- 1Key Laboratory of Freshwater Aquatic Genetic Resources, Ministry of Agriculture and Rural Affairs, Shanghai Ocean University, Shanghai, China
- 2Science and Technology Service Center, Shanghai MugaoBiotechnology co., Ltd., Shanghai, China
- 3Shanghai Engineering Research Center of Aquaculture, Shanghai Ocean University, Shanghai, China
Background: Shell and pearl formation in bivalves is a sophisticated biomineralization process that encompasses immunological and mineralization aspects, particularly during shell repair and the initial stages of pearl cultivation when a nucleus is inserted. Here, we describe a novel C-type lectin, HcLec1, isolated and characterized from the freshwater pearl mussel Hyriopsis cumingii Lea.
Methods: Immune challenge, RNA interference (RNAi) experiments, ELISA, and antibacterial assays were employed to investigate the role of HcLec1 in innate immunity. We also established shell damage repair and pearl nucleus insertion models to examine the impact of HcLec1 on the biomineralization process in Hyriopsis cumingii Lea. In vitro calcium carbonate crystallization assays were conducted to explore the direct role of HcLec1 in calcium carbonate crystal formation.
Results: The HcLec1 gene sequence is a full-length cDNA of 1552 bp, encoding 240 amino acids. HcLec1 comprises an N-terminal signal peptide and a carbohydrate-recognition domain (CRD), with QPD (Gln-Pro-Asp) and MND (Met-Asn-Asp) motifs for polysaccharide binding. Tissue expression analysis showed that HcLec1 is predominantly expressed in the gill tissue of Hyriopsis cumingii Lea under normal conditions, and its expression is significantly elevated in both gill and pearl sac tissues following nucleus insertion for pearl cultivation (P < 0.05). After immune stimulation with Aeromonas hydrophila and lipopolysaccharides (LPS), HcLec1 expression levels significantly increased in both cases (P < 0.01), indicating a role in bivalve innate immunity. RNA interference (RNAi)-mediated knockdown of HcLec1 led to a significant decrease in the expression levels of immune-related genes (WAP, α2m, and Lyso) and mineralization-related genes (CA, CHS, Nacrein, and Pif) (P < 0.05). In animal models for shell damage and nucleus insertion in pearl cultivation, HcLec1 showed a consistent expression pattern, with an initial significant decrease followed by a marked increase, peaking at day 14 (P < 0.05). This suggests a role for HcLec1 in pearl formation and shell repair. The recombinant HcLec1 protein demonstrated binding affinity to LPS and PGN, a robust ability to agglutinate Escherichia coli, Staphylococcus aureus, Aeromonas veronii, and Aeromonas hydrophila, and significantly inhibited bacterial growth (P < 0.05). Moreover, rHcLec1 promoted calcite crystal formation in saturated calcium carbonate solutions and altered crystal morphology.
Discussion: The HcLec1 gene plays a pivotal role in both innate immunity and biomineralization in the triangle sail mussel. This study enhances our understanding of the functional diversity of C-type lectins and provides a foundation for future studies on shell repair and pearl growth.
1 Introduction
Mollusks, inhabiting aquatic environments, are particularly susceptible to pathogen invasions (1, 2). As invertebrates, unlike fish, they rely predominantly on their innate immune system, which is distinguished by its non-specific, broad-spectrum, and swift response to pathogens (3, 4). This system targets conserved microbial elements termed pathogen-associated molecular patterns (PAMPs) rather than recognizing pathogens specifically. Upon infection, PAMPs are detected by pattern recognition receptors (PRRs), initiating the immune response (5–7). C-type lectins (CTLs), one of the most abundant families of PRRs, are particularly noteworthy in this context.
CTLs, alternatively known as C-type lectin receptors (CLRs), are calcium-dependent lectin proteins that bind carbohydrates and are ubiquitous in the animal kingdom (8). CTLs in vertebrates are notably conserved, whereas those in invertebrates exhibit considerable variation, indicative of their diverse functional roles (9–11). The CTL family is characterized by two principal polysaccharide-binding motifs: EPN (Glu-Pro-Asn)/QPD (Gln-Pro-Asp) and WND (Trp-Asn-Asp) (12). These motifs show substantial variation and are not uniformly conserved, owing to mutations. Studies indicate that Immulectin-2 from Manduca sexta and PcLec2 from Procambarus clarkii can activate the prophenoloxidase cascade, contributing to immune defense (13, 14). FcLec4, found in Fenneropenaeus chinensis, enhances the agglutination and elimination of Vibrio anguillarum, underscoring the role of CTLs as PRRs for PAMPs (15). Cflc-1, identified in Chlamys farreri, suppresses the proliferation of E. coli and Candida albicans and displays calcium-dependent agglutination of E. coli (16).
Hyriopsis cumingii Lea is a significant pearl-producing bivalve species, ranking first globally in total pearl production (17, 18). Both the shell and pearls are products of biomineralization in pearl oysters. The repair of shell damage and the nucleus implantation process during pearl cultivation involve two key biological phenomena: (1) resistance to waterborne pathogens and the immune response triggered by nucleus implantation, and (2) guidance of calcium carbonate crystal structure formation by matrix proteins in the mantle tissue. These proteins play a critical role in the biomineralization of newly formed shells and pearls following shell damage or nucleus implantation (19, 20). This process reflects the complex interaction and regulation between immune and biomineralization functions in H. cumingii Lea. Following nucleus implantation, the mussel undergoes stimulation, activating immune-related genes and proteins. This response persists until pearl sac formation (wound healing), at which point the expression of mineralization-related genes and proteins changes, initiating and enhancing biomineralization functions (21, 22).
A multitude of studies underscore the potential of CTLs in the process of biomineralization. CTLs broadly refer to any proteins containing one or more C-type lectin domains (CTL domain or CRD) (8). Wilt et al. identified a CRD in the spicule matrix protein SM50 from the sea urchin Strongylocentrotus purpuratus (23). They found that this CRD independently influences sea urchin mineralization and stabilizes amorphous calcium carbonate (ACC). Numerous cloned spicule matrix proteins in sea urchins contain the CRD (24). In earlier studies, a C-type lectin matrix protein, Perlucin, was isolated from the inner shell of abalone and shown to induce calcium carbonate crystal nucleation and guide crystal morphology (25). Purified Perlucin from Haliotis discus was also found to influence calcium carbonate crystal morphology, likely due to its high glycine and aspartic acid content, which affects the surface thermodynamics of crystal growth (26). These findings suggest that CTLs with CRDs function as matrix proteins in the biomineralization processes of sea urchins and abalones, participating in the organization and growth of calcium carbonate crystal structures. Currently, three types of CTLs have been identified in H. cumingii Lea. HcLec4, which contains four CRDs, binds to LPS and PGN, promoting early bacterial clearance through antimicrobial peptides (AMPs) (27). HcCUB-Lec, with a single CRD and a complement Uegf Bmp1 (CUB) domain, binds and agglutinates various bacteria, contributing to innate immunity (28). Perlucin, containing six conserved cysteine residues and a CRD, has been identified as a critical gene for nacre formation (29). However, at present, no studies have supported that CTLs in H. cumingii Lea possess both immune and biomineralization functions simultaneously.
This study identified and cloned a novel C-type lectin gene, HcLec1, from the mantle tissue, a critical biomineralization organ in the triangle sail mussel. HcLec1 encodes a single typical CRD and an N-terminal signal peptide. By employing gene expression profiling, constructing the HcLec1 protein expression vector, RNA interference (RNAi), Raman spectroscopy, and scanning electron microscopy, we explored the functional roles of this lectin in both innate immunity and biomineralization of shells and pearls. This research expands the molecular understanding of immunity and biomineralization processes in freshwater bivalves, providing valuable insights to improve shell repair and pearl production.
2 Materials and methods
2.1 Experimental animals and sources of materials
Triangle sail mussels were obtained from the Wuyi Aquaculture Base in Zhejiang, with an average length of 5–6 cm. They were kept in freshwater at room temperature (22–24°C) under laboratory conditions and fed Chlorella twice daily at 11:00 am and 11:00 pm.
Hemolymph was extracted from the hemocoel of the adductor muscle using a disposable syringe and centrifuged at 4°C and 700 × g for 10 minutes to separate the serum (supernatant) from the hemocytes (pellet). The mussel shells were opened by cutting the adductor muscle with a sterile scalpel, and tissue samples, including the mantle, gills, adductor muscle, foot, gonad, and hepatopancreas, were collected using sterile instruments. Samples were washed with sterile PBS, preserved in 1 mL of RNA preservation solution, rapidly frozen in liquid nitrogen, and stored at -80°C.
Aeromonas hydrophila was purchased from Beina Biological Co., while lipopolysaccharides (LPS) were obtained from Macklin Biochemical Co. Staphylococcus aureus, Aeromonas veronii, and Escherichia coli strains were sourced from our laboratory. Acid phosphatase assay kit and Alkaline phosphatase assay kit were purchased from Nanjing Jiancheng Bioengineering Institute.
2.2 Obtaining and bioinformatics analysis of full-length cDNA of HcLec1
The 3’ and 5’ UTRs of HcLec1 were amplified using the SMARTER® RACE 5’/3’ Kit (Takara Bio, USA) with primers listed in Table 1. The amplified product was purified using a gel extraction kit (Beyotime, China), then ligated into the pMD19-T vector (Takara, Japan) at 16°C for 4 h, followed by transformation into DH5α competent cells for positive clone selection. Sequencing was performed by GENEWIZ (Suzhou, China). The full-length cDNA sequence was obtained by aligning the 3’ and 5’ UTR sequences. Domain and signal peptide prediction, amino acid composition, and secondary structure prediction were carried out using the SMART platform (http://smart.embl-heidelberg.de/), the ExPASy website (https://web.expasy.org/protparam/), and the PRABI website (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma_f.html), respectively.
