- 1Department of Urology, Taizhou Central Hospital (Taizhou University Hospital), Taizhou, Zhejiang, China
- 2Department of Urology, First Affiliated Hospital of Gannan Medical University, Ganzhou, Jiangxi, China
- 3Department of Pathology, The Third Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
Urological malignant tumors pose a significant threat to human health, with a high incidence rate each year. Prostate cancer, bladder cancer, and renal cell carcinoma are among the most prevalent and extensively researched urological malignancies. Despite advancements in research, the prognosis for these tumors remains unfavorable due to late detection, postoperative recurrence, and treatment resistance. A thorough investigation into their pathogenesis is crucial for early diagnosis and treatment. Recent studies have highlighted the close association between microRNAs (miRNAs) and cancer progression. miRNAs are small non-coding RNAs composed of 19-23 nucleotides that regulate gene expression by binding to the 3’ untranslated region (3’UTR) of target mRNAs, impacting key cellular processes such as proliferation, differentiation, apoptosis, and migration. Dysregulation of miRNAs can disrupt the expression of oncogenes and tumor suppressor genes, contributing to cancer development. Among the various miRNAs studied, miR-152 has garnered attention for its role in urological malignancies. Several studies have indicated that dysregulation of miR-152 expression is significant in these cancers, warranting a comprehensive review of the evidence. This review focuses on the expression and function of miR-152 in prostate cancer, bladder cancer, and renal cell carcinoma, elucidating its mechanisms in cancer progression and exploring its potential as a therapeutic target and biomarker in urological malignancies.
Introduction
Urological malignancies encompass prostate cancer, bladder cancer, renal cell carcinoma, penile cancer, testicular cancer, and uroepithelial cancer. According to GLOBOCAN, there were approximately 2.5 million new cases of urological malignancies globally in 2022, representing 12.6% of all cancers. These cases resulted in 770,000 deaths, accounting for 8% of all cancer-related deaths (1). The number of new cases and deaths of urinary malignant tumors in 2022 has exceeded the number published by GLOBOCAN in 2020 (2). Prostate cancer, bladder cancer, and renal cell carcinoma are among the most prevalent and extensively researched urological malignancies. Prostate cancer, the second most common cancer in men, was responsible for 1,466,680 new cases and 396,792 deaths in 2022. The majority of patients with prostate cancer progress to desmoplasia-resistant prostate cancer, leading to a poor prognosis. Bladder cancer, the second most common urological malignancy, ranked ninth in terms of incidence in 2022 (1) Renal cell carcinoma, although less common than prostate and bladder cancer, saw 434,419 new cases and 155,702 deaths in 2022, ranking 14th among all cancers (1). The incidence of urological malignancies is expected to rise globally due to population growth and aging (3). These malignancies pose a significant burden on global health and finances, with the United States estimated to have spent $314.7 billion in 2020 on the prevention and treatment of urological malignancies (4).
Early urological malignancies are often treated with surgical resection, which boasts a high success rate. However, the lack of specific symptoms in the early stages leads to most patients being diagnosed at an advanced stage. Additionally, some non-invasive biomarkers, like prostate specific antigen (PSA), have limitations in the early diagnosis of prostate cancer, such as low specificity and overdiagnosis (5, 6). Urine cytological testing is even less sensitive for diagnosing bladder cancer, while cystoscopic biopsy is invasive and inconvenient (7). This highlights the urgent need for sensitive diagnostic methods to aid in early urological tumour diagnosis. For patients with advanced cancer, chemotherapy and radiotherapy are commonly used to slow disease progression and improve quality of life. Unfortunately, cancer often develops resistance to these treatments after several courses, posing a significant challenge (8). Despite recent advancements in immunotherapy and targeted therapies, challenges persist due to drug-resistant gene mutations post-targeted therapies, immune adverse effects post-immunotherapy, and limited use of targeted drugs due to high costs. Thus, there is a pressing need to develop new therapeutic strategies for urological tumours, necessitating a comprehensive understanding of their pathogenesis.
In the past two decades, the study of miRNAs has sparked a molecular revolution, with numerous studies demonstrating their significant role in cancer. miRNAs impact essential cellular processes like cell proliferation, differentiation, cell cycle regulation, invasion, metastasis, and angiogenesis by modulating the expression of target genes. Given the close connection between these processes and cancer, miRNAs are intricately linked to cancer development. Among the well-researched miRNAs in oncology, miR-152 stands out for its consistent oncogenic role across various cancer types, including colorectal (9, 10), gastric (11, 12), hepatocellular (13, 14), lung (15–17), breast (18, 19), ovarian (20, 21), cervical (22, 23), and glioma (24, 25). While studies on the relationship between miR-152 and urological tumors abound, a comprehensive review on this subject is lacking. This paper aims to fill this gap by examining the impact of miR-152 on biological behaviors of cancer cells in urological tumors, such as proliferation, invasion, metastasis, angiogenesis, and apoptosis. It also explores the potential of miR-152 as a biomarker for early diagnosis, treatment, and prognosis of urological tumors, as well as its therapeutic implications in miRNA-based cancer treatments. By delving into the role of miR-152, this review seeks to enhance our understanding of the pathogenesis and therapeutic strategies for urological malignancies.
Overview of miRNAs
miRNAs were initially discovered in 1993 in the nematode Caenorhabditis elegans (26), Since then, a growing number of miRNAs have been identified and characterized, with over 28,000 miRNAs currently known across a variety of organisms (27). These small RNAs, approximately 19-23 nucleotides in length, are evolutionarily conserved and play a crucial role in gene expression regulation. They typically bind to the 3’ untranslated region (3’ UTR) of target mRNA, leading to either degradation or inhibition of translation (28, 29). In some rare instances, miRNAs can enhance translation of target genes, contributing to post-transcriptional gene regulation (3, 30, 31). It is estimated that miRNAs regulate at least one-third of all genes (32). The biogenesis of miRNA is an extremely complex process, as shown in Figure 1.
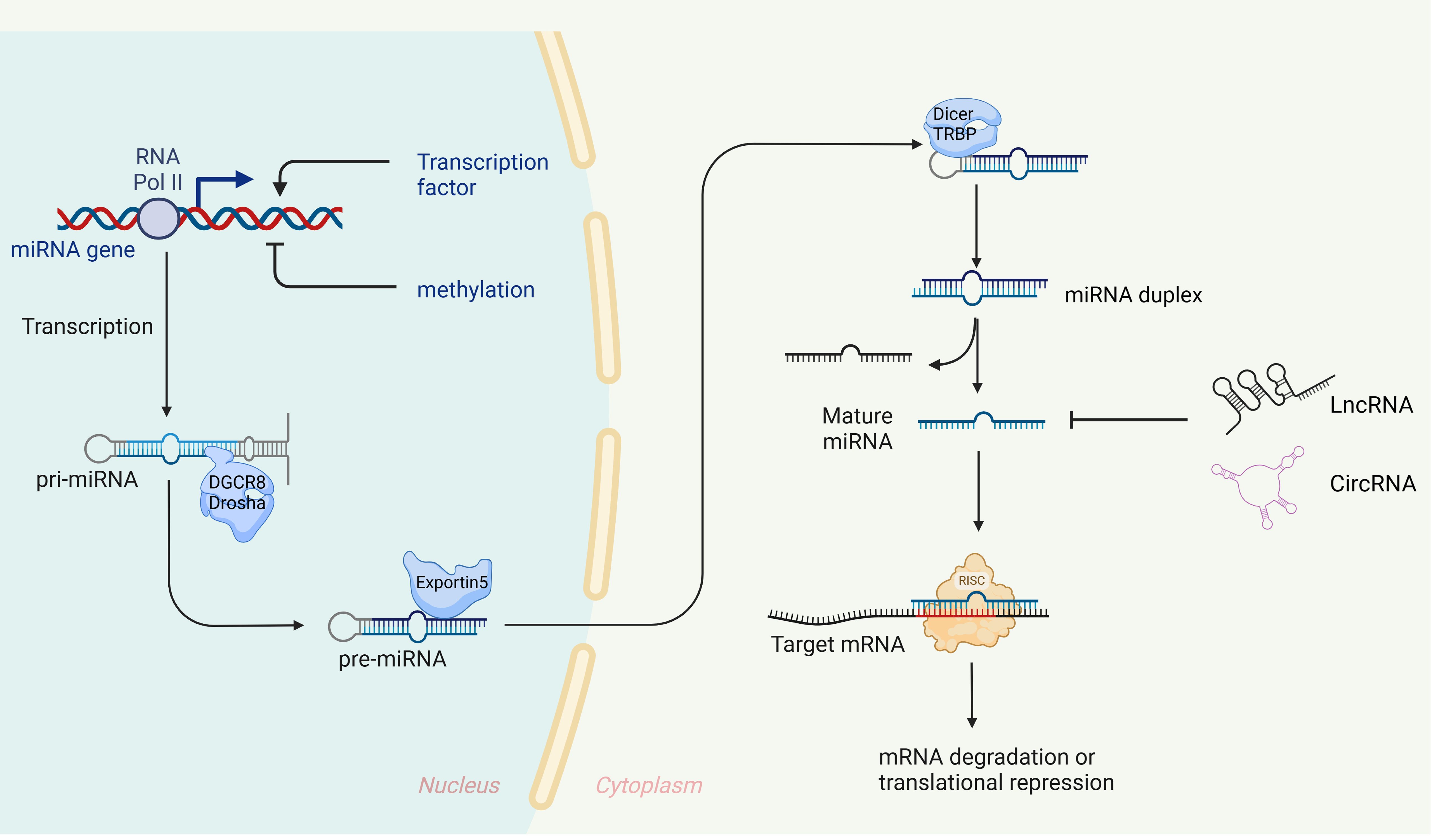
Figure 1. Biogenesis and expression regulation of miRNAs. In the nucleus, primary miRNA (pri-miRNA), which possesses specific structural characteristics, is transcribed by RNA polymerase II. These pri-miRNAs are then cleaved by a microprocessor complex, composed of Drosha and its cofactor DGCR8, to generate precursor miRNAs (pre-miRNAs) of a defined length (120–123), The pre-miRNAs are transported to the cytoplasm via Exportin-5 (124, 125), where Dicer and TRBP further process them into double-stranded RNA molecules (126, 127). These molecules then unwind to form mature single-stranded miRNAs, with one strand designated as ‘5p’ and the other as ‘3p’. Finally, these mature miRNAs are integrated into the RNA-induced silencing complex (RISC) with the assistance of argonaut 2 protein (AGO2) (128). miRNAs with nucleotide sequences 2-7, known as ‘seed’ sequences, are directed by AGO2 to recognize and bind the 3’UTR of target mRNA (129), When fully complementary, the target mRNA is degraded (130), when not fully complementary, translation is inhibited (32).
