- 1School of Clinical Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 2Dermatological Department, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, China
Exosome-derived microRNAs (miRNAs) are biomacromolecules and nanoscale extracellular vesicles originating from intracellular compartments that are secreted by most cells into the extracellular space. This review examines the formation and function of exosomal miRNAs in biological information transfer, explores the pathogenesis of vitiligo, and highlights the relationship between exosomal miRNAs and vitiligo. The aim is to deepen the understanding of how exosomal miRNAs influence immune imbalance, oxidative stress damage, melanocyte-keratinocyte interactions, and melanogenesis disorders in the development of vitiligo. This enhanced understanding may contribute to the development of potential diagnostic and therapeutic options for vitiligo.
1 Introduction
Vitiligo is recognized as an autoimmune disorder characterized by the progressive destruction of epidermal melanocytes (1, 2). It occurs due to a dynamic interaction between genetic and environmental factors, leading to autoimmune destruction of melanocytes. Defects in melanocyte adhesion and increased oxidative stress further augment the immune response in vitiligo (3).
Exosomes are extracellular vesicles found in various biofluids and tissues, ranging in diameter from 40 to 160 nm (averaging 100 nm).They are released by all cells as part of normal physiology and in response to abnormalities (4). Exosomes deliver bioactive cargoes, such as proteins, mRNAs, miRNAs, and lipids, to recipient cells (5–7). Exosomes are involved in regulating various physiological and pathological processes, such as immune regulation, cell growth, and differentiation (8, 9). Increasing research indicates that exosomes play a significant role in vitiligo (10).
MicroRNAs (miRNAs) are small non-coding RNA molecules (about 22 54 nucleotides) that are essential in regulating various biological processes (11, 12). Cells can selectively package miRNAs into exosomes, which are then secreted to nearby or distant targets (13). Evidence suggests that changes in the profiles of miRNAs delivered by exosomes are closely linked to the progression of vitiligo (10, 14). This feature renders miRNAs as highly interesting therapeutic tools to restore cell functions altered as part of a vitiligo phenotype (15). The present review aims to provide a deep insight into the relationship between exosome-delivered mRNAs and vitiligo and highlight the role of exosomes in immune regulation, oxidative stress, melanocyte-keratinocyte interactions, and melanogenesis. Further understanding of the role of exosomes in vitiligo may contribute to the development of potential diagnostic markers and therapeutic options for vitiligo.
2 Exosome-derived miRNAs
2.1 Biological functions of exosome
Exosomes are extracellular vesicles characterized by vesicle-like structures with a diameter of 30 ~ 150 nm, wrapped by a phospholipid bilayer (10, 16, 17). They are present in almost all types of prokaryotic and eukaryotic cells (18). Initially recognized in parasitic cells as debris and “cellular dust” in human platelets, they were believed to have no significant role in biology (19). Later, their role as carriers of bioactive substances was discovered (20). Previously, intercellular communication was thought to be mediated through soluble substances, including cytokines and interleukins. However, an increasing number of studies now suggest that exosomes are the primary channel for long-distance communication between various cell types (4, 21). Exosomes are thought to originate from multivesicular bodies (MVBs),which form intraluminal vesicles through dual invaginations of the plasma membrane (22). The plasma membrane primarily invaginates to form disc-shaped structures called early sorting endosomes, which contain phagocytic proteins and genetic material from the cytoplasm as well as cytosolic proteins produced by membrane invagination. Early sorting endosomes then mature into late sorting endosomes, which eventually form MVBs through secondary fusion of endosome-restricted membranes. The resulting MVBs are either degraded by autophagosomes or lysosomes, or are released as exosomes into the extracellular space after fusing with the plasma membrane (18, 23).
Exosomes have been reported to carry signaling molecules such as proteins, DNA, RNA, enzymes, and even organelles (24–27). They are present in a variety of biological fluids, including blood, urine, saliva, and breast milk (27, 28). Exosomes contain two types of proteins: conserved proteins and exosome-specific proteins (10). Conserved proteins are similar proteins that appear in each exosome and include cluster of differentiation (CD) 36, CD81, CD9, CD82 (29, 30). Heat shock protein (Hsp) 500, Hsp70, Hsp90, ALIX, and tumor susceptibility gene 101 (TSG101) (31–33),which serve as pan-markers for the common detection of exosomes (34). The composition of exosome-specific proteins depends on the cellular origin of the exosome and may change depending on the physiological changes and stimuli acting on the cell; for example, exosomes derived from mesenchymal stem cells or stromal cells (MSC) are endowed with immunosuppressive and tumor-homing capabilities (18).
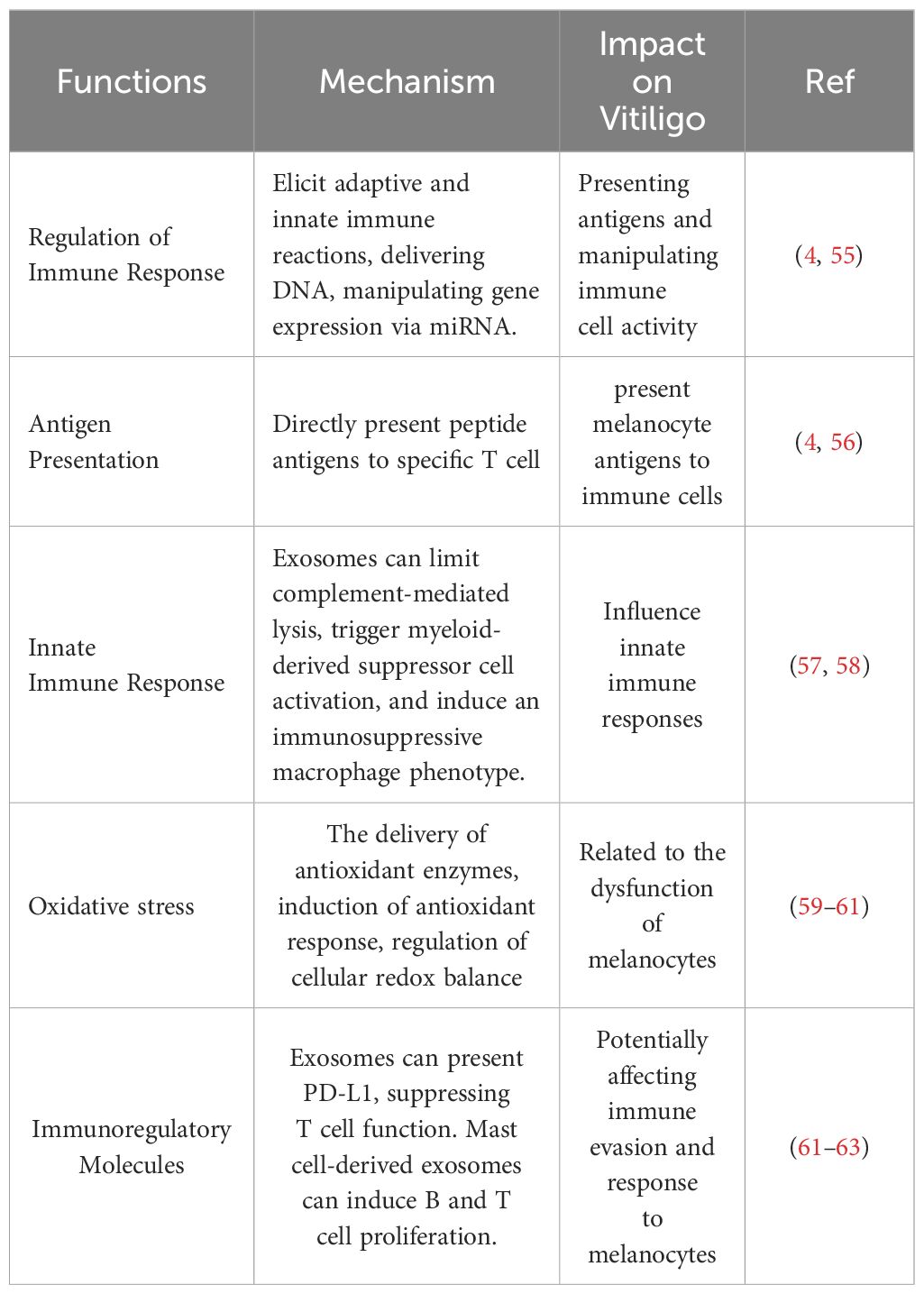
Notably, exosomes are not only involved in a variety of physiological processes such as inflammatory responses (35, 36)、skin trauma repair (37)、angiogenesis (38)、immune responses (39, 40) and immune surveillance (41), but also play an important role in the pathological state of several diseases. For example, exosomes are believed to contribute to skin immunity and melanogenesis, thereby supporting the maintenance of skin homeostasis (41, 42). In addition, they enhance the function of regulatory T cells and suppress the activity of CD8+ cells and natural killer (NK) cells (43, 44). They are closely associated with the pathogenesis of other diseases as well, such as autoimmune diseases (45) and skin diseases (4, 46). Also, the contents carried by exosomes reflect the physiological or pathological condition of their cellular origin. Many studies have reported that the RNA profile of exosomes of disease origin or pathological cells differs from that of exosomes of healthy cellular origin (47–49). Exosomal RNA has been used or has the potential to be used in the diagnosis and treatment of multiple diseases (50, 51). For example, a multicenter cohort study showed that a small RNA was specifically enriched in salivary exosomes, tissues and cells of patients with pancreatic ductal adenocarcinoma, and detection of a double signal composed of these small RNAs was able to diagnose patients with pancreatic ductal adenocarcinoma with high sensitivity (90.50%) and specificity (94.20%) (52). In melanoma patients, the levels of circulating exosomal PD-L1 are positively correlated with the levels of IFN-γ, and exosomal PD-L1 is also a marker of immune activation after the initiation of treatment with PD-1-blocking antibodies, (53, 54). The analysis of exosomes contributes to a better understanding of the biological mechanisms of disease and helps to develop new diagnostic and therapeutic approaches. Table 1 summarizes the role of exosomes in autoimmune diseases.
2.2 MiRNAs
MiRNAs are 18-25 nucleotide non-coding RNAs generated by nearly all cells in the body and primarily transcribed by RNA polymerase (64). They are initially transcribed into large precursors called primary miRNAs (pri-miRNAs), which are then transcribed in the nucleus by RNA polymerase II or RNA polymerase III and cleaved into pre-miRNAs by Drosha and its cofactors (65). These pre-miRNAs are exported to the cytoplasm and processed by Dicer to produce mature miRNA products (66, 67). MiRNAs have been shown to play various roles in numerous diseases and physiological states, both in vitro and in vivo, through their functional transfer via exosomes. They primarily regulate gene expression by recognizing homologous sequences and interfering with transcriptional, translational, or epigenetic processes (68). MiRNAs are highly conserved during evolution and participate in essential biological processes such as cell proliferation, differentiation, metabolism, apoptosis, development, and aging processes (66). Over 2600 mature human miRNAs have been reported to date (69) and are expressed in distinct spatial and temporal patterns during embryonic and postnatal development, as well as in adult tissues (70).
2.3 Exosome-derived miRNAs
Exosomes offer a unique mode of intercellular communication, where miRNAs produced and released by one cell are taken up by distant cells, affecting gene expression in health and disease (71–73). Exosome-derived miRNAs have several advantages over intracellular and cell-free blood sources of miRNA (74). For instance, exosomes can protect miRNAs from degradation in vivo and enable superior systemic retention compared to liposomes, allowing miRNAs to function at distant sites (75). Moreover, the miRNA profiles derived from exosomes differ significantly from those originating from their cells of origin, indicating that numerous miRNA species are selectively encapsulated into exosomes (76).
Currently, various attempts have been made to elucidate the mechanisms of loading and sorting miRNA into exosomes. Several pathways for loading miRNAs into exosomes have been described. The first pathway originates from the inherent structure of miRNA with a 3′ end adenylation and uridylation, which is essential for recognition by AGO2.miRNAs with an adenylated 3′ end are predominantly found in cells, while miRNAs with a uridylated 3′ end are sorted into exosomes, as demonstrated in RNA sequencing research on human B cells and their associated exosomes (77). The second sorting pathway involves sphingomyelinase 2-dependency, which is the first molecule identified to be associated with miRNA loading into exosomes. Overexpression of sphingomyelinase 2 promotes miRNA sorting into exosomes, while its inhibition produces the opposite result (78). The third sorting pathway involves the miRNA-induced silencing complex (miRISC) association. The miRISC mainly consists of miRNA, miRNA-repressible mRNA, GW182, and AGO2. The RNA-binding protein mediates the final sorting pathway. SYNCRIP selectively sorts miRNAs and has a 4-nucleotide motif near the 3′ end, independently of hnRNPA2B1 (79). The fourth pathway is the sumoylated heterogeneous nuclear ribonucleoprotein (hnRNP; primarily including hnRNPA2B1, hnRNPA1, and hnRNPC)-dependent pathway, which binds to miRNAs and facilitates their loading into exosomes (80). Y-box protein 1 can selectively sort miR-223 into exosomes in HEK-293T cells (81). Additionally, a growing number of studies clarify the mechanisms by which miRNA sorting occurs in exosomes or cells. Accumulating research evidence demonstrates that miRNAs possess sorting sequences that determine their exosomal secretion or cellular retention, and different cell types, including endothelium, brown and white adipocytes, muscle, and liver, preferentially use specific sorting sequences, thus determining the exosomal miRNA profile of that cell type. Insertion or knockdown of these CELLmotifs or EXOmotifs into a miRNA alters their retention in the cells that produce or secrete them into exosomes. Two RNA-binding proteins, Fus and Alyref, are involved in the export of miRNAs carrying one of the strongest EXOmotifs, CGGGAG. Increased miRNA delivery mediated by EXOmotifs leads to enhanced inhibition of receptor genes in distant cells (82, 83).