2.3 Immune challenge, RNA extraction, and cDNA synthesis
To examine the tissue expression profile of HcLec1 and its expression changes before and after nucleus insertion in the triangle sail mussel, six untreated mussels and six mussels 14 days post-insertion were randomly selected, and various tissues were collected. Sampling and preservation methods are described in Section 2.1. The mussels were divided into three groups: a PBS control group, an Aeromonas hydrophila injection group, and an LPS injection group. A. hydrophila was cultured overnight in broth at 37°C, centrifuged at 4000 rpm for 20 minutes, and resuspended in PBS. Approximately 1 × 107 cells were injected into the adductor muscle of each mussel. For the LPS group, 100 μL of LPS diluted in PBS to a concentration of 1 mg/mL was injected into the adductor muscle, while the PBS control group was injected with 100 μL of sterile PBS. At 0, 6, 12, 24, and 48 h post-injection, six mussels from each group were randomly selected, and hemocytes, mantle, and gill tissues were collected. Sampling and preservation methods are detailed in Section 2.1. RNA was extracted using the TRIzol method. RNA concentration was measured with a NanoDrop 2000c (Thermo Fisher Scientific, USA). The extracted RNA was reverse-transcribed into cDNA using the Evo M-MLV RT Mix Kit, followed by real-time quantitative PCR analysis.
2.4 Real-time quantitative PCR
Quantitative analysis was conducted using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara, Japan) on a Bio-Rad CFX96 system (Bio-Rad, Hercules, CA, USA) with a reaction volume of 20 μL. EF-1α was used as the reference gene, with primers listed in Table 1. A negative control was included for each primer pair. Relative expression level analysis was performed with 3–6 biological replicates and three technical replicates. The group with the lowest expression or the control group was used for normalization, and gene expression levels were evaluated using the 2-ΔΔCT method.
2.5 In vivo RNA interference assay
Three dsRNA-HcLec1 constructs with T7 promoters at the 5’ end and one dsRNA-pEGFP construct with a T7 promoter at the 5’ end were designed. These constructs were transcribed into dsRNA using the T7 High Efficiency Transcription Kit and diluted to a concentration of 300 ng/μL. The three interference strands were designated as Chain 1, Chain 2, and Chain 3. The triangle sail mussels were divided into a negative control (NC) group and a dsRNA-HcLec1 injection group. Injections of 100 μL EGFP and 100 μL dsRNA-HcLec1 were administered into the adductor muscle of the mussels. After 24, 48, and 72 h post-injection, mantle tissue samples were collected. qRT-PCR and the 2-ΔΔCT method were used to assess knockdown efficiency, with normalization against the negative control group.
2.6 Construction of recombinant plasmids and protein induction in vitro
The CDS region of HcLec1 was ligated into the pET-32a plasmid (without the thrombin site) (from our laboratory) using a seamless cloning kit via the EcoRI restriction site. The recombinant plasmid was transformed into competent Origami (DE3) cells, which were cultured at 37°C, 200 rpm until the OD600 reached 0.6–0.8. IPTG was then added to a final concentration of 0.5 mM, and the cells were induced overnight at 16°C, 180 rpm. SDS-PAGE gel electrophoresis was performed to assess the induction of recombinant protein expression. The induced protein was subsequently purified using a His-Tag Protein Purification Kit (Beyotime, China) and analyzed by SDS-PAGE. The recombinant HcLec1 proteins are referred to as rHcLec1 throughout the study.
2.7 Western blot
After SDS-PAGE, the protein bands were transferred onto a methanol-activated PVDF membrane using a semi-dry transfer method. The PVDF membrane was then blocked with 10% non-fat milk diluted in TBST at room temperature for 1 h, followed by incubation with a 1:1000 dilution of mouse anti-His tag monoclonal antibody (Beyotime, China) at 4°C overnight. After three washes with TBST, the membrane was incubated with a 1:1000 dilution of HRP-conjugated goat anti-mouse IgG secondary antibody at room temperature for 1 h. Following three additional washes with TBST, the membrane was developed, and detection was performed using a versatile gel imaging system (Bio-Rad, Hercules, CA, USA).
2.8 Detection of binding activity of rHcLec1 and PAMPs
The binding activity of rHcLec1 with LPS and peptidoglycan (PGN) was assessed using an ELISA kit. In a 96-well plate, 5 μL of LPS and PGN at a concentration of 80 μg/mL were added to each well and incubated at 37°C overnight. Subsequently, 50 μL of 2% BSA (1 mg/mL) was added to each well and incubated at room temperature for 2 h, followed by five washes with TBS. In the negative control group, 50 μL of rTrx (recombinant Thioredoxin) was added, while 50 μL of rHcLec1 protein was added to the experimental group, and incubation occurred at 37°C for 2 h. Each well was then treated with 100 μL of anti-His-tag mouse monoclonal antibody (1:1000 dilution) and 100 μL of HRP-conjugated sheep anti-mouse secondary antibody (1:1000 dilution), incubated at 37°C for 2 h, and washed five times with TBS. Color development was achieved by adding 100 μL of TMB substrate solution for 30 minutes, followed by 100 μL of TMB stop solution. Absorbance was measured at 450 nm. A result was considered positive if (P [sample] - B [blank])/(N [negative] - B [blank]) > 2.1. Each experiment was performed in triplicate.
2.9 Bacterial inhibition test
Staphylococcus aureus, Escherichia coli, Aeromonas veronii, and Aeromonas hydrophila were cultured to the logarithmic growth phase (15) and diluted to a concentration of 1 × 104 CFU/mL in TBS solution, followed by inoculation into a 96-well plate. For the experimental group, 50 μL of rHcLec1 (200 μg/mL) was added to 50 μL of bacterial solution, while the control group received 50 μL of TBS solution. The mixtures were incubated at 37°C for 2 h. Subsequently, 20 μL of each mixture was transferred to a nutrient broth medium for further incubation. OD600 absorbance values were measured at 12 h using a microplate reader.
2.10 Bacterial agglutination test
Bacteria in the logarithmic growth phase were collected for Giemsa staining, and the stained microorganisms were suspended in TBS buffer. A 10 μL aliquot of the microbial suspension was incubated with 25 μL of rHcLec1 (200 μg/mL) dissolved in TBS buffer or with 25 μL of TBS buffer (control group) at room temperature for 1 h. Observations were made using an optical microscope.
2.11 In Vitro growth of crystals in the presence of rHcLec1
A saturated calcium carbonate solution was prepared following the method of Zhenguang Yan et al. (30). With continuous stirring, 30 mL of 100 mM sodium bicarbonate was added dropwise to 120 mL of 40 mM calcium chloride solution until the solution became cloudy, at which point stirring was immediately stopped. The pH was adjusted to 8.2 with NaOH, and the solution was filtered through a 0.22 μm filter. To assess the effect of rHcLec1 on calcite crystal morphology in vitro, the recombinant protein was incubated with the prepared saturated calcium carbonate solution at 4°C for 48 h. Crystals were examined using scanning electron microscopy, and their crystal forms were analyzed by Raman spectroscopy. The experiment included a blank control group (300 μL saturated calcium carbonate solution only), a negative control group (300 μL saturated calcium carbonate solution with protein elution buffer), and an experimental group (300 μL saturated calcium carbonate solution with 50 μL of rHcLec1 at 1.0 mg/mL).
2.12 Statistical analysis
The graphs were plotted using GraphPad Prism 9. Data analysis was performed using ANOVA and the t-test in SPSS 26.0. Tukey’s test was used for multiple comparisons, with P < 0.05 indicating significant differences.
3 Results
3.1 Cloning and sequence analysis of HcLec1
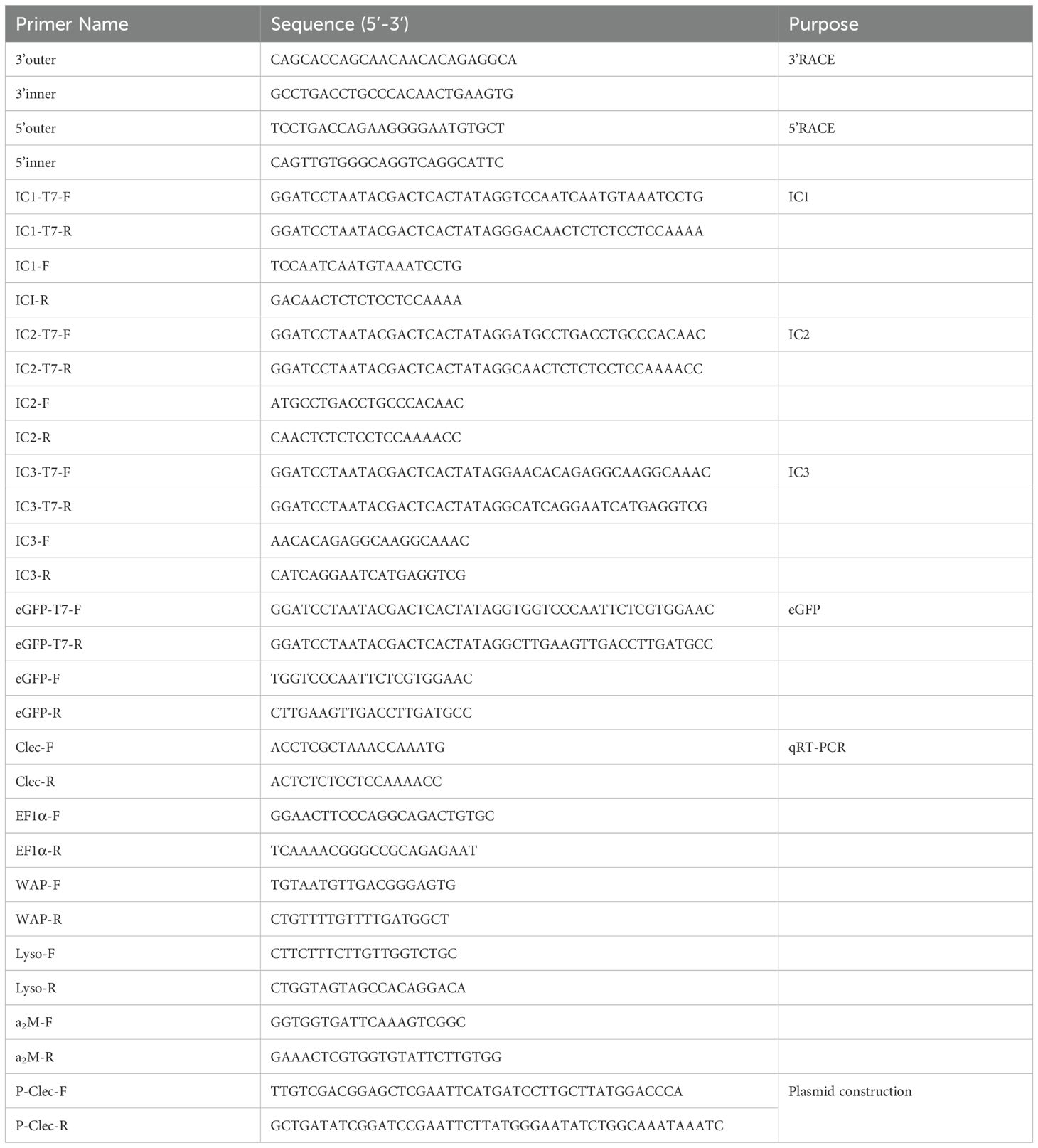
Cloning results revealed that the full-length cDNA of HcLec1 is 1552 bp (Figure 1A), with a 3’ UTR of 770 bp, a 5’ UTR of 59 bp, and a CDS of 723 bp, encoding 240 amino acids. Among these, Glycine (Gly) accounts for 8.3%, followed by Glutamic acid (Glu) and Leucine (Leu) at 7.9% each (Figure 1C). The structure of HcLec1 is different from that of previous CTLs in H. cumingii Lea. SMART analysis predicted that the HcLec1 protein contains an N-terminal signal peptide and a carbohydrate recognition domain (CRD) with QPD (Gln-Pro-Asp) and MND (Met-Asn-Asp) motifs for polysaccharide binding (Figure 1B). The predicted molecular weight of the HcLec1 protein is 27.03 kDa, with a secondary structure comprising 37.5% random coils and 35.83% alpha helices (Figure 1D). The GenBank accession number is PP056159.