miRNAs play a crucial role in regulating cellular processes, with their dysregulation linked to various diseases, including cancer. The connection between cancer and dysregulated miRNA expression was initially observed in patients with chronic lymphocytic leukaemia (CML), where the absence of chromosomal regions coding for miR15 and miR16 was noted (33). Cancer-associated miRNAs can be broadly categorized into oncogenic miRNAs (oncomiRs) and tumor suppressor miRNAs (miRsupps) (34, 35). OncomiRs promote cancer progression by influencing cell proliferation, apoptosis, and other cancer-related processes, typically exhibiting overexpression in cancers (36). Conversely, miRsupps impede malignant behaviors in cancer cells, thus hindering cancer progression, and are usually underexpressed in cancer (37). The roles of specific miRNAs can vary across different cancer types. For instance, miR-155 was found to be overexpressed in renal cell carcinoma, promoting proliferation and metastasis (38), while in malignant melanoma, its overexpression inhibited cancer progression (39). In addition, miR-137 functions as a tumor suppressor in various cancers, including lung cancer (40), gastric cancer (41), ovarian cancer (42), endometrial cancer (42), and nervous system tumors (43). However, some studies have indicated that miR-137 can also act as an oncomiR, promoting cancer progression (44, 45). Notably, the same miRNA may exhibit contradictory roles within the same cancer, potentially influenced by factors such as technological variations, diverse targets, and cancer subtypes.
Function and expression regulation of miR-152
miR-152, a member of the miR-148/152 family along with miR-148a and miR-148b, was initially discovered in the mouse colon through tissue-specific cloning (46), These three members share the same seed sequence, leading to the targeting of genes with immediate homology. Variations in reported miRNA targets across studies could be attributed to the focus on individual miRNAs within the family without considering others (47). Moreover, the 3’-terminal non-seeded sequence of miRNAs plays a role in stabilizing their binding to target genes (48), suggesting that the miR-148/152 family has the potential to target different mRNAs based on their distinct non-seeded sequences.
Aberrant expression of miR-152 has been observed in various oncological and non-oncological diseases. For instance, Nielsen et al. identified 12 upregulated human miRNAs, including miR-152, in the serum of patients with type 1 diabetes (49). Moreover, increased miR-152 expression has been detected in atheromatous plaque tissue and classical monocytes (50). Research has shown that miR-152 overexpression promotes lipid accumulation in preadipocytes, correlating with in vivo adipogenesis and intramuscular adipogenesis, indicating its potential relevance in obesity and obesity-related metabolic syndrome (51). Furthermore, up-regulation of miR-152-5p expression was noted in the mandible of a rat osteoporosis model, suggesting its involvement in regulating osteogenic differentiation by targeting the autophagy-related protein ATG14. These findings collectively demonstrate the diverse functions of miR-152 and contribute to our understanding of its significance in non-tumour diseases.
miR-152 has been extensively studied in various types of tumors, showing reduced expression and potential as a tumor suppressor miRNA. For instance, in hepatocellular carcinoma, miR-152-3p was significantly down-regulated in tumor tissues compared to non-tumor tissues. Overexpression of miR-152-3p inhibited ROBO1, blocking the malignant characteristics of hepatocellular carcinoma (14). Similarly, in gastric cancer, reduced expression of miR-152-3p led to inhibited proliferation, clone formation, migration, and induced apoptosis in cancer cells (52). In cholangiocarcinoma, miR-152 targeted DNMT1, with reduced expression in cancer cells (53). Notably, miR-152 was also found to be down-regulated in ovarian cancer and correlated with the malignant phenotype of ovarian cancer cells (54). Reduced miR-152 expression was associated with poorer survival outcomes in endometrial plasma adenocarcinoma (55). Furthermore, in lung cancer miR-152 targets TNS1 to regulate the Akt/mTOR/RhoA pathway, inhibiting the progression of non-small cell lung cancer (15). In breast cancer, miR-152 inhibits cell survival and promotes apoptosis by targeting EPAS1, enhancing the sensitivity of breast cancer cells to paclitaxel (56). Kong et al. found that miR-152 inhibits glioma progression and tumorigenesis by targeting FBXL7, and increases the cytotoxicity of temozolomide-induced glioma cells (24). Overall, miR-152 shows decreased expression in digestive system tumors, reproductive system tumors, lung cancer, breast cancer, and glioma, with this decreased expression correlating with the malignant phenotype of cancer cells. Restoring miR-152 expression inhibits cancer progression and increases the sensitivity of cancer cells to chemotherapeutic agents, indicating its role as a tumor-suppressor miRNA in various cancers. Therefore, miR-152 may serve as a potential target in cancer therapy.
The expression and activity of miR-152 are tightly regulated both temporally and spatially in normal physiological conditions. Disruption of this regulation is closely linked to various human diseases, such as cancer growth and metastasis (57). Several mechanisms can lead to dysregulation of miRNAs, including epigenetic changes, abnormal binding of transcription factors, interference with miRNA production, post-transcriptional modifications of RNA, and RNA degradation (37, 58). There is growing evidence of epigenetic connections between DNA methylation changes and miR-152 levels in cancer. miR-152 is often silenced in different types of cancer, like prostate (59), bladder (60), colorectal (61), breast (62) endometrial (63), and glioblastoma (25), due to DNA hypermethylation. This hypermethylation is driven by the increased activity of DNMT1, an enzyme responsible for maintaining DNA methylation levels. Interestingly, DNMT1 is a target of miR-152, and its levels are inversely related to miR-152 in many cancers (64, 65), suggesting a negative feedback loop between them. miR-152 expression is also influenced by various transcription factors, such as ELF1, which can enhance miR-152-3p levels by directly interacting with its promoter (17). In breast cancer, the activation of miR-152 expression is triggered by the translocation of the β-linker protein and PKM2 complex into the nucleus in response to IGF-1 (66). Moreover, the activity of miR-152 is regulated by competing endogenous RNAs (ceRNAs) that bind to miR-152 and reduce its inhibitory effects on downstream targets, primarily including certain lncRNAs, circRNAs, and pseudogene transcripts (67, 68).
miR-152 in prostate cancer
Among the studies focusing on miR-152 and urological tumors, prostate cancer has been the most extensively researched. Numerous studies have demonstrated that miR-152 is downregulated in prostate cancer tissues and cell lines, and its altered expression is intricately linked to the development of prostate cancer (Figure 2). Restoring miR-152 expression has been shown to impede the progression of prostate cancer by targeting and suppressing genes and signaling pathways that are crucial in cancer development. This inhibition ultimately hinders the advancement of prostate cancer. The subsequent discussion provides a detailed explanation of the role and mechanism of miR-152 in prostate cancer.