Following intracellular loading of miRNA into exosomes, binding to their target cells’ membrane regions, and through plasma membrane fusion, miRNA-containing exosomes are eventually released. Six possible methods of exosome miRNA uptake have been documented, and are illustrated in Figure 1: 1) Receptor-Mediated Endocytosis. Mechanisms involving clathrin- or caveolae-regulated endocytosis, phagocytosis, or macropinocytosis (84). 2) Clathrin-Coated Pits (CCPs).CCPs form as a result of the AP2 complex recognizing membrane and cargo, subsequently recruiting clathrin triskelia (85, 86). 3) Lipid Rafts. The role of lipid rafts in the intracellular mechanism of exosome entry has been investigated. Studies show that disrupting dendritic cell lipid rafts significantly reduces and blocks exosome uptake, indicating that lipid rafts are involved in this process. However, the precise mechanism of this pathway remains to be elucidated (85). 4) Receptor signaling. MiRNAs docked in exosomes can be assimilated by recipient cells by directly targeting corresponding cell membrane receptors, generating intracellular signaling, and subsequently activating or inhibiting related pathways such as the Ca2+/MEK/ERK signaling pathway (87, 88). 5) Direct Fusion. Targeting the recipient cells’ surface immediately and fusing with the cell membrane (89). 6) Caveolae. Exosomes have been shown to follow a different endocytic pathway than liposomes through caveolae internalization (90). Co-localization studies between fluorescently labeled extracellular vesicles (EVs) and cholera toxin B exhibit poor co-localization in recipient cells, suggesting that EVs utilize independent caveolin (91).
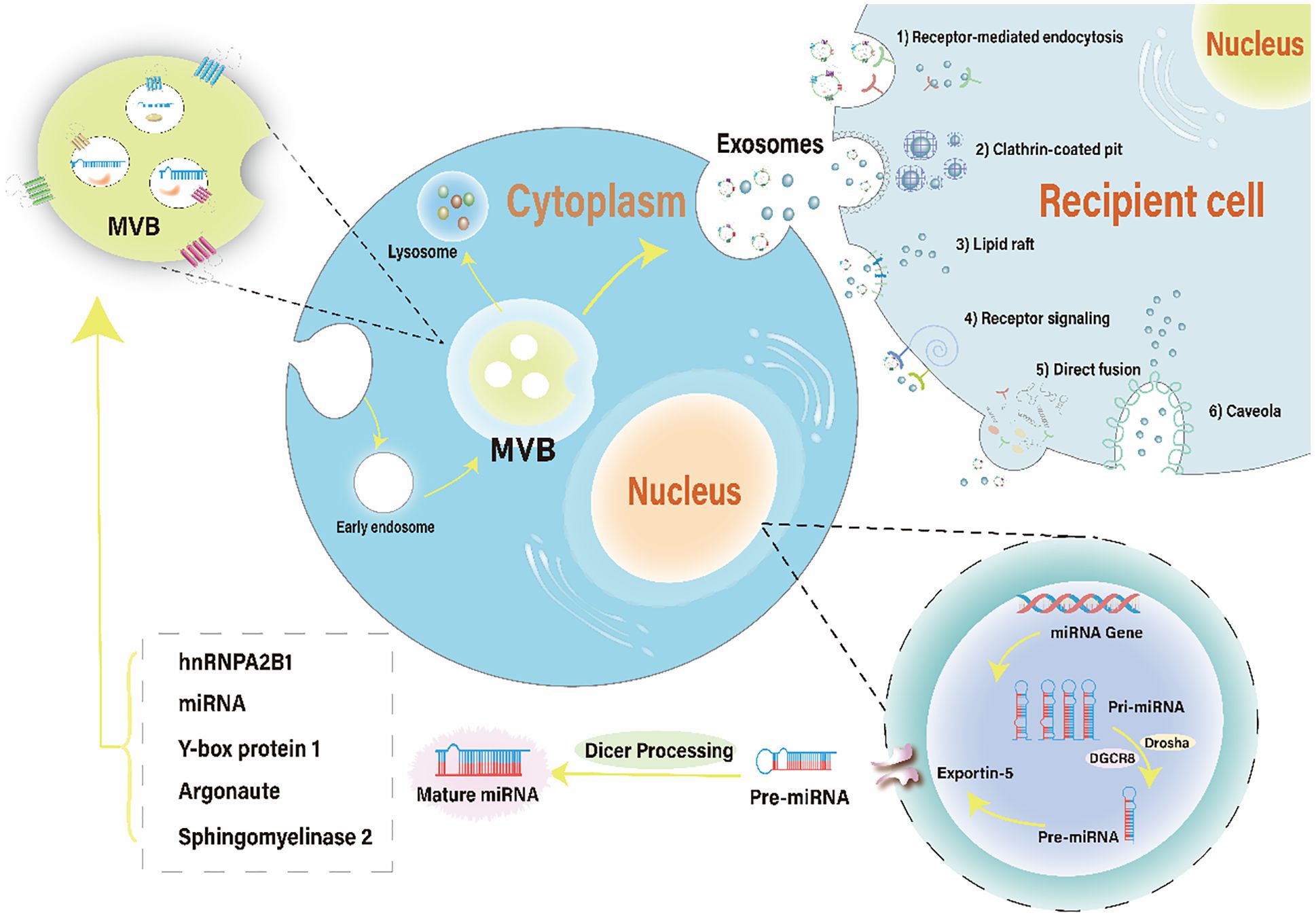
Figure 1 The brief mechanism of exosomal miRNA and uptake by recipient cells. The biogenesis of miRNA involves transcription of a pri-miRNA, formation of pre-miRNA, translocation to the cytoplasm, and maturation of the miRNA. miRNAs containing different RNA motifs can be loaded into MVBs via various RNA-binding proteins. MVBs can either follow a degradation pathway by fusing with lysosomes or release the intraluminal vesicles as exosomes into the extracellular space. Recipient cells can uptake exosomal miRNAs through six pathways: receptor-mediated endocytosis, clathrin-coated pit, lipid raft, receptor signaling, direct fusion, and caveola. MVBs, multivesicular bodies.
3 Pathogenesis of vitiligo
Vitiligo is an autoimmune disease (92), and its exact pathogenesis is not fully understood. Two main biological process have been reported to explain its pathogenesis.
3.1 Autoimmune
The immune system attacks and destroys melanocytes, leading to the development of white patches. A cross-sectional study by Gill L et al. showed that about 20% of vitiligo patients have at least one autoimmune disease in combination throughout the disease progression, suggesting that vitiligo’s occurrence is likely related to immune mechanisms (93). Immunosuppressive agents have shown better results in vitiligo treatment, further suggesting that autoimmunity can influence vitiligo development (94, 95). The immunological mechanism of vitiligo development is closely related to humoral and cellular immunity, especially cellular immunity, where many cytokines have been found to play a direct or indirect regulatory role. In cellular immunology, the main influences on vitiligo development are helper T cells (Th), cytotoxic T cells (CTL), and regulatory T cells (Treg). In the whole process of vitiligo disease, cellular immune function regulation is mainly acted upon by Th1 and Th17 cells, and interleukin-17A (IL-17A) is significantly increased in the cellular expression level of patients’ serum when Th1 and Th17 cells’ levels significantly increase in the peripheral blood (96).
CD8 cytotoxic T cells occupy a crucial position among the whole T lymphocytes and are capable of toxic killing of cells. These cells can specifically target and destroy melanocytes. As their numbers increase, their ability to kill melanocytes intensifies, leading to a greater extent of skin lesions in patients (97, 98). Activated CD8+ T cells secrete various cytokines, such as IFN-γ and TNF-α. These cytokines have multiple effects, including inducing melanocyte apoptosis, inhibiting melanocyte proliferation, and reducing pigment production (99).
Recent studies increasingly suggest that dysfunction of Treg cells significantly contributes to the onset of vitiligo (100, 101). In the lesional skin of vitiligo patients, a reduction in the number of Tregs is often observed. Similarly, flow cytometry analysis of circulating Tregs shows that the number of Tregs in the blood of vitiligo patients is reduced compared to healthy individuals (102–105). Treg cells are a subpopulation of T cells with tolerance to self-antigens, and they help prevent autoimmune diseases. When the number of Treg cells decreases or their function is impaired, they fail to inhibit the proliferation of CD8 and CD4 T cells. The increase in CD8 T cells may destroy melanocytes via the granzyme B or perforin pathways, or by releasing cytokines that promote melanocyte destruction. This ultimately leads to the loss of melanocytes through apoptosis (101, 102). Currently, studies based on single-cell RNA sequencing of human vitiligo patients indicate that CCL5-CCR5 cytokine signaling acts as a connecting axis between effector CD8 T cells and Treg cells (106).
Additionally, the differential ability of fibroblasts in various parts of the patient to respond to gamma interferon determines not only their capacity to recruit CD8+ T cells but also the preferred locations for vitiligo onset. This recruitment mechanism in vitiligo pathogenesis coordinates the accumulation of CD8+ T cells at damaged skin junctions. As a result, these cells can continuously attack melanocytes in healthy areas, causing a progressive expansion of depigmented regions (107). Secretion of interferon-γ (IFN-γ) by natural killer (NK) cells and innate lymphocytes (ILC) induces the expression of C-X-C motif chemokine receptor (CXCR) 3B on the melanocyte surface and the release of C-X-C motif chemokine ligand 9 (CXCL9), C-X-C motif chemokine ligand 10 from keratinocytes and melanocytes (CXCL10), and C-X-C motif chemokine ligand 11 (CXCL11). CXCL10 then activates CXCR3B and triggers apoptosis of melanocytes (108).
3.2 Oxidative stress
Multiple factors and mechanisms have been proposed for the etiopathogenesis of vitiligo, among which oxidative stress has been widely accepted as a key factor in initiating melanocyte loss. Oxidative stress is a reaction process caused by the accumulation of free radicals induced by an altered environment, including inflammation and mitochondrial dysfunction (109). Excessive reactive oxygen species (ROS) have been identified as essential free radicals that exacerbate oxidative stress and aggravate tissue dysfunction (110). However, when ROS production severely exceeds clearance capacity, ROS can accumulate and affect cells, leading to DNA damage, protein misfolding and chromosome instability (111). This theory suggests that increased oxidative stress caused by factors such as sun exposure, hormonal imbalance, or certain chemicals damages melanocytes and leads to vitiligo. The imbalance in redox status caused by oxidative stress, including excessive production of ROS and reduced activity of the antioxidant system in the skin, reduce the resistance of melanocytes to exogenous or endogenous stimuli, ultimately impairing normal defense mechanisms and leading to melanocyte deficiency (112). Low levels of catalase (CAT), glutathione peroxidase (GPx), glucose-6-phosphate dehydrogenase (G6PD), and superoxide dismutase (SOD) have been demonstrated in the epidermis and blood of vitiligo patients (113). These enzymes are closely associated with a state of chronic oxidative stress (OS), which may lead to melanocytes being programmed to become “senescent melanocytes.” They recruit immune cells to kill senescent cells by expressing various cytokines (114). In addition, excessive accumulation of reactive oxygen species (ROS) induces the production of damage-associated molecular patterns (DAMPs) and the release of melanosomal antigens that activate innate immunity (98).
There is mounting evidence revealing an association between oxidative stress and autoimmunity (115–117). ROS can activate immune macrophages and dendritic cells, leading to these cells releasing more cytokines (such as TNF-α, IL-6, IL-1β), thereby affecting melanogenesis (114, 118). Autoimmune responses can further exacerbate oxidative stress. For example, when T cells attack melanocytes, they produce more ROS, worsening the local oxidative stress environment (119).
Additionally, MiRNA plays a crucial role in immune regulation and oxidative stress in vitiligo, contributing to the process of melanocyte death (14). For example, the increased expression of miR-25 promotes melanocyte dysfunction and oxidative stress-induced damage (120). MiR-211 inhibits UVB-induced human melanocyte migration by suppressing MMP9 (matrix metalloproteinase 9) and p53 (121). Figure 2 depicts the abnormally expressed miRNAs involved in melanocyte destruction (adapted from reference (14)).
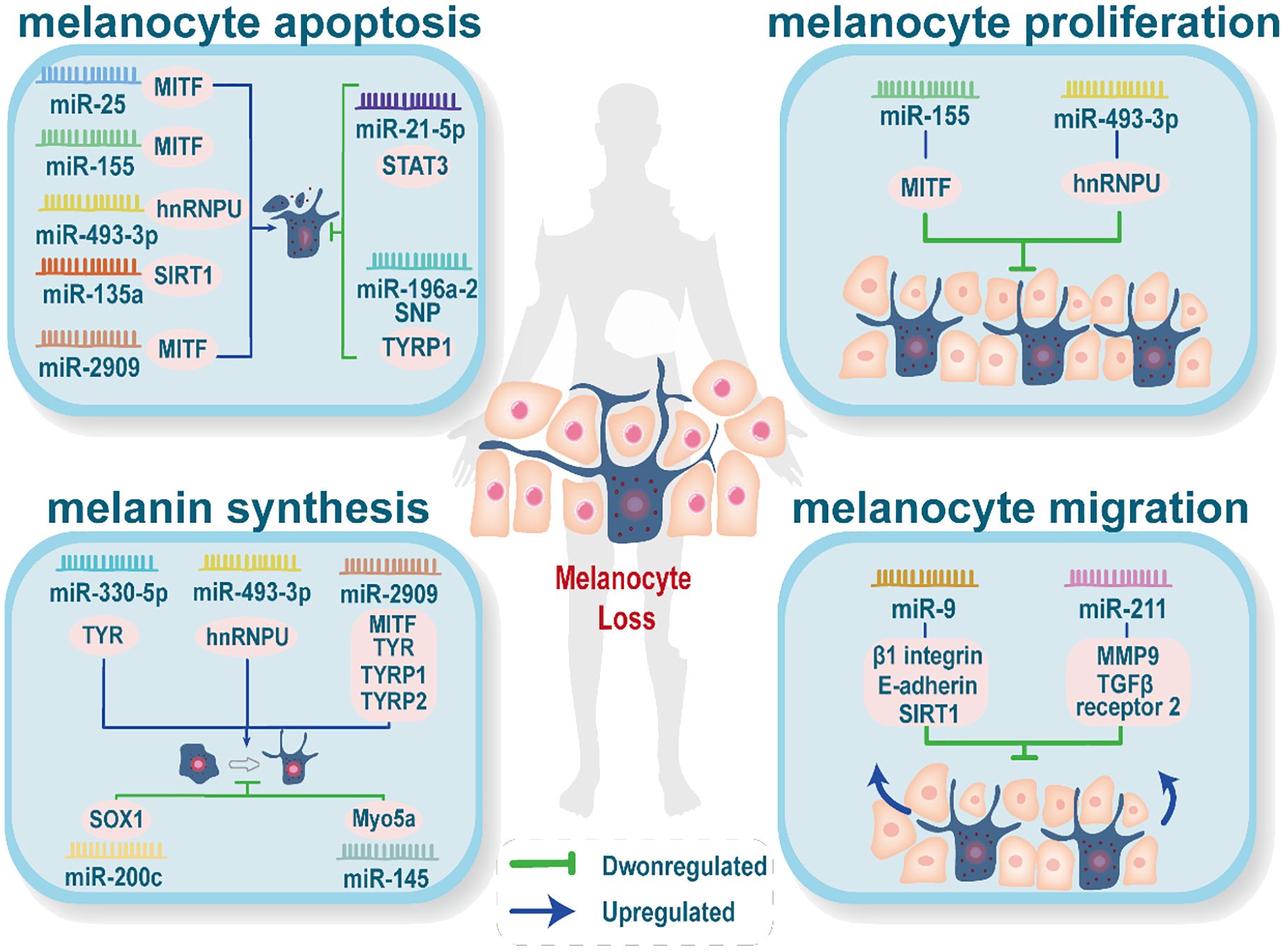
Figure 2 Abnormally expressed miRNAs affect melanin production. Abnormal expression of miRNAs reduces skin melanin through four processes: melanocyte apoptosis, melanocyte proliferation, melanin synthesis, and melanocyte migration. Each process may be influenced by several abnormally expressed miRNAs, and the same abnormally expressed miRNA may be involved in more than one process.