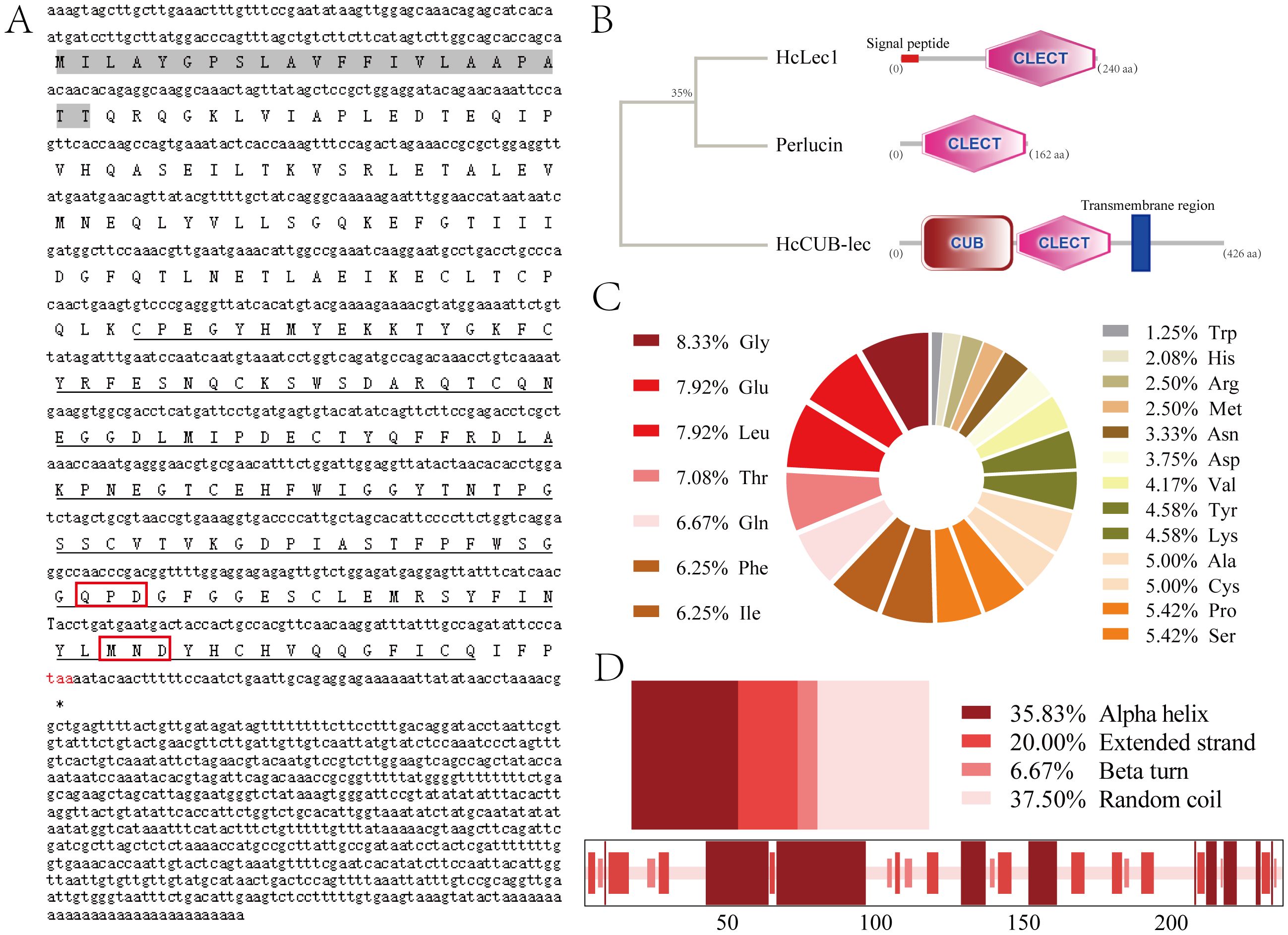
Figure 1. Bioinformatic analysis of HcLec1. (A) Full-length HcLec1 gene obtained by cloning. The gray background indicates the signal peptide, the black dashed line represents the conserved domains, the red box highlights the polysaccharide-binding motifs, and the red “taa” indicates the stop codon. (B) Structural domain prediction of HcLec1 and comparison of HcLec1 with other CTL sequences from H. cumingii Lea. The “CLECT” denotes the C-type lectin domain. (C) Amino acid composition of the HcLec1 protein. (D) Proportions of secondary structure elements in the HcLec1 protein.
3.2 Expression profile of HcLec1 in various tissues of H. cumingii Lea
The qPCR results demonstrated that HcLec1 is expressed in all tissues of H. cumingii Lea, with significantly higher relative expression levels observed in the gill tissues of untreated mussels compared to other tissues, followed by the mantle (P < 0.05, Figure 2B). Fourteen days after the nucleus implantation is a critical period for the formation of the pearl sac. During this time, the smooth muscle in the mantle undergoes specialization into columnar epithelial cells, forming a cavity (the pearl sac) that encloses the foreign object. The specialized pearl sac tissue at this stage is crucial for the study of biomineralization (Figure 2A). The quantitative analysis revealed that HcLec1 expression levels were significantly elevated (P < 0.01) in the gill tissue, mantle tissue, pearl sac (specialized mantle tissue), adductor muscle, and gonads (Figure 2B). Specifically, following nucleus implantation, the relative expression levels of HcLec1 in the mantle, gills, adductor muscle, and gonads increased approximately 4.6-, 10-, 17-, and 3-fold, respectively, with no significant changes detected in the hepatopancreas, foot, or hemocytes (P > 0.05). Although hemocyte-mediated immunity is a crucial component of innate immunity in mollusks, our results revealed that HcLec1 exhibited the lowest relative expression in hemocytes. These findings suggest that HcLec1 may primarily play an immune role in the gills, while exerting a biomineralization function in the mantle and pearl sac.
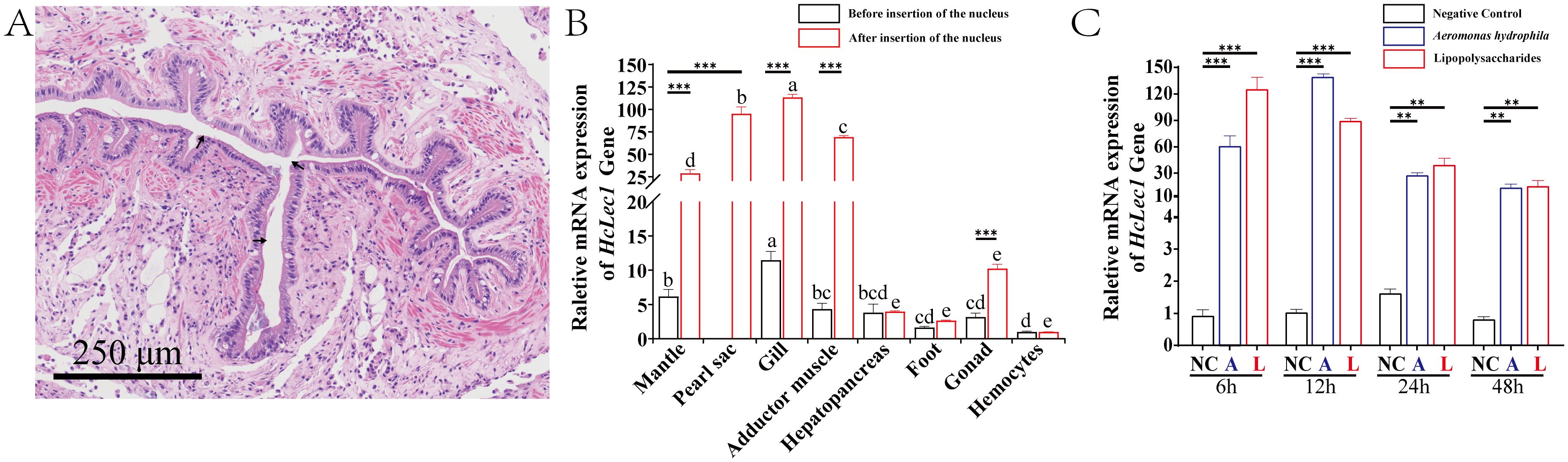
Figure 2. HcLec1 tissue expression profile and challenge assay. (A) HE-stained schematic showing pearl sac formation after nucleus implantation, with black arrows indicating the cavity formed by the pearl sac. (B) Relative expression levels of HcLec1 in various tissues of untreated and nucleus-implanted mussels. Different lowercase letters indicate significant differences between tissues within the same group (P < 0.05). (C) Relative expression levels of HcLec1 following bacterial challenge. NC represents the negative control group (injected with sterile PBS), while A and L represent the A. hydrophila and LPS injection groups, respectively. ** denotes highly significant differences (P < 0.01), and *** denotes extremely significant differences (P < 0.001).