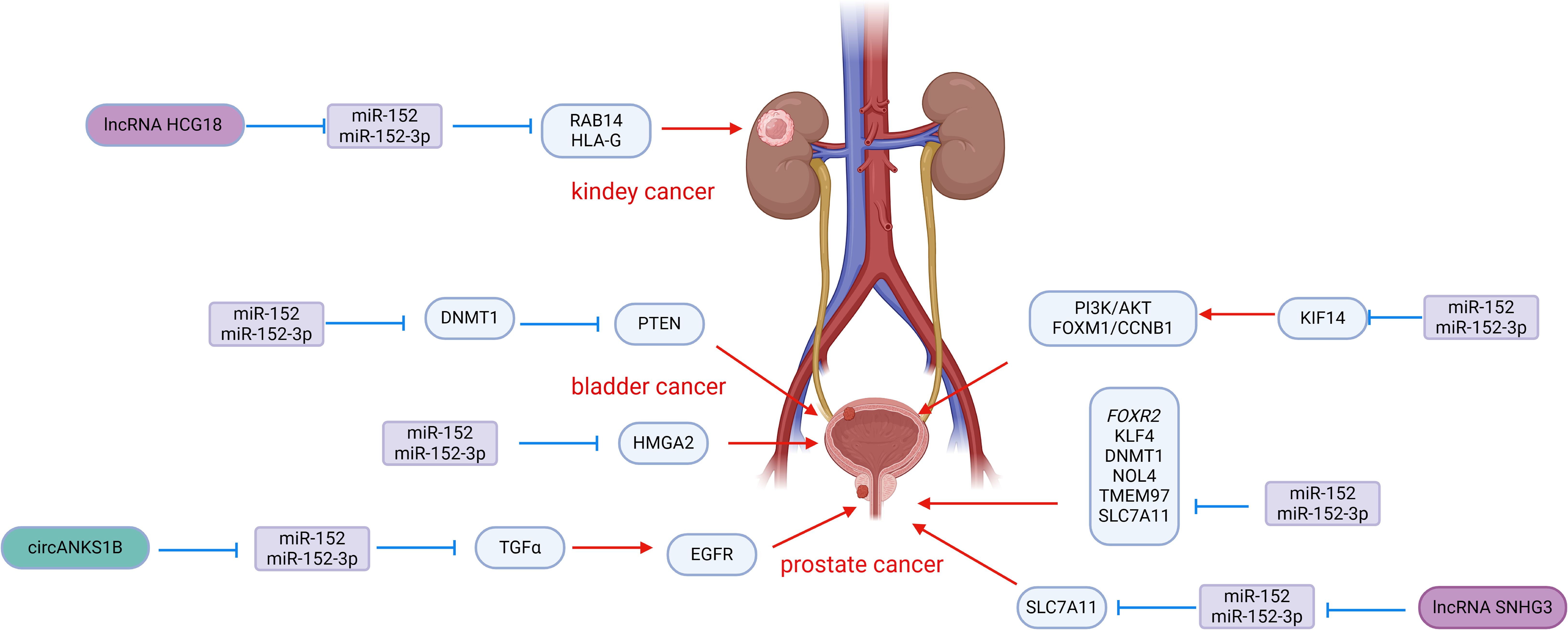
Figure 2. Roles of miR-152 and its targeted proteins and signaling cascades in the development of urologic malignancies.
Prostate cancer is known for its aggressive nature, with a strong invasive ability that allows it to infiltrate and metastasize to surrounding tissues, contributing to its poor prognosis and high rate of postoperative recurrence. Chen et al. conducted a study on miR-152 expression in 48 primary prostate cancers, finding a significant down-regulation compared to non-malignant control tissues. Specifically, patients with a Gleason score >7 showed lower levels of miR-152, which was also associated with the pathological stage of prostate cancer. Overexpression of miR-152 in PC-3 and DU145 cells effectively suppressed the invasive and migratory abilities of prostate cancer cells by targeting TGFα, a key player in EGFR signaling (69). The activation of EGFR is crucial for cell proliferation, angiogenesis, motility, and metastasis (70, 71), with previous research linking EGFR to invasion and metastasis in various tumors (72, 73). By inhibiting TGFα-EGFR signaling, miR-152 also downregulated downstream genes MMP2 and MMP9, further suppressing the invasive and migratory potential of prostate cancer cells in vitro (69). Interestingly, the tumor-suppressive effects of miR-152 could be counteracted by circANKS1B, which acts as a sponge for miR-152, binding to miR-152-3p and attenuating the inhibition of TGFα. This mechanism promotes the invasion and metastasis of prostate cancer cells, highlighting the intricate interplay between miR-152, TGFα, and circANKS1B in prostate cancer progression (74). Xu et al. reported that miR-152 targets the 3′UTR of FOXR2 in prostate cancer, leading to downregulation of FOXR2 expression, which in turn inhibits cell proliferation and induces apoptosis. Additionally, lncRNA HOTAIR was observed to interact with miR-152, resulting in upregulation of FOXR2 and further promoting prostate cancer progression (75).
Ramalho-Carvalho et al. employed a combined approach involving micro-RNA expression analysis and differential methylation localization to identify novel miRNAs downregulated by aberrant DNA methylation in prostate cancer. They discovered a transcription unit comprising COPZ2-miR-152-3p (76). Their study highlighted that the downregulation of miR-152-3p is a common characteristic of prostate cancer and plays a role in sustaining malignant traits and tumor growth. Functional assays conducted in vitro revealed that overexpression of miR-152-3p led to a significant decrease in cell viability in LNCaP and PC3 cells, along with an increase in cell accumulation in S and G2/M phases. Moreover, the overexpression of miR-152-3p resulted in the downregulation of several cell cycle regulators at the transcriptional level. Additionally, miR-152-3p overexpression suppressed the expression of epithelial-mesenchymal transition-related genes TWIST and VIM, leading to a notable reduction in the invasive capacity of PC3 cells. Conversely, miR-152-3p mimics induced apoptosis in both cell lines (76). Mechanistically, NOL4 (nucleolin 4) and TMEM97 (transmembrane protein 97) were identified as targets of miR-152-3p. TMEM97 has been found to be up-regulated in various malignant tumors, including prostate cancer (77, 78). miR-152-3p also down-regulates the expression of genes involved in the MAPK/ERK, TFG-Beta, JAK-STAT3, and EMT pathways, which are classical pathways in cancer.
Feng et al. discovered a synergistic effect of miR-148-3p and miR-152-3p in prostate cancer. Their study revealed a reduction in the expression of both miR-148-3p and miR-152-3p in prostate cancer cells. Functional assays conducted in vitro showed that the combined overexpression of miR-148-3p and miR-152-3p had a greater inhibitory impact on cell proliferation and apoptosis induction compared to individual overexpression. This suggests a synergistic relationship between miR-148-3p and miR-152-3p in inhibiting the growth of PC3 and LNCaP cells (79). Mechanistically, miR-148-3p and miR-152-3p, being part of the same family, share a common seed sequence and target the 3′ UTR of KLF4. By jointly inhibiting KLF4, miR-148-3p and miR-152-3p effectively impede the progression of prostate cancer. This synergistic effect was further validated in prostate cancer xenograft mice (79).
miR-152 is present in prostate cancer and is involved in a feedback pathway with DNMT1, serving as a target gene for miR-152. miR-152 functions by repressing DNMT1 expression post-transcriptionally. Conversely, increased expression of DNMT1 results in widespread hypermethylation of DNA, leading to a notable decrease in miR-152 expression (47). Theodore et al. observed hypermethylation of the promoter region in the LNCaP and PC-3 cell lines, resulting in the inactivation of miR-152 through hypermethylation in prostate cancer. They proposed that miR-152, which acts as a tumor suppressor, is inactivated by methylation, indicating that the epigenetic regulation of miR-152/DNMT1 may have a significant impact on the aggressiveness of prostate cancer (59). Similarly, Gurbuz et al. found a substantial increase in DNMT1 gene expression levels and a notable decrease in DNMT3b and PTEN expression in patients with prostate cancer and metastatic prostate cancer. Linear regression analysis revealed a significant relationship between miR-152 and DNMT1/DNMT3b/PTEN, although this study did not confirm a direct association of miR-152 with these genes (80).
Wang et al. found that lncRNA SNHG3 expression was markedly upregulated in prostate cancer tissues and cell lines, correlating with poor prognosis. Functional assays revealed that SNHG3 overexpression promoted proliferation, migration, and invasion of prostate cancer cells while inhibiting apoptosis (81). Mechanistically, SNHG3 acts as a molecular sponge for miR-152-3p, competitively modulating the inhibition of miR-152-3p on its target SLC7A11. The upregulation of SLC7A11 reduces methionine-dependence in cancer cells, a common feature among cancer cells (82, 83). Methionine dependence refers to the inability of cells to grow in media containing homocysteine instead of methionine, suggesting methionine restriction as a potential cancer treatment strategy (84, 85). This study highlights the role of the SNHG3/miR-152-3p/SLC7A11 axis in promoting prostate cancer progression by influencing methionine-dependence, underscoring the significance of miR-152-3p in inhibiting prostate cancer (81).
In summary, miR-152 is frequently downregulated in prostate cancer tissues and cell lines. Acting as a tumor suppressor miRNA, miR-152 targets and suppresses pro-oncogenic factors, leading to the inhibition of prostate cancer cell growth, movement, and invasion. It also triggers cell cycle arrest and apoptosis, ultimately impeding the progression and spread of prostate cancer. These findings indicate that miR-152 holds promise as a potential target for prostate cancer therapy.