4 Roles of exosomal miRNAs in vitiligo
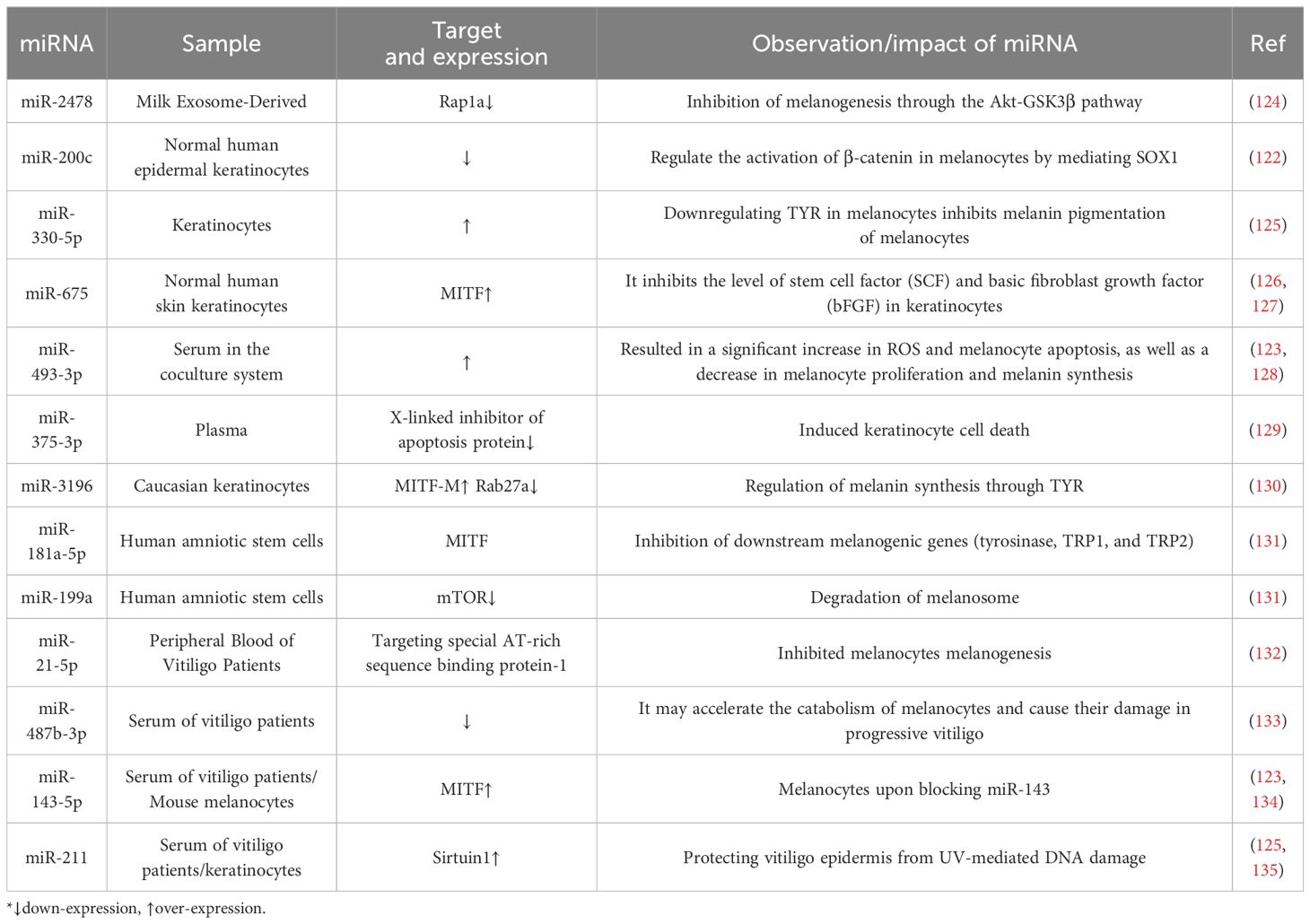
The vital role of miRNAs encapsulated in exosomes in vitiligo is increasingly recognized. High-throughput sequencing of clinical samples has shown that the exosomal miRNA expression profiles differ between patients with vitiligo and healthy individuals, with increased expression of miR-493-3p, miR-370-3p, and miR-143-5p, and decreased expression of miR-885-5p, miR-16-5p, miR-92a-3p, and miR-92b-3p (96). In addition, exosome MiR-200c released from keratinocytes in vitiligo patients was significantly downregulated (122). However, the expression of Exosomal MiR-493-3p was significantly increased in the blood and surrounding tissues of vitiligo patients (123). Table 2 summarizes the reported exosomal miRNAs that may be associated with vitiligo pathogenesis and their potential mechanisms of action.
Exosomal miRNAs play multiple roles in the pathogenesis of vitiligo, influencing the development and progression of the disease through various mechanisms such as regulating immune responses, participating in oxidative stress, and affecting melanogenesis. The following will detail the roles of exosomal miRNAs in the pathogenesis of vitiligo.
4.1 Immune imbalance
The immunomodulatory function of exosomal miRNAs has been widely reported (136, 137). Exosomes of immune cell origin (e.g., T cells, B cells, macrophages, etc.) have been shown to induce suppressive or active immune responses and to participate in immune regulation and immune stimulation (138). Studies have shown that antigen-presenting exosomes secreted by cells such as dendritic cells and T lymphocytes are enriched with co-stimulatory molecules, antigenic peptide-MHC class I, and antigenic peptide class II molecular complexes, which are presented to the corresponding T cells to activate the immune response (139). Researchers have also found that exosomes from normal immune, non-immune, or pathological cell types can influence immune homeostasis. For example, exosomes from dendritic cells express stimulatory molecules of MHC class I and class II T cells in tumor peptide culture, which subsequently activate CD8+ cytotoxic cells and produce cytotoxicity against mouse tumor cells (140). In addition, exosomes may induce the differentiation of type 1 T helper cells into type 2 T helper cells and reduce T cell differentiation into type 2 T helper cells. They also reduce T cell differentiation to interleukin 17-producing effector T cells (Th17 cells) and increase the number of Treg cells and the level of cytotoxic T lymphocyte-associated proteins (141). Thus, exosomal miRNAs, as intercellular mediators, are likely to be involved in the regulation of immune homeostasis.
Vitiligo and immune imbalance are closely related. The immune imbalance in vitiligo involves two main aspects of the adaptive immune response:
1) Role of CD8+ T cells. Activated CD8+ cytotoxic T lymphocytes (CTL) exhibit a strong skin-homing ability and cytotoxic effect; they kill melanocytes by secreting granzyme B and perforin, leading to skin pigmentation deficiency (142). Current studies have found that IFN-γ reduces mRNA levels of TYR, tyrosinase-related protein 1 (TRP1), and microphthalmia-associated transcription factor (MITF), inhibits melanocyte production, and reduces their number around skin lesions. IFN-γ is mainly derived from NK cells, CD4+ T lymphocytes, and activated CD8+ CTL; however, the main source of IFN-γ in vitiligo patients is CD8+ CTL (143, 144). In conclusion, CD8+ T cells not only kill melanocytes directly but also inhibit melanocyte function by secreting IFN-γ.
2) Imbalance between effector T cells. An imbalance occurs between Treg cells (reduced) and CD8+ CTL (significantly elevated) around the skin lesions of vitiligo patients. This imbalance leads to massive secretion of IFN-γ to kill melanocytes (145, 146). Notably, this imbalance is associated with the secretion of pro-inflammatory factors IL-17, IL-6, and TNF-α by differentiated Th17 cells from CD4+ T cells. These cytokines actively regulate inflammatory and autoimmune responses in vivo. Among them, IL-6 acts as a hub between Treg and Th17 cells (147, 148). Currently, miRNA regulation of transcription factor expression, thereby affecting the Treg/Th17 balance, is the most extensively studied aspect of the Treg/Teff balance (149). For example, miR-155 and miR-221-5p target SOCS1, inhibiting the activation of STAT5 (150, 151). SOCS3, a negative regulator of STAT3 activation, is a target of miR-384 and miR-206 (152, 153). In addition to Treg/Th17, the Treg/Th1 and Treg/Th2 balance can also be regulated by various miRNAs. For instance, miR-23a-3p and miR-155 disrupt the Th1/Treg balance by targeting Sirt17 (154, 155).
Exosomes are necessary for the regulation of CD8+ T, and Th17 cells (10). The immunosuppressive effect of exosomal miRNAs on CD8+ T cells has been frequently described in tumor lesions. Exosomes from malignant or immune cells can be taken up by tumor-infiltrating CD8+ T cells and modulate the tumor microenvironment to produce anti-tumor effects (156). Dokyung Jung generated interleukin-2-tethered sEVs (IL2-sEVs) from engineered Jurkat T cells expressing IL2 at the plasma membrane via a flexible linker to induce an autocrine effect. IL2-sEVs increased the anti-cancer ability of CD8+ T cells without affecting Treg and down-regulated cellular and exosomal PD-L1 expression in melanoma cells, causing their increased sensitivity to CD8+ T cell-mediated cytotoxicity. Its effect on CD8+ T and melanoma cells was mediated by several IL2-sEV-resident miRNAs, whose expressions were upregulated by the autocrine effects of IL2. Among the miRNAs, miR-181a-3p and miR-223-3p notably reduced the PD-L1 protein levels in melanoma cells. Interestingly, miR-181a-3p increased the activity of CD8+ T cells while suppressing Treg cell activity (137). In addition, exosomal miRNAs can also induce suppressors of CD8+ T cell activity; tumor-derived exosomes lead to a significant loss of CD27/CD28 and thus induce suppressors of CD8+ T cells (74).
Moreover, exosomes have an important role in the regulation of Treg cells. For example, human umbilical cord MSC-derived exosomes increased the proportion of CD4+CD25+Foxp3+ Treg cells and decreased the proportion of CD4+IL17A+ T cells (Th17 cells) (10). A study showed that MSC-derived exosomes have the capacity to decrease Th17/Treg imbalance in aplastic anemia through sphingosine kinase isoenzyme 1 (SphK1)-mediated exosomal sphingosine 1-phosphate enrichment (S1P) in both in vivo and in vitro experiments (157). In addition, bone marrow-derived mesenchymal stem cell (BMSC)-derived exosome miR-181a is essential for the maintenance of immune tolerance. It induced CD3+CD1+FOXP3+ Treg cells production via SIRT4/acetylation/FOXP3 (158). BMSC-derived exosomes can partially transfer miR-17a-6p to suppress IL-23 expression and thereby balance the Th17/Treg ratio in aplastic anemia (159).
Exosomes secreted by multiple immune cells can act on different types of immune cells and alter their functions (160). First, the unidirectional transfer of T cell exosomal miRNAs to antigen-presenting cells (APCs) plays a genetic regulatory role during immune synapse formation (161). In addition, it has been reported that Treg cells transfer miRNAs to various immune cells, including helper T cell 1 (Th1) cells, via exosomes to inhibit Th1 cell proliferation and cytokine secretion. The mechanism lies in the fact that miRNA-containing exosomes mediate the silencing of Th1 cell genes by Treg cells through non-cell-autonomous gene regulation. Exosomal miRNAs may cooperate with other Treg cell mechanisms to achieve optimal inhibition (162). This may be one of the reasons for the ability of Treg cells to regulate immunity. B cells release more exosomes upon stimulation with CD40 and IL-4 antigens, and these exosomes initiate dendritic cells (DCs) through transfer, which together with CD4+ T cells activate CD8+ T cells to trigger the killing response of CD8+ T cells (139). Immunosuppressive effects of B-cell-derived exosomes have also been reported, inducing apoptosis of CD4+ T cells (163). Classically activated macrophages secrete exosomes that deliver inflammatory signals to immature macrophages and DCs, stimulating cellular inflammation and releasing more Th1 cytokines (164).
Natural killer (NK) cells secrete exosomal miRNAs that also regulate T cell immune responses. miR-10b-5p, miR-92a-3p, and miR-155-5p, found in NK cell exosomes, specifically target molecules involved in the Th1 response, promote CD4+ T cell activation, and produce the cytokines IFN-γ and IL-2 (165). Increasing evidence suggests that NK cell-derived exosomes can induce IFN-γ secretion and increased T-bet expression, with exosomal miRNAs associated with this effect including miR-20a-5p, miR-25-3p, miR-10b-5p, miR-92a-3p, and miR-155. In addition, NK cell-derived exosomes can drive CD4+ T cell activation and may enhance antigen presentation and co-stimulatory abilities by increasing monocyte polarization toward moDCs. They also indirectly affect T cell responses by upregulating MHC-II and CD86 expression (165). Therefore, NK cell-derived exosomes may influence vitiligo development by affecting IFN-γ secretion and CD4+ T cell activation.