3.3 A. hydrophila and LPS stimulated HcLec1 expression
The relative expression of HcLec1 increased sharply and significantly following stimulation with A. hydrophila and LPS (P < 0.01), peaking at 6 h and 12 h post-stimulation. Expression levels significantly decreased at 24 h and 48 h but remained substantially higher than those of the control group (P < 0.01, Figure 2C). Under A. hydrophila and LPS stimulation, HcLec1 expression increased approximately 60-fold and 120-fold compared to the control group at 6 h and about 140-fold and 90-fold at 12 h. By 24 h, expression sharply decreased (P < 0.01), and at 48 h, expression levels further declined but remained 14–15 times higher than the NC group (P < 0.01). These findings suggest that HcLec1 is rapidly upregulated upon stimulation with A. hydrophila and LPS, with peak expression occurring earlier following LPS stimulation compared to A. hydrophila.
3.4 HcLec1 regulates immune and mineralization-related genes and enzyme activities
The results of HcLec1 relative expression after RNAi demonstrated that all three dsRNAs achieved significant knockdown efficiency at 48 h and 72 h (P < 0.001), with Chain 2 showing a 90% knockdown at 72 h (Figure 3A). Consequently, Chain 2 was selected for subsequent RNAi experiments. Interference with HcLec1 using Chain 2 significantly reduced the expression levels of immune-related genes WAP, α2m, and Lyso (P < 0.01, Figure 3B). CA, CHS, Nacrein, and Pif are key matrix proteins in H. cumingii Lea (30–32). After HcLec1 knockdown, the relative expression levels of mineralization-related genes CA, CHS, and Pif were significantly reduced by 10–20 fold (P < 0.01, Figure 3C), while Nacrein expression decreased by 30% (P < 0.05). ACP and ALP activities are crucial for biomineralization (33). This study revealed that ACP and ALP enzyme activities were significantly downregulated following HcLec1 interference (P < 0.05). ACP activity decreased by 35% in serum and 22% in hemocytes (Figure 3D), whereas ALP activity showed a significant 40% reduction in both serum and hemocytes (Figure 3E). These findings suggest that HcLec1 knockdown adversely impacts both immune and biomineralization processes in H. cumingii Lea.
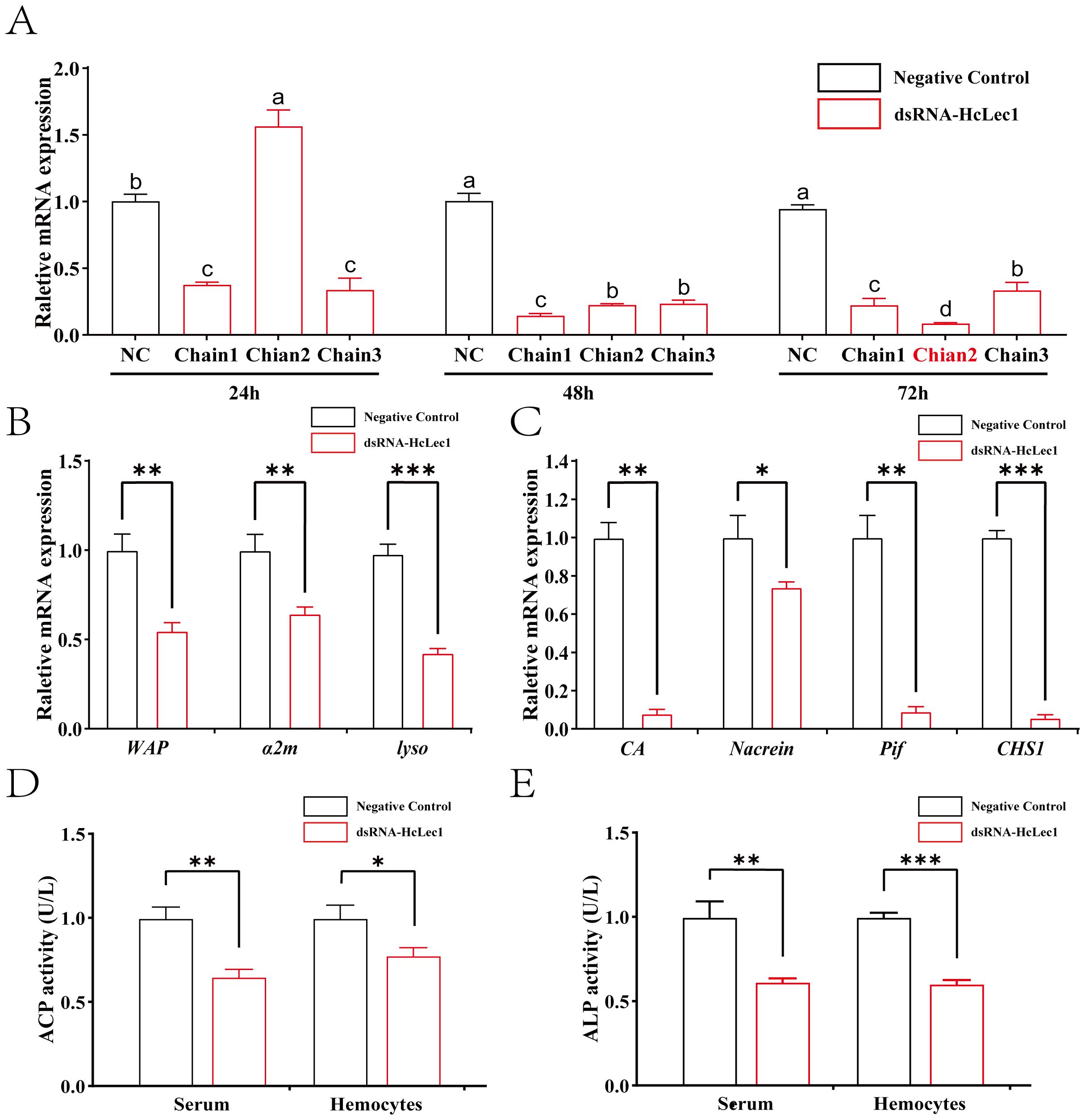
Figure 3. HcLec1 RNAi Experiment. (A) Effects of three dsRNA-HcLec1 sequences. NC represents the negative control group (injected with eGFP), while Chain1, Chain2, and Chain3 correspond to the experimental groups (each injected with one of the three dsRNA-HcLec1 sequences). Different lowercase letters indicate significant differences between the same groups at different times (P < 0.05). (B, C) display relative expression changes in immune- and mineralization-related genes following HcLec1 knockdown. (D, E) depict alterations in ACP and ALP enzyme activities after HcLec1 knockdown. Black bars represent the control group (injected with eGFP), and red bars represent the experimental group (injected with dsRNA-HcLec1). * indicates P < 0.05, ** indicates P < 0.01, and *** indicates P < 0.001.
3.5 HcLec1 involvement in shell and pearl mineralization processes in H. cumingii Lea
In the shell damage experiment, cracks in the shell of H. cumingii Lea gradually develop into a thin membrane (immature shell (34)) over time, which subsequently mineralizes and heals (Figure 4A). Upon inserting a small piece of mantle tissue from the donor mussel (known as “saibo”), the recipient mantle is stimulated to form a pearl sac, which envelops the tissue and secretes nacre, leading to pearl formation (Figure 4C). Results show that during both shell repair and “saibo” insertion, HcLec1 expression follows a similar pattern: it decreases significantly initially and then rises steadily to reach control levels (P < 0.05, Figure 4B, D). Compared to the control, HcLec1 expression was significantly reduced on day 1 and remained lower than control levels from days 1 to 7 (P < 0.05). By day 14, HcLec1 expression significantly increased to 1.5–1.6 times the control level. Days 10 to 15 represent the phase of membrane formation and subsequent hardening at the shell damage site, as well as the initial formation of the pearl sac at the insertion site. On days 21 and 24, HcLec1 expression levels declined and stabilized, showing no significant difference from the control group (P > 0.05). These findings suggest that HcLec1 plays a role in both shell repair and pearl sac formation processes in H. cumingii Lea.