miR-152 in bladder cancer
Bladder cancer is a prevalent malignancy among urological tumors, with approximately 70% of patients being diagnosed with non-muscle invasive bladder cancer. Out of these cases, 50%-70% face recurrence, and 10%-20% progress to muscle invasive bladder cancer with a poor prognosis (86) Despite recent advancements in understanding the mechanisms, diagnosis, and treatment of bladder cancer, the high recurrence rate and low survival rate of patients with muscle-invasive bladder cancer remain unresolved issues. Various studies have highlighted the significance of miR-152 in bladder cancer (Figure 2). Like in prostate cancer, miR-152 is often under-expressed in bladder cancer tissues and cell lines, functioning as a tumor suppressor miRNA. Zhang et al. observed significantly lower levels of miR-152 expression in bladder cancer cell lines T24 cells and UM-UC-3 cells compared to the normal urinary tract epithelial cell line SV-HUC-1. Moreover, miR-152 expression was notably lower in bladder cancer tissues compared to adjacent non-cancerous tissues. The reduced levels of miR-152 expression were found to be associated with the stage and grade of bladder cancer, and the overexpression of miR-152 effectively inhibited the growth of bladder cancer cells. The methylation rate of miR-152 promoter CpG islands was notably higher in bladder cancer cell lines and tissues compared to normal tissues. Mechanistic studies indicated that DNMT1-mediated hypermethylation was responsible for the decreased expression of miR-152 (60). Furthermore, DNMT1 was identified as a potential target of miR-152, with luciferase reporter gene assays showing direct binding of miR-152 to DNMT1 3′UTR, leading to its inhibition in bladder cancer cells (60). These findings suggest a reciprocal regulatory relationship between DNMT1 and miR-152 in bladder cancer, where increased DNMT1 levels cause hypermethylation of miR-152, resulting in its downregulation. Simultaneously, miR-152 suppresses DNMT1 expression by targeting its 3′-UTR. Additionally, research by Liu et al. revealed that DNMT1 promoted the promoter methylation of phosphatase and tensin homologue (PTEN), inhibiting PTEN expression. Knockdown of PTEN mitigated DNMT1’s effects on bladder cancer cell proliferation and migration, indicating that miR-152-3p inhibits DNMT1 expression, which in turn affects PTEN via DNA methylation regulation. The miR-152-3p/DNMT1/PTEN pathway plays a crucial role in bladder cancer development (65). These studies enhance our understanding of the intricate relationship between miR-152 and DNMT1 in bladder cancer. Kinesin family member 14 (KIF14) plays a crucial role in various biological processes. Studies have shown that overexpression of KIF14 is linked to the advancement and spread of different types of cancers (87, 88). Fang et al. reported that the overexpression of miR-152-3p and the knockdown of KIF14 inhibited the proliferation, migration, and invasion of bladder cancer cells while inducing apoptosis (89). Mechanistically, miR-152-3p directly targets KIF14, leading to a reduction in its expression, which in turn inhibits the PI3K/AKT and FOXM1/CCNB1 pathways, thereby impeding the progression of bladder cancer. The PI3K/AKT and FOXM1/CCNB1 pathways are well-established pathways involved in cancer development and cell cycle regulation (89). Moreover, miR-152-3p was found to target high mobility histone A2 (HMGA2), further inhibiting bladder cancer cell proliferation and invasion by suppressing HMGA2 expression (90). Additionally, Zhang et al. reported that lncRNA CCAT1 promotes the proliferation, migration, and invasion of bladder cancer cells. Further examination of its potential pathways revealed that miR-152-3p is associated with these functions. lncRNA CCAT1 may function as a sponge for miR-152-3p by binding to it, thereby weakening the activity of miR-152-3p and inhibiting its target genes, which in turn promotes the progression of bladder cancer (91).
miR-152 in renal cell carcinoma
Renal cell carcinoma (RCC) is the third most common urological tumor with a high mortality rate. Clear renal cell carcinoma (ccRCC) is a predominant subtype, constituting approximately 80% of RCC cases (2). Compared with prostate cancer, research on miR-152 in renal cell carcinoma is limited. Several studies have highlighted the significance of miR-152 in RCC (Figure 2). Yang et al. reported that LncRNA HCG18 is highly expressed in clear cell renal cell carcinoma (ccRCC) cells and tissues. Further research identified miR-152-3p as a downstream target of HCG18. HCG18 promotes the proliferation, migration, and growth of ccRCC by inhibiting the activity of miR-152-3p (92). Moreover, RAB14 was identified as a downstream target of miR-152-3p; overexpression of RAB14 mitigated the effects of HCG18 knockdown on cell viability and metastasis. This indicates that HCG18 positively regulates the expression of RAB14 by sponging miR-152-3p, thereby promoting the progression of ccRCC and highlighting the inhibitory effect of miR-152-3p on ccRCC (92). Additionally, Jasinski-Bergner et al. (93) demonstrated that miR-152 exhibits a strong affinity for the 3′-UTR of human leukocyte antigen G (HLA-G), a gene commonly overexpressed in RCC, thereby shielding tumor cells from immune cell-mediated cytotoxicity (94). This implies that miR-152 can suppress HLA-G, enhancing immune cell-mediated killing of tumor cells and impeding RCC progression.
The potential of miR-152 as a biomarker for urological malignancies
Biomarkers play a crucial role in providing valuable cancer-related information for the early screening, diagnosis, treatment, and prognosis of cancer. The traditional prostate cancer diagnostic indicator, blood PSA, has faced criticism in recent years due to its lack of specificity, false positives, and potential for overdiagnosis in benign prostate disorders (5), As PSA levels do not directly correlate with prostate cancer staging, there is a need for biomarkers with higher specificity and sensitivity in clinical management. Dysregulation of miRNA has been observed in various cancers (95, 96), with miRNA signatures proving to be more accurate than PSA in detecting different cancer types (97–99). The expression differences of miR-152 in prostate cancer compared to non-cancerous tissues, along with its correlation with Gleason score and pathological staging, suggest its potential as a diagnostic and prognostic biomarker for prostate cancer.
Liu et al. investigated the use of miR-146a and miR-152 in prostate cancer and their association with clinicopathological parameters. They found that the serum levels of miR-152 in prostate cancer patients were significantly lower compared to normal levels. The expression of miR-152 was strongly linked to clinical stage, presence of bone metastasis, and pathological stage. The diagnostic accuracy of prostate cancer using miR-152 was evaluated with an area under the curve (AUC) of 0.699 and a specificity of 94.64%. Furthermore, combined detection with miR-146a showed higher sensitivity than individual detection, suggesting that monitoring changes in the expression of miR-146a and miR-152 in serum could enhance the diagnostic precision of prostate cancer (100). Moya et al. also identified four significantly up-regulated miRNAs in the plasma of prostate cancer patients, including miR-152. These miRNAs exhibited high specificity and sensitivity in diagnosing prostate cancer. The AUC suggested that their diagnostic capability exceeded that of PSA, indicating that miRNA-152 in plasma holds promise as a diagnostic biomarker for prostate cancer (6, 101). Circulating miR-152 has shown promise in early detection of postoperative recurrence of prostate cancer. Chen et al. found that miR-152 levels in the serum of prostate cancer patients were significantly lower compared to controls, with even lower levels in patients experiencing postoperative recurrence. The ROC curve analysis demonstrated an AUC of 0.906, sensitivity of 91.4%, and specificity of 80.6%, indicating the potential of miR-152 as a biomarker for early prediction of postoperative recurrence (102). A MATE analysis also highlighted 14 down-regulated miRNAs, including miR-152, with diagnostic and predictive potential in distinguishing prostate cancer from benign prostatic hyperplasia (103). For metastasis detection, Lin et al. identified miR-152 as a potential marker through bioinformatics modelling, further confirming its role in prostate cancer metastasis (104). Additionally, Lichner et al. discovered 25 differentially expressed miRNAs associated with biochemical failure risk post-prostatectomy, with miR-152 interacting with ERBB3 and influencing cell proliferation to predict biochemical failure risk. These findings suggest the utility of miR-152 in predicting postoperative recurrence and metastatic features of prostate cancer, as well as in guiding postoperative adjuvant therapy decisions (105).
Early diagnosis of bladder cancer is crucial for reducing mortality rates. While urine cytology is a simple and non-invasive method, its sensitivity is limited. Cystoscopy-guided biopsy is the gold standard but is invasive and inconvenient for cancer screening. Therefore, the search for more sensitive and non-invasive biomarkers continues. Serum miRNA has emerged as a promising option for bladder cancer diagnosis and prognosis. A study on genome-wide miRNA analysis identified six miRNAs, including miR-152, in serum with high accuracy and sensitivity levels, outperforming urinary cytology. Notably, miR-152 was linked to NMIBC tumor recurrence, indicating its clinical significance (106). Additionally, miR-152’s tumor suppressor role was hindered by DNA hypermethylation, suggesting its potential as an epigenetic biomarker for bladder cancer. Köhler et al. found that miR-152 had low expression and hypermethylation in bladder cancer cell lines, proposing its use as an epigenetic biomarker for the disease (107).
In a bioinformatics analysis of potential therapeutic targets for hypertension-associated renal cell carcinoma (RCC), miR-152-3p was identified as significantly associated with hypertension-associated RCC (108). Furthermore, five miRNAs, including miR-152-3p, were found to be dysregulated in the urine of RCC patients both before and after surgery. These miRNAs exhibited a strong correlation, indicating their potential utility as post-operative disease status markers (109).
In summary, miR-152 in serum demonstrates high specificity and sensitivity in diagnosing prostate cancer, aiding in early detection. It is associated with the pathological stage and Gleason score of prostate cancer. Furthermore, miR-152 facilitates early monitoring of postoperative prostate cancer recurrence and metastasis risk, complementing the limitations of PSA. This suggests that miR-152 holds significant promise as a biomarker for the diagnosis, treatment, and prognosis of prostate cancer. In bladder cancer, miR-152, alongside other miRNAs, shows high sensitivity across various stages and offers clinical value in predicting postoperative recurrence. Studies also link miR-152 to renal cell carcinoma, indicating its potential as a biomarker for postoperative follow-up, although further research is necessary to fully understand its clinical significance.