4.2 The role of exosomal miRNAs derived from keratinocytes
Skin color is mainly determined by the content and composition of melanin in the epidermis, and the development of vitiligo is closely related to impaired melanin production and transport. Summarizing the available research results, keratinocytes have two roles in skin tone formation. On the one hand, they are melanin-receiving cells. Melanin is synthesized within the melanosomes of melanocytes and transported from its perinuclear region of origin to the cell periphery, where it will be further transferred to the adjacent keratinocytes, forming different skin tones (166). Reportedly, the melanosomes in the keratinocytes in the lesions of patients with stable vitiligo are single and large; however, in contrast, the melanosomes in active vitiligo skin are smaller and less dense (167). Currently, melanin transfer patterns from melanocytes to keratinocytes are described in four main categories:1) Cytophagy. where melanosome-containing melanocytes are phagocytosed by adjacent keratinocytes via dendrites or filamentous pseudopods (168). 2) Membrane fusion, where melanosomes are transferred directly through the membrane tunnel formed between the two cells (169). 3) Shedding and swallowing model, where membrane vesicles containing individual or aggregated melanosomes are released from melanocytes and then phagocytosed by keratin-forming cells (169). 4) Cytosolic-endocytosis. The melanosome membrane fuses with the plasma membrane and secretes naked melanin from the melanocytes (170). In addition, the role of vesicles in this mechanism has been demonstrated, and studies have shown that vesicles containing melanosomes exiting melanocytes are phagocytosed by keratinocytes (168, 171). Transport of melanosomes is also regulated by the small GTPase Rab27a (172), and the role of Rab27 in exosomes has also been reported (173). Thus, melanin transport may also be associated with exosomal miRNAs.
On the other hand, keratinocytes not only passively receive melanosomes transferred from melanocytes but also actively regulate melanocyte function. According to reports, extracellular vesicles released by keratinocytes carry miRNAs that target melanocytes and regulate their state by altering gene expression patterns and enzyme activity (125, 174–178). Studies have shown that exosome protein markers, such as TSG101, Alix, and CD63, were detected in the culture supernatant of keratinocytes (125, 130). Moreover, the expression of miRNAs in exosomes was not affected by RNase intervention in the keratinocyte culture supernatant, as they were encapsulated within the phospholipid bilayer of the exosomes (126). The process of exosomes transferring from keratinocytes to melanocytes has been recently confirmed. The work by Lo Cicero et al. utilized CD63-GFP to track the movement of exosomes in keratinocytes. Initially, CD63-positive regions were detected around the nuclei of keratinocytes, which then moved towards the plasma membrane. During co-culture with melanocytes, it was observed that exosome levels in melanocytes significantly increased after 24 hours of co-culture (130). Furthermore, exosomes derived from keratinocytes carrying miR-330-5p resulted in a significant increase in the expression of miR-330-5p in melanocytes, indicating that nucleic acids from keratinocytes are delivered to melanocytes via exosomes (125).
Furthermore, Hai-Xia Shi et al. studies showed that miRNAs were upregulated in EVs from keratinocytes. KEGG pathway analysis indicated that exosome-derived miRNAs can target one or more genes and regulate one or more signaling pathways that affect the biological activity of melanocytes, such as the MAPK signaling pathway, endocytosis, axon guidance, and neurotrophic factor signaling pathways. In addition, PKH67, a green fluorescent dye was used as a marker to study the internalization process of exosomes. The experimental results showed that scattered green fluorescent particles could be seen in melanocytes co-cultured with exosomes derived from keratinocytes. These findings suggest that exosomes secreted by keratinocytes are transported to melanocytes through certain pathways. Exosomes from keratinocytes not only promote the proliferation and dendritic morphology of melanocytes but also enhance their tyrosinase activity and melanin production (175). Another study investigated the effect of UVB radiation on melanogenesis. After irradiation, the number of exosomes released from keratinocytes increased almost twofold. When using keratinocyte exosome modulators D609 and GW4869, the number of melanocytes in the co-culture system was significantly reduced, indicating that the quantitative changes in keratinocyte exosomes are of significant importance in the regulation of human skin color development (179). Figure 3 summarizes the process of exosome miRNA formation from keratin-forming cells and its uptake by melanocytes, which affects melanin formation.
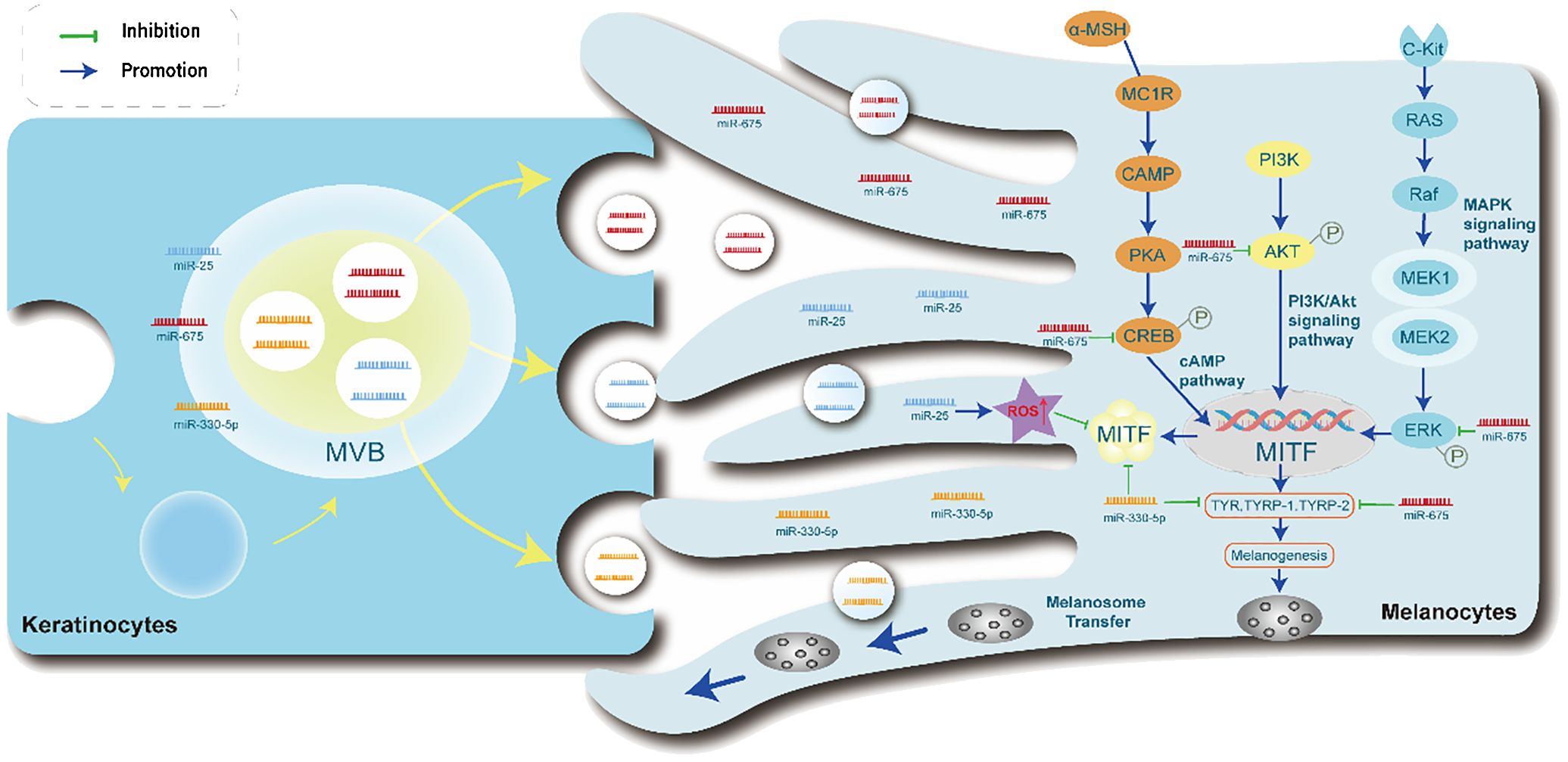
Figure 3 Exosome-derived miRNAs in keratinocytes modulate melanocyte Function. The miRNA-containing exosomes are taken up by melanocytes, followed by the transfer of miRNAs. Furthermore, exosomal miRNAs are involved in the PI3K/Akt, MAPK, and cAMP signaling pathways to regulate the transcription of MITF in melanogenesis. Exosomal miR-675 inhibits the phosphorylation of CREB, ERK, and AKT signaling molecules, subsequently causing the inhibition of TYR, TYRP1, and TYRP2 expression. Exosomal miR-25 promotes ROS increase, suppressing the expression of MITF. Exosomal miR-330-5p directly inhibits the expression of MITF protein, leading to the inhibition of TYR, TYRP1, and TYRP2 expression. PI3K/Akt, phosphatidylinositol 3-kinase/protein kinase B, MAPK, mitogen-activated protein kinase, cAMP, cyclic adenosine monophosphate.
Unfortunately, there have been limited reports on the study of secreted exosomes from melanocytes to keratinocytes. Current research indicates that UVA radiation is a necessary condition for human melanocytes to secrete exosomes. Under UVA irradiation, the exosome protein markers flotillin-1 and CD63 were detected in the conditioned medium of melanocytes (180, 181). It was also determined that exosomes were released from melanocytes and preferentially absorbed by keratinocytes without cell-to-cell contact, enhancing the proliferation of keratinocytes (182). Petra Wäster purified exosomes from UVA-exposed melanocytes and added them to keratinocytes, which resulted in enhanced anti-apoptotic signal transduction in the keratinocytes (183).
4.3 Abnormal melanin production
The influence of exosomes on melanogenesis pathways and their possible role in the pathogenesis of pigment deposition diseases have been proposed, but the mechanisms are not yet fully elucidated. There are four main pathways through which miRNAs regulate melanin production:1) miRNA may cause abnormal proliferation, differentiation, migration, and apoptosis of melanocytes, thereby affecting pigment deposition (14). For example, overexpression of miR-155 reduces the expression of melanogenesis-related genes in melanocytes and keratinocytes (184); MiR-211 inhibits UVB-induced human melanocyte migration by suppressing MMP9 (matrix metalloproteinase 9) and p53 (185). 2) miRNA targets proteins involved in melanogenesis pathways, including SOX5, β-catenin, cyclin-dependent kinase 2, and MITF (186). For example, exosomal miR-21-5p derived from peripheral blood of vitiligo patients inhibits melanogenesis in melanocytes by targeting special AT-rich sequence-binding protein-1 (SATB1) (107); MiRNA-141-3p and miRNA-200a-3p regulate melanogenesis stimulated by α-melanocyte-stimulating hormone through directly targeting MITF (187). 3) miRNA affects pigment deposition by inhibiting melanin transport.MiR-203 reduces melanosomes transport by targeting KIF5B and negatively regulating the CREB1/MITF/Rab27a pathway (188). 4) miRNA may be involved in the destruction of melanosomes. Human amniotic stem cell-derived exosomal miR-181a-5p and miR-199a inhibit melanogenesis in excessive skin pigmentation and promote melanosome degradation (131).
Exosomal miRNAs participate in melanogenesis through MAPK signaling pathways, Ras signaling pathways, Rap1 signaling pathways, cAMP signaling pathways, endocytosis, axon guidance, and Wnt signaling pathways (10). A large number of miRNAs carried by exosomes participate in intercellular communication and selectively target miRNAs in melanocytes to regulate melanin pigmentation by changing gene expression patterns and/or enzyme activity (189). Studies have shown that miR-27a-3p, miR-675, miR-145, miR-25, miR-340, miR-330-5p, miR-211, miR-155, miR-218, miR-21a-5p8, and other miRNAs affect melanogenesis through various mechanisms (175).
4.4 Oxidative stress-induced damage
Oxidative stress is commonly detected in vitiligo and is related to immune response and melanocyte death progression (190). The regulation of endogenous ROS production and its balance with the antioxidant system is dynamic and complex. Under physiological conditions, mild stress such as moderate sun exposure or physical exercise can induce ROS release, activating responses that maintain cellular defense barrier functions. Excessive ROS can cause the death of a small number of melanocytes by causing molecular and organelle dysfunction and exposing melanocyte-specific antigens (14, 191). Recent research has found that miRNAs regulate the response of oxidative stress in vitiligo. MiRNAs may promote the development of vitiligo by regulating the expression and function of oxidative stress-related genes in melanocytes.
Studies have shown that the expression of miR-493-3p in circulating exosomes and perilesional skin of SV patients is significantly increased. Overexpression of miR-493-3p in keratinocytes increases the concentration of dopamine in the culture supernatant, leading to a significant increase in ROS and melanocyte apoptosis, and a decrease in melanocyte proliferation and melanin synthesis in the co-culture system by targeting HNRNPU. The miR-493-3p/HNRNPU/COMT/dopamine axis may contribute to the dysregulation of melanocytes in the pathogenesis of SV (123). Oxidative stress also increases the expression of miR-25 in melanocytes and keratinocytes. Q Shi performed luciferase reporter assays, indicating that miR-25 directly regulates MITF expression by binding to the predicted target sites in the 3′UTR of its mRNAs (120). In studies of miR-25 on melanocytes, overexpression of miR-25 alone does not affect melanocyte apoptosis but can inhibit the antioxidant capacity of melanocytes by suppressing the MITF-apyrimidinic endonuclease (APE1) pathway, making melanocytes more susceptible to the effects of oxidative stress-induced apoptosis (120).
It has been reported that mesenchymal stem cell-derived exosomes can prevent H2O2-induced damage to keratinocytes, improve the antioxidant capacity of keratinocytes, and reduce the cellular response to oxidative stress (192). This provides new ideas and possibilities for the treatment of vitiligo.
5 Conclusions and perspective
Given the significant clinical implications of miRNAs in vitiligo, specifically targeting exosomal miRNAs as an in vitro strategy presents a promising approach for vitiligo treatment. In vitiligo patients, a large number of miRNAs are abnormally expressed, reflecting the activity and histological changes of the disease. These miRNAs are involved in oxidative stress responses (e.g., miR-9, miR-211), immune imbalance(e.g.,miR-21-5p,miR-133b, miR-224-3p), and melanin production (e.g.,miR-330-5p, miR-211,miR-21-5p,miR-200a-3p,hsa-miR-149-5p) (2, 163, 184). These miRNAs can reflect the activity and histological changes of vitiligo, which are considered biomarkers of vitiligo. Therefore, exosomal miRNAs become attractive diagnostic and therapeutic targets.