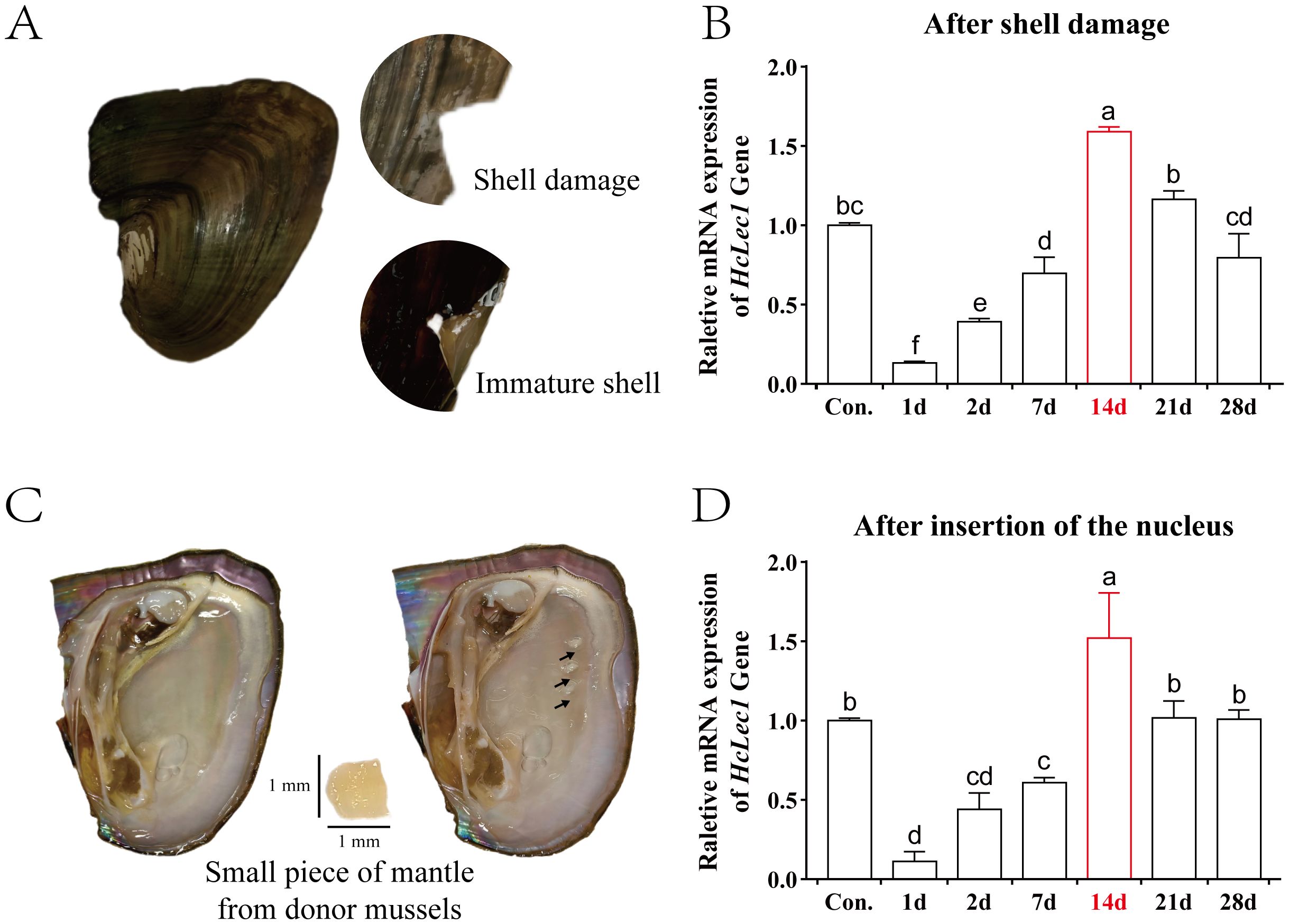
Figure 4. Relative Expression Changes of HcLec1 in H. cumingii Lea During Mineralization. (A) Procedure and observations of the shell damage experiment in H. cumingii Lea. (B) Relative expression trend of HcLec1 during shell repair following damage. (C) Nucleus insertion procedure in H. cumingii Lea, with the black arrow indicating the insertion site. (D) Relative expression trend of HcLec1 after nucleus insertion, where "Con." represents the control group at day 0. Different lowercase letters indicate significant differences between groups (P < 0.05).
3.6 Recombinant HcLec1 protein inhibits bacterial growth and agglutinates bacteria by recognizing PAMPs
To investigate the response of HcLec1 protein to bacteria in vitro, we successfully induced the recombinant HcLec1 protein (referred to as rHcLec1, Figure 5A). The results indicated that rHcLec1 can recognize and bind both LPS and PGN, with a higher binding affinity for PGN compared to LPS (Figure 5B). This finding supports the role of rHcLec1 as a PRR that binds to PAMPs. PGN, a key component of the cell walls in both Gram-positive and Gram-negative bacteria, suggests that rHcLec1 has broad-spectrum antibacterial activity, allowing it to recognize both bacterial types. Bacterial inhibition assays showed that rHcLec1 significantly suppressed the growth of E. coli, S. aureus, A. veronii, and A. hydrophila, reducing their growth rates by 43%, 53%, 18%, and 26%, respectively, when added to the growth medium (Figure 5C). Giemsa staining revealed a clear agglutination effect of rHcLec1 on these four bacterial species (Figure 5D). This suggests that, as a C-type lectin, rHcLec1 may inhibit bacterial growth not only by activating immune-related genes but also by directly inducing bacterial agglutination.
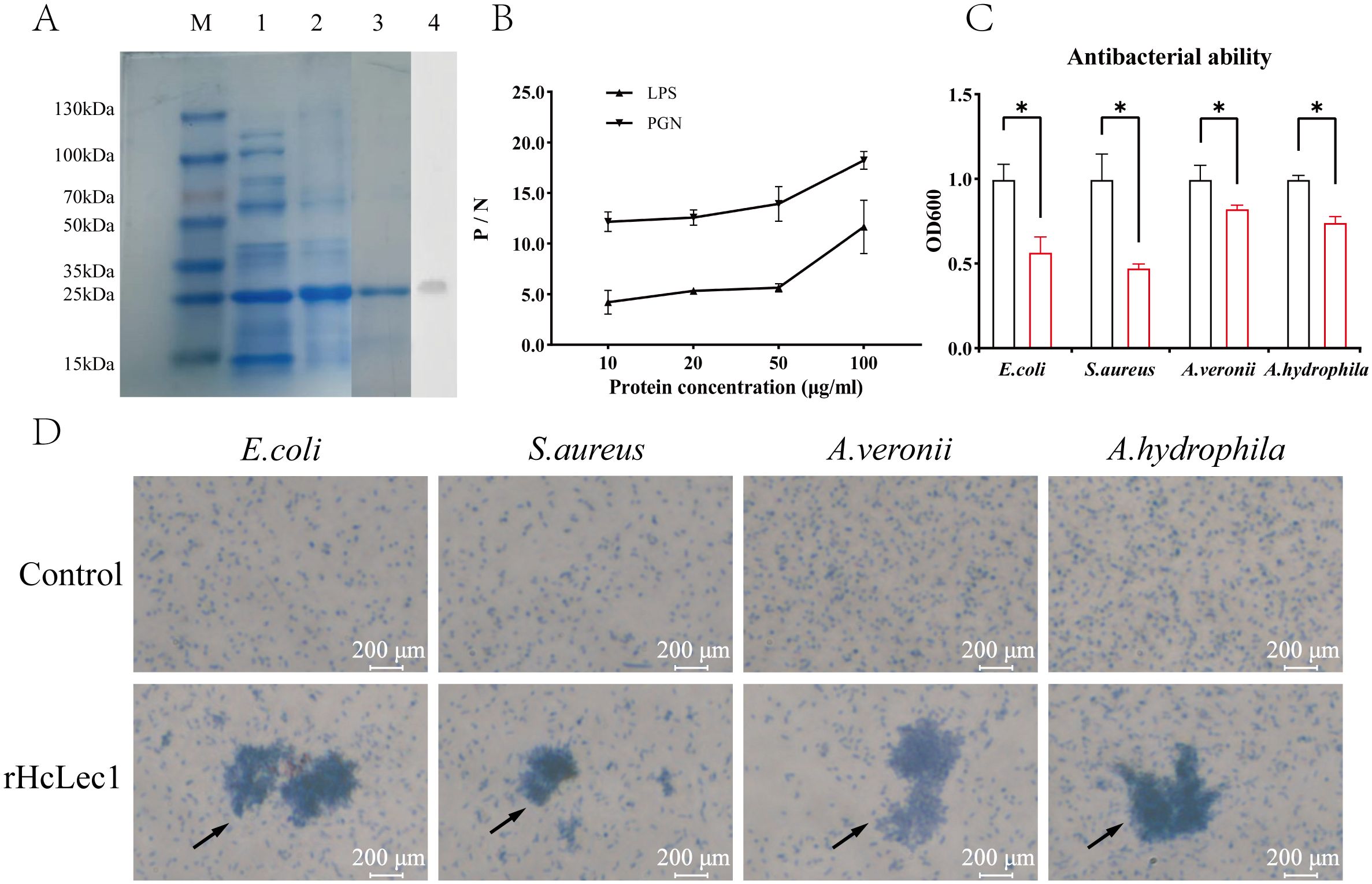
Figure 5. Prokaryotic Expression and Antibacterial Activity Analysis of rHcLec1. (A) SDS-PAGE and Western blot validation of rHcLec1. Lane M: Marker; Lane 1: Expression of rHcLec1 in the supernatant; Lane 2: Expression of rHcLec1 in the precipitate; Lane 3: Fourth elution of rHcLec1; Lane 4: Western blot validation results. (B) Binding activity of rHcLec1 to PGN and LPS. (C) Antibacterial effect of rHcLec1, with red bars representing the experimental group (200 μg/mL of rHcLec1) and black bars representing the control group (equivalent concentration of TBS). (D) Agglutination effect of rHcLec1 on S. aureus, E. coli, A. veronii, and A. hydrophila. The experimental group (rHcLec1, 200 μg/mL) is labeled as "rHcLec1," and the control group (TBS at the same concentration) as "Control." Black arrows indicate areas of significant agglutination. * indicates P < 0.05.
