- 1Department of Biomedical Science and Environmental Biology, PhD Program in Life Sciences, College of Life Science, Kaohsiung Medical University, Kaohsiung, Taiwan
- 2School of Dentistry, Taipei Medical University, Taipei, Taiwan
- 3Department of Oral and Maxillofacial Surgery, Chi-Mei Medical Center, Tainan, Taiwan
- 4School of Post-Baccalaureate Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan
- 5Department of Radiation Oncology, Kaohsiung Medical University Hospital, Kaohsiung Medical University, Kaohsiung, Taiwan
- 6Graduate Institute of Natural Products, Kaohsiung Medical University, Kaohsiung, Taiwan
- 7Department of Pharmacy and Master Program, College of Pharmacy and Health Care, Tajen University, Pingtung, Taiwan
- 8Department of Computer Science and Information Engineering, National Pingtung University, Pingtung, Taiwan
- 9Department of Urology, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan
- 10Department of Urology, Kaohsiung Gangshan Hospital, Kaohsiung Medical University, Kaohsiung, Taiwan
- 11Center for Cancer Research, Kaohsiung Medical University, Kaohsiung, Taiwan
- 12Department of Medical Research, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan
Immunogenic cell death (ICD) spatiotemporally regulates damage-associated molecular patterns (DAMPs) derived from dying cancer cells to signal the immune response. Intriguingly, these DAMPs and cytokines also induce cellular responses in non-immune cells, particularly cancer cells. Several ICD-modulating natural products and miRNAs have been reported to regulate the DAMP, cytokine, and cell death responses, but they lack systemic organization and connection. This review summarizes the impacts of natural products and miRNAs on the DAMP and cytokine responses and cancer cell death responses (apoptosis, autophagy, ferroptosis, necroptosis, and pyroptosis). We establish the rationale that ICD inducers of natural products have modulating effects on miRNAs, targeting DAMPs and cytokines for immune and cancer cell death responses. In conclusion, DAMP, cytokine, and cell death responses are intricately linked in cancer cells, and they are influenced by ICD-modulating natural products and miRNAs.
1 Introduction
1.1 Immunogenic cell death, damage-associated molecular patterns, and cytokines
ICD induces antitumor immunity by triggering several immune signals and damage-associated molecular patterns (DAMPs) involved in immune and cell death responses (1). DAMPs are molecules that induce intracellular responses but generate immunogenic responses in an extracellular environment (2). Generally, DAMPs are released by damaged or dying cells and tissues, triggering an innate immune response against these damaged and infected cells as well as cancer cells (3).
The danger signals from DAMPs include the cell surface exposure of calreticulin (CALR), the release of the high-mobility group box 1 (HMGB1) protein, and the secretion of ATP (4). Subsequently, they cooperatively activate dendritic cells and cytotoxic T lymphocytes (CTLs), causing them to kill cancer cells (5). LDL receptor-related protein-1 (CD91), P2X7 receptor (P2X7R), and Toll-like receptor 2 (TLR2) are located on the dendritic cell surface and recognize CALR, ATP, and HMGB1, respectively.
In addition to DAMPs, ICD inducers also promote the secretion of inflammatory cytokines from cancer cells. The DAMPs and cytokines activate dendritic and natural killer (NK) cells, promoting the secretion of effector cytokines. After ICD inducer treatment, the dying cancer cells promote DAMP responses, and, in turn, they release cytokines that activate the immune response, such as C-X-C motif chemokine ligand 10 (CXCL10; IL8) and interleukin 6 (IL6) (6, 7); additionally, IFNG (IFN-γ) is released by T helper 1 (Th1) cells and CTLs and interleukin 17A (IL17A; IL17) is released by Th17 cells (8). Moreover, activated dendritic cells secrete interleukin 12 (IL12), which causes NK cells to secrete interferon γ (IFNG; IFNγ) and tumor necrosis factor-alpha (TNF; TNFA; TNFα) (9). Furthermore, macrophages secrete interferon β (IFNB1; IFNβ1; IFNB), promoting apoptosis in neutrophils (10). Myeloid-derived suppressor cells (MDSCs) are responsible for the immune suppression activity of macrophages and dendritic cells (11). Therefore, ICD modulates DAMP and cytokine responses.
1.2 Some DAMPs and cytokines involved in ICD were selected as ICD gene candidates
As described above, ICD initiates spatiotemporal DAMP signals, such as the cell surface translocation of CALR and heat shock proteins (HSP70 and HSP90) and the release of ATP and HMGB1, leading to cell death (12). Consequently, DAMPs and cytokines are vital for ICD induction.
In this review, we selected DAMPs involved in ICD modulation, including HMGB1, CALR, heat shock protein family A (HSP70) member 1A (HSPA1A), HSP70 member 1B (HSPA1B), heat shock protein 90 alpha family class A member 1 (HSP90AA1), and heat shock protein 90 beta family member 1 (HSP90B1) (1, 3, 12, 13). The selected cytokines involved in ICD modulation include interleukin 6 (IL6), C-X-C motif chemokine ligand 8 (CXCL8; IL8), CXCL10, IL12A, IL12B, IL17A, IL23A (IL23), IFNB1, IFNG, and TNF (14, 15). The protein signaling pathway involving both DAMPs and selected cytokines in ICD modulation were shown (Figure 1) (15–21).
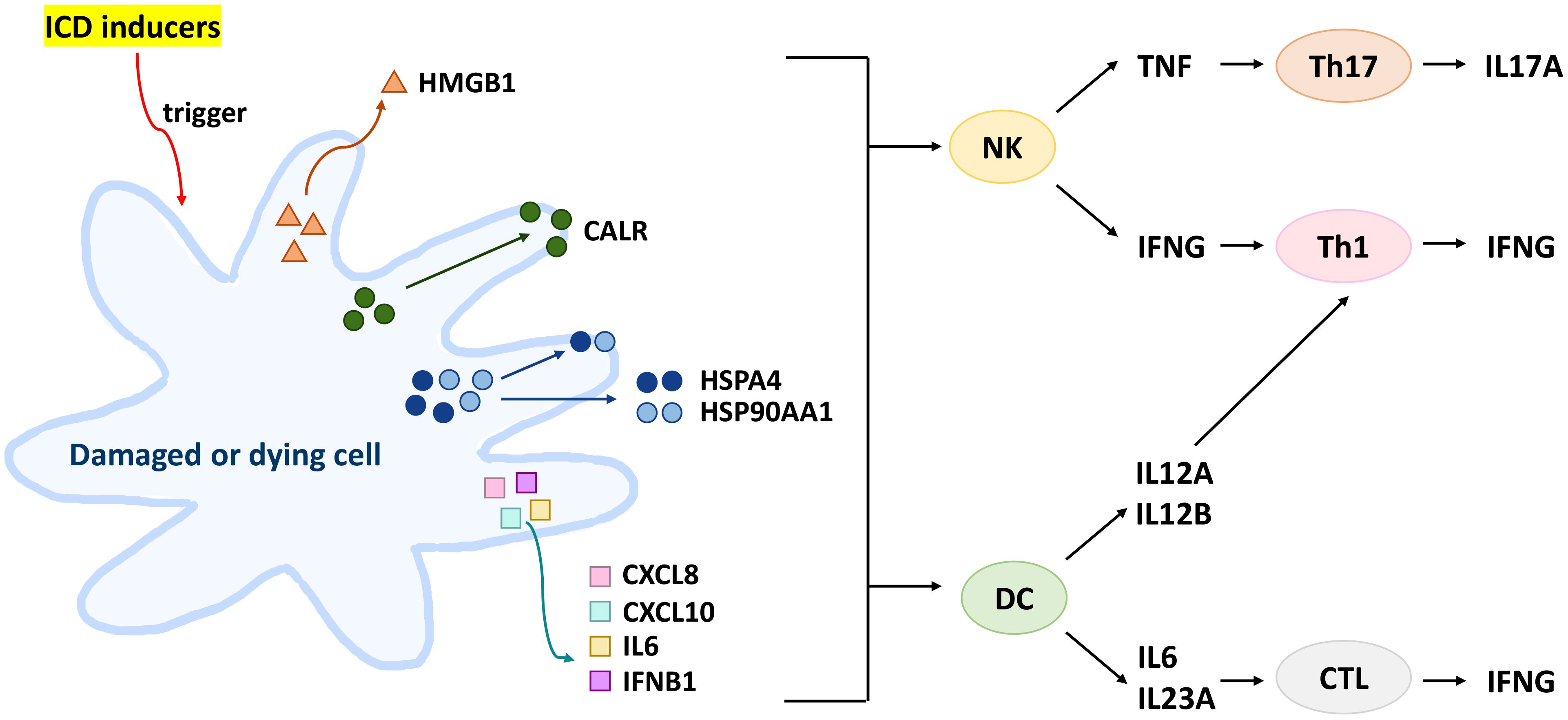
Figure 1. Protein signaling pathway involving both DAMPs and selected cytokines in ICD modulation. ICD inducer triggers HMGB1 secretion, CALR surface exposure, HSPA4/HSP90AA1 surface exposure and secretion, and inflammatory cytokine secretion from dying cancer cells or damaged cells. Subsequently, HSPA4/HSP90AA1 and inflammatory cytokines can activate both NK and DC cells. NK cells secrete INFG and TNF to activate Th1 and Th17 to secrete IFNG and IL17A, respectively. Moreover, DC cells secrete IL12A and IL12B to activate Th1 cells and secrete IL6 and IL23A to activate CTL cells for IFNG secretion.
1.3 miRNAs have ICD-modulating effects
miRNAs are short nucleotide (21-23 nts) molecules that can regulate genes both positively (22–25) and negatively (2). Several miRNAs exhibit tumor-promoting or suppressing effects. In the immune response, miRNAs may regulate the ICD-modulated expression, translocation, and secretion of some DAMPs (2). Moreover, miRNAs may regulate cytokine responses in a manner that modulates the immune response (26, 27). Consequently, DAMP-targeting miRNAs are potential regulators of immune-modulated responses and cell death in cancer cells (2).
1.4 ICD inducers of natural products
Drugs with the ability to induce ICD have anticancer effects by enhancing DAMP and cell death responses (28). Several natural products exhibiting ICD-inducing functions have been reported (28). However, the impact of miRNA regulation on natural ICD-modulating products remains unclear. Later, we will illustrate the organization between ICD, natural products, and miRNAs in detail.
1.5 Rationale for this review
Many natural products exhibit miRNA-modulating effects when used for anticancer treatment (29, 30). Moreover, many of these products have induced ICD (31, 32). Accordingly, natural products have the potential to regulate ICD through miRNAs, but they lack systemic organization.
This review illustrates the systemic connection between ICD (DAMP and cytokine responses), cell death responses (apoptosis, autophagy, ferroptosis, necroptosis, and pyroptosis), miRNAs, and natural products (Figure 2). With the help of bioinformatics, the potential ICD targets of ICD-modulating miRNAs in natural product studies were retrieved from miRDB (270). The detailed reports on ICD, natural products, and miRNAs from the literature were retrieved using Google Scholar. Finally, we propose the rationale that the ICD inducers among natural products modulate ICD targets that involve DAMPs and cytokines (Section 2). In this review, miRNAs that target DAMPs and cytokines were collected, and the immune and cell death responses were integrated (Sections 3 and 4). Moreover, the interaction between ICD-inducing natural products and ICD-modulating miRNAs is also explored (Section 5).
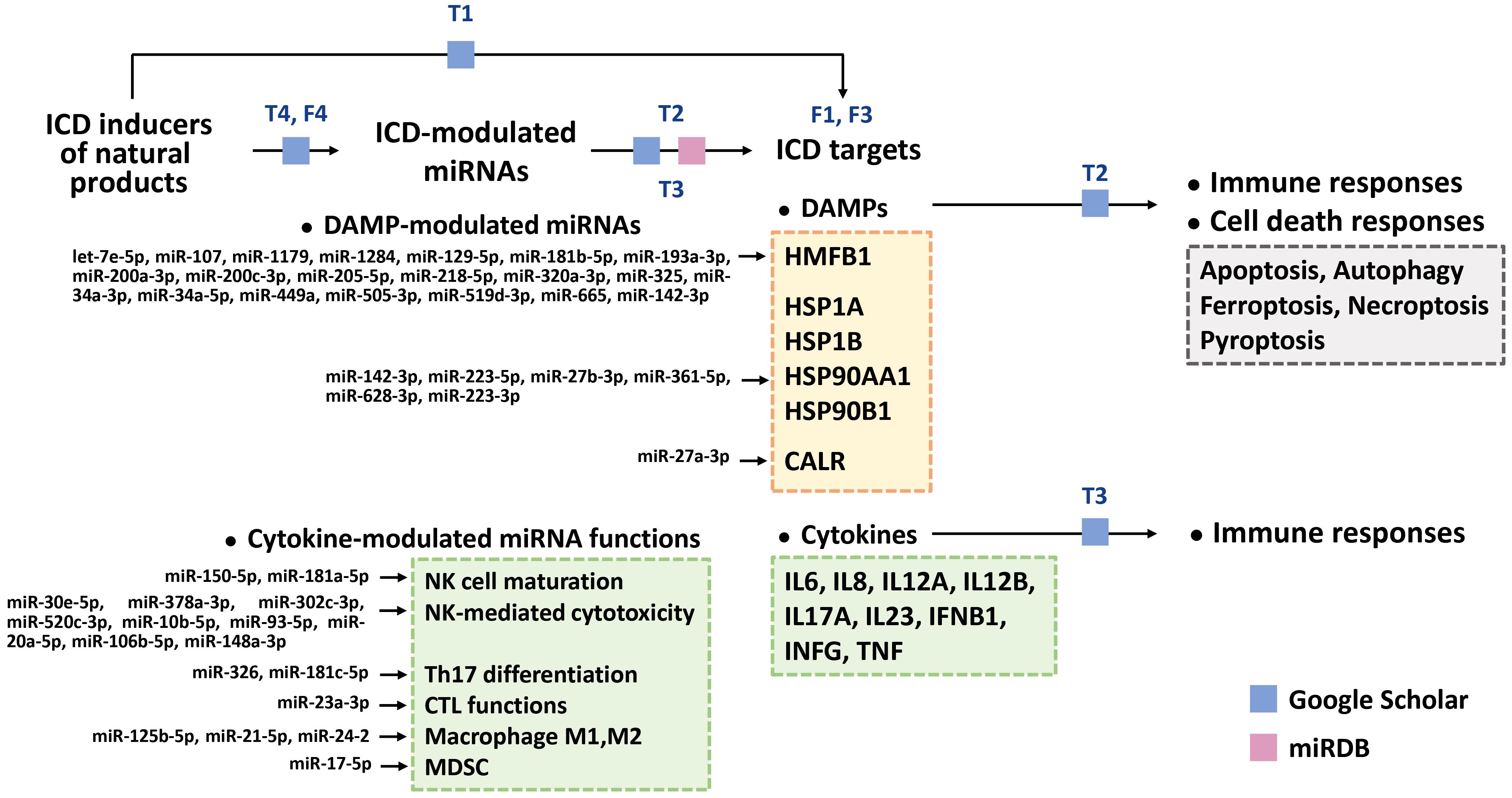
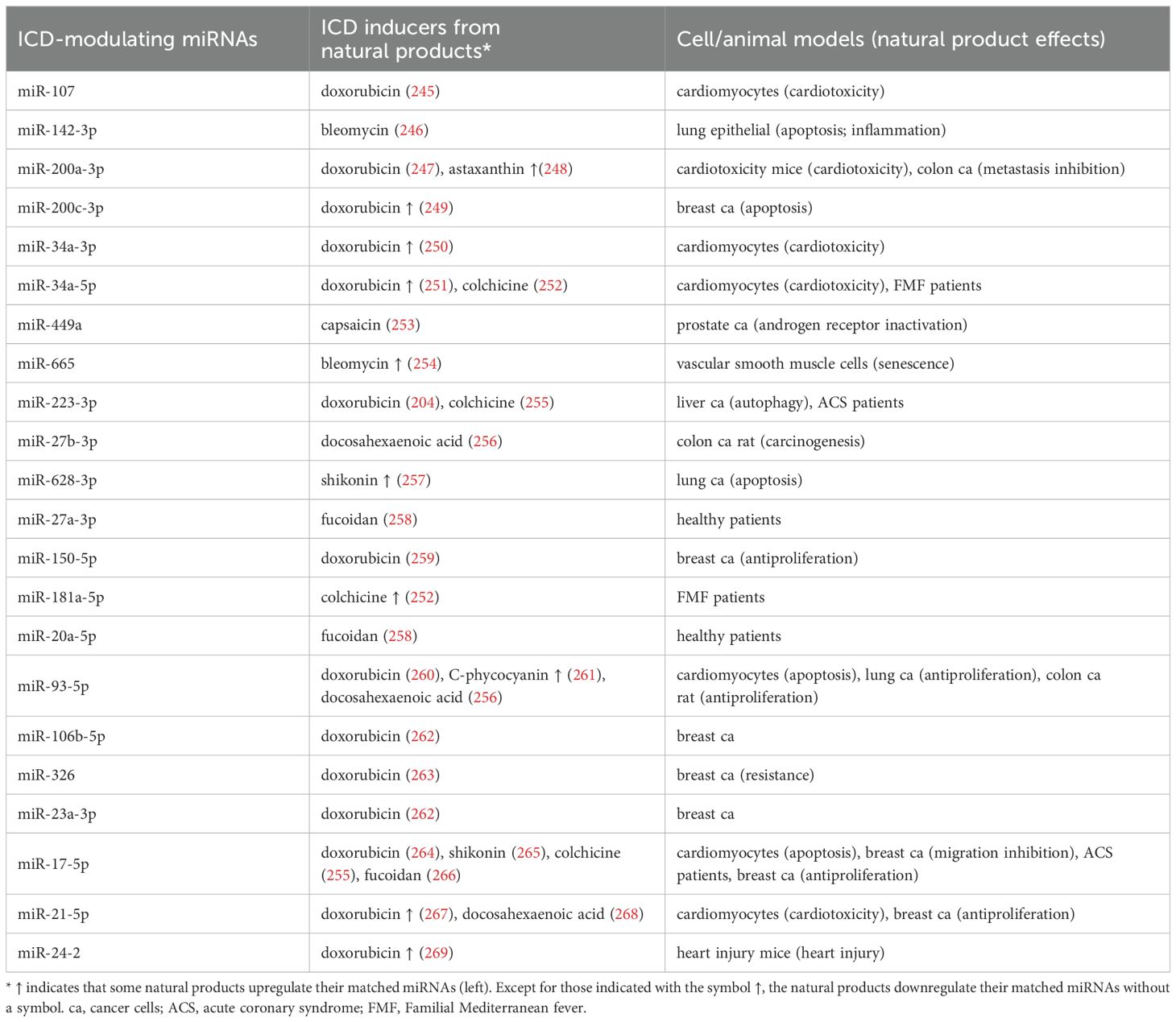
Figure 2. Arrangement of this review. Using Google Scholar, we organized natural products with ICD-inducing functions and their ICD targets, such as DAMPs and cytokines [Table 1 (T1)]. ICD-modulating miRNAs that regulate DAMPs and cytokines were also retrieved [Tables 2 and 3 (T2, T3)]. Based on our searches in Google Scholar and miRDB (270), the potential ICD targets (DAMPs (HMGB1, CALR, HSP1A, HSP1B, HSP90AA1, and HSP90B1) [Figure 3 (F3)] and cytokines [IL6, IL8, IL12A, IL12B, IL17A, IL23, IFNB1, INFG, and TNF)] for these miRNAs were collected (Tables 2, 3). Moreover, the DAMP-modulated miRNAs (Table 1) targeting DAMPs are shown. In comparison, the functions of cytokine-modulated miRNAs (Table 2) are presented. For DAMP-modulating miRNAs, the immune and cell death responses were categorized (Table 2). Cell death responses, such as apoptosis, autophagy, ferroptosis, necroptosis, and pyroptosis, were also retrieved. For cytokine-modulating miRNAs, their immune responses and expression levels in cancer cells were categorized [Table 3 (T3)]. Finally, we ascertained the relationship between ICD inducers of natural products and ICD-modulating miRNAs [Table 4 (T4) and Figure 4 (F4)].