Therapeutic strategies for tumours based on miRNA therapy
miRNAs play a significant role in cancer regulation and can be targeted for therapeutic purposes. Cancer-associated miRNAs, categorized as oncogenic or tumor-suppressor miRNAs, can be targeted through two mechanisms. For oncogenic miRNAs, miRNA inhibitors can be administered to patients to reduce or eliminate their activity by isolating and binding to the miRNAs (35). These inhibitors typically consist of reverse sequences that complement the miRNAs. Additionally, ceRNAs, such as lncRNAs, circRNAs, and pseudogene transcripts, act as natural miRNA inhibitors by binding to miRNAs and competing for their limited availability, thereby reducing their function. On the other hand, for tumor-suppressor miRNAs, miRNA mimics can be delivered to patients to replace the downregulated miRNAs (110). miRNA therapy for cancer may involve the use of miRNA mimics or inhibitors alone or in combination with chemotherapy, radiotherapy, and immunotherapy to enhance treatment efficacy (111).
Gene therapy, which involves the therapeutic delivery of nucleic acids into cancer cells, has been regarded as a promising approach for decades. Recently, the U.S. Food and Drug Administration (FDA) has approved several new therapies utilizing small interfering RNA (siRNA), signaling the onset of a new era in targeted therapy. In 2018, the FDA approved Patisiran, the first-ever siRNA-based drug, for the treatment of transthyretin-mediated amyloidosis, representing a significant milestone in the history of RNA interference (RNAi) technology (112). The following year, the FDA approved Givlaari, the second siRNA drug, which targets aminolevulinic acid synthase 1 (ALAS1) mRNA for the treatment of acute hepatic porphyria (113). In 2020, Lumasiran was approved by the FDA as a treatment for primary hyperoxaluria type 1 (PH1) (114). Furthermore, several studies investigating miRNA-based therapies for advanced cancers have progressed to phase II clinical trials, including TargomiR (a miR-16 mimic-based therapy) for mesothelioma (115), Cobomarsen (an anti-miR-155 therapy) (116, 117), and Miravirsen (an anti-miR-122 therapy) for individuals infected with hepatitis C (118). The number of siRNA-based drugs is anticipated to increase significantly in the future, as siRNA-based therapies emerge as a promising treatment option that could revolutionize the clinical management of numerous diseases due to their capacity to selectively modulate protein expression. Nevertheless, siRNA-based cancer treatments must still address several challenges. One major challenge is identifying the most suitable miRNA candidate or target for each type of cancer (119). Since a single miRNA can have multiple targets, including both oncogenes and tumor suppressor genes, miRNAs may have dual roles in cancer progression, either promoting tumor development or inhibiting tumor progression, depending on various factors such as the cumulative effect of all targets and the specific tumor microenvironment. Using miRNAs as therapeutic targets requires a delicate balance between their oncogenic and tumor-suppressing effects to ensure their clinical efficacy. Another challenge is the lack of tissue-specific targeting of miRNAs delivered to the body, which may lead to unintended uptake by other tissues and subsequent adverse reactions. To address this issue, researchers have developed vectors, such as viral particles and nanoparticles, to facilitate targeted delivery of miRNAs to specific tissues. Additionally, the degradation of oligonucleotides by serum and intracellular RNA enzymes upon entry into the body poses a problem. To mitigate this, scientists have modified the chemical properties of oligonucleotides by adding phosphorothioate groups to the nucleotide or RNA backbone, thereby preventing degradation. These delivery vectors not only aid in targeting but also provide protection by encapsulating the miRNA. Furthermore, the potential toxicity, immune response, and off-target effects of miRNAs and vectors post-administration into the body are important considerations that must be addressed.
Despite the existing challenges associated with miRNA therapy for cancer treatment, addressing these issues could lead to significant breakthroughs in the management of various diseases, including cancer. Notably, miR-152 has demonstrated considerable promise in the treatment of cancer, particularly in relation to urinary malignancies. Studies indicate that miR-152 functions as a tumor-suppressor miRNA with decreased expression in prostate, bladder, and renal cell carcinomas. In vitro experiments have shown that overexpression of miR-152 can inhibit cancer cell proliferation, migration, invasion, angiogenesis, and promote apoptosis. This tumor-suppressor effect has also been validated in vivo through tumor xenografts (76, 81). Consequently, miR-152 emerges as a strong candidate for miRNA-based therapy in urological malignancies, with in vivo delivery of miR-152 mimics presenting a viable therapeutic approach. Ongoing advancements in miRNA carriers and chemical modifications hold promise for addressing targeting and stability issues associated with miR-152 treatment. However, further preclinical studies are necessary to assess the safety and efficacy of this approach.
Conclusion and prospects
This review provides a comprehensive summary of the relationship between miR-152 and urological tumors (Table 1). Current research indicates that miR-152 acts as a tumor-suppressor miRNA in prostate, bladder, and renal cell carcinomas. By targeting downstream genes and signaling pathways associated with the biological behaviors of cancer cells, miR-152 inhibits tumor progression. Furthermore, studies demonstrate that miR-152 interacts with multiple genes and signaling cascades, forming a protein-protein interaction network that plays a role in the pathophysiological process of urological malignancies. These findings suggest that miR-152 could be a promising gene target for the treatment of urological malignancies. In the realm of urological malignancies, the expression of miR-152 exhibits notable variations and is closely linked to key pathological parameters such as cancer stage, grading, and metastasis. This suggests that miR-152 holds promise as a biomarker for the detection, treatment, and prognosis of urological malignancies. For instance, in prostate cancer, serum miR-152 demonstrates high sensitivity and specificity in diagnosing the disease, as well as in monitoring metastasis and postoperative recurrence. Similarly, in bladder cancer, serum miR-152 exhibits high sensitivity across different stages of the disease, surpassing the sensitivity of urocytology. This indicates that miR-152 could serve as a non-invasive biomarker with significant potential in the realm of urological malignancies. However, there are certain limitations in the current research on miR-152 in urological malignancies. Firstly, miR-152 remains relatively understudied in the urinary system, particularly in renal cell carcinoma, and the precise mechanism of miR-152 in urological malignancies remains poorly understood. Secondly, there is a lack of information on the stability of miR-152 and its targeting of tissues in vivo in current studies on miR-152 and urinary malignancies. Lastly, existing studies are predominantly focused on cellular and animal models, with a scarcity of clinical research. Numerous challenges need to be addressed before miR-152 can be effectively utilized in clinical settings. Once these challenges are overcome, miR-152 has the potential to emerge as an innovative therapeutic avenue for urological malignancies.
In conclusion, miR-152 has demonstrated significant promise as both a therapeutic target and biomarker for urological malignancies. However, there are unresolved issues that must be addressed before miR-152 can realize its full potential in the treatment of urological malignancies and other types of tumors. Further research is essential to expand our understanding in this field.
Author contributions
XL: Writing – original draft. BQ: Writing – review & editing. XC: Writing – original draft. MS: Writing – original draft. SZ: Writing – original draft. XZ: Writing – review & editing. JH: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by Zhejiang Provincial Natural Science Foundation of China under Grant (No. LGY23H050001), Science and Technology Planning Project of Taizhou City, Zhejiang Province (ID: 22ywa19), Zhejiang Medical and Health Science and Technology Program (No. 2023KY1328).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
PSA, prostate specific antigen; 3’ UTR, 3’untranslated region; pri-miRNAs, primary miRNAs; pre-miRNAs, precursor miRNAs; RISC, RNA-induced silencing complex; AGO2, assistance of argonaut 2 protein; CML, chronic lymphocytic leukaemia; oncomiRs, oncogenic miRNAs; miRsupps, tumor suppressor miRNAs; ATG14, autophagy-related protein 14; DNMT1, DNA methyltransferase 1; ELF1, ETS transcription factor 1; ceRNAs, competing endogenous RNAs; TGFα, transforming growth factor-alpha; NOL4, nucleolin 4; TMEM97, transmembrane protein 97; KLF4, Kruppel-like factor 4; PTEN, phosphate and tensin homolog; SNHG3, Small nucleolar RNA host gene 3; KIF14, Kinesin family member 14; HMGA2, high mobility histone A2; RCC, Renal cell carcinoma; HLA-G, human leukocyte antigen G; AUC, area under the curve; siRNA, small interfering RNA.