3.7 rHcLec1 promotes calcite formation in vitro
The mineralization capacity of bivalves is primarily reflected in calcium carbonate deposition and the regulation of the morphology and properties of calcium carbonate crystals. After co-culturing at 4°C for 48 h, rectangular crystals were observed in the solutions of both the blank and negative control groups (Figure 6A). Compared to the control group, crystal growth was significantly enhanced in the presence of rHcLec1. At the same magnification, crystals formed under the influence of rHcLec1 protein were several times larger than those in the control group and exhibited significant fusion. Raman spectroscopy analysis of the crystals indicated nearly identical spectral bands across all three groups (Figure 6B). Two main bands were identified at 279.34–281.16 and 1083.67–1085.50, along with two weaker bands at 154.34–159.36 and 708.68–712.31, which are characteristic of calcite (35–37). These results suggest that rHcLec1 can promote calcite formation and fusion.
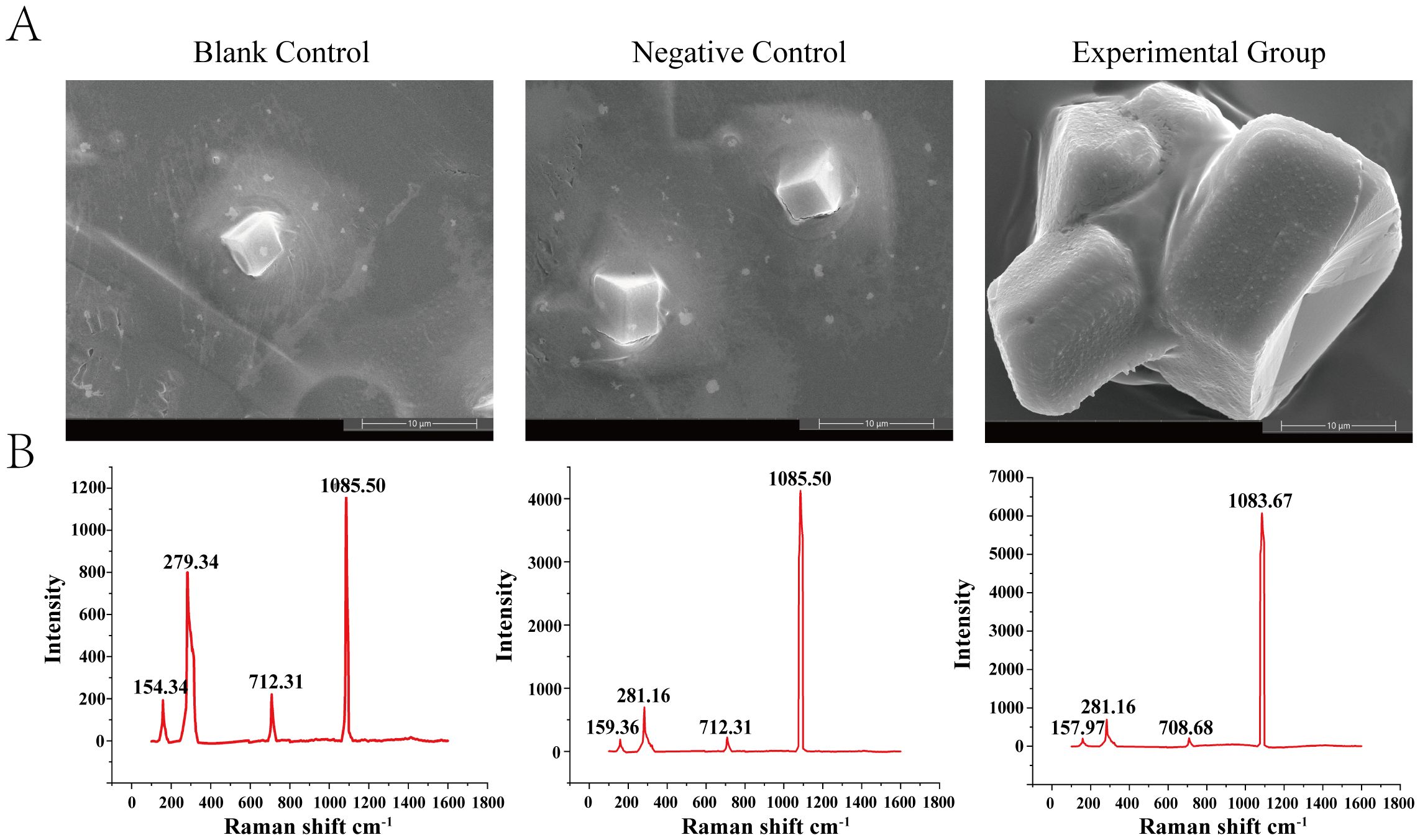
Figure 6. Role of rHcLec1 in the in vitro crystallization of calcium carbonate. (A) Results from scanning electron microscopy. (B) Corresponding Raman spectroscopy results. The Blank Control contains only a saturated calcium carbonate solution, the Negative Control includes protein elution buffer, and the Experimental Group includes 50 μL of rHcLec1 (1.0 mg/mL).
4 Discussion
C-type lectins are among the largest families of pattern recognition receptors (PRRs), activating the immune system against invading pathogens through the recognition of pathogen-associated molecular patterns (PAMPs). They are widely distributed in both vertebrates and invertebrates; however, in invertebrates, they are generally not conserved. This structural diversity has resulted in a wide array of functions within the CTL family in invertebrates. In mollusks, CTLs have been reported to serve various functions. Beyond their well-known roles in innate immunity (38), recognizing and binding to PAMPs (39), and agglutinating bacteria (40), they had also been shown to perform many other functions. In the echinoderm Strongylocentrotus purpuratus, it was found that sea urchin spicule matrix proteins, such as SpSM29, SpC-lectin, SM30, and PM27, contain CTL carbohydrate recognition domains (CRDs), which are involved in post-spicule formation (23, 24). In mollusks, a CTL named MeML was found in the labial palps of Crassostrea virginica and Mytilus edulis, and it may be involved in the selection of food particles in suspension-feeding bivalves (41, 42). In Ruditapes philippinarum, CTLs are thought to have potential functions in responding to cold stress (43). Additionally, Perlucin, a purified CTL from Haliotis discus and H. cumingii Lea, has been shown to play a role in altering the morphology of calcium carbonate crystals and in nacre formation (26, 29).
In this study, a C-type lectin (HcLec1) from the freshwater pearl mussel H. cumingii Lea was isolated, identified, and cloned. Structurally simple, HcLec1 contained only a signal peptide and a CRD, making it suitable for functional studies as a representative C-type lectin. The results of this study demonstrated that HcLec1 was expressed across various tissues, with the lowest expression in hemocytes. This finding was similar to those for Perlucin in Haliotis discus discus and FcLec4 in Fenneropenaeus chinensis (15, 26), suggesting tissue-specific expression of CTLs (44, 45). Huang et al. found that the mantle of Pinctada fucata contains a high concentration of CTLs, with more immune genes expressed in the mantle than in hemocytes, a pattern similar to that of HcLec1 (46). The gill tissue in H. cumingii Lea is a key site for exchange with the external water environment and for pathogen defense (16, 47, 48). Thus the high expression of HcLec1 in the gill implied its potential role in innate immunity.
The CRD of CTLs contained carbohydrate-binding motifs that were critical for recognizing and binding to PAMPs (29, 39, 49). The CRD of HcLec1 contained two carbohydrate-binding motifs (QPD and MND), and results from injection stimulation and ELISA indicated that these motifs responded to A. hydrophila and LPS and could bind to both LPS and PGN. LPS and PGN are representative PAMPs of Gram-negative and Gram-positive bacteria, respectively (7). Our findings also showed that rHcLec1 could directly agglutinate Gram-positive bacteria (S. aureus) and Gram-negative bacteria (E. coli, A. veronii, and A. hydrophila) in vitro, without the addition of Ca2+, indicating that rHcLec1’s agglutination function is calcium-independent. This result was consistent with the functional studies of SPL-1 in Saxidomus purpuratus and CfLec-2 in Chlamys farreri (50, 51). Additionally, HcLec1 was found to directly inhibit the growth of these four bacterial species in vitro, suggesting that HcLec1 not only enhanced bacterial phagocytosis through complement system activation in vivo but also directly bound to and agglutinates bacteria, affecting their proliferation.
The pearl formation process in H. cumingii Lea can be divided into three main stages: an initial immune rejection stage following the implantation of a small mantle tissue piece from the donor mussel into the recipient’s mantle, a stage where the connective tissues of both fuse and specialize into a pearl sac, and finally, a mineralization stage where nacre is secreted and deposited to form the pearl (52, 53). Throughout this process, both the mantle and pearl sac serve as critical mineralizing tissues in H. cumingii Lea, with the pearl sac being particularly essential for pearl cultivation (21, 54, 55). The significant expression of HcLec1 in both the mantle and pearl sac suggested that HcLec1 may play roles not only in immunity but also in mineralization. Our results indicated that the expression pattern of HcLec1 aligned with the mineralization process during nucleus implantation and shell self-repair. The 10–14 day period is critical for pearl sac and shell repair, as the pearl sac nearly forms, and new shell material is deposited, marking a peak in mineralization activity (29, 56, 57).
CTLs have been shown to participate in in vitro mineralization and guide crystal formation. The protein Perlucin from Haliotis discus discus and Haliotis laevigata has been demonstrated to regulate crystal formation by either promoting or inhibiting calcium carbonate crystal formation and controlling crystal morphology and orientation (25, 26, 58, 59). Our study corroborated these findings: rHcLec1 was observed to promote calcite formation in vitro and facilitate the fusion of multiple calcite crystals into a larger, more complex structure. Morphologically, the resulting crystals appeared as a larger aggregate formed by the fusion of several smaller crystals, with a connecting and fused region between them. Although we did not investigate further whether this fusion is due to rHcLec1 producing a glue-like substance or embedding multiple crystals together, previous research has shown that P60 in Pinctada fucata can bind crystals together, suggesting that rHcLec1 may have a similar function in crystal aggregation (60). The formation of pearls depends on the combined action of foreign materials, calcium carbonate, and proteins (61). Many proteins can randomly or directionally regulate the formation of crystal morphology and size. Currently, the cultivation and harvesting of pearls cannot be controlled artificially. Therefore, protein-mediated formation of high-quality pearls may be a future research direction (62, 63). rHcLec1, as a protein identified from H. cumingii Lea with dual functions in immunity and biomineralization, could provide new molecular materials and a foundation for forming high-quality pearls by influencing crystal formation.