2 ICD-inducing natural products that modulate DAMPs and cytokines
Many natural compounds can induce ICD (31, 32). Most natural product studies focus on cancer cells’ antiproliferative and immunomodulatory activity, while evidence for the induction of ICD in the immune system is limited. Consequently, we summarized the ICD-inducing natural products, focusing on the ICD-modulated responses to DAMPs and cytokines, particularly in cancer cells (Table 1). The ICD inducers of natural products are classified based on their targeting network, which is constructed by analyzing the STRING database (271) (Figure 3). The natural products are classified into five functions: cardiac glycosides, topoisomerase II inhibitors, anti-mitotic agents, antibiotics, and multiple functions (Table 1) (Sections 2.1-2.5).
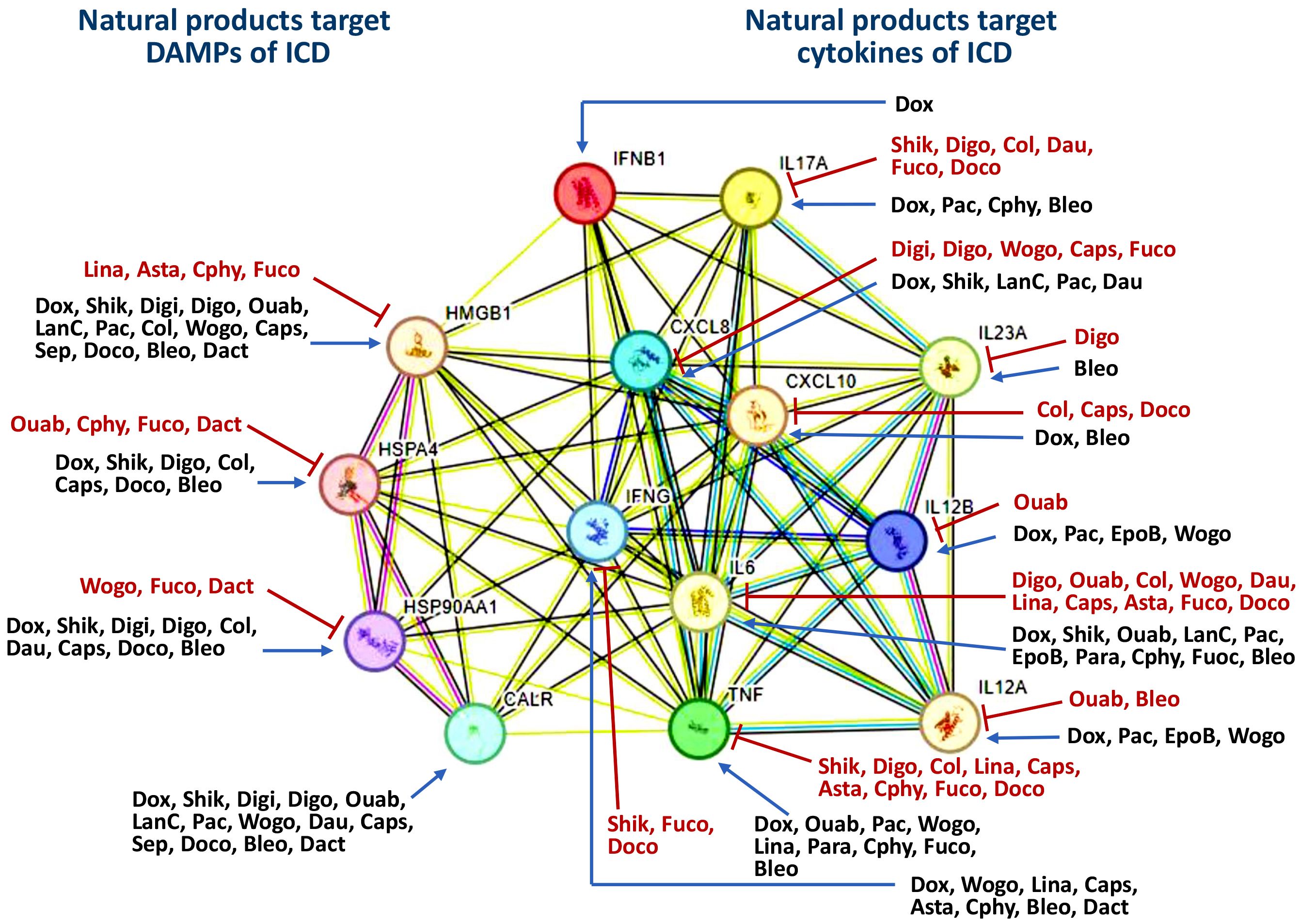
Figure 3. Classification of ICD inducers of natural products into different classes based on their targeting. The potential interaction for targets was analyzed by STRING database. All the information was derived from Table 1. The symbols of “T” and “arrow-line” indicate that natural products downregulate and upregulate their targets. HSP70 and HSP90 are labeled with HSPA4 and HSP90AA1 in the network, respectively. Dox, Doxorubicin; Shik, Shikonin; Digi, Digitoxin; Digo, Digoxin; Ouab, Ouabain; LanC, Lanatoside C; Pac, Paclitaxel; Col, Colchicine; EpoB, Epothilone B; Wogo, Wogonin; Dau, Daunorubicin; Lina, Linalool; Caps, Capsaicin; Asta, Astaxanthin; Para, Paramylon; Cphy, C-phycocyanin; Fuco, Fucoidan; Sep, Septacidin; Doco, Docosahexaenoic acid; Bleo, Bleomycin; Dact, Dactinomycin.
2.1 Cardiac glycosides
2.1.1 Digitoxin and digoxin
Digitoxin (33) and digoxin (35), foxglove (Digitalis purpurea)-derived cardenolides, upregulate ICD-related molecules such as DAMPs (HSP90, CALR, and HMGB1) in osteosarcoma cells (Table 1) (Figure 3). Digoxin also upregulates HSP70 levels in patients with chronic heart failure (36).
These natural products also induce ICD-modulated cytokine responses (Table 1) (Figure 3). Digitoxin downregulates CXCL8 in cultured lung epithelial cells (34). Similarly, digoxin downregulates various ICD-modulating cytokines, as well as CXCL8, IL6, and TNF, in peripheral blood mononuclear cells (37). Digoxin downregulates bortezomib-induced IL23A in brain vascular endothelial cells (38) and IL17A in colonic mucosa (39). Accordingly, digitoxin and digoxin regulate DAMPs’ and ICD-modulated cytokines’ functions.
2.1.2 Ouabain and lanatoside C
Ouabain and lanatoside C, the cardiac glycosides, induce DAMP responses (CALR exposure, ATP secretion, and HMGB1 release) (Table 1) (Figure 3) (40). Moreover, these natural products regulate ICD-modulating cytokines. For example, ouabain upregulates IL6 and TNF in peripheral blood mononuclear cells (42). In comparison, it downregulates IL6 signaling (43), TNF-induced IL12 (44), and IL17A (45) in cultured skeletal muscle cells, dendritic cells, and bronchial epithelial cells, respectively. Additionally, lanatoside C upregulates IL6 and CXCL8 in pericytes (46). Accordingly, ouabain and lanatoside C regulate DAMPs and ICD-modulated cytokines’ functions.
2.2 Topoisomerase II inhibitors
2.2.1 Doxorubicin
Doxorubicin upregulates DAMPs (CARL, HSP70, and HSP90), causing them to translocate to the cell surface and causing the release of HMGB1 in leukemia, ovarian, and prostate cancer cells (Table 1) (Figure 3) (14). In terms of ICD-modulating cytokines, doxorubicin upregulates the production of IL6 (50) and IFNG (272). IL17A enhances the doxorubicin sensitivity of breast cancer cells (52). Moreover, doxorubicin upregulates CXCL8 and TNF in lung cancer cells (48) and IL12-induced IFNG in xenografted breast cancer (51). Accordingly, it regulates DAMPs and ICD-modulating cytokines’ functions.
2.2.2 Daunorubicin
Daunorubicin, a Streptomyces peucetius-derived antibiotic, induces DAMPs (CARL exposure and the release of HSP70/HSP90) in acute myeloid leukemia cells (53) (Table 1) (Figure 3). It also modulates several ICD-modulating cytokines. Histone deacetylase 8 (HDAC8) is upregulated in daunorubicin-resistant AML cells. In contrast, the inhibition of HDAC8 promotes daunorubicin sensitivity by downregulating IL6 (54). Moreover, the upregulation of AKT and IL17A enhances the daunorubicin resistance of B cell acute lymphoblastic leukemia (ALL) cells (55), suggesting that daunorubicin downregulates IL17A.
2.2.3 Dactinomycin
Actinomycin D (dactinomycin), a natural chromopeptide, promotes the release of HMGB1 and IFNG and the exposure of CALR in osteosarcoma cells (Table 1) (Figure 3) (56). In chicken embryo cells, TGFβ upregulates HSPA4 and HSP90AA1, which are downregulated by dactinomycin (57).
Accordingly, bleomycin and dactinomycin modulate DAMPs and ICD-modulated cytokines’ effects.
2.3 Microtubule inhibitors
2.3.1 Paclitaxel
Paclitaxel is an ICD inducer whose activity is characterized by the release of HMGB1 in osteosarcoma cells (58) and upregulation of CALR in lung cancer cells (Table 1) (Figure 3) (59). Moreover, paclitaxel regulates ICD-modulating cytokines. For example, it upregulates CXCL8 and IL6 in ovarian cancer patients (60) and CXCL10 expression in lung cancer cells (61). In in vivo studies, paclitaxel upregulated the expression of IL12 in macrophages in fibrosarcoma-xenografted mice (62), of IL17A in solid Ehrlich carcinoma mouse models (63), and of TNF in hippocampus tissue (64).
2.3.2 Colchicine and epothilone B
Originally isolated from Colchicum autumnale, colchicine triggers dendritic cell maturation (65) and upregulates or downregulates DAMPs or ICD-modulating cytokines (Table 1) (Figure 3). It upregulates DAMPs (HMGB1, HSPA4, and HSP90AA1) in melanoma cells, for example (66). In comparison, colchicine downregulates CXCL10 (67). Moreover, it downregulates IL6 (68), IL17A (69), and LPS-induced TNF (70) expression in cardiac fibroblasts, atrial fibrillation patients, and macrophages, respectively. Additionally, epothilone B (patupilone), a Sorangium cellulosum-derived microtubular inhibitor, is an ICD inducer in ovarian cancer cells that upregulates IL12 and IL6 (71).
Accordingly, paclitaxel, colchicine, and epothilone B modulate DAMPs and ICD-modulated cytokines’ functions.
2.4 Antibiotics
Septacidin, an L-heptopyranose isolated from Streptomyces fibriatus, enhances CALR exposure and ATP and HGMB1 secretion from osteosarcoma cells (Table 1) (Figure 3) (72). Bleomycin, a Streptomyces verticillus-derived antibiotic, induces HMGB1, CALR, and IFNG expression in colon cancer cells (73). It upregulates HSPA4 in the lung epithelium (75) and HSP90AA1 in the interstitial lung fibroblasts (76) of mice. Moreover, bleomycin regulates several ICD-modulating cytokines. For example, it increases γδ T-cell populations and upregulates CXCL10, affecting inflammation (77). Bleomycin upregulates IL6 and TNF in the lung homogenates of CBA/J mice (78), as well as IL17A and IL23A in C57BL/6 mice (80). In comparison, the transcription factor Fli-1 is downregulated in systemic sclerosis. Bleomycin downregulates IL12A in Fli-1+/− mice, a skin fibrosis model (79).
2.5 Multiple functions
2.5.1 Shikonin and wogonin
Shikonin promotes the release of HSP70, HSP90, and HMGB1 in melanoma cells by enhancing immunogenic apoptosis (Table 1) (Figure 3) (81). It also upregulates DAMPs (HMGB1, HSP70, and CALR) in glioma cells (82). In comparison, it inhibits several ICD-modulating cytokines. For example, it suppresses TNF-induced IL6 and CXCL8 production in human periodontal ligament cells (273) and inhibits T cell proliferation by downregulating IFNG and IL17A (84). It also downregulates TNFA in rheumatoid arthritis-like cell models (85).
Wogonin, a Scutellaria baicalensis-derived natural product, induces DAMP responses (Table 1) (Figure 3). For example, it promotes ICD and ER stress in dendritic cells, causes CALR exposure on the cell surface, and triggers the release of HMGB1 and ATP (86). Subsequently, these released molecules cause dendritic cells to secrete cytokines (86). In breast cancer cells, wogonin inhibits proteins downstream of HSP90AA1 such as EGFR, Cdk4, and survivin (87).
Moreover, wogonin regulates ICD-modulating cytokines. For example, it suppresses IL-1β’s promotion of IL6 and CXCL8 expression in retinal pigment epithelial cells (88). It also induces IL12 and TNF expression in breast cancer cells (89) and downregulates IFNG generation in splenocytes (90).
Accordingly, shikonin and wogonin regulate DAMPs and ICD-modulated cytokines’ functions.
2.5.2 Linalool and capsaicin
Linalool improves Th1 cellular immunity in breast cancer cells by inducing the release of IFNG and TNF in lymphocytes (Table 1) (Figure 3) (91). It downregulates HMBG1, TNF, and IL6 in cisplatin-induced acute kidney injury in rat models (92).
Capsaicin, a red pepper-derived compound, suppresses the proliferation of many cancer cells (Table 1) (Figure 3) (93). It induces DAMPs in human bladder cancer cells by upregulating HMBG1, CALR, HSPA4, and HSP90AA1 (94). Regarding ICD-modulating cytokines, capsaicin downregulates TNF, IL6, and CXCL8 in monocytes (95). It has an anti-inflammatory effect in wound healing by downregulating TNF, IL6, and CXCL10 (96). In comparison, it upregulates IFNG in murine Peyer’s patch cells (97).
Accordingly, daunorubicin, linalool, and capsaicin regulate DAMPs and ICD-modulated cytokines’ functions.
2.5.3 Astaxanthin, paramylon, and C-phycocyanin
Astaxanthin, a carotenoid derived from the green alga Hematococcus pluvialis, induces ICD (Table 1) (Figure 3) (1). It attenuates spinal cord edema by downregulating HMGB1 in rat models (99). Moreover, it modulates several ICD-modulating cytokines, such as by enhancing immunity through inducing the release of IFNG in lymphocytes (98). Astaxanthin downregulates IL6 expression in activated microglia (100) and cerulein-/resistin-stimulated pancreatic acinar cells (101) and TNF expression in LPS-treated macrophages (102).
Paramylon, a Euglena gracilis-derived beta-(1–>3)-D-glucan, induces ICD (Table 1) (Figure 3) (274). Regarding cytokine regulation, paramylon nanofibers upregulate TNF and IL6 mRNA expression in lymphocytes (103).
Spirulina microalgae-derived C-phycocyanin induces ICD (104). It enhances the secretion of IL6 and TNF in murine macrophages (Table 1) (Figure 3) (104). HMGB1 induces ulcers, which, in turn, are suppressed by its downregulation. C-phycocyanin attenuates ethanol-induced gastric ulcers in rats by downregulating HMGB1 and TNF (105). Similarly, dietary C-phycocyanin increases the lifespan of Drosophila melanogaster by downregulating HSPA4, a member of the HSP70 family (106). In comparison, some cytokines are induced by C-phycocyanin, which upregulates IFNG and IL17A in BALB/c mice (107).
Accordingly, astaxanthin, paramylon, and C-phycocyanin can regulate DAMPs and ICD-modulated cytokines.
2.5.4 Fucoidan and docosahexaenoic acid
Fucoidan can modulate DAMPs and ICD-modulated cytokines (Table 1) (Figure 3). For example, it inhibits HSPA4 and HSP90AA1 protein expression in liver cancer cells (110), while downregulating HMGB1, IL6, and TNF levels in ischemia–reperfusion rats (109). It also induces IL6 and TNF expression, promoting the maturation of spleen dendritic cells (108). Fucoidan-rich Ascophyllum nodosum extract (Ascophyscient®) downregulates TNF, IL6, and CXCL8 in bronchial epithelial cells (111). Fucoidan downregulates IL17A and IFNG in T helper cells (Th1/Th2/Th17) (275).
Docosahexaenoic acid (DHA) enhances myeloma apoptosis by improving CALR exposure, HMGB1, and HSP90 secretion and activating dendritic cells (Table 1) (Figure 3) (112). DHA also upregulates HSPA4 in rainbow trout leukocytes (113). In comparison, it downregulates several ICD-modulating cytokines, such as Cxcl10 in the lupus flaring mouse model (114). DHA downregulates IL6 and TNF secretion in LPS-activated dendritic cells, preventing their maturation (115). It also downregulates IL17A in T cells in psoriatic skin models (116). Moreover, DHA intake downregulates IFNG production in mice (117).
Accordingly, fucoidan, septacidin, and DHA show a regulating effect on DAMP and ICD-modulated cytokines.
3 DAMP-modulating miRNAs
Several miRNAs that modulate DAMPs can regulate ICD and tumor proliferation (2). miRNAs exhibit tumor-promoting and tumor-suppressing effects by targeting various DAMPs, whose expression levels are changed in cancers (2). Therefore, the ICD response of the immune system and the cell death responses of ICD-modulating miRNAs need further investigation.
This section presents an overview of the impacts of miRNAs on immune and cell death responses. The immune responses affected by DAMP-modulating miRNAs (Section 3.1), and cell death responses affected by DAMP-modulating miRNAs (Section 3.2) and DAMP-targeting miRNAs (Section 3.3) are described (Table 2).
3.1 Immune responses affected by DAMP-modulating miRNAs
Some DAMP-targeting miRNAs modulate immune responses (Table 2). For example, miR-181b-5p upregulation in chronic lymphocytic leukemia B cells enhances cytotoxic T cell function, inhibiting tumor growth (131). miR-200c-3p inhibits the tumor-infiltrating function of macrophages (147), while miR-34a-5p enhances that of CD8+ T lymphocytes (169).
miR-142-3p is highly expressed in immune cells, such as CD3+ spleen lymphocytes derived from experimental autoimmune encephalomyelitis mice, compared to Complete Freund’s Adjuvant CD3+ (188) (Table 2). miR-223-3p enhances the differentiation of macrophages via M2 polarization (203).
miR-27b-3p enhances ammonia-triggered apoptosis by targeting TNF receptor-associated death domain (TRADD), Fas-associated death domain (FADD), and apoptotic protease activating factor-1 (APAF1) (Table 2). Ammonia induces apoptosis and immunosuppression by causing a T helper cell type 1 (Th1)/Th2 imbalance and regulatory T cell (Treg)/Th17 imbalance in chicken peripheral blood lymphocytes (196). Accordingly, miR-27b-3p promotes immunosuppression through Th1/Th2 and Treg/Th17 imbalances.
Notably, most of the miRNAs listed in Table 2 are rarely investigated in terms of their involvement in the immune response. In the future, the immune-regulating function of these ICD-modulating miRNAs should be carefully and more thoroughly investigated.
3.2 Cell death responses of DAMP-modulating miRNAs
ICD is a general term that includes various cell death responses, such as apoptosis, autophagy, ferroptosis, pyroptosis, and necroptosis (276, 277). These responses exert synergistic effects by enhancing or suppressing antitumor immune responses (277). Currently, the cell death responses of ICD-modulating miRNAs are rarely highlighted. To address this gap, we outline miRNAs that modulate ICD (Table 2) by regulating apoptosis, autophagy, ferroptosis, necroptosis, and pyroptosis in the following (Sections 3.3 to 3.5).
3.3 DAMP-targeting miRNAs
DAMPs, such as HMGB1, HSP70, HSP90, and CALR (Sections 3.3.1-3.3.4), are targeted by various miRNAs (Table 2). Based on a search of miRDB, we describe the potential targets of DAMPs and cytokines (270). The cell death responses they influence, including apoptosis, autophagy, ferroptosis, necroptosis, and pyroptosis, are summarized (Table 2) (Figure 2).
3.3.1 HMGB1-targeting miRNAs
A series of investigations have reported that many miRNAs have HMGB1-modulating functions. By targeting HMGB1, these miRNAs (let-7e-5p, miR-107, miR-1179, miR-1284, miR-129-5p, miR-181b-5p, miR-193a-3p, miR-200a-3p, miR-200c-3p, miR-205-5p, miR-218-5p, miR-320a-3p, miR-325, miR-34a-3p, miR-34a-5p, miR-449a, miR-505-3p, miR-519d-3p, miR-665, and miR-142-3p) exhibit anticancer effects associated with various cell death responses (Table 2) (Figure 2) (Sections 3.3.3.1-3.3.3.6).