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2024) 74:229–63. doi: 10.3322/caac.21834
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
3. Shen M, Chen T, Li X, Zhao S, Zhang X, Zheng L, et al. The role of miR-155 in urologic Malignancies. BioMed Pharmacother. (2024) 174:116412. doi: 10.1016/j.biopha.2024.116412
4. Mariotto AB, Yabroff KR, Shao Y, Feuer EJ, Brown ML. Projections of the cost of cancer care in the United States: 2010-2020. J Natl Cancer Inst. (2011) 103:117–28. doi: 10.1093/jnci/djq495
5. Bokhorst LP, Bangma CH, van Leenders GJ, Lous JJ, Moss SM, Schröder FH, et al. Prostate-specific antigen-based prostate cancer screening: reduction of prostate cancer mortality after correction for nonattendance and contamination in the Rotterdam section of the European Randomized Study of Screening for Prostate Cancer. Eur Urol. (2014) 65:329–36. doi: 10.1016/j.eururo.2013.08.005
6. Moya L, Meijer J, Schubert S, Matin F, Batra J. Assessment of miR-98-5p, miR-152-3p, miR-326 and miR-4289 Expression as Biomarker for Prostate Cancer Diagnosis. Int J Mol Sci. (2019) 20. doi: 10.3390/ijms20051154
7. Shefer HK, Masarwe I, Bejar J, Naamnih LH, Gueta-Milshtein K, Shalata A, et al. Performance of CellDetect for detection of bladder cancer: Comparison with urine cytology and UroVysion. Urol Oncol. (2023) 41:296.e1–8. doi: 10.1016/j.urolonc.2022.12.012
8. Chen T, Zheng L, Luo P, Zou J, Li W, Chen Q, et al. Crosstalk between m6A modification and autophagy in cancer. Cell Biosci. (2024) 14:44. doi: 10.1186/s13578-024-01225-5
9. Zhu X, Jin X, Li Z, Chen X, Zhao J. miR-152-3p facilitates cell adhesion and hepatic metastases in colorectal cancer via targeting AQP11. Pathol Res Pract. (2023) 244:154389. doi: 10.1016/j.prp.2023.154389
10. Sun LB, Zhao SF, Zhu JJ, Han Y, Shan TD. Long noncoding RNA UCID sponges miR−152−3p to promote colorectal cancer cell migration and invasion via the Wnt/β−catenin signaling pathway. Oncol Rep. (2020) 44:1194–205. doi: 10.3892/or.2020.7670
11. You W, Zhang X, Ji M, Yu Y, Chen C, Xiong Y, et al. MiR-152-5p as a microRNA passenger strand special functions in human gastric cancer cells. Int J Biol Sci. (2018) 14:644–53. doi: 10.7150/ijbs.25272
12. Wang X, Zhang Y, Li W, Liu X. Knockdown of cir_RNA PVT1 Elevates Gastric Cancer Cisplatin Sensitivity via Sponging miR-152-3p. J Surg Res. (2021) 261:185–95. doi: 10.1016/j.jss.2020.12.013
13. Chen K, Zhang L. LINC00339 regulates ROCK1 by miR-152 to promote cell proliferation and migration in hepatocellular carcinoma. J Cell Biochem. (2019) 120:14431–43. doi: 10.1002/jcb.v120.9
14. Yin T, Zhao H. miR-152-3p impedes the Malignant phenotypes of hepatocellular carcinoma by repressing roundabout guidance receptor 1. Cell Mol Biol Lett. (2022) 27:22. doi: 10.1186/s11658-022-00322-y
15. Duan J, Wang L, Shang L, Yang S, Wu H, Huang Y, et al. miR-152/TNS1 axis inhibits non-small cell lung cancer progression through Akt/mTOR/RhoA pathway. Biosci Rep. (2021) 41. doi: 10.1042/BSR20201539
16. Cheng Z, Ma R, Tan W, Zhang L. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp Mol Med. (2014) 46:e112. doi: 10.1038/emm.2014.51
17. Zhao L, Wu X, Zhang Z, Fang L, Yang B, Li Y. ELF1 suppresses autophagy to reduce cisplatin resistance via the miR-152-3p/NCAM1/ERK axis in lung cancer cells. Cancer Sci. (2023) 114:2650–63. doi: 10.1111/cas.v114.6
18. Chen MJ, Cheng YM, Chen CC, Chen YC, Shen CJ. MiR-148a and miR-152 reduce tamoxifen resistance in ER+ breast cancer via downregulating ALCAM. Biochem Biophys Res Commun. (2017) 483:840–6. doi: 10.1016/j.bbrc.2017.01.012
19. Jiang CF, Xie YX, Qian YC, Wang M, Liu LZ, Shu YQ, et al. TBX15/miR-152/KIF2C pathway regulates breast cancer doxorubicin resistance via promoting PKM2 ubiquitination. Cancer Cell Int. (2021) 21:542. doi: 10.1186/s12935-021-02235-w
20. Li LW, Xiao HQ, Ma R, Yang M, Li W, Lou G. miR-152 is involved in the proliferation and metastasis of ovarian cancer through repression of ERBB3. Int J Mol Med. (2018) 41:1529–35. doi: 10.3892/ijmm.2017.3324
21. Woo HH, László CF, Greco S, Chambers SK. Regulation of colony stimulating factor-1 expression and ovarian cancer cell behavior in vitro by miR-128 and miR-152. Mol Cancer. (2012) 11:58. doi: 10.1186/1476-4598-11-58
22. Wu J, Chi X, Yang Q, Li J. Expression of miR-152 in cervical cancer and its influence on cisplatin resistance. Afr Health Sci. (2023) 23:246–55. doi: 10.4314/ahs.v23i2.28
23. Zhou J, Lei N, Tian W, Guo R, Gao F, Fu H, et al. Hypoxic tumor cell-derived small extracellular vesicle miR-152-3p promotes cervical cancer radioresistance through KLF15 protein. Radiat Oncol. (2023) 18:183. doi: 10.1186/s13014-023-02369-3
24. Kong S, Fang Y, Wang B, Cao Y, He R, Zhao Z. miR-152-5p suppresses glioma progression and tumorigenesis and potentiates temozolomide sensitivity by targeting FBXL7. J Cell Mol Med. (2020) 24:4569–79. doi: 10.1111/jcmm.15114
25. Zhang P, Sun H, Yang B, Luo W, Liu Z, Wang J, et al. miR-152 regulated glioma cell proliferation and apoptosis via Runx2 mediated by DNMT1. BioMed Pharmacother. (2017) 92:690–5. doi: 10.1016/j.biopha.2017.05.096
26. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. (1993) 75:855–62. doi: 10.1016/0092-8674(93)90530-4
27. Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. (2011) 39:D152–7. doi: 10.1093/nar/gkq1027
28. Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. (2009) 21:452–60. doi: 10.1016/j.ceb.2009.04.009
29. Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. (2008) 20:214–21. doi: 10.1016/j.ceb.2008.01.006
30. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509–24. doi: 10.1038/nrm3838
31. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. (2009) 136:215–33. doi: 10.1016/j.cell.2009.01.002
32. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. (2008) 9:102–14. doi: 10.1038/nrg2290
33. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U.S.A. (2002) 99:15524–9. doi: 10.1073/pnas.242606799
34. Shen M, Li X, Qian B, Wang Q, Lin S, Wu W, et al. Crucial roles of microRNA-mediated autophagy in urologic Malignancies. Int J Biol Sci. (2021) 17:3356–68. doi: 10.7150/ijbs.61175
35. Mollaei H, Safaralizadeh R, Rostami Z. MicroRNA replacement therapy in cancer. J Cell Physiol. (2019) 234:12369–84. doi: 10.1002/jcp.v234.8
36. Liu S, Ruan Y, Chen X, He B, Chen Q. miR-137: a potential therapeutic target for lung cancer. Front Cell Dev Biol. (2024) 12:1427724. doi: 10.3389/fcell.2024.1427724
37. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of microRNA expression in cancer. Int J Mol Sci. (2020) 21. doi: 10.3390/ijms21051723
38. Kulkarni P, Dasgupta P, Hashimoto Y, Shiina M, Shahryari V, Tabatabai ZL, et al. A lncRNA TCL6-miR-155 interaction regulates the Src-Akt-EMT network to mediate kidney cancer progression and metastasis. Cancer Res. (2021) 81:1500–12. doi: 10.1158/0008-5472.CAN-20-0832
39. Li H, Song JB, Chen HX, Wang QQ, Meng LX, Li Y. MiR-155 inhibits proliferation, invasion and migration of melanoma via targeting CBL. Eur Rev Med Pharmacol Sci. (2019) 23:9525–34. doi: 10.26355/eurrev_201911_19447
40. Luo Y, Hu S, Wang F, Yang J, Gong D, Xu W, et al. miR-137 represses migration and cell motility by targeting COX-2 in non-small cell lung cancer. Transl Cancer Res. (2022) 11:3803–13. doi: 10.21037/tcr-22-2177
41. Chen Q, Chen X, Zhang M, Fan Q, Luo S, Cao X. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci. (2011) 56:2009–16. doi: 10.1007/s10620-010-1536-3
42. Sun J, Cai X, Yung MM, Zhou W, Li J, Zhang Y, et al. miR-137 mediates the functional link between c-Myc and EZH2 that regulates cisplatin resistance in ovarian cancer. Oncogene. (2019) 38:564–80. doi: 10.1038/s41388-018-0459-x
43. Wang Y, Chen R, Zhou X, Guo R, Yin J, Li Y, et al. miR-137: A novel therapeutic target for human glioma. Mol Ther Nucleic Acids. (2020) 21:614–22. doi: 10.1016/j.omtn.2020.06.028
44. Chang TH, Tsai MF, Gow CH, Wu SG, Liu YN, Chang YL, et al. Upregulation of microRNA-137 expression by Slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer Lett. (2017) 402:190–202. doi: 10.1016/j.canlet.2017.06.002
45. Su TJ, Ku WH, Chen HY, Hsu YC, Hong QS, Chang GC, et al. Oncogenic miR-137 contributes to cisplatin resistance via repressing CASP3 in lung adenocarcinoma. Am J Cancer Res. (2016) 6:1317–30.
46. Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. (2002) 12:735–9. doi: 10.1016/S0960-9822(02)00809-6
47. Friedrich M, Pracht K, Mashreghi MF, Jäck HM, Radbruch A, Seliger B. The role of the miR-148/-152 family in physiology and disease. Eur J Immunol. (2017) 47:2026–38. doi: 10.1002/eji.201747132
48. Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. (2007) 27:91–105. doi: 10.1016/j.molcel.2007.06.017
49. Nielsen LB, Wang C, Sørensen K, Bang-Berthelsen CH, Hansen L, Andersen ML, et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res. (2012) 2012:896362. doi: 10.1155/2012/896362
50. Bidzhekov K, Gan L, Denecke B, Rostalsky A, Hristov M, Koeppel TA, et al. microRNA expression signatures and parallels between monocyte subsets and atherosclerotic plaque in humans. Thromb Haemost. (2012) 107:619–25. doi: 10.1160/TH11-09-0607
51. Fan Y, Gan M, Tan Y, Chen L, Shen L, Niu L, et al. Mir-152 regulates 3T3-L1 preadipocyte proliferation and differentiation. Molecules. (2019) 24. doi: 10.3390/molecules24183379
52. Tan Z, Wang W, Peng J, Zhou Z, Pan J, Peng A, et al. Impact of Amarogentin on Gastric Carcinoma Cell Multiplication, Apoptosis and Migration via circKIF4A/miR-152-3p. J Immunol Res. (2022) 2022:2156204. doi: 10.1155/2022/2156204
53. Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human Malignant cholangiocytes. Hepatology. (2010) 51:881–90. doi: 10.1002/hep.23381
54. Zhou X, Zhao F, Wang ZN, Song YX, Chang H, Chiang Y, et al. Corrigendum] Altered expression of miR−152 and miR−148a in ovarian cancer is related to cell proliferation. Oncol Rep. (2022) 47. doi: 10.3892/or.2022.8285
55. Hiroki E, Akahira J, Suzuki F, Nagase S, Ito K, Suzuki T, et al. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. (2010) 101:241–9. doi: 10.1111/j.1349-7006.2009.01385.x
56. Song Y, Zhang M, Lu MM, Qu LY, Xu SG, Li YZ, et al. EPAS1 targeting by miR-152-3p in Paclitaxel-resistant Breast Cancer. J Cancer. (2020) 11:5822–30. doi: 10.7150/jca.46898
57. Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. (2014) 24:R762–76. doi: 10.1016/j.cub.2014.06.043
58. Lujambio A, Lowe SW. The microcosmos of cancer. Nature. (2012) 482:347–55. doi: 10.1038/nature10888
59. Theodore SC, Davis M, Zhao F, Wang H, Chen D, Rhim J, et al. MicroRNA profiling of novel African American and Caucasian Prostate Cancer cell lines reveals a reciprocal regulatory relationship of miR-152 and DNA methyltranferase 1. Oncotarget. (2014) 5:3512–25. doi: 10.18632/oncotarget.v5i11
60. Zhang H, Qi D, Li J, Peng T, Yang L, Yuan J, et al. A novel regulatory circuit of miR−152 and DNMT1 in human bladder cancer. Oncol Rep. (2018) 40:1803–12. doi: 10.3892/or.2018.6553
61. Wang C, Ma X, Zhang J, Jia X, Huang M. DNMT1 maintains the methylation of miR-152-3p to regulate TMSB10 expression, thereby affecting the biological characteristics of colorectal cancer cells. IUBMB Life. (2020) 72:2432–43. doi: 10.1002/iub.v72.11
62. Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. (2013) 5:3–13. doi: 10.1093/jmcb/mjs049
63. Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I, et al. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. (2011) 71:6450–62. doi: 10.1158/0008-5472.CAN-11-0364
64. Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. (2014) 33:378–86. doi: 10.1038/onc.2012.575
65. Liu P, Wu L, Chand H, Li C, Hu X, Li Y. Silencing of miR-152 contributes to DNMT1-mediated CpG methylation of the PTEN promoter in bladder cancer. Life Sci. (2020) 261:118311. doi: 10.1016/j.lfs.2020.118311
66. Wen YY, Liu WT, Sun HR, Ge X, Shi ZM, Wang M, et al. IGF-1-mediated PKM2/β-catenin/miR-152 regulatory circuit in breast cancer. Sci Rep. (2017) 7:15897. doi: 10.1038/s41598-017-15607-y
67. Shi J, Zhang Y, Qin B, Wang Y, Zhu X. Long non-coding RNA LINC00174 promotes glycolysis and tumor progression by regulating miR-152-3p/SLC2A1 axis in glioma. J Exp Clin Cancer Res. (2019) 38:395. doi: 10.1186/s13046-019-1390-x
68. Jin Y, Yang L, Li X, Liu F. Circular RNA KIF4A promotes cell migration, invasion and inhibits apoptosis through miR-152/ZEB1 axis in breast cancer. Diagn Pathol. (2020) 15:55. doi: 10.1186/s13000-020-00963-7
69. Zhu C, Li J, Ding Q, Cheng G, Zhou H, Tao L, et al. miR-152 controls migration and invasive potential by targeting TGFα in prostate cancer cell lines. Prostate. (2013) 73:1082–9. doi: 10.1002/pros.v73.10
70. Pospiech K, Orzechowska M, Nowakowska M, Anusewicz D, Płuciennik E, Kośla K, et al. TGFα-EGFR pathway in breast carcinogenesis, association with WWOX expression and estrogen activation. J Appl Genet. (2022) 63:339–59. doi: 10.1007/s13353-022-00690-3
71. Kidd M, Schimmack S, Lawrence B, Alaimo D, Modlin IM. EGFR/TGFα and TGFβ/CTGF signaling in neuroendocrine neoplasia: theoretical therapeutic targets. Neuroendocrinology. (2013) 97:35–44. doi: 10.1159/000334891
72. Guo G, Gong K, Beckley N, Zhang Y, Yang X, Chkheidze R, et al. EGFR ligand shifts the role of EGFR from oncogene to tumour suppressor in EGFR-amplified glioblastoma by suppressing invasion through BIN3 upregulation. Nat Cell Biol. (2022) 24:1291–305. doi: 10.1038/s41556-022-00962-4
73. Ying X, Liu B, Yuan Z, Huang Y, Chen C, Jiang X, et al. METTL1-m(7) G-EGFR/EFEMP1 axis promotes the bladder cancer development. Clin Transl Med. (2021) 11:e675. doi: 10.1002/ctm2.v11.12
74. Tao LJ, Pan XY, Wang JW, Zhang L, Tao LS, Liang CZ. Circular RNA circANKS1B acts as a sponge for miR-152-3p and promotes prostate cancer progression by upregulating TGF-α expression. Prostate. (2021) 81:271–8. doi: 10.1002/pros.24102
75. Xu WB, Hou JQ, Chang JK, Li S, Liu GC, Cao SQ. The mechanism of HOTAIR regulating the proliferation and apoptosis of prostate cancer cells by targeting down-regulation of miR-152 to improve the expression of FOXR2. Zhonghua Yi Xue Za Zhi. (2019) 99:1887–92. doi: 10.3760/cma.j.issn.0376-2491.2019.24.010
76. Ramalho-Carvalho J, Gonçalves CS, Graça I, Bidarra D, Pereira-Silva E, Salta S, et al. A multiplatform approach identifies miR-152-3p as a common epigenetically regulated onco-suppressor in prostate cancer targeting TMEM97. Clin Epigenet. (2018) 10:40. doi: 10.1186/s13148-018-0475-2
77. Mao D, Zhang X, Wang Z, Xu G, Zhang Y. TMEM97 is transcriptionally activated by YY1 and promotes colorectal cancer progression via the GSK-3β/β-catenin signaling pathway. Hum Cell. (2022) 35:1535–46. doi: 10.1007/s13577-022-00759-5
78. Schmit K, Michiels C. TMEM proteins in cancer: A review. Front Pharmacol. (2018) 9:1345. doi: 10.3389/fphar.2018.01345
79. Feng F, Liu H, Chen A, Xia Q, Zhao Y, Jin X, et al. miR-148-3p and miR-152-3p synergistically regulate prostate cancer progression via repressing KLF4. J Cell Biochem. (2019) 120:17228–39. doi: 10.1002/jcb.v120.10
80. Gurbuz V, Sozen S, Bilen CY, Konac E. miR-148a, miR-152 and miR-200b promote prostate cancer metastasis by targeting DNMT1 and PTEN expression. Oncol Lett. (2021) 22:805. doi: 10.3892/ol.2021.13066
81. Wang X, Song Y, Shi Y, Yang D, Li J, Yin B. SNHG3 could promote prostate cancer progression through reducing methionine dependence of PCa cells. Cell Mol Biol Lett. (2022) 27:13. doi: 10.1186/s11658-022-00313-z
82. Lien EC, Ghisolfi L, Geck RC, Asara JM, Toker A. Oncogenic PI3K promotes methionine dependency in breast cancer cells through the cystine-glutamate antiporter xCT. Sci Signal. (2017) 10:510. doi: 10.1126/scisignal.aao6604
83. Poirson-Bichat F, Gonfalone G, Bras-Gonçalves RA, Dutrillaux B, Poupon MF. Growth of methionine-dependent human prostate cancer (PC-3) is inhibited by ethionine combined with methionine starvation. Br J Cancer. (1997) 75:1605–12. doi: 10.1038/bjc.1997.274
84. Sorin M, Watkins D, Gilfix BM, Rosenblatt DS. Methionine dependence in tumor cells: The potential role of cobalamin and MMACHC. Mol Genet Metab. (2021) 132:155–61. doi: 10.1016/j.ymgme.2021.01.006
85. Chaturvedi S, Hoffman RM, Bertino JR. Exploiting methionine restriction for cancer treatment. Biochem Pharmacol. (2018) 154:170–3. doi: 10.1016/j.bcp.2018.05.003
86. Kaufman DS, Shipley WU, Feldman AS. Bladder cancer. Lancet. (2009) 374:239–49. doi: 10.1016/S0140-6736(09)60491-8
87. Yang Z, Li C, Yan C, Li J, Yan M, Liu B, et al. KIF14 promotes tumor progression and metastasis and is an independent predictor of poor prognosis in human gastric cancer. Biochim Biophys Acta Mol Basis Dis. (2019) 1865:181–92. doi: 10.1016/j.bbadis.2018.10.039
88. Li TF, Zeng HJ, Shan Z, Ye RY, Cheang TY, Zhang YJ, et al. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int. (2020) 20:123. doi: 10.1186/s12935-020-01191-1
89. Meng F, Zhang Z. MicroRNA-152 specifically targets kinesin family member 14 to suppress the advancement of bladder cancer cells via PI3K/AKT pathway. Biochem Biophys Res Commun. (2024) 692:149337. doi: 10.1016/j.bbrc.2023.149337
90. Li S, Pan M, Wu X, Bai Y, Qi M, Chen J, et al. miR-152-3p inhibits proliferation, invasion and autophagy in UMUC3 and TCCSUP bladder cancer cell lines via suppressing HMGA2 expression. Ann Clin Lab Sci. (2023) 53:607–18.