In summary, we demonstrated that HcLec1 in Hyriopsis cumingii Lea participated in innate immunity, could agglutinate and inhibit bacterial growth in vitro, promoted calcite crystal formation and fusion, and played a significant role in the biomineralization process of the mussel. This study may provide a molecular basis for shell repair and pearl growth, offering new materials and insights for biomineralization applications.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/2647031310, nuccore/2647031310.
Author contributions
XC: Investigation, Validation, Writing – original draft, Writing – review & editing. XS: Software, Writing – original draft, Writing – review & editing. SL: Conceptualization, Methodology, Writing – review & editing. WL: Data curation, Formal analysis, Investigation, Supervision, Validation, Writing – original draft, Writing – review & editing. HW: Formal analysis, Software, Visualization, Writing – review & editing. JL: Formal analysis, Funding acquisition, Resources, Supervision, Writing – review & editing. ZB: Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China 296 (2022YFD2400105), the earmarked fund for China Agriculture Research System of MOF and 297 MARA (CARS-49), the National Natural Science Foundation of China (No. 31201991).
Conflict of interest
Author SL was employed by the company Shanghai MugaoBiotechnology co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zhong L, Xu BH, Yan DF, Xiao TY, Liu QL. Pathogen isolation and pathologic observation on explosive epidemics of hyriopsis cumingii lea. Turkish J Fisheries Aquat Sci. (2016) 16:935–45. doi: 10.4194/1303-2712-v16_4_21
2. Bai ZY, Yin YX, Hu SN, Wang GL, Zhang XW, Li JL. Identification of genes involved in immune response, microsatellite, and SNP markers from expressed sequence tags generated from hemocytes of freshwater pearl mussel (Hyriopsis cumingii). Mar Biotechnol. (2009) 11:520–30. doi: 10.1007/s10126-008-9163-0
3. Bouallegui Y. Immunity in mussels: An overview of molecular components and mechanisms with a focus on the functional defenses. Fish Shellfish Immunol. (2019) 89:158–69. doi: 10.1016/j.fsi.2019.03.057
4. Burgos-Aceves MA, Faggio C. An approach to the study of the immunity functions of bivalve haemocytes: Physiology and molecular aspects. Fish Shellfish Immunol. (2017) 67:513–7. doi: 10.1016/j.fsi.2017.06.042
5. Rumpret M, von Richthofen HJ, Peperzak V, Meyaard L. Inhibitory pattern recognition receptors. J Exp Med. (2021) 219(1):e20211463. doi: 10.1084/jem.20211463
6. Teixeira MM, Almeida IC, Gazzinelli RT. Introduction: innate recognition of bacteria and protozoan parasites. Microbes Infection. (2002) 4:883–6. doi: 10.1016/s1286-4579(02)01625-8
7. Medzhitov R, Janeway CA. Decoding the patterns of self and nonself by the innate immune system. Science. (2002) 296:298–300. doi: 10.1126/science.1068883
8. Robinson MJ, Sancho D, Slack EC, LeibundGut-Landmann S, Sousa CRE. Myeloid C-type lectins in innate immunity. Nat Immunol. (2006) 7:1258–65. doi: 10.1038/ni1417
9. Richardson MB, Williams SJ. MCL and Mincle: C-type lectin receptors that sense damaged self and pathogen-associated molecular patterns. Front Immunol. (2014) 5:288. doi: 10.3389/fimmu.2014.00288
10. Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. (2005) 272:6179–217. doi: 10.1111/j.1742-4658.2005.05031.x
11. Wang XW, Wang JX. Diversity and multiple functions of lectins in shrimp immunity. Dev Comp Immunol. (2013) 39:27–38. doi: 10.1016/j.dci.2012.04.009
12. Zelensky AN, Gready JE. Comparative analysis of structural properties of the C-type-lectin-like domain (CTLD). Proteins-Structure Funct Genet. (2003) 52:466–77. doi: 10.1002/prot.10626
13. Yu XQ, Kanost MR. Immulectin-2, a lipopolysaccharide specific lectin from an insect, Manduca sexta, is induced in response to gram-negative bacteria. J Biol Chem. (2000) 275:37373–81. doi: 10.1074/jbc.M003021200
14. Wang XW, Zhang HW, Li XA, Zhao XF, Wang JX. Characterization of a C-type lectin (PcLec2) as an upstream detector in the prophenoloxidase activating system of red swamp crayfish. Fish Shellfish Immunol. (2011) 30:241–7. doi: 10.1016/j.fsi.2010.10.012
15. Wang XW, Zhang XW, Xu WT, Zhao XF, Wang JX. A novel C-type lectin (FcLec4) facilitates the clearance of Vibrio Anguillarum in vivo in Chinese white shrimp. Dev Comp Immunol. (2009) 33:1039–47. doi: 10.1016/j.dci.2009.05.004
16. Wang H, Song LS, Li CH, Zhao JM, Zhang H, Ni DJ, et al. Cloning and characterization of a novel C-type lectin from Zhikong scallop Chlamys farreri. Mol Immunol. (2007) 44:722–31. doi: 10.1016/j.molimm.2006.04.015
17. Shi L, Liu X, Mao J, Han X. Study of coloration mechanism of cultured freshwater pearls from mollusk hyriopsis cumingii. J Appl Spectrosc. (2014) 81:97–101. doi: 10.1007/s10812-014-9893-x
18. Li JL, Wang GL, Bai ZY. Genetic diversity of freshwater pearl mussel (Hyriopsis cumingii) in populations from the five largest lakes in China revealed by inter-simple sequence repeat (ISSR). Aquacult Int. (2009) 17:323–30. doi: 10.1007/s10499-008-9204-8
19. Wu HL, Yang CY, Hao RJ, Liao YS, Wang QH, Deng YW. Lipidomic insights into the immune response and pearl formation in transplanted pearl oyster Pinctada fucata martensii. Front Immunol. (2022) 13:1018423. doi: 10.3389/fimmu.2022.1018423
20. Yang CY, Yang JM, Hao RJ, Du XD, Deng YW. Molecular characterization of OSR1 in Pinctada fucata martensii and association of allelic variants with growth traits. Aquaculture. (2020) 516:734617. doi: 10.1016/j.aquaculture.2019.734617
21. Mariom, Take S, Igarashi Y, Yoshitake K, Asakawa S, Maeyama K, et al. Gene expression profiles at different stages for formation of pearl sac and pearl in the pearl oyster Pinctada fucata. BMC Genomics. (2019) 20:240. doi: 10.1186/s12864-019-5579-3
22. Li XA, Feng SL, Xuan XR, Wang H, Shen XY, Chen YG, et al. A proteomic approach reveals biomineralization and immune response for mantle to pearl sac in the freshwater pearl mussel (Hyriopsis cumingii). Fish Shellfish Immunol. (2022) 127:788–96. doi: 10.1016/j.fsi.2022.06.057
23. Wilt FH, Killian CE, Livingston BT. Development of calcareous skeletal elements in invertebrates. Differentiation. (2003) 71:237–50. doi: 10.1046/j.1432-0436.2003.7104501.x
24. Rao A, Seto J, Berg JK, Kreft SG, Scheffner M, Cölfen H. Roles of larval sea urchin spicule SM50 domains in organic matrix self-assembly and calcium carbonate mineralization. J Struct Biol. (2013) 183:205–15. doi: 10.1016/j.jsb.2013.06.004
25. Blank S, Arnoldi M, Khoshnavaz S, Treccani L, Kuntz M, Mann K, et al. The nacre protein perlucin nucleates growth of calcium carbonate crystals. J Microscopy-Oxford. (2003) 212:280–91. doi: 10.1111/j.1365-2818.2003.01263.x
26. Wang N, Lee YH, Lee JH. Recombinant perlucin nucleates the growth of calcium carbonate crystals:: Molecular cloning and characterization of perlucin from disk abalone, Haliotis discus discus. Comp Biochem Physiol B-Biochem Mol Biol. (2008) 149:354–61. doi: 10.1016/j.cbpb.2007.10.007
27. Zhao LL, Wang YQ, Dai YJ, Zhao LJ, Qin QW, Lin L, et al. A novel C-type lectin with four CRDs is involved in the regulation of antimicrobial peptide gene expression in Hyriopsis cumingii. Fish Shellfish Immunol. (2016) 55:339–47. doi: 10.1016/j.fsi.2016.06.007
28. Huang Y, Ren Q. HcCUB-Lec, a newly identified C-type lectin that contains a distinct CUB domain and participates in the immune defense of the triangle sail mussel Hyriopsis cumingii. Dev Comp Immunol. (2019) 93:66–77. doi: 10.1016/j.dci.2018.12.012
29. Lin JY, Ma KY, Bai ZY, Li JL. Molecular cloning and characterization of perlucin from the freshwater pearl mussel, Hyriopsis cumingii. Gene. (2013) 526:210–6. doi: 10.1016/j.gene.2013.05.029
30. Xiang L, Su JT, Zheng GL, Liang J, Zhang GY, Wang HZ, et al. Patterns of expression in the matrix proteins responsible for nucleation and growth of aragonite crystals in flat pearls of pinctada fucata. PloS One. (2013) 8(6):e66564. doi: 10.1371/journal.pone.0066564
31. Zhang WL, Wang YD, Andrew E, Martin G. Characterization of two carbonic anhydrase isoforms in the pulmonate snail (Lymnaea Stagnalis) and their involvement in Molluskan calcification. Comp Biochem Physiol B-Biochem Mol Biol. (2025) 275: 111028. doi: 10.1016/j.cbpb.2024.111028