There are two strategies for the application of miRNAs: 1) Anti-miRNAs. Anti-miRNAs can be used to counteract the over-activation of miRNAs. For example, Short tandem target mimic-miR-143-5p can upregulate the expression of MYO5A, which in turn increases the levels of MITF, promoting melanogenesis (134). 2) miRNA Replacement. This involves reintroducing a gene-suppressor miRNA mimic or using adeno-associated virus -mediated miRNA gain-of-function to modulate gene expression. For instance,miR-211 mimic can alter the migratory capacity of melanocytes through the p53-TRPM1/miR-211-MMP9 axis, enhancing melanin levels (2, 193).
The delivery of miRNA through exosomes has attracted widespread attention. For example, Kuang et al. pointed out that in C57BL/6 mice exposed to cerebral ischemia, natural MSC-derived exosomes (rather than exosomes obtained from MSCs pre-treated with anti-miR-25-3p) regulated oligonucleotide autophagy flux and cell death by modulating the p53-BNIP3 pathway (194). Exosomal miRNAs have several excellent properties for disease treatment. Exosomes have high stability, effectively protecting miRNAs from degradation by endogenous RNases. In addition, they are widely distributed in various body fluids such as milk, urine, blood, and saliva, making them easy to isolate and non-invasively obtain. Moreover, in vitiligo, miRNAs encapsulated in exosomes have been found to exert biological functions by regulating specific aspects such as immune response, oxidative stress, and regulation of keratinocyte and melanocyte function. Importantly, many exosomal miRNAs are involved in more than two of these processes and have good consistency in their mode of action.
Currently, there are 49 clinical trials with different statuses related to miRNAs encapsulated in exosomes registered at https://clinicaltrials.gov/. Of these 49 trials, 12 are completed, 16 are active and recruiting, 5 are active but not recruiting, 2 are not yet recruiting, and 14 are unknown. It is expected that the dawn of the exosomal miRNA era will arrive.
However, there are significant challenges to be addressed when applying exosomal miRNA for disease treatment in clinical settings.
1) Exosomes have low yields, mainly obtained in limited fluids (such as culture media). Traditional exosome isolation methods, like ultracentrifugation, require multiple steps and cause significant loss and damage to exosomes, reducing their quality and purity (195). MSCs have remarkable ex vivo proliferation abilities, making them a key source for therapeutic exosome production (196). Mendt et al. developed a scalable exosome extraction method based on GMP standards, which extracts exosomes from human bone marrow-derived MSCs with yields three times higher than those from human foreskin fibroblasts (197). MSCs are widely found in adipose tissue, human umbilical cords, and bone marrow (198). Adipose tissue can be obtained from the abdominal and gluteal fat of healthy adult donors, and MSCs can be screened and cultured using specific media. Additionally, human embryonic kidney cells(HEK293) are extensively used as exosome donors in various studies due to their high transfection efficiency, ease of culture, and ability to produce large quantities of exosomes (199). Importantly, HEK293 cells can produce proteins most similar to those synthesized by the human body, with low immunogenicity, making them an ideal tool for producing protein or peptide-based drugs (200, 201).
2) In exosome-based clinical research, the practicality, safety, and compliance of exosome-related drugs are crucial issues that need to be addressed. Firstly, the choice of donor cells for exosomes is important; whether to use autologous cells or allogeneic cells with low immunogenicity requires adherence to relevant regulations and ethical review requirements. Secondly, the method of drug loading and release must be determined, optimizing the interaction between exosomes and the drug to ensure the drug’s stability and proper release rate. Thirdly, improving the targeting and tissue localization capabilities of exosomes requires developing surface modification strategies suitable for clinical application, such as using targeting peptides, antibodies, or other molecules to allow exosomes to specifically bind to target cells or tissues.
Given these limitations, future research should focus on these directions. High-yield, efficient, safe, and standardizable production of exosomes for treating vitiligo remains to be explored and developed. New effective miRNA targets for vitiligo treatment can be further investigated, with exosome delivery as a potential approach. Currently, exosome-related therapies are primarily in the preclinical stage, and their safety and efficacy need to be demonstrated. Additionally, regulatory guidelines for exosome-based drugs need to be refined. Furthermore, exosomes modified through genetic and metabolic engineering have shown more effective, stable, and safe immunomodulatory effects in the progression of autoimmune diseases (202). Researchers should continue to explore the potential value of modified exosomes in vitiligo-related therapies, focusing on aspects such as targeted delivery, circulatory stability, and biocompatibility. This will help advance the clinical application of exosomes.
In summary, our review covers the biological functions of exosomes and the involvement of exosomal miRNAs in the pathological processes of vitiligo, providing new insights for the diagnosis and treatment of the disease. Future research should delve deeper into the mechanisms between exosomal miRNAs and vitiligo progression and address the challenges of using exosomes in the clinical treatment of vitiligo.
Author contributions
WL: Writing – original draft. YP: Writing – original draft. QH: Writing – original draft. ZS: Writing – original draft. XX: Writing – original draft. JZ: Writing – review & editing. JG: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (grant nos. 82074443); Chengdu University of Traditional Chinese Medicine “Xinglin Scholars” Discipline Talent Research Improvement Program, Project Number: QJJJ2021001; Chengdu University of Traditional Chinese Medicine “Foundation Thickening” Action Plan (2023-42); Chengdu University of Traditional Chinese Medicine Hospital’s “Hundred Person Plan” (21-L03 and 22-B09). “Young Qihuang” Scholars of the State Administration of Traditional Chinese Medicine (2022-256).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ezzedine K, Eleftheriadou V, Whitton M, vanGeel N. Vitiligo. Lancet. (2015) 386:74–84. doi: 10.1016/S0140-6736(14)60763-7
2. Feng Y, Lu Y. Advances in vitiligo: Update on therapeutic targets. Front Immunol. (2022) 13:986918. doi: 10.3389/fimmu.2022.986918
3. Bergqvist C, Ezzedine K. Vitiligo : A Rreview. Dermatology. (2020) 236:571–92. doi: 10.1159/000506103
4. Zhang R, Wei Y, Wang T, Nie X, Shi Z, Deng Y, et al. Exosomal miRNAs in autoimmune skin diseases. Front Immunol. (2023) 14:1307455. doi: 10.3389/fimmu.2023.1307455
5. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. (2019) 21:9–17. doi: 10.1038/s41556-018-0250-9
6. Xie F, Zhou X, Fang M, Li H, Su P, Tu Y, et al. Extracellular vesicles in cancer immune microenvironment and cancer immunotherapy. Advanced Sci. (2019) 6:1901779. doi: 10.1002/advs.201901779
7. Hussein GM, Mohammed SM, Faris M, Mohammed A, Kadhim MJ, Awadh SA, et al. Find new channel for overcoming chemoresistance in cancers: Role of stem cells-derived exosomal microRNAs. Int J Biol macromolecules. (2022) 219:530–7. doi: 10.1016/j.ijbiomac.2022.07.253
8. Sjoqvist S, Kasai Y, Shimura D, Ishikawa T, Ali N, Iwata T, et al. Oral keratinocyte-derived exosomes regulate proliferation of fibroblasts and epithelial cells. Biochem Biophys Res Commun. (2019) 514:706–12. doi: 10.1016/j.bbrc.2019.04.202
9. Fernández-Messina L, Rodríguez-Galán A, de Yébenes VG, Gutiérrez-Vázquez C, Tenreiro S, Seabra MC, et al. Transfer of extracellular vesicle-microRNA controls germinal center reaction and antibody production. EMBO Rep. (2020) 21:e48925. doi: 10.15252/embr.201948925
10. Wong PM, Yang L, Yang L, Wu H, Li W, Ma X, et al. New insight into the role of exosomes in vitiligo. Autoimmun Rev. (2020) 19:102664. doi: 10.1016/j.autrev.2020.102664
11. Wang W, Yue C, Gao S, Li S, Zhou J, Chen J, et al. Promising roles of exosomal microRNAs in systemic lupus erythematosus. Front Immunol. (2021) 12:757096. doi: 10.3389/fimmu.2021.757096
12. Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J Cell Physiol. (2019) 234:5451–65. doi: 10.1002/jcp.27486
13. Groot M, Lee H. Sorting mechanisms for microRNAs into extracellular vesicles and their associated diseases. Cells. (2020) 9:1044. doi: 10.3390/cells9041044
14. Yu X, Cui Y, Zhu X, Xu H, Li L, Gao G. MicroRNAs: Emerging players in the pathogenesis of vitiligo. Front Cell Dev Biol. (2022) 10:964982. doi: 10.3389/fcell.2022.964982
15. Diener C, Keller A, Meese E. Emerging concepts of miRNA therapeutics: from cells to clinic. Trends genetics: TIG. (2022) 38:613–26. doi: 10.1016/j.tig.2022.02.006
16. Gurunathan S, Kang MH, Jeyaraj M, Qasim M, Kim JH. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. (2019) 8:307. doi: 10.3390/cells8040307
17. Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J extracellular vesicles. (2015) 4:27031. doi: 10.3402/jev.v4.27031
18. Rehman FU, Liu Y, Zheng M, Shi B. Exosomes based strategies for brain drug delivery. Biomaterials. (2023) 293:121949. doi: 10.1016/j.biomaterials.2022.121949
19. Lee ST, Im W, Ban JJ, Lee M, Jung KH, Lee SK, et al. Exosome-based delivery of miR-124 in a Huntington's disease model. J movement Disord. (2017) 10:45–52. doi: 10.14802/jmd.16054
20. Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol therapy: J Am Soc Gene Ther. (2011) 19:1769–79. doi: 10.1038/mt.2011.164
21. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J extracellular vesicles. (2018) 7:1535750. doi: 10.1080/20013078.2018.1535750
22. Ren X, Zhao Y, Xue F, Zheng Y, Huang H, Wang W, et al. Exosomal DNA aptamer targeting α-synuclein aggregates reduced neuropathological deficits in a mouse Parkinson's disease model, molecular therapy. Nucleic Acids. (2019) 17:726–40. doi: 10.1016/j.omtn.2019.07.008
23. Izco M, Blesa J, Schleef M, Schmeer M, Porcari R, Al-Shawi R, et al. Systemic exosomal delivery of shRNA minicircles prevents parkinsonian pathology. Mol therapy: J Am Soc Gene Ther. (2019) 27:2111–22. doi: 10.1016/j.ymthe.2019.08.010
24. Liu H, Tian Y, Xue C, Niu Q, Chen C, Yan X. Analysis of extracellular vesicle DNA at the single-vesicle level by nano-flow cytometry. J extracellular vesicles. (2022) 11:e12206. doi: 10.1002/jev2.12206
25. Salomon C, Das S, Erdbrügger U, Kalluri R, Kiang Lim S, Olefsky JM, et al. Extracellular vesicles and their emerging roles as cellular messengers in endocrinology: an endocrine society scientific statement. Endocrine Rev. (2022) 43:441–68. doi: 10.1210/endrev/bnac009
26. Hade MD, Suire CN, Mossell J, Suo Z. Extracellular vesicles: Emerging frontiers in wound healing. Medicinal Res Rev. (2022) 42:2102–25. doi: 10.1002/med.21918
27. Singh A, Das K, Banerjee S, Sen P. Elucidation of the signalling pathways for enhanced exosome release from Mycobacterium-infected macrophages and subsequent induction of differentiation. Immunology. (2023) 168:63–82. doi: 10.1111/imm.13561
28. Pitzer CR, Paez HG, Alway SE. The contribution of tumor derived exosomes to cancer cachexia. Cells. (2023) 12:292. doi: 10.3390/cells12020292
29. Martínez-Greene JA, Hernández-Ortega K, Quiroz-Baez R, Resendis-Antonio O, Pichardo-Casas I, Sinclair DA, et al. Quantitative proteomic analysis of extracellular vesicle subgroups isolated by an optimized method combining polymer-based precipitation and size exclusion chromatography. J extracellular vesicles. (2021) 10:e12087. doi: 10.1002/jev2.12087
30. Wang M, Jiang Y, Fan ZL, Zhang N, Liang J, Zheng Z. Exosomal MEF2C's association with the progression of CRC in regulating CD36 transcription. Altern therapies Health Med. (2023) 29:198–209.