91. Zhang C, Wang W, Lin J, Xiao J, Tian Y. lncRNA CCAT1 promotes bladder cancer cell proliferation, migration and invasion. Int Braz J Urol. (2019) 45:549–59. doi: 10.1590/s1677-5538.ibju.2018.0450
92. Yang Y, Gong P, Yao D, Xue D, He X. LncRNA HCG18 promotes clear cell renal cell carcinoma progression by targeting miR-152-3p to upregulate RAB14. Cancer Manag Res. (2021) 13:2287–94. doi: 10.2147/CMAR.S298649
93. Jasinski-Bergner S, Stoehr C, Bukur J, Massa C, Braun J, Hüttelmaier S, et al. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology. (2015) 4:e1008805. doi: 10.1080/2162402X.2015.1008805
94. Bukur J, Seliger B. The role of HLA-G for protection of human renal cell-carcinoma cells from immune-mediated lysis: implications for immunotherapies. Semin Cancer Biol. (2003) 13:353–9. doi: 10.1016/S1044-579X(03)00026-9
95. Tumilson CA, Lea RW, Alder JE, Shaw L. Circulating microRNA biomarkers for glioma and predicting response to therapy. Mol Neurobiol. (2014) 50:545–58. doi: 10.1007/s12035-014-8679-8
96. Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. (2010) 23:1157–64. doi: 10.1038/modpathol.2010.111
97. Watahiki A, Macfarlane RJ, Gleave ME, Crea F, Wang Y, Helgason CD, et al. Plasma miRNAs as biomarkers to identify patients with castration-resistant metastatic prostate cancer. Int J Mol Sci. (2013) 14:7757–70. doi: 10.3390/ijms14047757
98. Endzeliņš E, Melne V, Kalniņa Z, Lietuvietis V, Riekstiņa U, Llorente A, et al. Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: a systematic review. Mol Cancer. (2016) 15:41. doi: 10.1186/s12943-016-0523-5
99. Sharova E, Grassi A, Marcer A, Ruggero K, Pinto F, Bassi P, et al. A circulating miRNA assay as a first-line test for prostate cancer screening. Br J Cancer. (2016) 114:1362–6. doi: 10.1038/bjc.2016.151
100. Liu Y, Gao S, Du Q, Shao M. miR-146a and miR-152 in prostate cancer and clinicopathological parameters. J buon. (2019) 24:1692–9.
101. Matin F, Jeet V, Moya L, Selth LA, Chambers S, Clements JA, et al. A plasma biomarker panel of four microRNAs for the diagnosis of prostate cancer. Sci Rep. (2018) 8:6653. doi: 10.1038/s41598-018-24424-w
102. Chen JF, Liao YF, Ma JB, Mao QF, Jia GC, Dong XJ. Circulating miR-152 helps early prediction of postoperative biochemical recurrence of prostate cancer. Zhonghua Nan Ke Xue. (2017) 23:603–8.
103. Song CJ, Chen H, Chen LZ, Ru GM, Guo JJ, Ding QN. The potential of microRNAs as human prostate cancer biomarkers: A meta-analysis of related studies. J Cell Biochem. (2018) 119:2763–86. doi: 10.1002/jcb.v119.3
104. Lin Y, Chen F, Shen L, Tang X, Du C, Sun Z, et al. Biomarker microRNAs for prostate cancer metastasis: screened with a network vulnerability analysis model. J Transl Med. (2018) 16:134. doi: 10.1186/s12967-018-1506-7
105. Lichner Z, Fendler A, Saleh C, Nasser AN, Boles D, Al-Haddad S, et al. MicroRNA signature helps distinguish early from late biochemical failure in prostate cancer. Clin Chem. (2013) 59:1595–603. doi: 10.1373/clinchem.2013.205450
106. Jiang X, Du L, Wang L, Li J, Liu Y, Zheng G, et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. (2015) 136:854–62. doi: 10.1002/ijc.v136.4
107. Köhler CU, Bryk O, Meier S, Lang K, Rozynek P, Brüning T, et al. Analyses in human urothelial cells identify methylation of miR-152, miR-200b and miR-10a genes as candidate bladder cancer biomarkers. Biochem Biophys Res Commun. (2013) 438:48–53. doi: 10.1016/j.bbrc.2013.07.021
108. Huang W, Wu K, Wu R, Chen Z, Zhai W, Zheng J. Bioinformatic gene analysis for possible biomarkers and therapeutic targets of hypertension-related renal cell carcinoma. Transl Androl Urol. (2020) 9:2675–87. doi: 10.21037/tau-20-817
109. Oto J, Herranz R, Plana E, Sánchez-González JV, Pérez-Ardavín J, Hervás D, et al. Identification of miR-20a-5p as Robust Normalizer for Urine microRNA Studies in Renal Cell Carcinoma and a Profile of Dysregulated microRNAs. Int J Mol Sci. (2021) 22. doi: 10.3390/ijms22157913
110. Hosseinahli N, Aghapour M, Duijf PHG, Baradaran B. Treating cancer with microRNA replacement therapy: A literature review. J Cell Physiol. (2018) 233:5574–88. doi: 10.1002/jcp.v233.8
111. He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi S, et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int J Biol Sci. (2020) 16:2628–47. doi: 10.7150/ijbs.47203
112. Garber K. Alnylam launches era of RNAi drugs. Nat Biotechnol. (2018) 36:777–8. doi: 10.1038/nbt0918-777
113. Yasuda M, Keel S, Balwani M. RNA interference therapy in acute hepatic porphyrias. Blood. (2023) 142:1589–99. doi: 10.1182/blood.2022018662
114. Scott LJ, Keam SJ. Lumasiran: first approval. Drugs. (2021) 81:277–82. doi: 10.1007/s40265-020-01463-0
115. van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent Malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. (2017) 18:1386–96. doi: 10.1016/S1470-2045(17)30621-6
116. Anastasiadou E, Seto AG, Beatty X, Hermreck M, Gilles ME, Stroopinsky D, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, slows DLBCL tumor cell growth in vitro and in vivo. Clin Cancer Res. (2021) 27:1139–49. doi: 10.1158/1078-0432.CCR-20-3139
117. Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M, et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol. (2018) 183:428–44. doi: 10.1111/bjh.2018.183.issue-3
118. van der Ree MH, van der Meer AJ, de Bruijne J, Maan R, van Vliet A, Welzel TM, et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. (2014) 111:53–9. doi: 10.1016/j.antiviral.2014.08.015
119. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. (2017) 16:203–22. doi: 10.1038/nrd.2016.246
120. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. (2004) 303:83–6. doi: 10.1126/science.1091903
121. Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. (2002) 21:4663–70. doi: 10.1093/emboj/cdf476
122. Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. (2003) 425:415–9. doi: 10.1038/nature01957
123. Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. (2004) 18:3016–27. doi: 10.1101/gad.1262504
124. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. (2004) 432:231–5. doi: 10.1038/nature03049
125. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. (2003) 17:3011–6. doi: 10.1101/gad.1158803
126. Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. (2001) 293:1146–50. doi: 10.1126/science.1064023
127. Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PloS Biol. (2005) 3:e235. doi: 10.1371/journal.pbio.0030235
128. Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. (2011) 59:671–81. doi: 10.1016/j.eururo.2011.01.044
129. Gorski SA, Vogel J, Doudna JA. RNA-based recognition and targeting: sowing the seeds of specificity. Nat Rev Mol Cell Biol. (2017) 18:215–28. doi: 10.1038/nrm.2016.174
Keywords: miR-152, urological malignancy, therapeutic target, biomarker, microRNA
Citation: Li X, Qian B, Chen X, Shen M, Zhao S, Zhang X and He J (2024) The role of miR-152 in urological tumors: potential biomarkers and therapeutic targets. Front. Immunol. 15:1464327. doi: 10.3389/fimmu.2024.1464327
Received: 13 July 2024; Accepted: 28 October 2024;
Published: 13 November 2024.
Edited by:
Giandomenico Roviello, University of Firenze, ItalyReviewed by:
Soumasree De, University of Bern, SwitzerlandYing Shi, Huazhong University of Science and Technology, China
Copyright © 2024 Li, Qian, Chen, Shen, Zhao, Zhang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinsheng Zhang, emhhbmd4czA2MjRAMTYzLmNvbQ==; Jian He, amlhbmhlMjAyMDA2MDZAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Xin Li1†
Xin Li1† Xu Chen
Xu Chen Jian He
Jian He