32. Suman S, Pavan-Kumar A, Saurabh S, Katneni VK, Prabhudas SK, Varshney S, et al. Transcriptome analysis of mantle and pearl sac of freshwater pearl mussel Lamellidens marginalis to identify biomineralisation-associated genes. Molluscan Res. (2024) 44:107–17. doi: 10.1080/13235818.2024.2324466
33. Zhao LQ, Lu YN, Yang F, Liang J, Deng YW. Transgenerational biochemical effects of seawater acidification on the Manila clam (Ruditapes philippinarum). Sci Total Environ. (2020) 710: 136420. doi: 10.1016/j.scitotenv.2019.136420
34. Li M, Tang JY, Yuan MQ, Huang BY, Liu YQ, Wei L, et al. Outer fold is sole effective tissue among three mantle folds with regard to oyster shell colour. Int J Biol Macromol. (2023) 241:124655. doi: 10.1016/j.ijbiomac.2023.124655
35. Kontoyannis CG, Orkoula MG, Koutsoukos PG. Quantitative analysis of sulfated calcium carbonates using Raman spectroscopy and X-ray powder diffraction. Analyst. (1997) 122:33–8. doi: 10.1039/a606167b
36. Fu PG, Zheng HF. Raman spectra of aragonite and calcite at high temperature and high pressure. Spectrosc Spectral Anal. (2013) 33:1557–61. doi: 10.3964/j.issn.1000-0593(2013)06-1557-05
37. Rivera-Perez C, Flores-Sánchez IA, de Areyano J, Posadas DIR, Hernández-Saavedra NY. A shell matrix protein of Pinctada mazatlanica produces nacre platelets in vitro. Sci Rep. (2020) 10: 20201. doi: 10.1038/s41598-020-77320-7
38. Zhu L, Song LS, Xu W, Qian PY. Molecular cloning and immune responsive expression of a novel C-type lectin gene from bay scallop Argopecten irradians. Fish Shellfish Immunol. (2008) 25:231–8. doi: 10.1016/j.fsi.2008.05.004
39. Yang JL, Huang MM, Zhang H, Wang LL, Wang H, Wang LL, et al. CfLec-3 from scallop: an entrance to non-self recognition mechanism of invertebrate C-type lectin. Sci Rep. (2015) 5:10068. doi: 10.1038/srep10068
40. Lan TY, Li Z, Peng MX, Niu DH, Li Y, Li JLA. four-CRD C-type lectin from razor clam Sinonovacula constricta mediates agglutination and phagocytosis. Gene. (2020) 728: 144287. doi: 10.1016/j.gene.2019.144287
41. Espinosa EP, Eckstein M, Allam B. Density of compatible ligands on the surface of food particles modulates sorting efficiency in the blue mussel mytilus edulis. Front Mar Sci. (2022) 9:882356. doi: 10.3389/fmars.2022.882356
42. Espinosa EP, Perrigault M, Ward JE, Shumway SE, Allam B. Lectins associated with the feeding organs of the oyster crassostrea virginica can mediate particle selection. Biol Bull. (2009) 217:130–41. doi: 10.1086/BBLv217n2p130
43. Li DD, Nie HT, Jahan K, Yan XW. Expression analyses of C-type lectins (CTLs) in Manila clam under cold stress provide insights for its potential function in cold resistance of Ruditapes philippinarum. Comp Biochem Physiol C-Toxicol Pharmacol. (2020) 230: 108708. doi: 10.1016/j.cbpc.2020.108708
44. Luo T, Zhang XB, Shao ZZ, Xu X. PmAV, a novel gene involved in virus resistance of shrimp Penaeus monodon. FEBS Lett. (2003) 551:53–7. doi: 10.1016/s0014-5793(03)00891-3
45. Sun YD, Fu LD, Jia YP, Du XJ, Wang Q, Wang YH, et al. hepatopancreas-specific C-type lectin from the Chinese shrimp Fenneropenaeus chinensis exhibits antimicrobial activity. Mol Immunol. (2008) 45:348–61. doi: 10.1016/j.molimm.2007.06.355
46. Huang JL, Li L, Jiang TF, Xie LP, Zhang RQ. Mantle tissue in the pearl oyster Pinctada fucata secretes immune components via vesicle transportation. Fish Shellfish Immunol. (2022) 121:116–23. doi: 10.1016/j.fsi.2022.01.001
47. Tassanakajon A, Somboonwiwat K, Supungul P, Tang S. Discovery of immune molecules and their crucial functions in shrimp immunity. Fish Shellfish Immunol. (2013) 34:954–67. doi: 10.1016/j.fsi.2012.09.021
48. Wang YY, Chen Y, Cao ML, Wang XQ, Wang GL, Li JL. Identification of wnt2 in the pearl mussel Hyriopsis cumingii and its role in innate immunity and gonadal development. Fish Shellfish Immunol. (2021) 118:85–93. doi: 10.1016/j.fsi.2021.08.022
49. Song KK, Li DF, Zhang MC, Yang HJ, Ruan LW, Xu X. Cloning and characterization of three novel WSSV recognizing lectins from shrimp Marsupenaeus japonicus. Fish Shellfish Immunol. (2010) 28:596–603. doi: 10.1016/j.fsi.2009.12.015
50. Zheng PL, Wang H, Zhao JM, Song LS, Qiu LM, Dong CH, et al. A lectin (CfLec-2) aggregating Staphylococcus haemolyticus from scallop Chlamys farreri. Fish Shellfish Immunol. (2008) 24:286–93. doi: 10.1016/j.fsi.2007.11.014
51. Unno H, Higuchi S, Goda S, Hatakeyama T. Novel carbohydrate-recognition mode of the invertebrate C-type lectin SPL-1 from Saxidomus purpuratus revealed by the GlcNAc-complex crystal in the presence of Ca 2+. Acta Crystallographica Section F-Structural Biol Commun. (2020) 76:271–7. doi: 10.1107/s2053230x20007256
52. Huang DD, Shen JX, Li JL, Bai ZY. Integrated transcriptome analysis of immunological responses in the pearl sac of the triangle sail mussel (Hyriopsis cumingii) after mantle implantation. Fish Shellfish Immunol. (2019) 90:385–94. doi: 10.1016/j.fsi.2019.05.012
53. Shen WY, Hun YW, He ZK, Xu ST, Xu XY, Zhang GF, et al. Histological and comparative transcriptome analyses provide insights into the immune response in pearl sac formation of hyriopsis cumingii. Front Mar Sci. (2020) 7:256. doi: 10.3389/fmars.2020.00256
54. Wang XY, Liu SF, Xie LP, Zhang RQ, Wang Z. Pinctada fucata mantle gene 3 (PFMG3) promotes differentiation in mouse osteoblasts (MC3T3-E1). Comp Biochem Physiol B-Biochem Mol Biol. (2011) 158:173–80. doi: 10.1016/j.cbpb.2010.11.004
55. Blay C, Planes S, Ky CL. Cultured pearl surface quality profiling by the shell matrix protein gene expression in the biomineralised pearl sac tissue of pinctada margaritifera. Mar Biotechnol. (2018) 20:490–501. doi: 10.1007/s10126-018-9811-y
56. Kishore P, Southgate PC. A detailed description of pearl-sac development in the black-lip pearl oyster, Pinctada margaritifera (Linnaeus 1758). Aquacult Res. (2016) 47:2215–26. doi: 10.1111/are.12674
57. Sillanpää JK, Ramesh K, Melzner F, Sundh H, Sundell K. Calcium mobilisation following shell damage in the Pacific oyster, Crassostrea gigas. Mar Genomics. (2016) 27:75–83. doi: 10.1016/j.margen.2016.03.001
58. Weiss IM, Kaufmann S, Mann K, Fritz M. Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevigata. Biochem Biophys Res Commun. (2000) 267:17–21. doi: 10.1006/bbrc.1999.1907
59. Weber E, Weiss IM, Cölfen H, Kellermeier M. Recombinant perlucin derivatives influence the nucleation of calcium carbonate. Crystengcomm. (2016) 18:8439–44. doi: 10.1039/c6ce01878e
60. Lao YX, Zhang XQ, Zhou J, Su WW, Chen RJ, Wang YG, et al. Characterization and in vitro mineralization function of a soluble protein complex P60 from the nacre of Pinctada fucata. Comp Biochem Physiol B-Biochem Mol Biol. (2007) 148:201–8. doi: 10.1016/j.cbpb.2007.05.010
61. Jin C, Zhao JY, Liu XJ, Li JL. Expressions of shell matrix protein genes in the pearl sac and its correlation with pearl weight in the first 6Months of pearl formation in hyriopsis cumingii. Mar Biotechnol. (2019) 21:240–9. doi: 10.1007/s10126-019-09876-z
62. Koleini MM, Raee MJ, Tamaddon AM, Salmanpour M. Mechanistic insights of amino acid binding to hydroxyapatite: molecular dynamics charts future directions in biomaterial design. Langmuir. (2024) 40:22136–44. doi: 10.1021/acs.langmuir.4c02537
Keywords: C-type lectin, immunity, biomineralization, calcite formation, bacterial agglutination, shell repair
Citation: Chen X, Shen X, Liu S, Li W, Wang H, Li J and Bai Z (2025) A c-type lectin HcLec1 with dual function of immunology and mineralization from the freshwater oyster (Hyriopsis cumingii Lea). Front. Immunol. 15:1530732. doi: 10.3389/fimmu.2024.1530732
Received: 19 November 2024; Accepted: 26 December 2024;
Published: 14 January 2025.
Edited by:
Guirong Wang, Upstate Medical University, United StatesReviewed by:
Zhi Liao, Zhejiang Ocean University, ChinaLiang Haiying, Guangdong Ocean University, China
Copyright © 2025 Chen, Shen, Liu, Li, Wang, Li and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjuan Li, d2psaUBzaG91LmVkdS5jbg==; Zhiyi Bai, enliYWlAc2hvdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Xiaofeng Chen
Xiaofeng Chen Xiaoya Shen
Xiaoya Shen Shijun Liu
Shijun Liu Wenjuan Li
Wenjuan Li He Wang
He Wang Jiale Li
Jiale Li Zhiyi Bai
Zhiyi Bai