3.3.1.1 let-7e-5p, miR-107, miR-1179, and miR-1284
let-7e-5p overexpression suppresses the proliferation and migration of thyroid cancer cells by targeting and downregulating HMGB1 (118) (Table 2). Regarding cell death responses, let-7e-5p downregulation triggers apoptosis but blocks autophagy in articular chondrocytes (119). miR-107 is underexpressed in breast cancer cell lines and tissues. Its overexpression inhibits proliferation, migration, and autophagy by downregulating HMGB1 (120), while its upregulation triggers apoptosis in glioma cells (121). Moreover, miR-107 enhances proliferation by downregulating the LPS-triggered pyroptosis of chondrocytes (122) (Table 2).
Gastric cancer shows low levels of miR-1179. The overexpression of miR-1179 inhibits gastric cell migration and proliferation and promotes apoptosis by targeting HMGB1 (123) (Table 2), while its downregulation promotes autophagy in oral cancer cells (124). Moreover, miR-1284 is underexpressed in cervical cancer cell lines and tissues. Its overexpression sensitizes cells to cisplatin and promotes apoptosis in cervical cancer cells by downregulating HMGB1 (125) (Table 2).
Accordingly, let-7e-5p, miR-107, miR-1179, and miR-1284 modulate cell death responses.
3.3.1.2 miR-129-5p, miR-181b-5p, and miR-193a-3p
miR-129-5p is less expressed and HMGB1 is more expressed in gastric cancer than in normal tissues (126) (Table 2). The former regulates several cell death responses. For example, its upregulation triggers apoptosis in gastric cancer cells by downregulating HMGB1. miR-129-5p improves the radiosensitization of colon cancer cells by downregulating autophagy (127). It blocks ferroptosis in intestinal epithelial cells (128), while a miR-129-5p antagomir attenuates LPS-triggered neuronal pyroptosis in rat pheochromocytoma cells (129) (Table 2).
miR-181b-5p is underexpressed in AML patients. Its overexpression sensitizes cells to doxorubicin, suppressing proliferation and inducing apoptosis in AML cells, by targeting HMGB1 (130) (Table 2). This miRNA also regulates several cell death responses. For example, it inhibits ginsenoside Rg3’s suppression of proliferation in gallbladder cancer cells by upregulating autophagy (132). miR-181b-5p is highly expressed in osteoarthritic cell models, while in osteoarthritic chondrocytes, its downregulation inhibits ferroptosis by upregulating GPX4 (133). Atrazine triggers inflammation and necroptosis in carp lymphocytes by downregulating miR-181-5p (134). The overexpression of the latter attenuates NLRP3 inflammasome-mediated pyroptosis in vascular endothelial cells (135) (Table 2).
miR-193a-3p is more downregulated in lung cancer than in normal cells (278). It inhibits the migration and proliferation of lung cancer cells and downregulates HMGB1 (136) (Table 2). miR-193a-3p modulates several cell death responses. For example, it suppresses proliferation and causes apoptosis in colon cancer cells (137). miR-193a-3p mimics downregulated autophagy in liver cancer cells, which was reverted by miR-193a-3p inhibitors (138). miR-193a-3p downregulation caused congenital heart disease by upregulating ferroptosis in rat cardiomyocytes (139) (Table 2).
Accordingly, miR-129-5p, miR-181b-5p, and miR-193a-3p modulate cell death responses.
3.3.1.3 miR-200a-3p, miR-200c-3p, and miR-205-5p
miR-200a-3p shows low expression in liver cancer, while HMGB1 is highly expressed (140) (Table 2). This miRNA inhibits liver cancer cell proliferation by downregulating HMGB1 and modulates several cell death responses. For example, miR-200a-3p upregulation suppresses proliferation and promotes apoptosis (141) in prostate cancer cells. Its upregulation also attenuates diabetic cardiomyopathy damage in mice by upregulating autophagy (142), while its downregulation suppresses hypoxia/reoxygenation (H/R)-triggered ferroptosis and protects the myocardial cells (143). Moreover, miR-200a-3p mimics suppress lipopolysaccharide-induced inflammation and necrosis in intestinal epithelial cells (144). miR-200a-3p’s downregulation suppresses pyroptosis in human aortic endothelial cells (145) (Table 2).
miR-200c-3p shows low expression in non-small cell lung cancer (NSCLC) (146). It suppresses epithelial–mesenchymal transition, invasion, and migration in lung cancer cells by downregulating HMGB1 (146) (Table 2). It modulates several cell death responses. For example, miR−200c−3p mimics promote proliferation and suppress apoptosis in trabecular meshwork cells (148). miR-200c-3p induces autophagy in prostate cancer cells (149), and its downregulation attenuates high glucose-induced pyroptosis in human retinal microvascular endothelial cells (HRMECs) (150), suggesting that miR-200c-3p has a pyroptosis-inducing function (Table 2).
miR-205-5p shows low expression in breast cancer cells, enhancing EMT and invasion (151). Its overexpression suppresses the migration and proliferation of breast cancer cells by targeting HMGB1 (Table 2). Similarly, miR-205-5p is underexpressed in gastric cancer. Its overexpression suppresses proliferation and metastasis and triggers apoptosis in gastric cancer cells (152). miR-205 inhibits autophagy, improving the cisplatin sensitivity of prostate cancer cells (153). lncAABR07025387.1, highly expressed in myocardial ischemia/reperfusion (MI/R) injury, sponges miR-205-5p and upregulates ferroptosis (154) (Table 2), suggesting that miR-205-5p downregulates ferroptosis.
Accordingly, miR-200a-3p, miR-200c-3p, and miR-205-5p modulate cell death responses.
3.3.1.4 miR-218-5p, miR-320a-3p, and miR-325-3p
miR-218-5p is underexpressed in NSCLC (279). The overexpression of miR-218-5p inhibits migration in lung cancer cells by targeting and downregulating HMGB1 (155) (Table 2). miR-218-5p modulates several cell death responses. For example, its upregulation promotes apoptosis in cervical cancer cells (156). miR-218-5p silencing suppresses proliferation by triggering apoptosis and autophagy in rheumatoid arthritis synovial fibroblasts (RASFs) (157). Moreover, necrotic and hypoxic mesenchymal glioblastomas exhibit low miR-218-5p levels (158), suggesting that miR-218-5p downregulates necrosis in mesenchymal glioblastoma cells.
miR-320a-3p is downregulated in liver cancer tissues (159). Its overexpression suppresses the invasion and metastasis of liver cancer cells by targeting HMGB1 (Table 2). miR-320a-3p modulates several cell death responses. For example, LINC00963 overexpression induces the apoptosis and autophagy of diffuse large B-cell lymphoma by downregulating miR-320a, which is reversed by miR-320a-3p mimics (160). LINC00460 downregulation inhibits proliferation but enhances ferroptosis in breast cancer cells by upregulating miR-320a-3p (161) (Table 2).
In lung cancer patients, miR-325-3p and HMGB1 are underexpressed and overexpressed, respectively (162) (Table 2). The overexpression of miR-325-3p inhibits the proliferation of lung cancer cells by targeting HMGB1. miR-325-3p modulates several cell death responses. For example, it inhibits the proliferation of gastric cancer cells by triggering apoptosis (163). Its downregulation suppresses myocardial ischemia/reperfusion-induced autophagy and cell death in cardiomyocytes (164) (Table 2).
Accordingly, miR-218-5p, miR-320a-3p, and miR-325-3p modulate cell death responses.
3.3.1.5 miR-34a-3p, miR-34a-5p, and miR-449a
miR-34a-3p is expressed at low levels in retinoblastoma cells (280). Its overexpression inhibits proliferation and autophagy and triggers apoptosis in retinoblastoma cells by targeting HMGB1 (165) (Table 2). It also modulates other cell death responses. For example, anti-miR-34a-3p inhibits late apoptosis and necrosis in meningioma cells (166). LncRNA SNHG7 suppresses pyroptosis in liver cancer cells by sponging miR-34a-3p (167). This suggests that miR-34a-3p promotes apoptosis, necrosis, and pyroptosis.
miR-34a-5p shows low expression in colon cancer tissues (168). Its upregulation in colon cancer cells inhibits proliferation and migration by targeting HMGB1 (Table 2). It modulates other cell death responses. For example, apigenin triggers apoptosis in lung cancer cells by upregulating miR-34a-5p (170). miR-34a-5p enhances autophagy in chicken ovarian granulosa cells (171), while its downregulation inhibits pyroptosis in doxorubicin-induced cardiomyopathy (172) (Table 2).
miR-449a is poorly expressed in lung cancer cells and tissues (173). Its upregulation suppresses the proliferation and migration of lung cancer cells by targeting HMGB1 (Table 2). miR-449a modulates several cell death responses, suppresses proliferation and promotes apoptosis in liver cancer cells (174), and suppresses autophagy in T lymphocytes (175) (Table 2).
Accordingly, miR-34a-3p, miR-34a-5p, and miR-449a modulate cell death responses.
3.3.1.6 miR-505-3p, miR-519d-3p, miR-665, and miR-142-3p
miR-505-3p is downregulated in liver cancer cells. Its overexpression inhibits the proliferation and invasion of liver cancer cells by targeting HMGB1 (176) (Table 2). miR-505-3p modulates other cell death responses. For example, its upregulation promotes apoptosis in lung cancer cells, which is reversed by its downregulation (177). miR-505-3p blocks autophagy in rat primary neurons by downregulating HMGB1 (178) (Table 2).
miR-519d-3p shows low expression in lung cancer tissues, promoting proliferation and migration by upregulating HMGB1 (179) (Table 2). Hence, it potentially targets HMGB1. Moreover, it triggers apoptosis and autophagy in liver cancer cells (180).
miR-665 is underexpressed in glioma (181). Its upregulation inhibits glioma cell proliferation, migration, and invasion by downregulating HMGB1 (Table 2). miR-665 modulates several cell death responses. For example, its mimics stimulate cell proliferation and suppress lung (281) and breast (183) cancer cell apoptosis. lncRNA MIAT promotes autophagy in Bacillus Calmette-Guerin (BCG)-infected macrophages by sponging miR-665 (184), suggesting that the latter inhibits macrophage autophagy. Moreover, dexmedetomidine downregulates miR-665, attenuating myocardial ischemia–reperfusion injury (MIRI) by inhibiting pyroptosis (185) (Table 2), suggesting that miR-665 promotes pyroptosis.
HMGB1 is a target of miR-142-3p (186) in breast cancer cells (Table 2). miR-142-3p is expressed at lower levels in breast cancer than in normal cells (282). Moreover, doxorubicin-resistant breast cancer cells exhibit lower levels of miR-142-3p than parental cells. miR-142-3p modulates several cell death responses. For example, its overexpression induces apoptosis and inhibits breast cancer cell autophagy, attenuating doxorubicin resistance by downregulating HMGB1 (186). miR-142-3p triggers ferroptosis in M1 macrophages, improving liver cancer development (189), and its upregulation attenuates coronary microembolization (CME)-induced pyroptosis in myocardial injury (190) (Table 2).
Accordingly, miR-505-3p, miR-519d-3p, miR-665, and miR-142-3p modulate cell death responses by targeting HMGB1.
3.3.2 HSP70 and HSP90-targeting miRNAs
Some members of the HSP70 family were chosen as DAMP targets in this review, including HSPA1B and HSPA1A (Section 3.3.2.1). Some members of the HSP90 family were also selected, including HSP90AA1 and HSP90B1 (Section 3.3.2.2). These are targeted by several ICD-modulating miRNAs (Table 2) (Figure 2), and these HSP70 and HSP90-targeting miRNAs exhibit anticancer effects that are associated with various cell death responses (Table 2) (Figure 2).
3.3.2.1 HSP70-targeted miRNAs
HSPA1B and HSPA1A are targeted by miR-223-5p and miR-142-3p (Table 2). Osteosarcoma overexpresses HSP70, which is downregulated by miR-223-5p (191). The overexpression of the latter enhances cisplatin sensitivity in osteosarcoma, while the upregulation of miR-142-3p in pancreatic cancer cells inhibits proliferation by targeting HSPA4 (HSP70) (187).
Regarding cell death responses, the apoptosis- and necroptosis-modulating effects of miR-223-5p are summarized (Table 2). miR-223-5p inhibitors suppress apoptosis signaling, such as caspase-3 in spinal cord injury rats (192), suggesting that miR-223-5p induces apoptosis. Moreover, miR-223-5p and miR-223-3p jointly inhibit necroptosis in ischemic/reperfused mouse hearts (194).
3.3.2.2 HSP90-targeted miRNAs
HSP90AA1 and HSP90B1 are targeted by miR-27b-3p/miR-361-5p/miR-628-3p and miR-223-3p, respectively (Table 2). miR-27b-3p is underexpressed in lung cancer (283). LncRNA KCNQ1OT1 enhances the proliferation of lung cancer cells by upregulating HSP90AA1 and downregulating miR-27b-3p (195). Accordingly, miR-27b-3p suppresses HSP90AA1 expression in lung cancer cells. Moreover, miR-27b-3p promotes tamoxifen-triggered apoptosis in breast cancer cells (197) (Table 2).
miR-361-5p shows lower levels in cervical cancer than in normal tissues and cells (198). Its upregulation suppresses EMT and the invasion of cervical cancer cells by targeting HSP90 (Table 2). miR-361-5p also triggers cell death responses. For example, ovarian cancer cells exhibit high levels of the miRNA, whereas, in these cells, its downregulation promotes apoptosis (199). Moreover, miR-361-5p inhibits autophagy’s ability to improve chemoresistance in gastric cancer cells (200). Sp1 knockdown inhibits prostate cell growth and hypoxia-triggered autophagy by upregulating miR-361-5p (193). In comparison, miR-628-3p mimics cause apoptosis and suppress the migration of lung cancer cells by targeting and downregulating HSP90AA1 (201) (Table 2).
By targeting HSP90B1, miR-223-3p can induce anticancer effects and cell death responses (Table 2). For example, osteosarcoma shows a low level of oncogenic heat shock protein 90 kDa beta member 1 (HSP90B1) (202), a member of the HSP90 family. The upregulation of miR-223-3p suppresses osteosarcoma cell proliferation and promotes apoptosis by downregulating HSP90B1.
miR-223-3p also triggers cell death responses (Table 2). For example, doxorubicin induces autophagy in liver cancer cells by downregulating miR-223-3p. The upregulation of the latter suppresses doxorubicin-triggered autophagy, improving the chemoresistance of liver cancer cells (204). miR-223-3p enhances pyroptosis in cardiomyocytes (207). In comparison, exosomal miR-223-3p isolated from mesenchymal stem cells inhibits HBV-X protein (HBx)-triggered ferroptosis in podocytes (205), as well as necroptosis in macrophage cell death (206) (Table 2).
3.3.3 CALR-targeting miRNAs
miR-27a-3p targets CALR (2). It is overexpressed in colon cancer cells and tissues, improving proliferation, suppressing apoptosis (208) (Table 2) (Figure 2), and triggering pre-osteoblast cell autophagy (209). SLC7A11, which is highly expressed in lung cancer patients, triggers the ferroptosis of lung cancer cells by downregulating miR-27a-3p (210), suggesting that miR-27a-3p inhibits ferroptosis.
Following treatment with ICD inducers (oxaliplatin and mitoxantrone), miR-27a-3p knockdown induces more cell surface CALR and HMGB1 secretion by colon cancer cells compared to that observed in cells overexpressing miR-27a-3p (284).
3.3.4 Potential targets for DAMPs and cytokines according to miRDB search
Based on a search of the miRDB, in addition to targeting HMGB1, miR-181b-5p, miR-320a-3p, and miR-34a-5p/miR-449a potentially target DAMPs (CALR/HSP90B1, HSPA4, and HSPA1B) (270) (Table 2) (Figure 2). let-7e-5p, miR-1284, miR-181b-5p, miR-200a-3p, miR-320a-3p, miR-34a-3p, miR-34a-5p, and miR-505-3p also potentially target ICD-modulating cytokines, such as IL6, IL12B, TNF, IL17A, IL12B, TNF, CXCL10, IFNG, and HSPA1B, which are co-targeted by HMGB1 according to a search of miRDB (270) (Table 2) (Figure 2).
4 Cytokine-modulating miRNAs
In addition to DAMPs, ICD inducers may promote the secretion of inflammatory cytokines such as IL6 and CXCL8 (IL8) from cancer cells. The DAMPs and cytokines activate dendritic cells and NK cells, which then release effector cytokines, stimulating CTL and Th1 cells to release IFNG and Th17 cells to release IL17 (15). Moreover, M0 macrophages are stimulated to differentiate into M1 or M2 macrophages by several cytokines. M1 macrophages regulate many inflammatory, cytotoxic, and tissue damage functions. In comparison, M2 macrophages inhibit inflammatory and immune functions and promote tissue repair (285). MDSCs are responsible for the immune suppression activity of macrophages and dendritic cells (11). Several miRNAs regulating the cytokines that modulate factors related to immune responses, such as NK cell maturation, NK-mediated cytotoxicity, Th17 differentiation, CTL function, macrophage M1/M2 polarization, and MDSC levels, have been summarized (Table 3) (Figure 2) (Sections 4.1-4.5).
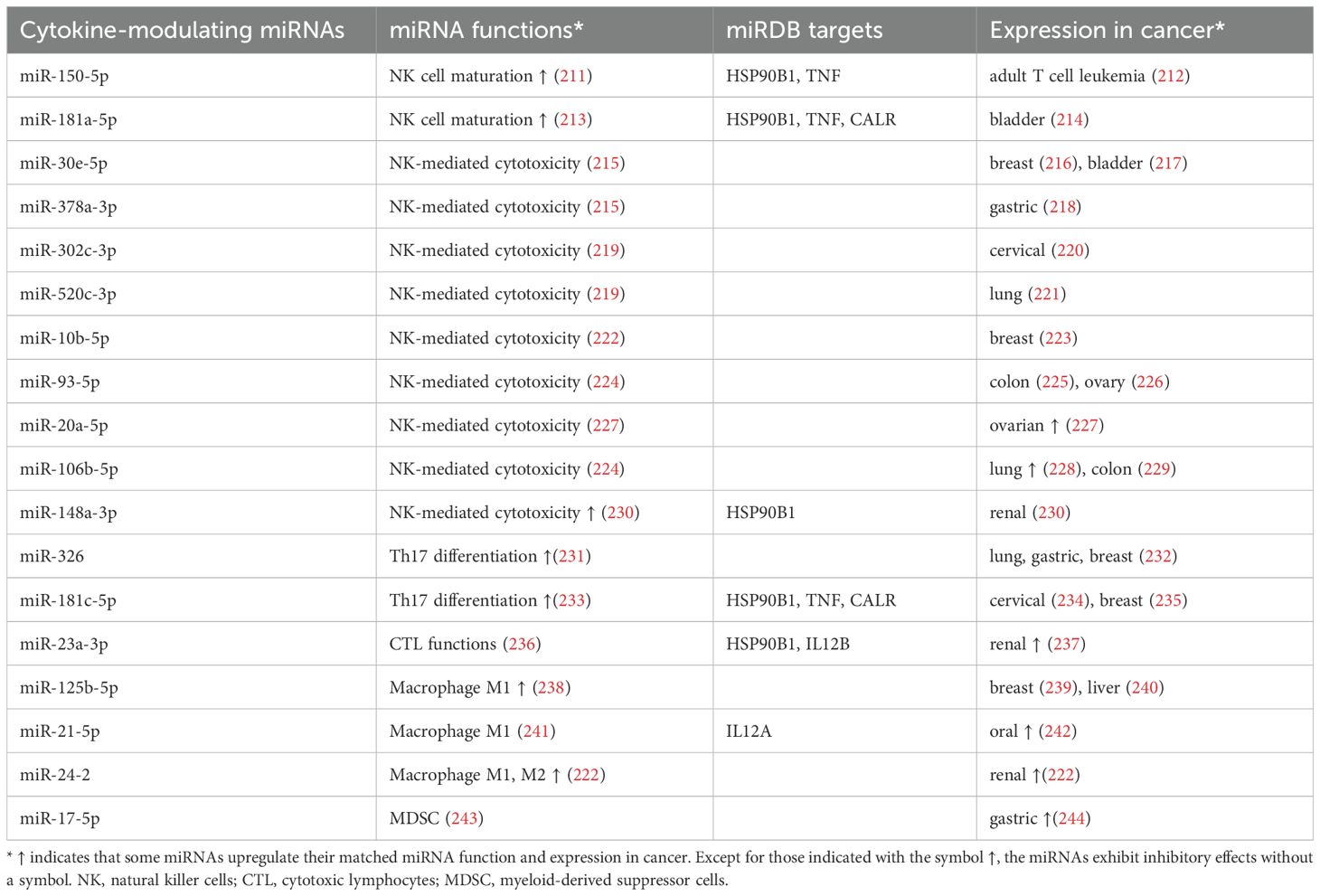
Table 3. Potential miRDB targets, immune function, and expression in cancer cells for cytokine-modulating miRNAs.