31. H Rashed M, Bayraktar E, K Helal G, Abd-Ellah MF, Amero P, Chavez-Reyes A, et al. Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci. (2017) 18:538. doi: 10.3390/ijms18030538
32. Song F, Wang C, Wang C, Wang J, Wu Y, Wang Y, et al. Multi-phenotypic exosome secretion profiling microfluidic platform for exploring single-cell heterogeneity. Small Methods. (2022) 6:e2200717. doi: 10.1002/smtd.202200717
33. Salvermoser L, Flisikowski K, Dressel-Böhm S, Nytko KJ, Rohrer Bley C, Schnieke A, et al. Elevated circulating Hsp70 levels are correlative for Malignancies in different mammalian species. Cell Stress chaperones. (2023) 28:105–18. doi: 10.1007/s12192-022-01311-y
34. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. (2020) 182:1044–1061.e18. doi: 10.1016/j.cell.2020.07.009
35. Xu D, Song M, Chai C, Wang J, Jin C, Wang X, et al. Exosome-encapsulated miR-6089 regulates inflammatory response via targeting TLR4. J Cell Physiol. (2019) 234:1502–11. doi: 10.1002/jcp.27014
36. Zeng X, Zhang YD, Ma RY, Chen YJ, Xiang XM, Hou DY, et al. Activated Drp1 regulates p62-mediated autophagic flux and aggravates inflammation in cerebral ischemia-reperfusion via the ROS-RIP1/RIP3-exosome axis. Military Med Res. (2022) 9:25. doi: 10.1186/s40779-022-00383-2
37. Davies OG, Williams S, Goldie K. The therapeutic and commercial landscape of stem cell vesicles in regenerative dermatology. J Controlled release: Off J Controlled Release Soc. (2023) 353:1096–106. doi: 10.1016/j.jconrel.2022.12.025
38. Li L, Mu J, Zhang Y, Zhang C, Ma T, Chen L, et al. Stimulation by exosomes from hypoxia preconditioned human umbilical vein endothelial cells facilitates mesenchymal stem cells angiogenic function for spinal cord repair. ACS nano. (2022) 16:10811–23. doi: 10.1021/acsnano.2c02898
39. Zeng B, Chen Y, Chen H, Zhao Q, Sun Z, Liu D, et al. Exosomal miR-211-5p regulates glucose metabolism, pyroptosis, and immune microenvironment of melanoma through GNA15. Pharmacol Res. (2023) 188:106660. doi: 10.1016/j.phrs.2023.106660
40. Mahmoudi F, Hanachi P. Extracellular vesicles of immune cells; immunomodulatory impacts and therapeutic potentials. Clin Immunol. (2023) 248:109237. doi: 10.1016/j.clim.2023.109237
41. Moloudizargari M, Asghari MH, Jørgensen MM, Reiter RJ, Kabelitz D. Editorial: Extracellular vesicles in cancer immunosurveillance. Front Immunol. (2022) 13:993967. doi: 10.3389/fimmu.2022.993967
42. Mirzaei R, Zamani F, Hajibaba M, Rasouli-Saravani A, Noroozbeygi M, Gorgani M, et al. The pathogenic, therapeutic and diagnostic role of exosomal microRNA in the autoimmune diseases. J neuroimmunology. (2021) 358:577640. doi: 10.1016/j.jneuroim.2021.577640
43. Guan L, Wu B, Li T, Beer LA, Sharma G, Li M, et al. HRS phosphorylation drives immunosuppressive exosome secretion and restricts CD8(+) T-cell infiltration into tumors. Nat Commun. (2022) 13:4078. doi: 10.1038/s41467-022-31713-6
44. Li X, Guo X, Huang J, Lin Q, Qin B, Jiang M, et al. Recruiting T cells and sensitizing tumors to NKG2D immune surveillance for robust antitumor immune response. J Controlled release: Off J Controlled Release Soc. (2023) 353:943–55. doi: 10.1016/j.jconrel.2022.12.032
45. Miao C, Wang X, Zhou W, Huang J. The emerging roles of exosomes in autoimmune diseases, with special emphasis on microRNAs in exosomes. Pharmacol Res. (2021) 169:105680. doi: 10.1016/j.phrs.2021.105680
46. Yu H, Feng H, Zeng H, Wu Y, Zhang Q, Yu J, et al. Exosomes: The emerging mechanisms and potential clinical applications in dermatology. Int J Biol Sci. (2024) 20:1778–95. doi: 10.7150/ijbs.92897
47. Perez-Hernandez J, Martinez-Arroyo O, Ortega A, Galera M, Solis-Salguero MA, Chaves FJ, et al. Urinary exosomal miR-146a as a marker of albuminuria, activity changes and disease flares in lupus nephritis. J Nephrol. (2021) 34:1157–67. doi: 10.1007/s40620-020-00832-y
48. Vázquez-Mera S, Martelo-Vidal L, Miguéns-Suárez P, Saavedra-Nieves P, Arias P, González-Fernández C, et al. Serum exosome inflamma-miRs are surrogate biomarkers for asthma phenotype and severity. Allergy. (2023) 78:141–55. doi: 10.1111/all.15480
49. Wermuth PJ, Piera-Velazquez S, Jimenez SA. Exosomes isolated from serum of systemic sclerosis patients display alterations in their content of profibrotic and antifibrotic microRNA and induce a profibrotic phenotype in cultured normal dermal fibroblasts. Clin Exp Rheumatol. (2017) 35 Suppl 106:21–30.
50. Jafari N, Llevenes P, Denis GV. Exosomes as novel biomarkers in metabolic disease and obesity-related cancers, Nature reviews. Endocrinology. (2022) 18:327–8. doi: 10.1038/s41574-022-00666-7
51. Sun JX, Xu JZ, An Y, Ma SY, Liu CQ, Zhang SH, et al. Future in precise surgery: Fluorescence-guided surgery using EVs derived fluorescence contrast agent. J Controlled release: Off J Controlled Release Soc. (2023) 353:832–41. doi: 10.1016/j.jconrel.2022.12.013
52. Li K, Lin Y, Luo Y, Xiong X, Wang L, Durante K, et al. A signature of saliva-derived exosomal small RNAs as predicting biomarker for esophageal carcinoma: a multicenter prospective study. Mol Cancer. (2022) 21:21. doi: 10.1186/s12943-022-01499-8
53. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. (2018) 560:382–6. doi: 10.1038/s41586-018-0392-8
54. Daassi D, Mahoney KM, Freeman GJ. The importance of exosomal PDL1 in tumour immune evasion, Nature reviews. Immunology. (2020) 20:209–15. doi: 10.1038/s41577-019-0264-y
55. Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles, Nature reviews. Immunology. (2014) 14:195–208. doi: 10.1038/nri3622
56. Serra P, Santamaria P. Antigen-specific therapeutic approaches for autoimmunity. Nat Biotechnol. (2019) 37:238–51. doi: 10.1038/s41587-019-0015-4
57. Takahashi A, Okada R, Nagao K, Kawamata Y, Hanyu A, Yoshimoto S, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun. (2017) 8:15287. doi: 10.1038/ncomms15287
58. Steinbichler TB, Dudás J, Skvortsov S, Ganswindt U, Riechelmann H, Skvortsova II. Therapy resistance mediated by exosomes. Mol Cancer. (2019) 18:58. doi: 10.1186/s12943-019-0970-x
59. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. (2007) 9:654–9. doi: 10.1038/ncb1596
60. Hessvik NP, Llorente A. Current knowledge on exosome biogenesis and release. Cell Mol Life sciences: CMLS. (2018) 75:193–208. doi: 10.1007/s00018-017-2595-9
61. Hazrati A, Soudi S, Malekpour K, Mahmoudi M, Rahimi A, Hashemi SM, et al. Immune cells-derived exosomes function as a double-edged sword: role in disease progression and their therapeutic applications. biomark Res. (2022) 10:30. doi: 10.1186/s40364-022-00374-4
62. Liu J, Peng X, Yang S, Li X, Huang M, Wei S, et al. Extracellular vesicle PD-L1 in reshaping tumor immune microenvironment: biological function and potential therapy strategies. Cell communication signaling: CCS. (2022) 20:14. doi: 10.1186/s12964-021-00816-w
63. Skokos D, Le Panse S, Villa I, Rousselle JC, Peronet R, David B, et al. and T lymphocyte activation is mediated by the secretion of immunologically active exosomes. J Immunol (Baltimore Md.: 1950). (2001) 166:868–76. doi: 10.4049/jimmunol.166.2.868
64. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. (2009) 136:642–55. doi: 10.1016/j.cell.2009.01.035
65. Tomari Y, Zamore PD. MicroRNA biogenesis: drosha can't cut it without a partner. Curr biology: CB. (2005) 15:R61–4. doi: 10.1016/j.cub.2004.12.057
66. Du S, Ling H, Guo Z, Cao Q, Song C. Roles of exosomal miRNA in vascular aging. Pharmacol Res. (2021) 165:105278. doi: 10.1016/j.phrs.2020.105278
67. Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. (2019) 30:656–73. doi: 10.1016/j.cmet.2019.07.011
68. Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G. Trends in the development of miRNA bioinformatics tools. Briefings Bioinf. (2019) 20:1836–52. doi: 10.1093/bib/bby054
69. Dragomir MP, Knutsen E, Calin GA. Classical and noncanonical functions of miRNAs in cancers. Trends genetics: TIG. (2022) 38:379–94. doi: 10.1016/j.tig.2021.10.002
70. Walz AL, Ooms A, Gadd S, Gerhard DS, Smith MA, Guidry Auvil JM, et al. Recurrent DGCR8, DROSHA, and SIX homeodomain mutations in favorable histology Wilms tumors. Cancer Cell. (2015) 27:286–97. doi: 10.1016/j.ccell.2015.01.003
71. Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. (2017) 542:450–5. doi: 10.1038/nature21365
72. Abels ER, Maas SLN, Nieland L, Wei Z, Cheah PS, Tai E, et al. Glioblastoma-associated microglia reprogramming is mediated by functional transfer of extracellular miR-21. Cell Rep. (2019) 28:3105–3119.e7. doi: 10.1016/j.celrep.2019.08.036
73. Crewe C, Joffin N, Rutkowski JM, Kim M, Zhang F, Towler DA, et al. An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell. (2018) 175:695–708.e13. doi: 10.1016/j.cell.2018.09.005
74. Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J extracellular vesicles. (2014) 3. doi: 10.3402/jev.v3.23743
75. Xin W, Qin Y, Lei P, Zhang J, Yang X, Wang Z. From cerebral ischemia towards myocardial, renal, and hepatic ischemia: Exosomal miRNAs as a general concept of intercellular communication in ischemia-reperfusion injury, Molecular therapy. Nucleic Acids. (2022) 29:900–22. doi: 10.1016/j.omtn.2022.08.032
76. O'Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications, Nature reviews. Mol Cell Biol. (2020) 21:585–606. doi: 10.1038/s41580-020-0251-y
77. Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MAJ, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. (2014) 8:1649–58. doi: 10.1016/j.celrep.2014.08.027
78. Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. (2013) 288:10849–59. doi: 10.1074/jbc.M112.446831
79. Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. (2016) 17:799–808. doi: 10.1016/j.celrep.2016.09.031
80. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. (2013) 4:2980. doi: 10.1038/ncomms3980
81. Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. -box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. (2016) 5:e19276. doi: 10.7554/eLife.19276
82. Garcia-Martin R, Wang G, Brandão BB, Zanotto TM, Shah S, Kumar Patel S, et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature. (2022) 601:446–51. doi: 10.1038/s41586-021-04234-3
83. Temoche-Diaz MM, Shurtleff MJ, Nottingham RM, Yao J, Fadadu RP, Lambowitz AM, et al. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. eLife. (2019) 8:e47544. doi: 10.7554/eLife.47544
84. Banizs AB, Huang T, Nakamoto RK, Shi W, He J. Endocytosis pathways of endothelial cell derived exosomes. Mol pharmaceutics. (2018) 15:5585–90. doi: 10.1021/acs.molpharmaceut.8b00765
85. Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J extracellular vesicles. (2014) 3:3. doi: 10.3402/jev.v3.24641
86. Smith SM, Larocque G, Wood KM, Morris KL, Roseman AM, Sessions RB, et al. Multi-modal adaptor-clathrin contacts drive coated vesicle assembly. EMBO J. (2021) 40:e108795. doi: 10.15252/embj.2021108795
87. Fan Q, Yang L, Zhang X, Peng X, Wei S, Su D, et al. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. (2018) 414:107–15. doi: 10.1016/j.canlet.2017.10.040
88. Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, et al. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer. (2022) 21:207. doi: 10.1186/s12943-022-01671-0
89. Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. (2017) 27:172–88. doi: 10.1016/j.tcb.2016.11.003
90. Le Saux S, Aarrass H, Lai-Kee-Him J, Bron P, Armengaud J, Miotello G, et al. Post-production modifications of murine mesenchymal stem cell (mMSC) derived extracellular vesicles (EVs) and impact on their cellular interaction. Biomaterials. (2020) 231:119675. doi: 10.1016/j.biomaterials.2019.119675
91. Boudna M, Campos AD, Vychytilova-Faltejskova P, Machackova T, Slaby O, Souckova K. Strategies for labelling of exogenous and endogenous extracellular vesicles and their application for in vitro and in vivo functional studies. Cell communication signaling: CCS. (2024) 22:171. doi: 10.1186/s12964-024-01548-3
92. Rodrigues M, Ezzedine K, Hamzavi I, Pandya AG, Harris JE. New discoveries in the pathogenesis and classification of vitiligo. J Am Acad Dermatol. (2017) 77:1–13. doi: 10.1016/j.jaad.2016.10.048
93. Gill L, Zarbo A, Isedeh P, Jacobsen G, Lim HW, Hamzavi I. Comorbid autoimmune diseases in patients with vitiligo: A cross-sectional study. J Am Acad Dermatol. (2016) 74:295–302. doi: 10.1016/j.jaad.2015.08.063
94. Karagaiah P, Valle Y, Sigova J, Zerbinati N, Vojvodic P, Parsad D, et al. Emerging drugs for the treatment of vitiligo. Expert Opin emerging Drugs. (2020) 25:7–24. doi: 10.1080/14728214.2020.1712358
