogensen M, Beck-Nielsen H, et al. Impaired insulin-induced site-specific phosphorylation of TBC1 domain family, member 4 (TBC1D4) in skeletal muscle of type 2 diabetes patients is restored by endurance exercise-training. Diabetologia. (2011) 54:157–67. doi: 10.1007/s00125-010-1924-4
88. Papageorgiou MV, Hadziyannis E, Tiniakos D, Georgiou A, Margariti A, Kostas A, et al. Serum levels of vascular endothelial growth factor in non-alcoholic fatty liver disease. Ann Gastroenterol. (2017) 30:209–16. doi: 10.20524/aog.2016.0107
89. de Oliveira Dos Santos AR, de Oliveira Zanuso B, Miola VFB, Barbalho SM, Santos Bueno PC, Flato UAP, et al. Adipokines, myokines, and hepatokines: crosstalk and metabolic repercussions. Int J Mol Sci. (2021) 22:2639. doi: 10.3390/ijms22052639
Keywords: oenocytes, Drosophila, lipid metabolism, tissue communication, hepatocyte, adipocyte, fat body cells, hydrocarbon
Citation: Huang K, Liu Y and Perrimon N (2022) Roles of Insect Oenocytes in Physiology and Their Relevance to Human Metabolic Diseases. Front. Insect Sci. 2:859847. doi: 10.3389/finsc.2022.859847
Received: 21 January 2022; Accepted: 14 February 2022;
Published: 17 March 2022.
Edited by:
Geoffrey Michael Attardo, University of California, Davis, United StatesReviewed by:
John Tower, University of Southern California, United StatesGareth Lycett, Liverpool School of Tropical Medicine, United Kingdom
Copyright © 2022 Huang, Liu and Perrimon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Norbert Perrimon, cGVycmltb24mI3gwMDA0MDtnZW5ldGljcy5tZWQuaGFydmFyZC5lZHU=
 Kerui Huang
Kerui Huang Ying Liu
Ying Liu Norbert Perrimon
Norbert Perrimon