4.1 NK cell maturation
NK cell maturation is enhanced by miR-150-5p and miR-181a-5p (Table 3) (Figure 2). By directly targeting c-Myb, miR-150-5p improves the development and maturation of NK cells (211). miR-181a-5p enhances NK cell maturation by downregulating nemo-like kinase (NLK), an inhibitor of Notch signaling (Table 3) (213). Moreover, miR-150-5p (212) and miR-181a-5p (214) are downregulated in adult T-cell leukemia (ATL) and bladder cancer cells. Accordingly, they both exhibit tumor-suppressive potential by improving NK cell maturation; however, this needs further validation.
4.2 NK-mediated cytotoxicity
Several miRNAs, such as miR-30e-5p, miR-378a-3p, miR-302c-3p, miR-520c-3p, miR-10b-5p, miR-20a-5p, miR-93-5p, and miR-106b-5p, have inhibitory effects on NK-mediated cytotoxicity, while miR-148a-3p has induces it (Table 3).
miR-30e-5p and miR-378a-3p inhibit NK cell cytotoxicity (215) (Table 3) (Figure 2). Moreover, these miRNAs also exhibit anticancer effects. Breast (216) and bladder (217) cancer tissues and cells show low levels of miR-30e-5p (Table 3). miR-378a-3p is underexpressed in gastric cancer. When it is upregulated, proliferation is inhibited and apoptosis is promoted in gastric cancer cells (218). Similarly, 1,25(OH)2D3 inhibits miR-302c-3p and miR-520c-3p expression, improving NK cell cytotoxicity in breast cancer cells (219), suggesting that miR-302c-3p and miR-520c-3p downregulate NK cell cytotoxicity. Moreover, miR-302c-3p (220) and miR-520c-3p (221) are downregulated in cervical and lung cancer cells. The overexpression of the former suppresses proliferation and induces apoptosis in cervical cancer cells.
MHC class I chain-related protein B (MICB) is a stress-induced ligand of the activating NK-cell receptor NKG2D. miR-10b-5p inhibits MICB expression, suppressing NK cell cytotoxicity and thereby promoting cancer immune evasion, suggesting that miR-10b-5p downregulates NK cell cytotoxicity (286) (Table 3). In comparison, and based on data from the TCGA database, breast cancer tissues express lower levels of miR-10b-5p than normal controls (223). Moreover, the overexpression of miR-93-5p and miR-106b-5p downregulates MICA expression (224), suppressing NK cell cytotoxicity (287). Colon (225) and ovarian (226) cancer tissues exhibit low levels of miR-93-5p (Table 3), the upregulation of which inhibits the proliferation of these two types of cancer. Accordingly, miR-30e-5p, miR-378a-3p, miR-302c-3p, miR-520c-3p, miR-10b-5p, and miR-93-5p have tumor-promoting potential because they suppress NK-mediated cytotoxicity, which warrants detailed study.
Furthermore, ovarian cancer tissues exhibit high levels of miR-20a-5p, which downregulates NK cell cytotoxicity in ovarian cancer cells (Table 3) and shows tumor-promoting effects (227). Hence, miR-20a-5p overexpression promotes the immune escape of ovarian cancer cells from NK cells. In comparison, miR-106b-5p has a dual role in tumor suppression and in promoting functionality. It is downregulated and upregulated in colon (229) and lung (228) cancer tissues (Table 3), respectively, enhancing lung cancer cell proliferation and suppressing colon cancer metastasis.
In contrast, some miRNAs exhibit NK-mediated cytotoxicity (Table 3) (Figure 2). Classical human leukocyte antigen G (HLA-G) is commonly expressed in renal cancer cells, inhibiting the cytotoxic activity of T and NK cells (230). The overexpression of miR-148a-3p downregulates HLA-G expression and induces cell death in renal cancer cells, activating NK cell cytotoxicity (230) (Table 3). Consequently, miR-148a-3p is downregulated in renal cancer cells and, accordingly, possesses a tumor-suppressive function, upregulating NK-mediated cytotoxicity. This warrants a detailed investigation.
4.3 Th17 differentiation and CTL function
Some miRNAs, such as miR-326 and miR-181c-5p, induce Th17 differentiation (Table 3) (Figure 2). Both enhance Th17 development by targeting the negative regulators ETS-1 (231) and SMAD7 (233). miR-326 suppresses lung tumor growth in mice by promoting T cell cytotoxicity (288). It also consistently suppresses immune escape and metastasis in lung cancer cells (288). Th17 cells are crucial for a host’s defense against specific bacteria and fungi and for their anticancer functions (289–291). Accordingly, these miRNAs have anticancer potential. miR-326 is underexpressed in lung, gastric, and breast cancers (232). Similarly, miR-181c-5p shows low expression in cervical (234) and breast (235) cancer cells (Table 3). Therefore, miR-326 and miR-181c-5p have anticancer effects, in addition to promoting Th17 differentiation.
Some miRNAs induce CTL cytotoxicity. miR-23a-3p inhibits the cytotoxicity of CD8+ CTLs, while miR-23a-3p knockdown attenuates TGF-β-induced immunosuppression (236) (Table 3) (Figure 2). Additionally, miR-23a-3p is highly expressed in renal cancer tissues and cells (237), whereas its knockdown inhibits cell proliferation in these cells. Accordingly, miR-23a-3p has regulatory effects on CTL and causes anticancer.
4.4 Macrophage M1/M2 polarization and MDSC level
M1 macrophage polarization exhibits pro-inflammatory and anti-tumor functions, whereas M2 macrophage polarization causes immunosuppression and promotes tumor formation (292). Some miRNAs have modulating effects on macrophages (Table 3) (Figure 2). miR-125b-5p is upregulated in M1 macrophages, enhancing antigen presentation for T-cell activation and inhibiting tumor growth (238). Moreover, miR-125b-5p is underexpressed in breast (239) and liver (240) cancer cells, while their proliferation and migration are inhibited by its overexpression. This suggests miR-125b-5p has tumor-suppressive effects.
In contrast, miR-21-5p and miR-24-2 are downregulated in M1 macrophages. miR-21-5p depletion promotes pro-inflammatory and tumoricidal macrophage (M1) polarization (293) (Table 3) (Figure 2), while its overexpression inhibits lymphocyte migration and enhances immunotherapeutic resistance to breast cancer (241). M1 macrophage stimulation downregulates miR-24-2 expression, but M2 macrophage stimulation has the opposite effect (294), suggesting that miR-24-2 induces M2 macrophages and promotes tumor growth. Moreover, miR-21-5p and miR-24-2 are highly expressed in oral cancer patients (242) and renal cancer tissues (295) (Table 3). Consequently, both have tumor-promoting effects.
Furthermore, MDSCs have immunosuppressive effects (296) and are modulated by miRNAs. miR-17-5p inhibits immune suppression in MDSCs derived from colon tumor-bearing mice (243) (Table 3) (Figure 2). Moreover, miR-17-5p is highly expressed in gastric cancer patients (244) and, consequently, has tumor-promoting effects.
4.5 Potential targets for cytokine-regulating miRNAs
As previously mentioned, target information for these cytokine-modulating miRNAs (Table 3) is rarely reported. By applying miRDB data mining (270), their potential targets can be described as follows: miR-150-5p, miR-181a-5p, miR-148a-3p, miR-181c-5p, and miR-23a-3p potentially target HSP90B1; miR-150-5p, miR-181a-5p, and miR-181c-5p can target TNF; and miR-181a-5p and miR-181c-5p potentially target CALR. Moreover, IL12A and IL12B are potentially targeted by miR-21-5p and miR-23a-3p, respectively. In the future, a detailed examination is warranted to explore the role of these targets in miRNA-regulating immune and anticancer activity.
5 Relationship between ICD-modulating natural products and miRNA
We analyzed the relationship between ICD-modulating natural products (Table 1) and miRNAs (Tables 2 and 3). From a miRNA-centric view (Table 4), different miRNAs may be regulated by various ICD inducers of natural products; meanwhile, different natural products may regulate the same miRNAs.
From the point of view of natural products, the regulation of miRNAs by doxorubicin, shikonin, colchicine, capsaicin, astaxanthin, C-phycocyanin, fucoidan, docosahexaenoic acid, and bleomycin is summarized in Figure 4.
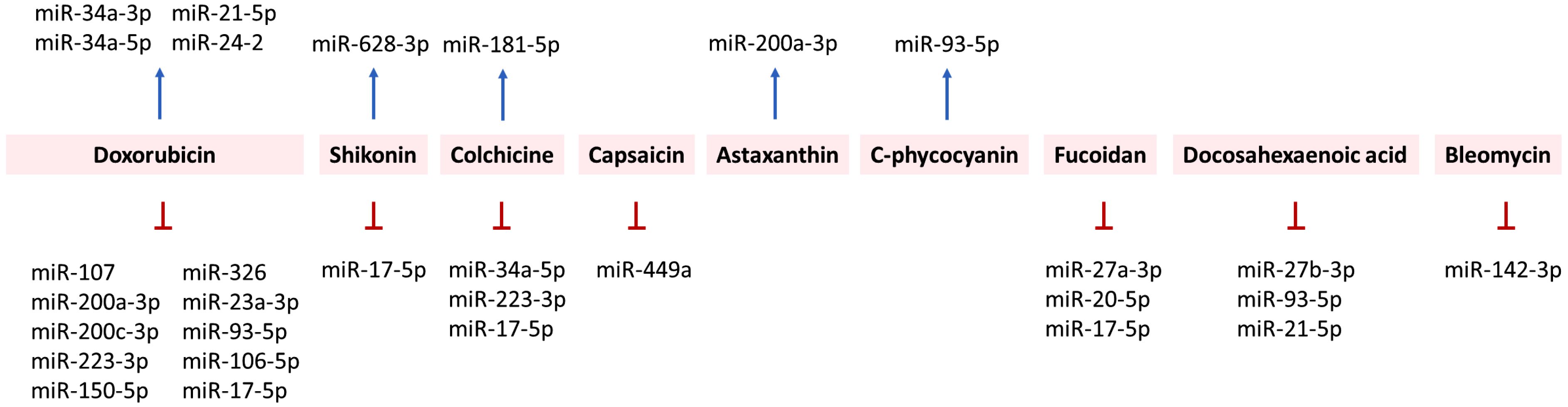
Figure 4. Relationship between ICD inducers of natural products and the regulation of ICD-modulating miRNAs.
5.1 Doxorubicin
In both non-cancer and cancer studies, oxorubicin has exhibited miRNA-modulating effects. In non-cancer studies (cardiomyocytes), doxorubicin has caused cardiotoxicity by upregulating miR-34a-3p (250), miR-34a-5p (251), and miR-21-5p (267). In comparison, doxorubicin downregulated miR-107 (245), miR-93-5p (260), and miR-17-5p (264) in cardiomyocytes. In animal models, doxorubicin induced cardiotoxicity in mice by upregulating miR-24-2 (269) and downregulating miR-200a-3p (247).
In cancer studies, doxorubicin may downregulate or upregulate miRNAs depending on the type of cancer cell. miR-200c-3p suppresses the migration of lung cancer cells by downregulating HMGB1 (146) but enhances the doxorubicin sensitivity of breast cancer cells (249). This finding suggests that doxorubicin may be associated with the upregulation of miR-200c. In comparison, doxorubicin downregulates miR-223-3p in liver cancer cells (204). Under short-term starvation, doxorubicin downregulates miR-23a-3p and miR-106b-5p in breast cancer cells (262). Moreover, miR-150-5p inhibitors suppress the migration and viability of doxorubicin-treated breast cancer cells, suggesting that doxorubicin inhibits breast cancer cell proliferation by downregulating miR-150-5p (259). Accordingly, doxorubicin may differentially regulate miRNAs in various cancer cells.
5.2 Shikonin, colchicine, capsaicin, astaxanthin, and C-phycocyanin
Shikonin promotes apoptosis in lung cancer cells by increasing miR-628-3p expression, which is reversed by its inhibition (257). Shikonin inhibits the migration of breast cancer cells by downregulating miR-17-5p, which is overexpressed in breast cancer (265). This suggests shikonin has miR-628-3p-upregulating and miR-17-5p-downregulating abilities.
Colchicine downregulates miR-17-5p and miR-223-3p in acute coronary syndrome (ACS) patients (255). Colchicine upregulates miR-181a-5p but downregulates miR-34a-5p in Familial Mediterranean Fever (FMF) patients (252). Capsaicin upregulates miR-449a in prostate cancer cells (253). Astaxanthin exhibits anti-metastatic effects on colon cancer cells by upregulating miR-200a-3p (248). @Moreover, C-phycocyanin induces miR-93-5p expression in lung cancer cells (297). Accordingly, these miRNAs are differentially regulated by these natural products.
5.3 Fucoidan, docosahexaenoic acid, and bleomycin
Fucoidan and docosahexaenoic acid exhibit inhibitory effects on various ICD-modulating miRNAs. For example, fucoidan downregulates human plasma miR-27a-3p and miR-20a-5p in healthy patients (258). It also suppresses the proliferation of breast cancer cells by downregulating miR-17-5p (266). Similarly, docosahexaenoic acid downregulates miR-27b-3p and miR-93-5p in azoxymethane-induced colon cancer in Sprague–Dawley rats (256). Docosahexaenoic acid downregulates miR-21-5p in breast cancer cells (268).
In comparison, bleomycin may exhibit a dual function in regulating ICD-modulating miRNAs. miR-142-3p upregulation attenuates bleomycin-induced injury in lung epithelial cells (246), suggesting that bleomycin downregulates miR-142-3p. In comparison, bleomycin promotes the senescence of vascular smooth muscle cells by upregulating miR-665 (254). Accordingly, these miRNAs are differentially regulated by these natural products.
6 Conclusion
ICD is the spatiotemporal immune cell death process caused by the exposure of DAMPs from damaged or dying cancer cells, which triggers the release of many cytokines involved in cancer cell killing. Different DAMPs may be either translocated or show altered secretion in a spatiotemporal manner. However, the functions of DAMPs and cytokines are not limited to the immune response.
DAMPs exhibit intracellular functions but generate extra immunogenic effects when responding to extracellular stimulation (2). Accordingly, they have anticancer and immune-modulating effects. Moreover, natural products and miRNAs have been reported to modulate the immune (DAMPs and cytokines) and cancer cell death responses.
However, there are gaps in the literature regarding the interplay between ICD inducers of natural products and ICD-modulating miRNAs, between natural products and ICD targets, between ICD-modulating miRNAs and ICD targets, and between ICD-modulating miRNAs and immune and cell death responses.
To address these gaps, we have provided an integrated view connecting ICD, cell death responses, miRNAs, and natural products. In this review, we organized reports from the literature regarding the impacts of natural products and miRNAs on the DAMP, cytokine, and cell death responses (apoptosis, autophagy, ferroptosis, necroptosis, and pyroptosis) in various cancer types. We collated and mapped out the potential DAMP and cytokine targets and responses of ICD-modulating miRNAs and natural products. This improvement proves the rationale that ICD inducers of natural products modulate miRNAs, and they, in turn, target DAMPs and cytokines, triggering immune and cancer cell death responses.
Notably, we used miRDB to process target retrieval. This is a comprehensive and reliable database constituting a vast array of experimental data, but the target information acquired may be derived from different cells. This target information needs to be further validated in future ICD studies. Moreover, the functions of DAMPs and miRNAs are context-dependent (2). For instance, HMGB1 exhibits pro- and anti-tumoral functions depending on its location in extracellular or intracellular environments. Besides its immune functions, HMGB1 exhibits tumor suppression and oncogenic functions in the context of receptors, targeted cells, and redox status (298). Moreover, miRNAs can regulate different immune responses and modulate death responses in cancer cells. A detailed investigation of the relationship between the natural products and miRNA-modulated immune and cell death responses needs to be conducted in the future.
Altogether, this review summarizes the changes in DAMPs and cytokines and cell death responses in cancer cells, linking these with natural products and miRNAs with ICD-modulating effects. This work sheds light on the anticancer effects of natural products and the mechanisms by which they modulate ICD with respect to miRNAs.