95. Pichler R, Sfetsos K, Badics B, Gutenbrunner S, Berg J, Auböck J. Lymphocyte imbalance in vitiligo patients indicated by elevated CD4+/CD8+ T-cell ratio. Wiener medizinische Wochenschrift (1946). (2009) 159:337–41. doi: 10.1007/s10354-009-0699-z
96. Riding RL, Harris JE. The role of memory CD8(+) T cells in vitiligo. J Immunol. (2019) 203:11–9. doi: 10.4049/jimmunol.1900027
97. Chang WL, Lee WR, Kuo YC, Huang YH. Vitiligo: an autoimmune skin disease and its immunomodulatory therapeutic intervention. Front Cell Dev Biol. (2021) 9:797026. doi: 10.3389/fcell.2021.797026
98. Richmond JM, Frisoli ML, Harris JE. Innate immune mechanisms in vitiligo: danger from within. Curr Opin Immunol. (2013) 25:676–82. doi: 10.1016/j.coi.2013.10.010
99. Chen J, Li S, Li C. Mechanisms of melanocyte death in vitiligo. Medicinal Res Rev. (2021) 41:1138–66. doi: 10.1002/med.21754
100. Das D, Akhtar S, Kurra S, Gupta S, Sharma A. Emerging role of immune cell network in autoimmune skin disorders: An update on pemphigus, vitiligo and psoriasis. Cytokine Growth factor Rev. (2019) 45:35–44. doi: 10.1016/j.cytogfr.2019.01.001
101. Zhang J, Hu W, Wang P, Ding Y, Wang H, Kang X. Research progress on targeted antioxidant therapy and vitiligo. Oxid Med Cell Longev. (2022) 2022:1821780. doi: 10.1155/2022/1821780
102. Lili Y, Yi W, Ji Y, Yue S, Weimin S, Ming L. Global activation of CD8+ cytotoxic T lymphocytes correlates with an impairment in regulatory T cells in patients with generalized vitiligo. PloS One. (2012) 7:e37513. doi: 10.1371/journal.pone.0037513
103. Chatterjee S, Eby JM, Al-Khami AA, Soloshchenko M, Kang HK, Kaur N, et al. A quantitative increase in regulatory T cells controls development of vitiligo. J Invest Dermatol. (2014) 134:1285–94. doi: 10.1038/jid.2013.540
104. Dwivedi M, Laddha NC, Arora P, Marfatia YS, Begum R. Decreased regulatory T-cells and CD4(+) /CD8(+) ratio correlate with disease onset and progression in patients with generalized vitiligo. Pigment Cell melanoma Res. (2013) 26:586–91. doi: 10.1111/pcmr.12105
105. Oparaugo NC, Ouyang K, Nguyen NPN, Nelson AM, Agak GW. Human regulatory T cells: understanding the role of tregs in select autoimmune skin diseases and post-transplant nonmelanoma skin cancers. Int J Mol Sci. (2023) 24:1527. doi: 10.3390/ijms24021527
106. Gellatly KJ, Strassner JP, Essien K, Refat MA, Murphy RL, Coffin-Schmitt A, et al. scRNA-seq of human vitiligo reveals complex networks of subclinical immune activation and a role for CCR5 in T(reg) function. Sci Trans Med. (2021) 13:eabd8995. doi: 10.1126/scitranslmed.abd8995
107. Xu Z, Chen D, Hu Y, Jiang K, Huang H, Du Y, et al. Anatomically distinct fibroblast subsets determine skin autoimmune patterns. Nature. (2022) 601:118–24. doi: 10.1038/s41586-021-04221-8
108. Tulic MK, Cavazza E, Cheli Y, Jacquel A, Luci C, Cardot-Leccia N, et al. Innate lymphocyte-induced CXCR3B-mediated melanocyte apoptosis is a potential initiator of T-cell autoreactivity in vitiligo. Nat Commun. (2019) 10:2178. doi: 10.1038/s41467-019-09963-8
109. Abdul-Muneer PM, Chandra N, Haorah J. Interactions of oxidative stress and neurovascular inflammation in the pathogenesis of traumatic brain injury. Mol Neurobiol. (2015) 51:966–79. doi: 10.1007/s12035-014-8752-3
110. Rastogi R, Geng X, Li F, Ding Y. NOX activation by subunit interaction and underlying mechanisms in disease. Front Cell Neurosci. (2016) 10:301. doi: 10.3389/fncel.2016.00301
111. Wang X, Zhou Y, Gao Q, Ping D, Wang Y, Wu W, et al. The role of exosomal microRNAs and oxidative stress in neurodegenerative diseases. Oxid Med Cell Longev. (2020) 2020:3232869. doi: 10.1155/2020/3232869
112. He S, Xu J, Wu J. The promising role of chemokines in vitiligo: from oxidative stress to the autoimmune response. Oxid Med Cell Longev. (2022) 2022:8796735. doi: 10.1155/2022/8796735
113. Lyu C, Sun Y. Immunometabolism in the pathogenesis of vitiligo. Front Immunol. (2022) 13:1055958. doi: 10.3389/fimmu.2022.1055958
114. Ohtani N. Deciphering the mechanism for induction of senescence-associated secretory phenotype (SASP) and its role in aging and cancer development. J Biochem. (2019). 166:289–5. doi: 10.1093/jb/mvz055
115. Ibáñez-Cabellos JS, Pallardó FV, García-Giménez JL, Seco-Cervera M. Oxidative stress and epigenetics: miRNA involvement in rare autoimmune diseases. Antioxidants (Basel Switzerland). (2023) 12:800. doi: 10.3390/antiox12040800
116. Wójcik P, Gęgotek A, Žarković N, Skrzydlewska E. Oxidative stress and lipid mediators modulate immune cell functions in autoimmune diseases. Int J Mol Sci. (2021) 22:723. doi: 10.3390/ijms22020723
117. Wigerblad G, Kaplan MJ. Neutrophil extracellular traps in systemic autoimmune and autoinflammatory diseases, Nature reviews. Immunology. (2023) 23:274–88. doi: 10.1038/s41577-022-00787-0
118. Nathan C, Ding A. Nonresolving inflammation. Cell. (2010) 140:871–82. doi: 10.1016/j.cell.2010.02.029
119. Vallée A, Lecarpentier Y. TGF-β in fibrosis by acting as a conductor for contractile properties of myofibroblasts. Cell bioscience. (2019) 9:98. doi: 10.1186/s13578-019-0362-3
120. Shi Q, Zhang W, Guo S, Jian Z, Li S, Li K, et al. Oxidative stress-induced overexpression of miR-25: the mechanism underlying the degeneration of melanocytes in vitiligo. Cell Death differentiation. (2016) 23:496–508. doi: 10.1038/cdd.2015.117
121. Sahoo A, Lee B, Boniface K, Seneschal J, Sahoo SK, Seki T, et al. MicroRNA-211 regulates oxidative phosphorylation and energy metabolism in human vitiligo. J Invest Dermatol. (2017) 137:1965–74. doi: 10.1016/j.jid.2017.04.025
122. Zhao C, Wang D, Wang X, Mao Y, Xu Z, Sun Y, et al. Down-regulation of exosomal miR-200c derived from keratinocytes in vitiligo lesions suppresses melanogenesis. J Cell Mol Med. (2020) 24:12164–75. doi: 10.1111/jcmm.15864
123. Li D, Zhou T, She Q, Nie X, Liu Z, Pan R, et al. Circulating exosomal miR-493-3p affects melanocyte survival and function by regulating epidermal dopamine concentration in segmental vitiligo. J Invest Dermatol. (2022) 142:3262–3273.e11. doi: 10.1016/j.jid.2022.05.1086
124. Bae IS, Kim SH. Milk exosome-derived microRNA-2478 suppresses melanogenesis through the akt-GSK3β Pathway. Cells. (2021) 10:2848. doi: 10.3390/cells10112848
125. Liu Y, Xue L, Gao H, Chang L, Yu X, Zhu Z, et al. Exosomal miRNA derived from keratinocytes regulates pigmentation in melanocytes. J Dermatol Sci. (2019) 93:159–67. doi: 10.1016/j.jdermsci.2019.02.001
126. Kim NH, Choi SH, Kim CH, Lee CH, Lee TR, Lee AY. Reduced MiR-675 in exosome in H19 RNA-related melanogenesis via MITF as a direct target. J Invest Dermatol. (2014) 134:1075–82. doi: 10.1038/jid.2013.478
127. Jin S, Chen L, Xu Z, Xing X, Zhang C, Xiang L. 585 nm light-emitting diodes inhibit melanogenesis through upregulating H19/miR-675 axis in LEDs-irradiated keratinocytes by paracrine effect. J Dermatol Sci. (2020) 98:102–8. doi: 10.1016/j.jdermsci.2020.03.002
128. Zhu Z, Chen B, Peng L, Gao S, Guo J, Zhu X. Blockade of LINC01605-enriched exosome generation in M2 macrophages impairs M2 macrophage-induced proliferation, migration, and invasion of human dermal fibroblasts. Int J immunopathology Pharmacol. (2021) 35:20587384211016724. doi: 10.1177/20587384211016724
129. Zhang C, Zhu Z, Gao J, Yang L, Dang E, Fang H, et al. Plasma exosomal miR-375-3p regulates mitochondria-dependent keratinocyte apoptosis by targeting XIAP in severe drug-induced skin reactions. Sci Trans Med. (2020) 12:eaaw6142. doi: 10.1126/scitranslmed.aaw6142
130. Lo Cicero A, Delevoye C, Gilles-Marsens F, Loew D, Dingli F, Guéré C, et al. Exosomes released by keratinocytes modulate melanocyte pigmentation. Nat Commun. (2015) 6:7506. doi: 10.1038/ncomms8506
131. Wang XY, Guan XH, Yu ZP, Wu J, Huang QM, Deng KY, et al. Human amniotic stem cells-derived exosmal miR-181a-5p and miR-199a inhibit melanogenesis and promote melanosome degradation in skin hyperpigmentation. respectively Stem Cell Res Ther. (2021) 12:501. doi: 10.1186/s13287-021-02570-9
132. Zhang C, Guo W, Wang S, Di Y, Wu D. Peripheral blood of vitiligo patients-derived exosomal miR-21-5p inhibits melanocytes melanogenesis via targeting SATB1. Iranian J Public Health. (2022) 51:2706–16. doi: 10.18502/ijph.v51i12.11461
133. Luo H, Xie B, Xu J, Zhu Y, Sun J, Shen Y, et al. Differential expression of serum exosomal hsa-miR-487b-3p in progressive vitiligo before and after systemic corticosteroid treatment. Clinical cosmetic investigational Dermatol. (2022) 15:1377–86. doi: 10.2147/CCID.S372112
134. Qi S, Liu B, Zhang J, Liu X, Dong C, Fan R. Knockdown of microRNA−143−5p by STTM technology affects eumelanin and pheomelanin production in melanocytes. Mol Med Rep. (2019) 20:2649–56. doi: 10.3892/mmr.2019.10492
135. Brahmbhatt HD, Gupta R, Gupta A, Rastogi S, Misri R, Mobeen A, et al. The long noncoding RNA MALAT1 suppresses miR-211 to confer protection from ultraviolet-mediated DNA damage in vitiligo epidermis by upregulating sirtuin 1. Br J Dermatol. (2021) 184:1132–42. doi: 10.1111/bjd.19666
136. Rezaie J, Etemadi T, Feghhi M. The distinct roles of exosomes in innate immune responses and therapeutic applications in cancer. Eur J Pharmacol. (2022) 933:175292. doi: 10.1016/j.ejphar.2022.175292
137. Jung D, Shin S, Kang SM, Jung I, Ryu S, Noh S, et al. Reprogramming of T cell-derived small extracellular vesicles using IL2 surface engineering induces potent anti-cancer effects through miRNA delivery. J extracellular vesicles. (2022) 11:e12287. doi: 10.1002/jev2.12287
138. Agarwal A, Fanelli G, Letizia M, Tung SL, Boardman D, Lechler R, et al. Regulatory T cell-derived exosomes: possible therapeutic and diagnostic tools in transplantation. Front Immunol. (2014) 5:555. doi: 10.3389/fimmu.2014.00555
139. Fang Y, Ni J, Wang YS, Zhao Y, Jiang LQ, Chen C, et al. Exosomes as biomarkers and therapeutic delivery for autoimmune diseases: Opportunities and challenges. Autoimmun Rev. (2023) 22:103260. doi: 10.1016/j.autrev.2022.103260
140. Zhang M, Hu S, Liu L, Dang P, Liu Y, Sun Z, et al. Engineered exosomes from different sources for cancer-targeted therapy. Signal transduction targeted Ther. (2023) 8:124. doi: 10.1038/s41392-023-01382-y
141. Chen W, Huang Y, Han J, Yu L, Li Y, Lu Z, et al. Immunomodulatory effects of mesenchymal stromal cells-derived exosome. Immunologic Res. (2016) 64:831–40. doi: 10.1007/s12026-016-8798-6
142. Zhang M, Xia T, Lin F, Yu J, Yang Y, Lei W, et al. Vitiligo: An immune disease and its emerging mesenchymal stem cell therapy paradigm. Transplant Immunol. (2023) 76:101766. doi: 10.1016/j.trim.2022.101766
143. Yang L, Wei Y, Sun Y, Shi W, Yang J, Zhu L, et al. Interferon-gamma inhibits melanogenesis and induces apoptosis in melanocytes: A pivotal role of CD8+ Cytotoxic T lymphocytes in vitiligo. Acta dermato-venereologica. (2015) 95:664–70. doi: 10.2340/00015555-2080
144. Vaish U, Kumar AA, Varshney S, Ghosh S, Sengupta S, Sood C, et al. Micro RNAs upregulated in Vitiligo skin play an important role in its aetiopathogenesis by altering TRP1 expression and keratinocyte-melanocytes cross-talk. Sci Rep. (2019) 9:10079. doi: 10.1038/s41598-019-46529-6
145. Ujiie H. Regulatory T cells in autoimmune skin diseases. Exp Dermatol. (2019) 28:642–6. doi: 10.1111/exd.13535
146. Hajam EY, Panikulam P, Chu CC, Jayaprakash H, Majumdar A, Jamora C. The expanding impact of T-regs in the skin. Front Immunol. (2022) 13:983700. doi: 10.3389/fimmu.2022.983700
147. Huo J, Liu T, Li F, Song X, Hou X. MicroRNA−21−5p protects melanocytes via targeting STAT3 and modulating Treg/Teff balance to alleviate vitiligo. Mol Med Rep. (2021) 23:51. doi: 10.3892/mmr.2020.11689
148. Zhou J, Ling J, Song J, Wang Y, Feng B, Ping F. Interleukin 10 protects primary melanocyte by activation of Stat-3 and PI3K/Akt/NF-κB signaling pathways. Cytokine. (2016) 83:275–81. doi: 10.1016/j.cyto.2016.05.013
149. Xu SJ, Chen JH, Chang S, Li HL. The role of miRNAs in T helper cell development, activation, fate decisions and tumor immunity. Front Immunol. (2023) 14:1320305. doi: 10.3389/fimmu.2023.1320305
150. Guan Y, Ma Y, Tang Y, Liu X, Zhao Y, An L. MiRNA-221-5p suppressed the Th17/Treg ratio in asthma via RORγt/Foxp3 by targeting SOCS1. Allergy asthma Clin immunology: Off J Can Soc Allergy Clin Immunol. (2021) 17:123. doi: 10.1186/s13223-021-00620-8
151. Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. (2009) 30:80–91. doi: 10.1016/j.immuni.2008.11.010
152. Ye X, Lu Q, Yang A, Rao J, Xie W, He C, et al. MiR-206 regulates the Th17/Treg ratio during osteoarthritis. Mol Med (Cambridge Mass.). (2021) 27:64. doi: 10.1186/s10020-021-00315-1
153. Qu X, Han J, Zhang Y, Wang Y, Zhou J, Fan H, et al. MiR-384 regulates the th17/treg ratio during experimental autoimmune encephalomyelitis pathogenesis. Front Cell Neurosci. (2017) 11:88. doi: 10.3389/fncel.2017.00088
154. Zhang D, Qiu X, Li J, Zheng S, Li L, Zhao H. MiR-23a-3p-regulated abnormal acetylation of FOXP3 induces regulatory T cell function defect in Graves' disease. Biol Chem. (2019) 400:639–50. doi: 10.1515/hsz-2018-0343
155. Zheng Y, Dong C, Yang J, Jin Y, Zheng W, Zhou Q, et al. Exosomal microRNA-155-5p from PDLSCs regulated Th17/Treg balance by targeting sirtuin-1 in chronic periodontitis. J Cell Physiol. (2019) 234:20662–74. doi: 10.1002/jcp.28671
156. Wang X, Shen H, He Q, Tian W, Xia A, Lu XJ. Exosomes derived from exhausted CD8+ T cells impaired the anticancer function of normal CD8+ T cells. J Med Genet. (2019) 56:29–31. doi: 10.1136/jmedgenet-2018-105439
157. Li Y, Wang F, Guo R, Zhang Y, Chen D, Li X, et al. Exosomal sphingosine 1-phosphate secreted by mesenchymal stem cells regulated Treg/Th17 balance in aplastic anemia. IUBMB Life. (2019) 71:1284–92. doi: 10.1002/iub.2035
158. Wang R, Li R, Li T, Zhu L, Qi Z, Yang X, et al. Bone Mesenchymal Stem Cell-Derived Exosome-Enclosed miR-181a Induces CD4(+)CD25(+)FOXP3(+) Regulatory T Cells via SIRT1/Acetylation-Mediated FOXP3 Stabilization. J Oncol. (2022) 2022:8890434. doi: 10.1155/2022/8890434
159. Shi QZ, Yu HM, Chen HM, Liu M, Cheng X. Exosomes derived from mesenchymal stem cells regulate Treg/Th17 balance in aplastic anemia by transferring miR-23a-3p. Clin Exp Med. (2021) 21:429–37. doi: 10.1007/s10238-021-00701-3
160. Lindenbergh MFS, Stoorvogel W. Antigen presentation by extracellular vesicles from professional antigen-presenting cells. Annu Rev Immunol. (2018) 36:435–59. doi: 10.1146/annurev-immunol-041015-055700
161. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González M, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. (2011) 2:282. doi: 10.1038/ncomms1285
162. Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, et al. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity. (2014) 41:89–103. doi: 10.1016/j.immuni.2014.05.019
163. Klinker MW, Lizzio V, Reed TJ, Fox DA, Lundy SK. Human B cell-derived lymphoblastoid cell lines constitutively produce fas ligand and secrete MHCII(+)FasL(+) killer exosomes. Front Immunol. (2014) 5:144. doi: 10.3389/fimmu.2014.00144
164. Cheng L, Wang Y, Huang L. Exosomes from M1-polarized macrophages potentiate the cancer vaccine by creating a pro-inflammatory microenvironment in the lymph node. Mol therapy: J Am Soc Gene Ther. (2017) 25:1665–75. doi: 10.1016/j.ymthe.2017.02.007
165. Dosil SG, Lopez-Cobo S, Rodriguez-Galan A, Fernandez-Delgado I, Ramirez-Huesca M, Milan-Rois P, et al. Natural killer (NK) cell-derived extracellular-vesicle shuttled microRNAs control T cell responses. eLife. (2022) 11:e76319. doi: 10.7554/eLife.76319
166. Tian X, Cui Z, Liu S, Zhou J, Cui R. Melanosome transport and regulation in development and disease. Pharmacol Ther. (2021) 219:107707. doi: 10.1016/j.pharmthera.2020.107707
167. Xiong XX, Ding GZ, Zhao WE, Li X, Ling YT, Sun L, et al. Differences in the melanosome distribution within the epidermal melanin units and its association with the impairing background of leukoderma in vitiligo and halo nevi: a retrospective study. Arch Dermatol Res. (2017) 309:323–33. doi: 10.1007/s00403-017-1730-7
168. Tadokoro R, Murai H, Sakai KI, Okui T, Yokota Y, Takahashi Y. Melanosome transfer to keratinocyte in the chicken embryonic skin is mediated by vesicle release associated with Rho-regulated membrane blebbing. Sci Rep. (2016) 6:38277. doi: 10.1038/srep38277
169. Moreiras H, Seabra MC, Barral DC. Melanin transfer in the epidermis: the pursuit of skin pigmentation control mechanisms. Int J Mol Sci. (2021) 22:4466. doi: 10.3390/ijms22094466
170. Zhu Y, Li D, Huang H. Activity and cytosolic Na(+) regulate synaptic vesicle endocytosis. J neuroscience: Off J Soc Neurosci. (2020) 40:6112–20. doi: 10.1523/JNEUROSCI.0119-20.2020
171. Ando H, Niki Y, Ito M, Akiyama K, Matsui MS, Yarosh DB, et al. Melanosomes are transferred from melanocytes to keratinocytes through the processes of packaging, release, uptake, and dispersion. J Invest Dermatol. (2012) 132:1222–9. doi: 10.1038/jid.2011.413
172. Ishida M, Arai SP, Ohbayashi N, Fukuda M. The GTPase-deficient Rab27A(Q78L) mutant inhibits melanosome transport in melanocytes through trapping of Rab27A effector protein Slac2-a/melanophilin in their cytosol: development of a novel melanosome-targetinG tag. J Biol Chem. (2014) 289:11059–67. doi: 10.1074/jbc.M114.552281
173. Liao Y, Chen X, Miller-Little W, Wang H, Willard B, Bulek K, et al. The Ras GTPase-activating-like protein IQGAP1 bridges Gasdermin D to the ESCRT system to promote IL-1β release via exosomes. EMBO J. (2023) 42:e110780. doi: 10.15252/embj.2022110780
174. Jella KK, Rani S, O'Driscoll L, McClean B, Byrne HJ, Lyng FM. Exosomes are involved in mediating radiation induced bystander signaling in human keratinocyte cells. Radiat Res. (2014) 181:138–45. doi: 10.1667/RR13337.1
175. Shi HX, Zhang RZ, Xiao L, Wang L. Effects of keratinocyte-derived and fibroblast-derived exosomes on human epidermal melanocytes. Indian J dermatology venereology leprology. (2022) 88:322–31. doi: 10.25259/IJDVL_1087_19
176. Nasiri G, Azarpira N, Alizadeh A, Goshtasbi S, Tayebi L. Shedding light on the role of keratinocyte-derived extracellular vesicles on skin-homing cells. Stem Cell Res Ther. (2020) 11:421. doi: 10.1186/s13287-020-01929-8
177. Prospéri MT, Giordano C, Gomez-Duro M, Hurbain I, Macé AS, Raposo G, et al. Extracellular vesicles released by keratinocytes regulate melanosome maturation, melanocyte dendricity, and pigment transfer. Proc Natl Acad Sci United States America. (2024) 121:e2321323121. doi: 10.1073/pnas.2321323121
178. Yoon JH, Jo CS, Hwang JS. Comprehensive analysis of exosomal microRNAs derived from UVB-irradiated keratinocytes as potential melanogenesis regulators. Int J Mol Sci. (2024) 25:3095. doi: 10.3390/ijms25063095
179. Takano K, Hachiya A, Murase D, Tanabe H, Kasamatsu S, Takahashi Y, et al. Quantitative changes in the secretion of exosomes from keratinocytes homeostatically regulate skin pigmentation in a paracrine manner. J Dermatol. (2020) 47:265–76. doi: 10.1111/1346-8138.15202
180. Gemel J, Kilkus J, Dawson G, Beyer EC. Connecting exosomes and connexins. Cancers. (2019) 11:475. doi: 10.3390/cancers11040476
181. Xie S, Zhang Q, Jiang L. Current knowledge on exosome biogenesis, cargo-sorting mechanism and therapeutic implications. Membranes. (2022) 12:498. doi: 10.3390/membranes12050498
182. Wäster P, Eriksson I, Vainikka L, Rosdahl I, Öllinger K. Extracellular vesicles are transferred from melanocytes to keratinocytes after UVA irradiation. Sci Rep. (2016) 6:27890. doi: 10.1038/srep27890
183. Wäster P, Eriksson I, Vainikka L, Öllinger K. Extracellular vesicles released by melanocytes after UVA irradiation promote intercellular signaling via miR21. Pigment Cell melanoma Res. (2020) 33:542–55. doi: 10.1111/pcmr.12860
184. Šahmatova L, Tankov S, Prans E, Aab A, Hermann H, Reemann P, et al. MicroRNA-155 is dysregulated in the skin of patients with vitiligo and inhibits melanogenesis-associated genes in melanocytes and keratinocytes. Acta dermato-venereologica. (2016) 96:742–7. doi: 10.2340/00015555-2394
185. Dai X, Rao C, Li H, Chen Y, Fan L, Geng H, et al. Regulation of pigmentation by microRNAs: MITF-dependent microRNA-211 targets TGF-β receptor 2. Pigment Cell melanoma Res. (2015) 28:217–22. doi: 10.1111/pcmr.12334
186. Aguennouz M, Guarneri F, Oteri R, Polito F, Giuffrida R, Cannavò SP. Serum levels of miRNA-21-5p in vitiligo patients and effects of miRNA-21-5p on SOX5, beta-catenin, CDK2 and MITF protein expression in normal human melanocytes. J Dermatol Sci. (2021) 101:22–9. doi: 10.1016/j.jdermsci.2020.10.014
187. Ji K, Zhang P, Zhang J, Fan R, Liu Y, Yang S, et al. MicroRNA 143-5p regulates alpaca melanocyte migration, proliferation and melanogenesis. Exp Dermatol. (2018) 27:166–71. doi: 10.1111/exd.13480
188. Noguchi S, Kumazaki M, Yasui Y, Mori T, Yamada N, Akao Y. MicroRNA-203 regulates melanosome transport and tyrosinase expression in melanoma cells by targeting kinesin superfamily protein 5b. J Invest Dermatol. (2014) 134:461–9. doi: 10.1038/jid.2013.310
189. Cai XW, Zhu R, Ran L, Li YQ, Huang K, Peng J, et al. A novel non−contact communication between human keratinocytes and T cells: Exosomes derived from keratinocytes support superantigen−induced proliferation of resting T cells. Mol Med Rep. (2017) 16:7032–8. doi: 10.3892/mmr.2017.7492
190. Di Dalmazi G, Hirshberg J, Lyle D, Freij JB, Caturegli P. Reactive oxygen species in organ-specific autoimmunity. Auto- Immun highlights. (2016) 7:11. doi: 10.1007/s13317-016-0083-0
191. Pang YB, Wu S, He YJ, Nian Q, Lei J, Yao YJ, et al. Plant-derived compounds as promising therapeutics for vitiligo. Front Pharmacol. (2021) 12:685116. doi: 10.3389/fphar.2021.685116
192. Wang T, Jian Z, Baskys A, Yang J, Li J, Guo H, et al. MSC-derived exosomes protect against oxidative stress-induced skin injury via adaptive regulation of the NRF2 defense system. Biomaterials. (2020) 257:120264. doi: 10.1016/j.biomaterials.2020.120264
193. Su M, Miao F, Jiang S, Shi Y, Luo L, He X, et al. Role of the p53−TRPM1/miR−211−MMP9 axis in UVB−induced human melanocyte migration and its potential in repigmentation. Int J Mol Med. (2020) 45:1017–26. doi: 10.3892/ijmm.2020.4478
194. Kuang Y, Zheng X, Zhang L, Ai X, Venkataramani V, Kilic E, et al. Adipose-derived mesenchymal stem cells reduce autophagy in stroke mice by extracellular vesicle transfer of miR-25. J extracellular vesicles. (2020) 10:e12024. doi: 10.1002/jev2.12024
195. Kim H, Lee JW, Han G, Kim K, Yang Y, Kim SH. Extracellular vesicles as potential theranostic platforms for skin diseases and aging. Pharmaceutics. (2021) 13:760. doi: 10.3390/pharmaceutics13050760
196. Janockova J, Slovinska L, Harvanova D, Spakova T, Rosocha J. New therapeutic approaches of mesenchymal stem cells-derived exosomes. J Biomed Sci. (2021) 28:39. doi: 10.1186/s12929-021-00736-4
197. Mendt M, Kamerkar S, Sugimoto H, McAndrews KM, Wu CC, Gagea M, et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. (2018) 3:e99263. doi: 10.1172/jci.insight.99263
198. Cai J, Wu J, Wang J, Li Y, Hu X, Luo S, et al. Extracellular vesicles derived from different sources of mesenchymal stem cells: therapeutic effects and translational potential. Cell bioscience. (2020) 10:69. doi: 10.1186/s13578-020-00427-x
199. Abaandou L, Quan D, Shiloach J. Affecting HEK293 cell growth and production performance by modifying the expression of specific genes. Cells. (2021) 10:1667. doi: 10.3390/cells10071667
200. Hu J, Han J, Li H, Zhang X, Liu LL, Chen F, et al. Human embryonic kidney 293 cells: A vehicle for biopharmaceutical manufacturing. Struct Biology Electrophysiology Cells tissues organs. (2018) 205:1–8. doi: 10.1159/000485501
201. Tan E, Chin CSH, Lim ZFS, Ng SK. HEK293 cell line as a platform to produce recombinant proteins and viral vectors. Front bioengineering Biotechnol. (2021) 9:796991. doi: 10.3389/fbioe.2021.796991
Keywords: exosome, microRNAs, vitiligo, autoimmunity, biomacromolecule, oxidative stress
Citation: li W, Pang Y, He Q, Song Z, Xie X, Zeng J and Guo J (2024) Exosome-derived microRNAs: emerging players in vitiligo. Front. Immunol. 15:1419660. doi: 10.3389/fimmu.2024.1419660
Received: 18 April 2024; Accepted: 24 June 2024;
Published: 08 July 2024.
Edited by:
Shahnawaz Jadeja, Trinity College Dublin, IrelandReviewed by:
Cristina Casalou, University College Dublin, IrelandPrashant Giri, St. Jude Children’s Research Hospital, United States
Nik T. Georgopoulos, Sheffield Hallam University, United Kingdom
Copyright © 2024 li, Pang, He, Song, Xie, Zeng and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinhao Zeng, emVuZ2ppbmhhb0BjZHV0Y20uZWR1LmNu; Jing Guo, Z3VvamluZzY2QGNkdXRjbS5lZHUuY24=
 Wenquan li
Wenquan li Yaobin Pang
Yaobin Pang Qingying He1,2
Qingying He1,2 Jinhao Zeng
Jinhao Zeng Jing Guo
Jing Guo