Author contributions
Y-TC: Conceptualization, Methodology, Writing – original draft. C-YY: Conceptualization, Methodology, Writing – original draft. J-YT: Methodology, Writing – original draft. F-RC: Methodology, Writing – original draft. Y-HT: Methodology, Writing – original draft. K-CW: Methodology, Writing – original draft. T-MC: Conceptualization, Methodology, Supervision, Writing – review & editing. H-WC: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was partly supported by funds from the Ministry of Science and Technology (MOST 111-2320-B-037-015-MY3 and NSC 113-2314-B-037-063), the Kaohsiung Medical University (KMU-DK(A)113003), and the Kaohsiung Medical University Research Center (KMU-TC113A04).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sansone C, Bruno A, Piscitelli C, Baci D, Fontana A, Brunet C, et al. Natural compounds of marine origin as inducers of immunogenic cell death (ICD): potential role for cancer interception and therapy. Cells. (2021) 10:231. doi: 10.3390/cells10020231
2. Lamberti MJ, Nigro A, Casolaro V, Rumie Vittar NB, Dal Col J. Damage-associated molecular patterns modulation by microRNA: Relevance on immunogenic cell death and cancer treatment outcome. Cancers (Basel). (2021) 13:2566. doi: 10.3390/cancers13112566
3. Roh JS, Sohn DH. Damage-associated molecular patterns in inflammatory diseases. Immune Netw. (2018) 18:e27. doi: 10.4110/in.2018.18.e27
4. Balsamo JA, Penton KE, Zhao Z, Hayes MJ, Lima SM, Irish JM, et al. An immunogenic cell injury module for the single-cell multiplexed activity metabolomics platform to identify promising anti-cancer natural products. J Biol Chem. (2022) 298:102300. doi: 10.1016/j.jbc.2022.102300
5. Radogna F, Diederich M. Stress-induced cellular responses in immunogenic cell death: Implications for cancer immunotherapy. Biochem Pharmacol. (2018) 153:12–23. doi: 10.1016/j.bcp.2018.02.006
6. Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. (2008) 14:6735–41. doi: 10.1158/1078-0432.CCR-07-4843
7. Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J Clin Invest. (2007) 117:3660–3. doi: 10.1172/JCI34237
9. Ross ME, Caligiuri MA. Cytokine-induced apoptosis of human natural killer cells identifies a novel mechanism to regulate the innate immune response. Blood. (1997) 89:910–8. doi: 10.1182/blood.V89.3.910
10. Kumaran Satyanarayanan S, El Kebir D, Soboh S, Butenko S, Sekheri M, Saadi J, et al. IFN-beta is a macrophage-derived effector cytokine facilitating the resolution of bacterial inflammation. Nat Commun. (2019) 10:3471. doi: 10.1038/s41467-019-10903-9
11. Ma T, Renz BW, Ilmer M, Koch D, Yang Y, Werner J, et al. Myeloid-derived suppressor cells in solid tumors. Cells. (2022) 11:310. doi: 10.3390/cells11020310
12. Andocs G, Meggyeshazi N, Balogh L, Spisak S, Maros ME, Balla P, et al. Upregulation of heat shock proteins and the promotion of damage-associated molecular pattern signals in a colorectal cancer model by modulated electrohyperthermia. Cell Stress Chaperones. (2015) 20:37–46. doi: 10.1007/s12192-014-0523-6
13. Tukaj S, Sitko K. Heat shock protein 90 (Hsp90) and Hsp70 as potential therapeutic targets in autoimmune skin diseases. Biomolecules. (2022) 12:1153. doi: 10.3390/biom12081153
14. Fucikova J, Kralikova P, Fialova A, Brtnicky T, Rob L, Bartunkova J, et al. Human tumor cells killed by anthracyclines induce a tumor-specific immune response. Cancer Res. (2011) 71:4821–33. doi: 10.1158/0008-5472.CAN-11-0950
15. Showalter A, Limaye A, Oyer JL, Igarashi R, Kittipatarin C, Copik AJ, et al. Cytokines in immunogenic cell death: Applications for cancer immunotherapy. Cytokine. (2017) 97:123–32. doi: 10.1016/j.cyto.2017.05.024
16. Ha SJ, Kim DJ, Baek KH, Yun YD, Sung YC. IL-23 induces stronger sustained CTL and Th1 immune responses than IL-12 in hepatitis C virus envelope protein 2 DNA immunization. J Immunol. (2004) 172:525–31. doi: 10.4049/jimmunol.172.1.525
17. Huseni MA, Wang L, Klementowicz JE, Yuen K, Breart B, Orr C, et al. CD8(+) T cell-intrinsic IL-6 signaling promotes resistance to anti-PD-L1 immunotherapy. Cell Rep Med. (2023) 4:100878. doi: 10.1016/j.xcrm.2022.100878
18. Alam MS, Otsuka S, Wong N, Abbasi A, Gaida MM, Fan Y, et al. TNF plays a crucial role in inflammation by signaling via T cell TNFR2. Proc Natl Acad Sci U.S.A. (2021) 118:e2109972118. doi: 10.1073/pnas.2109972118
19. Multhoff G. Hyperthermia classic commentary: Activation of natural killer (NK) cells by heat shock protein 70, Gabriele Multhoff. Int J Hyperthermia. (2002) 18:576–85. doi: 10.1080/02656730902835672
20. Xie ZX, Zhang HL, Wu XJ, Zhu J, Ma DH, Jin T. Role of the immunogenic and tolerogenic subsets of dendritic cells in multiple sclerosis. Mediators Inflammation. (2015) 2015:513295. doi: 10.1155/2015/513295
21. Yin SY, Yang NS, Lin TJ. Molecular basis of shikonin-induced immunogenic cell death: insights for developing cancer therapeutics. Receptors Clin Invest. (2016) 3:e1234. doi: 10.14800/rci.1234
22. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. (2007) 318:1931–4. doi: 10.1126/science.1149460
23. Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol. (2010) 42:1316–29. doi: 10.1016/j.biocel.2009.09.016
24. Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, et al. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep. (2012) 2:842. doi: 10.1038/srep00842
25. Ramchandran R, Chaluvally-Raghavan P. miRNA-mediated RNA activation in mammalian cells. Adv Exp Med Biol. (2017) 983:81–9. doi: 10.1007/978-981-10-4310-9_6
26. Flemming JP, Hill BL, Haque MW, Raad J, Bonder CS, Harshyne LA, et al. miRNA- and cytokine-associated extracellular vesicles mediate squamous cell carcinomas. J Extracell Vesicles. (2020) 9:1790159. doi: 10.1080/20013078.2020.1790159
27. Chakraborty C, Sharma AR, Sharma G, Lee SS. The interplay among miRNAs, major cytokines, and cancer-related inflammation. Mol Ther Nucleic Acids. (2020) 20:606–20. doi: 10.1016/j.omtn.2020.04.002
28. Diederich M. Natural compound inducers of immunogenic cell death. Arch Pharm Res. (2019) 42:629–45. doi: 10.1007/s12272-019-01150-z
29. Chuang YT, Tang JY, Shiau JP, Yen CY, Chang FR, Yang KH, et al. Modulating effects of cancer-derived exosomal miRNAs and exosomal processing by natural products. Cancers (Basel). (2023) 15:318. doi: 10.3390/cancers15010318
30. Shiau JP, Chuang YT, Yen CY, Chang FR, Yang KH, Hou MF, et al. Modulation of AKT pathway-targeting miRNAs for cancer cell treatment with natural products. Int J Mol Sci. (2023) 24:3688. doi: 10.3390/ijms24043688
31. Radogna F, Dicato M, Diederich M. Natural modulators of the hallmarks of immunogenic cell death. Biochem Pharmacol. (2019) 162:55–70. doi: 10.1016/j.bcp.2018.12.016
32. Deng LJ, Qi M, Li N, Lei YH, Zhang DM, Chen JX. Natural products and their derivatives: Promising modulators of tumor immunotherapy. J Leukoc Biol. (2020) 108:493–508. doi: 10.1002/JLB.3MR0320-444R
33. Menger L, Vacchelli E, Adjemian S, Martins I, Ma Y, Shen S, et al. Cardiac glycosides exert anticancer effects by inducing immunogenic cell death. Sci Transl Med. (2012) 4:143ra99. doi: 10.1126/scitranslmed.3003807
34. Srivastava M, Eidelman O, Zhang J, Paweletz C, Caohuy H, Yang Q, et al. Digitoxin mimics gene therapy with CFTR and suppresses hypersecretion of IL-8 from cystic fibrosis lung epithelial cells. Proc Natl Acad Sci U.S.A. (2004) 101:7693–8. doi: 10.1073/pnas.0402030101
35. Kepp O, Menger L, Vacchelli E, Adjemian S, Martins I, Ma Y, et al. Anticancer activity of cardiac glycosides: At the frontier between cell-autonomous and immunological effects. Oncoimmunology. (2012) 1:1640–2. doi: 10.4161/onci.21684
36. Kupper N, Gidron Y, Winter J, Denollet J. Association between type D personality, depression, and oxidative stress in patients with chronic heart failure. Psychosom Med. (2009) 71:973–80. doi: 10.1097/PSY.0b013e3181bee6dc
37. Ihenetu K, Espinosa R, de Leon R, Planas G, Perez-Pinero A, Waldbeser L. Digoxin and digoxin-like immunoreactive factors (DLIF) modulate the release of pro-inflammatory cytokines. Inflammation Res. (2008) 57:519–23. doi: 10.1007/s00011-008-7249-9
38. Lu Y, Chen X, Liu X, Shi Y, Wei Z, Feng L, et al. Endothelial TFEB signaling-mediated autophagic disturbance initiates microglial activation and cognitive dysfunction. Autophagy. (2023) 19:1803–20. doi: 10.1080/15548627.2022.2162244
39. Tani S, Takano R, Tamura S, Oishi S, Iwaizumi M, Hamaya Y, et al. Digoxin attenuates murine experimental colitis by downregulating Th17-related cytokines. Inflammation Bowel Dis. (2017) 23:728–38. doi: 10.1097/MIB.0000000000001096
40. Reddy D, Kumavath R, Barh D, Azevedo V, Ghosh P. Anticancer and antiviral properties of cardiac glycosides: A review to explore the mechanism of actions. Molecules. (2020) 25:3596. doi: 10.3390/molecules25163596
41. Zhang D, Ciciriello F, Anjos SM, Carissimo A, Liao J, Carlile GW, et al. Ouabain mimics low temperature rescue of F508del-CFTR in cystic fibrosis epithelial cells. Front Pharmacol. (2012) 3:176. doi: 10.3389/fphar.2012.00176
42. Matsumori A, Ono K, Nishio R, Igata H, Shioi T, Matsui S, et al. Modulation of cytokine production and protection against lethal endotoxemia by the cardiac glycoside ouabain. Circulation. (1997) 96:1501–6. doi: 10.1161/01.cir.96.5.1501
43. Pirkmajer S, Bezjak K, Matkovic U, Dolinar K, Jiang LQ, Mis K, et al. Ouabain suppresses IL-6/STAT3 signaling and promotes cytokine secretion in cultured skeletal muscle cells. Front Physiol. (2020) 11:566584. doi: 10.3389/fphys.2020.566584
44. Nascimento CR, Valente RC, Echevarria-Lima J, Fontes CF, de Araujo-Martins L, Araujo EG, et al. The influence of Ouabain on human dendritic cells maturation. Mediators Inflammation. (2014) 2014:494956. doi: 10.1155/2014/494956
45. Kreindler JL, Bertrand CA, Lee RJ, Karasic T, Aujla S, Pilewski JM, et al. Interleukin-17A induces bicarbonate secretion in normal human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. (2009) 296:L257–66. doi: 10.1152/ajplung.00344.2007
46. Jansson D, Dieriks VB, Rustenhoven J, Smyth LCD, Scotter E, Aalderink M, et al. Cardiac glycosides target barrier inflammation of the vasculature, meninges and choroid plexus. Commun Biol. (2021) 4:260. doi: 10.1038/s42003-021-01787-x
47. Ellrich JA, Ehlers SM. Field observations in pebble beach habitats link plastiglomerate to pyroplastic via pebble clasts. Mar pollut Bull. (2022) 174:113187. doi: 10.1016/j.marpolbul.2021.113187
48. Niiya M, Niiya K, Kiguchi T, Shibakura M, Asaumi N, Shinagawa K, et al. Induction of TNF-alpha, uPA, IL-8 and MCP-1 by doxorubicin in human lung carcinoma cells. Cancer Chemother Pharmacol. (2003) 52:391–8. doi: 10.1007/s00280-003-0665-1
49. Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nat Med. (2014) 20:1301–9. doi: 10.1038/nm.3708
50. Sauter KA, Wood LJ, Wong J, Iordanov M, Magun BE. Doxorubicin and daunorubicin induce processing and release of interleukin-1beta through activation of the NLRP3 inflammasome. Cancer Biol Ther. (2011) 11:1008–16. doi: 10.4161/cbt.11.12.15540
51. Zhu S, Waguespack M, Barker SA, Li S. Doxorubicin directs the accumulation of interleukin-12 induced IFN gamma into tumors for enhancing STAT1 dependent antitumor effect. Clin Cancer Res. (2007) 13:4252–60. doi: 10.1158/1078-0432.CCR-06-2894
52. Hum NR, Sebastian A, Martin KA, Rios-Arce ND, Gilmore SF, Gravano DM, et al. IL-17A increases doxorubicin efficacy in triple negative breast cancer. Front Oncol. (2022) 12:928474. doi: 10.3389/fonc.2022.928474
53. Fredly H, Ersvaer E, Gjertsen BT, Bruserud O. Immunogenic apoptosis in human acute myeloid leukemia (AML): primary human AML cells expose calreticulin and release heat shock protein (HSP) 70 and HSP90 during apoptosis. Oncol Rep. (2011) 25:1549–56. doi: 10.3892/or.2011.1229
54. Wu J, Zhang L, Feng Y, Khadka B, Fang Z, Liu J. HDAC8 promotes daunorubicin resistance of human acute myeloid leukemia cells via regulation of IL-6 and IL-8. Biol Chem. (2021) 402:461–8. doi: 10.1515/hsz-2020-0196
55. Bi L, Wu J, Ye A, Wu J, Yu K, Zhang S, et al. Increased Th17 cells and IL-17A exist in patients with B cell acute lymphoblastic leukemia and promote proliferation and resistance to daunorubicin through activation of Akt signaling. J Transl Med. (2016) 14:132. doi: 10.1186/s12967-016-0894-9
56. Humeau J, Sauvat A, Cerrato G, Xie W, Loos F, Iannantuoni F, et al. Inhibition of transcription by dactinomycin reveals a new characteristic of immunogenic cell stress. EMBO Mol Med. (2020) 12:e11622. doi: 10.15252/emmm.201911622
57. Takenaka IM, Hightower LE. Regulation of chicken Hsp70 and Hsp90 family gene expression by transforming growth factor-beta 1. J Cell Physiol. (1993) 155:54–62. doi: 10.1002/jcp.1041550108
58. Liu P, Zhao L, Loos F, Iribarren K, Lachkar S, Zhou H, et al. Identification of pharmacological agents that induce HMGB1 release. Sci Rep. (2017) 7:14915. doi: 10.1038/s41598-017-14848-1
59. Solari JIG, Filippi-Chiela E, Pilar ES, Nunes V, Gonzalez EA, Figueiro F, et al. Damage-associated molecular patterns (DAMPs) related to immunogenic cell death are differentially triggered by clinically relevant chemotherapeutics in lung adenocarcinoma cells. BMC Cancer. (2020) 20:474. doi: 10.1186/s12885-020-06964-5
60. Penson RT, Kronish K, Duan Z, Feller AJ, Stark P, Cook SE, et al. Cytokines IL-1beta, IL-2, IL-6, IL-8, MCP-1, GM-CSF and TNFalpha in patients with epithelial ovarian cancer and their relationship to treatment with paclitaxel. Int J Gynecol Cancer. (2000) 10:33–41. doi: 10.1046/j.1525-1438.2000.00003.x
61. Liu C, Zheng S, Wang Z, Wang S, Wang X, Yang L, et al. KRAS-G12D mutation drives immune suppression and the primary resistance of anti-PD-1/PD-L1 immunotherapy in non-small cell lung cancer. Cancer Commun (Lond). (2022) 42:828–47. doi: 10.1002/cac2.12327
62. Mullins DW, Burger CJ, Elgert KD. Paclitaxel enhances macrophage IL-12 production in tumor-bearing hosts through nitric oxide. J Immunol. (1999) 162:6811–8. doi: 10.4049/jimmunol.162.11.6811
63. Elsisi A, Sokar S, El-Mahrouk S, Abu-Risha S. Potentiation of paclitaxel antitumor activity by galloflavin or oxamate as lactate dehydrogenase inhibitors. J Adv Med Pharm Res. (2021) 2:64–76. doi: 10.21608/jampr.2021.98224.1020
64. Li Z, Zhao S, Zhang HL, Liu P, Liu FF, Guo YX, et al. Proinflammatory factors mediate paclitaxel-induced impairment of learning and memory. Mediators Inflammation. (2018) 2018:3941840. doi: 10.1155/2018/3941840
65. Mizumoto N, Gao J, Matsushima H, Ogawa Y, Tanaka H, Takashima A. Discovery of novel immunostimulants by dendritic-cell-based functional screening. Blood. (2005) 106:3082–9. doi: 10.1182/blood-2005-03-1161
66. Wen CC, Chen HM, Chen SS, Huang LT, Chang WT, Wei WC, et al. Specific microtubule-depolymerizing agents augment efficacy of dendritic cell-based cancer vaccines. J BioMed Sci. (2011) 18:44. doi: 10.1186/1423-0127-18-44
67. Aral CA, Aral K, Yay A, Ozcoban O, Berdeli A, Saraymen R. Effects of colchicine on gingival inflammation, apoptosis, and alveolar bone loss in experimental periodontitis. J Periodontol. (2018) 89:577–85. doi: 10.1002/JPER.17-0359
68. Wu Q, Liu H, Liao J, Zhao N, Tse G, Han B, et al. Colchicine prevents atrial fibrillation promotion by inhibiting IL-1beta-induced IL-6 release and atrial fibrosis in the rat sterile pericarditis model. BioMed Pharmacother. (2020) 129:110384. doi: 10.1016/j.biopha.2020.110384
69. Yue H, Liang W, Gu J, Zhao X, Zhang T, Qin X, et al. Comparative transcriptome analysis to elucidate the therapeutic mechanism of colchicine against atrial fibrillation. BioMed Pharmacother. (2019) 119:109422. doi: 10.1016/j.biopha.2019.109422
70. Li Z, Davis GS, Mohr C, Nain M, Gemsa D. Inhibition of LPS-induced tumor necrosis factor-alpha production by colchicine and other microtubule disrupting drugs. Immunobiology. (1996) 195:624–39. doi: 10.1016/s0171-2985(96)80027-1
71. Pellicciotta I, Yang CP, Goldberg GL, Shahabi S. Epothilone B enhances Class I HLA and HLA-A2 surface molecule expression in ovarian cancer cells. Gynecol Oncol. (2011) 122:625–31. doi: 10.1016/j.ygyno.2011.05.007
72. Sukkurwala AQ, Adjemian S, Senovilla L, Michaud M, Spaggiari S, Vacchelli E, et al. Screening of novel immunogenic cell death inducers within the NCI Mechanistic Diversity Set. Oncoimmunology. (2014) 3:e28473. doi: 10.4161/onci.28473
73. Bugaut H, Bruchard M, Berger H, Derangere V, Odoul L, Euvrard R, et al. Bleomycin exerts ambivalent antitumor immune effect by triggering both immunogenic cell death and proliferation of regulatory T cells. PloS One. (2013) 8:e65181. doi: 10.1371/journal.pone.0065181
74. Fucikova J, Kepp O, Kasikova L, Petroni G, Yamazaki T, Liu P, et al. Detection of immunogenic cell death and its relevance for cancer therapy. Cell Death Dis. (2020) 11:1013. doi: 10.1038/s41419-020-03221-2
75. Luzina IG, Kopach P, Lockatell V, Kang PH, Nagarsekar A, Burke AP, et al. Interleukin-33 potentiates bleomycin-induced lung injury. Am J Respir Cell Mol Biol. (2013) 49:999–1008. doi: 10.1165/rcmb.2013-0093OC
76. Sibinska Z, Tian X, Korfei M, Kojonazarov B, Kolb JS, Klepetko W, et al. Amplified canonical transforming growth factor-beta signalling via heat shock protein 90 in pulmonary fibrosis. Eur Respir J. (2017) 49:1501941. doi: 10.1183/13993003.01941-2015
77. Pociask DA, Chen K, Choi SM, Oury TD, Steele C, Kolls JK. gammadelta T cells attenuate bleomycin-induced fibrosis through the production of CXCL10. Am J Pathol. (2011) 178:1167–76. doi: 10.1016/j.ajpath.2010.11.055
78. Smith RE, Strieter RM, Phan SH, Lukacs N, Kunkel SL. TNF and IL-6 mediate MIP-1alpha expression in bleomycin-induced lung injury. J Leukoc Biol. (1998) 64:528–36. doi: 10.1002/jlb.64.4.528
79. Taniguchi T, Asano Y, Akamata K, Noda S, Takahashi T, Ichimura Y, et al. Fibrosis, vascular activation, and immune abnormalities resembling systemic sclerosis in bleomycin-treated Fli-1-haploinsufficient mice. Arthritis Rheumatol. (2015) 67:517–26. doi: 10.1002/art.38948
80. Sonnenberg GF, Nair MG, Kirn TJ, Zaph C, Fouser LA, Artis D. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J Exp Med. (2010) 207:1293–305. doi: 10.1084/jem.20092054
81. Chen HM, Wang PH, Chen SS, Wen CC, Chen YH, Yang WC, et al. Shikonin induces immunogenic cell death in tumor cells and enhances dendritic cell-based cancer vaccine. Cancer Immunol Immunother. (2012) 61:1989–2002. doi: 10.1007/s00262-012-1258-9
82. Zhou Z, Lu B, Wang C, Wang Z, Luo T, Piao M, et al. RIP1 and RIP3 contribute to shikonin-induced DNA double-strand breaks in glioma cells via increase of intracellular reactive oxygen species. Cancer Lett. (2017) 390:77–90. doi: 10.1016/j.canlet.2017.01.004
83. Diederich M, Muller F, Cerella C. Cardiac glycosides: From molecular targets to immunogenic cell death. Biochem Pharmacol. (2017) 125:1–11. doi: 10.1016/j.bcp.2016.08.017
84. Zeng Q, Qiu F, Chen Y, Liu C, Liu H, Liang CL, et al. Shikonin prolongs allograft survival via induction of CD4(+)FoxP3(+) regulatory T cells. Front Immunol. (2019) 10:652. doi: 10.3389/fimmu.2019.00652
85. Sun WX, Liu Y, Zhou W, Li HW, Yang J, Chen ZB. Shikonin inhibits TNF-alpha production through suppressing PKC-NF-kappaB-dependent decrease of IL-10 in rheumatoid arthritis-like cell model. J Nat Med. (2017) 71:349–56. doi: 10.1007/s11418-016-1064-3
86. Yang Y, Li XJ, Chen Z, Zhu XX, Wang J, Zhang LB, et al. Wogonin induced calreticulin/annexin A1 exposure dictates the immunogenicity of cancer cells in a PERK/AKT dependent manner. PloS One. (2012) 7:e50811. doi: 10.1371/journal.pone.0050811
87. Song X, Yao J, Wang F, Zhou M, Zhou Y, Wang H, et al. Wogonin inhibits tumor angiogenesis via degradation of HIF-1alpha protein. Toxicol Appl Pharmacol. (2013) 271:144–55. doi: 10.1016/j.taap.2013.04.031
88. Nakamura N, Hayasaka S, Zhang XY, Nagaki Y, Matsumoto M, Hayasaka Y, et al. Effects of baicalin, baicalein, and wogonin on interleukin-6 and interleukin-8 expression, and nuclear factor-kappab binding activities induced by interleukin-1beta in human retinal pigment epithelial cell line. Exp Eye Res. (2003) 77:195–202. doi: 10.1016/s0014-4835(03)00116-7
89. Yang D, Guo Q, Liang Y, Zhao Y, Tian X, Ye Y, et al. Wogonin induces cellular senescence in breast cancer via suppressing TXNRD2 expression. Arch Toxicol. (2020) 94:3433–47. doi: 10.1007/s00204-020-02842-y
90. Shin HS, Bae MJ, Choi DW, Shon DH. Skullcap (Scutellaria baicalensis) extract and its active compound, wogonin, inhibit ovalbumin-induced Th2-mediated response. Molecules. (2014) 19:2536–45. doi: 10.3390/molecules19022536
91. Chang MY, Shen YL. Linalool exhibits cytotoxic effects by activating antitumor immunity. Molecules. (2014) 19:6694–706. doi: 10.3390/molecules19056694
92. Mohamed ME, Abduldaium YS, Younis NS. Ameliorative effect of linalool in cisplatin-induced nephrotoxicity: the role of HMGB1/TLR4/NF-κB and Nrf2/HO1 pathways. Biomolecules. (2020) 10:1488. doi: 10.3390/biom10111488
93. Diaz-Laviada I, Rodriguez-Henche N. The potential antitumor effects of capsaicin. Prog Drug Res. (2014) 68:181–208. doi: 10.1007/978-3-0348-0828-6_8
94. D'Eliseo D, Manzi L, Velotti F. Capsaicin as an inducer of damage-associated molecular patterns (DAMPs) of immunogenic cell death (ICD) in human bladder cancer cells. Cell Stress Chaperones. (2013) 18:801–8. doi: 10.1007/s12192-013-0422-2
95. Belenahalli Shekarappa S, Kandagalla S, Malojirao VH, Pavan Kumar GS, Prabhakar BT, Hanumanthappa M. A systems biology approach to identify the key targets of curcumin and capsaicin that downregulate pro-inflammatory pathways in human monocytes. Comput Biol Chem. (2019) 83:107162. doi: 10.1016/j.compbiolchem.2019.107162
96. Huang CJ, Pu CM, Su SY, Lo SL, Lee CH, Yen YH. Improvement of wound healing by capsaicin through suppression of the inflammatory response and amelioration of the repair process. Mol Med Rep. (2023) 28:1–13. doi: 10.3892/mmr.2023.13042
97. Takano F, Yamaguchi M, Takada S, Shoda S, Yahagi N, Takahashi T, et al. Capsicum ethanol extracts and capsaicin enhance interleukin-2 and interferon-gamma production in cultured murine Peyer's patch cells ex vivo. Life Sci. (2007) 80:1553–63. doi: 10.1016/j.lfs.2007.01.031
98. Lin KH, Lin KC, Lu WJ, Thomas PA, Jayakumar T, Sheu JR. Astaxanthin, a carotenoid, stimulates immune responses by enhancing IFN-gamma and IL-2 secretion in primary cultured lymphocytes in vitro and ex vivo. Int J Mol Sci. (2015) 17:44. doi: 10.3390/ijms17010044
99. Abbaszadeh F, Jorjani M, Joghataei MT, Raminfard S, Mehrabi S. Astaxanthin ameliorates spinal cord edema and astrocyte activation via suppression of HMGB1/TLR4/NF-kappaB signaling pathway in a rat model of spinal cord injury. Naunyn Schmiedebergs Arch Pharmacol. (2023) 396:3075–86. doi: 10.1007/s00210-023-02512-7
100. Kim YH, Koh HK, Kim DS. Down-regulation of IL-6 production by astaxanthin via ERK-, MSK-, and NF-kappaB-mediated signals in activated microglia. Int Immunopharmacol. (2010) 10:1560–72. doi: 10.1016/j.intimp.2010.09.007
101. Kwak MS, Lim JW, Kim H. Astaxanthin inhibits interleukin-6 expression in cerulein/resistin-stimulated pancreatic acinar cells. Mediators Inflammation. (2021) 2021:5587297. doi: 10.1155/2021/5587297
102. Ohgami K, Shiratori K, Kotake S, Nishida T, Mizuki N, Yazawa K, et al. Effects of astaxanthin on lipopolysaccharide-induced inflammation. Vitro vivo. Invest Ophthalmol Vis Sci. (2003) 44:2694–701. doi: 10.1167/iovs.02-0822
103. Barsanti L, Gualtieri P. Paramylon, a potent immunomodulator from WZSL mutant of Euglena gracilis. Molecules. (2019) 24:3114. doi: 10.3390/molecules24173114
104. Chen HW, Yang TS, Chen MJ, Chang YC, Wang EIC, Ho CL, et al. Purification and immunomodulating activity of C-phycocyanin from Spirulina platensis cultured using power plant flue gas. Process Biochem. (2014) 49:1337–44. doi: 10.1016/j.procbio.2014.05.006
105. Alzokaky AA, Abdelkader EM, El-Dessouki AM, Khaleel SA, Raslan NA. C-phycocyanin protects against ethanol-induced gastric ulcers in rats: Role of HMGB1/NLRP3/NF-kappaB pathway. Basic Clin Pharmacol Toxicol. (2020) 127:265–77. doi: 10.1111/bcpt.13415
106. Kumar A, Christian PK, Panchal K, Guruprasad BR, Tiwari AK. Supplementation of spirulina (Arthrospira platensis) improves lifespan and locomotor activity in paraquat-sensitive DJ-1beta(Delta93) flies, a Parkinson's disease model in Drosophila melanogaster. J Diet Suppl. (2017) 14:573–88. doi: 10.1080/19390211.2016.1275917
107. Jang JY, Lee BM, Jun SY, Yang YJ, Shin HS. Attenuation of psoriasis symptoms following treatment with C-phycocyanin from Spirulina maxima in a mouse model. Biotechnol Bioprocess Eng. (2022) 27:407–14. doi: 10.1007/s12257-022-0022-z
108. Jin JO, Zhang W, Du JY, Wong KW, Oda T, Yu Q. Fucoidan can function as an adjuvant in vivo to enhance dendritic cell maturation and function and promote antigen-specific T cell immune responses. PloS One. (2014) 9:e99396. doi: 10.1371/journal.pone.0099396
109. Li C, Gao Y, Xing Y, Zhu H, Shen J, Tian J. Fucoidan, a sulfated polysaccharide from brown algae, against myocardial ischemia-reperfusion injury in rats via regulating the inflammation response. Food Chem Toxicol. (2011) 49:2090–5. doi: 10.1016/j.fct.2011.05.022
110. Geinguenaud F, Sainte-Catherine O, Poirier F, Besnard V, Haddad O, Chaubet F, et al. Iron oxide nanoparticles functionalized with fucoidan: A potential theranostic nanotool for hepatocellular carcinoma. Chembiochem. (2022) 23:e202200265. doi: 10.1002/cbic.202200265
111. Dutot M, Grassin-Delyle S, Salvator H, Brollo M, Rat P, Fagon R, et al. A marine-sourced fucoidan solution inhibits Toll-like-receptor-3-induced cytokine release by human bronchial epithelial cells. Int J Biol Macromol. (2019) 130:429–36. doi: 10.1016/j.ijbiomac.2019.02.113
112. D'Eliseo D, Di Renzo L, Santoni A, Velotti F. Docosahexaenoic acid (DHA) promotes immunogenic apoptosis in human multiple myeloma cells, induces autophagy and inhibits STAT3 in both tumor and dendritic cells. Genes Cancer. (2017) 8:426–37. doi: 10.18632/genesandcancer.131
113. Samples BL, Pool GL, Lumb RH. Polyunsaturated fatty acids enhance the heat induced stress response in rainbow trout (Oncorhynchus mykiss) leukocytes. Comp Biochem Physiol B Biochem Mol Biol. (1999) 123:389–97. doi: 10.1016/s0305-0491(99)00083-8
114. Benninghoff AD, Bates MA, Chauhan PS, Wierenga KA, Gilley KN, Holian A, et al. Docosahexaenoic acid consumption impedes early interferon-and chemokine-related gene expression while suppressing silica-triggered flaring of murine lupus. Front Immunol. (2019) 10:2851. doi: 10.3389/fimmu.2019.02851
115. Kong W, Yen JH, Vassiliou E, Adhikary S, Toscano MG, Ganea D. Docosahexaenoic acid prevents dendritic cell maturation and in vitro and in vivo expression of the IL-12 cytokine family. Lipids Health Dis. (2010) 9:12. doi: 10.1186/1476-511X-9-12
116. Morin S, Belanger S, Cortez Ghio S, Pouliot R. Eicosapentaenoic acid reduces the proportion of IL-17A-producing T cells in a 3D psoriatic skin model. J Lipid Res. (2023) 64:100428. doi: 10.1016/j.jlr.2023.100428
117. Fritsche KL, Anderson M, Feng C. Consumption of eicosapentaenoic acid and docosahexaenoic acid impair murine interleukin-12 and interferon-gamma production in vivo. J Infect Dis. (2000) 182 Suppl 1:S54–61. doi: 10.1086/315925
118. Ding C, Yu H, Shi C, Shi T, Qin H, Cui Y. MiR-let-7e inhibits invasion and magration and regulates HMGB1 expression in papillary thyroid carcinoma. BioMed Pharmacother. (2019) 110:528–36. doi: 10.1016/j.biopha.2018.11.057
119. Feng L, Feng C, Wang CX, Xu DY, Chen JJ, Huang JF, et al. Circulating microRNA let−7e is decreased in knee osteoarthritis, accompanied by elevated apoptosis and reduced autophagy. Int J Mol Med. (2020) 45:1464–76. doi: 10.3892/ijmm.2020.4534
120. Ai H, Zhou W, Wang Z, Qiong G, Chen Z, Deng S. microRNAs-107 inhibited autophagy, proliferation, and migration of breast cancer cells by targeting HMGB1. J Cell Biochem. (2019) 120:8696–705. doi: 10.1002/jcb.28157
121. He J, Zhang W, Zhou Q, Zhao T, Song Y, Chai L, et al. Low-expression of microRNA-107 inhibits cell apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell Biol. (2013) 45:1962–73. doi: 10.1016/j.biocel.2013.06.008
122. Qian J, Fu P, Li S, Li X, Chen Y, Lin Z. miR-107 affects cartilage matrix degradation in the pathogenesis of knee osteoarthritis by regulating caspase-1. J Orthop Surg Res. (2021) 16:40. doi: 10.1186/s13018-020-02121-7
123. Li Y, Qin C. MiR-1179 inhibits the proliferation of gastric cancer cells by targeting HMGB1. Hum Cell. (2019) 32:352–9. doi: 10.1007/s13577-019-00244-6
124. Gao Y, Xu H, Pu T. MicroRNA-1179 suppresses the proliferation and enhances vincristine sensitivity of oral cancer cells via induction of apoptosis and modulation of MEK/ERK and PI3K/AKT signalling pathways. AMB Express. (2020) 10:149. doi: 10.1186/s13568-020-01082-8
125. Chen J, Li G. MiR-1284 enhances sensitivity of cervical cancer cells to cisplatin via downregulating HMGB1. BioMed Pharmacother. (2018) 107:997–1003. doi: 10.1016/j.biopha.2018.08.059
126. Feng J, Guo J, Wang JP, Chai BF. MiR-129-5p inhibits proliferation of gastric cancer cells through targeted inhibition on HMGB1 expression. Eur Rev Med Pharmacol Sci. (2020) 24:3665–73. doi: 10.26355/eurrev_202004_20829
127. Xu JY, Zheng FC, Yu WS. MicroRNA-129-5p-mediated inhibition of autophagy enhanced the radiosensitivity of human colon cancer cells. Int J Clin Exp Pathol. (2016) 9:12179–87.
128. Wei Z, Hang S, Wiredu Ocansey DK, Zhang Z, Wang B, Zhang X, et al. Human umbilical cord mesenchymal stem cells derived exosome shuttling mir-129-5p attenuates inflammatory bowel disease by inhibiting ferroptosis. J Nanobiotechnology. (2023) 21:188. doi: 10.1186/s12951-023-01951-x
129. Wang F, Wang L, Sui G, Yang C, Guo M, Xiong X, et al. Inhibition of miR-129 improves neuronal pyroptosis and cognitive impairment through IGF-1/GSK3beta signaling pathway: An in vitro and in vivo study. J Mol Neurosci. (2021) 71:2299–309. doi: 10.1007/s12031-021-01794-x
130. Lu F, Zhang J, Ji M, Li P, Du Y, Wang H, et al. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int J Oncol. (2014) 45:383–92. doi: 10.3892/ijo.2014.2390
131. Di Marco M, Veschi S, Lanuti P, Ramassone A, Pacillo S, Pagotto S, et al. Enhanced expression of miR-181b in B cells of CLL improves the anti-tumor cytotoxic T cell response. Cancers (Basel). (2021) 13:257. doi: 10.3390/cancers13020257
132. Wu K, Huang J, Xu T, Ye Z, Jin F, Li N, et al. MicroRNA-181b blocks gensenoside Rg3-mediated tumor suppression of gallbladder carcinoma by promoting autophagy flux via CREBRF/CREB3 pathway. Am J Transl Res. (2019) 11:5776–87.
133. Wang D, Fang Y, Lin L, Long W, Wang L, Yu L, et al. Upregulating miR-181b promotes ferroptosis in osteoarthritic chondrocytes by inhibiting SLC7A11. BMC Musculoskelet Disord. (2023) 24:862. doi: 10.1186/s12891-023-07003-7
134. Cui Y, Yin K, Gong Y, Qu Y, Liu H, Lin H. Atrazine induces necroptosis by miR-181-5p targeting inflammation and glycometabolism in carp lymphocytes. Fish Shellfish Immunol. (2019) 94:730–8. doi: 10.1016/j.fsi.2019.09.068
135. Xu X, Yang Y, Wang G, Yin Y, Han S, Zheng D, et al. Low shear stress regulates vascular endothelial cell pyroptosis through miR-181b-5p/STAT-3 axis. J Cell Physiol. (2021) 236:318–27. doi: 10.1002/jcp.29844
136. Wu H, Zhou C. Long non-coding RNA UCA1 promotes lung cancer cell proliferation and migration via microRNA-193a/HMGB1 axis. Biochem Biophys Res Commun. (2018) 496:738–45. doi: 10.1016/j.bbrc.2018.01.097
137. Ma T, Li H, Yang W, Liu Q, Yan H. Over-expression of miR-193a-3p regulates the apoptosis of colorectal cancer cells by targeting PAK3. Am J Transl Res. (2022) 14:1361–75.
138. Qu L, Tian Y, Hong D, Wang F, Li Z. Mig-6 inhibits autophagy in HCC cell lines by modulating miR-193a-3p. Int J Med Sci. (2022) 19:338–51. doi: 10.7150/ijms.66040
139. Zhong L, Yang H, Zhu B, Zhao X, Xie M, Cao M, et al. The TBX1/miR-193a-3p/TGF-beta2 axis mediates CHD by promoting ferroptosis. Oxid Med Cell Longev. (2022) 2022:5130546. doi: 10.1155/2022/5130546
140. Li SL, Huang Y, Huang Y, Fu YM, Tang DL, Kang R, et al. The long non-coding RNA TP73-AS1 modulates HCC cell proliferation through miR-200a-dependent HMGB1/RAGE regulation. J Exp Clin Cancer Res. (2017) 36:51. doi: 10.1186/s13046-017-0519-z
141. Guan H, You Z, Wang C, Fang F, Peng R, Mao L, et al. MicroRNA-200a suppresses prostate cancer progression through BRD4/AR signaling pathway. Cancer Med. (2019) 8:1474–85. doi: 10.1002/cam4.2029
142. You P, Chen H, Han W, Deng J. miR-200a-3p overexpression alleviates diabetic cardiomyopathy injury in mice by regulating autophagy through the FOXO3/Mst1/Sirt3/AMPK axis. PeerJ. (2023) 11:e15840. doi: 10.7717/peerj.15840
143. Li S, Lv D, Lu Y, Zhang Y, Jia Y. High-throughput sequencing to investigate the expression and potential role of differentially expressed microRNAs in myocardial cells after ischemia-reperfusion injury. Front Biosci (Landmark Ed). (2024) 29:38. doi: 10.31083/j.fbl2901038
144. Liu Y, Wang Z, Huang H, Shou K. miR-200a-3p improves neonatal necrotizing enterocolitis by regulating RIPK1. Am J Transl Res. (2021) 13:12662–72.
145. Liu J, Yan Y, Zheng D, Zhang J, Wang J. Inhibiting microRNA-200a-3p attenuates pyroptosis via targeting the SIRT1/NF-kappaB/NLRP3 pathway in H(2)O(2)-induced HAEC. Aging (Albany NY). (2023) 15:11184–200. doi: 10.18632/aging.205121
146. Liu PL, Liu WL, Chang JM, Chen YH, Liu YP, Kuo HF, et al. MicroRNA-200c inhibits epithelial-mesenchymal transition, invasion, and migration of lung cancer by targeting HMGB1. PloS One. (2017) 12:e0180844. doi: 10.1371/journal.pone.0180844
147. Raue R, Frank AC, Fuhrmann DC, de la Cruz-Ojeda P, Rosser S, Bauer R, et al. MicroRNA-200c attenuates the tumor-infiltrating capacity of macrophages. Biol (Basel). (2022) 11:349. doi: 10.3390/biology11030349
148. Shen Y, Zhu Y, Rong F. miR−200c−3p regulates the proliferation and apoptosis of human trabecular meshwork cells by targeting PTEN. Mol Med Rep. (2020) 22:1605–12. doi: 10.3892/mmr.2020.11198
149. Sohn EJ. MicroRNA 200c-3p regulates autophagy via upregulation of endoplasmic reticulum stress in PC-3 cells. Cancer Cell Int. (2018) 18:2. doi: 10.1186/s12935-017-0500-0
150. Li W, Yang S, Chen G, He S. MiR-200c-3p regulates pyroptosis by targeting SLC30A7 in diabetic retinopathy. Hum Exp Toxicol. (2022) 41:9603271221099589. doi: 10.1177/09603271221099589
151. Wang L, Kang FB, Wang J, Yang C, He DW. Downregulation of miR-205 contributes to epithelial-mesenchymal transition and invasion in triple-negative breast cancer by targeting HMGB1-RAGE signaling pathway. Anti-Cancer Drugs. (2019) 30:225–32. doi: 10.1097/Cad.0000000000000705
152. Chen X, Zhang L, Geng J, Chen Z, Cui X. MiR-205-5p functions as a tumor suppressor in gastric cancer cells through downregulating FAM84B. J Oncol. (2022) 2022:8267891. doi: 10.1155/2022/8267891
153. Pennati M, Lopergolo A, Profumo V, De Cesare M, Sbarra S, Valdagni R, et al. miR-205 impairs the autophagic flux and enhances cisplatin cytotoxicity in castration-resistant prostate cancer cells. Biochem Pharmacol. (2014) 87:579–97. doi: 10.1016/j.bcp.2013.12.009
154. Sun W, Wu X, Yu P, Zhang Q, Shen L, Chen J, et al. LncAABR07025387.1 enhances myocardial ischemia/reperfusion injury via miR-205/ACSL4-mediated ferroptosis. Front Cell Dev Biol. (2022) 10:672391. doi: 10.3389/fcell.2022.672391
155. Zhang CL, Ge SL, Hu CL, Yang N, Zhang JR. MiRNA-218, a new regulator of HMGB1, suppresses cell migration and invasion in non-small cell lung cancer. Acta Biochim Et Biophys Sin. (2013) 45:1055–61. doi: 10.1093/abbs/gmt109
156. Zhang J, Li S, Li Y, Liu H, Zhang Y, Zhang Q. miRNA-218 regulates the proliferation and apoptosis of cervical cancer cells via targeting Gli3. Exp Ther Med. (2018) 16:2433–41. doi: 10.3892/etm.2018.6491
157. Chen M, Li M, Zhang N, Sun W, Wang H, Wei W. Mechanism of miR-218-5p in autophagy, apoptosis and oxidative stress in rheumatoid arthritis synovial fibroblasts is mediated by KLF9 and JAK/STAT3 pathways. J Investig Med. (2021) 69:824–32. doi: 10.1136/jim-2020-001437
158. Mathew LK, Skuli N, Mucaj V, Lee SS, Zinn PO, Sathyan P, et al. miR-218 opposes a critical RTK-HIF pathway in mesenchymal glioblastoma. Proc Natl Acad Sci U.S.A. (2014) 111:291–6. doi: 10.1073/pnas.1314341111
159. Lv G, Wu M, Wang M, Jiang X, Du J, Zhang K, et al. miR-320a regulates high mobility group box 1 expression and inhibits invasion and metastasis in hepatocellular carcinoma. Liver Int. (2017) 37:1354–64. doi: 10.1111/liv.13424
160. Cui Y, Xu H, Yang Y, Zhao D, Wen Y, Lv C, et al. The regulation of miR-320a/XBP1 axis through LINC00963 for endoplasmic reticulum stress and autophagy in diffuse large B-cell lymphoma. Cancer Cell Int. (2021) 21:305. doi: 10.1186/s12935-021-01992-y
161. Zhang C, Xu L, Li X, Chen Y, Shi T, Wang Q. LINC00460 facilitates cell proliferation and inhibits ferroptosis in breast cancer through the miR-320a/MAL2 axis. Technol Cancer Res Treat. (2023) 22:15330338231164359. doi: 10.1177/15330338231164359
162. Yao S, Zhao T, Jin H. Expression of MicroRNA-325-3p and its potential functions by targeting HMGB1 in non-small cell lung cancer. BioMed Pharmacother. (2015) 70:72–9. doi: 10.1016/j.biopha.2015.01.013
163. Huang Z, Luo Y, Chen C, Zhou C, Su Z, Cai C, et al. miR-325-3p reduces proliferation and promotes apoptosis of gastric cancer cells by inhibiting human antigen R. Can J Gastroenterol Hepatol. (2023) 2023:6882851. doi: 10.1155/2023/6882851
164. Bo L, Su-Ling D, Fang L, Lu-Yu Z, Tao A, Stefan D, et al. Autophagic program is regulated by miR-325. Cell Death Differ. (2014) 21:967–77. doi: 10.1038/cdd.2014.18
165. Liu K, Huang J, Xie M, Yu Y, Zhu S, Kang R, et al. MIR34A regulates autophagy and apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy. (2014) 10:442–52. doi: 10.4161/auto.27418
166. Werner TV, Hart M, Nickels R, Kim YJ, Menger MD, Bohle RM, et al. MiR-34a-3p alters proliferation and apoptosis of meningioma cells in vitro and is directly targeting SMAD4, FRAT1 and BCL2. Aging (Albany NY). (2017) 9:932–54. doi: 10.18632/aging.101201
167. Chen Z, He M, Chen J, Li C, Zhang Q. Long non-coding RNA SNHG7 inhibits NLRP3-dependent pyroptosis by targeting the miR-34a/SIRT1 axis in liver cancer. Oncol Lett. (2020) 20:893–901. doi: 10.3892/ol.2020.11635
168. Chandrasekaran KS, Sathyanarayanan A, Karunagaran D. Downregulation of HMGB1 by miR-34a is sufficient to suppress proliferation, migration and invasion of human cervical and colorectal cancer cells. Tumour Biol. (2016) 37:13155–66. doi: 10.1007/s13277-016-5261-1
169. Taheri F, Ebrahimi SO, Shareef S, Reiisi S. Regulatory and immunomodulatory role of miR-34a in T cell immunity. Life Sci. (2020) 262:118209. doi: 10.1016/j.lfs.2020.118209
170. Aida R, Hagiwara K, Okano K, Nakata K, Obata Y, Yamashita T, et al. miR-34a-5p might have an important role for inducing apoptosis by down-regulation of SNAI1 in apigenin-treated lung cancer cells. Mol Biol Rep. (2021) 48:2291–7. doi: 10.1007/s11033-021-06255-7
171. Han S, Zhao X, Zhang Y, Amevor FK, Tan B, Ma M, et al. MiR-34a-5p promotes autophagy and apoptosis of ovarian granulosa cells via the Hippo-YAP signaling pathway by targeting LEF1 in chicken. Poult Sci. (2023) 102:102374. doi: 10.1016/j.psj.2022.102374
172. Zhong Z, Gao Y, Zhou J, Wang F, Zhang P, Hu S, et al. Inhibiting mir-34a-5p regulates doxorubicin-induced autophagy disorder and alleviates myocardial pyroptosis by targeting Sirt3-AMPK pathway. BioMed Pharmacother. (2023) 168:115654. doi: 10.1016/j.biopha.2023.115654
173. Wu D, Liu J, Chen J, He H, Ma H, Lv X. miR-449a suppresses tumor growth, migration, and invasion in non-small cell lung cancer by targeting a HMGB1-mediated NF-kappaB signaling pathway. Oncol Res. (2019) 27:227–35. doi: 10.3727/096504018X15213089759999
174. Liu Y, Wang Y, Sun X, Mei C, Wang L, Li Z, et al. miR-449a promotes liver cancer cell apoptosis by downregulation of Calpain 6 and POU2F1. Oncotarget. (2016) 7:13491–501. doi: 10.18632/oncotarget.4821
175. Zhang N, Qiu L, Li T, Wang X, Deng R, Yi H, et al. MiR-449a attenuates autophagy of T-cell lymphoma cells by downregulating ATG4B expression. BMB Rep. (2020) 53:254–9. doi: 10.5483/BMBRep.2020.53.5.219
176. Lu L, Qiu C, Li D, Bai G, Liang J, Yang Q. MicroRNA-505 suppresses proliferation and invasion in hepatoma cells by directly targeting high-mobility group box 1. Life Sci. (2016) 157:12–8. doi: 10.1016/j.lfs.2016.05.039
177. Tang H, Lv W, Sun W, Bi Q, Hao Y. miR−505 inhibits cell growth and EMT by targeting MAP3K3 through the AKT−NFkappaB pathway in NSCLC cells. Int J Mol Med. (2019) 43:1203–16. doi: 10.3892/ijmm.2018.4041
178. Guo Y, Xu X, Tang T, Sun L, Zhang X, Shen X, et al. miR-505 inhibits replication of Borna disease virus 1 via inhibition of HMGB1-mediated autophagy. J Gen Virol. (2022) 103. doi: 10.1099/jgv.0.001713
179. Ye Y, Zhao L, Li Q, Xi C, Li Y, Li Z. circ_0007385 served as competing endogenous RNA for miR-519d-3p to suppress Malignant behaviors and cisplatin resistance of non-small cell lung cancer cells. Thorac Cancer. (2020) 11:2196–208. doi: 10.1111/1759-7714.13527
180. Zhang YJ, Pan Q, Yu Y, Zhong XP. microRNA-519d induces autophagy and apoptosis of human hepatocellular carcinoma cells through activation of the AMPK signaling pathway via Rab10. Cancer Manag Res. (2020) 12:2589–602. doi: 10.2147/CMAR.S207548
181. Shen H, Xu L, You C, Tang H, Wu H, Zhang Y, et al. miR-665 is downregulated in glioma and inhibits tumor cell proliferation, migration and invasion by targeting high mobility group box 1. Oncol Lett. (2021) 21:156. doi: 10.3892/ol.2020.12417
182. Chen TJ, Zheng Q, Gao F, Yang T, Ren H, Li Y, et al. MicroRNA-665 facilitates cell proliferation and represses apoptosis through modulating Wnt5a/beta-Catenin and Caspase-3 signaling pathways by targeting TRIM8 in LUSC. Cancer Cell Int. (2021) 21:215. doi: 10.1186/s12935-021-01913-z
183. Zhao XG, Hu JY, Tang J, Yi W, Zhang MY, Deng R, et al. miR-665 expression predicts poor survival and promotes tumor metastasis by targeting NR4A3 in breast cancer. Cell Death Dis. (2019) 10:479. doi: 10.1038/s41419-019-1705-z
184. Jiang F, Lou J, Zheng XM, Yang XY. LncRNA MIAT regulates autophagy and apoptosis of macrophage infected by Mycobacterium tuberculosis through the miR-665/ULK1 signaling axis. Mol Immunol. (2021) 139:42–9. doi: 10.1016/j.molimm.2021.07.023
185. Wang L, Liu J, Wang Z, Qian X, Zhao Y, Wang Q, et al. Dexmedetomidine abates myocardial ischemia reperfusion injury through inhibition of pyroptosis via regulation of miR-665/MEF2D/Nrf2 axis. BioMed Pharmacother. (2023) 165:115255. doi: 10.1016/j.biopha.2023.115255
186. Liang L, Fu J, Wang S, Cen H, Zhang L, Mandukhail SR, et al. MiR-142-3p enhances chemosensitivity of breast cancer cells and inhibits autophagy by targeting HMGB1. Acta Pharm Sin B. (2020) 10:1036–46. doi: 10.1016/j.apsb.2019.11.009
187. MacKenzie TN, Mujumdar N, Banerjee S, Sangwan V, Sarver A, Vickers S, et al. Triptolide induces the expression of miR-142-3p: a negative regulator of heat shock protein 70 and pancreatic cancer cell proliferation. Mol Cancer Ther. (2013) 12:1266–75. doi: 10.1158/1535-7163.MCT-12-1231
188. Mandolesi G, De Vito F, Musella A, Gentile A, Bullitta S, Fresegna D, et al. miR-142-3p is a key regulator of IL-1beta-dependent synaptopathy in neuroinflammation. J Neurosci. (2017) 37:546–61. doi: 10.1523/JNEUROSCI.0851-16.2016
189. Hu Z, Zhang H, Liu W, Yin Y, Jiang J, Yan C, et al. Mechanism of HBV-positive liver cancer cell exosomal miR-142-3p by inducing ferroptosis of M1 macrophages to promote liver cancer progression. Transl Cancer Res. (2022) 11:1173–87. doi: 10.21037/tcr-22-96
190. Lee S, Jia B, Liu J, Pham BP, Kwak JM, Xuan YH, et al. A 1-Cys peroxiredoxin from a thermophilic archaeon moonlights as a molecular chaperone to protect protein and DNA against stress-induced damage. PloS One. (2015) 10:e0125325. doi: 10.1371/journal.pone.0125325
191. Tang Q, Yuan Q, Li H, Wang W, Xie G, Zhu K, et al. miR-223/Hsp70/JNK/JUN/miR-223 feedback loop modulates the chemoresistance of osteosarcoma to cisplatin. Biochem Biophys Res Commun. (2018) 497:827–34. doi: 10.1016/j.bbrc.2018.02.091
192. Guan YZ, Sun C, Wang HL, Xia XL, Lu FZ, Song J, et al. MiR-223-5p inhibitor suppresses microglia inflammation and promotes Nrg-1 in rats of spinal cord injury. Eur Rev Med Pharmacol Sci. (2019) 23:9746–53. doi: 10.26355/eurrev_201911_19537
193. Ling Z, Liu D, Zhang G, Liang Q, Xiang P, Xu Y, et al. miR-361-5p modulates metabolism and autophagy via the Sp1-mediated regulation of PKM2 in prostate cancer. Oncol Rep. (2017) 38:1621–8. doi: 10.3892/or.2017.5852
194. Qin D, Wang X, Li Y, Yang L, Wang R, Peng J, et al. MicroRNA-223-5p and -3p cooperatively suppress necroptosis in ischemic/reperfused hearts. J Biol Chem. (2016) 291:20247–59. doi: 10.1074/jbc.M116.732735
195. Dong Z, Yang P, Qiu X, Liang S, Guan B, Yang H, et al. KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating miRNA-27b-3p/HSP90AA1 axis. J Cell Physiol. (2019) 234:11304–14. doi: 10.1002/jcp.27788
196. Zhang J, Cui J, Wang Y, Lin X, Teng X, Tang Y. Complex molecular mechanism of ammonia-induced apoptosis in chicken peripheral blood lymphocytes: miR-27b-3p, heat shock proteins, immunosuppression, death receptor pathway, and mitochondrial pathway. Ecotoxicol Environ Saf. (2022) 236:113471. doi: 10.1016/j.ecoenv.2022.113471
197. Zhu J, Zou Z, Nie P, Kou X, Wu B, Wang S, et al. Downregulation of microRNA-27b-3p enhances tamoxifen resistance in breast cancer by increasing NR5A2 and CREB1 expression. Cell Death Dis. (2016) 7:e2454. doi: 10.1038/cddis.2016.361
198. Xu D, Dong P, Xiong Y, Yue J, Konno Y, Ihira K, et al. MicroRNA-361-mediated inhibition of HSP90 expression and EMT in cervical cancer is counteracted by oncogenic lncRNA NEAT1. Cells. (2020) 9:632. doi: 10.3390/cells9030632
199. Ling J, He P. miR-361-5p regulates ovarian cancer cell proliferation and apoptosis by targeting TRAF3. Exp Ther Med. (2021) 21:199. doi: 10.3892/etm.2021.9632
200. Tian L, Zhao Z, Xie L, Zhu J. MiR-361-5p suppresses chemoresistance of gastric cancer cells by targeting FOXM1 via the PI3K/Akt/mTOR pathway. Oncotarget. (2018) 9:4886–96. doi: 10.18632/oncotarget.23513
201. Pan J, Jiang F, Zhou J, Wu D, Sheng Z, Li M. HSP90: A novel target gene of miRNA-628-3p in A549 cells. BioMed Res Int. (2018) 2018:4149707. doi: 10.1155/2018/4149707
202. Li GD, Cai M, Fu D, Chen K, Sun MX, Cai ZD, et al. Heat shock protein 90B1 plays an oncogenic role and is a target of microRNA-223 in human osteosarcoma. Cell Physiol Biochem. (2012) 30:1481–90. doi: 10.1159/000343336
203. Gu J, Xu H, Chen Y, Li N, Hou X. MiR-223 as a regulator and therapeutic target in liver diseases. Front Immunol. (2022) 13:860661. doi: 10.3389/fimmu.2022.860661
204. Zhou Y, Chen E, Tang Y, Mao J, Shen J, Zheng X, et al. miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. (2019) 10:843. doi: 10.1038/s41419-019-2053-8
205. Chen Y, Yang X, Feng M, Yu Y, Hu Y, Jiang W. Exosomal miR-223-3p from bone marrow mesenchymal stem cells targets HDAC2 to downregulate STAT3 phosphorylation to alleviate HBx-induced ferroptosis in podocytes. Front Pharmacol. (2024) 15:1327149. doi: 10.3389/fphar.2024.1327149
206. Jia Y, Cheng L, Yang J, Mao J, Xie Y, Yang X, et al. miR-223-3p prevents necroptotic macrophage death by targeting Ripk3 in a negative feedback loop and consequently ameliorates advanced atherosclerosis. Arterioscler Thromb Vasc Biol. (2024) 44:218–37. doi: 10.1161/ATVBAHA.123.319776
207. Zhao S, Tan Y, Qin J, Xu H, Liu L, Wan H, et al. MicroRNA-223-3p promotes pyroptosis of cardiomyocyte and release of inflammasome factors via downregulating the expression level of SPI1 (PU. 1). Toxicol. (2022) 476:153252. doi: 10.1016/j.tox.2022.153252
208. Su C, Huang DP, Liu JW, Liu WY, Cao YO. miR-27a-3p regulates proliferation and apoptosis of colon cancer cells by potentially targeting BTG1. Oncol Lett. (2019) 18:2825–34. doi: 10.3892/ol.2019.10629
209. Ren LR, Yao RB, Wang SY, Gong XD, Xu JT, Yang KS. MiR-27a-3p promotes the osteogenic differentiation by activating CRY2/ERK1/2 axis. Mol Med. (2021) 27:43. doi: 10.1186/s10020-021-00303-5
210. Lu X, Kang N, Ling X, Pan M, Du W, Gao S. MiR-27a-3p promotes non-small cell lung cancer through SLC7A11-mediated-ferroptosis. Front Oncol. (2021) 11:759346. doi: 10.3389/fonc.2021.759346
211. Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J Exp Med. (2011) 208:2717–31. doi: 10.1084/jem.20111386
212. Moles R, Bellon M, Nicot C. STAT1: A novel target of miR-150 and miR-223 is involved in the proliferation of HTLV-I-transformed and ATL cells. Neoplasia. (2015) 17:449–62. doi: 10.1016/j.neo.2015.04.005
213. Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, et al. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. (2011) 187:6171–5. doi: 10.4049/jimmunol.1100835
214. Mao W, Huang X, Wang L, Zhang Z, Liu M, Li Y, et al. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression. J Exp Clin Cancer Res. (2019) 38:169. doi: 10.1186/s13046-019-1136-9
215. Wang P, Gu Y, Zhang Q, Han Y, Hou J, Lin L, et al. Identification of resting and type I IFN-activated human NK cell miRNomes reveals microRNA-378 and microRNA-30e as negative regulators of NK cell cytotoxicity. J Immunol. (2012) 189:211–21. doi: 10.4049/jimmunol.1200609
216. Liu MM, Li Z, Han XD, Shi JH, Tu DY, Song W, et al. MiR-30e inhibits tumor growth and chemoresistance via targeting IRS1 in Breast Cancer. Sci Rep. (2017) 7:15929. doi: 10.1038/s41598-017-16175-x
217. Zhang Z, Qin H, Jiang B, Chen W, Cao W, Zhao X, et al. miR-30e-5p suppresses cell proliferation and migration in bladder cancer through regulating metadherin. J Cell Biochem. (2019) 120:15924–32. doi: 10.1002/jcb.28866
218. Xu X, Li Y, Liu G, Li K, Chen P, Gao Y, et al. MiR-378a-3p acts as a tumor suppressor in gastric cancer via directly targeting RAB31 and inhibiting the Hedgehog pathway proteins GLI1/2. Cancer Biol Med. (2022) 19:1662–82. doi: 10.20892/j.issn.2095-3941.2022.0337
219. Min D, Lv XB, Wang X, Zhang B, Meng W, Yu F, et al. Downregulation of miR-302c and miR-520c by 1,25(OH)2D3 treatment enhances the susceptibility of tumour cells to natural killer cell-mediated cytotoxicity. Br J Cancer. (2013) 109:723–30. doi: 10.1038/bjc.2013.337
220. Ding HM, Zhang H, Wang J, Zhou JH, Shen FR, Ji RN, et al. miR−302c−3p and miR−520a−3p suppress the proliferation of cervical carcinoma cells by targeting CXCL8. Mol Med Rep. (2021) 23:322. doi: 10.3892/mmr.2021.11961
221. Li X, Fu Q, Li H, Zhu L, Chen W, Ruan T, et al. MicroRNA-520c-3p functions as a novel tumor suppressor in lung adenocarcinoma. FEBS J. (2019) 286:2737–52. doi: 10.1111/febs.14835
222. Omar HA, El-Serafi AT, Hersi F, Arafa EA, Zaher DM, Madkour M, et al. Immunomodulatory microRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. (2019) 286:3540–57. doi: 10.1111/febs.15000
223. Wang J, Yan Y, Zhang Z, Li Y. Role of miR-10b-5p in the prognosis of breast cancer. PeerJ. (2019) 7:e7728. doi: 10.7717/peerj.7728
224. Kishikawa T, Otsuka M, Yoshikawa T, Ohno M, Takata A, Shibata C, et al. Regulation of the expression of the liver cancer susceptibility gene MICA by microRNAs. Sci Rep. (2013) 3:2739. doi: 10.1038/srep02739
225. Chen YL, Wang GX, Lin BA, Huang JS. MicroRNA-93-5p expression in tumor tissue and its tumor suppressor function via targeting programmed death ligand-1 in colorectal cancer. Cell Biol Int. (2020) 44:1224–36. doi: 10.1002/cbin.11323
226. Chen G, Yan Y, Qiu X, Ye C, Jiang X, Song S, et al. miR-93-5p suppresses ovarian cancer Malignancy and negatively regulate CCND2 by binding to its 3'UTR region. Discovery Oncol. (2022) 13:15. doi: 10.1007/s12672-022-00478-1
227. Xie J, Liu M, Li Y, Nie Y, Mi Q, Zhao S. Ovarian tumor-associated microRNA-20a decreases natural killer cell cytotoxicity by downregulating MICA/B expression. Cell Mol Immunol. (2014) 11:495–502. doi: 10.1038/cmi.2014.30
228. Wei K, Pan C, Yao G, Liu B, Ma T, Xia Y, et al. MiR-106b-5p promotes proliferation and inhibits apoptosis by regulating BTG3 in non-small cell lung cancer. Cell Physiol Biochem. (2017) 44:1545–58. doi: 10.1159/000485650
229. Ni S, Weng W, Xu M, Wang Q, Tan C, Sun H, et al. miR-106b-5p inhibits the invasion and metastasis of colorectal cancer by targeting CTSA. Onco Targets Ther. (2018) 11:3835–45. doi: 10.2147/OTT.S172887
230. Jasinski-Bergner S, Stoehr C, Bukur J, Massa C, Braun J, Huttelmaier S, et al. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology. (2015) 4:e1008805. doi: 10.1080/2162402X.2015.1008805
231. Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. (2009) 10:1252–9. doi: 10.1038/ni.1798
232. Pan YJ, Wan J, Wang CB. MiR-326: Promising biomarker for cancer. Cancer Manag Res. (2019) 11:10411–8. doi: 10.2147/CMAR.S223875
233. Zhang Z, Xue Z, Liu Y, Liu H, Guo X, Li Y, et al. MicroRNA-181c promotes Th17 cell differentiation and mediates experimental autoimmune encephalomyelitis. Brain Behav Immun. (2018) 70:305–14. doi: 10.1016/j.bbi.2018.03.011
234. Li N, Cheng C, Wang T. MiR-181c-5p mitigates tumorigenesis in cervical squamous cell carcinoma via targeting glycogen synthase kinase 3beta interaction protein (GSKIP). Onco Targets Ther. (2020) 13:4495–505. doi: 10.2147/OTT.S245254
235. Xie D, Li S, Wu T, Wang X, Fang L. MiR-181c suppresses triple-negative breast cancer tumorigenesis by targeting MAP4K4. Pathol Res Pract. (2022) 230:153763. doi: 10.1016/j.prp.2022.153763
236. Lin R, Chen L, Chen G, Hu C, Jiang S, Sevilla J, et al. Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J Clin Invest. (2014) 124:5352–67. doi: 10.1172/JCI76561
237. Quan J, Pan X, Li Y, Hu Y, Tao L, Li Z, et al. MiR-23a-3p acts as an oncogene and potential prognostic biomarker by targeting PNRC2 in RCC. BioMed Pharmacother. (2019) 110:656–66. doi: 10.1016/j.biopha.2018.11.065
238. Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O'Connell RM, et al. MicroRNA-125b potentiates macrophage activation. J Immunol. (2011) 187:5062–8. doi: 10.4049/jimmunol.1102001
239. Li Y, Wang Y, Fan H, Zhang Z, Li N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun. (2018) 504:277–82. doi: 10.1016/j.bbrc.2018.08.172
240. Hua S, Quan Y, Zhan M, Liao H, Li Y, Lu L. miR-125b-5p inhibits cell proliferation, migration, and invasion in hepatocellular carcinoma via targeting TXNRD1. Cancer Cell Int. (2019) 19:203. doi: 10.1186/s12935-019-0919-6
241. Wang Z, Han J, Cui Y, Zhou X, Fan K. miRNA-21 inhibition enhances RANTES and IP-10 release in MCF-7 via PIAS3 and STAT3 signalling and causes increased lymphocyte migration. Biochem Biophys Res Commun. (2013) 439:384–9. doi: 10.1016/j.bbrc.2013.08.072
242. Garajei A, Allameh A, Azadi M, Emami A, Atashbasteh M, Mostafavi M, et al. Evaluation of the expression levels of miR-21-5p and miR-429 genes in biopsy samples from patients with oral squamous cell carcinoma. Diagnostics (Basel). (2023) 13:1244. doi: 10.3390/diagnostics13071244
243. Zhang M, Liu Q, Mi S, Liang X, Zhang Z, Su X, et al. Both miR-17-5p and miR-20a alleviate suppressive potential of myeloid-derived suppressor cells by modulating STAT3 expression. J Immunol. (2011) 186:4716–24. doi: 10.4049/jimmunol.1002989
244. Yifei S, Chunxiao H, Dinuo L. MiR-17-5p inhibits the proliferation and metastasis of gastric cancer cells by targeting PTEN protein. Altern Ther Health Med. (2022) 28:23–9.
245. Li C, Zhang L, Bu X, Wang J, Li L, Yang Z. Circ-LTBP1 is involved in doxorubicin-induced intracellular toxicity in cardiomyocytes via miR-107/ADCY1 signal. Mol Cell Biochem. (2022) 477:1127–38. doi: 10.1007/s11010-022-04360-0
246. Guo F, Lin SC, Zhao MS, Yu B, Li XY, Gao Q, et al. microRNA-142-3p inhibits apoptosis and inflammation induced by bleomycin through down-regulation of Cox-2 in MLE-12 cells. Braz J Med Biol Res. (2017) 50:e5974. doi: 10.1590/1414-431X20175974
247. Hu X, Liu H, Wang Z, Hu Z, Li L. miR-200a attenuated doxorubicin-induced cardiotoxicity through upregulation of Nrf2 in mice. Oxid Med Cell Longev. (2019) 2019:1512326. doi: 10.1155/2019/1512326
248. Kim HY, Kim YM, Hong S. Astaxanthin suppresses the metastasis of colon cancer by inhibiting the MYC-mediated downregulation of microRNA-29a-3p and microRNA-200a. Sci Rep. (2019) 9:9457. doi: 10.1038/s41598-019-45924-3
249. Safaei S, Amini M, Najjary S, Mokhtarzadeh A, Bolandi N, Saeedi H, et al. miR-200c increases the sensitivity of breast cancer cells to Doxorubicin through downregulating MDR1 gene. Exp Mol Pathol. (2022) 125:104753. doi: 10.1016/j.yexmp.2022.104753
250. Chaudhari U, Nemade H, Gaspar JA, Hescheler J, Hengstler JG, Sachinidis A. MicroRNAs as early toxicity signatures of doxorubicin in human-induced pluripotent stem cell-derived cardiomyocytes. Arch Toxicol. (2016) 90:3087–98. doi: 10.1007/s00204-016-1668-0
251. Zhu JN, Fu YH, Hu ZQ, Li WY, Tang CM, Fei HW, et al. Activation of miR-34a-5p/Sirt1/p66shc pathway contributes to doxorubicin-induced cardiotoxicity. Sci Rep. (2017) 7:11879. doi: 10.1038/s41598-017-12192-y
252. Hortu HO, Karaca E, Sozeri B, Gulez N, Makay B, Gunduz C, et al. Evaluation of the effects of miRNAs in familial Mediterranean fever. Clin Rheumatol. (2019) 38:635–43. doi: 10.1007/s10067-017-3914-0
253. Zheng L, Chen J, Ma Z, Liu W, Yang F, Yang Z, et al. Capsaicin causes inactivation and degradation of the androgen receptor by inducing the restoration of miR-449a in prostate cancer. Oncol Rep. (2015) 34:1027–34. doi: 10.3892/or.2015.4055
254. Zhang Y, Liang Q, Zhang Y, Hong L, Lei D, Zhang L. Olmesartan alleviates bleomycin-mediated vascular smooth muscle cell senescence via the miR-665/SDC1 axis. Am J Transl Res. (2020) 12:5205–20.
255. Barraclough JY, Joglekar MV, Januszewski AS, Martinez G, Celermajer DS, Keech AC, et al. A microRNA signature in acute coronary syndrome patients and modulation by colchicine. J Cardiovasc Pharmacol Ther. (2020) 25:444–55. doi: 10.1177/1074248420922793
256. Shah MS, Schwartz SL, Zhao C, Davidson LA, Zhou B, Lupton JR, et al. Integrated microRNA and mRNA expression profiling in a rat colon carcinogenesis model: effect of a chemo-protective diet. Physiol Genomics. (2011) 43:640–54. doi: 10.1152/physiolgenomics.00213.2010
257. Pan J, Li M, Yu F, Zhu F, Wang L, Ning D, et al. Up-regulation of p53/miR-628-3p pathway, a novel mechanism of shikonin on inhibiting proliferation and inducing apoptosis of A549 and PC-9 non-small cell lung cancer cell lines. Front Pharmacol. (2021) 12:766165. doi: 10.3389/fphar.2021.766165
258. Gueven N, Spring KJ, Holmes S, Ahuja K, Eri R, Park AY, et al. Micro RNA expression after ingestion of fucoidan; a clinical study. Mar Drugs. (2020) 18:143. doi: 10.3390/md18030143
259. Sugita BM, Rodriguez Y, Fonseca AS, Nunes Souza E, Kallakury B, Cavalli IJ, et al. MiR-150-5p overexpression in triple-negative breast cancer contributes to the in vitro aggressiveness of this breast cancer subtype. Cancers (Basel). (2022) 14:2156. doi: 10.3390/cancers14092156
260. Lin X, Ma A. LncRNA SUMO1P3 aggravates doxorubicin-induced cardiomyocyte apoptosis by targeting miR-93-5p/Bin1. Res Square. (2021). doi: 10.21203/rs.3.rs-167434/v2
261. Hao S, Yang Q, Zhang W, Li Q, Li F, Cheng H, et al. LINC00852 participates in the phycocyanin-mediated restrained vitality of non-small cell lung cancer cells via miR-29a-3p/LAMTOR1 axis. J Funct Foods. (2023) 106:105588. doi: 10.1016/j.jff.2023.105588
262. Rizzo S, Cangemi A, Galvano A, Fanale D, Buscemi S, Ciaccio M, et al. Analysis of miRNA expression profile induced by short term starvation in breast cancer cells treated with doxorubicin. Oncotarget. (2017) 8:71924–32. doi: 10.18632/oncotarget.18028
263. Assidicky R, Tokat UM, Tarman IO, Saatci O, Ersan PG, Raza U, et al. Targeting HIF1-alpha/miR-326/ITGA5 axis potentiates chemotherapy response in triple-negative breast cancer. Breast Cancer Res Treat. (2022) 193:331–48. doi: 10.1007/s10549-022-06569-5
264. Yu X, Ruan Y, Shen T, Qiu Q, Yan M, Sun S, et al. Dexrazoxane protects cardiomyocyte from doxorubicin-induced apoptosis by modulating miR-17-5p. BioMed Res Int. (2020) 2020:5107193. doi: 10.1155/2020/5107193
265. Bao C, Liu T, Qian L, Xiao C, Zhou X, Ai H, et al. Shikonin inhibits migration and invasion of triple-negative breast cancer cells by suppressing epithelial-mesenchymal transition via miR-17-5p/PTEN/Akt pathway. J Cancer. (2021) 12:76–88. doi: 10.7150/jca.47553
266. Wu SY, Wu AT, Yuan KS, Liu SH. Brown seaweed fucoidan inhibits cancer progression by dual regulation of miR-29c/ADAM12 and miR-17-5p/PTEN axes in human breast cancer cells. J Cancer. (2016) 7:2408–19. doi: 10.7150/jca.15703
267. Wang Q, Jiang F, Zhao C, Song J, Hu M, Lv Y, et al. miR-21-5p prevents doxorubicin-induced cardiomyopathy by downregulating BTG2. Heliyon. (2023) 9:e15451. doi: 10.1016/j.heliyon.2023.e15451
268. Mandal CC, Ghosh-Choudhury T, Dey N, Choudhury GG, Ghosh-Choudhury N. miR-21 is targeted by omega-3 polyunsaturated fatty acid to regulate breast tumor CSF-1 expression. Carcinogenesis. (2012) 33:1897–908. doi: 10.1093/carcin/bgs198
269. Desai VG, CK J, Vijay V, Moland CL, Herman EH, Lee T, et al. Early biomarkers of doxorubicin-induced heart injury in a mouse model. Toxicol Appl Pharmacol. (2014) 281:221–9. doi: 10.1016/j.taap.2014.10.006
270. Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. (2020) 48:D127–D31. doi: 10.1093/nar/gkz757
271. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. (2023) 51:D638–D46. doi: 10.1093/nar/gkac1000
272. Mattarollo SR, Loi S, Duret H, Ma Y, Zitvogel L, Smyth MJ. Pivotal role of innate and adaptive immunity in anthracycline chemotherapy of established tumors. Cancer Res. (2011) 71:4809–20. doi: 10.1158/0008-5472.CAN-11-0753
273. Shindo S, Hosokawa Y, Hosokawa I, Ozaki K, Matsuo T. Shikonin inhibits inflammatory cytokine production in human periodontal ligament cells. Inflammation. (2016) 39:1124–9. doi: 10.1007/s10753-016-0344-0
274. Kondo Y, Kato A, Hojo H, Nozoe S, Takeuchi M, Ochi K. Cytokine-related immunopotentiating activities of paramylon, a beta-(1–>3)-D-glucan from Euglena gracilis. J Pharmacobiodyn. (1992) 15:617–21. doi: 10.1248/bpb1978.15.617
275. Sun T, Liang H, Xue M, Liu Y, Gong A, Jiang Y, et al. Protective effect and mechanism of fucoidan on intestinal mucosal barrier function in NOD mice. Food Agric Immunol. (2020) 31:939–53. doi: 10.1080/09540105.2020.1789071
276. Catanzaro E, Feron O, Skirtach AG, Krysko DV. Immunogenic cell death and role of nanomaterials serving as therapeutic vaccine for personalized cancer immunotherapy. Front Immunol. (2022) 13:925290. doi: 10.3389/fimmu.2022.925290
277. Gao W, Wang X, Zhou Y, Wang X, Yu Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct Target Ther. (2022) 7:196. doi: 10.1038/s41392-022-01046-3
278. Liang H, Liu M, Yan X, Zhou Y, Wang W, Wang X, et al. miR-193a-3p functions as a tumor suppressor in lung cancer by down-regulating ERBB4. J Biol Chem. (2015) 290:926–40. doi: 10.1074/jbc.M114.621409
279. Zhu KG, Ding HY, Wang WG, Liao ZC, Fu Z, Hong YT, et al. Tumor-suppressive miR-218-5p inhibits cancer cell proliferation and migration via EGFR in non-small cell lung cancer. Oncotarget. (2016) 7:28075–85. doi: 10.18632/oncotarget.8576
280. Yin W, Gao F, Zhang S. MicroRNA−34a inhibits the proliferation and promotes the chemosensitivity of retinoblastoma cells by downregulating Notch1 expression. Mol Med Rep. (2020) 22:1613–20. doi: 10.3892/mmr.2020.11238
281. Wang Z, Lin M, He L, Qi H, Shen J, Ying K. Exosomal lncRNA SCIRT/miR-665 transferring promotes lung cancer cell metastasis through the inhibition of HEYL. J Oncol. (2021) 2021:9813773. doi: 10.1155/2021/9813773
282. Xu T, He BS, Pan B, Pan YQ, Sun HL, Liu XX, et al. MiR-142-3p functions as a tumor suppressor by targeting RAC1/PAK1 pathway in breast cancer. J Cell Physiol. (2020) 235:4928–40. doi: 10.1002/jcp.29372
283. Sun Y, Xu T, Cao YW, Ding XQ. Antitumor effect of miR-27b-3p on lung cancer cells via targeting Fzd7. Eur Rev Med Pharmacol Sci. (2017) 21:4113–23.
284. Colangelo T, Polcaro G, Ziccardi P, Muccillo L, Galgani M, Pucci B, et al. The miR-27a-calreticulin axis affects drug-induced immunogenic cell death in human colorectal cancer cells. Cell Death Dis. (2016) 7:e2108. doi: 10.1038/cddis.2016.29
285. Ahmed I, Ismail N. M1 and M2 macrophages polarization via mTORC1 influences innate immunity and outcome of Ehrlichia infection. J Cell Immunol. (2020) 2:108–15. doi: 10.33696/immunology.2.029
286. Tsukerman P, Enk J, Mandelboim O. Metastamir-mediated immune evasion: miR-10b downregulates the stress-induced molecule MICB, hence avoid recognition by NKG2D receptor. Oncoimmunology. (2013) 2:e22245. doi: 10.4161/onci.22245
287. Codo P, Weller M, Meister G, Szabo E, Steinle A, Wolter M, et al. MicroRNA-mediated down-regulation of NKG2D ligands contributes to glioma immune escape. Oncotarget. (2014) 5:7651–62. doi: 10.18632/oncotarget.2287
288. Shao L, He Q, Wang J, He F, Lin S, Wu L, et al. MicroRNA-326 attenuates immune escape and prevents metastasis in lung adenocarcinoma by targeting PD-L1 and B7-H3. Cell Death Discovery. (2021) 7:145. doi: 10.1038/s41420-021-00527-8
289. Knochelmann HM, Dwyer CJ, Bailey SR, Amaya SM, Elston DM, Mazza-McCrann JM, et al. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell Mol Immunol. (2018) 15:458–69. doi: 10.1038/s41423-018-0004-4
290. Bailey SR, Nelson MH, Himes RA, Li Z, Mehrotra S, Paulos CM. Th17 cells in cancer: the ultimate identity crisis. Front Immunol. (2014) 5:276. doi: 10.3389/fimmu.2014.00276
291. Karpisheh V, Ahmadi M, Abbaszadeh-Goudarzi K, Mohammadpour Saray M, Barshidi A, Mohammadi H, et al. The role of Th17 cells in the pathogenesis and treatment of breast cancer. Cancer Cell Int. (2022) 22:108. doi: 10.1186/s12935-022-02528-8
292. Parisi L, Gini E, Baci D, Tremolati M, Fanuli M, Bassani B, et al. Macrophage polarization in chronic inflammatory diseases: Killers or builders? J Immunol Res. (2018) 2018:8917804. doi: 10.1155/2018/8917804
293. Xi J, Huang Q, Wang L, Ma X, Deng Q, Kumar M, et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene. (2018) 37:3151–65. doi: 10.1038/s41388-018-0178-3
294. Ma S, Liu M, Xu Z, Li Y, Guo H, Ge Y, et al. A double feedback loop mediated by microRNA-23a/27a/24-2 regulates M1 versus M2 macrophage polarization and thus regulates cancer progression. Oncotarget. (2016) 7:13502–19. doi: 10.18632/oncotarget.6284
295. Jin L, Li Y, Nie L, He T, Hu J, Liu J, et al. MicroRNA−24−2 is associated with cell proliferation, invasion, migration and apoptosis in renal cell carcinoma. Mol Med Rep. (2017) 16:9157–64. doi: 10.3892/mmr.2017.7705
296. Yaseen MM, Abuharfeil NM, Darmani H, Daoud A. Mechanisms of immune suppression by myeloid-derived suppressor cells: the role of interleukin-10 as a key immunoregulatory cytokine. Open Biol. (2020) 10:200111. doi: 10.1098/rsob.200111
297. Hao S, Liu Y, Li S, Wang J, Zhao L, Wang C, et al. Insight into the potential antineoplastic mechanism of phycocyanin in non-small cell lung carcinoma A549 cells based on micro-RNA sequencing. J Funct Foods. (2020) 74:104175. doi: 10.1016/j.jff.2020.104175
Keywords: ICD, DAMPs, cytokines, microRNAs, natural products, targets
Citation: Chuang Y-T, Yen C-Y, Tang J-Y, Chang F-R, Tsai Y-H, Wu K-C, Chien T-M and Chang H-W (2024) The modulation of immune cell death in connection to microRNAs and natural products. Front. Immunol. 15:1425602. doi: 10.3389/fimmu.2024.1425602
Received: 30 April 2024; Accepted: 27 November 2024;
Published: 20 December 2024.
Edited by:
Hui-Min David Wang, National Chung Hsing University, TaiwanReviewed by:
Alexander T. Sougiannis, The Ohio State University, United StatesJiunn-Kiang Ko, Chung Shan Medical University, Taiwan
Copyright © 2024 Chuang, Yen, Tang, Chang, Tsai, Wu, Chien and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tsu-Ming Chien, MTAyMDQzMUBrbXVoLm9yZy50dw==; Hsueh-Wei Chang, Y2hhbmdod0BrbXUuZWR1LnR3
†These authors have contributed equally to this work
 Ya-Ting Chuang1†
Ya-Ting Chuang1† Jen-Yang Tang
Jen-Yang Tang Fang-Rong Chang
Fang-Rong Chang Yi-Hong Tsai
Yi-Hong Tsai Hsueh-Wei Chang
Hsueh-Wei Chang